Abstract
Understanding molecular, cellular, genetic and functional heterogeneity of tumors at the single-cell level has become a major challenge for cancer research. The microfluidic technique has emerged as an important tool that offers advantages in analyzing single-cells with the capability to integrate time-consuming and labour-intensive experimental procedures such as single-cell capture into a single microdevice at ease and in a high-throughput fashion. Single-cell manipulation and analysis can be implemented within a multi-functional microfluidic device for various applications in cancer research. Here, we present recent advances of microfluidic devices for single-cell analysis pertaining to cancer biology, diagnostics, and therapeutics. We first concisely introduce various microfluidic platforms used for single-cell analysis, followed with different microfluidic techniques for single-cell manipulation. Then, we highlight their various applications in cancer research, with an emphasis on cancer biology, diagnosis, and therapy. Current limitations and prospective trends of microfluidic single-cell analysis are discussed at the end.
Keywords: Microfluidic Lab-on-a-chip, Single cell analysis, Cancer biology, Cancer diagnosis, Cancer therapy, Cancer research
1. Introduction
Single cell analysis has been gaining attention and popularity in recent years [1, 2]. Single-cell assays reveal heterogeneities in morphology, functions, composition, and genetic performance of seemingly identical cells. An accurate analysis of biomolecules in cancer cells (e.g. DNA mutation and protein biomarkers) can help to improve fundamental understanding of the development, the progression and the classification of tumor types at the cellular and molecular levels, and may further aid the development of new diagnostic and therapeutic approaches [1, 2]. The current knowledge of cancer biology is mostly based on information obtained from bulk experiments using traditional population-averaged approaches, such as Western blotting, proliferation assays, DNA sequencing or cytotoxicity assays which are performed in microtubes or microwells with the sample volume on a microliter scale, much larger (6 orders of magnitude) than the single cell volume of picoliters [3, 4]. By using these approaches, some key information, such as molecular distributions, functional variations, and drug-target interactions, may be hidden at the single-cell level. In addition, these traditional approaches always encounter difficulties in manipulating a single cell such as single-cell isolation and capture at low efficiency [1, 5]. Therefore, there is an urgent need to develop new tools to advance single-cell analysis.
The microfluidic lab-on-a-chip (LOC), a type of miniaturized devices mostly produced by the microfabrication technique, has developed rapidly in the last few decades, since its advent in the 1990s [6]. Microfluidic devices have been widely used in various applications, especially in bio-applications, thanks to their significant advantages (e.g. fast analysis, low reagent consumption, and high capillary electrophoresis (CE) separation efficiency) associated with their inherent miniaturization, integration, portability, and automation [7–16]. This technique has several advantages for cellular assays: (1) the micrometer-sized liquid channels or physical structures are compatible with the cell sizes, and this makes single cell manipulation or single-cell capture applicable and improves cell capture efficiency [17]; (2) Many traditional instruments like fluorescence-activated cell sorter (FACS) require a large amount of cells for their desired performance. However, microfluidic devices require only a small amount of samples, and this aspect is especially important in bioanalytical applications since biological samples are often limited in quantity; (3) the reagent consumption is low, which reduces the cost of assays; and (4) microfluidic technique can integrate multiple functions such as cell sampling, fluid control, single-cell capture, cell lysis, mixing, and detection on a single device. Among microfluidic cellular applications, single-cell analysis is preferred because the traditional bulk cellular analysis often overlooks cellular heterogeneity and does not provide information on cell-to-cell variations [18]. Additionally, physicochemical modeling of biological processes also demands single-cell data [19]. Therefore, microfluidic single-cell analysis has been widely used in numerous applications, including intracellular signaling [20], myocyte contraction [21], drug discovery [22], patch-clamp recording [23], multidrug resistance [24, 25], genetic analysis [26], protein analysis [27], and so on. Among those applications, particularly, considering that cancer cells show high levels of cellular heterogeneity and cancer originates from a small number of cells [19, 28, 29], microfluidic single-cell analysis plays a more and more important role in cancer research [19, 30–32], ranging from fundamental studies in cellular interactions to cancer diagnosis and cancer treatment such as multidrug resistance (MDR) modulation [18] and anticancer drug discovery [33, 34]. In addition, circulating tumor cells (CTCs) which are known as a liquid biopsy and exist at low concentrations in a complex mixture of biological samples, have been confirmed to have a high level of heterogeneity in the single cell analysis of CTCs using live single cell mass spectrometry (MS) integrated with microfluidic-based cell enrichment techniques [35]. Hence, cancer research at the single-cell level using microfluidics has become one of the most popular applications [1, 31, 36].
This article reviews most recent advances in microfluidic technologies for single-cell analysis in cancer research and practice. We first introduce microfluidic platforms for single-cell analysis. Then, we summarize microfluidic techniques for single cell isolation, single cell treatment, followed with various applications of microfluidics for single-cell analysis in cancer research, with an emphasis on cancer biology, diagnosis, and therapy. Finally, we discuss current limitations of microfluidic single cell analysis and perspectives.
2. Microfluidic platforms for single-cell analysis
A significant number of microfluidic platforms have been developed for single-cell analysis. In this section, based on fabrication materials, we separate these platforms into three major categories: polydimethylsiloxane (PDMS), glass, and paper.
2.1. PDMS-based microfluidic devices
PDMS, a polymer material used in microfluidic fabrication, is flexible, low-cost, and optically transparent down to 230 nm in UV light. PDMS is compatible with biological studies and becomes the most widely used chip substrate in single-cell analysis, because of its easy fabrication, low-cost, and O2 permeability properties. Although PDMS devices can be fabricated by multiple methods such as laser ablation [9, 37, 38], injection and so on, most PDMS devices are fabricated by the soft-lithography method using a mold or master to replicate patterns. These masters can be made of a variety of materials through the photolithography technique. One of the most popular master materials is SU-8, a UV-sensitive, high-contrast, epoxy-based negative tone photoresist designed for the lithography of ultra-thick resists [39]. To fabricate the PDMS replica, briefly, the mixture of PDMS precursor (liquid) and crosslinking curing agent (10:1, v/v) is firstly degassed then poured onto blank masters for curing (70 °C for 3 h, 80 °C for 2 h or 95 °C for 1 h) to form the top PDMS layer with patterns from a master mold, which is often fabricated with SU-8 on a wafer, and pattern mask covered UV exposure, as shown in Fig. 1A. Once the PDMS is hardened, it can be peeled off from the mold, and proceed with a bonding process after plasma surface treatment [40]. For instance, through a standard soft lithography method, Li et al. [41] fabricated a chip device by combining a single-cell-arrayed agarose layer with a microfluidics-based oxygen gradient-generating layer using a PDMS membrane. A top layer was designed for cell culture and the bottom layer for chemical reaction channels. The mixture of PDMS precursor and curing agent (10:1, v/v) was firstly degassed and poured onto a blank master for curing (70 °C for 3 h) to form the top layer with the thickness of 5 mm. Using tirapazamine (TPZ) and bleomycin (BLM) as model anticancer drugs, this PDMS based chip demonstrated single cell loading, cell cultivation, and subsequent drug treatment for the analysis of oxygen-dependent cytotoxicity and genotoxicity of anticancer drugs.
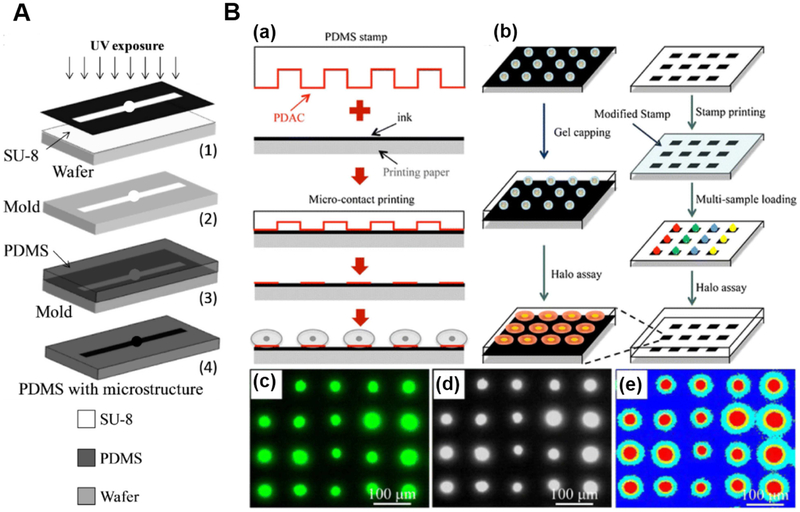
Fig. 1.
(A) Schematic diagram of SU-8 involved soft-lithography for PDMS-based microfluidic chip fabrication. Adapted with permission from Ref. [62]. Copyright 2014 Elsevier. (B) Schematic diagram of a paper-based HaloChip fabrication and the method of the single cell HaloChip assay. (a) The steps used to form single cell arrays on ink-covered paper using microcontact printing technique; (b) The method used to assess DNA damage on the ink-covered paper using HaloChip assay. (c) Original halo image; (d) Grayscale image; (e) Identifying halos and nuclei in an array. Adapted with permission from Ref. [58]. Copyright 2016 Springer.
It is easier to use PDMS to create high-aspect-ratio microstructures like micropillars than glass for cellular study in a three-dimensional (3D) environment, in combination with gels. Marasso et al. [42] presented a PDMS-based microfluidic platform for cell motility analysis in a 3D environment, in which injection of both single cells and cell spheroids were investigated under spatially and temporally controlled chemical stimuli. A chip layout based on a central microchannel confined by micropillars and two lateral reservoirs was selected as the most effective. The microfluidics had an internal height of 350 μm to accommodate cell spheroids of a considerable size. The chip was fabricated by obtaining the PDMS replica from a Si/SU-8 master. Then, the chip was bonded on a thin microscope cover glass slide with 170-μm thickness that allows high spatial resolution live microscopy. In order to allow both cost-effective and highly repeatable production of chips with high-aspect-ratio (5:1) micropillars, specific design and fabrication processes were optimized. This design permitted spatial confinement of the gel where cells were grown, the creation of a stable gel–liquid interface, and the formation of a diffusive gradient of a chemoattractant (> 48 h).
2.2. Glass-based microfluidic devices
Glass is another material commonly used for single-cell analysis, particularly in the first two decades of the microfluidic lab-on-a-chip, due to its excellent mechanical and optical properties and chemical resistance. Because glass is compatible to the traditional microfabrication technique, most glass devices are fabricated through the standard photolithography process, which involves substrate cleaning, photoresist spinning, alignment and UV exposure, photoresist developing, chrome etching, glass etching, stripping of the remaining photoresist and chrome, and thermal bonding [43]. Glass can be patterned by wet etching, dry etching as well as laser ablation techniques [39, 44]. The widely used wet etching for glass is normally an isotropic process with hydrofluoric acid (HF) based solutions. By manipulating the concentration of HF, or adding other strong acids like HCl, HNO3, H2SO4, or H3PO4 in the solution, the etch rate can be altered [39, 45, 46]. For example, using a wet etching process, Li et al. [19, 47] fabricated multiple glass based microfluidic chips for single cell analysis for cytotoxicity application and drug resistance tests. They combined a one-level microfabrication method with a post-etching process to create a dam structure for single-cell capture, which usually requires two-level microfabrication procedures [47]. Recently, Faigle et al. [48] fabricated an optical stretcher glass chip, involving the bonding of two asymmetrically etched glass plates, for sorting and measuring single cells in a heterogeneous population. The two slides of asymmetrically etched glass were achieved through a standard manufacturing process of wet etching with bulk glasses.
Other than wet etching, laser ablation has also been used to fabricate nano-sized channels on glass. Laser ablation is the process to remove materials from a solid surface by irradiating it with a laser beam. Yun et al. [49] presented a microfluidic chip with a nanoinjection system integrated with capillary electrophoresis (CE) for precise direct delivery of biomolecules into single cells. The nanoinjection structure was fabricated by using femtosecond-laser (fs-laser) ablation in a single solid glass layer. Nanochannels with a diameter of 800 nm in the nanoinjection structure were achieved by precise laser ablation with a higher resolution than soft lithography such as PDMS. The fluorescent results showed successful delivery of red fluorescent protein (RFP) and expression of plasmid DNA in several different types of cells.
2.3. Paper-based microfluidic devices
Recently, paper-based microfluidics has attracted increasing attention due to a number of merits such as low cost, ease of fabrication and surface modification, portability, and facile integration with other devices. Paper-based microfluidics devices have been widely used in point-of-care testing, public health, food quality control, environmental monitoring [8, 9, 13, 14, 50–52]. In addition, paper microfluidics has also been endowed with applications in cell/tissue culture for cellular analysis, due to its inherent porous and flexible micro/nanostructures. Paper substrates provide a natural 3D scaffold to mimic native cellular microenvironments, enabling different applications related to cellular biology in a 3D fashion at the single-cell level [53, 54]. A variety of methods have been developed for the paper-based device fabrication mostly using either cellulose chromatography paper or nitrocellulose membrane, including photolithography [55], wax printing [56], screen printing [57], micro-contact printing [58], and cutting [59, 60].
The porosity, surface chemistry as well as optical properties of paper have been incorporated into different microfluidic systems to engineer cell culture microenvironment for applications like cell differentiation, disease model construction, single cell analysis and drug screening [61]. With a micro-contact printing method, Ma et al.[58] developed a paper-based low-cost single cell HaloChip assay that can be used to assess drug- and radiation-induced DNA damage at the point of care. Instead of forming cell arrays on silicon or glass substrates, single cell arrays were patterned on normal printing paper, as shown in Fig. 1B. Instead of using labile chemicals for surface modification, multiple ink printing layers were used to modify paper surface to provide excellent water-repelling ability. A PDMS stamp treated with polydiallyldimethyl ammonium chloride (PDAC) was brought into contact with the ink-covered paper. Slight pressure was applied on the stamp manually to ensure the microcontact between a PDMS stamp and the ink-covered paper, and then the stamp was peeled off from the paper. Then, cells were seeded and incubated on the paper, and unattached cells were rinsed away with PBS. After gel solidification and X-ray exposure, cells were stained with SYBR green I and fluorescent images were analyzed to quantify DNA damage from drug efficacy and radiation condition.
3. Techniques of microfluidic single cell manipulation
Prior to single cell analysis, cells are generally processed via different manipulation techniques. In this section, techniques of single cell manipulation using microfluidics will be summarized and discussed based on two major purposes, single cell isolation and single cell treatment.
3.1. Single cell isolation
Single cell isolation is crucial to single-cell analysis in order to better understand the variations from cell to cell, which can provide valuable information for diagnostics and other biomedical applications. Early technologies to isolate single cells include serial dilution, density gradient centrifugation, membrane filtration, and manual cell picking or micromanipulation, which are simple and convenient to operate, while suffering from low purity of isolated cells and low-throughput [63–65]. Currently, new advanced techniques have been developed for single-cell isolation, such as optical tweezers [66, 67], laser-capture microdissection (LCM) [68, 69], fluorescence-activated cell sorter (FACS) [70], and magnetic-activated cell sorting (MACS) [71], which provide precise isolation and purity of cells and high specificity. However, the intrinsic limitations of these conventional methods including bulky and costly instruments, trained personnel needed, requirement of a large number of cells, low throughput (e.g. for optical tweezers), and time-consuming procedures, have hindered their broad applications.
Microfluidic technologies are advantageous in isolating individual cells by providing precise fluid control, facile automation, low sample consumption, and high throughput. Single cell isolation using microfluidic platforms allows for precise manipulation by trapping, transporting, and releasing single cells [4]. Numerous structural designs with different geometries have been developed to isolate single cells, such as microwells [72, 73], microdams [74, 75], and microvalves [76, 77]. For instance, high-throughput trapping of single cells was achieved by using U-shaped microdams [78]. Active methods such as dielectrophoresis (DEP) have also been developed to isolate single cells on different microfluidic platforms [79, 80]. In this section, we mainly focused on two main types of microfluidic trapping strategies for single cell isolation, mechanical traps, and microfluidic droplets.
3.1.1. Mechanical traps
Mechanical microfluidic traps are mainly characterized by microstructures designed as barriers or chambers that control the flow of cell suspension to capture a single cell in determined compartments such as microwells, microdams, and valves [4]. Additionally, computation fluid dynamics simulations have been employed to study flow behaviors on single-cell manipulation [81].
Microwells approaches for single cell isolation enhances long-term cell viability and minimizes cell stress in trapping areas. Various microwells structures with different sizes or shapes were designed to capture, and separate cells. For example, a dual-well microfluidic chip was designed with top and bottom layers, composed of 25-μm-diameter trapping wells and 285- and 485-μm-diameter culture wells, for proliferation and differentiation. Single-cells were captured by gravity from the microchannel stream into the 25 μm diameter layer placed at the bottom, while excess of cells were washed away, and release of trapped single-cells into larger wells was enabled by flipping the microfluidic device. A ~77% single-cell loading for in vitro cell culture of A549 and MDA-MB-435 cancer cells and KT98 stems cells was achieved [72]. Comparably, Tu et al. [73] reported a microfluidic combination of physical microwells and protein patterns, in which single-cells were captured by flipping a layered chip after protein binding. Within a triangular shape, HeLa and gall bladder carcinoma (SGC-996) cells were captured with 79% efficiency for cell patterning studies. To improve selectivity, a combination of microwells with a permanent magnet was developed to analyze immunomagnetic labeled (THP-1) Leukemia cells, achieving a 99.6% purity and 62% trapping efficiency for single-cell analysis [82].
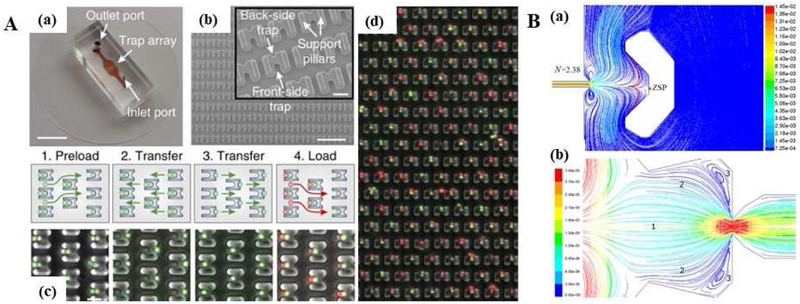
Similar to a ball-in-a-maze puzzle, microdams were designed with different geometries and holes to trap single cells. Semicircular structure microchip demonstrated a label-free and rapid single-cell sequential isolation of MDA-MB-231 human breast cancer cells [74]. It was reported that a microfluidic device with 50 identical semicircular microsieves with a 100 μm inner diameter and specific offsets and distances controlled the hydrodynamic trajectory of cells to achieve a 100% isolation yield and > 95% sequential isolation efficiency [74]. Moreover, to study adaptive immune response of lymphocytes and antigen-presenting cells (APCs) for cell-cell interactions, Dura et al. [75] designed a high-throughput microfluidic device with about 1,000 trapping sites for single cell pairing. Dynamic interactions of CD8 T cells with APCs, along with characterization of melanoma mouse CD8 T cells were evaluated. As shown in Fig. 2A, a PDMS microfluidic device composed with multiple semicircular back and front cell capture cups of 6–8 μm were filled by loading 1–5 μL of a cell suspension into the inlet reservoir. Systematically, a four-step back-forth loading process allowed cell capture at the front-side two-cell trap; next, an opposite flow direction temporarily held the desired cell at the back-side, while excess of cells were removed, and another flow reverse allowed cell pairing with a second cell population at the front-side two-cell trap. As a result, flow rates between 100 nL/min–2 μL/min reduced cell damage and restrained cell aggregation to obtain an 80% trapping efficiency and a > 95% fill factor within a density of 500–580 traps per mm2 in less than 40 s.
Fig. 2.
(A) A microfluidic device with ~1000 semicircular trapping sites for high-throughput single-cell capture. (a) Picture of the microfluidic device with channels and traps. (b) SEM image of single cells trapping structures. (c) Microfluidic cell pairing mechanism. (d) Fluorescence images of lymphocytes cells stained with Dil (red) and Dio (green) dyes. Adapted with permission of Ref. [75]. Copyright 2015 Springer Nature Publishing. (B) CFD simulations for single-cell capture. Streamline patterns in two different single-cell retention structures (a-b). Adapted with permission from Ref. [81] Copyright 2014 API Publishing.
Microfluidic valves structures are able to control fluids over trapping sites, facilitating selection of the desired single-cell capture, and have demonstrated high programmability for high throughput [83]. A flexible microfluidic V-Type valve system with a PDMS membrane was able to isolate prostate cancer (PC3) single-cells by opening and closing the valve to further push and trap cells within a microchamber [76]. Moreover, to analyze anticancer peptides, a microfluidic device with a combination of pneumatic valves and microchambers was designed to study drug response on hundreds of MCF-7 single-cells. It was found that 612 chambers distributed in 34 rows and 16 columns with a doughnut shape and a minimal chamber size (<100 pL) minimized the shear stress. However, only 200 chambers were filled with MCF-7 single cells, due to cell aggregation [77].
Furthermore, cell retention design affects dynamic flow behaviors and shear stress to cells. To improve single-cell capture results and minimize shear stress to captured cells, Li et al. [81] developed a method of computational fluid dynamics (CFD) simulations to compare the flow behaviors on single-cell manipulation and shear stress reduction in different cell retention structures (Fig. 2B). The CFD simulation results showed that the flow behaviors affected the single-cell capture locations (or zero-speed points (ZSP)) in the U-shaped structure from Chip V1 (Fig. 2B(a)). In addition, the flow distribution showed that a waist band configuration with a gap-leading channel at the back in Chip V2 had enhanced flow behaviors for single-cell capture (Fig. 2B(b)), which also minimized shear stress on cells mainly through the newly added leading channel. CFD simulations can provide useful guidelines for single-cell chip design and flow optimization.
3.1.2. Droplet traps
Droplet-based microfluidic technology has shown promising capabilities in single-cell isolation in a high-throughput fashion for widespread applications in biochemical and biomedical fields. There are numerous advantages of droplet-based microfluidic devices such as high throughput production, precise fluid control and stable manipulation of cells both spatially and temporally, and well-defined sizes and structures [84]. Individual cells can be trapped in each droplet, which acts as a discrete compartment to isolate cells as well as further storing, identifying, sorting, and transporting cells. Based on different techniques used to control the droplet formation [85], there are generally two categories of droplet-based microfluidic devices: passive and active.
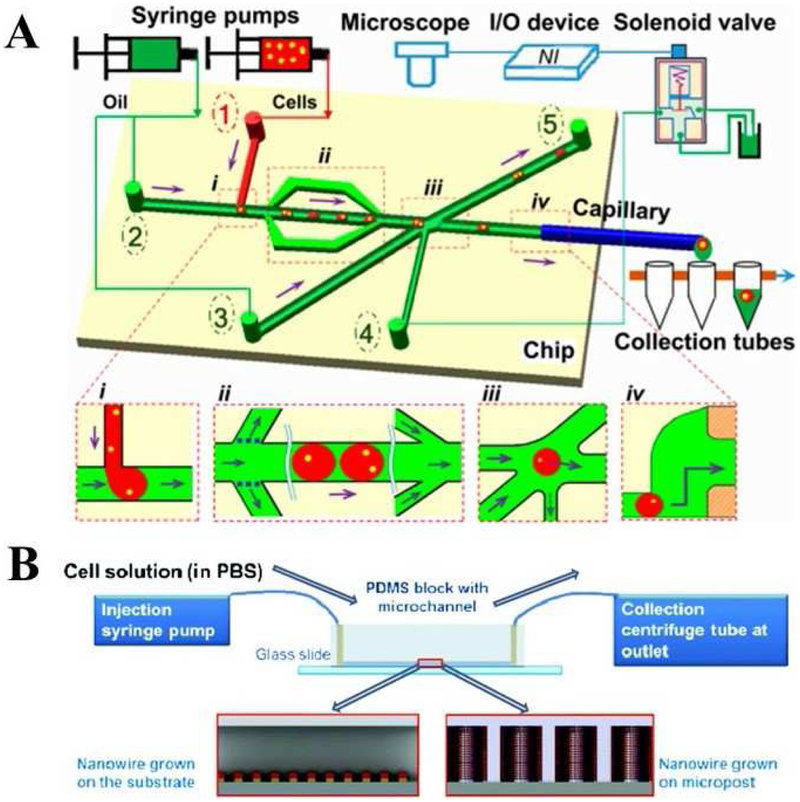
Droplets generation on passive microfluidic devices can be achieved commonly by applying T-junction or flow-focusing structures, where one dispersed phase fluid is introduced to another immiscible continuous fluid by syringes or pumps. Single cells can be q. [86] developed a highly-integrated system for single-cell genomic analysis of Escherichia coli, by integrating a programmable passive droplet-based microfluidic device with conventional protocols of gene-specific analyses. The droplet-based microfluidic device was fabricated mainly using PDMS and the droplets were generated in a passive mode. As shown in Fig. 3A(a), the chip consisted of a T-junction configuration to encapsulate cells in water-in-oil droplets (about 60 μm in diameter), and different branch-channel structures to isolate and export single cells. Syringe pumps were used to assist the cell and oil injection, while an optical microscope was applied to inspect the isolation workflow. Basically, four procedures were conducted on one chip for single-cell isolations in droplets, including droplet generation, droplet deceleration by the branch-channel structure, droplet sorting by solenoid valve suction, and droplet exporting into a collection tube through an embedded capillary interface. The success rate of single-cell droplet isolation was 94.4 ± 2.0% with an average collection throughput around 0.05 cells/s (i.e. 20 s/cell). The droplet-based microfluidic system was further applied in single-cell cultivation with a success rate of 80%, high-quality gene-specific analyses, and whole genome sequencing. The proposed method using microfluidic droplets improved the accessibility of single-cell analysis and provided a facile platform for downstream analyses. Another passive droplet-based microfluidic device using similar flowing-focus junction structures was developed to coencapsulate single cells and nanoparticles in individual droplets [17]. The droplets were tracked in a spectrally independent manner using fluorescence microscopy, enabling a robust quantification of single-cell dose response to drugs. The integration of nanoparticles on the droplet-based microfluidic platform brings out the possibility of dynamic cellular measurements and distinct subpopulations identification of cancer cells.
Fig. 3.
Microfluidic single-cell manipulation including cell isolation using droplet microfluidics (A) and cell lysis (B). (a) Schematics of a passive droplet-based microfluidic platform for single cell isolation, including (i) cell encapsulation, (ii) droplet deceleration, (iii) sorting of single-cell droplets, and (iv) export of single-cell droplets into tubes. Adapted with permission from Ref. [86]. Copyright 2017 Springer Nature Publishing. (B) Experimental setup of mechanical cell lysis using a nanowire integrated microfluidic device including: Injection of the cell solution through the microchannel by using the syringe pumps; Anchoring, stretching and bursting of the cells on the nanowire arrays; Centrifugation and collection of a cell solution. Adapted with permission from Ref. [87]. Copyright 2012 The Royal Society of Chemistry.
To meet the requirement of droplet formation with on-demand or tunable sizes, structures, formation rates, active droplet-based microfluidic devices have also been developed to isolate single cells. Different from passive droplet-based microfluidic devices, assorted external actuation techniques are used herein, including electrical, centrifugal, magnetic, mechanical, optical, thermal controls. For instance, single cell isolation of CTCs was conducted by applying magnetic controls on a microfluidic device [71]. The whole system contained a single cell isolation microfluidic chip (SIM-Chip), a lateral magnetophoretic microseparator, an electric impedance cytometer, and a single-cell microshooter. The PDMS SIM-Chip enabled the isolation of single cells from a heterogeneous cell mixture (whole blood samples) by applying a magnetic field using immunomagnetic nanobeads specifically bound to CTCs. The enriched cells were electrically measured using the impedance cytometer and identified by the size difference from normal blood cells. The CTCs were transferred individually to the microshooters and dispensed in a particular droplet along with the shooting buffer, allowing subsequent downstream analyses. The single-cell isolation throughput was 1.4 cells/s, while the efficiency was 82.4% with the purity of 92.45%. The recovery rate of the SIM-Chip was measured consistently around 99.78%, while the downstream fluorescence-based viability of isolated single cells was over 80%. This active microfluidic system could isolate extremely rare single cells from blood samples with no assist of expensive instruments and was compatible with conventional downstream analyses.
3.1.3. Other techniques
In addition to mechanical traps and droplet traps, there are other techniques to isolate single cells using microfluidic devices. For example, a bioelectronic chip was developed to manipulate cells via DEP due to different dielectric and conductive properties of cells and the cultivation medium [88]. The trapping of single cells could be achieved by using embedded electrodes and switching electrode polarities. The DEP-chip based assay allowed the same-single-cell analysis (SASCA) in the drug accumulation stage in order to study the multidrug resistance (MDR) activity in leukemic blast cells [89–91]. SASCA was reported for the first time by Li et al. to address the issue that positive MDR drug effects could be obscured due to cellular heterogeneity from a small amount of cancer cells [18, 19]. Another technique to isolate single cells was optofluidic microdevices, such as optical tweezers. For instance, a tightly focused laser beam with a specific wavelength was applied to trap individual living cells in a non-invasive manner [92]. By optimizing the parameters of applied infrared lasers, single cells were stably and quantitatively confined in optical traps and used for the studies of cell tolerance to phototoxic stress over a time period of several hours.
3.2. Single cell treatment
For single cell analysis, integration of cell treatment, especially cell lysis or fusion, into the microfluidic platform is of great importance. Single cell lysis is a significant step in the analysis of single cells that breaks the cell membrane to release DNA, proteins, and other components from the cell [93]. Single cell fusion is an important biological process that involves combining two membrane-bound entities into one. It is a critical cellular process that commonly occurs during differentiation, embryogenesis, and morphogenesis [4]. In the subsequent sections, advances in microfluidic single cell lysis and fusion are presented.
3.2.1. Single cell lysis
Single-cell lysis is a vital step in the molecular analysis of single cells. Chemical, electrical, and mechanical single cell lysis are the major microfluidics-based single cell lysis methods.
Owing to the long history of chemical cell lysis in the bulk analysis of cells, chemical cell lysis has become a popular technique in the field of single-cell lysis [2]. Sarkar et al. [94] presented a microfluidic probe that lysed single adherent cells from standard tissue culture and captured the lysate contents to perform single-cell biochemical assays. The probe was able to hydrodynamically confine the lysis buffer from the tip to an area on the scale of a single cell via a balanced surrounding inflow. The buffer was a complex mixture containing 1% commercial Triton X-100 detergent. Abate and coworkers developed a method based on alkaline lysis, which ruptured cell membranes and rapidly denatured endogenous nucleases and inhibitory proteins in a high pH buffer [95]. Several studies reported single lymphocyte capture together with chemical lysis using lithium dodecyl sulfate in microdroplets with a lysis efficiency of 100% [96–98].
Electrical lysis relies on the generation of small transient pores in cell membranes by applying an electric field to release intracellular molecules [4]. Jokilaakso et al. [99] reported a method to position and lyse individual cells on silicon nanowire and nanoribbon biological field effect transistors. In this reported method, HT-29 cancer cells were positioned on top of transistors by manipulating magnetic beads using external magnetic fields. Ultra-rapid cell lysis was subsequently performed by applying 600–900 mVpp at 10 MHz for as little as 2 ms across the transistor channel and the bulk substrate. Jen et al. [100] developed an electroosmotic-driven microfluidic chip with arrays of 30-μm-diameter microwells for single-cell electric lysis. The results of electrical lysis experiments at the single-cell level indicated that the cells were gradually lysed as the direct current (DC) voltage of 30 V was applied; and the cells were fully lysed after 25 s. Recently, de Lange et al. [101] demonstrated robust cell lysis without the use of detergents or other chemicals that combined applying an electric field and using lysozyme for lysis in droplets. In this method, cells were exposed to an electric field immediately before encapsulation in droplets, resulting in cell lysis. This approach was simple to integrate into microfluidic devices and compatible with high-throughput single-cell screening assays. The lysis efficiency was >90%, which was a significant improvement than using lysozyme alone.
Mechanical lysis can directly damage the cells to release the intracellular components through the application of mechanical forces [4]. Kim et al. [87] introduced an interesting microfluidic device integrated with patterned one-dimensional nanostructure arrays for facile and high-throughput mechanical cell lysis. The geometry of the hydrothermally grown ZnO nanowires, characterized by sharp tips and high aspect ratios, aided in anchoring the cell and tearing the plasma membrane, enabling simple and highly efficient extraction of cellular proteins and nucleic acids. The cell solution was flown through the microchannel using a pair of syringe pumps (one for injection at the inlet and another for withdrawal at the outlet) as described in Fig. 3B. Over time, the cells anchoring on the nanowire array was stretched by the shear force of the flow and then got burst. Cell lysis efficiency was enhanced using nanowires integrated in microchannels. Nanowire integration on microchannels resulted in a 41% increase in released protein and a 48% increase in the released nucleic acid. Hoefemann et al. [102] achieved single-cell lysis using a vapor bubble generation technique. Single-cell lysis was performed by bubble generators placed inside a flow channel directly below the cells’ path. Bubble generation pushed the cells towards the channel ceiling or the channel wall and cells were lysed due to the large shear force at an efficiency of 100%. Cycle times for sorting and lysis have experimentally been determined to be 5 ms and 20 ms, respectively. Recently, Cheng et al. [103] demonstrated a pump-on-a-chip microfluidic platform for mechanical cell lysis. A 50 μl cell sample was effectively lysed through on-chip multi-disruption in 36 s without introducing any chemical agent and suffering from clogging by cellular debris. After 30 cycles of circulating disruption, 80.6% and 90.5% cell disruption rates were achieved for HEK293 cell and human natural killer cell samples, respectively.
3.2.2. Single cell fusion
Cell fusion, in which several uninuclear cells merge into multinuclear cells, has been extensively investigated for analyzing gene expression, cancer immunotherapy, cell reprogramming, and tissue generation [4, 104, 105]. Hu et al. [106] designed a high-throughput cell electrofusion microfluidic chip on a silicon insulator wafer and tested for in vitro cell fusion under a low applied voltage. The chip consisted of six individual straight microchannels with a highly conductive doped Si layer (40 μm thickness) as the microchannel wall. An individual cell electrofusion process could complete within 10 min. Kimura et al. [107] presented a novel electrofusion device that enabled massive parallelism, using an electrically insulating sheet having a 2D micro‐orifice array. The sheet was sandwiched by a pair of micro‐chambers with immersed electrodes, and each chamber was filled with the suspensions of two types of cells to be fused. The 2D orifice arrangement at the pitch of 50 μm achieved simultaneous fusion of 6×103 cells by superimposing a pulse (10 V, 50 μs) on a 4 mm diameter chip, with the fusion yields of 78–90% among various sizes and types of cells. Additionally, Yang and coworkers designed a microfluidic device integrated with 3D thin film microelectrode arrays wrapped around serpentine-shaped microchannel walls to test cell electrofusion [108]. The integration of a soft protruding microelectrode array into the microfluidic device allowed for cell attachment onto the microchannel wall when applying an AC electric field, followed by electroporation and fusion of cells under DC electric pulses. The fusion efficiency was about 43.1% of total cells loaded into the device which was higher than the efficiency of the existing microfluidics-based electrofusion devices. The on-chip cell fusion thus provided an important tool for cell treatment at the single-cell level.
4. Applications of microfluidic single cell analysis in cancer biology, diagnosis and therapy
With those aforementioned microfluidic techniques for single-cell manipulation, single-cell analysis on microfluidic devices has found vast applications in cancer research owing to the remarkable capacity for single-cell manipulation, high-degree integration of multiple functions on a single device, simplified operation, high-throughput capacity, low sample and reagent consumption, and so on [109]. Herein, we review recent applications of microfluidic single-cell analysis in cancer research, with a focus on cancer biology, diagnosis, and therapy. We also summarize these different applications in Table 1.
Table 1.
Summarized applications of microfluidic single-cell analysis in cancer biology, diagnosis, and therapy
| App Category | Research Area | Substrates | Cancer Cells | Cell Capture Techniques | Detection Method | Study Focus | Ref |
|---|---|---|---|---|---|---|---|
| Cancer Biology | Cell-cell interaction | PDMS | Human leukemia cells | Droplet trap | Phasor-FLIM | Label-free singl- cell screening and interaction between cancer cells and fibroblasts | [113] |
| PDMS | PC3 cells | Mechanical trap (Electrolytic valve) | LIVE/DEAD fluorescence staining assay | Cell proliferation during coculture | [114] | ||
| Cell metastasis | PDMS | Breast cancer cells (MCF-7, MDA-MB- 231, SUM- 159) | Mechanical trap (Microhook) | Transwell migration assay | Cell migration regulated by overexpression of MCPIP1 | [118] | |
| PDMS | CTCs | Mechanical trap (Herringbone structure) | Three-color immunocytochemistry method | Heterogeneous secretion profile studies of IL-8 and VEGF in cancer patients’ blood samples | [119] | ||
| Cancer Diagnosis | Genetic Analysis | Glass | PD7591, PD483 and PD798 | Mechanical trap | Fluorescence detection | Successful gene sequencing | [123] |
| PDMS | Breast cancer cells (MCF-7 and MDA- MB-231) | Droplet trap | Fluorescence detection | miRNA over expression in cancer cells | [26] | ||
| PDMS | Raji B-lymphocyte cells | Droplet trap | Barcoding Hydrogel Beads | Generating genes sequence-ready libraries | [124] | ||
| PDMS | HT-29 | Droplet trap | Fluorescence detection | Detecting of CNVs and SNVs | [126] | ||
| Protein Analysis | PDMS | HepG2 | Mechanical trap | Fluorescence detection | Quantification of exosomes | [115] | |
| PDMS | HepG2 | Mechanical trap | Fluorescence detection | Diversity in the single-cell protease activity | [133] | ||
| Hydrogel | Breast cancer cells (MCF-7 and MDA-MB-231) | Droplet trap | Fluorescence detection | No cytokines secretions in MCF7, cytokines secretions in MDA-MB-231 | [27] | ||
| PDMS | U-937 | Mechanical trap | Fluorescence detection | Advance sensitivity of soluble proteins detection | [134] | ||
| PDMS | U-937 and HEK 293 | Mechanical trap | Fluorescence detection | Detection of low concentration of the enzyme GAPDH | [135] | ||
| Cancer Therapy | Drug Discovery | PDMS | CTCs | Mechanical trap | Immunofluorescence staining | CTCs expansion approach for drug screening and patient therapeutic monitoring | [144] |
| PDMS | MCF-7 | Droplet trap | LIVE/DEAD fluorescence staining assay | Quantitatively drug screening | [116] | ||
| Immunotherapy | PDMS | NK-92; RPMI-8226 | Droplet trap | fluorescence staining assay | Activity-based screening and molecular immunotherapy | [149] | |
| PDMS | Jurkat E6.1; K562; TCR T | Droplet trap | Fluorescence; PCR and sequencing | TCR T cell screening with TCR tracking for immunotherapy | [152] |
CNV: copy number variation; CTC: circulating tumor cell; FLIM: fluorescence lifetime imaging microscopy; GAPDH: glyceraldehyde 3-phosphate dehydrogenase; IL-8: interleukin-8; MCPIP1: monocyte chemotactic protein induced protein 1; PC: prostate cancer; PCR: polymerase chain reaction; PDMS: polydimethylsiloxane; SNV: single nucleotide variation; TCR: T cell receptor; VEGF: vascular endothelial growth factor.
4.1. Cancer biology
Cancer biology includes an extensive range of fundamental knowledge to understand what cancer is, how cancer develops, and which possible strategies to treat cancer. Research on the biology of cancer is undoubtedly critical to disease prevention and cure by studying basic processes, such as cell growth, cell metastasis, cell-cell interaction, cell division, cell metabolism, and cell apoptosis [110].
Microfluidic single-cell analysis has attracted sustained interest in cancer biology studies, as it provides precise control of microenvironments and detailed information. Current single cell analysis about cancer biology focuses on primordial mechanisms covering a broad range of scenarios, such as the biological identification of cancer cells from normal cells, the signaling pathways of cancer development and progress, while studying the interactions within cancer cells or with the host microenvironment and the metastasis of cancer cells has attracted tremendous attention [111]. This work lays the foundation of cancer prevention, diagnosis, and therapy. For instance, CTCs have played a vital role in cancer biology research, especially in the metastatic spread of carcinoma. Recent researches have confirmed a high level of heterogeneity in the single cell analysis of CTCs [35], which contributes to cancer progression and metastasis. Single cell analysis of CTCs could also provide a patient’s disease status by revealing metastasis and real-time information of cancer cells. However, one challenge remains in the analysis of CTCs due to natural paucity. To address this issue, characterization, separation, and enrichment of CTCs using microfluidic devices have become one of the hot research areas in the field of microfluidics [35].
4.1.1. Cell-Cell interaction
Interactions between cells are constitutive factors while studying the physiological properties and functions of organs and tissues, the signaling pathways, and the development and migration profiles of cancer. One imperative facet of cancer biology research is to understand cell-to-cell interactions between tumor cells and target cells, like endothelial and immune cells. On one aspect, cell-cell interactions can be investigated via cell motility and cell proliferation studies. On another aspect, cells can communicate directly by releasing and sending signaling molecules, which commonly diffuse to target cells. Hence, studying the mechanisms at the single cell level is necessary to understand the development and progression of cancer. Several microfluidic devices have been reported to spatially and temporally control, identify, pair single cells, as well as investigate signaling molecules released from single cells [112].
Ma et al. [113] reported a passive PDMS-based microfluidic device for single cancer cell dormancy studies. In this study, the droplet-based microfluidic technology was used for single cell isolation and combined with the phasor approach for fluorescence lifetime imaging microscopy (phasor-FLIM). Single cells were encapsulated in droplets and characterized by FLIM on the device which was integrated with a unique droplet collection chamber to prevent the droplet motion. Using the device, two kinds of human leukemia cells K562 erythromyeloid and Jurkat T-cell leukemia were first encapsulated one-to-one in the droplets and then distinguished by using phasor-FLIM. Furthermore, a label-free single-cell screening test was performed using human foreskin fibroblast model. The results showed that human foreskin fibroblasts treated with different types of serum starvation could be clearly distinguished from the normal condition treated-cells, mainly due to different fractions of free nicotinamide adenine dinucleotide (NAD(P)H) and bound NAD(P)H in two types of cells. A significant increase of free NAD(P)H was quantified with serum-free treatment, leading to the induction of cells in a quiescent state, while a significant shift of serum-starvation groups was observed from a high bound NAD(P)H fraction to a lower bound fraction. Moreover, it was suggested that a higher lactate production would occur as introduced by serum-starvation and create an acidic microenvironment for gene expressions regulation, which provided vital information in tumor progression and wound healing promotion. In addition to studying heterogeneity and metabolic differences of single cells, this proposed method demonstrated a feasible strategy for further understanding of basic mechanisms in the communication and interaction between tumor cells and fibroblasts.
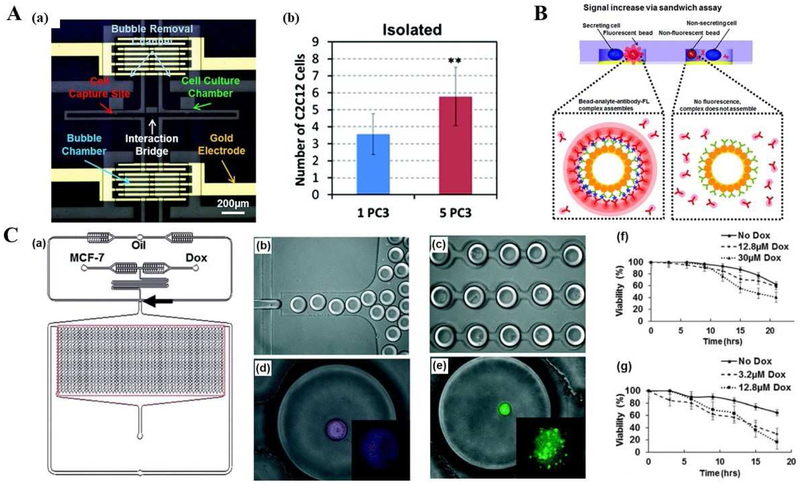
Cancer-stromal interaction was studied by using a PDMS-based microfluidic platform, in which an electrolytic cell isolation design was applied to control the co-control microenvironment [114]. The platform was composed of PDMS layers and a gold electrodes-patterned substrate, forming cell culture chambers, bubble chambers, a shallow interaction bridge (10 μm in width), microchannels (40 μm in depth), and capture sites, as shown in Fig. 4A(a). Each cell culture chamber was sandwiched between two bubble chambers, where a bubble was generated when applying a potential for electrolysis. The isolated cells captured on the chip could interact via the interaction bridge by diffusion of secreted factors, cytokines. Besides, the cell isolation can be controlled by simply applying negative pressure to the channels to remove bubbles. The capture rate of single cells was over 90%, and the pairing ratios of two types of cells could be controlled, varying from 1:1 to 5:1. The on-chip cell-cell interaction assay was conducted between prostate cancer (PC3) and myoblast (C2C12) cells. The results in Fig. 4A(b) indicated that with electrolytic isolation, the proliferation rate was enhanced, when C2C12 cells were co-cultured with PC3 cells at a high pairing ratio, which was mainly due to the accumulation of secreted factors in the isolated chambers. However, the pairing ratio would not affect the proliferation rates without the isolated chamber.
Fig. 4.
Single cell analysis applications using microfluidic platforms in cancer biology, diagnosis, and therapy. (A) Cell-cell interaction assay using a microfluidic platform. (a) Microphotograph of the fabricated device including culture chambers, an interaction bridge, bubble chambers, bubble removal channels, and a gold electrode for electrolysis. (b) Proliferation rates of C2C12 cells co-cultured with one PC3 and five PC3 cells with bubble isolation. Adapted with permission from Ref. [114]. Copyright 2014 Royal Society of Chemistry. (B) Principle of the reconfigurable microfluidic device for real-time monitoring of secreted proteins at the single-cell level. Each individual compartment consists of single cells and sensing beads surrounded by PE-labelled detection antibodies as reporter molecules. Once released by cells, target molecules diffused inside the chamber, bound to sensing bead, causing fluorescence to increase on the bead surface via a sandwich assay. Adapted with permission from Ref. [115]. Copyright 2016 The Royal Society of Chemistry. (C) Droplet array design, single cell encapsulation images and Dox cell viability tests with different MCF-7 cell lines. (a) Schematics of integrated microfluidic platform and droplet generation array. (b) Droplet generation. (c) Droplet docking in a microarray. (d) Image of live MCF-7S cell in a droplet after incubation with Cy-5 conjugated ABCB-1 mRNA. (e) Image of live Dox-resistant MCF-7R cell encapsulated in a droplet, Calcein AM was hydrolyzed after entering live cells. (f) Cumulative cell viability of MCF7 Dox-resistant (MCF-7R) cells (f) and MCF7 Dox-sensitive (MCF-7S) cells (g) treated with two concentrations of Dox. Adapted with permission from Ref. [116]. Copyright 2015 The Royal Society of Chemistry.
4.1.2. Cancer metastasis
Cancer metastasis is a complex process where cancer cells break away from the original sites of primary cancer, travel through the blood or lymph system, and spread to different parts of the body, generally forming new tumors. Metastasis is the main cause of the mortality of cancer, which primarily relies on cell motility. Microfluidic single-cell analysis can lead to key mechanisms and features of metastasis by directly investigating collective cell migration [117]. Cellular dynamics of the metastatic process can be inspected for a better understanding of cancer biology.
A PDMS single cell mobility analysis platform (SCM-Chip) was designed to study cancer metastasis by maintaining an independent microenvironment for single cells [118]. The SCM-Chip was applied to capture breast cancer cells (MCF-7, MDA-MB-231, and SUM-159) at the level of single cells, and the cell capture rates were tested to be over 80%. The migration of captured single cells was monitored in real time by analyzing the migration distance. The results showed that metastatic MDA-MB-231 and SUM-159 cells exhibited stronger migratory than nonmetastatic MCF-7 cells, which was consistent with the wound-healing assay and the transwell migration assay. Additionally, the post-translational mechanisms on tumor metastasis were studied and it was found that the overexpression of monocyte chemotactic protein induced protein 1 (MCPIP1) was associated with the decrease of cell mobility. Moreover, the inhibition of the transforming growth factor-β (TGF-β) pathway restored the levels of low-migratory cells back to comparable levels of cells with high MCPIP1 expression. This SCM-Chip provided a general platform to isolate single cancer cells and characterize cells with different migratory profiles, which could be adopted for the mechanistic study of key regulators for cancer metastasis and drug development.
Metastatic propensities of cancer cells have been also investigated by assessing functional secreted proteins at the single-cell level. One effort was made to identify the profiling of the functional proteins from whole blood at a single CTC resolution [119]. Specifically, a PDMS-based microvotex-generating herringbone chip was designed to capture CTCs via photocleavable single-stranded DNA-encoded antibody conjugates. Under UV irradiation, captured CTCs were then released and proceeded with a two-step purification process to remove red blood cells and white blood cells. CTCs with high purity were transported to a microfluidic chip coated with an enhanced poly-l-lysine barcode pattern, where single CTCs were recaptured into each microchamber. By using the single-cell barcode chip (SCBC), quantitative measurement of secreted proteins from individual CTCs was then achieved. The result has shown a highly heterogeneous secretion profile of two proteins, interleukin-8 (IL-8) and vascular endothelial growth factor (VEGF), which were present in the cancer patients’ blood samples.
4.2. Cancer diagnosis
Due to cellular heterogeneity, it is important to study individual cells to understand the complex biology of the heterogeneous population [120]. These minute differences in cellular activities at the single-cell level could be essential to the development of cancer diagnostic methods and CTCs are one of such examples [31]. Eventually, single-cell-based diagnostics methods can be highly beneficial and essential to precision medicine and personalized medicine.
Attributed to the remarkable capability of microfluidics, microfluidic single-cell analysis has found significant applications in cancer diagnosis [4, 121]. Herein, we summarize those recent applications into two categories based on the types of diagnostic assays, namely single-cell gene and protein analysis.
4.2.1. Single-cell genetic analysis
Many cancers originate from gene mutation of a small number of cells. Detection and analysis of DNA or RNA are fundamentally crucial for cancer diagnosis. Because the analysis of genes at the single-cell level allows understanding the genotypic characteristics of each molecule, it is particularly important in identifying abnormal genes. Microfluidic techniques based on single-cell genetic analysis have been widely used for cancer diagnosis [122]. Bhagwat et al. [123] developed a fully integrated glass-based microfluidic platform for flow cytometry-based isolation of CTCs and clusters from blood for whole transcriptome analysis or targeted RNA transcript quantification. A pre-enrichment platform was connected in-line with a BD Influx™ cell sorter. This developed platform utilized in-line magnetic particle-based leukocyte depletion and acoustic cell focusing and washing to achieve >98% reduction of blood cells and non-cellular debris, along with >1.5 log-fold enrichment of spiked tumor cells. Whole blood was labeled with antibodies against CTC markers as well as magnetic microparticles that bind unwanted blood cells. The sample then passed through a magnetic depletion step that removed unwanted blood cells. It was followed by an in-line acoustic focusing and washing step, which removed debris and concentrated the sample prior to cell sorting. 63 single CTCs from a genetically engineered pancreatic cancer mouse model (n = 12 mice) were isolated and transcriptionally characterized. PD798 cells were spiked into healthy mouse blood and individual cells were sorted to generate libraries from 11/13 single cells (85% success rate).
Because there are only picogram quantities of genes within a single cell, due to sensitivity limits, it is quite challenging to sequence and detect single cell genomes directly with high sensitivity without amplification methods. Thus, to obtain a sufficient quantity of material for sequencing and detection of genes in single cells, microfluidic single cell amplification is recently presented. Guo et al. [26] developed a rapid, PCR-free, single-cell miRNA assay on a continuous-flow PDMS-based microfluidic platform fabricated with soft lithography. A DNA hybridization chain reaction was employed in this assay to amplify the target miRNA signal. With a non-enzymatic hybridization chain reaction (HCR) of specifically designed hairpin DNA sequences, the novel enzyme-free isothermal amplification of the target miRNA was spontaneously triggered. It amplified the fluorescence signals within 20 minutes in droplets after single-cell lysis, enabling high-specificity measurements. A customized photomultiplier (PMT) detector allowed the continuous-flow droplet screening of single-cell miRNA with a throughput of 300–500 cells per minute for profiling of large-scale physiological samples. Three different breast cell samples, MCF-7 (cancer), MDA-MB-231 (cancer) and MCF-10A (healthy), were screened by using this system. Cancer cells (MCF-7 and MDA-MB-231) showing high miRNA expression levels were clearly distinguished from healthy cells (MCF-10A). Moreover, the aggressive cancer cells (MDA-MB-231) showed a higher miRNA expression level than the less aggressive cancer cells (MCF-7) in the statistical analysis. Pellegrino et al. [124] developed a PDMS based microfluidic device using a PCR method and barcoded amplified genomic DNA from thousands of individual cancer cells confined to droplets. This technique enabled the characterization of genetic heterogeneity in tumor cell populations. By using this approach, longitudinally collected acute myeloid leukemia (AML) tumor populations were sequenced from two patients and genotyped up to 62 disease relevant loci across more than 16,000 individual cells. Targeted single-cell sequencing was able to sensitively identify cells harboring pathogenic mutations during complete remission and uncover complex clonal evolution within AML tumors that were not observable with bulk sequencing. The developed microfluidic platform could generate sequence-ready libraries in less than 2 days through the use of picoliter volume droplets that consumes minimal reagent to barcode genomic DNA. These key features significantly lowered the barriers for performing single-cell DNA sequencing and had the potential for high-resolution analysis of clonal architectures within tumors.
Multiple displacement amplification (MDA) is another DNA amplification method that relies on a combination of random sequence primers and the strand-displacement properties of the Phi29 polymerase to isothermally amplify DNA [125]. MDA has been used for single cell whole-genome amplification (WGA). Fu et al. [126] reported a PDMS based microfluidic device for emulsion WGA (eWGA) to overcome the fluctuation of amplification yield, and avoid false-positive errors for single nucleotide variations (SNVs) identification. The proof-of-principle of eWGA was demonstrated by sequencing nine single HT-29 cancer cells expanded from a single clone with MDA. This easy-to-operate approach enabled simultaneous detection of copy number variations (CNVs) and SNVs in an individual human cell, exhibiting significantly improved amplification evenness and accuracy. CNVs at 250-kb size with 50-kb resolution and SNVs with error rate <2 × 10−5 were achieved.
4.2.2. Single-cell protein analysis
Heterogeneity of cancer is also characterized at the protein level. A number of proteins are important disease biomarkers such as prostate-specific antigen (PSA) for prostate cancer [127–130] and Epithelial Cell Adhesion Molecule (EpCAM), a specific cancer cell surface protein expressed in CTCs [131]. However, the low abundance of target cells in a biological sample and/or low copy numbers of protein markers and the inherent complexity of the proteome make protein analysis at the single-cell level a challenge for conventional assays [132]. Microfluidic methods with features of reduced sample consumption and increased sample throughput are well suited for measuring low levels of proteins as cancer biomarkers [1, 4].
Real-time monitoring of secreted proteins from an individual cell using microfluidic devices is one of the most popular research areas in the field. Son et al. [115] described a reconfigurable PDMS based microfluidic device for confining single cells along with antibody-modified sensing beads inside 20 picoliter (pL) microcompartments for monitoring cellular secretory activities. The principle of operation of the device is shown in Fig. 4B. An array of ~7000 microchambers fabricated in the roof of the reconfigurable microfluidic device could be raised or lowered by applying negative pressure. The floor of the device was micropatterned to contain cell attachment sites in the microcompartments. Using this set-up, the detection of inflammatory cytokine IFN-γ and exosomes from single immune cells and cancer cells were demonstrated, respectively. The detection scheme was similar in both cases: cells were first captured on the surface inside the microfluidic device, then sensing microbeads were introduced into the device so that, once the microcompartments were lowered, single cells and microbeads became confined together. A solution containing fluorescently-labeled secondary antibodies (Abs) bathed the beads and the cells inside the compartments. The cell-secreted molecules onto microbeads were captured, followed by binding with secondary antibodies, which caused microbeads to become fluorescent. The fluorescence intensity of the microbeads changed over time, providing dynamics of single cell secretory activities. The average quantity of exosomes secreted by single HepG2 cells was estimated over the course of 12 h using a calibration curve. The log–log model was applied to the calibration curve (R2 = 0.995). This quantity of exosomes was based on the protein content and was estimated to be 0.92 ± 0.29 fg. Wu et al. [133] developed a PDMS microfluidic platform to simultaneously monitor protease activity of many single cells in a time-dependent manner. This platform isolated individual microwells rapidly on demand and thus allowed single-cell activity measurement of both cell-surface and secreted proteases by confining individual cells with diffusive Forster resonance energy transfer (FRET)-based substrates. With this platform, it was observed that dose-dependent heterogeneous protease activation of HepG2 cells treated with phorbol 12-myristate 13-acetate (PMA). To study the temporal behavior of PMA-induced protease response, the pericellular protease activity of the same single cells was monitored during three different time periods and revealed the diversity in the dynamic patterns of single-cell protease activity profile upon PMA stimulation.
Microfluidic immunoassays, particularly enzyme-linked immunosorbent assays (ELISA), have been developed to enhance the detection sensitivity for the measurement of low-abundance proteins from single cells, because cancer biomarkers often exist in biological fluids at concentrations in the range of 10−12 −10−16 M [1]. Chen and coworkers developed an integrated platform consisting of smart hydrogel immunosensors for the sensitive detection of protein secretions [27]. A single cell and smart hydrogel microparticles were encapsulated within a droplet. After incubation, target secreted proteins from the cell were captured in the smart hydrogel particle for immunoassays. The temperature-induced volume phase transition of the hydrogel biosensor allowed the concentration of analytes within the gel matrix to increase, enabling high-sensitivity measurements. Distinct heterogeneity in low-abundance essential secretions from live cells was determined from 6000 cells within 1 h, including interleukin-6 (IL-6), IL-8, and monocyte chemoattractant protein-1 (MCP-1) secretions of both suspended cells (HL60) and adherent cells (MCF7 and MDA-MB-231). HL60 cells were observed to secrete predominantly IL-8 and MCP-1, and MDA-MB-231 cells secreted all three cytokines, whereas MCF7 showed almost no secretions. The in-droplet conditioning of the cells with 50 ng mL−1 soluble cytokine tumor necrosis factor alpha (TNFα), a well-known cytokine linked with tumor progression, elevated the single cellular secretion of these three cytokines, except for MCF7 cells which still maintained their low secretion activities. This platform was very flexible and could be used to simultaneously measure a wide range of clinically relevant cellular secretions. Herrera et al. [134] used quantum dots (QDs) in PDMS microwell arrays for a sandwich immunoassay to detect secretion of the soluble cytokine TNF-α from single cells. The detection sensitivity of QD sandwich immunoassays was advanced for the measurement of soluble proteins using single QD imaging and amplification of binding of QDs to each captured TNF-α molecule using bioorthogonal chemical reaction, reaching a lower threshold of 60 aM. The detection format used a simple sandwich immunoassay and a standard fluorescence microscope, and thus came with additional benefits such as assay speed, simplicity, large dynamic range, and the spatial resolution essential for single cell secretion studies. The QD-based detection method increased the number of single cells that could be interrogated for TNF-α secretion by 3 folds in comparison to an organic fluorophore, which was achieved by extending detection range down to nearly one molecule captured per microwell. In addition, Eyer et al. [135] combined a microfluidic device with the analytical strength of ELISA for single-cell studies to reliably identify intracellular proteins, secondary messengers, and metabolites. The microfluidic device allowed parallel single-cell trapping and isolation in 60 of 625-pL microchambers, repeated treatment and washing steps, and subsequent lysis and analysis by ELISA. Using a sandwich ELISA, the concentration of the enzyme GAPDH in the range of 1–4 attomole per cell was quantitatively determined in single U937 cells and HEK 293 cells. Furthermore, a competitive ELISA was performed to determine the concentration of the secondary messenger, cyclic adenosine monophosphate (cAMP), in murine leydig tumor (MLT) cells, in response to the hormone, lutropin. The half maximal effective concentration (EC50) of lutropin was determined to have an average value of 2.51 ± 0.44 ng/mL.
4.3. Cancer therapy
Although there are many types of cancer therapy such as surgery, radiation therapy, chemotherapy and so on, anticancer drug-related chemotherapy and targeted therapy play an important role in cancer treatment [136, 137]. Our current drug discovery mainly relies on bulk experiments using traditional drug screening approaches such as cell proliferation and cytotoxicity assays. While these approaches have been discovered for understanding treatment efficacy, other information such as normal tissue toxicity, molecular distributions, and drug-cell interactions, might be hidden at the level of single cell [1]. In the past few decades, microfluidic single-cell analysis has been widely used in a number of cancer therapeutic applications, such as anticancer drug screening, cytotoxic effects and cancer immune-targeted therapy [138–140].
4.3.1. Anticancer drug discovery & testing
It is known that in vitro drug research contributes to the evaluation of anticancer efficiency, drug resistance, and malignant potential in clinical practice. Cell-based assays develop fundamental practices in the drug screening process in early stages. The investigation of cellular and molecular events is of special importance not only in providing pharmaceutical information, but also in economizing workforce and costs for further clinical trials [141, 142]. The rapid development of microfluidic single-cell analysis technologies, in combination with the basic in vitro quantifiable assays, showed prominent feasibility for drug discovery with high throughput and sensitivity [138].
The development of personalized cancer therapy depends on a robust system to monitor the patient’s individual response from anticancer treatment. Anticancer drug efficacy has been tested on CTCs derived from patient blood samples. Current attempts to culture these primary cancer cells using a traditional approach require long-term maintenance under growth factor supplements into cell lines, which usually takes more than 6 months and results in a low CTC expansion efficiency below 20% [137, 143]. Recently, Khoo et al. [144] developed a microfluidics-based culture approach with 8000 microwells for high-throughput testing. This microfluidic approach required minimal preprocessing (~30 min) but not prior enrichment of CTCs or dependence on the use of growth factor supplements. The microfluidic device was fabricated via a standard SU8-based PDMS-replica-molding approach. After cultures of clinical cancer patient samples, tight and well-defined clusters containing CTCs were formed and clearly distinguished from cultures of healthy blood samples. Immunostaining of these clusters showed the presence of CD45–/Hoechst+ cells, which were indicative of CTCs. To test the performance of the assay, the method was characterized with commercially available breast cancer cell lines, from less aggressive (MCF-7) to more metastatic breast cancer subtypes (MDA-MB-231). Drug screening was carried out with a model anticancer drug Doxorubicin (Dox). They improved the throughput of drug screening by incorporating a tree-like gradient generator so that they could apply several drugs at various concentrations to cells at the same time. Among exposure to various concentrations of Dox, cell viability showed a sharp decrease to 30% at the concentration of 0.1 μM Dox. Similarly, quite a few previous studies demonstrated cytotoxic effects with various concentrations of different anticancer drugs through microfluidic single cell analysis [19, 47, 145, 146].
Cancer chemotherapy has been impeded by multidrug resistance developed from cancer patients, resulting in the intracellular drug concentration to be lower than the drug’s cytotoxic threshold within cancer cells and leading to the failure of many forms of chemotherapy [19]. Microfluidic same-single-cell analysis approaches have been developed to study MDR modulation by investigating drug uptake and efflux [18, 19]. Recently, microfluidic single-cell devices increased the throughput of drug testing by using droplet generators. Sarkar et al.[116] reported a droplet microfluidics approach to assess the dynamics of drug uptake, efflux, and cytotoxicity in drug-sensitive and drug-resistant breast cancer cells with Dox. An integrated droplet generation and docking microarray were generated by a PDMS-based microfluidic chip. The chip design and process to encapsulate MCF-7 single cells in droplet arrays were shown in Fig. 4C. Generated droplets were docked in a 1000-traping site microarray for high throughput functional assessment. Drug-sensitive cells (MCF-7S) showed more deaths in the presence of Dox compared to the drug-resistant cells (MCF-7R). MCF-7S cells showed greater viability in the absence of Dox compared to two doses of Dox treatment, as observed in bulk cell experiments (64% vs. 16–30%). Treatment of MCF-7R with 12.8 μM Dox showed no significant viability changes, when compared to untreated MCF-7R cells. However, the viability of drug-resistant cells was four folds higher than that of the drug-sensitive cell line MCF-7S. Moreover, MCF-7Sand MCF-7R exhibited different cell apoptosis processes in droplets. MCF-7S cells presented membrane blebbing and rupture after sustained Dox treatment, while MCF-7R cells appeared pronounced morphological deformation, with standard apoptosis hallmarks (such as condensed nuclear matter, cell shrinkage, extensive blebbing and cytoplasmic fragmentation).
4.3.2. Cancer immunotherapy
Cancer immunotherapy, also called biologic therapy, is a type of cancer treatment that generates enhancement of the body’s natural defenses for cancer fighting [147]. Cancer immunotherapies can be antibodies involved, vaccines involved, and T cell infusions [148]. Immune-targeted therapies that activate effector lymphocytes such as Natural Killer (NK) cells are currently being investigated for the treatment of Multiple myeloma (MM), the second most common form of hematological cancer. However, individual NK cells are highly heterogeneous in their cytolytic potential, leading difficulties in detection, quantification and correlation of the outcome of dynamic effector-target cell interactions at the single-cell resolution which can provide key information about cell-cell interactions such as contacting duration. With microfluidic devices, immune-interactions can be captured and analyzed at the single-cell level. Sarkar et al. [149] presented a PDMS-based microfluidic single-cell bioassay device applicable to activity-based screening and molecular immunotherapies, and meanwhile, identifying functional signatures from NK-MM cell interactions. The injection of an immunomodulatory drug lenalidomide triggered responses of NK-susceptible MM cells, while no response for NK-tolerant MM cells. Antitumor cytotoxicity against RPMI-8226 cells of the NK-mediated cellular increased along with the block of programmed cell death 1/programmed death ligand 1 (PD1/PDL1) axis as well as clinically relevant cell line NK92. Compared with primary NK cells, NK-mediated cells (NK92) showed shorter contact duration to form single contacts with RPMI-8226 (137 min to 46 min, respectively) and required less time to kill targets, with four folds higher number of NK92 cells (i.e. higher killing efficiency) which remained being conjugated with MM cells at the time of death (NK92: 31 ± 2%, primary NK: 7 ± 6%).
T cell receptor (TCR) T cell therapy has great potential for cancer immunotherapy with encouraging clinical results [148, 150, 151]. However, finding the right TCR T cell clone is a tedious, time-consuming, and costly process. Therefore, there is a pressing need for single-cell technologies to conduct fast and multiplexed functional analyses followed by recovery of the clone of interest. Segaliny et al. [152] reported a droplet microfluidics device for single TCR T cell analysis, with functional screening and real-time monitoring of activation of TCR T cell upon recognition of target tumor cells. Meanwhile, with validation from downstream single-cell reverse-transcription PCR and sequencing of TCR chains, the microfluidic platform could track each clone after a sorting procedure. As a result, NY-ESO-1 TCR T cells were specifically selected based on their high activation kinetics, using the Enhanced green fluorescent protein (EGFP) reporter expressed in the downstream TCR signaling pathway, which showed a feasible way to facilitate immunotherapeutic screening and development of T cell therapies.
5. Conclusions & Outlook
Because of many advantages of the microfluidic lab-on-chip technology over conventional methods, such as high throughput, miniaturization, and integration of multiple cellular-assay steps on a multi-functional single device, numerous microfluidic devices have been developed over the past decade for single-cell analysis in cancer research including cancer biology, diagnostics, and therapeutics. In this review, we present recent advances in microfluidic single-cell analysis for cancer biology, diagnosis, and therapy. Microfluidic single-cell manipulation techniques such as single-cell capture, lysis, and cell fusion lay a solid foundation for integrated and high-throughput single-cell analysis in those cancer-related applications. The microfluidic technologies will shape the future direction of single cancer cell research by providing an integrated, versatile and efficient analytical platform. Microfluidic single-cell analysis is becoming a powerful tool for cancer research, especially for cancer mechanistic studies and personalized diagnostics and medicine in the near future.
Despite the exciting progress in microfluidic single-cell analysis, there are still some limitations for its application in cancer research. First, the amount of analytes in a single cell is very limited, which requires ultra-high detection sensitivity, a challenge for many detection methods and instruments. Therefore, the combination of signal-amplification techniques (e.g. PCR, HCR and WGA) with microfluidics and the development of new ultra-sensitive phototransducers (e.g. Electron Multiplying Charge Coupled Device (EMCCD)) will significantly expand single-cell analysis applications. In the meantime, with the development of new ultra-sensitive detection techniques and phototransducers, single-cell analysis may gradually tend towards subcellular analysis. Second, current microfluidic platforms for single cell analysis focus on detection of a very limited number of contents such as DNA or RNA or proteins. However, to explore unknown cancer biology, it will require the combination of multiple characterizations and detection methods for post-capture analysis of multiple analytes simultaneously in a systematic manner, such as the combination of amplification techniques, CE separation, mass spectrometry (MS), fluorescence spectroscopy, and so on. Another limitation is that most work in microfluidic single-cell analysis just demonstrates proof of concept. There are several examples for clinical applications of microfluidic assays for single-cell analysis [119, 124, 144]. For instance, Pellegrino et al. [124] applied a microfluidic single-cell sequencing technology relying on cell-identifying molecular barcodes to detect acute AML tumor cells from patients’ bone marrow aspirate samples. 74.7% accuracy in mapping of the 23 most commonly mutated genes associated with AML progression was achieved. Despite these progresses in clinical applications of microfluidic assays for single-cell analysis, clinical validation of microfluidic single-cell studies and their correlations with real cancer biology may be one of the major focuses for the microfluidic single-cell analysis in the next or the next few decades.
Reviewed recent advances microfluidic single cell analysis in cancer research.
Introduced various microfluidic platforms for single cancer cell analysis.
Summarized different microfluidic techniques for single cancer cell manipulation.
Highlighted applications of microfluidics in cancer biology, diagnosis and therapy.
Discussed current limitation and future prospects.
Acknowledgment
The Li group would like to acknowledge the financial support from the National Institute of Allergy and Infectious Disease of the NIH (R21AI107415), the U.S. NSF-PREM program (DMR 1827745), the Philadelphia Foundation, and the Medical Center of the Americas Foundation. Financial support from the National Institute of General Medical Sciences of the NIH (SC2GM105584), the NIH RCMI Pilot grant, the University of Texas at El Paso for the IDR Program, and University of Texas (UT) System for the STARS award is also greatly acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interest
All the authors declare no conflict of interest.
References
- [1].Kang D-K, Ali MM, Zhang K, Pone EJ, Zhao W, TrAC Trends in Analytical Chemistry 58 (2014) 145. [Google Scholar]
- [2].Huang Q, Mao S, Khan M, Lin J-M, Analyst 144 (2019) 808. [DOI] [PubMed] [Google Scholar]
- [3].Altschuler SJ, Wu LF, Cell 141 (2010) 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lin L, Chen Q, Sun J, TrAC Trends in Analytical Chemistry 99 (2018) 66. [Google Scholar]
- [5].Armbrecht L, Dittrich PS, Analytical chemistry 89 (2016) 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Dou MW, Sanjay ST, Benhabib M, Xu F, Li XJ, Talanta 145 (2015) 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Dou M, Dominguez DC, Li X, Sanchez J, Scott G, Analytical chemistry 86 (2014) 7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Sanjay ST, Dou M, Sun J, Li X, Scientific reports 6 (2016) 30474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Dou M, Sanjay ST, Dominguez DC, Liu P, Xu F, Li X, Biosensors and Bioelectronics 87 (2017) 865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Liu P, Li X, Greenspoon SA, Scherer JR, Mathies RA, Lab on a Chip 11 (2011) 1041. [DOI] [PubMed] [Google Scholar]
- [11].Li X, Valadez AV, Zuo P, Nie Z, Bioanalysis 4 (2012) 1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zuo P, Li X, Dominguez DC, Ye BC, Lab Chip 13 (2013) 3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Dou M, Macias N, Shen F, Bard JD, Domínguez DC, Li X, EClinicalMedicine (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Dou M, Sanchez J, Tavakoli H, Gonzalez JE, Sun J, Bard JD, Li X, Analytica Chimica Acta (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Li XJ, Zhou Y, Microfluidic devices for biomedical applications, Elsevier, 2013. [Google Scholar]
- [16].Li X, Nie Z, Cheng C, Goodale A, Whitesides G, Proc. Micro Total Analysis Systems. 2010, pp. 1487. [Google Scholar]
- [17].Vaithiyanathan M, Bajgiran KR, Darapaneni P, Safa N, Dorman JA, Melvin AT, Analytical and Bioanalytical Chemistry 411 (2019) 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Li X, Chen Y, Li PC, Lab on a Chip 11 (2011) 1378. [DOI] [PubMed] [Google Scholar]
- [19].Li X, Ling V, Li PC, Analytical chemistry 80 (2008) 4095. [DOI] [PubMed] [Google Scholar]
- [20].Welkenhuysen N, Borgqvist J, Backman M, Bendrioua L, Goksör M, Adiels CB, Cvijovic M, Hohmann S, BMC systems biology 11 (2017) 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ionescu A, Zahavi EE, Gradus T, Ben-Yaakov K, Perlson E, European journal of cell biology 95 (2016) 69. [DOI] [PubMed] [Google Scholar]
- [22].Hochstetter A, Stellamanns E, Deshpande S, Uppaluri S, Engstler M, Pfohl T, Lab on a Chip 15 (2015) 1961. [DOI] [PubMed] [Google Scholar]
- [23].van den Hurk M, Erwin JA, Yeo GW, Gage FH, Bardy C, Frontiers in Molecular Neuroscience 11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Li YW, Chen DJ, Zhang YF, Liu C, Chen P, Wang Y, Feng XJ, Du W, Liu BF, Sensors and Actuators B-Chemical 225 (2016) 563. [Google Scholar]
- [25].Lin L, Lin X, Lin L, Feng Q, Kitamori T, Lin J-M, Sun J, Analytical chemistry 89 (2017) 10037. [DOI] [PubMed] [Google Scholar]
- [26].Guo S, Lin WN, Hu Y, Sun G, Phan D-T, Chen C-H, Lab on a Chip 18 (2018) 1914. [DOI] [PubMed] [Google Scholar]
- [27].Hsu MN, Wei SC, Guo S, Phan DT, Zhang Y, Chen CH, Small 14 (2018) 1802918. [DOI] [PubMed] [Google Scholar]
- [28].Kuboki Y, Fischer CG, Beleva Guthrie V, Huang W, Yu J, Chianchiano P, Hosoda W, Zhang H, Zheng L, Shao X, The Journal of pathology 247 (2019) 347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Sun D, Cao F, Cong L, Xu W, Chen Q, Shi W, Xu S, Lab on a Chip 19 (2019) 335. [DOI] [PubMed] [Google Scholar]
- [30].Sarkar S, Sabhachandani P, Stroopinsky D, Palmer K, Cohen N, Rosenblatt J, Avigan D, Konry T, Biomicrofluidics 10 (2016) 054115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Yeo T, Tan SJ, Lim CL, Lau DPX, Chua YW, Krisna SS, Iyer G, San Tan G, Lim TKH, Tan DS, Scientific reports 6 (2016) 22076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Antfolk M, Kim SH, Koizumi S, Fujii T, Laurell T, Scientific reports 7 (2017) 46507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Li X, Xue X, Li PC, Integrative Biology 1 (2009) 90. [DOI] [PubMed] [Google Scholar]
- [34].Khamenehfar A, CH Li P, Current pharmaceutical biotechnology 17 (2016) 810. [DOI] [PubMed] [Google Scholar]
- [35].Abouleila Y, Onidani K, Ali A, Shoji H, Kawai T, Lim CT, Kumar V, Okaya S, Kato K, Hiyama E, Yanagida T, Masujima T, Shimizu Y, Honda K, Cancer Sci (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kapanidis AN, Strick T, Trends in biochemical sciences 34 (2009) 234. [DOI] [PubMed] [Google Scholar]
- [37].Dou MW, Garcia JM, Zhan SH, Li XJ, Chemical Communications 52 (2016) 3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Dou MW, Dominguez DC, Li XJ, Sanchez J, Scott G, Analytical Chemistry 86 (2014) 7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Dietzel A, Microsystems for pharmatechnology, Springer, 2016. [Google Scholar]
- [40].MichelleáGrandin H, Lab on a Chip 7 (2007) 1074.17653351 [Google Scholar]
- [41].Li LL, Li YQ, Shao ZX, Luo GA, Ding MY, Liang QL, Analytical Chemistry 90 (2018) 11899. [DOI] [PubMed] [Google Scholar]
- [42].Marasso SL, Puliafito A, Mombello D, Benetto S, Primo L, Bussolino F, Pirri CF, Cocuzza M, Microfluidics and Nanofluidics 21 (2017). [Google Scholar]
- [43].Baker CA, Bulloch R, Roper MG, Analytical and bioanalytical chemistry 399 (2011) 1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Iliescu C, Informacije Midem-Journal of Microelectronics Electronic Components and Materials 36 (2006) 204. [Google Scholar]
- [45].Harrison DJ, Fluri K, Seiler K, Fan Z, Effenhauser CS, Manz A, Science 261 (1993) 895. [DOI] [PubMed] [Google Scholar]
- [46].Li PC, Microfluidic lab-on-a-chip for chemical and biological analysis and discovery, CRC press, 2005. [Google Scholar]
- [47].Li X, Huang J, Tibbits GF, Li PCH, Electrophoresis 28 (2007) 4723. [DOI] [PubMed] [Google Scholar]
- [48].Faigle C, Lautenschlager F, Whyte G, Homewood P, Martin-Badosa E, Guck J, Lab on a Chip 15 (2015) 1267. [DOI] [PubMed] [Google Scholar]
- [49].Yun CK, Hwang JW, Kwak TJ, Chang WJ, Ha S, Han K, Lee S, Choi YS, Lab on a Chip 19 (2019) 580. [DOI] [PubMed] [Google Scholar]
- [50].Dou M, Sanjay ST, Dominguez DC, Zhan S, Li X, Chemical Communications 53 (2017) 10886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Nilghaz A, Lu X, Analytica chimica acta 1046 (2019) 163. [DOI] [PubMed] [Google Scholar]
- [52].Shibata H, Hiruta Y, Citterio D, Analyst (2019). [DOI] [PubMed] [Google Scholar]
- [53].Ma J, Yan S, Miao C, Li L, Shi W, Liu X, Luo Y, Liu T, Lin B, Wu W, Lu Y, Advanced Healthcare Materials 8 (2019) 1801084. [DOI] [PubMed] [Google Scholar]
- [54].Martinez AW, Phillips ST, Butte MJ, Whitesides GM, Angewandte Chemie 119 (2007) 1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Martinez AW, Phillips ST, Butte MJ, Whitesides GM, Angewandte Chemie-International Edition 46 (2007) 1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Carrilho E, Martinez AW, Whitesides GM, Analytical chemistry 81 (2009) 7091. [DOI] [PubMed] [Google Scholar]
- [57].Dungchai W, Chailapakul O, Henry CS, Analyst 136 (2011) 77. [DOI] [PubMed] [Google Scholar]
- [58].Ma LY, Qiao Y, Jones R, Singh N, Su M, Analytical and Bioanalytical Chemistry 408 (2016) 7753. [DOI] [PubMed] [Google Scholar]
- [59].Giokas DL, Tsogas GZ, Vlessidis AG, Analytical chemistry 86 (2014) 6202. [DOI] [PubMed] [Google Scholar]
- [60].Thuo MM, Martinez RV, Lan W-J, Liu X, Barber J, Atkinson MB, Bandarage D, Bloch J-F, Whitesides GM, Chemistry of Materials 26 (2014) 4230. [Google Scholar]
- [61].Hu J, Wang S, Wang L, Li F, Pingguan-Murphy B, Lu TJ, Xu F, Biosensors and Bioelectronics 54 (2014) 585. [DOI] [PubMed] [Google Scholar]
- [62].Bhardwaj R, Lightson N, Ukita Y, Takamura Y, Sensors and Actuators B: Chemical 192 (2014) 818. [Google Scholar]
- [63].Hu P, Zhang W, Xin H, Deng G, Front Cell Dev Biol 4 (2016) 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Gross A, Schoendube J, Zimmermann S, Steeb M, Zengerle R, Koltay P, International Journal of Molecular Sciences 16 (2015) 16897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Bithi SS, Vanapalli SA, Scientific Reports 7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Stetciura IY, Yashchenok A, Masic A, Lyubin EV, Inozemtseva OA, Drozdova MG, Markvichova EA, Khlebtsov BN, Fedyanin AA, Sukhorukov GB, Gorin DA, Volodkin D, Analyst 140 (2015) 4981. [DOI] [PubMed] [Google Scholar]
- [67].Keloth A, Anderson O, Risbridger D, Paterson L, Micromachines 9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Yamada A, Umeki K, Saeki Y, Hashikura Y, Nomura H, Yamamoto I, Umekita K, Takajo I, Koshimoto C, Okayama A, Journal of Microbiological Methods 155 (2018) 42. [DOI] [PubMed] [Google Scholar]
- [69].Kummer S, Wilichowski E, Mitochondrion 43 (2018) 37. [DOI] [PubMed] [Google Scholar]
- [70].Kuo CT, Thompson AM, Gallina ME, Ye FM, Johnson ES, Sun W, Yu JB, Wu IC, Fujimoto B, DuFort CC, Carlson MA, Hingorani SR, Paguirigan AL, Radich JP, Chiu DT, Nature Communications 7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Kim J, Cho H, Han SI, Han KH, Anal Chem 88 (2016) 4857. [DOI] [PubMed] [Google Scholar]
- [72].Lin C-H, Hsiao Y-H, Chang H-C, Yeh C-F, He C-K, Salm EM, Chen C, Chiu M, Hsu C-H, Lab on a Chip 15 (2015) 2928. [DOI] [PubMed] [Google Scholar]
- [73].Tu C, Huang B, Zhou J, Liang Y, Tian J, Ji L, Liang X, Ye X, Micromachines 8 (2017) 1. [Google Scholar]
- [74].Tran QD, Kong TF, Hu D, Lam RH, Lab on a Chip 16 (2016) 2813. [DOI] [PubMed] [Google Scholar]
- [75].Dura B, Dougan SK, Barisa M, Hoehl MM, Lo CT, Ploegh HL, Voldman J, Nature Communications 6 (2015). [DOI] [PubMed] [Google Scholar]
- [76].Rho HS, Yang Y, Hanke AT, Ottens M, Terstappen LW, Gardeniers H, Lab on a Chip 16 (2016) 305. [DOI] [PubMed] [Google Scholar]
- [77].Armbrecht L, Gabernet G, Kurth F, Hiss JA, Schneider G, Dittrich PS, Lab on a Chip 17 (2017) 2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Kim HS, Devarenne TP, Han A, Lab on a Chip 15 (2015) 2467. [DOI] [PubMed] [Google Scholar]
- [79].Khamenehfar A, Beischlag TV, Russell PJ, Ling MTP, Nelson C, Li PCH, Biomicrofluidics 9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Sharifi HN, Soo M, Khamenehfar A, Li P, Electrophoresis (2019). [DOI] [PubMed] [Google Scholar]
- [81].Shen F, Li X, Li PC, Biomicrofluidics 8 (2014) 014109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Huang N-T, Hwong Y-J, Lai RL, Microfluidics and Nanofluidics 22 (2018) 16. [Google Scholar]
- [83].Streets AM, Zhang XN, Cao C, Pang YH, Wu XL, Xiong L, Yang L, Fu YS, Zhao L, Tang FC, Huang YY, Proceedings of the National Academy of Sciences of the United States of America 111 (2014) 7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Babahosseini H, Misteli T, DeVoe DL, Lab Chip 19 (2019) 493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Zhu P, Wang L, Lab Chip 17 (2016) 34. [DOI] [PubMed] [Google Scholar]
- [86].Zhang Q, Wang T, Zhou Q, Zhang P, Gong Y, Gou H, Xu J, Ma B, Sci Rep 7 (2017) 41192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Kim J, Hong JW, Kim DP, Shin JH, Park I, Lab on a chip 12 (2012) 2914. [DOI] [PubMed] [Google Scholar]
- [88].Khamenehfar A, Gandhi MK, Chen YC, Hogge DE, Li PCH, Analytical Chemistry 88 (2016) 5680. [DOI] [PubMed] [Google Scholar]
- [89].Khamenehfar A, Wan CPL, Li PC, Letchford K, Burt HM, Analytical and bioanalytical chemistry 406 (2014) 7071. [DOI] [PubMed] [Google Scholar]
- [90].Haiyan Wang AK, Wong Michael C.K., Wang James L.Z., Li Paul C.H., Zhou Da, Sarunic Marinko, Feng Feng, Cao Hairong and Leung Fung King, Can. J Pure & Appl. Sci 11 (2017) 4053. [Google Scholar]
- [91].Avid Khamenehfar P.C.L.a.E.L.L., Can. J Pure & Appl. Sci 12 (2018) 4375. [Google Scholar]
- [92].Pilat Z, Jonas A, Jezek J, Zemanek P, Sensors 17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Zare RN, Kim S, Annual review of biomedical engineering 12 (2010) 187. [DOI] [PubMed] [Google Scholar]
- [94].Sarkar A, Kolitz S, Lauffenburger DA, Han J, Nature communications 5 (2014) 3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Kim SC, Clark IC, Shahi P, Abate AR, Analytical chemistry 90 (2018) 1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].DeKosky BJ, Kojima T, Rodin A, Charab W, Ippolito GC, Ellington AD, Georgiou G, Nature medicine 21 (2015) 86. [DOI] [PubMed] [Google Scholar]
- [97].Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Cell 161 (2015) 1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW, Cell 161 (2015) 1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Jokilaakso N, Salm E, Chen A, Millet L, Guevara CD, Dorvel B, Reddy B, Karlstrom AE, Chen Y, Ji H, Lab on a Chip 13 (2013) 336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Jen C-P, Amstislavskaya TG, Liu Y-H, Hsiao J-H, Chen Y-H, Sensors 12 (2012) 6967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].De Lange N, Tran T, Abate A, Biomicrofluidics 10 (2016) 024114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Hoefemann H, Wadle S, Bakhtina N, Kondrashov V, Wangler N, Zengerle R, Sensors and Actuators B: Chemical 168 (2012) 442. [Google Scholar]
- [103].Cheng Y, Wang Y, Wang Z, Huang L, Bi M, Xu W, Wang W, Ye X, Biomicrofluidics 11 (2017) 024112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Sukhorukov VL, Reuss R, Endter JM, Fehrmann S, Katsen-Globa A, Geßner P, Steinbach A, Müller KJ, Karpas A, Zimmermann U, Biochemical and Biophysical Research Communications 346 (2006) 829. [DOI] [PubMed] [Google Scholar]
- [105].Vassilopoulos G, Wang P-R, Russell DW, Nature 422 (2003) 901. [DOI] [PubMed] [Google Scholar]
- [106].Hu N, Yang J, Yin ZQ, Ai Y, Qian S, Svir IB, Xia B, Yan JW, Hou WS, Zheng XL, Electrophoresis 32 (2011) 2488. [DOI] [PubMed] [Google Scholar]
- [107].Kimura Y, Gel M, Techaumnat B, Oana H, Kotera H, Washizu M, Electrophoresis 32 (2011) 2496. [DOI] [PubMed] [Google Scholar]
- [108].Skelley AM, Kirak O, Suh H, Jaenisch R, Voldman J, Nature methods 6 (2009) 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Yang Z, Yu B, Zhu J, Huang X, Xie J, Xu S, Yang X, Wang X, Yung BC, Lee LJ, Lee RJ, Teng L, Nanoscale 6 (2014) 9742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Vander Heiden MG, DeBerardinis RJ, Cell 168 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Wu H, Zhu J, Huang Y, Wu D, Sun J, Molecules 23 (2018). [Google Scholar]
- [112].Rothbauer M, Zirath H, Ertl P, Lab Chip 18 (2018) 249. [DOI] [PubMed] [Google Scholar]
- [113].Jin Q, Shen Y, Ma L, Pan Y, Zhu S, Zhang J, Zhou W, Wei X, Li X, Catalysis Today 327 (2019) 279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Chen YC, Ingram P, Yoon E, Analyst 139 (2014) 6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Son KJ, Rahimian A, Shin D-S, Siltanen C, Patel T, Revzin A, Analyst 141 (2016) 679. [DOI] [PubMed] [Google Scholar]
- [116].Sarkar S, Cohen N, Sabhachandani P, Konry T, Lab on a Chip 15 (2015) 4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Stuelten CH, Parent CA, Montell DJ, Nat Rev Cancer 18 (2018) 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Zhuang J, Wu Y, Chen L, Liang S, Wu M, Zhou L, Fan C, Zhang Y, Adv Sci (Weinh) 5 (2018) 1801158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Deng YL, Zhang Y, Sun S, Wang ZH, Wang MJ, Yu BQ, Czajkowsky DM, Liu BY, Li Y, Wei W, Shi QH, Scientific Reports 4 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Murphy TW, Zhang Q, Naler LB, Ma S, Lu C, Analyst 143 (2018) 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Lecault V, White AK, Singhal A, Hansen CL, Current opinion in chemical biology 16 (2012) 381. [DOI] [PubMed] [Google Scholar]
- [122].Zhong JF, Chen Y, Marcus JS, Scherer A, Quake SR, Taylor CR, Weiner LP, Lab on a Chip 8 (2008) 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Bhagwat N, Dulmage K, Pletcher CH, Wang L, DeMuth W, Sen M, Balli D, Yee SS, Sa S, Tong F, Scientific reports 8 (2018) 5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Pellegrino M, Sciambi A, Treusch S, Durruthy-Durruthy R, Gokhale K, Jacob J, Chen TX, Geis JA, Oldham W, Matthews J, Genome research 28 (2018) 1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Yu Z, Lu S, Huang Y, Analytical chemistry 86 (2014) 9386. [DOI] [PubMed] [Google Scholar]
- [126].Fu Y, Li C, Lu S, Zhou W, Tang F, Xie XS, Huang Y, Proceedings of the National Academy of Sciences 112 (2015) 11923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Fu G, Sanjay ST, Dou M, Li X, Nanoscale 8 (2016) 5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Wu G, Datar RH, Hansen KM, Thundat T, Cote RJ, Majumdar A, Nature biotechnology 19 (2001) 856. [DOI] [PubMed] [Google Scholar]
- [129].Fu G, Sanjay ST, Zhou W, Brekken RA, Kirken RA, Li X, Analytical Chemistry 90 (2018) 5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Fu G, Sanjay ST, Li X, Analyst 141 (2016) 3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Shi J, Lyu J, Tian F, Yang M, Biosensors and Bioelectronics 93 (2017) 182. [DOI] [PubMed] [Google Scholar]
- [132].Zhang L, Sun J, Wang Y, Wang J, Shi X, Hu G, Analytical chemistry 88 (2016) 7344. [DOI] [PubMed] [Google Scholar]
- [133].Wu L, Claas AM, Sarkar A, Lauffenburger DA, Han J, Integrative Biology 7 (2015) 513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Herrera V, Hsu S-CJ, Rahim MK, Chen C, Nguyen L, Liu WF, Haun JB, Analyst 144 (2019) 980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Eyer K, Stratz S, Kuhn P, Kuster S, Dittrich PS, Analytical chemistry 85 (2013) 3280. [DOI] [PubMed] [Google Scholar]
- [136].McQuade JL, Daniel CR, Hess KR, Mak C, Wang DY, Rai RR, Park JJ, Haydu LE, Spencer C, Wongchenko M, The Lancet Oncology 19 (2018) 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Maheswaran S, Haber DA, Cancer research 75 (2015) 2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Heath JR, Ribas A, Mischel PS, Nature reviews Drug discovery 15 (2016) 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Liadi I, Singh H, Romain G, Rey-Villamizar N, Merouane A, Adolacion JRT, Kebriaei P, Huls H, Qiu P, Roysam B, Cancer immunology research 3 (2015) 473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Elitas M, Brower K, Lu Y, Chen JJ, Fan R, Lab on a Chip 14 (2014) 3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Walsh JC, Lebedev A, Aten E, Madsen K, Marciano L, Kolb HC, Antioxidants & redox signaling 21 (2014) 1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Sinha S, Sarma P, Sehgal R, Medhi B, Frontiers in pharmacology 8 (2017) 754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Yu M, Bardia A, Aceto N, Bersani F, Madden MW, Donaldson MC, Desai R, Zhu H, Comaills V, Zheng Z, Science 345 (2014) 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Khoo BL, Grenci G, Lim YB, Lee SC, Han J, Lim CT, Nature Protocols 13 (2018) 34. [DOI] [PubMed] [Google Scholar]
- [145].Li X, Li PC, CANADIAN JOURNAL OF PURE AND APPLIED SCIENCES (2014) 2663. [Google Scholar]
- [146].Li X, Xue X, Li PC, Integrative Biology 1 (2008) 90. [DOI] [PubMed] [Google Scholar]
- [147].Drake CG, Lipson EJ, Brahmer JR, Nature reviews Clinical oncology 11 (2014) 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Weller M, Le Rhun E, Nature Reviews Clinical Oncology (2019) 1. [DOI] [PubMed] [Google Scholar]
- [149].Sarkar S, McKenney S, Sabhachandani P, Adler J, Hu XZ, Stroopinksy D, Rosenblatt J, Avigan D, Konry T, Sensors and Actuators B-Chemical 282 (2019) 580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].June CH, O’Connor RS, Kawalekar OU, Ghassemi S, Milone MC, Science 359 (2018) 1361. [DOI] [PubMed] [Google Scholar]
- [151].Adriani G, Pavesi A, Tan AT, Bertoletti A, Thiery JP, Kamm RD, Drug Discovery Today 21 (2016) 1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Segaliny AI, Li GD, Kong LS, Ren C, Chen XM, Wang JK, Baltimore D, Wu GK, Zhao WA, Lab on a Chip 18 (2018) 3733. [DOI] [PMC free article] [PubMed] [Google Scholar]