Summary
Cell-surface protein-protein interactions (PPIs) mediate cell-cell communication, recognition, and responses. We executed an interactome screen of 564 human cell-surface and secreted proteins, most of which are immunoglobulin superfamily (IgSF) proteins, using a high-throughput, automated ELISA-based screening platform employing a pooled-protein strategy to test all 318,096 PPI combinations. Screen results, augmented by phylogenetic homology analysis, revealed ∼380 previously unreported PPIs. We validated a subset using surface plasmon resonance and cell binding assays. Observed PPIs reveal a large and complex network of interactions both within and across biological systems. We identified new PPIs for receptors with well-characterized ligands and binding partners for “orphan” receptors. New PPIs include proteins expressed on multiple cell types and involved in diverse processes including immune and nervous system development and function, differentiation/proliferation, metabolism, vascularization, and reproduction. These PPIs provide a resource for further biological investigation into their functional relevance and may offer new therapeutic drug targets.
Keywords: cell-surface, protein-protein interaction screen, receptor, ligand, interactome, human IgSF, network
Graphical Abstract
Highlights
-
•
Human IgSF interactome reveals complex network of cell-surface protein interactions
-
•
Phylogenetic homology analysis predicts protein-protein interactions
-
•
∼380 previously unknown protein-protein interactions identified
-
•
Deorphanization of receptors and new binding partners for well-studied receptors
A high-throughput protein-protein interaction screen, carried out to map human cell-surface receptor-ligand interactions between proteins belonging to the immunoglobulin domain superfamily (IgSF), begins to unravel the complex network of cell-surface interactions that allows cells to recognize and respond to one another and their dynamically changing environment.
Introduction
Protein-protein interactions (PPIs) at the cell surface allow cells to respond to one another and their environment in a highly dynamic, context-dependent and spatiotemporal manner (Wood and Wright, 2019). The essential role played by cell-surface PPIs is exemplified by estimates that ∼20% of genes in the human genome encode cell-surface proteins and ∼10% encode secreted proteins (Fonseca et al., 2016).
At present, a comprehensive human cell-surface interactome is lacking. Mapping extracellular PPIs has proved challenging because most cell-surface proteins are refractory to classic biochemical screening techniques and cell-surface PPIs are underrepresented in affinity purification/mass spectrometry-based datasets (Huttlin et al., 2015, 2017). Additionally, cell surface PPIs often have fast binding kinetics spanning a broad range of affinities (low nM to hundreds of μM) (van der Merwe and Barclay, 1994), rendering them difficult to detect using standard biochemical assays.
In recent years, several assays have been developed that allow detection of low-affinity cell-surface PPIs by imparting avidity through clustering of secreted proteins and the extracellular domains (ECDs) of single transmembrane (STM) cell-surface proteins. Clustering is achieved using multimerization domains, and can increase binding signal up to 250-fold (Bushell et al., 2008). Experimental platforms that utilize clustering include several variations of ELISA-based binding assays (Wojtowicz et al., 2007; Bushell et al., 2008; Özkan et al., 2013), Bio-Plex beads (Li et al., 2017), protein microarrays (Sun et al., 2012), cell-signaling assays (Barrow et al., 2018), cell-surface staining microarrays (Turner et al., 2013), and bead-based assays (Husain et al., 2019). Multiple groups have shown that ELISA-based binding assays have a remarkably low false-positive rate (Wojtowicz et al., 2007; Bushell et al., 2008; Söllner and Wright, 2009; Martin et al., 2010; Crosnier et al., 2011; Özkan et al., 2013; Visser et al., 2015; Ranaivoson et al., 2019).
Previously, we conducted a screen for all ∼200 Drosophila cell-surface and secreted proteins containing three types of domains: immunoglobulin (Ig) and Ig-like, fibronectin type III (FN3), and leucine-rich repeats (LRRs) (Özkan et al., 2013). This screen reported over 80 new PPIs, including a previously unknown immunoglobulin superfamily (IgSF) PPI network between members of the Dpr and DIP subfamilies. Since we reported the Dpr-DIP network, functional studies have revealed that this network mediates neuronal wiring decisions in the fly brain and neuromuscular system (for review, see Honig and Shapiro, 2020; Sanes and Zipursky, 2020).
In humans, there are an estimated ∼4,000 secreted and STM proteins, totaling ∼8 M putative PPIs. Screening this vast number requires a high-throughput assay. Here, we developed a screening platform that combines a high-throughput version of the ELISA-based extracellular interactome assay (ECIA) (Özkan et al., 2013) with an automated pooled-protein strategy (apECIA). We performed a screen of human IgSF secreted and STM cell-surface proteins (excluding antibodies and T cell receptors), along with other select proteins of interest. The IgSF is the largest and most functionally diverse family in the cell-surface proteome. Members include receptor tyrosine kinases, phosphatases, co-stimulatory or co-inhibitory immune receptors, growth factor and adhesion receptors, among many others, and are present in most, if not all, cell types.
We produced 564 proteins, and screened every possible PPI (564 × 564 = 318,096). We observed 426 PPIs, of which 345 (81%) are previously unreported. New PPIs form a complex network and include PPIs between phylogenetically related proteins within a subfamily, different subfamilies, and distantly related proteins. Screen results were combined with phylogenetic homology analysis (PHA) to predict additional PPIs between subfamily members using a nearest-neighbor approach. We confirmed a subset of both screen and PHA predicted PPIs using surface plasmon resonance (SPR) and cell binding assays.
Results
Selection and Production of Proteins for PPI Screening
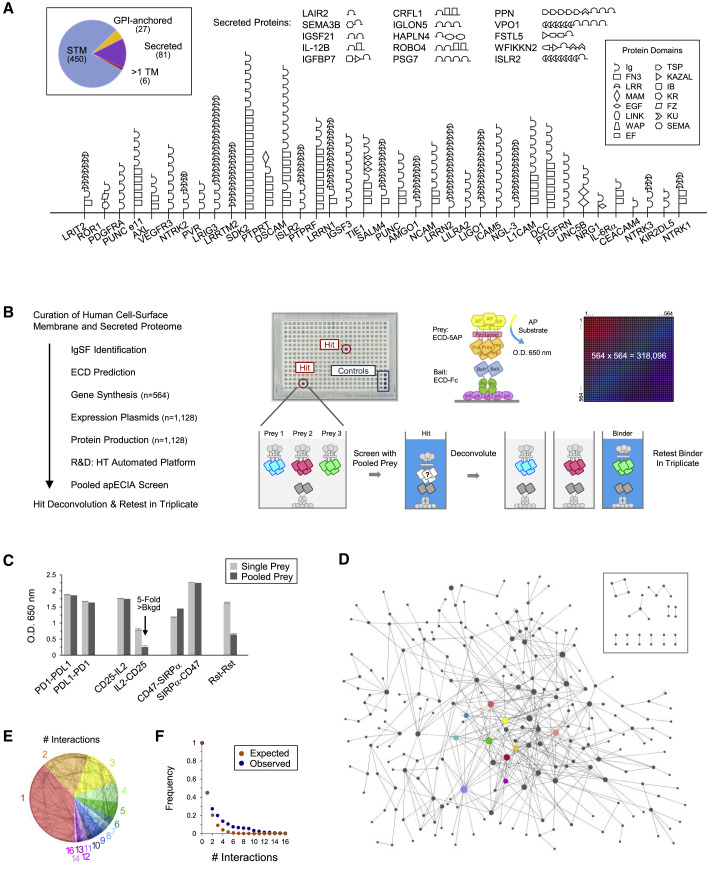
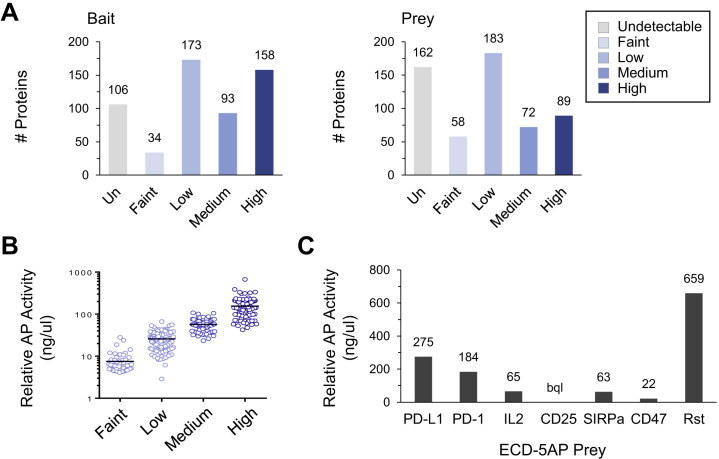
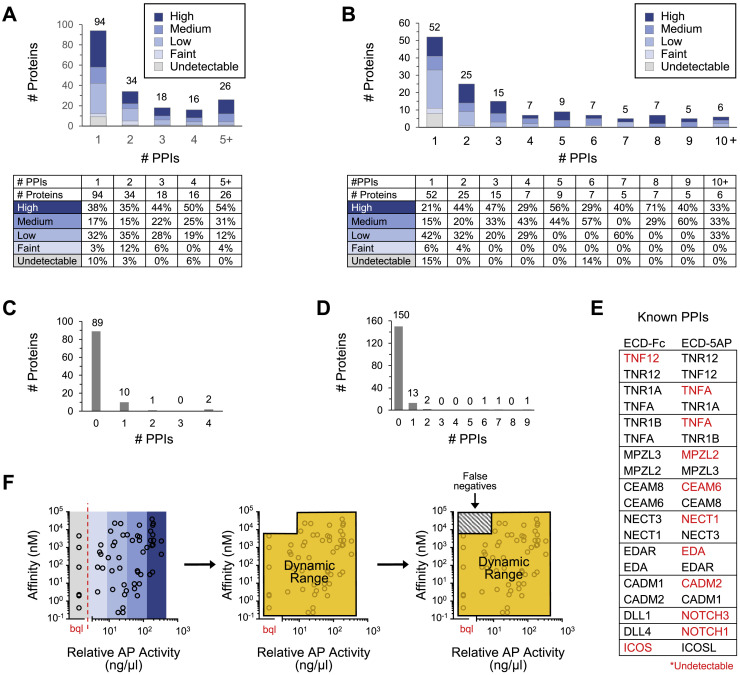
To identify human IgSF proteins, we utilized the HUGO Gene Nomenclature Committee (HGNC) (Yates et al., 2017), Human Protein Atlas (Uhlén et al., 2015), and UniProt (UniProt Consortium, 2019) databases. ECDs and secreted proteins for 458 IgSF and 106 non-IgSF proteins of interest were produced with “bait” and “prey” multimerization domains into cell supernatants (Figures 1A and 1B; Data S1 and S2) and expression was confirmed by western blot (Data S3). Westerns revealed detectable levels of protein for 82% of baits and 70% of preys. We and others have observed that PPIs can be detected in the ECIA and other ELISA-based binding assays even when proteins are present at levels below the limit of detection by western (Özkan et al., 2013; Visser et al., 2015; Ranaivoson et al., 2019). As such, all bait and prey were included in the screen regardless of whether protein was detected.
Figure 1.
apECIA Screen Details and Overview of Results
(A) Schematic depiction of a subset of proteins in library. Pie graph of library distribution. Full protein domain names at http://smart.embl.de/ (Sun et al., 2012).
(B) Left: flow chart of screen. HT, high-throughput. Right: example plate from screen. Schematic of ECIA and pooled-prey strategy. Illustration of screen matrix.
(C) ECIA of undiluted prey (single prey) versus 3-fold diluted prey (pooled prey). Background subtracted data are represented as ±SD. Bkgd, background.
(D) Network of PPIs observed in screen. Inset: the 18 PPIs that are not connected to the network. Node size is proportional to number of PPIs. Siglec subfamily nodes are highlighted in color.
(E) Pie graph of distribution of the number of binding partners observed in screen overlaid on the network in a degree-sorted circular layout.
(F) Observed versus expected frequency with which each protein will bind at least “x” number of proteins up to the maximally observed PPIs assuming random chance of interactions (p = 0.01; Kolmogorov-Smirnov [K-S] test).
See also Figure S1, Figure S2, Figure S3, and S4 and Data S1, S2, S3, and S4.
Development of an Automated Pooled-Prey ECIA Platform (apECIA)
ECIA and other ELISA-based assays allow bait and prey proteins to be tested for binding directly from conditioned media (Figure 1B). These assays test one bait-prey pair per well. To increase throughput, we pooled three preys and, following screening, deconvoluted positive wells to identify PPIs (Figure 1B). Pooling experiments with a panel of known PPIs showed binding for all 3-fold diluted prey (Figures 1C, S1 C, and data not shown). As bait-prey pairs are tested in both orientations, a false-negative resulting from pooling can be “rescued” by a positive result in the converse orientation. We reasoned that the advantages of pooling, which reduces the number of binding reactions, outweighed the potential increase in false negatives. To further improve throughput, we optimized a 384-well format and developed an automated workflow using liquid handling robots. The apECIA platform allows testing of 55,296 bait-prey combinations per week.
Figure S1.
Bait and Prey Levels in Conditioned Media, Related to Figure 1 and Data S3
(A) Qualitative levels of bait (left) and prey (right) proteins in conditioned media used for screen as assessed by western blot against C-terminal 6xHis and 9xHis tags, respectively. Levels were estimated by comparing band intensity of proteins to a purified ECD-Fc standard on each gel. Un; undetectable.
(B) Qualitative prey levels in conditioned media (panel A) versus quantitative relative AP activity (ng/μl), as determined by AP enzymatic assay. Black lines denote mean.
(C) Relative AP activity (ng/μl), as determined by AP enzymatic assay, for all prey used in pooling experiments. Prey levels span the entire spectrum, from the max AP levels obtained in conditioned media (Rst: 659 ng/μl) using our expression system down to levels below quantitation (bql).
PPI Screen Reveals a Complex Network of PPIs
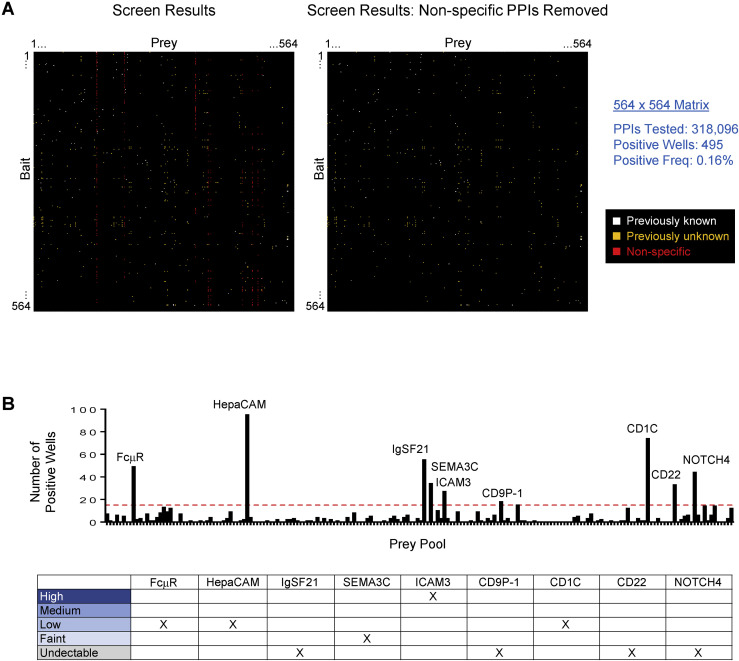
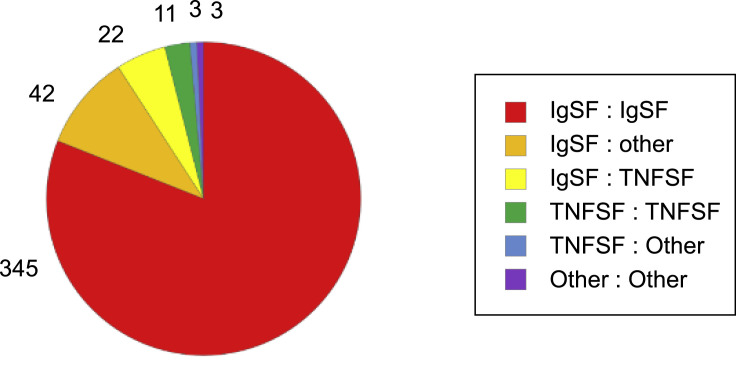
Following screening, deconvolution of positive wells (≥2 fold-over-background) was performed by re-testing each prey individually (Figure 1B). Nine prey gave rise to large numbers of wells with positive signals and were excluded as non-specific binders (Figure S2 ). Following removal of non-specific PPIs, deconvolution revealed 495 positive wells comprising 426 unique PPIs (Data S4). In each case, only one of the three deconvoluted prey yielded a positive signal. To confirm binding, the positive prey was re-tested against its cognate bait in triplicate. Eighty-one percent (345/426) of PPIs are between IgSF proteins. The remaining 19% include PPIs between IgSF and other proteins present in the screen (Figure S3 ).
Figure S2.
Identification of Non-specific PPIs in Screen, Related to Data S3 and S4
(A) Left. Screen results for 564 × 564 matrix showing positive wells (≥2-fold over background). White, previously known PPI; orange, previously unknown PPI; red, non-specific PPI. Right. Screen results following removal of non-specific PPIs. Non-specific PPIs may result from misfolded proteins. Freq, frequency.
(B) Number of positive wells (≥2-fold over background) in the screen for each of the 188 prey pools. Prey pools with > 16 positive wells (above red dotted line) were classified as “sticky,” potentially exhibiting non-specific binding. In the screen, 16 PPIs for Siglec-10 were observed, many of which were PPIs with other Siglec subfamily members. As many Siglec-Siglec PPIs were observed in the screen, Siglec-10’s 16 PPIs were determined to not be the result of “stickiness,” and 16 PPIs was selected as a conservative cut-off for non-specific “sticky” prey to minimize the possibility of including false positives in the PPI dataset. Each prey in the screen was individually tested against mCherry-Fc bait and mock controls (data not shown) and positive binding signals were used to identify the “sticky” prey in each pool which were excluded from the PPI dataset. Table shows the protein expression level of the nine “sticky” prey in the conditioned media used for the screen. Qualitative expression levels were assessed by western blot.
Figure S3.
Distribution of PPIs between Protein Families, Related to Figure 1 and Data S1 and S4
Pie graph showing the number of PPIs between proteins belonging to different families. Proteins categorized as “other” include non-IgSF DEATH domain receptors and cytokines, among others. IgSF; immunoglobulin superfamily. TNFSF; tumor necrosis factor superfamily.
Almost half of the proteins tested were involved in a PPI (254/564, 45%). Proteins not involved in PPIs may be misfolded, have binding partners not included in the screen, require co-receptors, or fall outside the dynamic range of the assay (false negatives) which is determined by PPI affinity and bait and prey concentrations (Figure S4 F). Confirming the sensitivity of the assay, many bait or prey proteins expressed at very low levels still engaged in one or two PPIs (Figures S4A and S4B). A small number of PPIs were observed with bait or prey proteins exhibiting undetectable levels in conditioned media (Figures S4C–S4E). To interrogate the dynamic range of the assay we plotted prey AP levels for PPIs with reported affinities (Figure S4F). These data suggest that, for very poorly expressed prey proteins (Figures S1A, S4A, and S4B), PPIs with KD >4.5 μM are likely to be missed (false negatives; Figure S4F).
Figure S4.
Distribution of Bait and Prey Expression and Dynamic Range, Related to Data S3 and S4
(A) Number of bait proteins engaging in one to > 5 PPIs by qualitative expression level as assessed by western blot.
(B) Number of prey proteins engaging in one to > 10 PPIs by qualitative expression level as assessed by western blot.
(C) Number of PPIs involving bait proteins that were undetectable in conditioned media by western blot.
(D) Number of PPIs involving prey proteins that were undetectable in conditioned media by western blot and quantitative AP enzymatic assay.
(E) Known PPIs involving bait or prey proteins that were undetectable in conditioned media by western blot (bait and prey) and quantitative AP enzymatic assay (prey). PPIs listed are the only PPIs observed in the screen for these undetectable bait and prey proteins. Red font, undetectable protein.
(F) Relative AP activity (ng/μl) versus binding affinity (nM) for 34 known PPIs observed in the screen with published KD values to illustrate the dynamic range of apECIA. PPIs that fall outside the dynamic range (orange perimeter), which is determined by the combined effect of PPI affinity and bait and prey concentrations, will likely result in false negatives (box with gray slashes). Blue shaded bars (colors as in panels A and B) represent qualitative prey protein levels as determined by western blot. Red dotted line segregates prey that were both undetectable by Western and below the quantitation limit (bql) using the AP enzymatic assay. The break in the x axis is to emphasize that data points to the left of the break (bql) could not be quantitated and, as such, are all positioned vertically in the center of the gray bar.
Of the 426 PPIs, 345 (81%), are previously unreported based on literature and PPI databases (Data S4). The majority of PPIs (408/426) form one large network comprising 226 proteins (Figure 1D). Only 28 proteins involving 18 PPIs are not connected to the network. Different regions of the network are shown in Figure 2 . Ninety-eight proteins (39%) had one PPI, 113 (44%) had two to five PPIs, and 43 (17%) had >5 PPIs (Figure 1E). Because 45% of proteins exhibit binding, we calculated the expected frequency with which each protein will bind at least “x” number of proteins up to the maximally observed 16 PPIs (Figure S2B) and compared the expected and observed frequencies (Figure 1F). The observed number of binding partners is significantly greater than predicted by random chance of PPIs for a network of this size.
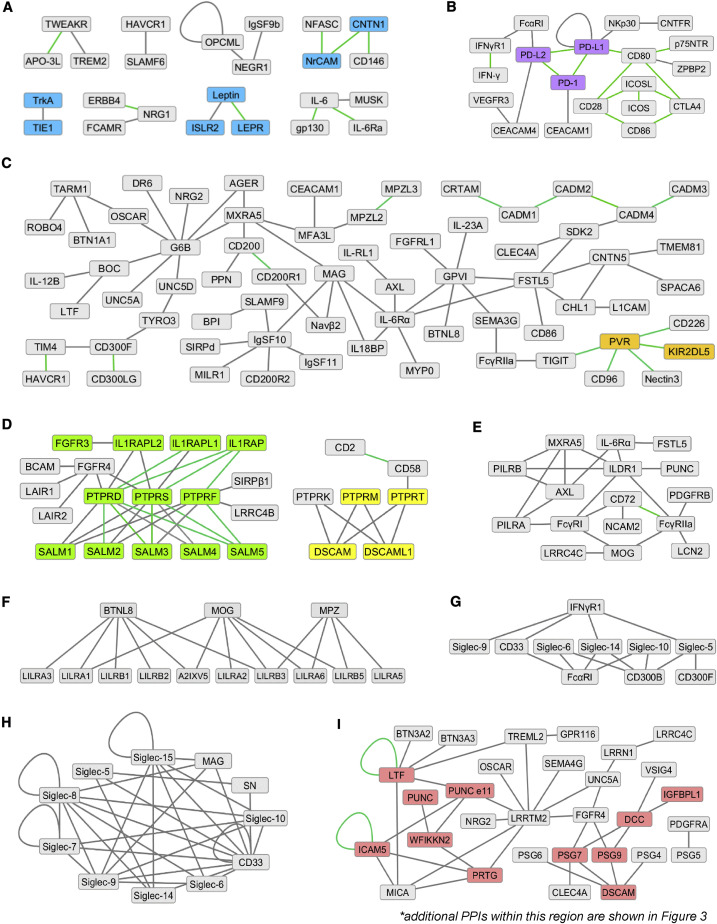
Figure 2.
Select Regions of the Complex PPI Network
(A) Select PPIs including four proteins not connected to the network (CD146, CNTN1, NFASC, and NrCAM).
(B) Region largely comprised of immune system proteins.
(C) Region comprising PPIs both within and across biological systems.
(D) Two regions highlighting subfamily-specific type IIA and type IIB PTPR PPIs. Type IIA PPIs with SALMs and IL1APs include PPIs observed in screen and PHA predicted PPIs validated by SPR (Figures 4 and S6).
(E) Region highlighting PPIs for “orphan” receptors ILDR1 and PUNC.
(F) Region showing a subset of LILR subfamily PPIs.
(G) Region showing a subset of Siglec PPIs with non-Siglecs (CD33/Siglec-3).
(H) Region showing Siglec-Siglec PPIs (CD33/Siglec-3; MAG/Siglec-4a; SN/Siglec-1).
(I) Region highlighting PPIs between nervous system proteins and with proteins in immune and reproductive systems. Within this region, multiple additional PHA predicted PPIs were validated by SPR (Figure 3; Table 1).
Because a network is composed of interconnected nodes, some linkage proteins are present in more than one panel. Colored nodes denote proteins and PPIs validated by additional experiments. Green line, previously reported PPI; gray line, previously unreported PPI.
See also Data S4.
Phylogenetic Homology Analysis (PHA) to Predict PPIs
PPIs often occur between phylogenetically related proteins both within and between subfamilies. We performed multiple sequence alignments to identify subfamily members within our library and generated phylogenetic trees. Using a combined approach, which we call apECIA-PHA, we analyzed screen data alongside the phylogenetic trees to predict additional PPIs between subfamily members that may have been missed in the screen.
PPI Validation by Surface Plasmon Resonance
We selected a subset of screen and PHA predicted PPIs to validate by SPR. Bona fide PPIs are expected to display distinct association and dissociation, which can be observed with high sensitivity by SPR. To increase avidity, and therefore sensitivity, Fc dimerized ECD analytes and ligands were used in most experiments (Figure S5 ). This increase in sensitivity prevented us from determining monomeric KD constants. Binding profiles characteristic of PPIs, exhibiting clear resonance signals above background (negative control ligand and receptor) and concentration-dependent binding curves, were deemed indicative of a specific ligand-analyte PPI. Non-specific PPIs generally exhibit deviations from this behavior, such as high background and non-linear concentration responses.
Figure S5.
Size-Exclusion Chromatography of ECD-Fc Bait Proteins, Related to Figures 3, 4, 5, and 6 and STAR Methods
FPLC traces of ECD-Fc proteins used for SPR and cell-surface binding experiments.
Twenty-four newly identified PPIs observed in our screen were tested by SPR. Of these, we observed 23 specific ligand-analyte interactions. We additionally tested 35 PHA predicted PPIs and observed PPIs for 33. Three additional PPIs were observed between homologous proteins in mouse and cross-species (human-mouse). In total, we SPR validated 59 newly identified PPIs (Table 1 ; Data S5).
Table 1.
SPR-Validated apECIA Screen and PHA Predicted PPIs
| Ligand-Fc | Analyte-Fc | apECIA | PHA |
|---|---|---|---|
| TIE1 | TrkA | + | N/A |
| hISLR2 | hLEPa | + | N/A |
| mLepa | N/A | + | |
| mIslr2 | hLEPa | N/A | + |
| mLepa | N/A | + | |
| SALM1 | PTPRD | N/A | + |
| PTPRF | N/A | + | |
| PTPRS | N/A | + | |
| SALM2 | PTPRD | known | N/A |
| PTPRF | + | N/A | |
| PTPRS | + | N/A | |
| SALM3 | PTPRD | known | N/A |
| PTPRF | + | N/A | |
| PTPRS | known | N/A | |
| SALM4 | PTPRD | N/A | + |
| PTPRS | N/A | + | |
| SALM5 | PTPRD | known | N/A |
| PTPRF | known | N/A | |
| PTPRS | known | N/A | |
| IL1RAPL1 | PTPRS | + | N/A |
| IL1RAPL2 | FGFR3 | + | N/A |
| PTPRS | + | N/A | |
| DSCAM-CT | PTPRM | + | N/A |
| PTPRT | + | N/A | |
| DSCAML1-CT | PTPRM | + | N/A |
| PTPRT | + | N/A | |
| KIR2DL5 | PVR | known | N/A |
| IL-6Rα | ISLR2 | + | N/A |
| PSG7 | DCC | + | N/A |
| DSCAM | + | N/A | |
| PSG9 | DSCAM | + | N/A |
| ICAM5 | DCC | N/A | + |
| IL-6Rα | N/A | + | |
| ISLR2 | N/A | + | |
| NEO1 | N/A | + | |
| PRTG | + | N/A | |
| PUNC | N/A | + | |
| PUNC e11 | + | N/A | |
| WFIKKN2 | DCC | N/A | + |
| IL-6Rα | N/A | + | |
| ISLR2 | N/A | + | |
| PRTG | + | N/A | |
| PUNC | + | N/A | |
| PUNC e11 | + | N/A | |
| PD-L1 | PD-L1 | N/A | + |
| PD-L2 | PD-L1 | known | N/A |
| LTF | DCC | N/A | + |
| IL-6Rα | N/A | + | |
| ISLR2 | N/A | + | |
| NEO1 | N/A | + | |
| PRTG | N/A | + | |
| PUNC | N/A | + | |
| PUNC e11 | + | N/A | |
| IGFBPL1 | DCC | + | N/A |
| IL-6Rα | N/A | + | |
| ISLR2 | N/A | + | |
| NEO1 | N/A | + | |
| PRTG | N/A | + | |
| PUNC | N/A | + | |
| PUNC e11 | N/A | + | |
| IGFBP7 | DCC | N/A | + |
| IL-6Rα | N/A | + | |
| ISLR2 | N/A | + | |
| NEO1 | N/A | + | |
| PRTG | N/A | + | |
| PUNC | N/A | + | |
| PUNC e11 | N/A | + |
Combined apECIA-PHA Approach Reveals Multiple DCC Subfamily PPIs
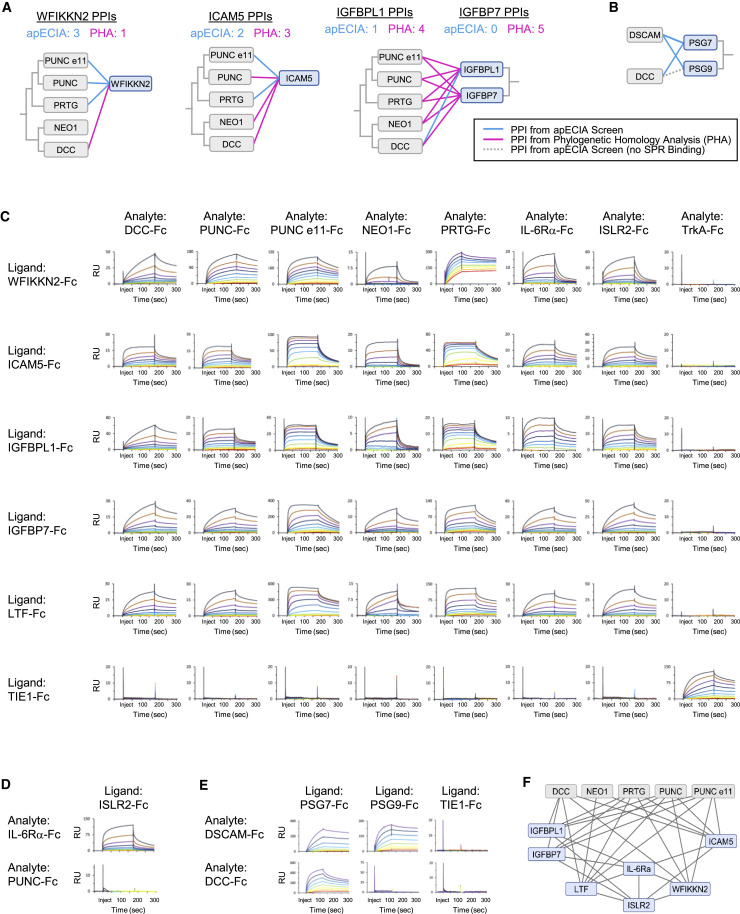
The netrin-1 receptor DCC has well-characterized functions in the nervous system (Finci et al., 2015). DCC is a dependence receptor and is implicated in colorectal and other cancers, but its roles in these cancers are not well understood (Goldschneider and Mehlen, 2010). We observed DCC binding to insulin-like growth factor-binding protein-like 1 (IGFBPL1), but not to insulin-like growth factor-binding protein 7 (IGFBP7) (Figures 2I and 3A). Our phylogenetic tree revealed IGFBPL1 and IGFBP7 reside in a cluster and share the highest amino acid sequence similarity (55%) among subfamily members. As such, we examined binding of DCC to both IGFBPL1 and IGFBP7 by SPR and observed binding for both (Figure 3C).
Figure 3.
SPR Validation of DCC Subfamily PPIs Identified by apECIA-PHA Approach
(A) Dendrograms of a subset of DCC subfamily PPIs showing the number of SPR-validated PPIs observed in the apECIA screen (turquoise) and predicted by PHA (magenta).
(B) Dendrogram of a subset of PSG PPIs.
(C) SPR sensorgrams for DCC subfamily, IL6-Rα, and ISLR2 analytes (2-fold dilutions; 2–2,048 nM; 512 nM for PRTG:WFIKKN2 due to incomplete chip regeneration at higher concentrations) binding to various ligands. TrkA and TIE1, negative controls.
(D) SPR sensorgram for IL-6Rα analyte (2-fold dilutions; 2–2,048 nM) binding to ISLR2 ligand. PUNC, negative control.
(E) SPR sensorgrams for DSCAM and DCC analytes (2-fold dilutions; 16–2,048 nM) binding to PSG ligands. TIE1, negative control.
(F) Subnetwork of SPR-validated PPIs.
RU, resonance units.
PHA pointed us to four proteins that cluster with DCC: PUNC, PUNC e11, neogenin (NEO1), and protogenin (PRTG) (Figure 3A). Together, these proteins play roles in diverse processes including nervous system development, myogenesis and angiogenesis, inflammation and tissue regeneration, leukocyte migration, neural tube and mammary gland formation, development of bone and connective tissues, and stem cell differentiation (Salbaum, 1998; Wilson and Key, 2007; Takahashi et al., 2010; Schievenbusch et al., 2012; Dakouane-Giudicelli et al., 2014). PUNC is an “orphan” receptor expressed in the developing nervous system (Salbaum, 1998). PUNC e11 and PRTG bound intercellular adhesion molecule 5 (ICAM5) (Figure 3A), a protein exclusively expressed in the brain that functions in synapse formation, stabilization, and refinement (Gahmberg et al., 2014). Cleaved ICAM5 ECD exhibits immunosuppressive functions through cytokine regulation and may play important roles in regulation of brain inflammation. We confirmed binding of PUNC e11 and PRTG to ICAM5 by SPR (Figure 3C). Although PPIs with ICAM5 were not detected in the screen for the remaining DCC subfamily members, we tested them by SPR and observed binding for all three (Figure 3C).
In our screen, one or more DCC subfamily members also bound to: (1) WFIKKN2, a secreted protein that binds transforming growth factor-beta subfamily members and modulates their presentation to cells (Monestier and Blanquet, 2016), (2) lactotransferrin (LTF), an iron-binding protein with antimicrobial activity (Hao et al., 2019), (3) interleukin-6 receptor subunit alpha (IL-6Rα), a cytokine receptor (Schaper and Rose-John, 2015), and (4) ISLR2/LINX, which functions in nervous system development (Mandai et al., 2014; Panza et al., 2015; Abudureyimu et al., 2018). We confirmed binding of all DCC subfamily members to these proteins by SPR and to other proteins observed in our screen (Figures 3C, 3D, and 3F). These results demonstrate the value of using a combined apECIA-PHA approach to identify additional PPIs beyond screen data, resulting in the elucidation of a more complete network (Figure 3F).
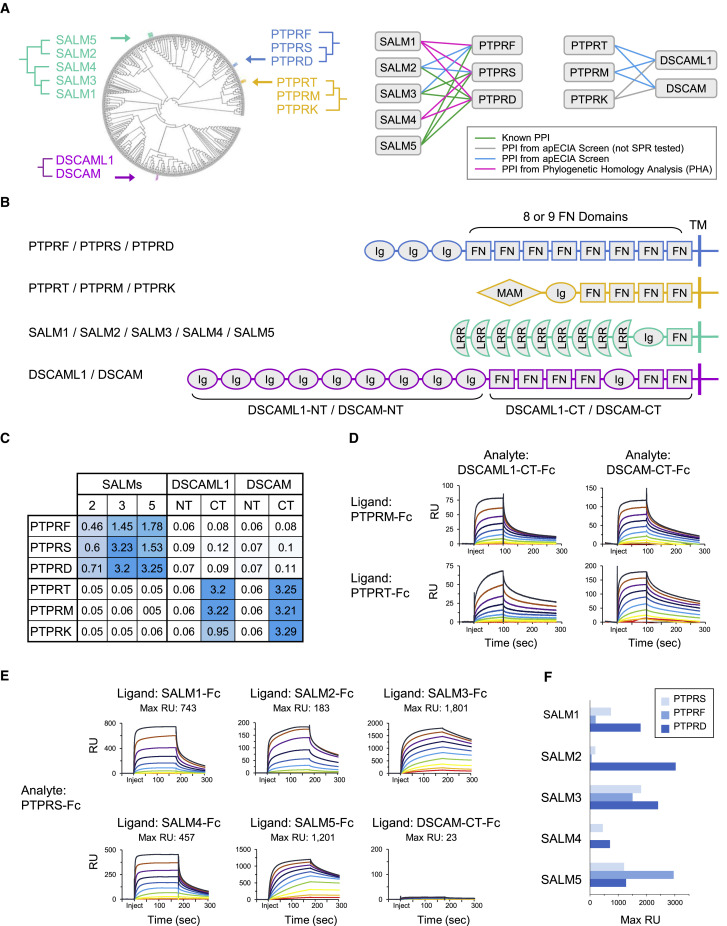
LAR-PTPR Subfamily PPIs with SALMs
The LAR-family of type IIA protein tyrosine phosphatase receptors (LAR-PTPRs) comprises PTPRF (also known as LAR), PTPRD, and PTPRS (Figures 4A and 4B). LAR-PTPRs play important roles in synaptic organization and function (Nam et al., 2011). Pre-synaptic LAR-PTPRs mediate trans-synaptic adhesion through PPIs with multiple post-synaptic ligands. Mouse mutants in LAR-PTPRs and their ligands exhibit defects in synapse structure and function (Lie et al., 2018).
Figure 4.
SPR Validation of Type IIA and Type IIB PTPRs with SALMs and DSCAMs
(A) Left: dendrogram highlighting phylogenetic clustering of SALMs, DSCAMs, type IIA PTPRs (PTPRF/S/D), and type IIB PTPRs (PTPRT/M/K). Right: SPR-validated PPIs: green line, previously known PPI; turquoise line, previously unknown PPI observed in screen; magenta line, previously unknown PHA predicted PPI. Gray line, screen PPI not SPR tested.
(B) Protein domain structures. Ig, immunoglobulin-like domain; FN, fibronectin type III domain; MAM, meprin, A-5 protein domain; LRR, leucine-rich repeat; TM, transmembrane; NT, N-terminal region of ECD; CT, C-terminal region of ECD.
(C) Screen data showing PPI specificity of PTPRF/S/D and PTPRT/M/K subfamilies. Background subtracted O.D. 650 nm data are represented as mean of triplicate wells.
(D) SPR sensorgrams for DSCAML1-CT and DSCAM-CT analytes (2-fold dilutions; 16–2,048 nM) binding to PTPRM and PTPRT ligands.
(E) SPR sensorgrams for PTPRS analyte (2-fold dilutions; 2–2,048 nM) binding to SALMs and DSCAM-CT ligands.
(F) SPR max RU values for every pairwise combination of PTPRF/S/D analytes with SALM ligands.
RU; resonance units.
See also Figures S5 and S6 and Data S5.
Several PPIs are known between specific LAR-PTPRs and SALM2/3/5 (Li et al., 2015; Choi et al., 2016; Goto-Ito et al., 2018) (Figure 4A). We observed these, as well as additional PPIs in our screen (Figures 2D, 4A, and 4C). No LAR-PTPR binding to SALM1 or SALM4 has been reported, and we did not observe binding in our screen. PHA led us to test binding of all LAR-PTPRs to all SALMs by SPR. With the exception of PTPRF-SALM4, we observed binding of all LAR-PTPRs to all SALMs (Figures 4E and S6 A). PTPR-SALM pairs exhibited differences in maximum response units (RU), a relative comparison of binding strength, suggesting a spectrum of binding affinities may exist among LAR-PTPRs and SALMs (Figure 4F).
Figure S6.
SPR Validation of LAR-PTPR, IL1AP, and PD-L2 PPIs, Related to Figure 4 and Table 1
(A) SPR sensorgrams for PTPRF-Fc and PTPRD-Fc analytes (2-fold dilutions; 2-2048 nM) binding to SALM1/2/3/4/5 ECD-Fc.
(B) SPR sensorgrams for PTPRF-Fc and PTPRS-Fc analytes (2-fold dilutions; 2-2048 nM) binding to IL1RAP-Fc, IL1RAPL1-Fc and IL1RAPL2-Fc ligands.
(C) SPR sensorgrams for PD-L1-Fc analyte (2-fold dilutions; 16-16000 nM) binding to PD-L1-Fc, PD-L2-Fc and PD-1-Fc ligands. TIE1-Fc ligand, negative control.
RU; resonance units.
LAR-PTPR Subfamily PPIs with Interleukin-1 Receptor Accessory Proteins (IL1APs)
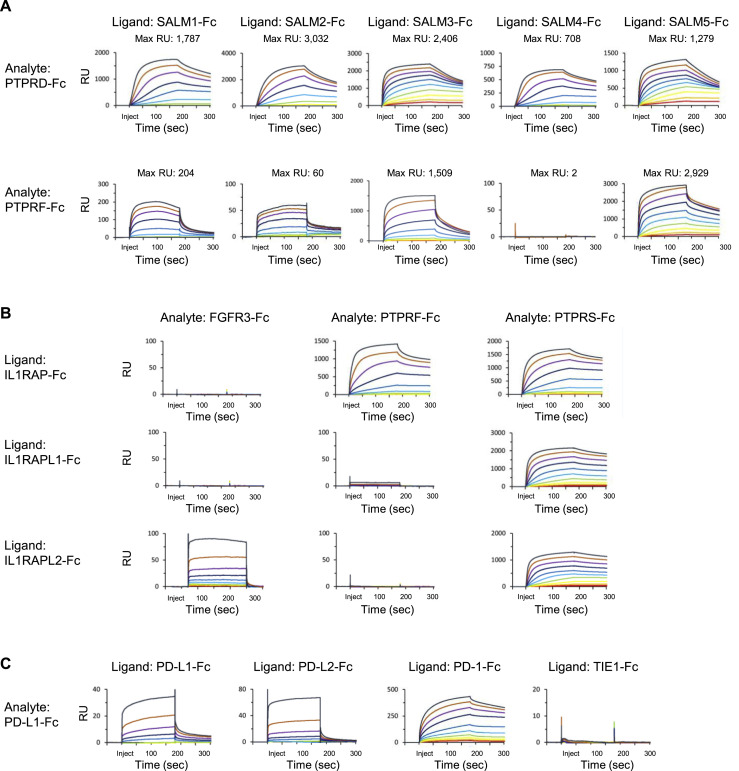
Trans-synaptic LAR-PTPR interactions with IL1RAP and IL1RAPL1 induce bidirectional pre- and post-synaptic differentiation (Yoshida et al., 2011, 2012). IL1RAP is known to interact with all three LAR-PTPRs and IL1RAPL1 with PTPRD. We observed these PPIs and, additionally, binding of IL1RAPL1 to PTPRS (Figure 2D). IL1RAPL1 shares 79% sequence similarity with IL1RAPL2, an orphan receptor expressed in the brain with no known biological function (Boraschi et al., 2018). In our screen, we detected binding of IL1RAPL2 to PTPRD and PTPRS (Figure 2D). We tested both known and previously unknown PPIs by SPR and observed binding for all (Figure S6B). IL1RAPL2 additionally interacted with fibroblast growth factor receptor 3 (FGFR3) in the screen and by SPR (Figure S6B).
Type IIB PTPR Subfamily PPIs with DSCAMs
Type IIB PTPRs are expressed in most tissues and regulate diverse processes including cell growth, migration, and differentiation; immune cell development; endothelial cell adhesion and migration; neuronal development and synapse formation; and oncogenic transformation (Lee, 2015; Stoker, 2005). The type IIB subfamily is composed of PTPRK, PTPRM, and PTPRT (Figures 4A and 4B), and multiple binding partners are known. We observed binding of all type IIB PTPRs with Down syndrome cell adhesion molecule (DSCAM) and Down syndrome cell adhesion molecule like-1 (DSCAML1) (Figures 4A and 4C), proteins that play various roles in nervous system development (Sanes and Zipursky, 2020).
The ECD of DSCAMs contains nine Ig-like domains followed by four FN3 domains, one Ig-like domain, and two FN3 domains (Figure 4B). The N-terminal region engages in homophilic binding and mediates binding to secreted chemoattractant and chemorepellent ligands, netrin-1 and slit-1, respectively (Wojtowicz et al., 2004; Ly et al., 2008; Alavi et al., 2016). No binding partner has been reported for the C-terminal ECD region. In our screen, we observed binding between the C-terminal regions of both DSCAMs (DSCAM-CT and DSCAML1-CT) and all three type IIA PTPRs (Figure 4C). We confirmed binding of DSCAM-CT and DSCAML1-CT to PTPRT and PTPRM by SPR (PTPRK was not expressed at sufficient levels) (Figure 4D).
New PPIs between Immune System Proteins
New PPIs were observed for well-studied immune system proteins, as well as orphan receptors (Figure 2B). Two PPIs were detected for the orphan granulocyte receptor CEACAM4: (1) vascular endothelial growth factor receptor 3 (VEGFR3), and (2) programmed cell death 1 ligand 2 (PD-L2). CEACAM4 is a member of the carcinoembryonic antigen-related cell adhesion molecule subfamily expressed in the immune system, epithelial and endothelial cells, and brain (Wakabayashi-Nakao et al., 2014; Zinn and Özkan, 2017). In the immune system, CEACAMs play roles in immunity and development. We also observed binding between CEACAM1 and the immune checkpoint receptor, programmed cell death protein 1 (PD-1).
Interactions of PD-1 with its ligands PD-L1 and PD-L2 inhibit T cell proliferation, cytokine production, and cytotoxic activity (Bardhan et al., 2016). PD-L2 competes with PD-L1 for binding to PD-1 and can be expressed on tumor cells where it may play a role in tumor evasion (Ghiotto et al., 2010; Cheng et al., 2013; Bardhan et al., 2016). Using SPR, we confirmed the known PPI between PD-L1 and PD-L2 (Lee et al., 2016) and observed homophilic binding of PD-L1, a previously unknown PPI (Figure S6C). By comparison with PD-L1 binding to PD-1, which approaches saturation at ∼400 nM, we infer that the affinity of PD-L1 homophilic binding is lower than binding to PD-1. This may explain why this PPI within the well-studied PD-1/PD-L1 axis has not been identified previously, and highlights the value of using multimerized proteins to detect low-affinity PPIs.
Homophilic and Heterophilic Siglec Subfamily PPIs
Eight of the proteins with the highest number of PPIs in our screen are members of the Siglec subfamily (14 members), proteins highly restricted to the immune system that have immune modulatory effects on Toll-like receptor (TLR) signaling and play important roles in self versus non-self discrimination (Macauley et al., 2014). Siglecs display differential expression on cells and exhibit a broad spectrum of Siglec-specific recognition of sialylated glycan profiles present on healthy cells, inflamed or malignant cells, and pathogens. We observed a network of homophilic and heterophilic PPIs among Siglecs (Figure 2H; Data S4), as well as PPIs with distantly related proteins (a subset are shown in Figure 2G; Data S4).
PPIs between Immune and Nervous System Proteins
The signaling lymphocytic activation molecule subfamily (SLAM; 9 members) is expressed on most immune cells (Dragovich and Mor, 2018). SLAMs function as both co-stimulatory and co-inhibitory molecules in innate and adaptive immunity, and play an integral role in autoimmune disorders. SLAMF9 is an orphan receptor.
We observed SLAMF9 binding to bactericidal permeability-increasing protein (BPI) and IGSF10 (Figure 2C). BPI is a neutrophil-derived antibiotic protein that participates in bacteria killing through its highly cationic N-terminal region (Bülow et al., 2018). The C-terminal region of BPI exhibits no bactericidal activity and is believed to interact with other proteins and function in different processes. IgSF10 is an orphan receptor involved in the migration of gonadotropin-releasing hormone expressing (GnRH) neurons (Howard et al., 2016). IgSF10 has no known function in the immune system, and SLAMF9 function in the nervous system is unknown, revealing a PPI between two orphan receptors from different biological systems.
PPIs with Pregnancy-Specific Glycoproteins (PSGs)
During pregnancy, PSGs (10 members) are the most abundant trophoblastic proteins in maternal blood and serve as markers for trophoblast quality and embryo viability (Moore and Dveksler, 2014). The mechanisms of PSG action in pregnancy are not well understood. Studies suggest PSGs also have immunoregulatory, pro-angiogenic, and anti-platelet aggregation functions. We observed several PSG binding partners including: (1) platelet-derived growth factor receptor alpha (PDGFRA), (2) fibroblast growth factor receptor 4 (FGFR4), (3) C-type lectin domain family 4 member A (CLEC4A), (4) DCC, and (5) DSCAM (Figures 2I and 3B).
Observed PSG interactions occur selectively and differentially to these binding partners (Figure 2I; Data S4). Using SPR, we examined binding of DCC and DSCAM to PSG7 and PSG9 (Figure 3E). We observed binding for DSCAM with PSG9, and both DCC and DSCAM with PSG7. Interestingly, DCC is expressed in human placenta (Dakouane-Giudicelli et al., 2014). These PPIs present new candidate receptors for studying the role of PSGs in pregnancy, immunoregulation, and angiogenesis.
LILR Subfamily PPIs with BTNL8 and Myelination Proteins
Leukocyte immunoglobulin-like receptors (LILRs; 11 members) are a subfamily of activating (LILRA) and inhibitory (LILRB) receptors that exhibit immunomodulatory activity and function in inflammation regulation, tolerance, and differentiation (Burshtyn and Morcos, 2016). We observed binding of multiple activating and inhibitory LILRs to butyrophilin-8 (BTNL8) (Figure 2F). BTNLs are members of the extended B7 family of molecules and function as co-stimulatory or co-inhibitory signals for T cell activation (Rhodes et al., 2016).
LILRs are also expressed on neurons and function in the regulation of development, synaptic plasticity, and axonal regeneration (Hirayasu and Arase, 2015). Myelin, the protective insulating layer around axons, inhibits neuronal regeneration following spinal injury (Monje, 2018). Three myelin proteins, Nogo, MAG, and OMgp, are known to interact with PirB in mouse, one of only two mouse LILRB orthologs (Atwal et al., 2008). We observed binding of multiple LILRs to two additional myelin proteins: myelin-oligodendrocyte glycoprotein (MOG) and myelin protein P0 (MPZ) (Figure 2F). These PPIs present MOG and MPZ PPIs with LILRs as new candidates for neuronal regeneration studies.
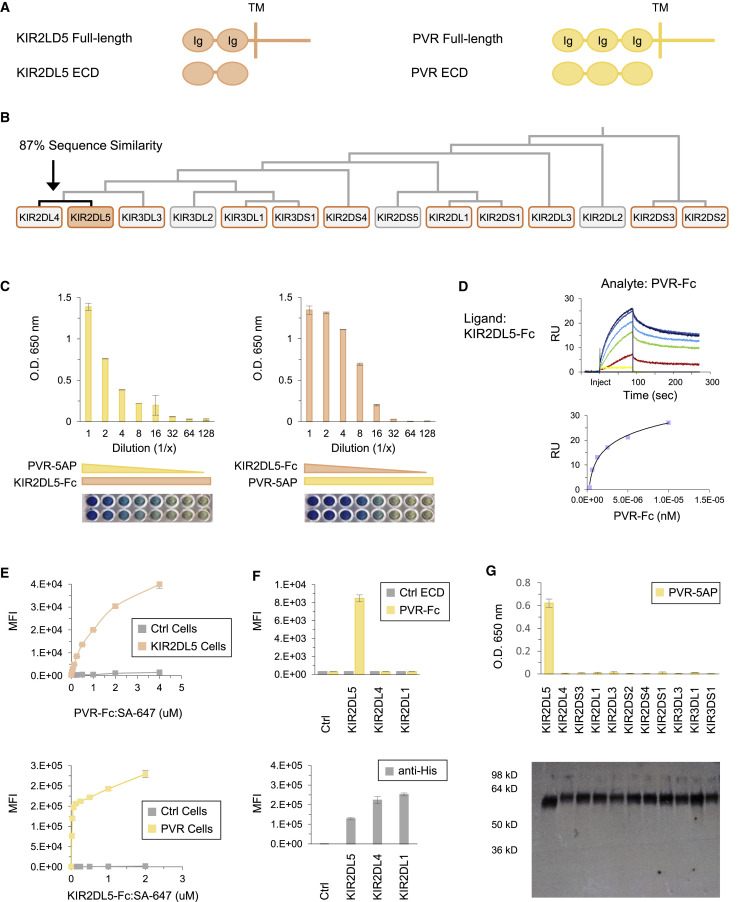
PVR Selectively Interacts with a Killer-Cell Immunoglobulin-like Receptor (KIR)
The polygenic Killer-cell immunoglobulin-like receptors (KIRs) are a highly polymorphic subfamily of activating and inhibitory proteins expressed on natural killer (NK) cells that regulate development, maturation, and activation (Pende et al., 2019). NK cells initially express a stochastic combination of KIRs that is refined during maturation to tune killing response threshold and ensure optimal discrimination of target cells from healthy cells. We observed binding of KIR2DL5 to poliovirus receptor (PVR) (Figures 2C and 5A), which validates a recently reported PPI (Husain et al., 2019). We confirmed KIR2DL5 binding to PVR by ECIA titration analysis and SPR (Figures 5C and 5D).
Figure 5.
PVR Specifically Interacts with KIR2DL5
(A) Protein domain structures of KIR2DL5 and PVR.
(B) Dendrogram showing KIR family members in screen. KIRs outlined in orange were tested in (G).
(C) ECIA titration of PVR-5AP on KIR2DL5-Fc (left) and KIR2DL5-Fc on PVR-5AP (right). Images are scanned ELISA plates. Background subtracted data are represented as mean ± SD.
(D) SPR sensorgram and steady-state curve for PVR analyte (2-fold dilutions; 156–10,000 nM) binding to KIR2DL5 ligand.
(E) Cell staining of full-length KIR2DL5-transfected and control cells with PVR-Fc:SA-647 (top) and full-length PVR-transfected and control cells with KIR2DL5-Fc:SA-647 (bottom) and analysis by flow cytometry. Data are represented as mean ± SD.
(F) Top: cell staining of full-length transfected KIR2DL5, KIR2DL4, KIR2DL1, and control cells with PVR-Fc:SA-647 and control protein and analysis by flow cytometry. Bottom: flow cytometry analysis of anti-His-647 stained cells. Data are represented as mean ± SD.
(G) Top: ECIA of PVR-5AP with 11 KIR ECD-Fc baits. Background subtracted data are represented as mean ± SD. Bottom: following ECIA, anti-His western blot analysis of KIR-Fc proteins on ELISA plate.
RU, resonance units; MFI, mean fluorescence intensity; Ig, immunoglobulin-like domain; TM, transmembrane; Ctrl, control.
To examine binding at the cell surface, fluorescent tetramers of PVR-Fc and KIR2DL5-Fc (ECD-Fc:SA-647) were incubated with full-length KIR2DL5- and PVR-transfected cells, respectively. Flow cytometry analysis revealed concentration-dependent binding of both ligands to cells expressing cognate full-length receptor, but not control cells (Figure 5E). PVR-Fc tetramers did not bind cells transfected with full-length KIR2DL4 and KIR2DL1, which share 87% and 69% sequence similarity with KIR2DL5, respectively (Figures 5B and 5F). To further examine the specificity of PVR for KIR2DL5, ECIA was performed using 11 KIRs. PVR bound specifically to KIR2DL5 (Figure 5G). Because KIR2DL5 has been associated with increased virus susceptibility and reduced anti-viral response to therapy, this specificity may have implications for the role of KIR2DL5 in immunity.
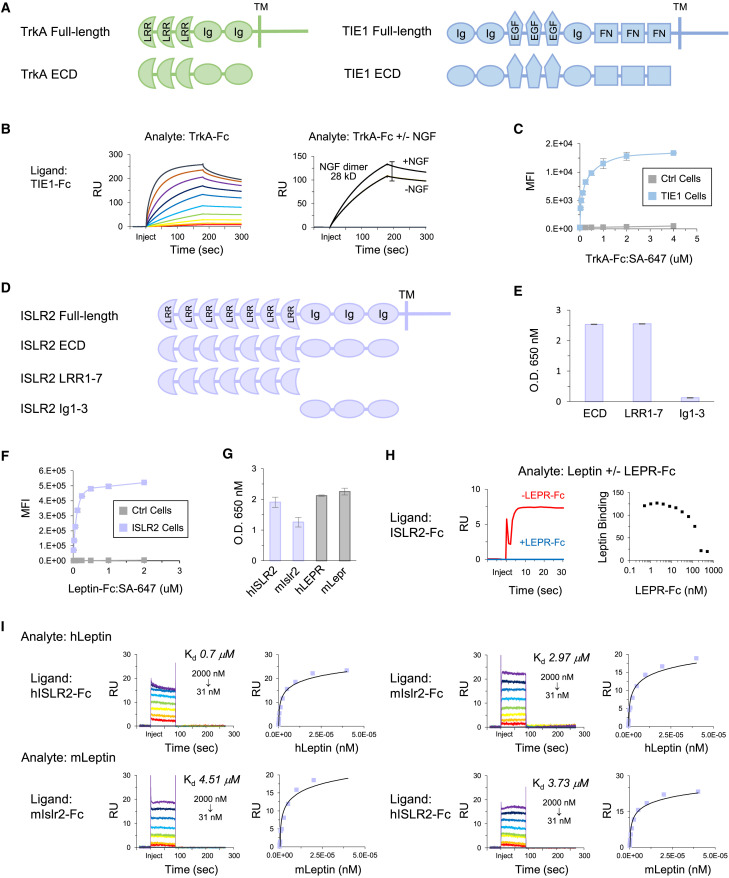
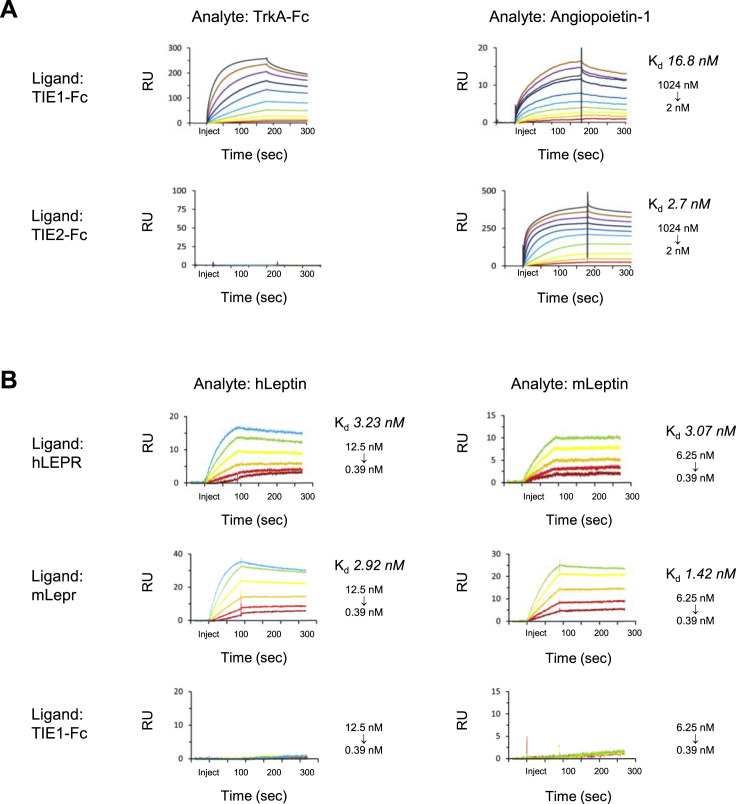
TrkA Selectively Interacts with TIE1
High-affinity nerve growth factor receptor (NTRK1), also known as TrkA, has multiple well-studied functions in the nervous system (Amatu et al., 2019). In the immune system, where its function is not well understood, TrkA is expressed on monocytes, macrophages, dendritic cells, resting and activated B cells and neutrophils, and erythroblasts (Minnone et al., 2017). In our screen, we observed TrkA binding to tyrosine-protein kinase receptor TIE1, but not to TIE2 (Figures 2A and 6A). TIE1 is expressed on endothelial cells, immature hematopoietic cells and platelets, and functions in complex with the angiopoietin-TIE2 pathway to inhibit angiogenesis (Eklund et al., 2017). We performed SPR and observed TrkA binding to TIE1, but not to TIE2 (Figures 6B and S7 A). As a positive control for TIE2, we confirmed binding to monomer angiopoietin-1 (Figure S7A) (Davis et al., 1996).
Figure 6.
TrkA Interacts with TIE1 and Leptin Interacts with ISLR2
(A) Protein domain structures of TrkA and TIE1.
(B) Left: SPR sensorgram for TrkA analyte (2-fold dilutions; 2–2,048 nM) binding to TIE ligand. Right: SPR sensorgram for 100 nM TrkA analyte ± 128 nM NGF binding to TIE1 ligand.
(C) Cell staining of full-length TIE1-transfected and control cells with TrkA-Fc:SA-647 and analysis by flow cytometry. Data are represented as mean ± SD.
(D) Protein domain structures of ISLR2 ECD and truncations.
(E) ECIA of leptin-5AP with ISLR2 ECD-Fc, LRR1-7-Fc, and Ig1-3-Fc. Background subtracted data are represented as mean ± SD.
(F) Cell staining of full-length ISLR2-transfected and control cells with leptin-Fc:SA-647 and analysis by flow cytometry. Data are represented as mean ± SD.
(G) ECIA of human leptin-5AP with human ISLR2-Fc and mouse Islr2-Fc. LEPR-Fc and Lepr-Fc; positive controls. Background subtracted data are represented as mean ± SD.
(H) Left: SPR sensorgram for 128 nM leptin ± 256 nM LEPR analyte binding to ISLR2 ligand. Right: SPR data showing 128 nM leptin analyte binding to ISLR2 ligand following pre-incubation with increasing concentrations of LEPR (2-fold dilutions; 0.5–512 nM).
(I) SPR sensorgrams and steady-state curves for monomer human and mouse leptin analytes (2-fold dilutions; 31–2,000 nM) binding to human ISLR2 and mouse Islr2 ligands.
RU, resonance units; MFI, mean fluorescence intensity; Ig, immunoglobulin-like domain; FN, fibronectin type III domain; LRR, leucine-rich repeat; EGF, epidermal growth factor repeat domain; TM, transmembrane; Ctrl, control.
See also Figures S5 and S7 and Data S5.
Figure S7.
Control SPR Experiments for TIE2 and Leptin-LEPR, Related to Figure 6
(A) SPR sensorgrams for Trk-Fc analyte (2-fold dilutions; 2-2048 nM) and monomer angiopoietin-1 analyte (2-fold dilutions; 2-1024 nM) binding to TIE1-Fc and TIE2-Fc ligands. As TIE2-Fc ligand showed no binding to TrkA-Fc analyte, monomer angiopoietin-1 analyte was included as a positive control for TIE2-Fc ligand.
(B) SPR sensorgrams for monomer human leptin analyte (2-fold dilutions; 0.39-12.5 nM) and monomer mouse leptin analyte (2-fold dilutions; 0.39-6.25 nM) binding to human LEPR-Fc and mouse Lepr-Fc ligands. Kd values are shown. TIE1-Fc ligand, negative control.
RU; resonance units.
We next investigated whether TrkA can bind NGF and TIE1 simultaneously. We pre-incubated TrkA with NGF and compared binding of TrkA ± NGF to TIE1 via SPR (Figure 6B). Compared with TrkA alone, TrkA:NGF binding to TIE1 exhibited an increase in max RU corresponding to the molecular weight of NGF dimers, demonstrating that NGF-bound TrkA can interact with TIE1. We tested TrkA-Fc tetramer binding to full-length TIE1-transfected cells and observed concentration-dependent binding to TIE1-transfected but not control cells (Figure 6C). TrkA interaction with TIE1 may play a role in angiogenesis and/or other biological processes.
Leptin Interacts with ISLR2
Leptin is a cytokine predominantly secreted by adipose cells and enterocytes in the small intestine and is required for the maintenance of energy homeostasis and body weight (Friedman, 2016). The function of leptin in this process is mediated by binding to the leptin receptor (LEPR) in a subtype of hypothalamic neurons. Leptin deficiency results in various metabolic disorders and rare genetic defects. The role of leptin in regulation of fat stores is its most highly studied function. However, leptin is secreted by many other types of cells and LEPR is expressed in other neuronal subtypes and non-neuronal cells suggesting leptin plays roles in additional processes.
We observed leptin binding to neuronally expressed ISLR2 (Figure 2A) (Mandai et al., 2014). The ECD of ISLR2 contains seven LRRs followed by three Ig-like domains (Figure 6D). To map the region of ISLR2 where leptin binds, we generated two ECD truncations comprising LRR1-7 and Ig1-3 and measured leptin binding using ECIA. Leptin bound equivalently to the entire ECD and LRR1-7. No binding was observed to Ig1-3 (Figure 6E). We tested leptin-Fc tetramer binding to full-length ISLR2-transfected cells and observed concentration-dependent binding to ISLR2-transfected but not control cells (Figure 6F).
As Lep−/− mice are a well-studied model of obesity (Friedman, 2016), we tested human leptin binding to mouse Islr2 using ECIA. Human leptin bound mouse Islr2 (Figure 6G). We then measured the binding affinities of monomer human and mouse leptin to both human ISLR2 and mouse Islr2 using SPR (Figure 6I). In controls, human and mouse leptin bound their cognate LEPR and Lepr with affinities in agreement with the range of published KD values (Verkerke et al., 2014) (Figure S7B). To investigate whether leptin can bind ISLR2 and LEPR simultaneously, we pre-incubated leptin with LEPR and compared binding of leptin ± LEPR to ISLR2 via SPR. Compared with leptin alone, leptin:LEPR exhibited no binding to ISLR2, suggesting that leptin cannot bind LEPR and ISLR2 simultaneously (Figure 6H). To assess whether this decrease in leptin binding to ISLR2 occurs in a LEPR concentration-dependent manner, we pre-incubated leptin with increasing concentrations of LEPR. Leptin binding to ISLR2 decreased as LEPR concentration increased (Figure 6H). ISLR2 interaction with leptin presents a candidate PPI for future studies into the function of leptin in the regulation of fat stores or other physiological processes, and in leptin deficiency diseases.
Discussion
Elucidation of the map of extracellular PPIs is essential to understanding the role of cell-surface PPIs in both healthy and disease tissues. Our screen revealed a highly connected and complex network of PPIs. This dataset provides a deep resource for further investigation. We anticipate it will provide candidates for the broader research community to interrogate the function of these PPIs in many biological systems. The complete list of PPIs (Data S4) contains many that have not been described in the Results. The approaches for selecting PPIs to investigate are multiple and varied. Below, we discuss the strategies that guided our exploration.
Homology-Guided Exploration of PPIs
Using a phylogenetic homology pipeline, we were able to expand upon our screen results by predicting and testing additional subfamily PPIs. This revealed an effective strategy to maximize the number of PPIs identified. Homology-guided identification of protein subfamilies was also important to correctly classify subfamily PPIs. For instance, in the absence of phylogenetic analysis, we may not have identified CD33, SN, and MAG as members of the Siglec subfamily. This demonstrates that phylogenetic analysis of PPIs is important to ensure completeness of networks.
Homology-guided exploration can reveal new pathways to investigate. As each PPI branches to include interactions with one or more proteins, homology-guided investigation results in a “domino effect,” revealing increasingly complex branching patterns of interconnected PPIs. We used this approach to explore interactions emanating from the DCC subfamily, interrogating new PPIs that exhibit one degree of separation. These revealed linked PPI pathways between proteins with two degrees of separation. Our findings illustrate that each newly identified PPI has the potential to create additional interconnected branches within the network.
A quick survey of screen data reveals multiple phylogenetic pathways that can be explored in the future. It must, however, be noted that some proteins engage in a highly specific PPI with only one subfamily member. Our finding that PVR specifically binds KIR2DL5, but none of the other KIRs examined, illustrates this point.
Functional-Guided Exploration of PPIs
Functional homology can be used to guide exploration in cases where PPIs are observed with proteins belonging to different, distantly related subfamilies. In this approach, proteins are clustered by function, and PPIs are selected for investigation based on shared biological processes. Using this approach, we identified several PPIs of interest. For instance, LILRs interact with MOG and MPZ, two proteins that do not share significant sequence homology but are both involved in the formation and maintenance of myelin sheath. In another example for future studies, the megakaryocyte and platelet inhibitory receptor (G6b) bound advanced glycosylation end product-specific receptor (AGER/RAGE) and IGFBP7. These three proteins share no significant sequence homology, but all play roles in inhibition of platelet activation and homeostasis and other vascular processes.
Expression-Guided Exploration of PPIs
Cell-type and tissue expression can be used to guide PPI exploration in cases where proteins share no sequence homology and where biological function within a system is unknown. In this approach, proteins are clustered by expression and PPIs are evaluated by shared expression, or by expression in cells that encounter one another. Using these criteria, respectively, we selected leptin-ISLR2 and TrkA-TIE1 for investigation. Expression-guided exploration of PPIs is limited by the availability and resolution of mRNA and protein expression data and broad expression across multiple tissues and cell types. As such, expression-guided exploration is a useful tool for proteins that exhibit more restricted expression patterns.
PPIs with Signaling-Competent Cell-Surface Proteins
Many PPIs in the screen were observed with signaling-competent proteins including receptor tyrosine kinases (RTKs), PTPRs, ITIM-, and ITAM-containing proteins and proteins with B30.2 domains (e.g., BTNL8). Interestingly, in some cases, both receptor and ligand exhibit signaling capacity. PVR and KIR2DL5 are both ITIM-containing inhibitory receptors. TrkA and TIE1 are both RTKs. VEGFR3 is an RTK that bound CEACAM4, an ITAM-containing activating receptor. In other cases, a ligand bound two receptors with the same signaling competence: PSGs bound two RTKs, PDGFRA and FGFR4. Together, these signaling receptors regulate a broad spectrum of pathways. As signaling receptors are relevant targets across multiple cancers, these PPIs may suggest new candidates for therapeutic investigation.
Therapeutic Relevance and Future Applications
In our screen, ∼60% of proteins with binding partners engaged in >1 PPI. Several of these proteins are targets of current drugs, including TrkA (cenegermin), Siglec-2 (Besponsa), PD-L1 (atezolizumab, avelumab, and durvalumab), PTPRS (Adrovance and Didronel), FGFR4 (pemigatinib), IL-6Rα (tocilizumab), and VEGFR3 (axitinib), among others. Newly identified PPIs for these targets suggest proteins that should be considered when investigating the mechanisms of drug action as they may contribute to off-target effects or unanticipated signaling mechanisms. In addition, previously unknown PPIs may lead to the discovery of new alternative drug targets for clinically proven pathways or mechanisms. Continued cell-surface PPI mapping is fundamental to understand the intricate networks that function in health and disease.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-His-647 | R&D Systems | Cat#IC0501R |
| Penta·His Antibody; BSA free | QIAGEN | Cat#34660; RRID:AB2619735 |
| Polyclonal Rabbit-Anti-Mouse Immunoglobulin HRP | Agilent | Cat#P0161; RRID:AB2687969 |
| Bacterial and Virus Strains | ||
| Mix & Go Competent Cells - Strain DH5 Alpha | Zymo Research | Cat#T3007 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 2-mercaptoethanol | Thermo Fisher Scientific | Cat#21985023 |
| 4-12% Bis-Tris NuAPGE Protein Gels | Thermo Fisher Scientific | Cat#NP0322 |
| 4–20% Criterion TGX Precast Midi Protein Gel | Bio-Rad | Cat#5671095 |
| 4–20% Mini-PROTEAN® TGX Precast Protein Gels | Bio-Rad | Cat#4561096 |
| Adenosine 5′-triphosphate disodium Salt Hydrate (ATP) | Sigma | Cat#A2383-10G |
| Alexa Fluor 647 NHS Ester (Succinimidyl Ester) | Thermo Fisher Scientific | Cat#A37573 |
| Alkaline Phosphatase | Thermo Fisher Scientific | Cat#31391 |
| Amersham ECL Prime Western Blotting Detection Reagent | Fisher Scientific | Cat#45-002-401 |
| Antibiotic-Antimycotic Solution (100x) | Fisher Scientific | Cat#15-240-096 |
| AscI Restriction Enzyme | New England Biolabs | Cat#R0558S |
| β-Nicotinamide adenine dinucleotide (NAD+) 50 mM solution | New England Biolabs | Cat#B9007S |
| Bicine | Sigma | Cat#B8660-100G |
| Bio-Safe Coomassie Stain | Bio-Rad | Cat#1610786 |
| BirA Ligase | Özkan et al., 2013 | N/A |
| Blotting-Grade Blocker | Bio-Rad | Cat#1706404 |
| Bovine Serum Albumin Fraction V | Roche | Cat#03116956001 |
| Bromophenol Blue | Fisher Scientific | Cat#BP115-25 |
| Carbenicillin, disodium salt | Sigma | Cat#C1389-10G |
| cOmplete, EDTA-free Protease Inhibitor Cocktail | Sigma | Cat#5056489001 |
| dNTP mix; 10 mM each | Genscript | Cat#C01582 |
| d-Biotin | Sigma | Cat#B4501-5G |
| DTT | Sigma | Cat#D0632-25G |
| Expi293 Expression Medium | Thermo Fisher Scientific | Cat#A14635 |
| ExpiFectamine | Thermo Fisher Scientific | Cat#A14635 |
| Fetal Bovine Serum, ultra-low IgG | Thermo Fisher Scientific | Cat#16250078 |
| FreeStyle 293 Expression Medium | Thermo Fisher Scientific | Cat#12338026 |
| Glycerol | Fisher Scientific | Cat#BP229-4 |
| HBS-P+ Buffer 10X | GE Healthcare | Cat#BR-1006-71 |
| HEPES solution, 1 M, pH 7.0-7.6 | Sigma | Cat#H0887 |
| Imidazole | Acros Organics | Cat#12202-5000 |
| InstantBlue Protein Stain | Expedeon | Cat#ISB1L |
| LB Agar, Miller (Powder), 2 kg | Fisher Scientific | Cat#BP1425-2 |
| Linear Polyethylenimine | Fisher Scientific | Cat#NC9197339 |
| Invitrogen UltraPure Agarose | Thermo Fisher Scientific | Cat#16500500 |
| Ethidium Bromide (1% Solution), 10 mL | Fisher Scientific | Cat#BP1302-10 |
| Luria-Bertani (LB) Broth (Miller) | Sigma | Cat#L3522-1KG |
| MACS buffer | Miltenyi | Cat#130-091-221 |
| Magnesium acetate, hexahydrate | Sigma | Cat#M5661-250G |
| Magnesium chloride hexahydrate | Sigma | Cat#M9272-100G |
| MOPS NuPAGE SDS Running Buffer (20X) | Thermo Fisher Scientific | Cat#NP0001 |
| Ni-NTA Agarose | QIAGEN | Cat#30250 |
| NotI-HF Restriction Enzyme | New England Biolabs | Cat#R3189L |
| Opti-MEM Reduced Serum | Thermo Fisher Scientific | Cat#31985062 |
| PEG-8000 | Sigma | Cat#81268-1KG |
| Phosphate Buffered Saline, pH 7.2 | GIBCO | Cat#20012-043 |
| Phusion HF DNA Polymerase | New England Biolabs | Cat#M0530L |
| Propidium iodide | Thermo Fisher Scientific | Cat#P3566 |
| Precision Plus Protein Dual Color Standards | Bio-Rad | Cat#1610374 |
| Recombinant Human Angiopoietin-1 Protein, CF | R&D Systems | Cat#923-AN-025/CF |
| Recombinant Human β-NGF | R&D Systems | Cat#256-GF/CF |
| Recombinant Human Leptin Protein, CF | R&D Systems | Cat#398-LP-01M |
| Recombinant Mouse Leptin Protein, CF | R&D Systems | Cat#498-OB-01M |
| Recombinant protein A | Thermo Fisher Scientific | Cat#21184 |
| SeeBlue® Plus2 Pre-stained Protein Standard | Thermo Fisher Scientific | Cat#LC5925 |
| Series S Sensor Chip SA | GE Healthcare | Cat#BR-1005-31 |
| Sodium bicarbonate | Fisher Scientific | Cat#BP328-500 |
| Sodium Chloride | Fisher Scientific | Cat#S271-3 |
| Streptavidin | Invitrogen | Cat#S888 |
| Superdex 200 Increase column | GE Healthcare | Cat#28990944 |
| Superose S6 column | GE Healthcare | Cat#17-5172-01 |
| T5 Exonuclease | New England Biolabs | Cat#M0363L |
| Taq Ligase | New England Biolabs | Cat#M0530L |
| Trans-Blot® Turbo RTA Midi PVDF Transfer Kit | Bio-Rad | Cat#1704273 |
| Tris base | Fisher Scientific | Cat#BP152-1 |
| Tris-HCl | Fisher Scientific | Cat#BP153-1 |
| Tween-20; Sigma | Sigma | Cat#P1379-500ML |
| Ultrapure SDS | Life Technologies | Cat#15525-017 |
| Critical Commercial Assays | ||
| BCA Kit | Pierce | Cat#23227 |
| KPL BluePhos Microwell Substrate Kit | VWR | Cat#95059-220 |
| QIAQuick Gel Extraction Kit | QIAGEN | Cat#28506 |
| QIAprep Spin Miniprep Kit | QIAGEN | Cat#27106 |
| Deposited Data | ||
| Dryad | This Study | https://doi.org/10.5061/dryad.xsj3tx9bd |
| Experimental Models: Cell Lines | ||
| Expi 293F cells | Thermo Fisher Scientific | Cat#A14635; RRID: CVCL_D615 |
| HEK293 Freestyle cells | Thermo Fisher Scientific | Cat#:R79007; RRID:CVCL_6642 |
| Oligonucleotides | ||
| ISLR2 LRR1-7 forward: 5′- CTGAGCTACATCTTC TGCCTGGTGTTCGCTGCGGCCGCCTCTTGTCC TGAACCTTGTGCCTGCG-3′ |
This Study | N/A |
| ISLR2 LRR1-7 reverse: 5′- CCAGGCCC TTGAAACAGAACTTCCAACCCCGGC GCGCCAGGAGCGCAAGGCAGAGCAG-3′ |
This Study | N/A |
| ISLR2 Ig1-3 forward: 5′-CTGAGCTACATCTTC TGCCTGGTGTTCGCTGCGGCCGCCCCTAGC GTTCACCTGTCTGCC-3′ |
This Study | N/A |
| ISLR2 Ig1-3 reverse: 5′-CCAGGCCCTTGAAAC AGAACTTCCAACCCCGGCGCGCCAGAGGGC AGCTCTTTCTTGGTGC-3′ |
This Study | N/A |
| pD649-HAsp-ECD-Fc(DAPA)-AviTag-6xHis (sequencing) forward:5′-ATTTAGGTGACACTATAG-3′ | This Study | N/A |
| pD649-HAsp-ECD-Fc(DAPA)-AviTag-6xHis (sequencing) reverse: 5′-CACGTACCAGTTGAACTTCACC-3′ | This Study | N/A |
| Recombinant DNA | ||
| pD649-HAsp-ECD-Fc(DAPA)-AviTag-6xHis | This study | Addgene |
| pD649-HAsp-ECD-COMP5AP-AviTag-9xHis | This study | Addgene |
| pTT3-SP-6XHis-NTRK1(FL)-FLAG | This study | Addgene |
| pTT3-SP-6XHis-TIE1(FL)-FLAG | This study | Addgene |
| pTT3-SP-6XHis-PVR(FL)-FLAG | This study | Addgene |
| pTT3-SP-6XHis-KIR2DL5(FL)-FLAG | This study | Addgene |
| pTT3-SP-6XHis-KIR2DL4(FL)-FLAG | This study | Addgene |
| pTT3-SP-6XHis-KIR2DL1(FL)-FLAG | This study | Addgene |
| pTT3-SP-6XHis-ISLR2(FL)-FLAG | This study | Addgene |
| Software and Algorithms | ||
| Biacore T200 Control Software (version 3.2) | GE Healthcare | https://www.biacore.com/lifesciences/service/downloads/downloads/index.html |
| BioGRID3.5 | Oughtred et al., 2019 | https://thebiogrid.org/ |
| CytExpert 2.3 | Beckman Coulter | https://www.beckman.com/flow-cytometry/instruments/cytoflex/software |
| FlowJo 10.4.2 | FlowJo | https://www.flowjo.com/ |
| HUGO Gene Nomenclature Committee (HGNC) | Yates et al., 2017 | https://www.genenames.org/ |
| Human Protein Atlas | Uhlén et al., 2015 | https://www.proteinatlas.org/ |
| IntAct | Orchard et al., 2014 | https://www.ebi.ac.uk/intact/ |
| Interactive Tree of Life (iTOL) (version 5.5) | Letunic and Bork, 2019 | https://itol.embl.de/ |
| MAFFT (Multiple Alignment using Fast Fourier Transform) | Kuraku et al., 2013; Katoh et al., 2019 | https://mafft.cbrc.jp/alignment/server/ |
| MUltiple Sequence Comparison by Log- Expectation (MUSCLE) | Edgar, 2004 | https://www.ebi.ac.uk/Tools/msa/muscle/ |
| PredGPI | Pierleoni et al., 2008 | http://gpcr.biocomp.unibo.it/predgpi/pred.htm |
| SignalP-5.0 | Almagro Armenteros et al., 2019 | http://www.cbs.dtu.dk/services/SignalP/ |
| SnapGene 5.1 | GSL Biotech LLC | https://www.snapgene.com/ |
| STRING (version 11.0) | Szklarczyk et al., 2019 | |
| SoftMax Pro | Molecular Devices | https://www.moleculardevices.com/products/microplate-readers/acquisition-and-analysis-software/softmax-pro-software |
| Freedom EVOware Standard (version 2.5) Service Pack 3, Build 2.5.21.0 | TECAN | https://lifesciences.tecan.com/software-freedom-evoware? |
| TMHMM2.0 | Krogh et al., 2001 | http://www.cbs.dtu.dk/services/TMHMM/ |
| UniProt | The UniProt Consortium | https://www.uniprot.org/ |
| Other | ||
| Econo-Pak Chromatography Column | Bio-Rad | Cat#7321011 |
| 12-Column Reservoirs | Agilent | Cat#201256-100 |
| 12-Column Reservoirs | Integra | Cat#6361 |
| 96-well deep well plates | USA Scientific | Cat#1896-2110 |
| 96-well round-bottom plate | Corning | Cat#3799 |
| AirOtop Enhanced Seal for Ultra Yield | Fisher Scientific | Cat#NC0063892 |
| Aluminum Foil Seals | Corning | Cat#7200684 |
| Amicon Ultra-4 Centrifugal Filter Unit | EMD Millipore | Cat#UFC805024 |
| Amicon Ultra-15 Centrifugal Filter Unit | EMD Millipore | Cat#UFC901024 |
| Hyperfilm ECL | Sigma | Cat#GE28-9068-39 |
| Nunc clear flat-bottom maxisorp 384-well plates | Thermo Fisher Scientific | Cat#464718 |
| Nunc clear flat-bottom maxisorp 384-well plates | Thermo Fisher Scientific | Cat#464718 |
| Nunc clear flat-bottom maxisorp 96-well plates | Thermo Fisher Scientific | Cat#442404 |
| PolarSeal | E&K Scientific | Cat#T592100 |
| Rapid-Flow Sterile Disposable Filter Units with PES Membrane; 250 ml (0.2 um) | Thermo Fisher Scientific | Cat#168-0045 |
| Rapid-Flow Sterile Disposable Filter Units with PES Membrane; 50 ml (0.2 um) | Thermo Fisher Scientific | Cat#564-0020 |
| Rapid-Flow Sterile Disposable Filter Units with PES Membrane; 1 L (0.2 um) | Thermo Fisher Scientific | Cat#567-0020 |
| Seahorse Single Cavity P.P. Pyramid Base Reservoirs | Agilent | Cat#201244-100 |
| TECAN TP unfiltered 200 ul tips; Zymark | MBP/Thermo Fisher Scientific | Cat#923-261 |
| TECAN TR unfiltered 100 ul tips; Zymark | MBP/Thermo Fisher Scientific | Cat#907-261 |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, K. Christopher Garcia (kcgarcia@stanford.edu).
Materials Availability
Plasmids generated in this study (1,144 plasmids) will be deposited to Addgene (Addgene preassigned plasmid #s: 156487-157630). Due to COVID-19 reduced Addgene lab capacity and the queue for accepting large deposits at this time, plasmids may not be available from Addgene until early 2021. Full plasmid sequences are included in Data S2.
Data and Code Availability
Information for all 564 proteins in screen including gene name, UniProt entry name, aliases, full-length protein sequence, ECD boundaries and sequence, superfamily, type (secreted, STM, multi-pass TM, and GPI-anchored), and predicted molecular weight of ECD-Fc and ECD-5AP proteins is included in Data S1. Full plasmid sequences for all bait and prey constructs are included in Data S2. Data for qualitative assessment of ECD-Fc and ECD-5AP levels in conditioned media by western blot and AP quantitation of ECD-5AP conditioned media (relative AP activity; ng/μl) are included in Data S3. Screen data and multiple sequence alignement (MSA) files have been deposited to Dryad (https://doi.org/10.5061/dryad.xsj3tx9bd) and are included in Data S4. SPR conditions for all ligand-analyte pairs tested including ligand RU, maximum analyte concentration, analyte RU at maximum concentration, number of analyte concentrations tested, injection time (seconds), injection rate (μl/minute), dissociation time (seconds), and regeneration conditions are included in Data S5.
Experimental Model and Subject Details
HEK293 Freestyle suspension cells were grown in FreeStyle 293 Expression Medium (Thermo Fisher Scientific) in a 37°C, 5% CO2 incubator with 130 rpm shaking. Expi293F suspension cells were grown in Expi293 Expression Medium (Thermo Fisher Scientific) in a 37°C, 5% CO2 incubator with 130 rpm shaking.
Method Details
Selection of proteins and ECD prediction
Two lists of human IgSF proteins were generated using the HUGO Gene Nomenclature Committee (HGNC) database (https://www.genenames.org/data/genegroup/#!/group/589) and the Human Protein Atlas (HPA) database (https://www.proteinatlas.org/humanproteome/tissue/secretome). Lists were checked for overlap and a master list was generated using the HPA majority decision-based method (MDM). Metadata for all master list proteins was extracted from UniProt, and extracted signal peptide (SP) and TM predictions were compared to the SIgnalP-5.0 (http://www.cbs.dtu.dk/services/SignalP/) (Almagro Armenteros et al., 2019) and TMHMM2.0 prediction servers (http://www.cbs.dtu.dk/services/TMHMM/) (Krogh et al., 2001). SP and TM predictions were used to map the ECD boundaries of cell-surface proteins. For glycosylphosphatidylinositol-anchored (GPI) proteins, PredGPI (http://gpcr.biocomp.unibo.it/predgpi/) (Pierleoni et al., 2008) and manual sequence analysis were used to predict the ECD boundary upstream of the phospholipase cleavage site. ECD sequences containing a Cys residue within the C-terminal 10 amino acids were manually analyzed to determine whether the Cys residue fell within the last protein domain, or within the linker region between the last protein domain and the TM. In cases where the Cys residue was predicted to reside within the linker region, the ECD boundary was adjusted to end at the residue preceding the Cys to prevent potential protein misfolding due to aberrant Cys-Cys disulfide bond formation between this putatively unpaired Cys and Cys residues in the Ig domains of the Fc. Following ECD domain boundary prediction, canonical protein sequences were extracted from UniProt and compiled for back translation and optimization by GeneArt/Life Sciences Technology for gene synthesis (see below).
Amino acid sequence homology analysis
Mutliple sequence alignment (MSA) was performed as follows. ECD amino acid sequences were submitted to the MUltiple Sequence Comparison by Log- Expectation (MUSCLE) (https://www.ebi.ac.uk/Tools/msa/muscle/) (Edgar, 2004) and Multiple Alignment using Fast Fourier Transform (MAFFT) (https://mafft.cbrc.jp/alignment/server/) (Kuraku et al., 2013; Katoh et al., 2019) online resources and analyzed using both first iteration and second iteration parameters (MUSCLE) and default parameters (MAFFT). Output files were submitted to the Interactive Tree of Life (iTOL) (https://itol.embl.de/) (Letunic and Bork, 2019), an agglomerative hierarchical clustering algorithm, to build a cluster hierarchy and generate phylogenetic trees. MSA files have been deposited to Dryad (https://doi.org/10.5061/dryad.xsj3tx9bd).
Bait and prey expression plasmids
Genes encoding curated ECD and secreted proteins were synthesized at GeneArt/Life Sciences Technologies and subcloned into both pD649-HAsp-ECD-Fc(DAPA)-AviTag-6xHis and pD649-HAsp-ECD-COMP5AP-AviTag-9xHis bait and prey expression vectors, respectively. Genes were subcloned in-frame with the upstream hemagglutinin signal peptide (HAsp) (MKTIIALSYIFCLVFA) and downstream Fc(DAPA)-AviTag-6xHis or COMP5AP-AviTag-9xHis modules via 5′ NotI and 3′ AscI sites. Bait expression vectors fuse genes in-frame with the Fc region of human IgG containing D265A and P329A mutations (DAPA), followed by an AviTag (GLNDIFEAQKIEWHE), and 6xHis. The DAPA mutation was used because it prevents binding of Fc receptors to the Fc. Prey expression vectors fuse the gene in-frame with a 3C enzyme cleavage site (LEVLFQGP), followed by the rat cartilage oligomeric matrix protein (COMP) pentamerization domain (Holler et al., 2000) fused to human placental alkaline phosphatase (5AP), an AviTag, and 9xHis. A MaxiPrep of plasmid DNA was provided by GeneArt/Life Sciences Technologies at 1 μg/ml in 20 mM Tris, pH 8.0. For complete plasmid sequences of all 564 bait and prey expression vectors, see Data S2.
ISLR2-Fc truncation expression plasmids
ECD-Fc expression vectors for two ISLR2 ECD truncations comprising: 1) the N-terminal seven LRRs (LRR1-7) and 2) the C-terminal three Ig domains (Ig1-3) were generated by PCR amplification of the regions encoding these domains from pD649-HAsp-ISLR2-Fc(DAPA)-AviTag-6xHis. Resulting PCR products were cloned into the NotI-AscI double-digested parental pD649-HAsp-ECD-Fc(DAPA)-AviTag-6xHis backbone by isothermal assembly. LRR1-7 comprises amino acids 19-232 and Ig1-3 comprises amino acids 233-589. Primer sequences are listed in the Key Resources Table. PCR products were run on an agarose gel (Thermo Fisher Scientific) stained with ethidium bromide (Fisher Scientific), and purified using a QIAQuick Gel Extraction Kit (QIAGEN). Isothermal assembly was performed using 1 μL digested plasmid DNA (∼50 ng/μl), 1-4 μL purified PCR product (∼80 ng/μl) and 15 μL isothermal assembly mix. Isothermal assembly mix contained 320 μL 5X ITA buffer (0.5 M Tris-HCl, pH 7.5, 50 mM MgCl2, 1 mM dNTP mix (Genscript), 50 mM DTT, 25% w/v PEG-8000, 0.33% w/v nicotinamide adenine dinucleotide (New England Biolabs)), 1 μL T5 exonuclease (New England Biolabs), 20 μL Phusion HF DNA polymerase (New England Biolabs), 160 μL Taq ligase (New England Biolabs) and 700 μL Ultrapure H2O (EMD Millipore). Reactions were incubated at 50°C for 1 hour, transformed into competent DH5α cells (Zymogen), plated on Luria-Bertani (LB) Broth agar (Fisher Scientific) plates containing 50 μg/ml carbenicillin (Sigma), and incubated overnight at 37°C. Colonies were picked into 8 mL LB (Sigma) containing 50 μg/ml carbenicillin and grown ∼18 hours at 37°C with 250 rpm shaking. Plasmid DNA was purified using a QIAprep Spin Miniprep Kit (QIAGEN) and absorbance at 260 nm was measured using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific) to determine plasmid DNA concentration. LRR1-7 and Ig1-3 sequences were confirmed by sequencing (MCLab) using primers listed in the Key Resources Table. For complete plasmid sequences, see Data S2.
Full-length protein expression plasmids
Genes encoding full-length proteins were synthesized at GeneArt/Life Sciences Technologies and subcloned into the pTT3 expression vector. Genes contain the endogenous signal peptide, a 6xHis tag (to facilitate cell-surface expression analysis), the remaining full-length coding region of the gene, and a FLAG tag, followed by a stop codon. Synthesized DNA contained upstream NotI and NheI and downstream MluI and XbaI restriction sites. Synthesized DNA was subcloned into pTT3 using 5′ NotI and 3′ XbaI sites. A MaxiPrep of plasmid DNA was provided by GeneArt/Life Sciences Technologies at 1 μg/ml in 20 mM Tris, pH 8.0. For transgene sequences, see Data S2.
Conditioned media for standard ECIA
ECD-Fc bait and ECD-5AP prey conditioned media were produced in Expi293F suspension cells grown in Expi293 Expression Medium (Thermo Fisher Scientific) in a 37°C, 5% CO2 incubator with 130 rpm shaking. Transfection was performed using Expifectamine as per manufacturer’s instructions (Thermo Fisher Scientific). Briefly, cells were grown to 4-5x106 cells/ml and diluted to 3x106 cells/ml. 1 μg of plasmid DNA was used per ml of culture. Plasmid DNA was mixed with 1/20 transfection volume of OptiMem (Thermo Fisher Scientific) and, in a separate tube, 2.7 μL Expifectamine/μg DNA was mixed with 1/20 transfection volume of OptiMEM. After 3 minutes incubation of Expifectamine with OptiMem at room temperature, solutions containing plasmid DNA and OptiMem were mixed and incubated for 5 minutes at room temperature before adding dropwise to cells. Enhancer 1 and Enhancer 2 were added ∼18 hours later according to the manufacturer’s instructions. Three days following addition of Enhancers, cell cultures were harvested, and spun 7 minutes at 1,294 x g to pellet cells. Conditioned media was supplemented to 10% ultra-low IgG Fetal Bovine Serum (Thermo Fisher Scientific) and 1X antibiotic-antimycotic solution (Fisher Scientific), and stored at 4°C.
Conditioned media for apECIA screen
ECD-Fc bait and ECD-5AP prey conditioned media were produced using the Protein Expression and Purification Platform (PEPP), a fully-automated suite of custom robotic platforms (Rue et al., 2019). PEPP is extensively described in a recent book chapter (Rue et al., 2019). Briefly, HEK293 Freestyle cells were grown in FreeStyle 293 Expression Medium (Thermo Fisher Scientific). Cells were cultured in custom-made flasks and transfected using linear polyethylenimine (PEI) (Fisher Scientific). Three days post-transfection, cells were spun down, conditioned media was supplemented to 10% ultra-low IgG Fetal Bovine Serum (Thermo Fisher Scientific), 1X antibiotic-antimycotic solution (Fisher Scientific) and cOmplete protease inhibitor (Sigma), 0.2 um filtered (Thermo Fisher Scientific), and stored at 4°C.
Western blot analysis of conditioned media
ECD-Fc bait and ECD-5AP prey conditioned media were analyzed by western blot against C-terminal 6xHis (bait) and 9xHis (prey) tags to confirm all full-length proteins were of the expected size. Westerns were performed as follows: 10 μL of conditioned media was combined with 2 μL of 6X SDS loading dye (Tris/SDS buffer (0.5 M Tris, pH 6.8, 0.4% SDS), 38% glycerol (weight/volume), 10% (weight/volume) SDS, 0.55 M 2-mercaptoethanol, 0.0024% bromophenol blue (weight/volume)). Samples were incubated at 95°C for 3 minutes and run on a 4%–20% Mini-PROTEAN gel (Bio-Rad). 10 μL of 1 ng/μl FPLC-purified IL2-Fc was run as a concentration standard on each gel. After running, gels were rinsed in dH2O and transferred to a PVDF membrane using the High MW setting on a Bio-Rad TransBlot Turbo system using the Midi PVDF Transfer Kit (Bio-Rad) according to manufacturer’s instructions. Membranes were blocked for 1-2 hours at room temperature with shaking in Western blocking buffer (20 mM Tris, pH 7.2, 0.5M NaCl, 0.2% Tween-20, 5% Blotting-Grade Blocker (Bio-Rad)). Penta-His antibody (QIAGEN) was added at 1:1000 and blots were incubated at 4°C overnight with shaking. Membranes were washed 10 times for 2 minutes on an orbital shaker at 300 rpm in Western blocking buffer and stained in Western blocking buffer + 1:5000 HRP-tagged polyclonal rabbit anti-mouse antibody (Agilent) for 1-1.5 hours at room temperature on an orbital shaker at 70 rpm. Membranes were incubated with 4 mL ECL Prime Western Blotting Detection Reagent (Fisher Scientific), exposed to Hyperfilm ECL, and developed on an All-Pro 100 Plus X-ray film processor (ALLPRO Imaging). Exposure times ranged from 5 s to 4 hours in order to adequately cover the wide range of expression levels of proteins present in conditioned media.
To qualitatively assess protein levels we included a known concentration of a purified His-tagged reference protein, IL2-Fc. Westerns were semiquantitatively analyzed by comparing the intensity of conditioned media bands with the IL2-Fc standard. If conditioned media bands were visible at an intensity which could be directly compared with the standard, an estimate of the relative concentration of protein in the conditioned media could be determined from 0.1X to 10X of the standard. In cases where conditioned media proteins were present at very high levels, in a short exposure where the standard was barely visible, it was scored as > 10X. In cases where the conditioned media was barely visible while the standard band was overexposed, it was scored as < 0.1x. In cases where the conditioned media band was barely visible at the longest exposure, it was denoted as “faint.” If a conditioned media failed to show any band at the longest exposure (4 hours), it was scored as “undetectable.” Levels are reported in Data S3.
AP quantitation of prey conditioned media
AP standards were prepared as follows: 2-fold dilutions (1.5625-800 pg/μl) of purified alkaline phosphatase (AP) (Thermo Fisher Scientific) were prepared using 1X TBS (20 mM Tris pH 7.4, 150 mM NaCl). AP standards (10 μl) and 1X TBS (10 μl; blank) were added to duplicate wells in clear, untreated 384-well plates. ECD-5AP conditioned media samples (10 μl) and conditioned media from mock transfections (10 μl; blank) were also added in duplicate. 50 μL of BluePhos AP substrate (VWR) was added to all wells. O.D. 650 nm was measured using a SpectraMax Paradigm plate reader (Molecular Devices) with SoftMax Pro software. Measurements were taken at multiple time points (4-60 minutes) to obtain values within the linear range of various regions of the AP standard curve for quantitation of ECD-5AP proteins exhibiting a broad spectrum of expression levels (relative AP activity: 5 ng/μl to ∼650 ng/μl). AP quantitation results were in good agreement with qualitative Western estimates (Figure S1B). Complete quantitation results are reported in Data S3.
6xHis tag purification of ECD-Fc protein
ECD-Fc proteins were produced in Expi293F cells using transfection conditions described above. Following harvesting of cell conditioned media, 1 M Tris, pH 8.0 was added to a final concentration of 20 mM. Ni-NTA Agarose (QIAGEN) was added to ∼5% conditioned media volume. 1 M sterile PBS, pH 7.2 (GIBCO) was added to ∼3X conditioned media volume. The mixture was stirred overnight at 4°C. Ni-NTA agarose beads were collected in a Buchner funnel and washed with ∼300 mL protein wash buffer (30 mM HEPES, pH 7.2, 150 mM NaCl, 20 mM imidazole). Beads were transferred to an Econo-Pak Chromatography column (Bio-Rad) and protein was eluted in 15 mL of elution buffer (30 mM HEPES, pH 7.2, 150 mM NaCl, 200 mM imidazole). Proteins were concentrated using Amicon Ultracel 10K filters (Millipore) to a volume of ∼0.5 mL and absorbance at 280 nm was measured using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific) to determine protein concentration.
Biotinylation and FPLC purification
Proteins were biotinylated as described previously (Özkan et al., 2013). Briefly, up to 10 mg of protein was incubated at 4° overnight in 2X Biomix A (0.5 M bicine buffer), 2X Biomix B (100 mM ATP, 100 mM MgOAc, 500 μM d-biotin), Bio200 (500 uM d-biotin) to a final concentration of 20 uM, and 60-80 units BirA ligase in a final volume of 1 ml. Proteins were further purified by size-exclusion chromatography using an S200 Increase or a Superose S6 column (GE Healthcare), depending on protein size, on an ÄKTA Pure FPLC (GE Healthcare). All proteins were eluted in 1X HBS (30 mM HEPES pH 7.2, 150 mM NaCl). Following elution, a sample from each protein fraction was run on a 4%–20% Mini-PROTEAN gel and gels were Coomassie stained to check for protein degradation and identify fractions containing intact protein for pooling of samples. 2 to 10 μL of peak sample fractions were mixed with 2 μL 6X loading dye (see above) in a final volume of 12 μL and incubated at 95°C for 5 minutes prior to running on a 4%–20% Mini-PROTEAN gel (Bio-Rad). Gels were briefly rinsed 3X in dH2O, incubated in warm dH2O for 10-15 minutes on an orbital shaker at 60 rpm to fully remove residual SDS, then incubated 1 hour to overnight at room temperature with shaking in Bio-Safe Coomassie Stain (Bio-Rad). Gels were briefly rinsed 3X in dH2O, then washed in dH2O with shaking at room temperature for several hours to overnight prior to imaging in a Bio-Rad Gel Doc EZ Imager (Bio-Rad). Fractions containing intact protein were pooled, concentrated using Amicon Ultracel 10K filters (Millipore) to a volume of 100-300 μL and absorbance at 280 nm was measured as above. SEC data are shown in Figure S5.
Strep-shift analysis of biotinylated proteins
1 to 10 μg of FPLC-purified protein was added to 3.7 μL 6X SDS loading dye (see above) in a final volume of 20 μl, incubated at 95°C for 5 minutes, then placed on ice for 10 minutes. Equal volumes of samples were split into two tubes. 4X molar excess of streptavidin (Invitrogen) was added to one tube, and an equivalent volume of 1X HBS (30 mM HEPES pH 7.2, 150 mM NaCl) was added to the other tube. Streptavidin was also added to an otherwise blank reaction containing 1X HBS and 6X SDS loading dye. Samples were incubated on ice for 20 minutes prior to running on a 4%–20% Mini-PROTEAN gel (Bio-Rad). Binding of streptavidin to biotinylated protein causes the molecular weight of the biotinylated protein to increase (shift) in molecular weight by ∼40 kD. The fraction of biotinylated protein in the sample was estimated by comparing the intensity of the band in the no-streptavidin lane with that of the shifted band in the plus-streptavidin lane.
Optimization of ECIA in 384-well plates
Early pooling experiments were performed in a 96-well plate format. We noticed that when we moved to a 384-well plate format, the sensitivity (maximum O.D. 650 nm signal) was ∼1.4-fold lower than observed in 96-well plates. We optimized three parameters to increase the sensitivity in 384-well plates. To optimize the volume of BluePhos substrate (VWR), we added 4 μL of a 2-fold dilution series (undiluted to 1/64 dilution) of ECD-5AP conditioned media or mock conditioned media to multiple rows of a 384-well plate, and added increasing volumes of BluePhos substrate (10 μL increments from 10 μL to 100 μl) (prepared according to manufacturer’s instructions). O.D. 650 nm was measured at multiple time points (20 minutes, 40 minutes, 1 hour, 1.5 hours, 2 hours, and 3 hours) using a SpectraMax Paradigm plate reader (Molecular Devices) with SoftMax Pro software. At low ECD-5AP concentrations, 40 μL and 60 μL of BluePhos substrate yielded maximum O.D. 650 nm values, while retaining minimal background signal (i.e., highest signal to background ratio). Follow-up experiments were performed to fine-tune the volume of BluePhos substrate and 50 μL was found to be the optimal volume for high signal to background. As the plate was read at multiple times following addition of BluePhos substrate, this experiment also allowed us to optimize BluePhos substrate incubation time. The 2 hour time point yielded the highest signal to background ratio (BluePhos oxidizes over time, thereby turning blue without recourse to enzymatic activity).
To optimize the volume of ECD-Fc bait and ECD-5AP prey conditioned media in 384-well plates, we tested 5 μl, 10 μL and 20 μL using conditioned media with different binding pairs exhibiting a range of binding levels from weak to high (O.D. 650 nm values, 2-fold to 40-fold over background signal). For the weakest binding pairs, 20 μL of bait and 20 μL of prey was required to obtain ≥ 2-fold over background signals (cutoff for categorizing a binding pair as a putative “hit”).
ECIA using pooled prey
Pooled prey experiments were carried out in a 384-well clear Nunc MaxiSorp plate (Thermo Fisher Scientific). Protein A was diluted to 10 μg/ml (from a stock concentration of 10 mg/ml in 1X PBS, pH 7.4) in 100 mM sodium bicarbonate buffer, pH 9.6 and sterile filtered (0.2 um). 20 μL of Protein A solution was added to each well and sealed plates (PolarSeal; E&K Scientific) were incubated at 4°C overnight. Protein A solution was removed and 90 μL blocking solution (1X TBST (20 m M Tris, pH 7.4, 150 mM NaCl, 0.05% w/v Tween-20), 3% BSA) was added. Sealed plates were blocked at room temperature for 3 hours. Blocking buffer was removed and plates were washed 3 times with 90 μL 1X TBST. 20 μL of ECD-Fc bait conditioned media (or mCherry-Fc control conditioned media) was added to wells and sealed plates were incubated at 4°C overnight. Plates were washed 3 times with 90 μL 1X TBST using an automated HydroSpeed plate washer (TECAN). 20 μL of undiluted or 1:3 diluted ECD-5AP prey conditioned media was added and sealed plates were incubated at room temperature overnight. Plates were washed 3 times with 90 μL 1X TBST and 50 μL of KPL BluePhos substrate (VWR) was added (prepared according to manufacturer’s instructions). Following incubation for 2 hours at room temperature, O.D. 650 nm was measured as above. Background was subtracted using the absorbance of mCherry-Fc control wells.
ECIA titration
Titration experiments were carried out in 96-well Nunc Maxisorp plates as above with the following modifications. Increased volumes were used including 100 μL 10 μg/ml Protein A solution, 200 μL 1X TBST wash buffer, 250 μL blocking solution, and 150 μL BluePhos substrate (VWR). For ECD-Fc bait and ECD5-AP prey, 100 μL of undiluted or a 2-fold dilution series of conditioned media was used (dilutions performed using mock conditioned media). As above, mCherry-Fc conditioned media was used for bait control wells.
ECIA testing of PVR binding to KIRs
ECIA in 96-well Nunc Maxisorp plates was carried out as above with the following modifications. Because specificity of PVR-5AP prey binding was tested to many KIR-Fc bait family members, it was essential to ensure that all KIR-Fc well surfaces were saturated to compare differences in binding to KIR-Fc proteins. To ensure KIR-Fc saturation, multiple applications of 150 μL of KIR-Fc conditioned media were applied sequentially. Applications were incubated for 3 hours at room temperature before removal and addition of the subsequent application. The last application was incubated at 4°C overnight. To confirm KIR-Fc saturation of all wells, following ECIA and O.D. 650 nm measurement, plates were washed 3 times with 200 μL 1X TBST. 50 μL 6X SDS loading dye (see above) was added to each well and plates were placed at −20°C. Plates were removed from −20°C, allowed to thaw to room temperature and placed on a 42°C heat block for 20 minutes to assist in releasing protein from the plate surface. 10 μL was run on a 4%–20% Mini-PROTEAN gel (Bio-Rad) and anti-His western blot was performed as above.
Design of robotic choreography for apECIA
Development of an automated platform required a design strategy that arranged bait and prey stock proteins in a format which allowed all bait and pooled prey pairs to be combined by choreographed transfer into 384-well microtiter reaction plates using a liquid handling system. 384-well plates are composed of 96 quadrants, each a 2x2 matrix. We utilized a 2x2 matrix scheme to test 384 unique combinations of bait and pooled prey in each plate. To achieve this, two bait proteins were arrayed in alternating rows. Pooled prey proteins were transferred from two stock plates (X and Y, each containing 94 unique prey pools) into alternating columns in two contiguous rows.
The 96-quadrant, 2x2 matrix layout was achieved using a customized robotic workstation. We developed a choreographed robotic workflow, and programmed robot methods to execute a two-step process for: 1) sample processing into distinct layouts to generate bait and prey conditioned media stock plates and 2) all steps of screening in ELISA plates. The automated platform utilized two pipetting robotic arms with user-defined aspiration and dispense parameters for liquid speed and acceleration and deceleration of plungers to ensure precision volume pipetting of samples with varying viscosity, and z axis control for non-contact pipetting. The first is an 8-channel, fixed-tip, positive pressure liquid displacement handling arm (LiHa) with programmable features for sample aspiration and dispense tracking, variable y-spacing, and individual channel control for x- and y-axis transfer of samples (“cherry-picking”). The second is an air displacement multichannel dispensing arm (MCA) with 96-tip head for user-defined multi-tip transfer (“stamping”). The workstation includes a 3-axis plate transport robotic manipulator arm (RoMA) and integrates a Power Washer PW384 plate washer (TECAN) and Infinite M1000 multimodal spectrophotometer plate reader (TECAN). A wavelength of 650 nm was used to detect the substrate absorbance of the BluePhos phosphatase substrate (KPL) used to detect protein interactions. To implement real-time pipetting volume verification after addition of each reagent into 384-well reaction plates, each plate was also read at 980 nm as liquid volume (absorbance pathlength) for non-opaque aqueous solutions correlates with optical density at 980 nm (McGown and Hafeman, 1998). Following both LiHa and MCA dispensing of every reagent into 384-reaction plates, we performed volume QC using automated spectrometry analysis. All automated liquid handling processes were carried out using TECAN Freedom Evo2 robotics workstations with in-house designed components for sample processing to allow larger volume sample handling.
apECIA screen
Studies using ECIA and other variations of this assay have demonstrated that results are highly reproducible. As such, we performed the pooled prey screen once and validated “hits” in triplicate. The screen was performed in 384-well plates using reagent volumes determined by optimization experiments (see above). Each week 48 plates were screened (47 test plates and one control plate). Test plates contained two ECD-Fc baits in alternating rows; as such, 94 baits were tested each week. The control plate contained alternating rows of mCherry-Fc conditioned media and mock conditioned media. As prey were tested in pools of three, the 564 prey proteins in the screen comprised 188 pools. Prey pools were distributed into two 96-deep well stock plates designated “X” and “Y,” each containing 94 prey pools. The remaining two wells contained positive control preys for testing to positive control baits located in eight wells in the bottom right corner of each plate (wells M23-24, N23-24, O23-24 and P23-24). Controls included four bait-prey pairs in duplicate wells including a non-binding, low binding, intermediate binding, and strong binding pair. Using a 96-tip stamping method, prey pools from plate “X” were transferred to 384-well ELISA plates in two subsequent rows within each of the odd-numbered columns. Prey pools from plate “Y” were similarly transferred into two subsequent rows within each of the even-numbered columns.
Each week 96 baits were tested (94 test baits, mCherry-Fc and mock) against all prey pools (and 4 prey controls) in 48 plates totaling 18,432 reaction wells. As 94 test baits were tested against all 564 preys, 53,016 bait-prey combinations were tested per week. We conducted the screen in 6 weeks testing 318,096 combinations (i.e., 53,016 × 6 = 318,096), the total number in a matrix of 564 proteins (i.e., 564 × 564 = 318,096). Screen data are available at Dryad (https://doi.org/10.5061/dryad.xsj3tx9bd) and Data S4.
apECIA screen deconvolution and validation
Nine of the 188 prey pools gave rise to large numbers of wells with positive signal (≥2-fold over background) and were excluded as non-specific PPIs. Following removal of wells representing non-specific PPIs, the 756 wells with the highest fold-over-background signal were selected for deconvolution. Background was calculated using the average signal of each prey pool against all 564 baits and controls (mCherry-Fc and mock). Deconvolution was performed for each well by re-testing bait with each of the three pool prey proteins in separate wells. The 756 wells selected for deconvolution included wells exhibiting the highest signal (> 50 fold-over-background) down to wells with a nominal signal (1.3 fold-over-background). We chose to deconvolute wells with such a low signal due to the possibility that actual PPIs may have yielded nominal signals as a result of decreased prey concentration brought about by pooling. In such cases, re-screening against undiluted prey may result in higher signal and identification of an otherwise missed PPI (false negative). Deconvolution revealed 495 wells with signal ≥ 2 fold-over-background. In each case, only one of the three deconvoluted prey yielded a positive binding signal, referred to as the ‘true-binding’ prey. To confirm binding, each ‘true-binding’ prey was re-tested against its partner bait in triplicate and against a negative control bait (mCherry-Fc). All re-tested bait-prey PPI pairs exhibited binding signal ≥ 2 fold-over-background. To determine whether PPIs were known, we searched the literature using PubMed and Google Scholar, and the PPI databases STRING (Szklarczyk et al., 2019), BioGRID3.5 (Oughtred et al., 2019), and IntAct (Orchard et al., 2014). In addition to human, we considered PPIs reported in mouse, rat, and zebrafish to be known.
Cell-surface binding assay
To examine PPIs at the cell surface, we performed cell-surface protein binding assays. Expi293F cells were transfected as above with expression plasmids encoding full-length proteins containing an N-terminal 6xHis tag (following the signal peptide, see above). Two days following transfection, cultures were harvested, cells were spun down for 4 min at 1600 rpm (∼400 x g), and resuspended in cold MACS buffer (Miltenyi) to a final density of ∼3×106 cells/ml. To generate tetramerized ECD-Fc ligands to test for binding to cells expressing full-length proteins, FPLC-purified biotinylated ECD-Fc proteins (see above) were incubated with streptavidin tetramers conjugated to Alexa Fluor 647 (SA-647) (Thermo Fisher Scientific) at a 4:1 molar ratio on ice for at least 15 minutes. To assess cell-surface expression of full-length proteins, 1:100 mouse anti-His-647 antibody (R&D Systems) staining of cells was also performed in parallel. Approximately 150,000 cells were incubated with ECD-Fc:SA-647 complexes or antibody in a final volume of 100 μL in 96-well round-bottom plates (Corning) for 1 hour at 4°C protected from light. Two-fold dilutions series of ECD-Fc:SA-647 complexes were performed. Depending on the experiment, final ECD-Fc:SA-647 complex concentrations (when incubated with cells) ranged from 31.25 nM to 4000 nM (PVR-Fc:SA-647, KIR2DL5-Fc:SA-647, TrkA-Fc:SA-647: 31-4000 nM; leptin-Fc:SA-647: 16-2000 nM). For staining of full-length KIR2DL5-, KIR2DL4- and KIR2DL1-transfected cells, 250 nM PVR-Fc:SA-647 was used. Mock transfected cells were used as a negative control and TCR/OTII-Fc protein was used as a negative control ligand for staining. Following incubation, cells were washed two times with 200 μL cold MACS buffer and resuspended in 120 μL cold MACS buffer with 1:200 propidium iodide (Thermo Fisher Scientific). Immunofluorescence staining was analyzed using a Cytoflex (Beckman Coulter) and data were collected for 20,000 cells. Data were analyzed using FlowJo v10.4.2 software. All data report median fluorescence intensity (MFI). Concentration-dependent binding of ECD-Fc:SA-647 to full-length receptor-expressing, but not mock control cells, was deemed indicative of cell-surface binding.
SPR experiments
SPR experiments were performed using a Biacore T200 instrument (GE Healthcare). Biotinylated ECD-Fc proteins (ligands) were sequentially purified using nickel-NTA and FPLC (see above) and captured on a Streptavidin (SA) Series S Sensor Chip (GE Healthcare). Chip capture was performed in HBS-P+ buffer (GE Healthcare) aiming for ∼300-700 ligand response units (RU). Flow cell 1 was left empty to use as a reference flow cell for on-line subtraction of bulk solution refractive index and for evaluation of non-specific binding of analyte to the chip surface using Biacore T200 Control Software (version 3.2) (GE Healthcare). In most cases, test ligands were captured in flow cells 2 and 3. A ligand for which binding was not expected was captured in flow cell 4 to serve as a negative control. Except where noted, FPLC-purified unbiotinylated ECD-Fc protein was used as analyte. Analytes were run in HBS-P+ buffer using 2-fold increasing protein concentrations to generate a series of sensorgrams. Since ECD-Fc analyte proteins were presented in a bivalent form (due to dimerization by the Fc-domain), sensorgrams followed a bivalent binding model which is not suitable for deriving a single binding constant for straightforward numerical quantification of the binding interaction. Nonetheless, these sensorgrams could be used to derive a qualitative assessment of binding since they represent an increasing response profile with increasing analyte concentrations, and below threshold response to the negative control ligand. The validation of interactions using bivalent analyte proteins allowed us to verify interactions that, in their monovalent form, may exhibit extremely fast kinetics and off-rates which would have been difficult, if not impossible, to capture without this avidity enhancement. Commercially-available monomer analytes used for SPR included recombinant mouse and human leptin (R&D Systems), recombinant human β-NGF (R&D Systems) and recombinant human angiopoietin-1 (R&D Systems). SPR conditions for each analyte-ligand pair tested including ligand RUs, analyte RUs, concentration range of 2-fold analyte dilutions, injection rate, injection and dissociation times and regeneration conditions are provided in Data S5.
Quantification and Statistical Analysis
Alkaline phosphatase enzymatic activity
Data from standard ECIA and apECIA in duplicate and triplicate reaction wells, respectively, are reported as mean of background subtracted values ± SD or as mean of fold-over-background ± SD (indicated in legends), where background was measured by testing prey binding to a negative control bait(s), mCherry-Fc and/or mock conditioned media (Figures 1C, 4C, 5C, 5G, 6E, and 6G; Data S4). Prey levels in conditioned media were quantitated from duplicate reaction wells against a standard curve of purified alkaline phosphatase and mean values from duplicate reaction wells are reported (Figures S1B, S1C, and S4F; Data S3).
Cell binding assay
Data were collected for 20,000 cells in duplicate reactions and analyzed using FlowJo v10.4.2 software. Values are reported as median fluorescence intensity (MFI) (Figures 5 and 6).
Statistical analysis of number of PPIs
Statistical analysis of the expected versus observed PPIs (Figure 1F) was performed using the Kolmogorov–Smirnov (K-S) test, a non-parametric and distribution-free test with no assumption of data distribution.
Acknowledgments
The authors thank Sean Parker and the Parker Foundation for a generous gift, the Howard Hughes Medical Institute (K.C.G.), the G. Harold and Leila Y. Mathers Charitable Foundation (K.C.G.), the National Institutes of Health (NIH R37 NS28182) (K.Z.), and the Caltech Beckman Institute (Caltech Protein Expression Center, J.V.) for support of this work. We thank Daisuke Hattori for help with data and statistical analyses.
Author Contributions
Conceptualization, K.C.G., W.M.W., J.V., R.A.F., D.H.S., and K.Z.; Methodology, W.M.W., J.V., R.A.F., D.H.S., C.L.E., and P.W.A.; Software, J.V.; Validation, W.M.W., J.V., R.A.F., D.H.S., and C.L.E.; Formal Analysis, W.M.W., J.V., R.A.F., and D.H.S.; Investigation, W.M.W., J.V., R.A.F., D.H.S., C.L.E., G.B.C., S.C., H.K., P.W.A., S.M.R., J.J.M., M.L.B., J.V., and A.W.L.; Resources, J.V., S.A.L., S.C., H.K., and P.W.A.; Data Curation, D.H.S. and K.Z.; Writing – Original Draft, W.M.W. and J.V.; Writing – Review & Editing, K.C.G., W.M.W., J.V., R.A.F., D.H.S., C.L.E., and K.Z.; Visualization, W.M.W., J.V., R.A.F., D.H.S., and C.L.E.; Supervision, K.C.G., W.M.W., J.V., K.Z., S.A.L., S.C., P.W.A., and S.M.R.; Project Administration, K.C.G., W.M.W., J.V., S.A.L., and S.C.; Funding Acquisition, K.C.G.
Declaration of Interests
The authors declare no competing interests.
Published: August 20, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2020.07.025.
Contributor Information
Woj M. Wojtowicz, Email: woj@stanford.edu.
K. Christopher Garcia, Email: kcgarcia@stanford.edu.
Supplemental Information
Information for all 564 proteins in screen including gene name, UniProt entry name, aliases, full-length protein sequence, ECD boundaries and sequence, superfamily, type (secreted, STM, multi-pass TM, and GPI-anchored), and predicted molecular weight of ECD-Fc and ECD-5AP proteins.
Complete sequences for all ECD-Fc bait and ECD-5AP prey in pD649 expression plasmids, and full-length gene sequences in pTT3 expression plasmids.
Qualitative assessment of ECD-Fc bait and ECD-5AP prey levels in conditioned media by western blot using a blue color scale. Values for AP quantitation of ECD-5AP prey conditioned media (relative AP activity; ng/μl).
Raw O.D. 650 nm data for all PPIs observed in screen, notation regarding whether PPI was observed in both orientations, and PPI status regarding whether or not the PPI was previously known. Following PubMed and PPI database searches, PPI status is designated as ‘previously reported’ or ‘not found in literature or PPI databases’. ‘Previously reported’ PPIs include PPIs reported in mouse, rat and zebrafish.
Table of SPR conditions for all ligand-analyte pairs tested including ligand RU, maximum analyte concentration, analyte RU at maximum concentration, number of analyte concentrations tested, injection time (seconds), injection rate (μl/minute), dissociation time (seconds), and regeneration conditions.
References
- Abudureyimu S., Asai N., Enomoto A., Weng L., Kobayashi H., Wang X., Chen C., Mii S., Takahashi M. Essential Role of Linx/Islr2 in the Development of the Forebrain Anterior Commissure. Sci. Rep. 2018;8:7292. doi: 10.1038/s41598-018-24064-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alavi M., Song M., King G.L.A., Gillis T., Propst R., Lamanuzzi M., Bousum A., Miller A., Allen R., Kidd T. Dscam1 Forms a Complex with Robo1 and the N-Terminal Fragment of Slit to Promote the Growth of Longitudinal Axons. PLoS Biol. 2016;14:e1002560. doi: 10.1371/journal.pbio.1002560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almagro Armenteros J.J., Tsirigos K.D., Sønderby C.K., Petersen T.N., Winther O., Brunak S., von Heijne G., Nielsen H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019;37:420–423. doi: 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- Amatu A., Sartore-Bianchi A., Bencardino K., Pizzutilo E.G., Tosi F., Siena S. Tropomyosin receptor kinase (TRK) biology and the role of NTRK gene fusions in cancer. Ann. Oncol. 2019;30:viii5–viii15. doi: 10.1093/annonc/mdz383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwal J.K., Pinkston-Gosse J., Syken J., Stawicki S., Wu Y., Shatz C., Tessier-Lavigne M. PirB is a functional receptor for myelin inhibitors of axonal regeneration. Science. 2008;322:967–970. doi: 10.1126/science.1161151. [DOI] [PubMed] [Google Scholar]
- Bardhan K., Anagnostou T., Boussiotis V.A. The PD1:PD-L1/2 Pathway from Discovery to Clinical Implementation. Front. Immunol. 2016;7:550. doi: 10.3389/fimmu.2016.00550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrow A.D., Edeling M.A., Trifonov V., Luo J., Goyal P., Bohl B., Bando J.K., Kim A.H., Walker J., Andahazy M. Natural Killer Cells Control Tumor Growth by Sensing a Growth Factor. Cell. 2018;172:534–548. doi: 10.1016/j.cell.2017.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boraschi D., Italiani P., Weil S., Martin M.U. The family of the interleukin-1 receptors. Immunol. Rev. 2018;281:197–232. doi: 10.1111/imr.12606. [DOI] [PubMed] [Google Scholar]
- Bülow S., Zeller L., Werner M., Toelge M., Holzinger J., Entzian C., Schubert T., Waldow F., Gisch N., Hammerschmidt S., Gessner A. Bactericidal/Permeability-Increasing Protein Is an Enhancer of Bacterial Lipoprotein Recognition. Front. Immunol. 2018;9:2768. doi: 10.3389/fimmu.2018.02768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burshtyn D.N., Morcos C. The Expanding Spectrum of Ligands for Leukocyte Ig-like Receptors. J. Immunol. 2016;196:947–955. doi: 10.4049/jimmunol.1501937. [DOI] [PubMed] [Google Scholar]
- Bushell K.M., Söllner C., Schuster-Boeckler B., Bateman A., Wright G.J. Large-scale screening for novel low-affinity extracellular protein interactions. Genome Res. 2008;18:622–630. doi: 10.1101/gr.7187808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X., Veverka V., Radhakrishnan A., Waters L.C., Muskett F.W., Morgan S.H., Huo J., Yu C., Evans E.J., Leslie A.J. Structure and interactions of the human programmed cell death 1 receptor. J. Biol. Chem. 2013;288:11771–11785. doi: 10.1074/jbc.M112.448126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y., Nam J., Whitcomb D.J., Song Y.S., Kim D., Jeon S., Um J.W., Lee S.-G., Woo J., Kwon S.-K. SALM5 trans-synaptically interacts with LAR-RPTPs in a splicing-dependent manner to regulate synapse development. Sci. Rep. 2016;6:26676. doi: 10.1038/srep26676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosnier C., Bustamante L.Y., Bartholdson S.J., Bei A.K., Theron M., Uchikawa M., Mboup S., Ndir O., Kwiatkowski D.P., Duraisingh M.T. Basigin is a receptor essential for erythrocyte invasion by Plasmodium falciparum. Nature. 2011;480:534–537. doi: 10.1038/nature10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dakouane-Giudicelli M., Alfaidy N., de Mazancourt P. Netrins and their roles in placental angiogenesis. BioMed Res. Int. 2014;2014:901941. doi: 10.1155/2014/901941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S., Aldrich T.H., Jones P.F., Acheson A., Compton D.L., Jain V., Ryan T.E., Bruno J., Radziejewski C., Maisonpierre P.C., Yancopoulos G.D. Isolation of angiopoietin-1, a ligand for the TIE2 receptor, by secretion-trap expression cloning. Cell. 1996;87:1161–1169. doi: 10.1016/s0092-8674(00)81812-7. [DOI] [PubMed] [Google Scholar]
- Dragovich M.A., Mor A. The SLAM family receptors: Potential therapeutic targets for inflammatory and autoimmune diseases. Autoimmun. Rev. 2018;17:674–682. doi: 10.1016/j.autrev.2018.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklund L., Kangas J., Saharinen P. Angiopoietin-Tie signalling in the cardiovascular and lymphatic systems. Clin. Sci. (Lond.) 2017;131:87–103. doi: 10.1042/CS20160129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finci L., Zhang Y., Meijers R., Wang J.-H. Signaling mechanism of the netrin-1 receptor DCC in axon guidance. Prog. Biophys. Mol. Biol. 2015;118:153–160. doi: 10.1016/j.pbiomolbio.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca A.L., da Silva V.L., da Fonsêca M.M., Meira I.T.J., da Silva T.E., Kroll J.E., Ribeiro-Dos-Santos A.M., Freitas C.R., Furtado R., de Souza J.E. Bioinformatics Analysis of the Human Surfaceome Reveals New Targets for a Variety of Tumor Types. Int. J. Genomics. 2016;2016:8346198. doi: 10.1155/2016/8346198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J. The long road to leptin. J. Clin. Invest. 2016;126:4727–4734. doi: 10.1172/JCI91578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahmberg C.G., Ning L., Paetau S. ICAM-5: A Neuronal Dendritic Adhesion Molecule Involved in Immune and Neuronal Functions. In: Berezin V., Walmod P.S., editors. Cell Adhesion Molecules: Implications in Neurological Diseases. Springer; New York, NY: 2014. pp. 117–132. [DOI] [PubMed] [Google Scholar]
- Ghiotto M., Gauthier L., Serriari N., Pastor S., Truneh A., Nunès J.A., Olive D. PD-L1 and PD-L2 differ in their molecular mechanisms of interaction with PD-1. Int. Immunol. 2010;22:651–660. doi: 10.1093/intimm/dxq049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschneider D., Mehlen P. Dependence receptors: a new paradigm in cell signaling and cancer therapy. Oncogene. 2010;29:1865–1882. doi: 10.1038/onc.2010.13. [DOI] [PubMed] [Google Scholar]
- Goto-Ito S., Yamagata A., Sato Y., Uemura T., Shiroshima T., Maeda A., Imai A., Mori H., Yoshida T., Fukai S. Structural basis of trans-synaptic interactions between PTPδ and SALMs for inducing synapse formation. Nat. Commun. 2018;9:269. doi: 10.1038/s41467-017-02417-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao L., Shan Q., Wei J., Ma F., Sun P. Lactoferrin: Major Physiological Functions and Applications. Curr. Protein Pept. Sci. 2019;20:139–144. doi: 10.2174/1389203719666180514150921. [DOI] [PubMed] [Google Scholar]
- Hirayasu K., Arase H. Functional and genetic diversity of leukocyte immunoglobulin-like receptor and implication for disease associations. J. Hum. Genet. 2015;60:703–708. doi: 10.1038/jhg.2015.64. [DOI] [PubMed] [Google Scholar]
- Holler N., Kataoka T., Bodmer J.L., Romero P., Romero J., Deperthes D., Engel J., Tschopp J., Schneider P. Development of improved soluble inhibitors of FasL and CD40L based on oligomerized receptors. J. Immunol. Methods. 2000;237:159–173. doi: 10.1016/s0022-1759(99)00239-2. [DOI] [PubMed] [Google Scholar]
- Honig B., Shapiro L. Adhesion Protein Structure, Molecular Affinities, and Principles of Cell-Cell Recognition. Cell. 2020;181:520–535. doi: 10.1016/j.cell.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard S.R., Guasti L., Ruiz-Babot G., Mancini A., David A., Storr H.L., Metherell L.A., Sternberg M.J., Cabrera C.P., Warren H.R. IGSF10 mutations dysregulate gonadotropin-releasing hormone neuronal migration resulting in delayed puberty. EMBO Mol. Med. 2016;8:626–642. doi: 10.15252/emmm.201606250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain B., Ramani S.R., Chiang E., Lehoux I., Paduchuri S., Arena T.A., Patel A., Wilson B., Chan P., Franke Y. A Platform for Extracellular Interactome Discovery Identifies Novel Functional Binding Partners for the Immune Receptors B7-H3/CD276 and PVR/CD155. Mol. Cell. Proteomics. 2019;18:2310–2323. doi: 10.1074/mcp.TIR119.001433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin E.L., Ting L., Bruckner R.J., Gebreab F., Gygi M.P., Szpyt J., Tam S., Zarraga G., Colby G., Baltier K. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell. 2015;162:425–440. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin E.L., Bruckner R.J., Paulo J.A., Cannon J.R., Ting L., Baltier K., Colby G., Gebreab F., Gygi M.P., Parzen H. Architecture of the human interactome defines protein communities and disease networks. Nature. 2017;545:505–509. doi: 10.1038/nature22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2019;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Kuraku S., Zmasek C.M., Nishimura O., Katoh K. aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res. 2013;41:W22–W28. doi: 10.1093/nar/gkt389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-R. Protein tyrosine phosphatase PTPRT as a regulator of synaptic formation and neuronal development. BMB Rep. 2015;48:249–255. doi: 10.5483/BMBRep.2015.48.5.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-R., Bechstein D.J.B., Ooi C.C., Patel A., Gaster R.S., Ng E., Gonzalez L.C., Wang S.X. Magneto-nanosensor platform for probing low-affinity protein-protein interactions and identification of a low-affinity PD-L1/PD-L2 interaction. Nat. Commun. 2016;7:12220. doi: 10.1038/ncomms12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47(W1):W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhang P., Choi T.-Y., Park S.K., Park H., Lee E.-J., Lee D., Roh J.D., Mah W., Kim R. Splicing-Dependent Trans-synaptic SALM3-LAR-RPTP Interactions Regulate Excitatory Synapse Development and Locomotion. Cell Rep. 2015;12:1618–1630. doi: 10.1016/j.celrep.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Watson A., Olechwier A., Anaya M., Sorooshyari S.K., Harnett D.P., Lee H.P., Vielmetter J., Fares M.A., Garcia K.C. Deconstruction of the beaten Path-Sidestep interaction network provides insights into neuromuscular system development. eLife. 2017;6:e28111. doi: 10.7554/eLife.28111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lie E., Li Y., Kim R., Kim E. SALM/Lrfn Family Synaptic Adhesion Molecules. Front. Mol. Neurosci. 2018;11:105. doi: 10.3389/fnmol.2018.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly A., Nikolaev A., Suresh G., Zheng Y., Tessier-Lavigne M., Stein E. DSCAM is a netrin receptor that collaborates with DCC in mediating turning responses to netrin-1. Cell. 2008;133:1241–1254. doi: 10.1016/j.cell.2008.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macauley M.S., Crocker P.R., Paulson J.C. Siglec-mediated regulation of immune cell function in disease. Nat. Rev. Immunol. 2014;14:653–666. doi: 10.1038/nri3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandai K., Reimert D.V., Ginty D.D. Linx mediates interaxonal interactions and formation of the internal capsule. Neuron. 2014;83:93–103. doi: 10.1016/j.neuron.2014.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S., Söllner C., Charoensawan V., Adryan B., Thisse B., Thisse C., Teichmann S., Wright G.J. Construction of a large extracellular protein interaction network and its resolution by spatiotemporal expression profiling. Mol. Cell. Proteomics. 2010;9:2654–2665. doi: 10.1074/mcp.M110.004119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGown E.L., Hafeman D.G. Multichannel pipettor performance verified by measuring pathlength of reagent dispensed into a microplate. Anal. Biochem. 1998;258:155–157. doi: 10.1006/abio.1998.2621. [DOI] [PubMed] [Google Scholar]
- Minnone G., De Benedetti F., Bracci-Laudiero L. NGF and Its Receptors in the Regulation of Inflammatory Response. Int. J. Mol. Sci. 2017;18:1028. doi: 10.3390/ijms18051028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monestier O., Blanquet V. WFIKKN1 and WFIKKN2: “Companion” proteins regulating TGFB activity. Cytokine Growth Factor Rev. 2016;32:75–84. doi: 10.1016/j.cytogfr.2016.06.003. [DOI] [PubMed] [Google Scholar]
- Monje M. Myelin Plasticity and Nervous System Function. Annu. Rev. Neurosci. 2018;41:61–76. doi: 10.1146/annurev-neuro-080317-061853. [DOI] [PubMed] [Google Scholar]
- Moore T., Dveksler G.S. Pregnancy-specific glycoproteins: complex gene families regulating maternal-fetal interactions. Int. J. Dev. Biol. 2014;58:273–280. doi: 10.1387/ijdb.130329gd. [DOI] [PubMed] [Google Scholar]
- Nam J., Mah W., Kim E. The SALM/Lrfn family of leucine-rich repeat-containing cell adhesion molecules. Semin. Cell Dev. Biol. 2011;22:492–498. doi: 10.1016/j.semcdb.2011.06.005. [DOI] [PubMed] [Google Scholar]
- Orchard S., Ammari M., Aranda B., Breuza L., Briganti L., Broackes-Carter F., Campbell N.H., Chavali G., Chen C., del-Toro N. The MIntAct project--IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014;42:D358–D363. doi: 10.1093/nar/gkt1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oughtred R., Stark C., Breitkreutz B.-J., Rust J., Boucher L., Chang C., Kolas N., O’Donnell L., Leung G., McAdam R. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 2019;47(D1):D529–D541. doi: 10.1093/nar/gky1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özkan E., Carrillo R.A., Eastman C.L., Weiszmann R., Waghray D., Johnson K.G., Zinn K., Celniker S.E., Garcia K.C. An extracellular interactome of immunoglobulin and LRR proteins reveals receptor-ligand networks. Cell. 2013;154:228–239. doi: 10.1016/j.cell.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panza P., Sitko A.A., Maischein H.-M., Koch I., Flötenmeyer M., Wright G.J., Mandai K., Mason C.A., Söllner C. The LRR receptor Islr2 is required for retinal axon routing at the vertebrate optic chiasm. Neural Dev. 2015;10:23. doi: 10.1186/s13064-015-0050-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pende D., Falco M., Vitale M., Cantoni C., Vitale C., Munari E., Bertaina A., Moretta F., Del Zotto G., Pietra G. Killer Ig-Like Receptors (KIRs): Their Role in NK Cell Modulation and Developments Leading to Their Clinical Exploitation. Front. Immunol. 2019;10:1179. doi: 10.3389/fimmu.2019.01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierleoni A., Martelli P.L., Casadio R. PredGPI: a GPI-anchor predictor. BMC Bioinformatics. 2008;9:392. doi: 10.1186/1471-2105-9-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranaivoson F.M., Turk L.S., Ozgul S., Kakehi S., von Daake S., Lopez N., Trobiani L., De Jaco A., Denissova N., Demeler B. A Proteomic Screen of Neuronal Cell-Surface Molecules Reveals IgLONs as Structurally Conserved Interaction Modules at the Synapse. Structure. 2019;27:893–906. doi: 10.1016/j.str.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D.A., Reith W., Trowsdale J. Regulation of Immunity by Butyrophilins. Annu. Rev. Immunol. 2016;34:151–172. doi: 10.1146/annurev-immunol-041015-055435. [DOI] [PubMed] [Google Scholar]
- Rue S.M., Anderson P.W., Gaylord M.R., Miller J.J., Glaser S.M., Lesley S.A. A High-Throughput System for Transient and Stable Protein Production in Mammalian Cells. Methods Mol. Biol. 2019;2025:93–142. doi: 10.1007/978-1-4939-9624-7_5. [DOI] [PubMed] [Google Scholar]
- Salbaum J.M. Punc, a novel mouse gene of the immunoglobulin superfamily, is expressed predominantly in the developing nervous system. Mech. Dev. 1998;71:201–204. doi: 10.1016/s0925-4773(98)00005-7. [DOI] [PubMed] [Google Scholar]
- Sanes J.R., Zipursky S.L. Synaptic Specificity, Recognition Molecules, and Assembly of Neural Circuits. Cell. 2020;181:536–556. doi: 10.1016/j.cell.2020.04.008. [DOI] [PubMed] [Google Scholar]
- Schaper F., Rose-John S. Interleukin-6: Biology, signaling and strategies of blockade. Cytokine Growth Factor Rev. 2015;26:475–487. doi: 10.1016/j.cytogfr.2015.07.004. [DOI] [PubMed] [Google Scholar]
- Schievenbusch S., Sauer E., Curth H.-M., Schulte S., Demir M., Toex U., Goeser T., Nierhoff D. Neighbor of Punc E 11: expression pattern of the new hepatic stem/progenitor cell marker during murine liver development. Stem Cells Dev. 2012;21:2656–2666. doi: 10.1089/scd.2011.0579. [DOI] [PubMed] [Google Scholar]
- Söllner C., Wright G.J. A cell surface interaction network of neural leucine-rich repeat receptors. Genome Biol. 2009;10:R99. doi: 10.1186/gb-2009-10-9-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoker A.W. Protein tyrosine phosphatases and signalling. J. Endocrinol. 2005;185:19–33. doi: 10.1677/joe.1.06069. [DOI] [PubMed] [Google Scholar]
- Sun Y., Gallagher-Jones M., Barker C., Wright G.J. A benchmarked protein microarray-based platform for the identification of novel low-affinity extracellular protein interactions. Anal. Biochem. 2012;424:45–53. doi: 10.1016/j.ab.2012.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K.F., Kiyoshima T., Kobayashi I., Xie M., Yamaza H., Fujiwara H., Ookuma Y., Nagata K., Wada H., Sakai T. Protogenin, a new member of the immunoglobulin superfamily, is implicated in the development of the mouse lower first molar. BMC Dev. Biol. 2010;10:115. doi: 10.1186/1471-213X-10-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner L., Lavstsen T., Berger S.S., Wang C.W., Petersen J.E.V., Avril M., Brazier A.J., Freeth J., Jespersen J.S., Nielsen M.A. Severe malaria is associated with parasite binding to endothelial protein C receptor. Nature. 2013;498:502–505. doi: 10.1038/nature12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- UniProt Consortium UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47(D1):D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Merwe P.A., Barclay A.N. Transient intercellular adhesion: the importance of weak protein-protein interactions. Trends Biochem. Sci. 1994;19:354–358. doi: 10.1016/0968-0004(94)90109-0. [DOI] [PubMed] [Google Scholar]
- Verkerke H., Naylor C., Zabeau L., Tavernier J., Petri W.A., Jr., Marie C. Kinetics of leptin binding to the Q223R leptin receptor. PLoS ONE. 2014;9:e94843. doi: 10.1371/journal.pone.0094843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser J.J., Cheng Y., Perry S.C., Chastain A.B., Parsa B., Masri S.S., Ray T.A., Kay J.N., Wojtowicz W.M. An extracellular biochemical screen reveals that FLRTs and Unc5s mediate neuronal subtype recognition in the retina. eLife. 2015;4:e08149. doi: 10.7554/eLife.08149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi-Nakao K., Hatakeyama K., Ohshima K., Ken Yamaguchi K., Mochizuki T. Carcinoembryonic antigen-related cell adhesion molecule 4 (CEACAM4) is specifically expressed in medullary thyroid carcinoma cells. Biomed. Res. 2014;35:237–242. doi: 10.2220/biomedres.35.237. [DOI] [PubMed] [Google Scholar]
- Wilson N.H., Key B. Neogenin: one receptor, many functions. Int. J. Biochem. Cell Biol. 2007;39:874–878. doi: 10.1016/j.biocel.2006.10.023. [DOI] [PubMed] [Google Scholar]
- Wojtowicz W.M., Flanagan J.J., Millard S.S., Zipursky S.L., Clemens J.C. Alternative splicing of Drosophila Dscam generates axon guidance receptors that exhibit isoform-specific homophilic binding. Cell. 2004;118:619–633. doi: 10.1016/j.cell.2004.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtowicz W.M., Wu W., Andre I., Qian B., Baker D., Zipursky S.L. A vast repertoire of Dscam binding specificities arises from modular interactions of variable Ig domains. Cell. 2007;130:1134–1145. doi: 10.1016/j.cell.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood L., Wright G.J. Approaches to identify extracellular receptor-ligand interactions. Curr. Opin. Struct. Biol. 2019;56:28–36. doi: 10.1016/j.sbi.2018.10.002. [DOI] [PubMed] [Google Scholar]
- Yates B., Braschi B., Gray K.A., Seal R.L., Tweedie S., Bruford E.A. Genenames.org: the HGNC and VGNC resources in 2017. Nucleic Acids Res. 2017;45(D1):D619–D625. doi: 10.1093/nar/gkw1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T., Yasumura M., Uemura T., Lee S.-J., Ra M., Taguchi R., Iwakura Y., Mishina M. IL-1 receptor accessory protein-like 1 associated with mental retardation and autism mediates synapse formation by trans-synaptic interaction with protein tyrosine phosphatase δ. J. Neurosci. 2011;31:13485–13499. doi: 10.1523/JNEUROSCI.2136-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T., Shiroshima T., Lee S.-J., Yasumura M., Uemura T., Chen X., Iwakura Y., Mishina M. Interleukin-1 receptor accessory protein organizes neuronal synaptogenesis as a cell adhesion molecule. J. Neurosci. 2012;32:2588–2600. doi: 10.1523/JNEUROSCI.4637-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinn K., Özkan E. Neural immunoglobulin superfamily interaction networks. Curr. Opin. Neurobiol. 2017;45:99–105. doi: 10.1016/j.conb.2017.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Information for all 564 proteins in screen including gene name, UniProt entry name, aliases, full-length protein sequence, ECD boundaries and sequence, superfamily, type (secreted, STM, multi-pass TM, and GPI-anchored), and predicted molecular weight of ECD-Fc and ECD-5AP proteins.
Complete sequences for all ECD-Fc bait and ECD-5AP prey in pD649 expression plasmids, and full-length gene sequences in pTT3 expression plasmids.
Qualitative assessment of ECD-Fc bait and ECD-5AP prey levels in conditioned media by western blot using a blue color scale. Values for AP quantitation of ECD-5AP prey conditioned media (relative AP activity; ng/μl).
Raw O.D. 650 nm data for all PPIs observed in screen, notation regarding whether PPI was observed in both orientations, and PPI status regarding whether or not the PPI was previously known. Following PubMed and PPI database searches, PPI status is designated as ‘previously reported’ or ‘not found in literature or PPI databases’. ‘Previously reported’ PPIs include PPIs reported in mouse, rat and zebrafish.
Table of SPR conditions for all ligand-analyte pairs tested including ligand RU, maximum analyte concentration, analyte RU at maximum concentration, number of analyte concentrations tested, injection time (seconds), injection rate (μl/minute), dissociation time (seconds), and regeneration conditions.
Data Availability Statement
Information for all 564 proteins in screen including gene name, UniProt entry name, aliases, full-length protein sequence, ECD boundaries and sequence, superfamily, type (secreted, STM, multi-pass TM, and GPI-anchored), and predicted molecular weight of ECD-Fc and ECD-5AP proteins is included in Data S1. Full plasmid sequences for all bait and prey constructs are included in Data S2. Data for qualitative assessment of ECD-Fc and ECD-5AP levels in conditioned media by western blot and AP quantitation of ECD-5AP conditioned media (relative AP activity; ng/μl) are included in Data S3. Screen data and multiple sequence alignement (MSA) files have been deposited to Dryad (https://doi.org/10.5061/dryad.xsj3tx9bd) and are included in Data S4. SPR conditions for all ligand-analyte pairs tested including ligand RU, maximum analyte concentration, analyte RU at maximum concentration, number of analyte concentrations tested, injection time (seconds), injection rate (μl/minute), dissociation time (seconds), and regeneration conditions are included in Data S5.