Significance
Virtually all animals are associated with symbiotic bacteria. How these associations are modulated across an animal’s life cycle is a key question in understanding animal–bacteria interactions, particularly in organisms that undergo metamorphosis during development. Here, we used the cereal weevil to show how symbiosis is reorganized during metamorphosis, a developmental process entailing drastic tissue rearrangements. In this insect, symbionts are housed within specialized host cells, the bacteriocytes, that form the bacteriome tissue. We show that the bacteriome is completely remodeled during metamorphosis through host–symbiont communication, involving adhesion and motility host proteins and a temporary symbiont infectious behavior. This interkingdom dialogue results in the adaptation of the bacteriome to adulthood, highlighting the intertwining of symbiosis with host development.
Keywords: endosymbiosis, bacteriome, metamorphosis, insect, dual RNA-seq
Abstract
Bacterial intracellular symbiosis (endosymbiosis) is widespread in nature and impacts many biological processes. In holometabolous symbiotic insects, metamorphosis entails a complete and abrupt internal reorganization that creates a constraint for endosymbiont transmission from larvae to adults. To assess how endosymbiosis copes—and potentially evolves—throughout this major host-tissue reorganization, we used the association between the cereal weevil Sitophilus oryzae and the bacterium Sodalis pierantonius as a model system. S. pierantonius are contained inside specialized host cells, the bacteriocytes, that group into an organ, the bacteriome. Cereal weevils require metabolic inputs from their endosymbiont, particularly during adult cuticle synthesis, when endosymbiont load increases dramatically. By combining dual RNA-sequencing analyses and cell imaging, we show that the larval bacteriome dissociates at the onset of metamorphosis and releases bacteriocytes that undergo endosymbiosis-dependent transcriptomic changes affecting cell motility, cell adhesion, and cytoskeleton organization. Remarkably, bacteriocytes turn into spindle cells and migrate along the midgut epithelium, thereby conveying endosymbionts to midgut sites where future mesenteric caeca will develop. Concomitantly, endosymbiont genes encoding a type III secretion system and a flagellum apparatus are transiently up-regulated while endosymbionts infect putative stem cells and enter their nuclei. Infected cells then turn into new differentiated bacteriocytes and form multiple new bacteriomes in adults. These findings show that endosymbiosis reorganization in a holometabolous insect relies on a synchronized host–symbiont molecular and cellular “choreography” and illustrates an adaptive feature that promotes bacteriome multiplication to match increased metabolic requirements in emerging adults.
Symbiotic associations between animals and bacteria are widespread in nature, impact organisms in different ways, and represent a driving force in evolution (1–4). Symbiosis is found in all ecological niches and occurs at different levels of host integration, culminating in intracellular symbiosis (endosymbiosis) in which the genomes of both partners are present within specific host cells, the bacteriocytes. Endosymbiosis is prevalent in insects thriving on nutritionally unbalanced diets, where endosymbionts provide nutritional complementation to their hosts and thereby increase their fitness and their adaptive capacities (5–7). Endosymbiosis occurs both in insects going through incomplete metamorphosis (Hemimetabola) and complete metamorphosis (Holometabola). Metamorphosis is a major postembryonic developmental process that represents a drastic anatomical remodeling of internal and external morphological organization (8). Such extreme changes allow the discrete life stages of a given species to adapt to distinct environmental niches, hence limiting intraspecies competition (8, 9). It also allows adaptive decoupling: for example, task specialization of growth at the larval stage versus reproduction at the adult stage (9, 10). However, metamorphosis is also associated with constraints, including higher susceptibility to predators and pathogens, as well as the conservation and adaptation of beneficial symbionts (9, 11). In hemimetabolous insects, the relative morphological stability associated with incomplete metamorphosis is believed to facilitate symbiont maintenance and transmission across developmental stages (11). On the other hand, complete metamorphosis creates a double challenge for holometabolous hosts: 1) Maintaining symbionts amid a drastic morphological reorganization and 2) adapting the symbiotic relationship to a new life stage that might have different metabolic needs. While the evolution of gut symbionts through metamorphosis has been addressed in several insects (12–14), very few insects harboring intracellular symbionts have been studied in this regard (15–17). One of the most extreme cases reported to date is the olive fly Bactrocera oleae, whose endosymbiont transitions from being intracellular in larvae to extracellular in adults (17), although the underlying mechanisms and associated advantages are yet to be established.
To address how endosymbiosis is shaped by a holometabolous lifestyle, we studied the cereal weevil Sitophilus oryzae. This coleopteran species houses its obligatory bacterial endosymbiont Sodalis pierantonius within specialized cells, the bacteriocytes, that group together into an organ, the bacteriome (18–21). S. pierantonius provides S. oryzae with vitamins and amino acids that are at very low abundance in cereals (22–25). While endosymbiont load remains relatively stable during the larval stages, it drastically and rapidly increases during 1 wk following the end of metamorphosis (22). It was shown that endosymbionts supply the host with large amounts of the aromatic amino acids, phenylalanine and tyrosine, which are necessary components for cuticle synthesis following final ecdysis (22). Once the cuticle is complete, endosymbionts are rapidly eliminated and recycled from the gut bacteriomes (22, 26). Hence, while cereal weevils remain in the same ecological niche as larvae and adults, host metabolic needs increase greatly following the end of metamorphosis, as does the endosymbiont metabolic contribution to those needs. In parallel with the endosymbiont dynamics, weevil metamorphosis is associated with a complete reshaping of the bacteriome organ, starting with the dissociation of the single larval bacteriome and ending with the formation of multiple new bacteriomes in pupae, at the apex of the midgut mesenteric caeca (27–29). These morphological changes could represent a morphological optimization for the symbiotic burst and increased bacterial metabolic input in young (i.e., less than 7-d-old) adults. In this study, we sought to understand the mechanistic basis driving the bacteriome reshaping in the S. oryzae–S. pierantonius symbiotic association, as well as the respective implications for each partner. We present an in depth histological characterization of the bacteriome remodeling during metamorphosis. By developing a dual RNA-sequencing (RNA-seq) methodology specifically tailored for S. oryzae and S. pierantonius, we unraveled a coordinated host–endosymbiont transcriptomic dialogue during this reorganization process. Our data suggest that both host and endosymbiont are actively involved in this process through a synchronized transcriptomic dialogue. Our results provide evidence on how symbiont–host coevolution has selected a complex interplay between metamorphosis and endosymbiosis that ensures both symbiont transmission to the adult stage and morphological modifications adapted to the insect’s evolving metabolic needs.
Results and Discussion
Weevil Metamorphosis Entails Bacteriome Localization Shift and Remodeling.
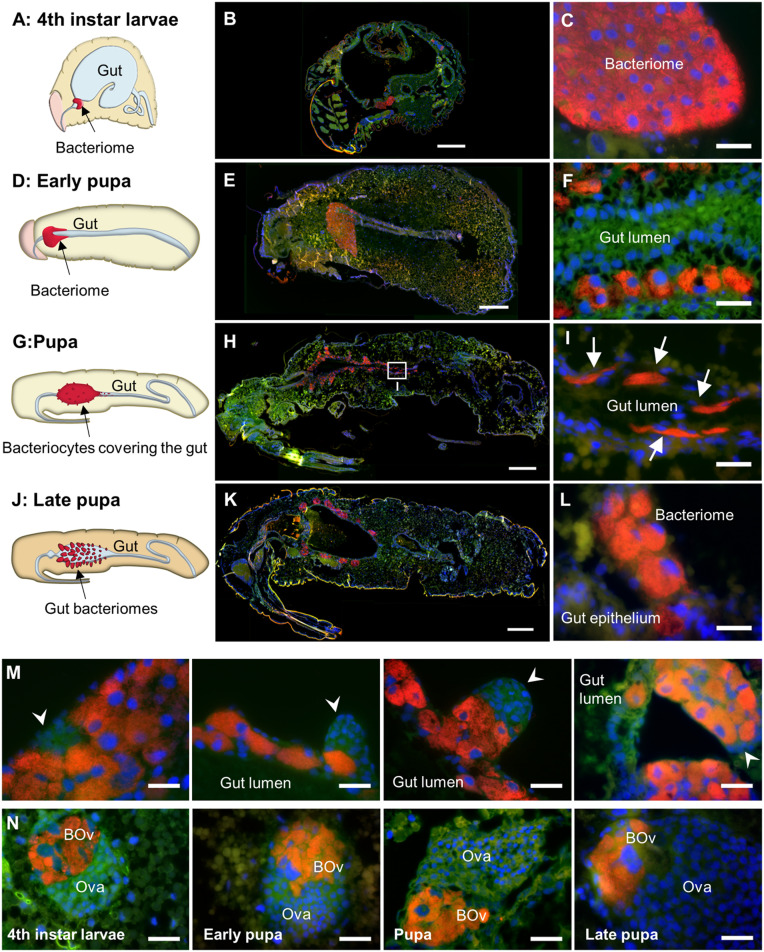
In the 1920s and 1930s, K. Mansour described a shift in bacteriocyte localization during cereal weevil metamorphosis (20, 28, 29). To refine these data with state-of-the-art imaging methods, we revisited this histological analysis during metamorphosis by performing FISH with an S. pierantonius-specific probe (Fig. 1 and SI Appendix, Fig. S1A). In early pupae, the bacteriome dissociates and releases bacteriocytes, which migrate along the midgut (Fig. 1 D–F). In pupae, once the larval bacteriome is fully dissociated, bacteriocytes cover the entire midgut but remain separated from the gut lumen by the intestinal epithelium, and also do not invade the fat body (Fig. 1 G–I). Groupings of bacteriocytes at repetitive specific sites along the midgut lead to the formation of multiple de novo bacteriomes, where mesenteric caeca develop (Fig. 1 J–M). In late pupae, bacteriomes are completely formed at the apex of the growing mesenteric caeca and no bacteriocytes are seen elsewhere (Fig. 1 J–L). In artificially symbiont-depleted (aposymbiotic) individuals, no bacteriome-like structure is observed at either larval or pupal stages (SI Appendix, Fig. S2), consistent with previous observations in aposymbiotic adults (22). Mesenteric caeca still form during metamorphosis of aposymbiotic individuals but contain only gut epithelial cells and no bacteriocyte (SI Appendix, Fig. S2). This suggests that in both larvae and pupae, bacteriocyte differentiation and bacteriome formation are bacteria-dependent, unlike what has been reported in the pea aphid Acyrthosiphon pisum (30). Interestingly, endosymbiont quantification by flow cytometry showed that endosymbiont density remains stable during metamorphosis and increases only once metamorphosis is achieved (SI Appendix, Fig. S1B), supporting the idea that the formation of multiple gut bacteriomes in pupae may be a prerequisite to the symbiotic burst required to fulfill the increased metabolic demands in young adults.
Fig. 1.
The bacteriome organ is drastically remodeled during weevil metamorphosis. (A–C) Endosymbiont localization in fourth-instar larvae schematized in a cartoon [A, adapted from ref. 31, with permission from Elsevier] and visualized by FISH (B and C). The bacteriome is a unique bilobed organ present at the foregut-midgut junction. (D–F) Endosymbiont localization in early pupae schematized in a cartoon (D) and visualized by FISH (E and F). Bacteriocytes are migrating from the larval bacteriome along the shrunken midgut, between a layer of epithelial cells and muscles. (G–I) Endosymbiont localization in pupae schematized in a cartoon (G) and visualized by FISH (H and I). I is a magnification from H. Bacteriocytes completely cover the midgut and start forming new bacteriomes. Arrows in I point to spindle-shaped bacteriocytes. (J–L) Endosymbiont localization in late pupae schematized in a cartoon (J) and visualized by FISH (K and L). Bacteriomes are completely formed and bacteriocytes are no longer present around the midgut. (M) Clusters of small cells (arrowheads) at the apex of forming bacteriomes in pupae (first three panels) and late pupae (last panel). (N) Ovarian bacteriome morphology during metamorphosis, observed by FISH. BOv, Ovarian bacteriome; Ova, Ovariole. Bacteriomes present at the apices of ovarioles in females do not go through any remodeling. All panels: Red: S. pierantonius; green: autofluorescence; blue: DAPI. (Scale bars, 250 μm for B, E, H, and K; 25 μm for C, F, I, L, M, and N.)
In the fourth and last nymphal instar of the hemimetabolous whitefly Bemisia tabaci, a few bacteriocytes were described to migrate from the bacteriome to the ovaries, where they are incorporated into developing oocytes (32). In contrast, we observed in the cereal weevil that ovarian bacteriomes, from which endosymbionts infect oocytes and are transmitted vertically through host generations, are already present in early larval stages (Fig. 1N). This is in line with previous observations that weevil endosymbionts permanently infect germ cells during early embryonic development (20, 33). Unlike gut bacteriomes, ovarian bacteriomes do not seem to go through any remodeling during metamorphosis (Fig. 1N). Hence, the two bacteriocyte populations appear to represent distinct developmental fates, from the larval to the adult stage. Whereas gut bacteriomes strongly increase their symbiont load in the first days of adult life before being eliminated by day 14, ovarian bacteriomes are maintained in the ovaries during the females’ entire lifespan with a relatively constant bacterial density (22).
Dual RNA-Seq Experiments Reveal Synchronized Host–Symbiont Transcriptomic Changes during Metamorphosis.
The fact that bacteriomes form only in symbiotic individuals strongly suggests that the bacteriocyte differentiation process relies on a host–symbiont molecular dialogue. To elucidate such a dialogue, we analyzed the transcriptomic changes from both host and endosymbiont simultaneously, using a dual RNA-seq approach (34–38). We sequenced 12 dual RNA-seq libraries from weevil bacteriocyte-containing guts dissected from fourth-instar larvae, early pupae, pupae, and late pupae (39). The raw reads were cleaned from adaptors and filtered for low quality and were subsequently mapped against the genomes of either S. oryzae (PRJNA431034) (40) or S. pierantonius (CP006568.1) (41) (SI Appendix, Table S1).
Dual RNA-seq libraries were successfully depleted of ribosomal RNA and yielded from 26 to 32 M reads mapped against the S. oryzae genome and from 0.8 to 3.2 M reads mapped against the S. pierantonius genome. Pairwise comparisons were conducted between consecutive time points to identify differentially expressed genes (DEG). In S. oryzae, we detected 2,062 DEG between early pupae vs. fourth-instar larvae, 724 for pupae vs. early pupae, and 1,192 for late pupae vs. pupae (Table 1 and Dataset S1). In S. pierantonius, we detected a total of 114 DEG in early pupae vs. fourth-instar larvae, 215 in pupae vs. early pupae, and 112 in late pupae vs. pupae (Table 1 and Dataset S2). Up- and down-regulated DEG were further analyzed by gene ontology (GO) enrichment analysis (42) and short time-series expression miner (STEM) profiling analysis (43) (Table 1, SI Appendix, Figs. S3–S5, and Datasets S3 and S4). Transcriptomic changes in the host included genes involved in lipid, carbohydrate, and chitin metabolism, and in extracellular matrix and cuticle synthesis (Table 1, SI Appendix, Fig. S3, and Dataset S3), in accordance with a classic metamorphosis process, as previously observed (44–49), and validating the quality of the sequencing data.
Table 1.
Main GO categories differentially expressed in both S. oryzae and S. pierantonius during host metamorphosis
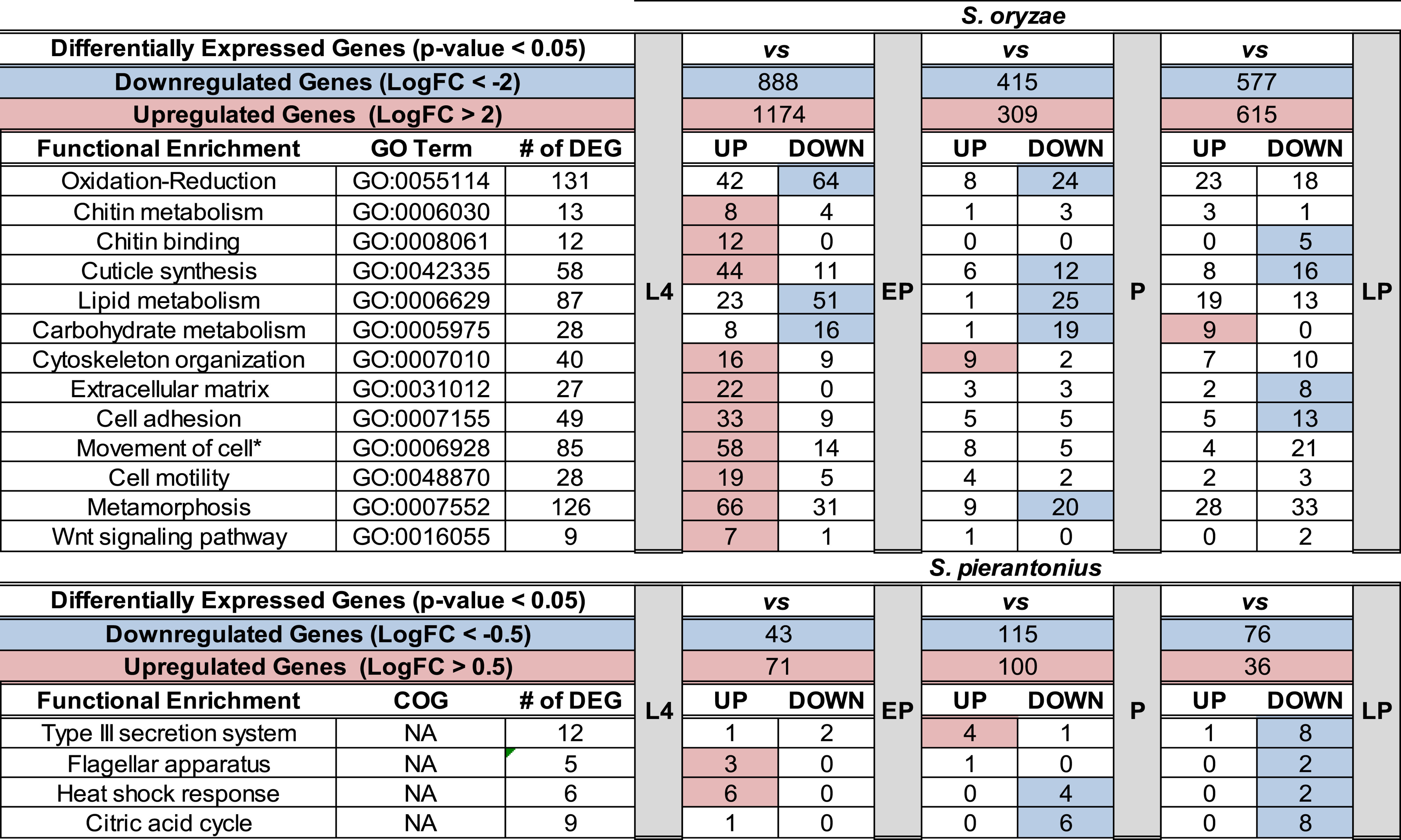 |
For each organism independently are represented: The total number of DEGs between two successive time points, either down- or up-regulated (first two lines); the main affected processes based on GO enrichment and manual analysis, and the number of genes either down- or up-regulated in each process between two successive time points. EP, early pupa; L4, fourth-instar larva; LP, late pupa; P, pupa.
GO 0006928. Movement of cell or subcellular component.
Symbiotic State Modulates the Expression of Host Genes Involved in Cell Adhesion and Migration during Metamorphosis.
In addition to the aforementioned transcriptomic changes, strong variations were observed in genes whose products are involved in cell adhesion, cell motility, movement of cell and organelles, and cytoskeleton organization (Table 1, SI Appendix, Figs. S3 and S5, and Datasets S1 and S3). For example, many genes associated with epithelial–mesenchymal transition (EMT) in other systems were found to be up-regulated at metamorphosis. EMT, well known to promote individual or collective cell migrations during embryogenesis or diseases (e.g., cancer metastasis) in many organisms, requires modulation of cell junctions, cell interactions with the extracellular matrix, cytoskeleton reorganization, and cellular shape modification (50, 51). Remarkably, around 70% of up-regulated genes homologous to markers of EMT cell adhesion and migration (52) were within the DEG we detected in our analysis (Dataset S5).
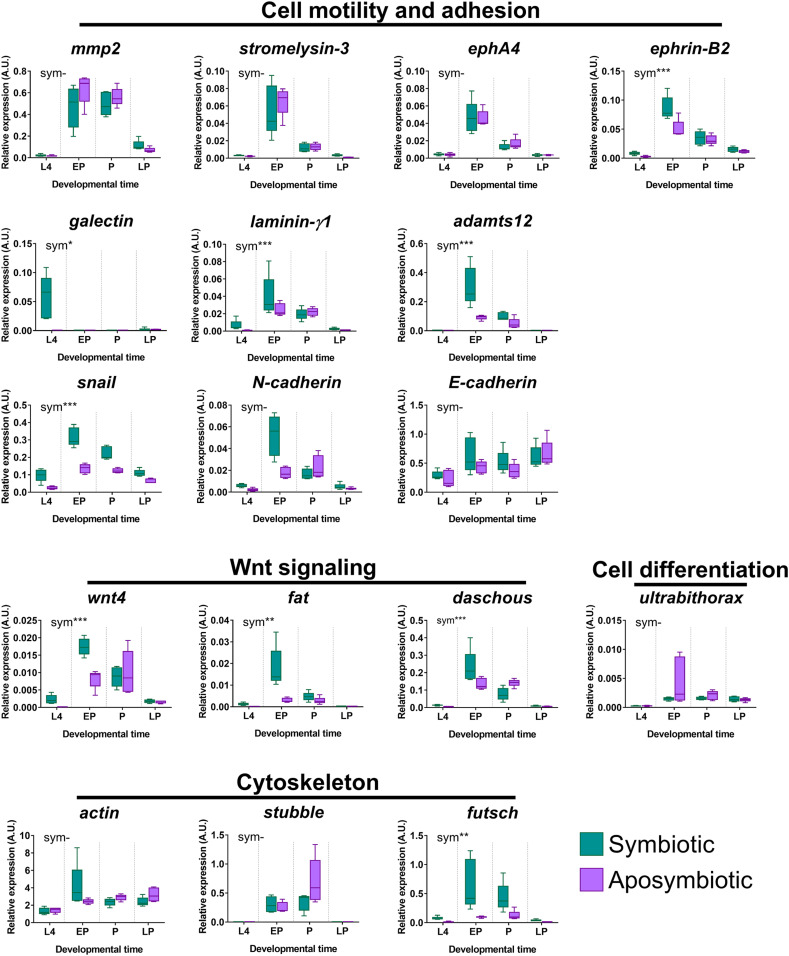
The transcriptomic changes observed could be directly linked either to metamorphosis processes of the gut per se or to changes in the bacteriocyte transcriptome underlying their change in behavior. To discriminate between these two possibilities, we selected 17 host DEG based on their documented functions in cell adhesion, cytoskeleton organization, or cell polarity in related organisms, and compared by qRT-PCR their expression in symbiotic and aposymbiotic metamorphosing weevils. Aposymbiotic individuals do not bear bacteriocytes but go through metamorphosis and gut remodeling (SI Appendix, Fig. S2) and possess the same genetic background as symbiotic individuals.
Eight of the tested genes (mmp2, stromelysin-3, ephA4, ultrabithorax, actin, stubble, E-cadherin, and N-cadherin) did not show significant differences between symbiotic and aposymbiotic individuals (Fig. 2). These genes are therefore likely involved in nonsymbiosis-related developmental processes entailed by metamorphosis. In contrast, expression profiles of the nine remaining genes were significantly up-regulated in symbiotic individuals as compared to their aposymbiotic counterparts (Fig. 2), indicating a bacteriocyte-specific modulation that could be involved in modifications of bacteriocyte cell behavior during metamorphosis. Among these genes, some have been described to be involved in cytoskeleton organization, cell polarity, cell adhesion, or cell motility, including EMT. The futsch gene, which encodes a microtubule-associated protein, promotes cytoskeleton rearrangement during Drosophila’s neuronal development (53, 54). Its up-regulation in symbiotic early pupae and pupae (Fig. 2) suggests it could participate in microtubule reorganization in the weevil’s migrating bacteriocytes. Fat and Daschous have been reported to play a role in the establishment of planar cell polarity in several Drosophila tissues, including the eyes and wings (55–58), by generating subcellular asymmetry. Modifications in cell polarity could participate in the bacteriocyte ability to migrate properly in weevil early pupae. Laminin-γ1 mediates cell adhesion (59, 60), Ephrin-B2 and Adamts12 promote cell invasion in mammals (61–63), and Wnt4 is required for proper cell migration during ovary and salivary-gland morphogenesis in Drosophila (64, 65). The expression of the genes encoding these proteins was increased between the larval and early pupal stages in symbiotic individuals (Fig. 2), suggesting they could participate in the bacteriocyte-invasive behavior and migration along the gut at this step of metamorphosis (Fig. 1 F and I). Nonetheless, we note that the expression levels of some of these genes—including wnt4, fat, and laminin-γ1—are relatively low, and differences between symbiotic and aposymbiotic individuals are subtle, albeit statistically significant. Determining whether these differences are biologically significant will require further functional analyses targeting those specific genes.
Fig. 2.
Specific host transcriptomic changes in genes affecting cell adhesion, motility, differentiation, polarity, and cytoskeleton organization underlie bacteriome remodeling. Expression kinetics of selected host genes were analyzed by qRT-PCR on dissected bacteriomes/guts during metamorphosis in symbiotic (green) and aposymbiotic (purple) weevils. A.U., arbitrary units. Boxes represent first quartile to third quartile for at least four independent replicates, the middle lines represent the medians, and the whiskers represent the minimum and maximum values. Asterisks indicate a significant impact of the symbiotic status (sym) based on the analysis of a generalized linear model (-P > 0.05; *P ≤ 0.05; **P < 0.01; ***P < 0.001). EP, early pupa; L4, fourth-instar larva; LP, late pupa; P, pupa. Gene IDs are available in SI Appendix, Table S3.
Expression of the EMT regulator snail was also significantly up-regulated in symbiotic relative to aposymbiotic individuals (Fig. 2). During metamorphosis, bacteriocytes change shape when initiating migration (Fig. 1 C, F, and I), adopting a spindle-like morphology typical of migrating cells. However, the expression of three other EMT markers [mmp2, involved in the composition of extracellular matrix (66), and E-cadherin and N-cadherin, which mediate cell–cell and cell–matrix junctions (67, 68)], did not exhibit symbiosis-specific expression profiles (Fig. 2). The expression profile of E-cadherin was even more puzzling, as it appeared to increase during metamorphosis, despite being known to be transcriptionally repressed by Snail in many models (69, 70). Taken together, these results indicate that, while Snail is probably involved in a symbiosis-related process during metamorphosis, its function might differ from the canonical Snail-mediated EMT mechanisms known in other models. Moreover, other non-EMT related, E-cadherin-independent, functions of Snail could be at work during symbiosis remodeling, such as roles in cell survival, stem cell biology, and regulation of immunity (71). Thus, a subtype-specific approach will be required to elucidate the exact functions of Snail in symbiosis remodeling during metamorphosis.
A galectin Gene Participates in Proper Bacteriocyte Migration toward Their Target Sites in Pupa.
One of the highest fold-changes from fourth-instar larvae to early pupa was observed for a galectin gene (down-regulated 4,000-fold in the dual RNA-seq data) (Dataset S1). We confirmed by qRT-PCR that galectin is specifically expressed in the bacteriome of fourth-instar larvae and displays very low transcript levels at all other developmental times (Fig. 2). galectin transcript levels were also very low at all time points in aposymbiotic individuals, strongly suggesting a function in bacteriocyte dissociation and migration. Galectins are lectins that bind surface β-galactosides through carbohydrate-recognition domains, through which they can mediate cell–cell and cell–matrix adhesion. They were shown to either increase or decrease cell motility in cases of tumor progression, depending on the context and tumor type (72, 73), and to promote EMT in several studies (74, 75). They are also involved in various ways in host-cell–bacteria interactions (76), and a Galectin-encoding gene was recently found to be significantly more expressed in the symbiotic organ of the tortoise leaf beetle Cassida rubiginosa when compared to the rest of the body (77). To further address galectin function in bacteriocyte migration, we used an RNA interference approach (SI Appendix, Fig. S6). Strikingly, galectin knockdown through double-stranded RNA (dsRNA)-injection at the third-larval instar resulted in isolated, circular-shaped bacteriocytes “left behind” in the fat body and the gut lumen in pupae (SI Appendix, Fig. S7 and Table S2). These data suggest that Galectin is essential for the proper migration of bacteriocytes.
Taken together, the above results reveal an endosymbiont-driven cell reprogramming at the onset of metamorphosis that parallels bacteriocyte behavioral changes. This symbiosis-specific program includes the transcriptomic modification of genes involved in cell adhesion, cell motility, cell polarity, and cytoskeleton organization, and it allows bacteriocytes to migrate and transport endosymbionts along the gut. Because of the complex, pleiotropic, and model-dependent functions controlled by many of the genes of interest, fine functional analyses will be required to address their specific roles in bacteriome remodeling during metamorphosis.
Endosymbiont Up-Regulates Type III Secretion System- and Flagellum-Encoding Genes during Metamorphosis.
Although changes in host gene expression during morphological rebuilding at metamorphosis were expected, our dual RNA-seq experiments showed that S. pierantonius gene expression is also impacted by metamorphosis (Table 1, SI Appendix, Figs. S4 and S5, and Datasets S2 and S4). This behavior contrasts markedly with that of Buchnera, the endosymbiont of the pea aphid A. pisum, whose genome lacks most gene regulators and shows limited transcriptional regulation (78–80). We observed a transient up-regulation (in early pupae) of genes coding for chaperones and heat-shock proteins, which could be a stress response to the wide cellular changes the bacteriocytes undergo. It is noteworthy that in Buchnera, heat-shock proteins have been found to be constitutively expressed (81), as opposed to their inducibility observed here in S. pierantonius. The citric-acid cycle was down-regulated (in pupae and late pupae) in S. pierantonius, suggesting a decrease in bacterial oxidative metabolism and a switch to fermentative metabolism. It was shown in Aedes aegypti that metamorphosis initiation was dependent on gut hypoxia (82), and a similar hypoxia during S. oryzae metamorphosis could explain this switch in bacterial metabolism.
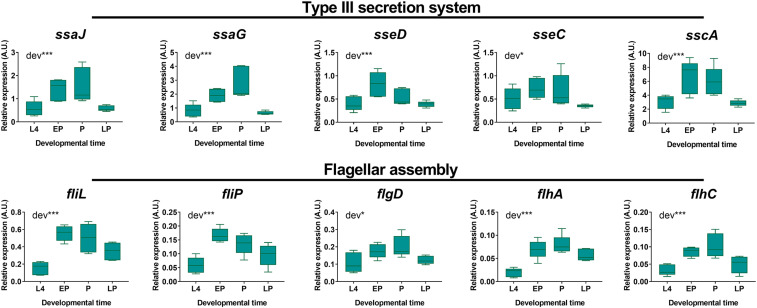
Importantly, genes encoding flagellum and type III secretion system (T3SS) components were up-regulated in early pupae and pupae and down-regulated in late pupae (Dataset S2), as further confirmed by qRT-PCR (Fig. 3). For many bacterial species, the flagellum is crucial during infections due to its motility and secretion functions (83). The T3SS forms a needle-like structure that enables bacteria to inject into the host cell cytosol effector molecules that can manipulate the cell and favor a subsequent infection (84). We detected both the structural genes of the T3SS apparatus, as well as effector genes, as differentially expressed. Hence, the up-regulation of flagellum- and T3SS-encoding genes might underlie a transient “infectious state” of the endosymbiont.
Fig. 3.
Genes encoding virulence factors are up-regulated in S. pierantonius during host metamorphosis. Expression kinetics of selected bacterial genes involved in synthesis of the T3SS or in flagellar assembly were analyzed by qRT-PCR on dissected bacteriomes/guts during metamorphosis in symbiotic (green) weevils. A.U., arbitrary units. Boxes represent the first quartile to third quartile for five independent replicates, the middle lines represent the medians, and the whiskers represent minimum and maximum values. Asterisks indicate a significant impact of the developmental time point (dev) based on a one-way ANOVA (*P < 0.05; ***P < 0.001). EP, early pupa; L4, fourth-instar larva; LP, late pupa; P, pupa.
This transcriptomic signature was previously reported in the sibling-species Sitophilus zeamais (85). The conservation of such virulence factors is usually limited to recently acquired symbionts, such as S. pierantonius and the secondary tsetse-fly symbiont Sodalis glossinidius (25), both of which have not suffered a drastic genome reduction (25, 86, 87) as observed in long-established endosymbionts (88). In the tsetse fly Glossina morsitans, S. glossinidius mutants lacking a functional T3SS are unable to infect host cells (89). T3SS-encoding genes were also shown to be essential in the formation of symbiotic nodules in the association between the legume Aeschynomene indica and the bacterium Bradyrhizobium (90). Since the T3SS from S. pierantonius is complete and predicted to be functional (25), it might play a similar proinfectious role here. Contrary to the T3SS, several genes involved in flagellum synthesis are lost or pseudogenized in S. pierantonius (25), including fliC, which encodes the flagellum filament. The flagellum of S. pierantonius would therefore presumably not support motility functions. Instead, we hypothesize it might serve as an additional secretion apparatus, as reported in Yersinia enterocolitica, Campylobacter jejuni, and Xenorhabdus nematophila, species in which the flagellum-export apparatus was shown to export nonflagellum-related proteins, including virulence factors (91–93). Interestingly, the genome of Buchnera also does not contain a fliC gene (94). However, other flagellum genes are among the most highly expressed genes in endosymbionts isolated from adult aphids (79, 95), and it was thus hypothesized that the flagellum could act as a secretion apparatus in this species (79, 95, 96). Hence, both the up-regulation of T3SS and flagellum-export apparatus genes in early pupae and pupae could allow S. pierantonius to switch to an infectious behavior during host metamorphosis.
S. pierantonius Infects Putative Stem Cells and Initiates the Formation of New Bacteriocytes.
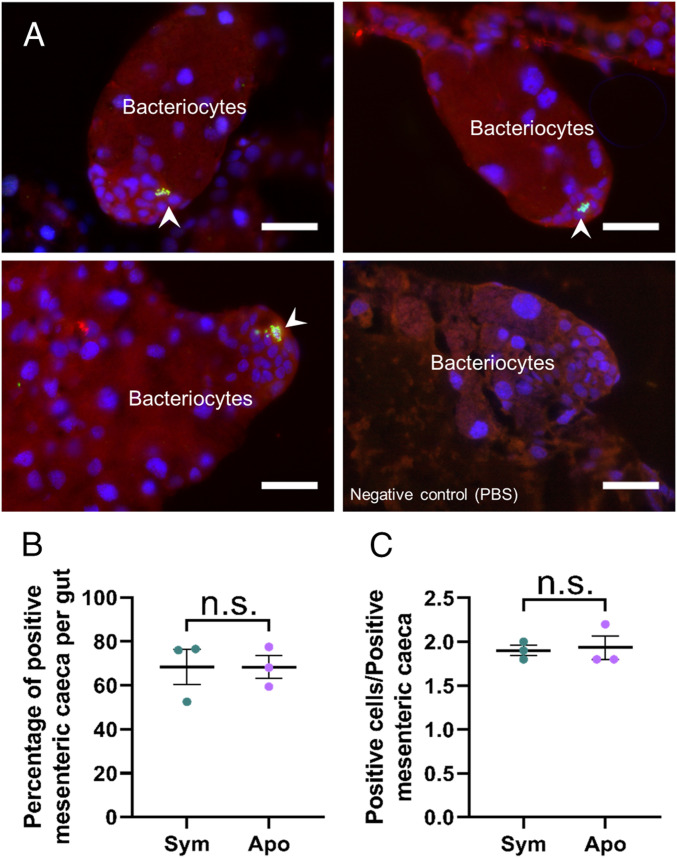
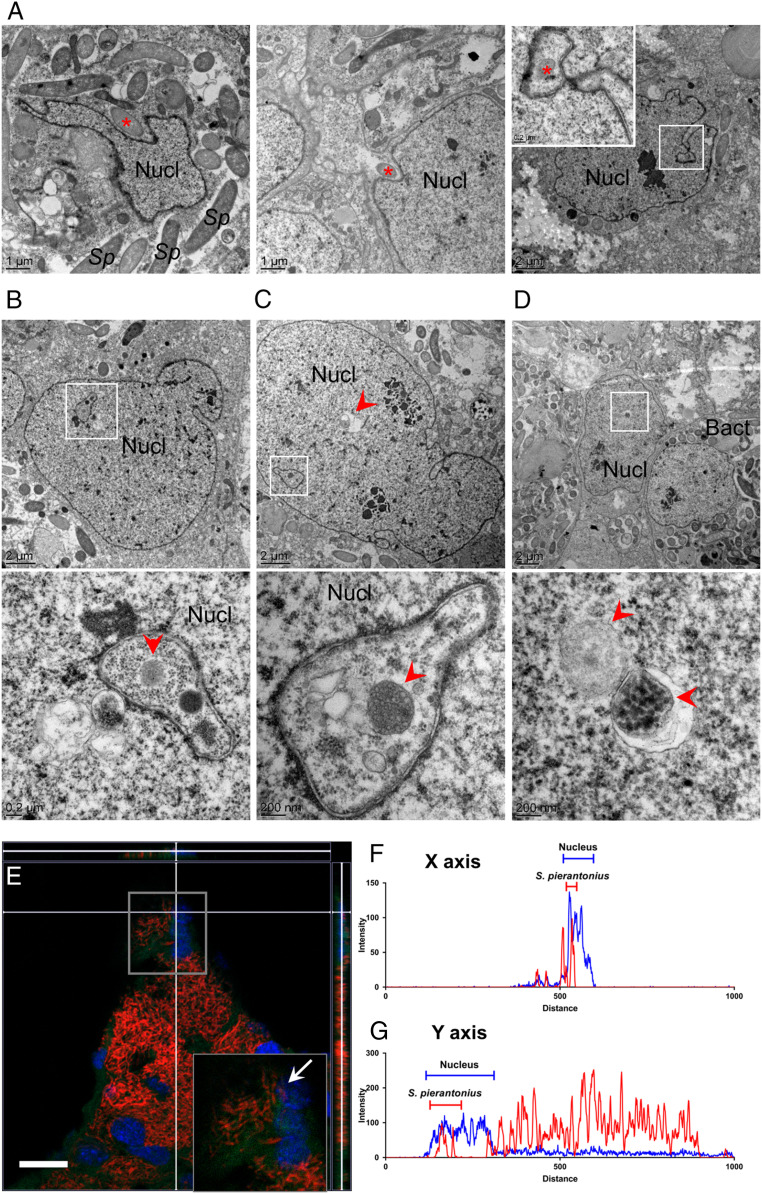
Histological analyses showed that migrating bacteriocytes end their migration next to clusters of small cells that are present at repetitive sites along the pupal midgut. During metamorphosis, these groups of cells continued to be clearly visible at the apex of forming bacteriomes (Fig. 1M). These cells showed a high nucleus:cytoplasm ratio and were often strongly stained after antiphosphorylated-histone-3 (PH3) immunostaining, indicating that they were actively dividing (Fig. 4A). Because these cells were also present at the extremity of each mesenteric cecum in aposymbiotic insects and showed a similar division activity (based on PH3-immunostaining) as in symbiotic insects (Fig. 4 B and C), we hypothesized that they are epithelial stem cells.
Fig. 4.
Putative epithelial stem cells are present at the apices of forming bacteriomes. (A) PH3 localization by immunostaining in forming bacteriomes in pupae. A negative control was included (Right, PBS). PH3 is specifically localized in clusters of small cells at the apices of forming bacteriomes (arrowheads), indicating that they might be stem cells. Red: autofluorescence; green: PH3; blue: DAPI. (Scale bars, 25 µm.) (B) Percentages of bacteriome containing PH3+ cells in whole guts from symbiotic and aposymbiotic pupae. The means and SEs for three independent replicates are represented. No significant difference was found between symbiotic and aposymbiotic individuals, based on a Welch t test. (C) Numbers of PH3+ cells per positive bacteriome in symbiotic and aposymbiotic pupae. The means and SEs for three independent replicates are represented. No significant difference was found between symbiotic and symbiotic individuals, based on a Welch t test; n.s., not significant.
FISH and transmission electron microscopy (TEM) experiments revealed the presence of isolated endosymbionts inside these putative epithelial stem cells (Fig. 5). Along with the endosymbiont’s transcriptomic up-regulation of its T3SS and flagellar export apparatus, this indicates that at this developmental stage, some S. pierantonius present in neighboring bacteriocytes following their migration manage to escape bacteriocytes of larval origin, activate an infectious state, and invade putative epithelial stem cells. Because no PH3-staining was observed in bacteriocytes from the larval bacteriome, and because the newly infected cells contained low numbers of bacteria, we hypothesize that the newly infected cells differentiate into bacteriocytes and contribute to the nascent adult bacteriomes. The presence of similar highly proliferative putative stem cells in aposymbiotic insects, in which mesenteric caeca form without any bacteriocyte/bacteriome formation (Fig. 4 B and C), suggests that in the absence of endosymbionts, these cells differentiate instead into intestinal cells, and that specific signals linked to endosymbiont infection trigger their differentiation into bacteriocytes. Similarly, it was shown in the flatworm Paracatenula galateia that bacteriocytes originate from noninfected pluripotent stem cells that also differentiate into other cell types, although the cell differentiation and endosymbiont infection mechanisms remain elusive (97). In endosymbiotic insects, the developmental origin of bacteriocytes is largely unknown and may vary among species. Intriguingly, genes that were previously shown to be involved in embryonic bacteriocyte formation in hemimetabolous insects [i.e., distal-less, engrailed, abdominal-A, and ultrabithorax (30, 98)] were not differentially expressed during S. oryzae’s metamorphosis, suggesting that other genes might be involved in this holometabolous insect.
Fig. 5.
Endosymbionts enter putative stem cells during bacteriome formation. (A and B) Localization of S. pierantonius cells by FISH in stem cells (arrowheads) at the apices of forming bacteriomes in pupae. Red: S. pierantonius; green: autofluorescence; blue: DAPI. These enlargements are cropped portions of the initial, full images that were acquired with a 60× objective. Enlargements were cropped and enlarged 1.7× (A) and 2.1× (B) compared to the original images. (Scale bars, 25 μm.) (C–E) TEM images showing stem cells (cell borders are highlighted by white dashed lines) infected with few endosymbionts (red arrows), contrasting with the high bacterial density in neighboring bacteriocytes. Bact: bacteriocytes; Nucl: nuclei.
Endosymbionts Ultimately Enter the Nucleus of Neoinfected Cells.
Unexpectedly, TEM revealed the presence of endosymbionts not only in the cytosol but also within the nuclei of putative stem cells (Fig. 6). In some cells, endosymbionts were observed surrounded by nuclear-membrane invaginations (Fig. 6A), suggesting that they were being internalized by the nucleus. In line with this, endosymbionts were also observed within vesicles inside nuclei (Fig. 6 B–D). These vesicles presented strong similarities with the nuclear membrane, supporting a nuclear origin. FISH experiments with a S. pierantonius probe observed by confocal microscopy confirmed the occurrence of S. pierantonius within stem-cell nuclei (Fig. 6 E–G). We also noticed a reduced size of endosymbionts that were present inside the nuclei, being on average half the size of cytoplasmic endosymbionts (SI Appendix, Fig. S8). We did not observe any nuclear endosymbionts past the pupal stage. These observations suggest that nuclear infection of stem cells by endosymbionts is a transient phenomenon that correlates with the transient up-regulation of T3SS- and flagellum-encoding genes and might be terminated by nuclear degradation of the endosymbionts.
Fig. 6.
Intranuclear occurrence of S. pierantonius in bacteriome stem cells. Dissected midguts from pupae were observed by TEM. (A) Nuclear membrane invaginations containing S. pierantonius (red asterisks). (B–D) S. pierantonius are observed inside vesicles (red arrowheads) within nuclei of cells that present low bacterial density. Bact, Bacteriocyte; Nucl, nuclei; Sp, S. pierantonius. (E) Z-stack representation of a pupa forming bacteriome as observed by confocal microscopy following FISH against S. pierantonius. Red: S. pierantonius; green: autofluorescence; blue: DAPI. This enlargement is a cropped portion of the initial, full image that was acquired with a 60× objective. The enlargement was cropped and enlarged 1.6× compared to the original image. (Scale bar, 10 μm.) (F and G) Superposition of blue (DAPI) and red (TAMRA-labeled S. pierantonius) signal intensities along the x (F) and y (G) axes of E. Capped lines on graphs show zones of blue and red signal colocalization, suggesting the nuclear presence of S. pierantonius. The gray square in E is an enlargement of the area where blue and red signals colocalize. The white arrow points at an apparent nuclear endosymbiont.
Although additional experiments are needed to ensure that the nuclear localization of S. pierantonius is biologically relevant and not a by-product of the putative stem-cell infection, the observation of intranuclear signal using an S. pierantonius-specific probe, the restriction of intranuclear localization to the putative stem cells, and the recent report of intranuclear Sodalis-related endosymbionts in the psyllid Bactericera trigonica (99) all support the hypothesis that S. pierantonius is transiently intranuclear during S. oryzae’s metamorphosis. Endonuclear bacteria have been described previously, in both pathogenic and mutualistic associations in protists, bivalves, and arthropods (100). Infection mechanisms remain elusive in most cases. In Paramecium, nuclear infection by Holospora is assumed to involve the so-called “infection tip,” a protein that would allow movement within the cell and nuclear infection (101, 102). In Euglena hemichromata, it was hypothesized that bacteria transit through the cytoplasm within phagocytotic vacuoles, which in turn fuse with the nuclear membrane, allowing for bacterial uptake by the nucleus (103). However, none of what has been characterized so far involves nuclear vesicles as seen in S. oryzae. A potential involvement of the T3SS and flagellar-export apparatus from S. pierantonius cannot be excluded. One striking difference from what we have observed in S. oryzae is that endonuclear bacteria in other systems usually persist within the nucleus. For example, in the leafhopper Nephotettix cincticeps, Rickettsia endosymbionts are able to permanently infect sperm nuclei, from which they are paternally transmitted to the offspring (104). In contrast, intranuclear occurrence of S. pierantonius is rare and transient, and the intranuclear bacteria rapidly decrease in size, suggesting bacterial degradation.
Nucleus-surface measurements showed that nuclei were significantly smaller in noninfected putative stem cells than in bacteriocytes (SI Appendix, Fig. S9), suggesting that bacteriocyte differentiation may entail a step of genome polyploidization, as has been proposed for several other insect bacteriocytes (29, 105–107). Intranuclear endosymbionts could be involved in triggering stem-cell differentiation into bacteriocytes, rather than into intestinal cells, as well as influencing genome polyploidization. The endosymbiont intranuclear localization could favor its interference with host gene expression. Several pathogenic, cytoplasmic bacteria were shown to interfere with host gene expression through the manipulation of epigenetic markers, such as histone modifications or chromatin rearrangements (108–111). The proximity of intranuclear S. pierantonius to its host genetic machinery at this stage makes epigenetic manipulation a strong possibility for bacteria-induced cell reprogramming, and especially genome polyploidization.
Conclusion
In summary, we show in this study how the host developmental program is intertwined with endosymbiosis, and how metamorphosis enables symbiont maintenance to adulthood and provides an opportunity to adapt a symbiotic organ to evolving metabolic needs. We provide a histological, molecular, and functional characterization of a remarkably regulated and timely reorganization of the bacteriome during weevil metamorphosis. The larval bacteriome dissociates, and bacteriocytes undergo migration along the gut epithelium, where they form de novo bacteriomes. We show that this process involves coordinated responses from both the host and the endosymbiont, including the up-regulation of T3SS- and flagellum-encoding genes. This activation of an infectious behavior allows the endosymbiont to invade the cytosol and, transiently, the nuclei of presumed epithelial stem cells, where endosymbionts might deliver a yet unknown signal that triggers stem-cell polyploidization and differentiation into bacteriocytes. We hypothesize that this bacteriome remodeling improves the trophic-interaction efficiency between S. oryzae and S. pierantonius at the onset of a short yet crucial period for the association, early adulthood, when symbiont-assisted cuticle synthesis takes place (22). We speculate that the increase in bacteriome number enhances productivity and that morphological rearrangements in close association with mesenteric caeca increase the exchange surface between bacteriocytes and the gut/fat body, likely optimizing metabolic flows. Hence, metamorphosis would allow a switch from a small bacteriome in larvae, when metabolic dependency on endosymbionts is lower, to multiple bacteriomes in adults, enabling massive symbiont growth in young adults and facilitating host–symbiont metabolic exchanges necessary for a rapid cuticle synthesis. Thus, this work shows how metamorphosis not only maintains endosymbionts, despite massive morphological rearrangements, but also enables adaptive decoupling with regards to endosymbiosis by restructuring the bacteriome and modulating host investment into endosymbiont housing according to metabolic needs.
Materials and Methods
Insect Rearing and Sampling.
S. oryzae weevils were reared on wheat grains at 27.5 °C and at 70% relative humidity. The Bouriz strain was chosen in this work because it is free of any facultative symbionts, including Wolbachia, and harbors only S. pierantonius. The full procedure for the generation of aposymbiotic insects through heat-treatment is described in the SI Appendix.
Sitophilus’ metamorphosis was divided as follows: Early pupae are more elongated than fourth-instar larvae, their color evolves from yellow to white, and they still possess a cephalic capsule. Pupae, on the other hand, lose the cephalic capsule, while late pupae have started cuticle sclerotization (SI Appendix, Fig. S1A).
Insect organs (bacteriomes, guts, ovaries) were dissected in diethylpyrocarbonate-treated Buffer A (25 mM KCl, 10 mM MgCl2, 250 mM Sucrose, 35 mM Tris/HCl, pH 7.5). For RNA extractions, at least 15 organs per condition were pooled and stored at −80 °C and each sampling was independently repeated five times.
Sample Preparation for Histology.
Whole individuals or dissected organs were prepared as described previously (112, 113).
Endosymbiont Localization by FISH.
FISH was performed as previously described (114). Probe sequence for S. pierantonius was: TAMRA-ACCCCCCTCTACGAGAC (10 μg/mL). Observations, image acquisition and treatment were performed as previously described (112, 113). Nuclei-surface measurements on acquired pictures were done using ImageJ.
Confocal microscopy and image acquisition were performed on a LSM800 confocal microscope (Zeiss, lasers 405, 488, 561 nm). Analyses, orthogonal projections and fluorescence-profile plots on acquired pictures were done using the Zen software (Zeiss), ImageJ, and GraphPad.
Endosymbiont Quantification by Flow Cytometry.
Bacteriomes/guts of fourth-instar larvae, early pupae, pupae, late pupae, and young adults (1-d-old) were dissected in PBS. For each developmental time point, five organs were pooled and each sampling was repeated three times. Organs were grinded in 300 μL PBS and centrifuged 2 min at 400 × g. The supernatant was recovered and centrifuged for 5 min at 13,800 × g. The pellet was resuspended in 1 mL PBS, filtered on a 50-μm nylon membrane, and centrifuged for 5 min at 13,800 × g. The pellet was resuspended in 300 μL PFA 4% and left overnight at 4 °C. Samples were then centrifuged for 5 min at 13,800 × g, washed with 1 mL PBS, and centrifuged for 5 min at 13,800 × g. Finally, samples were resuspended in PBS: 200 μL for larvae and pupae, 800 μL for adults. Samples were then incubated during 15 min at room temperature with Syto9 (0.5 μL/200 μL) and iodide propidium (0.5 μL/200 μL) from the Live/Dead BacLight Bacterial Viability Kit (ThermoFisher). Bacterial enumeration was performed with a C6 flow cytometer (BD Accuri), using a fluorescence gating for either Syto9 or iodide propidium.
RNA Isolation and Library Construction for Dual RNA-Seq.
Total RNA was extracted with TRIzol reagent (ThermoFisher Scientific) following the manufacturer’s instructions. Nucleic acids were then purified using the NucleoSpin RNA Clean up kit (Macherey Nagel). Genomic DNA was then removed from the samples with the DNA-free DNA removal kit (ThermoFisher Scientific). Total RNA concentration and quality were checked using the Qubit Fluorometer (ThermoFisher Scientific) and the Agilent 2200 TapeStation (Agilent Technologies). Ribo-depletion and dual RNA-seq libraries were then performed with 100 ng of total RNA using the Ovation Universal RNA-seq System (NuGEN) following the manufacturer’s instructions and sequenced on an Ion Torrent Proton platform (Life Technologies) at the sequencing platform of the Institut de Génomique Fonctionnelle de Lyon, Ecole Normale Supérieure de Lyon, France. Each replicate was sequenced on an independent run of ∼90 million reads for a total of more than one billion sequenced reads. Raw sequencing data from this study have been deposited at the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under PRJNA484327.
Differential Gene-Expression Analysis.
The raw reads were processed using CUTADAPT (115) to remove adapters and filter out reads shorter than 35 bp. The clean reads were mapped against the S. oryzae draft genome (GenBank: PRJNA431034) (40) with STAR (116) and S. pierantonius (GenBank: CP006568.1) (41) with Bowtie2 (117). Shared reads between the two genomes were filtered out with the aid of SAMTOOLS (118) and PICARD (available from https://broadinstitute.github.io/picard/). Gene counts were obtained with featureCounts method from the SUBREAD package (119) and tested for differential expression with DESEq2 package from R (120). Gene counts are available in Datasets S6 and S7. Genes were considered differentially expressed when the adjusted P value (P-adj) was lower than 0.05 and were selected for further analysis when log2 fold-change (LogFC) was greater than 2 (for up-regulated genes) or smaller than −2 (for down-regulated genes). Since bacterial RNA was present in smaller quantities than the insect’s RNA, P-adj, and LogFC were not as pronounced. Nevertheless, we were able to detect DEG when the LogFC threshold was set to 0.5 and −0.5. Genes were also clustered based on their expression profile using STEM (43) and these sets of genes were separately tested for functional analysis as well.
Functional Annotation.
The genomes of the insect and the endosymbiont were annotated for GO terms with the aid of EggNog (121). GO counts are available in Dataset S8. Enriched GO terms were detected using a custom R script with a Fischer test. After testing, the P values were adjusted with the Benjamini–Hochberg (122) correction for multiple testing. The GO terms were reduced to representative nonredundant terms with the use of the REVIGO tool (42).
qRT-PCR Transcript Quantification.
Total RNA was extracted and purified as described in the RNA isolation section described above. Reverse transcription and transcript quantification was performed as previously described (112, 113). The quantification was performed with a CFX Connect Real-Time PCR Detection System (Bio-Rad) using the LightCycler Fast Start DNA Master SYBR Green I kit (Roche Diagnostics). Host data were normalized using the ratio of the target cDNA concentration to that of three housekeeping genes: Ribosomal protein L29 (rpL29), glyceraldehyde 3-phosphate dehydrogenase (gapdh), and malate dehydrogenase (mdh). Symbiont data were normalized the same way, using two bacterial housekeeping genes: Ribosomal protein O (rplO) and ATP synthase-γ (atpG). Primers were designed to amplify fragments of ∼200 bp. Primers for rpL29, gapdh, and mdh were previously published (113). A complete list of other primers can be found in SI Appendix, Table S3. Based on the draft version of the genome, we have designed a specific qPCR primer for the galectin gene (Gene ID: LOC115886684). In the second assembly, the initial portion of the gene was not annotated as part of the transcript. We have manually verified that this portion was indeed transcribed, and we provide in SI Appendix, Sequence S1 the manually annotated sequence.
galectin Knockdown by RNAi.
dsRNA synthesis and injection were performed as described previously (112, 123). Primers used for dsRNA synthesis are listed in SI Appendix, Table S3. Injection of gfp dsRNA was used as a negative control. Six days following the injection of 50 ng of dsRNA in third-instar larvae, fourth-instar larvae were recovered and dissected to assess galectin knockdown. Ten days after dsRNA injection, early pupae and pupae were recovered for histological work.
Statistical Analyses.
Gene-expression kinetics for eukaryotic genes (Fig. 2) were analyzed by fitting a generalized linear model with a γ-transformation to the data. Developmental time points were considered as a four-level categorical variable. Symbiotic status was considered as a two-level categorical variable. The impact of symbiotic status was assessed by comparing with a t test the regression coefficients between symbiotic and aposymbiotic individuals.
Gene-expression kinetics for prokaryotic genes (Fig. 3) were analyzed by fitting a generalized linear model with a γ-transformation to the data. Developmental time points were considered as a four-level categorical variable. To confirm the dual RNA-seq data, the impact of developmental time was assessed with a one-way ANOVA with a χ2 test.
galectin knockdown (SI Appendix, Fig. S6) was analyzed doing pairwise comparisons using a Welch’s t test on the log-transformed gene-expression data.
PH3 Localization by Immunostaining.
Paraffin sections were dewaxed twice in methylcyclohexane for 10 min, rinsed in ethanol 100%, and rehydrated through an ethanol gradient to PBS. Afterward, slides were plunged during 20 min at 99 °C into an antigen retrieval buffer (anhydrous citric acid 1.8 mM; sodium citrate 8.2 mM), and rinsed with water for 10 min. The slides were incubated with 1% BSA in PBS for 30 min prior to primary antibody incubation, overnight at 4 °C. Rabbit anti-PH3 antibody (Abcam) was used as the primary antibody, diluted 1:200 in PBS containing 0.1% BSA. BSA 0.1% in PBS was used as a negative control. After primary antiserum incubation, sections were washed with PBS containing 0.2% Tween. Primary antibodies were detected with fluorescent donkey anti-rabbit IgG, labeled with Alexa Fluor 488 (ThermoFisher Scientific). This secondary antibody was applied for 1 h at room temperature, diluted at 1:600 in 0.1% BSA in PBS. They were washed with PBS-Tween, rinsed with PBS and washed several times with tap water. Sections were then dried and mounted using PermaFluor Aqueous Mounting Medium (ThermoFisher Scientific), together with 4DAPI (Sigma-Aldrich) for nuclear staining (3 μg/mL of medium). Images were acquired and treated as described above.
For whole-mount experiments, guts from symbiotic and aposymbiotic pupae were dissected in PBS and fixed in PFA 4% for 2 d. They were then washed twice in PBS for 10 min, and tissues were permeabilized for 10 min in Triton 0.1% in PBS. The same protocol used for the slides was then used for these organs.
Ultrastructural Observation of Midguts by TEM.
Sample preparation and embedding, sectioning, contrasting, and TEM were carried out as previously described (22). Observations and image acquisition were performed on a CM120 electron microscope (Philips) at the Centre Technologique des Microstructures (Université de Lyon). Bacterial diameter measurements on acquired pictures were done using ImageJ.
Supplementary Material
Acknowledgments
The authors thank Catherine Fiol, Sergio López-Madrigal, Rita Rebollo, and Carlos Vargas-Chávez for their valuable help on animal rearing and sampling; Fabien Filaire and Sandra Hamamjian for their help in microtomy; Ina Atrée and Eric Faudry for kindly reviewing our data on the type III secretion system and discussing with us the conservation of these bacterial genes in Sodalis; and the anonymous reviewers and editor for their constructive comments that helped improve the manuscript. Confocal and electron microscopy was performed at the Centre Technologique des Microstructures. Funding for this project was provided by Institut National de la Recherche Agronomique, Institut National des Sciences Appliquées-Lyon, the French ANR-13-BSV7-0016-01 (IMetSym), the French ANR-17_CE20_0031_01 (GREEN), the “Santé des Plantes et Environnement” Institut National de la Recherche Agronomique department, and a grant CMIRA from Région Rhône-Alpes.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Raw sequencing data from this study have been deposited at the National Center for Biotechnology Information Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra (accession no. PRJNA484327).
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2007151117/-/DCSupplemental.
Data Availability.
Raw sequencing data from this study have been deposited at the NCBI SRA under PRJNA484327. All other data are available in this paper.
References
- 1.Margulis L., Origins of species: Acquired genomes and individuality. Biosystems 31, 121–125 (1993). [DOI] [PubMed] [Google Scholar]
- 2.Moran N. A., Symbiosis. Curr. Biol. 16, R866–R871 (2006). [DOI] [PubMed] [Google Scholar]
- 3.McFall-Ngai M. et al., Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. U.S.A. 110, 3229–3236 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heddi A., Zaidman-Rémy A., Endosymbiosis as a source of immune innovation. C. R. Biol. 341, 290–296 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Buchner P., Endosymbiosis of Animals with Plant Microorganisms, (Interscience Publishers/John Wiley, New York, 1965). [Google Scholar]
- 6.Baumann P., Biology bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu. Rev. Microbiol. 59, 155–189 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Douglas A. E., Multiorganismal insects: Diversity and function of resident microorganisms. Annu. Rev. Entomol. 60, 17–34 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Truman J. W., Riddiford L. M., The origins of insect metamorphosis. Nature 401, 447–452 (1999). [DOI] [PubMed] [Google Scholar]
- 9.Rolff J., Johnston P. R., Reynolds S., Complete metamorphosis of insects. Philos. Trans. R. Soc. Lond. B Biol. Sci. 374, 20190063 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moran N. A., Adaptation and constraint in the complex cife cycles of animals. Annu. Rev. Ecol. Syst. 25, 573–600 (1994). [Google Scholar]
- 11.Hammer T. J., Moran N. A., Links between metamorphosis and symbiosis in holometabolous insects. Philos. Trans. R. Soc. Lond. B Biol. Sci. 374, 20190068 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parmentier A. et al., A different gut microbial community between larvae and adults of a wild bumblebee nest (Bombus pascuorum). Insect Sci. 25, 66–74 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Kim J. K. et al., Molting-associated suppression of symbiont population and up-regulation of antimicrobial activity in the midgut symbiotic organ of the Riptortus-Burkholderia symbiosis. Dev. Comp. Immunol. 43, 10–14 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Johnston P. R., Rolff J., Host and symbiont jointly control gut microbiota during complete metamorphosis. PLoS Pathog. 11, e1005246 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stoll S., Feldhaar H., Fraunholz M. J., Gross R., Bacteriocyte dynamics during development of a holometabolous insect, the carpenter ant Camponotus floridanus. BMC Microbiol. 10, 308 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oishi S., Moriyama M., Koga R., Fukatsu T., Morphogenesis and development of midgut symbiotic organ of the stinkbug Plautia stali (Hemiptera: Pentatomidae). Zoological Lett. 5, 16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Estes A. M., Hearn D. J., Bronstein J. L., Pierson E. A., The olive fly endosymbiont, “Candidatus Erwinia dacicola,” switches from an intracellular existence to an extracellular existence during host insect development. Appl. Environ. Microbiol. 75, 7097–7106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heddi A., Grenier A. M., Khatchadourian C., Charles H., Nardon P., Four intracellular genomes direct weevil biology: Nuclear, mitochondrial, principal endosymbiont, and Wolbachia. Proc. Natl. Acad. Sci. U.S.A. 96, 6814–6819 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heddi A., Charles H., Khatchadourian C., Bonnot G., Nardon P., Molecular characterization of the principal symbiotic bacteria of the weevil Sitophilus oryzae: A peculiar G + C content of an endocytobiotic DNA. J. Mol. Evol. 47, 52–61 (1998). [DOI] [PubMed] [Google Scholar]
- 20.Mansour K., Preliminary studies on the bacterial cell-mass (accessory cell-mass) of Calandra oryzae (Linn.): The rice weevil. Q. J. Microsc. Sci. 73, 421–435 (1930). [Google Scholar]
- 21.Heddi A., Charles H., Khatchadourian C., Intracellular bacterial symbiosis in the genus Sitophilus: The “biological individual” concept revisited. Res. Microbiol. 152, 431–437 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Vigneron A. et al., Insects recycle endosymbionts when the benefit is over. Curr. Biol. 24, 2267–2273 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Wicker C., Nardon P., Development responses of symbiotic and aposymbiotic weevils Sitophilus oryzae L. (Coleoptera, curculionidae) to a diet supplemented with aromatic amino acids. J. Insect Physiol. 28, 1021–1024 (1982). [Google Scholar]
- 24.Wicker C., Differential vitamin and choline requirements of symbiotic and aposymbiotic S. oryzae (Coleoptera: Curculionidae). Comp. Biochem. Physiol. Part A. Physiol. 76, 177–182 (1983). [Google Scholar]
- 25.Oakeson K. F. et al., Genome degeneration and adaptation in a nascent stage of symbiosis. Genome Biol. Evol. 6, 76–93 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masson F. et al., Weevil endosymbiont dynamics is associated with a clamping of immunity. BMC Genom. 16, 819 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murray F. V., Tiegs O. W., The metamorphosis of Calandra oryzae. J. Cell Sci. s2–s77, 405–495 (1935). [Google Scholar]
- 28.Mansour K., The development of the larval and adult mid-gut of Calandra oryzae (Linn.): The rice weevil. J. Cell Sci. s2–s71, 313–352 (1927). [Google Scholar]
- 29.Mansour K., On the so-called symbiotic relationship between coleopterous insects and intracellular micro-organisms. J. Cell Sci. s2–s77, 255–271 (1934). [Google Scholar]
- 30.Braendle C. et al., Developmental origin and evolution of bacteriocytes in the aphid-Buchnera symbiosis. PLoS Biol. 1, E21 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masson F., Zaidman-Rémy A., Heddi A., Antimicrobial peptides and cell processes tracking endosymbiont dynamics. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371, 403–411 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luan J.-B. et al., Cellular and molecular remodelling of a host cell for vertical transmission of bacterial symbionts. Proc. Biol. Sci. 283, 218–230 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nardon P., Grenier A. M., “Genetical and biochemical interactions between the host and its endocytobiotes in the weevils Sitophilus (Coleoptere, Curculionidae) and other related species” in Cell to Cell Signals in Plant, Animal and Microbial Symbiosis, Scannerini S., Smith D., Bonfanti-Fasolo P., Gianinazzi-Pearson V., Eds. (Springer Berlin Heidelberg, 1988), pp. 255–270. [Google Scholar]
- 34.Bing X. et al., Unravelling the relationship between the tsetse fly and its obligate symbiont Wigglesworthia: Transcriptomic and metabolomic landscapes reveal highly integrated physiological networks. Proc. Biol. Sci. 284, 20170360 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burns J. A., Zhang H., Hill E., Kim E., Kerney R., Transcriptome analysis illuminates the nature of the intracellular interaction in a vertebrate-algal symbiosis. eLife 6, e22054 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grote A. et al., Defining Brugia malayi and Wolbachia symbiosis by stage-specific dual RNA-seq. PLoS Negl. Trop. Dis. 11, e0005357 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Medina Munoz M., Pollio A. R., White H. L., Rio R. V. M., Into the wild: Parallel transcriptomics of the tsetse-Wigglesworthia tutualism within kenyan populations. Genome Biol. Evol. 9, 2276–2291 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mateus I. D. et al., Dual RNA-seq reveals large-scale non-conserved genotype × genotype-specific genetic reprograming and molecular crosstalk in the mycorrhizal symbiosis. ISME J. 13, 1226–1238 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maire J., et al. , Application of a dual RNA-seq strategy to unravel the symbiotic organ remodeling during metamorphosis in the cereal weevil Sitophilus oryzae. NCBI. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA484327/. Deposited 8 March 2018.
- 40.Parisot N., et al. , Data from “Sitophilus oryzae breed Bouriz, whole genome shotgun sequencing project.” GenBank. https://www.ncbi.nlm.nih.gov/nuccore/1735659643. Accessed 2 June 2020.
- 41.Oakeson K. F., et al. , Candidatus Sodalis pierantonius str. SOPE, complete genome. GenBank. https://www.ncbi.nlm.nih.gov/nuccore/CP006568.1/. Accessed 2 June 2020.
- 42.Supek F., Bošnjak M., Škunca N., Šmuc T., REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6, e21800 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ernst J., Bar-Joseph Z., STEM: A tool for the analysis of short time series gene expression data. BMC Bioinf. 7, 191 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ylla G., Piulachs M.-D., Belles X., Comparative transcriptomics in two extreme neopterans reveals general trends in the evolution of modern insects. iScience 4, 164–179 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Won H. I. et al., De novo assembly of the burying beetle Nicrophorus orbicollis (Coleoptera: Silphidae) transcriptome across developmental stages with identification of key immune transcripts. J Genomics 6, 41–52 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koutsos A. C. et al., Life cycle transcriptome of the malaria mosquito Anopheles gambiae and comparison with the fruitfly Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 104, 11304–11309 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kou F. et al., Temporal transcriptomic profiling of the ant-feeding assassin bug Acanthaspis cincticrus reveals a biased expression of genes associated with predation in nymphs. Sci. Rep. 7, 12691 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Si Q., Luo J.-Y., Hu Z., Zhang W., Zhou C.-F., De novo transcriptome of the mayfly Cloeon viridulum and transcriptional signatures of Prometabola. PLoS One 12, e0179083 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pesch Y. Y., Riedel D., Patil K. R., Loch G., Behr M., Chitinases and Imaginal disc growth factors organize the extracellular matrix formation at barrier tissues in insects. Sci. Rep. 6, 18340 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Revenu C., Gilmour D., EMT 2.0: Shaping epithelia through collective migration. Curr. Opin. Genet. Dev. 19, 338–342 (2009). [DOI] [PubMed] [Google Scholar]
- 51.Rørth P., Collective cell migration. Annu. Rev. Cell Dev. Biol. 25, 407–429 (2009). [DOI] [PubMed] [Google Scholar]
- 52.Gröger C. J., Grubinger M., Waldhör T., Vierlinger K., Mikulits W., Meta-analysis of gene expression signatures defining the epithelial to mesenchymal transition during cancer progression. PLoS One 7, e51136 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roos J., Hummel T., Ng N., Klämbt C., Davis G. W., Drosophila Futsch regulates synaptic microtubule organization and is necessary for synaptic growth. Neuron 26, 371–382 (2000). [DOI] [PubMed] [Google Scholar]
- 54.Hummel T., Krukkert K., Roos J., Davis G., Klämbt C., Drosophila Futsch/22C10 is a MAP1B-like protein required for dendritic and axonal development. Neuron 26, 357–370 (2000). [DOI] [PubMed] [Google Scholar]
- 55.Yang C. H., Axelrod J. D., Simon M. A., Regulation of Frizzled by fat-like cadherins during planar polarity signaling in the Drosophila compound eye. Cell 108, 675–688 (2002). [DOI] [PubMed] [Google Scholar]
- 56.Ma D., Yang C. H., McNeill H., Simon M. A., Axelrod J. D., Fidelity in planar cell polarity signalling. Nature 421, 543–547 (2003). [DOI] [PubMed] [Google Scholar]
- 57.Matakatsu H., Blair S. S., Interactions between Fat and Dachsous and the regulation of planar cell polarity in the Drosophila wing. Development 131, 3785–3794 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Simon M. A., Planar cell polarity in the Drosophila eye is directed by graded Four-jointed and Dachsous expression. Development 131, 6175–6184 (2004). [DOI] [PubMed] [Google Scholar]
- 59.Ishibashi M. et al., The cyclooxygenase inhibitor indomethacin modulates gene expression and represses the extracellular matrix protein laminin γ1 in human glioblastoma cells. Exp. Cell Res. 302, 244–252 (2005). [DOI] [PubMed] [Google Scholar]
- 60.Wolfstetter G., Holz A., The role of LamininB2 (LanB2) during mesoderm differentiation in Drosophila. Cell. Mol. Life Sci. 69, 267–282 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sentürk A., Pfennig S., Weiss A., Burk K., Acker-Palmer A., Ephrin Bs are essential components of the Reelin pathway to regulate neuronal migration. Nature 472, 356–360 (2011). [DOI] [PubMed] [Google Scholar]
- 62.Krusche B. et al., EphrinB2 drives perivascular invasion and proliferation of glioblastoma stem-like cells. eLife 5, e14845 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Beristain A. G., Zhu H., Leung P. C. K., Regulated expression of ADAMTS-12 in human trophoblastic cells: A role for ADAMTS-12 in epithelial cell invasion? PLoS One 6, e18473 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cohen E. D. et al., DWnt4 regulates cell movement and focal adhesion kinase during Drosophila ovarian morphogenesis. Dev. Cell 2, 437–448 (2002). [DOI] [PubMed] [Google Scholar]
- 65.Harris K. E., Beckendorf S. K., Different Wnt signals act through the Frizzled and RYK receptors during Drosophila salivary gland migration. Development 134, 2017–2025 (2007). [DOI] [PubMed] [Google Scholar]
- 66.Gialeli C., Theocharis A. D., Karamanos N. K., Roles of matrix metalloproteinases in cancer progression and their pharmacological targeting. FEBS J. 278, 16–27 (2011). [DOI] [PubMed] [Google Scholar]
- 67.Thiery J. P., Acloque H., Huang R. Y. J., Nieto M. A., Epithelial-mesenchymal transitions in development and disease. Cell 139, 871–890 (2009). [DOI] [PubMed] [Google Scholar]
- 68.Lamouille S., Xu J., Derynck R., Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15, 178–196 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cano A. et al., The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 2, 76–83 (2000). [DOI] [PubMed] [Google Scholar]
- 70.Peinado H., Ballestar E., Esteller M., Cano A., Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol. Cell. Biol. 24, 306–319 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu Y., Zhou B. P., Snail: More than EMT. Cell Adhes. Migr. 4, 199–203 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu F.-T., Rabinovich G. A., Galectins as modulators of tumour progression. Nat. Rev. Cancer 5, 29–41 (2005). [DOI] [PubMed] [Google Scholar]
- 73.Johannes L., Jacob R., Leffler H., Galectins at a glance. J. Cell Sci. 131, jcs208884 (2018). [DOI] [PubMed] [Google Scholar]
- 74.Bacigalupo M. L. et al., Galectin-1 triggers epithelial-mesenchymal transition in human hepatocellular carcinoma cells. J. Cell. Physiol. 230, 1298–1309 (2015). [DOI] [PubMed] [Google Scholar]
- 75.Zhu J. et al., Galectin-1 induces metastasis and epithelial-mesenchymal transition (EMT) in human ovarian cancer cells via activation of the MAPK JNK/p38 signalling pathway. Am. J. Transl. Res. 11, 3862–3878 (2019). [PMC free article] [PubMed] [Google Scholar]
- 76.Baum L. G., Garner O. B., Schaefer K., Lee B., Microbe-host interactions are positively and negatively regulated by galectin-glycan interactions. Front. Immunol. 5, 284 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bauer E., Kaltenpoth M., Salem H., Minimal fermentative metabolism fuels extracellular symbiont in a leaf beetle. ISME J. 14, 866–870 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Moran N. A., Dunbar H. E., Wilcox J. L., Regulation of transcription in a reduced bacterial genome: Nutrient-provisioning genes of the obligate symbiont Buchnera aphidicola. J. Bacteriol. 187, 4229–4237 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Reymond N. et al., Different levels of transcriptional regulation due to trophic constraints in the reduced genome of Buchnera aphidicola APS. Appl. Environ. Microbiol. 72, 7760–7766 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brinza L., Calevro F., Charles H., Genomic analysis of the regulatory elements and links with intrinsic DNA structural properties in the shrunken genome of Buchnera. BMC Genom. 14, 73 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wilcox J. L., Dunbar H. E., Wolfinger R. D., Moran N. A., Consequences of reductive evolution for gene expression in an obligate endosymbiont. Mol. Microbiol. 48, 1491–1500 (2003). [DOI] [PubMed] [Google Scholar]
- 82.Coon K. L. et al., Bacteria-mediated hypoxia functions as a signal for mosquito development. Proc. Natl. Acad. Sci. U.S.A. 114, E5362–E5369 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Khan S., Scholey J. M., Assembly, functions and evolution of archaella, flagella and cilia. Curr. Biol. 28, R278–R292 (2018). [DOI] [PubMed] [Google Scholar]
- 84.Galán J. E., Waksman G., Protein-injection machines in bacteria. Cell 172, 1306–1318 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dale C., Plague G. R., Wang B., Ochman H., Moran N. A., Type III secretion systems and the evolution of mutualistic endosymbiosis. Proc. Natl. Acad. Sci. U.S.A. 99, 12397–12402 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Clayton A. L. et al., A novel human-infection-derived bacterium provides insights into the evolutionary origins of mutualistic insect-bacterial symbioses. PLoS Genet. 8, e1002990 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Charles H., Condemine G., Nardon C., Nardon P., Genome size characterization of the principal endocellular symbiotic bacteria of the weevil Sitophilus oryzae, using pulsed field gel electrophoresis. Insect Biochem. Mol. Biol. 27, 345–350 (1997). [Google Scholar]
- 88.Moran N. A., Bennett G. M., The tiniest tiny genomes. Annu. Rev. Microbiol. 68, 195–215 (2014). [DOI] [PubMed] [Google Scholar]
- 89.Dale C., Young S. A., Haydon D. T., Welburn S. C., The insect endosymbiont Sodalis glossinidius utilizes a type III secretion system for cell invasion. Proc. Natl. Acad. Sci. U.S.A. 98, 1883–1888 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Teulet A. et al., The rhizobial type III effector ErnA confers the ability to form nodules in legumes. Proc. Natl. Acad. Sci. U.S.A. 116, 21758–21768 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Young G. M., Schmiel D. H., Miller V. L., A new pathway for the secretion of virulence factors by bacteria: The flagellar export apparatus functions as a protein-secretion system. Proc. Natl. Acad. Sci. U.S.A. 96, 6456–6461 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Konkel M. E. et al., Secretion of virulence proteins from Campylobacter jejuni is dependent on a functional flagellar export apparatus. J. Bacteriol. 186, 3296–3303 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Singh P., Park D., Forst S., Banerjee N., Xenocin export by the flagellar type III pathway in Xenorhabdus nematophila. J. Bacteriol. 195, 1400–1410 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shigenobu S., Watanabe H., Hattori M., Sakaki Y., Ishikawa H., Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407, 81–86 (2000). [DOI] [PubMed] [Google Scholar]
- 95.Viñuelas J. et al., Conservation of the links between gene transcription and chromosomal organization in the highly reduced genome of Buchnera aphidicola. BMC Genom. 8, 143 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Maezawa K. et al., Hundreds of flagellar basal bodies cover the cell surface of the endosymbiotic bacterium Buchnera aphidicola sp. strain APS. J. Bacteriol. 188, 6539–6543 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dirks U. et al., Bacterial symbiosis maintenance in the asexually reproducing and regenerating flatworm Paracatenula galateia. PLoS One 7, e34709 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Matsuura Y., Kikuchi Y., Miura T., Fukatsu T., Ultrabithorax is essential for bacteriocyte development. Proc. Natl. Acad. Sci. U.S.A. 112, 9376–9381 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ghosh S. et al., An intranuclear Sodalis-Like symbiont and Spiroplasma coinfect the carrot psyllid, Bactericera trigonica (Hemiptera, Psylloidea). Microorganisms 8, 692 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schulz F., Horn M., Intranuclear bacteria: Inside the cellular control center of eukaryotes. Trends Cell Biol. 25, 339–346 (2015). [DOI] [PubMed] [Google Scholar]
- 101.Iwatani K. et al., Translocation of an 89-kDa periplasmic protein is associated with Holospora infection. Biochem. Biophys. Res. Commun. 337, 1198–1205 (2005). [DOI] [PubMed] [Google Scholar]
- 102.Sabaneyeva E. V. et al., Actin-based mechanism of Holospora obtusa trafficking in Paramecium caudatum. Protist 160, 205–219 (2009). [DOI] [PubMed] [Google Scholar]
- 103.Shin W., Boo S. M., Fritz L., Endonuclear bacteria in Euglena hemichromata (Euglenophyceae): A proposed pathway to endonucleobiosis. Phycologia 42, 198–203 (2003). [Google Scholar]
- 104.Watanabe K., Yukuhiro F., Matsuura Y., Fukatsu T., Noda H., Intrasperm vertical symbiont transmission. Proc. Natl. Acad. Sci. U.S.A. 111, 7433–7437 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nardon P., Contribution a l’etude des symbiotes ovariens de Sitophilus sasakii : Localisation, histochimie et ultrastructure chez la femelle adulte. C. R. Acad. Sci. 272D, 2975–2978 (1971). [Google Scholar]
- 106.Nardon P., Louis C., Nicolas G., Kermarrec A., Mise en evidence et etude des bacteries symbiotiques chez deux charançons parasites du bananier : Cosmopolites sordidus (Germar) et Metamasius hemipterus (L.) (Col. Curculionidae). Ann. Soc. Entomol. Fr. 21, 245–258 (1985). [Google Scholar]
- 107.Luan J., Sun X., Fei Z., Douglas A. E., Maternal inheritance of a single somatic animal cell displayed by the bacteriocyte in the whitefly Bemisia tabaci. Curr. Biol. 28, 459–465.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pennini M. E., Perrinet S., Dautry-Varsat A., Subtil A., Histone methylation by NUE, a novel nuclear effector of the intracellular pathogen Chlamydia trachomatis. PLoS Pathog. 6, e1000995 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lebreton A. et al., A bacterial protein targets the BAHD1 chromatin complex to stimulate type III interferon response. Science 331, 1319–1321 (2011). [DOI] [PubMed] [Google Scholar]
- 110.Eskandarian H. A. et al., A role for SIRT2-dependent histone H3K18 deacetylation in bacterial infection. Science 341, 1238858 (2013). [DOI] [PubMed] [Google Scholar]
- 111.Rolando M. et al., Legionella pneumophila effector RomA uniquely modifies host chromatin to repress gene expression and promote intracellular bacterial replication. Cell Host Microbe 13, 395–405 (2013). [DOI] [PubMed] [Google Scholar]
- 112.Maire J. et al., Weevil pgrp-lb prevents endosymbiont TCT dissemination and chronic host systemic immune activation. Proc. Natl. Acad. Sci. U.S.A. 116, 5623–5632 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Maire J., Vincent-Monégat C., Masson F., Zaidman-Rémy A., Heddi A., An IMD-like pathway mediates both endosymbiont control and host immunity in the cereal weevil Sitophilus spp. Microbiome 6, 6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Balmand S., Lohs C., Aksoy S., Heddi A., Tissue distribution and transmission routes for the tsetse fly endosymbionts. J. Invertebr. Pathol. 112, S116–S122 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10 (2011). [Google Scholar]
- 116.Dobin A. et al., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li H. et al.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 120.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jensen L. J. et al., eggNOG: Automated construction and annotation of orthologous groups of genes. Nucleic Acids Res. 36, D250–D254 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Benjamini Y., Hochberg Y., Controlling the false discovery rate: A practical and powerful spproach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995). [Google Scholar]
- 123.Vallier A., Vincent-Monégat C., Laurençon A., Heddi A., RNAi in the cereal weevil Sitophilus spp: Systemic gene knockdown in the bacteriome tissue. BMC Biotechnol. 9, 44 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data from this study have been deposited at the NCBI SRA under PRJNA484327. All other data are available in this paper.