Coronaviruses typically express several accessory proteins. They vary in number and nature, and only one is conserved among most of the coronaviruses, pointing at an important biological function for this protein. PEDV is peculiar in that it expresses just this one accessory protein, termed the ORF3 protein. While its analogs in other coronaviruses have been studied to different extents, and these studies have indicated that they share an ion channel property, little is still known about the features and functions of the PEDV ORF3 protein except for its association with virulence. In this investigation, we studied the intracellular trafficking of the ORF3 protein both in infected cells and when expressed independently. In addition, we analyzed the effects of mutations in five sorting motifs in its C-terminal domain and investigated whether the protein, found to follow the same exocytic route by which the viral structural membrane proteins travel, is also incorporated into virions.
KEYWORDS: PEDV, coronavirus, accessory protein, YxxØ, intracellular traffic, exocytic secretion
ABSTRACT
Accessory genes occurring between the S and E genes of coronaviruses have been studied quite intensively during the last decades. In porcine epidemic diarrhea virus (PEDV), the only gene at this location, ORF3, encodes a 224-residue membrane protein shown to exhibit ion channel activity and to enhance virus production. However, little is known about its intracellular trafficking or about its function during PEDV infection. In this study, two recombinant PEDVs were rescued by targeted RNA recombination, one carrying the full-length ORF3 gene and one from which the gene had been deleted entirely. These viruses as well as a PEDV encoding a naturally truncated ORF3 protein were employed to study the ORF3 protein’s subcellular trafficking. In addition, ORF3 expression vectors were constructed to study the protein’s independent transport. Our results show that the ORF3 protein uses the exocytic pathway to move to and accumulate in the Golgi area of the cell similarly in infected and transfected cells. Like the S protein, but unlike the other structural proteins M and N, the ORF3 protein was additionally observed at the surface of PEDV-infected cells. In addition, the C-terminally truncated ORF3 protein entered the exocytic pathway but it was unable to leave the endoplasmic reticulum (ER) and ER-to-Golgi intermediate compartment (ERGIC). Consistently, a YxxØ motif essential for ER exit was identified in the C-terminal domain. Finally, despite the use of sensitive antibodies and assays no ORF3 protein could be detected in highly purified PEDV particles, indicating that the protein is not a structural virion component.
IMPORTANCE Coronaviruses typically express several accessory proteins. They vary in number and nature, and only one is conserved among most of the coronaviruses, pointing at an important biological function for this protein. PEDV is peculiar in that it expresses just this one accessory protein, termed the ORF3 protein. While its analogs in other coronaviruses have been studied to different extents, and these studies have indicated that they share an ion channel property, little is still known about the features and functions of the PEDV ORF3 protein except for its association with virulence. In this investigation, we studied the intracellular trafficking of the ORF3 protein both in infected cells and when expressed independently. In addition, we analyzed the effects of mutations in five sorting motifs in its C-terminal domain and investigated whether the protein, found to follow the same exocytic route by which the viral structural membrane proteins travel, is also incorporated into virions.
INTRODUCTION
Porcine epidemic diarrhea (PED) was first recognized as a severe enteric disease in feeder and fattening pigs in England in 1971. The etiological agent was isolated from pigs exhibiting diarrhea during a PED outbreak in Belgium in 1978 (1) and was named porcine epidemic diarrhea virus (PEDV). In the 1980s and 1990s, PED was relatively insignificant in Europe, with only a few outbreaks being reported during this period. However, in Asian countries PED became more and more epidemic, especially in China (2–5), where the disease caused huge economic losses to the country’s pig industry particularly since 2010 (6). In 2013, the disease spread to the United States and its neighboring countries, with similar great economic consequences (7).
PEDV is a positive-sense RNA virus classified in the genus Alphacoronavirus, family Coronaviridae. Its genome, about 28 kb in length, comprises at least 7 open reading frames (ORFs) which encode two nonstructural polyproteins, polyproteins 1a and 1ab (pp1a and pp1ab), and four structural proteins: the 180- to 220-kDa glycosylated spike (S) protein, 7-kDa envelope (E) protein, 25-kDa membrane (M) protein, and 58-kDa nucleocapsid (N) protein. The virus encodes only one known accessory protein, the ORF3 protein (8–10). Typically, the precursors pp1a and pp1ab are processed to yield several functional proteins that together direct PEDV genome replication and transcription. The S protein is glycosylated and forms trimers that constitute the typical spikes on the outside of the virion; the protein is responsible for attachment of the virus to the cell surface and for cell entry by mediating the fusion of the virus envelope with the cell membrane (11–13). The E protein plays a role in virion assembly and has ion channel activity (14). The M protein is essential in virus particle assembly; it interacts with all structural components and forms the thick lipoprotein virion envelope (15, 16). The N protein usually occurs in a phosphorylated form which, together with the genomic RNA, forms a helical nucleocapsid (RNP) (9).
While coronaviruses generally express various accessory proteins from the 3′ part of their genomes, the single accessory ORF occurring in PEDV is the only one conserved in all coronaviruses (17, 18). PEDV ORF3 encodes an anion channel protein of 224 amino acids (aa) with a theoretical molecular weight of 25 kDa, predicted to be a multispanning membrane protein (19). The protein is prone to undergoing deletion or mutation when the virus is adapted to growth in cell culture, e.g., by serial passaging. Thus, the coding region of the ORF3 gene of attenuated PEDV strains such as KPED-9 and P-5V has a 51-nucleotide (nt) internal deletion and encodes a predicted protein of 207 amino acids; the 17-residue deletion has been proposed to be responsible for the reduced pathogenicity of these viruses (20). Similarly, a 49-nt deletion in the ORF3 gene has been observed in the vaccine strains CV777 and DR13, the latter of which was used as the backbone for the recombinant PEDVs used in the present study. The removal of nt 245 to 293 leads to a premature translation stop at nt 274 to 276, giving rise to a naturally truncated ORF3 protein of 91 residues designated the ORF3trun protein (21). Field isolates with large deletions of ORF3 have also been documented (22).
As there are only a few reports on the characteristics of the ORF3 protein, little is actually known about its precise functioning during PEDV infection. In one study, the ORF3 protein was shown to exhibit ion channel activity when expressed in Xenopus laevis oocytes and yeast cells (19). In another study, Vero cells constitutively expressing the ORF3 were generated and the protein was found to localize in the cytoplasm, to prolong the cell cycle’s S phase, and to augment vesicle formation in the cells (23). In a more recent study, the ORF3 protein was observed to localize in the cytoplasm, partially in the endoplasmic reticulum (ER) and the Golgi apparatus (24). In contrast, very recently the protein was reported to localize in the ER, triggering an ER stress response and inducing cellular autophagy (25).
In view of the inconsistent literature on the ORF3 protein’s intracellular trafficking, we focused in the present study on this fundamental issue. Using a reverse genetics system based on strain DR13att (26), we generated two isogenic recombinant PEDVs (rPEDVs), one carrying an intact ORF3 gene and one lacking the ORF3 gene entirely. With these rPEDVs in combination with PEDV DR13att, which expresses the ORF3trun protein, we studied the protein’s transport and localization in infected cells. In addition, plasmids expressing the wild-type ORF3 protein and a C-terminally tagged form thereof were also constructed to investigate the intrinsic subcellular localization of the ORF3 protein in transfected cells. To identify signals involved in the intracellular sorting of the protein, we studied the effects of mutations in certain predicted targeting motifs. Finally, as we observed the ORF3 protein joining the viral structural membrane proteins in the exocytic pathway, we also analyzed whether the protein becomes integrated into the virions during virus assembly.
RESULTS
Construction and verification of the recombinant plasmids.
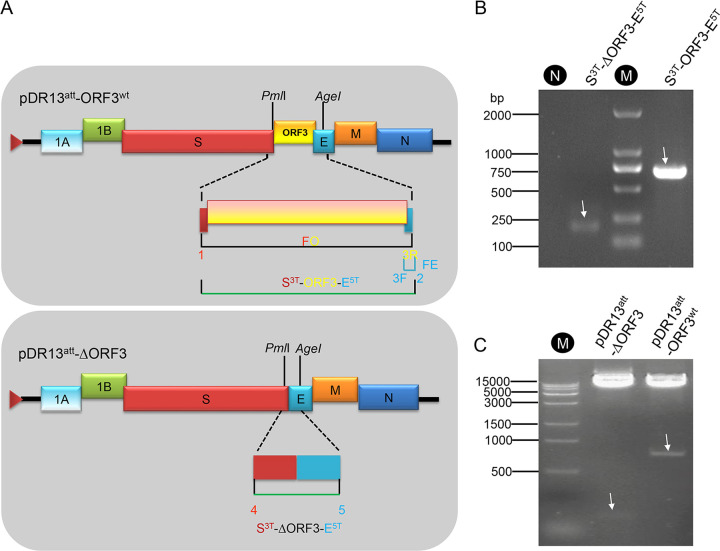
The wild-type PEDV DR13 ORF3 gene (ORF3wt; GenBank accession no. JQ023161) was generated by using a gene synthesis method based on overlap extension PCR and its sequence was confirmed. It was subsequently used to construct the recombinant plasmids pORF3wt and pDsRed-ORF3wt to express in eukaryotic cells the natural ORF3 protein and an ORF3 protein C-terminally extended with the red fluorescent protein DsRed2, respectively. The synthetic gene was also used to construct transfer vector pDR13att-ORF3wt (Fig. 1A), a tool required to generate recombinant virus rDR13att-ORF3wt using an RNA recombination method described earlier (26, 27). A similar transfer vector but lacking ORF3, pDR13att-ΔORF3 (Fig. 1A), was constructed to similarly generate the recombinant virus rDR13att-ΔORF3. The recombinant constructs pDR13att-ORF3wt and pDR13att-ΔORF3 were verified by PCR analysis of the relevant inserts (Fig. 1B) and by double endonuclease digestion analysis using PmlI and AgeI (Fig. 1C).
FIG 1.
Design of transfer vectors pDR13att-ORF3wt and pDR13att-ΔORF3 (A) and verification (B and C). (A) The pDR13att-ORF3wt transfer vector contains the genomic 5′-proximal 605 nt fused to the approximately 8-kb 3′ part of the PEDV-DR13 genome, in which the truncated ORF3 was replaced with ORF3wt (yellow box). In the pDR13att-ΔORF3 vector, the truncated ORF3 gene (ORF3att) was deleted completely. The triangle at the 5′ end of the vector represents the T7 promoter from which synthetic RNAs were made in vitro using T7 RNA polymerase. The numbers 1, 2, 3F, 3R, 4, and 5 refer to the primers listed in Table 3 and indicate their approximate positions. The green lines indicate the fragments amplified with the primers for the construction of the vectors. (B) PCR amplification of S3T-ΔORF3-E5T and S3T-ORF3-E5T from transfer vectors pDR13att-ΔORF3 and pDR13att-ORF3wt, respectively, using the primers mentioned for panel A and specified in Table 3. Lane N, negative control; lane M, DL 2000 DNA ladder. (C) Confirmation of recombinant transfer plasmids by double digestion with PmlI/AgeI. Lane M, Trans 15,000 (15K) DNA ladder (TransGen Biotech Inc., Beijing, China). Arrows point at the relevant bands, the predicted sizes of which are 191 nt and 725 nt for lanes 2 and 4 in panel B and 130 nt and 770 nt for lanes 2 and 3 in panel C, respectively.
Generation and characterization of recombinant PEDVs.
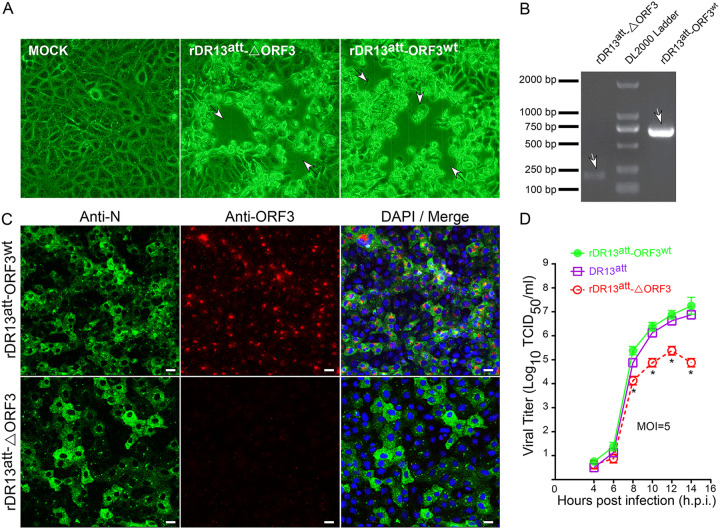
To generate recombinant PEDVs using targeted RNA recombination (26, 27), LR7 cells infected with mPEDV (a recombinant PEDV carrying most of the S gene of mouse hepatitis coronavirus) were electroporated with synthetic RNA prepared by in vitro transcription from pDR13att-ORF3wt and pDR13att-ΔORF3 and cocultured with Vero cells in which the intended recombinant PEDVs can proliferate. Cytopathic effect (CPE) appeared in the Vero cells at about 40 h postinfection (h.p.i.) (Fig. 2A). The recombinant viruses were purified by endpoint dilution and propagated for identification. Reverse transcription-PCR (RT-PCR) was employed to amplify specific segments of the RNA genome. Using primers mapping in the 3′ end of the S gene and the 5′ end of the E gene, DNA fragments of expected sizes were amplified; i.e., a 770-bp fragment was amplified from rDR13att-ORF3wt, while a 191-bp fragment was obtained from rDR13att-ΔORF3 (Fig. 2B). Sequencing of the fragments additionally confirmed their identity. An immunofluorescence analysis (IFA) of Vero cells infected with either of the two rescued recombinant PEDVs using a mouse anti-PEDV N monoclonal antibody revealed the wide spread cytoplasmic staining typical of coronavirus N proteins (Fig. 2C). To verify the differential expression of the ORF3 protein by the two viruses, the same cells were also probed with the ORF3-specific antibody P71-3. While the rDR13att-ΔORF3-infected Vero cells were completely negative, a typical punctate staining pattern was observed after infection with rDR13att-ORF3wt (Fig. 2C). Taken together, these results confirmed that the intended recombinant viruses rDR13att-ORF3wt and rDR13att-ΔORF3 were successfully rescued by the targeted RNA recombination procedure.
FIG 2.
Characterization of the recombinant PEDVs. (A) Cytopathic effect (arrows) in Vero cells infected with the recombinant viruses. Images were taken at 40 h.p.i. using an inverted microscope at a magnification of ×200. (B) RT-PCR verification of recombinant viruses rDR13att-ORF3wt and rDR13att-ΔORF3. RT-PCR was performed across the ORF3 region (primers 1/3R [Table 3]) using the isolated viral RNAs as the template, and the products were analyzed by agarose gel electrophoresis. Arrows point at the relevant bands, the predicted sizes of which are 191 nt and 725 nt for the left and right lanes, respectively. (C) Indirect immunofluorescence analysis of N protein and ORF3 protein expression in Vero cells infected with rDR13att-ORF3wt and rDR13att-ΔORF3. Cells were inoculated at an MOI of 0.1 and incubated for 24 h. They were then fixed and permeabilized, and an immunofluorescence analysis was performed to detect ORF3 (red) and N protein (green) with rabbit anti-ORF3 (P71-3; 1:50) and mouse anti PEDV N monoclonal antibody (3F-12; 1:100), respectively. Cell nuclei were stained with DAPI (blue). Separate images showing N protein expression, ORF3 protein expression, and a merger of both were acquired as indicated. Scale bar represents 50 μm. (D) One-step growth kinetics of rPEDVs and DR13att in Vero cells. Vero cells were infected with rDR13att-ORF3wt, rDR13att-ΔORF3, and DR13att at the indicated times postinfection (MOI = 5). Cells and culture media were harvested by three rounds of freezing and thawing, followed by centrifugation to remove cell debris. Supernatants were used to measure viral titers. Results are expressed as the mean values from at least three parallel tests, and error bars represent standard deviations. Statistical significance is indicated by asterisks as follows: *, P < 0.05.
To further characterize the viruses, we studied their growth kinetics in Vero cells in comparison with those of a natural PEDV strain, DR13att. As shown in Fig. 2D, after an initial period of similar growth, from about 8 h.p.i. rDR13att-ΔORF3 started to lag behind the two other viruses, which replicated similarly, reaching titers of around 107 50% tissue culture infective doses (TCID50)/ml, about 2 log units higher than rDR13att-ΔORF3. In conclusion, PEDV DR13att, which encodes a C-terminally truncated ORF3 protein, replicated indistinguishably from PEDV with intact ORF3, while propagation of the virus lacking ORF3 was significantly impaired.
Cellular localization of the ORF3 protein.
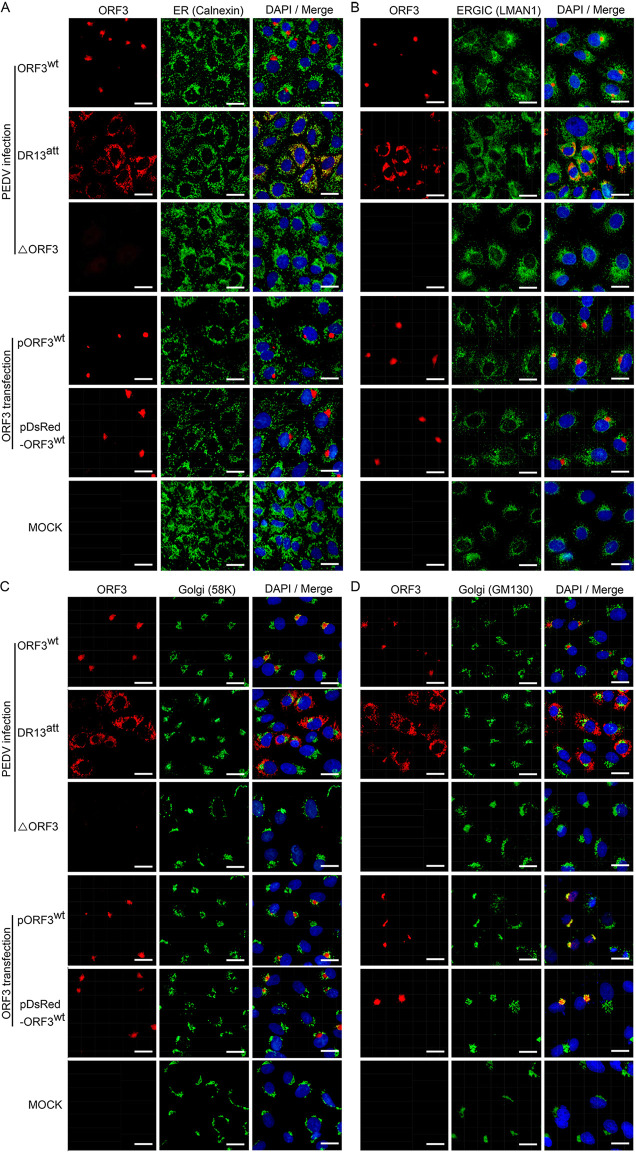
To try to elucidate the identity of the punctate structures in which the ORF3 protein appeared to accumulate (Fig. 2C), we performed costaining with antibodies against established marker proteins of different intracellular compartments: endoplasmic reticulum (ER), ER-Golgi intermediate compartment (ERGIC), and Golgi complex. Besides the viruses rDR13att-ORF3wt and rDR13att-ΔORF3, we again included in our analysis the parental virus DR13att.
As is clear in the results shown in Fig. 3 for different costaining experiments, the wild-type ORF3 protein exhibited a consistent punctate staining pattern both in infected cells and when expressed individually by transfection. A completely different pattern was observed for the PEDV DR13att-derived truncated ORF3 protein, which exhibited a more diffuse intracellular distribution. This protein appeared to stay in early exocytic compartments as it colocalized clearly with the ER marker calnexin and partly with the ERGIC marker LMAN1 (Fig. 3A and B). Wild-type ORF3 protein, on the other hand, did not colocalize with these markers. Rather, its staining pattern was indicative of a localization in the Golgi complex, as demonstrated by its clear costaining with two established Golgi marker proteins, 58K (Fig. 3C) and, more particularly, GM130, the appearance of which was generally somewhat patchier (Fig. 3D).
FIG 3.
Intracellular localization of ORF3 protein relative to organelle marker proteins in transfected and infected Vero cells. Vero cells were infected with rDR13att-ORF3wt, DR13att, or rDR13att-ΔORF3 at an MOI of 0.5, transfected with pORF3wt or pDsRed-ORF3wt, or mock infected. After 24 h (infection) or 36 h (transfection) of incubation, the cells were fixed, permeabilized, and processed for IFA using the ORF3 protein antibody P71-3 and appropriate organelle marker monoclonal antibodies, including mouse anti-calnexin (A), mouse anti-LMAN1 (B), mouse anti-58K Golgi protein (C), and mouse anti-Golgi GM130 polyclonal antibody (D) as primary antibodies; Alexa Fluor 647-conjugated goat anti-rabbit IgG (red) and Alexa Fluor 488-conjugated goat anti-mouse IgG (green) were used as secondary antibodies. Separate images showing ORF3 protein localization and organelle marker distribution are presented, as well as mergers of the two. In the merged images the yellow color indicates regions of protein colocalization. Cellular nuclei were stained with DAPI (blue). Scale bar represents 50 μm.
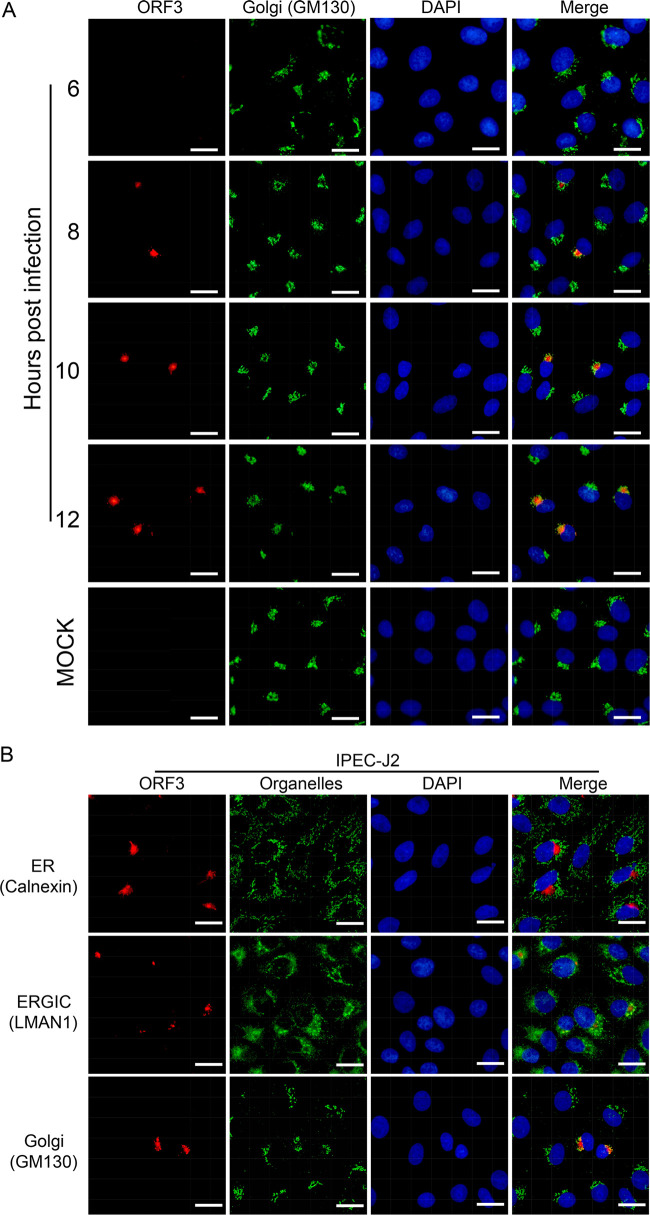
To confirm these observations in a different way, we performed a time course experiment in which we monitored ORF3 protein localization in the course of an infection. Immunofluorescence images were made at various times starting at 6 h.p.i., at which time the ORF3 protein was still undetectable (Fig. 4A). From 8 h.p.i. onwards the ORF3 protein could be detected and the Golgi-associated localization was the predominant one, showing the ORF3 protein colocalizing with the GM130 Golgi marker as observed before (Fig. 3D).
FIG 4.
Intracellular localization of ORF3 protein relative to GM130 Golgi marker protein in infected Vero and transfected IPEC-J2 cells. (A) Time course study of ORF3 protein expression in rDR13att-ORF3wt-infected Vero cells. Vero cells grown on coverslips were inoculated with rDR13att-ORF3wt at an MOI of 5. At different time points (6, 8, 10, and 12 h.p.i.), coverslips were harvested and processed for IFA under permeabilization conditions by using the P71-3 and mouse anti-Golgi GM130 polyclonal antibodies as primary antibodies. Alexa Fluor 647-conjugated goat anti-rabbit IgG (red) and Alexa Fluor 488-conjugated goat anti-mouse IgG (green) were used as secondary antibodies. Scale bar represents 50 μm. (B) ORF3 protein expression in transfected IPEC-J2 cells. IPEC-J2 cells were transfected with pORF3wt plasmids. At 36 h posttransfection, cells were fixed and permeabilized for IFA using the P71-3 antibody and appropriate mouse originated organelle marker antibodies as primary antibodies. Secondary antibodies were used as for panel A. Separate images showing ORF3 distribution, organelle marker distribution, and a merger of both were acquired as indicated. Cellular nuclei were stained with DAPI (blue). Scale bar represents 50 μm.
As Vero cells are of monkey origin, we checked the localization of the ORF3 protein also in porcine cells. IPEC-J2 cells, which are derived from porcine enterocytes, were transfected with the ORF3 expression vector pORF3wt. Also in these cells, the expressed ORF3 protein was found to colocalize with Golgi marker GM130 (Fig. 4B). Attempts to infect IPEC-J2 cells with the rPEDVs were not successful.
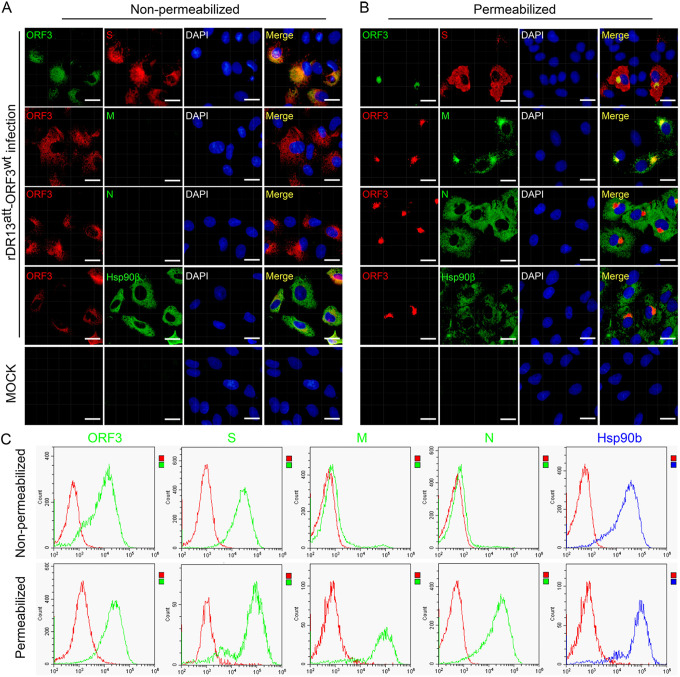
Finally, we also investigated whether the ORF3 protein was transported beyond the Golgi complex to the plasma membrane. Immunofluorescence staining of nonpermeabilized rDR13att-ORF3wt-infected Vero cells was performed using antibodies against the ORF3 protein and against the cell surface marker protein hsp90β as well as against the PEDV structural proteins N, M, and S, of which the last is known to be transported to the cell surface and to cause cell fusion. As Fig. 5A shows, the ORF3 protein was clearly observable at the plasma membrane together with the two surface markers, hsp90β and S. No surface exposure of the N and M proteins was detected, but these proteins became detectable upon permeabilization of the cells, which also revealed their typical intracellular localization in the cytoplasm and in the Golgi complex, the latter clearly coinciding with the intracellular localization of the ORF3 protein (Fig. 5B). These results were confirmed in an independent experiment in which we analyzed similarly stained infected cells by flow cytometry (Fig. 5C). The observations revealed the surface exposure specifically of the ORF3 and S proteins together with the hsp90β marker protein on the nonpermeabilized cells, while viral proteins M and N became detectable only after permeabilization.
FIG 5.
Detection of the ORF3 protein on the surfaces of PEDV-infected cells. (A and B) Cell surface and intracellular location of PEDV proteins as determined by IFA. Vero cells were infected with rDR13att-ORF3wt at an MOI of 0.5 or mock infected. After 24 h of incubation, the cells were fixed and processed for IFA under both nonpermeabilization (A) and permeabilization (B) conditions. PEDV proteins S, M, and N and ORF3 protein were detected by staining using rabbit anti-S antibody (1:1,000), mouse anti-M antibody (1:100), mouse anti-N antibody (1:100), mouse anti-ORF3 (aa 1 to 71) polyclonal antibody (1:50, top row), or P71-3 (1:50, rows 2 to 4) as primary antibodies. The hsp90β protein, which served as a cell surface marker, was detected by using mouse anti-hsp90β monoclonal antibody (1:100) as a primary antibody. Secondary antibodies were used as for Fig. 4. Cellular nuclei were stained with DAPI (blue). Scale bar represents 50 μm. (C) Cell surface and intracellular location of PEDV proteins as determined by flow cytometry. Cells cultured in 6-well plates were infected with rDR13att-ORF3wt at an MOI of 0.5 or mock infected. After 36 h of incubation, the cells were collected and the cell surface and intracellular proteins were stained under nonpermeabilization (top row) and permeabilization (bottom row) conditions as described for panels A and B, respectively. Flow cytometry was performed as described in Materials and Methods. Data for the PEDV proteins are shown in green and those for hsp90β in blue, while those for an isotype (negative) control are shown in red.
Sorting signal 170YLAI173 involved in ORF3 protein’s intracellular transport.
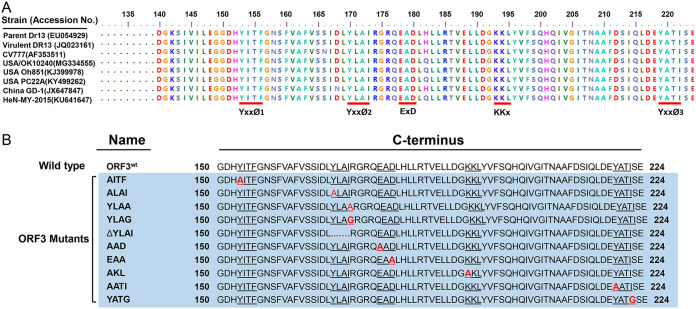
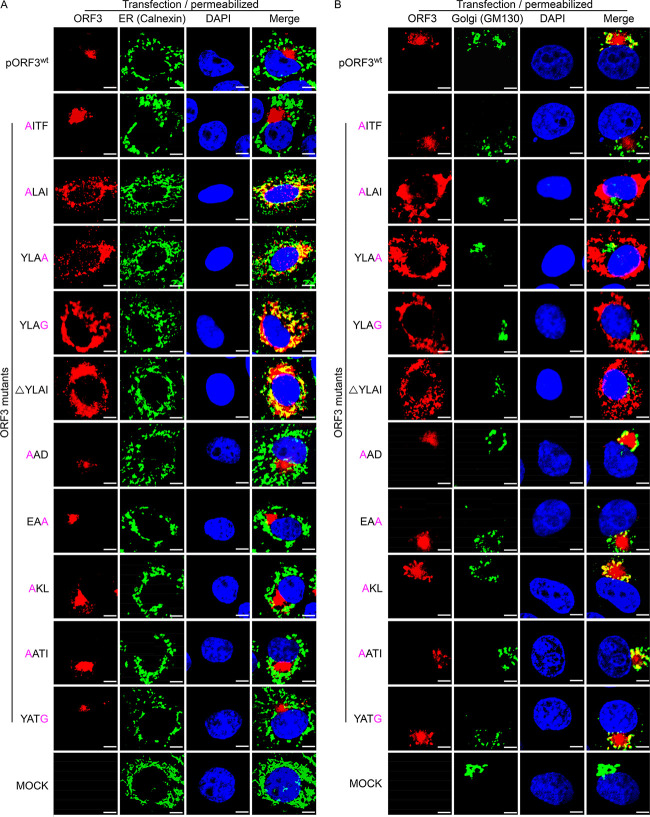
To deepen our understanding of the ORF3 protein’s trafficking behavior, we focused on the role of three kinds of well-established sorting motifs that occur in the protein’s C-terminal amino acid sequence: the tyrosine-based motif YxxØ, in which x can be any residue and Ø is a residue with a bulky hydrophobic side chain; the diacidic motif ExD; and the dibasic motif KKx. When comparing the amino acid sequences of PEDV ORF3 proteins from several representative isolates available in the NCBI database, we found that the C-terminal part contains 3 YxxØ motifs (residues 153 to 156, YITF; residues 170 to 173, YLAI; and residues 219 to 222, YATI), 1 EAD motif (residues 178 to 180), and 1 KKL motif (residues 193 to 195), which are conserved in all isolates studied (Fig. 6A). We applied single residue mutations and a motif deletion within ORF3 and obtained 10 expression vectors: AITF, ALAI, YLAA, YLAG, ΔYLAI, AAD, EAA, AKL, AATI, and YATG (Fig. 6B). The localization of these mutant proteins was compared with that of the ORF3wt protein by transfection and staining of Vero cells. The results show that the inactivating mutations in all motifs were without detectable effect except for the YLAI motif (YxxØ2 in Fig. 6A). Different substitutions of the key residues Y and I as well the entire deletion of the YLAI motif rendered the ORF3 protein apparently transport incompetent, as the mutant protein was found concentrated in the ER region (Fig. 7A) rather than in the Golgi area (Fig. 7B). The observations suggest that the YLAI motif mediates the protein’s export from the ER.
FIG 6.
Putative sorting motifs in the ORF3 protein’s C-terminal sequences (aa 140 to 224) and design of mutations. (A) Comparison of amino acid sequences in the C-terminal domain of ORF3 proteins of different PEDV isolates. Sequences were obtained from the National Center for Biotechnology Information (NCBI). The potential sorting motifs, YxxØ 1 to 3, ExD, and KKx, are underlined in red. (B) Mutations introduced into the motifs and designation of ORF3 mutants. The putative sorting motifs are underlined, the substituted amino acid in each motif is marked in red, and dots indicate residue deletions. All of the ORF3 mutant constructs are framed in light blue.
FIG 7.
Effects of mutations or deletion of sorting motifs on intracellular ORF3 protein transport. Vero cells grown on coverslips were transfected with pORF3wt and ORF3 mutant constructs including AITF, YLAG, YLAA, ALAI, AKL, AATI, YATG, EAA, AAD, and ΔYLAI. After 36 h of incubation, the cells were processed for IFA and stained for ORF3 protein, calnexin, and GM-130 as for Fig. 4. Scale bar represents 10 μm.
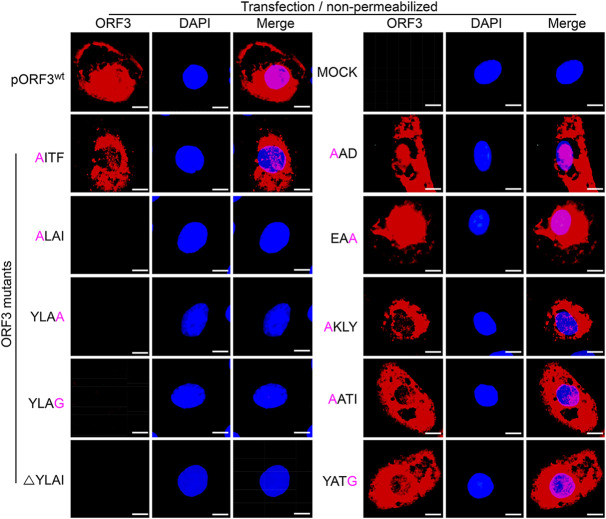
To investigate whether the mutations of the sorting motifs also affect ORF3 protein’s presence at the cell surface, we also performed immunofluorescence staining of nonpermeabilized cells. Except again for the mutations in the YLAI sequence, none of the mutations in the other motifs affected the fate of the expressed ORF3 protein, as judged by the clear cell surface appearance typical for the wild-type protein. In contrast, no such exposure was observed for any of the YLAI mutant proteins (Fig. 8). The results are consistent with the YLAI motif being an essential signal in the regulation of ORF3 protein’s intracellular transport. Taken together, our results indicate that the 170YLAI173 motif (YxxØ2 in Fig. 6A) is the functional motif involved in the ORF3 protein’s intracellular transport.
FIG 8.
Cell surface expression of ORF3 protein and its mutants. Vero cells grown on coverslips were transfected with pORF3wt and ORF3 mutant constructs including AITF, YLAG, YLAA, ALAI, AKL, AATI, YATG, EAA, AAD, and ΔYLAI. After 36 h of incubation, the cells were fixed and processed for IFA under nonpermeabilization conditions by using P71-3 as primary antibody and Alexa Fluor 647-conjugated goat anti-rabbit IgG (red) as secondary antibody. Cells transfected with empty vector served as a negative control (mock). Cell nuclei were stained with DAPI (blue). Separate images showing surface ORF3 protein expression and nuclear staining are presented, as well as mergers of the two. Scale bar represents 15 μm.
Occurrence of ORF3 protein in purified PEDV virions.
In view of its occurrence in the intracellular compartments where coronavirus assembly is known to take place, we wondered whether the ORF3 protein might become incorporated into virions. Hence, we purified our recombinant viruses rDR13att-ORF3wt, PEDV DR13att, and rDR13att-ΔORF3 and analyzed them by Western blotting, including in the analyses also samples of infected cells and their culture supernatants to verify the detection of the different viral proteins. Surprisingly, no trace of an ORF3 protein could be detected in the samples from the purified viruses rDR13att-ORF3wt and PEDV DR13att, indicating that the protein was not assembled into PEDV virions, as was the structural N protein used as a control (Fig. 9).
FIG 9.
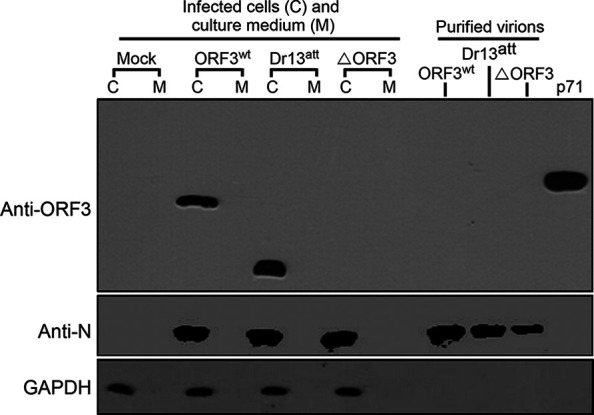
Occurrence of ORF3 protein in PEDV virions. Vero cells were mock infected or infected with PEDVs at an MOI of 2 in T225 flasks. After 2 days of incubation, the cells were harvested and viral particles in the culture medium were purified by sucrose gradient centrifugation. Both the cellular samples and purified virions were analyzed for their protein compositions in a separate SDS-10% polyacrylamide gel which was subsequently processed for immunoblot analysis with antibodies against ORF3 (rabbit anti-ORF3 polyclonal antibody, P71-3; 1:2,000) and N (mouse anti-PEDV N protein monoclonal antibody, 3F-12; 1:2,000) proteins; GAPDH served as a protein loading control. Lanes corresponding with the lysates of the cells (C), the corresponding culture media (M), and the purified virus particles are indicated. The rightmost lane, labeled p71, refers to the analysis of an ORF3-glutathione S-transferase (GST) positive-control protein, derived by prokaryotic expression of pGEX-4T-1-p71, which specifies a fusion protein consisting of ORF3 aa 1 to 71 linked to a GST tag (∼33.5 kDa).
Mass spectrometric characterization of PEDV component proteins.
To confirm this conclusion, we did a mass spectrometry (MS) analysis of cellular and virus samples. Cells were infected with rDR13att-ORF3wt or rDR13att-ΔORF3, after which cell lysates were prepared and virus was purified from the culture medium by pelleting through a sucrose cushion. Proteins from both specimens were separated by SDS-PAGE, and slices of the gel containing proteins of around 25 kDa were excised and subjected to MS analysis. ORF3 peptide fragments were identified in the rDR13att-ORF3wt-infected cell sample but not in that of rDR13att-ΔORF3-infected cells (Table 1). They were also not detected in the sample of rDR13att-ORF3wt virion particles, confirming our earlier conclusion. Interestingly, besides the expected peptides derived from the M protein, peptides corresponding to other viral structural proteins, including S and N, were also detected, indicative of the sensitivity of the analysis as well as perhaps of some degradation of these larger proteins during sample processing.
TABLE 1.
Mass spectroscopic identification of proteins from purified PEDV virions and from infected cells
| Group | M | N | S | ORF3 |
|---|---|---|---|---|
| rDR13att-ΔORF3 cells | + | + | + | − |
| rDR13att-ORF3wt cells | + | + | + | + |
| rDR13att-ORF3wt virus | + | + | + | − |
DISCUSSION
Coronavirus accessory proteins are generally dispensable for virus replication in cell culture, but their encoded proteins usually contribute to viral virulence (28). In the present study, we demonstrated that the PEDV ORF3 protein is indeed an accessory protein that is not essential for the viability of the virus in vitro but can promote virus replication. Using a reverse genetics system based on targeted RNA recombination, we successfully rescued 2 recombinant PEDVs having the same genetic background but differing in their ORF3 proteins and of which the virus entirely lacking this ORF was fully viable. The virus expressing an intact wild-type ORF3 and the attenuated DR13 with a truncated ORF3 were used to study the intracellular trafficking and location of the ORF3 protein. Similar to the structural membrane proteins of coronaviruses, the wild-type ORF3 protein was found to enter into and move through the exocytic pathway, accumulating in the Golgi area of the cell and being transported partly to the cell surface. In contrast, the truncated ORF3 protein was severely impaired in its transport and retained in the ER/ERGIC region.
Studies on the intracellular localization of the PEDV ORF3 protein are limited and inconsistent. Ye et al. expressed an N-terminal fluorescently tagged ORF3 protein in Vero cells (23). Though its localization was qualified as cytoplasmic by the authors, the concentrated appearance of the protein in their IF picture seems more consistent with an accumulation in the Golgi area, as was observed in the present work. Myc-tagged ORF3 protein transport was studied by Kaewborisuth et al., both in PEDV-infected cells and in cells individually expressing the protein (24). In both situations the IF staining appeared to be diffusely spread within the cells, without clearly specific localization, and it was also observed at the cell surface. Yet part of the intracellular protein was found to colocalize with markers for ER and Golgi complex. These authors also found the ORF3 protein to colocalize and interact with the viral spike protein, both intracellularly and at the plasma membrane. Finally, Zou et al. recently expressed an enhanced green fluorescent protein (EGFP)-ORF3 fusion protein in cells and found it to occur in an aggregated form in the ER, where it induced an ER stress reaction (25). It is unclear why the protein was not transport competent.
Though nothing is actually known about the functioning of the ORF3 protein during natural PEDV infection, an important role in viral pathogenicity is suggested by the attenuation that correlates with debilitating mutations in its gene. While complete deletion of the gene from an infectious clone did not seem to affect replication and infection by the virus, diarrheic scores in infected piglets were reduced, indicating the protein’s association with disease severity (29). Passaging of PEDV in culture cells often leads to deletions that are related with a reduction of virulence. An example is the deletion of 49 nucleotides that was observed in independently passaged isolates DR13 and CV777 and which resulted in viruses with severely truncated ORF3 proteins. When analyzing the intracellular location of the DR13att protein, we observed that the 91-residue-long polypeptide was unable to reach the Golgi complex but was retained in ER and ERGIC membranes. Various reasons may account for this early retention. One is that the truncated protein may not be able to fold into a conformation acceptable for the cellular quality control system to allow its exit from the ER. Such conformation might, for instance, require homo-oligomerization into tetramers, as was proposed to occur with the wild-type protein by Wang et al. on theoretical grounds (19). Another reason might be that specific trafficking signals contained in the lost C-terminal segment of the protein are required for efficient ER export and/or targeting to the Golgi complex (30–32).
Intracellular trafficking motifs are found in the C-terminal domains of many cellular and viral transmembrane proteins. These regulatory sequence motifs usually consist of 2 to 4 amino acids which are recognized by host adapter proteins of the cellular exo- and endocytic sorting pathways. The C-terminal domain of the PEDV ORF3 protein contains quite a number of such established motifs, and these are well conserved among various virus isolates. Thus, 3 tyrosine-based YxxØ motifs, an ExD diacidic motif, and a KKx dibasic motif occur; we evaluated their involvement in the protein’s intracellular transport. Only the 170YLAI173 (YxxØ) motif appeared to be critical for the protein’s efficient export from the ER. Signal-inactivating mutations of the key residues Y and I as well as deletion of the entire motif rendered the protein unable to exit the ER, thus preventing its transport through the Golgi complex to the plasma membrane. None of the mutations in the other sorting motifs affected ORF3 protein traffic, as judged from its efficient export from the ER and its appearance at the cell surface. The abundant presence of these conserved sorting motifs in the ORF3 protein is quite remarkable, and it is unclear whether they play any other roles in the functioning of the protein. Of note, many transmembrane proteins carry cytoplasmic domains that contain diacidic sequences on the C-terminal side of YxxØ motifs (33, 34). Thus, the ER export signal in vesicular stomatitis virus (VSV) G protein requires a YxxØ motif immediately proximal to a diacidic sequence (34, 35). Similarly, the combined deletion of the YxxØ and diacidic motifs from the severe acute respiratory syndrome coronavirus (SARS-CoV) U274 (3a) protein abolished the protein’s transport to the cell surface, suggesting their importance for exit from the ER and/or subsequent transport to the plasma membrane (36). In the ORF3 protein the juxtaposed 170YxxØ173 and 178EAD180 motifs do not appear to act together, as the separate inactivation of the EAD motif did not influence the protein’s trafficking behavior. It is interesting that the Tyr170 residue, besides being an essential element of the 170YxxØ173 transport motif, is also crucial for the ORF3 protein’s ion channel activity. Mutation of this residue (Y170A) was found to reduce the activity by about half (19). This may suggest that transport and channel activity somehow correlate and that the channel activity is most optimal in a transport-competent ORF3 protein. This might, for instance, be the case if the Y170A mutation would interfere with the protein’s oligomerization, which is likely to be important both for ER exit and for ion channel activity.
As already mentioned, many ORF3 analogs, particularly those in the Alphacoronavirus genus, have been demonstrated to encode proteins with ion channel properties (14). This is quite remarkable, as such function is also served by the E protein (37). Some ORF3-like proteins were also shown to enhance virus production and/or to be incorporated into virions (38, 39). In the present study, we also found the ORF3 protein to function in promoting virus production whether in intact or truncated form, consistent with another recent report (27). However, despite our use of sensitive antibodies and assays, we were unable to detect the ORF3 protein in purified PEDV particles. Hence, we conclude that the protein is not a structural virion component. This is consistent with its accumulation in the Golgi area, which is beyond the main cellular compartments where coronavirus assembly occurs, i.e., the ER and ERGIC (40).
MATERIALS AND METHODS
Cells, viruses, and antibodies.
Murine L (LR7) s (a L-2 murine fibroblast line stably expressing the murine hepatitis virus receptor, a gift of Peter Rottier, Utrecht University), African green monkey kidney cells (Vero; ATCC CCL-81), and porcine intestinal columnar epithelial cells (IPEC-J2, ACC-701; Leibnitz Institute DSMZ, German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany) were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS; Gibco, Invitrogen), penicillin (100 U/ml), and streptomycin (100 μg/ml) at 37°C in a humidified atmosphere with 5% CO2. The recombinant PEDV carrying the ORF3 gene of wild-type PEDV DR13 (GenBank accession no. JQ023161) (rDR13att-ORF3wt), the cell culture-adapted PEDV DR13 strain (DR13att), derived from a commercial vaccine of GreenCross, South Korea (GenBank accession no. JQ023162), and a recombinant PEDV without the ORF3 gene (rDR13att-ΔORF3) were propagated and titrated in Vero cells.
Rabbit anti-ORF3 polyclonal antibody (P71-3) was produced against a synthetic N-terminal 12-aa peptide (10IDTVVKDVSKSA21) of the ORF3 protein of PEDV DR13 (GenBank accession no. JQ023161). Mouse anti-ORF3 (aa 1 to 71) polyclonal antibody was produced against a recombinantly expressed N-terminal 71-aa peptide of the ORF3 protein of PEDV DR13 (GenBank accession no. JQ023161). Mouse anti-PEDV N protein monoclonal antibody 3F-12 was purchased from BioNote, Hwaseong-si, South Korea (catalog no. 9191). Rabbit anti-PEDV S polyclonal antibody was produced against a recombinantly expressed C-terminal 423-aa part of the spike protein (aa 961 to 1383) which was cloned from strain CV777 (GenBank accession no. AF353511). Mouse anti-PEDV M monoclonal antibody was kindly provided by Shaobo Xiao (Huazhong Agricultural University, Wuhan, China). Mouse anti-58K Golgi protein monoclonal antibody (catalog no. ab27043), mouse anti-GM130 Golgi protein polyclonal antibody (catalog no. ab169276), mouse anti-calnexin monoclonal antibody (catalog no. ab31290), mouse anti-LMAN1 monoclonal antibody (also known as ERGIC-53 antibody) (catalog no. ab118407) and mouse anti-hsp90β monoclonal antibody (catalog no. ab53497) were purchased from Abcam Biotech Inc., China. Horseradish peroxidase (HRP)-labeled goat anti-rabbit IgG (catalog no. D110058), HRP-labeled goat anti-mouse IgG (catalog no. D110087), and rabbit anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) polyclonal antibody (catalog no. D110016) were purchased from Sangon Biotech Inc., Shanghai, China. Alexa Fluor 647-conjugated goat anti-rabbit IgG (catalog no. A0468), Alexa Fluor 488-conjugated goat anti-mouse IgG (catalog no. A0428), and 4′,6-diamidino-2-phenylindole (DAPI) were purchased from Beyotime Biotech Inc., China.
Synthesis of full-length ORF3wt gene through overlapping-PCR.
The full-size 675-bp ORF3 gene of the wild-type PEDV strain DR13 (GenBank accession no. JQ023161) was constructed from overlapping synthetic cDNA fragments by PCR. Nine pairs of single-stranded and overlapping oligonucleotides (Table 2), each 59 nt in length, encompassing the entire ORF3wt gene, were used for the synthesis of the full-length ORF3wt gene, which were then cloned into pJET 1.2 blunt vector (Thermo Fisher, Carlsbad, CA). Recombinant plasmid containing the insert fragment was designated pJET 1.2-ORF3wt, and its insert was verified by sequencing.
TABLE 2.
Nucleotide sequences of oligonucleotide primers used for parental DR13 ORF3 (ORF3wt) gene synthesis
| Primer | Locationa | Sequence (5′–3′) |
|---|---|---|
| F1-F | 24785–24837 | GAATTCATGTTTCTTGGACTTTTTCAATACACGATTGACACAGTTGTCAAAGATGTCTC |
| F1-R | 24817–24875 | ACTCTTGGACAGCATCCAAAGACAAGTTGGCAGACTTTGAGACATCTTTGACAACTGTG |
| F2-F | 24855–24913 | CTTTGGATGCTGTCCAAGAGTTGGAGCTCAATGTAGTTCCAATTAGACAAGCTTCAAAT |
| F2-R | 24893–24951 | AAGTAGATAAAAACACTGGTGAAAAGAAAACCCGTAACATTTGAAGCTTGTCTAATTGG |
| F3-F | 24931–24989 | CACCAGTGTTTTTATCTACTTCTTTGCACTGTTTAAAGCGTCTTCTTTGCGGCGCAATT |
| F3-R | 24969–25027 | AAGAACGATGACAGCAAAACGCGCTGCCAACATAATATAATTGCGCCGCAAAGAAGACG |
| F4-F | 25007–25065 | CGTTTTGCTGTCATCGTTCTTTATTGCCCACTTTTATATTATTGTGGTGCATTTTTAGA |
| F4-R | 25045–25103 | AAAGCCTGCCAATAAGTGTGCAACAAATAATAGTTGCATCTAAAAATGCACCACAATAA |
| F5-F | 25083–25141 | GCACACTTATTGGCAGGCTTTGTTTAGTCTGCTTTTACTCCTGGCGCTATAAAAATGCG |
| F5-R | 25121–25179 | AGGAAAGAAAGTGTCGTAGTATTAAAGATAATAAAGAGCGCATTTTTATAGCGCCAGGA |
| F6-F | 25159–25217 | TACTACGACACTTTCTTTCCTCAATGGTAAAGCAGCTTATTATGACGGCAAATCCATTG |
| F6-R | 25197–25255 | GCCAAAAGTGATGTAATGGTCACCACCTTCTAGAATCACAATGGATTTGCCGTCATAAT |
| F7-F | 25235–25293 | GACCATTACATCACTTTTGGCAACTCTTTTGTTGCTTTCGTTAGTAGCATTGACTTGTA |
| F7-R | 25273–25331 | GATGTAGGTCAGCTTCTTGCCGCCCACGTATAGCTAGATACAAGTCAATGCTACTAACG |
| F8-F | 25311–25369 | GGCAAGAAGCTGACCTACATCTGTTGCGAACTGTTGAGCTTCTTGATGGCAAGAAGCTT |
| F8-R | 25349–25407 | TTAGTAATGCCGACAATTTGATGTTGCGAAAAGACATAAAGCTTCTTGCCATCAAGAAG |
| F9-F | 25387–25445 | TCAAATTGTCGGCATTACTAATGCCGCATTTGACTCAATTCAACTAGACGAGTATGCTA |
| F9-R | 25425–25477 | TTAACTACTAGACCATTACTCGAGTCATTCACTAATTGTAGCATACTCGTCTAGTTGAA |
Nucleotide positions are in accordance with the sequence of the virulent PEDV DR13 (GenBank accession no. JQ023161).
Recombinant plasmid constructions.
The construction of the pPEDV-DR13att vector, used as a donor plasmid in the targeted RNA recombination system for PEDV, has been described previously (26, 27). By replacing in this construct ORF3trun with its counterpart ORF3wt, the donor plasmid pDR13att-ORF3wt was generated (Fig. 1A). Briefly, a DNA fragment (FO) covering the ORF3wt gene and including a 20-bp 5′-terminal sequence of the E gene was PCR amplified using primers 1 and 3R (Table 3) and pJET 1.2-ORF3wt as a template. Another DNA fragment (FE; 203 bp) covering the E gene was PCR amplified with primers 3F and 2 and pPEDV-DR13att as a template. Then fragments FO and FE were fused into ORF3-E by PCR with primers 1 and 2. Finally, the PCR product was digested with PmlI and AgeI and cloned into the similarly digested pPEDV-DR13att to obtain pDR13att-ORF3wt. The acquired recombinant plasmid was verified by sequencing.
TABLE 3.
Primers used for plasmid constructions and rPEDV genome verification
| Primer | Location(s)a | Sequence (5′–3′)b |
|---|---|---|
| 1 | 24737–24786 | TGAAAAGGTCCACGTGCAGTGATGTTTCTTGGACTTTTTCAATACACGAT |
| 2 | 25513–25544 | GGTCAATTCGCATGTAAGACTTATAAACTCTA |
| 3F | 25342–25383 | GACTCAATTCAACTAGACGAGTATGCTACAATTAGTGAATGA |
| 3R | 25342–25383 | TCATTCACTAATTGTAGCATACTCGTCTAGTTGAATTGAGTC |
| 4 | 24726–24758 and 25343–25378 | TACGAAGCTTTTGAAAAGGTCCACGTGCAGTGAACTCAATTCAACTAGACGAGTATGCTACAATTAGTG |
| 5 | 25480–25500 | ATAGGTGTGTAAACTGCGCTA |
| 3F-XhoI | 24758–24779 | ATTCTCGAGATGTTTCTTGGACTTTTTCAAT |
| 3Rst-BamHI | 25362–25383 | ATTGGATCCCGTCATTCACTAATTGTAGCATAC |
| 3R-BamHI | 25362–25383 | ATTGGATCCCGTTCACTAATTGTAGCATAC |
The locations of primers are relative to the full genome sequence of the attenuated PEDV strain DR13 (GenBank accession no. JQ023162).
Restriction enzyme sites are marked by italic, bold letters. The stop codon is underlined.
For the construction of pDR13att-ΔORF3, a S-E-ΔORF3 DNA fragment was first amplified with primers 4 and 5 (Table 3) and pPEDV-DR13att as a template. The amplified fragment was then cloned into pPEDV-DR13att (as PmlI-AgeI fragment) to give pDR13att-ΔORF3.
To generate pDsRed-ORF3wt, a fragment corresponding to the full-size ORF3 but without the 3′ stop codon was generated by PCR using pJET 1.2-ORF3wt as a template and the 3F-XhoI/3R-BamHI primers (Table 3). The PCR product was inserted into pDsRed2-N1 (Clontech Laboratories, Inc.) using XhoI and BamHI restriction sites to yield the pDsRed-ORF3wt expression plasmid. pORF3wt was constructed similarly to pDsRed-ORF3wt except that the stop codon of ORF3 was recovered by using 3F-XhoI/3Rst-BamHI primers (Table 3).
To investigate the potential trafficking motifs in the C-terminal part of the ORF3 protein, ORF3s with mutant or deleted motifs were constructed by PCR-based mutagenesis of pORF3wt. The following ORF3 mutants were prepared: pORF3wt-AITF (AITF), pORF3wt-ALAI (ALAI), pORF3wt-YLAA (YLAA), pORF3wt-YLAG (YLAG), pORF3wt-ΔYLAI (ΔYLAI), pORF3wt-AAD (AAD), pORF3wt-EAA (EAA), pORF3wt-AKL (AKL), pORF3wt-AATI (AATI), and pORF3wt-YATG (YATG). All mutants were confirmed by DNA sequencing. The primer sequences, PCR conditions, and cloning details are available upon request.
Rescuing of recombinant PEDVs.
Generation and rescue of recombinant PEDVs were carried out by homologous RNA recombination as described previously (26, 27). Briefly, LR7 cells were grown to 90% confluence and infected at a multiplicity of infection (MOI) of 1.0 with mPEDV, a recombinant PEDV carrying most of the S gene of mouse hepatitis coronavirus, thereby enabling the virus to infect murine LR7 cells. When obvious cytopathic effect (CPE) started to appear around 8 h.p.i., cells were treated with trypsin to produce a single cell suspension and washed three times with phosphate-buffered saline (PBS). Capped runoff RNA transcripts (donor RNA) were synthesized from the PacI-linearized plasmids pDR13att-ΔORF3 and pDR13att-ORF3wt, using a T7 RNA polymerase kit (Ambion, Carlsbad, CA) as specified by the manufacturer. Then the donor RNAs were transferred into the above-mentioned mPEDV-infected LR7 cells by electroporation (300 V, 975 μF, two consecutive pulses) using a Gene Pulser apparatus (SCIENTZ-2C; Ningbo Scientz Biotech Inc., China). Finally, the electroporated cells were resuspended in DMEM supplemented with 2% FBS and cocultured in a 25-cm2 flask with 2 × 107 to 5 × 107 Vero cells. After 4 to 5 days of incubation at 37°C, progeny virus was harvested by freeze-thawing of the cultures 3 times, after which candidate recombinant viruses were purified by three rounds of endpoint dilutions on Vero cells as described below. The genotypes of the recombinant viruses were confirmed by RT-PCR and subsequent sequencing.
Growth curves of rPEDVs and DR13.
Vero cells grown in 6-well plates were inoculated in parallel with recombinant PEDVs and strain DR13 at an MOI of 5. After adsorption at 37°C for 60 min, the cells were fed with maintenance medium and incubated further at 37°C under 5% CO2. At various time points during the infections, the culture supernatant was harvested by freeze-thawing and candidate viruses were stored at –80°C, and the virus titers were determined according to previously described and expressed as 50% tissue culture infective dose (TCID50) per milliliter (27).
Purification of the recombinant virus by terminal dilution method.
The rescued viruses (passage 0 [p0]) were purified using the terminal dilution method. Briefly, the supernatant, clarified by centrifugation at 3,000 × g for 10 min at 4°C, was serially diluted in 2-fold increments and inoculated onto 90% confluent monolayers of Vero cells in 96-well plates using four duplicate wells per dilution. After 5 days of incubation, the cells exhibiting CPE at the highest dilution were harvested by three cycles of freeze-thawing, followed by removal of cell debris by centrifugation, and the supernatants were harvested for a further 2 rounds of similar purification. The final harvests were stored in aliquots as p3 stocks at –80°C.
Virus titration.
Viral titers were measured in 96-well plates using TCID50 assays. The culture supernatants were serially diluted 10-fold from 10−1 to 10−10, triplicate per dilution, and added onto a monolayer of Vero cells in 96-well culture plates. After 5 days of infection, the virus titers were determined according to the Reed and Muench method (41) and expressed as TCID50 per milliliter.
Transient expression of recombinant ORF3 proteins for localization studies.
Cells were seeded into 24-well plates in which coverslips had been placed. After culturing for approximately 1 day, cells were transfected by adding 1.0 μg of ORF3-expressing plasmid/well in Lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s recommendations and incubated at 37°C for approximately 24 h. Then cells were processed for indirect immunofluorescence microscopy as described below.
Indirect immunofluorescence assay.
For indirect immunofluorescence analysis (IFA), Vero cells on coverslips infected with the PEDVs or transfected with plasmids DNA were washed three times with PBS at 24 or 36 h after infection or transfection, respectively, fixed with 4% paraformaldehyde (PFA), and processed for indirect immunofluorescence analysis under nonpermeabilization or permeabilization conditions. For intracellular expression and localization purposes, cells were permeabilized with 0.1% Triton X-100. The cell sheet was blocked with 2% bovine serum albumin (BSA) for 1 h at room temperature. Cells were subsequently incubated for 1 h with rabbit anti-ORF3 polyclonal antibody (P71-3; 1:50) and antibodies to cellular marker proteins in blocking buffer at a dilution of 1:100. After three washings with PBS, corresponding Alexa Fluor-labeled secondary antibodies in blocking buffer at a dilution of 1:200 were added and the cells were further incubated for 1 h. Then the nuclei were stained with DAPI (1:1,000 dilution) at 37°C for 15 min. Images were viewed by using a fluorescence microscope (Axio Scope A1; Carl Zeiss, Germany) or a commercially available confocal scanning fluorescence microscope.
For analysis of cell surface expression, the PFA-fixed, nonpermeabilized Vero cells were blocked with 2% BSA, after which the exposed ORF3 protein was stained with mouse anti-ORF3 polyclonal antibody (aa 1 to 71; 1:50) or rabbit anti-ORF3 polyclonal antibody (P71-3; 1:50) under nonpermeabilization conditions. For comparison, PEDV proteins S, M, and N were probed by treatment with rabbit anti-S antibody (1:1,000), mouse anti-M antibody (1:100), and mouse anti-N antibody (1:100) as primary antibodies. The cellular hsp90β protein, which served as a cell surface marker, was detected by using mouse anti-hsp90β monoclonal antibody (1:100) as a primary antibody. Alexa Fluor 647-conjugated goat anti-rabbit IgG (1:100, red) and Alexa Fluor 488-conjugated goat anti-mouse IgG (1:200, green) were used as secondary antibodies and DAPI was used for staining of the cell nuclei. Images were viewed as described above. For cell surface detection of expressed ORF3 protein and its mutants, Vero cells were transfected with expression constructs for wild-type ORF3 protein (pORF3wt) or mutant ORF3 proteins (AITF, ALAI, YLAA, YLAG, ΔYLAI, AAD, EAA, AKL, AATI, and YATG). ORF3 protein expression on the cell surface was detected using rabbit-ORF3 (P71-3; 1:50) and Alexa Fluor 647-conjugated goat anti-rabbit antibodies (1:100, red) as primary and secondary antibodies, respectively, without permeabilization.
Flow cytometry.
Cell surface and intracellular ORF3 protein localization was additionally examined by flow cytometric analysis using a CytoFLEX flow cytometer (Beckman Coulter; 405/488-nm lasers) and Cytexpert software (2.3 version). Vero cells grown in 6-well plates were infected with PEDVs. After the desired time of incubation, the cells were trypsinized and harvested by centrifugation, after which they were washed twice by gentle resuspension in PBS and centrifugation. Half of the cells were fixed with 4% PFA and permeabilized for 30 min with ice-cold methanol. Both permeabilized and nonpermeabilized cells were resuspended in fluorescence-activated cell sorting (FACS) buffer (PBS containing 0.1% BSA) and subsequently stained with antibodies against the ORF3 protein as well as, for comparison, with antibodies against the PEDV structural proteins S, M, and N and with antibodies against cell surface marker hsp90β. Permeabilized and nonpermeabilized cells were stained with primary antibodies for 1 h at room temperature, washed twice with FACS buffer, and finally resuspended in 100 μl of the same buffer. The primary signal was detected using Alexa Fluor 647-conjugated goat anti-rabbit IgG (1:100) or Alexa Fluor 488-conjugated goat anti-mouse IgG (1:200). At least 5,000 events were acquired and FACS data were computed using Cytexpert software (2.3 version).
Virus purification by sucrose gradient ultracentrifugation.
For virus purification, confluent monolayers of Vero cells grown in T225 flasks (Nunc, Thermo Fisher, USA) were infected with rDR13att-ORF3wt, PEDV DR13att, and rDR13att-ΔORF3 at an MOI of 2. After infection for 2 days at 37°C in medium supplemented with 5% FCS, the culture supernatants were harvested and precleared by centrifugation at 1,500 × g for 10 min at 4°C to remove cell debris. Supernatants were then centrifuged at 28,000 rpm for 3 h at 4°C, and the pellets were resuspended in PBS. For additional purification of virus, the resuspended pellets were layered onto 20% to 60% (wt/wt) sucrose gradients (in PBS) and centrifuged for 6 h at 28,000 rpm. From each gradient, the virus particle band visible in the 40% to 50% zone of the sucrose gradient was collected and diluted with PBS. The viruses were finally concentrated by pelleting for 3 h at 28,000 rpm and dissolving in a small volume of PBS. The purified virus was analyzed by SDS-PAGE and Western blotting.
SDS-PAGE and Western blotting.
For sample preparation, purified virus was diluted in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 1 mM EDTA) containing protease inhibitors (Sigma-Aldrich, St. Louis, MO, USA) and taken up in SDS loading buffer (50 mM Tris-HCl [pH 6.8], 2% SDS, 10% glycerol, 1% β-mercaptoethanol, and 0.1% bromophenol blue). Cells were lysed by adding RIPA buffer onto the monolayers and incubated at 4°C for 30 min. Lysates were clarified by centrifugation at 15,000 × g for 20 min at 4°C, after which a sample of each supernatant was diluted in an equal volume of SDS loading buffer. The proteins in these samples were then separated by electrophoresis in 10% polyacrylamide gel and blotted onto a polyvinylidene difluoride (PVDF) membrane (Pall Corporation, East Hills, NY). Membranes were treated with 5% nonfat dry milk dissolved in TBST (Tris-HCl [pH 7.4], 150 mM NaCl, 0.1% Tween 20) for 1 h at room temperature and incubated overnight at 4°C with the specific primary antibodies. After a washing with TBS containing 0.1% Tween 20, the membranes were further incubated for 1 h with HRP-conjugated secondary antibodies (1:10,000 dilution). The immunolabeled proteins were visualized using Pierce ECL Western blotting substrate (Thermo Fisher, Carlsbad, CA) according to the manufacturer’s instructions.
MS characterization of proteins of PEDV virions and infected cells.
Subconfluent Vero cells were inoculated with rDR13att-ORF3wt or rDR13att-ΔORF3 at an MOI of 0.1 for 1 h, after which the inoculum was replaced with normal growth medium and incubation was continued. At 18 h.p.i., before the appearance of obvious CPE, the infected cells and culture medium were harvested separately. The culture medium from rDR13att-ORF3wt-infected cells was first centrifuged at 3,000 × g for 10 min to remove cell debris. The clarified supernatant (9 ml) was then placed in a centrifuge tube on top of a 2-ml 20% sucrose cushion and ultracentrifuged at 150,000 × g for 6 h at 4°C (Hitachi P40ST; Hitachi High-Technologies, Tokyo, Japan) to pellet the virus. Infected cell lysates were prepared as described above. For the identification of the proteins, the virus and cell samples obtained were processed by SDS-PAGE, after which the candidate protein bands around 25 to 28 kDa (about 1 cm2 of gel) were excised from each lane. The excised gel slices were digested with trypsin (in-gel digestion); the samples were desalted with a C18 ZipTip (Millipore Corporation, Bedford, MA) and then applied to analysis by nano-high-performance liquid chromatography-tandem mass spectrometry (nano-HPLC-MS/MS) (Q-Exactive mass spectrometer; Thermo Fisher, Carlsbad, CA) (42). Tandem mass spectra were processed by PEAKS Studio version 8.5 (Bioinformatics Solutions Inc., Waterloo, Canada).
ACKNOWLEDGMENTS
We thank Peter Rottier for critical readings of and helpful suggestions for the manuscript.
This study was partially supported by the National Natural Science Foundation of China (31572519), National Key Research and Development Program of China (no. 2016YFD0500101-03), International Science and Technology Cooperation Program of China (2013DFG32370), and Shanghai Key Project on Agricultural Development (no. 2015-6-1-9).
REFERENCES
- 1.Pensaert MB, de Bouck P. 1978. A new coronavirus-like particle associated with diarrhea in swine. Arch Virol 58:243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kusanagi K, Kuwahara H, Katoh T, Nunoya T, Ishikawa Y, Samejima T, Tajima M. 1992. Isolation and serial propagation of porcine epidemic diarrhea virus in cell cultures and partial characterization of the isolate. J Vet Med Sci 54:313–318. doi: 10.1292/jvms.54.313. [DOI] [PubMed] [Google Scholar]
- 3.Song D, Park B. 2012. Porcine epidemic diarrhoea virus: a comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Genes 44:167–175. doi: 10.1007/s11262-012-0713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takahashi K, Okada K, Ohshima K. 1983. An outbreak of swine diarrhea of a new-type associated with coronavirus-like particles in Japan. Nihon Juigaku Zasshi 45:829–832. doi: 10.1292/jvms1939.45.829. [DOI] [PubMed] [Google Scholar]
- 5.Jinghui F, Yijing L. 2005. Cloning and sequence analysis of the M gene of porcine epidemic diarrhea virus LJB/03. Virus Genes 30:69–73. doi: 10.1007/s11262-004-4583-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun RQ, Cai RJ, Chen YQ, Liang PS, Chen DK, Song CX. 2012. Outbreak of porcine epidemic diarrhea in suckling piglets, China. Emerg Infect Dis 18:161–163. doi: 10.3201/eid1801.111259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stevenson GW, Hoang H, Schwartz KJ, Burrough ER, Sun D, Madson D, Cooper VL, Pillatzki A, Gauger P, Schmitt BJ, Koster LG, Killian ML, Yoon KJ. 2013. Emergence of porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J Vet Diagn Invest 25:649–654. doi: 10.1177/1040638713501675. [DOI] [PubMed] [Google Scholar]
- 8.Kocherhans R, Bridgen A, Ackermann M, Tobler K. 2001. Completion of the porcine epidemic diarrhoea coronavirus (PEDV) genome sequence. Virus Genes 23:137–144. doi: 10.1023/a:1011831902219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee C. 2015. Porcine epidemic diarrhea virus: an emerging and re-emerging epizootic swine virus. Virol J 12:193. doi: 10.1186/s12985-015-0421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duarte M, Tobler K, Bridgen A, Rasschaert D, Ackermann M, Laude H. 1994. Sequence analysis of the porcine epidemic diarrhea virus genome between the nucleocapsid and spike protein genes reveals a polymorphic ORF. Virology 198:466–476. doi: 10.1006/viro.1994.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park SJ, Moon HJ, Yang JS, Lee CS, Song DS, Kang BK, Park BK. 2007. Sequence analysis of the partial spike glycoprotein gene of porcine epidemic diarrhea viruses isolated in Korea. Virus Genes 35:321–332. doi: 10.1007/s11262-007-0096-x. [DOI] [PubMed] [Google Scholar]
- 12.Bosch BJ, van der Zee R, de Haan CA, Rottier PJ. 2003. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol 77:8801–8811. doi: 10.1128/jvi.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Godet M, Grosclaude J, Delmas B, Laude H. 1994. Major receptor-binding and neutralization determinants are located within the same domain of the transmissible gastroenteritis virus (coronavirus) spike protein. J Virol 68:8008–8016. doi: 10.1128/JVI.68.12.8008-8016.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang K, Xie S, Sun B. 2011. Viral proteins function as ion channels. Biochim Biophys Acta 1808:510–515. doi: 10.1016/j.bbamem.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Haan CA, Kuo L, Masters PS, Vennema H, Rottier PJ. 1998. Coronavirus particle assembly: primary structure requirements of the membrane protein. J Virol 72:6838–6850. doi: 10.1128/JVI.72.8.6838-6850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen V-P, Hogue BG. 1997. Protein interactions during coronavirus assembly. J Virol 71:9278–9284. doi: 10.1128/JVI.71.12.9278-9284.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang XC, Zhang JX, Zhang SY, Wang P, Fan XH, Li LF, Li G, Dong BQ, Liu W, Cheung CL, Xu KM, Song WJ, Vijaykrishna D, Poon LL, Peiris JS, Smith GJ, Chen H, Guan Y. 2006. Prevalence and genetic diversity of coronaviruses in bats from China. J Virol 80:7481–7490. doi: 10.1128/JVI.00697-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zuñiga S, Pascual-Iglesias A, Sanchez CM, Sola I, Enjuanes L. 2016. Virulence factors in porcine coronaviruses and vaccine design. Virus Res 226:142–151. doi: 10.1016/j.virusres.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang K, Lu W, Chen J, Xie S, Shi H, Hsu H, Yu W, Xu K, Bian C, Fischer WB, Schwarz W, Feng L, Sun B. 2012. PEDV ORF3 encodes an ion channel protein and regulates virus production. FEBS Lett 586:384–391. doi: 10.1016/j.febslet.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park SJ, Moon HJ, Luo Y, Kim HK, Kim EM, Yang JS, Song DS, Kang BK, Lee CS, Park BK. 2008. Cloning and further sequence analysis of the ORF3 gene of wild- and attenuated-type porcine epidemic diarrhea viruses. Virus Genes 36:95–104. doi: 10.1007/s11262-007-0164-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J, Wang C, Shi H, Qiu H, Liu S, Chen X, Zhang Z, Feng L. 2010. Molecular epidemiology of porcine epidemic diarrhea virus in China. Arch Virol 155:1471–1476. doi: 10.1007/s00705-010-0720-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X, Zeng L, Yang J, Yu F, Ge J, Guo Q, Gao X, Song T. 2013. Sequence heterogeneity of the ORF3 gene of porcine epidemic diarrhea viruses field samples in Fujian, China, 2010–2012. Viruses 5:2375–2383. doi: 10.3390/v5102375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye S, Li Z, Chen F, Li W, Guo X, Hu H, He Q. 2015. Porcine epidemic diarrhea virus ORF3 gene prolongs S-phase, facilitates formation of vesicles and promotes the proliferation of attenuated PEDV. Virus Genes 51:385–392. doi: 10.1007/s11262-015-1257-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaewborisuth C, He Q, Jongkaewwattana A. 2018. The accessory protein ORF3 contributes to porcine epidemic diarrhea virus replication by direct binding to the spike protein. Viruses 10:399. doi: 10.3390/v10080399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zou D, Xu J, Duan X, Xu X, Li P, Cheng L, Zheng L, Li X, Zhang Y, Wang X, Wu X, Shen Y, Yao X, Wei J, Yao L, Li L, Song B, Ma J, Liu X, Wu Z, Zhang H, Cao H. 2019. Porcine epidemic diarrhea virus ORF3 protein causes endoplasmic reticulum stress to facilitate autophagy. Vet Microbiol 235:209–219. doi: 10.1016/j.vetmic.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li C, Li Z, Zou Y, Wicht O, van Kuppeveld FJ, Rottier PJ, Bosch BJ. 2013. Manipulation of the porcine epidemic diarrhea virus genome using targeted RNA recombination. PLoS One 8:e69997. doi: 10.1371/journal.pone.0069997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Si F, Hu X, Wang C, Chen B, Wang R, Dong S, Yu R, Li Z. 2020. Porcine epidemic diarrhea virus (PEDV) ORF3 enhances viral proliferation by inhibiting apoptosis of infected cells. Viruses 12:214. doi: 10.3390/v12020214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Haan CA, Masters PS, Shen X, Weiss S, Rottier PJ. 2002. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology 296:177–189. doi: 10.1006/viro.2002.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beall A, Yount B, Lin CM, Hou Y, Wang Q, Saif L, Baric R. 2016. Characterization of a pathogenic full-length cDNA clone and transmission model for porcine epidemic diarrhea virus strain PC22A. mBio 7:e01451-15. doi: 10.1128/mBio.01451-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonifacino JS, Traub LM. 2003. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem 72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- 31.Bonifacino JS, Glick BS. 2004. The mechanisms of vesicle budding and fusion. Cell 116:153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- 32.Beckers CJ, Block MR, Glick BS, Rothman JE, Balch WE. 1989. Vesicular transport between the endoplasmic reticulum and the Golgi stack requires the NEM-sensitive fusion protein. Nature 339:397–398. doi: 10.1038/339397a0. [DOI] [PubMed] [Google Scholar]
- 33.Bannykh SI, Balch WE. 1998. Selective transport of cargo between the endoplasmic reticulum and Golgi compartments. Histochem Cell Biol 109:463–475. doi: 10.1007/s004180050248. [DOI] [PubMed] [Google Scholar]
- 34.Sevier CS, Weisz OA, Davis M, Machamer CE. 2000. Efficient export of the vesicular stomatitis virus G protein from the endoplasmic reticulum requires a signal in the cytoplasmic tail that includes both tyrosine-based and di-acidic motifs. Mol Biol Cell 11:13–22. doi: 10.1091/mbc.11.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishimura N, Balch WE. 1997. A di-acidic signal required for selective export from the endoplasmic reticulum. Science 277:556–558. doi: 10.1126/science.277.5325.556. [DOI] [PubMed] [Google Scholar]
- 36.Tan YJ, Teng E, Shen S, Tan TH, Goh PY, Fielding BC, Ooi EE, Tan HC, Lim SG, Hong W. 2004. A novel severe acute respiratory syndrome coronavirus protein, U274, is transported to the cell surface and undergoes endocytosis. J Virol 78:6723–6734. doi: 10.1128/JVI.78.13.6723-6734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Castaño-Rodriguez C, Honrubia JM, Gutiérrez-Álvarez J, DeDiego ML, Nieto-Torres JL, Jimenez-Guardeño JM, Regla-Nava JA, Fernandez-Delgado R, Verdia-Báguena C, Queralt-Martín M, Kochan G, Perlman S, Aguilella VM, Sola I, Enjuanes L. 2018. Role of severe acute respiratory syndrome coronavirus viroporins E, 3a, and 8a in replication and pathogenesis. mBio 9:e02325-17. doi: 10.1128/mBio.02325-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ito N, Mossel EC, Narayanan K, Popov VL, Huang C, Inoue T, Peters CJ, Makino S. 2005. Severe acute respiratory syndrome coronavirus 3a protein is a viral structural protein. J Virol 79:3182–3186. doi: 10.1128/JVI.79.5.3182-3186.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang R, Wang K, Ping X, Yu W, Qian Z, Xiong S, Sun B. 2015. The ns12.9 accessory protein of human coronavirus OC43 is a viroporin involved in virion morphogenesis and pathogenesis. J Virol 89:11383–11395. doi: 10.1128/JVI.01986-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klumperman J, Locker JK, Meijer A, Horzinek MC, Geuze HJ, Rottier P. 1994. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J Virol 68:6523–6534. doi: 10.1128/JVI.68.10.6523-6534.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Hyg 27:493–497. [Google Scholar]
- 42.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. 2006. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]