Significance
Plant cells have a polysaccharide-based wall that maintains their structural and functional integrity and determines their shape. Reorganization of wall components is required to allow growth and differentiation. One matrix polysaccharide that is postulated to play an important role in this reorganization is xyloglucan (XyG). While the structure of XyG is well understood, its biosynthesis is not. Through genetic studies with Arabidopsis CSLC genes, we demonstrate that they are responsible for the synthesis of the XyG glucan backbone. A quintuple cslc mutant is able to grow and develop normally but lacks detectable XyG. These results raise important questions regarding cell wall structure and its reorganization during growth. The series of cslc mutants will be valuable tools for investigating these questions.
Keywords: Arabidopsis, cell wall, xyloglucan, glucan synthase, CSLC
Abstract
Xyloglucan (XyG) is an abundant component of the primary cell walls of most plants. While the structure of XyG has been well studied, much remains to be learned about its biosynthesis. Here we employed reverse genetics to investigate the role of Arabidopsis cellulose synthase like-C (CSLC) proteins in XyG biosynthesis. We found that single mutants containing a T-DNA in each of the five Arabidopsis CSLC genes had normal levels of XyG. However, higher-order cslc mutants had significantly reduced XyG levels, and a mutant with disruptions in all five CSLC genes had no detectable XyG. The higher-order mutants grew with mild tissue-specific phenotypes. Despite the apparent lack of XyG, the cslc quintuple mutant did not display significant alteration of gene expression at the whole-genome level, excluding transcriptional compensation. The quintuple mutant could be complemented by each of the five CSLC genes, supporting the conclusion that each of them encodes a XyG glucan synthase. Phylogenetic analyses indicated that the CSLC genes are widespread in the plant kingdom and evolved from an ancient family. These results establish the role of the CSLC genes in XyG biosynthesis, and the mutants described here provide valuable tools with which to study both the molecular details of XyG biosynthesis and the role of XyG in plant cell wall structure and function.
Xyloglucan (XyG) consists of a structurally complex collection of polysaccharides found in the cell walls of higher plants (1). The backbone of the polymers consists of β-1,4–linked glucosyl residues. Half to three fourths of the glucosyl residues are substituted on the 6-position by α-linked xylosyl residues, depending upon the plant species or the tissue, and many of these xylosyl residues are further substituted by other sugars, most often galactose and fucose (1). XyG biosynthesis occurs in the Golgi apparatus before transport of the polysaccharides to the cell surface where they are incorporated into the wall matrix (1, 2). During the past two decades, considerable progress has been made in identifying the genes and enzymes involved in the biosynthesis of this complex collection of polymers, although many important questions about the biosynthetic process remain to be answered.
Among the first genes and enzymes identified were those involved in adding the fucose and galactose side chains to XyG (3, 4). The genes and enzymes involved in adding the xylose side chains were identified about the same time (5, 6). Reverse genetics studies on the family of genes encoding XyG xylosyltransferases (XXT genes) revealed that double (xxt1 xxt2) and triple (xxt1 xxt2 xxt5) mutant plants were phenotypically relatively normal under standard growth conditions, but lacked detectable levels of XyG (7, 8). These observations provide strong evidence that the enzymes encoded by these genes are involved in XyG biosynthesis but also leave important questions unanswered. For example, how do plants adapt to a dramatic reduction in XyG levels or a complete loss of XyG? Do other cell wall polymers increase in abundance in the absence of XyG?
Cocuron et al. (9) provided evidence that CSLC genes encode the glucan synthases involved in making the backbone of XyG. They reported that a CSLC gene was highly expressed in developing nasturtium seeds that were depositing large quantities of XyG as a storage polymer. Both the nasturtium CSLC gene and an Arabidopsis CSLC4 gene produced short β-1,4–linked glucan chains when expressed in the yeast Pichia. Moreover, the coexpression of Arabidopsis CSLC4 and XXT1 in yeast cells resulted in the production of large quantities of long chains of β-1,4–linked glucan. However, to date, there is no in vivo evidence supporting a role of the CSLC family members as β-1,4–linked glucan synthases. Arabidopsis contains five putative CSLC genes, but their role in XyG biosynthesis and their importance in different tissues are still not clear. Based on the previous characterization of the XXT genes (7, 8), the loss of CSLC function may have similar phenotypes to xxt1 xxt2 mutant plants, but this is yet to be demonstrated. The diversification of the CSLC proteins suggests tissue or developmental specificity for the action of these proteins, but this remains to be shown.
Here we addressed these fundamental knowledge gaps using a reverse genetics strategy and isolated single mutants of each of the five Arabidopsis CSLC genes (CSLC4, CSLC5, CSLC6, CSLC8, and CSLC12). We then generated and characterized various double, triple, and quadruple mutant combinations plus a quintuple mutant. We established that the CSLC proteins share largely overlapping roles during development. Furthermore, we provide conclusive evidence that the concomitant disruption of the five CSLC genes reduces the levels of XyG below the limit of detection, while it does not ignite significant adaptive responses at a transcriptional level. We also show that the Arabidopsis CSLC proteins are responsible for XyG biosynthesis using complementation analyses. The similar phenotypes of the xxt1 xxt2 mutant and the cslc quintuple mutant raise important questions regarding the role of XyG in plant cell wall structure and function. The collection of mutants generated in this work will be crucial during future efforts to address these questions.
Results
CSLC Proteins Have Sequence Similarity, but CSLC Genes Show Different Patterns of Expression.
As sequence information became available for Arabidopsis and other plant genomes, families of genes closely related to the cellulose synthase (CESA) genes were identified and designated as cellulose synthase-like (CSL) genes (10). Early speculations suggested that these genes played a role in the synthesis of matrix polysaccharides of plant cell walls (10, 11), but providing experimental evidence to evaluate this attractive hypothesis has been difficult. Cocuron et al. (9) provided evidence that the Arabidopsis CSLC4 gene encodes a glucan synthase that is likely involved in making the backbone of XyG. Arabidopsis contains five members of the CSLC gene family: CSLC4, 5, 6, 8, and 12 (10, 12). A pairwise comparison of the amino acid sequences of the five CSLC proteins indicates significant sequence similarity and a similar number of putative transmembrane domains (SI Appendix, Fig. S1). Furthermore, all five CSLC proteins contain the canonical D, D, D, QxxRW motif found in GT2 glycan synthases (11, 13, 14) (SI Appendix, Fig. S1). These similarities support the hypothesis that all five of the proteins perform the same biochemical function as XyG glucan synthases.
To further investigate the function of the CSLC family members, we examined their patterns of expression. The first step was to mine the publicly available eFP Browser expression databases (SI Appendix, Fig. S2) (15). Examination of the browser data revealed that CSLC4 and CSLC8 are widely expressed in Arabidopsis, although CSLC8 is expressed at lower levels compared to CSLC4. The other three CSLC genes show more distinct patterns of expression, with CSLC5 highly expressed in developing seeds while CSLC6 and CSLC12 are highly expressed in pollen grains (SI Appendix, Fig. S2). In addition, CSLC4 and CSLC12 are both highly expressed in root hairs (SI Appendix, Fig. S2). Some of these expression patterns were confirmed by qRT-PCR analyses (SI Appendix, Fig. S3). Therefore, some CSLC genes have acquired tissue or organ-specific expression and possibly nonredundant functions at a tissue level.
Mutants Lacking the Five CSLC Genes Have Tissue-Specific Phenotypes.
We next aimed to test the hypothesis that CSLC genes could have branched into distinct functional groups in different organs or tissues by isolating homozygous T-DNA insertion lines for each of the five CSLC genes (Fig. 1). The single cslc lines were crossed to generate various double, triple, and quadruple mutant combinations as well as a quintuple mutant (Table 1). The presence of a T-DNA insert in each of the five CSLC genes was verified by PCR using genomic DNA for each of the alleles (Fig. 1) as well as transcriptomic analyses (RNA-seq) of the cslc456812 quintuple mutant (16) (hereafter called the cslc quintuple mutant; Fig. 2 and SI Appendix, Fig. S4). Using these approaches, we verified disruption of the individual genetic loci and the lack of full-length transcripts for each of the disrupted genes in the cslc quintuple mutant.
Fig. 1.
Isolation of cslc single mutants. (A) Gene models of all five cslc single mutants. Black rectangles, exons; black lines, introns; red triangles, T-DNA insertion sites; blue lines, noncoding sequences. Numbers in the red triangles indicate alleles. (B) The genotype of each cslc mutant was confirmed by PCR using gene- and T-DNA–specific primers. LP and RP primers were used to amplify a portion of the genomic sequence of each CSLC gene. LB is the specific primer on the T-DNA used to verify the presence of T-DNA. In each panel, DNA amplification is presented for the wild type (first lane) and three biological replicates for each single mutant.
Table 1.
Generation of higher-order cslc mutants
| Higher-order mutant line | Female parent | Male parent |
| Double mutants | ||
| cslc45-1 | cslc5-1 | cslc4-1 |
| cslc46-1 | cslc6-1 | cslc4-1 |
| cslc45-2 | cslc5-1 | cslc4-3 |
| cslc46-2 | cslc6-1 | cslc4-3 |
| cslc48 | cslc4-3 | cslc8 |
| cslc68 | cslc8 | cslc6-1 |
| cslc612 | cslc6-1 | cslc12-2 |
| Triple mutants | ||
| cslc456-1 | cslc45-1 | cslc46-1 |
| cslc456-2 | cslc45-2 | cslc46-2 |
| cslc458 | cslc48 | cslc45-2 |
| cslc468 | cslc46-2 | cslc48 |
| cslc4612 | cslc612 | cslc46-2 |
| cslc5612 | cslc56 | cslc612 |
| Quadruple mutants | ||
| cslc4568 | cslc458 | cslc468 |
| cslc45612 | cslc4612 | cslc5612 |
| Quintuple mutant | ||
| cslc456812 | cslc45612 | cslc4568 |
The table shows the parental lines used to produce the higher-order mutants used in this work. Two cslc456 triple mutants, cslc456-1 and cslc456-2, were generated with the cslc4-1 and cslc4-3 alleles, respectively. The cslc quadruple and quintuple mutants were generated with the cslc4-3 allele.
Fig. 2.
Confirmation of the lack of full-length transcripts in the cslc456812 quintuple mutant. Reads from RNA-seq analysis were mapped onto the Arabidopsis reference genome, and the read densities from the wild type and the cslc quintuple mutant are presented as a stacked density graph (wild type, dark blue; cslc quintuple mutant, light blue). Both 5′ and 3′ ends of genes are indicated in the graph. Arrows indicate T-DNA insertion sites. SI Appendix, Fig. S4, provides additional details of the T-DNA insertion sites.
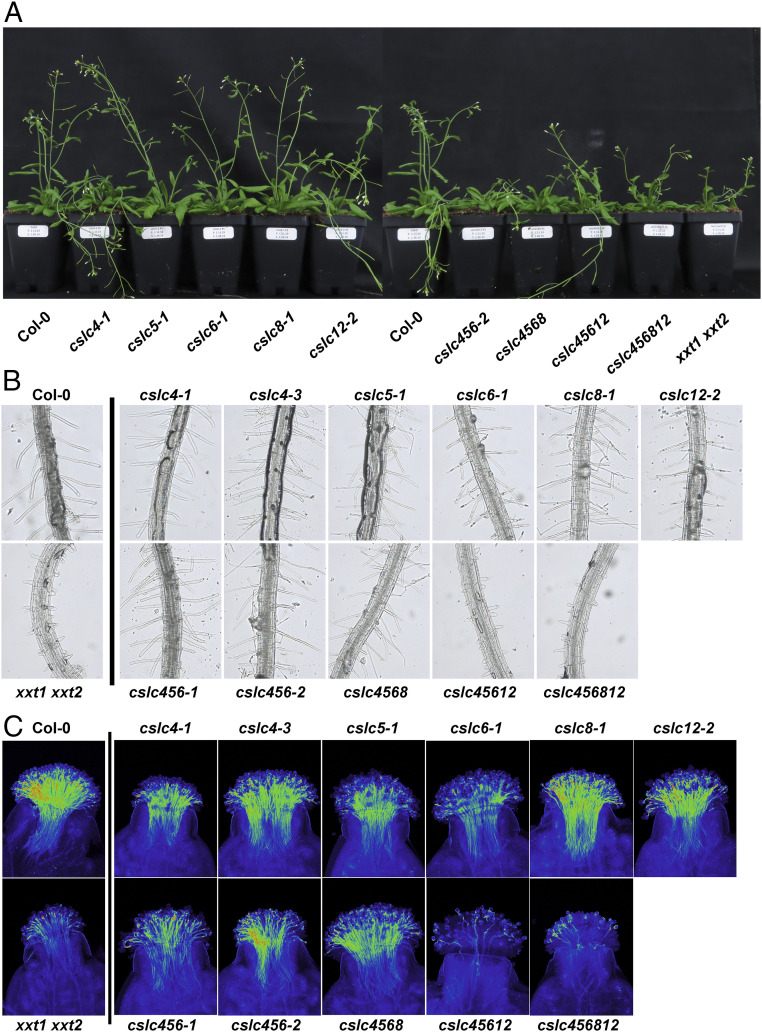
Next, the phenotypes of the various single and higher-order mutants were analyzed, focusing on the tissues or organs such as stem, root hairs, or pollen that exhibited prevalent expression of specific CSLCs (SI Appendix, Figs. S2 and S3). No significant differences in the rosette sizes or inflorescence stem heights of the single cslc mutants were found (SI Appendix, Fig. S5). However, higher-order cslc mutants exhibited smaller rosettes and shorter inflorescence stems compared to wild type (SI Appendix, Fig. S5). Mutant lines with a cslc4 allele showed bending of the inflorescence stems (Fig. 3A), possibly indicating weaker stems in these mutants. Also, the higher-order mutants with the cslc12 allele (i.e., cslc45612 and cslc456812) exhibited shorter root hairs and a reduced number of pollen tubes following 1-h pollinations compared to wild type and the other mutants (Fig. 3 B and C and SI Appendix, Fig. S6). These results, along with expression data (SI Appendix, Fig. S2), indicate that CSLC12, likely in combination with CSLC4, is required for proper growth of root hairs and interaction of pollen with the tissues of the pistil.
Fig. 3.
Organ- and tissue-specific roles of CSLCs. (A) Images of 6-wk-old plants of the indicated genotypes. (B) Root hairs of the primary roots of 1-wk-old seedlings of the indicated genotypes. (C) Wild-type pistils, highlighting the pollen tubes that formed 1 h after pollinating the stigmas with pollen from the indicated genotypes.
Taken together, these observations led us to conclude that the five CSLC proteins have some overlapping and tissue-specific roles during Arabidopsis development. The smaller rosettes and shorter inflorescence stems of the cslc higher-order mutants as well as the root hair defects of the cslc45612 quadruple and cslc456812 quintuple mutants were reminiscent of the phenotypes of the xxt1 xxt2 mutant, which lacked detectable levels of XyG (8) (Fig. 3 and SI Appendix, Fig. S5). Thus, we next sought to determine XyG levels in the various mutant plants.
CSLC Mutations Cause Reduced XyG Levels in the Cell Wall.
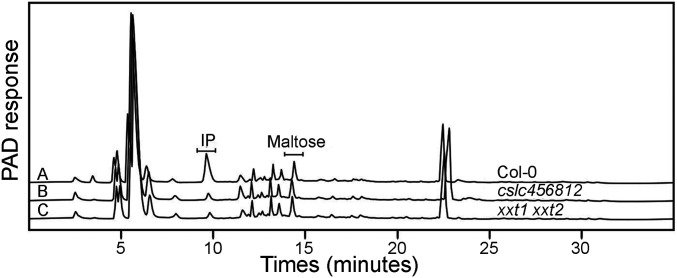
If the CSLC proteins are involved in XyG biosynthesis, as hypothesized previously (9), then mutations of the CSLC genes should cause a decrease in XyG levels in the cell wall. In addition, because earlier work on the xxt1 xxt2 mutant demonstrated that this mutant lacked detectable XyG, we hypothesized that the cslc quintuple mutant would be XyG-deficient, considering the similarity of phenotypes between the cslc quintuple mutants and the xxt1 xxt2 mutants (Fig. 3 and SI Appendix, Figs. S5 and S6). To test this hypothesis, we pursued multiple strategies to determine the XyG levels in the walls of the cslc mutants. The first strategy was to assay the levels of isoprimeverose (IP; xylose-α-1,6-glucose) as a proxy for the levels of XyG, as done earlier (8). To measure IP levels, alcohol-insoluble residue (AIR) from the hypocotyl of cslc mutants was digested using partially purified Driselase. This enzyme mixture degrades wall polysaccharides mainly to monosaccharides, except that it lacks α-xylosidase activity (17), thus releasing IP from XyG. The digested cell wall fractions were analyzed using high-performance anion exchange chromatography (HPAEC) with pulsed amperometric detection (PAD). IP was detected in all of the samples tested, with even the xxt1 xxt2 mutant and the cslc quintuple mutant showing a small peak eluting at the retention time of IP (Fig. 4, Table 2, and SI Appendix, Fig. S7). Because low levels of IP were not observed in a previous study of the xxt1 xxt2 mutant (8), we performed more detailed analyses to determine the nature of the compound in the mutants eluting at the IP retention time (∼10 min). This material was collected and subjected to linkage analysis by GC-MS to determine the sugar linkages present. Authentic IP and the material present in Driselase digests of wild-type cell walls contained terminal xylose (t-Xyl) and 6-linked glucose (6-Glc), as would be expected if IP were present (SI Appendix, Fig. S8B). When linkage analysis was performed on the small peak eluting near 10 min from cell wall Driselase digests of the xxt1 xxt2 mutant or the cslc quintuple mutant (Fig. 4), the GC traces did not have a peak near t-Xyl or 6-Glc (SI Appendix, Fig. S8A). Moreover, the spectra recorded from the appropriate retention time from the xxt1 xxt2 mutant and the cslc quintuple mutant did not have spectra consistent with t-Xyl or 6-Glc (SI Appendix, Fig. S8 C and D). Thus, although the identity of the material eluting near the retention time of IP is not known, we concluded that there was no detectable IP in the xxt1 xxt2 mutant, consistent with earlier reports (8). We also concluded that the cslc quintuple mutant lacked detectable XyG, as one would predict if the CSLC genes are responsible for making the backbone of XyG.
Fig. 4.
Xyloglucan content in cell wall preparations using isoprimeverose (IP) as a proxy. Determination of isoprimeverose (IP) content of the AIR prepared from etiolated hypocotyls of Arabidopsis Col-0 (graph A), clsc456812 (graph B), and xxt1 xxt2 (graph C). AIR was digested with Driselase, and the digested material was analyzed by high-performance anion exchange chromatography (HPAEC). The IP content in the sample was quantified using maltose as an internal standard.
Table 2.
Quantification of isoprimeverose (IP) after Driselase digestion of AIR of various Arabidopsis mutants
| Genotype | IP, µg/mg AIR | Relative abundance of IP peak, % |
| Col-0 | 8.23 ± 1.33 | 100 |
| xxt1 xxt2 | n.d. | n.d. |
| clsc456812 | n.d. | n.d. |
| cslc45612 | 1.00 ± 0.61* | 12.2 |
| cslc4568 | 1.02 ± 0.53* | 12.4 |
| cslc456-2 | 1.87 ± 0.55* | 22.7 |
| cslc456-1 | 3.40 ± 0.96* | 41.3 |
| clsc12-2 | 9.60 ± 2.38 | 116.6 |
| cslc8-1 | 10.11 ± 2.11 | 122.8 |
| cslc6-1 | 8.18 ± 0.87 | 99.4 |
| clsc5-1 | 7.54 ± 1.25 | 91.5 |
| cslc4-3 | 8.09 ± 2.47 | 98.3 |
The IP content was determined using HPAEC (Fig. 4). IP quantification (µg/mg AIR) was determined using maltose as an internal standard. The IP values were obtained from three biological replicates (n = 3, ±SD). Asterisks indicate statistically significant differences from Col-0, (Student’s t test, P < 0.05). The relative abundance of the IP peak in the samples was calculated with respect to Col-0. n.d., not detected.
A second strategy was to employ simplified glycan arrays (18, 19) with antibodies specific for XyG (LM15) (20) to monitor the levels of XyG in extracts from cell wall preparations derived from the hypocotyls of the mutants. Antibodies to pectic polysaccharides (LM6) (21) were used as a control. The results revealed that the cslc456-2 triple mutant had dramatically reduced levels of XyG, while both the cslc quintuple mutant and the xxt1 xxt2 mutant had undetectable levels of XyG (SI Appendix, Fig. S9A). The levels of pectic polysaccharides were unchanged in all of the plants tested (SI Appendix, Fig. S9B).
Still a third strategy was to evaluate XyG abundance and to investigate the substitution patterns of the residual XyG using oligosaccharide mass profiling (OLIMP) by MALDI-TOF mass spectrometry (22). Ion signals representing known XyG oligosaccharides in the wild-type walls were observed (SI Appendix, Fig. S10). In the triple mutant cslc456-1, all of these ions were also observed, albeit in lower quantities compared to the wild type. However, the cslc quintuple mutant walls lacked any of those ions representing XyG oligosaccharides, confirming a lack of XyG in the quintuple mutant or a lack of XyG that was accessible to the enzyme used (XyG specific endoglucanase). It is interesting to note that, despite reduced levels of XyG in the cslc456-1 triple mutant, the side chain substitution patterns remain unchanged in the hypocotyl. The important conclusion is that all three strategies provide compelling evidence that the CSLC genes are involved in XyG biosynthesis.
All CSLC Protein Family Members Have XyG Glucan Synthase Activity.
The information above provides compelling evidence that the CSLC genes are involved in XyG biosynthesis, most probably in synthesizing the glucan backbone of XyG. A related question is whether all of the proteins encoded by these genes are redundant in biochemical function with CSLC4, which was previously shown to have glucan synthase activity (9). To address this question, we generated complementation lines of the cslc quintuple mutant using each CSLC gene. For complementation with the CSLC4, CSLC5, or CSLC6 gene, expression was driven by the CSLC4 promoter. For complementation with the CSLC8 or CSLC12 gene, expression was driven by the 35S constitutive promoter (SI Appendix, Fig. S11). The XyG levels in complemented plants were analyzed by performing Driselase digestions on cell wall material from leaves and then determining IP levels using LC/Qtof MS. The IP levels, and therefore the XyG levels, increased significantly for each of the complemented lines (Table 3). In addition, each of the complementation lines showed longer root hairs than the cslc quintuple mutant. Although the extent of complementation was variable, there was good correlation between the two measures of complementation in that lines with more XyG had longer root hairs (Table 3). These results led us to conclude that each of the CSLC proteins has XyG glucan synthase activity.
Table 3.
Complementation of the cslc456812 mutant using each CSLC member
| Genotype (no. of replicates for IP analysis) | IP ± SEM, µg/mg of AIR | Relative abundance of IP | Root hair length ± SEM, µm |
| Col-0 (3) | 12.88 ± 2.16 | 100.00 | 426.7 ± 24.04a |
| cslc456812 (3) | 0.064 ± 0.01 | 0.49 | 78.16 ± 2.86b |
| CSLC4pro::CSLC4com-1 (3) | 0.42 ± 0.14 | 3.26 | 157.1 ± 8.65c |
| CSLC4pro::CSLC5com-1 (3) | 14.58 ± 7.39 | 113.22 | 295.7 ± 18.20d |
| CSLC4pro::CSLC6com-1 (3) | 17.09 ± 1.64 | 132.75 | 448.4 ± 20.74a |
| CSLC4pro::CSLC6com-2 (3) | 18.88 ± 1.28 | 146.64 | 466.3 ± 18.89a |
| 35Spro::CSLC8com-1 (1) | 1.54 | 11.95 | 206.2 ± 16.44c |
| 35Spro::CSLC8com-2 (1) | 2.32 | 18.01 | 179 ± 22.39c |
| 35Spro::CSLC8com-3 (1) | 4.46 | 34.63 | 187.3 ± 14.56c |
| 35Spro::CSLC12com-1 (1) | 4.73 | 36.72 | 147.9 ± 12.72bc |
| 35Spro::CSLC12com-2 (1) | 6.79 | 52.74 | 145.2 ± 13.78bc |
| 35Spro::CSLC12com-3 (1) | 12.04 | 93.46 | 323.9 ± 24.74d |
The levels of IP after Driselase digestion and lengths of root hairs (from 2-wk-old plants) from each complementation line. One-way ANOVA (Tukey’s test, α = 0.05, P < 0.05) was used to determine statistical differences in root hair lengths among genotypes. Each statistical group was labeled with lowercase letters (a, b, c, and d), which indicates that the length of root hair in the same group is not statistically different. Promoters (CSLC4pro or 35Spro) used to express each CSLC gene are specified.
Loss of XyG Does Not Impact Expression of Other Wall-Related Genes.
Because XyG is an abundant component in primary cell walls, it is possible that the loss of XyG would activate compensation mechanisms. To investigate the possibility that other wall components are elevated when XyG is missing, we performed sugar analyses of cell wall preparations from wild-type and cslc mutant plants. The xxt1 xxt2 mutant was analyzed as a control (Fig. 4, Table 2, and SI Appendix, Fig. S7) because earlier studies demonstrated no compensation in the walls of these plants lacking XyG (8). Analysis of the neutral sugar composition showed that the higher-order mutants had reduced levels of fucose, xylose, and noncellulosic glucose, as would be expected from reductions in XyG (SI Appendix, Table S1). More importantly, dramatic increases in other sugars were not observed (SI Appendix, Table S1), supporting the conclusion that the cslc mutants did not overproduce other wall polymers.
A more sensitive indicator of changes in the abundance of wall polymers is linkage analysis, which was performed on the cell walls from both cslc quintuple and xxt1 xxt2 mutant plants. The results revealed that both mutants had reduced levels of t-Xylp and 4,6-Glcp (SI Appendix, Table S2), as would be expected from reduced levels of XyG. However, relevant to the question of compensation, the linkages associated with other wall components, such as pectic or xylan polysaccharides, did not change significantly.
To further investigate possible compensation mechanisms, we hypothesized that, if genetic reprograming due to a lack of XyG occurred in the cslc quintuple mutant, then this mutant would exhibit significant changes in the expression of genes involved in cell wall metabolism and the transcriptional regulation of wall-associated genes. To test this possibility, we looked for changes in gene expression by comparing wild-type plants with the cslc456-2 triple and the cslc quintuple mutants by comparative transcript profiling. When DESeq2 (23) was used to identify differentially expressed genes (DEGs) in the cslc mutants, 67 of them were found in the cslc quintuple mutant and 60 in the cslc triple mutant, where expression levels changed at least twofold (|log2 fold change| > 1) after pairwise comparison with wild type (Dataset S1). Among these DEGs, 41 were differentially expressed in both mutant plants as compared to wild type. Among the common 41 DEGs, only 2 genes were expressed at a higher level compared to wild type, while 39 genes were expressed at lower levels in both cslc triple and quintuple mutants. We then analyzed for gene enrichment in specific biological processes using Gene Ontology (GO) term enrichment (24) focusing on the DEGs in the cslc triple and quintuple mutants. No significant GO term enrichments were found, supporting the conclusion that global transcriptional responses were not activated in specific biological processes due to XyG reductions in the cell wall. We next aimed to gain functional insights into the DEGs through an analysis of the subcellular localization of the protein products using the SUBA4 database (25). We hypothesized that gene expression changes would be coordinated by nuclear proteins, such as transcription factors. Fifteen DEGs were predicted to encode proteins localized in the nucleus, but all were expressed at extremely low levels (RPKM < 2), making their involvement in global genetic reprogramming processes unlikely (Dataset S1). In addition, we noticed that the expression levels of other genes involved in XyG, pectin, and cellulose synthesis were not greatly changed in terms of RPKM values (Dataset S2). Most genes showed log2 fold change lower than 1, suggesting that the genes involved in cell wall synthesis or modification are not strongly changed. Only XUT1, GAUT7, and RGXT1 showed meaningful increase (log2 fold change slightly higher than 1) in cslc quintuple mutant, but their expression levels were very low (Dataset S2). Together, these results led us to conclude that a lack of XyG in the cell wall does not trigger significant transcriptional responses.
The CSLC Genes Expanded from an Ancestral Family.
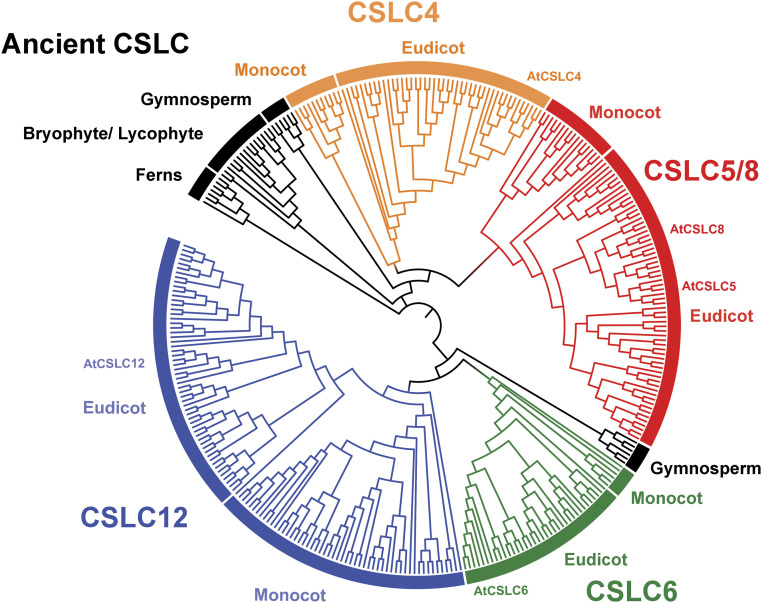
Having established that all CSLCs are XyG backbone synthases, we next investigated their phylogenetic relationship across plant genomes. Therefore, we performed cross-species phylogenetic analysis with CSLC protein sequences (>90% sequence coverage) obtained from publicly available plant genome databases. Although XyG-like polysaccharides were reported previously in charophytes (26), we did not include any putative CSLCs from charophytes due to the difficulty of differentiating CSLC from CSLA. After building a phylogenetic tree using the maximum-likelihood method, we verified an evolutionary branching of CSLC proteins into five major groups: an ancient CSLC-like group, a CSLC4-like group, a CSLC6-like group, a CSLC12-like group, and a cluster of CSLC5 and 8 groups (Fig. 5), similar to a previous report (27). Interestingly, bryophytes, early embryophytes such as Marchantia polymorpha, Physcomitrella patens, and Sphagnum fallax, as well as an ancient tracheohyte, Selaginella moellendorffii, and ferns, such as Azolla filiculoides and Salvinia cucullata, contain only the ancient form of CSLC-like proteins, supporting an ancestral phylogenetic position of these organisms and the presence of XyG in groups as early as bryophytes (28). No CSLC homologs were identified in chlorophyte sequences available in Phytozome, which is consistent with the absence of XyG-related genes in chlorophytes reported earlier (29). We observed a marked diversification of the CSLCs among seed plants, as supported by the emergence of CSLC6 in both gymnosperms and angiosperms, and the presence of both CSLC4 and CSLC12 in angiosperm lineages.
Fig. 5.
Phylogenetic analysis of the CSLC protein family. A total of 325 CSLC sequences from various plant groups were used (bryophytes/lycophytes, 15; ferns, 7; gymnosperms, 12; basal angiosperms, 3; monocots, 84; eudicots, 204). The phylogenetic tree was built using the MEGA X application with 1,000 replicates in the bootstrap test and the maximum-likelihood method. Orange, CSLC4 lineage; red, CSLC5/8 lineage; blue, CSLC12 lineage; green, CSLC6 lineage; black, ancient CSLC.
We also found that both monocots and eudicots contain CSLC4 proteins, but the structures of CSLC4 genes are different between the two groups. In eudicots, the CSLC4 genes are the only members of the CSLC family that have four exons and three introns (Fig. 1A). In contrast, the monocot CSLC4 genes are similar to the other CSLC genes in that they have five exons and four introns. Most eudicots have at least one member in CSLC4, CSLC5, CSLC6, and CSLC12 subfamilies, while the monocot lineages, especially the grasses, have at least one member of each of the CSLC4 and CSLC5 genes and duplicated copies of CSLC12. Amborella trichopoda, considered as a basal lineage of angiosperms, has CSLC5, 6, and 12 genes, suggesting the possible loss of CSLC6 and acquisition of extra CSLC12 genes during grass evolution. In Arabidopsis, because of high expression in pollen (SI Appendix, Figs. S2 and S3) along with their presence only in seed plants (Fig. 5), the CSLC6 and CSLC12 genes could be correlated with the evolution of reproductive systems in angiosperms. In summary, these bioinformatics analyses support an increased diversification of CSLC genes including the recent evolution of CSLC4 genes in eudicots, most likely from an ancestral CSLC group, and important roles of XyG in plant reproduction.
Discussion
Despite the abundance of XyG in plant cell walls, a molecular understanding of XyG biosynthesis has not yet been attained. In this work, we addressed this fundamental knowledge gap and showed that the five Arabidopsis CSLC family members are responsible for the synthesis of the glucan backbone of XyG in planta. The genetic experiments described here complement the biochemical evidence reported earlier that the Golgi-localized CSLC4 functions as a β-1,4 glucan synthase (9). The earlier observations led to the conclusion that the CSLC proteins are likely involved in XyG biosynthesis. Here we advanced these findings by providing direct evidence for a role of the five CSLCs in XyG biosynthesis in Arabidopsis and ascribe a degree of tissue specificity for the function of some of the CSLC proteins. By pursuing a reverse genetics approach based on loss-of-function alleles of all five of the Arabidopsis CSLC genes, we gathered evidence that, while each of the single mutants contained nearly normal levels of XyG (Fig. 4, Table 2, and SI Appendix, Fig. S7), the higher-order mutants had significantly reduced levels of XyG and the cslc quintuple mutant had undetectable levels of XyG, as determined with three different methods. The first measured levels of IP, a disaccharide that is derived exclusively from XyG (Fig. 4, Table 2, and SI Appendix, Fig. S8). The second method, used on the triple and quintuple mutants, employed simplified glycan arrays to directly estimate XyG levels (SI Appendix, Fig. S9). The third method, also used on the triple and quintuple mutants, was OLIMP analysis of oligosaccharides released from XyG (SI Appendix, Fig. S10). This last method also provides information on the side-chain substitution patterns in the residual XyG. While many questions remain about the functional significance of XyG substitution patterns, the observation that they remain unchanged in the hypocotyl during large reductions in XyG levels caused by elimination of three CSLC proteins implies that the substitution patterns are controlled in ways that are not fully understood. The important point is that all three methods led to the same conclusion, i.e., the triple mutant had significantly reduced levels of XyG, while XyG could not be detected in wall fractions from the quintuple mutant. This pattern is consistent with the conclusion that the CSLC genes are required for XyG biosynthesis. The complementation of the quintuple mutant with each of the CSLC genes further strengthens this conclusion and provides evidence that each of the CSLC genes encodes a XyG glucan synthase (Table 3). Furthermore, the observation that the loss of CSLC4 and CSLC12 causes visible plant phenotypes (Fig. 3) argues that the function of CSLC enzymes is only partially overlapping at a tissue level.
While the evidence presented here supports the conclusion that all members of the Arabidopsis CSLC gene family are involved in XyG biosynthesis, it remains to be determined the extent to which this conclusion can be extended to other plant species. Because both the XyG polymer and the CSLC genes are widely distributed in the plant kingdom (1, 26), it seems likely that the CSLCs have retained the same function in all plant species and are involved in XyG biosynthesis. Phylogenetic analyses of the CESA superfamily revealed that CSLC genes are closely related to CSLA genes (30–32). Little et al. (2018) suggested that the grouping of CSLA and CSLC genes has no direct phylogenetic connection with the other members of the CESA superfamily (32). Thus, they speculated that the CSLA/CSLC group arose via an independent evolutionary event. The close evolutionary relationship between the CSLC and CSLA gene families combined with a lack of connection to other members of the CESA superfamily (32) may have consequences for understanding their biochemical functions (12). The CSLA proteins synthesize a β-1,4–linked mannan backbone or a β-1,4–linked glucomannan backbone (33–37). The CSLA proteins interact with α-galactosyltransferases from the GT34 family that add galactosyl residues to the 6-position of the mannose residues in the backbone (38, 39) to produce galactomannan or galactoglucomannan. The CSLC proteins have striking functional similarities in that they synthesize β-1,4–linked backbones and interact with glycosyltransferases from the GT34 family to add sugars to the 6-position of the sugars in the backbone (12). In the case of CSLC proteins, they interact with α-xylosyltransferases that add xylose residues to the glucan backbone to form XyG (6, 40, 41). In contrast, the other members of the CESA superfamily, e.g., CSLF and CSLH that synthesize mixed-linkage glucan (42, 43) or CSLD that synthesizes a β-1,4–linked glucan (44), generate sugar backbones that lack side chains, meaning that there would be no need for interaction with a glycosyltransferase from the GT34 family. Thus, the functional differences between most members of the CESA superfamily and CSLA and CSLC proteins are consistent with different phylogenetic origins of CSLC and CSLA proteins and the rest of the CESA family.
One important unresolved question regarding the XyG-deficient mutants is how they are able to grow and develop relatively normally (Fig. 3 and SI Appendix, Fig. S5). Given the current hypotheses regarding the roles that XyG performs in plant cell wall structure and function (see below for more discussion of this topic), one possible answer is that the quantities of some wall components may be increased in the XyG-deficient plants, thereby providing compensation for the lack of XyG. However, we found no evidence for increased levels of known wall components (SI Appendix, Tables S1 and S2; see also tables 1 and 2 in Cavalier et al., 2008 [8]). Further evidence making this possibility unlikely is that comparative RNA-seq analyses on the quintuple mutant and wild type did not show significant changes in the levels of expression for genes responsible for the synthesis of known wall components (Datasets S1 and S2). Another possible explanation for the relatively normal growth and development of the mutant plants is that redundancy exists among wall components and that some wall component is able to substitute for XyG without an increase in its level. One possibility is pectin. Support for this hypothesis comes from solid-state NMR studies of the cell walls of wild type and the xxt1 xxt2 xxt5 triple mutant that lacks XyG (45). The results led these authors to suggest that the load-bearing network in Arabidopsis cell walls includes cellulose, pectin, and XyG and that the network can continue to function in the absence of XyG. Another possible component that may substitute for XyG is the glycoprotein extensin. Gille et al. (2009) observed that enzymatic degradation of XyG caused greater hypocotyl elongation in a mutant, xeg113, than in wild type (46). This mutant has defects in extensin glycosylation (46). The authors hypothesized that XyG and extensin have similar, or possibly redundant, functions. Thus, if their hypothesis is correct, extensin may functionally substitute for XyG in the mutants lacking XyG.
While Arabidopsis plants are able to grow and develop relatively normally in the absence of XyG, the presence of “dysfunctional” XyG in an XyG galactosyltransferase mutant, mur3-3, which lacks galactose and fucose side chains, causes more severe phenotypes. This mutation produces plants “that are dwarfed with curled rosette leaves, short petioles, and short inflorescence stems” (47, 48). Further studies have shown that elimination of the aberrant XyG by combining the mur3-3 mutant with the xxt1 xxt2 mutant eliminates the morphological phenotype of the mur3-3 mutant (47, 49). Molecular studies comparing changes in gene expression in the various mutants demonstrated several alterations in the mur3-3 mutant containing dysfunctional XyG, whereas the patterns of gene expression in the xxt1 xxt2 mutant lacking XyG are quite similar to wild-type plants (49). Correspondingly, we did not observe any remarkable changes in gene expression in the cslc quintuple mutant compared to wild type (Datasets S1 and S2). These observations provide further support for the hypothesis of Xu et al. (49) that the presence of abnormal XyG rather than absence of XyG in Arabidopsis elicits transcriptional responses that impact plant growth.
XyG is proposed to play an important role in the structure and function of primary plant cell walls (1, 50–54). In most models, XyG is thought to function as a tether between cellulose microfibrils forming a load-bearing network that needs to be reorganized to allow growth. Several studies have utilized the XyG-deficient mutants to investigate the impact on wall properties with a goal of evaluating such models. Anderson et al. (2010) reported that cellulose organization is significantly altered in the root cells of xxt1 xxt2 mutant plants (55). Park and Cosgrove (2012) examined the mechanical properties of cell walls in wild-type and mutant Arabidopsis plants and reported that “acid growth” and α-expansin–induced growth was reduced in the xxt1 xxt2 mutant (56). They also provided evidence that pectins and xylans may substitute for XyG in the petiole cell walls of the mutant plants. In addition to a possible role for pectins to compensate for the loss of XyG in the xxt1 xxt2 xxt5 mutant (45), Xiao et al. (2016) found that microtubule patterning and stability was altered, cellulose fibrils were more aligned, and the rate of cellulose synthase mobility was reduced in the xxt1 xxt2 mutant (57). Consistent with these observations is that XyG might represent a “spacer” molecule to limit interactions between cellulose fibrils and thus maintain their functionality. More recently, the xxt1 xxt2 mutant was used to determine that XyG and microtubules function synergistically to maintain meristem geometry and phyllotaxis in Arabidopsis (58).
As a result of these and other studies on the role of XyG in plant cell walls, new ideas for the organization of plant cell wall polymers have emerged. Rather than postulating that XyG forms the load-bearing tethers between cellulose microfibrils (50, 51), one newer idea suggests that highly localized “biomechanical hotspots” play an important role in controlling the mechanical properties of plant cell walls (52, 53). The role, if any, of XyG in the structure and function of these hotspots remains to be determined (59). The new series of cslc mutants provides additional tools with which to investigate some of the new ideas regarding cell wall organization and resolve some of the questions regarding the role of XyG in cell wall functions. They also provide the opportunity to test a role for XyG under stress conditions, which may be not apparent during standard conditions of growth as used here. Furthermore, because of the tissue-specific expression of the various CSLC genes, this series of mutants has the potential to allow the investigation of the role of XyG in the cells of different tissues throughout plant development.
Methods
Plant Material and Growth Condition.
Homozygous cslc4-1 (SALK_146718), cslc4-3 (SAIL_837B10), cslc5-1 (SAIL_187G09), cslc6-1 (SALK_088720-11), cslc8 (WiscDsLox_497-02H), and cslc12-2 (SAIL_168F02) T-DNA insertion lines were used in this study after confirmation by PCR-based genotyping with specific primers. Details are provided in SI Appendix, SI Materials and Methods. Seeds for the cslc456–2 triple mutant, the two quadruple mutants, and the quintuple mutant (Table 1) have been deposited in the ABRC at Ohio State University.
Transcriptomic and qRT-PCR Analysis.
In this study, RNA was extracted using a plant RNA extraction kit (https://www.mn-net.com/). After RNA isolation, total RNA from 7-d-old etiolated hypocotyls of wild type and cslc456-2 and cslc456812 mutant were sent to the Research Technology Support Facility at Michigan State University for generating RNA-seq libraries and sequencing the libraries using the Illumina platform (paired 125 bp). For qRT-PCR and RT-PCR of CSLC family genes, 1 µg of total RNA was used to investigate expression of CSLC genes during Arabidopsis development. Details are provided in SI Appendix, SI Materials and Methods.
Driselase Digestion of AIR and HPAEC and LC/Qtof MS Analyses.
The Driselase enzyme mix (https://www.sigmaaldrich.com) was partially purified as previously reported (60). AIR was prepared using 7-d-old etiolated hypocotyls as described earlier (22). Approximately 1.3 mg of AIR obtained from the etiolated hypocotyls were digested with 300 µL of 0.03% of Driselase enzyme mix (25 mM NaAc, pH 5.0) at 37 °C for 48 h, and the reaction was stopped by heating the samples at 95 °C for 5 min. Details are provided in SI Appendix, SI Materials and Methods.
Glycosidic Linkage of Isolated Isoprimeverose.
Eluting material at the retention time of IP (10 min, HPAEC separation of Driselase digest) was collected, pooled, and freeze-dried. To account for the significant reduction in IP in the Driselase digests derived from the cslc456812 and xxt1 xxt2 mutants compared to wild type (Fig. 4), 10 times more Driselase-digested material from the mutants was subjected to HPAEC-PAD via multiple injections compared to wild type. The collected material was air-dried and subjected to glycosidic linkage analysis as described previously (8).
Complementation of the cslc456812 Quintuple Mutant.
To generate constructs to complement the cslc456812 quintuple mutant, each CSLC gene was synthesized by Thermo Fisher (https://www.thermofisher.com/) and cloned into binary vectors containing the CSLC4 promoter or the constitutive 35S promoter to drive CSLC expression. Details are provided in SI Appendix, SI Materials and Methods.
Phylogenetic Tree Construction.
To identify CSLC homologs from diverse plant groups, the Arabidopsis CSLC5 amino acid sequence was used to identify CSLC homologs by BLASTp using sequences stored in Phytozome 12 (https://phytozome.jgi.doe.gov/), and sequences with over 90% sequence coverage were selected for further analysis. Due to a lack of gymnosperm and fern sequences in Phytozome, CSLC-like sequences from Ginkgo biloba, Picea abies, Pinus sylvestris, Pseudotsuga menziesii, Cycas micholitzii, Pinus pinaster, and two ferns, A. filiculoides and S. cucullata, were obtained from PLAZA (https://bioinformatics.psb.ugent.be/plaza/versions/gymno-plaza/) and FernBase (https://www.fernbase.org/). A total of 325 sequences from bryophytes, lycophytes, ferns, gymnosperms, and angiosperms were used to build a phylogenetic tree using the maximum-likelihood method and a JTT matrix-based model with a 1,000 bootstrap value using MEGA X (61) after sequence alignment using Clustal W. iTOL v5 was used to visualize the tree generated from MEGA X (62).
Supplementary Material
Acknowledgments
We thank Dr. David Cavalier, who was involved in early efforts to identify the cslc single mutants and in the preparation of some of the double and triple mutants. We also thank Cliff Foster (Great Lakes Bioenergy Research Center cell wall facility), Dr. A. Daniel Jones and Dr. Anthony Schilmiller (Michigan State University [MSU] Mass Spectrometry Facility), and Dr. Melinda Frame (MSU Center for Advanced Microscopy) for technical assistance with plant cell wall composition, IP analyses, and confocal microscopy, respectively. This work was funded primarily by the Department of Energy (DOE) Great Lakes Bioenergy Research Center (DOE Biological and Environmental Research Office of Science DE-FC02-07ER64494 and DE-SC0018409). We also acknowledge partial support from the Chemical Sciences, Geosciences and Biosciences Division, Office of Basic Energy Sciences, Office of Science, US Department of Energy (Award Number DE-FG02-91ER20021), NSF (MCB1727362), and AgBioResearch (MICL02598) to F.B. The contributions of M.P. and B.C. were funded by the Deutsche Forschungsgemeinschaft (German Research Foundation) under Germany’s Excellence Strategy, EXC-2048/1, Project 390686111.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2007245117/-/DCSupplemental.
Data Availability.
RNA-seq data have been deposited in the National Center for Biotechnology Information database (accession no. PRJNA642312).
References
- 1.Pauly M., Keegstra K., Biosynthesis of the plant cell wall matrix polysaccharide xyloglucan. Annu. Rev. Plant Biol. 67, 235–259 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Kim S.-J., Brandizzi F., The plant secretory pathway for the trafficking of cell wall polysaccharides and glycoproteins. Glycobiology 26, 940–949 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Perrin R. M. et al., Xyloglucan fucosyltransferase, an enzyme involved in plant cell wall biosynthesis. Science 284, 1976–1979 (1999). [DOI] [PubMed] [Google Scholar]
- 4.Madson M. et al., The MUR3 gene of Arabidopsis encodes a xyloglucan galactosyltransferase that is evolutionarily related to animal exostosins. Plant Cell 15, 1662–1670 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Faik A., Price N. J., Raikhel N. V., Keegstra K., An Arabidopsis gene encoding an α-xylosyltransferase involved in xyloglucan biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 99, 7797–7802 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavalier D. M., Keegstra K., Two xyloglucan xylosyltransferases catalyze the addition of multiple xylosyl residues to cellohexaose. J. Biol. Chem. 281, 34197–34207 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Zabotina O. A. et al., Mutations in multiple XXT genes of Arabidopsis reveal the complexity of xyloglucan biosynthesis. Plant Physiol. 159, 1367–1384 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cavalier D. M. et al., Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. Plant Cell 20, 1519–1537 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cocuron J.-C. et al., A gene from the cellulose synthase-like C family encodes a β-1,4 glucan synthase. Proc. Natl. Acad. Sci. U.S.A. 104, 8550–8555 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richmond T. A., Somerville C. R., The cellulose synthase superfamily. Plant Physiol. 124, 495–498 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richmond T. A., Somerville C. R., Integrative approaches to determining Csl function. Plant Mol. Biol. 47, 131–143 (2001). [PubMed] [Google Scholar]
- 12.Liepman A. H., Cavalier D. M., The CELLULOSE SYNTHASE-LIKE A and CELLULOSE SYNTHASE-LIKE C families: Recent advances and future perspectives. Front. Plant Sci. 3, 109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morgan J. L. W., Strumillo J., Zimmer J., Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature 493, 181–186 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sethaphong L. et al., Tertiary model of a plant cellulose synthase. Proc. Natl. Acad. Sci. U.S.A. 110, 7512–7517 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winter D. et al., An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2, e718 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim S.-J., Thrower N., Danhof L., Keegstra K., Brandizzi F., Sequence from Arabidopsis cslc mutants. Sequence Read Archive. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA642312. Deposited 27 June 2020.
- 17.Gardner S. L., Burrell M. M., Fry S. C., Screening of Arabidopsis thaliana stems for variation in cell wall polysaccharides. Phytochemistry 60, 241–254 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Moller I. et al., High-throughput mapping of cell-wall polymers within and between plants using novel microarrays. Plant J. 50, 1118–1128 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Jensen J. K. et al., The DUF579 domain containing proteins IRX15 and IRX15-L affect xylan synthesis in Arabidopsis. Plant J. 66, 387–400 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Marcus S. E. et al., Pectic homogalacturonan masks abundant sets of xyloglucan epitopes in plant cell walls. BMC Plant Biol. 8, 60 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Willats W. G. T., Marcus S. E., Knox J. P., Generation of monoclonal antibody specific to (1–>5)-α-L-arabinan. Carbohydr. Res. 308, 149–152 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Günl M., Kraemer F., Pauly M., “Oligosaccharide mass profiling (OLIMP) of cell wall polysaccharides by MALDI-TOF/MS” in The Plant Cell Wall: Methods and Protocols, Popper Z. A., Ed. (Humana Press, Totowa, NJ, 2011), pp. 43–54. [DOI] [PubMed] [Google Scholar]
- 23.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian T. et al., agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 45, W122–W129 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hooper C. M., Castleden I. R., Tanz S. K., Aryamanesh N., Millar A. H., SUBA4: The interactive data analysis centre for Arabidopsis subcellular protein locations. Nucleic Acids Res. 45, D1064–D1074 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Del-Bem L.-E., Xyloglucan evolution and the terrestrialization of green plants. New Phytol. 219, 1150–1153 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Dwivany F. M. et al., The CELLULOSE-SYNTHASE LIKE C (CSLC) family of barley includes members that are integral membrane proteins targeted to the plasma membrane. Mol. Plant 2, 1025–1039 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Popper Z. A., Evolution and diversity of green plant cell walls. Curr. Opin. Plant Biol. 11, 286–292 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Del Bem L. E. V., Vincentz M. G. A., Evolution of xyloglucan-related genes in green plants. BMC Evol. Biol. 10, 341 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yin Y., Huang J., Xu Y., The cellulose synthase superfamily in fully sequenced plants and algae. BMC Plant Biol. 9, 99 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yin Y., Johns M. A., Cao H., Rupani M., A survey of plant and algal genomes and transcriptomes reveals new insights into the evolution and function of the cellulose synthase superfamily. BMC Genom. 15, 260 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Little A. et al., Revised phylogeny of the cellulose synthase gene superfamily: Insights into cell wall evolution. Plant Physiol. 177, 1124–1141 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liepman A. H. et al., Functional genomic analysis supports conservation of function among cellulose synthase-like a gene family members and suggests diverse roles of mannans in plants. Plant Physiol. 143, 1881–1893 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dhugga K. S. et al., Guar seed β-mannan synthase is a member of the cellulose synthase super gene family. Science 303, 363–366 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Liepman A. H., Wilkerson C. G., Keegstra K., Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proc. Natl. Acad. Sci. U.S.A. 102, 2221–2226 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goubet F. et al., Cell wall glucomannan in Arabidopsis is synthesised by CSLA glycosyltransferases, and influences the progression of embryogenesis. Plant J. 60, 527–538 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Voiniciuc C., Dama M., Gawenda N., Stritt F., Pauly M., Mechanistic insights from plant heteromannan synthesis in yeast. Proc. Natl. Acad. Sci. U.S.A. 116, 522–527 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edwards M. E. et al., Molecular characterisation of a membrane-bound galactosyltransferase of plant cell wall matrix polysaccharide biosynthesis. Plant J. 19, 691–697 (1999). [DOI] [PubMed] [Google Scholar]
- 39.Edwards M. E. et al., The seeds of Lotus japonicus lines transformed with sense, antisense, and sense/antisense galactomannan galactosyltransferase constructs have structurally altered galactomannans in their endosperm cell walls. Plant Physiol. 134, 1153–1162 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chou Y.-H., Pogorelko G., Young Z. T., Zabotina O. A., Protein-protein interactions among xyloglucan-synthesizing enzymes and formation of Golgi-localized multiprotein complexes. Plant Cell Physiol. 56, 255–267 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Chou Y.-H., Pogorelko G., Zabotina O. A., Xyloglucan xylosyltransferases XXT1, XXT2, and XXT5 and the glucan synthase CSLC4 form Golgi-localized multiprotein complexes. Plant Physiol. 159, 1355–1366 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burton R. A. et al., Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3;1,4)-β-D-glucans. Science 311, 1940–1942 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Doblin M. S. et al., A barley cellulose synthase-like CSLH gene mediates (1,3;1,4)-β-D-glucan synthesis in transgenic Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 5996–6001 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang J. et al., Biochemical and genetic analysis identify CSLD3 as a beta-1,4-glucan synthase that functions during plant cell wall synthesis. Plant Cell 32, 1749–1767 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dick-Pérez M. et al., Structure and interactions of plant cell-wall polysaccharides by two- and three-dimensional magic-angle-spinning solid-state NMR. Biochemistry 50, 989–1000 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Gille S., Hänsel U., Ziemann M., Pauly M., Identification of plant cell wall mutants by means of a forward chemical genetic approach using hydrolases. Proc. Natl. Acad. Sci. U.S.A. 106, 14699–14704 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kong Y. et al., Galactose-depleted xyloglucan is dysfunctional and leads to dwarfism in Arabidopsis. Plant Physiol. 167, 1296–1306 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jensen J. K., Schultink A., Keegstra K., Wilkerson C. G., Pauly M., RNA-seq analysis of developing nasturtium seeds (Tropaeolum majus): Identification and characterization of an additional galactosyltransferase involved in xyloglucan biosynthesis. Mol. Plant 5, 984–992 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu Z. et al., DGE-seq analysis of MUR3-related Arabidopsis mutants provides insight into how dysfunctional xyloglucan affects cell elongation. Plant Sci. 258, 156–169 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Somerville C. et al., Toward a systems approach to understanding plant cell walls. Science 306, 2206–2211 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Hayashi T., Kaida R., Functions of xyloglucan in plant cells. Mol. Plant 4, 17–24 (2011). [DOI] [PubMed] [Google Scholar]
- 52.Park Y. B., Cosgrove D. J., Xyloglucan and its interactions with other components of the growing cell wall. Plant Cell Physiol. 56, 180–194 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Cosgrove D. J., Diffuse growth of plant cell walls. Plant Physiol. 176, 16–27 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Buckeridge M. S., The evolution of the glycomic codes of extracellular matrices. Biosystems 164, 112–120 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Anderson C. T., Carroll A., Akhmetova L., Somerville C., Real-time imaging of cellulose reorientation during cell wall expansion in Arabidopsis roots. Plant Physiol. 152, 787–796 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park Y. B., Cosgrove D. J., Changes in cell wall biomechanical properties in the xyloglucan-deficient xxt1/xxt2 mutant of Arabidopsis. Plant Physiol. 158, 465–475 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiao C., Zhang T., Zheng Y., Cosgrove D. J., Anderson C. T., Xyloglucan deficiency disrupts microtubule stability and cellulose biosynthesis in Arabidopsis, altering cell growth and morphogenesis. Plant Physiol. 170, 234–249 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao F. et al., Xyloglucans and microtubules synergistically maintain meristem geometry and phyllotaxis. Plant Physiol. 181, 1191–1206 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park Y. B., Cosgrove D. J., A revised architecture of primary cell walls based on biomechanical changes induced by substrate-specific endoglucanases. Plant Physiol. 158, 1933–1943 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fry S. C., The Growing Plant Cell wall: Chemical and Metabolic Analysis, (The Blackburn Press, Caldwell, NJ, 2000). [Google Scholar]
- 61.Kumar S., Stecher G., Li M., Knyaz C., Tamura K., MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Letunic I., Bork P., Interactive tree of life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 47, W256–W259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited in the National Center for Biotechnology Information database (accession no. PRJNA642312).