Abstract
Long noncoding RNAs (lncRNAs) have multiple functions in the regulation of cellular homeostasis. In recent years, numerous studies have shown that tumor-associated lncRNAs play key roles in promoting and maintaining tumor initiation and progression by shaping the tumor microenvironment through changing tumor cell intrinsic properties. Here, we focus on the roles of lncRN As in cancer immunology. In the first part, we provide an overview of the roles played by lncRNAs and their deregulation in cancer at the cancer cell- and tumor microenvironment-associated immune cell levels. We go on to describe preclinical strategies for targeting lncRNAs, particularly highlighting the effects on tumor microenvironments. We then discuss the possibility of combining lncRNA targeting and tumor immune checkpoint inhibitor antibodies to treat cancer.
Keywords: Long noncoding RNA, Tumor-cell-intrinsic factors, Tumor-cell-extrinsic factors, Cancer immunotherapy
1. Introduction
Long non-coding RNAs (lncRNAs) are defined as RNA transcripts that are longer than 200 nucleotides but exhibit low coding potential (Perkel, 2013). With the advancement of high-throughput technology, more and more lncRNAs have been identified. In recent years, in-depth studies of lncRNAs have identified characteristics that are distinct from protein coding RNAs (mRNAs), such as tissue specificity; 78% of lncRNAs are tissue-specific, compared to ~19% for mRNAs (Cabili, et al., 2011; Ravasi, et al., 2006). LncRNAs also show higher developmental stage specificity and cell subtype specificity (S. J. Liu, et al., 2016; Yan, et al., 2013). Due to these specific manifestations, lncRNAs have been found to be involved in the regulation of multiple physiological functions, including regulating cell cycles, cell growth, differentiation, apoptosis, motility and invasion, signal transduction, DNA damage regulation, survival, immune response, and pluripotency (Huarte, 2015; C. Lin & Yang, 2018). They also play essential roles in early embryonic development and regulating tissue homeostasis in adults (Perry & Ulitsky, 2016; Salviano-Silva, Lobo-Alves, Almeida, Malheiros, & Petzl-Erler, 2018). Recently, numerous studies demonstrated that aberrant expression of lncRNAs plays crucial roles in both the development of tumors and metastasis (Batista & Chang, 2013; Wu, et al., 2019). In addition to affecting cancer cells themselves, lncRNAs also influence the tumor microenvironment (TME) (Denaro, Merlano, & Lo Nigro, 2019). The role of lncRNAs as oncogenes or tumor suppressors at the cancer cell level is still a matter of debate due to their differential effects in different cancer types. Meanwhile, at the microenvironment level, lncRNAs contribute to mediating and controlling several immune and cancer cell interactions and important mechanisms of immune response (Y. Zhou, Zhu, Xie, & Ma, 2019). Thus, targeting lncRNAs as a cancer therapy may require more in-depth studies.
Gene alteration is considered an important cause of tumorigenesis. With the advancement of large-scale sequencing technology, numerous somatic mutations, copy number alteration, and cancer-related single-nucleotide polymorphisms (SNPs) have been discovered from clinical cancer patient samples. Interestingly, the majority of these gene alterations are in the noncoding region of the genome (Fatica & Bozzoni, 2014; Maurano, Wang, Kutyavin, & Stamatoyannopoulos, 2012; Melton, Reuter, Spacek, & Snyder, 2015; Reuter, Spacek, & Snyder, 2015). Among those non-coding genes, lncRNAs have emerged as essential new participants involved in tumorigenesis (Beroukhim, et al., 2010; Fatica & Bozzoni, 2014; Melton, et al., 2015). Dysregulation of numerous lncRNA targets has been reported to associate with the stage and prognosis of many tumor types (Jendrzejewski, et al., 2012), including breast cancer (A. Lin, et al., 2016; T. Zhang, et al., 2019), lung cancer (Loewen, Jayawickramarajah, Zhuo, & Shan, 2014; Lu, Wang, Chen, Liu, & Jiao, 2018; Tao, et al., 2016), and liver cancer (Mai, et al., 2019; Mehra & Chauhan, 2017; O’Brien, Zhou, Tan, Alpini, & Glaser, 2019), as well as linked to resistance against chemotherapy and targeted therapy (Chen & Xu, 2018; De Los Santos, Dragomir, & Calin, 2019; W. Jiang, et al., 2020; K. Liu, et al., 2020; Majidinia & Yousefi, 2016; Pan, Xie, Li, & Chen, 2015). Although much has been learned about the multiple functions of lncRNAs in tumor cell proliferation, apoptosis, migration, invasion, and maintenance of stemness during cancer development (Batista & Chang, 2013; Slack & Chinnaiyan, 2019), little is known about their potential role in regulating tumor immunity as tumor-cell-intrinsic factors or tumor-cell-extrinsic factors that affect the cancer immunotherapy.
In recent years, breakthroughs in the field of tumor immunotherapy have greatly improved the efficacy of tumor treatments. Nevertheless, only a small percentage of cancer patients benefit from tumor immunotherapy, and the majority of patients develop primary or secondary resistance following treatment (Corrales, Matson, Flood, Spranger, & Gajewski, 2017; Cristescu, et al., 2018; Farhood, Najafi, & Mortezaee, 2019; Jia, et al., 2017; Ribas, 2012; Sharma, Hu-Lieskovan, Wargo, & Ribas, 2017; Y. Zhou, et al., 2019). Further in-depth studies suggest that both tumor-cell-intrinsic and tumor-cell-extrinsic factors may be involved in cancer immunotherapy resistance mechanisms (Fares, Van Allen, Drake, Allison, & Hu-Lieskovan, 2019; Sharma, et al., 2017). Many clinical sample-based studies have shown that the dysregulation of the antigen presentation pathway is one of the major factors in driving tumor immunotherapy resistance. For example, studies suggested that tumor cell antigen loss leads to the tumor failing to respond to immune checkpoint therapy (Gubin, et al., 2014; Iorgulescu, Braun, Oliveira, Keskin, & Wu, 2018). Alternatively, cancer cells may express tumor antigens but develop mechanisms to avoid presenting them on the surface through the major histocompatibility complex (MHC) due to alterations in antigen-presenting machinery (such as proteasome subunits or transporters associated with antigen processing), beta-2-microglobulin (B2M), or the MHC itself (Q. Hu, et al., 2019; Marincola, Jaffee, Hicklin, & Ferrone, 2000; Sucker, et al., 2014). In addition to defects in antigen processing, some new dysregulated tumor-intrinsic signaling pathways have also been shown to associate with tumor immunotherapy resistance, such as the mitogen-activated protein kinase (MAPK)-PI3K and PTEN-PI3K signaling pathways, WNT/β-catenin signaling pathway, interferon-gamma (IFNγ) signaling pathways, and a lack of the immune checkpoint Programmed death-ligand 1 (PD-L1) expression (Sharma, et al., 2017; Stutvoet, et al., 2019). Although there are many studies focusing on the causes of tumor cell resistance, the detailed mechanisms of this phenomenon are still unclear. To better understand the mechanisms of tumor immunotherapy resistance, more tumor cell intrinsic factors need to be discovered.
2. Roles of lncRNAs as tumor-cell-intrinsic factors
Recently, numerous tumor-associated lncRNAs have been recognized as tumor cell intrinsic factors or tumor-cell-extrinsic factors that mediate tumor cell escape of immunosurveillance (Figure 1). They are involved in cancer immunity regulation as either oncogenic genes or tumor-suppressive genes (Table 1). These tumor-associated lncRNAs may play vital roles in immunotherapy resistance (Denaro, et al., 2019; Y. Zhou, et al., 2019).
Figure 1. Tumor-cell-intrinsic and tumor-cell-extrinsic roles of lncRNAs.
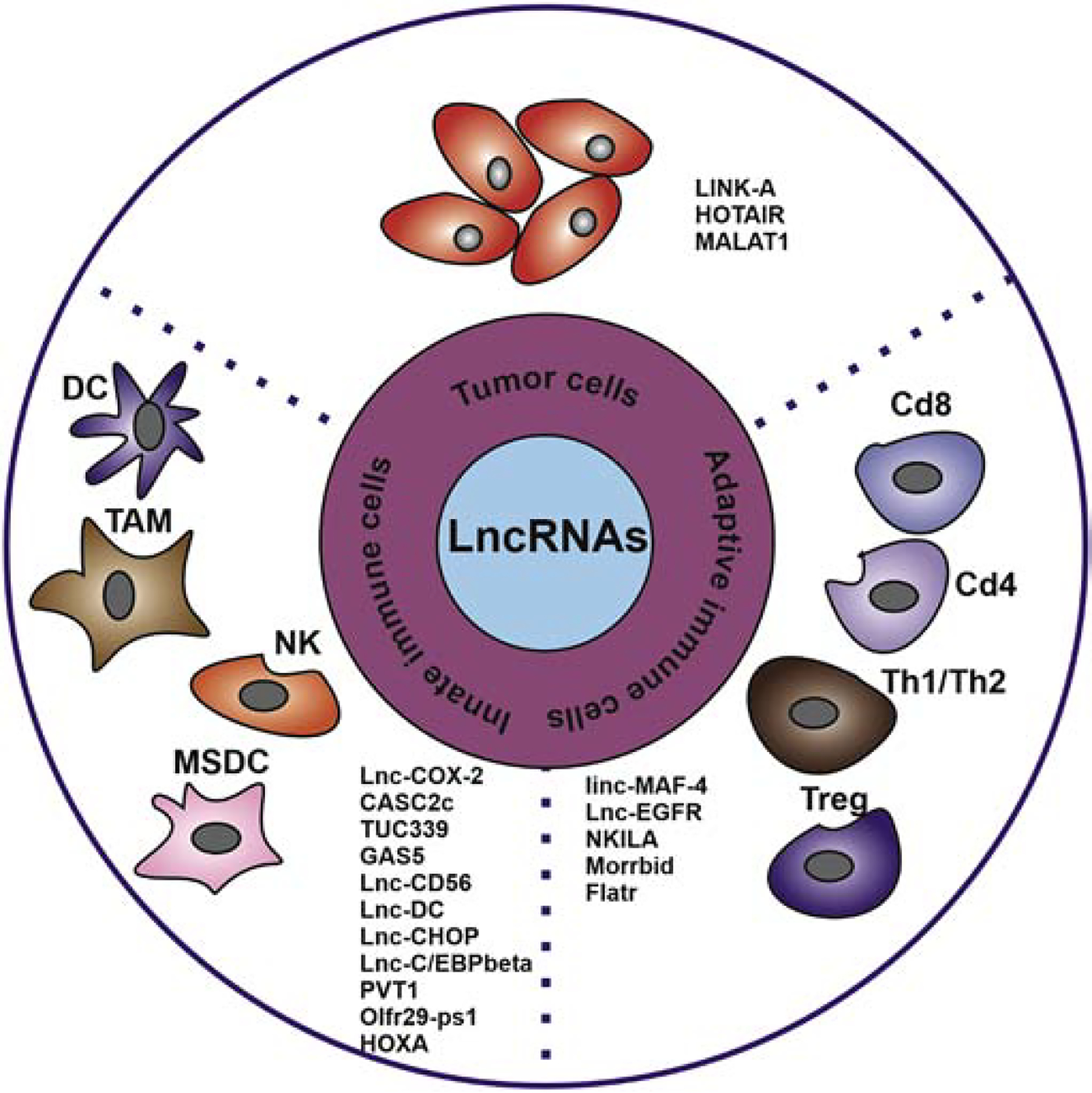
Intrinsic factor roles of lncRNAs regulate antigen processing machinery and constitutive PD-L1 expression in cancer cells; Extrinsic factors roles of lncRNAs regulate the activity of tumor microenvironment immune cells, such as TAM, CD8+, NK cells, Th1/Th2, DC, and MDSC.
Table 1.
Oncogenic or Tumor-Suppressive Long Non-coding RNAs involved in cancer immunity regulation
| LncRNA | O/T | Cell type | Localization | Immunity-Related Mechanisms | Reference |
|---|---|---|---|---|---|
| LINK-A | O | Tumor cell | Cytoplasm | Inhibits breast cancer cell antigen presentation and decrease CTL cells infiltration | (Q. Hu, et al., 2019) |
| HOTAIR | O | Tumor cell, macroph age and MDSC | Nuclear | Promotes gastric cancer cell (HLA)-G expression via inhibiting miR-152 and miR-148a, also facilitates macrophage and MDSC activity in liver cancer through transcriptional upregulation of chemokine (C-C motif) ligand 2 | (Fujisaka, et al., 2018; Song, et al., 2015) |
| MALAT1 | O | Tumor cell and MDSC | Nuclear | Increase the expression of PD-L1 by sponging miR-195 and modulating miR-200a, promotes infiltration of MDSC in lung cancer | (Q. M. Wang, et al., 2019; Wei, et al., 2019; Q. Zhou, et al., 2018) |
| LncRNA-C ox2 | O | Macrop hages | Nuclear | Regulates the inflammatory gene expression in the macrophages via interacts with heterogeneous nuclear ribonucleoprotein A/B and A2/B1, assembles into the switch/sucrose nonfermentable (SWI/SNF) complex and regulating the NF-κb signaling axis | (Bian, Yang, Zhang, Li, Wang, Jiang, Chen, Li, et al., 2019; Carpenter, et al., 2013; Elling, et al., 2018) |
| CASC2c | T | Macrop hages | Nuclear | Inhibits of M2 polarization and decreased proliferation of tumor cells via downregulating the expression and extracellular production of coagulation factor X (FX) | (Y. Zhang, et al., 2018) |
| TUC339 | O | Macrop hagy | Nuclear | Upregulates the expression of M2 markers after IFN-γ + LPS treatment | (X. Li, et al., 2018) |
| Morrbid | T | T cell | Nuclear | Controls the expansion, survival, and effector function of CD8+ T cells by regulating the expression of the pro-apoptotic factor Bcl2111 and the strength of the PI3K-AKT signaling pathway | (Kotzin, et al., 2019) |
| Lnc-CD56 | T | NK Cell | Nuclear | Promotes the expression of CD56 and differentiation of NK cells from human CD34 + hematopoietic progenitor cells | (R. Zhang, et al., 2016) |
| GAS5 | T | NK Cell | Nuclear | Enhances the killing effect of NK cell on liver cancer through regulating miR-544/RUNX3 | (Fang, et al., 2019) |
| Linc-MAF-4 | T | T Cell | Nuclear | Facilitates the differentiation of T cells toward the TH2 phenotype through regulating MAF transcriptional activities by recruiting chromatin modifiers | (Ranzani, et al., 2015) |
| Lnc-DC | T | DC Cell | Cytoplasm | Promotes the DC differentiation by activating the transcription factor STAT3 and increase the ability of DCs to stimulate T cell activation | (P. Wang, et al., 2014) |
| Flatr | O | T Cell | Nuclear | Modulates chromatin accessibility of the FoxP3 gene loci, leading to impaired Treg activity and autoimmunity | (Brajic, et al., 2018) |
| NKILA | T | T Cell | Regulates T cell sensitivity to AICD by inhibiting NF-κB activity | (Huang, et al., 2018) | |
| Lnc-EGFR | O | T Cell | Cytoplasm | Stimulates the differentiation of Treg cells, thereby inhibiting CTL activity in the tumor area and enhancing the growth of HCC | (R. Jiang, et al., 2017) |
| Lnc-CHOP | O | MDSC | Nuclear | Promotes MDSC differentiation by interacting with CHOP and C / EBPβ isoforms to encourage C / EBPβ activation and regulate a large number of target transcripts in MDSC | (Gao, Wang, et al., 2018) |
| Lnc-C/EBP beta | O | MDSC | Nuclear | Controls immune-suppressive function and differentiation of MDSCs by regulating a set of target transcripts, such as Arg-1, NOS2, NOX2, and COX2 | (Gao, Sun, et al., 2018) |
| PVT1 | O | MDSC | Nuclear | Promotes the immunosuppressive function of G-MDSCs in vitro and in vivo | (Zheng, et al., 2019) |
| Olfr29-ps1 | O | MDSC | Nuclear | Regulates the differentiation and function of MDSCs through a m6A-modified Olfr29-ps1/miR-214–3p/MyD88 regulatory network | (Shang, et al., 2019) |
| HOXA | T | MDSC | Nuclear | delay tumor progression and enhance the antitumor immune response by downregulating the immunosuppression of MDSCs | (Tian, et al., 2018) |
O/T represents the role of lncRNA as Oncogenic or Tumor-Suppressive Long Non-coding RNAs regulating cancer immunity, respectively.
2.1. LINK-A
Long intergenic non-coding RNA for kinase activation (LINK-A) was first identified to predominately reside in the cytoplasm and is highly expressed in triple-negative breast cancer (TNBC). Further studies showed that LINK-A-dependent signaling pathway activation promotes breast cancer metabolic reprogramming and tumorigenesis. The expression of LINK-A is correlated with advanced lymph node metastasis stages and shorter survival time for breast cancer patients (A. Lin, et al., 2017; A. Lin, et al., 2016). Transgenic expression of LINK-A in mouse mammary glands results in mammary gland malignancies and lung metastasis. Mechanistically, LINK-A facilitates the association between phosphatidylinositol (3,4,5)-trisphosphate (PIP3) and inhibitory G protein-coupled receptors (GPCRs), facilitating the cAMP-dependent protein kinase (PKA)-mediated phosphorylation of the E3 ligase TRIM71 (also known as LIN-41). As a consequence, TRIM71 catalyzes the K48-linked polyubiquitination and proteasome-mediated degradation of the peptide-loading complex (PLC) upon LINK-A expression (Figure 2). Treatment with LINK-A Locked Nucleic Acids (LNAs) led to stabilization of the PLC components and improved the infiltration and cytotoxicity of tumor-resident CD8+ T cells in vivo. Treatment with LINK-A LNAs in combination with immune checkpoint blockers exhibits synergistic efficacy in repressing breast tumor growth. Most importantly, human breast cancer tissues with high LINK-A expression are associated with decreased activated CD8+ T cell and antigen-presenting cell (APCs) infiltration in tumors. LINK-A upregulation and the downregulation of the PLC are correlated in TNBC patients whose tumors are resistant to anti-PD-1 immunotherapy. Hence, lncRNA-directed targeted therapy using LNAs could serve as a promising strategy to improve breast tumor antigenicity and sensitize TNBC patients to immunotherapy (Q. Hu, et al., 2019).
Figure 2. The mechanisms of how LINK-A regulates antigen processing machinery.
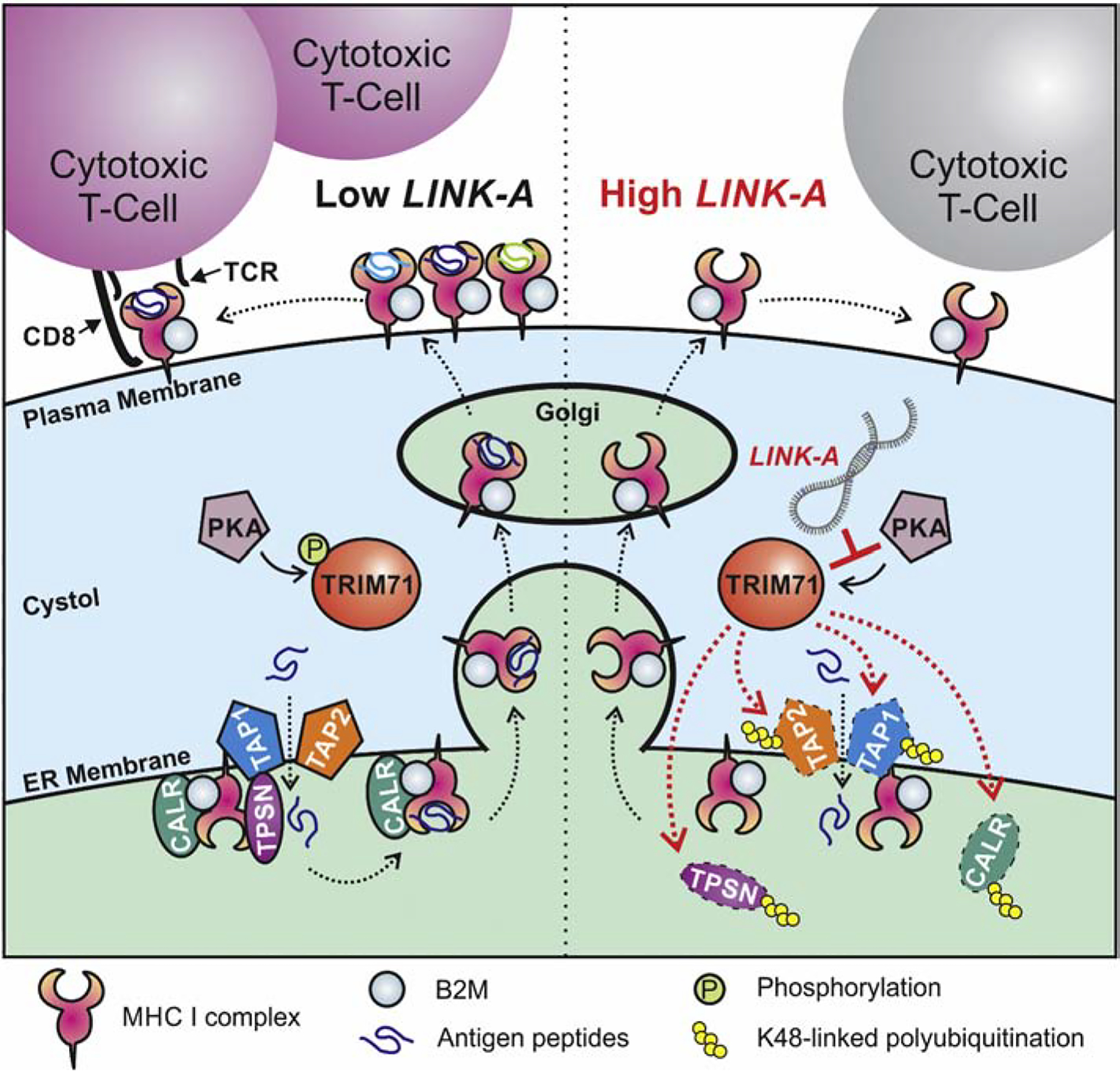
LINK-A-facilitated, TRIM71-mediated downregulation of antigen presentation machinery impairs antigen presentation during tumor initiation.
2.2. HOTAIR
HOX transcript antisense intergenic RNA (HOTAIR) has been identified at the HOXC locus and inhibits the transcription of the HOXD locus. HOTAIR binds to Polycomb Repressive Complex 2 (PRC2) and plays an important role in the occupation of PRC2 at the HOXD locus and modulation of histone H3 lysine 27 trimethylation (Rinn, et al., 2007; Tsai, et al., 2010). It has been reported that HOTAIR is highly expressed in numerous cancers, providing notable prognostic value (Cai, Song, Cai, & Zhang, 2014; Gupta, et al., 2010; Hajjari & Salavaty, 2015). The role of HOTAIR in transcriptional regulation in cancer cells has been extensively reviewed (Hajjari & Salavaty, 2015; Tang & Hann, 2018). Recent advances have suggested that HOTAIR may play important roles in antigen presentation and immunotherapy. Song et al. reported that HOTAIR overexpression in gastric cancer cells might also involve tumor cell immune escape mechanisms by influencing HLA-G upregulation via inhibition of miR-152 and miR-148a (Song, et al., 2015; Sun, et al., 2016). Recent studies indicated that HOTAIR facilitates macrophage and myeloid-derived suppressor cell activity in liver cancer through transcriptional upregulation of chemokine (C-C motif) ligand 2 (Fujisaka, et al., 2018).
2.3. MALAT1
The transcript of the MALAT1 gene, also known as NEAT2, represents nuclear-rich enriched transcript 2 and consists of >8000 nucleotides (Ji, et al., 2003). Although early reports provided evidence for its association with metastasis in patients with early non-small cell lung cancer (NSCLC), subsequent studies have reported that MALAT1 is abundant and highly conserved in 33 different mammals (Eissmann, et al., 2012). Computational and biochemical studies demonstrated the interaction between MALAT1 and the serine and arginine (SR) proteins involved in splicing regulation or interaction with spliceosome proteins (Yoshimoto, Mayeda, Yoshida, & Nakagawa, 2016). Recently, MALAT1 have been found to positively regulate the expression of PD-L1 by sponging miR-195, thus promoting cancer cell migration and immune escape by regulating the proliferation and apoptosis of CD8+ T cells (Q. M. Wang, Lian, Song, Huang, & Gong, 2019). MALAT1 also has been showed to modulate miR-200a and the expression of PD-L1, serving as one of the mechanisms of lung cancer progression (Wei, Wang, Huang, Zhao, & Zhao, 2019). In lung cancer, the expression of MALAT1 is correlated with infiltration of Myeloid-derived suppressor cells (MDSCs) and possibly modulates the patient’s response to immunotherapy (Q. Zhou, et al., 2018).
3. Roles of lncRNAs as tumor-cell-extrinsic factors
Recently, the functional roles of lncRNAs in innate immune response have emerged. Aberrant expression of lncRNAs has also been found to closely associate with innate and adaptive immune response regulation, which have important effects on cancer progression (Figure 1 and Table 1).
3.1. lncRNA-Cox2
Inducing gene expression is an important feature of host inflammatory response (Ahmed, Williams, & Hannigan, 2015; Mathy & Chen, 2017; Smale & Natoli, 2014; J. Wang, Blanchard, & Ernst, 2001). Recently, lincRNA-Cox2 has been found to be highly expressed in macrophages upon stimulation of Toll-like receptors (G. Hu, et al., 2016). Further studies found that it also mediates both the activation and repression of distinct classes of immune genes. Mechanistically, lincRNA-Cox2 interacts with heterogeneous nuclear ribonucleoprotein A/B and A2/B1 to inhibit the transcription of target genes (Carpenter, et al., 2013). To evaluate the in vivo functions of lincRNA-Cox2, Elling et al. developed a variety of lincRNA-Cox2-deficient mouse models. They found that the loss of LincRNA-Cox2 led to an inflammatory response in mouse macrophages and tissues (Elling, et al., 2018). In addition to interacting with heterogeneous nuclear ribonucleoprotein A/B and A2/B1, lincRNA-Cox2 also assembles into the switch/sucrose nonfermentable (SWI/SNF) complex in cells after Lipopolysaccharides (LPS) stimulation. Consequently, SWI / SNF-related chromatin remodeling leads to the activation of inflammatory gene expression under the influence of macrophages in the presence of a microbial challenge (G. Hu, et al., 2016). This lncRNA also plays important roles in regulating the NF-κb signaling axis and inflammatory response (Bian, Yang, Zhang, Li, Wang, Jiang, Chen, & Zeng, 2019).
3.2. CASC2c
Macrophages play an important role in innate immunity and TME (Hao, et al., 2012; Noy & Pollard, 2014; Pathria, Louis, & Varner, 2019). Macrophages can transform into the M1 type or M2 type when induced by LPS and IFN-γ or IL-4 (interleukin 4) and IL-13 (interleukin 13), respectively, and the two phenotypes can mutually transform (Martinez & Gordon, 2014). Expression of the lncRNA CASC2c has been shown to inhibit the expression and extracellular production of coagulation factor X (FX) (Y. Zhang, et al., 2018). The reduced production of FX leads to decreased phosphorylation and the inactivation of ERK1/2 and AKT in macrophages. As a consequence, the expression of CASC2c contributes to repression of M2 polarization and decreased proliferation of tumor cells (Y. Zhang, et al., 2018).
3.3. TUC339
The lncRNA TUC339 was first identified as a lncRNA that is highly-expressed in hepatocellular carcinoma (HCC) cell-derived extracellular vesicles (Kogure, Yan, Lin, & Patel, 2013). X Li et al. reported that cellular HCC-derived exosomes contain high levels of lncRNA TUC339, which are subsequently taken up by macrophages (X. Li, Lei, Wu, & Li, 2018). Depletion of TUC339 in macrophages results in increased production of pro-inflammatory cytokines, expression of costimulatory molecules, and phagocytosis. In addition, compared to M1 macrophages, elevated levels of TUC339 in M2 macrophages were detected. Depletion of TUC339 in the macrophages led to reduced expression of M2 markers after IL-4 treatment, while the overexpression of TUC339 in the macrophages enhanced the expression of M2 markers after IFN-γ + LPS treatment, indicating that TUC339 plays a key role in M1 / M2 polarization (X. Li, et al., 2018).
3.4. Morrbid
CD8+ T cells are key immune cells that eliminate cancer cells with MHC class I molecules (Janeway & Bottomly, 1994; Mueller, Jenkins, & Schwartz, 1989; N. Zhang & Bevan, 2011). To achieve this, cells must first be triggered by their primary interactions with dendritic cells (DCs), natural killer (NK) cells, and CD4+ T cells, with the cells themselves playing a key role in this initiation. The cells are then activated to form effector cytotoxic T lymphocytes (CTLs), which kill cancer cells by releasing particles or inducing FasL-mediated apoptosis (Farhood, et al., 2019; van der Leun, Thommen, & Schumacher, 2020). Recently, the Henao-Mejia J. group has identified a long non-coding RNA named Morrbid, which regulates the survival of neutrophils, eosinophils, and monocytes upon stimulation of mouse survival cytokines. To control the life span of these cells, Morrbid maintains gene equilibrium states by recruiting the PRC2 complex to the Bim promoter, thereby regulating the transcription of the neighboring pro-apoptotic gene Bim (Kotzin, et al., 2016). In a recent study, Kotzin JJ, et al reported that T cell receptor (TCR) and type I IFN stimulation specifically induces the transcription of lncRNA Morrbid during the early stages of acute and chronic lymphocytic choroidal meningitis virus (LCMV) infection. In response to type I IFN, Morrbid and its loci control the expansion, survival, and effector function of CD8+ T cells by regulating the expression of the pro-apoptotic factor Bcl2111 and the strength of the PI3K-AKT signaling pathway (Kotzin, et al., 2019).
3.5. Lnc-CD56
Natural killer (NK) cells are innate immune lymphocytes that play critical roles in host defense against viral infection and surveillance against malignant transformation (Chiossone, Dumas, Vienne, & Vivier, 2018; Smyth, Hayakawa, Takeda, & Yagita, 2002; Vivier, et al., 2011). NK cells exhibit cytotoxicity toward adjacent cells that express oncogenic transformation-associated surface markers (Shimasaki, Jain, & Campana, 2020). In addition, NK cells also enhance the response of antibodies and the activation of T cells, which proves that NK cells have play important role in tumor immunotherapy (Souza-Fonseca-Guimaraes, Cursons, & Huntington, 2019). Recently, R. Zhang et al. determined the expression profile of lncRNAs in human primary lymphocytes. They found that innovative NK-specific lncRNAs are closely related to the differentiation and function of NK cells (R. Zhang, et al., 2016). Among them, they found that the expression of a NK-specific lncRNA, lnc-CD56, is positively correlated with the expression of NK cell surface marker CD56. Further stud ies found that lnc-CD56 may play important roles in promoting the expression of CD56 and differentiation of NK cells from human CD34 + hematopoietic progenitor cells (R. Zhang, et al., 2016). Another study indicated that lncRNAs MEG3, GAS5, and numerous microRNAs modulate the killing efficacy of NK cells in a variety of cancer types (Fang, et al., 2019; W. Liu, et al., 2017; Yang, Shen, Feng, & Li, 2019).
3.6. Linc-MAF-4
Recently, the importance of stimulating CD4+ T helper cell (Th) response in cancer immunotherapy has been increasing (Bretscher, 2019; Cohen, et al., 2000; Knutson & Disis, 2005). By activating antigen-specific effector cells and recruiting cells of the innate immune system, Th cells are essential for the development of an immune response (Dong & Flavell, 2000; Hung, et al., 1998; Walker & McKenzie, 2018). There are two main Th cell subtypes: Th1 and Th2. Th1 cells are characterized by the secretion of IFN-γ and TNF-α (Tumour Necrosis Factor alpha), which are closely associated with cytotoxic T cell activation (Kalams & Walker, 1998). Th1/Th2 cells enhance the ability of APCs to influence antigen presentation and promote the cytotoxicity of CD8+ T cells (Fallarino, et al., 2000). LncRNAs have been suggested to regulate adaptive immunity by controlling the differentiation and function of different types of T cells (Atianand, Caffrey, & Fitzgerald, 2017). Ranzani et al. reported that the expression of linc-MAF-4 is negatively correlated with the expression of TH2-related transcription factor MAF (Ranzani, et al., 2015). The down-regulation of linc-MAF-4 facilitates the differentiation of T cells toward the TH2 phenotype (Ranzani, et al., 2015; F. Zhang, Liu, Wei, Gao, & Hao, 2017). They also indicated that linc-MAF-4 regulates MAF transcriptional activities by recruiting chromatin modifiers (Ranzani, et al., 2015).
3.7. Lnc-DC
Dendritic cells (DCs) are a special class of antigen-presenting cells that play a key role in the initiation and regulation of innate and adaptive immune response. Therefore, there is currently great interest in regulating DC function to improve the effectiveness of cancer immunotherapy (Bottcher & Reis, 2018; Demoulin, Herfs, Delvenne, & Hubert, 2013; Wculek, et al., 2020). In one pioneering study, the Cao group identified a new lncRNA named lnc-DC, which is expressed only in human conventional dendritic cells (DC) (P. Wang, et al., 2014). Knocking down lnc-DC impairs the DC differentiation of human monocytes and mouse bone marrow cells in vitro and reduces the ability of DCs to stimulate T cell activation (P. Wang, et al., 2014). lnc-DC mediates these effects by activating the transcription factor STAT3 (signal transducer and transcriptional activator 3) (P. Wang, et al., 2014). Recently, lnc-DC has been shown to regulate the immune response of dendritic cells upon hepatitis B virus (HBV) infection through a Toll-like receptor 9 (TLR9) and STAT3-mediated signaling axis (Zhuang, Tian, Zhang, Wang, & Huang, 2018).
3.8. Flatr
Regulatory T cells (Tregs) are a specialized subpopulation of CD4+ T cells and are essential for maintaining self-tolerance (Benoist & Mathis, 2012; Corthay, 2009; Vignali, Collison, & Workman, 2008). Treg dysfunctions are associated with severe autoimmune diseases such as inflammatory bowel disease, multiple sclerosis, rheumatoid arthritis, colitis, and type 1 diabetes (Alroqi & Chatila, 2016; Buckner, 2010). However, excessive activation of these cells is also harmful because it inhibits beneficial anti-pathogens and anti-tumor immunity (Dannull, et al., 2005; Franceschini, et al., 2009). Recently, the Adrian Liston group identified a new lncRNA named Flatr (Foxp3-specific lncRNA of Treg expected), which is highly conserved and enriched in activated Treg cells (Brajic, et al., 2018). Jiang et al. found that Treg cells in HCC tumor areas specifically express lnc-EGFR, and further research found that lnc-EGFR stimulates the differentiation of Treg cells, thereby inhibiting CTL activity in the tumor area and enhancing the growth of HCC. (R. Jiang, et al., 2017). Furthermore, the lncRNA Flicr has been shown to negatively regulate the expression of FoxP3, which is a master regulator of Treg (Rudensky, 2011). Flicr modulates chromatin accessibility of the FoxP3 gene loci, leading to impaired Treg activity and autoimmunity (Zemmour, Pratama, Loughhead, Mathis, & Benoist, 2017).
3.9. Lnc-CHOP
Myeloid suppressor cells (MDSCs) produced from bone marrow-like progenitor cells in the bone marrow are one of the major components of the tumor microenvironment (Bronte, et al., 2016; Gabrilovich, 2017; Talmadge & Gabrilovich, 2013). These cells have become key regulators of immune response in cancer and other pathological conditions (Bronte, et al., 2016; Kumar, Patel, Tcyganov, & Gabrilovich, 2016; Talmadge & Gabrilovich, 2013; Ugel, De Sanctis, Mandruzzato, & Bronte, 2015). Gao et. al identified an uncharacterized lncRNA named lnc-CHOP. They found that lnc-CHOP may interact with CHOP and C / EBPβ isoforms to encourage C / EBPβ activation and regulate a large number of target transcripts in MDSC to control immunosuppressive function and MDSC differentiation in inflammatory and tumor environments (Gao, Wang, Li, Zhang, & Yang, 2018). MicroRNAs and other lncRNAs, including lnc-C/EBPbeta, Olfr29-ps1, Pvt1, MALAT1, and HOXA regulate the immunosuppressive activity of MDSC (Gao, Sun, et al., 2018; Shang, Gao, Tang, Zhang, & Yang, 2019; Shang, et al., 2017; Tian, et al., 2018; Zheng, et al., 2019; Q. Zhou, et al., 2018).
4. LncRNAs as Potential Targets for Immunotherapies
The tumor-specific upregulation and important functional roles of lncRNAs as oncogenes brings attention to considering lncRNAs as promising therapeutic targets in cancer (Arun, Diermeier, & Spector, 2018; M. C. Jiang, Ni, Cui, Wang, & Zhuo, 2019; Lavorgna, et al., 2016). Moreover, as previously mentioned, lncRNAs also act as tumor suppresser genes that regulate immune cell function and immune response in the TME. With this in mind, there are several strategies currently available in preclinical studies to assess the role of lncRNAs as targets of immunotherapy, including downregulating the oncogenic role of lncRNAs through ASO-based strategies, siRNAs, small molecule inhibitors, and CRISPR inhibition (CRISPRi) strategies, and upregulating the tumor suppresser role of lncRNAs via CRISPR activation (CRISPRa) strategies. We also proposed a combinatorial therapy with immune checkpoint antibodies treatments, which need to be evaluated for their effects on cancer immunotherapy (Figure 3).
Figure 3. Targeting lncRNAs as a cancer therapy.
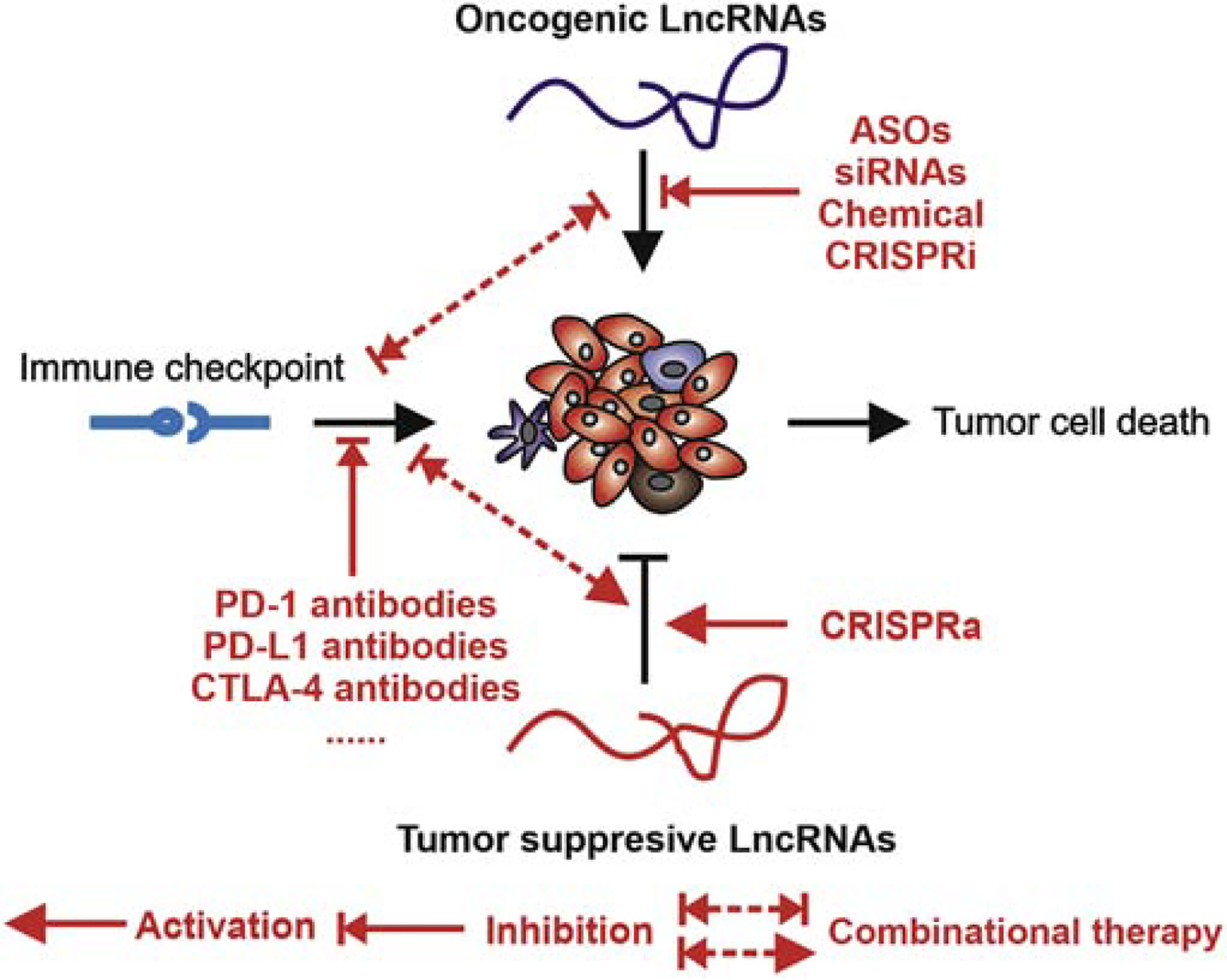
Targeting tumor-associated lncRNAs using ASO, siRNA, small molecular inhibitors, and gene editing methods.
4.1. ASO-based strategies
Anti-sense oligonucleotides (ASO), including ASO gapmers (Kole, Krainer, & Altman, 2012), duplex RNA (Williams & Fried, 1986), and locked nucleic acids (LNAs) (Vester & Wengel, 2004) bind to lncRNA transcripts via base-pairing. The RNA-DNA duplexes trigger RNase-H-dependent cleavage (Chan, Lim, & Wong, 2006). The new generation of ASOs utilizes chemical modification of the sugar backbone to improve binding affinity and stability in vivo (Geary, Norris, Yu, & Bennett, 2015), and ASOs have advanced to clinical trials (Hong, et al., 2015). The mixed LNA-DNA-LNA gapmers are base-paired to RNA targets and can be used to silence RNA targets in cell line-based experiments and animal models. The application of ASO to knock down lncRNAs in vivo has been investigated in a variety of cancer models and has been shown to have significant inhibitory effects on tumor growth and progression. For example, LNAs targeting PVT1 have been shown to significantly reduce cell proliferation, migration, and invasion (Iden, et al., 2016). A study found that lncARSR promotes the resistance of cancer cells to sunitinib and that administration of LNAs targeting lncARSR enhances the sensitivity of cancer cells to sunitinib (Qu, et al., 2016). Therapeutic delivery of locked nucleic acids (LNAs) targeting BCAR4, LINK-A, and MAYA strongly suppresses breast cancer growth and metastasis in mouse models (C. Li, et al., 2017; A. Lin, et al., 2016; Xing, et al., 2014). Systematic knockout of Malat1 using antisense oligonucleotides (ASO) in a MMTV (mouse mammary tumor virus) -PyMT mouse breast cancer model resulted in inhibited tumor growth with significantly increased cystic tumor differentiation and reduced metastasis (Arun, et al., 2016). In addition to its use in cancer, it has also been suggested that ASO could benefit patients with neuronal diseases and other human pathological conditions (Khorkova & Wahlestedt, 2017).
4.2. siRNAs
Using siRNAs targeting lncRNAs to inhibit the expression of these targets is a strategy that has been successfully applied to various preclinical models (Ozpolat, Sood, & Lopez-Berestein, 2010, 2014; Rupaimoole, et al., 2015). Recently, nanoliposomes based on oleylphosphatidylcholine (DOPC) have been developed for use with nucleotide-based therapeutics (siRNA, microRNA, lncRNA, antisense oligonucleotides, etc.) in vivo and clinical delivery (Ozpolat, et al., 2014). Studies have shown that DOPC-nanoliposomal siRNAs effectively inhibit the expression of target proteins in mouse tumors for about 3–5 days (Aslan, et al., 2015). DOPC nanoliposomes with an average size of 50 nm can be administered by a single intravenous or intraperitoneal injection to deliver the selected siRNA and anti-miR to tumor cells in vivo. This single administration significantly inhibits gene expression levels and reduces tumor size in both mouse models and preclinical models of human cancer, including subcutaneous xenografts and orthotopic tumor models (C. Lin & Yang, 2018; Ozpolat, et al., 2010, 2014). Other research has indicated that nanoparticle-encapsulated siRNAs deplete brain metastasis-associated lncRNAs in preclinical mouse models (S. Wang, et al., 2017).
4.3. Small Molecule Inhibitors
LncRNAs form complex tertiary structures (Jones & Sattler, 2019; Zampetaki, Albrecht, & Steinhofel, 2018). Whether the secondary or three-dimensional structure of lncRNAs is conserved among different species remains unclear (Rivas, Clements, & Eddy, 2017). RNA molecules are potential targets for small molecule inhibitors (Connelly, Moon, & Schneekloth, 2016). High-throughput screening has been used to identify small molecule compounds that may inhibit RNAs (Brustikova, et al., 2018; Lucas-Hourani, et al., 2014). For example, Howe et al. reported the discovery and characterization of riboprotein, which is a highly selective chemical regulator of bacterial riboflavin riboswitches and was identified in phenotypic screening as a natural ligand for Flavin single core. Structurally unique nucleotide mimics have been screened and characterized to inhibit bacterial growth and riboswitch-mediated ribB gene expression (Howe, et al., 2015). A platform has been established to screen and identify small molecule inhibitors that target non-coding RNAs; this platform will help with large-scale screening of drugs that target lncRNAs (Velagapudi, et al., 2017).
4.4. Gene editing to target lncRNAs
In recent years, with major breakthroughs in gene editing technology like CRISPR-Cas9, gene therapy has reignited people’s interest. New technologies continue to be used to treat animal models of disease, cell editing, and in vivo gene repair (Bortesi & Fischer, 2015; Nelson & Gersbach, 2016; H. Wang, La Russa, & Qi, 2016). In order to better utilize gene editing technology to treat human diseases, new delivery technologies are constantly being developed, such as liposomes, protein delivery, nanoparticles, adeno-associated virus delivery vectors, adenovirus delivery vectors, and lentivirus delivery vectors (Khan, 2019; Nelson & Gersbach, 2016). However, direct insertion / deletion terminated by a single double-strand break is unlikely to cause functional ablation of non-coding genes. Therefore, in terms of lncRNA function, a more comprehensive approach must be considered (Zhen & Li, 2019). Currently, nuclease-inactivated Cas9-based technologies have been reported to target the promoter sequence of a target gene to apply RNA-directed transcriptional regulation without the need to permanently modify the genome. Therefore, through activation (CRISPRa) or inhibition (CRISPRi), it could be possible to control lncRNA expression transiently or stably without altering the genomic sequence (Kampmann, 2018). For example, the CRISPRa-based method has been developed to screen important lncRNAs related to drug resistance. Through this method, it was found that a lncRNA named GAS6-AS2 promotes the activation of the GAS6/TAM pathway, which is a drug resistance mechanism for a variety of cancers, including AML (Bester, et al., 2018). To explore the function of lncRNAs in humans, a CRISPR interference (CRISPRi) platform has been developed to screen growth-regulating lncRNA genes in seven cell types (S. J. Liu, et al., 2017).
4.5. Combinational treatment
As previously mentioned, there is strong evidence that lncRNAs play an important role in cancer drug resistance and cancer immunotherapy resistance. Therefore, the use of targeted lncRNA drugs in combination with chemotherapeutic drugs or immunotherapy antibodies could be an effective strategy for treating cancer (Y. Zhou, et al., 2019). In recent years, breakthroughs in the field of tumor immunotherapy (mainly in the form of immune checkpoint drugs such as PD-1, PD-L1 and CTLA-4 antibodies) have greatly improved the effectiveness of tumor treatments. Nevertheless, only a small percentage of people benefit from tumor immunotherapy, and the majority of patients develop primary or secondary resistance following treatment. Recent studies suggest that many factors affect the efficiency of immune checkpoint therapy, including antigen presentation, tumor mutational burden, and T cell infiltration (Sharma, et al., 2017). Recently, some tumor-associated lncRNAs have been found to mediate tumor cell immune system evasion by inhibiting antigen presentation and T cell infiltration. For example, LINK-A affects breast cancer cell MHC-1 stability, and NKILA affects CTL infiltration (Huang, et al., 2018). Therefore, we believe that some lncRNAs that lead to immune checkpoint antibody therapy resistance can be effectively utilized in combinatorial therapies by utilizing drugs targeting the lncRNAs together with immune checkpoint drugs (particularly PD1, PD-L1, and CTLA-4 antibodies). For example, treatment with LINK-A LNAs in combination with immune checkpoint blockers exhibited synergistic efficacy in repressing breast tumor growth (Q. Hu, et al., 2019). It is possible that a combinatorial treatment that considers lncRNA ASOs or LNAs together with immune checkpoint inhibitors could exhibit synergistic effects on anti-tumor immunity.
Concluding Remarks and Future Perspectives
Ultimately, the role of lncRNAs as therapeutic targets against cancer requires further study. Although emerging evidence has indicated the importance of lncRNAs in regulating the immune escape of tumor cells, this is only the tip of the iceberg. How lncRNAs affect the tumor microenvironment and regulate the function of tumor immune cells needs more research. In terms of clinical application prospects, lncRNAs with tissue-specific expression features are potential targets for cancer immunotherapy.
Future studies of the regulatory roles of lncRNAs in cancer immunotherapy will define the future of the field. Although numerous lncRNAs have been identified so far, it has been challenging to demonstrate the functional relevance of lncRNAs in cancer immunotherapy. To answer this problem, single cell sequencing, CyToF, and cellular and xenograft models have been the commonly used to study the roles that lncRNAs play in cancer and are useful tools in cursory evaluations of their functions. However, conclusions that are more definitive will require representative in vivo models of cancer, such as genetic models and patient-Derived xenograft (PDX) models using humanized mice that better characterize the tumor microenvironment. It is critical to determine whether tissue-specific expression of lncRNAs modulates the immune microenvironment and whether organ-specific delivery of ASOs targeting lncRNAs demonstrates effective anti-tumor immunity.
We believe that with the rise of single-cell sequencing technology and the large number of clinical trials involving cancer immunotherapy, numerous lncRNAs with tissue- and cell-type specificities will be identified. Therefore, a clear understanding of the mechanisms of lncRNAs as tumor-cell-intrinsic and tumor-cell-extrinsic factors will be greatly beneficial to expanding current cancer therapy strategies. How to develop safe and effective targeted therapies against tumor-associated lncRNAs and effectively deliver these drugs to tumors will be the primary tasks of this field.
Acknowledgements
This work was supported by C.R.L.’s NIH R01 awards (1 R01 CA218025-01, 1R01CA231011-01), CPRIT individual investigator research award (180259), and DoD Breakthrough award BC180196, as well as L.Q.Y.’s NIH R01 award (1 R01 CA218036-01), DoD Breakthrough awards (BC151465, BC181384) grant, Andrew Sabin Family Foundation Fellows award, AACR-Bayer Innovation and Discovery Grant (18-80-44) and CPRIT individual investigator research award (200423).
Abbreviations
- LncRNAs
Long noncoding RNAs
- TME
tumor microenvironment
- SNPs
single-nucleotide polymorphisms
- MHC
major histocompatibility complex
- B2M
beta-2-microglobulin
- MAPK
mitogen-activated protein kinase
- IFNγ
interferon-gamma
- PD-L1
Programmed death-ligand 1
- LINK-A
Long intergenic non-coding RNA for kinase activation
- GPCRs
G protein-coupled receptors
- PLC
peptide-loading complex
- LNAs
Locked Nucleic Acids
- APCs
antigen-presenting cell
- TNBC
Triple-negative breast cancer
- HOTAIR
HOX transcript antisense intergenic RNA
- PRC2
Polycomb Repressive Complex 2
- MALAT1
Metastasis Associated Lung Adenocarcinoma Transcript 1
- MDSCs
Myeloid-derived suppressor cells
- SWI/SNF
switch/sucrose nonfermentable
- LPS
Lipopolysaccharides
- IL-4
interleukin 4
- IL-13
interleukin 13
- HCC
hepatocellular carcinoma
- DCs
dendritic cells
- NK
natural killer
- CTLs
cytotoxic T lymphocytes
- Morrbid
myeloid RNA repressor of BCL2L11 induced death
- Th
T helper cell
- STAT3
signal transducer and transcriptional activator 3
- Tregs
Regulatory T cells
- Flatr
Foxp3-specific lncRNA of Treg expected
- ASO
Anti-sense oligonucleotides
- DOPC
nanoliposomes based on oleylphosphatidylcholine
- PDX
patient-Derived xenograft.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors have no conflicts of interest to disclose.
References:
- Ahmed AU, Williams BR, & Hannigan GE (2015). Transcriptional Activation of Inflammatory Genes: Mechanistic Insight into Selectivity and Diversity. Biomolecules, 5, 3087–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alroqi FJ, & Chatila TA (2016). T Regulatory Cell Biology in Health and Disease. Curr Allergy Asthma Rep, 16, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arun G, Diermeier S, Akerman M, Chang KC, Wilkinson JE, Hearn S, Kim Y, MacLeod AR, Krainer AR, Norton L, Brogi E, Egeblad M, & Spector DL (2016). Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev, 30, 34–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arun G, Diermeier SD, & Spector DL (2018). Therapeutic Targeting of Long Non-Coding RNAs in Cancer. Trends Mol Med, 24, 257–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslan B, Monroig P, Hsu MC, Pena GA, Rodriguez-Aguayo C, Gonzalez-Villasana V, Rupaimoole R, Nagaraja AS, Mangala S, Han HD, Yuca E, Wu SY, Ivan C, Moss TJ, Ram PT, Wang H, Gol-Chambers A, Ozkayar O, Kanlikilicer P, Fuentes-Mattei E, Kahraman N, Pradeep S, Ozpolat B, Tucker S, Hung MC, Baggerly K, Bartholomeusz G, Calin G, Sood AK, & Lopez-Berestein G (2015). The ZNF304-integrin axis protects against anoikis in cancer. Nat Commun, 6, 7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atianand MK, Caffrey DR, & Fitzgerald KA (2017). Immunobiology of Long Noncoding RNAs. Annu Rev Immunol, 35, 177–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, & Chang HY (2013). Long noncoding RNAs: cellular address codes in development and disease. Cell, 152, 1298–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoist C, & Mathis D (2012). Treg cells, life history, and diversity. Cold Spring Harb Perspect Biol, 4, a007021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, Mc Henry KT, Pinchback RM, Ligon AH, Cho YJ, Haery L, Greulich H, Reich M, Winckler W, Lawrence MS, Weir BA, Tanaka KE, Chiang DY, Bass AJ, Loo A, Hoffman C, Prensner J, Liefeld T, Gao Q, Yecies D, Signoretti S, Maher E, Kaye FJ, Sasaki H, Tepper JE, Fletcher JA, Tabernero J, Baselga J, Tsao MS, Demichelis F, Rubin MA, Janne PA, Daly MJ, Nucera C, Levine RL, Ebert BL, Gabriel S, Rustgi AK, Antonescu CR, Ladanyi M, Letai A, Garraway LA, Loda M, Beer DG, True LD, Okamoto A, Pomeroy SL, Singer S, Golub TR, Lander ES, Getz G, Sellers WR, & Meyerson M (2010). The landscape of somatic copy-number alteration across human cancers. Nature, 463, 899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bester AC, Lee JD, Chavez A, Lee YR, Nachmani D, Vora S, Victor J, Sauvageau M, Monteleone E, Rinn JL, Provero P, Church GM, Clohessy JG, & Pandolfi PP (2018). An Integrated Genome-wide CRISPRa Approach to Functionalize lncRNAs in Drug Resistance. Cell, 173, 649–664 e620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian Y, Yang L, Zhang B, Li W, Wang S, Jiang S, Chen X, Li W, & Zeng L (2019). LincRNA Cox-2 Regulates Lipopolysaccharide-Induced Inflammatory Response of Human Peritoneal Mesothelial Cells via Modulating miR-21/NF-kappaB Axis. Mediators Inflamm, 2019, 8626703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian Y, Yang L, Zhang B, Li W, Wang S, Jiang S, Chen X, & Zeng L (2019). LincRNA Cox-2 Regulates Lipopolysaccharide-Induced Inflammatory Response of Human Peritoneal Mesothelial Cells via Modulating miR-21/NF-kappaB Axis. Mediators Inflamm, 2019, 8626703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortesi L, & Fischer R (2015). The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol Adv, 33, 41–52. [DOI] [PubMed] [Google Scholar]
- Bottcher JP, & Reis ESC (2018). The Role of Type 1 Conventional Dendritic Cells in Cancer Immunity. Trends Cancer, 4, 784–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brajic A, Franckaert D, Burton O, Bornschein S, Calvanese AL, Demeyer S, Cools J, Dooley J, Schlenner S, & Liston A (2018). The Long Non-coding RNA Flatr Anticipates Foxp3 Expression in Regulatory T Cells. Front Immunol, 9, 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher P (2019). On Analyzing How the Th1/Th2 Phenotype of an Immune Response Is Determined: Classical Observations Must Not Be Ignored. Front Immunol, 10, 1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronte V, Brandau S, Chen SH, Colombo MP, Frey AB, Greten TF, Mandruzzato S, Murray PJ, Ochoa A, Ostrand-Rosenberg S, Rodriguez PC, Sica A, Umansky V, Vonderheide RH, & Gabrilovich DI (2016). Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat Commun, 7, 12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brustikova K, Sedlak D, Kubikova J, Skuta C, Solcova K, Malik R, Bartunek P, & Svoboda P (2018). Cell-Based Reporter System for High-Throughput Screening of MicroRNA Pathway Inhibitors and Its Limitations Front Genet, 9, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner JH (2010). Mechanisms of impaired regulation by CD4(+)CD25(+)FOXP3(+) regulatory T cells in human autoimmune diseases. Nat Rev Immunol, 10, 849–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, & Rinn JL (2011). Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev, 25, 1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai B, Song XQ, Cai JP, & Zhang S (2014). HOTAIR: a cancer-related long non-coding RNA. Neoplasma, 61, 379–391. [DOI] [PubMed] [Google Scholar]
- Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, Byron M, Monks B, Henry-Bezy M, Lawrence JB, O’Neill LA, Moore MJ, Caffrey DR, & Fitzgerald KA (2013). A long noncoding RNA mediates both activation and repression of immune response genes. Science, 341, 789–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JH, Lim S, & Wong WS (2006). Antisense oligonucleotides: from design to therapeutic application. Clin Exp Pharmacol Physiol, 33, 533–540. [DOI] [PubMed] [Google Scholar]
- Chen DL, & Xu RH (2018). The emerging role of long non-coding RNAs in the drug resistance of colorectal cancer. Int J Clin Exp Pathol, 11, 4735–4743. [PMC free article] [PubMed] [Google Scholar]
- Chiossone L, Dumas PY, Vienne M, & Vivier E (2018). Natural killer cells and other innate lymphoid cells in cancer. Nat Rev Immunol, 18, 671–688. [DOI] [PubMed] [Google Scholar]
- Cohen PA, Peng L, Plautz GE, Kim JA, Weng DE, & Shu S (2000). CD4+ T cells i n adoptive immunotherapy and the indirect mechanism of tumor rejection. Crit Rev Immunol, 20, 17–56. [PubMed] [Google Scholar]
- Connelly CM, Moon MH, & Schneekloth JS Jr. (2016). The Emerging Role of RNA as a Therapeutic Target for Small Molecules. Cell Chem Biol, 23, 1077–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrales L, Matson V, Flood B, Spranger S, & Gajewski TF (2017). Innate immune signaling and regulation in cancer immunotherapy. Cell Res, 27, 96–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corthay A (2009). How do regulatory T cells work? Scand J Immunol, 70, 326–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristescu R, Mogg R, Ayers M, Albright A, Murphy E, Yearley J, Sher X, Liu XQ, Lu H, Nebozhyn M, Zhang C, Lunceford JK, Joe A, Cheng J, Webber AL, Ibrahim N, Plimack ER, Ott PA, Seiwert TY, Ribas A, McClanahan TK, Tomassini JE, Loboda A, & Kaufman D (2018). Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science, 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannull J, Su Z, Rizzieri D, Yang BK, Coleman D, Yancey D, Zhang A, Dahm P, Chao N, Gilboa E, & Vieweg J (2005). Enhancement of vaccine-mediated antitumor immunity in cancer patients after depletion of regulatory T cells. J Clin Invest, 115, 3623–3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Los Santos MC, Dragomir MP, & Calin GA (2019). The role of exosomal long non-coding RNAs in cancer drug resistance. Cancer Drug Resist, 2, 1178–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demoulin S, Herfs M, Delvenne P, & Hubert P (2013). Tumor microenvironment converts plasmacytoid dendritic cells into immunosuppressive/tolerogenic cells: insight into the molecular mechanisms. J Leukoc Biol, 93, 343–352. [DOI] [PubMed] [Google Scholar]
- Denaro N, Merlano MC, & Lo Nigro C (2019). Long noncoding RNAs as regulators of cancer i mmunity. Mol Oncol, 13, 61–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, & Flavell RA (2000). Cell fate decision: T-helper 1 and 2 subsets in immune responses. Arthritis Res, 2, 179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissmann M, Gutschner T, Hammerle M, Gunther S, Caudron-Herger M, Gross M, Schirmacher P, Rippe K, Braun T, Zornig M, & Diederichs S (2012). Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol, 9, 1076–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elling R, Robinson EK, Shapleigh B, Liapis SC, Covarrubias S, Katzman S, Groff AF, Jiang Z, Agarwal S, Motwani M, Chan J, Sharma S, Hennessy EJ, FitzGerald GA, McManus MT, Rinn JL, Fitzgerald KA, & Carpenter S (2018). Genetic Models Reveal cis and trans Immune-Regulatory Activities for lincRNA-Cox2. Cell Rep, 25, 1511–1524 e1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallarino F, Grohmann U, Bianchi R, Vacca C, Fioretti MC, & Puccetti P (2000). Th1 and Th2 cell clones to a poorly immunogenic tumor antigen initiate CD8+ T cell -dependent tumor eradication in vivo. J Immunol, 165, 5495–5501. [DOI] [PubMed] [Google Scholar]
- Fang P, Xiang L, Chen W, Li S, Huang S, Li J, Zhuge L, Jin L, Feng W, Chen Y, & Pan C (2019). LncRN A GAS5 enhanced the killing effect of NK cell on liver cancer through regulating miR-544/RUNX3. Innate Immun, 25, 99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares CM, Van Allen EM, Drake CG, Allison JP, & Hu-Lieskovan S (2019). Mechanisms of Resistance to Immune Checkpoint Blockade: Why Does Checkpoint Inhibitor Immunotherapy Not Work for All Patients? Am Soc Clin Oncol Educ Book, 39, 147–164. [DOI] [PubMed] [Google Scholar]
- Farhood B, Najafi M, & Mortezaee K (2019). CD8(+) cytotoxic T lymphocytes in cancer immunotherapy: A review. J Cell Physiol, 234, 8509–8521. [DOI] [PubMed] [Google Scholar]
- Fatica A, & Bozzoni I (2014). Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet, 15, 7–21. [DOI] [PubMed] [Google Scholar]
- Franceschini D, Paroli M, Francavilla V, Videtta M, Morrone S, Labbadia G, Cerino A, Mondelli MU, & Barnaba V (2009). PD-L1 negatively regulates CD4+CD25+Foxp3+ Tregs by limiting STAT-5 phosphorylation in patients chronically infected with HCV. J Clin Invest, 119, 551–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisaka Y, Iwata T, Tamai K, Nakamura M, Mochizuki M, Shibuya R, Yamaguchi K, Shimosegawa T, & Satoh K (2018). Long non-coding RNA HOTAIR up-regulates chemokine (C-C motif) ligand 2 and promotes proliferation of macrophages and myeloid-derived suppressor cells in hepatocellular carcinoma cell lines. Oncol Lett, 15, 509–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrilovich DI (2017). Myeloid-Derived Suppressor Cells. Cancer Immunol Res, 5, 3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Sun W, Shang W, Li Y, Zhang D, Wang T, Zhang X, Zhang S, Zhang Y, & Yang R (2018). Lnc-C/EBPbeta Negatively Regulates the Suppressive Function of Myeloid-Derived Suppressor Cells. Cancer Immunol Res, 6, 1352–1363. [DOI] [PubMed] [Google Scholar]
- Gao Y, Wang T, Li Y, Zhang Y, & Yang R (2018). Lnc-chop Promotes Immunosuppressive Function of Myeloid-Derived Suppressor Cells in Tumor and Inflammatory Environments. J Immunol, 200, 2603–2614. [DOI] [PubMed] [Google Scholar]
- Geary RS, Norris D, Yu R, & Bennett CF (2015). Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv Drug Deliv Rev, 87, 46–51. [DOI] [PubMed] [Google Scholar]
- Gubin MM, Zhang X, Schuster H, Caron E, Ward JP, Noguchi T, Ivanova Y, Hundal J, Arthur CD, Krebber WJ, Mulder GE, Toebes M, Vesely MD, Lam SS, Korman AJ, Allison JP, Freeman GJ, Sharpe AH, Pearce EL, Schumacher TN, Aebersold R, Rammensee HG, Melief CJ, Mardis ER, Gillanders WE, Artyomov MN, & Schreiber RD (2014). Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature, 515, 577–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, & Chang HY (2010). Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature, 464, 1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajjari M, & Salavaty A (2015). HOTAIR: an oncogenic long non-coding RNA in different cancers. Cancer Biol Med, 12, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao NB, Lu MH, Fan YH, Cao YL, Zhang ZR, & Yang SM (2012). Ma crophages in tumor microenvironments and the progression of tumors. Clin Dev Immunol, 2012, 948098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong D, Kurzrock R, Kim Y, Woessner R, Younes A, Nemunaitis J, Fowler N, Zhou T, Schmidt J, Jo M, Lee SJ, Yamashita M, Hughes SG, Fayad L, Piha-Paul S, Nadella MV, Mohseni M, Lawson D, Reimer C, Blakey DC, Xiao X, Hsu J, Revenko A, Monia BP, & MacLeod AR (2015). AZD9150, a next-generation antisense oligonucleotide inhibitor of STAT3 with early evidence of clinical activity in lymphoma and lung cancer. Sci Transl Med, 7, 314ra185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe JA, Wang H, Fischmann TO, Balibar CJ, Xiao L, Galgoci AM, Malinverni JC, Mayhood T, Villafania A, Nahvi A, Murgolo N, Barbieri CM, Mann PA, Carr D, Xia E, Zuck P, Riley D, Painter RE, Walker SS, Sherborne B, de Jesus R, Pan W, Plotkin MA, Wu J, Rindgen D, Cummings J, Garlisi CG, Zhang R, Sheth PR, Gill CJ, Tang H, & Roemer T (2015). Selective small-molecule inhibition of an RNA structural element. Nature, 526, 672–677. [DOI] [PubMed] [Google Scholar]
- Hu G, Gong AY, Wang Y, Ma S, Chen X, Chen J, Su CJ, Shibata A, Strauss-Soukup JK, Drescher KM, & Chen XM (2016). LincRNA-Cox2 Promotes Late Inflammatory Gene Transcription in Macrophages through Modulating SWI/SNF-Mediated Chromatin Remodeling. J Immunol, 196, 2799–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Ye Y, Chan LC, Li Y, Liang K, Lin A, Egranov SD, Zhang Y, Xia W, Gong J, Pan Y, Chatterjee SS, Yao J, Evans KW, Nguyen TK, Park PK, Liu J, Coarfa C, Donepudi SR, Putluri V, Putluri N, Sreekumar A, Ambati CR, Hawke DH, Marks JR, Gunaratne PH, Caudle AS, Sahin AA, Hortobagyi GN, Meric-Bernstam F, Chen L, Yu D, Hung MC, Curran MA, Han L, Lin C, & Yang L (2019). Oncogenic lncRNA downregulates cancer cell antigen presentation and intrinsic tumor suppression. Nat Immunol, 20, 835–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Chen J, Yang L, Ouyang Q, Li J, Lao L, Zhao J, Liu J, Lu Y, Xing Y, Chen F, Su F, Yao H, Liu Q, Su S, & Song E (2018). NKILA lncRNA promotes tumor immune evasion by sensitizing T cells to activation-induced cell death. Nat Immunol, 19, 1112–1125. [DOI] [PubMed] [Google Scholar]
- Huarte M (2015). The emerging role of lncRNAs in cancer. Nat Med, 21, 1253–1261. [DOI] [PubMed] [Google Scholar]
- Hung K, Hayashi R, Lafond-Walker A, Lowenstein C, Pardoll D, & Levitsky H (1998). The central role of CD4(+) T cells in the antitumor immune response. J Exp Med, 188, 2357–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iden M, Fye S, Li K, Chowdhury T, Ramchandran R, & Rader JS (2016). The lncRNA PVT1 Contributes to the Cervical Cancer Phenotype and Associates with Poor Patient Prognosis. PLoS One, 11, e0156274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorgulescu JB, Braun D, Oliveira G, Keskin DB, & Wu CJ (2018). Acquired mechanisms of immune escape in cancer following immunotherapy. Genome Med, 10, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeway CA Jr., & Bottomly K (1994). Signals and signs for lymphocyte responses. Cell, 76, 275–285. [DOI] [PubMed] [Google Scholar]
- Jendrzejewski J, He H, Radomska HS, Li W, Tomsic J, Liyanarachchi S, Davuluri RV, Nagy R, & de la Chapelle A (2012). The polymorphism rs944289 predisposes to papillary thyroid carcinoma through a large intergenic noncoding RNA gene of tumor suppressor type. Proc Natl Acad Sci U S A, 109, 8646–8651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, Thomas M, Berdel WE, Serve H, & Muller-Tidow C (2003). MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene, 22, 8031–8041. [DOI] [PubMed] [Google Scholar]
- Jia H, Truica CI, Wang B, Wang Y, Ren X, Harvey HA, Song J, & Yang JM (2017). Immunotherapy for triple-negative breast cancer: Existing challenges and exciting prospects. Drug Resist Updat, 32, 1–15. [DOI] [PubMed] [Google Scholar]
- Jiang MC, Ni JJ, Cui WY, Wang BY, & Zhuo W (2019). Emerging roles of lncRNA in cancer and therapeutic opportunities. Am J Cancer Res, 9, 1354–1366. [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Tang J, Chen Y, Deng L, Ji J, Xie Y, Wang K, Jia W, Chu WM, & Sun B (2017). The long noncoding RNA lnc-EGFR stimulates T-regulatory cells differentiation thus promoting hepatocellular carcinoma immune evasion. Nat Commun, 8, 15129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Xia J, Xie S, Zou R, Pan S, Wang ZW, Assaraf YG, & Zhu X (2020). Long non-coding RNAs as a determinant of cancer drug resistance: Towards the overcoming of chemoresistance via modulation of lncRNAs. Drug Resist Updat, 50, 100683. [DOI] [PubMed] [Google Scholar]
- Jones AN, & Sattler M (2019). Challenges and perspectives for structural biology of lncRNAs -the example of the Xist lncRNA A-repeats. J Mol Cell Biol, 11, 845–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalams SA, & Walker BD (1998). The critical need for CD4 help in maintaining effective cytotoxic T lymphocyte responses. J Exp Med, 188, 2199–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampmann M (2018). CRISPRi and CRISPRa Screens in Mammalian Cells for Precision Biology and Medicine. ACS Chem Biol, 13, 406–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan SH (2019). Genome-Editing Technologies: Concept, Pros, and Cons of Various Genome-Editing Techniques and Bioethical Concerns for Clinical Application. Mol Ther Nucleic Acids, 16, 326–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khorkova O, & Wahlestedt C (2017). Oligonucleotide therapies for disorders of the nervous system. Nat Biotechnol, 35, 249–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson KL, & Disis ML (2005). Tumor antigen-specific T helper cells in cancer immunity and immunotherapy. Cancer Immunol Immunother, 54, 721–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogure T, Yan IK, Lin WL, & Patel T (2013). Extracellular Vesicle-Mediated Transfer of a Novel Long Noncoding RNA TUC339: A Mechanism of Intercellular Signaling in Human Hepatocellular Cancer. Genes Cancer, 4, 261–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kole R, Krainer AR, & Altman S (2012). RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat Rev Drug Discov, 11, 125–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotzin JJ, Iseka F, Wright J, Basavappa MG, Clark ML, Ali MA, Abdel-Hakeem MS, Robertson TF, Mowel WK, Joannas L, Neal VD, Spencer SP, Syrett CM, Anguera MC, Williams A, Wherry EJ, & Henao-Mejia J (2019). The long noncoding RNA Morrbid regulates CD8 T cells in response to viral infection. Proc Natl Acad Sci U S A, 116, 11916–11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotzin JJ, Spencer SP, McCright SJ, Kumar DBU, Collet MA, Mowel WK, Elliott EN, Uyar A, Makiya MA, Dunagin MC, Harman CCD, Virtue AT, Zhu S, Bailis W, Stein J, Hughes C, Raj A, Wherry EJ, Goff LA, Klion AD, Rinn JL, Williams A, Flavell RA, & Henao-Mejia J (2016). The long non-coding RNA Morrbid regulates Bim and short-lived myeloid cell lifespan. Nature, 537, 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Patel S, Tcyganov E, & Gabrilovich DI (2016). Th e Nature of Myeloid-Derived Suppressor Cells in the Tumor Microenvironment. Trends Immunol, 37, 208–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavorgna G, Vago R, Sarmini M, Montorsi F, Salonia A, & Bellone M (2016). Long non-coding RNAs as novel therapeutic targets in cancer. Pharmacol Res, 110, 131–138. [DOI] [PubMed] [Google Scholar]
- Li C, Wang S, Xing Z, Lin A, Liang K, Song J, Hu Q, Yao J, Chen Z, Park PK, Hawke DH, Zhou J, Zhou Y, Zhang S, Liang H, Hung MC, Gallick GE, Han L, Lin C, & Yang L (2017). A ROR1-HER3-lncRNA signalling axis modulates the Hippo-YAP pathway to regulate bone metastasis. Nat Cell Biol, 19, 106–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Lei Y, Wu M, & Li N (2018). Regulation of Macrophage Activation and Polarization by HCC-Derived Exosomal lncRNA TUC339. Int J Mol Sci, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A, Hu Q, Li C, Xing Z, Ma G, Wang C, Li J, Ye Y, Yao J, Liang K, Wang S, Park PK, Marks JR, Zhou Y, Zhou J, Hung MC, Liang H, Hu Z, Shen H, Hawke DH, Han L, Zhou Y, Lin C, & Yang L (2017). The LINK-A lncRNA interacts with PtdIns(3,4,5)P3 to hyperactivate AKT and confer resistance to AKT inhibitors. Nat Cell Biol, 19, 238–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A, Li C, Xing Z, Hu Q, Liang K, Han L, Wang C, Hawke DH, Wang S, Zhang Y, Wei Y, Ma G, Park PK, Zhou J, Zhou Y, Hu Z, Zhou Y, Marks JR, Liang H, Hung MC, Lin C, & Yang L (2016). The LINK-A lncRNA activates normoxic HIF1alpha signalling in triple-negative breast cancer. Nat Cell Biol, 18, 213–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, & Yang L (2018). Long Noncoding RNA in Cancer: Wiring Signaling Circuitry. Trends Cell Biol, 28, 287–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Gao L, Ma X, Huang JJ, Chen J, Zeng L, Ashby CR Jr., Zou C, & Chen ZS (2020). Long non-coding RNAs regulate drug resistance in cancer. Mol Cancer, 19, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SJ, Horlbeck MA, Cho SW, Birk HS, Malatesta M, He D, Attenello FJ, Villalta JE, Cho MY, Chen Y, Mandegar MA, Olvera MP, Gilbert LA, Conklin BR, Chang HY, Weissman JS, & Lim DA (2017). CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science, 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SJ, Nowakowski TJ, Pollen AA, Lui JH, Horlbeck MA, Attenello FJ, He D, Weissman JS, Kriegstein AR, Diaz AA, & Lim DA (2016). Single-cell analysis of long non-coding RNAs in the developing human neocortex. Genome Biol, 17, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Liu X, Luo M, Luo Q, Tao H, Wu D, Lu S, Jin J, Zhao Y, & Zou L (2017). dNK derived IFN-gamma mediates VSMC migration and apoptosis via the induction of LncRNA MEG3: A role in uterovascular transformation. Placenta, 50, 32–39. [DOI] [PubMed] [Google Scholar]
- Loewen G, Jayawickramarajah J, Zhuo Y, & Shan B (2014). Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol, 7, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Wang Y, Chen D, Liu J, & Jiao W (2018). Potential clinical application of lncRNAs in non-small cell lung cancer. Onco Targets Ther, 11, 8045–8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas-Hourani M, Munier-Lehmann H, Helynck O, Komarova A, Despres P, Tangy F, & Vidalain PO (2014). High-throughput screening for broad-spectrum chemical inhibitors of RNA viruses. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai H, Zhou B, Liu L, Yang F, Conran C, Ji Y, Hou J, & Jiang D (2019). Molecular pattern of lncRNAs in hepatocellular carcinoma. J Exp Clin Cancer Res, 38, 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majidinia M, & Yousefi B (2016). Long non-coding RNAs in cancer drug resistance development. DNA Repair (Amst), 45, 25–33. [DOI] [PubMed] [Google Scholar]
- Marincola FM, Jaffee EM, Hicklin DJ, & Ferrone S (2000). Escape of human solid tumors from T-cell recognition: molecular mechanisms and functional significance. Adv Immunol, 74, 181–273. [DOI] [PubMed] [Google Scholar]
- Martinez FO, & Gordon S (2014). The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep, 6, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathy NW, & Chen XM (2017). Long non-coding RNAs (lncRNAs) and their transcriptional control of inflammatory responses. J Biol Chem, 292, 12375–12382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano MT, Wang H, Kutyavin T, & Stamatoyannopoulos JA (2012). Widespread site-dependent buffering of human regulatory polymorphism. PLoS Genet, 8, e1002599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehra M, & Chauhan R (2017). Long Noncoding RNAs as a Key Player in Hepatocellular Carcinoma. Biomark Cancer, 9, 1179299X17737301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton C, Reuter JA, Spacek DV, & Snyder M (2015). Recurrent somatic mutations in regulatory regions of human cancer genomes. Nat Genet, 47, 710–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller DL, Jenkins MK, & Schwartz RH (1989). Clonal expansion versus functional clonal inactivation: a costimulatory signalling pathway determines the outcome of T cell antigen receptor occupancy. Annu Rev Immunol, 7, 445–480. [DOI] [PubMed] [Google Scholar]
- Nelson CE, & Gersbach CA (2016). Engineering Delivery Vehicles for Genome Editing. Annu Rev Chem Biomol Eng, 7, 637–662. [DOI] [PubMed] [Google Scholar]
- Noy R, & Pollard JW (2014). Tumor-associated macrophages: from mechanisms to therapy. Immunity, 41, 49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien A, Zhou T, Tan C, Alpini G, & Glaser S (2019). Role of Non-Coding RNAs in the Progression of Liver Cancer: Evidence from Experimental Models. Cancers (Basel), 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozpolat B, Sood AK, & Lopez-Berestein G (2010). Nanomedicine based approaches for the delivery of siRNA in cancer. J Intern Med, 267, 44–53. [DOI] [PubMed] [Google Scholar]
- Ozpolat B, Sood AK, & Lopez-Berestein G (2014). Liposomal siRNA nanocarriers for cancer therapy. Adv Drug Deliv Rev, 66, 110–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan JJ, Xie XJ, Li X, & Chen W (2015). Long Non-coding RNAs and Drug Resistance. Asian Pac J Cancer Prev, 16, 8067–8073. [DOI] [PubMed] [Google Scholar]
- Pathria P, Louis TL, & Varner JA (2019). Targeting Tumor-Associated Macrophages in Cancer. Trends Immunol, 40, 310–327. [DOI] [PubMed] [Google Scholar]
- Perkel JM (2013). Visiting “noncodarnia”. Biotechniques, 54, 301, 303–304. [DOI] [PubMed] [Google Scholar]
- Perry RB, & Ulitsky I (2016). The functions of long noncoding RNAs in development and stem cells. Development, 143, 3882–3894. [DOI] [PubMed] [Google Scholar]
- Qu L, Ding J, Chen C, Wu ZJ, Liu B, Gao Y, Chen W, Liu F, Sun W, Li XF, Wang X, Wang Y, Xu ZY, Gao L, Yang Q, Xu B, Li YM, Fang ZY, Xu ZP, Bao Y, Wu DS, Miao X, Sun HY, Sun YH, Wang HY, & Wang LH (2016). Exosome-Transmitted lncARSR Promotes Sunitinib Resistance in Renal Cancer by Acting as a Competing Endogenous RNA. Cancer Cell, 29, 653–668. [DOI] [PubMed] [Google Scholar]
- Ranzani V, Rossetti G, Panzeri I, Arrigoni A, Bonnal RJ, Curti S, Gruarin P, Provasi E, Sugliano E, Marconi M, De Francesco R, Geginat J, Bodega B, Abrignani S, & Pagani M (2015). The long intergenic noncoding RNA landscape of human lymphocytes highlights the regulation of T cell differentiation by linc-MAF-4. Nat Immunol, 16, 318–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasi T, Suzuki H, Pang KC, Katayama S, Furuno M, Okunishi R, Fukuda S, Ru K, Frith MC, Gongora MM, Grimmond SM, Hume DA, Hayashizaki Y, & Mattick JS (2006). Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res, 16, 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter JA, Spacek DV, & Snyder MP (2015). High-throughput sequencing technologies. Mol Cell, 58, 586–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas A (2012). Tumor immunotherapy directed at PD-1. N Engl J Med, 366, 2517–2519. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, & Chang HY (2007). Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell, 129, 1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas E, Clements J, & Eddy SR (2017). A statistical test for conserved RNA structure shows lack of evidence for structure in lncRNAs. Nat Methods, 14, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudensky AY (2011). Regulatory T cells and Foxp3. Immunol Rev, 241, 260–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupaimoole R, Lee J, Haemmerle M, Ling H, Previs RA, Pradeep S, Wu SY, Ivan C, Ferracin M, Dennison JB, Millward NMZ, Nagaraja AS, Gharpure KM, McGuire M, Sam N, Armaiz-Pena GN, Sadaoui NC, Rodriguez-Aguayo C, Calin GA, Drapkin RI, Kovacs J, Mills GB, Zhang W, Lopez-Berestein G, Bhattacharya PK, & Sood AK (2015). Long Noncoding RNA Ceruloplasmin Promotes Cancer Growth by Altering Glycolysis. Cell Rep, 13, 2395–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salviano-Silva A, Lobo-Alves SC, Almeida RC, Malheiros D, & Petzl-Erler ML (2018). Besides Pathology: Long Non-Coding RNA in Cell and Tissue Homeostasis. Noncoding RNA, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang W, Gao Y, Tang Z, Zhang Y, & Yang R (2019). The Pseudogene Olfr29-ps1 Promotes the Suppressive Function and Differentiation of Monocytic MDSCs. Cancer Immunol Res, 7, 813–827. [DOI] [PubMed] [Google Scholar]
- Shang W, Tang Z, Gao Y, Qi H, Su X, Zhang Y, & Yang R (2017). LncRNA RNCR3 promotes Chop expression by sponging miR-185–5p during MDSC differentiation. Oncotarget, 8, 111754–111769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Hu-Lieskovan S, Wargo JA, & Ribas A (2017). Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell, 168, 707–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimasaki N, Jain A, & Campana D (2020). NK cells for cancer immunotherapy. Nat Rev Drug Discov, 19, 200–218. [DOI] [PubMed] [Google Scholar]
- Slack FJ, & Chinnaiyan AM (2019). The Role of Non-coding RNAs in Oncology. Cell, 179, 1033–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale ST, & Natoli G (2014). Transcriptional control of inflammatory responses. Cold Spring Harb Perspect Biol, 6, a016261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth MJ, Hayakawa Y, Takeda K, & Yagita H (2002). New aspects of natural -killer-cell surveillance and therapy of cancer. Nat Rev Cancer, 2, 850–861. [DOI] [PubMed] [Google Scholar]
- Song B, Guan Z, Liu F, Sun D, Wang K, & Qu H (2015). Long non-coding RNA HOTAIR promotes HLA-G expression via inhibiting miR-152 in gastric cancer cells. Biochem Biophys Res Commun, 464, 807–813. [DOI] [PubMed] [Google Scholar]
- Souza-Fonseca-Guimaraes F, Cursons J, & Huntington ND (2019). The Emergence of Natural Killer Cells as a Major Target in Cancer Immunotherapy. Trends Immunol, 40, 142–158. [DOI] [PubMed] [Google Scholar]
- Stutvoet TS, Kol A, de Vries EG, de Bruyn M, Fehrmann RS, Terwisscha van Scheltinga AG, & de Jong S (2019). MAPK pathway activity plays a key role in PD-L1 expression of lung adenocarcinoma cells. J Pathol, 249, 52–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucker A, Zhao F, Real B, Heeke C, Bielefeld N, Mabetaen S, Horn S, Moll I, Maltaner R, Horn PA, Schilling B, Sabbatino F, Lennerz V, Kloor M, Ferrone S, Schadendorf D, Falk CS, Griewank K, & Paschen A (2014). Genetic evolution of T-cell resistance in the course of melanoma progression. Clin Cancer Res, 20, 6593–6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Chu H, Ji J, Huo G, Song Q, & Zhang X (2016). Long non-coding RNA HOTAIR modulates HLA-G expression by absorbing miR-148a in human cervical cancer. Int J Oncol, 49, 943–952. [DOI] [PubMed] [Google Scholar]
- Talmadge JE, & Gabrilovich DI (2013). History of myeloid-derived suppressor cells. Nat Rev Cancer, 13, 739–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q, & Hann SS (2018). HOTAIR: An Oncogenic Long Non-Coding RNA in Human Cancer. Cell Physiol Biochem, 47, 893–913. [DOI] [PubMed] [Google Scholar]
- Tao H, Yang JJ, Zhou X, Deng ZY, Shi KH, & Li J (2016). Emerging role of long noncoding RNAs in lung cancer: Current status and future prospects. Respir Med, 110, 12–19. [DOI] [PubMed] [Google Scholar]
- Tian X, Ma J, Wang T, Tian J, Zhang Y, Mao L, Xu H, & Wang S (2018). Long Non-Coding RNA HOXA Transcript Antisense RNA Myeloid-Specific 1-HOXA1 Axis Downregulates the Immunosuppressive Activity of Myeloid-Derived Suppressor Cells in Lung Cancer. Front Immunol, 9, 473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, & Chang HY (2010). Long noncoding RNA as modular scaffold of histone modification complexes. Science, 329, 689–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugel S, De Sanctis F, Mandruzzato S, & Bronte V (2015). Tumor-induced myeloid deviation: when myeloid-derived suppressor cells meet tumor-associated macrophages. J Clin Invest, 125, 3365–3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Leun AM, Thommen DS, & Schumacher TN (2020). CD8(+) T cell states in human cancer: insights from single-cell analysis. Nat Rev Cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velagapudi SP, Luo Y, Tran T, Haniff HS, Nakai Y, Fallahi M, Martinez GJ, Childs-Disney JL, & Disney MD (2017). Defining RNA-Small Molecule Affinity Landscapes Enables Design of a Small Molecule Inhibitor of an Oncogenic Noncoding RNA. ACS Cent Sci, 3, 205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vester B, & Wengel J (2004). LNA (locked nucleic acid): high-affinity targeting of complementary RNA and DNA. Biochemistry, 43, 13233–13241. [DOI] [PubMed] [Google Scholar]
- Vignali DA, Collison LW, & Workman CJ (2008). How regulatory T cells work. Nat Rev Immunol, 8, 523–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, Yokoyama WM, & Ugolini S (2011). Innate or adaptive immunity? The example of natural killer cells. Science, 331, 44–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JA, & McKenzie ANJ (2018). TH2 cell development and function. Nat Rev Immunol, 18, 121–133. [DOI] [PubMed] [Google Scholar]
- Wang H, La Russa M, & Qi LS (2016). CRISPR/Cas9 in Genome Editing and Beyond. Annu Rev Biochem, 85, 227–264. [DOI] [PubMed] [Google Scholar]
- Wang J, Blanchard TG, & Ernst PB (2001). Host Inflammatory Response to Infection. [PubMed]
- Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, Jiang Z, Xu J, Liu Q, & Cao X (2014). The STAT3 -binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science, 344, 310–313. [DOI] [PubMed] [Google Scholar]
- Wang QM, Lian GY, Song Y, Huang YF, & Gong Y (2019). LncRNA MALAT1 promotes tumorigenesis and immune escape of diffuse large B cell lymphoma by sponging miR-195. Life Sci, 231, 116335. [DOI] [PubMed] [Google Scholar]
- Wang S, Liang K, Hu Q, Li P, Song J, Yang Y, Yao J, Mangala LS, Li C, Yang W, Park PK, Hawke DH, Zhou J, Zhou Y, Xia W, Hung MC, Marks JR, Gallick GE, Lopez-Berestein G, Flores ER, Sood AK, Huang S, Yu D, Yang L, & Lin C (2017). JAK2-binding long noncoding RNA promotes breast cancer brain metastasis. J Clin Invest, 127, 4498–4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wculek SK, Cueto FJ, Mujal AM, Melero I, Krummel MF, & Sancho D (2020). Dendritic cells in cancer immunology and immunotherapy. Nat Rev Immunol, 20, 7–24. [DOI] [PubMed] [Google Scholar]
- Wei S, Wang K, Huang X, Zhao Z, & Zhao Z (2019). LncRNA MALAT1 contributes to non-small cell lung cancer progression via modulating miR-200a-3p/programmed death-ligand 1 axis. Int J Immunopathol Pharmacol, 33, 2058738419859699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T, & Fried M (1986). A mouse locus at which transcription from both DNA strands produces mRNAs complementary at their 3’ ends. Nature, 322, 275–279. [DOI] [PubMed] [Google Scholar]
- Wu Y, Shao A, Wang L, Hu K, Yu C, Pan C, & Zhang S (2019). The Role of lncRNAs in the Distant Metastasis of Breast Cancer. Front Oncol, 9, 407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Z, Lin A, Li C, Liang K, Wang S, Liu Y, Park PK, Qin L, Wei Y, Hawke DH, Hung MC, Lin C, & Yang L (2014). lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell, 159, 1110–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Yang M, Guo H, Yang L, Wu J, Li R, Liu P, Lian Y, Zheng X, Yan J, Huang J, Li M, Wu X, Wen L, Lao K, Li R, Qiao J, & Tang F (2013). Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol, 20, 1131–1139. [DOI] [PubMed] [Google Scholar]
- Yang C, Shen C, Feng T, & Li H (2019). Noncoding RNA in NK cells. J Leukoc Biol, 105, 63–71. [DOI] [PubMed] [Google Scholar]
- Yoshimoto R, Mayeda A, Yoshida M, & Nakagawa S (2016). MALAT1 long non-coding RNA in cancer. Biochim Biophys Acta, 1859, 192–199. [DOI] [PubMed] [Google Scholar]
- Zampetaki A, Albrecht A, & Steinhofel K (2018). Long Non-coding RNA Structure and Function: Is There a Link? Front Physiol, 9, 1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemmour D, Pratama A, Loughhead SM, Mathis D, & Benoist C (2017). Flicr, a long noncoding RNA, modulates Foxp3 expression and autoimmunity. Proc Natl Acad Sci U S A, 114, E3472–E3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Liu G, Wei C, Gao C, & Hao J (2017). Linc-MAF-4 regulates Th1/Th2 differentiation and is associated with the pathogenesis of multiple sclerosis by targeting MAF. FASEB J, 31, 519–525. [DOI] [PubMed] [Google Scholar]
- Zhang N, & Bevan MJ (2011). CD8(+) T cells: foot soldiers of the immune system. Immunity, 35, 161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Ni F, Fu B, Wu Y, Sun R, Tian Z, & Wei H (2016). A long noncoding RNA positively regulates CD56 in human natural killer cells. Oncotarget, 7, 72546–72558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Hu H, Yan G, Wu T, Liu S, Chen W, Ning Y, & Lu Z (2019). Long Non-Coding RNA and Breast Cancer. Technol Cancer Res Treat, 18, 1533033819843889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Feng J, Fu H, Liu C, Yu Z, Sun Y, She X, Li P, Zhao C, Liu Y, Liu T, Liu Q, Liu Q, Li G, & Wu M (2018). Coagulation Factor X Regulated by CASC2c Recruited Macrophages and Induced M2 Polarization in Glioblastoma Multiforme. Front Immunol, 9, 1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen S, & Li X (2019). Application of CRISPR-Cas9 for Long Noncoding RNA Genes in Cancer Research. Hum Gene Ther, 30, 3–9. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Tian X, Wang T, Xia X, Cao F, Tian J, Xu P, Ma J, Xu H, & Wang S (2019). Long noncoding RNA Pvt1 regulates the immunosuppression activity of granulocytic myeloid-derived suppressor cells in tumor-bearing mice. Mol Cancer, 18, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Tang X, Tian X, Tian J, Zhang Y, Ma J, Xu H, & Wang S (2018). LncRNA MALAT1 negati vely regulates MDSCs in patients with lung cancer. J Cancer, 9, 2436–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhu Y, Xie Y, & Ma X (2019). The Role of Long Non-coding RNAs in Immunotherapy Resistance. Front Oncol, 9, 1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang L, Tian J, Zhang X, Wang H, & Huang C (2018). Lnc-DC regulates cellular turnover and the HBV-induced immune response by TLR9/STAT3 signaling in dendritic cells. Cell Mol Biol Lett, 23, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]


