Significance
Eukaryotic gelsolin superfamily proteins generally comprise three or more related domains. Here we characterize single- and double-domain gelsolins from Thorarchaeota (Thor). Similar domain architectures are present in Heimdall-, Loki-, and Odinarchaeota. Thor gelsolins are functional in regulating rabbit actin in in vitro assays, showing a range of activities including actin filament severing and bundling. These gelsolins bind to the eukaryotic gelsolin/cofilin-binding site on actin. Two-domain, but not one-domain, gelsolins are calcium regulated. Thor gelsolins appear to have the characteristics and structure consistent with primitive gelsolins/cofilins, suggesting that these single- and double-domain gelsolins are a record of a nascent preeukaryotic actin-regulation machinery.
Keywords: actin, gelsolin, Asgard archaea, eukaryogenesis, X-ray crystallography
Abstract
Asgard archaea genomes contain potential eukaryotic-like genes that provide intriguing insight for the evolution of eukaryotes. The eukaryotic actin polymerization/depolymerization cycle is critical for providing force and structure in many processes, including membrane remodeling. In general, Asgard genomes encode two classes of actin-regulating proteins from sequence analysis, profilins and gelsolins. Asgard profilins were demonstrated to regulate actin filament nucleation. Here, we identify actin filament severing, capping, annealing and bundling, and monomer sequestration activities by gelsolin proteins from Thorarchaeota (Thor), which complete a eukaryotic-like actin depolymerization cycle, and indicate complex actin cytoskeleton regulation in Asgard organisms. Thor gelsolins have homologs in other Asgard archaea and comprise one or two copies of the prototypical gelsolin domain. This appears to be a record of an initial preeukaryotic gene duplication event, since eukaryotic gelsolins are generally comprise three to six domains. X-ray structures of these proteins in complex with mammalian actin revealed similar interactions to the first domain of human gelsolin or cofilin with actin. Asgard two-domain, but not one-domain, gelsolins contain calcium-binding sites, which is manifested in calcium-controlled activities. Expression of two-domain gelsolins in mammalian cells enhanced actin filament disassembly on ionomycin-triggered calcium release. This functional demonstration, at the cellular level, provides evidence for a calcium-controlled Asgard actin cytoskeleton, indicating that the calcium-regulated actin cytoskeleton predates eukaryotes. In eukaryotes, dynamic bundled actin filaments are responsible for shaping filopodia and microvilli. By correlation, we hypothesize that the formation of the protrusions observed from Lokiarchaeota cell bodies may involve the gelsolin-regulated actin structures.
Asgard archaea are some of the most fascinating organisms on the planet since they possess eukaryotic-like genes (1, 2), which encode for functional proteins (3), providing evidence for the origins of the eukaryotic cell. In particular, Asgard genomes encode potential homologs for a regulated actin cytoskeleton (1, 2), which is critical for membrane remodeling in eukaryotes (4). It is not known whether this potential dynamic actin cytoskeleton is related to the membrane blebs, protrusions or vesicles observed in the first Asgard archaea to be isolated (5). All phyla of Asgard archaea possess actin and actin-related proteins (ARPs) (1, 2, 6–8). The actins have yet to be demonstrated to form filaments, and the functions of the ARPs are presently unknown. Asgard and eukaryotic profilins support barbed-end actin filament elongation and inhibit spontaneous actin nucleation (3). In addition, Asgard genomes also encode potential homologs for gelsolin domains, while some species of Heimdallarchaeota may contain a distant homolog of ARP2/3 subunit 4 (1, 2, 6–8). The presence of these genes raises the question to what extent these archaea are able to regulate their actin dynamics by this limited number of proteins, beyond the control afforded by profilin (3).
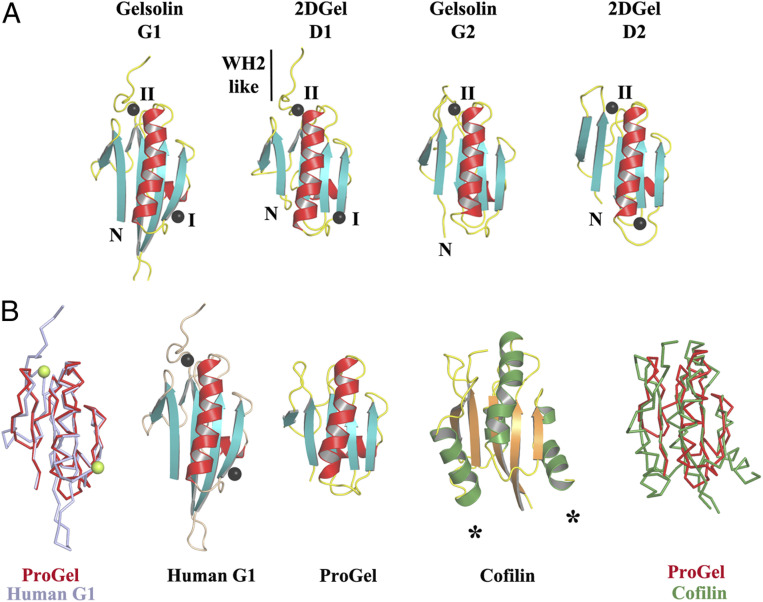
In eukaryotes, many actin-regulating activities, such as monomer sequestration and filament nucleation, elongation, annealing, bundling, capping and severing (9, 10), are elicited by the calcium-regulated multidomain gelsolin family of proteins (Fig. 1A) (11, 12). These are predicted to have arisen from serial gene multiplication events (12, 13). Sequence comparison of the domains indicates that two serial single domain gene multiplication events were followed by a third whole gene duplication to produce the 3 and 6 domain proteins, respectively (Fig. 1A) (12, 13). The last common ancestor of eukaryotes likely possessed a protein comprised of three gelsolin domains, and possibly a second protein of six gelsolin domains, since 3 and 6 domain gelsolins are predicted across a broad spectrum of eukaryotes from sequence databases. In addition, eukaryotes contain single domain ADF/cofilins and double cofilin domain twinfilins, some of which bind to the sides of filaments, sever actin filaments, sequester actin monomers, and share a core domain topology with the gelsolin domain (14–18).
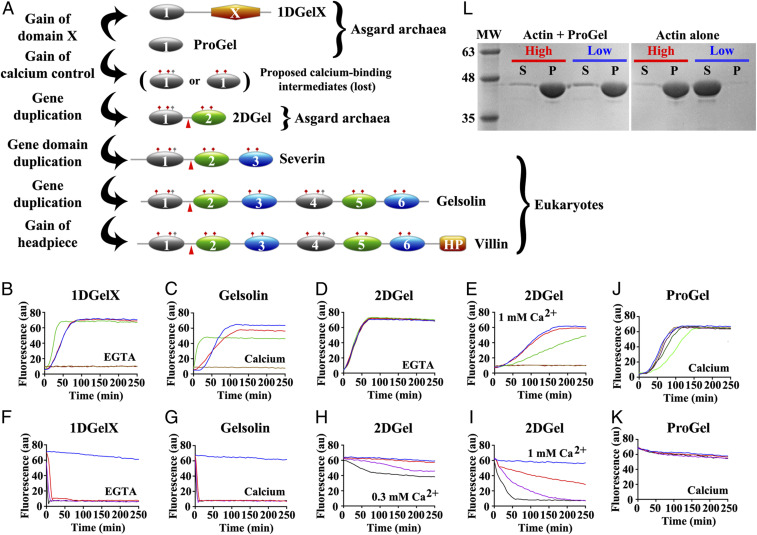
Fig. 1.
Thor gelsolins and actin regulation. (A) Schematic representation of the three Thor gelsolin architectures and the hypothetical evolution of the gelsolin family. Ovals depict gelsolin domains. Ticks indicate potential calcium-binding residues and red triangles denote a central WH2-like motif. Since Type II calcium-binding sites (48) (red ticks) are found in both domains of 2DGel, a calcium-binding single-domain protein likely existed in evolution that is not found in the current sequence databases. This is indicated by the “proposed calcium-binding intermediates.” The Type I site (48) (gray ticks) may have been present in this proposed calcium-binding intermediate, and later lost from domain two after the first gene duplication. Alternatively, the Type I site may have appeared in domain one after the first gene duplication. The architectures of typical eukaryotic gelsolin-like proteins are included for comparison. (B–E) Pyrene-actin polymerization profiles of 2 μM actin (blue) supplemented with (B) 1DGelX (1 mM EGTA), at 10 nM (red), 0.1 μM (green), or 2 μM (fawn) or 16 μM (dark brown), (C) supplemented with 5 nM (red), 0.05 μM (green), 2 μM (fawn) human gelsolin (0.3 mM CaCl2), or supplemented with (D) 2DGel (1 mM EGTA) or (E) 2DGel (1.0 mM CaCl2) at the concentrations in B. (F–I) Actin depolymerization profiles of 2 μM actin (blue), supplemented by (F) 1DGelX (1 mM EGTA), at 2 μM (red), 8 μM (lilac), or 32 μM (black), (G) human gelsolin in 0.3 mM CaCl2, concentrations as in C, (H) 2DGel (0.3 mM EGTA) or (I) 2DGel (1 mM CaCl2) at the concentrations in F. Two other 2DGel orthologs, 2DGel2 and 2DGel3, showed additional filament nucleation activity and more potent severing activity. All three 2DGel proteins were less active at 0.3 mM than at 1 mM Ca2+, and inactive in 1 mM EGTA in terms of severing activity (SI Appendix, Fig. S2 E–P). (J) Pyrene-actin polymerization profiles of 2 μM actin (blue) supplemented with 2 μM (fawn), 8 μM (lilac), 32 μM (dark green), or 128 μM (light green) ProGel in 0.3 mM Ca2+. (K) Pyrene-actin depolymerization profiles of 2 μM actin (blue) supplemented with 10 nM (red), 0.1 μM (green), 2 μM (beige), or 16 μM (dark brown) ProGel in 0.3 mM CaCl2. (L) SDS/PAGE analysis of actin filaments (8 μM) in the presence or absence of ProGel (256 μM). At 150,000 × g (high) filaments were pelleted in both conditions, whereas at 10,000 × g (low) actin was pelleted as bundles only in the presence of ProGel.
Here, we analyzed protein sequences from Asgard archaea genomes to search for proteins that contain gelsolin domains. We identified three gelsolin-like architectures from Thorarchaeota (Thor) sequence databases (Fig. 1A): sequences comprising only of the prototypical gelsolin/cofilin domain (ProGel); sequences comprising a single gelsolin/cofilin domain followed by an unknown domain X (1DGelX); and sequences comprising two-domain gelsolins (2DGel). Similar architectures are found in, and are unique to, other Asgard archaea, including Lokiarchaeota (SI Appendix, Fig. S1 A–D). We investigated these mini gelsolins to determine whether they provide a record of the initial gene duplication in this superfamily of proteins (Fig. 1A). Asgard gelsolins are uncharacterized at the protein level. To further investigate the actin regulation in Asgard archaea, we tested the properties of Thor gelsolins. We carried out in vitro biochemical experiments to demonstrate that Thor single-domain and double-domain gelsolins are functional with eukaryotic actin in order to establish that these proteins are genuine functionally-related homologs of eukaryotic gelsolin and cofilin. Variety in Thor gelsolin activities indicates a wide-ranging array of functions. Together these data provide insight in to the regulation of the early pre-eukaryotic actin cytoskeleton.
Results
Biochemical Analysis of Thor Gelsolins.
We expressed and purified five recombinant Thor gelsolins from E. coli (SI Appendix, Fig. S1E). ProGel, 1DGelX and 2DGel derive from the same species/assembly (SMTZ1-83), while 2DGel2 and 2DGel3 originate from different species/assemblies AB_25 and SMTZ1-45, respectively (SI Appendix, Fig. S1F). 2DGel, 2DGel2 and 2DGel3 share 76–89% identity (SI Appendix, Fig. S1G). These proteins displayed a range of binding affinities in interacting with rabbit muscle actin (rActin) as assessed by surface plasmon resonance studies: 1DGelX (Kd = 7.7 ± 0.3 μM), 2DGel2 (Kd = 54 ± 7 μM), 2DGel (Kd = 113 ± 16 μM), 2DGel3 (Kd = 135 ± 21 μM), with ProGel (Kd = 337 ± 70 μM) showing little or negligible binding (SI Appendix, Fig. S1H). In pyrene-actin assays, 1DGelX and 2DGel showed robust activities. In the actin assembly assay, low concentrations of 1DGelX (0.1 μM) reduced the lag phase of actin polymerization (2 μM) consistent with filament nucleation, while 1:1 ratios inhibited polymerization (Fig. 1B), similar to calcium-activated human gelsolin (hGelsolin) (Fig. 1C). The 1DGelX activity was similar in the presence of ethylene glycol-bis(2-aminoethylether)-N,N,N′-tetraacetic acid) (EGTA) or Ca2+ (SI Appendix, Fig. S2A), indicating an absence of calcium control. By contrast, 2DGel showed no activity in the assembly assay in the presence of EGTA (Fig. 1D). However, in the presence of Ca2+ (1 mM), a low concentration of 2DGel (0.1 μM) significantly slowed actin polymerization indicating filament capping (Fig. 1E), and at a 1:1 ratio, no increase in pyrene fluorescence was observed, indicative of actin monomer sequestration. The sequestering and capping effects of 2DGel were reduced at intermediate Ca2+ levels (0.3 mM, See SI Appendix, Fig. S2B).
In a pyrene-actin depolymerization assay, 1DGelX produced a rapid drop in fluorescence, indicative of robust severing (Fig. 1F), similar to calcium-activated gelsolin (Fig. 1G). 1DGelX’s severing activity was comparable in EGTA to Ca2+ (SI Appendix, Fig. S2C), revealing that this activity is also Ca2+ independent. 2DGel in 0.3 mM Ca2+ showed partial loss in fluorescence implying incomplete depolymerization (Fig. 1H). At higher calcium levels (1 mM) the effects were more dramatic. 2DGel induced F-actin depolymerization with a fast, initial loss followed by a slower decline, consistent with filament severing followed by monomer sequestration (Fig. 1I). These effects were lost in EGTA (SI Appendix, Fig. S2D), implying that the calcium signaling range in Thorarchaeota is likely to be higher than the micromolar range in some eukaryotes. ProGel had weak inhibitory effects on actin assembly and no effect on filament disassembly (Fig. 1 J and K and SI Appendix, Fig. S3). These pyrene assay data reveal that 1DGelX and 2DGel display diverse gelsolin-like activities, including monomer sequestration and filament nucleation, capping and severing, with 2DGel showing calcium regulation, as did the two 2DGel orthologs 2DGel2 and 2DGel3 (SI Appendix, Fig. S2 E–P).
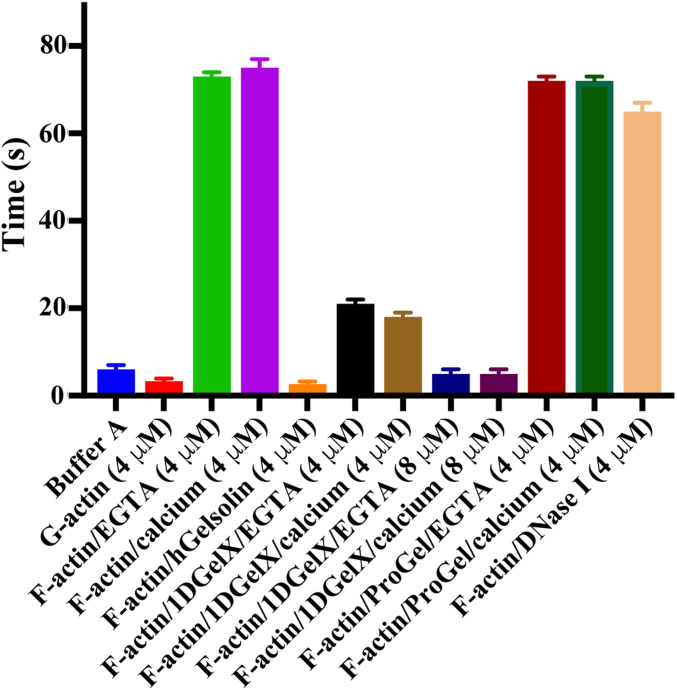
To further provide evidence of calcium-independent severing by 1DGelX we measured its effect on actin filament viscosity (Fig. 2). 1DGelX (4 μM and 8 μM) showed a titratable reduction in F-actin (4 μM) viscosity, consistent with severing, which was not dependent on calcium, reaching levels close to G-actin or to F-actin treated with calcium-activated hGelsolin (both 4 μM). By contrast, ProGel (4 μM) or DNase I (4 μM), an actin monomer sequestering protein, were unable to substantially change the viscosity of F-actin (4 μM) during the time course of the experiment (∼4 min).
Fig. 2.
F-actin severing by 1DGelX. Viscometry experiments demonstrate that 4 μM ProGel or DNase I (actin-sequestering protein control) do not significantly change the viscosity of F-actin (∼70 s) in the timeframe of the experiment, whereas, 4- and 8 μM 1DGelX lowers the F-actin viscosity levels close to G-actin (∼5 s) or to gelsolin (F-actin severing protein control). x axis indicates solution conditions.
Subsequently, we asked whether the Thor gelsolins could be pelleted with F-actin under slow and fast centrifugation speeds, suitable for sedimentation of actin filament bundles and actin filaments (including bundles), respectively. ProGel, at the high concentration (256 μM) that showed little effect in the pyrene assays, was able to cause F-actin (8 μM) to be pelleted at low speed, indicating that ProGel induced filament bundling (Fig. 1L and SI Appendix, Fig. S4A). Substantial amounts of ProGel were observed in the pellets, despite careful washing. This suggests that ProGel binds to the sides actin filaments causing bundling. At low concentration (1 μM) of 1DGelX, 2DGel, 2DGel2, or 2DGel3 the filaments (8 μM) largely remained intact, pelleting only at the highest speed, as assessed by electrophoresis, implying that the filaments remained single (SI Appendix, Fig. S4B). The 2DGel proteins were only observed in the soluble fraction, indicating that these proteins do not associate with the sides of actin filaments. The number of filament ends was likely insufficient to determine filament end binding in this experiment. 1DGelX migrated at a similar position to actin. Hence, its filament association properties were difficult to determine, however, some 1DGelX was observed in the soluble fractions of the high-speed spins.
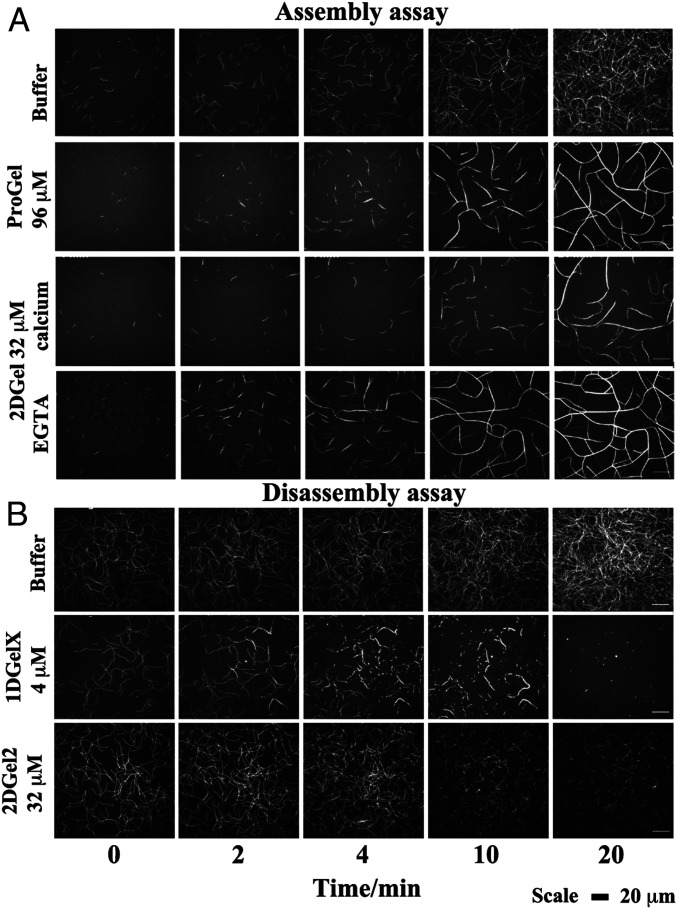
Next, we applied total internal reflection fluorescence (TIRF) microscopy to observe fluorescently-labeled actin filaments. Assembly of actin (1.5 μM) under high concentrations of ProGel (96 μM) confirmed an initial minor reduction in polymerization (0–4 min), but showed striking filament bundling and annealing at later time points under the assay’s molecular crowding conditions (0.25% methylcellulose, Fig. 3A and see SI Appendix, Fig. S5A and Movie S1). Bundling of actin filaments by ProGel was apparent in the sedimentation assay (Fig. 1L), but not in the pyrene actin assembly assay (Fig. 1J), which has been shown to be insensitive to bundling by villin (19). In the TIRF assembly assay, increasing concentrations of 1DGelX (100 nM to 4 μM) produced a decreased number of filaments and some bundling at the highest concentration (4 μM, and see SI Appendix, Fig. S5B and Movie S2). 2DGel (32 μM) in EGTA bundled filaments (Fig. 3A and Movie S3). Addition of Ca2+ to 2DGel reduced the number of actin filaments compared to the control, and by consequence, reduced the formation of bundles (Fig. 3A and Movie S3). The reduction in actin filament number is consistent with the decrease in pyrene fluorescence observed in the pyrene actin assembly assay (Fig. 1E). Filament bundling was not apparent in the sedimentation studies for either 1DGelX or 2DGel (SI Appendix, Fig. S4B), which suggests that bundling is aided by the molecular crowding conditions in the TIRF assay.
Fig. 3.
The regulation of actin assembly and disassembly by Thor gelsolins followed by TIRF microscopy. Time course of the (A) assembly and (B) disassembly of 1.5 μM actin in the presence of various concentrations of Thor gelsolins. (Scale bar, 20 μm.) Titrations of ProGel and 1DGelX in the assembly assay can be found in SI Appendix, Fig. S5, and comparison of 2DGel, 2DGel2, and 2DGel3 in the disassembly assay are found in SI Appendix, Fig. S6. Movies of the assembly/disassembly of Thor gelsolins are found in Movies S1–S5. ProGel and 1DGelX assays were carried out in 1 mM EGTA. The 2DGel assembly assays were in 0.3 mM CaCl2 or 1 mM EGTA, and the disassembly assay in 0.3 mM CaCl2.
Adding 1DGelX or the 2DGel orthologs to preformed actin filaments resulted in severing by different modes. 1DGelX (4 μM) disassembled F-actin in a complex pattern (Fig. 3B and Movie S4). 1DGelX initially severed filaments into short fragments, which associated into short bundles, and the bundles were further severed to complete dissociation of actin structures. In 0.3 mM Ca2+, 2DGel (32 μM) bundled, 2DGel2 (32 μM) severed single actin filaments with no bundling, and 2DGel3 (32 μM) initially severed single filaments, followed by the bundling and annealing of the fragments into larger structures (Fig. 3B, SI Appendix, Fig. S6, and Movie S5). These TIRF data reveal a complex regulation of the assembly and disassembly of actin filaments and bundles.
Structures of the Thor Gelsolin/rActin Complexes.
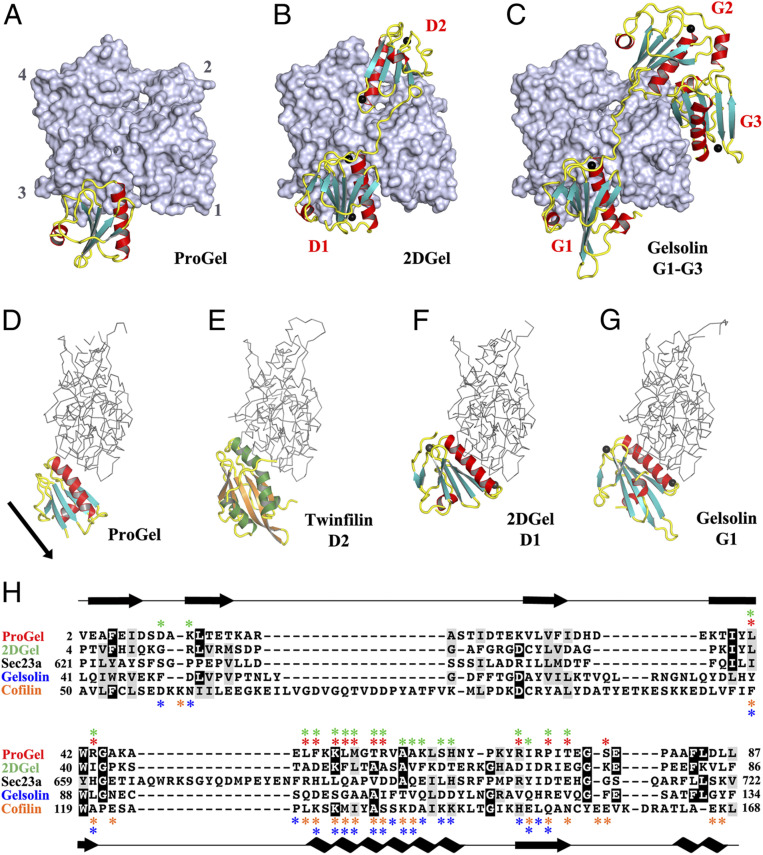
To compare the Asgard and eukaryotic gelsolin interactions with actin, we determined the X-ray structures of two types of Thor gelsolin/rActin complex, ProGel and 2DGel. Despite its weak interaction with G-actin (SI Appendix, Fig. S1H), ProGel bound to actin at a site that overlaps with gelsolin domain 1 (G1) (20), and with the cofilin family (21) (Fig. 4 A and D–H). Significantly, the protein is translated by one turn of the main helix relative to G1 and the calcium-binding sites are absent. The crystal structure of 2DGel bound to actin (Fig. 4B) revealed that domain 1 (D1) and the central WH2-like motif (LRRV) are similar to hGelsolin (Fig. 4C and SI Appendix, Fig. S7A), however, D2 is positioned differently (22). D1 of 2DGel packs tightly to the cleft between actin subdomains 1 and 3, similar to hGelsolin G1, which is a capping orientation for hGelsolin (23). ProGel more loosely associates with the same region of actin, in a similar orientation to the cofilin family, which for human cofilin allows interaction with the sides of actin filaments (24). The residues on rActin that interact with ProGel, 2DGel (D1), cofilin, and hGelsolin (G1) are largely conserved across Asgard actins (SI Appendix, Fig. S8A). Some substitutions in the 2DGel, 2DGel2, and 2DGel3 residues that contact actin may account for their functional differences (SI Appendix, Fig. S7B and S8B). ProGel and 2DGel contain the gelsolin DWG motif that is replaced by a WP motif in the cofilin family (SI Appendix, Fig. S9 A and B), implying that ProGel is related to both gelsolin and cofilin (Fig. 5). Next, we soaked the 2DGel crystals with 1 mM TbCl3 and examined the resultant anomalous electron density to confirm the cation-binding sites (SI Appendix, Fig. S9C) (25). Four Ca2+ bind to 2DGel, three sites in common with hGelsolin (Fig. 5A and SI Appendix, Fig. S8B). We speculate that gene duplication to produce 2DGel resulted in tighter calcium control and extra functionality, due to multiplication of the calcium and actin-binding sites (Fig. 1A). Moreover, attempts to crystallize 1DGelX failed. The 1DGelX gelsolin domain shares 35% identity with ProGel and is likely to similarly bind to actin (Fig. 4A and SI Appendix, Fig. S8 C and D). Domain X is predicted to form a coiled-coil structure with no homology to known actin-binding proteins (SI Appendix, Fig. S7C).
Fig. 4.
The structures of ProGel and 2DGel in complex with rActin. (A) The ProGel/rActin complex. rActin is shown as a surface and ProGel is in schematic representation. (B) The structure of the 2DGel/rActin complex. The four calcium ions associated with 2DGel are shown as black spheres. The crystal structure of 2DGel3/rActin complex is found in SI Appendix, Fig. S7A. (C) The structure of the first three domains of human gelsolin in complex with rActin for comparison (PDB ID code 1EQY). (D–G) Side views of (D) ProGel, (E) twinfilin domain 2 (D2), a cofilin-family member (PDB ID code 3DAW), (F) 2DGel domain 1 (D1), and (G) gelsolin domain G1, in complex with actin. Actin is shown as a trace. Similar representations for 2DGel3 is found in SI Appendix, Fig. S7B. The arrow indicates the displacement of ProGel relative to G1. Data collection and refinement statistics are found in SI Appendix, Table S1. (H) Structure-based sequence alignment of the core region of the gelsolin/cofilin domain of ProGel, 2DGel, Sec23a (which has not been shown to bind actin), hGelsolin, and cofilin. rActin interacting residues are indicated by stars. Asgard proteins above the alignment ProGel, red; 2DGel, green) and human proteins below (gelsolin, blue; cofilin, orange). The consensus secondary structure is shown in black.
Fig. 5.
Structural homology to gelsolin and cofilin. (A) The three conserved calcium-binding sites in 2DGel and 2DGel3 are shared with the first two domains of human gelsolin. (B) Structural comparisons and superimpositions of ProGel with human gelsolin G1 (PDB ID code 3FFN) and cofilin (PDB ID code 4KEE). Calcium ions are shown as lime or black spheres. Asterisks indicate additional helices in the cofilin fold relative to ProGel.
Localization and Calcium Control in a Cellular Context.
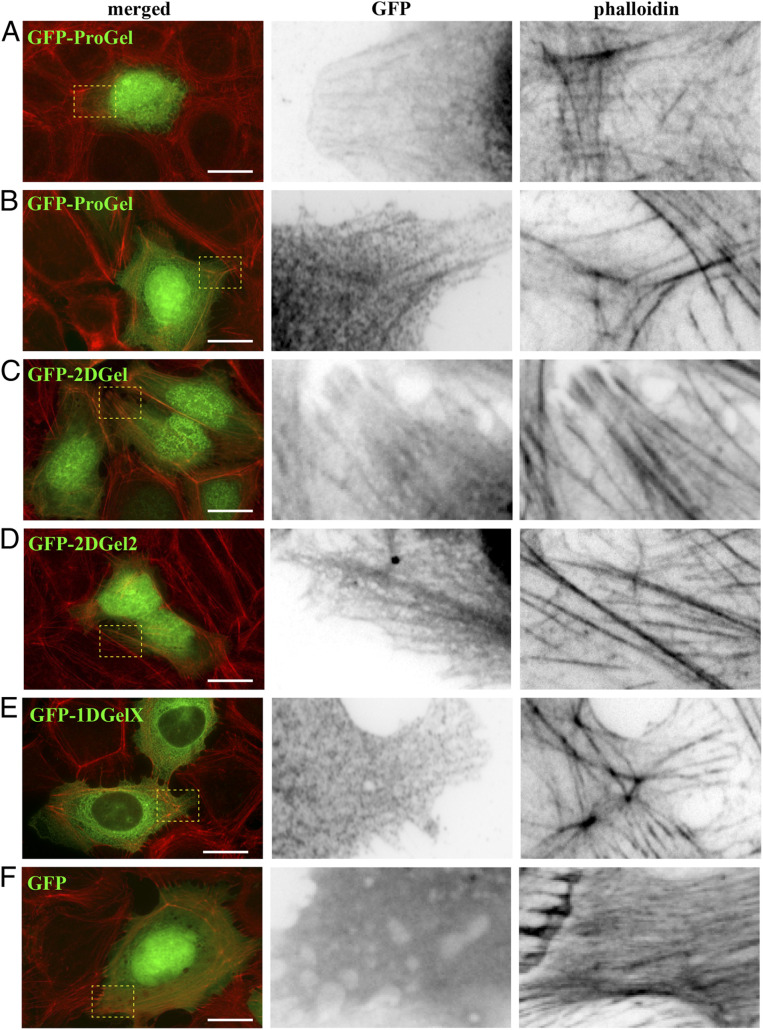
Next, we tested the localization of GFP-tagged Thor gelsolins in transfected human U2OS cells. The cells were treated with rhodamine-phalloidin, which stains actin filaments, to determine whether these proteins associate with eukaryotic cellular structures. GFP-positive cells showed typical cell morphologies and actin cytoskeleton arrangements for all constructs (Fig. 6). GFP-ProGel was present in the cytoplasm and nucleus (Fig. 6 A and B) similar to GFP alone (Fig. 6F). In the cytoplasm, GFP-ProGel was diffuse but also localized to filament structures, consistent with actin–filament binding (Fig. 6 A and B). GFP-2DGel and its ortholog GFP-2DGel2 were present in the nucleus and cytoplasm (Fig. 6 C and D), with both proteins appearing to be enriched in actin–filament structures.
Fig. 6.
Localization of GFP-Thor proteins in mammalian cells. Ectopically expressed Asgard gelsolins in human U2OS cells followed by fluorescence imaging. (A and B) Two examples of merged images of GFP-ProGel (green) and rhodamine-phalloidin staining of actin filament structures (red), followed by enlargements of the box regions with separated GFP and phalloidin channels. Similar representative images of (C) GFP-2DGel, (D) GFP-2DGel2, (E) GFP-1DGelX, and (F) GFP control.
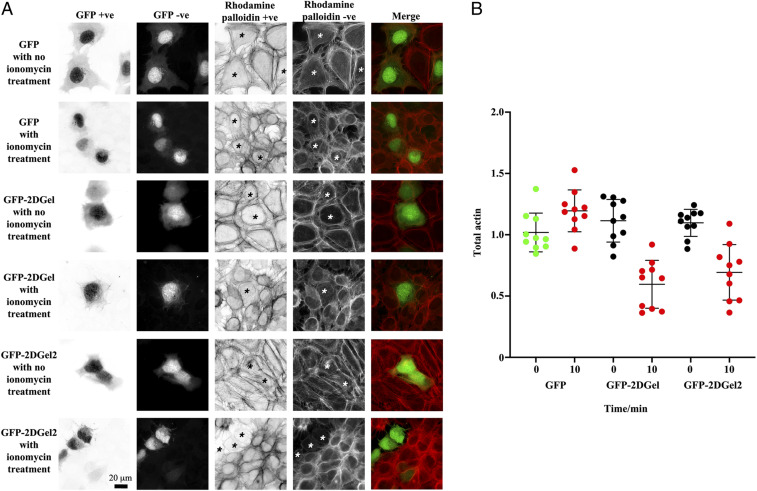
By contrast, 1DGelX was present throughout the cytoplasm but excluded from the nucleus, concentrating at the regions typical of the nuclear membrane and endoplasmic reticulum (Fig. 6E). The 1DGelX association with filaments was weak in comparison to ProGel, GFP-2DGel, and GFP-2DGel2. Finally, we tested whether GFP-2DGel and GFP-2DGel2 could respond to calcium signaling in a cellular context. Treatment of the cells with ionomycin (10 μM, 10 min), to release calcium from intracellular stores, led to a contraction of all cells. In the GFP-2DGel and GFP-2DGel2 expressing cells, but not GFP-expressing cells, a dramatic loss in rhodamine-phalloidin signal was observed (Fig. 7). This indicates that calcium signaling to 2DGel and 2DGel2 resulted in the loss of F-actin structures, demonstrating that these proteins are functional in the eukaryotic cellular environment.
Fig. 7.
Calcium signaling to ectopically expressed 2DGel proteins in human U2OS cells followed by fluorescence imaging. (A) Cells expressing GFP, GFP-2DGel, or GFP-2DGel2 are indicated by signal the GFP channel, columns 1 and 2, and highlighted with asterisks. Actin filaments and larger structures are observed in the rhodamine-phalloidin channel, columns 3 and 4. “+ve” refers to normal and “−ve” to the reversed image. Merged images of the GFP channel (green) and the rhodamine-phalloidin channel (red) are in the final column. Different cells were imaged immediately before, or 10 min after, treatment with ionomycin to release calcium. (B) Quantification of rhodamine-phalloidin fluorescence before and 10 min after treatment with ionomycin. Twelve-bit monochrome images of actin fluorescence intensity, of typical cells, were quantified as a ratio for GFP and adjacent non-GFP expressing cells, in three separate experiments.
Discussion
The combination of structural biology and biochemistry has a long history of illuminating evolutionary relationships for the regulated actin cytoskeleton, and has been particularly insightful in situations where relationships are not readily apparent from sequence analyses (18, 26). Protein sequence alignment-based phylogenetic analyses detect signatures of three-dimensional (3D) structural and functional conservation in proteins. We have directly determined these structural and functional signatures from ProGel and 2DGel, and we have compared them to the eukaryotic proteins, gelsolin and cofilin. The Asgard and eukaryotic proteins adopt similar core structures; and despite ∼2 billion y of divergence these proteins interact with eukaryotic actin in similar orientations, at a common binding site on eukaryotic actin. The most parsimonious explanation of these data is that the Thor gelsolins and eukaryotic gelsolins and cofilins are derived from a common ancestor gene. Here, we speculate on the implications for the evolution of the gelsolin and cofilin families of proteins that can be inferred from this analysis of single- and double-domain Thor gelsolins. The core structure of ProGel is similar to eukaryotic cofilin and more similar to eukaryotic gelsolin, yet the ProGel actin-binding mode is most similar to cofilin (Figs. 4 D–H and 5B and SI Appendix, Fig. S4A). This indicates that present-day eukaryotic cofilins and gelsolins likely evolved from a protein that had similar architecture and F-actin–binding capability to ProGel. The binding sites on actin are consistent with the Thor gelsolins competing with actin–actin interactions to elicit their effects (SI Appendix, Fig. S10). The acquisition of sequences outside of the core gelsolin/cofilin domain appears to be a critical factor in extending the range of functions. Eukaryotic cofilin has additional helices whereas eukaryotic gelsolins have duplications of the core domain (Figs. 1A and 5B), and both of these proteins bind F-actin. Comparison of the activities of ProGel with 1DGelX and 2DGel indicates how expansion of the core gelsolin domain leads to altered activities. We hypothesize that the C-terminal extension of 1DGelX contains an actin side-binding domain or motif. The F-actin binding functionality would locate 1DGelX to the side of a filament allowing the gelsolin domain to evolve to compete for actin:actin contacts to gain severing functionality. Thus, we propose that ProGel represents a record of an initial gelsolin/cofilin protein that originally emerged as a relatively simple actin regulator, which through gaining C-terminal domains, or expansion of the core, added additional functionalities. Duplication of the ProGel scaffold would have resulted in 2DGel, after it acquired calcium binding, which was present in the last common ancestor of Asgard archaea. We propose that the Asgard ancestor of eukaryotes would have possessed ProGel and 2DGel, which evolved into ADF/cofilin and three domain gelsolins, respectively, proteins which were present in the last common ancestor of eukaryotes. The variety of Asgard gelsolin-like sequences is consistent with this structure-function–based proposal of gelsolin/cofilin family evolution; however, the signatures of 3D structural and functional conservation in the gelsolin/cofilin core domain appear to be too weak to be reliably detected by phylogenetic analysis (SI Appendix, Fig. S11).
Localization of the Thor proteins to different eukaryotic cellular locations reveals the functional compatibility of the Asgard and eukaryotic actin regulation systems. ProGel showed clear actin filament localization (Fig. 6 A and B), while Asgard 2DGels were able to dismantle eukaryotic cellular F-actin in a calcium-dependent manner (Fig. 7). The sensitivity of the Thor 2DGels to calcium indicates that Asgard organisms are capable of calcium signaling, and that calcium signaling predates eukaryogenesis. Taken together, these data demonstrate that Asgard mini gelsolins possess equivalent actin-regulating properties (monomer sequestration, filament nucleation, bundling, and severing) to their larger eukaryotic counterparts (Fig. 1A) and to proteins that possess different architectures, such as capping protein and fascin. We speculate that the emergence of distinct actin regulators in eukaryotes allowed for greater and more diverse control of actin dynamics, enabling its incorporation into an expanded number of cell processes (27). However, many of the fundamental actin architectures were achievable in preeukaryotic organisms. We now have an emerging picture of a relatively complex regulated Asgard eukaryotic-like polymerization/depolymerization cycle that is executed by a limited number of proteins, yet has many of the characteristics of the actin dynamics in eukaryotes, where a large part of the functional output involves eliciting membrane perturbations, similar to those observed in Lokiarchaeota (5).
The presence of functional gelsolin-like proteins in Asgard archaea has several possible scenarios with regard to the emergence of the domains of life, which has been debated in relation to different phylogenetic analyses of Asgard genomes (1–3, 28–31). In the three-domain hypothesis, eukaryotes and archaea form separate clades. In this case, there are at least two possible explanations for the existence of Asgard gelsolins that are capable of interacting with eukaryotic actin. Firstly, gelsolins, profilins and actins may have been present in the last common ancestor of archaea, but later lost from most branches of archaea, with the exception of Asgard archaea. Such widespread loss of these genes in Archaea would seem unlikely given the prevalence of these proteins throughout eukaryotes, indicating their advantage for survival. Secondly, the genes may have been passed by horizontal gene transfer to stem Asgard archaea from a stem eukaryote(s) or vice versa. We specify stem organisms, since all Asgard phyla contain ProGel and 1DGelX, and since these genes are not found in present day eukaryotes, they would likely not be present in last common ancestor of eukaryotes. Given the abundance of eukaryotic-like protein sequences in Asgard archaea genomes, this would require a sizable transfer, or transfers, of genetic material. Alternatively, in the two-domain hypothesis, eukaryotes emerge from within the archaea domain. In this scenario, which is the most parsimonious explanation for the presence of the actin regulation system in Asgard archaea (3), the gelsolin, profilin, and actin proteins arose in a common ancestor of eukaryotes and Asgard archaea, within archaea. Further metagenomic sequencing of diverse Asgard archaea, followed by careful phylogenetic analysis, is required to enhance the useful phylogenetic signal in the Asgard ribosomal sequences, and categorically settle the debate on the relative positions of Asgard archaea and eukaryotes within the tree of life. Regardless of the ultimate phylogeny, the primitive gelsolins analyzed here represent a record of a nascent actin cytoskeleton regulation machinery that was likely a prerequisite for eukaryogenesis.
Materials and Methods
Protein Expression and Purification.
The Asgard gelsolin gene sequences were synthesized and codon optimized for Escherichia coli (GenScript) and placed in the pSY5 vector (3, 23). Proteins were expressed as described previously (3). Asgard gelsolin containing BL21 DE3 Rosetta cell pellets (10 g) were resuspended in binding buffer 50 mL (20 mM Hepes, pH 7.5, 500 mM NaCl, 20 mM imidazole, 1 mM tris(2-carboxyethyl)phosphine [TCEP], and 1 mM EGTA), supplemented with benzonase (2 μL of 10,000 U/μL, Merck), Protease Inhibitor mixture (Set III, EDTA-Free, Calbiochem), and Triton X-100 (0.01%). Cell lysis, Ni-nitrilotriacetic acid (NTA) affinity, and size-exclusion chromatography (16/60 Superdex 75 PG, GE Healthcare) in gel filtration buffer (20 mM Hepes, pH 7.5, 150 mM NaCl, 1 mM TCEP, and 1 mM EGTA) were performed as described previously (3). Asgard gelsolin fractions were pooled and concentrated (10,000 MWCO Vivaspin concentrator, Vivascience). Recombinant human gelsolin was purified as described previously (23). Freshly prepared rActin was purified including a final gel filtration step and was labeled with pyrene as described previously (3). For TIRF studies, actin was labeled on lysines by incubating actin filaments with Alexa-488 succinimidyl ester (Molecular Probes) (32). Human profilin was expressed in BL21 DE3 Rosetta cells and purified as reported (33). Attempts to express Thor actin and constructs comprising individual 1DGelX gelsolin and X domains failed.
Pyrene–Actin Assays.
Pyrene–actin assembly and disassembly assays were performed with 2-μM rabbit skeletal-muscle G-actin (10% pyrene labeled). Gelsolin polymerization and depolymerization assays were performed in the presence of Ca2+ or EGTA. In the pyrene–actin assembly assays, G-actin in buffer A (2 mM Tris.HCl, pH 7.4, 0.2 mM adenosine triphosphate [ATP], 0.5 mM dithiothreitol [DTT], 0.3 or 1 mM CaCl2, 1 mM Na azide) was mixed and incubated with a 20-fold dilution of 20× Mg-exchange buffer (1 mM MgCl2, 4 mM EGTA) for 2 min in order to preexchange the calcium ion for magnesium. Subsequently, actin polymerization was initiated by the addition 10 μL of 10× actin polymerization KMI buffer (500 mM KCl, 10 mM MgCl2, 100 mM imidazole-HCl, pH 7.5) supplemented by the appropriate concentrations of EGTA or CaCl2 in a total volume of 100 μL.
Pyrene–actin disassembly assays were carried out in presence of calcium (0.3 mM or 1 mM Ca2+) or EGTA (1 mM or 2 mM). For pyrene–actin disassembly assays, 2 μM G-actin (10% pyrene labeled) was mixed with buffer A. Actin polymerization was initiated by the addition of 10 μL of 10× KMI or KMEI (KMI with 1 mM EGTA) buffer. The polymerization was monitored around 2 h at room temperature in 96-well, black, flat-bottomed plates (Corning, Nunc) in total volume of 90 μL. Then, 10 μL of different concentrations of gelsolin proteins in buffer A were added to the preformed F-actin in the appropriate concentrations of CaCl2 or EGTA. The fluorescence intensities were monitored at wavelength 407 nm after excitation at 365 nm with a Safire2 fluorimeter (Tecan). Seeding-pyrene assays were performed as described (3).
Viscometry Experiments.
Relative viscosity was assessed using an uncalibrated Semi-Micro Viscometer (Cannon-Manning, size 50) which functions under gravity. The go-through time of UltraPure water, buffer A, G-actin, and F-actin were measured at room temperature for a total sample volume of 1,000 μL. Actin was either polymerized (4 μM) actin by adding 10× KMEI or KMI for 90 min. Subsequently, the test protein (50 μL) was added to the preformed filament solution and incubated for 2 min. Then, the time for the solution to travel through the viscometer was measured, which is relative to the viscosity of the solution. All experiments were performed three times independently.
Sedimentation Studies.
Thor proteins were tested for their ability to bind F-actin in the presence of calcium (1 mM) or EGTA (1 mM). All proteins were first centrifuged at 150,000 × g for 1 h to remove potential aggregates. Actin (8 μM) in buffer A was polymerized in the presence of the Thor proteins with addition of 10× KMEI or KMI for 90 min at room temperature. The samples were centrifuged using a TLA120.1 rotor in a Beckman Optima Max ultracentrifuge for 30 min at 10,000 × g (low speed for bundling assays) and 150, 000 × g (high speed for filament binding assays). Equal volumes of pellet and supernatant were analyzed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis.
Surface Plasmon Resonance.
Surface plasmon resonance experiments were performed with Biacore T100 (GE Healthcare) at 25 °C. Asgard gelsolins and actin were desalted into modified buffer A (2 mM Tris.HCl, pH 7.4, 0.2 mM ATP, 0.5 mM DTT, 1 mM CaCl2, 1 mM Na azide, 100 mM NaCI, and 0.01% Tween 20). All buffers were degassed prior to the experiment. Actin was immobilized to CM5 chips via amine coupling with 10 μL/min flow rate for 1,800 s as described in the Biacore manual. The immobilized levels were between 400 and 500 resonance units (RU). Asgard gelsolin proteins were captured to actin-immobilized CM5 flow cell with the flow rate of 30 μL/min to a level of ∼35 RU for 1DGelX, ∼20 RU for ProGel, ∼8 RU for 2DGel and 2DGel2, and ∼5 RU for 2DGel3. One flow cell was used with buffer as a reference surface. Gelsolin proteins in modified buffer A were injected for 60 s; the dissociation was monitored for 300 s. Final responses from target proteins were obtained upon the subtraction of responses from buffer reference. Kinetic analysis of the final responses were analyzed by global fitting using a 1:1 binding model. All experiments were repeated at least three times with similar results.
TIRF Assays.
Coverslips (20 × 20 mm2) and microscope slides (Agar Scientific) were extensively cleaned, oxidized with oxygen plasma (5 min at 80 W, Harrick Plasma), and incubated with 1 mg/mL of silane-PEG, MW 5K (Creative PEG Works) overnight. Actin assembly was initiated in polymerization chambers of 20 × 20 mm2 × 4.5-µm height or in a polydimethylsiloxane (PDMS) chamber with a reaction volume of 30 µL, by addition of the actin polymerization mix (2.6 mM ATP, 10 mM DTT, 1 mM EGTA, 50 mM KCl, 5 mM MgCl2, 10 mM Hepes, pH 7.5, 3 mg/mL glucose, 20 µg/mL catalase, 100 µg/mL glucose oxidase, 0.2% wt/vol bovine serum albumin [BSA], and 0.25% wt/vol methylcellulose) containing actin monomers (1.5 µM, 20% Alexa488-labeled). The polymerization chambers were constructed using a double-sided tape (70 µm height) between a glass coverslip and slide coated with silane-PEG. The PDMS chambers were prepared using the published protocol (34). To observe the effect on actin polymerization, Thor gelsolins were added at the desired concentration directly in the polymerization mix. To show effect on F-Actin, the PDMS chamber was used to reconstitute actin network during 5–10 min, then the gelsolin protein was added carefully and mixed. The controls were carried out using gelsolin storage buffer (20 mM Hepes, pH 7.5, 150 mM NaCl, 1 mM TCEP, and 1 mM EGTA) without gelsolin.
Crystallization.
Crystals of Asgard gelsolin/actin complexes of ProGel/rActin, 2DGel/rActin and 2DGel3/rActin were grown from 1:1 molar ratios (400 μM Asgard gelsolin: 400 μM rabbit actin) of the proteins were under the following conditions: ProGel/rActin, 0.1 M 2-(N-morpholino)ethanesulfonic acid (MES), pH 6.0, 0.2 M magnesium chloride hexahydrate, 20% wt/vol polyethylene glycol 6000, at 291 K; 2DGel/rActin, 0.1 M Bis-Tris, pH 5.5, 0.2 M magnesium chloride hexahydrate, 25% wt/vol polyethylene glycol 3350, at 291 K; and 2DGel3/rActin, 0.1 M N-cyclohexyl-2-aminoethanesulfonic acid, pH 9.5, 20% wt/vol polyethylene glycol 8000, at 291 K; CaCl2 was added to a final concentration of 1 mM prior to crystallization for 2DGel/rActin and 2DGel3/rActin. Latrunculin B (4 mM) was added to the ProGel/rActin complex prior to concentration and crystallization. All crystallization trials were performed at ∼10 mg/mL protein concentration using the sitting-drop or hanging-drop vapor-diffusion methods. Crystals were flash frozen in the crystallization buffer, which was supplemented by 25% glycerol for the 2DGel/rActin complex, prior to X-ray data collection. Crystallization screens for the Asgard gelsolins alone, or for the 1DGelX/rActin and 2DGel2/rActin complexes were negative.
Structure Determination, Model Building, and Refinement.
Native X-ray diffraction datasets from single crystals of 2DGel/rActin and 2DGel3/rActin on a RAYONIX MX-300 HS charge-coupled device (CCD) detector on beamline TPS 05A (NSRRC) controlled by BLU-ICE (version 5.1) at λ = 1.0 Å. Data were indexed, scaled, and merged in HKL2000 (version 715) (35) (SI Appendix, Table S1). A native-data X-ray diffraction dataset from a crystal of the ProGel/rActin complex was collected on beamline MX2 (Australian Synchrotron) on an Eiger16M detector at λ = 1.0 Å. Data were indexed, scaled, and merged in XDS (Version November 2016) (36) and ccp4-7.0 CCP4-7.0 AIMLESS (version 0.5.29) (SI Appendix, Table S1). Terbium anomalous diffraction data for 2DGel/rActin were collected and merged in the range 20.0–1.7 Å (Rmerge 0.056, Rpim 0.056, anomalous completeness 99.4%, redundancy 5.5) using the same protocols as for the native data set.
Molecular replacement using the ProGel/rActin and 2DGel/rActin datasets using the native actin (Protein Data Bank [PDB] ID code 3HBT) (37) as the search model was carried out in the PHENIX suite (Version 1.13–2998) (38) Phaser. The model for 2DGel/rActin was extended in AutoBuild (38), whereas the gelsolin chains for ProGel/rActin were built by hand. All manual adjustments to the models and refinement were carried out in Coot (Version 0.8.9 EL) (39). Final refinement for ProGel/rActin was carried out in CCP4-7.0 Refmac5 (40). The final model for 2DGel/rActin was used as the molecular replacement search model for 2DGel3/rActin. All final models were verified for good stereochemistry in PHENIX suite (Version 1.13–2998) (38) MolProbity (41) (SI Appendix, Table S1). The final ProGel/rActin models (two in the asymmetric unit) consist of ProGel residues 1–88 missing the final four residues (chain B and D) and rActin 5–42 and 51–374 (chain A) and rActin 5–41 and 51–374 (chain C). The actins are each associated with ATP, latrunculin B, and a magnesium ion. The final 2DGel/rActin model consists of 2DGel residues 2–197 (missing the final residue), associated with five calcium ions, and rActin 5–41 and 49–372. The final 2DGel3/rActin model consists of 2DGel3 residues 2–198, associated with five calcium ions, and rActin 6–39 and 51–372. The model of the structure of domain X from 1DGelX was generated by I-TASSER (42).
Sequence Analyses.
Protein domain homolog identification was carried in BLAST (43) using reported Asgard sequences (1, 2). Domain architectures of proteins were created in Prosite MyDomains (44). Structure-based sequence alignments were carried out in PROMALS3D (45). Phylogenetic analysis was carried out on the cofilin/gelsolin G1 core domain (Fig. 4H). Sequences were aligned in MUSCLE (46) and potential phylogenetic relationships calculated in Phylogeny.fr (47) using the WAG model with 100 bootstrap replicates.
Cell Culture and Transfection.
Protein constructs were synthesized with an N-terminus GFP in pcDNA3.1(+)-N-eGFP (GenScript). Actin staining was performed with rhodamine-phalloidin (TRITC) (ThermoFisher R415) at 1:200/500 dilution and nuclei stained with H33342 (ThermoFisher 62249) at 1 µg/mL U2OS (human osteosarcoma) cells were cultured in high glucose Dulbecco’s modified Eagle’s (DME) media with 4,500 mg/L glucose, supplemented with 10% fetal bovine serum (FBS) (HyClone). Cells were grown at 37 °C in an incubator filled with 5% CO2 and 99% humidity. U2OS cells seeded onto 22 × 22-mm glass coverslips in 35-mm culture dishes and grown to subconfluence. Each dish was then transfected with 0.5 μg of GFP control of fusion gelsolin construct plasmid DNA using the Mirus TransIT LT1 transfection reagent according to the manufacturer’s protocol, incubated at 37 °C and allowed to express for 24 h. Cells were then fixed with 3.7% formaldehyde in phosphate-buffered saline (PBS) at room temperature for 20 min.
Intracellular calcium induction.
Cells were transfected and observed to ensure ∼50% of cells were expressing GFP, appeared healthy, and of comparable density. The media was then changed to reduced serum DME (1% FBS, 1.8 mM calcium) with or without 10 μM ionomycin (calcium ionophore) and incubated at 37 °C for 10 min. Cells were then fixed with 3.7% formaldehyde in PBS at room temperature for 20 min.
Immunofluorescence and microscopy.
Fixed cells were washed with PBS and permeabilized in 0.2% TritonX-100 in PBS for 10 min and were then blocked with 10% serum for 10 min. They were then incubated in rhodamine-phalloidin, diluted with 5% BSA, 0.1% TritonX-100 in PBS for 1 h at room temperature, after which cells were washed with 0.1% TritonX-100 in PBS. Coverslips were mounted with fluorescent mounting medium (Thermo Scientific), images acquired using a Zeiss Axioplan2 microscope equipped with CoolSnap HQ cold CCD camera at 400× magnification for quantification and higher (630×) for representative images.
Quantification.
Eight-bit monochrome images of total actin fluorescence intensity, of typical cells, were quantified as a ratio for GFP and adjacent non-GFP expressing cells, in three separate experiments. At least 20 different cells were measured. Analyses were performed using ImageJ (NIH) software.
Statistics and Reproducibility.
All biochemical experiments were repeated three times with similar results.
Supplementary Material
Acknowledgments
We thank Agency for Science, Technology and Research, Singapore National Medical Research Council (NMRC Grant OFIRG/0067/2018), the Vidyasirimedhi Institute of Science and Technology, Research Institute for Interdisciplinary Science, and Japan Society for the Promotion of Science (KAKENHI Grant JP20H00476) for support, and William Burkholder for reagents. This work was supported by a grant from European Research Council (741773 [AAA]) awarded to L.B. We appreciate the experimental facility and the technical services provided by The Synchrotron Radiation Protein Crystallography Facility of the National Core Facility Program for Biotechnology, Ministry of Science and Technology and the National Synchrotron Radiation Research Center, a national user facility supported by the Ministry of Science and Technology, Taiwan, Republic of China; and by the Australian Synchrotron, part of ANSTO Australian Nuclear Science and Technology Organisation. We thank Professor Jian-Ren Shen and Professor Yuichiro Takahashi for use of reagents and access to equipment and Esra Balıkçı for technical support.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2009167117/-/DCSupplemental.
Data Availability.
The atomic coordinates and structure factors have been deposited in the PDB under accession codes 7C2F, 7C2G, and 7C2H (49–51).
References
- 1.Spang A. et al., Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521, 173–179 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zaremba-Niedzwiedzka K. et al., Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 541, 353–358 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Akıl C., Robinson R. C., Genomes of Asgard archaea encode profilins that regulate actin. Nature 562, 439–443 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Pollard T. D., Cooper J. A., Actin, a central player in cell shape and movement. Science 326, 1208–1212 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Imachi H. et al., Isolation of an archaeon at the prokaryote-eukaryote interface. Nature 577, 519–525 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seitz K. W. et al., Asgard archaea capable of anaerobic hydrocarbon cycling. Nat. Commun. 10, 1822 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai M. et al., Diverse Asgard archaea including the novel phylum Gerdarchaeota participate in organic matter degradation. Sci. China Life Sci. 63, 886–897 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Liu Y. et al., Comparative genomic inference suggests mixotrophic lifestyle for Thorarchaeota. ISME J. 12, 1021–1031 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pollard T. D., Actin and actin-binding proteins. Cold Spring Harb. Perspect. Biol. 8, a018226 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xue B., Robinson R. C., Guardians of the actin monomer. Eur. J. Cell Biol. 92, 316–332 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Nag S., Larsson M., Robinson R. C., Burtnick L. D., Gelsolin: The tail of a molecular gymnast. Cytoskeleton 70, 360–384 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Ghoshdastider U., Popp D., Burtnick L. D., Robinson R. C., The expanding superfamily of gelsolin homology domain proteins. Cytoskeleton 70, 775–795 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Ampe C., Vandekerckhove J., The F-actin capping proteins of Physarum polycephalum: cap42(a) is very similar, if not identical, to fragmin and is structurally and functionally very homologous to gelsolin; cap42(b) is Physarum actin. EMBO J. 6, 4149–4157 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blanchoin L., Pollard T. D., Mechanism of interaction of Acanthamoeba actophorin (ADF/Cofilin) with actin filaments. J. Biol. Chem. 274, 15538–15546 (1999). [DOI] [PubMed] [Google Scholar]
- 15.Goode B. L., Drubin D. G., Lappalainen P., Regulation of the cortical actin cytoskeleton in budding yeast by twinfilin, a ubiquitous actin monomer-sequestering protein. J. Cell Biol. 142, 723–733 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moseley J. B. et al., Twinfilin is an actin-filament-severing protein and promotes rapid turnover of actin structures in vivo. J. Cell Sci. 119, 1547–1557 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Paavilainen V. O. et al., Structural conservation between the actin monomer-binding sites of twinfilin and actin-depolymerizing factor (ADF)/cofilin. J. Biol. Chem. 277, 43089–43095 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Hatanaka H. et al., Tertiary structure of destrin and structural similarity between two actin-regulating protein families. Cell 85, 1047–1055 (1996). [DOI] [PubMed] [Google Scholar]
- 19.Huang S. et al., Arabidopsis VILLIN1 generates actin filament cables that are resistant to depolymerization. Plant Cell 17, 486–501 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McLaughlin P. J., Gooch J. T., Mannherz H. G., Weeds A. G., Structure of gelsolin segment 1-actin complex and the mechanism of filament severing. Nature 364, 685–692 (1993). [DOI] [PubMed] [Google Scholar]
- 21.Paavilainen V. O., Oksanen E., Goldman A., Lappalainen P., Structure of the actin-depolymerizing factor homology domain in complex with actin. J. Cell Biol. 182, 51–59 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xue B., Leyrat C., Grimes J. M., Robinson R. C., Structural basis of thymosin-β4/profilin exchange leading to actin filament polymerization. Proc. Natl. Acad. Sci. U.S.A. 111, E4596–E4605 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nag S. et al., Ca2+ binding by domain 2 plays a critical role in the activation and stabilization of gelsolin. Proc. Natl. Acad. Sci. U.S.A. 106, 13713–13718 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanaka K. et al., Structural basis for cofilin binding and actin filament disassembly. Nat. Commun. 9, 1860 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chumnarnsilpa S. et al., Calcium ion exchange in crystalline gelsolin. J. Mol. Biol. 357, 773–782 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Bork P., Sander C., Valencia A., An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc. Natl. Acad. Sci. U.S.A. 89, 7290–7294 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gunning P. W., Ghoshdastider U., Whitaker S., Popp D., Robinson R. C., The evolution of compositionally and functionally distinct actin filaments. J. Cell Sci. 128, 2009–2019 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Da Cunha V., Gaia M., Gadelle D., Nasir A., Forterre P., Lokiarchaea are close relatives of Euryarchaeota, not bridging the gap between prokaryotes and eukaryotes. PLoS Genet. 13, e1006810 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Da Cunha V., Gaia M., Nasir A., Forterre P., Asgard archaea do not close the debate about the universal tree of life topology. PLoS Genet. 14, e1007215 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eme L., Spang A., Lombard J., Stairs C. W., Ettema T. J. G., Archaea and the origin of eukaryotes. Nat. Rev. Microbiol. 15, 711–723 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Spang A. et al., Asgard archaea are the closest prokaryotic relatives of eukaryotes. PLoS Genet. 14, e1007080 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Isambert H. et al., Flexibility of actin filaments derived from thermal fluctuations. Effect of bound nucleotide, phalloidin, and muscle regulatory proteins. J. Biol. Chem. 270, 11437–11444 (1995). [DOI] [PubMed] [Google Scholar]
- 33.Almo S. C., Pollard T. D., Way M., Lattman E. E., Purification, characterization and crystallization of Acanthamoeba profilin expressed in Escherichia coli. J. Mol. Biol. 236, 950–952 (1994). [DOI] [PubMed] [Google Scholar]
- 34.Gressin L., Guillotin A., Guérin C., Blanchoin L., Michelot A., Architecture dependence of actin filament network disassembly. Curr. Biol. 25, 1437–1447 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Otwinowski Z., Minor W., Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 36.Kabsch W., XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang H., Robinson R. C., Burtnick L. D., The structure of native G-actin. Cytoskeleton 67, 456–465 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Adams P. D. et al., The Phenix software for automated determination of macromolecular structures. Methods 55, 94–106 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murshudov G. N., Vagin A. A., Dodson E. J., Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997). [DOI] [PubMed] [Google Scholar]
- 41.Chen V. B. et al., MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang J., Zhang Y., I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 43, W174–W181 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Madden T. L., Tatusov R. L., Zhang J., Applications of network BLAST server. Methods Enzymol. 266, 131–141 (1996). [DOI] [PubMed] [Google Scholar]
- 44.Hulo N. et al., The 20 years of PROSITE. Nucleic Acids Res. 36, D245–D249 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pei J., Grishin N. V., PROMALS3D: Multiple protein sequence alignment enhanced with evolutionary and three-dimensional structural information. Methods Mol. Biol. 1079, 263–271 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dereeper A. et al., Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, W465–W469 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Choe H. et al., The calcium activation of gelsolin: Insights from the 3A structure of the G4-G6/actin complex. J. Mol. Biol. 324, 691–702 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Robinson R. C., Akil C., Crystal Structure of the Thorarchaeaota ProGel/rabbit actin complex. Protein Data Bank. https://www.rcsb.org/. Deposited 7 May 2020.
- 50.Robinson R. C., Akil C., Crystal Structure of the Thorarchaeaota 2DGel/rabbit actin complex. Protein Data Bank. https://www.rcsb.org/. Deposited 7 May 2020.
- 51.Robinson R. C., Akil C., Crystal Structure of the Thorarchaeaota 2DGel3/rabbit actin complex. Protein Data Bank. https://www.rcsb.org/. Deposited 7 May 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structure factors have been deposited in the PDB under accession codes 7C2F, 7C2G, and 7C2H (49–51).