Significance
A specific ATP-binding cassette (ABC) transporter is generally viewed to function as either an exporter or an importer, but in principle ABC transporters can transport substrates in both directions across the membrane. Structural studies of the prokaryotic ABC exporter NaAtm1 demonstrate that progression through the transport cycle is accompanied by changes in transmembrane helix 6 (TM6) that modulate the binding cavity for transported substrate. Significantly, kinking of TM6 in a post-ATP hydrolysis state stabilized by MgADPVO4 eliminates the substrate-binding cavity. The presence of this cavity during the transition from the inward-facing to outward-facing conformational states, and its absence in the reverse direction, thereby provide an elegant and conceptually simple mechanism for enforcing the export directionality of transport.
Keywords: ATP-binding cassette transporters, ABC transporters, alternating access mechanism, glutathione transport, transport cycle
Abstract
The ATP-binding cassette (ABC) transporter of mitochondria (Atm1) mediates iron homeostasis in eukaryotes, while the prokaryotic homolog from Novosphingobium aromaticivorans (NaAtm1) can export glutathione derivatives and confer protection against heavy-metal toxicity. To establish the structural framework underlying the NaAtm1 transport mechanism, we determined eight structures by X-ray crystallography and single-particle cryo-electron microscopy in distinct conformational states, stabilized by individual disulfide crosslinks and nucleotides. As NaAtm1 progresses through the transport cycle, conformational changes in transmembrane helix 6 (TM6) alter the glutathione-binding site and the associated substrate-binding cavity. Significantly, kinking of TM6 in the post-ATP hydrolysis state stabilized by MgADPVO4 eliminates this cavity, precluding uptake of glutathione derivatives. The presence of this cavity during the transition from the inward-facing to outward-facing conformational states, and its absence in the reverse direction, thereby provide an elegant and conceptually simple mechanism for enforcing the export directionality of transport by NaAtm1. One of the disulfide crosslinked NaAtm1 variants characterized in this work retains significant glutathione transport activity, suggesting that ATP hydrolysis and substrate transport by Atm1 may involve a limited set of conformational states with minimal separation of the nucleotide-binding domains in the inward-facing conformation.
The translocation mechanism of ATP-binding cassette (ABC) transporters is generally described in terms of the alternating access model involving inward-facing, occluded, and outward-facing states, with the transitions between states coupled to the binding and hydrolysis of ATP, along with product release (1–5). ABC exporters constitute an important branch of ABC transporters found in all forms of life, exhibiting a broad range of activities from exporting macromolecular building blocks to serving as multidrug efflux pumps. Despite the name, it has generally not been experimentally demonstrated that exporters have a unique transport directionality, or how this directionality is maintained. An informative example is provided by the recent studies of mycobacterial ABC transporters with exporter folds that mediate the uptake of various water-soluble compounds (6, 7). The structural characterization of ABC exporters in distinct conformational states has been achieved primarily by controlling the nucleotide state, often in combination with the ATPase deficient “E-to-Q” mutant in the Walker-B motif (8). Other approaches to stabilizing specific conformational states include introduction of disulfide crosslinks between the nucleotide-binding domains (NBDs) (9, 10) or through the binding of state-specific nanobodies (11), inhibitors (10), or substrate (12). Detailed structural frameworks are now available for addressing the transport mechanism of different ABC exporter systems, including CFTR (13–15), MsbA (16, 17), TmrAB (18), PglK (19), and P-glycoprotein (10, 20–22) in distinct conformational states. These studies reinforce the view that significant conformational variability exists between different members of the ABC exporter family.
The ABC transporter of mitochondria (Atm1) is a homodimeric transporter involved in iron-sulfur cluster biogenesis in yeast (23) and has been shown to bind glutathione (24, 25); it is also a homolog to the human ABCB6 and ABCB7 transporters and the plant atm3 transporter that were all found to play a role in iron homeostasis (26–29). Previous analysis of the bacterial homolog from Novosphingobium aromaticivorans (NaAtm1) revealed a role in heavy-metal detoxification, plausibly by exporting metallated glutathione species (30). The initial characterization of NaAtm1 established the structure in an inward-facing conformation and defined the binding interactions for glutathione derivatives (30). To stabilize NaAtm1 in different conformational states, we introduced disulfide crosslinks at multiple positions within the NBD dimerization interface. In this work, we report the structures and functions of disulfide crosslinking variants, together with the “E-to-Q” ATP-hydrolysis-deficient mutant and wild-type NaAtm1 in distinct conformational states. From these structures, a framework for the transport cycle of NaAtm1 is delineated. A prominent feature of the transport cycle is the coupling between the conformation of transmembrane helix 6 (TM6) and the glutathione-binding site. During the transition from inward to outward conformation, a large binding cavity exists, but in the return process, as trapped by MgADPVO4 in a post-ATP hydrolysis state, kinking of TM6 eliminates this cavity. The net effect would be to restrict glutathione transport to the export direction, providing a mechanism for enforcing directionality of the transport mechanism.
Results
Disulfide Crosslinking.
To stabilize NaAtm1 in distinct conformational states, we introduced cysteine mutations at the dimerization interface between the NBDs of the natively cysteine-less homodimeric NaAtm1. Three residues identified through sequence alignments of NaAtm1 to other transporters with dimerized NBDs (9, 10, 31) are T525, S526, and A527, positioned near the Walker-B motif where disulfide bonds could potentially form between the two equivalent residues (SI Appendix, Fig. S1A). These residues were separately mutated to cysteine to generate three single-site variants: NaT525C, NaS526C, and NaA527C. Initial crosslinking tests with purified protein revealed similar crosslinking yields at ∼70% (SI Appendix, Fig. S1B). Despite extensive efforts, we were unable to achieve quantitative disulfide bond formation or separate disulfide-linked protein from the uncrosslinked protein. We crystallized and solved the structures of all three variants to assess the consequences of the disulfide crosslinks on the conformational states of NaAtm1.
Crystal Structures of Inward-Facing Occluded Conformations.
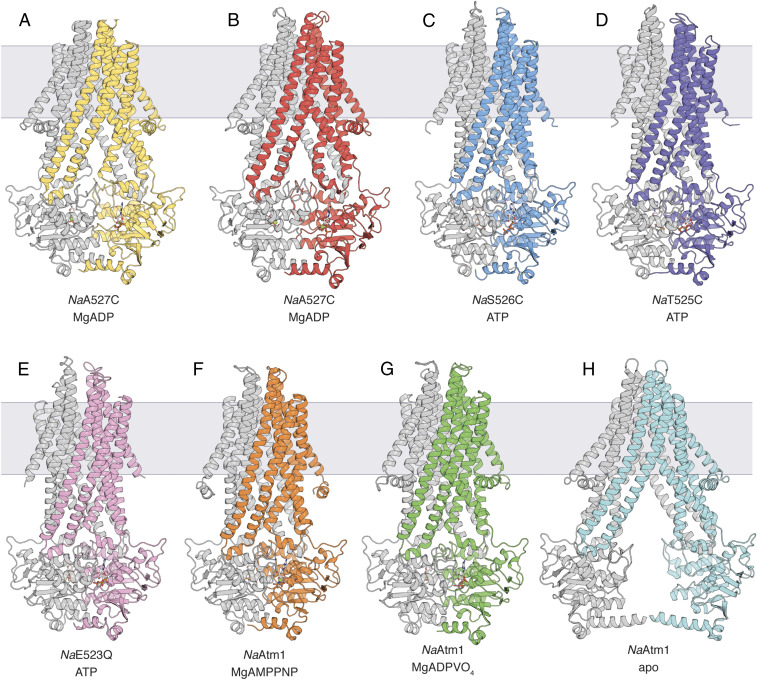
NaA527C crystallized in a MgADP-bound state with four transporters in the asymmetric unit. Despite the moderate anisotropic diffraction to 3.7-Å resolution, two distinguishable inward-facing occluded states, designated state #1 (Fig. 1A) and state #2 (Fig. 1B), were evident. The presence of two conformational states was confirmed from the positions of the selenium sites in selenomethionine-substituted protein crystals (SI Appendix, Fig. S2A). Clear electron density was observed for the disulfide bridges and the nucleotides in all four transporters (SI Appendix, Fig. S2 B and C). A structural alignment of the two states revealed that, while the transmembrane domains (TMDs) are structurally similar, sharing an overall rmsd of 1.7 Å (SI Appendix, Fig. S3A), the α-helical subdomains of the NBDs differ by a small rotation (∼10°) about the molecular two-fold axis of the transporter (SI Appendix, Fig. S3B). These two states also differ from the previous NaAtm1 inward-facing conformation with rmsds of 2.1 and 4.4 Å for state #1 and state #2, respectively (SI Appendix, Fig. S3 C and D); the primary differences are in the separation of the NBDs and changes near the coupling helices of the TMDs that interact with the NBDs.
Fig. 1.
NaAtm1 structure overview. Crystal structures of NaA527C in the inward-facing occluded conformation in (A) state #1 and (B) state #2, both with MgADP bound. Crystal structures of (C) NaS526C, (D) NaT525C, and (E) NaE523Q in the occluded conformation, all with ATP bound. (F) Crystal structure of wild-type NaAtm1 in an occluded conformation with MgAMPPNP bound. (G) Single-particle cryo-EM structure of NaAtm1 in a closed conformation with MgADPVO4 bound. (H) Single-particle cryo-EM structure of NaAtm1 in a wide-open inward-facing conformation. All structures are colored with one chain in gray and the second chain in yellow, red, blue, purple, pink, orange, green, and cyan, respectively. Nucleotides are shown as sticks with Mg2+ shown as green spheres.
Crystals of NaA527C were prepared either with or without 5 mM oxidized glutathione (GSSG). While weak density in the substrate-binding pocket was observed for crystals grown in the presence of GSSG, the moderate resolution was insufficient to unambiguously establish the presence of GSSG. To crystallographically assess the substrate binding capability of NaA527C, we cocrystallized NaA527C with a glutathione-mercury complex (GS-Hg) and collected data at the Hg absorption edge. Anomalous electron density peaks identifying Hg sites were found in all GSSG-binding sites (30) and were strongest in the two best-resolved transporters in the asymmetric unit (SI Appendix, Fig. S4). These sites were not observed in control studies using mercury compounds in the absence of glutathione, supporting the presence of GS-Hg and the substrate-binding capability of NaA527C.
Crystal Structures of the Prehydrolysis Occluded Conformations.
Crystals of NaS526C diffracted anisotropically to 3.4-Å resolution, and the transporter adopted an occluded conformation with bound ATP and a fully resolved disulfide bridge (Fig. 1C and SI Appendix, Fig. S5 A and B). Even though GSSG or GS-Hg were included during crystallization, the binding of substrate was not detected in the electron density maps. The periplasmic regions of the transporter (residues 60 to 82 and 284 to 300) were not well resolved in the electron density maps, possibly due to the lack of stabilizing crystal contacts. These regions were accordingly modeled based on the previously determined high-resolution crystal structure and refined with reduced occupancies.
Surprisingly, crystals of NaT525C were nearly isomorphous to those of NaS526C, and the structures were found to be quite similar (Fig. 1D). In contrast to the NaA527C and NaS526C structures, however, no disulfide bridge was observed between the two T525C residues (Fig. 1D and SI Appendix, Fig. S5 C and D). Instead, the two T525C residues were 13 Å apart, calculated from the corresponding Cα positions. Since no reductant was present, presumably crystallization of the crosslinked protein was not favored in this condition, and instead the uncrosslinked population crystallized.
Crystals of the ATP-hydrolysis–deficient NaE523Q diffracted anisotropically to 3.3-Å resolution, and the structure revealed an occluded conformation with ATP bound (Fig. 1E and SI Appendix, Fig. S5E). While the overall structure of NaE523Q was similar to that observed for NaS526C and NaT525C, including the flexible periplasmic loops, the crystal form was distinct. Overall, the three occluded structures are similar with rmsds in the range of 0.5 to 0.7 Å (SI Appendix, Fig. S5 F–H).
Wild-type NaAtm1 was crystallized in an occluded state with bound MgAMPPNP (Fig. 1F and SI Appendix, Fig. S6A). This occluded structure shared similar overall architecture to the other three mutant occluded structures with alignment rmsds of ∼1 Å (SI Appendix, Fig. S6 B–D); the major conformational differences are in the resolved periplasmic loop regions and the partially resolved elbow helices. Although GSSG was present in the crystallization conditions, bound substrate was not evident in the crystal structure, which is the same outcome observed for the other occluded structures.
Single-Particle Cryo-Electron Microscopy Structure of the Posthydrolysis Closed Conformation.
To capture the posthydrolysis state of the transporter, we utilized the posthydrolysis ATP analog MgADPVO4 (32) and determined a single-particle cryo-electron microscopy (cryo-EM) structure of NaAtm1 in a closed conformation at 3.0-Å resolution (Fig. 1G and SI Appendix, Fig. S7 A–C). For this analysis, NaAtm1 was reconstituted in nanodiscs formed by the membrane scaffolding protein MSP1D1 (33). The overall architecture of this single-particle structure has rmsds in the range of 1.7 to 2.1 Å with the crystal structures in the prehydrolysis occluded state (SI Appendix, Fig. S7 D–G). These structures all adopt similarly dimerized NBDs, but with pronounced differences in the TMDs. The kinked TM6 observed in the posthydrolysis MgADPVO4 occluded structure contrasts with the straightened conformation observed in the prehydrolysis occluded states; as a consequence of this structural change, the GSSG-binding cavity is eliminated, and so this form is designated as a “closed” state.
Single-Particle Structure of the Inward-Facing Conformation.
We determined an inward-facing conformation of NaAtm1 reconstituted into nanodiscs by single-particle cryo-EM in the absence of ligands. This reconstruction was obtained at 3.9-Å resolution (Fig. 1H and SI Appendix, Fig. S8 A and B) where large side chains are well resolved (SI Appendix, Fig. S8C). This structure is designated as a “wide-open” inward-facing conformation, since the NBDs are significantly farther apart in comparison to the original crystal structures of NaAtm1. The rmsds between this structure and other inward-facing conformation (30) and the inward-facing occluded structures are 5.8 to 9.3 Å, with the major difference in the separation of the two half-transporters (SI Appendix, Fig. S8 D–F).
ATPase Activities, Transport Activities, and Coupling Efficiencies of All Variants.
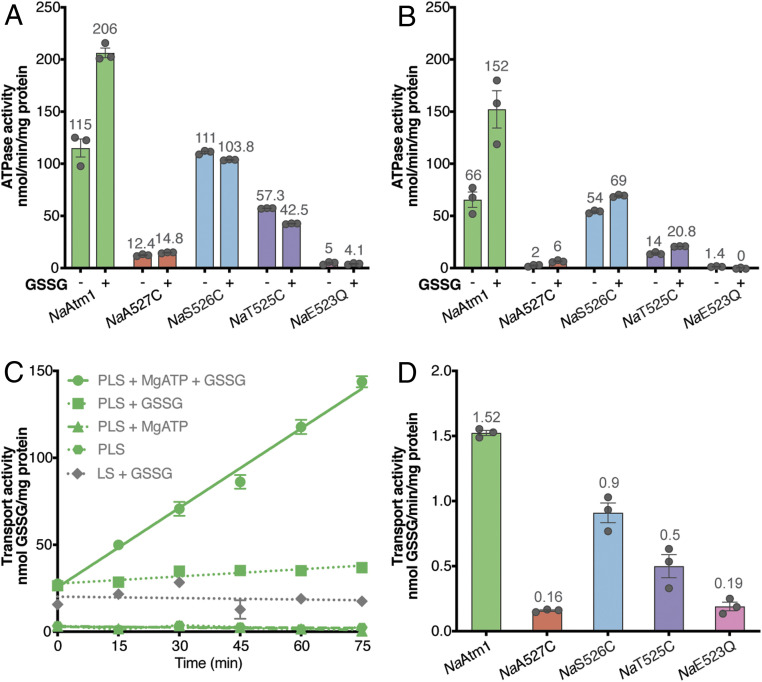
To establish the functional competence of the disulfide crosslinked variants and wild-type NaAtm1, their ATPase and transport activities were characterized (Fig. 2). ATPase activities were measured in both detergent and proteoliposomes (PLS) with 10 mM ATP and 2.5 mM GSSG, concentrations approximating those measured physiologically (34) (Fig. 2 A and B). Wild-type NaAtm1 in detergent showed a basal ATPase activity of 115 ± 15 nmol inorganic phosphate (Pi)⋅min−1⋅mg−1 transporter, and the addition of 2.5 mM GSSG stimulated the ATPase activity by nearly two-fold to 206 ± 8 nmol Pi⋅min−1⋅mg−1 transporter. NaAtm1 reconstituted in PLS showed an ATPase activity of 66 ± 13 nmol Pi⋅min−1⋅mg−1 transporter, again with an approximately two-fold stimulation in the presence of 2.5 mM GSSG to 152 ± 31 nmol Pi⋅min−1⋅mg−1 transporter. Based on the NaAtm1 molecular weight of 133 kDa (with 1 mg ∼ 7.5 nmol), neglecting orientation effects and assuming that all of the transporters are functionally active, an ATPase rate of 152 nmol Pi⋅min−1⋅mg−1 corresponds to a turnover rate of ∼20 ATP min−1. Unexpectedly, NaS526C had a similar basal ATPase activity to the wild-type protein, with no stimulation by GSSG in detergent, but a slight stimulation by GSSG in PLS. NaT525C exhibited reduced ATPase activities in detergent and PLS. The NaA527C and NaE523Q constructs exhibited little ATPase activity in either detergent or PLS.
Fig. 2.
NaAtm1 transport and ATPase activities. ATPase activities of wild-type NaAtm1 and variants at 10 mM MgATP in the absence and presence of 2.5 mM GSSG in (A) detergent and (B) PLS. (C) Wild-type NaAtm1 transport activities with various controls at 10 mM MgATP and 2.5 mM GSSG. (D) Transport activities of NaAtm1 and variants with 10 mM MgATP and 2.5 mM GSSG. Error bars represent the SEMs, and circles represent the results of individual measurements. All measurements were done in triplicate. The individual rate measurements for the ATPase and transport activities are presented in SI Appendix, Tables S2 and S3.
The transport activity was by necessity measured only in PLS, again using 10 mM MgATP and 2.5 mM GSSG (Fig. 2 C and D). Control experiments indicated that low levels of GSSG may stick to liposomes, complicating the measurement of very low levels of transport activities. For wild-type NaAtm1, the uptake of GSSG was linear with time, corresponding to a rate of 1.52 ± 0.03 nmol GSSG⋅min−1⋅mg−1 transporter, equivalent to ∼0.2 GSSG translocated per minute per transporter (Fig. 2C). Interestingly, NaS526C exhibited ∼60% of wild-type transport activity at a rate of 0.9 ± 0.1 nmol GSSG⋅min−1⋅mg−1 transporter (Fig. 2D), while NaT525C retained about 30% of the transport activity of wild-type NaAtm1. NaA527C and NaE523Q exhibited GSSG uptake rates ∼10% of the wild-type protein transport activity (Fig. 2D). A complicating feature in the interpretation of these results is that, since disulfide bond formation in these variants was not quantitative, we cannot eliminate some contribution by the uncrosslinked material to the observed ATPase and transport activities. If the uncrosslinked material exhibits the same activity as wild-type protein, then, in the absence of transport activity by the crosslinked protein, a residual ∼30% transport activity would be anticipated based on the estimate of 70% disulfide bond formation. As the transport activity measured for the NaS526C variant exceeds this threshold, NaAtm1 with a disulfide crosslink between the two Cys-526 residues is competent for GSSG transport. In contrast, the transport activity NaT525C variant is at the 30% threshold value, and hence the transport competence of this crosslinked variant cannot be confidently established.
From the ATPase and transport activities, the coupling efficiency—the number of ATP hydrolyzed per GSSG translocated—can be computed. For this calculation, we defined the coupling efficiency as the ratio of the total ATPase rate to the transport rate measured under the same conditions (in the presence of 2.5 mM substrate). For the wild-type protein, the coupling efficiency is measured to be 100 ± 20, compared to 76 ± 6 for the NaS526C variant. Although these coupling efficiencies may seem high relative to the canonical value of 2 typically envisioned for ABC transporters (35), they are in the range reported for other transporters (SI Appendix, Table S1), reflecting the generally high basal ATPase activity and coupling inefficiencies of ABC transporters.
Volumes of Substrate-Binding Cavities.
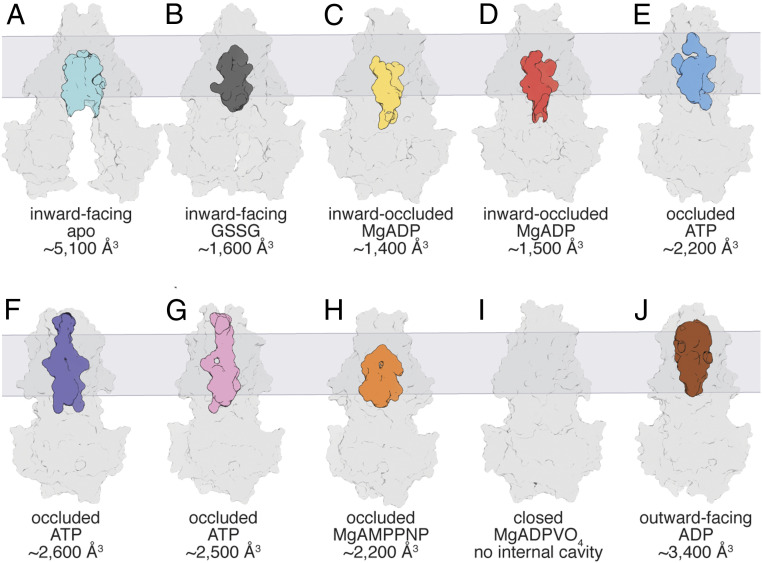
The volumes of the substrate-binding cavities in different structures were calculated using the program CastP with a 2.5 Å probe radius (36). For reference, the volume of a GSSG molecule (molecular weight 612.6 g⋅mol−1) is estimated to be ∼740 Å3, assuming a partial specific volume for GSSG typical of globular proteins (∼0.73 cm3⋅g−1) (37). With the exception of the MgADPVO4 stabilized state, the binding pockets in the various NaAtm1 structures were all found to be of sufficient size to accommodate GSSG (Fig. 3). Significantly, the binding pocket is no longer present in the “posthydrolysis” MgADPVO4 stabilized state (Fig. 3I). For reference, the outward-facing conformation of Sav1866 (31) has a cavity volume of ∼3,400 Å3 (Fig. 3J). As GSSG was not experimentally found to be present in the prehydrolysis occluded conformations, the absence of substrate in these structures suggests that they may represent lower affinity, posttranslocational states of the transporter when the empty transporter is transitioning back to the inward-facing conformation. The comparison with the posthydrolysis occluded structure suggests that a key conformational change after ATP hydrolysis is the generation of kinked TM6 helices, which eliminates the central cavity.
Fig. 3.
Comparison of the central cavities for the NaAtm1 structures. Surface representations of the overall structures and internal binding cavities for (A) NaAtm1 wide-open inward-facing conformation, (B) NaAtm1 inward-facing conformation (PDB ID: 4MRN), (C) NaA527C inward-facing occluded state #1, (D) NaA527C inward-facing occluded state #2, (E) NaS526C occluded conformation, (F) NaT525C occluded conformation, (G) NaE523Q occluded conformation, (H) NaAtm1 MgAMPPNP-bound occluded structure, and (I) NaAtm1 MgADPVO4-bound closed structure. (J) Surface representation of the overall structure and central binding cavity of Sav1866 in the outward-facing conformation (PDB ID: 2HYD). The cavity of the NaAtm1 wide-open inward-facing structure is shown in cyan, the NaAtm1 inward-facing structure is shown in gray, the NaA527C inward-occluded structure #1 in yellow, the NaA527C inward-occluded structure #2 in red, the NaS526C occluded structure in blue, the NaT525C occluded structure in purple, the NaE523Q occluded structure in pink, the NaAtm1 MgAMPPNP occluded structure in orange, and the Sav1866 outward-facing structure in brown. The volumes of the cavities were all calculated by CastP (36) with a probe radius of 2.5 Å.
Discussion
We have expanded the structurally characterized conformations of the ABC exporter NaAtm1 from the previously determined inward-facing conformation (30) to multiple occluded conformations, through introduction of disulfide bridges and the use of nucleotide analogs. From all available NaAtm1 structures, we have observed the inward-facing conformations (including the inward-facing occluded conformation) in either nucleotide-free or MgADP-bound forms, while the occluded conformations are stabilized by ATP or analogs (Fig. 1). Substrates have been observed to bind only to the inward-facing conformations (30), even though substrate-binding cavities of sufficient size to accommodate GSSG are present in the occluded conformations (Fig. 4). The most striking changes between these different conformational states occur in the TMDs, most notably in the positioning of TM4–5 described previously (30) and the straightening and kinking of TM6s.
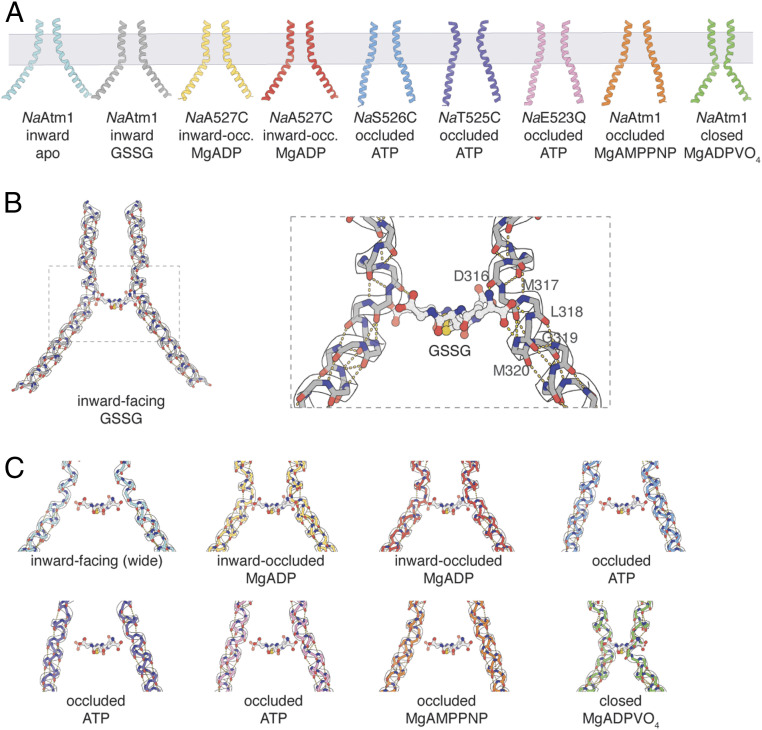
Fig. 4.
TM6 comparisons of NaAtm1. (A) TM6 (residues 300 to 340) conformations observed in different NaAtm1 structures. (B) Hydrogen-bonding interactions between GSSG and TM6 main chain groups from residues in the kink region of the primary binding site [residues 316 to 320 (PDB ID: 4MRS)]. (C) Potential interactions between GSSG and residues 316 to 320 in other NaAtm1 structures. The approximate location of GSSG is obtained from the structural alignments of TM6s in different structures to the TM6 of the inward-facing conformation of NaAtm1 (PDB ID: 4MRS). Hydrogen-bonding interactions are shown by yellow dashes and the GSSG positions are shown by ball and sticks in B and C. TM6s in the NaAtm1 wide-open inward-facing structure are shown in cyan, the NaAtm1 inward-facing structure (PDB ID: 4MRS) in gray, the NaA527C inward-occluded structure #1 in yellow, the NaA527C inward-occluded structure #2 in red, the NaS526C occluded structure in blue, the NaT525C occluded structure in purple, the NaE523Q occluded structure in pink, the NaAtm1 MgAMPPNP-bound occluded structure in orange, and the NaAtm1 MgADPVO4-bound closed structure in green.
The conformation of TM6 is directly coupled to the binding of glutathione by NaAtm1 and hence to the directionality of transport. The kinking of TM6 observed in the inward-facing, inward-facing occluded, and the posthydrolysis occluded structures (Fig. 4A) contrasts with the more subtly bent TM6 helices in the prehydrolysis occluded structures of NaAtm1 (Fig. 4A). The TM6 kink present in the inward-facing conformation occurs adjacent to a turn of the 310 helix at residues 314 to 317, such that the amide NH groups of residues 319 and 320 hydrogen bond to the α-carboxyl of the γ-Glu of GSSG, while the α-amino group of GSSG hydrogen bonds to the carbonyl CO of residue 316 (30) (Fig. 4B). Upon transition to the occluded conformation of NaAtm1, TM6 adopts a regular α-helical structure (Fig. 4C), accompanied by an increased separation between the symmetry-related TM6s, so that these peptide bond groups are no longer available to hydrogen-bond the transported substrate. As a consequence, as NaAtm1 transitions from the inward to outward-facing conformational states, the binding site for the transported substrate is restructured for substrate release, providing a mechanism for coupling protein conformation to the ligand-binding affinity. As NaAtm1 resets to the inward-facing conformation, kinking of TM6 in the MgADPVO4 stabilized form effectively closes the substrate-binding cavity. The inability to bind glutathione in the posthydrolysis state enforces the unidirectionality of the transport cycle, since the binding site exists only during the transition from the inward- to outward-facing conformations.
Other ABC transporter systems, such as the lipid-linked oligosaccharide flippase PglK, the multidrug resistance protein TmrAB, and the drug efflux pump ABCB1 (P-glycoprotein) also exhibit changes in TM6 kinking and binding between distinct conformational states (SI Appendix, Fig. S9). The detailed progression of changes in TM6 varies between the different transporters, however, and NaAtm1 is so far unique in the nearly straight TM6s present in the occluded structure. The coupling of TM6 to substrate binding observed with NaAtm1 has also been noted for other transporters. From the analysis of multiple TmrAB structures, TM6 of each subunit was identified as the gatekeeper controlling access to substrate-binding cavities (18), while in ABCB1 (10), TM6 and TM12 (equivalent to TM6 in a full ABC transporter) were found to be important in substrate binding, along with TM4 and TM10. Rather than a universal set of structural changes associated with the transport cycle, however, each system will have its own distinct structural features. Thus, detailed characterizations of the different intermediate states of a specific transporter are essential for addressing its transport mechanism.
The structural and functional characterization of the disulfide crosslinked variants revealed several unanticipated findings that reflect the apparent the plasticity of the NBD–NBD interface. The structures of the two disulfide crosslinking variants NaA527C and NaS526C determined in this study included three different conformational states, the two inward-occluded conformations of NaA527C and the occluded conformation of NaS526C. Unexpectedly, the disulfide crosslinked NaS526C variant not only hydrolyzed ATP, but also could transport substrate (Fig. 2). While the presence of a disulfide crosslink between the NBDs might be expected to inhibit the functionality by restricting relevant conformational changes, the observation that this variant is transport competent indicates that ATP hydrolysis and substrate transport involve a limited set of conformational states that do not require wide separation of the NBDs.
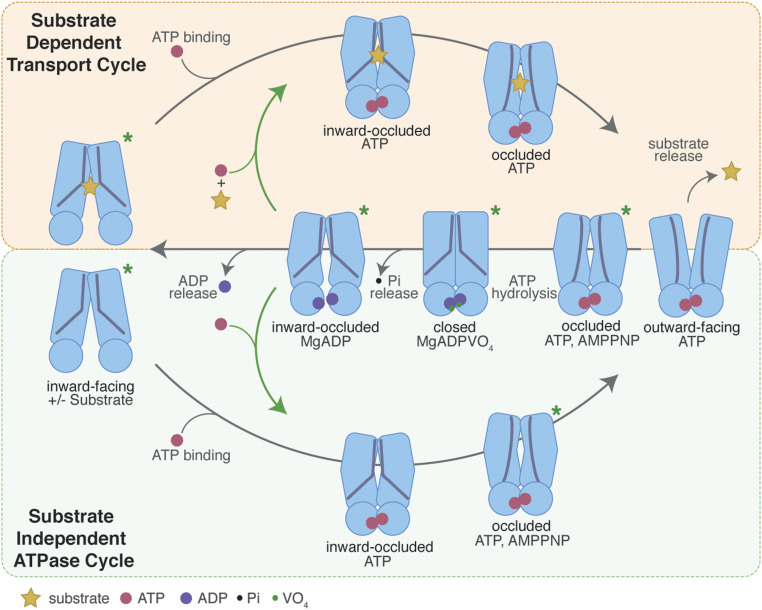
The characterization of NaAtm1 in multiple conformational states provides the opportunity to outline the structural changes that occur in the transport cycle. Since each structure represents a separate experiment determined under different conditions, it is not possible to unambiguously order the structures in a sequence along the transport cycle. With the introduction of several assumptions about the relationships between structures, however, a working model for a structure-based transport mechanism of NaAtm1 can be devised. These assumptions are the following: 1) the available NaAtm1 structures approximate on-path intermediates, 2) the transported ligand binds preferentially to the inward-facing conformation (25, 30), and 3) the outward-facing conformation is stabilized by ATP (see refs. 22, 31, 38), with return to the inward-facing conformation accompanying Pi dissociation (18). A schematic mechanism for substrate-dependent transport by NaAtm1 is presented in Fig. 5 (Top) with the main features summarized as follows. Substrate and MgATP can bind to the transporter in both the inward-facing and inward-facing occluded states. The transition from the inward-facing to outward-facing conformation through the occluded states is accompanied by ATP binding. The progression from the inward-facing to outward-facing conformations is reflected in the straightening of TM6, which alters the GSSG-binding site, creating the larger cavity of the occluded conformation and ultimately leading to dissociation from the outward-facing conformation. Although we were unable to capture an outward-facing conformation by either X-ray crystallography or electron microscopy, it is possible that the flexibility of the periplasmic loops observed in the occluded conformation may transiently open an exit pathway for glutathione release. On resetting to the inward conformation, regeneration of kinked TM6 helices creates a closed conformation by eliminating the substrate-binding cavity as trapped in the posthydrolysis MgADPVO4-trapped state. Although we have observed only symmetric NaAtm1 structures, we cannot exclude the presence of asymmetric states as observed for TmrAB (18). Upon Pi dissociation, the transporter returns back to the inward-facing and/or the inward-facing occluded states, and the transporter is ready for the next transport cycle.
Fig. 5.
Schematic representation of a structure-based transport mechanism for NaAtm1. The substrate-dependent transport cycle (Top cycle) and the substrate-independent ATPase pathways (Bottom) differ by the presence or absence of bound substrate, respectively, during the transition from the inward- to outward-facing conformation. Resetting the outward-facing to the inward-facing conformation involves a set of intermediates common to both pathways, including the closed, posthydrolysis conformation lacking a substrate-binding site. Ligand binding to disulfide crosslinked variants may occur in the inward-occluded conformations (green arrows), without accessing the fully inward-facing conformations. Inward-facing and inward-facing occluded structures have been determined in both apo and substrate-bound states (PDB IDs: 6VQU [wide-open], 4MRN [apo], 4MRS [GSSG bound], 4MRP [GSH bound], 4MRV [GS-Hg bound], and 6PAM [MgADP bound]). The inward-occluded state with both substrate and ATP present is modeled on the crystal structure of the inward-facing occluded structure with MgADP bound (PDB ID: 6PAM). The ATP-bound occluded state has been determined for the disulfide crosslinking mutants (NaS526C and NaT525C with PDB IDs 6PAN and 6PAO, separately), the ATP hydrolysis-deficient mutant (NaE523Q with PDB ID: 6PAQ), and the wild-type transporter with MgAMPPNP bound (PDB ID: 6PAR). The outward-facing conformation of NaAtm1 has not been structurally characterized, but its presence is essential for substrate release. The posthydrolysis closed state has been characterized with MgADPVO4 bound (PDB ID: 6VQT), while the ADP-bound occluded state after ATP hydrolysis has been solved with NaA527C (PDB ID: 6PAM). Asterisks denote the structurally characterized structures to date for NaAtm1.
A key question for ABC exporters centers on the role of the substrate in the transport cycle; given the observed basal ATPase activities, there is not an obligatory coupling between the ATPase activity and substrate binding/transport. This is a surprising situation given what is known for the transport cycles of well-coupled transporters such as lac permease (39) and P-type ATPases (40), where both the transported substrate and the energy source (H+ or ATP, respectively) must be present for translocation to proceed. As depicted in Fig. 5 (Bottom), the uncoupled activity could reflect the operation of a parallel, but distinct, pathway for ATP hydrolysis that occurs in the absence of the transported substrate, as proposed for certain ABC importers (41) and ABC exporters (18). A key intermediate for understanding the coupling mechanism of an ABC exporter is the occluded state with both substrate and ATP bound (Fig. 5); this state has not been captured for NaAtm1 or, to our knowledge, for any other ABC exporter. This particular species perhaps exists only transiently before the substrate dissociates from the outward-facing conformation [also likely a transiently occurring state (18, 42)], even in the transport-competent form with the disulfide-linked NBDs. Engineering a construct adopting an occluded conformation with both substrate and ATP bound will be a priority for future investigations to address the coupling mechanism of NaAtm1.
Materials and Methods
Mutagenesis and Protein Expression.
The gene encoding NaAtm1 (GenBank accession no. ABD27067) was previously cloned into a pJL-H6 ligation independent vector with 6-His tag on the carboxy-terminus (30) and deposited in Addgene (catalog #78308). All mutants were generated using Q5 Site-Directed Mutagenesis Kit (New England Biolabs). All proteins were overexpressed in Escherichia coli BL21-gold (DE3) cells (Agilent Technologies) using ZYM-5052 autoinduction media and selenomethionine substituted proteins were overexpressed in E. coli B834 (DE3) cells (Novagen) using PASM-5052 autoinduction media as described previously (30). Cells were collected by centrifugation and stored at −80 °C until use.
Purification and Crosslinking.
Frozen cell pellets of cysteine mutants were resuspended in lysis buffer containing 100 mM NaCl, 20 mM Tris, pH 7.5, 40 mM imidazole, pH 7.5, 5 mM β-mercaptoethanol (BME), 10 mM MgCl2, 0.5% (wt/vol) N-dodecyl-β-d-maltopyranoside (DDM) (Anatrace), 0.5% (wt/vol) octaethylene glycol monododecyl ether (C12E8) (Anatrace), lysozyme, DNase, and a protease inhibitor tablet. The resuspended cells were lysed either by stirring for 3 h at 4 °C or by using a M-110L pneumatic microfluidizer (Microfluidics). Unlysed cells and cell debris were removed by ultracentrifugation at ∼113,000 × g for 45 min at 4 °C. The supernatant was collected and loaded onto a prewashed NiNTA column with NiNTA wash buffer at 4 °C. NiNTA wash buffer contained 100 mM NaCl, 20 mM Tris, pH 7.5, 50 mM imidazole, pH 7.5, 5 mM BME, 0.05% DDM, and 0.05% C12E8. Elution was achieved using the same buffer containing 350 mM imidazole instead. The eluted protein was then buffer-exchanged to 100 mM NaCl, 20 mM Tris, pH 7.5, 0.05% DDM, and 0.05% C12E8 (size exclusion chromatography [SEC buffer]). Oxidation of engineered cysteines to form disulfide bonds was achieved by incubating buffer-exchanged protein with 1 mM Cu (II)(1,10-phenanthroline)3 for 1 h at 4 °C. A crosslinked sample was buffer-exchanged into SEC buffer to remove the oxidant and further purified by SEC using HiLoad 16/60 Superdex 200 (GE Healthcare). Fractions were pooled and concentrated to 20 to 35 mg/mL using an Amicon Ultra 15 concentrator (Millipore) with a molecular weight cutoff of 100 kDa. Wild-type NaAtm1 was solubilized in lysis buffer containing 1% DDM and purified in NiNTA and SEC buffers containing 0.1% DDM for crystallization in the occluded conformation with MgAMPPNP.
Crystallizations.
NaA527C was crystallized in MemGold (Molecular Dimensions) condition #68. Upon optimization of the crystallization conditions, including the use of additive screens (Hampton Research), the best crystals were grown from 100 mM NaCl, 100 mM Tris, pH 8.3, 25 mM MgCl2, and 28% polyethylene glycol monomethyl ether 2000 (PEG 2000 MME) with 20 mM ATP, pH 7.5, at 20 °C. The NaA527C crystallization sample was prepared at 20 mg/mL with 1 mM ATP, pH 7.5, 5 mM ethylenediaminetetraacetic acid (EDTA), pH 7.5, with or without 5 mM GSSG, pH 7.5. Crystals appeared in about 2 wk and lasted for about a month. Crystals were harvested in cryoprotectant solutions containing 100 mM NaCl, 100 mM Tris, pH 8.3, 25 mM MgCl2, 28% PEG 2000 MME with PEG 400 at 10, 15, and 20% before flash-freezing in liquid nitrogen.
NaS526C, NaT525C, and NaE523Q crystals were prepared under the same condition as NaA527C except MgCl2 was removed in the crystallization well solution. All crystallization samples were prepared with 1 mM ATP, pH 7.5, and 5 mM EDTA, pH 7.5. The crystallization condition of NaT525C was further optimized with 200 mM of NDSB-221 using the Additive Screen (Hampton Research). The crystallization condition of NaE523Q was further optimized with 10 mM DTT but without 20 mM ATP, pH 7.5, in the crystallization conditions. Crystals of all three constructs were harvested in cryoprotectant solutions containing 100 mM NaCl, 100 mM Tris, pH 8.3, 28% PEG 2000 MME with PEG 400 at 10, 15, and 20% before flash-freezing in liquid nitrogen.
NaAtm1 purified in DDM was crystallized in MemChannel (Molecular Dimensions) condition #29. The crystallization sample was prepared with 1 mM AMPPNP, pH 7.5, 2 mM MgCl2 with or without 5 mM GSSG, pH 7.5. The condition was further optimized to 50 mM ADA, pH 7.1, 8 to 10% PEG 1000 and 8 to 10% PEG 1500 at 20 °C with protein at 8 mg/mL. Crystals appeared within a week and were harvested in cryoprotectant solutions containing 50 mM ADA, pH 7.1, 10% PEG 1,000 and 10% PEG 1,500 with PEG 400 at 10, 15, and 20% before flash-freezing in liquid nitrogen.
X-Ray Data Collection and Structure Determinations.
X-ray datasets were collected at the Stanford Synchrotron Radiation Laboratory beamline 12–2 using a Pilatus 6M detector with Blu-Ice interface (43) and Advanced Photon Source GM/CA beamline 23ID-B using an Eiger 16M detector with JBluIce-EPICS interface (44). All datasets were processed and integrated with XDS (45) and scaled with Aimless (46). For the NaA527C crystal structure, the first three transporters in the asymmetric unit were identified by searching for multiple copies of the TMDs and NBDs using the inward-facing structure (PDB ID: 4MRN) with Phaser in Phenix (47). Due to the relatively poor electron density for the fourth transporter, the helices of the TMDs were first built using Find Helices and Strands in Phenix (47), and then a full transporter from the previously identified partial model was superposed onto the built helices in Coot (48), which resulted in the misplacement of one NBD. The misplaced NBD was removed from the model and was subsequently correctly placed using Molrep in CCP4 (46). For the NaS526C structure, molecular replacement was carried out using Sav1866 (PDB ID: 2HYD) with superposed NaAtm1 sequence as the input model for Phaser in Phenix (47). For the NaT525C, NaE523Q, and NaAtm1 structures, molecular replacement was carried out using the NaS526C structure as the input model for Phaser in Phenix (47). For all structures, experimental phase information was derived from SeMet datasets by MR-SAD using AutoSol in Phenix (47). Iterative refinement and model-building cycles were carried out with phenix.refine in Phenix (47), Refmac in CCP4 (46), and Coot (48). The final refinements were performed with phenix.refine in Phenix (47). Data collection and refinement statistics are presented in SI Appendix, Tables S4–S8.
Single-Particle Sample and Grid Preparation.
The expression plasmid for the membrane scaffolding protein (MSP1D1) was purchased from Addgene (plasmid #20061), and its expression and purification were carried out using published protocols (33). The reconstitutions were performed with 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC) at a ratio of NaAtm1:MSP1D1:POPC = 1:(2 to 4):130. For the structure with MgADPVO4 bound, MSP1D1 and POPC were added after incubating detergent-purified protein with 4 mM MgCl2, 4 mM ATP, pH 7.5, and 4 mM VO43- for 4 h to allow for ATP hydrolysis at 4 °C. VO43- was prepared from sodium orthovanadate following a published protocol (49). The reconstituted samples were incubated overnight at 4 °C with a BioBeads addition at 400 mg/mL for detergent removal. The samples were subjected to size exclusion chromatography with Superdex 200 Increase 10/300 (GE Healthcare). Peak fractions were either pooled and concentrated or directly used for grid preparation. Grids were prepared with protein concentrations between 0.6 and 4 mg/mL. Briefly, 3 µL of protein solution was applied to freshly glow-discharged UltrAuFoil 2/2 200 mesh grids (MgADPVO4 stabilized closed conformation) and QuantiFoil Au R1.2/1.3 200 mesh grids (wide-open inward-facing conformation) and blotted for 4 s with 0 blot force and 100% humidity at room temperature using Vitrobot Mark IV (FEI).
Single-Particle Data Collection, Processing, and Refinement.
For the MgADPVO4 stabilized closed conformation, two datasets were collected with a Gatan K3 direct electron detector on a 300 keV Titan Krios in the superresolution mode using SerialEM. For data collection, a total dosage of 60 e−/Å2 was utilized with a defocus range between −1.5 and −3.5 μm at the Caltech CryoEM facility and a total dosage of 48.6 e−/Å2 with a defocus range between −1.7 and −2.4 μm at the Stanford-SLAC Cryo-EM Center (S2C2). The two datasets were processed separately. For the inward-facing conformation, data were collected with a Gatan K2 Summit direct electron detector on a 300 keV Titan Krios in the superresolution mode using EPU with a total dosage of 36 e−/Å2 with a defocus range between −1.0 and −3.5 μm at the Caltech CryoEM facility.
Processing of all datasets was performed with cryoSPARC 2 (50), using patch motion for motion correction and estimating the contrast transfer function (CTF) parameters with CTFFIND (51). Particles were picked using a previous reconstruction of NaAtm1 or a blob as template and then extracted. Rounds of two-dimensional classifications were performed, leaving ∼170,000 particles for the MgADPVO4-stabilized closed conformation using data from the S2C2 collection and ∼100,000 particles for the inward-facing conformation for the three-dimensional (3D) reconstruction. The resulting 3D reconstructions were then refined with homogeneous and nonuniform refinements in cryoSPARC 2 (50). For the MgADPVO4 stabilized closed structure, local CTF refinement was carried out to refine per-particle defocus, and the final local refinement was performed using symmetry expanded (C2) particles in cryoSPARC 2 (50) along with a mask generated in Chimera (52) based on a fitted model.
Initial model fitting was carried out with phenix.dock_in_map in Phenix (47) using a single chain of the MgAMPPNP-bound occluded structure (PDB ID: 6PAR) as the starting model for the MgADPVO4-bound closed structure, and a single chain of the inward-facing conformation (PDB ID: 4MRN) for the wide open inward-facing conformation. Model building and ligand fitting were manually carried out in Coot (48), and the structures were iteratively refined using phenix.real_space_refine in Phenix (47). Data collection, refinement and validation statistics are presented in SI Appendix, Tables S9–S10.
Proteoliposome Preparation and Transport Assay.
PLS were prepared by following the published protocol for ABC transporter PLS reconstitution (53) with an additional step of a Biobeads addition at 40 mg/mL to ensure the complete removal of detergents. The transport assay was conducted with NaAtm1 reconstituted in PLS in a 1-mL format at 37 °C. The reaction mixture contained PLS at 5 mg/mL, 10 mM MgATP, pH 7.5, 2.5 mM GSSG, pH 7.5, transport buffer at 85 mM NaCl, and 17 mM Tris, pH 7.5. The different controls were also prepared in a similar fashion. Aliquots of 150 µL of the reaction mixture were taken every 15 min, added to 1 mL of cold transport buffer, and then ultracentrifuged at ∼203,000 × g in a TLA 100.3 rotor in a Beckman Ultima benchtop ultracentrifuge for 10 min at 4 °C. The pellets were washed 10 times with cold transport buffer and then resuspended to 100 µL with solubilization buffer (85 mM NaCl, 17 mM Tris, pH 7.5, and 2% sodium dodecanoyl sarcosine [Anatrace]). The samples were solubilized for 2 h until the solution clarified before spinning down in the TLA 100 rotor to remove bubbles. Samples of 10 µL were taken for GSSG quantification using the Glutathione Quantification Assay (Sigma-Aldrich). Reactions were done in triplicate, and the rates were not corrected for orientation of NaAtm1 in PLS.
ATPase Assay.
The ATPase activity was determined by the molybdate-based phosphate quantification method (54). All reactions were performed in a 250-µL scale with a final protein concentration of 0.05 mg/mL for both PLS and detergent at various concentrations of MgATP and GSSG, pH 7.5, at 37 °C. Fifty microliters of reactions were taken every 5 min for four times and mixed with 50 µL of 12% sodium dodecyl sulfate in a 96-well plate at room temperature. One hundred microliters of ascorbic acid/molybdate mix was added and incubated for 5 min before the addition of 150 µL of citric acid/arsenite/acetic acid solution. The reaction was then incubated for 20 min at room temperature before reading at 850 nm with a Tecan plate reader. Reactions were done in triplicate, the measurements were plotted against time, and the final linear rates were fitted with nonlinear regression fit using Prism 8. The rates were not corrected for orientation of NaAtm1 in PLS.
Data and Materials Availability.
Atomic coordinates and structure factors have been deposited in the Protein Data Bank with accession codes 6PAM (NaA527C-MgADP), 6PAN (NaS526C-ATP), 6PAO (NaT525C-ATP), 6PAQ (NaE523Q-ATP), 6PAR (NaAtm1-MgAMPPNP), 6VQT (NaAtm1-MgADPVO4), and 6VQU (NaAtm1). The two cryo-EM single-particle structures in nanodiscs have also been deposited in the Electron Microscopy Data Bank under accession codes EMD-21356 (NaAtm1-MgADPVO4) and EMD-21357 (NaAtm1). The raw data for ATPase and transport assays that support the findings in Fig. 2 are included in SI Appendix, Tables S2 and S3.
Supplementary Material
Acknowledgments
We thank the staffs of the Stanford Synchrotron Radiation Lightsource (SSRL) beamline 12-2 and of the Advanced Photon Source GM/CA beamline for support during X-ray diffraction data collection; Paul Adams, Kaspar Locher, Oded Lewinson, Gabriele Meloni, William Clemons, the organizers and speakers at the Cold Spring Harbor X-ray Method in Structural Biology Course (2018), the CCP4/APS School for Macromolecular Crystallography (2017), and the SBGrid/NE-CAT Phenix Workshop (2016) for discussions; Haoqing Wang, Andrey Malyutin, Songye Chen, and Megan Meyer for their support during single-particle cryo-EM data collection; and the Gordon and Betty Moore Foundation and the Beckman Institute for their generous support of the Molecular Observatory at Caltech. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the US Department of Energy (DOE), Office of Science, Office of Basic Energy Sciences, under contract no. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE of Biological and Environmental Research and by the NIH National Institute of General Medical Sciences (NIGMS) (Grant P41GM103393). GM/CA@APS was funded in whole or in part by federal funds from the National Cancer Institute (Grant ACB-12002) and the NIGMS (Grant AGM-12006). This research used resources of the Advanced Photon Source, a DOE Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under contract no. DE-AC02-06CH11357. The Eiger 16M detector was funded by NIH Office of Research Infrastructure Programs High-End Instrumentation Grant 1S10OD012289-01A1. The cryo-EM was performed in the Beckman Institute Resource Center for Transmission Electron Microscopy at Caltech and at the Stanford SLAC Cryo-EM Center (S2C2). The S2C2 is supported by the NIH Health Common Fund Transformative High Resolution Cryo-Electron Microscopy program.
Footnotes
The authors declare no competing interest.
Data deposition: Data for this article have been deposited in the RCSB Protein Data Bank under IDs 6PAM, 6PAN, 6PAO, 6PAQ, 6PAR, 6VQT, and 6VQU. The two cryo-EM single-particle structures in nanodiscs have also been deposited in the Electron Microscopy Data Bank under accession codes EMD-21356 (NaAtm1-MgADPVO4) and EMD-21357 (NaAtm1).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2006526117/-/DCSupplemental.
References
- 1.Higgins C. F., ABC transporters: From microorganisms to man. Annu. Rev. Cell Biol. 8, 67–113 (1992). [DOI] [PubMed] [Google Scholar]
- 2.Rees D. C., Johnson E., Lewinson O., ABC transporters: The power to change. Nat. Rev. Mol. Cell Biol. 10, 218–227 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Locher K. P., Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat. Struct. Mol. Biol. 23, 487–493 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Thomas C., Tampé R., Multifaceted structures and mechanisms of ABC transport systems in health and disease. Curr. Opin. Struct. Biol. 51, 116–128 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Davidson A. L., Dassa E., Orelle C., Chen J., Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 72, 317–364 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rempel S. et al., A mycobacterial ABC transporter mediates the uptake of hydrophilic compounds. Nature 580, 409–412 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Arnold F. M. et al., The ABC exporter IrtAB imports and reduces mycobacterial siderophores. Nature 580, 413–417 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moody J. E., Millen L., Binns D., Hunt J. F., Thomas P. J., Cooperative, ATP-dependent association of the nucleotide binding cassettes during the catalytic cycle of ATP-binding cassette transporters. J. Biol. Chem. 277, 21111–21114 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korkhov V. M., Mireku S. A., Locher K. P., Structure of AMP-PNP-bound vitamin B12 transporter BtuCD-F. Nature 490, 367–372 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Alam A. et al., Structure of a zosuquidar and UIC2-bound human-mouse chimeric ABCB1. Proc. Natl. Acad. Sci. U.S.A. 115, E1973–E1982 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alam A., Kowal J., Broude E., Roninson I., Locher K. P., Structural insight into substrate and inhibitor discrimination by human P-glycoprotein. Science 363, 753–756 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson Z. L., Chen J., Structural basis of substrate recognition by the multidrug resistance protein MRP1. Cell 168, 1075–1085.e9 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Liu F., Zhang Z., Csanady L., Gadsby D. C., Chen J., Molecular structure of the human CFTR ion channel. Cell 169, 85–95.e88 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Zhang Z., Liu F., Chen J., Conformational changes of CFTR upon phosphorylation and ATP binding. Cell 170, 483–491.e8 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Liu F. et al., Structural identification of a hotspot on CFTR for potentiation. Science 364, 1184–1188 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ward A., Reyes C. L., Yu J., Roth C. B., Chang G., Flexibility in the ABC transporter MsbA: Alternating access with a twist. Proc. Natl. Acad. Sci. U.S.A. 104, 19005–19010 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mi W. et al., Structural basis of MsbA-mediated lipopolysaccharide transport. Nature 549, 233–237 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hofmann S. et al., Conformation space of a heterodimeric ABC exporter under turnover conditions. Nature 571, 580–583 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perez C., Mehdipour A. R., Hummer G., Locher K. P., Structure of outward-facing PglK and molecular dynamics of lipid-linked oligosaccharide recognition and translocation. Structure 27, 669–678.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Aller S. G. et al., Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science 323, 1718–1722 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin M. S., Oldham M. L., Zhang Q., Chen J., Crystal structure of the multidrug transporter P-glycoprotein from Caenorhabditis elegans. Nature 490, 566–569 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim Y., Chen J., Molecular structure of human P-glycoprotein in the ATP-bound, outward-facing conformation. Science 359, 915–919 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Kispal G., Csere P., Guiard B., Lill R., The ABC transporter Atm1p is required for mitochondrial iron homeostasis. FEBS Lett. 418, 346–350 (1997). [DOI] [PubMed] [Google Scholar]
- 24.Kuhnke G., Neumann K., Mühlenhoff U., Lill R., Stimulation of the ATPase activity of the yeast mitochondrial ABC transporter Atm1p by thiol compounds. Mol. Membr. Biol. 23, 173–184 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Srinivasan V., Pierik A. J., Lill R., Crystal structures of nucleotide-free and glutathione-bound mitochondrial ABC transporter Atm1. Science 343, 1137–1140 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Lill R., Kispal G., Mitochondrial ABC transporters. Res. Microbiol. 152, 331–340 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Pondarré C. et al., The mitochondrial ATP-binding cassette transporter Abcb7 is essential in mice and participates in cytosolic iron-sulfur cluster biogenesis. Hum. Mol. Genet. 15, 953–964 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Cavadini P. et al., RNA silencing of the mitochondrial ABCB7 transporter in HeLa cells causes an iron-deficient phenotype with mitochondrial iron overload. Blood 109, 3552–3559 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Zuo J. et al., Mitochondrial ABC transporter ATM3 is essential for cytosolic iron-sulfur cluster assembly. Plant Physiol. 173, 2096–2109 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee J. Y., Yang J. G., Zhitnitsky D., Lewinson O., Rees D. C., Structural basis for heavy metal detoxification by an Atm1-type ABC exporter. Science 343, 1133–1136 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dawson R. J., Locher K. P., Structure of a bacterial multidrug ABC transporter. Nature 443, 180–185 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Goodno C. C., Myosin active-site trapping with vanadate ion. Methods Enzymol. 85 Pt B, 116–123 (1982). [DOI] [PubMed] [Google Scholar]
- 33.Ritchie T. K. et al., Chapter 11: Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 464, 211–231 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bennett B. D. et al., Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 5, 593–599 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patzlaff J. S., van der Heide T., Poolman B., The ATP/substrate stoichiometry of the ATP-binding cassette (ABC) transporter OpuA. J. Biol. Chem. 278, 29546–29551 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Tian W., Chen C., Lei X., Zhao J., Liang J., CASTp 3.0: Computed atlas of surface topography of proteins. Nucleic Acids Res. 46, W363–W367 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Erickson H. P., Size and shape of protein molecules at the nanometer level determined by sedimentation, gel filtration, and electron microscopy. Biol. Proced. Online 11, 32–51 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stefan E., Hofmann S., Tampé R., A single power stroke by ATP binding drives substrate translocation in a heterodimeric ABC transporter. eLife 9, e55943 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaback H. R., A chemiosmotic mechanism of symport. Proc. Natl. Acad. Sci. U.S.A. 112, 1259–1264 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palmgren M. G., Nissen P., P-type ATPases. Annu. Rev. Biophys. 40, 243–266 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Lewinson O., Livnat-Levanon N., Mechanism of action of ABC importers: Conservation, divergence, and physiological adaptations. J. Mol. Biol. 429, 606–619 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Grossmann N. et al., Mechanistic determinants of the directionality and energetics of active export by a heterodimeric ABC transporter. Nat. Commun. 5, 5419 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McPhillips T. M. et al., Blu-Ice and the Distributed Control System: Software for data acquisition and instrument control at macromolecular crystallography beamlines. J. Synchrotron Radiat. 9, 401–406 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Stepanov S. et al., JBluIce-EPICS control system for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 67, 176–188 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kabsch W., XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winn M. D. et al., Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adams P. D. et al., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oldham M. L., Chen J., Snapshots of the maltose transporter during ATP hydrolysis. Proc. Natl. Acad. Sci. U.S.A. 108, 15152–15156 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pettersen E. F. et al., UCSF Chimera: A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Geertsma E. R., Nik Mahmood N. A., Schuurman-Wolters G. K., Poolman B., Membrane reconstitution of ABC transporters and assays of translocator function. Nat. Protoc. 3, 256–266 (2008). [DOI] [PubMed] [Google Scholar]
- 54.Chifflet S., Torriglia A., Chiesa R., Tolosa S., A method for the determination of inorganic phosphate in the presence of labile organic phosphate and high concentrations of protein: Application to lens ATPases. Anal. Biochem. 168, 1–4 (1988). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.