Abstract
N-Linked glycans are critical to the infection cycle of HIV, and most neutralizing antibodies target the high-mannose glycans found on the surface envelope glycoprotein-120 (gp120). Carbohydrate-binding proteins, particularly mannose-binding lectins, have also been shown to bind these glycans. Despite their therapeutic potency, their ability to cause lymphocyte proliferation limits their application. In this study, we report one such lectin named horcolin (Hordeum vulgare lectin), seen to lack mitogenicity owing to the divergence in the residues at its carbohydrate-binding sites, which makes it a promising candidate for exploration as an anti-HIV agent. Extensive isothermal titration calorimetry experiments reveal that the lectin was sensitive to the length and branching of mannooligosaccharides and thereby the total valency. Modeling and simulation studies demonstrate two distinct modes of binding, a monovalent binding to shorter saccharides and a bivalent mode for higher glycans, involving simultaneous interactions of multiple glycan arms with the primary carbohydrate-binding sites. This multivalent mode of binding was further strengthened by interactions of core mannosyl residues with a secondary conserved site on the protein, leading to an exponential increase in affinity. Finally, we confirmed the interaction of horcolin with recombinant gp120 and gp140 with high affinity and inhibition of HIV infection at nanomolar concentrations without mitogenicity.
Keywords: HIV gp120, lectin, horcolin, anti-HIV agent, molecular dynamics, isothermal titration calorimetry (ITC), glycoprotein, antiviral agent
N-Linked glycans on glycoprotein-120 (gp120) are critical to the biology of HIV for its infectious cycle. They are mainly comprised of Man5–9-GlcNAc2-Asn sugars and display Manα1–2-Man residues at their nonreducing termini. These glycans are found in a cluster away from the receptor-binding sites and the trimeric interface of gp120 (1). The glycan distribution is in line with the “antigenic map” of gp120, which mainly constitutes the neutralizing face, the heavily glycosylated silent face, and the non-neutralizing face (2, 3). Most of the neutralizing antibodies are designed to target the neutralizing face of the glycoprotein as the glycans on gp120 are constrained there within tight clusters. It is therefore not surprising that all of the clades of HIV-1 are readily neutralized by some mannose-specific lectins. Moreover, Manα1-2-Man residues on gp120 are valuable as vaccine targets as they are antigenically conserved (4–6).
Mannose-specific jacalin-related lectins (mJRLs) constitute a large proportion of this class of proteins and are members of one of the most characterized lectin families. They are known to recognize the terminal and/or internal mannose moieties of N-glycans (7). mJRLs exhibit a common β-prism fold arrangement constituting three Greek motifs with the potential to harbor several sugar-binding sites in the same monomer (8, 9). Their combining sites are conducive for cooperative binding to enhance the binding affinities for additive effect. This cooperativity is especially important for the lectin-sugar interactions, as the binding affinity for a given binding site is usually relatively weak at 2–4 kcal/mol (10).
In this paper, we describe the characterization of a mannose-binding lectin from the coleoptile of Hordeum vulgare (barley), detailing the thermodynamic fingerprint of its interaction with mannose and mannooligosaccharides by isothermal titration calorimetry (ITC), which reveals subtle features of its recognition of bi- and triantennary glycans. The ITC data are further complemented by molecular dynamics simulation studies. The results obtained demonstrate that glycan length and flexibility serve as modulators of the thermodynamics of the protein-binding process. Furthermore, its high affinity for the recognition of glycans in gp120 and gp140 and its ability to neutralize HIV-1 infectivity, coupled with the absence of mitogenicity, highlight its potential as a desirable component of microbicides in our armamentarium against HIV.
Results and discussion
Homolog identification and bioinformatic analysis of horcolin
The 146-residue protein sequence retrieved from Uniprot (ID: Q5U9T2) was annotated to have the jacalin-like lectin domain by the Pfam database. Lectins of this family contain β-prism type I fold with three internal repeats, each being an antiparallel β-sheet (8, 11). A BLAST search of the nonredundant protein sequences with HR as the query, identified 50 target jacalin-like proteins as HR homologs with a query cover of 97–100% having sequence similarity of over 34% with lectins from Oryza sativa (orysata, a salt protein), calsepa lectin, and various banana lectins (BanLec), which were used for further analysis. Previous literature reports the presence of two binding sites in BanLec, a mitogenic lectin (12, 13), whereas orysata, with one active mannose-binding site, is nonmitogenic (14). The phylogenetic tree constructed using the maximum likelihood method positioned HR under the same clade as the orysata lectin with a 44% sequence identity between the sequences. Multiple-sequence alignment (MSA) identified two signature motifs: the Gly-Gly recognition loop and GXXXD regions for carbohydrate binding across the homologs, hinting at the presence of two binding pockets in HR similar to BanLec (Fig. 1). The sequence analyses of the individual primary binding sites had a higher percentage identity with BanLec protein (∼80%), whereas the common secondary binding site was similar to the orysata lectin (∼30%), revealing stark differences in the binding site residues between the three proteins, laying the foundation of this study to characterize its binding pockets and its potential in recognition of high-mannose N-glycans. The residues 134GAFLD138 will henceforth be called carbohydrate-binding site I (CBS I), and residues 35GAIVD39 will be called carbohydrate-binding site II (CBS II) (Fig. S1).
Figure 1.
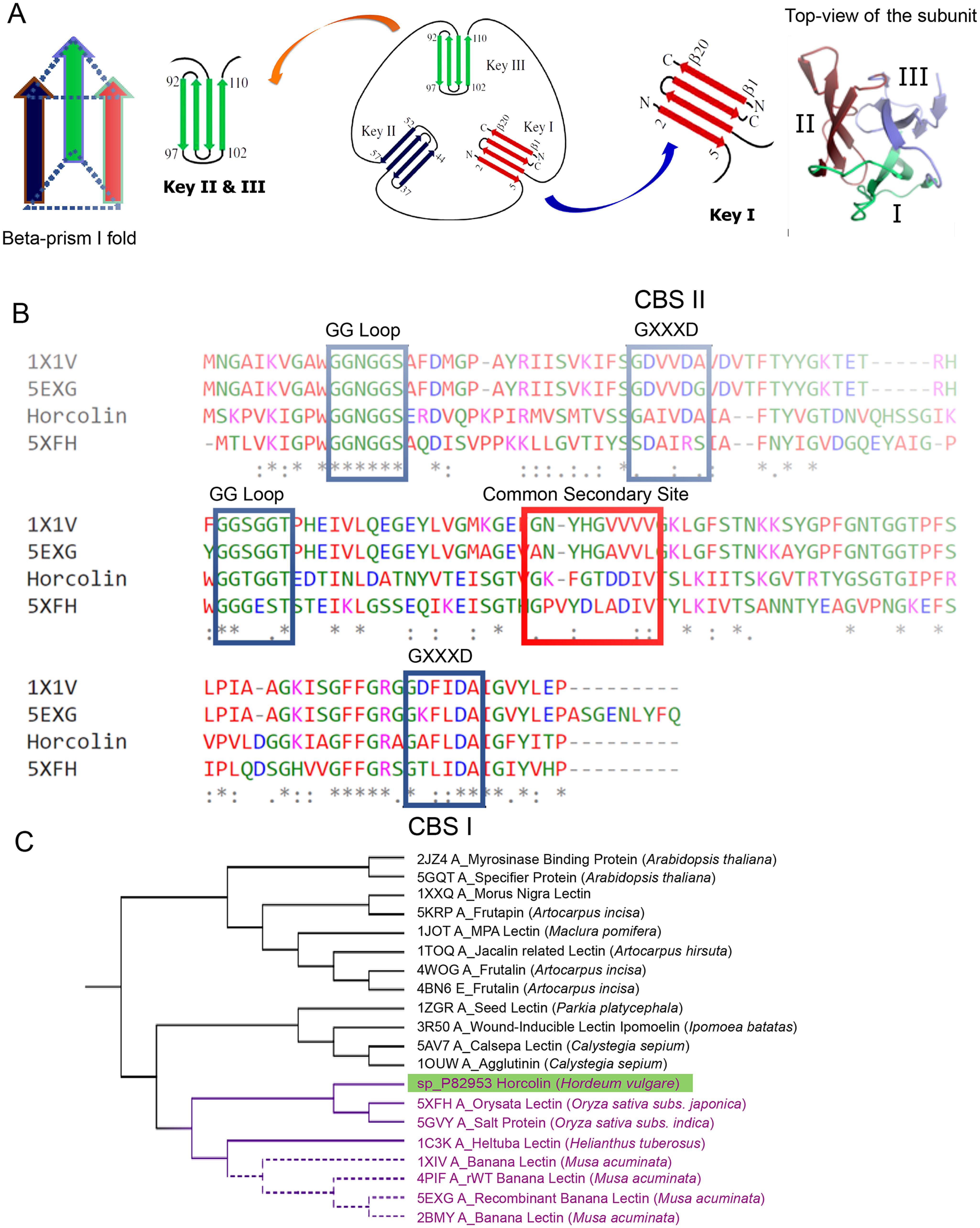
Homolog identification and bioinformatic analysis of horcolin. A, schematic representation of the jacalin fold indicating the four-stranded Greek key motif. Each monomer of the jacalin-related lectin is shown in a 3-fold axis with three Greek keys represented in different colors. B, multiple-sequence alignment of relevant mJRL homologs. The alignment of horcolin, BanLec, and orysata was obtained using Clustal Omega. The residues involved in mannose binding in CBS I and II are highlighted in blue, and the residues in the common secondary site are highlighted in red. C, molecular phylogenetic analysis by the maximum likelihood method. Horcolin was found to lie in the same clade as BanLec and orysata lectin, highlighted in purple.
Carbohydrate-binding specificity of horcolin
Protein expression, purification, and characterization of horcolin
HR was subcloned in a pET22b(+) vector and purified to homogeneity using affinity chromatography on a mannose-Sepharose column. HR appears as a single band of Mr; 15,000 (15% SDS-PAGE). A major peak of 14,992 Da observed on electrospray ionization-MS correlates well with the theoretical mass (15,122 Da (i.e. 14,9921130, the mass of methionine)). Experiments with size-exclusion chromatography coupled to multiangle light scattering (SEC-MALS) provided Mr 30,000, indicating HR to be a dimer. The CD spectra of the protein had a negative peak at 218 nm and a positive peak at 196 nm indicating it to be composed mostly of β-sheets. HR was thermally stable across the pH range of 5.5–7.4, and at pH 7.4, it exhibited maximum stability with a Tm of 79.12 ± 0.11 °C, whereas the His6-tagged construct had a Tm of 64.66 ± 0.22 °C. The intrinsic fluorescence profiles of HR tagged and tagless were consistent with the above (Fig. 2, A–F).
Figure 2.
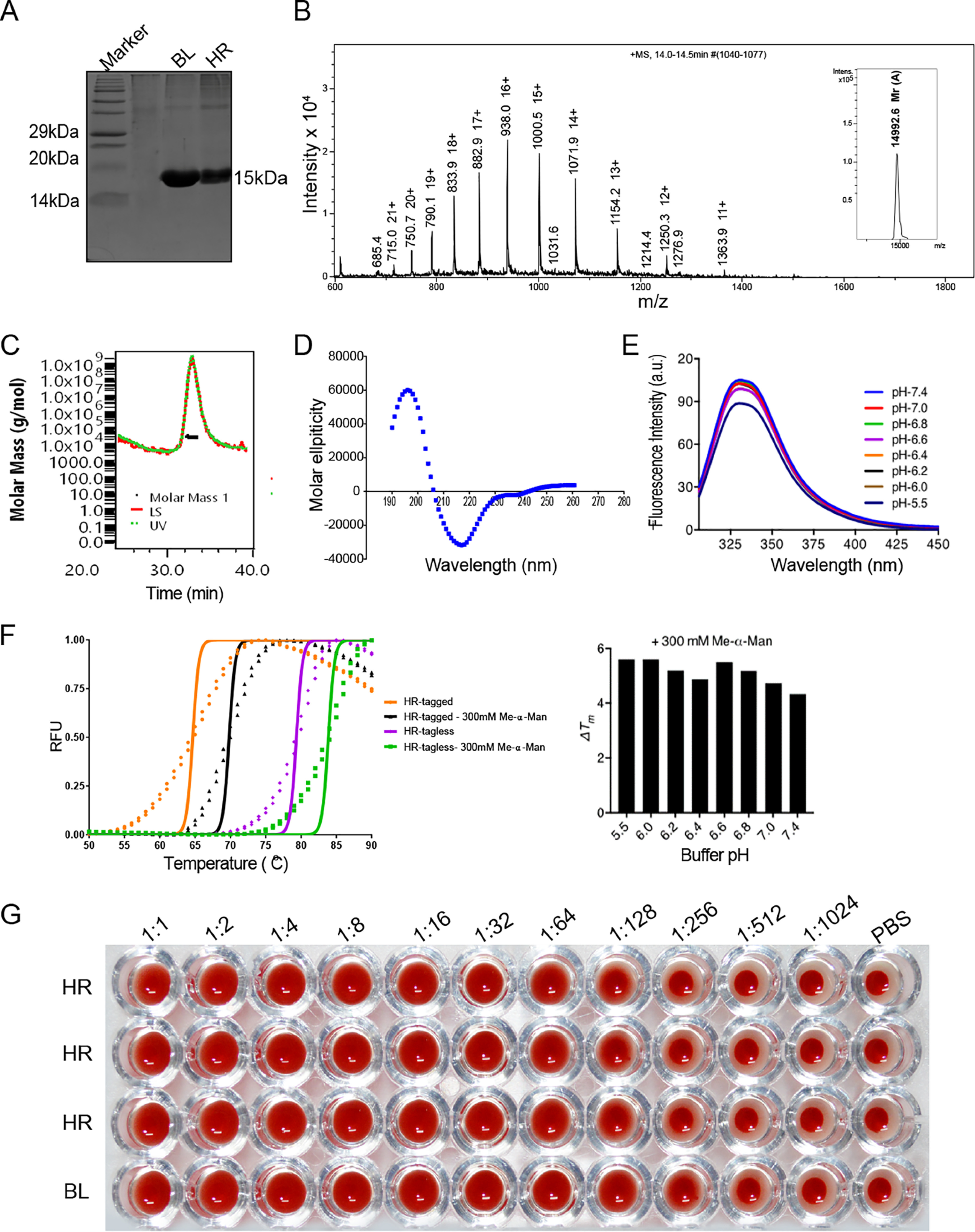
Protein expression, purification, and in vitro characterization of horcolin. A, 15% SDS-PAGE showing purity of the recombinant horcolin protein subsequent to purification with affinity chromatography. Lane 1, marker; lane 2, HR; lane 3, BanLec. HR appears as a single band with Mr ∼15 kDa in SDS–PAGE. B, spectrum of intensity versus mass and charge ratio, showing the observed experimental mass of the purified protein from electrospray ionization-MS (14,992 Da). C, SEC-MALS profile for the HR protein. The peak at 30 ml corresponds to Mr ∼28.9 kDa (D) far-UV CD spectra of the purified recombinant HR showing β-sheet composition. E, intrinsic fluorescence emission spectra of protein across pH range 5.5–7.4. F, thermal shift assay of horcolin (tagged and tagless) at varying pH conditions along with the ligand-induced stabilization (Me-α-Man) to determine the effect of pH on protein thermal stability. G, an initial concentration of 25 μg/ml was serially diluted (2-fold) to characterize the hemagglutination activity of the lectin. The experiment was done as triplicates for HR with BanLec (BL) as the positive control and 1× PBS as the negative control.
Assay for the hemagglutinating activity of the lectin
The minimum concentration of HR required for agglutinating activity was found to be 195 ng/ml (Fig. 2G). Methyl-α-mannopyranoside (Me-α-Man) and α-d-Mannose (Man) completely inhibited the hemagglutination activity of HR at 18.75 and 75 mm, respectively. Glucose (Glc) showed partial inhibition at 300 mm concentration, and Galactose (Gal), N-acetyl glucosamine (GlcNAc), and N-acetyl galactosamine (GalNAc) were ineffective (Fig. S2). Further, a thermal shift assay conducted to determine the ligand-induced stabilization of the protein revealed that Me-α-Man at 300 mm imparted maximum stability to the protein at pH 7.4 (Fig. 2F). Thus, horcolin was found to be mannose-specific, corroborating the previous literature on β prism–type 1 fold mannose-binding lectins (15–20).
Thermodynamics of horcolin-glycan interactions
ITC experiments were performed to determine the thermodynamics of protein-glycan interactions. Me-α-Man and Man, together with 11 manno-oligosaccharides were studied (Fig. 3A). Mannopentaose (Man5), mannoheptaoses (Man7D1/D3), and mannonanose (Man9) used in the study cover the most abundant differentially processed N-linked glycans on the HIV-1 gp120. The result of a typical ITC experiment is shown in Fig. 3B, where the top panel shows individual heats of binding for each injection, whereas the bottom panel gives a plot of the incremental heat released as a function of the oligosaccharide/HR monomer ratio. The results display a monotonic decline in exothermic heat of binding with successive injections of the ligand until saturation is achieved. A nonlinear least-squares fit of the data is consistent with the “one set of sites” binding model. The thermodynamic parameters associated with the binding (i.e. stoichiometry of binding (n), association constant (Ka), enthalpy of binding (ΔH°), and entropy of binding (ΔS°)) obtained at 25 °C are shown in Table 1 (Fig. 3C).
Figure 3.
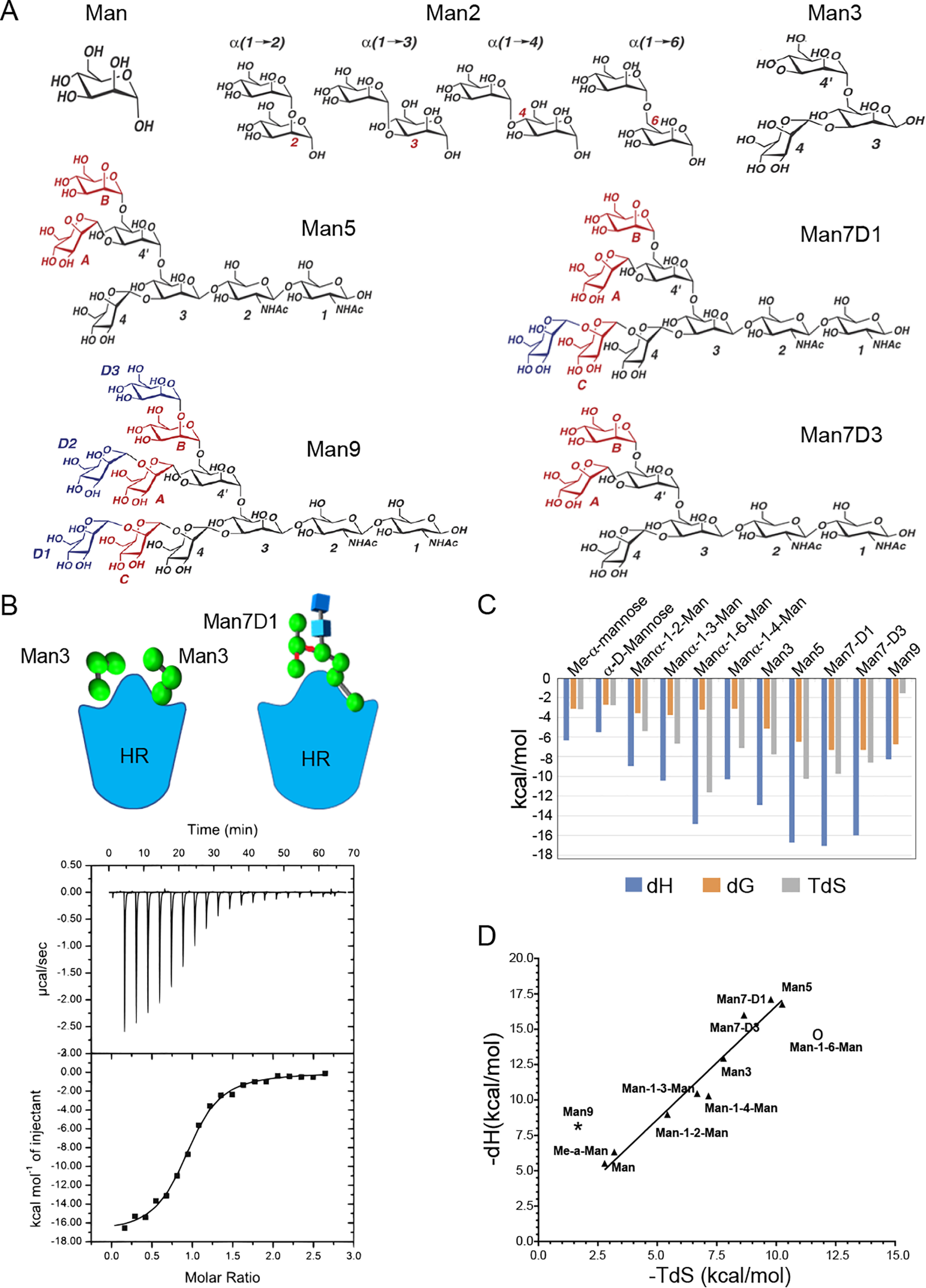
Thermodynamics of horcolin-glycan interactions. A, structures of the various manno-oligosaccharides used in the study. B, schematic representation of the HR complexed with Man3 and Man7D1. Shown are results of a typical isothermal titration calorimetry experiment. Top, exothermic microcalorimetric traces of Man7-D1 injections into horcolin solution (0.8 mm). Bottom, Wiseman plot of heat releases versus molar ratio of injectant/protein in the cell and a nonlinear fit of the binding isotherm for equivalent binding site(s). C, thermodynamic profile indicating binding free energy, enthalpy, and entropy for the 11-HR mannooligosaccharide interaction. D, enthalpy-entropy compensation plot of glycan binding to horcolin at 25 °C. Enthalpy and entropy values are from Table 2. Straight line, best least-squares fit of the data.
Table 1.
Thermodynamic parameters of manno-oligosaccharide binding to horcolin at 25 °C
n is stoichiometry coefficient (number of sugars bound/mol of horcolin monomer), ΔH is enthalpy, ΔS is entropy, and ΔG is reaction energy (calculated with the formula, ΔG = −RT ln Kb, where r = 1.987 cal/mol·K).
| Ligand | n | Ka | KD | ΔH | ΔG | TΔS |
|---|---|---|---|---|---|---|
| m−1 | kcal/mol | kcal/mol | kcal/mol | |||
| Me-α-mannopyranoside | 1.72 + 0.328 | 200 + 27.8 | 5.00 mm | −6.324 | −3.1354 | −3.1886 |
| α-d-Mannose | 1.90 + 0.521 | 101 + 12.5 | 9.90 mm | −5.517 | −2.73368 | −2.78332 |
| Manα1–2-Man | 1.946 + 0.4028 | 398.9 + 41.42 | 2.50 mm | −8.974 | −3.5504 | −5.4236 |
| Manα1–3-Man | 1.88 ± 0.0724 | 596 ± 14.6 | 1.67 mm | −10.46 | −3.7848 | −6.6752 |
| Manα1–6-Man | 1.78 ± 0.243 | 227 ± 10.3 | 4.40 mm | −14.88 | −3.2282 | −11.6518 |
| Manα1–4-Man | 1.77 ± 0.649 | 196 ± 21.2 | 5.10 mm | −10.28 | −3.128 | −7.152 |
| Man3 | 2.01 + 0.0275 | 6.10 E3 + 300 | 163 μm | −12.94 | −5.1622 | −7.7778 |
| Man5 | 0.774 ± 0.076 | 5.99 E4 ± 1.00 E5 | 16.69 μm | −16.76 | −6.5088 | −10.2512 |
| Man7-D1 | 0.925 ± 0.0117 | 2.61 E4 ± 2.87 E4 | 3.831 μm | −17.1 | −7.3256 | −9.7744 |
| Man7-D3 | 1.09 ± 0.00711 | 2.4 E5 ± 1.45 E4 | 4.16 μm | −15.99 | −7.348 | −8.642 |
| Man9 | 1.34 + 0.0185 | 8.60 E4 + 1.21 E4 | 11.6 μm | −8.277 | −6.73038 | −1.54662 |
The stoichiometries for binding to mono-, di-, and trisaccharides were close to 2, indicating that each protomer of HR contains two identical binding sites for these ligands. The close fit of the data to the identical site model shows that the ligand binds to each of the two sites independently with similar binding constants and enthalpies (1:2). Interestingly, the stoichiometries for the interaction of Man5, Man7, and Man9 with HR could be fitted to only one per protomer, indicating that both of the binding sites of the lectin interact with a single oligosaccharide simultaneously (1:1). The apparent affinities were found to be in the millimolar range for the monosaccharides (∼10 mm) and disaccharides (∼1.67 mm) and low micromolar range for the high-mannose oligosaccharides (4–16 μm), signifying a clear preference toward higher oligosaccharides. The order of binding affinities for the sugars used was found to be Man7D1 ≥ Man7D3 > Man9 > Man5 > Man3 > Man3 > Man2 > Me-α-Man > Man. A dramatic increase in the affinity for the higher glycans thus demonstrates that the length, the branching of the glycan, and hence the total carbohydrate-binding valency (multivalent effect) can be attributed to a favorable change in the free energies of interactions for them.
The Man2 affinity data suggested that HR prefers α1–3– and α1–2–linked mannobioses compared with their α1–6– or α1–4–linked counterparts. A better affinity of mannobioses over that of mannose is due to a decrease in their binding enthalpies, which suggests that their nonreducing mannosyl unit interacts with a complementary locus, the site adjacent to the two primary binding sites (viz. the common secondary binding site in the lectin). The possibility of a multivalent interaction between Man3 and the lectin can be ruled out not only because of n = 2 but also because the value of Ka obtained for Man3 was not found to be a product of the Ka for α-d-Man (101 m−1 × 101 m−1= 10,201) (viz. the ΔG value was not an additive function of the ΔG values (−2.7 + −2.7 = –5.4 kcal/mol) of the two primary binding sites). Differences in the ΔH values for the interaction of Man, Me-α-Man, Man2, and Man3 amount to ∼4 kcal/mol, indicating that, besides binding to the primary sites CBS I and II, Man2 and Man3 also interact with the common secondary site on the same monomer of HR via the Man-4 moiety. Trimannose had additional interactions of its nonreducing mannosyl residues in the central arm (Man-3, Man-4′) with the amino acids in the common secondary site.
A reduction in the stoichiometries for the binding of Man5, Man7D1 and Man7D3 (n = 1) appears to emanate from a simultaneous interaction between two of their nonreducing end mannosyl residues with the two primary binding sites in each monomer of HR. This can be envisaged as follows. While the D1 arm of Man5 and Man7 binds to the CBS I, a nonreducing terminal mannosyl residue from the D2/D3 arm of these saccharides interacts with the CBS II. This interpretation is further supported by the observation of the additive free energies of binding of two different nonreducing end mannosyl residues of the same oligosaccharide (Man5 and Man7-D1/3), implying additional interactions between Man-3 and Man-4′ of the central arm aided by the interactions of the terminal mannose moiety, Man-B, in the bisected glycans. An increment of 3.8 kcal/mol for the binding of Man5 over and above that observed for Man3 is indicative of the contribution of the additional interaction between the central mannosyl unit (Man-3) of the Man5 glycan and Phe-87 and Thr-89 residues in the lectin, which constitute the common secondary binding site. Likewise, an increase of 0.5 kcal for the binding of Man7-D1 and Man7-D3 indicates a consolidation of the interaction between “OH” of the terminal mannose in their D3 arm (Man-B or Man-D3) with Gly-88 and Lys-86 residues of the lectin. Thus, the residues distant from the primary binding sites modulate the binding thermodynamics. Additionally, both of the glycans showed similar binding affinities, suggesting an analogous mode of interaction with a similar number of contacts with the protein (Table 2). All of the sugars displayed a favorable enthalpic change associated with the binding process, suggesting the dominance of polar, hydrogen-bonding, and van der Waals interactions principally to stabilize these complexes. This favorable enthalpic change was seen to compensate for the loss in conformational entropy thus aiding the binding process. The trend for the binding enthalpies for each sugar with the protein depended on the number of mannose moieties except for Man9, the interpretation of which will be discussed in the subsequent section.
Table 2.
Enthalpy and entropy contributions for the complexes of HR with various manno-oligosaccharides
| Ligand | −TΔS | −ΔH |
|---|---|---|
| kcal/mol | kcal/mol | |
| Me-α-mannopyranoside | 3.1886 | 6.324 |
| α-d-Mannose | 2.783 | 5.517 |
| Manα1–2-Man | 5.4236 | 8.974 |
| Manα1–3-Man | 6.6752 | 10.46 |
| Manα1–6-Man | 11.651 | 14.88 |
| Manα1–4-Man | 7.152 | 10.28 |
| Man3 | 7.7778 | 12.94 |
| Man5 | 10.2512 | 16.76 |
| Man7-D1 | 9.7744 | 17.1 |
| Man7-D3 | 8.642 | 15.99 |
| Man9 | 1.54 | 8.27 |
The ITC experiments were also carried out at different temperatures, −5, 15, and 25 °C for mono-, tri-, and pentamannose, to calculate the heat capacity changes to characterize the nature of forces stabilizing these interactions. The HR–mannooligosaccharide binding reactions were found to be enthalpically driven with little change in the heat capacity (Cp) of binding (−0.01 < Cp < −0.08 kcal/mol/K). Further, the enthalpy-entropy compensation with a slope of TΔS of 1.357 was noted for the interaction. It was observed that as the size of the ligand increases there is a large favorable change in enthalpy, indicating that the interactions are dominated by hydrogen bonding that is compensated for by a corresponding loss in conformational entropy due to the restricted rotational and vibrational degrees of freedom (Table 2 and Fig. 3D). This is consistent with the earlier reports on lectin-sugar interactions (21–24). It is also to be noted that Manα1–6-Man and Man9 do not fit into this pattern. Although Manα1–6-Man exhibits considerable binding enthalpy, it suffers from unfavorable entropy owing to its greater degree of conformational flexibility. This is in agreement with the literature on the role of glycosidic linkages on conformational entropy in solution (25). On the other hand, Man9 lacks a corresponding favorable change in enthalpy (as in the case of Man5/7) and displays a relatively large unfavorable entropy, which can be attributed to a failure to achieve an optimal orientation of the three binding arms. The reorientation of the unbound arm results in an entropic penalty, which utilizes a portion of the enthalpic component in moving the unbound arm away from the binding site, which in turn leads to a decrease in the binding free energy. Results from the ITC experiments thus provided a thermodynamic model to explain the mode of bivalent interaction for binding of higher glycans to the lectin protomer with multiple subsites. This is consistent with a recent native-MS study that showed qualitatively a bidentate mode of interaction between BanLec and Man5–Man9 (26).
Furthermore, to validate the in silico characterization of the two primary binding sites of HR, we generated three mutants of HR-His–tagged protein by replacing Asp in the GXXXD motif with Ala in CBS I and II (i.e. D138A and D39A, respectively) as well as a double mutant that substituted Asp in both of the pockets (D138A/D39A). The mutants were expressed and purified using Ni-NTA resin, and their ability to bind to α-d-Man was explored with ITC. The mutants D138A and D39A displayed half the stoichiometries (i.e. n = 1) for binding to mannose as compared with their WT counterpart. The mutants did not show any appreciable change in their binding affinity. The double mutant was completely inactive and did not show any heat of binding when titrated with mannose in the ITC experiment (Fig. S3C). Mutation at D138A abrogated the CBS I, D39A abrogated CBS II, and the double mutant D39A/D138A abolished altogether the sugar-binding ability of HR and therefore completely inactivated it. Mutation studies thus corroborate the in silico prediction of the presence and the role of the two carbohydrate-binding sites in HR (Table S1).
Molecular modeling and MD simulation of glycan-binding modes of horcolin
The ITC experiments revealed the potential of high manno-oligosaccharides to undergo bivalent interaction. Due to the paucity of structural information for HR, comparative modeling using Modeller9v19 with BanLec (40% identity) as the template was used to predict the three-dimensional structure of the protein. The structure was subjected to scrutiny through algorithms in the Structural Analysis and Verification (SAVES) server, where Procheck revealed 92.8% residues to be in the allowed region of the Ramachandran plot. ERRAT results showed the quality factor to be 72.46%, indicating the goodness of the model compared with highly refined structures in the PDB (Fig. 4A). The structural superposition of the modeled protein with its homolog, BanLec and orysata, performed using MISTRAL revealed that the proteins were closely superposed across the β-sheets. However, the loop constituting the secondary site exhibited relatively higher deviation. The root mean square deviation (RMSD) of the corresponding 146 Cα atoms was 1.3 Å (Fig. 4B, Fig. S4 and supporting information).
Figure 4.
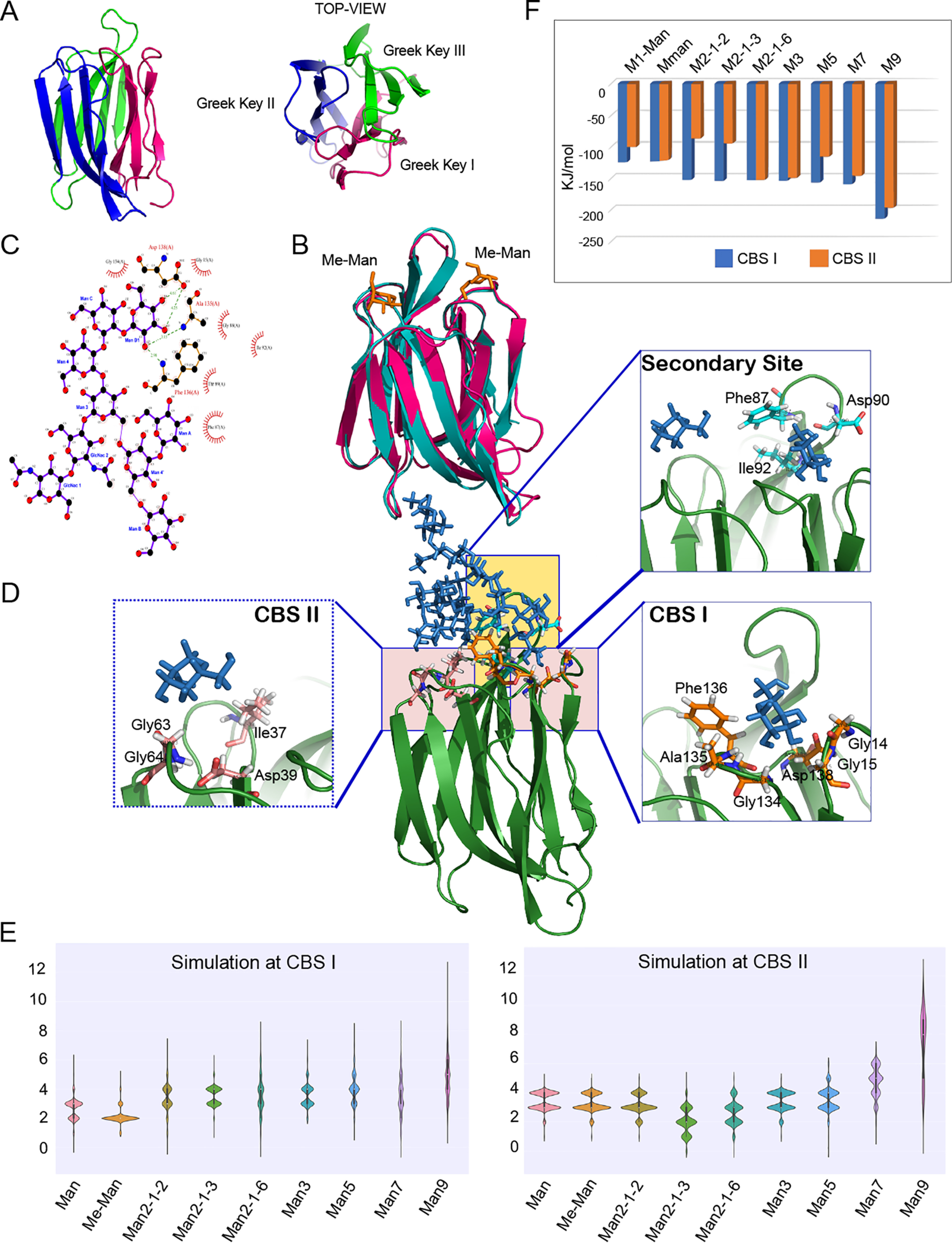
Molecular modeling and MD simulation of glycan-binding modes of horcolin. A, overall fold of horcolin, shown in a cartoon representation with the three Greek keys (β1–2 + β11–12; β3–6; β7–10) color-coded magenta, blue, and green, respectively, and the 12 β-strands numbered. B, superposition of horcolin and BanLec in complex with methyl-α-d-mannopyranoside (PDB ID 1X1V). horcolin is represented in a magenta cartoon, BanLec in teal, and Me-α-Man as orange sticks. C, results from Ligplot analysis of the HR-Man7 complex, where the dashed green lines indicate hydrogen bonds and the half-moon indicates van der Waals interactions. D, schematic representation of the interactions of the glycan Man7 with HR where the glycan is shown in stick representation and the protein in cartoon. Shown in the inset is the residue-level interaction of the glycan Man7 (ball and stick representation) with the residues in the CBS I and II and the common secondary site (stick representation). E, summary of the hydrogen bonds for the nine complexes at binding sites CBS I and CBS II shown as the average number of hydrogen bonds indicated as violin plots. F, results from MM/PBSA analysis for the complexes at binding sites CBS I and II shown in blue and orange, respectively.
We next focused on the specific interaction of the HR protein with the sugars studied by ITC. To this end, the time-averaged structure of the modeled protein was used for the docking studies using DOCK6. The two primary binding sites were identified by pocket prediction tools and were used for active-site sphere generation. The dock poses hence obtained were verified with the crystal structures of its homologs (Fig. 4C and Fig. S5). Simulation studies were conducted with the ligand bound individually at CBS I and II to preclude pocket-based bias. This was followed by studying the occupancies of the smaller ligands independently at both CBS I and II as observed from the ITC data (n = 2). The stability and flexibility were monitored throughout the 100-ns run. The structure of the glycan is represented by the orientation of the arms D1, D2, and D3 and the core chitobiose. The branched oligomannosides involving the Manα1–6-Man branch exhibited maximum flexibility in the D2/3 arms owing to this linkage driving the conformational space of the glycan in question (Fig. 4D). Root mean square fluctuation (RMSF) analysis revealed that the binding of higher sugars (Man5/7/9) simultaneously at both CBS I and CBS II diminished the fluctuations in the common secondary site, which facilitates the bidentate mode of binding as observed experimentally. This was not observed for smaller sugars, where the role of the secondary site is absent or negligible. Also, engagement of both CBS I and II pockets in bivalent binding mode by manno-oligosaccharides Man5/7, by their nonreducing terminal mannosyl residues at their D1 and D3 arms, respectively, potentiated the avidity of the interaction (Supporting information, Fig. S5–S8).
During the entire run of the simulation, there was a constant maintenance of hydrogen bonds from the primary binding sites. These hydrogen bonds are tabulated with respect to their percentage existence of the simulation time (Table S2 and supporting information). In CBS I, the residues 134GAFLD138 from the primary binding site were involved in hydrogen-bonding interactions along with the residues from the secondary binding site: Gly-85, Lys-86, Phe-87, and Asp-90. In the case of Man7-D1, both the D1 arm (Man-D1) and D3 arm (Man-B), respectively, were interacting simultaneously with both the primary binding pockets CBS I and CBS II, in the same subunit of HR. They also displayed strong interactions with the secondary binding site that serves as a bridge for both of the primary binding sites. The nonbinding arm D2 (Man-A) along with the chitobiosyl unit were seen to be solvent-exposed owing to the flexibility of its α1–6 linkage. It was observed that Asp-90 aids in binding of the oligosaccharides starting from mannobiose tethered to CBS I. The chitobiose unit was also seen to form hydrogen bonds in the case of Man5 glycan, where the α1–3–binding mode had interactions of the GlcNAc-2 (O6) with residues in the secondary binding site. Additionally, Phe-87 stabilizes the interaction of glycans through hydrophobic contact. In the case of CBS II, Asp-39 retained the hydrogen bond during the entire simulation, indicating that it was crucial for the anchorage of higher manno-oligosaccharides. The glycans tethered at CBS II also displayed interaction with the secondary binding site for a significant portion (>50%) of the simulation time (Gly-88, Thr-89, and Asp-90). Thus, the interactions at CBS II are relatively less extensive as compared with CBS I. Gly-63/64 of the recognition loop aided the binding via hydrogen bonds with the mannose in both of the pockets (Fig. 4, D and E).
We further employed molecular mechanics/Poisson–Boltzmann surface area (MM/PBSA) to utilize the enthalpy contributions of the interactions to identify the preference of HR and to calculate relative free energies of binding of the various high-mannose oligosaccharides. It is to be noted that the decomposition of the total free energy associated with the manno-oligosaccharide binding is semi-quantitative, allowing the prediction of the trend in binding preference rather than absolute values as the current methods suffer from the limitation in the decomposition of the binding free energy into total enthalpy and entropy parameters, consequently obviating a direct comparison with the data from ITC. The analysis was conducted independently for the snapshots of the ligands in the two primary binding pockets as well as different binding modes for Man5/7/9 glycans exhibiting higher flexibility owing to the α1–6 linkage (considering computational time and accuracy for large flexible ligands). The average ΔE for mannose was found to be −161.453 ± 7.385 kJ/mol, and the addition of single mannose entity in α1–6 linkage brought it to −152.418 ± 10.048 kJ/mol, whereas increased branching and the addition of eight mannose units in the case of Man9 had a favorable impact on it (ΔE = −214.058 ± 20.063 kJ/mol). This is in line with the ITC data, where the addition of mannose units was accompanied by a significant increase in binding affinity. A similar trend was observed for ligands interacting at CBS II. Despite having a similar interaction profile, CBS I had lower binding energy, implying higher affinity for the larger glycans compared with CBS II, indicating the dominance of the former binding pocket for interactions with Man5/7/9. These differences were marginal in the case of lower sugars that showed similar interaction with both of the pockets. It is to be noted that MM/PBSA analysis allows for addressing only the contributions from electrostatic and van derWaals interactions and does not capture the entropic component of the free energy calculation and thus accounts for the greater binding energy values displayed by Manα1–6-Man. Owing to this drawback, the results from these analyses should be considered only for a relative binding energy comparison for the various ligand complexes. It is interesting to note that in the case of Man9, with the D1 arm tethered at CBS I, the D2 arm had prominent interaction with the CBS II, whereas the D3 arm had a flipped conformation, leading to its interaction with the backbone residues of the common secondary site as noted from the cluster analysis. This, however, was not maintained across the simulation time as can be observed in RMSF analysis of the glycones and the residence time of the hydrogen bond interaction between the D2 arm and the protein (2–5% of the simulation time) (Fig. S9). This is consistent with the observation of relatively poor enthalpy for binding in the ITC data, implying a reduced number of hydrogen bonds and an increased entropy owing to the high flexibility of the arms of Man9. The ligand conformational entropy, an important contributor in glycan binding, is rather difficult to address through simulation. However, from the ITC data, it is apparent that the gain in the entropic parameter of Man9 led to a diminution in the enthalpic contribution for the binding reaction, reflecting a loss in hydrogen-bonding interactions.
Simulation studies supplement and validate the monovalent mode of binding for mono-, di-, and trisaccharide and the bivalent mode of interaction for Man5/7/9. This bidentate mode of interaction can be explained as a two-step process. First, the D1 arm randomly approaches CBS I and II. The initial tethering by the terminal mannose at CBS I is followed by potentiation of its retention at CBS I due to its lock-in of the D3 arm at the CBS II owing to the flexibility of its α1–6 linkage. These studies also show that the bidentate interaction in the reversed orientation is disallowed, as the placement of the D1 arm at CBS II precluded the reach of the D2/D3 arm to CBS I. Thus, the in silico studies indicated a possibility of one pocket offering better interaction than the other.
In vitro interaction of horcolin with the viral glycoproteins
As a part of the preliminary characterization of the potential of HR for anti-HIV activity, its interaction with the HIV envelope glycoprotein gp120 was analyzed to gain insight into its binding kinetics. The glycoprotein gp120 is known to contain nearly 20–30 N-linked high-mannose structures, which make up ∼50% of its molecular weight (27, 28). In vitro interaction with recombinant gp120 from the HIV-1YU2 strain was monitored using surface plasmon resonance. It was observed that immobilized HR, both tagged and tagless proteins (∼1,000 RU) dose-dependently bound to the gp120 in phosphate buffer containing 150 mm NaCl (1× PBS). The sensograms reveal effective binding with the glycoprotein; in particular, at 125 nm concentration, >170 RU were observed. From the kinetic analysis, the calculated dissociation constant (KD) was 14.4 nm, whereas for the His-tagged protein, the KD was 36.9 nm (Table 3). Inverting the assay and immobilizing gp120 on the surface yields similar affinity values for the HR-gp120 interaction (Table S3 and Fig. S10). Likewise, the binding interaction analysis was also conducted on gp140, a derivative of gp160, which lacks the transmembrane domain from the HIV-1YU2 strain. Sensogram analysis revealed a higher affinity with the trimeric glycoprotein, wherein it displayed very tight binding with minimal dissociation (KD = 2.51E−14 m). With altered NaCl concentration to 250 mm, we observed a KD of 5.53E−07 m (Fig. 5A and Table 3). Similar KD values were also observed for its interaction with the gp120 and gp140 from the HIV-116055 strain. Previous studies have reported the binding constant for cyanovirin with gp120 to be 8.65 nm (29), Galanthus nivalis (GNA) with HIV IIIB gp120 showed a KD of 0.33 nm (30), and griffithsin (GRFT) lectin was seen to exhibit a lower KD of 72.7 pm (31). On the other hand, HR exhibited strong interactions with the native-like trimeric gp140, in a stark contrast to the other mentioned well-studied antiviral lectins. A very tight binding of HR with a KD value of 0.02 pm was observed because of an extremely slow dissociation rate constant for the complex between HR and gp140. However, the KD value for the association of the trimeric gp140 with GRFT was in the range of 0.1–4.3 nm, for BanLec it was 5.2 nm, for CV-N it was 2.3 nm, and for GNA it was found to be 7.2 nm (32). Thus, a comparison of these KD values implies a possibility of differential modes of interaction by the antiviral lectins with the glycoproteins.
Table 3.
Rate and equilibrium constant for the binding of different forms of glycoprotein gp120 and gp140 to horcolin measured by SPR (Biacore)
ka and kd are rate constants for association and dissociation, respectively. KD is the equilibrium dissociation constant. Ka is the association constant.
| Concentration of analyte | ka | kd | Ka | KD |
|---|---|---|---|---|
| 1/m·s | 1/s | 1/m | m | |
| Immobilized horcolin-tagged with gp120 and gp140 as analyte | ||||
| gp120 | ||||
| 125 nm | 1.66E+05 | 5.91E−03 | 2.80E+07 | 3.57E−08 |
| 100 nm | 1.89E+05 | 4.08E−03 | 4.64E+07 | 2.16E−08 |
| 50 nm | 2.09E+05 | 2.05E−03 | 1.02E+08 | 9.83E−09 |
| 25 nm | 1.85E+05 | 7.74E−04 | 2.39E+08 | 4.19E−09 |
| 10 nm | 2.17E+05 | 1.93E−04 | 1.12E+09 | 8.90E−10 |
| gp140 | ||||
| 2000 nm | 1.35E+04 | 5.31E−12 | 2.55E+15 | 3.93E−16 |
| 1,000 nm | 1.82E+04 | 2.43E−10 | 7.49E+13 | 1.33E−14 |
| 500 nm | 2.92E+04 | 7.07E−12 | 4.14E+15 | 2.42E−16 |
| 250 nm | 2.41E+04 | 9.94E−10 | 2.42E+13 | 4.13E−14 |
| 100 nm | 3.60E+04 | 2.53E−09 | 1.42E+13 | 7.04E−14 |
| Immobilized horcolin-tagless with gp120 and gp140 as analyte | ||||
| gp120 | ||||
| 125 nm | 1.23E+05 | 8.87E−03 | 1.39E+07 | 7.19E−08 |
| 100 nm | 9.74E+04 | 7.03E−03 | 1.39E+07 | 7.22E−08 |
| 50 nm | 1.24E+05 | 3.83E−03 | 3.24E+07 | 3.08E−08 |
| 25 nm | 1.22E+05 | 1.14E−03 | 1.07E+08 | 9.37E−09 |
| 10 nm | 1.00E+05 | 3.38E−05 | 2.97E+09 | 3.37E−10 |
| gp140 | ||||
| 2000 nm | 1.72E+04 | 2.85E−12 | 6.05E+15 | 1.65E−16 |
| 1,000 nm | 2.30E+04 | 5.13E−10 | 4.48E+13 | 2.23E−14 |
| 500 nm | 3.55E+04 | 9.94E−13 | 3.57E+16 | 2.80E−17 |
| 250 nm | 2.62E+04 | 3.10E−12 | 8.46E+15 | 1.18E−16 |
| 100 nm | 4.62E+04 | 2.35E−10 | 1.97E+14 | 5.08E−15 |
Figure 5.
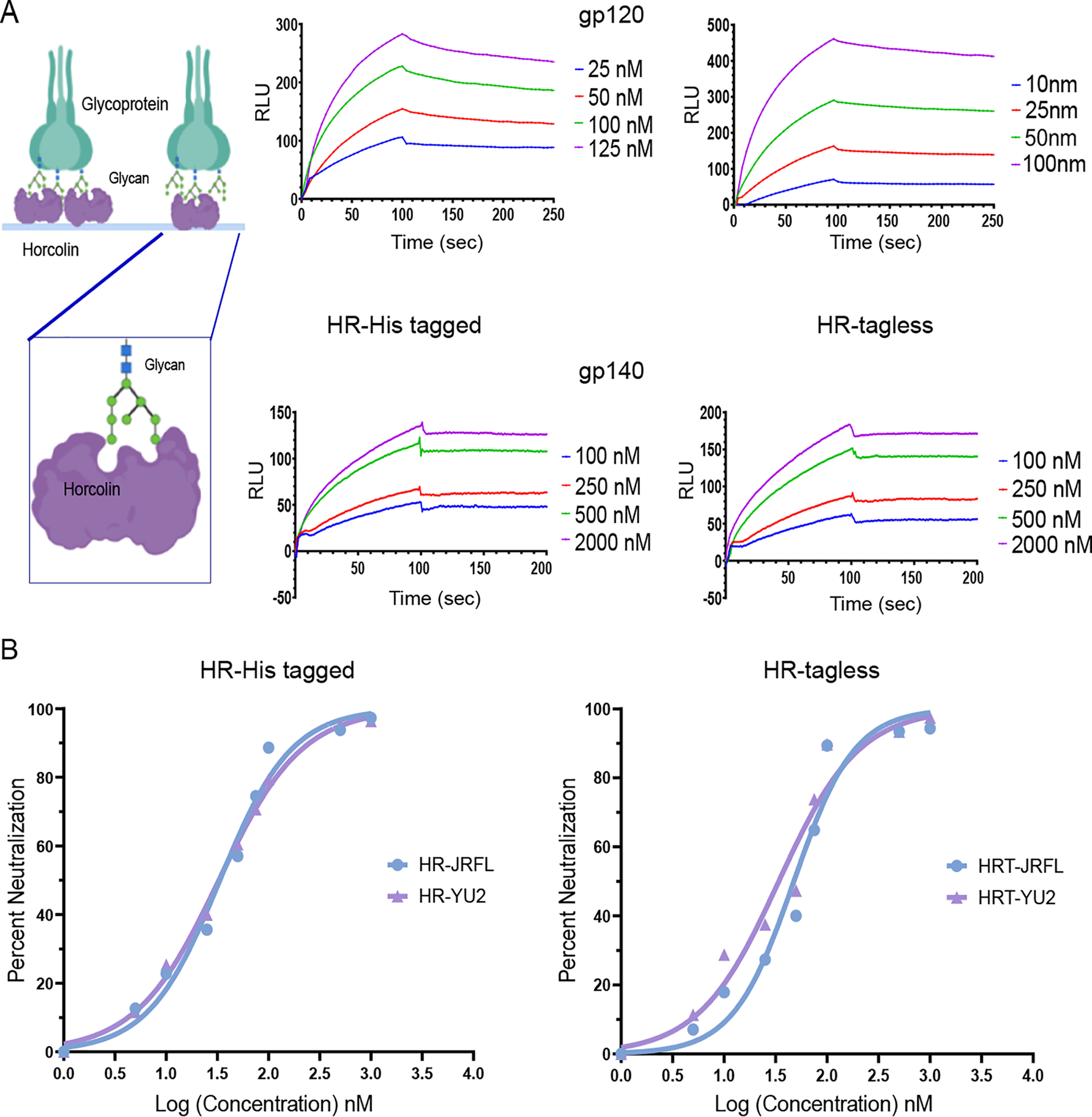
Analysis of the antiviral activity of HR tagged and tagless proteins using SPR and neutralization assay. A, schematic representation of immobilized horcolin interacting with the glycoprotein gp120 (shown on the left). SPR sensogram overlays are shown on the right for the interaction between immobilized gp120 and HR (top) and immobilized gp140 and HR (bottom). The concentration of analyte injected is indicated at the right of each sensogram overlay. Solid lines, global fitting of the data to a 1:1 Langmuir binding model (A + B ↔ AB, BIAevaluation 3.0). B, anti-HIV activity of HR tagged and tagless proteins against the two viral isolates, HIV-1JRFL (blue) and HIV-1YU2 (purple). The average of three biological replicates is shown, and the IC50 value was calculated using nonlinear regression using GraphPad Prism. RLU, relative luminescence units.
Neutralization of HIV infectivity by HR
We next characterized the neutralizing potential of HR on HIV-1 isolates from JRFL and YU2 strains (33, 34). Each experiment was performed by determining the anti-HIV activity of HR by a Luciferase-based reporter assay in TZM-bl cells (35) at noncytotoxic concentrations with b12 neutralizing antibody as the positive control. Our results indicate that both HR tagged and tagless proteins inhibit the entry and thereby infection of HIV-1 isolates in a dose-dependent manner. For the strain HIV-1YU2, the IC50 values of 32.67 and 33.93 nm, respectively, for tagged and tagless proteins were observed. For HIV-1JRFL, IC50 values of 33.39 and 47.51 nm for tagged and tagless proteins, respectively, were obtained (Fig. 5B and Fig. S10C). Additionally, HR displayed analogous inhibitory potential against both of the strains (HIV-1JRFL and HIV-1YU2). The results show a potency of HR comparable with that of the previously known anti-HIV lectins, where the calculated IC50 values were reported to be 34.3 nm for the snowdrop lectin, GNA, 3.18 nm for BanLec, and 0.42 nm for GRFT in a similar neutralization assay using the TZM-bl cells with the relative luminescence unit measurements (27). These results thus highlight the potential of horcolin's induction as an anti-HIV agent in microbicides.
Lack of mitogenicity and the absence of cytokine-inducing activity of horcolin
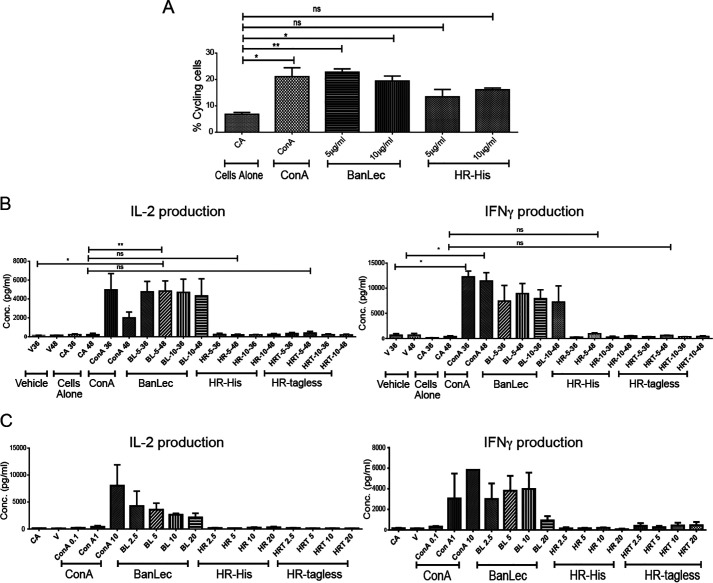
Next, we ventured to study whether HR's subtly different primary binding sites and a distinctly different secondary site influences its mitogenic behavior, highlighting its advantage over BanLec in its prospective application as a viricidal agent. The mitogenicity was initially studied by the changes in cytokine levels, where the binding of the lectin augments the production of various cytokines by the activated lymphocytes. Interleukin-2 (IL-2), IL-6, and interferon-γ (IFNγ) are important proliferative cytokines and serve as inducers for cell-mediated immune response. The IL-2 cytokine is known to be a T-cell growth factor that induces the clonal expansion of T-cells following antigen or lectin stimulation. It is also important for the differentiation of CD4+ T cells into Th1 and Th2 effector subsets and stimulates NK cells and B cells (36). The increase in the production of IL-2 and IFNγ by splenocytes was studied as a mitogenic response induced by the positive control lectins, ConA and BanLec. The initial time course experiments were conducted with two concentrations of the proteins (5 and 10 μg/ml). Both ConA and BanLec treatment showed a good amount of cycling cells at both of the time points. However, HR treatment even at 48 h induced a relatively smaller fraction of cycling cells, affirming its poor mitogenicity (Fig. 6A and Fig. S11). The activated splenocytes at the two time points, analyzed for their cytokine production ability using ELISA, were noted to be insignificant, although some amount of IFNγ was seen by HR-treated cells at the 48-h time point, which was deemed feeble compared with other stronger mitogens BanLec and ConA, indicative of the former being a very weak mitogen (Fig. 6B). This was followed by concentration-dependent experiments (2.5–20 μg/ml), where at 36 h, we observed an up-regulation of both IL-2 and IFNγ levels in ConA- and BanLec-treated cells. HR (tagged and tagless) treatment was incapable of producing increased IL-2 and IFNγ expression even at 20 μg/ml (Fig. 6C). Although HR is homologous to BanLec, differing from it by a few residues, HR's slightly different primary sites CBS I/II and a significant variation in the common secondary binding site is seen to provide a differential recognition of glycans on the cell surface and is likely to underlie its very weak mitogenicity. Thus, both the cytokine and cell-cycle analyses highlight HR's advantage over BanLec.
Figure 6.
Nonmitogenic activity and the absence of cytokine-inducing activity of HR. A, flow cytometry analysis of cell proliferation data of splenocytes treated with ConA, BanLec, and HR at 5 and 10 µg/ml at 36-h time points. FACS data show the amount of propidium iodide staining representing cells in different phases of the cell cycle. B, cytokine profile of the time-dependent (36- and 48-h) protein stimulation experiment by ConA, BanLec, and HR (tagged and tagless) at 10 µg/ml concentration on mouse splenocytes. Values shown are the mean from triplicate samples. C, cytokine profile of concentration-dependent stimulation (2.5–20 µg/ml) of mouse splenocytes by ConA, BanLec, and HR tagged and tagless after 48-h incubation. Values shown are the mean from triplicate samples.
In conclusion, we demonstrate that the bivalent mode of recognition of manno-oligosaccharides by horcolin is modulated by glycan length and flexibility. The differential mode of its interactions with glycans as compared with the mitogenic BanLec appears to underpin horcolin's lack of mitogenicity. The absence of mitogenicity together with the strong and specific recognition of HIV envelope glycoproteins (gp120/140) and the ability to neutralize HIV infection highlight horcolin's potential for inclusion as an anti-HIV agent in microbicides.
Experimental procedures
Materials
Chemicals
The enzymes used for cloning were obtained from New England Biolabs. The Ni-NTA metal affinity resin and isopropyl β-d-1-thiogalactopyranoside were procured from Novagen. Ampicillin, imidazole, methyl-α-mannopyranoside, mannose, glucose, and SDS-PAGE reagents were from Sigma. Me-α-Man and α-d-Man were from Sigma, and Man2, Man3, Man5, Man7, and Man9 were from Dextra Laboratories Ltd.
Strains and plasmids
Escherichia coli DH5α cells (Novagen) were used for cloning, and E. coli BL21 (λDE3) (Novagen) was used for expression.
Protein identification
Sequence characterization
The protein sequence of a lectin of H. vulgare was obtained from UniProtKB (accession number Q5U9T2) (37).The sequence was submitted to the Pfam webserver (38) to obtain the different domain information in the sequence. The ProtParam webserver (39) was used for the calculation of molecular weight, theoretical pI, and instability index.
Homolog identification
BLASTp (40) was performed against the PDB to obtain 50 target sequences using the BLOSUM62 scoring matrix.
Multiple-sequence analysis
The top sequence having a total score of over 90 and a sequence identity over 39% in the BLASTp result was utilized to perform MSA. The MSA was performed using default parameters in Clustal Omega (RRID:SCR_001591).
Phylogenetic tree construction
The top results having a query cover of 97–100% and a minimum of 37% of sequence similarity with query sequence were chosen. The phylogenetic tree of the respective sequences was prepared using the tree building algorithm in MEGA6 (41) and was visualized using iTOL (42).
In vitro characterization
Cloning
A codon-optimized DNA sequence for bacterial expression encoding HR was obtained from GeneArt (Thermo Fisher Scientific). The codon-optimized DNA sequence was subcloned in the pET-22b(+) vector using NdeI and XhoI restriction sites. A stop codon insertion at the end of the protein-encoding sequence allowed C-terminal His-tagged and tagless expression of recombinant HR. The restriction digestion of the native vector and the gene-containing vector was followed by gel extraction and ligation using T4 DNA ligase. The positive constructs were screened and selected.
Protein expression
E. coli (BL21-DE3) cells were transformed with the cloned plasmid, and a colony was inoculated for primary culture in Luria broth supplemented with ampicillin (100 μg/ml) and allowed to grow until the A600 reached 0.8. The secondary culture was then inoculated with 1% primary culture and allowed to grow at 37 °C until A600 reached 0.6. Protein production was induced by adding isopropyl β-d-1-thiogalactopyranoside (0.2 mm) and incubated at 18 °C for 18 h. The culture was centrifuged (4 °C, 5,000 rpm, 10 min) postinduction, and cells were washed with 1× PBS. The pellet was resuspended in 30 ml of lysis buffer (1× PBS, pH 7.4, and 1 mm phenylmethylsulfonyl fluoride) and sonicated at 28% amplitude for 30 min (2 s on and 6 s off).
Purification of tagless protein
The sonicated mixture was centrifuged (14,000 rpm, 45 min, 4 °C), and the supernatant was incubated with 5 ml of mannose-Sepharose beads prepared as indicated (43) for binding on an end-on rocker for 2 h at 4 °C. Flow-through was discarded, and beads were washed with 1,000 ml of 1× PBS wash buffer. Elution fractions of 5 ml each were collected (EB1: 100 mm mannose 1× PBS buffer; EB2: 200 mm mannose 1× PBS; and EB3: 300 mm mannose 1× PBS buffer). The purity of the eluted protein was checked on 15% SDS-PAGE. The pure protein fractions were dialyzed against 1× PBS. The protein concentration was determined by absorbance at 280 nm utilizing theoretical ε 16,500 m−1 cm−1. Our optimized protocol provided a yield of 1 mm, 15.21 mg/ml HR (1 ml) from 1.4 liter of the E. coli secondary culture.
Purification of His-tagged protein
The sonicated mixture was centrifuged (14,000 rpm, 45 min, 4 °C), and the supernatant was incubated with 2 ml of Ni-NTA beads for binding on an end-on rocker for 2 h at 4 °C. Flow-through was discarded, and beads were washed with 100 ml of wash buffer 1 (1× PBS buffer 1: 10 mm imidazole) and 150 ml of wash buffer 2 (1× PBS buffer 1: 20 mm imidazole). Elution fractions of 2 ml each were collected (EB1: 30 mm imidazole; EB2: 50 mm imidazole; EB3: 75 mm imidazole; and EB4: 200 mm imidazole). The purity of the eluted protein was checked on 15% SDS-PAGE. The pure protein fractions were dialyzed against 1× PBS. The protein concentration was determined by absorbance at 280 nm utilizing theoretical ε 16,500 m−1 cm−1.
Purification of His-tagged mutant protein
The above clone was used as a template to introduce D39A and D138A mutation by site-directed mutagenesis. In brief, the primers were designed using SiteFind with the mutations D39A and D138A. PCR was performed using the designed primers (Table S4) followed by digestion with DpnI restriction enzyme for cleaving the parental plasmid DNA. The digested mixture was transformed in E. coli DH5α strain (Promega), and the mutant colonies were screened by plasmid isolation followed by digestion with the respective restriction enzymes. The identity of the clone was confirmed by sequencing. The mutant protein was expressed, purified in the same way as the WT His-tagged protein, and used for ITC experiments.
SEC-MALS
The spectrum for SEC-MALS was obtained to check the oligomerization status of the protein. The Superdex 200 10/300 column was pre-equilibrated with 500 mm mannose, 1× PBS to prevent the binding of the protein to the column. The column was then connected to a Wyatt Treos multiangle light-scattering instrument with an inline refractive index detector (Waters 2414 RI detector) and a multiwavelength UV detector (Shimadzu SPD-10 A VP UV-visible detector). Approximately 100 μl of 16 mg/ml protein solution was injected onto the column. The molar mass and mass percentage of each fraction were calculated using the ASTRA program (Wyatt Technology).
Circular dichroism
The CD spectrum of the purified protein (15 μm) in 1× phosphate buffer without the saline (pH 7.4) was carried out in a 260 to 195-nm range to determine the secondary structural characteristics of the protein. The experiment was done on a JASCO-J715 polarimeter in a 0.2-cm pathlength cuvette, with a slit width of 1 nm, a response time of 4 s, and a scan speed of 50 nm/s. Each data point was an average of four accumulations. GraphPad Prism version 7 software was used to plot the CD spectrum. The analysis of the predicted CD plot was done using the K2D2 webserver.
Fluorescence spectrum measurement
Fluorescence spectrum measurement was performed using intrinsic fluorophore tryptophan. Fluorescence spectra of the purified recombinant protein (15 μm) were measured at different pH conditions in 1× PBS at room temperature. The emission spectrum was recorded by exciting the protein at 295 nm and recording the emission in a 300–400 nm range in a quartz cuvette on a Jasco FP spectrofluorometer. The excitation and emission slit widths were 5 nm. Each spectrum was an average of four consecutive scans, and all of the experiments were done in duplicates.
Hemagglutination
Preparation of rabbit erythrocytes
Rabbit blood (2-5 ml) was obtained from the Central Animal Facility at the Indian Institute of Science, Bangalore. The blood was washed with 1× PBS (pH 7.4) and centrifuged three times at 1,200 rpm at 4 °C for 10 min to isolate the erythrocytes from the blood sample obtained in the form of a pellet. The treated rabbit erythrocytes were then stored at 4 °C and diluted appropriately in 1× PBS for use in the hemagglutination experiment.
Hemagglutination assay
The hemagglutinating activity of the lectin was determined by a 2-fold serial dilution procedure in a microtiter U-plate using heparin-treated 4% rabbit erythrocytes in 1× PBS buffer. The results were read after 1 h, when the blank or the negative control had fully sedimented. The hemagglutination titer is defined as the reciprocal of the highest dilution still exhibiting hemagglutination, which was reckoned as one hemagglutination unit. Specific activity is the number of hemagglutination units/mg of the protein. The experiment was performed at different pH levels to determine the optimum pH for activity starting from the initial concentration of 25 μg/ml. The experiment was done in triplicate for HR with BanLec as the positive control and 1× PBS as the negative control.
For hemagglutination inhibition assay, 25 μl of 10 μg/ml HR was incubated with an equal volume of a serial 2-fold dilution of the carbohydrate being tested. After incubation at room temperature for 30 min, the mixture was mixed with 50 μl of a 4% suspension of rabbit erythrocytes. The minimal concentration of the carbohydrate in the final reaction mixture capable of completely inhibiting four hemagglutination units of the lectin was calculated from the results.
Thermal shift assay
A fluorescence microplate reader (iQ5, Bio-Rad iCycler Multicolor Real-Time PCR detection system) was used to monitor protein unfolding as a function of temperature. The detection involves an increase in fluorescence of a fluorophore, SYPRO Orange (Sigma, S5692), upon binding to the hydrophobic regions of the gradually unfolded protein. Protein samples were mixed with 1× PBS buffer (pH 7.4) consisting of 1× SYPRO Orange dye and appropriate concentrations of ligand over the range of severalfold. Samples (10 μm) in triplicates were kept in a 96-well PCR microplate (Bio-Rad) in the i-Cycler iQ5 Multicolor RT-PCR detection system. The samples were heated at 0.5 °C/min, ranging from 25 to 95 °C, and the fluorescence intensities were measured at an interval of 0.5 °C. A Cy5 filter with red-orange color intensity was selected for the SYPRO Orange detection. Appropriate buffer blanks, in triplicates, were kept, and the intensities were subtracted for the same in each set. The average fluorescence intensities from the samples kept in triplicates were plotted as a function of temperature using GraphPad Prism version 7 software, and the Tm of protein was determined using the sigmoidal dose-response function of GraphPad. These experiments were also carried out at different ionic strengths of the buffer, different pH levels, and varying ligand concentrations.
Isothermal titration calorimetry
HR protein with a 6× histidine tag at the C terminus was used mostly for ITC studies. An ITC-200 microcalorimeter (GE Healthcare) was used for data collection. The protein was dialyzed in 1× PBS at pH 7.4 for at least 24 h, with three buffer changes prior to the titration. All of the reactant solutions were degassed for 30 min before the titrations. After preparation, the solutions, including the reactants and buffer solutions used for rinsing the cells and for the reaction setup, were degassed for 30 min. HR (80–2000 μm) was loaded in the sample cell and titrated with 1.6–2 μl of ligand, prepared in the dialysis buffer until saturation in the exothermic heats was observed (Table S5). The volume of the first injection was 0.6 μl. Each injection had a duration of 20 s, and they were 200 s apart. A stirring speed of 1,000 rpm was used. The exothermic heats associated with the titration of the ligand into the buffer were subtracted from that of the titration of the ligand into the protein solution. A plot of the incremental heat evolved as a function of the molar concentration of the ligand to the molar concentration of the lectin monomer was least squares–fitted to a single set of sites to obtain binding affinity (Kb), stoichiometry (n), and change in enthalpy (ΔH) and entropy using Origin 7.0 software. The change in entropy (ΔS) was calculated using the following equations,
| (Eq. 1) |
| (Eq. 2) |
where R is the gas constant, 8.314 J mol−1 K−1, and T is the absolute temperature in kelvin. The results reported are the average of three or more independent experiments.
For most titrations, the protein concentration was in the acceptable range of the “c-value,” a unitless constant, defined by Ka × Mt(0), where Ka is the association constant and Mt(0) is the initial concentration of the HR monomers. For mono-, di-, and trimannoside, the monomer concentration was multiplied by 2, as HR has 2 binding sites/subunit, and the c-value achieved was found to be <1 for the Me-α-Man, Man, and mannobioses as higher concentrations (>2 mm) of HR could not be contained because of protein aggregation and precipitation. Man5, Man7D1/3, and Man9 bind simultaneously to both of the sites in each protomer of HR, and the c-value achieved was 12, 22, and 22, respectively, adding to the confidence in stoichiometric and thermodynamic parameters. Despite a c-value of 0.5 for mono- and disaccharides, the n and Ka values obtained can be considered to be reliable as evident from an agreement between the KD = 1/Ka values from ITC experiments and the hemagglutination inhibition studies, although the enthalpy values for them could be overestimated as they are extrapolated from the fit. There was no significant difference observed in the binding thermodynamics between HR tagged and its tagless counterpart (Fig. S3 and Table S6).
In silico structure generation and validation
Modeling of the protein and validation
To overcome the paucity of structural data, two modeling approaches—comparative modeling and threading—were employed. The protein was modeled using homology modeling by the Phyre2 web server (44) and Modeller9v19 and a threading approach using I-TASSER (45). For Modeller, the top 10 BLASTp hits were taken, and the structure was prepared using the alignment of these sequences and structure as a template. This was followed by loop optimization to cater to the loops adjoining the β-sheets. The structure was found to constitute 62.3% β-sheet, 12.3% bend, and 6.2% turn
Validation of model structures was performed by monitoring for stereochemical quality, including planarity, chirality, ϕ/ψ preferences, χ angles, nonbonded contact distances, and unsatisfied donors and acceptors. The validation of ϕ/ψ preference for the modeled structure and structure validation was performed using PROCHECK, Ramachandran plot with the most favored, additionally allowed, generously allowed, and disallowed regions according to observed frequencies in high-resolution structures in the PDB. The tool ERRAT (46) was used to determine the quality of the model. Because HR has a β-prism fold consisting of 12 β-sheets, the topology of the modeled structure was generated. The wiring plot with respect to the residues and topology diagram for the secondary structural elements of the structures were generated using the PDBsum web server (47). A simulation of 100 ns was performed to check for the stability of the structure and the maintenance of secondary structure by the modeled structure. Prior to docking and MD simulations, the stability and the flexibility of the structure were analyzed using RMSD and RMSF of the 100 ns run. The maintenance of the secondary structure throughout the simulations was analyzed using the dssp (database of secondary structure assignments) function.
Oligosaccharide preparation
The oligosaccharides were prepared using GLYCAM-WEB (RRID:SCR_018260). The Man9 structure was truncated to obtain the different shorter oligosaccharides for the docking and simulation analysis. Independent, unbound states of the glycans were also subjected to MD simulation to obtain minimum energy conformers (Fig. S5). Glycans are depicted using symbols for monosaccharides as presented in the current version of the Symbol Nomenclature for Glycans.
Docking studies
Web servers COACH (48) and metaPocket (49) were used to detect the cavities and binding sites in the modeled protein. For further analysis of the number of binding pockets in the modeled lectin, a pairwise alignment was performed using the sequence of BanLec. All docking calculations were performed using Dock6 (version 6.6) (RRID:SCR_000128) (50). The protein receptor was processed using the Dock Prep module of Chimera using AMBER parm99 partial charges and the consequent output in Mol2 format (57). Active sites were identified and prepared by selecting spheres at a distance of 1–5 Å from the binding regions. All of the input files required to define the negative image of the binding site were prepared to superpose the ligands using the programs present in the DOCK distribution. Ligands were protonated and assigned AM1-BCC charges and then docked. A standard flexible docking protocol was employed to sample internal degrees of freedom of the ligands. The important parameters were as follows: A minimum of five atoms were treated as rigid, the maximum number of anchor orientations attempted was set to 1,000, and all 1,000 orientations were retained using a pruning clustering coefficient of 100 combining conformer rank and RMSD. A simplex minimizer was employed with score convergence set to 0.1 kcal/mol, and translational, rotational, and torsional step sizes for the simplex minimizer were set to 1.0 Å, 0.1p radians, and 10.0, respectively. A single round of simplex minimizer constituted a maximum of 500 iterations for the anchor and 500 iterations for the partially grown molecule. A pruning score cut-off was maintained at 100 kcal/mol to reject the conformers greater than the cut-off after energy minimization. Grid scoring based on the intermolecular nonbonded terms (viz. van der Waals for steric and electrostatic interactions) was computed using AMBER force field ff99 to identify the best orientation of each ligand. Finally, the best-scored conformer was retained for scrutiny and further refinement and Amber-based scoring. The docked poses were verified with respect to the crystal structures to ensure the relative positions of the docked sugar Man-D1 vis-à-vis crystal structure, and the best dock pose among different iterations was chosen for each of the carbohydrate-binding sites. It is to be noted that for glycans, Man5, Man7-D1, and Man9, the D1 arm of the glycan was transposed onto the mannose structure of HR at both of the carbohydrate-binding sites.
Conventional molecular dynamic simulations
Initially, independent simulations were performed on the HR system to ensure the maintenance of its three β-sheets and on the oligosaccharides to obtain the various plausible conformations of the glycans. Further, to examine the protein with different high-mannose derivatives and the possibility of cooperativity in their binding, docking and simulation studies were conducted. For the high-mannose oligosaccharide recognition, we consider the role of the whole surface of the CBS, including the monosaccharide-binding pocket, along with the residues that make up the common secondary binding site between the two binding pockets to participate in the binding of the complex glycans as seen from the ITC data.
Simulation parameters
The docked mannose on the predicted structure of HR was used for all-atom molecular dynamics simulations using GROMACS version 5.1.5 (51). The simulation of the docked structure was carried out using the OPLS-AA (optimized potential for liquid simulation—all atoms) force field (modified appropriately for carbohydrates) (52) and TIP3P water model. A vacuum energy minimization step was performed by employing the steepest descent algorithm for 50,000 steps, and conjugate gradient minimization was performed for 5,000 steps. Sodium (Na+) and chloride (Cl−) counter ions were added at a physiological concentration to neutralize the system after solvent addition by assuming normal charge state of ionizable groups with reference to pH 7.0 and by adjusting the boundaries of the dodecahedron box by 10 Å in a unit cell. The docked structure system in the solvent was again subjected to energy minimization for 50,000 steps for stabilization. Position restraint and unrestrained dynamics simulations of the solvated system were performed, and the Parrinello–Rahman pressure-coupling bath was used for the equilibration step (NVT and NPT) of the system at temperature 300 K using a Nose–Hoover thermostat under pressure of 1 atm for 1 ns. For simulations, all bonds were constrained using the LINCS algorithm. The particle Mesh–Ewald method was used for electrostatic calculations by maintaining a cut-off distance of 1.4 nm for coulomb and Van der Waals interactions, and finally, the production run for 100 ns was performed for the simulations.
Trajectories obtained from simulations were analyzed using tools provided by GROMACS. PyMOL (53) and UCSF Chimera (57) were used for the visual analysis of the snapshots. GROMACS built-in programs were used for the calculation of RMSD. The hydrogen bonds being formed throughout the 100 ns trajectory were analyzed using the hbond built-in function. Xmgrace was used for the generation of graphs.
Binding energy calculation using MM/PBSA
The binding free energies of the lectin-oligosaccharide complexes were calculated using the MM/PBSA approach (54) to evaluate the relative stabilities of these different complexes. This method involves a combination of molecular mechanics energy with implicit solvation models to calculate binding free energies. In MM/PBSA, binding free energy (ΔGbind) between a ligand (L) and a target (T) to form a complex is calculated using the equations,
| (Eq. 3) |
| (Eq. 4) |
| (Eq. 5) |
| (Eq. 6) |
where ΔEMM, ΔGSol, and −TΔS are the changes of the gas-phase molecular mechanics energy, the solvation free energy, and the conformational entropy upon binding, respectively. ΔEMM comprises ΔEInternal (bond, angle, and dihedral energies), ΔEElectrostatic (electrostatic energies), and ΔEVdw (van der Waals energies). ΔGSolv is the sum of electrostatic solvation energy (polar contribution) ΔGPB and nonelectrostatic solvation component (nonpolar contribution) ΔGSA. The polar contribution is calculated using the Poisson–Boltzmann surface area model, whereas the nonpolar energy is estimated from the solvent-accessible surface area (55). Convergence analysis suggested the calculation of the free energies on the last 90 ns of the trajectory.
Interaction of horcolin with the viral glycoproteins by surface plasmon resonance (SPR)
SPR assays were performed on a BIAcore 3000 instrument (Biacore, Uppsala, Sweden) optical biosensor at 25 °C. The HR protein (tagged and tagless) was immobilized onto a CM5 dextran sensor surface through standard amine coupling techniques using 0.2 m EDC and 0.05 m NHS, based on manufacturer protocols and as described previously. Briefly, the carboxymethyl dextran was activated by injecting 35 μl of the EDC/NHS solution over the surface at 5 μl/min. The protein was diluted to 0.01 μg/μl in 10 mm sodium acetate, pH 4.5, and manually injected over the activated CM5 sensor surface until the appropriate amount of the HR protein (in response units or 1,000 RU) was immobilized onto the surface. The reactive surface was then quenched by injecting 35 μl of 1 m ethanolamine, pH 8.0, over the surface at 5 μl/min. A nonspecific reference surface was created in an empty channel. This reference surface, along with buffer injections (1× PBS, pH 7.4), was used to correct for nonspecific interactions and instrumental artifacts and served as a negative control for each binding interaction. Serial dilutions of gp120 and gp140 were made in 1× PBS and injected over the surface at 50 μl/min with a 100-s association and up to 200-s dissociation. Protein concentrations ranged from 10 to 125 nm for gp120 and 100 nm to 2 μm for gp140. The surfaces were then regenerated with a 5-s pulse of 2 m magnesium chloride, 50 μl/min. Each binding curve was corrected for nonspecific binding by subtracting the signal obtained from the negative empty flow cell. Kinetic analysis of gp120 and gp140 binding to HR protein was performed by fitting the binding curves to a Langmuir 1:1 model using the BIAevaluation 4.0 program, with low residuals and χ2. This analysis determined the average ka and kd values, which were then used to calculate the associated Ka and KD values.
Characterization of anti-HIV activity of horcolin
Preparation of infectious virus stocks
HEK-293T cells were seeded at a density of 2.5 × 106 cells/100-mm Petri dish containing 100 μl of DMEM (Gibco) supplemented with 10% FBS (Gibco) and 1% penicillin/streptomycin (Gibco) to avoid bacterial contamination. 12 h postseeding, the medium was replaced with fresh complete medium, and cells were transfected with molecular clones of HIV-1JRFL and HIV-1YU2 using the CalPhos mammalian transfection kit (Takara) according to the manufacturer's protocol. 36 h post-transfection, supernatant was collected and subjected to ultracentrifugation at 28,000 rpm for 2.5 h using an SW28Ti rotor (Beckman). The virus pellet was resuspended in serum-free RPMI 1640 (Gibco) and stored in small aliquots at −80 °C for further use.
Determination of infectivity of prepared virus stocks
TZM-bl cells were seeded at the density of 0.125 × 106 cells/well of a 24-well plate containing 500 μl of DMEM (Gibco) supplemented with 10% FBS (Gibco) and 1% penicillin/streptomycin (Gibco). 12 h postseeding, the medium from each well was replaced with fresh complete medium, and the cells were infected with different concentrations of HIV-1JRFL and HIV-1YU2 as described previously (56). 48 h postinfection, medium was removed, and infected cells were fixed with 0.1% formaldehyde. 15 min postfixation, cells were washed twice with 500 μl of 1× PBS and were stained with X-gal–based staining solution, and blue cells were counted in different fields of each well.
Neutralization assay
TZM-bl cells were seeded at a density of 10,000–12,000 cells/well of a 96-well plate containing 100 μl of DMEM (Gibco) supplemented with 10% FBS (Gibco) and 1% penicillin/streptomycin (Gibco) to avoid bacterial contaminations. 12 h postseeding, TZM-bl cells were infected for 4 h with a 0.1 multiplicity of infection of HIV-1JRFL and HIV-1YU2 in the presence or absence of HR proteins or controls independently. Untreated cells were considered as negative controls, and b12-treated cells were considered as positive control. 48 h postinfection, the medium was removed, and cells were washed twice with 100 μl of 1× PBS using a multichannel pipette. After washing, 50 μl of SteadyGlo substrate (Promega) was added to each well (1:1 with 1× PBS) and transferred to an opaque plate using a multichannel pipette. Luminescence was measured using a Molecular Devices M5 microplate reader.
Each experiment was performed in duplicate, and three biological replicates of the assay were conducted. The average of the three experiments was taken for the IC50 calculation by nonlinear regression using GraphPad Prism software.
Assay of mitogenic and cytokine-inducing activity
All of the animal experiments were performed strictly according to the guidelines of the animal ethics committee. Clearance to perform the animal experiments was obtained from the Institutional Animal Ethics Committee before the commencement of the experiments.
Splenocyte isolation
C57BL/6 male mice, aged 6–8 weeks, were sacrificed, and the spleen was isolated. Cells from the spleen were isolated by carrying out teasing using a pair of forceps. Splenocytes were isolated using the Percoll gradient centrifugation method and washed using 5% RPMI medium. Cells were counted using a hemocytometer and seeded at a density of 0.25 million cells/well in a 96-well plate. Cells were then activated with different concentrations of ConA, BanLec, and HR (tagged and tagless) and were analyzed at different time points (36 and 48 h) after activation.
Propidium iodide assay
At desired time points, cells were taken and fixed with 70% ethanol (chilled). It was added in a dropwise manner to avoid clumping. Cells were then fixed on ice for 15–30 min or kept at −20 °C for long storage. After fixing, the cells were washed with 1 ml of 1× PBS. 100 μg/ml RNase A was added to each tube. After this, 50 μg/ml propidium iodide was added and incubated at room temperature for 15–30 min. Cells were analyzed using the flow cytometer, and the percentage of cycling cells was calculated.
ELISA for the quantification of cytokines
Cell supernatant was used to measure cytokine levels. A 96-well plate was used for the coating of the desired antibody with the help of a coating buffer. The coating was carried by adding the antibody to each well and incubated at 4 °C overnight. Washing was then carried out, and 1× assay diluent or ELISA blocking buffer containing phosphate-buffered solution supplemented with fetal bovine serum as a source of animal serum proteins was added to each well. This was used for the blocking of nonspecific binding of ELISA plates. After 1 h of incubation, wells were washed using the washing buffer, 1× PBS containing 0.05% Tween 20. The sample and desired standards were added to the washed wells. The buffer used for samples and the standard preparation was 1× assay diluent made from 5× stock. The plate was then incubated at 37 °C for 2–3 h. The wells were washed, and the detection antibody diluted in blocking buffer was added to each well, making the volume in each well 100 μl. The plate was incubated for 1–2 h at room temperature. In the end, the antibody conjugated to the enzyme (avidin-horseradish peroxidase) was added at the appropriate concentration after washing. After 15 min of incubation at room temperature, the TMB substrate was added to each well. After the color was generated, the reaction was stopped by adding 50 μl of 1 m H2SO4 (stop solution), making the final volume in each well 150 μl. The absorbance level is detected using a plate reader using a 450-nm wavelength of light. Unknown concentrations were calculated using the standard graph. The antibodies used in coating and detection were procured from the eBioscience ELISA kit (250×/1,000×) diluted to 1× for the experiment.
Statistical analysis
All data were analyzed using nonparametric analysis of variance tests. To detect differences among groups, the Newman–Keuls multiple-comparison test was used for pairwise comparison. All results were expressed using mean values ± S.D. and were considered statistically significant when the p value was <0.05.
Data availability
All data are contained within the article.
Supplementary Material
Acknowledgments
Prof. Raghavan Varadarajan (Indian Institute of Science) is acknowledged for the molecular clones of HIV-1JRFL and b12 neutralizing antibody. Dr. Shailendra Kumar (Sterogene Bioseparations, Inc.) is acknowledged for helpful suggestions. Prof. Richard Wyatt (Scripps Research) is acknowledged for the kind gift of viral glycoproteins. We also thank MBU for the high-performance computing facility, Srilatha N. from the SPR facility; Sunitha Prakash from the proteomics facility, MBU; the Indian Institute of Science FACS facility and animal house facility; and a Grant for NanoMachines for the nano-ITC facility.
This article contains supporting information.
Author contributions—N. G. J., R. V., S. Y., S. P., J. T., N. J., D. N., D. M., and A. Surolia data curation; N. G. J., A. Singh, and A. Surolia formal analysis; N. G. J. validation; N. G. J. visualization; N. G. J. methodology; N. G. J. writing-original draft; N. J., D. N., D. M., and A. Surolia resources; N. J., D. N., D. M., and A. Surolia investigation; A. Surolia conceptualization; N. J., D. N., D. M., and A. Surolia supervision; A. Surolia funding acquisition; A. Surolia project administration; A. Surolia writing-review and editing.
Funding and additional information—This study was supported by Department of Biotechnology (DBT), Government of India, Grant BT/PR27659/BID/7/829/2018 (to A. Surolia) and in part by Science and Engineering Research Board (SERB), Department of Science and Technology, Government of India, Grant SB/DF – 003/2018 (to A. Surolia). N. G. J. is an SRF fellow in the DBT grant. A. S. is a SERB Distinguished Fellow, Government of India; D. M. is a JC Bose fellow of SERB, Government of India; N. G. J. received a Department of Science Technology, Government of India, INSPIRE Fellowship for initial training.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- HR
- horcolin
- CBS
- carbohydrate-binding site
- MD
- molecular dynamics
- EDC
- 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide
- NHS
- N-hydroxysuccinimide
- X-Gal
- 5-bromo-4-chloro-3-indolyl β-d-galactoside
- Man
- mannose
- mJRL
- mannose-specific jacalin-related lectin
- ITC
- isothermal titration calorimetry
- BanLec
- banana lectin(s)
- MSA
- multiple-sequence alignment
- SEC-MALS
- size-exclusion chromatography coupled to multiangle light scattering
- Me-α-Man
- methyl-α-mannopyranoside
- Ni-NTA
- nickel-nitrilotriacetic acid
- PDB
- Protein Data Bank
- RMSD
- root mean square deviation
- RMSF
- root mean square fluctuation
- RU
- response units
- GNA
- Galanthus nivalis agglutinin
- GRFT
- griffithsin
- IL
- interleukin
- ConA
- concanavalin A
- MM/PBSA
- molecular mechanics/Poisson–Boltzmann surface area
- SPR
- surface plasmon resonance
- DMEM
- Dulbecco's modified Eagle's medium
- gp120
- glycoprotein 120
- gp140
- glycoprotein 140
- BLAST
- Basic Local Alignment Search Tool
- GROMACS
- Groningen Machine for Chemical Simulations.
References
- 1. Doores K. J., Bonomelli C., Harvey D. J., Vasiljevic S., Dwek R. A., Burton D. R., Crispin M., and Scanlan C. N. (2010) Envelope glycans of immunodeficiency virions are almost entirely oligomannose antigens. Proc. Natl. Acad. Sci. U. S. A. 107, 13800–13805 10.1073/pnas.1006498107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pantophlet R., Ollmann Saphire E., Poignard P., Parren P. W. H. I., Wilson I. A., and Burton D. R. (2003) Fine mapping of the interaction of neutralizing and nonneutralizing monoclonal antibodies with the CD4 binding site of human immunodeficiency virus type 1 gp120. J. Virol. 77, 642–658 10.1128/jvi.77.1.642-658.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhou T., Zheng A., Baxa U., Chuang G.-Y., Georgiev I. S., Kong R., O'Dell S., Shahzad-Ul-Hussan S., Shen C.-H., Tsybovsky Y., Bailer R. T., Gift S. K., Louder M. K., McKee K., Rawi R., et al. (2018) A neutralizing antibody recognizing primarily N-linked glycan targets the silent face of the HIV envelope. Immunity 48, 500–513.e6 10.1016/j.immuni.2018.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boyd M. R., Gustafson K. R., McMahon J. B., Shoemaker R. H., O'Keefe B. R., Mori T., Gulakowski R. J., Wu L., Rivera M. I., Laurencot C. M., Currens M. J., Cardellina J. H., Buckheit R. W., Nara P. L., Pannell L. K., et al. (1997) Discovery of cyanovirin-N, a novel human immunodeficiency virus-inactivating protein that binds viral surface envelope glycoprotein gp120: potential applications to microbicide development. Antimicrob. Agents Chemother. 41, 1521–1530 10.1128/AAC.41.7.1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barrientos L. G., Matei E., Lasala F., Delgado R., and Gronenborn A. M. (2006) Dissecting carbohydrate-Cyanovirin-N binding by structure-guided mutagenesis: functional implications for viral entry inhibition. Protein Eng. Des. Sel. 19, 525–535 10.1093/protein/gzl040 [DOI] [PubMed] [Google Scholar]
- 6. Koharudin L. M. I., and Gronenborn A. M. (2014) Antiviral lectins as potential HIV microbicides. Curr. Opin. Virol. 7, 95–100 10.1016/j.coviro.2014.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jeyaprakash A. A., Srivastav A., Surolia A., and Vijayan M. (2004) Structural basis for the carbohydrate specificities of artocarpin: variation in the length of a loop as a strategy for generating ligand specificity. J. Mol. Biol. 338, 757–770 10.1016/j.jmb.2004.03.040 [DOI] [PubMed] [Google Scholar]
- 8. Sankaranarayanan R., Sekar K., Banerjee R., Sharma V., Surolia A., and Vijayan M. (1996) A novel mode of carbohydrate recognition in jacalin, a Moraceae plant lectin with a β-prism fold. Nat. Struct. Biol. 3, 596–603 10.1038/nsb0796-596 [DOI] [PubMed] [Google Scholar]
- 9. Singh D. D., Saikrishnan K., Kumar P., Surolia A., Sekar K., and Vijayan M. (2005) Unusual sugar specificity of banana lectin from Musa paradisiaca and its probable evolutionary origin: crystallographic and modelling studies. Glycobiology 15, 1025–1032 10.1093/glycob/cwi087 [DOI] [PubMed] [Google Scholar]
- 10. Johnson Q., Lindsay R., Petridis L., Shen T., Johnson Q. R., Lindsay R. J., Petridis L., and Shen T. (2015) Investigation of carbohydrate recognition via computer simulation. Molecules 20, 7700–7718 10.3390/molecules20057700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shimizu T., and Morikawa K. (1996) The β-prism: a new folding motif. Trends Biochem. Sci. 21, 3–6 10.1016/S0968-0004(06)80018-6 [DOI] [PubMed] [Google Scholar]
- 12. Goldstein I. J., Winter H. C., Mo H., Misaki A., Van Damme E. J. M., and Peumans W. J. (2001) Carbohydrate binding properties of banana (Musa acuminata) lectin. Eur. J. Biochem. 268, 2616–2619 10.1046/j.1432-1327.2001.02149.x [DOI] [PubMed] [Google Scholar]
- 13. Singh D. D., Saikrishnan K., Kumar P., Dauter Z., Sekar K., Surolia A., and Vijayan M. (2004) Purification, crystallization and preliminary X-ray structure analysis of the banana lectin from Musa paradisiaca. Acta Crystallogr. D Biol. Crystallogr. 60, 2104–2106 10.1107/S0907444904024114 [DOI] [PubMed] [Google Scholar]
- 14. Han N., and Mu Y. (2013) Plasticity of 150-loop in influenza neuraminidase explored by Hamiltonian replica exchange molecular dynamics simulations. PLoS ONE 8, e60995 10.1371/journal.pone.0060995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bourne Y., Zamboni V., Barre A., Peumans W. J., Van Damme E. J. M., and Rougé P. (1999) Helianthus tuberosus lectin reveals a widespread scaffold for mannose- binding lectins. Structure 7, 1473–1482 10.1016/S0969-2126(00)88338-0 [DOI] [PubMed] [Google Scholar]
- 16. Peumans W. J., Zhang W., Barre A., Houlès Astoul C., Balint-Kurti P. J., Rovira P., Rougé P., May G. D., Van Leuven F., Truffa-Bachi P., and Van Damme E. J. M. (2000) Fruit-specific lectins from banana and plantain. Planta 211, 546–554 10.1007/s004250000307 [DOI] [PubMed] [Google Scholar]
- 17. Pratap J., Jeyaprakash A. A., Rani P. G., Sekar K., Surolia A., and Vijayan M. (2002) Crystal structures of artocarpin, a Moraceae lectin with mannose specificity, and its complex with methyl-α-d-mannose: implications to the generation of carbohydrate specificity. J. Mol. Biol. 317, 237–247 10.1006/jmbi.2001.5432 [DOI] [PubMed] [Google Scholar]
- 18. Tateno H., Winter H. C., Petryniak J., and Goldstein I. J. (2003) Purification, characterization, molecular cloning, and expression of novel members of jacalin-related lectins from rhizomes of the true fern Phlebodium aureum (L) J. Smith (polypodiaceae). J. Biol. Chem. 278, 10891–10899 10.1074/jbc.M211840200 [DOI] [PubMed] [Google Scholar]
- 19. Grunwald I., Heinig I., Thole H. H., Neumann D., Kahmann U., Kloppstech K., and Gau A. E. (2007) Purification and characterisation of a jacalin-related, coleoptile specific lectin from Hordeum vulgare. Planta 226, 225–234 10.1007/s00425-006-0467-x [DOI] [PubMed] [Google Scholar]
- 20. Azarkan M., Feller G., Vandenameele J., Herman R., El Mahyaoui R., Sauvage E., Vanden Broeck A., Matagne A., Charlier P., and Kerff F. (2018) Biochemical and structural characterization of a mannose binding jacalin-related lectin with two-sugar binding sites from pineapple (Ananas comosus) stem. Sci. Rep. 8, 11508 10.1038/s41598-018-29439-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Khan M. I., Sastry M. V., and Surolia A. (1986) Thermodynamic and kinetic analysis of carbohydrate binding to the basic lectin from winged bean (Psophocarpus tetragonolobus). J. Biol. Chem. 261, 3013–3019 [PubMed] [Google Scholar]
- 22. Schwarz F. P., Puri K., and Surolia A. (1991) Thermodynamics of the binding of galactopyranoside derivatives to the basic lectin from winged bean (Psophocarpus tetrogonolobus). J. Biol. Chem. 266, 24344–24350 [PubMed] [Google Scholar]
- 23. Surolia A., Sharon N., and Schwarz F. P. (1996) Thermodynamics of monosaccharide and disaccharide binding to Erythrina corallodendron lectin. J. Biol. Chem. 271, 17697–17703 10.1074/jbc.271.30.17697 [DOI] [PubMed] [Google Scholar]
- 24. Bachhawat-Sikder K., Thomas C. J., and Surolia A. (2001) Thermodynamic analysis of the binding of galactose and poly-N-acetyllactosamine derivatives to human galectin-3. FEBS Lett. 500, 75–79 10.1016/S0014-5793(01)02586-8 [DOI] [PubMed] [Google Scholar]
- 25. Morris M. J., and Striegel A. M. (2014) Influence of glycosidic linkage on the solution conformational entropy of gluco- and mannobioses. Carbohydr. Res. 398, 31–35 10.1016/j.carres.2014.06.022 [DOI] [PubMed] [Google Scholar]
- 26. Hopper J. T. S., Ambrose S., Grant O. C., Krumm S. A., Allison T. M., Degiacomi M. T., Tully M. D., Pritchard L. K., Ozorowski G., Ward A. B., Crispin M., Doores K. J., Woods R. J., Benesch J. L. P., Robinson C. V., et al. (2017) The tetrameric plant lectin BanLec neutralizes HIV through bidentate binding to specific viral glycans. Structure. 25, 773–782.e5 10.1016/j.str.2017.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Swanson M. D., Winter H. C., Goldstein I. J., and Markovitz D. M. (2010) A lectin isolated from bananas is a potent inhibitor of HIV replication. J. Biol. Chem. 285, 8646–8655 10.1074/jbc.M109.034926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Al Atalah B., Fouquaert E., Vanderschaeghe D., Proost P., Balzarini J., Smith D. F., Rougé P., Lasanajak Y., Callewaert N., and Van Damme E. J. M. (2011) Expression analysis of the nucleocytoplasmic lectin “Orysata” from rice in Pichia pastoris. FEBS J. 278, 2064–2079 10.1111/j.1742-4658.2011.08122.x [DOI] [PubMed] [Google Scholar]
- 29. Parajuli B., Acharya K., Bach H. C., Parajuli B., Zhang S., Smith A. B., Abrams C. F., and Chaiken I. (2018) Restricted HIV-1 Env glycan engagement by lectin-reengineered DAVEI protein chimera is sufficient for lytic inactivation of the virus. Biochem. J. 475, 931–957 10.1042/BCJ20170662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hoorelbeke B., Van Damme E. J. M., Rougé P., Schols D., Van Laethem K., Fouquaert E., and Balzarini J. (2011) Differences in the mannose oligomer specificities of the closely related lectins from Galanthus nivalis and Zea mays strongly determine their eventual anti-HIV activity. Retrovirology 8, 10 10.1186/1742-4690-8-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hoorelbeke B., Montfort T., Van, Xue J., Liwang P. J., Tanaka H., Igarashi Y., Damme E. J. M., Van, Sanders R. W., and Balzarini J. (2013) HIV-1 envelope trimer has similar binding characteristics for carbohydrate-binding agents as monomeric gp120. FEBS Lett. 587, 860–866 10.1016/j.febslet.2013.02.037 [DOI] [PubMed] [Google Scholar]
- 32. Lusvarghi S., Bewley C., Lusvarghi S., and Bewley C. A. (2016) Griffithsin: an antiviral lectin with outstanding therapeutic potential. Viruses 8, 296 10.3390/v8100296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. O'Brien W. A., Koyanagi Y., Namazie A., Zhao J. Q., Diagne A., Idler K., Zack J. A., and Chen I. S. (1990) HIV-1 tropism for mononuclear phagocytes can be determined by regions of gp120 outside the CD4-binding domain. Nature 348, 69–73 10.1038/348069a0 [DOI] [PubMed] [Google Scholar]
- 34. Li Y., Hui H., Burgess C. J., Price R. W., Sharp P. M., Hahn B. H., and Shaw G. M. (1992) Complete nucleotide sequence, genome organization, and biological properties of human immunodeficiency virus type 1 in vivo: evidence for limited defectiveness and complementation. J. Virol. 66, 6587–6600 10.1128/JVI.66.11.6587-6600.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sarzotti-Kelsoe M., Bailer R. T., Turk E., Lin C., Bilska M., Greene K. M., Gao H., Todd C. A., Ozaki D. A., Seaman M. S., Mascola J. R., and Montefiori D. C. (2014) Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. J. Immunol. Methods 409, 131–146 10.1016/j.jim.2013.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Larsson E. L., and Coutinho A. (1979) The role of mitogenic lectins in T-cell triggering. Nature 280, 239–241 10.1038/280239a0 [DOI] [PubMed] [Google Scholar]
- 37. Bateman A., Martin M. J., O'Donovan C., Magrane M., Alpi E., Antunes R., Bely B., Bingley M., Bonilla C., Britto R., Bursteinas B., Bye-AJee H., Cowley A., Da Silva A., De Giorgi M., et al. (2017) UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45, D158–D169 10.1093/nar/gkw1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. El-Gebali S., Mistry J., Bateman A., Eddy S. R., Luciani A., Potter S. C., Qureshi M., Richardson L. J., Salazar G. A., Smart A., Sonnhammer E. L. L., Hirsh L., Paladin L., Piovesan D., Tosatto S. C. E., et al. (2019) The Pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432 10.1093/nar/gky995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wilkins M. R., Gasteiger E., Bairoch A., Sanchez J. C., Williams K. L., Appel R. D., and Hochstrasser D. F. (1999) Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112, 531–552 10.1385/1-59259-584-7:531 [DOI] [PubMed] [Google Scholar]
- 40. Altschul S. F., Gish W., Miller W., Myers E. W., and Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 41. Tamura K., Stecher G., Peterson D., Filipski A., and Kumar S. (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Letunic I., and Bork P. (2016) Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245 10.1093/nar/gkw290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Satish P. R., and Surolia A. (2002) Sugars as affinity ligands. in Methods for Affinity-based Separations of Enzymes and Proteins (Gupta M. N., ed) pp. 115–129, Birkhäuser, Basel, Switzerland [Google Scholar]
- 44. Kelley L. A., Mezulis S., Yates C. M., Wass M. N., and Sternberg M. J. E. (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yang J., Yan R., Roy A., Xu D., Poisson J., and Zhang Y. (2015) The I-TASSER suite: protein structure and function prediction. Nat. Methods 12, 7–8 10.1038/nmeth.3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Colovos C., and Yeates T. O. (1993) Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 2, 1511–1519 10.1002/pro.5560020916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Laskowski R. A. (2001) PDBsum: summaries and analyses of PDB structures. Nucleic Acids Res. 29, 221–222 10.1093/nar/29.1.221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wu Q., Peng Z., Zhang Y., and Yang J. (2018) COACH-D: Improved protein-ligand binding sites prediction with refined ligand-binding poses through molecular docking. Nucleic Acids Res. 46, W438–W442 10.1093/nar/gky439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Huang B. (2009) Metapocket: a meta approach to improve protein ligand binding site prediction. OMICS 13, 325–330 10.1089/omi.2009.0045 [DOI] [PubMed] [Google Scholar]
- 50. Allen W. J., Balius T. E., Mukherjee S., Brozell S. R., Moustakas D. T., Lang P. T., Case D. A., Kuntz I. D., and Rizzo R. C. (2015) DOCK 6: impact of new features and current docking performance. J. Comput. Chem. 36, 1132–1156 10.1002/jcc.23905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Abraham M. J., Murtola T., Schulz R., Páll S., Smith J. C., Hess B., and Lindah E. (2015) Gromacs: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2, 19–25 10.1016/j.softx.2015.06.001 [DOI] [Google Scholar]
- 52. Kony D., Damm W., Stoll S., and Van Gunsteren W. F. (2002) An improved OPLS-AA force field for carbohydrates. J. Comput. Chem. 23, 1416–1429 10.1002/jcc.10139 [DOI] [PubMed] [Google Scholar]
- 53. DeLano W. L. (2012) The PyMOL Molecular Graphics System, version 1.5.0.1, Schrödinger, LLC, New York [Google Scholar]
- 54. Kumari R., Kumar R., Lynn A., and Lynn A, Open Source Drug Discovery Consortium, (2014) g_mmpbsa—a GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 54, 1951–1962 10.1021/ci500020m [DOI] [PubMed] [Google Scholar]
- 55. Zhou R. (2004) Exploring the protein folding free energy landscape: coupling replica exchange method with P3ME/RESPA algorithm. J. Mol. Graph. Model. 22, 451–463 10.1016/j.jmgm.2003.12.011 [DOI] [PubMed] [Google Scholar]
- 56. Trivedi J., Alam A., Joshi S., Kumar T. P., Chippala V., Mainkar P. S., Chandrasekhar S., Chattopadhyay S., and Mitra D. (2020) A novel isothiocyanate derivative inhibits HIV-1 gene expression and replication by modulating the nuclear matrix associated protein SMAR1. Antiviral Res. 173, 104648 10.1016/j.antiviral.2019.104648 [DOI] [PubMed] [Google Scholar]
- 57. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article.