Abstract
Chronic lung diseases remain major healthcare burdens, for which the only curative treatment is lung transplantation. In vitro human models are promising platforms for identifying and testing novel compounds to potentially decrease this burden. Directed differentiation of pluripotent stem cells is an important strategy to generate lung cells to create such models. Current lung directed differentiation protocols are limited as they do not 1) recapitulate the diversity of respiratory epithelium, 2) generate consistent or sufficient cell numbers for drug discovery platforms, and 3) establish the histologic tissue-level organization critical for modeling lung function. In this review, we describe how lung development has formed the basis for directed differentiation protocols, and discuss the utility of available protocols for lung epithelial cell generation and drug development. We further highlight tissue engineering strategies for manipulating biophysical signals during directed differentiation such that future protocols can recapitulate both chemical and physical cues present during lung development.
Keywords: Pluripotent stem cells, Directed differentiation, In vitro disease modelling, Lung, Airway, Mechanical cues
Graphical abstract

1. Introduction
End-stage lung disease is the third leading cause of morbidity and mortality worldwide [1,2], and produces a significant burden on healthcare systems due to extensive resource expenditures for disease management and as lung transplantation is the only curative treatment option. Such diseases include acute respiratory distress syndrome, chronic obstructive pulmonary disease, cystic fibrosis, and pulmonary fibrosis. Chronic pulmonary diseases result in 3 million global deaths per year [3,4]. Patients who receive transplants face continued complications associated with chronic immunosuppression and graft rejection with the transplant survival rates at 5 and 10 years being 54% and 32%, respectively [5]. Furthermore, since lungs function as an important barrier between the internal and the external environments, they are a critical site for bacterial and viral infections and disease transmission; particularly relevant given the current COVID-19 pandemic. There is, therefore, a critical need to better elucidate the mechanisms of infection, disease progression, host response, and cellular repair in the lung to enable the development of novel targeted therapeutics for lung disease.
Tissue-engineered models have emerged as a technology to address this challenge and shown some success in drug identification and toxicology studies. For example, commercially available airway epithelial models such as EpiAirwayTM (MatTek Life Sciences) serve as convenient platforms, with air-liquid interface culture capabilities, for assessing the effect of chemical and physical stimuli [[6], [7], [8]]. Other examples include the Alveolus Lung-Chip and Airway Lung-Chip systems (Emulate Inc.), originally developed in the Ingber laboratory, which mimic the epithelial-endothelial interface of the airway and provide a more dynamic platform for testing new anti-inflammatory compounds in asthma [9] and new small molecule targets to decrease cancer-associated pulmonary edema [10]. More complex models have also been reported which involve self-assembly of heterogeneous progenitor cells into 3D structures, termed organoids [21]. These organoid models can recapitulate aspects of human lung development in terms of tissue structures and protein expression, and therefore present a promising opportunity for drug screening [11,12].
A challenge in developing such human in vitro lung models to screen for drugs, however, is the requirement for large batches of similar human cells as a starting population for tissue manufacturing to ensure minimal heterogeneity between test wells [13]. Achieving this is especially challenging when using primary human lung cells, which exhibit considerable heterogeneity across donors and have a limited ability to grow and differentiate reliably [14,15]. Furthermore, primary cells are often extracted from diseased donors, which is not ideal for conducting controlled studies due to the wide range of therapeutic and environmental factors these cells have already been exposed to. Directed differentiation of pluripotent populations has the potential to create vast numbers of cells, from either healthy or diseased patients. It allows introduction of specific disease-associated mutations via CRISPR/Cas9 gene editing to recapitulate and understand pathologies in a controlled manner. As such, directed differentiation enables the generation of an attractive cell source for drug screening platforms and personalized disease models that may provide insight into tissue regeneration mechanisms [[16], [17], [18]].
Directed differentiation protocols to manufacture specific cell populations from pluripotent stem cells (PSCs) have been developed to meet the need for a homogeneous human cell source. Older lung directed differentiation protocols from the late 2000s have been proven inefficient due to the non-standardized methods through which they derive lung endoderm from embryoid bodies [[19], [20], [21]]. A series of more standardized stepwise protocols have since emerged in the last decade that provide avenues for developing airway and lung epithelia, albeit with variable efficiencies [11,[22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33]]. The first influential directed differentiation protocol to produce lung epithelia used human PSCs in 2011 [23], which was further supported by two prominent studies conducted using mouse PSCs in 2012 [22,24]. These protocols have continued to be enhanced through adaptations related to the selection of growth factors and small molecules, the chronology of morphogen delivery, as well as innovations in enabling platforms such as cell sorting, 3D culture, and single-cell analyses to efficiently derive normal and diseased lung epithelia from human PSCs [11,22,25,26,28,32,[34], [35], [36], [37], [38]]. Despite such advancements, limitations pertaining to heterogeneity in the resulting populations still exist, which are likely attributed to variability across directed differentiation trials, PSC cell lines, or the persistence of contaminating cell populations belonging to other lineages. While protocols have progressed to some degree in differentiating proximal airway and distal alveolar epithelia, they remain limited. Overall, many unanswered questions remain with regards to the identity, maturity, and functionality of resulting cell types, as well as their utility for tissue engineering and drug testing approaches. Therefore, these protocols must be optimized further to reliably produce large numbers of spatially relevant and functional lung and airway epithelial cells that appropriately respond to both chemical and mechanical stimuli in the context of disease modeling and drug discovery.
In this review, we discuss the directed differentiation protocols that attempt to recapitulate lung development and disease and highlight possible opportunities to enhance these protocols in the future. We first describe development of native lung tissue and the patterning events that occur, that differentiation models attempt to mimic, and highlight how human lung embryology has served as the blueprint to create the common pathway of lung directed differentiation protocols. We then discuss the evolution of directed differentiation protocols to find opportunities for creating specific populations of airway and lung epithelia through targeted manipulation of key signaling pathways in 2D and 3D models. We further describe how these models have been used to recapitulate different airway and lung diseases. Finally, we discuss how tissue engineering and biophysical cues using biomaterials can be utilized during lung directed differentiation to mimic patterning cues present in development to augment current differentiation protocols.
2. Human embryology as a blueprint for lung directed differentiation
2.1. Overview of key developmental stages
Directed differentiation protocols have been designed to mimic in vivo human lung development [39]. Indeed, in vitro models of lung development have provided unique insight into human lung development [40]. As human lung development has been described at great length in earlier reviews, [41,42], we provide a brief overview as follows (schematically represented in Fig. 1 ). During early embryogenesis (at 14 days post fertilization), a process called gastrulation begins with the appearance of a structure called primitive streak, through which cells migrate to form the primary embryonic germ layers (definitive endoderm, mesoderm, and ectoderm) [[43], [44], [45]]. Definitive endoderm expands, thereby forming the primitive gut tube comprised of three endodermal regions: foregut, midgut, and hindgut [46,47]. This is when lung development begins, at approximately four weeks into embryonic life, with the outgrowth of foregut endoderm [48,49] and continues through eight years of post-natal life [50]. There are five stages to lung development:
-
1.
Embryonic (weeks 4–7): The future lung buds emerge from the ventral side of the primitive foregut endoderm into the surrounding mesenchyme and develop into embryonic lung buds with early trachea and bronchi [[51], [52], [53], [54], [55]].
-
2.
Pseudoglandular (weeks 5–16): Branching of the airway continues, leading to formation of conducting and terminal bronchioles, while the proximal airway epithelium begins to develop [50,56].
-
3.
Canalicular (weeks 16–25): Development of the respiratory, or gas-exchanging, airways is initiated, primitive alveoli form, and the future distal epithelium begins to thin as distal epithelial markers are expressed [41,57].
-
4.
Saccular (weeks 24–40): Emergence of sac-shaped distal airways, which develop crests with muscle and elastin to create indentations. These distal airways extend to form alveoli by 29 weeks [58]. The developing epithelium and vasculature within the future alveolus continue to merge closer together to facilitate future gas exchange and further differentiation of alveolar epithelial cells (AEC) I and II takes place.
-
5.
Alveolar periods (week 40–8 years): True alveoli are seen in week 36 and the majority of alveolarization takes place through saccule septation, a process by which the sac-shaped distal airways change their internal architecture and create thin walls intraluminally. Septation leads to an increase in surface area of the gas exchanging portion of the developing lung and prepares the fetus to breath air during this stage [50,59].
Fig. 1.
Schematic of human lung development from an epithelial perspective.
2.2. Lung organogenesis: molecularly defining lung fate in the embryo
During the embryonic period, early lung is genetically defined by the expression of transcription factor NK2 Homeobox 1 (NKX2.1) and Sry-box 2 (SOX2) [[60], [61], [62]]. During human lung development, it has been found that the lung buds and branches given off during the pseudoglandular period are mostly SOX2+SOX9+ [17,62,63]. Both SOX2 and SOX9 are individual markers of the early proximal or distal lineage, respectively [60,[64], [65], [66]]. Over the course of the canalicular and saccular periods of development (weeks 16–40), these double positive populations downregulate one SOX protein and maintain expression of the other as these cells mature towards proximal or distal lineages [62]. The proximal airway (closer to the mouth) is comprised of a pseudostratified columnar epithelium that is responsible for the conducting airway function: debris and pathogen removal (ciliated cells), mucus production (goblet cells), prevention of airway inflammation (club cells), and humidification of air as it passes through to the distal lung compartment [[67], [68], [69]]. The squamous distal epithelium, composed of alveolar epithelial cells (AEC I and II), facilitates the respiratory function of the lung as air in the epithelial compartment is brought into close apposition to blood from the pulmonary vasculature; it also secretes surfactants, which play an immunologic role and decrease the surface tension present at the air-liquid interface, thereby preventing alveolar collapse [70]. In humans, a number of cell types are found in the proximal airway, each identified with specific markers (Table 1 ). This includes basal cells (tumor protein p63/P63, keratin/KRT5, nerve growth factor receptor/NGFR, integrin α6/ITGA6, integrin β4/ITGB4), ciliated cells (Forkhead BoxJ1/FOXJ1, acetylated tubulin/AcTUB), goblet cells (mucin 5AC/MUC5AC, mucin 5B/MUC5B), club cells (club cell secretory protein/CCSP or SCGB1A1), and pulmonary neuroendocrine cells (PNECs; synaptophysin/SYP, chromogranin A/CHGA). On the other hand, homeodomain-only protein (HOPX) identifies the distal lung along with AEC I cells (T1α, podoplanin/PDPN, aquaporin 5/AQP5) while AEC II cells are recognized via surfactant protein B (SPC), pro-surfactant protein C (pro-SPC or SPC), and HT2-280 [71,72].
Table 1.
Epithelial populations in native human airways and lungs
| Region | Cell Type | Associated Markers for Cell Characterization | Cell Proportions in Native Lung |
|---|---|---|---|
| Proximal Airway | Ciliated Cell | FOXJ1, AcTUB | 48–70% [[122], [123]] |
| Goblet Cell | MUC5AC, MUC5B | 6–25% [[122], [124]] | |
| Club Cell | CCSP, SCGB1A1, SCGB3A2 | 7–11% [[122], [125]] | |
| Basal Cell | P63, KRT5, NGFR, ITGA6, ITGB4 | 12–30% [[122], [126]] | |
| Distal Lung | Alveolar epithelial cell type I (AEC I) | HOPX, PDPN, AQP5 | ~33% [127] |
| Alveolar epithelial cell type II (AEC II) | SPB, SPC, HT2-280 | ~66% [127] |
One mechanism by which lung epithelia begin to mature is based on chemokine secretions from the surrounding mesenchyme and the developing heart field which are well reviewed here [73]. Key players including fibroblast growth factors (FGFs) [64,[74], [75], [76], [77], [78], [79], [80]], WNTs [[81], [82], [83], [84]], and bone morphogenetic proteins (BMPs) [[85], [86], [87], [88], [89], [90]] are known to induce the differentiation of early lung progenitors in a controlled manner. For example, in mouse, it has been found that FGF10 plays a role in bud outgrowth [77] and drives lung progenitors towards a distal fate [78,79] through canonical WNT signaling [64,81,91]. Proximal epithelia develop because they are located further away from distally located FGF reservoirs in the mesenchyme, in a mechanism that appears dependent on concentration gradients [64]. BMP4 plays a key role in lung bud formation from foregut endoderm and establishment of both dorsoventral (back to front) and proximodistal (top to bottom) patterning in the nascent lung [88]. BMP4 is also present at high levels in distal bud tips and epithelia including AEC II cells [88,89], however, its inhibition promotes a proximal fate and, along with BMP2 inhibition, ciliated cell development [87,88,90].
2.3. Branching morphogenesis and other mechanical cues generated during lung development
While the cell fate of early proximal and distal lineages is directed through chemical signals, the lung epithelium itself undergoes marked changes in architecture, a process known as branching morphogenesis [79,92]. From the simple tube of the anterior foregut endoderm to the complex tubular structure of the adult, a highly stereotyped mechanism of branching morphogenesis facilitates the outgrowth, division, placement, and structure of lung airway [42]. Branching morphogenesis of the lung is driven by three simple and iteratively used processes: domain branching, planar and orthogonal bifurcation [93]. The first form of branching is domain branching: along a primary branch, buds form in a linear and sequential fashion, from proximal to distal. The next form of branching is planar bifurcation, in which the tip of the forming tube bifurcates to create two new tips, which subsequently elongate and bifurcate again, creating four tips. The last process of branching is known as orthogonal bifurcation. In this process, the initial planar bifurcation is followed by a rotation around the planar axis which creates two new tips through bifurcation. A critical gene in this process, Sprouty, has been found to attenuate Erk1/2 signaling, thereby altering the orientation of cell division and future tube elongation [94]. Other critical genes and regulatory networks associated with FGF signaling also contribute to controlling the periodicity of the branched network [95]. Although elements such as domain specification, bifurcation, rotation and branch generation remain largely undetermined [93,96], new technologies involving high-resolution live imaging, tension sensing, and force-mapping are opening paths to further explore and explain the branching morphogenesis phenomenon [97].
The early structure of the lung gives rise to a striking architectural separation of future SOX2+ proximal lineages and SOX9+ distal lineages, at least in mice [98]. The diameter of tube generated during branching morphogenesis in the pseudoglandular and canalicular stages has a small degree of variance within each stage as measured from electron micrograph sources of fetal human tissue [99]. This suggests that the branching program is rigorous in its control of lung structure and that tubes themselves may have instructive potential on the developing epithelia. Once the basic organ structure has formed, the lung continues to be exposed to mechanical cues as it continues to mature. In several cases, these cues have been shown to be essential for correct organ function. In utero, the fetal lung is a secretory organ that only converts to an absorptive one, to prepare for breathing after birth, through a change in the activity of chloride and sodium channels late in development. Fetal lung secretions result in a static fluid pressure of around 2.5 cmH2O in the developing terminal sacs of the fetus, which propels branching morphogenesis outwards into the developing thoracic cavity [100,101]. Lack of amniotic fluid in the developing lung alters the expression of distal epithelial markers and consequently results in the creation of smaller than normal lungs (pulmonary hypoplasia) [102], highlighting the importance of this mechanical pressure during lung development. In addition, cyclic strain is generated from fetal breathing movements (FBM) in utero that prime the airway for use after birth. FBM are detectable from the tenth week of pregnancy and begin as infrequent and erratic activity with long quiescent periods. As development continues, these quiescent periods decrease and sustained periods of fetal breathing occur. These breathing movements vary with the fetal sleep cycle and can be chemically tuned [57], and alter the volume of terminal sacs by around 5% [100,103], again highlighting the importance of mechanical signals influencing lung development. Finally, a novel FGF10/FGFR2-dependent tensional mechanism has been shown by which distal epithelial cells in the lung accumulate motor proteins at the apex of the cell, thereby becoming resistant to compression from increasing fluid pressure within the tube lumen. Cells under this tension are more likely to become AEC II cells, while those under compression become AEC I cells [102]. Interestingly, while the above examples highlight the importance of specific mechanical signals in the growth, development, and differentiation of the lung, PSC directed differentiation protocols of the lung are primarily based on mimicking the sequential chemical changes that occur during lung development.
3. Directed differentiation of lung epithelia inspired by embryology
Early attempts to create lung epithelia from PSCs began in mouse and did not attempt to mimic the stepwise changes in chemical signaling that occur during development. Rather, groups focused on applying lung-like physical cues such as air-liquid interface [21,104]. These protocols, while successful in generating NKX2.1+ positive populations, also produced contaminating cells expressing pluripotency markers (OCT4, NANOG, SSEA4, TRA1-60, TRA1-81). These early attempts solidified that further optimization, particularly related to the chemical cues applied, was needed to reliably create lung progenitors from pluripotent sources without remnant pluripotent contaminating cells. More successful directed differentiation protocols were rationalized from the detailed understanding of the chemical changes during lung embryology. In this section, we describe in detail the different differentiation protocols currently available that evolved from this approach.
3.1. Mouse embryonic stem cell derived lung epithelia
Although mouse models do not fully recapitulate human lung development, they have served as guides for earlier iterations of PSC directed differentiation protocols and have identified critical chemical cues for lung organogenesis. Broadly speaking, these protocols begin by driving stem cells towards a definitive endoderm fate (SOX17+ and FOXA2+; mimicking the pre-embryonic period of human lung development, weeks 0–2) through high doses of the nodal activating molecule, Activin A [[105], [106], [107], [108]]. Foregut endoderm is then induced via transforming growth factor beta (TGFβ) inhibition either alone [24] or with BMP inhibition [22] for a short period; a process called anteriorization (as during the embryonic period of human lung development, weeks 4–7). This foregut endoderm (FOXA2+SOX2+) is subsequently induced to generate NKX2.1+ cells (putative lung progenitors) by stimulating the retinoic acid (RA), BMP, WNT and FGF signaling pathways [109,110]. These lung progenitors are further matured, as demonstrated by increased NKX2.1 expression, through application of corticosteroids [22]. In brief, each protocol begins with PSCs guided through definitive endoderm, followed by anteriorization to foregut endoderm, and subsequent ventralization to generate NKX2.1+ cells [111]. These protocols formed the basis and backbone for the creation of human lung epithelia from PSCs.
Given the structural and cellular complexity of the lung, it is reasonable that the earliest protocols focused on mouse. However, there are subtle differences that highlight how human models are different in terms of structure, patterning, and differentiation. For example, the entire human conducting airway is comprised of a pseudostratified epithelium, even at diameters less than 0.5mm [112,113]. In contrast, conducting airways in mice only exhibit pseudostratified epithelium with accompanying submucosal glands and cartilage in the most proximal portion of the airway and transition directly into alveolar sacs [72]. This difference in histology affects the residing cell populations, as evidenced by the lack of basal cells in the lower portion of the proximal airways of mice [114,115]. Similarly, mouse models suggest that SOX2+SOX9+ progenitors are quite rare, and their cell fate is ambiguous [116]. However, evidence from directed differentiation of human lung epithelia [26,63], which has been confirmed in vivo [17,26,62,63], reveals that SOX2+SOX9+ progenitors are common in the developing lung buds and that branch tips of the pseudoglandular staged lung give rise to both proximal and distal epithelia [63]. Moreover, specific protein markers have been found to differ in both timing and location of expression between human and mouse models: pro-SPC in mouse is expressed early and throughout the developing mouse epithelium [117,118], while in human, pro-SPC is rarely detected early in development and is only robustly found later in distal epithelia [63]. These examples highlight that, while there are similarities, development and patterning of mouse and human lungs is different, and these differences require human models to be fully appreciated.
3.2. Human pluripotent stem cell-derived lung epithelia
Human PSC protocols have generally followed the same differentiation chronology as that of mouse directed differentiation, wherein definitive endoderm, anterior foregut endoderm, and NKX2.1+ lung progenitors are produced sequentially.
Different groups have adhered to their own methods of generating definitive endoderm, which primarily involves exposing PSCs to high concentrations of Activin A. Slight variations such as introducing WNT agonism (through WNT3a or CHIR99021) prior to [25] or alongside [11,30,32,33] Activin A, or additional exposure to BMP4 and FGF2 [23,25,29] during this stage exist across protocols for differentially inducing primitive streak and its anteriorization towards producing definitive endoderm. In addition, the use of embryoid bodies, which are limited by user experience and technique, has resulted in a wide range of production efficiencies for achieving this stage: from 45% CKIT+CXCR4+EPCAM+ cells [33] to >90% CKIT+ CXCR4+ cells [23,25]. Recent advances in commercial products have led to development of standardized 2D culture-based media (STEMdiff Definitive Endoderm Kit, STEMCELL Technologies) which allow reliable derivation of >95% of definitive endoderm [27,28,36,119].
Similarly, generation of both anterior foregut endoderm and ventralized lung progenitor populations has been subject to much investigation and modification. Earlier work suggested that SOX2+FOXA2+ (92 ± 2% in their case) anterior foregut endoderm can only be induced by subjecting definitive endoderm to TGFβ and BMP inhibition [23]. Subsequent studies, however, attempted to anteriorize definitive endoderm to foregut endoderm through TGFβ inhibition alone (50-60% SOX2+), a combination of endogenous WNT, TGFβ and BMP inhibition (not quantified) [25], and via Sonic Hedgehog (SHH) and FGF2 signaling (78% FOXA2+ EPCAM+) [11]. A comparison of the latter two strategies demonstrated that SHH and FGF2 are insufficient in producing reliable NKX2.1+ lung progenitors [25], possibly because FGF2 is involved in promoting thyroid lineages [120]. In general, TGFβ and BMP inhibition [23] is the basis for currently applied endoderm anteriorization strategies [27,[29], [30], [31],36,121].
Factors involved in early versions of ventralization in directed differentiation protocols included WNT3a, FGF7, FGF10, BMP4, epidermal growth factor (EGF), and RA have now been reduced based on elimination studies [25,120]. As such, CHIR99021 (CHIR; WNT agonist), BMP4, and RA are necessary and sufficient for producing lung progenitors from anterior foregut endoderm derived from both mouse and human PSCs [25,30,32,119]. Despite finding that FGF7 and FGF10 are non-essential for inducing NKX2.1 expression, they continue to be used for ventralization in some protocols [29]. Although each protocol differs in terms of the duration of each phase, NKX2.1+ lung progenitors are generally achieved by 15 days, with the exception of a study by de Carvalho et al., in which they maintained their cultures for an additional 10 days in FGF7, FGF10, and CHIR99021 to attain 90-98% NKX2.1+FOXA2+ lung progenitors. In all cases, these lung progenitors are then either sorted or directly guided towards proximal or distal progeny in 2D or 3D culture systems. Ideally, products of directed differentiation protocols should mimic the cell proportions present in human airways and lungs (Table 1), however current protocols have not progressed that far. While these protocols continue to be refined, the percentage of select cell populations generated from these protocols have been summarized in Table 2 .
Table 2.
Products of airway and lung directed differentiation protocols
| Definitive Endoderm | Anterior Foregut Endoderm | Lung Specification | Proximal Epithelia | Distal Epithelia | Contaminant Populations | |
|---|---|---|---|---|---|---|
| Green et al. 2011 | >90% CKIT+CXCR4+ | 92±2% FOXA2+ SOX2+ | 37±6% NKX2.1+ | Not quantified | Not quantified | None observed |
| Mou et al. 2012 | 85–90% FOXA2+SOX17+ | 50-60% FOXA2+ SOX2+ | 10–30% NKX2.1 | NKX2.1+P63+in vivo (Not quantified) |
Not quantified | None observed |
| Wong et al. 2012 | 87.5% CD117+CXCR4+ | 78% FOXA2+EPCAM+ | 32.1% NKX2.1 32.2% PanKRT+ 30.9% CFTR+ 53.9% P63+ |
42.8% CFTR+ panKRT+ 24.8% FOXJ1+ 14.6% CFTR+FOXJ1+ |
SOX9+, SPC+ (Not quantified) |
PDX1+ TG+ PAX9+ HNF4+ AFP+ |
| Huang et al. 2014 | >96% CKIT+ CXCR4+ | Not quantified | 86.4% NKX2.1+FOXA2+ | 2–5% SCGB1A1+ P63+, MUC5AC+, FOXJ1+ (Not quantified) |
50% SPB+ Surfactant metabolism |
TUJ1+ PAX6+ |
| Firth et al. 2014 | 64.8±4.2% FOXA2+SOX17+ ~45% EPCAM+ CXCR+CKIT+ |
45.9±8.4% NKX2.1+SOX2+ 55.6±5% FOXA2+NKX2.1+ |
Not Quantified | 27.3±3.7% CCSP+ 1–2% MUC5AC+ AcTUB+, CFTR+ (Not quantified) |
None observed | TG+ TUJ1+ |
| Gotoh et al. 2014 | ≥80% CXCR4+ | ≥88% FOXA2+SOX2+ | 57–77% FOXA2+NKX2.1+ | CCSP+ (Not quantified) |
3.82%±0.5% CPM+SPC+ SPB+ AQP5+ (Not quantified) |
None observed |
| Dye et al. 2015 | Not reported | FOXA2+SOX2+NKX2.1+ (Not quantified) |
57% NKX2.1+ | 39% P63+ 3% FOXJ1+ SCGB1A1+ (Not quantified) |
5% SPC+ 4% HOPX+ |
Vimentin+ SMA+ layer around airway-like structures |
| Konishi et al. 2016 | Not reported | Not reported | NKX2.1+SOX2+ (Not quantified) |
72–87% FOXJ1+ AcTUB+, CFTR+, MUC5AC+, KRT5+, SCGB1A1+, CHGA+, SYP+ (Not quantified) Ciliary beating |
None observed | None observed |
| Chen et al. 2017 | Not reported | 89.07±3.36% FOXA2+ 92.08±1.88% EPCAM+ |
51.26±4.37% NKX2.1+ | 23.78±5.21 SOX2+ | 76.75±6.89% SOX9+ SPC+, SPB+, MUC1+, HT2-280+ (Not quantified) Surfactant metabolism |
Vimentin+ SMA+ CD90+ PDGFRA+ PDGFRB+ |
| McCauley et al. 2017 | Not reported | Not reported | 35.2% NKX2.1-GFP+ | SOX2+, P63+, KRT5+, SCGB3A2+, AcTUB+ (Not quantified) |
SOX9+, SPC+, SPB+ (Not quantified) | None observed |
| Jacob et al. 2017 | Not reported | Not reported | >78.2% NKX2.1-GFP+ | None observed | 98.7% NKX2.1+SPC+ | None observed |
| Yamamoto et al. 2017 | Not reported | Not reported | 85.2%±5.6% NKX2.1+ | None observed | 51.2%±1.2% SPC+ SPB+, PDPN+, AQP5+ (Not quantified) |
None observed |
| de Carvalho et al. 2018 | Not reported | Not reported | 90-98% NKX2.1+ | 53.5±4.31% P63+ 45±10.5% ITGA6+ITGB4+ 16.5±5.85% EPCAM+NGFR+ ~15% of AcTUB+ <1% of MUC5B+ <1% of CC10+ |
<12.1±5.3% EPCAM+HT-280+ <2% PDPN+HOPX+ |
None observed |
| Miller et al. 2019 | Not reported | Not reported | SOX2+SOX9+ID2+NKX2.1+ (Not quantified) |
P63+, AcTUB+, FOXJ1+, MUC5AC+ (Not quantified) |
HOPX+, SPC+, SPB+ (Not quantified) | SMA+ layer around airway-like regions |
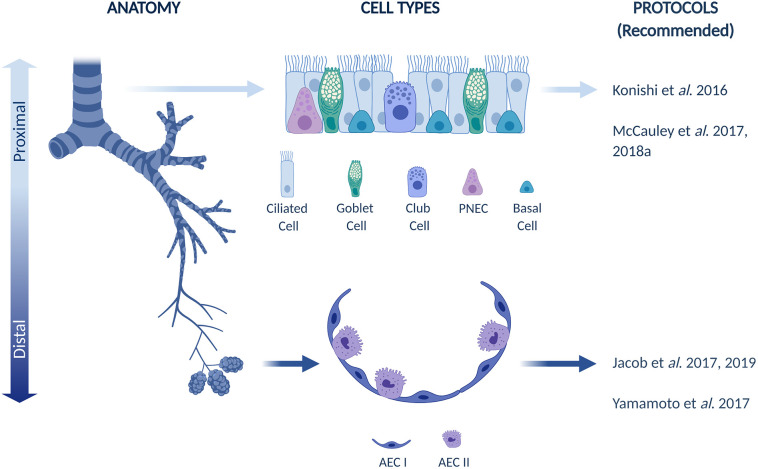
3.3. Creation of human proximal lung epithelia
Protocols to create proximal lung epithelia have focused on the production of the four major cells types present: ciliated, goblet, club, and basal cells (see Table 1 for a summary of markers for each cell type). Motivation for creating proximal epithelia in the field has primarily been to develop patient-specific cystic fibrosis (CF) models [11,24,27] and/or to produce epithelia with multi-ciliated cell populations for protocol validation [30,33]. A shift towards human PSC-derived CF models has been critical as mouse models do not accurately represent CF disease progression and phenotypes seen in humans [[128], [129], [130]]. As such, the first evidence of human PSC proximalization using CF patient-derived PSCs was shown by Mou et al., who exposed anterior foregut endoderm to BMP4, GSK3iXV (WNT agonist), FGF2, and RA-supplemented B27 to generate 10-30% NKX2.1+ cells by Day 12. Although contaminating neuroectodermal and distal lung NKX2.1+SOX9+ cells were present, day 12 populations included proximal NKX2.1+SOX2+ progenitors. Subcutaneous implantation of this population in immunodeficient mice for 30 days resulted in emergence of NKX2.1+P63+ cells, however no mature epithelial markers for ciliated, goblet, and club cells were found.
Wong et al. employed a longer, 2D differentiation approach to produce mature proximal airway epithelia in vitro. Through a process they called “proximal specification”, they generated day 15 lung progenitors via low levels of BMP4 (mimicking signaling gradients in the airway), FGF7, and FGF10 which began expressing proximal genes. Further culture with FGF7, FGF10, and FGF18 resulted in upregulated gene expression of KRT5, P63, FOXJ1, SOX17, cystic fibrosis transmembrane conductance regulator (CFTR), and SCGB1A1 to a lesser extent, along with low levels of distal SOX9 and SPC by day 19. Protein expression amounted to 32.1% NKX2.1+, 32.2% panKRT+, 53.9% P63+, 36% FOXJ1+ cells. These cells were subsequently matured in air-liquid interface (ALI) culture for 5 weeks (1 week of submerged culture with FGF18, followed by 4 weeks of ALI culture) to generate 42.8% CFTR+panKRT+, 24.8% FOXJ1+ with 14.6% co-expressing CFTR, and 19% CFTR+LHS28+ cells. The resulting epithelium ranged from being squamous to cuboidal with sparse pseudostratified regions, implying that this protocol lacked specific maturation cues. Contaminating thyroid (thyroglobulin and PAX9), liver (HNF4 and AFP), and pancreatic (PDX1) lineages were detected through quantitative PCR, while percentages of goblet, club, and basal cell populations (barring gene expression analysis) were not evaluated.
A similar 2D culture approach was employed by Firth et al. to generate proximal lung progenitors which were subsequently matured into multi-ciliated epithelia. They optimized lower concentrations of BMP4 required during the ventralization phase (day 9–17) to promote higher gene expression of FOXA2, SOX17, MUC1, MUC5AC, KRT5, P63, CFTR, and NGFR by the end of their differentiation protocol (day 45). Proximal lung progenitor efficiency was not reported at day 17, however earlier assessments at day 9 and 10 identified contaminating thyroid and neuroectodermal cells, with approximately 28% of total cells portraying mesenchymal characteristics based on CD90 protein expression. This is plausible given the low definitive endoderm efficiency (45% CKIT+CXCR4+EPCAM+). Regardless, day 17 cells underwent further maturation in ALI culture for 28 days, resulting in a cuboidal epithelial layer with an underlying mesenchymal layer; only 45-50% of these cells were EPCAM+. The resulting epithelium was devoid of multi-ciliated cells but comprised 27.3±3.7% CCSP+ club cells, 1–2% MUC5AC+ goblet cells, and some functional CFTR+ cells. Multi-ciliated cells were only achieved through NOTCH inhibition (DAPT) during ALI culture.
Konishi et al. were the first to introduce a 3D culture-based protocol to generate proximal epithelia from human PSCs. Post-ventralization, they sorted day 14 cells based on expression of carboxypeptidase M (CPM), an AEC I-specific marker which was used as a surrogate marker for identifying lung progenitors based on the finding that 92% of CPM+ cells reliably co-expressed NKX2.1 [32]. Inspired by the formation of tracheospheres from primary tracheal epithelia in earlier studies [131,132], they embedded CPM+ cells in Matrigel and cultured them for 14 days in the presence of FGF10 and CHIR. Resulting spheroids were NKX2.1+SOX2+ but devoid of multi-ciliated cells, therefore an additional 14 days of culture in PneumaCultTM-ALI media (Stem Cell Technologies) was added for further maturation. Day 42 spheroids depicted AcTUB+ and MUC5AC+ cells in the lumen with smaller numbers of KRT5+ SCGB1A1+, CHGA+ and SYP+ cells; no AQP5+ and SPC+ cells were detected. NOTCH inhibition between day 28 and 42 resulted in significant upregulation of FOXJ1, SYP, and CHGA genes. Meanwhile, SCGB1A1 was undetected and MUC5AC expression was 100-fold less than levels found in adult tracheae. Spheroid cells were dissociated and cultured under ALI conditions (with continued NOTCH inhibition) for 14 days to generate multi-ciliated cells (72±6.6% to 87.06±0.43% FOXJ1+ across cell lines) with ciliary beating frequency similar to that of primary bronchial epithelial cells; other proximal markers were not evaluated at this point. Overall, the use of 3D culture resulted in tri-lineage (ciliated, goblet, and club cell) differentiation of proximal airway epithelia, which was not observed in their 2D ALI cultures.
McCauley et al. developed a protocol [119] based on their findings that withdrawal of WNT signaling post lung progenitor specification at day 15 promotes a proximal fate (upregulated SOX2, P63, SCGB2A2) both in 2D and 3D culture [27]. They sorted day 15 lung progenitors, through an NKX2.1-GFP reporter or by selecting a CD47highCD26low population that identifies NKX2.1+ cells [34], and subsequently embedded them in Matrigel for 2 weeks of 3D culture in the presence of FGF2, FGF10, and corticosteroid agonists: dexamethasone, 8-bromo-cAMP and isobutylmethylxanthine (DCI). This led to significant upregulation of TP63, SCGB3A2, SCGB1A1, MUC5B, CFTR, FOXJ1, and SFTPB (AEC II marker) genes, along with protein expression of P63, SOX2, KRT5, MUC5AC, SCGB3A2, and SFTPB. Another study by this group reported that these cells can be divided into three clusters at this stage: 1) secretory cells (SCGB3A2+, SCGB1A1+), 2) non-secretory airway basal cells (TP63+, ITGA6+) and 3) non-lung hepatic cells (SERPINA1+, SOX9+, APOA2+, AFP+) [27]. Multi-ciliated cells were only observed within spheroids when NOTCH signaling was inhibited, at the expense of downregulated SCGB1A1 expression, or when spheroid cells were further cultured in ALI conditions. Other mature epithelial populations including club and goblet cells were not assessed post ALI culture. It is unclear whether 2D or 3D culture systems resulted in more representative proximal populations, although it is worth noting that the 3D spheroids could be manipulated to produce a variety of proximal epithelia ranging from progenitor to differentiated populations.
The most recent approach, described by de Carvalho et al., extended the lung specification phase by 10 days, while maintaining CHIR, FGF7 and FGF10, to achieve 90-98% NKX2.1+ cells by day 25. Subsequent withdrawal of CHIR during collagen I-embedded 3D culture resulted in differentiation of AEC I, mature AEC II, and proximal cells, with the latter representing 34.52±2.25% P63+, 24±12% ITGA6+ITGB4+, and <1% of AcTUB+, CC10+, and MUC5B+ cells at day 50. Prolonged culture until day 80 produced 16.5±5.85% mature EPCAM+NGFR+ basal cells, which formed a primarily squamous epithelium depicting abnormal AcTUB and limited MUC5B and CC10 expression after 6 weeks of ALI culture. The multilineage differentiation seen in CHIR withdrawal cultures, in discordance with McCauley et al., were attributed to GSK3β-associated cell cycling inhibition as opposed to canonical WNT signaling. In fact, WNT3a in absence of CHIR led to significant increase in P63+ (51.5±9.78%) and ITGA6+ITGB4+ (31.8±12.95%) cells by day 50. Employment of NOTCH inhibition, in addition to CHIR withdrawal, during 3D culture increased P63+ (53.5±4.31%), ITGA6+ITGB4+ (45±10.5%), and ciliated cell proportions, while repressing club and AEC II cell markers, indicating that NOTCH signaling plays a role in proximodistal specification. In general, this protocol produced cell types of both proximal and distal lineages, however, lacked in its ability to generate differentiated epithelia of appropriate proportions and maturity despite lengthy cultures.
3.4. Comparisons of proximal airway directed differentiation protocols
Overall, the aforementioned studies differed drastically from each other with regards to the timing and chemical modulation of each phase of differentiation towards proximal epithelia, and consequently produced variable results. While it is evident that 3D culture augments maturation, no protocol to date has been able to efficiently produce all functional epithelial populations present in the airway in proportions representative of those in vivo. Furthermore, these studies have not thoroughly elucidated the mechanisms of proximal patterning. Barring the application of FGF18 [11] (known to enhance proximal programming [133]), protocols have adopted growth factors based on trial and error without understanding why, for example, FGF10 signaling (which is known to favor distal lung development) promotes production of proximal progenitors [11,27,30,33,37,119]. As such, the quest for obtaining mature airway progenitors, such as NGFR+ cells, comes at the cost of elongated protocol lengths, heterogenous maturation levels of resulting populations, and missed opportunities for understanding why these populations do not result in a histologically appropriate epithelium [29].
It is apparent that the timing of signaling molecule delivery as well as the competence of subjected cell populations to respond to a given signaling molecule are of extreme importance. The spatiotemporal dynamics of cell signaling are non-linear, are more complex in vivo, and are not fully appreciated in the latter stages of current directed differentiation protocols. This may explain the incongruence amongst different protocols, primarily those assessing the effects of GSK3β and WNT signaling [27,29,30], all of which targeted populations at non-comparable protocol stages. Therefore, a deeper analysis is required to appropriately explain and mimic these dynamics in vitro. Furthermore, recreating these spatiotemporal signaling patterns during directed differentiation protocols may potentially require repurposing molecular delivery tools from other fields such as drug delivery and tissue engineering [[134], [135], [136]].
Interestingly, most existing protocols have been skewed towards generating multi-ciliated cells at the expense of goblet and club cells by subjecting airway progenitors to NOTCH inhibition, which is known to decrease goblet cell populations [137,138]. Goblet cells, in addition to club cells, have recently been discovered as a source for generating multi-ciliated cells in primary airway epithelia [139]. Club cells play a key role in epithelial injury, wherein they de-differentiate into basal cells in the absence of basal cells such that they can give rise to ciliated and club cell populations to repair a denuded epithelium [140]. Therefore, in the future it will be critical to identify protocols to create PSC-derived cultures containing these cell types, and not just multi-ciliated cells, in order to fully capture the dynamics of airway injury and repair for drug screening. Overall, based on current progress, the Konishi et al. and McCauley et al. protocols are considered the most relevant for generating functional airway epithelia.
3.5. Creation of human distal lung epithelia
The alveolar space in the distal lung is comprised of two epithelial cell types: AEC I and AEC II (see Table 1 for specific markers of each cell type). Distal lung directed differentiation protocols have progressed drastically in the last 5 years, with most of them employing a 3D organoid culture phase for maturation to acquire these progenies. Earlier work by Huang et al. focused on efficiently inducing lung progenitors in 2D culture by streamlining the ventralization phase of the protocol. Day 15 lung progenitors (86.4.1± 1.7% FOXA2+NKX2.1+) were multipotent, leading to proximal and distal lineages both in vitro and in vivo, albeit with contaminating non-lung and mesenchymal cells, respectively. Aggregate culture of these cells in the presence of CHIR, FGF7, FGF10, and DCI for 10 days resulted in proximal NKX2.1+SOX2+ and NKX2.1+P63+ cells by day 25. Interestingly, subsequent culture in the presence of DCI alone resulted in distal SPB+ cells (50%) by day 48, of which 52% could metabolize surfactant, a characteristic of AEC II cells.
A study by Gotoh et al. demonstrated that 3D co-culture with fetal lung fibroblast significantly augmented distal epithelial maturation. In this investigation, day 14 lung progenitors (57–77% FOXA2+NKX2.1+) were sorted based on CPM+ expression and cultured in 2D in the presence of FGF7 and DCI for 14 days. Resulting cells expressed SPB but not SPC (indicating a lack of AEC II cells), which remained unchanged even when CPM+ cells were extracted on day 23 instead of day 14; this implies that prolonged 2D culture does not promote AEC maturity, necessitating transition into a 3D culture phase. As such, when day 14 CPM+ cells were embedded with fetal lung fibroblasts in Matrigel for 3D culture, spheroids expressing distal epithelial markers formed by day 24, of which 3.82% ± 0.5% of extracted cells were SPC+. Therefore, 3D culture of lung progenitors rapidly induced a distal population in contrast to 2D culture of lung progenitor aggregates, by Huang et al., which required prolonged culture in different media conditions to achieve distal maturation.
Furthering this work, Yamamoto et al. developed a modified protocol, wherein day 14 lung progenitors were “preconditioned” for 7 days in CHIR, FGF7, FGF10, and DAPT. This additional culture phase enhanced the lung progenitor population, as compared to the older Gotoh et al. protocol, by resulting in 85.2% ± 5.6% NKX2.1+ by day 21 [12]. This preconditioning also positively influenced the 3D co-culture phase, wherein day 35 alveolar organoids expressed SPC (51.2% ± 1.2%), SPB and PDPN (AEC I marker) and depicted lamellar-body-like organelles reminiscent of AEC II cells. With an ability to self-renew across multiple passages, SPC+ populations gained maturity, while SPC-PDPN+AQP5+ AEC I-like cells also began appearing in later passages. The emergence of both alveolar populations in this protocol was especially promising. Additionally, these alveolar organoids enabled drug toxicology assessment as evidenced by appropriate lamellar body enlargement in response to GNE7915 and Amiodarone treatment.
A fibroblast-free alveolar organoid culture protocol was subsequently developed by Jacob et al. in which sorted NKX2.1-GFP+ or CD47highCD26low (>78.2%) lung progenitors underwent 3D culture for 14 days. Via elimination studies, it was found that CHIR and FGF7 (with DCI) were sufficient to promote distal patterning, with CHIR primarily being responsible for significantly increasing SPC expression (up to 33% by day 38), consistent with their other findings [27]. NKX2.1+ alveolar organoids were devoid of proximal markers, however, also negligibly expressed AEC I markers such as PDPN and AQP5. Lamellar body-like features were seen in alveolar organoids that were maintained across serial passages (up to 98.7% NKX2.1+SPC+ by passage 9) [36], while SPC+ cells synthesized and secreted surfactants according to lipidomic analyses. These alveolar organoids exhibited amenability for toxicology studies as they appropriately initiated an immune response by activating NK-κB signaling upon exposure to canonical ligands such as TNF-α and IL-1β.
Another 3D approach for generating distal lung epithelia was described by Dye et al [31]. Herein, free-floating endoderm clusters formed foregut spheroids (expressing SOX2, FOXA2, and NKX2.1), with mesenchymal contamination, in the presence of FGF4, NOGGIN (BMP inhibitor), SB431542 (TGFβ inhibitor), CHIR, and SAG (sonic hedgehog/SHH agonist) by day 10. Matrigel-embedded culture of these organoids with FGF10 signaling resulted in NKX2.1+ organoids with SOX2+ and SOX9+ domains. Prolonged culture (>60 days) led to development of immature lung organoids (57% NKX2.1+) comprising 14.5±0.6% proximal and 85.5±0.6% distal/mesenchymal structures. Specifically, 39% P63+ and 3% FOXJ1+ cells were present, however no indications of apical cilia, goblet cell, and club cell formation were found. Meanwhile, progenitor-like SOX9+SPC+ and SOX9+HOPX+ clusters were prominently present with minimal mature SPC+ (5%) and HOPX+ (4%) populations. Further refinement of this protocol bifurcated proximal “human lung” and distal “bud tip progenitor” organoid development by culturing foregut spheroids in FGF10 with 1% serum or FGF7, CHIR, and RA in serum-free media, respectively [121]. After 65 days of culture, the “human lung” organoids expressed P63, FOXJ1, and mesenchymal markers with no sign of mature epithelial features; some SPC and HOPX staining was also observed. Only after an 8 week-long in vivo implantation did mature ciliated AcTUB+ cells appear. “Bud tip progenitor” organoids also contained heterogenous MUC5AC+, HOPX+, SPB+, and SPC+ cells after 120 days. However, when seeded into naphthalene-injured mouse airways, they gave rise to AcTUB+ and MUC5AC+ cells. In general, this protocol diverged to produced lung organoids with heterogeneous populations of either predominantly proximal or distal epithelia, which required prolonged culture or in vivo implantation for maturation (limited in this case). A key aspect of the “human lung” organoids was their inclusion of a mesenchymal population to study epithelial-mesenchymal crosstalk during lung development.
Chen and colleagues described a similar approach wherein 3D branching projections of anteriorized foregut endoderm (89.07±3.36% FOXA2+ and 92.08±1.88% EPCAM+) were cultured in CHIR, BMP4, FGF7 and FGF10 for 25 days [26]. Resulting lung bud organoids (LBOs) were NKX2.1+ (51.26±4.37%), were surrounded by mesodermal cells, and did not express any mature lung or airway markers other than P63 (18.59±1.49%). When embedded in Matrigel and grown in the same media conditions, they rapidly branched, forming tubular structures. Day 70 LBOs expressed FOXA2 (95.17±1.54%), NKX2.1 (74.97±4.37%), and SOX9 (92.42 ± 3.81%), along with MUC1, SPC, and SPB; proximal markers were absent, except for minimal MUC5AC+ cells. Further growth of LBOs up to >170 days revealed saccule-like structures, depicting NKX2.1+ (84.86±5.21%), SOX9+ (76.75 ± 6.89%), and SOX2+ (23.78 ± 5.21%) cells. The presence of AEC II-like cells with lamellar bodies was evident, which were able to process surfactant. Meanwhile, except for HOPX, markers of AEC I were not present and proximal markers were limited to MUC5AC and SCGB3A2. Other mature airway and lung markers did not appear until after 7 months of in vivo implantation, with those of AEC I cells still absent. This work further confirmed that lung organoids produce immature, heterogeneous populations of proximal and distal epithelia which require extended 3D culture or in vivo implantation to promote maturation (incomplete in this case).
3.6. Comparisons of distal lung directed differentiation protocols
All described directed differentiation protocols for distal epithelia utilized a 3D culture approach in some format, however only those that established lung specification in 2D culture prior to a 3D transition demonstrated promising results. These protocols do not completely depend on spontaneous organoid assembly, are highly responsive to fine tuning with morphogens, and can therefore provide better insight into the cellular responses in lung development to generate therapeutic strategies accordingly.
Although the “human lung” organoid and “lung bud progenitor” organoid-based protocols may be useful for studying complex cellular interactions during disease progression or repair, they are currently under-developed, do not recapitulate proximodistal patterning adequately in vitro, and are limited in terms of protocol lengths and incomplete array of relevant cell types produced. Especially in the Miller et al. study, both organoid models stochastically led to production of proximal cells ex vivo or in vivo, thereby diluting the need for developing two approaches for generating either proximal (“human lung” organoids) or distal (“lung bud progenitor” organoids) epithelia. A potential factor contributing to complications in the Miller protocol was their method for determining signaling cues: they eliminated FGF7, FGF10, BMP4, CHIR, and RA one at a time while culturing bud tips isolated from mice [62]. In contrast to analysis done using mouse and human PSCs during directed differentiation in previous studies [25,120], they found that removal of BMP4 enhanced SOX9 gene expression. This was perhaps due to the differences in complexity of the aforementioned models studied, especially in relation to the mouse lung buds being equipped with a heterogeneous, potentially mesenchymal, cell population and its associated paracrine signaling that may compensate for presence or absence of growth factors. Therefore, it is important to elucidate essential cues in simpler models prior to progressing towards 3D models that involve complex signaling patterns.
The Yamamoto et al. and Jacob et al. protocols presented the strongest results for generating AEC II-like cells that expanded rapidly and matured with serial passaging. While the surfactant processing ability of differentiated AEC II cells was only exhibited by Jacob and co-authors, both protocols generated alveolar organoids (simple spheroids containing alveolar cells) that can mimic functional properties of AEC II cells by responding appropriately to pharmaceutical and molecular stimuli.
The presence of mature AEC I cell markers was only seen in the Yamamoto protocol, with a major difference being its dependence on a mesenchymal feeder population. Although feeder-free generation of alveolar organoids was possible, only 23.1% ±1.4% of collected cells expressed SPC. Evidently, the mesenchymal niche provided key maturation signals that require further investigation for advancing feeder-free protocols. While there is general consensus regarding the involvement of WNT signaling for distal pattering, RNA sequencing-associated gene ontology, in this study, suggested the involvement of Hippo and MAPK pathways as well. Furthermore, the employment of NOTCH inhibition during the “preconditioning” phase may have played a role in promoting both AEC I and AEC II populations, and therefore the aforementioned signaling pathways need to be collectively assessed. Inspiration can be sought from a recent study which described a computational modeling approach, based on single cell RNA sequencing, that predicted the optimal time point for CHIR withdrawal for maintaining a NKX2.1+SPC+ lung fate; its findings were further supported by empirical studies [35]. Employment of such techniques will prove essential for understanding fate choice and developing customized target lung populations.
3.7. Modeling airway and lung diseases for drug discovery
The study of airway and lung diseases is limited by animal models as they do not recapitulate human disease phenotypes and progression adequately. For example, existing mouse models of cystic fibrosis (CF) vary greatly in their ability to represent relevant organ pathologies and are deficient in developing spontaneous lung disease observed in humans [[141], [142], [143]]. Similarly, pulmonary fibrosis is most commonly studied in the bleomycin-induced lung fibrosis mouse model, which results in faster disease progression, eventual resolution of disease phenotype over time, and is obscured by the wide range of bleomycin doses administered for induction of injury [144]. Studies exploiting PSC-derived lung culture models to explore the effect of drugs in human lung cells are therefore beginning to emerge.
Culture models of proximal airway epithelia have been applied for drug discovery primarily in the context of CF. CF is an autosomal recessive genetic disorder caused by mutations in the epithelial chloride channel gene, CFTR, which consequently leads to accumulation of excess mucous and compromised mucociliary clearance [145], affecting multiple organs. CF phenotypes have been studied widely in primary or PSC-derived intestinal [[146], [147], [148]], rectal [[149], [150], [151]], pancreatic [152], and airway models [11,27,33,153,154].
In the context of airway models, ALI culture has been the gold-standard for studying primary airway epithelia derived from CF patients. In concordance with this method, Wong et al. attempted to differentiate CF patient-derived PSCs, albeit with great heterogeneity with regards to CFTR expression and activity across PSC lines and trials. Regardless, they used the models to explore mechanisms of drug action and provided evidence of CFTR activity in response to cyclic-AMP agonism through forskolin, isobutylmethyl xanthine, and genistein in select PSC lines. Additionally, upon treatment of a CF patient-derived PSC line with an analog of VX-809 (an FDA-approved drug for CF), partial restoration of CFTR localization in the plasma membrane was observed [11]. Similarly, Firth et al. demonstrated characteristic apical expression of CFTR in their differentiated PSC-derived ALI cultures along with appropriate CFTR activity upon exposure to forskolin and/or an CFTR inhibitor [33].
Organoid-based culture systems have also been used for drug exploration in CF. Based on a previously developed CFTR assay [151], McCauley et al. used an organoid-based approach to assess the application of PSC-derived airway epithelial cultures for CF disease modeling. Their protocol was able to reproducibly generate airway spheroids from normal PSC lines that were responsive to forskolin. In contrast, CF patient-derived spheroids were non-responsive, thereby replicating the expected CF disease phenotype. This effect was confirmed to be CFTR-dependent as CFTR-corrected PSC spheroids behaved similarly to normal spheroids [155]. While CFTR function can therefore be modeled in both 2D and 3D culture systems, the function of mucociliary clearance may only be appropriately modeled using the ALI culture system. Assessment of this specific airway function, of course, is dependent on the ability of the chosen directed differentiation protocol to produce all the relevant cell types involved (ciliated and goblet cells) using ALI culture. The presence of goblet cells, for example, was not confirmed by the Wong et al. protocol to recapitulate the mucociliary clearance function.
Distal lung organoid models have been applied to drug development in a broader range of diseases. For example, pulmonary fibrosis is a fatal disease which involves progressive and irreversible alveolar fibrosis, leading to lung failure [156,157]. A handful of reports have emerged using human PSC-derived epithelium to model pulmonary fibrosis. In one study, Chen et al. developed LBOs (complex organoids with branching structures) to model pulmonary fibrosis associated with an autosomal recessive disorder called Hermansky-Pudlak syndrome (HPS) by deleting the HPS1 gene in their PSC line [158,159]. These LBOs demonstrated diminished branching, contained a high proportion of proliferative mesenchymal cells, and showed increased deposition of ECM proteins (collagen I, collagen III, and fibronectin), consistent with a fibrotic state [26]. Similarly, using the Yamamoto et al. protocol, Korogi and colleagues modeled pulmonary fibrosis in alveolar organoids (simple spheroids containing alveolar cells) with an HPS2 mutation [160] and reported deviant size and distribution of lysosome-like organelles of lamellar bodies, as well as abnormal lamellar body enlargement and secretion. At the same time, a wider range of PSC lines was generated by Strikoudis et al., who mutated individual HPS genes (HPS1, HPS2, HPS4, and HPS8) and comprehensively corroborated the LBO fibrotic phenotype of each mutation with its clinical presentation [161]. This study implicated interleukin 11 (IL-11) in inducing fibrotic phenotypes (increased mesenchymal populations and ECM deposition), which was validated by IL-11 expression pulmonary fibrosis and HPS clinical patient samples. Deletion of IL-11 in fibrotic HPS4-/- LBOs rescued the wildtype phenotype, thereby identifying IL-11 as a therapeutic target for lung fibrotic disease [161]. In a concurrent study, Ng et al. confirmed the pro-fibrotic role of IL-11 and developed an IL-11 antibody which reversed late-stage lung fibrosis by significantly decreasing ECM deposition in an animal model [162]. Evaluation of this IL-11 antibody using PSC-derived lung organoid models can provide better insight into their applicability for human disease.
Distal lung organoids have also been applied to model respiratory viral infections caused by respiratory syncytial virus (RSV), human parainfluenza virus type 3 (HPIV3) [163,164], as well as the measles virus (MeV). Chen and colleagues infected LBOs with RSV, resulting in characteristic luminal shedding of epithelia [26], which leads to small airway obstruction and consequent bronchiolitis in clinical settings [165]. Interestingly, Sach et al. demonstrated that prior incubation with palivizumab (an antibody that prevents RSV from fusing with cells) prevented RVS from replicating in primary airway organoids [154], which would be valuable to assess in PSC-derived lung or airway organoids. Meanwhile, HPIV3 infected AEC II in LBOs and temporally reached peak infection similar to that in primary alveolar epithelia [164], confirming clinical data. HPIV3 infection did not result in either epithelial shedding or syncytium formation in the LBOs as did RSV [26] and MeV [164] infections, respectively, again confirming clinical phenotypes. This showed the ability of lung organoid models to not only demonstrate virus-specific infection, but also to recapitulate phenotypes observed in the clinic.
Another condition modeled by alveolar or lung organoids is SPB deficiency, a lethal neonatal autosomal recessive disease which necessitates lung transplantation for patient survival. Both Jacob et al. and Leibel et al. developed alveolar [28] (simple spheroids containing alveolar cells) and lung [166] (complex organoids with branching structures) organoids, respectively, from patient-derived PSCs with an SPB mutation. Despite major differences in their cellular composition, both organoid models similarly demonstrated diminished evidence of lamellar bodies and SPB protein expression. These models open avenues for drug development to manage SPB deficiency and consequently prolong neonatal survival.
Currently, little to no effective therapeutic options exist for many of the aforementioned diseases. Limitations of current pre-clinical disease models are particularly unveiled during clinical trials as drugs that seemingly ameliorate disease progression in standard models either fail or result in adverse effects in humans [167]. Specifically, at least 16 drugs have failed in Phase 2 or 3 of clinical trials for pulmonary fibrosis treatment in the last decade.[167] A detailed examination of pre-clinical models is required to assess their limitations in replicating drug efficacy results of clinical trials. Based on their ability to recapitulate human disease and genetic profiles, PSC-derived lung organoid models may augment existing pre-clinical models by determining drug efficacy more appropriately prior to clinical trials. The identification of IL-11 as a therapeutic target for IPF by Strikoudis et al. and Ng et al. is an excellent example of exploiting human PSC-derived lung organoid models to identify novel therapeutic targets. From this point forward, protocols will ideally be selected for drug discovery based on which organoid models best fit the disease of interest as no single protocol exists that 1) appropriately represents all relevant cell types and 2) has been shown to model a wide range of region-specific infections and diseases.
3.8. Limitations of current directed differentiation protocols
While there has been great progress in the establishment and maturation of lung epithelia from PSC populations, a number of limitations have emerged that will require optimization and augmentation of current protocols to create better developmental and disease models, and specific cell populations:
-
a.
Lack of control over which populations are produced - understanding or recapitulation of signaling pathways beyond proximodistal patterning is currently limited, as the ratio of AEC II versus AEC I cells; or club cells versus goblet cells cannot be reliably predicted. Furthermore, while development of reporter lines and identification of surface markers for sorting have [32,34] helped the advancement of distal lung protocols (for AEC II cells specifically) in the last few years [28,36], such techniques are limited in current proximal airway protocols[168].
-
b.
Lack of maturity and characterization - there is inconsistent maturation of resulting cell populations within and across available protocols with results ranging from first trimester to adulthood-like cells [25,26,28,29,35,37].
-
c.
Variability in cell production - the same protocol does not consistently and reliably produce the same types of cells in similar ratios. Further, inherent differences amongst PSC lines limits development of standardized protocols.
-
d.
Cell are not correctly patterned and organized - with respect to lung epithelial directed differentiation, proximal and distal populations are interspersed amongst each other and do not mimic the patterning seen in vivo. In organoid cultures, while separate populations of both proximal and distal epithelia have been created, these organoids do not accurately reproduce the patterning of the in vivo lung.
-
e.
Contamination from other organ cell types - lung protocols suffer from contamination of thyroid, gastrointestinal, and neural tissues. Protocols that did not employ a lung progenitor sorting step were often contaminated with thyroid, pancreas and liver contaminants. While thyroid contamination was decreased via streamlining of signaling cues, specifically elimination of FGF2 [120], liver contaminants (12.1%) continue to remain despite NKX2.1+ cell sorting [37]. Beyond the usual suspects, Huang et al. found undefined contaminants that cannot be categorized as any specific tissue type [25]. This issue is partially due to the stochastic nature of PSC-based directed differentiation and a lack of early definitive lung markers for selection.
-
f.
Minimal recapitulation of physiological conditions - current alveolar organoids are beginning to represent relevant cell types, however, they are embedded in Matrigel and grown in submerged culture. They, therefore, fail to provide an ALI environment, which is critical to in vivo functionality. As in proximal protocols, cell products often need to be dissociated and regrown on transwells, which allow ALI culture by exposing cells to media basally and air apically, for further assessment.
-
g.
High cost - associated with the growth factor and small molecules required for chemically directed stem cell differentiation, and the expertise required to reliably create lung epithelia with these protocols. The use of commercially available 2D based endoderm differentiation kits have greatly decreased the level of expertise required to achieve this early stage of differentiation.
Based on current advances, we have made recommendations for directed differentiation protocols that generate proximal and distal epithelia in Fig. 2 .
Fig. 2.
Organization of airway and lung epithelium with recommended directed differentiation protocols to generate specific populations.
4. Opportunities to exploit mechanical cues for improving directed differentiation protocols in the future
The timing of chemical signals present during lung development has been well mimicked in current differentiation protocols. The lung, however, develops in response to chemical signals within a highly dynamic mechanical environment of cyclic strain, pressure and a complex branching tubular architecture [169]. Indeed, it is well established that mechanical cues can impact progenitor cell fate [102,[170], [171], [172], [173], [174], [175], [176], [177], [178], [179], [180]] and emerging evidence suggests that the mechanical environment can be manipulated to produce predictable fate choices in stem and progenitor cells [[181], [182], [183], [184]]. For example, the importance of biophysical manipulation associated with tissue structure was beautifully articulated by von Erlach et al., who described a mechanistic link between cell geometry and lipid rafts within plasma membranes that play a key role in cell signalling and therefore, cell fate. Mediated by the actin cytoskeleton, mesenchymal stem cells (MSCs) constrained by geometry acquired higher contractility, an abundance of lipid raft structures, and consequently favoured differentiation towards osteogenic versus adipogenic lineages [182]. The use of mechanical cues that mimic those experienced during development, has been exploited only to a limited extent to augment and guide directed differentiation protocols to address some of the above limitations [170,[185], [186], [187]]. Organoid cultures, for instance, allow self-assembly of tissue-like structures and enable further maturation of proximal and distal epithelia [12,26,28,119]. In this section of the review, we highlight tools from tissue engineering that have been used to manipulate tissue structure and the resulting biophysical signals experienced during stem cell differentiation in both 2D and 3D (Fig. 3 and Table 3 ). Furthermore, we explore the opportunity to utilize such tools to engineer mechanical signaling as a strategy to augment and refine existing chemical differentiation protocols. Note, we do not consider the use of simply culturing differentiating cells on substrates or in 3D hydrogels with variable mechanical stiffness but point the reader to excellent reviews on this subject [188,189].
Fig. 3.
Potential applications of mechanical cues for enhancing lung directed differentiation protocols.
Table 3.
Highlights of mechanical cues influencing cell fate. *High throughput not shown in the paper but could be easily developed.
| Physical Manipulation Technique | Cellularity Level | Dimension | Physical Cue Modulated | Effect of Physical Cue Modulation | Throughput | Reference |
|---|---|---|---|---|---|---|
| Micropatterning | Single Cells | 2D | Cell size | Endothelial cell apoptosis is inversely related to cell spreading area. | High* | Chen et al. 1997 |
| Cell shape | Constraining MSC culture area within pentagons with concave lines or in shapes with high aspect ratios promotes osteogenic differentiation due to increased actomyosin contractility. | High* | Kilian et al. 2010 | |||
| Cell Colonies | 2D | Perimeter topology | Melanoma cells occupying larger arc angles at the tumour periphery demonstrate greater tumorigenicity due to hypoxia-induced mechanotransduction. | High* | Lee et al. 2017 | |
| Colony size | Colonies of larger diameters allow maintenance of human PSC pluripotency and can further be used as a platform to study early PSC fate in response to specific chemical cues. | High | Nazareth et al. 2013 | |||
| Mediated by colony boundary, not size, human PSCs organize into radially segregated germ layer regions in response of BMP4. | High | Warmflash et al. 2014 Tewary et al. 2017 |
||||
| Organoids | 3D | Organoid shape (surface area: volume ratio) using microfibres | Increased patterning and organization of cerebral organoids grown around microfibres lead to mature neuronal features. | Low | Lancaster et al. 2017 | |
| Cell position via micromoulding | Geometry of hollow tubules reliably predict branching patterns of mammary epithelia through mechanical stress gradients, and further reveal mechanisms of cellular rearrangement. | Low | Nelson et al. 2006, 2008 Mori et al. 2009 Gjorevski and Nelson 2010 |
|||
| Cell curvature through micromoulding | Tubular diameters exert differential cellular tension and thereby dictate cell fate of bipotent lung progenitors. | Low | Soleas et al. 2020 | |||
| Substrate Topography | Multiple cells | 2D | Cell position in response to undulation | Epidermis-inspired topography induces human keratinocytes to pattern into distinct regions of progenitor, differentiating, and proliferating cells, as seen in vivo. | Low- Medium | Viswanathan et al. 2016 |
| Single Cells/ Cell Patches | 2D | Cell shape via “TopoChip” topographies | Smaller feature size is most essential for maintaining human PSC pluripotency. | High | Unadkat et al. 2011 Reimer et al. 2016 |
|
| Single Cells/ Cell Clusters/ Monolayer | 2D | Cell shape via grooves (nano to micro scale) | Due to actomyosin contractility, neuronal differentiation is promoted on anisotropic nanoscale grooves in a highly expedited manner compared to standard protocols. | High | Ankam et al. 2013, 2015 | |
| Single Cells/ Cell Patches | 2D | Cell shape via grooves (micro scale) | Associated with cell morphology and focal adhesion formation, wider groove ridges promote adipogenic differentiation, while thinner ridges promote osteogenic differentiation of MSCs. | High* | Abagnale et al. 2015 | |
| Single Cells/ Cell Patches/ Cell Colonies | 2D | Colony shape via grooves (nano scale) | Linked to differential YAP and TAZ activity, grooves elongate human PSC colonies which maintain pluripotency and are highly responsive to morphogenic differentiation cues. | High* | Abagnale et al. 2017 |
4.1. Micropatterning in 2D
Micropatterning of the culture surface is one strategy that has been used to manipulate the physical organization of 2D stem cell colonies and the resulting mechanical environment individual cells experience within the cell sheet. Micropatterning entails deposition of extracellular matrix (ECM) protein islands with highly specific shapes and sizes on non-adhesive surfaces, via micro-contact printing (μCP) [[190], [191], [192], [193], [194]] or soft lithography [[195], [196], [197]]. Individual cells or cell populations are thereby restricted to the area of the surface where the adhesive protein islands are present. The shape of the island, therefore, geometrically constrains the shape of single and groups of cells in 2D, which determines the pattern of adhesive attachments between the cells and the underlying surface, and hence the mechanical state of the cells [198].
Chen and colleagues presented early evidence that geometric constraint affects cell fate by demonstrating that cell growth and apoptosis are directly related to ECM pattern size through its control of cell spreading [194,199]. Not only is the size of the ECM pattern important, but also the shape it holds, specifically in relation to its aspect ratio and subcellular curvature. As shown by Kilian et al., despite the presence of equipotential differentiation signals, osteogenic differentiation of heterogeneous mesenchymal stem cells (MSCs) was promoted by increasing the ECM pattern aspect ratio at the single cell level [181]. Further, in a pentagon-shaped design, the curvature of the lines connecting vertices was varied from convex to concave and was shown to guide cell differentiation choice from adipogenic to osteogenic, respectively, by manipulating subcellular myosin II polarization, tension, and integrin localization. Evidently, such micropatterned islands not only exert control over the growth and survival of cells, but also enable manipulation of cell differentiation through changes in intracellular tension. For example, by probing tension at the cell-boundary interface through confined 2D geometries, Lee et al. found that patterned melanoma cancer cells occupying larger arc angles, or smaller magnitudes of curvature, expressed higher cancer stem cell markers [195]. Furthermore, these markers were preferentially found at the edge of the micropattern, a consequence of perimeter tension acting through the p38-MAP kinase pathway. Interestingly, once removed from the defined geometric environment, the cells lost their activated cancer phenotype. These tools could provide an excellent platform for subtly manipulating self-organization of PSC populations to understand and influence their differentiation.
While not in the context of augmenting directed differentiation specifically, the use of micropatterning has been applied to explore pluripotency and fate choice during early development. Based on a previous finding that human PSC differentiation is dependent on colony size [200], Nazareth et al. developed a high throughput μCP platform, with optimized colony size, and probed early cell fates (pluripotent, neuroectoderm, primitive streak, and extraembryonic) in response to different media conditions and developmental factors [190]. Such micropatterned surfaces were also used by Warmflash and colleagues to investigate embryonic germ layer patterning of human PSCs [201]. They found that BMP4 treatment results in spatially segregated regions that delineate ectoderm, primitive streak and trophoblast-like tissues within the patterned colonies. This pattern was shown to be mediated by the colony edge, as opposed to colony size, with BMP4 signalling progressively being restricted to the edge of the colony. These findings were further confirmed by Tewary et al., who explained this effect to be caused by the emergence of a phosphorylated SMAD1 gradient. The establishment of this gradient is initially controlled via inhibition by Noggin, followed by a restriction of BMP4 sensitivity to the edge of the colony due to re-localization of BMP receptors throughout the rest of the colony [202]. Such micropatterned platforms have also been used to map fate choices made during mouse [203] and human [204] gastrulation events, and therefore are a powerful tool for elucidating fate choice during lung development. It is not clear however, how the mechanical state of the cells within these micropatterned islands impacts chemical cue secretion, and hence the local gradients of chemical signals that result in patterning of cell fate in these studies.
4.2. Substrate texture
Another method to manipulate the physical organization and biophysical cues in a differentiating cell sheet is through substrate texture [205]. Cellular behaviours including proliferation, adhesion, and differentiation have been linked to underlying substrate topographical cues [[205], [206], [207], [208], [209], [210]]. These cues are recognized by cellular protrusions called filopodia and lamellipodia, through integrin receptors and focal adhesions [[211], [212], [213], [214]], which in turn dynamically modify their shape and exert protrusive forces [[215], [216], [217], [218], [219]]. In this section, we will provide key examples of substrate topographies, as well as grooves, based on their relevance for lung epithelial organization. These topographies can be microfabricated using various techniques, such as etching, photolithography, soft lithography, and stereolithography, that are scalable, precise, and provide high fidelity [220].
4.2.1. Stem cell behaviour on substrate topographies
Stem cell fate choices have been shown to respond to topographical features [221,222]. For example, Viswanathan et al. assessed the ability of different topographies to mimic sinusoidal epidermal undulation to induce in vivo-like biophysical cues. Their screened undulating topography created β1 integrin patterning that is reminiscent of the human dermis and more differentiated cells were found localized to the troughs of their pattern in a highly repeatable fashion. These findings suggest that replicating the physical organization of the dermal microenvironment promotes tissue-level organization and alters the positioning of the epidermal stem cells towards the in vivo state [223]. This group further applied a screening platform called TopoChip [224], that incorporated features of varying sizes, roundedness, and distribution density, to assess human PSC proliferation and pluripotency in the absence of ECM coatings [225]. Topographies that ranked high in the screen not only supported PSC proliferation, but also allowed maintenance of OCT4 and SOX2-expressing pluripotent colonies. In conjunction with computational modeling, this platform was able to predict topographical features conducive to maintaining PSC pluripotency, thus demonstrating great promise for exploring how to use tissue organization patterning to control cell fate. Application of this platform for probing keratinocyte differentiation revealed that differentiation is linked to changes in cell morphology, which is influenced by substrate topography, and mediated by Rho kinase activity [226].
Based on these studies, it is evident that application of biophysical cues alone can impact differentiation. High-throughput technologies like TopoChip could be used in the future to understand and mimic cell fates of proximal or distal lung epithelia. Towards this goal of recapitulating the physiological morphology of distal lung alveoli, we recently developed larger-sized topographical features, specifically hemispherical cavities, that enabled seeding of multiple cells and further allowed maintenance of primary AEC I and AEC II cells [227]. The ability of this platform to promote PSC differentiation towards these cell types has yet to be explored, however.
4.2.1.1. Stem cell behaviour on grooves
Grooved topographical cues specifically, through their ability to modulate cytoskeletal alignment and cellular shape, have also demonstrated great promise in guiding cell fate [228,229]. In the context of neural differentiation, Ankam and colleagues generated a multi-architectural chip (MARC), that incorporated a range of isotropic and anisotropic topographies at both micro and nano scales, to differentiate PSCs towards neural progeny without the use of embryoid bodies. Anisotropic nanoscale grooves (250 nm) promoted neuronal differentiation with cell alignment and elongation, and isotropic pillars enhanced astrocyte differentiation with cellular branching within 7 days of culture. Meanwhile, conventional culture protocols were unable to induce these populations without additional culture steps and/or prolonged culture up to 30 days [230]. Neural differentiation on nanogrooves was attributed to actomyosin contractility via vinculin-associated focal adhesions [231]. MARC further enabled investigation of nuclear morphology and histone methylation [232], thereby exemplifying that such platforms can allow exploration of the mechanism of biophysical cues translating to DNA modulation during differentiation.
The influence of groove topography on differentiation has been highlighted in other contexts as well. Abagnale et al. developed a micro-grooved chip, incorporating systematic variation of groove widths and ridges, to study MSC differentiation towards adipogenic and osteogenic progeny. While wider ridges led to higher adipogenic differentiation with formation of fat droplets, thinner ridges enhanced osteogenic differentiation with calcium phosphate precipitation [233]. Interestingly, groove width had minimal impact on favouring differentiation towards either lineage. The ridge-mediated differentiation effect was associated with cell morphology and focal adhesion formation wherein wider ridges resulted in rounder cell morphology with many large focal adhesions, as compared to thinner ridges leading to cellular elongation with fewer and smaller sized focal adhesions. Nano-scale groove topography was also shown to alter the spatial conformation of PSC colonies by elongating them, consequently affecting cell fate [234]. This effect was particularly potent at the colony edges and controlled by separate and differential localization of YAP and TAZ during PSC maintenance and differentiation.
In general, grooved topography can provide great insights into PSC fate choice and potentially expedite differentiation protocols. The MARC platform illustrates the importance of combining biophysical and biochemical cues, especially as exposure to topography induced a higher yield of functional and mature progeny within a short time frame as compared to flat substrates or standard directed differentiation protocols [230,235]. This is of extreme importance as current directed differentiation protocols for airway and lung epithelia require longer than 60 days of culture to achieve functional cell types [12,27,28,30,36,119]. Although the inclusion of topography has not been investigated for promoting lung differentiation, we have explored grooved substrates for aligning airway epithelia during differentiation to achieve coordinated unidirectional ciliary beating [236]. Our unpublished data demonstrates that while primary human basal-derived epithelia lose their alignment on grooved topography over time, epithelia generated from human PSC-derived airway progenitors maintain their alignment throughout differentiation in ALI culture.
4.3. Micropatterning in 3D: organoid systems
While 2D cues have enabled the community to clearly demonstrate the capability of biophysical cues to manipulate cell fate, 2D approaches are limited in their biological applicability as most physiologic physical and mechanical cues occur in 3D environments [237]. This necessitates development of 3D platforms that can better recapitulate the developmental structures and biophysical environments observed during tissue development and specification. Some efforts have emerged to control the 3D structure of differentiating cells and assess the resulting fate patterning. For example, to dissect fate choice events during embryonic patterning and subsequent cardiac development, Ma et al. created highly reproducible, confined microchambers of varying diameters (200, 400 and 600 μm) that stimulated PSCs towards a mesodermal fate via WNT agonism. This platform enabled mimicry of mesenchymal condensation during epithelial-mesenchymal transition as cells at the periphery of larger geometrically constrained patterns (400 and 600 μm) expressed OCT4 and E-cadherin. As such, cells in the middle of the patterns differentiated into beating cardiomyocytes with structures mimicking the linear heart tube, while cells at the periphery expressed myofibroblast markers [186]. Further, these microchambers were validated for use in developmental drug toxicity screening, through which they were able to represent Thalidomide embryopathy by exhibiting diminished contractility, beating frequency, and size of cardiac chambers.
Geometric constraint was also applied to cerebral organoids by Lancaster and colleagues to minimize variability associated with neural induction efficiency. This entailed addition of a physical cue in the form of polymer microfibres, around which the organoid self-organized [170]. In conjunction with an established chemical protocol, these microfibres served to pattern developing organoids leading to recognizable neuronal features including a cortical plate, radial units, along with organized radial neuronal migration in a reproducible manner. This addition of a simple physical cue substantially increased the patterning and organization of brain organoids compared to those derived from protocols only relying on biochemical cues.
Another strategy to control cell and tissue geometry in 3D is the use of micromoulding to create defined mechanical microenvironments which in turn alter cell and tissue level organization and differentiation. This approach was first developed by Nelson et al. using 3D collagen moulds to study branching morphogenesis of mammary epithelial cells [238,239]. Seeded mammary epithelia conformed to the 3D architectures, forming hollow tubules, and demonstrated predictable branching patterns according to mould geometry and presence of inhibitory morphogens. This technique was further used to understand the mechanism of cellular rearrangement in mammary ducts [240] and exhibit that mechanical stress gradients control the pattern of branching morphogenesis [241]. Inspired by this method and its applicability for studying lung branching morphogenesis, we developed tubular constructs of physiologically relevant diameters to guide self-assembly of lung progenitors [242]. Using this approach, we demonstrated that specification of these bipotent SOX2+SOX9+ lung progenitors was dependent on geometry, wherein tubes of 100 μm diameter led to a distal SOX9+ fate, while 400 μm diameter tubes remained in a SOX2+SOX9+ lung progenitor state. The mechanism of this effect was dependent on canonical WNT signalling, and due to differences in cellular tension induced by patterning the progenitor cells into a 3D tube structure.
While the addition of mechanical cues influences fate choice, its role in inducing cell functionality, especially at the organoid level, needs to be elucidated. Currently, there is scant evidence of lung directed differentiation being manipulated in a 3D context [183,242,243]. Beyond our exploration of patterning early lung fates through micromoulding, Dye et al. have recently applied tissue engineering techniques during lung organoid formation. In their case, foregut endoderm spheroids cultured on highly degradable synthetic polymers demonstrated enhanced ability to differentiate into proximal airway epithelia after in vivo implantation [183]. Evidently, the field of lung directed differentiation is in its nascent stages for using biophysical manipulation to enhance differentiation during in vitro culture. The addition of simple mechanical cues such as polymer microfibres to already established lung directed differentiation protocols, as well as delving deeper into understanding cell fate in tubular constructs are excellent starting points for potentially enhancing 3D lung models.
5. Future outlook
In the future, human PSC-derived lung tissue models have the potential to enable exploration of infection, disease and regeneration mechanisms of action to impact drug discovery and drug development, and further inform patient-specific drug selection. While lung models remain in their infancy, the investment necessary to translate such models into practical use is worthwhile given that they offer a number of key advantages over primary cells or mouse models. Firstly, PSC-derived platforms enable modeling of human disease. Another major advantage of PSC-derived cells, specifically in the context of lung, is the potential to directly associate specific patient genetics and cell phenotypes with clinical conditions, as is underway in the field of lung cancer [244,245]. This will avoid complications associated with prior exposure of primary cell donors to a plethora of environmental (such as smoking) and pharmaceutical stimuli. Furthermore, establishing models specifically from PSC sources potentially enables generation of the large number of cells and cell types necessary for personalized disease modeling.
A number of challenges exist however, to translate these models into widespread use for drug discovery and development. One major challenge in the field, highlighted in this review, is the standardization of robust cell manufacturing protocols. Lung epithelial models require not only large cell numbers, but also the correct proportion of cell types. Additionally, for a variety of functional read-outs, these cell types must be appropriately spatially organized. Therefore, standardized protocols are needed to both manufacture lung cells and assemble these cells into reproducible and clinically representative “lung tissues” at the scales required for screening. This will be essential to enable the generation of in vitro lung test tissues with sufficiently low batch-to-batch and within-batch variation for screening with high reproducibility.
Towards this challenge, methods will continue to emerge to control PSC differentiation into the different proximal airway and distal lung cell types. For example, in this review, we have highlighted the emerging evidence that an opportunity exists to further improve differentiation control and disease modeling by mimicking mechanical cues experienced during development. Beyond the strategies described in previous sections, this concept could be expanded further in the future to mimic additional aspects of lung development. For example, the developing lung is exposed to variations in oxygenation [246,247], which is known to impact cell fate choices [[248], [249], [250]], therefore the use of optimized oxygenation levels could be an attractive and easily scalable strategy to further refine directed differentiation culture protocols. The developing lung is also subject to various other physical cues at the organ-scale including 1) pressure from amniotic fluid, which serves to expand the nascent alveolar compartment; 2) fetal breathing movements that provide a stretch-based physical cue which serves as a maturation signal; and 3) pulsatile flow from the extensive vascular network present throughout the organ. Techniques that mimic these forces to control lung cell fate are emerging. These include the use of shear to produce relatively homogenous populations of AEC I and II [251], the use of cyclic mechanical stretch [252], and the use of patterned hydrogels to enable perfusion of lung-type structures [253]. Scaling some of these complex mechanical setups to enable large scale efficient manufacturing, however, could be a challenge.
The challenge of assembling lung cells into reproducible arrays of “lung tissues” is starting to be addressed by emerging high-throughput techniques that seek to purify specific cell populations [[254], [255], [256]] that could be later mixed in controlled ratios to generate precise tissue compositions. Bioprinting [[257], [258], [259]] and cellular assembly efforts, including organoid fabrication through DNA programming [260], could also potentially prove useful to enabling complex tissue assembly in a manner that is adequately scalable and reproducible for screening. Many of these techniques exploit biomaterials as carriers to pattern cells into 3D structures. An opportunity, therefore, exists to design novel materials that both pattern cells and augment directed differentiation culture protocols. Such materials should consider both approaches to enable timed and patterned delivery of chemical cues and the design of appropriate mechanical properties and structures (such as topography) to mimic any biophysical cues that can augment chemical signaling in the system.
Beyond the manufacturing challenge, another significant challenge limiting translation of tissue models for drug discovery and development is the inadequate availability of human validation data sets to demonstrate the accuracy of the models in predicting drug response in corresponding clinical populations. Based on single-cell RNA sequencing, data sets pertaining to airway [139,261] and lung [35] cellular compositions have emerged, that can serve as references for identifying and validating [262] differentiation products. Similarly, some evidence profiling the inflammatory response associated with CF [263] and epithelial dysfunction during pulmonary fibrosis [264,265] exists, however, establishment of a comprehensive repository informing the identity of diseased cells and their response to specific drugs is important. Data sets correlating genetic profiles with drug responses are severely limited in the lung field. Establishing such “ground-truth” benchmarking data sets to validate the ability of in vitro models to distinguish both positive and negative hits will be absolutely critical to establish confidence in lung culture models and to ensure wider spread community adoption and impact.
Acknowledgements
This work was funded by a Canada First Research Excellence Fund (CFREF) Medicine by Design, New Ideas Grant to TKW and APM (NIS-2016-02), a McLaughlin Centre Award, CIHR Training Program in Regenerative Medicine Award, and a Henry White Kinnear Foundation Award to JS and a Henry White Kinnear Foundation Award to RV. All figures were created using BioRender (Toronto ON, Canada). The authors have no conflicts of interest to declare.
References
- 1.Murphy S.L., Xu J., Kochanek K.D., Arias E. Mortality in the United States. 2017, NCHS Data Brief. 2018:1–8. [PubMed] [Google Scholar]
- 2.Khaltaev N., Axelrod S. Chronic respiratory diseases global mortality trends, treatment guidelines, life style modifications, and air pollution: preliminary analysis. J Thorac Dis. 2019;11:2643–2655. doi: 10.21037/jtd.2019.06.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burney P.G., Patel J., Newson R., Minelli C., Naghavi M. Global and regional trends in COPD mortality, 1990-2010. Eur. Respir. J. 2015;45:1239–1247. doi: 10.1183/09031936.00142414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.G.B.D.C.O.D. Collaborators Global, regional, and national age-sex specific mortality for 264 causes of death, 1980-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2017;390:1151–1210. doi: 10.1016/S0140-6736(17)32152-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thabut G., Mal H. Outcomes after lung transplantation. J Thorac Dis. 2017;9:2684–2691. doi: 10.21037/jtd.2017.07.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freishtat R.J., Nino G., Tsegaye Y., Alcala S.E., Benton A.S., Watson A.M., Reeves E.K., Haider S.K., Damsker J.M. Pharmacologically-induced mitotic synchrony in airway epithelial cells as a mechanism of action of anti-inflammatory drugs. Respir. Res. 2015;16 doi: 10.1186/s12931-015-0293-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Czekala L., Simms L., Stevenson M., Tschierske N., Maione A.G., Walele T. Toxicological comparison of cigarette smoke and e-cigarette aerosol using a 3D in vitro human respiratory model. Regul. Toxicol. Pharmacol. 2019;103:314–324. doi: 10.1016/j.yrtph.2019.01.036. [DOI] [PubMed] [Google Scholar]
- 8.Kilic A., Ameli A., Park J.A., Kho A.T., Tantisira K., Santolini M., Cheng F., Mitchel J.A., McGill M., O'Sullivan M.J., De Marzio M., Sharma A., Randell S.H., Drazen J.M., Fredberg J.J., Weiss S.T. Mechanical forces induce an asthma gene signature in healthy airway epithelial cells. Sci. Rep. 2020;10:966. doi: 10.1038/s41598-020-57755-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benam K.H., Villenave R., Lucchesi C., Varone A., Hubeau C., Lee H.H., Alves S.E., Salmon M., Ferrante T.C., Weaver J.C., Bahinski A., Hamilton G.A., Ingber D.E. Small airway-on-a-chip enables analysis of human lung inflammation and drug responses in vitro. Nat. Methods. 2016;13:151–157. doi: 10.1038/nmeth.3697. [DOI] [PubMed] [Google Scholar]
- 10.Huh D., Leslie D.C., Matthews B.D., Fraser J.P., Jurek S., Hamilton G.A., Thorneloe K.S., McAlexander M.A., Ingber D.E. A human disease model of drug toxicity-induced pulmonary edema in a lung-on-a-chip microdevice. Sci. Transl. Med. 2012;4:159ra147. doi: 10.1126/scitranslmed.3004249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong A.P., Bear C.E., Chin S., Pasceri P., Thompson T.O., Huan L.J., Ratjen F., Ellis J., Rossant J. Directed differentiation of human pluripotent stem cells into mature airway epithelia expressing functional CFTR protein. Nat. Biotechnol. 2012;30:876–882. doi: 10.1038/nbt.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamamoto Y., Gotoh S., Korogi Y., Seki M., Konishi S., Ikeo S., Sone N., Nagasaki T., Matsumoto H., Muro S., Ito I., Hirai T., Kohno T., Suzuki Y., Mishima M. Long-term expansion of alveolar stem cells derived from human iPS cells in organoids. Nat. Methods. 2017;14:1097–1106. doi: 10.1038/nmeth.4448. [DOI] [PubMed] [Google Scholar]
- 13.Van Norman G.A. Limitations of Animal Studies for Predicting Toxicity in Clinical Trials: Part 2: Potential Alternatives to the Use of Animals in Preclinical Trials. JACC Basic Transl Sci. 2020;5:387–397. doi: 10.1016/j.jacbts.2020.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rayner R.E., Makena P., Prasad G.L., Cormet-Boyaka E. Optimization of Normal Human Bronchial Epithelial (NHBE) Cell 3D Cultures for in vitro Lung Model Studies. Sci. Rep. 2019;9 doi: 10.1038/s41598-018-36735-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van de Laar E., Clifford M., Hasenoeder S., Kim B.R., Wang D., Lee S., Paterson J., Vu N.M., Waddell T.K., Keshavjee S., Tsao M.S., Ailles L., Moghal N. Cell surface marker profiling of human tracheal basal cells reveals distinct subpopulations, identifies MST1/MSP as a mitogenic signal, and identifies new biomarkers for lung squamous cell carcinomas. Respir. Res. 2014;15 doi: 10.1186/s12931-014-0160-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirsch T., Rothoeft T., Teig N., Bauer J.W., Pellegrini G., De Rosa L., Scaglione D., Reichelt J., Klausegger A., Kneisz D., Romano O., Secone Seconetti A., Contin R., Enzo E., Jurman I., Carulli S., Jacobsen F., Luecke T., Lehnhardt M., Fischer M., Kueckelhaus M., Quaglino D., Morgante M., Bicciato S., Bondanza S., De Luca M. Regeneration of the entire human epidermis using transgenic stem cells. Nature. 2017;551:327–332. doi: 10.1038/nature24487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Danopoulos S., Alonso I., Thornton M.E., Grubbs B.H., Bellusci S., Warburton D., Al Alam D. Human lung branching morphogenesis is orchestrated by the spatiotemporal distribution of ACTA2, SOX2, and SOX9. Am. J. Phys. Lung Cell. Mol. Phys. 2018;314:L144–L149. doi: 10.1152/ajplung.00379.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Biehl J.K., Russell B. Introduction to stem cell therapy. J Cardiovasc Nurs. 2009;24:98–103. doi: 10.1097/JCN.0b013e318197a6a5. quiz 104-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang D., Haviland D.L., Burns A.R., Zsigmond E., Wetsel R.A. A pure population of lung alveolar epithelial type II cells derived from human embryonic stem cells. Proc. Natl. Acad. Sci. U. S. A. 2007;104:4449–4454. doi: 10.1073/pnas.0700052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Samadikuchaksaraei A., Cohen S., Isaac K., Rippon H.J., Polak J.M., Bielby R.C., Bishop A.E. Derivation of distal airway epithelium from human embryonic stem cells. Tissue Eng. 2006;12:867–875. doi: 10.1089/ten.2006.12.867. [DOI] [PubMed] [Google Scholar]
- 21.Van Haute L., De Block G., Liebaers I., Sermon K., De Rycke M. Generation of lung epithelial-like tissue from human embryonic stem cells. Respir. Res. 2009;10 doi: 10.1186/1465-9921-10-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Longmire T.A., Ikonomou L., Hawkins F., Christodoulou C., Cao Y., Jean J.C., Kwok L.W., Mou H., Rajagopal J., Shen S.S., Dowton A.A., Serra M., Weiss D.J., Green M.D., Snoeck H.W., Ramirez M.I., Kotton D.N. Efficient derivation of purified lung and thyroid progenitors from embryonic stem cells. Cell Stem Cell. 2012;10:398–411. doi: 10.1016/j.stem.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Green M.D., Chen A., Nostro M.C., d'Souza S.L., Schaniel C., Lemischka I.R., Gouon-Evans V., Keller G., Snoeck H.W. Generation of anterior foregut endoderm from human embryonic and induced pluripotent stem cells. Nat. Biotechnol. 2011;29:267–272. doi: 10.1038/nbt.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mou H., Zhao R., Sherwood R., Ahfeldt T., Lapey A., Wain J., Sicilian L., Izvolsky K., Musunuru K., Cowan C., Rajagopal J. Generation of multipotent lung and airway progenitors from mouse ESCs and patient-specific cystic fibrosis iPSCs. Cell Stem Cell. 2012;10:385–397. doi: 10.1016/j.stem.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang S.X., Islam M.N., O'Neill J., Hu Z., Yang Y.G., Chen Y.W., Mumau M., Green M.D., Vunjak-Novakovic G., Bhattacharya J., Snoeck H.W. Efficient generation of lung and airway epithelial cells from human pluripotent stem cells. Nat. Biotechnol. 2014;32:84–91. doi: 10.1038/nbt.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y.W., Huang S.X., de Carvalho A., Ho S.H., Islam M.N., Volpi S., Notarangelo L.D., Ciancanelli M., Casanova J.L., Bhattacharya J., Liang A.F., Palermo L.M., Porotto M., Moscona A., Snoeck H.W. A three-dimensional model of human lung development and disease from pluripotent stem cells. Nat. Cell Biol. 2017;19:542–549. doi: 10.1038/ncb3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCauley K.B., Hawkins F., Serra M., Thomas D.C., Jacob A., Kotton D.N. Efficient Derivation of Functional Human Airway Epithelium from Pluripotent Stem Cells via Temporal Regulation of Wnt Signaling. Cell Stem Cell. 2017;20:844–857. doi: 10.1016/j.stem.2017.03.001. e846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacob A., Morley M., Hawkins F., McCauley K.B., Jean J.C., Heins H., Na C.L., Weaver T.E., Vedaie M., Hurley K., Hinds A., Russo S.J., Kook S., Zacharias W., Ochs M., Traber K., Quinton L.J., Crane A., Davis B.R., White F.V., Wambach J., Whitsett J.A., Cole F.S., Morrisey E.E., Guttentag S.H., Beers M.F., Kotton D.N. Differentiation of Human Pluripotent Stem Cells into Functional Lung Alveolar Epithelial Cells. Cell Stem Cell. 2017;21:472–488. doi: 10.1016/j.stem.2017.08.014. e410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Carvalho A., Strikoudis A., Liu H.Y., Chen Y.W., Dantas T.J., Vallee R.B., Correia-Pinto J., Snoeck H.W. Glycogen synthase kinase 3 induces multilineage maturation of human pluripotent stem cell-derived lung progenitors in 3D culture. Development. 2019;146 doi: 10.1242/dev.171652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Konishi S., Gotoh S., Tateishi K., Yamamoto Y., Korogi Y., Nagasaki T., Matsumoto H., Muro S., Hirai T., Ito I., Tsukita S., Mishima M. Directed Induction of Functional Multi-ciliated Cells in Proximal Airway Epithelial Spheroids from Human Pluripotent Stem Cells. Stem Cell Reports. 2016;6:18–25. doi: 10.1016/j.stemcr.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dye B.R., Hill D.R., Ferguson M.A., Tsai Y.H., Nagy M.S., Dyal R., Wells J.M., Mayhew C.N., Nattiv R., Klein O.D., White E.S., Deutsch G.H., Spence J.R. In vitro generation of human pluripotent stem cell derived lung organoids. Elife. 2015;4 doi: 10.7554/eLife.05098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gotoh S., Ito I., Nagasaki T., Yamamoto Y., Konishi S., Korogi Y., Matsumoto H., Muro S., Hirai T., Funato M., Mae S., Toyoda T., Sato-Otsubo A., Ogawa S., Osafune K., Mishima M. Generation of alveolar epithelial spheroids via isolated progenitor cells from human pluripotent stem cells. Stem Cell Reports. 2014;3:394–403. doi: 10.1016/j.stemcr.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Firth A.L., Dargitz C.T., Qualls S.J., Menon T., Wright R., Singer O., Gage F.H., Khanna A., Verma I.M. Generation of multiciliated cells in functional airway epithelia from human induced pluripotent stem cells. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E1723–E1730. doi: 10.1073/pnas.1403470111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hawkins F., Kramer P., Jacob A., Driver I., Thomas D.C., McCauley K.B., Skvir N., Crane A.M., Kurmann A.A., Hollenberg A.N., Nguyen S., Wong B.G., Khalil A.S., Huang S.X., Guttentag S., Rock J.R., Shannon J.M., Davis B.R., Kotton D.N. Prospective isolation of NKX2-1-expressing human lung progenitors derived from pluripotent stem cells. J. Clin. Invest. 2017;127:2277–2294. doi: 10.1172/JCI89950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hurley K., Ding J., Villacorta-Martin C., Herriges M.J., Jacob A., Vedaie M., Alysandratos K.D., Sun Y.L., Lin C., Werder R.B., Huang J., Wilson A.A., Mithal A., Mostoslavsky G., Oglesby I., Caballero I.S., Guttentag S.H., Ahangari F., Kaminski N., Rodriguez-Fraticelli A., Camargo F., Bar-Joseph Z., Kotton D.N. Reconstructed Single-Cell Fate Trajectories Define Lineage Plasticity Windows during Differentiation of Human PSC-Derived Distal Lung Progenitors. Cell Stem Cell. 2020;26:593–608. doi: 10.1016/j.stem.2019.12.009. e598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jacob A., Vedaie M., Roberts D.A., Thomas D.C., Villacorta-Martin C., Alysandratos K.D., Hawkins F., Kotton D.N. Derivation of self-renewing lung alveolar epithelial type II cells from human pluripotent stem cells. Nat. Protoc. 2019;14:3303–3332. doi: 10.1038/s41596-019-0220-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCauley K.B., Alysandratos K.D., Jacob A., Hawkins F., Caballero I.S., Vedaie M., Yang W., Slovik K.J., Morley M., Carraro G., Kook S., Guttentag S.H., Stripp B.R., Morrisey E.E., Kotton D.N. Single-Cell Transcriptomic Profiling of Pluripotent Stem Cell-Derived SCGB3A2+ Airway Epithelium. Stem Cell Reports. 2018;10:1579–1595. doi: 10.1016/j.stemcr.2018.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Basil M.C., Katzen J., Engler A.E., Guo M., Herriges M.J., Kathiriya J.J., Windmueller R., Ysasi A.B., Zacharias W.J., Chapman H.A., Kotton D.N., Rock J.R., Snoeck H.W., Vunjak-Novakovic G., Whitsett J.A., Morrisey E.E. The Cellular and Physiological Basis for Lung Repair and Regeneration: Past, Present, and Future. Cell Stem Cell. 2020;26:482–502. doi: 10.1016/j.stem.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moodley Y., Thompson P., Warburton D. Stem cells: a recapitulation of development. Respirology. 2013;18:1167–1176. doi: 10.1111/resp.12186. [DOI] [PubMed] [Google Scholar]
- 40.Conway R.F., Frum T., Conchola A.S., Spence J.R. Understanding human lung development through in vitro model systems. Bioessays. 2020;42 doi: 10.1002/bies.202000006. e2000006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Warburton D., El-Hashash A., Carraro G., Tiozzo C., Sala F., Rogers O., De Langhe S., Kemp P.J., Riccardi D., Torday J., Bellusci S., Shi W., Lubkin S.R., Jesudason E. Lung organogenesis. Curr. Top. Dev. Biol. 2010;90:73–158. doi: 10.1016/S0070-2153(10)90003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herriges M., Morrisey E.E. Lung development: orchestrating the generation and regeneration of a complex organ. Development. 2014;141:502–513. doi: 10.1242/dev.098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williams K., Johnson M.H. Adapting the 14-day rule for embryo research to encompass evolving technologies. Reprod Biomed Soc Online. 2020;10:1–9. doi: 10.1016/j.rbms.2019.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pera M.F. Human embryo research and the 14-day rule. Development. 2017;144:1923–1925. doi: 10.1242/dev.151191. [DOI] [PubMed] [Google Scholar]
- 45.Tam P.P., Beddington R.S. Establishment and organization of germ layers in the gastrulating mouse embryo. CIBA Found. Symp. 1992;165:27–41. doi: 10.1002/9780470514221.ch3. discussion 42-29. [DOI] [PubMed] [Google Scholar]
- 46.Scheibner K., Bakhti M., Bastidas-Ponce A., Lickert H. Wnt signaling: implications in endoderm development and pancreas organogenesis. Curr. Opin. Cell Biol. 2019;61:48–55. doi: 10.1016/j.ceb.2019.07.002. [DOI] [PubMed] [Google Scholar]
- 47.Chin A.M., Hill D.R., Aurora M., Spence J.R. Morphogenesis and maturation of the embryonic and postnatal intestine. Semin. Cell Dev. Biol. 2017;66:81–93. doi: 10.1016/j.semcdb.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Domyan E.T., Sun X. Patterning and plasticity in development of the respiratory lineage. Dev. Dyn. 2011;240:477–485. doi: 10.1002/dvdy.22504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cardoso W.V., Lu J. Regulation of early lung morphogenesis: questions, facts and controversies. Development. 2006;133:1611–1624. doi: 10.1242/dev.02310. [DOI] [PubMed] [Google Scholar]
- 50.Sadler T.W., Langman J. 12th ed. Wolters Kluwer Health/Lippincott Williams & Wilkins; Philadelphia: 2012. Langman's medical embryology. [Google Scholar]
- 51.Hislop A. Developmental biology of the pulmonary circulation. Paediatr. Respir. Rev. 2005;6:35–43. doi: 10.1016/j.prrv.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 52.Tam P.P., Kanai-Azuma M., Kanai Y. Early endoderm development in vertebrates: lineage differentiation and morphogenetic function. Curr. Opin. Genet. Dev. 2003;13:393–400. doi: 10.1016/s0959-437x(03)00085-6. [DOI] [PubMed] [Google Scholar]
- 53.Ioannides A.S., Massa V., Ferraro E., Cecconi F., Spitz L., Henderson D.J., Copp A.J. Foregut separation and tracheo-oesophageal malformations: the role of tracheal outgrowth, dorso-ventral patterning and programmed cell death. Dev. Biol. 2010;337:351–362. doi: 10.1016/j.ydbio.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hall S.M., Hislop A.A., Haworth S.G. Origin, differentiation, and maturation of human pulmonary veins. Am. J. Respir. Cell Mol. Biol. 2002;26:333–340. doi: 10.1165/ajrcmb.26.3.4698. [DOI] [PubMed] [Google Scholar]
- 55.E.D. Congdon, Transformation of the aortic-arch system during the development of the human embryo.
- 56.Bucher U., Reid L. Development of the intrasegmental bronchial tree: the pattern of branching and development of cartilage at various stages of intra-uterine life. Thorax. 1961;16:207–218. doi: 10.1136/thx.16.3.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gleason C.A., Devaskar S.U., Avery M.E. 9th ed. Elsevier/Saunders; Philadelphia, PA: 2012. Avery's diseases of the newborn / [edited by] Christine A. Gleason, Sherin U. Devaskar. [Google Scholar]
- 58.Hislop A.A. Airway and blood vessel interaction during lung development. J. Anat. 2002;201:325–334. doi: 10.1046/j.1469-7580.2002.00097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maeda Y., Dave V., Whitsett J.A. Transcriptional control of lung morphogenesis. Physiol. Rev. 2007;87:219–244. doi: 10.1152/physrev.00028.2006. [DOI] [PubMed] [Google Scholar]
- 60.Que J., Luo X., Schwartz R.J., Hogan B.L. Multiple roles for Sox2 in the developing and adult mouse trachea. Development. 2009;136:1899–1907. doi: 10.1242/dev.034629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gontan C., de Munck A., Vermeij M., Grosveld F., Tibboel D., Rottier R. Sox2 is important for two crucial processes in lung development: branching morphogenesis and epithelial cell differentiation. Dev. Biol. 2008;317:296–309. doi: 10.1016/j.ydbio.2008.02.035. [DOI] [PubMed] [Google Scholar]
- 62.Miller A.J., Hill D.R., Nagy M.S., Aoki Y., Dye B.R., Chin A.M., Huang S., Zhu F., White E.S., Lama V., Spence J.R. In Vitro Induction and In Vivo Engraftment of Lung Bud Tip Progenitor Cells Derived from Human Pluripotent Stem Cells. Stem Cell Reports. 2018;10:101–119. doi: 10.1016/j.stemcr.2017.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nikolic M.Z., Caritg O., Jeng Q., Johnson J.A., Sun D., Howell K.J., Brady J.L., Laresgoiti U., Allen G., Butler R., Zilbauer M., Giangreco A., Rawlins E.L. Human embryonic lung epithelial tips are multipotent progenitors that can be expanded in vitro as long-term self-renewing organoids. Elife. 2017;6 doi: 10.7554/eLife.26575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Volckaert T., Campbell A., Dill E., Li C., Minoo P., De Langhe S. Localized Fgf10 expression is not required for lung branching morphogenesis but prevents differentiation of epithelial progenitors. Development. 2013;140:3731–3742. doi: 10.1242/dev.096560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rawlins E.L., Clark C.P., Xue Y., Hogan B.L. The Id2+ distal tip lung epithelium contains individual multipotent embryonic progenitor cells. Development. 2009;136:3741–3745. doi: 10.1242/dev.037317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rockich B.E., Hrycaj S.M., Shih H.P., Nagy M.S., Ferguson M.A., Kopp J.L., Sander M., Wellik D.M., Spence J.R. Sox9 plays multiple roles in the lung epithelium during branching morphogenesis. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E4456–E4464. doi: 10.1073/pnas.1311847110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heijink I.H., Nawijn M.C., Hackett T.L. Airway epithelial barrier function regulates the pathogenesis of allergic asthma. Clin. Exp. Allergy. 2014;44:620–630. doi: 10.1111/cea.12296. [DOI] [PubMed] [Google Scholar]
- 68.Bermbach S., Weinhold K., Roeder T., Petersen F., Kugler C., Goldmann T., Rupp J., Konig P. Mechanisms of cilia-driven transport in the airways in the absence of mucus. Am. J. Respir. Cell Mol. Biol. 2014;51:56–67. doi: 10.1165/rcmb.2012-0530OC. [DOI] [PubMed] [Google Scholar]
- 69.Hamilton N., Bullock A.J., Macneil S., Janes S.M., Birchall M. Tissue engineering airway mucosa: a systematic review. Laryngoscope. 2014;124:961–968. doi: 10.1002/lary.24469. [DOI] [PubMed] [Google Scholar]
- 70.Guillot L., Nathan N., Tabary O., Thouvenin G., Le Rouzic P., Corvol H., Amselem S., Clement A. Alveolar epithelial cells: master regulators of lung homeostasis. Int. J. Biochem. Cell Biol. 2013;45:2568–2573. doi: 10.1016/j.biocel.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 71.Barkauskas C.E., Chung M.I., Fioret B., Gao X., Katsura H., Hogan B.L. Lung organoids: current uses and future promise. Development. 2017;144:986–997. doi: 10.1242/dev.140103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tata P.R., Rajagopal J. Plasticity in the lung: making and breaking cell identity. Development. 2017;144:755–766. doi: 10.1242/dev.143784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Volckaert T., De Langhe S.P. Wnt and FGF mediated epithelial-mesenchymal crosstalk during lung development. Dev. Dyn. 2015;244:342–366. doi: 10.1002/dvdy.24234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Serls A.E., Doherty S., Parvatiyar P., Wells J.M., Deutsch G.H. Different thresholds of fibroblast growth factors pattern the ventral foregut into liver and lung. Development. 2005;132:35–47. doi: 10.1242/dev.01570. [DOI] [PubMed] [Google Scholar]
- 75.Nyeng P., Norgaard G.A., Kobberup S., Jensen J. FGF10 maintains distal lung bud epithelium and excessive signaling leads to progenitor state arrest, distalization, and goblet cell metaplasia. BMC Dev. Biol. 2008;8 doi: 10.1186/1471-213X-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Peters K., Werner S., Liao X., Wert S., Whitsett J., Williams L. Targeted expression of a dominant negative FGF receptor blocks branching morphogenesis and epithelial differentiation of the mouse lung. EMBO J. 1994;13:3296–3301. doi: 10.1002/j.1460-2075.1994.tb06631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bellusci S., Grindley J., Emoto H., Itoh N., Hogan B.L. Fibroblast growth factor 10 (FGF10) and branching morphogenesis in the embryonic mouse lung. Development. 1997;124:4867–4878. doi: 10.1242/dev.124.23.4867. [DOI] [PubMed] [Google Scholar]
- 78.Park W.Y., Miranda B., Lebeche D., Hashimoto G., Cardoso W.V. FGF-10 is a chemotactic factor for distal epithelial buds during lung development. Dev. Biol. 1998;201:125–134. doi: 10.1006/dbio.1998.8994. [DOI] [PubMed] [Google Scholar]
- 79.Hyatt B.A., Shangguan X., Shannon J.M. FGF-10 induces SP-C and Bmp4 and regulates proximal-distal patterning in embryonic tracheal epithelium. Am. J. Phys. Lung Cell. Mol. Phys. 2004;287:L1116–L1126. doi: 10.1152/ajplung.00033.2004. [DOI] [PubMed] [Google Scholar]
- 80.Lebeche D., Malpel S., Cardoso W.V. Fibroblast growth factor interactions in the developing lung. Mech. Dev. 1999;86:125–136. doi: 10.1016/s0925-4773(99)00124-0. [DOI] [PubMed] [Google Scholar]
- 81.Ostrin E.J., Little D.R., Gerner-Mauro K.N., Sumner E.A., Rios-Corzo R., Ambrosio E., Holt S.E., Forcioli-Conti N., Akiyama H., Hanash S.M., Kimura S., Huang S.X.L., Chen J. beta-Catenin maintains lung epithelial progenitors after lung specification. Development. 2018;145 doi: 10.1242/dev.160788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pongracz J.E., Stockley R.A. Wnt signalling in lung development and diseases. Respir. Res. 2006;7 doi: 10.1186/1465-9921-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Okubo T., Hogan B.L. Hyperactive Wnt signaling changes the developmental potential of embryonic lung endoderm. J. Biol. 2004;3:11. doi: 10.1186/jbiol3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shu W., Guttentag S., Wang Z., Andl T., Ballard P., Lu M.M., Piccolo S., Birchmeier W., Whitsett J.A., Millar S.E., Morrisey E.E. Wnt/beta-catenin signaling acts upstream of N-myc, BMP4, and FGF signaling to regulate proximal-distal patterning in the lung. Dev. Biol. 2005;283:226–239. doi: 10.1016/j.ydbio.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 85.Xu B., Chen C., Chen H., Zheng S.G., Bringas P., Jr., Xu M., Zhou X., Chen D., Umans L., Zwijsen A., Shi W. Smad1 and its target gene Wif1 coordinate BMP and Wnt signaling activities to regulate fetal lung development. Development. 2011;138:925–935. doi: 10.1242/dev.062687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sountoulidis A., Stavropoulos A., Giaglis S., Apostolou E., Monteiro R., Chuva de Sousa Lopes S.M., Chen H., Stripp B.R., Mummery C., Andreakos E., Sideras P. Activation of the canonical bone morphogenetic protein (BMP) pathway during lung morphogenesis and adult lung tissue repair. PLoS One. 2012;7 doi: 10.1371/journal.pone.0041460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cibois M., Luxardi G., Chevalier B., Thome V., Mercey O., Zaragosi L.E., Barbry P., Pasini A., Marcet B., Kodjabachian L. BMP signalling controls the construction of vertebrate mucociliary epithelia. Development. 2015;142:2352–2363. doi: 10.1242/dev.118679. [DOI] [PubMed] [Google Scholar]
- 88.Weaver M., Yingling J.M., Dunn N.R., Bellusci S., Hogan B.L. Bmp signaling regulates proximal-distal differentiation of endoderm in mouse lung development. Development. 1999;126:4005–4015. doi: 10.1242/dev.126.18.4005. [DOI] [PubMed] [Google Scholar]
- 89.Chung M.I., Bujnis M., Barkauskas C.E., Kobayashi Y., Hogan B.L.M. Niche-mediated BMP/SMAD signaling regulates lung alveolar stem cell proliferation and differentiation. Development. 2018;145 doi: 10.1242/dev.163014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bellusci S., Henderson R., Winnier G., Oikawa T., Hogan B.L. Evidence from normal expression and targeted misexpression that bone morphogenetic protein (Bmp-4) plays a role in mouse embryonic lung morphogenesis. Development. 1996;122:1693–1702. doi: 10.1242/dev.122.6.1693. [DOI] [PubMed] [Google Scholar]
- 91.Jones M.R., Dilai S., Lingampally A., Chao C.M., Danopoulos S., Carraro G., Mukhametshina R., Wilhelm J., Baumgart-Vogt E., Al Alam D., Chen C., Minoo P., Zhang J.S., Bellusci S. A Comprehensive Analysis of Fibroblast Growth Factor Receptor 2b Signaling on Epithelial Tip Progenitor Cells During Early Mouse Lung Branching Morphogenesis. Front. Genet. 2018;9:746. doi: 10.3389/fgene.2018.00746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Weaver M., Dunn N.R., Hogan B.L. Bmp4 and Fgf10 play opposing roles during lung bud morphogenesis. Development. 2000;127:2695–2704. doi: 10.1242/dev.127.12.2695. [DOI] [PubMed] [Google Scholar]
- 93.Metzger R.J., Klein O.D., Martin G.R., Krasnow M.A. The branching programme of mouse lung development. Nature. 2008;453:745–750. doi: 10.1038/nature07005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tang N., Marshall W.F., McMahon M., Metzger R.J., Martin G.R. Control of mitotic spindle angle by the RAS-regulated ERK1/2 pathway determines lung tube shape. Science. 2011;333:342–345. doi: 10.1126/science.1204831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Herriges J.C., Verheyden J.M., Zhang Z., Sui P., Zhang Y., Anderson M.J., Swing D.A., Zhang Y., Lewandoski M., Sun X. FGF-Regulated ETV Transcription Factors Control FGF-SHH Feedback Loop in Lung Branching. Dev. Cell. 2015;35:322–332. doi: 10.1016/j.devcel.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lang C., Conrad L., Michos O. Mathematical Approaches of Branching Morphogenesis. Front. Genet. 2018;9:673. doi: 10.3389/fgene.2018.00673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang S., Sekiguchi R., Daley W.P., Yamada K.M. Patterned cell and matrix dynamics in branching morphogenesis. J. Cell Biol. 2017;216:559–570. doi: 10.1083/jcb.201610048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Alanis D.M., Chang D.R., Akiyama H., Krasnow M.A., Chen J. Two nested developmental waves demarcate a compartment boundary in the mouse lung. Nat. Commun. 2014;5 doi: 10.1038/ncomms4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Campiche M.A., Gautier A., Hernandez E.I., Reymond A. An Electron Microscope Study of the Fetal Development of Human Lung. Pediatrics. 1963;32:976–994. [PubMed] [Google Scholar]
- 100.Kitterman J.A. The effects of mechanical forces on fetal lung growth. Clin. Perinatol. 1996;23:727–740. [PubMed] [Google Scholar]
- 101.Scarpelli E.M., Condorelli S., Cosmi E.V. Lamb fetal pulmonary fluid. I. Validation and significance of method for determination of volume and volume change. Pediatr. Res. 1975;9:190–195. doi: 10.1203/00006450-197504000-00010. [DOI] [PubMed] [Google Scholar]
- 102.Li J., Wang Z., Chu Q., Jiang K., Li J., Tang N. The Strength of Mechanical Forces Determines the Differentiation of Alveolar Epithelial Cells. Dev. Cell. 2018;44:297–312. doi: 10.1016/j.devcel.2018.01.008. e295. [DOI] [PubMed] [Google Scholar]
- 103.Harding R. Fetal pulmonary development: the role of respiratory movements. Equine Vet J Suppl. 1997:32–39. doi: 10.1111/j.2042-3306.1997.tb05076.x. [DOI] [PubMed] [Google Scholar]
- 104.Coraux C., Nawrocki-Raby B., Hinnrasky J., Kileztky C., Gaillard D., Dani C., Puchelle E. Embryonic stem cells generate airway epithelial tissue. Am. J. Respir. Cell Mol. Biol. 2005;32:87–92. doi: 10.1165/rcmb.2004-0079RC. [DOI] [PubMed] [Google Scholar]
- 105.Loh K.M., Ang L.T., Zhang J., Kumar V., Ang J., Auyeong J.Q., Lee K.L., Choo S.H., Lim C.Y., Nichane M., Tan J., Noghabi M.S., Azzola L., Ng E.S., Durruthy-Durruthy J., Sebastiano V., Poellinger L., Elefanty A.G., Stanley E.G., Chen Q., Prabhakar S., Weissman I.L., Lim B. Efficient endoderm induction from human pluripotent stem cells by logically directing signals controlling lineage bifurcations. Cell Stem Cell. 2014;14:237–252. doi: 10.1016/j.stem.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.D'Amour K.A., Agulnick A.D., Eliazer S., Kelly O.G., Kroon E., Baetge E.E. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat. Biotechnol. 2005;23:1534–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- 107.Gouon-Evans V., Boussemart L., Gadue P., Nierhoff D., Koehler C.I., Kubo A., Shafritz D.A., Keller G. BMP-4 is required for hepatic specification of mouse embryonic stem cell-derived definitive endoderm. Nat. Biotechnol. 2006;24:1402–1411. doi: 10.1038/nbt1258. [DOI] [PubMed] [Google Scholar]
- 108.Yasunaga M., Tada S., Torikai-Nishikawa S., Nakano Y., Okada M., Jakt L.M., Nishikawa S., Chiba T., Era T., Nishikawa S. Induction and monitoring of definitive and visceral endoderm differentiation of mouse ES cells. Nat. Biotechnol. 2005;23:1542–1550. doi: 10.1038/nbt1167. [DOI] [PubMed] [Google Scholar]
- 109.Duester G. Retinoic acid synthesis and signaling during early organogenesis. Cell. 2008;134:921–931. doi: 10.1016/j.cell.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Desai T.J., Chen F., Lu J., Qian J., Niederreither K., Dolle P., Chambon P., Cardoso W.V. Distinct roles for retinoic acid receptors alpha and beta in early lung morphogenesis. Dev. Biol. 2006;291:12–24. doi: 10.1016/j.ydbio.2005.10.045. [DOI] [PubMed] [Google Scholar]
- 111.Ikonomou L., Herriges M.J., Lewandowski S.L., Marsland R., 3rd, Villacorta-Martin C., Caballero I.S., Frank D.B., Sanghrajka R.M., Dame K., Kandula M.M., Hicks-Berthet J., Lawton M.L., Christodoulou C., Fabian A.J., Kolaczyk E., Varelas X., Morrisey E.E., Shannon J.M., Mehta P., Kotton D.N. The in vivo genetic program of murine primordial lung epithelial progenitors. Nat. Commun. 2020;11:635. doi: 10.1038/s41467-020-14348-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rock J.R., Hogan B.L. Epithelial progenitor cells in lung development, maintenance, repair, and disease. Annu. Rev. Cell Dev. Biol. 2011;27:493–512. doi: 10.1146/annurev-cellbio-100109-104040. [DOI] [PubMed] [Google Scholar]
- 113.Morrisey E.E., Hogan B.L. Preparing for the first breath: genetic and cellular mechanisms in lung development. Dev. Cell. 2010;18:8–23. doi: 10.1016/j.devcel.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rock J.R., Hogan B.L. Epithelial progenitor cells in lung development, maintenance, repair, and disease. Annu. Rev. Cell Dev. Biol. 2011;27:493–512. doi: 10.1146/annurev-cellbio-100109-104040. [DOI] [PubMed] [Google Scholar]
- 115.Rock J.R., Randell S.H., Hogan B.L. Airway basal stem cells: a perspective on their roles in epithelial homeostasis and remodeling. Dis. Model. Mech. 2010;3:545–556. doi: 10.1242/dmm.006031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mahoney J.E., Mori M., Szymaniak A.D., Varelas X., Cardoso W.V. The hippo pathway effector Yap controls patterning and differentiation of airway epithelial progenitors. Dev. Cell. 2014;30:137–150. doi: 10.1016/j.devcel.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Laresgoiti U., Nikolic M.Z., Rao C., Brady J.L., Richardson R.V., Batchen E.J., Chapman K.E., Rawlins E.L. Lung epithelial tip progenitors integrate glucocorticoid- and STAT3-mediated signals to control progeny fate. Development. 2016;143:3686–3699. doi: 10.1242/dev.134023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wuenschell C.W., Sunday M.E., Singh G., Minoo P., Slavkin H.C., Warburton D. Embryonic mouse lung epithelial progenitor cells co-express immunohistochemical markers of diverse mature cell lineages. J. Histochem. Cytochem. 1996;44:113–123. doi: 10.1177/44.2.8609367. [DOI] [PubMed] [Google Scholar]
- 119.McCauley K.B., Hawkins F., Kotton D.N. Derivation of Epithelial-Only Airway Organoids from Human Pluripotent Stem Cells. Curr Protoc Stem Cell Biol. 2018;45 doi: 10.1002/cpsc.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Serra M., Alysandratos K.D., Hawkins F., McCauley K.B., Jacob A., Choi J., Caballero I.S., Vedaie M., Kurmann A.A., Ikonomou L., Hollenberg A.N., Shannon J.M., Kotton D.N. Pluripotent stem cell differentiation reveals distinct developmental pathways regulating lung- versus thyroid-lineage specification. Development. 2017;144:3879–3893. doi: 10.1242/dev.150193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Miller A.J., Dye B.R., Ferrer-Torres D., Hill D.R., Overeem A.W., Shea L.D., Spence J.R. Generation of lung organoids from human pluripotent stem cells in vitro. Nat. Protoc. 2019;14:518–540. doi: 10.1038/s41596-018-0104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yang J., Zuo W.L., Fukui T., Chao I., Gomi K., Lee B., Staudt M.R., Kaner R.J., Strulovici-Barel Y., Salit J., Crystal R.G., Shaykhiev R. Smoking-Dependent Distal-to-Proximal Repatterning of the Adult Human Small Airway Epithelium. Am. J. Respir. Crit. Care Med. 2017;196:340–352. doi: 10.1164/rccm.201608-1672OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Schamberger A.C., Staab-Weijnitz C.A., Mise-Racek N., Eickelberg O. Cigarette smoke alters primary human bronchial epithelial cell differentiation at the air-liquid interface. Sci. Rep. 2015;5 doi: 10.1038/srep08163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rogers D.F. The airway goblet cell. Int. J. Biochem. Cell Biol. 2003;35:1–6. doi: 10.1016/s1357-2725(02)00083-3. [DOI] [PubMed] [Google Scholar]
- 125.Boers J.E., Ambergen A.W., Thunnissen F.B. Number and proliferation of clara cells in normal human airway epithelium. Am. J. Respir. Crit. Care Med. 1999;159:1585–1591. doi: 10.1164/ajrccm.159.5.9806044. [DOI] [PubMed] [Google Scholar]
- 126.Boers J.E., Ambergen A.W., Thunnissen F.B. Number and proliferation of basal and parabasal cells in normal human airway epithelium. Am. J. Respir. Crit. Care Med. 1998;157:2000–2006. doi: 10.1164/ajrccm.157.6.9707011. [DOI] [PubMed] [Google Scholar]
- 127.Crapo J.D., Young S.L., Fram E.K., Pinkerton K.E., Barry B.E., Crapo R.O. Morphometric characteristics of cells in the alveolar region of mammalian lungs. Am. Rev. Respir. Dis. 1983;128:S42–S46. doi: 10.1164/arrd.1983.128.2P2.S42. [DOI] [PubMed] [Google Scholar]
- 128.Guilbault C., Saeed Z., Downey G.P., Radzioch D. Cystic fibrosis mouse models. Am. J. Respir. Cell Mol. Biol. 2007;36:1–7. doi: 10.1165/rcmb.2006-0184TR. [DOI] [PubMed] [Google Scholar]
- 129.Clarke L.L., Grubb B.R., Gabriel S.E., Smithies O., Koller B.H., Boucher R.C. Defective epithelial chloride transport in a gene-targeted mouse model of cystic fibrosis. Science. 1992;257:1125–1128. doi: 10.1126/science.257.5073.1125. [DOI] [PubMed] [Google Scholar]
- 130.Snouwaert J.N., Brigman K.K., Latour A.M., Malouf N.N., Boucher R.C., Smithies O., Koller B.H. An animal model for cystic fibrosis made by gene targeting. Science. 1992;257:1083–1088. doi: 10.1126/science.257.5073.1083. [DOI] [PubMed] [Google Scholar]
- 131.Rock J.R., Onaitis M.W., Rawlins E.L., Lu Y., Clark C.P., Xue Y., Randell S.H., Hogan B.L. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc. Natl. Acad. Sci. U. S. A. 2009;106:12771–12775. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Rock J.R., Gao X., Xue Y., Randell S.H., Kong Y.Y., Hogan B.L. Notch-dependent differentiation of adult airway basal stem cells. Cell Stem Cell. 2011;8:639–648. doi: 10.1016/j.stem.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Whitsett J.A., Clark J.C., Picard L., Tichelaar J.W., Wert S.E., Itoh N., Perl A.K., Stahlman M.T. Fibroblast growth factor 18 influences proximal programming during lung morphogenesis. J. Biol. Chem. 2002;277:22743–22749. doi: 10.1074/jbc.M202253200. [DOI] [PubMed] [Google Scholar]
- 134.Rinker T.E., Philbrick B.D., Hettiaratchi M.H., Smalley D.M., McDevitt T.C., Temenoff J.S. Microparticle-mediated sequestration of cell-secreted proteins to modulate chondrocytic differentiation. Acta Biomater. 2018;68:125–136. doi: 10.1016/j.actbio.2017.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bratt-Leal A.M., Nguyen A.H., Hammersmith K.A., Singh A., McDevitt T.C. A microparticle approach to morphogen delivery within pluripotent stem cell aggregates. Biomaterials. 2013;34:7227–7235. doi: 10.1016/j.biomaterials.2013.05.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Carpenedo R.L., Bratt-Leal A.M., Marklein R.A., Seaman S.A., Bowen N.J., McDonald J.F., McDevitt T.C. Homogeneous and organized differentiation within embryoid bodies induced by microsphere-mediated delivery of small molecules. Biomaterials. 2009;30:2507–2515. doi: 10.1016/j.biomaterials.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gomi K., Arbelaez V., Crystal R.G., Walters M.S. Activation of NOTCH1 or NOTCH3 signaling skews human airway basal cell differentiation toward a secretory pathway. PLoS One. 2015;10 doi: 10.1371/journal.pone.0116507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gerovac B.J., Valencia M., Baumlin N., Salathe M., Conner G.E., Fregien N.L. Submersion and hypoxia inhibit ciliated cell differentiation in a notch-dependent manner. Am. J. Respir. Cell Mol. Biol. 2014;51:516–525. doi: 10.1165/rcmb.2013-0237OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ruiz Garcia S., Deprez M., Lebrigand K., Cavard A., Paquet A., Arguel M.J., Magnone V., Truchi M., Caballero I., Leroy S., Marquette C.H., Marcet B., Barbry P., Zaragosi L.E. Novel dynamics of human mucociliary differentiation revealed by single-cell RNA sequencing of nasal epithelial cultures. Development. 2019;146 doi: 10.1242/dev.177428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Tata P.R., Mou H., Pardo-Saganta A., Zhao R., Prabhu M., Law B.M., Vinarsky V., Cho J.L., Breton S., Sahay A., Medoff B.D., Rajagopal J. Dedifferentiation of committed epithelial cells into stem cells in vivo. Nature. 2013;503:218–223. doi: 10.1038/nature12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Semaniakou A., Croll R.P., Chappe V. Animal Models in the Pathophysiology of Cystic Fibrosis. Front. Pharmacol. 2018;9:1475. doi: 10.3389/fphar.2018.01475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.McCarron A., Donnelley M., Parsons D. Airway disease phenotypes in animal models of cystic fibrosis. Respir. Res. 2018;19 doi: 10.1186/s12931-018-0750-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Fisher J.T., Zhang Y., Engelhardt J.F. Comparative biology of cystic fibrosis animal models. Methods Mol. Biol. 2011;742:311–334. doi: 10.1007/978-1-61779-120-8_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Tashiro J., Rubio G.A., Limper A.H., Williams K., Elliot S.J., Ninou I., Aidinis V., Tzouvelekis A., Glassberg M.K. Exploring Animal Models That Resemble Idiopathic Pulmonary Fibrosis. Front Med (Lausanne) 2017;4:118. doi: 10.3389/fmed.2017.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mou H., Brazauskas K., Rajagopal J. Personalized medicine for cystic fibrosis: establishing human model systems. Pediatr. Pulmonol. 2015;50(Suppl. 40):S14–S23. doi: 10.1002/ppul.23233. [DOI] [PubMed] [Google Scholar]
- 146.Fleischer A., Vallejo-Diez S., Martin-Fernandez J.M., Sanchez-Gilabert A., Castresana M., Del Pozo A., Esquisabel A., Avila S., Castrillo J.L., Gainza E., Pedraz J.L., Vinas M., Bachiller D. iPSC-Derived Intestinal Organoids from Cystic Fibrosis Patients Acquire CFTR Activity upon TALEN-Mediated Repair of the p.F508del Mutation. Mol Ther Methods Clin Dev. 2020;17:858–870. doi: 10.1016/j.omtm.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Mithal A., Capilla A., Heinze D., Berical A., Villacorta-Martin C., Vedaie M., Jacob A., Abo K., Szymaniak A., Peasley M., Stuffer A., Mahoney J., Kotton D.N., Hawkins F., Mostoslavsky G. Generation of mesenchyme free intestinal organoids from human induced pluripotent stem cells. Nat. Commun. 2020;11:215. doi: 10.1038/s41467-019-13916-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hohwieler M., Perkhofer L., Liebau S., Seufferlein T., Muller M., Illing A., Kleger A. Stem cell-derived organoids to model gastrointestinal facets of cystic fibrosis. United European Gastroenterol J. 2017;5:609–624. doi: 10.1177/2050640616670565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Berkers G., van Mourik P., Vonk A.M., Kruisselbrink E., Dekkers J.F., de Winter-de Groot K.M., Arets H.G.M., Marck-van der Wilt R.E.P., Dijkema J.S., Vanderschuren M.M., Houwen R.H.J., Heijerman H.G.M., van de Graaf E.A., Elias S.G., Majoor C.J., Koppelman G.H., Roukema J., Bakker M., Janssens H.M., van der Meer R., Vries R.G.J., Clevers H.C., de Jonge H.R., Beekman J.M., van der Ent C.K. Rectal Organoids Enable Personalized Treatment of Cystic Fibrosis. Cell Rep, 26. 2019;e1703:1701–1708. doi: 10.1016/j.celrep.2019.01.068. [DOI] [PubMed] [Google Scholar]
- 150.Dekkers J.F., Berkers G., Kruisselbrink E., Vonk A., de Jonge H.R., Janssens H.M., Bronsveld I., van de Graaf E.A., Nieuwenhuis E.E., Houwen R.H., Vleggaar F.P., Escher J.C., de Rijke Y.B., Majoor C.J., Heijerman H.G., de Winter-de Groot K.M., Clevers H., van der Ent C.K., Beekman J.M. Characterizing responses to CFTR-modulating drugs using rectal organoids derived from subjects with cystic fibrosis. Sci. Transl. Med. 2016;8:344ra384. doi: 10.1126/scitranslmed.aad8278. [DOI] [PubMed] [Google Scholar]
- 151.Dekkers J.F., Wiegerinck C.L., de Jonge H.R., Bronsveld I., Janssens H.M., de Winter-de Groot K.M., Brandsma A.M., de Jong N.W., Bijvelds M.J., Scholte B.J., Nieuwenhuis E.E., van den Brink S., Clevers H., van der Ent C.K., Middendorp S., Beekman J.M. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat. Med. 2013;19:939–945. doi: 10.1038/nm.3201. [DOI] [PubMed] [Google Scholar]
- 152.Shik Mun K., Arora K., Huang Y., Yang F., Yarlagadda S., Ramananda Y., Abu-El-Haija M., Palermo J.J., Appakalai B.N., Nathan J.D., Naren A.P. Patient-derived pancreas-on-a-chip to model cystic fibrosis-related disorders. Nat. Commun. 2019;10 doi: 10.1038/s41467-019-11178-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Liu Z., Anderson J.D., Deng L., Mackay S., Bailey J., Kersh L., Rowe S.M., Guimbellot J.S. Human Nasal Epithelial Organoids for Therapeutic Development in Cystic Fibrosis. Genes (Basel) 2020;11 doi: 10.3390/genes11060603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sachs N., Papaspyropoulos A., Zomer-van Ommen D.D., Heo I., Bottinger L., Klay D., Weeber F., Huelsz-Prince G., Iakobachvili N., Amatngalim G.D., de Ligt J., van Hoeck A., Proost N., Viveen M.C., Lyubimova A., Teeven L., Derakhshan S., Korving J., Begthel H., Dekkers J.F., Kumawat K., Ramos E., van Oosterhout M.F., Offerhaus G.J., Wiener D.J., Olimpio E.P., Dijkstra K.K., Smit E.F., van der Linden M., Jaksani S., van de Ven M., Jonkers J., Rios A.C., Voest E.E., van Moorsel C.H., van der Ent C.K., Cuppen E., van Oudenaarden A., Coenjaerts F.E., Meyaard L., Bont L.J., Peters P.J., Tans S.J., van Zon J.S., Boj S.F., Vries R.G., Beekman J.M., Clevers H. Long-term expanding human airway organoids for disease modeling. EMBO J. 2019;38 doi: 10.15252/embj.2018100300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.(!!! INVALID CITATION !!! [26]).
- 156.Lederer D.J., Martinez F.J. Idiopathic Pulmonary Fibrosis. N. Engl. J. Med. 2018;379:797–798. doi: 10.1056/NEJMc1807508. [DOI] [PubMed] [Google Scholar]
- 157.Marchioni A., Tonelli R., Ball L., Fantini R., Castaniere I., Cerri S., Luppi F., Malerba M., Pelosi P., Clini E. Acute exacerbation of idiopathic pulmonary fibrosis: lessons learned from acute respiratory distress syndrome? Crit. Care. 2018;22 doi: 10.1186/s13054-018-2002-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.De Jesus Rojas W., Young L.R. Hermansky-Pudlak Syndrome. Semin Respir Crit Care Med. 2020;41:238–246. doi: 10.1055/s-0040-1708088. [DOI] [PubMed] [Google Scholar]
- 159.Huizing M., Malicdan M.C.V., Gochuico B.R., Gahl W.A. In: M.P. Adam H.H. Ardinger, Pagon R.A., Wallace S.E., Bean L.J.H., Stephens K., Amemiya A., editors. GeneReviews((R)); Seattle (WA): 1993. Hermansky-Pudlak Syndrome. [Google Scholar]
- 160.Korogi Y., Gotoh S., Ikeo S., Yamamoto Y., Sone N., Tamai K., Konishi S., Nagasaki T., Matsumoto H., Ito I., Chen-Yoshikawa T.F., Date H., Hagiwara M., Asaka I., Hotta A., Mishima M., Hirai T. In Vitro Disease Modeling of Hermansky-Pudlak Syndrome Type 2 Using Human Induced Pluripotent Stem Cell-Derived Alveolar Organoids. Stem Cell Reports. 2019;13:235. doi: 10.1016/j.stemcr.2019.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Strikoudis A., Cieslak A., Loffredo L., Chen Y.W., Patel N., Saqi A., Lederer D.J., Snoeck H.W. Modeling of Fibrotic Lung Disease Using 3D Organoids Derived from Human Pluripotent Stem Cells. Cell Rep. 2019;27:3709–3723. doi: 10.1016/j.celrep.2019.05.077. e3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ng B., Dong J., D'Agostino G., Viswanathan S., Widjaja A.A., Lim W.W., Ko N.S.J., Tan J., Chothani S.P., Huang B., Xie C., Pua C.J., Chacko A.M., Guimaraes-Camboa N., Evans S.M., Byrne A.J., Maher T.M., Liang J., Jiang D., Noble P.W., Schafer S., Cook S.A. 2019. Interleukin-11 is a therapeutic target in idiopathic pulmonary fibrosis, Sci Transl Med, 11. [DOI] [PubMed] [Google Scholar]
- 163.Efstathiou C., Abidi S.H., Harker J., Stevenson N.J. Revisiting respiratory syncytial virus's interaction with host immunity, towards novel therapeutics. Cell. Mol. Life Sci. 2020 doi: 10.1007/s00018-020-03557-0. ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Porotto M., Ferren M., Chen Y.W., Siu Y., Makhsous N., Rima B., Briese T., Greninger A.L., Snoeck H.W., Moscona A. Authentic Modeling of Human Respiratory Virus Infection in Human Pluripotent Stem Cell-Derived Lung Organoids. MBio. 2019;10 doi: 10.1128/mBio.00723-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Schweitzer J.W., Justice N.A. StatPearls; Treasure Island (FL): 2020. Respiratory Syncytial Virus Infection (RSV) [Google Scholar]
- 166.Leibel S.L., Winquist A., Tseu I., Wang J., Luo D., Shojaie S., Nathan N., Snyder E., Post M. Reversal of Surfactant Protein B Deficiency in Patient Specific Human Induced Pluripotent Stem Cell Derived Lung Organoids by Gene Therapy. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-49696-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Mora A.L., Rojas M., Pardo A., Selman M. Emerging therapies for idiopathic pulmonary fibrosis, a progressive age-related disease. Nat. Rev. Drug Discov. 2017;16:810. doi: 10.1038/nrd.2017.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Drick N., Sahabian A., Pongpamorn P., Merkert S., Gohring G., Welte T., Martin U., Olmer R. Generation of a NKX2.1 - p63 double transgenic knock-in reporter cell line from human induced pluripotent stem cells (MHHi006-A-4) Stem Cell Res. 2020;42 doi: 10.1016/j.scr.2019.101659. [DOI] [PubMed] [Google Scholar]
- 169.Liu M., Post M. Invited review: mechanochemical signal transduction in the fetal lung. J. Appl. Physiol. 2000;89:2078–2084. doi: 10.1152/jappl.2000.89.5.2078. [DOI] [PubMed] [Google Scholar]
- 170.Lancaster M.A., Corsini N.S., Wolfinger S., Gustafson E.H., Phillips A.W., Burkard T.R., Otani T., Livesey F.J., Knoblich J.A. Guided self-organization and cortical plate formation in human brain organoids. Nat. Biotechnol. 2017;35:659–666. doi: 10.1038/nbt.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Przybyla L., Lakins J.N., Weaver V.M. Tissue Mechanics Orchestrate Wnt-Dependent Human Embryonic Stem Cell Differentiation. Cell Stem Cell. 2016;19:462–475. doi: 10.1016/j.stem.2016.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Santos M., Nogueira-Silva C., Baptista M.J., Soares-Fernandes J., Moura R.S., Correia-Pinto J. Pulmonary epithelial cell differentiation in the nitrofen-induced congenital diaphragmatic hernia. J. Pediatr. Surg. 2007;42:1231–1237. doi: 10.1016/j.jpedsurg.2007.02.014. [DOI] [PubMed] [Google Scholar]
- 173.Takayasu H., Nakazawa N., Montedonico S., Sugimoto K., Sato H., Puri P. Impaired alveolar epithelial cell differentiation in the hypoplastic lung in nitrofen-induced congenital diaphragmatic hernia. Pediatr. Surg. Int. 2007;23:405–410. doi: 10.1007/s00383-006-1853-y. [DOI] [PubMed] [Google Scholar]
- 174.Kloxin A.M., Lewis K.J., DeForest C.A., Seedorf G., Tibbitt M.W., Balasubramaniam V., Anseth K.S. Responsive culture platform to examine the influence of microenvironmental geometry on cell function in 3D. Integr Biol (Camb) 2012;4:1540–1549. doi: 10.1039/c2ib20212c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lewis K.J., Tibbitt M.W., Zhao Y., Branchfield K., Sun X., Balasubramaniam V., Anseth K.S. In vitro model alveoli from photodegradable microsphere templates. Biomater Sci. 2015;3:821–832. doi: 10.1039/c5bm00034c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Moore K.A., Polte T., Huang S., Shi B., Alsberg E., Sunday M.E., Ingber D.E. Control of basement membrane remodeling and epithelial branching morphogenesis in embryonic lung by Rho and cytoskeletal tension. Developmental dynamics : an. 2005;232:268–281. doi: 10.1002/dvdy.20237. [DOI] [PubMed] [Google Scholar]
- 177.Hirashima T., Adachi T. Polarized cellular mechano-response system for maintaining radial size in developing epithelial tubes. Development. 2019;146 doi: 10.1242/dev.181206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Spurlin J.W., Siedlik M.J., Nerger B.A., Pang M.F., Jayaraman S., Zhang R., Nelson C.M. Mesenchymal proteases and tissue fluidity remodel the extracellular matrix during airway epithelial branching in the embryonic avian lung. Development. 2019;146 doi: 10.1242/dev.175257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Neumann N.M., Perrone M.C., Veldhuis J.H., Huebner R.J., Zhan H., Devreotes P.N., Brodland G.W., Ewald A.J. Coordination of Receptor Tyrosine Kinase Signaling and Interfacial Tension Dynamics Drives Radial Intercalation and Tube Elongation. Dev. Cell. 2018;45:67–82. doi: 10.1016/j.devcel.2018.03.011. e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Gao B., Ajima R., Yang W., Li C., Song H., Anderson M.J., Liu R.R., Lewandoski M.B., Yamaguchi T.P., Yang Y. Coordinated directional outgrowth and pattern formation by integration of Wnt5a and Fgf signaling in planar cell polarity. Development. 2018;145 doi: 10.1242/dev.163824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kilian K.A., Bugarija B., Lahn B.T., Mrksich M. Geometric cues for directing the differentiation of mesenchymal stem cells. Proc. Natl. Acad. Sci. U. S. A. 2010;107:4872–4877. doi: 10.1073/pnas.0903269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.von Erlach T.C., Bertazzo S., Wozniak M.A., Horejs C.M., Maynard S.A., Attwood S., Robinson B.K., Autefage H., Kallepitis C., Del Rio Hernandez A., Chen C.S., Goldoni S., Stevens M.M. Cell-geometry-dependent changes in plasma membrane order direct stem cell signalling and fate. Nat. Mater. 2018;17:237–242. doi: 10.1038/s41563-017-0014-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Dye B.R., Youngblood R.L., Oakes R.S., Kasputis T., Clough D.W., Spence J.R., Shea L.D. Human lung organoids develop into adult airway-like structures directed by physico-chemical biomaterial properties. Biomaterials. 2020;234:119757. doi: 10.1016/j.biomaterials.2020.119757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kawai N., Ouji Y., Sakagami M., Tojo T., Sawabata N., Yoshikawa M., Taniguchi S. Induction of lung-like cells from mouse embryonic stem cells by decellularized lung matrix. Biochem Biophys Rep. 2018;15:33–38. doi: 10.1016/j.bbrep.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Poling H.M., Wu D., Brown N., Baker M., Hausfeld T.A., Huynh N., Chaffron S., Dunn J.C.Y., Hogan S.P., Wells J.M., Helmrath M.A., Mahe M.M. Mechanically induced development and maturation of human intestinal organoids in vivo. Nat Biomed Eng. 2018;2:429–442. doi: 10.1038/s41551-018-0243-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Ma Z., Wang J., Loskill P., Huebsch N., Koo S., Svedlund F.L., Marks N.C., Hua E.W., Grigoropoulos C.P., Conklin B.R., Healy K.E. Self-organizing human cardiac microchambers mediated by geometric confinement. Nat. Commun. 2015;6 doi: 10.1038/ncomms8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Shao Y., Taniguchi K., Gurdziel K., Townshend R.F., Xue X., Yong K.M.A., Sang J., Spence J.R., Gumucio D.L., Fu J. Self-organized amniogenesis by human pluripotent stem cells in a biomimetic implantation-like niche. Nat. Mater. 2017;16:419–425. doi: 10.1038/nmat4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Vining K.H., Mooney D.J. Mechanical forces direct stem cell behaviour in development and regeneration. Nat. Rev. Mol. Cell Biol. 2017;18:728–742. doi: 10.1038/nrm.2017.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Murphy W.L., McDevitt T.C., Engler A.J. Materials as stem cell regulators. Nat. Mater. 2014;13:547–557. doi: 10.1038/nmat3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Nazareth E.J., Ostblom J.E., Lucker P.B., Shukla S., Alvarez M.M., Oh S.K., Yin T., Zandstra P.W. High-throughput fingerprinting of human pluripotent stem cell fate responses and lineage bias. Nat. Methods. 2013;10:1225–1231. doi: 10.1038/nmeth.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Peerani R., Bauwens C., Kumacheva E., Zandstra P.W. Patterning mouse and human embryonic stem cells using micro-contact printing. Methods Mol. Biol. 2009;482:21–33. doi: 10.1007/978-1-59745-060-7_2. [DOI] [PubMed] [Google Scholar]
- 192.Tan J.L., Liu W., Nelson C.M., Raghavan S., Chen C.S. Simple approach to micropattern cells on common culture substrates by tuning substrate wettability. Tissue Eng. 2004;10:865–872. doi: 10.1089/1076327041348365. [DOI] [PubMed] [Google Scholar]
- 193.Anderson D.G., Levenberg S., Langer R. Nanoliter-scale synthesis of arrayed biomaterials and application to human embryonic stem cells. Nat. Biotechnol. 2004;22:863–866. doi: 10.1038/nbt981. [DOI] [PubMed] [Google Scholar]
- 194.Chen C.S., Mrksich M., Huang S., Whitesides G.M., Ingber D.E. Geometric control of cell life and death. Science. 1997;276:1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 195.Lee J., Abdeen A.A., Wycislo K.L., Fan T.M., Kilian K.A. Interfacial geometry dictates cancer cell tumorigenicity. Nat. Mater. 2016;15:856–862. doi: 10.1038/nmat4610. [DOI] [PubMed] [Google Scholar]
- 196.Whitesides G.M., Ostuni E., Takayama S., Jiang X., Ingber D.E. Soft lithography in biology and biochemistry. Annu. Rev. Biomed. Eng. 2001;3:335–373. doi: 10.1146/annurev.bioeng.3.1.335. [DOI] [PubMed] [Google Scholar]
- 197.Kane R.S., Takayama S., Ostuni E., Ingber D.E., Whitesides G.M. Patterning proteins and cells using soft lithography. Biomaterials. 1999;20:2363–2376. doi: 10.1016/s0142-9612(99)00165-9. [DOI] [PubMed] [Google Scholar]
- 198.McBeath R., Pirone D.M., Nelson C.M., Bhadriraju K., Chen C.S. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev. Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 199.Ostuni E., Whitesides G.M., Ingber D.E., Chen C.S. Using self-assembled monolayers to pattern ECM proteins and cells on substrates. Methods Mol. Biol. 2009;522:183–194. doi: 10.1007/978-1-59745-413-1_12. [DOI] [PubMed] [Google Scholar]
- 200.Peerani R., Rao B.M., Bauwens C., Yin T., Wood G.A., Nagy A., Kumacheva E., Zandstra P.W. Niche-mediated control of human embryonic stem cell self-renewal and differentiation. EMBO J. 2007;26:4744–4755. doi: 10.1038/sj.emboj.7601896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Warmflash A., Sorre B., Etoc F., Siggia E.D., Brivanlou A.H. A method to recapitulate early embryonic spatial patterning in human embryonic stem cells. Nat. Methods. 2014;11:847–854. doi: 10.1038/nmeth.3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Tewary M., Ostblom J., Prochazka L., Zulueta-Coarasa T., Shakiba N., Fernandez-Gonzalez R., Zandstra P.W. A stepwise model of reaction-diffusion and positional information governs self-organized human peri-gastrulation-like patterning. Development. 2017;144:4298–4312. doi: 10.1242/dev.149658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Morgani S.M., Metzger J.J., Nichols J., Siggia E.D., Hadjantonakis A.K. Micropattern differentiation of mouse pluripotent stem cells recapitulates embryo regionalized cell fate patterning. Elife. 2018;7 doi: 10.7554/eLife.32839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Martyn I., Siggia E.D., Brivanlou A.H. Mapping cell migrations and fates in a gastruloid model to the human primitive streak. Development. 2019;146 doi: 10.1242/dev.179564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Krishna L., Dhamodaran K., Jayadev C., Chatterjee K., Shetty R., Khora S.S., Das D. Nanostructured scaffold as a determinant of stem cell fate. Stem Cell Res Ther. 2016;7 doi: 10.1186/s13287-016-0440-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Chen G.Y., Pang D.W., Hwang S.M., Tuan H.Y., Hu Y.C. A graphene-based platform for induced pluripotent stem cells culture and differentiation. Biomaterials. 2012;33:418–427. doi: 10.1016/j.biomaterials.2011.09.071. [DOI] [PubMed] [Google Scholar]
- 207.Bae D., Moon S.H., Park B.G., Park S.J., Jung T., Kim J.S., Lee K.B., Chung H.M. Nanotopographical control for maintaining undifferentiated human embryonic stem cell colonies in feeder free conditions. Biomaterials. 2014;35:916–928. doi: 10.1016/j.biomaterials.2013.10.031. [DOI] [PubMed] [Google Scholar]
- 208.Zanden C., Hellstrom Erkenstam N., Padel T., Wittgenstein J., Liu J., Kuhn H.G. Stem cell responses to plasma surface modified electrospun polyurethane scaffolds. Nanomedicine. 2014;10:949–958. doi: 10.1016/j.nano.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 209.Ko J.Y., Oh H.J., Lee J., Im G.I. Nanotopographic Influence on the In Vitro Behavior of Induced Pluripotent Stem Cells. Tissue Eng Part A. 2018;24:595–606. doi: 10.1089/ten.TEA.2017.0144. [DOI] [PubMed] [Google Scholar]
- 210.Huang J., Chen Y., Tang C., Fei Y., Wu H., Ruan D., Paul M.E., Chen X., Yin Z., Heng B.C., Chen W., Shen W. The relationship between substrate topography and stem cell differentiation in the musculoskeletal system. Cell. Mol. Life Sci. 2019;76:505–521. doi: 10.1007/s00018-018-2945-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Geiger B., Spatz J.P., Bershadsky A.D. Environmental sensing through focal adhesions. Nat. Rev. Mol. Cell Biol. 2009;10:21–33. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- 212.Yim E.K., Darling E.M., Kulangara K., Guilak F., Leong K.W. Nanotopography-induced changes in focal adhesions, cytoskeletal organization, and mechanical properties of human mesenchymal stem cells. Biomaterials. 2010;31:1299–1306. doi: 10.1016/j.biomaterials.2009.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Seo C.H., Furukawa K., Montagne K., Jeong H., Ushida T. The effect of substrate microtopography on focal adhesion maturation and actin organization via the RhoA/ROCK pathway. Biomaterials. 2011;32:9568–9575. doi: 10.1016/j.biomaterials.2011.08.077. [DOI] [PubMed] [Google Scholar]
- 214.Trappmann B., Gautrot J.E., Connelly J.T., Strange D.G., Li Y., Oyen M.L., Cohen Stuart M.A., Boehm H., Li B., Vogel V., Spatz J.P., Watt F.M., Huck W.T. Extracellular-matrix tethering regulates stem-cell fate. Nat. Mater. 2012;11:642–649. doi: 10.1038/nmat3339. [DOI] [PubMed] [Google Scholar]
- 215.Dalby M.J., Gadegaard N., Riehle M.O., Wilkinson C.D., Curtis A.S. Investigating filopodia sensing using arrays of defined nano-pits down to 35 nm diameter in size. Int. J. Biochem. Cell Biol. 2004;36:2005–2015. doi: 10.1016/j.biocel.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 216.Dalby M.J., Riehle M.O., Johnstone H., Affrossman S., Curtis A.S. Investigating the limits of filopodial sensing: a brief report using SEM to image the interaction between 10 nm high nano-topography and fibroblast filopodia. Cell Biol. Int. 2004;28:229–236. doi: 10.1016/j.cellbi.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 217.Cojoc D., Difato F., Ferrari E., Shahapure R.B., Laishram J., Righi M., Di Fabrizio E.M., Torre V. Properties of the force exerted by filopodia and lamellipodia and the involvement of cytoskeletal components. PLoS One. 2007;2 doi: 10.1371/journal.pone.0001072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Mattila P.K., Lappalainen P. Filopodia: molecular architecture and cellular functions. Nat. Rev. Mol. Cell Biol. 2008;9:446–454. doi: 10.1038/nrm2406. [DOI] [PubMed] [Google Scholar]
- 219.Bornschlogl T. How filopodia pull: what we know about the mechanics and dynamics of filopodia. Cytoskeleton (Hoboken) 2013;70:590–603. doi: 10.1002/cm.21130. [DOI] [PubMed] [Google Scholar]
- 220.Nikkhah M., Edalat F., Manoucheri S., Khademhosseini A. Engineering microscale topographies to control the cell-substrate interface. Biomaterials. 2012;33:5230–5246. doi: 10.1016/j.biomaterials.2012.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Oh S., Brammer K.S., Li Y.S., Teng D., Engler A.J., Chien S., Jin S. Stem cell fate dictated solely by altered nanotube dimension. Proc. Natl. Acad. Sci. U. S. A. 2009;106:2130–2135. doi: 10.1073/pnas.0813200106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Dalby M.J., Gadegaard N., Tare R., Andar A., Riehle M.O., Herzyk P., Wilkinson C.D., Oreffo R.O. The control of human mesenchymal cell differentiation using nanoscale symmetry and disorder. Nat. Mater. 2007;6:997–1003. doi: 10.1038/nmat2013. [DOI] [PubMed] [Google Scholar]
- 223.Viswanathan P., Guvendiren M., Chua W., Telerman S.B., Liakath-Ali K., Burdick J.A., Watt F.M. Mimicking the topography of the epidermal-dermal interface with elastomer substrates. Integr Biol (Camb) 2016;8:21–29. doi: 10.1039/c5ib00238a. [DOI] [PubMed] [Google Scholar]
- 224.Unadkat H.V., Hulsman M., Cornelissen K., Papenburg B.J., Truckenmuller R.K., Carpenter A.E., Wessling M., Post G.F., Uetz M., Reinders M.J., Stamatialis D., van Blitterswijk C.A., de Boer J. An algorithm-based topographical biomaterials library to instruct cell fate. Proc. Natl. Acad. Sci. U. S. A. 2011;108:16565–16570. doi: 10.1073/pnas.1109861108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Reimer A., Vasilevich A., Hulshof F., Viswanathan P., van Blitterswijk C.A., de Boer J., Watt F.M. Scalable topographies to support proliferation and Oct4 expression by human induced pluripotent stem cells. Sci. Rep. 2016;6 doi: 10.1038/srep18948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Zijl S., Vasilevich A.S., Viswanathan P., Helling A.L., Beijer N.R.M., Walko G., Chiappini C., de Boer J., Watt F.M. Micro-scaled topographies direct differentiation of human epidermal stem cells. Acta Biomater. 2019;84:133–145. doi: 10.1016/j.actbio.2018.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Poon J.C.H., Liao Z., Suzuki T., Carleton M.M., Soleas J.P., Aitchison J.S., Karoubi G., McGuigan A.P., Waddell T.K. Design of biomimetic substrates for long-term maintenance of alveolar epithelial cells. Biomater Sci. 2018;6:292–303. doi: 10.1039/c7bm00647k. [DOI] [PubMed] [Google Scholar]
- 228.Oakley C., Brunette D.M. The sequence of alignment of microtubules, focal contacts and actin filaments in fibroblasts spreading on smooth and grooved titanium substrata. J. Cell Sci. 1993;106(Pt 1):343–354. doi: 10.1242/jcs.106.1.343. [DOI] [PubMed] [Google Scholar]
- 229.Clark P., Connolly P., Curtis A.S., Dow J.A., Wilkinson C.D. Cell guidance by ultrafine topography in vitro. J. Cell Sci. 1991;99(Pt 1):73–77. doi: 10.1242/jcs.99.1.73. [DOI] [PubMed] [Google Scholar]
- 230.Ankam S., Suryana M., Chan L.Y., Moe A.A., Teo B.K., Law J.B., Sheetz M.P., Low H.Y., Yim E.K. Substrate topography and size determine the fate of human embryonic stem cells to neuronal or glial lineage. Acta Biomater. 2013;9:4535–4545. doi: 10.1016/j.actbio.2012.08.018. [DOI] [PubMed] [Google Scholar]
- 231.Ankam S., Lim C.K., Yim E.K. Actomyosin contractility plays a role in MAP2 expression during nanotopography-directed neuronal differentiation of human embryonic stem cells. Biomaterials. 2015;47:20–28. doi: 10.1016/j.biomaterials.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 232.Ankam S., Teo B.K.K., Pohan G., Ho S.W.L., Lim C.K., Yim E.K.F. Temporal Changes in Nucleus Morphology, Lamin A/C and Histone Methylation During Nanotopography-Induced Neuronal Differentiation of Stem Cells. Front Bioeng Biotechnol. 2018;6:69. doi: 10.3389/fbioe.2018.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Abagnale G., Steger M., Nguyen V.H., Hersch N., Sechi A., Joussen S., Denecke B., Merkel R., Hoffmann B., Dreser A., Schnakenberg U., Gillner A., Wagner W. Surface topography enhances differentiation of mesenchymal stem cells towards osteogenic and adipogenic lineages. Biomaterials. 2015;61:316–326. doi: 10.1016/j.biomaterials.2015.05.030. [DOI] [PubMed] [Google Scholar]
- 234.Abagnale G., Sechi A., Steger M., Zhou Q., Kuo C.C., Aydin G., Schalla C., Muller-Newen G., Zenke M., Costa I.G., van Rijn P., Gillner A., Wagner W. Surface Topography Guides Morphology and Spatial Patterning of Induced Pluripotent Stem Cell Colonies. Stem Cell Reports. 2017;9:654–666. doi: 10.1016/j.stemcr.2017.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Tan K.K.B., Lim W.W.M., Chai C., Kukumberg M., Lim K.L., Goh E.L.K., Yim E.K.F. Sequential Application of Discrete Topographical Patterns Enhances Derivation of Functional Mesencephalic Dopaminergic Neurons from Human Induced Pluripotent Stem Cells. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-27653-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Soleas J.P., Waddell T.K., McGuigan A.P. Topographically grooved gel inserts for aligning epithelial cells during air-liquid-interface culture. Biomater Sci. 2015;3:121–133. doi: 10.1039/c4bm00237g. [DOI] [PubMed] [Google Scholar]
- 237.Baker B.M., Chen C.S. Deconstructing the third dimension: how 3D culture microenvironments alter cellular cues. J. Cell Sci. 2012;125:3015–3024. doi: 10.1242/jcs.079509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Nelson C.M., Vanduijn M.M., Inman J.L., Fletcher D.A., Bissell M.J. Tissue geometry determines sites of mammary branching morphogenesis in organotypic cultures. Science. 2006;314:298–300. doi: 10.1126/science.1131000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Nelson C.M., Inman J.L., Bissell M.J. Three-dimensional lithographically defined organotypic tissue arrays for quantitative analysis of morphogenesis and neoplastic progression. Nat. Protoc. 2008;3:674–678. doi: 10.1038/nprot.2008.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Mori H., Gjorevski N., Inman J.L., Bissell M.J., Nelson C.M. Self-organization of engineered epithelial tubules by differential cellular motility. Proc. Natl. Acad. Sci. U. S. A. 2009;106:14890–14895. doi: 10.1073/pnas.0901269106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Gjorevski N., Nelson C.M. Endogenous patterns of mechanical stress are required for branching morphogenesis. Integr Biol (Camb) 2010;2:424–434. doi: 10.1039/c0ib00040j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Soleas J.P., D'Arcangelo E., Huang L., Karoubi G., Nostro M.C., McGuigan A.P., Waddell T.K. Assembly of lung progenitors into developmentally-inspired geometry drives differentiation via cellular tension. Biomaterials. 2020;254 doi: 10.1016/j.biomaterials.2020.120128. [DOI] [PubMed] [Google Scholar]
- 243.Wilkinson D.C., Alva-Ornelas J.A., Sucre J.M., Vijayaraj P., Durra A., Richardson W., Jonas S.J., Paul M.K., Karumbayaram S., Dunn B., Gomperts B.N. Development of a Three-Dimensional Bioengineering Technology to Generate Lung Tissue for Personalized Disease Modeling. Stem Cells Transl. Med. 2017;6:622–633. doi: 10.5966/sctm.2016-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Rios Velazquez E., Parmar C., Liu Y., Coroller T.P., Cruz G., Stringfield O., Ye Z., Makrigiorgos M., Fennessy F., Mak R.H., Gillies R., Quackenbush J., Aerts H. Somatic Mutations Drive Distinct Imaging Phenotypes in Lung Cancer. Cancer Res. 2017;77:3922–3930. doi: 10.1158/0008-5472.CAN-17-0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Yang Y., Yin W., He W., Jiang C., Zhou X., Song X., Zhu J., Fei K., Cao W., Jiang G. Phenotype-genotype correlation in multiple primary lung cancer patients in China. Sci. Rep. 2016;6 doi: 10.1038/srep36177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Lakshminrusimha S., Saugstad O.D. The fetal circulation, pathophysiology of hypoxemic respiratory failure and pulmonary hypertension in neonates, and the role of oxygen therapy. J. Perinatol. 2016;36(Suppl. 2):S3–S11. doi: 10.1038/jp.2016.43. [DOI] [PubMed] [Google Scholar]
- 247.Vogel E.R., Britt R.D., Jr., Trinidad M.C., Faksh A., Martin R.J., MacFarlane P.M., Pabelick C.M., Prakash Y.S. Perinatal oxygen in the developing lung. Can. J. Physiol. Pharmacol. 2015;93:119–127. doi: 10.1139/cjpp-2014-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Xi Y., Kim T., Brumwell A.N., Driver I.H., Wei Y., Tan V., Jackson J.R., Xu J., Lee D.K., Gotts J.E., Matthay M.A., Shannon J.M., Chapman H.A., Vaughan A.E. Local lung hypoxia determines epithelial fate decisions during alveolar regeneration. Nat. Cell Biol. 2017;19:904–914. doi: 10.1038/ncb3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Schmiedl A., Roolfs T., Tutdibi E., Gortner L., Monz D. Influence of prenatal hypoxia and postnatal hyperoxia on morphologic lung maturation in mice. PLoS One. 2017;12 doi: 10.1371/journal.pone.0175804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Polosukhin V.V., Cates J.M., Lawson W.E., Milstone A.P., Matafonov A.G., Massion P.P., Lee J.W., Randell S.H., Blackwell T.S. Hypoxia-inducible factor-1 signalling promotes goblet cell hyperplasia in airway epithelium. J. Pathol. 2011;224:203–211. doi: 10.1002/path.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Ghaedi M., Mendez J.J., Bove P.F., Sivarapatna A., Raredon M.S., Niklason L.E. Alveolar epithelial differentiation of human induced pluripotent stem cells in a rotating bioreactor. Biomaterials. 2013;35:699–710. doi: 10.1016/j.biomaterials.2013.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Sanchez-Esteban J., Wang Y., Gruppuso P.A., Rubin L.P. Mechanical stretch induces fetal type II cell differentiation via an epidermal growth factor receptor-extracellular-regulated protein kinase signaling pathway. Am. J. Respir. Cell Mol. Biol. 2004;30:76–83. doi: 10.1165/rcmb.2003-0121OC. [DOI] [PubMed] [Google Scholar]
- 253.Grigoryan B., Paulsen S.J., Corbett D.C., Sazer D.W., Fortin C.L., Zaita A.J., Greenfield P.T., Calafat N.J., Gounley J.P., Ta A.H., Johansson F., Randles A., Rosenkrantz J.E., Louis-Rosenberg J.D., Galie P.A., Stevens K.R., Miller J.S. Multivascular networks and functional intravascular topologies within biocompatible hydrogels. Science. 2019;364:458–464. doi: 10.1126/science.aav9750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Mishra A., Dubash T.D., Edd J.F., Jewett M.K., Garre S.G., Karabacak N.M., Rabe D.C., Mutlu B.R., Walsh J.R., Kapur R., Stott S.L., Maheswaran S., Haber D.A., Toner M. Ultrahigh-throughput magnetic sorting of large blood volumes for epitope-agnostic isolation of circulating tumor cells. Proc. Natl. Acad. Sci. U. S. A. 2020;117:16839–16847. doi: 10.1073/pnas.2006388117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Mair B., Aldridge P.M., Atwal R.S., Philpott D., Zhang M., Masud S.N., Labib M., Tong A.H.Y., Sargent E.H., Angers S., Moffat J., Kelley S.O. High-throughput genome-wide phenotypic screening via immunomagnetic cell sorting. Nat Biomed Eng. 2019;3:796–805. doi: 10.1038/s41551-019-0454-8. [DOI] [PubMed] [Google Scholar]
- 256.Nitta N., Sugimura T., Isozaki A., Mikami H., Hiraki K., Sakuma S., Iino T., Arai F., Endo T., Fujiwaki Y., Fukuzawa H., Hase M., Hayakawa T., Hiramatsu K., Hoshino Y., Inaba M., Ito T., Karakawa H., Kasai Y., Koizumi K., Lee S., Lei C., Li M., Maeno T., Matsusaka S., Murakami D., Nakagawa A., Oguchi Y., Oikawa M., Ota T., Shiba K., Shintaku H., Shirasaki Y., Suga K., Suzuki Y., Suzuki N., Tanaka Y., Tezuka H., Toyokawa C., Yalikun Y., Yamada M., Yamagishi M., Yamano T., Yasumoto A., Yatomi Y., Yazawa M., Di Carlo D., Hosokawa Y., Uemura S., Ozeki Y., Goda K. Intelligent Image-Activated Cell Sorting. Cell, 175. 2018;e213:266–276. doi: 10.1016/j.cell.2018.08.028. [DOI] [PubMed] [Google Scholar]
- 257.Kupfer M.E., Lin W.H., Ravikumar V., Qiu K., Wang L., Gao L., Bhuiyan D., Lenz M., Ai J., Mahutga R.R., Townsend D., Zhang J., McAlpine M.C., Tolkacheva E.G., Ogle B.M. In Situ Expansion, Differentiation and Electromechanical Coupling of Human Cardiac Muscle in a 3D Bioprinted, Chambered Organoid. Circ. Res. 2020 doi: 10.1161/CIRCRESAHA.119.316155. ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Sharma R., Smits I.P.M., De La Vega L., Lee C., Willerth S.M. 3D Bioprinting Pluripotent Stem Cell Derived Neural Tissues Using a Novel Fibrin Bioink Containing Drug Releasing Microspheres. Front Bioeng Biotechnol. 2020;8:57. doi: 10.3389/fbioe.2020.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Skylar-Scott M.A., Uzel S.G.M., Nam L.L., Ahrens J.H., Truby R.L., Damaraju S., Lewis J.A. Biomanufacturing of organ-specific tissues with high cellular density and embedded vascular channels. Sci. Adv. 2019;5 doi: 10.1126/sciadv.aaw2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Todhunter M.E., Jee N.Y., Hughes A.J., Coyle M.C., Cerchiari A., Farlow J., Garbe J.C., LaBarge M.A., Desai T.A., Gartner Z.J. Programmed synthesis of three-dimensional tissues. Nat. Methods. 2015;12:975–981. doi: 10.1038/nmeth.3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Plasschaert L.W., Zilionis R., Choo-Wing R., Savova V., Knehr J., Roma G., Klein A.M., Jaffe A.B. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature. 2018;560:377–381. doi: 10.1038/s41586-018-0394-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Cahan P., Li H., Morris S.A., Lummertz da Rocha E., Daley G.Q., Collins J.J. CellNet: network biology applied to stem cell engineering. Cell. 2014;158:903–915. doi: 10.1016/j.cell.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Schupp J.C., Khanal S., Gomez J.L., Sauler M., Adams T.S., Chupp G.L., Yan X., Poli S., Zhao Y., Montgomery R.R., Rosas I.O., Dela Cruz C.S., Bruscia E.M., Egan M.E., Kaminski N., Britto C.J. Single Cell Transcriptional Archetypes of Airway Inflammation in Cystic Fibrosis. Am. J. Respir. Crit. Care Med. 2020 doi: 10.1164/rccm.202004-0991OC. ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Xu Y., Mizuno T., Sridharan A., Du Y., Guo M., Tang J., Wikenheiser-Brokamp K.A., Perl A.T., Funari V.A., Gokey J.J., Stripp B.R., Whitsett J.A. Single-cell RNA sequencing identifies diverse roles of epithelial cells in idiopathic pulmonary fibrosis. JCI Insight. 2016;1 doi: 10.1172/jci.insight.90558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.McDonough J.E., Ahangari F., Li Q., Jain S., Verleden S.E., Herazo-Maya J., Vukmirovic M., DeIuliis G., Tzouvelekis A., Tanabe N., Chu F., Yan X., Verschakelen J., Homer R.J., Manatakis D.V., Zhang J., Ding J., Maes K., De Sadeleer L., Vos R., Neyrinck A., Benos P.V., Bar-Joseph Z., Tantin D., Hogg J.C., Vanaudenaerde B.M., Wuyts W.A., Kaminski N. Transcriptional regulatory model of fibrosis progression in the human lung. JCI Insight. 2019;4 doi: 10.1172/jci.insight.131597. [DOI] [PMC free article] [PubMed] [Google Scholar]