Abstract
As obligate intracellular parasites with limited coding capacity, RNA viruses rely on host cells to complete their multiplication cycle. Viral RNAs (vRNAs) are central to infection. They carry all the necessary information for a virus to synthesize its proteins, replicate and spread and could also play essential non-coding roles. Regardless of its origin or tropism, vRNA has by definition evolved in the presence of host RNA Binding Proteins (RBPs), which resulted in intricate and complicated interactions with these factors. While on one hand some host RBPs recognize vRNA as non-self and mobilize host antiviral defenses, vRNA must also co-opt other host RBPs to promote viral infection. Focusing on pathogenic RNA viruses, we will review important scenarios of RBP-vRNA interactions during which host RBPs recognize, modify or degrade vRNAs. We will then focus on how vRNA hijacks the largest ribonucleoprotein complex (RNP) in the cell, the ribosome, to selectively promote the synthesis of its proteins. We will finally reflect on how novel technologies are helping in deepening our understanding of vRNA-host RBPs interactions, which can be ultimately leveraged to combat everlasting viral threats.
Keywords: Virology, RNA viruses, RNA biology, Viral Translation, Viral RNA sensing, Innate Immunity, Technology
1. Introduction
Viruses are obligate intracellular parasites that strictly rely on host cells to translate and amplify their genomes [[1], [2], [3]]. Viral RNAs (vRNAs) play a central role during infection as they bear all the necessary information for a virus to express its proteins, replicate and spread. Unlike DNA viruses where the messenger RNA must be first transcribed for the synthesis of viral proteins, RNA viruses use their RNA molecules both for protein synthesis and as replication templates. Viruses encode an array of vRNA-binding proteins (vRBPs), essential for different steps in the viral lifecycle, comprising translation, synthesis, packaging of the viral genome and cell-to-cell spread. Importantly, vRNA is never really naked in the cellular milieu. Apart from its interactions with vRBPs, it has evolved to co-opt specific sets of host encoded RBPs. This is illustrated by the variety of described strategies that vRNAs use to hijack key cellular machineries and evade the host’s defense arsenal. Unlike vRBPs that promote infection, host encoded RBPs can either have pro- or antiviral functions. Indeed, a large body of work produced during the last decades describe the involvement of host encoded RBPs in almost every known stage of vRNA lifecycle.
Certain host RBPs, ordinarily functioning in cellular tasks, are repurposed by vRNAs to guide them through the viral lifecycle, including genome translation, synthesis, modification, localization and packaging. On the other hand, many host RBPs have evolved dedicated antiviral functions, ranging from the recognition of the invading vRNA to the restriction of viral replication. The ability of vRNA to either subvert or get antagonized by cellular RBPs determine the outcome of a viral infection. Indeed, vRNA-RBPs interactions could dictate the permissiveness of certain cell types to infection, host range, tissue tropism, viral evolution, efficiency of viral replication, pathology of infection and immune clearance [4]. vRNAs interact with numerous and very diverse RBPs during infection. This is illustrated by the rich literature on the subject that has yielded many discoveries in the last decades. For example, the study of intimate vRNA-RBP interactions shaped our understanding of how RNA interference (RNAi) functions as the main antiviral defense system in plants and invertebrates, and how bacteria defend themselves against invading phages through the CRISPR system [5,6].
Herein, we review the mechanisms at play when RNA viruses infect their animal hosts, taking a vRNA-centric view. Focusing on viruses that are important human pathogens, we will describe how cellular RBPs act on early steps after viral entry, driving mechanisms of vRNA recognition, degradation and modification to limit or to promote viral replication and spread. We will further describe a number of crucial mechanisms during which vRNA hijacks the ribosome to preferentially translate the viral program. Finally, we will reflect on how new emerging techniques are allowing to get a better grasp on the diversity of host RBPs-vRNA interactions. Elucidating the mechanisms by which RBP-vRNA interactions influence viral infection, is advancing our understanding of cell biology by revealing unforeseen biological knowledge and may also provide new targets for host-directed antiviral therapies [7]. Indeed, recent Corona, Ebola and Zika virus outbreaks remind us that new innovative antiviral approaches are clearly needed, particularly for emerging RNA viruses with important epidemic potential [[8], [9], [10]].
2. Detection and elimination of vRNA
Despite the fact that the molecular composition of RNA is universal throughout life kingdoms, host immune pathways are able to differentiate and recognize non-self nucleic acids based on specific features, structures and modifications. Moreover, despite the molecular mimicry set by RNA viruses to resemble cellular mRNAs and escape host recognition, the viral nucleic acid still needs to embark on a long journey through a hostile cell environment and must overcome the obstacles put in place by the host antiviral system in order to be translated and replicated.
2.1. Recognition of vRNA structure by host sensors
Cellular innate immunity comprises a rather sophisticated set of host factors that discriminate molecular alterations with a high specificity and restrict RNA virus infection through direct or indirect activity on the vRNA. In vertebrate animals, a variety of cellular sensors have evolved to detect foreign RNA sensing to activate the innate immune response, which involves transcriptional and post-transcriptional activation of genes of interferon-stimulated genes (ISGs). Pattern recognition receptors (PRRs) are host-encoded proteins that sense molecular features of vRNA molecules which are generally absent in the majority of cellular RNAs. Multiple molecular receptors are involved in the recognition of these so-called pathogen-associated molecular patterns (PAMPs) [11,12]. A major molecular signature and weakness of all RNA viruses is the long double stranded (ds) RNA molecules that are generated during replication. The sensing of dsRNA is believed to be sequence-independent and rather depends on the distinct molecular features of the viral dsRNA structure mostly absent in uninfected host transcriptome [13].
Among the first cellular proteins that detect the invading virus are the Toll-like receptors (TLRs) (Fig. 1 ), which are transmembrane glycoproteins that are constitutively expressed or pathogen-induced, located on the plasma membrane or intracellular endosomes which are preferential entry routes for vRNAs (reviewed in [14,15]). TLRs share strong similarities in their structures and organization. Their N-terminal region recognizes ligands thanks to its extracellular or luminal domains and is connected to the C-terminal cytoplasmic domains (CTDs) by a single membrane-spanning domain. Among the 10 TLRs identified in humans so far, TLR7 and TLR8 recognize ssRNAs and TLR3 recognizes dsRNAs [12,16,17]. In TLR3, the N-terminal region recognizes the ligand, dsRNA, in the lumen of endosomes whereas the C-terminal signalling domain resides in the cytoplasm, and upon ligand recognition, TLR3 initiates the signalling process that culminates in transcriptional induction of specific immune genes. TLR3 was believed to be a major sentinel against viral infections since it responds to a common by-product of viral replication ( Table 1 ). For instance, encephalomyocarditis virus (EMCV) infection leads to a TLR3-dependent innate stress response, which is involved in mediating protection against virus-induced myocardial injury [18]. Similarly, TLR3 recognizes dengue virus (DENV), limiting its replication [19], while Zika virus (ZIKV)-mediated TLR3 activation was shown to deplete neural progenitor cells [20]. Indeed, the protective versus pathogenic role of TLR3 in viral pathogenesis is debated [21].
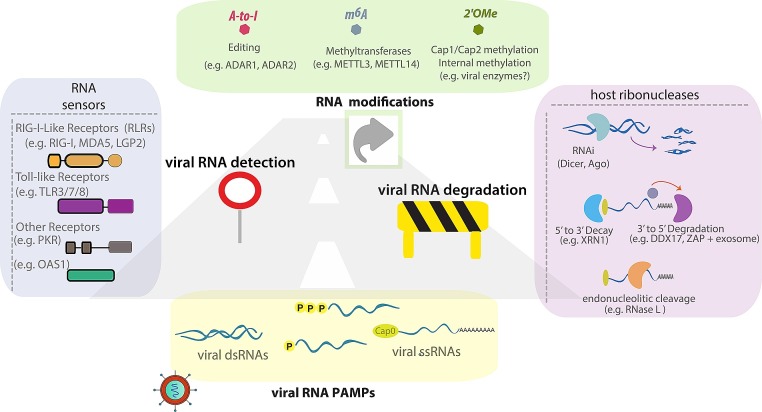
Fig. 1.
Detection, Modification and Degradation of vRNA by Cellular RBPs.
The figure depicts different vRNA structures recognized by host RBPs. vRNA is sensed by different families of receptors (e.g. RLRs, TLRs, PKR). vRNA is edited by ADARs and methylated by host METTL proteins (m6A) or viral methyl transferases (2′-O-me). vRNA can also be degraded by 5′-3′ or 3′-5′ exonucleases (e.g. XRN1 and RNA exosome respectively) or by endonucleases (e.g. Dicer).
Table 1.
Sensor RBPs involved in host responses to RNA viruses.
| Host RBPs | Recognized structure | Molecular function | Effect on viral infection | Examples of virus | References |
|---|---|---|---|---|---|
| SENSORS | |||||
| RIG-I | triphosphate (PPP) or diphosphate (PP) at the 5′-end of RNA molecules with partial base-pairing to a complementary strand | Induction of Type I IFN | Antiviral | EBOV, NiV, NDV, IAV, DENV, ZIKV, SeV, VSV, JEV |
[27,28,29,30] |
| MDA5 | uninterrupted RNA duplexes longer than a few hundred base pairs | Induction of Type I IFN | Antiviral | EMCV, CVB3, and other picornaviruses; DENV, WNV, MHV |
[28,36,238,239] |
| TLR3 | Long dsRNA | Induction antiviral and inflammatory response | Antiviral | EMCV, DENV, ZIKV | [18,19,20] |
| SENSORS/ EFFECTORS | |||||
| PKR | Long dsRNA (>30bp) |
Phosphorylation EIF2alpha and inhibition protein synthesis | Antiviral | Virtually all viruses generating a dsRNA intermediate | [44,45,240] |
| OAS and RNaseL | Long dsRNA | Production 2′,5′-oligoadenylates/ Degradation viral RNA | Antiviral | WNV, SINV, HCV | [83,241] |
| DICER | Long dsRNA | antiviral degradation and immunity mediated by small RNAs | Antiviral | EMCV, NoV, HEV71, IAV, ZIKV | [88,91,92,242] |
| ADAR | Long dsRNA | Adenosines to inosines editing; Suppression innate immunity |
Antiviral/ Proviral |
Virtually all viruses generating a dsRNA intermediate | [101] |
| METTL3, METTL14 | Unknown | N6-methyladenosine (m6A) writer; Suppression innate immunity |
Proviral | HCV, YFV, ZIKV, DENV, WNV, HMPV | [106,107] |
| IFIT1 | 5′ triphosphate RNAs and non-2′-O methylated capped transcripts |
Inhibition protein synthesis by out competing the cellular translation initiation factors | Antiviral | JEV, WNV, HCoV, VEEV | [97,116,118] |
Ebola virus, EBOV; Nipah virus, NiP; Newcastle disease virus, NDV; Hepatitis C virus, HCV; Influenza A virus, IAV; Encephalomyocarditis virus, EMCV; Vesicular stomatitis virus, VSV; Dengue virus, DENV; West Nile virus, WNV; Zika virus, ZIKV; Coxsackievirus B3, CVB3; Murine coronavirus mouse hepatitis virus, MHV; Nodamura virus, NoV ; Human enterovirus 71, HEV71. human coronavirus, HcoV ; Japanese Encephalitis virus, JEV ; Venezuelan Equine Encephalitis virus, VEEV ; Sindbis Virus, SINV ; Yellow Fever Vius, YFV ; human metapneumovirus (HMPV).
Another group of molecular sentinels that senses foreign RNA is represented by the IFN-induced intracellular cytosolic receptors, named Retinoid acid-inducible gene I (RIG-I)-like receptors (RLRs), which comprise DExD/H-box helicases such as RIG-I (or DDX58), melanoma differentiation-associated protein 5 (MDA5 or IFIH1) and laboratory of genetics and physiology 2 (LGP2 or DHX58) [22] (Fig. 1, Table 1). All RLRs possess a central DExD/H-box RNA helicase domain and a CTD which are necessary for detection of foreign RNAs. RIG-I and MDA5 have as well two N-terminal caspase activation and recruitment domains (CARDs), which mediate downstream signal transduction, while LGP2 lacks them. Although RIG-I and MDA5 share similar structural composition, their ability to detect vRNA is not redundant but specific [22] (Table 1). RIG-I monitors the 5′ ends of RNA molecules via its CTD and helicase domain in order to initiate cytokine production in response to a wide range of viruses. triphosphate (PPP) or diphosphate (PP) at the uncapped 5′-end of RNA molecules with partial base-pairing to a complementary strand of RNA molecules are recognized and required for RIG-I activation [[23], [24], [25], [26]]. The typical structure of the panhandle of negative-strand viral genomes confers full RIG-I ligand activity. RIG-I recognizes many single stranded negative RNA virus families, such as Paramyxoviridae, Rhabdoviridae, Orthomyxoviridae, Bunyaviridae and Filoviridae [[27], [28], [29]]. RIG-I also detects some positive-sense RNA viruses such as dengue and Zika viruses of the Flaviviridae family, by recognition of nascent viral genomes containing 5′-PPP groups prior to capping [30].
MDA5 senses both RNA length and secondary structure by recognizing uninterrupted RNA duplexes longer than a few hundred base pairs [31] and forming filaments around dsRNA to initiate signalling [32]. MDA5 is required for immunity against several classes of viruses, including single-stranded positive RNA viruses such as picornaviruses [28,33,34], flaviviruses [35] and coronaviruses [36] (Table 1).
The third RLR family member LGP2 lacks the amino-terminal tandem CARDs and thus lacks signal-transducing activity. Several studies indicate a disparate regulatory role for LGP2 in either triggering or dampening the innate immune signalling pathways following RNA virus infection [37]. On one hand, LGP2 can function as a feedback inhibitor of RIG-I by sequestrating double-stranded but not single-stranded RNAs. LGP2 negatively regulates Sendai Virus (SV)-induced IRF-3 or NF-κB signaling acting as a natural inhibitor of antiviral responses, and as a repressor of RIG-I signaling [38]. However, LGP2 has a positive effect on MDA5-mediated antiviral response. LGP2-deficient mice exhibited a defect in type I IFN production in response to infection by the encephalomyocarditis virus, the replication of which activates MDA5-dependent innate immune response [39]. Moreover, a recent study indicated that LGP2 can inhibit antiviral RNAi in mammals by competing with and preventing Dicer-mediated processing of dsRNAs into siRNAs both in vitro and in cells [40].
Interestingly, other DExD/H-box helicases, such as DDX17, DDX21, DDX6 or DDX56, are also emerging as important sentinels that influence viral sensing in multiple ways. These helicases have been shown to bind vRNAs, acting either as antiviral effectors or contributing to viral replication [41].
The presence and replication of viral nucleic acids in vertebrate cells can also induce the activation of other antiviral enzymes that are both sensors and effectors ( Table 1 ). These also recognize viral signatures; however, their main function is not necessarily to only induce a transcriptional immune response, but rather to directly attack vRNA by degrading it or inhibiting its translation. For this reason, these are not usually referred to as receptors [12,42,43]. Among them, double-stranded RNA (dsRNA) activated protein kinase R (PKR; also known as eIF2AK2) or adenosine deaminase and 2′-5′-oligoadenylate synthetase class of enzymes (OASs), can bind dsRNA in a sequence independent manner, recognizing the structure as a PRR (Fig. 1, Table 1) 43].
First, PKR is a serine-threonine kinase constitutively expressed in all tissues at a basal level as an inactive monomer. Viral dsRNA binding to the two N-terminal RNA binding motifs induces a conformational change that allows ATP binding to the C-terminal kinase domain. PKR can be activated by dsRNA from reoviruses [44] but can also recognize dsRNA from replication intermediates of + RNA viruses [45]] (Table 1). Moreover, it has been shown that highly structured RNA elements from retroviruses such as human immunodeficiency virus (HIV) transactivation-response region (TAR) RNA hairpins [46] or dsRNA formed by the antiparallel mRNA transcripts of some DNA viruses can also activate PKR [47]. Upon dsRNA binding PKR dimerizes and auto- phosphorylates threonine residues in its activation domains, thereby stabilizing the dimers and increasing kinase activity [48]. This in turn leads to a series of events that inhibit viral translation and replication, as will be detailed later on in this review. Interestingly, PKR is an interferon stimulated gene (ISG) itself and its transcription can be up-regulated upon type I interferon production by virus-infected cells, stimulating its accumulation in neighboring cells. PKR activation following infection of interferon-primed neighboring cells inhibits global protein synthesis and reduces viral spread, making this activation a key player in the innate response to viruses [49]. Therefore, PKR can be considered a major molecular sentinel of antiviral innate immunity, recognizing a diverse set of stress-inducing stimuli and mounting an appropriate antiviral state amongst which inhibition of translation initiation. Given its importance in antiviral defense, it is not surprising to note that viruses have evolved countermeasures against PKR activation. For example, some DNA viruses such as Adenoviruses or Epstein-Barr virus encode small RNA decoys (e.g. VA RNA or EBV EBERs) that can bind PKR but do not activate it. RNA elements present in RNA viruses such as hepatitis C virus (HCV) IRES or HIV-1 TAR-cis elements have also been shown to act as PKR pseudosubstrates [[50], [51], [52]]. Other viruses inhibit PKR function by directly targeting it for degradation. For instance, Rift Valley Fever Virus (RVFV) Non-structural S (NSs) protein recruits the E3 ligase SCF (SKP1-CUL-F-box) (FBXW11) to PKR and promotes its early degradation through the proteasome [53] (Fig. 3).
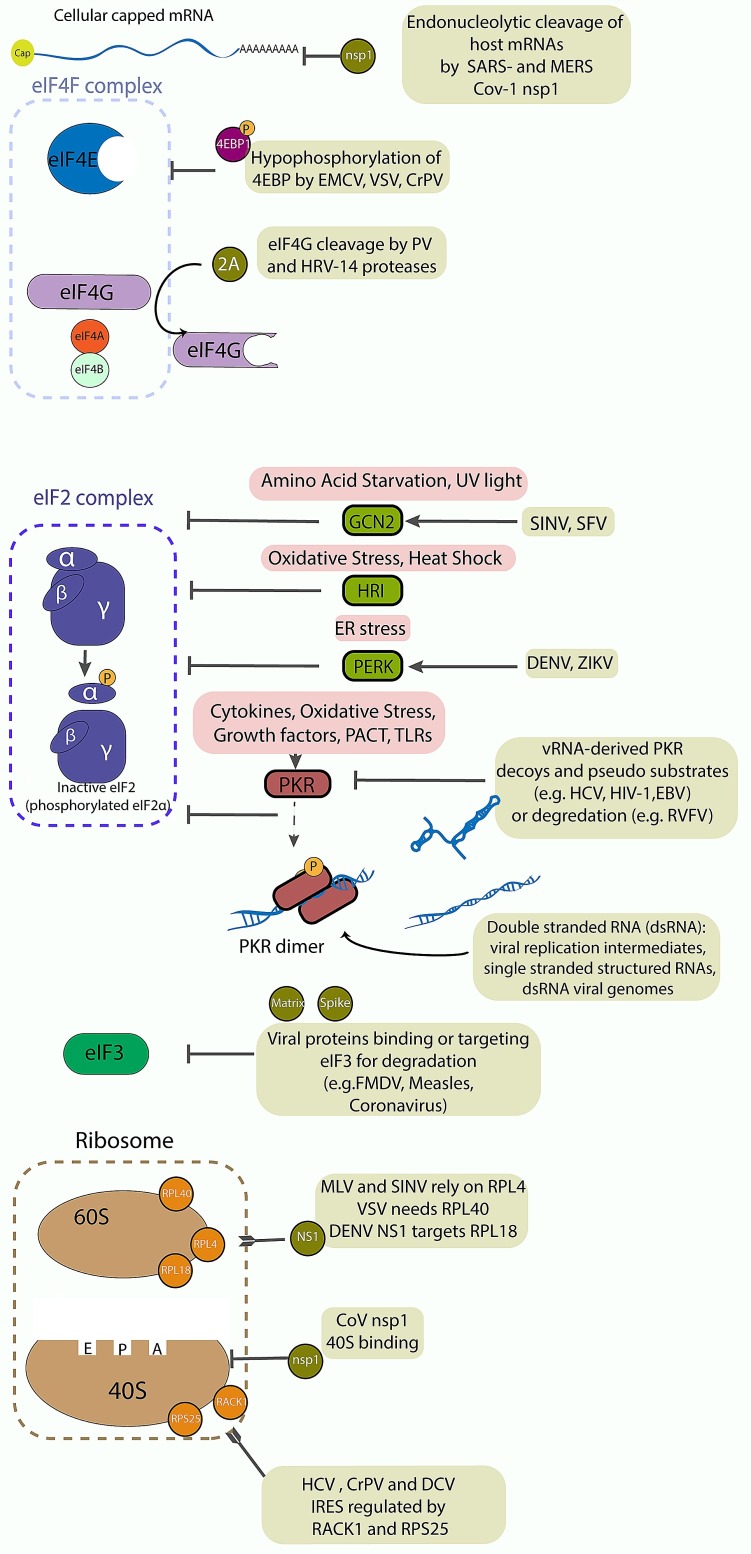
Fig. 3.
Cellular translation initiation arrest upon viral infection. The figure depicts different modes of translation initiation arrest caused by viral infection. In green, viral proteins or RNA structures that influence translation initiation are listed.
These can either act on endogenous mRNAs, eIF4F, eIF2, eIF3 or the ribosomal subunits. In red are listed the four known cellular eIF2alpha kinases: GCN2, HRI, PERK and PKR.
The second known class of dsRNA sensors and effectors is represented by the 2′,5′-oligoadenylate synthetases (OASes) (Fig. 1, Table 1), which also act as PRRs for the detection of many RNA viruses [54]. The four human OAS genes, OAS1, OAS2, OAS3 and OASL (OAS-like), are constitutively expressed at low levels in the cytoplasm and are activated in response to viral dsRNA. As an example, OAS1 protein accumulates in the cytoplasm as an inactive monomer [55] and following activation by viral dsRNA, it forms a tetramer that synthesizes 2′,5′- oligoadenylates (2-5 As). These 2-5As activate the ribonuclease L (RNaseL) to cleave cellular and viral RNAs and supress viral replication (see below). Overall, the functional redundancy, provided by multiple host proteins that recognize different features of vRNAs, is a successful strategy for the host to fight and contain the infection [56]. Similar to PKR, the OAS-RNaseL axis can be antagonized by some RNA viruses. For example, certain Corona and Rotaviruses encode proteins belonging to the phosphodiesterase family, able to cleave 2–5 A molecules, thereby preventing activation of RNaseL [57].
2.2. Antiviral RNA degradation by host ribonucleases
In concert with innate immune sensors linked to IFN activation, mammalian cells can also depend on intrinsic factors that participate in the direct antiviral response to non-self nucleic acids. Accumulating evidence suggests that the cellular RNA surveillance pathway restricts viral infection by modulating vRNA stability [58]. The presence of specific molecular features on the vRNA and the binding of trans-acting antiviral host RBPs can drive vRNA through the host mRNA quality control pathways (Fig. 1). For instance, in mammalian cells, the non-sense mediated decay (NMD) pathway has recently been shown to function in the restriction of positive-sense (+) RNA viruses such as alphaviruses in mammalian cells [59]. In fact, alphavirus RNA genome resembles cellular mRNAs and is translated into non-structural viral proteins (nsPs) but has an untranslated 3′ UTR, much longer than typical cellular mRNAs, which is detected and degraded by the NMD pathway [59]. The 5′ to 3′ RNA decay machinery can also directly target vRNA for degradation in the cytoplasm. In this pathway, the monomethyl guanosine (m7G) cap structure is cleaved from mRNAs by decapping enzymes exposing the 5′ monophosphorylated product to progressive 5′→3′ exoribonucleolytic degradation by Xrn1. The Hepatitis C virus (HCV) genomic RNA, which lacks a 5′ cap and is therefore susceptible to Xrn-1 mediated decay, uses an unusual mechanism to overcome this pathway [60]. Binding of the liver-abundant miR-122 to the 5′UTR of HCV in association with Argonaute-2 (Ago2) protein stabilizes the vRNA and slows Xrn1 decay of the viral genome in infected cells [60,61] (Fig. 1). Other Flaviviridae are able to take advantage of their susceptibility to Xrn-1 mediated vRNA decay. Dengue, West Nile, Yellow fever or Zika vRNAs contain a highly structured 300–700 bp-long non-coding RNA in their 3′end. An incomplete 5′-3′ degradation of the viral genome by the cellular XRN1 exonuclease produces the so called small flaviviral RNA (sfRNA). During RNA degradation, XRN1 is stalled at the 3′UTR extremity of flaviviral vRNAs, more precisely at stem-loops/pseudoknots, causing the accumulation of different species of sfRNA. Although, sfRNA does not seem to have a direct role in viral replication per se, it contributes to viral pathogenicity in vivo partly by interfering or evading innate immune responses, by sequestering cellular RBPs and inhibiting their endogenous functions [[62], [63], [64], [65], [66], [67], [68], [69], [70]]. Interestingly, sfRNA seems to function in a species-specific manner. While it facilitates replication and pathogenesis by inhibiting IFN-signalling in mammalian hosts, a recent report shows that ZIKV sfRNA possess an anti-apoptotic function that helps the virus to disseminate and reach saliva in mosquito hosts, thus promoting productive infection and transmission [71]. It has also been proposed that sfRNA could be a suppressor of RNAi [72].
In the 3′ decay pathway, deadenylated mRNA degradation is mediated by a cytoplasmic multisubunit 3′-to-5′ exoribonuclease complex, namely the RNA exosome (Fig. 1). This degradation pathway is emerging as a crucial effector for recognition and degradation of specific vRNAs [73]. The exosome interacts with a variety of RBPs, some of which are exported to the cytoplasm in response to viral infection and serve as adaptors to specifically target particular RNAs. Several antiviral RBPs have been found to interact with the exosome, suggesting that their mechanism of action may involve exosomal degradation. For instance, the conserved RNA helicase DDX17 translocates from the nucleus to the cytoplasm upon Rift Valley Fever Virus (RVFV) infection, binds a specific stem-loop structure in the vRNA and recruits the exosome to degrade the vRNA [74]. Another example, is the zinc-finger antiviral protein (ZAP), which binds vRNAs containing a ZAP response element (ZRE) and induces RNA degradation via interaction of its N-terminal domain with host decay machinery mediated [75] (Fig. 1). ZAP is constitutively expressed in many cell types and it is induced upon pathogen-sensing and in response to interferon. ZAP exhibits broad antiviral activity against a broad spectrum of viruses, such as alphaviruses, enteroviruses, filoviruses, influenza and porcine reproductive and respiratory virus [42] but also retroviruses [76]. For instance, ZAP binds regions of HIV-1 viral RNA containing CpGs and induce degradation of viral RNAs via interaction with the essential cellular cofactor KHNYN [77]. Moreover, ZAP antiviral activity has been linked to its subcellular localization in stress granules, which are induced upon infection by sindbis virus (SINV) for example [78]. Interestingly, TRIM25, an E3 ubiquitin ligase with no previous known role in RNA binding is emerging as a regulator of many antiviral factors, including ZAP [79,80].
Other ribonucleases can exhibit broad-spectrum antiviral effects through viral RNA binding and degradation. The ribonuclease MCPIP1 is involved in the suppression of Japanese encephalitis virus (JEV) and DENV, but also other positive-sense RNA viruses, such as SINV, HCV and EMCV, and negative-sense RNA virus, such as influenza virus [81,82].
Finally, two conserved endoribonucleases have been shown to play a major antiviral role in different animal species, namely RNase L and Dicer (Fig. 1, Table 1). RNase L is composed of three major domains: an N-terminal regulatory ankyrin repeat domain (ARD), a protein kinase (PK)-like domain, and a C-terminal ribonuclease domain (RNASE). In the absence of 2–5 A, the ARD represses the RNASE domain. Binding of 2-5As, generated by OASes, to the ARD alters the conformation of RNase L, thereby exposing protein–protein interaction domains and releasing the RNASE domain from internal inhibitory sequence, triggering its dimerization and activation. RNase L cleaves single-stranded RNAs at the UU/UA dinucleotides, which are relatively rare in the coding regions of cellular mRNAs compared to viral RNAs [55]. In the case of HCV infection, RNase L degradation of vRNA produces small RNA cleavage products with 3′-monophosphate (3′-p) and 5′-hydroxyl (5′-OH) at their termini, which fold back into potent PAMPs recognized by RIG-I, amplifying the innate immune responses [83]. Interestingly, some HCV genotypes escape RNaseL cleavage by accumulating silent mutations at UA and UU dinucleotides, which are preferentially targeted by the nuclease [84].
The RNase III endoribonuclease Dicer is active against different RNA viruses in both vertebrate and invertebrate animals (Table 1) [6,[85], [86], [87], [88], [89]]. RNA interference (RNAi) is a primordial form of antiviral immunity mediated by small RNAs and is the major immune defence in insects, plants and nematodes species [12,90]. In the antiviral RNAi pathway, Dicer is able to bind and cleave viral dsRNAs into ∼21 nt-long small interfering RNAs (siRNAs). The siRNAs are subsequently loaded into the Argonaute-containing RNA Induced Silencing Complex (RISC) and program RISC to target vRNA in a sequence-specific manner, inducing vRNA degradation or translational arrest. Although evidence of Dicer activity against certain RNA viruses have been reported in undifferentiated mouse stem cells [88], and in some other specific context [91,92], the physiological antiviral role of RNAi and its interplay with the interferon response in vertebrate animals is still an area of active debate [[93], [94], [95]].
3. Hide and seek: modifications on vRNA to trick the infected cell
In recent years, it has been revealed that the RNA code is much more complex than the primary RNA sequence. In particular, covalent RNA base and sugar modifications recently emerged as an additional layer in the regulation of gene expression. Such modifications can change the RNA structure and its interaction with other RNAs or proteins, thereby regulating important steps in RNA metabolism including splicing, RNA stability and translation. These RNA modifications are also utilized by host cells as a way to distinguish self-RNAs from vRNAs [[96], [97], [98]]. It is therefore not surprising that viruses evolved strategies to modify their own genomes and transcripts or hijack dedicated cellular enzymes responsible for these modifications. Here we briefly describe three RNA modifications known to be present on vRNAs (Adenosine-to-Inosine editing, N 6-Methyladenosine (m6A) and 2′-O-methylation) [[88], [89], [90]]. We expect that other RNA modifications may have the ability to also influence vRNA stability, translation and immune potential.
3.1. Adenosine -to-Inosine editing
Deamination of adenosines to inosines (A-to-I editing) is a covalent RNA modification occurring on dsRNA structures, by the adenosine deaminase acting on RNA (ADAR) enzymes (Fig. 1, Table 1). Among the three mammalian ADARs, ADAR1 is expressed in its constitutive nuclear form ADAR1p110 and the IFN-inducible cytoplasmic one, ADAR1p150. Both enzymes consist of a C-terminal deaminase domain, three consecutive dsRNA binding motifs and one or two copies of N-terminal Z-DNA binding domains [99]. The consequence of the RNA modification by ADAR1 is two-fold: first, if editing takes place in coding sequences, inosines will be misinterpreted by the translation machinery and will be read as guanosines. Second, A-to-I conversion have the capacity to destabilize dsRNA structures by changing the Watson-Crick base pairing and impairing recognition by cytoplasmic antiviral receptors, such as RLRs, PKR and OAS1. For these reasons, ADAR proteins play a pivotal role in masking self RNAs against innate immune detection by selective labelling [100].
Given the opposite effects of ADAR1 and antiviral sensors in the control of innate immune responses to endogenous duplex RNA, the A-to-I editing contribution in antiviral defence remains unclear. A bias toward A-to-G mutations dependent on ADAR has been described for a wide range of viruses [101]. This could potentially lead to the synthesis of dysfunctional viral proteins and destabilize key RNA structures such as dsRNA replication intermediates. Although several studies have reported high A-to-I editing levels indicative of a potential mutagenic and antiviral effect, ADAR activity seems to be rather proviral due to its dampening effects on the innate immune system [101]. One clear example where ADAR plays a proviral role is the case of Hepatitis D virus (HDV). HDV contains only one ORF responsible of expressing the HDV Delta antigen (HDAg), essential for HDV replication and spread. Two forms of HDAg are expressed: a short form S-HDAg and one that is 19 amino acid longer (L-HDag). HDV needs both to be expressed in order to replicate and spread and this depends on ADAR1. In fact, ADAR1 recognizes a specific structure on the rod-like RNA of HDV antigenomic RNA and converts adenosine to inosine. Replication of this mutated RNA leads to the substitution of guanosine in its genome, specifically leading to the replacement of an amber codon (UAG) for the small Delta antigen by a tryptophan codon (UGG) which produces a protein that is 19 amino acid longer. The synthesis of this longer protein is crucial for HDV spread, therefore ADAR1 is a proviral host RBP essential for HDV infection [102].
3.2. m6A-methylation
In mammalian cells, N 6-methyladenosine (m6A) is the most prevalent and dynamically modulated internal mRNA modification. Such a modification is reversible via the activity of two key enzyme classes: methyltransferases or “writers” (e.g. Methyltransferase Like 3 (METTL3) and 14 (METTL14)) and demethylases or “erasers” (e.g. (FTO) and ALKBH5) (Fig. 1, Table 1). The m6A writers and erasers can alter the cellular RNA fate by destabilizing local RNA structures or by affecting protein binding to modulate host RNA stability or translation efficiency. In particular, the m6A readers such as YTH domain-containing family proteins (YTHDF1–YTHDF3) play crucial roles [103]. Recent reports show that m6A could be a regulator of the immune system, playing a role in the discrimination between endogenous and exogenous RNAs [104]. Indeed, several studies have identified m6A modifications on a variety of vRNAs and this enhances global viral gene expression by increasing RNA stability and translation [96,105]. Interestingly, mapping of m6A on the RNA genomes of Flaviviridae, including dengue, Zika, yellow fever, and West Nile virus, identified conserved regions modified by m [6]A, suggesting that this modification is a conserved regulatory mark across Flaviviridae genomes [106]. Another case where m6A appears to function in innate immune discrimination between self and non-self RNAs is given by the human metapneumovirus (HMPV) [107]. m6A methylation of vRNA promotes HMPV replication and gene expression. In the absence of m6A residues on the vRNA, viral infection is compromised due to a RIG-I-dependent increase of type I interferon [107,108]. This notion is reinforced by the recent report of the effect of the loss of the m6A writer METTL3 in the mouse hematopoietic system. Indeed, METTL3 knock-out results in the upregulation of MDA5-RIG-I, PKR and OAS-RNase L pathways due to the aberrant accumulation of endogenous dsRNA [109]. Therefore, m6A modification appears to prevent formation of certain dsRNA, so it would be interesting in future work to check whether this has implications during viral infection.
3.3. 2′O-methylation
The 2′O- methylation (2′O-me) modification is a highly abundant chemical modification found in the RNA of virtually all eukaryotic species, whereby a methyl group is added to the 2′-OH of the ribose ring by cellular 2′-O-methyltransferases (2′O-MTase) (Fig. 1). 2′O-me is enriched in the first and second transcribed nucleotides next to the N7 methylated 5′ cap structure of higher eukaryote mRNAs. While N-7-methylation is important for stability and translation of the mRNA, the 2′- O-methylation has been shown to antagonize the innate immune response by allowing endogenous mRNAs to escape recognition by immune sensors [97,110]. Notably, 2′-O-me of the first (cap 1) and often the second (cap 2) nucleotide abolishes interaction of host mRNAs with RIG-I and MDA5 [97]. The functional importance of 5′-structures and modifications in mRNAs is supported by the fact that many cytoplasmic RNA viruses (including picornaviruses, flaviviruses and coronaviruses) have evolved alternative 5′ elements. These include small viral proteins linked to the 5′ end of genomic RNA, or encode functions associated with the formation of a 5′ cap that are homologous to those found in eukaryotic cells. Many viruses have evolved mechanisms to generate their own cap structures with methyl ribose in 2′-O position in addition to the one at the N7 position of the capped guanine to evade innate immune recognition. For instance, the coronavirus 2′-O-methyltransferase activity associated with the highly conserved viral nonstructural protein 16 (nsp16) is necessary to inhibit MDA5 recognition of vRNA and activation of IFN pathway [111]. 2′-O methylation of the 5′ cap of vRNA also subverts mammalian antiviral responses by evading restriction by interferon-induced proteins with tetratricopeptide repeats (IFITs). IFITs are cytoplasmic antiviral proteins which differ in specificity against RNA structures. Among them, IFIT1 binds 5′PPP RNAs [112] and non-2′-O methylated viral capped transcripts [113,114] to inhibit their translation by out-competing the cellular translation initiation apparatus [115] (Table 1). Members of the flaviviruses and coronaviruses are able to evade IFIT1 sensing by adding a 2′O-methyl cap structure to their own mRNA via viral proteins [116,117]. Interestingly, alphaviruses whose genomic RNA has a 5′ cap lacking 2′-O methylation can avoid IFIT1 immune restriction and efficiently replicate thanks to a secondary structure within their 5′ untranslated region (UTR) which alters IFIT1 binding and function [118].
4. The Translation battlefield
Once it has managed to escape host cell sensing and degradation pathways, vRNA must compete with endogenous mRNAs for accessing the same protein synthesis machinery. RNA viruses have evolved several mechanisms to bypass the highly regulated translation cycle, particularly at the initiation phase and most of these mechanisms rely on peculiar vRNA-host RBPs interactions. Importantly, changes in abundance, modification and activity of translation factors could regulate both vRNA and host mRNA translation [50]. Therefore, RNA viruses often employ strategies that favor the selective translation of their genomes. They can accomplish that by targeting, modifying or cleaving host translation factors and/or by usurping the identity of endogenous mRNAs to have a privileged access to the ribosome [119].
4.1. Cellular translation: the basics
Canonical cellular mRNAs are capped at their 5′end co-transcriptionally. The cap structure, consisting of a 7‑methylguanosine (m7G) linked to the 5′ nucleoside of the mRNA chain through a 5′–5′ triphosphate bridge, is crucial for mRNA stability and translatability. Specifically, the cap protects mRNA from 5′-3′ exonucleases and ensures recognition by the cap-binding complex eukaryotic translation initiation factors (eIF4F) [120] (Fig. 2 ). Interestingly, vRNA caps can be stolen (“cap-snatching”) from cellular mRNAs or synthesized using either a host- or virus-encoded capping apparatus, and these capping assemblies exhibit a wide diversity in organization, structure and mechanism [121].
Fig. 2.
Cap v/s IRES-dependent Translation Initiation. Upper panel: The figure depicts Cap-dependent translation initiation mechanism. Capped RNA is recognized by EIF4F complex recruiting the 43S complex to the 5′UTR of mRNA. This is followed by scanning until a Start codon in encountered, followed by the formation of 80S ribosomes before elongation. Lower panel: Depicted are different modes of IRES-dependent translation (I, II, III and IV). Note the minimal requirement for initiation factors depending on IRES class.
Recognition of the cap by the eIF4F subunit eIF4E is thought to prepare mRNAs for recruitment of the 43S complex containing multiple eIFs, the initiator methionine tRNA (Met-tRNAi Met) as a ternary complex with eIF2 and GTP (eIF2-TC), and the small 40S ribosomal subunit (Fig. 2). The polyadenylated 3′ mRNA end is recognized by a poly(A)-binding protein (PABP), which is bridged to the 5′ end via PABP interactions with the eIF4G subunit of eIF4F end. The assembled 43S complex scans the 5′UTR of the mRNA to locate the initiator start codon (AUG) (Fig. 2). After AUG recognition ribosomal 60S subunits joining trigger eIFs release to form the 80S ribosome where elongation can proceed for synthesis of the polypeptide chain.
Translation is one of the most energy-expensive processes in the cell and is therefore tightly regulated. Cells rapidly reprogram gene expression when subjected to stress to conserve energy and minimize damage [122]. The arrest of bulk protein synthesis is a classical reaction cells employ during viral infection and it is often characterized by a specific inhibition of translation initiation. In the never-ending arms-race, many viruses have however evolved mechanisms to overcome the cellular stress they induce.
4.2. Cellular translation shutoff during viral infection
Generally, translation initiation is inhibited either by the disruption of the formation of the eIF4F complex or by the inhibition of eIF2-TC recycling [122] (Fig. 3 ). One major cellular regulator of eIF4F complex formation is the Mechanistic Target of Rapamycin (mTOR) pathway and will not be discussed here [[122], [123], [124]]. The second major mechanism that causes translational arrest is the disruption of eIF2-TC recycling, caused by phosphorylation of the α subunit of eIF2. Mammalian cells encode four known eIF2α kinases among which PKR (Table 1) (see above). The other three eIF2α kinases: Haeme-regulated inhibitor (HRI), general control non-derepressible protein 2 (GCN2) and PKR-like endoplasmic reticulum (ER) kinase (PERK), respond to various cellular stresses including heat shock, amino acid deprivation, and ER stress [122] (Fig. 3). These cellular stress states can be indirect indicators of viral infections. For instance, flaviviruses like dengue and Zika viruses, which replicate in close association to ER-membranes, induce ER stress that results in PERK activation and eIF2α phosphorylation early during viral infection [125,126] (Fig. 3). In some cases, GCN2 can also be activated by vRNA to arrest translation. For example, GCN2 can recognize two regions of Sindbis virus (SINV) genomic RNA and inhibit vRNA replication by blocking early viral translation. Strikingly, mice lacking GCN2 are extremely susceptible to intranasal SINV infections, demonstrating high virus titers in the brain compared to similarly infected control animals [127]. Alphaviruses like SINV take advantage of eIF2 inactivation and can be totally insensitive to translational arrest by eIF2 phosphorylation. Subgenomic mRNAs of SINV and another alphavirus, Semliki Forest Virus (SFV), initiate translation in the presence of high levels of phosphorylated eIF2α, utilizing a highly stable RNA hairpin loop downstream of the AUG initiator codon that stalls the ribosomes on the correct site to initiate translation of their mRNAs, thus bypassing the requirement for a functional eIF2 [128].
As mentioned earlier, dsRNA is detected by and activates PKR for the rapid inhibition of translation initiation [129]. Following activation by dsRNA binding, dimeriziation and auto-phosphorylation, PKR phosphorylates eIF2α which locks it onto its guanine nucleotide exchange factor eIF2B, thereby preventing the regeneration of the eIF2-TC that is critical for canonical translation initiation [122] (Fig. 3). Although initially identified because of its ability to regulate translation in response to dsRNA, PKR can also be activated by other cellular stresses such as oxidative stress, cytokines or following the stimulation of TLRs [130,131]. Furthermore, upon activation, PKR not only inhibits translation initiation, but also modulates a plethora of different signal transduction pathways, including the activation of p38 mitogen-activated protein kinase (MAPK), c-Jun N-terminal kinase (JNK), signal transducer and activator of transcription 1 and 3 (STAT1 and 3) and the tumor suppressor p53 [132]. PKR can indirectly activate the nuclear factor κB (NFκB), which has among other functions an important role in type I IFN induction [132].
4.3. Selective vRNA translation during infection
Selective translation is a common strategy used by RNA viruses to maintain intact their protein synthesis during global translation shutoff. We will briefly describe mechanisms used by viruses to promote the selective translation of their genomes during global protein shutdown. Sequence, structure and chemical modifications present in the viral genome can determine the identity of the interacting RBPs serving as translation factors. Selective translation can therefore be achieved when vRNA shunts the lengthy and elaborate multi-step mechanisms that yield assembled and productive 80S ribosomes to gain a privileged access to the ribosome. Other strategies, such as targeting and degrading translation factors, ribosomal RNAs or messenger RNAs might also confer a selective advantage for vRNAs over endogenous ones. We will discuss mechanisms that target the translation initiation step, nevertheless it is important to note that viruses are also able to trick translation elongation or termination steps [133].
4.3.1. Tricking translation initiation
While some positive-sense RNA viruses like flavi or coronaviruses possess a cap-structure on the 5′end of their vRNA, some others like caliciviruses or picornaviruses (e.g. Poliovirus or Rhinovirus) decorate their 5ʹend with a covalently bound viral protein, VPg. In some cases, VPg proteins linked to the 5′ end of positive-strand RNA genomes (e.g. feline calicivirus (FCV) or human norovirus) plays the role of a cap structure, recruiting ribosomes through interaction with eIF3 [134]. In fact, modifications at the 5′ end of vRNAs largely dictate the translation initiation mode that could either be cap -dependent or -independent. Coronavirus mRNAs are thought to undergo a classical cap-dependent translation initiation mechanism. A small compound that prevents the formation of the eIF4F complex, a hallmark of cap-dependent translation, by blocking the interaction between eIF4E and eIF4G is able to significantly decrease human coronavirus 229E replication and infectious virus titers [135]. To favor the translation of their vRNAs over endogenous mRNAs, Severe Acute Respiratory Syndrome Coronaviruses (SARS-CoV) non-structural protein 1 (nsp1) is able to efficiently and selectively inhibit host mRNA translation by binding 40S subunits [136] (Fig. 3) and promote host mRNA degradation. For example, SARS-CoV-1 nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs, however viral mRNAs seem to be resistant to nsp1-induced RNA cleavage [137]. One mechanism permitting this selectivity have been shown for Middle East Respiratory Syndrome coronavirus (MERS-CoV) where nsp1 selectively target mRNAs transcribed in the nucleus and spares mRNAs of cytoplasmic origin, notably vRNAs [138].
Flaviviruses are also thought to undergo a canonical cap-dependent initiation of translation requiring both the eIF4F complex and PABP [[139], [140], [141]]. Interestingly, in contrast to most cellular mRNAs, flavivirus vRNA lacks a 3′ poly-A tail; however, PABP is still able to associate internally to DENV 3′UTR upstream of a conserved 3′stem-loop, PABP/3′UTR interaction mimicking the role of mRNA poly-A tail and presumably stimulating translation initiation 141]. Both UTR regions (5′ and 3′) in flaviviruses possesses secondary structures shown to stimulate viral translation [139,142,143]. Interestingly, under cellular conditions where translation factors are limiting or eIF4E is inhibited, DENV has been shown to alternate between canonical cap-dependent translation initiation and a noncanonical mechanism that does not require a functional m [7]G cap [144]. Although the mechanism by which flaviviruses undergo cap-independent translation initiation is not completely clear, it has been recently suggested that DENV and ZIKV 5′ Untranslated Regions could harbor Internal Ribosomal Entry Site (IRES) functions [145]. In fact, IRES-dependent initiation of translation is the other major mechanism used by many viruses to initiate their translation, dispensing them from cap-dependence.
IRES sequences are specialized cis-acting RNA elements present in viral 5′UTRs capable of bypassing the extensive requirement of translation initiation factors to recruit ribosomal complexes and drive protein synthesis. IRESs therefore confer a considerable competitive advantage to viral mRNAs, freeing them from host regulatory constraints and sustaining viral protein synthesis when canonical cap-dependent translation is impaired [119] (Fig. 2). There are four known types of viral IRESs (I, II, III and IV). Although IRESs perform a similar function, there are no known universal IRES sequences. In fact, IRES elements present in the genome of different families of RNA viruses lack overall conserved features [146,147].The classification of viral IRESs in four types stems from their structural organization, their respective dependence on sets of translation initiation factors, and whether they use scanning or instead directly recruit ribosomes to the start codon [148] (Fig. 2). Type I and II IRESes were first discovered in picornaviruses, such as poliovirus (type I) and encephalomyocarditis virus (type II). These two IRES classes interact with eIF4G C-terminal region which binds to eIF3 and eIF4A [149]. Both require eIF5B, eIF2 and Met-tRNAi and are stimulated by the activity of eIF1, eIF1A and eIF4B [150] (Fig. 2). Type III IRESs, exemplified those of pestiviruses and Hepatitis C Virus IRES, require even less translation initiation factors to recruit 40S ribosomes. In fact, the HCV IRES bypasses the requirement for eIF4F and the scanning step of translation initiation. HCV IRES is able to directly interact with eIF3 and the ribosomal 40S subunit, placing the start codon present in the vRNA in the ribosomal P-site [151] (Fig. 2). Finally, type IV IRESs are amongst the most remarkable as they completely obviate the need for canonical initiation factors. These IRESs are common in the dicistroviridae family of viruses such as Cricket Paralysis Virus (CrPV) or Drosophila C Virus (DCV), which infect insects. These viruses contain a single positive-sense RNA genome that encodes two non-overlapping open reading frames separated by a short intergenic region (IGR). The IGR region acts as an IRES to initiate translation by recruiting 80S ribosomes in the absence of initiator tRNAi Met or any canonical initiation factors, from a GCU alanine codon located in the A-site of the ribosome [152,153] (Fig. 2). Indeed, the IGR RNA structure mimics a tRNA, thus tricking the ribosome into translation initiation without the need of the eIF2–GTP–Met–tRNA complex.
4.3.2. Viral targeting of cellular mRNA translation
Using shortcuts for a privileged access to the translation machinery by tricking the ribosome is not the only strategy RNA viruses employ to selectively favor the synthesis of viral proteins. Another effective tactic is targeting and inhibiting the translation of cellular mRNAs, which frees up a large pool of ribosomes, making them readily available for vRNA translation. This is generally achieved by targeting host RBPs, specifically translation initiation factors or ribosomal proteins.
Viruses that use IRES-dependent translation for example, actively provoke the shut-off of host cap-dependent translation initiation, by targeting specific initiation factors. For instance, enteroviruses including Rhino and Polioviruses, encode proteases that cleave eIF4G, impeding the formation of the eIF4F complex and therefore cap-dependent host mRNA translation [154] (Fig. 3). Other picornaviruses, such as EMCV suppresses cap-dependent translation by activating the translational repressor 4EBP1 [155]. Hypophosphorylated 4EBP1 binds eIF4E preventing, eIF4E-eIF4G interaction, thus eIF4F assembly. In fact, 4EBP hypophosphorylation is a strategy common to many viruses to inhibit host mRNA translation (e.g., CrPV, VSV) [[156], [157], [158]] (Fig. 3). Targeting eIF4F-associated PABP proteins is also used by some viruses to influence host translation. For example, Rotavirus Nsp3 interacts with eIF4G and displaces PABP, therefore inhibiting host translation [159]. Likewise, enterovirus and calicivirus proteases cleave PABP, and the rubella virus capsid protein binds PABP to suppress cellular mRNA translation [119,154,159]. Other translation initiation factors like eIF3 can also be targeted by viral proteins. eIF3‑binding proteins from measles and rabies viruses inhibit host protein synthesis [160,161], whereas foot-and-mouth disease virus protease degrades eIF3a and eIF3b subunits [162,119]. Also, the coronavirus spike protein has been shown to interact with eIF3f influencing translational control [163] (Fig. 3). Although initiation factors are the privileged target of viral proteins to shut-off host mRNA translation, some examples of viruses targeting translation elongation factors are also reported. For instance, it has been shown that EF1A and eEF2 are respectively inactivated by SARS-CoV-1 N protein and avian reovirus p17 [164,165]. In many of these examples, how vRNA translation is able to proceed upon inactivation of these essential initiation or elongation factors remains unclear.
Viral RBPs can directly target the ribosome to promote translation of their genomes and viral RNA may also directly interact with ribosomal proteins to preferentially translate their genomes. DENV NS1 protein interacts with RPL18 to promote viral protein synthesis [166]. Conversely, vRNA can rely on specific nonessential ribosomal proteins, dispensable for translating most host mRNAs. Moloney leukemia virus and SINV stop codon readthrough and frameshifting efficiency, have been shown to depend on RPL4 [167]. VSV cap-dependent mRNA translation have been shown to be affected by RPL40 presence [168] (Fig. 3). Also, some IRES functions, like those found in polio, HCV or dicistroviridae, have been shown to be regulated by small ribosomal proteins such as RACK1 and RPS25 [[169], [170], [171], [172], [173]] (Fig. 3). Interestingly, RACK1 seem to be a privileged target of Poxviruses to promote selective translation of viral mRNAs [174]. Finally, RBPs that are not necessary translation factors or ribosomal can also positively influence viral translation. For example, Pelo has been shown to be a host factor needed for efficient translation of viral capsids of Drosophila C Virus (DCV) [175]. Vigilin and RRBP1, two Endoplasmic Reticulum localized RBPs have been shown to bind DENV and ZIKV vRNAs, promoting their translation, stability and replication [176].
5. Technology guiding discovery; the case of RBP-vRNA interactions
5.1. From classical genetics to structural biology
During the last decades, our knowledge of the intricate interactions of cellular RBPs with vRNAs have been a direct result of technical advances in modern molecular biology. When it comes to the study of RBPs that are viral sensors or antiviral effectors, many major discoveries in how TLRs, RLRs, Dicer or other RBPs recognize and/or antagonize vRNA, were made thanks to in vivo genetic screening technologies and innovations [177]. Both, hypothesis-driven reverse genetics approaches, and forward genetic techniques have been able to yield susceptibility or resistance phenotypes, revealing genes with unanticipated and important roles in recognizing and/or targeting vRNAs. The involvement of TLR 3, 9, MDA5 and RIG-I in the innate antiviral response wouldn’t have been possible without in vivo genetic manipulation and screening of mouse models [[178], [179], [180]]. Another classical example where genetics played a fundamental role in discovering important antiviral host RBPs, is the study of the RNAi pathway components. Argonaute and Dicer proteins’ involvement in antiviral defenses across several animal and plant species was only possible thanks to genetic manipulation and screening techniques in model organisms [85,[181], [182], [183]]. Importantly, biochemical and structural biology approaches including X-Ray crystallography and cryo-electron microscopy (Cryo-EM) guided these discoveries and were able to determine structurally how sensor and effector RBPs interact with vRNA [[184], [185], [186], [187], [188]].
Likewise, recent technical innovations and improvements in RNA isolation, manipulation and sequencing have enabled detection of new RNA species and RNA chemical modifications (e.g. m6A, 2′-O-me) in vRNAs, revealing an underappreciated role of these species and modifications in viral lifecycles [96,189,190]. Thus, the use of oxidative treatment on the RNA prior to its sequencing helped to unravel the existence of 2′-O-methylated viral small RNAs species that accumulate during SINV infection [191]. More recently, the use of single cell RNA sequencing allowed to determine at an unprecedented resolution the accumulation of viral transcripts in a dynamic manner [192]. While this has so far been used to study DNA viruses’ transcription, it might also prove useful to study replication of RNA viruses. We are still lacking a global picture of RNA modifications that are present in viral RNAs, and more importantly, we need to determine their exact contribution to the infectious cycle. To this end, it is crucial to develop and validate good antibodies targeting modified ribonucleotides, or to turn to emerging sequencing technologies, such as the Oxford Nanopore one, that allows direct RNA sequencing. This strategy has recently allowed the direct detection of m6A by sequencing of cellular RNAs [193,194].
Similarly, the last decades of research brought very detailed insights into how vRNAs co-opt host ribosomes. This is partly due to the history of research in the mRNA translation field. The ribosome being the most conserved and largest known RNP complex in living organisms, deciphering mRNA translation has been an endeavor pursued since the birth of modern molecular biology [195]. Important knowledge in our understanding of mRNA translation were made in the past thanks to technical innovations and improvements. For example, in vitro translation systems, reconstituted from rabbit reticulocytes, were instrumental to understand many aspects of endogenous mRNA translational control [195]. Interestingly, thanks to such in vitro systems, we know since the 1970′s the inhibitory role of viral dsRNA or eIF2 phosphorylation on translation initiation [196,197]. in vitro translation systems coupled to reporter assays were also crucial later on to decipher the different mechanisms by which IRES-dependent translation proceeds [50,152,198]. More recently, cryo-EM approaches were able to paint a more detailed picture of how different IRESes can interact and co-opt host ribosomes [150,199,200]. The rapid development and improvement of cryo-EM techniques is expected to generate further knowledge, surpassing IRES-Ribosome interactions. Indeed, cryo-EM techniques have been lately used to unravel the structural basis of flaviviral sfRNA production for example [63]. Most recently, cryo-EM structures of SARS-Cov2 Nsp1 with human 40S ribosomal subunit revealed that Nsp1 C-terminus binds to and obstructs the mRNA entry tunnel, explaining how this protein is able to shutdown endogenous mRNA translation [201]. Another very recent cryo-EM study was able to identify residues of Nsp1 crucial for mediating translation inhibition [202]. In the future, these approaches promise to aid structure-based drug design not only against SARS-CoV2 but also other emerging RNA viruses [201]. It is finally important to note that other novel and innovative techniques developed to study translation such as single-molecule approaches [203] or ribosome profiling by deep sequencing [204] are revealing the complexity of vRNA interaction with host ribosomes. For instance, single-molecule studies of IRES-mediated translation have revealed insights into the dynamics of HCV and CrPV IRES interaction with ribosomes and clarified decades of biochemical research, providing an outline of the conformational and compositional trajectory of the ribosome during initiation [151]. Likewise, ribosome profiling revealed strategies that some RNA viruses use to induce Programmed Ribosomal Frameshifting (PRF) that would have not been discovered otherwise. Frameshifting events are ‘programmed’ because they occur at specific sequences, stimulated by cis-acting elements generally present in the vRNA at rates that are orders of magnitude more frequent than nonprogrammed events [205]. Cardioviruses such as EMCV and its relative Theiler’s murine encephalomyelitis virus (TMEV) induce PRF but are atypical due to the absence of an appropriately positioned stimulatory RNA structure in their genomes and a failure to reconstitute PRF in vitro, outside of the context of virus infection. Remarkably, thanks to ribosome profiling, a recent study shows that EMCV frameshifting is trans-activated by viral protein 2A. As a result, the frameshifting efficiency increases from 0 to 70 % (one of the highest known in a mammalian system) over the course of infection, temporally regulating the expression levels of the viral structural and enzymatic proteins [206]. In the future, ribosome profiling techniques are expected to reveal more secrets of vRNA translation, especially for viruses that annexes organelles like the ER to complete their translation [207].
5.2. New frontiers in RBP-vRNA discovery
Each time a technological leap is achieved the scope of what we know about vRNAs interaction with cellular RBPs has expanded. Two global strategies have been used to study vRNA-RBP interactions; protein-centric or RNA-centric methods. Protein-centric methods start with a known RBP of interest and characterize its interaction with RNA. These approaches involve UV-crosslinking followed by antibody purification of the RBP of interest and identification of bound RNAs, often by deep sequencing, broadly termed HITS-CLIP (cross-linking immunoprecipitation followed by high-throughput sequencing) [208]. These methods have been used in the past in the context of viral infections to characterize the RNA species that bind either known host RBPs [176,[209], [210], [211]] or of viral proteins [212,213]. We will not focus further on these methods here but readers can refer to excellent reviews on the subject in the context of viral infections [[214], [215], [216]].
RNA-centric approaches are more recent and focus on purifying an RNA of interest to identify its associated proteins. A large number of the so-called “unconventional RBPs” have been identified as associated to RNAs thanks to these RNA-centric approaches. For example, RNA interactome capture (RNA-IC), have largely expanded the repertoire of known RBPs [217]. RNA-IC entails ultraviolet crosslinking of RBPs to RNA in live cells, followed by collective capture of RNPs with polyadenylated (poly(A)) RNA on oligo(dT) beads and identification of proteins by quantitative mass spectrometry (Q-MS) [217,218]. These methods discovered new RBPs that generally lack canonical RNA-binding domains and are conserved across species, suggesting the existence of previously unidentified modes of RNA binding and new biological functions for protein–RNA interactions [217]. This method was applied to study cellular RNA-interactome during SINV infection and was able to identify over 200 RBPs that interacted differentially with host RNA upon viral infection, amongst which, RNAs encoding proviral and restriction factors [219]. Although RNA-IC is able to identify RBPs that directly interact with RNAs, one limitation of this method is its reliance on oligo(dT) to pull-down poly(A) containing RNAs. Another novel method, Comprehensive Identification of RNA binding Proteins-Mass Spectrometry” (ChIRP-MS), addresses this limitation by using 20-mer oligonucleotide probes complimentary to the RNA of interest. ChIRP-MS uses formaldehyde (FA) cross-linking prior to pull-down with biotinylated probes, able to hybridize with an RNA of interest. Unlike UV-crosslinking, the use of FA for crosslinking doesn’t only identify direct RNA binders but also their associated protein complexes. Cross-linked RNP complexes are then captured with streptavidin beads prior to proteomic analysis [220]. Originally developed to identify RBPs associated with long non-coding RNAs (lncRNAs) [221], ChIRP-MS was recently applied to identify RBPs associated with DENV and ZIKV vRNAs [176]. ChIRP-MS identified viral non-structural proteins NS3 and NS5 as highly enriched with DENV and ZIKV vRNA. Furthermore, this method identified 464 high confidence hits from the human proteome that were specifically and reproducibly associated with the DENV or ZIKV vRNA [222]. Many of those proteins were localized to the ER, in line with the known biology of flaviviral RNA, being translated and replicated at specific ER sites [223]. Importantly, when viral infection is difficult to synchronize, ChIRP-MS is unable to differentiate at which step in the viral lifecycle vRNA-RBPs associations occur (e.g. entry, translation, replication, assembly). A recent approach addresses partly this limitation and is able to capture interactions with just the pre-replicated viral RNA genome [224]. Viral cross-linking and solid-phase purification (VIR-CLASP) relies on infection of unlabeled host cells with 4-thiouridine (4SU)-labeled viral genomes [224]. Irradiation of infected cells with 365 nm light generates covalent cross-links between 4SU-labeled viral genomes and interacting host or viral proteins. Solid-phase capture of these complexes with Solid-phase reversible immobilization (SPRI) beads leads to purification of total RNA but only proteins covalently crosslinked to the 4SU-labeled viral RNA. This purification precedes (LC–MS/MS) identification of the crosslinked proteins [224]. Using VIR-CLASP, the study identifies early interactomes of different viral genomes (e.g. CHIKV, IAV, ZIKV, VSV). The authors of the study find that incoming CHIKV vRNA binds both proviral and antiviral factors. While on one hand, CHIKV incoming vRNA hijacks the lipid-modifying enzyme fatty acid synthase (FASN) for pro-viral activity, it is on the other hand N6-methyladenosine modified, binding to YTHDF1 which suppresses its replication [224].
The aforementioned RNA-centric methods (RNA-IC, ChIRP-MS, VIR-CLASP) all use crosslinking (UV or FA) to identify in vivo interactions by purifying the RNA under denaturing conditions that remove noncovalent interactions, and subsequently extracting only the cross-linked proteins for identification [225]. Other approaches, that do not require crosslinking of RNPs, use proximity proteomics to identify RBP-RNA interactions in live cells. These approaches rely on ‘promiscuous’ biotin ligases that are able to label preferentially proteins that are within 20 nm of distance with biotin [226,227]. Originally developed to identify protein-protein interactions, this approach was recently adapted to identify RNA-RBPs interactions. The RNA-protein interaction detection (RaPID) method allows to use this spatial detection constraint to detect RBPs bound to RNA by tagging an RNA of interest with a BoxB aptamer to recruit a fusion protein of λ-N and a promiscuous biotin ligase [228,225]. The biotin sprayer binds the BoxB motif through its λ-N domain and labels proteins proximal to its bound RNA [225]. RaPID-MS was successfully used to identify RBPs that bind ZIKV UTR sequences. Interestingly, the identified RBPs show enriched expression in neural tissue, which is in line with the neurotropic nature of ZIKV [228]. RaPID pointed to a known RNA-binding protein, QKI, that is highly expressed in neural progenitor cells (NPCs) and whose depletion reduces ZIKV RNA levels by 90 % [228]. Importantly, because these RNA-centric methods rely on RNA sequence specificity they can be transposed and applied to identify RBPs associated with virtually any RNA virus. After these proof-of-principle studies, all of these novel RNA-centric approaches are expected to yield important fundamental knowledge about vRNA interactions with host RBPs, if applied to emerging or re-emerging RNA viruses with epidemic potential such as Ebola or SARS-Cov2 [229,230].
6. Conclusion and outlook
RNA-protein interactions are central to most cellular processes. It is thus not surprising that RNA viruses and cellular genes evolved intricate vRNA-RBP interactions that determine many aspects of the viral life cycle [4]. From the host cell perspective, the affinity of its RBPs to the invading vRNA, could be a double-edged sword. Many cellular genes have evolved to specifically recognize and/or target vRNAs and alert the immune system to mobilize cellular defenses. This is illustrated by the relative conservation of many vRNA sensors and anti-vRNA effectors across species, such as components of the RNAi pathway or vRNA sensors like RLRs and TLRs [12]. Likewise, many viral genes and RNA structures have specifically evolved to co-opt fundamental host cell machineries, without which the virus in unable to replicate. This is exemplified by the convergent evolution of many RNA elements in the viral genomes, with roles in hijacking ribosomes and the cellular machinery in general [133]. In fact, structured RNA elements, such as IRESs are shared amongst viruses from very different families, ranging from plant to animal viruses [133,148,231]. Also, viral enzymes that can interfere with endogenous mRNA lifecycles favoring vRNAs translation, are also found across a wide range of viral families [119,156].
During the last decades, since the rise of modern molecular biology we are witnessing a remarkable and accelerated development of tools and technologies that are enabling to delve deeper into fundamental cellular mechanisms. As mentioned in this review, each time a technical innovation emerged, our understanding of vRNA-RBP interactions expanded. For example, proteins identified by the novel aforementioned RNA-centric methods (e.g. ChIRP-MS, VIR-CLASP, RNA-IC) are highly enriched with classical RBPs as expected. However, hundreds of “unorthodox” RBPs have also been found to bind vRNAs [219,222]. Some of those unorthodox’ RBPs have already known cellular functions and weren’t expected to have an influence on viral lifecycles by binding vRNAs. For example, cellular transport motors like dyneins have been shown to have an RNA binding activity that might be responsible of RNAs trafficking, uncoating, reverse transcription and in some cases viral replication factory assembly [232]. Another eloquent example comes from the interaction of ER-resident multimolecular complexes, such as Sec61 translocon complex or the Oligosaccharyltransferase (OST) complex, with the lifecycle of flaviviruses [3,233]. Although these complexes have known and established roles in protein translocation and glycosylation in the ER, new unexpected RNA-related functions of these complexes are just emerging [234]. For example, flavivirus vRNAs that are translated and replicate in close proximity to ER membranes, seem to require “non-canonical” functions of the OST complex [3]. This might involve tethering viral RNA genomes to the ER for efficient translation and replication [176,207,232,234].
Future work will clarify which unconventional RBPs are truly important for the viral infectious cycle. In the meantime, these system-wide approaches are uncovering new host RBPs with unexpected new roles during viral lifecycles. Understanding mechanistically the relevance of these newly discovered interactions is crucial in the future. Alongside careful and rigorous biochemistry, this can be tackled with new complimentary cutting-edge methods (e.g. CLIP-seq, ribosome profiling) [204,206,210,222] to determine the footprints of RBPs on viral and host RNAs. Other new approaches to visualize, track, target or detect RNAs (e.g. CRISPR-Cas13), if applied to vRNAs, will clarify the role of the newly discovered host RBP-vRNA interactions [[235], [236], [237]]. Finally, there are many reasons to look at the future of the field with optimism. Indeed, continuing to dissect the intimate relationship between vRNAs and host RBPs with new tools, is expected to both expand our fundamental understanding of virology and cell biology, and to guide the development of much needed novel antiviral approaches.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgments
The authors would like to thank Dr. Alex G. Johnson for his thorough and critical reading of the manuscript. The authors apologize to colleagues whose work could not be cited due to space limitations.
Acknowledgments
Funding
This work has been supported by the H2020 Marie-Curie Actions MSCA-IF-792661-HipShot (KM). SP is supported by the European Research Council (ERC-CoG-647455 RegulRNA) and the LABEX: ANR-10-LABX-0036_NETRNA, which benefits from a funding from the state managed by the French National Research Agency as part of the Investments for the future program. EG has also received funding from the People Programme (Marie Curie Actions) of the European Union’s Seventh Framework Program (FP7/2007-2013) under REA grant agreement n° PCOFUND-GA-2013-609102, through the PRESTIGE program coordinated by Campus France. This work was also supported under the framework of the LABEX ANR-10-LABX-0028_HEPSYS (TB).
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.semcdb.2020.08.006.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Ahlquist P., Noueiry A.O., Lee W.-M., Kushner D.B., Dye B.T. Host factors in positive-strand RNA virus genome replication. J. Virol. 2003;77:8181–8186. doi: 10.1128/JVI.77.15.8181-8186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.König R. Human host factors required for influenza virus replication. Nature. 2010;463:813–817. doi: 10.1038/nature08699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marceau C.D. Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens. Nature. 2016;535:159–163. doi: 10.1038/nature18631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Z., Nagy P.D. Diverse roles of host RNA binding proteins in RNA virus replication. RNA Biol. 2011;8:305–315. doi: 10.4161/rna.8.2.15391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marraffini L.A. CRISPR-Cas immunity in prokaryotes. Nature. 2015;526:55–61. doi: 10.1038/nature15386. [DOI] [PubMed] [Google Scholar]
- 6.Ding S.W., Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bekerman E., Einav S. Infectious disease. Combating emerging viral threats. Science. 2015;348:282–283. doi: 10.1126/science.aaa3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhatt S. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lazear H.M., Diamond M.S.Zika. Virus: new clinical syndromes and its emergence in the western hemisphere. J. Virol. 2016;90:4864–4875. doi: 10.1128/JVI.00252-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poon L.L.M., Peiris M. Emergence of a novel human coronavirus threatening human health. Nat. Med. 2020;26:317–319. doi: 10.1038/s41591-020-0796-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jensen S., Thomsen A.R. Sensing of RNA viruses: a review of innate immune receptors involved in recognizing RNA virus invasion. J. Virol. 2012;86:2900–2910. doi: 10.1128/JVI.05738-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Majzoub K., Wrensch F., Baumert T.F. The innate antiviral response in animals: an evolutionary perspective from flagellates to humans. Viruses. 2019;11 doi: 10.3390/v11080758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schlee M., Hartmann G. Discriminating self from non-self in nucleic acid sensing. Nat. Rev. Immunol. 2016;16:566–580. doi: 10.1038/nri.2016.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iwasaki A., Medzhitov R. Toll-like receptor control of the adaptive immune responses. Nat. Immunol. 2004;5:987–995. doi: 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- 15.Lester S.N., Li K. Toll-like receptors in antiviral innate immunity. J. Mol. Biol. 2014;426:1246–1264. doi: 10.1016/j.jmb.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diebold S.S., Kaisho T., Hemmi H., Akira S., Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 17.Heil F. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 18.Hardarson H.S. Toll-like receptor 3 is an essential component of the innate stress response in virus-induced cardiac injury. Am. J. Physiol. Heart Circ. Physiol. 2007;292:H251–258. doi: 10.1152/ajpheart.00398.2006. [DOI] [PubMed] [Google Scholar]
- 19.Tsai Y.-T., Chang S.-Y., Lee C.-N., Kao C.-L. Human TLR3 recognizes dengue virus and modulates viral replication in vitro. Cell. Microbiol. 2009;11:604–615. doi: 10.1111/j.1462-5822.2008.01277.x. [DOI] [PubMed] [Google Scholar]
- 20.Dang J. Zika virus depletes neural progenitors in human cerebral organoids through activation of the innate immune receptor TLR3. Cell Stem Cell. 2016;19:258–265. doi: 10.1016/j.stem.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perales-Linares R., Navas-Martin S. Toll-like receptor 3 in viral pathogenesis: friend or foe? Immunology. 2013;140:153–167. doi: 10.1111/imm.12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rehwinkel J., Gack M.U. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat. Rev. Immunol. 2020 doi: 10.1038/s41577-020-0288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hornung V. 5’-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 24.Pichlmair A. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5’-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 25.Schlee M. Recognition of 5’ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt A. 5’-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc. Natl. Acad. Sci. U. S. A. 2009;106:12067–12072. doi: 10.1073/pnas.0900971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Habjan M. Processing of genome 5’ termini as a strategy of negative-strand RNA viruses to avoid RIG-I-dependent interferon induction. PLoS One. 2008;3:e2032. doi: 10.1371/journal.pone.0002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kato H. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 29.Kato H. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23:19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 30.Chazal M. RIG-I recognizes the 5’ region of dengue and Zika virus genomes. Cell Rep. 2018;24:320–328. doi: 10.1016/j.celrep.2018.06.047. [DOI] [PubMed] [Google Scholar]
- 31.Dias A.G., Jr., Sampaio N.G., Rehwinkel J.A. Balancing act: MDA5 in Antiviral Immunity and Autoinflammation. Trends Microbiol. 2019;27:75–85. doi: 10.1016/j.tim.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berke I.C., Yu X., Modis Y., Egelman E.H. MDA5 assembles into a polar helical filament on dsRNA. Proc Natl Acad Sci U. S. A. 2012;109:18437–18441. doi: 10.1073/pnas.1212186109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng Q. Enterovirus 2Apro targets MDA5 and MAVS in infected cells. J. Virol. 2014;88:3369–3378. doi: 10.1128/JVI.02712-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng Q. MDA5 detects the double-stranded RNA replicative form in picornavirus-infected cells. Cell Rep. 2012;2:1187–1196. doi: 10.1016/j.celrep.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muñoz-Jordán J.L., Fredericksen B.L. How flaviviruses activate and suppress the interferon response. Viruses. 2010;2:676–691. doi: 10.3390/v2020676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zalinger Z.B., Elliott R., Rose K.M., Weiss S.R. MDA5 is critical to host defense during infection with murine coronavirus. J. Virol. 2015;89:12330–12340. doi: 10.1128/JVI.01470-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Satoh T. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc. Natl. Acad. Sci. U. S. A. 2010;107:1512–1517. doi: 10.1073/pnas.0912986107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rothenfusser S. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J. Immunol. 2005;175:5260–5268. doi: 10.4049/jimmunol.175.8.5260. [DOI] [PubMed] [Google Scholar]
- 39.Venkataraman T. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J. Immunol. 2007;178:6444–6455. doi: 10.4049/jimmunol.178.10.6444. [DOI] [PubMed] [Google Scholar]
- 40.van der Veen A.G. The RIG-I-like receptor LGP2 inhibits Dicer-dependent processing of long double-stranded RNA and blocks RNA interference in mammalian cells. EMBO J. 2018;37 doi: 10.15252/embj.201797479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taschuk F., Cherry S. DEAD-box helicases: sensors, regulators, and effectors for antiviral defense. Viruses. 2020;12 doi: 10.3390/v12020181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.García-Sastre A., Biron C.A. Type 1 interferons and the virus-host relationship: a lesson in détente. Science. 2006;312:879–882. doi: 10.1126/science.1125676. [DOI] [PubMed] [Google Scholar]
- 43.Sadler A.J., Williams B.R.G. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith J.A., Schmechel S.C., Williams B.R.G., Silverman R.H., Schiff L.A. Involvement of the interferon-regulated antiviral proteins PKR and RNase L in reovirus-induced shutoff of cellular translation. J. Virol. 2005;79:2240–2250. doi: 10.1128/JVI.79.4.2240-2250.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weber F., Wagner V., Rasmussen S.B., Hartmann R., Paludan S.R. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 2006;80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heinicke L.A. RNA dimerization promotes PKR dimerization and activation. J. Mol. Biol. 2009;390:319–338. doi: 10.1016/j.jmb.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Willis K.L., Langland J.O., Shisler J.L. Viral double-stranded RNAs from vaccinia virus early or intermediate gene transcripts possess PKR activating function, resulting in NF-kappaB activation, when the K1 protein is absent or mutated. J. Biol. Chem. 2011;286:7765–7778. doi: 10.1074/jbc.M110.194704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nallagatla S.R., Toroney R., Bevilacqua P.C. Regulation of innate immunity through RNA structure and the protein kinase PKR. Curr. Opin. Struct. Biol. 2011;21:119–127. doi: 10.1016/j.sbi.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stern-Ginossar N., Thompson S.R., Mathews M.B., Mohr I. Translational control in virus-infected cells. Cold Spring Harb. Perspect. Biol. 2019;11 doi: 10.1101/cshperspect.a033001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jan E., Mohr I., Walsh D. A cap-to-Tail guide to mRNA translation strategies in virus-infected cells. Annu. Rev. Virol. 2016;3:283–307. doi: 10.1146/annurev-virology-100114-055014. [DOI] [PubMed] [Google Scholar]
- 51.Tycowski K.T. Viral noncoding RNAs: more surprises. Genes Dev. 2015;29:567–584. doi: 10.1101/gad.259077.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vyas J., Elia A., Clemens M.J. Inhibition of the protein kinase PKR by the internal ribosome entry site of hepatitis C virus genomic RNA. RNA N. Y. N. 2003;9:858–870. doi: 10.1261/rna.5330503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mudhasani R. Protein kinase r degradation is essential for rift valley fever virus infection and is regulated by SKP1-CUL1-F-box (SCF)FBXW11-NSs E3 ligase. PLoS Pathog. 2016;12:e1005437. doi: 10.1371/journal.ppat.1005437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Silverman R.H. Viral encounters with 2’,5’-oligoadenylate synthetase and RNase L during the interferon antiviral response. J. Virol. 2007;81:12720–12729. doi: 10.1128/JVI.01471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chakrabarti A., Jha B.K., Silverman R.H. New insights into the role of RNase L in innate immunity. J. Interferon Cytokine Res. Off. J. Int. Soc. Interferon Cytokine Res. 2011;31:49–57. doi: 10.1089/jir.2010.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rong E. Molecular mechanisms for the adaptive switching between the OAS/RNase l and OASL/RIG-I pathways in birds and mammals. Front. Immunol. 2018;9:1398. doi: 10.3389/fimmu.2018.01398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang R. Homologous 2’,5’-phosphodiesterases from disparate RNA viruses antagonize antiviral innate immunity. Proc. Natl. Acad. Sci. U. S. A. 2013;110:13114–13119. doi: 10.1073/pnas.1306917110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Molleston J.M., Cherry S. Attacked from all sides: RNA decay in antiviral defense. Viruses. 2017;9 doi: 10.3390/v9010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Balistreri G. The host nonsense-mediated mRNA decay pathway restricts Mammalian RNA virus replication. Cell Host Microbe. 2014;16:403–411. doi: 10.1016/j.chom.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 60.Li Y., Masaki T., Yamane D., McGivern D.R., Lemon S.M. Competing and noncompeting activities of miR-122 and the 5’ exonuclease Xrn1 in regulation of hepatitis C virus replication. Proc. Natl. Acad. Sci. U. S. A. 2013;110:1881–1886. doi: 10.1073/pnas.1213515110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shimakami T. Stabilization of hepatitis C virus RNA by an Ago2-miR-122 complex. Proc. Natl. Acad. Sci. U. S. A. 2012;109:941–946. doi: 10.1073/pnas.1112263109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Akiyama B.M. Zika virus produces noncoding RNAs using a multi-pseudoknot structure that confounds a cellular exonuclease. Science. 2016;354:1148–1152. doi: 10.1126/science.aah3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chapman E.G. The structural basis of pathogenic subgenomic flavivirus RNA (sfRNA) production. Science. 2014;344:307–310. doi: 10.1126/science.1250897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chapman E.G., Moon S.L., Wilusz J., Kieft J.S. RNA structures that resist degradation by Xrn1 produce a pathogenic Dengue virus RNA. eLife. 2014;3 doi: 10.7554/eLife.01892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Donald C.L. Full genome sequence and sfRNA interferon antagonist activity of zika virus from Recife, Brazil. PLoS Negl. Trop. Dis. 2016;10 doi: 10.1371/journal.pntd.0005048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Funk A. RNA structures required for production of subgenomic flavivirus RNA. J. Virol. 2010;84:11407–11417. doi: 10.1128/JVI.01159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Manokaran G. Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science. 2015;350:217–221. doi: 10.1126/science.aab3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moon S.L. A noncoding RNA produced by arthropod-borne flaviviruses inhibits the cellular exoribonuclease XRN1 and alters host mRNA stability. RNA N. Y. N. 2012;18:2029–2040. doi: 10.1261/rna.034330.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pijlman G.P. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe. 2008;4:579–591. doi: 10.1016/j.chom.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 70.Silva P.A.G.C., Pereira C.F., Dalebout T.J., Spaan W.J.M., Bredenbeek P.J. An RNA pseudoknot is required for production of yellow fever virus subgenomic RNA by the host nuclease XRN1. J. Virol. 2010;84:11395–11406. doi: 10.1128/JVI.01047-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Slonchak A. Zika virus noncoding RNA suppresses apoptosis and is required for virus transmission by mosquitoes. Nat. Commun. 2020;11:2205. doi: 10.1038/s41467-020-16086-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schnettler E. Noncoding flavivirus RNA displays RNA interference suppressor activity in insect and mammalian cells. J. Virol. 2012;86:13486–13500. doi: 10.1128/JVI.01104-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sokoloski K.J., Wilusz C.J., Wilusz J. Viruses: overturning RNA turnover. RNA Biol. 2006;3:140–144. doi: 10.4161/rna.3.4.4076. [DOI] [PubMed] [Google Scholar]
- 74.Moy R.H. Stem-loop recognition by DDX17 facilitates miRNA processing and antiviral defense. Cell. 2014;158:764–777. doi: 10.1016/j.cell.2014.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guo X., Ma J., Sun J., Gao G. The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA. Proc. Natl. Acad. Sci. U. S. A. 2007;104:151–156. doi: 10.1073/pnas.0607063104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Takata M.A. CG dinucleotide suppression enables antiviral defence targeting non-self RNA. Nature. 2017;550:124–127. doi: 10.1038/nature24039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ficarelli M. KHNYN is essential for the zinc finger antiviral protein (ZAP) to restrict HIV-1 containing clustered CpG dinucleotides. eLife. 2019;8 doi: 10.7554/eLife.46767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Law L.M.J. ZAP’s stress granule localization is correlated with its antiviral activity and induced by virus replication. PLoS Pathog. 2019;15 doi: 10.1371/journal.ppat.1007798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Choudhury N.R. RNA-binding activity of TRIM25 is mediated by its PRY/SPRY domain and is required for ubiquitination. BMC Biol. 2017;15:105. doi: 10.1186/s12915-017-0444-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li M.M.H. TRIM25 enhances the antiviral action of zinc-finger antiviral protein (ZAP) PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lin R.-J. MCPIP1 ribonuclease exhibits broad-spectrum antiviral effects through viral RNA binding and degradation. Nucleic Acids Res. 2013;41:3314–3326. doi: 10.1093/nar/gkt019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lin R.-J. MCPIP1 suppresses hepatitis C virus replication and negatively regulates virus-induced proinflammatory cytokine responses. J. Immunol. Baltim. Md. 1950;193:4159–4168. doi: 10.4049/jimmunol.1400337. 2014. [DOI] [PubMed] [Google Scholar]
- 83.Malathi K. RNase L releases a small RNA from HCV RNA that refolds into a potent PAMP. RNA N. Y. N. 2010;16:2108–2119. doi: 10.1261/rna.2244210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Han J.-Q., Barton D.J. Activation and evasion of the antiviral 2’-5’ oligoadenylate synthetase/ribonuclease L pathway by hepatitis C virus mRNA. RNA N. Y. N. 2002;8:512–525. doi: 10.1017/s1355838202020617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Galiana-Arnoux D., Dostert C., Schneemann A., Hoffmann J.A., Imler J.L. Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nat. Immunol. 2006;7:590–597. doi: 10.1038/ni1335. [DOI] [PubMed] [Google Scholar]
- 86.Girardi E. Cross-species comparative analysis of Dicer proteins during Sindbis virus infection. Sci. Rep. 2015;5:10693. doi: 10.1038/srep10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li Y., Lu J., Han Y., Fan X., Ding S.W. RNA interference functions as an antiviral immunity mechanism in mammals. Science. 2013;342:231–234. [Google Scholar]
- 88.Maillard P.V. Antiviral RNA interference in mammalian cells. Science. 2013;342:235–238. [Google Scholar]
- 89.Qiu Y. Flavivirus induces and antagonizes antiviral RNA interference in both mammals and mosquitoes. Sci. Adv. 2020;6 doi: 10.1126/sciadv.aax7989. eaax7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.tenOever B.R. The evolution of antiviral defense systems. Cell Host Microbe. 2016;19:142–149. doi: 10.1016/j.chom.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 91.Qiu Y. Human virus-derived small RNAs can confer antiviral immunity in mammals. Immunity. 2017;46:992–1004. doi: 10.1016/j.immuni.2017.05.006. e5. [DOI] [PubMed] [Google Scholar]
- 92.Li Y. Induction and suppression of antiviral RNA interference by influenza A virus in mammalian cells. Nat. Microbiol. 2016;2:16250. doi: 10.1038/nmicrobiol.2016.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cullen B.R. Viruses and RNA interference: issues and controversies. J. Virol. 2014;88:12934–12936. doi: 10.1128/JVI.01179-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schuster S., Overheul G.J., Bauer L., van Kuppeveld F.J.M., van Rij R.P. No evidence for viral small RNA production and antiviral function of Argonaute 2 in human cells. Sci. Rep. 2019;9:13752. doi: 10.1038/s41598-019-50287-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Aguado L.C. RNase III nucleases from diverse kingdoms serve as antiviral effectors. Nature. 2017;547:114–117. doi: 10.1038/nature22990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gonzales-van Horn S.R., Sarnow P. Making the mark: the role of adenosine modifications in the life cycle of RNA viruses. Cell Host Microbe. 2017;21:661–669. doi: 10.1016/j.chom.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hyde J.L., Diamond M.S. Innate immune restriction and antagonism of viral RNA lacking 2׳-O methylation. Virology. 2015;479–480:66–74. doi: 10.1016/j.virol.2015.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Frye M., Jaffrey S.R., Pan T., Rechavi G., Suzuki T. RNA modifications: what have we learned and where are we headed? Nat. Rev. Genet. 2016;17:365–372. doi: 10.1038/nrg.2016.47. [DOI] [PubMed] [Google Scholar]
- 99.Barraud P., Allain F.H.-T.A. DAR proteins: double-stranded RNA and Z-DNA binding domains. Curr. Top. Microbiol. Immunol. 2012;353:35–60. doi: 10.1007/82_2011_145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lamers M.M., van den Hoogen B.G., Haagmans B.L. ADAR1: ‘Editor-in-Chief’ of cytoplasmic innate immunity. Front. Immunol. 2019;10:1763. doi: 10.3389/fimmu.2019.01763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Samuel C.E. Adenosine deaminases acting on RNA (ADARs) are both antiviral and proviral. Virology. 2011;411:180–193. doi: 10.1016/j.virol.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Taylor J.M. Hepatitis d virus replication. Cold Spring Harb. Perspect. Med. 2015;5 doi: 10.1101/cshperspect.a021568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Huang H., Weng H., Chen J. The biogenesis and precise control of RNA m6A methylation. Trends Genet. TIG. 2020;36:44–52. doi: 10.1016/j.tig.2019.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shulman Z., Stern-Ginossar N. The RNA modification N6-methyladenosine as a novel regulator of the immune system. Nat. Immunol. 2020;21:501–512. doi: 10.1038/s41590-020-0650-4. [DOI] [PubMed] [Google Scholar]
- 105.Dang W. N6-methyladenosine and viral infection. Front. Microbiol. 2019;10:417. doi: 10.3389/fmicb.2019.00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gokhale N.S. N6-methyladenosine in flaviviridae viral RNA genomes regulates infection. Cell Host Microbe. 2016;20:654–665. doi: 10.1016/j.chom.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lu M. N6-methyladenosine modification enables viral RNA to escape recognition by RNA sensor RIG-I. Nat. Microbiol. 2020;5:584–598. doi: 10.1038/s41564-019-0653-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Thiel V. Viral RNA in an m6A disguise. Nat. Microbiol. 2020;5:531–532. doi: 10.1038/s41564-020-0689-x. [DOI] [PubMed] [Google Scholar]
- 109.Gao Y. m6A modification prevents formation of endogenous double-stranded RNAs and deleterious innate immune responses during hematopoietic development. Immunity. 2020;52:1007–1021. doi: 10.1016/j.immuni.2020.05.003. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Freund I., Eigenbrod T., Helm M., Dalpke A.H. RNA modifications modulate activation of innate toll-like receptors. Genes. 2019;10 doi: 10.3390/genes10020092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Züst R. Ribose 2’-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 2011;12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pichlmair A. IFIT1 is an antiviral protein that recognizes 5’-triphosphate RNA. Nat. Immunol. 2011;12:624–630. doi: 10.1038/ni.2048. [DOI] [PubMed] [Google Scholar]
- 113.Habjan M. Sequestration by IFIT1 impairs translation of 2’O-unmethylated capped RNA. PLoS Pathog. 2013;9:e1003663. doi: 10.1371/journal.ppat.1003663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kimura T. Ifit1 inhibits Japanese encephalitis virus replication through binding to 5’ capped 2’-O unmethylated RNA. J. Virol. 2013;87:9997–10003. doi: 10.1128/JVI.00883-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Diamond M.S. IFIT1: a dual sensor and effector molecule that detects non-2’-O methylated viral RNA and inhibits its translation. Cytokine Growth Factor Rev. 2014;25:543–550. doi: 10.1016/j.cytogfr.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Daffis S. 2’-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Menachery V.D. Attenuation and restoration of severe acute respiratory syndrome coronavirus mutant lacking 2’-o-methyltransferase activity. J. Virol. 2014;88:4251–4264. doi: 10.1128/JVI.03571-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hyde J.L. A viral RNA structural element alters host recognition of nonself RNA. Science. 2014;343:783–787. doi: 10.1126/science.1248465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Walsh D., Mohr I. Viral subversion of the host protein synthesis machinery. Nat. Rev. Microbiol. 2011;9:860–875. doi: 10.1038/nrmicro2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ramanathan A., Robb G.B., Chan S.-H. mRNA capping: biological functions and applications. Nucleic Acids Res. 2016;44:7511–7526. doi: 10.1093/nar/gkw551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Decroly E., Ferron F., Lescar J., Canard B. Conventional and unconventional mechanisms for capping viral mRNA. Nat. Rev. Microbiol. 2011;10:51–65. doi: 10.1038/nrmicro2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.McCormick C., Khaperskyy D.A. Translation inhibition and stress granules in the antiviral immune response. Nat. Rev. Immunol. 2017;17:647–660. doi: 10.1038/nri.2017.63. [DOI] [PubMed] [Google Scholar]
- 123.Zoncu R., Efeyan A., Sabatini D.M. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Le Sage V., Cinti A., Amorim R., Mouland A.J. Adapting the stress response: viral subversion of the mTOR signaling pathway. Viruses. 2016;8 doi: 10.3390/v8060152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Peña J., Harris E. Dengue virus modulates the unfolded protein response in a time-dependent manner. J. Biol. Chem. 2011;286:14226–14236. doi: 10.1074/jbc.M111.222703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Roth H. Flavivirus infection uncouples translation suppression from cellular stress responses. mBio. 2017;8 doi: 10.1128/mBio.02150-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Berlanga J.J. Antiviral effect of the mammalian translation initiation factor 2alpha kinase GCN2 against RNA viruses. EMBO J. 2006;25:1730–1740. doi: 10.1038/sj.emboj.7601073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ventoso I. Translational resistance of late alphavirus mRNA to eIF2alpha phosphorylation: a strategy to overcome the antiviral effect of protein kinase PKR. Genes Dev. 2006;20:87–100. doi: 10.1101/gad.357006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.García M.A., Meurs E.F., Esteban M. The dsRNA protein kinase PKR: virus and cell control. Biochimie. 2007;89:799–811. doi: 10.1016/j.biochi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 130.García M.A. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol. Mol. Biol. Rev. MMBR. 2006;70:1032–1060. doi: 10.1128/MMBR.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ruvolo P.P., Gao F., Blalock W.L., Deng X., May W.S. Ceramide regulates protein synthesis by a novel mechanism involving the cellular PKR activator RAX. J. Biol. Chem. 2001;276:11754–11758. doi: 10.1074/jbc.M011400200. [DOI] [PubMed] [Google Scholar]
- 132.Munir M., Berg M. The multiple faces of proteinkinase R in antiviral defense. Virulence. 2013;4:85–89. doi: 10.4161/viru.23134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jaafar Z.A., Kieft J.S. Viral RNA structure-based strategies to manipulate translation. Nat. Rev. Microbiol. 2019;17:110–123. doi: 10.1038/s41579-018-0117-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Daughenbaugh K.F., Fraser C.S., Hershey J.W.B., Hardy M.E. The genome-linked protein VPg of the Norwalk virus binds eIF3, suggesting its role in translation initiation complex recruitment. EMBO J. 2003;22:2852–2859. doi: 10.1093/emboj/cdg251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cencic R. Blocking eIF4E-eIF4G interaction as a strategy to impair coronavirus replication▿. J. Virol. 2011;85:6381–6389. doi: 10.1128/JVI.00078-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kamitani W., Huang C., Narayanan K., Lokugamage K.G., Makino S. A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat. Struct. Mol. Biol. 2009;16:1134–1140. doi: 10.1038/nsmb.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Huang C. SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: viral mRNAs are resistant to nsp1-induced RNA cleavage. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lokugamage K.G. Middle east respiratory syndrome coronavirus nsp1 inhibits host gene expression by selectively targeting mRNAs transcribed in the nucleus while sparing mRNAs of cytoplasmic origin. J. Virol. 2015;89:10970–10981. doi: 10.1128/JVI.01352-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Clyde K., Harris E. RNA secondary structure in the coding region of dengue virus type 2 directs translation start codon selection and is required for viral replication. J. Virol. 2006;80:2170–2182. doi: 10.1128/JVI.80.5.2170-2182.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Garcia-Blanco M.A., Vasudevan S.G., Bradrick S.S., Nicchitta C. Flavivirus RNA transactions from viral entry to genome replication. Antiviral Res. 2016;134:244–249. doi: 10.1016/j.antiviral.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 141.Polacek C., Friebe P., Harris E. Poly(A)-binding protein binds to the non-polyadenylated 3’ untranslated region of dengue virus and modulates translation efficiency. J. Gen. Virol. 2009;90:687–692. doi: 10.1099/vir.0.007021-0. [DOI] [PubMed] [Google Scholar]
- 142.Chiu W.-W., Kinney R.M., Dreher T.W. Control of translation by the 5’- and 3’-terminal regions of the dengue virus genome. J. Virol. 2005;79:8303–8315. doi: 10.1128/JVI.79.13.8303-8315.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Holden K.L., Harris E. Enhancement of dengue virus translation: role of the 3’ untranslated region and the terminal 3’ stem-loop domain. Virology. 2004;329:119–133. doi: 10.1016/j.virol.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 144.Edgil D., Polacek C., Harris E. Dengue virus utilizes a novel strategy for translation initiation when cap-dependent translation is inhibited. J. Virol. 2006;80:2976–2986. doi: 10.1128/JVI.80.6.2976-2986.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Song Y., Mugavero J., Stauft C.B., Wimmer E. Dengue and Zika virus 5’ untranslated regions harbor internal ribosomal entry site functions. mBio. 2019;10 doi: 10.1128/mBio.00459-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lozano G., Martínez-Salas E. Structural insights into viral IRES-dependent translation mechanisms. Curr. Opin. Virol. 2015;12:113–120. doi: 10.1016/j.coviro.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 147.Martinez-Salas E., Francisco-Velilla R., Fernandez-Chamorro J., Embarek A.M. Insights into structural and mechanistic features of viral IRES elements. Front. Microbiol. 2017;8:2629. doi: 10.3389/fmicb.2017.02629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mailliot J., Martin F. Viral internal ribosomal entry sites: four classes for one goal. Wiley Interdiscip. Rev. RNA. 2018;9 doi: 10.1002/wrna.1458. [DOI] [PubMed] [Google Scholar]
- 149.Hellen C.U.T. IRES-induced conformational changes in the ribosome and the mechanism of translation initiation by internal ribosomal entry. Biochim. Biophys. Acta. 2009;1789:558–570. doi: 10.1016/j.bbagrm.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Plank T.-D.M., Kieft J.S. The structures of nonprotein-coding RNAs that drive internal ribosome entry site function. WIREs RNA. 2012;3:195–212. doi: 10.1002/wrna.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Johnson A.G., Grosely R., Petrov A.N., Puglisi J.D. Dynamics of IRES-mediated translation. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 2017;372 doi: 10.1098/rstb.2016.0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Jan E., Sarnow P. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J. Mol. Biol. 2002;324:889–902. doi: 10.1016/s0022-2836(02)01099-9. [DOI] [PubMed] [Google Scholar]
- 153.Wilson J.E., Pestova T.V., Hellen C.U., Sarnow P. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000;102:511–520. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]
- 154.Lloyd R.E. Translational control by viral proteinases. Virus Res. 2006;119:76–88. doi: 10.1016/j.virusres.2005.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gingras A.C., Svitkin Y., Belsham G.J., Pause A., Sonenberg N. Activation of the translational suppressor 4E-BP1 following infection with encephalomyocarditis virus and poliovirus. Proc. Natl. Acad. Sci. U. S. A. 1996;93:5578–5583. doi: 10.1073/pnas.93.11.5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Walsh D., Mathews M.B., Mohr I. Tinkering with translation: protein synthesis in virus-infected cells. Cold Spring Harb. Perspect. Biol. 2013;5 doi: 10.1101/cshperspect.a012351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Connor J.H., Lyles D.S. Vesicular stomatitis virus infection alters the eIF4F translation initiation complex and causes dephosphorylation of the eIF4E binding protein 4E-BP1. J. Virol. 2002;76:10177–10187. doi: 10.1128/JVI.76.20.10177-10187.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Garrey J.L., Lee Y.-Y., Au H.H.T., Bushell M., Jan E. Host and viral translational mechanisms during cricket paralysis virus infection. J. Virol. 2010;84:1124–1138. doi: 10.1128/JVI.02006-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Smith R.W.P., Gray N.K. Poly(A)-binding protein (PABP): a common viral target. Biochem. J. 2010;426:1–12. doi: 10.1042/BJ20091571. [DOI] [PubMed] [Google Scholar]
- 160.Sato H. Measles virus N protein inhibits host translation by binding to eIF3-p40. J. Virol. 2007;81:11569–11576. doi: 10.1128/JVI.00570-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Komarova A.V. Rabies virus matrix protein interplay with eIF3, new insights into rabies virus pathogenesis. Nucleic Acids Res. 2007;35:1522–1532. doi: 10.1093/nar/gkl1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Rodríguez Pulido M., Serrano P., Sáiz M., Martínez-Salas E. Foot-and-mouth disease virus infection induces proteolytic cleavage of PTB, eIF3a,b, and PABP RNA-binding proteins. Virology. 2007;364:466–474. doi: 10.1016/j.virol.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 163.Xiao H., Xu L.H., Yamada Y., Liu D.X. Coronavirus spike protein inhibits host cell translation by interaction with eIF3f. PLoS One. 2008;3:e1494. doi: 10.1371/journal.pone.0001494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhou B. The nucleocapsid protein of severe acute respiratory syndrome coronavirus inhibits cell cytokinesis and proliferation by interacting with translation elongation factor 1alpha. J. Virol. 2008;82:6962–6971. doi: 10.1128/JVI.00133-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ji W.T., Wang L., Lin R.C., Huang W.R., Liu H.J. Avian reovirus influences phosphorylation of several factors involved in host protein translation including eukaryotic translation elongation factor 2 (eEF2) in Vero cells. Biochem. Biophys. Res. Commun. 2009;384:301–305. doi: 10.1016/j.bbrc.2009.04.116. [DOI] [PubMed] [Google Scholar]
- 166.Cervantes-Salazar M. Dengue virus NS1 protein interacts with the ribosomal protein RPL18: this interaction is required for viral translation and replication in Huh-7 cells. Virology. 2015;484:113–126. doi: 10.1016/j.virol.2015.05.017. [DOI] [PubMed] [Google Scholar]
- 167.Green L., Houck-Loomis B., Yueh A., Goff S.P. Large ribosomal protein 4 increases efficiency of viral recoding sequences. J. Virol. 2012;86:8949–8958. doi: 10.1128/JVI.01053-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Lee A.S.-Y., Burdeinick-Kerr R., Whelan S.P.J. A ribosome-specialized translation initiation pathway is required for cap-dependent translation of vesicular stomatitis virus mRNAs. Proc. Natl. Acad. Sci. U. S. A. 2013;110:324–329. doi: 10.1073/pnas.1216454109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Majzoub K. RACK1 controls IRES-mediated translation of viruses. Cell. 2014;159:1086–1095. doi: 10.1016/j.cell.2014.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hertz M.I., Landry D.M., Willis A.E., Luo G., Thompson S.R. Ribosomal protein S25 dependency reveals a common mechanism for diverse internal ribosome entry sites and ribosome shunting. Mol. Cell. Biol. 2013;33:1016–1026. doi: 10.1128/MCB.00879-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.LaFontaine E., Miller C.M., Permaul N., Martin E.T., Fuchs G. Ribosomal protein RACK1 enhances translation of poliovirus and other viral IRESs. Virology. 2020;545:53–62. doi: 10.1016/j.virol.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Johnson A.G. RACK1 on and off the ribosome. RNA. 2019;25:881–895. doi: 10.1261/rna.071217.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Neupane R., Pisareva V.P., Rodriguez C.F., Pisarev A.V., Fernández I.S. A complex IRES at the 5’-UTR of a viral mRNA assembles a functional 48S complex via an uAUG intermediate. eLife. 2020;9 doi: 10.7554/eLife.54575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Jha S. Trans-kingdom mimicry underlies ribosome customization by a poxvirus kinase. Nature. 2017;546:651–655. doi: 10.1038/nature22814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wu X. Pelo is required for high efficiency viral replication. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ooi Y.S. An RNA-centric dissection of host complexes controlling flavivirus infection. Nat. Microbiol. 2019;4:2369–2382. doi: 10.1038/s41564-019-0518-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Beutler B. Genetic analysis of resistance to viral infection. Nat. Rev. Immunol. 2007;7:753–766. doi: 10.1038/nri2174. [DOI] [PubMed] [Google Scholar]
- 178.Kato H. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 179.Gitlin L. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl. Acad. Sci. U. S. A. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Tabeta K. Toll-like receptors 9 and 3 as essential components of innate immune defense against mouse cytomegalovirus infection. Proc. Natl. Acad. Sci. U. S. A. 2004;101:3516–3521. doi: 10.1073/pnas.0400525101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Ding S.-W., Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Schott D.H., Cureton D.K., Whelan S.P., Hunter C.P. An antiviral role for the RNA interference machinery in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 2005;102:18420–18424. doi: 10.1073/pnas.0507123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.van Rij R.P. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 2006;20:2985–2995. doi: 10.1101/gad.1482006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Elkayam E. The structure of human Argonaute-2 in complex with miR-20a. Cell. 2012;150:100–110. doi: 10.1016/j.cell.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Botos I., Segal D.M., Davies D.R. The structural biology of toll-like receptors. Structure. 2011;19:447–459. doi: 10.1016/j.str.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wu B. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell. 2013;152:276–289. doi: 10.1016/j.cell.2012.11.048. [DOI] [PubMed] [Google Scholar]
- 187.Kolakofsky D., Kowalinski E., Cusack S. A structure-based model of RIG-I activation. RNA. 2012;18:2118–2127. doi: 10.1261/rna.035949.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Macrae I.J. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 189.Williams G.D., Gokhale N.S., Horner S.M. Regulation of viral infection by the RNA modification N6-Methyladenosine. Annu. Rev. Virol. 2019;6:235–253. doi: 10.1146/annurev-virology-092818-015559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.McIntyre W. Positive-sense RNA viruses reveal the complexity and dynamics of the cellular and viral epitranscriptomes during infection. Nucleic Acids Res. 2018;46:5776–5791. doi: 10.1093/nar/gky029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Girardi E., Chane-Woon-Ming B., Messmer M., Kaukinen P., Pfeffer S. Identification of RNase L-dependent, 3’-end-modified, viral small RNAs in Sindbis virus-infected mammalian cells. mBio. 2013;4 doi: 10.1128/mBio.00698-13. e00698-00613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Erhard F. scSLAM-seq reveals core features of transcription dynamics in single cells. Nature. 2019;571:419–423. doi: 10.1038/s41586-019-1369-y. [DOI] [PubMed] [Google Scholar]
- 193.Jenjaroenpun P. Decoding the epitranscriptional landscape from native RNA sequences. Nucleic Acids Res. 2020 doi: 10.1093/nar/gkaa620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Lorenz D.A., Sathe S., Einstein J.M., Yeo G.W. Direct RNA sequencing enables m6A detection in endogenous transcript isoforms at base-specific resolution. RNA N. Y. N. 2020;26:19–28. doi: 10.1261/rna.072785.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Tahmasebi S., Sonenberg N., Hershey J.W.B., Mathews M.B.Protein. Cold spring harbSynthesis and translational control: a historical perspective. Perspect. Biol. Med. 2019;11:a035584. doi: 10.1101/cshperspect.a035584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Ehrenfeld E., Hunt T. Double-stranded poliovirus RNA inhibits initiation of protein synthesis by reticulocyte lysates. Proc. Natl. Acad. Sci. 1971;68:1075–1078. doi: 10.1073/pnas.68.5.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Farrell P.J., Balkow K., Hunt T., Jackson R.J., Trachsel H. Phosphorylation of initiation factor elF-2 and the control of reticulocyte protein synthesis. Cell. 1977;11:187–200. doi: 10.1016/0092-8674(77)90330-0. [DOI] [PubMed] [Google Scholar]
- 198.Kaminski A., Belsham G.J., Jackson R.J. Translation of encephalomyocarditis virus RNA: parameters influencing the selection of the internal initiation site. EMBO J. 1994;13:1673–1681. doi: 10.1002/j.1460-2075.1994.tb06431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Kieft J.S. Viral IRES RNA structures and ribosome interactions. Trends Biochem. Sci. 2008;33:274–283. doi: 10.1016/j.tibs.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Spahn C.M. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science. 2001;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- 201.Thoms M. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science. 2020 doi: 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Schubert K. SARS-CoV-2 Nsp1 binds ribosomal mRNA channel to inhibit translation. bioRxiv. 2020 doi: 10.1101/2020.07.07.191676. 2020.07.07.191676. [DOI] [PubMed] [Google Scholar]
- 203.Khuperkar D. Quantification of mRNA translation in live cells using single-molecule imaging. Nat. Protoc. 2020;15:1371–1398. doi: 10.1038/s41596-019-0284-x. [DOI] [PubMed] [Google Scholar]
- 204.Ingolia N.T. Ribosome profiling: new views of translation, from single codons to genome scale. Nat. Rev. Genet. 2014;15:205–213. doi: 10.1038/nrg3645. [DOI] [PubMed] [Google Scholar]
- 205.Dinman J.D. Mechanisms and implications of programmed translational frameshifting. Wiley Interdiscip. Rev. RNA. 2012;3:661–673. doi: 10.1002/wrna.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Napthine S. Protein-directed ribosomal frameshifting temporally regulates gene expression. Nat. Commun. 2017;8:1–11. doi: 10.1038/ncomms15582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Reid D.W. Dengue virus selectively annexes endoplasmic reticulum-associated translation machinery as a strategy for Co-opting host cell protein synthesis. J. Virol. 2018;92 doi: 10.1128/JVI.01766-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Ule J. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 209.Scheel T.K.H. A broad RNA virus survey reveals both miRNA dependence and functional sequestration. Cell Host Microbe. 2016;19:409–423. doi: 10.1016/j.chom.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Flynn R.A. Dissecting noncoding and pathogen RNA-protein interactomes. RNA. 2015;21:135–143. doi: 10.1261/rna.047803.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Eckenfelder A. Argonaute proteins regulate HIV-1 multiply spliced RNA and viral production in a Dicer independent manner. Nucleic Acids Res. 2017;45:4158–4173. doi: 10.1093/nar/gkw1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Williams G.D. Nucleotide resolution mapping of influenza A virus nucleoprotein-RNA interactions reveals RNA features required for replication. Nat. Commun. 2018;9:465. doi: 10.1038/s41467-018-02886-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Sokoloski K.J. Identification of interactions between sindbis virus capsid protein and cytoplasmic vRNA as novel virulence determinants. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Bieniasz P.D., Kutluay S.B. CLIP-related methodologies and their application to retrovirology. Retrovirology. 2018;15 doi: 10.1186/s12977-018-0417-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Haddad C., Davila-Calderon J., Tolbert B.S. Integrated approaches to reveal mechanisms by which RNA viruses reprogram the cellular environment. Methods. 2020 doi: 10.1016/j.ymeth.2020.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Haecker I., Renne R. HITS-CLIP and PAR-CLIP advance viral miRNA targetome analysis. Crit. Rev. Eukaryot. Gene Expr. 2014;24:101–116. doi: 10.1615/critreveukaryotgeneexpr.2014006367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Hentze M.W., Castello A., Schwarzl T., Preiss T. A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 2018;19:327–341. doi: 10.1038/nrm.2017.130. [DOI] [PubMed] [Google Scholar]
- 218.Castello A. System-wide identification of RNA-binding proteins by interactome capture. Nat. Protoc. 2013;8:491–500. doi: 10.1038/nprot.2013.020. [DOI] [PubMed] [Google Scholar]
- 219.Garcia-Moreno M. System-wide profiling of RNA-binding proteins uncovers key regulators of virus infection. Mol. Cell. 2019;74:196–211. doi: 10.1016/j.molcel.2019.01.017. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Chu C., Chang H.Y. ChIRP-MS: RNA-directed proteomic discovery. Methods Mol. Biol. Clifton NJ. 2018;1861:37–45. doi: 10.1007/978-1-4939-8766-5_3. [DOI] [PubMed] [Google Scholar]
- 221.Chu C. Systematic discovery of Xist RNA binding proteins. Cell. 2015;161:404–416. doi: 10.1016/j.cell.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Ooi Y.S. An RNA-centric dissection of host complexes controlling flavivirus infection. Nat. Microbiol. 2019 doi: 10.1038/s41564-019-0518-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Neufeldt C.J., Cortese M., Acosta E.G., Bartenschlager R. Rewiring cellular networks by members of the Flaviviridae family. Nat. Rev. Microbiol. 2018;16:125–142. doi: 10.1038/nrmicro.2017.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Kim B. Discovery of widespread host protein interactions with the pre-replicated genome of CHIKV using VIR-CLASP. Mol. Cell. 2020;78:624–640. doi: 10.1016/j.molcel.2020.04.013. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Ramanathan M., Porter D.F., Khavari P.A. Methods to study RNA-protein interactions. Nat. Methods. 2019;16:225–234. doi: 10.1038/s41592-019-0330-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Kim D.I. An improved smaller biotin ligase for BioID proximity labeling. Mol. Biol. Cell. 2016;27:1188–1196. doi: 10.1091/mbc.E15-12-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Roux K.J., Kim D.I., Raida M., Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 2012;196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Ramanathan M. RNA-protein interaction detection in living cells. Nat. Methods. 2018;15:207–212. doi: 10.1038/nmeth.4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Jacob S.T. Ebola virus disease. Nat. Rev. Dis. Primer. 2020;6:1–31. doi: 10.1038/s41572-020-0147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Miras M., Miller W.A., Truniger V., Aranda M.A. Non-canonical translation in plant RNA viruses. Front. Plant Sci. 2017;8 doi: 10.3389/fpls.2017.00494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Garcia-Moreno M., Järvelin A.I., Castello A. Unconventional RNA-binding proteins step into the virus-host battlefront. Wiley Interdiscip. Rev. RNA. 2018;9:e1498. doi: 10.1002/wrna.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Zhang R. A CRISPR screen defines a signal peptide processing pathway required by flaviviruses. Nature. 2016;535:164–168. doi: 10.1038/nature18625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Jagannathan S. Multifunctional roles for the protein translocation machinery in RNA anchoring to the endoplasmic reticulum. J. Biol. Chem. 2014;289:25907–25924. doi: 10.1074/jbc.M114.580688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Freije C.A. Programmable inhibition and detection of RNA viruses using Cas13. Mol. Cell. 2019;76:826–837. doi: 10.1016/j.molcel.2019.09.013. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Abbott T.R. Development of CRISPR as an antiviral strategy to combat SARS-CoV-2 and influenza. Cell. 2020 doi: 10.1016/j.cell.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Cox D.B.T. RNA editing with CRISPR-Cas13. Science. 2017;358:1019–1027. doi: 10.1126/science.aaq0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Feng Q. Enterovirus 2Apro targets MDA5 and MAVS in infected cells. J. Virol. 2014;88:3369–3378. doi: 10.1128/JVI.02712-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Feng Q. MDA5 detects the double-stranded RNA replicative form in picornavirus-infected cells. Cell Rep. 2012;2:1187–1196. doi: 10.1016/j.celrep.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Dauber B., Wolff T. Activation of the antiviral kinase PKR and viral countermeasures. Viruses. 2009;1:523–544. doi: 10.3390/v1030523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Li Y. Activation of RNase L is dependent on OAS3 expression during infection with diverse human viruses. Proc. Natl. Acad. Sci. U. S. A. 2016;113:2241–2246. doi: 10.1073/pnas.1519657113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Xu Y.-P. Zika virus infection induces RNAi-mediated antiviral immunity in human neural progenitors and brain organoids. Cell Res. 2019;29:265–273. doi: 10.1038/s41422-019-0152-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.