In many natural and clinical settings, bacteria are associated with some type of biotic or abiotic surface that enables them to form biofilms, a multicellular lifestyle with bacteria embedded in an extracellular matrix. Staphylococcus aureus and Staphylococcus epidermidis, the most frequent causes of biofilm-associated infections on indwelling medical devices, can switch between an existence as single free-floating cells and multicellular biofilms. During biofilm formation, cells first attach to a surface and then multiply to form microcolonies.
KEYWORDS: Staphylococcus aureus, Staphylococcus epidermidis, biofilms, extracellular matrix, microbial communities, quorum sensing
SUMMARY
In many natural and clinical settings, bacteria are associated with some type of biotic or abiotic surface that enables them to form biofilms, a multicellular lifestyle with bacteria embedded in an extracellular matrix. Staphylococcus aureus and Staphylococcus epidermidis, the most frequent causes of biofilm-associated infections on indwelling medical devices, can switch between an existence as single free-floating cells and multicellular biofilms. During biofilm formation, cells first attach to a surface and then multiply to form microcolonies. They subsequently produce the extracellular matrix, a hallmark of biofilm formation, which consists of polysaccharides, proteins, and extracellular DNA. After biofilm maturation into three-dimensional structures, the biofilm community undergoes a disassembly process that leads to the dissemination of staphylococcal cells. As biofilms are dynamic and complex biological systems, staphylococci have evolved a vast network of regulatory mechanisms to modify and fine-tune biofilm development upon changes in environmental conditions. Thus, biofilm formation is used as a strategy for survival and persistence in the human host and can serve as a reservoir for spreading to new infection sites. Moreover, staphylococcal biofilms provide enhanced resilience toward antibiotics and the immune response and impose remarkable therapeutic challenges in clinics worldwide. This review provides an overview and an updated perspective on staphylococcal biofilms, describing the characteristic features of biofilm formation, the structural and functional properties of the biofilm matrix, and the most important mechanisms involved in the regulation of staphylococcal biofilm formation. Finally, we highlight promising strategies and technologies, including multitargeted or combinational therapies, to eradicate staphylococcal biofilms.
INTRODUCTION
The term bacterial biofilm was coined in 1978 by Costerton and colleagues, who described it as a structured microbial community that is attached to a surface and encased by an extracellular matrix (1). However, cell aggregates that form in the absence of any surface (2), as well as pellicles, floating biofilms that take shape at the air-liquid interface (3), are also often considered to be types of biofilms. Since the introduction of the biofilm model over 40 years ago, it has become clear that the majority of bacteria have the inherent capacity to grow in these self-generated ecosystems. Peters and colleagues demonstrated with early electron microscopy studies that staphylococcal cells adhere to central venous catheters, where they are embedded in a “slimy material,” the biofilm matrix (4, 5). The role of the self-produced biofilm matrix or extracellular polymeric substance (EPS), consisting mainly of polysaccharides, proteins, and extracellular DNA (eDNA), is to protect the cells from deleterious environmental factors, including antibiotics and the host immune system (6–9). Members of the Staphylococcus genus, including Staphylococcus aureus and Staphylococcus epidermidis, the most prominent member of the coagulase-negative staphylococci (CoNS), are opportunistic pathogens and both produce robust biofilms on abiotic and biotic surfaces. While nasal and skin colonization with these pathogens widely occurs in humans without any symptoms (10, 11), the switch between single free-floating cells, referred to as “planktonic state,” and a multicellular biofilm is a pivotal step for staphylococci to cause different types of infections, including infective endocarditis, osteomyelitis, and prosthetic joint infections (PJIs) (12–17). Whereas S. aureus has many mechanisms and virulence factors to evade the immune system, the pathogenic potential of S. epidermidis mainly depends on biofilm formation (18). Since implants, such as prosthetics, catheters, and other devices, provide an ideal surface for bacterial adhesion, S. epidermidis is remarkably well adapted to cause implant-associated infections (19). Approximately 30% to 40% of nosocomial bloodstream infections are caused by CoNS, such as S. epidermidis, and the majority of these bloodstream infections are the direct result of an intravascular catheter infection (ICI), a fact that strongly connects the biofilm-forming capacity of S. epidermidis to bloodstream infections. Besides that, CoNS are among the most frequent causes of prosthetic valve endocarditis (PVE) (15% to 40% of all PVE cases), cardiac pacemaker infections (incidence rates of 0.13% to 19.9%), and surgical site infections (20).
Since the biofilm lifestyle plays a central role in staphylococcal biology, the process of biofilm formation and disassembly is tightly controlled by several regulatory systems that integrate the physiological state of the cell, environmental cues, and the dynamics within the staphylococcal community. In this context, the best-studied regulatory system of staphylococcal biofilm formation is the accessory gene regulator (Agr) quorum sensing (QS) system, a cell-to-cell communication system that coordinates cellular behavior dependent on cell density (21–23). The Agr QS system most prominently regulates the production of proteases and phenol-soluble modulins (PSMs), which are main contributors of biofilm maturation and disassembly in S. aureus as well as S. epidermidis (24–26). Our understanding of staphylococcal biofilms has progressed tremendously as a consequence of advances in biochemical tools and new imaging technologies. Three-dimensional biofilm structure analysis in vitro revealed a high degree of complexity and spatial organization within staphylococcal biofilms. In addition, studies have shown that the staphylococcal EPS is highly variable in its composition, depending on nutrient availability, the host environment, and mechanical shear forces (27).
Although in the past researchers have extensively studied the molecular basis of staphylococcal biofilm formation in vitro, our knowledge about staphylococcal biofilm formation under in vivo conditions is still limited. In contrast to in vitro biofilm formation, in vivo staphylococci are subject to innate host defenses such as neutrophils, macrophages, and antimicrobial peptides (AMPs) (28, 29). During infection, staphylococcal biofilms confer protection against the host immune system as well as antibiotic treatment. Whereas biofilm recalcitrance against the immune response was long attributed to the biofilm microenvironment acting as a physical barrier for host immune cells (30), it has now become clear that biofilms shield bacterial cells from detection by the immune system by masking pathogen-associated molecular patterns (PAMPs) (13). Similarly, biofilms were originally postulated to prevent the diffusion of antibiotics, rendering the cells in the biofilm resistant to antibiotic treatment. However, new evidence suggests that cells residing inside biofilms have low metabolic activity, which increases their tolerance against antibiotics that primarily target metabolically active cells (31). Indeed, biofilm-associated cells with low metabolic activity share a high physiological similarity to persister cells and small-colony variants (SCV) (32, 33). The antibiotic tolerance of persister cells has been linked to low intracellular levels of ATP in S. aureus (34) as well as the Gram-negative bacterium Escherichia coli (35). Cells within a biofilm encounter low oxygen and nutrient availability, resulting in low metabolic cell activity and a drop in intracellular ATP levels, which presumably contribute to elevated antibiotic tolerance of the biofilm (32). Therefore, antibiofilm strategies that interfere with biofilm cells independent of their cellular activity, such as AMPs, surface modifications to prevent bacterial adhesion, nanoparticles with antimicrobial activity, and new technologies for physical biofilm removal, are considered to be highly attractive approaches (36).
This review seeks to give a comprehensive overview of staphylococcal biofilm formation, its regulation, and the resulting consequences for bacterial infections. The identification and development of antibiofilm agents has become a major research objective over the last decades, and strategies to interfere with biofilm formation and its regulation or to eradicate preexisting biofilms are discussed at the end of this review.
MOLECULAR MECHANISMS OF STAPHYLOCOCCAL BIOFILM FORMATION
Staphylococcal biofilm development is a complex process that can be divided into three main phases: (i) initial attachment, (ii) production of extracellular matrix and cell proliferation, and (iii) biofilm structuring and cell detachment. In the first phase, S. aureus attaches to a biotic or abiotic surface using a range of different factors and mechanisms, including sortase-attached surface adhesins (37, 38), wall teichoic acids (WTAs) (39, 40), changes in bacterial cell surface hydrophobicity (41), and eDNA production (42, 43). The attached bacteria then proliferate and build microcolonies on the surface. In the next stage, those microcolonies develop into distinct structures that establish the biofilm. During proliferation and biofilm maturation, the cells are held together by adhesive factors within the matrix. Remodeling of the biofilm occurs through disruptive factors, like nucleases and surfactants (PSMs), which are thought to be essential for the development of the three-dimensional structure of the mature staphylococcal biofilm with its distinctive towers and channels. Cell detachment during biofilm disassembly is then mainly caused by protease-driven degradation of the biofilm matrix and disruption of the biofilm by PSMs (24, 44, 45).
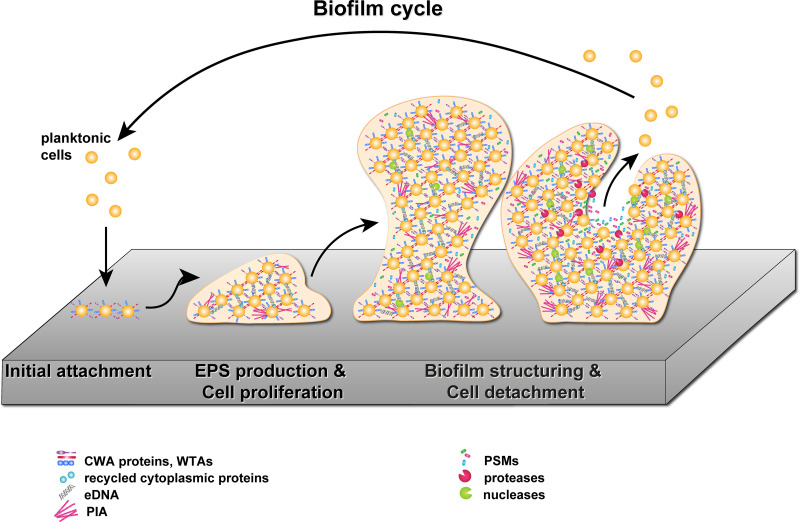
A further refined model of biofilm development has been postulated recently. This model expands the traditional biofilm model by two distinct new phases, referred to as the multiplication and the exodus phases, which take place after the initial attachment and before the maturation phase (46, 47). During the multiplication phase, the cells start to grow and divide and embed themselves in a matrix consisting of proteins and eDNA. The exodus phase is then triggered by the expression and secretion of the major nuclease Nuc1, which leads to degradation of the eDNA contained in the matrix, allowing the release of a subpopulation of cells from the biofilm. Cells remaining in the biofilm then undergo rapid growth, which leads to the formation of the characteristic tower-like structure of staphylococcal biofilms (46). Since the five-phase biofilm model has been postulated only recently and the current literature is almost exclusively based on the traditional three-phase biofilm model, this review was structured according to the traditional model. For a comprehensive overview of the five-phase biofilm model, we refer the reader to the review of Moormeier and Bayles (47). An overview of the above-described S. aureus biofilm formation steps is provided in the following subsections and is depicted in Fig. 1.
FIG 1.
Staphylococcal biofilm cycle. During the first step of biofilm formation, planktonic staphylococcal cells attach to a surface with CWA proteins and WTAs (initial attachment). The production of PIA, eDNA, and adhesins to establish intercellular interactions is upregulated and leads to the formation of microcolonies on the surface (EPS production and cell proliferation). Cells within the growing biofilm express nucleases and PSMs, which are responsible for the formation of channels and towers in the matured biofilm. In the last step, the biofilm is dispersed by the action of PSMs and proteases, allowing the bacterial cells to detach from the biofilm to colonize new niches (biofilm structuring and cell detachment). The depiction of matrix components was inspired by Fig. 5 from the work of Hobley and colleagues (9).
Initial Attachment
Surface attachment is the first stage of staphylococcal biofilm formation, at which planktonic staphylococcal cells attach to biotic or abiotic surfaces. During infection, staphylococci mainly encounter biotic surfaces composed of host matrix including fibrinogen, fibronectin, vitronectin, and collagen. Binding to these biotic surfaces is facilitated by cell wall-anchored (CWA) proteins, of which the best-characterized group is termed microbial surface components recognizing adhesive matrix molecules (MSCRAMMs). All MSCRAMMs contain a C-terminal cell wall-targeting motif (LPXTG motif) that covalently anchors the protein to the bacterial peptidoglycan by the enzyme sortase A (48). Several MSCRAMMs have been implicated to promote biofilm formation by facilitating the initial attachment to host matrix components. Among these are the fibronectin-binding proteins FnBPA and FnBPB (49–51), clumping factor A (ClfA) and clumping factor B (ClfB) (52–56), members of the serine-aspartate repeat family proteins (SdrC, SdrD, and SdrE) (57–60) and the bone sialoprotein-binding protein (Bbp) (61, 62). Similar to S. aureus, S. epidermidis also uses MSCRAMMs to bind to surfaces covered with host matrix proteins for primary attachment. Three main MSCRAMMs, SdrG, SdrF, and SdrH, all members of the serine-aspartate repeat family, have been identified in S. epidermidis (63). To date, no ligand has been identified for SdrH. On the other hand, several studies have shown that SdrG binds fibrinogen and SdrF binds the host matrix components collagen and keratin (38). For a more detailed overview of MSCRAMMs of Gram-positive cocci, the reader is referred to a recent review by Foster (38).
Binding to abiotic surfaces of catheters and implants, but also to the polystyrene surfaces of microtiter plates often used for in vitro biofilm experiments, is facilitated through hydrophobic and electrostatic interactions. The extracellular glycopolymers WTAs have been identified as the main drivers of these interactions in S. aureus (39) and in S. epidermidis (40). Besides their aforementioned interaction with host matrix components, FnBPs and SdrC can also drive attachment of S. aureus to abiotic surfaces (64, 65), which was also shown for the biofilm-associated protein (Bap) (66). In addition, the major autolytic enzymes AtlA in S. aureus (67–69) and the homologous protein AtlE in S. epidermidis (70) are major contributors to primary attachment to abiotic surfaces like polystyrene or glass. Although it is not known exactly how these hydrolytic enzymes contribute to surface attachment, it is hypothesized that the enzymatic activity results in major changes in bacterial surface hydrophobicity, resulting in enhanced surface attachment (41). Besides their involvement in primary attachment, AtlA and AtlE are key enzymes for the release of eDNA (42, 69), a known component of the staphylococcal in vitro biofilm matrix (43, 71, 72). Interestingly, addition of DNase I at the start of biofilm cultures prevented attachment of several different S. epidermidis strains, indicating an important role for eDNA not only as a component of the biofilm matrix but already during initial surface attachment (42). It is important to note that in contrast to the case with S. epidermidis, eDNA seems not to play an important role for S. aureus surface attachment, as DNase I treatment during the first 8 h does not have an apparent effect on biofilm formation (46, 73). Finally, the accumulation-associated protein (Aap) of S. epidermidis was shown to mediate attachment to polystyrene surfaces through its N-terminal A domain (74). Impaired surface attachment of an Aap mutant strain could be complemented by expression of the homologous protein S. aureus surface protein G (SasG), indicating that SasG could play a similar role in S. aureus surface attachment (74).
Production of Extracellular Matrix and Cell Proliferation
After initial attachment of single cells to a surface, these cells start to proliferate, resulting in the formation of so-called microcolonies. As part of the early biofilm maturation process, the bacteria also start to produce EPS, which eventually forms the biofilm matrix. EPS mainly consists of polysaccharides, proteins, teichoic acids, and eDNA, and dependent on the EPS composition, staphylococcal biofilms can be divided in two broad categories: biofilms consisting of a polysaccharide matrix and biofilms with a proteinaceous matrix (75, 76). In polysaccharide-dependent biofilms, the main EPS component is poly-β(1-6)-N-acetylglucosamine (PNAG), also referred to as polysaccharide intercellular adhesin (PIA) (77, 78). The production of PIA is dependent on the intracellular adhesion (ica) operon, which was first identified in an S. epidermidis transposon mutant screen for mutants deficient in biofilm formation (79) and later also in S. aureus (80). The ica operon comprises four genes arranged in the following order: icaA, icaD, icaB, and icaC (80, 81). In vitro assays using UDP-N-acetylglucosamine (UDP-GlcNAc) as the substrate showed that the transmembrane glycosyltransferase IcaA as well as IcaD is essential for PIA synthesis (82). Coexpression of the putative membrane protein IcaC is required to produce longer oligomer chains (82) and was therefore proposed to be important for translocation of the growing polysaccharide chain to the cell surface. Only recently has it become clear that IcaC is a predicted member of membrane-bound acyltransferases and presumably responsible for the succinylation of PIA (83). It is important to note that for a long time PIA has been described to be N-succinylated (84), but more recent evidence has supported the idea that PIA is indeed O-succinylated in staphylococci (85, 86). PIA is also partially deacetylated by the extracellular, cell surface-associated polysaccharide deacetylase IcaB (87), and deacetylated PIA is positively charged and localized on the bacterial surface in fibrous strands (87, 88). The transcription of the icaADBC operon is mainly regulated by IcaR, a repressor located upstream of the ica operon (89, 90). A more detailed description of ica regulation is given in “ica operon regulation” below.
Besides polysaccharide-dependent biofilms, staphylococci have also been shown to form biofilms with matrices mainly consisting of proteinaceous material. Such biofilms have been mainly observed for methicillin-resistant S. aureus (MRSA) isolates and S. epidermidis clinical isolates, while methicillin-sensitive S. aureus (MSSA) isolates are more often ica dependent (91–94). Importantly, the high prevalence of proteinaceous biofilms formed by S. epidermidis clinical isolates is likely connected with the absence of the ica operon in a high number of S. epidermidis isolates. Although the reported numbers vary, many studies have shown that only 20% to 60% of clinical S. epidermidis isolates carry the ica operon (95–99). In an early study, the surface protein Bap of S. aureus was identified as the main determinant for successful surface adherence as well as intercellular accumulation during biofilm development (66). A follow-up study by the same research group also showed that the ica operon, although present in the genome, did not contribute to biofilm formation of the S. aureus strain under investigation (100). Although the Bap protein is not widespread among S. aureus strains (66), a protein called Bap homologue protein (Bhp) is present in S. epidermidis (101). Furthermore, it has recently been shown that Bap proteins can build amyloid-like scaffolds that promote S. aureus biofilm formation (102). The role of several MSCRAMMs extends beyond initial attachment. More precisely, FnBPA, FnBPB, and SdrC all promote cell-cell interactions leading to increased biofilm formation (65, 103–105). One of the best-studied proteinaceous biofilm factors of S. epidermidis is the surface protein Aap (106). Originally thought to anchor PIA to the cell wall (106), Aap was recently shown to mediate biofilm formation after enzymatic cleavage by staphylococcal or host proteases (92, 107). More specifically, after cleavage the mature Aap protein accomplishes self-adhesion and intercellular adhesion via several B domain repeats (108, 109). Interestingly, the S. aureus Aap homologue SasG promotes biofilm formation by zinc-mediated dimerization and was shown to interact with Aap (110, 111).
A third type of staphylococcal biofilm, mainly dependent on eDNA, has attracted substantial interest. The initial discovery of this type of biofilm is supported by the observation that a mutant deficient in AtlA, the main autolysin in S. aureus responsible for the release of eDNA, showed a reduced biofilm formation phenotype in in vitro experiments (69), while mutation of the staphylococcal thermonuclease (Nuc) leads to increased biofilm formation (71). Finally, mature staphylococcal biofilms are sensitive to externally added DNase I, demonstrating that eDNA is a structural component of the biofilm matrix (43). Similar observations were made for biofilms of the Gram-negative bacterium Pseudomonas aeruginosa (112), indicating that the use of eDNA as a structural component of biofilms is conserved across different bacterial genera. There is also increasing evidence that cytoplasmic proteins associated with the outside of the cell via electrostatic interactions may be able to “moonlight” as eDNA-binding proteins in the biofilm matrix (113–115). Another recent study suggests that cell wall-associated lipoproteins contribute to S. aureus biofilm formation by linking bacterial cells together through noncovalent cross-links with high-molecular-weight eDNA found within the biofilm matrix (116). Additionally, investigation of the S. aureus biofilm proteome in a flow system demonstrated that secreted virulence factors and ribosomal proteins are trapped in the biofilm matrix and contribute to biofilm stability (115). In more detail, the biofilm environment is thought to be acidic, due to low oxygen levels and the release of fermentation products. The positive charge of alkaline virulence factors and ribosomal proteins in the acidic environment is then thought to mediate electrostatic interactions with surface components and eDNA, leading to biofilm stabilization (115). As described above, the biofilm matrix consists of different classes of molecules, namely, polysaccharides, proteins, and eDNA. While these are the principal building blocks of the biofilm matrix, the exact composition of the biofilm matrix can vary depending on substrate availability and physical factors. For example, S. epidermidis strains isolated from environments with high shear stress, such as found in the lumen of a catheter, rather produce PIA-dependent biofilms than isolates from low-shear environments (117). This exemplifies that environmental conditions, including the physical properties of the milieu, can have tremendous effects on staphylococcal biofilms.
Biofilm Structuring and Cell Detachment
Staphylococcal biofilms do not grow uniformly but rather consist of tower-like structures that are interspersed by fluid-filled channels that are developed during a process often referred to as biofilm maturation. The three-dimensional structure of biofilms is shaped by the activities of different enzymes and molecules that can degrade or interrupt the biofilm matrix (24, 118). As discussed before, the main components of the staphylococcal biofilm matrix are polysaccharides, proteins, and eDNA; accordingly, staphylococci have a vast array of exoenzymes that can target specific components of the biofilm matrix.
Protein biofilms are most susceptible to protease-mediated biofilm dispersal, and S. aureus produces and secretes 10 known proteases from different protease families: the metalloprotease aureolysin (Aur) (119), the two cysteine proteases SspB and ScpA (120, 121), and seven serine proteases known as SspA (also referred to as V8 protease) and SplA to -F (120, 122). The initial identification of staphylococcal proteases as biofilm-degrading agents was based on several studies investigating regulator mutants that were unable to form biofilms (103, 123, 124), a phenotype that could be correlated with elevated protease activity in these mutant strains (125–129). The importance of the individual proteases for biofilm degradation and dispersal is highly strain specific, and to date, the SspA, SspB, ScpA, and Aur proteases have been demonstrated to degrade S. aureus biofilms (103, 127, 129–131). The targets for two of the proteases have been identified. SspA degrades FnBPs and Bap (103, 127, 130), whereas aureolysin can degrade Bap as well as ClfB (127, 131). Although the proteins targeted by the staphopains SspB and ScpA have not been identified yet, their levels are decreased under biofilm-forming conditions, underlining their potential to inhibit biofilm formation (129). For S. epidermidis, three secreted proteases have been identified: the metalloprotease SepA (107, 132), the serine protease Esp, which was shown to degrade S. aureus biofilms (133–135), and the cysteine protease EcpA (136), all of which have low substrate specificity (137).
Another major component of the staphylococcal biofilm matrix, eDNA, is subject to degradation by nucleases. Two thermostable nucleases (Nuc1 and Nuc2), encoded by different open reading frames, are expressed by S. aureus (138). Transcription of nuc1 is at its maximum during the postexponential growth phase, whereas transcription of nuc2 is high during the early exponential phase and declines afterwards (139). Mutation of the nuc1 gene leads to increased staphylococcal biofilm formation, whereas overexpression of nuc1 leads to a reduction in biofilm formation (43, 71, 140). In accordance with the strong biofilm-degrading activity of Nuc1, its expression is highly repressed under in vitro biofilm growth conditions (141). It is interesting that eDNA needs to have a minimum size of 11 kDa in order to facilitate biofilm integrity (142), and the lack of nuclease activity is the driving factor for the preservation of high-molecular-weight eDNA (43, 71). In contrast to the well-established roles of Nuc1 during biofilm maturation and infection (141, 143), the role of Nuc2, a membrane-bound nuclease with an extracellular catalytic domain (144), has not been fully characterized yet.
Another group of molecules has emerged and been shown to participate in shaping staphylococcal biofilm structure. These molecules, so-called PSMs, are grouped in α-type (PSMα and PSMγ, which is also known as δ-toxin) and β-type (PSMβ) peptides and can disperse biofilms, most likely by disrupting noncovalent interactions between matrix components due to their pronounced amphipathic nature with potential surfactant-like properties (45, 118, 145). Indeed, external addition of PSMs to growing staphylococcal biofilms inhibits biofilm formation (45, 146). Periasamy and colleagues further determined that local differences of PSM expression in S. aureus biofilms are responsible for the formation of the characteristic fluid channels that are embedded in the biofilm matrix (45). In addition, PSM activity is also an important contributor to biofilm detachment and subsequent dissemination of staphylococcal cells in vivo (45). Whereas biofilm disassembly driven by soluble PSMs is well accepted, the biological role of polymerized PSMs in staphylococcal biofilms is contradictory. It was shown that PSMs of S. aureus not only can exist in their soluble form but also aggregate into amyloid-like fibrils that are able to stabilize the biofilm structure (147, 148). Zheng et al. confirmed that PSMs can form amyloid-like fibrils but proposed an alternative model in which PSMs, not in their amyloid form, protect eDNA from DNase-mediated degradation, which can then lead to enhanced S. aureus biofilm stability (149). PSMs are also important effectors for shaping the structure of S. epidermidis biofilms (150, 151). The surfactant-like β-type PSMs, first discovered in S. epidermidis (152), not only are involved in biofilm maturation and dispersal in vitro but also perform this function during catheter colonization in vivo (151). In addition, amyloid formation of S. epidermidis PSMs has not been observed yet, and this is in accordance with a recent study confirming a role of PSMs in biofilm structuring but not in its stabilization (26).
REGULATION OF STAPHYLOCOCCAL BIOFILM FORMATION
Biofilm formation is a social group behavior and every step, from the initial attachment to the dissemination of the mature biofilm, is tightly regulated (153). One of the main regulators of biofilm development is the QS system. In S. aureus, it is responsible for the switch from biofilm formation to its disassembly (22, 154, 155). Aside from QS, a vast number of environmental cues (e.g., pH, nutrient availability and fluid flow) as well as regulatory systems (e.g., Sae, SarA, Rot) are involved in the regulation of biofilm formation (23, 156, 157). The next section and Table 1 provide a comprehensive overview of the most important regulatory processes known to contribute to the formation, structuring, and disassembly of S. aureus biofilms. This review is mainly focused on protein-based regulatory systems. Nevertheless, there have been considerable advances in our knowledge of regulatory RNAs in staphylococci, and we refer the interested reader to two recent reviews (158, 159).
TABLE 1.
Major staphylococcal biofilm regulators
| Regulator | Functions | Involvement in biofilm development | References |
|---|---|---|---|
| Agr | Cell-cell communication system (quorum sensing), TCS with AIPs as signaling molecules; stimulation of protease production and PSMs; repression of adhesins | Important for initial attachment and biofilm disassembly | 21, 22, 45, 145, 175, 176, 178, 181, 182 |
| SaeRS | TCS; stimulation of adhesins and nuclease | Important for biofilm maturation; regulates the “exodus” event in the five-phase biofilm model | 46, 141, 217 |
| SarA | SarA family cytoplasmic regulator; repression of protease production and stimulation of PIA; regulation of adhesins | Important for biofilm formation mainly through repression of proteases; positive regulator of biofilm formation | 125, 226, 237, 242 |
| Rot | SarA family cytoplasmic regulator; main effector of the Agr QS; stimulation of adhesins and repression of proteases | Important for biofilm formation mainly through repression of proteases; positive regulator of biofilm formation. | 170, 179, 245, 249 |
| MgrA | SarA family cytoplasmic regulator; main effector of the ArlRS TCS; repression of adhesins | Important for biofilm formation mainly through repression of adhesins; negative regulator of biofilm formation | 250–252 |
| SigB | Alternative sigma factor for stress response; stimulation of adhesins and repression of proteases and nuclease; strain-dependent regulation of PIA | Important for initial attachment and biofilm disassembly | 71, 240, 255, 261, 263, 264, 269 |
| CodY | Cytoplasmic regulator for metabolic response; strain-dependent regulation of PIA; repression of proteases | Important for PIA-dependent biofilm formation | 276–279, 283–286 |
| LytSR | TCS regulating cell lysis; stimulation of LrgAB | Important for eDNA-dependent biofilm formation | 315–318 |
Quorum Sensing-Mediated Regulation of S. aureus Biofilm Development
QS enables bacteria to coordinate their behavior in a population density-dependent manner. This process involves the production of signaling molecules and their subsequent accumulation in the extracellular space surrounding the bacteria. When a certain threshold in concentration is reached, these signals are sensed by the bacteria and translated into alterations in their transcriptional profile and behavior. Therefore, QS allows bacteria to synchronize their behavior on a population-wide scale, enabling them to act like a multicellular organism. While the basic principles of QS are conserved among a wide variety of bacteria, the signaling molecules used for QS can differ greatly. The two best-studied systems are the acyl-homoserine lactone (AHL) QS system of Gram-negative bacteria and the (cyclic) peptide QS system of Gram-positive bacteria (160–162). S. aureus uses a QS system with autoinducing peptides (AIPs) as signaling molecules to regulate the expression of several virulence factors as well as the biosynthesis of AIPs as a function of cell population density (21, 22). The staphylococcal QS system, also known as the Agr system, is encoded by a 3.5-kb locus on the chromosome. The locus consists of the two transcriptional units, RNAII and RNAIII, driven by the two promoters P2 and P3, respectively. The RNAII transcript harbors the agrBDCA genes, which encode proteins required for AIP biosynthesis, transport, signal perception, and subsequent regulation of target genes (163, 164). The signaling cascade starts with the transcription and translation of AgrD, the propeptide precursor of AIP. AgrD is secreted and posttranslationally modified into the final peptide by AgrB, an integral membrane endopeptidase, and additionally processed via the type I signal peptidase SpsB (165, 166). When the AIP concentration in the environment reaches a certain threshold, known as a quorum, AIPs bind to the membrane-bound histidine kinase AgrC, which leads to autophosphorylation and initiation of the signal transduction cascade (21). AgrC then phosphorylates the response regulator AgrA, which, in turn, induces the expression of RNAIII from the P3 promoter (163, 167). RNAIII is the primary effector of the QS system that controls the upregulation of genes encoding secreted proteins, such as toxins and exoenzymes, and it also downregulates several genes encoding surface-associated adhesins (163, 168). Increased levels of RNAIII prevent the translation of the repressor of toxins (Rot) (169, 170), which is one of the main effectors of QS regulation. Although the majority of QS-regulated genes are regulated via RNAIII, some genes are directly regulated by AgrA. Most prominent among these is the direct regulation of RNAII from the P2 promoter, leading to elevated AIP production and a positive-feedback loop (171, 172). Additionally, the psmα and psmβ transcripts in S. aureus and S. epidermidis are directly regulated by AgrA (26, 145, 173) as well as the hld gene for δ-toxin, which is encoded within RNAIII (163). The psmα, psmβ, and hld transcripts encode peptides that belong to the family of PSMs that were already discussed above.
Although the basic principle of QS is the same in all bacteria, QS can have profoundly different impacts on biofilm formation in various bacterial species. P. aeruginosa, for example, uses its QS system in the early stages of biofilm development, where it initiates the differentiation of individual cells into a complex multicellular biofilm (174). The Agr QS system of S. aureus has the opposite effect on biofilm formation. Surface-associated proteins, such as FnBPs and SdrC, are important for cell attachment and subsequently the initiation of biofilm formation but are repressed by the Agr QS system (175, 176). Therefore, initiation of S. aureus biofilm formation requires conditions in which QS activity is low. Studies using fluorescent Agr reporter constructs have shown that the Agr QS system is not uniformly expressed in S. aureus biofilms, but only local patches of S. aureus cells show QS activity. Activation of the QS system in these local patches is followed by the detachment of these cells, creating voids in the biofilm that are filled by regrowing cells in these areas. Activation of QS in these cells at a later time point creates a periodic wave pattern of QS induction and cell detachment that is characteristic for S. aureus biofilms (177). One of the major forces driving the switch between biofilm formation and detachment is the degradation of the proteinaceous biofilm components by the Agr-regulated proteases (126, 127, 129, 178). Their upregulation by the Agr QS system is presumably a consequence of the negative regulation of Rot via the RNAIII effector (170, 179), whereas the core genome-encoded PSMs (PSMα, PSMβ, and δ-toxin) are directly regulated via AgrA (26, 145, 173). Therefore, activation of the Agr QS system leads to upregulation of PSMs and matrix-degrading enzymes, which are responsible for biofilm structuring and detachment of staphylococcal cells from the mature biofilm (45, 178, 180, 181). In agreement with these findings, inactivation of the Agr QS system enhances biofilm formation on certain surfaces, presumably by obstructing or delaying the dissemination process of the biofilm (182). Although the Agr QS system is an important regulator of S. aureus toxin production (183), the contribution of Agr to infections is also closely tied to its impact on biofilm development. The interconnections of Agr, biofilms, and infections are discussed below.
Impact of Agr on biofilm-associated infections.
Biofilm formation is a hallmark of chronic infections, and as outlined above, the Agr QS system is necessary for efficient S. aureus dissemination from a biofilm infection and subsequent spreading into neighboring tissues, a process that heavily relies on PSMs, nucleases, and proteases (24, 45, 184). Interestingly, agr mutants are defective in establishing osteomyelitis as well as endocarditis (185, 186). However, the beneficial role of the Agr QS system in staphylococcal infections is not always clear-cut. This point is emphasized by the fact that staphylococci adapt to the lifestyle of chronic infections by acquiring mutations in the agr operon. Naturally occurring agr mutants have been isolated from cystic fibrosis patients, persistent bacteremia, and biofilm-associated infections of indwelling medical devices (187–190). Similar results were obtained with four S. epidermidis isolates from indwelling catheter-associated infections, which all exhibited downregulation of RNAIII accompanied by enhanced initial attachment, eDNA release, and formation of thicker microcolonies (191). In addition, low Agr activity and low PSM production promote bacterial aggregation and biofilm formation in synovial fluid (192). An explanation for these observations might be the fact that activation of QS systems is assumed to display a high metabolic burden for the cell, leading to frequent spontaneous mutations within the QS system (193). The loss of the Agr QS system is tightly linked to a phenotype with elevated biofilm formation (146, 182, 190). While this can confer a fitness advantage in chronic infections (194, 195), it is also a potential maladaptation in the context of bacterial dissemination (196).
Modulators of Agr signaling and biofilm formation.
Bacterial cells constantly sense environmental cues and change their transcription profiles accordingly to maintain viability under diverse conditions. These environmental cues can be physicochemical parameters such as pH, temperature, and shear forces from fluid flow. Several of these parameters and nutrient availability can influence biofilm formation via the Agr QS system. Among the earliest environmental cues that has been shown to affect the Agr system are acidic metabolites derived from glucose catabolism (197). A subsequent study by Weinrick and colleagues showed that a decrease in pH in the absence of glucose was sufficient to inhibit agr transcription (198). In addition, glucose depletion of existing biofilms was shown to activate the Agr QS system, resulting in biofilm dispersal (178). In contrast to S. aureus, the addition of excess glucose is not necessary to promote biofilm formation in S. epidermidis, although it is not known whether this is related to Agr regulation (199). Another interesting link between glucose and biofilm formation was presented by Seidl and colleagues (200). The catabolite control protein A (CcpA), which regulates gene expression in response to different carbon sources, was shown to be important for biofilm growth in the presence of glucose. Moreover, the formed biofilm was susceptible to DNase, proteinase K, and sodium metaperiodate, showing that the biofilm matrix most likely consisted of proteins, eDNA, and PIA. In agreement with this observation, CcpA stimulated the upregulation of cidA, which encodes a holin involved in eDNA release, and icaA, a gene essential for PIA production (200). It is important to mention that an earlier study already established that ccpA deletion results in downregulation of RNAIII (201). Further studies will be needed to address this discrepancy. Since we have already established above that the Agr QS system is a negative regulator of biofilm formation, these described observations are a plausible explanation why the majority of biofilm studies with S. aureus are carried out using glucose-supplemented media (71, 177, 202).
Metals are essential micronutrients for bacteria, and many enzymes involved in vital bacterial processes rely on metals as cofactors. Within the host, S. aureus must compete for iron that is bound to iron-binding proteins such as hemoglobin, ferritin, lactoferrin, and transferrin (203, 204). In S. aureus, intracellular iron concentrations are regulated by the ferric uptake regulator (Fur), which is also able to interact with other metals such as cobalt, copper, manganese, and nickel (205). Biofilm formation in S. aureus is induced under iron-limiting conditions (206), and the increase of biofilm formation was demonstrated to be regulated by Fur, Agr, and the S. aureus exoprotein expression (Sae) regulator through the production of the two secreted proteins, the extracellular adhesion protein (Eap) and the extracellular matrix protein-binding protein (Emp) (207). Furthermore, Fur is required for elevated expression of Agr, Rot, and the Sae regulator under iron-limited conditions (208). These three regulatory systems play critical roles in S. aureus biofilm formation and are described in further detail elsewhere in this section. In contrast to the aforementioned studies, S. aureus strain SA113 showed reduced biofilm formation in the presence of an iron chelator, and the effect could be reversed by addition of iron to the growth medium (209). These contrasting results show that biofilm formation of S. aureus in response to various iron concentrations is likely strain dependent. For S. epidermidis, it was shown that iron excess as well as iron deficiency leads to impaired biomass accumulation and changes in the biofilm structure, ultimately leading to stunted biofilm formation (210).
The effect of physicochemical factors on staphylococcal biofilm formation have become a major research topic over the last couple of years. When colonizing indwelling medical devices such as catheters, staphylococci are subjected to mechanical shear forces from fluid flow, the local geometry, and surface chemistry of the medical device. Fluid flow can change the ability of bacterial cells to form biofilms (211, 212) by removing the QS signal molecules from the site of the producing bacteria (212, 213). Nonetheless, these signal molecules might then activate QS in cells located further downstream of the flow. This can lead to a relatively higher biofilm formation rate in regions located at sites on the upstream end of the flow than at sites further downstream. Even in environments with fluid flow, certain topographic regions, like small crevices, show a limited flow rate that allows the accumulation of QS molecules and subsequent activation of the S. aureus QS system (212). Another important factor regarding QS under flow conditions is biofilm thickness. Fluid flow is effective in removing QS signals only at the biofilm-liquid interface, while AIPs produced at the bottom of the biofilm are less affected by fluid flow. Therefore, AIPs can accumulate deep within the biofilm and activate the S. aureus QS system, subsequently leading to biofilm dissemination (212). Even though the interconnection of fluid flow, QS, and biofilm formation has not been directly studied for S. epidermidis, among a collection of over 100 clinical isolates, significantly more strains from high-shear environments either carried the ica operon or had other genetic determinants to increase biofilm formation (117). Altogether, these findings suggest that staphylococcal biofilm formation is increased in areas with high fluid flow due to the removal of the QS signal molecules in the case of S. aureus (212) and due to the presence of biofilm-enhancing determinants in S. epidermidis (117).
Regulatory Network of Staphylococcal Biofilm Formation
Several other regulatory systems are important during the biofilm development process, and some of these systems are interconnected with the Agr QS system. This network allows S. aureus to fine-tune its response to changing environmental conditions and modulate biofilm development. A simplified sketch of the Agr QS system and its regulatory network with the most important global regulators is depicted in Fig. 2 and described in the following section.
FIG 2.
Regulatory network of staphylococcal biofilm formation. Shown is a simplified overview of the Agr quorum sensing system, its regulatory interaction with the most important biofilm regulators (LytSR, SigB, CodY, SaeRS, MgrA, SarA, and Rot) and their influence on the molecular determinants of biofilm formation. Black arrows indicate stimulation, and red blunted arrows indicate repression.
The Sae two-component system.
The S. aureus exprotein expression (Sae) two-component system (TCS) consists of the SaeS sensor histidine kinase and the SaeR response regulator (214, 215). SaeS recognizes host signals such as human α-defensins and regulates the expression of various exoproteins important for pathogenesis and biofilm formation, among them α-hemolysin (Hla), the MSCRAMMs FnbA and FnbB, and the extracellular thermonuclease Nuc (141, 216–220). Fibronectin-binding proteins are known to be important for biofilm formation at the level of intercellular accumulation, allowing cells to bind together via homophilic interactions between surface proteins (103, 104), while Nuc was shown to cleave eDNA, a structural component of S. aureus biofilms (43, 71). An early biofilm dispersal event, termed exodus, that is part of the five-phase biofilm model, takes place before tower formation in S. aureus biofilm maturation and is independent of Agr QS (46). With a set of elegant experiments, it was shown that expression of Nuc preceded the exodus event, which likely led to degradation of eDNA in the biofilm matrix (46). The exodus event could directly be linked to the SaeRS system, as neither an sae nor a nuc mutant exhibited the exodus phenotype. Corroborating these results, Moormeier and colleagues showed that nuc transcription was regulated by the SaeRS system in their biofilm model (46). SaeS shows polymorphism across S. aureus strains (221). For example, a point mutation in SaeS (SaeSP) present in S. aureus Newman leads to hyperactivity of the SaeRS TCS, which was linked to the inability of this strain to form robust biofilms (221, 222). In a recent study, it was also determined that several Sae-regulated virulence genes in S. aureus are stochastically expressed in a subpopulation of cells in the nascent biofilm with an expression pattern matching the one of the thermonuclease Nuc (223). In S. epidermidis, mutation of saeRS leads to enhanced biofilm formation (224) concomitant with higher Aap expression and eDNA release (225).
The SarA protein family.
The staphylococcal accessory regulator SarA is a general transcription factor that binds to AT-rich sequences and activates or represses target gene expression (226, 227). SarA is transcribed on three different overlapping transcripts, initiated by three distinct upstream promoters (P1 to P3) (228). The SarA regulator directly modulates the expression of certain virulence factors and upregulates agr expression by bending the agr promoter region to enhance AgrA-dimer interaction (226, 229, 230). SarA also induces the expression of hla, fnbA, and fnbB but inhibits the transcription of protein A (spa) (216, 226, 231–234). The most important role of SarA regarding biofilm formation is the repression of extracellular proteases. A sarA mutant produces high levels of each protease (128, 235), has a decreased capacity to bind fibronectin (236), and is not able to form a static or flow cell biofilm (123). The observed absence of biofilm formation in sarA mutants could be restored only by concomitant inhibition of all three classes of proteinases (serine, cysteine, and metalloprotease) (125) or by simultaneous mutation of the four extracellular proteases Aur, ScpA, SspA, and SspB (237). SarA itself is subject to positive regulation by the alternative sigma factor SigB (σB) and consequently also by the σB regulator RsbU (238–241). These two regulators are discussed in more detail in the section below.
Although sarA mutation drastically decreases biofilm formation in most S. aureus strains, some exceptions exist. As described above, S. aureus strain Newman, for example, has a point mutation in SaeS which results in the constitutive activation of the SaeRS regulatory system and represses some protease production. Therefore, mutation of sarA in strain Newman is counterbalanced by the protease repression via the constitutively active SaeRS system, enabling this strain to form biofilms, even when sarA is mutated. These observations show that SaeRS and SarA synergistically repress protease production and drive biofilm formation in S. aureus (235). In S. epidermidis, SarA-mediated regulation of biofilm formation is highly strain dependent. It positively regulates the expression of the ica operon, and sarA mutations lead to biofilm-negative phenotypes in certain S. epidermidis strains (242). However, mutation of sarA in a PIA-negative S. epidermidis strain induced biofilm formation by overexpression of extracellular matrix-binding protein (Embp) and eDNA production via the upregulation of the metalloprotease SepA, which, in turn, processes the autolysin AtlE (243).
Besides SarA itself, the SarA regulator family also includes SarS, SarT, SarR, SarU, SarX, SarZ, MgrA, and Rot. The latter is known to promote the expression of surface proteins and immunomodulators and represses the production of extracellular toxins and enzymes (244–246). The regulatory activity of Rot is prevented by downregulation of rot translation via the QS effector RNAIII (169, 247), which, for example, leads to the activation of staphylococcal superantigen-like proteins (Ssl) in agr-negative S. aureus strains (248). Mutation of rot showed a significant decrease in biofilm formation in many clinical isolates (170). Similar to the observations made for SarA (described above), Rot was also shown to repress protease transcription, and the deletion of protease-encoding genes, as well as treatment with protease inhibitors, restored biofilm formation in a rot mutant. In contrast to the above-described sarA mutant, the general cysteine protease inhibitor E-64 or staphostatin SspC, a specific inhibitor of staphopain B, is necessary to restore biofilm formation (170). Further evidence suggests that Rot can potentially regulate all four protease loci, but the degree of regulation is highly medium dependent (170, 179, 245, 249). In summary, SarA and Rot, two members of the SarA regulator family, are important regulators of biofilm formation primarily through the repression of protease production. Another important member of the SarA protein family is the global regulator MgrA, which acts as the main effector of the ArlRS TCS (250, 251). MgrA acts as a repressor of eight cell wall-anchored proteins, and mutation of mgrA leads to loss of bacterial clumping but also to an increase in biofilm formation. The latter was attributed to the derepression of SasG, a surface protein that promotes intercellular interactions (250, 252) and was implicated in abiotic surface attachment (74).
The alternative sigma factor SigB.
The survival and successful adaptation of staphylococci under extreme environmental conditions are possible through the regulation of stress response genes via alternative sigma factors. Sigma factors direct the binding of the catalytic core of the RNA polymerase to specific promoter regions to induce gene transcription. The known S. aureus sigma factors are σA, σB, σH, and σS, with σA being the housekeeping sigma factor for transcription; σS seems to be important during starvation and exposure to elevated temperatures, and the alternative sigma factor σB acts as a transcription factor in response to different environmental stresses (240, 253–256). The recently identified sigma factor σH was indicated to play a role in staphylococcal natural competence regulation (257), but according to a more recent study, it does not regulate all components of the natural competence machinery and therefore can only induce low-level competence for natural genetic transformation (258). σB (sigB) is expressed from and regulated by the rsbUVW-sigB operon. The anti-sigma factor RsbW renders σB inactive when the cells are not under environmental stress. RsbU, which dephosphorylates RsbV (anti-anti-σB factor), is a positive regulator of σB and essential for the activation of σB (259). Donegan and Cheung showed that full σB activity is also dependent on the upstream mazE promoter (PmazE) of the MazEF toxin-antitoxin (TA) system, due to its transcriptional linkage with the downstream sigB operon (260). PmazE itself is positively regulated by SarA and downregulated by σB (260).
Among the >250 genes regulated by σB, genes for several adhesins for early stages of biofilm formation are upregulated (e.g., FnbA and ClfA) and various genes for exoproteins, known to be important for biofilm dispersal (e.g., nuclease and proteases), are repressed by σB (241, 255, 261–264). σB reduces the RNAIII levels of the Agr system, and it can also influence SarA expression depending on the strain background and environmental conditions (238, 265). Furthermore, σB was shown to downregulate the SaeRS TCS in certain strains (266).
It was also shown that σB promotes microcolony (267) and biofilm (268) formation. An S. aureus mucosal isolate was shown to form a biofilm in an in vitro static biofilm model induced by high-salt conditions. A mutation in the σB-encoding gene abolished the induction of biofilm formation by osmotic stress, showing that σB is crucial for stress-induced biofilm formation (268). In addition, sigB mutation has been demonstrated to influence PIA-dependent biofilm formation. It is important to mention that the available literature on PIA-dependent biofilm formation and its regulation by σB is contradictory (268–271). This phenomenon can most likely be explained by S. aureus strain-specific properties and the use of different media and biofilm systems across these studies. For example, mutation of sigB in the methicillin-sensitive mucosal isolate S. aureus MA12 correlated with reduced transcription of the ica operon (268). In contrast, the biofilm-negative phenotype in a sigB deletion mutant in S. aureus SH1000 and the USA300 strain LAC, a community-associated methicillin-resistant S. aureus (CA-MRSA) isolate, did not correlate with changes in the ica-dependent PIA levels but instead was caused by enhanced Agr activity and consequent increased protease activity (126). This might be explained by the fact that both strains have the capacity to form PIA-independent biofilms (178, 241, 272). In a recent study, Valle and colleagues evaluated the contribution of σB to biofilm formation in several genetically unrelated S. aureus strains. In this study, sigB mutants produced higher levels of PIA-dependent biofilms, which correlated with increased accumulation of the IcaC protein, indicating that σB acts as a repressor of PIA biosynthesis (271). Besides the aforementioned σB-dependent biofilm regulation via the ica operon and proteases, σB also negatively regulates the expression of the secreted thermonuclease Nuc. Cell-free supernatants of an USA300 sigB mutant were able to inhibit biofilm formation of different S. aureus strains. Subsequent fractionation and mass spectrometry analysis showed Nuc to be the active component in the supernatant responsible for biofilm inhibition. Deletion of the nuc gene in several S. aureus strains, including the USA300 lineage, led to enhanced biofilm formation, and mutation of nuc in a sigB background partially repaired the biofilm-negative phenotype of the sigB mutation (71). In S. epidermidis, mutation of rsbU as well as σB leads to decreased biofilm formation. This phenotype is explained by the upregulation of IcaR and consequent downregulation of the ica operon in the aforementioned mutants (273–275). In summary, the alternative sigma factor σB exerts regulatory control over several genes involved in the formation of different types of biofilms, including protein-, eDNA-, and PIA-containing biofilms.
The transcriptional repressor CodY.
Virulence factor production and biofilm formation can also be regulated by nutritional availability and the metabolic potential of the cell. Gram-positive bacteria can adapt to the stationary growth phase and starvation by regulating their gene expression via CodY, a GTP-sensing pleiotropic transcription repressor (276). More than 100 genes in S. aureus are directly or indirectly regulated by CodY (277, 278). Nutrient availability is sensed by CodY through the amount of the effector molecules, branched-chain amino acids (BCAAs; isoleucine, valine, and leucine), and GTP. Under sufficient nutrient conditions, CodY can interact with its effector molecules, leading to a conformational change of CodY, which promotes enhanced affinity of CodY to its DNA binding sites (276). CodY is thought to inhibit transcription by blocking RNA polymerase from binding to the promoter or acting as a roadblock for mRNA transcription (278). Under nutrient-limited conditions, the intracellular concentration of GTP and BCAAs decreases, CodY loses its DNA binding affinity, and CodY-repressed genes are activated. In S. aureus, CodY-regulated genes are involved in the primary metabolism, transport, and virulence (277, 278), and notably, CodY strongly represses the agr locus (279). In vitro binding assays confirmed interaction of CodY not with the P2 and P3 promoter regions of the Agr system but rather with a region within the agrC gene (278). A recent study confirmed the elevated expression of the agr locus and RNAIII in a codY mutant, as well as in vitro binding of CodY within the agrC gene. Nevertheless, the same study showed that this had no effect on agrA expression. In addition, Roux and colleagues observed in vitro binding of CodY to the P2 and P3 promoters, although with low affinity (280). Since RNAIII positively regulates transcription and translation of toxins and hemolysins (234, 281), CodY-dependent derepression of RNAIII in a codY mutant leads to higher hemolytic activity in culture supernatants (279). CodY was recently shown to repress rsaD, a small regulatory RNA (sRNA) that contributes to cell death regulation during weak acid stress (282), potentially leading to eDNA release and biofilm formation.
PIA-dependent biofilm formation is also regulated by CodY, and different biofilm phenotypes were shown in codY mutant strains. A transposon insertion in the codY gene in the clinical isolate S30 yielded reduced biofilm formation and PIA production (283). In contrast, a codY allelic replacement mutation in the two S. aureus clinical isolates SA564 and UAMS-1 showed an increase in biofilm formation, most likely resulting from the higher transcription of icaA and elevated PIA synthesis during in vitro growth (279). CodY-mediated regulation of the icaADBC locus is independent of IcaR (279), SarA, and RNAIII (278). In addition to the regulation of PIA-dependent biofilms, CodY represses two important modulators of biofilm formation and maturation, namely, extracellular proteases and nuc, the latter of which is indirectly regulated via the repression of the Sae TCS (284, 285). Most recent work from the Brinsmade lab investigated the contribution of CodY to biofilm formation in the clinical isolate SA564, a robust biofilm producer. Based on their results, they suggest that elevated PIA synthesis in cells with low CodY activity is the main contributor to biofilm formation. Moreover, in their model, PIA and eDNA can associate and have a synergistic contribution to biofilm formation rather than being mutually exclusive (286). Taken together, these findings show that the nutritional status of the cell has great influence on biofilm formation and is integrated into the regulatory network of S. aureus via CodY.
ica operon regulation.
PIA synthesis and regulation of the ica operon are governed by a complex regulatory network, the details of which are not completely understood. The icaADBC operon is negatively regulated by IcaR and TcaR (90). IcaR is a DNA-binding protein (287) located upstream of the icaADBC operon and acts as a repressor of icaA transcription in S. epidermidis (89) and S. aureus (90). A recent report showed that the 3′ untranslated region (3′ UTR) of icaR can base-pair with the Shine-Dalgarno sequence at the 5′ UTR of icaR, which interferes with translation, generates a double-stranded RNase III substrate, and consequently leads to reduced IcaR-mediated biofilm repression (288). Synthesis of IcaR is upregulated by the stress response regulator Spx (289) and indirectly downregulated by the AraC/XylS-type regulator Rbf via SarX (290, 291). TcaR, a transcriptional regulator of the teicoplanin-associated locus (292), was shown to act together with IcaR as a repressor of the ica operon in S. aureus and S. epidermidis (90, 293). In contrast, transcription of icaA is induced by the TCS SrrAB (staphylococcal respiratory response regulator) under anaerobic growth conditions (294, 295). Besides that, other environmental stresses, including sodium chloride (296, 297) and subinhibitory concentrations of certain protein synthesis inhibitors, such as tetracycline, were shown to upregulate the ica operon and induce PIA synthesis in staphylococci (298). In addition, ica operon expression was also shown to be activated by SarA (270) and partially by σB (268, 297). Going further, transcription of the ica locus is regulated by a specific 5-bp motif in the promoter region, and loss of this binding site causes a PIA-overproducing and biofilm-positive phenotype (287). Besides the described regulatory systems, PIA synthesis can be altered by phase variation via the insertion of the IS256 element into the ica operon of S. epidermidis and S. aureus (299, 300) or through mutation by slipped-strand mispairing in a tandem repeat in icaC (301).
An additional layer of regulatory elements are noncoding RNAs, of which several have been shown to participate in ica regulation in different staphylococcal species. One such noncoding RNA is icaZ, present only in ica-positive S. epidermidis strains and not in S. aureus. This regulatory molecule is a novel long noncoding RNA that was shown to downregulate icaR mRNA translation by targeting the 5′ UTR of icaR (302). Two additional RNA regulator molecules have been identified very recently. In contrast to icaZ, both are small noncoding RNAs, referred to as RsaI and RsaE. In S. aureus, RsaI is repressed by CcpA in the presence of high glucose concentrations and derepresses biofilm formation by binding to the 3′ UTR of icaR (303). RsaE has an interesting bifunctional role in S. epidermidis biofilm development, even though the effect is highly strain specific. The RNA molecule exists in a full-length 100-nucleotide form and a shorter processed form. Processed RsaE interacts with the 5′ UTR of icaR and leads to increased PIA production. The unprocessed form, on the other hand, interacts with the antiholin-encoding lrgA mRNA, leading to bacterial lysis and eDNA release, and potentially enhances biofilm matrix production (304). Given its unarguable importance for staphylococcal biofilm formation, it is not surprising that the ica operon and subsequent PIA biosynthesis are subject to regulation and control by a multitude of different regulatory systems that allow the bacterium to tightly control production of this polysaccharide.
The LytSR two-component system.
Cell death and lysis of S. aureus are regulated by the LytSR TCS (305) via a bacteriophage-like holin/antiholin system (306–308). The holin is encoded by cidA and facilitates cell lysis through increased extracellular murein hydrolase activity (306, 307). Confirming the role of CidA, a cidA mutant strain exhibited decreased lysis resulting in lower amounts of eDNA and impaired biofilm formation (202). The antiholin counteracts the activity of CidA and is encoded by lrgA (309). Mutation of the lrgAB operon results in the formation of more adherent biofilms with a higher eDNA content, demonstrating its role as a CidA antagonist (43). As the Cid/Lrg system controls such vital processes as lysis and cell death, its expression has to be tightly controlled. Several regulatory systems exert control over the Cid/Lrg system, including the LysR-type transcriptional regulator CidR (310), the alternative sigma factor σB (311), and, as already mentioned in the beginning of this section, the TCS LytSR (305). The lrgAB operon is also regulated by SarA and the Agr QS system (312), as well as by Rot (313). In addition, the presence of certain metabolites can alter the transcription of the Cid/Lrg system (314). The best-studied regulation of the Cid/Lrg system is exerted by the LytSR TCS, which positively regulates the lrgAB operon. LytSR is induced by the dissipation of the membrane potential (ΔΨ), and upregulation of lrgAB by alterations in ΔΨ is subsequently abolished in a lytS mutant. Furthermore, mutation of lytS decreases lrgAB transcription and leads to a thicker and more adherent biofilm with more eDNA in its matrix (315). These phenotypes clearly underscore the role of LytSR in regulation of the Cid/Lrg system and biofilm formation. Interestingly, expression of lrgAB is specifically upregulated in tower structures of S. aureus biofilms (316), and a small fraction of towers formed by a lytS mutant still exhibited strong lrgAB expression (317). This suggests that a LytS-independent pathway for LytR activation exists in these tower structures (317). The LytSR TCS is also present in S. epidermidis strains, and one study showed that mutation of lytSR in S. epidermidis strain 1457 leads to elevated biofilm formation (318), indicating that LytSR has an as-yet-unexplored role in S. epidermidis biofilm formation.
The LuxS/AI-2 system.
The LuxS/autoinducer-2 (AI-2) system, originally identified in Vibrio harveyi (319, 320), is thought to be an interspecies communication system due to its wide distribution in many bacterial species. LuxS synthesizes AI-2 (321), a furanosyl borate diester (322) that acts as the extracellular signaling molecule of the system. Sequence analysis showed that a functional LuxS/AI-2 system is conserved in different S. aureus strains (323). However, several studies have investigated the role of AI-2 in staphylococcal biofilm formation, with disparate results. Two early studies, one for S. aureus and one for S. epidermidis, identified LuxS as a negative regulator of biofilm formation, and external addition of AI-2 to a luxS mutant strain restored the wild-type phenotype (324, 325). These results were attributed to the activation of icaR transcription by AI-2, resulting in the downregulation of icaA (324). It is also important to note that an S. epidermidis luxS mutant was highly impaired in the production of PSMs, which are important for biofilm maturation and dispersal (326). The role of LuxS as a negative regulator of biofilm formation in S. aureus was corroborated in a recent study that showed increased PIA production in a LuxS-negative strain, and this phenotype was linked to increased expression of the positive biofilm regulator Rbf (327). In contrast to the aforementioned studies, Xue and colleagues observed downregulation of icaR and subsequent upregulation of the ica operon by AI-2 in S. epidermidis (328). In addition, upregulation of the ica operon was observed when the AI-2 precursor 4,5-dihydroxy-2,3-pentanedione (DPD) was added in micromolar concentrations (328). Yu and colleagues, on the other hand, observed changes in S. aureus biofilm formation only when DPD was added in the low nanomolar range; when this signal was provided at higher concentrations, no effect on biofilm formation was observed (324). This shows that despite the discussed role of AI-2 as a universal signaling molecule, the effect of AI-2 on each species might be very different and even concentration dependent.
The second messenger cyclic-di-AMP.
In bacteria, signaling nucleotides such as cyclic AMP (cAMP) and guanosine tetraphosphate (ppGpp) are involved in the regulation of carbon metabolism and the stringent response but have also been linked to the expression of virulence genes and biofilm formation (329). Two additional signaling nucleotides, cyclic-di-GMP and cyclic-di-AMP, have gained much attention in the scientific community. The second messenger c-di-GMP is predominantly found in proteobacteria, in which it was shown to regulate a multitude of different processes, including motility (330), cellulose production (331), and biofilm formation (332). In contrast, c-di-AMP signaling is mainly associated with Gram-positive bacteria, especially from the phylum Firmicutes (333). Production of c-di-AMP by S. aureus was first reported in 2011 by Corrigan and colleagues and is involved in the regulation of cell size, as well as membrane and cell wall stress (334). S. aureus synthesizes c-di-AMP from two molecules of ATP by the diadenylyl cyclase (DAC) enzyme DacA and degrades it with the phosphodiesterase (PDE) GdpP (334). Disruption of dacA in S. aureus was not possible in a screen for essential genes, indicating that c-di-AMP formation is an essential process for S. aureus (335). Intracellular c-di-AMP levels are sensed at least by the four receptor proteins KtrA, KdpD, PstA, and CpaA (336). Deletion of gdpP in S. aureus SEJ1 led to a 3-fold increase in biofilm formation (334), indicating that high c-di-AMP levels foster biofilm formation in this strain. It is not clear if this is a general phenotype of S. aureus, because neither S. aureus USA300 LAC nor its corresponding gdpP mutant was able to form a biofilm under the conditions used by Corrigan and colleagues (334). In another recent study, several homogenous oxacillin-resistant (HoR) S. aureus isolates with either single nucleotide polymorphisms (SNPs) or deletions in gdpP showed a decrease in icaADBC and agr expression and elevated expression of penicillin binding protein 2 (PBP2). These regulatory changes were accompanied by impaired polysaccharide-type biofilm development and a change to a more proteinaceous biofilm (272). Besides its influence on the polysaccharide and protein components of the biofilm matrix, c-di-AMP is also involved in eDNA release; more specifically, mutation of gdpP results in reduced levels of eDNA. In accordance with this observation, growth of S. aureus under biofilm-inducing conditions results in a GdpP-dependent drop in c-di-AMP levels (337). As different studies have shown different biofilm phenotypes for gdpP mutants, the influence of c-di-AMP on biofilm formation is likely to be strain dependent. Furthermore, dacA expression is elevated in tower structures of flow cell biofilms as well as in a static biofilm model, compared to planktonic growth. Surprisingly, between 30% and 50% of total c-di-AMP produced in biofilm cultures was found to be extracellular due to autolysis of S. aureus, and the extracellular c-di-AMP levels were sufficient to significantly elevate interferon beta (IFN-β) protein expression in an orthopedic implant biofilm infection model (338). In summary, these findings indicate that c-di-AMP levels might play a role in the biofilm matrix composition of staphylococci and could have an important impact on host defenses during chronic biofilm infections.
THE IMPORTANCE OF STAPHYLOCOCCAL BIOFILMS DURING INFECTION
Among the many types of infection that can be caused by staphylococcal species, biofilm-associated ones are the most challenging to treat owing to the increased resistance of staphylococci to antibiotic treatments and the host immune system (16). In adults, S. aureus is the major pathogen associated with osteomyelitis (339, 340). After initial biofilm formation, the host immune system attacks the invading bacteria, resulting in host tissue damage that further promotes biofilm development (16). Another common but serious problem is the formation of biofilms on indwelling medical devices (e.g., catheters) and implants (341–343). Usually, the indwelling device or implant readily becomes coated by host matrix, which eventually allows initial attachment and subsequent biofilm formation by S. aureus and other staphylococci (344). The major source of device-related infections is cross-contamination during surgery, and in this regard, colonization of the skin or other body sites of the host or health personnel is a major risk factor for implant-associated infections (343, 345, 346). Another type of infection that is very often associated with staphylococcal biofilms is a chronic wound. Staphylococci are among the pathogens most frequently isolated from chronic wounds (347), where they often reside as cellular aggregates, engulfed in a self-produced extracellular matrix (348). In that case, the wound healing process can be substantially prolonged due to secreted factors as well as the strong immune response to the bacteria present in the wound (349–351). According to recent studies, S. aureus has also become the leading cause of infective endocarditis (IE) worldwide (352–354). Whereas the role of biofilms in IE is obvious when IE is associated with an implanted cardiac device, involvement of biofilm formation in native valve IE is still under debate (355). Nevertheless, it has been shown that there is a correlation of in vitro biofilm formation and the ability to cause persistent bacteremia (356, 357), indirectly associating staphylococcal biofilm formation with IE.
Immune Evasion Mechanism
Staphylococcal biofilms can circumvent immune-mediated clearance by several mechanisms. First, it was proposed that the deficiency of phagocytes to kill staphylococci within a biofilm is mainly due to the physical diffusion barrier of a biofilm where immune cells are barely able to penetrate through the matrix (30). However, this is not supported by in vitro data, showing that human leukocytes are able to adhere and penetrate the biofilm matrix, even under conditions mimicking physiological shear (358). To evade the host immune system, S. aureus rather relies on directly attacking host immune cells with secreted factors (e.g., nucleases, PSMs, and hemolysins) and by skewing host immunity toward anti-inflammatory responses that favor staphylococcal persistence (13, 359). It was shown in an in vivo model that S. aureus biofilms are able to circumvent bacterial recognition pathways, such as Toll-like receptor (TLR) 2 and TLR9 signaling (360). Recent data by Bhattacharya and colleagues show that cells in an S. aureus biofilm secrete high concentrations of the leukocidins Panton-Valentin leukocidin (PVL) and γ-hemolysin and that either of these can induce NETosis to avoid clearance by neutrophils (361). NETosis, also referred to as neutrophil extracellular trap (NET) formation, is a mechanism employed by neutrophils that involves the controlled release of chromatin, modified histones, DNA, and granular proteins, resulting in the formation of web-like structures that can entrap and kill planktonic cells but are not able to clear cells within a biofilm (361, 362). In addition, modification of PIA by deacetylation of its poly-N-acetylglucosamine molecule is important for biofilm formation but was also shown to mediate resistance to neutrophil phagocytosis and AMPs (87). The immune response against S. epidermidis biofilms is generally less pronounced than for planktonic cells. This has been attributed to reduced activation and production of the transcription factor NF-κB and the cytokine interleukin-1β by macrophages (363), reduced antibody-mediated phagocytosis (364), and reduced deposition of the complement protein C3b and immunoglobulin G on the bacterial surface (365). These phenotypes have mainly been linked to the presence of PIA in the biofilm matrix (366), but the exact molecular mechanisms responsible for immune evasion of S. epidermidis biofilms are still to be determined.
Antibiotic Resistance
In general, it has been postulated that cells within a biofilm are 10 to 1,000 times more tolerant to antibiotics and AMPs than are planktonic cells (367). Although some antibiotics and AMPs show a partial decrease in their ability to penetrate the biofilm matrix (368–371), the high tolerance of biofilms to therapeutic agents is mainly attributed to the altered cell metabolism within a biofilm (372). Staphylococcal biofilms often show a high degree of cell heterogeneity with distinct specialized subpopulations of cells in the community. Cell heterogeneity can exist in staphylococcal biofilms due to the development of persister cells or different QS expression patterns or, for instance, by the acquisition of antibiotic resistance genes through horizontal gene transfer (373, 374). Indeed, high bacterial cell density within a biofilm community has been suggested to promote the transfer of resistance genes between bacterial cells (375). Besides the genetic diversity in biofilms, these communities are often a reservoir for antibiotic persister cells, which represent a subpopulation of cells that can survive antibiotic treatment without becoming resistant (376). In general, cells within a biofilm often show reduced proliferative and metabolic activity, leading to an increased tolerance to antibiotics targeting the bacterial cell wall, as well as DNA or protein synthesis inhibitors (31–33). Mapping of spatial patterns of DNA replication, protein biosynthesis, and oxygen concentrations in S. aureus and S. epidermidis biofilms allowed the distinction of four subpopulations: aerobically growing, fermentative growing, dormant, and dead cells (377).
The antibiotic rifampin has a high diffusion rate within biofilms, host cells, and bone tissue as well as bactericidal activity against S. aureus and S. epidermidis in the stationary phase of growth. Although rifampin is highly effective against staphylococcal biofilms and is often proposed for the treatment of staphylococcal PJIs, there is also a high risk of emergence of rifampin resistance during treatment, and therefore, it is administered only in combination with other antimicrobial agents (378–381). It is also generally assumed that bacterial cells within a biofilm are likely to encounter subinhibitory concentrations of antibiotics, which were shown to potentially stimulate biofilm production and alter the composition of the biofilm matrix (382–384).
ANTIBIOFILM STRATEGIES
The above-mentioned recalcitrance of staphylococcal cells within a biofilm makes it very difficult to effectively eradicate a biofilm once formed. Thus, implant-associated staphylococcal biofilm infections normally require surgical debridement and prolonged antimicrobial therapy (385). Besides these conventional therapeutic antibiofilm strategies, a broad range of substances with different mechanisms to prevent or eradicate staphylococcal biofilms have been identified. Those molecules either interfere with the Agr QS system (e.g., savirin and apicidin), degrade the biofilm matrix (e.g., recombinant DNase I and Dispersin B), or have bactericidal activity against bacterial cells within biofilms (e.g., lysostaphin and AMPs). Another highly promising strategy is to modify biomaterial surfaces such as catheters and implants to prevent staphylococcal adhesion and subsequent biofilm formation. In that regard, implants coated with surfactants, antibiotics, or antimicrobial peptides alone or in combination have shown promising effects (386–388). Another exciting new strategy to combat staphylococcal biofilms is the application of nanoparticles as delivery systems for conventional antibiotics or in the form of metal nanoparticles with antimicrobial activity. Poly(lactide-co-glycolide) (PLGA) nanoparticles, for example, have been successfully used as delivery systems for the antibiotics levofloxacin and ciprofloxacin to inhibit S. aureus biofilm formation (389, 390). Metal nanoparticles have been used in combination with near-infrared light irradiation for photothermal ablation of staphylococcal biofilms (391), and liposomes were shown to be promising delivery vehicles for antibiotics such as levofloxacin (392, 393) and vancomycin, with the latter having activity against mature biofilms (394).
Quorum-Sensing Modulators
Interference with bacterial communication, also called quorum quenching, has developed into a popular and productive research field over the last decades (395, 396). The idea is to identify molecules that specifically target the QS system, leading to attenuation of exoenzyme and toxin production while having no inhibitory effect on bacterial growth in order to limit resistance development. For example, a high-throughput screen was used to identify savirin (S. aureus virulence inhibitor), a small-molecule inhibitor of the Agr QS system that enhances host defense and S. aureus clearance in a murine skin infection model (397). QS inhibitory activity was also reported for natural products extracted from the leaves of the European chestnut (Castanea sativa) (398) and, recently, the fungus-derived apicidin (399). Parlet et al. showed that apicidin successfully prevents agr activation and efficiently attenuates MRSA virulence in apicidin-treated mice (399). Another promising strategy is the development of AIP mimetics to inhibit the AgrC receptor. This strategy is based on the fact that there are four Agr classes in S. aureus; each synthesizes a unique AIP molecule (AIP-I to AIP-IV) (395, 400), and except for AIP-I and AIP-IV, they show cross-inhibition, also known as Agr interference (22, 164, 401). A series of AIP mimetics based on the structure of AIP-III exhibited AgrC inhibition in all four Agr classes (402, 403). An additional study revealed the structural features important in AgrC-AIP interaction (404) that will eventually stimulate the development of new QS inhibitors. One consequence of Agr inhibition is the development of a thicker biofilm in vitro. This likely stems from the inhibition of Agr-regulated PSMs and protease biosynthesis, which are important for biofilm maturation but also foster dispersal of cells from the biofilm (45, 123, 126, 178). Therefore, QS inhibitors could be associated with an increase in S. aureus biofilm formation, raising concerns about their impact on chronic infections. Nonetheless, as already noted above, agr mutants have a defect in establishing infections like osteomyelitis and endocarditis (185, 186), and thus, QS inhibition might impact the biofilm structure in the infection but also lead to an overall less virulent phenotype. A possible approach to destroy mature S. aureus biofilms in vivo is the activation of the Agr QS system by the addition of exogenous AIPs, which was shown to lead to complete disassembly of mature biofilms and restoration of antibiotic susceptibility (178, 181).
Bioactive Molecules Interfering with Staphylococcal Biofilms
In general, there are two classes of bioactive molecules that can potentially be used to treat biofilm-associated infections. The first class are biofilm dispersal agents, which can degrade the biofilm matrix, and the second class are biofilm eradication agents, which are capable of killing the bacteria embedded in a biofilm, including metabolically inactive cells. Among the most promising classes of molecules to fight biofilm infections are biofilm-degrading enzymes. The usability of such enzymes is highly dependent on the composition of the biofilm matrix. For PIA-dependent biofilms, the PIAse Dispersin B was shown to be an effective agent (387, 405, 406), while eDNA-based biofilms can be dispersed by DNase I (43, 181) and protein-based biofilms are susceptible to proteinase K and different proteases secreted by staphylococci themselves, such as staphylococcal serine proteases, staphopains, and the S. epidermidis serine protease Esp (24, 133, 181). Importantly, the dispersal process of biofilms leads to the spreading of bacterial cells, which can then lead to a reinfection of the host (407). Therefore, biofilm dispersing agents need to be coadministered with antibiotics in order to effectively combat biofilms and prevent subsequent bacterial regrowth (387, 408). In addition, administration of proteases as biofilm dispersing agents could lead to degradation of nontarget proteins or peptides, resulting in host tissue damage and consequently reducing the therapeutic potential of proteases. For example, the human antimicrobial peptide LL-37 is cleaved by staphopain B (409) and aureolysin (410) and essentially all staphylococcal proteases have known host targets (411), including immune cells, immunoglobulins, and components of the complement system (412–415).
The second class of bioactive molecules to fight staphylococcal biofilms are the biofilm-eradicating molecules. Probably the most prominent member of this class of molecules is lysostaphin, a lysin produced by Staphylococcus simulans biovar staphylolyticus. It was originally discovered and proposed for the treatment of S. aureus infections over half a century ago (416, 417) and has recently garnered attention for its ability to combat staphylococcal biofilms, in combination with antibiotics, in in vitro studies and in a mouse catheter model (418–421). A very diverse group of biofilm-eradicating agents are natural and engineered AMPs, which were shown to have a plethora of different modes of action, ranging from pore formation and inhibition of DNA and protein synthesis to the disruption of the biofilm matrix (422, 423). An interesting AMP is the human cathelicidin peptide LL-37, because it not only shows activity against planktonic cells (424) but also exhibits strong antibiofilm activity against preformed S. aureus biofilms (425). Although AMPs are very promising candidates for antibiofilm therapy, cases of resistance development have been observed, including proteolytic degradation, alteration of the cell wall, and active secretion (426). Additionally, staphylococci use regulatory systems to increase their resistance to AMPs, one of which is the S. aureus TCS GraRS (427). A thoroughly studied class of AMPs are bacteriocins, with some of them showing increased activity against staphylococcal biofilms alone or in synergistic interactions with antibiotics (428–430). For a comprehensive review about different groups of bacteriocins with antibiofilm activity, the reader is referred to the work of Mathur et al. (431) and Newstead et al. (432).
Bacteriophage-Based Antibiofilm Strategies
An alternative strategy to fight biofilm-associated infections is the use of bacteriophages, viruses that specifically infect bacterial cells (433). Historically, bacteriophage therapy against bacterial infections had already been used over 100 years ago, but with the discovery of antibiotics, this form of treatment lost its popularity. However, the rise of multidrug-resistant bacteria has sparked new interest in the potential use of bacteriophages and their products to combat bacterial infections (434). Bacteriophages produce endolysins, which are able to enzymatically degrade the bacterial peptidoglycan, resulting in cell lysis and biofilm clearance (435, 436). There are three possible approaches to eradicate biofilms with bacteriophages: single-phage application, mixed-phage application with two or more phages, and phage-antibiotic combinations. An example that demonstrates the ability of a single phage to disperse staphylococcal biofilms was the recent isolation of phages UPMK_1 and UPMK_2. When applied separately, both phages have promising antibiofilm activity against pregrown S. aureus biofilms (437). The successful application of a bacteriophage mixture as an antibiofilm agent was demonstrated by Alvez and colleagues (438). They reported the isolation of a novel bacteriophage, DRA88, which in combination with bacteriophage K was demonstrated to strongly reduce the biofilm mass of S. aureus, including MRSA clones, and CoNS. In addition, combinatorial therapy with phages and antibiotics has shown promising results to eradicate biofilms of drug-resistant bacteria without the emergence of new antibiotic resistance (439–441). Another interesting approach is the use of phage-derived enzymes capable of dispersing bacterial biofilms. Recently, LysCSA13, an endolysin encoded by the bacteriophage CSA13, was shown to decrease staphylococcal biofilm mass up to 90% on several surfaces, including polystyrene, glass, and stainless steel (442, 443). Similar results were reported for the bacteriophage lysins LysGH15 and CF-301, and the latter also showed promising activity against bacterial mixed-species biofilms formed by S. aureus and S. epidermidis (444, 445). The two major advantages of phage therapy to treat biofilm-associated infections are the capability of phages to infect and kill biofilm embedded bacteria (433, 446) and their relatively narrow host range, which prevents the undesirable killing of bacterial commensals during treatment (447, 448). Nevertheless, a recent study determined that the amount of biofilm mass, strain susceptibility, initial phage titer, phage entrapment in the extracellular matrix, and phage inactivation are all factors contributing to successful biofilm penetration and need to be considered during the design of an effective phage-based therapy (446).
CONCLUDING REMARKS
Staphylococci, most prominently S. aureus and S. epidermidis, exist as multicellular communities or so-called biofilms in their natural habitats. When growing in a biofilm, the bacteria are encased in an extracellular matrix and display an altered metabolic state compared to that of planktonic cells (449), which leads to increased resistance to host defenses and antibiotic treatment (33). In order to coordinate the formation of biofilms, staphylococci rely on an extensive network of regulatory systems enabling the bacteria to sense and respond to an abundance of biotic and abiotic factors. The staphylococcal QS system is one of the best-studied regulatory systems and allows the bacteria to use the biofilm as a protected reservoir that serves as an initiating point for colonization of new body sites. As outlined in this review, the staphylococcal QS system is subject to regulation by many environmental factors and host components, in addition to physicochemical factors, like mechanical shear stress from fluid flow. Most interestingly, a recent study has shown that QS follows a heterogeneous activation pattern under flow conditions (212). Under stronger flow conditions, QS is essentially turned off, but the presence of areas of no or limited fluid flow, like small crevices or inside specific biofilm regions, allows the accumulation of AIP signals and subsequent activation of the staphylococcal QS system. An additional example of bimodal QS was recently published by García-Betancur and colleagues. They showed that S. aureus populations consist of two distinct subpopulations, one with QS turned on and one having it turned off. A high concentration of environmental magnesium triggers the downregulation of the QS system and most likely promotes S. aureus biofilm formation (450). These examples show that even though the connection between QS and biofilm formation has been studied extensively, there are still many aspects to investigate. For example, it is still not clear how different infection sites in the host can influence biofilm formation, either through the staphylococcal QS system or through other regulatory systems, and which environmental cues contribute to staphylococcal biofilm-associated infections.
In addition, multispecies biofilms more closely reflect the complex bacterial communities that are usually found in the host instead of single-species biofilms. For example, S. aureus and P. aeruginosa are frequently coisolated from chronic wound infections and cystic fibrosis lungs (347, 451–453), which represent types of infections associated with biofilm formation. Not surprisingly, the interaction between staphylococci and other microbial species has profound effects on antimicrobial resistance and the spatial organization within the biofilm. Several studies have shown that during mixed-species biofilm growth, although being present in the biofilm together, the participating species are spatially segregated and inhabit different niches within the biofilm (454, 455). Regarding alterations of antimicrobial resistance during mixed-species biofilm formation, S. aureus biofilms, when grown in the presence of cell-free P. aeruginosa supernatant, show increased sensitivity to cell wall-targeting antibiotics but also gain resistance to vancomycin (456, 457). In contrast to these observations, when S. aureus is grown with P. aeruginosa in a multispecies biofilm, S. aureus shows higher sensitivity to several classes of antibiotics (458). The ability of S. aureus to form interspecies biofilms is not limited to bacterial species but was also observed with the fungus Candida albicans. In these mixed-species biofilms, S. aureus forms microcolonies on top of the C. albicans biofilm and becomes more resistant to vancomycin (459–462). Considering that infections of the cystic fibrosis lung are always of a polymicrobial nature, it is reasonable to assume that S. aureus can also form mixed-species biofilms with several other bacterial and fungal species which have not been investigated yet.
Therefore, it will be crucial for future development of antibiofilm agents to incorporate mixed-species biofilm studies and to have a stronger focus on ex vivo and in vivo biofilm systems in order to understand which aspects of the already available knowledge on in vitro biofilms can be directly transferred to in vivo situations.
ACKNOWLEDGMENTS
Work in the authors’ laboratory is supported by Swiss National Science Foundation postdoctoral fellowship P2ZHP3_168570 (K.S.), by American Heart Association (AHA) postdoctoral fellowship 20POST35220011 (K.S.), by Merit Award I01 (I01 BX002711) (A.R.H.) from the Department of Veteran Affairs, and by NIH public health service grant AI083211 (A.R.H.).
Biographies

Katrin Schilcher is a postdoctoral fellow at the University of Colorado Anschutz Medical Campus. She obtained her B.Sc. and M.Sc. in molecular biology and molecular microbiology from the University of Graz, Austria. She then moved to Switzerland and received her Ph.D. from the University of Zurich in the laboratory of Dr. Annelies Zinkernagel, where she studied antibiotic-mediated modulation of Staphylococcus aureus biofilm formation and host-pathogen interaction. After completion of her Ph.D., Katrin joined the laboratory of Dr. Alexander Horswill to investigate the role of linear peptides in staphylococcal cell-cell signaling and host-pathogen interactions. She unraveled a new biosynthetic pathway for linear peptide processing and export from Staphylococcus aureus and is currently continuing her work on linear peptide signaling. Her research has been supported by an Early Postdoc Mobility fellowship from the Swiss National Science Foundation and a Postdoctoral Fellowship from the American Heart Association.

Alexander R. Horswill obtained his Ph.D. in 2001 from the University of Wisconsin-Madison studying Salmonella short-chain fatty acid metabolism under the direction of Professor Jorge Escalante-Semerena. From 2001 to 2005, Dr. Horswill was a Damon-Runyon postdoctoral fellow in the laboratory of Dr. Stephen Benkovic in the Department of Chemistry at Penn State University. In 2005, Dr. Horswill joined the Department of Microbiology at the University of Iowa as an Assistant Professor. He started a research program focused on mechanisms of Staphylococcus aureus quorum sensing, biofilm development, and host-pathogen interactions. In 2017, Dr. Horswill relocated his research group to the University of Colorado Anschutz Medical Campus, and currently he is a Full Professor in the Department of Immunology & Microbiology.
REFERENCES
- 1.Costerton JW, Geesey GG, Cheng KJ. 1978. How bacteria stick. Sci Am 238:86–95. doi: 10.1038/scientificamerican0178-86. [DOI] [PubMed] [Google Scholar]
- 2.Trunk T, Khalil HS, Leo JC. 2018. Bacterial autoaggregation. AIMS Microbiol 4:140–164. doi: 10.3934/microbiol.2018.1.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vaccari L, Molaei M, Niepa THR, Lee D, Leheny RL, Stebe KJ. 2017. Films of bacteria at interfaces. Adv Colloid Interface Sci 247:561–572. doi: 10.1016/j.cis.2017.07.016. [DOI] [PubMed] [Google Scholar]
- 4.Peters G, Locci R, Pulverer G. 1981. Microbial colonization of prosthetic devices. II. Scanning electron microscopy of naturally infected intravenous catheters. Zentralbl Bakteriol Mikrobiol Hyg B 173:293–299. [PubMed] [Google Scholar]
- 5.Peters G, Locci R, Pulverer G. 1982. Adherence and growth of coagulase-negative staphylococci on surfaces of intravenous catheters. J Infect Dis 146:479–482. doi: 10.1093/infdis/146.4.479. [DOI] [PubMed] [Google Scholar]
- 6.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 7.López D, Vlamakis H, Kolter R. 2010. Biofilms. Cold Spring Harb Perspect Biol 2:a000398. doi: 10.1101/cshperspect.a000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dufour D, Leung V, Lévesque CM. 2010. Bacterial biofilm: structure, function, and antimicrobial resistance. Endod Topics 22:2–16. doi: 10.1111/j.1601-1546.2012.00277.x. [DOI] [Google Scholar]
- 9.Hobley L, Harkins C, MacPhee CE, Stanley-Wall NR. 2015. Giving structure to the biofilm matrix: an overview of individual strategies and emerging common themes. FEMS Microbiol Rev 39:649–669. doi: 10.1093/femsre/fuv015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams RE. 1963. Healthy carriage of Staphylococcus aureus: its prevalence and importance. Bacteriol Rev 27:56–71. doi: 10.1128/MMBR.27.1.56-71.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kloos WE, Musselwhite MS. 1975. Distribution and persistence of Staphylococcus and Micrococcus species and other aerobic bacteria on human skin. Appl Microbiol 30:381–385. doi: 10.1128/AEM.30.3.381-395.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beenken KE, Dunman PM, McAleese F, Macapagal D, Murphy E, Projan SJ, Blevins JS, Smeltzer MS. 2004. Global gene expression in Staphylococcus aureus biofilms. J Bacteriol 186:4665–4684. doi: 10.1128/JB.186.14.4665-4684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scherr TD, Heim CE, Morrison JM, Kielian T. 2014. Hiding in plain sight: interplay between staphylococcal biofilms and host immunity. Front Immunol 5:37. doi: 10.3389/fimmu.2014.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paharik AE, Horswill AR. 2016. The staphylococcal biofilm: adhesins, regulation, and host response. Microbiol Spectrum 4:VMBF-0022-2015. doi: 10.1128/microbiolspec.VMBF-0022-2015. [DOI] [Google Scholar]
- 15.Vestby LK, Gronseth T, Simm R, Nesse LL. 2020. Bacterial biofilm and its role in the pathogenesis of disease. Antibiotics (Basel) 9:59. doi: 10.3390/antibiotics9020059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Archer NK, Mazaitis MJ, Costerton JW, Leid JG, Powers ME, Shirtliff ME. 2011. Staphylococcus aureus biofilms: properties, regulation, and roles in human disease. Virulence 2:445–459. doi: 10.4161/viru.2.5.17724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacqueline C, Caillon J. 2014. Impact of bacterial biofilm on the treatment of prosthetic joint infections. J Antimicrob Chemother 69:i37–i40. doi: 10.1093/jac/dku254. [DOI] [PubMed] [Google Scholar]
- 18.Foster TJ. 2005. Immune evasion by staphylococci. Nat Rev Microbiol 3:948–958. doi: 10.1038/nrmicro1289. [DOI] [PubMed] [Google Scholar]
- 19.Arciola CR, An YH, Campoccia D, Donati ME, Montanaro L. 2005. Etiology of implant orthopedic infections: a survey on 1027 clinical isolates. Int J Artif Organs 28:1091–1100. doi: 10.1177/039139880502801106. [DOI] [PubMed] [Google Scholar]
- 20.Rogers KL, Fey PD, Rupp ME. 2009. Coagulase-negative staphylococcal infections. Infect Dis Clin North Am 23:73–98. doi: 10.1016/j.idc.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 21.Novick RP, Geisinger E. 2008. Quorum sensing in staphylococci. Annu Rev Genet 42:541–564. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- 22.Thoendel M, Kavanaugh JS, Flack CE, Horswill AR. 2011. Peptide signaling in the staphylococci. Chem Rev 111:117–151. doi: 10.1021/cr100370n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kavanaugh JS, Horswill AR. 2016. Impact of environmental cues on staphylococcal quorum sensing and biofilm development. J Biol Chem 291:12556–12564. doi: 10.1074/jbc.R116.722710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lister JL, Horswill AR. 2014. Staphylococcus aureus biofilms: recent developments in biofilm dispersal. Front Cell Infect Microbiol 4:178. doi: 10.3389/fcimb.2014.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peschel A, Otto M. 2013. Phenol-soluble modulins and staphylococcal infection. Nat Rev Microbiol 11:667–673. doi: 10.1038/nrmicro3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le KY, Villaruz AE, Zheng Y, He L, Fisher EL, Nguyen TH, Ho TV, Yeh AJ, Joo HS, Cheung GYC, Otto M. 2019. Role of phenol-soluble modulins in Staphylococcus epidermidis biofilm formation and infection of indwelling medical devices. J Mol Biol 431:3015–3027. doi: 10.1016/j.jmb.2019.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flemming HC, Wingender J. 2010. The biofilm matrix. Nat Rev Microbiol 8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 28.Rigby KM, DeLeo FR. 2012. Neutrophils in innate host defense against Staphylococcus aureus infections. Semin Immunopathol 34:237–259. doi: 10.1007/s00281-011-0295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watters C, Fleming D, Bishop D, Rumbaugh KP. 2016. Host responses to biofilm. Prog Mol Biol Transl Sci 142:193–239. doi: 10.1016/bs.pmbts.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 30.Høiby N, Krogh Johansen H, Moser C, Song Z, Ciofu O, Kharazmi A. 2001. Pseudomonas aeruginosa and the in vitro and in vivo biofilm mode of growth. Microbes Infect 3:23–35. doi: 10.1016/s1286-4579(00)01349-6. [DOI] [PubMed] [Google Scholar]
- 31.Hogan D, Kolter R. 2002. Why are bacteria refractory to antimicrobials? Curr Opin Microbiol 5:472–477. doi: 10.1016/s1369-5274(02)00357-0. [DOI] [PubMed] [Google Scholar]
- 32.Waters EM, Rowe SE, O’Gara JP, Conlon BP. 2016. Convergence of Staphylococcus aureus persister and biofilm research: can biofilms be defined as communities of adherent persister cells? PLoS Pathog 12:e1006012. doi: 10.1371/journal.ppat.1006012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bui LM, Conlon BP, Kidd SP. 2017. Antibiotic tolerance and the alternative lifestyles of Staphylococcus aureus. Essays Biochem 61:71–79. doi: 10.1042/EBC20160061. [DOI] [PubMed] [Google Scholar]
- 34.Conlon BP, Rowe SE, Gandt AB, Nuxoll AS, Donegan NP, Zalis EA, Clair G, Adkins JN, Cheung AL, Lewis K. 2016. Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat Microbiol 1:16051. doi: 10.1038/nmicrobiol.2016.51. [DOI] [PubMed] [Google Scholar]
- 35.Shan Y, Brown Gandt A, Rowe SE, Deisinger JP, Conlon BP, Lewis K. 2017. ATP-dependent persister formation in Escherichia coli. mBio 8:e02267-16. doi: 10.1128/mBio.02267-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koo H, Allan RN, Howlin RP, Stoodley P, Hall-Stoodley L. 2017. Targeting microbial biofilms: current and prospective therapeutic strategies. Nat Rev Microbiol 15:740–755. doi: 10.1038/nrmicro.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Foster TJ, Geoghegan JA, Ganesh VK, Höök M. 2014. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat Rev Microbiol 12:49–62. doi: 10.1038/nrmicro3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Foster TJ. 2019. The MSCRAMM family of cell-wall-anchored surface proteins of Gram-positive cocci. Trends Microbiol 27:927–941. doi: 10.1016/j.tim.2019.06.007. [DOI] [PubMed] [Google Scholar]
- 39.Gross M, Cramton SE, Götz F, Peschel A. 2001. Key role of teichoic acid net charge in Staphylococcus aureus colonization of artificial surfaces. Infect Immun 69:3423–3426. doi: 10.1128/IAI.69.5.3423-3426.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holland LM, Conlon B, O'Gara JP. 2011. Mutation of tagO reveals an essential role for wall teichoic acids in Staphylococcus epidermidis biofilm development. Microbiology 157:408–418. doi: 10.1099/mic.0.042234-0. [DOI] [PubMed] [Google Scholar]
- 41.Otto M. 2014. Physical stress and bacterial colonization. FEMS Microbiol Rev 38:1250–1270. doi: 10.1111/1574-6976.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qin Z, Ou Y, Yang L, Zhu Y, Tolker-Nielsen T, Molin S, Qu D. 2007. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology 153:2083–2092. doi: 10.1099/mic.0.2007/006031-0. [DOI] [PubMed] [Google Scholar]
- 43.Mann EE, Rice KC, Boles BR, Endres JL, Ranjit D, Chandramohan L, Tsang LH, Smeltzer MS, Horswill AR, Bayles KW. 2009. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS One 4:e5822. doi: 10.1371/journal.pone.0005822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Joo HS, Otto M. 2012. Molecular basis of in vivo biofilm formation by bacterial pathogens. Chem Biol 19:1503–1513. doi: 10.1016/j.chembiol.2012.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Periasamy S, Joo HS, Duong AC, Bach TH, Tan VY, Chatterjee SS, Cheung GY, Otto M. 2012. How Staphylococcus aureus biofilms develop their characteristic structure. Proc Natl Acad Sci U S A 109:1281–1286. doi: 10.1073/pnas.1115006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moormeier DE, Bose JL, Horswill AR, Bayles KW. 2014. Temporal and stochastic control of Staphylococcus aureus biofilm development. mBio 5:e01341-14. doi: 10.1128/mBio.01341-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moormeier DE, Bayles KW. 2017. Staphylococcus aureus biofilm: a complex developmental organism. Mol Microbiol 104:365–376. doi: 10.1111/mmi.13634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mazmanian SK, Liu G, Ton-That H, Schneewind O. 1999. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285:760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- 49.Jönsson K, Signäs C, Muller HP, Lindberg M. 1991. Two different genes encode fibronectin binding proteins in Staphylococcus aureus. The complete nucleotide sequence and characterization of the second gene. Eur J Biochem 202:1041–1048. doi: 10.1111/j.1432-1033.1991.tb16468.x. [DOI] [PubMed] [Google Scholar]
- 50.Greene C, McDevitt D, Francois P, Vaudaux PE, Lew DP, Foster TJ. 1995. Adhesion properties of mutants of Staphylococcus aureus defective in fibronectin-binding proteins and studies on the expression of fnb genes. Mol Microbiol 17:1143–1152. doi: 10.1111/j.1365-2958.1995.mmi_17061143.x. [DOI] [PubMed] [Google Scholar]
- 51.Keane FM, Loughman A, Valtulina V, Brennan M, Speziale P, Foster TJ. 2007. Fibrinogen and elastin bind to the same region within the A domain of fibronectin binding protein A, an MSCRAMM of Staphylococcus aureus. Mol Microbiol 63:711–723. doi: 10.1111/j.1365-2958.2006.05552.x. [DOI] [PubMed] [Google Scholar]
- 52.McDevitt D, Francois P, Vaudaux P, Foster TJ. 1994. Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus. Mol Microbiol 11:237–248. doi: 10.1111/j.1365-2958.1994.tb00304.x. [DOI] [PubMed] [Google Scholar]
- 53.McDevitt D, Nanavaty T, House-Pompeo K, Bell E, Turner N, McIntire L, Foster T, Höök M. 1997. Characterization of the interaction between the Staphylococcus aureus clumping factor (ClfA) and fibrinogen. Eur J Biochem 247:416–424. doi: 10.1111/j.1432-1033.1997.00416.x. [DOI] [PubMed] [Google Scholar]
- 54.Ní Eidhin D, Perkins S, Francois P, Vaudaux P, Höök M, Foster TJ. 1998. Clumping factor B (ClfB), a new surface-located fibrinogen-binding adhesin of Staphylococcus aureus. Mol Microbiol 30:245–257. doi: 10.1046/j.1365-2958.1998.01050.x. [DOI] [PubMed] [Google Scholar]
- 55.Walsh EJ, Miajlovic H, Gorkun OV, Foster TJ. 2008. Identification of the Staphylococcus aureus MSCRAMM clumping factor B (ClfB) binding site in the alphaC-domain of human fibrinogen. Microbiology 154:550–558. doi: 10.1099/mic.0.2007/010868-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Geoghegan JA, Ganesh VK, Smeds E, Liang X, Höök M, Foster TJ. 2010. Molecular characterization of the interaction of staphylococcal microbial surface components recognizing adhesive matrix molecules (MSCRAMM) ClfA and Fbl with fibrinogen. J Biol Chem 285:6208–6216. doi: 10.1074/jbc.M109.062208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Josefsson E, McCrea KW, Ní Eidhin D, O’Connell D, Cox J, Höök M, Foster TJ. 1998. Three new members of the serine-aspartate repeat protein multigene family of Staphylococcus aureus. Microbiology 144:3387–3395. doi: 10.1099/00221287-144-12-3387. [DOI] [PubMed] [Google Scholar]
- 58.Barbu EM, Ganesh VK, Gurusiddappa S, Mackenzie RC, Foster TJ, Sudhof TC, Höök M. 2010. β-Neurexin is a ligand for the Staphylococcus aureus MSCRAMM SdrC. PLoS Pathog 6:e1000726. doi: 10.1371/journal.ppat.1000726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Askarian F, Uchiyama S, Valderrama JA, Ajayi C, Sollid JUE, van Sorge NM, Nizet V, van Strijp JAG, Johannessen M. 2017. Serine-aspartate repeat protein D increases Staphylococcus aureus virulence and survival in blood. Infect Immun 85:e00559-16. doi: 10.1128/IAI.00559-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Y, Wu M, Hang T, Wang C, Yang Y, Pan W, Zang J, Zhang M, Zhang X. 2017. Staphylococcus aureus SdrE captures complement factor H’s C-terminus via a novel ‘close, dock, lock and latch’ mechanism for complement evasion. Biochem J 474:1619–1631. doi: 10.1042/BCJ20170085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vazquez V, Liang X, Horndahl JK, Ganesh VK, Smeds E, Foster TJ, Höök M. 2011. Fibrinogen is a ligand for the Staphylococcus aureus microbial surface components recognizing adhesive matrix molecules (MSCRAMM) bone sialoprotein-binding protein (Bbp). J Biol Chem 286:29797–29805. doi: 10.1074/jbc.M110.214981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang X, Wu M, Zhuo W, Gu J, Zhang S, Ge J, Yang M. 2015. Crystal structures of Bbp from Staphylococcus aureus reveal the ligand binding mechanism with Fibrinogen alpha. Protein Cell 6:757–766. doi: 10.1007/s13238-015-0205-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McCrea KW, Hartford O, Davis S, Eidhin DN, Lina G, Speziale P, Foster TJ, Höök M. 2000. The serine-aspartate repeat (Sdr) protein family in Staphylococcus epidermidis. Microbiology 146:1535–1546. doi: 10.1099/00221287-146-7-1535. [DOI] [PubMed] [Google Scholar]
- 64.McCourt J, O’Halloran DP, McCarthy H, O’Gara JP, Geoghegan JA. 2014. Fibronectin-binding proteins are required for biofilm formation by community-associated methicillin-resistant Staphylococcus aureus strain LAC. FEMS Microbiol Lett 353:157–164. doi: 10.1111/1574-6968.12424. [DOI] [PubMed] [Google Scholar]
- 65.Feuillie C, Formosa-Dague C, Hays LMC, Vervaeck O, Derclaye S, Brennan MP, Foster TJ, Geoghegan JA, Dufrêne YF. 2017. Molecular interactions and inhibition of the staphylococcal biofilm-forming protein SdrC. Proc Natl Acad Sci U S A 114:3738–3743. doi: 10.1073/pnas.1616805114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cucarella C, Solano C, Valle J, Amorena B, Lasa I, Penadés JR. 2001. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J Bacteriol 183:2888–2896. doi: 10.1128/JB.183.9.2888-2896.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Biswas R, Voggu L, Simon UK, Hentschel P, Thumm G, Götz F. 2006. Activity of the major staphylococcal autolysin Atl. FEMS Microbiol Lett 259:260–268. doi: 10.1111/j.1574-6968.2006.00281.x. [DOI] [PubMed] [Google Scholar]
- 68.Houston P, Rowe SE, Pozzi C, Waters EM, O’Gara JP. 2011. Essential role for the major autolysin in the fibronectin-binding protein-mediated Staphylococcus aureus biofilm phenotype. Infect Immun 79:1153–1165. doi: 10.1128/IAI.00364-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bose JL, Lehman MK, Fey PD, Bayles KW. 2012. Contribution of the Staphylococcus aureus Atl AM and GL murein hydrolase activities in cell division, autolysis, and biofilm formation. PLoS One 7:e42244. doi: 10.1371/journal.pone.0042244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Heilmann C, Hussain M, Peters G, Götz F. 1997. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol Microbiol 24:1013–1024. doi: 10.1046/j.1365-2958.1997.4101774.x. [DOI] [PubMed] [Google Scholar]
- 71.Kiedrowski MR, Kavanaugh JS, Malone CL, Mootz JM, Voyich JM, Smeltzer MS, Bayles KW, Horswill AR. 2011. Nuclease modulates biofilm formation in community-associated methicillin-resistant Staphylococcus aureus. PLoS One 6:e26714. doi: 10.1371/journal.pone.0026714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Montanaro L, Poggi A, Visai L, Ravaioli S, Campoccia D, Speziale P, Arciola CR. 2011. Extracellular DNA in biofilms. Int J Artif Organs 34:824–831. doi: 10.5301/ijao.5000051. [DOI] [PubMed] [Google Scholar]
- 73.Grande R, Nistico L, Sambanthamoorthy K, Longwell M, Iannitelli A, Cellini L, Di Stefano A, Hall Stoodley L, Stoodley P. 2014. Temporal expression of agrB, cidA, and alsS in the early development of Staphylococcus aureus UAMS-1 biofilm formation and the structural role of extracellular DNA and carbohydrates. Pathog Dis 70:414–422. doi: 10.1111/2049-632X.12158. [DOI] [PubMed] [Google Scholar]
- 74.Conlon BP, Geoghegan JA, Waters EM, McCarthy H, Rowe SE, Davies JR, Schaeffer CR, Foster TJ, Fey PD, O’Gara JP. 2014. Role for the A domain of unprocessed accumulation-associated protein (Aap) in the attachment phase of the Staphylococcus epidermidis biofilm phenotype. J Bacteriol 196:4268–4275. doi: 10.1128/JB.01946-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.O’Gara JP. 2007. Ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol Lett 270:179–188. doi: 10.1111/j.1574-6968.2007.00688.x. [DOI] [PubMed] [Google Scholar]
- 76.Speziale P, Pietrocola G, Foster TJ, Geoghegan JA. 2014. Protein-based biofilm matrices in staphylococci. Front Cell Infect Microbiol 4:171. doi: 10.3389/fcimb.2014.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mack D, Fischer W, Krokotsch A, Leopold K, Hartmann R, Egge H, Laufs R. 1996. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: purification and structural analysis. J Bacteriol 178:175–183. doi: 10.1128/JB.178.1.175-183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maira-Litrán T, Kropec A, Abeygunawardana C, Joyce J, Mark G III, Goldmann DA, Pier GB. 2002. Immunochemical properties of the staphylococcal poly-N-acetylglucosamine surface polysaccharide. Infect Immun 70:4433–4440. doi: 10.1128/iai.70.8.4433-4440.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mack D, Nedelmann M, Krokotsch A, Schwarzkopf A, Heesemann J, Laufs R. 1994. Characterization of transposon mutants of biofilm-producing Staphylococcus epidermidis impaired in the accumulative phase of biofilm production: genetic identification of a hexosamine-containing polysaccharide intercellular adhesin. Infect Immun 62:3244–3253. doi: 10.1128/IAI.62.8.3244-3253.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cramton SE, Gerke C, Schnell NF, Nichols WW, Götz F. 1999. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect Immun 67:5427–5433. doi: 10.1128/IAI.67.10.5427-5433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Heilmann C, Schweitzer O, Gerke C, Vanittanakom N, Mack D, Götz F. 1996. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol Microbiol 20:1083–1091. doi: 10.1111/j.1365-2958.1996.tb02548.x. [DOI] [PubMed] [Google Scholar]
- 82.Gerke C, Kraft A, Sussmuth R, Schweitzer O, Götz F. 1998. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J Biol Chem 273:18586–18593. doi: 10.1074/jbc.273.29.18586. [DOI] [PubMed] [Google Scholar]
- 83.Atkin KE, MacDonald SJ, Brentnall AS, Potts JR, Thomas GH. 2014. A different path: revealing the function of staphylococcal proteins in biofilm formation. FEBS Lett 588:1869–1872. doi: 10.1016/j.febslet.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 84.McKenney D, Pouliot KL, Wang Y, Murthy V, Ulrich M, Döring G, Lee JC, Goldmann DA, Pier GB. 1999. Broadly protective vaccine for Staphylococcus aureus based on an in vivo-expressed antigen. Science 284:1523–1527. doi: 10.1126/science.284.5419.1523. [DOI] [PubMed] [Google Scholar]
- 85.Joyce JG, Abeygunawardana C, Xu Q, Cook JC, Hepler R, Przysiecki CT, Grimm KM, Roper K, Ip CC, Cope L, Montgomery D, Chang M, Campie S, Brown M, McNeely TB, Zorman J, Maira-Litran T, Pier GB, Keller PM, Jansen KU, Mark GE. 2003. Isolation, structural characterization, and immunological evaluation of a high-molecular-weight exopolysaccharide from Staphylococcus aureus. Carbohydr Res 338:903–922. doi: 10.1016/s0008-6215(03)00045-4. [DOI] [PubMed] [Google Scholar]
- 86.Sadovskaya I, Vinogradov E, Flahaut S, Kogan G, Jabbouri S. 2005. Extracellular carbohydrate-containing polymers of a model biofilm-producing strain, Staphylococcus epidermidis RP62A. Infect Immun 73:3007–3017. doi: 10.1128/IAI.73.5.3007-3017.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vuong C, Kocianova S, Voyich JM, Yao Y, Fischer ER, DeLeo FR, Otto M. 2004. A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J Biol Chem 279:54881–54886. doi: 10.1074/jbc.M411374200. [DOI] [PubMed] [Google Scholar]
- 88.Vuong C, Voyich JM, Fischer ER, Braughton KR, Whitney AR, DeLeo FR, Otto M. 2004. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell Microbiol 6:269–275. doi: 10.1046/j.1462-5822.2004.00367.x. [DOI] [PubMed] [Google Scholar]
- 89.Conlon KM, Humphreys H, O’Gara JP. 2002. icaR encodes a transcriptional repressor involved in environmental regulation of ica operon expression and biofilm formation in Staphylococcus epidermidis. J Bacteriol 184:4400–4408. doi: 10.1128/jb.184.16.4400-4408.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jefferson KK, Pier DB, Goldmann DA, Pier GB. 2004. The teicoplanin-associated locus regulator (TcaR) and the intercellular adhesin locus regulator (IcaR) are transcriptional inhibitors of the ica locus in Staphylococcus aureus. J Bacteriol 186:2449–2456. doi: 10.1128/jb.186.8.2449-2456.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fitzpatrick F, Humphreys H, O’Gara JP. 2005. Evidence for icaADBC-independent biofilm development mechanism in methicillin-resistant Staphylococcus aureus clinical isolates. J Clin Microbiol 43:1973–1976. doi: 10.1128/JCM.43.4.1973-1976.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rohde H, Burdelski C, Bartscht K, Hussain M, Buck F, Horstkotte MA, Knobloch JK, Heilmann C, Herrmann M, Mack D. 2005. Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol Microbiol 55:1883–1895. doi: 10.1111/j.1365-2958.2005.04515.x. [DOI] [PubMed] [Google Scholar]
- 93.Kogan G, Sadovskaya I, Chaignon P, Chokr A, Jabbouri S. 2006. Biofilms of clinical strains of Staphylococcus that do not contain polysaccharide intercellular adhesin. FEMS Microbiol Lett 255:11–16. doi: 10.1111/j.1574-6968.2005.00043.x. [DOI] [PubMed] [Google Scholar]
- 94.O’Neill E, Pozzi C, Houston P, Smyth D, Humphreys H, Robinson DA, O’Gara JP. 2007. Association between methicillin susceptibility and biofilm regulation in Staphylococcus aureus isolates from device-related infections. J Clin Microbiol 45:1379–1388. doi: 10.1128/JCM.02280-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cherifi S, Byl B, Deplano A, Nonhoff C, Denis O, Hallin M. 2013. Comparative epidemiology of Staphylococcus epidermidis isolates from patients with catheter-related bacteremia and from healthy volunteers. J Clin Microbiol 51:1541–1547. doi: 10.1128/JCM.03378-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Harris LG, Murray S, Pascoe B, Bray J, Meric G, Mageiros L, Wilkinson TS, Jeeves R, Rohde H, Schwarz S, de Lencastre H, Miragaia M, Rolo J, Bowden R, Jolley KA, Maiden MC, Mack D, Sheppard SK. 2016. Biofilm morphotypes and population structure among Staphylococcus epidermidis from commensal and clinical samples. PLoS One 11:e0151240. doi: 10.1371/journal.pone.0151240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Martínez-Meléndez A, Morfín-Otero R, Villarreal-Treviño L, Camacho-Ortíz A, González-González G, Llaca-Díaz J, Rodríguez-Noriega E, Garza-González E. 2016. Molecular epidemiology of coagulase-negative bloodstream isolates: detection of Staphylococcus epidermidis ST2, ST7 and linezolid-resistant ST23. Braz J Infect Dis 20:419–428. doi: 10.1016/j.bjid.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ahmed DM, Messih M, Ibrahim NH, Meabed MH, Abdel-Salam SM. 2019. Frequency of icaA and icaD determinants and biofilm formation among coagulase-negative staphylococci associated with nasal carriage in neonatal intensive care units. Germs 9:61–70. doi: 10.18683/germs.2019.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Farajzadeh Sheikh A, Asareh Zadegan Dezfuli A, Navidifar T, Fard SS, Dehdashtian M. 2019. Association between biofilm formation, structure and antibiotic resistance in Staphylococcus epidermidis isolated from neonatal septicemia in southwest Iran. Infect Drug Resist 12:1771–1782. doi: 10.2147/IDR.S204432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cucarella C, Tormo MA, Úbeda C, Trotonda MP, Monzón M, Peris C, Amorena B, Lasa I, Penadés JR. 2004. Role of biofilm-associated protein bap in the pathogenesis of bovine Staphylococcus aureus. Infect Immun 72:2177–2185. doi: 10.1128/iai.72.4.2177-2185.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bowden MG, Chen W, Singvall J, Xu Y, Peacock SJ, Valtulina V, Speziale P, Höök M. 2005. Identification and preliminary characterization of cell-wall-anchored proteins of Staphylococcus epidermidis. Microbiology 151:1453–1464. doi: 10.1099/mic.0.27534-0. [DOI] [PubMed] [Google Scholar]
- 102.Taglialegna A, Navarro S, Ventura S, Garnett JA, Matthews S, Penades JR, Lasa I, Valle J. 2016. Staphylococcal Bap proteins build amyloid scaffold biofilm matrices in response to environmental signals. PLoS Pathog 12:e1005711. doi: 10.1371/journal.ppat.1005711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.O’Neill E, Pozzi C, Houston P, Humphreys H, Robinson DA, Loughman A, Foster TJ, O’Gara JP. 2008. A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J Bacteriol 190:3835–3850. doi: 10.1128/JB.00167-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Herman-Bausier P, El-Kirat-Chatel S, Foster TJ, Geoghegan JA, Dufrêne YF. 2015. Staphylococcus aureus fibronectin-binding protein A mediates cell-cell adhesion through low-affinity homophilic bonds. mBio 6:e00413-15. doi: 10.1128/mBio.00413-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Barbu EM, Mackenzie C, Foster TJ, Höök M. 2014. SdrC induces staphylococcal biofilm formation through a homophilic interaction. Mol Microbiol 94:172–185. doi: 10.1111/mmi.12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hussain M, Herrmann M, von Eiff C, Perdreau-Remington F, Peters G. 1997. A 140-kilodalton extracellular protein is essential for the accumulation of Staphylococcus epidermidis strains on surfaces. Infect Immun 65:519–524. doi: 10.1128/IAI.65.2.519-524.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Paharik AE, Kotasinska M, Both A, Hoang TN, Büttner H, Roy P, Fey PD, Horswill AR, Rohde H. 2017. The metalloprotease SepA governs processing of accumulation-associated protein and shapes intercellular adhesive surface properties in Staphylococcus epidermidis. Mol Microbiol 103:860–874. doi: 10.1111/mmi.13594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Conrady DG, Brescia CC, Horii K, Weiss AA, Hassett DJ, Herr AB. 2008. A zinc-dependent adhesion module is responsible for intercellular adhesion in staphylococcal biofilms. Proc Natl Acad Sci U S A 105:19456–19461. doi: 10.1073/pnas.0807717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Conrady DG, Wilson JJ, Herr AB. 2013. Structural basis for Zn2+-dependent intercellular adhesion in staphylococcal biofilms. Proc Natl Acad Sci U S A 110:E202–E211. doi: 10.1073/pnas.1208134110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Geoghegan JA, Corrigan RM, Gruszka DT, Speziale P, O’Gara JP, Potts JR, Foster TJ. 2010. Role of surface protein SasG in biofilm formation by Staphylococcus aureus. J Bacteriol 192:5663–5673. doi: 10.1128/JB.00628-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Formosa-Dague C, Speziale P, Foster TJ, Geoghegan JA, Dufrêne YF. 2016. Zinc-dependent mechanical properties of Staphylococcus aureus biofilm-forming surface protein SasG. Proc Natl Acad Sci U S A 113:410–415. doi: 10.1073/pnas.1519265113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 113.Foulston L, Elsholz AK, DeFrancesco AS, Losick R. 2014. The extracellular matrix of Staphylococcus aureus biofilms comprises cytoplasmic proteins that associate with the cell surface in response to decreasing pH. mBio 5:e01667-14. doi: 10.1128/mBio.01667-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dengler V, Foulston L, DeFrancesco AS, Losick R. 2015. an electrostatic net model for the role of extracellular DNA in biofilm formation by Staphylococcus aureus. J Bacteriol 197:3779–3787. doi: 10.1128/JB.00726-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Graf AC, Leonard A, Schäuble M, Rieckmann LM, Hoyer J, Maass S, Lalk M, Becher D, Pané-Farré J, Riedel K. 2019. Virulence factors produced by Staphylococcus aureus biofilms have a moonlighting function contributing to biofilm integrity. Mol Cell Proteomics 18:1036–1053. doi: 10.1074/mcp.RA118.001120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kavanaugh JS, Flack CE, Lister J, Ricker EB, Ibberson CB, Jenul C, Moormeier DE, Delmain EA, Bayles KW, Horswill AR. 2019. Identification of extracellular DNA-binding proteins in the biofilm matrix. mBio 10:e01137-19. doi: 10.1128/mBio.01137-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schaeffer CR, Hoang TN, Sudbeck CM, Alawi M, Tolo IE, Robinson DA, Horswill AR, Rohde H, Fey PD. 2016. Versatility of biofilm matrix molecules in Staphylococcus epidermidis clinical isolates and importance of polysaccharide intercellular adhesin expression during high shear stress. mSphere 1:e00165-16. doi: 10.1128/mSphere.00165-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Otto M. 2013. Staphylococcal infections: mechanisms of biofilm maturation and detachment as critical determinants of pathogenicity. Annu Rev Med 64:175–188. doi: 10.1146/annurev-med-042711-140023. [DOI] [PubMed] [Google Scholar]
- 119.Banbula A, Potempa J, Travis J, Fernandez-Catalán C, Mann K, Huber R, Bode W, Medrano F. 1998. Amino-acid sequence and three-dimensional structure of the Staphylococcus aureus metalloproteinase at 1.72 A resolution. Structure 6:1185–1193. doi: 10.1016/s0969-2126(98)00118-x. [DOI] [PubMed] [Google Scholar]
- 120.Rice K, Peralta R, Bast D, de Azavedo J, McGavin MJ. 2001. Description of staphylococcus serine protease (ssp) operon in Staphylococcus aureus and nonpolar inactivation of sspA-encoded serine protease. Infect Immun 69:159–169. doi: 10.1128/IAI.69.1.159-169.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Golonka E, Filipek R, Sabat A, Sinczak A, Potempa J. 2004. Genetic characterization of staphopain genes in Staphylococcus aureus. Biol Chem 385:1059–1067. doi: 10.1515/BC.2004.137. [DOI] [PubMed] [Google Scholar]
- 122.Reed SB, Wesson CA, Liou LE, Trumble WR, Schlievert PM, Bohach GA, Bayles KW. 2001. Molecular characterization of a novel Staphylococcus aureus serine protease operon. Infect Immun 69:1521–1527. doi: 10.1128/IAI.69.3.1521-1527.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Beenken KE, Blevins JS, Smeltzer MS. 2003. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect Immun 71:4206–4211. doi: 10.1128/iai.71.7.4206-4211.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Trotonda MP, Manna AC, Cheung AL, Lasa I, Penadés JR. 2005. SarA positively controls Bap-dependent biofilm formation in Staphylococcus aureus. J Bacteriol 187:5790–5798. doi: 10.1128/JB.187.16.5790-5798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tsang LH, Cassat JE, Shaw LN, Beenken KE, Smeltzer MS. 2008. Factors contributing to the biofilm-deficient phenotype of Staphylococcus aureus sarA mutants. PLoS One 3:e3361. doi: 10.1371/journal.pone.0003361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lauderdale KJ, Boles BR, Cheung AL, Horswill AR. 2009. Interconnections between sigma B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation. Infect Immun 77:1623–1635. doi: 10.1128/IAI.01036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Martí M, Trotonda MP, Tormo-Más MA, Vergara-Irigaray M, Cheung AL, Lasa I, Penadés JR. 2010. Extracellular proteases inhibit protein-dependent biofilm formation in Staphylococcus aureus. Microbes Infect 12:55–64. doi: 10.1016/j.micinf.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 128.Zielinska AK, Beenken KE, Mrak LN, Spencer HJ, Post GR, Skinner RA, Tackett AJ, Horswill AR, Smeltzer MS. 2012. sarA-mediated repression of protease production plays a key role in the pathogenesis of Staphylococcus aureus USA300 isolates. Mol Microbiol 86:1183–1196. doi: 10.1111/mmi.12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mootz JM, Malone CL, Shaw LN, Horswill AR. 2013. Staphopains modulate Staphylococcus aureus biofilm integrity. Infect Immun 81:3227–3238. doi: 10.1128/IAI.00377-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.McGavin MJ, Zahradka C, Rice K, Scott JE. 1997. Modification of the Staphylococcus aureus fibronectin binding phenotype by V8 protease. Infect Immun 65:2621–2628. doi: 10.1128/IAI.65.7.2621-2628.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Abraham NM, Jefferson KK. 2012. Staphylococcus aureus clumping factor B mediates biofilm formation in the absence of calcium. Microbiology 158:1504–1512. doi: 10.1099/mic.0.057018-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Teufel P, Götz F. 1993. Characterization of an extracellular metalloprotease with elastase activity from Staphylococcus epidermidis. J Bacteriol 175:4218–4224. doi: 10.1128/jb.175.13.4218-4224.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Iwase T, Uehara Y, Shinji H, Tajima A, Seo H, Takada K, Agata T, Mizunoe Y. 2010. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 465:346–349. doi: 10.1038/nature09074. [DOI] [PubMed] [Google Scholar]
- 134.Sugimoto S, Iwamoto T, Takada K, Okuda K, Tajima A, Iwase T, Mizunoe Y. 2013. Staphylococcus epidermidis Esp degrades specific proteins associated with Staphylococcus aureus biofilm formation and host-pathogen interaction. J Bacteriol 195:1645–1655. doi: 10.1128/JB.01672-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chen C, Krishnan V, Macon K, Manne K, Narayana SV, Schneewind O. 2013. Secreted proteases control autolysin-mediated biofilm growth of Staphylococcus aureus. J Biol Chem 288:29440–29452. doi: 10.1074/jbc.M113.502039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dubin G, Chmiel D, Mak P, Rakwalska M, Rzychon M, Dubin A. 2001. Molecular cloning and biochemical characterisation of proteases from Staphylococcus epidermidis. Biol Chem 382:1575–1582. doi: 10.1515/BC.2001.192. [DOI] [PubMed] [Google Scholar]
- 137.Martínez-García S, Rodríguez-Martínez S, Cancino-Diaz ME, Cancino-Diaz JC. 2018. Extracellular proteases of Staphylococcus epidermidis: roles as virulence factors and their participation in biofilm. APMIS 126:177–185. doi: 10.1111/apm.12805. [DOI] [PubMed] [Google Scholar]
- 138.Tang J, Zhou R, Shi X, Kang M, Wang H, Chen H. 2008. Two thermostable nucleases coexisted in Staphylococcus aureus: evidence from mutagenesis and in vitro expression. FEMS Microbiol Lett 284:176–183. doi: 10.1111/j.1574-6968.2008.01194.x. [DOI] [PubMed] [Google Scholar]
- 139.Hu Y, Xie Y, Tang J, Shi X. 2012. Comparative expression analysis of two thermostable nuclease genes in Staphylococcus aureus. Foodborne Pathog Dis 9:265–271. doi: 10.1089/fpd.2011.1033. [DOI] [PubMed] [Google Scholar]
- 140.Beenken KE, Spencer H, Griffin LM, Smeltzer MS. 2012. Impact of extracellular nuclease production on the biofilm phenotype of Staphylococcus aureus under in vitro and in vivo conditions. Infect Immun 80:1634–1638. doi: 10.1128/IAI.06134-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Olson ME, Nygaard TK, Ackermann L, Watkins RL, Zurek OW, Pallister KB, Griffith S, Kiedrowski MR, Flack CE, Kavanaugh JS, Kreiswirth BN, Horswill AR, Voyich JM. 2013. Staphylococcus aureus nuclease is an SaeRS-dependent virulence factor. Infect Immun 81:1316–1324. doi: 10.1128/IAI.01242-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Izano EA, Amarante MA, Kher WB, Kaplan JB. 2008. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl Environ Microbiol 74:470–476. doi: 10.1128/AEM.02073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Berends ETM, Horswill AR, Haste NM, Monestier M, Nizet V, von Köckritz-Blickwede M. 2010. Nuclease expression by Staphylococcus aureus facilitates escape from neutrophil extracellular traps. J Innate Immun 2:576–586. doi: 10.1159/000319909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kiedrowski MR, Crosby HA, Hernandez FJ, Malone CL, McNamara JO II, Horswill AR. 2014. Staphylococcus aureus Nuc2 is a functional, surface-attached extracellular nuclease. PLoS One 9:e95574. doi: 10.1371/journal.pone.0095574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wang R, Braughton KR, Kretschmer D, Bach TH, Queck SY, Li M, Kennedy AD, Dorward DW, Klebanoff SJ, Peschel A, DeLeo FR, Otto M. 2007. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med 13:1510–1514. doi: 10.1038/nm1656. [DOI] [PubMed] [Google Scholar]
- 146.Vuong C, Gerke C, Somerville GA, Fischer ER, Otto M. 2003. Quorum-sensing control of biofilm factors in Staphylococcus epidermidis. J Infect Dis 188:706–718. doi: 10.1086/377239. [DOI] [PubMed] [Google Scholar]
- 147.Schwartz K, Syed AK, Stephenson RE, Rickard AH, Boles BR. 2012. Functional amyloids composed of phenol soluble modulins stabilize Staphylococcus aureus biofilms. PLoS Pathog 8:e1002744. doi: 10.1371/journal.ppat.1002744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Schwartz K, Ganesan M, Payne DE, Solomon MJ, Boles BR. 2016. Extracellular DNA facilitates the formation of functional amyloids in Staphylococcus aureus biofilms. Mol Microbiol 99:123–134. doi: 10.1111/mmi.13219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zheng Y, Joo HS, Nair V, Le KY, Otto M. 2018. Do amyloid structures formed by Staphylococcus aureus phenol-soluble modulins have a biological function? Int J Med Microbiol 308:675–682. doi: 10.1016/j.ijmm.2017.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Vuong C, Dürr M, Carmody AB, Peschel A, Klebanoff SJ, Otto M. 2004. Regulated expression of pathogen-associated molecular pattern molecules in Staphylococcus epidermidis: quorum-sensing determines pro-inflammatory capacity and production of phenol-soluble modulins. Cell Microbiol 6:753–759. doi: 10.1111/j.1462-5822.2004.00401.x. [DOI] [PubMed] [Google Scholar]
- 151.Wang R, Khan BA, Cheung GY, Bach TH, Jameson-Lee M, Kong KF, Queck SY, Otto M. 2011. Staphylococcus epidermidis surfactant peptides promote biofilm maturation and dissemination of biofilm-associated infection in mice. J Clin Invest 121:238–248. doi: 10.1172/JCI42520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mehlin C, Headley CM, Klebanoff SJ. 1999. An inflammatory polypeptide complex from Staphylococcus epidermidis: isolation and characterization. J Exp Med 189:907–918. doi: 10.1084/jem.189.6.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Parsek MR, Greenberg EP. 2005. Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol 13:27–33. doi: 10.1016/j.tim.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 154.Le KY, Otto M. 2015. Quorum-sensing regulation in staphylococci-an overview. Front Microbiol 6:1174. doi: 10.3389/fmicb.2015.01174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kong KF, Vuong C, Otto M. 2006. Staphylococcus quorum sensing in biofilm formation and infection. Int J Med Microbiol 296:133–139. doi: 10.1016/j.ijmm.2006.01.042. [DOI] [PubMed] [Google Scholar]
- 156.Otto M. 2008. Staphylococcal biofilms. Curr Top Microbiol Immunol 322:207–228. doi: 10.1007/978-3-540-75418-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wolska KI, Grudniak AM, Rudnicka Z, Markowska K. 2016. Genetic control of bacterial biofilms. J Appl Genet 57:225–238. doi: 10.1007/s13353-015-0309-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Fechter P, Caldelari I, Lioliou E, Romby P. 2014. Novel aspects of RNA regulation in Staphylococcus aureus. FEBS Lett 588:2523–2529. doi: 10.1016/j.febslet.2014.05.037. [DOI] [PubMed] [Google Scholar]
- 159.Desgranges E, Marzi S, Moreau K, Romby P, Caldelari I. 2019. Noncoding RNA. Microbiol Spectr 7:GPP3-0038-2018. doi: 10.1128/microbiolspec.GPP3-0038-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Waters CM, Bassler BL. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol 21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 161.Bassler BL. 2002. Small talk. Cell-to-cell communication in bacteria. Cell 109:421–424. doi: 10.1016/s0092-8674(02)00749-3. [DOI] [PubMed] [Google Scholar]
- 162.Fuqua C, Parsek MR, Greenberg EP. 2001. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu Rev Genet 35:439–468. doi: 10.1146/annurev.genet.35.102401.090913. [DOI] [PubMed] [Google Scholar]
- 163.Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, Moghazeh S. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J 12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ji G, Beavis R, Novick RP. 1997. Bacterial interference caused by autoinducing peptide variants. Science 276:2027–2030. doi: 10.1126/science.276.5321.2027. [DOI] [PubMed] [Google Scholar]
- 165.Zhang L, Gray L, Novick RP, Ji G. 2002. Transmembrane topology of AgrB, the protein involved in the post-translational modification of AgrD in Staphylococcus aureus. J Biol Chem 277:34736–34742. doi: 10.1074/jbc.M205367200. [DOI] [PubMed] [Google Scholar]
- 166.Kavanaugh JS, Thoendel M, Horswill AR. 2007. A role for type I signal peptidase in Staphylococcus aureus quorum sensing. Mol Microbiol 65:780–798. doi: 10.1111/j.1365-2958.2007.05830.x. [DOI] [PubMed] [Google Scholar]
- 167.Novick RP, Projan SJ, Kornblum J, Ross HF, Ji G, Kreiswirth B, Vandenesch F, Moghazeh S. 1995. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol Gen Genet 248:446–458. doi: 10.1007/BF02191645. [DOI] [PubMed] [Google Scholar]
- 168.Cheung GY, Wang R, Khan BA, Sturdevant DE, Otto M. 2011. Role of the accessory gene regulator agr in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. Infect Immun 79:1927–1935. doi: 10.1128/IAI.00046-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Geisinger E, Adhikari RP, Jin R, Ross HF, Novick RP. 2006. Inhibition of rot translation by RNAIII, a key feature of agr function. Mol Microbiol 61:1038–1048. doi: 10.1111/j.1365-2958.2006.05292.x. [DOI] [PubMed] [Google Scholar]
- 170.Mootz JM, Benson MA, Heim CE, Crosby HA, Kavanaugh JS, Dunman PM, Kielian T, Torres VJ, Horswill AR. 2015. Rot is a key regulator of Staphylococcus aureus biofilm formation. Mol Microbiol 96:388–404. doi: 10.1111/mmi.12943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ji G, Beavis RC, Novick RP. 1995. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc Natl Acad Sci U S A 92:12055–12059. doi: 10.1073/pnas.92.26.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Koenig RL, Ray JL, Maleki SJ, Smeltzer MS, Hurlburt BK. 2004. Staphylococcus aureus AgrA binding to the RNAIII-agr regulatory region. J Bacteriol 186:7549–7555. doi: 10.1128/JB.186.22.7549-7555.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Queck SY, Jameson-Lee M, Villaruz AE, Bach TH, Khan BA, Sturdevant DE, Ricklefs SM, Li M, Otto M. 2008. RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol Cell 32:150–158. doi: 10.1016/j.molcel.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 175.Saravia-Otten P, Müller HP, Arvidson S. 1997. Transcription of Staphylococcus aureus fibronectin binding protein genes is negatively regulated by agr and an agr-independent mechanism. J Bacteriol 179:5259–5263. doi: 10.1128/jb.179.17.5259-5263.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ythier M, Resch G, Waridel P, Panchaud A, Gfeller A, Majcherczyk P, Quadroni M, Moreillon P. 2012. Proteomic and transcriptomic profiling of Staphylococcus aureus surface LPXTG-proteins: correlation with agr genotypes and adherence phenotypes. Mol Cell Proteomics 11:1123–1139. doi: 10.1074/mcp.M111.014191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Yarwood JM, Bartels DJ, Volper EM, Greenberg EP. 2004. Quorum sensing in Staphylococcus aureus biofilms. J Bacteriol 186:1838–1850. doi: 10.1128/jb.186.6.1838-1850.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Boles BR, Horswill AR. 2008. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog 4:e1000052. doi: 10.1371/journal.ppat.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Oscarsson J, Tegmark-Wisell K, Arvidson S. 2006. Coordinated and differential control of aureolysin (aur) and serine protease (sspA) transcription in Staphylococcus aureus by sarA, rot and agr (RNAIII). Int J Med Microbiol 296:365–380. doi: 10.1016/j.ijmm.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 180.Novick RP. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol 48:1429–1449. doi: 10.1046/j.1365-2958.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- 181.Lauderdale KJ, Malone CL, Boles BR, Morcuende J, Horswill AR. 2010. Biofilm dispersal of community-associated methicillin-resistant Staphylococcus aureus on orthopedic implant material. J Orthop Res 28:55–61. doi: 10.1002/jor.20943. [DOI] [PubMed] [Google Scholar]
- 182.Vuong C, Saenz HL, Götz F, Otto M. 2000. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J Infect Dis 182:1688–1693. doi: 10.1086/317606. [DOI] [PubMed] [Google Scholar]
- 183.Jenul C, Horswill AR. 2018. Regulation of Staphylococcus aureus virulence. Microbiol Spectr 6:GPP3-0031-2018. doi: 10.1128/microbiolspec.GPP3-0031-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Le KY, Dastgheyb S, Ho TV, Otto M. 2014. Molecular determinants of staphylococcal biofilm dispersal and structuring. Front Cell Infect Microbiol 4:167. doi: 10.3389/fcimb.2014.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Cheung AL, Eberhardt KJ, Chung E, Yeaman MR, Sullam PM, Ramos M, Bayer AS. 1994. Diminished virulence of a sar-/agr- mutant of Staphylococcus aureus in the rabbit model of endocarditis. J Clin Invest 94:1815–1822. doi: 10.1172/JCI117530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Gillaspy AF, Hickmon SG, Skinner RA, Thomas JR, Nelson CL, Smeltzer MS. 1995. Role of the accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infect Immun 63:3373–3380. doi: 10.1128/IAI.63.9.3373-3380.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Fowler VG Jr, Sakoulas G, McIntyre LM, Meka VG, Arbeit RD, Cabell CH, Stryjewski ME, Eliopoulos GM, Reller LB, Corey GR, Jones T, Lucindo N, Yeaman MR, Bayer AS. 2004. Persistent bacteremia due to methicillin-resistant Staphylococcus aureus infection is associated with agr dysfunction and low-level in vitro resistance to thrombin-induced platelet microbicidal protein. J Infect Dis 190:1140–1149. doi: 10.1086/423145. [DOI] [PubMed] [Google Scholar]
- 188.Goerke C, Wolz C. 2010. Adaptation of Staphylococcus aureus to the cystic fibrosis lung. Int J Med Microbiol 300:520–525. doi: 10.1016/j.ijmm.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 189.Traber KE, Lee E, Benson S, Corrigan R, Cantera M, Shopsin B, Novick RP. 2008. agr function in clinical Staphylococcus aureus isolates. Microbiology 154:2265–2274. doi: 10.1099/mic.0.2007/011874-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Vuong C, Kocianova S, Yao Y, Carmody AB, Otto M. 2004. Increased colonization of indwelling medical devices by quorum-sensing mutants of Staphylococcus epidermidis in vivo. J Infect Dis 190:1498–1505. doi: 10.1086/424487. [DOI] [PubMed] [Google Scholar]
- 191.Dai L, Yang L, Parsons C, Findlay VJ, Molin S, Qin Z. 2012. Staphylococcus epidermidis recovered from indwelling catheters exhibit enhanced biofilm dispersal and “self-renewal” through downregulation of agr. BMC Microbiol 12:102. doi: 10.1186/1471-2180-12-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Dastgheyb SS, Villaruz AE, Le KY, Tan VY, Duong AC, Chatterjee SS, Cheung GY, Joo HS, Hickok NJ, Otto M. 2015. Role of phenol-soluble modulins in formation of Staphylococcus aureus biofilms in synovial fluid. Infect Immun 83:2966–2975. doi: 10.1128/IAI.00394-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Sandoz KM, Mitzimberg SM, Schuster M. 2007. Social cheating in Pseudomonas aeruginosa quorum sensing. Proc Natl Acad Sci U S A 104:15876–15881. doi: 10.1073/pnas.0705653104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Suligoy CM, Lattar SM, Noto Llana M, González CD, Alvarez LP, Robinson DA, Gómez MI, Buzzola FR, Sordelli DO. 2018. Mutation of Agr is associated with the adaptation of Staphylococcus aureus to the host during chronic osteomyelitis. Front Cell Infect Microbiol 8:18. doi: 10.3389/fcimb.2018.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Altman DR, Sullivan MJ, Chacko KI, Balasubramanian D, Pak TR, Sause WE, Kumar K, Sebra R, Deikus G, Attie O, Rose H, Lewis M, Fulmer Y, Bashir A, Kasarskis A, Schadt EE, Richardson AR, Torres VJ, Shopsin B, van Bakel H. 2018. Genome plasticity of agr-defective Staphylococcus aureus during clinical infection. Infect Immun 86:e00331-18. doi: 10.1128/IAI.00331-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Shopsin B, Eaton C, Wasserman GA, Mathema B, Adhikari RP, Agolory S, Altman DR, Holzman RS, Kreiswirth BN, Novick RP. 2010. Mutations in agr do not persist in natural populations of methicillin-resistant Staphylococcus aureus. J Infect Dis 202:1593–1599. doi: 10.1086/656915. [DOI] [PubMed] [Google Scholar]
- 197.Regassa LB, Novick RP, Betley MJ. 1992. Glucose and nonmaintained pH decrease expression of the accessory gene regulator (agr) in Staphylococcus aureus. Infect Immun 60:3381–3388. doi: 10.1128/IAI.60.8.3381-3388.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Weinrick B, Dunman PM, McAleese F, Murphy E, Projan SJ, Fang Y, Novick RP. 2004. Effect of mild acid on gene expression in Staphylococcus aureus. J Bacteriol 186:8407–8423. doi: 10.1128/JB.186.24.8407-8423.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Schaeffer CR, Woods KM, Longo GM, Kiedrowski MR, Paharik AE, Buttner H, Christner M, Boissy RJ, Horswill AR, Rohde H, Fey PD. 2015. Accumulation-associated protein enhances Staphylococcus epidermidis biofilm formation under dynamic conditions and is required for infection in a rat catheter model. Infect Immun 83:214–226. doi: 10.1128/IAI.02177-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Seidl K, Goerke C, Wolz C, Mack D, Berger-Bächi B, Bischoff M. 2008. Staphylococcus aureus CcpA affects biofilm formation. Infect Immun 76:2044–2050. doi: 10.1128/IAI.00035-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Seidl K, Stucki M, Ruegg M, Goerke C, Wolz C, Harris L, Berger-Bächi B, Bischoff M. 2006. Staphylococcus aureus CcpA affects virulence determinant production and antibiotic resistance. Antimicrob Agents Chemother 50:1183–1194. doi: 10.1128/AAC.50.4.1183-1194.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Rice KC, Mann EE, Endres JL, Weiss EC, Cassat JE, Smeltzer MS, Bayles KW. 2007. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Natl Acad Sci U S A 104:8113–8118. doi: 10.1073/pnas.0610226104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Bullen JJ. 1981. The significance of iron in infection. Rev Infect Dis 3:1127–1138. doi: 10.1093/clinids/3.6.1127. [DOI] [PubMed] [Google Scholar]
- 204.Parrow NL, Fleming RE, Minnick MF. 2013. Sequestration and scavenging of iron in infection. Infect Immun 81:3503–3514. doi: 10.1128/IAI.00602-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Xiong A, Singh VK, Cabrera G, Jayaswal RK. 2000. Molecular characterization of the ferric-uptake regulator, fur, from Staphylococcus aureus. Microbiology 146:659–668. doi: 10.1099/00221287-146-3-659. [DOI] [PubMed] [Google Scholar]
- 206.Johnson M, Cockayne A, Williams PH, Morrissey JA. 2005. Iron-responsive regulation of biofilm formation in Staphylococcus aureus involves Fur-dependent and Fur-independent mechanisms. J Bacteriol 187:8211–8215. doi: 10.1128/JB.187.23.8211-8215.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Johnson M, Cockayne A, Morrissey JA. 2008. Iron-regulated biofilm formation in Staphylococcus aureus Newman requires ica and the secreted protein Emp. Infect Immun 76:1756–1765. doi: 10.1128/IAI.01635-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Johnson M, Sengupta M, Purves J, Tarrant E, Williams PH, Cockayne A, Muthaiyan A, Stephenson R, Ledala N, Wilkinson BJ, Jayaswal RK, Morrissey JA. 2011. Fur is required for the activation of virulence gene expression through the induction of the sae regulatory system in Staphylococcus aureus. Int J Med Microbiol 301:44–52. doi: 10.1016/j.ijmm.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Lin MH, Shu JC, Huang HY, Cheng YC. 2012. Involvement of iron in biofilm formation by Staphylococcus aureus. PLoS One 7:e34388. doi: 10.1371/journal.pone.0034388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Oliveira F, França A, Cerca N. 2017. Staphylococcus epidermidis is largely dependent on iron availability to form biofilms. Int J Med Microbiol 307:552–563. doi: 10.1016/j.ijmm.2017.08.009. [DOI] [PubMed] [Google Scholar]
- 211.Purevdorj B, Costerton JW, Stoodley P. 2002. Influence of hydrodynamics and cell signaling on the structure and behavior of Pseudomonas aeruginosa biofilms. Appl Environ Microbiol 68:4457–4464. doi: 10.1128/AEM.68.9.4457-4464.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Kim MK, Ingremeau F, Zhao A, Bassler BL, Stone HA. 2016. Local and global consequences of flow on bacterial quorum sensing. Nat Microbiol 1:15005. doi: 10.1038/nmicrobiol.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Kirisits MJ, Margolis JJ, Purevdorj-Gage BL, Vaughan B, Chopp DL, Stoodley P, Parsek MR. 2007. Influence of the hydrodynamic environment on quorum sensing in Pseudomonas aeruginosa biofilms. J Bacteriol 189:8357–8360. doi: 10.1128/JB.01040-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Giraudo AT, Calzolari A, Cataldi AA, Bogni C, Nagel R. 1999. The sae locus of Staphylococcus aureus encodes a two-component regulatory system. FEMS Microbiol Lett 177:15–22. doi: 10.1111/j.1574-6968.1999.tb13707.x. [DOI] [PubMed] [Google Scholar]
- 215.Liu Q, Yeo WS, Bae T. 2016. The SaeRS two-component system of Staphylococcus aureus. Genes (Basel) 7:81. doi: 10.3390/genes7100081. [DOI] [Google Scholar]
- 216.Xiong YQ, Willard J, Yeaman MR, Cheung AL, Bayer AS. 2006. Regulation of Staphylococcus aureus alpha-toxin gene (hla) expression by agr, sarA, and sae in vitro and in experimental infective endocarditis. J Infect Dis 194:1267–1275. doi: 10.1086/508210. [DOI] [PubMed] [Google Scholar]
- 217.Liang X, Yu C, Sun J, Liu H, Landwehr C, Holmes D, Ji Y. 2006. Inactivation of a two-component signal transduction system, SaeRS, eliminates adherence and attenuates virulence of Staphylococcus aureus. Infect Immun 74:4655–4665. doi: 10.1128/IAI.00322-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Rogasch K, Rühmling V, Pané-Farré J, Hoper D, Weinberg C, Fuchs S, Schmudde M, Bröker BM, Wolz C, Hecker M, Engelmann S. 2006. Influence of the two-component system SaeRS on global gene expression in two different Staphylococcus aureus strains. J Bacteriol 188:7742–7758. doi: 10.1128/JB.00555-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Nygaard TK, Pallister KB, Ruzevich P, Griffith S, Vuong C, Voyich JM. 2010. SaeR binds a consensus sequence within virulence gene promoters to advance USA300 pathogenesis. J Infect Dis 201:241–254. doi: 10.1086/649570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Flack CE, Zurek OW, Meishery DD, Pallister KB, Malone CL, Horswill AR, Voyich JM. 2014. Differential regulation of staphylococcal virulence by the sensor kinase SaeS in response to neutrophil-derived stimuli. Proc Natl Acad Sci U S A 111:E2037–E2045. doi: 10.1073/pnas.1322125111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Mainiero M, Goerke C, Geiger T, Gonser C, Herbert S, Wolz C. 2010. Differential target gene activation by the Staphylococcus aureus two-component system saeRS. J Bacteriol 192:613–623. doi: 10.1128/JB.01242-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Cue D, Junecko JM, Lei MG, Blevins JS, Smeltzer MS, Lee CY. 2015. SaeRS-dependent inhibition of biofilm formation in Staphylococcus aureus Newman. PLoS One 10:e0123027. doi: 10.1371/journal.pone.0123027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.DelMain EA, Moormeier DE, Endres JL, Hodges RE, Sadykov MR, Horswill AR, Bayles KW. 2020. Stochastic expression of Sae-dependent virulence genes during Staphylococcus aureus biofilm development is dependent on SaeS. mBio 11:e03081-19. doi: 10.1128/mBio.03081-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Handke LD, Rogers KL, Olson ME, Somerville GA, Jerrells TJ, Rupp ME, Dunman PM, Fey PD. 2008. Staphylococcus epidermidis saeR is an effector of anaerobic growth and a mediator of acute inflammation. Infect Immun 76:141–152. doi: 10.1128/IAI.00556-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Lou Q, Zhu T, Hu J, Ben H, Yang J, Yu F, Liu J, Wu Y, Fischer A, Francois P, Schrenzel J, Qu D. 2011. Role of the SaeRS two-component regulatory system in Staphylococcus epidermidis autolysis and biofilm formation. BMC Microbiol 11:146. doi: 10.1186/1471-2180-11-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Chien Y, Manna AC, Projan SJ, Cheung AL. 1999. SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. J Biol Chem 274:37169–37176. doi: 10.1074/jbc.274.52.37169. [DOI] [PubMed] [Google Scholar]
- 227.Schumacher MA, Hurlburt BK, Brennan RG. 2001. Crystal structures of SarA, a pleiotropic regulator of virulence genes in S. aureus. Nature 409:215–219. doi: 10.1038/35051623. [DOI] [PubMed] [Google Scholar]
- 228.Bayer MG, Heinrichs JH, Cheung AL. 1996. The molecular architecture of the sar locus in Staphylococcus aureus. J Bacteriol 178:4563–4570. doi: 10.1128/jb.178.15.4563-4570.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Heinrichs JH, Bayer MG, Cheung AL. 1996. Characterization of the sar locus and its interaction with agr in Staphylococcus aureus. J Bacteriol 178:418–423. doi: 10.1128/jb.178.2.418-423.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Reyes D, Andrey DO, Monod A, Kelley WL, Zhang G, Cheung AL. 2011. Coordinated regulation by AgrA, SarA, and SarR to control agr expression in Staphylococcus aureus. J Bacteriol 193:6020–6031. doi: 10.1128/JB.05436-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Cheung AL, Ying P. 1994. Regulation of alpha- and beta-hemolysins by the sar locus of Staphylococcus aureus. J Bacteriol 176:580–585. doi: 10.1128/jb.176.3.580-585.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Chan PF, Foster SJ. 1998. Role of SarA in virulence determinant production and environmental signal transduction in Staphylococcus aureus. J Bacteriol 180:6232–6241. doi: 10.1128/JB.180.23.6232-6241.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Goerke C, Fluckiger U, Steinhuber A, Zimmerli W, Wolz C. 2001. Impact of the regulatory loci agr, sarA and sae of Staphylococcus aureus on the induction of alpha-toxin during device-related infection resolved by direct quantitative transcript analysis. Mol Microbiol 40:1439–1447. doi: 10.1046/j.1365-2958.2001.02494.x. [DOI] [PubMed] [Google Scholar]
- 234.Dunman PM, Murphy E, Haney S, Palacios D, Tucker-Kellogg G, Wu S, Brown EL, Zagursky RJ, Shlaes D, Projan SJ. 2001. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J Bacteriol 183:7341–7353. doi: 10.1128/JB.183.24.7341-7353.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Mrak LN, Zielinska AK, Beenken KE, Mrak IN, Atwood DN, Griffin LM, Lee CY, Smeltzer MS. 2012. saeRS and sarA act synergistically to repress protease production and promote biofilm formation in Staphylococcus aureus. PLoS One 7:e38453. doi: 10.1371/journal.pone.0038453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Blevins JS, Beenken KE, Elasri MO, Hurlburt BK, Smeltzer MS. 2002. Strain-dependent differences in the regulatory roles of sarA and agr in Staphylococcus aureus. Infect Immun 70:470–480. doi: 10.1128/iai.70.2.470-480.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Loughran AJ, Atwood DN, Anthony AC, Harik NS, Spencer HJ, Beenken KE, Smeltzer MS. 2014. Impact of individual extracellular proteases on Staphylococcus aureus biofilm formation in diverse clinical isolates and their isogenic sarA mutants. Microbiologyopen 3:897–909. doi: 10.1002/mbo3.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Bischoff M, Entenza JM, Giachino P. 2001. Influence of a functional sigB operon on the global regulators sar and agr in Staphylococcus aureus. J Bacteriol 183:5171–5179. doi: 10.1128/jb.183.17.5171-5179.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Cheung AL, Nishina KA, Trotonda MP, Tamber S. 2008. The SarA protein family of Staphylococcus aureus. Int J Biochem Cell Biol 40:355–361. doi: 10.1016/j.biocel.2007.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Pané-Farré J, Jonas B, Förstner K, Engelmann S, Hecker M. 2006. The sigmaB regulon in Staphylococcus aureus and its regulation. Int J Med Microbiol 296:237–258. doi: 10.1016/j.ijmm.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 241.Atwood DN, Loughran AJ, Courtney AP, Anthony AC, Meeker DG, Spencer HJ, Gupta RK, Lee CY, Beenken KE, Smeltzer MS. 2015. Comparative impact of diverse regulatory loci on Staphylococcus aureus biofilm formation. Microbiologyopen 4:436–451. doi: 10.1002/mbo3.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Tormo MA, Marti M, Valle J, Manna AC, Cheung AL, Lasa I, Penadés JR. 2005. SarA is an essential positive regulator of Staphylococcus epidermidis biofilm development. J Bacteriol 187:2348–2356. doi: 10.1128/JB.187.7.2348-2356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Christner M, Heinze C, Busch M, Franke G, Hentschke M, Bayard Dühring S, Büttner H, Kotasinska M, Wischnewski V, Kroll G, Buck F, Molin S, Otto M, Rohde H. 2012. sarA negatively regulates Staphylococcus epidermidis biofilm formation by modulating expression of 1 MDa extracellular matrix binding protein and autolysis-dependent release of eDNA. Mol Microbiol 86:394–410. doi: 10.1111/j.1365-2958.2012.08203.x. [DOI] [PubMed] [Google Scholar]
- 244.McNamara PJ, Milligan-Monroe KC, Khalili S, Proctor RA. 2000. Identification, cloning, and initial characterization of rot, a locus encoding a regulator of virulence factor expression in Staphylococcus aureus. J Bacteriol 182:3197–3203. doi: 10.1128/jb.182.11.3197-3203.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Saïd-Salim B, Dunman PM, McAleese FM, Macapagal D, Murphy E, McNamara PJ, Arvidson S, Foster TJ, Projan SJ, Kreiswirth BN. 2003. Global regulation of Staphylococcus aureus genes by Rot. J Bacteriol 185:610–619. doi: 10.1128/jb.185.2.610-619.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Tuffs SW, Herfst CA, Baroja ML, Podskalniy VA, DeJong EN, Coleman CEM, McCormick JK. 2019. Regulation of toxic shock syndrome toxin-1 by the accessory gene regulator in Staphylococcus aureus is mediated by the repressor of toxins. Mol Microbiol 112:1163–1177. doi: 10.1111/mmi.14353. [DOI] [PubMed] [Google Scholar]
- 247.Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, Chevalier C, Helfer AC, Benito Y, Jacquier A, Gaspin C, Vandenesch F, Romby P. 2007. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev 21:1353–1366. doi: 10.1101/gad.423507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Benson MA, Lilo S, Wasserman GA, Thoendel M, Smith A, Horswill AR, Fraser J, Novick RP, Shopsin B, Torres VJ. 2011. Staphylococcus aureus regulates the expression and production of the staphylococcal superantigen-like secreted proteins in a Rot-dependent manner. Mol Microbiol 81:659–675. doi: 10.1111/j.1365-2958.2011.07720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Gimza BD, Larias MI, Budny BG, Shaw LN. 2019. Mapping the global network of extracellular protease regulation in Staphylococcus aureus. mSphere 4:e00676-19. doi: 10.1128/mSphere.00676-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Crosby HA, Schlievert PM, Merriman JA, King JM, Salgado-Pabón W, Horswill AR. 2016. The Staphylococcus aureus global regulator MgrA modulates clumping and virulence by controlling surface protein expression. PLoS Pathog 12:e1005604. doi: 10.1371/journal.ppat.1005604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Crosby HA, Tiwari N, Kwiecinski JM, Xu Z, Dykstra A, Jenul C, Fuentes EJ, Horswill AR. 2020. The Staphylococcus aureus ArlRS two-component system regulates virulence factor expression through MgrA. Mol Microbiol 113:103–122. doi: 10.1111/mmi.14404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Kwiecinski JM, Crosby HA, Valotteau C, Hippensteel JA, Nayak MK, Chauhan AK, Schmidt EP, Dufrêne YF, Horswill AR. 2019. Staphylococcus aureus adhesion in endovascular infections is controlled by the ArlRS-MgrA signaling cascade. PLoS Pathog 15:e1007800. doi: 10.1371/journal.ppat.1007800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Deora R, Misra TK. 1995. Purification and characterization of DNA dependent RNA polymerase from Staphylococcus aureus. Biochem Biophys Res Commun 208:610–616. doi: 10.1006/bbrc.1995.1382. [DOI] [PubMed] [Google Scholar]
- 254.Deora R, Misra TK. 1996. Characterization of the primary sigma factor of Staphylococcus aureus. J Biol Chem 271:21828–21834. doi: 10.1074/jbc.271.36.21828. [DOI] [PubMed] [Google Scholar]
- 255.Kullik I, Giachino P, Fuchs T. 1998. Deletion of the alternative sigma factor sigmaB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J Bacteriol 180:4814–4820. doi: 10.1128/JB.180.18.4814-4820.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Shaw LN, Lindholm C, Prajsnar TK, Miller HK, Brown MC, Golonka E, Stewart GC, Tarkowski A, Potempa J. 2008. Identification and characterization of sigma, a novel component of the Staphylococcus aureus stress and virulence responses. PLoS One 3:e3844. doi: 10.1371/journal.pone.0003844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Morikawa K, Takemura AJ, Inose Y, Tsai M, Nguyen Thi Le T, Ohta T, Msadek T. 2012. Expression of a cryptic secondary sigma factor gene unveils natural competence for DNA transformation in Staphylococcus aureus. PLoS Pathog 8:e1003003. doi: 10.1371/journal.ppat.1003003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Fagerlund A, Granum PE, Håvarstein LS. 2014. Staphylococcus aureus competence genes: mapping of the SigH, ComK1 and ComK2 regulons by transcriptome sequencing. Mol Microbiol 94:557–579. doi: 10.1111/mmi.12767. [DOI] [PubMed] [Google Scholar]
- 259.Senn MM, Giachino P, Homerova D, Steinhuber A, Strassner J, Kormanec J, Flückiger U, Berger-Bächi B, Bischoff M. 2005. Molecular analysis and organization of the sigmaB operon in Staphylococcus aureus. J Bacteriol 187:8006–8019. doi: 10.1128/JB.187.23.8006-8019.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Donegan NP, Cheung AL. 2009. Regulation of the mazEF toxin-antitoxin module in Staphylococcus aureus and its impact on sigB expression. J Bacteriol 191:2795–2805. doi: 10.1128/JB.01713-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Bischoff M, Dunman P, Kormanec J, Macapagal D, Murphy E, Mounts W, Berger-Bächi B, Projan S. 2004. Microarray-based analysis of the Staphylococcus aureus sigmaB regulon. J Bacteriol 186:4085–4099. doi: 10.1128/JB.186.13.4085-4099.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Nair SP, Bischoff M, Senn MM, Berger-Bächi B. 2003. The sigma B regulon influences internalization of Staphylococcus aureus by osteoblasts. Infect Immun 71:4167–4170. doi: 10.1128/iai.71.7.4167-4170.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Ziebandt AK, Weber H, Rudolph J, Schmid R, Höper D, Engelmann S, Hecker M. 2001. Extracellular proteins of Staphylococcus aureus and the role of SarA and sigma B. Proteomics 1:480–493. doi:. [DOI] [PubMed] [Google Scholar]
- 264.Entenza JM, Moreillon P, Senn MM, Kormanec J, Dunman PM, Berger-Bächi B, Projan S, Bischoff M. 2005. Role of sigmaB in the expression of Staphylococcus aureus cell wall adhesins ClfA and FnbA and contribution to infectivity in a rat model of experimental endocarditis. Infect Immun 73:990–998. doi: 10.1128/IAI.73.2.990-998.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Horsburgh MJ, Aish JL, White IJ, Shaw L, Lithgow JK, Foster SJ. 2002. σB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J Bacteriol 184:5457–5467. doi: 10.1128/jb.184.19.5457-5467.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Geiger T, Goerke C, Mainiero M, Kraus D, Wolz C. 2008. The virulence regulator Sae of Staphylococcus aureus: promoter activities and response to phagocytosis-related signals. J Bacteriol 190:3419–3428. doi: 10.1128/JB.01927-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Bateman BT, Donegan NP, Jarry TM, Palma M, Cheung AL. 2001. Evaluation of a tetracycline-inducible promoter in Staphylococcus aureus in vitro and in vivo and its application in demonstrating the role of sigB in microcolony formation. Infect Immun 69:7851–7857. doi: 10.1128/IAI.69.12.7851-7857.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Rachid S, Ohlsen K, Wallner U, Hacker J, Hecker M, Ziebuhr W. 2000. Alternative transcription factor σB is involved in regulation of biofilm expression in a Staphylococcus aureus mucosal isolate. J Bacteriol 182:6824–6826. doi: 10.1128/jb.182.23.6824-6826.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Cerca N, Brooks JL, Jefferson KK. 2008. Regulation of the intercellular adhesin locus regulator (icaR) by SarA, sigmaB, and IcaR in Staphylococcus aureus. J Bacteriol 190:6530–6533. doi: 10.1128/JB.00482-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Valle J, Toledo-Arana A, Berasain C, Ghigo J-M, Amorena B, Penadés JR, Lasa I. 2003. SarA and not sigmaB is essential for biofilm development by Staphylococcus aureus. Mol Microbiol 48:1075–1087. doi: 10.1046/j.1365-2958.2003.03493.x. [DOI] [PubMed] [Google Scholar]
- 271.Valle J, Echeverz M, Lasa I. 2019. σB inhibits poly-N-acetylglucosamine exopolysaccharide synthesis and biofilm formation in Staphylococcus aureus. J Bacteriol 201:e0009819. doi: 10.1128/JB.00098-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Pozzi C, Waters EM, Rudkin JK, Schaeffer CR, Lohan AJ, Tong P, Loftus BJ, Pier GB, Fey PD, Massey RC, O’Gara JP. 2012. Methicillin resistance alters the biofilm phenotype and attenuates virulence in Staphylococcus aureus device-associated infections. PLoS Pathog 8:e1002626. doi: 10.1371/journal.ppat.1002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Knobloch JK, Jäger S, Horstkotte MA, Rohde H, Mack D. 2004. RsbU-dependent regulation of Staphylococcus epidermidis biofilm formation is mediated via the alternative sigma factor sigmaB by repression of the negative regulator gene icaR. Infect Immun 72:3838–3848. doi: 10.1128/IAI.72.7.3838-3848.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Handke LD, Slater SR, Conlon KM, O’Donnell ST, Olson ME, Bryant KA, Rupp ME, O’Gara JP, Fey PD. 2007. SigmaB and SarA independently regulate polysaccharide intercellular adhesin production in Staphylococcus epidermidis. Can J Microbiol 53:82–91. doi: 10.1139/w06-108. [DOI] [PubMed] [Google Scholar]
- 275.Jäger S, Jonas B, Pfanzelt D, Horstkotte MA, Rohde H, Mack D, Knobloch JK. 2009. Regulation of biofilm formation by sigma B is a common mechanism in Staphylococcus epidermidis and is not mediated by transcriptional regulation of sarA. Int J Artif Organs 32:584–591. doi: 10.1177/039139880903200907. [DOI] [PubMed] [Google Scholar]
- 276.Stenz L, Francois P, Whiteson K, Wolz C, Linder P, Schrenzel J. 2011. The CodY pleiotropic repressor controls virulence in gram-positive pathogens. FEMS Immunol Med Microbiol 62:123–139. doi: 10.1111/j.1574-695X.2011.00812.x. [DOI] [PubMed] [Google Scholar]
- 277.Pohl K, Francois P, Stenz L, Schlink F, Geiger T, Herbert S, Goerke C, Schrenzel J, Wolz C. 2009. CodY in Staphylococcus aureus: a regulatory link between metabolism and virulence gene expression. J Bacteriol 191:2953–2963. doi: 10.1128/JB.01492-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Majerczyk CD, Dunman PM, Luong TT, Lee CY, Sadykov MR, Somerville GA, Bodi K, Sonenshein AL. 2010. Direct targets of CodY in Staphylococcus aureus. J Bacteriol 192:2861–2877. doi: 10.1128/JB.00220-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Majerczyk CD, Sadykov MR, Luong TT, Lee C, Somerville GA, Sonenshein AL. 2008. Staphylococcus aureus CodY negatively regulates virulence gene expression. J Bacteriol 190:2257–2265. doi: 10.1128/JB.01545-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Roux A, Todd DA, Velázquez JV, Cech NB, Sonenshein AL. 2014. CodY-mediated regulation of the Staphylococcus aureus Agr system integrates nutritional and population density signals. J Bacteriol 196:1184–1196. doi: 10.1128/JB.00128-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Morfeldt E, Taylor D, von Gabain A, Arvidson S. 1995. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J 14:4569–4577. doi: 10.1002/j.1460-2075.1995.tb00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Augagneur Y, King AN, Germain-Amiot N, Sassi M, Fitzgerald JW, Sahukhal GS, Elasri MO, Felden B, Brinsmade SR. 2020. Analysis of the CodY RNome reveals RsaD as a stress-responsive riboregulator of overflow metabolism in Staphylococcus aureus. Mol Microbiol 113:309–325. doi: 10.1111/mmi.14418. [DOI] [PubMed] [Google Scholar]
- 283.Tu Quoc PH, Genevaux P, Pajunen M, Savilahti H, Georgopoulos C, Schrenzel J, Kelley WL. 2007. Isolation and characterization of biofilm formation-defective mutants of Staphylococcus aureus. Infect Immun 75:1079–1088. doi: 10.1128/IAI.01143-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Rom JS, Atwood DN, Beenken KE, Meeker DG, Loughran AJ, Spencer HJ, Lantz TL, Smeltzer MS. 2017. Impact of Staphylococcus aureus regulatory mutations that modulate biofilm formation in the USA300 strain LAC on virulence in a murine bacteremia model. Virulence 8:1776–1790. doi: 10.1080/21505594.2017.1373926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Waters NR, Samuels DJ, Behera RK, Livny J, Rhee KY, Sadykov MR, Brinsmade SR. 2016. A spectrum of CodY activities drives metabolic reorganization and virulence gene expression in Staphylococcus aureus. Mol Microbiol 101:495–514. doi: 10.1111/mmi.13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Mlynek KD, Bulock LL, Stone CJ, Curran LJ, Sadykov MR, Bayles KW, Brinsmade SR. 2020. Genetic and biochemical analysis of CodY-Mediated Cell aggregation in Staphylococcus aureus reveals an interaction between extracellular DNA and polysaccharide in the extracellular matrix. J Bacteriol 202:e00593-19. doi: 10.1128/JB.00593-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Jefferson KK, Cramton SE, Götz F, Pier GB. 2003. Identification of a 5-nucleotide sequence that controls expression of the ica locus in Staphylococcus aureus and characterization of the DNA-binding properties of IcaR. Mol Microbiol 48:889–899. doi: 10.1046/j.1365-2958.2003.03482.x. [DOI] [PubMed] [Google Scholar]
- 288.Ruiz de los Mozos I, Vergara-Irigaray M, Segura V, Villanueva M, Bitarte N, Saramago M, Domingues S, Arraiano CM, Fechter P, Romby P, Valle J, Solano C, Lasa I, Toledo-Arana A. 2013. Base pairing interaction between 5′- and 3′-UTRs controls icaR mRNA translation in Staphylococcus aureus. PLoS Genet 9:e1004001. doi: 10.1371/journal.pgen.1004001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Pamp SJ, Frees D, Engelmann S, Hecker M, Ingmer H. 2006. Spx is a global effector impacting stress tolerance and biofilm formation in Staphylococcus aureus. J Bacteriol 188:4861–4870. doi: 10.1128/JB.00194-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Cue D, Lei MG, Luong TT, Kuechenmeister L, Dunman PM, O’Donnell S, Rowe S, O’Gara JP, Lee CY. 2009. Rbf promotes biofilm formation by Staphylococcus aureus via repression of icaR, a negative regulator of icaADBC. J Bacteriol 191:6363–6373. doi: 10.1128/JB.00913-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Cue D, Lei MG, Lee CY. 2013. Activation of sarX by Rbf is required for biofilm formation and icaADBC expression in Staphylococcus aureus. J Bacteriol 195:1515–1524. doi: 10.1128/JB.00012-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Brandenberger M, Tschierske M, Giachino P, Wada A, Berger-Bächi B. 2000. Inactivation of a novel three-cistronic operon tcaR-tcaA-tcaB increases teicoplanin resistance in Staphylococcus aureus. Biochim Biophys Acta 1523:135–139. doi: 10.1016/s0304-4165(00)00133-1. [DOI] [PubMed] [Google Scholar]
- 293.Chang YM, Jeng WY, Ko TP, Yeh YJ, Chen CK, Wang AH. 2010. Structural study of TcaR and its complexes with multiple antibiotics from Staphylococcus epidermidis. Proc Natl Acad Sci U S A 107:8617–8622. doi: 10.1073/pnas.0913302107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Ulrich M, Bastian M, Cramton SE, Ziegler K, Pragman AA, Bragonzi A, Memmi G, Wolz C, Schlievert PM, Cheung A, Döring G. 2007. The staphylococcal respiratory response regulator SrrAB induces ica gene transcription and polysaccharide intercellular adhesin expression, protecting Staphylococcus aureus from neutrophil killing under anaerobic growth conditions. Mol Microbiol 65:1276–1287. doi: 10.1111/j.1365-2958.2007.05863.x. [DOI] [PubMed] [Google Scholar]
- 295.Wu Y, Wu Y, Zhu T, Han H, Liu H, Xu T, Francois P, Fischer A, Bai L, Götz F, Qu D. 2015. Staphylococcus epidermidis SrrAB regulates bacterial growth and biofilm formation differently under oxic and microaerobic conditions. J Bacteriol 197:459–476. doi: 10.1128/JB.02231-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Islam N, Ross JM, Marten MR. 2015. Proteome analyses of Staphylococcus aureus biofilm at elevated levels of NaCl. Clin Microbiol 4:219. doi: 10.4172/2327-5073.1000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Knobloch JK, Bartscht K, Sabottke A, Rohde H, Feucht HH, Mack D. 2001. Biofilm formation by Staphylococcus epidermidis depends on functional RsbU, an activator of the sigB operon: differential activation mechanisms due to ethanol and salt stress. J Bacteriol 183:2624–2633. doi: 10.1128/JB.183.8.2624-2633.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Rachid S, Ohlsen K, Witte W, Hacker J, Ziebuhr W. 2000. Effect of subinhibitory antibiotic concentrations on polysaccharide intercellular adhesin expression in biofilm-forming Staphylococcus epidermidis. Antimicrob Agents Chemother 44:3357–3363. doi: 10.1128/aac.44.12.3357-3363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Ziebuhr W, Krimmer V, Rachid S, Lössner I, Götz F, Hacker J. 1999. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol Microbiol 32:345–356. doi: 10.1046/j.1365-2958.1999.01353.x. [DOI] [PubMed] [Google Scholar]
- 300.Kiem S, Oh WS, Peck KR, Lee NY, Lee JY, Song JH, Hwang ES, Kim EC, Cha CY, Choe KW. 2004. Phase variation of biofilm formation in Staphylococcus aureus by IS 256 insertion and its impact on the capacity adhering to polyurethane surface. J Korean Med Sci 19:779–782. doi: 10.3346/jkms.2004.19.6.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Brooks JL, Jefferson KK. 2014. Phase variation of poly-N-acetylglucosamine expression in Staphylococcus aureus. PLoS Pathog 10:e1004292. doi: 10.1371/journal.ppat.1004292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Lerch MF, Schoenfelder SMK, Marincola G, Wencker FDR, Eckart M, Förstner KU, Sharma CM, Thormann KM, Kucklick M, Engelmann S, Ziebuhr W. 2019. A non-coding RNA from the intercellular adhesion (ica) locus of Staphylococcus epidermidis controls polysaccharide intercellular adhesion (PIA)-mediated biofilm formation. Mol Microbiol 111:1571–1591. doi: 10.1111/mmi.14238. [DOI] [PubMed] [Google Scholar]
- 303.Bronesky D, Desgranges E, Corvaglia A, François P, Caballero CJ, Prado L, Toledo-Arana A, Lasa I, Moreau K, Vandenesch F, Marzi S, Romby P, Caldelari I. 2019. A multifaceted small RNA modulates gene expression upon glucose limitation in Staphylococcus aureus. EMBO J 38:e99363. doi: 10.15252/embj.201899363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Schoenfelder SMK, Lange C, Prakash SA, Marincola G, Lerch MF, Wencker FDR, Förstner KU, Sharma CM, Ziebuhr W. 2019. The small non-coding RNA RsaE influences extracellular matrix composition in Staphylococcus epidermidis biofilm communities. PLoS Pathog 15:e1007618. doi: 10.1371/journal.ppat.1007618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Brunskill EW, Bayles KW. 1996. Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J Bacteriol 178:611–618. doi: 10.1128/jb.178.3.611-618.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Rice KC, Firek BA, Nelson JB, Yang SJ, Patton TG, Bayles KW. 2003. The Staphylococcus aureus cidAB operon: evaluation of its role in regulation of murein hydrolase activity and penicillin tolerance. J Bacteriol 185:2635–2643. doi: 10.1128/jb.185.8.2635-2643.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Ranjit DK, Endres JL, Bayles KW. 2011. Staphylococcus aureus CidA and LrgA proteins exhibit holin-like properties. J Bacteriol 193:2468–2476. doi: 10.1128/JB.01545-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Sadykov MR, Bayles KW. 2012. The control of death and lysis in staphylococcal biofilms: a coordination of physiological signals. Curr Opin Microbiol 15:211–215. doi: 10.1016/j.mib.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Groicher KH, Firek BA, Fujimoto DF, Bayles KW. 2000. The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J Bacteriol 182:1794–1801. doi: 10.1128/jb.182.7.1794-1801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Yang SJ, Rice KC, Brown RJ, Patton TG, Liou LE, Park YH, Bayles KW. 2005. A LysR-type regulator, CidR, is required for induction of the Staphylococcus aureus cidABC operon. J Bacteriol 187:5893–5900. doi: 10.1128/JB.187.17.5893-5900.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Rice KC, Patton T, Yang SJ, Dumoulin A, Bischoff M, Bayles KW. 2004. Transcription of the Staphylococcus aureus Cid and Lrg murein hydrolase regulators is affected by sigma factor B. J Bacteriol 186:3029–3037. doi: 10.1128/jb.186.10.3029-3037.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Fujimoto DF, Brunskill EW, Bayles KW. 2000. Analysis of genetic elements controlling Staphylococcus aureus lrgAB expression: potential role of DNA topology in SarA regulation. J Bacteriol 182:4822–4828. doi: 10.1128/jb.182.17.4822-4828.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Chu X, Xia R, He N, Fang Y. 2013. Role of Rot in bacterial autolysis regulation of Staphylococcus aureus NCTC8325. Res Microbiol 164:695–700. doi: 10.1016/j.resmic.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 314.Rice KC, Nelson JB, Patton TG, Yang SJ, Bayles KW. 2005. Acetic acid induces expression of the Staphylococcus aureus cidABC and lrgAB murein hydrolase regulator operons. J Bacteriol 187:813–821. doi: 10.1128/JB.187.3.813-821.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Sharma-Kuinkel BK, Mann EE, Ahn JS, Kuechenmeister LJ, Dunman PM, Bayles KW. 2009. The Staphylococcus aureus LytSR two-component regulatory system affects biofilm formation. J Bacteriol 191:4767–4775. doi: 10.1128/JB.00348-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Moormeier DE, Endres JL, Mann EE, Sadykov MR, Horswill AR, Rice KC, Fey PD, Bayles KW. 2013. Use of microfluidic technology to analyze gene expression during Staphylococcus aureus biofilm formation reveals distinct physiological niches. Appl Environ Microbiol 79:3413–3424. doi: 10.1128/AEM.00395-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Lehman MK, Bose JL, Sharma-Kuinkel BK, Moormeier DE, Endres JL, Sadykov MR, Biswas I, Bayles KW. 2015. Identification of the amino acids essential for LytSR-mediated signal transduction in Staphylococcus aureus and their roles in biofilm-specific gene expression. Mol Microbiol 95:723–737. doi: 10.1111/mmi.12902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Zhu T, Lou Q, Wu Y, Hu J, Yu F, Qu D. 2010. Impact of the Staphylococcus epidermidis LytSR two-component regulatory system on murein hydrolase activity, pyruvate utilization and global transcriptional profile. BMC Microbiol 10:287. doi: 10.1186/1471-2180-10-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Bassler BL, Wright M, Silverman MR. 1994. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol Microbiol 13:273–286. doi: 10.1111/j.1365-2958.1994.tb00422.x. [DOI] [PubMed] [Google Scholar]
- 320.Bassler BL, Wright M, Silverman MR. 1994. Sequence and function of LuxO, a negative regulator of luminescence in Vibrio harveyi. Mol Microbiol 12:403–412. doi: 10.1111/j.1365-2958.1994.tb01029.x. [DOI] [PubMed] [Google Scholar]
- 321.Schauder S, Shokat K, Surette MG, Bassler BL. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol Microbiol 41:463–476. doi: 10.1046/j.1365-2958.2001.02532.x. [DOI] [PubMed] [Google Scholar]
- 322.Chen X, Schauder S, Potier N, Van Dorsselaer A, Pelczer I, Bassler BL, Hughson FM. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545–549. doi: 10.1038/415545a. [DOI] [PubMed] [Google Scholar]
- 323.Doherty N, Holden MT, Qazi SN, Williams P, Winzer K. 2006. Functional analysis of luxS in Staphylococcus aureus reveals a role in metabolism but not quorum sensing. J Bacteriol 188:2885–2897. doi: 10.1128/JB.188.8.2885-2897.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Yu D, Zhao L, Xue T, Sun B. 2012. Staphylococcus aureus autoinducer-2 quorum sensing decreases biofilm formation in an icaR-dependent manner. BMC Microbiol 12:288. doi: 10.1186/1471-2180-12-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Xu L, Li H, Vuong C, Vadyvaloo V, Wang J, Yao Y, Otto M, Gao Q. 2006. Role of the luxS quorum-sensing system in biofilm formation and virulence of Staphylococcus epidermidis. Infect Immun 74:488–496. doi: 10.1128/IAI.74.1.488-496.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Li M, Villaruz AE, Vadyvaloo V, Sturdevant DE, Otto M. 2008. AI-2-dependent gene regulation in Staphylococcus epidermidis. BMC Microbiol 8:4. doi: 10.1186/1471-2180-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Ma R, Qiu S, Jiang Q, Sun H, Xue T, Cai G, Sun B. 2017. AI-2 quorum sensing negatively regulates rbf expression and biofilm formation in Staphylococcus aureus. Int J Med Microbiol 307:257–267. doi: 10.1016/j.ijmm.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 328.Xue T, Ni J, Shang F, Chen X, Zhang M. 2015. Autoinducer-2 increases biofilm formation via an ica- and bhp-dependent manner in Staphylococcus epidermidis RP62A. Microbes Infect 17:345–352. doi: 10.1016/j.micinf.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 329.McDonough KA, Rodriguez A. 2011. The myriad roles of cyclic AMP in microbial pathogens: from signal to sword. Nat Rev Microbiol 10:27–38. doi: 10.1038/nrmicro2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Ryjenkov DA, Simm R, Römling U, Gomelsky M. 2006. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J Biol Chem 281:30310–30314. doi: 10.1074/jbc.C600179200. [DOI] [PubMed] [Google Scholar]
- 331.Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, Mayer R, Braun S, de Vroom E, van der Marel GA, van Boom JH, Benziman M. 1987. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325:279–281. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- 332.Ha DG, O’Toole GA. 2015. c-di-GMP and its effects on biofilm formation and dispersion: a Pseudomonas Aeruginosa review. Microbiol Spectr 3:MB-0003-2014. doi: 10.1128/microbiolspec.MB-0003-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Corrigan RM, Gründling A. 2013. Cyclic di-AMP: another second messenger enters the fray. Nat Rev Microbiol 11:513–524. doi: 10.1038/nrmicro3069. [DOI] [PubMed] [Google Scholar]
- 334.Corrigan RM, Abbott JC, Burhenne H, Kaever V, Gründling A. 2011. c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog 7:e1002217. doi: 10.1371/journal.ppat.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Chaudhuri RR, Allen AG, Owen PJ, Shalom G, Stone K, Harrison M, Burgis TA, Lockyer M, Garcia-Lara J, Foster SJ, Pleasance SJ, Peters SE, Maskell DJ, Charles IG. 2009. Comprehensive identification of essential Staphylococcus aureus genes using transposon-mediated differential hybridisation (TMDH). BMC Genomics 10:291. doi: 10.1186/1471-2164-10-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Corrigan RM, Campeotto I, Jeganathan T, Roelofs KG, Lee VT, Gründling A. 2013. Systematic identification of conserved bacterial c-di-AMP receptor proteins. Proc Natl Acad Sci U S A 110:9084–9089. doi: 10.1073/pnas.1300595110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.DeFrancesco AS, Masloboeva N, Syed AK, DeLoughery A, Bradshaw N, Li GW, Gilmore MS, Walker S, Losick R. 2017. Genome-wide screen for genes involved in eDNA release during biofilm formation by Staphylococcus aureus. Proc Natl Acad Sci U S A 114:E5969–E5978. doi: 10.1073/pnas.1704544114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Gries CM, Bruger EL, Moormeier DE, Scherr TD, Waters CM, Kielian T. 2016. Cyclic di-AMP released from Staphylococcus aureus biofilm induces a macrophage type I interferon response. Infect Immun 84:3564–3574. doi: 10.1128/IAI.00447-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Lew DP, Waldvogel FA. 2004. Osteomyelitis. Lancet 364:369–379. doi: 10.1016/S0140-6736(04)16727-5. [DOI] [PubMed] [Google Scholar]
- 340.Muthukrishnan G, Masters EA, Daiss JL, Schwarz EM. 2019. Mechanisms of immune evasion and bone tissue colonization that make Staphylococcus aureus the primary pathogen in osteomyelitis. Curr Osteoporos Rep 17:395–404. doi: 10.1007/s11914-019-00548-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Trautner BW, Darouiche RO. 2004. Role of biofilm in catheter-associated urinary tract infection. Am J Infect Control 32:177–183. doi: 10.1016/j.ajic.2003.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Zimmerli W, Trampuz A, Ochsner PE. 2004. Prosthetic-joint infections. N Engl J Med 351:1645–1654. doi: 10.1056/NEJMra040181. [DOI] [PubMed] [Google Scholar]
- 343.Donlan RM. 2001. Biofilms and device-associated infections. Emerg Infect Dis 7:277–281. doi: 10.3201/eid0702.010226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Herrmann M, Vaudaux PE, Pittet D, Auckenthaler R, Lew PD, Schumacher-Perdreau F, Peters G, Waldvogel FA. 1988. Fibronectin, fibrinogen, and laminin act as mediators of adherence of clinical staphylococcal isolates to foreign material. J Infect Dis 158:693–701. doi: 10.1093/infdis/158.4.693. [DOI] [PubMed] [Google Scholar]
- 345.Datta R, Huang SS. 2008. Risk of infection and death due to methicillin-resistant Staphylococcus aureus in long-term carriers. Clin Infect Dis 47:176–181. doi: 10.1086/589241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Kwiecinski J, Kahlmeter G, Jin T. 2015. Biofilm formation by Staphylococcus aureus isolates from skin and soft tissue infections. Curr Microbiol 70:698–703. doi: 10.1007/s00284-014-0770-x. [DOI] [PubMed] [Google Scholar]
- 347.Gjødsbøl K, Christensen JJ, Karlsmark T, Jørgensen B, Klein BM, Krogfelt KA. 2006. Multiple bacterial species reside in chronic wounds: a longitudinal study. Int Wound J 3:225–231. doi: 10.1111/j.1742-481X.2006.00159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Fazli M, Bjarnsholt T, Kirketerp-Møller K, Jørgensen B, Andersen AS, Krogfelt KA, Givskov M, Tolker-Nielsen T. 2009. Nonrandom distribution of Pseudomonas aeruginosa and Staphylococcus aureus in chronic wounds. J Clin Microbiol 47:4084–4089. doi: 10.1128/JCM.01395-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Madsen SM, Westh H, Danielsen L, Rosdahl VT. 1996. Bacterial colonization and healing of venous leg ulcers. APMIS 104:895–899. doi: 10.1111/j.1699-0463.1996.tb04955.x. [DOI] [PubMed] [Google Scholar]
- 350.Bowling FL, Jude EB, Boulton AJ. 2009. MRSA and diabetic foot wounds: contaminating or infecting organisms? Curr Diab Rep 9:440–444. doi: 10.1007/s11892-009-0072-z. [DOI] [PubMed] [Google Scholar]
- 351.Lindsay S, Oates A, Bourdillon K. 2017. The detrimental impact of extracellular bacterial proteases on wound healing. Int Wound J 14:1237–1247. doi: 10.1111/iwj.12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Fowler VG Jr, Miro JM, Hoen B, Cabell CH, Abrutyn E, Rubinstein E, Corey GR, Spelman D, Bradley SF, Barsic B, Pappas PA, Anstrom KJ, Wray D, Fortes CQ, Anguera I, Athan E, Jones P, van der Meer JTM, Elliott TSJ, Levine DP, Bayer AS, ICE Investigators. 2005. Staphylococcus aureus endocarditis: a consequence of medical progress. JAMA 293:3012–3021. doi: 10.1001/jama.293.24.3012. [DOI] [PubMed] [Google Scholar]
- 353.Baddour LM, Wilson WR, Bayer AS, Fowler VG, Tleyjeh IM, Rybak MJ, Barsic B, Lockhart PB, Gewitz MH, Levison ME, Bolger AF, Steckelberg JM, Baltimore RS, Fink AM, O’Gara P, Taubert KA, American Heart Association Committee on Rheumatic Fever, Endocarditis, and Kawasaki Disease of the Council on Cardiovascular Disease in the Young, Council on Clinical Cardiology, Council on Cardiovascular Surgery and Anesthesia, and Stroke Council. 2015. Infective endocarditis in adults: diagnosis, antimicrobial therapy, and management of complications: a scientific statement for healthcare professionals from the American Heart Association. Circulation 132:1435–1486. doi: 10.1161/CIR.0000000000000296. [DOI] [PubMed] [Google Scholar]
- 354.Cresti A, Chiavarelli M, Scalese M, Nencioni C, Valentini S, Guerrini F, D’Aiello I, Picchi A, De Sensi F, Habib G. 2017. Epidemiological and mortality trends in infective endocarditis, a 17-year population-based prospective study. Cardiovasc Diagn Ther 7:27–35. doi: 10.21037/cdt.2016.08.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Holland TL, Baddour LM, Bayer AS, Hoen B, Miro JM, Fowler VG Jr. 2016. Infective endocarditis. Nat Rev Dis Primers 2:16059. doi: 10.1038/nrdp.2016.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Xiong YQ, Fowler VG, Yeaman MR, Perdreau-Remington F, Kreiswirth BN, Bayer AS. 2009. Phenotypic and genotypic characteristics of persistent methicillin-resistant Staphylococcus aureus bacteremia in vitro and in an experimental endocarditis model. J Infect Dis 199:201–208. doi: 10.1086/595738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Seidl K, Bayer AS, Fowler VG Jr, McKinnell JA, Abdel Hady W, Sakoulas G, Yeaman MR, Xiong YQ. 2011. Combinatorial phenotypic signatures distinguish persistent from resolving methicillin-resistant Staphylococcus aureus bacteremia isolates. Antimicrob Agents Chemother 55:575–582. doi: 10.1128/AAC.01028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Leid JG, Shirtliff ME, Costerton JW, Stoodley P. 2002. Human leukocytes adhere to, penetrate, and respond to Staphylococcus aureus biofilms. Infect Immun 70:6339–6345. doi: 10.1128/iai.70.11.6339-6345.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Ricciardi BF, Muthukrishnan G, Masters E, Ninomiya M, Lee CC, Schwarz EM. 2018. Staphylococcus aureus evasion of host immunity in the setting of prosthetic joint infection: biofilm and beyond. Curr Rev Musculoskelet Med 11:389–400. doi: 10.1007/s12178-018-9501-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Thurlow LR, Hanke ML, Fritz T, Angle A, Aldrich A, Williams SH, Engebretsen IL, Bayles KW, Horswill AR, Kielian T. 2011. Staphylococcus aureus biofilms prevent macrophage phagocytosis and attenuate inflammation in vivo. J Immunol 186:6585–6596. doi: 10.4049/jimmunol.1002794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Bhattacharya M, Berends ETM, Chan R, Schwab E, Roy S, Sen CK, Torres VJ, Wozniak DJ. 2018. Staphylococcus aureus biofilms release leukocidins to elicit extracellular trap formation and evade neutrophil-mediated killing. Proc Natl Acad Sci U S A 115:7416–7421. doi: 10.1073/pnas.1721949115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. 2004. Neutrophil extracellular traps kill bacteria. Science 303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 363.Schommer NN, Christner M, Hentschke M, Ruckdeschel K, Aepfelbacher M, Rohde H. 2011. Staphylococcus epidermidis uses distinct mechanisms of biofilm formation to interfere with phagocytosis and activation of mouse macrophage-like cells 774A.1. Infect Immun 79:2267–2276. doi: 10.1128/IAI.01142-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Cerca N, Jefferson KK, Oliveira R, Pier GB, Azeredo J. 2006. Comparative antibody-mediated phagocytosis of Staphylococcus epidermidis cells grown in a biofilm or in the planktonic state. Infect Immun 74:4849–4855. doi: 10.1128/IAI.00230-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Kristian SA, Birkenstock TA, Sauder U, Mack D, Götz F, Landmann R. 2008. Biofilm formation induces C3a release and protects Staphylococcus epidermidis from IgG and complement deposition and from neutrophil-dependent killing. J Infect Dis 197:1028–1035. doi: 10.1086/528992. [DOI] [PubMed] [Google Scholar]
- 366.Le KY, Park MD, Otto M. 2018. Immune evasion mechanisms of Staphylococcus epidermidis biofilm infection. Front Microbiol 9:359. doi: 10.3389/fmicb.2018.00359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Mah TF, O’Toole GA. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol 9:34–39. doi: 10.1016/S0966-842X(00)01913-2. [DOI] [PubMed] [Google Scholar]
- 368.Farber BF, Kaplan MH, Clogston AG. 1990. Staphylococcus epidermidis extracted slime inhibits the antimicrobial action of glycopeptide antibiotics. J Infect Dis 161:37–40. doi: 10.1093/infdis/161.1.37. [DOI] [PubMed] [Google Scholar]
- 369.Jefferson KK, Goldmann DA, Pier GB. 2005. Use of confocal microscopy to analyze the rate of vancomycin penetration through Staphylococcus aureus biofilms. Antimicrob Agents Chemother 49:2467–2473. doi: 10.1128/AAC.49.6.2467-2473.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Singh R, Ray P, Das A, Sharma M. 2010. Penetration of antibiotics through Staphylococcus aureus and Staphylococcus epidermidis biofilms. J Antimicrob Chemother 65:1955–1958. doi: 10.1093/jac/dkq257. [DOI] [PubMed] [Google Scholar]
- 371.Singh R, Sahore S, Kaur P, Rani A, Ray P. 2016. Penetration barrier contributes to bacterial biofilm-associated resistance against only select antibiotics, and exhibits genus-, strain- and antibiotic-specific differences. Pathog Dis 74:ftw056. doi: 10.1093/femspd/ftw056. [DOI] [PubMed] [Google Scholar]
- 372.Stewart PS. 2002. Mechanisms of antibiotic resistance in bacterial biofilms. Int J Med Microbiol 292:107–113. doi: 10.1078/1438-4221-00196. [DOI] [PubMed] [Google Scholar]
- 373.García-Betancur JC, Lopez D. 2019. Cell heterogeneity in staphylococcal communities. J Mol Biol 431:4699–4711. doi: 10.1016/j.jmb.2019.06.011. [DOI] [PubMed] [Google Scholar]
- 374.Águila-Arcos S, Álvarez-Rodríguez I, Garaiyurrebaso O, Garbisu C, Grohmann E, Alkorta I. 2017. Biofilm-forming clinical Staphylococcus isolates harbor horizontal transfer and antibiotic resistance genes. Front Microbiol 8:2018. doi: 10.3389/fmicb.2017.02018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Savage VJ, Chopra I, O’Neill AJ. 2013. Staphylococcus aureus biofilms promote horizontal transfer of antibiotic resistance. Antimicrob Agents Chemother 57:1968–1970. doi: 10.1128/AAC.02008-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Balaban NQ, Helaine S, Lewis K, Ackermann M, Aldridge B, Andersson DI, Brynildsen MP, Bumann D, Camilli A, Collins JJ, Dehio C, Fortune S, Ghigo JM, Hardt WD, Harms A, Heinemann M, Hung DT, Jenal U, Levin BR, Michiels J, Storz G, Tan MW, Tenson T, Van Melderen L, Zinkernagel A. 2019. Definitions and guidelines for research on antibiotic persistence. Nat Rev Microbiol 17:441–448. doi: 10.1038/s41579-019-0196-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Rani SA, Pitts B, Beyenal H, Veluchamy RA, Lewandowski Z, Davison WM, Buckingham-Meyer K, Stewart PS. 2007. Spatial patterns of DNA replication, protein synthesis, and oxygen concentration within bacterial biofilms reveal diverse physiological states. J Bacteriol 189:4223–4233. doi: 10.1128/JB.00107-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Coiffier G, Albert JD, Arvieux C, Guggenbuhl P. 2013. Optimizing combination rifampin therapy for staphylococcal osteoarticular infections. Joint Bone Spine 80:11–17. doi: 10.1016/j.jbspin.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 379.Raad I, Hanna H, Jiang Y, Dvorak T, Reitzel R, Chaiban G, Sherertz R, Hachem R. 2007. Comparative activities of daptomycin, linezolid, and tigecycline against catheter-related methicillin-resistant Staphylococcus bacteremic isolates embedded in biofilm. Antimicrob Agents Chemother 51:1656–1660. doi: 10.1128/AAC.00350-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Achermann Y, Eigenmann K, Ledergerber B, Derksen L, Rafeiner P, Clauss M, Nüesch R, Zellweger C, Vogt M, Zimmerli W. 2013. Factors associated with rifampin resistance in staphylococcal periprosthetic joint infections (PJI): a matched case-control study. Infection 41:431–437. doi: 10.1007/s15010-012-0325-7. [DOI] [PubMed] [Google Scholar]
- 381.Zimmerli W, Sendi P. 2019. Role of rifampin against staphylococcal biofilm infections in vitro, in animal models, and in orthopedic-device-related infections. Antimicrob Agents Chemother 63:e01746-18. doi: 10.1128/AAC.01746-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Mirani ZA, Jamil N. 2011. Effect of sub-lethal doses of vancomycin and oxacillin on biofilm formation by vancomycin intermediate resistant Staphylococcus aureus. J Basic Microbiol 51:191–195. doi: 10.1002/jobm.201000221. [DOI] [PubMed] [Google Scholar]
- 383.Kaplan JB, Izano EA, Gopal P, Karwacki MT, Kim S, Bose JL, Bayles KW, Horswill AR. 2012. Low levels of beta-lactam antibiotics induce extracellular DNA release and biofilm formation in Staphylococcus aureus. mBio 3:e00198-12. doi: 10.1128/mBio.00198-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Schilcher K, Andreoni F, Dengler Haunreiter V, Seidl K, Hasse B, Zinkernagel AS. 2016. Modulation of Staphylococcus aureus biofilm matrix by subinhibitory concentrations of clindamycin. Antimicrob Agents Chemother 60:5957–5967. doi: 10.1128/AAC.00463-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Zimmerli W, Moser C. 2012. Pathogenesis and treatment concepts of orthopaedic biofilm infections. FEMS Immunol Med Microbiol 65:158–168. doi: 10.1111/j.1574-695X.2012.00938.x. [DOI] [PubMed] [Google Scholar]
- 386.Bridgett MJ, Davies MC, Denyer SP. 1992. Control of staphylococcal adhesion to polystyrene surfaces by polymer surface modification with surfactants. Biomaterials 13:411–416. doi: 10.1016/0142-9612(92)90159-l. [DOI] [PubMed] [Google Scholar]
- 387.Donelli G, Francolini I, Romoli D, Guaglianone E, Piozzi A, Ragunath C, Kaplan JB. 2007. Synergistic activity of dispersin B and cefamandole nafate in inhibition of staphylococcal biofilm growth on polyurethanes. Antimicrob Agents Chemother 51:2733–2740. doi: 10.1128/AAC.01249-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Darouiche RO, Mansouri MD, Gawande PV, Madhyastha S. 2009. Antimicrobial and antibiofilm efficacy of triclosan and DispersinB combination. J Antimicrob Chemother 64:88–93. doi: 10.1093/jac/dkp158. [DOI] [PubMed] [Google Scholar]
- 389.Bastari K, Arshath M, Ng ZH, Chia JH, Yow ZX, Sana B, Tan MF, Lim S, Loo SC. 2014. A controlled release of antibiotics from calcium phosphate-coated poly(lactic-co-glycolic acid) particles and their in vitro efficacy against Staphylococcus aureus biofilm. J Mater Sci Mater Med 25:747–757. doi: 10.1007/s10856-013-5125-9. [DOI] [PubMed] [Google Scholar]
- 390.Thomas N, Thorn C, Richter K, Thierry B, Prestidge C. 2016. Efficacy of poly-lactic-co-glycolic acid micro- and nanoparticles of ciprofloxacin against bacterial biofilms. J Pharm Sci 105:3115–3122. doi: 10.1016/j.xphs.2016.06.022. [DOI] [PubMed] [Google Scholar]
- 391.Hu D, Li H, Wang B, Ye Z, Lei W, Jia F, Jin Q, Ren KF, Ji J. 2017. Surface-adaptive gold nanoparticles with effective adherence and enhanced photothermal ablation of methicillin-resistant Staphylococcus aureus biofilm. ACS Nano 11:9330–9339. doi: 10.1021/acsnano.7b04731. [DOI] [PubMed] [Google Scholar]
- 392.Gupta PV, Nirwane AM, Belubbi T, Nagarsenker MS. 2017. Pulmonary delivery of synergistic combination of fluoroquinolone antibiotic complemented with proteolytic enzyme: a novel antimicrobial and antibiofilm strategy. Nanomedicine (Lond) 13:2371–2384. doi: 10.1016/j.nano.2017.06.011. [DOI] [PubMed] [Google Scholar]
- 393.Gupta PV, Nirwane AM, Nagarsenker MS. 2018. Inhalable levofloxacin liposomes complemented with lysozyme for treatment of pulmonary infection in rats: effective antimicrobial and antibiofilm strategy. AAPS PharmSciTech 19:1454–1467. doi: 10.1208/s12249-017-0945-4. [DOI] [PubMed] [Google Scholar]
- 394.Scriboni AB, Couto VM, Ribeiro LNDM, Freires IA, Groppo FC, de Paula E, Franz-Montan M, Cogo-Müller K. 2019. Fusogenic liposomes increase the antimicrobial activity of vancomycin against Staphylococcus aureus biofilm. Front Pharmacol 10:1401. doi: 10.3389/fphar.2019.01401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Mayville P, Ji G, Beavis R, Yang H, Goger M, Novick RP, Muir TW. 1999. Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. Proc Natl Acad Sci U S A 96:1218–1223. doi: 10.1073/pnas.96.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Quave CL, Horswill AR. 2014. Flipping the switch: tools for detecting small molecule inhibitors of staphylococcal virulence. Front Microbiol 5:706. doi: 10.3389/fmicb.2014.00706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Sully EK, Malachowa N, Elmore BO, Alexander SM, Femling JK, Gray BM, DeLeo FR, Otto M, Cheung AL, Edwards BS, Sklar LA, Horswill AR, Hall PR, Gresham HD. 2014. Selective chemical inhibition of agr quorum sensing in Staphylococcus aureus promotes host defense with minimal impact on resistance. PLoS Pathog 10:e1004174. doi: 10.1371/journal.ppat.1004174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Quave CL, Lyles JT, Kavanaugh JS, Nelson K, Parlet CP, Crosby HA, Heilmann KP, Horswill AR. 2015. Castanea sativa (European chestnut) leaf extracts rich in ursene and oleanene derivatives block Staphylococcus aureus virulence and pathogenesis without detectable resistance. PLoS One 10:e0136486. doi: 10.1371/journal.pone.0136486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Parlet CP, Kavanaugh JS, Crosby HA, Raja HA, El-Elimat T, Todd DA, Pearce CJ, Cech NB, Oberlies NH, Horswill AR. 2019. Apicidin attenuates MRSA virulence through quorum-sensing inhibition and enhanced host defense. Cell Rep 27:187–198.e6. doi: 10.1016/j.celrep.2019.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.George EA, Muir TW. 2007. Molecular mechanisms of agr quorum sensing in virulent staphylococci. Chembiochem 8:847–855. doi: 10.1002/cbic.200700023. [DOI] [PubMed] [Google Scholar]
- 401.Jarraud S, Lyon GJ, Figueiredo AM, Lina G, Gérard L, Vandenesch F, Etienne J, Muir TW, Novick RP. 2000. Exfoliatin-producing strains define a fourth agr specificity group in Staphylococcus aureus. J Bacteriol 182:6517–6522. doi: 10.1128/jb.182.22.6517-6522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Tal-Gan Y, Stacy DM, Foegen MK, Koenig DW, Blackwell HE. 2013. Highly potent inhibitors of quorum sensing in Staphylococcus aureus revealed through a systematic synthetic study of the group-III autoinducing peptide. J Am Chem Soc 135:7869–7882. doi: 10.1021/ja3112115. [DOI] [PubMed] [Google Scholar]
- 403.Tal-Gan Y, Ivancic M, Cornilescu G, Yang T, Blackwell HE. 2016. Highly stable, amide-bridged autoinducing peptide analogues that strongly inhibit the AgrC quorum sensing receptor in Staphylococcus aureus. Angew Chem Int Ed Engl 55:8913–8917. doi: 10.1002/anie.201602974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Tal-Gan Y, Ivancic M, Cornilescu G, Cornilescu CC, Blackwell HE. 2013. Structural characterization of native autoinducing peptides and abiotic analogues reveals key features essential for activation and inhibition of an AgrC quorum sensing receptor in Staphylococcus aureus. J Am Chem Soc 135:18436–18444. doi: 10.1021/ja407533e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Kaplan JB, Velliyagounder K, Ragunath C, Rohde H, Mack D, Knobloch JK, Ramasubbu N. 2004. Genes involved in the synthesis and degradation of matrix polysaccharide in Actinobacillus actinomycetemcomitans and Actinobacillus pleuropneumoniae biofilms. J Bacteriol 186:8213–8220. doi: 10.1128/JB.186.24.8213-8220.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Pavlukhina SV, Kaplan JB, Xu L, Chang W, Yu X, Madhyastha S, Yakandawala N, Mentbayeva A, Khan B, Sukhishvili SA. 2012. Noneluting enzymatic antibiofilm coatings. ACS Appl Mater Interfaces 4:4708–4716. doi: 10.1021/am3010847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Fleming D, Rumbaugh K. 2018. The consequences of biofilm dispersal on the host. Sci Rep 8:10738. doi: 10.1038/s41598-018-29121-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Waryah CB, Wells K, Ulluwishewa D, Chen-Tan N, Gogoi-Tiwari J, Ravensdale J, Costantino P, Gökçen A, Vilcinskas A, Wiesner J, Mukkur T. 2017. In vitro antimicrobial efficacy of tobramycin against Staphylococcus aureus biofilms in combination with or without DNase I and/or Dispersin B: a preliminary investigation. Microb Drug Resist 23:384–390. doi: 10.1089/mdr.2016.0100. [DOI] [PubMed] [Google Scholar]
- 409.Sonesson A, Przybyszewska K, Eriksson S, Mörgelin M, Kjellstrom S, Davies J, Potempa J, Schmidtchen A. 2017. Identification of bacterial biofilm and the Staphylococcus aureus derived protease, staphopain, on the skin surface of patients with atopic dermatitis. Sci Rep 7:8689. doi: 10.1038/s41598-017-08046-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Sieprawska-Lupa M, Mydel P, Krawczyk K, Wójcik K, Puklo M, Lupa B, Suder P, Silberring J, Reed M, Pohl J, Shafer W, McAleese F, Foster T, Travis J, Potempa J. 2004. Degradation of human antimicrobial peptide LL-37 by Staphylococcus aureus-derived proteinases. Antimicrob Agents Chemother 48:4673–4679. doi: 10.1128/AAC.48.12.4673-4679.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Pietrocola G, Nobile G, Rindi S, Speziale P. 2017. Staphylococcus aureus manipulates innate immunity through own and host-expressed proteases. Front Cell Infect Microbiol 7:166. doi: 10.3389/fcimb.2017.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Smagur J, Guzik K, Bzowska M, Kuzak M, Zarebski M, Kantyka T, Walski M, Gajkowska B, Potempa J. 2009. Staphylococcal cysteine protease staphopain B (SspB) induces rapid engulfment of human neutrophils and monocytes by macrophages. Biol Chem 390:361–371. doi: 10.1515/BC.2009.042. [DOI] [PubMed] [Google Scholar]
- 413.Laarman AJ, Mijnheer G, Mootz JM, van Rooijen WJ, Ruyken M, Malone CL, Heezius EC, Ward R, Milligan G, van Strijp JA, de Haas CJ, Horswill AR, van Kessel KP, Rooijakkers SH. 2012. Staphylococcus aureus Staphopain A inhibits CXCR2-dependent neutrophil activation and chemotaxis. EMBO J 31:3607–3619. doi: 10.1038/emboj.2012.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Prokešová L, Potuẑníková B, Potempa J, Zikán J, Radl J, Hachová L, Baran K, Porwit-Bóbr Z, John C. 1992. Cleavage of human immunoglobulins by serine proteinase from Staphylococcus aureus. Immunol Lett 31:259–265. doi: 10.1016/0165-2478(92)90124-7. [DOI] [PubMed] [Google Scholar]
- 415.Laarman AJ, Ruyken M, Malone CL, van Strijp JA, Horswill AR, Rooijakkers SH. 2011. Staphylococcus aureus metalloprotease aureolysin cleaves complement C3 to mediate immune evasion. J Immunol 186:6445–6453. doi: 10.4049/jimmunol.1002948. [DOI] [PubMed] [Google Scholar]
- 416.Schindler CA, Schuhardt VT. 1964. Lysostaphin: a new bacteriolytic agent for the Staphylococcus. Proc Natl Acad Sci U S A 51:414–421. doi: 10.1073/pnas.51.3.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Schuhardt VT, Schindler CA. 1964. Lysostaphin therapy in mice infected with Staphylococcus aureus. J Bacteriol 88:815–816. doi: 10.1128/JB.88.3.815-816.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Hogan S, Zapotoczna M, Stevens NT, Humphreys H, O’Gara JP, O'Neill E. 2017. Potential use of targeted enzymatic agents in the treatment of Staphylococcus aureus biofilm-related infections. J Hosp Infect 96:177–182. doi: 10.1016/j.jhin.2017.02.008. [DOI] [PubMed] [Google Scholar]
- 419.Aguinaga A, Francés ML, Del Pozo JL, Alonso M, Serrera A, Lasa I, Leiva J. 2011. Lysostaphin and clarithromycin: a promising combination for the eradication of Staphylococcus aureus biofilms. Int J Antimicrob Agents 37:585–587. doi: 10.1016/j.ijantimicag.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 420.Wu JA, Kusuma C, Mond JJ, Kokai-Kun JF. 2003. Lysostaphin disrupts Staphylococcus aureus and Staphylococcus epidermidis biofilms on artificial surfaces. Antimicrob Agents Chemother 47:3407–3414. doi: 10.1128/aac.47.11.3407-3414.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Kokai-Kun JF, Chanturiya T, Mond JJ. 2009. Lysostaphin eradicates established Staphylococcus aureus biofilms in jugular vein catheterized mice. J Antimicrob Chemother 64:94–100. doi: 10.1093/jac/dkp145. [DOI] [PubMed] [Google Scholar]
- 422.Pletzer D, Coleman SR, Hancock RE. 2016. Anti-biofilm peptides as a new weapon in antimicrobial warfare. Curr Opin Microbiol 33:35–40. doi: 10.1016/j.mib.2016.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Jorge P, Lourenço A, Pereira MO. 2012. New trends in peptide-based anti-biofilm strategies: a review of recent achievements and bioinformatic approaches. Biofouling 28:1033–1061. doi: 10.1080/08927014.2012.728210. [DOI] [PubMed] [Google Scholar]
- 424.Turner J, Cho Y, Dinh NN, Waring AJ, Lehrer RI. 1998. Activities of LL-37, a cathelin-associated antimicrobial peptide of human neutrophils. Antimicrob Agents Chemother 42:2206–2214. doi: 10.1128/AAC.42.9.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Kang J, Dietz MJ, Li B. 2019. Antimicrobial peptide LL-37 is bactericidal against Staphylococcus aureus biofilms. PLoS One 14:e0216676. doi: 10.1371/journal.pone.0216676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Joo HS, Otto M. 2015. Mechanisms of resistance to antimicrobial peptides in staphylococci. Biochim Biophys Acta 1848:3055–3061. doi: 10.1016/j.bbamem.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Herbert S, Bera A, Nerz C, Kraus D, Peschel A, Goerke C, Meehl M, Cheung A, Götz F. 2007. Molecular basis of resistance to muramidase and cationic antimicrobial peptide activity of lysozyme in staphylococci. PLoS Pathog 3:e102. doi: 10.1371/journal.ppat.0030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Okuda K, Zendo T, Sugimoto S, Iwase T, Tajima A, Yamada S, Sonomoto K, Mizunoe Y. 2013. Effects of bacteriocins on methicillin-resistant Staphylococcus aureus biofilm. Antimicrob Agents Chemother 57:5572–5579. doi: 10.1128/AAC.00888-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Dosler S, Mataraci E. 2013. In vitro pharmacokinetics of antimicrobial cationic peptides alone and in combination with antibiotics against methicillin resistant Staphylococcus aureus biofilms. Peptides 49:53–58. doi: 10.1016/j.peptides.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 430.Saising J, Dube L, Ziebandt AK, Voravuthikunchai SP, Nega M, Götz F. 2012. Activity of gallidermin on Staphylococcus aureus and Staphylococcus epidermidis biofilms. Antimicrob Agents Chemother 56:5804–5810. doi: 10.1128/AAC.01296-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Mathur H, Field D, Rea MC, Cotter PD, Hill C, Ross RP. 2018. Fighting biofilms with lantibiotics and other groups of bacteriocins. NPJ Biofilms Microbiomes 4:9. doi: 10.1038/s41522-018-0053-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Newstead LL, Varjonen K, Nuttall T, Paterson GK. 2020. Staphylococcal-produced bacteriocins and antimicrobial peptides: their potential as alternative treatments for Staphylococcus aureus Infections. Antibiotics (Basel) 9:40. doi: 10.3390/antibiotics9020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Harper DR, Parracho HMRT, Walker J, Sharp R, Hughes G, Werthén M, Lehman S, Morales S. 2014. Bacteriophages and biofilms. Antibiotics (Basel) 3:270–284. doi: 10.3390/antibiotics3030270. [DOI] [Google Scholar]
- 434.Abdelkader K, Gerstmans H, Saafan A, Dishisha T, Briers Y. 2019. The preclinical and clinical progress of bacteriophages and their lytic enzymes: the parts are easier than the whole. Viruses 11:96. doi: 10.3390/v11020096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Schmelcher M, Shen Y, Nelson DC, Eugster MR, Eichenseher F, Hanke DC, Loessner MJ, Dong S, Pritchard DG, Lee JC, Becker SC, Foster-Frey J, Donovan DM. 2015. Evolutionarily distinct bacteriophage endolysins featuring conserved peptidoglycan cleavage sites protect mice from MRSA infection. J Antimicrob Chemother 70:1453–1465. doi: 10.1093/jac/dku552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Becker SC, Roach DR, Chauhan VS, Shen Y, Foster-Frey J, Powell AM, Bauchan G, Lease RA, Mohammadi H, Harty WJ, Simmons C, Schmelcher M, Camp M, Dong S, Baker JR, Sheen TR, Doran KS, Pritchard DG, Almeida RA, Nelson DC, Marriott I, Lee JC, Donovan DM. 2016. Triple-acting lytic enzyme treatment of drug-resistant and intracellular Staphylococcus aureus. Sci Rep 6:25063. doi: 10.1038/srep25063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Dakheel KH, Rahim RA, Neela VK, Al-Obaidi JR, Hun TG, Isa MNM, Yusoff K. 2019. Genomic analyses of two novel biofilm-degrading methicillin-resistant Staphylococcus aureus phages. BMC Microbiol 19:114. doi: 10.1186/s12866-019-1484-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Alves DR, Gaudion A, Bean JE, Perez Esteban P, Arnot TC, Harper DR, Kot W, Hansen LH, Enright MC, Jenkins AT. 2014. Combined use of bacteriophage K and a novel bacteriophage to reduce Staphylococcus aureus biofilm formation. Appl Environ Microbiol 80:6694–6703. doi: 10.1128/AEM.01789-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Jo A, Kim J, Ding T, Ahn J. 2016. Role of phage-antibiotic combination in reducing antibiotic resistance in Staphylococcus aureus. Food Sci Biotechnol 25:1211–1215. doi: 10.1007/s10068-016-0192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Tkhilaishvili T, Lombardi L, Klatt AB, Trampuz A, Di Luca M. 2018. Bacteriophage Sb-1 enhances antibiotic activity against biofilm, degrades exopolysaccharide matrix and targets persisters of Staphylococcus aureus. Int J Antimicrob Agents 52:842–853. doi: 10.1016/j.ijantimicag.2018.09.006. [DOI] [PubMed] [Google Scholar]
- 441.Dickey J, Perrot V. 2019. Adjunct phage treatment enhances the effectiveness of low antibiotic concentration against Staphylococcus aureus biofilms in vitro. PLoS One 14:e0209390. doi: 10.1371/journal.pone.0209390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Cha Y, Chun J, Son B, Ryu S. 2019. Characterization and genome analysis of Staphylococcus aureus podovirus CSA13 and its anti-biofilm capacity. Viruses 11:54. doi: 10.3390/v11010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Cha Y, Son B, Ryu S. 2019. Effective removal of staphylococcal biofilms on various food contact surfaces by Staphylococcus aureus phage endolysin LysCSA13. Food Microbiol 84:103245. doi: 10.1016/j.fm.2019.103245. [DOI] [PubMed] [Google Scholar]
- 444.Zhang Y, Cheng M, Zhang H, Dai J, Guo Z, Li X, Ji Y, Cai R, Xi H, Wang X, Xue Y, Sun C, Feng X, Lei L, Han W, Gu J. 2018. Antibacterial effects of phage lysin LysGH15 on planktonic cells and biofilms of diverse staphylococci. Appl Environ Microbiol 84:e00886-18. doi: 10.1128/AEM.00886-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Schuch R, Khan BK, Raz A, Rotolo JA, Wittekind M. 2017. Bacteriophage lysin CF-301, a potent antistaphylococcal biofilm agent. Antimicrob Agents Chemother 61:e02666-16. doi: 10.1128/AAC.02666-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.González S, Fernández L, Gutiérrez D, Campelo AB, Rodríguez A, García P. 2018. Analysis of different parameters affecting diffusion, propagation and survival of staphylophages in bacterial biofilms. Front Microbiol 9:2348. doi: 10.3389/fmicb.2018.02348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Lin DM, Koskella B, Lin HC. 2017. Phage therapy: an alternative to antibiotics in the age of multi-drug resistance. World J Gastrointest Pharmacol Ther 8:162–173. doi: 10.4292/wjgpt.v8.i3.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Moller AG, Lindsay JA, Read TD. 2019. Determinants of phage host range in Staphylococcus species. Appl Environ Microbiol 85:e00209-19. doi: 10.1128/AEM.00209-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Ammons MC, Tripet BP, Carlson RP, Kirker KR, Gross MA, Stanisich JJ, Copié V. 2014. Quantitative NMR metabolite profiling of methicillin-resistant and methicillin-susceptible Staphylococcus aureus discriminates between biofilm and planktonic phenotypes. J Proteome Res 13:2973–2985. doi: 10.1021/pr500120c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.García-Betancur JC, Goñi-Moreno A, Horger T, Schott M, Sharan M, Eikmeier J, Wohlmuth B, Zernecke A, Ohlsen K, Kuttler C, Lopez D. 2017. Cell differentiation defines acute and chronic infection cell types in Staphylococcus aureus. Elife 6:e28023. doi: 10.7554/eLife.28023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Frank DN, Wysocki A, Specht-Glick DD, Rooney A, Feldman RA, St Amand AL, Pace NR, Trent JD. 2009. Microbial diversity in chronic open wounds. Wound Repair Regen 17:163–172. doi: 10.1111/j.1524-475X.2009.00472.x. [DOI] [PubMed] [Google Scholar]
- 452.Harrison F. 2007. Microbial ecology of the cystic fibrosis lung. Microbiology 153:917–923. doi: 10.1099/mic.0.2006/004077-0. [DOI] [PubMed] [Google Scholar]
- 453.Hotterbeekx A, Kumar-Singh S, Goossens H, Malhotra-Kumar S. 2017. In vivo and in vitro interactions between Pseudomonas aeruginosa and Staphylococcus spp. Front Cell Infect Microbiol 7:106. doi: 10.3389/fcimb.2017.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Phalak P, Chen J, Carlson RP, Henson MA. 2016. Metabolic modeling of a chronic wound biofilm consortium predicts spatial partitioning of bacterial species. BMC Syst Biol 10:90. doi: 10.1186/s12918-016-0334-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Makovcova J, Babak V, Kulich P, Masek J, Slany M, Cincarova L. 2017. Dynamics of mono- and dual-species biofilm formation and interactions between Staphylococcus aureus and Gram-negative bacteria. Microb Biotechnol 10:819–832. doi: 10.1111/1751-7915.12705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Orazi G, O’Toole GA. 2017. Pseudomonas aeruginosa alters Staphylococcus aureus sensitivity to vancomycin in a biofilm model of cystic fibrosis infection. mBio 8:e00873-17. doi: 10.1128/mBio.00873-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Orazi G, Ruoff KL, O’Toole GA. 2019. Pseudomonas aeruginosa increases the sensitivity of biofilm-grown Staphylococcus aureus to membrane-targeting antiseptics and antibiotics. mBio 10:e01501-19. doi: 10.1128/mBio.01501-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Tavernier S, Crabbé A, Hacioglu M, Stuer L, Henry S, Rigole P, Dhondt I, Coenye T. 2017. Community composition determines activity of antibiotics against multispecies biofilms. Antimicrob Agents Chemother 61:e00302-17. doi: 10.1128/AAC.00302-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Harriott MM, Noverr MC. 2009. Candida albicans and Staphylococcus aureus form polymicrobial biofilms: effects on antimicrobial resistance. Antimicrob Agents Chemother 53:3914–3922. doi: 10.1128/AAC.00657-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Harriott MM, Noverr MC. 2010. Ability of Candida albicans mutants to induce Staphylococcus aureus vancomycin resistance during polymicrobial biofilm formation. Antimicrob Agents Chemother 54:3746–3755. doi: 10.1128/AAC.00573-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Kong EF, Tsui C, Kucharikova S, Andes D, Van Dijck P, Jabra-Rizk MA. 2016. Commensal protection of Staphylococcus aureus against antimicrobials by Candida albicans biofilm matrix. mBio 7:e01365-16. doi: 10.1128/mBio.01365-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Carolus H, Van Dyck K, Van Dijck P. 2019. Candida albicans and Staphylococcus species: a threatening twosome. Front Microbiol 10:2162. doi: 10.3389/fmicb.2019.02162. [DOI] [PMC free article] [PubMed] [Google Scholar]