Abstract
Contingent upon concentration, reactive oxygen species (ROS) influence cancer evolution in apparently contradictory ways, either initiating/stimulating tumorigenesis and supporting transformation/proliferation of cancer cells or causing cell death. To accommodate high ROS levels, tumor cells modify sulfur-based metabolism, NADPH generation, and the activity of antioxidant transcription factors. During initiation, genetic changes enable cell survival under high ROS levels by activating antioxidant transcription factors or increasing NADPH via the pentose phosphate pathway (PPP). During progression and metastasis, tumor cells adapt to oxidative stress by increasing NADPH in various ways, including activation of AMPK, the PPP, and reductive glutamine and folate metabolism.
Introduction
Oxidative stress, defined as a relative excess of reactive oxygen species (ROS) when compared with antioxidants, has been linked to neurodegenerative disease, cardiovascular disease, diabetes mellitus, and many other pathologies (Sies, 2015). These associations emphasize that a balance must be struck between the relative abundance of ROS and antioxidants. Cells possess complex biochemical and genetic mechanisms to maintain such a balance, and it is clear that their perturbation can have profound pathophysiological consequences.
Cancer cells exhibit aberrant redox homeostasis, but while ROS are pro-tumorigenic, high ROS levels are cytotoxic (Reczek et al., 2017). Specifically, hyperproliferation of tumor cells is accompanied by high ROS production, but they are adapted to thrive under conditions where this oxidative burden pushes redox balance away from a reduced state; tumor cells achieve this by increasing their antioxidant status to optimize ROS-driven proliferation, while at the same time avoiding ROS thresholds that would trigger senescence, apoptosis, or ferroptosis (Dodson et al., 2019; Redza-Dutordoir and Averill-Bates, 2016). Since realignment of redox underpins how tumor cells tolerate high ROS levels, emphasis has been placed in this review on the biochemistry of reduced glutathione ([GSH], L-γ-glutamyl-L-cysteinyl-glycine), thioredoxins (TXN1 and TXN2), and NADPH, and the mechanisms that govern their abundance under normal physiological conditions and during the initiation, progression and metastatic stages of cancer, as well as post-therapy recurrence. We also discuss heterogeneity inherent in the late stages of the evolution of cancer that may explain the apparently anomalous effects of antioxidants on the development and metastatic spread of different types of malignant disease.
Time-Dependent Cellular Adaptations to Oxidative Stress in Normal Cells
Cells generate ROS and reactive nitrogen species (RNS) as an unavoidable consequence of metabolism, and while they are potentially harmful these species are used as intracellular signaling molecules (see Box 1) (Halliwell and Gutteridge, 2015). To ensure ROS/RNS signaling processes are maintained and oxidative damage avoided, cells possess an array of antioxidant systems (Box 2). Besides those direct-acting antioxidants, cells are also equipped with indirect-acting antioxidant systems that either limit the formation of ROS/RNS or detoxify the reactive metabolites they generate (Box 3). A disproportionate increase in ROS/RNS relative to antioxidant capacity, is referred to as oxidative stress, and this is countered by the cell in various ways. In this context, GSH and TXN (Box 4) play central roles in countering oxidative stress, but their ability to do so is underpinned by NADPH which maintains both in a reduced state (Box 5).
Box 1. Chemical Properties and Sources of ROS and RNS.
Among different ROS (i.e., superoxide anion radical [O2●–]; hydrogen peroxide [H2O2]; hydroxyl radical [HO●]) and RNS (i.e., nitric oxide [NO●]; peroxynitrite anion [ONOO–]), each has distinct physicochemical properties and half-lives. Among these ROS, HO● is the most oxidizing, followed by O2●–, with H2O2 as a relatively weak oxidant (Buettner, 1993). While H2O2 and NO● are indispensable as signaling molecules, O2●– and ONOO– are highly reactive and can damage intracellular macromolecules, including polyunsaturated fatty acids (PUFAs) and nucleic acids. Oxidation of PUFAs by ROS leads to lipid peroxidation, where peroxidized PUFAs, together with their breakdown products (e.g., 4-hydroxy-2-nonenal [4-HNE]), can act as signaling molecules to stimulate inflammation, apoptosis, or ferroptosis (Breitzig et al., 2016; Dodson et al., 2019).
Principal intracellular sources of ROS include: leaking of electrons to O2 in mitochondria by reverse electron transport at complex I, or reaction between a semiquinone form of CoQ with O2 at complex III of the respiratory chain (Murphy, 2009); reduction of O2 to O2●– by NADPH oxidases (NOX1–5 and DUOX1/2) during growth factor signaling (Brown and Griendling, 2009); production of H2O2 during protein folding within the ER (Ye et al., 2017).
Less significant sources of ROS arise from the activities of cytochrome P450 (CYP), monoamine oxidase, xanthine oxidase, cyclooxygenase (COX), glycolate oxidase, hydroxyacid oxidase, aldehyde oxidase, and amino acid oxidase (Sies and Jones, 2020). Formation of the particularly reactive HO● from H2O2 occurs through Fenton chemistry that typically involves transition metal ions of iron, copper, or manganese. The principal RNS in the cell is the vasodilator NO●, produced by nitric oxide synthase (NOS) from L-arginine, with the inducible NOS2 isoenzyme largely responsible during inflammation. Reaction between NO● and O2●– yields ONOO– (Radi, 2018).
Box 2. Antioxidant Defences.
To prevent unrestrained accumulation of ROS and RNS, cells contain a spectrum of antioxidants. Non-catalytic small molecules that directly scavenge ROS and RNS include endogenously synthesized bilirubin, α-lipoic acid, melatonin, melanin, GSH, and uric acid, as well as exogenously derived vitamin E, vitamin C, β-carotene and plant polyphenols (Halliwell and Gutteridge, 2015). Among these, GSH is noteworthy because its synthesis is subject to homeostatic regulation (see below) and is often increased in a variety of cancer types (Gamcsik et al., 2012).
Catalytic antioxidants that scavenge O2●– include cytosolic copper/zinc superoxide dismutase (CuZnSOD, or SOD1), mitochondrial manganese MnSOD (SOD2), extracellular EC-SOD (SOD3), each of which catalyzes the conversion of O2●– to H2O2 and O2 (Sheng et al., 2014). SOD2 is of particular importance for viability as its knockout in mice results in perinatal death. Collectively, SOD isoenzymes can attenuate NOX-dependent redox signaling associated with activation of receptor tyrosine kinases and G-protein-coupled receptors, probably by facilitating production and maintenance of a diffusible H2O2 signal (Parascandolo and Laukkanen, 2019). Both SOD1 and SOD2 protect against spontaneous tumorigenesis, and while they have been referred to as tumor suppressors they may also be upregulated during tumorigenesis (Gill et al., 2016).
Enzymes that scavenge H2O2 include catalase (CAT), which converts H2O2 to H2O and O2 (Kirkman and Gaetani, 2007), as well as peroxiredoxins (PRDXs, also called PRXs) and glutathione peroxidases (GPXs), which reduce H2O2 to H2O (Brigelius-Flohe and Maiorino, 2013; Elko et al., 2019). In this context, the PRDXs are of particular physiological significance because they are abundant, have high catalytic activities, and reduce >90% of cellular peroxides; they are subdivided into “Typical” 2-Cys PRDX1–5, “Atypical” 2-Cys PRDX5, and 1-Cys PRDX6. Importantly, reduction of H2O2 by PRDX1–5 is at the expense of oxidizing TXN (simplified to thioredoxin-(SH)2), which results in condensation of two SH groups, and yields thioredoxin-S2 with an intramolecular disulfide bridge (Perkins et al., 2015). By contrast, reduction of H2O2 by the 1-Cys PRDX6 requires GSH rather than TXN (Fisher, 2017). Reduction of H2O2 by GPXs also requires GSH, but in this case it results in condensation between the Cys thiol (-SH) groups of two GSH molecules, yielding GSSG that contains an intermolecular disulfide bridge (Deponte, 2013). Besides limiting ROS levels, PRDXs and GPXs also counter the actions of RNS by contributing to the elimination of NO●, the reduction of ONOO– and also protein denitrosylation (Benhar, 2018).
In addition, sestrins (SESN1, 2, and 3) exert indirect antioxidant activity (Sanchez-Alvarez et al., 2019), in part by activation of transcription factor nuclear factor-erythroid 2 p45-related factor 2 (NRF2) and inhibition of mTORC1 (Rhee and Bae, 2015).
Box 3. ■■■.
-
Heme and iron-dependent proteins in antioxidant defences
Heme oxygenase isoenzymes (HO-1/2, encoded by HMOX1/2) serve as indirect antioxidants because they prevent free heme released from hemoproteins during oxidative stress from forming free radicals, and also because they participate in formation of bilirubin (Gozzelino et al., 2010). Proteins that sequester transition metals, or transport them from the cell, are also indirect-acting antioxidants because they suppress formation of HO● from H2O2 by Fenton chemistry: these include ferritin (comprising light FTL1 and heavy FTH1 subunits), ferroportin (FPN1/SLC40A1), metallothionein, and ceruloplasmin (Arosio et al., 2009; Pietrangelo, 2017). The ability of FTL1, FTH1, and FPN1 to limit intracellular levels of free intracellular iron helps prevent the redox cycling of Fe2+/Fe3+, which leads to lipid peroxidation and the likelihood of ferroptosis (Manz et al., 2016).
-
Drug-metabolizing enzymes in antioxidant defences
As certain xenobiotics are pro-oxidant, drug-metabolizing enzymes are often regarded as indirect-acting antioxidants. These include the phase 1 aldehyde dehydrogenase (ALDH) family, the aldo-keto reductase (AKR) family, carbonyl reductases (CBR), NAD(P)H:quinone oxidoreductase isoenzymes (NQO1 and 2) and short-chain dehydrogenases/reductases, as well as the phase 2 glutathione S-transferase (GST) family and the UDP-glucuronosyl transferase (UGT) family. Their actions prevent quinones and hydroquinones from redox cycling, and electrophiles and lipid peroxidation products from depleting GSH (Hayes et al., 2005; Jin and Penning, 2007; Oppermann, 2007; Rodriguez-Zavala et al., 2019; Rowland et al., 2013). Moreover, drug-metabolizing enzymes, along with GPX4, inactivate 4-HNE and other lipid-derived signaling molecules that can trigger apoptosis and ferroptosis (Breitzig et al., 2016; Dodson et al., 2019).
-
Sirtuin 3 as an indirect antioxidant
Sirtuin 3 (SIRT3) is a NAD+-dependent deacetylase involved in many aspects of mitochondrial quality control (Gomes et al., 2020; Meng et al., 2019). By catalyzing the deacetylation of SOD2, SIRT3 increases scavenging of O2●– in mitochondria, and by catalyzing the deacetylation of IDH2, SIRT3 increases generation of NADPH in mitochondria (Kincaid and Bossy-Wetzel, 2013). Furthermore, SIRT3 exerts multiple effects on mitochondrial metabolism by deacetylating acetyl-CoA synthase 2, long-chain acyl-CoA dehydrogenase and liver kinase B1 (Gomes et al., 2020).
Box 4. ■■■.
-
Glutathione synthesis and salvage
GSH homeostasis is achieved through de novo synthesis and salvage pathways. An ancillary tier of indirect-acting antioxidant proteins exists that supplies glutamate, cysteine, and glycine as building blocks for GSH synthesis. These include glutamine transporters, GLS1 and GLS2, the cystine-glutamate SLC7A11/xCT antiporter and the glycine transporter SLC6A9, as well as components of serine-glycine one-carbon metabolism that produce cysteine from serine and glycine (Ducker and Rabinowitz, 2017; Koppula et al., 2018). Also in this category, are the GSH biosynthetic enzymes glutamate-cysteine ligase ([GCL], previously called γ-glutamylcysteine synthetase [γGCS], comprising catalytic [GCLC, or γGCSh] and modifier [GCLM, or γGCSl] subunits), and glutathione synthetase (GSS), in which GCL catalyzes the initial rate-limiting step and GSS the second and final step (Lu, 2013). In cancer cells, expression of GCLC and GCLM are important determinants of sensitivity to both chemo- and radio-resistance (Lin et al., 2018; Liu et al., 2017), and high levels can be associated with poor prognosis after surgery (Sun et al., 2019). The γ-glutamyl cycle is responsible for the primary de novo synthesis of GSH, with the rate-limiting enzyme being GCL. Recycling of glutamate, cysteine, and glycine salvages GSH, a process accomplished by various enzymes, including the membrane-associated hydrolase GGT that acts on GSH, GSSG, and GS adducts. CHAC1 and CHAC2 as broad specificity γ-glutamyl cyclotransferases can degrade GSH: CHAC1 converts GSH to 5-oxoproline and cysteinylglycine (Oakley et al., 2008), and its expression may be dysregulated in breast and ovarian cancer (Goebel et al., 2012).
-
Thioredoxins in redox homeostasis
The “cytosolic” TXN1 and mitochondrial TXN2 are both small reductases that catalyze cysteine thiol-disulfide exchange reactions via a Cys-Gly-Pro-Cys motif in their active sites (Lu and Holmgren, 2014). The antioxidant function of TXNs is demonstrated principally through their abilities to transfer electrons to oxidized PRDX1–5, methionine sulfoxide reductases, and redox-sensitive transcription factors, thereby allowing their substrates to be reduced back to a more active state. This process results in oxidation of Cys residues in the active sites of TXN1 or TXN2, which can in turn be reduced by the selenoproteins TXNRD1 or TXNRD2, respectively, using NADPH as a cofactor. Thus, TXN1/2-mediated reduction of PRDXs, methionine sulfoxide reductases, and transcription factors is at the expense of NADPH and results in accumulation of NADP+. Similar to other redox pathways, the TXN antioxidant system is upregulated in a wide range of cancers (Jia et al., 2019) and is associated with poor prognosis (Leone et al., 2017). Overlap exists between the TXN system and GSH insofar as oxidized TXN1 can be reduced by glutaredoxins (GRX1 and 2), using GSH as a cofactor, and so under certain circumstances can replace TXNRD1 (Deponte, 2013; Ren et al., 2017). Furthermore, GSSG can be reduced by the TXN system, creating redundancy between the TXN- and GSH-based antioxidant systems. In tumors with compromised GSH homeostasis, inhibition of TXN/TXNRD induces synthetic lethality (Yan et al., 2019).
Box 5. NADPH-Based Redox Couples Are Central to Antioxidant Defences.
NADPH is a fundamentally important metabolite in the reductive biosynthesis of macromolecules, and is indispensable for cellular antioxidant defences. By binding to CAT, NADPH prevents inactivation of the enzyme by H2O2 and maintains its antioxidant capacity (Kirkman and Gaetani, 2007). Importantly, NADPH provides reducing equivalents that allow GSH to be generated from GSSG and thioredoxin-(SH)2 to be generated from thioredoxin-S2 by the catalytic actions of glutathione reductase (GSR) and thioredoxin reductases (TXNRD1 and TXNRD2), respectively (Ying, 2008). In turn, GSH and TXN enable GPXs and PRDXs to fulfill their antioxidant roles. To support proliferation, NADPH synthesis is frequently increased in tumor cells (Purohit et al., 2019).
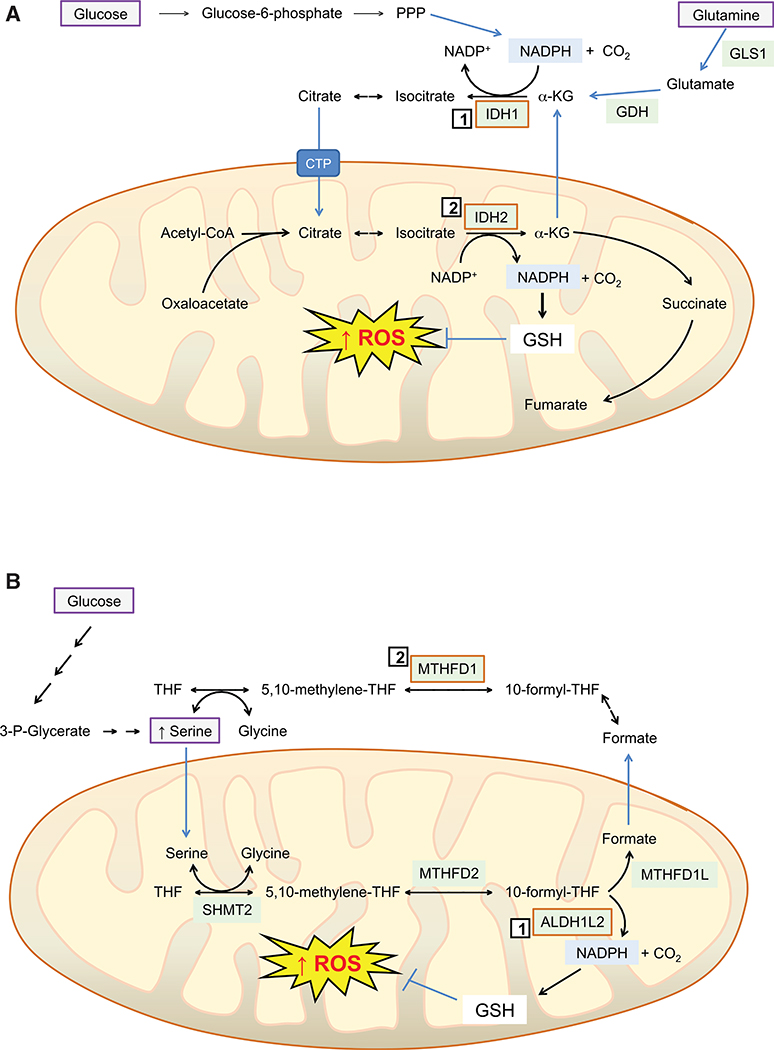
As NADPH exists as a redox couple with NADP+, and is constantly oxidized in aerobic conditions, it has to be continuously regenerated. This is primarily achieved by enzymes in the oxidative arm of the pentose phosphate pathway (PPP), glucose-6-phosphate dehydrogenase (G6PD), and 6-phosphogluconate dehydrogenase (6PGD) that provide ribose-5-phosphate, malic enzymes (ME1, 2, and 3) that provide pyruvate for the TCA cycle, and the serine-driven one-carbon metabolism enzymes methylenetetrahydrofolate dehydrogenase (MTHFD2), which forms the purine precursor 10-formyl-tetrahydrofolate, and aldehyde dehydrogenase 1L2 (ALDH1L2) that releases CO2 from 10-formyl-tetrahydrofolate (Ciccarese and Ciminale, 2017). Among these enzymes, ME1 has been reported to physically interact with, and activate, both 6PGD and G6PD, suggesting a level of coordinated regulation of these NADPH-generating enzymes that may be relevant in cancer cells (Yao et al., 2017). Under hypoxic conditions, the production of NADPH by MTHFD2 in MYC-transformed tumor cells can be enhanced by inducing serine hydroxymethyltransferase (SHMT2), which encodes an enzyme that catalyzes transfer of CH3 from serine to tetrahydrofolate and so increases substrate availability for MTHFD2 (Ye et al., 2014).
Other enzymes that generate NADPH include isocitrate dehydrogenase (IDH1/2), nicotinamide nucleotide transhydrogenase (NNT), and glutamate dehydrogenase (GDH1/2). The NADP+-dependent IDH1 and IDH2 catalyze reversible oxidative decarboxylation of isocitrate to yield α-ketoglutarate (α-KG) and CO2, whereas NNT transfers reducing equivalents from NADH to NADPH using the electron transport chain proton gradient (Purohit et al., 2019). Under hypoxic conditions, cancer cells can use IDH2 to reductively carboxylate α-KG obtained anaplerotically from glutamine and produce citrate at the expense of NADPH, which is compensated for by increased oxidation of α-KG to succinyl-CoA, which provides NADH, and is in turn used by NNT to produce NADPH (Mullen et al., 2014). GDH1 and GDH2 catalyze the reversible oxidative deamination of glutamate to α-KG using NAD(P)+ as cofactor (Plaitakis et al., 2017), but do not seem to contribute substantially to redox homeostasis (Fan et al., 2014b).
Somatic mutations in IDH1 and IDH2 have been reported in a range of solid and hematological malignancies arising early during tumorigenesis and consistent with events representing driver mutations; the mutations are heterozygous and neomorphic as they result in production of the oncometabolite 2-hydroxyglutarate (Dang and Su, 2017). In this instance, IDH1/2 encoded by the mutant gene acquires the ability to catalyze NADPH-dependent conversion of α-KG to 2-hydroxyglutarate as a consequence of substitutions that increases its affinity for NADPH and α-KG and causes loss of oxidative decarboxylation activity (Golub et al., 2019). The 2-hydroxyglutarate produced by mutant IDH1/2 inhibits competitively many 2-oxoglutarate-dependent dioxygenases, such as histone demethylases and DNA hydroxylases, and it is well established that this perturbs epigenetic mechanisms and thus cell differentiation. In addition, 2-hydroxyglutarate inhibits branched chain amino acid transaminases BCAT1 and BCAT2 that catalyze interconversion of 2-oxoglutarate with glutamate, resulting in a reduction in intracellular levels of glutamate and GSH in cells harboring mutant IDH1, which renders them sensitive to oxidative stress when treated with an inhibitor of glutaminase (GLS1 and GLS2) (McBrayer et al., 2018). As discussed later, inhibition of IDH mutants impacts ROS levels and has anticancer potential.
Cells adapt to oxidative stress in the short term by metabolic reprograming and in the longer term by genetic reprograming. Upon acute exposure to ROS, NADPH production by glucose-6-phosphate dehydrogenase (G6PD) plays a pivotal role in mitigating oxidative stress. Upon experiencing non-toxic threshold levels of H2O2, cells activate G6PD and reroute glucose metabolism from glycolysis through the oxidative arm of the pentose phosphate pathway (PPP) toward nucleotide synthesis, thereby allowing increased reduction of NADP+ to NADPH (Kuehne et al., 2015). This rapid metabolic rerouting is due to alleviation of the negative feedback regulation of G6PD activity exerted by NADPH, which occurs constitutively under non-stressed conditions, and is a consequence of acute depletion of NADPH caused by ROS (Dick and Ralser, 2015). In turn, the increase in NADPH enables GSR1 and TXNRD1/2 to augment the GSH- and TXN1/2-based antioxidant systems, subduing ROS to homeostatic levels.
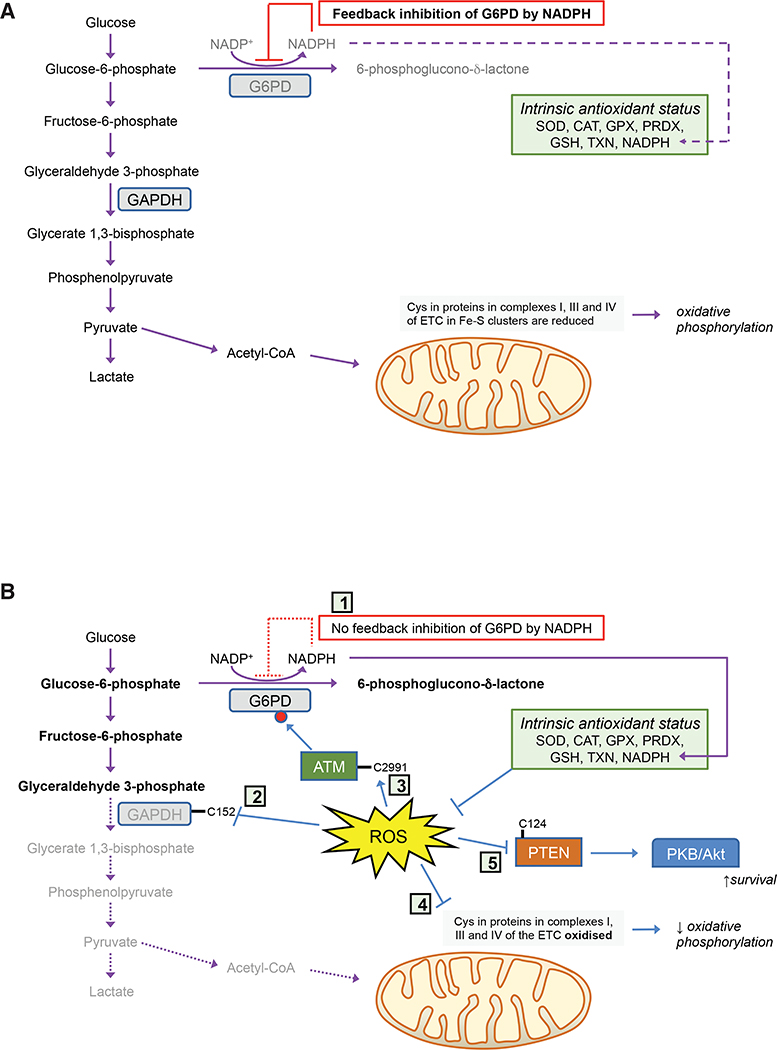
When exposed to non-toxic doses of H2O2 for modest periods (e.g., 15 min), cells use redox switches (see Box 6) in glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and pyruvate kinase M2 (PKM2) to block glycolysis and increase glucose catabolism via the PPP, causing accumulation of upper glycolysis intermediates, a spill-over of glucose-6-phosphate into the oxidative arm of the PPP, and increased production of NADPH by G6PD to ameliorate oxidative stress (Figure 1): the redox switch in GAPDH involves Cys-152, and in PKM2 involves Cys-358. Under these conditions, GAPDH activity can be further increased by phosphorylation by ataxia telangiectasia mutated (ATM), as a consequence of formation of an intermolecular disulfide bridge in ATM at Cys-2991, which also increases flux through the PPP (Cosentino et al., 2011). The effects of ROS on GAPDH, PKM2, and G6PD activities are likely coordinated with oxidation of Cys residues in at least six protein subunits within complexes I, III, and IV of the mitochondrial electron transport chain that contain Fe-S clusters, resulting in decreased O2 consumption and a decrease in ROS production (van der Reest et al., 2018). Acute oxidative stress may also inhibit phosphatase and tensin homolog (PTEN), by oxidizing Cys-124, and in so doing activate PKB/Akt by phosphatidylinositol 3-kinase (PI3K), upregulating antioxidant gene expression and increasing cell survival (Sakamoto et al., 2009; van der Reest et al., 2018).
Box 6. Protein Cysteines Provide Redox Switches.
Cellular responses to ROS predicate on regulatory proteins containing thiol-based redox switches that when oxidized cause the protein to adopt new or distinct functions (Holmstrom and Finkel, 2014). Critical to the reactivity of individual cysteines within redox switches, is the apparent pK (pKa) of their thiol moieties. Proteins which are susceptible to oxidative modification contain cysteines with low pKa values that exist in the thiolate anion state at physiological pH, caused by the presence of adjacent basic amino acids. The various oxidation states of the thiol (SH)-containing side chain of cysteine within proteins creates a circumstance where quantitative changes in intracellular ROS can regulate protein activity. The thiol of cysteine as a thiolate anion (−S–) or cysteinyl radical (−S●) is critical to its oxygen reactivity. Contingent upon the extent of oxidation, sulfenate (−SOH), sulfinate (−SO2H), sulfonate (−SO3H), or disulfide (−S–S−) species may be formed, with oxidation to sulfonate being essentially irreversible and requiring degradation of the protein to eliminate the modification. Also, S-glutathionylation (−SSG) and S-nitrosylation (−SNO) may result from exposure to ROS/RNS and sulfhydration/persulfidation (−SSH) from reaction with endogenous H2S (Yang et al., 2016a).
S-Glutathionylation may protect proteins from over-oxidation, or may alter their structure and function; thus, parallel to the kinase/phosphatase cycle is the S-glutathionylation cycle (Grek et al., 2013). The forward protein S-glutathionylation reaction can be driven by GST Pi 1–1 (GST P1–1), which demonstrates several mechanisms of pleiotropy (Paaby and Rockman, 2013). In particular, GST P1–1 engages in promiscuous interactions with target proteins, identifying them as substrates for S-glutathionylation. GST P1–1 has catalytic cysteines near the surface of the substrate channel that can act as GS– donors. By contrast, in GST Omega 1–1 (GST O1–1), which catalyzes deglutathionylation, the catalytic cysteine of each subunit is buried within the substrate channel, behaving as a GS– acceptor (Menon and Board, 2013). Deglutathionylation is also accomplished by GRX (Stroher and Millar, 2012) or SRXN1 (Findlay et al., 2006; Park et al., 2009).
With the exception of the sulfonate state, oxidized cysteines can be reduced, thereby recovering the protein’s homeostatic function. Oxidation of protein-S– to protein-SOH can be reduced by TXN and TXNRD, with NADPH as hydride donor. Similarly, protein-SSG and protein disulfide bridges formed from protein-SOH can be reduced by the actions of TXN, TXNRD, GSH, GSR, GST O1–1, GRX, and NADPH. At higher ROS levels, protein-SOH may be further oxidized to protein-SO2H, some of which can be reduced to protein-SOH by SRXN1 (Akter et al., 2018).
Figure 1. Metabolic Responses to Acute Oxidative Stress.
In cells under normal redox homeostatic conditions (A), glucose is principally oxidized by glycolysis to pyruvate, and via acetyl-CoA through the tricarboxylic acid cycle, with G6PD inhibited by NADPH and minimal flux through the PPP. However, upon acute oxidative stress (B), feedback inhibition of G6PD by NADPH is greatly diminished (1) and Cys residues in GAPDH (2), ATM (3), and complexes I, III, and IV of the electron transport chain (4) are oxidized, a combination of circumstances that result in inhibition of glycolysis, phosphorylation of G6PD, and increased metabolism through the PPP. Moreover, oxidation of Cys residues in PTEN (5) causes activation of PKB/Akt, resulting in increased cell survival.
An NO●-based protective mechanism against acute oxidative stress has been reported that entails increased flux through the PPP resulting from S-nitrosylation of PKM2 (Zhou et al., 2019). In this case, NOS3 activity increases in proximal tubule endothelial cells upon acute kidney injury, causing accumulation of S-nitroso coenzyme A (SNO-CoA) and S-nitrosylation of PKM2 at Cys-423 and Cys-424, which results in inhibition of the kinase, diversion of glucose metabolism through the PPP, increased NADPH production, and diminished ROS.
In contrast to acute oxidative stress, which can largely be resolved by metabolic rerouting, adaptation to chronic oxidative stress involves activation of genetic programs. In the short-to-medium time span, oxidative stress can alter the abundance and/or subcellular distribution of hypoxia-inducible factor 1α (HIF-1α), which leads to metabolic reprogramming. Traditionally this involves hypoxia and oxidation of Cys-326 in PHD2 that stabilizes HIF-1α and results in transcriptional changes that lead to a switch from glucose oxidation to glycolysis (Lee et al., 2016). Importantly, in chronic oxidative stress models that involve accumulation of endogenous electrophiles or depletion of GSH/TXN, adaptation entails upregulation of antioxidant genes (Blackburn et al., 2006; Chen et al., 2016; Patterson et al., 2013; Zheng et al., 2015).
Molecular Basis for Adaptation to Oxidative Stress
Transcription Factors that Control Intracellular Redox
It is established that many transcription factors, including activator protein 1 (AP-1), HIF-1α, heat shock factor 1 (HSF1), nuclear factor kB (NF-κB), nuclear factor-erythroid 2 p45-related factor 2 (NRF2), and tumor protein p53 (TP53 or Trp53 in mice), are activated by ROS and regulate the redox status of cells (reviewed in Marinho et al., 2014). While these transcription factors have all been implicated in carcinogenesis, the activities of forkhead box, class O (FOXO), and peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α), also increased by ROS (Brown and Webb, 2018; Guo et al., 2018), contribute to redox status and are implicated in carcinogenesis. Moreover, BTB and CNC homology 1 (BACH1), a repressor of the oxidative stress response that antagonizes NRF2 (Tan et al., 2013), is itself inhibited by pro-oxidants and also influences cancer evolution, including metastasis (Zhang et al., 2018a).
Stratified Responses of Antioxidant Transcription Factors
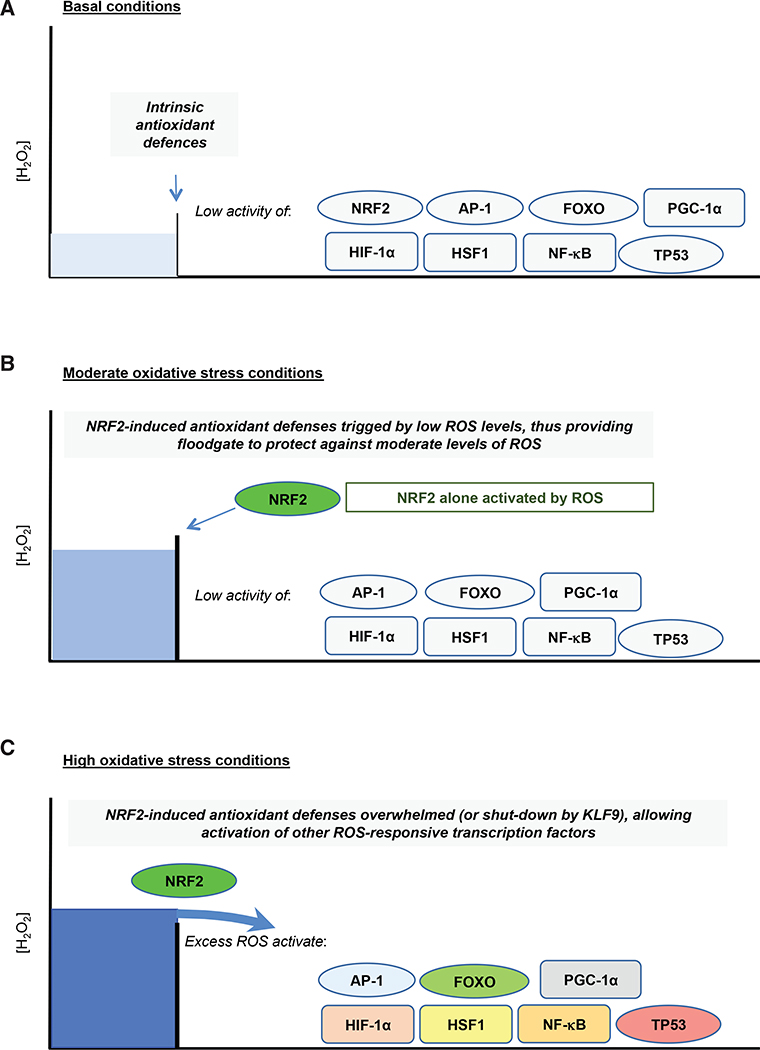
The extent to which individual members of the above network of antioxidant transcriptions are differentially activated by oxidative stress is uncertain, although it is improbable that all are activated simultaneously. Rather, different transcription factors likely respond to distinct threshold levels of ROS/RNS, in a concentration- and/or time-dependent manner that is probably attuned to the coexistence of metabolic stress, proteotoxic stress, hypoxia, inflammation, or DNA damage. Because it regulates a wide spectrum of antioxidant and detoxification genes, NRF2 provides a principal inducible defense against oxidative stress (Hayes and Dinkova-Kostova, 2014; Sies et al., 2017), but whether it is more readily activated by ROS than other redox-responsive transcription factors is unclear. Nevertheless, assuming that NRF2-directed gene expression provides an initial means to adapt to oxidative stress, it may offer a type of “floodgate” protection, analogous to that proposed for PRDXs (see Box 7), in which only once the antioxidant genes induced by NRF2 upon oxidative stress are overwhelmed at a particular ROS threshold are the other antioxidant transcription factors within the network activated (Figure 2). A modification of the floodgate model would include induction by NRF2 of Krüpple-like factor 9 (KLF9), which is a DNA-binding, transcriptional regulator that downregulates the antioxidant genes TXNRD2 and PRDX6 (Chhunchha et al., 2019; Zucker et al., 2014), and induction of KLF9 would shut-down antioxidant defences when ROS levels exceed a certain threshold or duration. In this scenario, other members of the network would only be activated when the antioxidant capacity of NRF2 target genes is exceeded—or when KLF9 is induced. Once NRF2-directed floodgate defences have been breached, the question of whether individual antioxidant transcription factors are activated in a stratified or coordinated manner is uncertain.
Box 7. Functioning of the Redox Switch in PRDX, and Associated “Floodgate” Hypothesis.
Antioxidant PRDX enzymes are a classic example of proteins regulated by redox. Each possesses a high-affinity H2O2-binding site, comprising a cysteine with a low pKa value that is readily oxidized, and therefore called a peroxidatic cysteine, or Cp (Perkins et al., 2015). In PRDX1–5, oxidation of their CP residues by H2O2 to a sulfenic acid derivative results in formation of an intermolecular disulfide between the CP residue and another cysteine, the resolving Cys, or CR, present in the other subunit within the PRDX dimer. In turn, the CP-CR disulfide is reduced by the TXN1/2-TXNRD1/2-NADPH system, thereby allowing regeneration of CP for reaction with H2O2, and completion of the catalytic cycle. For 1-Cys PRDX6, a monomeric enzyme with phospholipase activity that can reduce phospholipid hydroperoxides, its oxidized CP residue can be reduced by GSH, in a GST P1–1-catalyzed reaction (Fisher, 2017).
As a consequence of their CP residues being oxidized to sulfinic acid, PRDXs can be inactivated under conditions where H2O2 levels are relatively high. This allows H2O2 to accumulate and fulfill signaling functions, where PRDXs are posited to act through peroxide floodgates (Wood et al., 2003). Inactivation of PRDXs can also occur through phosphorylation, providing a classic example of crosstalk between kinase and redox signaling (Rhee et al., 2012). Because of their abundance, sensitivity to oxidation and ability to physically interact with other proteins, PRDXs have been proposed to transduce H2O2 signaling through physical associations with other proteins (Rhee and Kil, 2017).
Figure 2. NRF2 Provides an Inducible Floodgate Defense against Oxidative Stress.
Under non-stressed basal conditions (A), cellular redox homeostasis is maintained by constitutive expression of a battery of antioxidant genes. However, when exposed to acute oxidative stress (B), cells adapt to the increase in ROS levels by inducing genes encoding detoxification, GSH- and TXN-dependent antioxidants, and NADPH-generating enzymes that are regulated by NRF2. Should the capacity of the antioxidant systems that are induced by NRF2 become saturated and therefore insufficient to counter additional oxidative stress (C), or prolonged oxidative stress causes activation of KLF9 and downregulation of NRF2, the excess levels of ROS that are not countered by the NRF2-directed defences then trigger additional redox switches that activate other members of the antioxidant transcription factor network. When NRF2-orchestrated defences become saturated, activation of other members of the antioxidant transcription factor network may occur simultaneously, or they may be activated in a stratified manner with each transcription factor being activated at a distinct ROS threshold, which results in various cellular responses, including metabolic reprogramming, damage repair, cell-cycle arrest, senescence, and apoptosis.
A “hierarchical” or “rheostat” response to oxidative stress has been proposed that places NRF2 as a first-tier defense (activated by modest increases of ROS/RNS), with AP-1 and NF-κB as a second-tier defense (activated by higher ROS/RNS levels), and a third and final tier involving activation of apoptosis (Xiao et al., 2003). Within this model, it is plausible that FOXO, PGC-1α, and HIF-1α are activated by higher levels of ROS/RNS than are required to activate NRF2, and lower levels of ROS/RNS than required to stimulate apoptosis, but where they feature relative to AP-1 and NF-κB is a matter of conjecture. It can also be argued that TP53, which controls cell-cycle arrest, senescence, and apoptosis, provides a “final” response to excessive ROS levels.
Little has been reported about crosstalk between antioxidant transcription factors. Crosstalk between NRF2 and TP53 has been revealed using a bacterial artificial chromosome-GFP platform with quantitative high-throughput confocal microscopy to examine responses to oxidative stress caused by diethylmaleate and DNA damage caused by etoposide (Hiemstra et al., 2017). This is an approach that should be extended to define thresholds for various stressors that activate members of the antioxidant transcription factor network and tipping points for adaptation to stress. In this context, it should be noted that NRF2 regulates the expression of HSF1 (Paul et al., 2018) and overexpression of the NF-κB p65/RelA subunit antagonizes NRF2 by depriving it of CBP and recruiting histone deacetylase 3 (Liu et al., 2008) and that, when oxidative stress is sufficient to cause DNA damage, the ensuing activation of TP53 results in antagonism of NRF2, thereby heightening oxidative stress and facilitating apoptosis (Faraonio et al., 2006); together, these findings suggest that NRF2 is downregulated by oxidative stress sufficient to cause inflammation and pro-apoptotic signaling. It has also been reported that FOXO transcription factors can interact with PGC-1α and TP53 in a stimulus-dependent fashion (Eijkelenboom and Burgering, 2013).
The Antioxidant Transcription Factor Network
Members of this network each controls a distinct spectrum of antioxidant genes (Table 1), and each is implicated in tumorigenesis in different ways.
Table 1.
Redox Homeostasis-Associated Genes Regulated by Members of the Antioxidant Transcription Factor Network
| Transcription Factorsa | NRF2 | BACH1b | AP-1 | FOXO | PGC-1α | HIF-1α | HSF1 | NF-κB | TP53 |
|---|---|---|---|---|---|---|---|---|---|
| Genes for proteins that: | |||||||||
| Scavenge ROS | CAT | – | CAT | CAT | CAT | – | – | CAT | CAT |
| PRDX1 | – | – | PRDX1 | – | – | – | – | – | |
| – | – | – | PRDX3 | PRDX3 | – | – | – | – | |
| – | – | – | – | – | – | PRDX4 | – | – | |
| – | – | – | PRDX5 | PRDX5 | – | – | – | – | |
| PRDX6 | – | – | – | – | – | – | – | – | |
| – | – | – | – | GPX1 | – | – | GPX1 | GPX1 | |
| GPX2 | – | – | – | – | – | – | – | – | |
| – | – | SOD1 | SOD1 | – | – | – | SOD1 | – | |
| – | – | – | SOD2 | SOD2 | – | – | SOD2 | SOD2 | |
| Are auxiliary antioxidants | – | – | – | – | – | – | – | – | SESN1 |
| – | – | – | – | – | – | – | SESN2 | – | |
| – | – | – | SESN3 | – | – | – | – | – | |
| Synthesize GSH | GCLC | ↓GCLC | GCLC | – | – | – | – | GCLC | – |
| GCLM | ↓GCLM | GCLM | – | GCLM | GCLM | GCLM | GCLM | – | |
| – | – | – | – | – | GLS1 | GLS1 | – | – | |
| – | – | – | – | – | – | – | – | GLS2 | |
| GSR1 | – | – | – | – | – | – | – | – | |
| SLC6A9 | – | – | – | – | – | – | – | – | |
| SLC7A11 | ↓SLC7A11 | – | – | – | SLC7A11 | – | – | – | |
| Provide reduced TXN | TXN1 | – | – | – | – | – | – | TXN1 | – |
| – | – | – | TXN2 | TXN2 | – | TXN2 | TXN2 | – | |
| TXNRD1 | ↓TXNRD1 | – | – | – | – | – | – | – | |
| – | – | – | TXNRD2 | – | – | – | – | – | |
| Reduce sulfinic acid | SRXN1 | – | SRXN1 | – | – | – | – | – | – |
| Generate NADPH | G6PD | – | – | – | – | – | – | G6PD | – |
| IDH1 | – | – | – | – | – | – | – | – | |
| ME1 | – | – | – | – | – | – | – | – | |
| PGD | – | – | – | – | – | – | – | – | |
| – | – | – | – | – | – | – | – | TIGAR | |
| Metabolize heme/iron | FTH1 | ↓FTH1 | FTH1 | – | – | – | FTH1 | – | – |
| FTL1 | ↓FTL1 | – | – | – | – | – | FTL1 | – | |
| HMOX1 | ↓HMOX1 | HMOX1 | – | – | – | HMOX1 | HMOX1 | – | |
| SLC40A1 | ↓SLC40A1 | – | – | – | – | – | – | – | |
| Detoxify xenobiotics | AKR1B10 | – | – | – | – | – | – | – | AKR1B10 |
| AKR1C1 | – | – | – | – | – | – | AKR1C1 | – | |
| AKR1C3 | ↓AKR1C3 | – | – | – | – | – | – | – | |
| ALDH3A1 | – | – | – | – | – | – | – | – | |
| – | – | – | – | – | – | ALDH3B1 | – | – | |
| – | – | – | – | – | – | – | – | ALDH4 | |
| CBR1 | – | – | – | – | – | – | – | – | |
| EPHX1 | – | – | – | – | – | – | – | – | |
| GSTs | – | GSTP1 | – | – | – | – | GSTP1 | – | |
| NQO1 | – | NQO1 | – | – | – | – | NQO1 | – | |
| NQO2 | – | – | – | – | – | – | – | – | |
| PTGR1 | – | – | – | – | – | – | – | – | |
| UGT1A6 | – | – | – | – | – | – | – | – | |
| Deacetylase activity | – | – | – | – | SIRT3 | – | – | – | – |
Information about the target genes for each of the transcription factors was obtained as follows: for NRF2, from Hayes and Dinkova-Kostova (2014) and Hayes et al. (2016); for BACH1, from MacLeod et al. (2009) and Nishizawa et al. (2020); for AP-1, from Soriano et al. (2009) and Glorieux et al. (2016); for FOXO, from Klotz et al. (2015); for PGC-1α, from Valle et al. (2005), St-Pierre et al. (2006), Giralt et al. (2011), Guo et al. (2018), and Kaarniranta et al. (2018); for HIF-1α, from Budanov (2014) and Lu et al. (2015); for HSP1, from Kovács et al. (2019); for NF-κB, from Morgan and Liu (2011; http://www.bu.edu/nf-kb/generesourses/target-genes/); for TP53, from Maillet and Pervaiz (2012), Budanov (2014), and Nguyen et al. (2018).
Downward-pointing arrows signify antioxidant and detoxication genes that are repressed by BACH1, with repression of HMOX1 being most profound. All others are positively regulated by the transcription factors indicated at the top of the table.
NRF2 is the principal regulator of intracellular redox homeostasis and transactivates genes as a heterodimer with a small musculoaponeurotic fibrosarcoma (MAF) protein. As shown in Table 1, NRF2 transactivates a wide spectrum of antioxidant genes upon exposure to ROS or soft electrophiles (Hayes Dinkova-Kostova, 2014; Hayes JD; Yamamoto et al., 2018). An interesting feature of NRF2 is that it does not regulate SOD1 or SOD2, and so its activation will not directly quench O2●–-based redox signaling. It is not clear whether NRF2 can repress prooxidant genes, but it has been reported to downregulate NOX4 and, through its ability to repress the expression of genes encoding interleukin-1β (IL-1β) and IL-6, along with induction of the gene encoding peroxisome proliferator-activated receptor gamma (PPARγ), NRF2 dampens the levels of ROS formed during inflammation (Kobayashi et al., 2016). NRF2 mediates the protective effects of many cancer chemopreventive agents but it is also thought to support the latter stages of tumorigenesis (Rojo de la Vega et al., 2018). Its upregulation in cancer cells can increase serine biosynthesis through its ability to positively control ATF4-mediated induction of phosphoglycerate dehydrogenase, phosphohydroxythreonine aminotransferase, phosphoserine phosphatase, and SHMT2 that support GSH and nucleotide synthesis (DeNicola et al., 2015), although it should be recognized that ATF4 is activated by the integrated stress response that will likely be upregulated during tumorigenesis. Based on a computational method that analyses patterns of mutational signatures in cancer, Stephen Elledge and colleagues have classed the gene encoding NRF2 (i.e., NFE2L2) as an oncogene, and that encoding its principal repressor Kelch-like ECH-associated protein 1 (i.e., KEAP1) as a tumor suppressor (Davoli et al., 2013).
BACH1 binds DNA as a heterodimer with small MAF proteins, and is best known for its ability to repress HMOX1, and to a substantially lesser extent other antioxidant genes (MacLeod et al., 2009; Nishizawa et al., 2020). Upon binding heme and exposure to pro-oxidant agents, BACH1 is degraded, enabling NRF2 to transactivate genes it represses. Besides repression of antioxidant genes, BACH1 contributes positively to the expression of genes for metalloproteinase-1 (MMP-1) and CXCR4, and therefore supports metastatic disease (Liang et al., 2012). Moreover, BACH1 directs metabolic reprogramming by increasing expression of hexokinase 2 and Gapdh (Wiel et al., 2019) and suppresses expression of mitochondrial electron transport chain genes (Lee et al., 2019) increasing glycolysis and suppressing mitochondrial TCA metabolism, each a hallmark of cancer.
AP-1 represents a family of dimeric transcription factors comprising combinations of Jun (c-Jun, JunB, JunD), Fos (c-Fos, FosB, Fra-1, Fra-2), ATF (ATFa, ATF2, ATF3, ATF4), JDP (JDP-1, JDP-2), and MAF (c-MAF, MAFA, MAFB, MAFF, MAFG, MAFK) (Bejjani et al., 2019). Table 1 shows that they exert antioxidant effects through induction of genes that scavenge ROS, synthesize GSH, suppress levels of free iron, and metabolize pro-oxidant xenobiotics (Glorieux et al., 2016; Soriano et al., 2009). AP-1 members include oncogenes that can transform cells and are involved in cell proliferation, while others inhibit tumorigenesis, suggesting that their effects are context dependent (Eferl and Wagner, 2003; Shaulian and Karin, 2001).
FOXO forkhead motif-containing FOXO1, FOXO3, FOXO4, and FOXO6 support cellular homeostasis in various ways (Eijkelenboom and Burgering, 2013). They augment antioxidant status by inducing genes that eliminate ROS and improve mitochondrial redox and suppress levels of free transition metal ions by increasing levels of metallothionein and ceruloplasmin (Klotz et al., 2015). Besides responding to ROS, FOXOs also respond to nutrients and regulate genes involved in cell-cycle arrest, such as GADD45, and apoptosis (Eijkelenboom and Burgering, 2013). In cancer cells, FOXO can induce genes for MMP-9 and MMP-13 and may therefore aid epithelial-to-mesenchymal transition (EMT) required for metastatic spread (Liou and Storz, 2010).
PGC-1α transcriptional coactivator is a master regulator of mitochondrial biogenesis. It increases oxidative phosphorylation by activating PPARγ and nuclear respiratory factors 1 and 2, and also increases antioxidant capacity (Table 1) as well as decreasing mitochondrial production of ROS by activating uncoupling protein-1 and −2, and stimulating mitochondrial biogenesis (Guo et al., 2018; Kaarniranta et al., 2018; St-Pierre et al., 2006; Valle et al., 2005). In cancer, activation of PGC-1α can exert both positive and negative effects in that it supports survival and metabolic flexibility of tumor cells while also exhibiting anti-metastatic effects through inhibition of EMT (Gravel, 2018).
HIF-1α regulates expression of GSH-based antioxidant genes under hypoxic conditions (Lu et al., 2015; Stegen et al., 2016). It also exerts antioxidant effects by inducing genes for lactate dehydrogenase and pyruvate dehydrogenase kinase 1, the latter of which inhibits pyruvate dehydrogenase; together, these enzymes direct glucose catabolism from the TCA cycle to lactate, thereby decreasing mitochondrial ROS production (Samanta and Semenza, 2017). Activation of HIF-1α increases expression of proangiogenic genes, such as that for vascular endothelial growth factor, which is associated with metastatic disease (Semenza, 2012).
HSF1 responds to stressors that cause protein misfolding by inducing genes encoding heat shock protein chaperones. However, it also responds to ROS through Cys-35 and Cys-105 (Ahn and Thiele, 2003), and induces antioxidant genes (Table 1) (Kovács et al., 2019). HSF1 upregulation commonly occurs as a means of protecting tumor cells against diverse stresses (Dong et al., 2019).
NF-κB represents a family of transcription factors that comprise heterodimer or homodimer combinations of p50, p52, p65/RelA, RelB, and c-Rel subunits, which provide an inducible first-line defense against infection and other damaging agents (Perkins, 2007). Although NF-κB orchestrates an adaptive response to eradicate invading pathogens, by inducing expression of cytokines, chemokines, and receptors, and to repair tissue damage, it also regulates expression of antioxidant genes (Morgan and Liu, 2011). However, as NF-κB regulates expression of the pro-oxidant genes CYP2E1, NOX2, XOR, NOS2, COX2, ALOX5, and ALOX12 (Morgan and Liu, 2011; (http://www.bu.edu/nf-kb/gene-resourses/target-genes/) it may contribute little to adaptation to oxidative stress. Constitutive activation of NF-κB is observed in many cancers, where it promotes the survival, proliferation, and metastasis of tumor cells by increasing expression of anti-apoptotic genes, cyclins, MMPs, cell adhesion genes, and pro-angiogenic genes (Perkins, 2012). It also favors a metabolic switch to glycolysis and controls the tumor microenvironment by directing the pro-tumorigenic actions of immune cells (Taniguchi and Karin, 2018).
TP53 increases antioxidant status by transactivating genes encoding enzymes/proteins that scavenge ROS, support GSH synthesis, increase NADPH production, detoxify xenobiotics, and trans-repress genes for the pro-oxidant enzymes NOS2 and COX2 (Maillet and Pervaiz, 2012; Nguyen et al., 2018). The increase in production of NADPH affected by TP53 arises through upregulation of TP53-induced glycolysis and apoptosis regulator (TIGAR), which acts as a fructose-2,6-bisphosphatase and thereby decreases glycolysis and increases flux through the oxidative arm of the PPP (Cheung et al., 2013). Conversely, TP53 also exerts pro-oxidant effects by upregulating TP53-inducible genes (PIGs), including PIG3, which is a quinone oxidoreductase/ξ-crystallin producing ROS by redox cycling quinones and p67phox that activates the NOX2 complex (Italiano et al., 2012; Porte et al., 2009). TP53 protects against tumorigenesis primarily by controlling expression of genes involved in cell-cycle arrest, senescence, and apoptosis (Bieging et al., 2014).
Life and Death of Cells Is Heavily Influenced by Their Redox Status
Contributions of Redox to Physiology and Cell Signaling
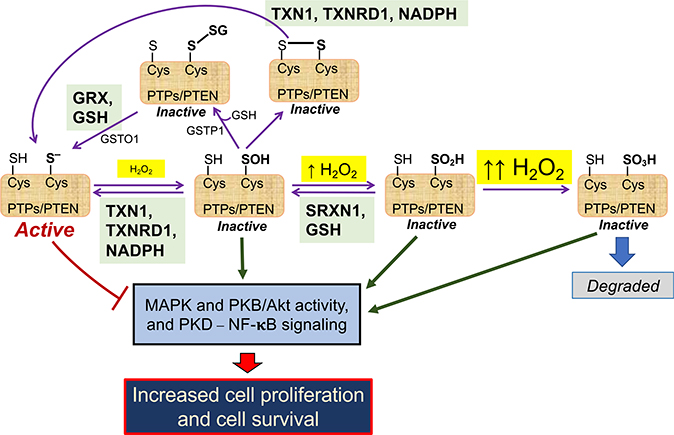
Typically, ROS signaling involves O2●– production by NOX or mitochondria, which, following reduction to H2O2, provides an effector molecule (Ushio-Fukai, 2009). Classic examples of ROS-mediated regulation include insulin/growth factor signaling (Tiganis, 2011) and transient receptor potential (TRP) channel-mediated cation signaling (Andersson et al., 2008). In addition, RAS proteins contain reactive Cys residues that are susceptible to modifications by ROS/RNS, which alter their signaling activities (Messina et al., 2019). In the case of insulin/growth factor signaling, ligand binding to the cognate receptor(s) stimulates production of O2●– by NOX enzymes that are closely juxta-positioned, with a resultant transient increase in H2O2 causing reversible inactivation of protein tyrosine phosphatases (e.g., PTP1B, PTPN2, and PTPN11) and the lipid phosphatase PTEN. Inactivation of these enzymes allows short-term de-repression of insulin/growth factor signaling and increased activity of various downstream mitogen-activated protein kinase (MAPK) enzymes, which leads to cell proliferation. Such inactivation of protein and lipid phosphatases occurs because H2O2 oxidizes their active-site cysteines to sulfenic, sulfinic, or S-glutathionylated forms (see Box 6), thereby inhibiting their activities until the oxidized active-site Cys is cycled back to its reduced form by the actions of TXN, TXNRD, SRXN, GRX, and NADPH (Figure 3). The duration and magnitude of the NOX-derived H2O2 redox signal is controlled by PRXN enzymes, which are themselves recruited to growth factor receptors and associated NOX shortly after receptor stimulation. Because the activity of PRXN can be suppressed by phosphorylation, and reactivated by phosphatases, they allow tight spatiotemporal control of redox signaling around growth factor receptors (Rhee et al., 2012).
Figure 3. Redox Regulation of PTP- and PTEN-Mediated Inhibition of Cell Proliferation and Survival.
Protein tyrosine phosphatases PTP1B, PTPN2, and PTPN11 and the lipid phosphatase PTEN suppress MAPK and PKB/Akt activity, as well as NF-κB signaling (bottom left). These phosphatases each possess an active-site Cys residue in a thiolate anion (S–) state that is susceptible to oxidation: as depicted across the center of the cartoon, the thiolate form of the active-site Cys can be oxidized to sulfenate (SOH), sulfinate (SO2H), or sulfonate (SO3H) states depending on the levels of H2O2 and duration of exposure to H2O2 (see Box 6). Alternatively, as shown at the top of the cartoon, the active-site Cys may form mixed disulfides by reacting with GSH (S-glutathionylated protein-SSG), which can be catalyzed by GST P1–1, or react with another thiol internally or in another protein to form a disulfide bridge (-S-S-). These oxidative modifications of the phosphatases result in their inactivation and therefore an increase in MAPK and PKB/Akt activity and NF-κB signaling. However, oxidative inactivation to sulfenate or sulfinate states can be reversed by the TXN1 or SRXN1 antioxidant systems (shown in green boxes), thereby allowing rescue of phosphatase activity and suppression of MAPK, PKB/Akt, and NF-κB activities. Similarly, active-site Cys that have been S-glutathionylated or have formed a disulfide bridge can be reversed by the GRX/GSH antioxidant system (green box), or de-glutathionylated by GST O1–1, thereby allowing recovery of phosphatase activity. Oxidation of the active-site Cys to a sulfonate state is irreversible (right), and the protein has to be eliminated.
For TRP channels, ROS and RNS activate TRPA1, TRPM2, TRPM7, TRPML1, and TRPML2, causing stimulation of Ca2+ signaling resulting in diverse responses, including chemosensitization/pain, inflammation, proliferation, cytoprotection, and cell death (Zhang et al., 2018b). TRPA1 is best understood in terms of its response to H2O2, 4-HNE, and 15d-PGJ2 (Andersson et al., 2008). Within human TRPA1, Cys-414, Cys-421, Cys-621, Cys-641, and Cys-665 contribute to regulation of the channel by electrophiles (Hinman et al., 2006; Macpherson et al., 2007). Cryoelectron microscopy has confirmed that Cys-621, Cys-641, and Cys-665 are highly reactive (Paulsen et al., 2015), but further experiments are required to determine how TRP channels are activated by ROS/RNS.
Oxidative Stress Activates Cell Death Pathways
ROS are potent stimulators of apoptosis and can activate the intrinsic mitochondrial pathway, the extrinsic death receptor pathway and the endoplasmic reticulum (ER) stress pathway (Redza-Dutordoir and Averill-Bates, 2016). In each of these instances, mitochondria are central to triggering apoptosis. ROS stimulate events that lead to loss of the inner mitochondrial membrane permeability and control of the mitochondrial permeability transition pore complex, disrupting membrane potential and resulting in release of cytochrome c and activation of caspase-3, −6, and −7 (Orrenius et al., 2015).
While under conditions of mild oxidative stress TP53 contributes to adaptation by inducing antioxidant genes, under more stringent conditions TP53 orchestrates apoptosis by stimulating production of ROS. TP53 target genes that stimulate apoptosis include PIG3 and PIG6, mitochondrial proline dehydrogenases that produce ROS indirectly by providing carbon for other mitochondrial dehydrogenases (Goncalves et al., 2014; Porte et al., 2009). Activation of TP53 by high levels of ROS induces genes for BAX, PUMA, and p66Shc, which perturb mitochondrial function and cause release of cytochrome c and increased ROS production (Liu and Xu, 2011). Also, when TP53 is activated under proapoptotic conditions, it represses the expression of SOD2 and several NRF2 target genes (Faraonio et al., 2006).
ROS can also trigger ferroptosis, an iron-dependent form of cell death, distinct from apoptosis, autophagy, and necrosis (Dixon et al., 2012), requiring redox cycling of Fe2+/Fe3+ that stimulates peroxidation of membrane phospholipids (Yang et al., 2014). Free intracellular redox-active iron allows increased ROS production via Fenton chemistry and/or increased lipoxygenase activity (Stockwell et al., 2017; Yang et al., 2016b). In turn, this results in oxidation of membrane phospholipid-polyunsaturated fatty acids, presumably altering membrane pores, integrity, or curvature (Dixon and Stockwell, 2019). Evidence suggests that GPX4 and maintenance of intracellular cysteine levels are each important in suppressing ferroptosis as the GPX4 inhibitor RSL-3, and the cystine/glutamate antiporter xCT inhibitor erastin, can markedly stimulate death in cancer cells harboring oncogenic RAS (Yang and Stockwell, 2008). Also, activation of the tumor suppressor p14ARF by ROS can stimulate ferroptosis by activating TP53 and suppressing the activity of NRF2, thereby downregulating SLC7A11 and diminishing xCT activity (Chen et al., 2017).
It is becoming increasingly apparent that redox signaling strongly influences apoptosis and ferroptosis (Benhar, 2020). Thus, the activities of the Fas receptor, BAX, cIAP, XIAP, caspase-9, and caspase-3 can be modified by S-nitrosylation, S-glutathionylation, S-persulfidation, or thiol oxidation. Ferroptosis is antagonized by ferroptosis suppressor protein 1 (FSP1) in a reaction in which ubiquinol (CoQH2) traps lipid peroxyl radicals and generates ubiquinone (CoQ), and FSP1 regenerates CoQH2 using NADPH. Future studies are required to determine the extent to which redox switches controlled by the GSH and/or TXN anti-oxidant systems suppress apoptosis and/or ferroptosis and the triggering of regulated cell death.
Oxidative Stress Increases Cancer Risk
It is recognized that chronic inflammation, with crosstalk between myeloid cell-derived ROS and tumor necrosis factor alpha (TNF-α)-mediated signaling, can lead to carcinogenesis (Canli et al., 2017). It is equally well known that sustained exposure to high ROS levels can damage DNA, and that a COSMIC mutation signature exists which is associated with oxidative DNA damage (Rose Li et al., 2020). The clearest evidence that ROS may increase the risk of cancer, when antioxidant defences are insufficient to provide protection against oxidative stress, comes from knockout of O2●–-scavenging enzymes. Thus, mice homozygous nulled for cytoplasmic Sod1, or heterozygous nulled for mitochondrial Sod2, exhibit evidence of severe oxidative damage and develop cancer spontaneously (Gill et al., 2016).
In the case of H2O2-scavenging enzymes, loss of certain genes encoding Prdx and selenium-dependent Gpx enzymes predispose to tumorigenesis. Specifically, Prdx1−/− and Prdx1+/− mice exhibit increased oxidative DNA damage, and the aging mutant mice exhibit a higher spontaneous incidence of hepatocellular carcinoma, fibrosarcoma, osteosarcoma, islet cell adenoma, lung adenocarcinoma, and breast adenocarcinoma than age-matched Prdx1+/+ mice (Neumann et al., 2003); by contrast, Prdx2−/−, Prdx4−/−, and Prdx6−/− mice do not spontaneously develop cancer (Hampton et al., 2018). Gpx1−/− and Gpx2−/− mice are healthy under normal laboratory conditions, but Gpx1/2 double knockout mice are susceptible to ileo-colitis upon weaning and develop ileal and colonic tumors at 6 months of age that are dependent on the presence of commensal microflora (Chu et al., 2004). Interestingly, Cat−/− mice show differential sensitivity to oxidants and peroxisomal function but do not suffer increased risk of cancer (Hwang et al., 2012).
Heightened production of HO● increases susceptibility to tumorigenesis by modifying DNA (Halliwell and Gutteridge, 2015). Consistent with this notion, patients with iron overload or hemochromatosis have a greater cancer risk (Torti et al., 2018). However, while ferritin-deficient mice have been reported, it is not known if iron-replete ferritin-knockout mice are more susceptible to cancer. In chronic inflammation, levels of ONOO– may increase substantially, which, following combination with CO2, leads to formation of nitrosoperoxycarbonate, which decomposes to CO3●– and ●NO2 and which in turn initiate selective oxidation and nitration of guanine in DNA, leading to guanine-thymidine crosslinks (Shafirovich and Geacintov, 2017).
As increases in steady-state levels of ROS are associated with heightened risk of tumorigenesis, it could be extrapolated that chronic loss of antioxidant status is associated with a similar risk. Evaluating a role for GSH in modulating the risk of carcinogenesis is confounded by the fact that knockout of Gclc or Gss is embryonically lethal. By contrast, knockout of Gclm has a less profound effect on GSH, with levels in liver, kidney, pancreas, and erythrocytes of mutant mice being ~15% of that in wild type (Yang et al., 2002). While fibroblasts prepared from Gclm−/− mice show increased amounts of ROS and DNA damage that is accompanied by upregulation of Tp53 and p21 (Chen et al., 2009), these mice do not develop tumors spontaneously.
Responses to Episodes of Oxidative Stress in Premalignant and Malignant Cells during the Multiple Stages of Tumor Development
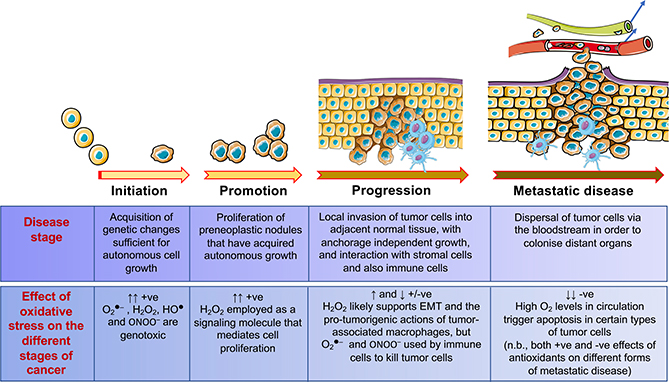
While ROS/RNS can initiate carcinogenesis, they support the proliferation of initiated cells during the promotion and progression stages of tumorigenesis (Figure 4). However, oxidative stress can stimulate senescence or apoptosis in neoplastic cells during all stages of tumorigenesis, including anchorage-independent growth and metastasis. It is also recognized that GSH levels and TXN and/or TXNRD are commonly increased in many cancer cells, presumably to combat the high ROS burden, and that this is associated with poor prognosis (Benhar et al., 2016).
Figure 4. An Ambiguous Role for Oxidative Stress in Tumorigenesis.
The cartoon depicts development of malignant disease from initiation through promotion and progression, until it acquires a highly malignant, invasive and metastatic phenotype. The contributions that excess levels of O2●–, H2O2, HO●, and ONOO– may make to the different stages of the disease are indicated in panel at the bottom.
ROS Production Is Heightened in Malignant Disease
Cancer cells contain higher levels of ROS than normal cells, and this sustains the phenotype. Sources of increased ROS production in tumor cells have been attributed to oncogene- and/or damage-stimulated production of (1) O2●– by mitochondria, associated with altered assembly of the electron transport chain, hypoxia, or anchorage-independent growth; (2) O2●– by NOX, associated with hyperproliferation or centrosome abnormalities; (3) H2O2 by 5-lipoxygenase, associated with cell mobility; and (4) H2O2 within the ER, associated with protein folding (Arnandis et al., 2018; Raimondi et al., 2020; Reczek et al., 2017; Zhou et al., 2014). Examples of oncogenes that increase ROS production include: (1) Ras that alters mitochondrial metabolism, decreases mitochondrial membrane potential, and activates NOX2 and/or NOX4; (2) Ras-related C3 botulinum toxin substrate 1 (Rac1) that activates NOX1; (3) STAT3 that alters mitochondrial metabolism and activates NOX4; (4) B cell lymphoma 2 (BCL-2) that alters mitochondrial function; and (5) MYC that downregulates PGC-1α and suppresses mitochondrial biogenesis (Chong et al., 2018; Igelmann et al., 2019; Liou et al., 2016; Marcar et al., 2019; Satoh et al., 2017). Increased ROS levels in tumor cells may also result from loss of tumor suppressor-mediated regulation of antioxidant genes, such as decreased expression of SOD2, GPX1, SESN1, and SESN2 following inactivation of TP53 (Gorrini et al., 2013), or by post-translational modifications, exemplified by acetylation of SOD2 (He et al., 2019), which confers upon antioxidant enzymes prooxidant properties. In addition, cancer cells may be stimulated to produce ROS by TNF-α secreted by immune cells, or they may encounter ROS generated by immune cells recruited to the tumor.
The increase in ROS in cancer cells stimulates proliferation and cell survival by blunting the activities of PTPs, PTEN (Figure 3), and MAPK phosphatases, thereby augmenting MAPK-ERK, PI3K-Akt, and PKD-NF-κB signaling cascades in a cell-specific manner (Moloney and Cotter, 2018). To support the proliferative benefits of high ROS levels while mitigating the risk of senescence/apoptosis, tumor cells upregulate antioxidant transcription factors and/or reprogram metabolism to increase NADPH and de novo synthesis of GSH by a variety of means (see below).
Besides augmenting antioxidant systems to ameliorate oxidative stress, tumor cells may stimulate anti-apoptotic and pro-survival pathways. For example, breast cancer cells can utilize the redox-sensitive TRPA1 channel to activate Ca2+ signaling and so stimulate ERK and PI3K-PKB/Akt pathways, which in turn activate MCL-1 contributing to oxidative stress tolerance and drug resistance; moreover, NRF2 regulates expression of TRPA1, reinforcing the link between redox homeostasis and Ca2+ signaling (Takahashi et al., 2018), and extending the influence of NRF2 beyond redox. Perhaps also relevant, downstream of these events, clusters of proteins that participate in Ca2+ homeostasis are particularly sensitive to S-glutathionylation (Grek et al., 2013).
ROS Production in Premalignant Cells Mandates Redox Adjustments Controlled by NRF2
The view that ROS are necessary for tumor promotion is supported by the report that their pharmacological suppression using N-acetylcysteine (NAC) or apocynin, attenuates lung adenocarcinoma in an oncogenic K-RASG12D-driven mouse model in which lung-specific knockout of Ikkα resulted in increased Nox2 expression and downregulation of Nrf2 (Song et al., 2018). Similarly, in an oncogenic B-RAFV600E-driven mouse melanoma model, knockout of Klf9 inhibited premalignant melanocyte hyperplasia because ROS production was diminished (Bagati et al., 2019), which is noteworthy because KLF9 suppresses NRF2 (Zucker et al., 2014). It is equally apparent that, for malignant lesions to develop, levels of ROS should not exceed an upper ceiling. Thus, loss of antioxidant capacity as a consequence of knockout of Tigar, which impairs generation of NADPH, decreases development of premalignant pancreatic intraepithelial neoplasia (PanIN) lesions in a variety of K-RASG12D-based mouse cancer models, including Trp53−/+ and Trp53R172H (Cheung et al., 2020).
Recognition that tumorigenesis requires maintenance of high intracellular ROS levels raises questions about how cells harboring mutations in oncogenes or tumor suppressor genes adjust their redox homeostasis so that the pro-tumorigenic effects of ROS are not quenched, while at the same time not succumbing to senescence or apoptosis. The Solt-Farber rat carcinogenesis model (induced by intraperitoneal [i.p.] treatments with diethylnitrosamine [DEN] to initiate carcinogenesis, followed 2 weeks later by two-thirds partial hepatectomy and dietary administration of 2-acetylaminofluorene to select growth of initiated cells) enables changes that accompany the different stages of cancer to be analyzed, with the appearance in rat liver of physically discernible preneoplastic nodules, comprising as few as 20 hepatocytes, before the appearance of preneoplastic nodules/adenomas, which in turn precede formation of hepatocellular carcinomas (Farber, 1984). Gene expression profiling revealed that GCLC and GSTP1 are induced in early rat liver preneoplastic foci obtained 4 weeks after initiation, and that robust induction of AKR7A1, GCLC, GCLM, GGT, GPX2, GSR, GSS, GSTA5, NQO1, TXNRD1, and UGT1A6 occurs in persistent nodules obtained 10–20 weeks after initiation (Petrelli et al., 2014). Besides induction of GCLC and GSTP1, early nodules increase both glycolysis and the PPP by augmenting activities/protein levels of glucose transporter 1 GLUT1 (SLC2A1), hexokinase II (HK2), monocarboxylate transporter 4 (MCT4, SLC16A3, for lactate), G6PD, and TIGAR, while at the same time suppressing OXPHOS by inducing the mitochondrial chaperone tumor necrosis factor receptor-associated protein 1 that binds complex II and IV of the electron transport chain and inhibits succinate dehydrogenase (Kowalik et al., 2016). Collectively, these findings suggest that antioxidant systems are upregulated and metabolism reconfigured in response to increases in ROS during the promotion and early progression stages of carcinogenesis. Interestingly, the changes in metabolism in rat liver preneoplastic nodules broadly resemble the biochemical responses to acute oxidative stress in Fh1loxP/loxP mouse kidney epithelial cells (van der Reest et al., 2018).
Many of the antioxidant genes upregulated in rat liver preneoplastic nodules are NRF2 target genes, suggesting that NRF2 mediates adaptation to oncogene-stimulated oxidative stress. This interpretation is supported by the fact that NRF2-null rats subject to a modified Solt-Farber protocol (i.e., DEN followed by a choline-devoid and methionine-deficient diet) do not form preneoplastic nodules (Orru et al., 2018). The mechanism by which NRF2 transactivation activity is increased in preneoplastic nodules provides an insight into the severity of stress that initiated cells have to withstand. Thus, while NRF2 can be upregulated either by ROS inhibiting its repressor KEAP1, an E3 ubiquitin ligase substrate adaptor that controls NRF2 protein stability (Suzuki et al., 2019), or by oncogenes transcriptionally increasing NRF2 mRNA production (DeNicola et al., 2011), neither of these mechanisms are of principal importance in hepatic preneoplastic nodules. Rather, these nodules frequently contain gain-of-function mutations in NFE2L2 (Zavattari et al., 2015), indicating that permanent activation of NRF2 is necessary to allow initiated cells to survive and proliferate, rather than succumb to oxidative stress-driven apoptosis (Figure 5A). Most remarkably, it was estimated that approximately 70% of early preneoplastic nodules harbored “hotspot” mutations in NFE2L2 encoding amino acids within NRF2 that physically engage with the Kelch-repeat domain of KEAP1 and are necessary for ubiquitylation of the transcription factor by CRLKEAP1 (Zavattari et al., 2015). Somatic mutations in NFE2L2 were also observed in approximately 60% of early and advanced hepatocellular carcinomas, respectively, suggesting that genetic activation of NRF2 is required throughout promotion and progression of liver tumorigenesis.
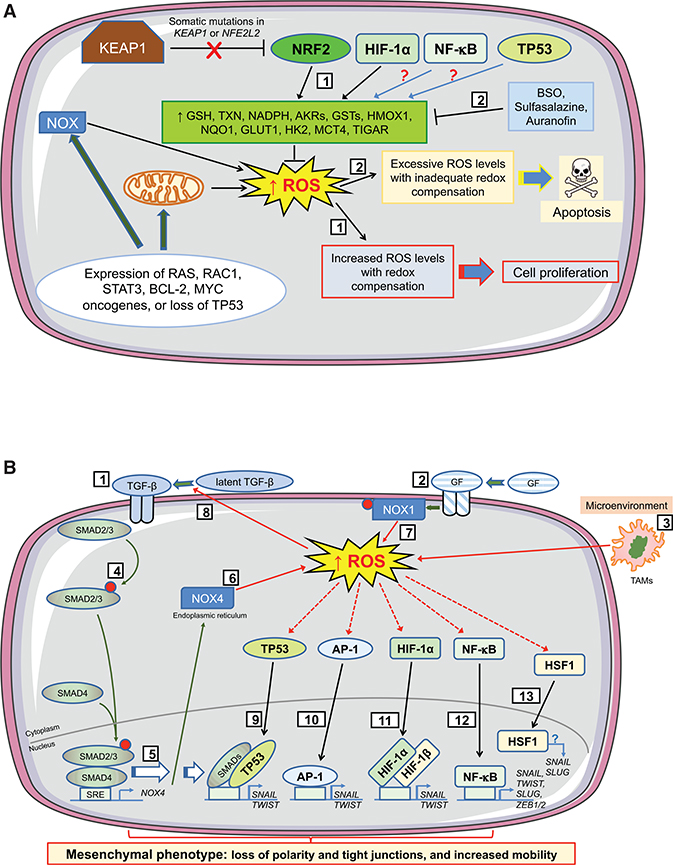
Figure 5. Influence of Oxidative Stress on Cell Fate during Early Stages of Tumorigenesis.
The cartoons depict how levels of ROS stimulate proliferation or apoptosis in preneoplastic cells during initiation of tumorigenesis, and how ROS support EMT during progression of tumorigenesis by altering TGF-β signaling and by activation of antioxidant transcription factors that control expression of EMT-TFs. During the earliest stages of tumorigenesis (A), activation of oncogenes, coupled with higher metabolic demands, results in an increase in intracellular ROS levels in early neoplastic lesions/adenomas. To benefit from the proliferative advantages associated with the increase in ROS, without succumbing to apoptosis, cells harboring activated oncogenes augment their antioxidant capacity by increasing transactivation of genes encoding GSH- and TXN-dependent enzymes along with antioxidant/detoxification enzymes (1). Often this readjustment of redox entails loss or blunting of repression of NRF2 by KEAP1 and induction of NRF2 target genes. In addition, HIF-1α increases expression of key metabolic proteins, such as GLUT1, HK2, and MCT4. Also, NF-κB and TP53 probably contribute to this adaptive process, although in lesions harboring mutant TP53 the latter is unlikely. Treatment of animals with BSO, which inhibits synthesis of GSH, before initiation of carcinogenesis will stimulate apoptosis of premalignant cells (2). However, once carcinogenesis has been initiated, stimulation of apoptosis in malignant cells requires inhibition of both the GSH-based (by BSO) and the TXN-based (by sulfasalazine or auranofin) antioxidant systems. During the progression stage of tumorigenesis (B), EMT is triggered by a variety of environmental factors, including those that alter intracellular redox. In this regard, TGF-β signaling (1), growth factor signaling (2), and tumor-associated macrophages (TAMs) in the microenvironment (3) will produce ROS. Binding of TGF-β to its cognate receptor causes phosphorylation of SMAD2/3 (4) and induction of NOX4 gene expression (5), which results in production of H2O2 at the ER (6). NOX4-generated ROS within the tumor cell is augmented by growth factor signaling causing phosphorylation and activation of NOX1 and production of O2●– at the plasma membrane (7), with increased ROS increasing processing of latent TGF-β (8). The increases in ROS from TGF-β and growth factor signaling, along with those generated by TAMs, activate TP53, which combines with SMAD proteins to induce transcription of genes encoding the EMT-TFs SNAIL and TWIST (9). Similarly, increased ROS levels activate AP-1 (10) and HIF-1α (11) and induce SNAIL and TWIST, whereas the activation by ROS of NF-κB (12) leads to induction of genes encoding SNAIL, TWIST, SLUG, ZEB1, and ZEB2. Together, SNAIL, TWIST, SLUG, ZEB1, and ZEB2 positively control expression of mesenchymal-associated genes and negatively control expression of epithelial-associated genes. ROS also activate HSF1 (13), whereas its downregulation decreases TGF-β-mediated expression of SNAIL and SLUG and inhibits EMT, although the precise mechanism(s) is not understood.
The hypothesis that formation of hepatic preneoplastic nodules in the rat requires NRF2 accords with mouse experiments in which livers from wild-type animals given a single i.p. dose of DEN all contained tumors after 9 months, compared with none from Nrf2-null mice (Ngo et al., 2017). This dependence on NRF2 also seems true of a number of other types of cancer. For example, the incidence of pancreatic cancer was markedly reduced in Nrf2-null mice relative to wild-type mice in a K-RasG12D-driven cancer model (DeNicola et al., 2011), and the incidence of lung tumors in Nrf2-null mice was also modestly reduced relative to wild-type mice in urethane-initiated cancer (Bauer et al., 2011; Satoh et al., 2013). Similarly, knockout of Keap1 in mice, leading to hyperactivation of Nrf2, accelerated cholangiocarcinoma driven by K-RasG12D and Trp53R172H (Nabeshima et al., 2020).
It is striking that the high frequency of somatic mutations in NFE2L2 observed in both rat liver preneoplastic nodules (Zavattari et al., 2015) and murine hepatoma (Ngo et al., 2017), which result in constitutive activation of NRF2, are clinically relevant as NFE2L2 or KEAP1 are frequently mutated in human tumors. In this context, it is imperative to recognize that there is no evidence that constitutive activation of NRF2, as for example observed in the Keap1FA/− mouse (in which the Keap1FA allele is hypomorphic [Taguchi et al., 2010]), is sufficient to trigger spontaneous tumorigenesis. Studies of Keap1FA/− mice have revealed that constitutive upregulation of Nrf2 increases proliferation of forestomach epithelium, but not cancer (Mitsuishi et al., 2012), and that Nrf2-mediated maximal transactivation of G6pd, Pgd, and Me1 requires activation of the PI3K-Akt signaling pathway, which likely results in inhibition of GSK-3 and the failure to form the phosphodegron in the Neh6 domain of NRF2 that is recognized by β-TrCP (Chowdhry et al., 2013). Moreover, constitutive activation of NRF2, by deleting its Neh2 KEAP1-interaction domain does not increase the rate of primary tumor formation in an MMTV-rtTA:TetO-Her2 mouse cancer model (Fox et al., 2020). Collectively, these results imply that, subsequent to a cancer initiating mutation(s), dysregulation of the NRF2-KEAP1 axis is a selective event under conditions of chronic oxidative stress, and so NRF2 activation per se is not sufficient to stimulate cancer initiation.
NRF2-Independent Antioxidant Mechanisms Can Also Support Early Tumorigenesis
The dependence on NRF2 for liver, pancreatic, and lung cancer observed in rodents may not be true for other malignancies. For example, the incidence of tumors in Nrf2-null mice was not diminished relative to wild-type mice in benzo[a]pyrene-initiated gastric cancer (Ramos-Gomez et al., 2001), 7,12-dimethylbenz [a]anthracene-initiated and 12-O-tetradecanoylphorbol-13-acetate-promoted skin cancer (Xu et al., 2006), or UV-initiated skin cancer (Knatko et al., 2015). In these cases, other members of the antioxidant transcription factor network may contribute to the putative resetting of redox homeostasis in early adenomas. Of possible relevance, NRF2-independent metabolic reprogramming occurs in rat preneoplastic nodules, as demonstrated by upregulation of GLUT1, HK2, MCT4, and TIGAR in the lesions (Kowalik et al., 2016): HIF-1α may be responsible for upregulation of GLUT1, HK2, and MCT4; NF-κB may be responsible for upregulation of GLUT1; TP53 may be responsible for upregulation of TIGAR.
In instances where NRF2 is not constitutively activated during the initiation of tumorigenesis, oncogenic tyrosine kinases may reprogram metabolism by phosphorylating the cancer-specific PKM2 isoform. In this scenario, phosphorylation of PKM2 causes it to adopt a dimeric rather than tetrameric quaternary structure, resulting in loss of pyruvate kinase activity and a relative block in the final steps of glycolysis (Li et al., 2014). In turn, spill-over of accumulated glucose-6-phosphate into the PPP increases NADPH generation by G6PDH, which enhances GSH- and TXN-based antioxidant defences (as indicated in Figure 1). Loss of pyruvate kinase activity also aids GSH synthesis. Specifically, increases in glyceraldehyde-3-phosphate, resulting from phosphorylation of PKM2, can be funneled through the serine synthesis pathway, with the resulting serine being available to form glycine (from serine by serine hydroxymethyltransferase within the one-carbon pathway) and cysteine (from cystathionine by cystathionine lyase within the trans-sulfuration pathway), two of the amino acids used to synthesize GSH (Yang and Vousden, 2016); the supply of cysteine can be augmented by the cystine/glutamate antiporter SLC7A11, the expression of which can be increased by oncogenic Ras (Lim et al., 2019). Besides glycine and cysteine, GSH biosynthesis requires a supply of glutamate, and this can be provided by the combined actions of the transporters ASCT2/SLC1A5 and SNAT2/SLC38A2, which import glutamine (Broer et al., 2019; Morotti et al., 2019) along with glutaminase, which converts glutamine to glutamate (Mates et al., 2020). Importantly, both the glutamine transporters and glutaminase are upregulated in tumor cells through oncogene activation or hypoxia (Bott et al., 2015; Lukey et al., 2016; Morotti et al., 2019).
Among 29 cancer types, significant positive selections for mutations in NFE2L2 and KEAP1 occur in bladder, endometrium, esophagus, head-neck, liver, and lung (Martincorena et al., 2017). Somatic mutations in NFE2L2 or KEAP1 are not, however, universal features of malignant disease, and so other mechanisms of NRF2 activation as well as NRF2-independent antioxidant systems likely contribute to adaptation to oxidative stress that accompanies tumorigenesis in certain types of cancer. Possibly, NRF2 is only genetically upregulated when certain oncogenes are activated or tumor suppressor genes lost.
Supply of Exogenous Antioxidants and Diminution of Endogenous Antioxidants Modulates Early Tumorigenesis
Using Cre-inducible Kras2LSL- and BrafCA-based lung cancer models, it has been found that administration of NAC in the drinking water, or vitamin E in the diet, 1 week after initiation of tumorigenesis by expressing K-RASG12D or B-RAFV600E, significantly increased tumor burden (Sayin et al., 2014). This was attributed to the ability of NAC and vitamin E to diminish oxidative stress, decrease DNA damage and blunt activation of wild-type TP53. However, it is noteworthy that many genes that were downregulated by the NAC and vitamin E treatments are NRF2 transcriptional targets, and the extent to which these antioxidants influenced the activity of other members of the antioxidant transcription factor network was not investigated. Nevertheless, it is interesting that NAC and vitamin E both support lung tumorigenesis, suggesting that maintenance of redox status throughout the cell is important during early tumor progression.
Experiments using the MMTV-PyMT spontaneous mammary tumor mouse model showed that in order for disease to proceed, cancer cells require GSH to maintain their appropriate redox status during the early stages of tumorigenesis, although this does not necessarily involve NRF2. Thus, immediately after weaning, administration of the glutamate-cysteine ligase (GCL) inhibitor buthionine-(S,R)-sulfoximine (BSO) to MMTV-PyMT mice greatly delayed tumor onset, suggesting that a critical threshold of GSH is required to support initiation of cancer (Harris et al., 2015). By contrast, when BSO was administered to MMTV-PyMT mice once mammary tumors had become physically evident, it was unable to delay tumor progression. Failure of BSO to inhibit later tumor progression is likely a consequence of malignant transformation causing overexpression of both CD44 and TXNRD1, which in turn allowed tumor cells to compensate for loss of GSH by increasing cystine uptake via xCT and by better utilizing TXN. Notably, when cancer cells were treated with a combination of BSO plus the xCT inhibitor sulfasalazine, or a combination of BSO plus the TXNRD1 inhibitor auranofin, a striking increase in cell death was observed, dependent on ROS. After tumors were established, treatment of MMTV-PyMT mice with BSO plus sulfasalazine markedly reduced tumor growth in vivo. Together, these results suggest that, during initiation of cancer, the GSH antioxidant system is principally responsible for attenuation of ROS that might trigger cell death, whereas after initiation, GSH and TXN synergistically support tumor cell survival. While NRF2 controls GSH homeostasis, other members of the antioxidant transcription factor network may also contribute to maintaining high GSH and TXN levels (see Table 1).
Influence of ROS and RNS on the Tumor Microenvironment during Progression of Disease
Cancer progression requires shaping the tumor microenvironment (TME). This involves reciprocal crosstalk between neoplastic cells and the TME, which includes ROS/RNS. The functions of cancer-associated fibroblasts (CAFs), tumor-associated macrophages (TAMs), and T cells are all affected by ROS/RNS within the TME. Thus, both CAFs and TAMs “cooperate” with neoplastic cells by contributing to a developmental program called EMT by helping to remodel the extracellular matrix (ECM), thereby stimulating tumor cell proliferation, tumor angiogenesis, immunosuppression and tumor invasion (Nieto et al., 2016). In addition, regulatory T (Treg) cells, which are critical for preventing autoimmunity, suppress effective tumor immunity; the presence of intratumoral Treg cells is often associated with poor prognosis (Beyer and Schultze, 2006). The TME also harbors specific cytotoxic CD8+ T cells. Although expected to support the immune destruction of tumor cells, CD8+ T cells often express co-inhibitory receptors, such as programmed death-1 (PD-1), and are considered terminally differentiated or “exhausted.” Conversely, the recently identified subset of stem cell-like tumor-infiltrating PD-1+ TCF1+ CD8+T cells contribute to limiting tumor progression in response to immunotherapy (Held et al., 2019).
In terms of the effects of ROS in the TME, chronic oxidative stress resulting from JunD inactivation has been shown to promote myofibroblast differentiation in stroma associated with mammary adenocarcinomas through activation of HIF-1α and increased production of the CXCL12 chemokine, which together stimulate tumor growth and vascular remodeling, and shorten survival (Toullec et al., 2010). Therefore, it is of interest that, in stromal remodeling associated with progression of prostate cancer, transforming growth factor β (TGF-β)-stimulated fibroblast-to-myofibroblast differentiation is preceded by induction of NOX4, increased production of ROS, and phosphorylation of JNK, and is accompanied by downregulation of the selenoproteins GPX3 and TXNRD1 (Sampson et al., 2011). By contrast, selenium supplementation decreases ROS levels and inhibits fibroblast differentiation into myofibroblasts, suggesting that ROS drive trans-differentiation of stromal cells. In addition, cancer cell-derived H2O2 causes metabolic changes in CAFs, including increased glucose uptake, decreased mitochondrial activity, and increased ROS production, whereas the presence of CAFs causes reciprocal metabolic changes in the adjacent cancer cells, such as decreased glucose uptake and increased mitochondrial activity (Martinez-Outschoorn et al., 2011). These metabolic alterations can be abrogated by addition of CAT, implicating H2O2 as the signaling molecule.
ROS also contribute to the pro-tumorigenic, anti-inflammatory and immunosuppressive properties of TAMs that favor tumor progression. In malignant melanoma, production by TAMs of mitochondrial ROS stimulates MAPK/ERK activity leading to secretion of TNF-α, which promotes tumor cell invasion (Lin et al., 2013). When exposed to cell-free tumor fluid, peritoneal macrophages that exist in the classically active pro-inflammatory (M1) phenotype adopt an alternatively activated (M2) phenotype associated with immunosuppression (Ghosh et al., 2015). Furthermore, ROS and RNS produced by TAMs suppress T cell functions (Ghosh et al., 2015; Hamilton et al., 2014). It has been shown that, in mice, O2●– generated by Nox2 stimulates Treg cells to dampen T cell-mediated inflammation (Kraaij et al., 2010). It is not clear if this process is recapitulated in TAMs, but if so it would further contribute to TAM-mediated immunosuppression. Notably, using oncogene-initiated zebrafish models of glioma, it was shown that TAMs engage in ATP-mediated interactions with preneoplastic cells at a very early stage of tumor development, and that depletion of TAMs or reducing the number of these interactions significantly impairs the proliferation of neoplastic cells (Chia et al., 2018, 2019). Whether or not ROS play a role in these interactions is currently unknown, although this is a distinct possibility considering that oncogene activation stimulates ROS production.
Myeloid-derived suppressor cells (MDSCs) induce antigen-specific CD8+ T cell tolerance, which constitutes a major mechanism of tumor escape from immune surveillance. Tumor-infiltrating MDSCs produce ONOO–, which nitrates Tyr residues in the T cell receptor-CD8 complex, thus disrupting binding of specific peptide-major histocompatibility complex (pMHC) dimers to the CD8+ T cell (Nagaraj et al., 2007); critically, use of a ONOO– scavenger abolished MDSC-induced T cell tolerance. Treatment of cancer cells with ONOO– inhibited binding of processed peptides to cancer cell-associated MHC, resulting in resistance to antigen-specific cytotoxic T cells, whereas inhibition of ONOO– production improved immunotherapy (Lu et al., 2011), implicating ONOO– as a critical regulator of the effects of cytotoxic T cells. Overall, in myeloid cells, mitochondria, NOX, arginase-1, and NOS2 all contribute to ROS production, where, in addition to O2●–, cooperation between arginase-1 and NOS2 results in the formation of ONOO–. The combined effect of ROS and RNS leads to T cell suppression, tolerance, and resistance to cytotoxic T cells.
In T cells, ROS play a dual role. Mitochondrial ROS are essential for T cell activation (Kaminski et al., 2012; Sena et al., 2013), but ROS in the TME can lead to T cell hypo-responsiveness (Cemerski et al., 2002). In tumor-infiltrating T cells, which are functionally impaired, mitochondrial activity is compromised (Scharping et al., 2016; Siska et al., 2017), but can be rescued by increasing mitochondrial biogenesis (Scharping et al., 2016), using mitochondrial ROS scavengers (Siska et al., 2017), or by overexpression of CAT (Ligtenberg et al., 2016). Critically, these manipulations also restore the antitumor activity of T cells, further demonstrating the importance of mitochondrial activity and balanced ROS production for T cell function. Interestingly, enhanced mitochondrial activity and ROS production synergize with the tumoricidal activity of PD-1 blockade by expansion of effector/memory T cells (Chamoto et al., 2017). In addition, production of ONOO– in the TME inhibits T cell migration into the tumor by nitrating and inactivating the chemokine CCL2 (Molon et al., 2011), which is a chemoattractant for myeloid cells, activated T cells, and natural killer cells.
Although information is limited, it appears that, unlike most other immune cells in the TME, neutrophils preserve their antitumor activity under oxidative stress conditions, which could be attributed to their high ROS-producing potential. Thus, tumor-associated neutrophils have been shown to suppress the expansion of the pro-tumorigenic IL-17+ γδ T cells via NOX2-mediated O2●– production (Mensurado et al., 2018). Moreover, IL-17-producing γδ T cells have lower GSH levels than their IFN-γ-producing γδ counterparts, providing an explanation for the exquisite sensitivity of IL-17+ γδ T cells to O2●– and H2O2. Importantly, Vδ1+ γδ T cells, the main γδ T cell subset that produces IL-17 in human tumors, also have low GSH levels in comparison with other human T cell subsets, and are sensitive to ROS, suggesting the clinical relevance of these findings.
These findings indicate that increases in ROS/RNS stimulate changes in the TME that support tumorigenesis by altering the functions of CAFs and TAMs, and at the same time they stimulate changes in T cells that might suppress immune responses to tumor cells.
EMT during Cancer Progression Entails Marked Changes in Redox Status and Tumor Cell Heterogeneity
For early neoplastic cells to progress to malignant carcinomas, they typically exploit EMT in order to increase mobility and invade adjacent stroma (Nieto et al., 2016), which can confer tumor-initiating potential and cancer stem cell properties, which enables them to form new foci when seeded elsewhere (San Juan et al., 2019). Activation of EMT is dictated by the tissue in which the neoplastic cell originated, as well as by CAFs and TAMs in their microenvironment (El-Kenawi et al., 2019), and thus provides major roles for Wnt, TGF-β, and Notch ligands, along with various growth factors (Chaffer et al., 2016) and ROS (Jiang et al., 2017), in initiating the program. It is a highly coordinated process during which early epithelial-like neoplastic cells acquire mesenchymal-like features, accompanied by restructuring of the composition of ECM proteins (Dongre and Weinberg, 2019).
Pioneering studies by Mina Bissell and colleagues revealed that initiation of EMT in murine SCp2 mammary epithelial cells upon exposure to MMP-3 involved expression of an alternatively spliced form of Rac1, called Rac1b, which by stimulating mitochondrial ROS production caused induction of vimentin, increased cell migration, and genomic instability (Radisky et al., 2005). These workers also discovered that the ability of MMP-3 to initiate EMT in SCp2 cells could be blocked by treatment with the antioxidant NAC or by ectopic expression of SOD2, but not by ectopic SOD1 or CAT, results suggesting that mitochondrially produced ROS are required during the very early stages of EMT. It is likely that SOD2 contributes to EMT, insofar as when it is overexpressed in MCF-7 cells (which exhibit an epithelial-like phenotype) it becomes acetylated on Lys-68, probably catalyzed by the GCN5L1 acetyltransferase, with the resulting post-translationally modified enzyme displaying pro-oxidant rather than antioxidant activity (He et al., 2019); it is currently envisaged that SOD2K68Ac increases ROS production, which in turn activates HIF-2α, producing a cancer stem cell-like phenotype that includes expression of Oct4 and Nanog.
EMT in neoplastic cells is directed by the master regulator EMT-activating transcription factors (EMT-TFs) SNAIL (also called SNAI1), SLUG (also called SNAI2), TWIST1, TWIST2, ZEB1, and ZEB2, which are considered key drivers of tumor progression as they transactivate genes associated with a mesenchymal state and repress genes associated with an epithelial state (Stemmler et al., 2019). Interestingly, EMT-TFs differ in their relative abilities to induce mesenchymal-associated genes and repress epithelial-associated genes. In some settings, SNAIL and ZEB1 only weakly induce mesenchymal-associated genes, while strongly repressing epithelial-associated genes, whereas TWIST1 exerts the opposite effects (Chaffer et al., 2016). Expression of SNAIL, SLUG, TWIST1, ZEB1, and ZEB2 is subject to complex control mechanisms, including transcriptional regulation by AP1, HIF-1α, NF-κB, and TP53 (Figure 5B), a fact that implies that the ability of EMT-TFs to induce mesenchymal-related genes and repress epithelial-related genes is controlled in part by redox signaling (Jiang et al., 2017); moreover, HSF1, which is also redox regulated, supports EMT (Powell et al., 2016; Xi et al., 2012). Consistent with the notion that redox influences EMT, this process can be initiated in SCp2 cells by MMP-3, which elicits an Rac1b-stimulated production of ROS that leads to activation of NF-κB, and in turn the upregulation of SNAIL (Cichon and Radisky, 2014).
As mentioned above, TGF-β regulates EMT-TFs in transformed cells and it is inextricably interrelated with redox. In this situation, binding of TGF-β to its cell-surface receptor, TGF-β receptor type 1 (TGFβR1), or TGFβR2, causes phosphorylation of SMAD2 and SMAD3 that, in turn, trimerize with SMAD4 and, following nuclear translocation, positively regulate SNAIL, SLUG, TWIST1, and ZEB1 (Massague, 2012). Induction of EMT-TFs by TGF-β signaling is heightened considerably by input from the RAS-MAPK pathway (David and Massague, 2018). Critically, from a redox perspective, activation of TGF-β signaling induces NOX4 and represses CAT, GCLC, GCLM, GPX3, and TXNRD1 (Liu et al., 2012; Sampson et al., 2011), with the resulting increase in ROS stimulating trans-differentiation and cell proliferation.
While ROS are necessary to initiate EMT, it is unclear to what extent they are required to maintain a mesenchymal phenotype. Consistent with ROS playing a role in initiation of EMT, treatment with the antioxidant NAC can antagonize stimulation of EMT by TGF-β in various cell types, including murine AML12 hepatocytes (Kim et al., 2019), human ARPE-19 adult retinal pigment epithelial cells (Yang et al., 2020), and human HCC4006 lung epithelial cells (Haley et al., 2019). Possibly more surprising is that, in a stably gefitinib-resistant human PC-9 non-small cell lung cancer cell line, treatment with NAC has been reported to increase an epithelial-phenotype and decrease the mesenchymal phenotype (Li et al., 2020), suggesting that a relative absence of ROS may stimulate mesenchymal-to-epithelial transition (MET). In contrast, oxidative stress in murine B16F10 melanoma cells, caused by inhibition of GSR1, has been reported to decrease EMT (Li et al., 2018), and in oral squamous cell carcinomas in which Aurora kinase A (AURKA) is aberrantly expressed, knockdown of AURKA increased ROS levels and inhibited EMT (Dawei et al., 2018). Collectively, these data suggest that the influence of ROS on a mesenchymal phenotype is dose and time dependent, with thresholds controlling the triggering of senescence/apoptosis being of critical importance.
One important question is whether oxidative stress contributes to the maintenance of the cancer mesenchymal cell phenotype. The fact that EMT is not a binary process, and is reversible (Chaffer et al., 2016), represents a confounding factor when interpreting the literature. Comparisons between epithelial-like human MCF-7 breast cancer cells and mesenchymal-like human MDA-MB-231 breast cancer cells revealed that the latter produce substantially higher levels of H2O2 than the former (Lunetti et al., 2019). Moreover, HCC4006, A549, and H538 non-small cell lung cancer cell lines treated with TGF-β to induce a mesenchymal phenotype produce substantially higher levels of ROS than their non-treated epithelial counterparts (Haley et al., 2019). By contrast, breast cancer stem cells (BCSCs) with an epithelial-type morphology (i.e., E-BCSCs) have been reported to generate high levels of ROS, exhibit metabolic flexibility, and possess heightened NRF2-regulated antioxidant defences necessary for survival, whereas BCSCs with a mesenchymal-type morphology (i.e., M-BCSCs) have low ROS levels, are dependent on glycolysis for energy, do not have high antioxidant defences, and do not require NRF2 for survival (Luo et al., 2018). By targeting these biochemical differences, it was found that M-BCSCs convert to E-BCSCs upon treatment with the glycolysis inhibitor 2-deoxyglucose (2-DG), which also increases ROS levels; the triggering of MET in M-BCSCs by 2-DG is thought to occur by activation of the AMPK-HIF-1α axis along with NRF2 (Luo et al., 2018). Remarkably, treatment of M-BCSCs with NAC antagonized the ability of 2-DG to stimulate MET, while treatment of E-BCSCs with 2-DG and the TXNRD inhibitor auranofin diminished their proliferation and/or self-renewal. Also, knockdown of NRF2 decreased E-BCSC numbers but had no effect on the number of M-BCSCs.
The above findings indicate that the effects of ROS on EMT are variable and probably reflect cancer cell-type specific and/or TME-dependent differences. It is plausible that this variability reflects heterogeneity in tumor cells, caused by factors controlling transition between epithelial and mesenchymal states. In this context, it is notable that NRF2 supports epithelial and hybrid epithelial/mesenchymal phenotypes, and knockout of NRF2 allows a mesenchymal phenotype to become established (Bocci et al., 2019; Zhou et al., 2016), although the degree to which this represents cause and effect is uncertain.
Adaptation to Oxidative Stress during Anchorage-Independent Growth
During the first stage of the metastatic cascade (see Nieto et al., 2016), resistance of tumor cells to death triggered by their detachment from the ECM and loss of integrin-mediated signaling, principally anoikis, is necessary for progression of tumorigenesis. Most importantly, ECM detachment results in catastrophic metabolic changes, including defective glucose uptake, attenuated PPP flux, and reduced ATP levels (Hawk and Schafer, 2018), which limits the metabolic flexibility of detached cells and renders them susceptible to oxidative stress. Indeed, detachment of non-transformed human MCF-10A breast epithelial cells increases ROS and decreases GSH due to diminished PPP flux (Schafer et al., 2009), while detachment of human A549 non-small cell lung cancer cells increases H2O2 and decreases NADPH levels (Jeon et al., 2012).
Cancer cells can use a wide variety of mechanisms to avoid anoikis, including triggering of EMT, integrin switching, growth factor overexpression, activating pro-survival signaling, activating oncogenes, and adapting metabolism to ameliorate stress (Paoli et al., 2013). In the last category, there is ample evidence that upregulation of antioxidant genes blunts anoikis and supports anchorage-independent growth. However, unlike adaptation to oxidative stress during initiation of tumorigenesis, which often involves somatic mutation of NFE2L2 or KEAP1, adaptation during anchorage-independent cell growth involves rerouting metabolism to increase generation of NADPH without resorting to constitutive activation of antioxidant genes. In 3D luminal-filling experiments, MCF-10A cells that lack matrix attachment are unable to form acinar structures because they succumb to anoikis, but this can be rescued by certain oncogenes, including ERBB2. By virtue of the fact it prevents downregulation of the epidermal growth factor receptor and maintains the PI3K pathway after matrix detachment, overexpression of ERBB2 enables MCF-10A cells to form acinar structures, thereby increasing glucose uptake and restoring ATP levels (Schafer et al., 2009). This increase in glucose uptake was found to be funneled down the PPP to enhance NADPH production by G6PD and increase GSH levels that might suppress ROS-induced cell death; this resembles, at least in part, adaptation to acute oxidative stress in normal cells (see Figure 1). Also of note, this increase in antioxidant status prevented ROS from inhibiting fatty acid oxidation and so maintained intracellular ATP levels sufficiently to avert metabolic crisis.
Matrix detachment produces energy stress that activates AMPK, which also contributes to increased production of NADPH (Jeon et al., 2012). In A549 cells, matrix detachment by phosphorylating and inhibiting acetyl-CoA carboxylase (ACC1 and ACC2), triggered AMPK activation to provide protection against oxidative stress, ultimately decreasing fatty acid synthesis and increasing fatty acid oxidation (Jeon et al., 2012). This blunting of fatty acid synthesis by inhibition of ACC1/2 conserves NADPH, making it available to support antioxidant defences, and so counter oxidative stress that would otherwise trigger cell death. Of note, knockdown of ACC1 in A549 cells blocked the increase in H2O2 and decreased the NADP+/NADPH ratio that occurs following matrix detachment, thus supporting such an interpretation.
Like MCF-10A and A549 cells, anchorage-independent growth of H460 lung cancer cells also requires increased generation of NADPH to mitigate oxidative stress that might limit tumor progression. In this case, however, ROS were found to accumulate primarily in mitochondria, and this was mitigated by increased generation of NADPH through reductive glutamine metabolism, which is normally only observed during hypoxia in an HIF-1α-dependent manner. Thus, in H460 spheroids, reductive carboxylation of α-ketoglutarate(α-KG) formed from glutamine is catalyzed by isocitrate dehydrogenase (IDH1) in the cytoplasm, independently of HIF-1α and hypoxia; this provides isocitrate, which is then imported into mitochondria where it is oxidized back to α-KG by IDH2, thereby providing reducing equivalents to fortify mitochondrial ROS defences (Jiang et al., 2016). During this process, cytoplasmic NADPH depletion resulting from reduction of α-KG by IDH1 is restored by the PPP, and so reducing equivalents from the PPP are transferred to mitochondria (Figure 6A). Clustering of detached tumor cells represents another strategy that aids anchorage-independent growth, because clustering stimulates HIF-1α-mediated mitophagy, which limits ROS production and in so doing increases dependence on glycolysis and reductive carboxylation of glutamine; this forced use of OXPHOS decreases survival of detached cells (Labuschagne et al., 2019).
Figure 6. Reductive Glutamine Metabolism and Serine-Driven Folate Metabolism Suppresses Mitochondrial ROS Accumulation to Support Anchorage-Independent Growth and/or Metastatic Disease.
(A) The increase in mitochondrial ROS that occurs when tumor cells are grown as spheroids can be mitigated by the concerted actions of IDH1 (1) and IDH2 (2), located in the cytoplasm and mitochondrion, respectively. Specifically, within the cytoplasm, IDH1 catalyzes the reductive carboxylation of α-ketoglutarate (α-KG), obtained from glutamine (by the sequential actions of GLS1 and GDH, see top right of cartoon), utilizing NADPH provided by the pentose phosphate pathway (PPP) (see top left and center of cartoon), to provide a supply of isocitrate and citrate. In turn, citrate in the cytoplasm is transferred to the mitochondrion via the citrate transporter protein (CPT, SLC25A1), before it is utilized by IDH2 to produce α-KG and NADPH. The latter is required to reduce GSSG and maintain high GSH levels, which ensures mitochondrial ROS levels are restrained and anoikis averted. Thus, IDH1 and IDH2 activities in the different subcellular compartments enable NADPH generated by the PPP, along with high glutamate/glutamine levels, to drive reductive carboxylation of α-KG in the cytoplasm, and so transfer reducing equivalents from the cytoplasm to the mitochondrion.
(B) The 10-formyl-tetrahydrofolate (THF) pathway represents a major source of NADPH for a variety of cell lines grown under in vitro cell culture conditions. For melanoma tumor cells to survive the relatively high O2 levels in the bloodstream and then colonize the liver, they increase production of NADPH by augmenting ALDH1L2 protein levels and maintaining levels of MTHFD1; ALDH1L2 (1) and MTHFD1 (2) are located in the mitochondrion and cytoplasm, respectively. The cartoon depicts how high de novo synthesis of serine, derived from glucose and 3-phosphoglycerate (top left-hand side), followed by transport into the mitochondrion, allows donation of a methyl group to THF, yielding 5,10-methylene-THF through a serine hydroxymethyl transferase (SHMT) 2-catalyzed reaction. In turn, 5,10-methylene-THF within the mitochondrion is converted to 10-formyl-THF by MTHFD2, which can in turn be utilized by ALDH1L2 to form NADPH (bottom right-hand side), which maintains GSH levels and prevents excess ROS from accumulating. Alternatively, 10-formyl-THF can be used by MTHFD1L to generate formate, which when transported out of the mitochondrion can be used in reversible MTHFD1-catalyzed reactions to generate 5,10-methylene-THF.
More recently it was observed that maintenance of fatty acid synthase (FASN) activity in mouse embryonic fibroblast (MEF) cells expressing oncogenic K-RASG12D, HER2(A775_G776insYVMA), or the PyMT antigen is an absolute requirement during the switch from 2D to 3D growth in colony-formation assays (Bueno et al., 2019). Pharmacological or genetic inhibition of FASN in MEF lines harboring these oncogenes prevented colony formation. Failure to grow upon inhibition/knockout of FASN was attributed to perturbation of glycolysis and mitochondrial function. In particular, inhibition/knockout of FASN caused a decrease in IDH1-catalyzed reductive carboxylation due to accumulation of citrate/isocitrate. This occurred because loss of FASN activity caused a buildup of acetyl-CoA, which inhibited ATP-citrate lyase, thereby increasing citrate. The inhibition of IDH1 by citrate/isocitrate resulted in an increase in mitochondrial ROS, decreased assembly of complex I of the electron transport chain into functional super-complexes, and a stalling of respiration. This is consistent with the notion that IDH1 is necessary for anchor-independent growth and tumorigenesis.
A common theme that emerges from studies into anchorage-independent cell growth is that treatment with antioxidants, such as NAC or Trolox (a water-soluble vitamin E derivative), can both rescue the metabolic defects caused by ECM deprivation and increase growth in soft agar (Bueno et al., 2019; Jeon et al., 2012; Schafer et al., 2009). This raises the question of whether antioxidants promote transformation of mammary epithelial cells. Consistent with this, upregulation of HO-1 protects against anoikis (Dey et al., 2015) and, conversely, knockdown of NQO1 renders non-small cell lung cancer cells more susceptible to anoikis (Madajewski et al., 2016).
Oxidative Stress Represents a Barrier to the Metastatic Spread of Cancer
For tumor cells to metastasize they need to undergo intravasation in order to enter the bloodstream, where they are exposed to a substantially higher O2 tension than their site of origin. They may also experience oxidative stress while migrating into the circulation, or before they extravasate to invade a distant organ (Nieto et al., 2016). This phase of tumorigenesis is inefficient, and its success requires plasticity on the part of the metastasizing cell. Experiments in mice involving subcutaneous or intravenous injection of efficient metastasizing melanomas revealed that the subcutaneous route of administration was substantially more effective than the latter in forming tumors (Piskounova et al., 2015). As ROS and GSSG levels were higher in circulating melanoma cells and metastatic melanoma cells than in subcutaneous tumors, and treatment with NAC increased the frequency of melanomas in the blood and the metastatic disease burden, it was reasoned the bloodstream is a pro-oxidant environment that provoked oxidative stress and impeded metastasis. Consistent with this view, metastatic nodules in the liver, pancreas, and lung from injected melanoma cells had higher levels of NADPH and NADP+ than did subcutaneous tumors, and this was associated with increased contribution of glucose to serine and glycine levels, augmented folate pathway activity, and an over-abundance of the mitochondrial ALDH1L2, and to a lesser extent cytoplasmic MTHFD1 (Figure 6B) (reviewed by Ducker and Rabinowitz, 2017). Thus, upregulation of these NADPH-generating enzymes may allow metastasizing tumor cells to survive in the bloodstream and invade distant organs (Piskounova et al., 2015). These authors also recognized that folate metabolism was not solely responsible for NADPH generation that drives metastasis, and that the PPP and malate pathways likely also contribute.
While high ROS levels limit metastatic spread of melanoma cells, those that are successful appear to be selected by their ability to use lactate as an energy source. This is possible because of the heterogeneity in expression of the lactate transporter MCT1 (SLC16A1) in tumor cells (Tasdogan et al., 2020). Antagonism of MCT1 with the selective inhibitor AZD3965, shRNA knockdown, or gene editing, has been shown to decrease the number of circulating melanoma cells and metastatic tumors arising from subcutaneous transplantation of melanoma cells, a process rescuable by treatment with NAC. Inhibition of MCT1 in melanoma cells increased ROS levels, decreased the abundance of GSH, NADPH, and flux through the oxidative arm of the PPP. It was concluded that inhibition of MCT1 would decrease flux through the oxidative arm of the PPP because lactate uptake is linked with proton flux, and so the increase in pH would increase glycolysis by activating phosphofructokinase and suppressing G6PD activity.
Evidence that ROS Have Diverse Effects on Different Forms of Metastatic Disease, and the Conundrum Posed by Antioxidants
Conflicting data exist relating to the effects of oxidative stress on metastasis. This likely reflects heterogeneity in redox thresholds for pathways controlling proliferation, mobility, and survival in different tumor cells. These are presumably dictated by the source of ROS and the subcellular compartments affected, and influenced by variables, including the oncogenes activated; the status of members of the antioxidant transcription factor network; and the extent to which neoplastic cells toggle between epithelial and mesenchymal phenotypes. In the latter case, oscillation between EMT and MET enables invading tumor cells to adapt to new environments (Dongre and Weinberg, 2019; Lu and Kang, 2019). It is becoming clear that the ability of cancer cells to adopt a hybrid epithelial/mesenchymal status is controlled by a number of “phenotypic stability factors,” such as grainyhead-like 2 (GRHL2), ovo-like zinc finger 2 (OVOL2), ΔNp63α, and NUMB, which stabilize such hybrid phenotypes (Aljagthmi et al., 2019; Bocci et al., 2017; He et al., 2020; Wu et al., 2017), and it could be hypothesized that the plasticity conferred by GRHL2, OVOL2, ΔNp63α, and NUMB is pivotal in determining the ability of tumor cells to withstand oxidative stress.
As discussed above, premalignant cells must adapt to the added ROS burden caused by activated oncogenes, which places extra demands on their antioxidant systems. Consistent with this, knockout of antioxidant defences provided by Nrf2 or Tigar markedly decreases development of PanIN lesions in K-RASG12D cancer models (Cheung et al., 2020; DeNicola et al., 2011). Counterintuitively, while high ROS levels can impede progression of tumorigenesis, there are examples where they actually enhance metastasis. Thus, in the K-RASG12D pancreatic cancer model, while knockout of Nrf2 or Tigar delays the appearance of PanIN lesions, their loss also increases metastasis to the lung and other organs, with resulting tumors having high ROS levels and exhibiting mesenchymal characteristics (Cheung et al., 2020). The increase in the metastatic potential of KrasG12D/+;Tigar−/− tumors was associated with activation of ERK signaling, attributed to downregulation of the dual-specificity phosphatase DUSP6. Ectopic expression of DUSP6 in Tigar-null cells was found to inhibit migration and the mesenchymal phenotype, as did treatment with NAC or the mitochondrially targeted antioxidant mito-TEMPO. In tail vein injection assays, treatment with NAC inhibited colonization of the lung by Tigar-null pancreatic tumor cells. Besides the ability of antioxidants to inhibit metastatic capabilities of Tigar-null pancreatic tumor cells, mito-TEMPO prevented lung metastasis of orthotopically injected MDA-MB-231 breast cancer cells in severe combined immunodeficiency mice (Porporato et al., 2014) and transgenic overexpression of mitochondrially targeted CAT diminished invasive breast cancer in MMTV-PyMT mice (Goh et al., 2011). Collectively, these findings suggest that antioxidants might trigger reductive stress in certain types of tumor cell.
In contrast to the observation that NAC and mito-TEMPO suppress metastatic spread of pancreatic tumors, an increasing literature indicates that antioxidants can support metastasis of melanoma and lung tumors. In mouse experiments, NAC and Trolox increase metastasis to the lung of melanoma cells harboring oncogenic B-RAFV600E with melanocyte-specific loss of Pten expression (Bagati et al., 2019; Le Gal et al., 2015) as well as human-derived metastasizing melanoma cells (Piskounova et al., 2015). Also, NAC and vitamin E increase metastasis to the liver, kidney, heart, and rib cage of lung cells harboring oncogenic K-RASG12D (Wiel et al., 2019).
The ability of NAC and vitamin E to increase metastasis of K-RASG12D-driven lung tumors was attributed to the ability of antioxidants to reduce the levels of ROS and free heme, thereby stabilizing BACH1, which in turn transcriptionally activated Hk2 and Gapdh and increased glucose uptake, glycolysis, and lactate secretion by Mct1 (Wiel et al., 2019). Importantly, BACH1 has been reported to be a master regulator of metastasis (Lee et al., 2013), and upregulates expression of metastasis-associated genes, such as MMP1, MMP3, CXCR4, CTGF, PGK2, and ROBO1 (Liang et al., 2012). Also, BACH1 can suppress mitochondrial activity by repressing expression of the electron transport chain genes ATP5D, COX15, UQCRC1, ATP5J, SLC25A22, and TIMM8B, and decrease glycolysis and the TCA cycle by increasing expression of pyruvate dehydrogenase kinase, thus inactivating phosphorylation of pyruvate dehydrogenase (Lee et al., 2019). Similar to the effects of antioxidants on K-RASG12D-driven lung tumorigenesis, it has been proposed that loss of Keap1 increases K-RASG12D-driven tumor metastasis via activation of Bach1. Thus, knockdown of Keap1 in KrasG12D/+;Trp53–/– tumors increased metastasis, but in this case the increase in Bach1 upon knockdown of Keap1 was due to Nrf2-mediated induction of Hmox1, which increased Bach1 stability by degrading heme (Lignitto et al., 2019). Similarly, in human HT1080 fibrosarcoma cells that harbor N-RAS and IDH1 mutations, ATF4-mediated induction of HMOX1 increased metastatic potential (Dey et al., 2015). Taken together, these observations suggest that administration of antioxidants can substantially affect the metastatic potential of cancer, but that this is probably influenced by tumor type and organ subject to colonization.
Dormant cancer cells that remain after therapy, and those that give rise to recurrent drug-resistant tumors during relapse, have to withstand oxidative stress resulting from their inability to utilize glucose as an energy source resulting in a switch to fatty acid β-oxidation. In particular, inhibitors of the receptor tyrosine kinase-RAS-MAPK pathway increase ROS (Krall et al., 2017), as does loss of oncogenic MYC, RAS, and Neu/Her2 signaling (Havas et al., 2017). In an MMTV-rtTA:TetO-Her2 mouse model, the small population of residual cells that survived in tumors after loss of oncogenic Her2 were found to upregulate Nrf2 in response to increased ROS (Fox et al., 2020). Moreover, constitutive activation of Nrf2 accelerated tumor recurrence, and knockdown of Nrf2 diminished recurrent tumor growth in vivo. Understandably, these are highly prescriptive models that for experimental reasons are limited to just several types of cancer. Because of the added ROS burden incurred during initiation, progression, and metastasis it is unclear how dormant and recurrent cancer cells with NRF2 already upregulated adapt to oxidative stress during and after chemotherapy.
Therapeutic Value of Manipulating Redox Status in Tumor Cells
Misunderstandings about how oxidative stress influences different stages of tumorigenesis is emphasized by the poor responses of at-risk patients given antioxidant therapy (Gill et al., 2016; Goodman et al., 2011). In animal carcinogenesis models, induction of endogenous antioxidant and detoxification systems by cancer chemopreventive agents confers protection against initiation of carcinogenesis caused by the subsequent exposure to chemical carcinogens (Wattenberg, 1985). However, administration of antioxidants per se may accelerate the later stages of certain types of cancer (Bagati et al., 2019; Wiel et al., 2019), results that support the notion that antioxidants have distinct effects on different stages of tumorigenesis and in a tumor type-specific fashion.
Like vitamin D, vitamin C sufficiency has been linked with innate and adaptive immune response to infective agents (Sorice et al., 2014). In this regard, preclinical studies have shown that high-dose vitamin C does exert anticancer effects in immunocompetent mice, where it can delay tumor growth in a T cell-dependent fashion, both through enhanced cytotoxicity of adoptively transferred T cells and cooperatively with immune checkpoint therapeutics (Magri et al., 2020). While a number of high-dose vitamin C clinical trials have been completed (Table 2), to date the results have not been encouraging. Perhaps design of future trials will benefit from the immunological approach, but for this tactic to reach general acceptance a number of remaining questions need to be addressed (e.g., Carr and Cook, 2018).
Table 2.
Overview of Drugs that Target Redox Platforms
| Drug | Mechanism of Action | Notes | Reference |
|---|---|---|---|
| Amifostine | aminothiol used to protect normal tissues against radiation toxicities | FDA approved in esophageal cancer | Shen et al. (2001) |
| MESNA | thiol used with alkylating agents to protect bladder | as an adjuvant, reduces incidence of hemorrhagic cystitis | Verschraagen et al. (2003) |
| NOV-002 | mimetic of GSSG; designed to interfere with cancer cell GSH homeostasis while protecting bone marrow | negative results in phase 3 clinical trials | Townsend et al. (2008) |
| Buthionine sulfoximine | inhibitor of de novo GSH biosynthesis | phase 1/2 clinical trials showed dose-limiting liver toxicity | O’Dwyer et al. (1992) |
| Sulforaphane | organosulfur isothiocyanate with reported chemoprevention properties | tested in a variety of preclinical and clinical settings | Hail et al. (2008) |
| Arsenic trioxide | crosslinks vicinal thiols in sensitive proteins | FDA approved in promyelocytic leukemia | Wang and Chen (2008) |
| Auranofin | non-specific binding, but inhibits thioredoxin reductase | plausible use in ovarian cancer | Fan et al. (2014a) |
| PX-12 | irreversible inhibitor of thioredoxin-1 | following drug administration, patients experienced strong, dose-limiting sulfurous odors | Baker et al. (2006) |
| Telcyta | Alkylating prodrug activated by GSTP | Negative results phase 3 clinical trials | Tew (2005) |
| Telintra | small-molecule inhibitor of GSTP | clinical benefit in myelodysplastic syndrome patients | Ruscoe et al. (2001) |
| Ethacrynic acid | Michael addition chemistry reacts with thiols | FDA approved as a diuretic; used in combination with alkylating agents; dose-limiting fluid imbalance in cancer patients | O’Dwyer et al. (1991) |
| PRIMA-1 or APR-246 | Michael addition chemistry with apparent specificity toward p53 | restores wild-type functions of p53; testing in progress | Ogiwara et al. (2019) |
| Erastin | small-molecule modulator of voltage-dependent anion channels and inhibitor of Gpx4 | analog development leads to phase 1 study of PRLX 93936 | Yang et al. (2014) |
| Ascorbate | high doses cause high H2O2 and deplete GSH and NADPH | numerous phase 1/2 trials but as yet, no consensus on efficacy | Chen et al. (2005) |
| CB-839 | inhibits glutaminase 1 (GLS1), limits glutamine, reduces glutamate and cysteine | evidence of cancer specificity based on salvage pathways; ongoing phase 1/2 trials in combinations | Romero et al. (2017) |
| Ivosidenib (AG-120) | inhibits mutant IDH1 variants with concomitant depletion of NADPH and GSH pools | numerous phase 1/2 trials; phase 3 with azacytidine in AML | Popovici-Muller et al. (2018) |
It can be argued that all types of cytotoxic cancer drugs cause direct, or indirect, oxidative stress. Nevertheless, a number of drugs have been developed using redox platforms to target various pathways. There are assorted categories that define their mechanisms of action: (1) adjuvants that protect normal tissues when given in combination with standard cancer drugs (amifostine, MESNA, NOV-002); (2) drugs that interfere directly with GSH homeostasis (BSO); (3) those that act as soft electrophiles in a chemoprevention setting by stimulating expression of ARE-regulated gene expression (sulforaphane); (4) hard electrophiles that covalently modify thiol groups in target proteins (arsenic trioxide, auranofin, PX-12, Telcyta); and (5) modification of target thiols through Michael addition (ethacrynic acid, PRIMA-1, APR-246). While this collection of drugs has diverse characteristics, the common theme is the nucleophilicity of cysteine thiol groups, whether within specific drug targets, or in the context of general stress response. Perhaps because of the critical importance of redox homeostasis in both normal and tumor cells and aspects of functional redundancy, limited clinical success has been an essential characteristic of this group of redox targeted drugs, nevertheless efforts to discover and develop new agents continue.
In context, many cancer cell lines are relatively resistant to death by BSO-initiated GSH depletion. This may be because TXN-based antioxidant systems compensate for depletion of GSH (Harris et al., 2015), or because, by upregulating HSF1 or increasing expression of deubiquitinases, tumor cells acquire tolerance of the effects of oxidative stress by attenuating their sensitivity to proteotoxicity and ER stress caused by protein misfolding (Harris et al., 2019). Together, these findings suggest that tumors can frequently circumvent redox targeting and that combinatorial approaches may be more successful. In fact, a number of combinatorial clinical trials are emerging with new redox-active agents. For example, erastin is a small-molecule modulator of voltage-dependent anion channels (VDAC2/VDAC3) (Yagoda et al., 2007) and of the GSH transporter xCT (Dixon et al., 2012). Treatment of cells with erastin causes GSH depletion, leading to inhibition of GPX4, which, in turn, increases ROS exposure (presumably through Fenton chemistry), triggering ferroptosis (Yang et al., 2014). Although erastin does not possess optimal therapeutic drug-like properties, analogs are under development with a view to clinical testing (Larraufie et al., 2015). Perhaps unexpectedly, GLS1 has also been shown to be a viable redox drug target for a new small molecule, CB839. NRF2 activation facilitates its antitumor effects, since in KEAP1 mutant lung cancer cells, GLS1 inhibition by CB839 reduces glutamine, thereby diminishing glutamate, which in turn limits cysteine and GSH levels (Sayin et al., 2017). Presently, lung cancers are a primary focus for clinical trials for CB839, but the effectiveness of GLS1 inhibition appears to be both tumor type and oncogene signaling dependent (Romero et al., 2017). Also, indirectly impacting redox homeostasis and inducing cell death through ROS, a new IDH1 inhibitor, Ivosidenib (AG-120), was shown to selectively target different mutants of IDH1 without apparent off-target effects on other dehydrogenases. Multiple clinical trials in IDH1 mutant cancers are underway using this inhibitor in combination with other anticancer drugs (Popovici-Muller et al., 2018).
There are examples where drugs can indirectly target redox pathways and homeostasis. For example, when solid cancers disseminate, metastases frequently have phenotypic characteristics that are distinct from the parent tumor cells. In melanomas, circulating cells that go on to form distant metastatic lesions have reversible adaptive changes that select for survival in a more biologically oxidative environment. Such adaptations include increased dependence on NADPH-generating enzymes in the folate pathway (Piskounova et al., 2015) and increased expression of the monocarboxylate transporter, MCT1 (Tasdogan et al., 2020). Since chemotherapy is one of the few approaches available for the treatment of metastases, interference with one-carbon metabolism with drugs, such as methotrexate, or impeding lactate/pyruvate metabolism and transport have potential as indirect means to interfere with redox regulation and restrict metastasis. Sulfasalazine and erastin are unrelated chemical structures that have promiscuous binding affinities for a number of intracellular targets. However, they do share the ability to inhibit the xCT glutamate cystine antiporter and, when used in combinations, have the potential to enhance the cytotoxic effects of other anticancer agents (Gout et al., 2001; Sayin et al., 2017). Moreover, combined inhibition of GSH (sulfasalazine) and TXN/TXNRD (auranofin) pathways has been found to lead to synergistic cancer cell death, implying roles for these complementary antioxidants in therapeutic intervention (Harris et al., 2015).
Radiation therapies cause release of ROS at levels contingent upon dose and time. There is a vast literature on stress responses consequent to radiation and general consensus that the nexus of TP53/APE1/NRF2 transcription factors contribute to a regulated response. Moreover, implicit in assessing tumor and normal tissue responses is the concept of dose versus time thresholds that determine the extent of response (Murray et al., 2018). As a translational biomarker, it has been possible to titrate the ROS caused by radiation exposure by measuring the occurrence of S-glutathionylated serine protease inhibitor (Serpin) in the blood of prostate cancer patients (Zhang et al., 2019). Interestingly, in a phase 1 clinical trial, pharmacological administration of vitamin C intravenously, in combination with gemcitabine and radiotherapy, has been found to be well tolerated and effective, suggesting that redox biology could be used to selectively target tumor tissue while protecting normal tissue (Alexander et al., 2018).
Concluding Comments
Throughout tumorigenesis, cancer cells have to endure oxidative stress at initiation, upon matrix detachment, during passage in the circulation, and when disease recurs after therapy. These episodes can be mitigated by tumor cells through various adaptive strategies, each of which ensures that ROS levels are restricted to a dynamic range that allows proliferation while avoiding cell death. During initiation, preneoplastic cells typically experience a high and sustained oncogene-generated ROS burden and they are therefore obliged to upregulate their antioxidant defences, which often entails overexpression of NRF2-regulated antioxidant genes, but can also involve oncogene-directed phosphorylation of PKM2 that increases NADPH production by diverting glucose metabolism down the PPP. During cancer progression, cooperation between neoplastic cells and CAFs and TAMs in their microenvironment results in ROS-stimulated migration and anchorage-independent growth of tumor cells, requiring them to reconfigure metabolism to increase NADPH generation via the actions of G6PD and/or IDH. During metastasis, circulating tumor cells experience high ROS levels after intravasation and existence in the blood, again requiring them to increase NADPH production, but in this case through increased use of the folate pathway (one-carbon metabolism) and upregulation of ALDH1L2, and to a lesser extent MTHFD1, while also ensuring glycolysis is maintained. In instances of post-therapy dormancy/recurrence, tumor cells depend on NRF2-directed gene expression to survive the switch to β-oxidation of fatty acids, rather than glycolysis, for energy.
Although overexpression of antioxidant genes, increased GSH synthesis, and increased NADPH generation will prevent oxidative stress, it is not certain that the “roll out” of the different tiers of oxidative stress defences during tumorigenesis and recurrence is orchestrated in precisely the order proposed above, and thus clarifying the stage-specific strategies used by tumor cells warrants further study. It may be more plastic than implied, and depend largely on the severity of the ROS burden preneoplastic cells have to counter at initiation of tumorigenesis, and also on the subcellular compartments where oxidative stress arises. For example, a high ROS burden at initiation may necessitate stochastic genetic changes, including somatic mutations in NFE2L2 or KEAP1 that are irreversible, whereas a lower ROS burden might be accommodated by reversible activation of members of the antioxidant transcription factor network and NADPH-generating pathways. It seems likely the adaptive mechanisms adopted at initiation will affect profoundly how tumor cells subsequently behave during cancer progression and to stimuli that dictate EMT and MET. It is striking that, as described above, adaptation to oxidative stress during cancer progression and metastasis entails bolstering NADPH-dependent antioxidant defences within the mitochondrion, which might not be so important during initiation and promotion of tumorigenesis. These observations suggest that mitochondrial function in cancer cells, and the dysregulation of particular mitochondrial metabolic pathways, may play a pivotal role in dictating which adaptive mechanisms are “available” in the later stages of tumorigenesis. It is also unclear the extent to which constitutive activation of NRF2 at initiation influences either the ability of tumor cells to adapt to oxidative stress during, and after therapy, or the necessity of tumor cells to adapt to oxidative stress during, and after therapy, and thus increases the likelihood of recurrent disease.
As we are only beginning to appreciate that redox homeostasis is of critical importance in cancer, there remain issues that need to be addressed. It is for example unclear how the antioxidant transcription factor network operates during different stages of tumorigenesis. In particular (1) the ROS thresholds that activate individual members of the network; (2) the extent to which thresholds for individual transcription factors change during the course of tumorigenesis, and upon constitutive activation of NRF2 or AP-1, or loss of TP53 function; (3) the degree of crosstalk between the factors during tumorigenesis; and (4) the impact of other forms of stress, such as metabolic stress, hypoxia, and inflammation, on the thresholds. An area that warrants further work is one that will determine the extent to which oxidative stress drives EMT, and the contribution of mitochondrial ROS to this process. Clarifying redox thresholds that influence EMT and MET will likely illuminate key biochemical pathways that influence the ability of different types of tumor cell to metastasize. It will also be important to provide a better understanding of factors controlling the acetylation of SOD2, and thus cause SOD2 to adopt pro-oxidant rather than antioxidant properties, during tumorigenesis.
Finally, the effects of antioxidants on the progression and metastatic stages of tumorigenesis are perplexing. It is hoped that research that provides a better understanding of how oscillations between EMT and TME are controlled will clarify the adaptive mechanisms used by different types of tumor cell in order to colonize distant tissues. The effects of antioxidants on ERK signaling in different tumor types should be further investigated, as should the possibility that antioxidants stimulate reductive stress in certain tumor types. Addressing these areas will improve therapeutic strategies.
ACKNOWLEDGMENTS
We thank the reviewers for their expert and constructive suggestions in fashioning this review. Also, we thank Professor Amedeo Columbano for critical and expert comments about the Solt-Farber model, and Dr Jean-Christophe Bourdon, Dr Sudhir Chowdhry, Dr Laureano de la Vega, Dr Dina Dikovskaya, Professor Stephen Keyse, Dr Holly Robertson, Professor Sonia Rocha, Dr Keiko Taguchi, and Professor Masayuki Yamamoto for valuable discussions. We gratefully acknowledge the Medical Research Council (MR/N009851/1, MR/T014644/1), Cancer Research UK (C20953/A18644), NIH (5P20GM103542; C06RR015455), and South Carolina Centers of Excellence program for funding.
Footnotes
DECLARATION OF INTERESTS
A.D.K. is on the Scientific Advisory Board of Evgen Pharma and is a consultant for Aclipse Therapeutics and Vividion Therapeutics.
REFERENCES
- Ahn SG, and Thiele DJ (2003). Redox regulation of mammalian heat shock factor 1 is essential for Hsp gene activation and protection from stress. Genes Dev. 17, 516–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akter S, Fu L, Jung Y, Conte ML, Lawson JR, Lowther WT, Sun R, Liu K, Yang J, and Carroll KS (2018). Chemical proteomics reveals new targets of cysteine sulfinic acid reductase. Nat. Chem. Biol. 14, 995–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander MS, Wilkes JG, Schroeder SR, Buettner GR, Wagner BA, Du J, Gibson-Corley K, O’Leary BR, Spitz DR, Buatti JM, et al. (2018). Pharmacologic ascorbate reduces radiation-induced normal tissue toxicity and enhances tumor radiosensitization in pancreatic cancer. Cancer Res. 78, 6838–6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aljagthmi AA, Hill NT, Cooke M, Kazanietz MG, Abba MC, Long W, and Kadakia MP (2019). DeltaNp63alpha suppresses cells invasion by downregulating PKCgamma/Rac1 signaling through miR-320a. Cell Death Dis. 10, 680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson DA, Gentry C, Moss S, and Bevan S (2008). Transient receptor potential A1 is a sensory receptor for multiple products of oxidative stress. J. Neurosci. 28, 2485–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnandis T, Monteiro P, Adams SD, Bridgeman VL, Rajeeve V, Gadaleta E, Marzec J, Chelala C, Malanchi I, Cutillas PR, and Godinho SA (2018). Oxidative stress in cells with extra centrosomes drives non-cell-autonomous invasion. Dev. Cell 47, 409–424 e409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arosio P, Ingrassia R, and Cavadini P (2009). Ferritins: a family of molecules for iron storage, antioxidation and more. Biochim. Biophys. Acta 1790, 589–599. [DOI] [PubMed] [Google Scholar]
- Bagati A, Moparthy S, Fink EE, Bianchi-Smiraglia A, Yun DH, Kolesnikova M, Udartseva OO, Wolff DW, Roll MV, Lipchick BC, et al. (2019). KLF9-dependent ROS regulate melanoma progression in stage-specific manner. Oncogene 38, 3585–3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker AF, Dragovich T, Tate WR, Ramanathan RK, Roe D, Hsu CH, Kirkpatrick DL, and Powis G (2006). The antitumor thioredoxin-1 inhibitor PX-12 (1-methylpropyl 2-imidazolyl disulfide) decreases thioredoxin-1 and VEGF levels in cancer patient plasma. J. Lab. Clin. Med. 147, 83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer AK, Cho HY, Miller-Degraff L, Walker C, Helms K, Fostel J, Yamamoto M, and Kleeberger SR (2011). Targeted deletion of Nrf2 reduces urethane-induced lung tumor development in mice. PLoS One 6, e26590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejjani F, Evanno E, Zibara K, Piechaczyk M, and Jariel-Encontre I (2019). The AP-1 transcriptional complex: local switch or remote command? Biochim. Biophys. Acta Rev. Cancer 1872, 11–23. [DOI] [PubMed] [Google Scholar]
- Benhar M (2018). Roles of mammalian glutathione peroxidase and thioredoxin reductase enzymes in the cellular response to nitrosative stress. Free Radic. Biol. Med. 127, 160–164. [DOI] [PubMed] [Google Scholar]
- Benhar M (2020). Oxidants, antioxidants and thiol redox switches in the control of regulated cell death pathways. Antioxidants (Basel) 9, 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhar M, Shytaj IL, Stamler JS, and Savarino A (2016). Dual targeting of the thioredoxin and glutathione systems in cancer and HIV. J. Clin. Invest. 126, 1630–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer M, and Schultze JL (2006). Regulatory T cells in cancer. Blood 108, 804–811. [DOI] [PubMed] [Google Scholar]
- Bieging KT, Mello SS, and Attardi LD (2014). Unravelling mechanisms of p53-mediated tumour suppression. Nat. Rev. Cancer 14, 359–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn AC, Matthaei KI, Lim C, Taylor MC, Cappello JY, Hayes JD, Anders MW, and Board PG (2006). Deficiency of glutathione transferase zeta causes oxidative stress and activation of antioxidant response pathways. Mol. Pharmacol. 69, 650–657. [DOI] [PubMed] [Google Scholar]
- Bocci F, Jolly MK, Tripathi SC, Aguilar M, Hanash SM, Levine H, and Onuchic JN (2017). Numb prevents a complete epithelial-mesenchymal transition by modulating Notch signalling. J. R. Soc. Interface 14, 20170512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocci F, Tripathi SC, Vilchez Mercedes SA, George JT, Casabar JP, Wong PK, Hanash SM, Levine H, Onuchic JN, and Jolly MK (2019). NRF2 activates a partial epithelial-mesenchymal transition and is maximally present in a hybrid epithelial/mesenchymal phenotype. Integr. Biol. (Camb.) 11, 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bott AJ, Peng IC, Fan Y, Faubert B, Zhao L, Li J, Neidler S, Sun Y, Jaber N, Krokowski D, et al. (2015). Oncogenic Myc induces expression of glutamine synthetase through promoter demethylation. Cell Metab. 22, 1068–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitzig M, Bhimineni C, Lockey R, and Kolliputi N (2016). 4-Hydroxy-2-nonenal: a critical target in oxidative stress? Am. J. Physiol. Cell Physiol. 311, C537–C543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigelius-Flohe R, and Maiorino M (2013). Glutathione peroxidases. Biochim. Biophys. Acta 1830, 3289–3303. [DOI] [PubMed] [Google Scholar]
- Broer A, Gauthier-Coles G, Rahimi F, van Geldermalsen M, Dorsch D, Wegener A, Holst J, and Broer S (2019). Ablation of the ASCT2 (SLC1A5) gene encoding a neutral amino acid transporter reveals transporter plasticity and redundancy in cancer cells. J. Biol. Chem. 294, 4012–4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AK, and Webb AE (2018). Regulation of FOXO factors inmammalian cells. Curr. Top Dev. Biol. 127, 165–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DI, and Griendling KK (2009). Nox proteins in signal transduction. Free Radic. Biol. Med. 47, 1239–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budanov AV (2014). The role of tumor suppressor p53 in the antioxidant defense and metabolism. Subcell Biochem. 85, 337–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno MJ, Jimenez-Renard V, Samino S, Capellades J, Junza A, Lopez-Rodriguez ML, Garcia-Carceles J, Lopez-Fabuel I, Bolanos JP, Chandel NS, et al. (2019). Essentiality of fatty acid synthase in the 2D to anchorage-independent growth transition in transforming cells. Nat. Commun. 10, 5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buettner GR (1993). The spin trapping of superoxide and hydroxyl free radicals withDMPO (5,5-dimethylpyrroline-N-oxide): more about iron. Free Radic. Res. Commun. 19 (Suppl 1 ), S79–S87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canli O, Nicolas AM, Gupta J, Finkelmeier F, Goncharova O, Pesic M, Neumann T, Horst D, Lower M, Sahin U, and Greten FR (2017). Myeloid cell-derived reactive oxygen species induce epithelial mutagenesis. Cancer Cell 32, 869–883.e5. [DOI] [PubMed] [Google Scholar]
- Carr AC, and Cook J (2018). Intravenous vitamin C for cancer therapy—identifying the current gaps in our knowledge. Front Physiol. 9, 1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cemerski S, Cantagrel A, Van Meerwijk JP, and Romagnoli P (2002). Reactive oxygen species differentially affect T cell receptor-signaling pathways. J. Biol. Chem. 277, 19585–19593. [DOI] [PubMed] [Google Scholar]
- Chaffer CL, San Juan BP, Lim E, and Weinberg RA (2016). EMT, cell plasticity and metastasis. Cancer Metastasis Rev. 35, 645–654. [DOI] [PubMed] [Google Scholar]
- Chamoto K, Chowdhury PS, Kumar A, Sonomura K, Matsuda F, Fagarasan S, and Honjo T (2017). Mitochondrial activation chemicals synergize with surface receptor PD-1 blockade for T cell-dependent antitumor activity. Proc. Natl. Acad. Sci. U S A 114, E761–E770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Tavana O, Chu B, Erber L, Chen Y, Baer R, and Gu W (2017). NRF2 is a major target of ARF in p53-independent tumor suppression. Mol. Cell 68, 224–232.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Espey MG, Krishna MC, Mitchell JB, Corpe CP, Buettner GR, Shacter E, and Levine M (2005). Pharmacologic ascorbic acid concentrations selectively kill cancer cells: action as a pro-drug to deliver hydrogen peroxide to tissues. Proc. Natl. Acad. Sci. U S A 102, 13604–13609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Johansson E, Fan Y, Shertzer HG, Vasiliou V, Nebert DW, and Dalton TP (2009). Early onset senescence occurs when fibroblasts lack the glutamate-cysteine ligase modifier subunit. Free Radic. Biol. Med. 47, 410–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Singh S, Matsumoto A, Manna SK, Abdelmegeed MA, Golla S, Murphy RC, Dong H, Song BJ, Gonzalez FJ, et al. (2016). Chronic glutathione depletion confers protection against alcohol-induced steatosis: implication for redox activation of AMP-activated protein kinase pathway. Sci. Rep. 6, 29743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung EC, Athineos D, Lee P, Ridgway RA, Lambie W, Nixon C, Strathdee D, Blyth K, Sansom OJ, and Vousden KH (2013). TIGAR is required for efficient intestinal regeneration and tumorigenesis. Dev. Cell 25, 463–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung EC, DeNicola GM, Nixon C, Blyth K, Labuschagne CF, Tuveson DA, and Vousden KH (2020). Dynamic ROS control by TIGAR regulates the initiation and progression of pancreatic cancer. Cancer Cell 37, 168–182.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhunchha B, Kubo E, and Singh DP (2019). Sulforaphane-induced Klf9/Prdx6 axis acts as a molecular switch to control redox signaling and determines fate of cells. Cells 8, 1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia K, Keatinge M, Mazzolini J, and Sieger D (2019). Brain tumours repurpose endogenous neuron to microglia signalling mechanisms to promote their own proliferation. eLife 8, e46912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia K, Mazzolini J, Mione M, and Sieger D (2018). Tumor initiating cells induce Cxcr4-mediated infiltration of pro-tumoral macrophages into the brain. eLife 7, e31918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong SJF, Lai JXH, Eu JQ, Bellot GL, and Pervaiz S (2018). Reactive oxygen species and oncoprotein signaling—a dangerous liaison. Antioxid. Redox Signal. 29, 1553–1588. [DOI] [PubMed] [Google Scholar]
- Chowdhry S, Zhang Y, McMahon M, Sutherland C, Cuadrado A, and Hayes JD (2013). Nrf2 is controlled by two distinct beta-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene 32, 3765–3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu FF, Esworthy RS, Chu PG, Longmate JA, Huycke MM, Wilczynski S, and Doroshow JH (2004). Bacteria-induced intestinal cancer in mice with disrupted Gpx1 and Gpx2 genes. Cancer Res. 64, 962–968. [DOI] [PubMed] [Google Scholar]
- Ciccarese F, and Ciminale V (2017). Escaping death: mitochondrial redox homeostasis in cancer cells. Front Oncol. 7, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichon MA, and Radisky DC (2014). ROS-induced epithelial-mesenchymal transition in mammary epithelial cells is mediated by NF-kB-dependent activation of Snail. Oncotarget 5, 2827–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosentino C, Grieco D, and Costanzo V (2011). ATMactivates the pentose phosphate pathway promoting anti-oxidant defence and DNA repair. EMBO J. 30, 546–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L, and Su SM (2017). Isocitrate dehydrogenase mutation and (R)-2-hydroxyglutarate: from basic discovery to therapeutics development. Annu. Rev. Biochem. 86, 305–331. [DOI] [PubMed] [Google Scholar]
- David CJ, and Massague J (2018). Contextual determinants of TGFbeta action in development, immunity and cancer. Nat. Rev. Mol. Cell Biol. 19, 419–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T, Xu AW, Mengwasser KE, Sack LM, Yoon JC, Park PJ, and Elledge SJ (2013). Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell 155, 948–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawei H, Honggang D, and Qian W (2018). AURKA contributes to the progression of oral squamous cell carcinoma (OSCC) through modulating epithelial-to-mesenchymal transition (EMT) and apoptosis via the regulation of ROS. Biochem. Biophys. Res. Commun. 507, 83–90. [DOI] [PubMed] [Google Scholar]
- DeNicola GM, Chen PH, Mullarky E, Sudderth JA, Hu Z, Wu D, Tang H, Xie Y, Asara JM, Huffman KE, et al. (2015). NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat. Genet. 47, 1475–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, Mangal D, Yu KH, Yeo CJ, Calhoun ES, et al. (2011). Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 475, 106–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deponte M (2013). Glutathione catalysis and the reaction mechanisms of glutathione-dependent enzymes. Biochim. Biophys. Acta 1830, 3217–3266. [DOI] [PubMed] [Google Scholar]
- Dey S, Sayers CM, Verginadis II, Lehman SL, Cheng Y, Cerniglia GJ, Tuttle SW, Feldman MD, Zhang PJ, Fuchs SY, et al. (2015). ATF4-dependent induction of heme oxygenase 1 prevents anoikis and promotes metastasis. J. Clin. Invest. 125, 2592–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick TP, and Ralser M (2015). Metabolic remodeling in times of stress: who shoots faster than his shadow? Mol. Cell 59, 519–521. [DOI] [PubMed] [Google Scholar]
- Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS, et al. (2012). Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 149, 1060–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon SJ, and Stockwell BR (2019). The hallmarks of ferroptosis. Annu. Rev. Cancer Biol. 3, 35–54. [Google Scholar]
- Dodson M, Castro-Portuguez R, and Zhang DD (2019). NRF2 plays a critical role in mitigating lipid peroxidation and ferroptosis. Redox Biol. 23, 101107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B, Jaeger AM, and Thiele DJ (2019). Inhibiting heat shock factor 1 in cancer: a unique therapeutic opportunity. Trends Pharmacol. Sci. 40, 986–1005. [DOI] [PubMed] [Google Scholar]
- Dongre A, and Weinberg RA (2019). New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 20, 69–84. [DOI] [PubMed] [Google Scholar]
- Ducker GS, and Rabinowitz JD (2017). One-carbon metabolism in health and disease. Cell Metab. 25, 27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eferl R, and Wagner EF (2003). AP-1: a double-edged sword in tumorigenesis. Nat. Rev. Cancer 3, 859–868. [DOI] [PubMed] [Google Scholar]
- Eijkelenboom A, and Burgering BM (2013). FOXOs: signalling integrators for homeostasis maintenance. Nat. Rev. Mol. Cell Biol. 14, 83–97. [DOI] [PubMed] [Google Scholar]
- El-Kenawi A, Hanggi K, and Ruffell B (2019). The immune microenvironment and cancer metastasis. Cold Spring Harb. Perspect. Med 10, a037424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elko EA, Cunniff B, Seward DJ, Chia SB, Aboushousha R, van de Wetering C, van der Velden J, Manuel A, Shukla A, Heintz NH, et al. (2019). Peroxiredoxins and beyond; redox systems regulating lung physiology and disease. Antioxid. Redox Signal. 31, 1070–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C, Zheng W, Fu X, Li X, Wong YS, and Chen T (2014a). Enhancement of auranofin-induced lung cancer cell apoptosis by selenocystine, a natural inhibitor of TrxR1 in vitro and in vivo. Cell Death Dis. 5, e1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Ye J, Kamphorst JJ, Shlomi T, Thompson CB, and Rabinowitz JD (2014b). Quantitative flux analysis reveals folate-dependent NADPH production. Nature 510, 298–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraonio R, Vergara P, Di Marzo D, Pierantoni MG, Napolitano M, Russo T, and Cimino F (2006). p53 suppresses the Nrf2-dependent transcription of antioxidant response genes. J. Biol. Chem. 281, 39776–39784. [DOI] [PubMed] [Google Scholar]
- Farber E (1984). The multistep nature of cancer development. Cancer Res. 44, 4217–4223. [PubMed] [Google Scholar]
- Findlay VJ, Townsend DM, Morris TE, Fraser JP, He L, and Tew KD (2006). A novel role for human sulfiredoxin in the reversal of glutathionylation. Cancer Res. 66, 6800–6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher AB (2017). Peroxiredoxin 6 in the repair of peroxidized cell membranes and cell signaling. Arch. Biochem. Biophys. 617, 68–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox DB, Garcia NMG, McKinney BJ, Lupo R, Noteware LC, Newcomb R, Liu J, Locasale JW, Hirschey MD, and Alvarez JV (2020). NRF2 activation promotes the recurrence of dormant tumour cells through regulation of redox and nucleotide metabolism. Nat. Metab. 2, 318–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamcsik MP, Kasibhatla MS, Teeter SD, and Colvin OM (2012). Glutathione levels in human tumors. Biomarkers 17, 671–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Mukherjee S, Choudhury S, Gupta P, Adhikary A, Baral R, and Chattopadhyay S (2015). Reactive oxygen species in the tumor niche triggers altered activation of macrophages and immunosuppression: role of fluoxetine. Cell Signal. 27, 1398–1412. [DOI] [PubMed] [Google Scholar]
- Gill JG, Piskounova E, and Morrison SJ (2016). Cancer, oxidative stress, and metastasis. Cold Spring Harb. Symp. Quant. Biol. 81, 163–175. [DOI] [PubMed] [Google Scholar]
- Giralt A, Hondares E, Villena JA, Ribas F, Diaz-Delfin J, Giralt M, Iglesias R, and Villarroya F (2011). Peroxisome proliferator-activated receptor-gamma coactivator-1alpha controls transcription of the Sirt3 gene, an essential component of the thermogenic brown adipocyte phenotype. J. Biol. Chem. 286, 16958–16966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glorieux C, Sandoval JM, Fattaccioli A, Dejeans N, Garbe JC, Dieu M, Verrax J, Renard P, Huang P, and Calderon PB (2016). Chromatin remodeling regulates catalase expression during cancer cells adaptation to chronic oxidative stress. Free Radic. Biol. Med. 99, 436–450. [DOI] [PubMed] [Google Scholar]
- Goebel G, Berger R, Strasak AM, Egle D, Muller-Holzner E, Schmidt S, Rainer J, Presul E, Parson W, Lang S, et al. (2012). Elevated mRNA expression of CHAC1 splicing variants is associated with poor outcome for breast and ovarian cancer patients. Br. J. Cancer 106, 189–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh J, Enns L, Fatemie S, Hopkins H, Morton J, Pettan-Brewer C, and Ladiges W (2011). Mitochondrial targeted catalase suppresses invasive breast cancer in mice. BMC Cancer 11, 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub D, Iyengar N, Dogra S, Wong T, Bready D, Tang K, Modrek AS, and Placantonakis DG (2019). Mutant isocitrate dehydrogenase inhibitors as targeted cancer therapeutics. Front Oncol. 9, 417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes P, Viana SD, Nunes S, Rolo AP, Palmeira CM, and Reis F (2020). The yin and yang faces of the mitochondrial deacetylase sirtuin 3 in age-related disorders. Ageing Res. Rev. 57, 100983. [DOI] [PubMed] [Google Scholar]
- Goncalves RL, Rothschild DE, Quinlan CL, Scott GK, Benz CC, and Brand MD (2014). Sources of superoxide/H2O2 during mitochondrial proline oxidation. Redox Biol. 2, 901–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman M, Bostick RM, Kucuk O, and Jones DP (2011). Clinical trials of antioxidants as cancer prevention agents: past, present, and future. Free Radic. Biol. Med. 51, 1068–1084. [DOI] [PubMed] [Google Scholar]
- Gorrini C, Harris IS, and Mak TW (2013). Modulation of oxidative stress as an anticancer strategy. Nat. Rev. Drug Discov. 12, 931–947. [DOI] [PubMed] [Google Scholar]
- Gout PW, Buckley AR, Simms CR, and Bruchovsky N (2001). Sulfasalazine, a potent suppressor of lymphoma growth by inhibition of the x(c)-cystine transporter: a new action for an old drug. Leukemia 15, 1633–1640. [DOI] [PubMed] [Google Scholar]
- Gozzelino R, Jeney V, and Soares MP (2010). Mechanisms of cell protection by heme oxygenase-1. Annu. Rev. Pharmacol. Toxicol. 50, 323–354. [DOI] [PubMed] [Google Scholar]
- Gravel SP (2018). Deciphering the Dichotomous effects of PGC-1alpha on tumorigenesis and metastasis. Front Oncol. 8, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grek CL, Zhang J, Manevich Y, Townsend DM, and Tew KD (2013). Causes and consequences of cysteine S-glutathionylation. J. Biol. Chem. 288, 26497–26504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Jiang Q, Tuccitto A, Chan D, Alqawlaq S, Won GJ, and Sivak JM (2018). The AMPK-PGC-1alpha signaling axis regulates the astrocyte glutathione system to protect against oxidative and metabolic injury. Neurobiol. Dis. 113, 59–69. [DOI] [PubMed] [Google Scholar]
- Hail N Jr., Cortes M, Drake EN, and Spallholz JE (2008). Cancer chemoprevention: a radical perspective. Free Radic. Biol. Med. 45, 97–110. [DOI] [PubMed] [Google Scholar]
- Haley JA, Ruiz CF, Montal ED, Wang D, Haley JD, and Girnun GD (2019). Decoupling of Nrf2 expression promotes mesenchymal state maintenance in non-small cell lung cancer. Cancers (Basel) 11, 10.3390/cancers11101488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B, and Gutteridge JMC (2015). Free Radicals in Biology and Medicine, Fifth Edition (Oxford University Press; ). [Google Scholar]
- Hamilton MJ, Bosiljcic M, Lepard NE, Halvorsen EC, Ho VW, Banath JP, Krystal G, and Bennewith KL (2014). Macrophages are more potent immune suppressors ex vivo than immature myeloid-derived suppressor cells induced by metastatic murine mammary carcinomas. J. Immunol. 192, 512–522. [DOI] [PubMed] [Google Scholar]
- Hampton MB, Vick KA, Skoko JJ, and Neumann CA (2018). Peroxiredoxin involvement in the initiation and progression of human cancer. Antioxid. Redox Signal. 28, 591–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris IS, Endress JE, Coloff JL, Selfors LM, McBrayer SK, Rosenbluth JM, Takahashi N, Dhakal S, Koduri V, Oser MG, et al. (2019). Deubiquitinases maintain protein homeostasis and survival of cancer cells upon glutathione depletion. Cell Metab. 29, 1166–1181.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris IS, Treloar AE, Inoue S, Sasaki M, Gorrini C, Lee KC, Yung KY, Brenner D, Knobbe-Thomsen CB, Cox MA, et al. (2015). Glutathione and thioredoxin antioxidant pathways synergize to drive cancer initiation and progression. Cancer Cell 27, 211–222. [DOI] [PubMed] [Google Scholar]
- Havas KM, Milchevskaya V, Radic K, Alladin A, Kafkia E, Garcia M, Stolte J, Klaus B, Rotmensz N, Gibson TJ, et al. (2017). Metabolic shifts in residual breast cancer drive tumor recurrence. J. Clin. Invest. 127, 2091–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawk MA, and Schafer ZT (2018). Mechanisms of redox metabolism and cancer cell survival during extracellular matrix detachment. J. Biol. Chem. 293, 7531–7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JD, and Dinkova-Kostova AT (2014). The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 39, 199–218. [DOI] [PubMed] [Google Scholar]
- Hayes JD, Ebisine K, Sharma RS, Chowdhry S, Dinkova-Kostova AT, and Sutherland C (2016). Regulation of the CNC-bZIP transcription factor Nrf2 by Keap1 and the axis between GSK-3 and -TrCP. Curr. Opin. Toxicol. 1, 92–103. [Google Scholar]
- Hayes JD, Flanagan JU, and Jowsey IR (2005). Glutathione transferases. Annu. Rev. Pharmacol. Toxicol. 45, 51–88. [DOI] [PubMed] [Google Scholar]
- He C, Danes JM, Hart PC, Zhu Y, Huang Y, de Abreu AL, O’Brien J, Mathison AJ, Tang B, Frasor JM, et al. (2019). SOD2 acetylation on lysine 68 promotes stem cell reprogramming in breast cancer. Proc. Natl. Acad. Sci. U S A 116, 23534–23541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Feng C, Zhu H, Wu S, Jin P, and Xu T (2020). Grainyhead-like 2 as a double-edged sword in development and cancer. Am. J. Transl. Res. 12, 310–331. [PMC free article] [PubMed] [Google Scholar]
- Held W, Siddiqui I, Schaeuble K, and Speiser DE (2019). Intratumoral CD8(+) T cells with stem cell-like properties: implications for cancer immunotherapy. Sci. Transl. Med. 11, eaay6863. [DOI] [PubMed] [Google Scholar]
- Hiemstra S, Niemeijer M, Koedoot E, Wink S, Klip JE, Vlasveld M, de Zeeuw E, van Os B, White A, and Water BV (2017). Comprehensive landscape of Nrf2 and p53 pathway activation dynamics by oxidative stress and DNA damage. Chem. Res. Toxicol. 30, 923–933. [DOI] [PubMed] [Google Scholar]
- Hinman A, Chuang HH, Bautista DM, and Julius D (2006). TRP channel activation by reversible covalent modification. Proc. Natl. Acad. Sci. U S A 103, 19564–19568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmstrom KM, and Finkel T (2014). Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat. Rev. Mol. Cell Biol. 15, 411–421. [DOI] [PubMed] [Google Scholar]
- Hwang I, Lee J, Huh JY, Park J, Lee HB, Ho YS, and Ha H (2012). Catalase deficiency accelerates diabetic renal injury through peroxisomal dysfunction. Diabetes 61, 728–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igelmann S, Neubauer HA, and Ferbeyre G (2019). STAT3 and STAT5 activation in solid cancers. Cancers (Basel) 11, 1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Italiano D, Lena AM, Melino G, and Candi E (2012). Identification of NCF2/p67phox as a novel p53 target gene. Cell Cycle 11, 4589–4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon SM, Chandel NS, and Hay N (2012). AMPK regulates NADPH homeostasis to promote tumour cell survival during energy stress. Nature 485, 661–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia JJ, Geng WS, Wang ZQ, Chen L, and Zeng XS (2019). The role of thioredoxin system in cancer: strategy for cancer therapy. Cancer Chemother. Pharmacol. 84, 453–470. [DOI] [PubMed] [Google Scholar]
- Jiang J, Wang K, Chen Y, Chen H, Nice EC, and Huang C (2017). Redox regulation in tumor cell epithelial-mesenchymal transition: molecular basis and therapeutic strategy. Signal. Transduct Target Ther. 2, 17036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Shestov AA, Swain P, Yang C, Parker SJ, Wang QA, Terada LS, Adams ND, McCabe MT, Pietrak B, et al. (2016). Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature 532, 255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, and Penning TM (2007). Aldo-keto reductases and bioactivation/detoxication. Annu. Rev. Pharmacol. Toxicol. 47, 263–292. [DOI] [PubMed] [Google Scholar]
- Kaarniranta K, Kajdanek J, Morawiec J, Pawlowska E, and Blasiak J (2018). PGC-1alpha protects RPE cells of the aging retina against oxidative stress-induced degeneration through the regulation of senescence and mitochondrial quality control. The significance for AMD pathogenesis. Int. J. Mol. Sci. 19, 2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski MM, Sauer SW, Kaminski M, Opp S, Ruppert T, Grigaravicius P, Grudnik P, Grone HJ, Krammer PH, and Gulow K (2012). T cell activation is driven by an ADP-dependent glucokinase linking enhanced glycolysis with mitochondrial reactive oxygen species generation. Cell Rep. 2, 1300–1315. [DOI] [PubMed] [Google Scholar]
- Kim JY, Park JH, Kim K, Leem J, and Park KK (2019). Melatonin inhibits transforming growth factor-beta1-induced epithelial-mesenchymal transition in AML12 hepatocytes. Biology (Basel) 8, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kincaid B, and Bossy-Wetzel E (2013). Forever young: SIRT3 a shield against mitochondrial meltdown, aging, and neurodegeneration. Front Aging Neurosci. 5, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkman HN, and Gaetani GF (2007). Mammalian catalase: a venerable enzyme with new mysteries. Trends Biochem. Sci. 32, 44–50. [DOI] [PubMed] [Google Scholar]
- Klotz LO, Sanchez-Ramos C, Prieto-Arroyo I, Urbanek P, Steinbrenner H, and Monsalve M (2015). Redox regulation of FoxO transcription factors. Redox Biol. 6, 51–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knatko EV, Ibbotson SH, Zhang Y, Higgins M, Fahey JW, Talalay P, Dawe RS, Ferguson J, Huang JT, Clarke R, et al. (2015). Nrf2 activation protects against solar-simulated ultraviolet radiation in mice and humans. Cancer Prev. Res. (Phila) 8, 475–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi EH, Suzuki T, Funayama R, Nagashima T, Hayashi M, Sekine H, Tanaka N, Moriguchi T, Motohashi H, Nakayama K, and Yamamoto M (2016). Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat. Commun. 7, 11624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppula P, Zhang Y, Zhuang L, and Gan B (2018). Amino acid transporter SLC7A11/xCT at the crossroads of regulating redox homeostasis and nutrient dependency of cancer. Cancer Commun. (Lond) 38, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovács D, Sigmond T, Hotzi B, Bohar B, Fazekas D, Deak V, Vellai T, and Barna J (2019). HSF1Base: a comprehensive database of HSF1 (heat shock factor 1) target genes. Int. J. Mol. Sci. 20, 5815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalik MA, Guzzo G, Morandi A, Perra A, Menegon S, Masgras I, Trevisan E, Angioni MM, Fornari F, Quagliata L, et al. (2016). Metabolic reprogramming identifies the most aggressive lesions at early phases of hepatic carcinogenesis. Oncotarget 7, 32375–32393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraaij MD, Savage ND, van der Kooij SW, Koekkoek K, Wang J, van den Berg JM, Ottenhoff TH, Kuijpers TW, Holmdahl R, van Kooten C, and Gelderman KA (2010). Induction of regulatory T cells by macrophages is dependent on production of reactive oxygen species. Proc. Natl. Acad. Sci. U S A 107, 17686–17691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krall EB, Wang B, Munoz DM, Ilic N, Raghavan S, Niederst MJ, Yu K, Ruddy DA, Aguirre AJ, Kim JW, et al. (2017). KEAP1 loss modulates sensitivity to kinase targeted therapy in lung cancer. eLife 6, e18970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehne A, Emmert H, Soehle J, Winnefeld M, Fischer F, Wenck H, Gallinat S, Terstegen L, Lucius R, Hildebrand J, and Zamboni N (2015). Acute activation of oxidative pentose phosphate pathway as first-line response to oxidative stress in human skin cells. Mol. Cell 59, 359–371. [DOI] [PubMed] [Google Scholar]
- Labuschagne CF, Cheung EC, Blagih J, Domart MC, and Vousden KH (2019). Cell clustering promotes a metabolic switch that supports metastatic colonization. Cell Metab. 30, 720–734.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larraufie MH, Yang WS, Jiang E, Thomas AG, Slusher BS, and Stockwell BR (2015). Incorporation of metabolically stable ketones into a small molecule probe to increase potency and water solubility. Bioorg. Med. Chem. Lett. 25, 4787–4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gal K, Ibrahim MX, Wiel C, Sayin VI, Akula MK, Karlsson C, Dalin MG, Akyurek LM, Lindahl P, Nilsson J, and Bergo MO (2015). Antioxidants can increase melanoma metastasis in mice. Sci. Transl. Med. 7, 308re8. [DOI] [PubMed] [Google Scholar]
- Lee G, Won HS, Lee YM, Choi JW, Oh TI, Jang JH, Choi DK, Lim BO, Kim YJ, Park JW, et al. (2016). Oxidative dimerization of PHD2 is responsible for its inactivation and contributes to metabolic reprogramming via HIF-1alpha activation. Sci. Rep. 6, 18928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Yesilkanal AE, Wynne JP, Frankenberger C, Liu J, Yan J, Elbaz M, Rabe DC, Rustandy FD, Tiwari P, et al. (2019). Effective breast cancer combination therapy targeting BACH1 and mitochondrial metabolism. Nature 568, 254–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee U, Frankenberger C, Yun J, Bevilacqua E, Caldas C, Chin SF, Rueda OM, Reinitz J, and Rosner MR (2013). A prognostic gene signature for metastasis-free survival of triple negative breast cancer patients. PLoS One 8, e82125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone A, Roca MS, Ciardiello C, Costantini S, and Budillon A (2017). Oxidative stress gene expression profile correlates with cancer patient poor prognosis: identification of crucial pathways might select novel therapeutic approaches. Oxid Med. Cell Longev. 2017, 2597581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang XH, Hu J, Shi M, Zhang L, and Chen H (2020). Combined treatment with N-acetylcysteine and gefitinib overcomes drug resistance to gefitinib in NSCLC cell line. Cancer Med. 9, 1495–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wu J, Zhang X, and Chen W (2018). Glutathione reductase-mediated thiol oxidative stress suppresses metastasis of murine melanoma cells. Free Radic. Biol. Med. 129, 256–267. [DOI] [PubMed] [Google Scholar]
- Li Z, Yang P, and Li Z (2014). The multifaceted regulation and functions of PKM2 in tumor progression. Biochim. Biophys. Acta 1846, 285–296. [DOI] [PubMed] [Google Scholar]
- Liang Y, Wu H, Lei R, Chong RA, Wei Y, Lu X, Tagkopoulos I, Kung SY, Yang Q, Hu G, and Kang Y (2012). Transcriptional network analysis identifies BACH1 as a master regulator of breast cancer bone metastasis. J. Biol. Chem. 287, 33533–33544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lignitto L, LeBoeuf SE, Homer H, Jiang S, Askenazi M, Karakousi TR, Pass HI, Bhutkar AJ, Tsirigos A, Ueberheide B, et al. (2019). Nrf2 activation promotes lung cancer metastasis by inhibiting the degradation of Bach1. Cell 178, 316–329.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligtenberg MA, Mougiakakos D, Mukhopadhyay M, Witt K, Lladser A, Chmielewski M, Riet T, Abken H, and Kiessling R (2016). Coexpressed catalase protects chimeric antigen receptor-redirected T cells as well as bystander cells from oxidative stress-induced loss of antitumor activity. J. Immunol. 196, 759–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JKM, Delaidelli A, Minaker SW, Zhang HF, Colovic M, Yang H, Negri GL, von Karstedt S, Lockwood WW, Schaffer P, et al. (2019). Cystine/glutamate antiporter xCT (SLC7A11) facilitates oncogenic RAS transformation by preserving intracellular redox balance. Proc. Natl. Acad. Sci. U S A 116, 9433–9442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin LC, Chen CF, Ho CT, Liu JJ, Liu TZ, and Chern CL (2018). gamma-Glutamylcysteine synthetase (gamma-GCS) as a target for overcoming chemo- and radio-resistance of human hepatocellular carcinoma cells. Life Sci. 198, 25–31. [DOI] [PubMed] [Google Scholar]
- Lin X, Zheng W, Liu J, Zhang Y, Qin H, Wu H, Xue B, Lu Y, and Shen P (2013). Oxidative stress in malignant melanoma enhances tumor necrosis factor-alpha secretion of tumor-associated macrophages that promote cancer cell invasion. Antioxid. Redox Signal. 19, 1337–1355. [DOI] [PubMed] [Google Scholar]
- Liou GY, Doppler H, DelGiorno KE, Zhang L, Leitges M, Crawford HC, Murphy MP, and Storz P (2016). Mutant KRas-induced mitochondrial oxidative stress in acinar cells upregulates EGFR signaling to drive formation of pancreatic precancerous lesions. Cell Rep. 14, 2325–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou GY, and Storz P (2010). Reactive oxygen species in cancer. Free Radic. Res. 44, 479–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CW, Hua KT, Li KC, Kao HF, Hong RL, Ko JY, Hsiao M, Kuo ML, and Tan CT (2017). Histone methyltransferase G9a drives chemotherapy resistance by regulating the glutamate-cysteine ligase catalytic subunit in head and neck squamous cell carcinoma. Mol. Cancer Ther. 16, 1421–1434. [DOI] [PubMed] [Google Scholar]
- Liu D, and Xu Y (2011). p53, oxidative stress, and aging. Antioxid. Redox Signal. 15, 1669–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu GH, Qu J, and Shen X (2008). NF-kappaB/p65 antagonizes Nrf2-ARE pathway by depriving CBP from Nrf2 and facilitating recruitment of HDAC3 to MafK. Biochim. Biophys. Acta 1783, 713–727. [DOI] [PubMed] [Google Scholar]
- Liu RM, Vayalil PK, Ballinger C, Dickinson DA, Huang WT, Wang S, Kavanagh TJ, Matthews QL, and Postlethwait EM (2012). Transforming growth factor beta suppresses glutamate-cysteine ligase gene expression and induces oxidative stress in a lung fibrosis model. Free Radic. Biol. Med. 53, 554–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Samanta D, Xiang L, Zhang H, Hu H, Chen I, Bullen JW, and Semenza GL (2015). Chemotherapy triggers HIF-1-dependent glutathione synthesis and copper chelation that induces the breast cancer stem cell phenotype. Proc. Natl. Acad. Sci. U S A 112, E4600–E4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, and Holmgren A (2014). The thioredoxin superfamily in oxidative protein folding. Antioxid. Redox Signal. 21, 457–470. [DOI] [PubMed] [Google Scholar]
- Lu SC (2013). Glutathione synthesis. Biochim. Biophys. Acta 1830, 3143–3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Ramakrishnan R, Altiok S, Youn JI, Cheng P, Celis E, Pisarev V, Sherman S, Sporn MB, and Gabrilovich D (2011). Tumor-infiltrating myeloid cells induce tumor cell resistance to cytotoxic T cells in mice. J. Clin. Invest. 121, 4015–4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, and Kang Y (2019). Epithelial-mesenchymal plasticity in cancer progression and metastasis. Dev. Cell 49, 361–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukey MJ, Greene KS, Erickson JW, Wilson KF, and Cerione RA (2016). The oncogenic transcription factor c-Jun regulates glutaminase expression and sensitizes cells to glutaminase-targeted therapy. Nat. Commun. 7, 11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunetti P, Di Giacomo M, Vergara D, De Domenico S, Maffia M, Zara V, Capobianco L, and Ferramosca A (2019). Metabolic reprogramming in breast cancer results in distinct mitochondrial bioenergetics between luminal and basal subtypes. FEBS J. 286, 688–709. [DOI] [PubMed] [Google Scholar]
- Luo M, Shang L, Brooks MD, Jiagge E, Zhu Y, Buschhaus JM,Conley S, Fath MA, Davis A, Gheordunescu E, et al. (2018). Targeting breast cancer stem cell state equilibrium through modulation of redox signaling. Cell Metab. 28, 69–86.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod AK, McMahon M, Plummer SM, Higgins LG, Penning TM, Igarashi K, and Hayes JD (2009). Characterization of the cancer chemopreventive NRF2-dependent gene battery in human keratinocytes: demonstration that the KEAP1-NRF2 pathway, and not the BACH1-NRF2 pathway, controls cytoprotection against electrophiles as well as redox-cycling compounds. Carcinogenesis 30, 1571–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson LJ, Dubin AE, Evans MJ, Marr F, Schultz PG, Cravatt BF, and Patapoutian A (2007). Noxious compounds activate TRPA1 ion channels through covalent modification of cysteines. Nature 445, 541–545. [DOI] [PubMed] [Google Scholar]
- Madajewski B, Boatman MA, Chakrabarti G, Boothman DA, and Bey EA (2016). Depleting tumor-NQO1 potentiates anoikis and inhibits growth of NSCLC. Mol. Cancer Res. 14, 14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magri A, Germano G, Lorenzato A, Lamba S, Chila R, Montone M, Amodio V, Ceruti T, Sassi F, Arena S, et al. (2020). High-dose vitamin C enhances cancer immunotherapy. Sci. Transl. Med. 12, eaay8707. [DOI] [PubMed] [Google Scholar]
- Maillet A, and Pervaiz S (2012). Redox regulation of p53, redox effectors regulated by p53: a subtle balance. Antioxid. Redox Signal. 16, 1285–1294. [DOI] [PubMed] [Google Scholar]
- Manz DH, Blanchette NL, Paul BT, Torti FM, and Torti SV (2016). Iron and cancer: recent insights. Ann. N Y Acad. Sci. 1368, 149–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcar L, Bardhan K, Gheorghiu L, Dinkelborg P, Pfaffle H, Liu Q, Wang M, Piotrowska Z, Sequist LV, Borgmann K, et al. (2019). Acquired resistance of EGFR-mutated lung cancer to tyrosine kinase inhibitor treatment promotes PARP inhibitor sensitivity. Cell Rep. 27, 3422–3432.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinho HS, Real C, Cyrne L, Soares H, and Antunes F (2014). Hydrogen peroxide sensing, signaling and regulation of transcription factors. Redox Biol. 2, 535–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincorena I, Raine KM, Gerstung M, Dawson KJ, Haase K, Van Loo P, Davies H, Stratton MR, and Campbell PJ (2017). Universal patterns of selection in cancer and somatic tissues. Cell 171, 1029–1041.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Outschoorn UE, Lin Z, Trimmer C, Flomenberg N, Wang C, Pavlides S, Pestell RG, Howell A, Sotgia F, and Lisanti MP (2011). Cancer cells metabolically “fertilize” the tumor microenvironment with hydrogen peroxide, driving the Warburg effect: implications for PET imaging of human tumors. Cell Cycle 10, 2504–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J (2012). TGFbeta signalling in context. Nat. Rev. Mol. Cell Biol. 13, 616–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mates JM, Di Paola FJ, Campos-Sandoval JA, Mazurek S, and Marquez J (2020). Therapeutic targeting of glutaminolysis as an essential strategy to combat cancer. Semin. Cell Dev. Biol. 98, 34–43. [DOI] [PubMed] [Google Scholar]
- McBrayer SK, Mayers JR, DiNatale GJ, Shi DD, Khanal J, Chakraborty AA, Sarosiek KA, Briggs KJ, Robbins AK, Sewastianik T, et al. (2018). Transaminase inhibition by 2-hydroxyglutarate impairs glutamate biosynthesis and redox homeostasis in glioma. Cell 175, 101–116.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng H, Yan WY, Lei YH, Wan Z, Hou YY, Sun LK, and Zhou JP (2019). SIRT3 regulation of mitochondrial quality control in neurodegenerative diseases. Front Aging Neurosci. 11, 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon D, and Board PG (2013). A role for glutathione transferase Omega 1 (GSTO1–1) in the glutathionylation cycle. J. Biol. Chem. 288, 25769–25779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mensurado S, Rei M, Lanca T, Ioannou M, Goncalves-Sousa N, Kubo H, Malissen M, Papayannopoulos V, Serre K, and Silva-Santos B (2018). Tumor-associated neutrophils suppress pro-tumoral IL-17+ gammadelta T cells through induction of oxidative stress. PLoS Biol. 16, e2004990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messina S, De Simone G, and Ascenzi P (2019). Cysteine-based regulation of redox-sensitive Ras small GTPases. Redox Biol. 26, 101282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuishi Y, Taguchi K, Kawatani Y, Shibata T, Nukiwa T, Aburatani H, Yamamoto M, and Motohashi H (2012). Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell 22, 66–79. [DOI] [PubMed] [Google Scholar]
- Molon B, Ugel S, Del Pozzo F, Soldani C, Zilio S, Avella D, De Palma A, Mauri P, Monegal A, Rescigno M, et al. (2011). Chemokine nitration prevents intratumoral infiltration of antigen-specific T cells. J. Exp. Med. 208, 1949–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moloney JN, and Cotter TG (2018). ROS signalling in the biology of cancer. Semin. Cell Dev. Biol. 80, 50–64. [DOI] [PubMed] [Google Scholar]
- Morgan MJ, and Liu ZG (2011). Crosstalk of reactive oxygen species and NF-kappaB signaling. Cell Res. 21, 103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morotti M, Bridges E, Valli A, Choudhry H, Sheldon H, Wigfield S, Gray N, Zois CE, Grimm F, Jones D, et al. (2019). Hypoxia-induced switch in SNAT2/SLC38A2 regulation generates endocrine resistance in breast cancer. Proc. Natl. Acad. Sci. U S A 116, 12452–12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen AR, Hu Z, Shi X, Jiang L, Boroughs LK, Kovacs Z, Boriack R, Rakheja D, Sullivan LB, Linehan WM, et al. (2014). Oxidation of alpha-ketoglutarate is required for reductive carboxylation in cancer cells with mitochondrial defects. Cell Rep. 7, 1679–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy MP (2009). How mitochondria produce reactive oxygen species. Biochem. J. 417, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray D, Mirzayans R, and McBride WH (2018). Defenses against prooxidant forces—maintenance of cellular and genomic integrity and longevity. Radiat. Res. 190, 331–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima T, Hamada S, Taguchi K, Tanaka Y, Matsumoto R, Yamamoto M, and Masamune A (2020). Keap1 deletion accelerates mutant Kras/p53-driven cholangiocarcinoma. Am. J. Physiol. Gastrointest. Liver Physiol. 318, G419–G427. [DOI] [PubMed] [Google Scholar]
- Nagaraj S, Gupta K, Pisarev V, Kinarsky L, Sherman S, Kang L, Herber DL, Schneck J, and Gabrilovich DI (2007). Altered recognition of antigen is a mechanism of CD8+ T cell tolerance in cancer. Nat. Med. 13, 828–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann CA, Krause DS, Carman CV, Das S, Dubey DP, Abraham JL, Bronson RT, Fujiwara Y, Orkin SH, and Van Etten RA (2003). Essential role for the peroxiredoxin Prdx1 in erythrocyte antioxidant defence and tumour suppression. Nature 424, 561–565. [DOI] [PubMed] [Google Scholar]
- Ngo HKC, Kim DH, Cha YN, Na HK, and Surh YJ (2017). Nrf2 mutagenic activation drives hepatocarcinogenesis. Cancer Res. 77, 4797–4808. [DOI] [PubMed] [Google Scholar]
- Nguyen TT, Grimm SA, Bushel PR, Li J, Li Y, Bennett BD, Lavender CA, Ward JM, Fargo DC, Anderson CW, et al. (2018). Revealing a human p53 universe. Nucleic Acids Res. 46, 8153–8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto MA, Huang RY, Jackson RA, and Thiery JP (2016). Emt: 2016. Cell 166, 21–45. [DOI] [PubMed] [Google Scholar]
- Nishizawa H, Matsumoto M, Shindo T, Saigusa D, Kato H, Suzuki K, Sato M, Ishii Y, Shimokawa H, and Igarashi K (2020). Ferroptosis is controlled by the coordinated transcriptional regulation of glutathione and labile iron metabolism by the transcription factor BACH1. J. Biol. Chem. 295, 69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Dwyer PJ, Hamilton TC, Young RC, LaCreta FP, Carp N, Tew KD, Padavic K, Comis RL, and Ozols RF (1992). Depletion of glutathione in normal and malignant human cells in vivo by buthionine sulfoximine: clinical and biochemical results. J. Natl. Cancer Inst. 84, 264–267. [DOI] [PubMed] [Google Scholar]
- O’Dwyer PJ, LaCreta F, Nash S, Tinsley PW, Schilder R, Clapper ML, Tew KD, Panting L, Litwin S, Comis RL, et al. (1991). Phase I study of thiotepa in combination with the glutathione transferase inhibitor ethacrynic acid. Cancer Res. 51, 6059–6065. [PubMed] [Google Scholar]
- Oakley AJ, Yamada T, Liu D, Coggan M, Clark AG, and Board PG (2008). The identification and structural characterization of C7orf24 as gamma-glutamyl cyclotransferase. An essential enzyme in the gamma-glutamyl cycle. J. Biol. Chem. 283, 22031–22042. [DOI] [PubMed] [Google Scholar]
- Ogiwara H, Takahashi K, Sasaki M, Kuroda T, Yoshida H, Watanabe R, Maruyama A, Makinoshima H, Chiwaki F, Sasaki H, et al. (2019). Targeting the vulnerability of glutathione metabolism in ARID1A-deficient cancers. Cancer Cell 35, 177–190.e8. [DOI] [PubMed] [Google Scholar]
- Oppermann U (2007). Carbonyl reductases: the complex relationships of mammalian carbonyl- and quinone-reducing enzymes and their role in physiology. Annu. Rev. Pharmacol. Toxicol. 47, 293–322. [DOI] [PubMed] [Google Scholar]
- Orrenius S, Gogvadze V, and Zhivotovsky B (2015). Calcium and mitochondria in the regulation of cell death. Biochem. Biophys. Res. Commun. 460, 72–81. [DOI] [PubMed] [Google Scholar]
- Orru C, Szydlowska M, Taguchi K, Zavattari P, Perra A, Yamamoto M, and Columbano A (2018). Genetic inactivation of Nrf2 prevents clonal expansion of initiated cells in a nutritional model of rat hepatocarcinogenesis. J. Hepatol. 69, 635–643. [DOI] [PubMed] [Google Scholar]
- Paaby AB, and Rockman MV (2013). The many faces of pleiotropy. Trends Genet. 29, 66–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoli P, Giannoni E, and Chiarugi P (2013). Anoikis molecular pathways and its role in cancer progression. Biochim. Biophys. Acta 1833, 3481–3498. [DOI] [PubMed] [Google Scholar]
- Parascandolo A, and Laukkanen MO (2019). Carcinogenesis and reactive oxygen species signaling: interaction of the NADPH oxidase NOX1–5 and superoxide dismutase 1–3 signal transduction pathways. Antioxid. Redox Signal. 30, 443–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JW, Mieyal JJ, Rhee SG, and Chock PB (2009). Deglutathionylation of 2-Cys peroxiredoxin is specifically catalyzed by sulfiredoxin. J. Biol. Chem. 284, 23364–23374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson AD, Carlson BA, Li F, Bonzo JA, Yoo MH, Krausz KW, Conrad M, Chen C, Gonzalez FJ, and Hatfield DL (2013). Disruption of thioredoxin reductase 1 protects mice from acute acetaminophen-induced hepatotoxicity through enhanced NRF2 activity. Chem. Res. Toxicol. 26, 1088–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Ghosh S, Mandal S, Sau S, and Pal M (2018). NRF2 transcriptionally activates the heat shock factor 1 promoter under oxidative stress and affects survival and migration potential of MCF7 cells. J. Biol. Chem. 293, 19303–19316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen CE, Armache JP, Gao Y, Cheng Y, and Julius D (2015). Structure of the TRPA1 ion channel suggests regulatory mechanisms. Nature 520, 511–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins A, Nelson KJ, Parsonage D, Poole LB, and Karplus PA (2015). Peroxiredoxins: guardians against oxidative stress and modulators of peroxide signaling. Trends Biochem. Sci. 40, 435–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins ND (2007). Integrating cell-signalling pathways with NF-kappaB and IKK function. Nat. Rev. Mol. Cell Biol. 8, 49–62. [DOI] [PubMed] [Google Scholar]
- Perkins ND (2012). The diverse and complex roles of NF-kappaB subunits in cancer. Nat. Rev. Cancer 12, 121–132. [DOI] [PubMed] [Google Scholar]
- Petrelli A, Perra A, Cora D, Sulas P, Menegon S, Manca C, Migliore C, Kowalik MA, Ledda-Columbano GM, Giordano S, and Columbano A (2014). MicroRNA/gene profiling unveils early molecular changes and nuclear factor erythroid related factor 2 (NRF2) activation in a rat model recapitulating human hepatocellular carcinoma (HCC). Hepatology 59, 228–241. [DOI] [PubMed] [Google Scholar]
- Pietrangelo A (2017). Ferroportin disease: pathogenesis, diagnosis and treatment. Haematologica 102, 1972–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskounova E, Agathocleous M, Murphy MM, Hu Z, Huddlestun SE, Zhao Z, Leitch AM, Johnson TM, DeBerardinis RJ, and Morrison SJ (2015). Oxidative stress inhibits distant metastasis by human melanoma cells. Nature 527, 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaitakis A, Kalef-Ezra E, Kotzamani D, Zaganas I, and Spanaki C (2017). The glutamate dehydrogenase pathway and its roles in cell and tissue biology in health and disease. Biology (Basel) 6, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovici-Muller J, Lemieux RM, Artin E, Saunders JO, Salituro FG, Travins J, Cianchetta G, Cai Z, Zhou D, Cui D, et al. (2018). Discovery of AG-120 (Ivosidenib): a first-in-class mutant IDH1 inhibitor for the treatment of IDH1 mutant cancers. ACS Med. Chem. Lett. 9, 300–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porporato PE, Payen VL, Perez-Escuredo J, De Saedeleer CJ, Danhier P, Copetti T, Dhup S, Tardy M, Vazeille T, Bouzin C, et al. (2014). A mitochondrial switch promotes tumor metastasis. Cell Rep. 8, 754–766. [DOI] [PubMed] [Google Scholar]
- Porte S, Valencia E, Yakovtseva EA, Borras E, Shafqat N, Debreczeny JE, Pike AC, Oppermann U, Farres J, Fita I, and Pares X (2009). Three-dimensional structure and enzymatic function of proapoptotic human p53-inducible quinone oxidoreductase PIG3. J. Biol. Chem. 284, 17194–17205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell CD, Paullin TR, Aoisa C, Menzie CJ, Ubaldini A, and Westerheide SD (2016). The heat shock transcription factor HSF1 induces ovarian cancer epithelial-mesenchymal transition in a 3D spheroid growth model. PLoS One 11, e0168389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purohit V, Simeone DM, and Lyssiotis CA (2019). Metabolic regulation of redox balance in cancer. Cancers (Basel) 11, 955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radi R (2018). Oxygen radicals, nitric oxide, and peroxynitrite: redox pathways in molecular medicine. Proc. Natl. Acad. Sci. U S A 115, 5839–5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radisky DC, Levy DD, Littlepage LE, Liu H, Nelson CM, Fata JE, Leake D, Godden EL, Albertson DG, Nieto MA, et al. (2005). Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Nature 436, 123–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimondi V, Ciccarese F, and Ciminale V (2020). Oncogenic pathways and the electron transport chain: a dangeROS liaison. Br. J. Cancer 122, 168–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Gomez M, Kwak MK, Dolan PM, Itoh K, Yamamoto M, Talalay P, and Kensler TW (2001). Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc. Natl. Acad. Sci. U S A 98, 3410–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reczek CR, Birsoy K, Kong H, Martinez-Reyes I, Wang T, Gao P, Sabatini DM, and Chandel NS (2017). A CRISPR screen identifies a pathway required for paraquat-induced cell death. Nat. Chem. Biol. 13, 1274–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redza-Dutordoir M, and Averill-Bates DA (2016). Activation of apoptosis signalling pathways by reactive oxygen species. Biochim. Biophys. Acta 1863, 2977–2992. [DOI] [PubMed] [Google Scholar]
- Ren X, Zou L, Zhang X, Branco V, Wang J, Carvalho C, Holmgren A, and Lu J (2017). Redox signaling mediated by thioredoxin and glutathione systems in the central nervous system. Antioxid. Redox Signal. 27, 989–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SG, and Bae SH (2015). The antioxidant function of sestrins is mediated by promotion of autophagic degradation of Keap1 and Nrf2 activation and by inhibition of mTORC1. Free Radic. Biol. Med. 88, 205–211. [DOI] [PubMed] [Google Scholar]
- Rhee SG, and Kil IS (2017). Multiple functions and regulation of mammalian peroxiredoxins. Annu. Rev. Biochem. 86, 749–775. [DOI] [PubMed] [Google Scholar]
- Rhee SG, Woo HA, Kil IS, and Bae SH (2012). Peroxiredoxin functions as a peroxidase and a regulator and sensor of local peroxides. J. Biol. Chem. 287, 4403–4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Zavala JS, Calleja LF, Moreno-Sanchez R, and Yoval-Sanchez B (2019). Role of aldehyde dehydrogenases in physiopathological processes. Chem. Res. Toxicol. 32, 405–420. [DOI] [PubMed] [Google Scholar]
- Rojo de la Vega M, Chapman E, and Zhang DD(2018). NRF2 and the hallmarks of cancer. Cancer Cell 34, 21–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero R, Sayin VI, Davidson SM, Bauer MR, Singh SX, LeBoeuf SE, Karakousi TR, Ellis DC, Bhutkar A, Sanchez-Rivera FJ, et al. (2017). Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat. Med. 23, 1362–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose Li Y, Halliwill KD, Adams CJ, Iyer V, Riva L, Mamunur R, Jen KY, Del Rosario R, Fredlund E, Hirst G, et al. (2020). Mutational signatures in tumours induced by high and low energy radiation in Trp53 deficient mice. Nat. Commun. 11, 394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland A, Miners JO, and Mackenzie PI (2013). The UDP-glucuronosyltransferases: their role in drug metabolism and detoxification. Int. J. Biochem. Cell Biol. 45, 1121–1132. [DOI] [PubMed] [Google Scholar]
- Ruscoe JE, Rosario LA, Wang T, Gate L, Arifoglu P, Wolf CR, Henderson CJ, Ronai Z, and Tew KD (2001). Pharmacologic or genetic manipulation of glutathione S-transferase P1–1 (GSTpi) influences cell proliferation pathways. J. Pharmacol. Exp. Ther. 298, 339–345. [PubMed] [Google Scholar]
- Sakamoto K, Iwasaki K, Sugiyama H, and Tsuji Y (2009). Role of the tumor suppressor PTEN in antioxidant responsive element-mediated transcription and associated histone modifications. Mol. Biol. Cell 20, 1606–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta D, and Semenza GL (2017). Maintenance of redox homeostasis by hypoxia-inducible factors. Redox Biol. 13, 331–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson N, Koziel R, Zenzmaier C, Bubendorf L, Plas E, Jansen-Durr P, and Berger P (2011). ROS signaling by NOX4 drives fibroblast-to-myofibroblast differentiation in the diseased prostatic stroma. Mol. Endocrinol. 25, 503–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Juan BP, Garcia-Leon MJ, Rangel L, Goetz JG, and Chaffer CL (2019). The complexities of metastasis. Cancers (Basel) 11, 1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Alvarez M, Strippoli R, Donadelli M, Bazhin AV, and Cordani M (2019). Sestrins as a therapeutic bridge between ROS and autophagy in cancer. Cancers (Basel) 11, 1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh H, Moriguchi T, Takai J, Ebina M, and Yamamoto M (2013). Nrf2 prevents initiation but accelerates progression through the Kras signaling pathway during lung carcinogenesis. Cancer Res. 73, 4158–4168. [DOI] [PubMed] [Google Scholar]
- Satoh K, Yachida S, Sugimoto M, Oshima M, Nakagawa T, Akamoto S, Tabata S, Saitoh K, Kato K, Sato S, et al. (2017). Global metabolic reprogramming of colorectal cancer occurs at adenoma stage and is induced by MYC. Proc. Natl. Acad. Sci. U S A 114, E7697–E7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayin VI, Ibrahim MX, Larsson E, Nilsson JA, Lindahl P, and Bergo MO (2014). Antioxidants accelerate lung cancer progression in mice. Sci. Transl. Med. 6, 221ra15. [DOI] [PubMed] [Google Scholar]
- Sayin VI, LeBoeuf SE, Singh SX, Davidson SM, Biancur D, Guzelhan BS, Alvarez SW, Wu WL, Karakousi TR, Zavitsanou AM, et al. (2017). Activation of the NRF2 antioxidant program generates an imbalance in central carbon metabolism in cancer. eLife 6, e28083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer ZT, Grassian AR, Song L, Jiang Z, Gerhart-Hines Z, Irie HY, Gao S, Puigserver P, and Brugge JS (2009). Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature 461, 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharping NE, Menk AV, Moreci RS, Whetstone RD, Dadey RE, Watkins SC, Ferris RL, and Delgoffe GM (2016). The tumor microenvironment represses T cell mitochondrial biogenesis to drive intratumoral T cell metabolic insufficiency and dysfunction. Immunity 45, 701–703. [DOI] [PubMed] [Google Scholar]
- Semenza GL (2012). Hypoxia-inducible factors in physiology and medicine. Cell 148, 399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena LA, Li S, Jairaman A, Prakriya M, Ezponda T, Hildeman DA, Wang CR, Schumacker PT, Licht JD, Perlman H, et al. (2013). Mitochondria are required for antigen-specific T cell activation through reactive oxygen species signaling. Immunity 38, 225–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafirovich V, and Geacintov NE (2017). Removal of oxidatively generated DNA damage by overlapping repair pathways. Free Radic. Biol. Med. 107, 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaulian E, and Karin M (2001). AP-1 in cell proliferation and survival. Oncogene 20, 2390–2400. [DOI] [PubMed] [Google Scholar]
- Shen H, Chen ZJ, Zilfou JT, Hopper E, Murphy M, and Tew KD (2001). Binding of the aminothiol WR-1065 to transcription factors influences cellular response to anticancer drugs. J. Pharmacol. Exp. Ther. 297, 1067–1073. [PubMed] [Google Scholar]
- Sheng Y, Abreu IA, Cabelli DE, Maroney MJ, Miller AF, Teixeira M, and Valentine JS (2014). Superoxide dismutases and superoxide reductases. Chem. Rev. 114, 3854–3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sies H (2015). Oxidative stress: a concept in redox biology and medicine. Redox Biol. 4, 180–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sies H, Berndt C, and Jones DP (2017). Oxidative stress. Annu. Rev. Biochem. 86, 715–748. [DOI] [PubMed] [Google Scholar]
- Sies H, and Jones DP (2020). Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat. Rev. Mol. Cell Biol. 10.1038/s41580-020-0230-3. [DOI] [PubMed] [Google Scholar]
- Siska PJ, Beckermann KE, Mason FM, Andrejeva G, Greenplate AR, Sendor AB, Chiang YJ, Corona AL, Gemta LF, Vincent BG, et al. (2017). Mitochondrial dysregulation and glycolytic insufficiency functionally impair CD8 T cells infiltrating human renal cell carcinoma. JCI Insight 2, e93411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song NY, Zhu F, Wang Z, Willette-Brown J, Xi S, Sun Z, Su L, Wu X, Ma B, Nussinov R, et al. (2018). IKKalpha inactivation promotes Kras-initiated lung adenocarcinoma development through disrupting major redox regulatory pathways. Proc. Natl. Acad. Sci. U S A 115, E812–E821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano FX, Baxter P, Murray LM, Sporn MB, Gillingwater TH, and Hardingham GE (2009). Transcriptional regulation of the AP-1 and Nrf2 target gene sulfiredoxin. Mol. Cell 27, 279–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorice A, Guerriero E, Capone F, Colonna G, Castello G, and Costantini S (2014). Ascorbic acid: its role in immune system and chronic inflammation diseases. Mini Rev. Med. Chem. 14, 444–452. [DOI] [PubMed] [Google Scholar]
- St-Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jager S, Handschin C, Zheng K, Lin J, Yang W, et al. (2006). Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell 127, 397–408. [DOI] [PubMed] [Google Scholar]
- Stegen S, van Gastel N, Eelen G, Ghesquiere B, D’Anna F, Thienpont B, Goveia J, Torrekens S, Van Looveren R, Luyten FP, et al. (2016). HIF-1alpha promotes glutamine-mediated redox homeostasis and glycogen-dependent bioenergetics to support postimplantation bone cell survival. Cell Metab. 23, 265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmler MP, Eccles RL, Brabletz S, and Brabletz T (2019). Non-redundant functions of EMT transcription factors. Nat. Cell Biol 21, 102–112. [DOI] [PubMed] [Google Scholar]
- Stockwell BR, Friedmann Angeli JP, Bayir H, Bush AI, Conrad M, Dixon SJ, Fulda S, Gascon S, Hatzios SK, Kagan VE, et al. (2017). Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell 171, 273–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroher E, and Millar AH (2012). The biological roles of glutaredoxins. Biochem. J. 446, 333–348. [DOI] [PubMed] [Google Scholar]
- Sun J, Zhou C, Ma Q, Chen W, Atyah M, Yin Y, Fu P, Liu S, Hu B, Ren N, and Zhou H (2019). High GCLC level in tumor tissues is associated with poor prognosis of hepatocellular carcinoma after curative resection. J. Cancer 10, 3333–3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Muramatsu A, Saito R, Iso T, Shibata T, Kuwata K, Kawaguchi SI, Iwawaki T, Adachi S, Suda H, et al. (2019). Molecular mechanism of cellular oxidative stress sensing by Keap1. Cell Rep. 28, 746–758 e744. [DOI] [PubMed] [Google Scholar]
- Taguchi K, Maher JM, Suzuki T, Kawatani Y, Motohashi H, and Yamamoto M (2010). Genetic analysis of cytoprotective functions supported by graded expression of Keap1. Mol. Cell. Biol. 30, 3016–3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Chen HY, Harris IS, Stover DG, Selfors LM, Bronson RT, Deraedt T, Cichowski K, Welm AL, Mori Y, et al. (2018). Cancer cells co-opt the neuronal redox-sensing channel TRPA1 to promote oxidativestress tolerance. Cancer Cell 33, 985–1003.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan MK, Lim HJ, Bennett EJ, Shi Y, and Harper JW (2013). Parallel SCF adaptor capture proteomics reveals a role for SCFFBXL17 in NRF2 activation via BACH1 repressor turnover. Mol. Cell 52, 9–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi K, and Karin M (2018). NF-kappaB, inflammation, immunity and cancer: coming of age. Nat. Rev. Immunol. 18, 309–324. [DOI] [PubMed] [Google Scholar]
- Tasdogan A, Faubert B, Ramesh V, Ubellacker JM, Shen B, Solmonson A, Murphy MM, Gu Z, Gu W, Martin M, et al. (2020). Metabolic heterogeneity confers differences in melanoma metastatic potential. Nature 577, 115–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tew KD (2005). TLK-286: a novel glutathione S-transferase-activated prodrug. Expert Opin. Investig. Drugs 14, 1047–1054. [DOI] [PubMed] [Google Scholar]
- Tiganis T (2011). Reactive oxygen species and insulin resistance: the good, the bad and the ugly. Trends Pharmacol. Sci. 32, 82–89. [DOI] [PubMed] [Google Scholar]
- Torti SV, Manz DH, Paul BT, Blanchette-Farra N, and Torti FM (2018). Iron and cancer. Annu. Rev. Nutr. 38, 97–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toullec A, Gerald D, Despouy G, Bourachot B, Cardon M, Lefort S, Richardson M, Rigaill G, Parrini MC, Lucchesi C, et al. (2010). Oxidative stress promotes myofibroblast differentiation and tumour spreading. EMBO Mol. Med. 2, 211–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend DM, He L, Hutchens S, Garrett TE, Pazoles CJ, and Tew KD (2008). NOV-002, a glutathione disulfide mimetic, as a modulator of cellular redox balance. Cancer Res. 68, 2870–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushio-Fukai M (2009). Compartmentalization of redox signaling through NADPH oxidase-derived ROS. Antioxid. Redox Signal. 11, 1289–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle I, Alvarez-Barrientos A, Arza E, Lamas S, and Monsalve M (2005). PGC-1alpha regulates the mitochondrial antioxidant defense system in vascular endothelial cells. Cardiovasc. Res. 66, 562–573. [DOI] [PubMed] [Google Scholar]
- van der Reest J, Lilla S, Zheng L, Zanivan S, and Gottlieb E (2018). Proteome-wide analysis of cysteine oxidation reveals metabolic sensitivity to redox stress. Nat. Commun. 9, 1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verschraagen M, Kedde MA, Hausheer FH, and Van Der Vijgh WJ (2003). The chemical reactivity of BNP7787 and its metabolite mesna with the cytostatic agent cisplatin: comparison with the nucleophiles thiosulfate, DDTC, glutathione and its disulfide GSSG. Cancer Chemother. Pharmacol. 51, 499–504. [DOI] [PubMed] [Google Scholar]
- Wang ZY, and Chen Z (2008). Acute promyelocytic leukemia: from highly fatal to highly curable. Blood 111, 2505–2515. [DOI] [PubMed] [Google Scholar]
- Wattenberg LW (1985). Chemoprevention of cancer. Cancer Res. 45, 1–8. [PubMed] [Google Scholar]
- Wiel C, Le Gal K, Ibrahim MX, Jahangir CA, Kashif M, Yao H, Ziegler DV, Xu X, Ghosh T, Mondal T, et al. (2019). BACH1 stabilization by antioxidants stimulates lung cancer metastasis. Cell 178, 330–345.e22. [DOI] [PubMed] [Google Scholar]
- Wood ZA, Poole LB, and Karplus PA (2003). Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science 300, 650–653. [DOI] [PubMed] [Google Scholar]
- Wu RS, Hong JJ, Wu JF, Yan S, Wu D, Liu N, Liu QF, Wu QW, Xie YY, Liu YJ, et al. (2017). OVOL2 antagonizes TGF-beta signaling to regulate epithelial to mesenchymal transition during mammary tumor metastasis. Oncotarget 8, 39401–39416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi C, Hu Y, Buckhaults P, Moskophidis D, and Mivechi NF (2012). Heat shock factor Hsf1 cooperates with ErbB2 (Her2/Neu) protein to promote mammary tumorigenesis and metastasis. J. Biol. Chem. 287, 35646–35657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao GG, Wang M, Li N, Loo JA, and Nel AE (2003). Use of proteomics to demonstrate a hierarchical oxidative stress response to diesel exhaust particle chemicals in a macrophage cell line. J. Biol. Chem. 278, 50781–50790. [DOI] [PubMed] [Google Scholar]
- Xu C, Huang MT, Shen G, Yuan X, Lin W, Khor TO, Conney AH, and Kong AN (2006). Inhibition of 7,12-dimethylbenz(a)anthracene-induced skin tumorigenesis in C57BL/6 mice by sulforaphane is mediated by nuclear factor E2-related factor 2. Cancer Res. 66, 8293–8296. [DOI] [PubMed] [Google Scholar]
- Yagoda N, von Rechenberg M, Zaganjor E, Bauer AJ, Yang WS, Fridman DJ, Wolpaw AJ, Smukste I, Peltier JM, Boniface JJ, et al. (2007). RAS-RAF-MEK-dependent oxidative cell death involving voltage-dependent anion channels. Nature 447, 864–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Kensler TW, and Motohashi H (2018). The KEAP1-NRF2 system: a thiol-based sensor-effector apparatus for maintaining redox homeostasis. Physiol. Rev. 98, 1169–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Zhang X, Wang L, Zhang R, Pu X, Wu S, Li L, Tong P, Wang J, Meng QH, et al. (2019). Inhibition of thioredoxin/thioredoxin reductase induces synthetic lethality in lung cancers with compromised glutathione homeostasis. Cancer Res. 79, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang IH, Lee JJ, Wu PC, Kuo HK, Kuo YH, and Huang HM (2020). Oxidative stress enhanced the transforming growth factor-beta2-induced epithelial-mesenchymal transition through chemokine ligand 1 on ARPE-19 cell. Sci. Rep. 10, 4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Carroll KS, and Liebler DC (2016a). The expanding landscape of the thiol redox proteome. Mol. Cell Proteomics 15, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, and Vousden KH (2016). Serine and one-carbon metabolism in cancer. Nat. Rev. Cancer 16, 650–662. [DOI] [PubMed] [Google Scholar]
- Yang WS, Kim KJ, Gaschler MM, Patel M, Shchepinov MS, and Stockwell BR (2016b). Peroxidation of polyunsaturated fatty acids by lipoxygenases drives ferroptosis. Proc. Natl. Acad. Sci. U S A 113, E4966–E4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WS, SriRamaratnam R, Welsch ME, Shimada K, Skouta R, Viswanathan VS, Cheah JH, Clemons PA, Shamji AF, Clish CB, et al. (2014). Regulation of ferroptotic cancer cell death by GPX4. Cell 156, 317–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WS, and Stockwell BR (2008). Synthetic lethal screening identifies compounds activating iron-dependent, nonapoptotic cell death in oncogenic-RAS-harboring cancer cells. Chem. Biol. 15, 234–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Dieter MZ, Chen Y, Shertzer HG, Nebert DW, and Dalton TP (2002). Initial characterization of the glutamate-cysteine ligase modifier subunit Gclm(−/−) knockout mouse. Novel model system for a severely compromised oxidative stress response. J. Biol. Chem. 277, 49446–49452. [DOI] [PubMed] [Google Scholar]
- Yao P, Sun H, Xu C, Chen T, Zou B, Jiang P, and Du W (2017). Evidence for a direct cross-talk between malic enzyme and the pentose phosphate pathway via structural interactions. J. Biol. Chem. 292, 17113–17120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Fan J, Venneti S, Wan YW, Pawel BR, Zhang J, Finley LW, Lu C, Lindsten T, Cross JR, et al. (2014). Serine catabolism regulates mitochondrial redox control during hypoxia. Cancer Discov. 4, 1406–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye ZW, Zhang J, Ancrum T, Manevich Y, Townsend DM, and Tew KD (2017). Glutathione S-transferase P-mediated protein S-glutathionylation of resident endoplasmic reticulum proteins influences sensitivity to drug-induced unfolded protein response. Antioxid. Redox Signal. 26, 247–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying W (2008). NAD+/NADH and NADP+/NADPH in cellular functions and cell death: regulation and biological consequences. Antioxid. Redox Signal. 10, 179–206. [DOI] [PubMed] [Google Scholar]
- Zavattari P, Perra A, Menegon S, Kowalik MA, Petrelli A, Angioni MM, Follenzi A, Quagliata L, Ledda-Columbano GM, Terracciano L, et al. (2015). Nrf2, but not beta-catenin, mutation represents an early event in rat hepatocarcinogenesis. Hepatology 62, 851–862. [DOI] [PubMed] [Google Scholar]
- Zhang L, Zhang J, Ye Z, Manevich Y, Townsend DM, Marshall DT, and Tew KD (2019). S-Glutathionylated serine proteinase inhibitors as biomarkers for radiation exposure in prostate cancer patients. Sci. Rep. 9, 13792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Guo J, Wei X, Niu C, Jia M, Li Q, and Meng D (2018a). Bach1: function, regulation, and involvement in disease. Oxid Med. Cell Longev. 2018, 1347969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Hu M, Yang Y, and Xu H (2018b). Organellar TRP channels. Nat. Struct. Mol. Biol. 25, 1009–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Cardaci S, Jerby L, MacKenzie ED, Sciacovelli M, Johnson TI, Gaude E, King A, Leach JD, Edrada-Ebel R, et al. (2015). Fumarate induces redox-dependent senescence by modifying glutathione metabolism. Nat. Commun. 6, 6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Shao L, and Spitz DR (2014). Reactive oxygen species in normal and tumor stem cells. Adv. Cancer Res. 122, 1–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HL, Zhang R, Anand P, Stomberski CT, Qian Z, Hausladen A, Wang L, Rhee EP, Parikh SM, Karumanchi SA, and Stamler JS (2019). Author Correction: metabolic reprogramming by the S-nitroso-CoA reductase system protects against kidney injury. Nature 570, E23. [DOI] [PubMed] [Google Scholar]
- Zhou W, Mo X, Cui W, Zhang Z, Li D, Li L, Xu L, Yao H, and Gao J (2016). Nrf2 inhibits epithelial-mesenchymal transition by suppressing snail expression during pulmonary fibrosis. Sci. Rep. 6, 38646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucker SN, Fink EE, Bagati A, Mannava S, Bianchi-Smiraglia A, Bogner PN, Wawrzyniak JA, Foley C, Leonova KI, Grimm MJ, et al. (2014). Nrf2 amplifies oxidative stress via induction of Klf9. Mol. Cell 53, 916–928. [DOI] [PMC free article] [PubMed] [Google Scholar]