Significance
Transposable elements (TEs) are genetic units that can selfishly propagate in a host genome. Despite often being considered “junk,” TEs can occasionally acquire useful functions such as regulating expression of nearby host genes. Here, we study gibbons, small apes whose genome contains a unique TE called LAVA (LINE-AluSz-VNTR-AluLIKE). We present evidence indicating that several LAVA insertions function as gene regulatory elements in the gibbon genome. Two of these insertions were also favored/preserved by natural selection, further indicating their functional importance for gibbons. Both of these LAVA elements were found inside genes that facilitate correct repair of DNA breaks, suggesting that LAVA’s incorporation in these genes may have influenced the regulation of biological processes that are crucial for maintaining genome integrity.
Keywords: transposable element, co-option, cis-regulatory element, transcription factor binding, DNA repair
Abstract
Co-option of transposable elements (TEs) to become part of existing or new enhancers is an important mechanism for evolution of gene regulation. However, contributions of lineage-specific TE insertions to recent regulatory adaptations remain poorly understood. Gibbons present a suitable model to study these contributions as they have evolved a lineage-specific TE called LAVA (LINE-AluSz-VNTR-AluLIKE), which is still active in the gibbon genome. The LAVA retrotransposon is thought to have played a role in the emergence of the highly rearranged structure of the gibbon genome by disrupting transcription of cell cycle genes. In this study, we investigated whether LAVA may have also contributed to the evolution of gene regulation by adopting enhancer function. We characterized fixed and polymorphic LAVA insertions across multiple gibbons and found 96 LAVA elements overlapping enhancer chromatin states. Moreover, LAVA was enriched in multiple transcription factor binding motifs, was bound by an important transcription factor (PU.1), and was associated with higher levels of gene expression in cis. We found gibbon-specific signatures of purifying/positive selection at 27 LAVA insertions. Two of these insertions were fixed in the gibbon lineage and overlapped with enhancer chromatin states, representing putative co-opted LAVA enhancers. These putative enhancers were located within genes encoding SETD2 and RAD9A, two proteins that facilitate accurate repair of DNA double-strand breaks and prevent chromosomal rearrangement mutations. Co-option of LAVA in these genes may have influenced regulation of processes that preserve genome integrity. Our findings highlight the importance of considering lineage-specific TEs in studying evolution of gene regulatory elements.
Transposable elements (TEs) comprise nearly half of mammalian genomes and provide a major source of genetic and epigenetic variation during evolution. While many studies have focused on the disruptive consequences of TE insertions, especially those that impact human health (1), growing evidence is revealing the widespread presence of advantageous TE insertions across lineages (2). Depending on their impact on the host, TEs face different evolutionary fates. Most TE insertions are neutral and therefore, drift randomly in the population, while disruptive insertions are actively selected against and removed from the population. The occasional adaptive TE insertions, however, are favored and preserved by selection, and they may ultimately become incorporated into the host genome, in a process known as “co-option” or “exaptation.” To date, several examples of co-opted TEs have been reported across vertebrates (reviewed in ref. 3).
Many co-opted TEs are capable of modifying gene expression in a tissue- or time-specific manner by forming new cis-regulatory elements (i.e., enhancers) or by being incorporated into already existing enhancers (4). Since regulatory TEs often contain transcription factor (TF) binding sites, their transposition in the genome can reshape entire gene regulatory networks by introducing similar regulatory modules near multiple genes (5). Furthermore, since the distribution of regulatory TE insertions often varies across lineages, co-option of these elements likely represents a major mechanism for evolution of lineage-specific gene regulation patterns (5–7). Indeed, a recent comparative study in primates demonstrated that nearly all human-specific regulatory elements overlapped TE sequences and that most TE families enriched at cis-regulatory regions were relatively young and lineage specific (7). These and other findings highlight young regulatory TEs as an important source for evolution of gene regulatory elements in primates (7–9). However, due to technical challenges associated with studying recent TE insertions (e.g., low mappability), their contributions to the evolution of gene regulatory adaptations still remain poorly understood, especially in nonhuman primates.
Among primates, the endangered gibbons (Hylobatidae) present an attractive model for exploring functional contributions of a lineage-specific TE. Gibbons (or small apes) occupy an important node in the primate phylogeny between Old World monkeys and great apes; they have an intriguing evolutionary history and have evolved many unique traits [e.g., locomotion via brachiating and monogamy (10)]. Most notably, the gibbon lineage has experienced drastic genomic rearrangements since its divergence from the common Hominidae ancestor ∼17 Mya (11). These evolutionary rearrangements are not only evident through comparisons with great ape genomes but also in the vastly different karyotypes of the four extant gibbon genera: Nomascus (2n = 52), Hylobates (2n = 44), Hoolock (2n = 38), and Siamang (2n = 50), which split almost instantaneously around 5 Mya. Factors leading to these evolutionary genome reorganizations are not fully understood, but a gibbon-specific retrotransposon called LAVA (LINE-AluSz-VNTR-AluLIKE) (Fig. 1A) may have played a role (12).
Fig. 1.
The LAVA element displays genus-specific expansion patterns. (A) The full-length composite LAVA element shares structural components with SVA. TSD, target site duplication; U, unique nonrepetitive sequence; CT, cytosine-thymine; L1ME, long interspersed nuclear element subfamily L1ME; SINE-R, short interspersed nuclear element subfamily R. (B) Representative species of the four extant gibbon genera shown along with our WGS sample sizes below each photo. (C) The number of LAVA insertion loci per individual varies greatly across genera. (D) Logarithmic PCA of LAVA genotypes groups individuals based on genus. PC, principal component. (E) Unsupervised hierarchical clustering of individuals, using LAVA genotype at 4,585 unfixed LAVA insertion loci, groups gibbons based on genus. In the heat map, dark gray indicates presence of LAVA, light gray indicates absence of LAVA, and white indicates missing data. Species abbreviations are as follows: Nomascus (Nomascus gabriellae [NGA]), Hylobates (Hylobates lar [HLA], Hylobates moloch [HMO], Hylobates muelleri [HMU], Hylobates pileatus [HPI]), Siamang (Siamang symphalangus [SSY]), and Hoolock (Hoolock [HLE]).
LAVA is a nonautonomous retrotransposon that relies on the L1 protein machinery for its retrotransposition via target primed reverse transcription (13, 14). The full-length ∼2-kb-long composite LAVA element is found only in gibbons, but its structure is composed of portions of TEs commonly found across all primate genomes, namely the 5′ portion of SVA (SINE-VNTR-AluLIKE; SINE, short interspersed nuclear element, VTNR, variable number tandem repeat), as well as pieces of AluSz and L1ME5 elements (Fig. 1A). Despite sharing much of their overall structure, SVA and LAVA elements have had drastically different propagation success in the gibbon lineage. The SVA element, which has thrived in all great apes, is only present in ∼30 copies in the gibbon genome (15, 16), whereas LAVA is estimated to be 20 to 40 times more abundant across gibbon genera (12, 13).
In the original analysis of the reference gibbon genome (Nleu3.0), which was derived from a northern white-cheeked gibbon (Nomascus leucogenys [NLE]), nearly half of the >1,000 identified LAVA insertions were located within or near genes, particularly genes involved in regulation of cell cycle and chromosome segregation (12). Since some intronic LAVA insertions are capable of terminating gene transcription prematurely, disruption of cell cycle genes by LAVA was considered a contributing cause for the abundant evolutionary genomic rearrangements in the gibbon lineage (12). However, LAVA’s successful and ongoing propagation in the gibbon genome (14), despite its disruptive effects, may have allowed a subset of insertions to adopt adaptive functions. In fact, TEs that are prevalent near genes have a stronger propensity for adopting enhancer function and modulating expression of adjacent genes (17). Thus, adaptive contributions from LAVA may have involved regulation of nearby genes and might have ultimately resulted in its co-option as a cis-regulatory element in the gibbon genome.
In this study, we characterized LAVA insertions across multiple gibbon genomes and used genomic, epigenetic, and evolutionary analyses to investigate evidence of LAVA’s functionality and co-option in the gibbon genome.
Results
Genome-Wide Identification and Genotyping of LAVA Insertions Across Gibbons Reveals Genus-Specific Expansion Patterns.
Since the LAVA element is still able to retrotranspose in the gibbon genome (14), its distribution is expected to vary across unrelated gibbons. To this end, we generated whole-genome sequencing (WGS) datasets from 23 unrelated gibbons across the four extant genera (Nomascus, Hylobates, Siamang, and Hoolock) (18) (Fig. 1B and Dataset S1). We selected the Mobile Element Locator Tool [MELT (19)] to identify and genotype LAVA insertion loci from our short-read data, as this software was found to effectively identify SVA elements from human WGS datasets (19, 20). MELT uses discrepancies in the alignment of WGS data to a reference genome to identify nonreference TE insertions and to detect absence of TE insertions that are represented in the reference genome (referred to as “deletions”). MELT can then combine this information across multiple datasets to characterize and genotype TE insertions (based on presence/absence) in a population.
Through in silico simulation analyses, we first validated MELT’s ability to identify insertions and deletions (indels) of the LAVA element relative to the reference gibbon genome sequence and showed that ≥10× WGS coverage is required for identifying ≥75% of LAVA indel sites within 10 bp of their true position with high sensitivity and specificity (SI Appendix, Fig. S1). We then used MELT to identify LAVA indels across our 23 WGS datasets, which all had >10× coverage (Dataset S1). Since all WGS datasets were aligned to the same gibbon reference genome [Nleu3.0, generated from the NLE species (12)], we were able to use MELT to identify and genotype syntenic LAVA insertion loci across genera. We identified an initial list of 20,734 nonreference LAVA insertion sites that we combined with 1,118 LAVA insertions previously identified in the Nleu3.0 assembly (12). To minimize false positives, we filtered these 21,852 LAVA insertions based on strict quality, length, and population frequency criteria, which reduced the total number of LAVA insertion loci to 5,490 high-confidence hits (Dataset S2). We found a significant effect of genus on the number of LAVA insertions identified (ANOVA, P < 0.0001) (Fig. 1C), as well as a significant effect of WGS coverage within each genus (P < 0.0001). The smallest numbers of LAVA insertions were found in the Nomascus genus, and the highest numbers were found in the Hoolock, the same genus in which LAVA was first discovered and found to form long centromeric expansions (13, 21).
Since LAVA is rarely deleted postinsertion (12), genotype differences at LAVA insertion loci are expected to mainly reflect differential insertions. Of the 5,490 LAVA insertion loci characterized in this study, 16.5% (905) appeared to be fixed in the gibbon lineage (i.e., homozygous for the presence of LAVA in all 23 gibbons), suggesting that LAVA insertion at these sites predated the genera split. In contrast, over half (2,888 or 52.6%) of LAVA insertions were genus specific (i.e., LAVA present in some or all individuals of a single genus), indicating that LAVA’s insertion at these sites likely occurred following the split of the four gibbon genera ∼5 Mya (12). The rest of LAVA insertion loci displayed presence/absence polymorphism both within and across genera, likely reflecting a mix of true evolutionary events, incomplete lineage sorting, and genotyping errors. Logarithmic principal component analysis (PCA) of LAVA genotypes at 4,585 unfixed insertion loci grouped individuals of the same genus together (Fig. 1D), and unsupervised clustering analysis organized gibbons in a pattern resembling a potential LAVA-based gibbon phylogeny (Fig. 1E). While this dendrogram recapitulates some of the published phylogenies obtained from gibbon mitochondrial DNA (22, 23), it does not match more recent phylogenies obtained based on nuclear genomes (24, 25). Therefore, it should be interpreted merely as a validation of our LAVA genotyping pipeline, rather than a true gibbon phylogeny, which still remains elusive.
LAVA Insertions Vary in Population Frequency and Are Unevenly Distributed in the Genome.
To reduce errors from cross-species alignment of WGS datasets, we focused all downstream analysis on the LAVA identified in NLE gibbons, the same species used to build the gibbon reference genome (12). Of the total of 2,266 LAVA insertion loci found among NLE individuals, 48% (1,095) had homozygous LAVA insertions in all 11 NLE individuals. These LAVA insertions, which have reached fixation/high frequency due to selection or drift, were called fixed LAVA. The remaining 1,171 LAVA insertions sites were present in lower population frequencies (<95%) and displayed presence/absence of polymorphism; hence, they were called polymorphic LINE-AluSz-VNTR-AluLIKE (poly-LAVA) (Fig. 2A and Dataset S2).
Fig. 2.
Fixed and poly-LAVA insertions have similar and uneven genomic distribution. (A) LAVA insertions present at 100% frequency in the population were classified as fixed LAVA (blue), and the rest were classified as poly-LAVA (red). Circos plot (Upper) shows the nonhomogeneous distribution of fixed (blue) and poly-LAVA (red) insertions across the gibbon chromosomes. Chromosome numbers are annotated on the outer circle, and 11 inner circles represent the genomes of the 11 NLE gibbons included in this study. (B) Proportion of LAVA observed in different positions with respect to genes is shown in bold, and mean ± SD expected percentages are reported below. *Significant deviation from expectation (permutation P < 0.01). UTR, untranslated region. (C) Correlations between gene density and LAVA density (counts per megabase) are shown across chromosomes for fixed (blue) and poly-LAVA (red).
In general, LAVA insertion loci were unevenly distributed across chromosomes [χ2fixed-LAVA(25,n=1,095) and χ2poly-LAVA(25,n=1,171), P < 0.001] (Fig. 2A and SI Appendix, Fig. S2A) and formed broad clusters within chromosomes (one-tailed permutation P < 0.001) (SI Appendix), likely due to genomic context such as repeat and gene density. Consistent with observations for other retrotransposons (26), LAVA insertions were enriched near repeats (permutation q < 0.05) (SI Appendix, Fig. S2B). Furthermore, both fixed and poly-LAVA insertion loci were significantly closer to genes than expected by random chance and were overrepresented in noncoding regions of genes (i.e., introns, promoters, or terminators; permutation P < 0.001) (Fig. 2B) (27). In line with these observations, we also found significant correlation between LAVA and gene density across chromosomes (R2fixed-LAVA = 0.23, P = 0.008 and R2poly-LAVA = 0.48, P = 0.0006) (Fig. 2C). Hence, both fixed and poly-LAVA insertion sites have uneven distribution in the genome and are overrepresented near genes, likely as a result of LAVA’s preferential insertion into open chromatin (28, 29), effects of postinsertion selection, or a combination of both.
Several LAVA Elements Show Chromatin Signatures of Enhancer Activity.
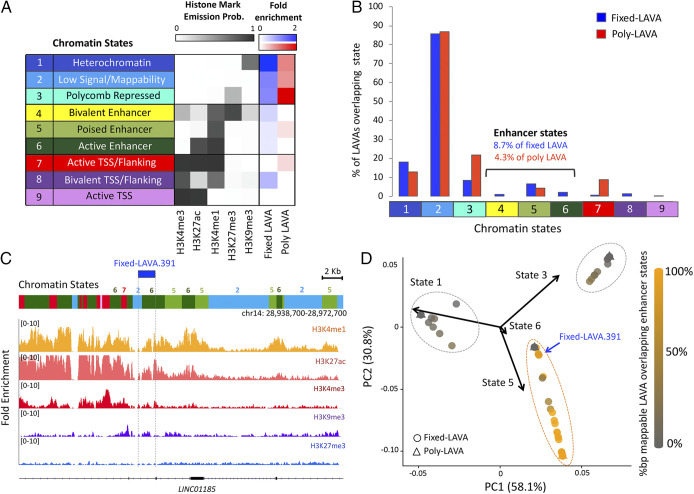
To investigate if LAVA elements have evolved regulatory function in the gibbon genome, we sought to identify LAVA elements displaying epigenetic hallmarks of enhancer activity. We performed chromatin immunoprecipitation sequencing (ChIP-seq) against three activating (H3K4me1, H3K27ac, and H3K4me3) and two repressing histone marks (H3K27me3 and H3K9me3), using Epstein-Barr Virus (EBV)-transformed lymphoblastoid cell lines (LCLs) established from three unrelated NLE individuals previously (12, 30) and in this study (Dataset S4) (18). We annotated the epigenetic landscape of the gibbon genome using nine chromatin states, each of which represented a different combination of histone marks (Fig. 3A). Since chromatin-state assignment is restricted to sequences present in the reference genome, we were only able to investigate and report chromatin states for the subset of LAVA insertions represented in the Nleu3.0 assembly (all 1,095 fixed LAVA and 23 poly-LAVA). We calculated “fold enrichment” of chromatin states overlapping these elements by accounting for the collective length and prevalence of LAVA and chromatin states in the genome. Of states overlapping fixed and poly-LAVA elements, constitutive “heterochromatin” and “polycomb-repressed” states had the highest fold enrichment, respectively (Fig. 3A). However, the chromatin state covering the most collective length (77% of fixed and 75% of poly-LAVA) (SI Appendix, Fig. S4A) and the greatest number of LAVA elements (86% of fixed and 87% of poly-LAVA) (Fig. 3B) was low signal/mappability, reflecting the difficulty in aligning short-read sequences to repetitive elements. While overlaps between LAVA and repressed chromatin states were expected, we were surprised to also find several LAVA elements overlapping enhancer chromatin states; specifically, 13 fixed LAVA overlapped bivalent enhancer chromatin, 72 fixed and 1 poly-LAVA colocalized with poised enhancer chromatin, and 23 fixed LAVA overlapped with active enhancer states (Fig. 3C). In total, 95 (8.7%) fixed LAVA insertions and 1 (4.3%) poly-LAVA element colocalized with at least one enhancer chromatin state. These overlaps ranged from 10 to 1,474 bp in length (median = 182 bp), consistent with the typical length of enhancers [10 to 1,000 bp (31)], and covered ∼3.5% of the collective length of LAVA elements in the reference genome (SI Appendix, Fig. S4A).
Fig. 3.
Several LAVA elements display epigenetic signatures of enhancers. (A) Chromatin states were characterized based on histone ChIP-seq signal. Fold enrichment (relative to random expectation) of states overlapping reference-present fixed (blue) and poly-LAVA (red) is shown. TSS, transcription start site. (B) Percentages of LAVA insertions with ≥10-bp overlap with each chromatin state are shown. Colors and numbers on the x axis correspond to chromatin states from A. (C) Example of a fixed LAVA that overlaps active enhancer chromatin state. ChIP-seq fold enrichment tracks (relative to input) are shown below the chromatin states. (D) PCA biplot analysis of LAVA elements based on their chromatin-state composition groups LAVA elements into three broad epigenetic groups (outlined with dashed circles). Shading elements based on their mappable percentage base pair overlap with enhancer states (state 4, 5, or 6) reveals one of these groups to be mostly composed of putatively functional LAVA (orange dashed circle). PC, principal component.
LAVA elements that overlapped enhancer states rarely colocalized with any additional states other than low signal/mappability. Similar patterns were observed for those elements overlapping silenced chromatin (SI Appendix, Fig. S4B). Consistently, PCA analysis of LAVA chromatin-state composition (excluding overlaps with low signal/mappability chromatin state and the 725 LAVA elements entirely covered by this state) indicated that three epigenetically distinct groups of LAVA may exist in the gibbon genome; one group consisted mostly of putatively functional LAVA overlapping enhancer states (Fig. 3D), while the other two groups consisted of putatively silenced LAVA elements that were almost exclusively within either constitutive heterochromatin or polycomb repressed chromatin (SI Appendix, Fig. S4 B and C).
LAVA Elements Provide TF Binding Motifs, and Some Elements Are Bound by PU.1.
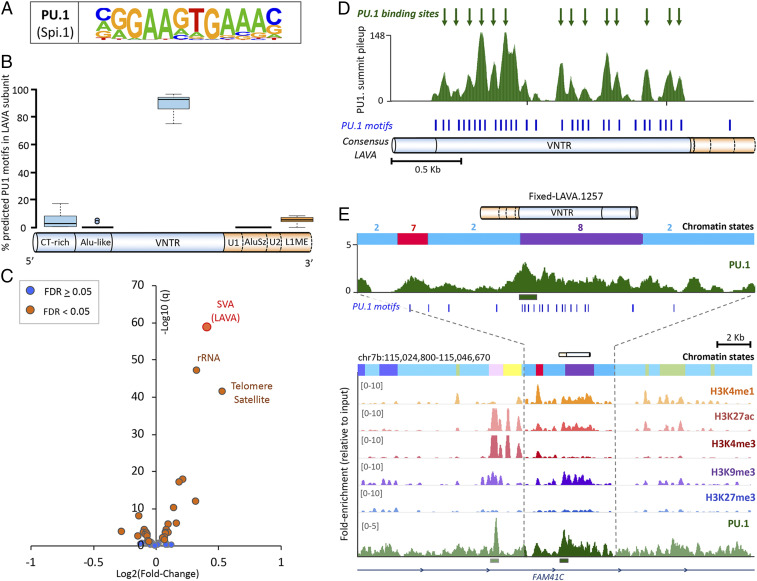
Regulatory TEs contribute roughly 20% of all TF binding sites in mammalian genomes (32). Using two different pipelines, we identified a conservative list of six TFs whose recognition motifs were significantly (q < 0.05) overrepresented in LAVA sequences and were predicted to bind LAVA with high affinity: PU.1 (encoded by SPI1), STAT3, SRF, SOX10, SOX17, and ZNF143 (SI Appendix, Fig. S5A). Since motif enrichment analysis on highly similar and repetitive sequences may lead to false positives, we sought to experimentally validate binding of one candidate TF to LAVA. We focused on PU.1, an important TF in the development of B-lymphoid cells (33), whose recognition motif was highly enriched in the VNTR (variable number of tandem repeats) of LAVA (Fig. 4 A and B). The LAVA VNTR is composed of variable numbers of 30- to 50-bp tandem repeat units, leading to most LAVA elements containing several closely spaced PU.1 binding motifs (12.8 ± 11; mean ± SD). To validate binding of PU.1 to LAVA, we performed ChIP-seq against PU.1 in two gibbon LCLs (Dataset S4) (18). We first used the RepEnrich2 software (34), which takes advantage of both unique and multimapping sequencing reads to assess overall enrichment of repeat families in ChIP-seq samples relative to input (indicating binding), without the need for high mappability or peak calling. Of the 16 repeat families significantly enriched in gibbon PU.1 ChIP-seq samples, the “SVA repeat family,” which is composed of SVA and LAVA elements, was the most significantly enriched (log fold change= 0.4, q = 1.52e-59) (Fig. 4C). It should be noted that in gibbons, enrichment of the SVA repeat family can be equated to enrichment of LAVA because SVA elements have only ∼30 insertions genome wide and make up a negligible proportion of this family (15, 16).
Fig. 4.
The PU.1 TF binds LAVA at the VNTR subunit. (A) PU.1 consensus binding motif from HOMER. (B) Proportions of LAVA’s predicted PU.1 binding motifs found in each subunit (based on 20 randomly selected full-length LAVA elements). U, unique nonrepetitive sequence. CT, cytosine-thymine; L1ME, long interspersed nuclear element subfamily L1ME. (C) Volcano plot displays enrichment of repeat families in gibbon PU.1 ChIP-seq. FDR, false discovery rate; rRNA, ribosomal RNA. (D) PU.1 ChIP-seq summit pileups are shown along the full-length LAVA consensus. Putative PU.1 binding sites (i.e., metasummits) are marked with green arrows. Blue ticks indicate predicted PU.1 binding motifs. (E) The fixed LAVA containing a significant PU.1 ChIP-seq peak (marked with green bar) and overlapping bivalent enhancer chromatin state (state 8 in purple). Predicted PU.1 binding motifs are marked with blue ticks. Chromatin-state colors and numbers match those in Fig. 3A.
To characterize sites of PU.1 binding within LAVA elements, we used multimapping and uniquely aligning reads to identify 22,264 peaks between our two PU.1 ChIP-seq replicates. Of these, the apex (i.e., summit) of 138 peaks overlapped LAVA elements in the reference gibbon genome. Using an approach similar to Fernandes et al. (35), we marked the positions of these overlapping summits within a consensus full-length LAVA element to generate a pileup of summit positions (Fig. 4D). Apexes of this pileup (i.e., metasummits), which represent putative PU.1 binding sites, were all located inside the VNTR subunit and were in overall agreement with the distribution of in silico predicted PU.1 recognition motifs (Fig. 4D). Lastly, to localize specific LAVA insertions bound by PU.1 in the gibbon genome, we performed peak calling using only uniquely mapping reads. We identified 13,920 unique peaks, which were collectively enriched for the consensus PU.1 recognition motif and co-occurred with active histone marks (SI Appendix, Fig. S5 C and D). In this approach, most overlaps between PU.1 peaks and LAVA are expected to be missed due to removal of multimapping reads. Nonetheless, we found significant PU.1 peaks inside the VNTR of two fixed LAVA elements. Based on our previous chromatin-state analysis, one of these elements (fixed LAVA.1257) overlapped bivalent enhancer chromatin state (Fig. 4E), while the other (fixed LAVA.1087) was entirely covered by low-signal/mappability chromatin state.
Since PU.1 appears to bind LAVA at the VNTR, a structure shared between LAVA and SVA elements (Fig. 1A), we investigated whether the ability to bind PU.1 was specific to LAVA or shared with SVA. After repeating our analysis on public human data from the Encyclopedia of DNA Elements (ENCODE), we did not detect any enrichment of the SVA repeat family in human PU.1 ChIP-seq datasets (q = 0.8) (SI Appendix, Fig. S6A) (36, 37). Moreover, there were no PU.1 ChIP-seq summits that mapped to the consensus human SVA sequence. Consistently, comparison of PU.1 recognition motifs between LAVA and SVA sequences revealed that 1) the consensus SVA sequence contained fewer predicted PU.1 recognition motifs compared with LAVA (5 vs. 32) (SI Appendix, Figs. S5B and S6B), 2) a smaller proportion of SVA repeats in the human reference genome contained PU.1 motifs (56% of SVAs vs. 41% of random size-matched background sequences) compared with LAVA in the gibbon genome (100% of LAVAs vs. 56% in background), and 3) the average density of PU.1 motifs in SVA repeats (2.5 ± 4.2; mean ± SD motifs per 1 kb) was lower than LAVA (8.6 ± 4.1; Mann–Whitney–Wilcoxon Test, P < 2.2e-16). Thus, despite the close relationship between LAVA and SVA, major differences exist in their ability to bind PU.1, and potentially other TFs, likely as a result of sequence differences that have evolved in the VNTR region since LAVA diverged from SVA in the gibbon lineage (38).
Genes Near LAVA Have Overall Higher Expression Compared with the Rest of the Genome.
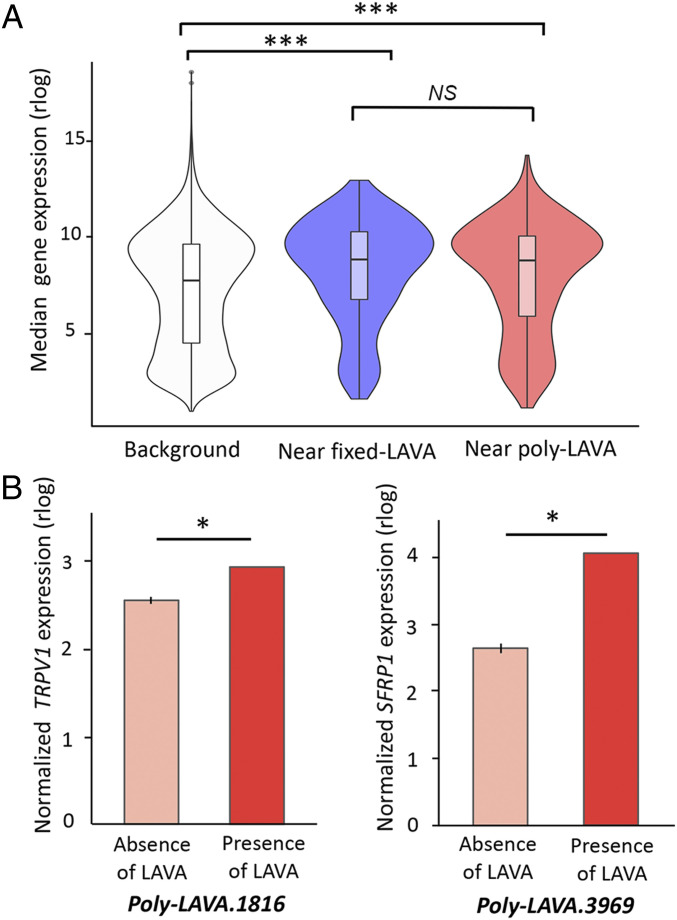
To investigate LAVA’s potential effect on expression of nearby genes, we generated RNA-sequencing (RNA-seq) data from nine NLE gibbon LCLs generated previously (12, 27, 30) and in this study. We considered genes with depth-normalized read counts (counts per million) higher than 0.5 in at least two of the nine gibbons to be “actively expressed.” Using this proxy, 72% of the nearest genes within 3 kb of fixed LAVA (448 of 620 genes) and 69% of the nearest genes within 3 kb of poly-LAVA (478 of 694) were considered actively expressed. These proportions were not significantly different from each other (two-tailed Fisher’s exact test, P = 0.58) but were both significantly higher than the 39% of genes (15,715 of 40,504) that were actively expressed genome wide (two-tailed χ2 test with Yates correction, P < 0.0001). Moreover, among actively expressed genes, those located near fixed and poly-LAVA had significantly higher median expression compared with the null distribution in the whole genome (two-tailed permutation test, P < 0.0001) (Fig. 5A).
Fig. 5.
LAVA is associated with higher expression of genes in cis. (A) Violin plots demonstrate distribution of median normalized gene expression for genes not located near LAVA insertions (white) vs. those ≤3 kb of fixed (blue) and poly-LAVA (red). ***Permutation P < 0.0001; NS, not significant. (B) Normalized expressions of TRPV1 (Left) and SFRP1 (Right) are shown against genotype at corresponding poly-LAVA. Bar heights represent mean normalized expression, and error bars are SD. All y axis are rlog-transformed gene counts normalized for gene length, GC (guanine-cytosine) content, and sequencing depth (i.e., log2 scale). *q < 0.05.
Next, we took advantage of the presence/absence of poly-LAVA insertions and examined correlation between LAVA genotype and the expression level of genes within 1 Mb of LAVA insertion loci. Despite being underpowered due to our small sample size of nine, we found two poly-LAVA insertions associated with significant increase in expression of a nearby gene (q < 0.05) (Fig. 5B). One of these genes was TRPV1 (transient receptor potential cation channel subfamily V member 1), whose expression was significantly higher in an individual with a LAVA insertion ∼300 kb downstream (P = 8.38e-07, q = 0.01) (Fig. 5 B, Left). In humans, TRPV1 is highly expressed in the central nervous system and has a human-specific SVA insertion hypothesized to have contributed to the evolution of human-specific behaviors (39). The other gene encodes the secreted frizzled related protein (SFRP1), a Wnt antagonist implicated in cell cycle regulation and senescence (40, 41), which was more highly expressed when a LAVA insertion was present ∼800 kb upstream (P = 3.8e-06, q = 0.04) (Fig. 5 B, Right). While our correlative analyses are consistent with these poly-LAVA elements functioning as cis-regulatory elements, neither insertion was present in the reference genome, and therefore, their chromatin state could not be determined. Also, changes in expression may be linked to other causal genetic variants located closer to the target genes.
Several LAVA Insertions Show Signatures of Gibbon-Specific Selective Constraint.
To investigate co-option of LAVA, we examined LAVA insertion loci for evidence of selection. We used Tajima’s D (42), a summary statistic that measures difference between two estimates of population genetic diversity (θ). Significantly negative Tajima’s D values indicate an excess of low-frequency alleles compared with expectation, a pattern of genetic variation often found surrounding sites that have been under strong recent positive selection (i.e., selective sweep) or around sites subjected to negative selection (i.e., background selection). We measured mean Tajima’s D in two 10-kb windows directly flanking each side of LAVA insertion sites and found that 11 of 808 poly-LAVA and 19 of 734 fixed LAVA elements included in our evolutionary analysis had Tajima’s D values within the 5% most negative values in the genome (i.e., P < 0.05 based on an empirical distribution of 10-kb pairs genome wide). These loci had Tajima’s D values <−2.0, compared with a genome-wide estimate of −0.95, suggestive of selection occurring on either side of, and presumably within, these LAVA elements. The most noteworthy was an ∼300-kb cluster of four fixed LAVA insertions on chromosome 18, which overlapped a major dip in Tajima’s D and displayed some of the lowest Tajima’s D values in the whole genome (SI Appendix, Fig. S7). We also fit a demographic model to the NLE allele frequency spectra at a set of putatively neutral loci using ∂a∂i (43) and generated coalescent simulations under neutrality, to estimate a simulation-based P value for each LAVA element. Except for two of the fixed LAVA loci, all LAVA insertions with P < 0.05 based on the empirical criteria described above also had P < 0.001 based on this simulation framework and P < 0.05 even under the most conservative simulation regime of no recombination, which artificially decreases null Tajima’s D values (44).
If the selection signals identified in gibbons were independent of LAVA (for example, if they merely reflected background purifying selection due to their proximity to genes), we may expect orthologous sequences in a sister taxon to show similar selection signals. We examined genetic diversity among humans within orthologous regions to the windows flanking LAVA elements. We successfully identified orthologous regions for 8 (of 11) poly- and 13 (of 17) fixed LAVA elements that displayed significant signatures of selection in gibbons, under both empirical and conservative simulation frameworks. Of these orthologous regions, none had significant Tajima’s D values in human, with the exception of one poly-LAVA. Therefore, in total we found 10 poly- and 17 fixed LAVA elements that showed significant signatures of positive/purifying selection in gibbon, but not in human (when orthologs were found) (Dataset S5), suggesting that they, or their immediately surrounding regions, have acquired gibbon-specific functional properties.
Enrichment and Potential Co-option of a Subset of LAVA Insertions Near DNA Repair Genes.
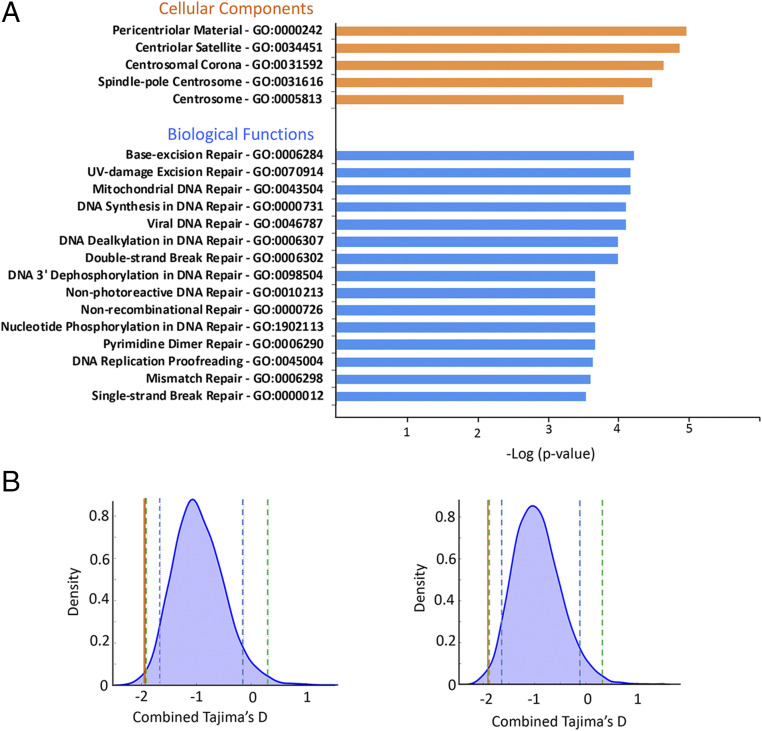
By investigating genes located near LAVA insertions, we can identify candidate biological processes influenced by LAVA. While no significant gene ontology (GO) term was enriched among genes near (≤3-kb) poly-LAVA, those near fixed LAVA insertions displayed significant enrichment (q < 0.1) for 15 biological functions, all related to DNA repair (e.g., double- and single-strand break repair), and 5 cellular components important in cell cycle (e.g., spindle-pole centrosome) (Fig. 6A and Dataset S6). Enrichment of these GO terms was validated using permutation analyses that accounted for gene length and LAVA’s preferential insertion near genes (two-tailed permutation P < 0.001) (SI Appendix). Overall, these results recapitulated the previously described association of LAVA with cell cycle and chromosome segregation genes (12). However, by characterizing LAVA insertions across multiple individuals and classifying them based on frequency in the population, we were able to also unravel an association between fixed LAVA and DNA repair pathways.
Fig. 6.
DNA repair genes are overrepresented near fixed LAVA and putatively co-opted elements. (A) Significant GO terms among genes ≤3 kb of fixed LAVA are shown. The −log P values reported by Enrichr are shown for GO terms with q < 0.1. UV, ultraviolet. (B) The (up-/downstream) combined Tajima’s D values of two putatively co-opted fixed LAVA enhancers are shown (orange lines) against the corresponding genome-wide null distributions (blue bell curves). Dashed blue and green lines mark the 95th and 99th percentiles of the null distribution (SI Appendix has details).
We then searched for co-opted regulatory LAVA insertions, which are expected to 1) be fixed in the gibbon genome, 2) show signatures of selection, and 3) overlap enhancer chromatin state. Based on these criteria, we found two putatively co-opted regulatory LAVA elements that were fixed across gibbon genera, had some of the smallest Tajima’s D values in the NLE genome (Fig. 6B), and overlapped poised enhancer state in gibbon LCL. These two LAVA insertions were located within introns of DNA repair genes encoding SETD2 (a histone methyltransferase) and RAD9A (a cell cycle checkpoint protein), which are specifically involved in facilitating accurate repair of DNA double-strand breaks (45–47). Considering the enrichment of several TF binding motifs in LAVA sequences (for example, that of STAT3 [SI Appendix, Fig. S5A], a major regulator of the DNA repair response), co-opted LAVA in these genes might be under selection to facilitate binding of TFs and alter regulation of DNA repair response globally or in a tissue-/time-specific context.
Of note, four additional LAVA elements under selection were located in genes implicated in the timely detection and precise repair of DNA lesions [TET3 (48, 49), KMT2C (50), NSD2 (51), and SMARCB1 (52)]. However, these LAVA elements did not overlap enhancer chromatin states in gibbon LCL and may perhaps function in other developmental/tissue contexts or via other mechanisms.
Discussion
In this study, we characterized insertions of the gibbon-specific LAVA retrotransposon across four extant gibbon genera using the largest set of whole-genome sequencing data generated from these endangered species to date. We found epigenetic and evolutionary evidence for the functionality of several LAVA insertions and showcased two putative co-opted LAVA enhancers within genes implicated in the accurate repair of DNA double-strand breaks.
We focused our investigation of LAVA’s functionality and co-option on the northern white-cheeked gibbon (NLE), as this is currently the only gibbon species with a published reference genome (12). Among the subset of fixed and poly-LAVA insertions that were present in the NLE reference genome, we found 96 unique elements overlapping active, poised, or bivalent enhancer chromatin states in gibbon LCLs. Consistent with known characteristics of enhancers (53), LAVA sequences were enriched in TF binding motifs including that of PU.1, an important regulator of gene expression in lymphocytes. After validating binding of PU.1 to LAVA in gibbon LCL, we showed that binding was mainly confined to the VNTR subunit, which harbors a tandem of PU.1 binding motifs. Although LAVA’s VNTR originates from SVA (Fig. 1A), PU.1 did not appear to bind SVA elements in the human genome. This finding was consistent with previous reports that LAVA’s VNTR has evolved distinct genetic features following its divergence from the SVA element (38). Furthermore, it indicated that LAVA has acquired unique epigenetic and functional properties, which might also be responsible for its successful propagation in the gibbon lineage (14, 16). Overall, we demonstrated that insertion of LAVA can introduce clusters of TF binding sites, which may alter the regulation of nearby genes (54, 55). Notably, genes near LAVA had overall higher expression compared with the rest of the genome. Also, despite low statistical power, we found two positive correlations between the presence of poly-LAVA loci and expression of a gene in cis. It should be noted, however, that these observations were correlational and may, at least partially, reflect LAVA’s preferential insertion into actively transcribed chromatin.
Since biochemical activity of TEs could reflect selfish strategies for their survival and propagation rather than adaptive function (29), we also investigated signatures of selection at LAVA insertion sites. We found significant signal of positive or purifying selection at 27 LAVA elements in gibbon, but not in any of the available orthologous regions in human, suggesting that the observed selection signal is specific to LAVA elements. Two of these evolutionarily significant fixed LAVA insertions also overlapped with poised enhancer chromatin state in gibbon LCL, representing two putative co-opted cis-regulator LAVA elements. Considering that these elements overlap with poised enhancer states and knowing that TE-derived enhancers often have tissue-specific activity (4), we predict that these putative co-opted LAVA enhancers are active in tissue/developmental contexts other than LCL. We also expect the true number of functional and co-opted LAVA insertions across gibbon tissues and genera to be larger than those identified here, considering all of the challenges that limited our analysis, including the lack of high-quality genome assemblies from all genera, exclusion of many LAVA elements from our epigenetic and evolutionary analyses, low mappability of short-read sequences to LAVA (56), and limited availability of tissues from the endangered gibbons.
While disruptive LAVA insertions are thought to have contributed to the emergence of genomic rearrangements in gibbons (12), evolutionary contributions of functional LAVA elements are not yet clear. Nonetheless, genes located near co-opted LAVA may provide insight. The two putative co-opted cis-regulatory LAVA elements identified in this study were located in the introns of SETD2 and RAD9A, both of which are crucial genes in maintaining genome integrity. SETD2 is an H3K36 histone methyltransferase that modifies chromatin at the site of DNA double-strand break to ensure its faithful repair via homologous recombination, rather than error-prone nonhomologous end joining (45, 57). RAD9A is a cell cycle checkpoint control protein that facilitates homologous recombination repair and prevents cell cycle progression before DNA double-strand breaks are repaired (46, 47). Regardless of how, or in which context, the co-opted LAVA insertions may alter regulation of these genes, any adaptive regulatory changes resulting in improvement of DNA repair and genome integrity would be favored/preserved by natural selection. As computational tools for studying TEs improve (58) and gibbon-induced pluripotent stem cells (iPSC) provide access to currently unavailable tissues, we will be able to further investigate functional roles of co-opted LAVA across tissues, particularly in the context of DNA double-strand repair. Insights from this and future studies will advance our understanding of how young TEs can contribute to lineage-specific evolution of gene regulatory novelty.
Materials and Methods
SI Appendix has further details on most of the sections described below.
Genome-Wide Identification, Genotyping, and Characterization of LAVA Insertions.
Genomic DNA from blood of 23 unrelated gibbons across the four extant genera (Nomascus = 13, Hylobates = 5, Hoolock = 3, Siamang = 2) was used to construct WGS libraries as described before (12). Libraries were sequenced paired end on Illumina HiSeq platforms. Reads were aligned to the gibbon reference genome (Nleu3.0) using BWA (59) with default settings. MELT v2.1.3 (19) was used to identify nonreference presence and absence of LAVA elements (referred to as insertion and deletion, respectively) and genotype all LAVA insertion sites from WGS alignments, similar to our simulation analysis (SI Appendix). LAVA elements were annotated based on their insertion position relative to the closest gene: exonic, intronic, promoter (<3 kb upstream of gene) or terminator (<3 kb downstream of gene), or intergenic. LAVA predictions were filtered to remove 1) low-quality inserts (as determined by MELT), 2) insertions present in only one copy in the population of 23 diploid genomes (i.e., heterozygous presence in only one genome), 3) insertions shorter than 290 bp (the minimum length required to discriminate a composite LAVA from its non-SVA subunits), and 4) inserts found on unplaced Nleu3.0 contigs. We generated binary LAVA genotype profiles for all individuals (heterozygous or homozygous LAVA insertion =1, absence of LAVA =0), performed hierarchical clustering using the hclust function with the ward D2 method in R 3.6.1, and visualized the results with heatmap.2 function in the ggplot2 package. Logistic PCA was carried out using the logisticPCA package in R (k = 2 and m = 4).
To reduce erroneous LAVA predictions due to cross-species WGS alignment, only LAVA insertions identified in the 11 NLE gibbons were used in downstream analysis. LAVA insertions found in two copies in all NLE individuals were called fixed LAVA, while the rest were called poly-LAVA. Due to sequence ambiguity and absence of many poly-LAVA insertions from the reference genome, we did not consider sequence polymorphism in our characterizations. We used permutation tests to characterize LAVA’s distribution across and within chromosomes and relative to repeats and genes (SI Appendix has details).
Histone ChIP-Seq and Chromatin-State Characterization.
ChIP-seq was performed on gibbon EBV transformed LCLs from three unrelated NLE individuals, as previously described (12) and outlined in SI Appendix. Raw reads were quality controlled with FastQC v0.11.5 (60). All reads were aligned to Nleu3.0 using BWA (59) with default single-end settings, and low-quality/multimapping read alignments (mapping quality < 30) were removed. ChIP-seq replicates displayed high correlation (Pearson correlation coefficient 75–96%) (SI Appendix, Fig. S3) (61) and were therefore combined (SI Appendix, Fig. S3). ChromHMM (62) was used to identify and characterize nine chromatin states based on the histone ChIP-seq alignments. Fold enrichment of each chromatin state at reference-present LAVA was calculated as (C/A)/(B/D), where: “A” is the genome-wide number of bases in the state, “B” is the collective length (base pairs) of LAVA elements in the genome, “C” is the collective length (base pairs) of overlaps between state and LAVA, and “D” is the total size of genome (base pairs). Lastly, we used BEDtools (63) to characterize overlap of chromatin states with reference-present LAVA elements, requiring each overlap to be ≥10-bp long. The chromatin composition of each LAVA was characterized by removing overlaps with low-signal/mappability state and calculating percentage length overlap of the remaining sequences with each chromatin states. The 725 LAVA elements that fully overlapped with low signal/mappability were removed. The chromatin composition matrix for the remaining 393 elements was used to perform PCA analysis in R and visualize epigenetically distinct groups of LAVA elements.
TF Motif Enrichment and PU.1 Binding to LAVA.
Sequences of all LAVA insertions represented on the assembled chromosomes of Nleu3.0 were used to perform motif enrichment analyses with the HOMER suite (64) and the Transcription factor Affinity Prediction [TRAP (65)] web tool. To meet length restrictions, 24 LAVA sequences that were longer than 3 kb were removed in TRAP analysis. P values were corrected using the Benjamini–Hochberg method (66), and only significant motif enrichments (q < 0.05) that agreed between the two methods were considered (Dataset S3). LAVA and SVA_A (SINE/VNTR/Alu composite subfamily A) consensus sequences were obtained as described in SI Appendix, and their PU.1 motifs were predicted using the HOMER suite (64). To assess distribution of PU.1 motifs across subunits of LAVA, we randomly selected 20 full-length LAVA elements, predicted their PU.1 motifs using the HOMER suite, and determined percentage of motifs found in each LAVA subunit. PU.1 ChIP-seq was performed on two gibbon LCLs, analyzed, and compared with public human LCL PU.1 ChIP-seq data from ENCODE [Gene Expression Omnibus (GEO) accession nos. GSM803531 and GSM803398 (36, 37)], as described in SI Appendix.
Characterization of Gene Expression Patterns Near LAVA.
RNA-seq gene count data were collected from two previously (12, 30) and seven newly established NLE LCLs and normalized as described in SI Appendix. We used the GraphPad tool (https://www.graphpad.com/quickcalcs/contingency1.cfm) to perform a two-tailed χ2 test with Yates correction and compare the proportion of actively expressed genes nearby fixed and poly-LAVA with the rest of the genome. We used custom R scripts described in SI Appendix to compare the (median of medians) expression level of genes near LAVA, and genes located elsewhere in the genome (27). Linear regressions between LAVA (presence/absence) genotypes at each poly-LAVA locus and expression of genes within 1 Mb were performed using Matrix-eQTL with default settings (67).
Assessing Selection Around LAVA Insertion Sites.
A detailed description of evolutionary analyses is available in SI Appendix. Briefly, we used ANGSD (68) to estimate folded allele frequency spectra and Tajima’s D in 10-kb windows. After filtering LAVA elements based on WGS coverage, we compared Tajima’s D in 10-kb windows upstream and downstream of these elements with randomly sampled loci in the genome and generated empirical P values. We considered a LAVA element under selection only if P was <0.05 for both upstream and downstream regions. In our simulation analyses, we generated simulated P values by comparing observed Tajima’s D in 20-kb windows centered at LAVA insertions with expectation under neutrality. To generate comparative human data, alignments from 20 Yoruba individuals from the 1000 Genomes Project (69) were used to calculate genome-wide Tajima’s D. We then used the multiz100way alignment to find orthologous human regions (Hg19) for the 10-kb windows flanking LAVA elements and assessed significance of Tajima’s D in these regions using the empirical framework used for gibbons.
GO Analysis of Genes Nearby Fixed and Poly-LAVA.
We used Enrichr (70) with GO Biological Process 2017b, GO Cellular Component 2017b, and GO Molecular Function 2017b libraries to test GO enrichment among the nearest genes (within 3 kb) of fixed and poly-LAVA. Significance of all GO terms that had P < 0.05 and q < 0.1 was validated using two permutation tests described in SI Appendix.
Supplementary Material
Acknowledgments
We thank the zoos (San Antonio Zoo and Aquarium, Point Defiance Zoo and Aquarium, Oregon Zoo, Gladys Porter Zoo, and Los Angeles Zoo) and the staff at the Gibbon Conservation Center (Santa Clarita, CA), especially the director Gabriella Skollar, who have provided us with opportunistic gibbon blood samples. We also thank Dr. Jeff Wall and Dr. Michael Hammer for their invaluable contributions to WGS data collection; Dr. Eugene Gardner for aiding in optimizing the MELT pipeline; Dr. Jessica Minnier for guidance in RNA-seq analysis; Dr. R. Alan Harris, Patty Langasek, and Christopher Klocke for their help with data analysis; and members of the laboratory of Dr. Shawn Chavez and L.C. for valuable feedback on the research. We acknowledge the ENCODE Consortium and Dr. Myers at HudsonAlpha Institute for Biotechnology, who generated the human PU.1 ChIP-seq data. Gibbon PU.1 ChIP-seq was performed by the Epigenetics Consortium at Knight Cardiovascular Institute of Oregon Health and Science University (OHSU). All ChIP-seq libraries were sequenced at the OHSU Massively Parallel Sequencing Shared Resource and the Genomics and Cell Characterization Core Facility at the University of Oregon. Data analyses were performed on the Exacloud super computer cluster at OHSU. This work was financially supported by a grant awarded to L.C. from the Leakey Foundation. R.J.O. and L.C. are also supported by National Science Foundation (NSF) Grant 1613856, N.A. and L.C. are supported by National Human Genome Research Institute (NHGRI) Grant R01HG010333, and L.C. is supported by National Insitute of Health Office of Directors (NIH/OD) Grant P51 OD011092 (to the Oregon National Primate Research Center). J.D.F. is supported by National Institute of General Medical Sciences (NIGMS) Grant F32GM125388, and S.R.S. is funded by NHGRI Grant 1R01HG010329.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: All WGS, ChIP-seq, and RNA-seq data are available at Gene Expression Omnibus (accession no. GSE136968). The gibbon gene annotation file is available on Dryad (https://doi.org/10.5061/dryad.7wm37pvq9).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2006038117/-/DCSupplemental.
Data Availability.
All WGS, ChIP-seq, and RNA-seq data are available at Gene Expression Omnibus (GEO accession no. GSE136968). The gibbon gene annotation file is available on Dryad (https://doi.org/10.5061/dryad.7wm37pvq9).
References
- 1.Konkel M. K., Batzer M. A., A mobile threat to genome stability: The impact of non-LTR retrotransposons upon the human genome. Semin. Cancer Biol. 20, 211–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schrader L., Schmitz J., The impact of transposable elements in adaptive evolution. Mol. Ecol. 28, 1537–1549 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Bourque G., Transposable elements in gene regulation and in the evolution of vertebrate genomes. Curr. Opin. Genet. Dev. 19, 607–612 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Trizzino M., Kapusta A., Brown C. D., Transposable elements generate regulatory novelty in a tissue-specific fashion. BMC Genomics 19, 468 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sundaram V. et al., Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res. 24, 1963–1976 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Warren I. A. et al., Evolutionary impact of transposable elements on genomic diversity and lineage-specific innovation in vertebrates. Chromosome Res. 23, 505–531 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Trizzino M. et al., Transposable elements are the primary source of novelty in primate gene regulation. Genome Res. 27, 1623–1633 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanco-Melo D., Gifford R. J., Bieniasz P. D., Co-option of an endogenous retrovirus envelope for host defense in hominid ancestors. eLife 6, e22519 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gombart A. F., Saito T., Koeffler H. P., Exaptation of an ancient Alu short interspersed element provides a highly conserved vitamin D-mediated innate immune response in humans and primates. BMC Genomics 10, 321 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cunningham C., Mootnick A., Gibbons. Curr. Biol. 19, R543–R544 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Carbone L. et al., A high-resolution map of synteny disruptions in gibbon and human genomes. PLoS Genet. 2, e223 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carbone L. et al., Gibbon genome and the fast karyotype evolution of small apes. Nature 513, 195–201 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carbone L. et al., Centromere remodeling in Hoolock leuconedys (Hylobatidae) by a new transposable element unique to the gibbons. Genome Biol. Evol. 4, 648–658 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyer T. J. et al., The flow of the gibbon LAVA element is facilitated by the LINE-1 retrotransposition machinery. Genome Biol. Evol. 8, 3209–3225 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang H. et al., SVA elements: A hominid-specific retroposon family. J. Mol. Biol. 354, 994–1007 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Ianc B., Ochis C., Persch R., Popescu O., Damert A., Hominoid composite non-LTR retrotransposons-variety, assembly, evolution, and structural determinants of mobilization. Mol. Biol. Evol. 31, 2847–2864 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Chuong E. B., Elde N. C., Feschotte C., Regulatory activities of transposable elements: From conflicts to benefits. Nat. Rev. Genet. 18, 71–86 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okhovat M., Carbone L., Data from “Co-option of the lineage-specific LAVA retrotransposon in the gibbon genome.” Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE136968. Deposited 5 September 2019.
- 19.Gardner E. J. et al.; 1000 Genomes Project Consortium , The mobile element locator tool (MELT): Population-scale mobile element discovery and biology. Genome Res. 27, 1916–1929 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sudmant P. H. et al.; 1000 Genomes Project Consortium , An integrated map of structural variation in 2,504 human genomes. Nature 526, 75–81 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hara T., Hirai Y., Jahan I., Hirai H., Koga A., Tandem repeat sequences evolutionarily related to SVA-type retrotransposons are expanded in the centromere region of the western hoolock gibbon, a small ape. J. Hum. Genet. 57, 760–765 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Chan Y.-C. et al., Mitochondrial genome sequences effectively reveal the phylogeny of Hylobates gibbons. PLoS One 5, e14419 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsudaira K., Ishida T., Phylogenetic relationships and divergence dates of the whole mitochondrial genome sequences among three gibbon genera. Mol. Phylogenet. Evol. 55, 454–459 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Veeramah K. R. et al., Examining phylogenetic relationships among gibbon genera using whole genome sequence data using an approximate bayesian computation approach. Genetics 200, 295–308 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi C.-M., Yang Z., Coalescent-based analyses of genomic sequence data provide a robust resolution of phylogenetic relationships among major groups of gibbons. Mol. Biol. Evol. 35, 159–179 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao C. et al., Characterization and functional annotation of nested transposable elements in eukaryotic genomes. Genomics 100, 222–230 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Okhovat M., et al. , Gibbon genome (Nleu3.0) custom gene annotation file, v3. Dryad. 10.5061/dryad.7wm37pvq9. Deposited 19 May 2020. [DOI]
- 28.Sultana T., Zamborlini A., Cristofari G., Lesage P., Integration site selection by retroviruses and transposable elements in eukaryotes. Nat. Rev. Genet. 18, 292–308 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Bourque G. et al., Ten things you should know about transposable elements. Genome Biol. 19, 199 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lazar N. H. et al., Epigenetic maintenance of topological domains in the highly rearranged gibbon genome. Genome Res. 28, 983–997 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li L., Wunderlich Z., An enhancer’s length and composition are shaped by its regulatory task. Front. Genet. 8, 63 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bourque G. et al., Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 18, 1752–1762 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rimmelé P. et al., Spi-1/PU.1 oncogene accelerates DNA replication fork elongation and promotes genetic instability in the absence of DNA breakage. Cancer Res. 70, 6757–6766 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Criscione S. W., Zhang Y., Thompson W., Sedivy J. M., Neretti N., Transcriptional landscape of repetitive elements in normal and cancer human cells. BMC Genomics 15, 583 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fernandes J. D. et al., The UCSC repeat browser allows discovery and visualization of evolutionary conflict across repeat families. Mob. DNA 11, 13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.ENCODE Project Consortium , An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis C. A. et al., The encyclopedia of DNA elements (ENCODE): Data portal update. Nucleic Acids Res. 46, D794–D801 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lupan I., Bulzu P., Popescu O., Damert A., Lineage specific evolution of the VNTR composite retrotransposon central domain and its role in retrotransposition of gibbon LAVA elements. BMC Genomics 16, 389 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gianfrancesco O., Bubb V. J., Quinn J. P., SVA retrotransposons as potential modulators of neuropeptide gene expression. Neuropeptides 64, 3–7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elzi D. J., Song M., Hakala K., Weintraub S. T., Shiio Y., Wnt antagonist SFRP1 functions as a secreted mediator of senescence. Mol. Cell. Biol. 32, 4388–4399 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Z., Wang J., Han X., Zhou J., Linder S., Up-regulation of human secreted frizzled homolog in apoptosis and its down-regulation in breast tumors. Int. J. Cancer 78, 95–99 (1998). [DOI] [PubMed] [Google Scholar]
- 42.Tajima F., Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gutenkunst R. N., Hernandez-Rodriguez R. D., Williamson S. H., Bustamante C. D., Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. 5, e1000695 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thornton K., Recombination and the properties of Tajima’s D in the context of approximate-likelihood calculation. Genetics 171, 2143–2148 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pfister S. X. et al., SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell Rep. 7, 2006–2018 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsai F.-L., Kai M., The checkpoint clamp protein Rad9 facilitates DNA-end resection and prevents alternative non-homologous end joining. Cell Cycle 13, 3460–3464 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pandita R. K. et al., Mammalian Rad9 plays a role in telomere stability, S- and G2-phase-specific cell survival, and homologous recombinational repair. Mol. Cell. Biol. 26, 1850–1864 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.An J. et al., Acute loss of TET function results in aggressive myeloid cancer in mice. Nat. Commun. 6, 10071 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang D., Wei S., Chen F., Zhang Y., Li J., TET3-mediated DNA oxidation promotes ATR-dependent DNA damage response. EMBO Rep. 18, 781–796 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rampias T. et al., The lysine-specific methyltransferase KMT2C/MLL3 regulates DNA repair components in cancer. EMBO Rep. 20, e46821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shah M. Y. et al., MMSET/WHSC1 enhances DNA damage repair leading to an increase in resistance to chemotherapeutic agents. Oncogene 35, 5905–5915 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim K. H., Roberts C. W. M., Mechanisms by which SMARCB1 loss drives rhabdoid tumor growth. Cancer Genet. 207, 365–372 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yáñez-Cuna J. O., Kvon E. Z., Stark A., Deciphering the transcriptional cis-regulatory code. Trends Genet. 29, 11–22 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Ridinger-Saison M. et al., Spi-1/PU.1 activates transcription through clustered DNA occupancy in erythroleukemia. Nucleic Acids Res. 40, 8927–8941 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ezer D., Zabet N. R., Adryan B., Homotypic clusters of transcription factor binding sites: A model system for understanding the physical mechanics of gene expression. Comput. Struct. Biotechnol. J. 10, 63–69 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ewing A. D., Transposable element detection from whole genome sequence data. Mob. DNA 6, 24 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carvalho S. et al., SETD2 is required for DNA double-strand break repair and activation of the p53-mediated checkpoint. eLife 3, e02482 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goerner-Potvin P., Bourque G., Computational tools to unmask transposable elements. Nat. Rev. Genet. 19, 688–704 (2018). [DOI] [PubMed] [Google Scholar]
- 59.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Andrews S., Fast Q. C., A quality control tool for high throughput sequence data (2010). www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed 6 January 2017.
- 61.Ramírez F., Dündar F., Diehl S., Grüning B. A., Manke T., deepTools: A flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–W191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ernst J., Kellis M., Chromatin-state discovery and genome annotation with ChromHMM. Nat. Protoc. 12, 2478–2492 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heinz S. et al., Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thomas-Chollier M. et al., Transcription factor binding predictions using TRAP for the analysis of ChIP-seq data and regulatory SNPs. Nat. Protoc. 6, 1860–1869 (2011). [DOI] [PubMed] [Google Scholar]
- 66.Benjamini Y. H. Y., Controlling the false fiscovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995). [Google Scholar]
- 67.Shabalin A. A., Matrix eQTL: Ultra fast eQTL analysis via large matrix operations. Bioinformatics 28, 1353–1358 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Korneliussen T. S., Albrechtsen A., Nielsen R., ANGSD: Analysis of next generation sequencing data. BMC Bioinformatics 15, 356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Auton A. et al.; 1000 Genomes Project Consortium , A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kuleshov M. V. et al., Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All WGS, ChIP-seq, and RNA-seq data are available at Gene Expression Omnibus (GEO accession no. GSE136968). The gibbon gene annotation file is available on Dryad (https://doi.org/10.5061/dryad.7wm37pvq9).