Abstract
Purpose
Retinoblastoma is a malignant tumor of the developing retina that mostly occurs in children. Our study aimed to investigate the effect of tripartite motif-containing protein 59 (TRIM59) on retinoblastoma growth and the underlying mechanisms.
Methods
We performed bioinformatic analysis of three datasets (GSE24673, GSE97508, and GSE110811) from the Gene Expression Omnibus database. Quantitative reverse-transcription PCR and immunoblotting of three retinoblastoma cell lines were conducted to verify TRIM59 as a differentially expressed gene. Specific siRNAs were used to inhibit TRIM59 expression in the HXO-Rb44 cell line. A lentiviral vector was transfected into the Y79 cell line to overexpress TRIM59. The effects of TRIM59 on retinoblastoma cell proliferation, cell cycling, and apoptosis were explored in vitro using the abovementioned cell lines. The effect of TRIM59 expression on retinoblastoma cell proliferation was evaluated in a mouse xenograft tumor model.
Results
TRIM59 expression in three retinoblastoma cell lines was remarkably elevated compared with normal control. Knocking down TRIM59 expression remarkably suppressed cell proliferation and growth and promoted cell apoptosis in HXO-Rb44 cells, whereas TRIM59 overexpression promoted tumor progression in Y79 cells. Silencing TRIM59 also markedly inhibited in vivo tumor growth in the xenograft model. Mechanistic studies revealed that TRIM59 upregulated phosphorylated p38, p-JNK1/2, p-ERK1/2, and p-c-JUN expression in retinoblastoma cells. Notably, the p38 inhibitor SB203580 attenuated the effects of TRIM59 on cell proliferation, apoptosis, and the G1/S phase transition.
Conclusions
TRIM59 plays an oncogenic role in retinoblastoma and exerts its tumor-promotive function by activating the p38–mitogen-activated protein kinase pathway.
Keywords: Retinoblastoma, TRIM59, p38–MAPK signaling pathway, biomarker
Retinoblastoma is an aggressive intraocular tumor that occurs in infancy or childhood, affecting one in every 18,000 newborns worldwide and accounting for 3% of all childhood cancers.1,2 The incidence of retinoblastoma is constant worldwide, at one case per 15,000 to 20,000 livebirths, which corresponds to about 9000 new cases annually.3 Tumor invasion is initiated from the retina to the sclera and postlaminar optic nerve and metastasizes to the central nervous system and distant organs, as well as the bones and bone marrow.4,5 The median time to diagnosis, the interval from symptom onset to diagnosis, ranges from 3 to 5 months and delayed diagnosis has been related to detrimental results on retinoblastoma sequelae, particularly with regard to increasing extraocular extension and mortality rates.1 Investigation of novel active proteins or signaling cascades involved in intraocular retinoblastoma might contribute to early diagnosis.
The tripartite motif (TRIM) protein family consists of nearly 80 members that are characterized by the presence of an N-terminal region, which contains a RING domain, B-box motifs, and a coiled-coil region.6 Updated evidence has revealed that TRIM family proteins are critical mediators involved in important biological activities, such as cell proliferation, cell-cycle progression, apoptosis, and natural viral immunity.7–9 Tripartite motif-containing protein 59 (TRIM59), mainly localized in the cytoplasm, is implicated in several tumors. The expression of TRIM59 is upregulated in gastric tumors and might promote tumor growth by accelerating p53 ubiquitination and degradation.10 In addition, TRIM59 can facilitate cell proliferation and migration in non-small-cell lung cancer and osteosarcoma.11,12 Knocking down TRIM59 expression can significantly decrease cellular growth and metastasis in cervical cancer.13 These reports indicate that TRIM59 exerts crucial roles in human cancers. Nonetheless, the relationship between TRIM59 and retinoblastoma remains unclear. In the present study, we aimed to explore TRIM59 expression in retinoblastoma and identify the function of TRIM59 in the progression of retinoblastoma.
Methods
Data Sources
Three microarray, gene expression datasets (GSE24673,14 GSE97508,15 and GSE11081116) were retrieved from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). Among these datasets, GSE24673 was based on the GPL6244 platform ([HuGene-1_0-st] Affymetrix Human Gene 1.0 ST Array); GSE97508 was based on the GPL15207 platform ([PrimeView] Affymetrix Human Gene Expression Array), and GSE110811 was based on the GPL16686 platform ([HuGene-2_0-st] Affymetrix Human Gene 2.0 ST Array). Nine retinoblastoma samples (GSM607938–GSM607946) and two normal healthy adult retinas (GSM607947 and GSM607948) were obtained from GSE24673. Six retinoblastoma (GSM2570469– GSM2570474) and three control tissues (GSM2570466–GSM2570468) were obtained from GSE97508. A total of 28 retinoblastoma samples (GSM3017123–GSM3017150) and three control tissues (GSM3017151–GSM3017153) were obtained from GSE110811.
After careful review, all samples in the GSE24673 dataset were considered to be satisfactory; however, three retinoblastoma samples (GSM2570472, GSM2570473, and GSM2570474) were eliminated from GSE97508, and one normal sample (GSM3017153) was removed from GSE110811 due to low sample quality.
Bioinformatic Analysis
Original .CEL files were analyzed and underwent background correction, quality control, and data standardization. Affy packages17 were used for the GSE24673 and GSE97508 datasets, and Oligo packages18 were used for the GSE110811 dataset. Principal component analysis was analyzed using the built-in princomp function in R 3.6.3 (R Foundation for Statistical Computing, Vienna, Austria), and plots were generated using the scatterplot3d package.19 Differentially expressed genes (DEGs) were screened using the limma package20 in R. The cutoff criteria for statistical significance were |logFC| (an absolute log2 value in the fold change of the expression of the genes) >1 and P < 0.05. Screened DEGs were used to create a heatmap using the built-in pheatmap function and a Venn diagram using the VennDiagram package in R.
Cell Culture
The human retinoblastoma cell lines Weri-Rb1, Y79, and HXO-Rb44 and the normal retinal epithelial cell line ARPE-19 were purchased from ATCC (Rockville, MD, USA). All of the cells were cultured in Dulbecco's Modified Eagle Medium/Nutrient Mixture F-12 (DMEM/F-12; Thermo Fisher Scientific, Waltham, MA, USA) supplemented with penicillin G, streptomycin, and 10% fetal bovine serum (Thermo Fisher Scientific). The cells were incubated in a humidified atmosphere with 5% CO2 at 37°C.
Lentiviral Transduction
For downregulation of TRIM59 expression, three human siRNA sequences (RNAi1, GGAAGCTGTTCTCCAGTAT [small interfering RNA of TRIM59-1, siTRIM59-1]; RNAi2, GAAGAGTCTCCACTTAAAT [siTRIM59-2]; and RNAi3, GAATGGAGCAGAACAGAAA [siTRIM59-3]) synthesized by Genewiz Company (Shanghai, China) were cloned into the pLKO.1 plasmid (Addgene, Cambridge, MA, USA). The coding DNA sequence region of TRIM59 (NM_173084.2), which was synthesized by Genewiz Company, was inserted into the EcoR I/BamH I restriction sites of the pLVX-Puro vector. The sequence of the constructed core plasmid was confirmed by DNA sequencing (Majorbio, Shanghai, China). Subsequently, pLKO.1-Puro-sh TRIM59 and pLVX-Puro-TRIM59 were cotransfected with the viral packaging plasmids psPAX2 and pMD2.G (Addgene) into 293T cells with Lipofectamine 2000 Transfection Reagent (Thermo Fisher Scientific). Forty-eight hours after infection, the lentiviral particles in the supernatants were collected. The HXO-Rb44 cell line was infected with the siRNA viral supernatants, and the Y79 cell line was infected with the overexpression viral supernatants. TRIM59 expression was examined by reverse-transcription quantitative polymerase chain reaction (qRT-PCR). TRIM59-overexpressing Y79 cells were treated in the absence or presence of the p38 inhibitor SB203580 (Cell Signaling Technology, Danvers, MA, USA) for 24 hours in subsequent experiments.
qRT-PCR
Total RNA was extracted from cells using TRIzol reagent (Thermo Fisher Scientific) according to the manufacturer's protocol. The extracted RNA was quantitated by spectrophotometry at 260 nm using a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). First-strand cDNA was synthesized using the QuantiTect Reverse Transcription Kit (Qiagen, Hilden, Germany). Later, RT-PCR with cDNA as the template was performed on the ABI PRISM 7000 Sequence Detection System (Thermo Fisher Scientific) using the QuantiFast SYBR Green PCR Kit (Qiagen). The thermal cycling conditions were 95°C for 10 minutes followed by an initial denaturation step at 95°C for 15 seconds, 40 cycles at 60°C for 45 seconds, and 60°C for 30 seconds. The experiments were carried out in triplicate for each data point. The primers for TRIM59 and β-actin were as follows: TRIM59 forward, 5′-TTGTCACCTGCCCTGAAC-3′; TRIM59 reverse, 5′-TCCTTATCGCCTTGGATC-3′; β-actin forward, 5′-TGGCATCCACGAAACTAC-3′; and β-actin reverse, 5′-CTTGATCTTCATGGTGCTG-3′. Quantifications were normalized by using β-actin as an internal reference21 and calculated by using the 2ΔΔCt method.
Immunoblotting
Cell lysates were prepared from cell lines using a RIPA Lysis and Extraction Buffer kit (Thermo Fisher Scientific), and the procedure was performed as described previously.22 Primary antibodies against target proteins, such as TRIM59, Bcl-2, cleaved caspase-3 (Abcam, Cambridge, UK), cyclin D1, phosphorylated p38 (p-p38), p38, p-JNK1/2, JNK1/2, p-ERK1/2, ERK1/2, p-c-JUN, c-JUN, and β-actin (Cell Signaling Technology), were diluted between 1:500 and 1:2000, and secondary antibodies (Beyotime Biotechnology, Shanghai, China) were diluted 1:1000.
Cell Proliferation
A Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Rockville, MD, USA) assay was applied to assess the cell proliferation rate. Approximately 3 × 103 HXO-Rb44 or Y79 cells were seeded in a 96-well plate and transfected with siRNA negative control (siNC), siTRIM59, vector, overexpression (oe)TRIM59, vehicle + vector, vehicle + oeTRIM59, SB203580 + vector, or SB203580 + oeTRIM59. At 0, 12, 24, 48, and 72 hours after transfection, CCK-8 reagent (10 µL) was added to each well and incubated for 2 hours at 37°C. The absorbance at 450 nm was measured using an ELISA microplate reader (Perlong, Beijing, China).
Cell-Cycle Analysis
HXO-Rb44 and Y79 cells were harvested and washed with ice-cold PBS. After fixation with 70% ethanol overnight at 4°C, the cells were washed twice with PBS and then treated with 1 mg/mL RNase A in 500 mL of PBS at 37°C for 30 minutes. Next, the cells were stained with 50 µg/mL propidium iodide (PI; Thermo Fisher Scientific) in the dark for 30 minutes. Flow cytometric analysis was performed using a FACScan flow cytometer (BD Biosciences, San Jose, CA, USA), and the results were analyzed using FlowJo 7.6 (FlowJo LLC, San Diego, CA, USA).
Apoptosis Assays
Cellular apoptosis was evaluated using an Annexin V-FITC Apoptosis Detection Kit (Beyotime Biotechnology). Cells were harvested using trypsin/EDTA and washed with PBS. Binding buffer was subsequently added to resuspend the cells. Next, 5 µL of the annexin V-FITC and PI solution were added, and samples were incubated for 15 minutes at room temperature. Finally, stained cells were analyzed using a CytoFLEX Flow Cytometer (Beckman Coulter, Indianapolis, IN, USA). Data were analyzed using the FlowJo 7.6 software. Apoptotic cells were defined by positive annexin V staining and negative PI staining. Normal, living cells were not stained by either annexin V-FITC or PI (cells in the third quadrant); early apoptotic cells were only stained by annexin V-FITC and were stained negative by PI (cells in the second quadrant); late apoptotic cells were simultaneously stained by both annexin V-FITC and PI (cells in the first quadrant); and necrotic cells were stained only by PI (cells in the fourth quadrant).
In Vivo Tumorigenicity Assay
Twelve 6-week-old male BALB/c nude mice weighing 18 to 20 g were obtained from Shanghai Experimental Animal Center (Shanghai, China) and bred in a specific pathogen-free animal facility. The animal protocol adhered to the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research and was approved by the Ethics Committee of the Second Affiliated Hospital of Nanchang University. Prior to injection, the mice were randomly divided into siTRIM59 and siNC groups (n = 6 for each group). A total of 5 × 105 HXO-Rb44 cells were subcutaneously injected into the left upper flank region. Tumor length and width were measured every three days. Tumor size was calculated as follows: volume (m3) = (length × width2) × 0.5. All mice were sacrificed 33 days later, and the tumor nodules were removed and weighed, and immunohistochemistry was then performed. The expression of TRIM59, cyclin D1, Bcl-2, and cleaved caspase-3 was determined by immunoblotting.
Immunohistochemistry
Paraffin-embedded tumor samples were sectioned into 4- to 7-µm-thick sections. Subsequent immunohistochemistry was performed according to the protocol in a previous report.22 An anti-TRIM59 primary antibody (Abcam) was used at a 1:600 dilution, and a secondary antibody (Abcam) was used at a 1:400 dilution. The slides were then stained with 3,3′-diaminobenzidine and counterstained with hematoxylin (Baso Diagnostics, Inc., Zhuhai, China) following the manufacturer's protocol. Immunohistochemical images were acquired using a Eclipse Ni microscope (Nikon Instruments, Inc., Melville, NY, USA). The sizes of positive areas were calculated using Nikon NIS-Elements, version 5.11.00.
Statistical Analysis
SPSS Statistics 23.0 (IBM, Chicago, IL, USA) was used for statistical analyses. In vitro experiments were performed in triplicate, and in vivo tests were repeated six times. Data are presented as the mean ± SD. Differences among multiple groups were analyzed by one-way ANOVA with Tukey's post hoc test, and the means of two groups were compared by Student's t-test. Two-way, repeated-measures ANOVA was used to analyze CCK-8 assay results. P < 0.05 was considered to indicate a statistically significant difference.
Results
Analysis of the GEO Database
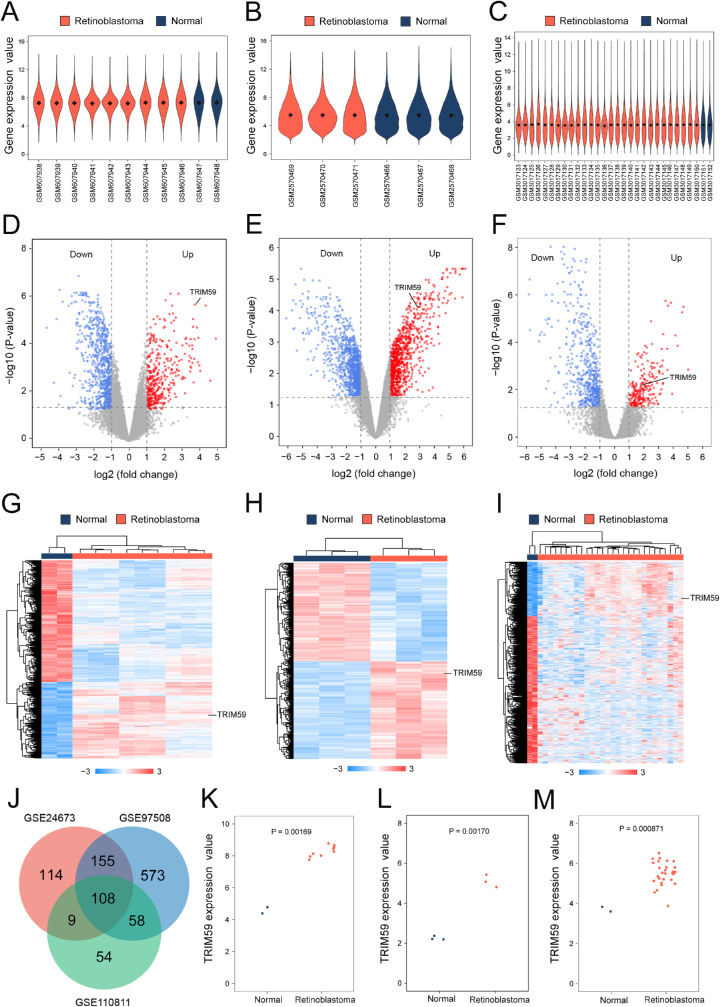
The mean values of gene expression for each sample in the GSE24673, GSE97508, and GSE110811 datasets were fundamentally the same after normalization, suggesting that the sample data sources were reliable and consistent for further downstream analyses (Figs. 1A–1C; Supplementary Fig. S1). After standardizing microarray results, 2758 DEGs were identified. Among these, 386 upregulated genes and 770 downregulated genes were screened in the GSE24673 dataset (Fig. 1D; Supplementary Table S1), 894 upregulated genes and 1007 downregulated genes were screened in the GSE97508 dataset (Fig. 1E; Supplementary Table S2), and 229 upregulated genes and 580 downregulated genes were screened in the GSE110811 dataset (Fig. 1F; Supplementary Table S3). The relative expressions of DEGs in each sample showed significant differences (Figs. 1G–1I).
Figure 1.
Bioinformatic analysis of TRIM59 expression in retinoblastoma samples from the GEO database. Violin plot of the number of genes in retinoblastoma patient and normal control samples from the GSE24673 (A), GSE97508 (B), and GSE110811 (C) datasets. Black dots represent mean values of gene expression after sample normalization. Volcano plot of the distribution of DEGs in the GSE24673 (D), GSE97508 (E), and GSE110811 (F) datasets, mapping upregulated genes (red dots) and downregulated genes (blue dots). Genes showing no significant changes are marked as gray dots. Heatmap using hierarchical clustering of DEGs in the GSE24673 (G), GSE97508 (H), and GSE110811 (I) datasets. (J) Venn diagram DEGs were selected with a fold change > 2 and P < 0.01 in the GSE24673, GSE97508, and GSE110811 datasets. All three datasets showed overlap of 243 genes. Scatterplot of TRIM59 expression among retinoblastoma patient samples and normal control samples in the GSE24673 (K), GSE97508 (L), and GSE110811 (M) datasets.
Furthermore, a Venn diagram revealed 108 overlapping upregulated genes in the GSE24673, GSE97508, and GSE110811 datasets (Fig. 1J). Compared with the normal control samples, the expression levels of TRIM59 in retinoblastoma samples were significantly increased in all three datasets (Figs. 1K–1M).
Upregulation of TRIM59 Expression in Retinoblastoma Cells
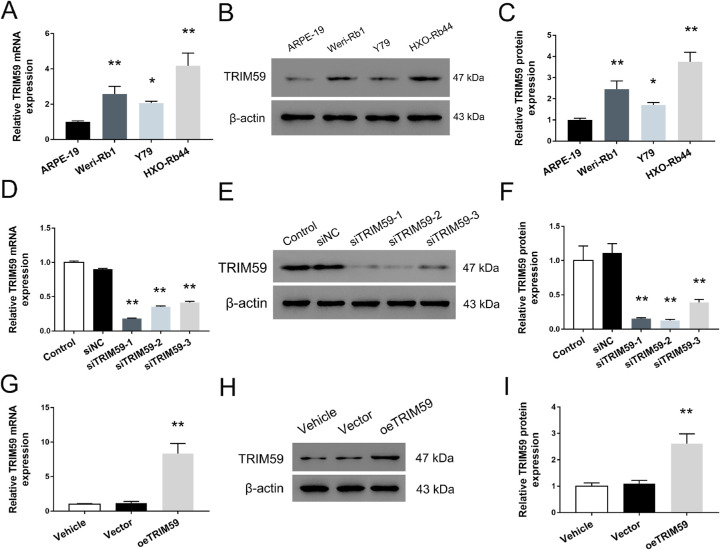
The expression levels of TRIM59 in the human retinoblastoma cell lines HXO-Rb44, Y79, and Weri-Rb1 were significantly higher than that in ARPE-19 cells (Fig. 2A). Meanwhile, the TRIM59 expression rate was highest in HXO-Rb44 cells and lowest in Y79 cells. Moreover, TRIM59 upregulation was further confirmed using immunoblotting, and the trend was consistent with the qRT-PCR data (Figs. 2B, 2C). To explore the biological function of TRIM59 in retinoblastoma cells, we used specific siRNAs to knock down the expression of TRIM59 in HXO-Rb44 cells, which had relatively high expression of TRIM59. In addition, Y79 cells with relatively low TRIM59 expression were used to generate a cell line with stable TRIM59 overexpression by infection with a lentivirus. As shown in Figures 2D to 2F, the mRNA and protein levels of TRIM59 in HXO-Rb44 cells were both significantly lower than those in control cells. Additionally, upregulated expression of TRIM59 in Y79 cells was observed by qRT-PCR (Fig. 2G) and immunoblotting (Fig. 2H, 2I).
Figure 2.
TRIM59 expression is upregulated in three retinoblastoma cell lines. Evaluation of mRNA expression using qRT-PCR (A) and protein expression using immunoblotting (B, C) of TRIM59 in the three human retinoblastoma cell lines, Weri-Rb1, Y79, and HXO-Rb44, as well as the normal retinal epithelial cell line ARPE-19. *P < 0.05, **P < 0.01 compared with ARPE-19 cells. Confirmation of knockdown performed by siRNAs targeting TRIM59 in HXO-Rb44 cells at the mRNA(D) and protein (E, F) levels. **P < 0.01 compared with the siNC group. Validation of upregulated mRNA (G) and protein expression (H, I) of TRIM59 in lentivirus transfected Y79 cells. **P < 0.01 compared with vector-transfected cells.
TRIM59 Accelerates the Cell Proliferation of Human Retinoblastoma Cells
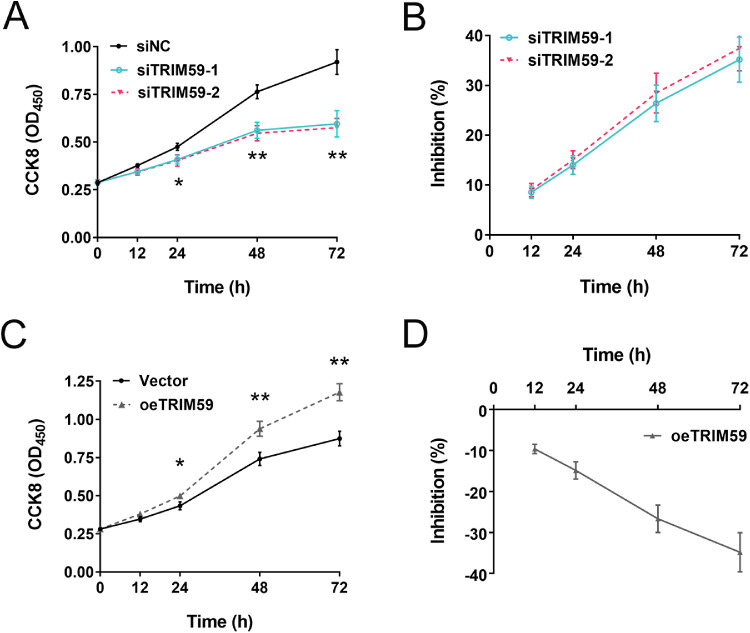
CCK-8 assays revealed that knocking down TRIM59 expression significantly suppressed cell proliferation in HXO-Rb44 cells compared with corresponding control cells (incubation time of 24 hours, P < 0.05; incubation times of 48 and 72 hours, P < 0.01) (Fig. 3A). There was no significant difference of inhibition rate between two siRNA-treated groups (P > 0.05) (Fig. 3B). In contrast, compared with control treatment, lentivirus-mediated overexpression of TRIM59 increased Y79 cell growth (Figs. 3C, 3D). Collectively, these data indicate that TRIM59 contributes to the proliferation of retinoblastoma cells and that knocking down TRIM59 expression significantly inhibits the proliferative potential of retinoblastoma cells.
Figure 3.
Effects of upregulating and downregulating TRIM59 expression on cell proliferation. (A, B) The growth curve and inhibition percentage of HXO-Rb44 cells from CCK8 assay, indicating that HXO-Rb44 cell growth was suppressed after silencing TRIM59. *P < 0.05, **P < 0.01 compared with the siNC group. (C, D) The growth curve and inhibition percentage of Y79 cells, suggesting that overexpression of TRIM59 promoted the cellular proliferation of Y79 cells. *P < 0.05, **P < 0.01 compared with vector-transfected Y79 cells.
TRIM59 Promotes Retinoblastoma Cell-Cycle Progression
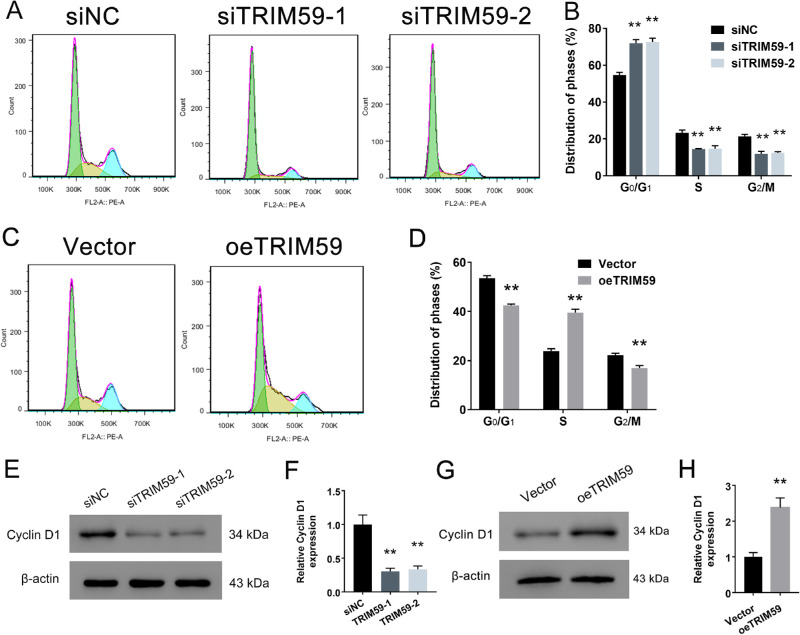
Flow cytometry analysis revealed that TRIM59 silencing significantly increased the proportion of cells in the G1 phase but reduced the proportion of cells in the S phase in HXO-Rb44 cells (Figs. 4A, 4B). In contrast, the opposite effects were observed in Y79 cells with TRIM59 overexpression (Figs. 4C, 4D). Furthermore, we examined the expression level of cyclin D1, a key cell-cycle-regulating protein, and found that the cyclin D1 expression levels in TRIM59-knockdown HXO-Rb44 cells were lower than those in control cells (Figs. 4E, 4F). Additionally, TRIM59-overexpressing cells had higher expression levels of the cyclin D1 protein than vector-transfected cells (Figs. 4G, 4H). All of these results suggest a positive correlation between the TRIM59 expression level and cell-cycle progression in retinoblastoma.
Figure 4.
Effects of TRIM59 on the cell-cycle distribution. (A, B) G1 phase arrest in HXO-Rb44 cells induced by TRIM59 expression downregulation was detected by flow cytometry. **P < 0.01 compared with the siNC group. (C, D) TRIM59 significantly promoted the G1/S phase transition in Y79 cells **P < 0.01 compared with vector-transfected cells. The cell-cycle-related protein cyclin D1 was detected in HXO-Rb44 cells (E, F) and Y79 cells (G, H) by immunoblotting. **P < 0.01 compared with the siNC and vector groups, respectively.
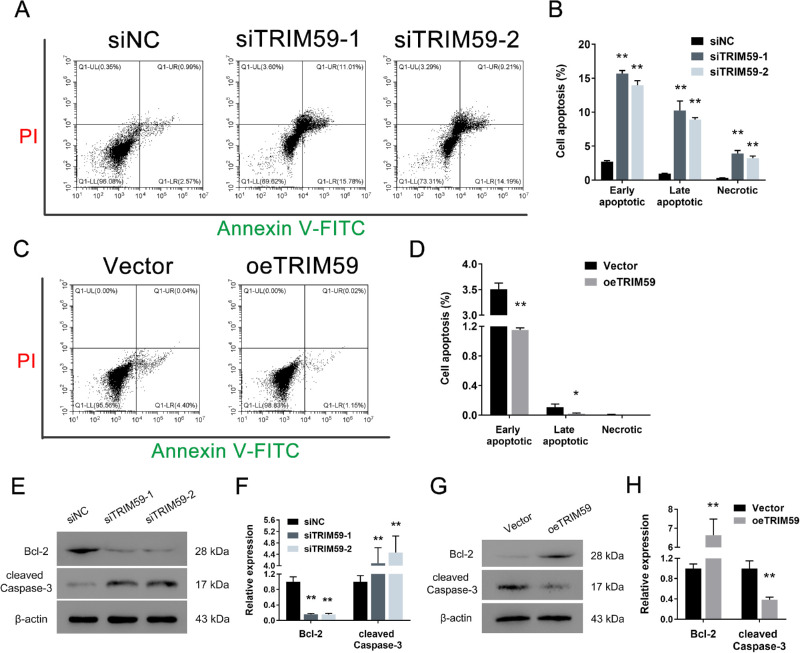
Downregulation of TRIM59 Expression Induces Retinoblastoma Cell Apoptosis
Compared with transfection of a negative control siRNA, TRIM59 depletion increased the percentage of apoptotic HXO-Rb44 cells (Figs. 5A, 5B). In contrast, compared with vector-transfected cells, overexpression of TRIM59 produced a remarkable decrease in the apoptotic cell percentage in Y79 cells (Figs. 5C, 5D). In addition, immunoblotting was used to detect the expression levels of two apoptosis-related biomarkers: Bcl-2 and cleaved caspase-3. As shown in Figures 5E and 5F, the results demonstrate that the expression of Bcl-2 in TRIM59-knockdown HXO-Rb44 cells was lower than that in control cells, and that of cleaved caspase-3 was higher than that in control cells. Additionally, TRIM59-overexpressing Y79 cells had a higher level of Bcl-2 and a lower level of cleaved caspase-3 than vector-transfected Y79 cells (Figs. 5G, 5H). Taken together, these data demonstrate that decreased expression of TRIM59 can increase retinoblastoma cell apoptosis.
Figure 5.
Silencing TRIM59 promotes cell apoptosis. Annexin V-FITC assay showed that increased apoptotic HXO-Rb44 cells (A, B) and decreased apoptotic Y79 (C, D) cells were detected by flow cytometry. *P < 0.05, **P < 0.01 compared with the siNC and vector groups, respectively. Immunoblotting was performed to measure the protein levels of Bcl-2 and cleaved caspase-3 in HXO-Rb44 cells (E, F) and Y79 cells (G, H). **P < 0.01 compared with the siNC and vector groups, respectively.
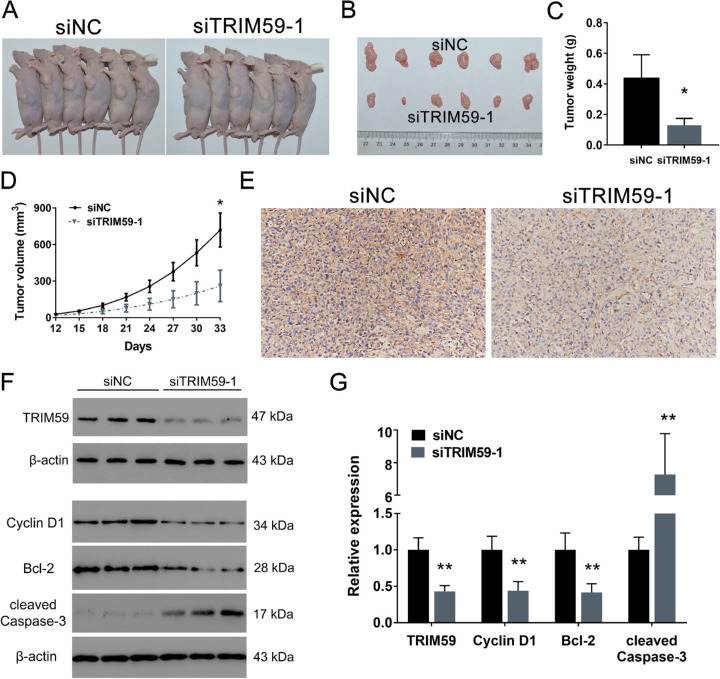
Silencing TRIM59 Suppresses the Tumorigenesis of Retinoblastoma Cells In Vivo
As shown in Figure 6A, all nude mice developed xenograft tumors at the injection site. However, tumor volume and weight were significantly lower in the mice injected with TRIM59-silenced cells than in those in the control group by approximately 69.96% and 63.73%, respectively (Figs. 6B–6D). TRIM59 knockdown was confirmed by immunohistochemical staining (Fig. 6E). Moreover, we also measured the protein levels of TRIM59, cyclin D1, Bcl-2, and cleaved caspase-3 and found that TRIM59-knockdown xenograft tumors had relatively low expression of TRIM59, cyclin D1, and Bcl-2 and relatively high expression of cleaved caspase-3 (Figs. 6F, 6G). Therefore, these results suggest that silencing TRIM59 can attenuate tumor growth and promote cell apoptosis.
Figure 6.
TRIM59 promotes retinoblastoma tumor growth in a xenograft model. (A, B) Representative images of subcutaneous tumors in mice receiving siTRIM59 or the negative control (siNC). (C) Quantification of tumor weight. *P < 0.05 compared with the siNC group. (D)Tumor growth curves comparing the group of mice receiving siTRIM59 with the control group. *P < 0.05 compared with the siNC group. (E) TRIM59 knockdown confirmed by immunohistochemical staining. Magnification, 200×. (F, G) Immunoblotting results indicating that, compared with control treatment, silencing TRIM59 led to reduced expression of TRIM59, cyclin D1, and Bcl-2 and increased expression of cleaved caspase-3. **P < 0.01 compared with the siNC group.
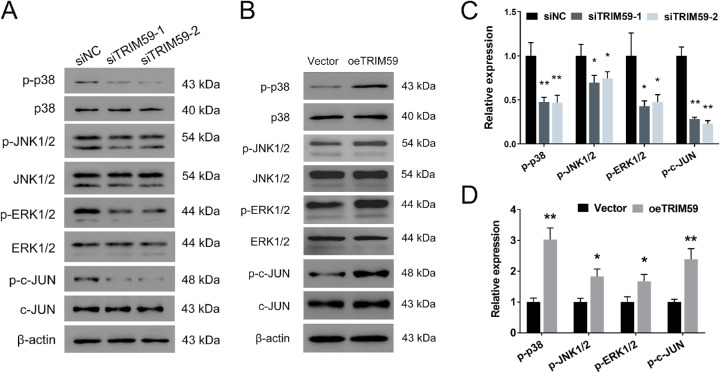
TRIM59 Regulates Retinoblastoma Cells Via the p38–Mitogen-Activated Protein Kinase Pathway
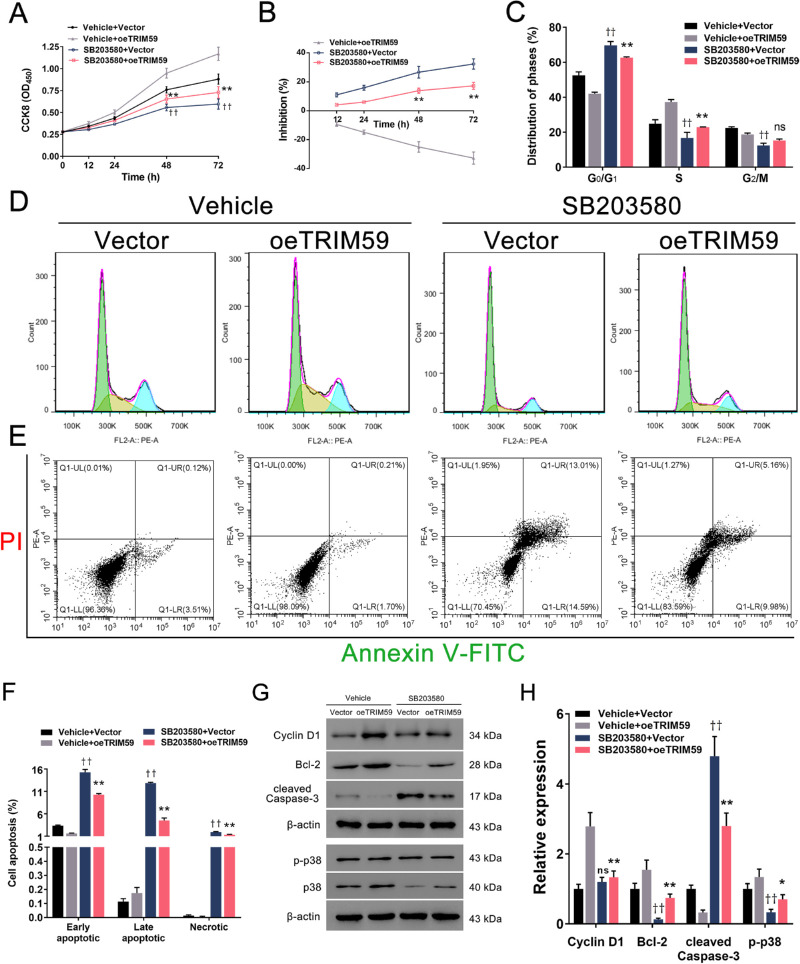
To elucidate the mechanism underlying the effects of TRIM59 on retinoblastoma cells, we evaluated the expression of several cell proliferation-associated genes upstream of cyclin D1. We found that TRIM59 knockdown decreased the phosphorylation of p38, JNK1/2, ERK1/2, and c-JUN (Figs. 7A, 7C), which are key components of the p38–mitogen-activated protein kinase (MAPK) pathway, in HXO-Rb44 cells, whereas TRIM59 overexpression increased the levels of the abovementioned proteins (Figs. 7B, 7D). Thus, we speculated that downregulation of TRIM59 expression leads to inactivation of the p38–MAPK pathway. Additionally, treatment of SB203580, an inhibitor specifically against p38, significantly suppressed cell proliferation (Figs. 8A, 8B) and G1/S phase transition (Figs. 8C, 8D) and promoted apoptosis (Figs. 8E, 8F) in TRIM59-overexpressing Y79 cells compared with controls. Immunoblotting revealed that cyclin D1, Bcl-2, and p-p38 expression levels were downregulated, whereas the expression of cleaved caspase-3 expression was upregulated (Figs. 8G, 8H) after SB203580 treatment. These data confirm that TRIM59 regulates retinoblastoma cells through the p38–MAPK pathway.
Figure 7.
TRIM59 downregulation inhibits the p38–MAPK signaling pathway. The expression of p-p38, p-JNK1/2, p-ERK1/2, and p-c-JUN was evaluated by immunoblotting in the HXO-Rb44 (A, C) and Y79 (B, D) cell lines. *P < 0.05, **P < 0.01 compared with the siNC and vector groups, respectively.
Figure 8.
p38 inhibition alleviates TRIM59-mediated retinoblastoma progression. The p38-specific inhibitor SB203580 effectively suppressed cell proliferation (A, B) and the G1/S phase transition (C, D) and promoted cell apoptosis (E, F) in TRIM59-overexpressing Y79 cells using the methods described above. **P < 0.01, “SB203580 + oeTRIM59” group compared with “vehicle + oeTRIM59” group; ††P < 0.01, “SB203580 + vector” group compared with “vehicle + vector” group. (G, H) Immunoblotting was performed to evaluate the expression of p-p38, cyclin D1, Bcl-2, and cleaved caspase-3. *P < 0.05, **P < 0.01, “SB203580 + oeTRIM59” group compared with “vehicle + oeTRIM59” group; ††P < 0.01, “SB203580 + vector” group compared with “vehicle + vector” group.
Discussion
Retinoblastoma is the most common intraocular neoplasm and a serious threat to vision and life in infants and young children.23 Although multidisciplinary treatments that include surgery, chemotherapy, and radiotherapy are currently available,24 the mortality rate of retinoblastoma patients is still approximately 70%, especially in the low- and middle-income countries where most affected children live.25 Even if patients receive enucleation or orbital exenteration, intracranial invasion, metastasis, or second nonocular neoplasm can lead to death.26 Therefore, investigating the molecular mechanism and promising biomarkers of intraocular retinoblastoma progression should be beneficial for early diagnosis and effective therapeutic target exploration.
Bioinformatic analysis is a well-established method that is widely used to help researchers identify potential biomarkers.27 Thus, we performed bioinformatic analysis of the GEO database to reveal that TRIM59 was one of the most upregulated genes in retinoblastoma samples, which was consistent among the three datasets analyzed. Through qRT-PCR and immunoblotting, we verified that the level of TRIM59 in retinoblastoma cells differed significantly from that in control cells. Previous studies have shown that TRIM59 appears to be an oncoprotein involved in various types of cancer.28,29 Elevated TRIM59 expression is correlated with a relatively poor prognosis and progression in breast cancer.30 Knocking down TRIM59 expression remarkably inhibits cell proliferation in neuroblastoma,31 hepatocellular carcinoma,32 and cholangiocarcinoma.33 However, to date, the role of TRIM59 in retinoblastoma has not been reported. Therefore, TRIM59 was selected for the present research. We have identified TRIM59 as a promising prognostic biomarker or therapeutic target in retinoblastoma. Moreover, gain- and loss-of-TRIM59 experiments with retinoblastoma cell lines suggested that TRIM59 promoted cell proliferation and growth and suppressed cell apoptosis. An in vivo assay further confirmed the tumorigenic function of TRIM59. Our findings suggest that TRIM59 plays an oncogenic role in the progression of retinoblastoma, which is consistent with the results previously observed in other cancers. According to the literature, TRIM59 can regulate colorectal cancer through the Akt pathway,34 regulate neuroblastoma through the Wnt pathway,31 and regulate bladder cancer through the TGF-β pathway.35 The intrinsic mechanism of TRIM59 involved in retinoblastoma needed to be identified.
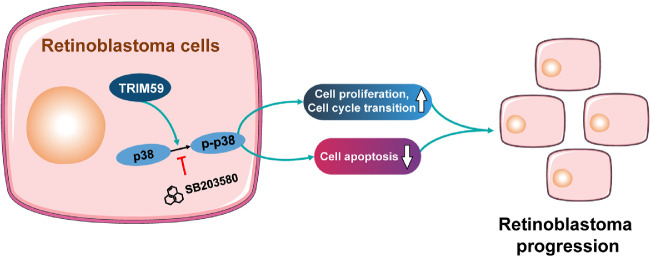
The p38–MAPK signaling pathway has been implicated in the inflammatory response, cell cycle regulation, cell death, differentiation, and tumor progression.36 Goldsmith et al.37 reported that inhibition of the p38–MAPK pathway led to decreased glioma cell invasion. Another study found that miR-3188 promoted cell proliferation and migration via the p38–MAPK pathway in breast cancer.38 Furthermore, miRNA-655 regulates retinoblastoma cell proliferation, invasion, and apoptosis via the p38–MAPK pathway.39 Our results showed that downregulation of TRIM59 expression significantly decreased the expression of the p38–MAPK pathway components p-p38, p-JNK1/2, p-ERK1/2, and p-c-JUN. The opposite results were observed in TRIM59-overexpressing Y79 cells. Moreover, inhibition of p38 with SB203580 abrogated the effects of TRIM59 on cell proliferation, apoptosis, and the G1/S phase transition. These results suggest involvement of the p38–MAPK signaling pathway in the functions of TRIM59 in retinoblastoma (Fig. 9).
Figure 9.
Schematic diagram indicates the proposed role of TRIM59 in retinoblastoma. Upregulation of TRIM59 promotes retinoblastoma cell proliferation and cell-cycle transition and suppresses cell apoptosis via p38–MAPK pathway activation.
These findings may be somewhat limited by the rarity of retinoblastoma patients, and we have not examined the level of TRIM59 in the metastatic samples. We also did not analyze the correlation between TRIM59 expression and survival in retinoblastoma patients. Therefore, the clinical significance of TRIM59 is worthy of further investigation.
In summary, to the best of our knowledge, our study shows for the first time that upregulated TRIM59 expression exerts a crucial role in the progression of retinoblastoma both in vitro and in vivo by activating the p38–MAPK pathway. Our findings offer insights into manipulating TRIM59 as a novel biomarker and target gene for retinoblastoma early diagnosis and treatment.
Supplementary Material
Acknowledgments
The authors thank Xin-Yuan Lao, PhD; Zhu-Jun Tan, PhD; and Cui-Xia Liang, PhD, who provided technical assistance with the experiments.
Supported by grants from the National Natural Science Foundation of China (81760176, 81960158), the Key Scientific Research Foundation of Jiangxi Education Department (GJJ190003), and the Youth Training Program of the Second Affiliated Hospital of Nanchang University (2019YNYX12002).
Disclosure: C. Wu, None; X.-Q. Shang, None; Z.-P. You, None; Q.-F. Jin, None; Y.-L. Zhang, None; Y. Zhou, None; Y.-Z. Zhang, None; K. Shi, None
References
- 1. Mattosinho CCS, Moura A, Oigman G, Ferman SE, Grigorovski N. Time to diagnosis of retinoblastoma in Latin America: a systematic review. Pediatr Hematol Oncol. 2019; 36: 55–72. [DOI] [PubMed] [Google Scholar]
- 2. Alkatan HM, Al Marek F, Elkhamary S. Demographics of pediatric orbital lesions: a tertiary eye center experience in Saudi Arabia. J Epidemiol Glob Health. 2019; 9: 3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kivela T. The epidemiological challenge of the most frequent eye cancer: retinoblastoma, an issue of birth and death. Br J Ophthalmol. 2009; 93: 1129–1131. [DOI] [PubMed] [Google Scholar]
- 4. Khosravi A, Shahrabi S, Shahjahani M, Saki N. The bone marrow metastasis niche in retinoblastoma. Cell Oncol (Dordr). 2015; 38: 253–263. [DOI] [PubMed] [Google Scholar]
- 5. Gunduz K, Muftuoglu O, Gunalp I, Unal E, Tacyildiz N. Metastatic retinoblastoma clinical features, treatment, and prognosis. Ophthalmology. 2006; 113: 1558–1566. [DOI] [PubMed] [Google Scholar]
- 6. Yamada Y, Takayama KI, Fujimura T, et al.. A novel prognostic factor TRIM44 promotes cell proliferation and migration, and inhibits apoptosis in testicular germ cell tumor. Cancer Sci. 2017; 108: 32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mirandola L, M JC, Cobos E, et al.. Cancer testis antigens: novel biomarkers and targetable proteins for ovarian cancer. Int Rev Immunol. 2011; 30: 127–137. [DOI] [PubMed] [Google Scholar]
- 8. Kimsa MW, Strzalka-Mrozik B, Kimsa MC, et al.. Differential expression of tripartite motif-containing family in normal human dermal fibroblasts in response to porcine endogenous retrovirus infection. Folia Biol (Praha). 2014; 60: 144–151. [DOI] [PubMed] [Google Scholar]
- 9. Raheja R, Liu Y, Hukkelhoven E, Yeh N, Koff A. The ability of TRIM3 to induce growth arrest depends on RING-dependent E3 ligase activity. Biochem J. 2014; 458: 537–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhou Z, Ji Z, Wang Y, et al.. TRIM59 is up-regulated in gastric tumors, promoting ubiquitination and degradation of p53. Gastroenterology. 2014; 147: 1043–1054. [DOI] [PubMed] [Google Scholar]
- 11. Hao L, Du B, Xi X. TRIM59 is a novel potential prognostic biomarker in patients with non-small cell lung cancer: a research based on bioinformatics analysis. Oncol Lett. 2017; 14: 2153–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liang J, Xing D, Li Z, Shen J, Zhao H, Li S. TRIM59 is upregulated and promotes cell proliferation and migration in human osteosarcoma. Mol Med Rep. 2016; 13: 5200–5206. [DOI] [PubMed] [Google Scholar]
- 13. Aierken G, Seyiti A, Alifu M, Kuerban G. Knockdown of tripartite-59 (TRIM59) inhibits cellular proliferation and migration in human cervical cancer cells. Oncol Res. 2017; 25: 381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nalini V, Segu R, Deepa PR, Khetan V, Vasudevan M, Krishnakumar S. Molecular insights on post-chemotherapy retinoblastoma by microarray gene expression analysis. Bioinform Biol Insights. 2013; 7: 289–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hudson LE, Mendoza P, Hudson WH, et al.. Distinct gene expression profiles define anaplastic grade in retinoblastoma. Am J Pathol. 2018; 188: 2328–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Danda R, Ganapathy K, Sathe G, et al.. Membrane proteome of invasive retinoblastoma: differential proteins and biomarkers. Proteomics Clin Appl. 2018; 12: e1700101. [DOI] [PubMed] [Google Scholar]
- 17. Gautier L, Cope L, Bolstad BM, Irizarry RA. affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004; 20: 307–315. [DOI] [PubMed] [Google Scholar]
- 18. Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010; 26: 2363–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ligges U, Mächler M. scatterplot3d - an R Package for visualizing multivariate data. J Stat Software. 2002; 8: 1–20. [Google Scholar]
- 20. Ritchie ME, Phipson B, Wu D, et al.. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li Z, Yang L, Wang J, et al.. β-Actin is a useful internal control for tissue-specific gene expression studies using quantitative real-time PCR in the half-smooth tongue sole Cynoglossus semilaevis challenged with LPS or Vibrio anguillarum. Fish Shellfish Immunol. 2010; 29: 89–93. [DOI] [PubMed] [Google Scholar]
- 22. You ZP, Zhang YL, Shi K, et al.. Suppression of diabetic retinopathy with GLUT1 siRNA. Sci Rep. 2017; 7: 7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Singh L, Kashyap S.. Update on pathology of retinoblastoma. Int J Ophthalmol. 2018; 11: 2011–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fabian ID, Onadim Z, Karaa E, et al.. The management of retinoblastoma. Oncogene. 2018; 37: 1551–1560. [DOI] [PubMed] [Google Scholar]
- 25. Dimaras H, Kimani K, Dimba EA, et al.. Retinoblastoma. Lancet. 2012; 379: 1436–1446. [DOI] [PubMed] [Google Scholar]
- 26. Kim JW, Kathpalia V, Dunkel IJ, Wong RK, Riedel E, Abramson DH. Orbital recurrence of retinoblastoma following enucleation. Br J Ophthalmol. 2009; 93: 463–467. [DOI] [PubMed] [Google Scholar]
- 27. You ZP, Zhang YL, Li BY, Zhu XG, Shi K. Bioinformatics analysis of weighted genes in diabetic retinopathy. Invest Ophthalmol Vis Sci. 2018; 59: 5558–5563. [DOI] [PubMed] [Google Scholar]
- 28. Wang Y, Zhou Z, Wang X, et al.. TRIM59 is a novel marker of poor prognosis and promotes malignant progression of ovarian cancer by inducing annexin A2 expression. Int J Biol Sci. 2018; 14: 2073–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhan W, Han T, Zhang C, et al.. TRIM59 promotes the proliferation and migration of non-small cell lung cancer cells by upregulating cell cycle related proteins. PLoS One. 2015; 10: e0142596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu Y, Dong Y, Zhao L, Su L, Diao K, Mi X. TRIM59 overexpression correlates with poor prognosis and contributes to breast cancer progression through AKT signaling pathway. Mol Carcinog. 2018; 57: 1792–1802. [DOI] [PubMed] [Google Scholar]
- 31. Chen G, Chen W, Ye M, Tan W, Jia B. TRIM59 knockdown inhibits cell proliferation by down-regulating the Wnt/beta-catenin signaling pathway in neuroblastoma. Biosci Rep. 2019; 39: BSR20181277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sun G, Sui X, Han D, Gao J, Liu Y, Zhou L. TRIM59 promotes cell proliferation, migration and invasion in human hepatocellular carcinoma cells. Pharmazie. 2017; 72: 674–679. [DOI] [PubMed] [Google Scholar]
- 33. Shen H, Zhang J, Zhang Y, et al.. Knockdown of tripartite motif 59 (TRIM59) inhibits proliferation in cholangiocarcinoma via the PI3K/AKT/mTOR signalling pathway. Gene. 2019; 698: 50–60. [DOI] [PubMed] [Google Scholar]
- 34. Sun Y, Ji B, Feng Y, et al.. TRIM59 facilitates the proliferation of colorectal cancer and promotes metastasis via the PI3K/AKT pathway. Oncol Rep. 2017; 38: 43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen W, Zhao K, Miao C, et al.. Silencing Trim59 inhibits invasion/migration and epithelial-to-mesenchymal transition via TGF-β/Smad2/3 signaling pathway in bladder cancer cells. Onco Targets Ther. 2017; 10: 1503–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Coulthard LR, White DE, Jones DL, McDermott MF, Burchill SA. p38(MAPK): stress responses from molecular mechanisms to therapeutics. Trends Mol Med. 2009; 15: 369–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Goldsmith CS, Kim SM, Karunarathna N, et al.. Inhibition of p38 MAPK activity leads to cell type-specific effects on the molecular circadian clock and time-dependent reduction of glioma cell invasiveness. BMC Cancer. 2018; 18: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen X, Chen J. miR-3188 regulates cell proliferation, apoptosis, and migration in breast cancer by targeting TUSC5 and regulating the p38 MAPK signaling pathway. Oncol Res. 2018; 26: 363–372. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39. Zhang M, Li Q, Pan Y, Wang H, Liu G, Yin H. MicroRNA-655 attenuates the malignant biological behaviours of retinoblastoma cells by directly targeting PAX6 and suppressing the ERK and p38 MAPK signalling pathways. Oncol Rep. 2018; 39: 2040–2050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.