Supplemental Digital Content is available in the text.
Keywords: Brugada syndrome, genetics, genotype, mutation, risk
Abstract
Background:
Brugada syndrome (BrS) is an oligogenic arrhythmic disease with increased risk of sudden cardiac arrest. Several BrS or ECG traits-related single-nucleotide polymorphisms (SNPs) were identified through previous genome-wide association studies in white patients. We aimed to validate these SNPs in BrS patients in the Taiwanese population, assessing the cumulative effect of risk alleles and the BrS-polygenic risk score in predicting cardiac events.
Methods:
We genotyped 190 unrelated BrS patients using the TWB Array, and Taiwan Biobank was used as controls. SNPs not included in the array were imputed by IMPUTE2. Cox proportional hazards model was used to evaluate the associations between each particular SNP, the collective BrS-polygenic risk score, and clinical outcomes.
Results:
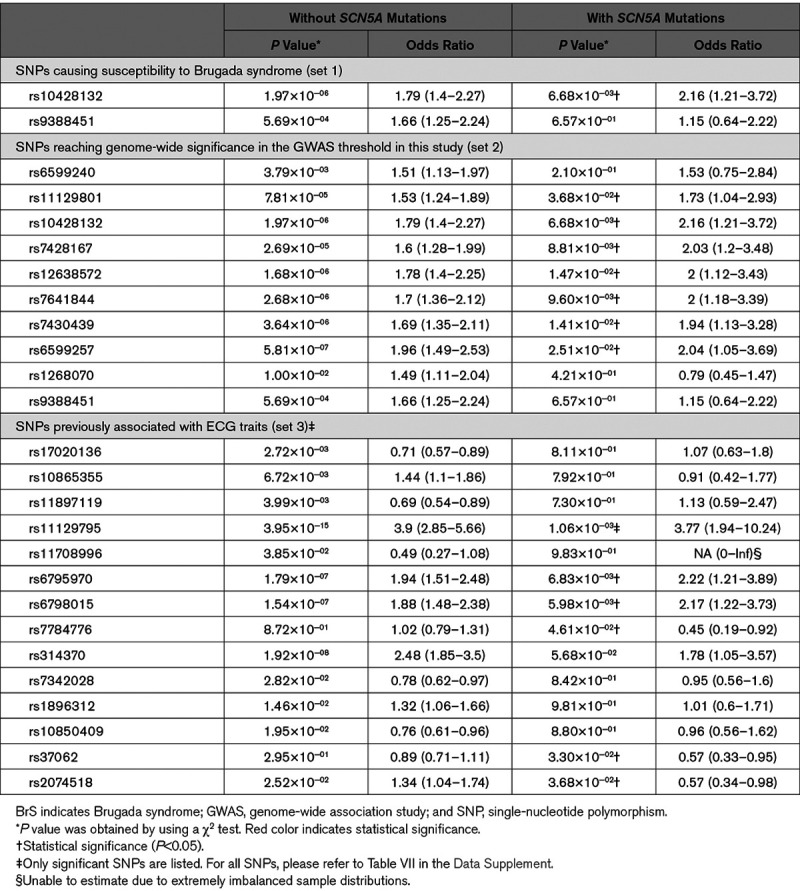
Of the 88 previously reported SNPs, 22 were validated in Taiwanese BrS patients (P<0.05). Of the 22 SNPs, 2 (rs10428132 and rs9388451) were linked with susceptibility to BrS, 10 were SNPs previously reaching genome-wide significance, and 10 were SNPs associated with ECG traits. For the 3 most commonly reported SNPs, disease risk increased consistently with the number of risk alleles (odds ratio, 3.54; Ptrend=1.38×10−9 for 5 risk alleles versus 1). Similar patterns were observed in both SCN5A mutation+ (odds ratio, 3.66; Ptrend=0.049) and SCN5A mutation− (odds ratio, 3.75; Ptrend=8.54×10−9) subgroups. Furthermore, BrS patients without SCN5A mutations had more risk alleles than BrS patients with SCN5A mutations regardless of the range of polygenic risk scores. Three SNPs (rs4687718, rs7784776, and rs2968863) showed significant associations with the composite outcome (sudden cardiac arrest plus syncope, hazard ratio, 2.13, 1.48, and 0.41; P=0.02, 0.006, and 0.008, respectively).
Conclusions:
Our findings suggested that some SNPs associated with BrS or ECG traits exist across multiple populations. The cumulative risk of the BrS-related SNPs is similar to that in white BrS patients, but it appears to correlate with the absence of SCN5A mutations.
Brugada syndrome (BrS), an oligogenic arrhythmic disease responsible for sudden cardiac arrest (SCA) in patients with structurally normal hearts, was first reported by Brugada and Brugada in 1992.1 BrS accounts for 4% of all sudden deaths and for up to 20% of sudden deaths in patients without structural cardiac disease.2 The prevalence of BrS is estimated to be 1 to 5 per 10 000 people in whites3 but is higher in Southeast Asians (12 per 10 000).4,5
In 1998, the SCN5A-encoded alpha-subunit of the voltage-gated Nav1.5 cardiac sodium channel was first associated with BrS.6 Although SCN5A is the most common BrS-susceptibility gene, it is responsible for only 20% of BrS cases in white populations7,8 and even less, 7.5% to 8% of BrS cases, in the Han Chinese population.9 BrS is generally considered a Mendelian disorder with autosomal dominant transmission and incomplete penetrance. Priori et al10 estimated that the overall disease penetrance across 4 small BrS families harboring mutations in the SCN5A gene was 16% (range, 12.5%–50%) based on their ECGs.11 In the past 2 decades, several BrS-associated genes and modifier genes have been reported, and most of these primarily encode sodium, potassium, and calcium channels or the proteins associated with these channels. However, disease-causing genes remain unknown in ≈80% to 85% of BrS patients. Additionally, the disease is sporadic in many patients.12,13 These observations suggest a more complex inheritance model.
To identify new genetic variants, Bezzina et al14 conducted a large-scale genome-wide association study (GWAS) with 312 BrS cases in the European populations and replicated the results in 594 cases from Europe and 208 cases from Japan. They reported 3 common single-nucleotide polymorphisms (SNPs) associated with BrS: rs11708996 in SCN5A, rs10428132 in SCN10A, and rs9388451 near HEY2. Furthermore, they analyzed the cumulative effect of these 3 SNPs on susceptibility to BrS with a BrS-polygenic risk score (BrS-PRS) and found that likelihood of BrS increased consistently with the number of risk alleles. However, it is unclear whether the previously identified SNPs are relevant in other racial groups such as the Taiwanese or whether population-specific SNPs influence BrS. In this study, we first aimed to validate the previously identified BrS-related SNPs in patients with BrS in Taiwan and assessed the cumulative risk of these SNPs on susceptibility to BrS. We then analyzed additional SNPs, including those reaching genome-wide significance in the GWAS from Bezzina et al and those associated with ECG traits (eg, PR interval, QRS duration, and QT interval). Since clinical outcomes of identified BrS-related common variants in the previous GWAS from Bezzina et al were not investigated, we further investigated the association between the BrS-PRS and clinical outcomes, and then compared them between BrS patients with and without pathogenic SCN5A variants.
Methods
This study was approved by the local ethical committee of National Taiwan University Hospital, and all participants gave informed consent before participating. The imputation workflow is illustrated in Figure 1, and the details of the methods are shown in the Data Supplement. The data that support the findings of this study are available from the corresponding author upon reasonable request.
Figure 1.
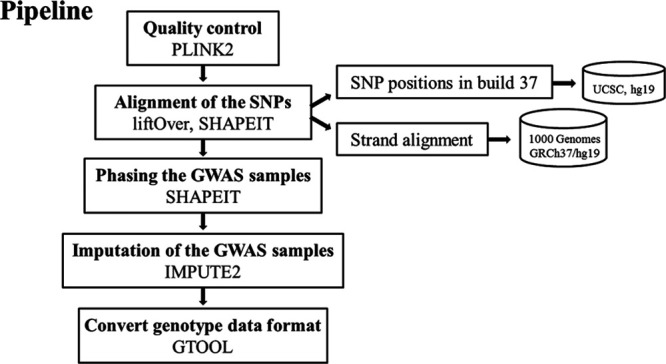
The workflow for the imputation approach. SNP indicates single-nucleotide polymorphism.
Results
Patient Population
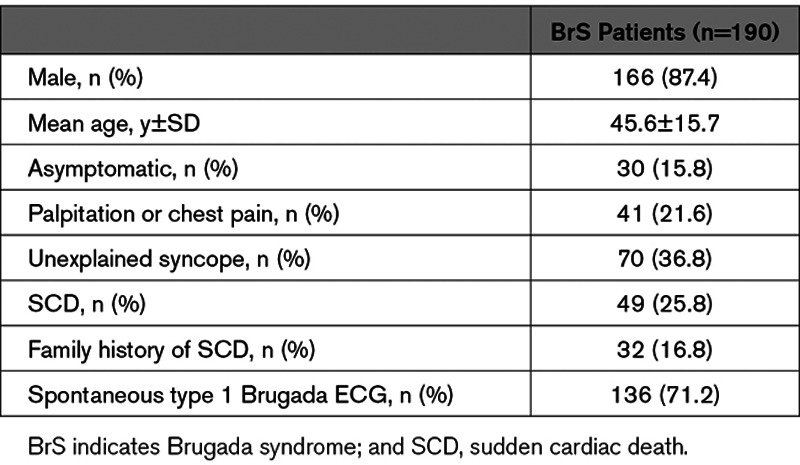
The basic demographic and clinical presentation of the 190 Taiwanese patients with BrS are shown in Table 1. The mean age of the patients with BrS was 45.6±15.7 years, and 87.4% of the patients were male. Regarding their symptoms, 62.6 % of the study patients were symptomatic (either SCA or unexplained syncope), and 71.2% presented with spontaneous type 1 Brugada ECG.
Table 1.
Demographic Data and Clinical Manifestations of the BrS Patients
Validation of the Imputation Method by In Silico Approaches and Direct Sequencing
To ensure the high quality of the imputed genotyping calls of the variants in this study, we verified the accuracy of the imputation method by using it on sequences for which microarray data were available. We found that the concordance of genotypes obtained by the imputation algorithm and the actual microarray was >90%. In addition to the in silico analyses, we performed direct sequencing to validate the genotyping calls obtained by imputation. We randomly selected 3 SNP loci in 20 samples for this validation, and all of the genotypes were the same as those obtained by using the imputation approach. These results indicate that imputation is a feasible and efficient approach to obtain genotyping calls for the SNP loci that were originally not detectable in the microarrays.
Comparisons of the Variants With Significance in BrS Patients Between White and Taiwanese Populations
Among the 3 SNPs previously shown to cause susceptibility to BrS (set 1), only rs9388451 was available on the Affymetrix TWB chip. As shown in Table 2, the allele frequency of rs9388451 was significantly higher in the Taiwanese BrS patients than the healthy controls (P=0.003). We obtained the genotyping calls of the other 2 SNPs (rs11708996 and rs10428132) by using the imputation approach, which was validated by Sanger sequencing. Only rs10428132 showed significant differences between BrS patients and controls (P=5.92×10−8).
Table 2.
Validation of the Previously Reported SNPs in the Taiwanese BrS Patients
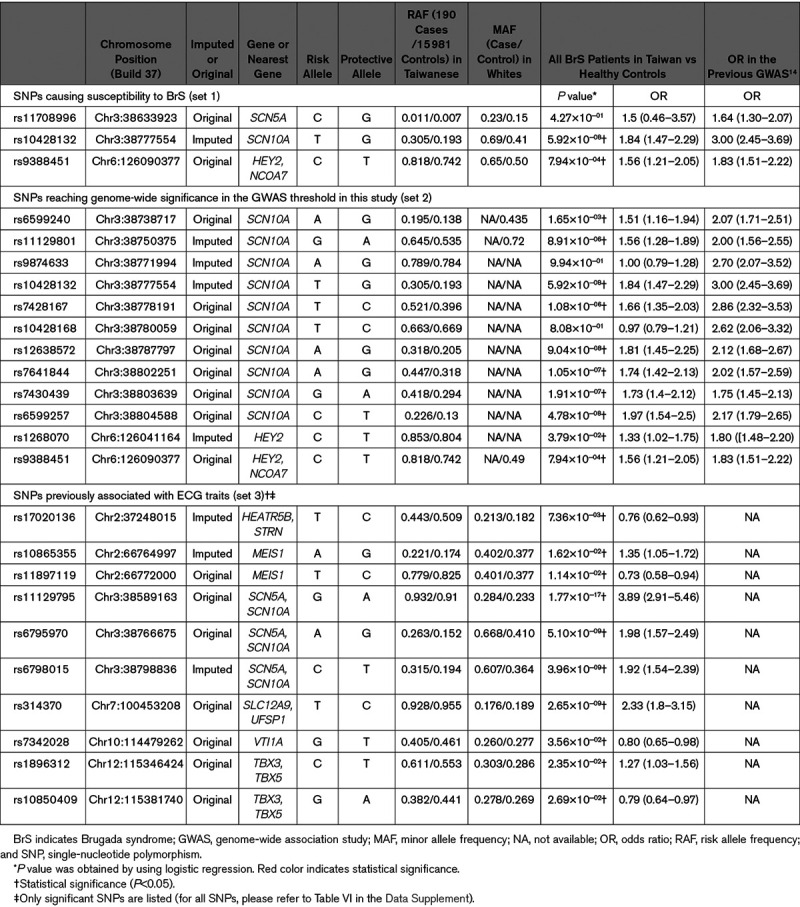
Among the 12 SNPs reaching genome-wide significance in the previous GWAS from Bezzina et al14 (set 2), most of them are located in SCN10A. As shown in Table 2, 10 out of the 12 SNPs had significantly higher allele frequencies in Taiwanese BrS patients than in the healthy controls. In general, the results were similar to those reported in the white BrS patients.
Among the 75 SNPs associated with ECG traits (set 3), 10 showed significant differences in Taiwanese BrS patients versus healthy controls (Table 2). Among the 10 SNPs, only 4—rs11129795, rs6795970, rs6798015, and rs314370—reached significance in the GWAS significance threshold (<5×10−8). In conclusion, the results suggested that the general patterns of the important SNPs in BrS patients were similar between white and Taiwanese populations, but the low number of replicated SNPs associated with ECG traits implied they were not important targets for BrS in Taiwanese patients.
Comparisons of the PRS Between BrS Patients and the Healthy Controls From Taiwan
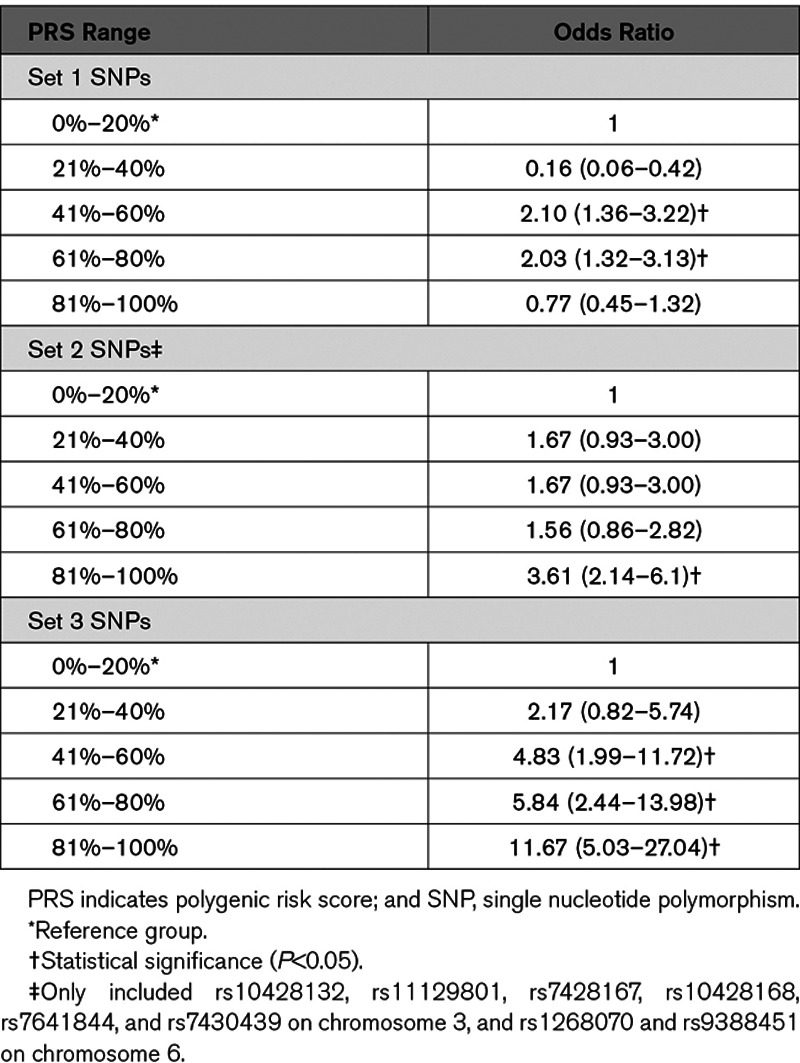
In addition to the single-marker tests, we developed PRS models using the 3 sets of SNPs. The weighting for the PRS models was obtained by using all BrS patients in Taiwan versus the healthy controls. The healthy controls were classified into 5 subgroups with equal PRS ranges. The group with the lowest PRS (0%–20%) was used as the reference to calculate the odds ratios (ORs) of disease risk. We calculated the ORs by dividing the number of BrS patients by the number of healthy controls in each subgroup. The results are summarized in Table 3. For the 3 SNPs in set 1 (BrS-PRS14), the BrS patients with the relatively higher PRS (61%–80%) had a significantly higher risk of disease (OR, 2.03) than the healthy controls, but this OR was not the highest. For the 12 SNPs reaching genome-wide significance (set 2), only rs10428132 and rs12638572 showed a high correlation (R2>0.7) in the LD calculation. We selected rs10428132 for further analyses because rs10428132 has lower P value in Table 2. After excluded rs12638572, other SNPs in set 2 were left for further analyses. To further address the issue of genomic inflation, we performed a condition analysis for each SNP set 2 by conditioning on rs10428132, which was the leading SNP reported by the previous GWAS. The results of the conditional analyses are summarized in Table I in the Data Supplement. Consequently, we used the 7 significant SNPs in Table I in the Data Supplement along with rs10428132 to develop the PRS model. As shown in Table 3, the BrS patients with the highest PRS (81%–100%) showed significantly higher disease risk (likelihood of BrS diagnosis) than controls (OR, 3.61 [2.14–6.10]). For the 75 SNPs associated with ECG traits (set 3), the BrS patients with relatively higher scores (41%–100%) reported significantly higher disease risk (ORs, 4.83–11.67). Similar to the second set, the BrS patients with the highest PRS (81%–100%) displayed the highest OR (11.67). In summary, the results of the 3 different PRS models demonstrated that the cumulative effects of the reported SNPs, as reflected in the PRSs, were effective markers in distinguishing Taiwanese BrS patients and healthy controls.
Table 3.
Comparisons of Disease Risk Using the PRS Models Generated by the 3 Sets of SNPs
Cumulative Effects of the 3 Major Risk Alleles (Set 1, BrS-PRS) on Susceptibility to BrS in Taiwanese BrS Patients
To compare the cumulative effect of the 3 previously reported major risk alleles in Taiwanese BrS patients, the allele frequencies of these 3 SNPs were evaluated between BrS patients and healthy controls.14 The result is illustrated in Figure 2. First, we compared the disease risk between all BrS patients and healthy controls and found that the risk increased consistently with the number of risk alleles present (Ptrend=1.38×10−9), with the estimated OR reaching 3.54 in the presence of >4 risk allele copies versus only 1 risk allele (Figure 2A). Second, we divided the BrS patients into 2 groups based on the presence or absence of SCN5A mutations. Similar patterns were observed in both SCN5A mutation+ (OR=3.66, Ptrend=0.049) and SCN5A mutation− (OR=3.75, Ptrend=8.54×10−9) subgroups (Figure 2B and 2C). However, some discrepancies of the ORs can be observed between the SCN5A mutation+ and SCN5A mutation− subgroups, suggesting some genetic differences may exist in the 2 subgroups.
Figure 2.
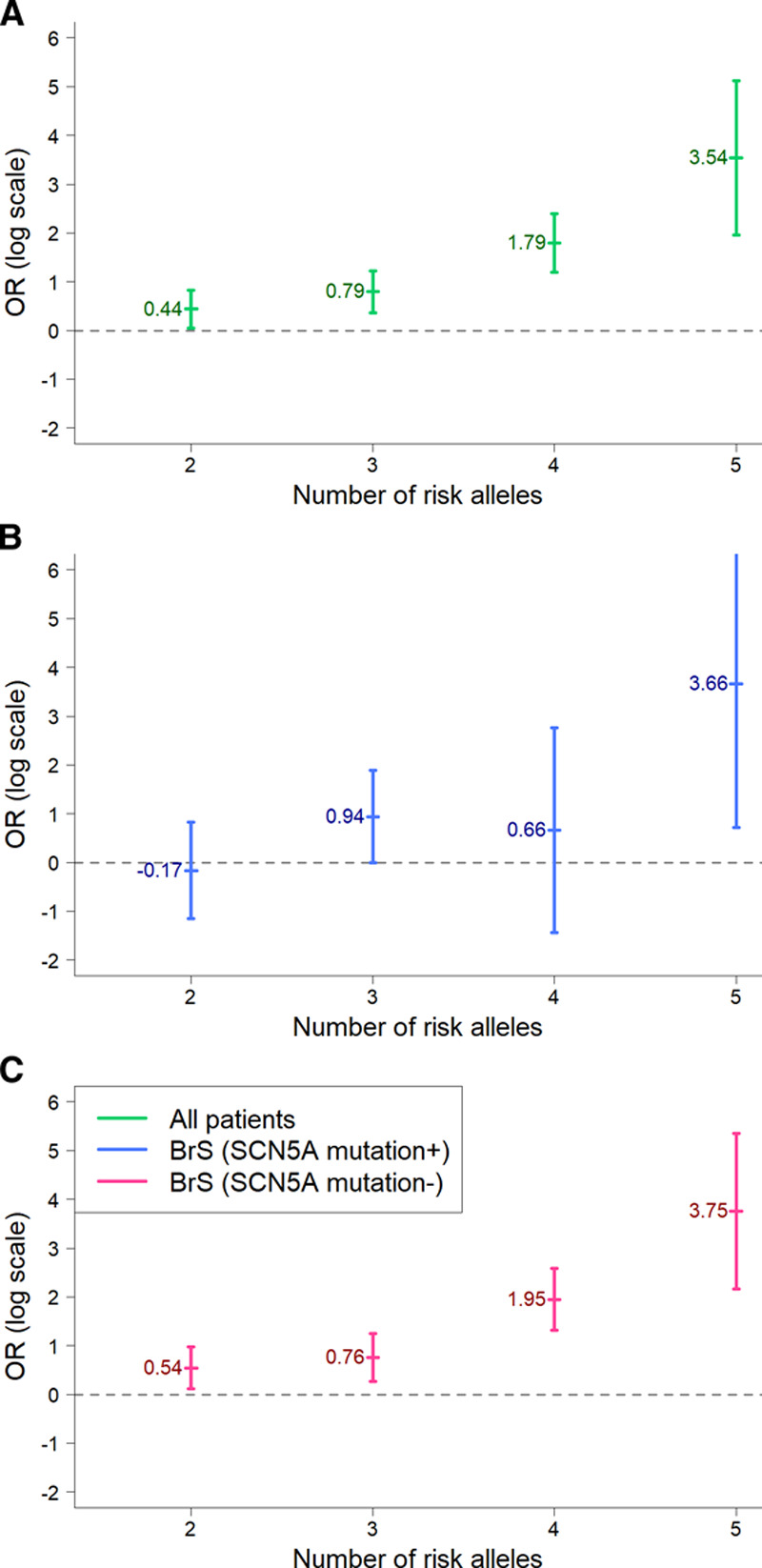
Cumulative effect of the 3 reported major risk alleles (set 1, Brugada syndrome [BrS]-polygenic risk score [PRS]14) on susceptibility to BrS in Taiwanese BrS patients. The x axis shows the number of risk alleles in one individual, whereas the y axis shows the odds ratios (ORs) of the BrS patients on a log scale. A, All BrS patients vs controls (Ptrend=1.38×10−9); (B) SCN5A mutation-positive BrS patients vs controls (Ptrend=0.049); and (C) SCN5A mutation-negative BrS patients vs controls (Ptrend=8.54×10−9).
Comparisons of the Previously Reported SNPs in BrS Patients With and Without SCN5A Mutations
Since SCN5A has been reported as the most dominant gene in BrS, albeit still accounting for only 20% of BrS in whites, we divided the BrS patients into 2 subgroups accordingly (SCN5A mutation+ versus SCN5A mutation−). The results are shown in Table 4. In set 1 SNPs, rs10428132 was significant in BrS patients in both subgroups (P<0.05). However, rs9388451 was only significant in the SCN5A mutation− BrS patients. In set 2 SNPs, the 10 SNPs which were significant in all combined BrS patients were still significant in the SCN5A mutation− BrS patients. Intriguingly, 3 SNPs (rs6599240, rs1268070, and rs9388451) were only significant in the SCN5A mutation− BrS patients but not in the SCN5A mutation+ BrS patients. Similar patterns were observed in the 75 SNPs associated with ECG traits (set 3 SNPs). Among the 10 significant SNPs of set 3 in Table 2, all of them were significant in the SCN5A mutation− BrS patients, but only 3 were significant in the SCN5A mutation+ BrS patients. Notably, 2 SNPs (rs37062 and rs7784776) were significant in the SCN5A mutation+ BrS patients alone, which were not in the 10 significant SNPs, and intriguingly they were both protective SNPs in the SCN5A mutation+ BrS patients. In addition, rs2074518 was not significant in all combined BrS patients, but it was significant in both BrS mutation+ and mutation− patients. However, rs2074518 was a risk allele in BrS SCN5A mutation− patients but a protective allele in BrS SCN5A mutation+ patients. Therefore, these differences between the SCN5A mutation+ and SCN5A mutation− BrS patients hinted that future investigations should classify the patients into subgroups instead of combining them into one unit.
Table 4.
Summary of the Significant Previously Reported SNPs in Taiwanese BrS Patients With or Without SCN5A Mutations
In addition to the single-marker tests, we examined the cumulative proportions of the 3 BrS-associated SNPs (set 1, BrS-PRS14) in the Taiwanese BrS patients after being stratified by the presence/absence of SCN5A pathogenic variants (Figure 3). Notably, the SCN5A mutation− BrS patients had more risk alleles (at least 4 risk alleles). Furthermore, we developed the PRS models using weighting schemes generated from the SCN5A mutation− BrS patients versus healthy controls. The results are summarized in Figure 4 and Table II in the Data Supplement. Using the PRS model generated from the 3 SNPs in set 1 (BrS-PRS14), the ORs of the disease risk showed differences in BrS patients with and without SCN5A variants (Figure 4A) in the 21% to 80% PRS groups. However, the results have negative ORs in the 21% to 40% PRS group whereas positive ORs in the 41% to 80% PRS groups. These fluctuating ORs may result from the low number of SNPs analyzed in the PRS model. Alternatively, obvious differences were observed in the ORs obtained from the PRS models generated from set 2 SNPs or set 3 SNPs (Figure 4B and 4C). In general, most ORs were higher in the SCN5A mutation− BrS patients than in the SCN5A mutation+ BrS patients, regardless of the high or low range of PRS. These results further indicated that the genetic markers differ between BrS1 patients with a pathogenic SCN5A variant and those patients diagnosed with BrS who are SCN5A negative.
Figure 3.
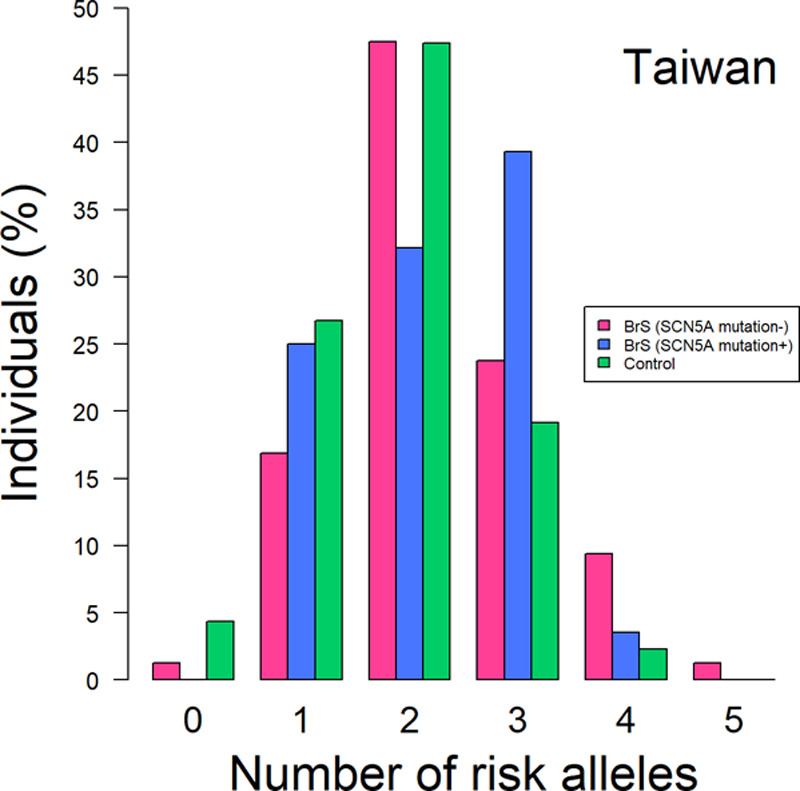
The distribution of the risk alleles in the Taiwanese Brugada syndrome (BrS) patients and healthy controls. The BrS patients were divided into 2 groups based on whether they possessed SCN5A mutations or not. The x axis shows the number of risk alleles in one individual, whereas the y axis shows the percentage of the people with the alleles.
Figure 4.
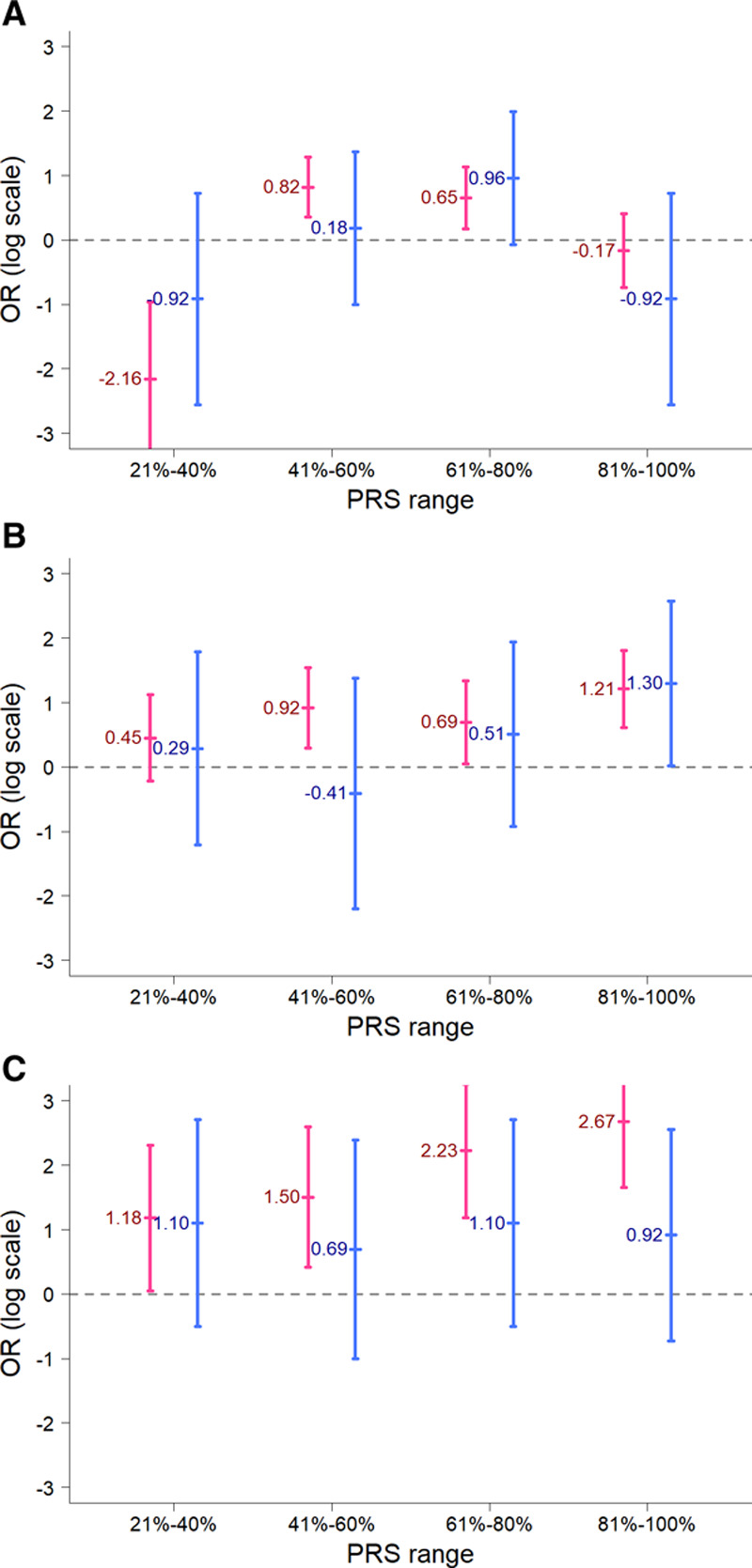
Comparisons of the odds ratios for the polygenic risk score (PRS) models between the Taiwanese Brugada syndrome (BrS) patients with and without SCN5A mutations. The x axis shows the ranges of PRS scores in the BrS patients, whereas the y axis shows the odds ratios on a log scale. A, The PRS model developed using the 3 single-nucleotide polymorphisms (SNPs; set 1, BrS-PRS14); (B) the PRS model was developed using the 8 SNPs from set 2 (rs6599257, rs11129801, rs10428132, rs7428167, rs7641844, and rs7430439 in chromosome 3 and rs1268070 and rs9388451 in chromosome 6); (C) the PRS model developed using the 75 SNPs (set 3). The BrS patients were divided into 2 groups based on whether they had pathogenic SCN5A mutations (blue color) or not (red color).
Associations of the Previously Reported SNPs and PRS With Clinical Outcomes
We evaluated whether these SNPs were able to predict clinical outcomes (SCA or unexplained syncope or both) in BrS patients. First, a single marker test was used for all previously reported SNPs (set 1–3) in Table III in the Data Supplement. Of these SNPs, 3 SNPs (rs4687718 in TKT, rs7784776 in IGFBP3, and rs2968863 in KCNH2) showed significant associations with the composite clinical outcome (SCA plus syncope; P<0.05, Table IV in the Data Supplement). Notably, rs7784776 was only significant in the SCN5A mutation− BrS patients, whereas rs4687718 was significant only in the SCN5A mutation+ BrS patients. Only the SNP, rs2968863, showed protective effects in all BrS patients no matter whether they had an SCN5A variant or not. We also examined whether the PRS models generated from the 3 sets of previously reported SNPs were able to predict the clinical outcomes or not. Unfortunately, none of the results of the 3 PRS models were significant, and thus they cannot serve as a predictor for the clinical outcomes in Taiwanese BrS patients.
Discussion
In this study, we successfully validated 22 of the 88 previously identified BrS- or ECG traits-related SNPs in patients with BrS in Taiwan. We assessed the cumulative effects of the SNPs on the risk of BrS and their association with clinical outcomes.
In 2013, Bezzina et al reported 3 important common SNPs associated with BrS, including rs11708996 in SCN5A, rs10428132 in SCN10A, and rs9388451 near HEY2,14 and found that disease risk (likelihood of BrS diagnosis) increased consistently with the number of risk alleles. In the current study, rs10428132 and rs9388451 were successfully validated in Taiwanese BrS patients, demonstrating that the same variants can affect BrS across different populations. Intriguingly, the MAF difference of rs9388451 between the cases and controls was only 0.08 in the Taiwanese population, which was lower than that in both the Japanese (0.11) and white populations (0.15),14 suggesting that racial differences should be taken into consideration because of the differing allele frequencies in the BrS patients with different ethnic backgrounds.
In addition, the cumulative effect of the 3 major risk alleles on susceptibility to BrS increased with the number of risk alleles present in the Taiwanese BrS patients, consistent with the results in white BrS patients.14 However, the cumulative effect was smaller in Taiwanese BrS patients than in white BrS patients; this might be caused by the smaller sample size of our study cohort or by ethnic differences.
Validation of the 12 SNPs Reaching Genome-Wide Significance in White BrS Patients
Among the 12 SNPs which reached genome-wide significance in white BrS patients (set 2), 10 were successfully validated in Taiwanese BrS patients, providing further evidence that SCN10A and HEY2 are highly associated with BrS. The P values were relatively higher in the Taiwanese population, which may have resulted from the limited sample size of BrS patients or from genetic differences between the 2 populations.
Comparisons of the Reported SNPs (Set 1, 2, and 3) and PRS Between the BrS Patients With SCN5A Mutation and the BrS Patients Without SCN5A Mutations
SCN5A is the major BrS-causing gene but is responsible for only 20% of BrS cases in white populations7,8 and even less in the BrS patients in the Japanese and Taiwanese population. (7.5%–8%),9,15 which might imply that the genetic background of BrS is partly different in different populations. In this study, we found that BrS patients without SCN5A mutations had more risk alleles (4 or 5 risk alleles) in the BrS-PRS14 than BrS patients with SCN5A mutations. In addition, the cumulative effect of the 3 common SNPs on susceptibility to BrS was larger in the BrS patients without SCN5A mutations than in BrS patients with SCN5A mutations. This may suggest that SCN5A mutations are dominant drivers of BrS whereas other minor variants/genes need higher quantities to display their effects. However, when the PRS generated by the 3 common SNPs was applied, there was no significant difference in disease risk between the 2 groups. This discrepancy might be explained by the limited number of the SNPs in this 3-SNP model or the assumption that the genetic effect of each SNP is the same. When the PRS generated by more SNPs (10 SNPs [set 2] or 75 SNPs [set 3], all ORs of disease risk were consistently higher in the BrS patients without SCN5A mutations than in the BrS patients with SCN5A mutations, regardless of the range of PRS. Furthermore, we found that more SNPs reaching genome-wide significance in the GWAS threshold in this study (set 2) and more SNPs associated with ECG traits (set 3) were validated in the BrS patients without SCN5A mutations than in the BrS patients with SCN5A mutations, although some of the SNPs were overlapped. These results indicated that it is necessary to consider the effects of the SNPs in BrS patients in the context of SCN5A variants, because the genetic architecture between BrS patients with SCN5A mutations and without SCN5A mutations may be different. In addition, the cumulative effects of SNP set 2 from Bezzina et al which were mostly located on chromosome 3, showed different ORs between SCN5A (+) and SCN5A (−) BrS patients (Figure 4B). This indicated that a SCN5A (−) BrS patient tends to have more risk alleles in SNP set 2. However, due to the limitations of the experimental techniques used in this study, we could only consider the effects of multiple SNPs by an additive model instead of a haplotype. Therefore, further investigations are warranted to explore whether chromosome 3 haplotype can provide synergetic effects in BrS patients.
Limitations
Some limitations exist in this study. First, although the sample size for all BrS patients was almost 200, it may still have lacked sufficient power to do the patient stratification by SCN5A mutations. It is well known that only around 20% of the BrS patients have pathogenic SCN5A variants, which resulted in <50 patients with SCN5A variants in this study. However, the current data indicated huge differences in MAFs between BrS patients with and without SCN5A mutations, suggest that it is worth doing stratification in future investigations. Second, we used the 1000 Genomes East Asian population as the reference in the imputation approach. This reference is perhaps not the best one, because the genetics of the Taiwanese population may be closer to the Han Chinese population. However, the sample size would be relatively limited if we used the Han Chinese population only, and thus we decided to use all East Asian populations to include more diverse haplotypes in the reference. Based on the high accuracy values validated by the imputation algorithm and the PCR experiment, we think this is not a critical issue in this study. In addition to the validations of DNA variants associated with BrS from a previous GWAS, we performed the analysis of all SNPs examined in the TWB chip in the current data set containing 190 BrS cases and around 16 000 healthy Taiwanese controls in Taiwan. The preliminary results of our primary GWAS were summarized in Table V in the Data Supplement. However, notably some of the P values were very low (Table V in the Data Supplement). We were afraid that these astounding findings may be too good to be true. These extremely low P values may be the result of the probe design, technical issues in the GWAS chip or the hugely imbalanced sample sizes in our GWAS (190 cases versus 16,000 controls). Therefore, validations of these DNA loci in independent cohorts are prerequisite to further apply them into advanced researchers and applications.
Conclusions
Of the 88 previously reported SNPs, 22 SNPs were validated in Taiwanese BrS patients, and 3 SNPs (rs4687718, rs7784776, and rs2968863) were associated with composite clinical outcomes (SCA plus syncope). The cumulative effect of the 3 major risk alleles on susceptibility to BrS was larger in BrS patients than in healthy controls. Furthermore, this effect was also larger in SCN5A genotype negative BrS patients than those with SCN5A-mediated BrS (BrS1). PRSs showed that all ORs of disease risk were consistently higher in BrS patients without SCN5A mutations than in those with SCN5A mutations, regardless of the range of PRS.
Acknowledgments
We are sincerely grateful to the staff of the Sixth Core Lab, Department of Medical Research, National Taiwan University Hospital and National Center of Genome Medicine for technical support in the genotyping, and all doctors participated the SADS-TW Brugada syndrome (BrS) registry (see Data Supplement). We thank Melissa Stauffer, of Scientific Editing Solutions, for editing the article.
Sources of Funding
Financial support for this research was provided partially through grants NTUH 106-S3469, NTUH106-S3458, NTUH 106-018, UN 103-018, and UN104-001 from National Taiwan University Hospital, Taiwan Health Foundation, and NSC 101-2314-B-002-168-MY2, NSC 101-2314-B-002-173-MY2, NSC 103-2314-B-002-148, MOST 104-2314-B-002-193-MY3, MOST 106-2314-B-002-047-MY3, MOST 106-2314-B-002 -134-MY2, MOST 106-2314-B-002-206, MOST 107-2314-B-002-261-MY3, MOST 108-2314-B-002-007 from the Ministry of Science and Technology, GTZ300 of the Center for Biotechnology, National Taiwan University, Taiwan. MJA is supported by the Mayo Clinic Windland Smith Rice Comprehensive Sudden Cardiac Death Program.
Disclosure
None.
Supplementary Material
Footnotes
Nonstandard Abbreviations and Acronyms
- BrS
- Brugada syndrome
- SCA
- sudden cardiac arrest
- PRS
- polygenic risk score
- SNP
- single-nucleotide polymorphism
For Sources of Funding and Disclosures, see page 210.
The Data Supplement is available at https://www.ahajournals.org/doi/suppl/10.1161/CIRCGEN.119.002797.
References
- 1.Brugada P, Brugada J. Right bundle branch block, persistent ST segment elevation and sudden cardiac death: a distinct clinical and electrocardiographic syndrome. A multicenter report. J Am Coll Cardiol. 1992;20:1391–1396. doi: 10.1016/0735-1097(92)90253-j. doi: 10.1016/0735-1097(92)90253-j. [DOI] [PubMed] [Google Scholar]
- 2.Antzelevitch C, Brugada P, Borggrefe M, Brugada J, Brugada R, Corrado D, Gussak I, LeMarec H, Nademanee K, Perez Riera AR, et al. Brugada syndrome: report of the second consensus conference: endorsed by the Heart Rhythm Society and the European Heart Rhythm Association. Circulation. 2005;111:659–670. doi: 10.1161/01.CIR.0000152479.54298.51. doi: 10.1161/01.CIR.0000152479.54298.51. [DOI] [PubMed] [Google Scholar]
- 3.Berne P, Brugada J. Brugada syndrome 2012. Circ J. 2012;76:1563–1571. doi: 10.1253/circj.cj-12-0717. doi: 10.1253/circj.cj-12-0717. [DOI] [PubMed] [Google Scholar]
- 4.Mizusawa Y, Wilde AA. Brugada syndrome. Circ Arrhythm Electrophysiol. 2012;5:606–616. doi: 10.1161/CIRCEP.111.964577. doi: 10.1161/CIRCEP.111.964577. [DOI] [PubMed] [Google Scholar]
- 5.Juang JM, Chen CY, Chen YH, Wu IC, Hsu CC, Chen LN, Tang FC, Wang CC, Juan CC, Chiu HC, et al. Prevalence and prognosis of brugada electrocardiogram patterns in an elderly han Chinese population: a nation-wide community-based study (halst cohort). Europace. 2015;17(suppl 2):ii54–62. doi: 10.1093/europace/euv141. [DOI] [PubMed] [Google Scholar]
- 6.Chen Q, Kirsch GE, Zhang D, Brugada R, Brugada J, Brugada P, Potenza D, Moya A, Borggrefe M, Breithardt G, et al. Genetic basis and molecular mechanism for idiopathic ventricular fibrillation. Nature. 1998;392:293–296. doi: 10.1038/32675. doi: 10.1038/32675. [DOI] [PubMed] [Google Scholar]
- 7.Kapplinger JD, Tester DJ, Alders M, Benito B, Berthet M, Brugada J, Brugada P, Fressart V, Guerchicoff A, Harris-Kerr C, et al. An international compendium of mutations in the SCN5A-encoded cardiac sodium channel in patients referred for Brugada syndrome genetic testing. Heart Rhythm. 2010;7:33–46. doi: 10.1016/j.hrthm.2009.09.069. doi: 10.1016/j.hrthm.2009.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crotti L, Marcou CA, Tester DJ, Castelletti S, Giudicessi JR, Torchio M, Medeiros-Domingo A, Simone S, Will ML, Dagradi F, et al. Spectrum and prevalence of mutations involving BrS1- through BrS12-susceptibility genes in a cohort of unrelated patients referred for Brugada syndrome genetic testing: implications for genetic testing. J Am Coll Cardiol. 2012;60:1410–1418. doi: 10.1016/j.jacc.2012.04.037. doi: 10.1016/j.jacc.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Juang JM, Tsai CT, Lin LY, Liu YB, Yu CC, Hwang JJ, Chen JJ, Chiu FC, Chen WJ, Tseng CD, et al. Unique clinical characteristics and SCN5A mutations in patients with Brugada syndrome in Taiwan. J Formos Med Assoc. 2015;114:620–626. doi: 10.1016/j.jfma.2013.02.002. doi: 10.1016/j.jfma.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 10.Priori SG, Napolitano C, Gasparini M, Pappone C, Della Bella P, Brignole M, Giordano U, Giovannini T, Menozzi C, Bloise R, et al. Clinical and genetic heterogeneity of right bundle branch block and ST-segment elevation syndrome: A prospective evaluation of 52 families. Circulation. 2000;102:2509–2515. doi: 10.1161/01.cir.102.20.2509. doi: 10.1161/01.cir.102.20.2509. [DOI] [PubMed] [Google Scholar]
- 11.Juang JJ, Horie M. Genetics of Brugada syndrome. J Arrhythm. 2016;32:418–425. doi: 10.1016/j.joa.2016.07.012. doi: 10.1016/j.joa.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schulze-Bahr E, Eckardt L, Breithardt G, Seidl K, Wichter T, Wolpert C, Borggrefe M, Haverkamp W. Sodium channel gene (SCN5A) mutations in 44 index patients with Brugada syndrome: different incidences in familial and sporadic disease. Hum Mutat. 2003;21:651–652. doi: 10.1002/humu.9144. doi: 10.1002/humu.9144. [DOI] [PubMed] [Google Scholar]
- 13.Hermida JS, Dassonvalle E, Six I, Amant C, Coviaux F, Clerc J, Herent D, Hermida A, Rochette J, Jarry G. Prospective evaluation of the familial prevalence of the Brugada syndrome. Am J Cardiol. 2010;106:1758–1762. doi: 10.1016/j.amjcard.2010.07.049. doi: 10.1016/j.amjcard.2010.07.049. [DOI] [PubMed] [Google Scholar]
- 14.Bezzina CR, Barc J, Mizusawa Y, Remme CA, Gourraud JB, Simonet F, Verkerk AO, Schwartz PJ, Crotti L, Dagradi F, et al. Common variants at SCN5A-SCN10A and HEY2 are associated with Brugada syndrome, a rare disease with high risk of sudden cardiac death. Nat Genet. 2013;45:1044–1049. doi: 10.1038/ng.2712. doi: 10.1038/ng.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamagata K, Horie M, Aiba T, Ogawa S, Aizawa Y, Ohe T, Yamagishi M, Makita N, Sakurada H, Tanaka T, et al. Genotype-phenotype correlation of SCN5A mutation for the clinical and electrocardiographic characteristics of probands with Brugada syndrome: a Japanese Multicenter Registry. Circulation. 2017;135:2255–2270. doi: 10.1161/CIRCULATIONAHA.117.027983. doi: 10.1161/CIRCULATIONAHA.117.027983. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


