Significance
t(8;21) acute myelogenous leukemia (AML) often presents different degrees of aberrant myeloid differentiation. Although most patients respond to current chemotherapy regimens, a sizeable number of patients are prone to relapse. Thus, the cellular heterogeneity and disease outcome may need exploration. Here, by integrating high-throughput DNA/RNA sequencing with phenotypic analysis, heterogeneous leukemic cell populations were identified. CD34+CD117dim myeloblasts had the characteristics of granulocyte-monocyte progenitors and might be responsible for drug-resistance. CD34+CD117bri and AM cells were blocked later than CD34+CD117dim cells, and AM cells were sensitive to chemotherapy. t(8;21) AML patients with a high proportion of CD34+CD117dim cells had inferior outcomes. The identification of the CD34+CD117dim proportion as a potential prognostic factor may lead to new tools for future tailored therapeutic strategies.
Keywords: t(8;21)(q22;q22) AML, cellular heterogeneity, gene expression profiling, single-cell RNA sequencing, cellular/clonal evolution
Abstract
t(8;21)(q22;q22) acute myelogenous leukemia (AML) is morphologically characterized by a continuum of heterogeneous leukemia cells from myeloblasts to differentiated myeloid elements. Thus, t(8;21) AML is an excellent model for studying heterogeneous cell populations and cellular evolution during disease progression. Using integrative analyses of immunophenotype, RNA-sequencing (RNA-seq), and single-cell RNA-sequencing (scRNA-seq), we identified three distinct intrapatient leukemic cell populations that were arrested at different stages of myeloid differentiation: CD34+CD117dim blasts, CD34+CD117bri blasts, and abnormal myeloid cells with partial maturation (AM). CD117 is also known as c-KIT protein. CD34+CD117dim cells were blocked in the G0/G1 phase at disease onset, presenting with the regular morphology of myeloblasts showing features of granulocyte-monocyte progenitors (GMP), and were drug-resistant to chemotherapy. Genes associated with cell migration and adhesion (LGALS1, EMP3, and ANXA2) were highly expressed in the CD34+CD117dim population. CD34+CD117bri blasts were blocked a bit later than the CD34+CD117dim population in the hematopoietic differentiation stage and displayed high proliferation ability. AM cells, which bear abnormal myelocyte morphology, especially overexpressed granule genes AZU1, ELANE, and PRTN3 and were sensitive to chemotherapy. scRNA-seq at different time points identified CD34+CD117dim blasts as an important leukemic cluster that expanded at postrelapse refractory stage after several cycles of chemotherapy. Patients with t(8;21) AML with a higher proportion of CD34+CD117dim cells had significantly worse clinical outcomes than those with a lower CD34+CD117dim proportion. Univariate and multivariate analyses identified CD34+CD117dim proportion as an independent factor for poor disease outcome. Our study provides evidence for the multidimensional heterogeneity of t(8;21)AML and may offer new tools for future disease stratification.
t(8;21)(q22;q22) acute myelogenous leukemia (AML) is a malignant hematologic disease that accounts for 10 to 15% of cases of adult de novo AML, most of the cases belonging to AML-M2b subtype in FAB nomenclature (1–4). The translocation t(8;21)(q22;q22) results in a RUNX1-RUNX1T1 fusion gene (also known as AML1-ETO). RUNX1-RUNX1T1 AML, along with inv(16)(p13q22)/t(16, 16)(p13;q22) AML, are core-binding factor (CBF) AML subtypes. A well-known morphological feature of t(8;21) AML is the presence of heterogeneous leukemia cells from myeloblasts to differentiated myeloid elements that are arrested at different myeloid stages, including myelocytes and metamyelocytes. The leukemic blasts are usually nucleolated and contain varying amounts of cytoplasm, often with numerous azurophil granules. Among the abnormal myeloid cells with partial maturation (AM), a group of cells described by some studies as “abnormal myelocytes” is remarkable (4–7). These cells are often observed with such characteristics as unsynchronized development of the nucleus and the cytoplasm, some nucleoli in the relatively condensed nucleus, and abundant cytoplasm.
t(8;21) AML is considered a favorable subtype, with a 5-y survival rate of ∼50% (2, 8). However, a high relapse rate after achieving complete remission (CR) has been reported in China and some other countries (9, 10). This unexpectedly high recurrence rate might be explained in part by differences in treatment strategy, such as the unavailability of gemtuzmab ozogamicin (11, 12) for most patients in China, or ethnic genetic background differences. Patients with t(8;21) AML with mutation in the KIT gene (encoding c-KIT protein, also known as CD117) are considered intermediate risk and are at greater risk of relapse (13, 14). RUNX1-RUNX1T19a, an alternatively spliced isoform of RUNX1-RUNX1T1, was also associated with poor outcome in our previous study (6).
The advent of single-cell RNA sequencing (scRNA-seq) made it possible to gain deep insights into the transcriptional features of individual cell types. To date, scRNA-seq studies in various tumors, including glioblastoma, melanoma, and lung cancer (15–18), have made great progresses in assessing the heterogeneity of cancer cells at a single-cell level. On the other hand, some recent studies described the dynamic patterns of clonal evolution during disease progression in paired diagnosis/relapse t(8;21) AML samples (19). However, there is limited information about intrapatient cellular heterogeneity in t(8;21) AML. The relationship between cellular evolution and treatment response also needs to be further addressed to refine the current model of disease evolution in t(8;21) AML.
In the present study, we used several approaches, including immunophenotypic and morphological analyses, RNA-seq, scRNA-seq, and functional experiments, to investigate the biological and molecular characteristics of the distinct leukemic cell populations arrested at different myeloid stages. Furthermore, we carried out scRNA-seq in a patient with t(8;21) AML to gain insight into the dynamic changes in the heterogeneous cell populations at diagnosis, CR, relapse, and postrelapse refractory disease stages. The cellular evolutionary pattern was validated in an extended cohort with clinical flow cytometry data. In addition, whole-exome sequencing (WES) was performed to study the acquired/selected or lost gene mutations that occurred during cellular evolution at different time points. Finally, we compared the transcriptional profiles and clinical outcome of the t(8;21) AML patients with different myeloblast population ratios. The relationships between distinct myeloblast populations and poor prognosis might provide ideas for the risk stratification and treatment of t(8;21) AML.
Results
Clinical Immunophenotypic Characteristics and Morphological Features of Distinct Leukemic Cell Populations in Patients with t(8;21) AML.
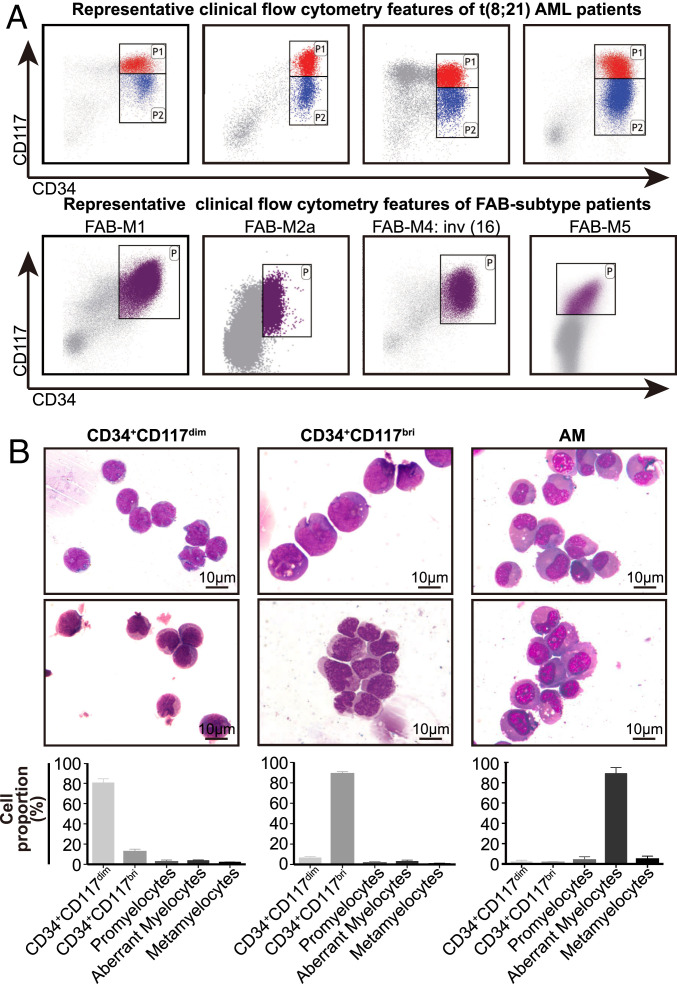
The study cohort included 101 patients with de novo t(8;21) AML, with a median age of 42 y. The patients’ clinical information is provided in Dataset S1. To study the immunophenotypic characteristics of the leukemia cell populations that are blocked at different hematopoietic stages, and to isolate the corresponding cell clusters through fluorescence-activated cell sorting (FACS) for further study, we retrospectively reviewed the clinical flow cytometry data of the 101 patients at Ruijin Hospital. Combined with careful analysis of our clinical flow cytometry data, we found that the combination of CD34 and CD117 could divide CD34+ myeloblasts into two groups: CD34+CD117dim and CD34+CD117bright(bri) populations (Fig. 1A). The forward-scatter (FSC) and side-scatter (SSC) values were lower in the CD34+CD117dim cells compared with the CD34+CD117bri cells, suggesting slight differences in morphology between these two cell groups (SI Appendix, Fig. S1A). Therefore, we sorted these two populations based on the marker combination using FACS.
Fig. 1.
Clinical immunophenotypic characteristics and morphological features of distinct leukemic cell populations in t(8;21) AML patients. (A) Representative clinical flow cytometry data showing the distribution of CD34+ myeloblasts by antigen CD34 and CD117 in four t(8;21) AML patients at diagnosis (Upper) and four AML patients with other FAB subtypes (except for M3) at diagnosis (Lower). (B, Upper) Representative Wright Giemsa-stained cytospin preparations of isolated CD34+CD117dim, CD34+CD117bri, and AM cell populations from the BM of t(8;21) AML patients. The AM population was isolated using the marker combination of CD34−CD117−HLA-DR−CD15+CD11b−. (B, Lower) Differential counts of the isolated cell populations (mean ± SD; n = 4), including CD34+CD117dim, CD34+CD117bri, and AM populations.
Differential counts of these two purified populations clearly demonstrated a significant, but not exclusive, enrichment of either population (Fig. 1B). Morphologically, CD34+CD117dim cells had the morphology of regular myeloblasts, whereas CD34+CD117bri cells had sunk nuclei that were relatively larger than those of CD34+CD117dim cells. Some CD34+CD117bri cells bore a promyelocyte-like morphology. These observations indicated that CD34+CD117dim cells could be blocked at an earlier stage of differentiation than CD34+CD117bri cells (Fig. 1B). It is worth noting that the characteristic immunophenotype could be specifically identified in t(8;21) AML and was seldom observed in other subtypes of AML, including inv(16) AML (Fig. 1A).
As the AM cells are a characteristic cell type of t(8;21) AML and could be observed in the bone marrow (BM) smear in most (98 of 101) patients with t(8;21) AML (Dataset S1 and SI Appendix, Fig. S1B), we attempted to analyze the immunophenotype of the AM cells using different combinations of myeloid differentiation antigens, including CD34, CD117, HLA-DR, CD15, CD11b, CD13, and CD33. The use of CD13 and CD33 could not distinguish AM cells well; however, the combination of CD34−CD117−HLA-DR−CD15+CD11b− could achieve a relatively satisfactory enrichment of AM cells, as shown by morphological analysis (Fig. 1B), although a few AM cells were left in other marker combinations (SI Appendix, Fig. S1 C and D).
Gene Expression Profiles and Biological Characteristics of CD34+CD117dim, CD34+CD117bri, and AM Populations.
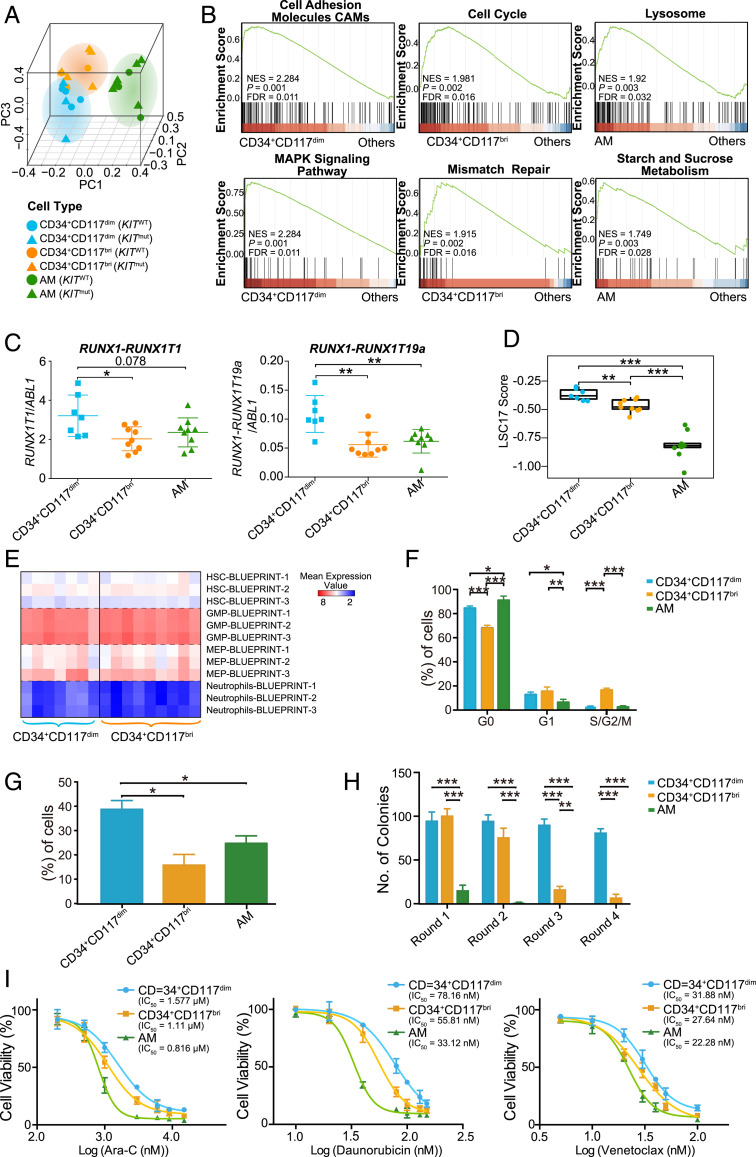
To explore the molecular basis underlying the distinct phenotypic characteristics of these three cell populations, we performed RNA-seq in sorted CD34+CD117dim (n = 7), CD34+CD117bri (n = 9), and AM (n = 9) cells from t(8;21) AML patients. Principal component analysis (PCA) showed that the cell populations clustered in consistence to their immunophenotypes rather than to KIT mutation status (Fig. 2A). Gene Ontology revealed that the up-regulated genes in CD34+CD117dim were enriched in cell migration, adhesion, and several important signaling pathways (e.g., MAPK cascade and GTPase activity). Cell cycle- and DNA repair-related gene sets were highly enriched in CD34+CD117bri populations, while biological processes involving the autophagy-lysosomal pathway and metabolism were activated in AM populations. Gene set enrichment analysis (GSEA) further confirmed these results (Fig. 2B and SI Appendix, Fig. S2A). Consistent with the pathway enrichment results, CD34+CD117dim cells overexpressed genes associated with cell migration and adhesion, such as LGALS1, EMP1, and ANXA2, whereas CD34+CD117bri overexpressed cell cycle regulators, including CCND1 and CCNG1 (SI Appendix, Fig. S2B). Both CD34+CD117dim and CD34+CD117bri cells expressed significantly higher levels of some drug-resistance genes than AM cells, with the highest levels seen in the CD34+CD117dim population (SI Appendix, Fig. S2C). We found that AM cells highly expressed the granule genes AZU1 and ELANE, in agreement with previous reports indicating that RUNX1-RUNX1T1 fusion could enhance the expression of these genes in granulocytic cells (20–22) (SI Appendix, Fig. S2B). The expression levels of RUNX1-RUNX1T1 and RUNX1-RUNX1T19a transcripts were also compared among these three populations using RNA-seq data. CD34+CD117dim cells showed significantly higher expression levels of RUNX1-RUNX1T1 and RUNX1-RUNX1T19a transcripts compared with CD34+CD117bri and AM cells (Fig. 2C and SI Appendix, Fig. S2D), as confirmed by droplet digital PCR results (SI Appendix, Fig. S2E).
Fig. 2.
Gene expression profiles and biological characteristics of CD34+CD117dim, CD34+CD117bri, and AM cell populations. (A) PCA plot of isolated CD34+CD117dim, CD34+CD117bri, and AM populations. Triangles and circles represent cells with KIT mutation (KITmut) and KIT wild-type (KITWT) status, respectively. (B) Representative GSEA plots showing the activated pathways in CD34+CD117dim, CD34+CD117bri, and AM cell populations. Normalized enrichment score (NES) values, nominal P values, and false discovery rate (FDR) values are given. (C) Relative expression levels of RUNX1-RUNX1T1 and RUNX1-RUNX1T19a transcripts in isolated CD34+CD117dim, CD34+CD117bri, and AM cell populations based on the RNA-seq data after normalization to the ABL1 read counts (internal reference). (D) LSC17 score of the CD34+CD117dim, CD34+CD117bri, and AM populations. Statistical significance was determined using a two-sided Wilcoxon test. (E) Comparison of the overexpressed gene sets of CD34+CD117dim and CD34+CD117bri myeloblasts with the Blueprint project gene sets. (F) Cell cycle analysis of the isolated CD34+CD117dim, CD34+CD117bri, and AM cell populations. Cell frequencies in G0, G1, and S/G2/M are shown (mean ± SD; n = 3 with duplicates). (G) Transwell migration of isolated CD34+CD117dim, CD34+CD117bri, and AM populations (mean ± SD; n = 3 with duplicates). (H) Colony-forming unit (CFU) assay of isolated CD34+CD117dim, CD34+CD117bri, and AM populations (mean ± SD; n = 3 with duplicates). (I) Cell viability of isolated CD34+CD117dim, CD34+CD117bri, and AM populations on exposure to cytarabine, daunorubicin, and venetoclax (n = 3 with triplicate). *P < 0.05; **P < 0.01; ***P < 0.001, two-sided Student’s t test.
We also compared the leukemia stem cell gene expression signatures (LSC17 score) (23) in the CD34+CD117dim, CD34+CD117bri, and AM populations. CD34+CD117dim showed a higher LSC17 score compared with the CD34+CD117bri and AM populations (Fig. 2D). To further investigate the hematopoietic properties of CD34+CD117dim and CD34+CD117bri myeloblasts, we compared the overexpressed gene sets of these two myeloblast populations with the Blueprint project gene sets (24). These two populations both had a noticeable positive correlation with granulocyte-monocyte progenitors (GMPs) and expressed GMP markers (Fig. 2E). This observation was supported by flow cytometry results (SI Appendix, Fig. S2F) and consistent with a previous study showing high GMP-like scores in AML patients harboring RUNX1-RUNX1T1 fusions (18).
We next compared the biological properties of the CD34+CD117dim, CD34+CD117bri, and AM cell populations. On cell cycle assays, CD34+CD117bri had a higher proportion of cells in S and G2/M phases, whereas CD34+CD117dim cells were arrested in G0/G1 phase (Fig. 2F). An in vitro transwell migration assay (25, 26) revealed greater migration toward the lower chamber in CD34+CD117dim cells compared with CD34+CD117bri and AM cells (Fig. 2G). In a colony-forming unit assay performed to evaluate the division/differentiation potential of these isolated populations, CD34+CD117dim cells formed significantly more colonies and maintained a longer clonogenic growth compared with CD34+CD117bri and AM cells, suggesting that CD34+CD117dim cells have a greater hematopoietic self-renewal/differentiation capacity (Fig. 2H). In addition, we treated the isolated cell populations with cytarabine, daunorubicin, venetoclax, and dasatinib and found that CD34+CD117dim cells were more resistant to cytarabine, daunorubicin, and venetoclax than CD34+CD117bri and AM cells (Fig. 2I). Overall, the distinctive biological features of each population suggest that patients with different composition of cell populations might have different disease parameters and show different treatment responses in t(8;21) AML.
Single-Cell Transcriptomic Analysis of t(8;21) AML at Diagnosis.
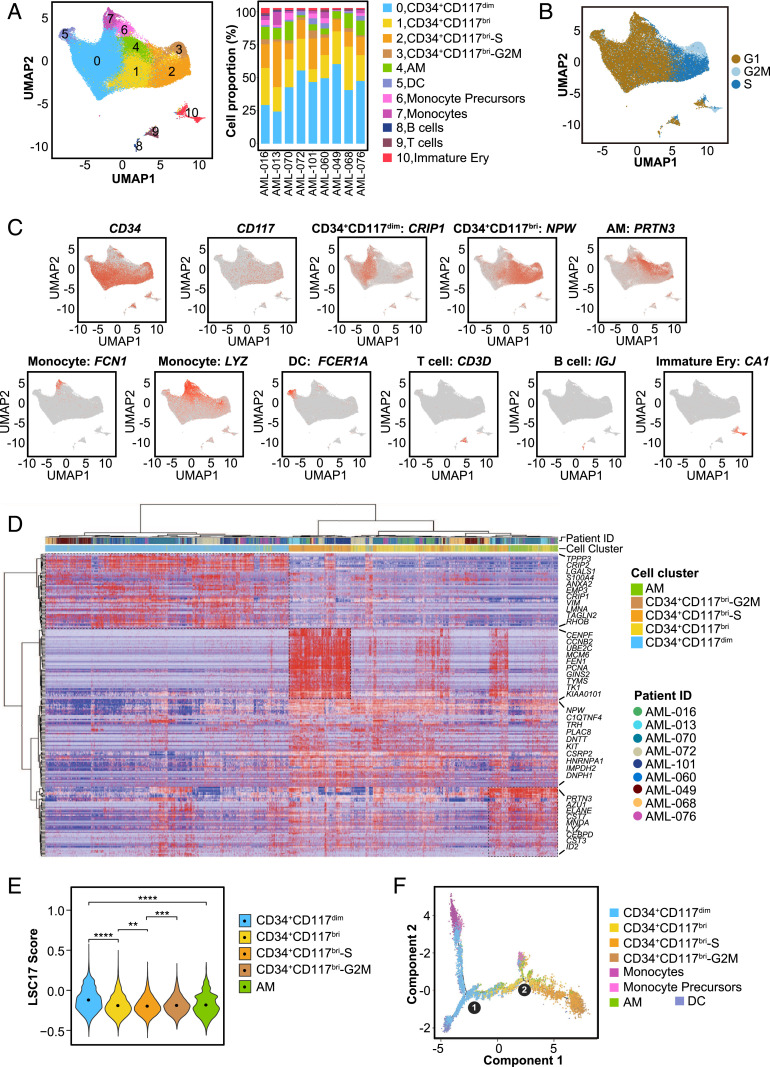
To obtain a comprehensive view of the cellular heterogeneity of the leukemic cells in t(8;21) AML and to gain deeper insight into the CD34+CD117dim, CD34+CD117bri, and AM populations, we performed scRNA-seq in nine t(8;21) AML patients at diagnosis using the droplet-based 10X Genomics platform. The morphological examination of BM smears revealed that the proportion of leukemic cells (blasts + AM) exceeded 60% for each patient (range, 63.5% to 93.5%; median, 87.0%; Dataset S1). After isolation of the BM mononuclear cells (BMMCs) by density-gradient centrifugation, the leukemic cells were enriched to >80% (SI Appendix, Fig. S3 A and B). After initial quality control, a total of 83,021 cells were analyzed from the nine BMMC samples, and the mean number of recovered cells from each sample was 9,225, with a median unique molecular identifier count of 6,675 and a median of 1,790 genes per cell (SI Appendix, Table S1). Cells were partitioned into several clusters using graph-based clustering and visualized by uniform manifold approximation and projection (UMAP) (Fig. 3A and SI Appendix, Fig. S3C). Annotation of cell cluster identities was determined using specific marker genes (SI Appendix, Table S2); expression patterns for the representative genes are displayed in Fig. 3C. The proportions of these identified cell clusters were consistent with the morphological analysis of the BMMCs cytospun from each sample (SI Appendix, Fig. S3D), and these BMMCs were composed mainly of malignant cells.
Fig. 3.
Single-cell transcriptomic analysis of t(8;21) AML at diagnosis. (A) UMAP analysis of BMMCs from nine primary t(8;21) AML patients after removing batch effects. Each dot represents a cell, and the colors represent different cell clusters. The right stacked column chart shows the percentage of each cell cluster in each patient. DC, dendritic cells; Immature Ery, immature erythroid cells. (B) UMAP plot of cell cycle state of cells in each cell cluster according to the expression level of cell cycle-specific gene sets. (C) UMAP plots displaying the expression patterns of CD34, CD117, and the representative markers for CD34+CD117dim, CD34+CD117bri, AM, monocytes, DCs, T cells, B cells, and immature erythroid cells. (D) Heatmap of the highly expressed genes in CD34+CD117dim, CD34+CD117bri, CD34+CD117bri-S, CD34+CD117bri-G2M, and AM cell populations from nine t(8;21) AML patients. The relative expression level of genes (rows) across cells (columns) is shown. (E) Violin plot showing the LSC17 score of CD34+CD117dim, CD34+CD117bri, CD34+CD117bri-S, CD34+CD117bri-G2M, and AM clusters. **P < 0.01; ***P < 0.001; ****P < 0.0001, two-sided Wilcoxon test. (F) Trajectory analysis using Monocle 2, with each dot representing an individual cell and colors representing different cell types. Circle 1 represents the beginning (the root), and circle 2 represents the branch point of the trajectory.
Four clusters with distinct characteristics were identified as myeloblasts based on the high expression levels of CD34 transcript (SI Appendix, Fig. S3E). KIT, which encodes CD117 protein, was identified as one of the top marker genes on the list of differentially expressed genes and was specifically overexpressed in CD34+CD117bri clusters (SI Appendix, Fig. S3E). According to the cell cycle score (16), CD34+CD117bri cells could be further classified as CD34+CD117bri, CD34+CD117bri-S, and CD34+CD117bri-G2M groups (Fig. 3B). CD34+CD117bri-S and CD34+CD117bri-G2M clusters highly expressed gene sets of S and G2/M phase, respectively, while CD34+CD117bri cells expressing neither of these markers were in G1 phase.
To compare the gene signatures of the CD34+CD117dim, CD34+CD117bri, and AM clusters identified on scRNA-seq with those identified on RNA-seq, we leveraged the highly expressed gene-sets of the isolated cell populations to construct reference gene-sets (SI Appendix, Table S3), and then compared the overexpressed genes identified in scRNA-seq with the reference gene sets. The highly expressed genes of each clusters identified on scRNA-seq could be well matched to their corresponding bulk gene sets in isolated populations, revealing that the clusters identified on scRNA-seq and the isolated populations had similar gene signatures (SI Appendix, Fig. S3F). Consistent with the RNA-seq data in isolated cell populations, CD34+CD117dim cells overexpressed cell migration and adhesion genes, such as LGALS1, ANXA2, EMP3, and CRIP1. KIT gene transcript was highly expressed in CD34+CD117bri cells. Genes participating in cell cycle and DNA replication (e.g., CCNB2, UBE2C, PCNA, TYMS, TK1) were overexpressed in CD34+CD117bri-S and CD34+CD117bri-G2/M populations. AM cells had high expression levels of genes encoding granule proteins, such as PRTN3, AZU1, and ELANE (Fig. 3D). Ingenuity pathway analysis further confirmed the characteristics of these populations (SI Appendix, Fig. S3G).
We also compared the LSC17 scores of CD34+CD117dim, CD34+CD117bri, and AM clusters. CD34+CD117dim showed higher LSC17 score compared with CD34+CD117bri and AM populations, which was consistent with the RNA-seq result (Fig. 3E). Furthermore, we compared the differentiation trajectories of these cell clusters using the Monocle 2 algorithm (27). CD34+CD117dim and CD34+CD117bri clusters were located at different nodes of the monocle tree (Fig. 3F). Compared with other cell clusters, CD34+CD117dim cells were located at the earliest stage of differentiation, while CD34+CD117bri cells were located after CD34+CD117dim cells, demonstrating the distinct and ordered states of these two myeloblast populations in terms of myeloid differentiation.
Single-Cell Transcriptomic Analysis and Clonal Evolution Analysis of t(8;21) AML along Disease Progression.
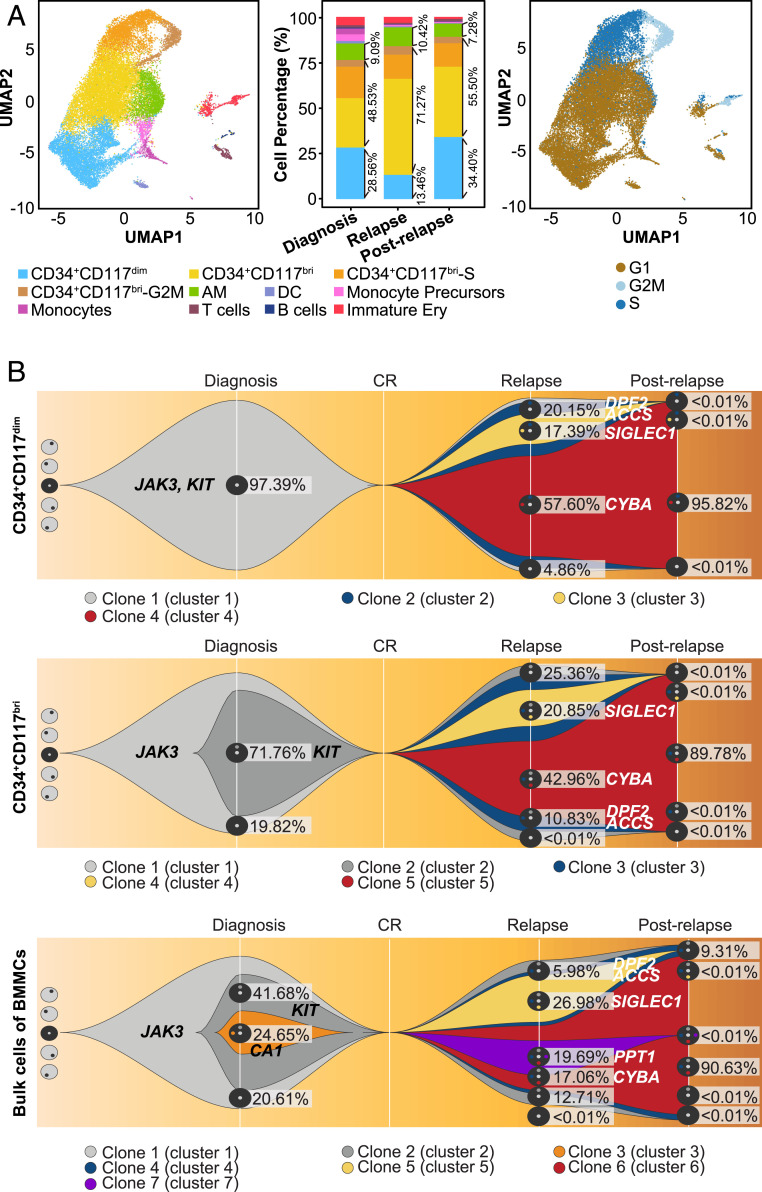
To investigate the dynamic changes in cell populations during leukemia progression, we performed scRNA-seq in BMMC samples obtained from a representative t(8;21) AML patient (patient AML-016) at four key time points: diagnosis, CR, relapse, and postrelapse refractory disease stages (Fig. 4A). The leukemic cells were >80% after BMMC enrichment (SI Appendix, Fig. S4A). The clinical features and detailed treatment histories at the different time points are summarized in SI Appendix, Table S4. The proportions of the cell populations varied during disease progression. At the CR stage, the proportion of the normal myeloblasts accounted for <5% of the BMMCs, while the proportions of lymphocytes and erythroid cells increased significantly, similar to the healthy controls (28) (SI Appendix, Fig. S4B).
Fig. 4.
Single-cell transcriptomic analysis and clonal evolution analysis of t(8;21) AML in disease progression. (A) UMAP analysis of BMMCs from three different time points (diagnosis, relapse, and postrelapse) of patient AML-016 during disease progression. Cells are colored by cell cluster (Left). The stacked column chart (Middle) shows the percentage of each cell cluster across different time points. The proportion of the leukemic blast cells for each immunophenotypic myeloblasts populations was >90%, consistent with the actual proportion of the leukemic blast cells in each population. (Right) UMAP plot showing the cell cycle states of cells in each cell cluster according to the expression level of cell cycle-specific gene sets. (B) Fish plots showing the clonal evolution landscapes for CD34+CD117dim cells (Upper), CD34+CD117bri cells (Middle), and bulk BMMCs (Lower) based on the median value of variant allele frequencies in patient AML-016.
Different leukemic cell populations still retained their characteristic gene expression patterns during the disease course (e.g., LGALS1, EMP3, ANXA2, and CRIP1 in CD34+CD117dim; TYMS, TK1, PCNA, UBE2C, TRH, and PLAC8 in CD34+CD117bri; ELANE, PRTN3, AZU1, and CST7 in AM), but there were slight differences in the gene expression profiles at different time points (SI Appendix, Fig. S4C). Genes participating in ribosome synthesis, such as RPL37, RPS21, and RPL36, were overexpressed at relapse, while genes involved in several biological pathways (e.g., MAPK signaling pathway and dasatinib drug resistance) were activated at postrelapse stage (SI Appendix, Fig. S4D). The number of differentially expressed genes among distinct populations decreased during relapse and postrelapse, indicating that the cells tended to be homogenized under the selection pressure after several cycles of chemotherapy. Indeed, the leukemic blasts showed higher stemness, as demonstrated by the higher LSC17 score at relapse and postrelapse stages (SI Appendix, Fig. S4E).
We further compared the expression levels of the CD34 gene transcript at different time points. The CD34+CD117dim population highly expressed CD34 at different time points during disease progression. A subset of AM cells started to express CD34 at relapse and postrelapse stages (SI Appendix, Fig. S4F), suggesting that these “atypical” AMs were arrested at an earlier phase of differentiation at these two time points, although their gene expression profile was still dominated by granule genes.
In patient AML-016, the proportion of CD34+CD117dim cells was significantly higher at postrelapse stage than at relapse stage (Fig. 4A). We validated this evolutionary pattern in an extended cohort of nine patients with clinical flow cytometry data at different time points. All patients exhibited similar patterns as patient AML-016 and had a significantly higher proportion of CD34+CD117dim cells at postrelapse stage compared with relapse (median increase in proportion of CD34+CD117dim cells, 21.73% (range, 10.69% to 52.64%) (SI Appendix, Fig. S4G).
To further examine the molecular mechanism of cellular evolution during disease progression, we also performed WES (∼219.67×) of CD34+CD117dim, CD34+CD117bri, and bulk cells obtained from patient AML-016 at different time points. In the CD34+CD117dim population, mutations including KIT and JAK3 constituted the cluster 1 mutations (clone 1 in Fig. 4B). Clone 1 evolved into clone 2 and clone 3 by gaining the relapse-specific mutations DPF2 and ACCS in clone 2 and DPF2, ACCS, and SIGLEC1 in clone 3. Clone 2 then evolved into clone 4 by gaining the CYBA mutation. At postrelapse stage with refractory disease, clone 4 had survived chemotherapy and expanded dramatically to become the dominant component of the CD34+CD117dim population (Fig. 4B). Compared with the CD34+CD117dim population, clone 2 including the KIT mutation on the basis of the cluster 1 mutations was identified at diagnosis, and this clone expanded at relapse and postrelapse refractory disease stages in the CD34+CD117bri population. The genetic background in the bulk cells was more complicated. Clone 3 (with CA1 mutation) and clone 7 (with PPT1 mutation) disappeared at relapse stage and postrelapse refractory disease stage, respectively. These minor subclones were not detected in CD34+CD117dim and CD34+CD117bri populations, indicating that these mutations in the minor subclones were gained in more mature myeloid cells at a late myeloid differentiation stage (Fig. 4B and Dataset S2). The clonal evolutionary patterns suggested that CD34+CD117dim population is located at an earlier stage of myeloid differentiation than CD34+CD117bri population, although both populations harbored same gene mutations. Different cell populations had their own clonal characteristics and they obtained clone components at different time during disease progression.
Gene Expression Profiles and Clinical Features of t(8;21) AML Patients with Different CD34+CD117dim Cell Proportions.
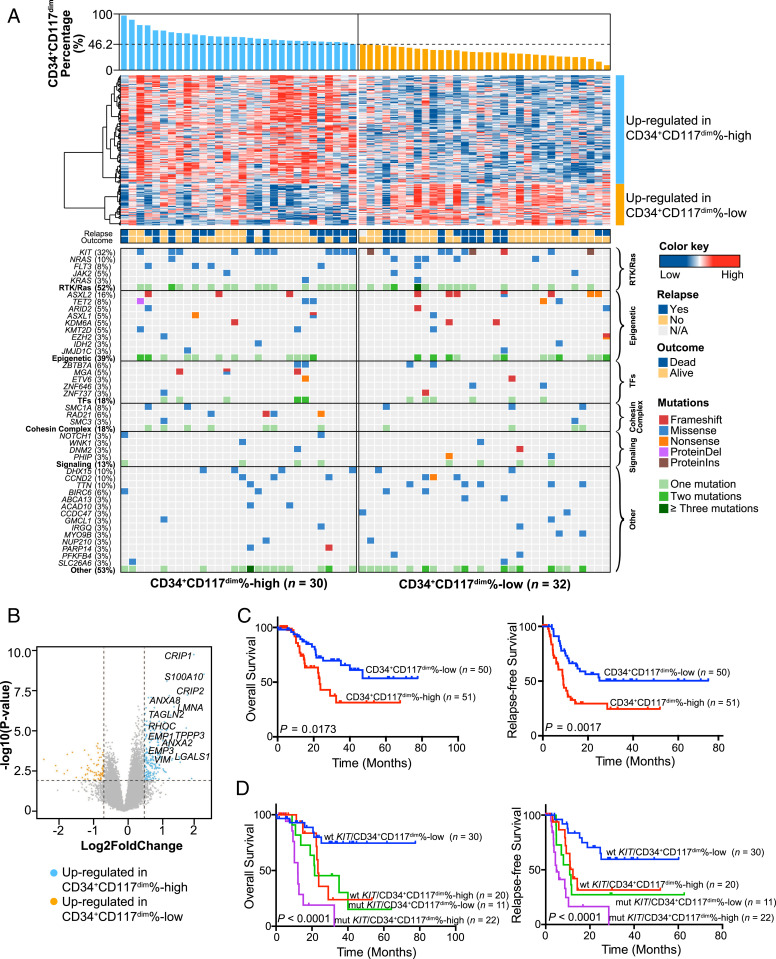
Since CD34+CD117dim cells have their distinct biological properties and play important roles at the postrelapse refractory disease stage through leukemia evolution, we wondered whether t(8;21) AML patients with different CD34+CD117dim ratios would have distinct gene expression profiles and clinical characteristics. We compared the transcriptomes of patients with different proportion of CD34+CD117dim cells to explore the molecular basis of the cellular heterogeneity in the t(8;21) AML cohort. Samples from 62 patients with good quality and sufficient quantity were subjected to RNA-seq and divided into two groups—CD34+CD117dim%-high and CD34+CD117dim%-low—according to the proportion of CD34+CD117dim in CD34+ myeloblasts. The cutoff point was determined by the median value of the CD34+CD117dim proportion identified in the clinical flow cytometry data from 101 patients in our cohort as well as the receiver operating characteristic (ROC) curve for the CD34+CD117dim proportion at 46.2% (Dataset S1, Fig. 5A, and SI Appendix, Fig. S5A). A total of 215 genes were highly expressed in the CD34+CD117dim%-high group, and 77 genes were highly expressed in the CD34+CD117dim%-low group (Dataset S3). Among the 215 up-regulated genes in the CD34+CD117dim%-high group, many were reported to be implicated in cell migration and adhesion (e.g., LGALS1, EMP3, ANXA2, CRIP1), and these highly expressed genes were also identified in our scRNA-seq and cell population-based RNA-seq data (Fig. 5B). GSEA revealed the significant activation of the gene sets associated with IL-6/JAK/STAT3, p53, WNT/β-catenin, NOTCH, and VEGF signaling pathways in the CD34+CD117dim%-high group compared with the CD34+CD117dim%-low group (SI Appendix, Fig. S5B).
Fig. 5.
Gene expression profiles and clinical features of t(8;21) AML patients with different CD34+CD117dim cell proportions. (A) Integrated molecular pattern of 62 t(8;21) AML patients subjected to RNA-seq. (Upper) The CD34+CD117dim proportion arranged from high to low according to the clinical flow cytometry data (Dataset S1), a heatmap showing the gene expression patterns in the CD34+CD117dim%-high and CD34+CD117dim%-low groups, and the outcomes and relapse status of the patients. (Lower) Mutations identified based on the RNA-seq data. Different colors represent distinct mutation types. (B) Volcano plot showing differentially expressed genes. The vertical axis shows the −log10 (P value), and the horizontal axis shows the log2 fold change. Up-regulated genes in the CD34+CD117dim%-high group are shown in blue (fold change >1.5; P < 0.01). (C) OS and RFS in 101 patients with t(8;21) AML according to the proportion of CD34+CD117dim cells among CD34+ myeloblasts. Survival curves were estimated with the Kaplan–Meier method and compared using a log-rank test. (D) Risk stratification combining KIT mutation status and CD34+CD117dim proportion. Survival curves were estimated with the Kaplan–Meier method and compared using a log-rank test.
Gene mutation detection was also performed based on the RNA-seq data in all 62 patients using the pipeline with a highly stringent procedure (29–31). In total, 40 genes were found to be mutated at least twice (Fig. 5A and Dataset S4). KIT, ASXL2, NRAS, DHX15, and CCND2 were identified as the most frequently mutated genes, consistent with previous reports (19).
Given the distinctive genetic characteristics of these two groups, we further analyzed the clinical features and outcomes of the 101 t(8;21) AML patients in our cohort. Patients in the CD34+CD117dim%-high group had a significantly higher white blood cell count than those in the CD34+CD117dim%-low group (SI Appendix, Table S5). Patients with a higher CD34+CD117dim proportion showed significantly worse overall survival (OS) and relapse-free survival (RFS) compared with the CD34+CD117dim%-low group (Fig. 5C). After excluding patients who underwent hematopoietic stem cell transplantation (HSCT), the CD34+CD117dim%-high group still had poorer OS and RFS than the CD34+CD117dim%-low group (SI Appendix, Fig. S5C). Interestingly, t(8;21) AML cases in the Gene Expression Omnibus (GSE37642) and AML cases in The Cancer Genome Atlas AML project with the highly expressed gene set of CD34+CD117dim%-high group also showed significantly worse OS than those with low expression of CD34+CD117dim gene set (SI Appendix, Fig. S5D).
Univariate and multivariate analyses for RFS and OS identified the proportion of CD34+CD117dim cells as an independent factor associated with RFS and OS, and this association was independent of KIT mutation and minimal residual disease status, which were also independent markers for poor outcome in multivariate analysis (Table 1). Furthermore, after combining KIT mutations with the CD34+CD117dim proportion and excluding the impact of HSCT, we could further stratify the t(8;21) AML patients into four groups with different prognoses (Fig. 5D): a low-risk subgroup (patients with WT KIT and a low CD34+CD117dim ratio), two intermediate-risk subgroups (patients with KIT mutations and a low CD34+CD117dim ratio and patients with WT KIT and a high CD34+CD117dim ratio), and a high-risk subgroup (patients with KIT mutations and a high CD34+CD117dim ratio). Although our sample size is small, our data suggest that compared with chemotherapy, HSCT could improve survival for patients with a higher proportion of CD34+CD117dim cells and/or with KIT mutations (SI Appendix, Fig. S5E). Overall, these results suggest that the identification of distinct leukemic cell populations might be an important approach for improving clinical outcomes in patients with t(8;21) AML.
Table 1.
Univariate and multivariate analyses for clinical characteristics of RFS and OS in 101 t(8;21) AML patients
| Factor | RFS | OS | ||||||
| Univariate | Multivariate | Univariate | Multivariate | |||||
| HR (95% CI) | P value | HR (95%CI) | P value | HR (95% CI) | P value | HR (95%CI) | P value | |
| Age, y | 1.010 (0.990–1.030) | 0.327 | 1.027 (1.003–1.052) | 0.025 | NS | |||
| Sex, male vs. female | 1.478 (0.833–2.623) | 0.181 | 0.955 (0.486–1.878) | 0.895 | ||||
| Marrow blasts, % | 1.011 (0.998–1.024) | 0.102 | 1.002 (0.986–1.017) | 0.852 | ||||
| AM, % | 0.991 (0.967–1.016) | 0.493 | 0.996 (0.968–1.026) | 0.811 | ||||
| White blood cells, × 109/L | 1.017 (1.002–1.032) | 0.022 | 1.021 (1.004–1.037) | 0.013 | 1.006 (0.985–1.027) | 0.599 | ||
| CD56, positive/negative | 1.507 (0.731–3.107) | 0.267 | 2.190 (0.847–5.667) | 0.106 | ||||
| CD19, positive/negative | 0.573 (0.328–0.999) | 0.050 | NS | 0.548 (0.279–1.076) | 0.080 | NS | ||
| t(8;21), alone vs. additional change | 1.052 (0.562–1.971) | 0.874 | 1.223 (0.578–2.586) | 0.598 | ||||
| Loss of sex chromosomes | 0.871 (0.478–1.587) | 0.651 | 0.738 (0.357–1.527) | 0.413 | ||||
| CD34+CD117dim proportion, high vs. low | 2.502 (1.410–4.439) | 0.002 | 2.369 (1.320–4.251) | 0.004 | 2.252 (1.135–4.468) | 0.020 | 2.649 (1.280–5.479) | 0.009 |
| KIT mutation | 2.628 (1.492–4.630) | 0.001 | 2.699 (1.495–4.875) | 0.001 | 3.466 (1.753–6.853) | <0.001 | 3.563 (1.754–7.238) | <0.001 |
| Minimal residual disease (achieving MMR vs. not achieving MMR)* | 0.389 (0.214–0.708) | 0.002 | 0.443 (0.242–0.813) | 0.009 | 0.326 (0.155–0.687) | 0.003 | 0.269 (0.120–0.603) | 0.001 |
| Transplantation status | 0.763 (0.369–1.576) | 0.465 | 0.252 (0.076–0.829) | 0.023 | 0.214 (0.062–0.739) | 0.015 | ||
Factors with P < 0.10 in the univariate analyses were subjected to multivariate analysis.
NS, not significant.
Major molecular remission (MMR) was based on the RUNX1-RUNX1T1 transcript level.
Discussion
In this study, we examined the heterogeneity of leukemic cells in t(8;21) AML patients via multidimensional analyses of immunophenotype, morphology, gene expression profiles, biological properties, scRNA-seq findings, gene mutations, and clinical features. In particular, we focused on the heterogeneous leukemic cell populations that were arrested at different hematopoietic stages in t(8;21) AML.
Intrapatient cellular heterogeneity is a hallmark of leukemia. In the continuously differentiated cell populations in t(8;21) AML, CD34+CD117dim cells seemed to locate at the earliest stage of myeloid differentiation. These cells exhibited high expression of GMP markers and a higher LSC17 score than CD34+CD117bri cells. The RUNX1-RUNX1T1 transcript level was higher in CD34+CD117dim cells than in the other cell types. Gene sets of several important signaling pathways, such as IL-6/JAK/STAT3, WNT/β-catenin, and VEGF, were significantly up-regulated in CD34+CD117dim%-high patients, and these patients had poorer clinical outcomes compared with CD34+CD117dim%-low patients. Further studies are warranted to elucidate the molecular regulatory circuits involved in CD34+CD117dim cells. Therapeutic drugs targeting the products of overexpressed genes in CD34+CD117dim cells also merit further investigation.
We tracked the complex cellular evolutionary process in an individual patient during leukemia progression. CD34+CD117dim cells expanded remarkably at the postrelapse refractory disease stage after several cycles of chemotherapy, implying that CD34+CD117dim cells evaded therapeutics and were selected in response to chemotherapeutic drugs, leading to the increased proportion and the patient’s refractory state. WES data suggest that clonal evolution could reshape the composition of the malignant clusters. It has been reported that chemotherapy does not induce mutations leading to the emergence of new clones, but rather selects for subclones that were already drug-resistant (32). Our results suggest the existence of both possibilities. The clonal evolutionary patterns reveal that different cell populations have their own clonal characteristics and obtain clone components at different time during disease progression. The combination of scRNA-seq and WES offers important insights into the cellular complexity and clonal architecture of AML.
We stratified t(8;21) AML patients based on their CD34+CD117dim ratio and demonstrated this ratio’s value in predicting the prognosis in t(8;21) AML. The higher LSC17 score in CD34+CD117dim cells indicated that this population might contain more LSCs, which are reportedly drug-resistant and responsible for disease relapse (33–36). Our clinical analysis was consistent with this and showed poorer outcomes in patients with a higher CD34+CD117dim proportion. KIT mutation is a well-established prognostic factor in t(8;21) AML. For patients without a KIT mutation and who did not receive HSCT, the CD34+CD117dim proportion could further stratify these patients into two distinct risk groups. The combination of KIT mutations with the CD34+CD117dim proportion could further refine the current prognostic stratification system for t(8;21) AML and could provide prognostic guidance for patients with this AML subtype, although a larger sample size is needed to confirm these observations.
To date, in addition to classical chemotherapy using anthracycline/cytarabine, different therapeutic strategies, such as targeted therapies, anti-CD33 monoclonal antibodies, and HSCT, have been reported in several clinical trials in CBF AML (9, 11, 12, 37, 38). Considering the cellular heterogeneity among leukemic cells during disease progression in t(8;21) AML, stratified therapeutic strategies tailored at the individual level of patients are needed to improve their survival. Zhu et al. (9) reported that in high-risk patients, HSCT was associated with reduced relapse and improved survival compared with chemotherapy. Our observations support those findings, albeit in a small cohort.
In summary, our data provide a comprehensive overview of the cellular heterogeneity in t(8;21) AML. Our evaluation of the molecular basis of diverse cell types and investigation of cellular/clonal evolution reveal that the heterogeneous cell populations have unique characteristics that may continue to evolve during disease progression. Our data also provide clinical prognostic markers to allow for better disease stratification as well as risk-adapted treatment guidance.
Materials and Methods
Patients.
A total of 101 patients with newly diagnosed t(8;21) AML seen at Shanghai Ruijin Hospital affiliated to Shanghai Jiao Tong University School of Medicine were enrolled in this study. The study was approved by the Ruijin Hospital Review Board, and informed consent was obtained from all patients in accordance with the Declaration of Helsinki. Details of the patients and treatments are provided in SI Appendix, Materials and Methods and Dataset S1.
Morphologic, Immunophenotypic, Biological, and Next-Generation Sequencing Analysis.
Details of these analysis are provided in SI Appendix, Materials and Methods.
Statistical Analysis.
The statistical analyses are described in detail in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by the Overseas Expertise Introduction Project for Discipline Innovation (111 Project; B17029), the Shanghai Collaborative Innovation Program on Regenerative Medicine and Stem Cell Research (2019CXJQ01), the Shanghai Clinical Medical Center for Hematological Disease (2017ZZ01002), the National Natural Science Foundation of China (81861148030, 81800147, and 81890994), the National Key Research and Development Plan of China (2018YFA0107800 and 2017YFA0506200), the Shanghai Sailing Program (18YF1413700), the Innovative Research Team of High-level Local Universities in Shanghai, the Samuel Waxman Cancer Research Foundation, and the Shanghai Guangci Translational Medical Research Development Foundation.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2003900117/-/DCSupplemental.
Data Availability.
Data for this study have been deposited at the National Omics Data Encyclopedia (NODE) (http://www.biosino.org/node/project/detail/OEP000629).
References
- 1.Döhner H. et al., Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 129, 424–447 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Papaemmanuil E. et al., Genomic classification and prognosis in acute myeloid leukemia. N. Engl. J. Med. 374, 2209–2221 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Faber Z. J. et al., The genomic landscape of core-binding factor acute myeloid leukemias. Nat. Genet. 48, 1551–1556 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yushu H., Shougeng B., Zhijian X., Yingchang M., Chao H. Z., Acute myeloid leukemia M2b. Haematologica 84, 193–194 (1999). [PubMed] [Google Scholar]
- 5.Xiao Z., Hao Y., Bian S., Acute myeloid leukemia M2B (subacute myeloid leukemia) in China. Leuk. Res. 21, 351–352 (1997). [DOI] [PubMed] [Google Scholar]
- 6.Jiao B. et al., AML1-ETO9a is correlated with C-KIT overexpression/mutations and indicates poor disease outcome in t(8;21) acute myeloid leukemia-M2. Leukemia 23, 1598–1604 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Bennett J. M. et al., Proposals for the classification of the acute leukaemias. French-American-British (FAB) co-operative group. Br. J. Haematol. 33, 451–458 (1976). [DOI] [PubMed] [Google Scholar]
- 8.Hospital M. A. et al., Core-binding factor acute myeloid leukemia in first relapse: A retrospective study from the French AML intergroup. Blood 124, 1312–1319 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Zhu H. H. et al., MRD-directed risk stratification treatment may improve outcomes of t(8;21) AML in the first complete remission: Results from the AML05 multicenter trial. Blood 121, 4056–4062 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Miyawaki S. et al., A randomized comparison of 4 courses of standard-dose multiagent chemotherapy versus 3 courses of high-dose cytarabine alone in postremission therapy for acute myeloid leukemia in adults: The JALSG AML201 study. Blood 117, 2366–2372 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Burnett A. K. et al., Identification of patients with acute myeloblastic leukemia who benefit from the addition of gemtuzumab ozogamicin: Results of the MRC AML15 trial. J. Clin. Oncol. 29, 369–377 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Hills R. K. et al., Addition of gemtuzumab ozogamicin to induction chemotherapy in adult patients with acute myeloid leukaemia: A meta-analysis of individual patient data from randomised controlled trials. Lancet Oncol. 15, 986–996 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cairoli R. et al., Prognostic impact of c-KIT mutations in core binding factor leukemias: An Italian retrospective study. Blood 107, 3463–3468 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Paschka P. et al.; Cancer and Leukemia Group B , Adverse prognostic significance of KIT mutations in adult acute myeloid leukemia with inv(16) and t(8;21): A cancer and leukemia group B study. J. Clin. Oncol. 24, 3904–3911 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Patel A. P. et al., Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344, 1396–1401 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tirosh I. et al., Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lambrechts D. et al., Phenotype molding of stromal cells in the lung tumor microenvironment. Nat. Med. 24, 1277–1289 (2018). [DOI] [PubMed] [Google Scholar]
- 18.van Galen P. et al., Single-cell RNA-seq reveals AML hierarchies relevant to disease progression and immunity. Cell 176, 1265–1281.e24 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christen F. et al., Genomic landscape and clonal evolution of acute myeloid leukemia with t(8;21): An international study on 331 patients. Blood 133, 1140–1151 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Shimada H., Ichikawa H., Ohki M., Potential involvement of the AML1-MTG8 fusion protein in the granulocytic maturation characteristic of the t(8;21) acute myelogenous leukemia revealed by microarray analysis. Leukemia 16, 874–885 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Dengler R. et al., Immunocytochemical and flow cytometric detection of proteinase 3 (myeloblastin) in normal and leukaemic myeloid cells. Br. J. Haematol. 89, 250–257 (1995). [DOI] [PubMed] [Google Scholar]
- 22.Lausen J., Liu S., Fliegauf M., Lübbert M., Werner M. H., ELA2 is regulated by hematopoietic transcription factors, but not repressed by AML1-ETO. Oncogene 25, 1349–1357 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Ng S. W. et al., A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature 540, 433–437 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Aran D., Hu Z., Butte A. J., xCell: Digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 18, 220 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zampini M. et al., Epigenetic heterogeneity affects the risk of relapse in children with t(8;21)RUNX1-RUNX1T1-rearranged AML. Leukemia 32, 1124–1134 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Stavropoulou V. et al., MLL-AF9 expression in hematopoietic stem cells drives a highly invasive AML expressing EMT-related genes linked to poor outcome. Cancer Cell 30, 43–58 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Trapnell C. et al., The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng G. X. et al., Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyer J. A. et al., Relapse-specific mutations in NT5C2 in childhood acute lymphoblastic leukemia. Nat. Genet. 45, 290–294 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lilljebjörn H. et al., Identification of ETV6-RUNX1-like and DUX4-rearranged subtypes in paediatric B-cell precursor acute lymphoblastic leukaemia. Nat. Commun. 7, 11790 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen B. et al., Identification of fusion genes and characterization of transcriptome features in T-cell acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. U.S.A. 115, 373–378 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shlush L. I. et al., Tracing the origins of relapse in acute myeloid leukaemia to stem cells. Nature 547, 104–108 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Thomas D., Majeti R., Biology and relevance of human acute myeloid leukemia stem cells. Blood 129, 1577–1585 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pollyea D. A., Jordan C. T., Therapeutic targeting of acute myeloid leukemia stem cells. Blood 129, 1627–1635 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Buccisano F. et al., Prognostic and therapeutic implications of minimal residual disease detection in acute myeloid leukemia. Blood 119, 332–341 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Lane S. W., Scadden D. T., Gilliland D. G., The leukemic stem cell niche: Current concepts and therapeutic opportunities. Blood 114, 1150–1157 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paschka P. et al., Adding dasatinib to intensive treatment in core-binding factor acute myeloid leukemia: Results of the AMLSG 11-08 trial. Leukemia 32, 1621–1630 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Boissel N. et al., Dasatinib in high-risk core binding factor acute myeloid leukemia in first complete remission: A French Acute Myeloid Leukemia Intergroup trial. Haematologica 100, 780–785 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data for this study have been deposited at the National Omics Data Encyclopedia (NODE) (http://www.biosino.org/node/project/detail/OEP000629).