Abstract
Recent findings have highlighted an interconnection between intestinal microbiota and the brain, referred to as microbiota–gut–brain axis, and suggested that alterations in microbiota composition might affect brain functioning, also in Alzheimer’s disease. To investigate microbiota–gut–brain axis biochemical pathways, in this work we developed an innovative device to be used as modular unit in an engineered multi-organ-on-a-chip platform recapitulating in vitro the main players of the microbiota–gut–brain axis, and an innovative three-dimensional model of brain cells based on collagen/hyaluronic acid or collagen/poly(ethylene glycol) semi-interpenetrating polymer networks and β-amyloid precursor protein-Swedish mutant-expressing H4 cells, to simulate the pathological scenario of Alzheimer’s disease. We set up the culturing conditions, assessed cell response, scaled down the three-dimensional models to be hosted in the organ-on-a-chip device, and cultured them both in static and in dynamic conditions. The results suggest that the device and three-dimensional models are exploitable for advanced engineered models representing brain features also in Alzheimer’s disease scenario.
Keywords: Hydrogel, in vitro brain models, Alzheimer’s disease, three-dimensional culture, organ-on-a-chip
Introduction
The intriguing hypotheses of a bidirectional functional relationship between intestinal microbiota and the brain, referred to as microbiota–gut–brain axis (MGBA), and the potential role of gut microbiota in pathological pathways, including Alzheimer’s disease (AD), the most common neurodegenerative disorder, have opened new scenarios and perspectives in neuroscience.1,2 The development of an engineered multi-organ-on-a-chip platform representing in vitro the main players of the MGBA, that is, the microbiota, the gut, the immune system, the blood–brain barrier, and the brain, can speed up the investigation of the impact of intestinal microbiota on brain functionality.2 The rationale of this approach is to couple the high technological features of organ-on-a-chip devices with the potential of advanced cell-based models to represent in vitro the key features of the biological systems involved in microbiota–brain interactions, such as mechanical stimuli, including physiologically relevant fluid shear stress conditions, and three-dimensional (3D) spatial architecture.
Organ-on-a-chip technology has dramatically boomed for its potential to revolutionize the healthcare system,2–5 also by reducing animal studies, in agreement with the 3Rs principle, while several studies in different contexts have evidenced that 3D cell models are more representative of in vivo conditions than two-dimensional (2D) monolayers.6–10 However, the possibility to represent in vitro the key features of the brain in both physiological and pathological conditions is still a challenge.
Choi et al.11 investigated the effect of oligomeric amyloid β (Aβ) on neural progenitor cells in 2D conditions by a microfluidic chip and recapitulated an in vivo–like microenvironment by flow rates in the range of the interstitial flow in brain tissues (0.1–0.3 mL/min g). In a successive work under static conditions, the setting up of an innovative, in vitro 3D model of brain cells was reported.12 ReNcell® cells expressing familial AD mutations in β-amyloid precursor protein (APP) and presenilin 1 were embedded in Matrigel. This culture model recapitulated the key hallmarks of AD. In particular, the presence of the hydrogel matrix acted as a physical barrier by limiting Aβ diffusion in culture medium and promoting its accumulation with time and toxicity. To develop a microfluidic model of a 3D neural circuit, Bang et al.13 modified an existing device and patterned the extracellular matrix (ECM) components of Matrigel by applying a stable hydrostatic pressure during gelation. Then, they plated rat cortical neurons on the gel surface and studied axon bundles. However, a miniaturized system suitable for the interstitial perfusion of 3D models of brain cells based on hydrogels of millimeter scale is still missing.
In the present work, we focused on two main goals: (1) the development of a new, miniaturized, and optically accessible microfluidic device as modular unit of a multi-organ-on-a-chip platform representing the main players of the MGBA and (2) an innovative 3D model of brain cells to be perfused in the aforementioned device, capable of hosting human cells overexpressing APP and suitable to promote extracellular accumulation of amyloid fragments, as required for a representative AD in vitro model. Starting from a prototypal device previously investigated in our laboratories for the interstitial perfusion of 3D cell constructs,14 to reach the first goal we developed an innovative microfluidic device and assessed its suitability for cell culture by computational fluid dynamics (CFD) simulations. To fulfill the second aim, we exploited two hydrogel matrices obtained by promoting collagen (COLL) fibrillogenesis in the presence of (1) poly(ethylene glycol) (PEG) and (2) ultrapure pharma-grade hyaluronic acid (HA). Their preparation was performed under physiological conditions without a crosslinking agent, thus reducing the risk of cytotoxicity. Both hydrogels were previously developed and tested for biocompatibility with brain cells.15,16 To obtain finely tunable matrices with enhanced controllability and reproducibility,15,16 both hydrogels were conceived as semi-interpenetrating polymer networks (semi-IPNs).
For hydrogel biological characterization, APP-Swedish double mutant (K595N/M596L)-expressing H4 (H4-SW) cells were selected as cell model. This mutation in the APP gene increases the levels of Aβ42, whose aberrant formation is peculiar of both sporadic and familial AD, through abnormal cleavage of cellular APP by γ-secretase.17 To set up the best cell-loading conditions, we considered two 3D cell models: (1) layered-based model, where a layer of hydrogel covers H4-SW cells attached to the microplate, and (2) embedded-based model, where H4-SW cells are mixed with the polymer solutions during hydrogel preparation.
To investigate the possibility to host the 3D models in the culture chambers of the organ-on-a-chip device, we studied COLL fibrillogenesis as a function of hydrogel thickness and cell loading/culturing density. Only for the embedded condition, we also examined the effect of embedding protocols, in particular the volumetric ratio between cell suspension and polymer solutions, on the key properties of the gels. Then, we continued the investigations of each 3D cell model with the cell density selected. To study the capability of both 3D culture systems to entrap soluble amyloid fragments, we evaluated H4-SW cell metabolic activity with time up to 21 days. After finding the peak, meaning the time point at which the cells showed the best performance in terms of possible protein production, we performed immunofluorescence assays and Western blotting. Finally, we loaded the thinnest hydrogel matrix with H4-SW cells according to the best performing culture condition, cultured it in the organ-on-a-chip device, and preliminarily assessed its potential as an in vitro brain model in AD pathological scenario.
Materials and methods
We purchased neuroglioma H4 cells (ATCC® code HTB-148) from LGC Standards (Sesto San Giovanni, Italy). We obtained plasticware from Corning (NY, USA) and reagents for cell culture from Thermo Fisher Scientific (Waltham, MA, USA). If not differently stated, we purchased other reagents from Sigma-Aldrich (St. Louis, MO, USA).
Microfluidic device
Starting from the microfluidic and optically accessible prototypal bioreactors,14,18 we completely re-designed and set up an innovative, versatile, and 3D printable device (Figure 1). We designed the device to be connected in series to obtain the multi-organ-on-a-chip platform representing the key players of the MGBA.
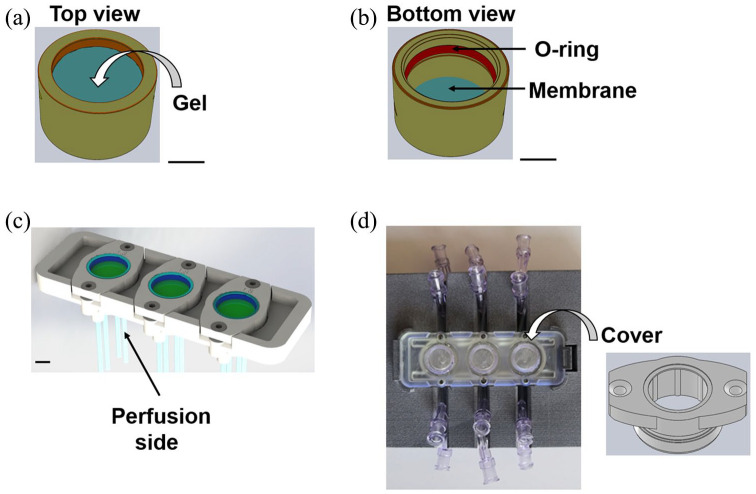
Figure 1.
Technical sketches (AutoCAD® Software; Autodesk, San Rafael, CA, USA) and views of the organ-on-a-chip device. Scale bars = 0.5 mm. (a) Technical sketch of the hemi-chamber hosting 3D culture models. (b) Technical sketch of the opposite hemi-chamber. (c) Technical sketch showing the main body of the device, with the culture chambers. Each hemi-chamber has an independent perfusion system. Perfusion is possible in both concurrent and counter-current configuration. (d) Top view showing the culture chambers and the main body of the device. Top view of the cover, ready to close a culture chamber.
To perform cell culture in triplicate, the microfluidic device hosts three identical culture chambers, suitable for suspended cells, 2D and 3D cell culture. The device design is compatible with housings for both optical and confocal microscopes, and the chambers are made up of optically transparent materials. For a rapid comparison with static culture conditions, they are round-shaped and their surface area is comparable to that of a culture well in 48-well plates. Luer lock connectors connect the chambers to a hydraulic circuit for the perfusion of culture medium, and sampling points are placed at both the inlet and the outlet of the culture chambers. Perfusion is possible in both concurrent and counter-current configuration, the latter to maximize diffusive phenomena.
Each chamber is divided into two hemi-chambers (about 2 mm high) by a transparent, semipermeable membrane that acts as a support for 2D cell cultures and 3D hydrogel-based cultures and it is suitable for cell culture on both sides. It has a pore diameter of 0.4 µm and a pore density of 2 × 106 pores/cm2 (Greiner Bio-One, Kremsmünster, Austria). It allows the diffusion of soluble factors and/or metabolites; thus, it can be exploited to study the crosstalk between different cell populations.
Stainless steel screws and an O-ring system bond the chambers to the main body of the bioreactor and guarantee both hydraulic sealing and sterility.
In this study, we applied the device to host an innovative model of brain cells and tested as described below. In the configuration tested, one hemi-chamber hosted the selected 3D model of brain cells, while the opposite one was empty and culture medium could freely perfuse the opposite side of the 3D construct. We sterilized the device by hydrogen peroxide gas plasma (STERRAD® 100S, ASP; Johnson & Johnson, Irvine, CA, USA).
Hydrogel preparation
We prepared COLL-based semi-IPNs by promoting COLL fibrillogenesis in the presence of ultrapure pharma-grade HA (Mw = 1×105 Da; Altergon, Morra de Sanctis, Italy) or PEG. We used PEG with two different molecular weights (Mw): 1.945×103 Da for COLL-PEG2000 gels and 3.270×103 Da for COLL-PEG3350 gels.
We mixed 8 parts (v/v) ultrapure type I COLL from bovine skin (3 mg/mL, batch SLBS7030) with 1 part (v/v) 10× phosphate-buffered saline solution (PBS) and 1 part (v/v) 0.1 N NaOH.
For COLL-HA gels, we dissolved HA in MilliQ water (5 mg/mL) and autoclaved (121°C for 15 min or with an equivalent time F0 equal to 13). We mixed COLL and HA solutions to final concentrations of 1.2 and 2.5 mg/mL, respectively.
For COLL-PEG gels, we dissolved PEG in saline (2.4 mg/mL) and autoclaved. We mixed COLL and PEG solutions to final concentrations of 1.8 and 0.6 mg/mL, respectively.
If not differently stated, samples were prepared in 96-well plates. To promote COLL fibrillogenesis, we incubated the solutions at 37°C.
Cell model
We cultured H4-SW cells at 37°C and 5% CO2 in high-glucose Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 100 U/mL penicillin, 100 µg/mL streptomycin sulfate, 300 μg/mL hygromycin B, and 10 μg/mL blasticidin S. We changed the medium every 3 days and split the cells twice a week.
We examined two 3D cell models:
(1) Layered-based model: we plated H4-SW cells. The following day, we removed the culture medium and covered the cells with the polymer solutions (supplemented with culture medium, 1:10 v/v);
(2) Embedded-based model: we mixed H4-SW cells with the polymer solutions and plated.
After covering or mixing cells with the polymer solutions, we incubated the samples at 37°C to promote COLL fibrillogenesis.
Numerical evaluation of the microfluidic device with 3D cell models
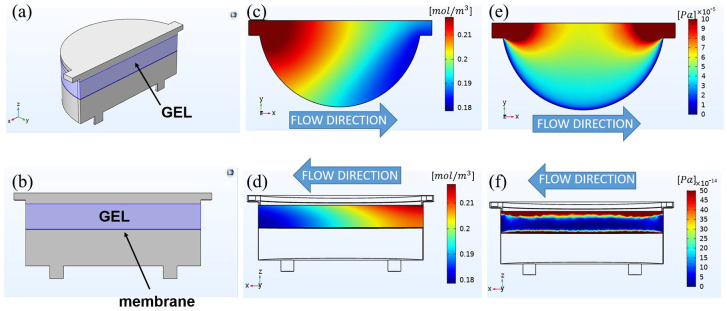
To assess the suitability of the microfluidic device for 3D cell culture, we performed CFD simulations with the software COMSOL Multiphysics® (Burlington, MA, USA), release 5.5. To design the geometry of the model (Figure 2(a) and (b)), we considered the internal space of the culture chambers, that is the fluidic pathway of culture medium in the hydrogel hemi-chamber, the hydrogel itself, the membrane, and the fluidic pathway of culture medium through the opposite hemi-chamber. We performed the simulation with perfusion in counter-current configuration and computed the velocity field, pressure drops, and oxygen distribution inside one chamber. We showed the results (Figure 2(c)–(f)) in the case of 3.125 × 105 cells/cm2 embedded (1:10 v/v) in 1.25-mm-thick COLL-PEG2000 gels (see hereinafter) and reported the numerical values of the parameters for the simulation in Table 1. For the reaction terms, we assumed a homogeneous cell distribution within the hydrogel.
Figure 2.
Technical sketches and color maps of the numerical characterization of the chambers hosting 1.25-mm-thick COLL-PEG2000 gels (embedded condition, 3.125 × 105 cells/cm2, 1:10 v/v). We reported the values of the parameters used for the simulation in Table 1. (a) Overall and (b) side view of the geometry. From top to bottom, it comprised four domains: the upper perfusion channel of the hydrogel chamber, the hydrogel (light blue), the membrane, and the other perfusion channel. (c) Top view and (d) side view of the color map showing the distribution of oxygen concentration. (e) Top view and (f) side view of the color map showing the distribution of shear stresses.
Table 1.
Numerical values of the parameters used for simulation.
| Property | Value |
|---|---|
| Atmospheric pressure (outlet) | 0 Pa |
| Condition at the walls | No-slip condition |
| Density of culture medium (at 37°C), ρ | 1 × 103 kg/m3 |
| Diffusion coefficient of oxygen, D | 2 × 10−9 m2/s |
| Dynamic viscosity of culture medium (at 37°C), µ | 8.1 × 10−4 Pa·s |
| COLL-PEG2000 hydrogel permeability | 1.868 × 10−16 m2 |
| COLL-PEG2000 hydrogel porosity, ε | 9.3 × 10−1 (Song et al.19) |
| Inlet velocity in the hydrogel chamber, v
(flow rate, 0.75 µL/min) |
4.16 × 10−5 m/s |
| Inlet velocity in the opposite chamber, v
(flow rate, 0.25 µL/min) |
1.54 × 10−5 m/s |
| Membrane porosity | 2.5 × 10−3 |
| Membrane permeability | 7.8 × 10−19 m2 |
| Oxygen concentration at the inlets | 2.2 × 10−1 mol/m3 |
| Maximum oxygen consumption rate Vmax
(for cells embedded in COLL-PEG2000 hydrogels) |
1.49×10−5 mol/(m3 s) (Berger et al.20) |
| Michaelis–Menten constant, Km | 0.15 mol/m3 (Cioffi et al.21) |
COLL: collagen; PEG: poly(ethylene glycol); HA: hyaluronic acid.
List of the properties and their values used for the computational fluid dynamics (CFD) model. We computed the velocity field, pressure drops, and oxygen distribution inside one chamber, in the presence of cells embedded in 1.25-mm-thick COLL-PEG2000 hydrogels. We estimated hydrogel permeability from the mesh size. We calculated this parameter by fitting the rheological data15 with the Generalized Maxwell Model,22 obtaining a value of 80 nm. We estimated a mesh size of 116 nm for COLL-HA and a mesh size of 75 nm for COLL-PEG3350 gels.
We determined the fluid velocity vector u, using the Brinkmann equation in stationary conditions
| (1) |
and a mass-balance equation
| (2) |
where ρ is the fluid density, u is the velocity field vector, p is the fluid pressure, I is the identity matrix, K is the permeability tensor of the porous media, and F is the volume force vector.
The permeability vector is defined as
| (3) |
where μ is the dynamic viscosity of the fluid and ε is the porosity.
We estimated oxygen distribution with the transport equation for diffusion and convection
| (4) |
where J is the mass flux vector, c is the concentration, R is the oxygen volumetric consumption rate, and S is the mass source.
The mass flux vector is defined by Fick’s law
| (5) |
where D is the diffusion coefficient of oxygen in the fluid.
We assumed that the reaction term R was a function of the local oxygen concentration according to the Michaelis–Menten kinetics
| (6) |
where Vmax is the maximum molar consumption rate in the hydrogel construct, c is the local oxygen concentration function, and Km is the Michaelis–Menten constant, which corresponds to the oxygen concentration at which the consumption is half of Vmax.
We obtained the shear stress τ with the formula
| (7) |
where ux is the velocity component vector parallel to the perfusion direction and z is the direction perpendicular to the hydrogel. To run the simulation, we chose the “Fine” element size for mesh building. The resulting mesh had 3.135.622 degrees of freedom to solve.
Effect of hydrogel thickness
To investigate the applicability of the semi-IPNs in the microfluidic device, improve optical accessibility, and increase the representativeness of the sample volume obtained from 3D reconstructions, we reduced hydrogel thickness from 5.10 mm, the condition already tested in our laboratories,15,16 to 1.25 mm. A further decrease does not ensure a uniform covering of the surface and leads to samples with non-homogeneous thickness. We studied COLL fibrillogenesis:
(1) In the absence of dilution of the polymer solutions with culture medium;
(2) After diluting the polymer solutions with culture medium (1:10 v/v). This condition represents the hydrogels covering H4-SW cells in the layered conditions.
For both conditions, we prepared 1.25-, 2.50-, 3.95-, and 5.10-mm-thick samples. We measured the optical density at 400 nm over time (Spark® Multimode Microplate Reader; Tecan, Männedorf, Switzerland). We set the following parameters:
Temperature: (37.0 ± 0.5°C);
Time interval between two consecutive measurements: 1 min;
Total duration of the measurements: 130 min.
We calculated the nucleation time, Tlag, that is the time before the onset of the turbidity increase, the slope of the turbidity curve in the growth phase, and the maximum value of the optical density (K400 max). We defined Tlag as the intersection between the baseline and the tangent to the sigmoidal part of the turbidity curve at the estimated inflection point. The slope is representative of fibril growth rate because of the lateral incorporation of freely diffusing monomers, while K400 max is representative of fibril diameter.
For 1.25- and 5.10-mm-thick gels, we estimated fibril diameter by transmission electron microscopy (TEM; CM 200 TEM/STEM; Philips, Eindhoven, the Netherlands), without any fixation or staining. We placed a small amount of sample on a TEM grid without carbon coating and dried in air. We clamped the grid between two glass coverslips and observed. We measured the diameter of collagen fibrils with the software ImageJ. For each condition, we acquired at least eight images. For each image, we considered sample variability by examining at least five fibrils. For non-homogeneous fibrils, we measured the diameter in two or three points of the same fibril.
Effect of cell density
To investigate the applicability of the 3D cell models in the microfluidic device and set up suitable preparation conditions, we assessed the effect of cell density on COLL fibrillogenesis. For the layered condition, we plated 2.34 × 104, 4.68 × 104, or 7.02 × 104 H4-SW cells/cm2 and covered with the polymer solutions, supplemented with culture medium, 1:10 v/v. We prepared 1.25-, 2.50-, and 3.95-mm-thick samples and studied COLL fibrillogenesis. As a control, we performed the study in the absence of H4-SW cells (0 cells). For the embedded condition, we mixed 1 part (v/v) cell suspension (3.125 × 105 or 6.250 × 105 H4-SW cells/cm2, that is, 1 × 105 or 2 × 105 cells/sample, in 96-well plates) to 9 parts (v/v) polymer solutions. We prepared 1.25-, 2.50-, 3.95-, and 5.10-mm-thick samples and studied COLL fibrillogenesis.
Effect of embedding protocols
For the embedded condition, we investigated the most suitable cell suspension/polymer solution ratio by fixing the cell density at 1 × 105 cells/sample and varying the volumetric ratio between cell suspension and polymer solutions. We prepared 1.25-, 2.50-, 3.95-, and 5.10-mm-thick samples by mixing 1 part (v/v) cell suspension (1 × 105 H4-SW cells/well) with 9 parts (v/v), 7 parts (v/v), 5 parts (v/v), 3 parts (v/v), or 1 part (v/v) polymer solutions and studied COLL fibrillogenesis.
Cell metabolic activity with time as a function of hydrogel thickness
After selecting the most adequate preparation conditions, we investigated cell metabolic activity to find the time point at which the 3D cell models showed the best performance in terms of possible protein production.
For the layered condition, we plated 2.34 × 104 H4-SW cells/cm2 and prepared 1.25-, 2.50-, and 3.95-mm-thick samples. As a control, we performed the study with H4-SW cells in 2D conditions (0 mm). For the embedded condition, we mixed 1 part (v/v) cell suspension (1 × 105 cells/sample) with 9 parts (v/v) polymer solutions and prepared 1.25-, 2.50-, 3.95-, and 5.10-mm-thick samples. For both conditions, we measured cell metabolic activity by MTS (Promega, Madison, WI, USA) assay up to 21 days.
Entrapment of soluble amyloid fragments by immunofluorescence and Western blotting
To investigate the capability of the 3D culture system to entrap amyloid fragments, we prepared 1.25- and 2.50-mm-thick samples in Lab-Tek™ II Chamber Slide™ systems, as described (1.325 × 105 cells/cm2). On day 10, we prepared for immunofluorescence, with minor modifications to Choi et al.12 We incubated with anti-Aβ, 1–16 antibody (6E10, 1:250; Biolegend, San Diego, CA, USA) and Alexa Fluor® 488 secondary antibody (1:500; Abcam, Cambridge, UK). After staining cell nuclei with Hoechst 33342 and cell membranes with lipophilic tracer (Vybrant™ DiI Cell-Labeling Solution; Thermo Fisher Scientific), we observed the samples by confocal microscopy (FV10i; Olympus, Shinjuku, Tokyo, Japan) with z-stacks of 1-μm-thick and performed 3D reconstructions with FV10-ASW 4.2 software. As a control, we performed the study with H4-SW cells in 2D conditions.
To validate the results from immunofluorescence, we prepared COLL-PEG2000 semi-IPNs in 48-well plates and analyzed amyloid protein content by Western blotting. For the layered condition, we plated 2.34 × 104 cells/cm2 and covered with 1.25-mm-thick gels. For the embedded condition, we prepared 1.25-mm-thick samples by mixing 1 part (v/v) cell suspension (1.325 × 105 cells/cm2) with 9 parts (v/v) polymer solution. As a control, we performed the study with H4-SW cells in 2D conditions. On day 10, we lysed the samples at 4°C with lysis buffer for 1 h and stored at −80°C. Before Western blotting, we centrifuged the lysates at 4°C for 10 min at 13,000 r/min. For 3D samples, we repeated until removal of hydrogel residues. Only for controls, we evaluated protein content with BCA (Bicinchoninic Acid) protein assay kit (PierceTM; Thermo Fisher Scientific) and loaded 20 μg protein in an 8% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). For 3D samples, we optimized the loading volume according to the results of a preliminary test. We transferred the electrophoresis gel to a nitrocellulose membrane (BioRad Laboratories, Hercules, CA, USA). We incubated the membrane with 6E10 monoclonal primary antibody (1:1000) overnight at 4°C, with horseradish peroxidase (HRP)-conjugated anti-mouse IgG antibody (1:15,000; Jackson ImmunoResearch, West Grove, PA, USA), and then used enhanced chemiluminescence (ECL) as the detection system (Millipore, Billerica, MA, USA). We developed the immunoreactive bands with Firereader V10 PLUS 26M Imaging system (Uvitec Ltd, Cambridge, UK) and quantified by ImageJ software.
Dynamic culture of the 3D models of brain cells in the microfluidic device
To investigate the potential of the microfluidic device for the dynamic culture of 3D cell systems of millimeter scale, we prepared 1.25-mm-thick samples by mixing 1 part (v/v) cell suspension (1.325 × 105 cells/cm2) with 9 parts (v/v) COLL-PEG2000 polymer solution. After COLL fibrillogenesis, we moved the culture chambers to a 24-well plate and flooded with culture medium. After 3 days in static conditions, we assembled the chambers into the device and perfused (PHD ULTRA™ Syringe Pumps; Harvard Apparatus, Holliston, MA, USA) in counter-current configuration (0.75 μL/min in the hydrogel chamber and 0.25 μL/min in the opposite chamber; the same flow rates applied for CFD simulations). As a control, we cultured the samples in static conditions. After 3 h, we lysed the samples and assessed the amyloid protein content by Western blotting, as reported. Finally, we repeated the analysis by extending the dynamic culturing time to 3 days.
Statistical analysis
We performed the experiments at least in triplicate on separate occasions and reported the results as mean ± standard deviation (SD). We analyzed the data with GraphPad Prism software (GraphPad Software, Inc.). We used one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparisons test for comparisons among the groups. We set the significance level at 0.05.
Results
Numerical evaluation of the microfluidic device with 3D cell models
The numerical evaluation allowed assessing the effect of the design of the microfluidic device for oxygen consumption and shear stress profiles in the 3D cell culture. Oxygen concentration (Figure 2(c)) showed a linear decay along the direction of perfusion. In the hydrogel construct, it varied from 0.220 to 0.179 mol/m3 at the outlet of the hydrogel chamber. In particular, the cells closer to the membrane were exposed to a lower oxygen concentration with respect to the cells close to the opposite face of the hydrogel (Figure 2(d)).
The plot of shear stress distribution was coherent with the values of the horizontal components (perfusion direction) of the velocity vector. On the top surface of the gel, we computed a maximum value of 1.0 × 10−4 Pa, which reduced to values close to zero on the chamber walls (Figure 2(e)). On the interior part of the hydrogel, the shear stress rapidly decreased while approaching the center: it was maximum (1.64 × 10−10 Pa) on both the superior (perfusion side) and the inferior part of the hydrogel (membrane side), while it was minimum in the central part. For these zones, we computed values of the shear stress close to zero (Figure 2(f)), thus indicating a shielding effect of the hydrogel matrix for embedded cells.
Effect of hydrogel thickness
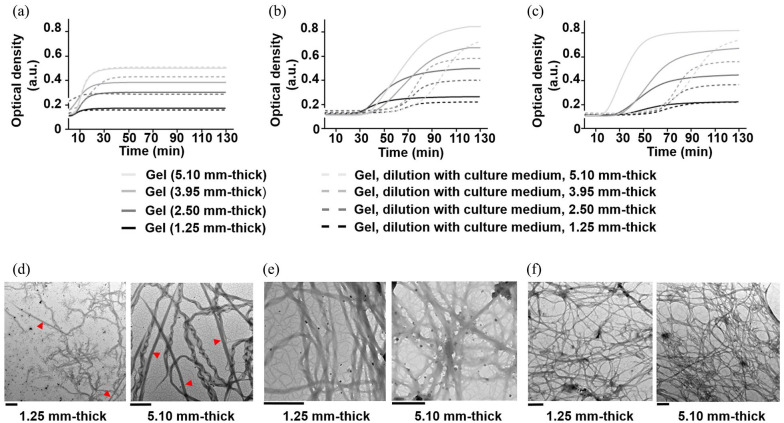
In the range of thickness examined, COLL-HA showed a short lag phase (Tlag < 6 min), which extended to 15–35 min for COLL-PEG3350 and 25–45 min for COLL-PEG2000 semi-IPNs (Figure 3(a)–(c), straight lines). For these materials, Tlag reduced when decreasing the thickness. For all the gels, the slope and K400 max decreased when reducing the thickness, and this trend was statistically significant. The scaling down of the COLL-based semi-IPNs from 5.10 to 1.25 mm thick is feasible, but in 1.25-mm-thick gels, fibril growth rate is slower and fibril diameter is smaller than in 5.10-mm-thick ones.
Figure 3.
Turbidity curves of type I collagen fiber self-assembly in vitro as a function of thickness for (a) COLL-HA, (b) COLL-PEG2000, and (c) COLL-PEG3350 semi-IPNs (straight lines) and (a) COLL-HA, (b) COLL-PEG2000, and (c) COLL-PEG3350 gels after diluting 9 parts (v/v) polymer solutions with 1 part (v/v) culture medium (dashed lines). We averaged six replicates/group and showed the mean curves. TEM images of (d) COLL-HA, (e) COLL-PEG2000, (f) COLL-PEG3350 semi-IPNs, for 1.25- and 5.10-mm-thick samples. The red triangles indicate the peculiar D-periodicity of collagen. Scale bars = 1 μm.
When diluting COLL-HA solution (Figure 3(a), dashed lines), the lag phase was absent for the lower thicknesses (1.25 and 2.50 mm). In these cases, we found that the slope reduced with respect to undiluted conditions, while we did not observe differences in K400 max. The preparation of COLL-HA gels of millimeter thickness supplemented with culture medium is feasible, but fibril growth rate is slower than in the absence of culture medium.
When diluting PEG-based solutions (Figure 3(b) and (c), dashed lines), Tlag extended to 55–75 min for COLL-PEG2000 and about 50–60 min for COLL-PEG3350 matrices. For both gels, the slope was similar, while K400 max decreased with respect to undiluted conditions. Only for 1.25-mm-thick COLL-PEG3350 samples, we did not find differences in K400 max between undiluted and diluted conditions. The preparation of PEG-based semi-IPNs of millimeter thickness supplemented with culture medium is feasible, but the duration of the lag phase is relevant. Only for COLL-PEG2000 gels, fibril diameter is smaller than in the absence of culture medium.
To compare the results related to fibril diameter obtained from turbidity curves in undiluted conditions, we observed 1.25 and 5.10-mm-thick samples by TEM (Figure 3(d)). Regardless of the thickness, we clearly observed the peculiar D-periodicity of COLL only for COLL-HA gels. When scaling down COLL-HA semi-IPNs from 5.10 to 1.25 mm thick, the mean fibril diameter decreased from 152 to 85 nm (p < 0.0001). The observation of a decrease agrees with turbidity curves.
For COLL-PEG2000 gels, the mean fibril diameter varied from 94 (5.10 mm thick) to 96 nm (1.25 mm thick). Oppositely from turbidity curves, TEM analysis indicated that the values were comparable (p > 0.05).
When scaling down COLL-PEG3350 semi-IPNs, the mean fibril diameter decreased from 75 to 68 nm. The decrease was not statistically significant; however, the observation of a decrease agrees with turbidity curves.
When reducing the thickness, we observed a decrease in the density of COLL fibrils in the network. This effect was apparent for COLL-HA and especially for COLL-PEG3350 matrices, while it appeared negligible for COLL-PEG2000 semi-IPNs. This result could indicate that the thermodynamic conditions controlling the formation of COLL-PEG2000 gels lead to the formation of a more stable network.
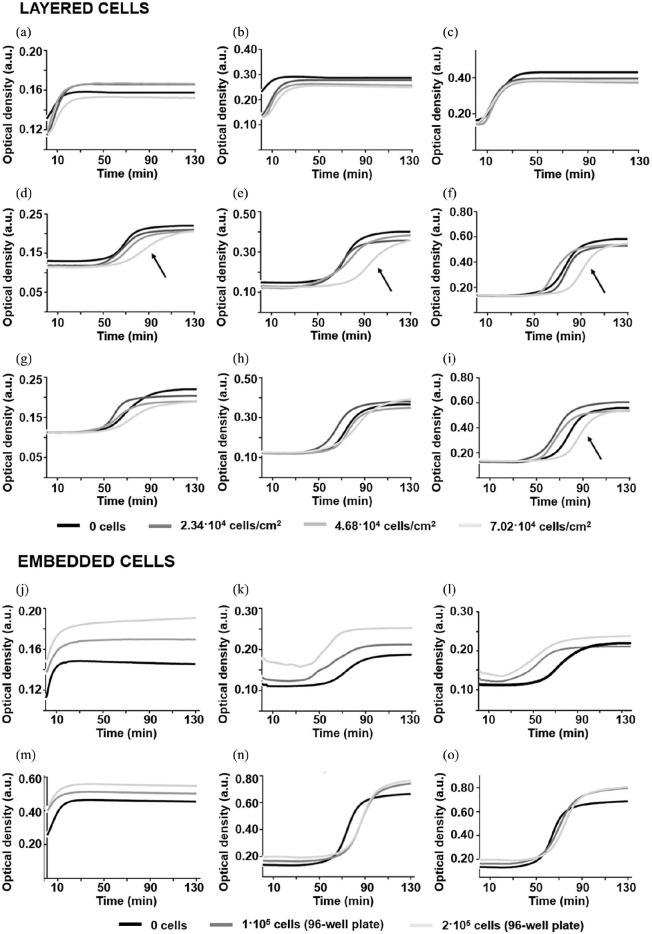
Effect of cell density
For a fixed thickness in layered conditions, the turbidity curves of COLL-HA matrices (Figure 4(a)–(c)) did not vary significantly with cell density. For PEG-based gels, only the slope and K400 max were independent of cell density. For COLL-PEG2000 ones (Figure 4(d)–(f)), the presence of 7.02 × 104 cells/cm2 increased Tlag with respect to the values estimated for the other cell densities, while for COLL-PEG3350 gels (Figure 4(g)–(i)), we observed this increase only for the thickest samples. Since AD is a chronic disease and time is an important parameter in its modeling, we aimed at studying 3D culture systems able to ensure cell survival for long periods. For this reason, we selected the lowest cell density (2.34 × 104 cells/cm2) for the evaluation of cell metabolic activity in layered conditions.
Figure 4.
Turbidity curves of type I collagen fiber self-assembly in vitro as a function of thickness and cell density for COLL-HA (a) 1.25, (b) 2.50, (c) 3.95-mm-thick; COLL-PEG2000 (d) 1.25, (e) 2.50, (f) 3.95-mm-thick; and COLL-PEG3350 (g) 1.25, (h) 2.50, (i) 3.95-mm-thick semi-IPNs in layered conditions and COLL-HA (j) 1.25, (m) 5.10-mm-thick; COLL-PEG2000 (k) 1.25, (n) 5.10-mm-thick; and COLL-PEG3350 (l) 1.25, (o) 5.10-mm-thick semi-IPNs in embedded conditions. We averaged six replicates/group and showed the mean curves. The black arrow indicates the increase in Tlag in the presence of 7.02 × 104 cells/cm2 for 3.95-mm-thick COLL-PEG samples. For the layered condition, we plated 2.34 × 104, 4.68 × 104, and 7.02 × 104 H4-SW cells/cm2 and covered with 1.25-, 2.50-, or 3.95-mm-thick gels, prepared by diluting 9 parts (v/v) polymer solutions with 1 part (v/v) culture medium. As a control, we investigated COLL fibrillogenesis in the absence of cells. For the embedded condition, we diluted 9 parts (v/v) polymer solutions with 1 part (v/v) cell suspension (1 × 105 or 2 × 105 H4-SW cells/sample) and prepared 1.25- or 5.10-mm-thick samples. As a control, we investigated COLL fibrillogenesis for samples prepared by diluting the polymer solutions with culture medium.
To study the effect of cell density independently from embedding protocols, we fixed the embedding procedure (mixing of 1 part (v/v) cell suspension with 9 parts (v/v) polymer solutions) and varied the cell density. As an example, we showed the results for 1.25- and 5.10-mm-thick samples. For COLL-HA gels (Figure 4(j) and (m)), the lag phase was absent. For PEG-based gels (COLL-PEG2000: Figure 4(k) and (n); COLL-PEG3350: Figure 4(i) and (o)), Tlag was not negligible. For COLL-PEG2000 samples, it increased with cell density. At a given cell density, it was shorter (45 min < Tlag < 65 min) for 1.25-mm-thick samples. For 1.25-mm-thick COLL-PEG3350 matrices, it reduced to about 30 min. However, when assessing the entrapment of soluble amyloid forms by confocal microscopy, we verified that the values of Tlag calculated for PEG-based semi-IPNs do not affect the uniformity of cell distribution within the matrices. In fact, only a small fraction of embedded cells adhered to the bottom of the microplate. For all the gels, the slope varied when embedding the cells. Generally, we observed a decrease, suggesting a slowdown of the process. For all the thicknesses and matrices, K400 max increased with cell density, suggesting an increase in fibril diameter and a reinforcement of the tensile strength of the gels.23
As optical microscopy observations indicated a more homogeneous cell distribution, we selected 3.125 × 105 cells/cm2 (1×105 cells/sample, in 96-well plates) as embedding density for further analyses.
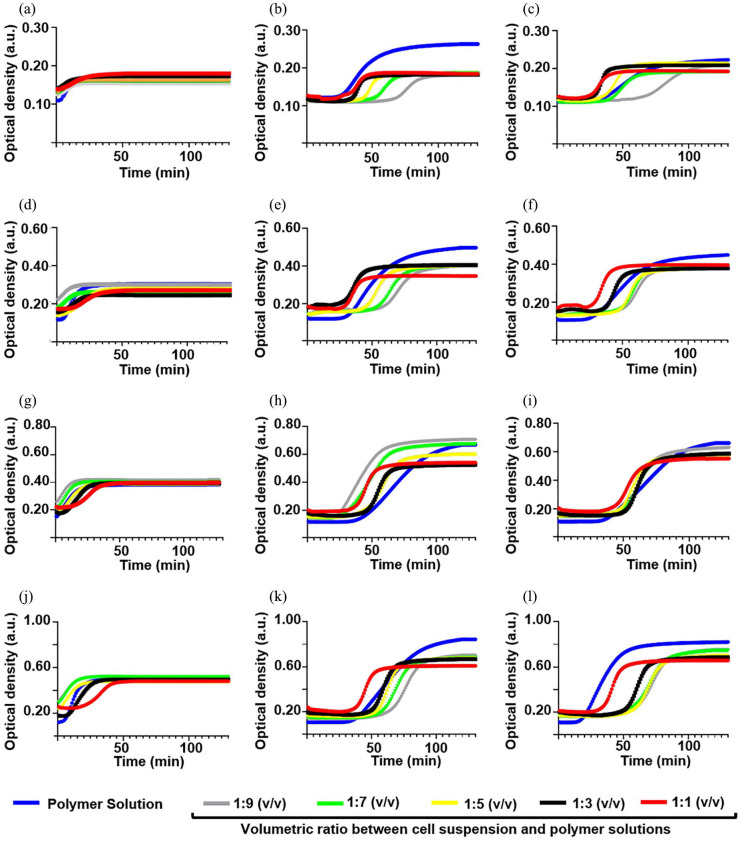
Effect of embedding protocols
The ideal embedding protocol should show the lowest values of Tlag and the highest values of slope and K400 max. With respect to Tlag and slope, we obtained (Figure 5) opposite results for COLL-HA and PEG-based semi-IPNs. For COLL-HA, we observed a lag phase from the mixing of 1 part (v/v) cell suspension with 5 parts (v/v) polymer solutions, while for PEG-based gels, Tlag decreased while increasing the dilution of the polymer solutions. For COLL-HA, we found a decrease in the slope from relevant dilution ratios (1:3 or 1:1), while for PEG-based gels, the slope increased while increasing the dilution of the polymer solutions. Generally, for all the conditions, K400 max decreased while increasing the dilution ratio.
Figure 5.
Turbidity curves of type I collagen fiber self-assembly in vitro for embedded conditions (1 × 105 cells/sample) as a function of thickness and volumetric ratio between cell suspension and polymer solutions. We averaged six replicates/group and showed the mean curves. Curves for 1.25-mm-thick (a) COLL-HA, (b) COLL-PEG2000, and (c) COLL-PEG3350 gels; curves for 2.50-mm-thick (d) COLL-HA, (e) COLL-PEG2000, and (f) COLL-PEG3350 gels; curves for 3.95-mm-thick (g) COLL-HA, (h) COLL-PEG2000, and (i) COLL-PEG3350 gels; curves for 5.10 mm-thick (j) COLL-HA, (k) COLL-PEG2000, and (l) COLL-PEG3350 gels.
For COLL-HA semi-IPNs, the results indicate 1:9 (v/v) as preferable dilution ratio. For PEG-based gels, we did not find a common dilution ratio capable to satisfy all the initial criteria. Since for a given COLL-based gel, K400 max is related to the size of COLL fibrils and, in turn, their diameter influences the tensile strength,23 we privileged the condition with the highest value of this parameter, that is, mixing 1 part (v/v) cell suspension with 9 parts (v/v) polymer solutions. Given the low thickness required to adapt to the miniaturized device, the use of 3D culture systems with greater mechanical strength, for a given formulation, could be useful to counteract material degradation while in dynamic culture, offering the opportunity for prolonged experimental time.
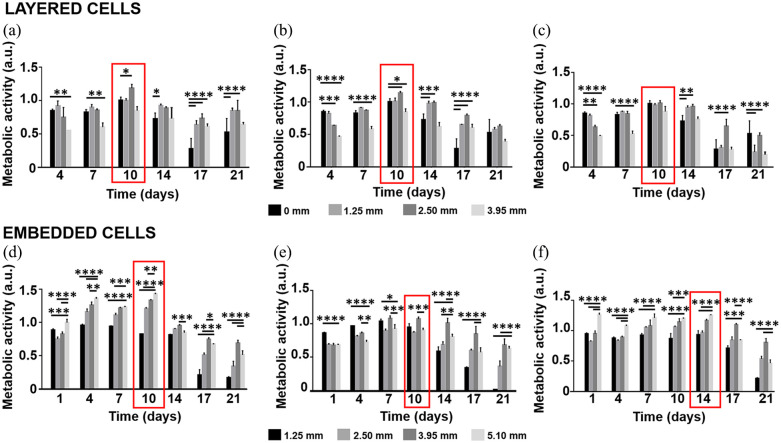
Cell metabolic activity with time as a function of hydrogel thickness
In layered conditions, cell metabolic activity was maximum on day 10 (Figure 6(a)–(c)); therefore, we studied the entrapment of soluble amyloid forms at this time point. Generally, cell metabolic activity was lower for the thickest matrices, while for 1.25- and 2.50-mm-thick samples, it was comparable or greater than that of 2D controls.
Figure 6.
Metabolic activity with time for H4-SW cells as a function of hydrogel thickness. We reported the results as mean ± SD, four replicates/group. The red rectangles highlight the conditions in which cell metabolic activity reached its maximum. For both conditions, we analyzed the results with one-way ANOVA followed by Tukey’s multiple comparisons test. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. For layered cells, we compared the results to controls (0 mm). For embedded cells, we compared the results to those from 5.1-mm-thick samples. For the layered condition, we plated 2.34 × 104 H4-SW cells/cm2 and covered with 1.25-, 2.50-, or 3.95-mm-thick (a) COLL-HA, (b) COLL-PEG2000, and (c) COLL-PEG3350 semi-IPNs. As a control, we performed the study with H4-SW cells in 2D conditions. For the embedded condition, we diluted 9 parts (v/v) polymer solutions with 1 part (v/v) cell suspension (1 × 105 H4-SW cells/sample) and prepared 1.25-, 2.50-, 3.95-, or 5.10-mm-thick (d) COLL-HA, (e) COLL-PEG2000, and (f) COLL-PEG3350 semi-IPNs.
In embedded conditions (Figure 6(d)–(f)), cell metabolic activity was maximum from day 4 to 10 for COLL-HA, from day 7 to 10 for COLL-PEG2000, and from day 7 to 14 for COLL-PEG3350 matrices. Therefore, also for this condition, we assessed the entrapment of soluble amyloid forms on day 10. Generally, for COLL-HA and COLL-PEG3350 gels, the lowest values were measured for the thinner samples (1.25 mm thick), while the greatest values were observed for the thicker ones (3.95 and 5.10 mm thick). Oppositely, for COLL-PEG2000 semi-IPNs, up to day 10 we obtained better results for the thinner gel. However, on longer time points, we confirmed the better performance of thicker gels.
Entrapment of soluble amyloid fragments by immunofluorescence and Western blotting
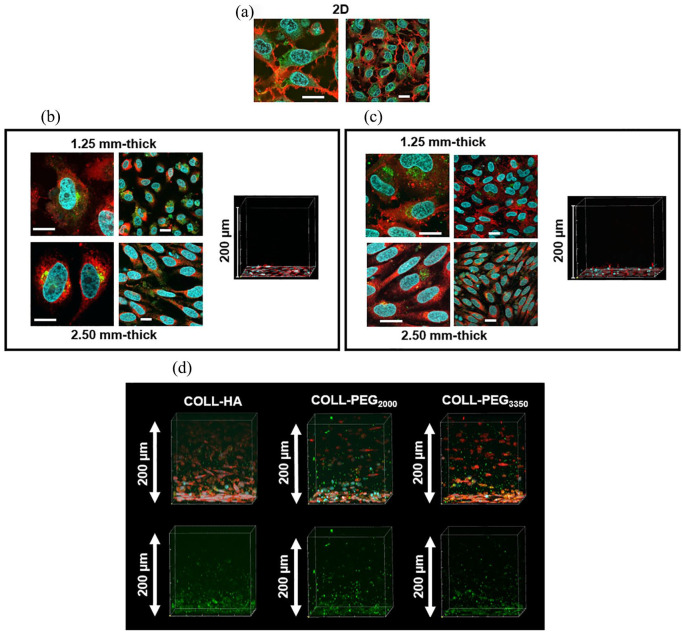
Immunofluorescence images for COLL-HA and COLL-PEG3350 in layered conditions are shown in Figure 7(b) and (c), but those for COLL-PEG2000 semi-IPNs are similar. In 2D conditions, amyloid fragments (stained green) were significantly present in the intracellular space. Nuclei (stained blue) were roundish and cells spread on the substrates. In the presence of a hydrogel layer, cell shape changed with hydrogel thickness: in the thicker samples (2.50 mm), cells stretched along preferred directions, while in the thinner ones (1.25 mm), they showed a more random distribution and a more rounded morphology. The content of soluble amyloids reduced with respect to 2D conditions, but their signal was still confined in the intracellular space, close to the nucleus. Because of the limited optical accessibility of confocal microscopy (about 200 μm from the cellular plane), we considered a representative volume to observe their spatial distribution in the hydrogel layer and we also tested layered samples not incubated with 6E10 primary antibody to assess whether the signal was specifically attributable to soluble amyloids. We detected immunoreactivity against amyloid protein in the hydrogel layers of both samples, but when projecting on the cellular plane we detected signals in the intracellular space only for the samples incubated with primary antibody (data not shown). These results suggest that the selected layered condition (2.34 × 104 cells/cm2) could not be effective in ensuring a sufficient production of soluble amyloid forms or their entrapment. Therefore, it could not to be appropriate for in vitro modeling of AD-related neurodegeneration.
Figure 7.
Immunofluorescence analysis by confocal microscopy for H4-SW cells in 2D, layered, or embedded conditions. For the layered condition, we plated 2.34 × 104 H4-SW cells/cm2 and covered with 1.25- or 2.50-mm-thick gels, prepared by diluting 9 parts (v/v) polymer solutions with 1 part (v/v) culture medium. For the embedded condition, we diluted 9 parts (v/v) polymer solutions with 1 part (v/v) cell suspension (3.125 × 105 H4-SW cells/ cm2) and prepared 2.50-mm-thick samples. We stained cell nuclei with Hoechst 33342 (blue), cell membranes with a lipophilic tracer (red), and soluble amyloid fragments with 6E10 primary antibody (green). (a) Control cells, (b) cells covered by a 1.25-mm or 2.50-mm-thick layer of COLL-HA semi-IPNs, (c) cells covered by a 1.25-mm or 2.50-mm-thick layer of COLL-PEG3350 gels, and (d) cells embedded in 2.50 mm-thick COLL-HA, COLL-PEG2000, or COLL-PEG3350. For (b) and (c), the panel of images on the left represents the projection on the cellular plane of the signal from the secondary antibody detected in the z-axis (200 μm) of the representative volume of hydrogel samples, while the square on the right shows the representative volume of hydrogel samples. Scale bars = 20 μm. For (d), the upper panel shows the complete staining, while in the lower one we omitted the signals from cell membranes and nuclei to highlight the amyloid signal. The z-axis for the representative volume of hydrogel samples was 200 μm.
Immunofluorescence images for 2.50-mm-thick gels in embedded conditions are shown in Figure 7(d), while those from 1.25-mm-thick matrices are reported in Supplementary Figure 1. For all the semi-IPNs, we detected immunoreactivity against amyloid protein in the representative volumes. We also noted evident bright inclusions, probably nonspecific aggregates. Even though much less diffused, we detected a non-negligible level of signal in samples not incubated with 6E10 primary antibody (data not shown). This observation needs further experiments for a confirmation, but it is possible that the more diffused signal found in samples is indicative of the presence of soluble fragments in the extracellular space. These results suggest that the embedded condition is preferable for in vitro models of AD-related neurodegeneration.
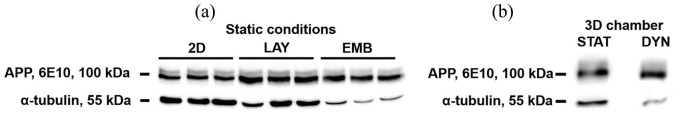
We also assessed APP content by Western blotting. The results (Figure 8(a)) indicated similar values of APP/α-tubulin ratio for controls in 2D conditions and layered H4-SW cells, but greater values for embedded cells. They suggested that, in contrast to the embedded condition, the layered one does not ensure an efficient entrapment of soluble amyloids forms. For this reason, together with the fact that it exploits a greater number of H4-SW cells (and, it maximizes the production source of soluble amyloid forms), we selected the embedded configuration for dynamic cell culture.
Figure 8.
(a) Western blot analysis to detect APP and α-tubulin in the lysates from 2D controls (2D; 2.34 × 104 cells/cm2), layered cells (LAY; 2.34 × 104 cells/cm2 covered with 1.25-mm-thick COLL-PEG2000 gels), and embedded cells (EMB; 3.125 × 105 cells/cm2, 1.25-mm-thick samples. We prepared the samples by mixing 1 part (v/v) cell suspension with 9 parts (v/v) COLL-PEG2000 solution). We prepared the samples in 48-well plates and cultured in static conditions for 10 days. (b) Western blot analysis to detect APP and α-tubulin in the lysates from embedded cells, cultured in static conditions for 3 days and then in static (STAT) or dynamic (DYN) conditions for 3 h.
Dynamic culture of 3D models of brain cells in the microfluidic device
As a preliminary assessment to investigate the suitability of the device for the dynamic culture of 3D culture systems of millimeter scale, we cultured 1.25-mm-thick COLL-PEG2000 gels in dynamic conditions for 3 h. The results from Western blotting (Figure 8(b)) indicated the capability of the selected 3D system to ensure cell viability and the accumulation of amyloid proteins also in the presence of a continuous, counter-current flow of medium. When we extended the duration of dynamic culture to 3 days, we observed a decrease in cell viability, but confirmed the presence of amyloid proteins (data not shown).
Discussion
The potential role of gut microbiota in brain-related pathological pathways is a new, exciting hypothesis in neuroscience.24,25 It represents a potential breakthrough in the field because, if verified, it would shift the investigation of the potential causes of brain-centered pathologies from the brain to the body periphery. In this scenario, the development of a reliable, engineered multi-organ-on-a-chip platform recapitulating in vitro the main players of the MGBA is a challenge, but also a great opportunity to speed up the investigations2—for instance, in the context of AD-related neurodegeneration, whose figures are rapidly growing worldwide. In fact, AD affects one out of nine people aged 65 and older, but this figure increases to one out of three for people aged 85 and older.26
In the present work, we finalized the first key steps to develop an in vitro 3D model of AD based on brain cells loaded in a suitable hydrogel matrix and cultured in an innovative microfluidic device under perfusion with flow rates in the range of the interstitial flow in brain tissues. More in detail, we focused on the development of (1) an innovative microfluidic device that represents the basic unit of the above-mentioned multi-organ platform and (b) a hydrogel-based 3D model of brain cells suitable to be hosted in the device and promote APP expression and accumulation of amyloid fragments as in AD pathological scenario. As main features, this new device is optically accessible and able to host suspended, 2D and 3D cell-based models, cultured alone or interconnected with other organ-on-a-chip units to also represent biological barriers, such as the blood–brain barrier. To design our brain cell models, we selected human neuroglioma H4-SW cells for their simplicity and capability to produce high levels of toxic Aβ. We considered two 3D conditions, referred to as layered- or embedded-based. In the first condition, cells are allowed to adhere to the microplate before hydrogel covering, while the second one guarantees a uniform cell distribution within the hydrogels.
In particular, in the context of AD in vitro models, Choi et al.11 cultured neural progenitor cells in a microfluidic chip under continuous flow conditions and a gradient of oligomeric assemblies of Aβ, but the cells were not in 3D conditions. The fascinating study by Choi et al.12 supported the amyloid hypothesis of AD, but it was performed in static conditions and the chemically undefined composition of Matrigel could influence the variability and reproducibility of the results. To overcome this limitation and tune mechanical cues independently on biomolecular functionalization, Papadimitriou et al.27 proposed star-shaped PEG and heparin gels. However, their study also was conducted under static conditions. Taking advantage of the same cell model as Choi et al., Park et al.28 modeled neuroinflammation activity and neural–glial interactions by a microfluidic device with an annular compartment surrounding a central chamber. Their system allowed to control the spatial and temporal distribution of cell microenvironment and to host 3D gels (100–600 μm), but perfusion was not included. Also in other contexts, the evolution of organ-on-a-chip devices to perfuse hydrogel-based 3D culture systems is just at the beginning. For applications in chemotaxis and migration, Uzel et al.29 proposed a device (300-µm thick) generating simultaneous or time-dependent linear gradients within the gel region. However, it was tested for short times (few hours) and with flow rates (4 µL/min, after a few minutes at 100 µL/min to purge the channels) above the physiological range of brain tissues.11 To obtain a tumor-endothelial co-culture model, Zheng et al.30 applied a microfluidic platform to generate and culture U87-MG cells and human umbilical vein endothelial cells (HUVEC) embedded in alginate microspheres. However, the use of microfluidic devices to perfuse millimeter-thick constructs, as those examined in this study, is not reported.
Since the brain is a highly oxygen-consuming organ, oxygen level is an important parameter for in vitro models of brain tissue. In accordance with the literature, we assumed that in mammalian cells oxygen consumption is governed by the Michaelis–Menten equation.31 However, the measurement of the average cellular consumption rate (oxygen consumption rate (OCR)) in 3D constructs is difficult, because tools for the precise quantification of oxygen concentration in space and time in tissues are lacking. It is a constant neither for a given cell type nor for a given tissue or construct. Moreover, for a construct with specific dimensions, Km and Vmax depend on cell number.32 We therefore assumed these parameters as constant in space and time. Previous computational studies measured the percentage of cells exposed to hypoxic and/or anoxic conditions or evaluated the zones at major risk.33 Cioffi et al.21 quantified oxygen transport in human chondrocytes in a 3D scaffold (a reconstructed cube of 1.25 mm per edge) in a perfusion bioreactor. They considered the very low threshold of 1% oxygen tension (0.02 mol/m3) and demonstrated that at the lower flow rate, about 6% of the cell construct was exposed to anoxic levels. In the context of Parkinson’s disease, Berger et al.20 examined midbrain organoids (1.2 mm thick) under perfusion and estimated that the “dead-core” zone, that is, the zone below the threshold of 0.04 mol/m3, corresponded to 1.8% of cell volume. Coherently with this study, we considered a critical oxygen threshold of 0.040 mol/m3 and computed a minimum oxygen concentration of 0.179 mol/m3 within hydrogel samples. Our results suggest that the culture chambers of the device ensure optimal oxygenation within all the sample thickness. Their design counteracts the potential negative effects of static conditions, where a strong oxygen gradient can expose the cells in the central part of the constructs to hypoxia.34 The numerical characterization also showed that in the center of the gels cultured in the device, the shear stress was close to zero, indicating a shielding effect of the matrix for embedded cells. The shear stress reached its maximum on the superior and inferior surface of the hydrogel, given their proximity to perfusion drag forces. However, the values remained several orders of magnitude lower than the critical ones (on average, in the order of 1–2 Pa).35 It is worth noting that on the inferior surface, the membrane works as a further shielding barrier against shear stresses for embedded cells.
To obtain a 3D model of brain cells to be hosted in the device and able to produce high levels of amyloid fragments, thus favoring their accumulation, we exploited COLL-based semi-IPNs already investigated in our lab15,16 and studied their scaling down from 5.10 to 1.25 mm thick. Because of its design, the thinnest condition is the most suitable to be hosted in the device, also to ensure an optimal optical accessibility during perfusion. For all the conditions in both layered and embedded configurations, we evaluated COLL fibrillogenesis as a function of hydrogel thickness by assessing cell density and cell loading protocols. We found that the setting up of 1.25-mm-thick culture systems is feasible, but the parameters describing COLL fibrillogenesis and cell metabolic activity are modified, and also some differences between COLL-PEG2000 and COLL-PEG3350 exist. This result supports the initial choice of hydrogels belonging to the class of semi-IPNs, whose features are easily tunable to obtain final properties suitable for applications in several contexts.
Despite the turbidity curves in undiluted conditions, TEM analysis showed a significant reduction in fibril diameter only for COLL-HA hydrogels, suggesting structural changes in the polymer network and a possible influence on biological performances.36 Starting from the fitting of rheological data, the Generalized Maxwell Model22 estimated that the mesh size decreased in this order: COLL-HA (116 nm) > COLL-PEG2000 (80 nm) > COLL-PEG3350 (75 nm). Since for similar concentrations of type I COLL, thin fibrils form networks with smaller pore sizes than thick fibers,37 the trends indicated by the Generalized Maxwell Model and TEM analysis agree.
At a given pH, an increase in COLL concentration speeds up the fibrillogenesis and increases fibril diameter because of the greater number of COLL molecules available to form the nucleus center and promote self-assembly.38 When 9 parts (v/v) polymer solutions were diluted with 1 part (v/v) culture medium, the results suggested an influence of the properties of the second polymer in solution. For COLL-HA gels, COLL concentration decreased from 1.2 to 1.08 mg/mL, but we did not estimate differences in K400 max. For PEG-based ones, it reduced from 1.8 to 1.62 mg/mL, leading to an increase in Tlag and a reduction of both slope and K400 max. Interestingly, for all the matrices at a given thickness, generally we estimated similar values of the slope. When we investigated the influence of cell density on COLL fibrillogenesis in layered conditions, we observed an effect for the highest condition tested (7.02 × 104 cells/cm2). In this case, adherent cells could form a more irregular surface, which hindered the direct interactions between COLL monomers during the nucleation phase.
The study of cell loading protocols hides the simultaneous variation of two parameters: COLL concentration, that decreasing while increasing the dilution ratio, and the pH of the final polymer suspension, that increases with the dilution ratio, because turbidity curves were acquired without carbon dioxide supply but with medium containing phenol red. While studying the self-assembly of COLL from pollock skin, Yan et al.38 found that for a given COLL concentration, an increase in pH (up to 7.2) accelerated the fibrillogenesis. This effect was due to the approaching of the isoelectric point, where repulsions among COLL molecules are minimal. In COLL-HA gels, the nucleation and growth phases were modified only for relevant dilution ratios, probably because they considerably decrease COLL concentration. On the contrary, in PEG-based matrices, we observed two effects. As expected from the reduction of COLL concentration, the increase in the dilution ratio decreased fibril diameter and nucleation times with respect to the undiluted condition, while the increase in pH could improve the conditions for COLL fibrillogenesis, increasing the growth rate and reducing the nucleation time when increasing the dilution.
In layered conditions, cell metabolic activity was greater for the lowest thicknesses. This result suggests that the gels act as a physical barrier, even though the results from immunofluorescence analysis did not confirm their efficiency to entrap amyloid fragments. If thickness is above 3.95 mm, the hydrogels may limit the exchange of nutrients, leading to a reduced cell metabolic activity, and eventually to cell suffering or death. By comparing PEG-based gels, we measured a better performance for COLL-PEG2000 in the last time points. For embedded conditions, we found a greater metabolic activity for the highest thicknesses (3.95 and 5.10 mm). This result suggests that a greater hydrogel volume provides better structural support to embedded cells, in accordance with the observation that 3D systems promote cell activity in terms of differentiation and maturation by providing a higher surface area for growth.12,28 Only for COLL-PEG2000 semi-IPNs in the first time points, we measured a better performance for 1.25-mm-thick gels. This was one of the reasons for selecting COLL-PEG2000 for dynamic culture.
On day 10, we tested the capability of our 3D models to allow the accumulation of amyloid fragments. When increasing thickness in layered conditions, H4-SW cells assumed a more elongated morphology with respect to controls in 2D conditions and oriented along specific directions. It might be due to the influence of the directional organization of COLL fibrils, which significantly influences cell adhesion dynamics.36 For both 2D controls and layered cells, the signal from soluble amyloids was mainly intracellular, but it seemed to decrease in the presence of the gels. Since on day 10 the MTS assay indicated that controls had a metabolic activity comparable to that of the cells covered with 1.25-mm-thick COLL-PEG2000 semi-IPNs (p > 0.05), we hypothesized that part of the secreted amyloid fragments were entrapped in the gel. To explore this hypothesis, we analyzed a representative hydrogel volume and assessed the entrapment of soluble amyloid fragments by immunofluorescence assays. The analysis of the representative volume did not provide a conclusive answer, both for the presence of a nonspecific background and the fact that after projecting the signal on the cellular plane, we observed that most of the signal from soluble amyloids was located close to cell nuclei. The analysis of protein content confirmed that the layered condition is probably ineffective to ensure Aβ accumulation. In embedded conditions, we also noticed a nonspecific signal within the representative volume. Bright spots were evident; however, the presence of a diffused amyloid signal in the hydrogel matrix was clear, and Western blot analysis confirmed the presence of amyloid proteins. It is worth noting that a key advantage of the embedded condition was the use of a greater cell number. This allowed increasing Aβ production and concentration, essential to trigger its pathological aggregation.39 This was another reason for selecting the embedded condition for dynamic culture.
Results from dynamic cell culture in the developed organ-on-a-chip device after both 3 h and 3 days suggested the suitability of the proposed device for the culture of 3D hydrogel-based models of brain cells in dynamic conditions. Future works should focus on increasing the complexity of the brain model—for instance, by co-culturing neurons and glial cells and assessing cell viability at long-term time points, as required to investigate microbiota–brain crosstalk in physiological and AD pathological scenario.
Conclusion
In this study, we developed a new, microfluidic and optically accessible organ-on-a-chip device for the dynamic culture of 3D hydrogel-based models of brain cells. Its extreme versatility makes this device exploitable in several contexts, such as the application in multi-organ-on-a-chip platforms to study the crosstalk between different cell populations. We assessed the best culture and cell loading conditions for an innovative 3D hydrogel-based model of brain cells to be hosted in the developed microfluidic device and proposed a 1.25-mm-thick culture system able to recapitulate the AD-related accumulation of amyloid proteins in both static and dynamic conditions. In conclusion, our results indicate that the coupling of the organ-on-a-chip device with the 3D culture system is exploitable for the development of more representative models of brain tissue in physiological conditions and AD pathological scenario.
Supplemental Material
Supplemental material, NEW_Supplementary_S1_Tunesi_et_al for A miniaturized hydrogel-based in vitro model for dynamic culturing of human cells overexpressing beta-amyloid precursor protein by Marta Tunesi, Luca Izzo, Ilaria Raimondi, Diego Albani and Carmen Giordano in Journal of Tissue Engineering
Acknowledgments
The authors are very grateful to Chiara Maffeis, Veronica Caddeo, and Andrea Serafini (Politecnico di Milano) for the technical support; Matteo Laganà (GemmaPrototipi, Longone al Segrino, Como) for microfluidic devices; Lorenzo Sardelli (Politecnico di Milano) for fitting rheological data with the Generalized Maxwell Model; and Felice Volpe (Altergon Italia srl, Morra De Sanctis, Italy; currently at IBSA Institut Biochimique SA, Lugano, Switzerland) for suggestions about HA handling and processing. The authors would like to specially thank Paola Petrini for critical suggestions and discussion.
Footnotes
Author contributions: M.T., D.A., and C.G. conceived the study. M.T. carried out the experiments and wrote the manuscript with support from L.I. and I.R. L.I. performed the computational fluid dynamics simulation. D.A. and C.G. supervised the present manuscript and the related project activities. All authors discussed the results and approved the final manuscript.
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was funded by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (G.A. 724734-MINERVA) to C.G. The results reflect only the authors’ views and the Agency is not responsible for any use that may be made of the information contained.
ORCID iD: Marta Tunesi  https://orcid.org/0000-0003-2507-6186
https://orcid.org/0000-0003-2507-6186
Supplemental material: Supplemental material for this article is available online.
References
- 1. Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behavior. Nat Rev Neurosci 2012; 13: 701–712. [DOI] [PubMed] [Google Scholar]
- 2. Raimondi I, Izzo L, Tunesi M, et al. Organ-on-a-chip in vitro models of the brain and the blood-brain barrier and their value to study the microbiota-gut-brain axis in neurodegeneration. Front Bioeng Biotechnol 2020; 7: 435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bhatia SN, Ingber DE. Microfluidic organs-on-chips. Nat Biotechnol 2014; 32: 760–772. [DOI] [PubMed] [Google Scholar]
- 4. Ingber DE. Reverse engineering human pathophysiology with organs-on-chips. Cell 2016; 164: 1105–1109. [DOI] [PubMed] [Google Scholar]
- 5. Rothbauer M, Rosser JM, Zirath H, et al. Tomorrow today: organ-on-a-chip advances towards clinically relevant pharmaceutical and medical in vitro models. Curr Opin Biotechnol 2019; 55: 81–86. [DOI] [PubMed] [Google Scholar]
- 6. Marchetto MC, Carromeu C, Acab A, et al. A model for neural development and treatment of rett syndrome using human induced pluripotent stem cells. Cell 2010; 143: 527–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dai X, Ma C, Lan Q, et al. 3D bioprinted glioma stem cells for brain tumor model and applications of drug susceptibility. Biofabrication 2016; 8: 045005. [DOI] [PubMed] [Google Scholar]
- 8. Mabry KM, Payne SZ, Anseth KS. Microarray analyses to quantify advantages of 2D and 3D hydrogel culture systems in maintaining the native valvular interstitial cell phenotype. Biomaterials 2016; 74: 31–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Duval K, Grover H, Han LH, et al. Modeling physiological events in 2D vs. 3D cell culture. Physiology (Bethesda) 2017; 32(4): 266–277. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 10. Bodgi L, Bahmad HF, Araji T, et al. Assessing radiosensitivity of bladder cancer in vitro: a 2D vs. 3D Approach Front Oncol 2019; 9: 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Choi YJ, Chae S, Kim JH, et al. Neurotoxic amyloid beta oligomeric assemblies recreated in microfluidic platform with interstitial level of slow flow. Sci Rep 2013; 3: 1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Choi SH, Kim YH, Hebisch M, et al. A three-dimensional human neural cell culture model of Alzheimer’s disease. Nature 2014; 515: 274–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bang S, Na S, Jang JM, et al. Engineering-aligned 3D neural circuit in microfluidic device. Adv Healthc Mater 2016; 5: 159–166. [DOI] [PubMed] [Google Scholar]
- 14. Izzo L, Tunesi M, Boeri L, et al. Influence of the static magnetic field on cell response in a miniaturized optically accessible bioreactor for 3D cell culture. Biomed Microdevices 2019; 21: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tunesi M, Batelli S, Rodilossi S, et al. Development and analysis of semi-interpenetrating polymer networks for brain injection in neurodegenerative disorders. Int J Artif Organs 2013; 36(11): 762–774. [DOI] [PubMed] [Google Scholar]
- 16. Tunesi M, Raimondi I, Russo T, et al. Hydrogel-based delivery of Tat-fused protein Hsp70 protects dopaminergic cells in vitro and in a mouse model of Parkinson’s disease. NPG Asia Mater 2019; 11: 28. [Google Scholar]
- 17. Czvitkovich S, Duller S, Mathiesen E, et al. Comparison of pharmacological modulation of APP metabolism in primary chicken telencephalic neurons and in a human neuroglioma cell line. J Mol Neurosci 2011; 43(3): 257–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tunesi M, Fusco F, Fiordaliso F, et al. Optimization of a 3D dynamic culturing system for in vitro modeling of frontotemporal neurodegeneration-relevant pathologic features. Front Aging Neurosci 2016; 8: 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Song X, Zhu C, Fan D, et al. A novel human-like collagen hydrogel scaffold with porous structure and sponge-like properties. Polymers 2017; 9: 638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Berger E, Magliaro C, Paczia N, et al. Millifluidic culture improves human midbrain organoid vitality and differentiation. Lab Chip 2018; 18: 3172–3183. [DOI] [PubMed] [Google Scholar]
- 21. Cioffi M, Kueffer S, Stroebel J, et al. Computational evaluation of oxygen and shear stress distributions in 3D perfusion culture systems: macroscale and micro-structured models. J Biomech 2008; 41: 2918–2925. [DOI] [PubMed] [Google Scholar]
- 22. Pacheco DP, Butnarasu CS, Briatico Vangosa F, et al. Disassembling the complexity of mucus barriers to develop a fast screening tool for early drug discovery. J Mater Chem B 2019; 7: 4940–4952. [DOI] [PubMed] [Google Scholar]
- 23. Doillon CJ, Dunn MG, Bender E, et al. Collagen fiber formation in repair tissue: development of strength and toughness. Coll Relat Res 1985; 5(6): 481–492. [DOI] [PubMed] [Google Scholar]
- 24. Kowalski K, Mulak A. Brain-gut-microbiota axis in Alzheimer’s disease. J Neurogastroenterol Motil 2019; 25: 48–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sarkar SR, Banerjee S. Gut microbiota in neurodegenerative disorders. J Neuroimmunol 2019; 328: 98–104. [DOI] [PubMed] [Google Scholar]
- 26. Dragomir A, Vrahatis A, Bezerianos A. A network-based perspective in Alzheimer’s disease: current state and an integrative framework. IEEE J Biomed Health Inform 2019; 23(1): 14–25. [DOI] [PubMed] [Google Scholar]
- 27. Papadimitriou C, Celikkaya H, Cosacak MI, et al. 3D culture method for Alzheimer’s disease modeling reveals interleukin-4 rescues Aβ42-induced loss of human neural stem cell plasticity. Dev Cell 2018; 46: 85–101. [DOI] [PubMed] [Google Scholar]
- 28. Park J, Wetzel I, Marriott I, et al. A 3D human triculture system modeling neurodegeneration and neuroinflammation in Alzheimer’s disease. Nat Neurosci 2018; 21(7): 941–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Uzel SGM, Amadi OC, Pearl TM, et al. Simultaneous or sequential orthogonal gradient formation in a 3D cell culture microfluidic platform. Small 2016; 12: 612–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zheng Y, Wu Z, Khan M, et al. Multifunctional regulation of 3D cell-laden microsphere culture on an integrated microfluidic device. Anal Chem 2019; 91: 12283–12289. [DOI] [PubMed] [Google Scholar]
- 31. Ahluwalia A. Allometric scaling in-vitro. Sci Rep 2017; 7: 42113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Magliaro C, Mattei G, Iacoangeli F, et al. Oxygen consumption characteristics in 3D constructs depend on cell density. Front Bioeng Biotechnol 2019; 7: 251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sharifi F, Firoozabadi B, Firoozbakhsh K. Numerical investigations of hepatic spheroids metabolic reactions in a perfusion bioreactor. Front Bioeng Biotechnol 2019; 7: 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lesher-Pérez S, Kim G, Kuo C, et al. Dispersible oxygen microsensors map oxygen gradients in three-dimensional cell cultures. Biomater Sci 2017; 5: 2106–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kretzmer G. Influence of stress on adherent cells. In: Schügerl K, Kretzmer G. (Eds) Influence of stress on cell growth and product formation, Berlin Heidelberg: Springer-Verlag, 2000, pp. 123–137. [Google Scholar]
- 36. Muthusubramaniam L, Peng L, Zaitseva T, et al. Collagen fibril diameter and alignment promote the quiescent keratocyte phenotype. J Biomed Mater Res A 2012; 100(3): 613–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kalbitzer L, Pompe T. Fibril growth kinetics link buffer conditions and topology of 3D collagen I networks. Acta Biomater 2018; 67: 206–214. [DOI] [PubMed] [Google Scholar]
- 38. Yan M, Bafang L, Zhao S, et al. Effect of concentration, pH and ionic strength on the kinetic self-assembly of acid-soluble collagen from walleye pollock (Theragra chalcogramma) skin. Food Hydrocoll 2012; 29: 199–204. [Google Scholar]
- 39. D’Avanzo C, Aronson J, Kim YH, et al. Alzheimer’s in 3D culture: challenges and perspectives. Bioessays 2015; 37(10): 1139–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, NEW_Supplementary_S1_Tunesi_et_al for A miniaturized hydrogel-based in vitro model for dynamic culturing of human cells overexpressing beta-amyloid precursor protein by Marta Tunesi, Luca Izzo, Ilaria Raimondi, Diego Albani and Carmen Giordano in Journal of Tissue Engineering