Abstract
Background:
MicroRNAs (miRNAs) are small, non-coding RNAs that play a key role in gene expression. Accumulating evidence suggests that aberrant miRNA expression contributes to the heart failure (HF) phenotype, however, the underlying molecular mechanisms are not well understood. A better understanding of the mechanisms of action miRNAs could potentially lead to targeted therapies that could halt the progression or even reverse HF.
Methods and Results:
We identified that miR-152 expression was upregulated in the failing human heart and experimental animal models of HF. Transgenic mice with cardiomyocyte-specific miR-152 overexpression developed impaired systolic function (mean difference, −38.74% [95% CI: −45.73% to −31.74%]; P<0.001) and dilated cardiomyopathy. At the cellular level, miR-152 overexpression induced aberrant mitochondrial ultrastructure and dysregulation of key genes involved in cardiomyocyte metabolism and inflammation. Mechanistically, we identified monothiol Glutaredoxin 5 (Glrx5), a critical regulator of mitochondrial iron and iron-sulfur cluster synthesis, as a direct miR-152 target. Finally, a proof-of-concept of the therapeutic efficacy of targeting miR-152 in vivo was obtained by utilizing a locked nucleic acid (LNA)-based inhibitor of miR-152 (LNA-152) in a murine model of heart failure subjected to transverse aortic constriction. We demonstrated that animals treated with LNA-152 (n=10) showed preservation of systolic function when compared to LNA-control treated animals (n=9) (mean difference, −18.25% [95% CI: −25.10% to – 11.39%] P<0.001).
Conclusion:
The upregulation of miR-152 expression in the failing myocardium contributes to the HF pathophysiology. Preclinical evidence suggests that miR-152 inhibition preserves cardiac function in a model of pressure overload-induced HF. These findings offer new insights into the pathophysiology of HF and point to miR-152 as a potential novel therapeutic target.
INTRODUCTION
MicroRNAs (miRNAs) are small, 18-22 nucleotide, non-coding RNA molecules that regulate gene expression post-transcriptionally by inhibiting the translation of protein-coding mRNA transcripts1. Recent evidence suggests that miRNAs are key regulators of different cellular processes underlying cardiovascular physiology and pathophysiology2, and several miRNAs have been associated with pathological cardiac remodeling preceding heart failure (HF)3-5. However, our understanding of the contribution of miRNAs to HF phenotype and their mechanism of action in the heart remains incomplete. As individual miRNAs often regulate the expression of multiple genes, targeting of a single miRNA can, in principle, influence an entire gene network and potentially ameliorate complex disease phenotypes6. Hence, elucidating the function of miRNAs in the heart could enable the discovery of novel mechanisms of disease and potentially lead to the development of miRNA-mediated therapies to prevent or treat HF7,8.
In this study, we identified that miR-152 expression was significantly increased in failing human myocardium. The miR-152 family of miRNAs, which includes miR-148a and miR148b, has been implicated in processes such as immuno-modulation, cell growth, proliferation, and cancer.9 As the role of miR-152 within the cardiac myocyte remains poorly understood, we sought to determine whether the upregulation of miR-152 contributes to HF. We report that overexpression of miR-152 in cardiomyocytes induces cardiac remodeling, systolic dysfunction, and dilated cardiomyopathy in vivo, and alters the expression of genes involved in cardiomyocyte metabolism and mitochondrial homeostasis. Mechanistically, we identified the mitochondrial monothiol Glutaredoxin 5 (Glrx5), a critical regulator of mitochondrial iron and iron-sulfur cluster synthesis, as a direct miR-152 target. Additionally, Anti-miR-152 therapy using a locked nucleic acid (LNA)-based antagomir prevented the functional deterioration of systolic function in a mouse model of pressure overload-induced HF. In conclusion, our data reveal a molecular link between miR-152 and the HF phenotype, providing new insight into how miRNAs contribute to the complex pathophysiology of HF and suggesting that modulation of miR-152 levels may represent a new effective therapeutic target.
METHODS
The data, methods used in the analysis, and materials used to conduct the research are available to any researcher for purposes of reproducing the results or replicating the procedure upon request. Additional detailed experimental methods used in the study are presented in the Supplementary Methods in the Supplementary Information.
Animal Care.
Animals were handled in accordance with the Guidelines for the Care and Use of Laboratory Animals published by the National Institutes of Health. The Icahn School of Medicine at Mount Sinai Institutional Animal Care and Use Committee, and by the Stanford University Animal Care and Use Committee approved the procedures.
Generation of conditional-miR-152 transgenic animals.
Cardiac-specific miR-152 expression was achieved using a ‘gene switch’ strategy as previously described10. Briefly, the human miR-152 precursor was PCR amplified from genomic DNA and inserted into the EcoRV site of the pMHC-flox vector (a kind gift from Dr. Y Wang) to generate a conditional “floxed” miR-152 expression vector under the control of the cardiac myocyte-specific α-myosin heavy chain promoter (Myh6). Tg(Myh6-miR-152) transgenic mice were generated by intra-nuclear injection of the KpnI/SstI fragment isolated from the pMHC-flox-miR-152 vector into fertilized eggs from B6C3 mice, and transgenic integration was confirmed by PCR. Following mating to Tg(Myh6-cre/Esr1*) mice (Jackson Laboratories), Cre-mediated miR-152 expression in mice 8 weeks of age was induced by oral delivery of Tamoxifen (Sigma-Aldrich) for four consecutive days of the same dose (20 μg/g reconstituted in peanut oil). All analyses were performed on heterozygote male Tg(Myh6-cre/Esr1*);Tg(Myh6-miR-152) and Tg(Myh6-cre/Esr1*) control littermates that were treated with tamoxifen.
TAC model and LNA antagomir treatment.
Twelve week old C57BL/6 mice (Jackson Laboratories) were subjected to transverse aortic constriction (TAC) using microsurgical techniques as previously described11. Briefly, the chest cavity was entered via the second intercostal space at the left upper sternal border through a small incision, and aortic constriction was performed by tying a 7-0 nylon suture ligature against a 27-gauge needle between the innominate and left common carotid arteries. Antagomirs were synthesized as fully phosphorothiolated oligonucleotides with perfect sequence complementary to the mature mmu-miR-152-3p sequence (16mer) or a scrambled sequence (control). Cardiac function was assessed 2 weeks post-surgery using pulse waved doppler analysis, and treatment started at that time in mice with a significant, hemodynamic relevant, constriction (velocity > 3200mm/s). Mice were administered a single daily intraperitoneal injection of LNA-control or LNA-anti-miR-152 (20mg/kg/day) for 3 subsequent doses (day-1, day-3, and day-7). Cardiac function was assessed by echocardiography at 2- and 3-weeks post treatment (4 weeks and 5 weeks post-surgery), followed by tissue collection.
Echocardiography.
Short axis two-dimensional images and M-mode tracings were recorded at the level of the papillary muscle to determine fractional shortening and left ventricular (LV) dimensions as previously described12. In the transgenic animal model, echocardiography was performed using a GE Vision Ultrasound system at designated time points under sedation with ketamine (50-80 mg/kg) optimized to maintain a heart rate >450 beats per minute. For the TAC model and LNA antagomir treatment experiments, echocardiography (two-dimensional M-mode) was performed in anesthetized mice (1.8% isoflurane) utilizing a VisualSonics Vevo 2100 Ultrasound system. The operator was blinded to the experimental conditions in all measurements.
miRNA expression profiling.
An enriched miRNA fraction was isolated from the tissue specimens using the mirVana miRNA Isolation Kit according to the manufacturer’s protocol (Ambion). The miRNA fraction was labeled and hybridized onto custom miRNA microarrays (TGmiRV1; Affymetrix) as previously described13. All the arrays were processed on an Affymetrix Fluidics Station and scanned on an Affymetrix G7 scanner. Estimates of miRNA expression were extracted using the Partek Genomic Suite (Partek), and the signal from replicate probes were median polished and normalized using cyclic loess. The resulting data were then analyzed for differential expression with SAM using its default parameters14,15, and miRNA genes with a false discovery rate <5% were considered differentially expressed.
Statistics.
All statistical analyses were performed using GraphPad Prism Software (San Diego, CA). The underlying assumption of normal distribution was investigated by performing a Kolmogorov–Smirnoff normality and normal probability plot test. Unpaired two-tailed Student’s t tests were used to determine the significance between two groups, assuming significance at P < 0.05. Analyses between multiple groups were performed using one-way ANOVA following Tukey multiple comparison post-test to determine significance. Echocardiography and PV loop measurements were analyzed by 2-way ANOVA or by 2-way ANOVA repeated measures test when the same animal was used over the course of the experiments. P < 0.05 was considered statistically significant.
RESULTS
MiR-152 gene expression levels are increased in human and experimental models of HF
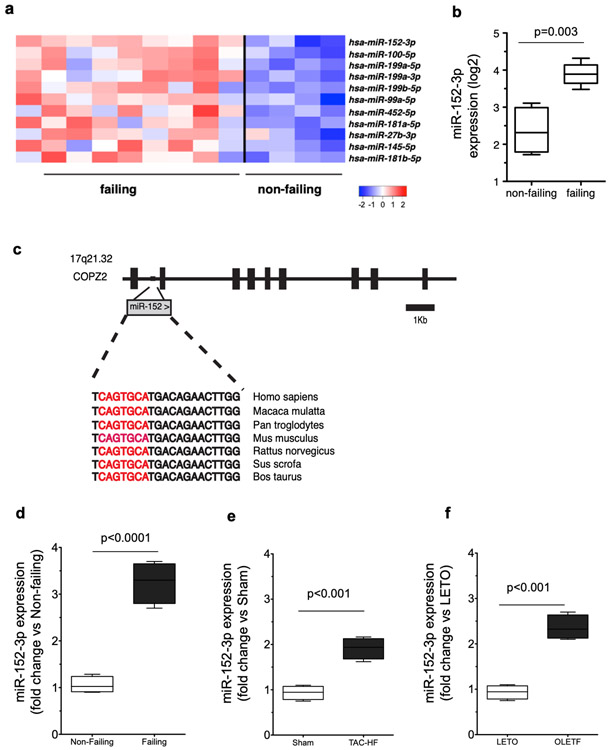
We conducted expression profiling of 462 known human miRNAs in LV tissue explanted from patients with end-stage HF (NYHA classification IV; n=9) and from control non-failing patients (n=4). We identified 11 miRNAs that were significantly upregulated in failing hearts, including a number of previously described miRNAs that have been associated with HF, such as miR-14516, miR-181b16, miR-10017, miR-199b18, and miR-199a19 (Figure 1a). Interestingly, miR-152-3p (hereafter referred to as miR-152) was the miRNA most increased in expression (4-fold, q < 0.05) in failing compared to non-failing samples (Figure 1b). MiR-152 is a member of the broadly conserved miR-148/152 family that also includes miR-148a and miR-148b and that is highly expressed in the mouse20 and human heart compared to other organs, such as brain, kidney and testis21. However, the role of miR-152 in the myocardium is currently unknown. The human miR-152 resides between exons 1 and 2 of the Coatomer Protein Complex Subunit Zeta-2 gene (COPZ2) and is evolutionarily conserved in vertebrates (Figure 1c). By quantitative real-time PCR we confirmed that the expression of miR-152 was significantly increased in the failing human (3.25-fold) when compared to non-failing LV heart tissue (Figure 1d). Additionally, the expression of miR-152 in rodent LV tissue was markedly increased in both pressure overload-induced (1.91-fold) and diabetes-induced (2.25-fold) models of HF when compared to non-failing controls (Figures 1e and 1f, respectively). These data were consistent with previous studies showing that miR-152 was upregulated in the later stages of the pressure overload-induced HF mouse model20, in rat models of catecholamine-induced cardiac stress22, and in the cardiac Gαq-overexpressing mice, a genetic model of cardiomyopathy that recapitulates seminal aspects of pressure-overload hypertrophy23. Together, these data demonstrate that miR-152 is significantly upregulated in both human end-stage heart disease and in experimental animal models of HF, suggesting a potential role for miR-152 in cardiac pathophysiology and HF.
Figure 1. miR-152 expression levels are increased in HF.
a) Hierarchical clustering of the dysregulated miRNAs in end-stage human HF. b) miR-152 was the top ranked miRNA that was significantly upregulated in human non-failing (n=4) versus failing hearts (n=9) (q < 0.05). c) Schematic representation of the structure of the human COPZ2 gene containing the evolutionarily conserved miR-152 gene embedded in intron 1. d) Real-time PCR expression analysis of miR-152 expression in human non-failing versus failing LV tissue samples (n=4 per group). e) Real-time PCR expression analysis of miR-152 gene expression in a mouse model of heart failure induced by trans-aortic constriction (TAC). Mean ± se TAC vs. Sham-operated animals (n=6 per group). f) Real-time PCR expression analysis miR-152 gene expression in a rat model of diabetes-induced heart failure. OLETF vs. LETO animals (n=4 per group). Box-and-whisker plots show the minimum, the 25th percentile, the median, the 75th percentile, and the maximum.*P < 0.001.
Cardiac myocyte-specific miR-152 overexpression in the adult mouse heart induces systolic dysfunction
To examine the potential role of miR152 in the myocardium we performed gain-of-function analyses of miR-152 in vitro. We observed that adenoviral-mediated miR-152 overexpression in adult rat cardiomyocytes causes a significant decrease in sarcomere shortening and aberrant intracellular Ca2+ cycling, such as elevated diastolic intracellular Ca2+ and slower relaxation kinetics in vitro (Supplementary Figure 1).
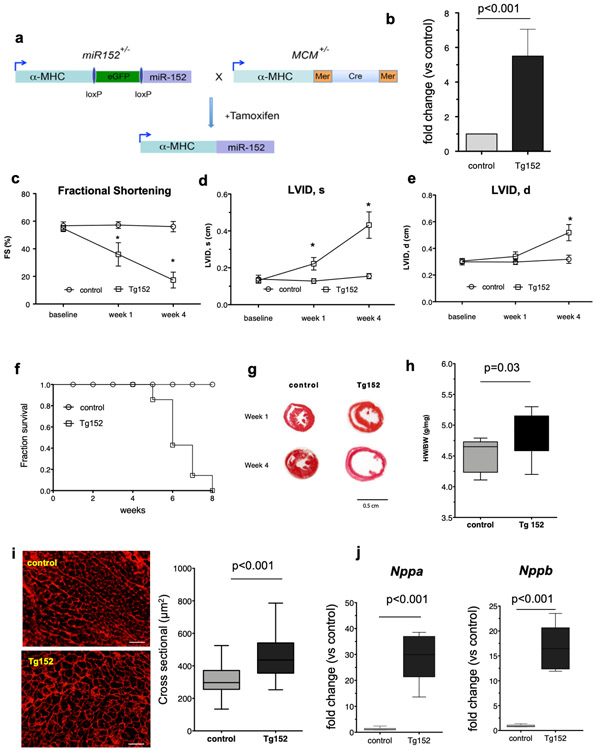
To further investigate the role of miR-152 in vivo, we engineering an inducible cardiac myocyte-specific miR-152 overexpression mouse model, using a gene-switch transgenic strategy10 (Figure 2a). We generated transgenic mice harboring a loxP-flanked genomic fragment of the miR-152 precursor expressed under the control of the α-myosin heavy chain promoter (Myh6). These mice were bred with Tg(Myh6-cre/Esr1*) mice harboring another Myh6 transgene expressing the “MerCreMer” tamoxifen-inducible estrogen receptor-cre recombinase fusion protein24. Upon tamoxifen administration to doubly transgenic mice (hereafter referred to as Tg152), cardiac miR-152 expression was significantly increased (5.5 – fold, P < 0.001) when compared to sex- and age-matched tamoxifen-treated Tg(Myh6-cre/Esr1*) littermate controls (hereafter referred to as Control; Figure 2b).
Figure 2. Forced miR-152 expression in cadiomyocytes induces cardiac dilatation and overt cardiac dysfunction in mice.
a) Schematic diagram of the Cre-LoxP–mediated gene-switch strategy to establish transgenic animals with temporally regulated gene overexpression of miR-152 in the myocardium. Tamoxifen-induced gene switch in the hearts of double transgenic animals carrying both miR-152 and MCM alleles. MCM: MerCreMer; a-MHC: α-myosin heavy chain. b) Real-time PCR expression analysis of miR-152 gene expression in the myocardium of bi-trangenic mice (Tg152) and littermate MCM control mice (n=4 per group). Comparison by paired Student’s t-test. c-e) Echocardiography analysis of cardiac function and dimensions of Tg152 mice and MCM control littermates prior to and at 1- and 4-weeks post tamoxifen treatment (n=6 per group at each time point). Data represent mean *P < 0.001 by 2-way ANOVA repeated measures test. f) Survival curve of Tg152 and control animals following tamoxifen administration. g) Representative Hematoxylin-Eosin staining of heart sections at 1 week and 4 weeks post tamoxifen treatment. h) Heart-weight-to-body weight ratios (n=9 animals per group). i) Assessment of cardiomyocyte diameters by wheat germ agglutinin staining (n=250 cardiomyocytes per group). Scale bar = 100 μm. j) Real-time PCR expression analysis of Nppb and Nppa genes in the myocardium of bi-trangenic mice (Tg152) and littermate MCM control mice (n=6 per group). All Box-and-whisker plots show the minimum, the 25th percentile, the median, the 75th percentile, and the maximum. FS=Fractional Shortening; LVID,s=Left ventricular internal diameter at end systole; LVID,d = Left ventricular internal diameter at end diastole.
By echocardiography, induction of miR-152 expression was observed to induce a rapidly progressive dilated cardiomyopathy characterized by a significant decrease in the fractional shortening and a significant increase in the LV dimensions (Figure 2c-e and Supplementary Table 1) by 4 weeks post tamoxifen (FS mean difference −38.74%, 95% CI: −45.73% to −31.74%; LVIDd mean difference 0.20, 95% CI: 0.15 to 0.25; LVIDs mean difference 0.28, 95% CI: 0.23 to 0.33). Hemodynamic analysis by LV catheterization in vivo confirmed the LV dilatation and reduced contractile function associated with miR-152 overexpression (Supplementary Figure 2a-b). Importantly, end-systolic pressure-volume relationship (ESPVR) and pre-load recruitable stroke work (PRSW), load-independent parameters for systolic function, were significantly decreased in Tg152 mice compared to controls (ESPVR mean difference −4.37, 95% CI: −6.62 to – 2.11; PRSW mean difference 43.46, 95% CI: −73.31 to −13.61) at 4 weeks post transgene induction (Supplementary Figure 2c and Supplementary Table 2). Consequently, the Tg152 mice died from HF within 8 weeks of tamoxifen treatment (Figure 2f). Consistent with the observed cardiac remodeling in vivo, post-mortem analysis at 1-week post tamoxifen treatment showed evidence of cardiac hypertrophy by gross histology (Figure 2g), gravimetric analysis (increased heart to body weight ratio; Figure 2h), and histologic assay of LV cardiomyocyte cross-sectional area (Figure 2i). Furthermore, we observed a significant increase in mRNA expression for brain natriuretic peptide (Nppb) and atrial natriuretic peptide (Nppa), two biomarkers for stressed myocardium25, in Tg152 mice when compared to controls at one week post-tamoxifen administration (Figure 2j).
Notably, the miR-152-induced systolic dysfunction was not accompanied by any increase in myocardial fibrosis or apoptosis (Supplementary Figure 3b and 3d, respectively). In addition, in contrast to what is often found in ischemic heart disease, the protein expression of sarcoplasmic reticulum calcium (Ca2+) ATPase 2a (Serca2a) and phospholamban (Pln), two critical determinants of active relaxation in diastole26, was unaltered in Tg152 mice (Supplementary Figure 4a-c). Instead, there was a small but significant increase in the phosphorylation of Pln Ser-16, consistent with a compensatory increase in Serca2a activity in response to contractile impairment. Finally, the expression and activation levels of the ryanodine receptors (Ryr2) were similar between control and Tg152 hearts (Supplementary Figure 4d). Together, these findings suggested that diastolic function was not impaired by miR-152 expression. In agreement, the LV end-diastolic pressure-volume relationship (EDPVR), an index of diastolic LV compliance, was similar in Tg152 and control mice (Supplementary Table 2). Taken together, these findings indicate that cardiac myocyte-specific miR-152 expression induces dilated cardiomyopathy with early mortality, characterized by progressive systolic function independent of any defect in diastolic function.
Global mRNA Transcriptional Changes in the Tg152 hearts
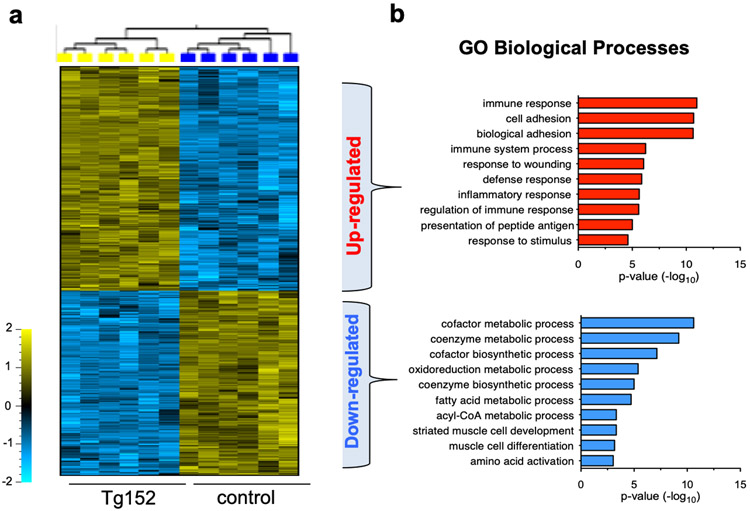
To elucidate the underlying molecular mechanisms that underlie the development systolic dysfunction induced by miR-152 overexpression, we performed genome-wide transcriptional profiling of the Tg152 hearts to reveal the steady-state transcript levels27. We identified 2277 transcripts that were differentially expressed (1370 up- and 907 down-regulated; q < 0.05) in Tg152 versus control hearts within 1 week of miR-152 overexpression (Figure 3a). The expression of a subset of these genes was validated by real-time quantitative PCR (Supplementary Figure 5). Gene Ontology (GO) analysis28 of mRNAs increased in expression showed a significant enrichment for immune responses (P = 1.3 x 10−29) (Figure 3b), including multiple mRNAs that encode toll like receptor 2 and 4 (Tlr2 and Tlr4), pro–inflammatory cytokines such as Interleukin 18 (Il18), and the tumor necrosis factor superfamily cytokine receptors (Tnfrsf1a and Tnfrsf1b), indicating a prevalence of immune system modulation is in the setting of the Tg152 heart29,30. The downregulated genes were enriched for metabolic processes (Figure 3b) including genes involved in fatty acid (FA) transport capacity (Cpt2, Slc25a20) and oxidation (Acadm, Acadv, Acot1, Acot2, Hahb). These changes are consistent with the downregulation of FA enzymes and the shift in metabolism and the activation of the innate immune system that triggers inflammatory responses in the failing heart30-34. Together, these data indicate that the immune system response and changes in metabolic pathways are potential contributors to the pathophysiological state associated with miR-152 overexpression.
Figure 3. Forced expression of miR152 induces global mRNA changes in the heart.
a) Heatmap of the top 500 differentially expressed genes between Tg152 and control hearts at 1-week post tamoxifen treatment. b) Top 10 GO biological processes enrichment analysis of the upregulated genes (upper panel) and the top 10 GO biological processes enrichment analysis of the downregulated genes (lower panel).
Glrx5 is a direct miR-152 target
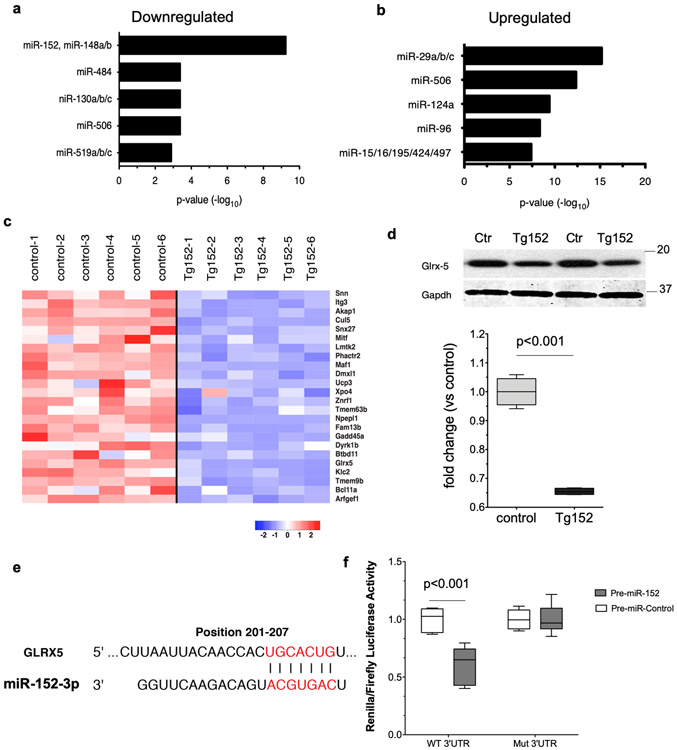
Changes in LV mRNA levels induced by miR-152 overexpression might be due to indirect mechanisms or driven by a direct interaction of miR-152 with target transcripts. To identify mRNAs altered in steady-state expression that might be direct miRNA targets, we analyzed the 3’ untranslated regions (3’-UTRs) of the mRNAs for the presence of miRNA recognition sequences. The miR-152 seed sequence CAGTGCA corresponding to nucleotides 2–7 of miR-15235,36 was the most significantly overrepresented motif (log P = 10−11) among the downregulated gene set (Figure 4a). This finding is consistent with the expected inverse correlation of miRNA expression and the expression of direct gene targets. In contrast, the up-regulated genes showed a significant enrichment for miRNA target motifs, such as miR-29 and miR-506 (P = 10−15 and 10−12, respectively; Figure 4b).
Figure 4. The mitochondrial Glrx5 gene is a direct miR-152 target.
a-b) Enrichment analysis of predicted miR152 ‘seed sequence’ motifs in the downregulated gene pool and upregulated gene pools. c) mRNA expression levels of 23 putative miR-152 targets represented as a heatmap. d) Representative Western blot analysis of miR-152 putative target Glrx-5 and densitometry analysis of the protein expression levels. Comparison by upaired student’s t-test; n=4 per group. Gapdh was used as a loading control. Numbers indicate the molecular weight marker size (kDa). e) Sequence complementarity between miR152 ‘seed sequence’ and Glrx5 3’-UTR. f) Pre-miR-152 mimic or pre-miR-control mimics were co-transfected with Luciferase reporter vector carrying the Glrx5 3′-UTR or a vector without the predicted binding site of miR-152. n=3 independent experiments. Comparison by 2-way ANOVA. All box-and-whisker plots show the minimum, the 25th percentile, the median, the 75th percentile, and the maximum.
Computational prediction of targets of miR-152 identified a putative miR-152 ‘seed sequence’ in the 3’-UTR of twenty-three mRNAs in the downregulated gene pool (Figure 4c). Among these putative mRNA-152 target genes, Glutaredoxin 5 (Glrx5) is one of the top predicted miR-152 target genes by Targetscan, a computational algorithm for miRNA target prediction37. The Glrx5 gene encodes monothiol glutaredoxin, an evolutionarily conserved protein that is essential for the biogenesis of iron-sulfur (Fe-S) clusters and regulates mitochondrial iron homoeostasis and function38,39. In line with the diminished expression at the mRNA level, Glrx5 protein levels were significantly repressed in Tg152 hearts (Figures 4d). To experimentally validate the link between Glrx5 mRNA repression and miR-152, the Glrx5 3’UTR harboring the evolutionarily conserved miR-152 ‘seed sequence’ (Figure 4e) was inserted into a luciferase reporter plasmid. We observed that the luciferase reporter activity was significantly inhibited by co-expression of a synthetic pre-miR-152 when compared to a control pre-miR mimic. In contrast, the luciferase activity of a control reporter construct lacking the miR-152 target 3-UTR motif was unaffected by expression of the pre-miR-152 mimic (Figure 4f).
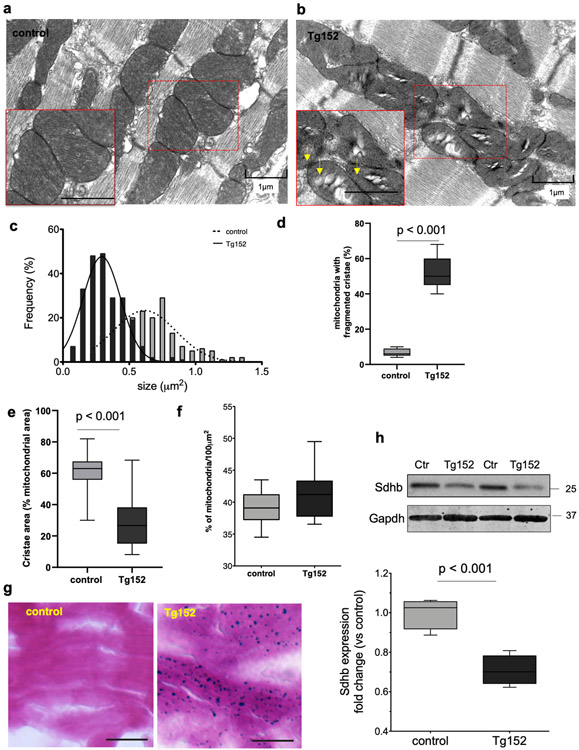
As Glrx5 is a critical regulator of the maturation of all cellular Fe/S proteins, cellular iron regulation and mitochondria homeostasis40, we assessed the mitochondria abundance and ultrastructure in Tg152 cardiomyocytes by electron microscopy. We observed that Tg152 cardiomyocytes displayed randomly distributed intermyofibrillar mitochondria characterized by reduced mitochondrial matrix electrodensity, external membrane disorganization, (Figure 5a-b). Furthermore, morphometric analysis of electron micrographs showed a significant decrease in cardiomyocyte mitochondria size in Tg152 compared to control hearts (Figure 5c), disrupted internal cristae with reduced density (Figure 5d-e), while the total mitochondria content was similar between control and Tg152 cardiomyocytes (Figure 5f). Importantly, Tg152 cardiomyocytes contained electron dense granules suggestive of intra-mitochondrial iron deposits, which were also evident by histological iron staining (Figure 5g). In line with these findings, Tg152 hearts showed a significant reduction in the expression levels of succinate dehydrogenase subunit (Sdhb), a Fe-S cluster-containing component of the mitochondrial respiratory chain complex II (Figure 5h). Together, these data demonstrate that Glrx5 is a direct target of miR-152, and the reduced expression of Glrx5 correlates with aberrations in cardiomyocyte mitochondrial morphology, Fe-S cluster deficit and iron accumulation in the Tg152 hearts.
Figure 5. Mitochondria abnormalities in Tg152 hearts.
a-b) Representative electron microscopy images of longitudinal LV tissue sections showing the myofilament and mitochondria ultrastructures in control and Tg152 cardiomyocytes. Arrows indicate electron dense particles in mitochondria. (n=3 animals per group). c) Morphometric analysis of mitochondrial size from the electron microscopy images (n=225 mitochondria, 3 hearts per group). d-e) Morphometric analysis of mitochondrial cristae quality and density in Tg152 and control cardiomyocytes. f) Quantification of mitochondria content in control and Tg152 cardiomyocytes expressed as percentage (%) of mitochondria occupied surface area per 100μm2. Comparison by unpaired student’s t-test; n=3 animals per group. g) Histological assessment of iron accumulation in heart LV cryosections in control and Tg152 by Prussian blue staining. Scale bar = 25 μm (n=3 animals in each group). h) Representative Western blot analysis of Sdhb and densitometry analysis of the protein expression levels. Comparison by unpaired Student’s t-test; n=4 animals per group. Gapdh was used as a loading control. Number indicate the molecular weight marker size (kDa). Box-and-whisker plots show the minimum, the 25th percentile, the median, the 75th percentile, and the maximum.
Silencing of miR-152 preserves systolic function in pressure overload-induced HF
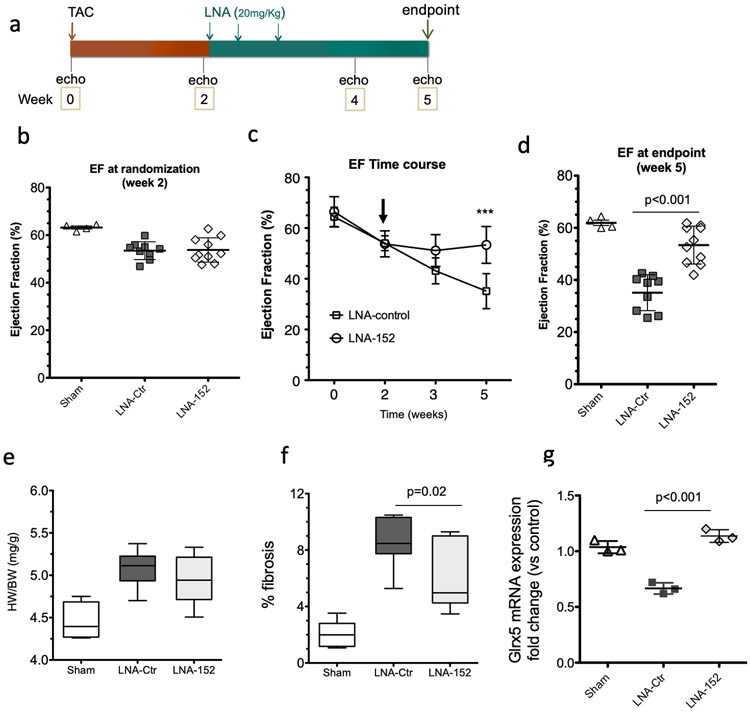
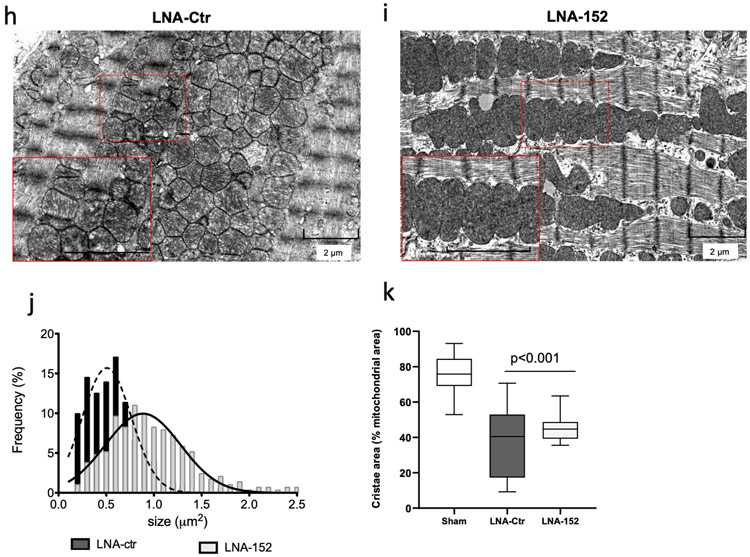
Anti-sense oligonucleotides that inhibit the expression of miRNAs are currently under development as therapeutic modalities for a diverse set of diseases associated with miRNA gain-of-function. We next considered whether pharmacological inhibition of miR-152 in the failing heart could have a beneficial effect in mitigating ventricular dysfunction. An LNA-based antimir (LNA-152) was synthesized41,42 to be complementary to nucleotides 2-16 of the mature miR-152 sequence (Supplementary Figure 6a). In control experiments, LNA-152 suppressed the expression of a Renilla luciferase-based miR-152 reporter vector in a dose response manner, while the control (LNA-control) had no effect on the reporter activity (Supplementary Figure 6b-c). To evaluate the therapeutic potential of silencing miR-152 in vivo, adult mice were subjected to chronic pressure overload by TAC surgery, a model for HF with reduced ejection fraction (EF). Consistent we previous studies, we observed a significant upregulation of miR-152 expression (Figure 1e). After two weeks of pressure overload, mice were randomly assigned to receive either LNA-152 or LNA-control for three consecutive days of therapy to test whether inhibition of miR-152 might suppress the deterioration of cardiac dysfunction associated with the long-term pressure overload (Figure 6a). At randomization, the LV ejection fractions (EF) were similar between controls and test groups (Figure 6b and Supplementary Table 3). At three weeks post treatment, mice treated with LNA-152 demonstrated preserved systolic function when compared to LNA control-treated animals as assessed by EF (mean difference −18.25%, 95% CI: −24.89% to −11.61%) (Figure 6c-d). The LNA-152 treatment also prevented ventricular dilatation as evident by the preservation of LV diameter at systole in LNA-152-treated mice (mean difference 0.65, 95 % CI: 0.24 to 1.07) (Supplementary Table 3). In contrast, treatment with LNA-152 did not attenuate cardiac hypertrophy at endpoint (heart weight-to-body-weight ratio) (Figure 6e). However, we observed reduced interstitial myocardial fibrosis in the LNA-152 treated hearts (Figure 6f). Consistent with the known role of miRNAs in tuning gene dosage by lowering target mRNA levels, the mRNA expression levels of Glrx5 were significantly increased in LNA-152 compared to LNA-control treated animals (Figure 6i). Finally, LNA-152-treated mice exhibited preserved mitochondria ultrastructure and significantly larger mitochondria with higher cristae density when compared to LNA-control animals as assessed by electron microscopy (Figure 6h-k). . Taken together, these data revealed that LNA-mediated silencing of miR-152 in vivo can attenuate the pathological cardiac remodeling and systolic dysfunction induced by chronic pressure overload in the mouse.
Figure 6. LNA-mediated inhibition of miR-152 in a TAC model of HF.
a) Schematic representation of the anti-miR-152 therapy protocol. At 2 weeks post TAC operation, mice were dosed at 20mg/Kg every three days for 1 week by intraperitoneal injections of the anti-miR oligonucleotides. b) Echocardiographic analysis of cardiac ejection fraction (EF) in TAC operated mice at randomization before receiving either anti-miR152 (LNA-152, n = 9) or control (LNA-control, n = 10). Sham operated animals (Sham, n=4) are shown for comparison. c) Time course assessment of cardiac EF. Box-and-whisker plots show the minimum, the 25th percentile, the median, the 75th percentile, and the maximum. ***P < 0.001 LNA-152 vs LNA control by 2-way ANOVA repeated measures. Arrow indicates the time of LNA administration. Sham, n =4; LNA-152, n = 9; LNA-control, n = 10. d) Cardiac EF at the endpoint. Values represent mean ± sd. Comparison by 1-way Anova. Sham, n =4; LNA-152, n = 9; LNA-control, n = 10. e) Heart weight-to-body weight ratios. Sham, n=4; LNA-152, n = 9; LNA-control, n = 10. Box-and-whisker plots show the minimum, the 25th percentile, the median, the 75th percentile, and the maximum. f) Assessment of cardiac fibrosis at the endpoint. Box-and-whisker plots show the minimum, the 25th percentile, the median, the 75th percentile, and the maximum. n=3 hearts per group. Comparison by one-way Anova: *P < 0.05 LNA-ctr vs LNA-152. g) Expression levels of Glrx5 at the endpoint. Comparison by 1-way ANOVA. ***P < 0.001 LNA-ctr vs LNA-152. n=3 hearts per group h-i) Representative electron microscopy images of mitochondria ultrastructures in treated animals. Inset scale bar = 2 μm. j-k) Morphometric analysis of mitochondrial size and cristae density at the endpoint. n=300 mitochondria from 2 hearts per group.
DISCUSSION
HF with reduced LV EF is highly prevalent and is associated with a 50% mortality in the 5 years after diagnosis, despite current therapy. This study implicates miR-152 as an important regulator of cardiac systolic function and a potential therapeutic target for HF with reduced EF that is associated with common diseases, such as ischemic heart disease and hypertension, and dilated cardiomyopathy. We determined that the miR-152 expression levels were increased in human and experimental animal models of HF. Upon overexpression of miR-152 in the murine myocardium at levels similar to those observed in human HF, the transgenic animals developed fulminant cardiomyopathy characterized by a systolic dysfunction and ventricular dilatation. Conversely, therapeutic miR-152 silencing attenuated the progression of cardiac dysfunction in a mouse model of pressure overload-induced HF in vivo.
The deleterious effects of miR-152 overexpression were closely associated with disturbances of metabolic gene networks. This is consistent with the cardiac adaptation in HF away from FA metabolism43. Similar to the Tg152 hearts, the failing heart is characterized by the downregulation of mitochondria FA oxidation enzymes and a decrease capacity of the heart for FA oxidation33,43,44. Studies in transgenic mouse models strongly support that the reduction in the capacity to oxidize FAs in the myocardium has deleterious effects possibly explaining the early onset of the pathological remodeling and cardiac dysfunction in the Tg152 mice45. Given the diverse effects of miRNAs in post-transcriptional gene regulation, we theorize that miR-152 acts as a pleiotropic modulator of lipid metabolism and metabolic processes in the heart.
Mechanistically, we have identified a direct target of miR-152, Glrx5, an evolutionary conserved protein localized in the mitochondria that is essential for Fe-S cluster biosynthesis and mitochondrial iron homeostasis38,39. We observed that the downregulation of Glrx5 correlates with alterations in the cardiomyocyte mitochondria, such as fragmentation, cristae remodeling, and iron overload. Consistent with our observations in the Tg152 hearts, the loss-of-function mutation of Glrx5 leads to mitochondrial iron overload in human fibroblasts46. Although there is a paucity of information regarding the role of Glrx5 in cardiovascular function, Fe-S protein biogenesis in mitochondria is a conserved and essential process of oxidative phosphorylation. It is plausible that the miR-152 plays an important role in mitochondrial iron homeostasis in the heart by modulating Glrx5. Since mitochondrial iron overload appears to be caused by a failure of Fe–S cluster biogenesis, it is likely that mitochondrial iron overload further impairs mitochondria structure and function by oxidatively damaging mitochondria, further contributing to the phenotype of the Tg152 mice. In support of this hypothesis, loss-of-function mutations in the frataxin gene (FXN), both in animal models47 and in patients diagnosed with Freidreich’s Ataxia (FRDA)48, disrupt the Fe-S complex biosynthesis, leading to iron overload in cardiac mitochondria, impairment of bioenergetics and HF. Considering the fundamental role of iron metabolism on cardiac energetics, these results suggest that modulation of Glrx5 levels by miR-152 may impact signaling networks playing an important role in mitochondrial homeostasis. However, we have identified that mir-152 interferes with multiple genes, although their specific contribution to HF pathogenesis remains to be elucidated. Recent studies suggest that miR-152 targets phosphatase and tensin homolog (PTEN) in the rat myoblast cell line H9C2 in vitro and hepatocytes in vivo49,50. However, we did not observe any significant changes in PTEN expression in our Tg152 transgenic animal model, suggesting that miRNA-target interactions are regulated in a context-dependent and cell-type-specific manner. The cellular context of where the miRNA of interest performs its physiological or pathological functions should be considered when establishing the repertoire of the miRNA target genes.
Finally, our proof-of-concept studies in a murine model of HF suggests that pharmacological silencing of miR-152 activity may protect against pathological cardiac remodeling and improve systolic function in patients with HF. LNA biochemistry enables the design of truncated LNA-modified anti-miRs, leading to high affinities toward their cognate miRNA targets in the heart in vivo51-54. Thus, pharmacological silencing of miR-152 activity has the potential for clinical translation. However, major challenges remain for development of oligonucleotide-based therapeutics, including bioavailability, specificity, and delivery. Future therapeutic approaches may include adeno-associated virus-based (AAV) anti-miR gene delivery engineered to enhance transduction specificity to cardiac myocytes within patients.
In summary, our findings show that miR-152 contributes to the heart failure phenotype, highlighting the pivotal roles played by miRNAs in mediating changes in gene expression and cardiomyocyte function. We demonstrated that pharmacological silencing of miR-152 attenuated the progression of systolic dysfunction in vivo, suggesting that modulation of miR-152 may represent a novel therapeutic approach for HF, and warrants further investigation of the role of miR-152 in HF pathophysiology.
Supplementary Material
WHAT IS NEW?
MicroRNA-152 (Mir-152) is a novel micro-RNA upregulated in human heart failure.
Cardiac myocyte specific overexpression of miR-152 in the murine heart leads to systolic dysfunction and dilated cardiomyopathy
Pharmacological inhibition of miR-152 utilizing a locked nucleic acid (LNA) improves cardiac function in a murine model pressure overload-induced of heart failure
We identified a new direct target of miR-152, Glutaredoxin-5 (GLRX-5), a mitochondrial iron-handling protein essential for iron-sulfur cluster synthesis and iron homeostasis.
WHAT ARE THE CLINICAL IMPLICATIONS?
Elucidating the role of miRNAs in heart function could offer a better understanding of the pathophysiological mechanisms that contribute the heart failure phenotype, and possibly the development of new diagnostic and therapeutic tools.
We provide proof-of-concept that modulation of miR-152 expression in the setting of pre-existing cardiac dysfunction could be a promising therapeutic strategy for heart failure
Acknowledgments
We acknowledge the support of Dr. Andrew Feber and Dr. Tony Godfrey for providing the miRNA arrays and assisting with the miRNA profiling assays. We also sincerely thank Dr. Michael Kapiloff for critical reading of the manuscript, and Amanda Chase for assistance in preparing the manuscript. We are grateful to Dr. Roger Hajjar and Dr. Djamel Lebeche for their support.
Sources of Funding
Funding support was provided by National Institutes of Health (NIH) R00 HL104002 (to I.K.), a German Research Foundation fellowship ( to T.S.), a fellowship from the Spanish Economy and Competitiveness Ministry (to I.P-G), and in part, by the ARRA Award Number 1S10RR026780-01 from the National Center for Research Resources (NCRR), and the Division of Critical Care Medicine at Lucile Packard Children’s Hospital (T.J.L).
Non-standard Abbreviations and Acronyms
- MerCreMer
tamoxifen-inducible Cre recombinase
- MHC
a-myosin heavy chain
- TAC
transverse aortic constriction
- LNA
locked nucleic acid
Footnotes
Conflict of Interest Disclosures
The authors declare no competing financial interests.
REFERENCES
- 1.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mrnas are conserved targets of micrornas. Genome Res. 2009;19:92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Verjans R, Derks WJA, Korn K, Sonnichsen B, van Leeuwen REW, Schroen B, van Bilsen M, Heymans S. Functional screening identifies micrornas as multi-cellular regulators of heart failure. Sci Rep. 2019;9:6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Small EM, Olson EN. Pervasive roles of micrornas in cardiovascular biology. Nature. 2011;469:336–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thum T, Condorelli G. Long noncoding rnas and micrornas in cardiovascular pathophysiology. Circ Res. 2015;116:751–762 [DOI] [PubMed] [Google Scholar]
- 5.Boon RA, Dimmeler S. Micrornas in myocardial infarction. Nat Rev Cardiol. 2015;12:135–142 [DOI] [PubMed] [Google Scholar]
- 6.Ebert MS, Sharp PA. Roles for micrornas in conferring robustness to biological processes. Cell. 2012;149:515–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lucas T, Bonauer A, Dimmeler S. Rna therapeutics in cardiovascular disease. Circ Res. 2018;123:205–220 [DOI] [PubMed] [Google Scholar]
- 8.Vegter EL, van der Meer P, de Windt LJ, Pinto YM, Voors AA. Micrornas in heart failure: From biomarker to target for therapy. Eur J Heart Fail. 2016;18:457–468 [DOI] [PubMed] [Google Scholar]
- 9.Liu X, Li J, Qin F, Dai S. Mir-152 as a tumor suppressor microrna: Target recognition and regulation in cancer. Oncol Lett. 2016;11:3911–3916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liao P, Georgakopoulos D, Kovacs A, Zheng M, Lerner D, Pu H, Saffitz J, Chien K, Xiao RP, Kass DA, Wang Y. The in vivo role of p38 map kinases in cardiac remodeling and restrictive cardiomyopathy. Proc Natl Acad Sci U S A. 2001;98:12283–12288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rockman HA, Ross RS, Harris AN, Knowlton KU, Steinhelper ME, Field LJ, Ross J Jr., Chien KR. Segregation of atrial-specific and inducible expression of an atrial natriuretic factor transgene in an in vivo murine model of cardiac hypertrophy. Proc Natl Acad Sci U S A. 1991;88:8277–8281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larocca TJ, Jeong D, Kohlbrenner E, Lee A, Chen J, Hajjar RJ, Tarzami ST. Cxcr4 gene transfer prevents pressure overload induced heart failure. J Mol Cell Cardiol. 2012;53:223–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feber A, Xi L, Pennathur A, Gooding WE, Bandla S, Wu M, Luketich JD, Godfrey TE, Litle VR. Microrna prognostic signature for nodal metastases and survival in esophageal adenocarcinoma. Ann Thorac Surg. 2011;91:1523–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tibshirani R, Chu G, Narasimhan B, Li J. Significance analysis of microarrays. R package version 2.0 http://CRAN.R-project.org/package=samr. 2011 [Google Scholar]
- 16.Naga Prasad SV, Duan ZH, Gupta MK, Surampudi VS, Volinia S, Calin GA, Liu CG, Kotwal A, Moravec CS, Starling RC, Perez DM, Sen S, Wu Q, Plow EF, Croce CM, Karnik S. Unique microrna profile in end-stage heart failure indicates alterations in specific cardiovascular signaling networks. J Biol Chem. 2009;284:27487–27499 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Sucharov C, Bristow MR, Port JD. Mirna expression in the failing human heart: Functional correlates. J Mol Cell Cardiol. 2008;45:185–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.da Costa Martins PA, Salic K, Gladka MM, Armand AS, Leptidis S, el Azzouzi H, Hansen A, Coenen-de Roo CJ, Bierhuizen MF, van der Nagel R, van Kuik J, de Weger R, de Bruin A, Condorelli G, Arbones ML, Eschenhagen T, De Windt LJ. Microrna-199b targets the nuclear kinase dyrk1a in an auto-amplification loop promoting calcineurin/nfat signalling. Nat Cell Biol. 2010;12:1220–1227 [DOI] [PubMed] [Google Scholar]
- 19.van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-responsive micrornas that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A. 2006;103:18255–18260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng Y, Ji R, Yue J, Yang J, Liu X, Chen H, Dean DB, Zhang C. Micrornas are aberrantly expressed in hypertrophic heart: Do they play a role in cardiac hypertrophy? Am J Pathol. 2007;170:1831–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meunier J, Lemoine F, Soumillon M, Liechti A, Weier M, Guschanski K, Hu H, Khaitovich P, Kaessmann H. Birth and expression evolution of mammalian microrna genes. Genome Res. 2013;23:34–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ali T, Mushtaq I, Maryam S, Farhan A, Saba K, Jan MI, Sultan A, Anees M, Duygu B, Hamera S, Tabassum S, Javed Q, da Costa Martins PA, Murtaza I. Interplay of n acetyl cysteine and melatonin in regulating oxidative stress-induced cardiac hypertrophic factors and micrornas. Arch Biochem Biophys. 2018;661:56–65 [DOI] [PubMed] [Google Scholar]
- 23.Matkovich SJ, Hu Y, Eschenbacher WH, Dorn LE, Dorn GW, 2nd. Direct and indirect involvement of microrna-499 in clinical and experimental cardiomyopathy. Circ Res. 2012;111:521–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sohal DS, Nghiem M, Crackower MA, Witt SA, Kimball TR, Tymitz KM, Penninger JM, Molkentin JD. Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible cre protein. Circ Res. 2001;89:20–25 [DOI] [PubMed] [Google Scholar]
- 25.Bernardo BC, Weeks KL, Pretorius L, McMullen JR. Molecular distinction between physiological and pathological cardiac hypertrophy: Experimental findings and therapeutic strategies. Pharmacol Ther. 2010;128:191–227 [DOI] [PubMed] [Google Scholar]
- 26.Bers DM. Cardiac excitation-contraction coupling. Nature. 2002;415:198–205 [DOI] [PubMed] [Google Scholar]
- 27.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian micrornas predominantly act to decrease target mrna levels. Nature. 2010;466:835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using david bioinformatics resources. Nat Protoc. 2009;4:44–57 [DOI] [PubMed] [Google Scholar]
- 29.Epelman S, Liu PP, Mann DL. Role of innate and adaptive immune mechanisms in cardiac injury and repair. Nat Rev Immunol. 2015;15:117–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mann DL. Innate immunity and the failing heart: The cytokine hypothesis revisited. Circ Res. 2015;116:1254–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doenst T, Nguyen TD, Abel ED. Cardiac metabolism in heart failure: Implications beyond atp production. Circ Res. 2013;113:709–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lopaschuk GD, Ussher JR, Folmes CD, Jaswal JS, Stanley WC. Myocardial fatty acid metabolism in health and disease. Physiol Rev. 2010;90:207–258 [DOI] [PubMed] [Google Scholar]
- 33.Sack MN, Rader TA, Park S, Bastin J, McCune SA, Kelly DP. Fatty acid oxidation enzyme gene expression is downregulated in the failing heart. Circulation. 1996;94:2837–2842 [DOI] [PubMed] [Google Scholar]
- 34.Stanley WC, Recchia FA, Lopaschuk GD. Myocardial substrate metabolism in the normal and failing heart. Physiol Rev. 2005;85:1093–1129 [DOI] [PubMed] [Google Scholar]
- 35.Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3’ utrs by comparison of several mammals. Nature. 2005;434:338–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microrna targets. Cell. 2003;115:787–798 [DOI] [PubMed] [Google Scholar]
- 37.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microrna target sites in mammalian mrnas. Elife. 2015;4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wingert RA, Galloway JL, Barut B, Foott H, Fraenkel P, Axe JL, Weber GJ, Dooley K, Davidson AJ, Schmid B, Paw BH, Shaw GC, Kingsley P, Palis J, Schubert H, Chen O, Kaplan J, Zon LI, Tubingen Screen C. Deficiency of glutaredoxin 5 reveals fe-s clusters are required for vertebrate haem synthesis. Nature. 2005;436:1035–1039 [DOI] [PubMed] [Google Scholar]
- 39.Rodriguez-Manzaneque MT, Tamarit J, Belli G, Ros J, Herrero E. Grx5 is a mitochondrial glutaredoxin required for the activity of iron/sulfur enzymes. Mol Biol Cell. 2002;13:1109–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stehling O, Lill R. The role of mitochondria in cellular iron-sulfur protein biogenesis: Mechanisms, connected processes, and diseases. Cold Spring Harb Perspect Biol. 2013;5:a011312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of micrornas in vivo with ‘antagomirs’. Nature. 2005;438:685–689 [DOI] [PubMed] [Google Scholar]
- 42.Krutzfeldt J, Kuwajima S, Braich R, Rajeev KG, Pena J, Tuschl T, Manoharan M, Stoffel M. Specificity, duplex degradation and subcellular localization of antagomirs. Nucleic Acids Res. 2007;35:2885–2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gibb AA, Hill BG. Metabolic coordination of physiological and pathological cardiac remodeling. Circ Res. 2018;123:107–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Razeghi P, Young ME, Alcorn JL, Moravec CS, Frazier OH, Taegtmeyer H. Metabolic gene expression in fetal and failing human heart. Circulation. 2001;104:2923–2931 [DOI] [PubMed] [Google Scholar]
- 45.Cheng L, Ding G, Qin Q, Huang Y, Lewis W, He N, Evans RM, Schneider MD, Brako FA, Xiao Y, Chen YE, Yang Q. Cardiomyocyte-restricted peroxisome proliferator-activated receptor-delta deletion perturbs myocardial fatty acid oxidation and leads to cardiomyopathy. Nat Med. 2004;10:1245–1250 [DOI] [PubMed] [Google Scholar]
- 46.Ye H, Jeong SY, Ghosh MC, Kovtunovych G, Silvestri L, Ortillo D, Uchida N, Tisdale J, Camaschella C, Rouault TA. Glutaredoxin 5 deficiency causes sideroblastic anemia by specifically impairing heme biosynthesis and depleting cytosolic iron in human erythroblasts. J Clin Invest. 2010;120:1749–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Puccio H, Simon D, Cossee M, Criqui-Filipe P, Tiziano F, Melki J, Hindelang C, Matyas R, Rustin P, Koenig M. Mouse models for friedreich ataxia exhibit cardiomyopathy, sensory nerve defect and fe-s enzyme deficiency followed by intramitochondrial iron deposits. Nat Genet. 2001;27:181–186 [DOI] [PubMed] [Google Scholar]
- 48.Campuzano V, Montermini L, Molto MD, Pianese L, Cossee M, Cavalcanti F, Monros E, Rodius F, Duclos F, Monticelli A, Zara F, Canizares J, Koutnikova H, Bidichandani SI, Gellera C, Brice A, Trouillas P, De Michele G, Filla A, De Frutos R, Palau F, Patel PI, Di Donato S, Mandel JL, Cocozza S, Koenig M, Pandolfo M. Friedreich’s ataxia: Autosomal recessive disease caused by an intronic gaa triplet repeat expansion. Science. 1996;271:1423–1427 [DOI] [PubMed] [Google Scholar]
- 49.Wang S, Wang L, Dou L, Guo J, Fang W, Li M, Meng X, Man Y, Shen T, Huang X, Li J. Microrna 152 regulates hepatic glycogenesis by targeting pten. FEBS J. 2016;283:1935–1946 [DOI] [PubMed] [Google Scholar]
- 50.Zhang Z, Li Y, Sheng C, Yang C, Chen L, Sun J. Tanshinone iia inhibits apoptosis in the myocardium by inducing microrna-152-3p expression and thereby downregulating pten. Am J Transl Res. 2016;8:3124–3132 [PMC free article] [PubMed] [Google Scholar]
- 51.Boon RA, Seeger T, Heydt S, Fischer A, Hergenreider E, Horrevoets AJ, Vinciguerra M, Rosenthal N, Sciacca S, Pilato M, van Heijningen P, Essers J, Brandes RP, Zeiher AM, Dimmeler S. Microrna-29 in aortic dilation: Implications for aneurysm formation. Circ Res. 2011;109:1115–1119 [DOI] [PubMed] [Google Scholar]
- 52.Montgomery RL, Hullinger TG, Semus HM, Dickinson BA, Seto AG, Lynch JM, Stack C, Latimer PA, Olson EN, van Rooij E. Therapeutic inhibition of mir-208a improves cardiac function and survival during heart failure. Circulation. 2011;124:1537–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Porrello ER, Johnson BA, Aurora AB, Simpson E, Nam YJ, Matkovich SJ, Dorn GW 2nd, van Rooij E, Olson EN. Mir-15 family regulates postnatal mitotic arrest of cardiomyocytes. Circ Res. 2011;109:670–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bernardo BC, Gao XM, Winbanks CE, Boey EJ, Tham YK, Kiriazis H, Gregorevic P, Obad S, Kauppinen S, Du XJ, Lin RC, McMullen JR. Therapeutic inhibition of the mir-34 family attenuates pathological cardiac remodeling and improves heart function. Proc Natl Acad Sci U S A. 2012;109:17615–17620 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.