A(H9N2) influenza viruses are widespread in poultry in many parts of the world and for over 20 years have sporadically jumped species barriers to cause human infection. As these viruses continue to diversify genetically and antigenically, it is critical to closely monitor viruses responsible for human infections, to ascertain if A(H9N2) viruses are acquiring properties that make them better suited to infect and spread among humans. In this study, we describe an active poultry surveillance system established in Vietnam to identify the scope of influenza viruses present in live bird markets and the threat they pose to human health. Assessment of a recent A(H9N2) virus isolated from an individual in China in 2018 is also reported, and it was found to exhibit properties of adaptation to humans and, importantly, it shows similarities to strains isolated from the live bird markets of Vietnam.
KEYWORDS: H9N2, influenza, surveillance studies, transmission
ABSTRACT
Low-pathogenicity avian influenza A(H9N2) viruses, enzootic in poultry populations in Asia, are associated with fewer confirmed human infections but higher rates of seropositivity compared to A(H5) or A(H7) subtype viruses. Cocirculation of A(H5) and A(H7) viruses leads to the generation of reassortant viruses bearing A(H9N2) internal genes with markers of mammalian adaptation, warranting continued surveillance in both avian and human populations. Here, we describe active surveillance efforts in live poultry markets in Vietnam in 2018 and compare representative viruses to G1 and Y280 lineage viruses that have infected humans. Receptor binding properties, pH thresholds for HA activation, in vitro replication in human respiratory tract cells, and in vivo mammalian pathogenicity and transmissibility were investigated. While A(H9N2) viruses from both poultry and humans exhibited features associated with mammalian adaptation, one human isolate from 2018, A/Anhui-Lujiang/39/2018, exhibited increased capacity for replication and transmission, demonstrating the pandemic potential of A(H9N2) viruses.
IMPORTANCE A(H9N2) influenza viruses are widespread in poultry in many parts of the world and for over 20 years have sporadically jumped species barriers to cause human infection. As these viruses continue to diversify genetically and antigenically, it is critical to closely monitor viruses responsible for human infections, to ascertain if A(H9N2) viruses are acquiring properties that make them better suited to infect and spread among humans. In this study, we describe an active poultry surveillance system established in Vietnam to identify the scope of influenza viruses present in live bird markets and the threat they pose to human health. Assessment of a recent A(H9N2) virus isolated from an individual in China in 2018 is also reported, and it was found to exhibit properties of adaptation to humans and, importantly, it shows similarities to strains isolated from the live bird markets of Vietnam.
INTRODUCTION
Widespread detection of low-pathogenicity avian influenza (LPAI) A(H9N2) viruses in gallinaceous poultry (i.e., domestic and game birds) has been reported over the past 20 years in Asia, the Middle East, and regions of Africa, with periodic detection in Europe and the Americas (1). There is extensive genetic diversity present among A(H9N2) viruses, with over a hundred distinct genotypes and several discrete lineages currently identified (2). Documented A(H9N2) human cases or evidence of human A(H9N2) virus infection or exposure via serologic studies have been reported in over a dozen countries (1, 3), highlighting the capacity for this virus subtype to cause mild and/or asymptomatic infections in humans. To date, confirmed human cases have been limited to the G1 and Y280 lineages (also known as the BJ94 or G9 lineages based on the A/chicken/Beijing/1/1994 and A/chicken/Hong Kong/G9/1997 prototype viruses, respectively).
While human-to-human transmission of A(H9N2) viruses has not been reported, viruses from both lineages have caused human infection and display enhanced binding to human-like α2,6-linked sialic acid receptors (4, 5). Multiple influenza virus subtypes associated with human infection, notably A(H7N9) viruses, possess internal genes derived from A(H9N2) viruses (6, 7), indicating that reassortment of avian influenza viruses with A(H9N2) viruses in poultry may enhance their capacity to adapt to mammalian hosts. A(H9N2) influenza viruses also possess heterogeneity in their threshold pH for fusion, with select viruses exhibiting pH stability similar to human transmissible viruses (8). Moreover, enhanced transmissibility following mammalian adaptation has been observed using the ferret model (4, 9). Finally, the lack of immunity to A(H9N2) subtype viruses in the general human population, coupled with their increasing genetic and antigenic diversity, adds to their public health threat and pandemic potential and the need for prepandemic candidate vaccine virus (CVV) assessment and development (1, 3).
Live bird markets (LBMs) can facilitate the circulation and reassortment of avian influenza viruses and are an ideal interface for human exposure to infected poultry and emergence of novel viruses. Approximately 70% of persons with confirmed A(H9N2) virus infection have reported poultry exposure prior to infection (3). In Vietnam, LBMs are common throughout the country, connecting farms with distributors, and play a substantial role in the poultry industry. As such, these markets are often sentinel locations for the identification of endemic and emerging avian influenza viruses and serve as critical sites from which to conduct active surveillance. Collection of samples from infected birds across national surveillance networks in Vietnam results in the testing of thousands of avian influenza viruses each year, allowing for their genetic and antigenic analysis as well as additional virologic characterization required for virus risk assessment.
Here, we examined genetically and antigenically diverse poultry viruses from Vietnam and compared them to contemporary LPAI A(H9N2) viruses inclusive of both G1 and Y280 lineages isolated from recent human infections. Genetic, antigenic, and receptor binding site characterization identified further divergence of recently detected A(H9N2) viruses and the need for continued CVV development. We found that A(H9N2) viruses were capable of high-titer replication in human respiratory tract cells and throughout the respiratory tract of both mice and ferrets. Transmission was efficient between cohoused ferrets, while a recent virus isolated following a human infection in China was capable of robust transmission through the air.
RESULTS
LBM surveillance and HA gene diversity.
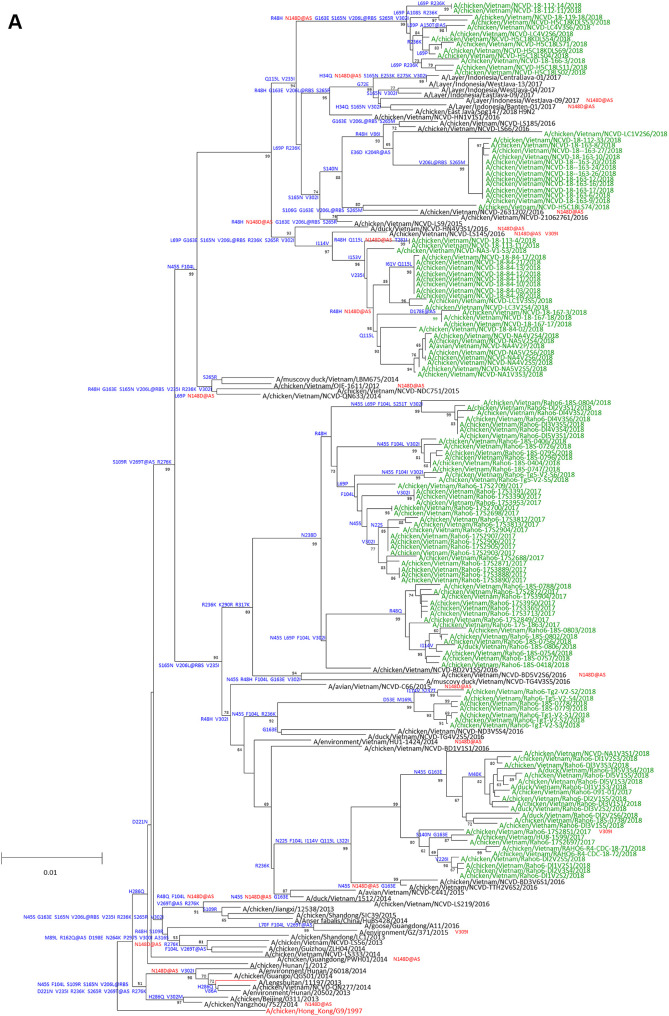
Surveillance in LBMs in Vietnam from September 2017 to September 2018 detected 163 A(H9N2) positive oropharyngeal swabs and environmental samples. A(H9N2) viruses made up 36% of the total avian influenza viruses detected. Other subtypes identified included H2N3, H3N2, H3N8, H5N1, H5N6, H6N2, H6N6, H7N6, H10N3, and H11N9. Codon complete genomic sequencing of viral RNAs was successful for 55 new Y280-lineage sequences from Vietnam. Viruses clustered with previously described Y280 lineage viruses, including the Y280 CVV, A/Hong Kong/308/2014 (HK/308). Viruses from Vietnam exhibited extensive genetic diversity clustering in multiple small groups and two larger overarching lineages classified as the A Y280 and B Y280 lineages (Fig. 1A and B). The A Y280 lineage viruses were collected from both northern, central and southern Vietnamese provinces, while B Y280 viruses were collected almost exclusively from central and northern provinces (see Table S1 in the supplemental material). Some clusters from both lineages contained previously identified A(H9N2) viruses from Vietnam and likely represented endemically circulating strains found in Vietnam since at least 2016. Other groups had no close ancestors to previous Vietnam viruses, suggesting recent emergence or introduction into Vietnam. One cluster of viruses in the A Y280 lineage was collected in northern provinces bordering China (i.e., Lang Son, Lao Cai, and Quang Ninh provinces) (Fig. 1A). The nearest ancestors were viruses detected in China in 2017 and 2018, suggesting introduction via shorter-distance poultry trade across porous borders. Three clusters of viruses belonging to the B Y280 lineage were identified with no known ancestors previously detected in Vietnam (Fig. 1B). One group contained viruses from northern border provinces with close phylogenetic proximity to viruses collected in Guangdong and Fujian provinces, China. Another group in the B Y280 lineage (collected in the northern border province of Lang Son) was closely related to viruses from Myanmar detected in 2018, suggesting more distant but rapid dispersal of these viruses in southeast Asia (Fig. 1B). Finally, a third cluster, also collected from Lang Son province, was related to poultry viruses from Guangdong province that caused a human infection in the same province in 2018. One A(H9N2)-positive environmental sample from Vinh Long province (Mekong Delta region of Vietnam) encoded HA viral RNA that belonged to the Eurasian/American wild bird lineage and was genetically distinct from all recommended A(H9N2) CVVs (data not shown).
FIG 1.
Evolution of A(H9N2) HA genes belonging to the Y280 lineages A and B. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches. (A and B) The A Y280 (A) and B Y280 (B) viruses tested in this study are shown in green. Candidate vaccine viruses are shown in red. Amino acid differences relative to the closest candidate vaccine viruses are shown in blue on each tree branch. Mutations found in the genetic changes inventory are shown in red. Bold lettering indicates a mutation at a putative antigenic site.
Molecular analysis of LPAI A(H9N2) viruses.
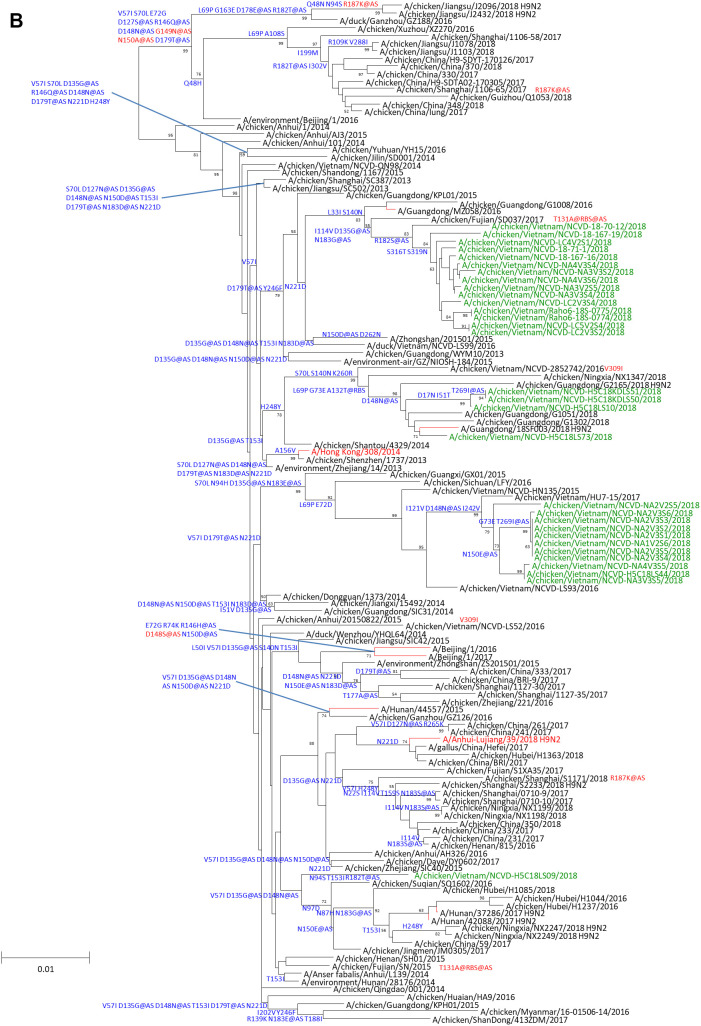
Representative H9N2 viruses isolated from LBM surveillance in Vietnam (inclusive of multiple lineages and genetic diversity) were chosen for subsequent analyses, contextualized with viruses isolated from recent human cases from both G1 and Y280 lineages. Viruses isolated from humans and tested in this study were collected through routine diagnostic detections of A(H9N2) infections reported by national authorities in Bangladesh, Hong Kong, and China. The majority of cases were detected in children with mild illness. Most cases reported direct contact with poultry prior to illness onset, and all cases recovered from infection with no evidence of further human-to-human transmission (10–12).
A Y280 viruses from Vietnam were genetically close to the A/chicken/Hong Kong/G9/97 CVV but possessed 23 to 26 amino acid differences in HA1 (Fig. 1A). Conserved amino acid differences were found in antigenic sites B (N148D), C (V269T), D (R162Q and Q217M) and E (R48H), but no differences resulted in changes in glycosylation status. Compared to the currently recommended CVV, A/Anhui-Lujiang/39/2018 (A-L/39, a human isolate from 2018), the B Y280 Vietnam viruses had 9 to 17 amino acid changes in the HA1. Three conserved amino acid substitutions in the HA1 of these viruses were in putative antigenic sites A (D135G) and B (D179T and N183G/E) but also did not result in glycosylation changes. Compared to HK/308 virus, B Y280 Vietnam viruses had between 7 to 23 amino acid differences in the HA1 protein (Fig. 1B). Despite the phylogenetic diversity of A and B Y280 HA genes, the viruses had similar amino acid residues at positions associated with receptor specificity and markers of mammalian adaptation potential and shared the same receptor specificity residues (i.e., Leu-Met-Gly at positions 226 to 228 by H3 numbering), indicating potential for dual-receptor specificity. This motif was shared by the Y280 human viruses tested in this study, while the G1 lineage human viruses had various combinations (Table 1). A Y280 viruses had predominately Ala at position 190, while B Y280 avian and human viruses had Thr, suggesting a potential difference in receptor specificity determined by the 190 helical domain. Both avian and human Y280 viruses had an Asn at position 183 associated with altered receptor binding specificity (Table 1).
TABLE 1.
Key molecular features and threshold pH of fusion of A(H9N2) viruses used in this study
| Virus name | Abbreviation | Lineage | HAa |
NA | PB2 |
PB1 | PA | pH of fusion | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 183 (173) | 190 (180) | 226 (216) | 227 (217) | 228 (218) | del62-64 | 627 | 701 | 368 | 185 | ||||
| A/Hong Kong/1073/1999 | HK/1073 | G1 | H | E | L | Q | G | no | E | D | I | K | 5.8 |
| A/Hong Kong/33982/2009 | HK/33982 | G1 | H | D | Q | Q | G | no | E | N | I | K | 5.5–5.8 |
| A/Bangladesh/0994/2011 | Bang/0994 | G1 | H | A | L | I | G | no | E | D | I | K | 5.6–5.7 |
| A/Hong Kong/308/2014 | HK/308 | B Y280 | N | T | L | M | G | yes | E | D | V | R | 5.4–5.5 |
| A/Anhui-Lujiang/39/2018 | A-L/39 | B Y280 | N | T | L | M | G | yes | V | D | V | R | 5.4 |
| A/ck/Vietnam/NCVD-18-70-12/2018 | ck/VN/70 | B Y280 | N | T | L | M | G | no | E | D | V | R | 5.5 |
| A/ck/Vietnam/NCVD-NA4V3S5/2018 | ck/VN/NA4 | B Y280 | N | T | L | M | G | yes | E | D | V | R | 5.3–5.4 |
| A/ck/Vietnam/NCVD-18-119-18/2018 | ck/VN/119 | A Y280 | N | A | L | M | G | no | E | D | V | R | 5.3–5.4 |
| A/ck/Vietnam/NCVD-18-167-18/2018 | ck/VN/167 | A Y280 | N | A | L | M | G | no | E | D | V | R | 5.4–5.5 |
For HA, H3 numbering is shown, with H9 numbering provided in parentheses.
The neuraminidase (NA) genes clustered with topologies similar to the HA genes (data not shown). At the amino acid level, only a small number of B Y280 viruses (n = 26) had a deletion of three amino acids (NA 62 to 64), indicative of adaptation to terrestrial poultry. This deletion was found in both of the Y280 human viruses characterized (Table 1). The presence of 294R among all H9N2 viruses evaluated here predicts sensitivity to the NA inhibitor oseltamivir. With the exception of A/Hong Kong/33982/2009, none of the PB2 proteins of the viruses characterized had 627K or 701N but instead possessed avian-consensus amino acids 627E/V and 701D. Additionally, while the G1 lineage human viruses tested contained an Ile at position 368 of the PB1 protein, the majority of Y280 viruses, including the human viruses, had a Val at this position. Finally, the PA proteins differed between the G1 and Y280 lineage viruses with the later lineage possessing Arg at position 185 (Table 1). GiRaF reassortment analysis did not identify reassortment within gene segments of the A(H9N2) viruses sequenced for this study (data not shown). In addition, there were no reassortment events confirmed among the A(H9N2) human-origin viruses analyzed in this study.
Antigenic characteristics of LPAI A(H9N2) viruses.
Hemagglutination inhibition (HI) testing of 14 recent A(H9N2) Y280 lineage viruses from Vietnam was performed (Table 2). Ferret antiserum raised to the A/chicken/Hong Kong/G9/1997-like CVV (IBCDC-RG2) inhibited hemagglutination of seven A Y280 viruses at titers that were equivalent or within 2-fold of the homologous virus titer. However, this antiserum had no cross-reactive titers with B Y280 viruses. In contrast, ferret antisera raised to wild-type A-L/39 virus had heterologous titers against six B Y280 Vietnam viruses that were within 4-fold of the homologous virus titer but significantly reduced heterologous titers (16- to 32-fold) to the A Y280 viruses (Table 2). In addition, ferret antisera raised to the Y280 wild-type virus, A/Hong Kong/308/2014, had heterologous titers that were, on average, 8-fold or greater reduced with both A and B Y280 Vietnam viruses compared to the homologous virus titer, indicating recent antigenic drift among viruses from both Y280 lineages.
TABLE 2.
Hemagglutination inhibition assay using representative A(H9N2) viruses
| Antigena | Lineage | G1 | G1 | A Y280 | B Y280 |
Date collectedc | |
|---|---|---|---|---|---|---|---|
| Bang/0994 | RG-31 | RG-2 | HK/308 | A-L/39 | |||
| Reference antigens | |||||||
| A/Bangladesh/0994/2011 | G1 | 5120b | 2,560 | 40 | 10 | 40 | 2/26/2011 |
| A/Bangladesh/0994/2011 IDCDC RG-31 | G1 | 5,120 | 2560 | 80 | 10 | 40 | NA |
| A/chicken/Hong Kong/G9/1997 IBCDC RG-2 | A Y280 | 640 | 20 | 320 | 10 | <10 | NA |
| A/Hong Kong/308/2014 | B Y280 | 40 | 10 | <10 | 1280 | 640 | 12/28/2013 |
| A/Anhui-Lujiang/39/2018 | B Y280 | 80 | <10 | <10 | 320 | 5120 | 1/2/2018 |
| Test Antigens | |||||||
| A/chicken/Vietnam/NCVD-18-119-18/2018 | A Y280 | 640 | 80 | 160 | 80 | 160 | 3/11/2018 |
| A/chicken/Vietnam/NCVD-H5C18LS02/2018 | A Y280 | 640 | 80 | 160 | 80 | 160 | 2/11/2018 |
| A/chicken/Vietnam/NCVD-H5C18KDLS54/2018 | A Y280 | 1280 | 160 | 320 | 160 | 320 | 3/15/2018 |
| A/chicken/Vietnam/NCVD-18–163-16/2018 | A Y280 | 640 | 80 | 160 | 80 | 160 | 6/4/2018 |
| A/chicken/Vietnam/NCVD-18-84-11/2018 | A Y280 | 1280 | 80 | 160 | 80 | 160 | 8/2/2018 |
| A/chicken/Vietnam/NCVD-18-113-11/2018 | A Y280 | 640 | 80 | 160 | 80 | 160 | 3/11/2018 |
| A/chicken/Vietnam/NCVD-18-167-18/2018 | A Y280 | 640 | 80 | 320 | 160 | 320 | 9/4/2018 |
| A/chicken/Vietnam/NCVD-NA4V2S6/2018 | A Y280 | 640 | 80 | 160 | 80 | 160 | 3/7/2018 |
| A/chicken/Vietnam/Raho6-18S-0775/2018 | B Y280 | 80 | <10 | <10 | 40 | 1280 | 4/5/2018 |
| A/chicken/Vietnam/NCVD-LC5V2S4/2018 | B Y280 | 80 | <10 | <10 | 320 | 5120 | 8/3/2018 |
| A/chicken/Vietnam/NCVD-18-70-12/2018 | B Y280 | 80 | <10 | <10 | 40 | 1280 | 3/4/2018 |
| A/chicken/Vietnam/NCVD-H5C18KDLS50/2018 | B Y280 | 40 | <10 | <10 | 320 | 2560 | 3/15/2018 |
| A/chicken/Vietnam/NCVD-NA4V3S5/2018 | B Y280 | 40 | <10 | <10 | 640 | 1280 | 5/4/2018 |
| A/chicken/Vietnam/NCVD-H5C18LS09/2018 | B Y280 | 20 | <10 | <10 | 160 | 2560 | 2/11/2018 |
A/Bangladesh/0994/2011 (Bang/0994); A/Bangladesh/0994/2011 IDCDC RG-31 (RG-31); A/chicken/Hong Kong/G9/1997 IBCDC RG-2 (RG-2); A/Hong Kong/308/2014 (HK/308); A/Anhui-Lujiang/39/2018 (A-L/39).
Hemagglutination inhibition titers represent the reciprocal of the titer that inhibited complete hemagglutination with a starting dilution of 1:10. Homologous hemagglutination titers are indicated in bold and underlined.
NA, not applicable.
A(H9N2) viruses exhibit a range of pH thresholds for HA activation.
An increasing body of evidence has supported the requirement for an acid-stable hemagglutinin for efficient airborne transmission of influenza viruses in mammalian models, as this allows survival of the virus in acidic milieu of the upper airways (13). Previous studies have indicated that most transmissible human influenza viruses have a fusion threshold pH of <5.5, while most A(H5) and A(H7) subtype avian influenza viruses generally fuse at a pH of >5.5 (14). Accordingly, we examined the ability of A(H9N2) viruses to induce syncytium formation in Vero cells upon exposure to low-pH fusion buffers (15). We found that the fusion threshold pH, as defined as the highest pH value to induce at least 50% or more syncytium, was at or above 5.5 (range of 5.5 to 5.8; Table 1) for all G1 lineage viruses tested in this study. In contrast, the fusion thresholds for all Y280 lineage viruses tested were equal to or below pH 5.5 (range of 5.3 to 5.5; Table 1), including viruses isolated from LBMs in Vietnam. The lower threshold pH for fusion exhibited by these A(H9N2) viruses is characteristic of the acid stability displayed by human seasonal influenza viruses.
Receptor binding properties of A(H9N2) viruses.
Human influenza viruses typically display a binding preference for α2,6-linked sialic acid receptors, while avian influenza viruses predominately bind to α2,3-linked sialic acids. Enhanced binding to human-like α2,6-linked sialic acids has been reported previously among human and avian A(H9N2) viruses (4, 5). To investigate whether recently isolated A(H9N2) viruses have maintained this capacity, we first employed a hemagglutination assay using differentially sialylated turkey red blood cells (RBCs). Comparable to the first-wave LPAI A(H7N9) virus, A/Shanghai/1/2013 (Shanghai/1), all G1 and B Y280 viruses bound to turkey RBCs sialylated with either α2,3- or α2,6-linked sialosides (Table 3), with all HA titers within a 2-fold difference. Interestingly, the two A Y280 A(H9N2) viruses bound only to α2,6-linked sialosides in this assay. These results support the notion that contemporary A(H9N2) viruses possess an ability to bind to α2,6-linked sialic acids, though strain-specific differences exist.
TABLE 3.
Hemagglutination of A(H9N2) viruses using resialylated red blood cells
| Virus | Subtype | HA titera
|
|||
|---|---|---|---|---|---|
| TRBC | α2-6 SA TRBC | α2-3 SA TRBC | Desialyated TRBC | ||
| HK/1073 | A(H9N2) | 2,048 | 4,096 | 8,192 | <2 |
| HK/33982 | A(H9N2) | 4,096 | 4,096 | 8,192 | <2 |
| Bang/0994 | A(H9N2) | 2,048 | 4,096 | 4,096 | <2 |
| HK/308 | A(H9N2) | 4,096 | 8,192 | 4,096 | <2 |
| A-L/39 | A(H9N2) | 4,096 | 8,192 | 4,096 | <2 |
| ck/VN/70 | A(H9N2) | 1,024 | 2,048 | 1,024 | <2 |
| ck/VN/NA4 | A(H9N2) | 2,048 | 2,048 | 2,048 | <2 |
| ck/VN/119 | A(H9N2) | 1,024 | 1,024 | <2 | <2 |
| ck/VN/167 | A(H9N2) | 2,048 | 2,048 | <2 | <2 |
| Shanghai/1 | A(H7N9) | 2,048 | 4,096 | 4,096 | <2 |
| Cal/7 | A(H1N1) | 32 | 64 | <2 | <2 |
| Switz/13 | A(H3N2) | 1,024 | 1,024 | 512 | <2 |
HA titers were measured with untreated turkey red blood cells (TRBC), Vibrio cholerae-treated TRBCs (desialylated), or α2,6 or α2,3 resialylated TRBCs. HA titer is representative of duplicate experiments and is the reciprocal of the highest dilution that results in agglutination of TRBC.
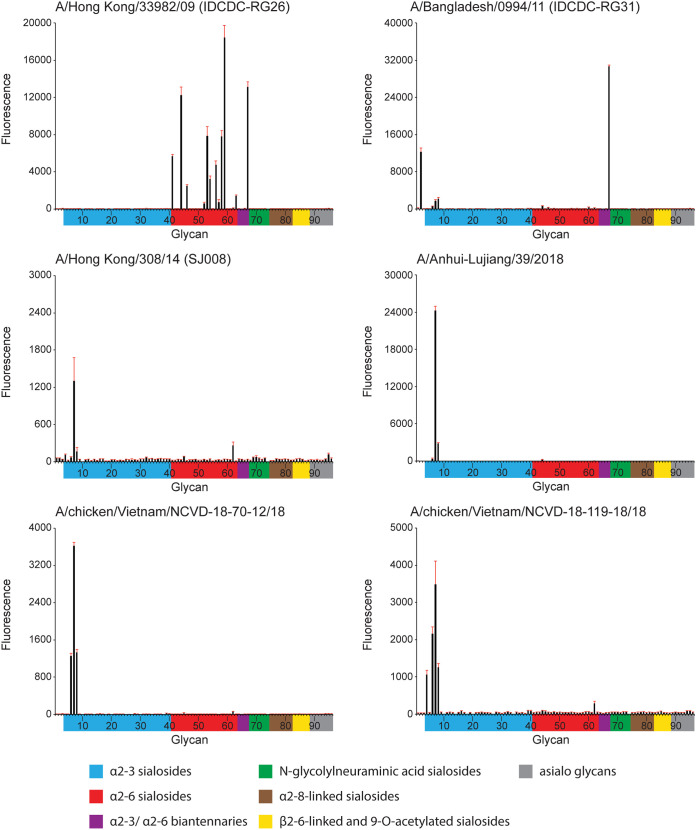
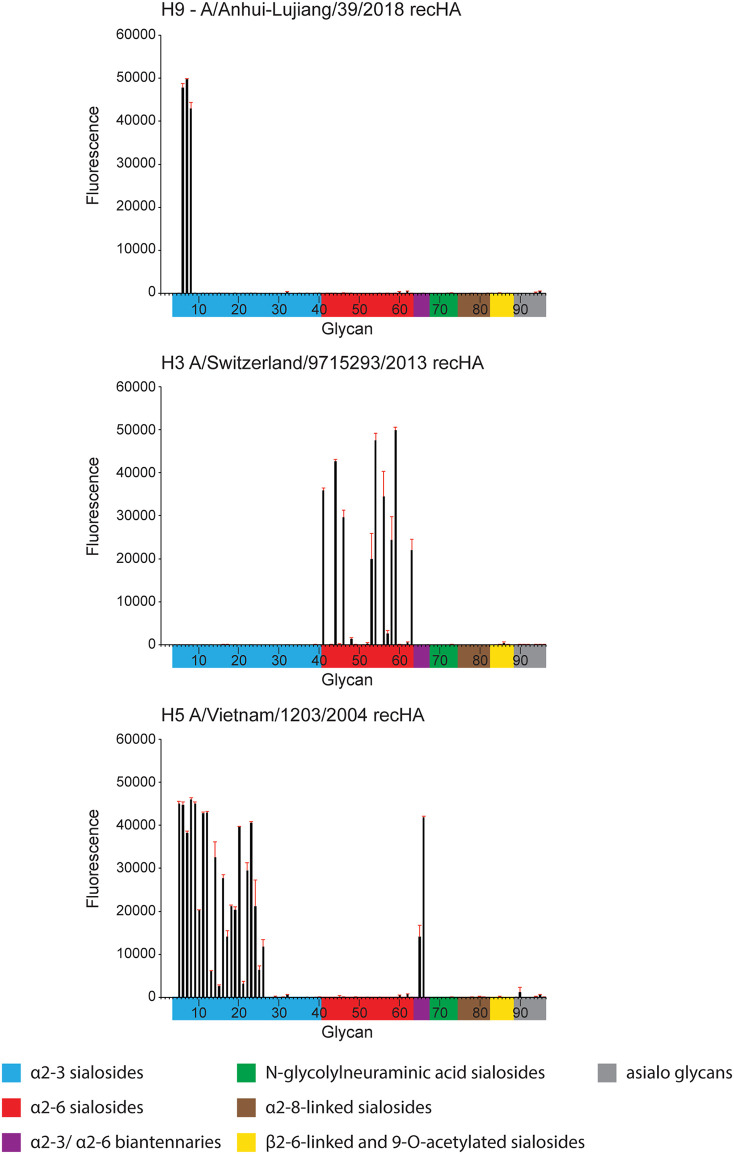
Next, a panel of recently isolated A(H9N2) viruses were subjected to glycan microarray analysis (Fig. 2) (16). In contrast to the RBC assay, which employs live virus and species-specific avian erythrocytes, glycan microarrays capture a complex profile of natural glycans from different species and tissue types. The G1 lineage virus HK/33982 bound well to α2,6 glycans, while Bang/0994 bound to a single α2,3/α2,6 di-sialoside (glycan number 67). In contrast, A and B Y280 lineage viruses revealed only limited binding to α2,3 sulfated sialosides (glycan numbers 6 to 8). Glycan microarray binding was also performed using a recombinant A-L/39 virus and avian Viet04 and seasonal Switz/13 HAs as controls; data were consistent with the A-L/39 virus results (Fig. 3).
FIG 2.
Glycan microarray analysis of representative A(H9N2) influenza viruses. Colored bars represent glycans that contain α-2,3 sialic acid (SA) (blue), α-2,6 SA (red), α-2,3/α-2,6 mixed SA (purple), N-glycolyl SA (green), α-2,8 SA (brown), β-2,6 and 9-O-acetyl SA (yellow), and non-SA (gray). Error bars reflect the standard error (SE) in the signal for 6 independent replicates on the array. Antisera for glycan array analysis were raised against homologous virus, with the exception of ck/VN/119 and ck/VN/70, for which antisera were raised against A-L/39 virus.
FIG 3.
Glycan microarray analysis of recombinant HA controls. A-L/39, avian Viet04, and seasonal Switz/13 virus HAs were analyzed. Colored bars represent glycans that contain α-2,3 sialic acid (SA) (blue), α-2,6 SA (red), α-2,3/α-2,6 mixed SA (purple), N-glycolyl SA (green), α-2,8 SA (brown), β-2,6 and 9-O-acetyl SA (yellow), and non-SA (gray). Error bars reflect SE in the signal for 6 independent replicates on the array.
A(H9N2) virus replication in human and ferret respiratory tract cells.
Previous studies have identified that A(H9N2) influenza viruses are capable of high-titer replication in human cultured respiratory tract cells and in ex vivo respiratory tract tissue samples (17, 18). However, recently isolated B Y280 viruses from human infections have not been extensively studied. To assess the capacity for contemporary A(H9N2) viruses from humans or chickens to replicate in a representative human respiratory tract cell line, Calu-3 cell cultures were inoculated with a panel of influenza A viruses and cultured at 37°C or 33°C, temperatures which emulate those of the human lower and upper respiratory tracts, respectively (Fig. 4).
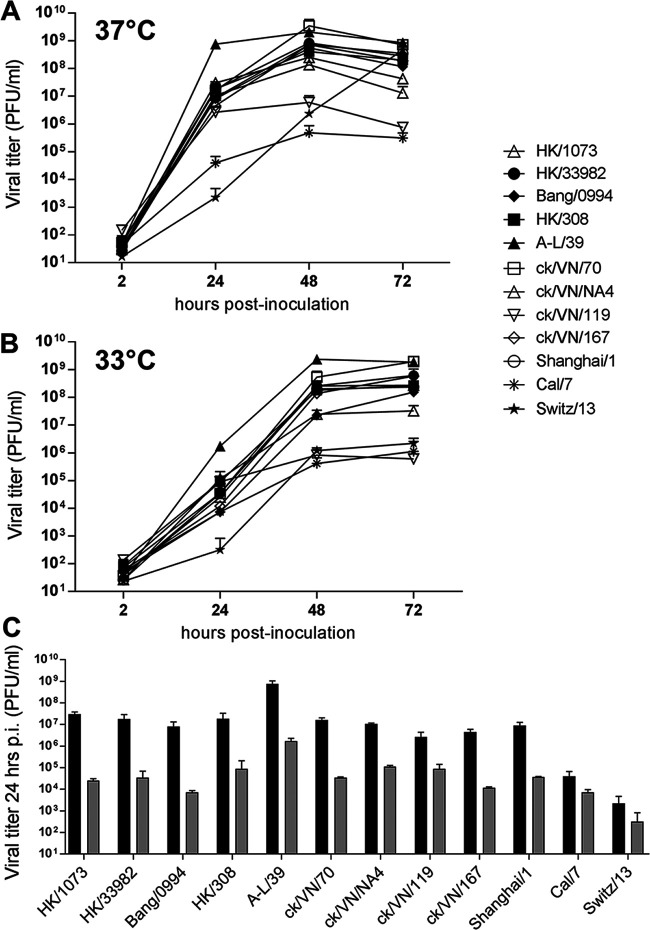
FIG 4.
Replication of A(H9N2) viruses in Calu-3 cells. Human bronchial epithelial (Calu-3) cells were grown on transwell inserts and infected apically in triplicate at an MOI of 0.01 with the indicated A(H9N2), A(H7N9) (Shanghai/1), 2009 pandemic A(H1N1) (Cal/7), or seasonal A(H3N2) (Switz/13) viruses. (A and B) Cells were incubated at 37°C (A) or 33°C (B). Culture supernatants were sampled at the indicated times p.i. and titrated in MDCK cells by standard plaque assay. (C) Comparison of infectious titers collected at 24 h p.i. from 37°C (black bars) and 33°C (gray bars) cultures is shown. The limit of virus detection was 10 PFU/ml.
All A(H9N2) viruses tested reached mean titers of >106 PFU/ml by 24 h postinfection (p.i.) at 37°C (Fig. 4A), demonstrating a high capacity for replication in this cell type, comparable to LPAI A(H7N9) viruses (represented here by the first-wave Shanghai/1 virus). Notably, A-L/39 virus replicated to significantly higher titer at 37°C than all other viruses examined at 24 h p.i. (P < 0.05). All A(H9N2) viruses reached mean peak titers by 48 h p.i. of >108 PFU/ml, with the exception of ck/VN/119 virus. In contrast, mean peak titers of the human 2009 pandemic A(H1N1) virus, A/California/7/2009 (Cal/7), were <106 PFU/ml at 48 h p.i., and the seasonal A(H3N2) virus, A/Switzerland/9715293/2013 (Switz/13), did not reach mean peak titers of >108 PFU/ml until 72 h p.i.
Avian influenza viruses typically replicate with reduced and delayed kinetics when incubated at 33°C, in contrast to human influenza viruses, which are often better suited for replication at the lower temperatures of the human upper airways (Fig. 4B) (19). In support of this, viral titers from cells infected by human A(H1N1) or A(H3N2) viruses cultured at 33°C were reduced by <1 log at 24 h p.i. compared with cells cultured at 37°C, while all A(H9N2) and A(H7N9) viruses tested exhibited titer reductions between 1.4 and 3.1 logs at this time point (Fig. 4C). Accordingly, culturing inoculated Calu-3 cells at 33°C resulted in delays in reaching mean peak titers for all viruses examined from 48 to 72 h (with two exceptions, A-L/39 and ck/VN/119). However, mean peak titers were within a log of one another for all A(H9N2) viruses tested at both temperatures by 72 h p.i. Taken together, all A(H9N2) viruses replicated efficiently in human bronchial epithelial cells, with the most recent A(H9N2) virus human isolate, A-L/39 virus, reaching the highest titers (>109 PFU/ml) at both culture temperatures.
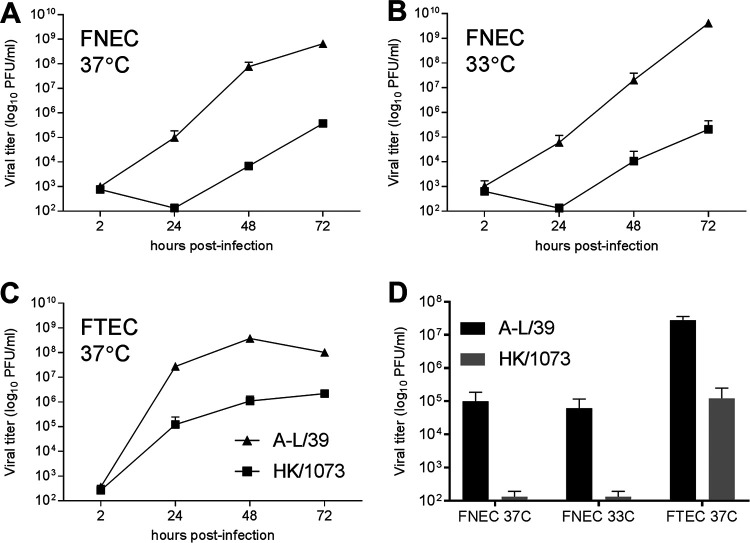
To evaluate the capacity of A(H9N2) viruses associated with human infection to replicate in the respiratory epithelia of another mammalian species, we inoculated cultures of differentiated primary ferret nasal epithelial cells (FNEC) or tracheal epithelial cells (FTEC) with either A-L/39 or HK/1073 virus (Fig. 5). In agreement with data from Calu-3 cells, A-L/39 virus replicated to significantly higher titers (P < 0.01) in either FNEC or FTEC cultured at 37°C compared with HK/1073 virus. Comparable virus replication in primary FNEC cultures was detected at 37°C or 33°C. Collectively, these data support a robust capacity of A(H9N2) viruses in general, and A-L/39 virus in particular, to replicate to high titer in the mammalian upper respiratory tract.
FIG 5.
Replication of A(H9N2) viruses in primary ferret cell cultures. (A to C) Ferret nasal epithelial cells (FNEC) (A and B) or ferret tracheal epithelial cells (FTEC) (C) were isolated independently from three naive ferrets, grown on transwell inserts, and inoculated apically at an MOI of 0.01 with either A-L/39 or HK/1073 A(H9N2) virus. Cells were incubated at 37°C or 33°C as indicated. At each indicated time point, 200 μl of medium was added apically and collected after 10 min; all samples were titrated in MDCK cells by standard plaque assay. (D) Comparison of infectious titers collected at 24 h p.i. from all cultures is shown. The limit of virus detection was 10 PFU.
Mammalian pathogenicity of H9N2 viruses.
Infection with A(H9N2) influenza viruses has been reported in numerous mammalian species (1); these viruses are typically associated with mild to moderate disease in laboratory mammalian models, with occasional exceptions (4, 18). Selected human and avian A(H9N2) influenza viruses were examined in the BALB/c mouse model to determine their capacity for efficient virus replication in vivo. All A(H9N2) viruses isolated from humans and tested here (inclusive of both G1 and B Y280 lineages) were capable of infection and replication in this species (Table 4). However, only HK/1073 virus infection caused moderate morbidity (17% weight loss) following high-dose (106 50% egg infective does [EID50]) inoculation, and it was the only virus tested that was capable of productive replication in the lungs of mice after delivery of a lower dose (103 EID50). Interestingly, mean viral titers in the nose remained elevated above 103 EID50 through day 6 p.i. for all A(H9N2) human isolates examined. In contrast, the two chicken A(H9N2) viruses isolated in 2018 did not cause weight loss, were detected in fewer mice on day 3 p.i., and were cleared in all mice by day 6 p.i.
TABLE 4.
A(H9N2) virus pathogenicity in mice
| Virus and dosea | Wt lossb | Viral titer (log10 EID50/ml) for:c
|
|||
|---|---|---|---|---|---|
| Day 3 lung | Day 3 nose | Day 6 lung | Day 6 nose | ||
| HK/1073 | |||||
| 106 | 17% (7) | 7.6 ± 0.1 | 3.1 ± 1.0 | 6.2 ± 0.4 | 3.4 ± 0.2 (2/3) |
| 103 | 6.4 ± 1.7 | 6.8 ± 0.5 | |||
| Bang/0994 | |||||
| 106 | 2.7% (6) | 7.4 ± 0.1 | 4.2 ± 0.9 | 6.3 ± 0.1 | 3.7 ± 0.2 |
| 103 | <1.5 (0/3) | <1.5 (0/3) | |||
| HK/308 | |||||
| 106 | 2.9% (7) | 6.6 ± 1.0 | 4.3 ± 0.5 | 6.5 ± 1.4 (2/3) | 3.2 ± 0.6 |
| 103 | <1.5 (0/3) | <1.5 (0/3) | |||
| A-L/39 | |||||
| 106 | 1.9% (8) | 5.3 ± 0.5 | 4.6 ± 0.1 | 2.0 (1/3) | 4.1 ± 0.9 (2/3) |
| 103 | 2.3 (1/3) | 2.3 (1/3) | |||
| ck/VN/70 | |||||
| 106 | None | 2.4 ± 0.5 (2/3) | 3.4 ± 1.2 (2/3) | <1.5 (0/3) | <1.5 (0/3) |
| 103 | <1.5 (0/3) | <1.5 (0/3) | |||
| ck/VN/119 | |||||
| 106 | None | <1.5 (0/3) | <1.5 (0/3) | <1.5 (0/3) | <1.5 (0/3) |
| 103 | <1.5 (0/3) | <1.5 (0/3) | |||
BALB/c mice were intranasally inoculated with 106 or 103 50% egg infectious doses (EID50) of virus in 50 μl as indicated.
Mean maximum percentage weight loss during days 1 to 14 p.i. following inoculation of mice with 106 EID50 of virus. Day maximum percentage recorded in parentheses. None, viral infection did not lead to weight loss >1%.
Mean viral titer ± standard deviation for all samples with detectable infectious virus (n = 3 mice/group unless specified in parentheses). Limit of detection was 1.5 log10 EID50/ml.
We next examined the capacity for selected A(H9N2) human isolates to cause disease in the ferret model, as the presentation of clinical signs of infection in ferrets is similar to humans, and humans and ferrets possess a high degree of similarity in respiratory tract tissues as well (20). Infection by either G1 (HK1073) or B Y280 (HK308 and A-L/39) lineage A(H9N2) viruses caused mild to moderate weight loss and transient fever in inoculated animals (Table 5); all ferrets survived the infection. Virus was detected at high titer in nasal wash (NW) specimens from all inoculated animals (≥106.8 EID50/ml). Interestingly, the most recently isolated virus (A-L/39) caused the highest mean maximum weight loss (11.2%), highest mean peak titers in NW samples (108.2 EID50/ml), and the highest frequency of nasal discharge/sneezing in inoculated ferrets.
TABLE 5.
Pathogenicity and transmissibility of A(H9N2) viruses in ferrets
| Experimental group | Wt lossa | Tempb | Nas disc | Peak NWd | Transe | Serof |
|---|---|---|---|---|---|---|
| HK/1073 | ||||||
| Inoculated | 6.2 (2/6) | 1.7 (6/6) | 3/6 | 6.8 (6/6) | NA | 320–640 (3/3) |
| DC contact | 5.0 (1/3) | 1.5 (3/3) | 0/3 | 6.5 (3/3) | 2/3 | 320 (2/3) |
| RD contact | None (0/3) | 1.3 (2/3) | 0/3 | <1.5 (0/3) | 0/3 | <10 (0/3) |
| HK/308 | ||||||
| Inoculated | 8.4 (4/6) | 1.4 (6/6) | 1/6 | 7.8 (6/6) | NA | 640–2,560 (6/6) |
| DC contact | None (0/3) | 1.0 (1/3) | 0/3 | 7.8 (3/3) | 3/3 | 640–2,560 (3/3) |
| RD contact | 16.8 (1/3) | None (0/3) | 0/3 | 3.0 (3/3) | 2/3 | 20–40 (2/3) |
| A-L/39 | ||||||
| Inoculated | 11.2 (9/9) | 1.6 (9/9) | 7/9 | 8.2 (9/9) | NA | 2,560–10,240 (9/9) |
| DC contact | 10.9 (3/3) | 2.3 (3/3) | 3/3 | 8.8 (3/3) | 3/3 | 5,120 (3/3) |
| RD contact | 7.8 (3/6) | 1.6 (5/6) | 0/6 | 8.0 (5/6) | 5/6 | 2,560–10,240 (5/6) |
Percentage mean maximum weight loss during days 1 to 14 p.i./p.c. following inoculation with 106 EID50/ml of virus. No. of ferrets with weight loss ≥5%/total no. of ferrets is in parentheses.
Mean maximum rise in degrees Celsius above baseline temperature (37.8 to 39.1°C) during days 1 to 14 p.i./p.c. No. of ferrets with temperature rise >1°C/total no. of ferrets in parentheses.
No. of ferrets with detected nasal discharge or sneezing during days 1 to 14 p.i./p.c./total no. of ferrets.
Mean maximum viral titer in ferret nasal wash (NW) specimens expressed as log10 EID50/ml. No. of ferrets with detectable virus/total no. of ferrets is in parentheses; limit of detection is 101.5 EID50/ml.
Transmission (Trans) is shown as the number of contact ferrets with viral titers in NW and seroconversion to homologous virus/total no. of ferrets. NA, not applicable. DC, direct contact; RD, respiratory droplets.
Seroconversion range to homologous virus as determined by HI assay. Titers represent the reciprocal of the dilution that inhibited hemagglutination. No. of ferrets with seroconversion/total no. of ferrets in parentheses. Serum was collected days 17–21 p.i./p.c.
Spread of A(H9N2) virus throughout the ferret respiratory tract and to extrapulmonary tissues was evaluated in ferrets 3 days after they received a 106 EID50 intranasal dose of virus (Fig. 6). All tested A(H9N2) viruses were detected throughout the ferret respiratory tract. A-L/39 virus displayed exceptional replicative ability and was the only virus detected at titers of >105 EID50/g (or ml) in all of the respiratory tract tissues examined. Virus detection in the olfactory bulb and brain was sporadic, potentially influenced by high titers of virus in the nasal passages (21). A-L/39 virus was detected in both rectal swab (RS) specimens (peak titers of 101.8 to 102.3 EID50/ml up to 5 days p.i.) and intestinal samples, while HK/308 and HK/1073 viruses were detected in RS (peak titers of 102.0 to 104.5 EID50/ml) but not in intestinal tissues. Infectious virus was not detected in the liver, spleen, kidneys, or blood of any ferret examined (data not shown).
FIG 6.
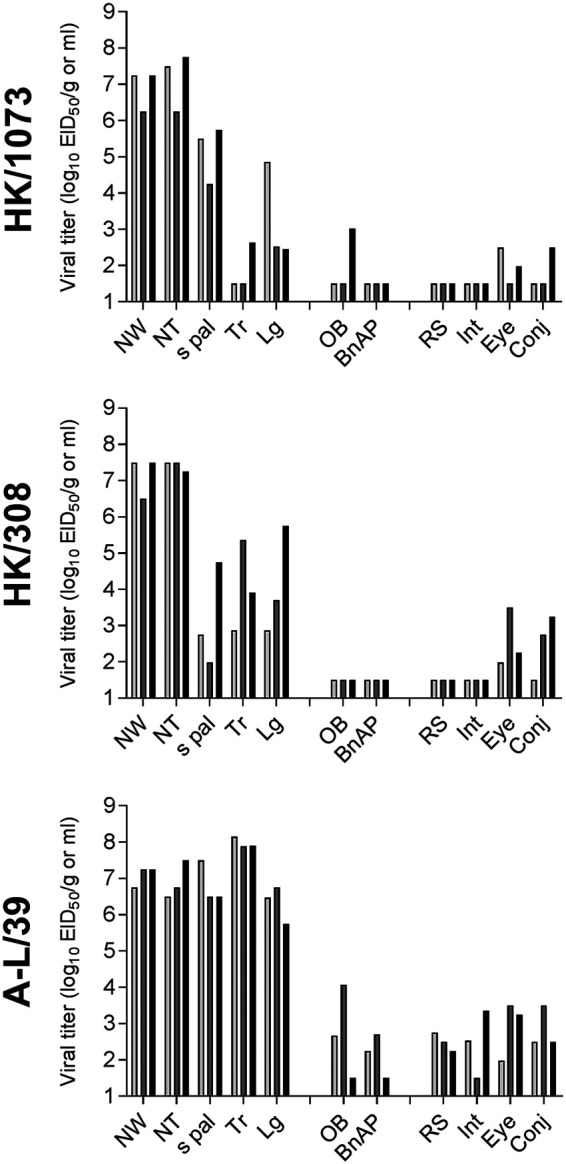
Pathogenicity of A(H9N2) viruses in ferrets. Three ferrets were inoculated intranasally with 106 EID50 of each virus indicated, and tissues and specimens were collected day 3 p.i. NW, nasal wash; NT, nasal turbinates; s pal, soft palate; Tr, trachea; Lg, lung; OB, olfactory bulb; BnAP, brain (pooled anterior and posterior); RS, rectal swab; Int, intestine (pooled duodenum, jejunoileum, and descending colon); Conj, surrounding conjunctiva. NW, NT, s pal, RS, Eye, and Conj are expressed as EID50/ml of sample or tissue homogenate, and Tr, Lg, OB, BnAP, and Int are expressed as EID50/g. The limit of virus detection was 101.5 EID50/g or ml.
Multiple subtypes of human and avian influenza viruses have demonstrated the capacity to use ocular exposure as an entry route or have been associated with conjunctivitis or other ocular complications following infection (22). While A(H9N2) viruses have not been previously linked with ocular disease in humans, multiple human ocular cell types have been shown to support virus replication with this subtype (23). All ferrets inoculated with A(H9N2) virus in this study had infectious virus detected in conjunctival wash (CW) specimens up to 5 days p.i., with peak titers of 103.3 to 104.5 EID50/ml (A-L/39 virus), 101.0 to 103.3 EID50/ml (HK/308 virus), and 102.5 to 103.5 EID50/ml (HK/1073 virus). Furthermore, infectious virus was detected in both the eye and surrounding conjunctiva on day 3 p.i. for all A(H9N2) viruses tested (Fig. 6). Compared with the G1 lineage HK/1073 virus, both B Y280 lineage viruses were associated with the highest frequency of virus detection in these tissues and the highest mean titers (>102.5 EID50/ml). However, mice inoculated ocularly with A-L/39 virus did not become productively infected (data not shown), demonstrating differences in susceptibility among mammalian models to A(H9N2) virus infection via the ocular route.
Ferret transmissibility of A(H9N2) viruses.
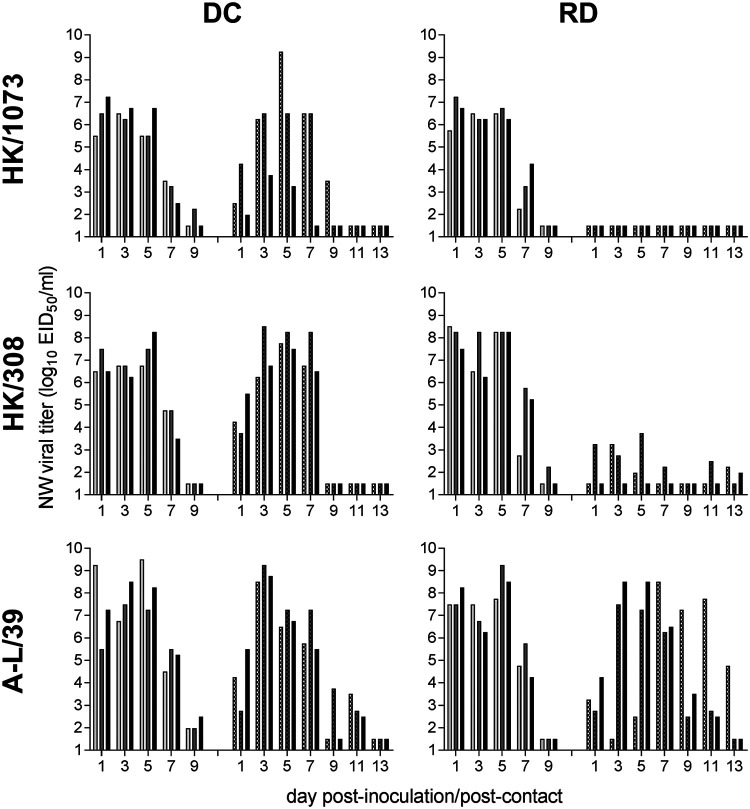
A(H9N2) G1 lineage influenza viruses have been previously shown to readily transmit among cohoused ferrets, but not in airborne transmission models (18, 24, 25). In contrast, selected Y280 viruses have demonstrated a capacity for transmission through the air, though these studies have largely been limited to poultry viruses and have not included viruses isolated from humans (4, 26). Here, we assessed the ability of a G1 lineage (HK/1073) and two B Y280 lineage (HK/308, A-L/39) A(H9N2) viruses (inclusive of a range of pH thresholds for HA activation, Table 1) to transmit between ferret pairs using a direct contact (DC) transmission model, which does not restrict the level of exposure between animals, and a respiratory droplet (RD) transmission model, which allows transmission to occur only through the air (Fig. 7). All A(H9N2) viruses that were tested displayed a capacity for transmission between ferret pairs when they were housed together in the same cage (2/3 for HK/1073 virus, 3/3 for HK/308 and A-L/39 viruses). NW samples collected from contact animals were positive for infectious virus as early as 1 day after exposure to inoculated cage mates, and titers reached levels comparable to those for directly inoculated ferrets (Table 5).
FIG 7.
Transmissibility of A(H9N2) viruses between ferrets. Six ferrets were inoculated intranasally with 106 EID50 of each virus indicated. At 24 h p.i., a naive ferret was placed in the same cage as an inoculated ferret (direct contact [DC] transmission model, left column) or in an adjacent cage with perforated side walls to an inoculated ferret (respiratory droplet [RD] transmission model, right column). Nasal washes (NW) were collected from inoculated (left sets of bars) or contact ferrets (right sets of bars) on alternate days p.i./p.c. Bars represent individual ferrets. The limit of virus detection was 101.5 EID50/ml.
During experiments using the more restrictive RD transmission model, HK/1073 virus did not transmit to any of the contact animals (Fig. 7), consistent with previous findings (18). In contrast, A-L/39 transmitted to 3/3 contact ferrets in the initial experiment (Fig. 7) and to 2/3 contact ferrets in a duplicate experiment (summarized in Table 5), resulting in a total of 5/6 contact ferrets shedding infectious virus and with confirmatory seroconversion to homologous virus. HK/308 virus was able to transmit to 2/3 contact ferrets in the RD transmission experiment. Both animals shed detectable virus in NW samples and seroconverted to HK/308 virus, although peak titers were >4 logs lower than those observed in inoculated animals or in the contact animals from the DC transmission experiment (Table 5). Serological titers in convalescent RD contact ferrets (20 to 40) were reduced as well compared to those for the other HK/308 virus-infected animals (640 to 2,560). The third animal in this experiment had trace levels of virus detected in an NW sample 13 days postcontact (p.c.) but did not display seroconversion.
To determine if mammalian adaptive mutations emerged during transmission experiments with A-L/39 virus, NW specimens collected from each transmission pair were analyzed by deep sequencing. Virus collected from experimentally inoculated ferrets possessed comparable levels of variants as the stock inoculum. Sequence deviations were not consistently identified among infected contact ferrets, though some selection was detected in the contact ferrets in RD transmission experiments (at positions PA-L532S, HA1-A39S, and HA1-L78R) that was not observed in the DC transmission experiments (Table 6). Collectively, these findings indicate that A-L/39 virus possesses an enhanced capacity for mammalian transmissibility compared with other B Y280 or G1 A(H9N2) influenza viruses, while, for the most part, maintaining genetic stability during a ferret-to-ferret transmission event.
TABLE 6.
Amino acid substitutions and frequencies of minor variants observed in ferret nasal wash samples collected during transmission experiments using A/Anhui-Lujiang/39/2018 virus
| Experiment | Animal sourceb |
Amino acid substitutions or variability (>5% cutoff threshold)a
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PB2 | PB1 | PA |
NP | NS1 | HA1c
|
HA2 |
||||||
| 563Q | 617D | 532L | 466N | 253I | 115L/I (84/16) | 39A | 78L | 224N/K (85/15) | 75T/N (90/10) | 215S | ||
| Respiratory droplet transmission experiments | ||||||||||||
| Ferret pair 1 | Inoculated (1 dpi) | Q/R (94/6) | D | L | N | I | L/I (89/11) | A | L | N/K (92/8) | T/N (89/11) | S |
| Ferret pair 1 | Contact (7 dpc) | Q | D | L | N | I | L | A | L | N | T | S |
| Ferret pair 2 | Inoculated (1 dpi) | Q | D | L | N | I | L/I (89/10) | A | L | N/K (81/19) | T/N (88/11) | F |
| Ferret pair 2 | Contact (3 dpc) | Q | D | L | N | I | L | A | L | N | T | S |
| Ferret pair 3 | Inoculated (1 dpi) | Q | D | L | N | I | L/I (88/12) | A | L | N/K (92/8) | T/N (86/14) | S |
| Ferret pair 3 | Contact (3 dpc) | Q | D | L | N | I | L | A | L | K/N (53/46) | T | S |
| Ferret pair 4 | Inoculated (1 dpi) | Q | D | L | N | I/T (94/6) | L/I (88/11) | A | L | N/K (84/16) | T/N (88/12) | S |
| Ferret pair 4 | Contact | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| Ferret pair 5 | Inoculated (1 dpi) | Q | D | L | N | I | L | A | L/R (68/31) | N/K (79/21) | T/N (86/14) | S |
| Ferret pair 5 | Contact (5 dpc) | Q | D | S | N | I | L | A | R | N | T | S |
| Ferret pair 6 | Inoculated (1 dpi) | Q | D | L | N | I | L/I (84/16) | A | L | N/K (91/9) | T/N (84/15) | S |
| Ferret pair 6 | Contact (5 dpc) | Q | D | L | N | I | L | S | L | N | T | S |
| Direct contact transmission experiments | ||||||||||||
| Ferret pair 1 | Inoculated (1 dpi) | Q | D | L | N | I | L/I (83/17) | A | L | N/K (87/13) | T/N (88/12) | S |
| Ferret pair 1 | Contact (3 dpc) | Q | D | L | N | I | L/I (82/18) | A | L | N/K (93/7) | T | S |
| Ferret pair 2 | Inoculated (3 dpi) | Q | D | L | N | I | L/I (91/8) | A | L | N/K (74/26) | T/N (92/8) | S |
| Ferret pair 2 | Contact (3 dpc) | Q | D | L | N | I | L/I (92/7) | A | L | N/K (94/6) | T/N (59/40) | S |
| Ferret pair 3 | Inoculated (3 dpi) | Q | D | L | N | I | L/I (88/12) | A | L | N/K (78/21) | T/N (88/11) | S |
| Ferret pair 3 | Contact (3 dpc) | Q | D/G (94/6) | L | N/S (94/6) | I | L | A | L/R (76/23) | K/N (54/45) | T/N (95/5) | S |
This table presents nonsynonymous changes compared to the inoculum. No amino acid changes or variability was detected for PB1-F2, PA-X, NA, MP, and NEP. Nasal wash samples representing peak titers were chosen for analysis. When mixed populations were detected, the percentage of each amino acid present in the sample is specified in parentheses. ND means that the virus was not detected in this animal during the course of the experiment (limit of detection was 101.5 EID50/ml).
dpi, days postinoculation; dpc, days postcontact.
Amino acid positions for HA1 are in H3 numbering.
DISCUSSION
The continued circulation of A(H9N2) viruses in poultry on multiple continents provides sustained opportunities for the virus to evolve genetically and antigenically, reassort with other avian influenza viruses, and acquire mutations that could enhance mammalian adaptation. Surveillance efforts, such as the active LBM surveillance program in Vietnam described here, play a key role in monitoring contemporary viruses and comparing these poultry isolates to viruses that have jumped the species barrier to cause human infection. While genomic data are essential to initiate viral risk assessment, additional phenotypic studies with isolates of these novel and emerging influenza viruses are crucial. As such, here we have performed a series of in vitro and in vivo studies to better understand the impact of virus circulation at the animal-human interface and the consequences of A(H9N2) evolution on key factors, including antigenic drift, receptor binding specificity, replication kinetics, pH stability, and mammalian pathogenicity and transmissibility.
The HA genes of A(H9N2) viruses continue to evolve within the Y280 lineage and exhibit considerable genetic diversity among the viruses detected. Importantly, the antigenic drift identified among recent virus isolates from both Y280 lineages warrants additional study to ensure appropriate selection of CVVs and pandemic preparedness (Table 2). Phylogenetic data show that, similar to studies of HPAI A(H5) viruses (27), A(H9N2) viruses detected in northern border provinces of Vietnam are closely related to those detected in China and indicate that spread of novel viruses from China into Vietnam continues to occur at ports of entry via importation of infected poultry. Still, genetic data from other viruses detected in Vietnam, strikingly similar to viruses detected in China and Myanmar, suggest that some viruses are transported via infected poultry across longer distances. Continued monitoring of borders and ports of entry will help to reduce the introduction and possible emergence of these A(H9N2) viruses into the country. Interestingly, A(H9N2) viruses that have caused human disease in China are genetically very similar to the B Y280 viruses isolated from LBMs in Vietnam, suggesting that viral introductions of these groups of A(H9N2) viruses may also pose a threat to human health in Vietnam. Furthermore, evidence of cross-border spread of a LPAI virus into Vietnam is concerning given the proximity of Vietnam to southern Chinese provinces where LPAI and HPAI A(H7N9) viruses have been detected in previous years. While surveillance for A(H7N9) viruses in Vietnamese LBMs has not detected these viruses, the data in this study suggest that infected poultry and their movements have the potential to introduce other avian influenza subtypes.
Influenza HA protein residues 226 and 228 (mature protein numbering 216 and 218) have been identified as host adaptation sites in A(H2) and A(H3) viruses. While human A(H3) viruses have an L226/S228 combination, avian A(H3) viruses typically have Q226/G228. With the exception of HK/33982 virus, all A(H9) viruses studied here bear the hybrid L216/G218 motif, similar to LPAI A(H7N9) viruses detected in China (L217/G219 by H7 numbering). Three HA residues (180, 216, 217; H3 numbering = 190, 226, 227) have also been the focus of analysis in recent receptor binding studies; for A-L/39 virus, these three positions are T180, L216, and M217. When aspartic acid or glutamic acid were present at position 180, H9 viruses were reported to bind human receptors (8). Alanine and threonine at this position, seen in more recent A(H9) viruses, were also reported to increase binding to human receptors (28). Interestingly, glycan array data for the A-L/39 virus highlighted poor binding to α2,3 and α2,6 glycans. Only the sulfated α2,3 glycan signals were apparent, which is in line with previous reports for A(H9) viruses with these glycans (28). This result was confirmed with recombinant HA. However, conditions in the resialylated RBC assays appeared to overcome the weak binding observed in the glycan microarray experiments. HA interactions with its ligands in glycan-based assays are sensitive to the types of assays employed and experimental conditions. It has been suggested that while real-time binding and enzyme-linked immunosorbent assays (ELISAs) are more sensitive for detecting subtle changes in binding avidity to α2,6 human-type receptors, glycan microarray is a more stringent test of binding specificity (29). The results presented here underscore the diversity of receptor binding capacities of different A(H9N2) viruses associated with human infection and the limitations of any given assay in achieving a complete, comprehensive understanding of this phenotype.
The efficient replication of A(H9N2) viruses in Calu-3 cells observed in this study is in agreement with previous studies employing cells derived from the human respiratory tract. Primary normal human bronchial epithelial (NHBE) cells, related human respiratory cell types, and human bronchus and lung organ cultures support high-titer replication of A(H9N2) viruses, isolated from either humans or avian species, when cultured at 37°C (17, 18, 24, 30), with sustained titers above those of a 2009 A(H1N1) pandemic virus (18, 23). Recent evaluation of A(H5N1) and A(H9N2) reassortant viruses in Calu-3 cells revealed that the presence of A(H9N2) gene segments (notably NS or PB2) conferred a replication advantage compared with the parental A(H5N1) virus, further supporting the high capacity of A(H9N2) viruses to replicate in mammalian cells (31). Similar to other avian influenza virus subtypes, including A(H5N1) and A(H7N9) viruses, all A(H9N2) viruses tested here demonstrated a delay in peak viral titers when Calu-3 cells were cultured at 33°C (19, 32). The robust replication of A-L/39 virus in both human and ferret cells at either 33°C or 37°C was striking, especially as this virus does not possess a lysine at PB2 position 627, which is frequently associated with enhanced replication in vitro and in vivo (33). In the case of A-L/39 virus, it is possible that the presence of valine at this position, identified previously as a potential intermediate between 627E and 627K for A(H7N9) viruses, contributes to this phenotype (34, 35). Interestingly, in this experiment a similar effect was not detected for HK/33982 virus, the only virus in this study to bear the compensatory mutation 701N in PB2 (36). Considering differences in replication kinetics observed between selected viruses, further study evaluating polymerase activity in the context of replication capacity would be warranted. As it was difficult to predict in vivo replication and transmission of A(H9N2) viruses based on in vitro phenotypes and replication efficiency (17), subsequent in vivo evaluation of these viruses was necessary.
While LPAI A(H7N9) viruses are capable of causing severe and fatal infection in mice, A(H9N2) virus infection, in general, manifests similarly to other LPAI viruses, in that mild to moderate weight loss (<15%) at high challenge doses is typically observed (18, 24, 37–40). The enhanced capacity of HK/1073 virus to cause disease and replicate efficiently in the murine lung is consistent with these prior studies; notably, the B Y280 lineage viruses that were isolated from humans only caused mild disease in mice (Table 4). A(H9N2) virus replication in nasal washes or nose tissue has been reported previously (4, 37) and supports the frequent detection of selected viruses in this tissue reported here. Reduced infection at lower inoculum doses (103 EID50) of recently isolated A(H9N2) viruses is in contrast with many Eurasian avian influenza viruses of the A(H5) and A(H7) subtypes (41, 42). Position 190 in the HA has been implicated in modulating viral replication of A(H9N2) virus in the murine lung, and the presence of threonine or alanine has been shown to diminish virus binding to mouse lung tissue (43), possibly contributing to the reduced replication of some of the Y280 viruses evaluated here. The lack of virus replication in mice following inoculation with ck/VN/119 virus is in line with the poor binding to α2,3-linked sialic acids detected by the hemagglutination assay (Tables 3 and 4). While it appears that several molecular determinants of virulence among A(H9N2) viruses are shared by other virus subtypes (44), additional work is required to better understand the capacity for certain A(H9N2) viruses to cause severe disease in this species (45).
Similar to other avian influenza virus subtypes, A(H9N2) viruses are capable of replication throughout the respiratory tracts of numerous mammalian species following high-dose inoculation (18, 46–49). All A(H9N2) viruses included in this study were detected throughout the ferret respiratory tract on day 3 p.i., consistent with prior studies, albeit showing strain-specific variability in replication and capacity to cause pathological lesions (4, 25, 50). With mean titers of >106 EID50 in nasal turbinates, soft palate, and lung tissues, and mean titers of >107 EID50 in NW and trachea samples at day 3 p.i., A-L/39 virus displayed the most efficient, high-titer replication among all three A(H9N2) viruses examined in ferrets. Furthermore, A-L/39 virus displayed an increased capacity for replication outside the respiratory tract, with higher titers and frequency of detection in brain (including olfactory bulb [OB]), intestinal, and ocular tissues compared with older G1 and B Y280 lineage viruses, as shown here and compared with previous studies (4, 25, 50).
A(H9N2) viruses isolated from mammals are generally capable of transmitting between cohoused ferrets (18, 23, 24), while virus transmission through the air is reportedly rare (4). A-L/39 virus displayed airborne transmission to 5 of 6 animals, mostly within the first day of exposure, despite lacking 627K and 701N in the PB2 protein. Influenza virus transmission is a multifactorial trait. In addition to enhanced polymerase activity, efficient binding to human receptors and a balanced HA and NA, most aerosol-transmissible influenza viruses boast an acid stable HA with a fusion pH threshold below 5.5 (14). It has been speculated that the relatively high fusion threshold (>5.5) for most avian influenza viruses may contribute to the lack of efficient airborne transmission in mammals. Interestingly, we found that the A(H9N2) viruses tested here exhibit a wide pH range for fusion. Many contemporary A(H9N2) viruses, including A-L/39 virus and poultry viruses detected in Vietnamese LBMs, possess a fusion threshold pH typical for human seasonal influenza viruses. As A(H9N2) viruses continue to accumulate molecular changes that further adapt them to the human host, the likelihood of the emergence of this virus subtype in susceptible populations increases. Our findings show the importance of surveillance activities in both poultry populations and humans to detect the emergence of zoonotic influenza viruses that pose a threat to public health. Continued study of A(H9N2) human viruses and the identification of genetic and phenotypic similarities they share with circulating avian viruses is critical for early detection of influenza viruses capable of zoonotic infections and potential pandemics.
MATERIALS AND METHODS
Surveillance.
LBM surveillance performed by the Department of Animal Health, Vietnam (Ministry of Agriculture and Rural Development), was conducted from September 2017 to September 2018 in 12 provinces. Provinces met specific criteria for LBM sampling, including reporting previous poultry outbreaks and having avian influenza prevalence in LBMs of >25% during the prior 2-year period, sharing a border with another country, having a high poultry population density, and high demand for poultry consumption. Five markets were selected per province. Samples were collected monthly per market from 6 poultry vendors selected at random; 5 birds were randomly swabbed (oropharyngeal) from each vendor and pooled (6 pooled samples/market). Environmental samples were also collected from each market with 5 fecal samples pooled into 1 sample and 5 water samples pooled into 1 sample. In total, 5,760 samples were collected in the 1-year period. Influenza A-positive samples, as determined by universal M gene real-time reverse transcription-PCR (rRT-PCR), were subjected to genomic sequencing using previously described methods (51).
Viruses.
Influenza A(H9N2) viruses identified in Vietnam LBMs are listed in Table S1, and those phenotypically characterized in this study are listed in Table 1. LPAI A(H7N9) A/Shanghai/1/2013 (Shanghai/1), seasonal A(H3N2) A/Switzerland/9715293/2013 (Switz/13), and 2009 pandemic A(H1N1) A/California/7/2009 (Cal/7) were also included in selected assays. All A(H9N2) and A(H7N9) viruses were propagated in the allantoic cavity of 10- to 11-day-old embryonated chicken eggs (Hyline) at 37°C for 48 h as described previously (42). A(H1) and A(H3) viruses were propagated in Madin-Darby canine kidney (MDCK) cells as described previously (52). Pooled allantoic fluid or cell culture supernatant was clarified by centrifugation and frozen in aliquots at −80°C until use, and stock 50% egg infectious dose (EID50) or PFU titers were determined. All experiments were conducted under biosafety level 3 conditions including enhancements required by the U.S. Department of Agriculture (53). GISAID accession numbers of the viruses sequenced for this study are included in Table S1, with viruses chosen for in vitro/in vivo analysis in boldface type.
Phylogenetic tree construction and molecular analysis.
A total of 832 A(H9N2) HA sequences from GISAID/GenBank databases were used to construct HA trees. Data were aligned via MUSCLE (54), and sequences were trimmed to the beginning of the mature HA1 protein gene sequence using BioEdit v7.0. The neighbor-joining method (55) with the Jukes-Cantor model in the MEGA 7.0 software package (56) were used to construct the final tree. The percentage of replicate trees in which the associated taxa clustered together was measured using the bootstrap test (1,000 replicates), and bootstrap values are shown next to the branches. Nucleotide sequence alignments used for the HA trees, as well as NA and internal gene alignments, were translated to amino acid protein sequences to compare the amino acid sequences among the virus.
Reassortment analysis.
Full genome sequences of A(H9N2) influenza viruses were used to identify potential reassortment events by aligning individual gene segments from this study to the top 100 BLAST hits in the GISAID database using Clustal W (57). Phylogenetic trees of all 8 segments were built using the MrBayes software (58) and screened for reassortment using the GiRaF version 1.09 software (59).
Antigenic characterization.
A(H9N2)-positive samples were inoculated into 10- to 11-day-old embryonated chicken eggs or MDCK cells, and allantoic fluid or supernatants were harvested 48 h postinoculation. Isolation-positive samples (n = 34) were identified by hemagglutination assay using turkey red blood cells. Antigenic characterization of virus isolates was performed using the hemagglutination inhibition assay with polyclonal ferret immune sera raised against representative viruses and CVVs, as previously described (60).
Virus-induced syncytium formation assay for HA fusion pH measurement.
Infection of Vero cells (ATCC) with influenza A viruses was used to determine the highest pH value at which syncytia were observed. As described previously (15), infected cell cultures were incubated with fusion buffer adjusted to a 5.2 to 6.0 range of pH values in 0.1-unit increments and were fixed and stained using anti-NP antibody 3 h after fusion induction. The fusion pH threshold is defined as the highest pH value at which 50% or more syncytia can be achieved.
HA receptor binding specificity.
Hemagglutination assays using resialylated turkey red blood cells were performed as previously described (61). Control viruses for this assay included beta-propiolactone (BPL)-inactivated A/mallard/Netherlands/12/2000 A(H7N7) (ATCC) (α2,3 binding control), A/Washington/74/2009 A(H1N1) (α2,6 binding control), and A/pheasant/New Jersey/1355/1998-PR8 A(H5N2) (ATCC) (dual α2,3 and α2,6 binding control).
Recombinant HA cloning, expression, and purification.
The cDNA encoding the mature ectodomain of A-L/39 virus was synthesized as codon-optimized construct (Genscript, Inc.) and subcloned into the baculovirus transfer vector pAcGP67-B (BD Biosciences) in frame with an N-terminal baculovirus GP67 signal peptide and a T4 fibritin sequence for generating functional trimers, and a His tag to aid purification (62). Transfection and virus amplification were carried out using AB Vector’s (San Diego, CA) linearized baculovirus vector DNA and their transfection protocols. Secreted soluble recombinant HA protein was recovered from the cell culture supernatant by tangential flow filtration through a 30-kDa-molecular-weight-cutoff membrane, metal affinity chromatography, and gel filtration chromatography. Recombinant HA of A/Vietnam/1203/2004 (Viet04) and A/Switzerland/9715293/2013 has been described elsewhere (63, 64).
Glycan binding studies.
Glycan microarray slides were produced under contract from the Centers for Disease Control and Prevention by James Paulson at The Scripps Research Institute (La Jolla, CA) (see Table S2 for glycans present on the array). BPL-inactivated viruses were diluted in phosphate-buffered saline (PBS) with 2% wt/vol bovine serum albumin (PBS-BSA) to an HA titer of 128. Viruses were applied to the slides and incubated at 4°C with gentle agitation for 1.5 h. Slides were rinsed with PBS with 0.05% Tween 20 (PBS-T) and PBS to remove unbound virus. The slides were then incubated sequentially with ferret serum (90 min; 1/1,000 dilution with PBS) raised against A(H9) viruses (sera used with each virus specified in Fig. 2), a biotinylated goat anti-ferret IgG antibody (30 min; 1/200 dilution with PBS) (Rockland), and with streptavidin-Alexa Fluor488 conjugate (30 min; ½,000 dilution with PBS) (Thermo Fisher, USA), with brief PBS-T/PBS washes being performed after each incubation. After the final PBS-T/PBS wash, the slides were briefly washed in deionized water, dried by a gentle steam of nitrogen gas, and immediately subjected to imaging. Fluorescence intensities were detected using an Innoscan 1100AL scanner (Innopsys, USA). Image analyses were carried out using ImaGene 9 image analysis software (BioDiscovery, Inc., USA).
Recombinant HA glycan microarray analyses were performed as described previously (21, 52). Briefly, recombinant HA-antibody complexes were prepared by mixing rHA, mouse anti-penta-His-Alexa Fluor 488 (Qiagen), and anti-mouse-IgG-Alexa Fluor 488 (Thermo Fisher Scientific) in a molar ratio of 4:2:1, respectively. Prepared complexes were incubated for 1 h at 4°C, diluted to 0.5 ml with PBS-BSA, and incubated on the microarray slide in a 4°C humidified chamber for 1.5 h. Slides were subsequently washed by successive rinses in PBS-T, PBS, and deionized water and then immediately subjected to imaging. Fluorescence intensities were detected using an Innoscan 1100AL scanner (Innopsys, USA). Image analyses were carried out using ImaGene 9 image analysis software (BioDiscovery, Inc., USA). The glycans used in these experiments are listed in Table S2.
In vitro experiments.
The human bronchial epithelial cell line Calu-3 (ATCC) was propagated and cultured as described previously (19). Cells were seeded on 12-well semipermeable inserts (Corning) and cultured with apical and basolateral media. Following washing, Calu-3 cells were inoculated apically at a multiplicity of infection (MOI) of 0.01 (based on stock PFU titer) in serum-free medium, and incubated for 1 h before washing and subsequent culture at either 37°C or 33°C. Culture supernatant aliquots were collected at the indicated times p.i. and frozen at –80°C until titration in MDCK cells for determination of infectious titer (PFU/ml).
FNEC and FTEC were isolated and cultured as previously described (65, 66). Cells were grown on 12-well inserts at air-liquid interface (ALI) conditions and were inoculated apically at an MOI of 0.01 in serum-free medium and incubated for 1 h before washing and subsequent culture at either 37°C or 33°C. Cells remained under ALI conditions for the duration of the experiment, with basolateral medium changed every-other-day. For sample collection, 200 μl medium was added apically for a 10-min incubation prior to collection and freezing at –80°C until titration in MDCK cells for determination of infectious titer (PFU/ml). The statistical significance of all viral replication kinetics experiments was assessed using two-way analysis of variance with a Tukey posttest.
Ethics statement.
All animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of the Centers for Disease Control and Prevention and were conducted in an Association for Assessment and Accreditation of Laboratory Animal Care International-accredited facility.
Mouse experiments.
Female BALB/c mice (Jackson Laboratories), 6 weeks of age, were anesthetized with vaporized isoflurane and inoculated by the intranasal route with 106 or 103 EID50 of the indicated A(H9N2) viruses in a 50-μl volume. Five mice per group inoculated with 106 EID50 of virus were observed daily for morbidity and mortality. Three mice per group inoculated with either dose were euthanized at day 3 or 6 p.i. for collection of nose and lung tissue for determination of virus replication by titration of homogenized tissues in eggs (limit of detection, 101.5 EID50/ml). Eight additional mice were anesthetized as described above and inoculated by the ocular route with 106 EID50 of A-L/39 virus in a 5-μl volume as described previously (67); four mice were euthanized at day 3 or 6 p.i. for collection of eye, nose, and lung tissue for determination of virus replication (limit of detection, 100.8 EID50/ml).
Ferret experiments.
Male Fitch ferrets (Triple F Farms), 6 to 11 months of age and serologically negative by a standard hemagglutination assay for currently circulating A(H1N1), A(H3N2), and B influenza viruses, were employed in this study. Ferrets were housed in a Duo-Flo Bioclean mobile environmental enclosure (Lab Products) for the duration of each experiment. Ferrets were anesthetized intramuscularly with a ketamine cocktail and inoculated intranasally with 106 EID50 of each A(H9N2) virus diluted in PBS in a 1-ml volume. Three ferrets per group were observed daily for clinical signs of infection, with nasal washes, conjunctival wash, and rectal swabs collected on alternate days p.i. for determination of virus shedding as previously described (68). Three additional ferrets per group were euthanized on day 3 p.i. for assessment of virus replication and systemic spread of virus, with sample titration of tissue specimens performed as described above.
Virus transmissibility was assessed by placing a serologically naive ferret in the same cage as an inoculated ferret (to assess virus transmission in the presence of direct contact [DC]) or in an adjacent cage with perforated side walls to allow air exchange in the absence of direct or indirect contact between animals (to assess virus transmission by respiratory droplets [RD]) as previously described (69). Note that the DC transmission model provides the opportunity for transmission to occur via contact or inhalation of airborne viruses, while the RD transmission model restricts transmission events to those occurring through the air. Serum was collected on days 18 to 21 p.i./postcontact (p.c.) to measure seroconversion against homologous virus by standard hemagglutinin inhibition assay with 0.5% turkey red blood cells.
Next-generation sequencing.
Viral RNA from ferret nasal wash specimens (the collection day associated with highest infectious viral load per ferret) was extracted with a QIAamp viral RNA minikit (Qiagen) per the manufacturer’s instructions. The codon complete influenza virus genomes were amplified and deep sequenced on an Illumina MiSeq instrument as described previously (70).
Supplementary Material
ACKNOWLEDGMENTS
We thank the staff of the Comparative Medicine Branch of the CDC for excellent care of animals during this study and Angie Foust and Jaber Hossain for access to embryonated eggs.
N.B. was supported by Chickasaw Nation Industries. H.D. and H.M.C. were supported by the Oak Ridge Institute for Science and Education. P.W.C. was supported by the Association of Public Health Laboratories. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention/Agency for Toxic Substances and Disease Registry.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Peacock THP, James J, Sealy JE, Iqbal M. 2019. A global perspective on H9N2 avian influenza virus. Viruses 11:620. doi: 10.3390/v11070620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li C, Wang S, Bing G, Carter RA, Wang Z, Wang J, Wang C, Wang L, Wu G, Webster RG, Wang Y, Sun H, Sun Y, Liu J, Pu J. 2017. Genetic evolution of influenza H9N2 viruses isolated from various hosts in China from 1994 to 2013. Emerg Microbes Infect 6:e106. doi: 10.1038/emi.2017.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pusch EA, Suarez DL. 2018. The multifaceted zoonotic risk of H9N2 avian influenza. Vet Sci 5:82. doi: 10.3390/vetsci5040082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X, Shi J, Guo J, Deng G, Zhang Q, Wang J, He X, Wang K, Chen J, Li Y, Fan J, Kong H, Gu C, Guan Y, Suzuki Y, Kawaoka Y, Liu L, Jiang Y, Tian G, Li Y, Bu Z, Chen H. 2014. Genetics, receptor binding property, and transmissibility in mammals of naturally isolated H9N2 avian influenza viruses. PLoS Pathog 10:e1004508. doi: 10.1371/journal.ppat.1004508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Obadan AO, Santos J, Ferreri L, Thompson AJ, Carnaccini S, Geiger G, Gonzalez Reiche AS, Rajao DS, Paulson JC, Perez DR. 2018. Flexibility in vitro of amino acid 226 in the receptor-binding site of an H9 subtype influenza A virus and its effect in vivo on virus replication, tropism, and transmission. J Virol 93:e02011-18. doi: 10.1128/JVI.02011-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu D, Shi W, Gao GF. 2014. Poultry carrying H9N2 act as incubators for novel human avian influenza viruses. Lancet 383:869. doi: 10.1016/S0140-6736(14)60386-X. [DOI] [PubMed] [Google Scholar]
- 7.Pu J, Wang S, Yin Y, Zhang G, Carter RA, Wang J, Xu G, Sun H, Wang M, Wen C, Wei Y, Wang D, Zhu B, Lemmon G, Jiao Y, Duan S, Wang Q, Du Q, Sun M, Bao J, Sun Y, Zhao J, Zhang H, Wu G, Liu J, Webster RG. 2015. Evolution of the H9N2 influenza genotype that facilitated the genesis of the novel H7N9 virus. Proc Natl Acad Sci U S A 112:548–553. doi: 10.1073/pnas.1422456112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peacock TP, Benton DJ, Sadeyen JR, Chang P, Sealy JE, Bryant JE, Martin SR, Shelton H, McCauley JW, Barclay WS, Iqbal M. 2017. Variability in H9N2 haemagglutinin receptor-binding preference and the pH of fusion. Emerg Microbes Infect 6:e11. doi: 10.1038/emi.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sorrell EM, Wan H, Araya Y, Song H, Perez DR. 2009. Minimal molecular constraints for respiratory droplet transmission of an avian-human H9N2 influenza A virus. Proc Natl Acad Sci U S A 106:7565–7570. doi: 10.1073/pnas.0900877106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peiris M, Yuen KY, Leung CW, Chan KH, Ip PL, Lai RW, Orr WK, Shortridge KF. 1999. Human infection with influenza H9N2. Lancet 354:916–917. doi: 10.1016/s0140-6736(99)03311-5. [DOI] [PubMed] [Google Scholar]
- 11.Cheng VC, Chan JF, Wen X, Wu WL, Que TL, Chen H, Chan KH, Yuen KY. 2011. Infection of immunocompromised patients by avian H9N2 influenza A virus. J Infect 62:394–399. doi: 10.1016/j.jinf.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 12.International Centre For Diarrhoeal Disease Research. 2011. Outbreak of mild respiratory disease caused by H5N1 and H9N2 infections among young children in Dhaka, Bangladesh, 2011. Health Sci Bull 9:5–12. [Google Scholar]
- 13.Russier M, Yang G, Rehg JE, Wong SS, Mostafa HH, Fabrizio TP, Barman S, Krauss S, Webster RG, Webby RJ, Russell CJ. 2016. Molecular requirements for a pandemic influenza virus: an acid-stable hemagglutinin protein. Proc Natl Acad Sci U S A 113:1636–1641. doi: 10.1073/pnas.1524384113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Russell CJ, Hu M, Okda FA. 2018. Influenza hemagglutinin protein stability, activation, and pandemic risk. Trends Microbiol 26:841–853. doi: 10.1016/j.tim.2018.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun X, Belser JA, Pappas C, Pulit-Penaloza JA, Brock N, Zeng H, Creager HM, Le S, Wilson M, Lewis A, Stark TJ, Shieh WJ, Barnes J, Tumpey TM, Maines TR. 2019. Risk assessment of fifth-wave H7N9 influenza A viruses in mammalian models. J Virol 93. doi: 10.1128/JVI.01740-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blixt O, Head S, Mondala T, Scanlan C, Huflejt ME, Alvarez R, Bryan MC, Fazio F, Calarese D, Stevens J, Razi N, Stevens DJ, Skehel JJ, van Die I, Burton DR, Wilson IA, Cummings R, Bovin N, Wong CH, Paulson JC. 2004. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc Natl Acad Sci U S A 101:17033–17038. doi: 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan RWY, Chan LLY, Mok CKP, Lai J, Tao KP, Obadan A, Chan MCW, Perez DR, Peiris JSM, Nicholls JM. 2017. Replication of H9 influenza viruses in the human ex vivo respiratory tract, and the influence of neuraminidase on virus release. Sci Rep 7:6208. doi: 10.1038/s41598-017-05853-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.SJCEIRS H9 Working Group. 2013. Assessing the fitness of distinct clades of influenza A (H9N2) viruses. Emerg Microbes Infect 2:e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeng H, Goldsmith C, Thawatsupha P, Chittaganpitch M, Waicharoen S, Zaki S, Tumpey TM, Katz JM. 2007. Highly pathogenic avian influenza H5N1 viruses elicit an attenuated type I interferon response in polarized human bronchial epithelial cells. J Virol 81:12439–12449. doi: 10.1128/JVI.01134-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belser JA, Katz JM, Tumpey TM. 2011. The ferret as a model organism to study influenza A virus infection. Dis Model Mech 4:575–579. doi: 10.1242/dmm.007823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zitzow LA, Rowe T, Morken T, Shieh WJ, Zaki S, Katz JM. 2002. Pathogenesis of avian influenza A (H5N1) viruses in ferrets. J Virol 76:4420–4429. doi: 10.1128/jvi.76.9.4420-4429.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Belser JA, Lash RR, Garg S, Tumpey TM, Maines TR. 2018. The eyes have it: influenza virus infection beyond the respiratory tract. Lancet Infect Dis 18:e220–e227. doi: 10.1016/S1473-3099(18)30102-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lenny BJ, Shanmuganatham K, Sonnberg S, Feeroz MM, Alam SM, Hasan MK, Jones-Engel L, McKenzie P, Krauss S, Webster RG, Jones JC. 2015. Replication capacity of avian influenza A(H9N2) virus in pet birds and mammals, Bangladesh. Emerg Infect Dis 21:2174–2177. doi: 10.3201/eid2112.151152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shanmuganatham KK, Jones JC, Marathe BM, Feeroz MM, Jones-Engel L, Walker D, Turner J, Rabiul Alam SM, Kamrul Hasan M, Akhtar S, Seiler P, McKenzie P, Krauss S, Webby RJ, Webster RG. 2016. The replication of Bangladeshi H9N2 avian influenza viruses carrying genes from H7N3 in mammals. Emerg Microbes Infect 5:e35. doi: 10.1038/emi.2016.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wan H, Sorrell EM, Song H, Hossain MJ, Ramirez-Nieto G, Monne I, Stevens J, Cattoli G, Capua I, Chen LM, Donis RO, Busch J, Paulson JC, Brockwell C, Webby R, Blanco J, Al-Natour MQ, Perez DR. 2008. Replication and transmission of H9N2 influenza viruses in ferrets: evaluation of pandemic potential. PLoS One 3:e2923. doi: 10.1371/journal.pone.0002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan J, Xu L, Bao L, Yao Y, Deng W, Li F, Lv Q, Gu S, Wei Q, Qin C. 2015. Characterization of an H9N2 avian influenza virus from a Fringilla montifringilla brambling in northern China. Virology 476:289–297. doi: 10.1016/j.virol.2014.12.021. [DOI] [PubMed] [Google Scholar]
- 27.Nguyen T, Davis CT, Stembridge W, Shu B, Balish A, Inui K, Do HT, Ngo HT, Wan XF, McCarron M, Lindstrom SE, Cox NJ, Nguyen CV, Klimov AI, Donis RO. 2009. Characterization of a highly pathogenic avian influenza H5N1 virus sublineage in poultry seized at ports of entry into Vietnam. Virology 387:250–256. doi: 10.1016/j.virol.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 28.Sealy JE, Yaqub T, Peacock TP, Chang P, Ermetal B, Clements A, Sadeyen JR, Mehboob A, Shelton H, Bryant JE, Daniels RS, McCauley JW, Iqbal M. 2018. Association of increased receptor-binding avidity of influenza A(H9N2) viruses with escape from antibody-based immunity and enhanced zoonotic potential. Emerg Infect Dis 25:63–72. doi: 10.3201/eid2501.180616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen LM, Blixt O, Stevens J, Lipatov AS, Davis CT, Collins BE, Cox NJ, Paulson JC, Donis RO. 2012. In vitro evolution of H5N1 avian influenza virus toward human-type receptor specificity. Virology 422:105–113. doi: 10.1016/j.virol.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xing Z, Harper R, Anunciacion J, Yang Z, Gao W, Qu B, Guan Y, Cardona CJ. 2011. Host immune and apoptotic responses to avian influenza virus H9N2 in human tracheobronchial epithelial cells. Am J Respir Cell Mol Biol 44–33. doi: 10.1165/rcmb.2009-0120OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arai Y, Ibrahim MS, Elgendy EM, Daidoji T, Ono T, Suzuki Y, Nakaya T, Matsumoto K, Watanabe Y. 2018. Genetic compatibility of reassortants between avian H5N1 and H9N2 influenza viruses with higher pathogenicity in mammals. J Virol 93:e01969-18. doi: 10.1128/JVI.01969-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeng H, Belser JA, Goldsmith CS, Gustin KM, Veguilla V, Katz JM, Tumpey TM. 2015. A(H7N9) virus results in early induction of proinflammatory cytokine responses in both human lung epithelial and endothelial cells and shows increased human adaptation compared with avian H5N1 virus. J Virol 89:4655–4667. doi: 10.1128/JVI.03095-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Subbarao EK, London W, Murphy BR. 1993. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J Virol 67:1761–1764. doi: 10.1128/JVI.67.4.1761-1764.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luk GS, Leung CY, Sia SF, Choy KT, Zhou J, Ho CC, Cheung PP, Lee EF, Wai CK, Li PC, Ip SM, Poon LL, Lindsley WG, Peiris M, Yen HL. 2015. Transmission of H7N9 influenza viruses with a polymorphism at PB2 residue 627 in chickens and ferrets. J Virol 89:9939–9951. doi: 10.1128/JVI.01444-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arai Y, Kawashita N, Ibrahim MS, Elgendy EM, Daidoji T, Ono T, Takagi T, Nakaya T, Matsumoto K, Watanabe Y. 2019. PB2 mutations arising during H9N2 influenza evolution in the Middle East confer enhanced replication and growth in mammals. PLoS Pathog 15:e1007919. doi: 10.1371/journal.ppat.1007919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou B, Pearce MB, Li Y, Wang J, Mason RJ, Tumpey TM, Wentworth DE. 2013. Asparagine substitution at PB2 residue 701 enhances the replication, pathogenicity, and transmission of the 2009 pandemic H1N1 influenza A virus. PLoS One 8:e67616. doi: 10.1371/journal.pone.0067616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mok CK, Lee HH, Chan MC, Sia SF, Lestra M, Nicholls JM, Zhu H, Guan Y, Peiris JM. 2013. Pathogenicity of the novel A/H7N9 influenza virus in mice. mBio 4:e00362-13. doi: 10.1128/mBio.00362-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Belser JA, Gustin KM, Pearce MB, Maines TR, Zeng H, Pappas C, Sun X, Carney PJ, Villanueva JM, Stevens J, Katz JM, Tumpey TM. 2013. Pathogenesis and transmission of avian influenza A (H7N9) virus in ferrets and mice. Nature 501:556–559. doi: 10.1038/nature12391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bi Y, Lu L, Li J, Yin Y, Zhang Y, Gao H, Qin Z, Zeshan B, Liu J, Sun L, Liu W. 2011. Novel genetic reassortants in H9N2 influenza A viruses and their diverse pathogenicity to mice. Virol J 8:505. doi: 10.1186/1743-422X-8-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi YK, Ozaki H, Webby RJ, Webster RG, Peiris JS, Poon L, Butt C, Leung YH, Guan Y. 2004. Continuing evolution of H9N2 influenza viruses in Southeastern China. J Virol 78:8609–8614. doi: 10.1128/JVI.78.16.8609-8614.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Belser JA, Lu X, Maines TR, Smith C, Li Y, Donis RO, Katz JM, Tumpey TM. 2007. Pathogenesis of avian influenza (H7) virus infection in mice and ferrets: enhanced virulence of Eurasian H7N7 viruses isolated from humans. J Virol 81:11139–11147. doi: 10.1128/JVI.01235-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maines TR, Lu XH, Erb SM, Edwards L, Guarner J, Greer PW, Nguyen DC, Szretter KJ, Chen LM, Thawatsupha P, Chittaganpitch M, Waicharoen S, Nguyen DT, Nguyen T, Nguyen HH, Kim JH, Hoang LT, Kang C, Phuong LS, Lim W, Zaki S, Donis RO, Cox NJ, Katz JM, Tumpey TM. 2005. Avian influenza (H5N1) viruses isolated from humans in Asia in 2004 exhibit increased virulence in mammals. J Virol 79:11788–11800. doi: 10.1128/JVI.79.18.11788-11800.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teng Q, Xu D, Shen W, Liu Q, Rong G, Li X, Yan L, Yang J, Chen H, Yu H, Ma W, Li Z. 2016. A single mutation at position 190 in hemagglutinin enhances binding affinity for human type sialic acid receptor and replication of H9N2 avian influenza virus in mice. J Virol 90:9806–9825. doi: 10.1128/JVI.01141-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li X, Qi W, He J, Ning Z, Hu Y, Tian J, Jiao P, Xu C, Chen J, Richt J, Ma W, Liao M. 2012. Molecular basis of efficient replication and pathogenicity of H9N2 avian influenza viruses in mice. PLoS One 7:e40118. doi: 10.1371/journal.pone.0040118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li C, Yu K, Tian G, Yu D, Liu L, Jing B, Ping J, Chen H. 2005. Evolution of H9N2 influenza viruses from domestic poultry in Mainland China. Virology 340:70–83. doi: 10.1016/j.virol.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 46.Wang J, Wu M, Hong W, Fan X, Chen R, Zheng Z, Zeng Y, Huang R, Zhang Y, Lam TT, Smith DK, Zhu H, Guan Y. 2016. Infectivity and transmissibility of avian H9N2 influenza viruses in pigs. J Virol 90:3506–3514. doi: 10.1128/JVI.02605-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang K, Xu W, Zhang Z, Wang T, Sang X, Cheng K, Yu Z, Zheng X, Wang H, Zhao Y, Huang G, Yang S, Qin C, Gao Y, Xia X. 2013. Experimental infection of non-human primates with avian influenza virus (H9N2). Arch Virol 158:2127–2134. doi: 10.1007/s00705-013-1721-8. [DOI] [PubMed] [Google Scholar]
- 48.Zhang C, Xuan Y, Shan H, Yang H, Wang J, Wang K, Li G, Qiao J. 2015. Avian influenza virus H9N2 infections in farmed minks. Virol J 12:180. doi: 10.1186/s12985-015-0411-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun Y, Bi Y, Pu J, Hu Y, Wang J, Gao H, Liu L, Xu Q, Tan Y, Liu M, Guo X, Yang H, Liu J. 2010. Guinea pig model for evaluating the potential public health risk of swine and avian influenza viruses. PLoS One 5:e15537. doi: 10.1371/journal.pone.0015537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao R, Bai T, Li X, Xiong Y, Huang Y, Pan M, Zhang Y, Bo H, Zou S, Shu Y. 2016. The comparison of pathology in ferrets infected by H9N2 avian influenza viruses with different genomic features. Virology 488:149–155. doi: 10.1016/j.virol.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 51.Yang G, Chowdury S, Hodges E, Rahman MZ, Jang Y, Hossain ME, Jones J, Stark TJ, Di H, Cook PW, Ghosh S, Azziz-Baumgartner E, Barnes JR, Wentworth DE, Kennedy E, Davis CT. 2019. Detection of highly pathogenic avian influenza A(H5N6) viruses in waterfowl in Bangladesh. Virology 534:36–44. doi: 10.1016/j.virol.2019.05.011. [DOI] [PubMed] [Google Scholar]
- 52.Maines TR, Jayaraman A, Belser JA, Wadford DA, Pappas C, Zeng H, Gustin KM, Pearce MB, Viswanathan K, Shriver ZH, Raman R, Cox NJ, Sasisekharan R, Katz JM, Tumpey TM. 2009. Transmission and pathogenesis of swine-origin 2009 A(H1N1) influenza viruses in ferrets and mice. Science 325:484–487. doi: 10.1126/science.1177238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chosewood LC, Wilson DE, Centers for Disease Control and Prevention (U.S.), National Institutes of Health (U.S.). 2009. Biosafety in microbiological and biomedical laboratories, 5th ed U.S. Dept. of Health and Human Services, Public Health Service, Centers for Disease Control and Prevention, National Institutes of Health, Washington, DC. [Google Scholar]
- 54.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 56.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 58.Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 59.Nagarajan N, Kingsford C. 2011. GiRaF: robust, computational identification of influenza reassortments via graph mining. Nucleic Acids Res 39:e34. doi: 10.1093/nar/gkq1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Balish AL, Davis CT, Saad MD, El-Sayed N, Esmat H, Tjaden JA, Earhart KC, Ahmed LE, Abd El-Halem M, Ali AH, Nassif SA, El-Ebiary EA, Taha M, Aly MM, Arafa A, O’Neill E, Xiyan X, Cox NJ, Donis RO, Klimov AI. 2010. Antigenic and genetic diversity of highly pathogenic avian influenza A (H5N1) viruses isolated in Egypt. Avian Dis 54:329–334. doi: 10.1637/8903-042909-Reg.1. [DOI] [PubMed] [Google Scholar]
- 61.Pulit-Penaloza JA, Pappas C, Belser JA, Sun X, Brock N, Zeng H, Tumpey TM, Maines TR. 2018. Comparative in vitro and in vivo analysis of H1N1 and H1N2 variant influenza viruses isolated from humans between 2011 and 2016. J Virol 92:e01444-18. doi: 10.1128/JVI.01444-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stevens J, Corper AL, Basler CF, Taubenberger JK, Palese P, Wilson IA. 2004. Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science 303:1866–1870. doi: 10.1126/science.1093373. [DOI] [PubMed] [Google Scholar]
- 63.Stevens J, Blixt O, Tumpey TM, Taubenberger JK, Paulson JC, Wilson IA. 2006. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science 312:404–410. doi: 10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]
- 64.Yang H, Carney PJ, Chang JC, Guo Z, Stevens J. 2018. Structural and molecular characterization of the hemagglutinin from the fifth-epidemic-wave A(H7N9) influenza viruses. J Virol 92:e00375-18. doi: 10.1128/JVI.00375-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zeng H, Goldsmith CS, Kumar A, Belser JA, Sun X, Pappas C, Brock N, Bai Y, Levine M, Tumpey TM, Maines TR. 2019. Tropism and infectivity of a seasonal A(H1N1) and a highly pathogenic avian A(H5N1) influenza virus in primary differentiated ferret nasal epithelial cell cultures. J Virol 93:e00080-19. doi: 10.1128/JVI.00080-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zeng H, Goldsmith CS, Maines TR, Belser JA, Gustin KM, Pekosz A, Zaki SR, Katz JM, Tumpey TM. 2013. Tropism and infectivity of influenza virus, including highly pathogenic avian H5N1 virus, in ferret tracheal differentiated primary epithelial cell cultures. J Virol 87:2597–2607. doi: 10.1128/JVI.02885-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Belser JA, Wadford DA, Xu J, Katz JM, Tumpey TM. 2009. Ocular infection of mice with influenza A (H7) viruses: a site of primary replication and spread to the respiratory tract. J Virol 83:7075–7084. doi: 10.1128/JVI.00535-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Belser JA, Gustin KM, Maines TR, Pantin-Jackwood MJ, Katz JM, Tumpey TM. 2012. Influenza virus respiratory infection and transmission following ocular inoculation in ferrets. PLoS Pathog 8:e1002569. doi: 10.1371/journal.ppat.1002569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maines TR, Chen LM, Matsuoka Y, Chen H, Rowe T, Ortin J, Falcon A, Nguyen TH, Mai Le Q, Sedyaningsih ER, Harun S, Tumpey TM, Donis RO, Cox NJ, Subbarao K, Katz JM. 2006. Lack of transmission of H5N1 avian-human reassortant influenza viruses in a ferret model. Proc Natl Acad Sci U S A 103:12121–12126. doi: 10.1073/pnas.0605134103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wilson JR, Belser JA, DaSilva J, Guo Z, Sun X, Gansebom S, Bai Y, Stark TJ, Chang J, Carney P, Levine MZ, Barnes J, Stevens J, Maines TR, Tumpey TM, York IA. 2017. An influenza A virus (H7N9) anti-neuraminidase monoclonal antibody protects mice from morbidity without interfering with the development of protective immunity to subsequent homologous challenge. Virology 511:214–221. doi: 10.1016/j.virol.2017.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.