Abstract
Forkhead box M1 (FOXM1) and NF-κB are oncogenic drivers in liver cancer that positively regulate each other. We showed that methionine adenosyltransferase 1A (MAT1A) is a tumor suppressor in the liver and it inhibits NF-κB activity. Here we examined the interplay between FOXM1/NF-kB and MAT1A in liver cancer. We examined gene and protein expression, effects on promoter activities and binding of proteins to promoter regions, effects of FOXM1 inhibitors T0901317 (T0) and FDI-6 in vitro and in xenograft and syngeneic models of liver cancer. We found in both hepatocellular carcinoma (HCC) and cholangiocarcinoma (CCA) an induction in FOXM1 and NF-κB expression is accompanied by fall in MATα1 (protein encoded by MAT1A). The TCGA dataset confirmed the inverse correlation between FOXM1 and MAT1A. Interestingly, FOXM1 directly interact with MATα1 and they negatively regulate each other. In contrast, FOXM1 positively regulates p50 and p65 expression via MATα1, as the effect is lost in its absence. FOXM1, MATα1 and NF-κB all bind to the FOX-binding sites in the FOXM1 and MAT1A promoters. However, binding of FOXM1 and NF-κB repressed MAT1A promoter activity but activated FOXM1 promoter. In contrast, binding of MATα1 repressed FOXM1 promoter. MATα1 also binds and represses the NF-κB element in the presence of p65 or p50. Inhibiting FOXM1 with either T0 or FDI-6 inhibited liver cancer cell growth in vitro and in vivo. However, inhibiting FOXM1 had minimal effects in liver cancer cells that do not express MAT1A.
Conclusion:
We have unveiled a novel crosstalk between FOXM1/NF-κB and MAT1A. Upregulation in FOXM1 lowers MAT1A but raises NF-κB expression and this is a feed forward loop that enhances tumorigenesis.
Keywords: Hepatocellular carcinoma, cholangiocarcinoma, TO901317, FDI-6, FOX-binding site
Forkhead box M1 (FOXM1) is a member of the forkhead box family of transcription factors that regulates cell cycle progression and mitosis.(1) Increased expression of FOXM1 has been reported in numerous cancers, including hepatocellular carcinoma (HCC),(1,2) and has been identified as an initiating factor in oncogenesis.(3) High expression of FOXM1 correlates with poor prognosis and has also been implicated in HCC progression through epithelial-mesenchymal transition (EMT) and metastasis.(4) A number of studies have examined targeting FOXM1 in cancer treatment. For instance, Forkhead Domain Inhibitory-6 (FDI-6), a small molecule that selectively targets the DNA binding domain of FOXM1, has been shown to repress transcription of FOXM1-target genes.(5) 9R-P201, a peptide that reduces the expression of FOXM1, reduced cell viability and induced apoptosis in HepG2 cells.(6) T0901317 (T0), an agonist for Liver X Receptor α (LXRα), suppressed FOXM1 transcription leading to HCC growth arrest in vitro.(7) Collectively these studies suggest targeting FOXM1 may be an attractive therapeutic strategy in HCC.
The human NF-κB family is composed of five cellular DNA-binding subunits: p50, p52, cRel, p65 (also known as RelA) and RelB, encoded by NF-κB1, NF-κB2, REL, RELA and RELB, respectively. The heterodimer p50/p65 is the most common form of NF-κB and a key driver in liver cancer.(8,9) FOXM1 promoter region contains a functional NF-κB element and is transcriptionally activated upon NF-κB binding in chronic myelogenous leukemia cells.(10) FOXM1 in return positively regulates NF-κB activity in lung cancer cells.(11) Interestingly, FOXM1 co-occupies with NF-κB (p65) on nearly half of the NF-κB binding sites in lymphoblastoid B cells.(12) Recently we showed that methionine adenosyltransferase 1A (MAT1A) is a tumor suppressor in the liver that inhibits NF-κB activity.(13) MAT1A is mainly expressed in normal hepatocytes and bile duct epithelial cells, encodes the enzyme MAT that is responsible for the biosynthesis of S-adenosylmethionine, the principle biological methyl donor. MAT1A expression falls in HCC and cholangiocarcinoma (CCA).(14,15) Downregulation of MAT1A expression in liver cancer has been attributed to promoter and coding region methylation, histone H4 deacetylation, and interactions with AUF1 (AU rich RNA binding factor 1).(15–17) In the current study we investigated the interplay between FOXM1/NF-κB and MAT1A in liver cancer and unveiled a novel feedforward loop triggered by higher FOXM1 and/or lower MAT1A expression in hepatocarcinogenesis.
EXPERIMENTAL PROCEDURES
Materials and reagents
Antibodies used for western blotting, immunohistochemistry (IHC), chromatin immunoprecipitation (ChIP), and sequential-ChIP (Seq-ChIP) for FOXM1, MATα1 (protein encoded by MAT1A), p65, p50, prohibitin 1 (PHB1), hepatocyte nuclear factor 4 (HNF4), alpha-fetoprotein (AFP), cytochrome P450 2E1 (CYP2E1), β-ACTIN, and IgG were purchased from Abcam (Cambridge, MA). MAT1A(18) and RELA (p65)(19) expression vectors were previously described. Please see Supplemental Experimental Procedures for sources of other reagents.
Source of normal liver, human HCC and CCA with adjacent non-tumorous tissues, primary biliary cholangitis (PBC) and primary sclerosing cholangitis (PSC)
Please see Supplemental Experimental Procedures for detailed description of the human liver specimens included in this study. The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki as reflected in a priori approval by the Institutional Review Boards of Cedars-Sinai Medical Center and the Medical Ethical Committee of Xiangya Hospital Central South University. Both tumor and normal tissues were evaluated histologically to confirm presence or absence of cancer.
Bioinformatics
Public genomic data from BaseSpace Correlation Engine (https://www.illumina.com/products/by-type/informatics-products/basespace-correlation-engine.html) was analyzed with the purpose of identifying the differential expression patterns of MAT1A, FOXM1, p65 and p50 at the mRNA level in liver cancer. Graphs showing oncoprint and survival analysis were generated using The Cancer Genome Atlas (TCGA) dataset from the cBio Cancer Genomics Portal (http://cbioportal.org).(20,21)
Ingenuity pathway analysis
Please see Supplemental Experimental Procedures for ingenuity pathway analysis.
Establishment of malignant HCC cell line from Mat1a knockout (KO) mice
Mat1a KO mice were described previously.(22) Please see Supplemental Experimental Procedures for detailed methodology. We named this cell line MATα1-D (D for deficient).
Human hepatocytes, cell lines and treatments
Primary human hepatocytes were obtained from ThermoFisher Scientific (Waltham, MA). MzChA-1 (human biliary adenocarcinoma), HepG2 (human hepatoblastoma), Hep3B (human hepatocellular carcinoma), H69 (human normal biliary epithelial cells),(14) SAMe-D (HCC cell line from Mat1a KO mouse),(23) and OKER cells (HCC cell line from glycine N-methyltransferase (Gnmt) KO mice)(24) were cultured and treated as described in Supplemental Experimental Procedures.
Treatment with T0 or FDI-6 in xenograft and syngeneic models
Details of T0 and FDI-6 treatments in the xenograft and syngeneic models are described in Supplemental Experimental Procedures. For xenografts, treatments began when the tumor reached 80 mm3 in size. In the syngeneic model treatment with FDI-6 was started at day seven and animals were sacrificed at day 11.
All procedure protocols, use, and the care of the animals were reviewed and approved by the Institutional Animal Care and Use Committee of University of South China (Hengyang, China) and Cedars-Sinai Medical Center (Los Angeles, CA).
Murine CCA model
Murine CCA model was previously described.(25) Frozen tissues from CCA and normal control livers were used in western blotting.
Promoter constructs and luciferase assay
The human MAT1A promoter was described previously.(26) The human 1.3kb FOXM1 promoter was purchased from Genecopoeia (NM_001243088). The construct containing multimerized NF-κB element (TGGGGACTTTCCGC)X5 was previously described.(13) Please see Supplemental Experimental Procedures for the details of mutagenesis where FOX binding sites were mutated in MAT1A and FOXM1 promoters.
Histology and immunohistochemistry (IHC)
Please see Supplemental Experimental Procedures for detailed methods.
RNA isolation and gene expression analysis
Please see Supplemental Experimental Procedures for details of RNA isolation and gene expression analysis.
Western blot analysis
Total protein extracts from cells expressing varying amounts of FOXM1, MATα1, p65 and p50 were subjected to western blot analysis as described.(14) To ensure equal loading, membranes were stripped and re-probed with anti-ACTIN antibodies. Blots were developed by enhanced chemiluminescence (Millipore Corporation, Temecula, CA).
Electrophoretic mobility shift assay (EMSA) and supershift
One probe was 32P-end-labeled double-stranded three FOX elements, corresponding to −810 to −818, −752 to −762 and −719 to −728 (relative to the transcriptional start codon of human MAT1A (NM-000429). Another probe was 32P-end-labeled double-stranded two NF-κB elements, corresponding to −664 to −679 and −30 to −45 (relative to the transcriptional start codon of human FOXM1 (NM_001243088). EMSA and supershift were done as previously described using recombinant proteins and lysates from HCC, CCA and their adjacent non-tumorous tissues.(27,28)
Chromatin immunoprecipitation (ChIP) and sequential-ChIP (Seq-ChIP) assay
ChIP was done to examine changes in protein binding to the FOX binding region of the human MAT1A and FOXM1 promoters using the manufacturer’s suggested protocol from the EpiTect ChIP OneDay kit (Qiagen, Germantown, MD). Please see Supplemental Experimental Procedures for details.
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay
Cells were seeded in 96-well plates at 5×103 cells per well. Upon reaching 70%−80% confluence cells were treated with different concentrations of FDI-6. Thereafter, 20μl of MTT (5g/mL) was added to the cells for four hours before MTT was removed. The value of optical density was measured at 490nm using a CLARIOSTAR (BMG Labtech, Cary, NC) while the cells were suspended in 150μl of dimethyl sulfoxide.
Migration and invasion assays
Migration and invasion assays were performed as described.(29)
MATα1 and FOXM1 interaction
Recombinant human MATα1 (CAT#: enz-493-b) was purchased from PROSPEC (East Brunswick NJ) and FOXM1 (CAT#: TP76157 was from OriGENE (Rockville, MD). Two μg of MATα1 or FOXM1 protein was immobilized to agarose beads by their respective antibody (Abcam). After washing, beads were mixed with 1 μg MATα1 or FOXM1 protein and rotated for 4 hours at 4°C. Beads were then washed 6 times, boiled in SDS sample buffer and proteins separated on SDS-PAGE.
Statistical analysis
Data are given as mean ± standard error of the mean. Statistical analysis was performed using analysis of variance followed by Fisher’s test for multiple comparisons, and two-tailed Student’s t-test for paired comparisons. For changes in mRNA and protein levels, ratios of genes or proteins to housekeeping genes or proteins densitometric values were compared. Student’s T-test was performed for statistical analysis of Pearson correlation. For survival analysis, log-rank test was used to compare the survival ratio differences between samples with high versus low MAT1A and FOXM1 expressions. Significance was defined by p<0.05.
RESULTS
FOXM1/NF-κB/MAT1A expression profiles in liver cancer
To analyze expression profiles of these genes, we retrieved FOXM1/NF-κB/MAT1A expression from databases of human liver cancers from BaseSpace CORRELATION ENGINE and GEO databases. We found MAT1A mRNA levels are lower while FOXM1/NF-κB (p50, p65) mRNA levels higher in HCC and CCA databases (Supplemental Tables 1–3). We also investigated mRNA levels of MAT1A and FOXM1 from 143 HCC tumor tissues and FOXM1 mRNA levels from seven CCA tumors and respective adjacent non-tumor liver tissues. Our CCA specimens, which have much lower MAT1A expression,(14) have markedly increased FOXM1 and MMP-7 mRNA levels (Fig. 1A). The same inverse relationship between MAT1A and FOXM1 mRNA levels is observed in our HCC specimens (Fig. 1B and C), in the TCGA dataset (Fig. 1D), and in numerous GEO datasets for HCC (Supplemental Fig. 1A) and CCA (Supplemental Fig. 1B). MAT1A mRNA levels are also inversely correlated with NF-κB1 (encodes for p50) and RELA (encodes for p65) mRNA levels in HCC, while FOXM1 mRNA levels positively correlated with NF-κB1 mRNA levels (Supplemental Fig. 2). In the TCGA database, HCC patients with higher FOXM1 (mainly amplification and mRNA up-regulation) and lower MAT1A mRNA levels had worse survival (Fig. 1E and 1F).
Figure 1. FOXM1 and MAT1A mRNA levels in HCC and CCA, and their influences on survival curves in HCC.
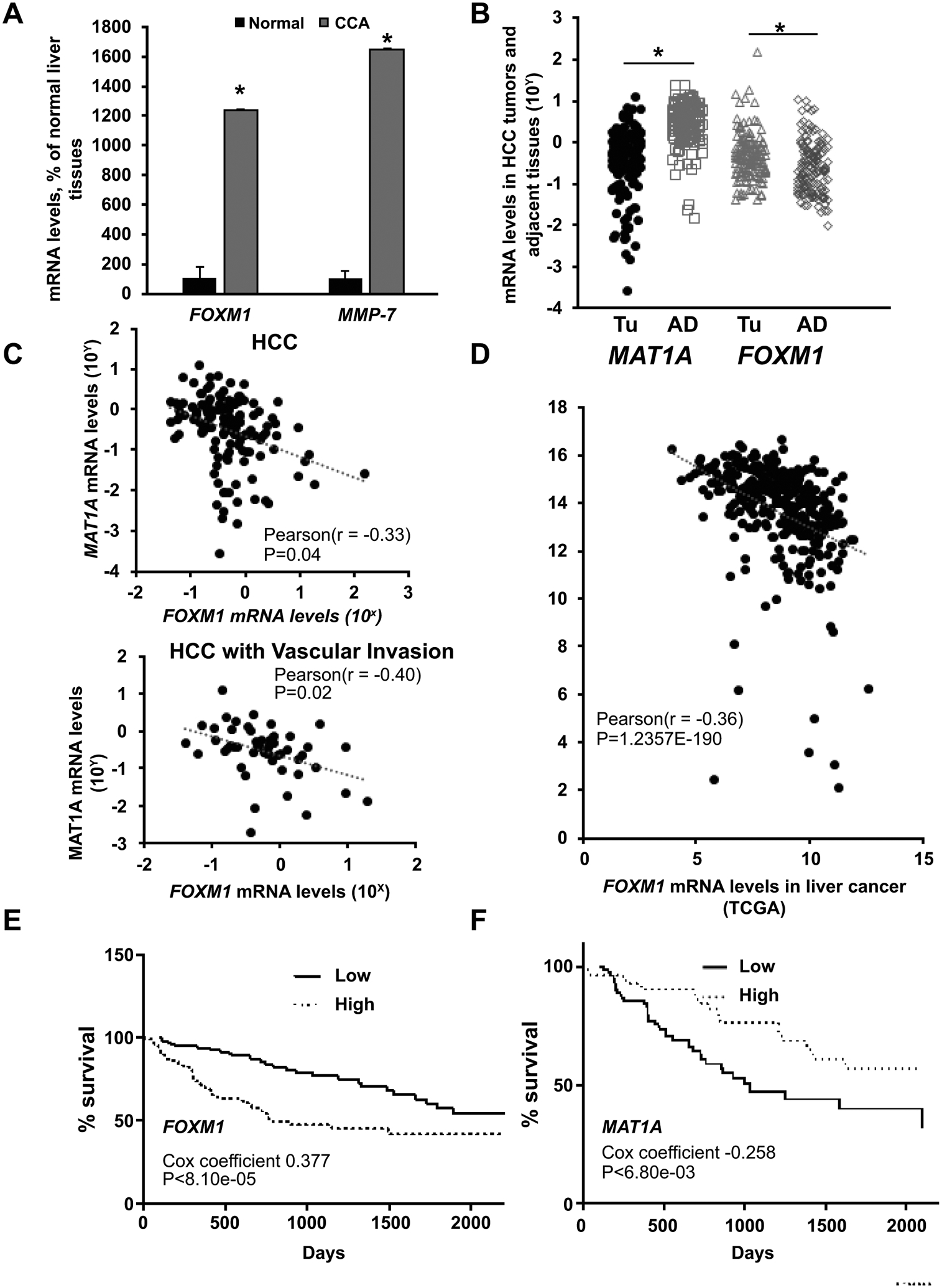
A) Relative FOXM1 and MMP-7 mRNA levels from seven patients with CCA compared to normal liver tissues. *p < 0.05 vs. normal human liver tissues. B) Relative MAT1A and FOXM1 mRNA levels from 143 patients with HCC (TU) compared to adjacent non-tumorous tissues (AD). *p < 0.05 vs. AD. C) Pearson correlation analysis of MAT1A and FOXM1 mRNA levels in 143 HCC specimens and in 52 HCCs with vascular invasion. D) Pearson correlation analysis of FOXM1 and MAT1A mRNA levels in HCC Samples (N=366) from the Cancer Genome Atlas (TCGA) dataset. E and F) Kaplan-Meier survival curves of HCC patients from the TCGA dataset stratified into high (upper 33rd percentile, n=118) or low (lower 33rd percentile, n=118) FOXM1 (E) and MAT1A (F) mRNA levels.
Consistent with changes in mRNA levels, MATα1 (encoded by MAT1A) protein levels are lower while FOXM1, p50, and p65 protein levels are higher in HCC and CCA compared to adjacent non-tumor tissues and normal livers on IHC (Fig. 2A) and western blots (Fig. 2B and 2C). On higher magnification, these proteins are present in the cytosolic and nuclear compartments (Fig. 2A). Downregulation in MATα1 and upregulation in FOXM1, p50, and p65 protein levels are also observed in chronic cholestatic liver diseases such as primary biliary cholangitis and primary sclerosing cholangitis (Supplemental Fig. 3).
Figure 2. Protein expression of MATα1, FOXM1, p50, and p65 from HCC and CCA, adjacent and normal liver tissues.
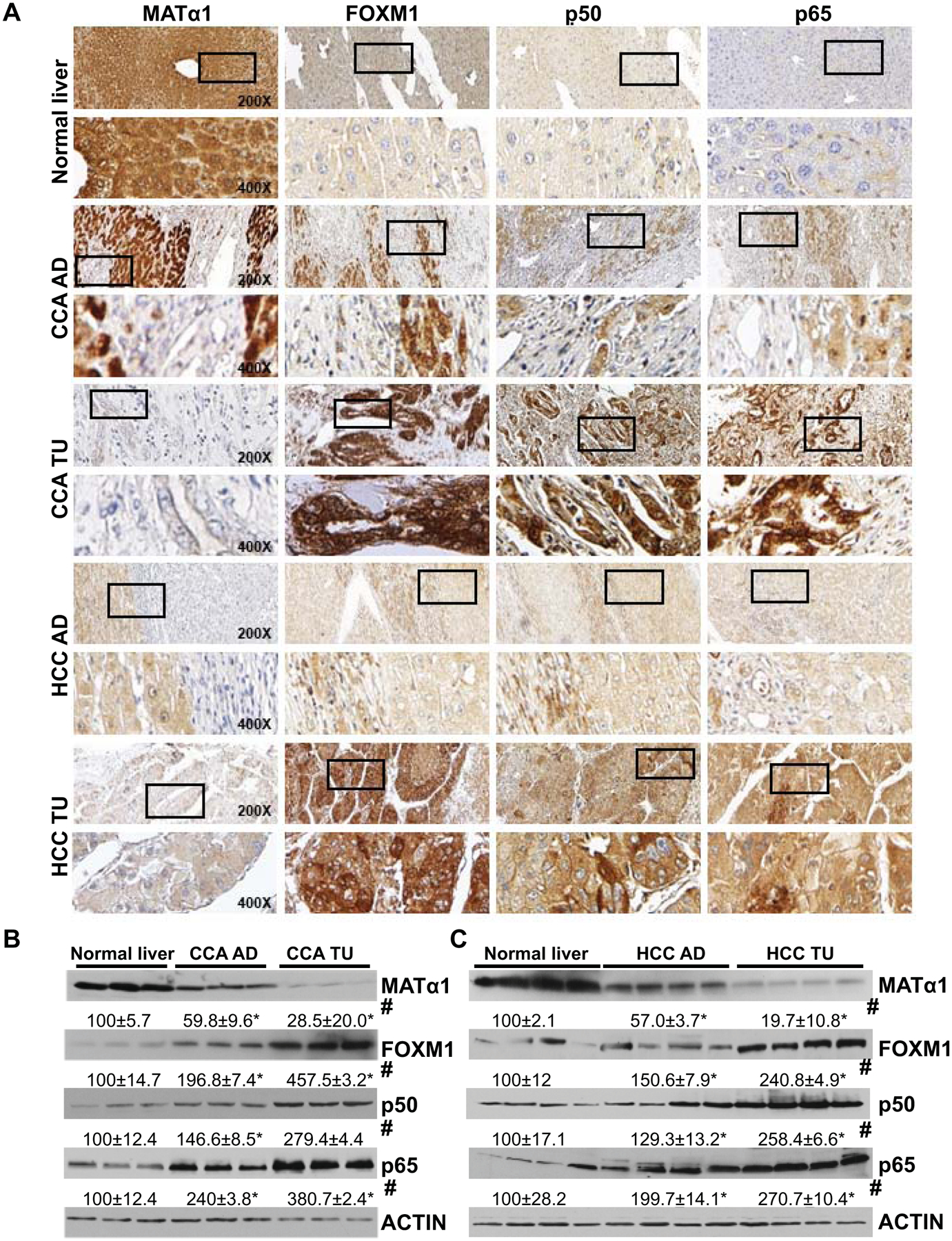
A) Representative immunohistochemistry (IHC) staining of MATα1, FOXM1, p50, and p65 from normal liver, non-tumorous tissue adjacent to CCA (CCA-AD), CCA (CCA-TU), HCC-AD, and HCC-TU. All are at 200X magnification. Higher magnification (400X) of boxed areas is shown below for each IHC. B) MATα1, FOXM1, p50, and p65 protein levels from three pairs of CCA and adjacent tissues (CCA-AD), and three normal liver specimens on western blotting. Numbers below the blots are densitometric values, expressed as percent of normal liver. *p < 0.05. vs. normal liver; #p < 0.05 vs. adjacent tissues. C) MATα1, FOXM1, p50, and p65 protein levels in four pairs of HCC and adjacent tissues (HCC-AD), and four normal liver specimens on western blotting. *p < 0.05 vs. normal liver tissues; #p < 0.05 vs. adjacent tissues.
Reciprocal Regulation Between FOXM1/NF-kB and MAT1A
Hepatocytes in culture rapidly dedifferentiate and one of the changes is a rapid fall in MAT1A mRNA level.(30) Using this model, we examined the relationship between MAT1A and FOXM1/NF-κB (p50/p65). We found MATα1 level decreased while FOXM1 and p65 levels increased as early as 30 minutes after plating, and this was followed closely by an increase in p50 levels. These changes coincided with loss of differentiated markers such as CYP2E1 and HNF4 and gain of de-differentiation marker AFP (Fig. 3A). To determine if there is a reciprocal regulation between FOXM1, NF-κB and MAT1A, we varied the expression of MAT1A or FOXM1 in HepG2 and SAMe-D cell lines using siRNA and overexpression vectors. In HepG2 cells, MAT1A knockdown raised the protein level of FOXM1, p50, and p65, while the opposite occurred with MAT1A overexpression (Fig. 3B). Interestingly, overexpressing the catalytic mutant of MAT1A (can only form MATα1 monomer, which is catalytically inactive) failed to suppress FOXM1, p50 and p65 expression (Fig. 3B). In return, FOXM1 overexpression lowered the protein level of MATα1 and raised the protein levels of p50 and p65, while the opposite occurred with FOXM1 knockdown (Fig. 3C). However, in SAMe-D cells, varying FOXM1 expression had no effect on p50 and p65 levels unless they were also transfected with MAT1A overexpression vector (Fig. 3D), suggesting regulation of p50 and p65 expression by FOXM1 required MATα1. To make sure the lack of effect of FOXM1 on NF-κB (p50/p65) expression is not unique to SAMe-D cells, we also examined MATα1-D cells. Similar to SAMe-D cells, varying FOXM1 expression did not affect p50 or p65 protein levels (Supplemental Fig. 4A and 4B). Although both SAMe-D and MATα1-D cells are from HCCs of Mat1a KOs, SAMe-D cells grew much slower in comparison to MATα1-D cells (Supplemental Fig. 4C).
Figure 3. Reciprocal regulation between FOXM1/NF-κB and MAT1A.
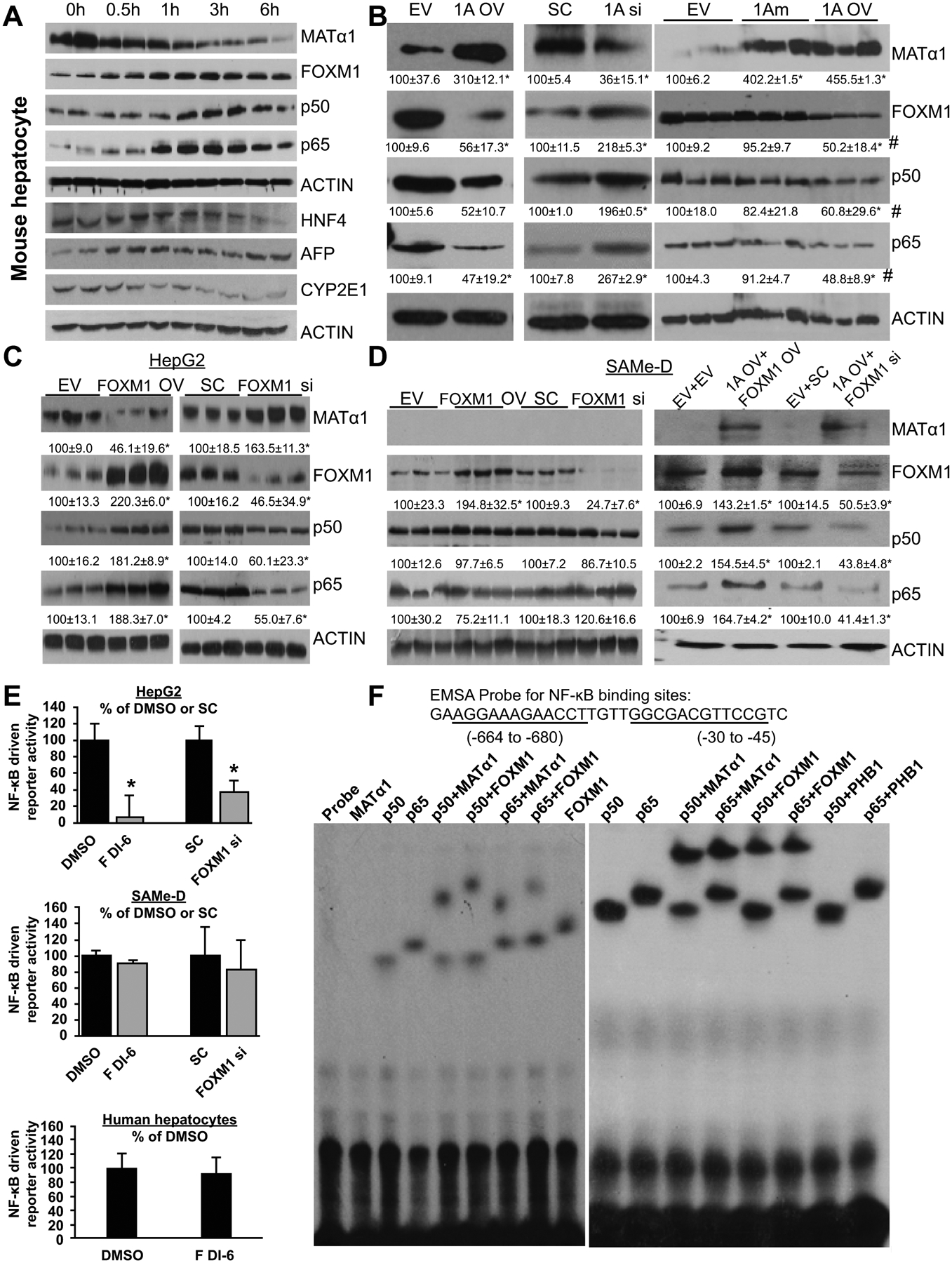
A) Protein levels of MATα1, FOXM1, p50, p65, HNF4, AFP, CYP2E1, and ACTIN were measured using western blotting in primary mouse hepatocytes at the time of isolation (0 hour = 0h) and up to 6 hours (6h) after plating. B) Protein levels of MATα1, FOXM1, p50 and p65 after overexpressing wild type MAT1A (1A OV), catalytic mutant of MAT1A (1Am) or siRNA knockdown (1A si) as compared to empty vector (EV) or scramble (SC) controls, respectively, in HepG2 cells. Numbers below the blots are densitometric values, expressed as percent of EV or SC in mean ± SEM from three experiments. *p < 0.05 vs. EV or SC; #p<0.05 vs. 1Am. C-D) Protein levels of MATα1, FOXM1, p50, and p65 after FOXM1 expression (FOXM1 OV) or siRNA knockdown (FOXM1 si) in HepG2 (C) and SAMe-D cells, which were also co-transfected with EV or MAT1A (D). Numbers below the blots are densitometric values, expressed as percent of EV or SC. Results are expressed as mean percent of control ± SEM from three experiments. *p < 0.05 vs. respective controls. E) Effect of FOXM1 siRNA or FDI-6 on NF-κB-driven promoter activity after 24h treatment in HepG2, SAMe-D cells, and human hepatocytes. *p < 0.05 FDI-6 vs. DMSO or FOXM1 si vs. SC. F) EMSA was performed by using a 32-bp double-stranded synthetic DNA containing two NF-κB motifs of the FOXM1 promoter and recombinant MATα1, p50, p65, FOXM1, PHB1 (all 100ng) alone or combined. The probe only served as a negative control. Results represent a total of at least 3 independent experiments.
FDI-6 is a known direct inhibitor of FOXM1 that blocks its transcriptional activity.(5) We reported MAT1A suppresses NF-κB-Luc activity.(13) We next examined if FDI-6 and FOXM1 regulates NF-κB-Luc activity and whether this required MAT1A. We found that both FDI-6 treatment and FOXM1 knockdown reduced NF-κB-Luc activity in HepG2 cells but not in SAMe-D cells (Fig. 3E) or MATα1-D cells (Supplemental Fig. 4D), suggesting FOXM1’s effect on NF-κB reporter activity is via MATα1. FDI-6 also had no effect on NF-κB-Luc activity in normal human hepatocytes (Fig. 3E bottom). Intrigued by the requirement of MATα1 in FOXM1-mediated regulation of NF-κB reporter activity we examined whether MATα1 and FOXM1 can bind to the consensus NF-κB element using recombinant proteins. Figure 3F shows that FOXM1, p50, and p65 alone (but not MATα1) can bind to the NF-κB element. Interestingly, MATα1 and FOXM1 can heterodimerize with either p50 or p65 to bind to the NF-κB element (Fig. 3F). In contrast, PHB1 was unable to bind to p50 or p65 to shift the band’s position (Fig. 3F). This suggests FOXM1’s inductive effect may be in part via suppressing MATα1’s expression and binding to the NF-κB element.
Direct interaction between FOXM1 and MATα1 and binding to the FOX element
Besides binding to the NF-κB element as heterodimers with p50 and p65 (Fig. 3F), MATα1 is also able to form heterodimer with transcription factors like MAX to bind to the E-box.(28) This raises the possibility that it may also interact with FOXM1 at the FOX binding site. Using co-immunoprecipitation (co-IP) assays with purified recombinant proteins we found direct interaction between FOXM1 and MATα1 (Fig. 4A). These two proteins also interact in HCC and mice CCA tissues (Fig. 4B, 4C). We next used EMSA and recombinant proteins to address binding of these proteins to the FOX element in the MAT1A promoter. MATα1, p50 or p65 individually could not bind to the FOX element, however, each was able to do so in the presence of FOXM1 (Fig. 5A, far left). In contrast, PHB1 was unable to displace FOXM1’s band, which assures interaction and binding are specific. Both HCC and CCA nuclear proteins exhibit increased binding to the FOX element and supershift analysis confirmed presence of MATα1, p50, p65, and FOXM1 in the protein complex that is bound to the FOX element (Fig. 5A, far right).
Figure 4. Interaction between FOXM1 and MATα1.
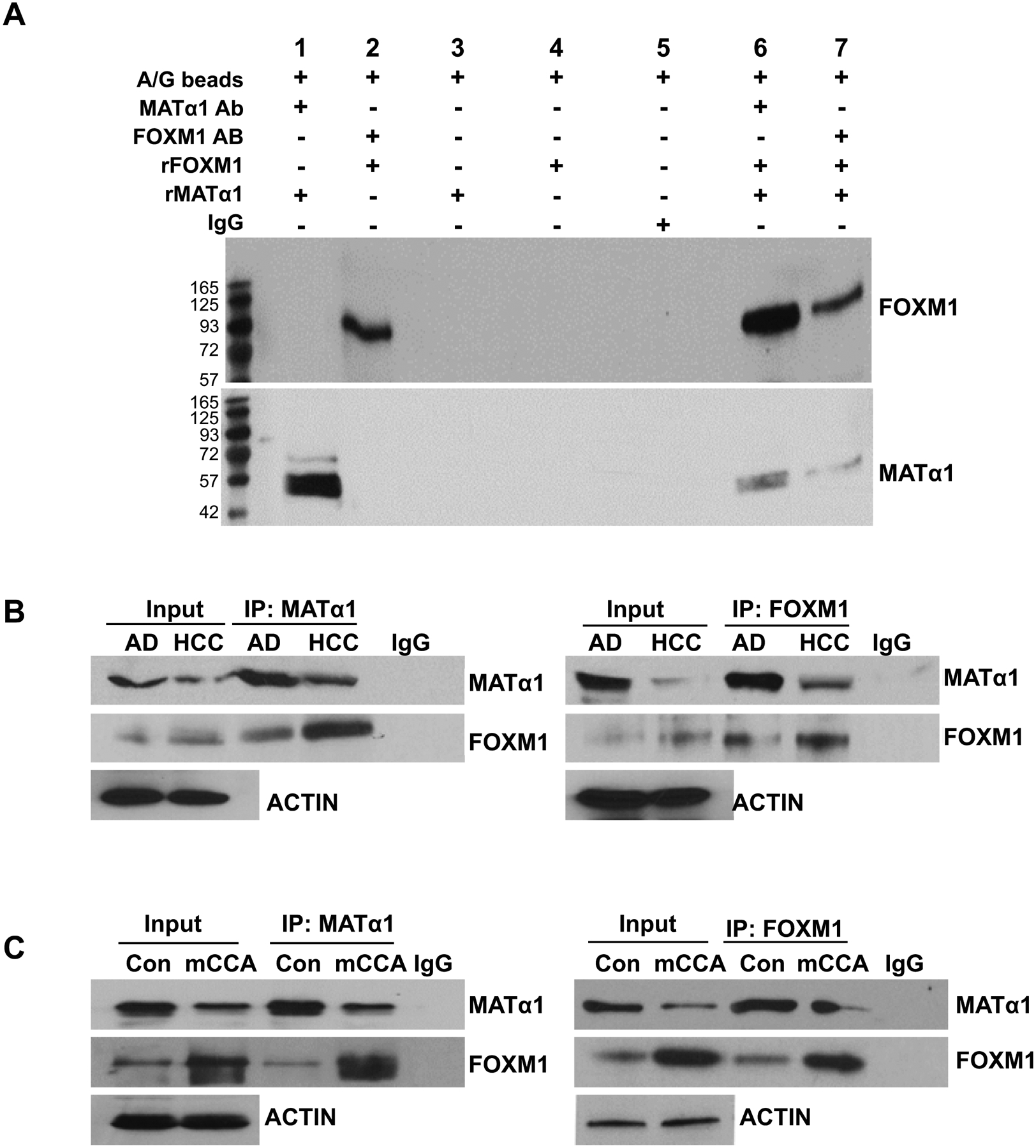
A) In vitro pull-down assay using immobilized recombinant MATα1 or FOXM1 and Immunoblot (IB) for MATα1 and FOXM1. B) Western blotting and co-immunoprecipitation (Co-IP) were used to detect MATα1 and FOXM1 protein levels and their interactions in human HCC and adjacent tissues (AD). C) MATα1 and FOXM1 protein levels and their interactions in mouse CCA and control livers.
Figure 5. FOXM1 and NF-κB regulate MAT1A expression through FOX binding sites.
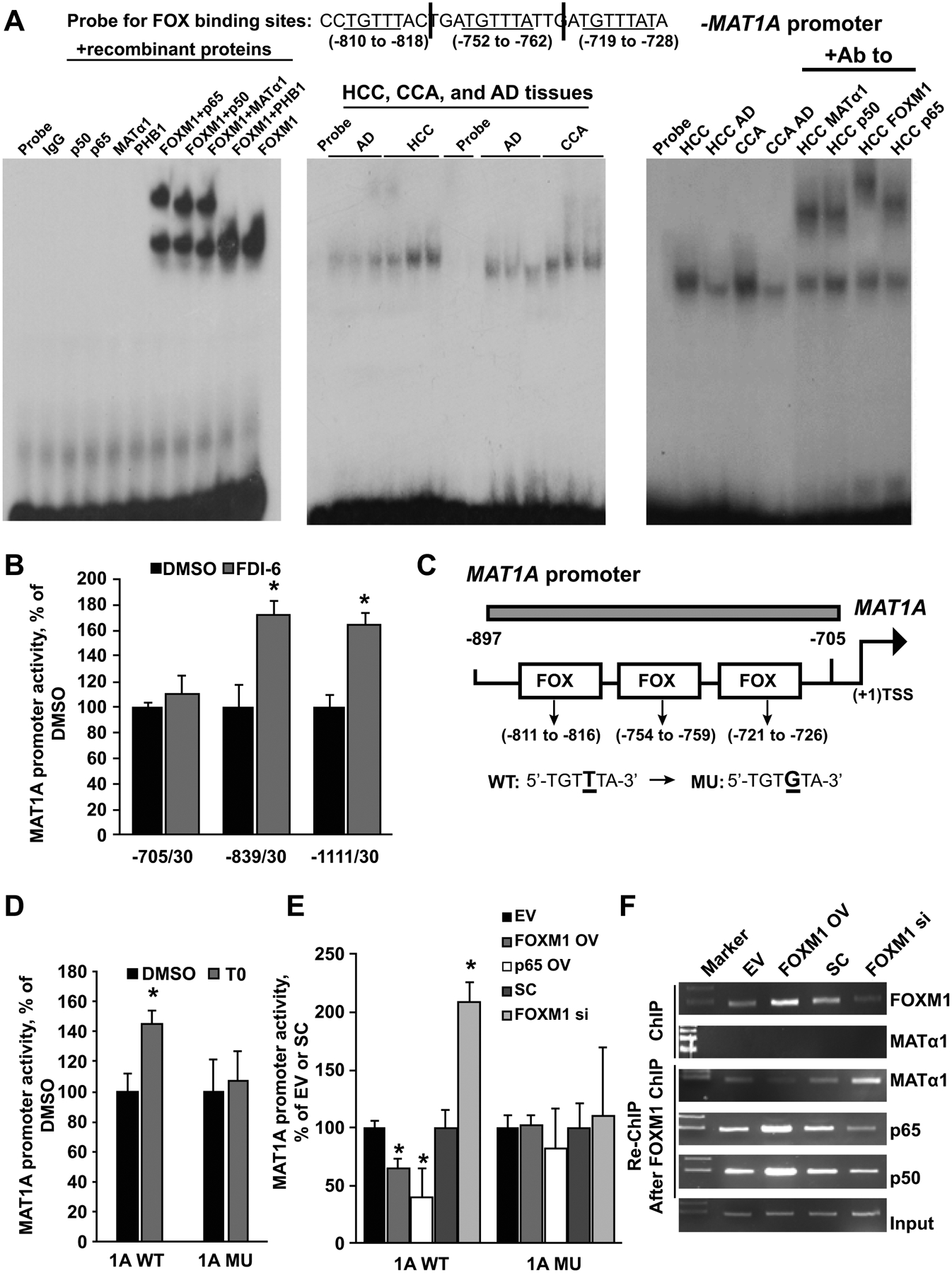
A) EMSA was done using labeled probes containing three FOX binding motifs of the MAT1A promoter as shown above and 100ng of recombinant FOXM1, MATα1, p50, p65, PHB1 alone or combined (left panel), nuclear proteins (0.2μg) from HCC, CCA, and respective adjacent non-tumorous tissues (AD) (middle panel), and with supershift using antibodies to MATα1, p50, FOXM1, and p65 (right panel). Results represent three or more independent experiments. Probe and IgG only served as negative controls. B) MAT1A promoter activity was measured in HepG2 cells following transient transfection with serial deletion constructs. Cells were treated during the last 24 hours of the transfection with DMSO or FDI-6 (5μM). Results represent mean ± SEM from four experiments done in triplicates, *p < 0.05 vs. DMSO. C) FOX binding sites and their mutants in the human MAT1A promoter were created as described in Materials and Methods. D). Effect of T0 (5μM) on the wild type MAT1A promoter (−839/30, 1A WT) or MAT1A promoter mutated at the FOX binding sites (1A MU) in HepG2 cells. Results represent mean ± SEM from four experiments done in triplicates, *p < 0.05 vs. DMSO. E) Activities of wild-type (WT) and FOX binding site mutants (MU) of the MAT1A promoter after FOXM1 OV, p65 OV, or FOXM1 si treatment for 24 hours in HepG2 cells. Results represent mean ± SEM from four experiments done in triplicates, *p < 0.05 vs. EV or SC. F) ChIP analysis spanning three FOX regions with anti-FOXM1 or anti-MATα1 antibody, and ChIP with anti-FOXM1 antibody followed by re-ChIP with MATα1, p65, or p50 antibodies spanning the FOX regions as shown in C) in HepG2 cells after FOXM1 OV or siRNA treatment for 24 hours.
FOXM1 regulates MAT1A directly at the FOX binding sites
There are multiple FOX binding sites in the human MAT1A promoter region (Supplemental Fig. 5A). To delineate sequences that FOXM1 regulates, three fragments of the human MAT1A promoter −1111/+30, −839/+30 and −705/+30 were cloned into the pGL3 vector and the effect of FOXM1 was measured by reporter activity. Figure 5B shows that treatment with FDI-6 (5μM), an inhibitor of FOXM1,(5) significantly increased the activity driven by the construct −839/+30 but not −705/+30, indicating the region between −839 to −705 contains key elements that are regulated by FOXM1 activity (Fig. 5B and 5C). T0, a potent liver X receptor (LXR) agonist known to inhibit FOXM1 protein expression,(7) also raised MAT1A −839/+30 promoter activity, but not if the three FOX binding sites were mutated (Figs. 5C&D). Both FDI-6 and T0 also induced MAT1A −839/+30 promoter activity in MzChA-1 cells (Supplemental Fig. 5B). To ensure that the effects of FDI-6 and T0 are due to FOXM1 inhibition, we varied the expression of FOXM1 in wild type (WT) MAT1A promoter compared to MAT1A promoter mutated at the three FOX binding sites. In both HepG2 and MzChA-1 cells FOXM1 overexpression lowered, while silencing FOXM1 increased MAT1A promoter activity (Fig. 5E, Supplemental Fig. 5C). However, when the MAT1A promoter was mutated at the FOX binding site, FOXM1 expression no longer exerted any influence.
We next examined whether FOXM1, MATα1, and NF-κB (p50, p65) interact at the FOX binding sites of the MAT1A promoter using ChIP and Seq-ChIP. We found that FOXM1 but not MATα1 can bind to the DNA by itself, consistent with result from EMSA (Fig. 5F). However, in the presence of FOXM1, MATα1 and NF-κB co-occupy this region on Seq-ChIP (Fig. 5F). FOXM1 overexpression led to an increase in FOXM1 and NF-κB binding but lowered that of MATα1, and the opposite was true with FOXM1 knockdown (Fig. 5F). To see if increased NF-κB binding also affects the MAT1A promoter activity, we overexpressed p65 and found that this lowered the MAT1A promoter activity by 60% but not if the FOX binding sites were mutated (Fig. 5E). Collectively these results show that FOXM1-NF-κB complex binds to the FOX elements of the MAT1A promoter and represses its activity.
MATα1 is a co-repressor at the FOX binding sites of the FOXM1 promoter
There are also FOX binding motifs within the FOXM1 promoter (Fig. 6A, Supplemental Fig. 6A). To see if FOXM1, MATα1, and NF-κB bind to the region containing the FOX binding sites, ChIP and Seq-ChIP were performed. Similar to the MAT1A promoter, MATα1 was able to bind to the region containing FOX binding sites of the FOXM1 promoter in the presence of FOXM1 (Fig. 6A). Overexpressing MAT1A lowered FOXM1 and NF-κB binding, whereas silencing MAT1A had the opposite effect. In contrast, FOXM1 overexpression increased FOXM1 and NF-κB binding while lowering MATα1 binding to this region, whereas silencing FOXM1 had the opposite effect (Fig. 6A). This suggests MATα1 acts as a corepressor of the FOXM1-NF-κB bound site. To confirm this possibility, we varied the expression of MAT1A and measured the effect on FOXM1-driven promoter activity where the binding site is mutated. In both HepG2 and MzChA-1 cells MAT1A overexpression lowered the FOXM1 promoter activity while silencing MAT1A had the opposite effect (Fig. 6B, Supplemental Fig. 6B). Importantly, when the promoter was mutated at the FOX binding sites, MAT1A expression no longer exerted any influence (Fig. 6B, Supplemental Fig. 6B). Thus, MAT1A and FOXM1 exert negative reciprocal regulation against each other via the FOX binding sites. The effect of NF-κB on FOXM1 promoter activity was also evaluated by overexpressing p65. We found that overexpressing p65 markedly induced FOXM1 promoter activity, but this was blunted if the FOX binding sites were mutated (Fig. 6C). This suggests p65 induces FOXM1 expression via both FOX element and likely NF-κB elements.
Figure 6. Effects of MAT1A and p65 on the FOXM1 promoter.
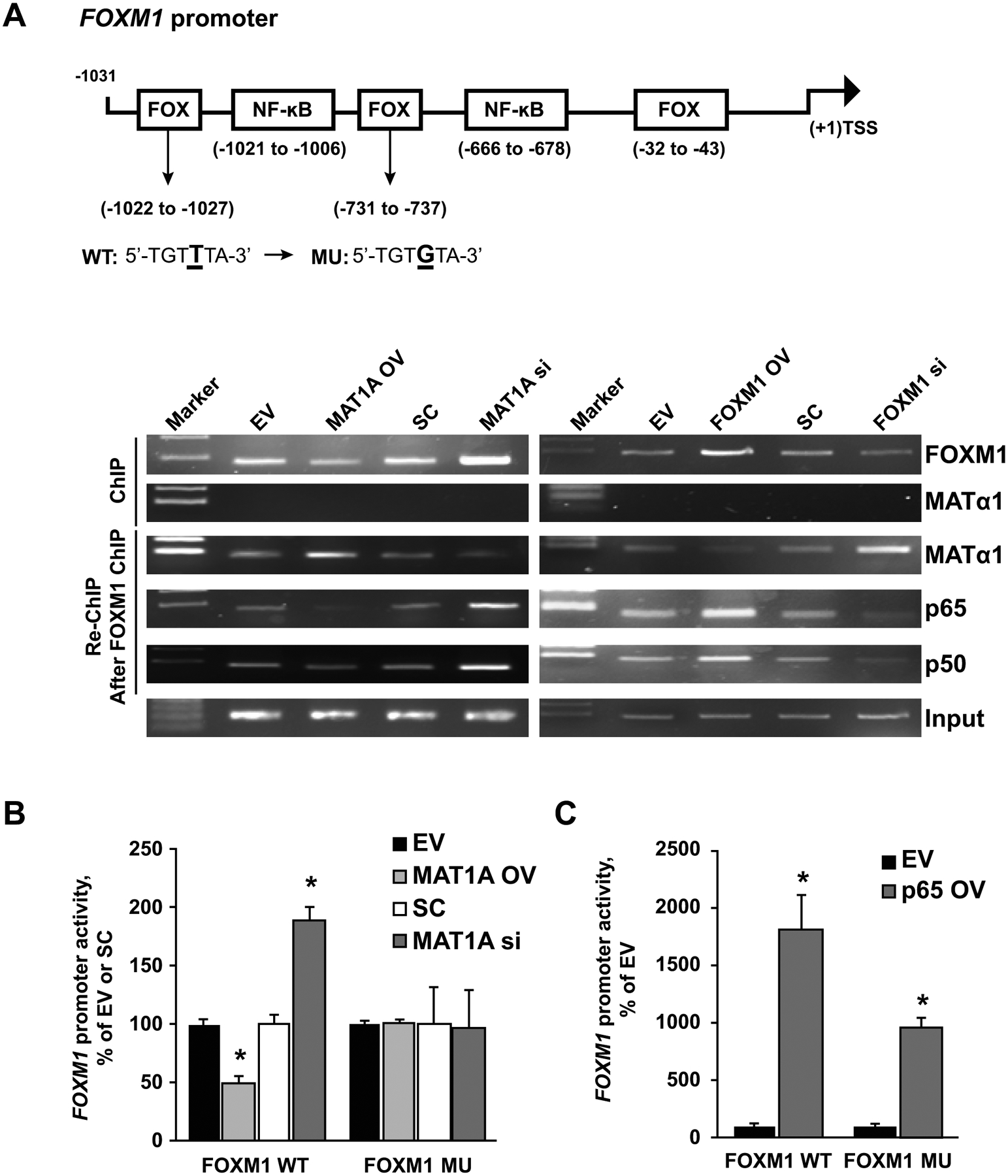
A) HepG2 cells were treated with MAT1A OV or si, or FOXM1 OV or si, and respective controls for 24 hours. ChIP analysis with anti-FOXM1 or MATα1 antibody, and then re-ChIP with anti-MATα1, p65, p50 antibodies after FOXM1 ChIP were performed as described in Materials and Methods. Representative results from three experiments are shown. B) Activities of the wild-type (WT) and FOX binding site mutants (MU) of the FOXM1 promoter after MAT1A OV and si treatment for 24 hours in HepG2 cell. Results are expressed as mean % of EV or SC ± SEM from three experiments done in duplicates *p < 0.05 vs. EV or. SC. C) Promoter activities of the WT and FOX binding site MU of the FOXM1 promoter after p65 OV for 24 hours in the HepG2 cell. Results are expressed as mean % of EV ± SEM from three experiments done in duplicates. *p < 0.05 vs. EV. Results represent at least three independent experiments done in duplicate.
To confirm MAT1A’s ability to negatively regulate FOXM1 and p65/50 in vivo, we examined their expression in the Mat1a KO mice livers that developed HCC. We found higher expression of FOXM1, p65/p50 in the HCCs of the Mat1a KO mice at the mRNA and protein levels (Supplemental Fig. 7).
We also examined whether this reciprocal regulation occurs in normal human hepatocytes and biliary epithelial cells. We did not observe the same reciprocal negative regulation between MAT1A and FOXM1/ NF-κB (Supplemental Fig. 8), suggesting this interplay occurs only after there has been a premalignant change.
FOXM1 promotes HCC and CCA cell proliferation, migration and invasion in part via inhibiting MAT1A
To examine the effect of MAT1A and FOXM1 on cell migration, HepG2 cells were first treated with overexpression vector or siRNA knockdown of FOXM1 or MAT1A for 24 hours. We found FOXM1 overexpression or MAT1A knockdown significantly increased cell migration in HepG2 cells, whereas combining FOXM1 overexpression with MAT1A overexpression eliminated FOXM1’s migration inductive effect. Similarly, when FOXM1 was silenced, the effect of MAT1A knockdown on migration was significantly attenuated. (Fig. 7A). The same findings were also observed in Hep3B cells (Supplemental Fig. 9).
Figure 7. Effects of MAT1A and FOXM1 on cell proliferation, migration and invasion.
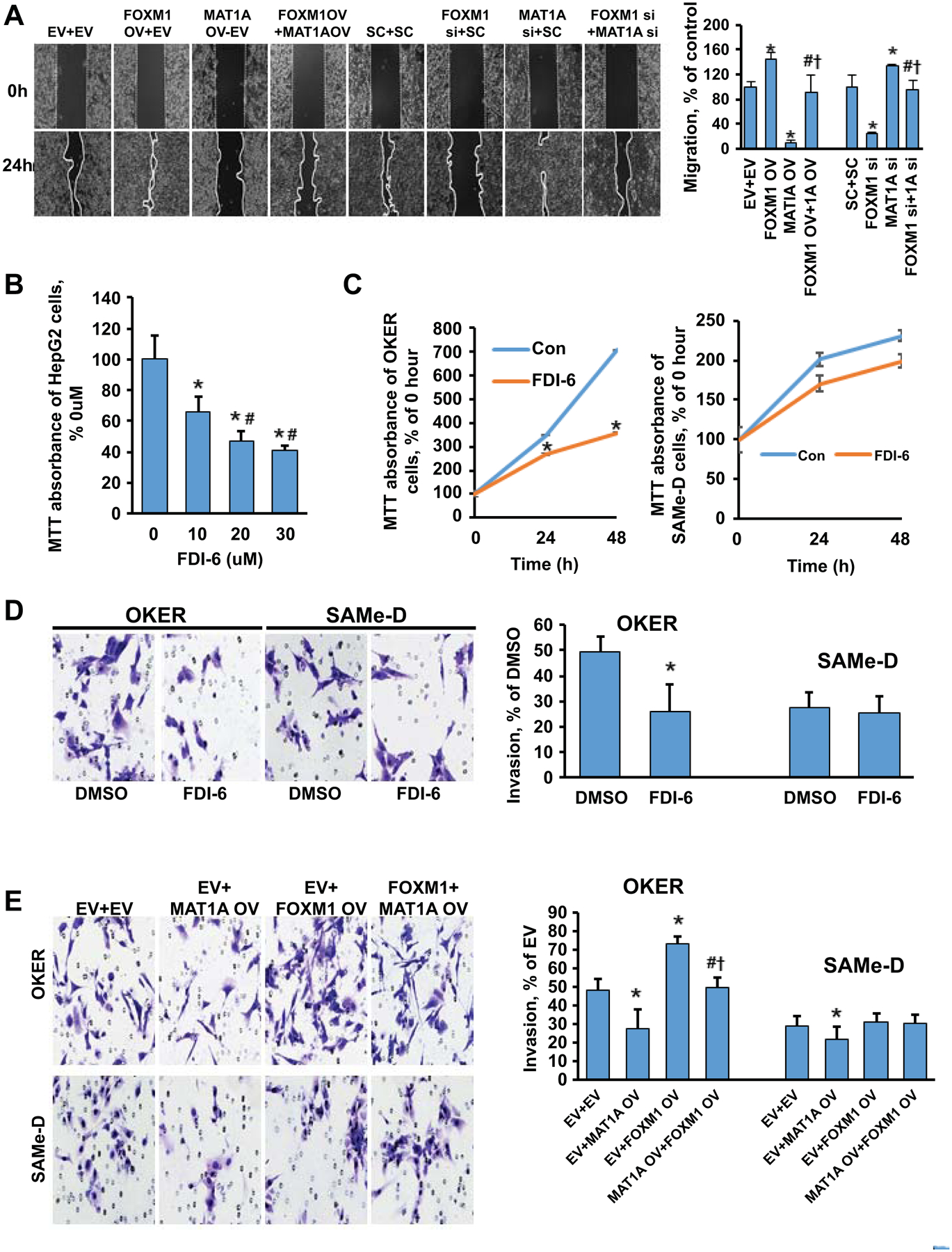
A) Effects of varying MAT1A and FOXM1 expressions on HepG2 cell migration. Quantitative values are summarized in the graph to the right. Results are expressed as mean % of respective controls ± SEM from three experiments done in duplicates. *p < 0.05 vs. EV+EV or SC+SC; #p < 0.05 vs. FOXM1 OV or siRNA; †p < 0.05 vs. MAT1A OV or siRNA. B) Dose-response effect of FDI-6 treatment on the MTT assay in HepG2 cells. Results are expressed as mean % of DMSO ± SEM from three experiments done in duplicates. *p < 0.05 vs. DMSO; #p < 0.05 vs FDI-6 (10uM). C) Time-dependent effect of FDI-6 (5uM) treatment on the MTT assay in OKER and SAMe-D cells. Results are expressed as mean % of DMSO ± SEM from three experiments done in duplicates. *p < 0.05 vs. DMSO. D) Representative invasion images of OKER and SAMe-D cells after FDI-6 (5uM) treatment. Quantitative analysis is summarized in the graph to the right. Results are expressed as mean % of DMSO ± SEM from three experiments done in duplicates. *p < 0.05 vs. DMSO. E) Effects of varied MAT1A and FOXM1 expressions on OKER (Top) and SAMe-D (Bottom) cell invasion. Quantitative values are summarized to the right of the invasion images. Results are expressed as mean % of EV ± SEM from three experiments done in duplicates. *p < 0.05 vs. EV+EV; #p < 0.05 vs. EV+MAT1A OV; †p<0.05 vs. EV+FOXM1 OV.
FDI-6 treatment repressed cell growth in a dose-dependent manner in HepG2 and MzChA-1 cells (Fig. 7B, Supplemental Fig. 10A). Similarly, FDI-6 (5μM) suppressed OKER cell growth and invasion but it had a much smaller inhibitory effect on growth and no effect on invasion in SAMe-D cells (Fig. 7C–D). Overexpressing MAT1A reduced invasion in both OKER and SAMe-D cells and attenuated the inductive effect of FOXM1 overexpression in OKER cells (Fig. 5E). Interestingly, FOXM1 overexpression had no influence on invasion in SAMe-D cells (Fig. 5E). Taken together, these results suggest MAT1A is a key downstream target for FOXM1.
Therapeutic targeting of FOXM1 reduced tumor growth, increased MATα1 but lowered p65/50 expression
We next examined the effects of targeting FOXM1 in liver cancer growth. T0 treatment in HepG2 and MzChA-1 cells raised MATα1 but lowered FOXM1 and p65/p50 protein levels (Fig. 8A, Supplemental Fig. 10B). To see if this was true in vivo, we injected subcutaneously HepG2 cell into the right flank and treated mice with T0 or vehicle control starting when the tumor size reached 80mm3 every 2 days. At the end of the 28 days, T0 treated mice had a 67% lower tumor volume compared to DMSO treated control mice (Fig. 8B). T0 lowered FOXM1 but increased MAT1A mRNA levels in the tumor (Fig. 8C). This translated to a similar change on the protein level shown by IHC (Fig. 8D). Similarly, FDI-6 treatment raised MATα1 but lowered FOXM1 and p65/p50 protein levels in HepG2 and MzChA-1 cells (Fig. 6E, Supplemental Fig. 10C). Both T0 and FDI-6 were equally effective in inhibiting tumor growth in the xenograft model from MzChA-1 cells, with similar changes on raising MATα1 while lowering FOXM1 and p65/p50 protein levels (Supplemental Fig. 11). Finally, we examined the effect of FDI-6 in immunocompetent syngeneic tumorigenesis model by injecting either OKER, SAMe-D, or MATα1-D cells in the flanks of C57/B6 WT mice. SAMe-D cells failed to grow in this model but OKER and MATα1-D cells grew up to 11 days. FDI-6 treatment was started at day seven and it reduced the tumor size from then on to day 11 (Fig. 8F). However, the effect of FDI-6 was blunted in MATα1-D cells (Supplemental Fig. 4E and 4F). IHCs showed FDI-6 increased MATα1 but lowered FOXM1 and p65/p50 protein levels (Fig. 8G).
Figure 8. Effects of T0 and FDI-6 on expression of MATα1, FOXM1, p50 and p65, and tumor growth.
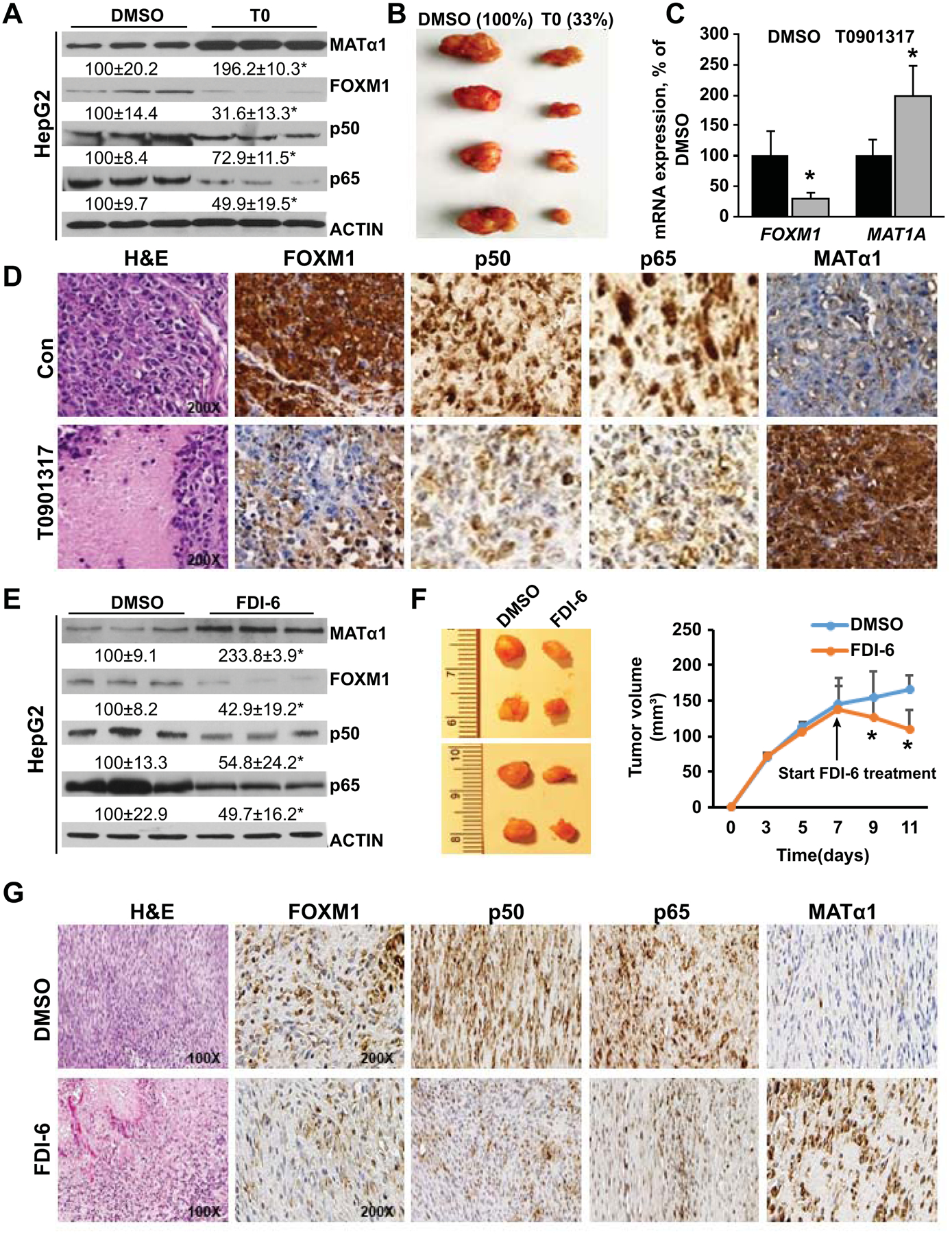
A) MATα1, FOXM1, p50, and p65 levels after T0 treatment (5μM) in HepG2 cells. Results are expressed as mean % of DMSO control ± SEM from three experiments done in duplicates. *p<0.05 vs. DMSO. B) Representative pictures of liver xenograft tumors at day 28 after injection of HepG2 cells in DMSO (left) and T0 treatment (right) groups. Results are expressed as mean % of DMSO from n=8 per group. C) FOXM1 and MAT1A mRNA levels from T0 (25 mg/kg/d) or DMSO treated xenograft tumor tissues. Results are expressed as mean % of control ± SEM from eight mice/group. D) Representative H&E and IHC pictures are shown from n = 8 each. Original magnification for H&E and IHCs is X200. E) Protein expression of MATα1, FOXM1, p50, and p65 after FDI-6 treatment in HepG2 cells. Results are expressed as mean % of control ± SEM from three experiments done in triplicates. *p < 0.05 vs. DMSO. F) Representative pictures of liver syngeneic tumors at day 11 after injection of OKER cells in DMSO (left) and FDI-6 treatment (right, 25 mg/kg/d started on day 7) groups. Tumor volumes are summarized in the graph to the right. Results are expressed as mean of control ± SEM from eight mice/group. *p < 0.05 vs. DMSO. G) Representative H&E and IHC of syngeneic tumors treated with DMSO or FDI-6 from day 7 after injection and sacrificed at day 11. Original magnification for H&E is X100, and X200 for all IHC.
Ingenuity pathway analysis (IPA)
We use IPA to analyze the canonical pathways associated with FOXM1, MAT1A, and NF-κb in CCA and HCC. They are functionally related to: 1. Wnt/β-catenin signaling; 2. ERK/MAPK signaling; 3. Hypoxia-inducible factor 1α (HIF1α) signaling, and 4. PI3K/AKT signaling. The network of these pathways is shown for CCA (Supplemental Fig. 12A) and HCC (Supplemental Fig. 12B).
DISCUSSION
FOXM1 is one of the most frequently upregulated genes in human cancers that plays a key role in tumor initiation and progression.(1) FOXM1-regulatory network has been shown to be a major predictor of adverse outcomes in 18,000 cancer patients across 39 human malignancies,(31) confirming its importance in human cancers. The mechanisms of FOXM1’s oncogenic action include activating the transcription of its targets, which include many cell cycle related genes, as well as through influencing proteins that it interacts with to enhance oncogenesis.(32) Although FOXM1 is overexpressed in HCC(4) and CCA,(33) how it influences liver tumorigenesis is incompletely understood. MAT1A is a tumor suppressor gene in HCC and CCA and its encoded protein acts as a co-repressor of E-box to inhibit the expression of several oncogenes such as c-MYC, MAFG, and c-MAF.(14) In addition, MAT1A negatively regulates NF-κB activity but the mechanism is unclear.(13) NF-κB is the link between chronic liver injury, fibrosis, and cancer and its role in both HCC and CCA is well-recognized.(9) We focused on p65/p50 because this is the most common heterodimer of NF-κB.(8,9) Interestingly, FOXM1 and NF-κB have been shown to reciprocally regulate each other positively and FOXM1 physically interacts and co-occupies with NF-κB on nearly half of the NF-κB binding sites in lymphoblastoid B cells.(11,12) In this work we examined whether there is an interplay between FOXM1/NF-κB and MAT1A in liver cancers. We included both HCC and CCA in our investigation because MAT1A expression is downregulated in both types of liver cancers.
In both human HCC and CCA, publicly available datasets and our own data show FOXM1, p50, and p65 are upregulated while MAT1A is downregulated at the mRNA and protein levels. A significant inverse correlation exists between MAT1A and FOXM1/NF-κB mRNA levels in HCC. This is due to the fact that MAT1A and FOXM1 can directly regulate each other negatively. In fact, MATα1 and FOXM1 can directly interact with each other and bind to the FOX binding motif as a complex with NF-κB (Fig. 5A). We narrowed down the region between −839 to −705 to be the FOXM1-responsive region for the MAT1A promoter, a region that contains multiple FOX binding sites (5′- TGTTTA-3′) (Fig. 5C). We found that binding of FOXM1/NF-κB to the FOX binding sites in the MAT1A promoter inhibited promoter activity (Fig. 5E). FOXM1 promoter also contains FOX binding sites and one of them is in the region −745 to −738 that was shown to be required for the auto-regulatory activation of FOXM1.(34) NF-κB activated FOXM1 promoter through the FOX and NF-κB binding sites (Fig. 6C), whereas MATα1’s interaction with FOXM1 at the FOX binding sites repressed FOXM1 promoter activity (Fig. 6B). Thus, by this mechanism FOXM1 and MAT1A are able to exert a negative reciprocal regulation against each other. We reported previously that MAT1A negatively regulates NF-κB-driven reporter activity(13) and here we found NF-κB negatively regulates MAT1A promoter activity in return. This could have happened because NF-κB induces FOXM1 expression and the complex FOXM1-NF-κB is a more potent repressor of MAT1A promoter. FOXM1 is known to interact with many other proteins such as β-catenin and p65 to further enhance their oncogenic activity.(32) Here we add MATα1 to the FOXM1 interactome and targets and provide a novel mechanism of how either FOXM1/NF-κB induction or MAT1A downregulation can be the nidus of a feedforward loop to accelerate tumorigenesis.
FOXM1 was reported to activate NF-κB in lung cancer.(11) FOXM1 can bind to the promoter region of Nfkb2 to activate its expression, whereas its stimulatory effect on Nfkb1 (p50) is indirect (no binding detected).(11) p50 has also been shown to auto-regulate via a functional NF-κB element in its promoter region.(35) We found overexpressing FOXM1 raised p65/p50 expression in most liver cancer cells. However, to our surprise FOXM1 had no influence on p65/p50 expression in SAMe-D and MATα1-D cells, which lack MAT1A. This suggests FOXM1 regulates p65/p50 expression via MAT1A. This notion was further supported by the observation that FOXM1 was able to positively regulate p65/p50 expression in SAMe-D cells expressing MAT1A (Fig. 3D). The finding that MATα1 and FOXM1 can heterodimerize with p50 and p65 and bind to the NF-κB element (Fig. 3F) suggests a scenario where FOXM1 and MATα1 compete with each other to exert opposing effects on NF-κB-driven reporter activity, with the former activating and the latter repressing. Thus, in the absence of MATα1 there is no competition for FOXM1 so its effect on p65/p50 is already at a maximum. This could explain why there is no induction on the expression of p65/p50 or the NF-κB-driven reporter activity when FOXM1 is overexpressed. However, FOXM1 knockdown also had no effect on p65/p50 and NF-κB-driven reporter activity in the absence of MATα1. This suggests induction of MATα1 during FOXM1 knockdown is a key mechanism for suppression of p65/p50 expression. It should be noted that the interplay between MAT1A and FOXM1/ NF-κB was not observed in normal human hepatocytes or biliary epithelial cells but was seen in the Mat1a KO livers as well as livers from patients with primary biliary cholangitis and primary sclerosing cholangitis. This suggests that this interplay only becomes activated when there are already changes in the premalignant environment. This is a subject that will require further investigation.
In addition to auto-regulation via FOX binding site, FOXM1 promoter can also be activated by CCCTC-binding factor (CTCF), cAMP responsive element-binding protein (CREB), and HIF1α but inhibited by liver X receptor α (LXRα) in HCC cells.(36) Consistently, treatment with LXRα agonist T0 and FOXM1 inhibitor FDI-6 lowered FOXM1 but raised MAT1A expression in multiple models in vitro and in vivo. The MAT1A promoter that is responsive to the effect of LXRα agonist does not have any LXR response element and the response is lost when FOX binding sites were mutated. These data suggest the effect of LXRα agonist on MAT1A expression is indirect via FOXM1.
To gain further insight on how the FOXM1-MAT1A interplay affects liver cancer tumorigenesis, we examined the effects of overexpression or knockdown of FOXM1 and MAT1A on cell growth, cell migration, and invasion. Our results demonstrated that upregulation of FOXM1 or downregulation of MAT1A significantly increased HepG2 and Hep3B cell migration. Whereas combining FOXM1 with MAT1A overexpression largely eliminated each other’s effects, supporting an important role for this interplay in liver cancer. This was further examined using FDI-6, a selective inhibitor of FOXM1. FDI-6 dose-dependently reduced the growth of HepG2 and MzChA-1 cells. However, FDI-6 exerted a very different effect in SAMe-D cells as compared to OKER cells, both murine HCC cells. While FDI-6 inhibited growth and invasion in OKER cells, it had minimal effect on growth and no effect on invasion in SAMe-D cells. This is not because SAMe-D cells do not respond since invasion was significantly reduced when MAT1A was overexpressed. One possibility may be related to lack of NF-κB regulation by FOXM1 in SAMe-D cells.
To further confirm MAT1A’s importance in FOXM1 biology, we measured tumor growth in response to FDI-6 treatment in a syngeneic model where OKER or MATα1-D cells were injected into the flanks. FDI-6 treatment exerted a much more inhibitory effect in OKER cells as compared to MATα1-D cells, further supporting the notion that in liver cancer cells the effect of FOXM1 is mediated in a large part by its influence on MAT1A expression.
In summary, we have unveiled a novel reciprocal negative regulation between FOXM1/NF-κB and MAT1A in HCC and CCA. These proteins all interact at the FOX and NF-κB binding sites, with FOXM1/NF-κB activating FOXM1 but inhibiting MAT1A transcription. MATα1 is a new target and interacting protein with FOXM1 that acts as a co-repressor of FOXM1 transcription. Unexpectedly, FOXM1’s positive regulation on NF-κB’s expression and its dependent reporter activity is lost in the absence of MAT1A. Taken together, treatments that raise MAT1A expression might be the most effective in targeting this feedforward interplay.
A schematic diagram that summarizes the key findings and interplays is shown in Supplemental Fig. 13.
Supplementary Material
Financial Support:
This work was supported by NIH grants R01CA172086 (HP Yang, JM Mato, and SC Lu) and P01CA233452 (HP Yang, E Seki and SC Lu), Plan Nacional of I+D SAF2017-88041-R (JM Mato). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations used (alphabetical order):
- CCA
cholangiocarcinoma
- ChIP
chromatin immunoprecipitation
- EMSA
electrophoretic mobility shift assay
- EMT
epithelial-mesenchymal transition
- FBS
fetal bovine serum
- FDI-6
Forkhead Domain Inhibitory-6
- FOXM1
forkhead box M1
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- H&E
hematoxylin and eosin
- HCC
hepatocellular carcinoma
- IHC
immunohistochemistry
- MAT
methionine adenosyltransferase
- miRNAs
MicroRNAs
- NF-κB
nuclear factor kappa B
- PCR
polymerase chain reaction
- PHB1
prohibitin 1
- PBC
primary biliary cholangitis
- PSC
primary sclerosing cholangitis
- SAMe
S-adenosylmethionine
- SC
scramble siRNA
- Seq-ChIP
sequential ChIP
- siRNA
small interfering RNA
- T0 (T0901317)
a potent liver X receptor (LXR) agonist
- HNF4
hepatocyte nuclear factor 4
- AFP
alpha-fetoprotein A
- CYP2E1
cytochrome P450 2E1
- MMP-7
matrix metalloproteinase-7
REFERENCES
- 1.Wierstra I FOXM1 (Forkhead box M1) in tumorigenesis: overexpression in human cancer, implication in tumorigenesis, oncogenic functions, tumor-suppressive properties, and target of anticancer therapy. Adv Cancer Res. 2013;119:191–419. [DOI] [PubMed] [Google Scholar]
- 2.Yu M, Tang Z, Meng F, Tai M, Zhang J, Wang R, et al. Elevated expression of FoxM1 promotes the tumor cell proliferation in hepatocellular carcinoma. Tumour Biol. 2016;37:1289–97. [DOI] [PubMed] [Google Scholar]
- 3.Koo CY, Muir KW, Lam EW. FOXM1: From cancer initiation to progression and treatment. Biochim Biophys Acta. 2012;1819:28–37. [DOI] [PubMed] [Google Scholar]
- 4.Meng FD, Wei JC, Qu K, Wang ZX, Wu QF, Tai MH, et al. FoxM1 overexpression promotes epithelial-mesenchymal transition and metastasis of hepatocellular carcinoma. World J Gastroenterol. 2015;21:196–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gormally MV, Dexheimer TS, Marsico G, Sanders DA, Lowe C, Matak-Vinković D, et al. Suppression of the FOXM1 transcriptional programme via novel small molecule inhibition. Nat Commun. 2014;5:5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bi Z, Liu W, Ding R, Wu Y, Dou R, Zhang W, et al. A novel peptide, 9R-P201, strongly inhibits the viability, proliferation and migration of liver cancer HepG2 cells and induces apoptosis by down-regulation of FoxM1 expression. Eur J Pharmacol. 2017;796:175–189. [DOI] [PubMed] [Google Scholar]
- 7.Hu C, Liu D, Zhang Y, Lou G, Huang G, Chen B, et al. LXRα-mediated downregulation of FOXM1 suppresses the proliferation of hepatocellular carcinoma cells. Oncogene 2014;33:2888–2897. [DOI] [PubMed] [Google Scholar]
- 8.He G, Karin M. NF-κB and STAT3 - key players in liver inflammation and cancer. Cell Res. 2011;21:159–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luedde T, Schwabe RF. NF-κB in the liver--linking injury, fibrosis and hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2011;8:108–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin B, Wang C, Li J, Du X, Ding K, Pan J. Anthelmintic Niclosamide Disrupts the Interplay of p65 and FOXM1/β-catenin and eradicates leukemia stem cells in chronic myelogenous leukemia. Clin Cancer Res. 2017;23:789–803. [DOI] [PubMed] [Google Scholar]
- 11.Wang IC, Ustiyan V, Zhang Y, Cai Y, Kalin TV, Kalinichenko VV. Foxm1 transcription factor is required for the initiation of lung tumorigenesis by oncogenic Kras(G12D). Oncogene 2014;33:5391–5396. [DOI] [PubMed] [Google Scholar]
- 12.Zhao B, Barrera LA, Ersing I, Willox B, Schmidt SC, Greenfeld H, et al. The NF-κB genomic landscape in lymphoblastoid B cells. Cell Rep. 2014;8(5):1595–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu T, Yang H, Fan W, Tu J, Li TWH, Wang J, et al. Mechanisms of MAFG Dysregulation in Cholestatic Liver Injury and Development of Liver Cancer. Gastroenterology 2018;155:557–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang H, Liu T, Wang J, Li TW, Fan W, Peng H, et al. Deregulated methionine adenosyltransferase α1, c-Myc, and Maf proteins together promote cholangiocarcinoma growth in mice and humans. Hepatology 2016;64:439–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Torres L, Avila MA, Carretero MV, Latasa MU, Caballería J, López-Rodas G, et al. Liver-specific methionine adenosyltransferase MAT1A gene expression is associated with a specific pattern of promoter methylation and histone acetylation: implications for MAT1A silencing during transformation. FASEB J. 2000;14:95–102. [DOI] [PubMed] [Google Scholar]
- 16.Tomasi ML, Li TWH, Li M, Mato JM, Lu SC. Inhibition of methionine adenosyltransferase 1A transcription by coding region methylation. J. Cell Physiol 2012;227:1583–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vázquez M, Fernández D, Embade N, Woodhoo A, Martinez N, Varela-Rey M, et al. HuR/Methylated-HuR and AUF1 regulate the expression of methionine adenosyltransferase during liver proliferation, differentiation and carcinogenesis. Gastroenterology 2010;138:1943–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, Ramani K, Sun Z, Zee C, Grant EG, Yang HP, et al. Forced expression of methionine adenosyltransferase 1A in human hepatoma cells suppresses in vivo tumorigenesis in mice. Am. J. Pathol 2010;176:2456–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang HP, Iglesias Ara A, Magilnick N, Xia M, Ramani K, Chen H, et al. Expression pattern, regulation and function of methionine adenosyltransferase 2beta alternative splicing variants in hepatoma cells. Gastroenterology 2008;134:281–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:l1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu SC, Alvarez L, Huang ZZ, Chen LX, An W, Corrales FJ, et al. Methionine adenosyltransferase 1A knockout mice are predisposed to liver injury and exhibit increased expression of genes involved in proliferation. Proc Natl Acad Sci USA 2001;98:5560–5565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martínez-López N, Varela-Rey M, Fernández-Ramos D, Woodhoo A, Vázquez-Chantada M, Embade N, et al. Activation of LKB1-Akt pathway independent of phosphoinositide 3-kinase plays a critical role in the proliferation of hepatocellular carcinoma from nonalcoholic steatohepatitis. Hepatology 2010;52:1621–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martínez-López N, García-Rodríguez JL, Varela-Rey M, Gutiérrez V, Fernández-Ramos D, Beraza N et al. Hepatoma cells from mice deficient in glycine N-methyltransferase have increased RAS signaling and activation of liver kinase B1. Gastroenterology 2012;143(30:787–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang HP, Li TWH, Peng J, Tang X, Ko KS, Xia M, et al. A mouse model of cholestasis-associated cholangiocarcinoma and transcription factors involved in progression. Gastroenterology 2011;141:378–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeng ZH, Huang ZZ, Chen CJ, Yang HP, Mao Z, Lu SC. Molecular cloning and functional characterization of the 5’-flanking region of human methionine adenosyltransferase 1 A gene. 2000;346:475–482. [PMC free article] [PubMed] [Google Scholar]
- 27.Yang HP, Li TWH, Ko KS, Xia M, and Lu SC. Switch from Mnt-Max to Myc-Max induces p53 and cyclin D1 expression and apoptosis during cholestasis in mice and human hepatocytes. Hepatology 2009;49:860–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fan W, Yang H, Liu T, Wang J, Li TWH, Mavila N, et al. Prohibitin 1 suppresses liver cancers tumorigenesis in mice and humans. Hepatology 2017;65(4):1249–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang JW, Murray B, Barbier-Torres L, Liu T, Liu Z, Yang HP, et al. The mitochondrial chaperone Prohibitin 1 negatively regulates interleukin-8 in human liver cancers. J. Biol. Chem 2019;294(6):1984–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomasi ML, Iglesias Ara A, Yang H, Ramani K, Feo F, Pascale MR, et al. S-adenosylmethionine Regulates Apurinic/Apyrimidinic Endonuclease 1 Stability: Implication in Hepatocarcinogenesis. Gastroenterology 2009;136:1025–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21(8):938–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gartel AL. FOXM1 in Cancer: Interactions and Vulnerabilities. Cancer Res. 2017;77:3135–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu L, Wu J, Guo Y, Xie W, Chen B, Zhang Y, et al. Overexpression of FoxM1 predicts poor prognosis of intrahepatic cholangiocarcinoma. Aging (Albany NY) 2018;10(12):4120–4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Halasi M, Gartel AL. A novel mode of FoxM1 regulation: positive auto-regulatory loop. Cell Cycle 2009;8(12):1966–1967. [DOI] [PubMed] [Google Scholar]
- 35.Cogswell PC, Scheinman RI, Baldwin AS Jr. Promoter of the human NF-kappa B p50/p105 gene. Regulation by NF-kappa B subunits and by c-REL. J Immunol. 1993;150(7):2794–2804. [PubMed] [Google Scholar]
- 36.Liao GB, Li XZ, Zeng S, Liu C, Yang SM, Yang L, et al. Regulation of the master regulator FOXM1 in cancer. Cell Commun. and Signaling 2018;16:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


