Summary
The health emergency caused by the recent Covid‐19 pandemic highlights the need to identify effective treatments against the virus causing this disease (SARS‐CoV‐2). The first clinical trials have been testing repurposed drugs that show promising anti‐SARS‐CoV‐2 effects in cultured cells. Although more than 2400 clinical trials are already under way, the actual number of tested compounds is still limited to approximately 20, alone or in combination. In addition, knowledge on their mode of action (MoA) is currently insufficient. Their first results reveal some inconsistencies and contradictory results and suggest that cohort size and quality of the control arm are two key issues for obtaining rigorous and conclusive results. Moreover, the observed discrepancies might also result from differences in the clinical inclusion criteria, including the possibility of early treatment that may be essential for therapy efficacy in patients with Covid‐19. Importantly, efforts should also be made to test new compounds with a documented MoA against SARS‐CoV‐2 in clinical trials. Successful treatment will probably be based on multitherapies with antiviral compounds that target different steps of the virus life cycle. Moreover, a multidisciplinary approach that combines artificial intelligence, compound docking, and robust in vitro and in vivo assays will accelerate the development of new antiviral molecules. Finally, large retrospective studies on hospitalized patients are needed to evaluate the different treatments with robust statistical tools and to identify the best treatment for each Covid‐19 stage. This review describes different candidate antiviral strategies for Covid‐19, by focusing on their mechanism of action.
Highlights
SARS‐CoV‐2 is a major threat to public health in the absence of drugs and vaccines.
The development of antiviral agents is urgently needed to treat patients with Covid‐19 and limit SARS‐CoV‐2 dissemination.
Several studies identified compounds that limit SARS‐CoV‐2 replication in vitro and determined the mode of action of some promising antiviral drugs.
Drug repurposing for Covid‐19 treatment is in progress, and the mode of action of these compounds needs to be clarified.
Abbreviations
- 3CLpro
chymotrypsin‐like 3
- Abs
antibodies
- ACE2
angiotensin‐converting enzyme 2
- CoV
coronaviruses
- E
envelope protein
- ExoN
exonuclease
- FDA
Food and Drug Administration
- HCQ
hydroxychloroquine
- HIV
human immunodeficiency virus
- IFN
interferon
- IL
interleukin
- M
membrane protein
- mAbs
monoclonal Abs
- MERS
Middle East respiratory syndrome
- MoA
mode of action
- N
nucleoprotein
- nsp
nonstructural protein
- PLpro
papain‐like
- RdRp
RNA‐dependent RNA polymerase
- S
spike protein
- SARS
severe acute respiratory syndrome
- TMPRSS2
transmembrane protease serine 2
1. INTRODUCTION TO SARS‐CORONAVIRUS 2
Over the last 20 years, three coronaviruses (CoVs) that cause severe pulmonary infections in humans have crossed the species barrier. 1 , 2 The last of these CoVs, named SARS‐CoV‐2, emerged in the Hubei province (China) in December 2019, and rapidly spread worldwide becoming a major public health threat. 3 In approximately 20% of patients, the disease progresses to severe pneumonia, respiratory and multi‐visceral failure, often leading to death of patients with comorbidity. 4 This worsening is associated with a deregulated immune response, including exacerbated production of pro‐inflammatory cytokines. 5 Therefore, specific and effective antiviral therapies are urgently needed, due to the absence of a vaccine.
Coronaviruses belong to the Nidovirales order, a group of enveloped viruses with genomic RNA of positive polarity. Their 27 to 34 kb genome encodes 16 nonstructural proteins (nsps) and 4 structural proteins (spike protein [S], envelope protein [E], membrane protein [M], and nucleoprotein [N]). 6 The virus life cycle, described in Figure 1, begins by the attachment of the viral particle to the angiotensin‐converting enzyme 2 (ACE2) cell surface receptor, mediated by the S protein. 7 , 8 Virus entry is achieved by endocytosis and/or direct fusion of the cell and viral membranes. This step requires S protein activation. The S protein is synthesized as an inactive precursor that is inserted in the viral membrane, and requires two subsequent cleavages by cellular proteases to become functionally active. Different cellular proteases, such as furin‐like enzymes and the transmembrane protease serine 2 (TMPRSS2), may cleave the S protein in two subunits, S1 and S2, in a process called “priming.” 9 , 10 A recent study showed that this furin‐mediated cleavage is important for virus entry in lung cells. 11 In addition, for virus entry, a second proteolytic cleavage is required at the S2′ site localized immediately upstream of the fusion peptide, 12 , 13 and seems to involve at least TMPRSS2. 14 The viral genome is then released into the cytoplasm of the infected cells. This allows the translation of the viral mRNA into two polyprotein precursors, pp1a and pp1ab, controlled by a −1 ribosomal frameshift. These polyprotein precursors are then cleaved by two viral proteases, chymotrypsin‐like 3 (3CLpro) and papain‐like (PLpro), to generate the 16 nsps (nsp1 to 16). Many of these proteins participate in the formation of the replication and transcription complex that orchestrates genome replication, mRNA synthesis and capping. At the final stage of viral infection, the N protein assembles with the neo‐synthesized viral genome to form the nucleocapsid that associates with the viral structural proteins to generate new virions released by exocytosis. 6
FIGURE 1.
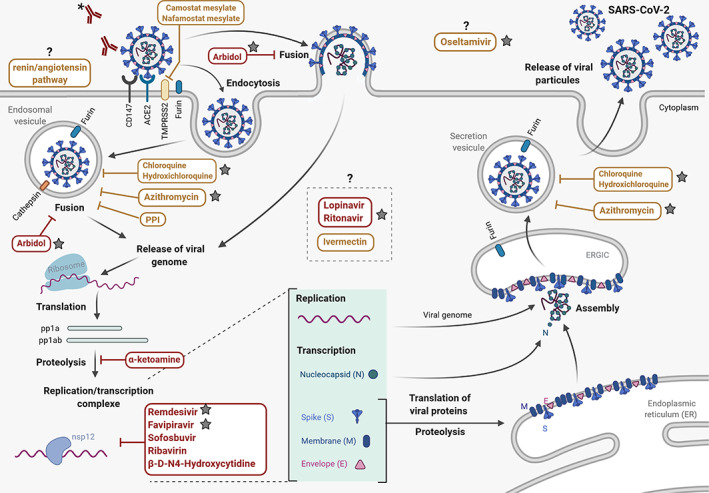
SARS‐CoV‐2 life cycle and potential mechanisms of action of currently evaluated therapeutics. SARS‐CoV‐2 life cycle and the presumed mechanisms of action (MoA) of the main SARS‐CoV‐2 replication inhibitors. Different compounds are assessed to find compounds targeting the different steps of SARS‐CoV‐2 life cycle. Molecules currently in clinical trials are indicated by a gray star. Therapeutic strategies based on antiviral compounds are indicated in red, and approved drugs used for other diseases or selected by virtual screening are indicated in yellow. The black question marks indicate unknown or elusive MoA in the context of CoV infection. *Corresponding to antibody (Ab) strategies including monoclonal antibodies and plasma from convalescent patient. The Biorender website was used to generate this figure
The ongoing Covid‐19 pandemic has already caused at least 4 million confirmed cases and more than 250 000 deaths. In response to this global health emergency, public health measures to control the virus spread have been put in place and efforts to identify potential antiviral molecules and to develop a vaccine have been intensified. The explored antiviral strategies are mainly based on existing drugs that were developed to treat viral infection or other diseases. Moreover, the many different clinical trials currently in progress (more than 2400) are testing a limited set of drugs, alone or in combination. 15 Although the drugs used in these therapeutic assays have shown some inhibitory effect in SARS‐CoV‐2‐infected cells in vitro, the molecular bases of their antiviral activity are often poorly understood, except for nucleoside analogs (eg, favipiravir for influenza virus) and fusion inhibitors (eg, arbidol). In addition, while waiting for the final results of these clinical studies, important concerns have arisen, such as the risk of inappropriate self‐treatment, the potential toxicity or adverse effects of some of these compounds, and the risk of depletion of pharmacy stocks needed for the treatment of other pathologies. On the other hand, the emergence of SARS‐CoV and Middle East respiratory syndrome coronavirus (MERS‐CoV) has led to a growing number of publications that describe new CoV inhibitors and precisely characterize their mode of action (MoA). In this review, we summarize promising candidate molecules that could be repurposed and anti‐CoV‐specific drugs derived from ongoing research on CoVs, for which we provide additional information on their potential MoA based on the current knowledge on SARS‐COV‐2 life cycle. In this review, we do not describe vaccine strategies, and supporting treatments, although they are also important for Covid‐19 management.
2. CLASSICAL ANTIVIRAL APPROACHES
In the absence of specific therapeutics, supportive care (eg, oxygen therapy or mechanical ventilation) remains the only option for managing severe Covid‐19 symptoms. 16 To limit virus proliferation in the early disease stages and reduce its severity, one important strategy is to develop treatments based on existing antiviral compounds. Following the rapid emergence and spread of SARS‐CoV in 2003 and MERS‐CoV in 2012, important efforts were made to identify drugs that target specific steps of the coronavirus life cycle (Figure 1) including (a) the interaction with ACE2 receptors, (b) enzymes that catalyze S protein cleavage, (c) viral entry, (d) polyprotein processing, and (e) replication/transcription complex. These compounds have been rapidly screened in SARS‐COV‐2‐infected cells, and many of them are now assessed in small animal models and/or clinical trials.
2.1. Antibody neutralization
A useful therapeutic strategy consists in interfering with the first step of the viral cycle by targeting the interaction of the viral S protein with the cell surface receptors. It was recently demonstrated that sera from patients with Covid‐19 contain neutralizing antibodies (Abs) that can limit SARS‐CoV‐2 viral infection in vitro. 17 Moreover, administration of plasma from convalescent patients has shown some efficacy in patients with respiratory distress, suggesting that passive immunity might help limit SARS‐CoV‐2 infection. 18 , 19 , 20 Clinical trials using plasmapheresis are under way to evaluate the efficacy of this strategy. 21 , 22 Nevertheless, recent randomized clinical trial have not shown significant improve for patient under convalescent plasma therapy. 23 The development of monoclonal Abs (mAbs) against the viral S protein is also an option. 24 This approach was used in the latest Ebola outbreak and led to the development of ZMapp. 25 Neutralizing mAbs against the SARS‐ and MERS‐CoV S protein have antiviral effect in cultured cells and animals infected with SARS‐CoV or MERS‐CoV. 26 , 27 , 28 , 29 , 30 Thus, the development of mAbs targeting the S protein is currently in progress. 31 The development of mAbs against ACE2 is also an interesting option to reduce viral entry, 32 , 33 but could be more risky due to potential interference with the physiological function of the ACE2 receptor, which is involved in angiotensin maturation. Another possibility is to use human recombinant soluble ACE2 to block SARS‐CoV‐2 entry into the cell. 34 This strategy seems to limit SARS‐CoV‐2 infection in cell culture, but the stability of the soluble receptor in the serum of infected patients and the potential adverse effects in patients remain to be determined. Notably, it has also been shown that ACE2 therapy protects mice from lung injury that can be observed during SARS‐CoV infection. 35 Interestingly, mAbs against other cellular receptors, such as CD147, also can block viral infection in cell culture, and the direct interaction between CD147 and the S protein has been demonstrated by coimmunoprecipitation. 36 These first results need be confirmed, but they suggest that SARS‐CoV‐2 could use an alternative entry receptor in some cell types. A clinical trial using an anti‐CD147 antibody (meplazumab) is currently ongoing in China 37 ; however, its antiviral effect is uncertain because the virus binds with higher affinity to the ACE2 receptor. 33
2.2. Antiviral molecules
Although SARS‐CoV‐2 has spread very recently, several antiviral compounds previously tested against other pathogenic CoVs have been already tried patients with Covid‐19 (Table 1). The first strategy is to interfere with virus entry using molecules that block virus‐cell fusion. This includes umifenovir (Arbidol), an antiviral molecule initially developed for the treatment of influenza infection. 62 Interestingly, umifenovir has a broad spectrum antiviral activity by inhibiting the entry of other viruses 63 and stimulating the immune response. 38 Moreover, umifenovir inhibits SARS‐CoV‐2 infection in vitro with an IC50 of 10 μM. 39 Although not yet approved by the FDA, this broad‐spectrum antiviral molecule is used in China and Russia for influenza treatment, and therefore was rapidly included in several clinical trials alone or in combination with other compounds. The first comparative study performed in a small group of patients showed a significant decrease of the viral load in the arm treated with umifenovir in combination with the protease inhibitors lopinavir/ritonavir (n = 16) compared with the arm treated with lopinavir/ritonavir alone (n = 17). 41 Nevertheless, one of the limitations of this study is the small‐sample size and investigation including large cohort need to be performed.
TABLE 1.
Main antiviral molecules currently being tested against SARS‐CoV‐2 infection
| Compounds | Target/MoA | Tests in vitro (CoV) | Clinical trials SARS‐CoV‐2 |
|---|---|---|---|
| Arbidol hydrochloride | Endocytosis/inhibition membrane fusion | (38, 39, 40) | NCT04252885 (41, 42) |
| Ribavirin | RNA polymerase/inhibition | (43, 44) | No data |
| Remdesivir | RNA polymerase/inhibition | (45, 46, 47, 48) | (49, 50) |
| Favipiravir | RNA polymerase/inhibition | No data | ChiCTR2000030894, NCT04280705, NCT04292730, NCT04292899, NCT04315948, WHO Solidarity Trial (42, 51) |
| β‐d‐N4‐hydroxycytidine | RNA polymerase/inhibition | (52, 53) | No data |
| Sofosbuvir | RNA polymerase/inhibition | (54) | No data |
| α‐ketoamide inhibitor | Proteases/inhibition | (55) | No data |
| Ritonavir | Proteases/inhibition | (56) | NCT04252885 (41, 49, 57, 58, 59) |
| Lopinavir | Proteases/inhibition | (56, 60) | NCT04252885 (41, 49, 57, 58, 59) |
| Nelfinavir | Proteases/inhibition | (61) | No data |
| Dolutegravir | Proteases/inhibition | (39) | No data |
Abbreviations: CoV, coronaviruses; MoA, mode of action.
After virus entry, the polyproteins pp1a and pp1ab are processed into 16 nsps by the viral proteases 3CLpro and PLpro. These cleavage events play a key role in the virus life cycle because they lead to the generation, among others, of the RNA‐dependent RNA polymerase (RdRp) required for viral RNA replication and transcription. Due to the high conservation of the cleavage sites and of the protease structures, these proteases are an optimal antiviral target. A recent study described the X‐ray structures of SARS‐CoV‐2 3CLpro alone and in the presence of an α‐ketoamide inhibitor. 55 Based on these findings, the α‐ketoamide inhibitor was optimized and showed antiviral activity against SARS‐CoV‐2 at concentrations lower than 10 μM in Calu3 lung cells. Protease inhibitors used in human immunodeficiency virus (HIV) therapy, such as lopinavir/ritonavir (a combination known as Kaletra), nelfinavir, also limit SARS‐CoV‐2 propagation in infected cells, 39 , 57 , 64 and they are being tested in several ongoing clinical trials. 56 , 60 , 61 However, the molecular basis of their inhibition mechanism has not been elucidated yet due to the fact that HIV and CoV proteases belong to different protease classes. Unfortunately, the first clinical trials reported that lopinavir, combined with the ritonavir that boosts lopinavir levels by interfering with cytochrome P450 metabolism, does not have any significant effect on SARS‐CoV‐2 infection. 58
Historically, the most attractive antiviral compounds are those that block virus replication by inhibiting RdRp activity. Nucleotide and nucleoside analogs are among the most promising groups of RdRp inhibitors. Their antiviral effect can be attributed to three nonmutually exclusive mechanisms. First, incorporation of nucleotide analogs (NAs) in the viral RNA by the error‐prone polymerase can induce early chain termination in an obligate (immediate) or nonobligate fashion, resulting in incomplete, noninfectious viral RNA. The second mechanism, named error catastrophe, is associated with the insertion and extension of NAs throughout the viral RNA that result in many errors during RNA synthesis. 65 , 66 Third, several NAs deplete the cytoplasmic levels of their equivalent native nucleotides, causing nucleotide pool imbalances that affect RdRp fidelity. 67 While nucleoside analogs have been successfully used for the treatment of other viral diseases, the situation is complicated in CoVs due to the presence of a viral exonuclease (nsp14 ExoN) with proof‐reading activity that can reduce NA antiviral effect. 68 , 69 Indeed, in vitro studies show that SARS‐CoV polymerase can incorporate ribavirin triphosphate during replication, but this purine NA is detected by nsp14 ExoN and eliminated by the repair mechanism. 68 This might partly explain the poor antiviral effect of ribavirin in SARS‐ and MERS‐CoV infection. 43 , 70 On the other hand, molecules such as remdesivir, initially developed against Ebola virus, 45 appear to have an inhibitory effect against SARS‐CoV and MERS‐CoV in vitro at the submicro‐ to micromolar range, depending on the infected cell type. 46 , 71 , 72 Remdesivir evaluation in a primate model infected by SARS‐CoV showed that the antiviral activity is dependent on early treatment after infection. 47 Remdesivir is incorporated into nascent RNA by the SARS‐CoV‐2 RdRp complex. 54 Several clinical trials, such as the European DISCOVERY clinical program, 49 are assessing remdesivir safety and efficacy in hospitalized patients with Covid‐19. The first results of these compassionate‐use studies showed clinical improvement in 68% of patients treated with remdesivir in a small cohort (n = 53). Notably however, these studies were not randomized and furthermore did not report the change in viral load following treatment. 50 While some adverse effects were reported, including abnormal liver function, diarrhea, rashes, renal impairment, and hypotension, remdesivir was licensed in May 2020 as an emergence treatment against SARS‐CoV‐2.
The cytidine analog β‐d‐N4‐hydroxycytidine (NHC) also is a potential anti‐CoV compound. NHC inhibits MERS‐CoV, SARS‐CoV, and SARS‐CoV‐2 replication in vitro in the micromolar range (0.09‐0.3 μM), without apparent interference from the viral proofreading activity of nsp14. 52 In infected cells, decreased viral replication was associated with increased mutation frequency, supporting a mechanism of lethal mutagenesis. Moreover, the orally bioavailable β‐D‐N4‐hydroxycytidine‐5′‐isopropyl ester improved pulmonary function and reduced virus titer and body weight loss in mice infected with SARS‐ and MERS‐CoV. These promising results suggest that NHC molecules might be considered for Covid‐19 treatment, 53 , 72 and should be assessed in clinical trials. Another potential treatment is the guanine analog favipiravir (T‐705) that was initially developed for influenza virus, 73 but has a broad‐spectrum activity, with antiviral effects against flaviviruses, 74 , 75 alphaviruses, 76 noroviruses, 77 and Ebola virus. 78 , 79 , 80 Favipiravir was approved for influenza treatment in Japan in 2014, and is currently assessed for Covid‐19 treatment in several clinical trials. 42 , 51 The preliminary results suggest a faster viral clearance time in the favipiravir arm compared with the control arm. 51 More recently, in vitro studies demonstrate that favipiravir exerts an antiviral effect as a nucleotide analogue through a combination of chain termination, slowed RNA synthesis and lethal mutagenesis. The use of favipiravir in infected cells induces C‐to‐U and G‐to‐A changes. 81 A last nucleoside analog potentially active against SARS‐CoV‐2 is sofosbuvir, a broad‐acting antiviral approved for hepatitis C virus infection management. A recent study showed that sofosbuvir triphosphate is incorporated by recombinant SARS‐CoV‐2 RdRp during RNA elongation. 54 More work is needed to confirm its antiviral effect in infected cells and animal models before moving to clinical trials.
3. DRUG REPOSITIONING STRATEGIES FOR COVID‐19 TREATMENT
The fastest option for the treatment of Covid‐19 is the identification of already approved drugs that were developed for other diseases, but that show inhibitory activity against SARS‐CoV‐2 in infected cells. The main advantages of this strategy are that these drugs are available on the market, and their safety and toxicity profiles are already documented. However, their MoA against the virus is often speculative, and the efficacy and clinical doses required for treatment in patients with Covid‐19 are unknown, due to the absence of large‐scale clinical studies. Moreover, it is important to consider the risk of overdose and of unexpected increase of viral load or symptoms. Several teams have already set up platforms to screen FDA‐approved libraries using SARS‐CoV‐2 infected cells. 39 , 64 , 82 Although the chemical libraries and experimental conditions used in these studies are quite different, a comparative analysis of their results reveals that some compounds, such as chloroquine derivatives, have been identified in different experimental conditions. Moreover, several groups reported the antiviral activity of FDA‐approved compounds, and some of them are already tested in clinical trials. 15 In this review, we cannot discuss all drugs identified by this strategy. Therefore, we selected the key compounds that are already in clinical trials and the most interesting classes of compounds, based on their potential MoA on CoV life cycle inferred from their known activity.
3.1. Immunomodulators
As some patients with Covid‐19 show an inefficient antiviral response, it has been suggested that immunomodulators might restore the immune system homeostasis. For instance, interferon (IFN) could be used during the early stages of infection to boost the innate immune response and promote viral clearance. IFN treatments are being tested in several clinical trials, but results are not available yet. However, as SARS‐CoV‐2 infection may also be accompanied by a dysregulated immune response leading to a massive production of pro‐inflammatory cytokines, IFN treatment could have unexpected effects. 5 , 83 , 84 , 85 , 86 Indeed, IFN triggers overexpression of ACE2, even in cell lines with low basal expression level, leading to a larger dissemination of SARS‐CoV‐2. 87 Conversely, in the second phase of the disease, the use of molecules that limit the effect of the cytokine storm could be advantageous. For instance, the mAbs mepolizumab against interleukin (IL)‐5, tocilizumab against IL‐6 receptor, 88 as well as anakinra an interleukin 1 receptor antagonist 89 could control the immune response, and be of therapeutic interest for managing the production of pro‐inflammatory cytokines in patients with severe infection. 90 Finally, Dexamethasone, used to reduce inflammation, has been tested in RECOVERY clinical trial and shown first promising results. Indeed, treatment with Dexamethasone reduces death by one‐third to one‐fifth. 91
3.2. Repositioned drugs that interfere with the viral life cycle
Several therapeutic molecules (Table 2), identified in antiviral screenings performed in infected cells, target the renin/angiotensin pathway. 39 , 100 , 101 The MoA of these class of inhibitors is speculative. Interestingly, ACE2 belongs to this pathway. An in vivo study has shown that inhibition of the renin/angiotensin pathway upregulates of ACE2 which can increase susceptibility to SARS‐CoV‐2 infection. 102 More recently, it has been shown that treatment target this pathway is not associated with higher severity of COVID19 103 Additional work is needed to understand how these compounds inhibit the virus before planning clinical trials.
TABLE 2.
Main repositioning molecules currently being tested against SARS‐CoV‐2 infection
| Compounds | Target/MoA | Tests in vitro (CoV) | Clinical trials SARS‐CoV‐2 |
|---|---|---|---|
| Antivirals | |||
| Nafamostat mesylate | TMPRSS2/inhibitor | (92) | No data |
| Camostat mesylate | TMPRSS2/inhibitor | (39, 92) | No data |
|
Chloroquine Hydroxychloroquine |
pH increases in endosomal compartment | (39, 48, 93, 94, 95, 96) | “Solidarity,” “Discovery” NCT04358068 (49, 97, 98) |
| Azithromycin (antibiotic) | pH increases in endosomal compartment/immunomodulator | (39) | NCT04358068 (98) |
| Omeprazole | PPI | (39) | No data |
| Vonoprazan | PPI | (39) | No data |
| Ivermectin | Limits viral infection | (99) | No data |
| Oseltamivir | Neuraminidase inhibitor | No data | No data |
Note: Clinical trials web site: https://clinicaltrials.gov/ct2/results?cond=SARS‐CoV2&term=&cntry=&state=&city=&dist=.
Abbreviations: CoV, coronaviruses; MoA, mode of action; PPI, proton pump inhibitor; TMPRSS2, transmembrane protease serine 2.
Once the virus binds to the target cell, the S protein is cleaved by cellular proteases into the S1 and S2 subunits (priming), and also upstream of the fusion peptide (S2’ cleavage site). The serine protease TMPRSS2, which is strongly expressed in lung cells, can cleave a variety of CoV S proteins, 12 , 13 and seems to play a key role in virus entry. 14 Camostat mesylate, a TMPRSS2 inhibitor approved for clinical use, blocks S protein processing in vitro. 92 Moreover, camostat mesylate and its derivative nafamostat mesylate can inhibit (in the nM to μM range) the entry of vesicular stomatitis virus pseudotypes carrying the SARS‐CoV‐2 S protein in different cell types that overexpress TMPRSS2. 92 , 104 This observation suggests that inhibition of S protein priming blocks ACE2‐mediated entry. However, the inhibition observed with camostat mesylate and nafamostat mesylate in VeroE6 cells infected by SARS‐CoV‐2 is lower and probably not enough for a therapeutic utilization of camostat mesylate. The picture is further complicated because other cellular proteases could be implicated in S protein processing. Indeed, cathepsin L 105 and furin 9 might be involved in S protein activation, depending on the infected cell type. Consequently, inhibitors of these different enzymes might limit virus propagation in some cell types, but their efficacy in the clinic could be limited. Perhaps, the best option is to combine furin and TMPRSS2 inhibitors to limit virus propagation. 14
Interestingly, several compounds identified in different screens of infected cells are implicated in endosomal acidification. 39 , 100 , 101 Increasing the endosomal pH might limit viral/endosomal membrane fusion, which is necessary for the release of the viral genome into the cytoplasm of the host cell. Two recent studies showed that proton pump inhibitors, such as omeprazole and vonoprazan, reduce the infection of cells by SARS‐CoV‐2. 39 , 64 In addition, chloroquine and hydroxychloroquine (HCQ), which have been extensively used for the treatment of malaria with known safety and efficacy, also limit acidification of endosomes, Golgi vesicles, and lysosomes. 106 These molecules significantly reduce SARS‐CoV and SARS‐CoV‐2 infection in vitro. 48 , 93 , 94 , 95 , 106 , 107 However, a recent study reported that HCQ does not have any effect on viral load levels and does not protect macaques against SARS‐CoV infection. 108 Moreover, chloroquine derivatives have immunosuppressive effects and inhibit the activation of innate immune receptors, such as toll‐like receptors 109 and have been associated with some adverse cardiac effects. Many clinical studies are currently testing these molecules, 97 , 98 and their initial results are controversial. 110 Randomized, double‐blind studies with a larger number of patients should bring robust conclusions. Recent in vitro studies have also suggested the therapeutic effect of the HCQ‐azithromycin combination. 98 Azithromycin is an antibiotic that has also antiviral activity in respiratory viral infections. 111 Interestingly, the MoA of azithromycin and HCQ may be related to the modulation of the endosomal and trans‐Golgi network pH. 112 , 113 In addition, azithromycin plays a role in regulating interleukin production that might help to control the immune response and prevent symptom worsening. 114 , 115 , 116 Azithromycin may also inhibit viral invasion by interfering with the CD147‐mediated recognition mechanism. 117 Nevertheless, both the FDA and the World Health Organization (WHO) have removed their support for the use of HCQ and chloroquine for COVID‐19, consequently the status of ongoing trials must be questioned.
More recently, it has been reported that ivermectin inhibits SARS‐CoV‐2 with an ~5000‐fold reduction in viral RNA after 48 hours. 99 Ivermectin is a FDA‐approved antiparasitic molecule, initially identified as an inhibitor of HIV protein nuclear import. 118 Its broad‐spectrum antiviral activity was documented in vitro. 119 , 120 Ivermectin perturbs nuclear import through IMPα/β1; however, the role of this machinery during the CoV life cycle has not been elucidated yet. Nevertheless, an observational study reported survival benefits during hospitalization, 121 but no data on viral load, highlighting the necessity to validate this observation in clinical trials. Recent review presents the pharmacokinetic properties of ivermectin, and highlights that the dose currently used for parasitic disease do not effective against SARS infection. 122
Finally, oseltamivir, a neuraminidase inhibitor that prevents influenza viral particle release, is also investigated. 123 Like for ritonavir/lopinavir, there is no molecular basis to support these trials because to the best of our knowledge, CoVs (unlike influenza viruses) do not rely on neuraminidases during their life cycle. If antiviral effect are observed, the off‐target mechanisms will have to be elucidated.
CONFLICT OF INTEREST
The authors have no competing interest.
AUTHOR CONTRIBUTIONS
Coralie Valle, Baptiste Martin, Bruno Coutard, and Etienne Decroly were major contributors to writing the manuscript. Coralie Valle, Baptiste Martin, Franck Touret, Ashleigh Shannon, Bruno Canard, Jean Claude Guillemot, Bruno Coutard, and Etienne Decroly checked and revised the manuscript. All authors read and approved the final version.
ACKNOWLEDGMENTS
This work was partly supported by Inserm through the REACTing initiative (REsearch and ACTion targeting emerging infectious diseases), a multidisciplinary collaborative network of French research institutions working on emerging infectious diseases, which aims to prepare and respond to epidemics (B. Canard, B. Coutard, E. D.), and by the Fondation pour la Recherche Médicale (Aide aux équipes, B. Canard, E. D.). The SCORE project has received funding from the European Union's Horizon 2020 research and innovation programme under grant agreement No 101003627.
Valle C, Martin B, Touret F, et al. Drugs against SARS‐CoV‐2: What do we know about their mode of action? Rev Med Virol. 2020;30:e2143. 10.1002/rmv.2143
Funding information European Union's Horizon 2020 research and innovation programme, Grant/Award Number: 101003627; SCORE project H2020 SC1‐PHE‐Coronavirus194 2020, Grant/Award Number: 101003627; Fondation pour la Recherche Médicale; Inserm
Contributor Information
Bruno Coutard, Email: bruno.coutard@univ-amu.fr.
Etienne Decroly, Email: etienne.decroly@afmb.univ-mrs.fr.
REFERENCES
- 1. Ksiazek TG, Erdman D, Goldsmith CS, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348(20):1953‐1966. [DOI] [PubMed] [Google Scholar]
- 2. Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus ADME, RAM F. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367(19):1814‐1820. [DOI] [PubMed] [Google Scholar]
- 3. Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265‐269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID‐19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054‐1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14(8):523‐534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li F. Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J Virol. 2015;89(4):1954‐1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade‐long structural studies of SARS coronavirus. J Virol. 2020;94(7):120–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E. The spike glycoprotein of the new coronavirus 2019‐nCoV contains a furin‐like cleavage site absent in CoV of the same clade. Antiviral Res. 2020;176:104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Matsuyama S, Nagata N, Shirato K, Kawase M, Takeda M, Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J Virol. 2010;84(24):12658‐12664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Markus H, Kleine‐Weber H, Pöhlmann S. A multibasic cleavage site in the spike protein of SARS‐CoV‐2 is essential for infection of human lung cells. Mol Cell. 2020;78(4):779–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Belouzard S, Chu VC, Whittaker GR. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc Natl Acad Sci U S A. 2009;106(14):5871‐5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Millet JK, Whittaker GR. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120‐134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bestle D, Heindl MR, Limburg H, et al. TMPRSS2 and furin are both essential for proteolytic activation and spread of SARS‐CoV‐2 in human airway epithelial cells and provide promising drug targets. bioRxiv;2020.04.15.042085. https://www.biorxiv.org/content/10.1101/2020.04.15.042085v1.abstract. [Accessed May 1, 2020]. [DOI] [PMC free article] [PubMed]
- 15. Thorlund K, Dron L, Park J, Hsu G, Forrest JI, Mills EJ. A real‐time dashboard of clinical trials for COVID‐19. Lancet Digit Heal. 2020;2:286–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clinical Management of Severe Acute Respiratory Infection When COVID‐19 Is Suspected. https://www.who.int/publications/i/item/clinical-management-of-covid-19. Accessed May 27, 2020.
- 17. Zhou G, Zhao Q. Perspectives on therapeutic neutralizing antibodies against the Novel Coronavirus SARS‐CoV‐2. Int J Biol Sci. 2020;16(10):1718‐1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cheng Y, Wong R, Soo YOY, et al. Use of convalescent plasma therapy in SARS patients in Hong Kong. Eur J Clin Microbiol Infect Dis. 2005;24(1):44‐46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mair‐Jenkins J, Saavedra‐Campos M, Baillie JK, et al. The effectiveness of convalescent plasma and hyperimmune immunoglobulin for the treatment of severe acute respiratory infections of viral etiology: a systematic review and exploratory meta‐analysis. J Infect Dis. 2015;211(1):80‐90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yeh K‐M, Chiueh T‐S, Siu LK, et al. Experience of using convalescent plasma for severe acute respiratory syndrome among healthcare workers in a Taiwan hospital. J Antimicrob Chemother. 2005;56(5):919‐922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen L, Xiong J, Bao L, Shi Y. Convalescent plasma as a potential therapy for COVID‐19. Lancet Infect Dis. 2020;20(4):398‐400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shen C, Wang Z, Zhao F, et al. Treatment of 5 critically ill patients with COVID‐19 with convalescent plasma. JAMA. 2020;323:1582–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li L, Zhang W, Hu Y, et al. Effect of convalescent plasma therapy on time to clinical improvement in patients with severe and life‐threatening COVID‐19. JAMA. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Berry JD. Rational monoclonal antibody development to emerging pathogens, biothreat agents and agents of foreign animal disease: the antigen scale. Vet J. 2005;170(2):193‐211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hoenen T, Groseth A, Feldmann H. Therapeutic strategies to target the Ebola virus life cycle. Nat Rev Microbiol. 2019;17(10):593‐606. [DOI] [PubMed] [Google Scholar]
- 26. Wang L, Shi W, Chappell JD, et al. Importance of neutralizing monoclonal antibodies targeting multiple antigenic sites on the Middle East respiratory syndrome coronavirus spike glycoprotein to avoid neutralization escape. J Virol. 2018;92(10):2002–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hofmann H, Hattermann K, Marzi A, et al. S protein of severe acute respiratory syndrome‐associated coronavirus mediates entry into hepatoma cell lines and is targeted by neutralizing antibodies in infected patients. J Virol. 2004;78(12):6134‐6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. He Y, Lu H, Siddiqui P, Zhou Y, Jiang S. Receptor‐binding domain of severe acute respiratory syndrome coronavirus spike protein contains multiple conformation‐dependent epitopes that induce highly potent neutralizing antibodies. J Immunol. 2005;174(8):4908‐4915. [DOI] [PubMed] [Google Scholar]
- 29. Sui J, Li W, Murakami A, et al. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc Natl Acad Sci U S A. 2004;101(8):2536‐2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Greenough TC, Babcock GJ, Roberts A, et al. Development and characterization of a severe acute respiratory syndrome–associated coronavirus–neutralizing human monoclonal antibody that provides effective immunoprophylaxis in mice. J Infect Dis. 2005;191(4):507‐514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wrapp D, De Vlieger D, Corbett KS, et al. Structural basis for potent neutralization of betacoronaviruses by single‐domain camelid antibodies. Cell. 2020;181(5):1004–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhu Z, Chakraborti S, He Y, et al. Potent cross‐reactive neutralization of SARS coronavirus isolates by human monoclonal antibodies. Proc Natl Acad Sci U S A. 2007;104(29):12123‐12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Walls AC, Park Y‐J, Tortorici MA, Wall A, AT MG, Veesler D. Structure, function, and antigenicity of the SARS‐CoV‐2 spike glycoprotein. Cell. 2020;181:281–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Monteil V, Kwon H, Prado P, et al. Inhibition of SARS‐CoV‐2 infections in engineered human tissues using clinical‐grade soluble human ACE2. Cell. 2020;181:905–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Imai Y, Kuba K, Penninger JM. The discovery of angiotensin‐converting enzyme 2 and its role in acute lung injury in mice. Exp Physiol. 2008;93:543‐548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang K, Chen W, Zhou YS, et al. SARS‐CoV‐2 invades host cells via a novel route: CD147‐spike protein. bioRxiv. 2020. https://www.biorxiv.org/content/10.1101/2020.03.14.988345v1. Accessed April 5, 2020.
- 37. Bian H, Zheng Z‐H, Wei D, et al. Meplazumab treats COVID‐19 pneumonia: an open‐labelled, concurrent controlled add‐on clinical trial. medRxiv. 2020. https://www.medrxiv.org/content/10.1101/2020.03.21.20040691v1. Accessed April 4, 2020.
- 38. Boriskin YS, Leneva IA, Pécheur E‐I, Polyak SJ. Arbidol: a broad‐spectrum antiviral compound that blocks viral fusion. Curr Med Chem. 2008;15(10):997‐1005. [DOI] [PubMed] [Google Scholar]
- 39. Touret F, Gilles M, Barral K, et al. in vitro screening of a FDA approved chemical library reveals potential inhibitors of SARS‐CoV‐2 replication. bioRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 40. Khamitov RA, Loginova SI, Shchukina VN, Borisevich SV, Maksimov VA, Shuster AM. Antiviral activity of arbidol and its derivatives against the pathogen of severe acute respiratory syndrome in the cell cultures. Vopr Virusol. 2008;53(4):9‐13. [PubMed] [Google Scholar]
- 41. Deng L, Li C, Zeng Q, et al. Arbidol combined with LPV/r versus LPV/r alone against Corona Virus Disease 2019: a retrospective cohort study. J Infect. 2020;81:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen C, Huang J, Cheng Z, et al. Favipiravir versus arbidol for COVID‐19: a randomized clinical trial. medRxiv. 2020. https://www.medrxiv.org/content/10.1101/2020.03.17.20037432v2. Accessed April 4, 2020.
- 43. Falzarano D, de Wit E, Martellaro C, Callison J, Munster VJ, Feldmann H. Inhibition of novel β coronavirus replication by a combination of interferon‐α2b and ribavirin. Sci Rep. 2013;3(1):1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Barnard DL, Day CW, Bailey K, et al. Enhancement of the infectivity of SARS‐CoV in BALB/c mice by IMP dehydrogenase inhibitors, including ribavirin. Antiviral Res. 2006;71(1):53‐63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Warren TK, Jordan R, Lo MK, et al. Therapeutic efficacy of the small molecule GS‐5734 against Ebola virus in rhesus monkeys. Nature. 2016;531(7594):381‐385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Agostini ML, Andres EL, Sims AC, et al. Coronavirus susceptibility to the antiviral remdesivir (GS‐5734) is mediated by the viral polymerase and the proofreading exoribonuclease. MBio. 2018;9(2):e00221–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. de Wit E, Feldmann F, Cronin J, et al. Prophylactic and therapeutic remdesivir (GS‐5734) treatment in the rhesus macaque model of MERS‐CoV infection. Proc Natl Acad Sci U S A. 2020;117(12):6771‐6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang M, Cao R, Zhang L, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019‐nCoV) in vitro. Cell Res. 2020;30(3):269‐271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Launch of a European Clinical Trial Against COVID‐19 | Newsroom | INSERM. https://presse.inserm.fr/en/launch-of-a-european-clinical-trial-against-covid-19/38737/. Accessed April 6, 2020.
- 50. Grein J, Ohmagari N, Shin D, et al. Compassionate use of remdesivir for patients with severe Covid‐19. N Engl J Med. 2020;382:2327–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cai Q, Yang M, Liu D, et al. Experimental treatment with favipiravir for COVID‐19: an open‐label control study. Engineering, in press. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sheahan TP, Sims AC, Zhou S, et al. An orally bioavailable broad‐spectrum antiviral inhibits SARS‐CoV‐2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci Transl Med. 2020;12:eabb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Agostini ML, Pruijssers AJ, Chappell JD, et al. Small‐molecule antiviral β‐D‐ N 4 ‐hydroxycytidine inhibits a proofreading‐intact coronavirus with a high genetic barrier to resistance. J Virol. 2019;93(24):e01348–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gordon CJ, Tchesnokov EP, Woolner E, et al. Remdesivir is a direct‐acting antiviral that inhibits RNA‐dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J Biol Chem. 2020;295:6785–6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang L, Lin D, Sun X, et al. Crystal structure of SARS‐CoV‐2 main protease provides a basis for design of improved α‐ketoamide inhibitors. Science. 2020;368:409–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chu CM, Cheng VCC, Hung IFN, et al. Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax. 2004;59(3):252‐256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bhatnagar T, Murhekar MV, Soneja M, et al. Lopinavir/ritonavir combination therapy amongst symptomatic coronavirus disease 2019 patients in India: protocol for restricted public health emergency use. Indian J Med Res. 2020;151:184–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cao B, Wang Y, Wen D, et al. A trial of lopinavir‐ritonavir in adults hospitalized with severe Covid‐19. N Engl J Med. 2020;382:1787–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lim J, Jeon S, Shin HY, et al. Case of the index patient who caused tertiary transmission of COVID‐19 infection in Korea: the application of lopinavir/ritonavir for the treatment of COVID‐19 infected pneumonia monitored by quantitative RT‐PCR. J Korean Med Sci. 2020;35(6):e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. de Wilde AH, Jochmans D, Posthuma CC, et al. Screening of an FDA‐approved compound library identifies four small‐molecule inhibitors of Middle East respiratory syndrome coronavirus replication in cell culture. Antimicrob Agents Chemother. 2014;58(8):4875‐4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yamamoto N, Yang R, Yoshinaka Y, et al. HIV protease inhibitor nelfinavir inhibits replication of SARS‐associated coronavirus. Biochem Biophys Res Commun. 2004;318(3):719‐725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kadam RU, Wilson IA. Structural basis of influenza virus fusion inhibition by the antiviral drug Arbidol. Proc Natl Acad Sci U S A. 2017;114(2):206‐214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Vigant F, Santos NC, Lee B. Broad‐spectrum antivirals against viral fusion. Nat Rev Microbiol. 2015;13(7):426‐437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Riva L, Yuan S, Yin X, et al. A large‐scale drug repositioning survey for SARS‐CoV‐2 antivirals. bioRxiv. 2020. https://www.biorxiv.org/content/10.1101/2020.04.16.044016v1. Accessed April 27, 2020.
- 65. Crotty S, Maag D, Arnold JJ, et al. The broad‐spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat Med. 2000;6(12):1375‐1379. [DOI] [PubMed] [Google Scholar]
- 66. Pruijssers AJ, Denison MR. Nucleoside analogues for the treatment of coronavirus infections. Curr Opin Virol. 2019;35:57‐62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Feld JJ, Hoofnagle JH. Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature. 2005;436(7053):967‐972. [DOI] [PubMed] [Google Scholar]
- 68. Ferron F, Subissi L, Silveira De Morais AT, et al. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc Natl Acad Sci U S A. 2018;115(2):E162‐E171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Smith EC, Blanc H, Surdel MC, Vignuzzi M, Denison MR. Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: evidence for proofreading and potential therapeutics. PLoS Pathog. 2013;9(8):e1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ströher U, DiCaro A, Li Y, et al. Severe acute respiratory syndrome–related coronavirus is inhibited by interferon‐α. J Infect Dis. 2004;189(7):1164‐1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sheahan TP, Sims AC, Graham RL, et al. Broad‐spectrum antiviral GS‐5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med. 2017;9(396):eaal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sheahan TP, Sims AC, Leist SR, et al. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS‐CoV. Nat Commun. 2020;11(1):222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Furuta Y, Takahashi K, Fukuda Y, et al. In vitro and in vivo activities of anti‐influenza virus compound T‐705. Antimicrob Agents Chemother. 2002;46(4):977‐981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kim J‐A, Seong R‐K, Kumar M, Shin OS. Favipiravir and ribavirin inhibit replication of Asian and African strains of Zika virus in different cell models. Viruses. 2018;10(2):72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zmurko J, Marques RE, Schols D, Verbeken E, SJF K, Neyts J. The viral polymerase inhibitor 7‐deaza‐2’‐C‐methyladenosine is a potent inhibitor of in vitro Zika virus replication and delays disease progression in a robust mouse infection model. PLoS Negl Trop Dis. 2016;10(5):e0004695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Delang L, Segura Guerrero N, Tas A, et al. Mutations in the chikungunya virus non‐structural proteins cause resistance to favipiravir (T‐705), a broad‐spectrum antiviral. J Antimicrob Chemother. 2014;69(10):2770‐2784. [DOI] [PubMed] [Google Scholar]
- 77. Rocha‐Pereira J, Jochmans D, Dallmeier K, Leyssen P, Nascimento MSJ, Neyts J. Favipiravir (T‐705) inhibits in vitro norovirus replication. Biochem Biophys Res Commun. 2012;424(4):777‐780. [DOI] [PubMed] [Google Scholar]
- 78. Guedj J, Piorkowski G, Jacquot F, et al. Antiviral efficacy of favipiravir against Ebola virus: a translational study in cynomolgus macaques. PLoS Med. 2018;15(3):e1002535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Oestereich L, Lüdtke A, Wurr S, Rieger T, Muñoz‐Fontela C, Günther S. Successful treatment of advanced Ebola virus infection with T‐705 (favipiravir) in a small animal model. Antiviral Res. 2014;105:17‐21. [DOI] [PubMed] [Google Scholar]
- 80. Smither SJ, Eastaugh LS, Steward JA, Nelson M, Lenk RP, Lever MS. Post‐exposure efficacy of oral T‐705 (favipiravir) against inhalational Ebola virus infection in a mouse model. Antiviral Res. 2014;104:153‐155. [DOI] [PubMed] [Google Scholar]
- 81. Shannon A, Selisko B, Le TTN , et al. Favipiravir strikes the SARS‐CoV‐2 at its Achilles heel, the RNA polymerase. Nature Com, in press. 2020.
- 82. Jeon S, Ko M, Lee J, et al. Identification of antiviral drug candidates against SARS‐CoV‐2 from FDA‐approved drugs. bioRxiv. 2020. https://www.biorxiv.org/content/10.1101/2020.03.20.999730v3. Accessed April 27, 2020. [DOI] [PMC free article] [PubMed]
- 83. Cinatl J, Morgenstern B, Bauer G, Chandra P, Rabenau H, Doerr HW. Treatment of SARS with human interferons. Lancet. 2003;362(9380):293‐294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Hensley LE, Fritz EA, Jahrling PB, Karp C, Huggins JW, Geisbert TW. Interferon‐β 1a and SARS coronavirus replication. Emerg Infect Dis. 2004;10(2):317‐319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Paragas J, Blatt L, Hartmann C, Huggins J, Endy T. Interferon alfacon1 is an inhibitor of SARS‐corona virus in cell‐based models. Antiviral Res. 2005;66(2‐3):99‐102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Totura AL, Whitmore A, Agnihothram S, et al. Toll‐like receptor 3 signaling via TRIF contributes to a protective innate immune response to severe acute respiratory syndrome coronavirus infection. MBio. 2015;6(3):e00638‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ziegler CGK, Allon SJ, Nyquist SK, et al. SARS‐CoV‐2 receptor ACE2 is an interferon‐stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zhang C, Wu Z, Li J‐W, Zhao H, Wang GQ. The cytokine release syndrome (CRS) of severe COVID‐19 and Interleukin‐6 receptor (IL‐6R) antagonist Tocilizumab may be the key to reduce the mortality. Int J Antimicrob Agents. 2020;55:105954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Conti P, Gallenga CE, Tetè G, et al. How to reduce the likelihood of coronavirus‐19 (CoV‐19 or SARS‐CoV‐2) infection and lung inflammation mediated by IL‐1. J Biol Regul Homeost Agents. 2020;34(2):11–16. [DOI] [PubMed] [Google Scholar]
- 90. Conti P, Ronconi G, Caraffa A, et al. Induction of pro‐inflammatory cytokines (IL‐1 and IL‐6) and lung inflammation by Coronavirus‐19 (COVI‐19 or SARS‐CoV‐2): anti‐inflammatory strategies. J Biol Regul Homeost Agents. 2020;34(2). [DOI] [PubMed] [Google Scholar]
- 91. Ledford H. Coronavirus breakthrough: dexamethasone is first drug shown to save lives. Nature. 2020;582:469 [DOI] [PubMed] [Google Scholar]
- 92. Hoffmann M, Schroeder S, Kleine‐Weber H, Müller MA, Drosten C, Pöhlmann S. Nafamostat Mesylate Blocks Activation of SARS‐CoV‐2: New Treatment Option. The Currently Unfolding Coronavirus Pandemic Threatens Health Systems and Economies. 2020. http://aac.asm.org/. Accessed April 27, 2020.
- 93. Vincent MJ, Bergeron E, Benjannet S, et al. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol J. 2005;2(1):69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Keyaerts E, Vijgen L, Maes P, Neyts J, Van Ranst M. In vitro inhibition of severe acute respiratory syndrome coronavirus by chloroquine. Biochem Biophys Res Commun. 2004;323(1):264‐268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Yao X, Ye F, Zhang M, et al. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). Clin Infect Dis. 2020. [Online ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Liu J, Cao R, Xu M, et al. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS‐CoV‐2 infection in vitro. Cell Discov. 2020;6(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Gao J, Tian Z, Yang X. Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID‐19 associated pneumonia in clinical studies. Biosci Trends. 2020;14(1):72‐73. [DOI] [PubMed] [Google Scholar]
- 98. Gautret P, Lagier J‐C, Parola P, et al. Hydroxychloroquine and azithromycin as a treatment of COVID‐19: results of an open‐label non‐randomized clinical trial. Int J Antimicrob Agents. 2020;105949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Caly L, Druce JD, Catton MG, Jans DA, Wagstaff KM. The FDA‐approved Drug Ivermectin inhibits the replication of SARS‐CoV‐2 in vitro. Antiviral Res. 2020;178:104787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Kuster GM, Pfister O, Burkard T, et al. SARS‐CoV2: should inhibitors of the renin–angiotensin system be withdrawn in patients with COVID‐19? Eur Heart J. 2020;41:1801–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Meng J, Xiao G, Zhang J, et al. Renin‐angiotensin system inhibitors improve the clinical outcomes of COVID‐19 patients with hypertension. Emerg Microbes Infect. 2020;9(1):757‐760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Sukumaran V, Tsuchimochi H, Tatsumi E, Shirai M, Pearson JT. Azilsartan ameliorates diabetic cardiomyopathy in young db/db mice through the modulation of ACE‐2/ANG 1–7/Mas receptor cascade. Biochem Pharmacol. 2017;144:90‐99. [DOI] [PubMed] [Google Scholar]
- 103. Fosbøl EL, Butt JH, Østergaard L, et al. Association of angiotensin‐converting enzyme inhibitor or angiotensin receptor blocker use with COVID‐19 diagnosis and mortality. JAMA. 2020;126:1671–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Hoffmann M, Schroeder S, Kleine‐Weber H, Müller MA, Drosten C, Pöhlmann S. Nafamostat mesylate blocks activation of SARS‐CoV‐2: new treatment option for COVID‐19. Antimicrob Agents Chemother. 2020;64:754–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Hoffmann M, Kleine‐Weber H, Schroeder S, et al. SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Savarino A, Boelaert JR, Cassone A, Majori G, Cauda R. Effects of chloroquine on viral infections: an old drug against today's diseases. Lancet Infect Dis. 2003;3(11):722‐727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Pantoliano MW, Petrella EC, Kwasnoski JD, et al. High‐density miniaturized thermal shift assays as a general strategy for drug discovery. J Biomol Screen. 2001;6(6):429‐440. [DOI] [PubMed] [Google Scholar]
- 108. Tripathy S, Dassarma B, Roy S, Chabalala H, Matsabisa MG. A review on possible modes of action of chloroquine/hydroxychloroquine: repurposing against SAR‐CoV‐2 (COVID‐19) pandemic. Int J Antimicrob Agents. 2020;106028. http://dx.doi/10.1016/j.ijantimicag.2020.106028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Schrezenmeier E, Dörner T. Mechanisms of action of hydroxychloroquine and chloroquine: implications for rheumatology. Nat Rev Rheumatol. 2020;16(3):155‐166. [DOI] [PubMed] [Google Scholar]
- 110.Statement on IJAA Paper. International Society of Antimicrobial Chemotherapy. https://www.isac.world/news-and-publications/official-isac-statement. Accessed April 30, 2020.
- 111. Gielen V, Johnston SL, Edwards MR. Azithromycin induces anti‐viral responses in bronchial epithelial cells. Eur Respir J. 2010;36(3):646‐654. [DOI] [PubMed] [Google Scholar]
- 112. Deretic V, Timmins GS. Azithromycin and ciprofloxacin have a chloroquine‐like effect on respiratory epithelial cells. bioRxiv. 2020. https://www.biorxiv.org/content/10.1101/2020.03.29.008631v1. Accessed April 5, 2020.
- 113. Pani A, Lauriola M, Romandini A, Scaglione F. Macrolides and viral infections: focus on azithromycin in COVID‐19 pathology. Int J Antimicrob Agents. 2020;106053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Aghai ZH, Kode A, Saslow JG, et al. Azithromycin suppresses activation of nuclear factor‐kappa B and synthesis of pro‐inflammatory cytokines in tracheal aspirate cells from premature infants. Pediatr Res. 2007;62(4):483‐488. [DOI] [PubMed] [Google Scholar]
- 115. Taylor SP, Sellers E, Taylor BT. Azithromycin for the prevention of COPD exacerbations: the good, bad, and ugly. Am J Med. 2015;128(12):1362. [DOI] [PubMed] [Google Scholar]
- 116. Bouwman JJ, Visseren FL, Bouter PK, Diepersloot RJ. Azithromycin inhibits interleukin‐6 but not fibrinogen production in hepatocytes infected with cytomegalovirus and chlamydia pneumoniae. J Lab Clin Med. 2004;144(1):18‐26. [DOI] [PubMed] [Google Scholar]
- 117. Ulrich H, Pillat MM. CD147 as a target for COVID‐19 treatment: suggested effects of azithromycin and stem cell engagement. Stem Cell Rev Reports. 2020;16:434–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Wagstaff KM, Sivakumaran H, Heaton SM, Harrich D, Jans DA. Ivermectin is a specific inhibitor of importin α/β‐mediated nuclear import able to inhibit replication of HIV‐1 and dengue virus. Biochem J. 2012;443(3):851‐856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Lundberg L, Pinkham C, Baer A, et al. Nuclear import and export inhibitors alter capsid protein distribution in mammalian cells and reduce Venezuelan Equine Encephalitis Virus replication. Antiviral Res. 2013;100(3):662‐672. [DOI] [PubMed] [Google Scholar]
- 120. Tay MYF, Fraser JE, Chan WKK, et al. Nuclear localization of dengue virus (DENV) 1–4 non‐structural protein 5; protection against all 4 DENV serotypes by the inhibitor Ivermectin. Antiviral Res. 2013;99(3):301‐306. [DOI] [PubMed] [Google Scholar]
- 121. Patel A. Usefulness of Ivermectin in COVID‐19 Illness. 2020. https://papers.ssrn.com/sol3/papers.cfm?abstract_id=3580524. Accessed April 27, 2020.
- 122. Momekov G, Momekova D. Ivermectin as a potential COVID‐19 treatment from the pharmacokinetic point of view: antiviral levels are not likely attainable with known dosing regimens. Biotechnol Biotechnol Equip. 2020;34(1):469–474. 10.1101/2020.04.11.20061804. [DOI] [Google Scholar]
- 123. Gubareva LV, Kaiser L, Hayden FG. Influenza virus neuraminidase inhibitors. Lancet. 2000;355(9206):827‐835. [DOI] [PubMed] [Google Scholar]


