To the Editor:
Severe acute respiratory syndrome‐coronavirus‐2 (SARS‐CoV‐2) uses the SARS‐CoV receptor angiotensin‐converting enzyme 2 (ACE2) for entry to the cell and the transmembrane serine protease 2 (TMPRSS2) for S protein priming. 1 , 2 Higher ACE2 expression was recently reported in nasal compared to throat tissue. 3 In fact, higher SARS‐CoV‐2 viral load was detected in nasal compared to throat swabs obtained from COVID‐19–infected patients. This was attributed to the difference in ACE2 expression between both tissues. 4 Recently, we have also shown that the upper airway expresses more SARS‐CoV‐2 entry genes, ACE2 and TMPRSS2, compared to the lower airway. 5 Moreover, Hou et al. 6 have recently established that multiciliated cells are the main cell types expressing ACE2 in nasal tissue and infected with SARS‐CoV‐2. Moreover, Sungnak et al., 7 by analyzing data of single‐cell RNA‐sequencing from healthy human nasal epithelial cells, showed that ACE2 and TMPRSS2 are co‐expressed in nasal epithelium with genes involved in host innate immunity, referring to the potential role of these cells in initiating SARS‐CoV‐2 infection. Therefore, the level of SARS‐CoV‐2 receptors in nasal tissue may determine the level of viral infectivity because these receptors are not upregulated following infection. 1 With that in mind, we decided to investigate potential factors that may affect the expression of SARS‐CoV‐2 receptors and hence the risk of infectivity with COVID‐19 in various phenotypes of sinonasal inflammation.
To achieve this, gene‐expression data sets were used to determine the expression pattern of SARS‐CoV‐2 cell entry genes, ACE2 and TMPRSS2, in the inflamed uncinate process from patients with chronic rhinosinusitis without nasal polyps (CRSsNPs) and nasal polyps from patients with CRS with nasal polyps (CRSwNP) (see Supplementary Methods). A reduction in ACE2 expression was observed in the inflamed uncinate tissue of CRSsNP patients but not to a significant level (Fig. 1A). Interestingly, a significant reduction in the expression of ACE2 and TMPRSS2 was observed in the nasal polyps of CRSwNP patients compared to healthy controls (Fig. 1A). This data implies that patients with CRSwNP might have a lower risk of SARS‐CoV‐2 infection due to lower expression levels of SARS‐CoV‐2 cell entry genes.
FIGURE 1.
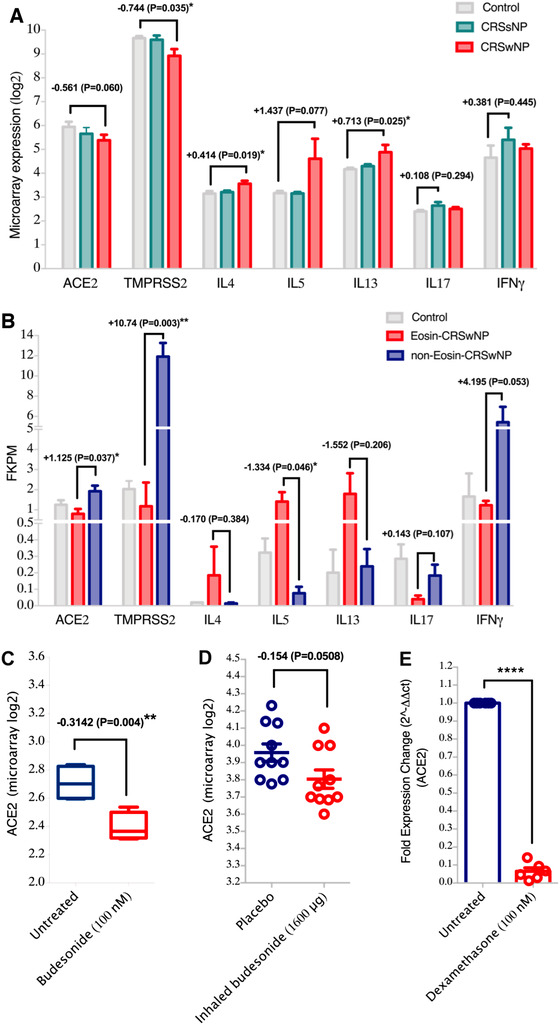
Reduced expression of SARS‐CoV‐2 host entry genes, TMPRSS2 and ACE2, in the nasal polyps of CRSwNP patients. (A) Gene expression of ACE2 and TMPRSS2 as well as proinflammatory cytokines in the inflamed uncinate tissues of CRSsNP patients (n = 3), nasal polyps of CRSwNP patients (n = 3), and uncinate tissues of healthy controls (n = 3). The data presented shows lower expression of ACE2 and TMPRSS2 in the nasal polyps of CRSwNP patients compared to normal uncinate tissue. Accordingly, gene expression levels of type 2 cytokines, IL4, IL‐5, and IL13 were higher in the nasal polyps of CRSwNP patients; whereas IL17 and IFNγ cytokines were more elevated in the uncinate tissue of CRSsNP patients. (B) Gene expression of ACE2 and TMPRSS2 as well as proinflammatory cytokines in nasal polyps of eosinophilic (n = 3), nasal polyps of non‐eosinophilic CRSwNP patients (n = 3), and sphenoid sinus mucosa of healthy controls (n = 3). The data presented shows decreased level of ACE2 and TMPRSS2 expression in the nasal polyps of eosinophilic compared to non‐eosinophilic CRSwNP patients. Additionally, it shows increased gene expression levels of type 2 cytokines, IL4, IL5, and IL13 in the nasal polyps of eosinophilic compared to non‐eosinophilic CRSwNP patients. In other hand, the gene expression levels of IL17 and IFNγ were higher in the nasal polyps of non‐eosinophilic compared to eosinophilic CRSwNP patients. (C) Gene expression level of ACE2 in the human bronchial airway epithelial BEAS‐2B cells untreated or treated with budesonide (100nM) for 18 hours. The data shows that treatment with budesonide downregulated ACE2 expression levels in the treated compared to untreated BEAS‐2B cells. (D) Gene expression level of ACE2 in bronchial biopsies from healthy male, non‐smoker, non‐allergic volunteers treated with placebo or budesonide (1600 μg). The data shows that treatment with budesonide downregulated ACE2 expression levels in the budesonide‐treated compared to placebo‐treated subjects. (E) Gene expression level of ACE2 in the primary bronchial fibroblasts untreated or treated with dexamethasone (100nM) for 24 hours. The data shows that treatment with dexamethasone downregulated ACE2 expression levels in the treated compared to untreated fibroblasts. ACE2 = angiotensin‐converting enzyme 2; CRSsNP = chronic rhinosinusitis without nasal polyps; CRSwNP = chronic rhinosinusitis with nasal polyps; IFN = interferon; IL = interleukin; TMPRSS2 = transmembrane serine protease 2.
To our knowledge, the impact of allergic inflammation on the level of expression of ACE2 and TMPRSS2 remains poorly understood. In an in vitro study, interleukin 4 (IL4) has been shown to reduce ACE2 gene expression levels in SARS‐CoV–infected cells. 8 Moreover, ACE2 was shown to be less activated in eosinophilic and allergic airway inflammation conditions, 9 which may indicate a potential regulatory effect of such conditions on these receptors. In our study, we investigated the expression levels of interferon γ (IFNγ), IL17, and type 2 cytokines in the tissues of these CRS patients. Significantly higher levels of IL4, IL5, and IL13 cytokines were observed in nasal polyp tissue from CRSwNP patients, whereas IFNγ and IL17 cytokines were more elevated in the uncinate tissue (Fig. 1A). This suggests that type 2 inflammation may downregulate ACE2 and TMPRSS2 expression.
To further investigate, we used another data set of nasal polyp tissue from eosinophilic and non‐eosinophilic CRSwNP patients. Interestingly, a reduction in expression of ACE2 and TMPRSS2 receptors was observed in the eosinophilic nasal polyp tissue compared to healthy controls. In contrast, an increase was observed in both receptors in non‐eosinophilic nasal polyp tissue compared to controls (Fig. 1B). In addition, ACE2 and TMPRSS2 expression in non‐eosinophilic polyp tissue was significantly higher than in eosinophilic tissue (p = 0.0378 and p = 0.0039, respectively).
We then assessed the level of expression of IFNγ, IL17, and type 2 cytokines in eosinophilic vs non‐eosinophilic tissue from CRSwNP patients. As expected, eosinophilic polyp tissue had higher levels of expression of IL4, IL5, and IL13 cytokines, whereas higher levels of IFNγ and IL‐17 were observed in non‐eosinophilic polyp tissue (Fig. 1B). It was also observed that ACE2 has positive correlation with IFNγ (r = 0.86, p = 0.003), but not with IL17. ACE2 levels negatively correlated with IL5 (r = 0.709, p = 0.033) and IL13 (r = 0.682, p = 0.043) (Supplementary Fig. 1A‐C). No significant correlation was found between ACE2 and IL4 levels. Correlations of TMPRSS2 with the abovementioned cytokines IFNγ, IL17, IL4, IL5, and IL13 were not significant.
Next, the differential expression of ACE2 and TMPRSS2 was assessed in datasets of human ethmoid sinus cells treated independently with IL4, IFNγ, or IL17. 6 IFNγ and IL17 increased the expression of both ACE2 and TMPRSS2 in these cells (Supplementary Fig. 2A,B), whereas IL4 suppressed their expression levels (Supplementary Fig. 2C). We further evaluated the expression of ACE2 and TMPRESS2 in a data set of IL13‐treated human nasal epithelial cells isolated from nasal turbinates. IL13 also suppressed ACE2, but increased the expression of TMPRSS2 in these cells (Supplementary Fig. 2D), consistent with recent reports. 6 , 10
This data suggest that eosinophilic inflammation and the associated type 2 cytokines downregulate the expression of ACE2 in nasal tissue of CRS patients and thus may have a protective role against COVID‐19 infection. In contrast, our data indicates that IFNγ increases ACE2 expression, in line with previously published data. 11 This may suggest that neutrophilic inflammation and the associated cytokines such as IFNγ and IL17 may upregulate COVID‐19 receptors levels and thus facilitate COVID‐19 infectivity. This data is also in line with our previous finding 5 indicating that patients with chronic obstructive pulmonary disease, characterized with neutrophilic inflammation, have significantly elevated ACE2 and TMPRSS2 lung tissue levels.
CRS patients are usually under prolonged exposure to corticosteroids, which may regulate ACE2 expression. A recent report suggested that in asthmatic subjects, sputum ACE2 expression is significantly lower in patients who are on inhaled corticosteroids compared to those who are not on inhaled corticosteroids. 12 Here, we hypothesized that corticosteroids may regulate ACE2 gene expression. To test this hypothesis, we examined the expression of ACE2 in relation to steroid treatment in 2 publicly available data sets of gene expression data from airway epithelial cells as well as bronchial biopsies. Treatment with budesonide downregulated the expression of ACE2 in both bronchial epithelial cells (Fig. 1C) and bronchial biopsies (Fig. 1D). To confirm this observation, we treated healthy primary bronchial fibroblasts with dexamethasone (100nM for 24 hours) and analyzed ACE2 expression using real‐time polymerase chain reaction (RT‐PCR). Dexamethasone significantly suppressed ACE2 expression in these cells (Fig. 1E). This suggests that prolonged exposure to corticosteroids may suppresses ACE2 gene expression levels in nasal tissue of CRS patients. Because the effect of corticosteroids on COVID‐19 patients is still highly debatable, more research is required to confirm our findings and observations.
Moreover, a recent study by Jackson et al. 13 suggested that atopic asthmatics might be at lower risk of COVID‐19–related illness because respiratory allergen exposure and type 2 cytokine IL13 reduces ACE2 gene expression in both nasal and bronchial epithelial cells. The analysis from our study provides further evidence that type 2 or eosinophilic inflammation is biased more toward lowering ACE2 gene expression, in the context of CRS as well, and shows for the first time that steroids may have a protective effect by downregulating ACE2 expression in the nasal tissues of CRS.
The limitation of our study is that although the difference in type 2 cytokines observed between polyp and uncinate tissue may contribute to the regulation of these receptors, this does not exclude the possibility that the difference in receptors expression may reflect tissue differences rather than endotype.
In conclusion, as presented in Figure 2, our data suggest that the type of inflammation underlying CRS, as well as corticosteroid treatment, may modulate ACE2 and TMPRSS2 gene expression levels in the nasal polyps of CRSwNP patients.
FIGURE 2.
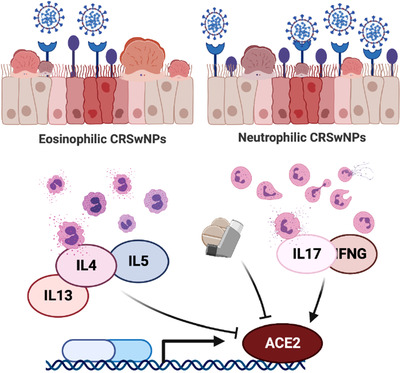
The effect of corticosteroids and pro‐inflammatory cytokines on the ACE2 gene expression level in the nasal polyps of eosinophilic and non‐eosinophilic CRSwNP patients. ACE2 = angiotensin‐converting enzyme 2; CRSwNP = chronic rhinosinusitis with nasal polyps.
Supporting information
Supplementary Table 1. General characteristics of CRSsNP and CRSwNP cohorts (GSE36830).
Supplementary Table 2. General characteristics of eosinophilic CRSwNP and non‐eosinophilic CRSwNP cohorts (GSE72713).
Supplementary Figure 1. (A) Positive correlation between ACE2 and IFN‐g (r=0.86, P=0.003) and negative correlation between ACE2 and (B) IL5 (r=0.709, P=033) and (C) IL13 (r=0.682, P=043).
Supplementary Figure 2. The effect of cytokines on the gene expression levels of ACE2 and TMPRSS2 in nasal cells. (A‐C) Gene expression level of ACE2 and TMPRSS2 in human ethmoid sinus cells treated or not with IFNγ, IL17, and IL4. The data shows that treatment with IFNγ or IL17 upregulated ACE2 and TMPRSS2 expression in these cells; while IL4 suppressed their expression levels. (D) Gene expression level of ACE2 and TMPRSS2 in human nasal epithelial cells treated or not with IL13. The data shows that treatment with IL13 downregulated ACE2 expression, but increased TMPRSS2 expression in these cells.
Supplementary Figure 3. (A) The gene expression levels of extracellular matrix depositions such as α‐SMA (ACTA2), Fibronectin (FN1), and Collagen type I (COL1A1) in the uncinate tissue of CRSsNP compared to nasal polyps of CRSwNP. The presented data shows that ECM depositions are non‐significant between uncinate tissues of CRSsNP and nasal polyps of CRSwNP.
Acknowledgements
This research was financially supported by Tissue Injury and Repair (TIR) group operational grant (Grant code: 150317), COVID‐19 research grant, and by a seed grant to RH (Grant code: 2001090275), University of Sharjah, UAE; and by Prince Abdullah Ben Khalid Celiac Disease Research Chair, under the Vice Deanship of Research Chairs, King Saud University, Riyadh, Kingdom of Saudi Arabia. The authors extend their appreciation to the Deputyship for Research & Innovation, Ministry of Education in Saudi Arabia for funding this research work through the project number (DRI‐KSU‐928). We gratefully acknowledge Zakaria Ratemi, College of Medicine, UDEM, Montreal, Canada, for his help in critically reviewing the manuscript.
Funding sources for the study: Tissue Injury and Repair (TIR) Group operational grant (150317); University of Sharjah, COVID‐19 research grant to R.H.; University of Sharjah, Seed grant (2001090275) to R.H.; University of Sharjah, Collaborative research grant (Grant code: 2001090278) to R.H.; Sandooq Al Watan Applied Research & Development grant to R.H.; Prince Abdullah Ben Khalid Celiac Disease Research Chair, under the Vice Deanship of Research Chairs, King Saud University, Riyadh, Kingdom of Saudi Arabia; and Deputyship for Research & Innovation, Ministry of Education in Saudi Arabia (DRI‐KSU‐928).
Potential conflict of interest: None provided.
References
- 1. Hoffmann M., Kleine‐Weber H, Schroeder S. SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271‐280.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270‐273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wu C, Zheng M. Single‐cell RNA expression profiling shows that ACE2, the putative receptor of Wuhan 2019‐nCoV, has significant expression in the nasal, mouth, lung and colon tissues, and tends to be co‐expressed with HLA‐DRB1 in the four tissues. Preprints. 2020;2020020247. https://www.preprints.org/manuscript/202002.0247/v1. Accessed August 13, 2020. [Google Scholar]
- 4. Zou L, Ruan F, Huang M, et al. SARS‐CoV‐2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177‐1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sharif‐Askari NS, Saheb Sharif‐Askari F, Alabed M, et al. Airways expression of SARS‐CoV‐2 receptor, ACE2, and TMPRSS2 is lower in children than adults and increases due to smoking and COPD. Mol Ther Methods Clin Dev. 2020;18:1‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hou YJ, Okuda K, Edwards CE, et al. SARS‐CoV‐2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell. 2020;182:429‐446.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sungnak W, Huang N, Bécavin C, et al. SARS‐CoV‐2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med. 2020;26:681‐687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Lang A, Osterhaus AD, Haagmans BL, Interferon‐γ and interleukin‐4 downregulate expression of the SARS coronavirus receptor ACE2 in Vero E6 cells. Virology. 2006;353:474‐481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dhawale VS, Amara VR, Karpe PA, et al. Activation of angiotensin‐converting enzyme 2 (ACE2) attenuates allergic airway inflammation in rat asthma model. Toxicol Appl Pharmacol. 2016;306:17‐26. [DOI] [PubMed] [Google Scholar]
- 10. Kimura H, Francisco D, Conway F, et al. Type 2 inflammation modulates ACE2 and TMPRSS2 in airway epithelial cells. J Allergy Clin Immunol. 2020;146:80‐88.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ziegler CGK, Allon SJ, Nyquist SK, et al. SARS‐CoV‐2 receptor ACE2 is an interferon‐stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016‐1035.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peters MC, Sajuthi S, Deford P, et al. COVID‐19 related genes in sputum cells in asthma. Relationship to demographic features and corticosteroids. Am J Respir Crit Care Med. 2020;202:83‐90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jackson DJ, Busse WW, Bacharier LB, et al. Association of respiratory allergy, asthma and expression of the SARS‐CoV‐2 receptor, ACE2. J Allergy Clin Immunol. 2020;146:203‐206.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. General characteristics of CRSsNP and CRSwNP cohorts (GSE36830).
Supplementary Table 2. General characteristics of eosinophilic CRSwNP and non‐eosinophilic CRSwNP cohorts (GSE72713).
Supplementary Figure 1. (A) Positive correlation between ACE2 and IFN‐g (r=0.86, P=0.003) and negative correlation between ACE2 and (B) IL5 (r=0.709, P=033) and (C) IL13 (r=0.682, P=043).
Supplementary Figure 2. The effect of cytokines on the gene expression levels of ACE2 and TMPRSS2 in nasal cells. (A‐C) Gene expression level of ACE2 and TMPRSS2 in human ethmoid sinus cells treated or not with IFNγ, IL17, and IL4. The data shows that treatment with IFNγ or IL17 upregulated ACE2 and TMPRSS2 expression in these cells; while IL4 suppressed their expression levels. (D) Gene expression level of ACE2 and TMPRSS2 in human nasal epithelial cells treated or not with IL13. The data shows that treatment with IL13 downregulated ACE2 expression, but increased TMPRSS2 expression in these cells.
Supplementary Figure 3. (A) The gene expression levels of extracellular matrix depositions such as α‐SMA (ACTA2), Fibronectin (FN1), and Collagen type I (COL1A1) in the uncinate tissue of CRSsNP compared to nasal polyps of CRSwNP. The presented data shows that ECM depositions are non‐significant between uncinate tissues of CRSsNP and nasal polyps of CRSwNP.


