Abstract
Protein loops make up a large portion of the secondary structure in nature. But very little is known concerning loop closure dynamics and the effects of loop composition on fold stability. We have designed a small system with stable β-sheet structures, including features that allow us to probe these questions. Using paired Trp residues that form aromatic clusters on folding, we are able to stabilize two β-strands connected by varying loop lengths and composition (an example sequence: RWITVTI – loop – KKIRVWE). Using NMR and CD, both fold stability and folding dynamics can be investigated for these systems. With the 16 residue loop peptide (sequence: RWITVTI-(GGGGKK)2GGGG-KKIRVWE) remaining folded (ΔGU = 1.6 kJ/mol at 295K). To increase stability and extend the series to longer loops, we added an additional Trp/Trp pair in the loop flanking position. With this addition to the strands, the 16 residue loop (sequence: RWITVRIW-(GGGGKK)2GGGG-WKTIRVWE) supports a remarkably stable β-sheet (ΔGU = 6.3 kJ/mol at 295K, Tm = ~55 °C). Given the abundance of loops in binding motifs and between secondary structures, these constructs can be powerful tools for peptide chemists to study loop effects; with the Trp/Trp pair providing spectroscopic probes for assessing both stability and dynamics by NMR.
Graphical Abstract
Introduction
Many proteins have relatively long loops with no defined secondary structure; in some cases, these segments are so flexible that they do not even appear in structures derived by X–ray crystallography. Variable length loops have a major role, connecting the secondary and tertiary structures within proteins, and even though such loops are flexible they are often important parts of bio-recognition binding sites1.
Loops makeup the majority of the binding interfaces for antibodies2,3, connecting portions of the β-sheets, corresponding to the variable regions modified to provide specific antigen binding. Because of this, stabilizing loops has been of interest in the creation of small synthetic vaccines and antibodies4,5, such as “monobodies” that mutate these binding loops into the smaller structure of Fibronectin type III6,7. These efforts have, to date, relied heavily on disulfide linkages and synthetic scaffolds8,9. For example, Hijnen9 et al. demonstrated that a synthetic vaccine, made from three discontinuous loops of the epitope of the pertactin protein of Bordetella pertussis, is able to give an antibody response.
Hard to target protein-protein interfaces are also commonly covered in loops, because of the large areas that the interactions take place over. The protein complex of vascular endothelial growth factors (VEGF) and receptor was found to be mediated by a 13 residue loop contained on an exterior β-hairpin of the VEGF. Diana10 et al., found that the hairpin of placental growth factor (HPLW of PlGF), when excised from the protein, still exhibited 32 μM affinity to the receptor. In addition to acting as segments participating in binding, loop sequences also play a crucial role as gatekeeper to catalytic sites deep within proteins11–13, with changes in the motion of such loops modulating the turnover rates of the enzyme.
Loops also act as a tuning mechanism for the overall dynamics of the protein. When a loop is attached to a region of a protein structure, the extensive segmental motion within the loop produces added flexibility near the anchor points in secondary structure elements, β-strands or helices, connected by the loop14. It was found that nature varies the loop length of proteins depending on the different environmental conditions15, with longer loops found in cold environments (psychrophile) and shorter loops found in warmer conditions (thermophiles). This has led protein engineers to use loop truncation as a common method of producing hyperthermostable proteins from mesophilic counterparts16,17.
β-Sheets are most vulnerable to unfolding at their termini. In a protein, this constitutes the connecting turns or loops, in a β-hairpin this includes the N and C termini and the reversing turn. With the addition of β-capping18 and turn optimization19,20, stable β-hairpins which include close matches to the sequence and structure of specific β-strand pairs can be created quite routinely. In such cases, β-capping prevents terminal fraying and loop shortening or turn optimization prevents residual segmental motion from pulling the strands apart from within. However, this could create a problem if the bioactive segment is contained within the loop.
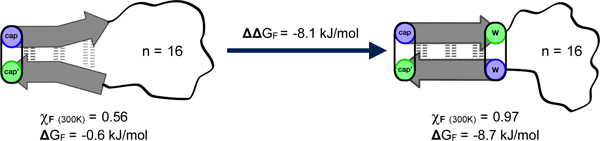
We set as our aim, the construction of β-strand—loop—β-strand motifs with minimalist β-strands, but still capable of forming stable folded structures that can incorporate quite long and variable loop sequences. We reported the first reasonably successful version of this motif, Ac-WITVTI-G3GKKGG3-KKIRVWTG-NH2, in 201018. This example of a β-sheet capped loop displayed 2-state folding with ΔGU280 = 2 kJ/mol (a folded state population, χF = 0.70). A key feature that provides stability to this small antiparallel β-sheet is a cross-strand indole/indole cluster (Figure 1) of defined geometry that also requires ancillary coulombic and/or H-bonding interactions21. Further developments have led to improved β-sheet caps that can be used with a wider variety of loops including ones as long as 16-residues22. The more recently developed β-caps22,23 include examples with the following terminal units - RW / WE, KW / WE, HW / WD, RW / WD – which correspond to W/W interactions combined with favorable π-cation and Coulombic interactions. Our studies18,21–24 indicate that W/W containing β-caps provide 9 – 12 kJ/mol of fold stabilization.
Figure 1.
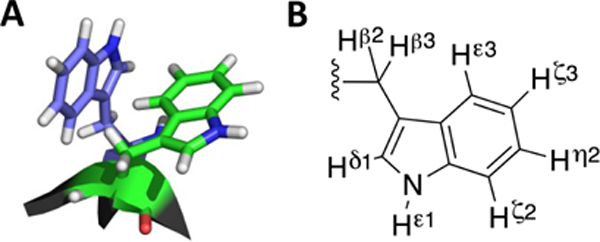
(A) Example of a Trp/Trp edge-to-face (EtF) aromatic cluster. (B) The protons of the Tryptophan sidechain.
Both turn-flanking and strand-terminal Trp/Trp pairs in capping units result in an exciton couplet (with [θ]228 on the order of +400,000° in Molar units21) and provide ring current shifted aryl-H signals that can serve as dynamics probes22,25–27, see Figure 1. A recently prepared construct with a loop length of 28 residues presents a surprisingly stable folded state; at 300K the folded state mole fraction is greater than 0.77. Here we provide the steps that went into the development of such stable β-sheet capped loops and details regarding the fold stabilities and 2-state folding behavior of the constructs examined along the way.
Materials and Methods
Peptide Synthesis
All peptides were synthesized using standard automated FMOC-based solid phase peptide synthesis, on a CEM Liberty blue microwave assisted synthesizer. Preloaded Wang resin was used for peptides with acid termini, while peptides with amidated C-termini were made on rink amide resin. Synthesis conditions used DIC/Oxyma for coupling and Piperidine for deprotection at a 5X excess. Peptides were cleaved from resin with a trifluoroacetic acid (TFA) cocktail using triisopropylsilane (TIPS) and water as radical scavangers (TFA:TIPS:H2O, 38:1:1). The crude peptides were washed 3 times in cold diethyl ether and purified using RP-HPLC, to ≥ 97% purity, with a gradient from 10–50% acetonitrile in water (with 0.1% TFA) over 25 mins. Purity and composition (mass and sequence) were confirmed using an Esquire Ion Trap ESI mass spectrometer (Bruker Daltonics). The purified peptides were lyophilized before use in experiments.
Spectroscopic Characterization
NMR samples were made by dissolving lyophilized peptide (~1 mM) in 20 mM potassium phosphate buffer with 10% D2O as the lock solvent and an internal standard of sodium 4,4-dimethyl-4-silapentane-1-sulfonate (DSS). Each peptide was fully assigned using backbone connectivities from 2D 1H TOCSY and NOESY (Bruker DRX 500, AV 700, or AV 800) spectra. The resulting NMR data has been deposited into our CSDb database (http://andersenlab.chem.washington.edu/CSDb). Chemical shift deviations (CSDs) were calculate by the subtraction of the known chemical shift at random coil from the observed shift using an in-house algorithm accounting for neighboring inductive effects and HN temperature and pH fluctuations, it is available within the CSDb20,28–30. Molar fraction folded (χF) values were calculated using CSDs from a number of diagnostic probes both within the β-strands and aromatic protons of the capping aromatic clusters, specifics for each system may be found in the supporting information. The excellent agreement (at χF = 0.40 – 0.90) between χF-values derived from widely spaced probes is a particularly strong validation of 2-state melting behavior.
CD samples were made by dissolving lyophilized peptide in 20 mM potassium phosphate buffer creating a stock solution of ~200 μM. The concentration was calculated based on the UV absorption of the peptide at 280 nm (ε280nm = 5690 and 1280 M−1 cm−1 for Trp and Tyr respectively). The sample was then diluted to 30 μM for measurements in 0.1 cm path length cells. Spectra were collected on a JASCO J-720 spectrophotometer from 190–270 nm and processed as previously described20,31.
Context and Results
Cross-strand indole/indole clusters in anti-parallel β-sheet structures provide significant fold stabilization when properly placed at non-H-bonded sites18,21. As previously noted, greater β-sheet stabilization is achieved with a W/W interaction involving Trp residues placed at the most terminal non-H-bonded sites of the associated β-strand; we designate such systems as “β-caps”18. It became apparent that the initial-developed alkanoyl-W/WTG cap did not provide optimal stabilization but it does present a unique feature for peptide conjugation and oligoimerization; the alkanoyl unit can be a small molecule with multiple –CO2H units24. We have been able to do this with numerous di-, tri- and even tetracarboxylic acids resulting in structured hairpins becoming linked to each carboxyl unit. This is one of the key discoveries that set the stage for our exploration of “Peptide Origami”32. The W/W interactions in β-caps were first examined as chemical shift probes for measuring the extent of folding at the termini and as probes for NMR relaxation-based dynamics measurements26. The present account focuses on β-capping development and how to improve fold stability in systems containing long loops.
The successful circular permutation of a WW domain22 provides an additional example of the fold-stabilizing effect of a W/W containing β-cap. In that case we employed a RW / WD cap, which was at the extreme ends of the sequence. This unit replaced the turn of the fold-nucleating, first hairpin of the WT Pin1 WW domain. The RW / WD cap is another example of a Coulombic β-cap. The edge-to-face geometry of the two Trp’s are clearly evident in the NMR structure in the derived NMR structure as it appears in Fig. 2 (panel B).
Figure 2.
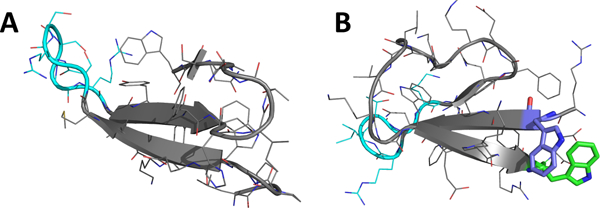
Structural Comparisons: (A) Wild-type Pin1 WW domain (from x-ray structure 3TCZ37. (B) Circular permutant cpPin1(−1)v3 made using a RW/WD β-cap (shown in purple and green, from NMR structure 2MDU22 ).
Turning to the stages in the development of β−caps, including the improvement that are reported for the first time herein, these can be conveniently summarized, as shown in Table 1. We are including the reference species in which the longer loop is replaced by short sequences that are known to favor hairpin formation (entries 3, 7, 9, and 14). Since these stable structures could be fully characterized by NMR, they provide expectation values for chemical shifts and exciton couplet magitudes corresponding to a fully-folded β-sheet state. Table 1 includes a number of reference species and stages in β-cap development that have not been reported previously.
Table 1.
The β-cap optimization process as it applies to loop closure.
| Entry # | System | Loop Length | Sequence cap – β-strand – loop – β-strand – cap’ | ΔGU (kJ/mol) | Fraction Folded (χF)c | |
|---|---|---|---|---|---|---|
| 280 K | 300K | 320K | ||||
| 1 | 10 | Ac-WVTI - G3GKKGG3 - KKIWTG-NH2 | − 2.2 | -- | <0.10 | |
| 2 | 10 | + 3.2 | -- | 0.49 | ||
| 3 | B2 | 2 | Ac-WITVTI - GG - KKIRVWTG-NH2 | + 9.0 | 0.96 | 0.91 |
| 4 | B2 | 4 | Ac-WITVTI – GGGG - KKIRVWTG-NH2 | + 2.9 | 0.72 | 0.58 |
| 5 | B2 | 10 | Ac-WITVTI - G3GKKGG3 - KKIRVWTG-NH2 | + 2.1 | 0.57 | 0.35 |
| 6 | B4 | 10 | Ac-WITVRIW-G3GKKGG3-WKTIRVWTG-NH2 | + 7.3 | 0.93 | 0.85 |
| 7 | B4 | 4a | Ac-WITVRIW - SNGK - WKTIRVWTG-NH2 | > 10 | 0.99 | 0.99 |
| 8 | C2 R1H | 10 | HWITVTI - G3GKKGG3 – KKIRVWE | + 3.8 | 0.75 | 0.57 |
| 9 | C2 | 2 | RWITVTI - GG - KKIRVWE | > 11 | 0.99 | 0.99 |
| 10 | C2 | 4 | RWITVTI - GGGG - KKIRVWE | + 5.8 | 0.88 | 0.74 |
| 11 | C2 | 10 | RWITVTI - (G3GKK)GG3 - KKIRVWE | + 5.0 | 0.84 | 0.59 |
| 12 | C2 | 16 | RWITVTI - (G3GKK)2GG3 - KKIRVWE | + 2.1 | 0.56 | 0.38 |
| 13 | C2 | 22 | RWITVTI - (G3GKK)3GG3 - KKIRVWE | − 1.7 | 0.29 | 0.25 |
| 14 | C4 | 4a | RWITVRIW - IGGK - WKTIRVWE | > 11 | 0.99 | 0.99 |
| 15 | C4 | 10 | RWITVRIW - G3GKKGG3 - WKTIRVWE | > 10 | 0.98 | 0.95 |
| 16 | C4 | 16 | RWITVRIW- (G3GKK)2GG3 -WKTIRVWE | + 8.6 | 0.97 | 0.92 |
| 16Y | 16 | RWITVRIY- (G3GKK)2GG3 -WKTIRVWE | + 7.1 | 0.96 | 0.89 | |
| 17 | C4 | 22 | RWITVRIW- (G3GKK)3GG3 -WKTIRVWE | + 5.5b | 0.84b | 0.61b |
| 18 | C4 | 28 | RWITVRIW- (G3GKK)4GG3 -WKTIRVWE | + 4.0b | 0.77b | 0.45b |
Loop size could be considered 4 or 2 residues since the outer residues of a turn are part of the β-strand.
Fraction folded values estimated by CD, values by NMR not possible due to line broadening.
Fraction folded values represent an average of multiple CSDs (further explained in the Supporting Information) the agreement of these values provides an error of ~0.04 when the mole fraction-folded was in the χF = 0.90 range. In the χF = 0.35 – 0.90 range, the agreement over the multiple probes provides an error of ~0.08.
Variable length loops were incorporated into four systems, two using a hydrophobic cap -- system B2 = Ac-WITVTI – loop – KKIRVWTG-NH2, and B4 = Ac-WITVRIW – loop – WKTIRVWTG-NH2 -- and two using a Coulombic cap -- C2 = RWITVTI – loop – KKIRVWE, and C4 = RWITVRIW – loop – WKTIRVWE. Fold stability and the melting behavior of these peptides was monitored using NMR and CD. The NMR data for multiple probes throughout the β-strands and in the cap, confirm β-strand association (and 2-state melting/folding); whereas CD monitors only the exciton couplet created by Trp/Trp EtF interactions. In the case of constructs with very long loops, CD became the most valuable tool due to significant broadening of the shifted HN and aromatic proton signals in the NMR spectra.
In the initial versions of our “capped loop” models for β-strand association, a tight hydrophobic interaction (containing an indole/indole cluster, a W/W interaction, and some buried H-bonds) between cap and cap’ as well as at least 4 cross-strand H-bonds, between the two β-strands are required to achieve a stable folded state. Each system is constructed similar to a β-hairpin, with a capped β-strand at each terminus, connected by a flexible linker. (Figure 3 demonstrates the system using peptide 11)
Figure 3.
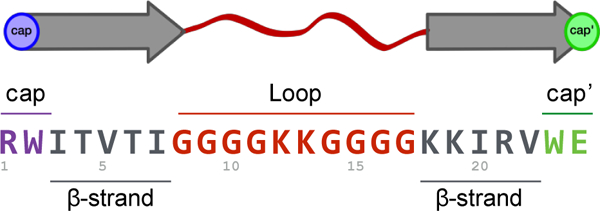
Each peptide’s sequence can be broken down into cap (purple) - β-strand (grey) - loop (red) – β-strand (grey) – cap’ (green).
The requirement for a >4-residue β-strand length can be seen in entries 1, 2 and 5 (comparisons in Table 1). The shorter strand system (1) had to be stapled with a cross-strand disulfide to form a moderately stable β-sheet fold (entry 2). The addition of two-residues to the β-strand, increases fold stability by 4 kJ/mol in this case. The construct of entry 518 served as the first validated long-loop AcW / WTG β-capping motif; entry 3 is the stable hairpin with the same strands that provides the chemical shift and CD measures that served as standards for the expectations for the 100%-folded state. In 2014, this end-capping motif was improved with better terminal and sidechain coulombic interactions23, an RW / WE cap, resulting in a variant of 3, that was in excess of 99% folded from 280–320K, 9. We have now extended these capped loop designs in several ways and produced a significant body of fold stability data (Table 1).
With entries 3, 7, 9, and 14 (which form stable hairpin structures, see Supporting Fig. S1) as “fully-folded” controls for chemical shift based measurement of the extent of folding, we can report χF-values (the mole faction of the folded state) for the longer loop species. The ΔG and χF values refer to β-sheet (and W/W interaction) formation. Individual χF values from strand versus cap proton sites are essentially identical; β-strand alignment and aryl cluster formation in the cap reflect the same two-state folding equilibrium. The nearly identical melting profiles along the β-strands of system C4 with the 16-residue loop, peptide 16, (Figure 4) serves as an example of this 2-state folding behavior, cooperative melting.
Figure 4.
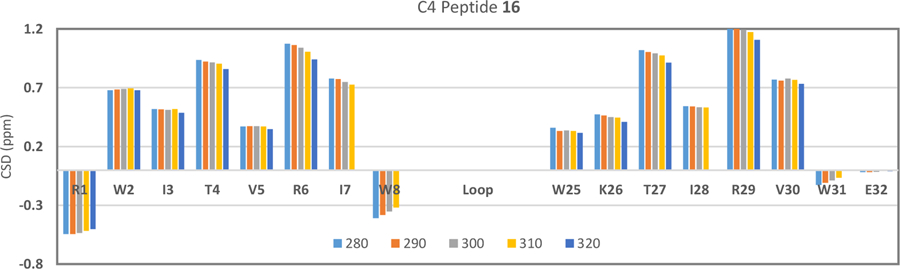
Thermal stability of the β-sheet at the ends of a 16-residue flexible loop as evident in an NMR structuring shift melt. The backbone Hα shifts of peptide 16 are shown as CSDs.
As expected, the signals of ring-current-shifted indole protons within the aryl clusters show some exchange broadening26,33, an initial qualitative analysis suggests25 that three systems with the same –G3GKKGG3- loop (entries 5, 6, 8) display nearly the same folding times, even though they have quite different fold stabilities. These constructs display chemical shifts for most of the loop residues, -GGGGKKGGGG-, that indicate rapid sampling of multiple conformations resulting in fully random chemical shift values (± 0.04 ppm). This validates our assumption and a key feature of our model: that the loop region remains a flexible linker in both the unfolded and folded state. The fully-flexible linker statement also applies to even longer loops (entries 12, 13 and 16 - 18). With loop lengths ≥10 there should, in principle, be no additional entropic penalty to achieving the folding transition state as the loop length increases. In contrast, the loop elongation from 2 to 4 residues, entries 3-4 and 9-10 (for systems B2 and C2 respectively), show a decrease in 5–6 kJ/mol in stability at 280K. This is much greater than expected for a 2 amino acid increase. Since the loop remains short, there are only a finite number of conformational solutions to reversing the direction of the backbone, leading to a large entropic cost. As the loop is extended the number of solutions increases: the entropic cost for each individual glycine addition is offset. This process is evident as the loop is increased from 4–10 residues. There is only a 0.8 kJ/mol loss between loop size 4 and 10 in both systems (peptides 4–5 and 9–11 in systems B2 and C2 respectively).
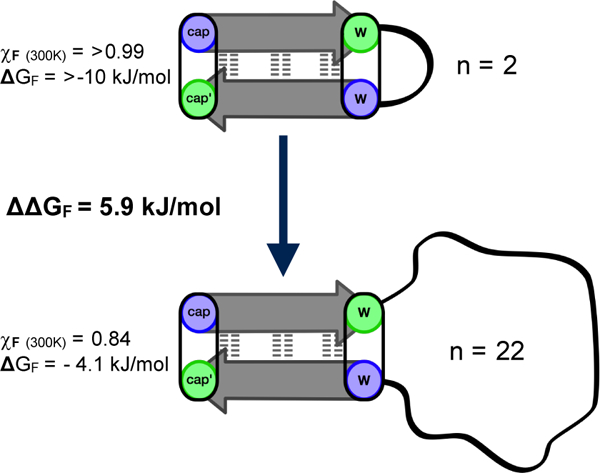
As expected, upon lengthening the loop, some fold stability is gradually lost. The stability loss upon going from a hairpin structure to a 10-residue-capped β-structure could be measured (~7 kJ/mol) with some accuracy only for system B2, the less effective cap. In system C2, the effect appears to be 6 kJ/mol, but might be greater because the overwhelming stability of 9 can only be estimated as >11 kJ/mol. With this somewhat more stable β cap, it was possible to extend the loop length to 16 and even 22 residues; although, at the 22-residue length the fractional population of the folded state dropped below 0.5 even at 280K (χF = 0.29 at 300K). Peptide 12 represents the point at which the elongated loop has become larger than the structured region of the peptide, with 14 residues of β-structure and 16 residues of unstructured loop. Surprisingly, 12 remains 71% folded at 280K, but drops to 38% at 320K. Most of this loss in stability is from the excess motion caused by the large loop, evident from the loss in CSD at different 1H sites, upon loop elongation. Figure 5 shows the change in CSD at the diagnostic Hζ3 site of the C-terminal Trp, and I7 HN for peptides 9, 11 and 12 (loop size = 2, 10 and 16 residues). It shows that although peptide 12 still remains 82% folded at cap’ (Trp Hζ3), it only appears to be ~60% folded at an H-bonded site next to the loop.
Figure 5.
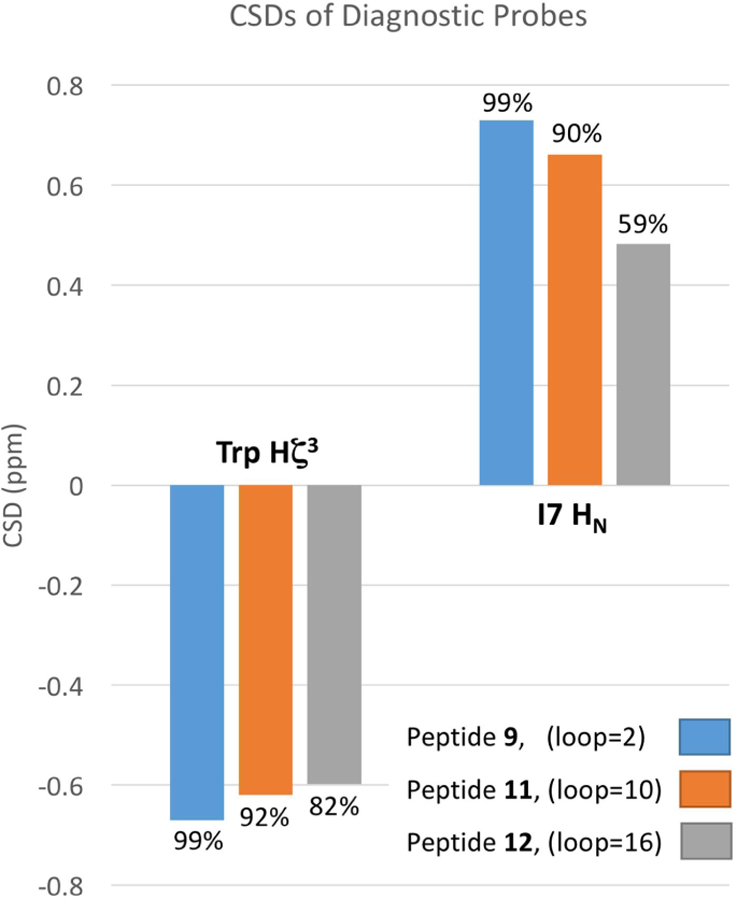
Graph of CSD change of End capping position vs. loop flanking position. Calculated site specific χF are shown as percentages above each bar.
Adding a Loop Flanking Trp/Trp
Inserting an additional Trp/Trp EtF aromatic cluster at the loop flanking position proved to be the most effective method of preventing the segmental motion of the loop from pulling apart the strands. To maintain the register required for EtF cluster formation at both sites, a two-residue insertion was required. The added, loop-flanking Trp’s in entry 6 displayed all of the classic diagnostics of an EtF W/W. As in W/W flanked turns21, the indole of the Trp at the N-terminus of the loop was the “edge” aryl ring in an “edge-to-face” (EtF) interaction, in contrast to “face-to-edge” (FtE) geometry of the end-capping motif. This is also in analogy to observations for W/W-flanked turns in β-hairpins31. This addition seems to add an energetic barrier to β-strand fraying near the loop and it was possible to make systems with longer loops. The addition of the loop-flanking aryl cluster (e.g., entry 5 versus 6 and entry 12 versus 16) resulted in a 5.2 – 6.5 kJ/mol stabilization which is at least 1.5 kJ/mol greater than the stabilization observed for just a 2-residue strand extension, but the stabilization associated with the W/W pairing is significantly less than the 5 – 10 kJ/mol values observed for W/W-flanking turns in β-hairpins21.
With both the improved Coulombic cap (ΔΔGU ≈ 3 kJ/mol) and chain extension with an additional loop-flanking W/W interaction, system C4, the 16 loop construct only shows (Fig. 4) a slight amount of melting (280 – 320K), with χF dropping from ≥0.98 to 0.92. Figure 4 shows this and the typical alternating pattern in the deshielding of Hα sites found in β-sheets. The changes in melting behavior with loop size are shown in Figure 6.
Figure 6.
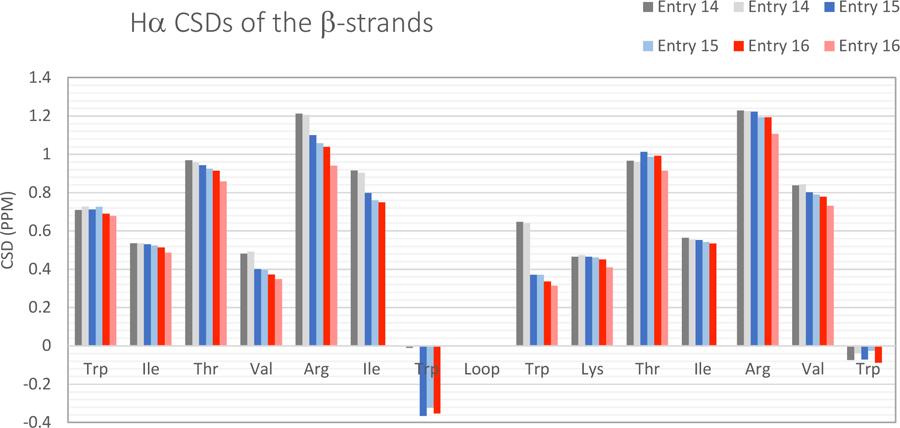
CSDs at 280 and 320K for system C4 loop sized 2/4, 10, and 16 residues (entries 14, 15, and 16 respectively).
Fold stability could also be modulated by manipulation of the loop-flanking aryl cluster. We recently reported21 ΔΔGF values for a W to Y mutation at the “edge” site of EtF W/W clusters: +3.6 and 2.1 kJ/mol, the smaller value applying to a turn-flanking location. A W8Y mutation was made to peptide 16, 16Y, resulting in only a +1.5 kJ/mol drop in stability at 280K: another indication that a loop-flanking EtF interaction is not as favorable as the same interaction flanking a well-defined turn sequence.
Although this system is dramatically more folded than the previous end-capped peptides, segmental motion of the loop is still seen at the Trp residues in the interior. When the CSDs of the aromatic protons on the edge Trp of these two aromatic clusters are compared, there is a different degree of folding at the two sites, as the loop is elongated. Between the end-capping and loop-flanking sites there is a 13% decrease in the apparent extent of folding in the 10 residue loop species, 15, and a 20% in the 16-loop, 16, at 280K (Supporting Fig. S2).
The C4 system was also employed for the longest 22 and 28 residue loops (entries 17 and 18), but many of the 1H NMR probes required for fold population analysis are broadened past visibility. As a result, peptide fold stability was primarily monitored using circular dichroism (CD). An advantage of the CD method, monitoring the Trp/Trp exciton couplet (at 228 nm, 5–95 °C), is that the entire melting range can be seen for each peptide. Figure 7 compares exciton melts for C2 and C4 series. For hairpin forms (the smallest loop size), the exciton couplet maximum for C4 is close to twice that of C2, reflecting the additional W/W interaction flanking the turn. It is apparent, given that the exciton couplet of C2 (reflecting only the β-cap) does not decrease on going from the hairpin to the 10-loop, we conclude that the loss of ellipticity observed for C4 with the same change reflect a greatly diminished exciton couplet magnitude for a loop-flanking versus turn-flanking W/W unit.
Figure 7.
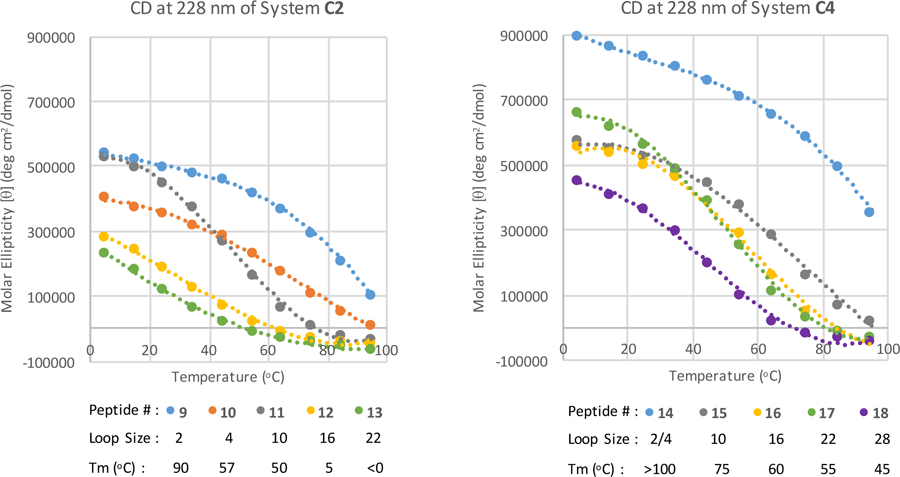
CD maximum at 228nm recorded from 5–95 °C. System C2 peptides 9, 10, 11, 12 and 13 (loop size 2, 4, 10, 16 and 22 residues). System C4 peptides 14, 15, 16, 17 and 18 (loop size 2(4), 10, 16, 22 and 28).
When there is only an end-capping Trp/Trp pair (8–11), a very small initial decrease in the intensity is found (2–10 loop) followed by a large decrease from 10–16 residues. The drop in the low temperature [θ]228-value reflects a lower population of the folded state (consistent with the NMR CSDs). The rapid loss of fold stability on further loop lengthening is also reflected in the Tm significantly decreasing, with an 85 °C drop from 2–16 residues (peptides 9–12).
In the spectra of the C4 peptide series (14–18), a major drop is only seen between the 2 and 10 residue loops. From 10 to 22 residues, minor changes in the melting temperature is observe. This is most likely due to a larger increase in flexibility of the loop flanking Trp/Trp EtF interaction from a 2–10 residue loop increase than from 10–22 residues. Since the increase between 2–10 residues includes this turn to loop transition, it can be thought of as a change from structured to random coil. In contrast, the 10 – 28 residue loop lengthening is already in the unstructured loop realm. It is assumed that peptide 18 is >80% folded at ≤ 25 °C, based on the trend of the CD melting curve (Fig 7). Some of the changes in the low temperature maximum are likely due to impurities from the synthesis, it was found to be difficult to purify peptides with so many repeating glycines and lysines. The 28 residue loop was at the limit of our purification capability. By comparing the CD spectra of peptides with similar sized loops, the drastic effect this additional Trp/Trp pair has on the thermostability of each peptide is evident. At the 16 residue loop size, a Tm increase of 55 °C was found (systems C2 to C4, peptides 12 to 16).
In order to verify that the spectroscopic results seen in these experiments are not due to a higher order assembly of the peptides, CD spectra at various concentrations were taken of the C4 22 loop, peptide 15. With the maximum, at 25 °C, remaining at an average [θ] = +252,000 ± 17,000° at maximum of the exciton couplet from 6.5 – 100 μM (Supporting Fig. S3) confirming the absence of dimers or higher order oligomers. As a further confirmation of the absence of aggregation effects, 1D 1H spectra were taken at 30 μM and 700 μM, with the spectra showing the same peak positions throughout (aromatic and shifted HN regions shown in supporting Figure S4).
Loop Composition Effects on Stability
Loops contained in the peptides mentioned thus far were designed to be as flexible as possible, this exaggerates the effects of natural loops. It is assumed that the effects of the loop are created by the motion caused by the large conformational entropy of each residue within a flexible loop34. If this intrinsic entropy is changed, the effects are found to be perpetuated throughout the system. This was probed by placing a turn-forming sequence at the center of the loop. Ile-(D-Pro)-Gly-Lys (IpGK) is a known turn nucleating sequence35, and was placed into the middle of three different-sized loops (6, 10 and 16 residues). A proline addition introduces “stiffness” to the loop independent of any specific local structuring effect, changing the folding landscape by decreasing the entropy of the unfolded state34,36.
The flexible nature of the loops discussed so far evolves from the conformational openness of the glycine residues in the loop. This leads to segmental motion, which in turn pulls the β-strands apart. Addition of a local area of stiffness, reduces this segmental motion thus creating a more stable system. It was found that in both the B2 and C2 systems there is a larger gain in stability in the smaller loops, and that this stability is more exaggerated at higher temperatures. Insertion of this IpGK segment into a 6 residue loop, 20 and 24, results in a ΔΔGU increase of 4.7 and 3.5 kJ/mol for the B2 and C2 systems respectively, at 300K. Most noticeably, there is a large decrease in melting; peptide 24 remaining fully folded at 320K. In the middle of the 10 and 16 residue loops there is still a large effect, with a free energy gain of 2.9 and 3.5 kJ/mol for the B2 and C2 systems with 10 residue loops, and a 2.4 kJ/mol stability gain in the 16 residue looped C2, 29 (all at 300K). It was also found that another turn sequence, NPATGK, which has a somewhat smaller “turn propensity” in hairpin contexts, has a similar effect. Within the 16 residue loop, 31 remains 63% folded at 320K. Table 2 collects the data for “turn” insertions in flexible loops.
Table 2:
Turn insertion into unstructured loops.
| Entry # | System | Loop Length | Sequence cap – β-strand – loop – β-strand – cap’ | ΔGU (kJ/mol) | Fraction Folded (χF)a | |
|---|---|---|---|---|---|---|
| 280 K | 300K | 320K | ||||
| 19 | B2 | 6 | Ac-WITVTI-GGGGGG-KKIRVWTG-NH2 | + 1.6 | 0.64 | 0.50 |
| 20 | B2 | 6 | Ac-WITVTI-(GIpGKG)-KKIRVWTG-NH2 | + 7.3 | 0.92 | 0.86 |
| 21 | B2 | 8 | Ac-WITVTI-(GNPATGKG)-KKIRVWTG-NH2 | + 2.2 | 0.71 | 0.54 |
| 5 | B2 | 10 | Ac-WITVTI-(G3GKK)GG3-KKIRVWTG-NH2 | + 2.1 | 0.57 | 0.35 |
| 22 | B2 | 10 | Ac-WITVTI-(G3IpGKG3)-KKIRVWTG-NH2 | + 4.7 | 0.81 | 0.63 |
| 23 | C2 | 6 | RWITVTI-GGGGGG-KKIRVWE | + 5.8 | 0.88 | 0.77 |
| 24 | C2 | 6 | RWITVTI - GIpGKG – KKIRVWE | + 6.9 | 0.97 | 0.98 |
| 25 | C2 | 6 | RWITVTI - GIPGKG – KKIRVWE | + 5.9 | 0.92 | 0.85 |
| 26 | C2 | 8 | RWITVTI - GNPATGKG – KKIRVWE | + 5.7 | 0.90 | 0.80 |
| 11 | C2 | 10 | RWITVTI-(G3GKK)GG3-KKIRVWE | + 5.0 | 0.84 | 0.59 |
| 27 | C2 | 10 | RWITVTI - G3IpGKG3 – KKIRVWE | + 6.4 | 0.93 | 0.86 |
| 28 | C2 | 10 | RWITVTI - G3IPGKG3 – KKIRVWE | + 5.1 | 0.90 | 0.72 |
| 12 | C2 | 16 | RWITVTI-(G3GKK)2GG3-KKIRVWE | + 2.1 | 0.56 | 0.38 |
| 29 | C2 | 16 | RWITVTI - G6IpGKG6 – KKIRVWE | + 4.2 | 0.77 | 0.57 |
| 30 | C2 | 16 | RWITVTI - G6IPGKG6 – KKIRVWE | + 3.4 | 0.69 | 0.45 |
| 31 | C2 | 16 | RWITVTI – G5NPATGKG5 – KKIRVWE | + 4.2 | 0.79 | 0.63 |
Fraction folded values represent an average of multiple CSDs (further explained in Supporting Information). Uncertainty in the fully-folded CSD values represents a circa 0.03 fraction-folded error in the χF = 0.90 range. In the χF = 0.35 – 0.90 range, the agreement over the multiple probes provides an error of ~0.09.
This increase in ΔGU for systems with loops containing an IpGK unit, is not just from a reduction in the conformational entropy of the four residues. There is also a gain in stabilization of the folded state by the turn propensity of IpGK to reverse the strands. This was tested using an I(L-Pro)GK turn within the loops (entries 25, 28 and 30). IPGK is, compared to IpGK, significantly less well folded as a turn, although the individual residues have the same conformational space restriction. From the data in Table 2 it is apparent that IPGK addition to loops has a less dramatic stabilizing effect. While insertion of IpGK into a 16 residue loop results in a 2.4 kJ/mol gain in stability, only a 1.4 kJ/mol gain is observed for the corresponding IPGK turn insertion (peptides 29 and 30 respectively, at 300K). While IPGK should yield the same increase in driving force for folding due to decreasing the conformational entropy of the unfolded state, IpGK has an added effect: stabilizing the folded state by ~1–2 kJ/mol.
In hairpin contexts the structuring of the IpGK β-turn is evidenced by the diagnostic HN chemical shifts within the turn, a downfield shifted Ile (S-1 position) and an upfield shifted Lys (T2 position). Smaller but still significant CSDs are observed for position I14 and K16 in peptide 19, 0.23 and −0.42 ppm respectively, even though the turn has 6 glycines on each side. In the L-Pro equivalent (peptide 30) these CSDs are reduced to 0.03 and −0.16 ppm. Additional data appears in Supporting Fig. S5. Based on this chemical shift analysis, these IpGK turns are 30–40% folded, and the extent of β-turn formation appears to be independent of the extent of β-strand association (the ΔGU of the folded state).
Discussion
The stability of a β-sheet is clearly related to the strength of cross-strand H-bonding and hydrophobic interactions, but the loops that connect the strands and the termini are also a consideration. Flexible loops are found to fray the strands internally as the loop size is increased due to the segmental motion of the longer loops. It was found that the addition of Trp/Trp cross-strand aromatic clusters were useful in preventing this motion from propagating into the strands. Using this, small stable β-hairpin-like systems could be created containing long loops. Two main β-capping styles were compared, one using hydrophobic Ac-W, WTG-NH2 (systems B2 and B4) termini18 and a hydrophilic RW, WE termini with additional coulombic interactions23 (systems C2 and C4). It was found that the coulombic capping system had a broader range of use, especially as the loop length increased. With peptides containing 10 residue flexible loops, system B2 (peptide 5) had a χF of 0.57 at 300K, while system C2 had a χF = 0.84 at 300K. Using the C2 system the loop could be elongated to 22 residues before the χF dropped off to a range below 50% folded. In order to remedy this, two methods were found to increase the ΔGU of folded states containing long loops. Surprisingly, insertion of a turn nucleating sequence into the middle of the 16 residue looped C2 system had a substantial effect on stability, ΔΔGU = 3.8 kJ/mol. This is a result of increased chain stiffness and reversal of the backbone direction associated with turns.
The most effective way to stabilize a loop containing system of any size was found to be the inserting of an additional Trp/Trp aromatic cluster at the loop flanking positions (Figure 7). This was found to decrease the effect of the segmental motion of the loop, keeping the β-strands associated all the way to the loop junctions. With the addition of this, both the hydrophobic and Coulombic systems (B4 and C4) became more stable, allowing the study to be extended to a peptide with an equal number of amino acids within the β-strands and in the loop attached to it. Regardless of size, near fully folded stability was observed by CD (Figure 6).
Conclusions
With relatively unstructured loops so pervasive in protein structures, binding motifs, gating of active sites, and at protein-protein interfaces, there is a need for research into their effects on the protein folding landscape. This and their use in protein design has been limited by a lack of smaller peptide systems that isolate and stabilize such loops. We have demonstrated the ability to created stable systems containing a variety of loop lengths. It was found that β-strands connected to these flexible chains were subjected to the segmental motion caused by the loops unstructured nature. To combat this, the strands can be stabilized by the addition of a loop flanking Trp/Trp aromatic cluster. With both a Trp/Trp β-cap and a loop flanking Trp/Trp cluster, a stable (χF = 0.77 at 300K from CD) system containing 28 residues of loop and only 16 residues of β-strand could be realized.
It was also found that the flexibility of the loop could also be reduced by the addition of structured turn regions ( -IpGK- or -NPATGK- sequences) into the middle of the loop. A 2.1 kJ/mol stability gain was possible by this strategy within a loop of 16 residues (peptide 12 verses 29 or 31). Native loops commonly have residues to promote a turn, but also may have specific hydrophobic and H-bonding contacts that would influence folding. Using a combination of loop size, composition and modulation of aromatic clusters, the extent and rigidity of the folding of regions in these small folded peptide motifs can be tuned. This offers a vast amount of control in the rational design of models of important loops within proteins.
Protein engineering has led to great advances, with loops playing an essential role in many functions. As the drive to produce smaller antibody and vaccine biologics increases, an ability to create small stable looped structures can serve as another answer. In addition to the direct benefits towards binding motifs, these systems will provide a way to model how protein loops effect the folding landscape of proteins.
Supplementary Material
Figure 8.
Example of the minimal effects of loop elongation on system C4.
Acknowledgments
We would like to acknowledge the funding that has made this work possible. National Institutes of Health (GM-059658 & -099889) and the National Science Foundation (CHE-0650318 & -1152218, DBI-0439063).
References
- (1).Fox DA; Larsson P; Lo RH; Kroncke BM; Kasson PM; Columbus LJ Am. Chem. Soc 2014, 136 (28), 9938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Ekiert DC; Kashyap AK; Steel J; Rubrum A; Bhabha G; Khayat R; Lee JH; Dillon MA; O’Neil RE; Faynboym AM; Horowitz M; Horowitz L; Ward AB; Palese P; Webby R; Lerner RA; Bhatt RR; Wilson IA Nature 2012, 489 (7417), 526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Sharon M; Kessler N; Levy R; Zolla-pazner S; Go M; Anglister J Structure 2003, 11 (3), 225. [DOI] [PubMed] [Google Scholar]
- (4).Zhai W; Glanville J; Fuhrmann M; Mei L; Ni I; Sundar PD; Van Blarcom T; Abdiche Y; Lindquist K; Strohner R; Telman D; Cappuccilli G; Finlay WJJ; Van Den Brulle J; Cox DR; Pons J; Rajpal AJ Mol. Biol 2011, 412 (1), 55. [DOI] [PubMed] [Google Scholar]
- (5).Skwarczynski M; Toth I Chem. Sci 2016, 7, 842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Batori V; Koide A; Koide S Protein Eng 2002, 15 (12), 1015. [DOI] [PubMed] [Google Scholar]
- (7).Koide A; Wojcik J; Gilbreth RN; Hoey RJ; Koide SJ Mol. Biol 2012, 415 (2), 393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Werkhoven PR; Van De Langemheen H; Van Der Wal S; Kruijtzer JAW; Liskamp RMJ J. Pept. Sci 2014, 20 (4), 235. [DOI] [PubMed] [Google Scholar]
- (9).Hijnen M; van Zoelen DJ; Chamorro C; van Gageldonk P; Mooi FR; Berbers G; Liskamp RMJ Vaccine 2007, 25 (37–38), 6807. [DOI] [PubMed] [Google Scholar]
- (10).Diana D; Basile A; De Rosa L; Di Stasi R; Auriemma S; Arra C; Pedone C; Turco MC; Fattorusso R; D’Andrea LD J. Biol. Chem 2011, 286 (48), 41680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Malabanan MM; Amyes TL; Richard JP Curr. Opin. Struct. Biol 2010, 20 (6), 702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Derreumaux P; Schlick T Biophys. J 1998, 74 (1), 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Chen M; Khalid S; Sansom MSP; Bayley H Proc. Natl. Acad. Sci. U. S. A 2008, 105 (17), 6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Nagi AD; Regan L Fold. Des 1997, 2 (1), 67. [DOI] [PubMed] [Google Scholar]
- (15).Balasco N; Esposito L; De Simone A; Vitagliano L Protein Sci 2013, 22 (7), 1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Villbrandt B; Sagner G; Schomburg D Protein Eng 1997, 10 (11), 1281. [DOI] [PubMed] [Google Scholar]
- (17).Vieille C; Zeikus GJ Microbiol. Mol. Biol. Rev 2001, 65 (1), 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Kier BL; Shu I; Eidenschink LA; Andersen NH Proc. Natl. Acad. Sci. U. S. A 2010, 107 (23), 10466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Haque TS; Gellman SH J. Am. Chem. Soc 1997, 7863 (15), 2303. [Google Scholar]
- (20).Fesinmeyer RM; Hudson FM; Andersen NH J. Am. Chem. Soc 2004, 126 (13), 7238. [DOI] [PubMed] [Google Scholar]
- (21).Anderson JM; Kier BL; Jurban B; Byrne A; Shu I; Eidenschink LA; Shcherbakov AA; Hudson M; Fesinmeyer RM; Andersen NH Biopolymers 2016, 105 (6), 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Kier BL; Anderson JM; Andersen NH J. Am. Chem. Soc 2014, 136 (2), 741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Anderson JM; Kier BL; Shcherbakov AA; Andersen NH FEBS Lett 2014, 588 (24), 4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Kier BL; Andersen NH J. Pept. Sci 2014, 20 (9), 704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Kier BL; Shu I; Kellock JK; Andersen NH In Proc. of the 31st Eur. Peptide Symposium; 2010; pp 508–509.
- (26).Scian M; Shu I; Olsen KA; Hassam K; Andersen NH Biochemistry 2013, 52 (15), 2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Kier BL; Anderson JM; Andersen NH In Proc. of the 23rd American Peptide Symposium; 2013; pp 144–145.
- (28).Andersen NH; Cao B; Chen C Biochem. Biophys. Res. Commun 1992, 184 (2), 1008. [DOI] [PubMed] [Google Scholar]
- (29).Andersen NH; Tong H Protein Sci 1997, 6, 1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Eidenschink L; Kier BL; Huggins KNL; Andersen NH Proteins Struct. Funct. Bioinforma 2009, 75 (2), 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Andersen NH; Olsen KA; Fesinmeyer RM; Tan X; Hudson FM; Eidenschink LA; Farazi SR J. Am. Chem. Soc 2006, 128 (18), 6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Kier BL; Andersen NH J. Pept. Sci 2014, 20 (S1), S71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Olsen KA; Fesinmeyer RM; Stewart JM; Andersen NH Proc. Natl. Acad. Sci. U. S. A 2005, 102 (43), 15483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Matthews BW; Nicholson H; Becktel WJ Proc. Natl. Acad. Sci. U. S. A 1987, 84 (October), 6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Stanger HE; Gellman SH J. Am. Chem. Soc 1998, 120 (7), 4236. [Google Scholar]
- (36).Sriprapundh D; Vieille C; Zeikus JG Protein Eng 2000, 13 (4), 259. [DOI] [PubMed] [Google Scholar]
- (37).Zhang Y; Daum S; Wildermann D; Zhou XZ; Verdecia MA; Bowman ME; Lucke C; Hunter T; Lu KP; Fischer G; Noel JP ACS Chem. Biol 2007, 2 (5), 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


