Significance
Bone remodeling requires a balanced interplay of osteoblasts and osteoclasts. While the intercellular signaling that triggers bone cell differentiation is well understood, it remains unclear how bone progenitor cells are recruited to remodeling sites. Various chemokines are upregulated under osteoporotic conditions. However, whether they are involved in progenitor recruitment or instead have inflammatory roles is unknown. Here we used a medaka fish osteoporosis model to identify the chemokine ligand Cxcl9l and receptor Cxcr3.2 as essential to control osteoclast progenitor recruitment and differentiation at bone resorption sites. Cxcr3.2 activity can be blocked by small-molecule inhibitors that protect bone from osteoporotic insult. Our study demonstrates the potential of fish for osteoporosis drug discovery and opens avenues for future osteoporosis therapy.
Keywords: osteoporosis, bone homeostasis, osteoblast, osteoclast, chemokine
Abstract
Bone homeostasis requires continuous remodeling of bone matrix to maintain structural integrity. This involves extensive communication between bone-forming osteoblasts and bone-resorbing osteoclasts to orchestrate balanced progenitor cell recruitment and activation. Only a few mediators controlling progenitor activation are known to date and have been targeted for intervention of bone disorders such as osteoporosis. To identify druggable pathways, we generated a medaka (Oryzias latipes) osteoporosis model, where inducible expression of receptor-activator of nuclear factor kappa-Β ligand (Rankl) leads to ectopic formation of osteoclasts and excessive bone resorption, which can be assessed by live imaging. Here we show that upon Rankl induction, osteoblast progenitors up-regulate expression of the chemokine ligand Cxcl9l. Ectopic expression of Cxcl9l recruits mpeg1-positive macrophages to bone matrix and triggers their differentiation into osteoclasts. We also demonstrate that the chemokine receptor Cxcr3.2 is expressed in a distinct subset of macrophages in the aorta-gonad-mesonephros (AGM). Live imaging revealed that upon Rankl induction, Cxcr3.2-positive macrophages get activated, migrate to bone matrix, and differentiate into osteoclasts. Importantly, mutations in cxcr3.2 prevent macrophage recruitment and osteoclast differentiation. Furthermore, Cxcr3.2 inhibition by the chemical antagonists AMG487 and NBI-74330 also reduced osteoclast recruitment and protected bone integrity against osteoporotic insult. Our data identify a mechanism for progenitor recruitment to bone resorption sites and Cxcl9l and Cxcr3.2 as potential druggable regulators of bone homeostasis and osteoporosis.
During bone remodeling, which is needed to maintain skeletal rigidity and stability, osteoblasts and osteoclasts form a functional unit to achieve a balance of bone resorption and formation (1). Deficiencies in this bone cell coupling, e.g., by excess osteoclast activity, lead to reduced bone mineral density and increased bone fracture risk as observed in osteoporosis patients (2, 3). Also, physiological age-related bone loss is associated with significant changes in bone remodeling characterized by reduced bone cell coupling and decreased bone formation relative to bone resorption, resulting in elevated bone fracture risk. How bone cells communicate with each other to coordinate progenitor cell recruitment to sites of bone remodeling in order to achieve homeostasis remains poorly understood. Well-characterized factors produced by bone-forming osteoblasts to activate osteoclast progenitors, which are cells of the macrophage/monocyte lineage, include receptor-activator of NF-kβ ligand (RANKL) and macrophage colony stimulating factor (M-CSF) (4, 5). Recently, it was shown that RANKL forward signaling in osteoblast progenitors delays osteoblast differentiation, which is later released by RANKL reverse signaling through vesicular RANK receptors secreted from osteoclasts demonstrating RANKL’s important role as a coupling factor (6–8). However, more coupling factors remain to be identified as osteoclasts also form in a RANKL-independent manner (9).
Zebrafish and medaka have become popular models for human skeletal disorders (10). Both species are amenable to advanced forward and reversed genetics and genome modification and uniquely suited for live bioimaging, which makes them ideal for bone research. Many cellular and molecular features of bone are highly similar if not identical in teleost fish and mammals. This includes mechanisms of bone formation (i.e., chondral and intramembranous bone formation), cellular phenotypes and cell biological characteristics of osteoblasts and osteoclasts, and most importantly the genetic networks that control bone cell differentiation (11). Fish mutants with skeletal defects uncovered new bone-relevant genes and a better understanding of bone formation and maintenance (12, 13). We earlier established bone reporter lines in medaka to monitor the dynamics and differentiation of bone cells during bone resorption and repair (14–16). To simulate osteoporosis-like conditions, we generated transgenic fish that express medaka Rankl under control of an inducible promoter (15). Results showed that upon Rankl induction, ectopic osteoclasts trigger excessive bone resorption, which leads to bone mineralization defects that can be prevented by treatment with bisphosphonates (17), similar to the situation in human osteoporosis patients.
In the present study, we performed transcriptome profiling of different bone cell types purified from Rankl-induced medaka to identify factors involved in cell recruitment to bone resorption sites. We identified members of the family of CXC motif chemokines (CXCL) and receptors (CXCR), which are known to be involved in the attraction and differentiation of immune cells. In vitro studies had earlier implicated individual CXCL members, including CXCL7, CXCL9, and CXCL10, in osteoclastogenesis (18–20), but their exact roles in progenitor recruitment in vivo to sites of bone resorption remained unclear. Here we used live imaging in medaka to demonstrate that osteoblast-derived Cxcl9l is required and sufficient to trigger osteoclast differentiation. We also show that the receptor Cxcr3.2 is expressed in macrophages, which are recruited to the mineralized matrix, where they differentiate into osteoclasts. Our mutant and inhibitor studies demonstrate that Cxcr3.2 is essential for macrophage recruitment to bone resorption sites. This establishes osteoblast-derived Cxcl9l and the macrophage receptor Cxcr3.2 as druggable components in bone cell coupling, thus presenting a mechanism to control osteoclast progenitor recruitment to sites of bone resorption.
Results
Up-Regulation of cxcl9l in Medaka Osteoblasts Under Osteoporotic Conditions.
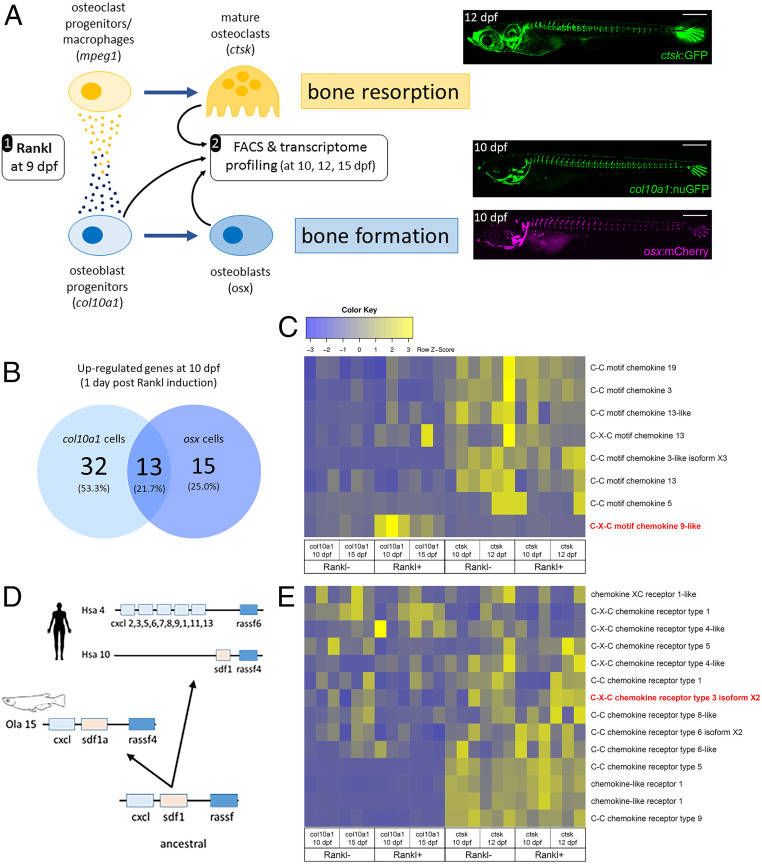
To identify factors involved in osteoblast–osteoclast cell communication, we performed RNAseq analysis in a medaka in vivo osteoporosis model (15). In this model, heat shock induction of transgenic Rankl expression at 9 d postfertilization (dpf) leads to ectopic differentiation of osteoclasts and excessive resorption of mineralized matrix of the vertebral bodies (17). Subsequently, at 2 d post-heat shock (dphs), col10a1-positive osteoblast progenitors accumulate at lesion sites, differentiate into premature osteoblasts, and contribute to remineralization of the lesioned matrix (14). To identify factors responsible for recruiting osteoclast and osteoblast progenitor cells to bone lesion sites, we purified osteoblast progenitors (col10a1), premature osteoblasts (osx), and osteoclasts (ctsk) from Rankl-induced medaka larvae by fluorescence activated cell sorting (FACS) during onset (at 10 and 12 dpf) and repair (at 15 dpf) of osteoporotic bone lesions and performed RNA sequencing (Fig. 1A). For FACS purification, 15 to 60 larvae were used per sample, leading to 10,000 to 270,000 isolated cells depending on strain and time point. Three independent biological repeats were sequenced per sample (for details, see ref. 21). Rankl:HSE:cfp nontransgenic siblings after heat shock were used as control. Bioinformatic analysis revealed that 45 genes were significantly up-regulated in osteoblast progenitors (col10a1 cells; baseMean > 10; FC > 2; P < 0.05) 1 d after Rankl induction, and 13 of these genes were also up-regulated in premature osteoblasts (osx cells; Fig. 1B). Among the genes up-regulated in col10a1 cells, we noted several genes with previously known functions in bone homeostasis, such as igfbp5, mmp9, mmp13 (collagenase 3), and tnfa (complete gene lists in SI Appendix, Tables S1–S4). In addition, we identified one medaka Cxcl chemokine, previously not associated with bone cells, as the only chemokine ligand to be significantly up-regulated in osteoblast progenitors upon Rankl induction (Fig. 1C). Phylogenetic and synteny analysis revealed that this medaka Cxcl (annotated as C-X-C motif chemokine 2 in ENSEMBL; ENSORLT00000011381) represents the basic vertebrate progenitor of the gene clusters of CXC chemokines on human chromosome 4 (Fig. 1D). By protein sequence, it is most closely related to CXCL9. We therefore refer to this gene as medaka cxcl9-like (cxcl9l, Olacxcl9l). Other chemokine ligands were expressed in osteoblasts and osteoclasts but not significantly regulated upon Rankl induction (Fig. 1C). Analyzing the regulation of chemokine receptors, we found the chemokine receptor gene cxcr3.2 up-regulated in Rankl-induced ctsk expressing osteoclasts at 12 dpf (Fig. 1E). Transcript-level (baseMean) analysis showed that cxcl9l and cxcr3.2 are expressed in osteoblast progenitors and osteoclasts, respectively, and qPCR validated the regulation of both genes in these cell types (SI Appendix, Fig. S1).
Fig. 1.
Transcriptome profiling of bone cells in a medaka osteoporosis model. (A) Experimental design: Medaka transgenic for rankl:HSE:cfp and carrying a reporter for macrophages (e.g., mpeg1:mCherry), osteoclasts (e.g., ctsk:GFP; Top Right), osteoblast progenitors (e.g., col10a1:nuGFP; Middle Right), and premature osteoblasts (e.g., osx:mCherry; Bottom Right) were heat-shocked at 9 dpf. This induced rankl expression, which in turn activated osteoblast (col10a1) and osteoclast (mpeg1) progenitor cells. Signaling between progenitors, as indicated by small circles, was suggested to trigger macrophage differentiation into osteoclasts (ctsk) in the vertebral column (image) and increased bone resorption, as well as differentiation of osteoblasts (osx) for bone formation. Larvae were dissociated to obtain col10a1 (at 10 and 15 dpf), osx (10 dpf), and ctsk (10 and 12 dpf) cells by FACS. Rankl nontransgenic reporter larvae after heat shock were used as controls. For details, see ref. 21. (B) Venn diagram showing number of significantly up-regulated genes in osteoblast progenitors and osteoblasts, at 10 dpf, 1 d after Rankl induction. (C) Heat map showing RNAseq-based expression levels of chemokine ligands in triplicates of osteoblast progenitors (col10a1, at 10 and 15 dpf) and osteoclasts (ctsk, at 10 and 12 dpf) without and with Rankl induction at 9 dpf. Note Rankl-induced up-regulation of cxcl9l (red) exclusively in col10a1 cells. (D) Inferred evolutionary history of C-X-C chemokine ligands. Deduced genomic organization of cxcl9l and neighboring genes on ancestral, medaka, and human chromosomes. (E) Expression levels of selected chemokine receptors. Cxcr3.2 is highlighted in red.
Cxcr3.2 Is Expressed in a Subset of Macrophages that Differentiates into Osteoclasts upon Rankl Induction.
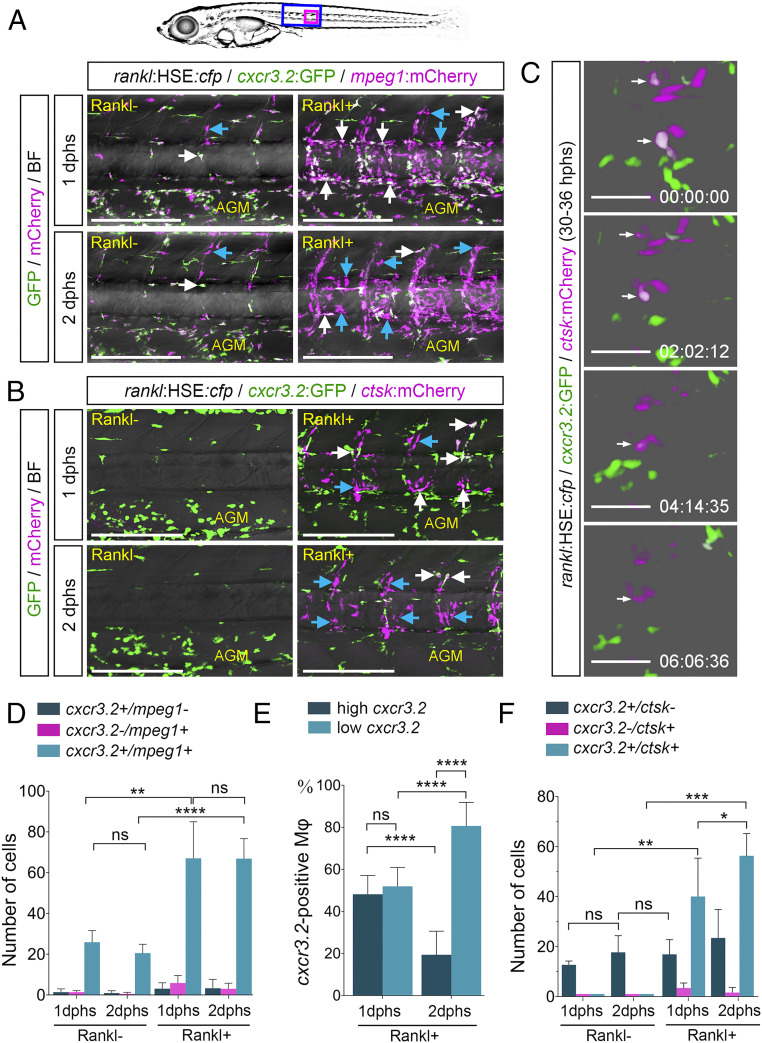
To address the function of cxcl9l and cxcr3.2 in bone homeostasis, we first determined the dynamics of cxcr3.2-positive macrophages after Rankl induction. For this, we used a reporter line that expresses GFP under control of the medaka cxcr3.2 promoter (formerly named cxcr3a; ref. 22). cxcr3.2:GFP fish were crossed to a line that expresses mCherry in macrophages under control of the medaka mpeg1 promoter (mpeg1:mCherry), as well as an osx:mCherry line that expresses mCherry in osteoblasts under control of an osx/sp7 promoter (16). We did not detect any expression of the cxcr3.2 reporter in osteoblasts but tight interactions of recruited cxcr3.2 cells with osx-expressing osteoblasts (SI Appendix, Fig. S2, blue arrows). On the other hand, control larvae without Rankl induction exhibited a significant portion of mpeg1 macrophages that coexpressed the cxcr3.2 reporter in the aorta-gonad-mesonephros (AGM) region, but only a few cells were detectable in the vertebral column (Fig. 2A, white arrow, and Movie S1). This changed dramatically after Rankl induction: the number of mpeg1 macrophages in the vertebral centra increased significantly at 1 dphs, with the vast majority of them being cxcr3.2-positive (Fig. 2 A and D and Movie S2). At 2 dphs, the number of mpeg1/cxcr3.2 double-positive macrophages remained constant, but expression of cxcr3.2 in these macrophages declined (Fig. 2 A, D, and E). Simultaneously, the number of ctsk-positive osteoclasts in the vertebral centra increased after Rankl induction (Fig. 2B), and the recruited cxcr3.2-positive cells started expressing ctsk:mCherry (Fig. 2 B, C, and F, white arrows, and Movie S3). The expression of cxcr3.2:GFP in osteoclasts decreased as they matured (Fig. 2C and Movie S4). These data suggest that cxcr3.2 is expressed in a subset of mpeg1 macrophages that becomes activated after Rankl induction and is recruited to the vertebral column. There macrophages differentiate into osteoclasts that gradually lose their cxcr3.2 expression.
Fig. 2.
cxcr3.2:GFP is expressed in a subset of macrophages and osteoclasts. (A) Expression of cxcr3.2:GFP in a subset of mpeg1:mCherry-positive macrophages without and with Rankl induction, at 1 or 2 dphs. Coexpressing cells are labeled with white arrows, and blue arrows mark cxcr3.2:GFP-negative macrophages. (B) Expression of cxcr3.2:GFP in a subset of ctsk:mCherry-positive osteoclasts. Coexpressing cells are labeled with white arrows, and blue arrows mark cxcr3.2:GFP-negative osteoclasts. (C) Reduction of cxcr3.2:GFP expression during osteoclast maturation. Still images are taken from Movie S4. (D) Quantification of cxcr3.2 and mpeg1 single and double-positive cells. (E) Quantification of cxcr3.2:GFP expressing macrophages with low or high GFP expression. (F) Quantification of cxcr3.2 and ctsk single and double-positive cells. Error bars show mean value ± SD, Brown–Forsythe ANOVA with Games–Howell’s multiple comparisons test (D), one-way ANOVA with Tukey’s multiple comparisons test (E), and Mann–Whitney U test (F); ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05; ns, nonsignificant; ncxcr3.2/mpeg1 = 34 fish, n cxcr3.2/ctsk = 7 fish. Time in C is hh:mm:ss. (Scale bars, 200 µm in A and B and 50 µm in C.)
Macrophage Recruitment Is Impaired Under Cxcr3.2-Deficient Conditions.
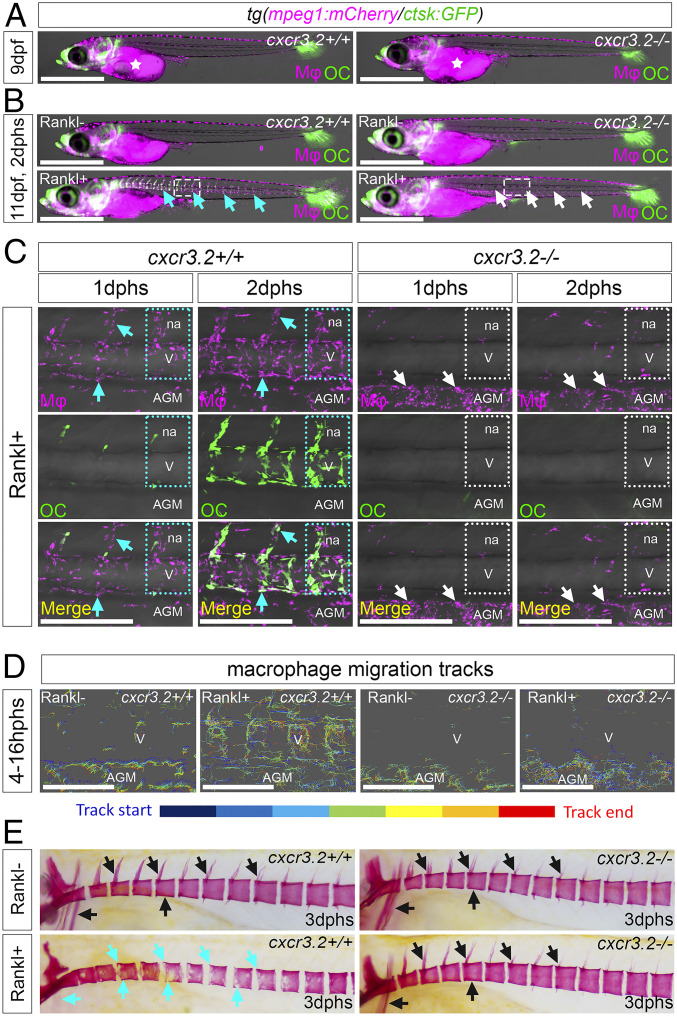
To determine whether Cxcr3.2 is required for macrophage activation and directed migration, we generated medaka cxcr3.2 mutants by CRISPR/Cas9 (SI Appendix, Fig. S3). Homozygous mutants developed normally without obvious malformations (Fig. 3 A and B). Next, we used time-lapse confocal imaging to track the dynamics of macrophages under cxcr3.2-deficient conditions (Movies S5–S8). In cxcr3.2 mutants without Rankl induction, macrophages were less migratory than in their wild-type siblings, which was evident from a significant reduction in directed displacement and speed of individually tracked cells (Fig. 3 B and D and SI Appendix, Fig. S4 and Movies S5 and S7). Upon Rankl induction, speed and migration distance were increased in mutants but were still significantly lower compared to wild-type cells (Fig. 3 B–D and SI Appendix, Fig. S4 and Movies S6 and S8). Although mutant macrophages increased their motility, they mainly persisted in the AGM and showed reduced directionality toward the centra of the vertebral column (Fig. 3 B–D; quantification in SI Appendix, Fig. S4F). This suggests that a Cxcr3.2 deficiency affects the general migratory behavior of macrophages and particularly interferes with their directed migration from the AGM to mineralized matrix in the vertebral column.
Fig. 3.
Impaired macrophage recruitment and osteoclast formation in cxcr3.2 mutants. (A) Normal distribution of mpeg1:mCherry macrophages and ctsk:GFP osteoclasts in cxcr3.2+/+ and cxcr3.2−/− siblings without Rankl induction. (B) Macrophage and osteoclast distribution in cxcr3.2+/+ and cxcr3.2−/− siblings at 11 dpf, 2 d after heat shock to induce Rankl expression (i.e., 2 dphs) in Rankl+ and Rankl− siblings. (C) Higher-magnification views of regions boxed in B. In cxcr3.2+/+, macrophages are found in neural arches and vertebral bodies at 1 dphs. Macrophage numbers are increased at 2 dphs, and macrophages start to differentiate into ctsk osteoclasts. In cxcr3.2−/−mutants, in contrast, macrophages remain in AGM and fail to migrate to vertebral bodies. Dotted lines demarcate individual vertebral bodies, cyan arrows mark macrophages and newly formed osteoclasts, and white arrows mark macrophages retained in the AGM. (D) Tracking of macrophage migration paths in cxcr3.2+/+ and cxcr3.2−/− siblings without and with Rankl induction. Time-lapse videos were taken from 4 to 16 hphs (Movies S5–S8). Upon Rankl induction, wild-type macrophages migrate dorsally toward vertebral bodies (V), while mutant macrophages have increased motility but exhibit lateral migration restricted to the AGM. (E) Alizarin Red staining of mineralized matrix in cxcr3.2+/+ and cxcr3.2−/− siblings without and with Rankl induction. Black arrows mark intact neural arches, centra and cleithrum, and cyan arrows indicate absent arches and severe lesions in centra and cleithrum. Mφ, macrophage; OC, osteoclast; na, neural arch. (Scale bars, 1 mm in A and B and 200 µm in C and D.)
Furthermore, after Rankl induction, macrophage to osteoclast differentiation was strongly impaired in cxcr3.2 mutants, and no ctsk-positive osteoclasts were detectable at 2 dphs, when wild-type siblings showed abundant osteoclasts (Fig. 3 B and C). Consequently, in the absence of ectopic osteoclasts, the integrity of mineralized matrix in vertebral bodies including neural and hemal arches was intact in cxcr3.2 mutants at 3 dphs, similar to the situation in Rankl− larvae (Fig. 3E). This is also consistent with the observation that osteoblasts formed normally in cxcr3.2 mutants (SI Appendix, Fig. S2).
Together, our data suggest that the chemokine receptor Cxcr3.2 is required for macrophage activation upon Rankl induction to recruit them to the mineralized matrix of the vertebral bodies, where they differentiate into osteoclasts. Importantly, other macrophage functions such as bacterial phagocytosis appeared not affected by cxcr3.2 deficiency. Escherichia coli injected into larvae were efficiently cleared by recruited cxcr3.2-deficient macrophages, similar to the situation in heterozygous and wild-type siblings (Movies S9–S11). Thus, Cxcr3.2 seems to be required particularly for those macrophages that are destined to differentiate into osteoclasts.
Chemical Inhibition of Cxcr3.2 Blocks Osteoclast Formation and Protects Bone Integrity.
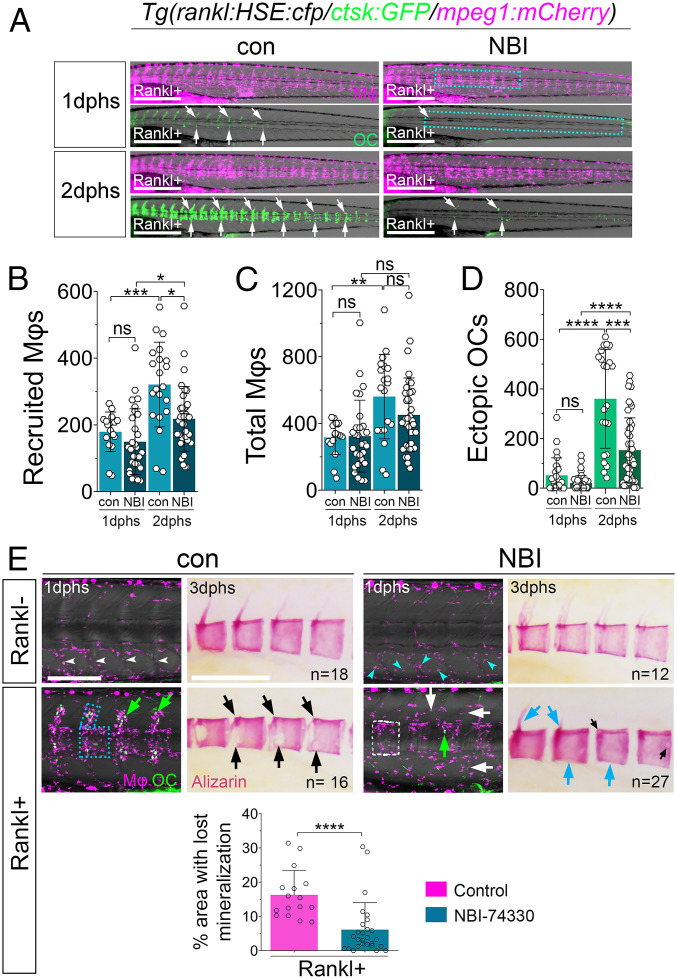
NBI-74330 and AMG487 are two structurally related antagonists of human CXCR3, with slightly different binding affinities (23–25). They have been pharmacologically tested previously in the context of atherosclerotic plaque formation and psoriasis (26, 27). To test whether human CXCR3 inhibitors interfere with Cxcr3.2 function in medaka and block osteoclast formation, we treated ctsk:GFP/mpeg1:mCherry/rankl:HSE:cfp transgenic medaka with different doses of these antagonists. Upon Rankl induction, control larvae treated with DMSO showed macrophage accumulation and osteoclast formation in neural arches and centra at 1 dphs (Fig. 4 A and E, blue box and green arrows), and osteoclast numbers increased at 2 dphs (Fig. 4 A and D). In NBI-74330–treated larvae at 1 dphs, the number of initially recruited macrophages was not significantly different from controls (Fig. 4 A and B); however, fewer macrophages were detectable in the region of the vertebral arches (Fig. 4E, white arrows). In contrast, at 2 dphs, when macrophage numbers increased strongly in controls, they remained at significantly lower levels in NBI-74330–treated larvae (Fig. 4B). Importantly, the total number of macrophages, including those in the AGM and tail region, was not significantly altered (Fig. 4C). Also osteoclast formation was strongly suppressed in NBI-74330–treated larvae (Fig. 4 A, D, and E). As a consequence, treatment with NBI-74330 protected bone integrity upon Rankl induction (Fig. 4E). In DMSO controls, Rankl induction resulted in a resorption of vertebral neural arches and a severe loss of mineralized matrix in the centra (Fig. 4E). In contrast, NBI-74330–treated larvae showed a significant reduction of mineralization defects and were almost indistinguishable from Rankl-negative larvae (Fig. 4E). Similar results were also obtained with AMG487 (SI Appendix, Fig. S5). In conclusion, chemical inhibitor experiments confirmed the findings in cxcr3.2 mutants that Cxcr3.2 is required for macrophage recruitment and osteoclast formation upon Rankl induction.
Fig. 4.
Cxcr3.2 inhibitor NBI-74330 interferes with macrophage recruitment and osteoclast differentiation. (A) Trunk regions of triple transgenic rankl:HSE:cfp/ctsk:GFP/mpeg1:mCherry larvae treated with DMSO (con) or NBI-74330 (NBI) for 3 h, followed by heat shock to induce Rankl and imaging at 1 and 2 dphs. ctsk osteoclasts are labeled with white arrows. (B–D) Quantification of recruited macrophages (B), total number of macrophages (C), and ectopic osteoclasts (D) in areas indicated by boxes in A. Data are represented as mean number of cells ± SD, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns, nonsignificant, Brown–Forsythe ANOVA with Tukey’s multiple comparisons test, 17 ≤ NLarvae ≤ 36, from three independent experiments. (E) Confocal images of macrophages (Mφ; magenta) and osteoclasts (OC; green) in vertebral bodies. Without Rankl induction, macrophages are confined to myosepta in AGM of control larvae (con; white arrowheads), while they are dispersed in AGM of NBI-treated larvae (NBI; cyan arrowheads). After Rankl induction, macrophages and osteoclasts are located along neural arches and vertebral centra in control larvae (blue box and green arrows). In NBI-treated larvae, macrophages are located in centra but less confined to neural arches (white box and white arrows), and fewer osteoclasts are detectable (green arrow). Alizarin Red staining shows severe lesions of mineralized matrix in majority of control larvae after Rankl induction, with neural arches resorbed and large lesions in centra (black arrows). In contrast, mineralization defects were strongly reduced in NBI-treated larvae with persistent mineralized neural arches and intact centra (cyan arrows). Areas with absent mineralization in centra were quantified (SI Appendix, Fig. S7) and are depicted in bar graph at bottom; Mann–Whitney U test, ****P < 0.0001. (Scale bars, 500 μm in A and 200 μm in E.)
Osteoblast-Derived Cxcl9l Triggers Macrophage Recruitment and Osteoclast Differentiation in the Absence of Rankl.
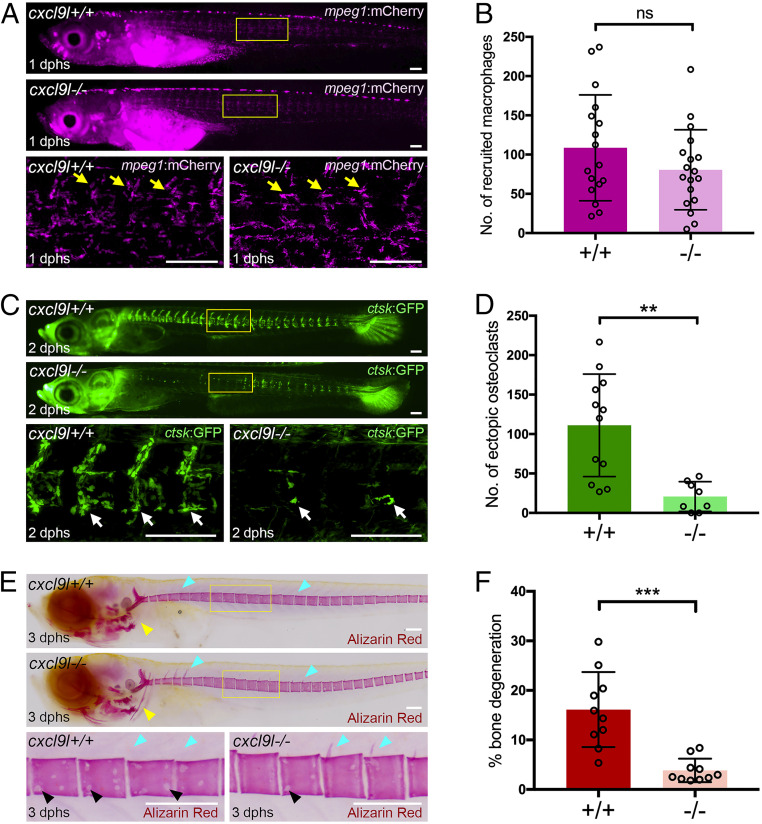
Chemokine receptors control cell migratory behavior in response to gradients of chemokine ligands. To address the source and nature of the chemokine responsible for macrophage recruitment to bone, we investigated whether the ligand Cxcl9l, up-regulated in osteoblast progenitors upon Rankl induction, is involved in this process. For this, we generated cxcl9l mutants by CRISPR/Cas9 (SI Appendix, Fig. S3) and tracked macrophage dynamics by time-lapse imaging. Without Rankl induction, no changes in macrophage distribution were observed in cxcl9l mutants (SI Appendix, Fig. S6A). After Rankl induction, the number of macrophages at 1 dphs recruited to the vertebral column was not significantly reduced (Fig. 5 A and B). However, osteoclast formation was severely reduced at 2 dphs, and only single ctsk-positive osteoclasts were detectable in the vertebral column of cxcl9l mutants, while wild-type siblings had abundant osteoclasts (Fig. 5 C and D). At 3 dphs, the severity of Rankl-induced mineralized matrix lesions, evident as resorbed neural arches and presence of nonmineralized lesions in centra of wild-type siblings, became significantly alleviated in Rankl-induced cxcl9l mutants (Fig. 5 E and F and SI Appendix, Fig. S7). Importantly, osteoblast numbers and distribution were not affected in cxcl9l mutants (SI Appendix, Fig. S8). Together, these data suggest that osteoblast-derived Cxcl9l is required for osteoclast formation but may be dispensable for macrophage recruitment.
Fig. 5.
Impaired osteoclast formation and reduced bone lesions in cxcl9l mutants after Rankl induction. (A) Overview and high-magnification confocal images of cxcl9l+/+ and cxcl9l−/− siblings in mpeg1:mCherry/rankl:HSE:cfp transgenic background at 1 dphs (10 dpf). Similar recruitment of macrophages to centra (yellow arrows) in cxcl9l+/+ and cxcl9l−/− siblings at 1 dphs. (B) Quantification of mpeg1:mCherry macrophage numbers in vertebral column (region marked as boxes in A) in cxcl9l+/+ and cxcl9l−/− siblings at 1 dphs. (C) Overview and confocal images of ctsk:GFP-expressing osteoclasts (white arrows) in in cxcl9l+/+ and cxcl9l−/− siblings at 2 dphs. (D) Quantification of ctsk:GFP osteoclast numbers in vertebral column (region marked as boxes in C). (E) Overview and high-magnification views of Alizarin Red stained mineralized matrix in in cxcl9l+/+ and cxcl9l−/− siblings at 12 dpf (3 dphs). Wild-type siblings but not mutants show enhanced resorption of mineralized matrix at cleithrum (yellow arrowheads), neural arches (cyan arrowheads), and vertebral bodies (black arrowheads). (F) Quantification of bone lesions in vertebral column (position indicated by boxes in E; for method, see SI Appendix, Fig. S7). Error bars indicate mean ± SD, **P < 0.01, ***P < 0.001; ns, not significant, unpaired Student’s two-tailed t test. (Scale bars, 100 μm.)
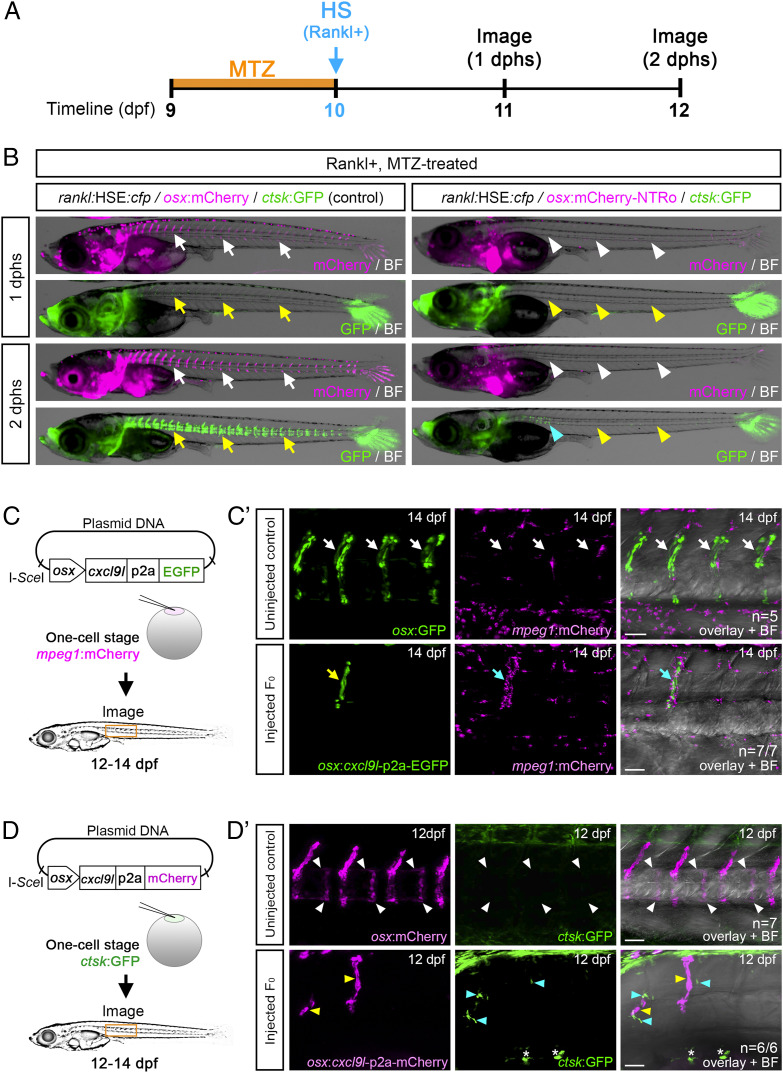
Upon Rankl induction, cxcl9l is up-regulated in osteoblasts. We therefore tested whether osteoblasts are the sole source for Cxcl9l required for osteoclast recruitment. As cxcl9l was up-regulated in col10a1 osteoblast progenitors as well as in premature osx-positive osteoblasts (SI Appendix, Tables S1 and S3), we made use of an established osx:mCherry-NTRo line to genetically ablate premature osteoblasts using a Nitroreductase (NTRo) approach as described earlier (28, 29) and quantified the formation of ctsk osteoclasts after Rankl induction. Metronidazole (Mtz) treatment of osx:mCherry-NTRo/rankl:HSE:cfp/ctsk:GFP transgenic medaka at 9 dpf for 24 h resulted in an almost complete ablation of osteoblasts from the vertebral column under both Rankl− and Rankl+ conditions (Fig. 6 A and B, white arrowheads, and SI Appendix, Fig. S9). Upon Rankl induction at 10 dpf, the number of ctsk osteoclasts was strongly reduced (by approximately 90%), while control treated larvae without ablation showed normal osteoclast induction (Fig. 6B and SI Appendix, Fig. S9). Importantly, the integrity of mineralized matrix was maintained in osteoblast-ablated larvae at 3 dphs, further confirming the absence of osteoclast activity in these fish (SI Appendix, Fig. S9). These ablation experiments strongly suggest that osteoblasts are the exclusive source of signals required for ectopic osteoclast formation upon Rankl induction.
Fig. 6.
Osteoblast-derived Cxcl9l is sufficient for macrophage recruitment and osteoclast differentiation in the absence of Rankl induction. (A) Experimental timeline for osteoblast ablation. osx:mCherry-NTRo larvae were treated with Metronidazole (MTZ) at 9 dpf for 24 h, and heat shock was applied at 10 dpf to induce Rankl expression. (B) Osteoblasts (magenta) and osteoclasts (green) were imaged at 1 and 2 dphs. osx:mCherry larvae treated with Mtz were used as controls. Osteoblasts and osteoclasts form normally in controls (white and yellow arrows, respectively) but are absent in osteoblast-ablated larvae (white and yellow arrowheads; blue arrowhead highlights remaining osteoclasts). (C) Strategy for macrophage analysis after ectopic cxcl9l expression in osx osteoblasts. mpeg1:mCherry transgenic embryos were injected with osx:cxcl9l-p2a-EGFP plasmid and I-SceI meganuclease at one-cell stage, raised to 12 to 14 dpf, and screened for mosaic osx:cxcl9l-p2a-EGFP expression in osteoblasts located at vertebral bodies. (C') Confocal images showing single macrophages present at neural arches of noninjected osx:GFP/mpeg1:mCherry control larvae (Top; white arrows, n = 5 fish). Enhanced recruitment of macrophages (cyan arrows) toward cxcl9l-p2a-EGFP expressing cells (yellow arrow) in neural arch of osx:cxcl9-p2a-EGFP injected larva (Bottom; n = 7/7 fish). (D) Strategy for osteoclast analysis upon ectopic cxcl9l expression in osx-expressing osteoblasts. ctsk:GFP transgenic embryos were injected with osx:cxcl9l-p2a-mCherry plasmid and I-SceI meganuclease at one-cell stage, raised to 12 to 14 dpf, and screened for mosaic osx:cxcl9l-p2a-mCherry expression in osteoblasts at vertebral bodies. (D') Confocal images showing absence of osteoclast formation (white arrowheads) in the vertebral bodies of uninjected osx:mCherry/ctsk:GFP larvae (Top; control, n = 7 fish). Ectopic formation of ctsk:GFP-expressing osteoclasts (cyan arrowheads) in vertebral bodies upon mosaic cxcl9l-p2a-mCherry expression in osx cells (yellow arrowheads; Bottom; n = 6/6 fish; 4/6 fish showed ectopic osteoclast formation both close to and distant from Cxcl9l-expressing cells, and 2/6 fish showed ectopic osteoclast formation at a distance from Cxcl9l-expressing cells). Asterisks indicate autofluorescent pigment cells at yolk region. (Scale bars, 50 μm.)
Finally, to test whether Cxcl9l alone is sufficient to trigger macrophage recruitment in the absence of Rankl induction, we ectopically expressed Cxcl9l in premature osteoblasts using a transient transgenic approach. For this, we injected an osx promoter construct driving expression of Cxcl9l fused to EGFP or mCherry via a self-cleaving p2a peptide into medaka embryos (Fig. 6 C and D). Larvae with mosaic osx:cxcl9l-p2a-mCherry or osx:cxcl9l-p2a-EGFP expression in osteoblasts on the vertebral column surface were selected for analysis at 10 to 14 dpf (Fig. 6 C' and D' and SI Appendix, Fig. S6 B and C). We noted a considerable accumulation of mpeg1:mCherry labeled macrophages directly adjacent to the cxcl9l-p2a-EGFP labeled cells, while the remaining vertebral column showed macrophages at a similarly low extent as noninjected controls (Fig. 6C'; for quantification, see SI Appendix, Fig. S6B). In a second approach, we analyzed whether ctsk-positive osteoclasts can form under conditions of ectopic Cxcl9l expression. Noninjected control larvae at 12 dpf never showed ctsk osteoclasts in the vertebral column under Rankl− conditions (Fig. 6D', Top; also see ref. 15). In contrast, larvae ectopically expressing Cxcl9l exhibited several ctsk-positive cells in the vertebral bodies (19 ctsk cells in 6 analyzed larvae; Fig. 6D'). Ten of the 19 identified ctsk cells were found positioned close to Cxcl9l-producing cells, while 9 cells were found farther away (SI Appendix, Fig. S6C). This shows that Cxcl9l is sufficient to induce macrophage migration and differentiation into osteoclasts, even in the absence of Rankl expression. Together, our results demonstrate that Cxcl9l is up-regulated in osteoblasts upon Rankl induction and mediates Rankl effects by controlling osteoclast recruitment and differentiation at the mineralized bone matrix.
Discussion
Bone homeostasis requires the coordinated recruitment of osteoclasts and osteoblasts to sites of remodeling. To achieve homeostasis, bone cells communicate with each other to balance cell numbers and activities. Rankl is well established as an osteoblast-derived inducer of osteoclast formation and activity (30). It has recently also been identified as an important coupling factor to control osteoblast differentiation by reverse signaling (6). However, the factors that control recruitment of osteoclast and osteoblast progenitors to sites of bone remodeling remain largely unknown. In this study using a medaka model, we identified the chemokine Cxcl9l and a chemokine receptor Cxcr3.2, which are up-regulated in osteoblasts and osteoclasts, respectively, under Rankl-induced osteoporotic conditions. We demonstrate that Cxcl9l is sufficient to recruit macrophages to bone resorption sites and drive their differentiation into osteoclasts independent of Rankl induction.
Medaka cxcl9l Encodes the Ancestral Gene of Human CXCL9.
The human genome encodes more than 40 chemokine ligands and 20 chemokine receptors. In contrast, the repertoire in teleost fish is much smaller. This suggests an expansion of chemokines and their cognate receptors in the tetrapod lineage related to additional and lineage-specific new functions of the more advanced immune system.
In the current medaka genome assembly, the cxcl transcript identified by our RNAseq analysis is annotated as C-X-C motif chemokine 2. However, our phylogenetic and synteny analysis revealed that this gene is the common precursor of two CXCL gene clusters on human chromosome 4, including CXCL 1, 2, 3, 4a and b (syn PF4 and PF4V1), 5, 6, 7 (syn PPBP), and 8 at 4q13.3 and CXCL 9, 10, 11, and 13 at 4q21.1. Such clusters evolve by local gene duplications and subsequent functional divergence (neofunctionalization or subfunction partition). The highest aa identity of OlaCxcl9l to human CXCL9 suggests that human CXCL9 shares the ancestral function with the identified medaka gene.
In mice, CXCL9 is produced in osteoblasts and functions as bone angiostatic and osteogenic factor (31). In addition, cell culture studies have shown that blocking CXCL9 or CXCL10 function in vitro impairs differentiation of osteoclast progenitors (19, 20, 32); however, their role for bone homeostasis in vivo is unclear. In humans, recent genome-wide association studies interestingly identified CXCL9 as a potential candidate locus for periodontitis, an inflammatory disease that destroys tissues surrounding and supporting teeth, leading to bone loss (33). However, whether and how CXCL9 acts to recruit osteoclasts to sites of bone resorption remained unknown.
Cxcl9l and Rankl Signaling.
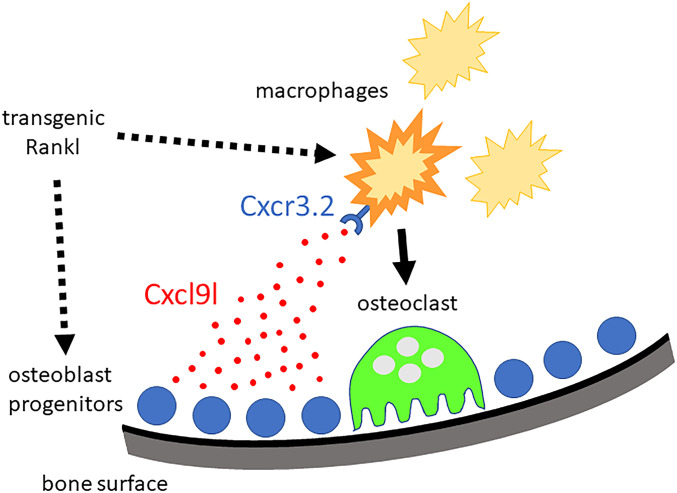
We found that upon Rankl induction, cxcl9l was up-regulated in col10a1 osteoblast progenitors. It is presently unclear whether this reflects a direct effect of Rankl activating a Rank receptor on col10a1 cells or happened indirectly as a consequence of factors released from activated osteoclasts. Consistent with the latter, we identified several up-regulated transcripts in Rankl-induced osteoclasts, including cxcr3.2, opening the possibility that factors released from Rankl-induced osteoclasts indirectly trigger up-regulation of cxcl9l in osteoblasts. On the other hand, however, it is known that at least in mammals, Rank receptor is expressed in mesenchymal stem cells (MSCs; ref. 7), which can differentiate into osteoblasts, as well as in osteoclast progenitors (34). In MSCs, forward Rank signaling blocks their differentiation into osteoblasts (7). Subsequently, Rankl-activated osteoclasts produce extracellular vesicles containing Rank receptors that bind to Rankl on MSCs and thereby induce reverse Rankl signaling to allow osteoblast differentiation (6). Hence, in mammals, Rankl has two major origins, from MSCs/osteoblast progenitors to block osteoblastogenesis and from osteocytes to promote osteoclastogenesis. Medaka have acellular bone that completely lacks osteocytes (35) but contains col10a1-positive osteoblast progenitors that differentiate into osx expressing premature osteoblasts (14). We hypothesize that Rankl forward signaling also occurs in medaka osteoblast progenitors. Transgenic Rankl expression might therefore activate Rank receptors on osteoblast progenitors to produce and release Cxcl9l. Cxcl9l then forms a chemokine gradient to attract macrophages to mineralized matrix where they differentiate into osteoclasts (Fig. 7). Whether these osteoclasts produce vesicular Rank to later promote osteoblast differentiation, similar to in mammals, remains to be investigated.
Fig. 7.
Chemokine control of osteoclast recruitment. Under osteoporotic conditions, Rankl acts on osteoblast progenitors located at the mineralized matrix of vertebral bodies and on macrophages as osteoclast progenitors. In osteoblast progenitors, Rankl either directly or indirectly induces the production and release of Cxcl9l. Freely diffusing Cxcl9l then activates macrophages that express the Cxcr3.2 receptor. This triggers the activation and recruitment of macrophages toward mineralized matrix, where they differentiate into osteoclasts and contribute to bone resorption.
The Chemokine Receptor Cxcr3.2 Marks a Subset of Macrophages Destined to Become Osteoclasts.
We found that cxcl9l and cxcr3.2 are expressed in medaka osteoblast progenitors and osteoclasts, respectively. Without Rankl induction, no osteoclasts appear in the vertebral column of medaka larvae at the analyzed stages (9 to 12 dpf), but ctsk-positive cells are present in head and fins (15). The up-regulation of cxcr3.2 transcription detected in FAC-sorted ctsk cells after Rankl induction thus likely reflects a de novo formation of osteoclasts in the vertebral column, rather than cxcr3.2 up-regulation in preexisting osteoclasts. This was confirmed by time-lapse analysis of a cxcr3.2:GFP reporter, which was initially expressed in a subset of mpeg1-positive/ctsk-negative macrophages. Later, the reporter persisted in differentiating osteoclasts that became ctsk-positive, but eventually, cxcr3.2 expression diminished in maturing osteoclasts. Together, our data suggest that Cxcr3.2 is expressed in macrophages that are destined to become osteoclasts. Once osteoclasts have reached their destination and become resorptive, cxcr3.2 gets down-regulated. Importantly, Cxcr3.2 is not required for macrophage migration or recruitment to clear inflammatory insult (Movies S9–S11 and ref. 36). Hence, we speculate that this receptor marks a distinct subset of macrophages, which differentiates into osteoclasts at mineralized bone matrix.
Cxcl9l Drives Progenitor Recruitment to Sites of Bone Resorption in Medaka.
Factors that control progenitor cell recruitment to bone matrix under physiological or pathological conditions gain increasing interest, mainly for therapeutic reasons. Various inflammatory cytokines and chemokines have been shown to promote osteoclastogenesis and modulate bone remodeling (37). Chemokines, including CXCL10, are mainly produced by immune cells and are implicated in inflammatory bone loss such as in rheumatoid arthritis or bone metastasis (32, 38). In the latter, chemokines activate osteoclasts to enhance bone resorption and facilitate colonization of bone marrow by metastasizing cancer cells (39). Expression in osteoblasts, on the other hand, has been reported for several chemokines (such as CCL2, CCL3, CXCL2, and CXCL12, among others) and linked, e.g., to tooth eruption (40), as well as fracture healing and bone repair (41, 42). However, while the immunomodulatory effects of chemokines are well characterized, the physiological roles of osteoblast-derived chemokines are much less understood. Consistent with reports in mammals, our study shows that medaka osteoblast progenitors produce several chemokines, but we found only Cxcl9l to be significantly up-regulated upon Rankl induction in vivo. Importantly, ectopic expression of Cxcl9l was sufficient to drive osteoclast progenitor recruitment in larvae even in the absence of Rankl induction. This may indicate that at least in larvae, Cxcl9l acts in a paracrine manner and independently or downstream of Rankl to recruit osteoclasts to bone matrix. Interestingly, we detected a considerable number of induced osteoclasts at a distance from ectopic Cxcl9l-producing cells. This could simply reflect the remarkable migratory dynamics of newly formed osteoclasts in medaka (see ref. 15). Alternatively, we cannot exclude that Cxcl9l controls osteoblasts in an autocrine manner and that consequently, other osteoblast-derived factors act at a distance to trigger osteoclast differentiation remotely.
The role of CXCL9–CXCR3 signaling in immune cell activation and migration in development and pathological processes has been well documented (43–45). It has, however, also been shown that CXCR3 acts independently of CXCL9, via CXCL4, CXCL10, or CXCL11 (46–48). Likewise, CXCL9 can interact with other receptors than CXCR3, such as CCR3 (49). Here the finding that cxcl9l and cxcr3.2 medaka mutants share a similar osteoclast phenotype proposes that Cxcl9l acts through Cxcr3.2 to control osteoclast recruitment to bone matrix. In contrast to cxcr3.2 mutants, however, cxcl9l mutants showed normal macrophage distribution upon Rankl induction suggesting that Cxcl9l is dispensable for cell recruitment. This could be due to the presence of additional chemokines produced by col10a1 cells, as described above. Importantly, deletion of cxcl9l blocked differentiation of macrophages into osteoclasts almost entirely, indicating an essential role in this process. Together with the fact that transgene-driven ectopic expression of cxcl9l promoted macrophage recruitment, this therefore strongly suggests that Cxcl9l is a critical chemotactic cue recognized by cxcr3.2-positive macrophages.
Interestingly, we observed that cxcr3.2-deficient macrophages exhibit increased motility upon Rankl induction but failed to directionally migrate toward bone matrix. We speculate that macrophages, in this case, were directly stimulated by Rankl binding to Rank receptors on these cells, which triggered dynamic migratory behavior in the absence of a chemotactic guidance cue. At present, however, we cannot exclude that Rankl-induced expression of other chemokines that bound to receptors other than Cxcr3.2 and stimulated cell movement, albeit not directed toward bone matrix.
Without Rankl induction, the cxcl9l and cxcr3.2 mutants generated in our study developed apparently normally with no obvious signs of osteoclast deficiency or osteopetrosis. We thus hypothesize that Cxcl9l and Cxcr3.2 are especially important under pathological conditions of severe Rankl overexpression, such as seen in rheumatoid arthritis and osteoporosis. Under nonpathological conditions, other chemokines and their receptors might be at play to recruit osteoclast precursors.
Chemical Inhibition of Cxcr3.2 Protects Bone Integrity in Medaka.
Fish models, such as medaka and zebrafish, are ideally suited for drug screening. Testable compounds can be directly added to small volumes of medium, and skeletal defects can be efficiently analyzed by live imaging in the translucent larvae (50). In the past, several chemical inhibitors for chemokine receptors have been developed and clinically tested, especially in the context of cancer, immune deficiencies, and inflammation (51–53). These include inhibitors for CXCR3 (54). NBI-74330 (24) and AMG487 (25) are two structurally related CXCR3 antagonists that have been tested in preclinical and clinical trials. AMG487 entered a phase II clinical trial for psoriasis that was stopped for lack of efficacy (overview in ref. 27). In an LPS-induced periodontitis mouse model, treatment with AMG487 resulted in a significant reduction of bone loss and decreased osteoclast numbers after LPS injections (33). Therefore, CXCR3 inhibitors have been highly effective in the context of several bone inflammatory diseases, but their role and efficacy in an osteoporosis context remained unclear. In our osteoporosis setting in medaka, NBI-74330 and AMG487 efficiently blocked osteoclast formation upon Rankl induction. Our in vivo experiments thus suggest that the chemokine-controlled process of osteoclast recruitment is druggable and that CXCR3 inhibitors can be used to modulate bone homeostasis. Targeting recruitment rather than activation of osteoclast progenitors opens new avenues for pharmacological treatment of a disease that affects millions of new patients every year and places extensive burden on families and health care systems.
Materials and Methods
Transgenic and Mutant Fish Lines.
All experiments were performed according to protocols approved by the Institutional Animal Care and Use Committee of the National University of Singapore (protocol numbers R14-293, R18-0562, and BR15-0119). For generation of mpeg1:mCherry transgenic medaka, a 2.1-kb sequence of the mpeg1 promoter including the endogenous ATG was amplified from genomic DNA of wild-type medaka using primers mpeg1.2F and mpeg1.2R (sequences in SI Appendix, Table S5). The PCR product was digested by NotI and BamHI and ligated in frame to farnesylated mCherry in a pI-SceI plasmid. The plasmid was injected into one-cell stage medaka embryos together with I-SceI meganuclease to obtain stable lines. Cxcr3.2:EGFP transgenic medaka were generated using a plasmid described earlier (22). To generate osx:cxcl9l-p2a-mCherry and osx:cxcl9l-p2a-EGFP plasmids for Cxcl9l overexpression, the medaka osx promoter (16) was cloned upstream of the cxcl9l coding sequence amplified from medaka cDNA using primers cxcl9lF and cxcl9lR (SI Appendix, Table S5). For osx:cxcl9l-p2a-mCherry, the cxcl9l stop codon was replaced with p2a-mCherry amplified from plasmid col10a1:creERT2-p2a-mCherry (29). For osx:cxcl9l-p2a-EGFP, mCherry from osx:cxcl9l-p2a-mCherry was replaced with EGFP. The plasmids were injected into one-cell stage medaka embryos together with I-SceI meganuclease. All other transgenic lines have been described before (15) and are available from the Medaka Stock Center, Laboratory of Bioresource, National Institute for Basic Biology, Okazaki, Japan (https://shigen.nig.ac.jp/medaka/). Staging of larvae, screening, and live imaging of transgenic medaka were performed as described before (50).
For CRISPR/Cas9 genome editing, guide RNAs targeting cxcl9l and cxcr3.2 (SI Appendix, Table S5) were designed using CRISPR/Cas9 target online prediction (CCTop) (55) and generated by integrated DNA technologies (IDT) as crRNAs. crRNAs (36 ng/μL each) were injected into one-cell stage medaka embryos together with tracrRNA (67 ng/μL, IDT) and Cas9 nuclease (0.25 μg/μL, IDT). Primers used for genotyping (cxcl9lgenoF/R and cxcr3.2F/R) are listed in SI Appendix, Table S5.
RNA-Seq Library Preparation of FACS Purified Bone Cells and Sequencing.
RNA libraries were prepared from FACS purified osteoblast progenitors (col10a1-positive), premature osteoblasts (osx/sp7), and osteoclasts (ctsk) as described previously (21). Briefly, medaka Rankl+ (rankl:HSE:cfp) and Rankl− control larvae expressing osteoblast (col10a1:nuGFP; osx:mCherry) or osteoclast (ctsk:mCherry) reporters were heat-shocked at 9 dpf for 2 h at 39 °C to induce Rankl expression. Larvae were dissociated for FACS using a collagenase/trypsin-based protocol for purification of osteoblasts at 10 dpf (col10a1, osx) or 15 dpf (col10a1) and osteoclasts (ctsk) at 10 or 12 dpf. RNAs were isolated using a total RNA isolation reagent (TRIzol)-based method and purified using the PureLink RNA Micro Kit (Invitrogen) following manufacturer’s instructions. cDNA was synthesized from 1 to 10 ng total RNA using the SMARTer Ultra Low Input RNA for Illumina Sequencing Kit and the Advantage2 polymerase mix (Clontech) for second strand synthesis. cDNAs were analyzed using an Agilent 2100 BioAnalyzer and a High Sensitivity DNA Chip. A Covaris AFA system was used to produce short cDNA fragments of 100 to 300 bp, and DNA concentration was determined using a Qubit dsDNA HS Assay Kit and Qubit2.0 Fluorometer. cDNA libraries were generated using the NEBNext Ultra DNA Library Prep Kit for Illumina and NEBNext Multiplex Oligos for Illumina (Index primer set 1). After adapter ligation, DNAs were PCR amplified with NEBNext high-fidelity 2× PCR master mix using Index primers (primers 1 to 12). Libraries were multiplexed, and paired-end sequencing (75 to 100 bp read lengths) was performed on an Illumina HiSeq platform at the Genome Institute of Singapore with sequencing depths between 50 and 100 million reads.
Bioinformatics Analysis.
For differential gene expression analysis, reads were aligned to the Japanese medaka Hd-rR strain genome (ASM223467v1) using spliced transcripts alignment to a reference (STAR; quantMode GeneCounts) (56). Differentially expressed genes were detected using the Bioconductor/R package DESeq2 (57). For further analyses a gene was considered to be differentially expressed at a baseMean ≥ 10, log2FC > 1, and adjusted p.value < 0.05. For functional clustering of genes involved in chemokine signaling, the Database for Annotation, Visualization and Integrated Discovery (https://david.ncifcrf.gov/home.jsp) was used based on human homologs.
To infer homology of medaka genes, orthology and paralogy relationships were established by using the BLAST/BLAT tools of ENSEMBL (http://asia.ensembl.org/Multi/Tools/Blast) with default parameters and conserved synteny analysis with GENOMICUS version 96.01 (https://www.genomicus.biologie.ens.fr/genomicus-96.01/cgi-bin/search.pl).
Quantitative PCR.
RNA was isolated from FACS purified osteoblast progenitors, osteoblasts, and osteoclasts derived from 15 to 20 larvae per sample using the PurelinkTM RNA micro kit (Invitrogen) as described previously (21). cDNA was synthesized and amplified using the PreAmp and Reverse Transcription Master Mix Kit (Fluidigm). qPCR was performed with PowerUp SYBR Green Master Mix (Applied Biosystems). Primers for qPCR are listed in SI Appendix, Table S5; β-actin was used for normalization. Comparisons of transcript levels in Rankl-expressing relative to heat-shocked Rankl-negative larvae were performed with three biological replicates and three technical replicates each. Samples for RNAseq analysis and qPCR validation were obtained from independent experiments. Data were analyzed using Bio-Rad CFX Maestro 1.0 software and Student’s t tests were performed for statistical analysis.
Genetic Ablation of Osteoblasts.
Transgenic lines used for nitroreductase-mediated cell ablation are described elsewhere (29). Osteoblasts were ablated by treating osx:mCherry-NTRo larvae with 8 mM metronidazole (Mtz) for 24 h at 9 dpf. Mtz-treated osx:mCherry larvae were used as controls.
CXCR3 Inhibitor Treatment.
The CXCR3 inhibitors AMG487 and NBI-74330 (Tocris Biosciences) were dissolved in DMSO and 100% ethanol to make 10 and 30 mM stocks, respectively. Stocks were added to 50% hydroxypropyl-β-cyclodextrin (Sigma) solution, vortexed vigorously, and further diluted with sterile water and 30% Danieau’s solution [19.3 mM NaCl, 0.23 mM KCl, 0.13 mM MgSO4, 0.2 mM Ca(NO3)2, 1.7 mM Hepes, pH 7.0] to 20 and 30 μM working concentrations, respectively. AMG487 solution was injected into the yolk at 3 h prior to heat shock. Thereafter, larvae were kept in fish medium containing 20 μM AMG487 for the course of the experiment. For NBI-74330, larvae were kept in fish medium containing 30 μM NBI-74330 for 3 h prior to heat shock, changed to the pure fish medium during heat shock, and then reverted to drug solution for the course of the experiment. Drug solutions were freshly made and changed daily.
Bone Staining.
For live bone staining, larvae were incubated in 0.01% Calcein (Sigma) in the dark for 1 h, washed with fish medium twice, and mounted for imaging. Alizarin Red bone staining was performed as previously described (16).
Imaging.
For live fluorescence imaging, medaka larvae were anesthetized with 0.016% Tricaine (MS222, Sigma), mounted in 1.2% low melting point agarose on a glass-bottom Petri dish, and imaged using an Olympus FluoView FV3000 confocal microscope or a Nikon SMZ18 stereomicroscope equipped with the NIS-Elements BR 3.0 software. Time-lapse imaging was performed with an Olympus FluoView FV3000 confocal microscope. To image fixed Alizarin Red stained samples, a Nikon Eclipse 90i upright microscope equipped with NIS-Elements BR 3.0 software was used. Images and time-lapse movies were processed with Fiji ImageJ, Bitplane Imaris, and Adobe Photoshop.
Quantification of Cells and Statistical Analysis.
Cell numbers (mpeg1:mCherry, ctsk:GFP, ctsk:mCherry, and cxcr3.2:GFP) were quantified as previously described (58) with slight modifications. Using Fiji, fluorescent images were converted to greyscale and thresholded, and the area of a cell (averaged from 15 cells from three larvae) as well as the total area of cells in a selected region of interest were obtained. Cell numbers were determined by dividing the total area of cells by the average cell area. ANOVA with multiple comparisons, Mann–Whitney U, and Student’s t tests were conducted in Prism (Graphpad) for statistical analysis. Details are indicated in corresponding figure legends.
Bacterial Infection.
Nonpathogenic E. coli K12 strains expressing a GFP reporter were prepared as described (59). Bacterial suspensions were microinjected into the muscle of 10 dpf larvae at ∼2,500 CFU per larva.
Supplementary Material
Acknowledgments
We thank Georges Lutfalla (Université Montpellier) for sharing fluorescent E. coli and pTol2-mCherry-F plasmid and Baubak Bajoghli (University of Tuebingen) for providing the cxcr3.2-GFP plasmid. We also thank the Centre for Bioimaging Sciences (CBIS) confocal unit and the fish facility at Department of Biological Sciences (DBS), National University of Singapore for continued support. This project is supported by grants from the Singapore Ministry of Education (MOE2016-T2-2-086) and the National Research Foundation Singapore (NRF2017-NRF-ISF002-2671).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2006093117/-/DCSupplemental.
Data Availability.
All data generated in this study are included in the paper and SI Appendix. All reagents described in this study are available from the corresponding author upon request.
References
- 1.Charles J. F., Aliprantis A. O., Osteoclasts: More than “bone eaters”. Trends Mol. Med. 20, 449–459 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vega E. et al., Bone mineral density and bone size in men with primary osteoporosis and vertebral fractures. Calcif. Tissue Int. 62, 465–469 (1998). [DOI] [PubMed] [Google Scholar]
- 3.Feng X., McDonald J. M., Disorders of bone remodeling. Annu. Rev. Pathol. 6, 121–145 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng X., Teitelbaum S. L., Osteoclasts: New insights. Bone Res. 1, 11–26 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyce B. F., Advances in osteoclast biology reveal potential new drug targets and new roles for osteoclasts. J. Bone Miner. Res. 28, 711–722 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ikebuchi Y. et al., Coupling of bone resorption and formation by RANKL reverse signalling. Nature 561, 195–200 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Chen X., Zhi X., Wang J., Su J., RANKL signaling in bone marrow mesenchymal stem cells negatively regulates osteoblastic bone formation. Bone Res. 6, 34 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang S. et al., Osteoclast regulation of osteoblasts via RANK-RANKL reverse signal transduction in vitro. Mol. Med. Rep. 16, 3994–4000 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Brien W. et al., RANK-independent osteoclast formation and bone erosion in inflammatory arthritis. Arthritis Rheumatol. 68, 2889–2900 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lleras-Forero L., Winkler C., Schulte-Merker S., Zebrafish and medaka as models for biomedical research of bone diseases. Dev. Biol. 457, 191–205 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Witten P. E., Harris M. P., Huysseune A., Winkler C., Small teleost fish provide new insights into human skeletal diseases. Methods Cell Biol. 138, 321–346 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Huitema L. F. et al., Entpd5 is essential for skeletal mineralization and regulates phosphate homeostasis in zebrafish. Proc. Natl. Acad. Sci. U.S.A. 109, 21372–21377 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spoorendonk K. M. et al., Retinoic acid and Cyp26b1 are critical regulators of osteogenesis in the axial skeleton. Development 135, 3765–3774 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Renn J., Büttner A., To T. T., Chan S. J., Winkler C., A col10a1:nlGFP transgenic line displays putative osteoblast precursors at the medaka notochordal sheath prior to mineralization. Dev. Biol. 381, 134–143 (2013). [DOI] [PubMed] [Google Scholar]
- 15.To T. T. et al., Rankl-induced osteoclastogenesis leads to loss of mineralization in a medaka osteoporosis model. Development 139, 141–150 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Renn J., Winkler C., Osterix-mCherry transgenic medaka for in vivo imaging of bone formation. Dev. Dyn. 238, 241–248 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Yu T. et al., Live imaging of osteoclast inhibition by bisphosphonates in a medaka osteoporosis model. Dis. Model. Mech. 9, 155–163 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goto Y. et al., CXCR4+ CD45- cells are niche forming for osteoclastogenesis via the SDF-1, CXCL7, and CX3CL1 signaling pathways in bone marrow. Stem Cells 34, 2733–2743 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Liu P. et al., Loss of menin in osteoblast lineage affects osteocyte-osteoclast crosstalk causing osteoporosis. Cell Death Differ. 24, 672–682 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwak H. B. et al., Monokine induced by interferon-gamma is induced by receptor activator of nuclear factor kappa B ligand and is involved in osteoclast adhesion and migration. Blood 105, 2963–2969 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Buettner A. et al., Fluorescence-activated cell sorting (FACS) of osteoblasts and osteoclasts for RNA sequencing in a medaka, Oryzias latipes (Temming & Schlegel, 1846), osteoporosis model. J. Appl. Ichthyology 34, 481–488 (2018). [Google Scholar]
- 22.Aghaallaei N., Bajoghli B., Schwarz H., Schorpp M., Boehm T., Characterization of mononuclear phagocytic cells in medaka fish transgenic for a cxcr3a:gfp reporter. Proc. Natl. Acad. Sci. U.S.A. 107, 18079–18084 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mladic M. et al., Metabolic profiling of ligands for the chemokine receptor CXCR3 by liquid chromatography-mass spectrometry coupled to bioaffinity assessment. Anal. Bioanal. Chem. 407, 7067–7081 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heise C. E. et al., Pharmacological characterization of CXC chemokine receptor 3 ligands and a small molecule antagonist. J. Pharmacol. Exp. Ther. 313, 1263–1271 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Johnson M. et al., Discovery and optimization of a series of quinazolinone-derived antagonists of CXCR3. Bioorg. Med. Chem. Lett. 17, 3339–3343 (2007). [DOI] [PubMed] [Google Scholar]
- 26.van Wanrooij E. J. et al., CXCR3 antagonist NBI-74330 attenuates atherosclerotic plaque formation in LDL receptor-deficient mice. Arterioscler. Thromb. Vasc. Biol. 28, 251–257 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Wijtmans M., Verzijl D., Leurs R., de Esch I. J., Smit M. J., Towards small-molecule CXCR3 ligands with clinical potential. ChemMedChem 3, 861–872 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Willems B. et al., Conditional ablation of osteoblasts in medaka. Dev. Biol. 364, 128–137 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Dasyani M. et al., Lineage tracing of col10a1 cells identifies distinct progenitor populations for osteoblasts and joint cells in the regenerating fin of medaka (Oryzias latipes). Dev. Biol. 455, 85–99 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Suda T. et al., Modulation of osteoclast differentiation and function by the new members of the tumor necrosis factor receptor and ligand families. Endocr. Rev. 20, 345–357 (1999). [DOI] [PubMed] [Google Scholar]
- 31.Huang B. et al., Osteoblasts secrete Cxcl9 to regulate angiogenesis in bone. Nat. Commun. 7, 13885 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benigni G. et al., CXCR3/CXCL10 axis regulates neutrophil-NK cell cross-talk determining the severity of experimental osteoarthritis. J. Immunol. 198, 2115–2124 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Hiyari S. et al., Genomewide association study identifies Cxcl family members as partial mediators of LPS-induced periodontitis. J. Bone Miner. Res. 33, 1450–1463 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J. et al., RANK is the intrinsic hematopoietic cell surface receptor that controls osteoclastogenesis and regulation of bone mass and calcium metabolism. Proc. Natl. Acad. Sci. U.S.A. 97, 1566–1571 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ofer L., Dumont M., Rack A., Zaslansky P., Shahar R., New insights into the process of osteogenesis of anosteocytic bone. Bone 125, 61–73 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Sommer F., Torraca V., Kamel S. M., Lombardi A., Meijer A. H., Frontline Science: Antagonism between regular and atypical Cxcr3 receptors regulates macrophage migration during infection and injury in zebrafish. J. Leukoc. Biol. 107, 185–203 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brylka L. J., Schinke T., Chemokines in physiological and pathological bone remodeling. Front. Immunol. 10, 2182 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun W. et al., B cells inhibit bone formation in rheumatoid arthritis by suppressing osteoblast differentiation. Nat. Commun. 9, 5127 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Esposito M., Guise T., Kang Y., The biology of bone metastasis. Cold Spring Harb. Perspect. Med. 8, a031252 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wise G. E., Que B. G., Huang H., Synthesis and secretion of MCP-1 by dental follicle cells—Implications for tooth eruption. J. Dent. Res. 78, 1677–1681 (1999). [DOI] [PubMed] [Google Scholar]
- 41.Wu A. C., Morrison N. A., Kelly W. L., Forwood M. R., MCP-1 expression is specifically regulated during activation of skeletal repair and remodeling. Calcif. Tissue Int. 92, 566–575 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Myers T. J. et al., BMP2 regulation of CXCL12 cellular, temporal, and spatial expression is essential during fracture repair. J. Bone Miner. Res. 30, 2014–2027 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Groom J. R., Luster A. D., CXCR3 ligands: Redundant, collaborative and antagonistic functions. Immunol. Cell Biol. 89, 207–215 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reynders N. et al., The distinct roles of CXCR3 variants and their ligands in the tumor microenvironment. Cells 8, 613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tokunaga R. et al., CXCL9, CXCL10, CXCL11/CXCR3 axis for immune activation—A target for novel cancer therapy. Cancer Treat. Rev. 63, 40–47 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deng S. et al., Non-platelet-derived CXCL4 differentially regulates cytotoxic and regulatory T cells through CXCR3 to suppress the immune response to colon cancer. Cancer Lett. 443, 1–12 (2019). [DOI] [PubMed] [Google Scholar]
- 47.Lee J. H. et al., Pathogenic roles of CXCL10 signaling through CXCR3 and TLR4 in macrophages and T cells: Relevance for arthritis. Arthritis Res. Ther. 19, 163 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Puchert M., Obst J., Koch C., Zieger K., Engele J., CXCL11 promotes tumor progression by the biased use of the chemokine receptors CXCR3 and CXCR7. Cytokine 125, 154809 (2020). [DOI] [PubMed] [Google Scholar]
- 49.Fulkerson P. C., Zhu H., Williams D. A., Zimmermann N., Rothenberg M. E., CXCL9 inhibits eosinophil responses by a CCR3- and Rac2-dependent mechanism. Blood 106, 436–443 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu T., Winkler C., Drug treatment and in vivo imaging of osteoblast-osteoclast interactions in a medaka fish osteoporosis model. J. Vis. Exp., 10.3791/55025 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rettig M. P., Ansstas G., DiPersio J. F., Mobilization of hematopoietic stem and progenitor cells using inhibitors of CXCR4 and VLA-4. Leukemia 26, 34–53 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lecavalier-Barsoum M. et al., Targeting the CXCL12/CXCR4 pathway and myeloid cells to improve radiation treatment of locally advanced cervical cancer. Int. J. Cancer 143, 1017–1028 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Pusic I., DiPersio J. F., Update on clinical experience with AMD3100, an SDF-1/CXCL12-CXCR4 inhibitor, in mobilization of hematopoietic stem and progenitor cells. Curr. Opin. Hematol. 17, 319–326 (2010). [DOI] [PubMed] [Google Scholar]
- 54.Andrews S. P., Cox R. J., Small molecule CXCR3 antagonists. J. Med. Chem. 59, 2894–2917 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Stemmer M., Thumberger T., Del Sol Keyer M., Wittbrodt J., Mateo J. L., CCTop: An intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS One 10, e0124633 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dobin A. et al., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Phan Q. T. et al., Neutrophils use superoxide to control bacterial infection at a distance. PLoS Pathog. 14, e1007157 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nguyen-Chi M. et al., Transient infection of the zebrafish notochord with E. coli induces chronic inflammation. Dis. Model. Mech. 7, 871–882 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated in this study are included in the paper and SI Appendix. All reagents described in this study are available from the corresponding author upon request.