Abstract
Paralytic shellfish poisoning (PSP) poses a serious health threat in Alaska and prevents effective utilization of shellfish resources by subsistence and recreational harvesters. Substantial economic losses also affect shellfish growers during PSP events. The toxins responsible for PSP are produced by dinoflagellates in the genus Alexandrium. Despite the persistent threat posed by PSP and the long history of shellfish toxicity research, there is still confusion concerning the Alexandrium species that cause PSP in Alaska. The primary objective of this study was to identify the toxic Alexandrium species present in Alaska and to develop polymerase chain reaction (PCR) assays for use in screening phytoplankton and sediment samples. Before developing the PCR assays for this study, we evaluated published assays and many were not adequate because of primer dimer formation or because of cross-reactivity. Rather than continue to grapple with the uncertainty and inadequacy of published assays, we developed new assays for the Alexandrium species most likely to be present in Alaska. Only Alexandrium fundyense Group I and A. ostenfeldii were identified from four sampling regions from southeast Alaska to Kodiak Island, indicating that these two species are widely distributed. PCR assays for these two species were converted to quantitative (q)PCR format for use in monitoring programs. During the course of this study, we realized that a systematic evaluation of all published (~150) Alexandrium species-specific assays would be of benefit. Toward this objective, we collated published Alexandrium PCR, qPCR, and in situ hybridization assay primers and probes that targeted the small-subunit (SSU), internal transcribed spacer (ITS/5.8S), or D1-D3 large-subunit (LSU) (SSU/ITS/LSU) ribosomal DNA genes. Each individual primer or probe was screened against the GenBank database and Alexandrium gene sequence alignments constructed as part of this study. These data were used to identify a suite of species-specific Alexandrium assays that can be recommended for evaluation by the global harmful algal bloom community.
Keywords: Alexandrium catenella, Alexandrium minutum, Alexandrium monilatum, Alexandrium pacificum, Alexandrium tamarense, Alexandrium monitoring, Gulf of Alaska, Paralytic shellfish poisoning (PSP), qPCR assay, Saxitoxin
INTRODUCTION
Paralytic shellfish poisoning (PSP) is a persistent health threat in Alaska caused by consumption of bivalves and other shellfish containing saxitoxins (STXs) produced by dinoflagellate species in the genus Alexandrium Halim (Smith 2010). STXs interact with voltage-gated sodium, potassium, and calcium channels and modulate the flux of these ions into various cell types. This results in disruption of nervous system function (Cusick & Sayler 2013), which may be fatal in severe cases. Documented cases of PSP in Alaska date back centuries to Captain George Vancouver’s survey of the Pacific coast in the early 1790s (Quayle 1969; Horner et al. 1997). PSP is sufficiently prevalent in Alaska that it has been designated as a public health emergency and all cases are required to be reported to the state’s Division of Epidemiology (Castrodale 2015). The remote locations of Alaska’s coastal communities and the high cost of shipping and testing make it impractical for the state to routinely monitor shellfish resources utilized by subsistence and recreational harvesters. only commercially harvested shellfish are tested by Alaska’s Division of Environmental Health for STXs before they are considered safe for sale or consumption. Despite the persistent PSP health threat, Alaskans harvest shellfish for personal use year round. Monitoring Alexandrium at key recreational shellfish harvesting beaches would help identify periods of elevated PSP risk that could be conveyed to the public before harvesting (R. RaLonde, personal observations).
The historical record of harmful algal bloom (HAB) literature for Alaska was described as “sparse” by Horner et al. (1997), with reports of possible Alexandrium species from Jack Bay in Prince William Sound, Tenakee Harbor, near Ketchikan, and other southeast Alaska sites. Hall (1982) collected samples containing Alexandrium (Protogonyaulax F.J.R.Taylor) from Dutch Harbor to Ketchikan and analyzed cellular toxin content. More recently, Trainer (2002) reported three species of Alexandrium in Alaska: A. catenella (Whedon & Kofoid) Balech, which was broadly distributed, A. fundyense (Balech) D.M.Anderson from Porpoise Island, and A. tamarense (M.Lebour) Balech (John et al. 2014a) from the Gulf of Alaska. Orlova et al. (2007) reported that seven species of Alexandrium, A. insuetum Balech, A. margalefii Balech, A. ostenfeldii (Paulsen) Balech & Tangen, A. catenella, A. pseudogonyaulax (Biecheler) Horiguchi ex Yuki & Fukuyo, A. tamarense, and A. tamutum Montresor, Beran & John, occur in the far eastern seas of Russia, including isolates collected from the Bering Sea in proximity to Alaskan waters. Most recently, Natsuike et al. (2013) assayed cysts collected from sediments at numerous stations in the Bering and Chukchi seas using species-specific polymerase chain reaction (PCR) assays for A. affine (H.Inoue & Y.Fukuyo) Balech, A. fraterculus Balech, A. catenella (Group IV, subsequently defined as A. pacificum by John et al. 2014a), A. pseudogonyaulax, A. tamarense Group III, and A. tamiyavanichi Balech. In their study, cyst densities ranged from 102 to 104 cm−3 of sediment. Despite the high cyst densities, only A. fundyense Group I was detected. With the exception of results from the Natsuike et al. (2013) study, the other Alexandrium species identifications previously reported were based solely on morphology. Unfortunately, morphological identification without molecular confirmation is unreliable for many Alexandrium species (John et al. 2014a; Kremp et al. 2014). This is particularly true for the A. tamarense species complex, which originally contained three broadly distributed, morphologically defined species, A. catenella, A. fundyense, and A. tamarense (John et al. 2014a). Molecular phylogenies based on ribosomal (r)DNA genes have consistently shown that the A. tamarense species complex is actually comprised of five distinct ribotype clades (Lilly et al. 2007; John et al. 2014a). Recently, John et al. (2014a) formally assigned species designations to A. fundyense (Group I), A. mediterraneum (Group II), A. tamarense (Group III), A. pacificum (Group IV), and A. australiense (Group V) and proposed formal rejection of the name Gonyaulax catenella (A. catenella) in John et al. (2014b). The John et al. (2014a) nomenclature is used throughout this study and references to the A. tamarense species complex are accompanied by a Groups I–V designation. This enables unambiguous species identification no matter the species name used in previous studies.
Taking into account recent taxonomic revisions within Alexandrium (John et al. 2014a; Kremp et al. 2014), the goals of this study were to develop and use quantitative, species-specific PCR (qPCR) assays that targeted the ribosomal gene complex to detect and quantify cells of Alexandrium collected from Alaska coastal sites. Six Alexandrium species were selected as subjects for assay development on the basis of the distributional data presented in Balech (1995). Initially, these assays were used qualitatively to detect Alexandrium species in surface water and sediment samples. After the presence of Alexandrium species was confirmed, species-specific assays were converted into a quantitative (qPCR) format. Alexandrium cysts from four geographically dispersed regions including Ketchikan, Juneau, Kachemak Bay, and Kodiak Island were isolated from sediment samples and propagated to establish cultures. This material was used to validate qPCR assays and to investigate the possible existence of other Alexandrium species.
Before developing the new species-specific assays we tested several published qPCR assays (data not shown). The results obtained in our laboratory revealed varying levels of primer dimer formation, occasional cross-reactivity with other Alexandrium species, and variable dynamic ranges. Consequently, we elected to develop our own species-specific qPCR assays. This approach has been used in the past by numerous researchers because of uncertainty surrounding the specificity and efficacy of published assays. Having developed molecular assays for other HAB projects (Litaker et al. 2002, 2003; Vandersea et al. 2006, 2012), we were conscious of the time and effort required to validate molecular assays. Therefore to aid other researchers in selecting creditable Alexandrium molecular assays, we opted to systematically evaluate existing Alexandrium assays and make recommendations to the scientific community. As a first step in this evaluation process, we collated published Alexandrium PCR, qPCR, and in situ hybridization assay primers and probes that target the small-subunit/internal transcribed spacer/large-subunit (SSU/ITS/LSU) rDNA genes. These data facilitated identification of validated, species-specific Alexandrium assays for monitoring and ecological studies worldwide.
MATERIAL AND METHODS
This study used two methods to identify the Alexandrium species present in Alaska. The first was to isolate cells from environmental samples and confirm their identity by molecular cloning and sequencing of diagnostic rDNA regions. The second was to develop PCR assays for species that were likely to be found in Alaska on the basis of previous reports and for which sufficient sequence data were available. Once species from these samples were identified using species-specific PCR assays, the assays were converted into a quantitative (qPCR) format. The PCR assays were used to confirm the identity of the isolates established from field material as well as to screen for species that were not easily cultured, or those present in such low numbers that a representative cell isolate could not be established.
Collection of phytoplankton and sediment samples
Live phytoplankton from nearshore locations in Ketchikan, Juneau, Kachemak Bay, and Kodiak Island (Figs 1–5) were collected via surface-water net tows using a 20-μm-mesh plankton net (model 9000, 1-litre cod end, Sea-Gear Corp., Melbourne, Florida, USA). The cod end from each tow was gently shaken to homogenize the sample and a 100-ml aliquot was preserved in neutral Lugol’s solution for screening using species-specific molecular assays. In this study sediment samples were collected from embayments and beaches possessing fine sediments and salinities ranging from 20 to 25. Surface sediment samples (upper 1–2 cm) were collected at low tide or by scuba divers using 50-ml screw-cap tubes. Samples were stored at 4°C in the dark until they could be processed for cyst germination or DNA extraction and PCR assay. Unpreserved whole-water and sediment samples were shipped with ice packs to the National Oceanic and Atmospheric Administration (NOAA) Center for Coastal Fisheries and Habitat Research in Beaufort, North Carolina, USA for single-cell isolations. Upon arrival the water samples were filtered sequentially through 500-μm and 200-μm mesh to remove larger fauna and debris. One-hundred-millilitre aliquots of each sieved sample were placed in vented, polystyrene tissue culture flasks and supplemented with ~20 ml of modified K medium (no Tris, no Cu, no Si) (Keller et al. 1987) and incubated for several days at 14°C under ~50 μmol photons·m−2·s−1 of light (light:dark = 12:12 h). Alexandrium cultures were then established from this incubated material via single-cell isolations (see methods below).
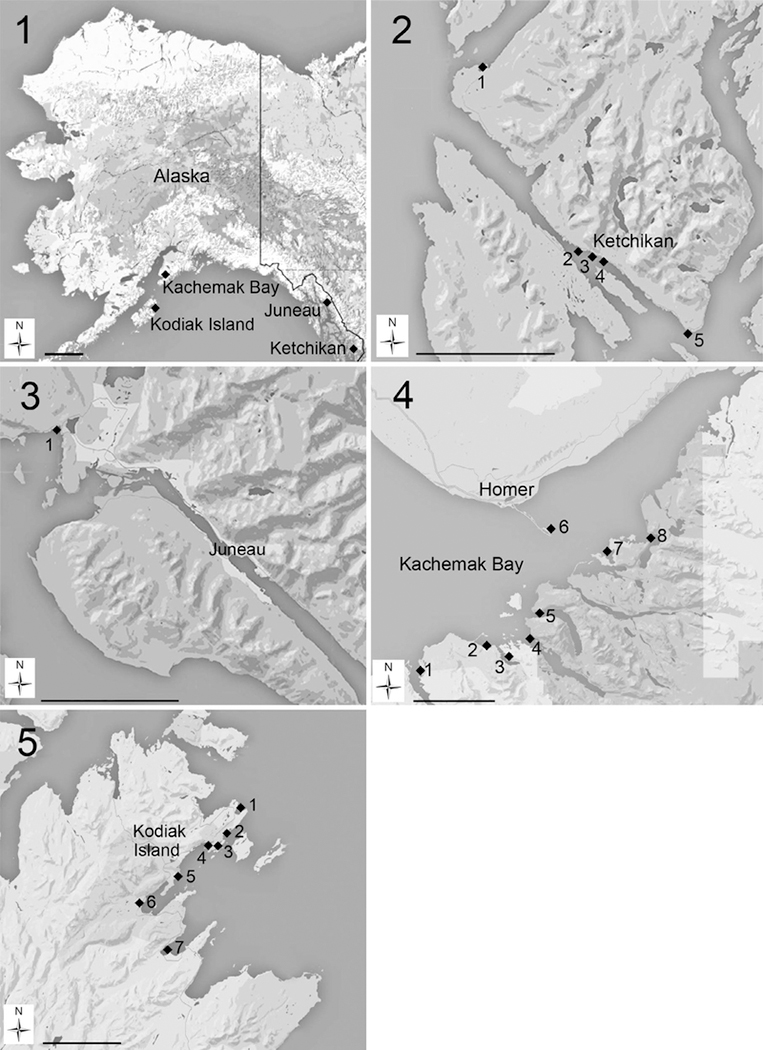
Figs 1–5. Locations of sampling sites.
Fig. 1: Location of coastal Alaska sampling sites. Scale bar = 250 km.
Fig. 2: Location of sampling sites in Ketchikan, Alaska. (1) Knudson Cove, (2) Bar Harbor Ramp, (3) City Float, (4) Thomas Basin, (5) Mountain Point. Scale bar = 10 km.
Fig. 3: Location of the Auke Bay sampling site in Juneau, Alaska. Scale bar = 10 km.
Fig. 4: Location of sampling sites in Kachemak Bay, Alaska. (1) Seldovia Harbor, (2) Kasitsna Bay NOAA Laboratory, (3) Jakolof Bay, (4) Tutka Bay, (5) Sadie Cove, (6) Homer Harbor, (7) Peterson Bay, (8) Halibut Cove. Scale bar = 10 km.
Fig. 5: Location of sampling sites in Kodiak Island, Alaska. (1) Mill Bay, (2) Mission Beach, (3) Trident Basin, (4) U.S. Coast Guard Station, (5) Buskin River, (6) Sargent Creek, (7) Womens Bay.
Sediment processing, cyst germination, and single-cell isolates
Sediment samples proved to be the best source of Alexandrium cells for single-cell isolation. Sediment samples were removed from storage at 4°C and rinsed through a 53-μm mesh screen with filtered seawater to remove large particulates. The 53-μm mesh screens were cleaned in 5% bleach between samples and were rinsed thoroughly with deionized (d)H2O to eliminate cross-contamination of Alexandrium DNA and other remaining debris. Approximately 0.5–3 ml of the resulting < 53-μm sediment slurry was added to three replicate 100 × 15 mm polystyrene Petri dishes containing 30 ml of filtered seawater at a salinity of 25. Next, 60 μl of NaNO3 stock (1.76 × 10−3 M), 30 μl of Na2 β-glycerophosphate stock (1.00 × 10−5 M), and 60 μl of germanium dioxide stock (1.23 × 10−4 M) were added to each dish. The dishes were then gently swirled to mix the contents and were incubated using the same conditions as above. The plates were checked for Alexandrium cells over the next 7 days using a stereoscope at ×40 magnification. Parallel 0.5-ml aliquots of sediment slurry were also filtered onto 47-mm, 8-μm pore size polycarbonate filters and subjected to DNA extraction as described below. Once motile Alexandrium cells were observed in the dishes they were collected with a pipette and transferred to a 55-mm Petri dish for isolation. A single cell was picked from the Petri dish using a micropipette and sequentially transferred through three to four drops of sterile medium. After each transfer, the drop was examined to ensure that only a single cell was present. After the final transfer, each isolated cell was placed into a separate well of a sterile 6- or 24-well tissue culture plate with a small volume of modified K medium (salinity 25). Isolates were then incubated as described above. When sufficient cell density was achieved, the clonal cultures were transferred to 50-ml glass Erlenmeyer flasks containing modified K medium and were eventually transferred to 250-ml glass Erlenmeyer flasks. Additional cultures were obtained from the National Center for Marine Algae and Microbiota (NCMA), reisolated, and maintained as described above to supplement the material isolated from Alaska. The genetic identities of clonal cultures established from sediment samples and from isolates established from NCMA were confirmed by sequencing the D1-D3 region of the LSU ribosomal gene as described below to ensure that they matched published sequences in GenBank. The Alexandrium cultures used in this study are listed in Table 1.
Table 1.
Alexandrium cultures used in this study.
| Strain | Species | Location |
|---|---|---|
| CCMP 112 | A. affine | Ria de Vigo, Spain |
| CCMP 113 | A. minutum | Ria de Vigo, Spain |
| CCMP 115 | A. tamarense Group III | Tamar Estuary, Plymouth, UK |
| CCMP 116 | A. affine | Ria de Vigo, Spain |
| CCMP 1598 | A. pacificum Group IV | Da-ya Bay Guan-dong (Canton) Province, bay west of Hong Kong Island, China |
| CCMP 1718 | A. andersoni | Town Cove, Eastham, Massachusetts, USA |
| CCMP 1719 | A. fundyense Group I | Portsmouth, New Hampshire, USA |
| CCMP 1771 | A. tamarense Group III | Tamar Estuary, Plymouth, UK |
| CCMP 1773 | A. ostenfeldii | Lim Fjorden, Denmark |
| CCMP 1888 | A. minutum | Laguna Obidos, Portugal |
| CCMP 1908 | A. fundyense Group I | East Sound, Washington, USA |
| CCMP 2082 | A. insuetum | Uchiumi Bay, Kagawa, Japan |
| CCMP 2215 | A. hiranoi | Jogashima, Misaki, Kanagawa, Japan |
| CCMP 2955 | A. leei | Singapore Strait, Singapore |
| Amon Miss VIMS | A. monilatum | Gulfport, Mississippi, USA |
| Amon YK VIMS | A. monilatum | York River, Virginia, USA |
| AoF0933 | A. ostenfeldii | Baltic Sea, Åland, Finland |
| AoNOR4 | A. ostenfeldii | Oslofjorden, Norway |
| AoTVA4 | A. ostenfeldii | Baltic Sea, Åland Finland |
| Ap002/B2NR | A. ostenfeldii | New River, North Carolina, USA |
| Ap003/A4NR | A. ostenfeldii | New River, North Carolina, USA |
| Ap004/A2NR | A. ostenfeldii | New River, North Carolina, USA |
| Ap006/A3NR | A. ostenfeldii | New River, North Carolina, USA |
| Auke Bay 1 | A. fundyense Group I | Juneau, Alaska, USA |
| Auke Bay 4 | A. fundyense Group I | Juneau, Alaska, USA |
| Auke Bay 9 | A. fundyense Group I | Juneau, Alaska, USA |
| Dentist Dock 1 | A. fundyense Group I | Ketchikan, Alaska, USA |
| Dentist Dock 2 | A. fundyense Group I | Ketchikan, Alaska, USA |
| Knudson Cove | A. fundyense Group I | Ketchikan, Alaska, USA |
| Mill Bay 3 | A. fundyense Group I | Kodiak Island, Alaska, USA |
| Mission Beach | A. fundyense Group I | Kodiak Island, Alaska, USA |
| Sargent Creek 1 | A. fundyense Group I | Kodiak Island, Alaska, USA |
| Sargent Creek 5 | A. fundyense Group I | Kodiak Island, Alaska, USA |
| Sargent Creek 6 | A. fundyense Group I | Kodiak Island, Alaska, USA |
| Sargent Creek 8 | A. fundyense Group I | Kodiak Island, Alaska, USA |
| Thomas Basin 1 | A. fundyense Group I | Ketchikan, Alaska, USA |
| Thomas Basin 2 | A. fundyense Group I | Ketchikan, Alaska, USA |
| Trident 1 | A. fundyense Group I | Kodiak Island, Alaska, USA |
| USCG 2 | A. fundyense Group I | Kodiak Island, Alaska, USA |
| Womens Bay 1 | A. fundyense Group I | Kodiak Island, Alaska, USA |
DNA extraction and sequencing protocols
DNA extraction, PCR amplification of the 3′ SSU through the 5′ LSU rDNA region, subcloning, and sequencing were performed using reagents and methods adapted from Vandersea et al. (2012). Approximately 10 ml of each Alexandrium culture (Table 1) at 1 × 103 cells·ml−1 were concentrated by filtration onto a 47-mm, 8-μm pore size polycarbonate filter (Whatman Nucleopore™, GE Healthcare Bio-sciences, Pittsburgh, Pennsylvania, USA). DNA was extracted from each filter using the Mo Bio Laboratories Power® Soil DNA isolation kit (Mo Bio Laboratories, Solana Beach, California, USA) following the manufacturer’s protocol except that 350 μl of cell lysate rather than the prescribed 450 μl was processed. The DNA extracts were eluted from the minicolumns using 50 μl of elution buffer. Spectrophotometric analysis (260 nm/280 nm) of the DNA was conducted to assess DNA concentration and purity. The DNA extracts were stored at 4°C. A ~1700-base pair (bp) product containing the 3′ SSU, ITS, and 5′ LSU rDNA domains was PCR amplified using the Alex1600F and LSUB primers (Table 2). The PCR amplification reaction mixtures contained 20 mM Tris-HCl, pH 8.4, 3 mM MgCl2, 50 mM KCl, 25 pmol of each primer, 2.5 mM of each deoxynucleoside triphosphate, 0.2 units of Platinum Taq DNA polymerase (Invitrogen™ Life Technologies, Carlsbad, California, USA), and 10 ng of genomic DNA in a total reaction volume of 50 μl. A Bio Rad PTC-100 Peltier thermal cycler (Bio Rad, Hercules, California, USA) was used to conduct the PCR with the following cycling conditions: 2 min at 95°C, 35× (30-s denaturation at 95°C, 30-s annealing temperature at 60°C, and an extension of 1.5 min at 72°C), and a final 5-min extension at 72°C. A 5-μl aliquot of each PCR reaction was checked for the presence of a specific amplification product by agarose gel electrophoresis (2% Tris-acetate/EDTA, 100 V) and ethidium bromide staining. The PCR products were cloned into the plasmid vector pCR2.1® (Invitrogen Life Technologies) using the TOPO TA cloning® kit following the manufacturer’s protocol. Plasmids were isolated and purified using the Promega Wizard® Plus Minipreps DNA purification system (Promega, Madison, Wisconsin, USA) and sequenced using an ABI3730xl DNA sequencer using the Deoxy™ Terminator Cycle sequencing kit (Applied Biosystems - ABI, Foster City, California, USA). DNA templates were sequenced completely in both directions using the primers listed in Table 2. The 3′ SSU to 5′ LSU sequence was assembled using the Vector NTI Advance™ 11 program (Invitrogen Life Technologies) and the resulting sequences compared with those in GenBank using basic local alignment search tool (BLAST) searches (Altschul et al. 1997) to establish the species identity of each isolate. The gene sequences obtained in this study were deposited in GenBank. (GenBank acession numbers: A. affine KX599339; KX599340, A. andersoni KX599343, A. fundyense KX599344; KX599345; KJ127879-KJ127911, A. minutum KX599341; KX599348, A. pacificum KX599346; KX599347, A. tamarense KX599342).
Table 2.
Oligonucleotide primers used in this study to amplify and sequence Alexandrium SSU, ITS1, 5.8S, ITS2, and the D1–D3 domain of the LSU.
| Primer name | Sequence (5′−3′) | Melting temperature, °C |
|---|---|---|
| PCR amplification primers | ||
| Alex1600F | GCYTGAGTCATCAGCTTGTGC | 64 |
| LSUB | ACGAACGATTTGCACGTCAG | 60 |
| Sequencing primers | ||
| 5.8SF | CATTGTGAATTGCAGAATTCC | 58 |
| Alex5.8SR | GCTCACGGAATTCTGCAATTC | 62 |
| AlexLSURl | CCACCCACTTTGCATTC | 52 |
| D1R | ACCCGCTGAATTTAAGCATA (Scholin et al. 1994) | 56 |
| M13F | GTAAAACGACGGCCAG | 50 |
| M13R | CAGGAAACAGCTATGAC | 50 |
Screening of environmental samples using the species-specific PCR assays
Water samples, sediment, and cell isolates from Ketchikan, Juneau, Kachemak Bay, and Kodiak Island were processed as described above. Genomic DNA was extracted from the phytoplankton or sediment samples collected at all four sampling locations (Figs 1–5) according to the methods described above. The resulting DNA extracts were used as the template for the species-specific PCR assays.
Design and testing of species-specific Alexandrium PCR assays
The SSU/ITS/LSU rDNA sequences of Alexandrium species obtained in this study were aligned with corresponding SSU, ITS, and D1-D3 LSU rDNA sequences from other Alexandrium species available from GenBank using the ClustalX program (Thompson et al. 1997) and sorted into species groups using the phylogenetic analyses methods described in John et al. (2014a). The species included in the alignments were A. affine (Inoue & Fukuyo) Balech, A. andersoni Balech, A. hiranoi Kita & Fukuyo, A. insuetum, A. minutum Halim, A. monilatum (J.F. Howell) Balech, A. cohorticula (Balech) Balech, A. tamiyavanichi Balech, A. fraterculus (Balech) Balech, A. leei Balech, A. margalefii, A. ostenfeldii, A. pseudogonyaulax, A. satoanum Yuki & Fukuyo, A. tamutum Montresor, A. taylori Balech, A. tropicale Balech, A. fundyense Group I, A. tamarense Group III, A. pacificum Group IV, and A. mediterraneum Group II John (John et al. 2014a). The D1-D3 LSU alignments were used for developing species-specific assays for A.fundyense Group I, A. minutum, A. monilatum, A. ostenfeldii, A. pacificum Group IV, and A. tamarense Group III. Unique forward and reverse species-specific primer sites were visually identified within the alignments for each species. The primer pairs were selected with the object of limiting the PCR amplicons to less than 200 bp in length. Each primer was also subjected to a BLAST search using the National Center for Biotechnology Information tool to determine potential cross-reactivity with non-Alexandrium species (Altschul et al. 1997). Primers that exhibited cross-reactivity with other Alexandrium species or with other dinoflagellates were eliminated from consideration. The primers were then tested systematically for secondary PCR products using genomic DNA extracted from the intended target species using the amplification conditions described below. Successful candidate primer pairs were analyzed for species specificity and cross-reactivity using genomic DNA extracted from other Alexandrium species (Table 1). The primer pairs chosen for final PCR assay validation are listed in Tables 3 and S1 (Vandersea et al. this study).
Table 3.
Excerpt from a literature survey completed in 2015 of the various probes available for detecting Alexandrium species. The complete literature survey is presented as supplemental material in Table S1. The literature citation, rDNA target domain, the original species the probe was designed to target, the probe sequences, and the actual species the probes will detect on the basis of current taxonomy are listed. The reverse primers and in situ hybridization probes target the noncoding strand. The probes are all listed in 5′–3′ orientation. The ribosomal gene locations of the primers and probes listed in this table are also identified in Figs 14 and S7–S9 on the species coding strand in 5′–3′ orientation.
| PCR primers |
Reporter probe/Detects |
Forward probe |
Reverse probe |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Reference | Domain | Target species | Assay type | Forward primer coding (sense) strand | Reverse primer/in situ probe noncoding (antisense) strand | Coding strand | BLAST search results | BLAST search results | |
| 56 | Vandersea et al., this study | LSU D1-D2 | A. fundyense | qPCR SYBR green | GATAAGTCTCCTGT GGGGGG | AAGCACAGGAACACA CACATA | A. fundyense | A. fundyense | |
| LSU D1-D2 | A. pacificum | PCR | CTTGCTTTGTGTGCCAGTTTT | ACCTCAAGGACAAGGACACAA | A. pacificum | A. pacificum | |||
| LSU D1-D2 | A. minutum | PCR | TTGTGCTTACTCTATCATTTATAT | AAATTTCACCAAACACATGCGT | A. minutum | A. minutum | |||
| LSU D1-D2 | A. monilatum | PCR | TGTTTGCATTTATGTGTTGAACA | CACAGGCACCTTACCTTTCAT | A. monolatum | A. monolatum | |||
| LSU D1-D2 | A. monilatum | qPCR TaqMan | TGAAAGGTAAGGTGCCTGTG | GCAGAAACATGTTGCCAAAG | TGCAAGCACAAGCAA CCCAGC A. monilatum | A. monolatum | A. monolatum | ||
| LSU D1-D2 | A. ostenfeldii | qPCR SYBR green | TGAGATTGTTGCGTCCACTTGT | AGGGAGGAGAGCCCTCCC | A. ostenfeldii | A. ostenfeldii | |||
| LSU D1-D2 | A. tamarense | PCR | TGAGATTGTTGCGTCCACTTGT | AGGGAGGAGAGCCCTCCC | A. tamarense | A. tamarense | |||
| 57 | Zhang & Li 2012 | 5.8S | Alexandrium genus | qPCR SYBR green | GATGAAGAATGCAGCAAAATG | CAAACCTTCAAGAATATCC | Alexandrium genus |
A.
fundyense A. pacificum A. tamarense |
|
| 58 | Zhen et al. 2011 | SSU | A. catenella | nuclease protection - sandwich hybridization | ATTTGGCACAGCCTGAGCATTTATC capture probe |
A.
australiense A. pacificum A. tamarense A. fundyense A. tamiyavanichii |
|||
| SSU | CACACCACACAGTCAAGTGCAGTTGTGCTTTCAAGATAAATGCTCAGGCTGTGCCAAAT nuclease protection probe |
A. affine A. cohorticula A. fundyense A. pacificum A. tamarense |
|||||||
| SSU | ACAACTGCACTTGACTGTGTGGTGTG signal probe | A. tamiyavanichii Alexandrium genus | |||||||
To further confirm species specificity, a mixture of DNA from seven Alexandrium species (A. affine, A. andersoni, A. fundyense Group I, A. minutum, A. ostenfeldii, A. pacificum Group IV, A. tamarense Group III) was PCR amplified using each of the PCR assay primer sets. The PCR products were purified using the Invitrogen Pure Link™ PCR purification kit following the manufacturer’s protocol, cloned using the TOPO TA cloning kit, and sequenced using an ABI3730xl DNA sequencer. The sequences were aligned and checked to ensure that they corresponded to the intended target species.
PCR assay cycling conditions
PCR assays were performed using an Eppendorf Master-cycler® ep realplex 4 system with white Eppendorf real-time tube strips (Eppendorf North America, Inc., Westbury, New York, USA) and a total reaction volume of 10.5 μl per tube. Each PCR reaction mixture contained 4.5 μl of 5 Prime RealMasterMix SYBR ROX 2.5× [0.05 units μl−1 Taq DNA polymerase, 10 mM Mg(CH3COO)2, 1.0 mM deoxynucleotide triphosphates, 20× SYBR® Green solution], each primer at a concentration of 0.15 μM, 4.7 μl of sterile deionized water, and 1 μl of template DNA. Thermal cycling conditions included denaturation at 95°C for 2 min followed by 40 cycles at 95°C for 10 s and annealing for 15 s at 60°C, with a subsequent extension at 68°C for 20 s. The fluorescence threshold was determined by the Eppendorf realplex 4 analytical software, and the PCR cycle during which fluorescence crossed the threshold was designated the quantification cycle (Cq) as defined by Bustin et al. (2009). A melting-curve analysis was performed after thermal cycling to check the specificity of the PCR reactions. The melting-curve profile consisted of denaturation at 95°C for 15 s followed by an annealing step for 15 s at 60°C. The fluorescence was continuously monitored during a steady 20-min temperature ramp from 60 to 95°C that was held at 95°C for 15 s. The melting-curve analysis was conducted by comparing the melting-temperature peak of positive control DNA to other experimental DNA samples. A limit of ±0.5°C for melting-temperature peak shift was set as the cutoff for species-specific amplifications.
Conversion of PCR assays into qPCR assays, qPCR standard curves, amplification efficiency, and limits of detection
The PCR assays were converted into qPCR assays using methods adapted from Vandersea et al. (2012). To construct cell-based standard curves, multiple strains (n = 5) of A. fundyense Group I cultures in log-phase growth were sieved through 50-μm mesh to remove cell debris and multicell aggregates. The sieved cells were gently mixed to ensure homogenization and then cell concentrations were measured in triplicate using a Beckman Coulter Multisizer™ 3 fitted with a 280-μm aperture tube (Beckman Coulter Inc., Brea, California, USA).
Individual cell dilutions ranging from 100 to 150,000 cells were prepared in triplicate using sterile filtered seawater at a salinity of 25. For each replicate sample 100 ml were concentrated by filtration onto 47-mm, 8-μm pore size polycarbonate filters as above. To obtain accurate results for low-range cell concentrations (10–100 cells), the cells were manually pipetted onto each polycarbonate filter and counted using a stereomicroscope. Using a range of cell concentrations to construct standard curves provides a more accurate calculation of qPCR cell concentration than amplifying DNA diluted from a single known cell concentration, as the latter method can greatly overestimate or underestimate actual cell concentrations from field samples. Genomic DNA extraction and qPCR amplification were carried out as described above. Each assay incorporated 1 μl of the 50-μl DNA extract in the qPCR reaction mix. Standard curves were constructed by plotting the triplicate Cq values against the log-transformed cell concentrations. A regression analysis of the Cq vs log-transformed cell concentrations was performed to calculate the slope values and confirm the linearity of the standard curves. The qPCR assay limit of detection (LOD) for the cell-based standard curves was the lowest cell concentration, ranging over four orders of magnitude, that amplified and yielded linear Cq values.
PCR amplicons were serially diluted and used to construct standard curves after the PCR amplicon copy numbers were determined. First, rDNA fragments for each species were PCR amplified using the Alex1600F and LSUB primers that flanked the qPCR assay target sites. The rDNA fragments were then cloned into the plasmid vector pCR2.1 using the TOPO TA cloning kit. Plasmid DNAs were isolated and purified using the Promega Wizard Plus Minipreps DNA purification system and stored at −20°C. The plasmid DNAs served as templates for producing the PCR amplicons used to construct the standard curves. When PCR amplicons were needed to construct standard curves, stock plasmids were thawed, diluted 1:3000 in sterile deionized water, and PCR amplified using the M13F and M13R primers. The PCR amplification reaction mixtures contained 20 mM Tris-HCl, pH 8.4, 3 mM MgCl2, 50 mM KCl, 25 pmol of each primer, 2.5 mM each deoxynucleoside triphosphate, 0.2 units of Platinum Taq DNA polymerase, and 10 ng of genomic DNA in a total reaction volume of 50 μl. The PCR amplification profile used to amplify ~ 1700-bp insert was as follows: 2 min at 95°C, 35× (30 s at 95°C, 30 s at 50°C, and 1.5 min at 72°C), and a final extension of 5 min at 72°C. The PCR reactions were checked for the presence of a specific amplification product by agarose gel electrophoresis (2% Tris-acetate/EDTA, 100 V) and ethidium bromide staining. PCR reactions containing specific amplicons were purified using the Invitrogen Pure Link PCR purification kit (Invitrogen Life Technologies) following the manufacturer’s protocol. The DNA concentration of the purified amplicons was estimated spectrophoto-metrically by measuring ultraviolet absorbance at 260 nm. The number of target copies per microlitre of stock solution for each species-specific PCR amplicon was determined as described in Vandersea et al. (2012).
To construct standard curves, the purified amplicons were serially diluted 1:10 in dH2O and encompassed six orders of magnitude. Each serial dilution was qPCR amplified for 40 cycles using the qPCR assay primers designed to amplify each species, followed by a melting-curve analysis to check for secondary qPCR products and primer dimers. Only Cq values from qPCR reactions that produced a specific product within 35 amplification cycles were included in the standard curves. For each standard curve, the Cq values generated from the 10-fold serial dilutions were plotted against the log-transformed copy numbers to obtain regression equations. A regression analysis was performed to calculate the slope values and confirm the linearity of the standard curves. The qPCR assay LOD for the diluted PCR amplicon-based standard curves was the lowest copy number concentration ranging over six orders of magnitude that amplified yielding linear Cq values. Regression equations derived from the standard curves were used to estimate the number of PCR amplicons per cell as described below.
Estimating cell numbers using qPCR standard curves
To obtain qPCR cell number estimates, the ratio of extractable PCR amplicons per cell was determined. The number of extractable PCR amplicons per cell was calculated by solving the regression equations derived from the diluted PCR amplicon-based standard curves using Cq values acquired from qPCR amplification of known numbers of Alexandrium cells. These values represented a functional ratio of the numbers of copies extracted per cell and cannot be used to calculate the number of rDNA copies per cell. An essential criterion for using this approach is that the slope values of the diluted PCR amplicon standard curves and cell-based standard curves must be almost equivalent (i.e. differ by ≤ 0.1). Adhering to this criterion ensures that the qPCR efficiency of both standard curves is nearly equivalent, facilitating a valid conversion of extractable PCR amplicons per cell to cell numbers (Bustin et al. 2009).
qPCR assay controls
To assess potential DNA contamination and PCR inhibitors in the extracted field samples, each qPCR assay included a positive control, a negative DNA control, a blank extraction control, and two supplemented DNA controls. The positive control contained a known amount of target DNA in the qPCR reaction mixture and ensured that the qPCR reagents were properly assembled and the DNA Taq polymerase was functional. The negative control included the addition of 1 μl of reaction buffer to a subset of reaction mixes to test for contaminated reagents or cross-contamination between samples. The blank controls were incorporated during the DNA extractions of the field samples to test for potential DNA contamination during the extraction process. Supplemented controls consisted of adding target DNA to a subset of the field samples to determine if PCR inhibitors were present.
Experimental analysis of qPCR accuracy
To check the accuracy and sensitivity of the qPCR assays, known concentrations of multiple Alexandrium isolates were mixed together and the live cells were enumerated using a Beckman Coulter Multisizer 3 fitted with a 280-μm aperture tube. Dilution series containing multiple strains of either A. fundyense Group I or A. ostenfeldii at concentrations ranging from ~0.10 to 9800 cells ml−1 were made in triplicate in natural seawater collected from the Center for Coastal Fisheries and Habitat Research’s dock in Beaufort, North Carolina. The diluted cells were concentrated onto 47-mm polycarbonate filters and the DNAs were extracted and qPCR assayed. Standard curves constructed from diluted PCR amplicons were run at the same time. The accuracy of the assays was determined by correlating qPCR cell estimates to the cell dilutions that were based on the multisizer cell counts.
Literature review of species-specific assays for Alexandrium species
To facilitate future molecular studies of Alexandrium species, we surveyed the literature for species-specific molecular assays that targeted the SSU through D1-D3 LSU rDNA region. A comprehensive table was assembled that included the author citation, the targeted ribosomal gene, the target species as published, the type of assay, the sequences of the primers or probes, and their species specificities (Tables 3 and S1). Each of the species-specific primer and probe sequences was manually mapped onto consensus SSU, ITS/5.8S, and D1-D3 alignments. The alignments were prepared as described in John et al. (2014a) and used to analyze the gene regions with the greatest concentration of species-specific sequences, and the relative intra- vs interspecific sequence variation. Analogous alignments were constructed for the ITS and SSU regions.
RESULTS
PCR assay of environmental samples
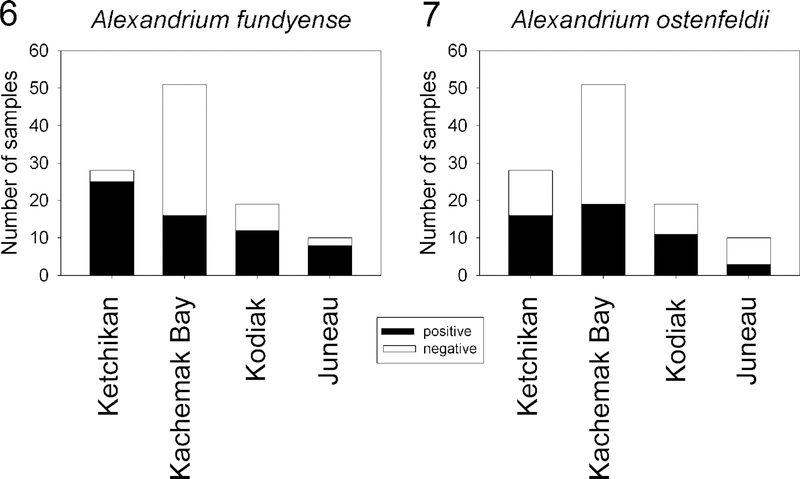
The species-specific PCR assays for Alexandrium fundyense Group I, A. pacificum Group IV, A. tamarense Group III, A. ostenfeldii, (Kremp et al. 2014) A. minutum, and A. monilatum [=Gonyaulax monilatum (Howell) Loeblich III] developed in this study did not cross-react with nontarget species as determined by melting-curve and sequencing analysis of PCR products (Figs S1–S6). They were successfully used to screen 108 water and sediment samples as well as 17 cell isolates collected from 10 stations at four coastal sites in Ketchikan, Juneau, Kachemak Bay, and Kodiak Island from September 2009 to November 2011 (Figs 1–5). Of the six species assayed, only A. fundyense Group I and A. ostenfeldii were detected in the four geographically isolated regions (Figs 6, 7). All 17 of the cell isolates were identified genetically as A. fundyense Group I (Table 1). Fifty-three percent of the environmental samples were positive for A. fundyense Group I, whereas 47% were positive for A. ostenfeldii. Alexandrium fundyense Group I was present in the majority of the samples from Ketchikan, Juneau, and Kodiak Island, but in only 31% of the samples from Kachemak Bay (Fig. 6). Alexandrium ostenfeldii was present in 58% of the samples from Ketchikan and Kodiak Island, in 37% of the Kachemak Bay samples, and in 30% of the Juneau samples (Fig. 7). Alexandrium fundyense Group I and A. ostenfeldii occurred together in 57% of the positive samples. Attempts to establish isolates of A. ostenfeldii from either phytoplankton or sediment samples were not successful. Whether this was due to low numbers of cells or unfavourable culture conditions was not determined.
Figs 6, 7.
Six PCR assays were developed in this study and used to screen 108 water and sediment samples collected from coastal sites in Ketchikan, Kachemak Bay, Juneau, and Kodiak Island during September 2009-November 2011. The graphs show the proportion of PCR-positive and -negative samples for each sampling area. The analysis combined data obtained from isolates, sediment, and water samples. Alexandrium fundyense Group I (Fig. 6) and A. ostenfeldii (Fig. 7) were the only species detected.
Assay sensitivity and amplification efficiency
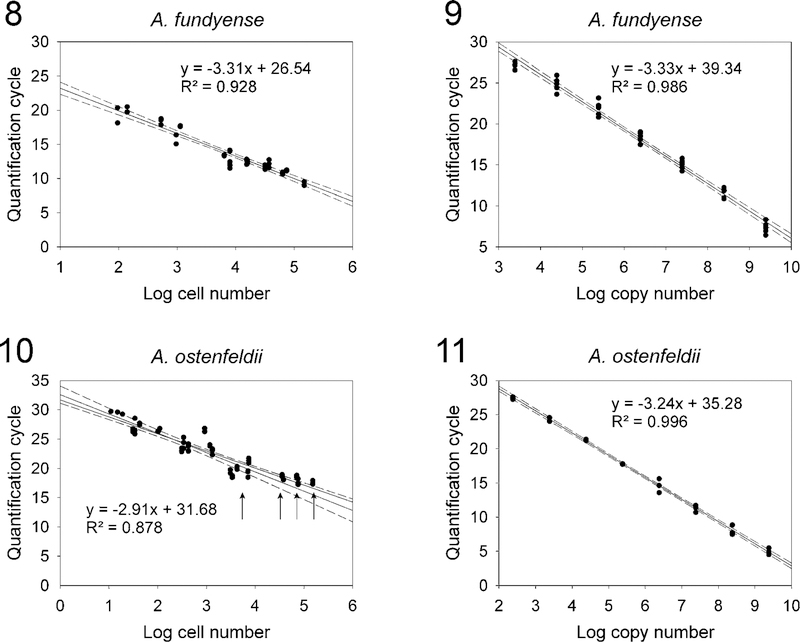
The A. fundyense Group I qPCR assay sensitivity was tested using cell concentrations ranging from ~0.01 to 14,800 cells·mL−1 diluted in local seawater. The A. ostenfeldii qPCR assay sensitivity was tested using cell concentrations ranging from 0.01 to 18,967 cells·ml−1. Both assays were capable of detecting less than 10 cells in a 1-litre sample, but the A. fundyense Group I Cq values were inconsistent below 0.09 cells·ml−1, whereas the A. ostenfeldii Cq values were inconsistent below 0.01 cells·ml−1 and fell outside the linear ranges of the standard curves. The assay sensitivities were also tested using serial dilutions of PCR amplicons that contained appropriate target sequence. The LOD for A. fundyense Group I assay was ~2440 copies and the LOD for A. ostenfeldii assay was ~241 copies. The standard curves were linear over at least six orders of magnitude. Amplification efficiencies were calculated using typical slope values of the regression lines for Cq vs log-transformed cell numbers and Cq vs log-transformed copy numbers using the equation E = 10(−1/slope) – 1. For the A. fundyense Group I assay, typical slope values approached the theoretical optimum of −3.3 and the amplification efficiencies ranged from 0.996 to 1.005. For the A. ostenfeldii assay typical slope values of the regression for Cq vs log-transformed copy number also approached −3.3, with an amplification efficiency of 0.979. In contrast, the slope of Cq vs log-transformed cell numbers was −2.91, with an amplication efficiency of 1.206. The differences in the slopes of the cell-based standard curves and diluted PCR amplicon standard curves were compared. The A. fundyense Group I assay varied by less than 0.1, whereas the A. ostenfeldii assay varied by 0.39. The linear relationships and representative data from the standard curves that were used to calculate the regressions are shown (Figs 8–11). The empirical relationship between PCR amplicons and cell numbers for A. fundyense Group I and A. ostenfeldii was derived by solving the regression equations for the PCR amplicon and cell-based standard curves using the methods described in Vandersea et al. (2012). The relationship between PCR amplicon number and cell number for A. fundyense Group I was determined to be 11,107 amplicons per cell and 12 amplicons per cell for A. ostenfeldii.
Figs 8–11. Alexandrium fundyense and Alexandrium ostenfeldii qPCR standard curves.
Fig. 8. Representative Alexandrium fundyense Group I qPCR assay standard curve constructed using a range of known cell concentrations. The solid line indicates the regression of log cell numbers vs quantification cycle (Cq). The 95% confidence interval is represented by the dashed lines.
Fig. 9. An Alexandrium fundyense qPCR assay standard curve constructed using 10-fold serial dilutions of a purified PCR amplicon containing the A. fundyense qPCR assay target domain. The regression of log copy numbers vs quantification cycle was plotted and dashed lines show the 95% confidence interval.
Fig. 10. Same as Fig. 8 except that Alexandrium ostenfeldii cells were used. The arrows indicate where Cq values obtained from cell concentrations ranging from ~5600 to ~ 151,000 cells plateaued at roughly 18 cycles and signaled the assay’s upper limit of detection.
Fig. 11. Same as Fig. 9 except the PCR amplicon contained the A. ostenfeldii qPCR assay target domain.
Assay accuracy
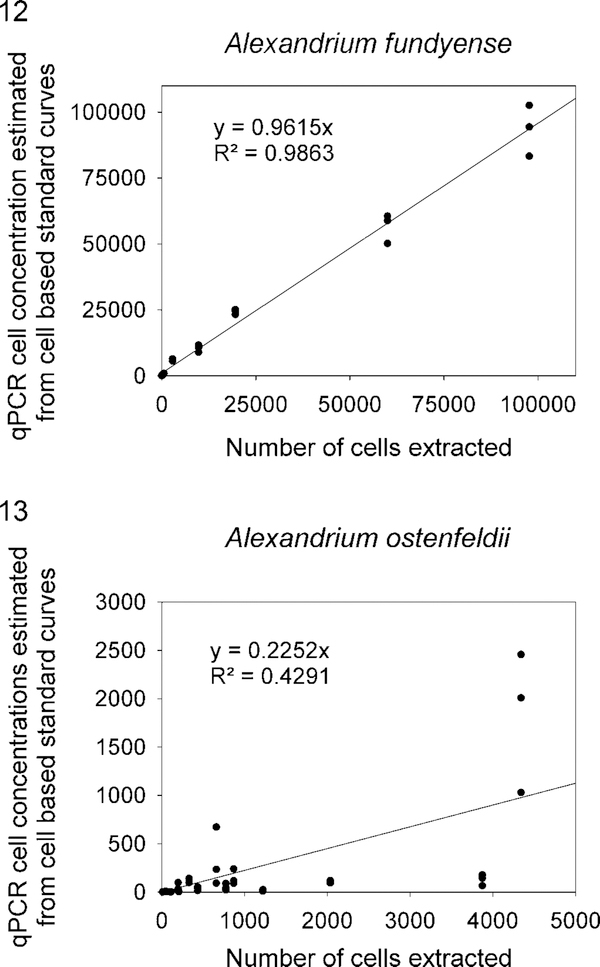
Five experiments using known concentrations of cells that were enumerated using a Beckman Coulter Multisizer were conducted to assess the accuracy of the A. fundyense Group I and A. ostenfeldii qPCR assays. Known numbers of cells were mixed with 100 ml of seawater from Beaufort, North Carolina. Each experiment contained three replicates of multiple strains of each species ranging from 10 to 9800 cells·mE−1 as well as standard curves constructed using serially diluted PCR amplicons. The qPCR assay cell concentration estimates for A. fundyense Group I exhibited a strong correlation with the multisizer cell counts (Fig. 12). In contrast, the qPCR assay cell concentration estimates for A. ostenfeldii did not correlate well with the multisizer cell counts (Fig. 13).
Figs 12, 13.
The relationship between known concentrations of cells of Alexandrium fundyense or A. ostenfeldii spiked into seawater samples and cell numbers estimated using qPCR.
Evaluation of the specificity of published Alexandrium molecular assays as a basis for developing standard assays
Alexandrium molecular assays were listed alphabetically by first author in Tables 3, 4 and S1. Each of the primers or probes was checked for cross-reactivity with other species using the National Center for Biotechnology Information (NCBI) nucleotide BLAST tool (Altschul et al. 1997). To convey a visual presentation of the BLAST results, the molecular probe and primer locations were manually mapped onto Alexandrium species consensus sequence alignments of the SSU/ITS/LSU rDNA genes (Figs 14 and S7–S9).
Table 4.
Additional references cited in a literature survey of Alexandrium molecular assays presented in supplemental materials Table S1 and Figs S7–S9.
Fig. 14.
Consensus sequences for Alexandrium species were aligned and used to map the location of hybridization sites in the LSU D1-D2 rDNA region. The figure shows one subsection of the D1-D2 region. The entire data set is presented in Fig. S7. Sequences from many species clustered into distinct subgroups. Identification of the subgroups including GenBank accession numbers is described in the Material and Methods section and the supplementary data in John et al. (2014a). To better illustrate differences among species, only variations from the consensus sequence are shown. A space in a position above the consensus sequence indicates that the base pair in that position is identical to the consensus sequence. A dash in a given position indicates that there is no corresponding base pair in the consensus sequence (i.e. missing nucleotide). Note: To simplify the figure, only coding strand sequences are displayed, including the reverse PCR primers and probes. All primers and probe locations are identified on the coding strand in 5′–3′ orientation. The sequences for the noncoding strand primers and probes are listed in Tables 3 and S1. The numbers in parentheses next to the primers and probe sequences correspond to authors listed in Tables 3 and S1.
DISCUSSION
Two toxic Alexandrium species, A. fundyense Group I and A. ostenfeldii, were found in sediment and water samples obtained from coastal sites in Ketchikan, Juneau, Kachemak Bay, and Kodiak (Figs 1–5). Alexandrium pacificum Group IV, A. tamarense Group III, A. minutum, and A. monilatum were not detected. These findings are significant because they are the first unambiguous report of Alexandrium species distribution for Alaskan coastal waters. They are consistent with the molecular results of Natsuike et al. (2013), who detected A. fundyense Group I cysts in sediments collected from the Bering and Chukchi Seas, often at extremely high densities. Even though our sampling efforts and those of Natsuike et al. (2013) suggest low Alexandrium diversity in this region, it is possible that with more intense sampling efforts other Alexandrium species will be identified in Alaskan waters using molecular methods.
The original reports of A. catenella occurring in Alaskan coastal waters (Horner et al. 1997; Trainer 2002) were based primarily on the observation that cells of A. catenella formed chains, which was previously considered a morphologically diagnostic feature for the species (Balech 1995). However, the review conducted by John et al. (2014a) demonstrated that chain formation is not a reliable species characteristic. Indeed, the only A. tamarense complex species identified from Alaska in our study was A. fundyense Group 1, which is also known to form chains. Similarly, A. fundyense Group I and A. tamarense Group III have been shown to be morphologically indistinguishable (John et al. 2014a), indicating that the report of A. tamarense in Alaskan waters by Trainer (2002) may have been due to misidentification of A. fundyense Group I cells, as the molecular screening conducted in this study found no A. tamarense Group III cells.
Interestingly, A. fundyense Group I is the dominant species found in eastern Canada and the Gulf of Maine, where it co-occurs with A. ostenfeldii (McGillicuddy et al. 2014). Though not conclusive, the similarity in the Alexandrium species composition between Alaska and eastern Canada and the Gulf of Maine is consistent with the hypothesis of Scholin et al. (1995), that eastern and western populations of Alexandrium may have originated from the same parental stocks. Given that A. fundyense Group I is the principal source of STXs in the Bay of Fundy and Gulf of Maine (Martin & Richard 1996; Anderson et al. 2005a, 2014; Martin et al. 2008; McGillicuddy et al. 2014), the presence of this species in the Gulf of Alaska likely accounts for the persistent occurrences of PSP in Alaska. This conclusion is supported by the identification of A. fundyense Group I in samples collected in this study (Thomas Basin, Mountain Point; Fig. 2) during a significant PSP event near Ketchikan, Alaska where 21 cases of PSP occurred in May-June 2011 (Porter et al. 2011).
The extent to which A. ostenfeldii contributes to the toxicity of shellfish in Alaska is unknown. Some strains of A. ostenfeldii have been reported to exhibit low toxicity, whereas others are known to be very toxic (Hansen et al. 1992; Mackenzie et al. 1996). Borkman et al. (2012) found STX production in A. ostenfeldii isolates obtained from Narragansett Bay, Rhode Island, as did Brown et al. (2010) from Scottish isolates. Toxicity testing of A. ostenfeldii isolates from Alaska is needed to address this question.
Historically, shellfish toxicity testing has been used to extrapolate Alexandrium species distributions and cell concentrations in Alaska (Hall 1982; Horner et al. 1997). Though indicative of where PSP problems are most severe, this type of monitoring does not provide crucial, real-time warning capability to identify the early stages of bloom development that can precede the onset of shellfish toxicity by days to weeks (Neale 1967; Matweyou 2003; Lefebvre et al. 2008). Initially, the A. fundyense Group I and A. ostenfeldii assays were used qualitatively in this study for species detection. After the presence of these species was confirmed in the environmental samples, the species-specific assays were converted into a qPCR format that could be used in near real time to determine Alexandrium distributions and relative abundances of cells. The feasibility of using the assays for these purposes was tested by adding known concentrations of A. fundyense and A. ostenfeldii into local filtered seawater and then comparing qPCR cell estimations to cell counts. We used this approach because the environmental samples collected in Alaska for this study were not collected quantitatively. Unfortunately, this negated our ability to make quantitative estimates using qPCR methods on the samples. Figs 12 and 13 show the qPCR assay cell estimates of supplemented seawater plotted against the cell numbers included in the samples. The A. fundyense qPCR cell estimates exhibited a strong positive correlation with Coulter counter-based cell counts and confirmed that the qPCR assay was quantitative (R2 = 0.987, Fig. 12). In contrast, the A. ostenfeldii qPCR cell estimates did not correlate with Coulter counter-based cell counts and was not quantitative (R2 = 0.437, Fig. 13), leading to the obvious question of why the A. fundyense qPCR assay was quantitative and the A. ostenfeldii was nonquantitative. One possibility we explored was qPCR inhibition. To test this, A. ostenfeldii DNA extracts were diluted 1:10 and 1:100 and PCR amplified. The results did not indicate qPCR inhibition. Supplemented control assays were also conducted in which concentrated genomic A. ostenfeldii DNA was added into replicate A. fundyense qPCR reactions and the Cq values of nonsupplemented A. fundyense qPCR reactions were compared with supplemented A. fundyense Cq values. On the basis of the equivalent Cq values of the supplemented and nonsupplemented A. fundyense assays, there was no sign of qPCR inhibition.
Inconsistent cell lysis could also account for the results, but microscopic inspection of the experimental extracts revealed no intact cells, indicating that this is not likely the primary cause of qPCR variability. It is more probable that an unidentified compound contained in A. ostenfeldii cells affected DNA extraction efficiency. It is known from molecular studies of plants and kelps that polyphenols, mucilaginous polysaccharides, and carbohydrates can interfere with the recovery of nucleic acids from certain species (Fox & Swanson 2007; Souza et al. 2012; Maeda et al. 2013; Ramosa et al. 2014). Alexandrium ostenfeldii may contain a compound that interfered with DNA purification. During our experiments DNA was purified from A. ostenfeldii cell concentrations that scaled over two orders of magnitude. Spectrophotometric determination of DNA concentrations revealed only a twofold difference between the highest and lowest cell concentrations. In comparison, DNA purified from A. fundyese cell concentrations, which also scaled over two orders of magnitude, yielded DNA concentrations that differed by 15-fold between the highest and lowest cell concentrations. This was a strong indication of interference in the DNA purification of the A. ostenfeldii cells. This premise is further supported by the limited dynamic range of the A. ostenfeldii cell-based standard curves (Fig. 10). The arrows in Fig. 10 indicate that the Cq values obtained from cell concentrations ranging from ~5600 to ~151,000 cells plateaued at roughly 18 cycles. These results signaled the assay’s upper limit of detection and that the DNA extraction method was saturated. The A. ostenfeldii cell-based standard curves exhibited more variability than the A. fundyense cell-based standard curves (R2 = 0.878 vs R2 = 0.928, respectively; Fig. 10). In contrast, typical diluted PCR amplicon standard curves for A. ostenfeldii were almost always linear over seven orders of magnitude (R2 = 0.996), demonstrating that the qPCR thermocycling, reagents, and reaction mixes were appropriately administered (Fig. 10). In conclusion, the A. fundyense Group I qPCR assay was robust and can be used to accurately assess a wide range of cell concentrations from field samples. More research, however, is needed to establish a DNA purification method for A. ostenfeldii before the assay can be used to quantify concentrations of this species in natural populations.
Review of species-specific molecular assays for identifying Alexandrium species
The earliest Alexandrium molecular studies were conducted by Scholin et al. (1993) and Scholin & Anderson (1994), who sequenced the A. fundyense SSU and the LSU D1-D2 rDNA genes and by Adachi et al. (1994), who conducted restricted fragment length polymorphism analysis of the ITS-5.8S regions of A. catenella, A. tamarense, A. insuetum, and A. pseudogonyaulax. During the intervening two decades many other investigators have conducted molecular studies and designed species-specific assays for numerous Alexandrium species. The majority of the assays are redundant and target identical species, particularly those known to produce STXs. One factor driving the development of seemingly duplicative assays is that no systematic compilation and evaluation of existing molecular assays has been carried out. In an effort to reduce the development of redundant species-specific assays we have collated and mapped published primer and probe locations shown in Figures 14 and S7–S9. These primer and probe locations targeted SSU/ITS/LSU rDNA gene regions. Analysis of the locations revealed that a hypervariable gene region in the LSU D1-D2 domain, from bp 420 and extending to bp 740, was most commonly targeted. Assays developed in our study also targeted this variable region. Further scrutiny revealed that probes and primers developed by different investigators for identical species often overlapped and were shifted several bps up or down from each other (Fig. S7). Similarly, the ITS/5.8S region contained numerous species-specific polymorphisms that were targeted for assay development and that tended to be evenly distributed over the entire region (Fig. S8). Molecular studies of Alexandrium have targeted the SSU rDNA gene regions less often because the SSU genes are more conserved than either the LSU D1-D2 or ITS1–5.8S-ITS2 gene regions and contain proportionately fewer polymorphic sites that can be targeted for molecular assay development. The most commonly targeted SSU gene region was located in a hypervariable region at the 5′ end of the gene between bps 140 and 260 (Fig. S9).
A systematic evaluation of published Alexandrium molecular assays is presented in Tables 3 and S1. The authors are listed in alphabetical order followed by the targeted rDNA domain, the intended target species, type of assay, probe and primer sequences, and species specificity as determined by NCBI BLAST search and by visual confirmation within the alignments (Figs 14 and S7–S9). The molecular analyses indicated that the majority of the assays were well designed and incorporated species-specific gene sequences as their targets (unshaded sections of Table S1). However, the analyses also revealed that some of the assays exhibited potential flaws due to a variety of reasons (shaded sections of Table S1). For example, numerous assays were not specific for their intended target species. The most apparent reason for this was that early investigations did not have access to the quantity and diversity of ribosomal gene sequence data that are available today. Consequently, determining cross-reactivity of molecular assays, which can be readily accomplished now, was not possible previously. Other sources of assay inaccuracies included mismatched gene sequence where the primer or probe sequence did not match the intended target sequence and assays that targeted polymorphic loci without incorporating degenerate bases in the probe design. Other assays were species specific, but only for a subset of the total sequence variants contained in GenBank. This could have the detrimental effect of reducing assay signal strength and efficiency as well as yielding false-negative results. Thorough assessment of within-species sequence variation is critical for optimal assay design and requires alignment of numerous gene sequences to identify unique sites that are conserved among alleles. This is particularly true for some Alexandrium species because sequence variation among ribosomal gene copies is high in some species but quite low in others (Scholin et al. 1993; Kim et al. 2004b; Miranda et al. 2012).
Recommendations
It is clear that a majority of the species-specific assays listed in Tables 3 and S1 (unshaded) are useful. However, at least for the toxic species, it would be beneficial if standard assays were adopted for worldwide monitoring. An immediate positive benefit of such an approach is that it would make implementation of the assays easier and more affordable. A logical starting point would be to evaluate and promote assays that have been used successfully in established monitoring programs and that have large linear dynamic ranges in the number of cells that can be detected in environmental samples. Examples include the qPCR assays used by the Cawthron Institute (Nelson, New Zealand) for several years to monitor A. australiense Group V, A. fundyense Group I, A. minutum, and A. pacificum Group IV (Harlow et al. 2005) and the one validated for A. fundyense Group I in this study. Other assays include those for A. andersoni and A. taylori developed by Penna et al. (2007). Similarly, the microarray sites identified in Galluzzi et al. (2011) for monitoring population densities of A. andersoni, A. mediterraneum Group II, A. minutum, A. pacificum Group IV, A. pseudogonyaulax, A. tamarense Group III, and A. taylori have been well vetted and may also prove useful as in situ probes. Representative in situ and sandwich hybridization assays include those for A. affine (Kim et al. 2005), A. andersoni (Touzet & Raine 2007), A. fraterculus (Kim et al. 2005), A. fundyense Group I (Scholin et al. 1996; Anderson et al. 1999; 2005b; John et al. 2003; Sako et al. 2004), A. mediterraneum Group II (John et al. 2005), A. minutum (Diercks et al. 2008; Touzet et al. 2008), A. ostenfeldii (Metfies et al. 2005), A. pacificum Group IV (Adachi et al. 1996a; Sako et al. 2004), A. tamarense Group III (John et al. 2005; Touzet et al. 2008), and A. tamiyavanichi (Kim et al. 2004a). Vetting and selecting representative assays from this group or others listed in Tables 3 and S1 will reduce future duplication of effort and facilitate investigation of this ecologically and toxicologically important genus. It will also widen the appeal of molecular identification methods to resource managers, public health officials, and commercial shellfish operators as well as improve public health protection and utilization of valuable shellfish resources.
Supplementary Material
ACKNOWLEDGEMENTS
We thank two anonymous reviewers who provided constructive comments. Reference to trade names do not imply product endorsement by the National Ocean Service (NOS), NOAA. Funding was provided by the National Center for Coastal Ocean Science (NCCOS), NOAA and North Pacific Research Board Project 1118. This is contribution #5231 from University of Maryland Center for Environmental Science (UMCES), #16-179 for University of Maryland Institute of Marine and Environmental Technology (IMET), and #ECO867 from the Ecology and Oceanography of Harmful Algal Blooms (ECOHAB) program. This research was also funded in part by grants from Oceans and Human Health National Institutes of Health R01ES021949-01/NSFOCE1313888 and NOAA-NOS- NCCOS-2012-2002987 to ARP.
Footnotes
SUPPLEMENTARY DATA
Supplementary data associated with this article can be found online at http://dx.doi.org/10.2216/16-41.1.s1.
REFERENCES
- Adachi M, Sako Y & Ishida Y 1994. Restriction-fragment length polymorphism of ribosomal DNA internal transcribed spacer and 5.8S regions in Japanese Alexandrium species (Dinophyceae). Journal of Phycology 30: 857–863. [Google Scholar]
- Adachi M, Sako Y & Ishida Y 1996a. Identification of the toxic dinoflagellates Alexandrium catenella and A. tamarense (Dinophyceae) using DNA probes and whole-cell hybridization. Journal of Phycology 32: 1049–1052. [Google Scholar]
- Adachi M, Sako Y & Ishida Y 1996b. Analysis of Alexandrium (Dinophyceae) species using sequences of the 5.8S ribosomal DNA and internal transcribed spacer regions. Journal of Phycology 32: 424–432. [Google Scholar]
- Altschul SF, Madden TL, SchÄffer AA, Zhang J, Zhang Z, Miller W & Lipman DJ 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DM, Kulis DM, Keafer BA & Berdalet E 1999. Detection of the toxic dinoflagellate Alexandrium fundyense (Dinophyceae) with oligonucleotide and antibody probes: variability in labeling intensity with physiological condition. Journal of Phycology 35: 870–883. [Google Scholar]
- Anderson DM, Stock CA, Keafer B, Nelson AB, Thompson B, McGillicuddy DJ, Keller M, Matrai PA & Martin JL 2005a. Alexandrium fundyense cyst dynamics in the Gulf of Maine. Deep-Sea Research Part II: Topical Studies in Oceanography 52: 2522–2542. [Google Scholar]
- Anderson DM, Kulis DM, Keafer BA, Gribble KE, Marin R & Scholin CA 2005b. Identification and enumeration of Alexandrium spp. from the Gulf of Maine using molecular probes. Deep-Sea Research Part II: Topical Studies in Oceanography 52: 2467–2490. [Google Scholar]
- Anderson DM, Keafer BA, Kleindinst JL, McGillicuddy DJ, Martin JL, Norton K, Pilskaln CH, Smith JL, Sherwood CR & Butman B 2014. Alexandrium fundyense cysts in the Gulf of Maine: long-term time series of abundance and distribution, and linkages to past and future blooms. Deep-Sea Research Part II: Topical Studies in Oceanography 103: 6–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balech E 1995. The genus Alexandrium Halim (Dinoflagellata). Sherkin Island Marine Station, Sherkin Island, Ireland: 151 pp. [Google Scholar]
- Borkman DG, Smayda TJ, Tomas CR, York R, Strangman W & Wright JLC 2012. Toxic Alexandrium peruvianum (Balech and de Mendiola) Balech and Tangen in Narragansett Bay, Rhode Island (USA). Harmful Algae 19: 92–100. [Google Scholar]
- Brosnahan ML, Kulis DM, Solow AR, Erdner DL, Percy L, Lewis J & Anderson DM 2010. Outbreeding lethality between toxic Group I and nontoxic Group III Alexandrium tamarense spp. isolates: predominance of heterotypic encystment and implications for mating interactions and biogeography. Deep- Sea Research Part II: Topical Studies in Oceanography 57: 175–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown L, Bresnan E, Graham J, Lacaze JP, Turrell E & Collins C 2010. Distribution, diversity and toxin composition of the genus Alexandrium (Dinophyceae) in Scottish waters. European Journal of Phycology 45: 375–393. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J & Wittwer CT 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry 55: 611–622. [DOI] [PubMed] [Google Scholar]
- Castrodale L 2015. State of Alaska epidemiology bulletin. Paralytic shellfish poisoning - Alaska, 1993– 2014. http://www.epi.alaska.gov/bulletins/docs/b2015_01.pdf;searchedon17March2016.
- Cusick KD & Sayler GS 2013. An overview on the marine neurotoxin, saxitoxin: genetics, molecular targets, methods of detection and ecological functions. Marine Drugs 11: 991–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diercks S, Medlin LK & Metfies K 2008. Colorimetric detection of the toxic dinoflagellate Alexandrium minutum using sandwich hybridization in a microtiter plate assay. Harmful Algae 7: 137–145. [Google Scholar]
- Diercks-Horn S, Metfies K, Jackel S & Medlin LK 2011. The ALGADEC device: a semi-automated rRNA biosensor for the detection of toxic algae. Harmful Algae 10: 395–401. [Google Scholar]
- Dyhrman ST, Erdner D, La Du J, Galac M & Anderson DM 2006. Molecular quantification of toxic Alexandrium fundyense in the Gulf of Maine using real-time PcR. Harmful Algae 5: 242–250. [DOI] [PubMed] [Google Scholar]
- Dyhrman ST, Haley ST, Borchert JA, Lona B, Kollars N & Erdner DL 2010. Parallel analyses of Alexandrium catenella cell concentrations and shellfish toxicity in the Puget sound. Applied and Environmental Microbiology 76: 4647–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdner DL, Percy L, Keafer B, Lewis J & Anderson DM 2010. A quantitative real-time PCR assay for the identification and enumeration of Alexandrium cysts in marine sediments. Deep- Sea Research Part II: Topical Studies in Oceanography 57: 279–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox CH & Swanson AK 2007. Nested PCR detection of microscopic life-stages of laminarian macroalgae and comparison with adult forms along intertidal height gradients. Marine Ecology Progress Series 332: 1–10. [Google Scholar]
- Galluzzi L, Penna A, Bertozzini E, Vila M, Garces E & Magnani M 2004. Development of a real-time PCR assay for rapid detection and quantification of Alexandrium minutum (a dinoflagellate). Applied and Environmental Microbiology 70: 1199–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Penna A, Bertozzini E, Giacobbe MG, Vila M, Garces E, Prioli S & Magnani M 2005. Development of a qualitative PCR method for Alexandrium spp. (Dinophyceae) detection in contaminated mussels (Mytilus galloprovincialis). Harmful Algae 4: 973–983. [Google Scholar]
- Galluzzi L, Bertozzini E, del Campo A, Penna A, Bruce IJ & Magnani M 2006. Capture probe conjugated to paramagnetic nanoparticles for purification of Alexandrium species (Dinophyceae) DNA from environmental samples. Journal of Applied Microbiology 101: 36–43. [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Bertozzini E, Penna A, Perini F, Garces E & Magnani M 2010. Analysis of rRNA gene content in the Mediterranean dinoflagellate Alexandrium catenella and Alexandrium taylori: implications for the quantitative real-time PCR-based monitoring methods. Journal of Applied Phycology 22: 1–9. [Google Scholar]
- Galluzzi L, Cegna A, Casabianca S, Penna A, Saunders N & Magnani M 2011. Development of an oligonucleotide microarray for the detection and monitoring of marine dinoflagellates. Journal of Microbiological Methods 84: 234–242. [DOI] [PubMed] [Google Scholar]
- Garneau ME, Schnetzer A, Countway PD, Jones AC, Seubert EL & Caron DA 2011. Examination of the seasonal dynamics of the toxic dinoflagellate Alexandrium catenella at Redondo Beach, california, by quantitative PcR. Applied and Environmental Microbiology 77: 7669–7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovesi B, Shin-Grzebyk MS, Grzebyk D, Laabir M, Gagnaire PA, Vaquer A, Pastoureaud A, Lasserre B, Collos Y, Berrebi P & Masseret E 2011. Assessment of cryptic species diversity within blooms and cyst bank of the Alexandrium tamarense complex (Dinophyceae) in a Mediterranean lagoon facilitated by semi-multiplex PcR. Journal of Plankton Research 33: 405–414. [Google Scholar]
- Gescher C, Metfies K & Medlin LK 2008. The ALEX CHIP – development of a DNA chip for identification and monitoring of Alexandrium. Harmful Algae 7: 485–494. [Google Scholar]
- Giacobbe MG, Penna A, Gangemi E, Maso M, Garces E, Fraga S, Bravo I, Azzaro F & Penna N 2007. Recurrent high-biomass blooms of Alexandrium taylori (Dinophyceae), a HAB species expanding in the Mediterranean. Hydrobiologia 580: 125–133. [Google Scholar]
- Godhe A, Otta SK, Rehnstam-Holm AS, Karunasagar I & Karunasagar I 2001. Polymerase chain reaction in detection of Gymnodinium mikimotoi and Alexandrium minutum in field samples from Southwest India. Marine Biotechnology 3: 152–162. [DOI] [PubMed] [Google Scholar]
- Gribble KE, Keafer BA, Quilliam MA, Cembella AD, Kulis DM, Manahan A & Anderson DM 2005. Distribution and toxicity of Alexandrium ostenfeldii (Dinophyceae) in the Gulf of Maine, USA. Deep-Sea Research Part II: Topical Studies in Oceanography 52: 2745–2763. [Google Scholar]
- Guillou L, Nezan E, Cueff V, Denn EEL, Cambon-Bonavita MA, Gentien P & Barbier G 2002. Genetic diversity and molecular detection of three toxic dinoflagellate genera (Alexandrium, Dinophysis, and Karenia) from French coasts. Protist 153: 223–238. [DOI] [PubMed] [Google Scholar]
- Haley ST, Cavender JF & Murray TE 1999. Detection of Alexandrium tamarensis by rapid PCR analysis. Biotechniques 26: 88–91. [DOI] [PubMed] [Google Scholar]
- Hall S 1982. Toxins and toxicity of Protogonyaulax from the northeast Pacific. PhD thesis University of Alaska Fairbanks. 392 pp. [Google Scholar]
- Hansen PJ, Cembella AD & Moestrup O 1992. The marine dinoflagellate Alexandrium ostenfeldii - paralytic shellfish toxin concentration, composition, and toxicity to a tintinnid ciliate. Journal of Phycology 28: 597–603. [Google Scholar]
- Harlow L, Rasmussen P, Bernard C, de Salas M & Hallegraeff G 2005. The development of real-time PCR detection methods for toxic Alexandrium dinoflagellate species in ship ballast water. University of Tasmania and the Australian Water Quality Centre, Hobart, Tasmania, Australia: 18 pp. [Google Scholar]
- Horner RA, Garrison DL & Plumley FG 1997. Harmful algal blooms and red tide problems on the U.S. west coast. Limnology and Oceanography 42: 1076–1088. [Google Scholar]
- Hosoi-Tanabe S & Sako Y 2005a. Species-specific detection and quantification of toxic marine dinoflagellates Alexandrium tamarense and Alexandrium catenella by real-time PCR assay. Marine Biotechnology 7: 506–514. [DOI] [PubMed] [Google Scholar]
- Hosoi-Tanabe S & Sako Y 2005b. Rapid detection of natural cells of Alexandrium tamarense and Alexandrium catenella (Dinophyceae) by fluorescence in situ hybridization. Harmful Algae 4: 319–328. [Google Scholar]
- Hosoi-Tanabe S & Sako Y 2006. Development and application of fluorescence in situ hybridization (FisH) method for simple and rapid identification of the toxic dinoflagellates Alexandrium tamarense and Alexandrium catenella in cultured and natural seawater. Fisheries Science 72: 77–82. [Google Scholar]
- Howell JF 1953. Gonyaulax monilata sp. nov. the causative dinoflagellate of a red tide in the east coast of Florida in August-September 1951. Transactions of the American Microscopical Society 72: 153–156. [Google Scholar]
- John U, Cembella A, Hummert C, Elebrachter M, Groben R & Medlin L 2003. Discrimination of the toxigenic dinoflagellates Alexandrium tamarense and A. ostenfeldii in co-occurring natural populations from scottish coastal waters. European Journal of Phycology 38: 25–40. [Google Scholar]
- John U, Medlin LK & Groben R 2005. Development of specific rRNA probes to distinguish between geographic clades of the Alexandrium tamarense species complex. Journal of Plankton Research 27: 199–204. [Google Scholar]
- John U, Litaker RW, Montresor M, Murray S, Brosnahan ML & Anderson DM 2014a. Formal revision of the Alexandrium tamarense species complex (Dinophyceae) taxonomy: the introduction of five species with emphasis on molecular-based (rDNA) classification. Protist 165: 779–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John U, Litaker RW, Montresor M, Murray S, Brosnahan ML & Anderson DM 2014b. (2032) Proposal to reject the name Gonyaulax catenella (Alexandrium catenella) (Dinophyceae). Taxon 63: 932–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamikawa R, Hosoi-Tanabe S, Nagai S, Itakura S & Sako Y 2005. Development of a quantification assay for the cysts of the toxic dinoflagellate Alexandrium tamarense using real-time polymerase chain reaction. Fisheries Science 71: 987–991. [Google Scholar]
- Kamikawa R, Nagai S, Hosoi-Tanabe S, Itakura S, Yamaguchi M, Uchida Y, Baba T & Sako Y 2007. Application of real-time PCR assay for detection and quantification of Alexandrium tamarense and Alexandrium catenella cysts from marine sediments. Harmful Algae 6: 413–420. [Google Scholar]
- Keller MD, Selvin RC, Claus W & Guillard RRL 1987. Media for the culture of oceanic ultraphytoplankton. Journal of Phycology 23: 633–638. [Google Scholar]
- Ki JS & Han MS 2006. A low-density oligonucleotide array study for parallel detection of harmful algal species using hybridization of consensus PCR products of LSU rDNA D2 domain. Biosensors & Bioelectronics 21: 1812–1821. [DOI] [PubMed] [Google Scholar]
- Kim CJ & Sako Y 2005. Molecular identification of toxic Alexandrium tamiyavanichii (Dinophyceae) using two DNA probes. Harmful Algae 4: 984–991. [Google Scholar]
- Kim CJ, Yoshimatsu SA, Sako Y & Kim CH 2004a. Molecular identification of the toxic Alexandrium tamiyavanichii (Dinophyceae) by the whole-cell FISH method. Journal of Fisheries Science and Technology 7: 175–183. [Google Scholar]
- Kim CJ, Yoshihiko S, Aritsune U & Kim CH 2004b. Molecular phylogenetic relationships within the genus Alexandrium (Dinophyceae) based on the nuclear-encoded SSU and LSU rDNA D1- D2 sequences. Journal of the Korean Society of Oceanography 39: 172–185. [Google Scholar]
- Kim CJ, Kim CH & Sako Y 2005. Development of molecular identification method for genus Alexandrium (Dinophyceae) using whole-cell FISH. Marine Biotechnology 7: 215–222. [DOI] [PubMed] [Google Scholar]
- Kremp A, Tahvanainen P, Litaker W, Krock B, Suikkanen S, Leaw CP & Tomas C 2014. Phylogenetic relationships, morphological variation, and toxin patterns in the Alexandrium Ostenfeldii (Dinophyceae) complex: implications for species boundaries and identities. Journal of Phycology 50: 81–100. [DOI] [PubMed] [Google Scholar]
- Lazerges M, Perrot H, Antoine E, Defontaine A & Compere C 2006. Oligonucleotide quartz crystal microbalance sensor for the microalgae Alexandrium minutum (Dinophyceae). Biosensors & Bioelectronics 21: 1355–1358. [DOI] [PubMed] [Google Scholar]
- Lefebvre KA, Bill BD, Erickson A, Baugh KA, O’Rourke L, Costa PR, Nance S & Trainer VL 2008. Characterization of intracellular and extracellular saxitoxin levels in both field and cultured Alexandrium spp. samples from Sequim Bay, Washington. Marine Drugs 6: 103–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardi M, Azzaro F, Galletta M, Giacobbe M, Maso M & Penna A 2006. Time-series evolution of toxic organisms and related environmental factors in a brackish ecosystem of the Mediterranean Sea. Hydrobiologia 555: 299–305. [Google Scholar]
- Lilly EL, Halanych KM & Anderson DM 2007. Species boundaries and global biogeography of the Alexandrium tamarense complex (Dinophyceae). Journal of Phycology 43: 1329–1338. [Google Scholar]
- Litaker RW, Vandersea MW, Kibler SR, Madden VJ, Noga EJ & Tester PA 2002. Life cycle of the heterotrophic dinoflagellate Pfiesteria piscicida (Dinophyceae). Journal of Phycology 38: 442–463. [Google Scholar]
- Litaker RW, Vandersea MW, Kibler SR, Reece KS, Stokes NA, Steidinger KA, Millie DF, Bendis BJ, Pigg RJ & Tester PA 2003. Identification of Pfiesteria piscicida (Dinophyceae) and Pfiesteria-like organisms using internal transcribed spacer-specific PCR assays. Journal of Phycology 39: 754–761. [Google Scholar]
- Mackenzie L, White D, Oshima Y & Kapa J 1996. The resting cyst and toxicity of Alexandrium ostenfeldii (Dinophyceae) in New Zealand. Phycologia 35: 148–155. [Google Scholar]
- Maeda T, Kawai T, Nakaoka M & Yotsukura N 2013. Effective DNA extraction method for fragment analysis using capillary sequencer of the kelp, Saccharina. Journal of Applied Phycology 25: 337–347. [Google Scholar]
- Martin JL & Richard D 1996. Shellfish toxicity from the Bay of Fundy, eastern Canada: 50 years in retrospect In: Harmful and toxic blooms (Ed. by Yasumoto T, Oshima Y & Fukuyo Y), pp. 3–6. UNESCO, Paris. [Google Scholar]
- Martin JL, LeGresley MM, Hanke AR & Page FH 2008. Alexandrium fundyense - red tides, PsP shellfish toxicity, salmon mortalities and human illnesses in 2003–2004 - before and after. In: Proceedings of the 12th International Conference on Harmful Algae, International Society for the Study of Harmful Algae and Intergovernmental Oceanographic Commission of UNESCO (Ed. by Moestrup O), pp. 206–208. UNESCO, Copenhagen. [Google Scholar]
- Masseret E, Enquebecq M, Laabir M, Genovesi B, Vaquer A & Avarre JC 2010. A simple and innovative method for species identification of phytoplankton cells on minute quantities of DNA. Environmental Microbiology Reports 2: 715–719. [DOI] [PubMed] [Google Scholar]
- Matweyou JA 2003. Paralytic shellfish poisoning: The relationship between Alexandrium abundance and PSP toxins on Kodiak Island, Alaska. MS thesis University of Alaska Fairbanks. 117 pp. [Google Scholar]
- McGillicuddy DJ, Brosnahan ML, Couture DA, He R, Keafer BA, Manning JP, Martin JL, Pilskaln CH, Townsend DW & Anderson DM 2014. A red tide of Alexandrium fundyense in the Gulf of Maine. Deep-Sea Research Part II: Topical Studies in Oceanography 103: 174–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metfies K, Huljic S, Lange M & Medlin LK 2005. Electrochemical detection of the toxic dinoflagellate Alexandrium ostenfeldii with a DNA-biosensor. Biosensors & Bioelectronics 20: 1349–1357. [DOI] [PubMed] [Google Scholar]
- Miranda LN, Zhuang YY, Zhang H & Lin S 2012. Phylogenetic analysis guided by intragenomic ssu rDNA polymorphism refines classification of “Alexandrium tamarense” species complex. Harmful Algae 16: 35–48. [Google Scholar]
- Nagai S 2011. Development of a multiplex PCR assay for simultaneous detection of six Alexandrium species (Dinophyceae). Journal of Phycology 47: 703–708. [DOI] [PubMed] [Google Scholar]
- Natsuike M, Nagai S, Matsuno K, Saito R, Tsukazaki C, Yamaguchi A & Imai I 2013. Abundance and distribution of toxic Alexandrium tamarense resting cysts in the sediments of the Chukchi Sea and the eastern Bering Sea. Harmful Algae 27: 52–59. [Google Scholar]
- Neale RA 1967. Fluctuations in the levels of paralytic shellfish toxin in four species of lamellibranch molluscs near Ketchikan, Alaska 1963–1965. PhD thesis University of Washington, Seattle. 149 pp. [Google Scholar]
- Orlova TY, Selina MS, Lilly EL, Kulis DM & Anderson DM 2007. Morphogenetic and toxin composition variability of Alexandrium tamarense (Dinophyceae) from the east coast of Russia. Phycologia 46: 534–548. [Google Scholar]
- Penna A & Magnani M 1999. Identification of Alexandrium (Dinophyceae) species using PCR and rDNA-targeted probes. Journal of Phycology 35: 615–621. [Google Scholar]
- Penna A & Magnani M 2000. A PCR method immunoassay for the detection of Alexandrium (Dinophyceae) species. Journal of Phycology 36: 1183–1186. [Google Scholar]
- Penna A, Bertozzini E, Battocchi C, Galluzzi L, Giacobbe MG, Vila M, Garces E, Luglie A & Magnani M 2007. Monitoring of HAB species in the Mediterranean Sea through molecular methods. Journal of Plankton Research 29: 19–38. [Google Scholar]
- Penna A, Battocchi C, Garces E, Angles S, Cucchiari E, Totti C, Kremp A, Satta C, Giacobbe MG, Bravo I & Bastianini M 2010. Detection of microalgal resting cysts in European coastal sediments using a PCR-based assay. Deep-Sea Research Part II: Topical Studies in Oceanography 57: 288–300. [Google Scholar]
- Porter K, Fearey D & Esposito T 2011. State of Alaska epidemiology bulletin. http://www.epi.alaska.gov/bulletins/docs/b2011_17.pdf; searched on 17 March 2016.
- Quayle DB 1969. Paralytic shellfish poisoning in British Columbia. Bulletin of the Fisheries Research Board of Canada 168: 1–69. [Google Scholar]
- Ramosa SNM, Salazarb MM, Pereira GAG & Efraima P 2014. Plant and metagenomic DNA extraction of mucilaginous seeds. MethodsX 1: 225–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sako Y, Hosoi-Tanabe S & Uchida A 2004. Fluorescence in situ hybridization using rRNA-targeted probes for simple and rapid identification of the toxic dinoflagellates Alexandrium tamarense and Alexandrium catenella. Journal of Phycology 40: 598–605. [Google Scholar]
- Scholin CA & Anderson DM 1994. Identification of group-specific and strain-specific genetic-markers for globally distributed Alexandrium (Dinophyceae) 1. RFLP analysis of SSU ribosomal-RNA genes. Journal of Phycology 30: 744–754. [Google Scholar]
- Scholin CA, Anderson DM & Sogin ML 1993. Two distinct small-subunit ribosomal -RNA genes in the North American toxic dinoflagellate Alexandrium fundyense (Dinophyceae). Journal of Phycology 29: 209–216. [Google Scholar]
- Scholin CA, Herzog M, Sogin M & Anderson DM 1994. identification of group-specific and strain-specific genetic markers for globally distributed Alexandrium (Dinophyceae) 2. sequence-analysis of a fragment of the LSU ribosomal-RNA gene. Journal of Phycology 30: 999–1011. [Google Scholar]
- Scholin CA, Hallegraeff G & Anderson DM 1995. Molecular evolution of the Alexandrium tamarense species complex: dispersal in the North American and West Pacific regions. Phycologia 34: 472–485. [Google Scholar]
- Scholin CA, Buck KR, Britschgi T, Cangelosi G & Chavez FP 1996. Identification of Pseudo-nitzschia australis (Bacillariophyceae) using rRNA-targeted probes in whole cell and sandwich hybridization formats. Phycologia 35: 190–197. [Google Scholar]
- Smith B 2010. State of Alaska epidemiology bulletin. Parlytic shellfish poisoning in Juneau, Kodiak, and Haines June 2010. http://www.epi.alaska.gov/bulletins/docs/b2010_17.pdf;searchedon17March2016.
- Souza HAV, Muller LAC, BrandÃo RL & Lovato MB 2012. Isolation of high quality and polysaccharide-free DNA from leaves of Dimorphandra mollis (Leguminosae), a tree from the Brazilian Cerrado. Genetics and Molecular Research 11: 756–764. [DOI] [PubMed] [Google Scholar]
- Tang XH, Yu RC, Zhou MJ & Yu ZG 2012. Application of rRNA probes and fluorescence in situ hybridization for rapid detection of the toxic dinoflagellate Alexandrium minutum. Chinese Journal of Oceanology and Limnology 30: 256–263. [Google Scholar]
- Taylor JD, Kegel JU, Lewis JM & Medlin LK 2014. Validation of the detection of Alexandrium species using specific RNA probes tested in a microarray format: calibration of signal using variability of RNA content with environmental conditions Harmful Algae 37: 207–207. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F & Higgins DG 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 24: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toebe K, Alpermann TJ, Tillmann U, Krock B, Cembella A & John U 2013. Molecular discrimination of toxic and non-toxic Alexandrium species (Dinophyta) in natural phytoplankton assemblages from the scottish coast of the North sea. European Journal of Phycology 48: 12–26. [Google Scholar]
- Touzet N & Raine R 2007. Discrimination of Alexandrium andersoni and A. minutum (Dinophyceae) using LSU rRNA-targeted oligonucleotide probes and fluorescent whole-cell hybridization. Phycologia 46: 168–177. [Google Scholar]
- Touzet N, Franco JM & Raine R 2007. Characterization of nontoxic and toxin-producing strains of Alexandrium minutum (Dinophyceae) in irish coastal waters. Applied and Environmental Microbiology 73: 3333–3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touzet N, Franco JM & Raine R 2008. PSP toxin analysis and discrimination of the naturally co-occurring Alexandrium tamarense and A. minutum (Dinophyceae) in Cork Harbour, Ireland. Aquatic Microbial Ecology 51: 285–299. [Google Scholar]
- Touzet N, Keady E, Raine R & Maher M 2009. Evaluation of taxa-specific real-time PCR, whole-cell FISH and morphotaxonomy analyses for the detection and quantification of the toxic microalgae Alexandrium minutum (Dinophyceae), Global Clade ribotype. FEMS Microbiology Ecology 67: 329–341. [DOI] [PubMed] [Google Scholar]
- Touzet N, Farrell H, Rathaille AN, Rodriguez P, Alfonso A, Botana LM & Raine R 2010. Dynamics of co-occurring Alexandrium minutum (Global Clade) and A. tamarense (West European) (Dinophyceae) during a summer bloom in Cork Harbour, Ireland (2006). Deep-Sea Research Part II: Topical Studies in Oceanography 57: 268–278. [Google Scholar]
- Trainer VL 2002. Harmful algal blooms on the U.S. west coast In: Harmful algal blooms in the PICES region of the North Pacific (Ed. by Taylor FJ & Trainer VL), PICES; Scientific Report no. 23. pp. 89–118. [Google Scholar]
- Vandersea MW, Litaker RW, Yonnish B, Sosa E, Landsberg JH, Pullinger C, Moon-Butzin P, Green J, Morris JA, Kator H, Noga EJ & Tester PA 2006. Molecular assays for detecting Aphanomyces invadans in ulcerative mycotic fish lesions. Applied Environmental Microbiology 72: 1551–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandersea MW, Kibler SR, Holland WC, Tester PA, Schultz TF, Faust MA, Holmes MJ, Chinain M & Litaker RW 2012. Development of semi-quantitative PCR assays for the detection and enumeration of Gambierdiscus species (Gonyaulacales, Dinophyceae). Journal of Phycology 48: 902–915. [DOI] [PubMed] [Google Scholar]
- Zhang FL & Li ZY 2012. Detection and quantification of cultured marine Alexandrium species by real-time PCR. World Journal of Microbiology & Biotechnology 28: 3255–3260. [DOI] [PubMed] [Google Scholar]
- Zhen Y, Mi TZ & Yu ZG 2011. Quantification methods for Alexandrium catenella, a toxic dinoflagellate: Comparison of competitive enzyme-linked immunosorbent assay and sandwich hybridization integrated with a nuclease protection assay. Harmful Algae 10: 589–597. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.