Significance
The formation of apocarotenoids by carotenoid cleavage dioxygenases (CCDs) is a critical process for several biological signaling systems. However, the active site determinants directing CCDs to cleave a specific alkene bond within the polyene backbone of carotenoids have remained unclear. Through analysis of the previously uncharacterized group of archaeal CCDs, we identified an enzyme, closely related to animal CCDs, that could be isolated in complex with its apocarotenoid product. Its crystal structure revealed the precise molecular interactions governing the enzyme’s unique regioselectivity. These insights pave the way toward control of CCD activity in carotenoid/retinoid metabolism through rational design of small-molecule modulators or targeted mutagenesis.
Keywords: β-apo-14′-carotenal, regioselectivity, RPE65, nonheme iron, apocarotenoid
Abstract
Apocarotenoids are important signaling molecules generated from carotenoids through the action of carotenoid cleavage dioxygenases (CCDs). These enzymes have a remarkable ability to cleave carotenoids at specific alkene bonds while leaving chemically similar sites within the polyene intact. Although several bacterial and eukaryotic CCDs have been characterized, the long-standing goal of experimentally visualizing a CCD–carotenoid complex at high resolution to explain this exquisite regioselectivity remains unfulfilled. CCD genes are also present in some archaeal genomes, but the encoded enzymes remain uninvestigated. Here, we address this knowledge gap through analysis of a metazoan-like archaeal CCD from Candidatus Nitrosotalea devanaterra (NdCCD). NdCCD was active toward β-apocarotenoids but did not cleave bicyclic carotenoids. It exhibited an unusual regiospecificity, cleaving apocarotenoids solely at the C14′–C13′ alkene bond to produce β-apo-14′-carotenals. The structure of NdCCD revealed a tapered active site cavity markedly different from the broad active site observed for the retinal-forming Synechocystis apocarotenoid oxygenase (SynACO) but similar to the vertebrate retinoid isomerase RPE65. The structure of NdCCD in complex with its apocarotenoid product demonstrated that the site of cleavage is defined by interactions along the substrate binding cleft as well as selective stabilization of reaction intermediates at the scissile alkene. These data on the molecular basis of CCD catalysis shed light on the origins of the varied catalytic activities found in metazoan CCDs, opening the possibility of modifying their activity through rational chemical or genetic approaches.
Carotenoids are a familiar part of our visual experience. These tetraterpenoid polyenes are light-absorbing pigments responsible for many of the brilliant colors observed in nature and are well-known for their antioxidant properties (1). Living organisms also transform carotenoids to generate a series of apocarotenoid metabolites. In both prokaryotes and eukaryotes, an apocarotenoid known as retinaldehyde serves as the universal chromophore of photosensory opsins such as bacteriorhodopsin and visual pigments (2), placing this isoprenoid among life’s oldest light-sensing compounds (3). Apocarotenoids are also used as hormones. In plants, abscisic acid and strigolactones influence diverse processes such as seed dormancy, morphogenesis, and environmental adaptation (4). In animals, retinoic acid regulates gene expression throughout life (5) and is the main effector molecule responsible for the nonvisual actions of vitamin A.
In general, apocarotenoids are generated from carotenoid precursors in vivo through the action of carotenoid cleavage dioxygenases (CCDs) (6). These enzymes are widely distributed in nature, consistent with their known biological importance (7). CCDs catalyze oxygenolysis of target alkene bonds of carotenoid (or apocarotenoid) substrates using a nonheme FeII prosthetic group that is coordinated by four highly conserved His residues (reviewed in ref. 8). Several CCDs from bacteria and eukaryotes have been characterized in terms of their substrate specificity, regioselectivity, and physiological functions. The first CCD to be cloned and functionally characterized was an enzyme from Zea mays known as viviparous-14 (VP14) (9). This enzyme cleaves 9-cis-epoxycarotenoids specifically at their C11–C12 double bond to generate xanthoxin, the precursor of abscisic acid. Vertebrate genomes typically encode two alkene-splitting CCDs. One of these is β-carotene oxygenase (BCO) 1, which cleaves carotenoids and apocarotenoids at the C15–C15′ position to generate the opsin chromophore and retinoic acid precursor retinaldehyde (10). The second enzyme, BCO2, prefers hydroxylated carotenoids as substrates and cleaves them asymmetrically at the C9–C10 (and/or C10′–C9′) double bond, an activity critical for cellular carotenoid homeostasis (11, 12). All of these examples illustrate the general rule that CCDs (with a few exceptions) cleave carotenoids at specific double bond positions with high fidelity despite the presence of several chemically similar alternative sites within the polyene structure.
The molecular basis of this remarkably regioselective carotenoid cleavage activity has been the subject of structure–function studies involving targeted mutagenesis, chemical biology, and crystallographic analyses of bacterial, plant, and animal CCDs. The first CCD to have its three-dimensional structure determined was that of an apocarotenoid oxygenase (ACO) from Synechocystis sp. PCC6803 (SynACO) (13). The structure revealed the basic CCD fold consisting of a seven-bladed β-propeller capped on one face by a cluster of α-helical and loop segments forming a dome that covers the bound iron within the active site. In that study, SynACO was crystallized in a metal-free form and then incubated with iron and 3-hydroxy-β-apo-8′-carotenol in an effort to observe the binding of this substrate to the active site. The structure showed a doubly bent electron density feature in the active site that the investigators suggested could represent a substrate molecule that had undergone isomerization from an all-trans to a 13,13′ di-cis configuration, implying that SynACO could have a secondary isomerase activity. These results were admitted to be inconclusive owing to incomplete electron density support for the bound ligand (13). Later, the electron density attributed to a bound carotenoid was shown to derive instead from a noncarotenoid molecule, likely the linear polyoxyethylene detergent used for SynACO crystallization (14). Failed attempts to trap CCD–carotenoid complexes for structural studies were also reported for VP14 (15). Thus, the structure of a CCD in complex with an (apo)carotenoid has remained elusive. A major contributing factor to the difficulty of such studies is the water-insolubility of carotenoids, which limits the formation of high-occupancy complexes in aqueous solutions. Despite these challenges, determining the molecular basis of CCD regioselectivity remains an important goal, as such information could to allow targeted mutagenesis to produce novel or rare apocarotenoids and enable structure-based design of small-molecule modulators to control CCD activity in various natural processes.
The molecular and physiological functions of metazoan CCDs have long been of interest. However, structural analysis of these proteins has been hampered by their limited capacity for heterologous expression and purification, leading to a gap in our understanding of their enzymology. In the present study, we examined a group of CCDs encoded in the genomes of archaea, a domain of life previously thought to lack CCDs (16). In doing so, we identified a metazoan-like CCD enzyme from an ammonia-oxidizing archaeon, Candidatus Nitrosotalea devanaterra, belonging to the phylum Thaumarchaeota (17), that was amenable to detailed characterization. Here, we describe the biochemical features and structure of this enzyme in complex with its apocarotenoid product, providing high-resolution insights into the mechanisms of carotenoid cleavage that are likely applicable to the catalytically diverse and biologically important group of metazoan CCDs.
Results
Distribution of CCDs in Archaea.
Our study began with a BLAST search of the NCBI nonredundant protein database for archaeal CCDs using bovine RPE65 and SynACO as query sequences. CCDs were most commonly found in halophilic Euryarchaeota, which have a known ability to synthesize carotenoids including bacterioruberin (SI Appendix, Fig. S1) (18). No examples of CCDs were found among DPANN group archaea, which are thought to form an early branching archaeal lineage (19). Among members of the TACK superphylum, CCDs were found only in Thaumarchaeota, the top scoring hit to RPE65 being from the nitrogen-fixing, acidophilic soil archaeon Ca. Nitrosotalea devanaterra (17). CCD sequences were also found in the metagenomes of some Asgard archaea such as Candidatus Odinarchaeota archaeon LCB_4. Notably, these TACK and Asgard archaea appear not to be carotenogenic, as judged from the absence of encoded phytoene synthases in their genomes, and also apparently do not express opsin proteins (SI Appendix, Fig. S1).
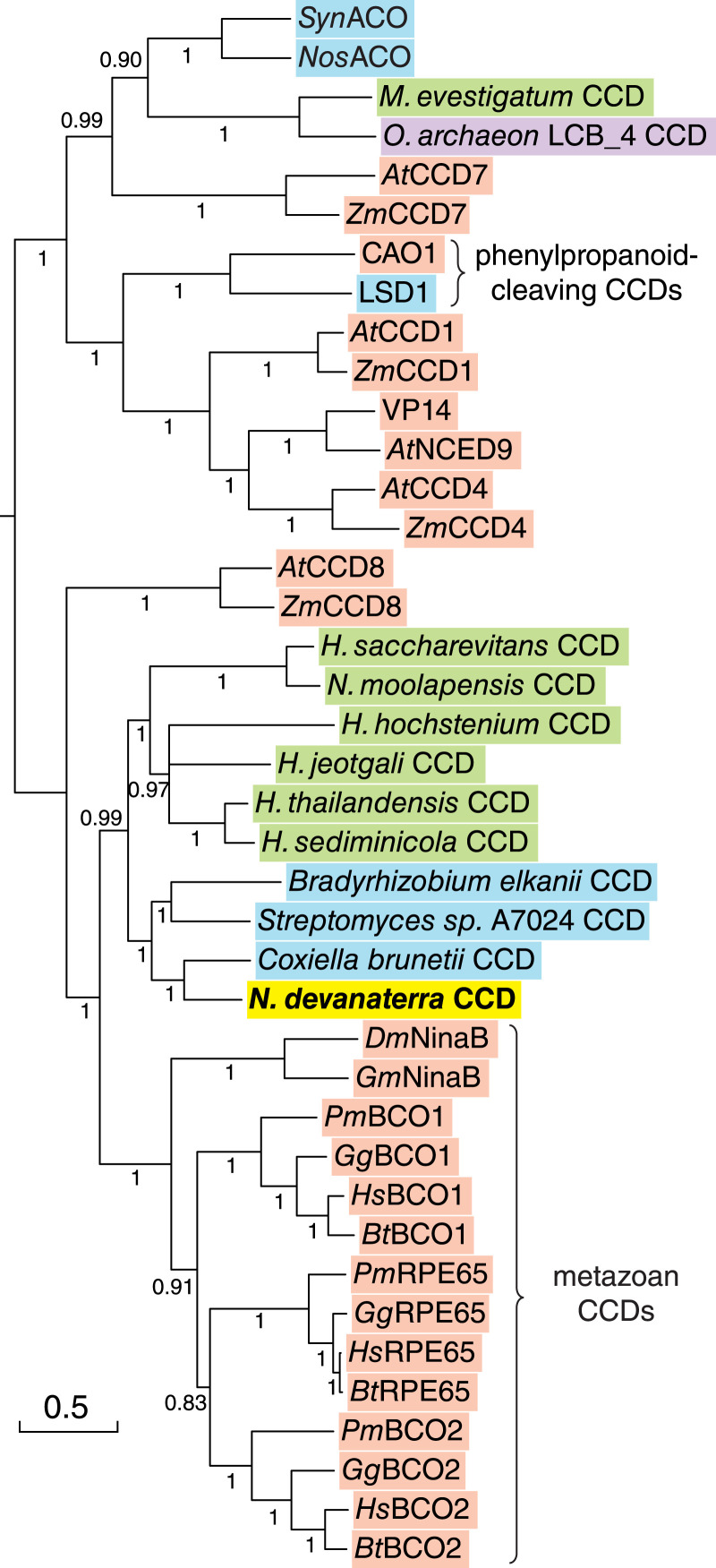
Archaea of the TACK (20) and Asgard (21) superphyla have been identified as close prokaryotic relatives of Eukarya, suggesting that CCDs from these archaea and eukaryotes might also have close phylogenetic relatedness. To test this hypothesis, we constructed a CCD phylogeny based on aligned amino acid sequences including top matching archaeal CCDs from the BLAST results (Fig. 1). The tree was topologically similar to previously reported phylogenies of bacterial and eukaryotic CCDs (22, 23), with archaeal CCDs appearing at two distinct locations. CCDs from Methanohalobium evestigatum (a euryarchaeote) and Ca. Odinarchaeota archaeon LCB_4 formed a sister group to cyanobacterial CCDs from Synechocystis and Nostoc, whereas CCDs from haloarchaea and N. devanaterra were found on the opposite side of the tree together with metazoan CCDs. N. devanaterra CCD (NdCCD) was located outside the haloarchaeal CCD cluster embedded within a group of CCDs from Coxiella burnetii and soil bacteria. Interestingly, nearly all of the closest-matching prokaryotic BLAST hits to NdCCD were from bacteria rather than archaea despite the current availability of genome sequences for over 124 archaea in the TACK superphylum. Analysis of the N. devanaterra genome with the DarkHorse algorithm (24) revealed an exceptionally low lineage probability index for NdCCD (SI Appendix, Fig. S2), demonstrating that NdCCD is highly atypical relative to most other N. devanaterra proteins. Together, these findings suggest that NdCCD may have been acquired by horizontal gene transfer from a bacterium. This mixed archaeal/bacterial CCD cluster formed a well-supported sister group to metazoan CCDs, with NdCCD being least diverged from the common ancestor of the two clades. Inspection of the NdCCD sequence revealed regions of striking similarity to metazoan CCDs, including a partially conserved “PDPCK” motif (“-DPCR” in NdCCD), a Val126 residue (Val or Ile in metazoan CCDs) that is known to influence ligand coordination by the iron center, as well as several residues predicted to line the substrate binding cleft (SI Appendix, Fig. S3). NdCCD shares 35% and 27% identity with RPE65 and SynACO, respectively. Owing to the excellent heterologous expression profile of NdCCD (SI Appendix, Fig. S4 and Table S1) and its similarity to metazoan CCDs of interest, we focused on this enzyme for further characterization.
Fig. 1.
Phylogenetic analysis of archaeal CCDs. The figure displays a CCD phylogeny illustrating the relationship of archaeal CCDs to those found in bacteria (blue background) and eukaryotes (salmon-colored background). Euryarchaeota are marked by a green background, while an Asgard CCD is shown on a purple background. The Thaumarchaeota CCD from N. devanaterra, which is the focus of this study, is shown on a yellow background. The majority-rule consensus tree was computed using MrBayes (25). Posterior probabilities are displayed along each bipartition. One bipartition with a posterior probability of <0.5 was collapsed. The tree scale indicates the number of substitutions per site. At, Arabidopsis thaliana; Bt, Bos taurus; CAO1, Neurospora crassa carotenoid oxygenase 1; Dm, Drosophila melanogaster; Gg, Gallus gallus; Gm, Galleria mellonella; Hs, Homo sapiens; LSD1, Sphingomonas paucimobilis TMY1009 lignostilbene α;β-dioxygenase 1; NinaB, neither inactivation nor afterpotential B; Nos, Nostoc; Pm, Petromyzon marinus; Syn, Synechocystis; Zm, Z. mays.
Expression and Enzymatic Characterization of NdCCD.
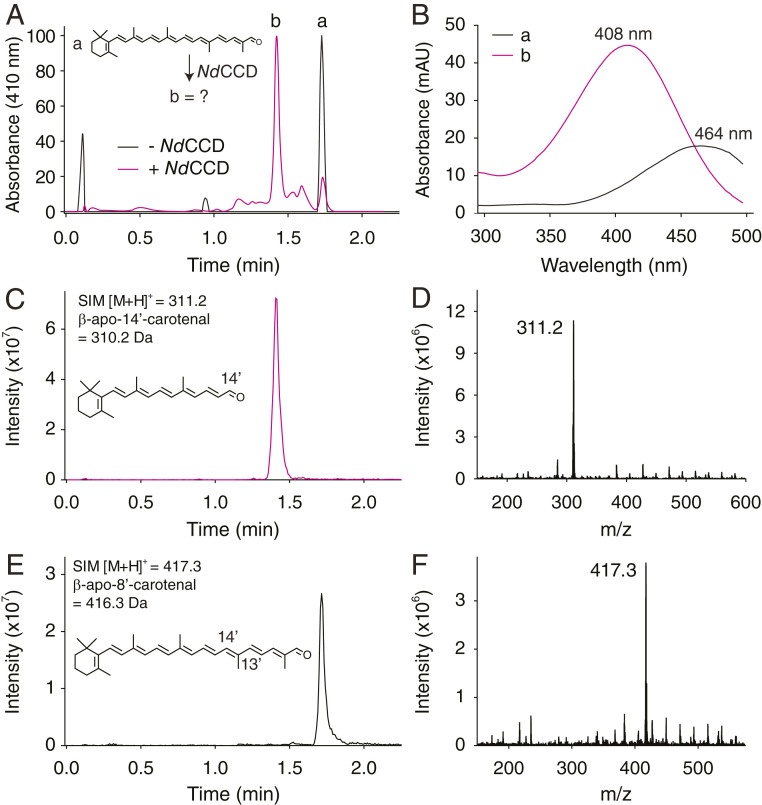
We expressed NdCCD in Escherichia coli and purified the protein in the absence of detergents by anion exchange and gel filtration chromatography (SI Appendix, Fig. S5). The purified sample was essentially homogenous (SI Appendix, Table S2) and contained nearly stoichiometric amounts of iron (SI Appendix, Table S3). We tested the ability of NdCCD to cleave β-apo-8′-carotenal and β-carotene, which are substrates of metazoan CCDs (26, 27). Whereas β-carotene was not accepted as a substrate (SI Appendix, Fig. S6A), NdCCD converted β-apo-8′-carotenal into a new compound with a λmax of ∼408 nm in polar solvent (Fig. 2 A and B). This value is slightly greater than the absorbance for retinal (λmax ∼ 383 nm in EtOH; ref. 28), but less than the 426-nm λmax known for β-apo-12′-carotenal in MeOH (29), suggesting a product with seven conjugated double bonds, namely β-apo-14′-carotenal. To confirm this identity, we further analyzed the product by LC-MS. We observed a single 311.2 m/z peak, corresponding to mass of protonated β-apo-14′-carotenal, with a retention time matching the λmax = 408 nm peak observed by UV/Vis spectroscopy (Fig. 2 C and D). Residual substrate was also detected as a 417.3 m/z peak whose elution time coincided with the UV/Vis peak observed for the substrate (Fig. 2 E and F). These data demonstrated that NdCCD cleaves β-apo-8′-carotenal specifically at the C14′–C13′ double bond.
Fig. 2.
Mass spectrometry analysis of the NdCCD reaction products demonstrates C14′–C13′ cleavage selectivity. (A) Reversed-phase HPLC analysis of β-apo-8′-carotenal (a) cleavage by NdCCD to form cleavage product (b). Absorbance for each trace is normalized to 100. The apocarotenoids were eluted with a 5 to 95% acetonitrile gradient in the presence of 0.1% formic acid. (B) UV/Vis absorbance spectra of peaks a and b from A. (C) Selective ion monitoring (SIM) at m/z = 311.2, corresponding to the molecular ion of β-apo-14′-carotenal, showed a peak with a retention time identical to that of peak b in A. (D) Mass spectrum of the SIM peak in C showing a dominance of the m/z = 311.2 species. (E) SIM at m/z = 417.3, corresponding to the molecular ion of β-apo-8′-carotenal, showed a peak with a retention time identical to that of peak a in A. (F) Mass spectrum of the SIM peak in E showing a dominance of the m/z = 417.3 species.
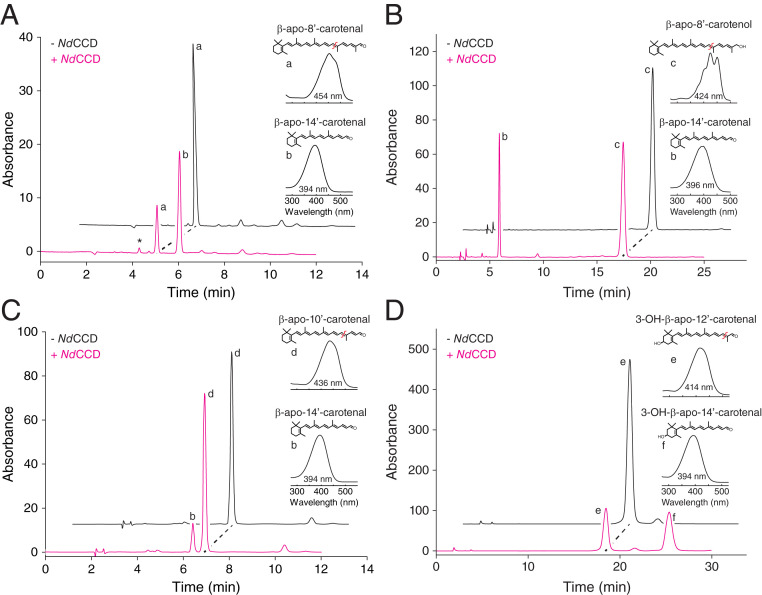
Next, we investigated the range of substrates accepted by NdCCD using apocarotenoids of various lengths as well as the C40 carotenoids lycopene and zeaxanthin. In addition to β-apo-8′-carotenal (Fig. 3A), NdCCD was able to cleave both β-apo-8′-carotenol and β-apo-10′-carotenal to produce β-apo-14′-carotenal (Fig. 3 B and C), demonstrating that the distal region of the apocarotenoid does not influence the enzyme’s regioselectivity. However, such changes did affect catalytic efficiency, as kcat and Km differed substantially between β-apo-8′-carotenal and β-apo-8′-carotenol (SI Appendix, Fig. S7). NdCCD also cleaved 3-hydroxy-β-apo-12′-carotenal, specifically at the C14′–C13′ position, demonstrating that substrate ring hydroxylation was also tolerated by the enzyme (Fig. 3D). By contrast, we did not detect cleavage of zeaxanthin, lycopene, or retinal (SI Appendix, Fig. S6 B–D). We also tested the ability of NdCCD to cleave resveratrol and isoeugenol, but neither compound was accepted as a substrate (SI Appendix, Fig. S6 E and F), consistent with the position of NdCCD outside of the phenylpropanoid-cleaving CCD clade (Fig. 1). Together, these results show that NdCCD can cleave a variety of apocarotenoids and does so specifically at their C14′–C13′ double bond. Such an activity has not been previously observed among molecularly characterized CCDs, although some can generate apo-14′-carotenals as side products (30–32). Interestingly, enzymatic β-apo-14′-carotenal production (EC 1.13.11.67) was reported previously from lysates of rabbit and rat intestinal mucosa, although the responsible enzyme was never molecularly identified (33).
Fig. 3.
Cleavage activity of NdCCD toward apocarotenoid substrates. (A) Normal-phase HPLC analysis demonstrating the cleavage of β-apo-8′-carotenal (a) by NdCCD to form β-apo-14′-carotenal (b) with an absorbance maximum of 394 nm. The asterisk indicates an isomer of β-apo-14′-carotenal that was generated by photoisomerization following the cleavage reaction. (B) NdCCD was similarly able to generate β-apo-14′-carotenal (b) from β-apo-8′-carotenol (c), indicating that the terminal polar group does not influence the site of cleavage. (C) NdCCD cleaved the shorter apocarotenoid, β-apo-10′-carotenal (d), to generate β-apo-14′-carotenal (b), indicating that cleavage specificity is not determined by the length of the distal end of the apocarotenoid substrate. (D) NdCCD also cleaved the polar carotenoid 3-hydroxy-β-apo-12′-carotenal (e) to form a product (f) with a greater retention time but similar absorbance spectrum compared to β-apo-14′-carotenal, indicating it represents 3-hydroxy-β-apo-14′-carotenal. Apocarotenoids were eluted with 90:10 hexane/ethyl acetate in A–C and 80:20 hexane/ethyl acetate in D.
Based on the close phylogenetic relationship between NdCCD and C. burnetii CCD (CbCCD), we hypothesized that these enzymes would exhibit the same C14′–C13′ cleavage regioselectivity. To test this hypothesis, we expressed CbCCD in E. coli and measured its activity directly from the supernatant fraction (SI Appendix, Fig. S8A and Table S1). As predicted, CbCCD cleaved β-apo-8′-carotenal specifically at the C14′–C13′ bond but did not accept β-carotene as a substrate (SI Appendix, Fig. S8 B and C). These findings indicate that CCDs with C14′–C13′ apocarotenoid cleavage activity could be widespread in both archaea and bacteria.
Structure of NdCCD.
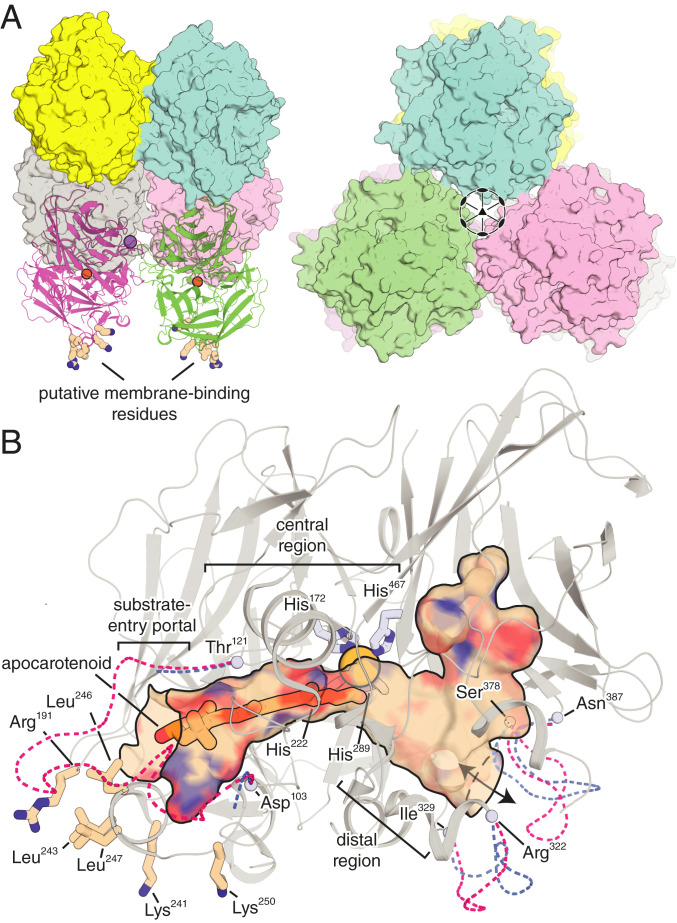
To gain insights into the structural basis of NdCCD’s regioselective cleavage activity, we crystallized NdCCD and determined its structure by X-ray diffraction analysis (SI Appendix, Table S4). The asymmetric unit of the crystals contained six NdCCD monomers in a D3-symmetric arrangement (Fig. 4A). Interactions between monomers related by threefold symmetry were sparse and unlikely to be stable in solution. Conversely, interactions between chains related by twofold symmetry were more extensive, burying ∼1,000 Å2 per dimer pair (Fig. 4A). This dimer interaction is distinct from the dimers observed for RPE65 (34) and stilbene-cleaving CCDs (35). The interaction surface was moderately hydrophobic according to ΔiG values computed in PISA (36), but also involved electrostatic interactions including sodium ion coordination by residues from each member of the dimer pair. A sequence alignment of NdCCD with metazoan CCDs shows that residues contributing to dimer formation are poorly conserved between the different proteins and reside in indel-prone areas (SI Appendix, Fig. S3). This dimer structure could promote NdCCD binding to membranes as a result of its parallel arrangement of hydrophobic surface residues (described later; Fig. 4A).
Fig. 4.
Crystal structure of NdCCD. (A) Side (Left) and front (Right) views of the D3-symmetric NdCCD hexamer showing interactions between monomers related by the twofold operator and threefold operator, respectively. The side-view image shows a parallel arrangement of putative membrane-interacting residues (shown as sticks) on the dimer surface. The active-site iron ions for one of the dimers are shown as brown spheres, and a sodium ion bound at the dimer interface is shown as a lavender sphere. (B) A composite view of NdCCD structures reported in this study. The active-site tunnel (shown in surface representation) is composed of a substrate entry portal located in proximity to the membrane binding patch, a central region that includes the iron prosthetic group, and a distal region that connects with the protein exterior as well as a hydrophilic cavity. The apocarotenoid studied in this work (described in detail later) is bound in the region of the tunnel closest to the membrane binding patch. The dashed lines near the tunnel openings show the various conformations observed for these segments (i.e., residues 103 to 121, 322 to 329, and 378 to 387) in the different crystal structures reported in this study. The metal center (iron or cobalt) is shown as a yellow-orange sphere coordinated by four His residues conserved throughout the CCD superfamily.
NdCCD exhibits the classic CCD fold described above for SynACO. Compared to CCDs of known structure, NdCCD was most similar to RPE65, with an rms difference (rmsd) of 1.6 Å over 445 matched Cα positions, consistent with these two proteins also having the highest amino acid sequence identity (SI Appendix, Figs. S3 and S9A). In light of its apocarotenoid oxygenase activity, NdCCD was also compared to SynACO, giving a superposition rmsd of 2.2 Å. CCDs typically possess hydrophobic patches on their surfaces, often surrounded by cationic residues (8), that allow them to interact with membranes where their carotenoid substrates are dissolved (1). Such a patch was also observed on the surface of NdCCD consisting of Leu residues 243, 246, and 247 together with Arg191, Lys241, and Lys250 (Fig. 4 A and B). This patch is notably smaller than the patches on both RPE65 and SynACO, a finding that is consistent with the soluble protein behavior of NdCCD, in contrast to RPE65 and SynACO, which require detergents for purification and/or crystallization. Nevertheless, the patch could confer an ability to transiently dip into lipid bilayers for substrate access. As mentioned above, NdCCD features a partially conserved PDPCK motif, which is positioned on the N-terminal side of a loop region thought to be involved in membrane interactions of RPE65 and other metazoan CCDs (37). Indeed, the Cys residue of this motif is palmitoylated in RPE65 (38) and other metazoan CCDs (39), findings consistent with its proximity to the lipid bilayer. However, this region has never been fully resolved in RPE65 crystal structures (34). Interestingly, the corresponding loop in chains E and F of NdCCD exhibited relatively clear electron density that allowed construction of a model for a majority of the sequence (SI Appendix, Fig. S9B). The structure of this region is mainly irregular, with only three β-turn elements in contrast to the α-helical structure that has been predicted for the corresponding region of RPE65 (37, 40). In NdCCD, this loop folds back against the helical cap region of the protein, partially occluding the main opening to the enzyme’s active site: a conformation that appears incompetent for substrate uptake (Fig. 4B). The loop is hydrophobic overall, with a few positively charged residues that may enhance the ability of NdCCD to interact with membranes, possibly triggering a conformational change to allow substrate entry (SI Appendix, Fig. S9B). Notably, residues 106 to 118 are entirely disordered in chains A through D, indicating that this region of NdCCD is also prone to being unstructured, similar to RPE65 (34). In the case of NdCCD, this disorder cannot be attributed to detergent-related destabilization.
By analogy to RPE65 (41), the active site of NdCCD can be divided into three regions: the substrate entry portal, the central active site including the metal center, and an expanded distal region that also connects with the protein exterior (Fig. 4B). The tunnel as a whole, which spans a length of ∼30 Å, is lined predominantly by nonpolar residues but includes a polar region in the central active site in which two water molecules are stably bound. Beginning with the relatively wide substrate entry portal, the active site tunnel narrows as it passes by the iron center before expanding and ramifying within the interior of the protein. In chains A and D, this interior pocket is nearly closed off with only a ∼2.3-Å-wide circular opening to the protein exterior (tunnel opening widths are given with respect to the Connolly surface). In chains B and C, a conformational difference in residues 322 to 328 and 378 to 386 produced a ∼4.5-by-7.5-Å elliptical opening in the protein structure that is partially covered by Ala382 (Fig. 4B). This wider conformation was associated with sodium ion coordination by residues near the opening. The distal segment also connects with a hydrophilic cavity of unclear functional significance that contains several buried water molecules. Extending from one opening to the other, the active site tunnel exhibits a shallow V-like shape with the bend located near the metal center (Fig. 4B).
The structure of the NdCCD active site largely resembled that of RPE65 in terms of amino acid conservation and overall shape, but differed markedly from that of SynACO despite the fact that these proteins have similar substrate specificities (SI Appendix, Fig. S9C). Prior studies on RPE65 have revealed that its substrate entry portal and central active site regions are responsible for binding of the retinoid, whereas the fatty acid acyl chain is accommodated in the inner pocket (41). The structure of the central active site is strikingly well conserved between NdCCD and RPE65, despite the fact that these proteins bind different (but related) substrates and catalyze distinct chemistry (SI Appendix, Fig. S9C). This conservation includes residues known to be critical for enforcing RPE65 isomerization regioselectivity (42), namely Phe61, Phe103, and Thr147, which are homologous to Phe58, Phe100, and Thr139 in NdCCD (SI Appendix, Fig. S3). The distal region of the active site pocket is more variable in terms of shape and sequence conservation, consistent with this pocket serving different functions in the two enzymes. In RPE65, this pocket exhibits a notable bend that is primarily due to the side chain of Phe418 protruding into the pocket. NdCCD, on the contrary, contains a less bulky Leu residue at the equivalent position, giving the pocket a more open shape. In both proteins, the distal pocket connects to both a hydrophilic cavity as well as an opening to the protein exterior, although the opening is much narrower and positioned slightly differently in RPE65 (SI Appendix, Fig. S9C).
The iron center is bound by inner sphere His residues 172, 222, 289, and 467, with the later three residues forming hydrogen bonding interactions with outer sphere Glu residues 140, 357, and 411 (SI Appendix, Fig. S9C). Strong, but heterogeneous, electron density was also observed in the primary sphere trans to His-172, indicative of a diffusible ligand. In chain D, the density could be explained by a single solvent molecule bound to the iron ion at a distance of ∼3 Å, whereas, in chain A, we observed an elongated density feature extending into the distal active site cavity that was not readily interpretable. The residual density in chains B and C was not adequately quenched by a single solvent alone, nor an O2 molecule. Based on the density appearance and the known binding of carboxylate ligands to the iron center of RPE65 (41), we modeled a bicarbonate anion in chains B and C, although the ligand identity could not be conclusively determined.
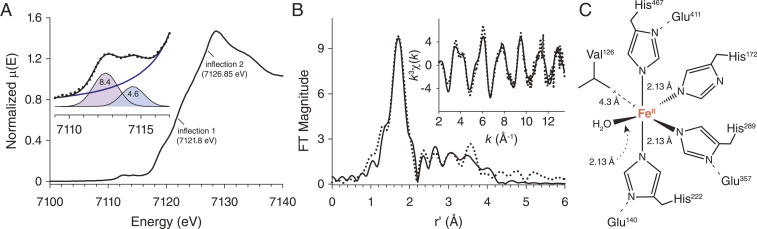
To clarify the resting state iron coordination, we measured iron K-edge X-ray absorption spectra on concentrated solutions of NdCCD. XAS provides coordination number and high-precision bond lengths for the primary metal coordination sphere and, importantly, can be carried out in simple buffered solutions closer to those used for activity studies. The XANES spectrum of NdCCD was similar to those of other CCDs, with a bimodal pre-edge feature exhibiting peaks at 7,112.5 and 7,114.4 eV with areas of 8.4 and 4.6 units, respectively (Fig. 5A). These peaks originate from 1s → 3d electron transitions whose intensity is related to the metal coordination number. The total pre-edge peak intensity (13 area units) and energy splitting (1.9 eV) were consistent with a five-coordinate FeII center. The area of the higher energy peak is greater than that reported for C4v-symmetric high-spin FeII model complexes suggesting D3h-symmetric character for the NdCCD iron center (43). The spectrum featured edge inflection points at 7,121.8 and 7,126.85 eV consistent with a ferrous iron state. Quantitative analysis of Fourier-filtered extended X-ray absorption fine structure (EXAFS) data incorporating multiple scattering paths (Fig. 5B and SI Appendix, Table S5) allowed construction of a model in which the iron is coordinated by four imidazole ligands and one O/N atom all at a distance of 2.13 Å (Fig. 5C). The absence of a prominent peak at r′ = 2.5 Å in the Fourier-transformed EXAFS spectrum together with the five-coordinate state supported by the pre-edge analysis argued against bidentate binding of a carboxylate ligand to the iron center as suggested by the crystallographic data. Instead, the EXAFS data were most simply explained by a single aquo ligand bound to the iron in addition to the four conserved His residues (Fig. 5C). Thus, the iron ligand heterogeneity we observed in crystallo may originate from bound components of the crystallization mixture or possibly arise from pH differences between the two conditions.
Fig. 5.
X-ray absorption spectroscopy analysis of the NdCCD iron center. (A) The near-edge spectrum shows a bimodal pre-edge absorption feature with an energy splitting of 1.9 eV and peak areas (Inset) as well as edge inflections at 7,121.8 and 7,126.85 eV. (B) Fourier-transformed (FT) EXAFS data (dotted black line) and best-fit simulated data (solid black line) for the model shown in SI Appendix, Table S5. (Inset) k3-weighted EXAFS data (dotted black line) and best-fit simulated data (solid black line). (C) Model of the NdCCD iron center derived from both XAS and crystallographic information. The FeII center is five-coordinate with a distorted D3h-symmetric ligand arrangement. The metal–ligand bond lengths are identical within the error of the data at 2.13 Å. The five-coordinate structure is promoted by the presence of a Val side chain that occludes one of the potential ligand binding sites in the coordination sphere.
Experimental Visualization of a CCD–Product Complex.
To gain atomic-level insights into the regioselectivity of NdCCD, we aimed to determine its structure in complex with an apocarotenoid substrate. The ability of this enzyme to be isolated and crystallized in the absence of detergent, together with its spatially restricted active site, appeared to make it an excellent candidate for such studies. To facilitate isolation the complex, we generated a cobalt-substituted version of NdCCD based on prior data showing that cobalt does not support CCD catalytic activity (25, 26). NdCCD was readily expressed in minimal media and incorporated cobalt in a near stoichiometric fashion as demonstrated by quantitative metal analysis (SI Appendix, Table S3). Concentrated Co-NdCCD samples had a pale pink color and optical absorbance peaks at 524, 541, and 562 nm, attributable to CoII d-d electronic transitions (SI Appendix, Fig. S10A). Co-NdCCD crystals were isomorphous to those of the native iron enzyme but routinely diffracted to higher resolution (SI Appendix, Table S4). Diffraction data collected above and below the cobalt K absorption edge allowed calculation of imaginary log-likelihood gradient maps (45) that demonstrated specific occupancy of cobalt at the active site of NdCCD (SI Appendix, Fig. S10B). The structure of Co-NdCCD was largely similar to that of the native enzyme as evidenced by the average 0.47-Å rmsd between equivalent chains. Electron density near the cobalt ion was uniform and supported modeling of a single solvent trans to His172 at a distance of ∼2 Å. The cobalt center had a distorted D3h symmetric structure, consistent with the symmetry of the NdCCD iron center derived from the XAS measurements described earlier. Structural variability was observed at the opening to the distal cavity (Fig. 4B). Specifically, residue Phe385 adopted a different conformation in chains A and D as compared to the corresponding chains in the Fe-NdCCD structure, resulting in a ∼3.2-by-5.8-Å elliptical opening. A polymorphic electron density feature was present in the distal cavity that could not be adequately modeled with known components of the crystallization mixture. Taken together, the Co-NdCCD structure closely mimicked native, iron-bound NdCCD.
Next, we isolated Co-NdCCD in complex with 3-hydroxy-β-apo-12′-carotenal, which was selected based on its relatively high aqueous solubility. The apocarotenal was added to the enzyme sample prior to each step of chromatography, and the protein and apocarotenoid absorbances were monitored at 280 and 420 nm, respectively. For each of the three steps of purification, we observed coelution of apocarotenoid with Co-NdCCD by UV/Vis spectroscopy (SI Appendix, Fig. S11). Based on the 280- and 420-nm peak areas and the extinction coefficients for NdCCD (74,688 M−1·cm−1) and 3-hydroxy-β-apo-12′-carotenal (75,600 M−1·cm−1; estimated from ref. 29), the final purified sample had an apocarotenoid to NdCCD molar stoichiometry of ∼0.7. We subjected a portion of the purified complex to hexane extraction and LC-MS analysis to assess the apocarotenoid composition of the final sample (SI Appendix, Fig. S12). Surprisingly, we found that a bulk of the apocarotenoid in the final preparation was the 3-hydroxy-β-apo-14′-carotenal product of catalysis instead of 3-hydroxy-β-apo-12′-carotenal, indicating that the substrate was cleaved during protein isolation. This result was unexpected given that cobalt substitution in similar CCDs was shown to eliminate catalytic activity (35, 44). It is likely that unavoidable low-level iron contamination (SI Appendix, Table S3) could have produced sufficient activity to turn over a majority of the copurified substrate, although cobalt may allow slow catalytic activity that was not detectable in conventional assays.
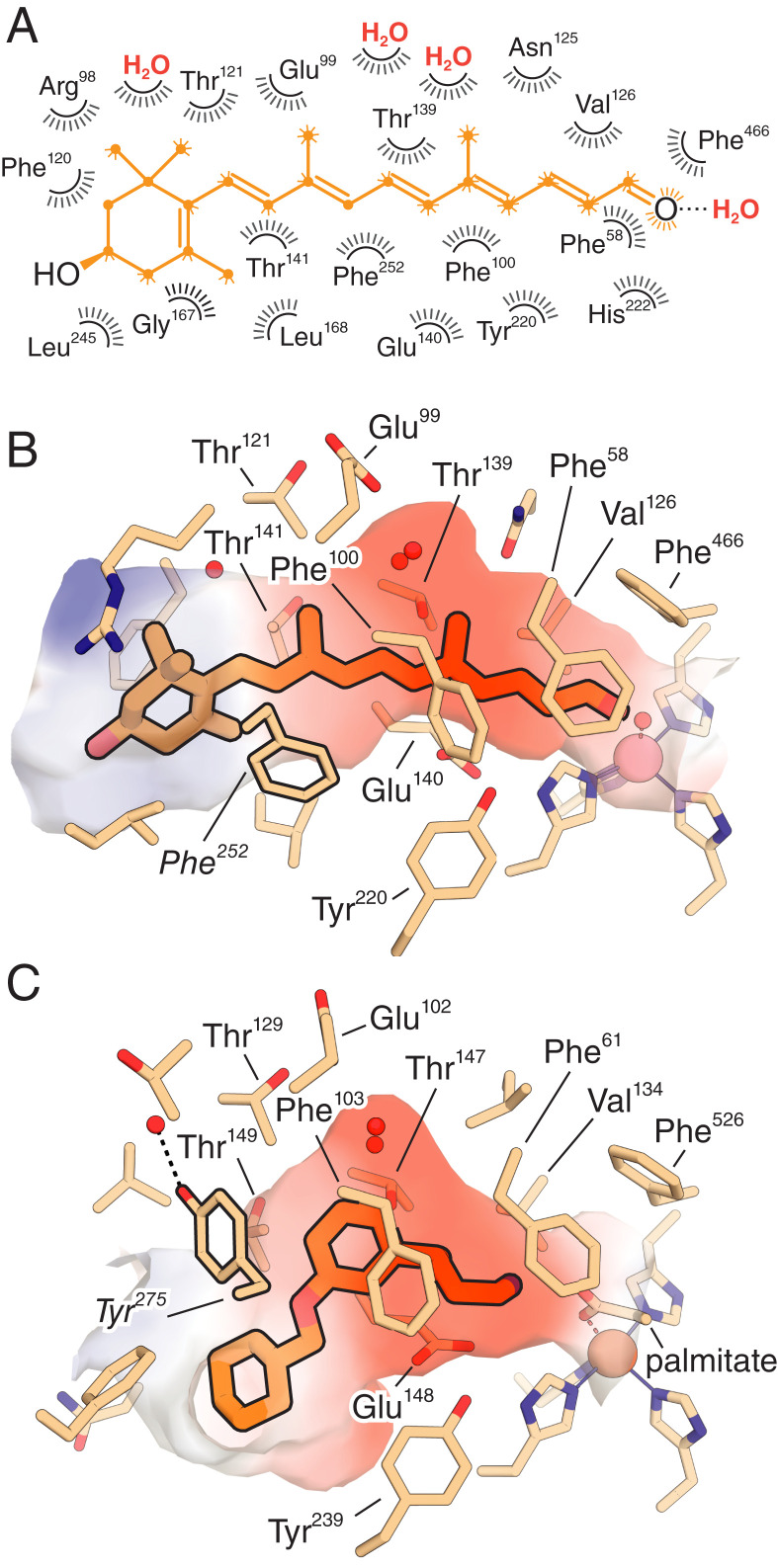
We proceeded to crystallize the Co-NdCCD–product complex under the same conditions used for unliganded Co-NdCCD. The orange-colored crystals obtained (SI Appendix, Fig. S11C) were isomorphous to the original Co-NdCCD crystals, with similar diffraction properties (SI Appendix, Table S4). Following rigid-body refinement, we observed residual electron density in the substrate entry portal and central active site suggestive of a bound ligand (SI Appendix, Fig. S13A). This was confirmed by calculation of an isomorphous-difference map, which revealed an excess of continuous positive density (SI Appendix, Fig. S13B). The entire apocarotenoid structure was represented in the electron density map with characteristic features for the 3-hydroxy-β-ionone ring and polyene methyl groups clearly present (SI Appendix, Fig. S13C). The molecule is snugly bound within the active site in an extended all-trans geometry with the C6–C7 bond in an s-cis-conformation (Fig. 6 A and B), which is enforced by the shape of the substrate-entry portal. The 3-hydroxy-β-ionone ring is partially solvent-exposed and does not engage in polar interactions with the protein involving its hydroxyl moiety (Fig. 4B). The polyene region of the molecule is completely isolated from the bulk solvent, residing in the tapered central active site. This active site region exhibits a negative electrostatic potential positioned over the central region of the apocarotenoid that originates from the main chain or side chain groups of Phe58, Glu99, Asn125, Thr139, Glu140, and Glu99 (Fig. 6B). Notably, the polyene binding site of RPE65 also features a negative electrostatic potential, and the responsible residues are conserved between the two proteins (Fig. 6C and SI Appendix, Fig. S3). The RPE65- and CCD-catalyzed reactions are both thought to proceed through cationic intermediates that would likely benefit from electrostatic stabilization (46). The electrostatic similarity between these evolutionarily divergent proteins indicates that anionic polyene binding sites are present in most metazoan CCDs and points to a unifying aspect of their catalytic functions. Besides these conserved residues, the polyene-binding site features two buried water molecules straddling the apocarotenoid C19 and C20 methyl groups that superimpose with equivalent water-binding sites found in RPE65 (41) (Fig. 6 A–C). These waters contribute to the cavity geometry by generating a surface that is complementary to the shape of the central region of the polyene. The C14′–O bond, which is equivalent to the scissile bond in apocarotenoid substrates of NdCCD, is positioned next to the metal center with the carbonyl oxygen forming a short hydrogen bond (∼2.4 Å bond length) with the Co-bound solvent (Fig. 6 A and B). The carbonyl group is additionally held in place by two orthogonal Phe side chains (residues 58 and 466) that interact with it from the top and side (Fig. 6B). Few active site changes were observed upon ligand complexation, the most notable being a ∼30° rotation of Glu140, which was required to avoid a clash with the polyene chain. Numerous van der Waals contacts made between the protein and its bound waters with the apocarotenoid (Fig. 6A) serve to enforce a strict mode of binding that leads to the high degree of regioselectivity we observed in NdCCD activity assays.
Fig. 6.
Mode of apocarotenoid binding to the NdCCD active site and its comparison to RPE65. (A) A two-dimensional representation of the NdCCD–β-apo-14′-carotenal complex showing numerous van der Waals contacts (radial lines surrounding atoms of the ligand) with both the protein as well as waters buried in the active site cavity that together enforce the observed mode of binding. The terminal carbonyl oxygen forms a strong hydrogen bond (dashed line) with a solvent molecule that is bound to the metal center. (B) Three-dimensional view of the NdCCD–product complex. Residues within 4.5 Å of the bound apocarotenoid (orange sticks) are shown as wheat-color sticks. The cobalt ion is shown as a salmon-colored sphere, and waters in proximity to the apocarotenoid are shown as red spheres. The electrostatic surface, calculated with APBS (47), in the vicinity of the bound apocarotenoid is shown, with red and blue representing negative and positive electrostatic potential, respectively. (C) Three-dimensional view of the RPE65 in complex with the 11-cis-retinoid mimetic emixustat (PDB accession code 4RSC). Residues within 4.5 Å of the emixustat molecule (orange sticks) are shown as wheat-colored sticks. The iron ion is shown as a brown sphere, and waters are shown as in A. A fragment of the iron-bound palmitate ligand is shown as wheat-colored sticks. Note the conserved negative electrostatic potential found in the central active site regions of both proteins as well as the presence of buried water molecules in the central region of both active sites. Also note the conformational difference between Phe252 in NdCCD and Tyr275 in RPE65 (equivalent positions in the CCD alignment as shown in SI Appendix, Fig. S3) that produces significantly different shapes near the substrate entrance. Other residues labeled in B and C are identical or highly conserved between NdCCD, RPE65, and other metazoan CCDs.
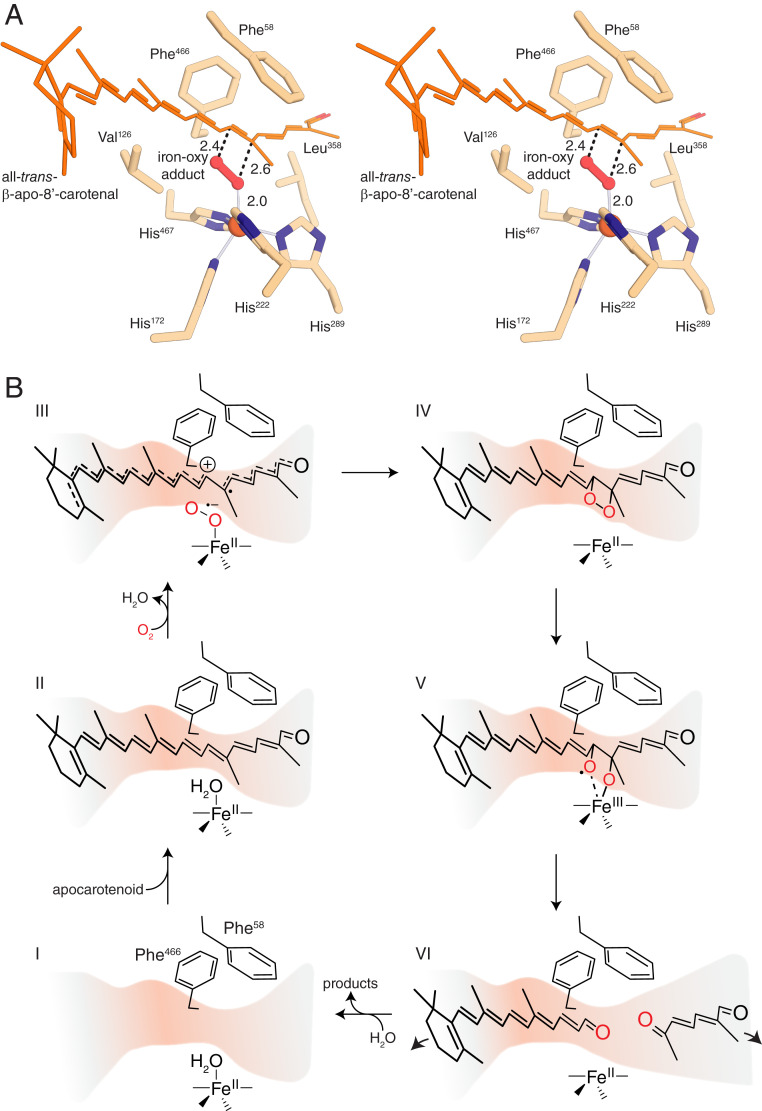
We further investigated the issue of regioselectivity in this enzyme by modeling β-apo-8′-carotenal in the active site using the bound 3-hydroxy-β-apo-14′-carotenal as a guide (Fig. 7A). The extra polyene structure was readily accommodated within the distal segment of the active site, although it must adopt a slight bend at the 12′–13′ single bond to avoid clashing with Leu358. The scissile bond is held in position by the phenyl rings of Phe58 and Phe466, the former residue being highly conserved in functionally characterized CCDs as part of the “FDG” motif (48) and the latter partially conserved among metazoan CCDs (SI Appendix, Fig. S3). These residues appear critical not only for constraining the position of the labile double bond but also likely help stabilize reaction intermediates via electrostatic interactions or π-electron donation. Together with the His222 Cε1 atom, these residues form a 3.8-Å constriction that would hinder or prevent passage of an ∼8-Å-wide β-ionone ring in the absence of conformational changes. This finding may partially explain the inability of NdCCD to cleave β,β-carotenoids. However, the β,β-carotenoid–cleaving enzyme BCO2 also has conserved Phe residues at these positions, indicating that other factors also determine whether bicyclic carotenoids are accepted or excluded. The scissile bond is oriented with respect to the metal center such that an end-on bound O2 molecule, modeled as suggested from computational studies (49, 50), would be well aligned with the C14′–C13′ bond for dioxetane formation (Fig. 7 A and B). To further illustrate how the active site selects this mode of substrate binding, we register-shifted the substrate by one double bond inward or outward to place the C15–C15′ or C12′–C11′ bonds in position to be cleaved (SI Appendix, Fig. S14). However, these modes of binding resulted in severe steric clashes in the central active site and substrate entry portal involving residues Phe120, Tyr220, His222, Val126, Phe252, and Glu140 as well as the buried water molecules. Our results indicate that the regioselectivity of NdCCD is enforced by multiple sites within the entry portal and central regions of the active site cavity. This conclusion is further supported by homology modeling of CbCCD, which, as demonstrated above, exhibits a regioselectivity identical to that of NdCCD. Inspection of the CbCCD model shows that residues forming the central active site are perfectly conserved between CbCCD and NdCCD (SI Appendix, Fig. S15A). By contrast, significant differences occur at the β-ionone ring binding site, in addition to smaller differences in the distal cavity. Most notably, Phe and Arg residues at positions 120 and 98 in NdCCD are replaced by smaller Pro and Ser residues in CbCCD, which reduces side chain bulk that conceivably could be involved in bottleneck formation and control of regioselectivity (SI Appendix, Fig. S15B). Despite this change in the substrate entry portal structure, C14′–C13′ regioselectivity is maintained in CbCCD. Taken together, the crystal structure and modeling experiments thoroughly explain the cleavage specificity of NdCCD for the C14′–C13′ double bond of β-apocarotenoid substrates.
Fig. 7.
Proposed mechanism for apocarotenoid cleavage by NdCCD. (A) A composite model of NdCCD in complex with β-apo-8′-carotenal and O2 shown in walleye stereo view. The model was generated by manually overlaying the substrate onto the experimentally determined structure of the bound product with a ∼30° rotation to the C13′–C12′ bond made to avoid a clash with Leu358. The geometry of the metal center and orientation of the scissile bond suggest that an end-on binding mode for O2 would be best suited for appropriate reactivity. (B) Proposed catalytic cycle for NdCCD. I, Resting state structure with the iron in a five-coordinate state. II, Apocarotenoid binds with little conformation change to the active site structure. Dissociation of the aquo ligand may be promoted by its close interaction with the apocarotenoid. III, The aquo ligand is replaced by O2, with simultaneous electron transfer from the apocarotenoid to the iron–oxy complex, an intermediate suggested by computational studies (49, 50). The negative surface potential (red color) may help to stabilize the cationic intermediate formed during this step. IV, End-on binding of O2 positions it appropriately for dioxetane formation with the target C14′–C13′ alkene. V, Decomposition of the dioxetane is facilitated by temporary oxidation of the FeII center to the +3 state as suggested by the computational studies referenced in step III. VI, The cleavage products dissociate, possibly through the two distinct openings, and water rebinds to the iron, restoring the active site to the resting state.
Discussion
The foregoing results expand our understanding of CCD enzymology from a number of perspectives. Specifically, we have demonstrated that 1) carotenoid-cleaving CCDs exist in the domain archaea; 2) the archaeal enzyme focused upon in this work, NdCCD, has a close phylogenetic, structural, and enzymatic relationship to metazoan as well as certain bacterial CCDs; 3) NdCCD catalyzes C14′–C13′ oxidative cleavage of apocarotenoids, a previously described enzymatic activity that was never molecularly linked to a specific gene but results in products of biological importance; and 4) this regioselective cleavage is governed by the geometry of both the entry portal and central region of the NdCCD active site tunnel—a finding that is likely broadly applicable to the biologically important metazoan CCD clade. Below we expand upon the implications of these results for understanding CCD structure and function.
NdCCD Illuminates the Determinants of Substrate Specificity, Catalytic Activity, and Regioselectivity within Metazoan CCDs.
We have shown that the overall architecture and physicochemical properties of the NdCCD and RPE65 active sites are remarkably similar given the vast evolutionary distance that separates them. Specifically, we have established that the central region of the active site, which encompasses most of the residues interacting with the apocarotenoid polyene chain and those surrounding the metal center, is highly conserved between these two proteins. This conservation indicates that apocarotenoid substrates of metazoan CCDs capable of cleaving such compounds are bound in a manner similar to that observed for NdCCD. A possible exception is the C9–C10 cleavage of apocarotenoids by some BCO2 orthologs (51). Preservation of the apocarotenoid orientation in this case would require stable binding of the substrate β-ionone ring within the narrow central active site close to the iron center, which seems unlikely given the strong sequence and structural conservation in this region. Instead, the β-ionone ring is more likely accommodated in the distal pocket with the end region of the polyene bound within the central active site. Not all BCO2 proteins are active toward apocarotenoids (52), so this potential difference is likely an exception rather than the rule. Moreover, our demonstration that the central active site region of NdCCD helps enforce regioselectivity suggests that this functional role is conserved throughout metazoan CCDs. Prior studies on SynACO posited that regioselectivity in this enzyme was controlled through a “bottleneck” mechanism whereby the depth of apocarotenoid insertion into the active site is controlled primarily by a constriction at the substrate entry portal that prevents passage of the bulky β-ionone ring of the substrate (13). However, subsequent mutagenesis of candidate residues forming the proposed bottleneck revealed that multiple sites within the substrate binding cleft must jointly contribute to holding the substrate in proper position, as the site of cleavage was maintained in all mutants examined (53). The work presented here supports the idea that regioselectivity is similarly determined by multiple substrate–enzyme interaction points in NdCCD and, by extension, metazoan CCDs. This idea is buttressed by our finding that C14′–C13′ regioselectivity is maintained in an NdCCD homolog from C. burnetii that differs substantially from NdCCD in the structure of its substrate entry portal. In light of the high level of structural conservation between NdCCD and metazoan CCDs, we expect that the orientation of the scissile alkene bond with respect to the metal center as determined from the NdCCD–product complex serves as an accurate model for animal CCDs.
Given the conservation of central active site residues in direct contact with the carotenoid/retinoid substrate at both the primary and tertiary structural levels, it is clear that sequence changes altering substrate specificity or catalytic activity must occur primarily at the active site entrance, the distal pocket, or residues that more subtly alter the conformations of residues lining the central active site. Indeed, mutagenesis (23, 51), phylogenetic (54, 55), and structural (41) studies are consistent with all of these mechanisms being operative in different metazoan CCDs. As an illustration of this point, residues thought to be responsible for stabilization of a catalytically critical retinyl cation intermediate in RPE65, namely Phe103 and Thr147 (41), are structurally conserved in NdCCD. Additionally, both proteins organize negative electrostatic surface potentials in the vicinity of the polyene that could help stabilize cationic reaction intermediates (Fig. 6 B and C). However, differences between these two enzymes at their substrate entry portal where the β-ionone ring is positioned result in markedly different cavity shapes that likely help determine whether or not the polyene is isomerically remodeled during catalysis. A difference of particular importance occurs at position 252 in NdCCD, where a Phe is substituted with a Tyr (position 275) in RPE65. These residues adopt two different conformations that contribute to the bent shape of the RPE65 binding pocket as opposed to the linear geometry observed for NdCCD (Fig. 6 B and C). This structural difference is reflected in the conformations of the bound ligands in these proteins. Specifically, emixustat adopts a bent conformation thought to mimic the structure of an 11-cis-retinoid (41), whereas 3-hydroxy-β-apo-14′-carotenal binds in an extended all-trans configuration. In addition to these unique features of the substrate entry portal, static or dynamic variations within the distal binding pocket and at the central active site constriction must also contribute to the ability of CCDs to accommodate their particular substrates. Although the NdCCD active site constriction in the vicinity of the metal center would appear prima facie to prevent passage of a β-ionone ring, it is important to note that the residues responsible for the constriction are conserved in some β,β-carotenoid cleaving enzymes, including BCO2. In such enzymes, these side chains likely exhibit dynamics not observed in our NdCCD crystal structure that allow passage of the β-ionone ring. Instead, variations within the distal binding pocket must determine the particularities and specificities of structures that can be accommodated in a given enzyme.
Recently, crystal structures of CCDs belonging to a subfamily whose members cleave resveratrol and related phenylpropanoids rather than carotenoids have been obtained in complex with substrates and products (35, 56). The orientation of the scissile bond in the NdCCD–apocarotenoid structure differs substantially from that of these stilbenoid-cleaving CCDs (35), where the labile double bond is shifted by ∼2 Å as a result of dramatically different active-site architecture (SI Appendix, Fig. S16). Knowledge of this positional shift will be important for future computational and experimental studies aimed at elucidating the nature of reactive carotenoid and oxygen intermediates generated during the catalytic cycles of metazoan CCDs.
Relationship of NdCCD to Bacterial and Eukaryotic CCDs Implicates Horizontal Gene Transfer as a Mechanism for CCD Acquisition.
An interesting observation from our analysis of NdCCD is that this enzyme bears the highest sequence similarity and phylogenetic relatedness to a variety of bacterial CCDs instead of archaea homologs. This apparent phyletic discrepancy suggests that NdCCD was acquired within the lineage leading to N. devanaterra by horizontal gene transfer instead of by vertical descent from the last archaeal common ancestor. This hypothesis is supported by several additional lines of evidence. First, CCDs are rarely found in members of the TACK superphylum so that a vertical mode of transmission would require assumption of several gene losses. Additionally, domain-wide phylogenetic analysis of the archaea has suggested that the ancestor of TACK archaea was anaerobic (19), which would appear to limit a survival benefit from expression of a dioxygenase enzyme. On the contrary, bacteria to archaea HGT is a well-established phenomenon (20), in particular the transfer of metabolic genes from bacteria to mesophilic archaea such as N. devanaterra (57). Moreover, several of the CCDs most similar to NdCCD are from bacteria that occupy the same ecological niche as N. devanaterra within agricultural soil (17). Collectively, these findings indicate that our HGT hypothesis is mechanistically easily envisioned.
Recent studies have indicated that Eukarya emerged from a common ancestor with Asgard archaea (21). It is thus interesting that CCDs are sparsely found within Asgard superphylum metagenomes reported to date and are absent in known Heimdallarchaeota, the closest known prokaryotic relatives of eukaryotes (58). Based on these observations and the occurrence of metazoan-like CCDs in the more distantly related TACK superphylum and bacteria, it is tempting to speculate that CCDs found in present-day eukaryotes did not derive from a single gene within the first common ancestor of eukaryotes (59). Instead, it seems more likely that CCDs were acquired by eukaryote lineages via nuclear gene transfer from internalized endosymbionts or potentially from external organisms. However, this hypothesis requires further detailed study to be confirmed or refuted and may become more or less likely as our knowledge of archaeal diversity and physiology becomes more complete.
Health and Environmental Implications of C14′–C13′ Apocarotenoid Cleavage Activity.
As noted earlier, the C14′–C13′ apocarotenoid cleavage enzymatic activity possessed by NdCCD was first described over 20 y ago during investigation of carotenoid cleavage in rat and rabbit intestinal lysates (33). Similarly, an enzyme that cleaves β-carotene specifically at the C14′–C13′ (C13–C14) bond was also reported from vertebrate intestinal mucosa lysates (60). However, subsequent cloning of vertebrate CCDs did not reveal any enzymes with C14′–C13′ cleavage activity, and this eccentric cleavage pathway has remained an orphan enzymatic activity. Our discovery of this activity being associated with a prokaryotic CCD gene that appears to be widely distributed among archaea and bacteria, including pathogens and common resident organisms of the intestinal lumen, provides a plausible explanation to reconcile these enzymatic and genomic data. Specifically, the intestinal mucosa used for the cleavage experiments are likely to have contained substantial amounts of microbiota, some of which could have expressed NdCCD-related proteins giving rise to the observed C14′–C13′ carotenoid cleavage activity. If correct, the production of β-apo-14′-apocarotenal by resident microbes including gut-associated archaea (e.g., halophiles [61]) and bacteria or pathogens such as C. burnetii could have implications for animal physiology and health, since this compound is known to modulate the activity of nuclear hormone receptors, including retinoic acid receptors (29) and peroxisome proliferator-activated receptors (62; reviewed in ref. 63). Because retinoic acid signaling is known to play a critical role in immune system function, including the induction of lymphocyte homing to the gut (64), β-apo-14′-carotenal production by resident or pathogenic prokaryotes could potentially have a profound impact on gastrointestinal physiology. Disruption of host carotenoid/retinoid metabolism was also proposed for a CCD from Mycobacterium tuberculosis that produces β-apo-14′-carotenal as one of several products (30). It will be of interest to further verify the phylogenetically inferred activity of these NdCCD/CbCCD-related prokaryotic CCDs and examine the impact of genetic or pharmacologic CCD inhibition on the physiology and pathogenicity of the organisms that harbor them.
Materials and Methods
Details regarding phylogenetic analysis, protein expression and purification, enzymatic assays, spectroscopic methods, and crystal structure determination are provided in the SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
This study was supported by funding from the National Institutes of Health (R01EY009339, P.D.K.; R01EY020551, J.v.L.), the Department of Veterans Affairs (I01BX004939, P.D.K.), and an unrestricted grant from Research to Prevent Blindness. This work is based in part upon research conducted at the APS Northeastern Collaborative Access Team beamline 24ID-E supported by grants GM103403, RR029205, and DE-AC02-06CH11357. Use of SSRL beamlines 7-3 and 12-2 was made possible by the US DOE Office of Science under Contract DE-AC02-76SF00515 to SLAC National Accelerator Laboratory and NIH Grant P41-GM-103393 (SSRL Structural Molecular Biology Program). Data for this study were measured at beamlines 17-ID-1 (AMX) and 17-ID-2 (FMX) of the National Synchrotron Light Source-II, which are supported by NIH Grant GM111244 and the DOE Office of Biological and Environmental Research KP1605010. The NSLS-II is supported in part by the DOE Office of Science, Office of Basic Energy Sciences Program under contract number DE-SC0012704 (KC0401040). We thank Dr. Adrian Wyss (DSM Nutrition) for kindly providing 3-hydroxy-β-apo-12′-carotenal for this study.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. P.S.B. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2004116117/-/DCSupplemental.
Data Availability.
Crystallographic coordinates and structure factor amplitudes have been deposited in the Protein Data Bank under accession codes 6VCF (Fe-NdCCD), 6VCG (Co-NdCCD), and 6VCH (Co-NdCCD-product complex). All other data are included in the manuscript or SI Appendix.
References
- 1.Britton G., Structure and properties of carotenoids in relation to function. FASEB J. 9, 1551–1558 (1995). [PubMed] [Google Scholar]
- 2.Spudich J. L., Yang C. S., Jung K. H., Spudich E. N., Retinylidene proteins: Structures and functions from archaea to humans. Annu. Rev. Cell Dev. Biol. 16, 365–392 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Zhong M., Kawaguchi R., Kassai M., Sun H., Retina, retinol, retinal and the natural history of vitamin A as a light sensor. Nutrients 4, 2069–2096 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Auldridge M. E., McCarty D. R., Klee H. J., Plant carotenoid cleavage oxygenases and their apocarotenoid products. Curr. Opin. Plant Biol. 9, 315–321 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Harrison E. H., Quadro L., Apocarotenoids: Emerging roles in mammals. Annu. Rev. Nutr. 38, 153–172 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giuliano G., Al-Babili S., von Lintig J., Carotenoid oxygenases: Cleave it or leave it. Trends Plant Sci. 8, 145–149 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Wyss A., Carotene oxygenases: A new family of double bond cleavage enzymes. J. Nutr. 134, 246S–250S (2004). [DOI] [PubMed] [Google Scholar]
- 8.Daruwalla A., Kiser P. D., Structural and mechanistic aspects of carotenoid cleavage dioxygenases (CCDs). Biochim. Biophys. Acta Mol. Cell Biol. Lipids, 158590 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwartz S. H., Tan B. C., Gage D. A., Zeevaart J. A., McCarty D. R., Specific oxidative cleavage of carotenoids by VP14 of maize. Science 276, 1872–1874 (1997). [DOI] [PubMed] [Google Scholar]
- 10.Wyss A. et al., Cloning and expression of beta,beta-carotene 15,15′-dioxygenase. Biochem. Biophys. Res. Commun. 271, 334–336 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Kiefer C. et al., Identification and characterization of a mammalian enzyme catalyzing the asymmetric oxidative cleavage of provitamin A. J. Biol. Chem. 276, 14110–14116 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Amengual J. et al., A mitochondrial enzyme degrades carotenoids and protects against oxidative stress. FASEB J. 25, 948–959 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kloer D. P., Ruch S., Al-Babili S., Beyer P., Schulz G. E., The structure of a retinal-forming carotenoid oxygenase. Science 308, 267–269 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Sui X. et al., Analysis of carotenoid isomerase activity in a prototypical carotenoid cleavage enzyme, apocarotenoid oxygenase (ACO). J. Biol. Chem. 289, 12286–12299 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Messing S. A. et al., Structural insights into maize viviparous14, a key enzyme in the biosynthesis of the phytohormone abscisic acid. Plant Cell 22, 2970–2980 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sui X., Kiser P. D., Lintig Jv., Palczewski K., Structural basis of carotenoid cleavage: From bacteria to mammals. Arch. Biochem. Biophys. 539, 203–213 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lehtovirta-Morley L. E., Stoecker K., Vilcinskas A., Prosser J. I., Nicol G. W., Cultivation of an obligate acidophilic ammonia oxidizer from a nitrifying acid soil. Proc. Natl. Acad. Sci. U.S.A. 108, 15892–15897 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dummer A. M. et al., Bacterioopsin-mediated regulation of bacterioruberin biosynthesis in Halobacterium salinarum. J. Bacteriol. 193, 5658–5667 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams T. A. et al., Integrative modeling of gene and genome evolution roots the archaeal tree of life. Proc. Natl. Acad. Sci. U.S.A. 114, E4602–E4611 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koonin E. V., Origin of eukaryotes from within archaea, archaeal eukaryome and bursts of gene gain: eukaryogenesis just made easier? Philos. Trans. R. Soc. B Biol. Sci. 370, 20140333 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zaremba-Niedzwiedzka K. et al., Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 541, 353–358 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Sui X. et al., Utilization of dioxygen by carotenoid cleavage oxygenases. J. Biol. Chem. 290, 30212–30223 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poliakov E., Soucy J., Gentleman S., Rogozin I. B., Redmond T. M., Phylogenetic analysis of the metazoan carotenoid oxygenase superfamily: A new ancestral gene assemblage of BCO-like (BCOL) proteins. Sci. Rep. 7, 13192 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Podell S., Gaasterland T., DarkHorse: A method for genome-wide prediction of horizontal gene transfer. Genome Biol. 8, R16 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ronquist F., Huelsenbeck J. P., MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (2003). [DOI] [PubMed] [Google Scholar]
- 26.von Lintig J., Dreher A., Kiefer C., Wernet M. F., Vogt K., Analysis of the blind Drosophila mutant ninaB identifies the gene encoding the key enzyme for vitamin A formation invivo. Proc. Natl. Acad. Sci. U.S.A. 98, 1130–1135 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.von Lintig J., Wyss A., Molecular analysis of vitamin A formation: Cloning and characterization of beta-carotene 15,15′-dioxygenases. Arch. Biochem. Biophys. 385, 47–52 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Garwin G. G., Saari J. C., High-performance liquid chromatography analysis of visual cycle retinoids. Methods Enzymol. 316, 313–324 (2000). [DOI] [PubMed] [Google Scholar]
- 29.Eroglu A. et al., Naturally occurring eccentric cleavage products of provitamin A β-carotene function as antagonists of retinoic acid receptors. J. Biol. Chem. 287, 15886–15895 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scherzinger D., Scheffer E., Bär C., Ernst H., Al-Babili S., The Mycobacterium tuberculosis ORF Rv0654 encodes a carotenoid oxygenase mediating central and excentric cleavage of conventional and aromatic carotenoids. FEBS J. 277, 4662–4673 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Heo J., Kim S. H., Lee P. C., New insight into the cleavage reaction of Nostoc sp. strain PCC 7120 carotenoid cleavage dioxygenase in natural and nonnatural carotenoids. Appl. Environ. Microbiol. 79, 3336–3345 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoffmann J., Bóna-Lovász J., Beuttler H., Altenbuchner J., In vivo and in vitro studies on the carotenoid cleavage oxygenases from Sphingopyxis alaskensis RB2256 and Plesiocystis pacifica SIR-1 revealed their substrate specificities and non-retinal-forming cleavage activities. FEBS J. 279, 3911–3924 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Dmitrovskii A. A., Gessler N. N., Gomboeva S. B., YuV Ershov, VYa Bykhovsky, Enzymatic oxidation of beta-apo-8′-carotenol to beta-apo-14′-carotenal by an enzyme different from beta-carotene-15,15′-dioxygenase. Biochemistry 62, 787–792 (1997). [PubMed] [Google Scholar]
- 34.Kiser P. D. et al., Structure of RPE65 isomerase in a lipidic matrix reveals roles for phospholipids and iron in catalysis. Proc. Natl. Acad. Sci. U.S.A. 109, E2747–E2756 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sui X. et al., Structure and spectroscopy of alkene-cleaving dioxygenases containing an atypically coordinated non-heme iron center. Biochemistry 56, 2836–2852 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krissinel E., Henrick K., Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Hamel C. P. et al., Molecular cloning and expression of RPE65, a novel retinal pigment epithelium-specific microsomal protein that is post-transcriptionally regulated in vitro. J. Biol. Chem. 268, 15751–15757 (1993). [PubMed] [Google Scholar]
- 38.Takahashi Y. et al., Identification of a novel palmitylation site essential for membrane association and isomerohydrolase activity of RPE65. J. Biol. Chem. 284, 3211–3218 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uppal S., Rogozin I. B., Redmond T. M., Poliakov E., Palmitoylation of metazoan carotenoid oxygenases. Molecules 25, E1942 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kiser P. D., Golczak M., Lodowski D. T., Chance M. R., Palczewski K., Crystal structure of native RPE65, the retinoid isomerase of the visual cycle. Proc. Natl. Acad. Sci. U.S.A. 106, 17325–17330 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kiser P. D. et al., Catalytic mechanism of a retinoid isomerase essential for vertebrate vision. Nat. Chem. Biol. 11, 409–415 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chander P., Gentleman S., Poliakov E., Redmond T. M., Aromatic residues in the substrate cleft of RPE65 protein govern retinol isomerization and modulate its progression. J. Biol. Chem. 287, 30552–30559 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Westre T. E. et al., A multiplet analysis of Fe K-edge 1s->3d pre-edge features of iron complexes. J. Am. Chem. Soc. 119, 6297–6314 (1997). [Google Scholar]
- 44.Sui X. et al., Preparation and characterization of metal-substituted carotenoid cleavage oxygenases. J. Biol. Inorg. Chem. 23, 887–901 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Read R. J., McCoy A. J., Using SAD data in Phaser. Acta Crystallogr. D Biol. Crystallogr. 67, 338–344 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fried S. D., Boxer S. G., Electric fields and enzyme catalysis. Annu. Rev. Biochem. 86, 387–415 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A., Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poliakov E. et al., Biochemical evidence for the tyrosine involvement in cationic intermediate stabilization in mouse beta-carotene 15, 15′-monooxygenase. BMC Biochem. 10, 31 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bai J., Hou Q. Q., Zhu W. Y., Liu Y. J., Mechanical insights into the oxidative cleavage of resveratrol catalyzed by dioxygenase NOV1 from Novosphingobium aromaticivorans: Confirmation of dioxygenase mechanism by QM/MM calculations. Catal. Sci. Technol. 9, 444–455 (2019). [Google Scholar]
- 50.Borowski T., Blomberg M. R., Siegbahn P. E., Reaction mechanism of apocarotenoid oxygenase (ACO): A DFT study. Chemistry 14, 2264–2276 (2008). [DOI] [PubMed] [Google Scholar]
- 51.Kelly M. E. et al., The biochemical basis of vitamin A production from the asymmetric carotenoid β-cryptoxanthin. ACS Chem. Biol. 13, 2121–2129 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dela Seña C. et al., Substrate specificity of purified recombinant chicken β-carotene 9′,10′-oxygenase (BCO2). J. Biol. Chem. 291, 14609–14619 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sui X., Zhang J., Golczak M., Palczewski K., Kiser P. D., Key residues for catalytic function and metal coordination in a carotenoid cleavage dioxygenase. J. Biol. Chem. 291, 19401–19412 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Poliakov E. et al., Origin and evolution of retinoid isomerization machinery in vertebrate visual cycle: Hint from jawless vertebrates. PLoS One 7, e49975 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Albalat R., Evolution of the genetic machinery of the visual cycle: A novelty of the vertebrate eye? Mol. Biol. Evol. 29, 1461–1469 (2012). [DOI] [PubMed] [Google Scholar]
- 56.McAndrew R. P. et al., Structure and mechanism of NOV1, a resveratrol-cleaving dioxygenase. Proc. Natl. Acad. Sci. U.S.A. 113, 14324–14329 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.López-García P., Zivanovic Y., Deschamps P., Moreira D., Bacterial gene import and mesophilic adaptation in archaea. Nat. Rev. Microbiol. 13, 447–456 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Williams T. A., Cox C. J., Foster P. G., Szöllősi G. J., Embley T. M., Phylogenomics provides robust support for a two-domains tree of life. Nat. Ecol. Evol. 4, 138–147 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eme L., Spang A., Lombard J., Stairs C. W., Ettema T. J. G., Archaea and the origin of eukaryotes. Nat. Rev. Microbiol. 15, 711–723 (2017). [DOI] [PubMed] [Google Scholar]
- 60.Tang G. W., Wang X. D., Russell R. M., Krinsky N. I., Characterization of beta-apo-13-carotenone and beta-apo-14′-carotenal as enzymatic products of the excentric cleavage of beta-carotene. Biochemistry 30, 9829–9834 (1991). [DOI] [PubMed] [Google Scholar]
- 61.Nkamga V. D., Henrissat B., Drancourt M., Archaea: Essential inhabitants of the human digestive microbiota. Hum. Micro J. 3, 1–8 (2017). [Google Scholar]
- 62.Ziouzenkova O. et al., Asymmetric cleavage of beta-carotene yields a transcriptional repressor of retinoid X receptor and peroxisome proliferator-activated receptor responses. Mol. Endocrinol. 21, 77–88 (2007). [DOI] [PubMed] [Google Scholar]
- 63.Eroglu A., Harrison E. H., Carotenoid metabolism in mammals, including man: Formation, occurrence, and function of apocarotenoids. J. Lipid Res. 54, 1719–1730 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Erkelens M. N., Mebius R. E., Retinoic acid and immune homeostasis: A balancing act. Trends Immunol. 38, 168–180 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Crystallographic coordinates and structure factor amplitudes have been deposited in the Protein Data Bank under accession codes 6VCF (Fe-NdCCD), 6VCG (Co-NdCCD), and 6VCH (Co-NdCCD-product complex). All other data are included in the manuscript or SI Appendix.