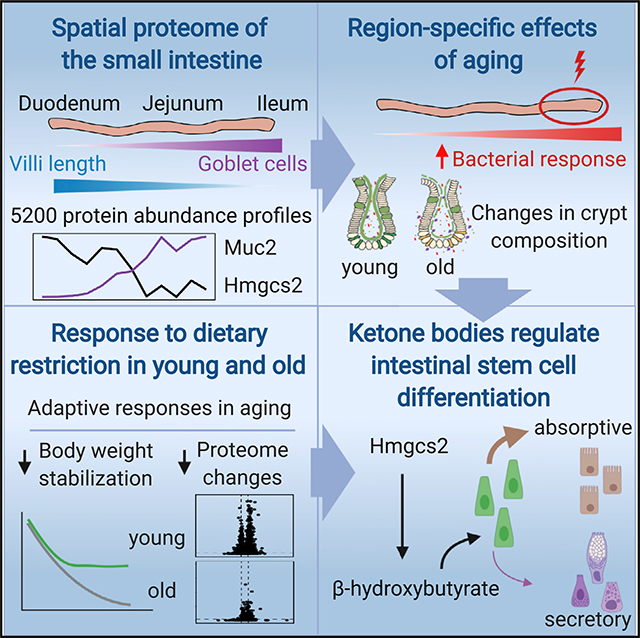
SUMMARY
The small intestine is responsible for nutrient absorption and one of the most important interfaces between the environment and the body. During aging, changes of the epithelium lead to food malabsorption and reduced barrier function, thus increasing disease risk. The drivers of these alterations remain poorly understood. Here, we compare the proteomes of intestinal crypts from mice across different anatomical regions and ages. We find that aging alters epithelial immunity, metabolism, and cell proliferation and is accompanied by region-dependent skewing in the cellular composition of the epithelium. Of note, short-term dietary restriction followed by refeeding partially restores the epithelium by promoting stem cell differentiation toward the secretory lineage. We identify Hmgcs2 (3-hydroxy-3-methylglutaryl-coenzyme A [CoA] synthetase 2), the rate-limiting enzyme for ketogenesis, as a modulator of stem cell differentiation that responds to dietary changes, and we provide an atlas of region- and age-dependent proteome changes of the small intestine.
In Brief
Using proteomics, Gebert et al. find that aging has region-specific effects on the small intestine epithelium of mice. These effects can be partially reversed by modulating ketone body signaling in intestinal stem cells via dietary interventions.
Graphical Abstract
INTRODUCTION
Aging is accompanied by a loss of structure and functionality of many tissues and organs. In the small intestine (SI), aging of the epithelium leads to reduced absorption of nutrients that contributes to malnutrition in elderly people (Nagaratnam, 2018). In animal models, the aging-related overgrowth of bacteria and bacterial dysbiosis in combination with a reduction of the barrier function facilitates crossover of the gut lumen contents, e.g., bacterial products and bacteria, into the body (Biteau et al., 2010; Thevaranjan et al., 2017). These changes contribute to increased tissue-level and systemic inflammation, a hallmark of human aging (Franceschi et al., 2007). During an entire lifespan, the intestinal epithelium shows a high cellular turnover that enables a complete self-renewal of the epithelial layer within 3–5 days (Barker, 2014). The continuous renewal of the intestinal epithelium is necessary to protect its integrity from mechanical insults and pathogens (Gehart and Clevers, 2019). The renewal of the intestinal epithelium is dependent on highly proliferative intestinal stem cells (ISCs) that give rise to multiple differentiated cell types of the absorptive and secretory lineages, e.g., enterocytes and goblet cells. Proliferation and differentiation of ISCs are tightly controlled by a combination of cell-intrinsic and extrinsic cues derived from the stem cell niche to ensure correct output of differentiation (i.e., to maintain the correct proportion between different lineages) and prevent overproliferation (i.e., cancer formation). A number of key pathways, including Wnt, Notch, Bmp, and Egf, have been implicated in controlling ISCs maintenance and fate specification (reviewed in Gehart and Clevers, 2019). In the aging intestine, stem cells lose their ability to maintain organ homeostasis, and, as a consequence, the integrity of the intestinal epithelium is disrupted. While in flies the aging phenotype of ISCs is characterized by overproliferation linked to dysbiosis (Biteau et al., 2010), in the aging mouse SI, ISCs show reduced proliferation (Nalapareddy et al., 2017), decreased number (Mihaylova et al., 2018), and regenerative capacity both in vitro (Mihaylova et al., 2018) and in vivo (Nalapareddy et al., 2017; Pentinmikko et al., 2019). Despite well-established anatomical differences between regions of the SI, no study has so far investigated region-specific effects of aging on the intestinal epithelium.
Dietary restriction (DR) is a lifespan-extending intervention conserved across species (Fontana and Partridge, 2015), and it was shown to restore the structure of the intestinal epithelium leading to a lifespan extension in fruit flies (Regan et al., 2016). ISCs are responsive to changes of diet, including DR and fasting, both of which promote ISC regenerative capacity (Beyaz et al., 2016; Mihaylova et al., 2018; Yilmaz et al., 2012). Recent work in a mouse model for colitis showed beneficial effects of a short fasting period followed by normal diet on inflammation levels, stem cell proliferation, and tissue maintenance in the SI (Rangan et al., 2019). These data consistently point to a beneficial effect of DR on the maintenance of the intestinal epithelium. However, the influence of DR on physiological tissue maintenance in older animals and, importantly, the mechanisms linking ISC fate to changes in food intake remain to be investigated.
Here, we use mass-spectrometry-based proteomics to investigate the spatial proteome of intestinal crypts along the SI. We compare the proteomes between animals of different age groups to reveal age- and region-specific differences in the cellular composition of intestinal crypts. Furthermore, we investigate how young and old mice respond to dietary changes and show that 1 month of DR followed by refeeding partially restores a youthful crypt composition in old animals. Finally, we demonstrate a mechanistic link between diet-induced ketogenic metabolism in ISCs and their differentiation output.
RESULTS
The Proteome of Intestinal Crypts Varies along the SI
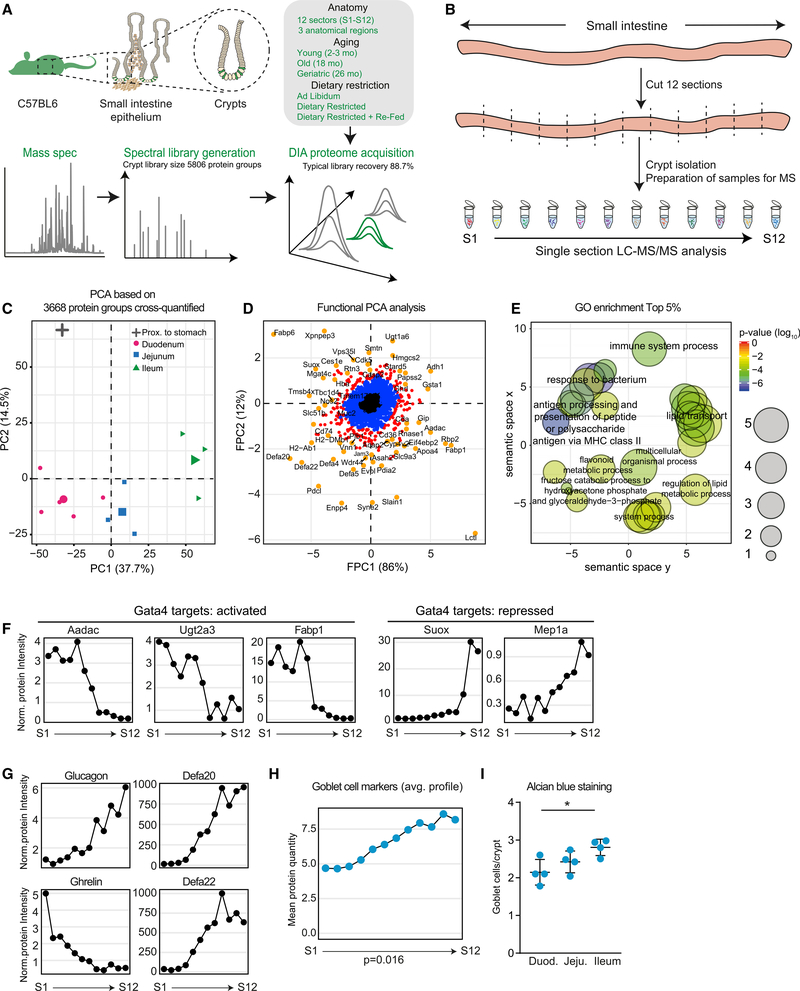
Using a quantitative proteomic workflow based on data-independent acquisition (DIA) mass spectrometry (Muntel et al., 2019), we monitored proteome changes across different regions of the SI. We decided to focus on intestinal crypts, since these anatomical structures beneath the absorptive epithelium are the location of ISCs and differentiated cell types responsible for the secretion of antimicrobial peptides (Paneth cells), mucus (goblet cells), and hormones (enteroendocrine [EE] cells) (Figure 1A). We isolated the SIs from three young C57BL6 mice, sectioned each in 12 pieces of 2–3 cm in length (S1 [proximal] to S12 [distal]) and isolated crypts from each section (Figure 1B; see STAR Methods). All crypt samples were analyzed by DIA mass spectrometry. We identified in total 5,762 proteins, of which 3,668 were quantified across all the 36 samples measured (Table S1). Principal- component analysis (PCA) of the obtained proteome profiles recapitulated the known anatomical organization of the SI with samples from duodenum, jejunum, and ileum forming clearly distinguishable clusters (Figure 1C). Next, we employed functional PCA (FPCA; Yao et al., 2005) to identify proteins whose abundances vary along the SI (Figure 1D). Therefore, we calculated two eigenfunctions: one describing abundance differences between proximal (duodenum) and distal (ileum) SI (FPC1; Figure S1A, top panel) and a second describing higher abundance in the jejunum but similar abundance in duodenum and ileum (FPC2; Figure S1A, bottom panel). Among the most variable proteins showing region-specific abundances (200 proteins, 95–100 percentile), we found an enrichment for Gene Ontology (GO) terms related to defense responses of the mucosal barrier, epidermis development, and positive regulation of transmembrane transport (Figure 1E; Table S1). Our data recapitulated region-dependent differences in abundance of proteins regulated by Gata4, a master regulator of jejunal identity (Thompson et al., 2017; Figure 1F), hormones such as ghrelin and glucagon, and a specific subset of antimicrobial peptides (e.g., Defa20 and Defa22; Figure 1G). In addition, the bile salt transporter Abcba1 and Fam3d, a protein known to be differentially expressed along the SI (de Wit et al., 2012), also displayed higher abundance toward the ileum (Figure S1A). We also noted regional differences in the abundance of rate-limiting enzymes of key metabolic pathways, in particular synthesis of ketone bodies and gluconeogenesis, suggesting that the metabolism of the intestinal epithelial cells varies along the SI (Figure S1B).
Figure 1. Anatomical Variation of the Small Intestine (SI) Crypt Proteome.
(A) Workflow for the mass spectrometry data acquisition and overview of samples analyzed in this study.
(B) Analysis of the regional proteome along the SI. Crypts were isolated from 12 sections (each 2–3 cm long) of three young SIs and single section proteomes were measured by DIA mass spectrometry (S1 [proximal] to S12 [distal]).
(C) Principal-component analysis of 12 crypt samples from different anatomical regions (average of n = 3).
(D) Functional principal-component analysis of protein expression profiles. Distributions of proteins were compared to an average distribution based on the expression pattern along the SI (Table S1).
(E) GO enrichment analysis among proteins showing region-specific abundance. Enrichment analysis was performed using GOrilla (Eden et al., 2009). The complete list of enriched GO terms is reported in Table S1.
(F and G) Abundance profiles of Gata4 target proteins (F; Thompson et al., 2017), hormones (G, left panels, glucagon and ghrelin), and defensins (G, right panels, Defa20 and Defa22) along the SI. Protein intensities were normalized to the median expression across all sections.
(H) Average abundance of marker proteins for differentiated goblet cells along the SI. Significance was tested between S1 and S12 using a Welch t test.
(I) Number of goblet cells per crypt in duodenum, jejunum, and ileum of young mice detected by Alcian blue staining (n = 4) represented as mean ± SD.
Using cell-type-specific markers defined by single-cell RNA sequencing (RNA-seq) (Haber et al., 2017; Table S1), we asked whether the cellular composition of intestinal crypts changes between different anatomical regions. Our data show that while the composition of intestinal crypts appears to be overall anatomically stable (Figure S1D), there are notable exceptions, including changes in subtypes of Paneth cells and enterocytes (Figure S1E), as previously suggested by single-cell analyses (Haber et al., 2017). In addition, we detected significant increase in the level of goblet cells markers toward the more distal part of the SI (Figures 1H and S1F). We validated this finding by counting the number of goblet cells per crypt using Alcian-blue-stained tissue sections obtained from duodenum, jejunum, and ileum. Consistent with the proteomics data, we found an increase in the number of goblet cells per crypt along the SIs that reaches an average of 3 goblet cells per crypt in the ileum (Figure 1I). Taken together, these data quantitatively characterize spatial variation of protein abundance along the SI, identify regional differences in cellular composition of intestinal crypts, and suggest region-specific metabolism of epithelial cells. Histological measurements and previous observations obtained by single-cell RNA-seq independently validated our crypt proteome analysis.
The Effect of Aging on the Crypt Proteome Mimics a Bacterial Infection
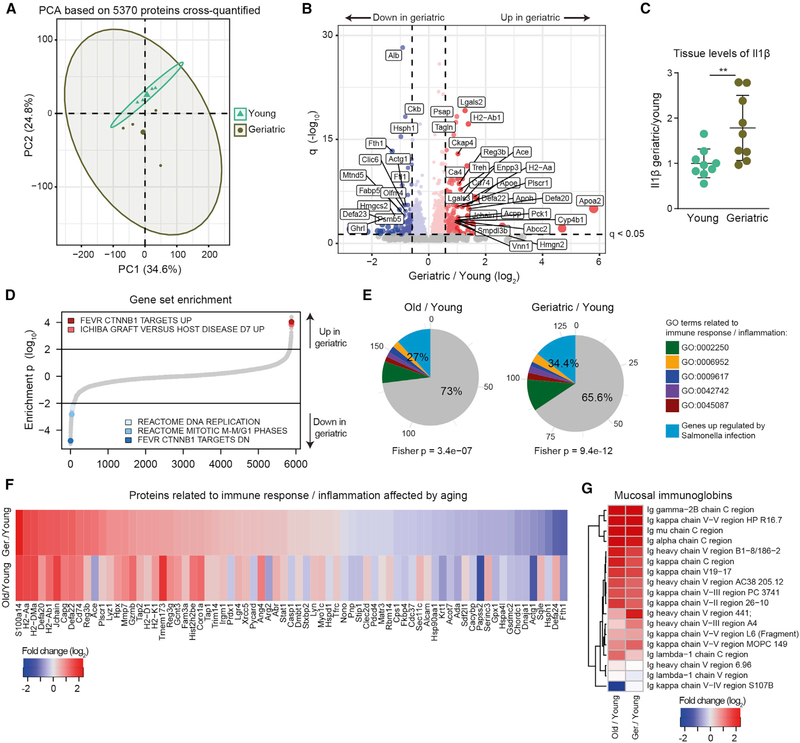
Next, we investigated the impact of aging on intestinal crypts isolated from the whole SI. We compared the abundance of over 5,000 proteins between young, old, and geriatric mice (Table S2). PCA revealed that the intestinal crypt proteome is significantly affected by aging (Figure 2A and S2A). We also noted increased variability among older animals indicated by a higher dispersion in the principal component (PC) space (Figure 2A and S2A). Among the 251 proteins that we identified as affected by aging in the geriatric, we found increased levels of proteins related to the immune response to pathogens, including antimicrobial peptides (e.g., Defa20 and Defa22) and other Paneth cell markers (Nalapareddy et al., 2017), major histocompatibility complex (MHC) proteins (e.g., H2-Aa and H2-Ab1) and lectins (e.g., regenerating islet-derived protein 3-beta [Reg3b]) (Figures 2B and S2B), as well as inflammatory cytokines known to be increased with aging (Franceschi et al., 2000), such as interleukin-1β (Il1β; Figure 2C). Consistent with a decrease in epithelium regenerative capacity in old age, we found decreased level of stem cell markers (Olfm4; Figure 2B) and proteins involved in DNA replication and cell-cycle progression, as revealed by gene set enrichment analysis (Figure 2D). In addition, we found perturbed levels of a group of proteins known to be affected in intestinal crypt cells upon deletion of β-catenin (Ctnnb1) (Fevr et al., 2007; Figure 2D), consistent with a decrease in canonical Wnt signaling in old crypts (Nalapareddy et al., 2017). Pathways related to DNA replication, cell cycle, and targets of Ctnnb1 were similarly affected already at 18 months of age (Figure S2C; Table S2). Intrigued by the increased levels of immune response-related proteins, we compared our aging proteome profiles to single-cell RNA-seq data describing the response of the intestinal epithelium to bacterial infection by Salmonella enterica (Haber et al., 2017). We found that genes upregulated in response to bacteria or annotated to GO categories related to immune response could explain a significant proportion of all the proteins upregulated in old (25.7%, Fisher test p = 2.4e-6) and geriatric (32.6%, Fisher test p = 2.1e-10) animals (Figure 2E). Importantly, most of the changes in inflammation-related proteins, as well as an increase in mucosal immunoglobulin (Ig), were already present in old animals (Figures 2F and 2G).
Figure 2. Proteome Changes in Intestinal Crypts during Aging.
(A) Principal-component analysis based on protein intensities measured by DIA mass spectrometry (n = 4).
(B) Volcano plot depicting proteins that significantly increase (red) or decrease (blue) abundance with aging (Table S2). Proteins not affected are shown in gray. Horizontal dashed line indicates a significance cutoff of q < 0.05 and vertical dashed lines an absolute fold change (log2 > 0.58.
(C) Ilβ levels measured by ELISA in the jejunum of young and geriatric animals (n = 9), represented as mean ± SD.
(D) Gene set enrichment analysis. Gene sets are plotted according to the log10 value of the calculated enrichment score. Positive and negative values are used for gene sets showing higher and lower abundance in old crypts, respectively. Selected significantly affected gene sets (q < 0.25) are highlighted. The complete list of enriched gene sets is reported in Table S2.
(E) Overlap between significantly regulated genes involved in response to bacterial infection (Haber et al., 2017, FDR < 0.25) and proteins significantly upregulated in aging. Fisher test was performed comparing significantly upregulated genes versus all other genes.
(F) Fold changes (log2) of significantly affected proteins related to bacterial/immune response.
(G) Heatmap showing age-related changes in abundance of mucosal-specific immunoglobulins during aging.
Taken together our data describe a distinct response of the intestinal crypt proteome to the aging process. A significant proportion of the age-related changes can be explained by an activation of bacterial response pathways that is detectable already in old mice and that goes hand in hand with a reduction of stem cell activity.
Region-Specific Effects of Aging on the Intestinal Crypt Proteome
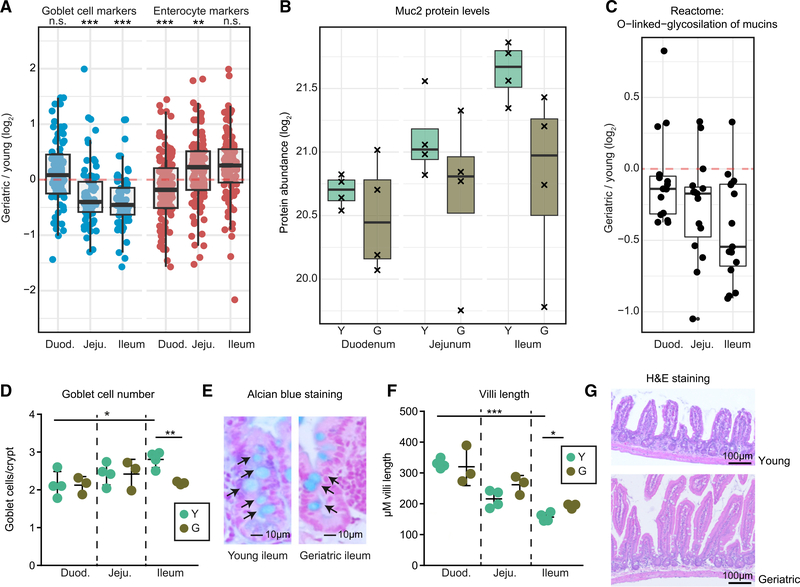
We used our spatial proteome data (Figure 1D) to analyze the distribution of aging-affected proteins along the SI. We found that proteins affected by aging tend to display region-specific abundance, as indicated by generally higher FPCA scores compared to non-affected proteins (Figure S3A), suggesting that different regions of the intestine might be differentially affected by aging. To further investigate this hypothesis, we repeated our proteome analysis on crypts isolated separately from duodenum, jejunum, and ileum from four young and four geriatric mice (Table S3). When we analyzed the effect of aging on markers for differentiated cell types, we found an age-dependent decrease in the abundance of goblet cell markers that manifested exclusively in the distal part of the SI. This was accompanied by a progressive and age-dependent increase in the abundance of enterocyte markers along the SI (Figure 3A). The decrease in goblet cells markers meant a close to 2-fold reduction in the levels of Mucin 2 (Muc2; Figure 3B), the major protein component of the mucosal layer, and of a group of enzymes responsible for the synthesis of the glycan component of mucins (Figure 3C). In order to validate our findings, we stained sections of the SI using Alcian blue and counted the number of goblet cells per crypt in different regions of the SI. We confirmed an age-dependent reduction in the number of goblet cells per crypt exclusively in the ileum of geriatric mice (Figures 3D and 3E) that was not dependent on changes in crypt length (Figure S3B). Notably, the reduction in goblet cells was already present in old mice (Figure S3C). These results confirm that the observed decrease of Muc2 protein levels derives from a reduction in the number of goblet cells that is specific to the ileum. In addition, we observed a co-occurring increase in villi length in old (Figure S3D) and geriatric animals (Figures 3F and 3G) that is also more pronounced in the ileum. Based on these data, we hypothesize that during aging local or systemic cues drive ISCs to differentiate preferentially toward enterocytes instead of goblet cells, leading to a change in crypt composition and macroscopic alterations of the small intestinal epithelium structure.
Figure 3. Specific Reduction of Mucus-Producing Goblet Cells in the Geriatric Ileum.
(A) Abundance changes of marker proteins for enterocytes and goblet cells in duodenum, jejunum, and ileum during aging analyzed by quantitative mass spectrometry (Wilcoxon rank sum test with continuity correction).
(B) Abundance levels for Muc2 in crypts from duodenum, jejunum, and ileum of young and geriatric animals (n = 4).
(C) Differential expressions of protein involved in O-glycosylation of mucins (Reactome: R-MMU-913709) showing an age-related decrease of the whole pathway toward the more distal part of the intestine.
(D) Quantification of goblet cells per crypt in sections stained with Alcian blue from duodenum, jejunum, and ileum of young (n = 4) and geriatric animals (n = 3), represented as mean ± SD.
(E) Representative pictures of Alcian blue stained sections from ileum of young and geriatric animals.
(F) Villi length in the ileum of young (n = 4) and geriatric animals (n = 3), represented as mean ± SD. Sections were stained with hematoxylin and eosin.
(G) Representative pictures of ileum sections from young and geriatric mice.
Old Mice Have Reduced Ability to Adapt to Changes of Diet
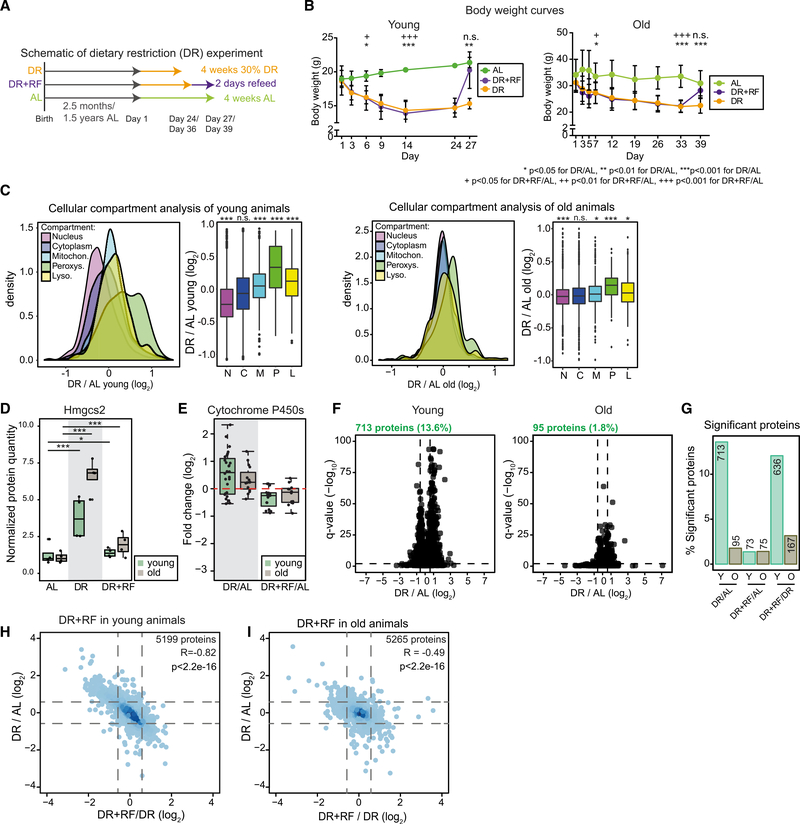
In order to investigate whether aging is affecting the capability of the intestinal epithelium to adapt to environmental changes, we investigated the effect of a short-term DR followed by refeeding on the SI of young and old mice. We compared mice fed a standard ad libitum (AL) diet to age-matched mice that underwent a 30% DR for a period of 4 weeks. A third group of mice was subjected to 2 days of AL refeeding following DR (DR+RF; Figure 4A; see STAR Methods for details). We chose this duration of refeeding because it was sufficient to restore body weight following DR in young mice (Figure 4B) and, additionally, it approximately represents the time of turnover of the intestinal epithelium in rodents (Darwich et al., 2014). Both young and old DR animals showed a reduction in body weight as compared to AL animals that was more pronounced in the old animals (−17.7% and −33.2%; Figures 4B and S4A). Of note, young DR animals were able to stabilize their body weight after 14 days of treatment whereas old DR animals did not show stabilization up until 39 days of treatment (Figure 4B). Following refeeding, both young and old animals were able to compensate their body weight loss within 2 days. Together, these data indicate a reduced ability of old animals to quickly adapt to sudden changes of nutrient availability. In order to gain molecular insights onto these differences, we analyzed the proteome of crypts from young and old animals that underwent changes of diet. DR had prominent effects on the organelle composition of cells and the abundance of metabolic enzymes in both young and old mice. We observed increased levels of peroxisomal proteins (Figure 4C; Table S4) and enzymes involved in lipid and fatty acid metabolism, including Hmgcs2 (3-hydroxy-3-methylglutaryl-coenzyme A [CoA] synthetase 2; Figure 4D), the key enzyme for production of ketone bodies. Both these observations are consistent with activation of Ppard signaling by DR (Mihaylova et al., 2018). Similarly, we found detoxifying enzymes to be increased by DR in young and old animals, e.g., proteins of the cytochrome P450 family (Figures 4E and S4B). Importantly, all of these changes were transient and they reverted to the baseline of AL-fed animals following refeeding (Figures 4E and S4B). Although the activation of nutrient sensing pathways and metabolic adaptations to DR appeared to be preserved in both young and old animals, we noted that the effects of DR were generally more pronounced in young mice with more proteins being significantly regulated by DR (Figures 4F and 4G). In addition, refeeding led to an almost complete reversion of the proteome changes induced by DR in young animals, while this effect was reduced in old animals, as indicated by PCA and a lower correlation between changes induced by DR and DR+RF (compare Figures 4H and 4I with Figures S4C and S4D). Together, these data indicate that old animals have a reduced ability to compensate for sudden changes of diet. This manifests as a reduced capability to stabilize body weight, and it correlates with a dampened response of the intestinal crypt proteome to DR.
Figure 4. Comparison of the Effect of Dietary Restriction (DR) in Young and Old Mice.
(A and B) Schematic of short-term DR experiment (A) and body weights of treated young (n = 4) and old (n = 6–8) animals (B) shown as mean ± SD. *p < 0.05 for DR/AL, **p < 0.01 for DR/AL, ***p < 0.001 for DR/AL, +p < 0.05 for DR+RF/AL, ++p < 0.01 for DR+RF/AL, +++p < 0.001 for DR+RF/AL, one-way ANOVA for multiple comparisons with Tukey’s correction.
(C) Cellular compartment analysis (Parca et al., 2018) of crypt proteome upon DR of young and old animals (Wilcoxon rank sum test with continuity correction).
(D) Protein abundance of Hmgcs2 in crypts of young and old animals (n = 4). Abundance level is shown as normalized protein intensity (mean of AL-fed mice = 1).
(E) Abundance of significant (q < 0.05) enzymes of the cytochrome P450 superfamily shown as fold changes (log2) of the comparison of DR and DR+RF animals to AL.
(F) Volcano plots showing all proteins changed by the DR treatment in young (left) and old (right) animals, horizontal dashed line indicates a significance cutoff of q < 0.05 and vertical dashed lines an absolute fold change (log2) > 0.58 (n = 4).
(G) Percentage of significantly affected proteins in crypts from young and old AL-, DR-, and DR+RF-treated animals, shown as percentage of all identified proteins in the indicated comparison. Numbers above bars are total numbers of significant proteins.
(H and I) Comparison of protein changes induced in young (H) and old (I) animals by DR and DR+RF. All proteins quantified in both comparisons are shown.
Partial Restoration of Cellular Composition of Intestinal Crypts upon Refeeding of Old Mice
Given previous data that showed beneficial effects of DR in promoting ISC function (Yilmaz et al., 2012), we investigated the effects of DR applied to old animals in relation to the aging phenotypes. We compared proteome changes induced by either DR or DR+RF in old animals to the aging signature of geriatric animals (Figures 5A and 5B). We focused exclusively on proteins affected by both aging and DR (Figure 5A) or aging and DR+RF (Figure 5B). We found an unexpected positive correlation between age-related and DR-induced changes (Pearson R = 0.38, p < 2.2e-16), with the majority of the commonly affected proteins (348 out of 465 [74.8%]) showing the same direction of abundance change in both comparisons. These proteins were enriched in GO terms related to metabolism and included members of cytochrome P450 family and rate-limiting metabolic enzymes such as Idh1 (tricarboxylic acid [TCA] cycle) and Fpb1 (gluconeogenesis; Figure 5C). Only for a small subset of proteins (117 out of 465 [25.2%]), the change of abundance induced by aging was reverted by DR treatment. Interestingly, these proteins were enriched in factors involved in defense response against bacterial infection (e.g., Defa20 and Defa22) and correlated with a reduction of tissue levels of Il1β following DR (Figure S5A). On the other hand, the majority of the protein changes induced by DR+RF were anti-correlated with age-related changes (244 out of 402 [60.7%]). GO analysis revealed decreased levels of proteins related to immune system and inflammation following DR+RF and increased levels of proteins related to cell cycle and DNA replication, suggesting a potential restoration of ISC activity (Figure 5B). Intrigued by this finding, we further investigated the impact of dietary interventions on the cellular composition of intestinal crypts. Using proteomics, we found that markers for differentiated secretory Paneth and goblet cells were elevated whereas enterocyte markers were decreased upon DR+RF of young and old animals (Figures 5D and S5B), a phenotype that is opposite to the one observed in aging (Figure 3A). In addition, levels of Muc2 were slightly increased in the old DR+RF animals (Figure S5C). We could validate the restoration of a higher number of goblet cells per crypt in the ileum using histology (Figures 5E and 5F), but we did not observe a significant effect of DR on the length of villi (Figure S5D). Together, these observations indicate that DR applied to old animals leads to proteome changes that are similar to the ones induced by aging, affecting primarily metabolic enzymes. However, old animals treated with a period of DR followed by refeeding displayed increased levels of cell cycle related proteins, a downregulation of immune response genes, and a partial restoration of cellular composition of intestinal crypts suggesting potential anti-aging effects of this dietary intervention.
Figure 5. Comparison of Aging and DR.
(A and B) Comparison of protein fold changes induced by aging and DR (A) and aging and DR+RF (B) in intestinal crypts from the whole SI. In (A) and (B), proteins were grouped according to the sign of the fold change in the compared experiments and GO terms enrichment assessed using GOrilla (Eden et al., 2009). Significantly enriched terms (false discovery rate [FDR] < 0.05) are shown.
(C) Abundance changes of rate-limiting enzymes for key metabolic pathways shown as log2 fold changes in the comparisons as indicated on the right (red indicates upregulation, blue downregulation, and gray indicates not detected for that comparison) (Table S4).
(D) Abundance changes of markers for differentiated cells in crypts from treated old animals (n = 4). See also STAR Methods.
(E) Goblet cell numbers per crypt in the ileum of treated old animals (averages of 50 crypts; n = 4) represented as mean ± SD.
(F) Representative pictures of crypts in the ileum of treated old animals. Scale bar, 50 μm.
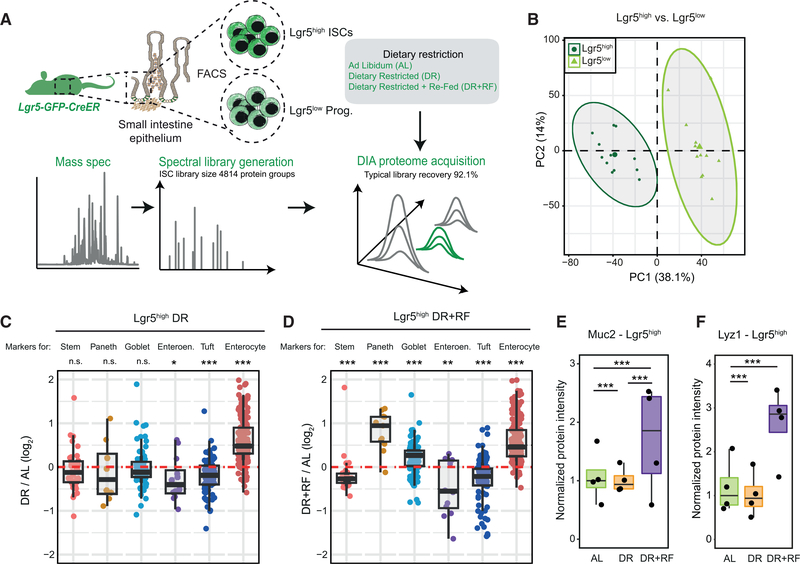
DR Affects the Protein Level of Differentiation Markers in Intestinal Stem and Progenitor Cells
Based on our finding that dietary changes appear to influence both cell proliferation and differentiation in intestinal crypts, we tested how DR and DR+RF influence ISCs. For this, we employed the Lgr5-eGFP-CreER (Barker et al., 2007) mouse model and developed a mass-spectrometry-based workflow to analyze the proteome of freshly isolated ISCs (Lgr5high) and progenitor cells (Lgr5low, Figure 6A). We tested the reproducibility of our workflow by directly comparing the proteome of Lgr5high and Lgr5low cells obtained from the same mouse for a large number of animals (n = 12; Figure 6B), and we could show the expected abundance pattern for stem cell markers (higher in Lgr5high ISCs) and differentiated cell markers (higher in Lgr5low progenitor cells; Figure S6A; Table S6). Next, we treated old Lgr5-eGFP-CreER mice with the same short-term DR+RF intervention, as described above (Figure 4A), and analyzed the proteome of both Lgr5high and Lgr5low cells from AL, DR, and DR+RF mice using DIA quantitative mass spectrometry. We were able to quantify on average 4,787 proteins from Lgr5high ISCs isolated from a single mouse and identified over 200 proteins affected by DR (Figure S6B; Table S5). Overall, the proteome signature induced in Lgr5high ISCs by DR that we measured in this study was similar to the signature induced by fasting on RNA level (Mihaylova et al., 2018; Figure S6F). In order to investigate potential diet-induced changes of ISC differentiation, we analyzed the abundance of differentiated cell markers in Lgr5high ISCs. ISCs and other adult stem cells, including muscle stem cells (van Velthoven et al., 2017), have been shown to express detectable levels of markers of differentiated cells, and the levels of these markers are indicative of the stem cell differentiation potential toward different lineages (Kim et al., 2016). Markers of differentiated cell markers were generally reduced upon DR with the exception of enterocyte markers (Figure 6C). In line with the crypt data, we found increased levels of Muc2 and other goblet cell markers in Lgr5high ISCs only in the DR+RF animals (Figures 6D and 6E). The levels of lysozyme and other Paneth cells markers were also increased in Lgr5high cells (Figures 6D and 6F). Together, our data indicate that short-term DR treatment followed by refeeding promotes the differentiation of Lgr5high cells toward the secretory lineage (Paneth and goblet cells) and thereby partially restores crypt composition in old mice (Figure 5D–F).
Figure 6. DR Alters the Polarization of Intestinal Stem Cells (ISCs) toward Different Lineages.
(A) Intestinal stem and progenitor cells were isolated from the whole SI of Lgr5-eGFP-CreER mice and analyzed by quantitative proteomics.
(B) Principal-component analysis of Lgr5high ISCs and Lgr5low progenitor cells based on protein intensities measured by DIA mass spectrometry (n = 12).
(C and D) Abundance changes for markers of stem cells and differentiated cells in Lgr5high ISCs from old animals that underwent DR (C) or DR+RF (D) (n = 4).
(E and F) Abundance of Muc2 (E) and Lyz1 (F) in Lgr5high ISCs of AL, DR, and DR+RF animals shown as normalized protein intensity (n = 4, mean of AL = 1).
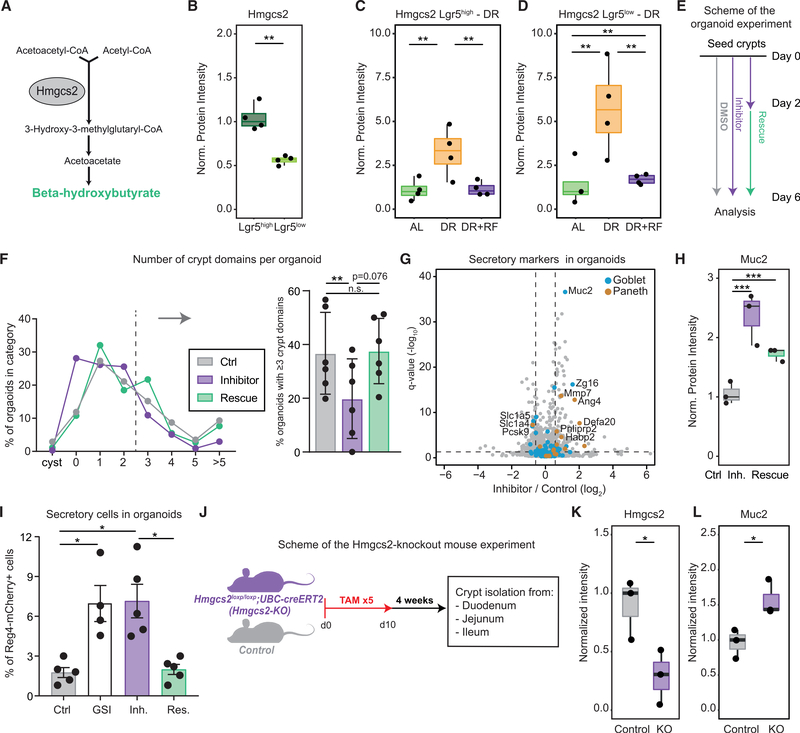
The Activity of Hmgcs2 Modulates the Differentiation of ISCs toward the Secretory Lineage
In order to investigate the mechanism influencing ISC differentiation following dietary changes, we turned our attention toward Hmgcs2. Hmgcs2 is the rate-late limiting enzyme in the synthesis of ketone bodies (Figure 7A). It is known to be highly abundant in ISCs (Muñoz et al., 2012; Figure 7B) and highly expressed in a subpopulation of ISCs showing increased quiescence (Basak et al., 2017). Importantly, Hmgcs2 levels are directly regulated by Mtor and Ppar signaling and robustly react to changes in diet both in crypts and ISCs (Figures 4D and 5C; Mihaylova et al., 2018; Sengupta et al., 2010). As expected, we found Hmgcs2 levels to be higher in old DR animals compared to AL, and DR+RF treatment reduced Hmgcs2 back to normal levels in both cell types (Figures 7C and 7D). In order to investigate whether modulating Hmgcs2 activity is sufficient to influence ISC differentiation, we treated intestinal crypts isolated from young mice with hymeglusin, an inhibitor for Hmgcs1/2 (Tomoda et al., 2004), and assessed organoid-forming capacity in vitro (Figure 7E). We found that inhibitor treatment leads to reduced survival (Figure S7A) and a reduced number of crypt domains per organoid, indicating a reduction of proliferation and differentiation capacity upon Hmgcs1/2 inhibition (Figure 7F). The effect on cell differentiation was partially rescued by the addition of β-hydroxybutyrate (OHB), the major metabolite downstream of Hmgcs2 (Figure 7F), while reduced survival was not (Figure S7A). In order to gain an unbiased view on the effect of Hmgcs1/2 inhibition, we analyzed the proteomes of organoids treated with hymeglusin and found a subset of markers for secretory cells (Paneth and goblet cells), including Muc2, to be significantly increased upon Hmgcs1/2 inhibition (Figures 7G and 7H; Table S6). To test whether Hmgcs2 regulates the differentiation of ISCs into secretory cells, we treated organoids carrying a reporter gene for Reg4+ secretory cells (Sasaki et al., 2016) with hymeglusin, hymeglusin in combination with OHB, and an inhibitor of Notch signaling (gamma secretase inhibitor [GSI]) as a positive control. We quantified the frequency of Reg4-mCherry+ secretory cells in organoids by flow cytometry. The inhibition of Hmgcs2 activity led to an increased number of secretory cells per organoid (Figure 7I), as it occurs upon Notch inhibition (Ohlstein and Spradling, 2007). Importantly, the addition of OHB restored the number of secretory cells per organoid (Figure 7I), highlighting a key role for this metabolite in instructing ISC fate decisions. Finally, we investigated the in vivo influence of Hmgcs2 activity on the epithelial composition using an Hmgcs2 knockout mouse model (Hmgcs2-KO; see STAR Methods; Figure 7J). We isolated crypts from the duodenum, jejunum, and ileum of Hmgcs2-KO and wild-type control mice that received repeated tamoxifen (TAM) injections. First, we verified the reduction of Hmgcs2 in duodenal crypts following TAM using parallel reaction monitoring (PRM) mass spectrometry (Figure 7K). Next, we analyzed the abundance of differentiated cell markers in Hmgcs2-KO and control mice using DIA mass spectrometry. Consistent with in vitro data, this analysis showed increased levels of both Paneth and goblet cells markers in crypts from Hmgcs2-KO mice (Figure S7C; Table S6). We verified increased levels of Muc2 following Hmgcs2 depletion using PRM (Figure 7L). Interestingly, the effect of Hmgcs2 depletion was more pronounced in the duodenum compared to the ileum (Figure S7C), consistent with higher abundance of Hmgcs2 in the proximal part of the SI (Figure S1C). Together, these data demonstrate that the activity of Hmgcs2 regulates ISC regeneration and differentiation in vitro and that reducing the activity of Hmgcs2 promotes the expression of secretory markers both in vitro and in vivo.
Figure 7. The Activity of Hmgcs2 Modulates the Regenerative Capacity and Differentiation of ISCs.
(A) Schematic representation of ketone bodies synthesis pathway. Hmgcs2 is the rate-limiting enzyme.
(B) Abundance of Hmgcs2 quantified by mass spectrometry in Lgr5high ISCs and Lgr5low progenitor cells of young mice (n = 4) shown as normalized protein intensity (mean of Lgr5high = 1).
(C) Abundance of Hmgcs2 in Lgr5high ISCs of ad libitum (AL)-fed, dietary restricted (DR), and dietary restricted and then refed (DR+RF) animals (n = 4). Protein levels are shown as normalized intensity (mean of AL = 1).
(D) Abundance of Hmgcs2 in Lgr5low progenitor cells of AL-fed, DR, and DR+RF animals (n = 4). Protein levels are shown as normalized intensity (mean of AL = 1).
(E) Scheme for the organoid experiment. Freshly isolated crypts were seeded on day 0 and treated with DMSO (control) or Hmgcs1/2 inhibitor (inhibitor) for 6 days or with Hmgcs1/2 inhibitor for 2 days followed by treatment with Hmgcs1/2 inhibitor in combination with β-hydroxybutyrate (rescue) until day 6.
(F) Crypt domain formation in organoids treated with Hmgcs1/2 inhibitor. The distribution is based on the number of crypt domains counted for each organoid after 6 days in culture. Displayed values are averages of n = 6 per condition. The barplot shows the percentage of organoids with ≥3 crypt domains per organoid (n = 6). For each replicate, 3–6 wells were counted and the values averaged. Error bars show mean ± SD.
(G) Volcano plot showing protein changes of organoids treated with DMSO (control) or Hmgcs1/2 inhibitor for 6 days. Horizontal dashed line indicates a significance cutoff of q < 0.05 and vertical dashed lines an absolute fold change (log2) > 0.58.
(H) Abundance of Muc2 in treated organoids shown as normalized protein intensity (mean of control = 1).
(I) Percentage of Reg4+ cells per organoid after treatment with gamma secretase inhibitor (GSI; n = 4), Hmgcs1/2 inhibitor (Inh.; n = 5), and Hmgcs1/2 inhibitor in combination with β-hydroxybutyrate (rescue [Res.], n = 5). Data are shown as mean ± SD (Holm Sidak’s multiple comparisons test).
(J) Scheme of the Hmgcs2-KO mouse experiment. Hmgcs2loxp/loxp;UBC-creERT2 mice (Hmgcs2-KO) were injected with tamoxifen (TAM) five times and kept after induction for 4 weeks before crypt isolation from duodenum, jejunum, and ileum.
(K and L) Parallel reaction monitoring data represented as mean intensity of selected peptides normalized to the median of control animals for Hmgcs2 (K) and Muc2 (L) (n = 3; Welch t test).
DISCUSSION
Here, we provide an atlas of the spatial organization of the SI crypt proteome in mouse. We show that hundreds of proteins have a tightly regulated abundance along the SI that underlies both changes in the cellular composition of the crypt (e.g., increase of goblet cells in the ileum), as well as changes in the proteome of specific cell types (e.g., different subtypes of Paneth and EE cells). Most of the protein differences appear to be continuous and progressive along the SI, suggesting the existence of cues derived from either the stem cell niche or the intestinal lumen (e.g., the microbiome) that are responsible for actively maintaining these specific protein expression programs. Two of such examples are the drastic increase of bacterial load that characterizes the more distal part of the SI (Sender et al., 2016) and the gradient of basal Wnt activity along the intestinal tract (Leedham et al., 2013).
Aging is known to affect different organs at different rates (Ori et al., 2015; Pisco et al., 2019), and differences have been observed also within the same organ, as previously shown for skeletal muscle (Messa et al., 2019). Similarly, in the SI, different anatomical regions show different aging phenotypes. For instance, we show that while in the duodenum there is a slight increase in the number of goblet cells per crypt, the more distal part of the SI and, in particular, the ileum is affected by a decreased number of goblet cells per crypt. Furthermore, the age-dependent increase in villi length is significantly more pronounced in the distal part of the SI. Together, the reduction in goblet cells and the increase in villi length in the ileum appear to equalize differences in the epithelial structure between duodenum and ileum of old animals that are present in young animals. This points to a dysregulation of epithelial maintenance during aging that is specific to the ileum and consistent with a more pronounced aging phenotype in this region of the intestine (Martin et al., 1998; Nalapareddy et al., 2017).
The anatomical differences that we describe might underlie some of the apparent discrepancies among previously reported data on mouse SI aging. Nalapareddy et al. reported an increase in Paneth cell number per crypt and goblet cell number per crypt-villus axis in old animals (Nalapareddy et al., 2017). Moorefield et al. investigated morphometric changes in the jejunum of old mice and reported increase of Paneth cells but no changes in goblet cell number per crypt (Moorefield et al., 2017). Finally, Mihaylova and colleagues confirmed the increase in Paneth cell number per crypt in the proximal and distal part of the SI; however, and in line with our data, they showed that the number of goblet cells per crypt is stable in the proximal and middle part of the SI, while it declines in the more distal region (Mihaylova et al., 2018). Given intrinsic differences in immune responses, metabolism, and exposure to environmental cues, our study clearly highlights the need to investigate different regions of the SI separately to study physiological (e.g., aging) and genetic responses.
We studied the ability of young and old animals to adapt to dietary changes to functionally assess the impact of aging on the plasticity of the intestinal epithelium. Although both young and old animals respond to DR, the response of old animals appears to be diminished both at the organismal level, as indicated by a reduced capability to stabilize body weight, as well as at the intestinal level, as indicated by a reduced response of the crypt proteome. The reduced adaptive response of the old intestinal epithelium that we describe corroborates previous observations from model organisms and humans pointing to an age-related reduced ability to adapt to stress and environmental changes (Pomatto and Davies, 2017), and it might drastically influence the way old individuals respond to changes in nutrient availability, microbiome changes, and drug treatment.
Unexpectedly, we found that DR induced a proteome signature in both young and old animals that was similar to the one that we observed during aging. This suggests that DR of old animals is potentially aggravating aging-related changes, an observation that was recently made also in other tissues (Hahn et al., 2019). The reversal of aging-related changes of protein abundance and epithelial composition was partially achieved when DR was followed by refeeding. Dietary interventions based on intermitting fasting or transient periods of fasting mimicking diet can have beneficial effects on human health parameters (Stekovic et al., 2019) and during recovery from colitis in an injury mouse model (Rangan et al., 2019). Both these types of interventions include cycles of transient DR followed by refeeding that resemble our DR+RF cycle. Our data indicate that the alternation between the fasted and fed state, rather than DR per se, promotes a restoration of epithelium composition and a partial rejuvenation of the crypt proteome when applied to old animals. It is tempting to speculate that these effects might contribute, at least in part, to the health-promoting effects of dietary interventions based on intermitted fasting. However, further investigations are required to demonstrate a causal link between this type of intervention and improvement of intestinal functions.
In addition to the already known effects of DR on ISCs regenerative capacity (Igarashi and Guarente, 2016; Mihaylova et al., 2018; Yilmaz et al., 2012), we show that changes of diet can also influence the differentiation of ISCs. Mechanistically, we directly link diet-induced changes in the levels of the mitochondrial enzyme Hmgcs2 and its downstream metabolite, OHB, to ISC differentiation. Hmgcs2 has been previously shown to influence the differentiation of colorectal cancer cell lines in vitro (Wang et al., 2017) and differentiation behavior of ISCs in vivo in young animals that underwent a ketogenic diet (Cheng et al., 2019). Here, we show that treatment of mice with DR leads to increased levels of Hmgcs2 in ISCs and a co-occurring reduction of all differentiation markers, with the exception of enterocyte markers. Subsequent refeeding of mice that underwent DR rapidly reduces Hmgcs2 protein levels in ISCs and progenitor cells and increases the expression of Paneth and goblet cell markers in ISCs, leading to restoration of goblet cell numbers in ileal crypts. Using organoid cultures, we confirmed that pharmacological inhibition of Hmgcs2 is sufficient to reduce organoid-forming capacity and branching and to promote the differentiation toward the secretory lineage. These effects were comparable to the one induced by gamma secretase inhibition in vitro, reverted by OHB supplementation, and recapitulated in vivo using a genetic model of Hmgcs2 depletion. Recently, Cheng et al. demonstrated that the effect of Hmgcs2 on ISCs is mediated by epigenetic modulation of Notch signaling via the histone deacetylase inhibitor activity of OHB (Cheng et al., 2019). Our work demonstrates that this metabolite-mediated mechanism contributes to the adaptation of the SI to changes of diet under physiological conditions. Similar mechanisms linking metabolic changes to modulation of developmental pathways via epigenetic alterations might extend beyond the SI and contribute to aging phenotypes in other stem cell compartments (Ermolaeva et al., 2018).
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to the Lead Contact, Alessandro Ori (alessandro.ori@leibniz-fli.de).
Materials Availability
This study did not generate new unique reagents. Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Alessandro Ori (alessandro.ori@leibniz-fli.de).
Data and Code Availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository with the dataset identifier PRIDE: PXD014311.
In addition, the proteomics data presented in this manuscript are available via a R shiny web server: https://orilab.shinyapps.io/IntestineAging/.
The codes supporting the study are available from the Lead Contact on request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All animal experiments were carried out with the approval of the state government of Thuringia (license no. FLI-17–006) or the Committee for Animal Care at MIT and under supervision of the Department of Comparative Medicine at MIT.
Mice
All wild-type mice were C57BL/6J obtained from Janvier or from internal breeding at the Leibniz Institute on Aging – Fritz Lipmann Institute (FLI). Lgr5-ki-eGFP-creERT2ki/+ mice (Barker et al., 2007) were bred at FLI and maintained on a C57BL/6J background. Hmgcs2loxp/loxp;UBC-creERT2 (Hmgcs2-KO) and Hmgcs2 wild-type (Control) mice were bread at MIT and previously described (Cheng et al., 2019). All animals were kept in a specific pathogen-free animal facility with a 12 h light/dark cycle. Young mice were aged 2–3 months, old mice were aged 18–20 months and geriatric animals were aged 24–26 months. For crypt spatial analysis and aging comparisons, male mice were used. Dietary restriction experiments were performed using female mice. The isolation of Lgr5 positive cells was performed from both male and female mice. For dietary restriction experiments, animals were randomly allocated to the different experimental groups.
Primary organoids
Crypts were isolated from mice as described in “Crypt and ISC isolation” below. Isolated crypts were counted and embedded in 70% v/v Matrigel™ (Corning #356231 growth factor reduced, enriched with Y-27632 dihydrochloridemonohydrate 10 μM Abcam #ab120129 and Jagged-1 1 μM Anaspec #ANA61298) mixed with organoid medium at 15–20 crypts/μL organoid medium which consists of Advanced DMEM F12 (Life Technologies #12634–010) supplemented with Egf 50ng/mL (Peprotech #315–09), Noggin 100 ng/mL (Peprotech #250–38), R-spondin 1 μg/mL (Peprotech #120–38), N-acetyl-L-cysteine 1 μM (Sigma #A9165), N2 1X (Life Technologies #17502048), B27 1X (Life Technologies #12587010). Crypts were plated onto a flat bottom 24-well plate (VWR #734–2325) and allowed to solidify for 10 min in a 37°C incubator. Matrigel droplets were overlaid with 350 μL organoid medium and medium was changed every three days. Organoids were cultured at 37°C in fully humidified chambers containing 5% CO2. Treatment with Hmgcs1/2 inhibitor hymeglusin (Sigma #SML0301) was started directly after seeding at a concentration of 0.25–0.3 μM, the control organoids were treated with DMSO. Organoids were rescued after 2 days in culture by adding 1 mM β-hydroxybutyrate to the culture medium. The number of surviving organoids and crypt domain formation was assessed on day 6 after seeding. For Reg4-mCherry reporter line, organoids were treated with 10 μM γ-secretase inhibitor MK-0752 (Cayman Chemical Company #471905–41-6), hymeglusin (0.25 μM) and β-hydroxybutyrate (100 μM, Sigma #54965). The percentage of Reg4+ cells in organoids was enumerated by flow cytometry based on the frequency of mCherry+ cells in the DAPI- live cells from the Reg4-mCherry reporter organoids.
Organoids for MS analysis were collected and washed 3x with cold PBS and collected at 70× g. After washing, organoids were incubated for 2 min with 500 μL TrypLE (Life Technologies #12604021) in 37°C and washed with cold PBS twice before lysis.
METHOD DETAILS
Dietary restriction
Mice were separated to single cages one week prior to the dietary treatment. Food weight and body weight were measured directly after separating the animals and before the dietary treatment. The daily food intake per animal was calculated and used for calculating the amount that refers to 70% for every animal dependent on its body weight. Food (ssniff #V1524–786) was given during the morning once per day to the DR and the DR+RF animals during the DR period and the animals were eating all the food they obtained. The AL animals had unlimited access to food during the experiment, their food intake was not measured. Animals of the DR+RF cohort underwent the same dietary treatment as DR animals and received ad libitum access to food for 2–3 days after the restriction.
Hmgcs2 deletion
Hmgcs2 deletion was induced as previously described (Cheng et al., 2019). Briefly, adult Hmgcs2loxp/loxp;UBC-creERT2 (Hmgcs2-KO) and Hmgcs2 wild-type (Control) mice received intraperitoneal tamoxifen injections (10mg/ml, 250 μl per 25 g of body weight) every other day, up to ten days for a total of five doses. Four weeks after the last tamoxifen injection, intestinal crypts were harvested as described in “Crypt and ISC isolation”. Samples were snap-frozen and stored in −80°C before “Crypt lysis and digestion for mass spectrometry (MS) analysis”.
Alcian Blue Staining
Small intestines were cleaned and cut into duodenum, jejunum and ileum. Samples were fixed in 4% formaldehyde (Roth 4979.1), dehydrated and embedded in paraffin for sectioning. Sections were rehydrated and Alcian Blue staining was performed according to manufacturer’s instructions (Vector Laboratories H-3501).
Hematoxylin and Eosin Staining
Small intestines were cleaned and cut into duodenum, jejunum and ileum. Samples were fixed in 4% formaldehyde (Roth 4979.1), dehydrated and embedded in paraffin for sectioning. Sections were rehydrated and hematoxyl (Merck, HX68296675) and eosin (Thermo, 71304) staining was performed.
Microscopy
Images of sections stained with either Alcian Blue or Hematoxylin and Eosin were acquired using Axio Imager or Axio Scan imaging systems (both from Zeiss) and the ZEN blue software v2 (Zeiss). Images of sections were acquired as individual tiles using 10x objectives after selection of 9–15 focus points per section. Tiles were automatically stitched together by ZEN. For further image analysis, the graphics tools for counting and measuring of the ZEN software were used.
Crypt and ISC isolation
Mice were euthanized with CO2 and the small intestine was isolated and cleaned with cold PBS. Crypts were separated by two rounds of incubation on ice in 5 mM EDTA for 15–20 min. Crypt suspensions were combined and filtered through a 70 μm mesh into a 50 mL conical tube to remove villus material and tissue fragments. Isolated crypts were counted, aliquoted and snap-frozen before storage in −80°C. For collection of ISCs, isolated crypts were digested to single cells for 20 min in a 37°C water bath with crypt digestion buffer (0.4 U/μL DNaseI from Sigma #4716728001, 5 mM MgCl2 from Roth #KK36.2, 80 μM Y-27632 dihydrochloridemonohydrate (Abcam #ab120129), 70% v/v TryplE (Life technologies #12604021). Cells were washed with cold PBS containing 80 μM Y-27632 dihydrochloridemonohydrate and filtered through 70 μm mesh directly before sorting. GFP-positive, DAPI-negative cells were sorted on a FACS Aria III into 2x lysis buffer for mass spectrometry (4% SDS from Sigma #75746, 100 mM HEPES from Sigma #H3375, 20 mM DTT from Roth #6908.3 in HPLC-H2O from Sigma #270733) and stored at −80°C.
Crypt lysis and digestion for mass spectrometry (MS) analysis
Approximately 20k crypts were resuspended in 0.4% RapiGest™ SF Surfactant (Waters #186002122, in 5 mM ammonium bicarbonate) and heat treated (95°C, 5 min). After a short cool-down 10M (Sigma #U6504) urea in 250 mM ammonium bicarbonate was added to a final concentration of 4 M urea (Sigma #U6504) and crypt lysates were solubilized by sonication in a Bioruptor Plus (Diagenode) for 10 cycles (1 min on and 30 s off) with high intensity at 20°C. Lysates were clarified by centrifugation for 5 min at 20800× g. For reduction and alkylation of cysteines, the clarified lysates were incubated at 37°C for 30 min with 10 mM DTT and subsequently treated with 15 mM iodacetamide (Sigma #I1148) for 30 min in the dark. Proteins were digested for 4 h at 37°C using 1:100 (w/w) LysC (Wako Chemicals GmbH #125–05061). Then, samples were diluted to 1.5 M urea with milliQ water and digested with 1:100 (w/w) trypsin (Promega #V5111) for 16 h at 37°C. Digested peptide solutions were then acidified with 10% trifluoroacetic acid and then desalted with Waters Oasis® HLB μElution Plate 30 μm (Waters 186001828BA) in the presence of a slow vacuum. In this process, the columns were conditioned with 3× 100 μL solvent B (80% acetonitrile from Biosolve #06914143) and equilibrated with 3× 100 μL solvent A (0.05% formic acid in milliQ water). Samples were loaded, washed 3 times with 100 μL solvent A, and eluted into PCR tubes with 50 μL solvent B. Eluates were dried with a speed vacuum centrifuge and dissolved in 5% acetonitrile, 0.1% formic acid to a peptide concentration of 1 μg/μL. 20 μL were transferred to an MS vial and 0.25 μL of HRM peptides (Biognosys AG #42896) were spiked into each sample prior to analysis by LC-MS.
ISC and organoids lysis and digestion for MS
Organoid pellets were lysed in lysis buffer (2% SDS (w/v), 100mM HEPES, 10 mM DTT in milliQ water) or cells sorted in 2x concentrated lysis buffer (4% SDS (w/v), 200 mM HEPES, 20 mM DTT in milliQ water) were thawed (after storage at −80°C) and sonicated in a Bioruptor Plus (Diagenode), as described above. Samples were then heated at 95°C for 10 min and sonicated again. The lysates were clarified and debris precipitated by centrifugation at 20800×g for 1 min, then incubated with 15 mM iodacetamide at room temperature, in the dark for 30 min. Each sample was treated with 8 volumes ice cold acetone (Biosolve #010306) and left overnight at −20°C to precipitate the proteins. The samples were then centrifuged at 20800× g for 1 h at 4°C. After removal of the supernatant, the precipitates were washed twice with ice cold 80% acetone. The pellets were then allowed to air-dry before being dissolved in digestion buffer at 0.25–0.5 μg/μL using 3 M urea in 100 mM HEPES (Sigma #H3375), pH 8. To facilitate the resuspension of the protein pellet, the samples were subjected to 3 rounds of sonication in the Bioruptor, as described above. Samples were then subjected to protein digestion, peptide desalting and preparation for LC-MS, as described above.
Data Independent Acquisition (DIA) for crypt and ISC samples
For spectral library generation, approx. 1 μg of reconstituted peptides was analyzed using Data Dependent Acquisition (DDA) using the nanoAcquity UPLC system (Waters) fitted with a trapping (nanoAcquity Symmetry C18, 5μm, 180 μm x 20 mm) and an analytical column (nanoAcquity BEH C18, 1.7 μm, 75 μm × 250 mm). The outlet of the analytical column was coupled directly to an Orbitrap Fusion Lumos (Thermo Fisher Scientific) using the Proxeon nanospray source. Solvent A was water, 0.1% (v/v) formic acid and solvent B was acetonitrile, 0.1% (v/v) formic acid. Peptides were eluted via a non-linear gradient from 0% to 40% B in 120 min. Total runtime was 145 min, including clean-up and column re-equilibration. The RF lens was set to 30%.
For crypt spectral library generation, 12 individual crypt samples and 4 pooled crypt samples were measured in DDA mode. The conditions for DDA were as follows: Full scan MS spectra with mass range 350–1650 m/z were acquired in profile mode in the Orbitrap with resolution of 60,000 FWHM. The filling time was set at maximum of 50 ms with limitation of 2× 105 ions. The “Top Speed” method was employed to take the maximum number of precursor ions (with an intensity threshold of 5× 104) from the full scan MS for fragmentation (using HCD collision energy, 30%) and quadrupole isolation (1.4 Da window) and measurement in the Orbitrap (resolution 15,000 FWHM, fixed first mass 120 m/z), with a cycle time of 3 s. The MIPS (monoisotopic precursor selection) peptide algorithm was employed but with relaxed restrictions when too few precursors meeting the criteria were found. The fragmentation was performed after accumulation of 2× 105 ions or after filling time of 22 ms for each precursor ion (whichever occurred first). MS/MS data were acquired in centroid mode. Only multiply charged (2+ – 7+) precursor ions were selected for MS/MS. Dynamic exclusion was employed with maximum retention period of 15 s and relative mass window of 10 ppm. Isotopes were excluded. For data acquisition and processing Tune version 2.1 was employed.
For the DIA, 3 μg of reconstituted peptides were loaded and the same gradient conditions were applied to the LC as for the DDA. The MS conditions were varied as follows: Full scan MS spectra with mass range 350–1650 m/z were acquired in profile mode in the Orbitrap with resolution of 120,000 FWHM. The filling time was set at maximum of 20 ms with limitation of 5× 105 ions. DIA scans were acquired with 34 mass window segments of differing widths across the MS1 mass range with a cycle time of 3 s. HCD fragmentation (30% collision energy) was applied and MS/MS spectra were acquired in the Orbitrap at a resolution of 30,000 FWHM over the mass range 200–2000 m/z after accumulation of 2× 105 ions or after filling time of 70 ms (whichever occurred first). Ions were injected for all available parallelizable time. Data were acquired in profile mode.
Data Independent Acquisition (DIA) for organoid and Hmgcs2-KO samples
For spectral library generation, approx. 1 μg of reconstituted peptides was analyzed using Data Dependent Acquisition (DDA) using the UltiMate 3000 UPLC system (Thermo Fisher Scientific) fitted with a trapping (Waters nanoEase M/Z Symmetry C18, 5 μm, 180 μm x 20 mm) and an analytical column (Waters nanoEase M/Z Peptide C18, 1.7 μm, 75 μm x 250 mm). The outlet of the analytical column was coupled directly to an Q exactive HF (Thermo Fisher Scientific) using the Proxeon nanospray source. Solvent A was water, 0.1% FA and solvent B was 80% (v/v) acetonitrile, 0.08% FA. Peptides were eluted via a non-linear gradient from 1% to 62.5% B in 131 min. Total runtime was 150 min, including clean-up and column re-equilibration. The S-lens RF value was set to 60. For crypt spectral library generation, 6 pooled crypt samples were measured in DDA mode. The conditions for DDA were as follows: Full scan MS spectra with mass range 350–1650 m/z were acquired in the Orbitrap with resolution of 60,000 FWHM. The filling time was set at maximum of 20 ms with an AGC target of 3× 106 ions. A Top15 method was employed to select precursor ions from the full scan MS for fragmentation (minimum AGC target of 1× 103 ions, normalized collision energy of 27%), quadrupole isolation (1.6 m/z) and measurement in the Orbitrap (resolution 15,000 FWHM, fixed first mass 120 m/z). The fragmentation was performed after accumulation of 2× 105 ions or after filling time of 25 ms for each precursor ion (whichever occurred first). Only multiply charged (2+ – 7+) precursor ions were selected for MS/MS. Dynamic exclusion was employed with maximum retention period of 20 s. Isotopes were excluded.
For DIA, 1 μg of reconstituted peptides were loaded using the same setup and LC conditions used for DDA. MS conditions were modified as follows: Full scan MS spectra with mass range 350–1650 m/z were acquired in profile mode in the Orbitrap with resolution of 120,000 FWHM. The filling time was set at maximum of 60 ms with an AGC target of 3× 106 ions. DIA scans were acquired with 40 mass window segments of differing widths across the MS1 mass range. The default charge state was set to 3+. HCD fragmentation (stepped normalized collision energy; 25.5, 27, 30%) was applied and MS/MS spectra were acquired with a resolution of 30,000 FWHM with a fixed first mass of 200 m/z after accumulation of 3× 106 ions or after filling time of 35 ms (whichever occurred first). Data were acquired in profile mode. For data acquisition and processing Tune version 2.9 and Xcalibur 4.1 were employed.
QUANTIFICATION AND STATISTICAL ANALYSIS
MS data analysis
For intestinal crypt and ISCs dataset, the DIA spectral library was generated using DDA data searched with MaxQuant (version 1.5.3.30) (Cox and Mann, 2008). The data were searched against a species-specific UniProt database (Mus musculus, entry only, release 2016_01) with a list of common contaminants appended using the Andromeda search engine (Cox et al., 2011). The data were searched with the following modifications: Carbamidomethyl (C) (fixed) and Oxidation (M) and Acetyl (Protein N-term) (variable). The mass error tolerance for the full scan MS spectra was set at 20 ppm and for the MS/MS spectra at 0.5 Da. A maximum of 2 missed cleavages were allowed. The library search was set to 1% false discovery rate (FDR) at both protein and peptide levels. The spectral library for crypt DIA analysis was generated using Spectronaut v9 (Biognosys AG) using the MaxQuant output. The library contained 98,394 precursors, corresponding to 5,759 protein groups using Spectronaut protein inference. The spectral library for ISC DIA analysis was generated using Spectronaut v10 using the corresponding MaxQuant output. The library contained 59,328 precursors, corresponding to 4,814 protein groups using Spectronaut protein inference.
For organoids and Hmgcs2-KO samples, the spectral libraries were generated using Pulsar in Spectronaut Professional v11 searching both DDA and DIA data. The data were searched against the mouse UniProt database (Mus musculus, entry only, release 2016_01) with a list of common contaminants appended. The data were searched with the following modifications: Carbamidomethyl (C) (Fixed), and Oxidation (M) and Acetyl (Protein N-term) (Variable). A maximum of 2 missed cleavages for trypsin were allowed. The identifications were filtered to satisfy FDR of 1% on peptide and protein level. The organoid library contained 111,625 precursors, corresponding to 6,272 protein groups using Spectronaut protein inference. The Hmgcs2-KO library contained 113,620 precursors, corresponding to 6,281 protein groups using Spectronaut protein inference.
DIA data were analyzed and searched against the specific spectral libraries using Spectronaut v9/10/11/13. Relative quantification was performed in Spectronaut for each pairwise comparison using the replicate samples from each condition. The data (candidate table) and protein quantity data reports were then exported and further data analyses and visualization were performed using R. Proteins with q < 0.05 and absolute log2 fold change > 0.58 were considered as significantly affected, unless otherwise stated. Displayed q-values are indicated as: *q < 0.05, **q < 0.01, ***q < 0.001, n.s. = not significant, unless otherwise stated.
Parallel Reaction Monitoring for quantification of Hmgcs2 and Muc2
For quantification of Muc2 and Hmgcs2 in crypts isolated Hmgcs2-KO and control mice, the same peptides samples as used for DIA analysis were used. Approx. 1μg of reconstituted peptides were analyzed using the UltiMate 3000 UPLC system (Thermo Fisher Scientific) fitted with a trapping (Waters nanoEase M/Z Symmetry C18, 5 μm, 180 μm x 20 mm) and an analytical column (Waters nanoEase M/Z Peptide C18, 1.7 μm, 75 μm x 250 mm). The outlet of the analytical column was coupled directly to a Q exactive HF (Thermo Fisher Scientific) using the Proxeon nanospray source. Solvent A was water, 0.1% (v/v) FA and solvent B was 80% (v/v) acetonitrile, 0.08% FA. Peptides were eluted via a non-linear gradient from 1% to 63% B in 40 min. Total runtime was 60 min, including clean-up and column re-equilibration. Parallel Reaction Monitoring (PRM) acquisition was performed in an unscheduled fashion for the duration of the entire gradient using the “DIA” node with the following settings: resolution 30,000 FWHM, AGC target 3e6, maximum injection time (IT) “auto,” isolation window 1 m/z. For each cycle, a “full MS” scan was acquired with the following settings: resolution 120,000 FWHM, AGC target 3× 106, maximum injection time (IT) 10 ms, scan range 350 to 1650 m/z.
For each protein, 2 or 3 proteotypic peptides were chosen and at least 5 fragment ions of the “y” series were used for quantification:
Peak group identification and quantification was performed using SpectroDive v9 (Biognosys). Quantification was performed using a label-free approach. Thereby, the summed height of all the identified transition was used to estimate the quantity of each peptide. Peptides quantities were normalized across samples by dividing for the integrated intensity of the Base Peak Chromatogram extracted for each sample from “full MS” scans using Xcalibur v4.1. Protein quantities were derived from the median value of the normalized peptide quantities. Changes of protein abundance between samples were assessed using a Welch t test performed on protein quantities.
Functional Principal Component Analysis (FPCA) of spatial crypt proteome data
Functional PCA analysis was implemented using the R package fdapace (https://cran.r-project.org/web/packages/fdapace/index.html). Briefly, log2 normalized protein abundance profiles along the 12 intestine sections were mean centered, and given as input to the FPCA function. Outlier proteins were identified with the package function CreateOutliersPlot using K = 3 and the ‘KDE’ as method for outlier detection. Normalized protein quantities, FPCA scores and outlier classification are reported in Table S1.
Analysis of markers for differentiated cell types
Definitions of marker proteins for different cell types composing the small intestine were taken from Haber et al. (2017) and they are reported in Table S1. For comparative analysis, the log2 fold changes of marker proteins for different cell types were used. Only marker proteins with q < 0.25 were selected. From the organoid data, proteins detected with more than 3 protein-specific peptides in the Matrigel, were removed. Significance was assessed by Wilcoxon rank sum test with continuity correction comparing individual groups of marker proteins against all proteins with q < 0.25. Statistical significance was tested using Wilcoxon rank sum test with continuity correction and indicated as: *p < 0.05, **p < 0.01, ***p < 0.001, n.s. = not significant, unless otherwise stated.
Histological analysis
Goblet cells were counted for 50 crypts per mouse and the average number of goblet cells per crypt was calculated. Villi length was measured for 50 villi per mouse and the average villi length was calculated. Averaged data were analyzed using GraphPad Prism 8.3. Significance was tested using unpaired two-tailed Student’s t test after testing for normal distribution using the Shapiro Wilk normality test, and indicated as: *p < 0.05, **p < 0.01, ***p < 0.001, n.s. = not significant, unless otherwise stated.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| Sodium beta-hydroxybutyrate | Sigma | Cat#54965 |
| Matrigel™ | Corning | Cat#356231 |
| DMEM/F12 | Life Technologies | Cat#12634-010 |
| Murine Egf | Peprotech | Cat#315-09 |
| R-Spondin | Peprotech | Cat#120-38 |
| Murine Noggin | Peprotech | Cat #250-38 |
| N-acetyl-L-cysteine | Sigma | Cat #A9165 |
| N2 | Life Technologies | Cat #17502048 |
| B27 | Life Technologies | Cat #12587010 |
| Y-27632 | Abcam | Cat #ab120129 |
| Jagged-1 | Anaspec | Cat #ANA61298 |
| γ-secretase inhibitor MK-0752 | Cayman Chemical Company | Cat#471905-41-6 |
| Hymeglusin | Sigma | Cat#SML0301 |
| Critical Commercial Assays | ||
| Alcian Blue stain kit | Vector Laboratories | Cat#H-3501 |
| II1β ELISA | R&D Systems | Cat#DY401 |
| HRM peptides | Biognosys AG | Cat#42896 |
| Deposited Data | ||
| Proteomics datasets | ProteomeXchange | PXD014311 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Lgr5-ki-eGFP-creERT2 | K.L. Rudolph, FLI | Barker et al., 2007 |
| Mouse: Hmgcs2loxp/loxp;UBC-creERT2 | Ö.H. Yilmaz, MIT | Cheng et al., 2019 |
| Software and Algorithms | ||
| Spectronaut™ | Biognosys AG | Sw-3001 |
| Xcalibur v4.1 | ThermoFisher Scientific | RRID:SCR_014593 |
| Prism v8.3 | GraphPad | RRID: SCR_002798 |
| RStudio v1.1.4 | RStudio, Inc. | RRID: SCR_000432 |
| ZEN Blue v2.6 | Zeiss | RRID:SCR_013672 |
| Other | ||
| R shiny web app for proteomics data | this paper | https://orilab.shinyapps.io/IntestineAging/ |
Highlights.
The effects of aging on the small intestine are region specific
Reduced plasticity of the old intestine to changes of diet
Dietary restriction followed by refeeding restores goblet cells in old mice
Ketone bodies prevent intestinal stem cells from differentiation into secretory cells
ACKNOWLEDGMENTS
The authors gratefully acknowledge support from the FLI Core Facilities Proteomics, Imaging, and Histology and the Mouse Facility. We thank Francesco Neri for critical reading of the manuscript; Omid Omrani for helpful discussion and advice; and Sonja Schätzlein, Antonina Klippert, Jennifer Freymann, and Anja Baar for support with animal experiments. A.O. acknowledges funding from the German Research Foundation (Deutsche Forschungsgemeinschaft [DFG]) via the Research Training Group ProMoAge (GRK 2155), the Else Kröner-Fresenius-Stiftung (award number 2019_A79), and the Deutsches Zentrum für Herz-Kreislaufforschung (award number 81X2800193). The FLI is a member of the Leibniz Association and is financially supported by the Federal Government of Germany and the State of Thuringia. Ö.H.Y. is supported by NIH grants R00 AG045144, R01CA211184, R01CA034992, and U54CA224068. C.-W.C. acknowledges funding from NIH grant K99 DK123407-01. O.W. is supported by the German Research Foundation (DFG grant SFB1127 ChemBioSys). K.L.R. is supported by the DFG within the collaborative research center “PolyTarget.”
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.107565.
REFERENCES
- Barker N (2014). Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat. Rev. Mol. Cell Biol. 15, 19–33. [DOI] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, and Clevers H (2007). Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007. [DOI] [PubMed] [Google Scholar]
- Basak O, Beumer J, Wiebrands K, Seno H, van Oudenaarden A, and Clevers H (2017). Induced quiescence of Lgr5+ stem cells in intestinal organoids enables differentiation of hormone-producing enteroendocrine cells. Cell Stem Cell 20, 177–190.e4. [DOI] [PubMed] [Google Scholar]
- Beyaz S, Mana MD, Roper J, Kedrin D, Saadatpour A, Hong SJ, Bauer-Rowe KE, Xifaras ME, Akkad A, Arias E, et al. (2016). High-fat diet enhances stemness and tumorigenicity of intestinal progenitors. Nature 531, 53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B, Karpac J, Supoyo S, Degennaro M, Lehmann R, and Jasper H (2010). Lifespan extension by preserving proliferative homeostasis in Drosophila. PLoS Genet. 6, e1001159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C-W, Biton M, Haber AL, Gunduz N, Eng G, Gaynor LT, Tripathi S, Calibasi-Kocal G, Rickelt S, Butty VL, et al. (2019). Ketone body signaling mediates intestinal stem cell homeostasis and adaptation to diet. Cell 178, 1115–1131.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, and Mann M (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372. [DOI] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, and Mann M (2011). Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805. [DOI] [PubMed] [Google Scholar]
- Darwich AS, Aslam U, Ashcroft DM, and Rostami-Hodjegan A (2014). Meta-analysis of the turnover of intestinal epithelia in preclinical animal species and humans. Drug Metab. Dispos. 42, 2016–2022. [DOI] [PubMed] [Google Scholar]
- de Wit NJW, IJssennagger N, Oosterink E, Keshtkar S, Hooiveld GJEJ, Mensink RP, Hammer S, Smit JWA, Müller M, and van der Meer R (2012). Oit1/Fam3D, a gut-secreted protein displaying nutritional status-dependent regulation. J. Nutr. Biochem. 23, 1425–1433. [DOI] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, and Yakhini Z (2009). GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolaeva M, Neri F, Ori A, and Rudolph KL (2018). Cellular and epigenetic drivers of stem cell ageing. Nat. Rev. Mol. Cell Biol. 19, 594–610. [DOI] [PubMed] [Google Scholar]
- Fevr T, Robine S, Louvard D, and Huelsken J (2007). Wnt/beta-catenin is essential for intestinal homeostasis and maintenance of intestinal stem cells. Mol. Cell. Biol. 27, 7551–7559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana L, and Partridge L (2015). Promoting health and longevity through diet: from model organisms to humans. Cell 161, 106–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi C, Bonafè M, Valensin S, Olivieri F, De Luca M, Ottaviani E, and De Benedictis G (2000). Inflamm-aging. An evolutionary perspective on immunosenescence. Ann. N Y Acad. Sci. 908, 244–254. [DOI] [PubMed] [Google Scholar]
- Franceschi C, Capri M, Monti D, Giunta S, Olivieri F, Sevini F, Panourgia MP, Invidia L, Celani L, Scurti M, et al. (2007). Inflammaging and anti-inflammaging: a systemic perspective on aging and longevity emerged from studies in humans. Mech. Ageing Dev. 128, 92–105. [DOI] [PubMed] [Google Scholar]
- Gehart H, and Clevers H (2019). Tales from the crypt: new insights into intestinal stem cells. Nat. Rev. Gastroenterol. Hepatol. 16, 19–34. [DOI] [PubMed] [Google Scholar]
- Haber AL, Biton M, Rogel N, Herbst RH, Shekhar K, Smillie C, Burgin G, Delorey TM, Howitt MR, Katz Y, et al. (2017). A single-cell survey of the small intestinal epithelium. Nature 551, 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn O, Drews LF, Nguyen A, Tatsuta T, Gkioni L, Hendrich O, Zhang Q, Langer T, Pletcher S, Wakelam MJO, et al. (2019). A nutritional memory effect counteracts benefits of dietary restriction in old mice. Nat. Metab. 1, 1059–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi M, and Guarente L (2016). mTORC1 and SIRT1 cooperate to foster expansion of gut adult stem cells during calorie restriction. Cell 166, 436–450. [DOI] [PubMed] [Google Scholar]
- Kim TH, Saadatpour A, Guo G, Saxena M, Cavazza A, Desai N, Jadhav U, Jiang L, Rivera MN, Orkin SH, et al. (2016). Single-cell transcript profiles reveal multilineage priming in early progenitors derived from Lgr5(+) intestinal stem cells. Cell Rep. 16, 2053–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leedham SJ, Rodenas-Cuadrado P, Howarth K, Lewis A, Mallappa S, Segditsas S, Davis H, Jeffery R, Rodriguez-Justo M, Keshav S, et al. (2013). A basal gradient of Wnt and stem-cell number influences regional tumour distribution in human and mouse intestinal tracts. Gut 62, 83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K, Kirkwood TBL, and Potten CS (1998). Age changes in stem cells of murine small intestinal crypts. Exp. Cell Res. 241, 316–323. [DOI] [PubMed] [Google Scholar]
- Messa GAM, Piasecki M, Hill C, McPhee JS, Tallis J, and Degens H (2019). Morphological alterations of mouse skeletal muscles during early ageing are muscle specific. Exp. Gerontol. 125, 110684. [DOI] [PubMed] [Google Scholar]
- Mihaylova MM, Cheng CW, Cao AQ, Tripathi S, Mana MD, Bauer-Rowe KE, Abu-Remaileh M, Clavain L, Erdemir A, Lewis CA, et al. (2018). Fasting activates fatty acid oxidation to enhance intestinal stem cell function during homeostasis and aging. Cell Stem Cell 22, 769–778.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorefield EC, Andres SF, Blue RE, Landeghem L Van Amanda, T., Santoro MA, and Ding S (2017). Aging effects on intestinal homeostasis associated with expansion and dysfunction of intestinal epithelial stem cells. Aging 9, 1898–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz J, Stange DE, Schepers AG, van de Wetering M, Koo B-K, Itzkovitz S, Volckmann R, Kung KS, Koster J, Radulescu S, et al. (2012). The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘+4′ cell markers. EMBO J. 31, 3079–3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muntel J, Kirkpatrick J, Bruderer R, Huang T, Vitek O, Ori A, and Reiter L (2019). Comparison of protein quantification in a complex background by DIA and TMT workflows with fixed instrument time. J. Proteome Res. 18, 1340–1351. [DOI] [PubMed] [Google Scholar]
- Nagaratnam N (2018). Malnutrition and malabsorption in the elderly In Advanced Age Geriatric Care, Nagaratnam N, Nagaratnam K, and Cheuk G, eds. (Springer; ), pp. 225–233. [Google Scholar]
- Nalapareddy K, Nattamai KJ, Kumar RS, Karns R, Wikenheiser-Brokamp KA, Sampson LL, Mahe MM, Sundaram N, Yacyshyn MB, Yacyshyn B, et al. (2017). Canonical Wnt Signaling Ameliorates Aging of Intestinal Stem Cells. Cell Rep. 18, 2608–2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein B, and Spradling A (2007). Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential notch signaling. Science 315, 988–993. [DOI] [PubMed] [Google Scholar]
- Ori A, Toyama BH, Harris MS, Bock T, Iskar M, Bork P, Ingolia NT, Hetzer MW, and Beck M (2015). Integrated transcriptome and proteome analyses reveal organ- specific proteome deterioration in old rats. Cell Syst. 1, 224–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parca L, Beck M, Bork P, and Ori A (2018). Quantifying compartment-associated variations of protein abundance in proteomics data. Mol. Syst. Biol. 14, e8131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pentinmikko N, Iqbal S, Mana M, Andersson S, Cognetta AB 3rd, Suciu RM, Roper J, Luopajärvi K, Markelin E, Gopalakrishnan S, et al. (2019). Notum produced by Paneth cells attenuates regeneration of aged intestinal epithelium. Nature 571, 398–02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, et al. (2019). The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47 (D1), D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisco AO, Schaum N, McGeever A, Karkanias J, Neff NF, Darmanis S, et al. (2019). A Single Cell Transcriptomic Atlas Characterizes Aging Tissues in the Mouse. bioRxiv 10.1101/661728. [DOI] [Google Scholar]
- Pomatto LCD, and Davies KJA (2017). The role of declining adaptive homeostasis in ageing. J. Physiol. 595, 7275–7309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangan P, Choi I, Wei M, Navarrete G, Guen E, Brandhorst S, Enyati N, Pasia G, Maesincee D, Ocon V, et al. (2019). Fasting-mimicking diet modulates microbiota and promotes intestinal regeneration to reduce inflammatory bowel disease pathology. Cell Rep. 26, 2704–2719.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan JC, Khericha M, Dobson AJ, Bolukbasi E, Rattanavirotkul N, Partridge L, Abad-Díez J, Calderón-Larrañaga A, Poncel-Falcó A, Poblador-Plou B, et al. (2016). Sex difference in pathology of the ageing gut mediates the greater response of female lifespan to dietary restriction. eLife 5, e10956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki N, Sachs N, Wiebrands K, Ellenbroek SIJ, Fumagalli A, Lyubimova A, Begthel H, van den Born M, van Es JH, Karthaus WR, et al. (2016). Reg4+ deep crypt secretory cells function as epithelial niche for Lgr5+ stem cells in colon. Proc. Natl. Acad. Sci. USA 113, E5399–E5407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sender R, Fuchs S, and Milo R (2016). Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14, e1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Peterson TR, Laplante M, Oh S, and Sabatini DM (2010). mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature 468, 1100–1104. [DOI] [PubMed] [Google Scholar]
- Stekovic S, Hofer SJ,Tripolt N, Aon MA, Royer P, Pein L, Stadler JT, Pendl T, Prietl B, Url J, et al. (2019). Alternate Day Fasting Improves Physiological and Molecular Markers of Aging in Healthy, Non-obese Humans. Cell Metab. 30, 462–76.e6. [DOI] [PubMed] [Google Scholar]
- Thevaranjan N, Puchta A, Schulz C, Naidoo A, Szamosi JC, Verschoor CP, Loukov D, Schenck LP, Jury J, Foley KP, et al. (2017). Age-associated microbial dysbiosis promotes intestinal permeability, systemic inflammation, and macrophage dysfunction. Cell Host Microbe 21, 455–466.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CA, Wojta K, Pulakanti K, Rao S, Dawson P, and Battle MA (2017). GATA4 is sufficient to establish jejunal versus ileal identity in the small intestine. Cell. Mol. Gastroenterol. Hepatol. 3, 422–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoda H, Ohbayashi N, Morikawa Y, Kumagai H, and Omura S (2004). Binding site for fungal beta-lactone hymeglusin on cytosolic 3-hydroxy-3-methylglutaryl coenzyme A synthase. Biochim. Biophys. Acta 1636, 22–28. [DOI] [PubMed] [Google Scholar]
- van Velthoven CTJ, de Morree A, Egner IM, Brett JO, and Rando TA (2017). Transcriptional profiling of quiescent muscle stem cells in vivo. Cell Rep. 21, 1994–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zhou Y, Rychahou P, Fan TW-M, Lane AN, Weiss HL, and Evers BM (2017). Ketogenesis contributes to intestinal cell differentiation. Cell Death Differ. 24, 458–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao F, Müller HG, and Wang JL (2005). Functional data analysis for sparse longitudinal data. J. Am. Stat. Assoc. 100, 577–590. [Google Scholar]
- Yilmaz ÖH, Katajisto P, Lamming DW, Gültekin Y, Bauer-Rowe KE, Sengupta S, Birsoy K, Dursun A, Yilmaz VO, Selig M, et al. (2012). mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature 486, 490–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository with the dataset identifier PRIDE: PXD014311.
In addition, the proteomics data presented in this manuscript are available via a R shiny web server: https://orilab.shinyapps.io/IntestineAging/.
The codes supporting the study are available from the Lead Contact on request.