Abstract
Methylome-wide association studies (MWASs) are promising complements to sequence variation studies. We used existing sequencing-based methylation data, which assayed the majority of all 28 million CpGs in the human genome, to perform an MWAS for schizophrenia in blood, while controlling for cell-type heterogeneity with a recently generated platform-specific reference panel. Next, we compared the MWAS results with findings from 3 existing large-scale array-based schizophrenia methylation studies in blood that assayed up to ~450 000 CpGs. Our MWAS identified 22 highly significant loci (P < 5 × 10−8) and 852 suggestively significant loci (P < 1 × 10−5). The top finding (P = 5.62 × 10−11, q = 0.001) was located in MFN2, which encodes mitofusin-2 that regulates Ca2+ transfer from the endoplasmic reticulum to mitochondria in cooperation with DISC1. The second-most significant site (P = 1.38 × 10−9, q = 0.013) was located in ALDH1A2, which encodes an enzyme for astrocyte-derived retinoic acid—a key neuronal morphogen with relevance for schizophrenia. Although the most significant MWAS findings were not assayed on the arrays, we observed significant enrichment of overlapping findings with 2 of the 3 array datasets (P = 0.0315, 0.0045, 0.1946). Overrepresentation analysis of Gene Ontology terms for the genes in the significant overlaps suggested high similarity in the biological functions detected by the different datasets. Top terms were related to immune and/or stress responses, cell adhesion and motility, and a broad range of processes essential for neurodevelopment.
Keywords: DNA methylation, MBD-seq, methylome-wide association study (MWAS), schizophrenia
Introduction
DNA methylation is a dynamic epigenetic modification that provides the cellular phenotype both stability and diversity. Investigations of these modifications provide promising complements to schizophrenia studies of sequence variation. In 2014, Aberg et al1 performed the first large-scale methylome-wide association study (MWAS) for schizophrenia, using methyl-binding domain sequencing (MBD-seq), which assays nearly all ~28 million CpGs in the human genome.2,3 Since then, 2 independent research teams, Hannon et al4 and Montano et al5 have reported large-scale array-based methylation studies for schizophrenia, which assay up to ~450 000 CpGs that also accounted for cell-type proportions.
In the current investigation, we used a recently generated MBD-seq specific leukocyte reference panel6 that allows us to perform MWAS for schizophrenia with the MBD-seq data while accounting for cell-type heterogeneity.7 Next, we used a multi-marker approach to investigate the cumulative MWAS signal across the top methylation sites. Furthermore, we use a permutation-based test to examine whether overlap between the new MBD-seq MWAS and the array-based methylation studies demonstrated enrichment. To the best of our knowledge, this is the first time that the results from multiple large-scale methylation datasets for schizophrenia have been compared in this manner.
Methods
Two Approaches for Methylome-Wide Association Studies
Whole genome bisulfite sequencing (WGB-seq), where bisulfite converted genomic DNA is sequenced to assess the methylation status of each site across the genome, is often considered the “gold standard” in DNA methylation research. However, WGB-seq remains too costly for large-scale projects. Other approaches, such as MBD-seq and the microarray-based Infinium Human Methylation 450K and MethylationEPIC assays, have been more frequently used for MWAS. Although both MBD-seq and array approaches are suitable for MWAS they have different advantages and limitations.
In MBD-seq, genomic DNA is fragmented into short ~150 bp fragments and a protein with high affinity for methylated CpGs is used to capture methylated fragments, which is then sequenced.8 With this approach the larger portion of the genome that lacks CpGs, and is not methylated, is not sequenced. In practice, this allows for assaying the majority of all 28 million CpGs in the genome.3,9 However, it is often challenging to distinguish between neighboring CpGs in regions where multiple CpGs are closely packed.
The methylation arrays use bisulfite converted DNA like WGB-seq, but instead of sequencing the entire converted genome, assay selected sites by hybridization to a microarray. This allows for methylation at single CpGs to be distinguished. However, current arrays are limited in scope and only assay a small percentage (~3%–4%) CpGs.
In this study, we use four existing large-scale datasets for schizophrenia that were generated with either MBD-seq or array-based technology.
Existing Methylation Sequencing Data
Our primary study sample included existing methylome-wide sequencing data from a Swedish schizophrenia cohort.1 The methylomes of DNA extracted from blood were assayed using MBD-seq as previously described.1,10 In the current study, sequencing reads were realigned (hg19/GRCh37) using an improved aligner Cushaw3.11 Data were then processed using the RaMWAS Bioconductor package,12 which is an efficient analysis tool designed for large-scale methylation studies. A description of the Swedish cohort and data processing is presented in the supplementary materials and supplementary table S1. After quality control, methylation data from 1448 subjects (744 schizophrenia and 704 controls) for 18 793 496 CpGs was available for statistical analysis.
Methylome-Wide Association Study Controlling for Cell Type Proportions
To test each CpG for association with schizophrenia status, we used RaMWAS12 to perform multiple regression analyses while controlling for age, sex, and assay-related variables like sample batch, and peak location.2 In this relatively homogenous sample, self-reported ethnicity (Caucasian with non-Finnish Nordic parents vs with Finnish parents) and ancestry estimated from genotype data did not substantially contribute to methylation variation, and thus were not included as covariates in the MWAS. To avoid confounding due to cell-type heterogeneity, we also included covariates for blood cell-type proportions estimated from the methylation data7 using reference methylomes.6 These reference methylomes were generated with MBD-seq after isolating all common cell types in blood as previously described.6 Finally, we used principal component analysis (PCA) as implemented in RaMWAS to capture major unmeasured sources of variation remaining after accounting for the measured covariates. The PCA is comparable with surrogate variable analysis and also uses latent variables, but allows for the >18 million CpG sites to be investigated simultaneously.6 Using a scree test (supplementary figure S1) we selected the first principal component that explained 1.17% of the remaining methylation variation for inclusion in the MWAS.
Determining the Significance of the Cumulative MWAS Signal by Resampling
To study the significance of the combined MWAS signals from associated methylation sites, we used the “ramwas7riskScoreCV” function in RaMWAS. This function uses elastic-nets13–15 in combination with 10-fold cross-validation16 to obtain unbiased predictions of case–control status for each individual. By testing whether these predictions are significantly correlated with actual schizophrenia status, we performed an “in sample replication” of the cumulative MWAS signal. Additional details are provided in the supplementary materials.
Permutation-Based Enrichment Test of Overlap and Gene Ontology Analyses
To further study the robustness and potential biological relevance of the observed associations we used the shiftR R-package, described previously,17 with 2 thresholds (top 1% and top 5% of results) to perform permutation-based enrichment tests of the overlap between our MWAS top results and other enrichment datasets. To account for “multiple testing”, the same thresholds are used in the permutations where the test statistic distribution under the null hypothesis is generated from the most significant (combination of) thresholds. The enrichment datasets included 3 previous large-scale methylation studies for schizophrenia (described later) for which we used 1 million permutations and genetic features such as genes, exons, introns, and promoters for which we used 100 000 permutations (supplementary table S2). To further test for robustness, we tested for enrichment of overlap with our previous analysis1 of the Swedish dataset (1 million permutations).
To test for enrichment of Gene Ontology (GO) terms among findings, we performed permutation-based tests that account for gene size and controls the family-wise error rate at the 0.05 level. BECon18 was used to identify CpGs among the overlapping top sites between the MBD-seq and array-based MWAS results that showed interindividual concordance (r ≥ |0.2|) between blood and brain. Further details are given in the supplementary materials.
Overlap Between Methylation Studies for Schizophrenia
In addition to our sequencing-based MWAS, 3 array-based large-scale methylation datasets for schizophrenia, here referred to as Montano,5 Hannon-1,4 and Hannon-24 have been reported. Details about these methylation study cohorts are given in the supplementary materials. Using the permutation-based enrichment tests we studied the overlap between these array-based datasets and our primary MWAS.
Results
Methylome-Wide Association Study Accounting for Cell Type Proportions
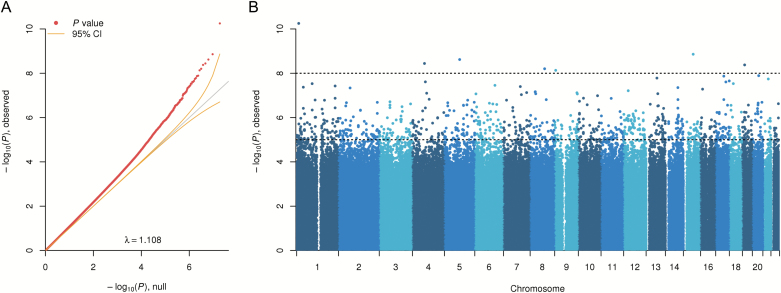
The quantile-quantile plot in figure 1A, and the lambda of 1.108, suggested many associated CpGs with relatively modest-to-large effects. To test if the lambda observed for the MBD-seq MWAS was caused by associations to the outcome variable, or if it was caused by deviation in the test statistic distribution from the theoretical null, we performed 100 MWAS where we permuted the schizophrenia case–control status and recorded the lambdas. Both the mean and median lambda from the permuted MWAS of this dataset were 1.00, with a 95% confidence interval of 0.98–1.02 (supplementary figure S2). Thus, the observed lambda in the MBD-seq MWAS likely reflects true associations.
Fig. 1.
Cell-type-corrected MWAS results. (A) Quantile-quantile plot. Observed methylome-wide association study (MWAS) −log10 (P values) is plotted against expected values (diagonal line) under the null hypothesis that no CpGs have an effect. Curved lines indicate the 95% CI. (B) Manhattan plot. Displays MWAS −log10 (P values) by chromosomal location.
When testing for enrichment of overlap between our new cell-type-corrected MWAS and our previous analysis without this correction1 we observed robust enrichment (P < 1.0 × 10−06, odds ratio = 16.05). Although we observe a very large overlap between the results, the highly significant top findings detected in the current analysis revealed newly associated sites.
The Manhattan plot in figure 1B and table 1 show 22 highly significant loci (P < 5 × 10−8). The top finding (P = 5.62 × 10−11, q = 0.001) was located in the MFN2 gene on chromosome 1. MFN2 encodes mitofusin-2, which regulates Ca2+ transfer from the endoplasmic reticulum to mitochondria in cooperation with DISC1.20 Deficits in mitofusin-2 result in impairments in neurogenesis and synaptogenesis,21 and degeneration of midbrain dopaminergic neurons22 as a result of mitochondrial dysfunction. The second top site (P = 1.38 × 10−9, q = 0.013) was found within ALDH1A2, which has been associated with schizophrenia in a previous candidate gene study.23ALDH1A2 encodes retinaldehyde dehydrogenase 2 (RALDH2) that is a key enzyme in production of astrocyte-derived retinoic acid in the brain.24,25 Retinoids are potent morphogens that are critical for neurodevelopment and have been broadly implicated in schizophrenia.26,27
Table 1.
Top Results (P < 5 × 10−8) From Methyl-Binding Domain Sequencing (MBD-seq) Methylome-Wide Association Study (MWAS)
| Chr. | Locationa | Geneb | MBD-seq MWAS | Extended Gene Annotationsc | |||
|---|---|---|---|---|---|---|---|
| Start (bp) | t stat | P value | q value | ||||
| 1 | 12058089 | MFN2 | 6.6 | 5.62E-11 | 0.0011 | TNFRSF8 (−65344) | MFN2 (+17721) |
| 15 | 58324541 | ALDH1A2 | 6.1 | 1.38E-09 | 0.013 | ALDH1A2 (+34074) | POLR2M (+325715) |
| 5 | 90591701 | 6.01 | 2.40E-09 | 0.015 | ARRDC3 (+87474) | GPR98 (+737085) | |
| 4 | 71851479 | MOB1B | 5.94 | 3.61E-09 | 0.0158 | DCK (−7676) | MOB1B (+83385) |
| 19 | 13279260 | 5.91 | 4.20E-09 | 0.0158 | IER2 (+16472) | CACNA1A (+337777) | |
| 8 | 86334323 | CA13 | 5.85 | 6.26E-09 | 0.0196 | CA1 (−43081) | CA3 (−16732) |
| 9 | 6138595 | 5.82 | 7.38E-09 | 0.0198 | RANBP6 (−122978) | IL33 (−103086) | |
| 20 | 37997060 | 5.72 | 1.29E-08 | 0.0282 | DHX35 (+406083) | ||
| 17 | 48260796 | COL1A1 | 5.71 | 1.35E-08 | 0.0282 | SGCA (+17431) | COL1A1 (+18196) |
| 13 | 64494841 | 5.68 | 1.65E-08 | 0.031 | No genes in this location | ||
| 21 | 32303327 | 5.66 | 1.81E-08 | 0.031 | KRTAP11-1 (−49454) | KRTAP19-8 (+107467) | |
| 17 | 80736726 | TBCD | 5.63 | 2.22E-08 | 0.0331 | TBCD (+26787) | ZNF750 (+61727) |
| 4 | 77169212 | FAM47E | 5.61 | 2.44E-08 | 0.0331 | FAM47E-STBD1 (−57966) | SCARB2 (−34167) |
| 17 | 60515203 | METTL2A | 5.61 | 2.47E-08 | 0.0331 | TLK2 (−41182) | METTL2A (+13976) |
| 18 | 23782139 | 5.58 | 2.94E-08 | 0.0346 | TAF4B (−23760) | PSMA8 (+68299) | |
| 1 | 92209862 | TGFBR3 | 5.58 | 2.95E-08 | 0.0346 | TGFBR3 (+141924) | CDC7 (+243198) |
| 6 | 120220770 | 5.54 | 3.51E-08 | 0.0388 | MAN1A1 (−549845) | ||
| 1 | 229239777 | 5.53 | 3.75E-08 | 0.0391 | TMEM78 (−145605) | RHOU (+368954) | |
| 7 | 100534233 | 5.52 | 3.99E-08 | 0.0393 | ACHE (−40693) | MUC3A (−17016) | |
| 1 | 39502674 | 5.51 | 4.26E-08 | 0.0393 | MACF1 (−46359) | NDUFS5 (+10685) | |
| 14 | 74385562 | ZNF410 | 5.5 | 4.45E-08 | 0.0393 | COQ6 (−31433) | ZNF410 (+31989) |
| 2 | 69225225 | 5.5 | 4.61E-08 | 0.0393 | ANTXR1 (−15084) | GKN1 (+23521) |
Note: Chr., chromosome; bp, base pair; stat, statistic.
aThe start location of the locus covering the CpGs investigated.
bUCSC Genes (gene symbol) within 2000 bp upstream (potential promoter region) or directly overlapping with the location of the locus.
cExtended gene annotation was performed with GREAT19 (v. 3.0.0) using a Basal Plus Extension that allowed for 5000 bp upstream, 1000 bp downstream, and a maximum extension of 1 000 000 bp. Annotated genes and the upstream/downstream (−/+) distance to the transcription start site is indicated.
In addition to the 22 highly significant loci, 852 suggestively significant (P < 1x10−5) CpGs were detected. The location of all 874 associated loci were enriched for a number of genomic features (supplementary table S2), including, genes (P < 1.00 × 10−5, odds ratio = 1.15) and potential promoter regions defined as 8 kb upstream of genes (P < 1.00 × 10−5, odds ratio = 1.18). The associated loci involved 388 genes that were enriched (P < 0.01, q < 0.25, minimum 3 gene overlap) for 6 GO terms that segregated into 3 clusters (see supplementary table S3a for full statistics). The first cluster (supplementary figure S3, red) involved immune responses and was primarily driven by top findings in toll-like receptor (TLR) genes. The second cluster of enriched GO terms (supplementary figure S3, yellow) involved integrin complex and collagen-activated signaling pathway.
The Significance of the Cumulative MWAS Signal
Encouraged by the MWAS findings that suggested many associated CpGs, we explored the cumulative effect of these signals. Results show that many markers together contribute more to the disease association than any single marker individually. When screening the top 100 000 markers, we detected a Pearson correlation of 0.252 (P = 8.6 × 10−23) between methylation-predicted status and actual disease status. Thus, the cumulative variation in methylation of these markers explain 6.4% of the schizophrenia disease status. The use of 100 000 markers corresponds to the point at which the correlation reaches a stable plateau (supplementary figure S4) suggesting this selection contains the majority of methylation markers with effect. However, it should be noted that not all these markers have an effect, but rather suggests that the majority of markers with an effect are located among the top 100 000.
Consistency in Findings Across Independent Methylation Studies
To study the robustness of the association findings we compared the results from the cell-type-corrected MBD-seq MWAS with the results from 3 array-based schizophrenia methylation studies of independent cohorts. The primary MBD-seq MWAS, presented earlier, included >99% of the 420 463–456 513 CpGs that were assayed via array. MBD-seq signals implicate loci that are up to the size of the sequenced DNA fragments. Therefore, we allowed for a 150 bp flank when matching the MBD-seq results to the array results. Similar to excluding single nucleotide polymorphisms with low minor allele frequencies, non-methylated CpGs are excluded in the MBD-seq quality control procedure. A substantial portion of these excluded sites were assayed by the arrays. Thus, using the 2 independent array datasets from Hannon et al, we cross-checked the methylation levels of the excluded sites. The array data also indicated that the methylation levels of these excluded CpGs were relatively low across all subjects (median/mean = 0.09/0.21 and 0.08/0.19, for Hannon-1 and Hannon-2, respectively) compared to the non-excluded sites in the same datasets (median/mean = 0.80/0.70 and 0.78/0.68, respectively). Thus, after excluding these uninformative sites and applying the flank 1 343 696–1 519 457 CpGs from the MBD-seq data could be mapped to 253 680–281 651 CpGs from the array data. The number of mapped sites for each dataset are given in table 2.
Table 2.
Enrichment Testing of Overlapping Top Sites Between the Methyl-Binding Domain Sequencing (MBD-seq) Methylome-Wide Association Study (MWAS) and Each of the Array-Based Methylation Studies
| Array Dataset | Sample Size | Number of Assayed CpGs | Number of MBD-seq CpGs Within 150 bp | Number of Unique CpGs Mapped | Odds Ratio | P |
|---|---|---|---|---|---|---|
| Montano | 1,334 | 456 219 | 1 519 457 | 281 651 | 1.1244 | 0.0315 |
| Hannon-1 | 675 | 420 463 | 1 344 100 | 253 771 | 1.0810 | 0.1946 |
| Hannon-2 | 847 | 420 374 | 1 343 696 | 253 680 | 1.0767 | 0.0045 |
Using only the mapped sites described in the previous paragraph, a significantly larger overlap than expected by chance was observed between top findings of the MBD-seq MWAS and 2 of the 3 array-based datasets (table 2). That is, the top 1% of findings in the study by Montano et al5 were more frequently observed (P = 0.0315, odds ratio = 1.12) among the top 5% of findings from the MBD-seq MWAS. Similarly, the top 5% of findings in the larger dataset (Hannon-2) from the study by Hannon et al4 showed significant enrichment of overlap (P = 0.0045, odds ratio = 1.08) among the top 5% of MBD-seq MWAS findings. Although the point estimate suggested enrichment, the smaller dataset (Hannon-1) from Hanon et al4 did not reach significance with the tested thresholds (P = 0.1946, odds ratio = 1.08).
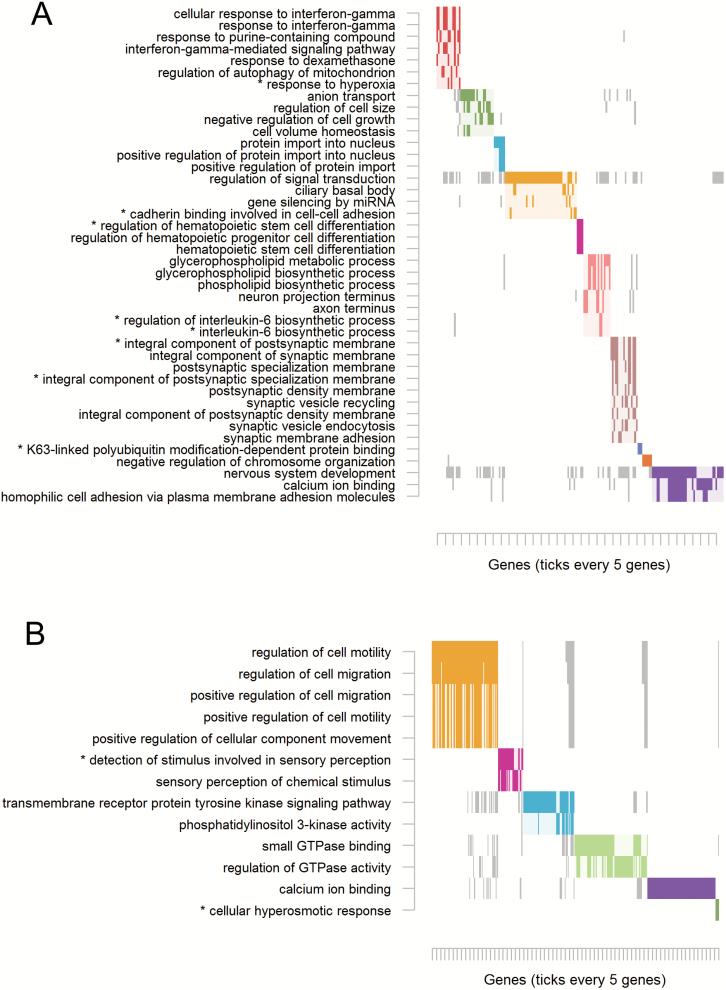
Focusing on the significantly overlapping datasets, we identified 457 and 2211 array CpGs overlapping top MBD-seq MWAS CpGs that mapped to 387 and 1613 genes for Montano and Hannon-2, respectively. Enrichment for 42 GO terms was observed for Montano that segregated into 10 clusters (supplementary table S3b). Most notably, overlapping results with Montano yielded a large cluster of genes involved with synaptic membrane function and structure (figure 2A, cluster 7, brown). Also implicated were nervous system development and calcium ion binding. Other notable clusters involved immune and/or stress responses (figure 2A, cluster 1, red), as well as regulation of signal transduction and cadherin binding involved in cell-cell adhesionfigure 2A, cluster 4, yellow).
Fig. 2.
Cluster plot of significantly enriched Gene Ontology terms for overlapping sites. (A) Overlap between methyl-binding domain sequencing (MBD-seq) methylome-wide association study (MWAS) and Montano. (B) Overlap between MBD-seq MWAS and Hannon-2. As pathways often share genes, the plot visualizes the clustering of pathways (y-axis) determined on the basis of their overlapping genes (x-axis). The solid rectangles indicate genes that were among the top MWAS results and belonged to the listed term. Only genes among the top MWAS results are plotted rather than all possible term members. Only terms containing ≥3 overlapping genes and those passing P < 0.01 and Q ≤ 0.25 are retained. Asterisks (*) indicate significance at an empirically determined family-wide error rate of 0.05. Complete overlaps and statistics are presented in supplementary table S2.
Results for the overlap with Hannon-2 yielded enrichment for 13 GO terms that formed 6 clusters (supplementary table S3c). The first and largest cluster of terms involved cell migration and motility (figure 2B, yellow). Many olfactory receptor genes also drove enrichment of terms for detection of stimulus involved in sensory perception and sensory perception of chemical stimulus (figure 2B, cluster 2, magenta). Finally, calcium ion binding, a term enriched for Montano, was again supported by the overlap with Hannon-2.
Discussion
In contrast to our previous MWAS for schizophrenia of the MBD-seq data,1 there is now an MBD-seq-specific reference panel available6 that allows for control of cell-type heterogeneity. Our new cell-type-corrected MWAS revealed several highly significant findings and the resampling approach further showed that the significance of the cumulative MWAS signal replicated. With the exception of the locus on chromosome 20, which reached suggestive significance in our previous study, the highly significant sites detected in this study were not detected by our previous analysis.1 Similarly, the replicating top loci located in FAM63B previously reported1 did not reach significance in the current analysis, emphasizing the importance of correcting for cell-type proportions in methylation studies. However, although our new MWAS revealed a different set of findings in the absolute top, we also observed a significant enrichment of overlapping top findings with the results from our previous analysis1 performed without correction for cell types. A site-by-site comparison of the significant findings detected in the current and previous study is reported in supplementary table S4.
We also observed significant enrichment of findings between the new MWAS and recent independent array-based methylation association studies for schizophrenia. Interestingly, overrepresentation analysis of the genes located in the top MWAS results and in the significant overlaps suggested similarity in the biological functions detected by the different datasets.
The inclusion of cell-type proportions as covariates in the MWAS minimizes the risks that methylation associations were caused by different distributions of cell-type proportions between cases and controls. However, it is still possible that other confounding factors influenced the results. One such factor is smoking, which is more frequent among schizophrenia patients than controls. Smoking status was not available for all participants in the MBD-seq MWAS and could not be included as a covariate in the analysis. However, none of the 22 highly significant or 852 suggestively significant top findings overlapped with any loci previously associated with smoking in methylation studies.28 Moreover, it should be noted that the studies by Montano et al5 and Hannon et al,4 both controlled for smoking status by either excluding sites with known smoking associations or estimating smoking status from the methylation profiles for use as a covariate. Thus, the observed enrichment between datasets is likely independent of smoking status in the participants. Taken together, these results suggest that the set of loci with schizophrenia associations detected by large-scale methylations studies is robust with potential biological relevance for schizophrenia.
Replicable Findings Across Studies
The majority (>99%) of all CpGs assayed on the array were also assayed in the MBD-seq data. In contrast, many markers assayed in the MBD-seq data were not assayed with the arrays. Therefore, the methylation array data cannot be used as a direct replication for most of the top findings detected in the primary MWAS. However, even though the array-based studies lacked information (mainly because the sites were not assayed) from the most significant schizophrenia associations detected in the MBD-seq MWAS, a significant enrichment of findings among the sites that were assayed with both platforms was detected. This further supports the notion that many loci are involved.
The significant overlap in findings between the MBD-seq MWAS and the results in the Montano and Hannon-2 datasets provided novel evidence supporting the robustness of findings in large-scale methylation association studies for schizophrenia. Critically, the use of 2 different approaches among the datasets mitigate artificial associations caused by platform-specific technical artifacts. Moreover, although the analysis for the different datasets followed standards used in the field, each dataset was generated by 3 independent research teams. Thus, artifacts caused by undetected errors in data processing and analysis are unlikely. In addition, the different datasets represent independent participants from different geographical locations who belonged to cohorts of different demographics. Therefore, findings support that schizophrenia associations are robust and reproducible in independent study samples.
Consistency in Biological Themes
The genes located in the overlapping loci between datasets and from the top MBD-seq MWAS findings showed overrepresentation among many GO terms with similar biological themes or those related to schizophrenia. Immune and/or stress-related pathways were featured among top MBD-seq MWAS findings and overlap with Montano. Genes for TLRs and those related to interferon-γ and interleukin signaling drove enrichment of immune-related GO terms. Although the TLRs implicated in our results, TLR1 and TLR6, have not been previously implicated in schizophrenia, enhanced TLR2-mediated cytokine release has been shown in psychosis.29 However, TLR1 and TLR6 serve as co-receptors and modulators of TLR2 activity.30 Overall, these results are consistent with the hypothesis of innate immune system dysregulation in schizophrenia.31,32
A second recurrent theme that emerged among enriched GO terms were those related to cell–cell adhesion and/or cell motility. Among the results for the new MBD-seq MWAS were genes enriched for integrin and collagen signaling. Integrins are key receptors for extracellular collagen and are essential for cell adhesion and motility.33 Integrins also interact with schizophrenia candidate gene RELN that affects neuronal migration.34,35 Interestingly, schizophrenia patient cells display altered focal adhesion complexes and cell motility36 as well as increased integrin expression.37 Overlap results for Montano further implicated cadherin binding and cell motility terms for Hannon-2. These apparent defects in cell migration pathways in blood may provide clues that similar events may occur in the brain of schizophrenia patients.
We also found support for altered methylation at genes important for calcium ion binding in the overlap vs Montano and in the overlap vs Hannon-2. Calcium signaling regulates essential features of neuronal and glial biology where its dysregulation contributes to numerous neuropsychiatric disorders.38,39 The endoplasmic reticulum and mitochondria interact to serve as sensors and buffers of intracellular calcium.40 A key regulator of mitochondrial fusion, MFN2, contained the top result of the MBD-seq MWAS. Numerous studies have demonstrated impaired mitochondrial respiration in postmortem brain,41 as well as in neurons from peripherally derived induced pluripotent stem cells of schizophrenia patients.42 Thus, mitochondriopathy in schizophrenia may be linked to deficits in calcium homeostasis that affects both central and peripheral tissues.
Findings of Potential Clinical Value
The 4 methylation datasets used in this work were generated from blood samples. The number of themes emerging from the overlapping datasets that were related to neurobiology was notable considering that previous reports indicate that few CpGs (~10%) show interindividual concordance between blood and brain.18,43–45 Therefore, we checked whether enriched GO terms were driven by CpGs known to have correlation between blood and brain. Overall, 52.1/23.1% of genes implicated by overlap with Montano/Hannon-2, respectively, were driven by at least 1 CpG with evidence of modest or better interindividual correlation (r ≥ |0.2|) between blood and brain. Although many top findings are indeed likely to be blood-specific, these results indicate that blood may still provide substantive information about methylation in brain for a good number of important sites.
For example, overlap of results between the current MWAS study and those of Hannon-2 implicated detection of stimulus involved in sensory perception. In particular, enrichment of this term was driven by a number of results found within clusters of olfactory receptor genes. This is curious considering that deficits in olfaction are observed in both drug-naïve schizophrenia46 and in premorbid schizophrenia.47 Therefore, epigenetic markers that may predict disease features or risk feasibly exist in blood.
Conclusion
Our reanalyzed MWAS identified highly significant loci associated with schizophrenia in genes such as MFN2 and ALDH1A2 with potentially critical biological relevance for disease etiology. These results significantly overlapped with those of other recent large-scale methylation studies for schizophrenia and identified consistent biological themes related to disease. Taken together, these studies have produced robust and replicable findings with importance for schizophrenia research.
Funding
National Institute of Mental Health (R03MH102723 and R01MH109525 to Dr Aberg and RC2MH089996 to Dr van den Oord); UK Medical Research Council (MR/K013807/1 and MR/R005176/1 to Dr Mill).
Supplementary Material
Acknowledgments
The MBD-seq dataset by Aberg et al was generated in collaboration with the Swedish Schizophrenia Consortia. Full association results from the Montano et al study were used with permission from the principal investigators of the Consortium on the Genetics of Endophenotypes in Schizophrenia (COGS) (PI David Braff, MD), the Project among African Americans To Explore Risks for Schizophrenia (PAARTNERS) (PIs Rodney C. Go, PhD, Rodney T. Perry, PhD), and the Multiplex Multigenerational Family Study of Schizophrenia (MGI) (PIs Raquel E. Gur, MD, PhD; Vishwajit Nimgaonkar, MD, PhD), that generated the data. The sponsors had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; or preparation, review, or approval of the manuscript.The authors declare no competing financial interests.
References
- 1. Aberg KA, McClay JL, Nerella S, et al. Methylome-wide association study of schizophrenia: identifying blood biomarker signatures of environmental insults. JAMA Psychiatry. 2014;71(3):255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aberg KA, Chan RF, Shabalin AA, et al. A MBD-seq protocol for large-scale methylome-wide studies with (very) low amounts of DNA. Epigenetics. 2017;12(9):743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chan RF, Shabalin AA, Xie LY, et al. Enrichment methods provide a feasible approach to comprehensive and adequately powered investigations of the brain methylome. Nucleic Acids Res. 2017;45(11):e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hannon E, Dempster E, Viana J, et al. An integrated genetic-epigenetic analysis of schizophrenia: evidence for co-localization of genetic associations and differential DNA methylation. Genome Biol. 2016;17(1):176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Montano C, Taub MA, Jaffe A, et al. Association of DNA Methylation Differences With Schizophrenia in an Epigenome-Wide Association Study. JAMA Psychiatry. 2016;73(5):506–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hattab MW, Shabalin AA, Clark SL, et al. Correcting for cell-type effects in DNA methylation studies: reference-based method outperforms latent variable approaches in empirical studies. Genome Biol. 2017;18(1):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinf. 2012;13:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Serre D, Lee BH, Ting AH. MBD-isolated Genome Sequencing provides a high-throughput and comprehensive survey of DNA methylation in the human genome. Nucleic Acids Res. 2010;38(2):391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aberg KA, Xie L, Chan RF, et al. Evaluation of Methyl-Binding domain based enrichment approaches revisited. PLoS One. 2015;10(7):e0132205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Aberg KA, McClay JL, Nerella S, et al. ; Swedish Schizophrenia Consortium MBD-seq as a cost-effective approach for methylome-wide association studies: demonstration in 1500 case–control samples. Epigenomics. 2012;4(6):605–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu Y, Popp B, Schmidt B. CUSHAW3: sensitive and accurate base-space and color-space short-read alignment with hybrid seeding. PLoS One. 2014;9(1):e86869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shabalin AA, Hattab MW, Aberg KA, Clark SL, Chan RF, Kumar G and van den Oord EJCG. RaMWAS: Fast Methylome-Wide Association Study Pipeline for Enrichment Platforms Bioinformatics. 2018;34(13):2283–2285.http://bioconductor.org/packages/release/bioc/html/ramwas.html. Accessed May 17, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33(1):1–22. [PMC free article] [PubMed] [Google Scholar]
- 14. Simon N, Friedman J, Hastie T, Tibshirani R. Regularization paths for Cox’s proportional hazards model via coordinate descent. J Stat Softw. 2011;39(5):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tibshirani R, Bien J, Friedman J, et al. Strong rules for discarding predictors in lasso-type problems. J R Stat Soc Series B Stat Methodol. 2012;74(2):245–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hastie T, Tibshirani R, Friedman J.. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. New York, NY: Springer Verlag; 2001. [Google Scholar]
- 17. Aberg KA, Dean B, Shabalin AA, Chan RF, et al. Methylome-wide association findings for major depressive disorder overlap in blood and brain and replicate in independent brain samples [published online ahead of print 2018]. Mol Psychiatry. doi: 10.1038/s41380-018-0247-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Edgar RD, Jones MJ, Meaney MJ, Turecki G, Kobor MS. BECon: a tool for interpreting DNA methylation findings from blood in the context of brain. Transl Psychiatry. 2017;7(8):e1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McLean CY, Bristor D, Hiller M, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28(5):495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Park SJ, Lee SB, Suh Y, et al. DISC1 modulates neuronal stress responses by Gate-Keeping ER-Mitochondria Ca2+ Transfer through the MAM. Cell Rep. 2017;21(10):2748–2759. [DOI] [PubMed] [Google Scholar]
- 21. Fang D, Yan S, Yu Q, Chen D, Yan SS. Mfn2 is required for mitochondrial development and synapse formation in human induced pluripotent stem Cells/hiPSC derived cortical neurons. Sci Rep. 2016;6:31462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pham AH, Meng S, Chu QN, Chan DC. Loss of Mfn2 results in progressive, retrograde degeneration of dopaminergic neurons in the nigrostriatal circuit. Hum Mol Genet. 2012;21(22):4817–4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wan C, Shi Y, Zhao X, et al. Positive association between ALDH1A2 and schizophrenia in the Chinese population. Prog Neuropsychopharmacol Biol Psychiatry. 2009;33(8):1491–1495. [DOI] [PubMed] [Google Scholar]
- 24. Mizee MR, Nijland PG, van der Pol SM, et al. Astrocyte-derived retinoic acid: a novel regulator of blood-brain barrier function in multiple sclerosis. Acta Neuropathol. 2014;128(5):691–703. [DOI] [PubMed] [Google Scholar]
- 25. Shearer KD, Fragoso YD, Clagett-Dame M, McCaffery PJ. Astrocytes as a regulated source of retinoic acid for the brain. Glia. 2012;60(12):1964–1976. [DOI] [PubMed] [Google Scholar]
- 26. Goodman AB. Three independent lines of evidence suggest retinoids as causal to schizophrenia. Proc Natl Acad Sci USA. 1998;95(13):7240–7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Palha JA, Goodman AB. Thyroid hormones and retinoids: a possible link between genes and environment in schizophrenia. Brain Res Rev. 2006;51(1):61–71. [DOI] [PubMed] [Google Scholar]
- 28. Gao X, Jia M, Zhang Y, Breitling LP, Brenner H. DNA methylation changes of whole blood cells in response to active smoking exposure in adults: a systematic review of DNA methylation studies. Clin Epigenetics. 2015;7:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McKernan DP, Dennison U, Gaszner G, Cryan JF, Dinan TG. Enhanced peripheral toll-like receptor responses in psychosis: further evidence of a pro-inflammatory phenotype. Transl Psychiatry. 2011;1:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee CC, Avalos AM, Ploegh HL. Accessory molecules for Toll-like receptors and their function. Nat Rev Immunol. 2012;12(3):168–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Müller N, Riedel M, Gruber R, Ackenheil M, Schwarz MJ. The immune system and schizophrenia. An integrative view. Ann N Y Acad Sci. 2000;917:456–467. [DOI] [PubMed] [Google Scholar]
- 32. Patterson PH. Immune involvement in schizophrenia and autism: etiology, pathology and animal models. Behav Brain Res. 2009;204(2):313–321. [DOI] [PubMed] [Google Scholar]
- 33. Heino J. Cellular signaling by collagen-binding integrins. In: Gullberg D, ed. I Domain Integrins. Dordrecht, the Netherlands: Springer; 2014:143–155. [DOI] [PubMed] [Google Scholar]
- 34. Dulabon L, Olson EC, Taglienti MG, et al. Reelin binds alpha3beta1 integrin and inhibits neuronal migration. Neuron. 2000;27(1):33–44. [DOI] [PubMed] [Google Scholar]
- 35. Guidotti A, Auta J, Davis JM, et al. Decrease in reelin and glutamic acid decarboxylase67 (GAD67) expression in schizophrenia and bipolar disorder: a postmortem brain study. Arch Gen Psychiatry. 2000;57(11):1061–1069. [DOI] [PubMed] [Google Scholar]
- 36. Fan Y, Abrahamsen G, Mills R, et al. Focal adhesion dynamics are altered in schizophrenia. Biol Psychiatry. 2013;74(6):418–426. [DOI] [PubMed] [Google Scholar]
- 37. Walsh MT, Ryan M, Hillmann A, et al. Elevated expression of integrin alpha(IIb) beta(IIIa) in drug-naïve, first-episode schizophrenic patients. Biol Psychiatry. 2002;52(9):874–879. [DOI] [PubMed] [Google Scholar]
- 38. Nanou E, Catterall WA. Calcium channels, synaptic plasticity, and neuropsychiatric disease. Neuron. 2018;98(3):466–481. [DOI] [PubMed] [Google Scholar]
- 39. Bazargani N, Attwell D. Astrocyte calcium signaling: the third wave. Nat Neurosci. 2016;19(2):182–189. [DOI] [PubMed] [Google Scholar]
- 40. Rizzuto R, De Stefani D, Raffaello A, Mammucari C. Mitochondria as sensors and regulators of calcium signalling. Nat Rev Mol Cell Biol. 2012;13(9):566–578. [DOI] [PubMed] [Google Scholar]
- 41. Flippo KH, Strack S. An emerging role for mitochondrial dynamics in schizophrenia. Schizophr Res. 2017;187:26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Robicsek O, Karry R, Petit I, et al. Abnormal neuronal differentiation and mitochondrial dysfunction in hair follicle-derived induced pluripotent stem cells of schizophrenia patients. Mol Psychiatry. 2013;18(10):1067–1076. [DOI] [PubMed] [Google Scholar]
- 43. Hannon E, Lunnon K, Schalkwyk L, Mill J. Interindividual methylomic variation across blood, cortex, and cerebellum: implications for epigenetic studies of neurological and neuropsychiatric phenotypes. Epigenetics. 2015;10(11):1024–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Davies MN, Volta M, Pidsley R, et al. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol. 2012;13(6):R43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bernardoni F, Roessner V, Walton E, et al. Correspondence of DNA methylation between blood and brain tissue and its application to schizophrenia research. Schizophr Bull. 2015;42(2):406–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Moberg PJ, Agrin R, Gur RE, Gur RC, Turetsky BI, Doty RL. Olfactory dysfunction in schizophrenia: a qualitative and quantitative review. Neuropsychopharmacology. 1999;21(3):325–340. [DOI] [PubMed] [Google Scholar]
- 47. Brewer WJ, Wood SJ, McGorry PD, et al. Impairment of olfactory identification ability in individuals at ultra-high risk for psychosis who later develop schizophrenia. Am J Psychiatry. 2003;160(10):1790–1794. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.