Summary
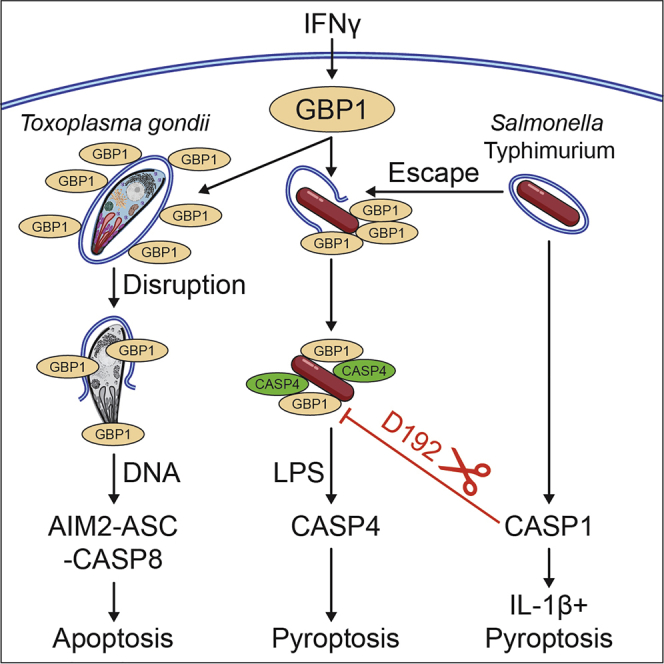
Interferon-inducible guanylate-binding proteins (GBPs) promote cell-intrinsic defense through host cell death. GBPs target pathogens and pathogen-containing vacuoles and promote membrane disruption for release of microbial molecules that activate inflammasomes. GBP1 mediates pyroptosis or atypical apoptosis of Salmonella Typhimurium (STm)- or Toxoplasma gondii (Tg)- infected human macrophages, respectively. The pathogen-proximal detection-mechanisms of GBP1 remain poorly understood, as humans lack functional immunity-related GTPases (IRGs) that assist murine Gbps. Here, we establish that GBP1 promotes the lysis of Tg-containing vacuoles and parasite plasma membranes, releasing Tg-DNA. In contrast, we show GBP1 targets cytosolic STm and recruits caspase-4 to the bacterial surface for its activation by lipopolysaccharide (LPS), but does not contribute to bacterial vacuole escape. Caspase-1 cleaves and inactivates GBP1, and a cleavage-deficient GBP1D192E mutant increases caspase-4-driven pyroptosis due to the absence of feedback inhibition. Our studies elucidate microbe-specific roles of GBP1 in infection detection and its triggering of the assembly of divergent caspase signaling platforms.
Keywords: apoptosis, caspases, GBP1, inflammasomes, pyroptosis, Salmonella enterica Typhimurium, Toxoplasma gondii
Graphical Abstract
Highlights
-
•
Development of two microscopy assays for microbe/microbe-containing vacuole lysis
-
•
Human GBP1 is essential for the lysis of Toxoplasma gondii vacuoles and parasites
-
•
Caspase-4 recruitment, but not cytosolic escape of Salmonella, is GBP1 dependent
-
•
Caspase-1 cleaves and inactivates GBP1 and suppresses caspase-4-driven pyroptosis
Fisch et al. find that GBP1 targets Toxoplasma vacuolar and parasite membranes for disruption of both membranes. In contrast, appearance of cytosolic Salmonella is GBP1 independent, but caspase-4 recruitment to bacteria and activation is GBP1 dependent. In a negative feedback loop, caspase-1 cleaves GBP1 and suppresses caspase-4-driven pyroptosis during Salmonella infection.
Introduction
Most nucleated cells can defend themselves against infection by viruses, bacteria, and eukaryotic parasites in a process called cell-intrinsic immunity. These defense programs respond to the detection of pathogens by membrane-bound or cytosolic pattern recognition receptors (PRRs) (Jorgensen et al., 2017; MacMicking, 2012; Mostowy and Shenoy, 2015; Randow et al., 2013). In addition to antimicrobial molecules that restrict or kill pathogens, host cell-death is a destructive yet effective mechanism of defense since it removes replicative niches and traps intracellular pathogens within cell remnants (Jorgensen et al., 2016). Antimicrobial immunity and cell death are enhanced by the type-II interferon (IFNγ), which induces the expression of up to 2,000 IFN-stimulated genes (ISGs) (MacMicking, 2012; Schoggins, 2019). The guanylate-binding protein (GBP) family of GTPases, which are highly abundant in cells exposed to IFNγ, consists of seven members in the humans and eleven members in mice (Kresse et al., 2008; Olszewski et al., 2006; Shenoy et al., 2007, 2012). GBPs target intracellular pathogens and mediate host-defense via autophagy, oxidative responses, inflammasomes, and cell death (Costa Franco et al., 2018; Feeley et al., 2017; Foltz et al., 2017; Gomes et al., 2019; Haldar et al., 2013, 2014, 2015; Kim et al., 2011, 2012; Li et al., 2017; Lindenberg et al., 2017; Liu et al., 2018; Man et al., 2015, 2017; Meunier et al., 2014, 2015; Piro et al., 2017; Santos et al., 2018; Shenoy et al., 2012; Tripal et al., 2007; Wallet et al., 2017; Wandel et al., 2017; Zwack et al., 2017).
Once GBPs translocate to a pathogen vacuole or the pathogen itself, they are thought to disrupt these membranes by an as yet uncharacterized mechanism (Kravets et al., 2016; Meunier et al., 2014; Selleck et al., 2013; Yamamoto et al., 2012). Disruption of barrier membranes leads to pathogen growth control and release of pathogen-associated molecular patterns (PAMPs), which are sensed by PRRs that can trigger host cell death. Whether GBPs directly recognize pathogen vacuolar membranes or PAMPs is an important question that has not yet been answered (Fisch et al., 2019a; Lagrange et al., 2018; Meunier et al., 2014; Pilla et al., 2014; Santos et al., 2018).
A large body of work on GBPs has been carried out in murine cells, wherein these proteins closely collaborate with members of a second family of IFN-induced GTPases, comprising 23 members, the p47 immunity-related GTPases (IRGs) (Haldar et al., 2014; Hunn et al., 2008; Khaminets et al., 2010; Miyairi et al., 2007; Shenoy et al., 2007; Singh et al., 2006, 2010; Tiwari et al., 2009). For instance, mouse Irgb10 targets bacteria following mGbp recruitment and contributes to the release of bacterial LPS and DNA, and mouse Irgm1 and Irgm3 are essential regulators of mGbp-targeting of some pathogen-containing vacuoles (Haldar et al., 2015; Man et al., 2016; Meunier et al., 2014; Singh et al., 2010). However, only one constitutively expressed, truncated IRG, called “IRGM,” is present in the human genome (Bekpen et al., 2005, 2010). Therefore, how human GBPs target intracellular pathogens remains unknown. In addition, some PRRs are unique to humans, for example caspase-4 and caspase-5 (Casson et al., 2015; Ding and Shao, 2017; Kayagaki et al., 2011, 2013; Shi et al., 2014), which enable human, but not mouse cells, to respond to tetra-acylated LPS (Lagrange et al., 2018). The mechanisms underlying GBP-mediated detection of pathogens and stimulation of human macrophage death therefore need to be investigated further.
All seven human GBPs have a conserved structure with an N-terminal globular GTPase domain and a C-terminal helical domain. GBP1, GBP2, and GBP5 can be isoprenylated at their C-terminal CaaX-box, allowing membrane anchoring (Britzen-Laurent et al., 2010; Nantais et al., 1996; Olszewski et al., 2006; Tripal et al., 2007). Differences in pathogen-targeting have been noted depending on the pathogen and cell type. We previously showed that human GBP1 fails to target the apicomplexan parasite Toxoplasma gondii (Tg) and two intracellular bacterial pathogens, Chlamydia trachomatis and Salmonella enterica subsp. enterica serovar Typhimurium (STm), in human A549 epithelial cells; however, GBP1 is required for the restriction of parasite growth, but not the bacterial pathogens (Johnston et al., 2016). On the other hand, in human macrophages GBP1 localizes to Tg, Chlamydia, and STm, but whether it can disrupt membranes that enclose these pathogens is not known (Al-Zeer et al., 2013; Fisch et al., 2019a).
During Tg or STm infection-induced death of human macrophages, GBP1 targeting to pathogens is necessary, even though downstream mechanisms of cell death are distinct. Since Tg induces the loss of several inflammasome proteins, including NLRP3 (NOD, leucine rich repeat and pyrin domain containing protein 3) and caspase-1, human macrophages undergo atypical apoptosis through the assembly of AIM2 (absent in melanoma 2) -ASC (apoptotic-associated speck-like protein with a CARD)-caspase-8 complexes. In contrast, GBP1 promotes activation of caspase-4 following its recruitment to STm, resulting in enhanced pyroptosis (Fisch et al., 2019a). Although our previous work suggested that GBP1 is involved in PAMP release for detection by these PRRs during natural infection, the underlying mechanisms involved in liberating microbial ligands was not investigated (Fisch et al., 2019a). In this study we show that GBP1 contributes to the lysis of parasite-containing vacuoles and the plasma membrane of Tg by employing two assays. We also show that GBP1 exclusively targets STm that are already cytosolic and does not contribute to their ability to reach the cytosol of human macrophages. In contrast, during STm infection, caspase-1 cleaves and inactivates GBP1, and thereby reduces its ability to recruit caspase-4. These studies reveal the feedback inhibition of GBP1-caspase-4-driven pyroptosis during STm infection and its dual membrane-disruptive actions during Tg infection.
Results
GBP1 Contributes to Toxoplasma Parasite and Vacuole Disruption and Infection Control
As GBP1 elicits divergent host cell death programs in response to Tg and STm, we sought to investigate the upstream mechanisms of GBP1 during infection by these two unrelated pathogens. We previously correlated GBP1 recruitment to Tg parasitophorous vacuoles (PVs) to the activation of AIM2-caspase-8 and recognition of parasite DNA (Fisch et al., 2019a). We hypothesized that, like some murine Gbps (Degrandi et al., 2013; Kravets et al., 2016; Selleck et al., 2013; Yamamoto et al., 2012), human GBP1 promotes PV opening and cytosolic access to intravacuolar pathogens.
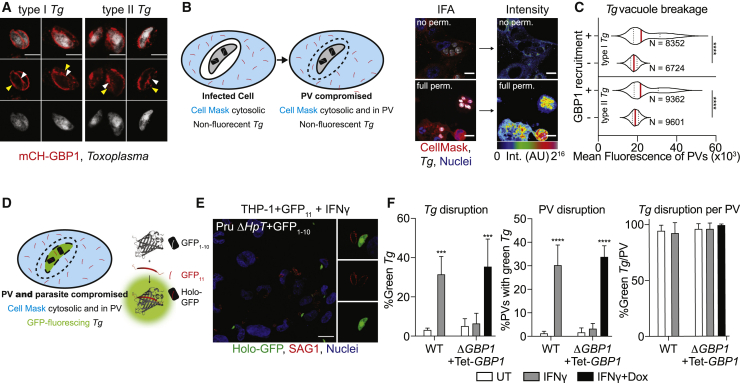
Extending our previous finding of GBP1 recruiting to the PV, we also localized GBP1 directly to the surface of Tg using AiryScan super-resolution microscopy (Figure 1A). To test whether GBP1 can disrupt Tg PVs, we used the cytosolic dye CellMask, which is excluded from PVs but enters once the PV membrane (PVM) is disrupted (Figure 1B). As positive control for this assay, PVs were chemically disrupted by detergent-mediated permeabilization, resulting in higher fluorescence within the vacuoles as compared to untreated cells (Figure 1B). Increased CellMask dye intensity within naturally disrupted PVs could be reliably quantified using our artificial intelligence-based high-throughput image analysis workflow HRMAn (Fisch et al., 2019b), which enabled us to enumerate dye access within thousands of PVs upon infection of type-I and type-II Tg strains. Analysis of CellMask fluorescence within PVs in IFNγ-primed THP-1 wild-type (WT) cells revealed increased intensities, indicating their disruption (Figure S1A). However, analysis of IFNγ-primed THP-1 ΔGBP1 cells showed that Tg vacuoles were not disrupted, as seen by the exclusion of CellMask dye (Figure S1A). Doxycycline (Dox) induced re-expression of GBP1 (THP-1 ΔGBP1+Tet-GBP1 cells) rescued vacuole breakage; as controls, empty-vector transduced cells (THP-1 ΔGBP1+Tet-EV) behaved like ΔGBP1 cells (Figure S1A). We next used Dox-induced expression of mCherry-GBP1 (THP-1 ΔGBP1+Tet-mCH-GBP1 cells) to allow quantification of GBP1-recruitment to Tg and stratify data on whether PVs that were decorated with mCH-GBP1 lost their integrity. Indeed, a population of GBP1+ PVs was unable to exclude CellMask dye, clearly indicating loss of membrane integrity (Figure 1C). Taken together, we concluded that GBP1 contributes to the opening of PVs, and GBP1-targeted vacuoles preferentially undergo loss of membrane integrity.
Figure 1.
GBP1 Disrupts Toxoplasma Vacuoles and Parasite Membrane
(A) AiryScan immunofluorescence images of type I or type II Toxoplasma gondii (Tg) decorated with mCH-GBP1 in IFNγ- and Doxycycline (Dox)-treated THP-1 ΔGBP1+Tet-mCH-GBP1 cells. Red: mCH-GBP1; white: Tg. White arrowhead indicates GBP1 on the parasite; yellow arrowhead indicates GBP1 on the vacuole membrane. Scale bar, 4 μm.
(B) Left: illustration of the high-throughput imaging assay to measure parasitophorous vacuole (PV) integrity by CellMask flooding. Right: representative immunofluorescence images from proof-of-principle experiment using THP-1 WT infected with type I Tg for 18 h and stained with CellMask but not permeabilized (no perm.; top) or fully permeabilized with saponin (full perm.; bottom) and corresponding rainbow intensity diagram to illustrate the CellMask signal from images used for quantification; AU: arbitrary units for fluorescence intensity values. Red: CellMask; gray: Tg; blue: nuclei. Scale bars, 10 μm.
(C) Representative quantification of CellMask fluorescence intensities within vacuoles (PV) of type I or type II Tg infected THP-1 ΔGBP1+Tet-mCH-GBP1 pre-treated with IFNγ and Dox to induce GBP1 expression. Plotted depending on whether PVs were decorated with GBP1 (+) or not (−). N = number of vacuoles.
(D) Illustration of the high-throughput imaging assays to measure Tg integrity with the split-GFP system.
(E and F) Example immunofluorescence image (E) and quantification (F) of disrupted and thus green-fluorescing type II Tg parasites expressing GFP1-10 fragment (Pru ΔHpt+GFP1-10) infecting IFNγ-primed THP-1+GFP11 or IFNγ and Dox-primed THP-1 ΔGBP1+Tet-GBP1+GFP11 cells stained for all Tg using anti-surface-antigen 1 (SAG1). Data plotted as proportion of all parasites (left), proportion of all PVs containing at least one green parasite (middle), or proportion of green parasites within the same PV (right). Red: SAG1; green: holo-GFP; blue: nuclei. Scale bar, 20 μm.
Graphs in (F) show mean ± SEM from n = 3 independent experiments. Graphs in (C) representative of n = 3 independent experiments. ∗∗∗p ≤ 0.001 or ∗∗∗∗p ≤ 0.0001 in (C) from nested Student’s t test comparing GBP1+ to GBP1− objects from the three experiments following adjustment for multiple comparisons and in (F) from two-way ANOVA comparing to untreated (UT) condition.
See also Figures S1 and S2.
Similar to our finding of GBP1 recruitment directly to the parasite surface, presumably within broken vacuoles, previous work localized murine Gbps directly onto the surface of the Tg plasma membrane (Kravets et al., 2016). Whether direct recruitment of a GBP to a Tg parasite leads to disruption of the parasite plasma membrane has not been studied. We developed a second assay that measures parasite membrane integrity (Figure 1D). In a split-GFP complementation approach, Tg parasites only fluoresce upon access of a GFP11 fragment expressed in the host-cell cytosol (Figure S1B) to the GFP1-10 fragment expressed in the Tg cytosol (Figure S1C); neither fragment is fluorescent on its own (Romei and Boxer, 2019). If the PV and the Tg membranes are both disrupted, the fragments can assemble to form fluorescent GFP holo-protein (Figure 1D). Indeed, we could observe GFP-fluorescing parasites in IFNγ-primed THP-1+GFP11 cells (Figure 1E) and quantify the proportion of parasites with GFP fluorescence (Figure 1F). Tg only became disrupted in the presence of GBP1 and all parasites within the same vacuole were disrupted, suggesting that once PV integrity is lost, the Tgs within them are susceptible to membrane damage (Figure 1F). Imaging results were confirmed using flow cytometry analysis of Tg from infected THP-1 cells (Figure S1D). Plaque assay of sorted parasites confirmed that green-fluorescing Tg were not viable (Figure S1E).
We validated our PV disruption assays by examining the ultrastructure of the vacuole membranes using correlative light and electron microscopy, which revealed ruffled and broken vacuole membranes in cells expressing GBP1 (Figure S2). In THP-1 ΔGBP1, most PVs analyzed by electron microscopy did not show structural defects or loss of membrane integrity (Figure S2). Together, our assays indicated that GBP1 contributes to the disruption of both the PV membrane and the Tg plasma membrane.
GBP1 Does Not Participate in Salmonella Vacuolar Escape but Targets Cytosolic Bacteria
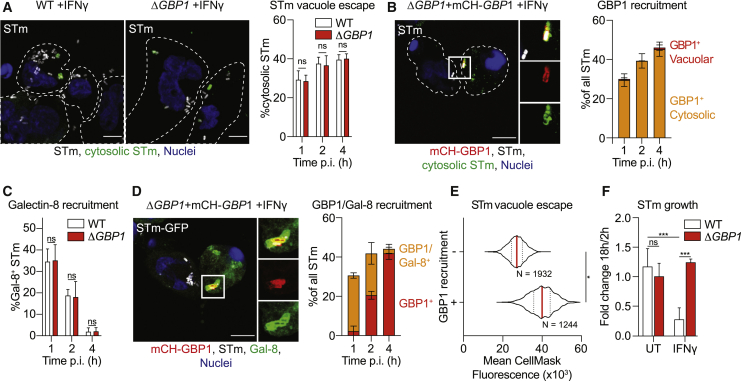
Having established an indispensable role for GBP1 in disrupting Tg PVs and parasites, we wanted to test if GBP1 also contributed to the escape of STm from Salmonella-containing vacuoles (SCVs). In murine cells, Gbps have been found to disrupt SCVs as well as directly recognize bacterial LPS in the cytoplasm (Meunier et al., 2014; Pilla et al., 2014). We used differential permeabilization (Meunier and Broz, 2015; Meunier et al., 2014) to determine whether the escape of STm from its vacuole into the cytosol required GBP1. Similar numbers of cytosolic bacteria were detected in WT and ΔGBP1 cells, suggesting that GBP1 is dispensable for vacuole escape of STm (Figure 2A) and indicating a microbe-specific role for GBP1 in disruption of pathogen compartments. Importantly, differential permeabilization revealed that GBP1 was exclusively recruited to cytosolic STm at all time points (Figure 2B). Therefore, as a second independent assay to analyze the ability of GBP1 to open STm vacuoles, we used galectin-8 (Gal-8) as a marker for cytosolic bacteria, which is recruited to disrupted SCVs in human epithelial cells, and promotes bacterial xenophagy and growth-restriction (Thurston et al., 2012). Consistent with the previously observed lack of a role for GBP1 in cytosolic escape of STm, similar proportions of STm were decorated with Gal-8 in WT and ΔGBP1 cells (Figure 2C). Temporal studies showed that SCVs were rapidly disrupted (became Gal-8+), but lost this marker over time (Figure 2C), as has been shown before in epithelial cells (Thurston et al., 2012). At later time points as the proportion of Gal-8+ vacuoles decreased, cytosolic STm retained GBP1 coating (Figure 2D). These single-cell assays revealed that unlike during Tg infection, GBP1 does not contribute to vacuole escape of STm, but recruits directly to cytosolic STm. We next used our CellMask dye influx assay and measured fluorescence in the vicinity of intracellular STm to determine if bacteria were vacuolar or in broken vacuoles/cytosolic (Figure S3A). Consistent with previous results, the assay confirmed that bacterial vacuole escape was similar in WT or ΔGBP1+Tet-GBP1 macrophages left untreated, treated with IFNγ, or treated with IFNγ+Dox (Figure S3B). Similarly, cytosolic STm that were decorated with mCH-GBP1 showed a higher CellMask fluorescence in their vicinity as compared to vacuolar STm that had not been targeted by GBP1 (Figure 2E). To assess the impact of GBP1 targeting on bacterial replication, we performed gentamicin-protection assays. IFNγ-treatment reduced bacterial replication by ∼70% (Figure 2F). This remarkable restriction of STm, however, was lost in ΔGBP1 cells (Figure 2F). Altogether, these results established that GBP1 does not contribute to STm vacuole escape; it targets cytosolic bacteria and reduces bacterial survival in macrophages.
Figure 2.
GBP1 Only Targets Already Cytosolic Salmonella and Recruits Caspase-4
(A) Representative immunofluorescence images at 2 h p.i. and quantification of the proportion of cytosolic Salmonella Typhimurium (STm) from differentially permeabilized, IFNγ-primed THP-1 WT or ΔGBP1 cells infected with STm SL1344 (MOI = 30) at indicated time p.i. Cells are outlined by the white, dashed line. Gray: STm; green: pseudo-colored cytosolic and extracellular STm; blue: nuclei. Scale bar, 10 μm.
(B) Representative immunofluorescence images at 2 h p.i., and quantification of GBP1 recruitment to cytosolic and intra-vacuolar STm in IFNγ-primed and Dox-treated THP-1 ΔGBP1+Tet-mCH-GBP1, infected with STm SL1344 (MOI = 30) at indicated times p.i. from differentially permeabilized cells stained for cytosolic STm and total STm. Cells are outlined by the white, dashed line. Red: mCH-GBP1; gray: STm; green: cytosolic STm; blue: nuclei. Scale bar, 10 μm.
(C) Quantification of galectin-8 (Gal-8) recruitment to STm SL1344-GFP in IFNγ-treated THP-1 WT or ΔGBP1 at the indicated times post infection.
(D) Representative immunofluorescence images at 1 h and quantification of Gal-8 recruitment to STm in IFNγ- and Dox-treated THP-1 ΔGBP1+Tet-mCH-GBP1 infected with STm SL1344-GFP (MOI = 30) at the indicated time post infection. Red: mCH-GBP1; gray: STm; green: Gal-8; blue: nuclei. Scale bar, 10 μm.
(E) Representative quantification of CellMask fluorescence intensities surrounding STm in infected THP-1 ΔGBP1+Tet-mCH-GBP1 pre-treated with IFNγ and Dox to induce GBP1 expression at 4 h p.i. plotted depending on whether STm were decorated with GBP1 (+) or not (−). N = number of STm quantified.
(F) Fold-change of intracellular STm colony forming units (CFUs) at 18 h p.i. normalized to 2 h p.i. of IFNγ-primed THP-1 WT or ΔGBP1 cells measured using gentamicin infection-protection assays.
Graphs in (A), (B) (C), (D), and (F) show mean ± SEM from n = 3 independent experiments and in (E) representative of n = 2 experiments. ∗p ≤ 0.05 or ∗∗∗p ≤ 0.001 in (E) from nested Student’s t test comparing GBP1+ to GBP1− objects from the two experiments following adjustment for multiple comparisons and in (A), (C), and (F) from two-way ANOVA following adjustment for multiple comparisons; ns, not significant.
See also Figure S3.
GBP1 Promotes Access to PAMPs for Cytosolic Host Defense and Interacts with Caspase-4 on the Surface of Salmonella
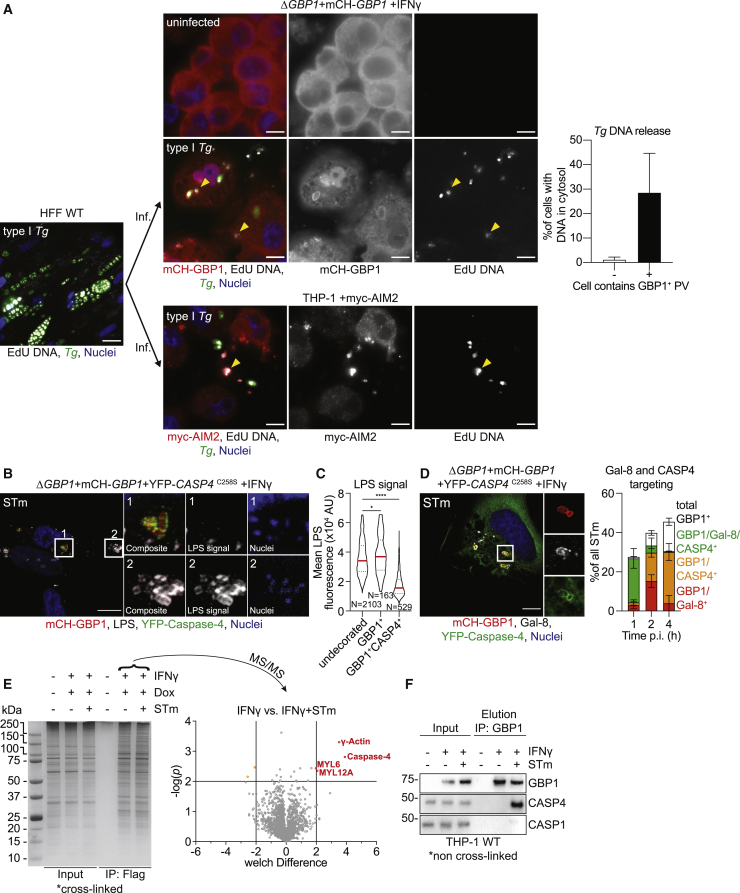
As Tg infection activates the DNA sensor AIM2 and we demonstrated that GBP1 promotes PV and Tg plasma membrane disruption, we wanted to visualize release of Tg-DNA into the cytoplasm of infected macrophages and subsequent recognition by AIM2. To this end, we labeled Tg-DNAs with EdU (5-ethynyl-2′-deoxyuridine) by growing them in human foreskin fibroblasts (HFFs), whose DNA remains unlabeled as they do not replicate due to contact-dependent growth inhibition. Following infection of macrophages with EdU-labeled Tg, we visualized Tg-DNA with Alexa Fluor 647 dye using click-chemistry and quantified macrophages containing cytosolic Tg-DNA (Figure 3A). Approximately 35% of infected macrophages that had at least one PV targeted by GBP1 (GBP1+) contained Tg-DNA in their cytosol at 6 h p.i. while uninfected macrophages or infected macrophages without targeted PVs did not show this phenotype (Figure 3A). Furthermore, infection of myc-AIM2-expressing THP-1 macrophages showed the association of the AIM2 receptor with EdU-labeled Tg-DNA in the cytosol (Figure 3A). Taken together, these results corroborated the model that human GBP1 actively ruptures the Tg PV and parasites and releases Tg-DNA into the cytosol for downstream detection by AIM2.
Figure 3.
GBP1 Mediates Access to PAMPs During Toxoplasma and Salmonella Infection
(A) Left: representative immunofluorescence image of type I Tg grown in human foreskin fibroblasts (HFF WTs) in the presence of EdU DNA label. Labeled type I Tg were harvested from the HFFs and used to infect (inf.) THP-1 ΔGBP1+Tet-mCH-GBP1 or THP-1 +myc-AIM2 for 6 h. THP-1 cells were pre-treated with IFNγ and Dox to induce mCH-GBP1 expression Middle: parasite DNA released into the cytoplasm was visualized by click-chemistry to label the incorporated EdU. Right: quantification of proportion of infected cells with cytosolic Tg-DNA based grouped based on whether the cells contain a GBP1-targeted (GBP1+) Tg PV. Red: mCH-GBP1 or immune-stained myc-AIM2; white: EdU-DNA; green: Tg; blue: nuclei. Some released, cytosolic (and additionally AIM2-bound, lower panels) Tg-DNA indicated by yellow arrowheads. Scale bar, 10 μm.
(B and C) Representative immunofluorescence images (B) and quantification (C) of LPS staining intensity of STm from IFNγ and Dox-treated THP-1 ΔGBP1+Tet-mCH-GBP1+YFP-CASP4C258S cells infected with STm SL1344 (MOI = 30) for 2 h. Red: mCH-GBP1; gray: STm-LPS; green: YFP-caspase-4; blue: nuclei. Scale bar, 10 μm. Contrast enhanced in the nuclei inset, to visualize STm-DNA used for detection of bacteria that do not stain for LPS.
(D) Representative immunofluorescence images at 1 h p.i. and quantification of Gal-8 and caspase-4 (CASP4) recruitment to STm in IFNγ-primed and Dox-treated THP-1 ΔGBP1+Tet-mCH-GBP1+YFP-CASP4C258S infected with STm SL1344 (MOI = 30) at indicated times p.i. Red: mCH-GBP1; gray: Gal-8; green: YFP-caspase-4; blue: nuclei. Scale bar, 10 μm.
(E) Left: silver stain of SDS-PAGE gel showing immunoprecipitation (IP) of FLAG-GBP1 from THP-1 ΔGBP1+Tet-FLAG-GBP1 treated with IFNγ and Dox and infected with STm for 2 h, left UT. Right: volcano plot of mass spectrometry hits comparing IFNγ treated cells with IFNγ treated and STm-infected cells. Plotted as Welch difference of mass spectrometry intensities versus −log10(p). Significant hits shown in orange/red circles.
(F) Representative immunoblots of IP of endogenous GBP1 from IFNγ-primed or naive THP-1 WT infected with STm for 2 h as indicated, showing co-precipitation of endogenous caspase-4 identified as a hit using mass spectrometry and no stable interaction with caspase-1.
Graphs in (A) and (D) show mean ± SEM from n = 3 independent experiments. Graph in (C) representative of n = 3 independent experiments. ∗p ≤ 0.05 and ∗∗∗∗p ≤ 0.0001 in (C) from nested one-way ANOVA comparing to undecorated vacuoles for the means of the n = 3 independent experiments; ns, not significant.
GBP1 thus promotes the sensing of PAMPs and formation of cytosolic signaling platforms also known as supramolecular organizing centers (SMOCs) (Kagan et al., 2014). We investigated the structure of caspase activation SMOCs promoted by GBP1 actions using structured illumination microscopy (SIM). Upon Tg infection, AIM2 detects Tg-DNA and nucleates the formation of an inflammasome containing ASC and caspase-8. Using SIM, we found that these atypical inflammasome complexes appear similar to previously described inflammasomes containing ASC and caspase-1 or caspase-8 (Man et al., 2013, 2014). We found a “donut”-like ASC ring enclosing caspase-8 in the center (Figures S4A and S4B). As we could not detect endogenous AIM2 by immunofluorescence microscopy, we resorted to using THP-1 cells expressing myc-AIM2 (Figure S4C), which revealed AIM2 recruitment to ASC specks in Tg-infected macrophages (Figures S4D and S4E). Altogether, these studies confirm that Tg-DNA is present within the macrophage cytosol as a result of GBP1-mediated disruption of the PVM and Tg membrane resulting in AIM2 activation.
We next decided to contrast GBP1 actions during STm infection, where we previously showed that caspase-4 is targeted exclusively to GBP1+ STm (Fisch et al., 2019a). The question was whether there is an interaction between GBP1 and caspase-4, which leads to recruitment of the LPS-sensor, and whether caspase-4 is recruited directly onto the surface of STm. Indeed, 3D-rendered SIM imaging demonstrated GBP1 recruited caspase-4 directly to the surface of cytosolic STm (Figure S4F). Bacteria were completely covered in GBP1, with a high degree of colocalization with YFP-CASP4C258S (Figure S4F). Interestingly, immunofluorescence staining of Salmonella-LPS using a monoclonal antibody revealed that GBP1+-CASP4+ bacteria stained either not at all or poorly, suggesting access to the epitope was blocked (Figures 3B and 3C). STm that were undecorated or CASP4–-GBP1+, however, were stained with anti-LPS antibody (Figure 3B). As caspase-4 can directly bind LPS through its CARD (Shi et al., 2014); this finding is consistent with the possibility that caspase-4 on the bacterial surface precludes antibody-mediated staining of LPS (Figure 3C). In addition, the monoclonal anti-LPS antibody stained bacteria in THP-1 ΔCASP4 cells (Figure S4G), pointing toward epitope occlusion by caspase-4.
In further agreement with targeting of cytosolic bacteria, the majority of Gal-8+-GBP1+ STm were also positive for caspase-4. Notably, GBP1 and caspase-4 were retained on STm over time even though Gal-8 staining had reduced (Figure 3D), which suggested that GBP1-caspase-4 are present on cytosolic STm longer during infection.
Our previous work showed that the translocation of GBP1 to STm and enhanced pyroptosis requires its GTPase function and isoprenylation (Fisch et al., 2019a), but did not determine whether these functions contributed to caspase-4 targeting to STm. THP-1 ΔGBP1 cells reconstituted with Dox-inducible variants of GBP1 that lacked GTPase activity (GBP1K51A) or isoprenylation sites (GBP1C589A or GBP1Δ589-592; Figure S4H) revealed that none of these variants supported the recruitment of caspase-4 (Figures S4I and S4J). Taken together, through single-cell comparative analyses we established that GBP1-targeting to Tg promotes the release of parasite DNA into the cytosol, whereas GBP1-targeting to STm enables caspase-4 recruitment to cytosolic bacteria. The reduced LPS staining on bacteria further suggest that GBP1 facilitates access of bacterial LPS ligand to caspase-4.
We additionally decided to use an unbiased proteomics approach to identify GBP1 binding-partners and other proteins recruited to GBP1-caspase-4 SMOCs on cytosolic STm. For this, we immunoprecipitated Dox-inducible FLAG-GBP1 from STm-infected THP-1 ΔGBP1+Tet-FLAG-GBP1 cells following protein cross linking (Figures 3E and S4K). Comparing infected to uninfected cells and correcting for non-specific binding of proteins to the FLAG-beads, we identified several proteins that were enriched in infected samples above the significance cut-off (p ≤ 0.01, Figure 3E; Data S1). Some of these proteins are known GBP1-interacting proteins such as γ-actin (ACTG1), myosin light polypeptide 6 (MYL6), and myosin regulatory light chain 12a (MYL12A; Forster et al., 2014; Ostler et al., 2014). The most prominent infection-specific GBP1 interaction partner we detected was caspase-4, which supported results from microscopy. To establish that this interaction is physiologically relevant during infection, we repeated immunoprecipitation experiments using antibodies against endogenous GBP1 from THP-1 WT cells (this time without prior cross linking). In agreement with our proteomics results, endogenous GBP1 interacted with caspase-4 only during STm infection, pointing toward its specific and crucial role in enabling LPS-sensing by caspase-4 (Figure 3F). In contrast, no stable interaction with caspase-1 was observed upon immunoprecipitation of endogenous GBP1 without or with infection (Figure 3F).
Taken together, these results indicated that GBP1 has two modes of assembling caspase-containing complexes depending on the infecting pathogen: (1) by proxy through Tg vacuole and parasite membrane disruption and release of Tg-DNA into the cytosol to trigger activation of the AIM2 inflammasome, and (2) by direct recruitment and interaction with caspase-4 on the surface of STm.
Caspase-1, but Not Caspase-4, Can Cleave GBP1
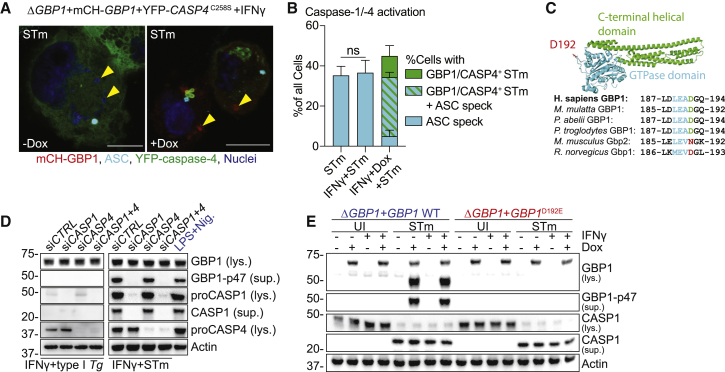
The noncanonical inflammasome in mouse macrophages involves sequential activation of caspase-4/11 and caspase-1, wherein caspase-4/11 activation precedes caspase-1. As both caspases are independently activated in IFNγ-stimulated human macrophages infected with STm (Fisch et al., 2019a), we wanted to investigate whether a crosstalk existed between the two pathways. This was also pertinent given a previous report of caspase-1-mediated cleavage of GBP1 in human umbilical vein endothelial cells (HUVECs) (Naschberger et al., 2017); however, the functional consequences of GBP1 proteolysis during infection were not investigated in that study. Noncanonical inflammasome activation during LPS-transfection is a cell-intrinsic process that involves K+ efflux-mediated activation of caspase-1 (Kayagaki et al., 2011; Rühl and Broz, 2015). We therefore first wanted to verify that caspase-1 and caspase-4 are activated within the same STm-infected macrophage. Our results showed a perfect correlation between bacterial targeting by GBP1-CASP4 and pyroptosis, and we indirectly quantified caspase-1 activation by measuring ASC speck formation. Indeed, single-cell microscopy confirmed that 80% of cells with GBP1+-CASP4+ STm (indicating active caspase-4), also had ASC specks (active caspase-1) (Figure 4A). Notably, caspase-4 was not recruited to ASC specks, which is consistent with previous work (Thurston et al., 2016)(Figures 4A and 4B). As these results suggested dual activation of caspase-1 and caspase-4 in the same cell, we investigated whether and how GBP1 proteolysis might affect caspase-4 recruitment to STm.
Figure 4.
Caspase-1, but Not Caspase-4, Cleaves GBP1 at Asp192 during Salmonella Infection
(A and B) Representative immunofluorescence images (A) and quantification (B) of ASC speck formation and GBP1+caspase-4 recruitment to STm from IFNγ-primed THP-1 ΔGBP1+Tet-mCH-GBP1+YFP-CASP4C258S infected with STm SL1344 (MOI = 30) for 2 h. Cells were treated with Dox to induce GBP1 expression or left UT. Yellow arrowheads indicate position of some STm within cells (DNA-staining dye). Red: mCH-GBP1; cyan: ASC; green: YFP-caspase-4; blue: nuclei. Scale bar, 10 μm.
(C) Crystal structure of human GBP1 (PDB: 1F5N) with GTPase domain highlighted in cyan, C-terminal helical domain in green and surface-exposed aspartate D192 in red (top). Multiple sequence alignment of human, primate, and rodent GBP1 orthologs depicting the caspase-1 cleavage site (cyan) surrounding Asp192 of human GBP1.
(D) Representative immunoblots from lysates (lys.) or culture supernatants (sup.) of THP-1 WT, transfected with the indicated siRNA and infected with type I Tg for 6 h, STm SL1344 for 4 h, and treated with LPS and Nigericin for 90 min.
(E) Representative immunoblots from lys. or culture sup. of THP-1 ΔGBP1+Tet-GBP1 WT or GBP1D192E cells treated with IFNγ and Dox as indicated and infected with STm SL1344 for 4 h or left uninfected (UI).
Graph in (B) shows mean ± SEM of n = 3 independent experiments. p values in in (B) from two-way ANOVA following adjustment for multiple comparisons; ns, not significant.
See also Figure S5.
We therefore examined the impact of caspase-1-mediated cleavage of GBP1 at the surface-exposed Asp192 residue that generates a stable p47 GBP1 C-terminal fragment (Figure 4C). Of note, phylogenetic analysis of representative GBPs (Shenoy et al., 2012) revealed that the Asp residue required for caspase-1 cleavage-site was present in all primates and absent in most rodents, including mice (Figure 4C). Infection of THP-1 with STm indeed confirmed that GBP1 is cleaved into a ∼47-kDa fragment that is prominently detected in cell culture supernatants. GBP1 proteolysis could be prevented by silencing caspase-1, but not caspase-4, confirming the dominant role of caspase-1 in the process (Figure 4D). LPS+Nigericin treatment for chemical activation of caspase-1 served as a positive control and also led to p47 GBP1 production. As expected with the lack of caspase-1 activation during Tg infection (Fisch et al., 2019a), GBP1 proteolysis could not be detected in Tg-infected THP-1 cells (Figure 4D).
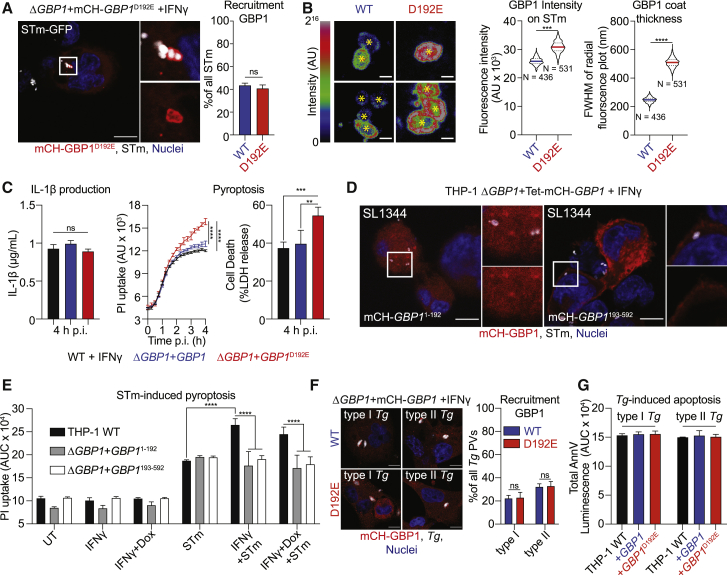
To confirm proteolysis of GBP1 at the Asp192 residue, we used a non-cleavable (D192E) variant. We created THP-1 ΔGBP1 cells expressing the non-cleavable GBP1D192E mutant without or with an mCherry tag (THP-1 ΔGBP1+Tet-GBP1D192E and THP-1 ΔGBP1+Tet-mCH-GBP1D192E cells; Figure S5A). Immunoblotting of GBP1 from STm-infected IFNγ-primed macrophages revealed caspase-1 activation and formation of p47 GBP1 from cells expressing WT GBP1 but not GBP1D192E (Figure 4E). Together, these results point toward the specificity of caspase-1 in cleaving GBP1, and that neither caspase-4 nor caspase-8 (active during Tg infection) can replace its role.
Caspase-1-Cleaved GBP1 Fragments Cannot Traffic to Microbial Vacuoles or Mediate Cell Death
As GBP1 can be cleaved by caspase-1, we wanted to investigate how this affects the pathogen-proximal activities of GBP1 in enabling PAMP access and triggering cell death. We infected mCH-GBP1D192E-expressing cells with STm and quantified GBP1 recruitment to bacteria. Consistent with a lack of role for GBP1 in SCV lysis, the proportion of GBP1+ STm was similar in cells expressing GBP1 WT and GBP1D192E (Figure 5A). However, the mean fluorescence intensity and the thickness of the protein coat of mCH-GBP1 around individual, decorated STm was markedly higher in cells expressing the GBP1D192E variant (Figure 5B), even though the expression and fluorescence of GBP1 WT and GBP1D192E was comparable in uninfected cells (Figure S5B). In agreement with increased amounts of GBP1 covering cytosolic bacteria, more STm-infected GBP1D192E cells underwent pyroptosis, as measured with propidium iodide (PI) uptake and lactate dehydrogenase (LDH) release assays, than WT cells, but released similar levels of IL-1β (Figure 5B). This finding is consistent with a major role for GBP1 in promoting caspase-4-driven pyroptosis, but not canonical caspase-1 activation, which is responsible for IL-1β production (Kortmann et al., 2015; Reyes Ruiz et al., 2017). These results led us to speculate that cleavage of GBP1 reduces the cellular pool of functional full-length GBP1, and its cleaved fragments do not support cell-death-related roles. Indeed, ΔGBP1 cells reconstituted with GBP11-192 or GBP1193-592 with or without mCherry-tag using our Dox-inducible system (Figure S5C) revealed that neither fragment was recruited to STm (Figure 5C) nor supported enhanced pyroptosis (Figure 5D). As caspase-1 is not activated during Tg infection, we anticipated that Tg targeting and apoptosis would be similar in cells expressing GBP1 WT or GBP1D192E. Indeed, the proportion of Tg-PVs decorated with GBP1 WT and GBP1D192E was similar (Figure 5E) and apoptosis remained unaffected (Figure 5F).
Figure 5.
Caspase-1-Driven GBP1 Proteolysis Regulates Cell Death during Salmonella, but Not Toxoplasma, Infection
(A) Representative immunofluorescence images and quantification of recruitment of GBP1 to STm in IFNγ-primed and Dox-treated THP-1 ΔGBP1+Tet-mCH-GBP1 or mCH-GBP1D192E cells infected with STm SL1344-GFP (MOI = 30) for 2 h. Red: mCH-GBP1; Gray: STm; Blue: Nuclei. Scale bar, 10 μm.
(B) Representative immunofluorescence images as rainbow intensity diagram of GBP1 WT or D192E recruitment to STm SL1344-GFP at 2 h p.i. (left) and quantification of fluorescence intensity and coat thickness measured as full-width half maximum (FWHM) of the radial intensity distribution surrounding the STm centroid (yellow asterisks; right). Scale bar, 1 μm. N = number of quantified GBP1+ bacteria in the respective condition.
(C) Left: IL-1β ELISA from the indicated THP-1 cells primed with IFNγ and Dox and infected with STm SL1344 (MOI = 30) at 4 h post-infection. Middle: real-time propidium iodide (PI) uptake assay from IFNγ-primed THP-1 cells of the indicated genotypes and infected with STm SL1344 for 4 h. Right: LDH release assay to measure cell death at 4 h p.i. with STm SL1344.
(D) Representative immunofluorescence image of mCherry-tagged GBP1 fragments in IFNγ- and Dox-primed THP-1 ΔGBP1+Tet cells expressing the indicated GBP1 fragment infected with STm SL1344 (MOI = 30) for 2 h. Red: mCH-GBP1; white: STm; blue: nuclei. Scale bar, 10 μm.
(E) Area under the curve (AUC) from 4 h live PI uptake cell death assay in THP-1 WT or THP-1 ΔGBP1+Tet-GBP1 cells expressing either GBP1 fragment 1-192 or 193-592, pre-stimulated with IFNγ only, with IFNγ and Dox to induce GBP1 expression or left UT and infected with STm SL1344 (MOI = 30).
(F) Representative immunofluorescence images and quantification of recruitment of GBP1 in IFNγ-primed and Dox-treated THP-1 ΔGBP1+Tet-mCH-GBP1 or mCH-GBP1D192E cells infected with type I or type II Tg for 6 h. Red: mCH-GBP1; gray: Tg; blue: nuclei. Scale bar, 10 μm.
(G) AnnV-Glo assay of THP-1 WT and ΔGBP1 cells stably reconstituted with Tet-GBP1 WT or GBP1D192E as indicated, infected with type I or type II Tg for 18 h. Plotted as AUC from real-time assays.
Graphs in (A), (C), (E), (F), and (G) show mean ± SEM from n = 3 independent experiments and in (B) representative of n = 3 independent experiments. ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001 for indicated comparisons in (B) from nested Student’s t test comparing means of the n = 3 experiments, in (A) from one-way ANOVA and in (C), (E), and (F) from two-way ANOVA following adjustment for multiple comparisons; ns, not significant.
See also Figure S5.
In summary, these results suggested that active caspase-1 cleaves a portion of the cellular GBP1 pool and generates protein fragments that cannot (1) target cytosolic STm, (2) recruit caspase-4, or (3) enhance pyroptosis induction. Because IL-1β maturation was not affected by GBP1D192E mutation, we speculate that this caspase-1-driven feedback mechanism balances caspase-1 and -4-driven cell death and caspase-1-driven IL-1β maturation during STm infection. Moreover, as caspase-1 does not contribute to cell death during Tg-infection, this proteolytic feedback regulation is pathogen specific.
Discussion
IFNγ-inducible GBPs have emerged as important proteins in host defense against a range of pathogens (Kutsch et al., 2020; Man et al., 2017; Meunier and Broz, 2016; Pilla-Moffett et al., 2016; Saeij and Frickel, 2017; Santos et al., 2020; Tretina et al., 2019; Wandel et al., 2020; Xavier et al., 2020). In this study, we have established that human GBP1 is essential for the breakdown of PVMs and Tg parasites through the use of two single-cell assays, combined with the artificial intelligence-driven image analysis pipeline HRMAn, that are adaptable for other pathogens (Fisch et al., 2019b). In contrast to Tg, GBP1 only decorates cytosolic STm and forms a complex with caspase-4, which it recruits onto the surface of bacteria. Caspase-1, but not caspase-4, also cleaves GBP1 at Asp192 to limit pyroptosis. These findings provide important insights on this key GTPase in human macrophages.
As the forerunner of the human GBP family, GBP1 has been extensively studied structurally and biochemically. We add the role of recruiting caspase-4 to STm dependent on functional GTPase activity and isoprenylation, which is in line with previous findings on mouse and human GBPs in vitro and in cellulo (Britzen-Laurent et al., 2010; Kohler et al., 2019; Nantais et al., 1996; Piro et al., 2017; Prakash et al., 2000; Shydlovskyi et al., 2017; Stickney and Buss, 2000). Recent work also points towards a role for GMP production in NLRP3 activation during C. trachomatis infection (Xavier et al., 2020).
Targeting of Tg vacuoles by murine Gbps and their interplay with the IRG proteins has been extensively studied (Degrandi et al., 2007; Haldar et al., 2013, 2014; Hunn et al., 2008; Kim et al., 2012; Miyairi et al., 2007; Shenoy et al., 2007; Singh et al., 2006, 2010; Tiwari et al., 2009; Traver et al., 2011; Virreira Winter et al., 2011). Uniquely in mice, two chromosomal loci each encode members of the Gbp (∼11 genes on Chr 3 and Chr 5) and IRG (∼23 genes on Chr 11 and Chr 18) families. Deletion of all mGbps on Chr3 (ΔGbpChr3) abrogates Tg vacuole rupture in macrophages and these mice are highly susceptible to Tg infection (Yamamoto et al., 2012). Single deletion of mGbp1 (Selleck et al., 2013) or mGbp2 (Degrandi et al., 2013) also results in enhanced susceptibility to Tg in vivo and in vitro. mGbp2 can homodimerize or form heterodimers with mGbp1 or mGbp5 before recruitment and attack of Tg vacuoles (Kravets et al., 2016). However, in the mouse, the hierarchical recruitment of IRG family GTPases to Tg vacuoles precedes the recruitment of Gbp family members. No Gbps are recruited to Tg in Irgm1/Irgm3−/− murine cells, pointing to their pivotal role in this process (Haldar et al., 2013). In addition to the absence of IRGs in humans, a direct role for individual GBPs in Tg vacuole disruption has not been demonstrated for GBPs before, even though mGbp2 has been found to localize inside Tg (Kravets et al., 2016), and vacuolar membrane integrity is not compromised in the absence of mGbp1, mGbp2 or all Gbps on chromosome 3 (Degrandi et al., 2013; Selleck et al., 2013; Yamamoto et al., 2012).
Our finding that GBP1 only targets bacteria already in the cytosol are consistent with bacterial staining with Gal-8, a marker for damaged endomembranes (Thurston et al., 2012). Furthermore, mouse Gbp recruitment is reduced in macrophages lacking Gal-3, which normally labels Legionella (Creasey and Isberg, 2012; Feeley et al., 2017; Liu et al., 2018; Pilla et al., 2014) or Yersinia (Feeley et al., 2017) expressing secretion systems that trigger damage of bacterial-containing vacuoles. Work with bacterial mutants that readily access the cytosol, such as Legionella pneumophila ΔsdhA and STm ΔsifA, also revealed no differences in cytosolic bacteria in mouse ΔGbpChr3 macrophages (Pilla et al., 2014). Similarly, release of Francisella novicida into the cytosol was shown to be independent of mouse Gbps (Man et al., 2015; Meunier et al., 2015). It is plausible that in human macrophages GBP1 is dispensable for release of STm into the cytosol, even though Gbps encoded at mouse Chr3 and mGbp2 have previously been implicated in this process in murine cells (Meunier et al., 2014). Indeed, STm escape to the cytosol requires its SPI-1 T3SS (Knodler et al., 2014; Malik-Kale et al., 2012; Radtke et al., 2007; Stévenin et al., 2019), and it will be interesting to test whether human GBP1 recruitment differs when macrophages are infected with STm that are deficient for SPI-1. Furthermore, it is tempting to speculate that human GBP1 recruitment to vacuolar STm is prevented by a bacterial virulence factor. Indeed, anti-GBP1 bacterial effectors have been identified in Shigella flexneri (Li et al., 2017; Piro et al., 2017; Wandel et al., 2017). Importantly, deletion of GBP1 drastically increased the survival of STm in IFNγ-primed human macrophages. However, these results should be interpreted with caution, as early gasdermin D (GSDMD)-pore formation prior to full-blown pyroptosis may enable the entry of gentamicin into cells and affect the viability of bacteria in such assays. Further work should also investigate whether other human GBPs also assemble alongside or assist GBP1 during STm infection.
Our work also shows unique GBP1 action during infection by these diverse pathogens whose distinct PAMPs are recognized by downstream innate immune pathways. Click-chemistry revealed that Tg-DNA is present in the cytoplasm of GBP1-expressing macrophages that subsequently induces the assembly of the atypical AIM2-ASC-caspase-8 SMOC and apoptosis. Super-resolution imaging structure of a caspase-8 containing AIM2 inflammasome closely resembles previously published structures of caspase-8 in NLRP3/NLRC4 inflammasomes (Man et al., 2013, 2014), revealing donut-like ASC rings enclosing AIM2 and caspase-8. Super-resolution microscopy during STm infection showed that GBP1 and caspase-4 formed a dense coat on STm, which reduced bacterial staining with anti-LPS antibody. Whether this reduced antibody access was due to the dense GBP1/caspase-4 coat or blocking of the LPS epitope by caspase-4 cannot be definitively distinguished; however, GBP1 alone (staining in ΔCASP4 cells) could not block access to the epitope. As caspase-4 by itself could not recruit to bacteria (Fisch et al., 2019a), we speculate that GBP1 is involved in exposing parts of the LPS that are buried deeper within the membrane. A direct interaction of GBP1 with LPS has been reported recently (Kutsch et al., 2020; Santos et al., 2020) and additionally suggested for mGbp5 (Santos et al., 2018). Our findings therefore support the hypothesis that isoprenylated, GTPase-activity competent GBP1 has a “detergent-like” function to open the bacterial outer membrane for caspase-4 to gain access to the otherwise-hidden lipid A of STm-LPS, which is consistent with recent reports (Kutsch et al., 2020).
We also uncovered a physiological role for GBP1-proteolysis by caspase-1 that was previous reported in vitro using HUVECs and in vivo from cerebrospinal fluid of meningitis patients (Naschberger et al., 2017), which we confirmed during natural infection of macrophages with STm. Notably, despite the 40% to 98% sequence similarity between human and mouse GBPs (Kim et al., 2011; Shenoy et al., 2007), and the conservation of Asp192 in other primate GBP1 sequences, Asp192 is absent in the closest murine homolog, mGbp2 (Olszewski et al., 2006), which is therefore unlikely to be regulated in this manner. Intriguingly, this finding mirrors our recent identification of the proteolysis of human, but not mouse, SQSTM1/p62 by caspase-8 at a conserved residue found in other mammalian SQSTM1/p62 sequences (Sanchez-Garrido et al., 2018). During STm infection, caspase-1 plays a dominant role in IL-1β maturation whereas IFNγ-induced GBP1 exclusively enhances caspase-4-driven pyroptosis. As a result, caspase-1-dependent proteolysis of GBP1 impaired GBP1-caspase-4-driven pyroptosis, but not caspase-1-driven IL-1β maturation. Thus, besides directly aiding the release or access to PAMPs for detection by caspases, GBP1 itself is a target of caspase-1 and a key regulatory hub that modulates host cell death. This contrasts our discovery of the ubiquitin-conjugating enzyme UBE2L3 as an indirect target of caspase-1 that specifically controls IL-1β production, but not pyroptosis (Eldridge et al., 2017). At the whole organism level, these mechanisms potentially enable differential responses based on the strength of the activating stimulus. Enhanced IL-1β production for adaptive immunity may be balanced by cell death that could enable pathogen uptake by other cell types such as neutrophils. Studies on cellular targets of caspases may therefore provide insights on homeostasis and disease.
Common themes also emerge from work on human and mouse GBPs. For instance, human GBP1 and mouse Gbp2 accumulate on vesicles generated through sterile damage, which suggests they could detect endogenous luminal ligands in the cytosol, such as in endogenous sulfated lipids (Bradfield, 2016; Feeley et al., 2017; Piro et al., 2017). The presence of Gal-3/Gal-8 and GBPs at sites of damaged membranes suggests GBPs may be assisted in sensing damage by other proteins, including other IFN-induced genes. Undoubtedly, future work in the area will focus on finding how human GBPs are targeted to diverse microbes, the ligands they sense and how they are regulated.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-AIM2 | Cell Signaling Technologies | Cat#12948; RRID:AB_2798067 |
| Rabbit polyclonal anti-ASC | Adipogen | Cat#AG-25B-006 |
| Mouse monoclonal anti-c-myc (Clone 9E10) | Merck | Cat#MABE282; RRID:AB_11213164 |
| Mouse monoclonal anti-Caspase-1 (Clone Bally-1) | Adipogen | Cat#AG-20B-0048; RRID:AB_2490257 |
| Mouse monoclonal anti-Caspase-8 | Cell Signaling Technologies | Cat#9746; RRID:AB_2275120 |
| Mouse monoclonal anti-Flag M2 | Sigma | Cat#F3165; RRID:AB_259529 |
| Goat polyclonal anti-Galectin-8 | R&D Systems | Cat#AF1305; RRID:AB_2137229 |
| Mouse monoclonal anti-GBP1 | Home-made (Frickel lab) | N/A |
| Rabbit polyclonal anti-mCherry | Abcam | Cat#ab167453; RRID:AB_2571870 |
| Rabbit polyclonal anti-panGBP | Home-made (Frickel lab) | N/A |
| Rabbit polyclonal anti-Salmonella | Abcam | Cat#ab35156; RRID:AB_777811 |
| Mouse monoclonal anti-Salmonella Typhimurium LPS (clone 1E6) | Abcam | Cat#ab8274; RRID:AB_306423 |
| Rabbit polyclonal anti-GFP/YFP | Abcam | Cat#ab6556; RRID:AB_305564 |
| Mouse monoclonal anti-β-Actin | Sigma | Cat#A5316; RRID:AB_476743 |
| Bacterial and Virus Strains | ||
| Salmonella Typhimurium SL1344 WT | Hoiseth and Stocker (1981), a kind gift from Jorge Galan (Yale University) | N/A |
| Salmonella Typhimurium SL1344-GFP | Fisch et al., 2019a | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Recombinant human IFNγ | R&D Systems | Cat#285-IF |
| Doxycycline | Sigma | Cat#D9891 |
| Nigericin | Invitrogen | Cat#N1495 |
| LPS-Sm | Adipogen | Cat#IAX-100-011 |
| 3xFlag peptide | Sigma | Cat#F4799 |
| CellMask Deep Red | Invitrogen | Cat#H32721 |
| Flag(M2)-agarose beads | Sigma | Cat#A2220; RRID:AB_10063035 |
| Critical Commercial Assays | ||
| RealTime-Glo Annexin V Apoptosis Assay | Promega | Cat#JA1001 |
| IL-1 beta Human Uncoated ELISA Kit | Invitrogen | Cat#88-7261; RRID:AB_2575052 |
| Pierce BCA protein assay kit | Thermo Scientific | Cat#23225 |
| Silver Stain Plus Kit | Biorad | Cat#1610449 |
| High-capacity cDNA synthesis kit | Applied Biosystems | Cat#4368813 |
| PowerUP SYBR green | Applied Biosystems | Cat#A25742 |
| Click-iT EdU Cell Proliferation Kit for Imaging, Alexa Fluor 647 dye | Invitrogen | Cat#C10340 |
| Alexa Fluor 647 Protein Labeling Kit | Invitrogen | Cat#A20173 |
| Deposited Data | ||
| Raw and analyzed data | This paper | https://dx.doi.org/10.17632/gkgrk7d8hg.1 |
| Experimental Models: Cell Lines | ||
| HEK293T | The Francis Crick Institute, Cell Services | RRID:CVCL_0063 |
| HFF | The Francis Crick Institute, Cell Services | RRID:CVCL_XB54 |
| THP-1 ΔCASP4 | Schmid-Burgk et al., 2015 | N/A |
| THP-1 ΔGBP1 | Fisch et al., 2019a | N/A |
| THP-1 ΔGBP1+Tet-EV | Fisch et al., 2019a | N/A |
| THP-1 ΔGBP1+Tet-GBP1 | Fisch et al., 2019a | N/A |
| THP-1 ΔGBP1+Tet-GBP1+GFP11 | This study | N/A |
| THP-1 ΔGBP1+Tet-GBP11-192 | This study | N/A |
| THP-1 ΔGBP1+Tet-GBP1193-592 | This study | N/A |
| THP-1 ΔGBP1+Tet-GBP1D192E | This study | N/A |
| THP-1 ΔGBP1+Tet-mCH-GBP1 | Fisch et al., 2019a | N/A |
| THP-1 ΔGBP1+Tet-mCH-GBP1 + YFP-CASP4C258S | Fisch et al., 2019a | N/A |
| THP-1 ΔGBP1+Tet-mCH-GBP1Δ589-592 +YFP-CASP4C258S | This study | N/A |
| THP-1 ΔGBP1+Tet-mCH-GBP11-192 | This study | N/A |
| THP-1 ΔGBP1+Tet-mCH-GBP1193-592 | This study | N/A |
| THP-1 ΔGBP1+Tet-mCH-GBP1C589A +YFP-CASP4C258S | This study | N/A |
| THP-1 ΔGBP1+Tet-mCH-GBP1D192E | This study | N/A |
| THP-1 ΔGBP1+Tet-mCH-GBP1K51A +YFP-CASP4C258S | This study | N/A |
| THP-1 WT | ATCC | Cat#TIB-202; RRID:CVCL_0006 |
| THP-1 WT+GFP11 | This study | N/A |
| THP-1 WT+Tet-CASP8-Flag | This study | N/A |
| THP-1 WT+Tet-CASP8-Flag + myc-AIM2 | This study | N/A |
| Experimental Models: Organisms/Strains | ||
| Type I (RH) Toxoplasma gondii-GFP-Luc | Kim et al., 2007 | N/A |
| Type II (Prugniaud) Toxoplasma gondii-GFP-Luc | Kim et al., 2007 | N/A |
| Type II (Prugniaud) Toxoplasma gondii ΔHpt | Moritz Treeck, The Francis Crick Institute, London, UK | N/A |
| Type II (Prugniaud) Toxoplasma gondii ΔHpt+GFP1-10 | This study | N/A |
| Oligonucleotides | ||
| Oligonucleotide primers for molecular cloning | This study | Table S1 |
| Oligonucleotide primers for qPCR | This study | Table S1 |
| siRNA for human CASP1 | Dharmacon | Cat#L-004401 |
| siRNA for human CASP4 | Dharmacon | Cat#L-004404 |
| siRNA for human CASP5 | Dharmacon | Cat#L-004405 |
| siRNA for human GSDMD | Dharmacon | Cat#L-016207 |
| Negative control siRNA | Dharmacon | Cat#D-001810 |
| Recombinant DNA | ||
| Plasmid pcDNA3-CASP8 | Stennicke and Salvesen, 1997 | Addgene #11817; RRID:Addgene_11817 |
| Plasmid LentiCRISPRv2 | Sanjana et al., 2014 | Addgene #52961; RRID:Addgene_52961 |
| Plasmid pLenti-Tet-CASP8-3xFlag | This study | N/A |
| Plasmid pLenti-Tet- 3xFlag-GBP1 | This study | N/A |
| Plasmid pGene-GBP1 | Home-made (Frickel lab) | N/A |
| Plasmid pLenti-Tet | Fisch et al., 2019a | N/A |
| Plasmid pLenti-Tet-mCH-GBP1 | Fisch et al., 2019a | N/A |
| Plasmid pLenti-Tet-GBP1 | Fisch et al., 2019a | N/A |
| Plasmid pMX-CMV-YFP-CASP4C258S | Fisch et al., 2019a | N/A |
| Plasmid pcDNA3-myc-AIM2 | Khare et al., 2014 | Addgene #73958; RRID:Addgene_73958 |
| Plasmid pLEX-MCS-ASC-GFP | de Almeida et al., 2015 | Addgene #73957; RRID:Addgene_73957 |
| Plasmid pLEX-MCS-myc-AIM2 | This study | N/A |
| Plasmid pLenti-P2A-Puro | This study | N/A |
| Plasmid pEGFP-C1 | Clontech | N/A |
| Plasmid pGRA-HA-HPT | Coppens et al., 2006 | N/A |
| Software and Algorithms | ||
| FlowJo version 10.3 | FlowJo, LLC | https://www.flowjo.com/ |
| Fiji | Schindelin et al., 2012 | https://fiji.sc/ |
| MaxQuant version 1.6.0.13 | MPI of Biochemistry, Martinsried, Germany | https://www.maxquant.org/ |
| LAS-AF software | Leica Microsystems | N/A |
| DeltaVision | GE Healthcare Life Sciences (Cytiva) | N/A |
| Imaris version 8.3.1. | Oxford Instruments | https://imaris.oxinst.com/ |
| TrakEM2 | Cardona et al., 2012 | https://imagej.net/TrakEM2 |
| KNIME Analytics Platform version 4.1.2 | Berthold et al., 2008 | https://www.knime.com/ |
| HRMAn | Fisch et al., 2019b | https://hrman.org/ |
| Prism version 8.1.1 | GraphPad Inc. | https://www.graphpad.com/ |
| MacPymol version 1.74. | Schrödinger, Inc. | https://pymol.org/2/ |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Eva-Maria Frickel (E.frickel@bham.ac.uk).
Materials Availability
All plasmids and cell lines generated and used in this study are available from the lead contact on request.
Data and Code Availability
The Mass spectrometry dataset is included in this study (Data S1). Quantification datasets are available from Mendeley data (https://doi.org/10.17632/gkgrk7d8hg.1). Any other data supporting the current study are available from the lead contact.
Experimental Model and Subject Details
THP-1 (TIB-202, ATCC, Male cell line, RRID:CVCL_0006) were maintained in RPMI with GlutaMAX (GIBCO) supplemented with 10% heat-inactivated FBS (Sigma), at 37°C in 5% CO2 atmosphere. THP-1 cells were differentiated with 50 ng/mL phorbol 12-myristate 13-acetate (PMA, P1585, Sigma) for 3 days followed by a rested for 2 days in complete medium without PMA. Cells were not used beyond passage 20. HEK293T (Female cell line, RRID:CVCL_0063) and human foreskin fibroblasts (HFF, Male cell line, RRID:CVCL_XB54) were maintained in DMEM with GlutaMAX (GIBCO) supplemented with 10% FBS at 37°C in 5% CO2 atmosphere. Tg expressing luciferase/eGFP (RH type I and Prugniaud (Pru) type II) were maintained by serial passage on monolayers of HFF cells. All cell culture was performed without addition of antibiotics unless otherwise indicated. Cell lines were routinely tested for mycoplasma contamination by PCR and agar test. Please refer to the Key Resources Table for an overview of all cell lines made/ used in this.
STm SL1344-GFP (with pFPV25.1 plasmid) was maintained under Ampicillin selection (11593027, GIBCO). STm SL1344 WT strain was maintained in the presence of Streptomycin (11860038, GIBCO) selection.
Method Details
Cell treatments
Cells were stimulated for 16 h prior to infection in complete medium at 37°C with addition of 50 IU/mL human IFNγ (285-IF, R&D Systems). Induction of Doxycycline-inducible cells was performed with 200 ng/mL Doxycycline overnight (D9891, Sigma). To chemically activate caspase-1, cells were treated with 10 μM Nigericin (N1495, Invitrogen) and 100 μg/mL LPS-Sm (IAX-100-011, Adipogen).
Creation of cell lines
Inducible GBP1 and caspase-8 cell lines
THP-1 ΔGBP1+Tet-GBP1 and THP-1 ΔGBP1+Tet-mCH-GBP1 have been published before and the THP-1 WT+Tet-CASP8-Flag cells were created identically using Lentiviral transductions (Fisch et al., 2019a).
To create the caspase-8-3xFlag expressing Dox-inducible plasmid (pLenti-Tet-CASP8-3xFlag), the empty vector backbone was digested with BamHI, CASP8 ORF was amplified from pcDNA3-CASP8 by PCR (Addgene #11817, a gift from Guy Salvesen) (Stennicke and Salvesen, 1997), 3xFlag was amplified from LentiCRISPRv2 (Addgene #52961, a gift from Feng Zhang) (Sanjana et al., 2014) and all fragments assembled with a Gibson assembly. Similarly, to create the 3xFlag-GBP1 expressing Dox-inducible plasmid (pLenti-Tet- 3xFlag-GBP1), the empty vector backbone was digested with BamHI, GBP1 ORF was amplified from pGene-GBP1 by PCR (Frickel lab), 3xFlag was amplified from LentiCRISPRv2 (Addgene #52961, a gift from Feng Zhang) (Sanjana et al., 2014) and all fragments assembled with a Gibson assembly. In the same way, GBP1 fragments 1-192 and 193-592 were amplified from pGene-GBP1 (Frickel lab) and Gibson assembled into BamHI digested pLenti-Tet (Fisch et al., 2019a) with and without addition of a mCherry tag. The obtained plasmids were then transduced into THP-1 ΔGBP1+Tet cells (Fisch et al., 2019a) using lentiviral particles.
To make the cells expressing GBP1D192E, the pLenti-Tet-mCH-GBP1 and pLenti-Tet-GBP1 plasmids (Fisch et al., 2019a) were mutated using site-directed mutagenesis and transduced into the THP-1 ΔGBP1+Tet target cells using lentiviral transduction as described before (Fisch et al., 2019a). To make cells expressing YFP-CASP4C258S and mutated GBP1 versions, THP-1 ΔGBP1+Tet-mCH-GBP1K51A, +Tet-mCH-GBP1C589A or +Tet-mCH-GBP1Δ589-592 (Fisch et al., 2019a) were transduced with pMX-CMV-YFP-CASP4C258S (Fisch et al., 2019a) using lentiviral particles. Please refer to the Key Resources Table and Table S1 for an overview of all oligonucleotide primers and plasmids used for cloning PCRs.
myc-AIM2 expressing cell line
To create a lentiviral vector for constitutive expression of myc-AIM2, the ORF was amplified from pcDNA3-myc-AIM2 (Addgene #73958, a gift from Christian Stehlik) (Khare et al., 2014) and Gibson assembled into BstBI and BsrGI digested pLEX-MCS-ASC-GFP (Addgene #73957, a gift from Christian Stehlik) (de Almeida et al., 2015) to create pLEX-MCS-myc-AIM2. The vector was then transduced into THP-1+Tet-CASP8-Flag cells to create THP-1+Tet-CASP8 + myc-AIM2 cells using lentiviral transduction as described above.
GFP11 expressing cell lines
To create an lentiviral vector for constitutive expression of GFP11, the sgRNA cassette from LentiCRISPRv2 (Addgene #52961, a gift from Feng Zhang) (Sanjana et al., 2014) was removed by digestion with KpnI and EcoRI and the plasmid re-ligated using annealed repair oligo pair 1 (see Key Resources Table and Table S1) and Quick Ligation Kit (M2200L, NEB). Next, the Cas9-ORF was removed by digestion with XbaI and BamHI and again the vector re-ligated using annealed repair oligo pair 2 (see Key Resources Table and Table S1), also adding a multiple cloning site, which created the vector pLenti-P2A-Puro. Next, the GFP11 ORF was amplified from pEGFP-C1 (Clontech) and ligated into BamHI and XbaI digested pLenti-P2A-Puro, to have the GFP11-ORF in frame with the P2A-Puro cassette, for Puromycin-selectable, constitutive expression of GFP11. The newly made vector was then transduced into THP-1 WT and THP-1 ΔGBP1+Tet-GBP1 cells using Lentiviral transduction as described above.
Creation of transgenic Toxoplasma gondii
To create Tg lines that constitutively express non-fluorescent GFP1-10 fragment, the GFP1-10 ORF was amplified from pEGFP-C1 (Clontech) and Gibson-assembled into NsiI and PacI digested pGRA-HA-HPT (a gift from Moritz Treeck) (Coppens et al., 2006), to have expression of the ORF under control of the TgGRA1 promoter.
Next the plasmid was transfected into type II (Pru) Tg ΔHpt (a gift from Moritz Treeck) using nucleofection as established by Young et al. (2019): The plasmid was linearized using PsiI-V2 and purified using phenol-chloroform precipitation and re-suspended in P3 solution (Lonza). Successful linearization was confirmed using agarose-gel electrophoresis. Next, type II (Pru) Tg ΔHpt were harvested from HFFs by syringe lysis and washed with PBS twice and then 5 × 106 parasites resuspended in P3 solution. Prior to nucleofection, 25 μg linearized DNA were added to the parasites and then nucleofected using 4D-Nucleofector™ (Lonza) with setting EO-115. Transfected parasites were then incubated for 12 minutes at room temperature, followed by platting onto fresh HFF cells into a T25 tissue culture flask. The next day, medium was replaced with complete DMEM containing 50 μg/mL xanthine and mycophenolic acid (MPA) each for selection. The selection medium was replaced every two days and the parasites passaged normally for two weeks when all Tg in the untransfected control had died. Successful integration of the plasmid and expression of GFP1-10 was confirmed by immunofluorescence and immunoblotting.
Toxoplasma gondii infection
Parasite were passaged the day before infection. Tg tachyzoites were harvested from HFFs by scraping and syringe lysing the cells through a 25 G needle. The Tg suspension was cleared by centrifugation at 50 x g for 5 min and then the parasites were pelleted by centrifugation at 550 x g for 7 min from the supernatant, washed with complete medium, and finally re-suspended in fresh medium. Viable parasites were counted with trypan blue and used for infection at a multiplicity of infection (MOI) of 3 for most experiments or 1 for immunofluorescence imaging. Infection was synchronized by centrifugation at 500 x g for 5 min. Two hours after infection, extracellular parasites were removed with three PBS washes.
Flow cytometry and sorting
For flow cytometry analysis of GFP-fluorescence, Tg ΔHpt+GFP1-10 were harvested from host cells by syringe lysis, washed twice with warm PBS and then re-suspended in PBS + 1% BSA. Parasites were analyzed on a LSR Fortessa (BD Biosciences) and data were processed using FlowJo version 10.3 (FlowJo, LLC). For viability determination of GFP-fluorescing versus non-fluorescing Tg the parasites were harvested and prepared identically, sorted on a FACSAria III (BD Biosciences) based on their GFP-fluorescence and then plated onto HFFs grown confluent in wells of a 24-well plate. 5 days post infection of the HFFs, cells were fixed with ice-cold methanol and stained with crystal violet. Following 5 washes with PBS, plaques were imaged on a GelCount Colony Counter (Oxford Optronix) and cell covered area determined using FIJI ImageJ.
Salmonella Typhimurium infection
One day before infection bacteria from a single colony were inoculated into 9 mL LB and grown overnight at 37°C. The overnight culture was diluted 1:50 into LB + 300 mM NaCl (746398, Sigma) and grown shaking in a closed container until an OD600 of 0.9. Bacteria were harvested by centrifugation at 1000 x g for 5 min, washed with serum-free cell culture medium twice and re-suspended in 1 mL medium. Cells were infected with STm at an MOI of 30 and infections were synchronized by centrifugation at 750 x g for 10 min. Infected cells were washed 30 min post-infection three times with warm PBS (806552, Sigma) to remove extracellular bacteria and fresh, serum-free medium containing 100 μg/mL Gentamicin (15750060, GIBCO) was added for 1 h. Medium was then replaced with medium containing 10 μg/mL Gentamicin and the infection continued for indicated times. Bacterial MOI used for infections were confirmed by plating on LB agar plates. For Gentamicin infection-protection assays, cells were lysed with 1 mL of PBS + 0.1% Triton X-100 (T8787, Sigma) for 5 mins including a brief vortex to ensure complete host cell-disruption at 2 h and 18 h p.i., suspensions diluted 1:1,000 to 1:10,000 in PBS, plated on LB-agar plates and incubated at 37°C overnight. Colony forming units (CFUs) were counted the next morning and replication calculated as fold-change normalized to the CFU at 2 h.
Real-time cell death assays and IL-1β ELISA
To measure live kinetics of cell death, 60,000 cells were seeded per well of a black-wall, clear-bottom 96-well plate (Corning) for differentiation with PMA, treated and infected as described above. Medium was replaced with phenol-red-free RPMI supplemented with 5 μg/mL propidium iodide (P3566, Invitrogen). The plate was sealed with a clear, adhesive optical plate seal (Applied Biosystems) and placed in a plate reader (Fluostar Omega, BMG Labtech) pre-heated to 37°C. PI fluorescence was recorded with top optics every 15 min for times as indicated.
Apoptosis kinetics were analyzed using the RealTime-Glo Annexin V Apoptosis Assay (JA1001, Promega) according to the manufacturer’s instruction. Simultaneously with infection, detection reagent was added. Luminescence was measured using a Fluostar Omega plate reader (BMG Labtech). No-cell, medium-only controls were used for background correction.
For IL-1β ELISA, the cell culture supernatant was harvested, cleared by centrifugation at 2000 x g for 5 minutes and diluted in the buffer provided with the ELISA kit. ELISA was performed according to the manufacturer’s instruction. IL-1β ELISA kit was from Invitrogen (#88-7261, detection range 2 - 150 pg/mL).
Immunoblotting and gel staining
For immunoblotting, 0.5 × 106 cells were seeded per well of a 48-well plate, differentiated with PMA, pre-treated and infected. Cells were washed with ice-cold PBS and lysed for 5 min on ice in 50 μL RIPA buffer supplemented with protease inhibitors (Protease Inhibitor Cocktail set III, EDTA free, Merck) and phosphatase inhibitors (PhosSTOP, Roche). Lysates were cleared by centrifugation at full speed for 15 min at 4°C. BCA assay (Pierce BCA protein assay kit, 23225, Thermo Scientific) was performed to determine protein concentration. 10 μg of total protein per sample were run on Bis-Tris gels (Novex, Invitrogen) in MOPS running buffer and transferred on Nitrocellulose membranes using iBlot transfer system (Invitrogen). Membranes were blocked with either 5% BSA (A2058, Sigma) or 5% dry-milk (M7409, Sigma) in TBS-T for at least 1 h at room temperature. Incubation with primary antibodies was performed at 4°C overnight (Please refer to the Key Resources Table for an overview of all antibodies used in this study). Blots were developed by washing the membranes with TBS-T, probed with 1:5000 diluted secondary antibodies in 5% BSA in TBS-T and washed again. Finally, the membranes were incubated for 2 minutes with ECL (Immobilon Western, WBKLS0500, Millipore) and luminescence was recorded on a ChemiDoc MP imaging system (Biorad). For silver staining of protein gels, following SDS-PAGE, the gels were washed in ddH2O and then silver stained following the manufacturers instruction (Silver Stain Plus Kit, 1610449, Biorad).
For immunoblots of culture supernatants, cells were treated in OptiMEM (1105802, GIBCO) without serum. Proteins in the supernatants were precipitated with 4x volume cold acetone (V800023, Sigma) overnight at −20°C, and pelleted by centrifugation. Pellets were air-dried and re-suspended in 40 μL 2x Laemmli loading dye.
Identification of GBP1-interacting proteins
Sample preparation
10 × 106 THP-1 ΔGBP1+Tet-Flag-GBP1 cells were seeded in 6-well plates and differentiated, pre-treated with IFNγ and Doxycycline and infected with STm as described before. 2 hours p.i. the interacting proteins were crosslinked with 1% formaldehyde (28906, Thermo Scientific) for 10 minutes at room temperature and the reaction quenched by addition of 125 mM glycine (Sigma). Cell were washed in ice-cold PBS and scraped from the plates. Cells were then pelleted by centrifugation and washed in PBS. Whole-cell lysates were prepared by adding 500 uL lysis buffer (1% Triton X-100, 20 mM Tris–HCl [pH 8], 130 mM NaCl, 1 mM dithiothreitol [DTT], 10 mM sodium fluoride, protease inhibitors (Protease Inhibitor Cocktail set III, EDTA free, Merck), phosphatase inhibitor cocktails (PhosSTOP, Roche)) and incubation for 15 minutes on ice. Lysates were cleared by centrifugation and then added to Flag(M2)-agarose beads (A2220, Sigma) washed three times with lysis buffer. Flag-GBP1 was captured by incubation on a rotator overnight at 4°C. Beads were then washed once with lysis buffer, three times with lysis buffer containing 260 mM NaCl and then again twice with lysis buffer. Proteins were eluted using 200 μg/mL 3xFlag peptide (F4799, Sigma) in lysis buffer by incubation on an orbital shaker (1400 rpm) for 2 hours at room temperature. Samples were then prepared by adding loading dye containing 5% β-Mercaptoethanol (Sigma) to reverse crosslinking and run on a 12% Bis-Tris polyacrylamide gel until the running front had entered the gel roughly 5 mm.
Trypsin digestion
Samples on the SDS-PAGE were excised as three vertical lanes each. The excised gel pieces were de-stained with 50% acetonitrile/50 mM ammonium bicarbonate, reduced with 10 mM DTT, and alkylated with 55 mM iodoacetamide. After alkylation, the proteins were digested with 250 ng of trypsin overnight at 37°C. The resulting peptides were extracted in 2% formic acid, 1% acetonitrile and speed vacuum dried. Prior to analysis the peptides were reconstituted in 50 μl of 0.1% TFA.
Mass spectrometry
The peptides were loaded on a 50 cm EASY-Spray column (75 μm inner diameter, 2 μm particle size, Thermo Fisher Scientific), equipped with an integrated electrospray emitter. Reverse phase chromatography was performed using the RSLC nano U3000 (Thermo Fisher Scientific) with a binary buffer system at a flow rate of 275 nl/min. Solvent A was 0.1% formic acid, 5% DMSO, and solvent B was 80% acetonitrile, 0.1% formic acid, 5% DMSO. The in-gel digested samples were run on a linear gradient of solvent B (2 - 30%) in 95.5 min, total run time of 120 min including column conditioning. The nano LC was coupled to an Orbitrap Fusion Lumos mass spectrometer using an EASY-Spray nano source (Thermo Fisher Scientific). The Orbitrap Fusion Lumos was operated in data-dependent acquisition mode acquiring MS1 scan (R = 120,000) in the Orbitrap, followed by HCD MS2 scans in the Ion Trap. The number of selected precursor ions for fragmentation was determined by the “Top Speed” acquisition algorithm with a cycle time set at 3 s. The dynamic exclusion was set at 30 s. For ion accumulation the MS1 target was set to 4 × 105 ions and the MS2 target to 2 × 103 ions. The maximum ion injection time utilized for MS1 scans was 50 ms and for MS2 scans was 300 ms. The HCD normalized collision energy was set at 28 and the ability to inject ions for all available parallelizable time was set to “true.”
Data processing and analysis
Orbitrap .RAW files were analyzed by MaxQuant (version 1.6.0.13), using Andromeda for peptide search. For identification, peptide length was set to 7 amino acids, match between runs was enabled and settings were kept as default. Parent ion and tandem mass spectra were searched against UniprotKB Homo sapiens and Salmonella typhimurium databases. For the search the enzyme specificity was set to trypsin with maximum of two missed cleavages. The precursor mass tolerance was set to 20 ppm for the first search (used for mass re-calibration) and to 6 ppm for the main search. Product mass tolerance was set to 20 ppm. Carbamidomethylation of cysteines was specified as fixed modification, oxidized methionines and N-terminal protein acetylation were searched as variable modifications. The datasets were filtered on posterior error probability to achieve 1% false discovery rate on protein level. Quantification was performed with the LFQ algorithm in MaxQuant using three replicate measurements per experiment.
Quantitative RT-PCR
RNA was extracted from 0.25 × 106 cells using Trizol reagent (15596026, Invitrogen). RNA (1 μg) was reverse transcribed using high-capacity cDNA synthesis kit (4368813, Applied Biosystems). qPCR used PowerUP SYBR green (A25742, Applied Biosystems) kit, 20 ng cDNA in a 20 μL reaction and primers (Please refer to the Key Resources Table and Table S1 for an overview of all oligonucleotide primers used for qPCR) at 1 μM final concentration on a QuantStudio 12K Flex Real-Time PCR System (Applied Biosystems). Recorded Ct values were normalized to Ct of human HPRT1 and data plotted as ΔCt (Relative expression).
siRNA transfection
Cells were transfected with siRNAs two days prior to infection, at the same time the THP-1 differentiation medium was replaced with medium without PMA. All siRNAs were used at a final concentration of 30 nM. For transfection, a 10x mix was prepared in OptiMEM containing siRNA(s) and TransIT-X2 transfection reagent (MIR 600x, Mirus) in a 1:2 stoichiometry. Please refer to the Key Resources Table for an overview of siRNAs used in this study.
Fixed immunofluorescence microscopy
For confocal imaging 0.25 × 106 THP-1 cells were seeded on gelatin-coated (G1890, Sigma) coverslips in 24-well plates. Following differentiation, treatments and infection, cells were washed three times with warm PBS prior to fixation to remove any uninvaded pathogens and then fixed with 4% methanol-free formaldehyde (28906, Thermo Scientific) for 15 min at room temperature. Following fixation, cells were washed again with PBS and kept at 4°C overnight to quench any unreacted formaldehyde. Fixed specimens were permeabilized with PermQuench buffer (0.2% (w/v) BSA and 0.02% (w/v) saponin in PBS) for 30 minutes at room temperature and then stained with primary antibodies for one hour at room temperature. After three washes with PBS, cells were incubated with the appropriated secondary antibody and 1 μg/mL Hoechst 33342 (H3570, Invitrogen) diluted in PermQuench buffer for 1 hour at room temperature. Cells were washed with PBS five times and mounted using 5 μL Mowiol.
Specimens were imaged on a Leica SP5-inverted confocal microscope using 100x magnification and analyzed using LAS-AF software. For structured-illumination super-resolution imaging, specimens were imaged on a GE Healthcare Lifesciences DeltaVision OMX SR imaging system and images reconstructed using the DeltaVision software. All images were further formatted using FIJI software. 3D rendering of image stacks and distance measurements of AIM2-ASC-CASP8 inflammasome specks was performed using Imaris 8.3.1.
Correlative light and electron microscopy
1.25 × 106 THP-1 cells were seeded in a μ-Dish35 mm, high Glass Bottom Grid-500 (81168, ibidi) and differentiated with PMA as described before. Cells were then pre-stimulated with IFNγ overnight and infected with type II (Pru) Tg at an MOI of one for 6 hours. One hour prior to fixation, 1 μg/mL CellMask Deep Red (H32721, Invitrogen) and 20 μM Hoechst 33342 (H3570, Invitrogen) was added to the culture medium to label the cells for detection in fluorescence microscopy. Cells were fixed by adding warm 8% (v/v) formaldehyde (Taab Laboratory Equipment Ltd) in 0.2 M phosphate buffer (PB) pH 7.4 directly to the cell culture medium (1:1) for 15min. The samples were then washed and imaged in PB using a Zeiss AiryScan LSM 880 confocal microscope. Samples were then processed using a Pelco BioWave Pro+ microwave (Ted Pella Inc) and following a protocol adapted from the National Centre for Microscopy and Imaging Research protocol (Deerinck et al., 2010) (See Table S2 for full BioWave program details). Each step was performed in the Biowave, except for the PB and water wash steps, which consisted of two washes followed by two washes in the Biowave without vacuum (at 250 W for 40 s). All chemical incubations were performed in the Biowave for 14 min under vacuum in 2 min cycles alternating with/without 100 W power. The SteadyTemp plate was set to 21°C unless otherwise stated. In brief, the samples were fixed again in 2.5% (v/v) glutaraldehyde (Taab) / 4% (v/v) formaldehyde in 0.1 M PB. The cells were then stained with 2% (v/v) osmium tetroxide (Taab) / 1.5% (v/v) potassium ferricyanide (Sigma), incubated in 1% (w/v) thiocarbohydrazide (Sigma) with SteadyTemp plate set to 40°C, and further stained with 2% osmium tetroxide in ddH2O (w/v). The cells were then incubated in 1% aqueous uranyl acetate (Agar Scientific), and then washed in dH2O with SteadyTemp set to 40°C for both steps. Samples were then stained with Walton’s lead aspartate with SteadyTemp set to 50°C and dehydrated in a graded ethanol series (70%, 90%, and 100%, twice each), at 250 W for 40 s without vacuum. Exchange into Durcupan ACM® resin (Sigma) was performed in 50% resin in ethanol, followed by 4 pure Durcupan steps, at 250 W for 3 min, with vacuum cycling (on/off at 30 s intervals), before embedding at 60°C for 48 h. Blocks were serial sectioned using a UC7 ultramicrotome (Leica Microsystems) and 70 nm sections were picked up on Formvar-coated slot copper grids (Gilder Grids Ltd). Consecutive sections were viewed using a 120 kV Tecnai G2 Spirit transmission electron microscope (Thermo Fischer Scientific) and images were captured using an Orius CCD camera (Gatan Inc). Individual TEM images of ∼25-30 consecutive sections per Tg parasite were converted as Tiff in Digital Micrograph (Gatan Inc.) and aligned using TrakEM2, a plugin of the FIJI framework (Cardona et al., 2012). The stacks were used to check the integrity of the PV and for coarse alignment with the AiryScan data.
Quantification of protein recruitment to vacuoles and ASC speck formation
Specimens were prepared as described above. Images were acquired using a Ti-E Nikon microscope equipped with LED-illumination and an Orca-Flash4 camera using a 60x magnification. All intracellular parasites/bacteria of 100 fields of view were automatically counted based on whether they showed recruitment of the protein of interest using HRMAn high-content image analysis (Fisch et al., 2019b). Further, the analysis pipeline was used to measure the fluorescence intensity of GBP1 on STm vacuoles using the radial intensity measurement implemented in HRMAn. The coat thickness of decorated vacuoles was determined by calculating the full width half maximum (FWHM) of the radial intensity measurement following a Gaussian curve fit of the raw fluorescence intensity data distribution measured starting from the centroid of the respective pathogen vacuole.
For quantification of ASC speck formation, 100 Tg-infected cells were manually counted per condition using a Ti-E Nikon microscope equipped with LED-illumination using 60x magnification based on whether they contain an ASC speck and whether STm was decorated with GBP1/CASP4. The experiment was repeated independently three times.
Visualization of Tg-DNA release
Type I (RH) Tg were grown in fully confluent and non-replicating HFFs for 3 days in the presence of 20 μM EdU to incorporate the nucleotide into their DNA. Labeled parasites were then harvested and used for infection as described above. 6 hours p.i. cells were fixed and EdU incorporated into Tg-DNA visualized by staining the specimens using Click-iT EdU Cell Proliferation Kit for Imaging, Alexa Fluor 647 dye (C10340, Invitrogen) according to the manufacturers instruction. Coverslips were further stained with Hoechst and mounted before imaging on a Ti-E Nikon microscope equipped with LED-illumination and an Orca-Flash4 camera using a 100x magnification.
Vacuole breakage assay - HRMAn
For quantification of Tg or STm vacuole integrity, cells seeded in black-wall 96-well imaging plates were infected and treated as described before. One hour prior to fixation, 1 μg/mL CellMask Deep Red (H32721, Invitrogen) was added to the culture medium to load the cytosol of host cells with this fluorescent dye. Following fixation and staining with Hoechst (H3570, Invitrogen), plates were imaged at 40x magnification on a Cell Insight CX7 High-Content Screening (HCS) Platform (Thermo Scientific) and 25 fields of view per well were recorded. For STm, plates were imaged at 60x magnification on a confocal Opera Phenix High Content Screening System (Perkin Elmer) and 100 fields of view per well were recorded with 8 z-slices each. Fluorescence of the dye within detected Tg vacuoles or STm was then analyzed using HRMAn (Fisch et al., 2019b). Additionally, HRMAn was used to classify Tg vacuoles and STm based on recruitment of mCH-GBP1 using the implemented neural network and the dataset stratified into decorated and non-decorated vacuoles or bacteria.
Differential stain for cytosolic STm
To distinguish between STm contained in vacuoles and bacteria that had escaped into the cytosol of infected macrophages, cells were differentially permeabilized using 25 μg/mL digitonin for one minute at room temperature as has been described before (Meunier and Broz, 2015). Cytosolic STm were then stained using anti-Salmonella antibody (ab35156, Abcam) that has been pre-labeled using Alexa Fluor 647 Protein Labeling Kit (A20173, Invitrogen) for 15 minutes at 37°C, prior to immediate fixation with 4% formaldehyde. Following fixation cells were permeabilized as described above and all STm were stained using the same but unlabeled antibody and corresponding Alexa 488 labeled secondary antibody. Cells were further stained with Hoechst (H3570, Invitrogen) and imaged on a Leica SP5-inverted confocal microscope using 100x magnification. For quantification, 100 fields of view per coverslip (typically > 1000 individual STm overall) were acquired using a Ti-E Nikon microscope equipped with LED-illumination and an Orca-Flash4 camera at 60x magnification and analyzed with HRMAn (Fisch et al., 2019b) for colocalization of fluorescent signal of all and cytosolic STm.
Quantification and Statistical Analysis
Data analysis used nested t test, one-way ANOVA or two-way ANOVA. Groups that were compared are indicated in the figure legends. Benjamini, Krieger and Yekutieli false-discovery rate (Q = 5%) based correction for multiple comparisons as implemented in Prism was used when making more than 3 comparisons. Graphs were plotted using Prism 8.1.1 (GraphPad Inc.) and presented as means of n = 3 experiments (with usually 3 technical repeats within each experiment) with error bars representing SEM, unless stated otherwise. Structure image of GBP1 was created using MacPymol v.1.74 (Schrödinger, Inc.).
Acknowledgments
We thank Matt Renshaw from the Crick Advanced Light Microscopy (CALM) STP for help with super-resolution SIM imaging, Julia Sanchez-Garrido for help in optimizing immunoblots and advice on reagents, Michael Howell from the Crick High-throughput screening (HTS) STP for help in performing automated imaging experiments, the Crick Genomics and Equipment Park STP for performing Sanger sequencing and DNA minipreps for cloning, Debipriya Das from the Crick Flow Cytometry STP for sorting Tg parasites, and Caia Dominicus, Jeanette Wagener, and Joanna Young from Moritz Treeck’s lab for help with creation of new transgenic Tg lines. We thank all members of the Frickel and the Shenoy labs for productive discussion and assistance in the project. This work was supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001076 to E.-M.F. and FC001999 to L.M.C. and A.P.S.), the UK Medical Research Council (FC001076 to E.-M.F. and FC001999 to L.M.C. and A.P.S.), and the Wellcome Trust (FC001076 to E.-M.F. and FC001999 to L.M.C. and A.P.S.). E.-M.F. was supported by a Wellcome Trust Career Development Fellowship (091664/B/10/Z) and a Wellcome Trust Senior Fellowship (217202/Z/19/Z). D.F. was supported by a Boehringer Ingelheim Fonds PhD fellowship. A.R.S. acknowledges support from the MRC (MR/P022138/1) and Wellcome Trust (108246/Z/15/Z). M.Y. was supported by the Research Program on Emerging and Re-emerging Infectious Diseases (JP18fk0108047) and the Japanese Initiative for Progress of Research on Infectious Diseases for Global Epidemic (JP18fk0108046) from the Agency for Medical Research and Development (AMED). H.B. was supported by a Grant-in-Aid for Scientific Research on Innovative Areas (17K15677) from the Ministry of Education, Culture, Sports, Science and Technology.
Author Contributions
D.F., A.R.S., and E.-M.F. conceived the idea for the study; D.F., B.C., M.-C.D., and V.E. performed experiments; H.B. and M.Y. provided essential reagents; L.M.C. and A.R.S. provided essential expertise and equipment; D.F., A.R.S., and E.-M.F. analyzed and interpreted the data and wrote the manuscript; and all authors revised the manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: August 11, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108008.
Contributor Information
Avinash R. Shenoy, Email: a.shenoy@imperial.ac.uk.
Eva-Maria Frickel, Email: e.frickel@bham.ac.uk.
Supplemental Information
References
- Al-Zeer M.A., Al-Younes H.M., Lauster D., Abu Lubad M., Meyer T.F. Autophagy restricts Chlamydia trachomatis growth in human macrophages via IFNG-inducible guanylate binding proteins. Autophagy. 2013;9:50–62. doi: 10.4161/auto.22482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekpen C., Hunn J.P., Rohde C., Parvanova I., Guethlein L., Dunn D.M., Glowalla E., Leptin M., Howard J.C. The interferon-inducible p47 (IRG) GTPases in vertebrates: loss of the cell autonomous resistance mechanism in the human lineage. Genome Biol. 2005;6:R92. doi: 10.1186/gb-2005-6-11-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekpen C., Xavier R.J., Eichler E.E. Human IRGM gene “to be or not to be”. Semin. Immunopathol. 2010;32:437–444. doi: 10.1007/s00281-010-0224-x. [DOI] [PubMed] [Google Scholar]
- Berthold M.R., Cebron N., Dill F., Gabriel T.R., Kötter T., Meinl T., Ohl P., Sieb C., Thiel K., Wiswedel B. Data Analysis, Machine Learning and Applications. Studies in Classification, Data Analysis, and Knowledge Organization. Springer; 2008. KNIME: the Konstanz Information Miner; pp. 319–326. [Google Scholar]
- Bradfield C. Yale University; 2016. Sulfated DAMPs Mobilize Human GBPs for Cell-Autonomous Immunity against Bacterial Pathogens. [Google Scholar]
- Britzen-Laurent N., Bauer M., Berton V., Fischer N., Syguda A., Reipschläger S., Naschberger E., Herrmann C., Stürzl M. Intracellular trafficking of guanylate-binding proteins is regulated by heterodimerization in a hierarchical manner. PLoS ONE. 2010;5:e14246. doi: 10.1371/journal.pone.0014246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardona A., Saalfeld S., Schindelin J., Arganda-Carreras I., Preibisch S., Longair M., Tomancak P., Hartenstein V., Douglas R.J. TrakEM2 software for neural circuit reconstruction. PLoS ONE. 2012;7:e38011. doi: 10.1371/journal.pone.0038011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson C.N., Yu J., Reyes V.M., Taschuk F.O., Yadav A., Copenhaver A.M., Nguyen H.T., Collman R.G., Shin S. Human caspase-4 mediates noncanonical inflammasome activation against gram-negative bacterial pathogens. Proc. Natl. Acad. Sci. USA. 2015;112:6688–6693. doi: 10.1073/pnas.1421699112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppens I., Dunn J.D., Romano J.D., Pypaert M., Zhang H., Boothroyd J.C., Joiner K.A. Toxoplasma gondii sequesters lysosomes from mammalian hosts in the vacuolar space. Cell. 2006;125:261–274. doi: 10.1016/j.cell.2006.01.056. [DOI] [PubMed] [Google Scholar]
- Costa Franco M.M., Marim F., Guimarães E.S., Assis N.R.G., Cerqueira D.M., Alves-Silva J., Harms J., Splitter G., Smith J., Kanneganti T.-D. Brucella abortus triggers a cGAS-independent STING pathway to induce host protection that involves guanylate-binding proteins and inflammasome activation. J. Immunol. 2018;200:607–622. doi: 10.4049/jimmunol.1700725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creasey E.A., Isberg R.R. The protein SdhA maintains the integrity of the Legionella-containing vacuole. Proc. Natl. Acad. Sci. USA. 2012;109:3481–3486. doi: 10.1073/pnas.1121286109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida L., Khare S., Misharin A.V., Patel R., Ratsimandresy R.A., Wallin M.C., Perlman H., Greaves D.R., Hoffman H.M., Dorfleutner A., Stehlik C. The PYRIN domain-only protein POP1 inhibits inflammasome assembly and ameliorates inflammatory disease. Immunity. 2015;43:264–276. doi: 10.1016/j.immuni.2015.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deerinck T.J., Bushong E., Thor A., Ellisman M. NCMIR methods for 3D EM: A new protocol for preparation of biological specimens for serial block face scanning electron microscopy. NCMIR. 2010 https://ncmir.ucsd.edu/sbem-protocol [Google Scholar]
- Degrandi D., Konermann C., Beuter-Gunia C., Kresse A., Würthner J., Kurig S., Beer S., Pfeffer K. Extensive characterization of IFN-induced GTPases mGBP1 to mGBP10 involved in host defense. J. Immunol. 2007;179:7729–7740. doi: 10.4049/jimmunol.179.11.7729. [DOI] [PubMed] [Google Scholar]
- Degrandi D., Kravets E., Konermann C., Beuter-Gunia C., Klümpers V., Lahme S., Wischmann E., Mausberg A.K., Beer-Hammer S., Pfeffer K. Murine guanylate binding protein 2 (mGBP2) controls Toxoplasma gondii replication. Proc. Natl. Acad. Sci. USA. 2013;110:294–299. doi: 10.1073/pnas.1205635110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J., Shao F. SnapShot: the noncanonical inflammasome. Cell. 2017;168:544–544.e1. doi: 10.1016/j.cell.2017.01.008. [DOI] [PubMed] [Google Scholar]
- Eldridge M.J.G., Sanchez-Garrido J., Hoben G.F., Goddard P.J., Shenoy A.R. The atypical ubiquitin E2 conjugase UBE2L3 is an indirect caspase-1 target and controls IL-1β secretion by inflammasomes. Cell Rep. 2017;18:1285–1297. doi: 10.1016/j.celrep.2017.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeley E.M., Pilla-Moffett D.M., Zwack E.E., Piro A.S., Finethy R., Kolb J.P., Martinez J., Brodsky I.E., Coers J. Galectin-3 directs antimicrobial guanylate binding proteins to vacuoles furnished with bacterial secretion systems. Proc. Natl. Acad. Sci. USA. 2017;114:E1698–E1706. doi: 10.1073/pnas.1615771114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisch D., Bando H., Clough B., Hornung V., Yamamoto M., Shenoy A.R., Frickel E.M. Human GBP1 is a microbe-specific gatekeeper of macrophage apoptosis and pyroptosis. EMBO J. 2019;38:e100926. doi: 10.15252/embj.2018100926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisch D., Yakimovich A., Clough B., Wright J., Bunyan M., Howell M., Mercer J., Frickel E. Defining host-pathogen interactions employing an artificial intelligence workflow. eLife. 2019;8:e40560. doi: 10.7554/eLife.40560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz C., Napolitano A., Khan R., Clough B., Hirst E.M., Frickel E.-M. TRIM21 is critical for survival of Toxoplasma gondii infection and localises to GBP-positive parasite vacuoles. Sci. Rep. 2017;7:5209. doi: 10.1038/s41598-017-05487-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster F., Paster W., Supper V., Schatzlmaier P., Sunzenauer S., Ostler N., Saliba A., Eckerstorfer P., Britzen-Laurent N., Schütz G. Guanylate binding protein 1-mediated interaction of T cell antigen receptor signaling with the cytoskeleton. J. Immunol. 2014;192:771–781. doi: 10.4049/jimmunol.1300377. [DOI] [PubMed] [Google Scholar]
- Gomes M.T.R., Cerqueira D.M., Guimarães E.S., Campos P.C., Oliveira S.C. Guanylate-binding proteins at the crossroad of noncanonical inflammasome activation during bacterial infections. J. Leukoc. Biol. 2019;106:553–562. doi: 10.1002/JLB.4MR0119-013R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar A.K., Saka H.A., Piro A.S., Dunn J.D., Henry S.C., Taylor G.A., Frickel E.M., Valdivia R.H., Coers J. IRG and GBP host resistance factors target aberrant, “non-self” vacuoles characterized by the missing of “self” IRGM proteins. PLoS Pathog. 2013;9:e1003414. doi: 10.1371/journal.ppat.1003414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar A.K., Piro A.S., Pilla D.M., Yamamoto M., Coers J. The E2-like conjugation enzyme Atg3 promotes binding of IRG and Gbp proteins to Chlamydia- and Toxoplasma-containing vacuoles and host resistance. PLoS ONE. 2014;9:e86684. doi: 10.1371/journal.pone.0086684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar A.K., Foltz C., Finethy R., Piro A.S., Feeley E.M., Pilla-Moffett D.M., Komatsu M., Frickel E.-M., Coers J. Ubiquitin systems mark pathogen-containing vacuoles as targets for host defense by guanylate binding proteins. Proc. Natl. Acad. Sci. USA. 2015;112:E5628–E5637. doi: 10.1073/pnas.1515966112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoiseth S.K., Stocker B.A.D. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- Hunn J.P., Koenen-Waisman S., Papic N., Schroeder N., Pawlowski N., Lange R., Kaiser F., Zerrahn J., Martens S., Howard J.C. Regulatory interactions between IRG resistance GTPases in the cellular response to Toxoplasma gondii. EMBO J. 2008;27:2495–2509. doi: 10.1038/emboj.2008.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston A.C., Piro A., Clough B., Siew M., Virreira Winter S., Coers J., Frickel E.-M. Human GBP1 does not localize to pathogen vacuoles but restricts Toxoplasma gondii. Cell. Microbiol. 2016;18:1056–1064. doi: 10.1111/cmi.12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen I., Zhang Y., Krantz B.A., Miao E.A. Pyroptosis triggers pore-induced intracellular traps (PITs) that capture bacteria and lead to their clearance by efferocytosis. J. Exp. Med. 2016;213:2113–2128. doi: 10.1084/jem.20151613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen I., Rayamajhi M., Miao E.A. Programmed cell death as a defence against infection. Nat. Rev. Immunol. 2017;17:151–164. doi: 10.1038/nri.2016.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan J.C., Magupalli V.G., Wu H. SMOCs: supramolecular organizing centres that control innate immunity. Nat. Rev. Immunol. 2014;14:821–826. doi: 10.1038/nri3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayagaki N., Warming S., Lamkanfi M., Vande Walle L., Louie S., Dong J., Newton K., Qu Y., Liu J., Heldens S. Non-canonical inflammasome activation targets caspase-11. Nature. 2011;479:117–121. doi: 10.1038/nature10558. [DOI] [PubMed] [Google Scholar]
- Kayagaki N., Wong M.T., Stowe I.B., Ramani S.R., Gonzalez L.C., Akashi-Takamura S., Miyake K., Zhang J., Lee W.P., Muszyński A. Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science. 2013;341:1246–1249. doi: 10.1126/science.1240248. [DOI] [PubMed] [Google Scholar]
- Khaminets A., Hunn J.P., Könen-Waisman S., Zhao Y.O., Preukschat D., Coers J., Boyle J.P., Ong Y.-C.C., Boothroyd J.C., Reichmann G., Howard J.C. Coordinated loading of IRG resistance GTPases on to the Toxoplasma gondii parasitophorous vacuole. Cell. Microbiol. 2010;12:939–961. doi: 10.1111/j.1462-5822.2010.01443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare S., Ratsimandresy R.A., de Almeida L., Cuda C.M., Rellick S.L., Misharin A.V., Wallin M.C., Gangopadhyay A., Forte E., Gottwein E. The PYRIN domain-only protein POP3 inhibits ALR inflammasomes and regulates responses to infection with DNA viruses. Nat. Immunol. 2014;15:343–353. doi: 10.1038/ni.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.-K., Karasov A., Boothroyd J.C. Bradyzoite-specific surface antigen SRS9 plays a role in maintaining Toxoplasma gondii persistence in the brain and in host control of parasite replication in the intestine. Infect. Immun. 2007;75:1626–1634. doi: 10.1128/IAI.01862-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B.-H., Shenoy A.R., Kumar P., Das R., Tiwari S., MacMicking J.D. A family of IFN-γ-inducible 65-kD GTPases protects against bacterial infection. Science. 2011;332:717–721. [Google Scholar]
- Kim B.-H., Shenoy A.R., Kumar P., Bradfield C.J., MacMicking J.D. IFN-inducible GTPases in host cell defense. Cell Host Microbe. 2012;12:432–444. doi: 10.1016/j.chom.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knodler L.A., Nair V., Steele-Mortimer O. Quantitative assessment of cytosolic Salmonella in epithelial cells. PLoS ONE. 2014;9:e84681. doi: 10.1371/journal.pone.0084681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler K.M., Kutsch M., Piro A.S., Wallace G., Coers J., Barber M.F. A rapidly evolving polybasic motif modulates bacterial detection by guanylate binding proteins. bioRxiv. 2019:689554. doi: 10.1128/mBio.00340-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortmann J., Brubaker S.W., Monack D.M. Cutting edge: inflammasome activation in primary human macrophages is dependent on flagellin. J. Immunol. 2015;195:815–819. doi: 10.4049/jimmunol.1403100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravets E., Degrandi D., Ma Q., Peulen T.-O.O., Klümpers V., Felekyan S., Kühnemuth R., Weidtkamp-Peters S., Seidel C.A., Pfeffer K. Guanylate binding proteins directly attack Toxoplasma gondii via supramolecular complexes. eLife. 2016;5:e11479. doi: 10.7554/eLife.11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kresse A., Konermann C., Degrandi D., Beuter-Gunia C., Wuerthner J., Pfeffer K., Beer S. Analyses of murine GBP homology clusters based on in silico, in vitro and in vivo studies. BMC Genomics. 2008;9:158–170. doi: 10.1186/1471-2164-9-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutsch M., Sistemich L., Lesser C.F., Goldberg M.B., Herrmann C., Coers J. Direct binding of polymeric GBP1 to LPS disrupts bacterial cell envelope functions. EMBO J. 2020;39:e104926. doi: 10.15252/embj.2020104926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagrange B., Benaoudia S., Wallet P., Magnotti F., Provost A., Michal F., Martin A., Di Lorenzo F., Py B.F., Molinaro A., Henry T. Human caspase-4 detects tetra-acylated LPS and cytosolic Francisella and functions differently from murine caspase-11. Nat. Commun. 2018;9:242. doi: 10.1038/s41467-017-02682-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Jiang W., Yu Q., Liu W., Zhou P., Li J., Xu J., Xu B., Wang F., Shao F. Ubiquitination and degradation of GBPs by a Shigella effector to suppress host defence. Nature. 2017;551:378–383. doi: 10.1038/nature24467. [DOI] [PubMed] [Google Scholar]
- Lindenberg V., Mölleken K., Kravets E., Stallmann S., Hegemann J.H., Degrandi D., Pfeffer K. Broad recruitment of mGBP family members to Chlamydia trachomatis inclusions. PLoS ONE. 2017;12:e0185273. doi: 10.1371/journal.pone.0185273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B.C., Sarhan J., Panda A., Muendlein H.I., Ilyukha V., Coers J., Yamamoto M., Isberg R.R., Poltorak A. Constitutive interferon maintains GBP expression required for release of bacterial components upstream of pyroptosis and anti-DNA responses. Cell Rep. 2018;24:155–168.e5. doi: 10.1016/j.celrep.2018.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMicking J.D. Interferon-inducible effector mechanisms in cell-autonomous immunity. Nat. Rev. Immunol. 2012;12:367–382. doi: 10.1038/nri3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik-Kale P., Winfree S., Steele-Mortimer O. The bimodal lifestyle of intracellular Salmonella in epithelial cells: replication in the cytosol obscures defects in vacuolar replication. PLoS ONE. 2012;7:e38732. doi: 10.1371/journal.pone.0038732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man S.M., Tourlomousis P., Hopkins L., Monie T.P., Fitzgerald K.A., Bryant C.E. Salmonella infection induces recruitment of Caspase-8 to the inflammasome to modulate IL-1β production. J. Immunol. 2013;191:5239–5246. doi: 10.4049/jimmunol.1301581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man S.M., Hopkins L.J., Nugent E., Cox S., Glück I.M., Tourlomousis P., Wright J.A., Cicuta P., Monie T.P., Bryant C.E. Inflammasome activation causes dual recruitment of NLRC4 and NLRP3 to the same macromolecular complex. Proc. Natl. Acad. Sci. USA. 2014;111:7403–7408. doi: 10.1073/pnas.1402911111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man S.M., Karki R., Malireddi R.K.S., Neale G., Vogel P., Yamamoto M., Lamkanfi M., Kanneganti T.D. The transcription factor IRF1 and guanylate-binding proteins target activation of the AIM2 inflammasome by Francisella infection. Nat. Immunol. 2015;16:467–475. doi: 10.1038/ni.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man S.M., Karki R., Sasai M., Place D.E., Kesavardhana S., Temirov J., Frase S., Zhu Q., Malireddi R.K.S., Kuriakose T. IRGB10 liberates bacterial ligands for sensing by the AIM2 and caspase-11-NLRP3 inflammasomes. Cell. 2016;167:382–396.e17. doi: 10.1016/j.cell.2016.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man S.M., Place D.E., Kuriakose T., Kanneganti T.-D. Interferon-inducible guanylate-binding proteins at the interface of cell-autonomous immunity and inflammasome activation. J. Leukoc. Biol. 2017;101:143–150. doi: 10.1189/jlb.4MR0516-223R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier E., Broz P. Quantification of cytosolic vs. vacuolar salmonella in primary macrophages by differential permeabilization. J. Vis. Exp. 2015;Jul 28:e52960. doi: 10.3791/52960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier E., Broz P. Interferon-inducible GTPases in cell autonomous and innate immunity. Cell. Microbiol. 2016;18:168–180. doi: 10.1111/cmi.12546. [DOI] [PubMed] [Google Scholar]
- Meunier E., Dick M.S., Dreier R.F., Schürmann N., Kenzelmann Broz D., Warming S., Roose-Girma M., Bumann D., Kayagaki N., Takeda K. Caspase-11 activation requires lysis of pathogen-containing vacuoles by IFN-induced GTPases. Nature. 2014;509:366–370. doi: 10.1038/nature13157. [DOI] [PubMed] [Google Scholar]
- Meunier E., Wallet P., Dreier R.F., Costanzo S., Anton L., Rühl S., Dussurgey S., Dick M.S., Kistner A., Rigard M. Guanylate-binding proteins promote activation of the AIM2 inflammasome during infection with Francisella novicida. Nat. Immunol. 2015;16:476–484. doi: 10.1038/ni.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyairi I., Tatireddigari V.R.R.A., Mahdi O.S., Rose L.A., Belland R.J., Lu L., Williams R.W., Byrne G.I. The p47 GTPases Iigp2 and Irgb10 regulate innate immunity and inflammation to murine Chlamydia psittaci infection. J. Immunol. 2007;179:1814–1824. doi: 10.4049/jimmunol.179.3.1814. [DOI] [PubMed] [Google Scholar]
- Mostowy S., Shenoy A.R. The cytoskeleton in cell-autonomous immunity: structural determinants of host defence. Nat. Rev. Immunol. 2015;15:559–573. doi: 10.1038/nri3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nantais D.E., Schwemmle M., Stickney J.T., Vestal D.J., Buss J.E. Prenylation of an interferon-γ-induced GTP-binding protein: the human guanylate binding protein, huGBP1. J. Leukoc. Biol. 1996;60:423–431. doi: 10.1002/jlb.60.3.423. [DOI] [PubMed] [Google Scholar]
- Naschberger E., Geißdörfer W., Bogdan C., Tripal P., Kremmer E., Stürzl M., Britzen-Laurent N. Processing and secretion of guanylate binding protein-1 depend on inflammatory caspase activity. J. Cell. Mol. Med. 2017;21:1954–1966. doi: 10.1111/jcmm.13116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski M.A., Gray J., Vestal D.J. In silico genomic analysis of the human and murine guanylate-binding protein (GBP) gene clusters. J. Interf. Cytokine. 2006;352:328–352. doi: 10.1089/jir.2006.26.328. [DOI] [PubMed] [Google Scholar]
- Ostler N., Britzen-Laurent N., Liebl A., Naschberger E., Lochnit G., Ostler M., Forster F., Kunzelmann P., Ince S., Supper V. Gamma interferon-induced guanylate binding protein 1 is a novel actin cytoskeleton remodeling factor. Mol. Cell. Biol. 2014;34:196–209. doi: 10.1128/MCB.00664-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilla D.M., Hagar J.A., Haldar A.K., Mason A.K., Degrandi D., Pfeffer K., Ernst R.K., Yamamoto M., Miao E.A., Coers J. Guanylate binding proteins promote caspase-11-dependent pyroptosis in response to cytoplasmic LPS. Proc. Natl. Acad. Sci. USA. 2014;111:6046–6051. doi: 10.1073/pnas.1321700111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilla-Moffett D., Barber M.F., Taylor G.A., Coers J. Interferon-inducible GTPases in host resistance, inflammation and disease. J. Mol. Biol. 2016;428:3495–3513. doi: 10.1016/j.jmb.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piro A.S., Hernandez D., Luoma S., Feeley E.M., Finethy R., Yirga A., Frickel E.M., Lesser C.F., Coers J. Detection of cytosolic Shigella flexneri via a C-terminal triple-arginine motif of GBP1 inhibits actin-based motility. MBio. 2017;8:e01979-17. doi: 10.1128/mBio.01979-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash B., Praefcke G.J.K., Renault L., Wittinghofer A., Herrmann C. Structure of human guanylate-binding protein 1 representing a unique class of GTP-binding proteins. Nature. 2000;403:567–571. doi: 10.1038/35000617. [DOI] [PubMed] [Google Scholar]
- Radtke A.L., Delbridge L.M., Balachandran S., Barber G.N., O’Riordan M.X.D. TBK1 protects vacuolar integrity during intracellular bacterial infection. PLoS Pathog. 2007;3:e29. doi: 10.1371/journal.ppat.0030029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randow F., MacMicking J.D., James L.C. Cellular self-defense: how cell-autonomous immunity protects against pathogens. Science. 2013;340:701–706. doi: 10.1126/science.1233028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes Ruiz V.M., Ramirez J., Naseer N., Palacio N.M., Siddarthan I.J., Yan B.M., Boyer M.A., Pensinger D.A., Sauer J.-D., Shin S. Broad detection of bacterial type III secretion system and flagellin proteins by the human NAIP/NLRC4 inflammasome. Proc. Natl. Acad. Sci. USA. 2017;114:13242–13247. doi: 10.1073/pnas.1710433114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romei M.G., Boxer S.G. Split green fluorescent proteins: scope, limitations, and outlook. Annu. Rev. Biophys. 2019;48:19–44. doi: 10.1146/annurev-biophys-051013-022846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rühl S., Broz P. Caspase-11 activates a canonical NLRP3 inflammasome by promoting K(+) efflux. Eur. J. Immunol. 2015;45:2927–2936. doi: 10.1002/eji.201545772. [DOI] [PubMed] [Google Scholar]
- Saeij J.P., Frickel E.-M. Exposing Toxoplasma gondii hiding inside the vacuole: a role for GBPs, autophagy and host cell death. Curr. Opin. Microbiol. 2017;40:72–80. doi: 10.1016/j.mib.2017.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Garrido J., Sancho-Shimizu V., Shenoy A.R. Regulated proteolysis of p62/SQSTM1 enables differential control of autophagy and nutrient sensing. Sci. Signal. 2018;11:eaat6903. doi: 10.1126/scisignal.aat6903. [DOI] [PubMed] [Google Scholar]
- Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos J.C., Dick M.S., Lagrange B., Degrandi D., Pfeffer K., Yamamoto M., Meunier E., Pelczar P., Henry T., Broz P. LPS targets host guanylate-binding proteins to the bacterial outer membrane for non-canonical inflammasome activation. EMBO J. 2018;37:e98089. doi: 10.15252/embj.201798089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos J.C., Boucher D., Schneider L.K., Demarco B., Dilucca M., Shkarina K., Heilig R., Chen K.W., Lim R.Y.H., Broz P. Human GBP1 binds LPS to initiate assembly of a caspase-4 activating platform on cytosolic bacteria. Nat. Commun. 2020;11:3276. doi: 10.1038/s41467-020-16889-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid-Burgk J.L., Gaidt M.M., Schmidt T., Ebert T.S., Bartok E., Hornung V. Caspase-4 mediates non-canonical activation of the NLRP3 inflammasome in human myeloid cells. Eur. J. Immunol. 2015;45:2911–2917. doi: 10.1002/eji.201545523. [DOI] [PubMed] [Google Scholar]
- Schoggins J.W. Interferon-stimulated genes: what do they all do? Annu. Rev. Virol. 2019;6:567–584. doi: 10.1146/annurev-virology-092818-015756. [DOI] [PubMed] [Google Scholar]
- Selleck E.M., Fentress S.J., Beatty W.L., Degrandi D., Pfeffer K., Virgin H.W., 4th, Macmicking J.D., Sibley L.D. Guanylate-binding protein 1 (Gbp1) contributes to cell-autonomous immunity against Toxoplasma gondii. PLoS Pathog. 2013;9:e1003320. doi: 10.1371/journal.ppat.1003320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy A.R., Kim B.H., Choi H.P., Matsuzawa T., Tiwari S., MacMicking J.D. Emerging themes in IFN-γ-induced macrophage immunity by the p47 and p65 GTPase families. Immunobiology. 2007;212:771–784. doi: 10.1016/j.imbio.2007.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy A.R., Wellington D.A., Kumar P., Kassa H., Booth C.J., Cresswell P., MacMicking J.D. GBP5 promotes NLRP3 inflammasome assembly and immunity in mammals. Science. 2012;336:481–485. doi: 10.1126/science.1217141. [DOI] [PubMed] [Google Scholar]
- Shi J., Zhao Y., Wang Y., Gao W., Ding J., Li P., Hu L., Shao F. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature. 2014;514:187–192. doi: 10.1038/nature13683. [DOI] [PubMed] [Google Scholar]
- Shydlovskyi S., Zienert A.Y., Ince S., Dovengerds C., Hohendahl A., Dargazanli J.M., Blum A., Günther S.D., Kladt N., Stürzl M. Nucleotide-dependent farnesyl switch orchestrates polymerization and membrane binding of human guanylate-binding protein 1. Proc. Natl. Acad. Sci. USA. 2017;114:E5559–E5568. doi: 10.1073/pnas.1620959114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S.B., Davis A.S., Taylor G.A., Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- Singh S.B., Ornatowski W., Vergne I., Naylor J., Delgado M., Roberts E., Ponpuak M., Master S., Pilli M., White E. Human IRGM regulates autophagy and cell-autonomous immunity functions through mitochondria. Nat. Cell Biol. 2010;12:1154–1165. doi: 10.1038/ncb2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stennicke H.R., Salvesen G.S. Biochemical characteristics of caspases-3, -6, -7, and -8. J. Biol. Chem. 1997;272:25719–25723. doi: 10.1074/jbc.272.41.25719. [DOI] [PubMed] [Google Scholar]
- Stévenin V., Chang Y.Y., Le Toquin Y., Duchateau M., Gianetto Q.G., Luk C.H., Salles A., Sohst V., Matondo M., Reiling N., Enninga J. Dynamic growth and shrinkage of the salmonella-containing vacuole determines the intracellular pathogen niche. Cell Rep. 2019;29:3958–3973.e7. doi: 10.1016/j.celrep.2019.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stickney J.T., Buss J.E. Murine guanylate-binding protein: incomplete geranylgeranyl isoprenoid modification of an interferon-γ-inducible guanosine triphosphate-binding protein. Mol. Biol. Cell. 2000;11:2191–2200. doi: 10.1091/mbc.11.7.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurston T.L.M.M., Wandel M.P., von Muhlinen N., Foeglein A., Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012;482:414–418. doi: 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurston T.L.M.M., Matthews S.A., Jennings E., Alix E., Shao F., Shenoy A.R., Birrell M.A., Holden D.W. Growth inhibition of cytosolic salmonella by caspase-1 and caspase-11 precedes host cell death. Nat. Commun. 2016;7:13292. doi: 10.1038/ncomms13292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S., Choi H.P., Matsuzawa T., Pypaert M., MacMicking J.D. Targeting of the GTPase Irgm1 to the phagosomal membrane via PtdIns(3,4)P(2) and PtdIns(3,4,5)P(3) promotes immunity to mycobacteria. Nat. Immunol. 2009;10:907–917. doi: 10.1038/ni.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traver M.K., Henry S.C., Cantillana V., Oliver T., Hunn J.P., Howard J.C., Beer S., Pfeffer K., Coers J., Taylor G.A. Immunity-related GTPase M (IRGM) proteins influence the localization of guanylate-binding protein 2 (GBP2) by modulating macroautophagy. J. Biol. Chem. 2011;286:30471–30480. doi: 10.1074/jbc.M111.251967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretina K., Park E.S., Maminska A., MacMicking J.D. Interferon-induced guanylate-binding proteins: guardians of host defense in health and disease. J. Exp. Med. 2019;216:482–500. doi: 10.1084/jem.20182031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripal P., Bauer M., Naschberger E., Mörtinger T., Hohenadl C., Cornali E., Thurau M., Stürzl M. Unique features of different members of the human guanylate-binding protein family. J. Interf. Cytokine Res. 2007;27:44–52. doi: 10.1089/jir.2007.0086. [DOI] [PubMed] [Google Scholar]
- Virreira Winter S., Niedelman W., Jensen K.D., Rosowski E.E., Julien L., Spooner E., Caradonna K., Burleigh B.A., Saeij J.P.J.J., Ploegh H.L., Frickel E.M. Determinants of GBP recruitment to Toxoplasma gondii vacuoles and the parasitic factors that control it. PLoS ONE. 2011;6:e24434. doi: 10.1371/journal.pone.0024434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallet P., Benaoudia S., Mosnier A., Lagrange B., Martin A., Lindgren H., Golovliov I., Michal F., Basso P., Djebali S. IFN-γ extends the immune functions of Guanylate Binding Proteins to inflammasome-independent antibacterial activities during Francisella novicida infection. PLoS Pathog. 2017;13:e1006630. doi: 10.1371/journal.ppat.1006630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandel M.P., Pathe C., Werner E.I., Ellison C.J., Boyle K.B., von der Malsburg A., Rohde J., Randow F. GBPs inhibit motility of Shigella flexneri but are targeted for degradation by the bacterial ubiquitin ligase IpaH9.8. Cell Host Microbe. 2017;22:507–518.e5. doi: 10.1016/j.chom.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandel M.P., Kim B.H., Park E.S., Boyle K.B., Nayak K., Lagrange B., Herod A., Henry T., Zilbauer M., Rohde J. Guanylate-binding proteins convert cytosolic bacteria into caspase-4 signaling platforms. Nat. Immunol. 2020:10.1038. doi: 10.1038/s41590-020-0697-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier A., Al-Zeer M.A., Meyer T.F., Daumke O. hGBP1 xoordinates Chlamydia restriction and inflammasome activation through sequential GTP hydrolysis. Cell Rep. 2020;31:107667. doi: 10.1016/j.celrep.2020.107667. [DOI] [PubMed] [Google Scholar]
- Yamamoto M., Okuyama M., Ma J.S., Kimura T., Kamiyama N., Saiga H., Ohshima J., Sasai M., Kayama H., Okamoto T. A cluster of interferon-γ-inducible p65 GTPases plays a critical role in host defense against Toxoplasma gondii. Immunity. 2012;37:302–313. doi: 10.1016/j.immuni.2012.06.009. [DOI] [PubMed] [Google Scholar]
- Young J., Dominicus C., Wagener J., Butterworth S., Ye X., Kelly G., Ordan M., Saunders B., Instrell R., Howell M. A CRISPR platform for targeted in vivo screens identifies Toxoplasma gondii virulence factors in mice. Nat. Commun. 2019;10:3963. doi: 10.1038/s41467-019-11855-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwack E.E., Feeley E.M., Burton A.R., Hu B., Yamamoto M., Kanneganti T.-D., Bliska J.B., Coers J., Brodsky I.E. Guanylate binding proteins regulate inflammasome activation in response to hyperinjected Yersinia translocon components. Infect. Immun. 2017;85:e00778-e16. doi: 10.1128/IAI.00778-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Mass spectrometry dataset is included in this study (Data S1). Quantification datasets are available from Mendeley data (https://doi.org/10.17632/gkgrk7d8hg.1). Any other data supporting the current study are available from the lead contact.