Fodrin and its erythroid cell-specific isoform spectrin are actin-associated fibrous proteins that play crucial roles in the maintenance of structural integrity in mammalian cells, which is necessary for proper cell function. Normal cell morphology is altered in diseases such as various cancers and certain neuronal disorders. Fodrin and spectrin are two-chain (αβ) molecules that are encoded by paralogous genes and share many features but also demonstrate certain differences. Fodrin (in humans, typically a heterodimer of the products of the SPTAN1 and SPTBN1 genes) is expressed in nearly all cell types and is especially abundant in neuronal tissues, whereas spectrin (in humans, a heterodimer of the products of the SPTA1 and SPTB1 genes) is expressed almost exclusively in erythrocytes.
KEYWORDS: apoptosis, cell signaling, cytoskeleton, drug interactions, mitosis
ABSTRACT
Fodrin and its erythroid cell-specific isoform spectrin are actin-associated fibrous proteins that play crucial roles in the maintenance of structural integrity in mammalian cells, which is necessary for proper cell function. Normal cell morphology is altered in diseases such as various cancers and certain neuronal disorders. Fodrin and spectrin are two-chain (αβ) molecules that are encoded by paralogous genes and share many features but also demonstrate certain differences. Fodrin (in humans, typically a heterodimer of the products of the SPTAN1 and SPTBN1 genes) is expressed in nearly all cell types and is especially abundant in neuronal tissues, whereas spectrin (in humans, a heterodimer of the products of the SPTA1 and SPTB1 genes) is expressed almost exclusively in erythrocytes. To fulfill a role in such a variety of different cell types, it was anticipated that fodrin would need to be a more versatile scaffold than spectrin. Indeed, as summarized here, domains unique to fodrin and its regulation by Ca2+, calmodulin, and a variety of posttranslational modifications (PTMs) endow fodrin with additional specific functions. However, how fodrin structural variations and misregulated PTMs may contribute to the etiology of various cancers and neurodegenerative diseases needs to be further investigated.
INTRODUCTION
Eukaryotic cells have a plasma membrane-bound cytoskeleton for the maintenance of their strength and structural integrity. The cytoskeleton is a dynamic proteinaceous network that provides the shape of the cell and holds onto the inner organelles of the cell in their proper position. Several filamentous structures such as microtubules, microfilaments, intermediate filaments, and spectrin filaments form the cytoskeleton, and they are in constant touch with each other to provide the proper functioning of the cytoskeleton. Red blood cells are a special example of cells that flow through the arteries and veins into constricted capillaries for gaseous exchange and thus vehemently need to maintain their cellular integrity. Spectrin filaments are especially important in red blood cells for structural maintenance. In other tissues, especially in neuronal tissues, nonerythroid spectrin or fodrin performs the duty of maintaining cellular integrity in association with other proteins.
Spectrins belong to the spectrin superfamily of proteins. Members such as spectrin, nonerythroid spectrin/fodrin, α-actinin, dystrophin, ABP-280, ABP-120, and fimbrin all carry a few to several units of spectrin repeats, characteristic of this family (1). Spectrins have two subunits, the α- and β-subunits. The α-subunit has two isoforms, αI-spectrin and αII-spectrin. αI-spectrin is expressed from the SPTA1 (spectrin alpha erythrocytic 1) gene exclusively in erythrocytes. However, SPTAN1 expresses the more ubiquitous and abundant αII-spectrin. This is the nonerythroid homologue of αI-spectrin, which is hence called nonerythroid α-spectrin or, more commonly, α-fodrin (2). The β-subunit has five isoforms, βI, βII, βIII, βIV, and βV, expressed by the SPTB, SPTBN1, SPTBN2, SPTBN4, and SPTBN5 genes, respectively. βI is expressed in erythrocytes and, to a certain extent, in lymphocytes. βIII and βIV are expressed in neurons, and βV is found associated with photoreceptors in rods and cones. βII is ubiquitous, as it is expressed in all nucleated cells. It is also called β-fodrin (2–7). In this review, we intend to focus on the structural differences in nonerythroid spectrin/fodrin (αIIβII) compared to erythroid spectrin (αIβI) and the emerging new functions of the nonerythroid isoform in cells that are likely to originate due to these structural differences. For ease of presentation, in this review, we refer to nonerythroid spectrin as fodrin and its two subunits as α-fodrin and β-fodrin to differentiate them from erythroid spectrin.
FODRIN
Fodrin was characterized initially in a group of proteins that translocated through axons (8). Additionally, it formed an undercoat near the plasma membrane in cells. In view of this localization pattern, it was termed fodrin, derived from the Greek word fodros, meaning lining (9). Certain reports also term this protein calspectin (calmodulin binding spectrin-like protein) or brain spectrin (10). Overlapping cDNA analysis of human lung fibroblasts revealed that the SPTAN1 gene, corresponding to α-human lung fibroblasts revealed that thefodrin, is mapped to chromosome 9. The coding sequence is a long stretch of 7,787 nucleotides producing a protein of 2,472 amino acids with a predicted molecular weight of 284 kDa. The SPTBN1 gene, encoding β-fodrin, is located in chromosome 2 in humans. It has a predicted molecular weight of 274 kDa (2, 11).
KEY STRUCTURAL ASPECTS OF SPECTRIN AND FODRIN
Spectrin repeats and other domains.
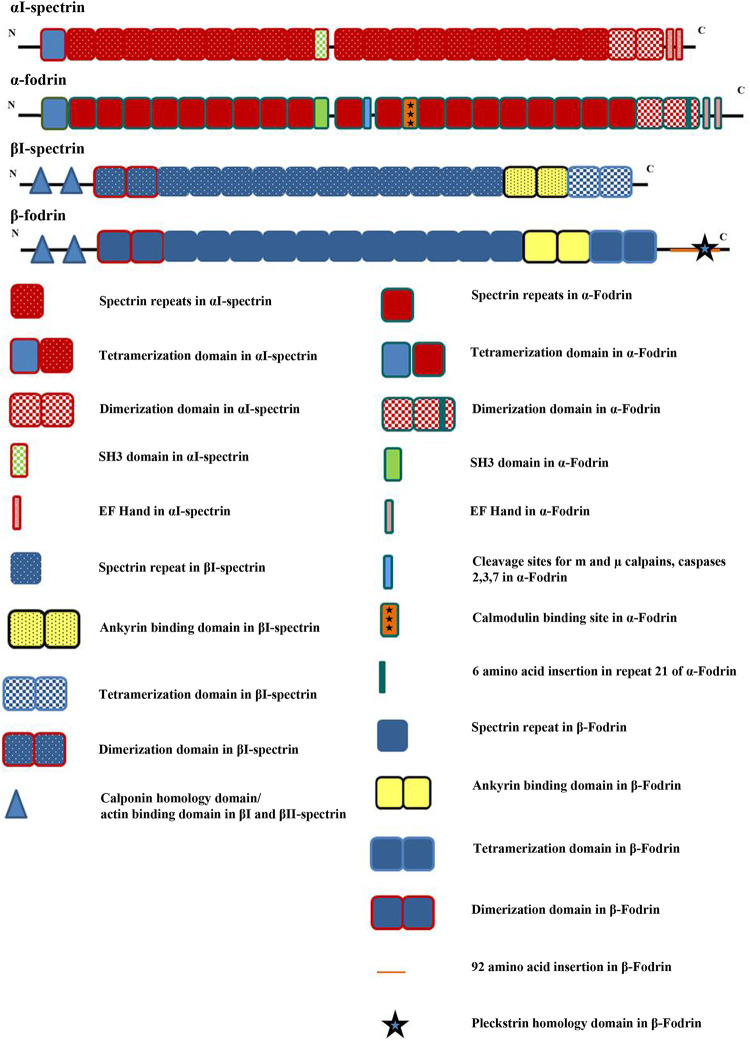
Spectrin and fodrin are characterized by spectrin domains or spectrin repeats. Each repeat, nearly 106 amino acids long (molecular weight, 12,000 Da), forms a characteristic triple-helical structure with intermittent unstructured loops facilitating the folding of the domain into a helix-loop-helix structure. The hydrophobic charged amino acids in these domains are arranged in a heptad periodic pattern enabling the structure to fold into an antiparallel coiled coil (12). The α-subunits of both comprise a tandem arrangement of 21 spectrin repeats, all of which show the above-mentioned triple-helical structure, except for domain 10 (3, 13). They also carry an SH3 (Src homology 3) domain in the central part of the molecule enabling involvement in signal transduction pathways (discussed below). α-Fodrin specifically inhabits calpain and caspase cleavage sites and a calmodulin binding domain. The significance of these sites is discussed below. The β-subunits have 17 such repeats in both molecules. Both βI-spectrin and β-fodrin have a calponin homology domain/actin binding domain (CH domain) at the N terminus and an ankyrin binding domain at the C terminus. Interestingly, β-fodrin exclusively harbors a pleckstrin homology domain toward the C terminus (Fig. 1) to facilitate binding with phospholipids (13).
FIG 1.
Different structural domains of spectrin and fodrin. This diagram illustrates the variations between spectrin and fodrin. The top portion is a schematic of the polypeptide chains of the α- and β-subunits of both molecules. A detailed description of the artwork used is given below.
Formation of the tertiary structure.
Spectrin and fodrin undergo regulated dimerization and tetramerization to form functionally relevant molecules. The α- and β-subunits associate in an antiparallel fashion to form a flexible and elastic heterodimer (12). The lock-and-key model explains this dimerization. Repeats 20 and 21 at the C terminus of the α-subunit and repeats 1 and 2 at the N terminus of the β-subunit (Fig. 1) serve as nucleation sites that associate in a complementary manner to initiate the formation of the heterodimer. The two subunits associate laterally at specific sites throughout the contour of the molecules (14). Constructing an operational tetramer from a heterodimer requires a head-to-head association of two dimer molecules at a specific site called the tetramerization domain. Repeats 16 and 17 at the C terminus of the β-subunit and repeats 0 and 1 at the N terminus of the α-subunit are involved in tetramerization. Ipsaro et al. were able to obtain a 2.8-Å crystal structure of the α0-1/β16-17 complex, showing that these repeats associate to recapitulate a spectrin repeat-like triple-helical bundle resulting in tetramerization (15).
A comparative account of the primary structures of spectrin and fodrin.
Peptide mapping has helped considerably in understanding the structural relatedness of spectrin and fodrin throughout evolution. The α-subunits of these molecules share a maximum of 52% and 54% homology in repeats 16 and 19, respectively, although the whole molecule shares less sequence identity (16). However, the β-subunits share 60% homology (2). Fodrin, being ubiquitous, is more conserved across species through evolution because mammalian α-fodrin shares 96% sequence identity with the avian homologue, whereas spectrin is much more divergent across species (17).
The linker region between spectrin repeats 10 and 11 of α-fodrin houses a proteolytically hypersensitive site. This site is recognized by m and μ calpains and caspases 2, 3, and 7. Calpains are calcium-dependent neutral proteases that digest proteins in a calcium-dependent manner. Caspases are apoptotic proteases that perform specific cleavage of proteins to execute apoptosis (18). α-Fodrin is processed by these proteases in a highly regulated manner to accomplish functions diverse from those of spectrin. Another major point of contention between spectrin and fodrin is the presence of a calmodulin binding site in the α-subunit of the latter. Calmodulin is a ubiquitous eukaryotic protein, functioning as a Ca2+-dependent messenger in various signal transduction pathways. Calmodulin’s Ca2+-dependent activity is particularly implicated in the synaptic transmission of nerve impulses. The presence of a calmodulin binding site in α-fodrin highlights its significance in neuronal transmission. Another notable variation is the functionality of EF (E helix and F helix of the parvalbumin protein that was first reported to possess EF hands) hands (detailed below). This difference is more qualitative than structural. EF hands are Ca2+ binding domains present at the C termini of the α-subunits of both spectrin and fodrin. However, the EF hands in spectrin are reportedly vestigial and do not bind to Ca2+ except under nonphysiological conditions. Contrarily, EF hands in fodrin show strong binding to Ca2+ in cells (Fig. 1) (19).
Evolutionary history.
Multiple-sequence alignment and phylogenetic analyses have revealed that all members of the spectrin family evolved from a common ancestor, α-actinin, an actin cross-linking protein with four spectrin repeats (ac1, ac2, ac3, and ac4). ac1 and ac2 show a high degree of sequence homology with the first two repeats of β-spectrin (β1 and β2). Interestingly, ac3 and ac4 are highly identical to the C-terminal spectrin repeats of α-spectrin (α19 and α20). α-Actinin occurs inside the cells as dimers. β1 and β2 of β-spectrin and α19 and α20 of α-spectrin are also involved in dimerization. Such sequence identity reveals that the dimerization property of these molecules was inherited from α-actinin and has undergone little change since then. Bootstrap analyses of the phylogenetic trees revealed that the α-actinin gene underwent an insertional event resulting in an ancestral block of 7 spectrin repeats between ac2 and ac3. This block was later duplicated by intragenic duplication followed by the insertion of a domain that is reminiscent of the tetramerization domain. This resulted in the ancestral α- and β-spectrin genes. The present-day β-spectrin gene evolved from this ancestral gene that underwent elongations and insertions. The α-spectrin gene, however, again encountered another duplication event and subsequent elongations to form the present-day α-spectrin (reviewed in references 10 and 11). Very little is known about the events that led to the distinction of spectrin and fodrin. However, it is speculated that vertebrates went through two whole-genome duplications that resulted in βI-spectrin and β-fodrin. Multiple events of alternative splicing led to the present variety of spectrin isoforms (reviewed in reference 12). A. J. Baines hypothesizes that the evolution of spectrin from fodrin is a process of neofunctionalization to allow the molecule to sustain mechanical shear forces and to confer a certain degree of elasticity to the membrane by the rapid “make and break” of the tetramers (20).
CALCIUM DEPENDENCE OF FODRIN
Calcium functions through multiple modes to affect fodrin and its operations. Below is a brief detailing of these modes and the biological relevance of each mode.
Direct binding of Ca2+ ions.
The association of Ca2+ ions directly with proteins is largely mediated through specialized motifs termed EF hands. These motifs have a helix-loop-helix structure providing accessibility to Ca2+. Investigation of the primary structure of α-fodrin revealed that it carries two such EF hands in its C terminus (3). 45Ca autoradiography experiments revealed the characteristics of the binding of calcium to fodrin. It possesses eight binding sites for Ca2+ per tetramer, that is, four in each dimer. Two of these sites are in the above-mentioned EF hands. Another site localizes between repeat 11 and repeat 12 of α-fodrin. The fourth site is in β-fodrin, which is about 25 kDa from the amino terminus. The EF hands bind to Ca2+ with high affinity (Kd [dissociation constant] = 2 × 10−8 to 30 × 10−8 M). The above-mentioned segment of β-fodrin also binds to Ca2+ with a quite high Kd of 1 × 10−6 to 3 × 10−6 M. The hypersensitive site between repeat 11 and repeat 12 of α-fodrin has a low affinity for Ca2+, with a Kd of 1 × 10−4 to 2 × 10−4 M (19). Magnesium inversely affects this binding. Barring the hypersensitive site in α-fodrin, the parameters of the binding of Ca2+ to α- and β-fodrins are comparable to those of calmodulin and calcium-dependent protease I. The physiological significance of this binding is evident during events of neuronal depolymerization. This process witnesses an upsurge in Ca2+. Ca2+ then binds with a high affinity to α-fodrin in a manner similar to calmodulin and calcium-dependent protease I. The binding of fodrin to actin and protein 4.1 to form a ternary complex is stimulated by Ca2+ binding. This binding may also expose the hydrophobic patches required for the binding of calmodulin (2, 19).
Ca2+-dependent binding of calmodulin to α-fodrin.
The presence of a calmodulin binding site in α-fodrin was revealed through ferritin-avidin labeling of biotinylated calmodulin bound to α-fodrin. There are two calmodulin binding sites per tetramer, that is, one per dimer (21). Studies using recombinant peptides spanning different segments of α-fodrin revealed the presence of a 24-amino-acid segment, KTASPWKSARLMVHTVATFNSIKE, housed in the 11th repeat. The charged and hydrophobic amino acids in this patch give rise to an amphipathic α-helical conformation. The Kd value for the binding reaction between this stretch and calmodulin is <10−7, equivalent to the binding affinities of other calmodulin binding partners. Moreover, binding could be abolished by Ca2+ chelators such as EGTA (22).
The physiological significance of the interaction of calmodulin and α-fodrin has been explained using various in vitro techniques. The direct linkage of α-fodrin to synaptosomal membrane sites is competitively inhibited by calmodulin, with a Ki of 1.3 μM. Furthermore, Ca2+ could stimulate this inhibition. However, this was not a consequence of the proteolysis or degradation of α-fodrin but of the inactivation or denaturation of the molecule, because the effect could be reversed by the action of calmodulin inhibitors and Ca2+ chelators. The inhibition induced on α-fodrin membrane interactions by calmodulin is considered to be significant in exposing secretory vesicles and in exocytosis (23).
A COMPARATIVE ACCOUNT OF THE POSTTRANSLATIONAL MODIFICATIONS IN SPECTRIN AND FODRIN
Both spectrin and fodrin undergo various modifications after synthesis, as shown in Table 1. These modifications affect the structural folding and flexibility of the molecules, their interactions with other binding partners, and, consequently, their functionality. Table 1 shows the posttranslational modifications (PTMs) reported in both molecules (24). Proteomic analysis revealed that the SH3 domain in α-fodrin is heavily loaded with modifying moieties, whereas αI-spectrin shows only two phosphorylations. The calmodulin binding site of α-fodrin reportedly shows various sites for phosphorylation and ubiquitination. These sites might be responsible for the efficient binding of α-fodrin to calmodulin and also for its specific cleavage by caspases and calpains. Comparison of the EF hands shows that the one in α-fodrin has many phosphorylations, ubiquitinations, and acetylations, but the equivalent in αI-spectrin has limited phosphorylation. This could explain why the EF hands in αI-spectrin are vestigial and bind to calcium only under nonphysiological conditions. The CH domain in the β-subunits of both molecules is differently modified but does not largely affect the actin binding property of these proteins. However, β-fodrin is quite different from βI-spectrin by the presence of heavy modifications, such as phosphorylations, acetylation, and ubiquitination in the pleckstrin homology domain in the former.
TABLE 1.
PTMs reported in select domains of fodrin and spectrina
| Type of PTM | Domain | Subunit | Position(s) (reference[s]) |
|---|---|---|---|
| Phosphorylation | SH3 | α-Fodrin | Y976-p (91), Y978-p (92), S982-p (93–97), T995-p (98), S999-p (93), T1000-p (99), Y1020-p, S1029-p (98, 100–103) |
| α-Spectrin | Y986-p, S992-p (95, 104, 105) | ||
| CaM binding domain | α-Fodrin | Y1176-p (91, 106–111), T1184-p, S1186-p (112), T1188-p (112, 113), S1190-p (93, 94, 98, 104, 108, 112, 114–117), S1194-p (94, 118), T1204-p (95, 101), S1207-p (101), S1217-p (119), S1226-p (95, 98), S1231-p (94, 95, 98, 101) | |
| EF hands | α-Fodrin | S2411-p, T2425-p, T2434-p, Y2423-p (120), Y2430-p, Y2440-p (121) | |
| α-Spectrin | T2288-p (98), Y2333-p, S2335-p (95), S2405-p, Y2407-p | ||
| CH domain | β-Fodrin | S163-p (93), S228-p (93, 95), S257-p (112), S278-p, S96-p (108), T80-p (92), Y274-p, Y79-p (92) | |
| β-Spectrin | S96-p (122), T104-p (104), T152-p, T76-p (123) | ||
| PH domain | β-Fodrin | S2221-p, S2251-p, S2303-p, T2297-p, Y2249-p (110), Y2268-p (124), Y2285-p (92) | |
| Acetylation | SH3 | α-Fodrin | K1022-ac (125), K1023-ac |
| EF hands | α-Fodrin | K2421-ac (125), K2426-ac (125) | |
| CH domain | β-Fodrin | K124-ac (125), K90-ac (125) | |
| β-Spectrin | K80-ac | ||
| PH domain | β-Fodrin | K2207-ac, K2269-ac, K2270-ac | |
| Ubiquitination | SH3 | α-Fodrin | K990-ub (126, 127) |
| CaM binding domain | α-Fodrin | K1187-ub (128), K1193-ub (127, 129), K1209-ub (129) | |
| EF hands | α-Fodrin | K2404-ub (126), K2421-ub (126), K2426-ub (129, 130) | |
| CH domain | β-Fodrin | K214-ub (126), K227-ub (127), K249-ub (126), K62-ub (129, 130), K118-ub (126, 127), K124-ub (118) | |
| β-Spectrin | K102-ub, K107-ub, K118-ub (126, 127), K124-ac (125, 131), K124-ub (118), K62-ub (130) | ||
| PH domain | β-Fodrin | K2207-ub (126), K2241-ub (129), K2276-ub (127) | |
| Sumoylation | CH domain | β-Fodrin | K214-sm (132) |
CaM, calmodulin. p, phosphorylation; ac, acetylation; ub, ubiquitination; sm, sumoylation; CH, calponin homology domain; PH, pleckstrin homology domain.
Despite the apparent diversity, both spectrin and fodrin share the function of supporting the plasma membrane. This was first evident from the outstanding electron micrographs of the membrane skeleton of erythrocytes shown by Byers and Branton in 1985. They showed polygonal networks containing 200-nm-long spectrin tetramers interlinked at junctional complexes rich in actin (25). However, such early studies were based on experiments involving high-shear-stress conditions; hence, the density of the subplasmalemmal network was underestimated. Through electron microscopy and computational biology, Brown et al. showed that the biological functional length of spectrin tetramers is ∼55 to 65 nm (26). But Xu et al. point out by stochastic optical reconstruction microscopy (STORM) analysis of the neuronal axons that the actin filaments form concentric rings along the circumference of the axonal shaft and that these are uniformly and periodically spaced to ∼180 nm by fodrin tetramers running parallel to the axonal axis (27). Spectrin in erythrocytes is probably required to remain compact functionally; however, the long axonal shafts require fodrin to be stretched out to maintain the actin network. This points to the structural flexibility of these molecules and the ability to evolve in accordance with the requirement of the cellular system that they decorate.
Fodrin not only stains the inner aspect of the plasma membrane in nonerythroid cells but also is present in a diffused pattern in the cell cytoplasm and, to a certain extent, in the nucleus (6, 28). Fodrin’s presence in both the periphery and the interior of the cells indicates the static as well as dynamic functions performed by this molecule.
FUNCTIONS COMMON TO SPECTRIN AND FODRIN
Structural support.
The spectrin-actin network on erythrocytes is significant in providing integrity and deformability to the cell membrane, as is evident in cases of hemolytic anemia and hereditary spherocytosis. Mutations in both subunits of spectrin result in brittle membranes with reduced elasticity (29, 30). After Levine and Willard (9), various groups worked out the characteristics of the fodrin-actin interaction and found that the former could result in gelation and cross-linking of actin. This is significant in maintaining the plasma membrane-associated microfilament network and also a probable regulatory step in cell motility over solid surfaces. This cross-linking of actin occurs at low intracellular Ca2+ concentrations. An upsurge in Ca2+ levels causes fodrin and F-actin to dissociate, resulting in the local dissolution of the cortex (31). Studies showed that the subplasmalemmal cortical network important for the capping of T lymphocytes contains fodrin in association with actin, myosin, and α-actinin (32). Fodrin also forms a ternary complex with actin and protein 4.1, another major cortical cytoskeletal protein (1, 33). This complex is important for maintaining the integrity and elasticity of the plasma membrane. In this context, it is important that fodrin also binds to microtubules and causes them to form bundled structures (33). This support is crucial for the anchoring of various receptors on the membrane and the relaying of signals into the cell. In the nucleus, α-fodrin exists to associate with proteins such as lamin and emerin to strengthen the nucleolaminar structure (34). Prime evidence to establish the significance of fodrin as a key structural support protein is its crucial association with other established structural factors. Unpublished data from our laboratory show that a deficiency of the same could result in neural cells with a disorganized cytoskeleton and a rounded morphology.
Role in activation of transmembrane proteins.
Research into the membrane protein organization led to the understanding that the spectrin cytoskeletal network is indispensable for the spatial arrangement and anchorage of these proteins. The interaction between spectrin and membrane proteins is mostly concentrated at two major sites, the ankyrin-band 3 junction and the protein 4.1R-MPP1-glycophorin C junction. Ankyrin binds to the C terminus of βI-spectrins, whereas protein 4.1R binds to the N terminus. Protein 4.1R is crucial for the spectrin-actin network (35). The protein 4.1R-MPP1-glycophorin C junctional complex contains other factors such as adducin, protein 4.2, and transmembrane proteins such as the AE1 dimer and GLUT1 (glucose transporter protein 1) (36). The association of ankyrin with βI-spectrin is essential to mediate the latter’s interaction with CD45 (cluster of differentiation 45). The surface sequestration of CD45 is a crucial step in T-cell receptor activation (37).
β-Fodrin has a specialized pleckstrin homology domain that mediates its ankyrin-regulated association with the membrane phospholipids. During neuronal development, the interactions of β-fodrin with immunoglobulin superfamily cell adhesion molecule family members, namely, L1, CHL1, neurofascin, NgCAM (neuron-glia cell adhesion molecule), NrCAM (neuron-glia-related cell adhesion molecule), and neuroglian, are considered crucial (36, 38). The actin- and spectrin-based membrane-associated periodic skeleton (MPS) of neuronal cells is reported to be important in NCAM1-mediated extracellular signal-regulated kinase (ERK) signaling. As revealed by STORM analysis, the reduction of β-fodrin causes the disruption of MPS accompanied by reduced ERK signaling (39). N-Methyl-d-aspartate (NMDA) receptors are glutamate-gated cation channels with high Ca2+ conductance that facilitate the transmission and plasticity of the excitatory synapses. α-Fodrin associates with NR2, a cytosolic subunit of NMDA, and this association is probably responsible for the modulation of membrane morphology during synaptic activity and plasticity (40, 41). Conditions such as spinocerebellar ataxia are associated with β-fodrin mutations. α-Fodrin mutations have been reported in the rare epileptic disease West syndrome (also known as infantile spasms) and epileptic encephalopathy (36). Ankyrin and spectrin/fodrin are key players in the spatial assembly and maintenance of physiologically important domains on the surfaces of diverse cells.
Role in actin dynamics and cell adhesion.
The α-subunits of spectrin and fodrin play a regulatory role in actin dynamics and consequently affect cell adhesion and spreading. The SH3 domain in spectrins is important in the integrin-based signaling pathway aiding cells to adhere, extend lamellipodia, migrate, proliferate, undergo cell death, or associate with the extracellular matrix. These processes require extensive actin reorganization. Migratory assays on integrin ligand surfaces reveal that such a stimulus leads to the clustering of integrin complexes. The SH3 domain of cleaved spectrin activates Rac, a small GTPase of the Rho GTPase family. Rac activation is upstream of actin polymerization and lamellipodium formation. The overexpression of the SH3 domain of spectrin results in reduced Rac activation (42). Thus, the association of spectrin with the initial integrin clusters and spectrin cleavage are crucial in regulating actin dynamics.
There is mounting evidence of the dependence of the actin network on the presence of functional fodrin. The depletion of α-fodrin in WM-266 human melanoma cells resulted in a reduced population of adherent cells with modified focal contacts, reduced expression of integrins, and loss of actin stress fibers (43).
Vascular permeability, a tool used by pathogens to mount an infection, is regulated by cortical actin assembly-mediated intercellular adhesion. VASP (vasodilator-stimulated phosphoprotein) is important in actin stress fiber formation for cell-to-cell adhesion. α-Fodrin associates with VASP, through its SH3 domain, forming an α-fodrin-VASP complex that helps in initiating cortical actin fiber formation, thus stabilizing intracellular contacts and reducing vascular permeability (44). Additionally, proteins that regulate actin dynamics and polymerization, such as EVL (Ena/VASP-like), WASP (Wiskott-Aldrich syndrome protein) subfamily proteins, WAVE (WASP family verpolin-homologous protein 1) proteins, and Abi1 (ABL interactor 1), relay their effects on actin through their association with spectrin (45, 46). Reports also indicate that βI-spectrin influences the actin network and cortical tension in cells by regulating the Hippo-signaling pathway. βI-spectrin mutations lead to the inhibition of this signaling and disturb the cortical actin organization (47, 48).
FUNCTIONS EXCLUSIVE TO FODRIN
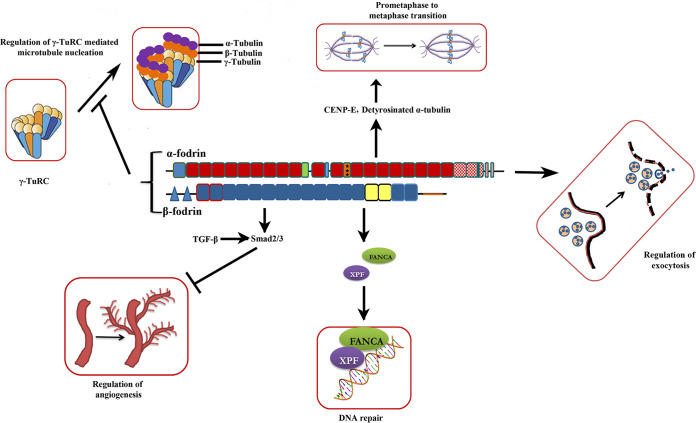
The functions exclusive to fodrin are summarized in Fig. 2.
FIG 2.
Newer functions of fodrin. The illustration focuses on the functions of fodrin that arise probably due to the additional domains and posttranslational modifications.
Role in exocytosis.
Exocytosis has been most extensively studied in the context of neurotransmitter release in neuronal cells. This is a strictly regulated and conserved mechanism. The first understanding of the involvement of fodrin in this mechanism comes from classic work by Perrin et al. By immunofluorescence, they demonstrate the change in the localization pattern of α-fodrin upon the stimulation of chromaffin cells with secretagogues. Unprimed cells display continuous rings of α-fodrin subjacent to the plasma membrane; however, stimulation by high concentrations of K+ ions or nicotine results in a patchy sublamellar pattern and an enhanced secretion of catecholamines by these cells. They also showed that this process was heavily dependent on the intracellular availability of Ca2+ (49). Through measurements of radioactive [3H]noradrenaline, this group also showed that α-fodrin antibody inhibited the process of secretion by permeabilized chromaffin cells (50). By way of elegant immunoelectron microscopy, it was shown that fodrin is concentrated at the presynaptic membrane region of neurons, which is also a site for the increased deposition of secretory granules (51). These secretory granules are populated with synapsin 1. By utilizing radioactively labeled secretory vesicles and purified immobilized fodrin, it was demonstrated that synapsin 1 and fodrin displayed a single-site binding association with a significant Kd value (52). Synapsin 1 is a phosphorylation target of CaMKII (Ca2+/calmodulin-dependent protein kinase II) (53). It is therefore proposed that the influx of Ca2+ in response to a stimulus results in the activation of the CaMKII-dependent phosphorylation of synapsin 1. This modification causes the release of secretory vesicles bound to the fodrin-actin network, facilitating the docking and fusion of these vesicles with the presynaptic membrane and the subsequent release of neurotransmitters into the synapse (54).
DNA repair.
Reports on the nuclear population of α-fodrin have highlighted its significance in DNA repair pathways and the maintenance of chromosome integrity. Fanconi anemia (FA), a hereditary bone marrow disorder occurring due to chromosome instability and cellular hypersensitivity to DNA interstrand cross-linking (ICL) agents, is frequently associated with reduced levels of α-fodrin. Evidence indicates that α-fodrin has a substantial affinity for interstrand cross-linked DNA, and it showcases a recruitment site at these locations for specific repair proteins. This is further corroborated by the increment of at least 5-fold in chromosomal aberrations upon a 40% reduction in α-fodrin expression in FA-A cells (55). Elegant immunological staining studies show that genotoxic overload in cells results in α-fodrin forming punctae that colocalize with FANCA (Fanconi anemia repair protein A) and XPF (xeroderma pigmentosum group F-complementing protein), which is an endonuclease important in the ICL repair pathway (56). Domain mapping studies of α-fodrin show that the SH3 domain is crucial for the docking of many repair proteins such as FANCG (57). The present consensus is that once α-fodrin binds to an ICL site, it encourages the binding of FANCG, XPF, and SLX4-SLX1 (structure-specific endonucleases 4 to 1). SLX4-SLX1 are nucleases that cause the cleavage and unhooking of ICL followed by translesion synthesis, homologous recombination, and nucleotide excision repair (34). In the nucleus, however, α-fodrin also associates with various nuclear skeletal components, indicating its structural roles apart from the DNA repair function. Thorough experimentation is required to decipher these dual characteristics and to discretely map the domains responsible for each.
Cellular proliferation.
α-Fodrin is evidently important for the G1/S transition. The reduced expression of α-fodrin results in a reduction in p21 expression, instigating the cells to be arrested in G1/S (43). Our laboratory has reported that α-fodrin is crucial for mitotic progression because α-fodrin-deficient cells showed a delayed prophase-to-metaphase transition with a sustained spindle assembly checkpoint (SAC). The depletion of α-fodrin resulted in a disorganized microtubule network and chromosomal misalignment in cells. This chromosome derangement was due to a reduced localization of the microtubule motor CENP-E (centromere protein E) at the kinetochore. CENP-E, a mitotic motor, carries chromosomes through the spindle with help from detyrosinated tubulin. We reported that α-fodrin-deficient cells displayed reduced detyrosinated tubulin (58). This could have important implications for the involvement of α-fodrin in the progression of cancer. Recent in vitro experiments from our laboratory reveal that fodrin directly binds to γ-tubulin and results in the negative regulation of γ-tubulin ring complex-mediated microtubule nucleation in the brain (59). Previous experiments by our group showed that fodrin is a component of γ-TuRC (γ-tubulin ring complex) and colocalizes with γ-tubulin at the centrosome of brain-derived cells (60, 61) in an actin-dependent manner. Detailed binding experiments revealed that this interaction with γ-tubulin occurs through a GRIP2 (γ-TuRC-interacting protein 2)-like motif in the C terminus of α-fodrin (59).
Reports also fortify the role of β-fodrin in cell cycle regulation. It regulates the phosphorylation status of retinoblastoma proteins by affecting the activity of cyclin-dependent kinase 4 (CDK4). CDK4, activated by the association with cyclin D1, hyperphosphorylates pRb (retinoblastoma protein), releasing bound E2F. E2F, a key transcription factor, causes the transcription of genes involved in the G1/S transition. The overexpression of β-fodrin reduces CDK4 activity, preventing the disassembly of the pRb-E2F complex, with subsequent G1/S arrest (62). The downregulation of β-fodrin increases the levels of p53 accompanying cellular stress. p53, being an important regulator of the G2/M checkpoint, results in G2/M arrest upon β-fodrin reduction (63). This evidence advocates for a significant contribution of β-fodrin to cell cycle progression. Although the indications are strong enough to tell that this homologue is important at various crucial points of the cell cycle, its importance is also reiterated by the modulation of αβ-subunits during tumorigenesis (discussed below).
Regulation of angiogenesis.
Angiogenesis is a bona fide hallmark of cancer. Invasive tumors possess a precocious ability to undergo neovascularization for a sustained supply of nutrients and oxygen and the continual evacuation of metabolic waste and carbon dioxide (64). The inhibition of the transforming growth factor β (TGF-β)/Smad pathway leads to the attenuation of angiogenesis (65). β-Fodrin, also termed ELF (embryonic liver fodrin), is an important regulator of angiogenesis. It acts as an adaptor for Smad3 and Smad4 of the TGF-β signaling pathway. ELF associates with receptor-induced endogenous Smad3. This complex then associates with Smad4, resulting in the translocation of the complex into the nucleus (66). β-Fodrin heterozygous null mice (elf+/−) exhibited a higher incidence of hepatocellular carcinoma associated with increased aberrant endovasculature, a noticeable amplification of progenitor endothelial cells, and a reduction in endothelial differentiation, followed by lethality (67). These results further cement the hypothesis that both α-fodrin and β-fodrin are important regulators of angiogenesis. However, it would be interesting to understand the upstream and downstream factors of this pathway to better comprehend angiogenesis and, consequently, tumor progression.
Organ development during embryogenesis.
Evidence justifying the involvement of fodrin in organ development, especially in vertebrates, has emerged only recently, largely due to advancements in targeted downregulation techniques. Experiments on C57BL/6 mice show that heterozygous knockout of the mouse Span2 gene that expresses mouse α-fodrin (Span2+/−) shows no peculiar phenotype. However, homozygous null mice (Span2−/−) undergo intrauterine death by embryonic day 12.5 (E12.5) to E16.5 accompanying craniofacial abnormalities, cardiac dilation, abnormal cardiac shape, incomplete neural tube closure, and thinning of the myocardium. Analysis of primary fibroblast cultures from E14.5 mice indicates that such cells possess sporadic lamellipodia with negligible cortical actin (68). This fortifies the belief that the fodrin-ankyrin-actin network is primal in tissue patterning and vertebrate development.
Like α-fodrin, the homozygous null mutant of β-fodrin is also embryonically lethal. Such embryos undergo intrauterine death at E16 due to the reduced size of the heart and compromised neural development. Tissue analysis of these organs revealed reduced cardiac muscle differentiation, reduced cardiac vasculature, and abnormal distributions of tropomyosin and dystrophin. These result in underdeveloped cardiomyocytes with reduced excitability and contractility (69). The cardiac cell-specific depletion of β-fodrin results in pronounced arrhythmia because of an altered association of ankyrin with β-fodrin (70). The data collected and analyzed so far delegate a profound role to fodrin in the context of organ development. However, the inherent pathways involved in this process are still quite elusive.
Apoptosis.
Programmed cell death or apoptosis warrants a regulated life span for each cell. Numerous data suggest that during the early stages of apoptosis, the proteolytic cleavage of actin and other actin-associated proteins results in a loss of adherence in cells. The cleavage of α-fodrin into fragments of 150 kDa and 120 kDa is associated with apoptosis in A1.1 T-cell hybridoma cells (18). Evidently, tumor necrosis factor (TNF) causes the calpain-independent and caspase 3-induced cleavage of α-fodrin into 150-kDa, 115-kDa, and 110-kDa fragments (71). The apoptosis-inducing agent staurosporine resulted in the simultaneous degradation of both α- and β-subunits in SHSY5Y neuroblastoma cells (72). As fodrin acts as a scaffold tethering several other proteins to keep the cell architecture intact, its proteolytic degradation could result in cytoskeletal dissolution followed by membrane blebbing and cell disintegration. It is important to note at this point that the cleavage of α-fodrin by m and μ calpains explicitly occurs in neuronal cells during synaptic transmission and during platelet cell activation (49, 73). The exact mechanism of discretion between calpain- and caspase-dependent cleavage in different cells is, however, a matter of study.
Role in neuronal diseases.
Various groups have looked into the status of fodrin in neuronal disease in adults. West syndrome or early infantile epileptic encephalopathy, where mutations cluster in spectrin repeats 21 and 22 of α-fodrin, results in the improper formation of the nodes of Ranvier and the sequestering of voltage-gated channels in myelinated neurons (74, 75). These events eventually lead to hypomyelination and neuronal atrophy. Experiments in squid axoplasms and Caenorhabditis elegans have shown independently that fodrin is crucial for dynein-dynactin-based retrograde transport, which is important for proper axonal maintenance and homeostasis (76, 77). Cholinergic degeneration in the forebrain is a prominent feature of Alzheimer’s disease (AD). An impairment of Ca2+ homeostasis in mouse models caused increased calpain-based cleavage of α-fodrin, accumulating large and stable breakdown products at the neurite plaques (78). The derailment of Ca2+ homeostasis, followed by unregulated α-fodrin cleavage, could act as an early marker for the diagnosis of neurodegenerative disorders.
Role in cancer.
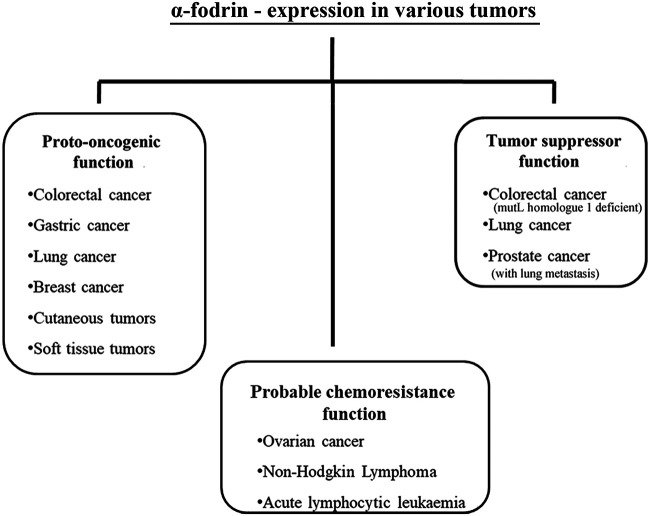
Cancer is one of the leading causes of global morbidities. α-Fodrin has been studied by many groups in a fair number of tumors, but the results are not coherent (Fig. 3). In certain tumors, it functions as a tumor suppressor, whereas in a few others, its gene functions as a proto-oncogene (79). Colorectal (80), gastric (81), lung (82), breast (83), and cutaneous (84) tumors exhibit a supranormal expression of α-fodrin. As fodrin is crucial for cellular integrity, anchoring the growth receptors at the cell membrane, and facilitating the epithelial-to-mesenchymal transition, the upregulation of this protein would promote tumor development. Interestingly, the plausible role of α-fodrin in enabling DNA repair mechanisms and its association with other cytoskeletal elements make it a good candidate for a tumor suppressor protein. This is evident in the case of prostate cancer with lung metastasis (85) and in specific cases of lung (86) and colorectal (87) cancers. This could be because of the augmenting load of genomic insults in cells in the absence of functional α-fodrin. However, there are a few tumors, such as ovarian cancer (88), non-Hodgkin lymphoma, and acute lymphoblastic leukemia (89), where the expression of α-fodrin appears to rise after chemotherapy, indicating a possible chemoresisting function. The genesis of such a multivalent role of α-fodrin in tumors still needs to find a basis through intense experimentation and research.
FIG 3.
Schematic representation of the relative expression of α-fodrin in different tumor types. α-Fodrin functions as a tumor suppressor and the gene functions as a proto-oncogene in different tumors, as indicated. In some, it also plays a probable role in chemoresistance.
There is limited evidence for the role of β-fodrin in tumorigenesis. Haploinsufficient mice (βIISp+/−) spontaneously developed hepatocellular carcinoma (90). These preliminary indications require further research. Nevertheless, it can be safely concluded that fodrin undergoes changes in its localization pattern and expression level in response to pathological stresses during cancer. This characteristic could be utilized to serve as a preliminary marker for neoplasia.
CONCLUSION
A robust cortical cytoskeleton is crucial for the survival of a eukaryotic cell. Both spectrin and fodrin help achieve this goal in different tissue systems. The canonical structure of fodrin provides for a flexible yet strong structure, permitting engagement in a plethora of functions, namely, cell signaling, DNA repair pathways, microtubule nucleation, cell proliferation, angiogenesis regulation, and apoptotic mechanisms. Considerable variation from this structure produces spectrin with focused and specific functionality. However, studies concerning both proteins agree about the indispensable nature of these isoforms toward structural support. Interestingly, the diverse functions of fodrin have only recently come to light. Studies conducted on various disease models for cancer and neurodegenerative disorders highlight the significance of this molecule in the regulation of these diseases. Modulating the spatial and temporal availability of fodrin appears to be a common hallmark of various disease conditions. The underlying mechanism and its regulation are still the thrust areas of research. It is expected that through this review that underlines the apparent diversity of these molecules and the various cellular functions performed by them, the focus will now shift toward understanding the detailed mechanisms that define the parameters of this diversity. This will thus help us solve certain key questions regarding the development of diseases.
ACKNOWLEDGMENTS
We acknowledge the Department of Science and Technology-Science Education and Research Board, Government of India, for extramural funding and the Department of Biotechnology, Rajiv Gandhi Center for Biotechnology, India, for core funding. J.S.S. and D.D. were provided fellowships by the University Grants Commission, Government of India, and the Indian Council for Medical Research, Government of India.
J.S.S. and S.S. were involved in writing the manuscript, data analysis, and conceptualization. R.J., D.D., and R.K.N. contributed to writing, data analysis, and critical reading. S.S. supervised and provided the funding. All authors approved the final form.
We declare that there is no conflict of interest.
REFERENCES
- 1.Bennett V, Gilligan DM. 1993. The spectrin-based membrane skeleton and micron-scale organization of the plasma membrane. Annu Rev Cell Biol 9:27–66. doi: 10.1146/annurev.cb.09.110193.000331. [DOI] [PubMed] [Google Scholar]
- 2.Winkelmann JC, Forget BG. 1993. Erythroid and nonerythroid spectrins. Blood 81:3173–3185. doi: 10.1182/blood.V81.12.3173.3173. [DOI] [PubMed] [Google Scholar]
- 3.Bennett V, Baines AJ. 2001. Spectrin and ankyrin-based pathways: metazoan inventions for integrating cells into tissues. Physiol Rev 81:1353–1392. doi: 10.1152/physrev.2001.81.3.1353. [DOI] [PubMed] [Google Scholar]
- 4.Berghs S, Aggujaro D, Dirkx R, Maksimova E, Stabach P, Hermel JM, Zhang JP, Philbrick W, Slepnev V, Ort T, Solimena M. 2000. βIV spectrin, a new spectrin localized at axon initial segments and nodes of Ranvier in the central and peripheral nervous system. J Cell Biol 151:985–1001. doi: 10.1083/jcb.151.5.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stabach PR, Morrow JS. 2000. Identification and characterization of βV spectrin, a mammalian ortholog of Drosophila β(H) spectrin. J Biol Chem 275:21385–21395. doi: 10.1074/jbc.C000159200. [DOI] [PubMed] [Google Scholar]
- 6.Stankewich MC, Tse WT, Peters LL, Ch’ng Y, John KM, Stabach PR, Devarajan P, Morrow JS, Lux SE. 1998. A widely expressed βIII spectrin associated with Golgi and cytoplasmic vesicles. Proc Natl Acad Sci U S A 95:14158–14163. doi: 10.1073/pnas.95.24.14158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong SY, Haack H, Kissil JL, Barry M, Bronson RT, Shen SS, Whittaker CA, Crowley D, Hynes RO. 2007. Protein 4.1B suppresses prostate cancer progression and metastasis. Proc Natl Acad Sci U S A 104:12784–12789. doi: 10.1073/pnas.0705499104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lorenz T, Willard M. 1978. Subcellular fractionation of intra-axonally transported polypeptides in the rabbit visual system. Proc Natl Acad Sci U S A 75:505–509. doi: 10.1073/pnas.75.1.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levine J, Willard M. 1981. Fodrin: axonally transported polypeptides associated with the internal periphery of many cells. J Cell Biol 90:631–642. doi: 10.1083/jcb.90.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanda K, Tanaka T, Sobue K. 1986. Calspectin (fodrin or nonerythroid spectrin)-actin interaction: a possible involvement of 4.1-related protein. Biochem Biophys Res Commun 140:1051–1058. doi: 10.1016/0006-291x(86)90741-2. [DOI] [PubMed] [Google Scholar]
- 11.Moon RT, McMahon AP. 1990. Generation of diversity in nonerythroid spectrins. J Biol Chem 265:4427–4433. [PubMed] [Google Scholar]
- 12.Grum VL, Li D, MacDonald RI, Mondragón A. 1999. Structures of two repeats of spectrin suggest models of flexibility. Cell 98:523–535. doi: 10.1016/s0092-8674(00)81980-7. [DOI] [PubMed] [Google Scholar]
- 13.Broderick MJF, Winder SJ. 2005. Spectrin, α-actinin, and dystrophin. Adv Protein Chem 70:203–246. doi: 10.1016/S0065-3233(05)70007-3. [DOI] [PubMed] [Google Scholar]
- 14.Speicher DW, Weglarz L, DeSilva TM. 1992. Properties of human red cell spectrin heterodimer (side-to-side) assembly and identification of an essential nucleation site. J Biol Chem 267:14775–14782. [PubMed] [Google Scholar]
- 15.Ipsaro JJ, Harper SL, Messick TE, Marmorstein R, Mondragón A, Speicher DW. 2010. Crystal structure and functional interpretation of the erythrocyte spectrin tetramerization domain complex. Blood 115:4843–4852. doi: 10.1182/blood-2010-01-261396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leto TL, Fortugno-Erikson D, Barton D, Yang-Feng TL, Francke U, Harris AS, Morrow JS, Marchesi VT, Benz EJ Jr.. 1988. Comparison of nonerythroid alpha-spectrin genes reveals strict homology among diverse species. Mol Cell Biol 8:1–9. doi: 10.1128/mcb.8.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glenney JR, Glenney P. 1984. Comparison of spectrin isolated from erythroid and non‐erythroid sources. Eur J Biochem 144:529–539. doi: 10.1111/j.1432-1033.1984.tb08498.x. [DOI] [PubMed] [Google Scholar]
- 18.Martin SJ, O’Brien GA, Nishioka WK, McGahon AJ, Mahboubi A, Saido TC, Green DR. 1995. Proteolysis of fodrin (non-erythroid spectrin) during apoptosis. J Biol Chem 270:6425–6428. doi: 10.1074/jbc.270.12.6425. [DOI] [PubMed] [Google Scholar]
- 19.Wallis CJ, Wenegieme EF, Babitch JA. 1992. Characterization of calcium binding to brain spectrin. J Biol Chem 267:4333–4337. [PubMed] [Google Scholar]
- 20.Baines AJ. 2009. Evolution of spectrin function in cytoskeletal and membrane networks. Biochem Soc Trans 37:796–803. doi: 10.1042/BST0370796. [DOI] [PubMed] [Google Scholar]
- 21.Tsukita S, Tsukita S, Ishikawa H, Kurokawa M, Morimoto K, Sobue K, Kakiuchi S. 1983. Binding sites of calmodulin and actin on the brain spectrin, calspectin. J Cell Biol 97:574–578. doi: 10.1083/jcb.97.2.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leto TL, Pleasic S, Forget BG, Benz EJ Jr, Marchesi VT. 1989. Characterization of the calmodulin-binding site of nonerythroid alpha spectrin. J Biol Chem 264:5826–5830. [PubMed] [Google Scholar]
- 23.Steiner JP, Walke HT, Bennett V. 1989. Calcium/calmodulin inhibits direct binding of spectrin to synaptosomal membranes. J Biol Chem 264:2783–2791. [PubMed] [Google Scholar]
- 24.Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E. 2015. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res 43:D512–D520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Byers TJ, Branton D. 1985. Visualization of the protein association in the erythrocyte membrane skeleton. Proc Natl Acad Sci U S A 82:6153–6157. doi: 10.1073/pnas.82.18.6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown JW, Bullitt E, Sriswasdi S, Harper S, Speicher DW, McKnight CJ. 2015. The physiological molecular shape of spectrin: a compact supercoil resembling a Chinese finger trap. PLoS Comput Biol 11:e1004302. doi: 10.1371/journal.pcbi.1004302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu K, Zhong G, Zhuang X. 2013. Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science 339:452–456. doi: 10.1126/science.1232251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sridharan DM, McMahon LW, Lambert MW. 2006. αII-spectrin interacts with five groups of functionally important proteins in the nucleus. Cell Biol Int 30:866–878. doi: 10.1016/j.cellbi.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 29.Delaunay J. 2007. The molecular basis of hereditary red cell membrane disorders. Blood Rev 21:1–20. doi: 10.1016/j.blre.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 30.Perrotta S, Gallagher PG, Mohandas N. 2008. Hereditary spherocytosis. Lancet 372:1411–1426. doi: 10.1016/S0140-6736(08)61588-3. [DOI] [PubMed] [Google Scholar]
- 31.Glenney JR, Glenney P, Weber K. 1982. F-actin-binding and cross-linking properties of porcine brain fodrin, a spectrin-related molecule. J Biol Chem 257:9781–9787. [PubMed] [Google Scholar]
- 32.Levine J, Willard M. 1983. Redistribution of fodrin (a component of the cortical cytoplasm) accompanying capping of cell surface molecules. Proc Natl Acad Sci U S A 80:191–195. doi: 10.1073/pnas.80.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burns NR, Ohanian V, Gratzer WB. 1983. Properties of brain spectrin (fodrin). FEBS Lett 153:165–168. doi: 10.1016/0014-5793(83)80140-9. [DOI] [PubMed] [Google Scholar]
- 34.Lambert MW. 2018. Spectrin and its interacting partners in nuclear structure and function. Exp Biol Med (Maywood) 243:507–524. doi: 10.1177/1535370218763563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karinch AM, Zimmer WE, Goodman SR. 1990. The identification and sequence of the actin-binding domain of human red blood cell β-spectrin. J Biol Chem 265:11833–11840. [PubMed] [Google Scholar]
- 36.Machnicka B, Czogalla A, Hryniewicz-Jankowska A, Bogusławska DM, Grochowalska R, Heger E, Sikorski AF. 2014. Spectrins: a structural platform for stabilization and activation of membrane channels, receptors and transporters. Biochim Biophys Acta 1838:620–634. doi: 10.1016/j.bbamem.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 37.Pradhan D, Morrow JS. 2002. The spectrin-ankyrin skeleton controls CD45 surface display and interleukin-2 production. Immunity 17:303–315. doi: 10.1016/S1074-7613(02)00396-5. [DOI] [PubMed] [Google Scholar]
- 38.Hortsch M, Nagaraj K, Godenschwege TA. 2009. The interaction between L1-type proteins and ankyrins—a master switch for L1-type CAM function. Cell Mol Biol Lett 14:57–69. doi: 10.2478/s11658-008-0035-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou R, Han B, Xia C, Zhuang X. 2019. Membrane-associated periodic skeleton is a signaling platform for RTK transactivation in neurons. Science 365:929–934. doi: 10.1126/science.aaw5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bennett V, Healy J. 2009. Membrane domains based on ankyrin and spectrin associated with cell-cell interactions. Cold Spring Harb Perspect Biol 1:a003012. doi: 10.1101/cshperspect.a003012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wechsler A, Teichberg VI. 1998. Brain spectrin binding to the NMDA receptor is regulated by phosphorylation, calcium and calmodulin. EMBO J 17:3931–3939. doi: 10.1093/emboj/17.14.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bialkowska K, Saido TC, Fox JEB. 2005. SH3 domain of spectrin participates in the activation of Rac in specialized calpain-induced integrin signaling complexes. J Cell Sci 118:381–395. doi: 10.1242/jcs.01625. [DOI] [PubMed] [Google Scholar]
- 43.Metral S, Machnicka B, Bigot S, Colin Y, Dhermy D, Lecomte M-C. 2009. AlphaII-spectrin is critical for cell adhesion and cell cycle. J Biol Chem 284:2409–2418. doi: 10.1074/jbc.M801324200. [DOI] [PubMed] [Google Scholar]
- 44.Derry JMJ, Ochs HD, Francke U. 1994. Isolation of a novel gene mutated in Wiskott-Aldrich syndrome. Cell 78:635–644. doi: 10.1016/0092-8674(94)90528-2. [DOI] [PubMed] [Google Scholar]
- 45.Dubielecka PM, Ladwein KI, Xiong X, Migeotte I, Chorzalska A, Anderson KV, Sawicki JA, Rottner K, Stradal TE, Kotula L. 2011. Essential role for Abi1 in embryonic survival and WAVE2 complex integrity. Proc Natl Acad Sci U S A 108:7022–7027. doi: 10.1073/pnas.1016811108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rotter B, Bournier O, Nicolas G, Dhermy D, Lecomte MC. 2005. αII-spectrin interacts with Tes and EVL, two actin-binding proteins located at cell contacts. Biochem J 388:631–638. doi: 10.1042/BJ20041502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wong KKL, Li W, An Y, Duan Y, Li Z, Kang Y, Yan Y. 2015. β-Spectrin regulates the Hippo signaling pathway and modulates the basal actin network. J Biol Chem 290:6397–6407. doi: 10.1074/jbc.M114.629493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deng H, Yang L, Wen P, Lei H, Blount P, Pan D. 2020. Spectrin couples cell shape, cortical tension, and Hippo signaling in retinal epithelial morphogenesis. J Cell Biol 219:e201907018. doi: 10.1083/jcb.201907018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perrin D, Aunis D. 1985. Reorganization of α-fodrin induced by stimulation in secretory cells. Nature 315:589–592. doi: 10.1038/315589a0. [DOI] [PubMed] [Google Scholar]
- 50.Perrin D, Langley OK, Aunis D. 1987. Anti-α-fodrin inhibits secretion from permeabilized chromaffin cells. Nature 326:498–501. doi: 10.1038/326498a0. [DOI] [PubMed] [Google Scholar]
- 51.Zagon I, Higbee R, Riederer B, Goodman S. 1986. Spectrin subtypes in mammalian brain: an immunoelectron microscopic study. J Neurosci 6:2977–2986. doi: 10.1523/JNEUROSCI.06-10-02977.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sikorski AF, Terlecki G, Zagon IS, Goodman SR. 1991. Synapsin I-mediated interaction of brain spectrin with synaptic vesicles. J Cell Biol 114:313–318. doi: 10.1083/jcb.114.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benfenati F, Valtorta F, Rubenstein JL, Gorelick FS, Greengard P, Czernik AJ. 1992. Synaptic vesicle-associated Ca2+/calmodulin-dependent protein kinase II is a binding protein for synapsin I. Nature 359:417–420. doi: 10.1038/359417a0. [DOI] [PubMed] [Google Scholar]
- 54.Benfenati F, Valtorta F, Chieregatti E, Greengard P. 1992. Interaction of free and synaptic vesicle-bound synapsin I with F-actin. Neuron 8:377–386. doi: 10.1016/0896-6273(92)90303-u. [DOI] [PubMed] [Google Scholar]
- 55.McMahon LW, Zhang P, Sridharan DM, Lefferts JA, Lambert MW. 2009. Knockdown of alphaII spectrin in normal human cells by siRNA leads to chromosomal instability and decreased DNA interstrand cross-link repair. Biochem Biophys Res Commun 381:288–293. doi: 10.1016/j.bbrc.2009.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang C, Lambert MW. 2010. The Fanconi anemia protein, FANCG, binds to the ERCC1-XPF endonuclease via its tetratricopeptide repeats and the central domain of ERCC1. Biochemistry 49:5560–5569. doi: 10.1021/bi100584c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lefferts JA, Wang C, Sridharan D, Baralt M, Lambert MW. 2009. The SH3 domain of αII spectrin is a target for the Fanconi anemia protein, FANCG. Biochemistry 48:254–263. doi: 10.1021/bi801483u. [DOI] [PubMed] [Google Scholar]
- 58.Nellikka RK, Sreeja JS, Dharmapal D, John R, Monteiro A, Macedo JC, Conde C, Logarinho E, Sunkel CE, Sengupta S. 2019. α-Fodrin is required for the organization of functional microtubules during mitosis. Cell Cycle 18:2713–2726. doi: 10.1080/15384101.2019.1656476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sreeja JS, Nellikka RK, John R, Sivakumar KC, Sreekumar E, Sengupta S. 2019. Binding of alpha‐fodrin to gamma‐tubulin accounts for its role in the inhibition of microtubule nucleation. FEBS Lett 593:1154–1165. doi: 10.1002/1873-3468.13425. [DOI] [PubMed] [Google Scholar]
- 60.Thomas NE, Shashikala S, Sengupta S. 2010. Cytoplasmic gamma-tubulin complex from brain contains nonerythroid spectrin. J Cell Biochem 110:1334–1341. doi: 10.1002/jcb.22647. [DOI] [PubMed] [Google Scholar]
- 61.Shashikala S, Kumar R, Thomas NE, Sivadasan D, James J, Sengupta S. 2013. Fodrin in centrosomes: implication of a role of fodrin in the transport of gamma-tubulin complex in brain. PLoS One 8:e76613. doi: 10.1371/journal.pone.0076613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baek HJ, Pishvaian MJ, Tang Y, Kim TH, Yang S, El Zouhairi M, Mendelson J, Shetty K, Kallakury B, Berry DL, Shin KH, Mishra B, Reddy EP, Kim SS, Mishra L. 2011. Transforming growth factor-β adaptor, β2-spectrin, modulates cyclin dependent kinase 4 to reduce development of hepatocellular cancer. Hepatology 53:1676–1684. doi: 10.1002/hep.24128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thenappan A, Shukla V, Khalek FJA, Li Y, Shetty K, Liu P, Li L, Johnson RL, Johnson L, Mishra L. 2011. Loss of transforming growth factor β adaptor protein β-2 spectrin leads to delayed liver regeneration in mice. Hepatology 53:1641–1650. doi: 10.1002/hep.24111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hanahan D, Weinberg RA. 2000. The hallmarks of cancer. Cell 100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 65.Lin S, Xie J, Gong T, Shi S, Zhang T, Fu N, Lin Y. 2016. Smad signal pathway regulates angiogenesis via endothelial cell in an adipose-derived stromal cell/endothelial cell co-culture, 3D gel model. Mol Cell Biochem 412:281–288. doi: 10.1007/s11010-015-2634-5. [DOI] [PubMed] [Google Scholar]
- 66.Tang Y, Katuri V, Dillner A, Mishra B, Deng C-X, Mishra L. 2003. Disruption of transforming growth factor-beta signaling in ELF beta-spectrin-deficient mice. Science 299:574–577. doi: 10.1126/science.1075994. [DOI] [PubMed] [Google Scholar]
- 67.Baek HJ, Lim SC, Kitisin K, Jogunoori W, Tang Y, Marshall MB, Mishra B, Kim TH, Cho KH, Kim SS, Mishra L. 2008. Hepatocellular cancer arises from loss of transforming growth factor beta signaling adaptor protein embryonic liver fodrin through abnormal angiogenesis. Hepatology 48:1128–1137. doi: 10.1002/hep.22460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stankewich MC, Cianci CD, Stabach PR, Ji L, Nath A, Morrow JS. 2011. Cell organization, growth, and neural and cardiac development require αII-spectrin. J Cell Sci 124:3956–3966. doi: 10.1242/jcs.080374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Derbala MH, Guo AS, Mohler PJ, Smith SA. 2018. The role of βII spectrin in cardiac health and disease. Life Sci 192:278–285. doi: 10.1016/j.lfs.2017.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Unudurthi SD, Greer-Short A, Patel N, Nassal D, Hund TJ. 2018. Spectrin-based pathways underlying electrical and mechanical dysfunction in cardiac disease. Expert Rev Cardiovasc Ther 16:59–65. doi: 10.1080/14779072.2018.1418664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Janicke RU, Ng P, Sprengart ML, Porter AG. 1998. Caspase-3 is required for alpha-fodrin cleavage but dispensable for cleavage of other death substrates in apoptosis. J Biol Chem 273:15540–15545. doi: 10.1074/jbc.273.25.15540. [DOI] [PubMed] [Google Scholar]
- 72.Wang KKW, Posmantur R, Nath R, McGinnis K, Whitton M, Talanian RV, Glantz SB, Morrow JS. 1998. Simultaneous degradation of αII- and βII-spectrin by caspase 3 (CPP32) in apoptotic cells. J Biol Chem 273:22490–22497. doi: 10.1074/jbc.273.35.22490. [DOI] [PubMed] [Google Scholar]
- 73.Fox JE, Reynolds CC, Morrow JS, Phillips DR. 1987. Spectrin is associated with membrane-bound actin filaments in platelets and is hydrolyzed by the Ca2+-dependent protease during platelet activation. Blood 69:537–545. doi: 10.1182/blood.V69.2.537.537. [DOI] [PubMed] [Google Scholar]
- 74.Saitsu H, Tohyama J, Kumada T, Egawa K, Hamada K, Okada I, Mizuguchi T, Osaka H, Miyata R, Furukawa T, Haginoya K, Hoshino H, Goto T, Hachiya Y, Yamagata T, Saitoh S, Nagai T, Nishiyama K, Nishimura A, Miyake N, Komada M, Hayashi K, Hirai S, Ogata K, Kato M, Fukuda A, Matsumoto N. 2010. Dominant-negative mutations in α-II spectrin cause West syndrome with severe cerebral hypomyelination, spastic quadriplegia, and developmental delay. Am J Hum Genet 86:881–891. doi: 10.1016/j.ajhg.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nonoda Y, Saito Y, Nagai S, Sasaki M, Iwasaki T, Matsumoto N, Ishii M, Saitsu H. 2013. Progressive diffuse brain atrophy in West syndrome with marked hypomyelination due to SPTAN1 gene mutation. Brain Dev 35:280–283. doi: 10.1016/j.braindev.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 76.Koushika SP, Schaefer AM, Vincent R, Willis JH, Bowerman B, Nonet ML. 2004. Mutations in Caenorhabditis elegans cytoplasmic dynein components reveal specificity of neuronal retrograde cargo. J Neurosci 24:3907–3916. doi: 10.1523/JNEUROSCI.5039-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Muresan V, Stankewich MC, Steffen W, Morrow JS, Holzbaur ELF, Schnapp BJ. 2001. Dynactin-dependent, dynein-driven vesicle transport in the absence of membrane proteins. Mol Cell 7:173–183. doi: 10.1016/S1097-2765(01)00165-4. [DOI] [PubMed] [Google Scholar]
- 78.Fernández-Shaw C, Marina A, Cazorla P, Valdivieso F, Vázquez J. 1997. Anti-brain spectrin immunoreactivity in Alzheimer’s disease: degradation of spectrin in an animal model of cholinergic degeneration. J Neuroimmunol 77:91–98. doi: 10.1016/S0165-5728(97)00066-0. [DOI] [PubMed] [Google Scholar]
- 79.Ackermann A, Brieger A. 2019. The role of nonerythroid spectrin αII in cancer. J Oncol 2019:7079604. doi: 10.1155/2019/7079604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Younes M, Harris AS, Morrow JS. 1989. Fodrin as a differentiation marker. Redistributions in colonic neoplasia. Am J Pathol 135:1197–1212. [PMC free article] [PubMed] [Google Scholar]
- 81.Lee S, Baek M, Yang H, Bang Y-J, Kim WH, Ha J-H, Kim D-K, Jeoung D-I. 2002. Identification of genes differentially expressed between gastric cancers and normal gastric mucosa with cDNA microarrays. Cancer Lett 184:197–206. doi: 10.1016/S0304-3835(02)00197-0. [DOI] [PubMed] [Google Scholar]
- 82.Sormunen R, Pääkkö P, Palovuori R, Soini Y, Lehto VP. 1994. Fodrin and actin in the normal, metaplastic, and dysplastic respiratory epithelium and in lung carcinoma. Am J Respir Cell Mol Biol 11:75–84. doi: 10.1165/ajrcmb.11.1.8018340. [DOI] [PubMed] [Google Scholar]
- 83.Simpson JF, Page DL. 1992. Altered expression of a structural protein (fodrin) within epithelial proliferative disease of the breast. Am J Pathol 141:285–289. [PMC free article] [PubMed] [Google Scholar]
- 84.Tuominen H, Sormunen R, Kallioinen M. 1996. Non-erythroid spectrin (fodrin) in cutaneous tumours: diminished in cell membranes, increased in the cytoplasm. Br J Dermatol 135:576–580. doi: 10.1111/j.1365-2133.1996.tb03834.x. [DOI] [PubMed] [Google Scholar]
- 85.Bii VM, Collins CP, Hocum JD, Trobridge GD. 2018. Replication-incompetent gammaretroviral and lentiviral vector-based insertional mutagenesis screens identify prostate cancer progression genes. Oncotarget 9:15451–15463. doi: 10.18632/oncotarget.24503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sun Z, Wang L, Eckloff BW, Deng B, Wang Y, Wampfler JA, Jang J, Wieben ED, Jen J, You M, Yang P. 2014. Conserved recurrent gene mutations correlate with pathway deregulation and clinical outcomes of lung adenocarcinoma in never-smokers. BMC Med Genomics 7:32. doi: 10.1186/1755-8794-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hinrichsen I, Ernst BP, Nuber F, Passmann S, Schäfer D, Steinke V, Friedrichs N, Plotz G, Zeuzem S, Brieger A. 2014. Reduced migration of MLH1 deficient colon cancer cells depends on SPTAN1. Mol Cancer 13:11. doi: 10.1186/1476-4598-13-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.L’Espérance S, Popa I, Bachvarova M, Plante M, Patten N, Wu L, Têtu B, Bachvarov D. 2006. Gene expression profiling of paired ovarian tumors obtained prior to and following adjuvant chemotherapy: molecular signatures of chemoresistant tumors. Int J Oncol 29:5–24. doi: 10.3892/ijo.29.1.5. [DOI] [PubMed] [Google Scholar]
- 89.Dubielecka PM, Jaźwiec B, Potoczek S, Wróbel T, Miłoszewska J, Haus O, Kuliczkowski K, Sikorski AF. 2005. Changes in spectrin organisation in leukaemic and lymphoid cells upon chemotherapy. Biochem Pharmacol 69:73–85. doi: 10.1016/j.bcp.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 90.Zhi X, Lin L, Yang S, Bhuvaneshwar K, Wang H, Gusev Y, Lee MH, Kallakury B, Shivapurkar N, Cahn K, Tian X, Marshall JL, Byers SW, He AR. 2015. βII-spectrin (SPTBN1) suppresses progression of hepatocellular carcinoma and Wnt signaling by regulation of Wnt inhibitor kallistatin. Hepatology 61:598–612. doi: 10.1002/hep.27558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mayya V, Lundgren DH, Hwang S-I, Rezaul K, Wu L, Eng JK, Rodionov V, Han DK. 2009. Quantitative phosphoproteomic analysis of T cell receptor signaling reveals system-wide modulation of protein-protein interactions. Sci Signal 2:ra46. doi: 10.1126/scisignal.2000007. [DOI] [PubMed] [Google Scholar]
- 92.Tsai C-F, Wang Y-T, Yen H-Y, Tsou C-C, Ku W-C, Lin P-Y, Chen H-Y, Nesvizhskii AI, Ishihama Y, Chen Y-J. 2015. Large-scale determination of absolute phosphorylation stoichiometries in human cells by motif-targeting quantitative proteomics. Nat Commun 6:6622. doi: 10.1038/ncomms7622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, Brunak S, Mann M. 2010. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal 3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 94.Mertins P, Yang F, Liu T, Mani DR, Petyuk VA, Gillette MA, Clauser KR, Qiao JW, Gritsenko MA, Moore RJ, Levine DA, Townsend R, Erdmann-Gilmore P, Snider JE, Davies SR, Ruggles KV, Fenyo D, Kitchens RT, Li S, Olvera N, Dao F, Rodriguez H, Chan DW, Liebler D, White F, Rodland KD, Mills GB, Smith RD, Paulovich AG, Ellis M, Carr SA. 2014. Ischemia in tumors induces early and sustained phosphorylation changes in stress kinase pathways but does not affect global protein levels. Mol Cell Proteomics 13:1690–1704. doi: 10.1074/mcp.M113.036392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mertins P, Mani DR, Ruggles KV, Gillette MA, Clauser KR, Wang P, Wang X, Qiao JW, Cao S, Petralia F, Kawaler E, Mundt F, Krug K, Tu Z, Lei JT, Gatza ML, Wilkerson M, Perou CM, Yellapantula V, Huang K, Lin C, McLellan MD, Yan P, Davies SR, Townsend RR, Skates SJ, Wang J, Zhang B, Kinsinger CR, Mesri M, Rodriguez H, Ding L, Paulovich AG, Fenyö D, Ellis MJ, Carr SA, NCI CPTAC . 2016. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 534:55–62. doi: 10.1038/nature18003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yi T, Zhai B, Yu Y, Kiyotsugu Y, Raschle T, Etzkorn M, Seo H-C, Nagiec M, Luna RE, Reinherz EL, Blenis J, Gygi SP, Wagner G. 2014. Quantitative phosphoproteomic analysis reveals system-wide signaling pathways downstream of SDF-1/CXCR4 in breast cancer stem cells. Proc Natl Acad Sci U S A 111:E2182–E2190. doi: 10.1073/pnas.1404943111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kettenbach AN, Schweppe DK, Faherty BK, Pechenick D, Pletnev AA, Gerber SA. 2011. Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci Signal 4:rs5. doi: 10.1126/scisignal.2001497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shiromizu T, Adachi J, Watanabe S, Murakami T, Kuga T, Muraoka S, Tomonaga T. 2013. Identification of missing proteins in the neXtProt database and unregistered phosphopeptides in the PhosphoSitePlus database as part of the chromosome-centric Human Proteome Project. J Proteome Res 12:2414–2421. doi: 10.1021/pr300825v. [DOI] [PubMed] [Google Scholar]
- 99.Sharma K, D’Souza RCJ, Tyanova S, Schaab C, Wiśniewski JR, Cox J, Mann M. 2014. Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep 8:1583–1594. doi: 10.1016/j.celrep.2014.07.036. [DOI] [PubMed] [Google Scholar]
- 100.Carrier M, Joint M, Lutzing R, Page A, Rochette-Egly C. 2016. Phosphoproteome and transcriptome of RA-responsive and RA-resistant breast cancer cell lines. PLoS One 11:e0157290. doi: 10.1371/journal.pone.0157290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bian Y, Song C, Cheng K, Dong M, Wang F, Huang J, Sun D, Wang L, Ye M, Zou H. 2014. An enzyme assisted RP-RPLC approach for in-depth analysis of human liver phosphoproteome. J Proteomics 96:253–262. doi: 10.1016/j.jprot.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 102.Franz-Wachtel M, Eisler SA, Krug K, Wahl S, Carpy A, Nordheim A, Pfizenmaier K, Hausser A, Macek B. 2012. Global detection of protein kinase D-dependent phosphorylation events in nocodazole-treated human cells. Mol Cell Proteomics 11:160–170. doi: 10.1074/mcp.M111.016014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Han G, Ye M, Liu H, Song C, Sun D, Wu Y, Jiang X, Chen R, Wang C, Wang L, Zou H. 2010. Phosphoproteome analysis of human liver tissue by long-gradient nanoflow LC coupled with multiple stage MS analysis. Electrophoresis 31:1080–1089. doi: 10.1002/elps.200900493. [DOI] [PubMed] [Google Scholar]
- 104.Zhou H, Di Palma S, Preisinger C, Peng M, Polat AN, Heck AJR, Mohammed S. 2013. Toward a comprehensive characterization of a human cancer cell phosphoproteome. J Proteome Res 12:260–271. doi: 10.1021/pr300630k. [DOI] [PubMed] [Google Scholar]
- 105.Lundby A, Secher A, Lage K, Nordsborg NB, Dmytriyev A, Lundby C, Olsen JV. 2012. Quantitative maps of protein phosphorylation sites across 14 different rat organs and tissues. Nat Commun 3:876. doi: 10.1038/ncomms1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Palacios-Moreno J, Foltz L, Guo A, Stokes MP, Kuehn ED, George L, Comb M, Grimes ML. 2015. Neuroblastoma tyrosine kinase signaling networks involve FYN and LYN in endosomes and lipid rafts. PLoS Comput Biol 11:e1004130. doi: 10.1371/journal.pcbi.1004130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Johnson H, Lescarbeau RS, Gutierrez JA, White FM. 2013. Phosphotyrosine profiling of NSCLC cells in response to EGF and HGF reveals network specific mediators of invasion. J Proteome Res 12:1856–1867. doi: 10.1021/pr301192t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rigbolt KTG, Prokhorova TA, Akimov V, Henningsen J, Johansen PT, Kratchmarova I, Kassem M, Mann M, Olsen JV, Blagoev B. 2011. System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci Signal 4:rs3. doi: 10.1126/scisignal.2001570. [DOI] [PubMed] [Google Scholar]
- 109.Iliuk AB, Martin VA, Alicie BM, Geahlen RL, Tao WA. 2010. In-depth analyses of kinase-dependent tyrosine phosphoproteomes based on metal ion-functionalized soluble nanopolymers. Mol Cell Proteomics 9:2162–2172. doi: 10.1074/mcp.M110.000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Moritz A, Li Y, Guo A, Villen J, Wang Y, MacNeill J, Kornhauser J, Sprott K, Zhou J, Possemato A, Ren JM, Hornbeck P, Cantley LC, Gygi SP, Rush J, Comb MJ. 2010. Akt-RSK-S6 kinase signaling networks activated by oncogenic receptor tyrosine kinases. Sci Signal 3:ra64. doi: 10.1126/scisignal.2000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nedrelow JH, Cianci CD, Morrow JS. 2003. c-Src binds alpha II spectrin’s Src homology 3 (SH3) domain and blocks calpain susceptibility by phosphorylating Tyr1176. J Biol Chem 278:7735–7741. doi: 10.1074/jbc.M210988200. [DOI] [PubMed] [Google Scholar]
- 112.Klammer M, Kaminski M, Zedler A, Oppermann F, Blencke S, Marx S, Muller S, Tebbe A, Godl K, Schaab C. 2012. Phosphosignature predicts dasatinib response in non-small cell lung cancer. Mol Cell Proteomics 11:651–668. doi: 10.1074/mcp.M111.016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chen Y, Choong L-Y, Lin Q, Philp R, Wong C-H, Ang B-K, Tan Y-L, Loh M-C-S, Hew C-L, Shah N, Druker BJ, Chong P-K, Lim Y-P. 2007. Differential expression of novel tyrosine kinase substrates during breast cancer development. Mol Cell Proteomics 6:2072–2087. doi: 10.1074/mcp.M700395-MCP200. [DOI] [PubMed] [Google Scholar]
- 114.Schweppe DK, Rigas JR, Gerber SA. 2013. Quantitative phosphoproteomic profiling of human non-small cell lung cancer tumors. J Proteomics 91:286–296. doi: 10.1016/j.jprot.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kim J-Y, Welsh EA, Oguz U, Fang B, Bai Y, Kinose F, Bronk C, Remsing Rix LL, Beg AA, Rix U, Eschrich SA, Koomen JM, Haura EB. 2013. Dissection of TBK1 signaling via phosphoproteomics in lung cancer cells. Proc Natl Acad Sci U S A 110:12414–12419. doi: 10.1073/pnas.1220674110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Santamaria A, Wang B, Elowe S, Malik R, Zhang F, Bauer M, Schmidt A, Silljé HHW, Körner R, Nigg EA. 2011. The Plk1-dependent phosphoproteome of the early mitotic spindle. Mol Cell Proteomics 10:M110.004457. doi: 10.1074/mcp.M110.004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Christensen GL, Kelstrup CD, Lyngso C, Sarwar U, Bogebo R, Sheikh SP, Gammeltoft S, Olsen JV, Hansen JL. 2010. Quantitative phosphoproteomics dissection of seven-transmembrane receptor signaling using full and biased agonists. Mol Cell Proteomics 9:1540–1553. doi: 10.1074/mcp.M900550-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mertins P, Qiao JW, Patel J, Udeshi ND, Clauser KR, Mani DR, Burgess MW, Gillette MA, Jaffe JD, Carr SA. 2013. Integrated proteomic analysis of post-translational modifications by serial enrichment. Nat Methods 10:634–637. doi: 10.1038/nmeth.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. 2006. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 120.Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, Nardone J, Lee K, Reeves C, Li Y, Hu Y, Tan Z, Stokes M, Sullivan L, Mitchell J, Wetzel R, MacNeill J, Ren JM, Yuan J, Bakalarski CE, Villen J, Kornhauser JM, Smith B, Li D, Zhou X, Gygi SP, Gu T-L, Polakiewicz RD, Rush J, Comb MJ. 2007. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 121.Gu T-L, Deng X, Huang F, Tucker M, Crosby K, Rimkunas V, Wang Y, Deng G, Zhu L, Tan Z, Hu Y, Wu C, Nardone J, MacNeill J, Ren J, Reeves C, Innocenti G, Norris B, Yuan J, Yu J, Haack H, Shen B, Peng C, Li H, Zhou X, Liu X, Rush J, Comb MJ. 2011. Survey of tyrosine kinase signaling reveals ROS kinase fusions in human cholangiocarcinoma. PLoS One 6:e15640. doi: 10.1371/journal.pone.0015640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ruse CI, McClatchy DB, Lu B, Cociorva D, Motoyama A, Park SK, Yates JR III. 2008. Motif-specific sampling of phosphoproteomes. J Proteome Res 7:2140–2150. doi: 10.1021/pr800147u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Casado P, Alcolea MP, Iorio F, Rodriguez-Prados J-C, Vanhaesebroeck B, Saez-Rodriguez J, Joel S, Cutillas PR. 2013. Phosphoproteomics data classify hematological cancer cell lines according to tumor type and sensitivity to kinase inhibitors. Genome Biol 14:R37. doi: 10.1186/gb-2013-14-4-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Johnson H, White FM. 2014. Quantitative analysis of signaling networks across differentially embedded tumors highlights interpatient heterogeneity in human glioblastoma. J Proteome Res 13:4581–4593. doi: 10.1021/pr500418w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. 2009. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 126.Akimov V, Barrio-Hernandez I, Hansen SVF, Hallenborg P, Pedersen A-K, Bekker-Jensen DB, Puglia M, Christensen SDK, Vanselow JT, Nielsen MM, Kratchmarova I, Kelstrup CD, Olsen JV, Blagoev B. 2018. UbiSite approach for comprehensive mapping of lysine and N-terminal ubiquitination sites. Nat Struct Mol Biol 25:631–640. doi: 10.1038/s41594-018-0084-y. [DOI] [PubMed] [Google Scholar]
- 127.Udeshi ND, Svinkina T, Mertins P, Kuhn E, Mani DR, Qiao JW, Carr SA. 2013. Refined preparation and use of anti-diglycine remnant (K-epsilon-GG) antibody enables routine quantification of 10,000s of ubiquitination sites in single proteomics experiments. Mol Cell Proteomics 12:825–831. doi: 10.1074/mcp.O112.027094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, Harper JW, Gygi SP. 2011. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell 44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Povlsen LK, Beli P, Wagner SA, Poulsen SL, Sylvestersen KB, Poulsen JW, Nielsen ML, Bekker-Jensen S, Mailand N, Choudhary C. 2012. Systems-wide analysis of ubiquitylation dynamics reveals a key role for PAF15 ubiquitylation in DNA-damage bypass. Nat Cell Biol 14:1089–1098. doi: 10.1038/ncb2579. [DOI] [PubMed] [Google Scholar]
- 130.Wagner SA, Beli P, Weinert BT, Nielsen ML, Cox J, Mann M, Choudhary C. 2011. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol Cell Proteomics 10:M111.013284. doi: 10.1074/mcp.M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Beli P, Lukashchuk N, Wagner SA, Weinert BT, Olsen JV, Baskcomb L, Mann M, Jackson SP, Choudhary C. 2012. Proteomic investigations reveal a role for RNA processing factor THRAP3 in the DNA damage response. Mol Cell 46:212–225. doi: 10.1016/j.molcel.2012.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lumpkin RJ, Gu H, Zhu Y, Leonard M, Ahmad AS, Clauser KR, Meyer JG, Bennett EJ, Komives EA. 2017. Site-specific identification and quantitation of endogenous SUMO modifications under native conditions. Nat Commun 8:1171. doi: 10.1038/s41467-017-01271-3. [DOI] [PMC free article] [PubMed] [Google Scholar]