Abstract
miR-155-5p is a well-known oncogenic microRNA, showing frequent overexpression in human malignancies, including breast cancer. Here, we show that high miR-155-5p levels are associated with unfavorable prognostic factors in two independent breast cancer cohorts (CSS cohort, n = 283; and TCGA-BRCA dataset, n = 1,095). Consistently, miR-155-5p results as differentially expressed in the breast cancer subgroups identified by the surrogate molecular classification in the CSS cohort and the PAM50 classifier in TCGA-BRCA dataset, with the TNBC and HER2-amplified tumors carrying the highest levels. Since the analysis of TCGA-BC dataset also demonstrated a significant association between miR-155-5p levels and the presence of mutations in homologous recombination (HR) genes, we hypothesized that miR-155-5p might affect cell response to the PARP-1 inhibitor Olaparib. As expected, miR-155-5p ectopic overexpression followed by Olaparib administration resulted in a greater reduction of cell viability as compared to Olaparib administration alone, suggesting that miR-155-5p might induce a synthetic lethal effect in cancer cells when coupled with PARP-1-inhibition. Overall, our data point to a role of miR-155-5p in homologous recombination deficiency and suggest miR-155-5p might be useful in predicting response to PARP1 inhibitors in the clinical setting.
Keywords: breast cancer, hsa-miR-155-5p, homologous recombination, PARP-1 inhibitors, Olaparib
Introduction
Breast cancer (BC) is the most frequent cancer among women worldwide, accounting for 13% of all cancer-related deaths (1). Breast cancer is a heterogeneous disease that includes different histological and molecular entities, clinical presentations, and behaviors, which vary in prognosis and response to therapy. To date, Oestrogen Receptor (ER), Progesterone Receptor (PgR), human epidermal growth factor receptor 2 (HER2), and Ki67, together with age, tumor size, histological grade, and lymph node engagement still represent the most reliable markers that provide prognostic information (2). Lately, microarray-based gene expression studies have identified intrinsic breast cancer subtypes of biological and, more importantly, clinical relevance consisting of two oestrogen receptor positive (ER+) subtypes, characterized by a relatively low (luminal A) and high (luminal B) expression of proliferation-related genes, a subtype enriched for HER2-amplified tumors (HER2-enriched), a subtype characterized by the absence of ER, PgR expression and HER2 amplification (Basal-Like) and a subtype ER +/– and negative for, PgR, HER2, claudin 3, claudin 4, claudin 7, and E-cadherin (Claudin-low) (3–6). This information has helped develop risk scores based on differential breast cancer molecular profiles that are currently entering the clinical practice to identify low-risk breast cancer patients who may avoid adjuvant treatment (7). Nevertheless, even these classification systems do not account for all the reported pathological and clinical heterogeneity of breast cancer.
MicroRNAs (miRNAs) are small endogenous non coding RNAs that fine-tune gene expression by post-transcriptional silencing of target mRNAs. Among the plethora of miRNAs that have been linked to human cancers, miR-155-5p (hsa-miR-155-5p) stands out as prominent oncomiR showing frequent overexpression in several hematological and solid tumors (8–12). In breast cancer, miR-155-5p has been found mostly upregulated, and associated with high-grade tumors, advanced stages, and lymph node metastases as well as worse disease-free and overall survival (13–22). At functional level, miR-155-5p validated target genes potentially place miR-155-5p within several cancer-related pathways encompassing cell proliferation, block of differentiation, epithelial-mesenchymal transition (EMT), and even DNA damage repair (DDR) (12, 23–26). However, these relationships between miR-155-5p and breast cancer clinical markers, such as ER and PgR status and tumor subtype, along with its causal role in breast cancer development remain controversial, likely due to limited patient sample sizes and discrepancies among studies in terms of methodologies and experimental models.
Herein, we report the results from the expression analysis of hsa-miR-155-5p we performed in a large cohort of 283 breast cancer patients with a complete median follow-up of 81 months that likely enabled us to define robust relationships between miR-155-5p and clinical parameters. We confirm that miR-155-5p expression is associated with unfavorable prognostic indicators in both our cohort and the TCGA-breast cancer dataset (TCGA-BRCA). We also found that higher miR-155-5p levels correlate with the basal-like subgroup followed by HER2-enriched tumors, whereas lower levels characterize the Luminal A and Luminal B tumors. Moreover, high miR-155-5p expression was found in breast cancer tumors from the TCGA dataset carrying mutations in HR genes. Lastly, we propose that miR-155-5p might play a role in the response to the poly(ADP-ribose) polymerase-1 (PARP-1) inhibitor Olaparib (AZD2281) which have been receiving much attention as promising therapeutic strategy beyond BRCA-mutant tumors, such as those with BRCAness and homologous recombination deficiency (HRD).
Materials and Methods
Study Design
We evaluated hsa-miR-155-5p expression in a retrospective (prospectively collected) cohort of 283 breast cancer cases with a median follow-up of 81-months (“Casa Sollievo della Sofferenza”, hereafter CSS, cohort). The study was conducted according to the REporting of tumor MARKer Studies (REMARK) guidelines (27). Breast cancer samples were collected at the Breast-Unit, IRCCS “Casa Sollievo della Sofferenza.” Upon receipt from surgery, tissues from the bulk of the tumor were sampled by the pathologist, then snap-frozen in liquid nitrogen and stored at −80°C. For legal reasons, only one tumor specimen (~50–100 mg of frozen tissue in weight) could be collected from each patient. Only women older than 18 years of age in were included in the study. All methods were carried out following the international Helsinki Declaration (7th rev, 2013, EU Directive 2004/23/EC) and Italian (D. Lgs. 30/06/2003, n. 196) regulations for research on patients. The experimental procedures of this study were approved by the Ethical Committee of the IRCCS “Casa Sollievo della Sofferenza” (Prot. N° 140/CE). Prior written informed consent was obtained from all patients in agreement with the experimental protocol approved by the Ethical Committee. All patients received either breast-conserving surgery or total mastectomy, plus sentinel node biopsy, or complete axillary dissection. The post-surgery treatments were performed according to the following guidelines: AIOM (Associazione Italiana Oncologia Medica), St Gallen, National Comprehensive Cancer Network (NCCN) and American Society of Clinical Oncology (ASCO). Progression was defined as evidence of loco-regional (recurrence) and/or distant disease over 4 months from diagnosis and after curative-intent surgical treatment. Clinical data were collected at each of the scheduled follow-up times.
Clinicopathological Data
Pathological assessment consists of evaluating the histological type, grade and stage. ER, PgR, Ki-67 labeling index, and HER2 status were determined by immunohistochemistry (IHC). Hormone receptor positive BCs, referred to as cases expressing oestrogen (ER) or progesterone (PgR) receptors in ≥1% of neoplastic cells by international guidelines (28), and HER2-amplified BCs were established according to standard recommendations (29). Of the 283 patients, 106 (38%) were lymph node negative, and 177 (62%) lymph node positive. Supplemental Table 1 summarizes descriptive statistics for the 283 cases from the CSS cohort analysed in this study. The median age of the study population was 59 years (range, 29–89), and the median tumor size was 2.5 cm (range, 0.5–11.0). Metastases at diagnosis were found in 15 cases whereas among non-metastatic patients (N = 268), 55 experienced disease progression (Incidence Rate, IR 3.25 events per 100 PY), and 30 of them (IR 1.65 events per 100 PY) died for the disease.
Cell Lines, Culture Conditions, and Reagents
In vitro assays were performed in a series of four breast cancer cell lines, encompassing the molecular subtypes of TNBC, which were as follows: MDA-MB-231 (claudin-low), MDA-MB-436 (basal-like), MDA-MB-468 (basal-like), and MDA-MB-453 (LAR (Luminal-Androgen-Receptor) subtype). MDA-MB-436 cell line was used as experimental model of BRCA1-mutant cells as carrying the pathogenic variant 5396+1G>A in the splice donor site of exon 20 resulting in a BRCA1 truncated protein (30). MDA-MB-436 (HTB-130, lot#63048503, p19), MDA-MB-468 (HTB-132, lot#63226339, p340), and MDA-MB-453 (HTB-131, lot#62959336, p348) cell lines were purchased from ATCC (American Type Culture Collection). MDA-MB-231 (Cat No. 92020424, lot#11D011, p40) cell line was purchased from ECACC (European Collection of Authenticated Cell Cultures). Culture conditions were as follows: MDA-MB-231, MDA-MB-468, and MDA-MB-453 were maintained in MEM w/o L-Gln, 10% FBS, 2 mM L-Gln, 1XNEAA, whereas MDA-MB-436 were maintained in MEM w/o L-Gln 10% FBS, 2 mM L-Gln, 1XNEAA, 5 mM NaP*, Human Insulin 10 μg/mL, GSH 16 μg/Ml. All cell lines were grown in a 5% CO2 humidified incubator at 37 °C. All cell lines were mycoplasma tested by Hoechst DNA staining and PCR by using N-GARDe Mycoplasma PCR kit (Euroclone).
The PARP-1 inhibitor Olaparib (AZD2281) was kindly provided by AstraZeneca, and dissolved in DMSO as a stock solution of 10 mg/mL.
RNA Isolation and Quantitative Reverse Transcription Polymerase Chain Reaction (RT-qPCR) Analysis
Total RNA from tissues and cells was isolated according to standard TRIzolTM protocol (Invitrogen, Thermo Fisher Sc). RNA samples from CSS cohort were selected as previously described (31). To assess miR-155-5p expression levels in our cohort of breast cancers, we applied a quantification method with standard curve. Briefly, for both miR-155-5p (ID 002623, Thermo Fisher Sc) and RNU48 (ID 001006, Thermo Fisher Sc) endogenous control, standard curves were constructed by plotting the threshold cycle (Ct) values against logarithm10 of the copy number and fitting by linear least square regression. For each sample, real-time PCR reactions were run in triplicate on ABI PRISM 7900 HT Sequence Detection System (Thermo Fis Sc). For tissue samples, miR-155-5p expression levels were determined as the ratio of the miR-155-5p copy number to the RNU48 copy number and then multiplied by 1000 for easier tabulation [i.e., (hsa-miR-155-5p/RNU48) × 1,000] (31).
Statistical and Bioinformatics Analyses
Patients' baseline characteristics were reported as median along with interquartile range (IQR, i.e., first-third quartiles) or frequencies and percentages for continuous and categorical variables, respectively. Comparisons between miR-155-5p levels and clinical pathological characteristics were assessed by Pearson correlation with continuous variables and by the two-sample t-test (or ANOVA model as appropriate) or Mann–Whitney U-test (or Kruskal–Wallis test when appropriate). In case of ANOVA model, pairwise comparisons were assessed from statistical contrasts (defined within the model) and computed p-values were adjusted following the step-up Bonferroni method due to Hochberg. In the case of the Kruskal–Wallis test, Dunn's post-hoc tests were carried out on each pair of groups and p-values were adjusted using the Bonferroni method. The assumption of normality distribution was checked by means of Q–Q plots and Shapiro–Wilks test. As the distribution of miR-155-5p expression levels in the CSS cohort was log-normal, all statistical analyses that involved this cohort were performed on their log-transformed values. Comparisons between distributions of categorical variables were assessed by Chi-Square test. Time-to-event analysis was performed in patients without metastases at diagnosis by univariable proportional hazards Cox regression models. Overall Survival (OS) was defined as the time between the enrollment date and cancer-related death. Progression Free Survival (PFS) was defined as the time between the enrollment date and the tumor progression. Metastasis Free Survival (MFS) was defined as the time between the enrollment date and the development of distant metastases. For patients who did not develop the event of interest, the endpoints (i.e., OS, PFS, and MFS) were defined as the time between the enrollment date and the date of the last available follow-up control. Incidence rates (IR, i.e., mortality and disease progression rates) were reported as number of events per 100 person-years (PY). A two-sided p < 0.05 was considered for statistical significance. All statistical analyses were performed using SAS Release 9.4 (SAS Institute, Cary, NC, USA) and plots were produced using R Foundation for Statistical Computing version 3.6. The PRISM software (version 5, GraphPad, Inc.) was used to make graphs and statistical analysis of in vitro assays. Single nucleotide variations (missense, frameshift, nonsense, and splicing SNV), copy number variations (CNV), gene, and miRNA expression data and clinical information concerning the TCGA-BRCA cohort, divided into tumor without synchronous metastases and healthy individuals, were retrieved from the NCI Genomic Data Commons (GDC, https://portal.gdc.cancer.gov/) data portal as pre-processed raw files. Germline point mutations in BRCA1 and BRCA2 genes were retrieved from cBioPortal (TCGA PanCancer Atlas subset, https://www.cbioportal.org/). Individuals were classified into Prediction Analysis of Microarray 50 (PAM50) subgroups by means of the R/Bioconductor TCGAbiolinks package. Files and data were manipulated using the R Foundation for Statistical Computing version 3.6. Clinicopathological characteristics of these individuals were summarized in Supplemental Table 2. The median age of the study population was 59 years (range, 27–90) and the median follow up was 12 months (IQR 3–35 months). Of the 1,095 patients, 991 (90%) were alive, and 104 (10%) died for the disease.
Determination of Short-Term Olaparib IC50 (IC5072 h) by PrestoBlueTM Viability Reagent
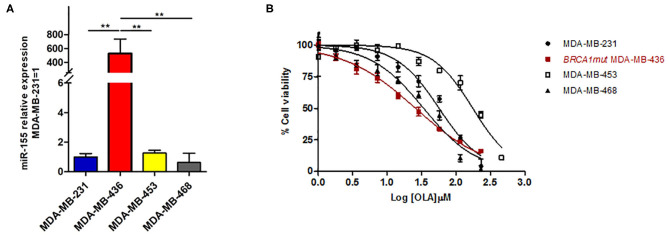
BC cells were seeded at 14,000 cells per well onto 96-well-plates and treated with a series of 2-fold Olaparib serial dilutions ranging from 230 to 1.8 μM, or vehicle (DMSO), in a final volume of 90 μL. For each treatment and vehicle condition, six technical replicates were prepared. Moreover, for each cell line, determination of Olaparib IC5072h was repeated in three independent 96-well-plates at different times. Cell viability was then evaluated after 72 h of treatment by PrestoBlue viability reagent (Thermo Fisher Sc). Briefly, 10 μL of the ready-to-use PrestoBlue Reagent were added directly to the medium. The 96-well-plates were incubated at 37°C for 2 h, and fluorescence was measured on a plate reader (Synergy HT Microplate Reader, BioTek) at 560 nm. Cell viability was calculated as the percentage of cells inhibited by the treatment as measured by the ratio in fluorescence between Olaparib-treated and vehicle-treated (DMSO) cells. The percentage of viable cells was plotted against Log concentrations of Olaparib. Individual data points on dose-response curves are shown as mean ± SD of n = 3 biological replicates. The Olaparib IC5072h for BC cell lines, i.e., the concentration of Olaparib causing a 50% reduction in cell viability in a time-interval of 72 h, were as followed: MDA-MB-436 IC5072h 25.8 μM (95%CI: 24.52–27.15), MDA-MB-231 IC5072h 56.8 μM (95%CI: 53.71–59.98), MDA-MB-468 IC5072h 33.81 μM (95%CI: 31.24–36.60), and MDA-MB-453 IC5072h 175 μM (95%CI: 160.8–190.6).
In vitro Transient Transfection of miR-155-5p Mimic/Inhibitor and Olaparib Treatment
To achieve transient overexpression or inhibition of miR-155-5p before Olaparib treatment, BC cells were first seeded onto 96-well-plates at 14,000 cells/well and 6-well-plates at 4.1 × 104 cells/well. Next, BC cell lines were transfected by using 50 nM mirVana miR-155-5p mimic (MC12601, Thermo Fisher Sc.) or mirVana miR-155-5p inhibitor (MH12601, Thermo Fisher Sc.), and correspondent negative controls (mirVana™ miRNA Mimic, Negative Control #1 and mirVana™ miRNA Inhibitor Negative Control #1, respectively, Thermo Fisher Sc.) according to Lipofectamine RNAiMAX forward transfection protocol (Thermo Fisher Sc). For each condition, eight and two technical replicates were prepared in each 96-well and 6-well-plate.
After 24 h from transfection, cell lines were treated with correspondent Olaparib IC5072h, except for MDA-MB-453 cell line, which was treated with a lower Olaparib dose (i.e., IC4072h, 60 μM) because showing higher sensitivity, in terms of cell tolerance, to liposomes-mediated transfection. Next, cell viability upon miR-155 modulation followed by 72-h exposure to Olaparib was measured by PrestoBlueTM viability reagent as described above.
To evaluate the true effects of miR-155-5p and Olaparib combination on cell viability and rule out those related to vehicle- and transfection procedures, experimental results are presented as comparison between miR-155 overexpressing/downregulated cells and control c(-) cells, treated with Olaparib or vehicle (DMSO). Data are shown as mean ± SD of n = 4 biological replicates (Student's t-test, *p < 0.05, **p < 0.01; ***p < 0.001).
To assess the extent of miR-155-5p overexpression/inhibition by RT-qPCR, adherent cells were harvested by both 96-well-plates and 6-well-plates, and processed for RNA isolation as described above. For each cell line, miR-155-5p levels in mimic/inhibitor-transfected cells were calculated as fold change (2−ΔΔCT) to correspondent negative control c(–)-transfected cells.
Western Blotting Analysis
Whole protein extracts from 6-well-plate seeded-cells were obtained by RIPA buffer (Tris-HCl 10 mM pH 7.5, NaCl 140 mM, Sodium deoxycholate 1%, sodium orthovanadate 1 mM, SDS 0.1%, Sodium fluoride 1 mM, EDTA 1 mM, Triton X-100 1%, 1X Protease inhibitor), resolved by SDS-PAGE, and transferred onto PVDF membranes. Primary antibodies for the bone-fide miR-155-5p target C/EBPβ (sc150, Santa Cruz Biotechnology) and normalizer Vinculin (#13901, Cell Signaling Tech) were detected by species-specific secondary HRP-linked antibodies, and revealed by using Pierce ECL 2 Substrate (Thermo Fisher Sc.). Image acquisition was performed on ChemiDoc XRS (Bio-Rad, Richmond, CA).
Results
Surrogate Molecular Classification of the CSS Cohort
Several genome-wide expression analyses have tried to identify clinically relevant molecular breast cancer subtypes (3–6). Among them, the PAM50 (4, 5) distinguishes five intrinsic subtypes: luminal A, luminal B, basal-like, HER2-enriched, and normal breast-like, characterized by different clinical behaviors. However, due to a lack of reimbursement, multigene tests are not readily available for all patients in many countries including Italy. Consequently, the use of immunohistochemistry (IHC) based biomarkers such as Ki-67 together with HER2 status has been proposed to distinguish ER-positive breast cancer cases at high risk of disease progression (i.e., bona fide Luminal B tumors) from those at low risk of disease progression (i.e., bona fide Luminal A tumors) (7). Instead, it is well established that HER2-amplification or intense overexpression (IHC 3+) is associated with worse prognosis in ER-positive BC patients. It is more difficult to classify the risk of ER-positive HER2-negative tumors because the prognostic performance of Ki-67 IHC staining shows a high variability among studies and laboratories (7). In our cohort, we evaluated two possible Ki-67 cut-offs: a 20% cut-off, as proposed by AIOM Guidelines (https://www.aiom.it/wp-content/uploads/2018/11/2018_LG_AIOM_Mammella.pdf), and the median cut off value of Ki-67 calculated in our study population (30%). As shown in Supplemental Table 3, only a Ki-67 cut-off of <30% was able to distinguish patients at low risk from patients at higher risk of disease progression (PFS HR = 0.38; 95%CI = 0.16–0.90; P = 0.029), metastases development (MFS HR = 0.34; 95%CI = 0.14–0.79; P = 0.012) and death (OS HR = 0.11; 95%CI = 0.01–0.87; P = 0.037) within the ER-positive subgroup. Thus, we defined Luminal A (LUMA) tumors as ER-positive/PgR positive, and HER2-negative with a low Ki-67 assessment (<30%), and Luminal B (LUMB) tumors as ER/PgR positive either HER2-positive or with a Ki-67 ≥ 30%. Overall, 94 cases (36%) were classified as Luminal A, 98 cases (37%) as Luminal B along with 34 HER2-amplified cases (13%), and 36 Triple Negative Breast Cancer cases (TNBC) (14%). The 21 remaining cases were not classified because HER2 or Ki-67 statuses were unknown.
miR-155-5p Expression Is Associated With Unfavorable Prognostic Indicators in Both CSS Cohort and TCGA-BRCA Dataset
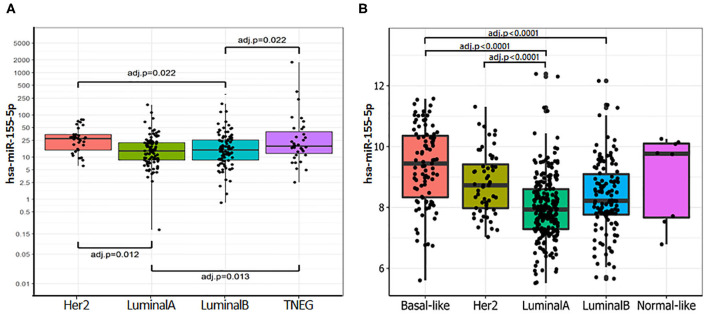
The expression of miR-155-5p [(hsa-miR-155-5p/RNU48) × 1,000] was evaluated in 283 primary breast tumors. As shown in Supplemental Table 4, miR-155 expression was associated with unfavorable prognostic indicators including high tumor grade (overall P = 0.0007; G2 vs. G3 P = 0.0053), reduced expression of ER (r = −0.243; P = 0.0002) and PgR (r = −0.240; P < 0.0001), and high Ki-67 expression (r = 0.215; P = 0.0005). Overall, these data suggest that miR-155 increased expression is associated with hormone receptor independency, high proliferation rate, and poor differentiation. However, no significant association was found with the pathological stage, lymph node status, and metastases development. These results were confirmed by the analysis of the TCGA-BRCA dataset (Supplemental Table 5). Then, we evaluated the expression of miR-155-5p across the surrogate molecular classification groups of the CSS cohort (n = 262) and the PAM50 subgroups for the TCGA-BRCA dataset (n = 505). In both cohorts, miR-155-5p was differentially expressed among the subgroups (CSS cohort P = 0.0008; TCGA-BRCA dataset P = 0.0005). In particular, higher miR-155 expression was found in HER2-amplified and TNBC subgroups, as compared with both Luminal A (P = 0.012 and 0.013, respectively) and Luminal B (both P = 0.0219) in the CSS cohort (Figure 1A). Accordingly, in the TCGA-BRCA dataset, the highest level of miR-155 was found in the basal-like subgroup followed by HER2-enriched tumors, whereas lower levels were detected in the Luminal A and Luminal B tumors. Pairwise comparison among groups evidenced statistically significant differences in the Basal-like subtype as compared with both Luminal A (P = 0.0001) and Luminal B subtypes (P = 0.0001), whereas the HER2-enriched subgroup showed significant differences only with the Luminal A subtype (P = 0.0001; Figure 1B). No significant associations were found between miR-155 expression and HER2 amplification/overexpression in neither CSS cohort nor TCGA-BRCA dataset.
Figure 1.
miR-155-5p is differentially expressed within the molecular breast cancer subgroups. (A) miR-155-5p expression within the surrogate molecular classification in CSS-cohort. High miR-155-5p expression was found in TNBC and HER2-amplified subgroups as compared with LUMA and LUMB tumors. (B) miR-155-5p expression within molecular subgroups identified by PAM50 in TCGA-BRCA dataset. High miR-155-5p levels were found in the Basal-like subgroup as compared with both Luminal A and Luminal B tumors, and in HER2-enriched subgroup as compared with Luminal A subgroup.
When we sought to determine putative associations of miR-155 with clinical outcomes, no statistically significant associations between miR-155 levels and patients' survival emerged from univariable Cox Regression analyses, in either the total population or molecular subgroups of both CSS cohort and TCGA-BRCA dataset. Moreover, no association was found in the subgroups of CSS patients stratified according to therapeutic regimens (chemotherapy, radiotherapy, and anti-HER2 monoclonal antibodies; data not shown).
miR-155-5p Is Differentially Expressed in Breast Cancer From TCGA-BRCA Dataset According to the Mutational Status of Homologous Recombination Genes
miR-155-5p has been found to be epigenetically repressed by wild type BRCA1 through its association with HDAC2 at MIR155 gene promoter and such a regulation is impaired by the R1699Q variant of BRCA1 (32). This, together with the findings that high levels of miR-155-5p associate with the Basal Like phenotype, a common feature of BRCA- mutated tumors, spurred us to investigate whether miR-155-5p may be associated with Homologous Recombination Deficiency (HRD), and thus represents a typical trait of BRCAness. To test this hypothesis, we selected those breast cancer cases for which the mutational status of 24 genes involved in the Homologous recombination pathway (Table 1) was available (n = 1,095 plus 104 normal controls) from the TCGA-BRCA dataset. Of those tumors, 801 carried at least one mutation (indel, missense or CNV) in one of the homologous recombination genes. Information about the expression levels of miR155-5p was available for 793 out of 801 mutated tumors and 291 out of the 295 cases without mutation in these genes (wt). Of the 793 mutated cases, 26 patients carried germline mutations in BRCA1 or BRCA2 (germMut), while 763 carried somatic mutations in homologous recombination genes (mutHR). Overall, 3,144 mutations were reported of which 91% were CNV and 9% were SNP or indels (Supplemental Table 6). The list of SNPs and indel mutations, together with their putative functional relevance, is reported in Supplemental Table 7. As shown in Figure 2A, miR-155 was increased in tumor samples (Median 302.94, IQR = 184.62–536.70) as compared with controls (n = 104) (Median = 129.96, IQR = 103.57–179.24; P = 0.0001). Higher miR-155 expression was found in the mutHR subgroup (Median = 446.600, IQR = 198.82–596.07) as compared with the wtHR subgroup (Median = 246.55, IQR = 148.43–419.62; P < 0.0001). The fact that no statistically significant differences were found for the germBRCA subgroup (Median = 446.60, IQR = 186.68–656.97) as compared with both wt (P = 0.134) and mutHR (P = 1.00), might be ascribed to the low number of germMut cases (n = 26; Figure 2A). Furthermore, the distribution of PAM50 subtypes differed significantly among the groups, with Basal-like and HER2-amplified tumors being more frequent in mutHR (83 and 55, respectively) as compared with wt (8 and 1, respectively; P = 0.0001). In the mutHR subgroup (Figure 2B), miR-155 was differentially expressed among the intrinsic molecular subtypes with higher expression in the Basal-Like (Median = 660.07, IQR = 296.79–1,262.98) and HER2-enriched (Median = 423.58, IQR = 251.10–653.04) tumors as compared with Luminal subtypes (LUMA Median = 252.87, IQR = 178.77–435.31, and LUMB Median = 292.89, IQR = 211.76–511.78; P < 0.0001; Figure 1B). Overall, these data indicate that miR-155 expression may represent a hallmark of homologous recombination deficiency (HRD).
Table 1.
Summary of mutations detected in the 24 homologous recombination genes evaluated in the TCGA-BRCA dataset.
| Gene symbol | Ensembl ID | Cytoband | Copy Number Variation | Frameshift mutation | SNP | ||||
|---|---|---|---|---|---|---|---|---|---|
| Deletions | Amplifications | Deletion | Insertion | Missense | Non sense | Splice site | |||
| ARID1A | ENSG00000117713 | 1p36.11 | 172 | 19 | 10 | 3 | 13 | 14 | 3 |
| ATM | ENSG00000149311 | 11q22.3 | 157 | 34 | 1 | 0 | 15 | 5 | 2 |
| ATRX | ENSG00000085224 | Xq21.1 | 21 | 18 | 2 | 0 | 39 | 3 | 1 |
| BAP1 | ENSG00000163930 | 3p21.1 | 140 | 11 | 1 | 1 | 9 | 1 | 0 |
| BARD1 | ENSG00000138376 | 2q35 | 47 | 36 | 0 | 0 | 0 | 0 | 0 |
| BLM | ENSG00000197299 | 15q26.1 | 14 | 92 | 1 | 0 | 15 | 1 | 0 |
| BRCA1 | ENSG00000012048 | 17q21.31 | 89 | 40 | 4 | 0 | 12 | 5 | 2 |
| BRCA2 | ENSG00000139618 | 13q13.1 | 86 | 73 | 5 | 0 | 18 | 3 | 1 |
| BRIP1 | ENSG00000136492 | 17q23.2 | 11 | 233 | 0 | 1 | 9 | 1 | 1 |
| CHEK1 | ENSG00000149554 | 11q24.2 | 157 | 25 | 0 | 0 | 2 | 0 | 0 |
| CHEK2 | ENSG00000183765 | 22q12.1 | 24 | 39 | 0 | 0 | 6 | 1 | 0 |
| FANCA | ENSG00000187741 | 16q24.3 | 64 | 40 | 0 | 0 | 8 | 0 | 0 |
| FANCD2 | ENSG00000144554 | 3p25.3 | 25 | 97 | 0 | 1 | 8 | 2 | 0 |
| FANCE | ENSG00000112039 | 6p21.31 | 31 | 66 | 0 | 0 | 3 | 0 | 0 |
| FANCF | ENSG00000183161 | 11p14.3 | 24 | 56 | 0 | 1 | 2 | 0 | 0 |
| FANCG | ENSG00000221829 | 9p13.3 | 18 | 56 | 0 | 0 | 3 | 0 | 0 |
| FANCL | ENSG00000115392 | 2p16.1 | 7 | 59 | 0 | 0 | 1 | 0 | 1 |
| MRE11 | ENSG00000020922 | 11q21 | 89 | 54 | 0 | 0 | 0 | 0 | 0 |
| NBN | ENSG00000104320 | 8q21.3 | 19 | 150 | 0 | 0 | 5 | 1 | 0 |
| PALB2 | ENSG00000083093 | 16p12.2 | 12 | 42 | 0 | 0 | 5 | 1 | 0 |
| RAD50 | ENSG00000113522 | 5q31.1 | 37 | 31 | 0 | 1 | 8 | 1 | 0 |
| RAD51 | ENSG00000051180 | 15q15.1 | 100 | 21 | 0 | 0 | 2 | 0 | 0 |
| RAD51B | ENSG00000182185 | 14q24.1 | 89 | 52 | 0 | 0 | 2 | 0 | 1 |
| WRN | ENSG00000165392 | 8p12 | 51 | 39 | 1 | 0 | 8 | 0 | 0 |
Figure 2.
miR-155-5p expression is associated with mutations in HR genes and basal-like phenotype. (A) Higher miR-155-5p levels were detected in tumors carrying mutations in HR genes. Statistically significant differences were found among controls with either germline BRCA1/2 mutated tumors (germBRCA), HR mutated (mutHR) or non HR mutated tumors (wt). High miR-155-5p expression was found in both germBRCA and mutHR tumors as compared with the wild type (wt) subgroups. (B) In mutHR tumors, miR-155-5p is differentially expressed according to PAM50 classification. Higher miR-155-5p levels were found in Basal-like and HER2 tumors as compared with the Luminal A subtype, and in the HER2-amplified tumors as compared with Luminal A subtype.
miR-155-5p May Indirectly Affect Cell Response to PARP-1 Inhibitor Olaparib
PARP-1 inhibitor Olaparib has been initially approved as monotherapy for the maintenance treatment of adult patients with platinum-sensitive relapsed BRCA-mutated (germline or somatic) high-grade ovarian cancer (33). More recently, results from the OlympiAD Phase 3 clinical trial indicate that Olaparib is also effective in BRCA1 or BRCA2 mutated HER2-negative metastatic breast cancer (34, 35). Our findings that high levels of miR-155-5p associate with the presence of mutations at HR genes prompted us to investigate whether miR-155-5p may affect the response to the PARP inhibitor Olaparib to some extent.
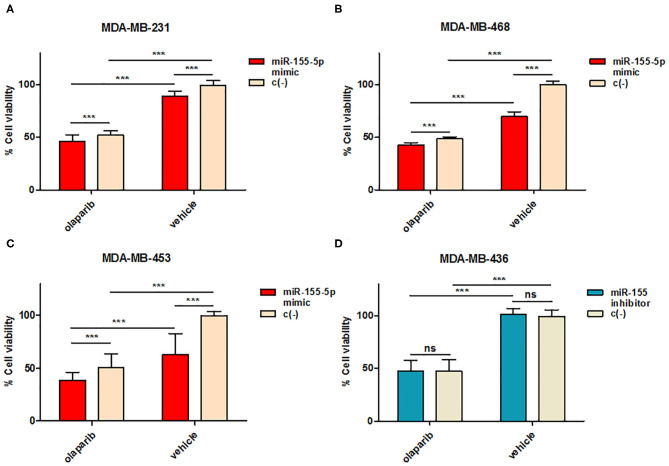
To this aim, we selected four breast cancer cell lines of TNBC, including the BRCA1 mutant cell line MDA-MB-436 (basal-like), and the BRCA1 wild type lines with BRCA1 allelic loss but normal transcript levels MDA-MB-468 (basal-like), MDA-MB-231 (claudin-low) and MDA-MB-453 (LAR-subtype). We did find that miR-155-5p endogenous levels were significantly elevated in mutBRCA1 MDA-MB-436 cells compared to the other cell lines (Figure 3A), suggesting the role of this BRCA1 pathogenic variant in abrogating negative regulation of miR-155-5p (30, 32). Interestingly, we also observed that, when we tested a wide range (see section Materials and Methods) of Olaparib concentrations that reduced cell viability significantly in a short time-interval of 72 h (IC5072), both basal-like TNBC cell lines encountered a 50% reduction of viability upon Olaparib treatment at similar doses independently of BRCA1 status (MDA-MB-436, 25.80 μM, 95%CI = 24.52–27.15; MDA-MB-468 33.81 μM 95%CI = 31.24–36.60) (Figure 3B). Next, to test the hypothesis that miR-155-5p might affect response to Olaparib, we ectopically overexpressed miR-155-5p in the less-responsive wt BRCA1 cell lines (MDA-MB-453, MDA-MB-231, and MDA-MB-468), and treated them at correspondent IC5072h, with the exception of MDA-MB-453 cell line, for which we chose a lower Olaparib dose (i.e., IC4072 h, 60 μM) because showing reduced tolerance to transfection procedures compared to the other cell lines. The ectopic overexpression of miR-155-5p itself induced a very significant decrease in cell viability in all cell lines compared to control (c(–)) cells. Moreover, when we treated both miR-155-5p overexpressing and control cells with Olaparib compound, we did observe a further reduction of cell viability, indicating that Olaparib and miR-155-5p may cooperate, and severely impair the survival of cancer cells (Figures 4A–C). Nevertheless, no alterations in Olaparib cell sensitivity were observed upon miR-155-5p inhibition in MDA-MB-436 (Figure 4D), suggesting that miR-155-5p regulatory role may be necessary but not sufficient for determining Olaparib sensitivity. The efficacy, as well as functionality of miR-155-5p overexpression/inhibition, was checked by both RT-qPCR and western blot analyses of its bone-fide target C/EBPβ protein levels (Figures 5A–D, Supplemental Figure 1).
Figure 3.
(A) miR-155-5p endogenous expression levels are significantly increased in the BRCA1mut MDA-MB-436 cell line as compared to the other wtBRCA-TNBC cell lines. miR-155-5p was quantified by RT-qPCR and normalized using RNU48 as endogenous control. Data are presented as fold increase over the expression levels of MDA-MB-231 and were derived from 6 biological replicates (Student's t-test, **p < 0.01). (B) Olaparib similarly affects cell viability of basal-like TNBC cell lines independently of BRCA-status. Growth curves of MDA-MB-231, MDA-MB-436, MDA-MB-468, and MDA-MB-453 (TNBC cell lines) were construed following 72 h of treatment with a range of 2-fold serial dilutions of Olaparib compound (230–1.8 μM) and cell viability assessment by PrestoBlueTM Reagent. The percentage of viable cells was plotted against Log-transformed Olaparib concentrations. Data are presented as mean (±SD) of three independent experiments.
Figure 4.
miR-155-5p effects on cell sensitivity to Olaparib. Cell viability was assessed by PrestoBlueTM reagent after induced overexpression in (A) MDA-MB-231, (B) MDA-MB-468, and (C) MDA-MB-453 or inhibition in (D) MDA-MB-436 of miR-155-5p by mirVana miR-155-5p mimic or inhibitor followed by Olaparib administration for 72 h. Data are presented as percentage of viable cells calculated in each condition (Olaparib vs. Vehicle) with respect to vehicle-treated c(–)-transfected cells, and represent the mean (±SD) of four independent experiments (Student's t-test, ***p < 0.001, ns = not significant).
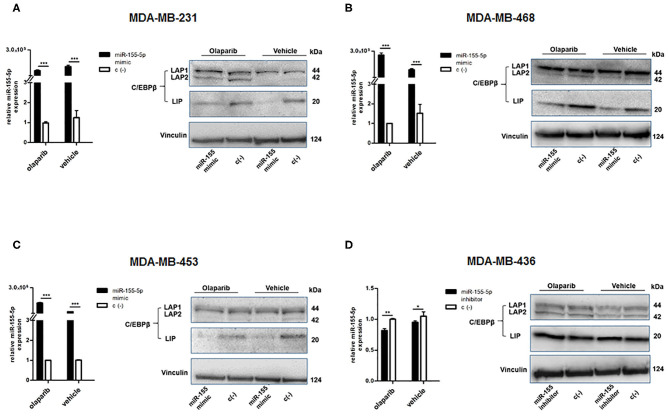
Figure 5.
Evaluation of efficacy and functionality of miR-155-5p overexpression/inhibition. miR-155-5p expression levels and western blot analyses of C/EBPβ protein levels in (A) MDA-MB-231, (B) MDA-MB-468, (C) MDA-MB-453, and (D) MDA-MB-436. miR-155-5p expression levels are presented as fold increase (2−ΔΔCT) with respect to correspondent negative control c(–)-transfected cells, in the presence of Olaparib or vehicle, and represent the mean (±SD) of four independent experiments (Student's t-test, *p < 0.05, **p < 0.01; ***p < 0.001).
Alterations of C/EBPβ Isoforms Abundance May Mediate the Growth Inhibitory Effect of miR-155-5p Coupled With Olaparib
As mentioned above, we evaluated the protein levels of miR-155-5p bona-fide target C/EBPβ (36) by western blot to ensure that ectopic modulation of miR-155-5p endogenous levels in the transfected-cell lines was functional. C/EBPβ (CCAAT/enhancer-binding protein) is a well-known transcription factor playing a central role in controlling the growth and differentiation of normal mammary gland, and it is produced in three isoforms through alternative usage of different start codons: the transcriptional activators liver activating protein 1 and 2 (LAP1 and LAP2), and the transcriptional inhibitor LIP, which can inhibit LAPs-mediated gene activation (36). The analyses of C/EBPβ protein levels in our cell lines showed a decreased level of the LIP isoform in miR-155-5p overexpressing cells compared to control cells, in both Olaparib- and vehicle-treated cells (Figures 5A–C), demonstrating the functionality of enforced miR-155-5p overexpression, and likely explaining the decrease in cell viability we observed after miR-155-5p overexpression. More interestingly, we noticed that, together with the miR-155-5p-mediated LIP reduction, a marked increase in LAP2 isoform in the claudin-low MDA-MB-231 cell line was induced specifically by Olaparib administration (Figure 5A). Since the smaller LAP2 isoform is considered to be the most transcriptionally active of the C/EBPβ isoforms, it is likely that its overexpression coupled with LIP downregulation, resulting in a LIP:LAP imbalance, severely impairs cell proliferation. We could not appreciate significant differences in MDA-MB-436 cell line, although transfection appeared to be successful, probably due to high C/EBPβ basal levels in this cell line, as shown by negative control (c(–))-transfected cells (Figure 5D).
Discussion
miR-155-5p is one of the best conserved and multifunctional miRNA involved in several physiological processes, such as proliferation, cell cycle, apoptosis, and differentiation (8–12). Altered expression of miR-155-5p has been found to be associated with hematopoietic lineage differentiation, immune response, inflammation, and tumorigenesis (8–12). Several studies suggest a role of miR-155-5p in breast cancer development and progression. However, its putative role as a biomarker is still controversial, likely due to limited patient sample sizes and discrepancies among studies in terms of methodologies and experimental models (13–22).
The primary aim of our study was the evaluation of miR-155-5p expression in a large cohort of breast cancer cases with a median follow up of 81 months. Our results were then validated in the TCGA-BRCA dataset. Consistently with previous reports, miR-155-5p expression was associated with negative prognostic factors including reduced expression of hormone receptor, high histological grade, and proliferation index measured by Ki67 expression in both our cohort and TCGA-BRCA dataset. Indeed, when we evaluated miR-155-5p expression within the subgroups identified by the surrogate molecular classification in the CSS cohort, and the intrinsic molecular classification in TCGA-BRCA dataset, the highest levels of expression were found in the TNBC and Basal-like subgroups followed by HER2-amplified tumors and the lowest expression in the Luminal subgroups. Overall, these data support the oncogenic role of miR-155-5p in deregulating cell proliferation and differentiation within the hormone-independent breast cancer subtypes. No associations were found with lymph node status and metastases development (Supplemental Tables 4, 5), as well as miR-155-5p did not correlate with patient's survival in univariable Cox regression analyses in both cohorts. These data rule against the role of miR155 as prognostic biomarkers in breast cancer. Nevertheless, the small number of TNBC (n = 34) and HER2 (n = 36) cases in our cohort, and the absence of information about PFS and MFS in the publicly available TCGA-BRCA dataset cannot completely exclude such a role in these breast cancer subgroups. Thus, this result should be further evaluated in retrospective or prospective studies with a specific focus on these subgroups.
Breast cancers with loss-of-function mutations in BRCA1 or BRCA2 genes are deficient in the HR (Homologous Recombination) pathway that manages the repair of DNA DSBs (Double Strand Breaks); hence, they are exquisitely sensitive to poly(ADP ribose) polymerase (PARP) inhibitors (37). This sensitivity to PARP inhibition derives from the synthetic lethality of cells with defective homologous recombination-mediated DNA repair to toxic replication intermediates generated on chromatin by PARP “trapping” (38). Preclinical studies and clinical trials in breast, ovarian cancer, and other cancers have shown the PARP inhibitors efficacy in BRCA1- and BRCA2-mutant patients (38–40). This led in 2014 to the approval by drug regulatory agencies of the PARP inhibitor Olaparib for the treatment of patients with recurrent ovarian cancer and BRCAs mutations (41). Encouraging results with Olaparib have also been obtained in patients with metastatic breast cancer-bearing germline BRCAs mutations (40–42). The randomized, phase 3 OlympiAD trial showed that among germline BRCA mutated patients affected by ER-positive HER2-negative metastatic TNBC, median progression-free survival was significantly longer with oral Olaparib monotherapy than with standard chemotherapy (34). This has finally led to Olaparib FDA-approval in January 2018 for gBRCA1/2+ HER2–breast cancers in the metastatic setting (35).
Although germline BRCA (germBRCA) mutations still remain the best clinical biomarkers for response to PARP inhibitor therapy (34, 43), genetic and epigenetic inactivation of other components of the homologous recombination apparatus can lead to HRD in sporadic cancers, broadly termed BRCAness (44). These alterations can occur in the form of germline mutations in HRR-associated genes, such as PALB2, FANCM, CHECK2, and RAD51C/D (44) or somatic mutations including ATM, BAP1, CDK12 (44) so that numerous studies are currently testing the possibility to apply mutational signatures to the prediction of PARP-inhibitors responsiveness (45, 46). BRCAness and HRD also involve methylation of the BRCA-promoter, which appears to be most common in TNBCs (47, 48). Breast tumors with BRCA1 methylation also show the higher histological grade, like that of BRCA1-mutated tumors (49). Additional contributors to HRD involve copy number alterations (CNA), and early research has revealed that the basal-like subtype of breast cancer has higher numbers of gains/losses, while the Luminal B subtype has a more frequent high-level DNA amplification (50).
Thus far, a few miRNAs have emerged as determining the BRCAness phenotype and, therefore, the response to PARP inhibition-based therapy (51, 52). miR-155-5p has been previously found to increase the tumor mutation load and promote the so-called “mutator phenotype” in inflammation-driven tumors due to the combined targeting of cell-cycle regulators and DNA repair enzymes (25, 26). Moreover, miR-155-5p expression has been found to be epigenetically controlled by BRCA1 through the recruitment of histone deacetylase 2 (HDAC2) at the MIR155 gene promoter (32) that ultimately leads to transcriptional silencing. In human cell lines, BRCA1-deficient cells showed 50-fold higher miR-155-5p levels compared with those with functional BRCA1. Finally, the transient overexpression of BRCA1 protein reduces the expression of miR-155-5p and, in clinical samples, miR-155-5p levels were 2- to 6-fold higher in BRCA1-mutant tumors compared to wt BRCA1 tumors.
For these reasons, we wondered whether miR-155-5p might correlate with defects in HR genes and represent a putative trait of BRCAness in the breast cancer of Triple Negative subtype (TNBC). Overall, we found that defects in the homologous recombination genes are a common feature of breast cancer, with 73% of cases in the TCGA-BRCA dataset carrying at least one germline or somatic mutation in HR genes. Almost all Basal Like and HER2-associated tumors were HR mutated. When we tested the putative association of miR-155-5p with the presence of mutations in Homologous Recombination genes, we were not surprised to find that miR-155-5p did show higher expression levels in the mutHR subgroup rather than in the wtHR subgroups in the TCGA Breast cancer dataset.
Next, we assessed the capability of miR-155-5p to alter the response to the first-in-class PARP inhibitor Olaparib (AZD2281) of four BC cell lines, differing in both molecular subtype and BRCA1 status. As expected, the MDA-MB-436 cells, representing a BRCA1-mutant basal-like TNBC cell line, showed a higher sensitivity to a short-term (72 h) treatment with Olaparib alone (IC5072h 25.80 μM, 95%CI = 24.52–27.15) compared to the other cell lines. In addition, we also observed that, independently of BRCA-mutational status, the second basal-like TNBC cell line MDA-MB-468, carrying normal levels of BRCA1 transcript despite allelic loss, and low expression of ATM, did show a high sensitivity to Olaparib (IC5072h 33.81 μM, 95%CI = 31.24–36.60), consistent with previous findings supporting this cell's BRCAness phenotype (49), and confirming that HR deficiency might be exploited to get a cancer cell response beyond BRCA-mutations. Then, when we enforced miR-155-5p overexpression in MDA-MB-231, MDA-MB-468, and MDA-MB-453 cell lines, we observed a significant decrease in cell viability following miR-155-5p overexpression alone in all cell lines. Interestingly, miR-155-5p-induced overexpression, followed by Olaparib exposure, resulted in an increased sensitivity to PARP inhibition and a further reduction in cell viability.
In order to determine the functionality of modulated miR-155-5p in our in vitro models, we analysed the protein expression levels of the transcriptional factor C/EBPβ, known to be one of bone-fide targets of miR-155-5p that plays an important role in regulating the balance between cell differentiation and proliferation in the mammary gland, according to the differential expression pattern of its isoforms. In particular, C/EBPβ has been recognized as core factor of the TGFβ cytostatic program, being essential for the induction of p15INK4b and the repression of c-MYC (53), and mediator of Ras-induced senescence (54). The association of C/EBPβ defects with breast cancer progression has been mostly attributed to aberrant levels of LIP isoform that, once elevated, may antagonize the transcriptional activities of LAPs and thus confer a growth advantage to cancer cells (55–58). Consistently with these data, ectopic overexpression of miR-155-5p in our cell lines showed to reduce the levels of the inhibitory LIP isoform in miR-155-5p-overexpressing cells with respect to control-transfected cells in both Olaparib- and vehicle-treated cells, thus demonstrating the functionality of synthetic oligos, and likely explaining the decrease in cell viability we observed after miR-155-5p mimic transfection (Figures 5A–C). Interestingly, we could detect a significant increase in the C/EBPβ LAP2 isoform protein level as a specific pattern of Olaparib treatment compared to vehicle in claudin-low MDA-MB-231 cells (Figure 5A). We might speculate that the decreased cell viability achieved in this cell line by miR-155-5p overexpression and Olaparib combination was a result of an enforced cell growth inhibitory effect exerted by LAP2 regulated genes and PARP inhibition. Moreover, since increased LAP2 was observed specifically in Olaparib-treated cells, we cannot rule out that the specific Olaparib action may go beyond PARP enzymes inhibition, and stimulate additional pathways toward growth inhibition. Hence, these data raise the question of whether increasing instead of inhibiting miR-155-5p in different cancer settings may enhance PARPi efficacy. Indeed, previous reports have already demonstrated the capability of miR-155-5p of boosting the anticancer immune response by targeting anti-inflammatory factors and immune checkpoints, including CTLA4 and PD-L1 [reviewed in (59)], something that may be of relevance for putative PARPi and Immunocheckpoints Inhibitors (ICI) combined therapies (60).
However, our results as well as the in vitro evidence that inhibition of miR-155-5p in BRCA1mut MDA-MB-436 cell line did not affect the higher sensitivity to PARP inhibition support the idea that several mechanisms underlie sensitivity to PARPi, and the understanding of the BRCAness as well as considering the specific BC subtype context warrants further investigation.
In conclusion, the analysis of two independent breast cancer cohorts corroborates the oncogenic role for miR-155-5p. Indeed, increased miR-155-5p expression was associated with loss of hormone receptor, reduced differentiation, and high proliferation rate. Consistently, miR-155-5p was differentially expressed in breast cancer molecular subgroups, with the highest expression in basal-like and HER2-related tumors. In addition, higher miR-155-5p expression was found in breast cancer cases from the TCGA dataset carrying mutations in HR genes. Lastly, we propose that a novel mechanism of synthetic lethality mediated by PARP inhibition and miR-155-5p may promote cancer cell death, and suggest miR-155-5p as an additional trait of the BRCAness phenotype.
Currently, several scoring systems aimed to identify HRD tumors are under investigation. These systems showed some predictive value in metastatic breast cancer patients treated with platinum derivatives (61). By using a high-depth whole-genome sequencing approach, Davies H. et al. developed a tool, they called HRDetect, as a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures (62). Such a technology was able to identify six different mutational signatures and classify BRCA1/2-deficient tumors correctly with 98.7% sensitivity. In addition, in a cohort of 560 individuals with BC, HRDetect identified 22 tumors with somatic loss-of-BRCA1 or BRCA2 and 47 tumors with functional BRCA1/2 deficiency, none of which had mutations detected with standard analysis (61). However, to date, HRDetect has not been correlated yet with therapeutic responses to PARPi. Another recent study evaluated a novel gene expression signature-generating algorithm to predict therapeutic response to PARPi (63). Our results indicate that, together with mutation and gene expression signatures, miRNA expression analysis may aid in evaluating the competency of homologous recombination, and thus eventually increase the ability to predict response to PARPi, in order to ultimately identify additional breast cancer patients eligible for PARP-inhibition including regimens.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The studies involving human participants were reviewed and approved by Fondazione IRCCS Casa Sollievo della Sofferenza. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
BP, RB, and PP: substantial contribution to conception and design of the study. BP, RB, MD, MR, SB, and SR: performed analytical procedures. AF and MC: performed statistical analyses. TB and TM: performed bioinformatics analyses. VV, MM, EM, and RM: collected clinical follow up data and reviewed the manuscript. PG: pathological evaluation of specimens. PP, BP, RB, MD, AF, MC, TB, and TM: analysis and interpretation of data. PP, BP, RB, AF, MC, and TM: manuscript writing. VF and ME: critical review of the manuscript. All authors: reviewed the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank AstraZeneca for providing Olaparib (AZD2281) free of charge under a grant agreement (NCR-16-12037).
Footnotes
Funding. Associazione Italiana Ricerca sul Cancro (AIRC) Project Code: 16747. Italian Ministry of Health (MoH) TRANSCAN Joint Transnational Call (JTC) 2013 co-funded by the European Regional Development Fund, A way of making Europe RRC-2014-2354565; Italian Ministry of Health (MoH) Ricerca Corrente Program and 5 × 1,000 voluntary contributions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.01415/full#supplementary-material
miR-155-5p expression levels following enforced overexpression/inhibition of miR-155-5p. miR-155-5p levels were quantified by RT-qPCR. Relative expression levels of miR-155-5p mimic/inhibitor-transfected cells were normalized to correspondent negative control c(–)-transfected cells (2−ΔΔCT) to rule out effects due to transfection procedures. Normalized data are presented as fold increase over the expression levels of MDA-MB-231 and represent the mean (±SD) of four independent experiments.
Clinicopathological characteristics of the CSS patient cohort (n = 283).
Clinicopathological characteristics of the TCGA-BRCA dataset (n = 1,095).
Univariable Cox Regression survival analysis evaluating two different KI-67 cut-off to estimate di risk of disease progression, metastases development, and death in ER positive breast cancer patients.
Associations between tumor clinicopathological characteristics and miR-155-5p expression in the CSS-cohort.
Associations between tumor clinicopathological characteristics and miR-155-5p expression in the TCGA-BRCA dataset.
Number of SNPs and indel detected according to gene and variant type.
List of SNPs and indel mutation detected in homologous recombination genes and their putative functional consequences.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2018) 68:394–424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Curigliano G, Burstein HJ, P Winer E, Gnant M, Dubsky P, Loibl S, et al. De-escalating and escalating treatments for early-stage breast cancer: the St. Gallen International Expert Consensus Conference on the Primary Therapy of Early Breast Cancer 2017. Ann Oncol. (2019) 30:1181. 10.1093/annonc/mdy537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumors. Nature. (2000) 406:747–52. 10.1038/35021093 [DOI] [PubMed] [Google Scholar]
- 4.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. (2003) 100:8418–23. 10.1073/pnas.0932692100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. (2002) 415:530–6. 10.1038/415530a [DOI] [PubMed] [Google Scholar]
- 6.Fougner C, Bergholtz H, Norum JH, Sørlie T. Re-definition of claudin-low as a breast cancer phenotype. Nat Commun. (2020) 11:1787. 10.1038/s41467-020-15574-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giuliano AE, Edge SB, Hortobagyi GN. Eighth edition of the AJCC cancer staging manual: breast cancer. Ann Surg Oncol. (2018) 25:1783–5. 10.1245/s10434-018-6486-6 [DOI] [PubMed] [Google Scholar]
- 8.Ferrajoli A, Shanafelt TD, Ivan C, Shimizu M, Rabe KG, Nouraee N, et al. Prognostic value of miR-155 in individuals with monoclonal B-cell lymphocytosis and patients with B chronic lymphocytic leukemia. Blood. (2013) 122:1891–9. 10.1182/blood-2013-01-478222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marcucci G, Maharry KS, Metzeler KH, Volinia S, Wu YZ, Mrozek K, et al. Clinical role of microRNAs in cytogenetically normal acute myeloid leukemia: miR-155 upregulation independently identifies high-risk patients. J Clin Oncol. (2013) 31:2086–93. 10.1200/JCO.2012.45.6228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. (2006) 103:2257–61. 10.1073/pnas.0510565103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He J, Zhang F, Wu Y, Zhang W, Zhu X, He X, et al. Prognostic role of microRNA-155 in various carcinomas: results from a meta-analysis. Dis Markers. (2013) 34:379–86. 10.1155/2013/856750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kong W, Yang H, He L, Zhao JJ, Coppola D, Dalton WS, et al. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol Cell Biol. (2008) 28:6773–84. 10.1128/MCB.00941-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. (2005) 65:7065–70. 10.1158/0008-5472.CAN-05-1783 [DOI] [PubMed] [Google Scholar]
- 14.Blenkiron C, Goldstein LD, Thorne NP, Spiteri I, Chin SF, Dunning MJ, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. (2007) 8:R214. 10.1186/gb-2007-8-10-r214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hui AB, Shi W, Boutros PC, Miller N, Pintilie M, Fyles T, et al. Robust global micro-RNA profiling with formalin-fixed paraffin-embedded breast cancer tissues. Lab Invest. (2009) 89:597–606. 10.1038/labinvest.2009.12 [DOI] [PubMed] [Google Scholar]
- 16.Zhu W, Qin W, Atasoy U, Sauter ER. Circulating microRNAs in breast cancer and healthy subjects. BMC Res Notes. (2009) 2:89. 10.1186/1756-0500-2-89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang F, Zheng Z, Guo J, Ding X. Correlation and quantitation of microRNA aberrant expression in tissues and sera from patients with breast tumor. Gynecol Oncol. (2010) 119:586–93. 10.1016/j.ygyno.2010.07.021 [DOI] [PubMed] [Google Scholar]
- 18.Chen J, Wang BC, Tang JH. Clinical significance of MicoRNA-155 expression in human breast cancer. J Surg Oncol. (2012) 106:260–6. 10.1002/jso.22153 [DOI] [PubMed] [Google Scholar]
- 19.Baffa R, Fassan M, Volinia S, O'Hara B, Liu CG, Palazzo JP, et al. MicroRNA expression profiling of human metastatic cancers identifies cancer gene targets. J Pathol. (2009) 219:214–21. 10.1002/path.2586 [DOI] [PubMed] [Google Scholar]
- 20.Kong W, He L, Richards EJ, Challa S, Xu C-X, Permuth-Wey J, et al. Upregulation of miRNA 155 promotes tumour angiogenesis by targeting VHL and is associated with poor prognosis and triple-negative breast cancer. Oncogene. (2014) 33:679–89. 10.1038/onc.2012.636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calvano Filho CM, Calvano-Mendes DC, Carvalho KC, Maciel GA, Ricci MD, Torres AP, et al. Triple-negative and luminal A breast tumors: differential expression of miR-18a-5p, miR-17-5p, and miR-20a-5p. Tumour Biol. (2014) 35:7733–41. 10.1007/s13277-014-2025-7 [DOI] [PubMed] [Google Scholar]
- 22.Ouyang M, Li Y, Ye S, Ma J, Lu L, Lv W, et al. MicroRNA profiling implies new markers of chemoresistance of triple-negative breast cancer. PLoS One. (2014) 9:e96228. 10.1371/journal.pone.0096228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willimott S, Wagner SD. MiR-125b and miR-155 contribute to BCL2 repression and proliferation in response to CD40 ligand (CD154) in human leukemic B-cells. J Biol Chem. (2011) 287:2608–17. 10.1074/jbc.M111.285718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang HQ, Yu XD, Liu ZH, Cheng X, Samartzis D, Jia LT, et al. Deregulated miR-155 promotes Fas-mediated apoptosis in human intervertebral disc degeneration by targeting FADD and caspase-3. J Pathol. (2011) 225:232–42. 10.1002/path.2931 [DOI] [PubMed] [Google Scholar]
- 25.Valeri N, Gasparini P, Fabbri M, Braconi C, Veronese A, Lovat F, et al. Modulation of mismatch repair and genomic stability by miR-155. Proc Natl Acad Sci U S A. (2010) 107:6982–7. 10.1073/pnas.1002472107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tili E, Michaille J-J, Wernicke D, Alder H, Costinean S, Volinia S, et al. Mutator activity induced by microRNA-155 (miR-155) links inflammation and cancer. Proc Natl Acad Sci U S A. (2011) 108:4908–13. 10.1073/pnas.1101795108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM, et al. REporting recommendations for tumor MARKer prognostic studies (REMARK). Br J Cancer. (2005). 93:387–91. 10.1038/sj.bjc.6602678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, et al. American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. (2010) 28:2784–95. 10.1043/1543-2165-134.7.e48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. (2013) 31:3997–4013. 10.1200/JCO.2013.50.9984 [DOI] [PubMed] [Google Scholar]
- 30.Elstrodt F, Hollestelle A, Nagel JH, Gorin M, Wasielewski M, van den Ouweland A, et al. BRCA1 mutation analysis of 41 human breast cancer cell lines reveals three new deleterious mutants. Cancer Res. (2006) 66:41–5. 10.1158/0008-5472.CAN-05-2853 [DOI] [PubMed] [Google Scholar]
- 31.Barbano R, Pasculli B, Rendina M, Fontana A, Fusilli C, Copetti M, et al. Stepwise analysis of MIR9 loci identifies miR-9-5p to be involved in Oestrogen regulated pathways in breast cancer patients. Sci Rep. (2017) 7:45283. 10.1038/srep45283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang S, Wang RH, Akagi K, Kim KA, Martin BK, Cavallone L, et al. Tumor suppressor BRCA1 epigenetically controls oncogenic micro-RNA-155. Nat Med. (2011) 17:1275–82. 10.1038/nm.2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pujade-Lauraine E, Ledermann JA, Selle F, Gebski V, Penson RT, Oza AM, et al. Olaparib tablets as maintenance therapy in patients with platinum-sensitive, relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-Ov21): a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. (2017) 18:1274–84. 10.1016/S1470-2045(17)30469-2 [DOI] [PubMed] [Google Scholar]
- 34.Robson M, Im SA, Senkus E, Xu B, Domchek SM, Masuda N, et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med. (2017) 377:523–33. 22. 10.1056/NEJMoa1706450 [DOI] [PubMed] [Google Scholar]
- 35.Robson ME, Tung N, Conte P, Im SA, Senkus E, Xu B, et al. OlympiAD final overall survival and tolerability results: Olaparib versus chemotherapy treatment of physician's choice in patients with a germline BRCA mutation and HER2-negative metastatic breast cancer. Ann Oncol. (2019) 30:558–66. 10.1093/annonc/mdz012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johansson J, Berg T, Kurzejamska E, Pang M-F, Tabor V, Jansson M, et al. MiR-155-mediated loss of C/EBPb shifts the TGF-b response from growth inhibition to epithelial-mesenchymal transition, invasion and metastasis in breast cancer. Oncogene. (2013) 32:5614–24. 10.1038/onc.2013.322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Venkitaraman AR. Linking the cellular functions of BRCA genes to cancer pathogenesis and treatment. Annu Rev Pathol. (2009) 4:461–87. 10.1146/annurev.pathol.3.121806.151422 [DOI] [PubMed] [Google Scholar]
- 38.Helleday T. The underlying mechanism for the PARP and BRCA synthetic lethality: clearing up the misunderstandings. Mol Oncol. (2011) 5:387–93. 10.1016/j.molonc.2011.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. (2005) 434:917–21. 10.1038/nature03445 [DOI] [PubMed] [Google Scholar]
- 40.Kaufman B, Shapira-Frommer R, Schmutzler RK, Audeh MW, Friedlander M, Balmaña J, et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J Clin Oncol. (2015) 33:244–250. 10.1200/JCO.2014.56.2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tutt A, Robson M, Garber JE, Domchek SM, Audeh MW, Weitzel JN, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet. (2010) 376:235–44. 10.1016/S0140-6736(10)60892-6 [DOI] [PubMed] [Google Scholar]
- 42.Ledermann JA, Harter P, Gourley C, Friedlander M, Vergote I, Rustin G, et al. Overall survival in patients with platinum-sensitive recurrent serous ovarian cancer receiving olaparib maintenance monotherapy: an updated analysis from a randomised, placebo-controlled, double-blind, phase 2 trial. Lancet Oncol. (2016). 17:1579–89. 10.1016/S1470-2045(16)30376-X [DOI] [PubMed] [Google Scholar]
- 43.Gelmon KA, Tischkowitz M, Mackay H, Swenerton K, Robidoux A, Tonkin K, et al. Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple negative breast cancer: a phase 2, multicentre, open-label, non-randomised study. Lancet Oncol. (2011) 12:852–61. 10.1016/S1470-2045(11)70214-5 [DOI] [PubMed] [Google Scholar]
- 44.Lord CJ, Ashworth A. BRCAness revisited. Nat Rev Cancer. (2016) 16:110–20. 10.1038/nrc.2015.21 [DOI] [PubMed] [Google Scholar]
- 45.Riaz N, Blecua P, Lim RS, et al. Pan-cancer analysis of bi-allelic alterations in homologous recombination DNA repair genes. Nat Commun. (2017) 8:857. 10.1038/s41467-017-00921-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Polak P., Kim J, Braunstein LZ, Karlic R, Haradhavala NJ, Tiao G, et al. A mutational signature reveals alterations underlying deficient homologous recombination repair in breast cancer. Nat. Genet. (2017) 49:1476. 10.1038/ng.3934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lips E, Mulder L, Oonk A, Van Der Kolk L, Hogervorst F, Imholz A, et al. Triple-negative breast cancer: BRCAness and concordance of clinical features with BRCA1-mutation carriers. Br J Cancer. (2013). 108:2172. 10.1038/bjc.2013.144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Timms KM, Abkevich V, Hughes E, Neff C, Reid J, Morris B, et al. Association of BRCA1/2 defects with genomic scores predictive of DNA damage repair deficiency among breast cancer subtypes. Breast Cancer Res. (2014) 16:475. 10.1186/s13058-014-0475-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roig B, Rodríguez-Balada M, Samino S, Lam EW, Guaita-Esteruelas S, Gomes AR, et al. Metabolomics reveals novel blood plasma biomarkers associated to the BRCA1-mutated phenotype of human breast cancer. Sci Rep. (2017) 7:17831. 10.1038/s41598-017-17897-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bergamaschi A, Kim YH, Wang P, Sørlie T, Hernandez-Boussard T, Lonning PE, et al. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosomes Cancer. (2006) 45:1033–40. 10.1002/gcc.20366 [DOI] [PubMed] [Google Scholar]
- 51.Moskwa P, Buffa FM, Pan Y, Panchakshari R, Gottipati P, Muschel RJ, et al. miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol Cell. (2011) 41:210–20. 10.1016/j.molcel.2010.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tommasi S, Pinto R, Danza K, Pilato B, Palumbo O, Micale L, et al. miR-151-5p, targeting chromatin remodeler SMARCA5, as a marker for the BRCAness phenotype. Oncotarget. (2016) 7:80363. 10.18632/oncotarget.10345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zahnow CA, Cardiff RD, Laucirica R, Medina D, Rosen JM. A Role for CCAAT/enhancer binding protein b-liver-enriched inhibitory protein in mammary epithelial cell proliferation. Cancer Res. (2001) 61:261–9. Available online at: https://cancerres.aacrjournals.org/content/61/1/261 [PubMed] [Google Scholar]
- 54.Gomis RR, Alarcon C, Nadal C, Van Poznak C, Massague J. C/EBPbeta at the core of the TGFβ cytostatic response and its evasion in metastatic breast cancer cells. Cancer Cell. (2006) 10:203–14. 10.1016/j.ccr.2006.07.019 [DOI] [PubMed] [Google Scholar]
- 55.Sebastian T, Malik R, Thomas S, Sage J, Johnson PF. C/EBPβ cooperates with RB:E2F to implement Ras(V12)-induced cellular senescence. EMBO J. (2005) 24:3301–12. 10.1038/sj.emboj.7600789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zahnow CA, Younes P, Laucirica R, Rosen J.M. Overexpression of C/EBPb-LIP, a naturally occurring, dominant-negative transcription factor, in human breast cancer. J Natl Cancer Inst. (1997) 89:1887–91. 10.1093/jnci/89.24.1887 [DOI] [PubMed] [Google Scholar]
- 57.Milde-Langosch K, Loning T, Bamberger A.M. Expression of the CCAAT/enhancer-binding proteins C/EBPa, C/EBPb and C/EBPd in breast cancer: correlations with clinicopathologic parameters and cell-cycle regulatory proteins. Breast Cancer Res Treat. (2003) 79:175–85. 10.1023/A:1023929504884 [DOI] [PubMed] [Google Scholar]
- 58.Raught B, Gingras AC, James A, Medina D, Sonenberg N, Rosen JM. Expression of a translationally regulated, dominant-negative CCAAT/enhancer-binding protein b isoform and up-regulation of the eukaryotic translation initiation factor 2a are correlated with neoplastic transformation of mammary epithelial cells. Cancer Res. (1996) 56:4382–6. [PubMed] [Google Scholar]
- 59.Michaille JJ, Awad H, Fortman EC, Efanov AA, Tili A. miR-155 expression in antitumor immunity: the higher the better? Genes Chromosomes Cancer. (2019) 58:208–18. 10.1002/gcc.22698 [DOI] [PubMed] [Google Scholar]
- 60.Peyraud F, Italiano A. Combined PARP inhibition and immune checkpoint therapy in solid tumors. Cancers. (2020) 12:1502. 10.3390/cancers12061502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Davies H, Glodzik D, Morganella S, Yates LR, Staaf J, Zou X, et al. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat Med. (2017) 23:517–25. 10.1038/nm.4292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McGrail DJ, Lin CC, Garnett J, Liu Q, Mo W, Dai H, et al. Improved prediction of PARP inhibitor response and identification of synergizing agents through use of a novel gene expression signature generation algorithm. NPJ Syst Biol Appl. (2017) 3:8. 10.1038/s41540-017-0011-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
miR-155-5p expression levels following enforced overexpression/inhibition of miR-155-5p. miR-155-5p levels were quantified by RT-qPCR. Relative expression levels of miR-155-5p mimic/inhibitor-transfected cells were normalized to correspondent negative control c(–)-transfected cells (2−ΔΔCT) to rule out effects due to transfection procedures. Normalized data are presented as fold increase over the expression levels of MDA-MB-231 and represent the mean (±SD) of four independent experiments.
Clinicopathological characteristics of the CSS patient cohort (n = 283).
Clinicopathological characteristics of the TCGA-BRCA dataset (n = 1,095).
Univariable Cox Regression survival analysis evaluating two different KI-67 cut-off to estimate di risk of disease progression, metastases development, and death in ER positive breast cancer patients.
Associations between tumor clinicopathological characteristics and miR-155-5p expression in the CSS-cohort.
Associations between tumor clinicopathological characteristics and miR-155-5p expression in the TCGA-BRCA dataset.
Number of SNPs and indel detected according to gene and variant type.
List of SNPs and indel mutation detected in homologous recombination genes and their putative functional consequences.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.