Abstract
Background: Primary colorectal cancer (PCRC) is a common digestive tract cancer in the elderly. However, the treatment effect of PCRC is still limited, and the long-term survival rate is low. Therefore, further exploring the pathogenesis of PCRC, and searching for specific molecular targets for diagnosis are the development trends of precise medical treatment, which have important clinical significance.
Methods: The public data were downloaded from Gene Expression Omnibus (GEO) database. Verification for repeatability of intra-group data was performed by Pearson’s correlation test and principal component analysis. Differentially expressed genes (DEGs) between normal and PCRC were identified, and the protein–protein interaction (PPI) network was constructed. Significant module and hub genes were found in the PPI network. A total of 192 PCRC patients were recruited between 2010 and 2019 from the Fourth Hospital of Hebei Medical University. RT-PCR was used to measure the relative expression of CLCA4 and MS4A12. Furthermore, the study explored the effect of expression of CLCA4 and MS4A12 for overall survival.
Results: A total of 53 DEGs were identified between PCRC and normal colorectal tissues. Ten hub genes concerned to PCRC were screened, namely CLCA4, GUCA2A, GCG, SST, MS4A12, PLP1, CHGA, PYY, VIP, and GUCA2B. The PCRC patients with low expression of CLCA4 and MS4A12 has a worse overall survival than high expression of CLCA4 and MS4A12 (P<0.05).
Conclusion: The research of DEGs in PCRC (53 DEGs, 10 hub genes, especially CLCA4 and MS4A12) and related signaling pathways is conducive to the differential analysis of the molecular mechanism of PCRC.
Keywords: bioinformatics, biomarker, hub genes, overall survival, primary colorectal cancer
Introduction
Primary colorectal cancer (PCRC) is a common digestive tract cancer in the elderly. The primary lesion can be seen in the left colon, the right colon, the upper or lower rectum [1,2]. PCRC is the second most commonly diagnosed cancer in women and the third most commonly diagnosed cancer in men, and the prevalence of male is higher than that of female in most areas [3,4]. With the social environment, lifestyle, and dietary structure changes, the incidence of PCRC is on the rise, and there is a trend of rejuvenation. This is a social issue worthy of attention [5]. At present, there is controversy about the pathogenesis of PCRC. It is generally believed that smoking, drinking, greasy diet, obesity, lack of exercise, colorectal inflammation, and genetic factors are all involved in the onset of cancer. But these factors are also the cause of many other tumors. Therefore, the specific etiological mechanism of PCRC has not yet been elucidated [6,7]. Some scholars believe that some genes or molecules are involved in the development of PCRC. These findings promote the research and treatment of PCRC [8–11]. At present, the treatment of PCRC includes traditional surgery, chemotherapy, radiotherapy, emerging immunotherapy, molecular targeted therapy, etc. Clinically, the single or combination therapy that best suits the condition is usually selected according to the actual situation of the patient [12,13]. However, the treatment effect of PCRC is still limited, and the long-term survival rate is low. The early prognosis of patients with early diagnosis is often better [14]. Therefore, further exploring the pathogenesis of PCRC, searching for specific molecular targets for diagnosis and treatment, realizing early diagnosis, targeted treatment and individualized treatment are the development trends of precise medical treatment, which have important clinical significance.
Personalized medicine refers to the treatment of existing diseases based on the information of each person’s disease genome [15]. It is now widely believed that majority of individual differences in drug response are due to genetic factors. Personalized medicine is a discipline that emphasizes studying the effect of genetic factors on a drug [16]. Recently, due to the smooth implementation of the human genome project and the rapid development of bioinformatics, personalized medicine has been strongly promoted, and the concept has been gradually developed [17].
Bioinformatics is a method to process and analyze biological data by combining biological knowledge with information processing technology. It is commonly used in high-throughput data analysis such as gene and proteomics. As a frontier interdisciplinary subject, bioinformatics analysis technology can realize the biological analysis of the structure and function of histological data, find the genes or proteins most relevant to diseases, and further analysis may find the molecules most relevant to diseases and can be used as disease markers [18,19]. At present, a large number of scholars have applied this technology to tumor research, that is, processing gene sequence or omics data by bioinformatics analysis technology to find genes or molecular markers most relevant to tumors [19–21].
Therefore, the present study aimed to use the bioinformatics to identify the hub genes of PCRC, and to verify their role on the overall survival of patients with PCRC based on the clinical data. And the research would provide novel insights for the personalized medicine on the treatment of patients with PCRC.
Material and methods
Lease start with dates and time, location of study, and the recruitments of patients
The present study recruited a total of 192 PCRC patients between 2010 and 2019 from the Fourth Hospital of Hebei Medical University, Shijiazhuang of Hebei province. Clinical and histopathological characteristics and follow-up and survival information were available for all patients, and were collected retrospectively from medical records. Patients aged 30–100 years old, histologically confirmed as PCRC, not received tumor treatment, and no history of gastrointestinal surgery will be screened for inclusion criteria. Exclusion criteria included: age <30 years old or >100 years old, combined with other malignant tumors, operation time more than one month after the last examination, and severs heart disease.
Ethical clearance and informed content
The research conformed to the Declaration of Helsinki and was authorized by the Human Ethics and Research Ethics Committees of the Fourth Hospital of Hebei Medical University. The written informed consents were obtained from all participates.
Download public data
The Gene Expression Omnibus (GEO) database [22] (http://www.ncbi.nlm.nih.gov/geo) is the largest, most comprehensive, and publicly available source of gene expression data. It contains information about the expression levels of multiple genes in different groups of clinical samples, such as the differences in gene expression between tumor tissues and normal tissues. GSE41258 (GPL96 [HG-U133A] Affymetrix Human Genome U133A Array) and GSE81558 (GPL15207 [PrimeView] Affymetrix Human Gene Expression Array) were obtained from the GEO database. A total of 240 samples, including 186 tumor colorectum tissues from PCRC patients and 54 normal colorectum tissues, were selected from GSE41258. A total of 32 samples, including 23 tumor colorectum tissues from PCRC patients and 9 normal colorectum tissues, were selected from GSE81558.
Verification for repeatability of intra-group data
First, repeatability of intra-group data were verified by the Pearson’s correlation test. The heatmap was drew via the R language environment, and presented the correlation among intra-group data. Second, principal component analysis (PCA) was the general method for sample clustering, and is commonly performed for diversity analysis, resequencing, gene expression, and other sample clustering based on various variable information. The verification for repeatability of intra-group data was executed by PCA.
Differentially expressed genes (DEGs) between normal and PCRC
GEO2R [23] (http://www.ncbi.nlm.nih.gov/geo/geo2r) could import data of the GEO database into the R language and perform differential analysis, essentially through the following two R packages, including limma packages and GEOquery. Therefore, through the GEO2R tool, DEGs were identified between normal and PCRC group. The adjusted P-values (adj. P) <0.05, and the fold change (FC) ≥ 1.5 or ≤ −1.5 were defined as significance. SangerBox (https://shengxin.ren), one open tool, was used to draw volcano maps [24]. Venn diagrams were delineated using an online Venn tool (http://bioinformatics.psb.ugent.be/webtools/Venn/), which would visualize common DEGs shared between GSE41258 and GSE81558.
Protein–protein interaction (PPI) network
The common DEGs, shared between GSE41258 and GSE81558, were converted into differently expressed proteins. The STRING (Search Tool for the Retrieval of Interacting Genes) online database (http://string-db.org) could construct PPI network, which was visualized by Cytoscape (version 2.8) [25].
GO and KEGG analysis via DAVID tool
One online tool, DAVID (https://david.ncifcrf.gov/home.jsp) (version 6.8, Maryland, America), was applied to carried out the functional annotation for DEGs. Gene Ontology (GO) [26] generally perform enrichment analysis of genomes. And there are mainly cellular components (CC), biological processes (BP), and molecular functions (MF) in the GO analysis. Kyoto Encyclopedia of Genes and Genomes (KEGG) (https://www.kegg.jp/) [27] is a comprehensive database of genomic, chemical, and systemic functional information. Therefore, DAVID was used to make analysis of GO and KEGG.
Significant module and hub genes
Molecular Complex Detection tool (MCODE) (version 1.5.1) [28], an open plug-in of Cytoscape, was performed to identify tested most significant module from the PPI network, and the criteria was that the maximum depth = 100, MCODE scores >5, cut-off = 2, k-score = 2, and node score cut-off = 0.2.
Then, cytoHubba [29], a free plug-in of Cytoscape, was applied to authorize the hub genes, when the degree ≥10. Chia-Hao Chin’s [29] research introduce a novel Cytoscape plugin cytoHubba for ranking nodes in a network by their network features. CytoHubba provide a user-friendly interface to explore important nodes in biological networks. When the degree ≥10 in the cytoHubba, the 10 hub genes would be obtained. And in the former publications [30–32], numerous researchers chose 10 hub genes out of the DEGs. Therefore, the present study chose 10 hub genes out of 53 DEGs.
Interaction between the hub genes
Pearson’s correlation analysis was also performed to present the interaction between the hub genes. The cBioPortal (http://www.cbioportal.org) [33], one online software, constructed the co-expression network of these hub genes. Simultaneously, the co-expression network of hub genes in the field of PCRC was also analyzed via Coexpedia, a free and open online tool(http://www.coexpedia.org/) [34].
Expression analysis of hub genes
UCSC Xena (https://xena.ucsc.edu/welcome-to-ucsc-xena/) could integrate the public genomic data sets to analyze and visualize the expression level of hub genes. Then, the clustering analysis of expression level of hub genes was performed using heatmaps based on the GSE41258 and GSE81558. Also, the expression profiles of hub genes in the human different organs were displayed with Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn/) [35]. In order to compare the expression of hub genes in the various tumors, GEPIA was used. And the expression profiles of hub genes in the PCRC and normal groups were analyzed using GEPIA.
Effect of hub gene expression for pathological stage and overall survival
Effect of hub gene expression for pathological stage and overall survival was analyzed by the GEPIA. Finally, the correlation and linear regression analysis between PCRC and hub gene expression were performed. And the receiver operator characteristic (ROC) curve analysis was performed to test the sensitivity and specificity of hub gene expression for diagnose PCRC. The SPSS software (version 21.0; IBM; New York; America) was used to conduct all the statistical analysis. A P-value <0.05 was defined as statistical significance.
RT-qPCR assay
Total RNA was extracted from tumor colorectum tissues from PCRC patients and adjacent normal colorectum tissues by the RNAiso Plus (Trizol) kit (Thermofisher, Massachusetts, America), and reverse transcribed to cDNA. RT-qPCR was performed using a Light Cycler® 4800 System with specific primers for the ten hub genes. Table 1 presents the primer sequences used in the experiments. The RQ values (2−ΔΔCt, where Ct is the threshold cycle) of each sample were calculated, and are presented as fold change in gene expression relative to the control group. GAPDH was used as an endogenous control. The expression level of CLCA4 and MS4A12 in PCRC patients was measured by RT-qPCR.
Table 1. Summaries for the function of 10 hub genes.
| No. | Gene symbol | Full name | Function |
|---|---|---|---|
| 1 | CLCA4 | Chloride channel accessory 4 | May be involved in mediating calcium-activated chloride conductance |
| 2 | GUCA2A | Guanylate cyclase activator 2A | Endogenous activator of intestinal guanylate cyclase. It stimulates this enzyme through the same receptor binding region as the heat-stable enterotoxins |
| 3 | GCG | Glucagon | Regulates blood glucose by increasing gluconeogenesis and decreasing glycolysis. GLP-1 is a potent stimulator of glucose-dependent insulin release. GLP-2 stimulates intestinal growth, concomitant with increased crypt cell proliferation |
| 4 | SST | Somatostatin | Somatostatin inhibits the release of somatotropin. This hormone is an important regulator of the endocrine system through its interactions with pituitary growth hormone, thyroid stimulating hormone, and most hormones of the gastrointestinal tract |
| 5 | MS4A12 | Membrane spanning 4-domains A12 | May be involved in signal transduction as a component of a multimeric receptor complex. Silencing of this gene in colon cancer cells inhibits the proliferation, cell motility, and chemotactic invasion of cells |
| 6 | PLP1 | Proteolipid protein 1 | This is the major myelin protein from the central nervous system. It plays an important role in the formation or maintenance of the multilamellar structure of myelin |
| 7 | CHGA | Chromogranin A | This gene product is a precursor to three biologically active peptides; vasostatin, pancreastatin, and parastatin |
| 8 | PYY | Peptide YY | This gut peptide inhibits exocrine pancreatic secretion, has a vasoconstrictory action, and inhibitis jejunal and colonic mobility |
| 9 | VIP | Vasoactive intestinal peptide | VIP causes vasodilation, lowers arterial blood pressure, stimulates myocardial contractility, increases glycogenolysis and relaxes the smooth muscle of trachea, stomach and gallbladder |
| 10 | GUCA2B | Guanylate cyclase activator 2B | May be a potent physiological regulator of intestinal fluid and electrolyte transport. May be an autocrine/paracrine regulator of intestinal salt and water transport |
Overall survival analysis of the PCRC
The Kaplan–Meier method was performed to analyze the overall survival. All statistical analyses were conducted using SPSS software (version 21.0), and P<0.05 was considered statistically significant.
Results
High repeatability of data
There exist strong correlations among samples in the PCRC group, and also strong correlations among samples in the control group in the GSE41258 via the Pearson’s correlation test (Supplementary Figure S1A). And there also exist strong correlations among samples in the PCRC group, and also strong correlations among samples in the control group in the GSE81558 via the Pearson’s correlation test (Supplementary Figure S1B). Furthermore, PCA was performed to verify the repeatability of data. Through the PCA, the repeatability of the data in GSE41258 was fine. The distances between per samples in the PCRC group were close, and the distances between per samples in the control group were also close in the dimension of PC1 (Supplementary Figure S1C). Through the PCA, the repeatability of the data in GSE81558 was fine. The distances between per samples in the PCRC group were close, and the distances between per samples in the control group were also close in the dimension of PC1 (Supplementary Figure S1D).
DEGs between control and PCRC
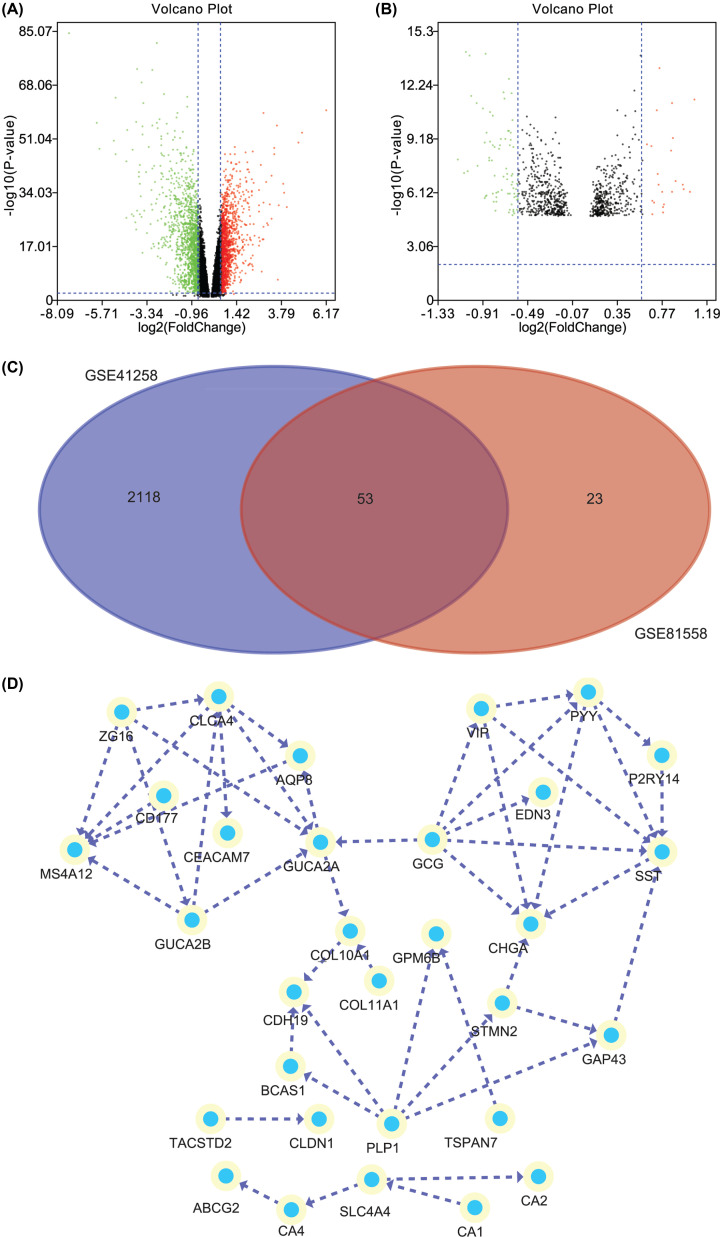
There are plenty of DEGs on the all chromosomes between PCRC and control samples (Supplementary Figure S1E). One volcano plot presents the DEGs in the GSE41258 (Figure 1A), and another volcano plot presents the DEGs in the GSE81558 (Figure 1B). In the volcano plots, the green nodes indicate the down-regulated DEGs, and the red nodes indicate the up-regulated DEGs. The Venn diagram manifested that a total of 53 DEGs were exist in the two datasets (GSE41258 and GSE81558) simultaneously (Figure 1C). After construction of PPI network for the common DEGs, there are 31 nodes and 46 edges in the PPI network (Figure 1D).
Figure 1. The differently expressed genes and PPI network.
(A) One volcano plot presents the DEGs in the GSE41258. (B) Another volcano plot presents the DEGs in the GSE81558. In the volcano plots, the green nodes indicate the down-regulated DEGs, and the red nodes indicate the up-regulated DEGs. (C) The Venn diagram manifested that a total of 53 DEGs were exist in the two datasets (GSE41258 and GSE81558) simultaneously. (D) The PPI network of the common DEGs.
The functional enrichment analysis of DEGs via GO and KEGG
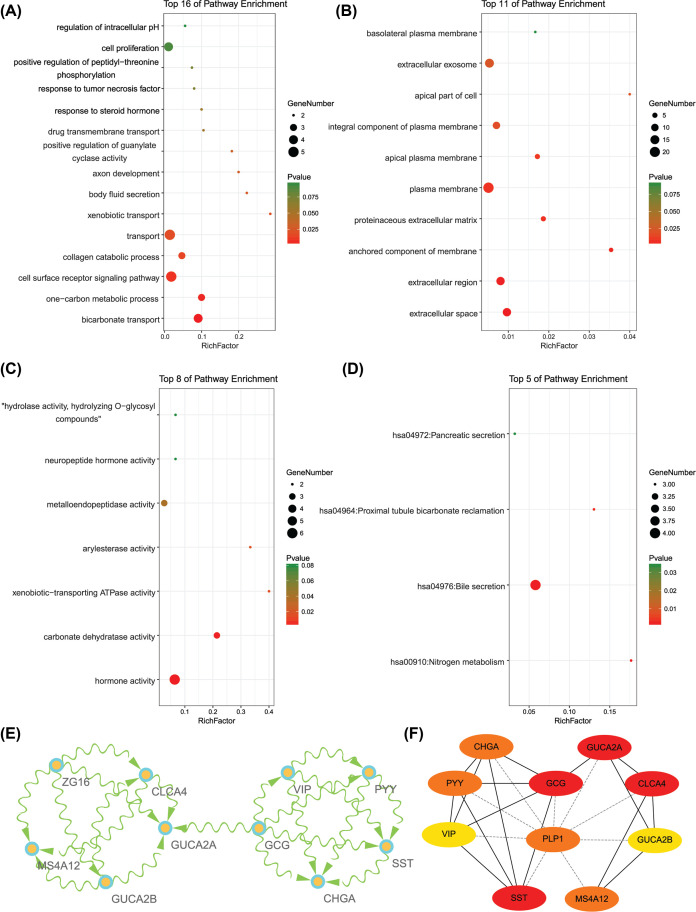
GO analysis manifested that variations in DEGs related with biological processes (BP) were significantly enriched in bicarbonate transport, one-carbon metabolic process, cell surface receptor signaling pathway, collagen catabolic process, transport, xenobiotic transport, body fluid secretion, axon development, positive regulation of guanylate cyclase activity, drug transmembrane transport, response to steroid hormone, response to tumor necrosis factor, positive regulation of peptidyl–threonine phosphorylation, cell proliferation, and regulation of intracellular pH (Figure 2A). The variations in DEGs related with cellular components (CC) were significantly enriched in extracellular space, extracellular region, anchored component of membrane, proteinaceous extracellular matrix, plasma membrane, apical plasma membrane, integral component of plasma membrane, apical part of cell, extracellular exosome, and basolateral plasma membrane (Figure 2B). The variations in DEGs related with molecular functions (MF) were significantly enriched in hormone activity, carbonate dehydratase activity, xenobiotic-transporting ATPase activity, arylesterase activity, metalloendopeptidase activity, neuropeptide hormone activity, and ‘hydrolase activity, hydrolyzing O-glycosyl compounds’ (Figure 2C). KEGG pathway enrichment analysis showed that the top pathways related with DEGs were nitrogen metabolism, bile secretion, proximal tubule bicarbonate reclamation, and pancreatic secretion (Figure 2D).
Figure 2. The enrichment analysis for DEGs and the identification of hub genes.
(A) Detailed information relating to changes in the biological processes (BP) of DEGs in PCRC and control colorectal samples. (B) Detailed information relating to changes in the cellular components (CC) of DEGs in PCRC and control colorectal samples. (C) Detailed information relating to changes in the molecular functions (MF) of DEGs in PCRC and control colorectal samples. (D) KEGG pathway analysis for DEGs. (E) The significant module of the PPI network. (F) The hub genes identified from the PPI network.
Significant module network and identification of hub genes
A significant module was screened from the PPI network, and the module network consisted of 10 nodes and 20 edges (Figure 2E). And ten hub genes were identified, including CLCA4, GUCA2A, GCG, SST, MS4A12, PLP1, CHGA, PYY, VIP, and GUCA2B (Figure 2F). The function of 10 hub genes were summarized in the Table 1.
Strong interaction among the hub genes
Through the Pearson’s correlation test, heat maps manifested that there were strong correlations among hub genes in the GSE41258 (Supplementary Figure S2A) and GSE81558 (Supplementary Figure S2B) datasets. PYY, SST, GCG, and VIP existed simultaneously in the co-expression network via cBioPortal (Supplementary Figure S2C). And through the analysis of Coexpedia, there were strong interactions among PYY, SST, GCG, CHGA, CLCA4, GUCA2B, and MS4A12 (Supplementary Figure S2D).
Difference of expression of hub genes between PCRC and control samples
Heat map showed that the expressions of all the hub genes were lower in the PCRC samples than the control samples (Supplementary Figure S2E). Hierarchical clustering allowed for simple differentiation of PCRC tissues from normal colorectal tissues via the expression levels of hub genes in the GSE41258 (Supplementary Figure S3A) and GSE81558 (Supplementary Figure S3B) datasets. The expressions of all the hub genes were lower in the PCRC group than the control group.
The analysis of expression level of hub genes
The hub genes in the human different organs were expressed in the Supplementary Figure S3C. The pink presents the tumor individuals, and the green presents the normal individuals. The expression of hub genes in the colorectum was higher in the normal individuals compared with the tumor samples (Supplementary Figure S3C). When we compared the expression of hub genes in the various tumors, the all hub genes were down-regulated in the PCRC samples, also named colon adenocarcinoma (COAD) (Supplementary Figure S3D). Through GEPIA analysis, the expressions of hub genes in the PCRC patients were lower than the normal individuals (Supplementary Figure S4A).
Association between hub gene expression, pathological stage, and overall survival
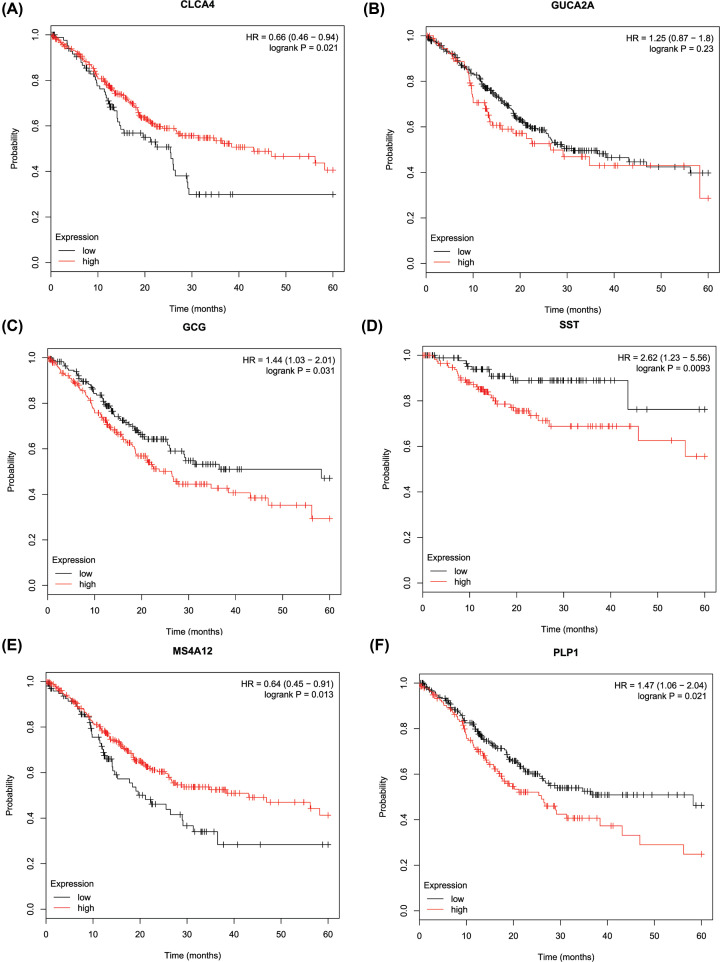
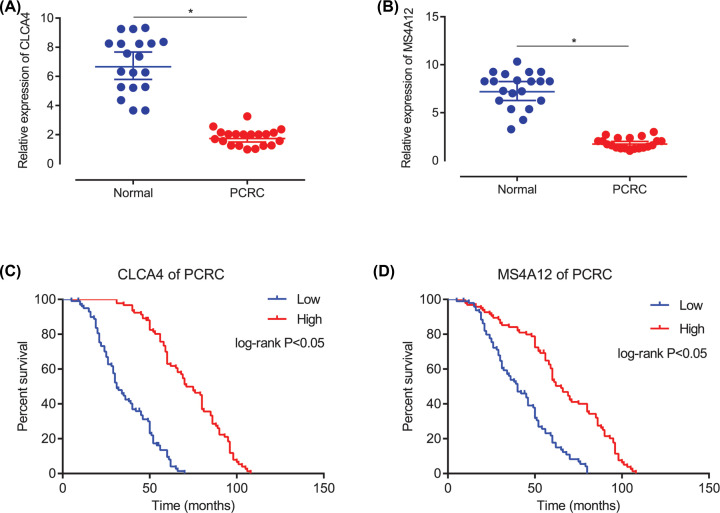
The results of GEPIA manifested that the expression of VIP was significantly positively related with pathological stage (P=0.027), while the expression of CLCA4, GUCA2A, GCG, SST, MS4A12, PLP1, CHGA, PYY, and GUCA2B was not as (Supplementary Figure S4B). Kaplan–Meier analysis showed that PCRC patients with low expression levels of CLCA4, and MS4A12 had poorer overall survival times than those with high expression levels (P<0.05; Figure 3A,E). PCRC patients with high expression levels of GCG, SST, PLP1, and CHGA had poorer overall survival times than those with low expression levels (P<0.05; Figure 3C,D,F; Supplementary Figure S5A). However, there was no statistically significant effect on OS associated with the expression of GUCA2A, PYY, VIP, and GUCA2B (P>0.05; Figure 3B, Supplementary Figure S5B–D).
Figure 3. The overall survival Kaplan–Meier of six hub genes.
(A) CLCA4, (B) GUCA2A, (C) GCG, (D) SST, (E) MS4A12, (F) PLP1.
Correlation, linear regression, and ROC analysis
The Pearson’s correlation coefficient was used in the correlation analysis, and CLCA4 (ρ = −0.868, P<0.001), GUCA2A (ρ = −0.837, P<0.001), GCG (ρ = −0.726, P<0.001), SST (ρ = −0.616, P=0.001), MS4A12 (ρ = −0.793, P<0.001), PLP1 (ρ = −0.763, P<0.001), CHGA (ρ = −0.634, P<0.001), PYY (ρ = −0.610, P<0.001), VIP (ρ = −0.600, P<0.001), and GUCA2B (ρ = −0.688, P<0.001) were significantly correlated with PCRC (Table 2). In the multivariate linear regression model, holding all other variables at a fixed value, the natural logarithmic DN remained associated with CLCA4, GUCA2A, SST, MS4A12, PLP1, CHGA, PYY, and GUCA2B (P<0.05) (Table 2).
Table 2. The correlation and linear regression analysis between PCRC and relevant gene expression.
| PCRC | ||||
|---|---|---|---|---|
| Gene symbol | Pearson’s correlation coefficient | Multiple linear regression | ||
| ρa | P-value | P-value | VIF | |
| CLCA4 | −0.868 | <0.001*** | <0.001*** | 6.959 |
| GUCA2A | −0.837 | <0.001*** | <0.001*** | 8.947 |
| GCG | −0.726 | <0.001*** | 0.087 | 4.944 |
| SST | −0.616 | 0.001** | 0.017* | 3.755 |
| MS4A12 | −0.793 | <0.001*** | 0.039* | 4.685 |
| PLP1 | −0.763 | <0.001*** | <0.001*** | 2.363 |
| CHGA | −0.634 | <0.001*** | 0.001** | 2.290 |
| PYY | −0.610 | <0.001*** | 0.004** | 2.195 |
| VIP | −0.600 | <0.001*** | 0.271 | 2.157 |
| GUCA2B | −0.688 | <0.001*** | 0.017* | 4.144 |
Pearson’s correlation coefficient between PCRC and relevant characteristics; ρ: Pearson’s correlation coefficient.
Multiple linear regression analysis; PCRC: primary colorectal cancer.
: P<0.05;
: P<0.01;
: P<0.001
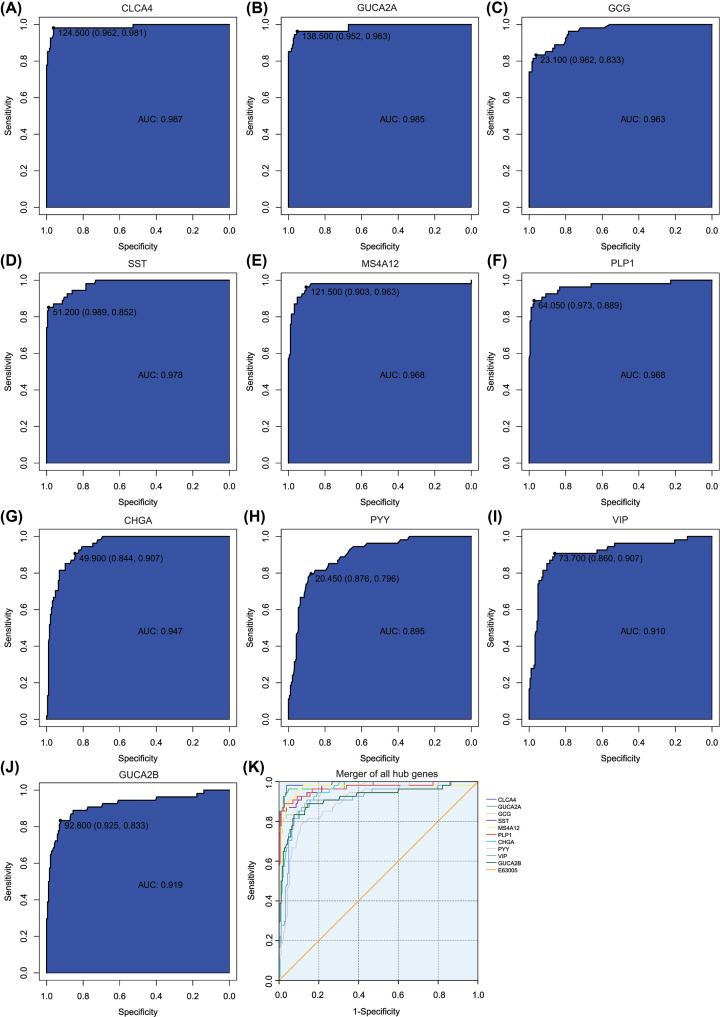
To identify accurate thresholds for hub genes to predict PCRC, we constructed ROC. The expression of all hub genes was associated with a diagnosis of PCRC (0.890 < AUC < 1, P-value<0.001) (Table 3 and Figure 4). The ROC curve of CLCA4 was shown in Figure 4A, and the area under curve of CLCA4 was maximal. The ROC curve of GUCA2A was shown in Figure 4B. The ROC curve of GCG was shown in Figure 4C. The ROC curve of SST was shown in Figure 4D. The ROC curve of MS4A12 was shown in Figure 4E. The ROC curve of PLP1 was shown in Figure 4F. The ROC curve of CHGA was shown in Figure 4G. The ROC curve of PYY was shown in Figure 4H. The ROC curve of VIP was shown in Figure 4I. The ROC curve of GUCA2B was shown in Figure 4J. The ROC curves of per hub genes are shown in Figure 4K.
Table 3. Receiver operator characteristic curve analysis of hub gene expression for PCRC.
| Gene symbol | PCRC | |||
|---|---|---|---|---|
| AUC | P-value | 95%CI | ODT | |
| CLCA4 | 0.987max | <0.001*** | 0.962–0.981 | 124.500 |
| GUCA2A | 0.985 | <0.001*** | 0.952–0.963 | 138.500 |
| GCG | 0.963 | <0.001*** | 0.833–0.962 | 23.100 |
| SST | 0.978 | <0.001*** | 0.852–0.989 | 51.200 |
| MS4A12 | 0.968 | <0.001*** | 0.903–0.963 | 121.500 |
| PLP1 | 0.968 | <0.001*** | 0.889–0.973 | 64.050 |
| CHGA | 0.947 | <0.001*** | 0.844–0.907 | 49.900 |
| PYY | 0.895 | <0.001*** | 0.796–0.876 | 20.450 |
| VIP | 0.910 | <0.001*** | 0.860–0.907 | 73.700 |
| GUCA2B | 0.919 | <0.001*** | 0.833–0.925 | 92.800 |
AUC: area under curve; max the maximum of AUC; *Significant variables; ODT: Optimal diagnostic threshold;
PCRC: primary colorectal cancer.***: P<0.001.
Figure 4. ROC curves of hub genes for PCRC.
(A) CLCA4, (B) GUCA2A, (C) GCG, (D) SST, (E) MS4A12, (F) PLP1, (G) CHGA, (H) PYY, (I) VIP, (J) GUCA2B, (K) ROC curves of all hub genes.
Basic information of PCRC patients
Patients’ basic information were presented in Table 4. The mean patient age was 66 years old (range, 35–96 years), and the median OS was 52 months (range, 5–108 months).
Table 4. Clinicopathological variables and the expression status of CLCA4 and MS4A12.
| CLCA4 | P | MS4A12 | P | ||||
|---|---|---|---|---|---|---|---|
| Low (%) | High (%) | Low (%) | High (%) | ||||
| Sex | |||||||
| Male | 161 | 77(40.1%) | 84(43.8%) | 0.007* | 73(38.0%) | 88(45.8%) | 0.001* |
| Female | 31 | 23(12.0%) | 8(4.2%) | 24(12.5%) | 7(3.6%) | ||
| Age | |||||||
| <65 years | 88 | 44(22.9%) | 44(22.9%) | 0.595 | 38(19.8%) | 50(26.0%) | 0.061 |
| ≥65 years | 104 | 56(29.2%) | 48(25.0%) | 59(30.7%) | 45(23.4%) | ||
| Overall survival | |||||||
| <60 months | 122 | 90(46.9%) | 32(16.7%) | <0.001* | 81(42.2%) | 41(21.4%) | <0.001* |
| ≥60 months | 70 | 10(5.2%) | 60(31.3%) | 16(8.3%) | 54(28.1%) | ||
Pearson’s chi-squared test was used.
P<0.05
RT-qPCR analysis validation of hub genes
As presented in the result, CLCA4 (P<0.05, Figure 5A) and MS4A12 (P<0.05, Figure 5B) were markedly down-regulated in PCRC samples, when compared with the adjacent normal colorectum tissues. It should be noted that the expression situation of CLCA4 and MS4A12 were consistent in above results of bioinformatics.
Figure 5. The verification of expression and overall survival analysis for CLCA4 and MS4A12.
(A) The relative expression of CLCA4 based on PCR. (B) The relative expression of MS4A12 based on PCR. (C) The overall survival of PCRC based the expression of CLCA4. (D) The overall survival of PCRC based the expression of MS4A12.
Low expression of CLCA4 and MS4A12 in PCRC patients were independent prognostic factors for the poor overall survival
The Kaplan–Meier OS curves were analyzed. Low expression of CLCA4 was predictive of a shorter OS in the PCRC patients (P<0.05, Figure 5C). Low expression of MS4A12 was predictive of a shorter OS in the PCRC patients (P<0.05, Figure 5D).
Discussion
PCRC is a common digestive tract cancer, which seriously affects the life expectancy and quality of life of patients. In recent years, the survey results show that the morbidity and mortality rate are on the rise [36]. The clinical manifestations of patients with PCRC are related to the location and pathological type of the tumor. The most common type of pathology is adenocarcinoma. The primary lesion located in the colon often causes diarrhea, obstruction, bleeding in the rectum, anemia, and cachexia in the later stage of cancer patients [37]. The current treatment is mainly surgery combined with chemotherapy or radiotherapy, while advocated exercise to enhance the body’s immunity and prevent infection [12]. Gavrilas et al. found that combination of dietary preparations such as curcumin and resveratrol with chemotherapeutic drugs contributed to the prognosis of PCRC [38]. Clinical application benefit and safety of epidermal growth factor EGFR-related targeted therapy and PD-1/PD-L1 immunotherapy are still to be further studied [39,40]. The investigation found that the cost of PCRC treatment is high, and it takes up a lot of medical resources, and the prognosis of patients is not necessarily proportional to the input. The early treatment of early treatment patients has a relatively low total cost of treatment and a good prognosis [41,42]. Therefore, to further explore the pathogenesis of PCRC, to find possible therapeutic targets, to achieve early diagnosis, targeted therapy, individualized treatment has important clinical value and market prospects.
Bioinformatics has been widely used as a new means of exploring disease mechanism and searching for disease-related genetic molecules. Zhang et al. found genes related to hepatocellular carcinoma by bioinformatics analysis. Further analysis confirmed the correlation between these differential genes and diseases, suggesting that these molecules may be used as molecular targets for early diagnosis and treatment [20]. Zhang et al. found the most relevant molecules of gastric cancer, miR-19b-3p and miR-16-5p, by analyzing the genome-wide miRNA microarray data of gastric cancer patients, which provided a new idea for the diagnosis and treatment of gastric cancer [43]. Sun found molecules related to the pathogenesis of colorectal cancer by screening from public databases. Further analysis showed that differentially expressed genes such as PPBP, CCL28, and CXCL12 are likely to be involved in the development of colorectal cancer and may be potential diagnostic and therapeutic targets [44]. We found 10 genes that were differentially expressed in patients with PCRC by bioinformatics analysis. Low expression of PLP1, VIP, SST, GCG, PYY, MS4A12, CLCA4, GUCA2A, CHGA, and GUCA2B in tumor patients compared with normal subjects. At the same time, we performed survival analysis on patients with PCRC. The results showed that CLCA4, GUCA2A, GCG, SST, MS4A12, and PLP1 genes were significantly associated with the survival of patients with PCRC.
CLCA4 is the chloride channel accessory 4. CLCA4 is a member of the calcium-sensitive chloride-transporting protein family involved in intracellular ion channel activity, chloride ion transmembrane transport, and proteolysis. Members of the calcium activated chloride channel (CLCA) gene family are thought to have multiple functions, including cell adhesion and tumor suppression. Ye et al. found that CLCA4 is low expressed in patients with oral tongue squamous cell carcinoma through genome-wide transcriptional mapping, which provides ideas for diagnosis and targeted therapy [45]. Bundela also found multiple differentially expressed genes in oral cancer patients in India, and suggested that CLCA4 may be a potential therapeutic target [46]. Yu et al. found that CLCA4 is low expressed in breast cancer patients. Further analysis revealed that CLCA4 is a marker of breast epithelial differentiation and may be involved in tumor proliferation and metastasis. Clinical data analysis showed that patients with breast cancer with low expression of CLCA4 had lower recurrence-free survival rate, suggesting that it may serve as a diagnostic and therapeutic target [47]. Hu found that CLCA4 was low expressed in bladder cancer tissues. Further analysis revealed that CLCA4 may be involved in the proliferation and invasion of bladder cancer through PI3K/AKT signal transduction, suggesting that CLCA4 may be a target for diagnosis and treatment [48]. Liu found that CLCA4 may inhibit epithelial–mesenchymal transition (EMT) by affecting PI3K/ATK phosphorylation, thereby inhibiting cell migration and invasion of hepatoma cells [49]. Yang found that patients with colorectal cancer CRC had low expression of CLCA1 and CLCA4, and further experiments confirmed that CLCA1 is involved in tumor proliferation and invasion [50]. Zhao found that CLCA4 was low expressed in colorectal patients [51]. Chen also found that CLCA4 was low expressed in patients with colorectal cancer, and believed that CLCA4 inhibited the epithelial–mesenchymal transformation of colorectal cancer through PI3K/ATK signaling pathway, thus participating in the proliferation and invasion of tumors, and may be used as a marker for diagnosis and judgment [52]. Consistent with the above results, we found that CLCA4 was lowly expressed in primary colorectal patients by bioinformatics analysis, and survival analysis of primary colorectal patients found that patients with low expression of CLCA4 had a worse prognosis in survival, suggesting the protective effect of CLCA4 on PCRC patients and its inhibitory effect on cancer. We speculated that CLCA4 affects epithelial–mesenchymal transition and intercellular communication via PI3K/ATK and participates in the development of primary colorectal patients, which may be potential diagnostic and therapeutic targets.
MS4A12 is membrane spanning 4-domains A12. As a cell protein, MS4A12 participates in cell membrane composition, cell differentiation, proliferation, and cell cycle regulation. Members of the MS4A family are likely to be part of the oligomeric cell surface complex, which has different signal transduction functions [53]. MS4A12 may promote the proliferation and invasion of colorectal cancer cells by influencing epidermal growth factor receptor. Further studies found that intestinal specific transcription factor CDX2 mediated by RNA interference (RNAi) is a trans-activator of growth-promoting gene expression in colorectal cancer, suggesting that MS4A12 may be a potential therapeutic target for colorectal cancer [54]. There was heterogeneity in cancer cells, and found that MS4A12 and other genes could be used to predict cancer patients, suggesting that MS4A12 might be a potential diagnostic target [55]. Multiple genes can be used as molecular markers to distinguish between colon adenomatous polyps and cancer, and that MS4A12 can be used as an early diagnostic target for colorectal cancer [56]. He found that MS4A12 participated in the differentiation of colon cancer cells. Survival analysis showed that patients with low expression of MS4A12 had a poor survival, suggesting that MS4A12 might be a molecular marker for diagnosis and prognosis [57]. We found that MS4A12 was low expressed in PCRC patients by bioinformatics analysis. Meanwhile, survival analysis of PCRC patients showed that patients with low expression of MS4A12 had worse survival than those with high expression of MS4A12, suggesting that MS4A12 was involved in the occurrence and development of PCRC and could inhibit the progression of cancer. We speculate that MS4A12 may be involved in the development of PCRC by affecting cell proliferation, differentiation and signaling pathway transduction, and may be a potential target for diagnosis and treatment.
Personalized medicine is based on the individual’s genetic structure to select the appropriate drug type and dose for patients to significantly improve the efficacy of drugs and reduce drug toxicity [15]. The research found that the expression of CLCA4 and MS4A12 were lower in the patients with PCRC than the normal individual, and overall survival analysis manifested that the PCRC patients with lower expression of CLCA4 and MS4A12 had poorer overall survivals. Therefore, CLCA4 and MS4A12 might be cancer suppressors which could be beneficial to the progression of patients with PCRC. Based on the principle of personalized medicine, the researchers could develop and popularize the drugs targeted to the CLCA4 and MS4A12 to suppress the occurrence and development of the PCRC. Detection of CLCA4 and MS4A12 expression might also help to identify risk factors with poor prognosis in patients with PCRC [58]. Therefore, assessing the level of CLCA4 and MS4A12 could help to accurately predict the prognosis, recurrence and the potential for secondary surgery, so that each patient can be treated individually and the therapeutic effect could be optimized [59]. Currently, the European and American countries have translated the experimental results of pharmacogenomics into clinical applications [60].
Limitations
Despite the rigorous analysis process, there are still some deficiencies in this study. First, there are no animal experiments to comprehensively verify the accuracy of the results. Second, the genetic information that can be used to guide personalized medicine still needs to be enriched, and the basic research needs to be strengthened. Finally, a successful genetic test for personalized medicine meets two criteria for clinical application: safety and effectiveness. However, the bioinformatics in this research focuses on gene screening, and does not verify its safety and effectiveness.
Conclusion
CLCA4 and MS4A12 might play a significant role in the development and survival of PCRC, and might eventually become biomarkers to the treatment of patients with PCRC. Detection of CLCA4 and MS4A12 in the clinical practice might provide the better evidence to guide the early diagnosis and treatment of PCRC. In the future, diagnostic reagent kit of CLCA4 and MS4A12 and the corresponding drugs should be developed to perform the multicenter randomized controlled clinical trial, which will provide new evidence and insights for exploring the early diagnosis of PCRC and personalized medicine.
Supplementary Material
Acknowledgements
We are thankful to Ling-bing Meng for his assistance and support during the submission process.
Abbreviations
- BP
biological processes
- DEG
differentially expressed gene
- EMT
epithelial–mesenchymal transition
- GEO
Gene Expression Omnibus
- MF
molecular functions
- PCRC
primary colorectal cancer
- PPI
protein–protein interaction
- RNAi
RNA interference
Data Availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
The authors declare that there are no sources of funding to be acknowledged.
Author Contribution
Jing Han performed the experiment, and was major contributor in writing. Xue Zhang was involved in critically revising manuscript for important intellectual content. Li Feng made substantial contributions to research conception, and designed the draft of the research process. Yan Liu analyzed the clinical data regarding PCRC. Li Jing and Yi-bing Liu were major contributors in submitting the manuscript, and gave the technical support in the bioinformatics. All authors read and approved the final manuscript.
References
- 1.Lee G.H., Malietzis G., Askari A., Bernardo D., Al-Hassi H.O. and Clark S.K. (2015) Is right-sided colon cancer different to left-sided colorectal cancer? - a systematic review Eur. J. Surg. Oncol. 41, 300–308 10.1016/j.ejso.2014.11.001 [DOI] [PubMed] [Google Scholar]
- 2.Bae J.M., Kim J.H. and Kang G.H. (2016) Molecular Subtypes of Colorectal Cancer and Their Clinicopathologic Features, With an Emphasis on the Serrated Neoplasia Pathway. Arch. Pathol. Lab. Med. 140, 406–412 10.5858/arpa.2015-0310-RA [DOI] [PubMed] [Google Scholar]
- 3.Mattiuzzi C. and Lippi G. (2019) Cancer statistics: a comparison between World Health Organization (WHO) and Global Burden of Disease (GBD). Eur. J. Public Health 10.1093/eurpub/ckz216 [DOI] [PubMed] [Google Scholar]
- 4.Cai Z. and Liu Q. (2019) Understanding the Global Cancer Statistics 2018: implications for cancer control. Sci. China Life Sci. 10.1007/s11427-019-9816-1 [DOI] [PubMed] [Google Scholar]
- 5.Teng A., Nelson D.W., AUID-Oho et al. (2019) Colon cancer as a subsequent malignant neoplasm in young adults. Cancer 125, 3749–3754 10.1002/cncr.32325 [DOI] [PubMed] [Google Scholar]
- 6.Zhang Q.L., Zhao L.G., Li H.L., Gao J., Yang G., Wang J. et al. (2018) The joint effects of major lifestyle factors on colorectal cancer risk among Chinese men: A prospective cohort study. Int. J. Cancer 142, 1093–1101 10.1002/ijc.31126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taheri Z., Asadzadeh A.H., Irani S., Modarressi M.H. and Noormohammadi Z. (2019) Clinical Correlation of miR-200c/141 Cluster DNA Methylation and miR-141 Expression with the Clinicopathological Features of Colorectal Primary Lesions/Tumors. Rep. Biochem. Mol. Biol. 8, 208–215 [PMC free article] [PubMed] [Google Scholar]
- 8.Inamoto S., Itatani Y., Yamamoto T. et al. (2016) Loss of SMAD4 Promotes Colorectal Cancer Progression by Accumulation of Myeloid-Derived Suppressor Cells through the CCL15-CCR1 Chemokine Axis. Clin. Cancer Res. 22, 492–501 10.1158/1078-0432.CCR-15-0726 [DOI] [PubMed] [Google Scholar]
- 9.Tang X.J., Wang W. and Hann S.S. (2019) Interactions among lncRNAs, miRNAs and mRNA in colorectal cancer. Biochimie 163, 58–72 10.1016/j.biochi.2019.05.010 [DOI] [PubMed] [Google Scholar]
- 10.Pallem P.V.S.P., Bodiga S. and Bodiga V.L. (2020) Dietary phytate lowers K-ras mutational frequency, decreases DNA-adduct and hydroxyl radical formation in azoxymethane-induced colon cancer. Iran J. Basic Med. Sci. 23, 20–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sameer A.S. (2013) Colorectal cancer: molecular mutations and polymorphisms. Front. Oncol. 3, 114 10.3389/fonc.2013.00114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marmol I., Sanchez-de-Diego C., Pradilla D.A., Cerrada E. and Rodriguez Y.M.J. (2017) Colorectal Carcinoma: A General Overview and Future Perspectives in Colorectal Cancer. Int. J. Mol. Sci. 18, 197 10.3390/ijms18010197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ciardiello D., Vitiello P.P., Cardone C. et al. (2019) Immunotherapy of colorectal cancer: Challenges for therapeutic efficacy. Cancer Treat. Rev. 76, 22–32 10.1016/j.ctrv.2019.04.003 [DOI] [PubMed] [Google Scholar]
- 14.Brenner H. and Chen C. (2018) The colorectal cancer epidemic: challenges and opportunities for primary, secondary and tertiary prevention. Br. J. Cancer 119, 785–792 10.1038/s41416-018-0264-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goetz L.H. and Schork N.J. (2018) Personalized medicine: motivation, challenges, and progress. Fertil. Steril. 109, 952–963 10.1016/j.fertnstert.2018.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Di S.M., Cipolloni L., Borro M., La Russa R., Santurro A., Scopetti M. et al. (2017) Clinical Applications of Personalized Medicine: A New Paradigm and Challenge. Curr. Pharm. Biotechnol. 18, 194–203 [DOI] [PubMed] [Google Scholar]
- 17.Abul-Husn N.S. and Kenny E.E. (2019) Personalized Medicine and the Power of Electronic Health Records. Cell 177, 58–69 10.1016/j.cell.2019.02.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nobile M.S., Cazzaniga P., Tangherloni A. and Besozzi D. (2017) Graphics processing units in bioinformatics, computational biology and systems biology. Brief. Bioinform. 18, 870–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malgerud L., Lindberg J. et al. (2017) Bioinformatory-assisted analysis of next-generation sequencing data for precision medicine in pancreatic cancer. Mol. Oncol. 11, 1413–1429, 0000-0002-1691-0880 AO 10.1002/1878-0261.12108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang C., Peng L., Zhang Y. et al. (2017) The identification of key genes and pathways in hepatocellular carcinoma by bioinformatics analysis of high-throughput data. Med. Oncol. 34, 101 10.1007/s12032-017-0963-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang T., Guo J., Gu J. et al. (2019) Identifying the key genes and microRNAs in colorectal cancer liver metastasis by bioinformatics analysis and in vitro experiments. Oncol. Rep. 41, 279–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clough E. and Barrett T. (2016) The Gene Expression Omnibus Database. Methods Mol. Biol. 1418, 93–110 10.1007/978-1-4939-3578-9_5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng H., Gu Z.Y., Li Q., Liu Q.H., Yang X.Y. and Zhang J.J. (2019) Identification of significant genes with poor prognosis in ovarian cancer via bioinformatical analysis. J. Ovarian Res. 12, 35 10.1186/s13048-019-0508-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meng L.B., Shan M.J., Qiu Y., Qi R., Yu Z.M., Guo P. et al. (2019) TPM2 as a potential predictive biomarker for atherosclerosis. Aging (Albany N.Y.) 11, 6960–6982 10.18632/aging.102231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calatroni A. and Wildfire J.J. (2017) Graphic depiction of bioinformatics data. J. Allergy Clin. Immunol. 140, 1519–1522 10.1016/j.jaci.2017.05.043 [DOI] [PubMed] [Google Scholar]
- 26.Kumata R., Ito J., Takahashi K., Suzuki T. and Sato K. (2020) A tissue level atlas of the healthy human virome. BMC Biol. 18, 55, 0000-0003-4431-1380, A.O. 10.1186/s12915-020-00785-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong S., Men W., Yang S. and Xu S. (2020) Identification of lung adenocarcinoma biomarkers based on bioinformatic analysis and human samples. Oncol. Rep. 43, 1437–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He T. and Chan K.C.C. (2018) Evolutionary Graph Clustering for Protein Complex Identification. IEEE/ACM Trans. Comput. Biol. Bioinform. 15, 892–904 10.1109/TCBB.2016.2642107 [DOI] [PubMed] [Google Scholar]
- 29.Chin C.H., Chen S.H., Wu H.H., Ho C.W., Ko M.T. and Lin C.Y. (2014) cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 8, S11 10.1186/1752-0509-8-S4-S11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y.R., Meng L.B., Su F. et al. (2020) Insights regarding novel biomarkers and the pathogenesis of primary colorectal carcinoma based on bioinformatic analysis. Comput. Biol. Chem. 85, 107229 10.1016/j.compbiolchem.2020.107229 [DOI] [PubMed] [Google Scholar]
- 31.Zou Y.F., Meng L.B., He Z.K. et al. (2019) Screening and authentication of molecular markers in malignant glioblastoma based on gene expression profiles. Oncol. Lett. 18, 4593–4604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu C., Meng L.B., Duan Y.C. et al. (2019) Screening and identification of biomarkers for systemic sclerosis via microarray technology. Int. J. Mol. Med. 44, 1753–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Torcivia-Rodriguez J., Dingerdissen H., Chang T.C. and Mazumder R. (2019) A Primer for Access to Repositories of Cancer-Related Genomic Big Data. Methods Mol. Biol. 1878, 1–37 10.1007/978-1-4939-8868-6_1 [DOI] [PubMed] [Google Scholar]
- 34.Yang S., Kim C.Y., Hwang S. et al. (2017) COEXPEDIA: exploring biomedical hypotheses via co-expressions associated with medical subject headings (MeSH). Nucleic Acids Res. 45, D389–D396 10.1093/nar/gkw868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang Z., Li C., Kang B., Gao G., Li C. and Zhang Z. (2017) GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45, W98–W102 10.1093/nar/gkx247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yin J., Bai Z., Zhang J. et al. (2019) Burden of colorectal cancer in China, 1990-2017: Findings from the Global Burden of Disease Study 2017. Chin. J. Cancer Res. 31, 489–498 10.21147/j.issn.1000-9604.2019.03.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lv X., Yu H., Gao P. et al. (2019) A nomogram for predicting bowel obstruction in preoperative colorectal cancer patients with clinical characteristics. World J. Surg. Oncol. 17, 21 10.1186/s12957-019-1562-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gavrilas L.I., Cruceriu D., Ionescu C., Miere D. and Balacescu O. (2019) Pro-apoptotic genes as new targets for single and combinatorial treatments with resveratrol and curcumin in colorectal cancer. Food Funct. 10, 3717–3726 10.1039/C9FO01014A [DOI] [PubMed] [Google Scholar]
- 39.Sepulveda A.R., Hamilton S.R., Allegra C.J. et al. (2017) Molecular Biomarkers for the Evaluation of Colorectal Cancer: Guideline From the American Society for Clinical Pathology, College of American Pathologists, Association for Molecular Pathology, and the American Society of Clinical Oncology. J. Clin. Oncol. 35, 1453–1486 10.1200/JCO.2016.71.9807 [DOI] [PubMed] [Google Scholar]
- 40.Yaghoubi N., Soltani A., Ghazvini K., Hassanian S.M. and Hashemy S.I. (2019) PD-1/ PD-L1 blockade as a novel treatment for colorectal cancer. Biomed. Pharmacother. 110, 312–318 10.1016/j.biopha.2018.11.105 [DOI] [PubMed] [Google Scholar]
- 41.Huang H.Y., Shi J.F., Guo L.W. et al. (2017) Expenditure and financial burden for the diagnosis and treatment of colorectal cancer in China: a hospital-based, multicenter, cross-sectional survey. Chin. J. Cancer 36, 41 10.1186/s40880-017-0209-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gani F., Cerullo M., Canner J.K. et al. (2017) Defining payments associated with the treatment of colorectal cancer. J. Surg. Res. 220, 284–292 10.1016/j.jss.2017.07.021 [DOI] [PubMed] [Google Scholar]
- 43.Zhang J., Song Y., Zhang C. et al. (2015) Circulating MiR-16-5p and MiR-19b-3p as Two Novel Potential Biomarkers to Indicate Progression of Gastric Cancer. Theranostics 5, 733–745 10.7150/thno.10305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun G., Li Y., Peng Y. et al. (2019) Identification of differentially expressed genes and biological characteristics of colorectal cancer by integrated bioinformatics analysis. J. Cell. Physiol. 234, 15215–15224 10.1002/jcp.28163 [DOI] [PubMed] [Google Scholar]
- 45.Hussein A.A., Forouzanfar T., Bloemena E., de Visscher J., Brakenhoff R.H., Leemans C.R. et al. (2018) A review of the most promising biomarkers for early diagnosis and prognosis prediction of tongue squamous cell carcinoma. Br. J. Cancer 119, 724–736 10.1038/s41416-018-0233-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bundela S., Sharma A. and Bisen P.S. (2014) Potential therapeutic targets for oral cancer: ADM, TP53, EGFR, LYN, CTLA4, SKIL, CTGF, CD70. PLoS ONE 9, e102610 10.1371/journal.pone.0102610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu Y., Walia V. and Elble R.C. (2013) Loss of CLCA4 promotes epithelial-to-mesenchymal transition in breast cancer cells. PLoS ONE 8, e83943 10.1371/journal.pone.0083943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hou T., Zhou L., Wang L., Kazobinka G., Zhang X. and Chen Z. (2017) CLCA4 inhibits bladder cancer cell proliferation, migration, and invasion by suppressing the PI3K/AKT pathway. Oncotarget 8, 93001–93013 10.18632/oncotarget.21724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Z., Chen M., Xie L.K. et al. (2018) CLCA4 inhibits cell proliferation and invasion of hepatocellular carcinoma by suppressing epithelial-mesenchymal transition via PI3K/AKT signaling. Aging (Albany N.Y.) 10, 2570–2584 10.18632/aging.101571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang B., Cao L., Liu B., McCaig C.D. and Pu J. (2013) The transition from proliferation to differentiation in colorectal cancer is regulated by the calcium activated chloride channel A1. PLoS ONE 8, e60861 10.1371/journal.pone.0060861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao Z.W., Fan X.X., Yang L.L. et al. (2019) The identification of a common different gene expression signature in patients with colorectal cancer. Math Biosci. Eng. 16, 2942–2958 [DOI] [PubMed] [Google Scholar]
- 52.Chen H., Liu Y., Jiang C.J., Chen Y.M., Li H. and Liu Q.A. (2019) Calcium-Activated Chloride Channel A4 (CLCA4) Plays Inhibitory Roles in Invasion and Migration Through Suppressing Epithelial-Mesenchymal Transition via PI3K/AKT Signaling in Colorectal Cancer. Med. Sci. Monit. 25, 4176–4185 10.12659/MSM.914195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eon K.L., Leffler M., Mackay G.A. and Hulett M.D. (2016) The MS4A family: counting past 1, 2 and 3. Immunol. Cell Biol. 94, 11–23 [DOI] [PubMed] [Google Scholar]
- 54.Xu H., Ma Y., Zhang J. et al. (2020) Identification and Verification of Core Genes in Colorectal Cancer. Biomed. Res. Int. 2020, 8082697, 0000-0003-4301-1781, A.O. 0000-0001-6291-2812, A.O. 0000-0002-8840-8283, A.O. 10.1155/2020/8082697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilbrey-Clark A., Roberts K. and Teichmann S.A. (2020) Cell Atlas technologies and insights into tissue architecture. Biochem. J. 477, 1427–1442 10.1042/BCJ20190341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qiu X., Cheng S.H., Xu F., Yin J.W., Wang L.Y. and Zhang X.Y. (2020) Weighted gene co-expression network analysis identified MYL9 and CNN1 are associated with recurrence in colorectal cancer. J. Cancer 11, 2348–2359 10.7150/jca.39723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He L., Deng H.Y. and Wang X.C. (2017) Decreased expression of MS4A12 inhibits differentiation and predicts early stage survival in colon cancer. Neoplasma 64, 65–73 10.4149/neo_2017_108 [DOI] [PubMed] [Google Scholar]
- 58.Wang Q., Yuan L.L. and Fan W.T. (2019) Analysis of the expressions of oncogene INHBA and anti-oncogene CLCA4 and CA4 in colorectal cancer based on GEO and TCGA databases. Zhongguo Ying Yong Sheng Li Xue Za Zhi 35, 279–282 [DOI] [PubMed] [Google Scholar]
- 59.Gao X. and Yang J. (2020) Identification of Genes Related to Clinicopathological Characteristics and Prognosis of Patients with Colorectal Cancer. DNA Cell Biol. 39, 690–699 10.1089/dna.2019.5088 [DOI] [PubMed] [Google Scholar]
- 60.Kalman L.V., Agúndez J., Appell M.L., Black J.L., Bell G.C., Boukouvala S. et al. (2016) Pharmacogenetic allele nomenclature: International workgroup recommendations for test result reporting. Clin. Pharmacol. Ther. 99, 172–185 10.1002/cpt.280 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.