Abstract.
Purpose: This study aims to develop and compare human-engineered radiomics methodologies that use multiparametric magnetic resonance imaging (mpMRI) to diagnose breast cancer.
Approach: The dataset comprises clinical multiparametric MR images of 852 unique lesions from 612 patients. Each MR study included a dynamic contrast-enhanced (DCE)-MRI sequence and a T2-weighted (T2w) MRI sequence, and a subset of 389 lesions were also imaged with a diffusion-weighted imaging (DWI) sequence. Lesions were automatically segmented using the fuzzy C-means algorithm. Radiomic features were extracted from each MRI sequence. Two approaches, feature fusion and classifier fusion, to utilizing multiparametric information were investigated. A support vector machine classifier was trained for each method to differentiate between benign and malignant lesions. Area under the receiver operating characteristic curve (AUC) was used to evaluate and compare diagnostic performance. Analyses were first performed on the entire dataset and then on the subset that was imaged using the three-sequence protocol.
Results: When using the full dataset, the single-parametric classifiers yielded the following AUCs and 95% confidence intervals: [0.82, 0.87], [0.80, 0.86], and [0.62, 0.75]. The two multiparametric classifiers both yielded AUCs of 0.87 [0.84, 0.89] and significantly outperformed all single-parametric methods classifiers. When using the three-sequence subset, the mpMRI classifiers’ performances significantly decreased.
Conclusions: The proposed mpMRI radiomics methods can improve the performance of computer-aided diagnostics for breast cancer and handle missing sequences in the imaging protocol.
Keywords: breast cancer, computer-aided diagnosis, radiomics, machine learning, multiparametric magnetic resonance imaging
1. Introduction
Breast magnetic resonance imaging (MRI) has shown high sensitivity for breast cancer detection and characterization.1 Although the main sequence in breast MRI exams, the dynamic contrast-enhanced (DCE)-MRI, offers morphological and functional lesion information and provides excellent sensitivity for breast cancer diagnosis, its moderate specificity may lead to unnecessary secondary patient management and anxiety.2 To overcome this limitation and assess additional functional data, multiparametric MRI (mpMRI) has been implemented in the routine clinical interpretation of breast MRI exams.2,3 T2-weighted (T2w) and diffusion-weighted MRI (DWI) are two commonly used sequences in mpMRI examined alongside the DCE sequence. Studies have shown that the incorporation of T2w and DWI sequences during interpretation is useful in the differential diagnosis of benign and malignant lesions.4–9 For example, fibroadenomas, a type of benign lesion that can exhibit similar contrast agent enhancement to that of malignant lesions on T1-weighted DCE-MRI, are usually hyperintense on T2w images, while malignant lesions are usually iso- or hypointense.4 DWI quantifies the random movement of water molecules in tissue, which is influenced by tissue microstructure and cell density. Cancers show decreased water diffusion because of increased cell density, which leads to higher signal intensity at DWI and lower signal intensity on the derived apparent diffusion coefficient (ADC) maps.7–9
As MRI is increasingly used for screening high-risk patients for breast cancer as well as in therapy response monitoring, the ability to distinguish benign from malignant lesions on breast MRI is increasingly important. Computer-aided diagnosis (CADx)/radiomics systems, which extract human-engineered features designed to characterize lesions in terms of intuitive characteristics, continue to be developed to enable artificial intelligence-assisted image interpretation for radiologists and potentially improve diagnostic performance.10–13 As MRI technology advances, multiparametric radiomics methods using multiple MRI sequences have also started to be explored.14–17 In this study, we propose and evaluate the performance of two multiparametric radiomics methods that utilize DCE, T2w, and DWI MRI sequences and show that the complementary information provided in them can improve the diagnostic performance in the task of distinguishing between benign and malignant breast lesions. In addition, we also examine the effect of dataset size and demonstrate the value of handling variability in mpMRI protocols in CADx systems as in clinical settings.
In our machine learning methodology, radiomic features were designed for and extracted from each MRI sequence, and classification was performed using a support vector machines (SVMs). Information from different mpMRI sequences was integrated at two different levels of the classification framework, namely (i) at the feature level by concatenating radiomic features extracted from multiple sequences (feature fusion) and (ii) at the classifier output level by aggregating the outputs from the single-parametric SVMs (classifier fusion). Our methodologies demonstrate strong potential in leveraging multiparametric information from three mpMRI sequences to predict the probability of breast lesion malignancy without the need for preprocessing, image registration, large datasets, or long training times.
2. Materials and Methods
2.1. Database
The database was retrospectively collected under Health Insurance Portability and Accountability Act-compliant Institutional Review Board protocols. All clinical information and images in this study were deidentified to the investigators, and hence consent from the participants was waived. The MRI exams in the database were consecutively acquired from 2007 to 2013 and imaged at a single institution. MRI studies that did not exhibit a visible lesion, lesions that did not have validation of the final diagnosis, or lesions whose DCE time intervals were unknown were excluded. In total, the database used in this study consisted of 852 unique breast lesions from 612 women (mean age, years; age range, 23 to 89 years).
Images in the database were acquired using either 1.5 T (66%) or 3 T (34%) Philips Achieva scanners. Each MR study contained a DCE-MRI sequence and a T2w MRI sequence acquired during the same exam, and exams for a subset of 389 lesions from 299 patients also included a DWI sequence. The scanning sequences for DCE, T2w, and DWI were a T1-weighted spoiled gradient sequence with fat saturation, a T2-w fast spin echo sequence with flow compensation, and a diffusion-weighted fast spin echo sequence with fat saturation, respectively. The DWI sequence contained various degrees (ranging from two to five) of diffusion weighting as measured by the -value. In-plane resolution and slice thickness also varied across the dataset.
Clinical characteristics of the dataset are detailed in Table 1. Of all lesions, 195 were benign (23%) and 657 were malignant (77%) as obtained from pathology and radiology reports. For all lesions categorized at MRI as breast imaging reporting and data system (BI-RADS) category 4, 5, or 6, diagnosis validation was achieved by histopathologic analysis. For all lesions categorized at MRI as BI-RADS category 2 or 3, diagnosis validation was obtained by MRI follow-up of at least 24 months.
Table 1.
Clinical characteristics of the dataset. Patient age is summarized on a patient basis, and lesion information (malignancy status and subtypes) is summarized on a lesion basis. The full set is a mixture of cases imaged using either two or three sequences, and the DWI subset contains cases imaged using three sequences.
| Full set () | DWI subset () | |
|---|---|---|
| Benign/malignant prevalence | Benign: 195 (22.9) | Benign: 66 (17.0) |
| Malignant: 657 (77.1) | Malignant: 323 (83.0) | |
| Age (years): |
||
| Unknown: 96 |
Unknown: 12 |
|
| Benign lesion characteristics | ||
| Lesion subtypes |
Fibroadenoma: 60 (30.8) | Fibroadenoma: 18 (27.3) |
| Columnar change: 15 (7.7) | Columnar change: 5 (7.6) | |
| Papilloma: 13 (6.7) | Papilloma: 6 (9.1) | |
| Parenchyma tissue: 11 (5.6) | Parenchyma tissue: 8 (12.1) | |
| Fibrotic tissue: 10 (5.1) | Fibrotic tissue: 5 (7.6) | |
| Hyperplasia: 8 (4.1) | Hyperplasia: 5 (7.6) | |
| Cystic change: 6 (3.1) | Cystic change: 3 (4.5) | |
| Fat necrosis: 4 (2.1) | Fat necrosis: 3 (4.5) | |
| Other: 26 (13.3) | Other: 12 (18.2) | |
| Unknown: 42 (21.5) |
Unknown: 1 (1.5) |
|
| Malignant lesion characteristics | ||
| Lesion subtypes | IDC: 133 (20.2) | IDC: 71 (22.0) |
| DCIS: 118 (18.0) | DCIS: 20 (6.2) | |
| IDC + DCIS: 316 (48.1) | IDC + DCIS: 197 (61.0) | |
| ILC: 27 (4.1) | ILC: 15 (4.6) | |
| ILC + LCIS: 24 (3.7) | ILC + LCIS: 5 (1.5) | |
| Other: 28 (4.3) | Other: 15 (4.6) | |
| Unknown: 11 (1.7) | ||
| Estrogen receptor status | Positive: 408 (62.1) | Positive: 235 (72.8) |
| Negative: 127 (19.3) | Negative: 83 (25.7) | |
| Unknown: 122 (18.6) | Unknown: 5 (1.5) | |
| Progesterone receptor status | Positive: 350 (53.3) | Positive: 209 (64.7) |
| Negative: 183 (27.9) | Negative: 108 (33.4) | |
| Unknown: 124 (18.9) | Unknown: 6 (1.9) | |
| HER-2 status | Positive: 87 (13.2) | Positive: 54 (16.7) |
| Negative: 401 (61.0) | Negative: 240 (74.3) | |
| Equivocal: 5 (0.8) | Equivocal: 2 (0.6) | |
| Unknown: 164 (25.0) | Unknown: 27 (8.4) | |
Note: Numbers in parentheses are percentages. For some subjects, only the decade of age was available (e.g., 60 s) as part of the patient information deidentification process. In these situations, the middle of the decade was used for the calculation of the mean subject age. DCE, dynamic contrast-enhanced sequence; T2w, T2-weighted sequence; DWI, diffusion-weighted imaging sequence; std, standard deviation; IDC, invasive ductal carcinoma; DCIS, ductal carcinoma in situ; ILC, invasive lobular carcinoma; and HER-2, human epidermal growth factor receptor 2.
2.2. Single-Parametric Methods
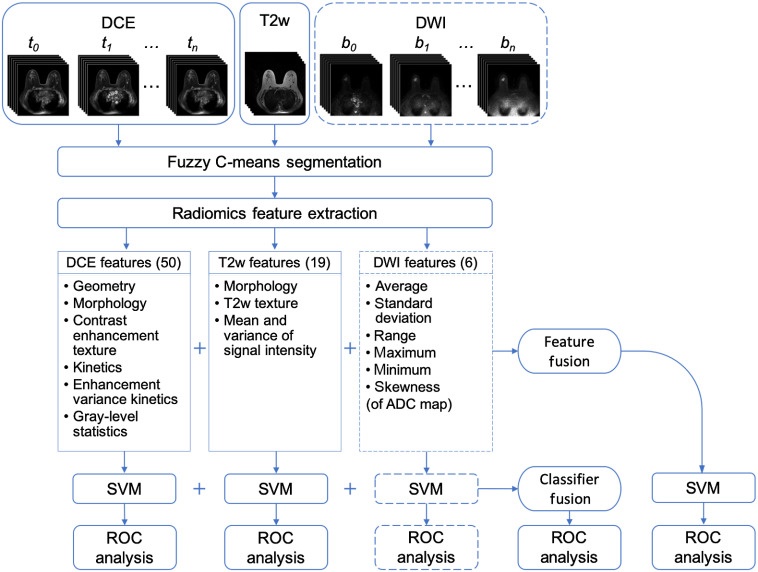
Figure 1 illustrates the human-engineered radiomic features extraction, machine learning classification, and evaluation process for both single-parametric and mpMRI approaches.
Fig. 1.
Lesion classification pipeline based on diagnostic images. Radiomic features were extracted from DCE, T2w, and DWI sequences. The mpMRI information was incorporated in two different ways: feature fusion, i.e., merging radiomic features extracted from all sequences to train an SVM classifier and classifier fusion, i.e., aggregating the PM output from all single-parametric classifiers via soft voting. Parentheses contain the numbers of features extracted from each sequence. The dashed lines for DWI indicate that the DWI sequence was only included in the classification process when it was available, while the DCE and T2w sequences were available for all lesions and thus were always included. ADC, apparent diffusion coefficient and ROC, receiver operating characteristic.
Lesions were segmented separately from each sequence using a fuzzy C-means method requiring only the manual indication of a seed point.18 Radiomic features were designed based on the biological phenotypes of lesions. Fifty radiomic features that characterize lesions in terms of their size, shape, morphology, enhancement texture, kinetics, and kinetics variance were extracted from DCE images.19–24 Likewise, three morphological features and 14 texture features as well as the mean and the variance of the signal intensity were extracted from T2w images.14 In addition, six first-order radiomic features were extracted from the ADC maps of DWI images.25 Morphological or texture features were not calculated from DWI due to its coarse resolution. Radiomic features related to contrast enhancement on DCE sequence were calculated in 4-D, and all other features were calculated in 3-D across the entire lesion. A complete list of radiomic features and their descriptions is included in the Appendix.
SVM classifiers with Gaussian radial basis function kernel were trained on the extracted radiomic features to differentiate between benign and malignant lesions (Python Version 3.7, Python Software Foundation).26 SVM was chosen over other classification methods due to its relative robustness to correlated data, which is an attribute of the radiomic features. Each SVM classifier was trained and evaluated using nested fivefold cross validation, where the inner cross validation was used for model development and the outer cross validation was used for testing. Within each training fold in the outer cross-validation loop, two SVM hyperparameters, namely the scaling parameter and the regularization parameter , were optimized on a grid search with an internal fivefold cross validation.27 Predictions on the five test folds in the outer cross-validation loop were aggregated for classification performance evaluation. Splitting was performed by patient, keeping all lesions from a patient in the same fold to eliminate the bias due to using correlated lesions for training and testing. Class prevalence was held constant across all cross-validation folds. Each training set was standardized to zero mean and unit variance, and the corresponding test set was standardized using the statistics of the training set. To address the problem of class imbalance, a misclassification penalty for cases in each class was assigned to be inversely proportional to its prevalence in the training data.
2.3. Multiparametric Methods
We investigated integrating information from the three MRI sequences at two different levels of the classification framework, as illustrated in Fig. 1. The two mpMRI approaches are referred to as feature fusion and classifier fusion. For the feature fusion approach, radiomic features extracted from each sequence separately were concatenated to form an ensemble of features, which was then input to an SVM classifier. The classifier training process then followed the single-parametric methods. For the classifier fusion approach, probability of malignancy (PM) outputs from the single-parametric SVM classifiers were aggregated via soft voting. That is, the PM outputs were averaged across all single-parametric classifiers to yield prediction scores.
2.4. Evaluation and Statistical Analysis
Classifier performances were evaluated using receiver operating characteristic (ROC) curve analysis, with area under the ROC curve (AUC) serving as the figure of merit.28,29 The 95% confidence intervals (CIs) of the AUCs were calculated by bootstrapping the posterior PMs (2000 bootstrap samples).30 Sensitivity and specificity, calculated at the optimal operating point on the ROC curve that minimizes , were also reported for each classifier.15
Classification performances of the mpMRI approaches were compared with those of the single-parametric classifiers using the DeLong test.31,32 Bonferroni–Holm corrections were used to account for multiple comparisons,33 and a corrected was considered to indicate a statistically significant difference in performance.
2.5. Protocol Variability
Missing data are a common challenge in multimodality imaging studies. Conventional methods typically discard modality-incomplete subjects, which reduces the subjects that can be used to train a diagnosis model and hence may degrade the diagnostic performance. To mimic potential clinical situations where radiologists perform assessments based on MRI exams acquired using different imaging protocols that contain various number of sequences, the analyses were first performed on the entire dataset of 852 lesions, in which exams contained either two or three sequences. For the feature fusion approach, an SVM classifier was trained on features extracted from three sequences for the subset of lesions for which all three sequences were acquired during their MRI exams, and another SVM classifier was trained on features extracted only from DCE and T2w sequences for the remaining lesions for which DWI was not acquired. For the classifier fusion approach, output PMs from all applicable single-parametric SVM classifiers were aggregated via soft voting, and subsequently input to ROC analysis and sensitivity/specificity calculations.
The same analyses were then performed on the subset of 389 lesions whose mpMRI protocol contained three sequences, discarding the modality-incomplete subset. The performances of mpMRI classifiers trained on this subset were compared with those trained on the full dataset to demonstrate the effect of the dataset size and the benefit of using all available data even when a subset contains missing sequences.
3. Results
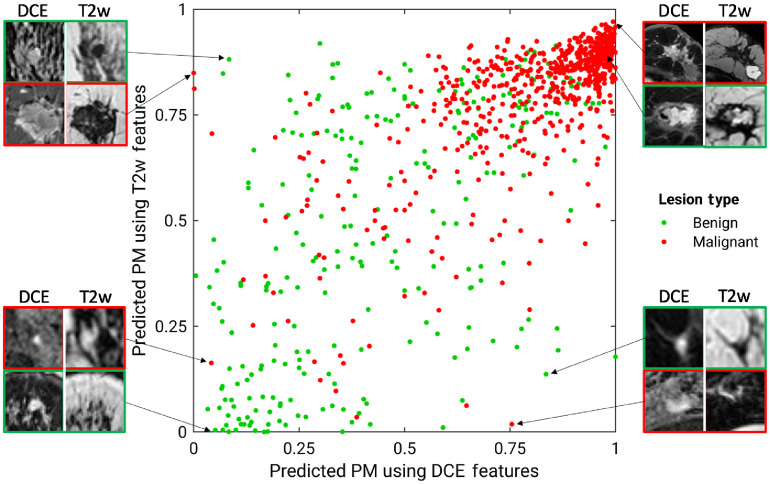
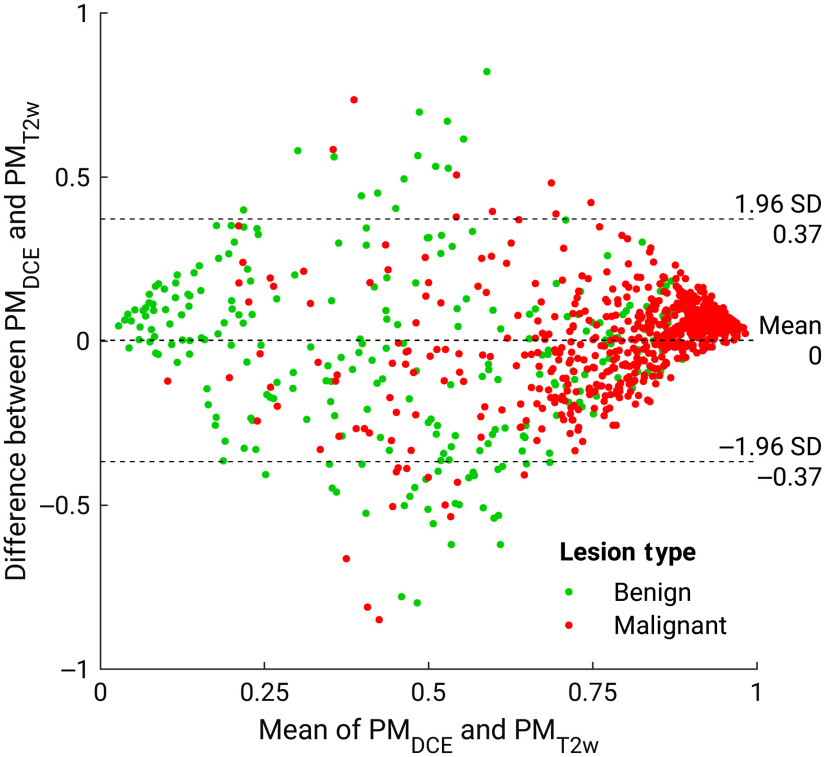
Figures 2 and 3 show the comparison between the PMs predicted by the single-parametric classifiers using DCE and T2w features. Although the majority of benign and malignant classes are separated from each other, there exists notable disagreement between the two single-parametric classifiers, suggesting that a fusion technique for features extracted from various mpMRI sequences may improve the predictive performance. Figure 2 also shows example lesions upon which these two classifiers agree or disagree, with their lesion types noted in the caption. For example, the benign papilloma lesion on the lower right was inaccurately predicted to have a high PM score using DCE features, but more accurately assigned with a low PM score when using T2w features, providing an example where combining features from mpMRI sequences would be beneficial.
Fig. 2.
Diagonal classifier agreement plot between the T2w and DCE single-parametric classifiers. The axis and axis denote the PM scores predicted by the classifiers using DCE and T2w features, respectively. Each point represents a lesion for which predictions were made. Points along or near the diagonal from bottom left to top right correspond to high classifier agreement; points far from the diagonal correspond to low agreement. Examples of lesions on which the two classifiers were in extreme agreement/disagreement are also included. Disagreement: lower right benign: papilloma; lower right malignant: mixture of invasive ductal carcinoma and ductal carcinoma in situ, HER-2 enriched; upper left benign: fibroadenoma; upper left malignant: mixture of invasive ductal carcinoma and ductal carcinoma in situ, luminal A. Agreement (both incorrect): upper right benign: hyalinized stromal fibrosis; lower left malignant: ductal carcinoma in situ. Agreement (both correct): upper right malignant: mixture of invasive ductal carcinoma and ductal carcinoma in situ, triple negative, very large; and lower left benign: fibroadenoma.
Fig. 3.
Bland–Altman plot illustrating classifier agreement between the single-parametric classifiers trained on DCE features and T2w features. The axis shows the difference between the SVM output scores of the two classifiers; the axis shows the mean of two classifiers’ outputs.
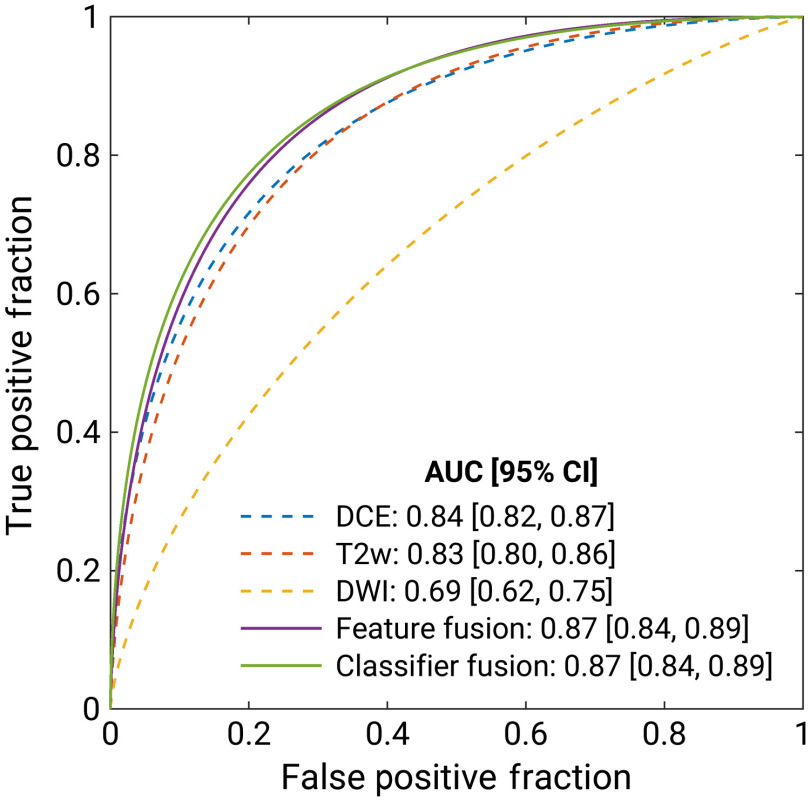
Figure 4 and Table 2 present the classification performances of the five classification models trained on the full dataset of 852 lesions imaged using either two- or three-sequence mpMRI protocols. Table 3 summarizes the -values and the 95% CIs for the comparisons between the multiparametric and single-parametric classifiers’ AUCs. Both mpMRI classification approaches significantly outperformed all single-parametric classifiers.
Fig. 4.
Fitted binomial receiver operating characteristic (ROC) curves for single-parametric (dashed line) and mpMRI classifiers (solid line) trained on the full set. The three single-parametric classifiers were trained separately on (i) DCE, (ii) T2w, and (iii) DWI features. The mpMRI models (iv) were trained on the ensemble of features extracted from all available sequences, and (v) aggregated the PM from the single-parametric classifiers via soft voting. The legend gives the AUC with the 95% CI for each classifier.
Table 2.
Sensitivity, specificity, and AUC along with the 95% CI of AUC for each classifier trained on the full set. Sensitivity and specificity presented are for the optimal operating point determined using a metric for cut-off value that minimizes . Because all lesions were referred for biopsy, the sensitivity and specificity of the data set were not calculated for clinical assessment.
| Classifier | DCE | T2w | DWI | Feature fusion | Classifier fusion |
|---|---|---|---|---|---|
| AUC [95% CI] | 0.84 [0.82, 0.87] | 0.83 [0.80, 0.86] | 0.69 [0.62, 0.75] | 0.87 [0.84, 0.89] | 0.87 [0.84, 0.89] |
| Sensitivity (%) | 75.7 | 76.3 | 61.4 | 79.1 | 79.0 |
| Specificity (%) | 76.3 | 74.5 | 62.9 | 77.2 | 78.4 |
Table 3.
Performance comparison for the six classification methods when classifiers were trained on the full set. The classifier names are shown in the first column (single-parametric) and first row (multiparametric). -value and 95% CI of the difference in AUCs for each comparison are presented, where each multiparametric classifier was compared with each single-parametric classifier using the DeLong test. -values were corrected for multiple comparisons using Bonferroni–Holm corrections.
| Classifier | Compared with feature fusion | Compared with classifier fusion |
|---|---|---|
| DCE | * | * |
| 95% CI | 95% CI | |
| T2w | * | * |
| 95% CI | 95% CI | |
| DWI | * | * |
| 95% CI | 95% CI |
Significance () after accounting for multiple comparisons.
When only including the subset imaged using the three-sequence protocol and discarding the subset, in which DWI was missing, the feature fusion and classifier fusion mpMRI approaches yielded AUCs [95% CIs] of 0.80 [0.73, 0.85] and 0.80 [0.74, 0.86], respectively, both significantly lower than their corresponding classifiers’ performances when the full set was used (95% CI of for both approaches). The results demonstrated that with the proposed method for handling exams acquired using different imaging protocols that contained inconsistent sequences, it would be beneficial to utilize the full dataset despite its incompleteness.
4. Discussion
The proposed radiomics methods that take advantage of the complimentary information provided by DCE, T2w, and DWI sequences in mpMRI demonstrated potential to improve performance over single-parametric CADx in the task of distinguishing between benign and malignant breast lesions. Two mpMRI approaches were examined: a feature fusion method that concatenated radiomic features extracted from mpMRI sequences at the classifier input and a classifier fusion method that aggregated the PM outputs from the single-parametric classifiers via soft voting. When trained on the entire clinical dataset of 852 lesions, where some exams did not include the DWI sequence, both mpMRI methods significantly outperformed all the single-parametric classifiers. When trained on the subset of 389 lesions that were imaged using the three-sequence mpMRI protocol, both mpMRI classifiers yielded reduced performances.
We believe that this is the first comprehensive study that investigated two human-engineered radiomics approaches of leveraging mpMRI information from three sequences for breast lesion classification. Previous studies were largely focused on using the DCE sequence alone.12,13,34–36 A few previous studies developed mpMRI frameworks to distinguish between malignant and benign lesions using radiomic features, but either only included DCE and T2w sequences or only investigated methods similar to the feature fusion approach in our study, and reported lower performance than the results achieved by our methods.14,15 The findings in our work demonstrate superiority of the mpMRI approaches, which can improve the currently available breast cancer CADx systems based on DCE alone.
Our study has a few limitations. First of all, an ideal model tuning procedure would involve a single training set, validation set, and held-out test set. However, we chose to use nested fivefold cross validation since no prior feature selection or parameter optimization was needed. The nested cross-validation scheme resulted in an 80%/20% split into independent development and test sets within one partition in the outer cross-validation loop, and thus did not lead to overfitting due to data leakage. Using cross validation as the evaluation technique allowed us to more efficiently use the data by reporting an overall score across five test sets instead of a single test set.
Moreover, only six first-order radiomic features were extracted from ADC maps. Other radiomic features were not calculated because we did not think high-order features, such as texture features, would be informative given the coarse resolution of DWI. Also feature selection was not included in our approach because our preliminary investigation of several feature selection and dimension reduction methods, including stepwise feature selection, recursive feature selection, principal component analysis, and -distributed stochastic neighbor embedding, showed that none of these methods resulted in an improved classification performance. It is worth noting that our approach was to extract radiomic features that are clinically or physiologically relevant to the diagnosis of breast cancer, rather than extracting as many features as possible and then select a subset based on statistical importance. A total of 75 features were extracted from three modalities, which was a reasonable size for SVM classifiers without feature selection, especially given the fairly large size of the database.
In addition, MRI exams used in this study were collected over the span of eight years, during which imaging technology advanced and some acquisition parameters did not remain constant. We ensured that no extreme imbalance that would potentially bias the results was present, e.g., the field strengths distribution was similar between the benign and malignant class. Among the 195 benign lesions, 141 (72%) of them were imaged with 1.5 T scanners and 54 were imaged with 3 T (28%) scanners; among the 657 malignant lesions, 422 (64%) were imaged with 1.5 T scanners and 235 were imaged with 3 T (36%) scanners. Although the use of such a retrospectively collected dataset provided us with an estimate of the robustness of our radiomics models to heterogeneous data, we plan to investigate harmonization of differences in acquisition parameters in future studies.
Common alternative approaches for handling missing modalities in multiparametric imaging studies include image imputation and feature imputation. Image imputation methods are task-specific, and while developing a satisfactory image imputation method for diagnosing breast cancer on mpMRI is an interesting topic for future investigation, it is beyond the scope of this study. As for feature imputation, a comparative experiment was performed in which the missing DWI radiomic features were imputed using a regression-based multivariate iterative feature imputation method and the classification results were compared with those from our original approach. The performance for all classifiers that utilized DWI features, namely the DWI single-parametric classifier, the feature fusion mpMRI classifier, and the classifier fusion mpMRI classifier, slightly decreased. Their AUCs [95% CIs] were 0.66 [0.62, 0.70], 0.85 [0.82, 0.88], and 0.86 [0.84, 0.89], indicating that the imputed DWI features did not benefit the classification performance. In addition to the classification performance, the advantages of our original approach also include its computational efficiency as it eliminates the imputation step, and its close analogy to the clinical diagnostic process, i.e., radiologists basing their assessment on either two or three sequences available in mpMRI exam for a particular case.
5. Conclusions
In conclusion, our study proposed two mpMRI approaches that both significantly outperformed single-parametric CADx in the task of distinguishing between benign and malignant breast lesions. Our methodology is highly automated, computationally efficient, and handles the common problem of missing modalities among clinical multiparametric imaging datasets. Future work will focus on understanding the disagreement between radiomics and deep learning approaches and taking advantage of the strengths from both. Furthermore, we plan to perform validation on an independent, external dataset to assess the robustness of the system.
6. Appendix
The following tables provide a complete list of radiomic features included in this study and their descriptions. Tables 4–6 list radiomic features extracted from DCE, T2w, and DWI sequences, respectively.
Table 4.
Radiomic features extracted from DCE sequence and their descriptions.
| Category | Feature name (unit) | Feature description |
|---|---|---|
| Geometry19 | Volume () | Volume of lesion |
| Effective diameter (mm) | Greatest dimension of a sphere with the same volume as the lesion | |
| Surface area () | Lesion surface area | |
| Maximum diameter (mm) | Maximum distance between any two voxels in the lesion | |
| Sphericity | Similarity of the lesion shape to a sphere | |
| Irregularity | Deviation of the lesion surface from the surface of a sphere | |
| Surface area/volume (1/mm) | Ratio of surface area to volume | |
| Morphology19 | Margin sharpness | Mean of the image gradient at the lesion margin |
| Variance of margin sharpness | Variance of the image gradient at the lesion margin | |
| Variance of radial gradient histogram | Degree to which the enhancement structure extends in a radial pattern originating from the center of the lesion | |
| Texture22 | Contrast | Location image variations |
| Correlation | Image linearity | |
| Difference entropy | Randomness of the difference of neighboring voxels’ gray levels | |
| Difference variance | Variations of difference of gray level between voxel pairs | |
| Angular second moment (energy) | Image homogeneity | |
| Entropy | Randomness of the gray levels | |
| Inverse difference moment (homogeneity) | Image homogeneity | |
| Information measure of correlation 1 | Nonlinear gray-level dependence | |
| Information measure of correlation 2 | Nonlinear gray-level dependence | |
| Maximum correlation coefficient | Nonlinear gray-level dependence | |
| Sum average | Overall brightness | |
| Sum entropy | Randomness of the sum of gray-level dependence | |
| Sum variance | Spread in the sum of the gray levels of neighboring voxels | |
| Sum of squares (variance) | Spread in the gray-level distribution | |
| Kinetics21 | Maximum enhancement | Maximum contrast enhancement |
| Time to peak (s) | Time at which the maximum enhancement occurs | |
| Uptake rate (1/s) | Uptake speed of the contrast enhancement | |
| Washout rate (1/s) | Washout speed of the contrast enhancement | |
| Curve shape index | Difference between late and early enhancement | |
| Enhancement at first postcontrast time point | Enhancement at first postcontrast time point | |
| Signal enhancement ratio | Ratio of initial enhancement to overall enhancement | |
| Volume of most enhancing voxels () | Volume of the most enhancing voxels | |
| Total rate variation () | How rapidly the contrast will enter and exit from the lesion | |
| Normalized total rate variation () | How rapidly the contrast will enter and exit from the lesion | |
| Enhancement-variance kinetics20 | Maximum enhancement-variance | Maximum spatial variance of contrast enhancement over time |
| Enhancement-variance time to peak (s) | Time at which the maximum variance occurs | |
| Enhancement-variance increasing rate (1/s) | Rate of increase of the enhancement-variance during uptake | |
| Enhancement-variance decreasing rate (1/s) | Rate of decrease of the enhancement-variance during washout | |
| Gray-level statistics24 | Mean voxel value precontrast | Average gray-level intensity within the lesion prior to contrast injection |
| Mean voxel value postcontrast injection | Average gray-level intensity within the lesion at first postcontrast injection time point | |
| Standard deviation of voxel value distribution precontrast | Variation in gray-level intensity within the lesion prior to contrast injection | |
| Standard deviation of voxel value distribution postcontrast | Variation in gray-level intensity within the lesion at first postcontrast injection time point | |
| Maximum voxel value precontrast | Maximum gray-level intensity within the lesion prior to contrast injection | |
| Maximum voxel value postcontrast | Maximum gray-level intensity within the lesion at first postcontrast injection time point | |
| Minimum voxel value precontrast | Minimum gray-level intensity within the lesion prior to contrast injection | |
| Minimum voxel value postcontrast | Minimum gray-level intensity within the lesion at first postcontrast injection time point | |
| Kurtosis of voxel value distribution precontrast | Tailedness of gray-level intensity distribution within the lesion prior to contrast injection | |
| Kurtosis of voxel value distribution postcontrast | Tailedness of gray-level intensity distribution within the lesion at first postcontrast injection time point | |
| Skewness of voxel value distribution precontrast | Asymmetry of gray-level intensity distribution about the mean within the lesion prior to contrast injection | |
| Skewness of voxel value distribution postcontrast | Asymmetry of gray-level intensity distribution about the mean within the lesion at first postcontrast injection time point |
Table 5.
Radiomic features extracted from T2w sequence and their descriptions.
| Category | Feature name | Feature description |
|---|---|---|
| Morphology14,19 | Margin sharpness | Mean of the image gradient at the lesion margin |
| Variance of margin sharpness | Variance of the image gradient at the lesion margin | |
| Variance of radial gradient histogram | Degree to which the enhancement structure extends in a radial pattern originating from the center of the lesion | |
| Texture14,22 | Contrast | Location image variations |
| Correlation | Image linearity | |
| Difference entropy | Randomness of the difference of neighboring voxels’ gray levels | |
| Difference variance | Variations of difference of gray level between voxel pairs | |
| Angular second moment (energy) | Image homogeneity | |
| Entropy | Randomness of the gray levels | |
| Inverse difference moment (homogeneity) | Image homogeneity | |
| Information measure of correlation 1 | Nonlinear gray-level dependence | |
| Information measure of correlation 2 | Nonlinear gray-level dependence | |
| Maximum correlation coefficient | Nonlinear gray-level dependence | |
| Sum average | Overall brightness | |
| Sum entropy | Randomness of the sum of gray-level dependence | |
| Sum variance | Spread in the sum of the gray-levels of neighboring voxels | |
| Sum of squares (variance) | Spread in the gray-level distribution | |
| Gray-level statistics14 | Mean voxel value | Average gray-level intensity within the lesion |
| Variance of voxel value | Variation in gray-level intensity within the lesion |
Table 6.
Radiomic features extracted from the ADC map derived from diffusion-weighted sequence and their descriptions.
| Category | Feature name | Feature description |
|---|---|---|
| ADC map statistics25 | Mean ADC | Average ADC within the lesion |
| Standard deviation of ADC distribution | Variation in ADC within the lesion | |
| Maximum ADC | Maximum ADC within the lesion | |
| Minimum ADC | Minimum ADC within the lesion | |
| Range of ADC distribution | Range of ADC distribution within the lesion | |
| Skewness of ADC distribution | Asymmetry of ADC distribution about the mean within the lesion |
Acknowledgments
The authors acknowledge other lab members, including Karen Drukker, PhD, MBA; Alexandra Edwards, MA; Hui Li, PhD; and John Papaioannou, MS, Department of Radiology, The University of Chicago, Chicago, IL, for their contributions to the datasets and discussions. This work was supported in part by the National Institutes of Health National Cancer Institute (NIH NCI) under Grant U01CA195564 and Grant R15 CA227948, the RSNA/AAPM Graduate Fellowship, and the University of Chicago Comprehensive Cancer Center Dancing with Chicago Celebrities Fund.
Biographies
Qiyuan Hu received her BA degrees in physics and mathematics from Carleton College in 2017. She is a PhD student in medical physics at the University of Chicago. Her research interests include radiomics and deep learning methodologies for computer-aided diagnosis. She is a student member of SPIE and an officer of the University of Chicago SPIE Student Chapter.
Heather M. Whitney is an associate professor of physics at Wheaton College and a visiting scholar in the Department of Radiology at the University of Chicago. Her experience in quantitative medical imaging has ranged from polymer gel dosimetry to radiation damping in nuclear magnetic resonance to now focusing on radiomics of breast cancer imaging. She is interested in investigating the effects of the physical basis of imaging on radiomics as well as the repeatability and robustness of radiomics. She is a member of SPIE.
Maryellen L. Giger is the A. N. Pritzker Professor of Radiology, Committee on Medical Physics, and the College at the University of Chicago. She has conducted research on computer-aided diagnosis, quantitative image analysis (radiomics), and deep learning in the areas of breast cancer, lung cancer, prostate cancer, and bone diseases. She is a fellow of SPIE and the 2018 SPIE President.
Disclosures
Q. H. declares no conflicts of interest. H. M. W. declares no conflicts of interest. M. L. G. is a stockholder in R2 technology/Hologic and QView, receives royalties from Hologic, GE Medical Systems, MEDIAN Technologies, Riverain Medical, Mitsubishi, and Toshiba, and is a cofounder of and equity holder in Quantitative Insights (now Qlarity Imaging). It is the University of Chicago Conflict of Interest Policy that investigators disclose publicly actual or potential significant financial interest that would reasonably appear to be directly and significantly affected by the research activities.
Contributor Information
Qiyuan Hu, Email: qhu@uchicago.edu.
Heather M. Whitney, Email: heather.whitney@wheaton.edu.
Maryellen L. Giger, Email: m-giger@uchicago.edu.
References
- 1.Kuhl C. K., et al. , “Mammography, breast ultrasound, and magnetic resonance imaging for surveillance of women at high familial risk for breast cancer,” J. Clin. Oncol. 23(33), 8469–8476 (2005). 10.1200/JCO.2004.00.4960 [DOI] [PubMed] [Google Scholar]
- 2.Leithner D., et al. , “Clinical role of breast MRI now and going forward,” Clin. Radiol. 73(8), 700–714 (2018). 10.1016/j.crad.2017.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Menezes G. L. G., et al. , “Magnetic resonance imaging in breast cancer: a literature review and future perspectives,” World J. Clin. Oncol. 5(2), 61 (2014). 10.5306/wjco.v5.i2.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuhl C. K., et al. , “Do T2-weighted pulse sequences help with the differential diagnosis of enhancing lesions in dynamic breast MRI?” J. Magn. Reson. Imaging 9(2), 187–196 (1999). [DOI] [PubMed] [Google Scholar]
- 5.Santamaría G., et al. , “Radiologic and pathologic findings in breast tumors with high signal intensity on T2-weighted MR images,” Radiographics 30(2), 533–548 (2010). 10.1148/rg.302095044 [DOI] [PubMed] [Google Scholar]
- 6.Westra C., et al. , “Using T2-weighted sequences to more accurately characterize breast masses seen on MRI,” Am. J. Roentgenol. 202(3), W183–W190 (2014). 10.2214/AJR.13.11266 [DOI] [PubMed] [Google Scholar]
- 7.Shi R., et al. , “Breast lesions: diagnosis using diffusion weighted imaging at 1.5 T and 3.0 T—systematic review and meta-analysis,” Clin. Breast Cancer 18(3), e305–e320 (2018). 10.1016/j.clbc.2017.06.011 [DOI] [PubMed] [Google Scholar]
- 8.Partridge S. C., et al. , “Diffusion-weighted MRI findings predict pathologic response in neoadjuvant treatment of breast cancer: the ACRIN 6698 multicenter trial,” Radiology 289(3), 618–627 (2018). 10.1148/radiol.2018180273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mann R. M., Cho N., Moy L., “Breast MRI: state of the art,” Radiology 292(3), 520–536 (2019). 10.1148/radiol.2019182947 [DOI] [PubMed] [Google Scholar]
- 10.Giger M. L., Chan H.-P., Boone J., “Anniversary paper: history and status of CAD and quantitative image analysis: the role of medical physics and AAPM,” Med. Phys. 35(12), 5799–5820 (2008). 10.1118/1.3013555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giger M. L., Karssemeijer N., Schnabel J. A., “Breast image analysis for risk assessment, detection, diagnosis, and treatment of cancer,” Annu. Rev. Biomed. Eng. 15, 327–357 (2013). 10.1146/annurev-bioeng-071812-152416 [DOI] [PubMed] [Google Scholar]
- 12.Whitney H. M., et al. , “Comparison of breast MRI tumor classification using human-engineered radiomics, transfer learning from deep convolutional neural networks, and fusion method,” Proc. IEEE 108(1), 163–177 (2020). 10.1109/JPROC.2019.2950187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu Q., Whitney H. M., Giger M. L., “Transfer learning in 4D for breast cancer diagnosis using dynamic contrast-enhanced magnetic resonance imaging,” arXiv1911.03022 (2019).
- 14.Bhooshan N., et al. , “Combined use of T2-weighted MRI and T1-weighted dynamic contrast—enhanced MRI in the automated analysis of breast lesions,” Magn. Reson. Med. 66(2), 555–564 (2011). 10.1002/mrm.22800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Truhn D., et al. , “Radiomic versus convolutional neural networks analysis for classification of contrast-enhancing lesions at multiparametric breast MRI,” Radiology 290(2), 290–297 (2019). 10.1148/radiol.2018181352 [DOI] [PubMed] [Google Scholar]
- 16.Dalmis M. U., et al. , “Artificial intelligence-based classification of breast lesions imaged with a multiparametric breast MRI protocol with ultrafast DCE-MRI, T2, and DWI,” Invest. Radiol. 54(6), 325–332 (2019). 10.1097/RLI.0000000000000544 [DOI] [PubMed] [Google Scholar]
- 17.Hu Q., Whitney H. M., Giger M. L., “A deep learning methodology for improved breast cancer diagnosis using multiparametric MRI,” Sci. Rep. 10(1), 1–11 (2020). 10.1038/s41598-020-67441-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen W., Giger M. L., Bick U., “A fuzzy c-means (FCM)-based approach for computerized segmentation of breast lesions in dynamic contrast-enhanced MR images,” Acad. Radiol. 13(1), 63–72 (2006). 10.1016/j.acra.2005.08.035 [DOI] [PubMed] [Google Scholar]
- 19.Gilhuijs K. G. A., Giger M. L., Bick U., “Computerized analysis of breast lesions in three dimensions using dynamic magnetic-resonance imaging,” Med. Phys. 25(9), 1647–1654 (1998). 10.1118/1.598345 [DOI] [PubMed] [Google Scholar]
- 20.Chen W., et al. , “Computerized interpretation of breast MRI: investigation of enhancement-variance dynamics,” Med. Phys. 31(5), 1076–1082 (2004). 10.1118/1.1695652 [DOI] [PubMed] [Google Scholar]
- 21.Chen W., et al. , “Automatic identification and classification of characteristic kinetic curves of breast lesions on DCE-MRI,” Med. Phys. 33(8), 2878–2887 (2006). 10.1118/1.2210568 [DOI] [PubMed] [Google Scholar]
- 22.Chen W., et al. , “Volumetric texture analysis of breast lesions on contrast-enhanced magnetic resonance images,” Magn. Reson. Med. 58(3), 562–571 (2007). 10.1002/mrm.21347 [DOI] [PubMed] [Google Scholar]
- 23.Chen W., et al. , “Computerized assessment of breast lesion malignancy using DCE-MRI: robustness study on two independent clinical datasets from two manufacturers,” Acad. Radiol. 17(7), 822–829 (2010). 10.1016/j.acra.2010.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drukker K., et al. , “Breast MRI radiomics for the pretreatment prediction of response to neoadjuvant chemotherapy in node-positive breast cancer patients,” J. Med. Imaging 6(3), 034502 (2019). 10.1117/1.JMI.6.3.034502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu Q., et al. , “Radiomics and deep learning of diffusion-weighted MRI in the diagnosis of breast cancer,” Proc. SPIE 10950, 109504A (2019). 10.1117/12.2512626 [DOI] [Google Scholar]
- 26.Scholkopf B., Smola A. J., Learning with Kernels: Support Vector Machines, Regularization, Optimization, and Beyond, MIT Press, Cambridge, Massachusetts: (2001). [Google Scholar]
- 27.Shawe-Taylor J., Sun S., “A review of optimization methodologies in support vector machines,” Neurocomput. 74(17), 3609–3618 (2011). 10.1016/j.neucom.2011.06.026 [DOI] [Google Scholar]
- 28.Metz C. E., Herman B. A., Shen J. H., “Maximum likelihood estimation of receiver operating characteristic (ROC) curves from continuously-distributed data,” Stat. Med. 17(9), 1033–1053 (1998). [DOI] [PubMed] [Google Scholar]
- 29.Metz C. E., Pan X., “‘Proper’ binormal ROC curves: theory and maximum-likelihood estimation,” J. Math. Psychol. 43(1), 1–33 (1999). 10.1006/jmps.1998.1218 [DOI] [PubMed] [Google Scholar]
- 30.Efron B., “Better bootstrap confidence intervals,” J. Am. Stat. Assoc. 82(397), 171–185 (1987). 10.1080/01621459.1987.10478410 [DOI] [Google Scholar]
- 31.DeLong E. R., DeLong D. M., Clarke-Pearson D. L., “Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach,” Biometrics 44(3), 837–845 (1988). 10.2307/2531595 [DOI] [PubMed] [Google Scholar]
- 32.Sun X., Xu W., “Fast implementation of DeLong’s algorithm for comparing the areas under correlated receiver operating characteristic curves,” IEEE Signal Process. Lett. 21(11), 1389–1393 (2014). 10.1109/LSP.2014.2337313 [DOI] [Google Scholar]
- 33.Holm S., “A simple sequentially rejective multiple test procedure,” Scand. J. Stat. 6(2), 65–70 (1979). 10.2307/4615733 [DOI] [Google Scholar]
- 34.Antropova N., Huynh B. Q., Giger M. L., “A deep feature fusion methodology for breast cancer diagnosis demonstrated on three imaging modality datasets,” Med. Phys. 44(10), 5162–5171 (2017). 10.1002/mp.12453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Antropova N., Abe H., Giger M. L., “Use of clinical MRI maximum intensity projections for improved breast lesion classification with deep convolutional neural networks,” J. Med. Imaging 5(1), 014503 (2018). 10.1117/1.JMI.5.1.014503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ji Y., et al. , “Independent validation of machine learning in diagnosing breast Cancer on magnetic resonance imaging within a single institution,” Cancer Imaging 19, 64 (2019). 10.1186/s40644-019-0252-2 [DOI] [PMC free article] [PubMed] [Google Scholar]