Abstract
Purpose
The clinical use of BRAF inhibitors in patients with melanoma is limited by intrinsic and acquired resistance. We asked whether next-generation sequencing of pretreatment tumors could identify coaltered genes that predict for intrinsic resistance to BRAF inhibitor therapy in patients with melanoma as a prelude to rational combination strategies.
Patients and Methods
We analyzed 66 tumors from patients with metastatic BRAF-mutant melanoma collected before treatment with BRAF inhibitors. Tumors were analyzed for > 250 cancer-associated genes using a capture-based next-generation sequencing platform. Antitumor responses were correlated with clinical features and genomic profiles with the goal of identifying a molecular signature predictive of intrinsic resistance to RAF pathway inhibition.
Results
Among the 66 patients analyzed, 11 received a combination of BRAF and MEK inhibitors for the treatment of melanoma. Among the 55 patients treated with BRAF inhibitor monotherapy, objective responses, as assessed by Response Evaluation Criteria in Solid Tumors (RECIST), were observed in 30 patients (55%), with five (9%) achieving a complete response. We identified a significant association between alterations in PTEN that would be predicted to result in loss of function and reduced progression-free survival, overall survival, and response grade, a metric that combines tumor regression and duration of treatment response. Patients with melanoma who achieved an excellent response grade were more likely to have an elevated BRAF-mutant allele fraction.
Conclusion
These results provide a rationale for cotargeting BRAF and the PI3K/AKT pathway in patients with BRAF-mutant melanoma when tumors have concurrent loss-of-function mutations in PTEN. Future studies should explore whether gain of the mutant BRAF allele and/or loss of the wild-type allele is a predictive marker of BRAFi sensitivity.
INTRODUCTION
BRAF inhibitors (BRAFis) and RAF/MEK inhibitor combinations are standard-of-care therapies for patients with BRAF V600E/K melanoma. Despite the profound clinical activity of BRAFis in this setting, the degree and durability of response are highly variable.1-6 Prior studies have identified mechanisms of acquired resistance to BRAFis, many of which have been validated using tumor tissues collected at the time of disease progression. These resistance mechanisms can be divided into two classes. Class 1 alterations confer drug resistance by abrogating the ability of the drugs to inhibit ERK signaling and include mutations in NRAS, NF1, MEK1/MEK2, and BRAF splice variants.7-14 Class 2 alterations result in reduced dependence on RAF signaling and include alterations that activate parallel pathways such as inactivating PTEN mutations15-17 and alterations in genes that dysregulate the downstream machinery that mediates cell cycle progression (eg, RB1, CDKN2A, CCND1)17-19 or apoptosis (eg, TP53, MCL1).20 In addition, alterations that overcome BRAF-mediated feedback-induced suppression of upstream receptor tyrosine kinases likely function by both mechanisms.8,21-27
The molecular basis of intrinsic resistance is less well characterized. Here, we used next-generation sequencing methods to determine whether genomic alterations present in pretreatment tumor tissue are predictive of intrinsic resistance to BRAFi therapy in patients with BRAF-mutant melanoma.
PATIENTS AND METHODS
Patient Clinical Characteristics
This study was conducted after institutional review board approval. Patients had stage IV or unresectable stage III BRAF V600E/K melanoma. Response was assessed using both Response Evaluation Criteria in Solid Tumors version 1.1 (RECIST) and response grade, a composite measure of the average of the percentage of lesion shrinkage and the duration of response.28 For response grade, patients were segregated into the following three classes: excellent (≥ 50% tumor shrinkage for ≥ 7 months or any shrinkage for ≥ 12 months), poor (tumor growth, any new lesions, or ≤ 50% shrinkage for < 4 months), or intermediate (neither excellent nor poor). Patients who died or were lost to follow-up before the first assessment or who had incomplete imaging were graded as not evaluable. Progression-free survival (PFS) was defined as the time from the start of BRAFi therapy until the date of progression on BRAFi, date of death, or the date of last follow-up; patients who were switched to immune checkpoint blockade were censored at the date of withdrawal from BRAFi therapy. Overall survival (OS) was measured as the time from the start of BRAFi therapy until the date of death; patients alive at last follow-up were censored at the date of last documented contact. The cutoff date was March 2016, with a median follow-up time for the entire cohort of 14.6 months.
Exon Capture Sequencing
DNA from tumor and blood was analyzed using a custom, exon capture DNA sequencing assay designed to capture all protein-coding exons and selected introns of > 250 cancer-associated genes.29,30 See Data Supplement for gene lists. Single nucleotide variants were detected using MuTect,31 and indels were detected using the SomaticIndelDetector tool in GATK. All candidate mutations were reviewed manually using the Integrative Genomics Viewer.32,33 Mean sequence coverage was calculated using the DepthOfCoverage tool in GATK and used to compute copy number.29,30 The FACETS algorithm was used to estimate tumor purity, ploidy, and allele-specific copy number.34 For correlations with response and outcome, we restricted the analysis to a subset of 56 genes selected based on their mutation frequency in melanoma and their ability to activate MAPK and/or PI3K signaling (Data Supplement). All clinical and genomic data are available through the Memorial Sloan Kettering cBio Portal for Cancer Genomics (http://cbioportal.org/).
Biostatistics
A Kruskal-Wallis test and one-way analysis of variance were used to assess the association between mutations and response. Fisher’s exact t test was used to compare the percentage of mutations for each designated gene between The Cancer Genome Atlas (TCGA) and Memorial Sloan Kettering Cancer Center/Vanderbilt-Ingram Cancer Center cohorts. BRAF-mutant allele frequencies were z score normalized relative to allele frequencies of other mutations in the same sample; deviation from the null distribution was computed using a one-sided Mann-Whitney U test. A Cox proportional hazards model was fit to obtain hazard ratios (HRs) and P values for PTEN mutation (Wald test) from univariable and multivariable models. Survival curves (OS and PFS) were obtained with Prism (GraphPad, La Jolla, CA), and a P value from a log-rank test is presented.
RESULTS
Patient Characteristics and BRAFi Response
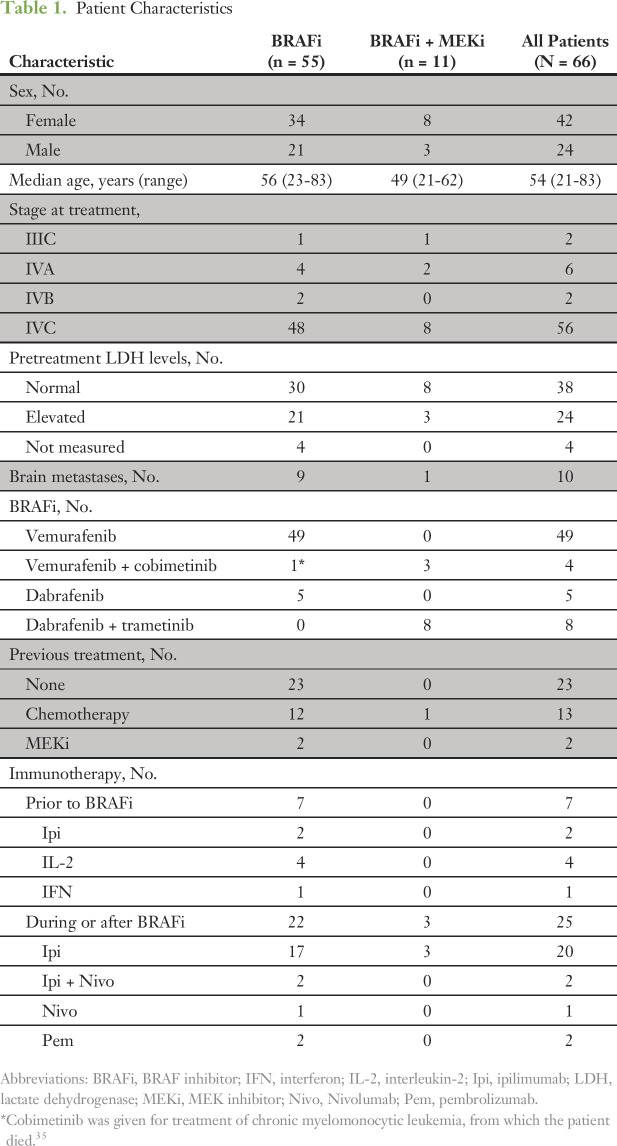
We analyzed pretreatment tumors from 66 patients with metastatic BRAF-mutant melanoma. Forty-nine patients received vemurafenib, five dabrafenib, and 11 a BRAFi plus MEK inhibitor (MEKi) combination. Notably, in the BRAFi therapy group, one patient received cobimetinib (MEKi) for the treatment of chronic myelomonocytic leukemia 75 weeks after the start of BRAFi therapy.35 For this patient, the clinical response to BRAFi monotherapy was only assessable before the administration of the MEKi. The clinical characteristics of the two cohorts of patients are listed in Table 1. Approximately 50% of the patients also received immunotherapy during their treatment course (29 of 55 BRAFi patients and three of 11 BRAFi plus MEKi patients), whereas 19.7% of patients received chemotherapy. Two (3%) of 66 patients had previously received the MEKi selumetinib before treatment with a BRAFi (Table 1).
Table 1.
Patient Characteristics
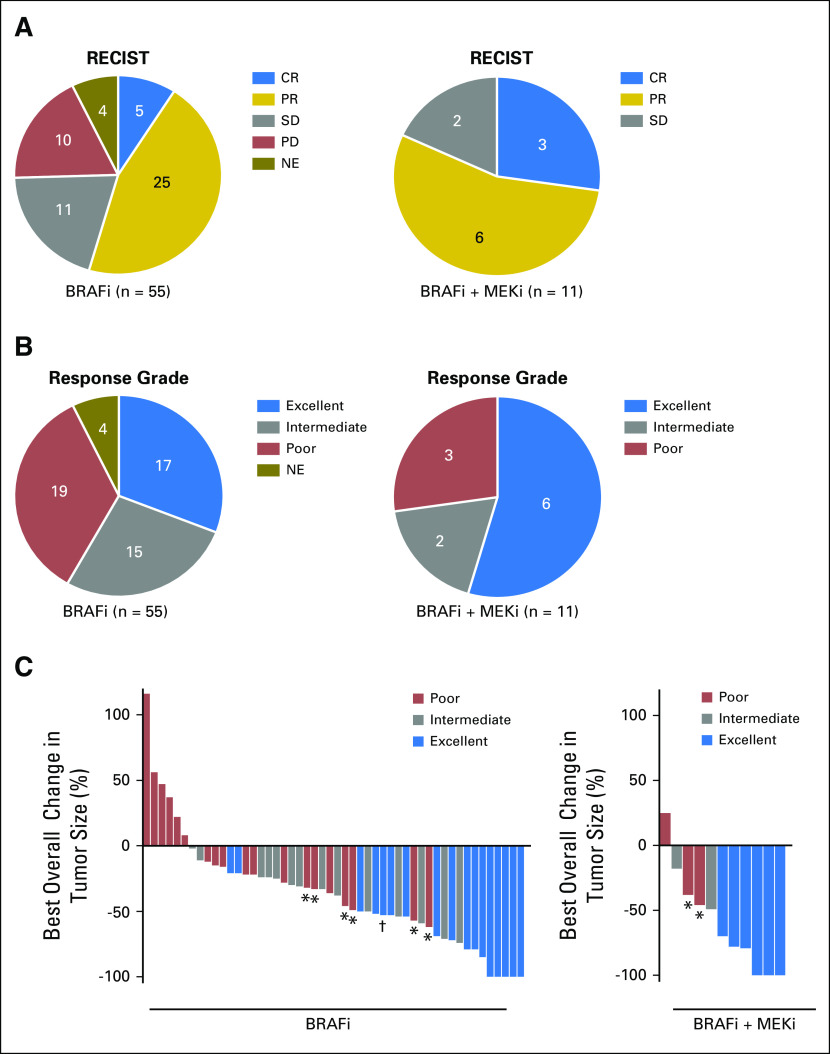
Objective responses, as assessed by RECIST criteria, to BRAFi monotherapy were observed in 30 (55%) of 55 patients, with five (9%) of 55 patients achieving a complete response and 25 (45%) of 55 patients achieving a partial response. Among the patients who achieved a complete response, three remained alive, with two being disease free 4.9 and 4.8 years after initiating therapy. Stable disease was observed in 11 (20%) of 55 patients, whereas 10 (18%) of 55 patients experienced primary progressive disease. Four (7%) of 55 patients were not evaluable for response as a result of rapid clinical deterioration (Fig 1A, left panel). In addition to RECIST, patients were stratified using a response grade classification that incorporates measures of both lesion shrinkage and response duration. Seventeen (31%) of 55 patients achieved an excellent response grade, and 19 (35%) of 55 patients had a poor response, whereas for 15 (27%) of 55 patients, the response was scored as intermediate. For four (7%) of 55 patients, the response grade was not evaluable (Fig 1B, left panel). PFS and OS in the three classes of responders are shown in the Data Supplement. As highlighted in Figure 1C, RECIST 1.1 and response grade classifications were not always concordant, with eight of 66 patients classified as partial responders on the basis of RECIST criteria but as poor responders on the basis of response grade because their responses to BRAFi therapy were of short duration.
Fig 1.
Response and response grade of patients with melanoma to BRAF-targeted therapy. (A) Classification of patients according to Response Evaluation Criteria in Solid Tumors (RECIST). (B) Patients were also stratified accordingly to response grade, which incorporates both tumor regression and response duration. (A and B) Left panel: patients treated with BRAF inhibitor (BRAFi) monotherapy; right panel: patients treated with the BRAFi plus MEK inhibitor (MEKi) combinations. (C) Waterfall plot showing best overall response. Left panel: BRAFi cohort (50 patients; in one patient, best overall response was not evaluable as a result of rapid clinical deterioration); right panel: BRAFi + MEKi (11 patients). Each bar represents an individual patient. CR, complete response; NE, not evaluable; PD, progression of disease; PR, partial response; SD, stable disease. (*) Indicates patients who had a PR according to RECIST but a poor response grade. (†) Indicates patient was treated with vemurafenib plus cobimetinib for chronic myelomonocytic leukemia.
Pattern of Genes Co-Mutated With BRAF
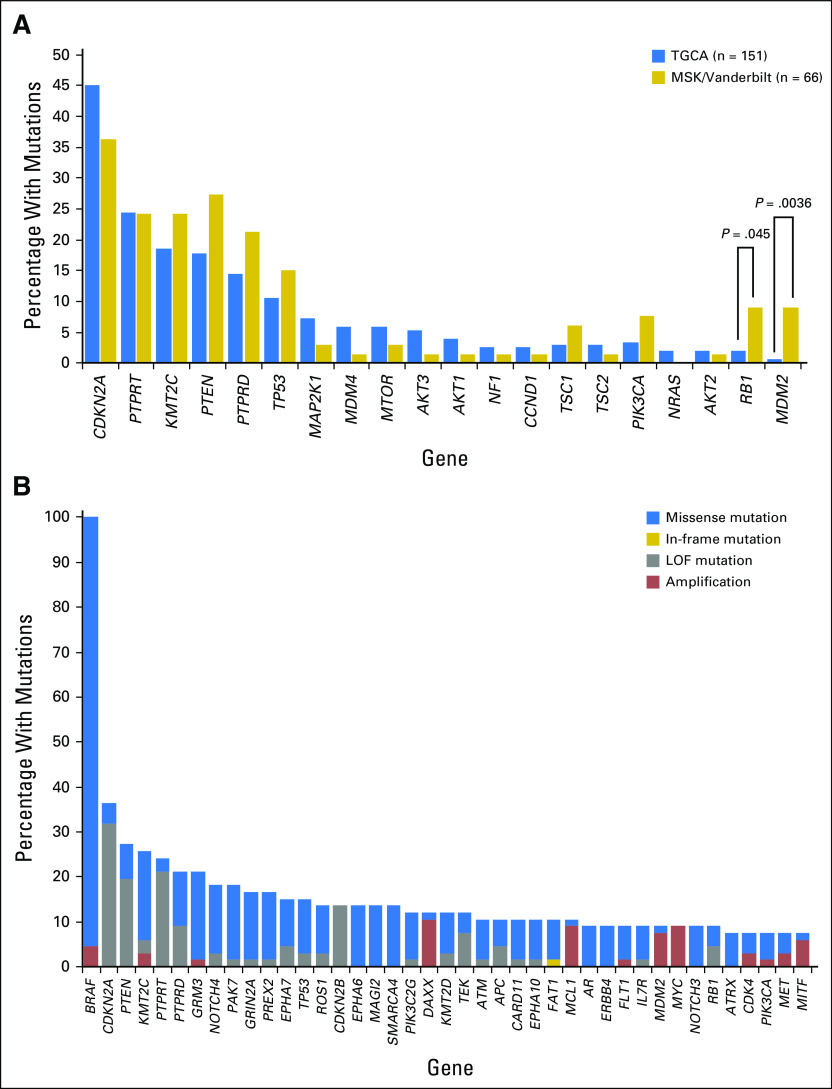
The primary objective of this study was to determine whether the pattern of genes co-mutated with BRAF was predictive of the clinical benefit from BRAFi treatment. Samples were sequenced as outlined in Patients and Methods to a mean coverage of 622-fold, identifying on average nine single nucleotide variants or indels per tumor (range, two to 39 variants or indels; Data Supplement). We confirmed the presence of a BRAF V600E/K mutation in all tumors. In contrast to patients treated with immune checkpoint blockade,36 no correlation was observed between the number of somatic mutations and treatment response (Data Supplement). BRAF amplification was observed in three tumors, all with intermediate responses. Although BRAF amplification has been associated with acquired resistance to BRAFis,37 these data indicate that BRAF amplification does not preclude BRAFi response. An increase in BRAF-mutant allele fraction relative to the mean allele fraction of other variants identified in a sample could indicate allelic imbalance resulting from selective gain of the mutated BRAF allele or loss of the wild-type allele. Significantly elevated BRAF-mutant allele fractions (more than twice the median of all variants detected in the sample) were observed in a total of nine patients (four excellent responders, four intermediate responders, and one poor responder; Data Supplement) in the BRAFi cohort. Normalizing relative to the mean in each case to account for differences in tumor content or purity, we observed a significant enrichment for elevated BRAF V600–mutant allele fraction in excellent responders (P < .001) compared with poor responders (P = .02). Quantitative estimates of tumor ploidy and purity could be calculated for a subset of patients and confirmed allelic imbalance in two excellent responders (Data Supplement), suggesting that selective amplification of the mutant allele or loss of the wild-type allele may be a sensitizing event in these tumors.
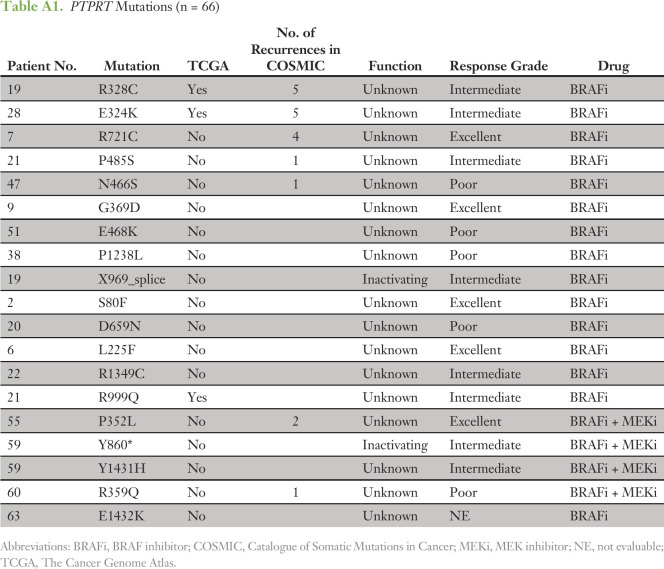
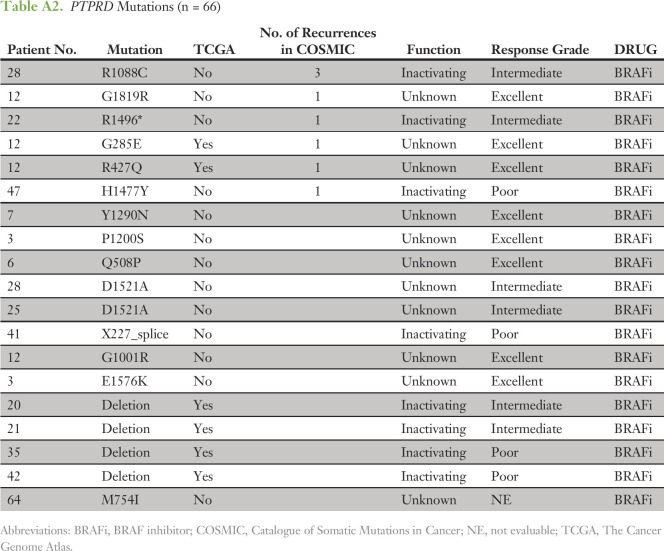
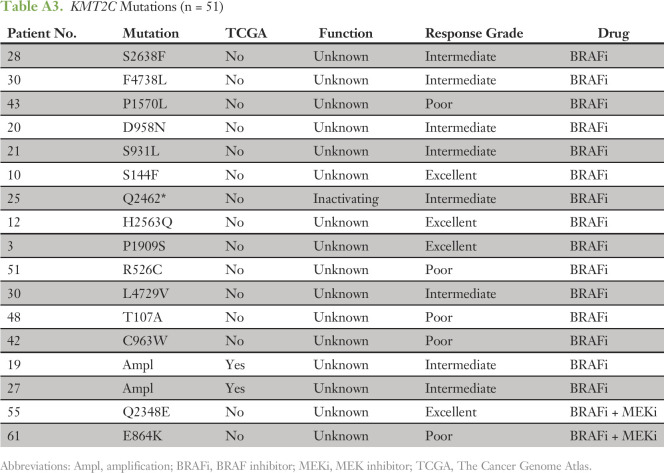
To confirm that the tumors in our cohort had a co-mutation pattern consistent with the genomic landscape of BRAF-mutated melanomas reported in other studies, we compared the 66 tumors analyzed here to the 151 BRAF V600E/K–mutated tumors from TCGA. In both cohorts, CDKN2A and PTEN were among the genes most commonly co-mutated with BRAF (Fig 2). A minority of tumors also harbored coalterations in a second RAS/MAPK pathway gene (NRAS, NF1, or MAP2K1) or in the PI3K/AKT/mTOR axis (PI3KCA, AKT1/2/3, PTEN, TSC1/2, or MTOR). We did observe enrichment for alterations in the RB1 and MDM2 genes, which may reflect the more aggressive clinical profiles of the patients in this study versus the TCGA (Fig 2A). The phosphatases PTPRT and PTRTD and the lysine methyltransferase KMT2C were among the genes mostly commonly coaltered with BRAF (Fig 2). Loss-of-function mutations have been reported in each of these tumor suppressor genes.35,36 In melanoma, the alterations were primarily missense variants of unknown significance (Appendix Tables A1-A3). Given the large size of these genes, the high mutation rate of melanomas and the distribution of the variants throughout the genes, most of the variants observed were likely passenger events (Data Supplement).
Fig 2.
Landscape of most frequently mutated genes across the 66 patients with melanoma treated with BRAF inhibitors. (A) Each bar represents the total percentage of alterations found in each gene in either The Cancer Genome Atlas (TCGA) or Memorial Sloan Kettering/Vanderbilt cohort. P < .05 was considered significant. (B) BRAF mutations were found in all samples (100%), with three of 66 exhibiting coamplification of BRAF. Loss-of-function (LOF) mutations included deep deletions, nonsense mutations, and frameshift alterations predicted to result in early truncation and loss of expression.
Genomic Predictors of Sensitivity to BRAFi Therapy
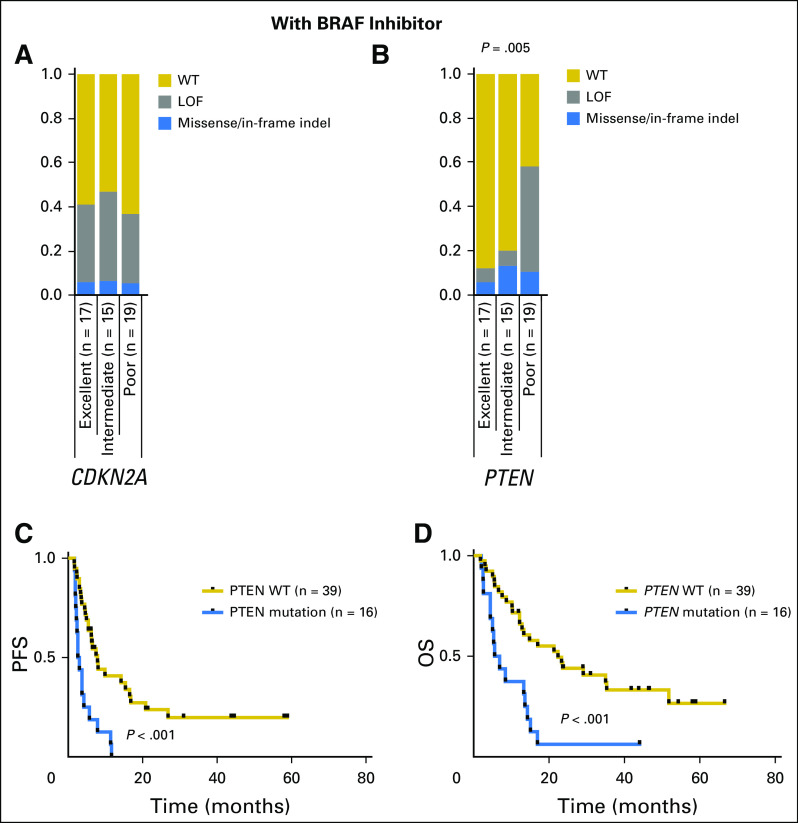
The co-mutational pattern identified in individual patients is shown as an OncoPrint in Figure 3. A prior report had suggested an association between CDKN2A alteration and BRAFi response.18 Twenty-one (41%) of 51 patients receiving BRAFi monotherapy had alterations in CDKN2A (Fig 3, left panel), of which 19 were putative loss-of-function alterations.38 Contrary to previous studies,7,18,39 mutation or homozygous deletion of CDKN2A did not correlate with response grade (Fig 4A).
Fig 3.

OncoPrint of key pathways altered in the three groups of responders. The OncoPrint highlights the pattern of comutation of 20 genes stratified by response grade into excellent (Exc), intermediate (Int), or poor cohorts. The red arrow identifies the patient tumor treated with the MEK inhibitor for chronic myelomonocytic leukemia. Orange dots indicate complete response and still alive; yellow dots indicate complete response and dead or disease progression. Dark green indicates recurrent and/or activating missense mutation; a cutoff of six recurrences or more reported in the Catalogue of Somatic Mutations in Cancer (COSMIC) in at least two tumor types was used to distinguish a recurrent mutation from a rare event. Light green indicates nonrecurrent missense variant of unknown significance; red indicates amplification; blue indicates deep deletion; black indicates truncating deletion; and orange indicates in-frame mutation.
Fig 4.
CDKN2A and PTEN mutation status as a function of response. Percentage of excellent, intermediate, and poor responders with wild-type (WT), loss-of-function (LOF), or missense mutations in (A) CDKN2A or (B) PTEN. Gold indicates WT; blue indicates missense mutation; gray indicates LOF mutation. (C) Progression-free survival (PFS) and (D) overall survival (OS) for patients with PTEN-mutant (gold line) and PTEN WT (blue line) melanomas.
Alterations in PTEN were significantly more common in patients with poor response grade treated with BRAFi monotherapy (11 alterations in patients with poor response, three in patients with intermediate response, and two in patients with excellent response; Fig 4B). Of the 11 PTEN alterations identified in the poor response grade cohort, 10 were likely inactivating, including deep deletions consistent with homozygous loss (n = 5), truncating mutations (n = 4), and one missense mutation (P204S) located in the C2 domain of the protein and known to affect its stability and catalytic activity.40 The one variant of unknown significance (P38S) was located in the phosphatase domain. Patients whose tumors had alterations in PTEN had shorter PFS (HR, 3.46; 95% CI, 1.79 to 6.71; P < .001) and reduced OS (HR, 3.10; 95% CI, 1.59 to 6.05; P < .001; Figs 4C and 4D). The association of PTEN mutation with PFS and OS remained statistically significant after controlling for stage and Eastern Cooperative Oncology Group performance status (PFS: HR, 3.30; 95% CI, 1.70 to 6.43; P < .001; OS: HR, 3.37; 95% CI, 1.70 to 6.70; P < .001). Despite the association between alterations in PTEN and response grade, PTEN alterations were not exclusive to the poor response grade cohort. Specifically, two excellent and three intermediate responders harbored either deep deletions or recurrent missense variants in PTEN (R15S, G132C, and Y177H; Fig 3).41
Given the association between PTEN alteration and BRAFi response, we assessed additional nodes in the PI3K/AKT/mTOR pathway. We detected four missense mutations in PIK3CA, which encodes for the catalytic subunit of PI3K, three of which are hotpot mutations (V344G, E545K, and H1047R). These mutations were not exclusive to patients with a poor response grade (Fig 3 and Data Supplement). In fact, two excellent responders harbored missense mutations in PIK3CA, one of which was an activating hotspot mutation (H1047R).42,43 No hotspot mutations in AKT were observed in the 51 patients treated with BRAFi alone (Fig 3, left panel). A hotspot E17K AKT3 mutation was observed in one poor responder treated with the RAF/MEK inhibitor combination (Fig 3, right panel). Nonrecurrent missense variants of unknown significance in TSC1, TSC2, and MTOR were rare and present in both excellent and poor responders (Fig 3, left panel).
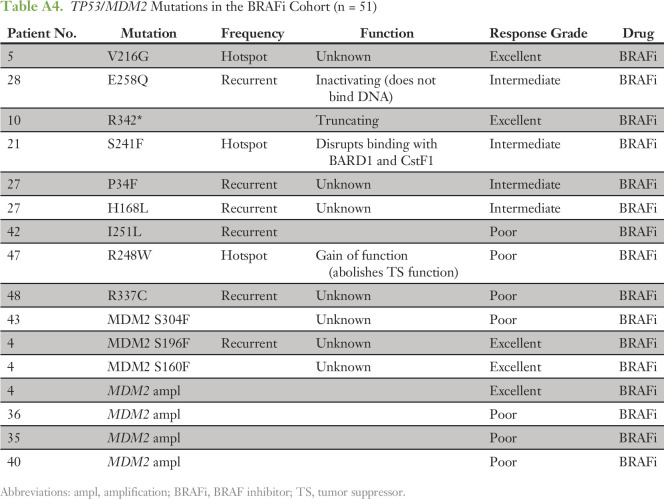
We observed that 25% of the patients (13 of 51 patients) harbored likely functional alterations in the TP53 or MDM2 genes. TP53 was mutated in eight patients (Appendix Table A4), and MDM2 was amplified in four patients and mutated in one patient (Appendix Table A4 and Data Supplement). When analyzed together, alterations in TP53 and MDM2 did not correlate with response grade or shorter PFS and OS (Fig 3, left panel, and Data Supplement).
Alterations that abrogate the ability of BRAFis to inhibit ERK activation (NRAS, MAP2K1, and NF1 mutations) have been shown to be common mechanisms of acquired resistance to BRAFis. None of the 66 pretreatment tumors analyzed harbored an RAS mutation (Fig 3 and Data Supplement). One patient had an alteration in NF1 that would be predicted to result in loss of expression (X2441_splice). Consistent with preclinical data indicating that loss of NF1 confers resistance to BRAFis,11,44 this patient had a poor response grade. MAP2K1 mutations were rare; only one patient, an intermediate responder, harbored an MEK1 mutation (P124S; Fig 3, left panel). A second patient, an intermediate responder treated with a BRAFi plus MEKi combination, had a known activating mutation in MEK1 (Q56P) with an allele frequency of 0.04 (compared with 0.19 for the BRAF mutation), consistent with the presence of a subclonal population. Both the MEK1 P124S and Q56P mutations have previously been shown to be associated with context-dependent resistance to both MEKis and BRAFis.12
We observed mutations in RB1 or amplifications in CCND1 in 11% of patients treated with BRAFi monotherapy; however, no association with response was observed (Data Supplement). Three patients had amplification of MITF, and consistent with a prior report,15 MITF amplifications were observed exclusively in poor or intermediate responders. However, given the rarity of this event, this association did not reach statistical significance (Data Supplement). Finally, four mutations in KDR (kinase insert domain receptor, VEGFR2) were identified, all in patients who exhibited a poor response grade (Fig 3, left panel, and Data Supplement). Of these mutations, only one (VEGFR2 R1032Q) is a hotspot,45 whereas the others are variants of unknown significance. Both OS and PFS were significantly shorter in patients with KDR mutations (Data Supplement). Although this association was statistically significant, given the small number of mutational events and the lack of functional data regarding the significance of these events, this association should be interpreted with caution.
DISCUSSION
BRAFis and MEKis are now US Food and Drug Administration–approved treatments for BRAF-mutant melanoma. Prior studies of small cohorts of patients who initially responded to BRAFis but later developed acquired resistance have identified molecular alterations that underlie acquired drug resistance.7-14,46,47 In this study, we sought to determine whether pre-existent co-occurring alterations predict for intrinsic resistance to BRAFis. As the benefit with anticancer agents has been shown to associate with both depth of response and response duration, we also correlated the genomic findings with response grade, a composite measure of tumor regression and duration of response. We found that loss-of-function alterations of PTEN correlate with poor response to BRAFis in BRAF-mutant melanoma. Furthermore, we observed that patients with PTEN alterations had shorter PFS and OS compared with the PTEN wild-type cohort.
Notably, PTEN and other PI3K pathway alterations were identified in a small number of excellent and intermediate BRAFi responders. This observation is consistent with prior clinical and laboratory studies that suggested that alterations in PTEN do not preclude an antitumor response to BRAFis.9,17,18 The data also suggest that additional comolecular events likely cooperate with PTEN loss to confer BRAFi resistance in patients who derive no clinical benefit from these agents. In sum, the results support the testing of PI3K inhibitors in combination with BRAFi and MEKi alone or in combination in patients with concurrent BRAF and PTEN alterations.48,49 Because complete responses were infrequent even in PTEN wild-type patients, coadministration of a PI3K inhibitor could also result in further incremental tumor regression or a longer duration of treatment response in patients who lack PI3K pathway mutations.
A notable observation from this study was a trend toward increased BRAF-mutant allele fraction in excellent responders. This allelic imbalance is consistent with a relative gain of the mutant BRAF allele. Because increased expression of wild-type BRAF would be predicted to confer resistance to vemurafenib, it is biologically plausible that loss of the wild-type allele leading to a decrease in the fraction of RAF dimers that contain a wild-type RAF protein could increase BRAFi sensitivity.50 The results support broader whole-exome or whole-genome sequencing of BRAF-mutant tumors to explore whether pre-existent BRAF allelic imbalance is a predictive marker of BRAFi sensitivity.
There were several limitations inherent to the sample set analyzed and methodology used in this study. First, the size of the cohort was too small to define associations between response and alterations in genes that are infrequently altered in melanoma. In addition, the current standard of care for BRAF-mutant melanoma has been shifting toward coadministration of BRAFi and MEKi, and PTEN alterations may have less effect in patients treated with the combination. A second limitation is that the approach used could not detect changes in the expression of genes such as PTEN resulting from epigenetic mechanisms. There was also variability in the duration between collection of the pretreatment sample and the initiation of BRAFi therapy, and the molecular status of some genes may have changed during this interval. In addition, only a single pretreatment tumor site was profiled, and thus, some alterations may not have been detected as a result of intratumoral or lesion-to-lesion tumor heterogeneity. Given these latter challenges to the use of pretreatment tumors, we are now exploring the analysis of tumor-derived DNA in plasma, which may better represent the spectrum of molecular alteration present in individual patients. Finally, broader sequencing methodologies such as whole-genome sequencing may identify predictive biomarkers in genes not included in the capture-based approach used in this study.
In summary, our results suggest that pre-existent mutations in PTEN are associated with poor BRAFi response and shorter survival in patients with BRAF-mutant melanoma. Additional studies using broader analysis methods and cell-free DNA may identify additional molecular signatures of intrinsic BRAFi resistance that could be used to guide first-line combinatorial strategies.
ACKNOWLEDGMENT
We thank Efsevia Vakiani, Gopakumar Iyer, and Tara Soumerai for technical assistance; Cyrus Hedvat for directing the core facility that performed the BRAF and NRAS typing; Armando Sanchez and Sherie Mar-Chaim for management of the clinical data; Nikolaus Schultz for the uploading of the data on the cBio Cancer Genomics Portal; and Valerio Donato for critical discussion and review of the article.
Appendix
Table A1.
PTPRT Mutations (n = 66)
Table A2.
PTPRD Mutations (n = 66)
Table A3.
KMT2C Mutations (n = 51)
Table A4.
TP53/MDM2 Mutations in the BRAFi Cohort (n = 51)
Footnotes
Supported by the Melanoma Research Alliance, the National Institutes of Health (NIH), Cycle for Survival, and the Marie-Josée and Henry R. Kravis Center for Molecular Oncology. Also supported in part by the Sloan Kettering Institute for Cancer Research Cancer Center Support Grant No. P30CA008748 and NIH Grant No. K23 CA204726 (to D.B.J.). J.J.H. was supported by a Young Investigator Award from the Conquer Cancer Foundation.
AUTHOR CONTRIBUTIONS
Conception and design: Federica Catalanotti, Donavan T. Cheng, Douglas B. Johnson, James J. Harding, Taha Merghoub, Jeffrey A. Sosman, Michael F. Berger, Paul B. Chapman, David B. Solit
Financial support: Michael F. Berger, David B. Solit
Provision of study materials or patients: Jeffrey A. Sosman, Paul B. Chapman, David B. Solit
Collection and assembly of data: Federica Catalanotti, Donavan T. Cheng, Alexander N. Shoushtari, Douglas B. Johnson, Parisa Momtaz, Catherine Higham, James J. Harding, Taha Merghoub, Michael F. Berger, Paul B. Chapman, David B. Solit
Data analysis and interpretation: Federica Catalanotti, Donavan T. Cheng, Alexander N. Shoushtari, Douglas B. Johnson, Katherine S. Panageas, Helen H. Won, James J. Harding, Neal Rosen, Michael F. Berger, Paul B. Chapman, David B. Solit
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
PTEN Loss-of-Function Alterations Are Associated With Intrinsic Resistance to BRAF Inhibitors in Metastatic Melanoma
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or po.ascopubs.org/site/ifc.
Federica Catalanotti
No relationship to disclose
Donavan T. Cheng
Employment: Illumina, Gilead Sciences (I)
Stock and Other Ownership Interests: Illumina, Gilead Sciences (I)
Alexander N. Shoushtari
Consulting or Advisory Role: Vaccinex, Castle Biosciences, Immunocore
Research Funding: Bristol-Myers Squibb, Immunocore
Travel, Accommodations, Expenses: Bristol-Myers Squibb
Douglas B. Johnson
Consulting or Advisory Role: Bristol-Myers Squibb, Genoptix, Merck
Research Funding: Incyte
Patents, Royalties, Other Intellectual Property: Intellectual property and patents pending surrounding use of MHC-II and response to immune therapy
Katherine S. Panageas
No relationship to disclose
Parisa Momtaz
No relationship to disclose
Catherine Higham
No relationship to disclose
Helen H. Won
No relationship to disclose
James J. Harding
No relationship to disclose
Taha Merghoub
No relationship to disclose
Neal Rosen
Stock and Other Ownership Interests: Beigene, Wellspring, Kura
Honoraria: Novartis, Eli Lilly, Bayer
Consulting or Advisory Role: AstraZeneca, Takeda-Millennium, Daiichi Sankyo, Chugai Pharma
Research Funding: Wellspring/Araxes, Chugai Pharma
Travel, Accommodations, Expenses: Plexxikon
Jeffrey A. Sosman
Honoraria: Amgen, Merck, Array BioPharma
Consulting or Advisory Role: Amgen, Merck, Array BioPharma
Michael F. Berger
Consulting or Advisory Role: Cancer Genetics, Sequenom
Paul B. Chapman
Honoraria: Bristol-Myers Squibb, GlaxoSmithKline, Genentech, Provectus, Momenta Pharmaceuticals, Daiichi Sankyo
Consulting or Advisory Role: Bristol-Myers Squibb, GlaxoSmithKline, Genentech, Daiichi Sankyo, Provectus, Momenta Pharmaceuticals
Research Funding: GlaxoSmithKline, Genentech, Bristol-Myers Squibb, Pfizer
Travel, Accommodations, Expenses: Bristol-Myers Squibb
David B. Solit
Honoraria: Loxo, Pfizer
Consulting or Advisory Role: Pfizer, Loxo
REFERENCES
- 1.Ascierto PA, Minor D, Ribas A, et al. : Phase II trial (BREAK-2) of the BRAF inhibitor dabrafenib (GSK2118436) in patients with metastatic melanoma. J Clin Oncol 31:3205-3211, 2013 [DOI] [PubMed] [Google Scholar]
- 2. McArthur GA, Chapman PB, Robert C, et al: Safety and efficacy of vemurafenib in BRAFV600E and BRAFV600K mutation-positive melanoma (BRIM-3): Extended follow-up of a phase 3, randomised, open-label study. Lancet Oncol 15:323-332, 2014. [DOI] [PMC free article] [PubMed]
- 3.Grob JJ, Amonkar MM, Martin-Algarra S, et al. : Patient perception of the benefit of a BRAF inhibitor in metastatic melanoma: Quality-of-life analyses of the BREAK-3 study comparing dabrafenib with dacarbazine. Ann Oncol 25:1428-1436, 2014 [DOI] [PubMed] [Google Scholar]
- 4.Hodi FS, O’Day SJ, McDermott DF, et al. : Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 363:711-723, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smalley KS: PLX-4032, a small-molecule B-Raf inhibitor for the potential treatment of malignant melanoma. Curr Opin Investig Drugs 11:699-706, 2010 [PubMed] [Google Scholar]
- 6.Flaherty KT, Infante JR, Daud A, et al. : Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med 367:1694-1703, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi H, Hugo W, Kong X, et al. : Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov 4:80-93, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nazarian R, Shi H, Wang Q, et al. : Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature 468:973-977, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trunzer K, Pavlick AC, Schuchter L, et al. : Pharmacodynamic effects and mechanisms of resistance to vemurafenib in patients with metastatic melanoma. J Clin Oncol 31:1767-1774, 2013 [DOI] [PubMed] [Google Scholar]
- 10.Whittaker SR, Theurillat JP, Van Allen E, et al. : A genome-scale RNA interference screen implicates NF1 loss in resistance to RAF inhibition. Cancer Discov 3:350-362, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nissan MH, Pratilas CA, Jones AM, et al. : Loss of NF1 in cutaneous melanoma is associated with RAS activation and MEK dependence. Cancer Res 74:2340-2350, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emery CM, Vijayendran KG, Zipser MC, et al. : MEK1 mutations confer resistance to MEK and B-RAF inhibition. Proc Natl Acad Sci USA 106:20411-20416, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. doi: 10.1016/j.celrep.2013.08.023. Villaneauva J, Infante JR, Krepler C, et al: Concurrent MEK2 mutation and BRAF amplification confer resistance to BRAF and MEK inhibitors in melanoma. Cell Rep 4:1090-1099, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poulikakos PI, Persaud Y, Janakiraman M, et al. : RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature 480:387-390, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Allen EM, Wagle N, Sucker A, et al. : The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov 4:94-109, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paraiso KHT, Xiang Y, Rebecca VW, et al. : PTEN loss confers BRAF inhibitor resistance to melanoma cells through the suppression of BIM expression. Cancer Res 71:2750-2760, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xing F, Persaud Y, Pratilas CA, et al. : Concurrent loss of the PTEN and RB1 tumor suppressors attenuates RAF dependence in melanomas harboring (V600E)BRAF. Oncogene 31:446-457, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nathanson KL, Martin AM, Wubbenhorst B, et al. : Tumor genetic analyses of patients with metastatic melanoma treated with the BRAF inhibitor dabrafenib (GSK2118436). Clin Cancer Res 19:4868-4878, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smalley KSM, Lioni M, Dalla Palma M, et al. : Increased cyclin D1 expression can mediate BRAF inhibitor resistance in BRAF V600E-mutated melanomas. Mol Cancer Ther 7:2876-2883, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fofaria NM, Frederick DT, Sullivan RJ et al: Overexpression of Mcl-1 confers resistance to BRAF V600E inhibitors alone and in combination with MEK1/2 inhibitors in melanoma. Oncotarget 6:40535-40556, 2015. [DOI] [PMC free article] [PubMed]
- 21.Straussman R, Morikawa T, Shee K, et al. : Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature 487:500-504, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson TR, Fridlyand J, Yan Y, et al. : Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature 487:505-509, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yadav V, Zhang X, Liu J, et al. : Reactivation of mitogen-activated protein kinase (MAPK) pathway by FGF receptor 3 (FGFR3)/Ras mediates resistance to vemurafenib in human B-RAF V600E mutant melanoma. J Biol Chem 287:28087-28098, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lito P, Rosen N, Solit DB: Tumor adaptation and resistance to RAF inhibitors. Nat Med 19:1401-1409, 2013 [DOI] [PubMed] [Google Scholar]
- 25.Villanueva J, Vultur A, Lee JT, et al. : Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell 18:683-695, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Basile KJ, Abel EV, Aplin AE: Adaptive upregulation of FOXD3 and resistance to PLX4032/4720-induced cell death in mutant B-RAF melanoma cells. Oncogene 31:2471-2479, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun C, Wang L, Huang S, et al. : Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature 508:118-122, 2014 [DOI] [PubMed] [Google Scholar]
- 28.Harding JJ, Catalanotti F, Munhoz RR, et al. : A retrospective evaluation of vemurafenib as treatment for BRAF-mutant melanoma brain metastases. Oncologist 20:789-797, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Catalanotti F, Solit DB, Pulitzer MP, et al. : Phase II trial of MEK inhibitor selumetinib (AZD6244, ARRY-142886) in patients with BRAFV600E/K-mutated melanoma. Clin Cancer Res 19:2257-2264, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wagle N, Berger MF, Davis MJ, et al. : High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov 2:82-93, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cibulskis K, Lawrence MS, Carter SL, et al. : Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 31:213-219, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thorvaldsdóttir H, Robinson JT, Mesirov JP: Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief Bioinform 14:178-192, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson JT, Thorvaldsdóttir H, Winckler W, et al. : Integrative Genomics Viewer. Nat Biotechnol 29:24-26, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen R, Seshan VE: FACETS: Allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res 44:e131, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abdel-Wahab O, Klimek VM, Gaskell AA, et al. : Efficacy of intermittent combined RAF and MEK inhibition in a patient with concurrent BRAF- and NRAS-mutant malignancies. Cancer Discov 4:538-545, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Snyder A, Makarov V, Merghoub T, et al. : Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 371:2189-2199, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi H, Moriceau G, Kong X, et al. : Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nat Commun 3:724, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yarbrough WG, Buckmire RA, Bessho M, et al. : Biologic and biochemical analyses of p16(INK4a) mutations from primary tumors. J Natl Cancer Inst 91:1569-1574, 1999 [DOI] [PubMed] [Google Scholar]
- 39.Moriceau G, Hugo W, Hong A, et al. : Tunable-combinatorial mechanisms of acquired resistance limit the efficacy of BRAF/MEK cotargeting but result in melanoma drug addiction. Cancer Cell 27:240-256, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Georgescu M-M, Kirsch KH, Kaloudis P, et al. : Stabilization and productive positioning roles of the C2 domain of PTEN tumor suppressor. Cancer Res 60:7033-7038, 2000 [PubMed] [Google Scholar]
- 41.Furnari FB, Huang H-JS, Cavenee WK: The phosphoinositol phosphatase activity of PTEN mediates a serum-sensitive G1 growth arrest in glioma cells. Cancer Res 58:5002-5008, 1998 [PubMed] [Google Scholar]
- 42.Bader AG, Kang S, Vogt PK: Cancer-specific mutations in PIK3CA are oncogenic in vivo. Proc Natl Acad Sci USA 103:1475-1479, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Isakoff SJ, Engelman JA, Irie HY, et al. : Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res 65:10992-11000, 2005 [DOI] [PubMed] [Google Scholar]
- 44.Krauthammer M, Kong Y, Bacchiocchi A, et al. : Exome sequencing identifies recurrent mutations in NF1 and RASopathy genes in sun-exposed melanomas. Nat Genet 47:996-1002, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cancer Genome Atlas Network : Comprehensive molecular characterization of human colon and rectal cancer. Nature 487:330-337, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Solit DB, Rosen N: Resistance to BRAF inhibition in melanomas. N Engl J Med 364:772-774, 2011 [DOI] [PubMed] [Google Scholar]
- 47.Manzano JL, Layos L, Bugés C, et al. : Resistant mechanisms to BRAF inhibitors in melanoma. Ann Transl Med 4:237, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marsh Durban V, Deuker MM, Bosenberg MW, et al. : Differential AKT dependency displayed by mouse models of BRAFV600E-initiated melanoma. J Clin Invest 123:5104-5118, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deuker MM, Marsh Durban V, Phillips WA, et al. : PI3′-kinase inhibition forestalls the onset of MEK1/2 inhibitor resistance in BRAF-mutated melanoma. Cancer Discov 5:143-153, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yao Z, Torres NM, Tao A, et al. : BRAF mutants evade ERK-dependent feedback by different mechanisms that determine their sensitivity to pharmacologic inhibition. Cancer Cell 28:370-383, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]