Abstract
Avian coronavirus infectious bronchitis virus (IBV) is an important pathogen threatening poultry production worldwide. Here, two recombinant IBVs (rYN-1a-aYN and rYN-1b-aYN) were generated in which ORF1a or ORF1b of the virulent YN genome were replaced by the corresponding regions from the attenuated strain aYN. The pathogenicity and virulence of rIBVs were evaluated in ovo and in vivo. The results revealed that mutations in the ORF1a gene during passage in embryonated eggs caused the decreased pathogenicity of virulent IBV YN strain, proven by determination of virus replication in ECEs and CEK cells, the observation of clinical signs, gross lesions, microscopic lesions, tracheal ciliary activity and virus distribution in chickens following exposure to rIBVs. However, mutations in ORF1b had no obvious effect on virus replication in both ECEs and CEK cells, or pathogenicity in chickens. Our findings demonstrate that the replicase 1a gene of avian coronavirus IBV is a determinant of pathogenicity.
Keywords: Coronavirus, Infections bronchitis virus, Replicase gene, Pathogenicity
Highlights
-
•
Avian coronavirus IBV is an important pathogen of chickens.
-
•
The effect of the replicase 1 ab gene on IBV pathogenicity were evaluated.
-
•
The replicase 1a gene is a determinant of IBV virulence.
1. Introduction
Coronaviridae are members of the Nidovirales order, a family of positive-sense RNA (+ssRNA) viruses that infect a wide range of host species. Formally, the Coronaviridae can be divided into alpha-, beta-, gamma-, and deltacoronavirus genera (Weiss and Leibowitz, 2011). The α-coronaviruses (CoVs) include transmissible gastroenteritis virus (TGEV), porcine epidemic diarrhea virus (PEDV) and feline infectious peritonitis virus (FIPV) (Perlman and Netland, 2009). Three human CoVs, severe acute respiratory syndrome coronavirus (SARS-CoV), middle east respiratory syndrome coronavirus (MERS-CoV) and a novel coronavirus (COVID-19), which belong to the β-CoVs, lately emerged from wild animals into the human population and caused severe respiratory disease with high morbidity and mortality rates (de Wit et al., 2016; Fehr et al., 2017; Hui et al., 2020). Infectious bronchitis virus (IBV) belongs to the γ-CoV genus and causes a contagious disease of chickens that negatively impacts on poultry production worldwide (Cavanagh, 2007; Cook et al., 2012). δ-CoVs were identified primarily in multiple songbird species and in leopard cats, and porcine δ-CoV was initially detected in 2009 in fecal samples from pigs in Asia (Woo et al., 2012; Zhang, 2016).
IBV, like the other CoVs, has one of the largest known positive-stranded RNA genomes, approximately 27.6 kb in size. The 5ʹ-proximal two-thirds of the CoV genome comprise two open reading frames (ORFs) that together constitute the replicase gene, ORF1a and ORF1b. ORF1a translation yields polyprotein 1a (pp1a; approximately 4000–4500 amino acid [aa] residues), the polyprotein encoded by ORF1b is generated by an a-1 ribosomal frameshift to yield pp1ab (6700–7200 aa residues in total). The pp1a and pp1ab polyproteins are proteolytically processed by two virally-encoded proteinases to generate 15 mature proteins that drive viral genome replication and subgenomic mRNA synthesis (Fang et al., 2010; Snijder et al., 2016). A virulent IBV field isolate can be attenuated by multiple passages in specific-pathogen-free (SPF) embryonated eggs due to the accumulation of amino acid mutations in the genome (Bande et al., 2015; Yan et al., 2018). Comparison of the genome sequences between virulent and attenuated avian IBV strains revealed that attenuated strain aYN shared 99.57% nucleotide (nt) sequence identity with the parental strain YN, and only eight sense mutations were found in the replicase gene ORF1a and two sense mutations in the ORF1b gene (Zhao et al., 2019a, Zhao et al., 2019b). It is not known whether these mutations are responsible for the attenuation and replication of virulent IBV, similar to the S gene and 5a that have been identified previously (Zhao et al., 2019a, Zhao et al., 2019b). The mutations in the replicase gene (ORF1a and ORF1b) associated with the attenuation of pathogenicity in chickens remain unknown.
In this study, we used an IBV reverse genetics system based on the virulent YN strain to generate recombinant IBVs (rIBVs) in which ORF1a or ORF1b of the YN genome was replaced by the corresponding regions from the attenuated strain aYN. The pathogenicity and virulence of recombinant rescued viruses were investigated in ovo and in vivo. The aim of this study was to determine whether the replicase gene 1a or 1b play a role in pathogenesis of IBV, to highlight viral components critical for efficient replication and pathogenicity of γ-CoV.
2. Materials and methods
2.1. Viruses and cells
Vaccinia virus strains vNotI/tk, CV-1 and D980R, and BHK-21 cells were kindly provided by Dr. Volker Thiel (University of Bern, Switzerland) and were used to assemble the full-length genome of mutant IBV. vNotI/tk-IBV rYN recombinant virus was constructed previously (Zhao et al., 2019a, Zhao et al., 2019b). Strains CV-1 and D980R were cultured in minimum essential medium (MEM; Thermo Fisher Scientific, Waltham, MA, USA) with 10% fetal bovine serum (FBS; Gibco, Grand Island, NY, USA). BHK cells were used for electroporation to rescue the rYN strain with ORF1a and ORF1b of aYN strain. Chicken embryonic kidney (CEK) cells were prepared from 18-day-old SPF chicken embryos and were cultured in Dulbecco's modified Eagle's medium (DMEM) (Hyclone, Logan, UT, USA) supplemented with 10% FBS, 100 U/ml penicillin and 100 mg/ml streptomycin (Gillette, 1973).
2.2. Animals and ethics statement
All SPF chickens and SPF embryonated eggs were purchased from Beijing Boehringer Ingelheim Vital Biotechnology Co. Ltd. (Beijing, China). The chickens were maintained in isolators throughout the experiments. The animal experimental protocols were approved by the Beijing Administration Committee of Laboratory Animals under the auspices of the Beijing Association for Science and Technology (approval ID SYXK [Jing] 2018-0038) and Ethical Censor Committee at China Agricultural University (CAU approval No. 19125).
2.3. Generation of recombinant virus
Recombinant rYN-1a-aYN and rYN-1b-aYN viruses were generated containing a replacement of aYN ORF1a or ORF1b. Briefly, the obtained positive plasmids, pGPT-ORF1a-positive and pGPT-ORF1b-positive, were integrated into the vaccinia virus genome by homologous recombination, to which only recombinant vaccinia viruses with the Ecogpt gene show resistance. The verified viruses were reconstructed with the obtained negative plasmids, pGPT-ORF1a-negative and pGPT-ORF1b-negative, which originate from the attenuated IBV strain aYN. The amino acid mutations were introduced into the rYN cDNA by homologous recombination using the transient dominant selection system (Zhao et al., 2019a, Zhao et al., 2019b). After screening and transcription in vitro, the full-length RNAs of rYN-ORF1a-aYN and rYN-ORF1b-aYN were electroporated into BHK cells, cells and supernatant were freeze-thawed three times, then inoculated into 10-day-old SPF embryonated chicken eggs (ECEs). The successful rescued viruses were passaged in 10-day-old ECEs until the stable appearance of dwarf embryos, the recombinant viruses were sequenced to make sure no extra mutations existed.
2.4. In vitro and in ovo growth kinetics of rIBVs
To compare the in vitro replication of the recombinant viruses and wild-type virus rYN on CEK cells, 200 μl of serum-free media containing 102.0 of the 50% tissue culture infectious dose (TCID50) of rYN, rYN-1a-aYN or rYN-1b-aYN virus were inoculated onto CEK cells in 24-well plates. Cell culture supernatants were collected at the indicated time points (6, 12, 24, 36, 48 and 60 h) for a real-time PCR detection assay for IBV N gene as described previously (Zhao et al., 2015). To compare the in ovo replication of the recombinant viruses and wild-type virus rYN in ECEs, 200 μl of serum-free media containing 102.0 of the 50% egg infectious dose (EID50) of rYN, rYN-1a-aYN or rYN-1b-aYN virus were inoculated into the allantoic cavities of 10-day-old ECEs. The allantoic fluid of five eggs from each group was harvested at the indicated time points (12, 24, 36, 48, 60 and 72 h) and pooled for a real-time PCR detection assay. The virus copy numbers at each time point were detected in triplicate and calculated according to a standard curve.
2.5. Comparison of the in vivo pathogenicity of rIBVs
One hundred 1-day-old SPF chickens were randomly divided into four groups (A, B, C, and D), with each group containing 22 birds being subdivided into 12 and 10 chickens for clinical sampling and observation, respectively. Chickens were inoculated via eye-drop with 100 μl containing 106.0 EID50 of IBV rYN (A), rYN-1a-aYN (B), rYN-1b-aYN (C) or PBS as the negative control (D). Chickens were observed daily for 14 days and their clinical signs and mortality were recorded. Clinical signs, consisted of sneezing, tracheal rale, dyspnea and depression, were given daily clinical scores: 0 for normal, 1 for mild depression, 2 for severe depression, 3 for paralysis/prostration and 4 for death.
At 3, 5, 7, 10 and 14 days post-infection (dpi), two birds from each sampled group were euthanized and necropsied. Gross lesions were noted and tissue samples from the trachea, kidney and lung were collected for virus detection by a real-time PCR assay or histopathological examination, meanwhile the tracheas of the necropsied chickens were evaluated for trachea ciliary activity.
2.6. Histopathology test
Tissue samples from the trachea, lung and kidney were fixed in 10% neutral buffered formalin for 24 h at room temperature. Fixed samples were embedded in paraffin wax. Sections were stained with hematoxylin and eosin (HE) and were observed using standard light microscopy for lesions due to IBV infection.
2.7. IBV detection by real-time RT-qPCR
The total RNA of sampled tissues, cell-culture supernatants and allantoic fluid was extracted using the Total RNA Purification Kit (GeneMarkbio, Taiwan, China) according to the manufacturer's instructions. Then, 500 ng of RNA was transcribed using PrimeScript™ RT Master Mix (TaKaRa, Shiga, Japan), and the cDNA was used as a template for SYBR Green I qPCR to detect the viral load of the IBV 5′UTR gene as described previously (Xu et al., 2018). All qPCR reactions were repeated at least twice, and expression of the IBV gene was calculated via a standard curve.
2.8. Inhibition of ciliary activity
To evaluate tracheal ciliostasis, three sections each of the upper, middle and lower part of the trachea were analyzed. Nine tracheal rings per bird were placed in a 96-well plate containing DMEM. The degree of integrity and preservation of the ciliary movement of the tracheal epithelial cells were observed under an inverted light microscope at a magnification of 400 × . The average ciliostasis score was calculated according to the following criteria: 0 if the cilia in the complete tracheal section showed movement; 1 if the cilia of 75%–100% of the tracheal section showed movement; 2 if the cilia of 50%–75% of the trachea section showed movement; 3 if the cilia of 25%–50% of the trachea section showed movement; and 4 if the cilia of less than 25% of the trachea section showed movement or there was no movement at all. The average ciliostasis score of each group was calculated.
2.9. Statistical analysis
Statistical analysis was performed using GraphPad Prism version 6.01 (GraphPad Software Inc., San Diego, CA, USA). Viral copies and ciliary activity were statistically evaluated by one-way or two-way ANOVA, followed by Bonferroni's multiple comparison tests. The following notations are used to indicate significant differences between groups: *p < 0.05; **p < 0.01; ***p < 0.001.
3. Results
3.1. Generation of recombinant IBVs
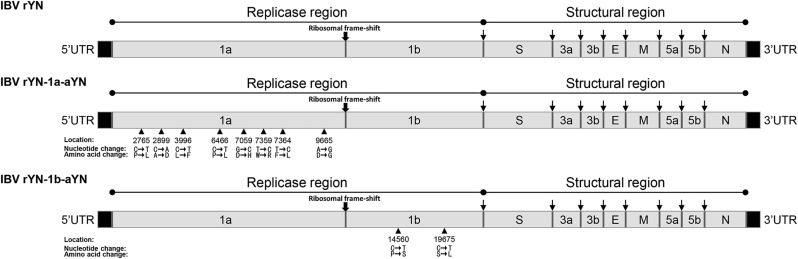
On comparing the ORF1ab amino acid sequence of attenuated virus aYN with the parental strain rYN, 10 sense mutations were detected in the replicase gene 1 ab, including eight mutations in ORF1a and two mutations in ORF1b. To determine whether these mutations in ORF1ab influenced the replication ability and virulence of IBV, two rIBVs were generated in which ORF1a or ORF1b were replaced with the corresponding regions of attenuated strain aYN, designated rYN-1a-aYN and rYN-1b-aYN (Fig. 1 ). The rIBVs were rescued and then passaged 10 times in SPF chicken embryos and confirmed by sequencing. The results demonstrate that rIBVs was obtained successfully and that the viruses remained stable upon passages. The 10th virus stocks were titrated and used for further characterization.
Fig. 1.
Schematic overview of the recombinant virus construction used in the present study. The nucleotide location was determined according to the IBV wild-type YN strain derived genome sequence.
3.2. Growth characteristics of rIBVs
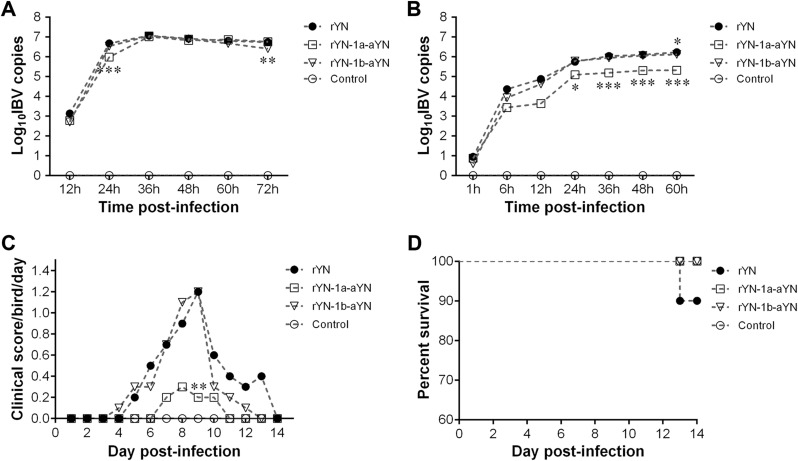
The growth characteristics of the rIBVs, rYN-1a-aYN and rYN-1b-aYN, were compared with those of the rescued wild-type virus rYN in vitro and in ovo (Fig. 2 ). Both rIBVs (rYN-1a-aYN and rYN-1b-aYN) showed growth profiles similar to that of rYN in ECEs, with maximum titers of 107 virus copies/200 μl by 36 h post-infection (hpi). No significant differences were observed in the growth kinetics of the rIBVs, with the exception of rYN-1a-aYN at 24 hpi and rYN-1b-aYN at 72 dpi, for which the titers were between 5.1-fold (p < 0.001) and 2.5-fold (p < 0.01) less than the titers observed for rYN at the corresponding time point (Fig. 2A).
Fig. 2.
Growth characteristics of IBV rYN, rYN-1a-aYN or rYN-1b-aYN, and clinical scores and survival rates of chickens inoculated with these IBV strains. (A) Growth characteristics in SPF embryonated eggs. (B) Growth characteristics in CEK cells. (C) Clinical signs consisted of sneezing, tracheal rale, dyspnea and depression, and were ascribed daily clinical scores. (D) Survival rates. Viral loads were determined by measuring the expression of the N gene in cell lysates using absolute RT-qPCR. Two-way ANOVA was used for the analysis of differences in GraphPad Prism 6.01 and significance levels were set as follows: significant at p < 0.05 (*), highly significant at p < 0.01 (**) and extremely significant at p < 0.001 (***).
In CEK cells, IBV rYN-1b-aYN showed similar growth kinetics to parental virus rYN, except that the replication titer was about 1.3-fold (p < 0.05) lower than that of the parental IBV rYN (Fig. 2B). However, the growth profile of IBV rYN-1a-aYN showed different traits, for which all of the titers were above 1 log 10 unit at all of the indicated time points, with the exception of 1 hpi. This indicated that there was less effective replication or release of rIBV following replacing with the mutations in ORF1a from aYN. These results revealed that the notable mutations in ORF1a during passage in embryonated eggs had a minor detrimental effect on virus replication in ECEs and a significant detrimental effect on virus replication in CEK cells. However, mutations in ORF1b had no obvious effect on virus replication in either ECEs or CEK cells.
3.3. Clinical signs in chickens following exposure to rIBVs
To understand the different clinical symptoms caused by the recombinant rIBVs, clinical scores were calculated for 10 chickens from each experiment group (Fig. 2C). The chickens infected with rIBVs began to show clinical signs at 4 dpi. The most prominent clinical signs were characterized by ruffled feathers, gasping, sneezing, tracheal rales and depression. Chickens infected with rYN-1b-aYN showed similar serious clinical symptoms to those infected with parental stain rYN during the 14 day observation period. In the rYN-1a-aYN challenged group, the clinical signs were milder and shorter in duration than those observed in the rYN-1b-aYN and rYN challenged groups, and the control group showed no clinical signs (Fig. 2C). Of the 10 chicks, one bird died at 13 dpi, giving a mortality rate of 10% in the rYN challenged group. No deaths attributable to IBV were observed in the rYN-1a-aYN or rYN-1b-aYN challenged groups or the control group (Fig. 2D).
3.4. Gross lesions in chickens following exposure to rIBVs
Gross lesions detected in the respiratory tract and kidney of chickens infected with rIBVs included tracheal exudate or hemorrhages, laryngeal hemorrhages, and swollen or pale kidneys with urate deposition in the organs and ureters. In the rYN and rYN-1b-aYN challenged groups, chickens showed more serious lesions than those in the rYN-1a-aYN group, including punctate hemorrhages and catarrhal exudates in the throat and trachea at 3, 5, 7, 10 and 14 dpi, congestion and edema in the lungs at 5 and 10 dpi, and swelling with urate deposits in the tubules and ureters of the kidney at 10 dpi. No gross lesions were observed in the trachea, lung or kidney in the control group. The detailed results are summarized in Table 1 .
Table 1.
Gross lesions in SPF chickens inoculated with IBV strains rYN-1a-aYN, rYN-1b-aYN and rYN.
| Strains | Lesions | Days post-challenge (dpi) |
||||
|---|---|---|---|---|---|---|
| 3 | 5 | 7 | 10 | 14 | ||
| rYN-1a-aYN | Laryngeal hemorrhage | 1/2 a | 1/2 | 1/2 | 1/2 | 2/7 b |
| Tracheal exudate/hemorrhage | 0/2 | 0/2 | 0/2 | 1/2 | 0/7 | |
| Renal enlargement/urate | 0/2 | 0/2 | 0/2 | 0/2 | 0/7 | |
| rYN-1b-aYN | Laryngeal hemorrhage | 2/2 | 2/2 | 2/2 | 2/2 | 4/5 c |
| Tracheal exudate/hemorrhage | 1/2 | 1/2 | 1/2 | 2/2 | 2/5 | |
| Renal enlargement/urate | 0/2 | 0/2 | 0/2 | 0/2 | 0/5 | |
| rYN | Laryngeal hemorrhage | 1/2 | 2/2 | 2/2 | 2/2 | 5/7 |
| Tracheal exudate/hemorrhage | 1/2 | 2/2 | 1/2 | 2/2 | 4/7 | |
| Renal enlargement/urate | 0/2 | 0/2 | 0/2 | 1/2 | 0/7 | |
| PBS | Laryngeal hemorrhage | 0/2 | 0/2 | 0/2 | 0/2 | 0/7 |
| Tracheal exudate/hemorrhage | 0/2 | 0/2 | 0/2 | 0/2 | 0/7 | |
| Renal enlargement/urate | 0/2 | 0/2 | 0/2 | 0/2 | 0/7 | |
Number of chickens exhibiting gross lesions out of the examined number of birds.
End of the observation period at 14 dpi.
Two chickens died during the observation period.
3.5. Microscopic lesions in chickens following exposure to rIBVs
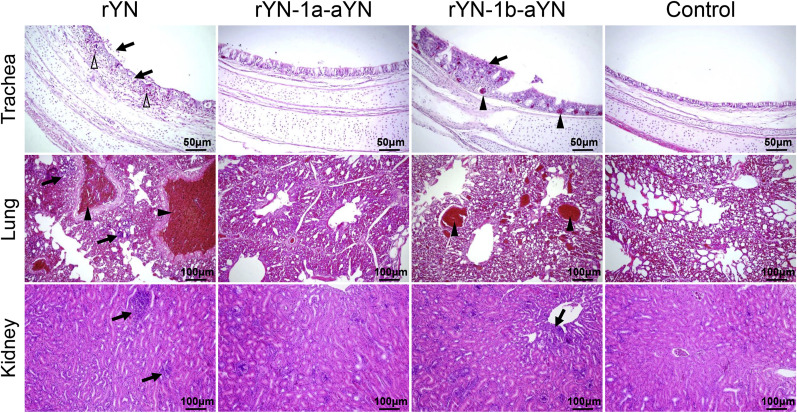
Microscopic lesions in the trachea, lung and kidney in all groups were also analyzed by histopathological examination and the HE staining results were consistent with the gross lesions described above. More severe histologic lesions were observed in the rYN and rYN-1b-aYN challenged group than those in the rYN-1a-aYN infected groups, with the most serious lesions appearing around 5, 7 and 10 dpi. The microscopic lesions of chickens at 10 dpi were showed in Fig. 3 . In the trachea of chickens in group rYN and rYN-1b-aYN, lesions included loss of cilia and epithelial cells (black arrows), hemorrhages (empty triangles), congestion (black triangles) and lymphoid cell infiltration (black arrows) in tracheal mucosa lamina propria. While congestion (black triangles) and infiltration of lymphoid cells (black arrows) into the lung, interstitial infiltration of lymphoid cells (black arrows) in the kidney were recorded in most infected chickens in group rYN and rYN-1b-aYN. No obvious lesions were observed in the rYN-1a-aYN challenged and control groups. These results showed that the lesions in the rYN and rYN-1b-aYN infected groups more significant than those in the rYN-1a-aYN infected groups, which indicated that the mutations in ORF1a have a significant influence on the pathogenicity of IBV, while the mutations in ORF1b have no obvious impact on pathogenicity.
Fig. 3.
Histopathologic changes in the trachea, lung and kidney at 10 dpi of chickens inoculated with IBV rYN, rYN-1a-aYN and rYN-1b-aYN strains. Different symbols indicate typical lesions detected in the tissue. Scale bar: 50 μm for trachea, 100 μm for lung and kidney.
3.6. Tracheal ciliary activity in chickens following exposure to rIBVs
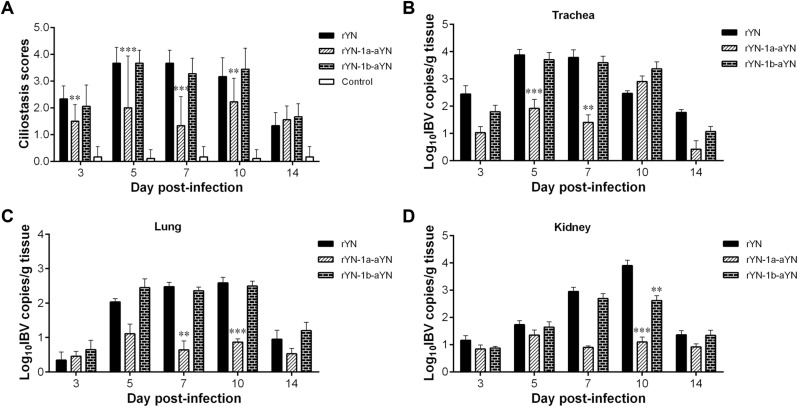
Inhibition of ciliary activity was observed at 3, 5, 7, 10 and 14 dpi in the trachea (Fig. 4 A). On comparing the damage to ciliary activity caused by rIBVs, the rYN-1a-aYN challenged group showed lower damage with a highly significant difference at 3 and 10 dpi (p < 0.01) and an extremely significant difference at 5 and 7 days post-challenge (p < 0.001) compared with the rYN group. However, the rYN-1b-aYN infected group showed similar inhibition of ciliary activity compared with the parental strain rYN challenged group, leading to severe damage of the trachea cilia. The results indicated that the amino acid mutations in ORF1a obviously improved ciliary activity (Fig. 4A).
Fig. 4.
Tracheal ciliostasis scores (A) and IBV viral loads in different tissues (B–D) of chickens inoculated with IBV rYN, rYN-1a-aYN and rYN-1b-aYN strains. Statistical significance was considered as follows: highly significant at p < 0.01 (**) and extremely significant at p < 0.001 (***).
3.7. Virus distribution in the tissues of chickens following exposure to rIBVs
We detected the number of viral RNA copies in the trachea, lung and kidney using an SYBR Green I RT-qPCR assay (Fig. 4B–D). The number of viral RNA copies varied in different organs between the different infected groups. The viral RNA levels in the tracheas of the rYN-1a-aYN infected group were significantly lower than those of the rYN group at 5 and 7 dpi (p < 0.01 or 0.001) (Fig. 4B). In the lungs, the virus copy numbers in the rYN-1a-aYN infected group were also significantly lower than those of the rYN group at 7 and 10 dpi (p < 0.01 or 0.001) (Fig. 4C). The numbers of viral RNA copies in the kidneys of the rYN-1a-aYN infected group were lower than in the rYN group at 10 dpi (p < 0.001) (Fig. 4D). There were no differences between the rYN-1b-aYN and rYN groups with regard to the viral titers in other tissues. All of the results indicated that the replacement of attenuated strain ORF1a limited viral replication in chicken tissues.
4. Discussion
IBV is a highly infectious pathogen in domestic fowl that replicates mainly in the respiratory tract but also in epithelial cells of other organs, including the kidney, oviduct and gut (Britton et al., 2012; Cavanagh, 2003; Cavanagh et al., 2007), causing infectious bronchitis (IB) that is responsible for economic losses in the poultry industry throughout the world. It is extremely important to identify viral components critical for efficient replication and pathogenicity of coronavirus, as this knowledge will be beneficial for vaccine development and disease prevention.
IB is mainly controlled by live attenuated vaccines derived from virulent viruses that have undergone multiple (usually >50) serial passages (Cavanagh, 2007; Sjaak et al., 2011). As a consequence, the mutations associated with attenuation of pathogenicity in chickens exist in structural, accessory and replicase genes. Our previous research compared the genome sequences of virulent IBV YN and attenuated IBV aYN and revealed 10 sense mutations in the replicase gene 1 ab, 10 mutations in the S gene region, and an 81-nt deletion in the inter region (IR) between M and ORF 5a, providing evidence that the S gene and 5a accessory gene are responsible for the attenuation of virulent IBV (Zhao et al., 2015, 2019). Many studies focused on the structural protein and the accessory protein have demonstrated their important role in the replication and pathogenicity of coronavirus (Casais et al., 2005; Shan et al., 2018; Laconi et al., 2018; van Beurden et al., 2018)). By contrast, little is known about the replicase gene, which accounts for two-thirds of the coronavirus genome. Researchers used the IBV Beaudette strain as a backbone to construct a recombinant virus containing the replicase gene from strain M41; however, virulence was not restored in this recombinant virus (Keep et al., 2018; Armesto et al., 2009). This may indicate that avirulent strain Beaudette was not the ideal background in which to study the role of IBV-specific mutations in pathogenicity. We suspect that mutations in the replicase gene may play an important role in attenuation of IBV.
In this study, we used an IBV reverse genetics system based on virulent strain YN of IBV to generate rIBVs in which the replicase gene (ORF 1a and ORF1b) of the YN genome were replaced with the corresponding regions from attenuated YN strain aYN. Successfully rescued viruses were characterized and subsequently investigated for pathogenicity and virulence in ovo and in vivo. According to the replication kinetic results on CEK cells and in SPF chicken embryos, the notable mutations in ORF1a had a minor detrimental effect on virus replication in ECEs but a significant detrimental effect on virus replication in CEK cells. Mutations in ORF1b had no effect on virus replication in both ECEs and CEK cells. The eight mutations in ORF1a had a significant effect on virus replication especially in CEK cells, unlike the S and 5a genes for which no differences were detected after replacement (Zhao et al., 2019a, Zhao et al., 2019b). The replicase gene plays a vital role in the coronavirus lifecycle and it can be translated and cleaved into nsp 1–nps16, that assemble into the replicase-transcriptase complex, which is instrumental in RNA synthesis, replication and transcription of sub-genomic RNAs (Mielech et al., 2014; Ziebuhr et al., 2000). We propose that the eight amino acid mutations affect or damage the function of one or more nsps, impacting on the replication of IBV; however, this requires further study.
The pathogenicity of 1-day-old SPF chickens provided further evidence of the decrease in virulence of rYN-1a-aYN. Compared with wild-type rYN, no mortality or clinical signs were observed during the observation period. Gross and pathological lesions in the trachea, lung and kidney were also negligible. The ciliary activity was improved and the viral loads in the trachea, lung and kidney were significantly decreased. According to these results, rYN-1a-aYN was attenuated after replacement of eight mutations from the replicase gene ORF1a. The replicase gene of IBV encodes 15 nsps, some of which have known enzymatic functions (Ziebuhr, 2008). How these proteins function in pathogenesis is still not well understood; however, mutation of some of the nsps of other coronaviruses has been associated with the loss of virulence. For example, the mutation of TGEV nsp 1 does not affect virus replication in cell culture but significantly reduces TGEV pathogenicity in pigs (Shen et al., 2019). The macrodomain and PLP2 mutation of MHV nsp3 triggers the production of type I interferon in vitro and causes attenuation in mice (Deng et al., 2019). For the nsp14 mutant of MHV, replication was attenuated in wild-type bone marrow-derived macrophages (BMMs) and nsp14 was required for resistance to the innate immune response (Case et al., 2018). The loss of nsp15 activity resulted in greatly attenuated disease in mice and stimulated a protective immune response, indicating that nsp15 is critical for evasion of host dsRNA sensors in macrophages, and that modulating nsp15 stability and activity is a strategy for generating live attenuated vaccines (Deng et al., 2017). An nsp 16 mutant of MERS-CoV demonstrated significant attenuation in both primary human airway cell cultures and in vivo (Menachery et al., 2017). Overall most of the nsps appear to serve important functions and have a major impact on virus replication and pathogenicity. That may explain why ORF1a affected virus replication in CEK cells and caused attenuation in SPF chickens.
In conclusion, our results demonstrate that the mutations in the ORF1a gene that result from the passage of IBV in ECEs are important and account for the decreased pathogenicity of virulent IBV and the detrimental effects on virus replication. However, mutations in ORF1b had no obvious effect on virus replication in ECEs or CEK cells, or on pathogenicity in chickens. It remains unclear which of the eight mutations in ORF1a had the most significant impact and whether this mutation affects nsp functions. Further studies are needed to address these questions.
CRediT authorship contribution statement
Jing Zhao: Conceptualization, Methodology, Software, Writing - original draft. Keran Zhang: Methodology, Formal analysis, Software. Jinlong Cheng: Methodology, Software, Visualization. Wenfeng Jia: Methodology. Ye Zhao: Supervision, Validation. Guozhong Zhang: Supervision, Writing - review & editing, Funding acquisition.
Declaration of competing interest
The authors have declared no conflicts of interest.
Acknowledgements
This study was supported by the National Key Research and Development Program of China (2017YFD0500700). We thank Kate Fox, DPhil, from Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
References
- Armesto M., Cavanagh D., Britton P. The replicase gene of avian coronavirus infectious bronchitis virus is a determinant of pathogenicity. PLoS One. 2009;4 doi: 10.1371/journal.pone.0007384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bande F., Arshad S.S., Bejo M.H., Moeini H., Omar A.R. Progress and challenges toward the development of vaccines against avian infectious bronchitis. J. Immunol. Res. 2015;2015:424860. doi: 10.1155/2015/424860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton P., Armesto M., Cavanagh D., Keep S. Modification of the avian coronavirus infectious bronchitis virus for vaccine development. Bioeng. Bugs. 2012;3:114–119. doi: 10.4161/bbug.18983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casais R., Davies M., Cavanagh D., Britton P. Gene 5 of the avian coronavirus infectious bronchitis virus is not essential for replication. J. Virol. 2005;79:8065–8078. doi: 10.1128/JVI.79.13.8065-8078.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case J.B., Li Y., Elliott R., Lu X., Graepel K.W., Sexton N.R., Smith E.C., Weiss S.R., Denison M.R. Murine hepatitis virus nsp 14 exoribonuclease activity is required for resistance to innate immunity. J. Virol. 2018;92 doi: 10.1128/JVI.01531-17. e01531-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D. Severe acute respiratory syndrome vaccine development: experiences of vaccination against avian infectious bronchitis coronavirus. Avian Pathol. 2003;32:567–582. doi: 10.1080/03079450310001621198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet. Res. 2007;38:281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Casais R., Armesto M., Hodgson T., Izadkhasti S., Davies M., Lin F., Tarpey I., Britton P. Manipulation of the infectious bronchitis coronavirus genome for vaccine development and analysis of the accessory proteins. Vaccine. 2007;25:5558–5562. doi: 10.1016/j.vaccine.2007.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook J.K., Jackwood M., Jones R.C. The long view: 40 years of infectious bronchitis research. Avian Pathol. 2012;41:239–250. doi: 10.1080/03079457.2012.680432. [DOI] [PubMed] [Google Scholar]
- de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Hackbart M., Mettelman R.C., O'Brien A., Mielech A.M., Yi G., Kao C.C., Baker S.C. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc. Natl. Acad. Sci. U.S.A. 2017;114:E4251–E4260. doi: 10.1073/pnas.1618310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Mettelman R.C., O'Brien A., Thompson J.A., O'Brien T.E., Baker S.C. Analysis of coronavirus temperature-sensitive mutants reveals an interplay between the macrodomain and papain-like protease impacting replication and pathogenesis. J. Virol. 2019;93 doi: 10.1128/JVI.02140-18. e02140-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S., Shen H., Wang J., Tay F.P., Liu D.X. Functional and genetic studies of the substrate specificity of coronavirus infectious bronchitis virus 3C-like proteinase. J. Virol. 2010;84:7325–7336. doi: 10.1128/JVI.02490-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Channappanavar R., Perlman S. Middle east respiratory syndrome: emergence of a pathogenic human coronavirus. Annu. Rev. Med. 2017;68:387–399. doi: 10.1146/annurev-med-051215-031152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillette K.G. Plaque formation by infectious bronchitis virus in chicken embryo kidney cell cultures. Avian Dis. 1973;17:369–378. [PubMed] [Google Scholar]
- Hui D.S., I Azhar E., Madani T.A., Ntoumi F., Kock R., Dar O., Ippolito G., Mchugh T.D., Memish Z.A., Drosten C., Zumla A., Petersen E. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health - the latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keep S., Bickerton E., Armesto M., Britton P. The ADRP domain from a virulent strain of infectious bronchitis virus is not sufficient to confer a pathogenic phenotype to the attenuated Beaudette strain. J. Gen. Virol. 2018;99:1097–1102. doi: 10.1099/jgv.0.001098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laconi A., van Beurden S.J., Berends A.J., Kramer-Kuhl A., Jansen C.A., Spekreijse D., Chenard G., Philipp H.C., Mundt E., Rottier P., Helene V.M. Deletion of accessory genes 3a, 3b, 5a or 5b from avian coronavirus infectious bronchitis virus induces an attenuated phenotype both in vitro and in vivo. J. Gen. Virol. 2018 doi: 10.1099/jgv.0.001130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Gralinski L.E., Mitchell H.D., Dinnon K.R., Leist S.R., Yount B.J., Graham R.L., McAnarney E.T., Stratton K.G., Cockrell A.S., Debbink K., Sims A.C., Waters K.M., Baric R.S. Middle east respiratory syndrome coronavirus nonstructural protein 16 is necessary for interferon resistance and viral pathogenesis. mSphere. 2017;2 doi: 10.1128/mSphere.00346-17. e00346-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielech A.M., Chen Y., Mesecar A.D., Baker S.C. Nidovirus papain-like proteases: multifunctional enzymes with protease, deubiquitinating and deISGylating activities. Virus Res. 2014;194:184–190. doi: 10.1016/j.virusres.2014.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan D., Fang S., Han Z., Ai H., Zhao W., Chen Y., Jiang L., Liu S. Effects of hypervariable regions in spike protein on pathogenicity, tropism, and serotypes of infectious bronchitis virus. Virus Res. 2018;250:104–113. doi: 10.1016/j.virusres.2018.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z., Wang G., Yang Y., Shi J., Fang L., Li F., Xiao S., Fu Z.F., Peng G. A conserved region of nonstructural protein 1 from alphacoronaviruses inhibits host gene expression and is critical for viral virulence. J. Biol. Chem. 2019;294:13606–13618. doi: 10.1074/jbc.RA119.009713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjaak D.W.J., Cook J.K., van der Heijden H.M. Infectious bronchitis virus variants: a review of the history, current situation and control measures. Avian Pathol. 2011;40:223–235. doi: 10.1080/03079457.2011.566260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Decroly E., Ziebuhr J. Chapter three - the nonstructural proteins directing coronavirus RNA synthesis and processing. In: Ziebuhr J., editor. vol. 96. Academic Press; 2016. pp. 59–126. (Advances in Virus Research). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Beurden S.J., Berends A.J., Kramer-Kuhl A., Spekreijse D., Chenard G., Philipp H.C., Mundt E., Rottier P., Verheije M.H. Recombinant live attenuated avian coronavirus vaccines with deletions in the accessory genes 3ab and/or 5ab protect against infectious bronchitis in chickens. Vaccine. 2018;36:1085–1092. doi: 10.1016/j.vaccine.2018.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S.R., Leibowitz J.L. Coronavirus pathogenesis. Adv. Virus Res. 2011;81:85–164. doi: 10.1016/B978-0-12-385885-6.00009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M., Zheng B.J., Chan K.H., Yuen K.Y. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G., Cheng J., Ma S., Jia W., Yan S., Zhang G. Pathogenicity differences between a newly emerged TW-like strain and a prevalent QX-like strain of infectious bronchitis virus. Vet. Microbiol. 2018;227:20–28. doi: 10.1016/j.vetmic.2018.10.019. [DOI] [PubMed] [Google Scholar]
- Yan S., Zhao J., Xie D., Huang X., Cheng J., Guo Y., Liu C., Ma Z., Yang H., Zhang G. Attenuation, safety, and efficacy of a QX-like infectious bronchitis virus serotype vaccine. Vaccine. 2018;36:1880–1886. doi: 10.1016/j.vaccine.2018.02.053. [DOI] [PubMed] [Google Scholar]
- Zhang J. Porcine deltacoronavirus: overview of infection dynamics, diagnostic methods, prevalence and genetic evolution. Virus Res. 2016;226:71–84. doi: 10.1016/j.virusres.2016.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Cheng J.L., Liu X.Y., Zhao J., Hu Y.X., Zhang G.Z. Safety and efficacy of an attenuated Chinese QX-like infectious bronchitis virus strain as a candidate vaccine. Vet. Microbiol. 2015;180:49–58. doi: 10.1016/j.vetmic.2015.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Cheng J., Xu G., Thiel V., Zhang G. Successful establishment of a reverse genetic system for QX-type infectious bronchitis virus and technical improvement of the rescue procedure. Virus Res. 2019;272:197726. doi: 10.1016/j.virusres.2019.197726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Cheng J., Yan S., Jia W., Zhang K., Zhang G. S gene and 5a accessory gene are responsible for the attenuation of virulent infectious bronchitis coronavirus. Virology. 2019;533:12–20. doi: 10.1016/j.virol.2019.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J. Coronavirus replicative proteins. In: Perlman S., Gallagher T., Snijder E.J., editors. Nidoviruses. ASM Press; Washington, DC: 2008. pp. 65–81. [Google Scholar]
- Ziebuhr J., Snijder E.J., Gorbalenya A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]