Abstract
Every mammalian species harbours a gut microbiota, and variation in the gut microbiota within mammalian species can have profound effects on host phenotypes. In this review, we summarize recent evidence that gut microbiotas have influenced the course of mammalian adaptation and diversification. Associations with gut microbiotas have: (i) promoted the diversification of mammalian species by enabling dietary transitions onto difficult-to-digest carbon sources and toxic food items; (ii) shaped the evolution of adaptive phenotypic plasticity in mammalian species through the amplification of signals from the external environment and from postnatal developmental processes; and (iii) generated selection for host mechanisms, including innate and adaptive immune mechanisms, to control the gut microbiota for the benefit of host fitness. The stability of specific gut microbiotas within host species lineages varies substantially across the mammalian phylogeny, and this variation may alter the ultimate evolutionary outcomes of relationships with gut microbiotas in different mammalian clades. In some mammalian species, including humans, relationships with host species-specific gut microbiotas appear to have led to the evolution of host dependence on the gut microbiota for certain functions. These studies implicate the gut microbiota as a significant environmental factor and selective agent shaping the adaptive evolution of mammalian diet, phenotypic plasticity, gastrointestinal morphology and immunity.
This article is part of the theme issue ‘The role of the microbiome in host evolution’.
Keywords: microbiome, host–microbe interactions, population genetics, holobiont, metagenome
1. Introduction
Animals, plants and fungi are colonized by microbial communities [1], but in few places do these communities reach densities approaching those found in the mammalian gastrointestinal tract. Mammalian guts are colonized by trillions of microorganisms representing hundreds of species, and bacterial cell counts can reach levels of 1012 cells ml−1 [2]. The evolutionary radiation of mammalian species has enabled the assembly of novel combinations of microorganisms into compositionally diverse gut microbiotas [3–7] and promoted the diversification of gut microbial lineages, some of which have co-diversified with host species and populations [8,9]. Today, mammalian guts represent one of the major reservoirs of microbial biodiversity on Earth [10].
Although every mammalian species harbours a gut microbiota, the consequences of these gut microbiotas for mammalian evolution and diversification remain largely unexplored. The question is particularly compelling, because natural history studies have indicated that most mammalian species harbour compositionally distinct gut microbiotas that reflect the phylogenetic histories of their hosts [3–7]. Moreover, experimental studies have shown that gut microbiotas are in many cases deeply intertwined with mammalian phenotypes spanning neuroendocrine, immune and metabolic systems [11–13], including traits related to fitness and adaptive differences between species.
Understanding the consequences of gut microbiotas in mammalian evolution first requires testable hypotheses regarding the specific modes by which they alter the adaptive landscapes experienced by hosts. The task is complicated by the immense diversity of microbes inhabiting the gut, and the variability in their identity and abundance over time: the gut microbiota within a host is likely to comprise both resident and transient microbes [14], with varying rates of transmission from other habitats and across generations within the host species. Nevertheless, recent reviews have proposed several complementary ways in which associating with a gut microbiota might influence the evolution of hosts. Microbiotas may influence host evolution by: (i) expanding host dietary niches, thereby altering future host adaptive trajectories [15,16]; (ii) influencing the evolution of host phenotypic plasticity in response to environmental change and during development [15–17]; and (iii) generating selection on hosts for traits that control the host's microbiota composition in ways that benefit host fitness [18]. Although these evolutionary effects have been well established in certain relatively simple animal–microbe systems, such as insects and bacterial endosymbionts [19], their influences on mammalian evolution are only beginning to be understood.
In this perspective, we critically evaluate the evidence for influences of the gut microbiota on mammalian evolution and diversification. Recent studies have provided unprecedented insights into how gut microbiotas have enabled the dietary diversification of mammalian species, including transitions to novel carbon sources and specialization on specific food items. In addition, gut microbiotas have shaped the evolution of mammalian phenotypic plasticity by serving as amplifiers of signals from the external environment and developmental processes. These evolutionary effects of the microbiota appear to have played out rapidly between closely related mammalian species and populations, and may have even affected recent human evolution. Similarly, the gut microbiota has contributed to the adaptive diversification of mammalian gut morphologies designed to harbour beneficial microorganisms. The gut microbiota probably also favoured the evolution of innate and adaptive mammalian immune systems. However, there remains minimal evidence for adaptive differences between mammalian species in immune-system traits that serve to control the microbiota for the benefit of host fitness, and there is a need for experiments capable of testing this hypothesis. Over long evolutionary time periods, associations between mammalian species and gut microbial lineages may inevitably lead to dependence of host development and function on the gut microbiota, a possibility with important implications for human health.
2. The diversification of mammalian dietary niches was enabled by gut microbiota
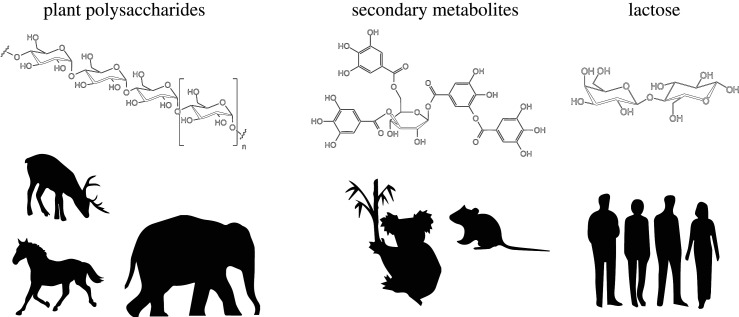
A major category of benefits that mammalian hosts derive from the gut microbiota is the expanded metabolic potential that gut microbial genomes provide relative to their hosts' genomes. Whereas the typical mammalian genome contains on the order of 10 000 genes, a typical mammalian gut microbiota can contain several million evolutionarily distinct gene families [20], some of which allow hosts to access foodstuffs that would be inaccessible otherwise. The functional repertoires contained within mammalian gut microbiotas appear to have enabled the evolution and diversification of herbivory and chitin-eating, the specialization of mammalian species and populations on toxic dietary items, and perhaps even recent dietary transitions in human evolution (figure 1).
Figure 1.
Dietary niche expansions mediated by the gut microbiota. The presence of a gut microbiota during mammalian evolution has had profound effects on mammalian dietary diversification. As herbivory has evolved multiple times independently throughout mammalian evolution, distantly related mammalian lineages have converged on gut microbiota compositions required for digesting complex plant polysaccharides (left). Dietary specialization on plants that produce toxic secondary metabolites, such as koala specialization on eucalyptus and woodrat specialization on creosote, has probably been facilitated by gut microbiota plasticity (centre). Gut microbiota-mediated metabolism of lactose in adulthood may have potentiated the recent evolution of lactase persistence phenotype in some human populations (right).
The most well-established example of a microbially provided dietary niche in mammals is herbivory. Plants contain complex polysaccharides that are indigestible by mammalian enzymes alone, but microorganisms in the gut can ferment these compounds, generating short-chain fatty acids and other metabolites that are readily accessible to their hosts [21]. Gut microbiotas are essential for herbivores. For example, early experiments in germ-free sheep found that hosts were unable to live long past weaning off milk and the transition to a solid diet [22]. Across the mammalian phylogeny, herbivory has evolved multiple times, and there has been widespread convergence of the composition and functional capacity of gut microbiotas between distantly related species of herbivores [3,4], further suggesting the adaptive value of these microbiotas. Bacterial lineages specific to herbivore guts, in general, arose much earlier in evolutionary history than those specific to carnivores, most likely before the diversification of vertebrates altogether [5], suggesting that their presence in the herbivore gut represents a co-option of pre-existing microbial potential rather than functionality that arose within the symbiosis. Once initiated, though, microbial lineages within herbivore guts, particularly those associated with the foregut-fermenting cetartiodactyla, seem to show an especially high degree of conservation within their hosts [23]. The expansion of dietary niche afforded by the gut microbiota has had enormous evolutionary consequences: diversification rate analyses have revealed that dietary transitions to herbivory are associated with accelerated diversification rates in mammalian clades, yielding diversification rates faster than those of any other mammalian dietary strategy [24]. These observations suggest that the acquisition and maintenance of a herbivory-associated microbiota has been a key innovation during mammalian evolution, comparable in its effects to other major evolutionary transitions in vertebrates [25].
Within herbivorous mammalian species, the gut microbiota also appears to have potentiated the dietary specialization on plant species containing toxic secondary metabolites. A clear example comes from the woodrat Neotoma lepida. Populations of this species have recently diversified and specialized on different dietary plants, including creosote, juniper and mesquite [26], each of which presents its own set of plant secondary metabolites. For example, creosote produces a phenolic-rich resin that is toxic to most woodrat populations, but consumable by woodrats with a history of feeding on creosote. An elegant series of experiments have demonstrated that the gut microbiota directly confers tolerance and digestibility of creosote secondary metabolites to woodrat hosts. Antibiotic treatment reduced woodrat ability to consume creosote, and transplanting the gut microbiota of woodrats from a population with a history of feeding on creosote into woodrats without such history led to improved performance of the recipient population on creosote [27]. More recently, further bacterial transplant experiments into germ-free rats have shown that bacteria from woodrats consuming tannin-rich diets improve rat performance on tannin-rich diets [28]. These results highlight how plasticity in gut microbiota composition in combination with the vast metabolic repertoire of microorganisms, particularly bacteria, can enable rapid host transitions to new dietary niches.
In addition to dietary specialization in woodrats, the gut microbiota may have facilitated koalas’ dietary specialization on Eucalyptus. Recent comparisons of the gut microbiotas of koalas and their sister species wombats have revealed that koalas harbour higher abundances of bacterial functional pathways related to the digestion of plant secondary metabolites [29]. Moreover, the gut microbiota of koalas specializing on different Eucalyptus species differs both in identity and predicted gene content [30], consistent with functional differences in the microbiome corresponding to phenotypic consequences for the host. A recent experiment using faecal transplant inocula indicates that these functional differences can be transferred between hosts, and may even support the presence of behavioural feedbacks from the microbiome: naive koalas ingest more of a novel Eucalyptus species after being inoculated with gut microbes from koalas specialized on it [31]. Therefore, variation in the gut microbial component of the koala's biotic environment alters the potential fitness impacts of another aspect of its biotic environment, namely the digestibility of different food species. This results in a modified adaptive landscape, and changes the fitness consequences of variation in a host phenotype: namely, the propensity to consume different food species. (Indeed, variation in individual preference for the novel food species was noted in the experiment, which if heritable would provide a pathway for adaptive evolution [31].) Together, these results suggest that the koala gut microbiota may have enabled koala specialization on Eucalyptus, and that koala dietary preferences may be influenced by the gut microbiota.
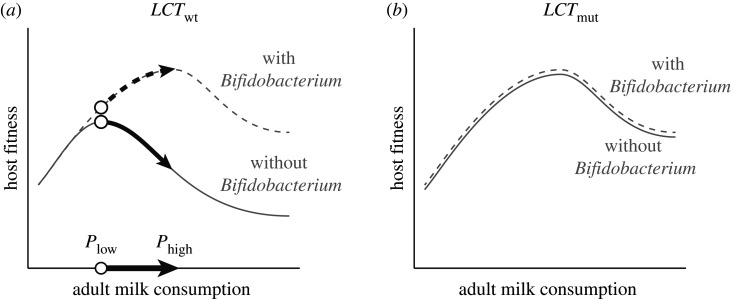
By altering the host's adaptive landscape, the microbiota might also potentiate host adaptation through phenotypes that ultimately remove reliance on microbial metabolism. The transition to milk consumption in adulthood in modern humans may be just such a case. Humans in certain populations began drinking milk in adulthood at least 6000 years ago [32], and this dietary transition has driven the rapid adaptive evolution of lactase persistence in the gut into adulthood in multiple human populations through mutations in the LCT gene [33]. Non-human mammals typically do not consume milk into adulthood, nor do they continue to express lactase (the enzyme that digests lactose in milk) in the gut after weaning. However, all mammals, including humans, contain gut bacteria, such as Bifidobacterium, that are capable of fermenting lactose, generating products that are accessible to their hosts' metabolisms. Interestingly, the alleles in the LCT gene responsible for lactase persistence are more significantly associated with gut microbiota composition than any other loci in the human genome [34]. Specifically, lactase persistence LCT alleles in historically dairy-consuming populations have been associated with reduced relative abundances of Bifidobacterium, consistent with reduced lactose availability for Bifidobacterium in lactase-persistent humans. These observations suggest the possibility that the presence of populations of Bifidobacterium (and potentially other lactose-fermenting microorganisms) in ancestral human populations [9] may have enabled the initial transition to milk-drinking in adulthood by providing access to sufficient energy from milk to justify the behavioural strategy. Under this model, the fitness advantages of milk consumption in adulthood provided by lactose-fermenting gut microorganisms were further refined through the evolution of lactase persistence via the LCT gene (figure 2).
Figure 2.
Gut microbes as potentiators of host dietary shifts. Graphs depict fitness surfaces, or the fitness outcomes of hosts (vertical axis) as a function of variation in a host phenotype value (horizontal axis), in this case, the degree of milk consumption as an adult. Presume that acquisition and consumption of milk has increasing fitness costs to the host, and fitness rewards that plateau at a certain point. Presume also that ability to digest lactose, either through secretion of endogenous host enzymes or via Bifidobacterium, increases the point at which that fitness reward plateaus. In a background population without adult production of lactase (LCTwt allele; (a)), a change in phenotype that increased milk acquisition and consumption (Plow → Phigh) might lead to a fitness decrease in the absence of Bifidobacterium in the gut, but a fitness increase in its presence. After fixation of the lactase-production allele (LCTmut) in the population (b), the host may no longer be sensitive to the presence of the lactose-degrading bacterium in the gut. In this way, the evolution of the host diet could be potentiated by gut bacteria, even without ultimately depending on the bacteria.
However, the association between gut microbiota and mammalian diet is not always straightforward. If the evolution of lactase persistence in some human populations was indeed potentiated by the activity of lactose-degrading bacteria in the gut, the microbial phenotype only served an intermediary role, and could in principle be lost once lactase expression became encoded in the host genome (figure 2b). Similar processes may help to explain inconsistencies in the correlation between diet and gut microbiome that have been observed elsewhere in the mammalian phylogeny. For example, despite the evidence for extensive convergence in the gut microbiota of the major groups of leaf-eating herbivores [3,4], the trend is not universal. Panda bears, for example, host a carnivore-like gut microbiota that does not appear to efficiently digest a diet composed almost entirely of bamboo [35]. In primates, leaf-chewing herbivory has evolved five times, with only subtle evidence of convergent changes in the gut microbiomes of folivorous groups relative to their omnivorous relatives [36]. At least two lineages of folivorous primates do seem to have independently evolved convergent changes to a duplicated copy of the hosts’ pancreatic RNASE1 enzyme, which is thought to be involved in the use of microbial fermentation products [37]. One possibility is that the lack of gut microbiota response to folivory in these species has increased the strength of selection for the adaptive evolution of digestive enzymes.
Another example of the complex interplay between the gut microbiota and the evolution of mammalian diet is myrmecophagy (i.e. specialization on ants and termites), which has evolved multiple times in mammals. This dietary transition has been associated with the compositional convergence in the gut microbiota across multiple distinct mammalian orders [38]. In some cases, this gut microbiota convergence has proceeded rapidly on an evolutionary timescale. For example, the gut microbiota of the myrmecophagous aardwolf, a close relative of carnivorous hyenas, has converged with the gut microbiotas of other myrmecophagous species in fewer than 10 Myr. In most of the myrmecophagous and otherwise invertebrate-specialized mammalian lineages, the hosts have also retained functional versions of most of the five copies of the CHIA chitinase genes inferred to be present in the genome of the most recent common ancestor of mammals, perhaps to assist with the degradation of the chitinous exoskeletons of their invertebrate prey [39]. By contrast, baleen whales eat a diet rich in chitinous invertebrates like krill and copepods, but inherited just a single functional copy of CHIA from the ancestor they share with their largely herbivorous relatives, the artiodactyls. Instead, whales may be using a gut microbiota that shares some high-level taxonomic similarities with herbivores to digest chitin in a fermentative process [40]. Baleen whales, unlike the terrestrial myrmecophages, have a complex multichambered foregut, itself probably remnant from a distant herbivorous ancestor. Although the precise metabolic contributions of host and microbial enzymes to chitin metabolism have yet to be determined for any of these taxa, it is clear that the particular evolutionary path taken by any given mammal lineage is the result of a complex interplay between the acquisition of metabolically relevant microbes and adaptation by the host.
In addition to enabling dietary shifts over evolutionary timescales, the gut microbiota may also aid mammals in coping with seasonal shifts in the diet that occur within individual hosts. For example, brown bears exhibit seasonal shifts in diet and gut microbiota composition, and transplantation of the summer-associated bear microbiota into germ-free mice induced adiposity in the recipient mice, an adaptive trait in bears before hibernation [41]. These results suggest that the seasonal compositional shifts in the bear gut microbiota provide metabolic benefits to their hosts. The gut microbiota has also been shown to vary seasonally in other mammalian species, such as mice [42] and humans [43], in ways that probably contribute to host metabolic needs in the context of seasonal dietary shifts. As in cases of mammalian dietary transitions over evolutionary time, the presence of a flexible gut microbiota during seasonal changes in diets may initially reduce selection pressures on hosts. However, by promoting mammalian fitness and enabling host-population persistence in variable environments, these flexible gut microbiotas can alter the future evolutionary trajectories of their host lineages.
3. Gut microbiotas shape mammalian evolution of phenotypic plasticity
One way in which biotic and abiotic environmental variables can affect host fitness is through influences on phenotypic plasticity and development. The modulation of phenotype using external cues can help to maximize fitness by matching organismal function to environmental conditions, but can also serve as a potential point of maladaptation, as seems to be happening at a global scale as a result of climate change [44]. The gut microbiota of mammals, as an important element of the mammalian biotic environment, is likely to be a source of similar effects for the host.
Recent evidence suggests that mammalian species have evolved to use their gut microbiotas as signals to initiate adaptive phenotypic plasticity in response to changes in the external environment as well as during key events in postnatal development. The composition of the mammalian gut microbiota is highly responsive to environmental variables, both from its host and from its host's external environment. The gut microbiota is also highly complex and multifaceted in terms of the signals it provides to its host. From the point of view of mammalian hosts, these features make the gut microbiota an excellent candidate transducer and amplifier of signals from both the external environment and developmental processes.
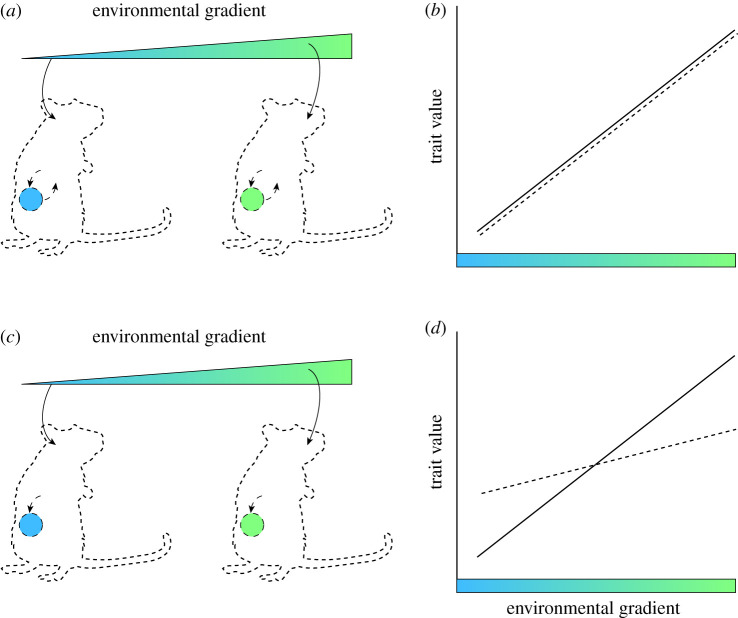
There is increasingly abundant experimental evidence that mammals have evolved to use the gut microbiota as a cue to initiate adaptive plastic responses to changing conditions in the external environment. For example, mouse experiments have shown that exposure to cold temperature caused a shift in the composition of the gut microbiota [45]. This shift was probably not driven by cold temperature directly, but rather by cold-induced changes to host physiology. Remarkably, transplantation of this cold-associated microbiota into germ-free house mice at room temperature induced plastic phenotypic responses in recipient hosts that are known to be adaptive in the context of cold exposure, including the browning of white fat. These results strongly suggest that house mice have evolved to use compositional shifts in the gut microbiota as cues to promote adaptive plastic responses to cold ambient temperatures. These results suggest that shifts in the gut microbiota caused by variation in the external environment may work in combination with the variation in the external environment itself to drive the evolution of adaptive plastic responses in mammalian species (figure 3). Under this view, the compositional plasticity of the gut microbiota serves as a signal amplification mechanism during the adaptive evolution of environmentally induced phenotypic plasticity in mammals.
Figure 3.
The gut microbiota as an amplifier of environmental signals for adaptive phenotypic plasticity in mammals. (a) An environmental gradient (coloured triangle) affects hosts (mouse cartoons), which in turn affects the gut microbiota (coloured circles). The shifts in the microbiota induced by host responses to the environmental gradient feed back to further affect host responses. (b) The combined direct and microbiota-mediated effects of the environmental gradient in (a) generate a host reaction norm (dashed line) that closely matches the fitness optimal reaction norm (solid line). (c) An environmental gradient (coloured triangle) affects hosts (mouse cartoons) and their microbiota (coloured circles), but the host does not respond to the shifts in the microbiota. (d) Under this scenario, the slope of the host reaction norm (dashed line) is less than the fitness optimal reaction norm (solid line). By serving as a signal amplification mechanism in this way, the microbiota could speed up adaptation during host transition to a new environmental gradient by allowing natural selection to act on host genetic variation in both direct and microbiota-mediated host phenotypic responses to the environmental gradient, rather than on direct host phenotypic responses alone. Alternatively, hosts may come to rely on the microbiota as a signal amplification mechanism through neutral processes, even if the host has previously evolved the optimal reaction norm through direct responses to the environmental gradient alone. Specifically, genetic drift may lead to the replacement of host direct responses to the environmental gradient by host responses to shifts in the microbiota caused by the gradient.
There is also a growing body of evidence that mammals have evolved to rely on host species-specific gut microbiotas for cues during the processes of postnatal development and function. For example, house mice (Mus musculus domesticus) colonized with the gut microbiota of rats or humans failed to develop fully differentiated T cell repertoires compared to house mice colonized with the gut microbiota of other house mice [46]. These results suggest that immunological development in house mice has evolved to integrate aspects of the house mouse-specific gut microbiota since the divergence of mice from rats. Similarly, germ-free house mice inoculated with the gut microbiotas of divergent host species within the genus Mus (i.e. Mus spretus and Mus pahari) displayed reduced growth rates, enlarged livers and less sexually dimorphic body compositions compared to germ-free house mice inoculated with the endogenous house mouse gut microbiota [47]. These results suggest that house mice have adapted to integrate the house mouse-specific gut microbiota into postnatal development since the divergence of house mice from other members of the genus Mus. Similarly, the wild mouse gut microbiota has been shown to promote fitness and improve disease resistance in laboratory mice [48] relative to the altered, laboratory-associated gut microbiota typically found in laboratory mice. In addition, Peromyscus polionotus mice inoculated with the gut microbiotas of different Peromyscus species displayed reduced digestive efficiency compared to P. polionotus mice inoculated with their endogenous gut microbiota [49]. This observation is consistent with the evolutionary integration of the species-specific gut microbiota into P. polionotus digestive physiology. Together, these results provide evidence that mammals adapt to features of their host species-specific gut microbiotas, suggesting a role for gut microbiota divergence in the evolutionary divergence of postnatal development and metabolic processes between closely related mammalian species.
Recent human studies have also indicated that the gut microbiota has been evolutionarily integrated into aspects of human postnatal development. For example, the caesarean section has been shown to affect the composition of the gut microbiota, and babies born by caesarean section tend to receive less microbial diversity from the mother than babies born vaginally [50]. Consistent with the hypothesis that the development of the human immune system has evolved to integrate the presence of a gut microbiota from birth, humans born by caesarean section also tend to display higher rates of certain autoimmune diseases, such as asthma, in adulthood [51]. More recent studies of the effects of caesarean section on human gut microbiota have provided conflicting evidence regarding the longevity of caesarean-induced microbiota differences. For example, one study found that the gut microbiota of caesarean-born babies is indistinguishable from vaginally born babies a few months after delivery [52]. Therefore, if lack of specific gut microbial lineages contributes to the development of autoimmune diseases in adulthood, these contributions may occur during critical windows of postnatal development.
Humans’ ancient associations with the gut microbiota are also being disrupted by other modern lifestyle practices, such as antibiotic use and reduced consumption of dietary fibre. Both of these environmental factors have been shown to reduce levels of gut microbiota diversity in laboratory animals [53] and in captive primates [54]. Lack of dietary fibre, in particular, has been shown in mice to lead to compounding losses of gut microbiota diversity over host generations [53], suggesting that dietary practices of current humans may have consequences for the gut microbiota of future generations. Consistent with this possibility, humans living in industrialized societies on average harbour lower levels of gut microbial diversity than any other wild-living primate [55,56]. These results motivate the need for further studies regarding the developmental consequences of the ongoing loss of ancestral gut microbiota diversity from human populations.
4. The adaptive evolution of mammalian mechanisms for controlling the gut microbiota
The gut microbiota may also affect mammalian evolution by generating selection pressures for host traits that control or reshape the microbiota for the benefit of host fitness. While many mammals modulate their external environment through ecosystem engineering [57], the compartmentalization of the gut microbiota within the digestive tract gives the host additional capacity for control. For gut microorganisms that make key contributions to host fitness, for example, the provisioning of essential nutrients from an otherwise inadequate diet, selection for host traits that control these microorganisms should be especially important [18].
Selection for host control of the gut microbiota has undoubtedly held influence during mammals' evolutionary past. For example, multiple herbivorous mammalian lineages have independently evolved foreguts to house microbiota capable of digesting complex plant polysaccharides. In addition, the innate and adaptive immune systems of mammals represent evolutionary innovations that probably were driven at least in part by a need to regulate the composition of the gut microbiota in ways that benefit host fitness [58,59]. The immune system provides means for identifying and eliminating microorganisms with pathogenic qualities while tolerating commensal or beneficial microorganisms. Experiments have shown that the absence of immune components from the body can lead to consequences for gut microbiota composition that are deleterious to hosts. For example, the deletion of Toll-like receptors from the host genome have been shown to lead to disrupted gut microbiota composition in mice that alters host energy harvest and metabolism in ways that are probably maladaptive [60]. Similarly, RAG1−/− mice, which lack adaptive immune systems, display altered gut microbiota compositions [61].
Despite the elaborate mechanisms that mammalian ancestors have evolved to control the gut microbiota, the influences of selection on hosts for control of the gut microbiota during the adaptive divergence of mammalian species remain poorly understood. Immune genes are among the most rapidly evolving protein-coding genes in mammalian genomes [62], but this pattern has historically been interpreted as evidence of red-queen coevolutionary dynamics with pathogens. The degree to which divergence in the gut microbiota between mammalian species contributes to the genetic divergence in immune genes has not been established. Continuously adapting to control the microbiota may be difficult for mammalian hosts, because microorganisms have the potential to evolve much more quickly in the context of an evolutionary arms race (although members of the microbiota must also evolve to compete with one another; [18]). Moreover, in contrast to evolving host control of pathogens of large fitness effect, controlling the gut microbiota requires the orchestration of potentially hundreds of microbial lineages, many of which are likely to have relatively small effects on host fitness.
One exciting possibility is that changes in the gut microbiota during host evolution have driven adaptive diversification of genes underlying traits that control the composition of the gut microbiota. This hypothesis generates predictions that remain untested to our knowledge. For example, given a mixture of microorganisms from a host species’ native environment, the host species when reared germ-free and inoculated with the mixture should be able to select a set of microorganisms that have greater beneficial effects on host fitness than would a random sampling of microorganisms in the mixture. The fitness effects of the gut microbiota sampled from the mixture by the gnotobiotic host could be assessed by further transplanting the gut microbiota of the gnotobiotic host into a new germ-free host. If the expected fitness effects on hosts are observed, it would suggest that hosts have evolved mechanisms to promote the growth of beneficial microorganisms, or that hosts have evolved to integrate whichever microorganisms are most fit within their guts into aspects of fitness. Differentiating between these alternatives could be accomplished by editing derived alleles in candidate host genes and repeating the experiments: if the genetically modified hosts are no longer able to select beneficial microorganisms from the microbial mixture, this result would support that selection for the ability to control of the microbiota contributed to the fixation of the alleles within the host species. With the growing availability of germ-free mammalian experimental systems, culturomics of gut microorganisms [63] and gene-editing approaches, such experiments are now feasible.
5. Ultimate evolutionary outcomes of mammalian associations with gut microbiotas
Gut microbiotas provide enormous benefits for mammalian species: they can enable host transitions to novel dietary niches and subsequent diversification (figures 1 and 2), they can act as amplifiers of environmental signals important for host fitness (figure 3), and they may contribute to the canalization of host developmental processes. However, gut microbiotas may also come with long-term evolutionary costs. One potential cost for animal hosts that has been proposed is ‘evolutionary addiction’ [16,64], wherein hosts become unable to survive and reproduce, or at least less fit, without the presence of their symbionts.
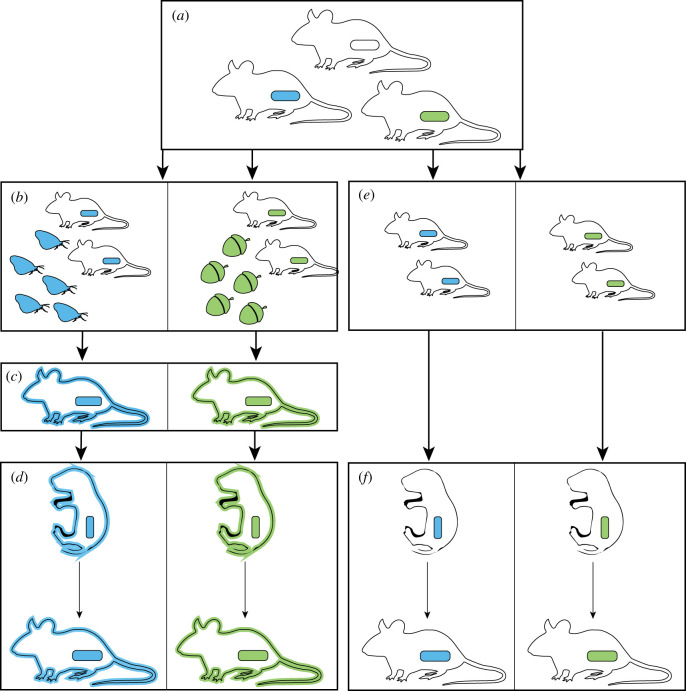
The effects of gut microbiotas on mammalian evolution suggest possible routes by which mammals may become evolutionarily addicted to specific gut microorganisms (figure 4). First, the presence of a specific gut microorganism or gut microbiota composition may enable transition onto a new dietary niche. Subsequent selection on hosts may lead to the evolution of host mechanisms that ensure the maintenance of the ecologically beneficial microorganisms. Further evolution of host development in the presence of this microbiota may lead to its integration into host developmental processes and evolutionary dependence. Although this process of ecologically beneficial symbionts leading to host evolutionary dependence has been previously described in other animal taxa, such as sap-feeding insects that rely entirely on microbial symbionts for the generation of essential amino acids [65], whether it occurs in mammals remains unclear.
Figure 4.
Potential pathways towards evolutionary dependence on a specific gut microbiota. Initial variation in gut microbiota composition within a species, denoted by coloured capsules (a), may generate differential fitness of lineages within the species in the presence of different food resources, denoted by coloured moths and acorns (b). Selection to retain ecological benefits of microbial associations may lead to the evolution of adaptive host mechanisms designed to maintain specific microbiotas, denoted by matching colours of host outlines and capsules (c). As the associations persist over evolutionary time, postnatal developmental processes may evolve to integrate signals from the specific microbiotas (d). Alternatively, host-lineage specific gut microbiotas can be evolutionarily integrated into host development even if the gut microbiotas have no effect on host fitness originally (e,f). Both pathways (a–d) and (a,e,f) can lead to deleterious effects on hosts if the symbiotic associations are disrupted.
Alternatively, the integration of features of the gut microbiota into host development could arise even if the features of the gut microbiota are not ecologically relevant for their hosts (figure 4e,f). For example, evidence for the integration of host species-specific gut microbiota into postnatal development has been observed in house mice [46,47], but it is likely that integration of house mouse-specific gut microbiota into house mouse development has been based on features of the gut microbiota that were not originally adaptive for hosts. Most differences between the gut microbiotas of house mice and other rodent species were probably neutral or deleterious when they first arose. However, the features of house mouse gut microbiotas that underlie the observed developmental effects have not been identified. Similarly, there is a need to identify the specific gut microorganisms on which human development has evolved to rely. The degree to which ‘evolutionary addiction’ to the gut microbiota in mammals centres around members of the microbiota with mutualistic, commensal or pathogenic properties remains an exciting area for future research.
Although every mammalian species harbours a gut microbiota, it is likely that the evolutionary outcomes of associating with a gut microbiota manifest differently among mammalian taxa. Recent analyses have shown that mammalian orders display differential levels of concordance between gut microbiota composition and host phylogenetic history (i.e. ‘phylosymbiosis’) [7,66]. The gut microbiotas of most mammalian orders show strong evidence of phylogenetic signal, but those of some orders, such as bats, show only weak associations with host phylogeny. The absence of a species-specific gut microbiota may also free mammalian species from the possibility of evolutionary dependence on a specific gut microbiota. Under these conditions, hosts may evolve only to integrate signals from non-specific or environmental microorganisms into development, rather than signals from specific microbes or sets of microbes. Alternatively, hosts may lose developmental reliance on microorganisms altogether. These hypotheses motivate the need for manipulative experiments in a wider diversity of mammalian species with varying degrees of phylogenetic signal in their gut microbiota.
6. Conclusion
The mammalian gut microbiota contains key aspects of the hosts' overall biotic environment, but it is also uniquely susceptible to host influence. Associating with gut microbiotas has reshaped the adaptive landscape experienced by mammals, facilitating the diversification of mammalian dietary niches and leading to the integration of the gut microbiota into host phenotypic plasticity as an amplifier of signals from the external environment and internal developmental processes. Similarly, the gut microbiota has driven the evolution of complex phenotypes in mammals that serve to control the gut microbiota for the benefit of host fitness, such as sophisticated gut compartments and immune mechanisms. However, the degree to which the gut microbiota has contributed to adaptive variation among the immune systems of mammalian species remains unclear, and this hypothesis represents an exciting area for future research. Moreover, many outstanding questions remain regarding how evolutionary impacts of gut microbiotas manifest across the mammalian phylogeny, how these evolutionary impacts are influenced by and contribute to the specificity of relationships between hosts and gut microbial lineages, and whether any mammalian species have overcome or avoided reliance on gut microbiota for metabolism or signals to initiate adaptive phenotypic plasticity.
Data accessibility
This article has no additional data.
Authors' contributions
A.H.M. and J.G.S. wrote the manuscript.
Competing interests
We declare we have no competing interests.
Funding
We received no funding for this study.
References
- 1.McFall-Ngai M, et al. 2013. Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl Acad. Sci. USA 110, 3229–3236. ( 10.1073/pnas.1218525110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sender R, Fuchs S, Milo R. 2016. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14, e1002533 ( 10.1371/journal.pbio.1002533) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ley RE, et al. 2008. Evolution of mammals and their gut microbes. Science 320, 1647–1651. ( 10.1126/science.1155725) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muegge BD, Kuczynski J, Knights D, Clemente JC, González A, Fontana L, Henrissat B, Knight R, Gordon JI. 2011. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332, 970–974. ( 10.1126/science.1198719) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Groussin M, Mazel F, Sanders JG, Smillie CS, Lavergne S, Thuiller W, Alm EJ. 2017. Unraveling the processes shaping mammalian gut microbiomes over evolutionary time. Nat. Commun. 8, 1–12. ( 10.1038/ncomms14319) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moeller AH, Suzuki TA, Lin D, Lacey EA, Wasser SK, Nachman MW. 2017. Dispersal limitation promotes the diversification of the mammalian gut microbiota. Proc. Natl Acad. Sci. USA 114, 13 768–13 773. ( 10.1073/pnas.1700122114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song SJ, et al. 2020. Comparative analyses of vertebrate gut microbiomes reveal convergence between birds and bats. MBio 11, e02901-19 ( 10.1128/mBio.02901-19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanders JG, Powell S, Kronauer DJC, Vasconcelos HL, Frederickson ME, Pierce NE. 2014. Stability and phylogenetic correlation in gut microbiota: lessons from ants and apes. Mol. Ecol. 23, 1268–1283. ( 10.1111/mec.12611) [DOI] [PubMed] [Google Scholar]
- 9.Moeller AH, et al. 2016. Cospeciation of gut microbiota with hominids. Science 353, 380–382. ( 10.1126/science.aaf3951) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson LR, et al. 2017. A communal catalogue reveals Earth's multiscale microbial diversity. Nature 551, 457–463. ( 10.1038/nature24621) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carabotti M, Scirocco A, Maselli MA, Severi C. 2015. The gut-brain axis: interactions between enteric microbiota, central and enteric nervous systems. Ann. Gastroenterol. Hepatol. 28, 203–209. [PMC free article] [PubMed] [Google Scholar]
- 12.Hooper LV, Littman DR, Macpherson AJ. 2012. Interactions between the microbiota and the immune system. Science 336, 1268–1273. ( 10.1126/science.1223490) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. 2012. Host-gut microbiota metabolic interactions. Science 336, 1262–1267. ( 10.1126/science.1223813) [DOI] [PubMed] [Google Scholar]
- 14.Shapira M. 2016. Gut microbiotas and host evolution: scaling up symbiosis. Trends Ecol. Evol. 31, 539–549. ( 10.1016/j.tree.2016.03.006) [DOI] [PubMed] [Google Scholar]
- 15.Alberdi A, Aizpurua O, Bohmann K, Zepeda-Mendoza ML, Gilbert MTP. 2016. Do vertebrate gut metagenomes confer rapid ecological adaptation? Trends Ecol. Evol. 31, 689–699. ( 10.1016/j.tree.2016.06.008) [DOI] [PubMed] [Google Scholar]
- 16.Moran NA, Ochman H, Hammer TJ. 2019. Evolutionary and ecological consequences of gut microbial communities. Ann. Rev. Ecol. Evol. Syst. 50, 451–475. ( 10.1146/annurev-ecolsys-110617-062453) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilbert SF, Bosch TCG, Ledón-Rettig C. 2015. Eco-Evo-Devo: developmental symbiosis and developmental plasticity as evolutionary agents. Nat. Rev. Genet. 16, 611–622. ( 10.1038/nrg3982) [DOI] [PubMed] [Google Scholar]
- 18.Foster KR, Schluter J, Coyte KZ, Rakoff-Nahoum S. 2017. The evolution of the host microbiome as an ecosystem on a leash. Nature 548, 43–51. ( 10.1038/nature23292) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moran NA. 2007. Symbiosis as an adaptive process and source of phenotypic complexity. Proc. Natl Acad. Sci. USA 104(Suppl. 1), 8627–8633. ( 10.1073/pnas.0611659104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao L, et al. 2015. A catalog of the mouse gut metagenome. Nat. Biotechnol. 33, 1103–1108. ( 10.1038/nbt.3353) [DOI] [PubMed] [Google Scholar]
- 21.Salyers AA, West SE, Vercellotti JR, Wilkins TD. 1977. Fermentation of mucins and plant polysaccharides by anaerobic bacteria from the human colon. Appl. Environ. Microbiol. 34, 529–533. ( 10.1128/AEM.34.5.529-533.1977) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soares JH, Leffel EC, Larsen RK. 1970. Neonatal lambs in a gnotobiotic environment. J. Anim. Sci. 31, 733–740. ( 10.2527/jas1970.314733x) [DOI] [PubMed] [Google Scholar]
- 23.Youngblut ND, Reischer GH, Walters W, Schuster N, Walzer C, Stalder G, Ley RE, Farnleitner AH. 2019. Host diet and evolutionary history explain different aspects of gut microbiome diversity among vertebrate clades. Nat. Commun. 10, 2200 ( 10.1038/s41467-019-10191-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Price SA, Hopkins SSB, Smith KK, Roth VL. 2012. Tempo of trophic evolution and its impact on mammalian diversification. Proc. Natl Acad. Sci. USA 109, 7008–7012. ( 10.1073/pnas.1117133109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heard SB, Hauser DL. 1995. Key evolutionary innovations and their ecological mechanisms. Hist. Biol. 10, 151–173. ( 10.1080/10292389509380518) [DOI] [Google Scholar]
- 26.Kohl KD, Dearing MD. 2016. The woodrat gut microbiota as an experimental system for understanding microbial metabolism of dietary toxins. Front. Microbiol. 7, 1165 ( 10.3389/fmicb.2016.01165) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kohl KD, Weiss RB, Cox J, Dale C, Dearing MD. 2014. Gut microbes of mammalian herbivores facilitate intake of plant toxins. Ecol. Lett. 17, 1238–1246. ( 10.1111/ele.12329) [DOI] [PubMed] [Google Scholar]
- 28.Kohl KD, Stengel A, Dearing MD. 2016. Inoculation of tannin-degrading bacteria into novel hosts increases performance on tannin-rich diets. Environ. Microbiol. 18, 1720–1729. ( 10.1111/1462-2920.12841) [DOI] [PubMed] [Google Scholar]
- 29.Shiffman ME, Soo RM, Dennis PG, Morrison M, Tyson GW, Hugenholtz P. 2017. Gene and genome-centric analyses of koala and wombat fecal microbiomes point to metabolic specialization for digestion. PeerJ 5, e4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brice KL, Trivedi P, Jeffries TC, Blyton MDJ, Mitchell C, Singh BK, Moore BD. 2019. The koala (Phascolarctos cinereus) faecal microbiome differs with diet in a wild population. PeerJ 7, e6534 ( 10.7717/peerj.6534) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blyton MDJ, Soo RM, Whisson D, Marsh KJ, Pascoe J, Le Pla M, Foley W, Hugenholtz P, Moore BD.. 2019. Faecal inoculations alter the gastrointestinal microbiome and allow dietary expansion in a wild specialist herbivore, the koala. Anim. Microbiome 1, 6 ( 10.1186/s42523-019-0008-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Charlton S, Ramsøe A, Collins M, Craig OE, Fischer R, Alexander M, Speller CF. 2019. New insights into Neolithic milk consumption through proteomic analysis of dental calculus. Archaeol. Anthropol. Sci. 11, 6183–6196. ( 10.1007/s12520-019-00911-7) [DOI] [Google Scholar]
- 33.Fan S, Hansen MEB, Lo Y, Tishkoff SA. 2016. Going global by adapting local: a review of recent human adaptation. Science 354, 54–59. ( 10.1126/science.aaf5098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goodrich JK, et al. 2016. Genetic determinants of the gut microbiome in UK twins. Cell Host Microbe 19, 731–743. ( 10.1016/j.chom.2016.04.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xue Z, et al. 2015. The bamboo-eating giant panda harbors a carnivore-like gut microbiota, with excessive seasonal variations. MBio 6, e00022-15 ( 10.1128/mBio.00022-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amato KR, et al. 2019. Evolutionary trends in host physiology outweigh dietary niche in structuring primate gut microbiomes. ISME J. 13, 576–587. ( 10.1038/s41396-018-0175-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Janiak MC, Burrell AS, Orkin JD, Disotell TR. 2019. Duplication and parallel evolution of the pancreatic ribonuclease gene (RNASE1) in folivorous non-colobine primates, the howler monkeys (Alouatta spp.). Sci. Rep. 9, 20366 ( 10.1038/s41598-019-56941-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Delsuc F, Metcalf JL, Wegener Parfrey L, Song SJ, González A, Knight R. 2014. Convergence of gut microbiomes in myrmecophagous mammals. Mol. Ecol. 23, 1301–1317. ( 10.1111/mec.12501) [DOI] [PubMed] [Google Scholar]
- 39.Emerling CA, Delsuc F, Nachman MW. 2018. Chitinase genes (CHIAs) provide genomic footprints of a post-Cretaceous dietary radiation in placental mammals. Sci. Adv. 4, eaar6478 ( 10.1126/sciadv.aar6478) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanders JG, Beichman AC, Roman J, Scott JJ, Emerson D, McCarthy JJ, Girguis PR. 2015. Baleen whales host a unique gut microbiome with similarities to both carnivores and herbivores. Nat. Commun. 6, 8285 ( 10.1038/ncomms9285) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sommer F, Ståhlman M, Ilkayeva O, Arnemo JM, Kindberg J, Josefsson J, Newgard CB, Fröbert O, Bäckhed F. 2016. The gut microbiota modulates energy metabolism in the hibernating brown bear Ursus arctos. Cell Rep. 14, 1655–1661. ( 10.1016/j.celrep.2016.01.026) [DOI] [PubMed] [Google Scholar]
- 42.Maurice CF, Knowles SCL, Ladau J, Pollard KS, Fenton A, Pedersen AB, Turnbaugh PJ. 2015. Marked seasonal variation in the wild mouse gut microbiota. ISME J. 9, 2423–2434. ( 10.1038/ismej.2015.53) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smits SA, et al. 2017. Seasonal cycling in the gut microbiome of the Hadza hunter-gatherers of Tanzania. Science 357, 802–806. ( 10.1126/science.aan4834) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mawdsley JR, O'malley R, Ojima DS. 2009. A review of climate-change adaptation strategies for wildlife management and biodiversity conservation. Conserv. Biol. 23, 1080–1089. ( 10.1111/j.1523-1739.2009.01264.x) [DOI] [PubMed] [Google Scholar]
- 45.Chevalier C, et al. 2015. Gut microbiota orchestrates energy homeostasis during cold. Cell 163, 1360–1374. ( 10.1016/j.cell.2015.11.004) [DOI] [PubMed] [Google Scholar]
- 46.Chung H, et al. 2012. Gut immune maturation depends on colonization with a host-specific microbiota. Cell 149, 1578–1593. ( 10.1016/j.cell.2012.04.037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moeller AH, Gomes-Neto JC, Mantz S, Kittana H, Segura Munoz RR, Schmaltz RJ, Ramer-Tait AE, Nachman MW. 2019. Experimental evidence for adaptation to species-specific gut microbiota in house mice. mSphere 4, e00387-19 ( 10.1128/mSphere.00387-19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosshart SP, et al. 2017. Wild mouse gut microbiota promotes host fitness and improves disease resistance. Cell 171, 1015–1028. ( 10.1016/j.cell.2017.09.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brooks AW, Kohl KD, Brucker RM, van Opstal EJ, Bordenstein SR.. 2016. Phylosymbiosis: relationships and functional effects of microbial communities across host evolutionary history. PLoS Biol. 14, e2000225 ( 10.1371/journal.pbio.2000225) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mueller NT, Bakacs E, Combellick J, Grigoryan Z, Dominguez-Bello MG. 2015. The infant microbiome development: mom matters. Trends Mol. Med. 21, 109–117. ( 10.1016/j.molmed.2014.12.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tamburini S, Shen N, Wu HC, Clemente JC. 2016. The microbiome in early life: implications for health outcomes. Nat. Med. 22, 713–722. ( 10.1038/nm.4142) [DOI] [PubMed] [Google Scholar]
- 52.Chu DM, Ma J, Prince AL, Antony KM, Seferovic MD, Aagaard KM. 2017. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat. Med. 23, 314–326. ( 10.1038/nm.4272) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK, Wingreen NS, Sonnenburg JL. 2016. Diet-induced extinctions in the gut microbiota compound over generations. Nature 529, 212–215. ( 10.1038/nature16504) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clayton JB, et al. 2016. Captivity humanizes the primate microbiome. Proc. Natl Acad. Sci. USA 113, 10 376–10 381. ( 10.1073/pnas.1521835113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moeller AH. 2017. The shrinking human gut microbiome. Curr. Opin. Microbiol. 38, 30–35. ( 10.1016/j.mib.2017.04.002) [DOI] [PubMed] [Google Scholar]
- 56.Moeller AH, Li Y, Mpoudi Ngole E, Ahuka-Mundeke S, Lonsdorf EV, Pusey AE, Peeters M, Hahn BH, Ochman H. 2014. Rapid changes in the gut microbiome during human evolution. Proc. Natl Acad. Sci. USA 111, 16 431–16 435. ( 10.1073/pnas.1419136111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davies GTO, Kirkpatrick JB, Cameron EZ, Carver S, Johnson CN. 2019. Ecosystem engineering by digging mammals: effects on soil fertility and condition in Tasmanian temperate woodland. R. Soc. Open Sci. 6, 180621 ( 10.1098/rsos.180621) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McFall-Ngai M. 2007. Adaptive immunity: care for the community. Nature 445, 153 ( 10.1038/445153a) [DOI] [PubMed] [Google Scholar]
- 59.Lee YK, Mazmanian SK. 2010. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science 330, 1768–1773. ( 10.1126/science.1195568) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vijay-Kumar M, et al. 2010. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 328, 228–231. ( 10.1126/science.1179721) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martínez I, et al. 2018. Experimental evaluation of the importance of colonization history in early-life gut microbiota assembly. eLife 7, e36521 ( 10.7554/eLife.36521) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meyerson NR, Sawyer SL. 2011. Two-stepping through time: mammals and viruses. Trends Microbiol. 19, 286–294. ( 10.1016/j.tim.2011.03.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lagier J-C, et al. 2016. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat. Microbiol. 1, 16203 ( 10.1038/nmicrobiol.2016.203) [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 64.Douglas AE. 2010. The symbiotic habit. Princeton, NJ: Princeton University Press. [Google Scholar]
- 65.Baumann P. 2005. Biology bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu. Rev. Microbiol. 59, 155–189. ( 10.1146/annurev.micro.59.030804.121041) [DOI] [PubMed] [Google Scholar]
- 66.Nishida AH, Ochman H. 2018. Rates of gut microbiome divergence in mammals. Mol. Ecol. 27, 1884–1897. ( 10.1111/mec.14473) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.