Abstract
The cell envelope of Gram-negative bacteria is a multilayered structure essential for bacterial viability; the peptidoglycan cell wall provides shape and osmotic protection to the cell, and the outer membrane serves as a permeability barrier against noxious compounds in the external environment. Assembling the envelope properly and maintaining its integrity are matters of life and death for bacteria. Our understanding of the mechanisms of envelope assembly and maintenance has increased tremendously over the past two decades. Here, we review the major achievements made during this time, giving central stage to the amino acid cysteine, one of the least abundant amino acid residues in proteins, whose unique chemical and physical properties often critically support biological processes. First, we review how cysteines contribute to envelope homeostasis by forming stabilizing disulfides in crucial bacterial assembly factors (LptD, BamA, and FtsN) and stress sensors (RcsF and NlpE). Second, we highlight the emerging role of enzymes that use cysteine residues to catalyze reactions that are necessary for proper envelope assembly, and we also explain how these enzymes are protected from oxidative inactivation. Finally, we suggest future areas of investigation, including a discussion of how cysteine residues could contribute to envelope homeostasis by functioning as redox switches. By highlighting the redox pathways that are active in the envelope of Escherichia coli, we provide a timely overview of the assembly of a cellular compartment that is the hallmark of Gram-negative bacteria.
Keywords: DsbA; DsbC; DsbD; BamA; LptD; LdtA; YbiS; sulfenic acid; FtsN; RcsF; NlpE; Rcs system; Cpx; l,d-transpeptidase; peptidoglycan; outer membrane; bacterial signal transduction; Gram-negative bacteria; disulfide; thioredoxin; lipopolysaccharide; oxidative stress
The cell envelope of Gram-negative bacteria is a complex macromolecular structure that consists of an inner membrane surrounding the cytoplasm and an outer membrane that separates the cell from the environment. While the inner membrane is a classic phospholipid bilayer, the outer membrane is asymmetric, with phospholipids in the inner leaflet and lipopolysaccharides in the outer leaflet (1). The two membranes are separated by the periplasm, a viscous compartment that represents 10–20% of the total cell volume (2) and contains a thin layer of peptidoglycan. The peptidoglycan, also referred to as the cell wall, is a polymer made of repeating units of a disaccharide (GlcNAc-N-acetylmuramic acid) cross-linked by short peptides (3, 4). In enterobacteria, the outer membrane and the peptidoglycan are covalently attached by protein tethers (5, 6). In the model bacterium Escherichia coli, approximately one-third of cellular proteins are destined for the cell envelope (7). Soluble proteins are present in the periplasm, where they engage in a variety of functions, including peptidoglycan assembly, protein folding, and nutrient import. Integral membrane proteins are present in both membranes. While inner membrane proteins cross the lipid bilayer via hydrophobic α-helices, proteins inserted in the outer membrane contain amphipathic β-strands that are arranged in a linear antiparallel β-sheet; this β-sheet folds into a barrel by establishing hydrogen bonds between the first and last β-strands (1, 8). Some of these so-called β-barrels function as passive diffusion channels, allowing small hydrophilic molecules to enter the cell, when others, connected to energy sources in the inner membrane, actively import specific compounds (9). Other important envelope proteins are the lipoproteins, globular proteins anchored to a membrane by a lipid moiety. Although some lipoproteins remain in the inner membrane, most of them are targeted to the outer membrane (10, 11).
Each envelope layer is essential for viability; the outer membrane serves as a permeability barrier against toxic compounds present in the surroundings (8), the peptidoglycan provides shape and osmotic protection to the cell (3), and the inner membrane delimits the cytoplasm and hosts many vital cellular processes, including respiratory systems. The crucial importance of the envelope is nicely illustrated by the fact that several antibiotics (3) and antibacterial toxins (12) target the mechanisms of peptidoglycan assembly while others, such as colistin (a last resort antibiotic), destabilize the outer membrane. Assembling the envelope properly is challenging, in part because the complex machineries involved in the biogenesis of its different layers need to coordinate and to adapt their activities to the growth rate. In addition, most of the envelope building blocks necessary for synthesis, being produced in the cytoplasm or in the inner membrane, need to be transported to their final destination in an assembly-competent state and correctly integrated in the construction despite the lack of an obvious energy source (there is no ATP in the periplasm (13)). In the case of envelope proteins, the large majority of which enter the periplasm in an unfolded state that is prone to aggregation (14), correct folding often involves the formation of one or more disulfide bonds.
The understanding of the mechanisms of envelope assembly and maintenance has increased tremendously during the past two decades. For instance, the machineries involved in the biogenesis of the outer membrane have been identified and their characterization has been initiated. Further, elegant mechanisms used by cells to monitor the integrity of their envelopes have started to be elucidated. These major achievements are reviewed here; however, they are discussed from an unusual perspective. Indeed, we have chosen to give central stage to the amino acid cysteine, one of the least abundant amino acid residues in proteins, whose unique chemical and physical properties make it often critical in biological processes. Putting cysteine residues under the spotlight brings to the surface often overlooked connections between essential cellular processes; it also highlights the important role played by redox-dependent mechanisms in cell envelope homeostasis. In the envelope, cysteine residues have two major functions that are reviewed here. First, they are involved in the formation of disulfide bonds that give envelope proteins further stability. Second, through their ability to function as nucleophiles in enzymatic reactions, cysteine residues are central to the activity of enzymes that are required for proper envelope assembly. Using nucleophilic cysteines comes with a price, however; because cysteines are highly vulnerable to oxidizing molecules that target bacteria during infection, cysteine-based enzymes are susceptible to irreversible inactivation and therefore need specific protection mechanisms. This is true both in the cytoplasm and in the periplasm. However, the reducing equivalents used by rescuing systems often originate from the cytoplasmic pool of NADPH (15). As a result, the protection of cysteine-based enzymes functioning in the envelope offers an additional challenge, as it involves transporting electrons across the inner membrane. Here, after briefly introducing the pathways of disulfide formation, we discuss the importance of cysteine residues in the folding of three essential assembly factors (FtsN, LptD, and BamA) and two proteins that cells use to monitor envelope integrity (RcsF and NlpE). Next, we focus on enzymes that utilize cysteine side chains as part of their catalytic machinery, and we address how these proteins are maintained as active in the oxidizing environment of the periplasm.
How disulfides are formed in the periplasm: a brief overview
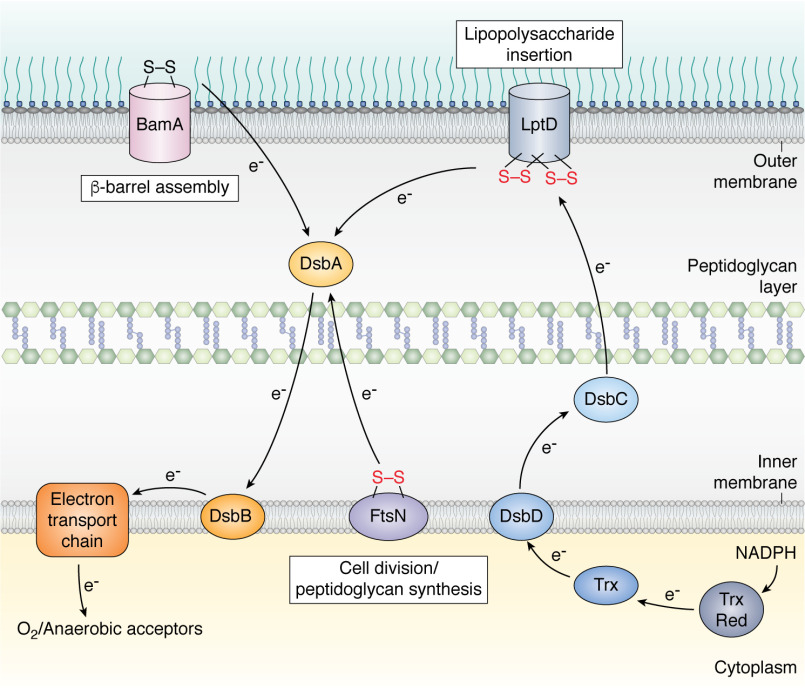
The formation of a disulfide bond between two cysteine residues stabilizes a protein structure, mainly by decreasing the conformational entropy of the denatured state. This stabilizing effect can be up to ∼4 kcal/mol per disulfide formed (16, 17). Disulfide bond formation is vital for the stability of many secreted proteins, both in bacteria and in eukaryotes. Proteins that are secreted to extracytoplasmic compartments such as the cell envelope or to the extracellular milieu benefit from stabilizing disulfides to remain folded in environments that lack ATP-dependent chaperones and often are rich in proteases and destabilizing compounds. Although disulfide bonds can form spontaneously in the presence of molecular oxygen, the process is rather slow and needs to be catalyzed in vivo. The first catalyst of disulfide bond formation identified in bacteria was E. coli DsbA (for Disulfide bond), a small (23-kDa) soluble periplasmic protein with a thioredoxin (Trx) fold. The biochemical characterization of DsbA established this protein as a highly oxidizing oxidoreductase (redox potential of −119 mV) (18) with a CXXC catalytic motif, mostly found oxidized in vivo. The oxidizing power of DsbA comes from the fact that reduction of the CXXC motif increases the stability of DsbA, thereby favoring the transfer of its catalytic disulfide to newly synthesized proteins entering the periplasm (19). The search for a protein capable of reoxidizing inactive DsbA led to the discovery of a small (20-kDa) inner membrane protein, DsbB (20) (Fig. 1 and Table 1). DsbB has two pairs of essential cysteine residues that mediate the transfer of electrons from DsbA to the electron transport chain and ultimately to molecular oxygen (21). Under anaerobic conditions, electrons flow from DsbB to anaerobic electron acceptors, such as nitrate and fumarate (21).
Figure 1.
Proteins that are essential for envelope assembly contain disulfide bonds in their native conformation. Formation of these disulfides is required for two of them, FtsN, an inner membrane protein involved in peptidoglycan synthesis during cell division, and LptD, an outer membrane β-barrel that inserts lipopolysaccharide molecules in the outer leaflet of the outer membrane. Essential disulfides are in red. In contrast, disulfide formation is not required, at least under the tested conditions, for the folding of BamA, the core component of the machinery that introduces β-barrel proteins into the outer membrane. Disulfide bond formation in envelope proteins is catalyzed by DsbA, which is recycled by transferring the electrons received from the substrate to the inner membrane protein DsbB. DsbB then shuttles electrons to the electron transport chain and ultimately to molecular oxygen or anaerobic acceptors. Several envelope proteins, like LptD, contain disulfides between cysteines that are not consecutive in the sequence. In this case, their folding also involves DsbC, a protein disulfide isomerase that corrects the errors of DsbA. DsbC is kept reduced by the inner membrane protein DsbD, which receives reducing equivalents from the cytoplasmic Trx system at the expense of NADPH. The Trx system consists of Trx and Trx reductase (Trx Red). The black arrows show the electron flow.
Table 1.
Key actors in cysteine-mediated envelope homeostasis in E. coli
| Enzyme | Function |
|---|---|
| Envelope oxidoreductases | |
| Disulfide bond formation | |
| DsbA | Catalyzes disulfide bond formation in the periplasm |
| DsbB | Inner membrane protein that recycles DsbA |
| Disulfide bond isomerization | |
| DsbC | Catalyzes disulfide bond isomerization in the periplasm |
| DsbD | Inner membrane protein that recycles DsbC |
| Cysteine protection | |
| DsbG | Rescues periplasmic single cysteine residues from oxidative damage |
| DsbD | Inner membrane protein that recycles DsbG |
| Essential envelope assembly factors with structural disulfide bonds | |
| LptD | Outer membrane protein that inserts lipopolysaccharide molecules into the membrane; two nonconsecutive disulfides; formation of at least one disulfide is essential |
| BamA | Outer membrane protein that inserts β-barrel proteins into the outer membrane; one nonessential disulfide bond |
| FtsN | Inner membrane protein with a large periplasmic domain regulating peptidoglycan synthesis; one essential disulfide bond in the periplasmic domain. |
| Disulfide-containing stress sensors monitoring envelope integrity | |
| RcsF | Outer membrane lipoprotein monitoring the integrity of the peptidoglycan and of the outer membrane; two nonconsecutive disulfides required for folding; induces the Rcs phosphorelay pathway under stress |
| NlpE | Outer membrane lipoprotein monitoring lipoprotein trafficking to the outer membrane; two consecutive disulfides required for folding; induces the Cpx system when lipoprotein transport is perturbed |
| Envelope assembly enzymes with a catalytic cysteine residue | |
| LdtA, LdtB, and LdtC | l,d-Transpeptidases catalyzing the attachment of the Braun lipoprotein Lpp to the peptidoglycan |
| LdtD and LdtE | l,d-Transpeptidases catalyzing the formation of 3-3 cross-links between two meso-diaminopimelic acid residues of adjacent stem peptides |
DsbA catalyzes disulfide bond formation as cysteines in its substrates enter the periplasm. Therefore, when disulfides need to be formed between cysteine residues that are nonconsecutive in the substrate sequence, DsbA often catalyzes the formation of nonnative disulfides, causing protein misfolding, aggregation, and/or degradation. DsbC, a V-shaped dimeric (47-kDa) oxidoreductase of the Trx family, was identified as a disulfide isomerase that corrects the errors of DsbA (22) (Fig. 1). DsbC harbors a CXXC catalytic motif, which, in contrast to DsbA, is found reduced in the periplasm (23), and displays an extended cleft whose inner surface is patched with uncharged and hydrophobic residues (24). These two features allow DsbC to recognize misfolded substrates; first, the N-terminal cysteine of the DsbC active site attacks a nonnative disulfide in the substrate, which results in the formation of an unstable mixed-disulfide complex. Next, the mixed disulfide is resolved either by the attack of another cysteine from the misfolded protein or by the C-terminal cysteine of DsbC itself. In the first case, DsbC acts as an isomerase that catalyzes the reshuffling of the disulfide in the substrate. In the second case, DsbC functions as a reductase, giving DsbA another chance to oxidize the substrate protein. Either way, the active site of DsbC needs to be kept reduced and active, which is the function of DsbD (Fig. 1 and Table 1) (23, 25), a 59-kDa protein with three domains; two domains, DsbDα and DsbDγ, are located in the periplasm, and the third domain, DsbDβ, is embedded in the inner membrane. DsbD uniquely transfers electrons across the membrane, from the cytoplasmic Trx system and NADPH (26) to DsbC (23, 25), via a cascade of thiol-disulfide exchange reactions (27, 28). The actual mechanism by which DsbD transfers electrons between cytoplasmic and periplasmic oxidoreductases is not fully understood but likely involves major conformational changes within DsbDβ, as suggested by structural studies with CcdA, a DsbDβ homolog (29, 30).
Three essential assembly factors have disulfide-bonded cysteines in their native conformations
There is strong bias against cysteine residues in envelope proteins; in a bacterium like E. coli, only ∼40% of envelope proteins have cysteines, compared with ∼85% of cytoplasmic proteins (31). Remarkably, about 70% of cysteine-containing envelope proteins have even numbers of cysteines, which has been shown to be a marker of disulfide bond formation (31). In cells lacking DsbA, these proteins do not fold properly and are subjected to proteolysis (32), which impairs cellular processes. For instance, cells lacking DsbA are nonmotile because FlgI, a flagellum component and a DsbA substrate, is not properly oxidized (33). In the same line, deleting dsbA decreases virulence in pathogenic strains of E. coli and other Gram-negative bacteria due to the misfolding of virulence factors involved in adhesion, secretion, and toxicity (34). For instance, in Pseudomonas aeruginosa, cells lacking DsbA fail to fold the protease LasB (35), the pilus component PilA (36), and other proteins important for pathogenicity (37). Thus, the proteins involved in disulfide formation are attractive targets for the design of innovative antivirulence strategies (38). Note, however, that the degree of protein misfolding observed in the absence of DsbA varies from protein to protein; while some can barely be detected in cells lacking dsbA (39), others, like the β-barrel OmpA, appear to be stable (40).
Strikingly, of the ∼40 essential proteins belonging to the machineries that assemble the peptidoglycan and the outer membrane, only 3 contain cysteine residues in their sequences (or in their periplasmic segments, in the case of inner membrane proteins). In all 3 cases, these cysteines are involved in disulfide formation. This remarkably small number of cysteine groups in the components of the assembly complexes suggests a negative selection for cysteine residues (31), potentially to protect these machineries from oxidative inactivation.
One of the assembly proteins with two cysteines is the inner membrane protein FtsN (Fig. 1 and Table 1), an essential constituent of the large protein complex that mediates cell division (41). Although the exact function of FtsN remains elusive, this protein likely regulates peptidoglycan synthesis during cytokinesis (septation) (42). The two cysteines of FtsN have been shown to form a disulfide in the large (∼30-kDa) periplasmic SPOR (sporulation-related repeat) domain of the protein (41), a region that is important for binding to denuded peptidoglycan (glycan strands lacking the stem peptides that normally cross-link the glycan polymers) (43). Mutation of the two cysteines of the SPOR domain decreases intracellular FtsN levels and causes cells to grow as filaments, a phenotype indicative of an impaired cell division process. Thus, the disulfide bond of the SPOR domain of FtsN stabilizes the structure of this protein and is important for function (41).
A second essential assembly protein with cysteine residues is LptD (Fig. 1 and Table 1), one of the components of the Lpt (Lipopolysaccharide transport) system that transports lipopolysaccharide molecules across the cell envelope (44). The biosynthesis of lipopolysaccharide, a glycolipid made of three distinct moieties (the lipid A anchor, the core oligosaccharides, and the O-antigen, a sugar polymer of variable composition (45)), takes place in the cytoplasm and in the inner membrane; lipopolysaccharide molecules are then extracted from the inner membrane by the LptB2FGC ABC transporter, transferred to the periplasmic protein LptA, and finally delivered to the outer membrane translocon, made of the large β-barrel LptD and the lipoprotein LptE. Together, the Lpt proteins form a membrane-to-membrane bridge for the unidirectional transport of lipopolysaccharides (46); ATP hydrolysis by LptB2FGC powers the entire process (47).
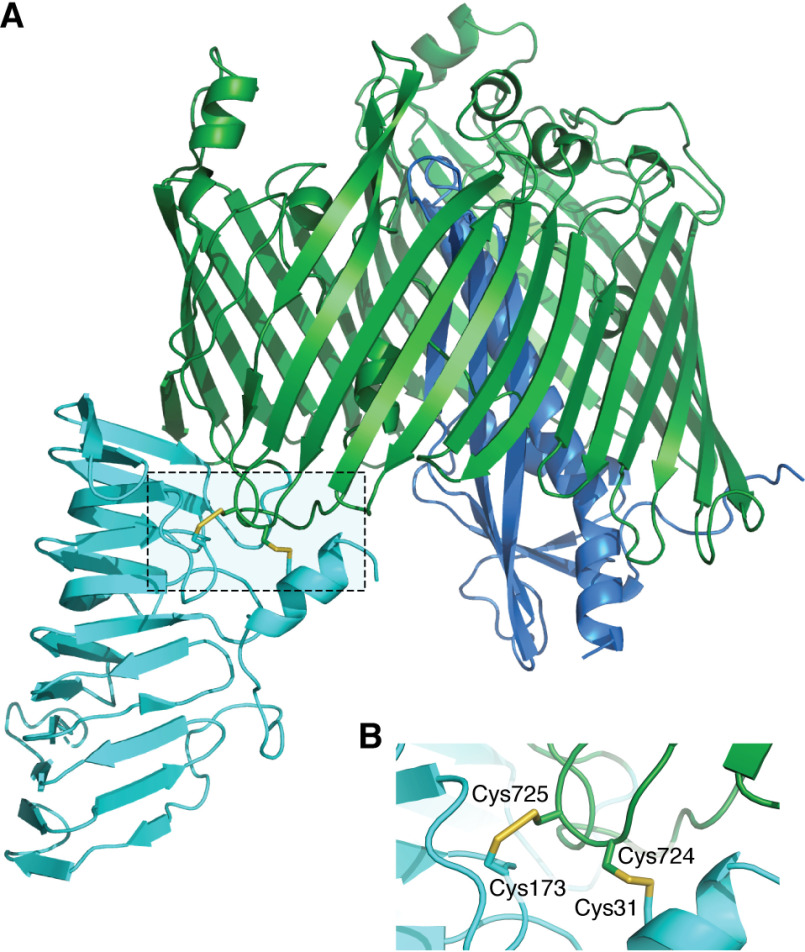
LptD contains four cysteine residues that form two nonconsecutive disulfides (Cys1-Cys3 and Cys2-Cys4) connecting the C-terminal β-barrel, in which LptE forms a plug (48, 49), to an N-terminal domain present in the periplasm (50, 51) (Fig. 2). Formation of at least one of these disulfides is required for function (50). The folding pathway of LptD is particularly complex; following translocation of the nascent protein into the periplasm, a first disulfide is introduced by DsbA between the first and second cysteines. Subsequent rearrangement of this disulfide into a Cys2-Cys4 bridge involves LptE (50, 51) and the protein disulfide isomerase DsbC (52). Formation of the second disulfide by DsbA can then occur. Given the essential function of LptD, one would expect ΔdsbA cells not to be viable. It is indeed the case, but only under anaerobic conditions in which LptD accumulates in a reduced, inactive form (53). In the presence of oxygen, however, ΔdsbA cells grow like WT cells, presumably because oxygen-dependent background oxidation, catalyzed by low-molecular-weight thiol-oxidizing compounds present in the periplasm, is sufficient for survival.
Figure 2.
Structure of E. coli LptD. A, cartoon representation of E. coli LptD (the N-terminal periplasmic domain is in cyan and the C-terminal β-barrel is in green) in complex with the lipoprotein LptE (dark blue). B, the two nonconsecutive disulfide bonds (represented as sticks) of LptD link the N-terminal domain to the C-terminal domain. Formation of at least one of them is required for folding and activity. This is a homology model generated using MODELLER v9.22 (108) and the PDB structures 4RHB (partial E. coli LptD (residues 230–784) in complex with LptE) and 4Q35 (full-length Shigella flexneri LptD in complex with LptE) (49) as templates.
The third assembly protein with two cysteines forming a disulfide is the outer membrane protein BamA, the core component of the β-barrel assembly machinery (BAM) that assembles β-barrel proteins in the outer membrane (Fig. 1 and Table 1). β-Barrel precursors are synthesized in the cytoplasm with an N-terminal signal peptide that targets them to the Sec translocon for transport across the inner membrane. Note that it was recently shown that the mature domains of the proteins destined for secretion contain multiple, degenerate, interchangeable hydrophobic stretches that also play a role in targeting to the translocase machinery (54). Upon emerging from the translocon on the periplasmic side of the inner membrane, the signal peptide is cleaved off and unfolded β-barrels interact with periplasmic chaperones for transport across the periplasm and delivery to BAM (14, 55).
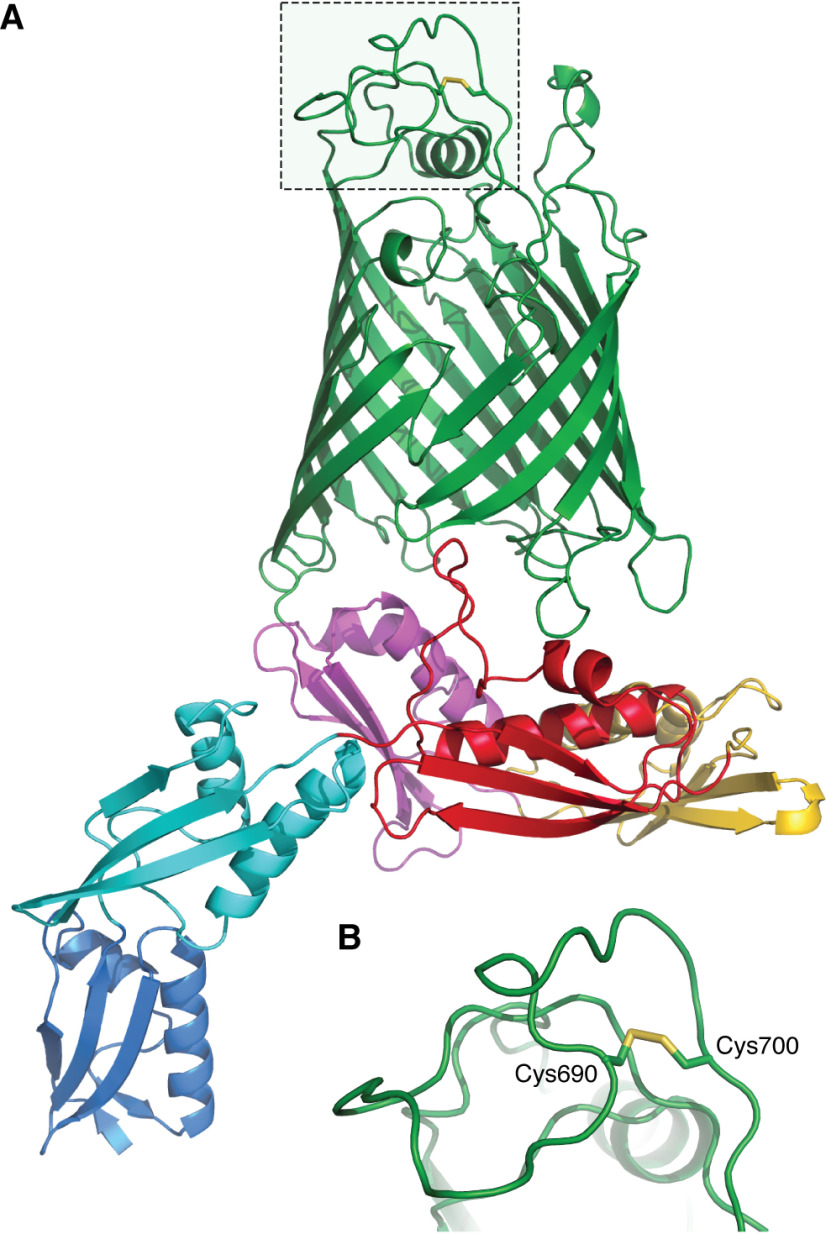
BamA is made of a 16-stranded C-terminal β-barrel embedded in the membrane and a large N-terminal periplasmic extension consisting of five POTRA (polypeptide transport-associated) domains (56) (Fig. 3). Structures of BAM have shown that BamA can adopt two conformations, namely, an outward-open conformation (57–59) in which the β-barrel domain opens between the first and last β-strands, opening a lateral gate to the membrane, and an inward-open conformation (59, 60) in which the lateral gate is closed while a periplasmic entry pore to the barrel lumen is open. In addition to BamA, BAM includes four accessory lipoproteins (BamB, BamC, BamD, and BamE) (61). All components are required for efficient β-barrel assembly, but only BamA and BamD are essential (56, 62). The disulfide bond of BamA is present in an extracellular loop (loop 6 in Fig. 3) of the β-barrel domain (63). Although this loop contains the most highly conserved segment of BamA and has been shown to be important for β-barrel assembly (64, 65), the two cysteine residues are not well conserved among BamA homologs (Iorga B. I., unpublished results), calling into question their functional importance. Accordingly, their mutation has no impact on BAM function (66). Despite important structural and functional insights, crucial questions remain unresolved regarding the mechanism of BAM (56, 67–69).
Figure 3.
Structure of E. coli BamA. A, cartoon representation of E. coli BamA from PDB structure 5D0O (59). The transmembrane β-barrel domain of BamA is colored green and the five periplasmic domains POTRA1, POTRA2, POTRA3, POTRA4, and POTRA5 in blue, cyan, red, yellow, and magenta, respectively. B, the disulfide bond present in extracellular loop 6 of BamA (represented as sticks) is dispensable for folding and activity.
Finally, DsbA was shown to catalyze the formation of disulfides in several BAM substrates (39). For instance, CirA, the outer membrane colicin 1 receptor protein, and FhuA, a β-barrel that functions as a ferrichrome iron receptor, contain one and two disulfide bonds, respectively. When and where these disulfides are formed (before or after BAM folding) remain to be established.
Disulfide bond formation is required for the folding of two important envelope stress sensors
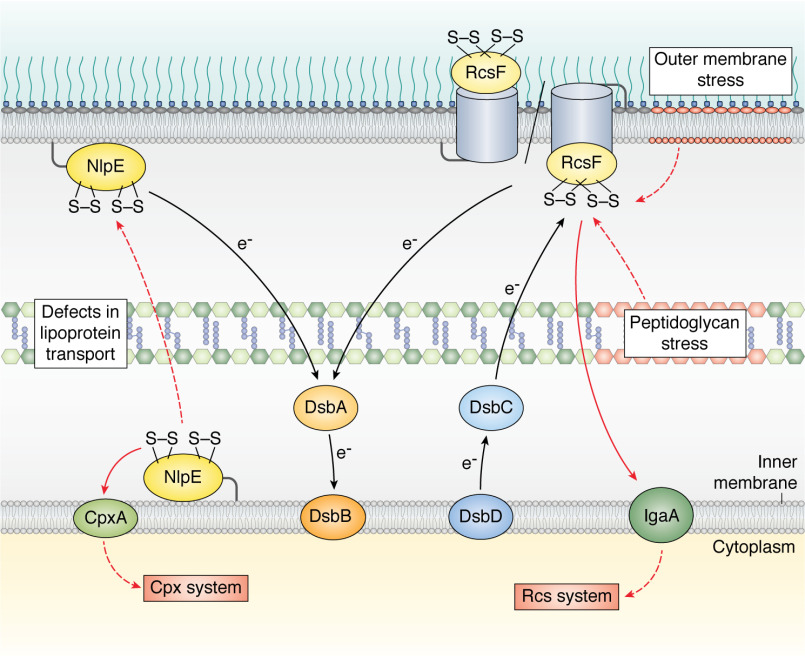
Bacteria evolve in always-changing environments in which they can be exposed to molecules or conditions that alter envelope integrity. Given the vital importance of this compartment, bacteria rely on stress sensor proteins to detect perturbations in their envelope and to respond in a fast and adequate manner to inflicted damage. In E. coli, two major envelope stress sensors, the outer membrane lipoproteins RcsF and NlpE (Fig. 4 and Table 1), both contain disulfide-linked cysteine residues in their native conformations.
Figure 4.
RcsF and NlpE, two outer membrane lipoproteins that monitor the integrity of the E. coli cell envelope, contain disulfide bonds in their native conformations. RcsF detects damage in the peptidoglycan and in the outer membrane. Upon stress, it activates the Rcs system by interacting with the inner membrane protein IgaA (109). NlpE detects perturbations in the transport of lipoproteins to the outer membrane; it accumulates in the inner membrane where it interacts with CpxA, activating the Cpx system. Disulfide bond formation in NlpE and RcsF is catalyzed by the DsbA-DsbB system (see the legend to Fig. 1). RcsF contains disulfides between cysteines that are not consecutive in the sequence; its folding involves the DsbC-DsbD isomerization system (see the legend to Fig. 1). The black arrows show the electron flow. The plain red arrows indicate proteins that directly interact under stress. The dotted arrows indicate processes that occur under stress.
RcsF is a small (11-kDa) surface-exposed lipoprotein that monitors the integrity of the outer part of the envelope, i.e. the outer membrane and the peptidoglycan, in enterobacteria (70). In particular, RcsF detects alterations caused by exposure to polymyxin B (71), a cationic antimicrobial peptide that disrupts the lipopolysaccharide leaflet, or to mecillinam (72), a β-lactam that interferes with peptidoglycan synthesis by inhibiting the essential transpeptidase penicillin-binding protein 2 (PBP2). As a result, RcsF triggers a complex signaling cascade known as the Rcs phosphorelay pathway, which tries to contain the inflicted damage by modulating the expression of dozens of genes, including those producing capsular oligosaccharides (70). It is remarkable that the folding of RcsF, a protein required to sense most Rcs-inducing cues, critically depends on two nonconsecutive disulfide bonds (Fig. 4); in cells impaired in disulfide formation (ΔdsbA or ΔdsbB) or disulfide isomerization (ΔdsbC or ΔdsbD), RcsF does not fold (40, 73) and is degraded by periplasmic proteases. Interestingly, one of the two RcsF disulfides connects two adjacent antiparallel β-strands (73), which is rarely seen in proteins (74). This unusual feature led to the proposal that this disulfide might function as a redox switch controlling the ability of the protein to detect stress (73). However, no evidence supporting this hypothesis has been reported so far. Instead, although the exact mechanism by which RcsF monitors envelope integrity is still a matter of debate (75–77), it is clear that the ability of RcsF to sense stress is linked to the unusual presence of this protein on the cell surface (the general view is that E. coli outer membrane lipoproteins face the periplasm). Interestingly, the export of RcsF to the surface is mediated by BAM via the assembly of complexes between RcsF and abundant β-barrel proteins, such as OmpC and OmpF (Fig. 4). The structure of a BamA-RcsF complex, which forms as an intermediate in the assembly of the complexes between RcsF and its β-barrel partners, was solved recently. In this complex, RcsF is lodged deep inside the lumen of the BamA barrel, which is observed in the inward-open conformation (78). Introduction of artificial disulfides again proved useful in revealing that RcsF does not bind to BamA when it is locked in the outward-open conformation (78).
A second envelope stress sensor with disulfide-bonded cysteines in its native conformation is the outer membrane lipoprotein NlpE (Fig. 4), which activates the Cpx stress response when lipoprotein trafficking to the outer membrane is perturbed (79, 80). In this case, NlpE accumulates in the inner membrane, where it can physically interact with the inner membrane sensor histidine kinase CpxA, triggering Cpx (79, 80). As a result, CpxA autophosphorylates and transfers its phosphoryl group to the cytoplasmic response regulator CpxR, which then binds DNA to regulate the expression of a large set of genes (81, 82).
NlpE consists of two distinct structural domains, namely, an N-terminal domain, which interacts with CpxA (80) and is homologous to the lipocalin Blc, a bacterial lipoprotein that binds hydrophobic ligands, and a C-terminal domain, which adopts an oligonucleotide/oligosaccharide-binding fold (83). Each domain of NlpE contains a disulfide bond that is introduced by DsbA (80) (Fig. 4). Interestingly, failure to form the C-terminal disulfide causes NlpE to induce Cpx (80), thus suggesting that the C-terminal disulfide functions as a molecular sensor for redox perturbations. Because dsbA is a Cpx regulon member, this sensing would establish a neat feedback loop. The molecular mechanism of this redox-regulated Cpx induction remains to be determined, however. It is noteworthy that, whereas RcsF occupies a critical position in Rcs, NlpE is not central for Cpx function. Indeed, most Cpx-inducing cues, such as accumulation of misfolded proteins in the periplasm, inner membrane stress, and cell wall perturbations (81, 82), are NlpE independent.
The activity of a family of enzymes important for envelope integrity depends on a single reduced cysteine residue
In E. coli, most envelope proteins either do not have any cysteine residues or have cysteine residues that are involved in disulfides. In the previous paragraphs, we discussed the importance of forming correct disulfide bonds in proteins that are required for envelope biogenesis and protection. In the following, we focus instead on the important role played by cysteine residues that are part of catalytic machineries and therefore need to remain reduced in the envelope. In fact, only a small group of enzymes use cysteine-based chemistry in the E. coli envelope, and these enzymes all belong to the l,d-transpeptidase family.
E. coli expresses six l,d-transpeptidases, but other bacteria, such as Bdellovibrio bacteriovorus, express more than 20 (84). Three of the E. coli l,d-transpeptidases, i.e. LdtA, LdtB, and LdtC, attach the C-terminal lysine residue of the outer membrane lipoprotein Lpp, the numerically most abundant protein in E. coli (also known as the Braun lipoprotein), to a diaminopimelic acid residue in the peptide stems of the peptidoglycan (6, 85) (Fig. 5 and Table 1). This reaction, which provides the only covalent connection between the outer membrane and the peptidoglycan, is required for envelope stiffness (86) and stress sensing (87). Furthermore, in cells lacking LdtA, LdtB, and LdtC, the architecture of the envelope is compromised; the intermembrane distance is modified (88), which impairs Rcs functioning and the ability to detect and to respond to envelope defects (87). The other l,d-transpeptidases expressed by E. coli have a different function. LdtD and LdtE catalyze the formation of 3-3 cross-links between two meso-diaminopimelic acid residues of adjacent stem peptides during peptidoglycan synthesis (89, 90) (Fig. 5 and Table 1), while the enzymatic activity of LdtF (YafK) remains unknown. In E. coli, there are only 2–10% 3-3 cross-links, with the majority of cross-links being between d-Ala and meso-diaminopimelic acid residues (4-3 cross-links) (91). Formation of 3-3 cross-links increases, however, when cells enter stationary phase and when the transport of lipopolysaccharides to the outer membrane is impaired (90, 91).
Figure 5.
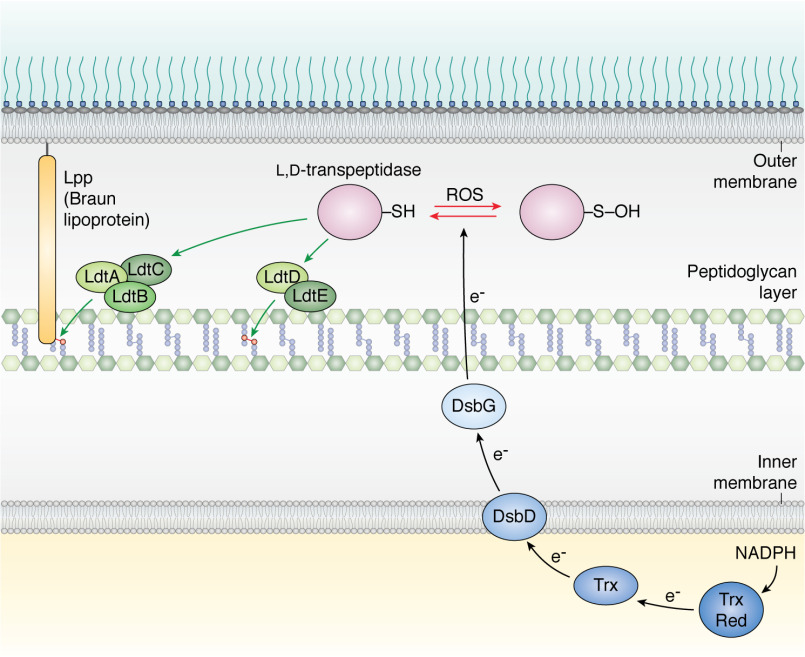
A family of l,d-transpeptidases catalyze reactions that are crucial for envelope assembly using a catalytic cysteine residue. Three l,d-transpeptidases (LdtA, LdtB, and LdtC) catalyze the covalent attachment of the Braun lipoprotein Lpp, the numerically most abundant protein in E. coli, to the peptidoglycan. Two other l,d-transpeptidases (LdtD and LdtE) catalyze the formation of 3-3 cross-links between two meso-diaminopimelic acid residues of adjacent stem peptides of the peptidoglycan. The catalytic cysteine residue of l,d-transpeptidases is prone to oxidation to a sulfenic acid (-SOH) when exposed to reactive oxygen species (ROS). Sulfenic acids can be further oxidized to sulfinic and sulfonic acids (not shown), two irreversible modifications. l,d-Transpeptidases are maintained as reduced and active in the periplasm by DsbG. Electrons are delivered to DsbG by the inner membrane protein DsbD. Electrons are delivered to DsbD as explained in the legend to Fig. 1. The black arrows show the electron flow.
The functional importance of l,d-transpeptidases in the assembly of the envelope is beginning to be fully appreciated, not only in E. coli but also in a large number of bacteria, including mycobacteria, where they play a major role in peptidoglycan assembly by catalyzing abundant 3-3 cross-links (92). These enzymes have an Achilles' heel, however; their activity involves a catalytic cysteine residue that needs to be kept reduced (93) (Fig. 5), which is challenging, given that cysteine residues are particularly sensitive to oxidation because of the electron-rich sulfur atom in their side chain (15). When exposed to oxidants produced by phagocytic cells (94), the thiol side chains of cysteine residues are indeed oxidized to sulfenic acids (-SOH), which are highly reactive and can be irreversibly oxidized to sulfinic acids (-SO2H) and sulfonic acids (-SO3H) (15). These latter two modifications are often detrimental for protein function and inactivate l,d-transpeptidases. Accordingly, it was recently shown that exposure of E. coli to copper, a redox-active metal that is able to catalyze cysteine oxidation in the presence of oxygen (95), inhibits l,d-transpeptidases, compromising Lpp attachment and 3-3 cross-link formation (96). Protecting the thiol functional group of l,d-transpeptidases from oxidation is the function of DsbG (93) (Fig. 5 and Table 1), a periplasmic dimeric oxidoreductase with a Trx-like domain and a CXXC catalytic motif. As in DsbC, this CXXC motif is maintained as reduced by electrons provided by DsbD (97) (Fig. 5). Thus, intracellular metabolism (DsbD is recycled at the expense of NADPH) provides the reducing equivalents to keep cysteine-based envelope enzymes functional, thus maintaining envelope integrity.
Conclusions and perspectives
Since the discovery of DsbA in 1991, an impressive body of research has revealed the critical role played by cysteine residues in the biogenesis and maintenance of the bacterial cell envelope. In the previous sections, we discussed the importance of disulfide bond formation for the folding and stability of envelope proteins, including crucial assembly factors and stress sensors, and we highlighted the functional relevance of enzymes that use cysteine-based chemistry to build the cell envelope properly. It is likely that additional examples of envelope assembly factors with cysteine residues important for folding and/or activity will be identified in the future, in E. coli or in other Gram-negative bacteria. In addition, future research will probably identify novel antioxidant factors protecting envelope proteins from oxidation, an area that has been less well explored than that of oxidative protein folding.
As we reach the end of this review, we would like to suggest the hypothesis that cysteine residues may play an additional role in the envelope by serving as regulatory switches controlling processes necessary for envelope homeostasis. Given their ability to undergo reversible redox modifications, cysteine residues indeed often act as powerful molecular switches allowing organisms to adapt to changes in the environment, as has been extensively described for the bacterial cytoplasm and for higher organisms (98, 99). To our knowledge, no such example has been described so far for the mechanisms that participate in envelope biogenesis. However, several envelope proteins display features that hint at potential redox regulation. For instance, as discussed above, the uncommon presence of a disulfide bond between two adjacent β-strands in RcsF (73) is intriguing and suggests that a layer of redox regulation remains to be discovered for this protein. In addition, both the stress sensor NlpE and PBP1a (an enzyme required for peptidoglycan synthesis) have a disulfide bond between two cysteine residues that are found in a CXXC motif. The fact that CXXC motifs can function as redox switches (74) suggests that these two proteins also may undergo redox regulation. Note that, in the case of NlpE, the redox state of the CXXC motif does not affect Cpx activation (79). Another intriguing case is the abundant outer membrane protein OmpA, which is important for envelope integrity. OmpA is composed of an N-terminal 8-stranded β-barrel and a C-terminal periplasmic domain binding to the peptidoglycan (100, 101). Although this two-domain conformation is well established, some studies have proposed that OmpA can also fold into a 16-stranded β-barrel with a large central pore (102, 103). Interestingly, a disulfide bond present in the C-terminal domain might function as a redox switch controlling the OmpA conformation, as suggested by work in Salmonella enterica serovar Typhymurium (104). Finally, a third envelope protein that could potentially be redox regulated is PBP5, a d,d-carboxypeptidase that cleaves the terminal d-alanine from the pentapeptide side chains in peptidoglycan (3). This enzyme has a single cysteine residing close to the active site but without being involved in catalysis (105). In an old study, however, it was shown that cysteine-modifying reagents inhibit PBP5 (106), strongly suggesting that PBP5 (as well as PBP6a and PBP6b, two E. coli paralogs in which the cysteine residue is conserved (3)) might be subject to redox regulation. The recent development of specific probes designed to monitor the redox state of cysteine residues (107) will facilitate further exploration of the versatile function and crucial roles of the amino acid cysteine in the bacterial cell envelope. In the same line, the fact that the cell envelope is an environment in which disulfides can be formed will prove very useful in studying the mechanism of crucial assembly and surveillance processes. Indeed, in addition to their native roles, disulfides can be artificially introduced into proteins to affect their structures, allowing inference of their function. In the case of BamA, for instance, using artificial disulfides has led to major mechanistic insights by demonstrating the importance of BamA cycling between an outward-open conformation and an inward-open conformation (57–59).
Acknowledgments
We thank Pauline Leverrier for providing insightful comments on the manuscript.
Author contributions—J.-F. C. and C. V. G. writing-original draft; J.-F. C., S.-H. C., B. I. I., and C. V. G. writing-review and editing.
Funding and additional information—This work was supported, in part, by grants from the Fonds de la Recherche Scientifique, from the CNRS, from the FRFS-WELBIO (Grant WELBIO-CR-2019-03), from the EOS Excellence in Research Program of the FWO and FRS-FNRS (Grant G0G0818N), and from the Fédération Wallonie-Bruxelles (Grant ARC 17/22-087).
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- Trx
- thioredoxin
- PBP
- penicillin-binding protein
- BAM
- β-barrel assembly machinery.
References
- 1. Silhavy T. J., Kahne D., and Walker S. (2010) The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2, a000414 10.1101/cshperspect.a000414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vollmer W., and Bertsche U. (2008) Murein (peptidoglycan) structure, architecture and biosynthesis in Escherichia coli. Biochim. Biophys. Acta 1778, 1714–1734 10.1016/j.bbamem.2007.06.007 [DOI] [PubMed] [Google Scholar]
- 3. Typas A., Banzhaf M., Gross C. A., and Vollmer W. (2011) From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 10, 123–136 10.1038/nrmicro2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Egan A. J., Errington J., and Vollmer W. (2020) Regulation of peptidoglycan synthesis and remodelling. Nat. Rev. Microbiol. 10.1038/s41579-020-0366-3 [DOI] [PubMed] [Google Scholar]
- 5. Braun V., and Hantke K. (2019) Lipoproteins: structure, function, biosynthesis. Subcell. Biochem. 92, 39–77 10.1007/978-3-030-18768-2_3 [DOI] [PubMed] [Google Scholar]
- 6. Asmar A. T., and Collet J. F. (2018) Lpp, the Braun lipoprotein, turns 50: major achievements and remaining issues. FEMS Microbiol. Lett. 365, fny199 10.1093/femsle/fny199 [DOI] [PubMed] [Google Scholar]
- 7. Leverrier P., Vertommen D., and Collet J. F. (2010) Contribution of proteomics toward solving the fascinating mysteries of the biogenesis of the envelope of Escherichia coli. Proteomics 10, 771–784 10.1002/pmic.200900461 [DOI] [PubMed] [Google Scholar]
- 8. Konovalova A., Kahne D. E., and Silhavy T. J. (2017) Outer membrane biogenesis. Annu. Rev. Microbiol. 71, 539–556 10.1146/annurev-micro-090816-093754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Noinaj N., Guillier M., Barnard T. J., and Buchanan S. K. (2010) TonB-dependent transporters: regulation, structure, and function. Annu. Rev. Microbiol. 64, 43–60 10.1146/annurev.micro.112408.134247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Okuda S., and Tokuda H. (2011) Lipoprotein sorting in bacteria. Annu. Rev. Microbiol. 65, 239–259 10.1146/annurev-micro-090110-102859 [DOI] [PubMed] [Google Scholar]
- 11. Szewczyk J., and Collet J. F. (2016) The journey of lipoproteins through the cell: one birthplace, multiple destinations. Adv. Microb. Physiol. 69, 1–50 10.1016/bs.ampbs.2016.07.003 [DOI] [PubMed] [Google Scholar]
- 12. Braun V., Helbig S., Patzer S. I., Pramanik A., and Römer C. (2015) Import and export of bacterial protein toxins. Int. J. Med. Microbiol. 305, 238–242 10.1016/j.ijmm.2014.12.006 [DOI] [PubMed] [Google Scholar]
- 13. Wulfing C., and Pluckthun A. (1994) Protein folding in the periplasm of Escherichia coli. Mol. Microbiol. 12, 685–692 [DOI] [PubMed] [Google Scholar]
- 14. De Geyter J., Tsirigotaki A., Orfanoudaki G., Zorzini V., Economou A., and Karamanou S. (2016) Protein folding in the cell envelope of Escherichia coli. Nat. Microbiol. 1, 16107 10.1038/nmicrobiol.2016.107 [DOI] [PubMed] [Google Scholar]
- 15. Ezraty B., Gennaris A., Barras F., and Collet J. F. (2017) Oxidative stress, protein damage and repair in bacteria. Nat. Rev. Microbiol. 15, 385–396 10.1038/nrmicro.2017.26 [DOI] [PubMed] [Google Scholar]
- 16. Clarke J., and Fersht A. R. (1993) Engineered disulfide bonds as probes of the folding pathway of barnase: increasing the stability of proteins against the rate of denaturation. Biochemistry 32, 4322–4329 10.1021/bi00067a022 [DOI] [PubMed] [Google Scholar]
- 17. Pantoliano M. W., Ladner R. C., Bryan P. N., Rollence M. L., Wood J. F., and Poulos T. L. (1987) Protein engineering of subtilisin BPNʹ: enhanced stabilization through the introduction of two cysteines to form a disulfide bond. Biochemistry 26, 2077–2082 10.1021/bi00382a002 [DOI] [PubMed] [Google Scholar]
- 18. Zapun A., Bardwell J. C., and Creighton T. E. (1993) The reactive and destabilizing disulfide bond of DsbA, a protein required for protein disulfide bond formation in vivo. Biochemistry 32, 5083–5092 10.1021/bi00070a016 [DOI] [PubMed] [Google Scholar]
- 19. Grauschopf U., Winther J. R., Korber P., Zander T., Dallinger P., and Bardwell J. C. (1995) Why is DsbA such an oxidizing disulfide catalyst? Cell 83, 947–955 10.1016/0092-8674(95)90210-4 [DOI] [PubMed] [Google Scholar]
- 20. Bardwell J. C., Lee J. O., Jander G., Martin N., Belin D., and Beckwith J. (1993) A pathway for disulfide bond formation in vivo. Proc. Natl. Acad. Sci. U.S.A. 90, 1038–1042 10.1073/pnas.90.3.1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bader M., Muse W., Ballou D. P., Gassner C., and Bardwell J. C. (1999) Oxidative protein folding is driven by the electron transport system. Cell 98, 217–227 10.1016/S0092-8674(00)81016-8 [DOI] [PubMed] [Google Scholar]
- 22. Zapun A., Missiakas D., Raina S., and Creighton T. E. (1995) Structural and functional characterization of DsbC, a protein involved in disulfide bond formation in Escherichia coli. Biochemistry 34, 5075–5089 10.1021/bi00015a019 [DOI] [PubMed] [Google Scholar]
- 23. Rietsch A., Bessette P., Georgiou G., and Beckwith J. (1997) Reduction of the periplasmic disulfide bond isomerase, DsbC, occurs by passage of electrons from cytoplasmic thioredoxin. J. Bacteriol. 179, 6602–6608 10.1128/jb.179.21.6602-6608.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McCarthy A. A., Haebel P. W., Torronen A., Rybin V., Baker E. N., and Metcalf P. (2000) Crystal structure of the protein disulfide bond isomerase, DsbC, from Escherichia coli. Nat. Struct. Biol. 7, 196–199 10.1038/73295 [DOI] [PubMed] [Google Scholar]
- 25. Rietsch A., Belin D., Martin N., and Beckwith J. (1996) An in vivo pathway for disulfide bond isomerization in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 93, 13048–13053 10.1073/pnas.93.23.13048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Collet J. F., and Messens J. (2010) Structure, function, and mechanism of thioredoxin proteins. Antioxid. Redox Signal. 13, 1205–1216 10.1089/ars.2010.3114 [DOI] [PubMed] [Google Scholar]
- 27. Katzen F., and Beckwith J. (2000) Transmembrane electron transfer by the membrane protein DsbD occurs via a disulfide bond cascade. Cell 103, 769–779 10.1016/S0092-8674(00)00180-X [DOI] [PubMed] [Google Scholar]
- 28. Collet J. F., Riemer J., Bader M. W., and Bardwell J. C. (2002) Reconstitution of a disulfide isomerization system. J. Biol. Chem. 277, 26886–26892 10.1074/jbc.M203028200 [DOI] [PubMed] [Google Scholar]
- 29. Williamson J. A., Cho S. H., Ye J., Collet J. F., Beckwith J. R., and Chou J. J. (2015) Structure and multistate function of the transmembrane electron transporter CcdA. Nat. Struct. Mol. Biol. 22, 809–814 10.1038/nsmb.3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhou Y., and Bushweller J. H. (2018) Solution structure and elevator mechanism of the membrane electron transporter CcdA. Nat. Struct. Mol. Biol. 25, 163–169 10.1038/s41594-018-0022-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dutton R. J., Boyd D., Berkmen M., and Beckwith J. (2008) Bacterial species exhibit diversity in their mechanisms and capacity for protein disulfide bond formation. Proc. Natl. Acad. Sci. U.S.A. 105, 11933–11938 10.1073/pnas.0804621105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Depuydt M., Messens J., and Collet J. F. (2011) How proteins form disulfide bonds. Antioxid. Redox Signal. 15, 49–66 10.1089/ars.2010.3575 [DOI] [PubMed] [Google Scholar]
- 33. Dailey F. E., and Berg H. C. (1993) Mutants in disulfide bond formation that disrupt flagellar assembly in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 90, 1043–1047 10.1073/pnas.90.3.1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Heras B., Shouldice S. R., Totsika M., Scanlon M. J., Schembri M. A., and Martin J. L. (2009) DSB proteins and bacterial pathogenicity. Nat. Rev. Microbiol. 7, 215–225 10.1038/nrmicro2087 [DOI] [PubMed] [Google Scholar]
- 35. Braun P., Ockhuijsen C., Eppens E., Koster M., Bitter W., and Tommassen J. (2001) Maturation of Pseudomonas aeruginosa elastase: formation of the disulfide bonds. J. Biol. Chem. 276, 26030–26035 10.1074/jbc.M007122200 [DOI] [PubMed] [Google Scholar]
- 36. Ha U.-H., Wang Y., and Jin S. (2003) DsbA of Pseudomonas aeruginosa is essential for multiple virulence factors. Infect. Immun. 71, 1590–1595 10.1128/iai.71.3.1590-1595.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Arts I. S., Ball G., Leverrier P., Garvis S., Nicolaes V., Vertommen D., Ize B., Tamu Dufe V., Messens J., Voulhoux R., and Collet J. F. (2013) Dissecting the machinery that introduces disulfide bonds in Pseudomonas aeruginosa. mBio 4, e00912–13 10.1128/mBio.00912-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Smith R. P., Paxman J. J., Scanlon M. J., and Heras B. (2016) Targeting bacterial Dsb proteins for the development of anti-virulence agents. Molecules 21, 811 10.3390/molecules21070811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vertommen D., Depuydt M., Pan J., Leverrier P., Knoops L., Szikora J. P., Messens J., Bardwell J. C., and Collet J. F. (2008) The disulphide isomerase DsbC cooperates with the oxidase DsbA in a DsbD-independent manner. Mol. Microbiol. 67, 336–349 10.1111/j.1365-2958.2007.06030.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kadokura H., Tian H., Zander T., Bardwell J. C., and Beckwith J. (2004) Snapshots of DsbA in action: detection of proteins in the process of oxidative folding. Science 303, 534–537 10.1126/science.1091724 [DOI] [PubMed] [Google Scholar]
- 41. Meehan B. M., Landeta C., Boyd D., and Beckwith J. (2017) The essential cell division protein FtsN contains a critical disulfide bond in a non-essential domain. Mol. Microbiol. 103, 413–422 10.1111/mmi.13565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Du S., and Lutkenhaus J. (2019) At the heart of bacterial cytokinesis: the Z ring. Trends Microbiol. 27, 781–791 10.1016/j.tim.2019.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ursinus A., van den Ent F., Brechtel S., de Pedro M., Holtje J. V., Lowe J., and Vollmer W. (2004) Murein (peptidoglycan) binding property of the essential cell division protein FtsN from Escherichia coli. J. Bacteriol. 186, 6728–6737 10.1128/JB.186.20.6728-6737.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sperandeo P., Martorana A. M., and Polissi A. (2019) The Lpt ABC transporter for lipopolysaccharide export to the cell surface. Res. Microbiol. 170, 366–373 10.1016/j.resmic.2019.07.005 [DOI] [PubMed] [Google Scholar]
- 45. Whitfield C., and Trent M. S. (2014) Biosynthesis and export of bacterial lipopolysaccharides. Annu. Rev. Biochem. 83, 99–128 10.1146/annurev-biochem-060713-035600 [DOI] [PubMed] [Google Scholar]
- 46. Sherman D. J., Xie R., Taylor R. J., George A. H., Okuda S., Foster P. J., Needleman D. J., and Kahne D. (2018) Lipopolysaccharide is transported to the cell surface by a membrane-to-membrane protein bridge. Science 359, 798–801 10.1126/science.aar1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xie R., Taylor R. J., and Kahne D. (2018) Outer membrane translocon communicates with inner membrane ATPase to stop lipopolysaccharide transport. J. Am. Chem. Soc. 140, 12691–12694 10.1021/jacs.8b07656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Freinkman E., Chng S. S., and Kahne D. (2011) The complex that inserts lipopolysaccharide into the bacterial outer membrane forms a two-protein plug-and-barrel. Proc. Natl. Acad. Sci. U.S.A. 108, 2486–2491 10.1073/pnas.1015617108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Qiao S., Luo Q., Zhao Y., Zhang X. C., and Huang Y. (2014) Structural basis for lipopolysaccharide insertion in the bacterial outer membrane. Nature 511, 108–111 10.1038/nature13484 [DOI] [PubMed] [Google Scholar]
- 50. Ruiz N., Chng S. S., Hiniker A., Kahne D., and Silhavy T. J. (2010) Nonconsecutive disulfide bond formation in an essential integral outer membrane protein. Proc. Natl. Acad. Sci. U.S.A. 107, 12245–12250 10.1073/pnas.1007319107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chng S. S., Xue M., Garner R. A., Kadokura H., Boyd D., Beckwith J., and Kahne D. (2012) Disulfide rearrangement triggered by translocon assembly controls lipopolysaccharide export. Science 337, 1665–1668 10.1126/science.1227215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Denoncin K., Vertommen D., Paek E., and Collet J. F. (2010) The protein-disulfide isomerase DsbC cooperates with SurA and DsbA in the assembly of the essential β-barrel protein LptD. J. Biol. Chem. 285, 29425–29433 10.1074/jbc.M110.119321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Meehan B. M., Landeta C., Boyd D., and Beckwith J. (2017) The disulfide bond formation pathway is essential for anaerobic growth of Escherichia coli. J. Bacteriol. 199, e00120–17 10.1128/JB.00120-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chatzi K. E., Sardis M. F., Tsirigotaki A., Koukaki M., Sostaric N., Konijnenberg A., Sobott F., Kalodimos C. G., Karamanou S., and Economou A. (2017) Preprotein mature domains contain translocase targeting signals that are essential for secretion. J. Cell Biol. 216, 1357–1369 10.1083/jcb.201609022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Goemans C., Denoncin K., and Collet J. F. (2014) Folding mechanisms of periplasmic proteins. Biochim. Biophys. Acta 1843, 1517–1528 10.1016/j.bbamcr.2013.10.014 [DOI] [PubMed] [Google Scholar]
- 56. Noinaj N., Gumbart J. C., and Buchanan S. K. (2017) The β-barrel assembly machinery in motion. Nat. Rev. Microbiol. 15, 197–204 10.1038/nrmicro.2016.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Iadanza M. G., Higgins A. J., Schiffrin B., Calabrese A. N., Brockwell D. J., Ashcroft A. E., Radford S. E., and Ranson N. A. (2016) Lateral opening in the intact β-barrel assembly machinery captured by cryo-EM. Nat. Commun. 7, 12865 10.1038/ncomms12865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bakelar J., Buchanan S. K., and Noinaj N. (2016) The structure of the β-barrel assembly machinery complex. Science 351, 180–186 10.1126/science.aad3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gu Y., Li H., Dong H., Zeng Y., Zhang Z., Paterson N. G., Stansfeld P. J., Wang Z., Zhang Y., Wang W., and Dong C. (2016) Structural basis of outer membrane protein insertion by the BAM complex. Nature 531, 64–69 10.1038/nature17199 [DOI] [PubMed] [Google Scholar]
- 60. Han L., Zheng J., Wang Y., Yang X., Liu Y., Sun C., Cao B., Zhou H., Ni D., Lou J., Zhao Y., and Huang Y. (2016) Structure of the BAM complex and its implications for biogenesis of outer-membrane proteins. Nat. Struct. Mol. Biol. 23, 192–196 10.1038/nsmb.3181 [DOI] [PubMed] [Google Scholar]
- 61. Wu T., Malinverni J., Ruiz N., Kim S., Silhavy T. J., and Kahne D. (2005) Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121, 235–245 10.1016/j.cell.2005.02.015 [DOI] [PubMed] [Google Scholar]
- 62. Hagan C. L., Silhavy T. J., and Kahne D. (2011) β-Barrel membrane protein assembly by the Bam complex. Annu. Rev. Biochem. 80, 189–210 10.1146/annurev-biochem-061408-144611 [DOI] [PubMed] [Google Scholar]
- 63. Noinaj N., Kuszak A. J., Gumbart J. C., Lukacik P., Chang H., Easley N. C., Lithgow T., and Buchanan S. K. (2013) Structural insight into the biogenesis of β-barrel membrane proteins. Nature 501, 385–390 10.1038/nature12521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Noinaj N., Kuszak A. J., Balusek C., Gumbart J. C., and Buchanan S. K. (2014) Lateral opening and exit pore formation are required for BamA function. Structure 22, 1055–1062 10.1016/j.str.2014.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Leonard-Rivera M., and Misra R. (2012) Conserved residues of the putative L6 loop of Escherichia coli BamA play a critical role in the assembly of β-barrel outer membrane proteins, including that of BamA itself. J. Bacteriol. 194, 4662–4668 10.1128/JB.00825-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rigel N. W., Ricci D. P., and Silhavy T. J. (2013) Conformation-specific labeling of BamA and suppressor analysis suggest a cyclic mechanism for β-barrel assembly in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 110, 5151–5156 10.1073/pnas.1302662110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lee J., Tomasek D., Santos T. M., May M. D., Meuskens I., and Kahne D. (2019) Formation of a β-barrel membrane protein is catalyzed by the interior surface of the assembly machine protein BamA. Elife 8, e49787 10.7554/eLife.49787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Doerner P. A., and Sousa M. C. (2017) Extreme dynamics in the BamA β-barrel seam. Biochemistry 56, 3142–3149 10.1021/acs.biochem.7b00281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Doyle M. T., and Bernstein H. D. (2019) Bacterial outer membrane proteins assemble via asymmetric interactions with the BamA β-barrel. Nat. Commun. 10, 3358 10.1038/s41467-019-11230-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wall E., Majdalani N., and Gottesman S. (2018) The complex Rcs regulatory cascade. Annu. Rev. Microbiol. 72, 111–139 10.1146/annurev-micro-090817-062640 [DOI] [PubMed] [Google Scholar]
- 71. Farris C., Sanowar S., Bader M. W., Pfuetzner R., and Miller S. I. (2010) Antimicrobial peptides activate the Rcs regulon through the outer membrane lipoprotein RcsF. J. Bacteriol. 192, 4894–4903 10.1128/JB.00505-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Laubacher M. E., and Ades S. E. (2008) The Rcs phosphorelay is a cell envelope stress response activated by peptidoglycan stress and contributes to intrinsic antibiotic resistance. J. Bacteriol. 190, 2065–2074 10.1128/JB.01740-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Leverrier P., Declercq J. P., Denoncin K., Vertommen D., Hiniker A., Cho S. H., and Collet J. F. (2011) Crystal structure of the outer membrane protein RcsF, a new substrate for the periplasmic protein-disulfide isomerase DsbC. J. Biol. Chem. 286, 16734–16742 10.1074/jbc.M111.224865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wouters M. A., Fan S. W., and Haworth N. L. (2010) Disulfides as redox switches: from molecular mechanisms to functional significance. Antioxid. Redox Signal. 12, 53–91 10.1089/ars.2009.2510 [DOI] [PubMed] [Google Scholar]
- 75. Cho S. H., Szewczyk J., Pesavento C., Zietek M., Banzhaf M., Roszczenko P., Asmar A., Laloux G., Hov A. K., Leverrier P., Van der Henst C., Vertommen D., Typas A., and Collet J. F. (2014) Detecting envelope stress by monitoring β-barrel assembly. Cell 159, 1652–1664 10.1016/j.cell.2014.11.045 [DOI] [PubMed] [Google Scholar]
- 76. Konovalova A., Perlman D. H., Cowles C. E., and Silhavy T. J. (2014) Transmembrane domain of surface-exposed outer membrane lipoprotein RcsF is threaded through the lumen of β-barrel proteins. Proc. Natl. Acad. Sci. U.S.A. 111, E4350–E4358 10.1073/pnas.1417138111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Konovalova A., Mitchell A. M., and Silhavy T. J. (2016) A lipoprotein/β-barrel complex monitors lipopolysaccharide integrity transducing information across the outer membrane. Elife 5, e15276 10.7554/eLife.15276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Létoquart J., Rodríguez-Alonso R., Louis G., Calabrese A. N., Radford S. E., Cho S.-H., Remaut H., and Collet J.-F. (2019) Structural insight into the formation of lipoprotein-β-barrel complexes by the β-barrel assembly machinery. bioRxiv 823146 10.1101/823146 [DOI] [PMC free article] [PubMed]
- 79. May K. L., Lehman K. M., Mitchell A. M., and Grabowicz M. (2019) A stress response monitoring lipoprotein trafficking to the outer membrane. mBio 10, e00618–19 10.1128/mBio.00618-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Delhaye A., Laloux G., and Collet J. F. (2019) The lipoprotein NlpE is a Cpx sensor that serves as a sentinel for protein sorting and folding defects in the Escherichia coli envelope. J. Bacteriol. 201, e00611–18 10.1128/JB.00611-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Raivio T. L. (2014) Everything old is new again: an update on current research on the Cpx envelope stress response. Biochim. Biophys. Acta 1843, 1529–1541 10.1016/j.bbamcr.2013.10.018 [DOI] [PubMed] [Google Scholar]
- 82. Laloux G., and Collet J. F. (2017) Major Tom to ground control: how lipoproteins communicate extracytoplasmic stress to the decision center of the cell. J. Bacteriol. 199, e00216–17 10.1128/JB.00216-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Hirano Y., Hossain M. M., Takeda K., Tokuda H., and Miki K. (2007) Structural studies of the Cpx pathway activator NlpE on the outer membrane of Escherichia coli. Structure 15, 963–976 10.1016/j.str.2007.06.014 [DOI] [PubMed] [Google Scholar]
- 84. Kuru E., Lambert C., Rittichier J., Till R., Ducret A., Derouaux A., Gray J., Biboy J., Vollmer W., VanNieuwenhze M., Brun Y. V., and Sockett R. E. (2017) Fluorescent d-amino-acids reveal bi-cellular cell wall modifications important for Bdellovibrio bacteriovorus predation. Nat. Microbiol. 2, 1648–1657 10.1038/s41564-017-0029-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Magnet S., Bellais S., Dubost L., Fourgeaud M., Mainardi J.-L., Petit-Frèe S., Marie A., Mengin-Lecreulx D., Arthur M., and Gutmann L. (2007) Identification of the l,d-transpeptidases responsible for attachment of the Braun lipoprotein to Escherichia coli peptidoglycan. J. Bacteriol. 189, 3927–3931 10.1128/JB.00084-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Mathelié-Guinlet M., Asmar A. T., Collet J.-F., and Dufrêne Y. F. (2020) Lipoprotein Lpp regulates the mechanical properties of the E. coli cell envelope. Nat. Commun. 11, 1789 10.1038/s41467-020-15489-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Asmar A. T., Ferreira J. L., Cohen E. J., Cho S. H., Beeby M., Hughes K. T., and Collet J. F. (2017) Communication across the bacterial cell envelope depends on the size of the periplasm. PLoS Biol. 15, e2004303 10.1371/journal.pbio.2004303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Cohen E. J., Ferreira J. L., Ladinsky M. S., Beeby M., and Hughes K. T. (2017) Nanoscale-length control of the flagellar driveshaft requires hitting the tethered outer membrane. Science 356, 197–200 10.1126/science.aam6512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Magnet S., Dubost L., Marie A., Arthur M., and Gutmann L. (2008) Identification of the l,d-transpeptidases for peptidoglycan cross-linking in Escherichia coli. J. Bacteriol. 190, 4782–4785 10.1128/JB.00025-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. More N., Martorana A. M., Biboy J., Otten C., Winkle M., Serrano C. K. G., Monton Silva A., Atkinson L., Yau H., Breukink E., den Blaauwen T., Vollmer W., and Polissi A. (2019) Peptidoglycan remodeling enables Escherichia coli to survive severe outer membrane assembly defect. mBio 10, e02729–18 10.1128/mBio.02729-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Glauner B., Holtje J. V., and Schwarz U. (1988) The composition of the murein of Escherichia coli. J. Biol. Chem. 263, 10088–10095 [PubMed] [Google Scholar]
- 92. Lavollay M., Arthur M., Fourgeaud M., Dubost L., Marie A., Veziris N., Blanot D., Gutmann L., and Mainardi J. L. (2008) The peptidoglycan of stationary-phase Mycobacterium tuberculosis predominantly contains cross-links generated by l,d-transpeptidation. J. Bacteriol. 190, 4360–4366 10.1128/JB.00239-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Depuydt M., Leonard S. E., Vertommen D., Denoncin K., Morsomme P., Wahni K., Messens J., Carroll K. S., and Collet J. F. (2009) A periplasmic reducing system protects single cysteine residues from oxidation. Science 326, 1109–1111 10.1126/science.1179557 [DOI] [PubMed] [Google Scholar]
- 94. Winterbourn C. C., and Kettle A. J. (2013) Redox reactions and microbial killing in the neutrophil phagosome. Antioxid. Redox Signal. 18, 642–660 10.1089/ars.2012.4827 [DOI] [PubMed] [Google Scholar]
- 95. Hiniker A., Collet J. F., and Bardwell J. C. (2005) Copper stress causes an in vivo requirement for the Escherichia coli disulfide isomerase DsbC. J. Biol. Chem. 280, 33785–33791 10.1074/jbc.M505742200 [DOI] [PubMed] [Google Scholar]
- 96. Peters K., Pazos M., Edoo Z., Hugonnet J. E., Martorana A. M., Polissi A., VanNieuwenhze M. S., Arthur M., and Vollmer W. (2018) Copper inhibits peptidoglycan ld-transpeptidases suppressing β-lactam resistance due to bypass of penicillin-binding proteins. Proc. Natl. Acad. Sci. U.S.A. 115, 10786–10791 10.1073/pnas.1809285115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Bessette P. H., Cotto J. J., Gilbert H. F., and Georgiou G. (1999) In vivo and in vitro function of the Escherichia coli periplasmic cysteine oxidoreductase DsbG. J. Biol. Chem. 274, 7784–7792 10.1074/jbc.274.12.7784 [DOI] [PubMed] [Google Scholar]
- 98. Fra A., Yoboue E. D., and Sitia R. (2017) Cysteines as redox molecular switches and targets of disease. Front. Mol. Neurosci. 10, 167 10.3389/fnmol.2017.00167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Groitl B., and Jakob U. (2014) Thiol-based redox switches. Biochim. Biophys. Acta 1844, 1335–1343 10.1016/j.bbapap.2014.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Pautsch A., and Schulz G. E. (1998) Structure of the outer membrane protein A transmembrane domain. Nat. Struct. Biol. 5, 1013–1017 10.1038/2983 [DOI] [PubMed] [Google Scholar]
- 101. Park J. S., Lee W. C., Yeo K. J., Ryu K. S., Kumarasiri M., Hesek D., Lee M., Mobashery S., Song J. H., Kim S. I., Lee J. C., Cheong C., Jeon Y. H., and Kim H. Y. (2012) Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the Gram-negative bacterial outer membrane. FASEB J. 26, 219–228 10.1096/fj.11-188425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Stathopoulos C. (1996) An alternative topological model for Escherichia coli OmpA. Protein Sci. 5, 170–173 10.1002/pro.5560050122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Reusch R. N. (2012) Insights into the structure and assembly of Escherichia coli outer membrane protein A. FEBS J. 279, 894–909 10.1111/j.1742-4658.2012.08484.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. van der Heijden J., Reynolds L. A., Deng W., Mills A., Scholz R., Imami K., Foster L. J., Duong F., and Finlay B. B. (2016) Salmonella rapidly regulates membrane permeability to survive oxidative stress. mBio 7, e01238–16 10.1128/mBio.01238-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Nicholas R., and Strominger J. (1988) Site-directed mutants of a soluble form of penicillin-binding protein 5 from Escherichia coli and their catalytic properties. J. Biol. Chem. 263, 2034–2040 [PubMed] [Google Scholar]
- 106. Curtis S. J., and Strominger J. (1978) Effects of sulfhydryl reagents on the binding and release of penicillin G by d-alanine carboxypeptidase IA of Escherichia coli. J. Biol. Chem. 253, 2584–2588 [PubMed] [Google Scholar]
- 107. Yang J., Carroll K. S., and Liebler D. C. (2016) The expanding landscape of the thiol redox proteome. Mol. Cell. Proteomics 15, 1–11 10.1074/mcp.O115.056051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Sali A., and Blundell T. L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 10.1006/jmbi.1993.1626 [DOI] [PubMed] [Google Scholar]
- 109. Hussein N. A., Cho S. H., Laloux G., Siam R., and Collet J. F. (2018) Distinct domains of Escherichia coli IgaA connect envelope stress sensing and down-regulation of the Rcs phosphorelay across subcellular compartments. PLoS Genet. 14, e1007398 10.1371/journal.pgen.1007398 [DOI] [PMC free article] [PubMed] [Google Scholar]