Abstract
Computer vision systems (CVS) have been shown to be a powerful tool for the measurement of live pig body weight (BW) with no animal stress. With advances in precision farming, it is now possible to evaluate the growth performance of individual pigs more accurately. However, important traits such as muscle and fat deposition can still be evaluated only via ultrasound, computed tomography, or dual-energy x-ray absorptiometry. Therefore, the objectives of this study were: 1) to develop a CVS for prediction of live BW, muscle depth (MD), and back fat (BF) from top view 3D images of finishing pigs and 2) to compare the predictive ability of different approaches, such as traditional multiple linear regression, partial least squares, and machine learning techniques, including elastic networks, artificial neural networks, and deep learning (DL). A dataset containing over 12,000 images from 557 finishing pigs (average BW of 120 ± 12 kg) was split into training and testing sets using a 5-fold cross-validation (CV) technique so that 80% and 20% of the dataset were used for training and testing in each fold. Several image features, such as volume, area, length, widths, heights, polar image descriptors, and polar Fourier transforms, were extracted from the images and used as predictor variables in the different approaches evaluated. In addition, DL image encoders that take raw 3D images as input were also tested. This latter method achieved the best overall performance, with the lowest mean absolute scaled error (MASE) and root mean square error for all traits, and the highest predictive squared correlation (R2). The median predicted MASE achieved by this method was 2.69, 5.02, and 13.56, and R2 of 0.86, 0.50, and 0.45, for BW, MD, and BF, respectively. In conclusion, it was demonstrated that it is possible to successfully predict BW, MD, and BF via CVS on a fully automated setting using 3D images collected in farm conditions. Moreover, DL algorithms simplified and optimized the data analytics workflow, with raw 3D images used as direct inputs, without requiring prior image processing.
Keywords: body composition, image analysis, machine learning, precision livestock farming, ultrasound
Introduction
The world human population is forecasted to reach 8.6 billion people by 2030 and, following current trends, the overall consumption of meat will increase substantially as low-income countries have more than tripled their overall meat consumption in recent years (FAO, 2018). To meet this expanding demand, farmers are faced with the challenge of increasing productivity while maintaining adequate animal welfare and reducing the use of human-edible crops as well as the waste produced (FAO, 2017). In this context, changes to the current livestock production systems for more efficient monitoring animal welfare and productivity are highly desirable.
Precision livestock farming proposes to address this challenge via the use of sensors, such as activity trackers and cameras (Berckmans, 2017; Benjamin and Yik, 2019). The use of computer vision systems (CVS) is particularly appealing since they provide nondisruptive monitoring of the animals and high-throughput measurements of traits of interest. In pig production, the literature presents applications for tracking behavior (Kashiha et al., 2013; Maselyne et al., 2017), identification of leg and back disorders (Stavrakakis et al., 2014), measurement of pig weight (Kongsro, 2014; Pezzuolo et al., 2018; Fernandes et al., 2019), among others.
Automatic measurement of weight on live pigs can be an extremely useful management tool for assessment of individual growth and to monitor group homogeneity, ultimately helping farmers with more efficient problem detection and optimal management decision. Another important application of monitoring pig growth is for precision feeding, i.e., precision adjustment of feed composition and quantity delivered for a group or individual pigs (Pomar and Remus, 2019). With precision feeding, farmers can reduce production costs, protein intake, excretion, and greenhouse gas emissions.
Nevertheless, for an optimal balance of rations, measurements of animal body composition can be even more important than just live body weight (BW). Currently, traits related to body composition, such as lean muscle and fat deposition, are occasionally measured via ultrasound, dual-energy x-ray absorptiometry (DXA), or computed tomography (Scholz et al., 2015; Carabús et al., 2016). Even though DXA and computed tomography produce the most accurate measurements and bring more information on body composition (Font-i-Furnols et al., 2015; Lucas et al., 2017), these methods are generally expensive and require the animal to be anesthetized, making them impractical in commercial operations. On the other hand, ultrasound scanners are portable and do not require pigs to be anesthetized. Therefore, DXA and computed tomography are mainly used for research purposes, while ultrasound is used mostly by breeding companies, as a mean to evaluate candidate breeders.
CVS has also been proposed for the estimation of body composition of live animals, with the advantage of allowing indirect measurements of the pigs. However, there are only a few studies on the use of CVS for measurement of muscle and fat composition in live pigs, using either features extracted from 2D images or reconstruction of the body shape via computer stereo vision (Wu et al., 2004; Doeschl-Wilson et al., 2005). From these early studies, only Doeschl-Wilson et al. (2005) reported results for prediction of body composition, with predictive squared correlation (R2) ranging from 0.19 to 0.31 for fat, and from 0.04 to 0.18 for lean muscle. Regarding muscle deposition, recently, Alsahaf et al. (2019) used Kinect cameras for the estimation of muscle score, which is an indirect and subjective measurement of pig muscularity traditionally scored by a trained evaluator on a scale from 1 to 5. By using a gradient tree boosting classifier, the authors achieved an accuracy ranging from 0.30 to 0.58. Even though these are promising results, better predictions should be achieved before CVS can be successfully used for the prediction of muscle and fat deposition on commercial pig farms.
In this context, the objectives of this study were: 1) to develop a CVS for prediction of live BW, muscle depth (MD), and back fat (BF) from top view 3D images of finishing pigs and 2) to compare different predictive approaches, including multiple linear (ML) regression, partial least squares (PLS), and machine learning techniques, such as elastic networks (EN), artificial neural networks (ANN), and deep learning (DL) image encoder.
Material and Methods
The datasets of video recordings and measurements of BW, MD, and BF were supplied by the Pig Improvement Company (PIC, a Genus company, Hendersonville, TN). The data collection followed rigorous animal-handling procedures that comply with federal and institutional regulations regarding proper animal care practices (FASS, 2010).
Animals and data acquisition
A total of 557 finishing pigs were used, including boars and gilts from three different PIC commercial lines with average BW of 120 kg (± 12.4 kg) and an average age of 151 ± 3 d, all on the same multiplier farm and raised under standard commercial settings. Pigs from the same pen were moved to a specific area for evaluation, where they had their identification tag read using an RFID (radio frequency identification) sensor, followed by measurements of BW, MD, BF, and videos recorded for each pig. For the measurement of the traits of interest, groups of pigs that were housed together were moved to a management area that was usually used for the collection of BW and ultrasound measurements. The video recordings from the top view of the pigs (Figure 1A) were acquired using a Microsoft Kinect V2 (Microsoft, Redmond, WA, USA) and automatically processed in MATLAB (The MathWorks, 2017) for feature extraction, as previously described in Fernandes et al. (2019). Measurements of BW were taken with an EziWeigh5i (Tru-Test Inc., Mineral Wells, TX, USA) electronic scale that has a measured standard error of ±1% of the weight load. The MD (mm) and BF (mm) were measured using an Aloka SSD 500 ultrasound device (Hitachi-Aloka, Tokyo, Japan) equipped with a 3.5-MHz, 12-cm linear probe. The measurements were collected by placing the probe perpendicular to the loin at the 10th intercostal space. The data acquisition was performed in the course of 3 mo, from February to April 2016, and all data were collected by the same team of trained professionals; Table 1 presents the descriptive statistics of the traits measured.
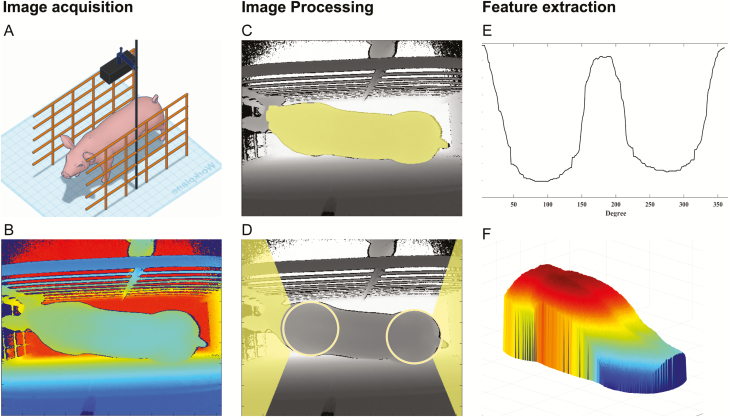
Figure 1.
Representative fluxogram of the image acquisition and processing pipeline: (A) setup of the 3D camera; (B) unprocessed depth image; (C) thresholded depth image, selected pig area highlighted; (D) identification of pig shoulder and rump, and removal of head and tail; (E) the PSD of the pig contour; and (F) processed pig body for measurements extraction.
Table 1.
Descriptive statistics for BF, MD, and live BW of the 618 finishing pigs evaluated
| Trait | Mean | Median | Standard deviation | Min | Max | Interquartile interval | Coefficient of variation |
|---|---|---|---|---|---|---|---|
| BF, mm | 6.03 | 5.60 | 1.47 | 3.00 | 10.90 | 1.60 | 0.24 |
| MD, mm | 65.07 | 65.00 | 6.19 | 48.20 | 89.40 | 8.00 | 0.10 |
| BW, kg | 119.97 | 119.50 | 12.43 | 83.00 | 156.50 | 16.62 | 0.10 |
Image processing and feature extraction
From each video recording, images from the pigs were extracted along with their respective features. This process was completely automated with the video recordings emulated on a virtual Kinect camera via the Kinect for Windows SDK v2.0 (Microsoft, 2014) and a CVS for processing the video feed initialized on an independent MATLAB routine. The connection between both instances was performed using a custom encapsulation of the Kinect for Windows SDK on a MATLAB MEX following the directions in the Kin2 toolbox (Terven and Córdova-Esparza, 2016). The CVS used for video processing and extraction of suitable images and their respective image features corresponds to a library of custom codes developed by the authors in MATLAB (Release 2017b; The MathWorks, 2017). This CVS is a combination of image processing and segmentation steps for the selection of frames with a well-positioned pig and subsequent removal of its head and tail as well as background noise from the images (Figure 1C and D). The algorithm then saves the original selected images, along with the segmented ones and the respective extracted features; a more detailed description of this algorithm can be found in Fernandes et al. (2019). Since the amount of time each pig stayed under the 3D camera was not the same and the images were selected automatically, multiple images were selected for each animal resulting in multiple image measurements, while only one direct measurement of BW, MD, and BF was acquired for each animal.
In the current study, the features extracted from the images were: 1) body measurements from the pigs, including apparent volume (V), surface area (A), length (L), heights (H), and widths (W) collected at 11 equidistant points along the animal’s back, and eccentricity (E); 2) 360 equidistant measurements from the polar shape contour of the top view image; and 3) the corresponding 360 Fourier descriptor features from the same polar shape contour. The body measurements were extracted from the 3D images (which is a map of the pixel distance to the camera) and were converted to the values on the metric scale by using the intrinsic focal length (f) from the Kinect depth camera used. The f was used for estimation of the image magnification factor ( in each pixel as , where is the distance in mm from the point to the camera plane. From , the pixel area () in mm was calculated as , and the total area of the pig calculated as the sum of . Similarly, the volume of a pixel is then , where is the pig height at the pixel p on the back of the pig. The pig volume was calculated as the sum of pixels’ volumes. The pig eccentricity was estimated as the ratio between the foci and the major axis of the ellipsis that has the same major and minor axis as the pig area. The polar shape descriptors (PSD) were measured as the distance from the centroid of the pig to points on its boundary contour as , where the point is the coordinate of the pig boundary at the degree t (Figure 1 E). The polar Fourier descriptors (PFD) were the real values from the Fourier transform of the pig contour PSD (Ostermeier et al., 2001; Zhang and Lu, 2002).
Statistical analysis
Alternative approaches for the prediction of BW, MD, and BF were evaluated. The predictive performance of each approach was assessed using a 5-fold cross-validation (CV) technique. Thus, the processes described below were repeated five times, in which the dataset was split into training (80% of the pigs) and validation (20% of the pigs) sets in each run of the CV.
The criteria used to compare these models were the mean absolute error (MAE), mean absolute scaled error (MASE), root mean square error (RMSE), and the R2. The models evaluated can be divided into two groups: 1) models that used as predictor variables the metrics extracted from the image processing step (ML regression, PLS, EN regression, and ANN) and 2) DL image encoder models, which used the raw 3D images as model input. A description of each of these modeling approaches and the specific predictive techniques considered is provided below.
Approaches using metrics from images as predictor variables
A total of 746 metrics were extracted from the images, including 26 biometrics, such as volume (V), area (A), length (L), eccentricity (E), 11 heights (H), and 11 widths (W), in addition to 360 PSD and 360 PFD image descriptors. These features, with and without information on pig sex and genetic line, were tested as predictor variables on the models that follow, in order to evaluate the importance of including sex and line information along with the image features. Since each pig had several images acquired but only one value of BW, MD, and BF, the truncated median at the third quartile of each predictor variable was calculated, as described in Fernandes et al. (2019). Thus, the final dataset used in this section had just one value (the truncated median) per pig for each of the features extracted from the available images.
Multiple linear regression
The ML regression models were implemented in R (R Core Team, 2017) using the MASS package (Venables and Ripley, 2002). To avoid overfitting a stepwise approach was used for variable selection using the stepAIC function from the MASS package, with the Bayesian information criteria for model selection. For each fold of the CV, the ML models were fitted on the training set and had their predictive performance evaluated on the validation set.
Partial least squares
The PLS models were implemented using the pls package in R (Mevik and Wehrens, 2007). For this analysis, a 10-fold CV on the training data was performed for selection of the most significant latent variables; therefore, it was performed five times, once for each training set of the original 5-fold CV. The selection of the best set of latent variables was performed using the randomization strategy (a.k.a. the van der Voet test) with a 5% significance level. This strategy removes latent variables from the model that reached the absolute optimum at the CV until there is no significant performance deterioration at the specified significance level.
Elastic network regression
The EN were implemented using the glmnet package in R (Friedman et al., 2010). In the current work, the EN evaluated were ML regressions with a mixture of the L1 (lasso) and L2 (ridge) penalties, in which the mixture of the two regularizations is controlled by the hyperparameter α, which ranges from 0 (a full ridge penalty) to 1 (lasso penalty). A grid of six equidistant values of α from 0 to 1 was tested. Apart from α, the EN has an additional parameter λ that controls the shrinkage magnitude of the regression coefficients. Thus, similarly to what was performed for the PLS analysis, for each value of α, a 10-fold CV was performed on the training set to obtain the value of λ that minimized the CV mean squared error (λ min). The search for λ min was done according to Friedman et al. (2010), using a pathwise coordinate descent with a warm start to generate a set of 100 values of lambda to be evaluated.
Artificial neural networks
ANN were implemented using the H2O platform for machine learning available as an R package (LeDell et al., 2019). ANN, also known as a multilayer perceptron, is a class of machine learning algorithms with an arbitrary number of features that have the purpose of learning nonlinear combinations of the input variables in order to predict a given output (Murphy, 2012). In ANN models, there are many possible parameters to be controlled and tuned, such as the number of layers and nodes in each layer, activation function, loss function, regularization parameters (such as L1 and L2 penalizations), and dropout rate. Therefore, one of the biggest challenges in fitting an ANN is to define its architecture. In the present study, the ANN models evaluated differed in: 1) the number of hidden layers and nodes, from a single-layer perceptron to at most three hidden layers with the number of nodes in each layer ranging from 5 to 100; 2) the activation function, which could be a rectified linear unit (ReLU) or max-out (Goodfellow et al., 2013), both with a constraint on the squared sum of the incoming weights that could be from 5 to 100 and a dropout rate of the input and hidden layer nodes from 20% to 80% of the nodes in each layer; 3) the loss functions evaluated were either a Gaussian loss (in this case, the mean squared error) or the Huber loss as a robust alternative (Hastie et al., 2009); 4) the inclusion of the L1 and L2 regularizations with their specific shrinkage parameters; and 5) the learning rate and time decay of the AdaDelta adaptive learning algorithm.
The search on the model space for possible architectures was conducted using a random discrete search on 500 candidate models, from which the 10 best candidate models were later fine-tuned. All the models were fitted on the training set and subsequently had their predictive performance evaluated on the validation set.
Deep learning image encoder
The DL image encoder models were implemented in Python (version 3.7) using the TensorFlow machine learning library (Abadi et al., 2015). The basic architecture of the image encoder models was a feed-forward multilayer perceptron. This architecture is similar to the encoder block of state of the art segmentation convolution neural networks (Poudel et al., 2019), which has been shown to be fast and has good predictive performance for image segmentation tasks.
The developed network architectures were composed of an input layer, encoder blocks, fully connected layers, and an output layer. The input layer was an array containing the 3D image of a pig and the focal length of the camera that generated the image (no information on pig sex and line was included). Each encoder block was composed of a convolutional block, followed by a max-pooling layer with a 2 by 2 window and a strider of the same size. Each convolutional block comprised of a convolutional layer with a 3 by 3 window, a batch normalization layer, and a ReLU activation function layer. The fully connected layers had L1 and L2 regularization (λ of 0.01), a dropout rate of 50%, and leaky ReLU activation function (α = 0.1).
The DL architectures tested diverged mainly on the size of the input image (ranging from 0.1 to 0.5 of the original image size), the number of encoder blocks (ranging from 1 to 5), and the number of nodes on the fully connected layers (ranging from 4 to 64).
Results
Descriptive statistics of image features
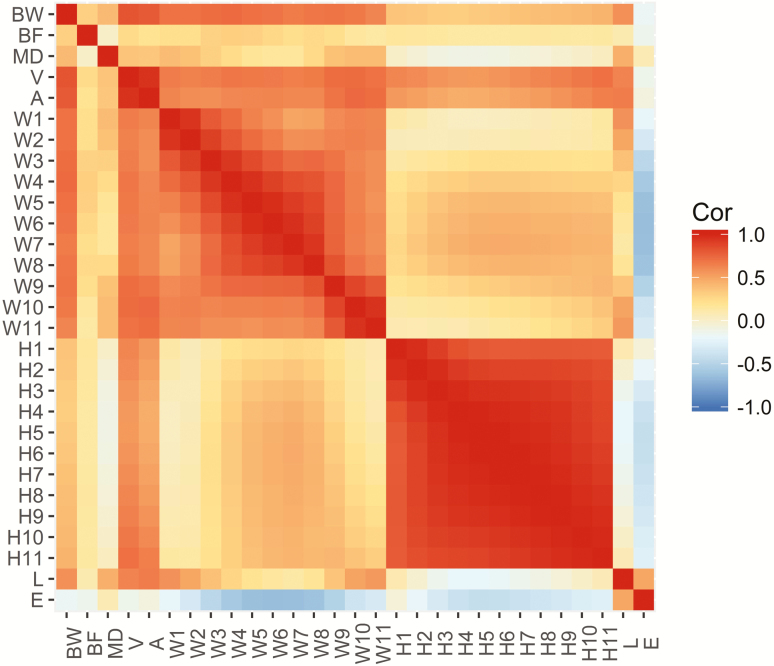
Figure 2 shows the correlations between biometric measurements and the three traits of interest (BW, BF, and MD). Overall, BW was found to be highly correlated with volume and area and positively correlated with BF and MD and most of the biometric measurements, except for eccentricity, with which it showed a low negative correlation. MD presented a low positive correlation with most of the other traits, with the highest values being with BW, V, and A. BF also presented low positive correlations with most of the other traits, with the highest value being with BW. In addition, the biometric measurements were mostly positively correlated with each other; notably, there was a higher correlation within widths, and within heights. The exception was for eccentricity, which presented mostly negative correlations with the other measurements and traits, except with MD and length.
Figure 2.
Correlations between BW, BF, MD, and the extracted biometric traits, such as volume (V), area (A), widths (W), heights (H), length (L), and eccentricity (E).
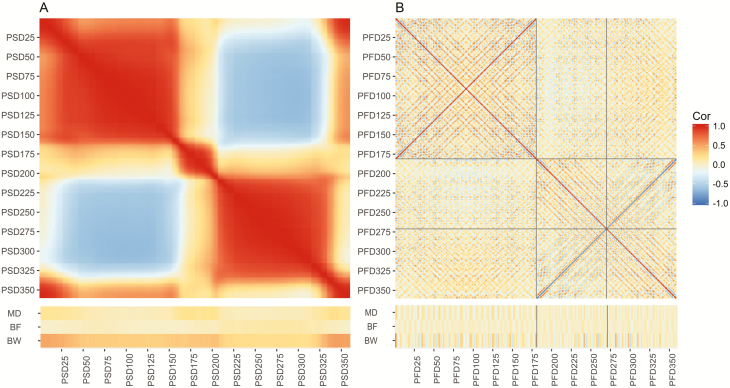
The correlations between BW, BF, MD, and the PSD are presented in Figure 3A. PSD presented, in general, higher correlations with BW than with MD or BF. Also, the correlation between PSD and BW or MD followed the same pattern, with higher correlations with PSD 1 to 25, 160 to 200, and 335 to 360. However, for the correlations between PSD and BF, this pattern was shifted. Overall, there was a high correlation between neighboring PSD with two big blocks of high correlated PSD, one from 30 to 160 degrees, and another from 210 to 335, and two small blocks of high positively correlated PSD, one from 165 to 209 degrees and another from 345 going back to 20 degrees. Regarding the correlations between the PFD and BW, MD, and BF (Figure 3B), there was a more complex pattern with shorter wavelength if compared with PSD, with some PFD having a high positive correlation with the traits of interest. Also, there was a similar trend for the correlations between PFD and both MD and BW, and this trend was inverted for BF.
Figure 3.
Correlations between BW, BF, MD, and the PSD extracted as the distances of the pig contour to the centroid at each degree (A). Correlations between BW, BF, MD, and the PFD transform of the PSD (B).
Model performance for prediction of BW, MD, and BF
Table 2 presents the results of MAE, RMSE, and R2 for the prediction of BW, MD, and BF for the various modeling strategies evaluated. For BW, DL presented better predictive performance with an MAE of 3.26 kg, which is on average 30% smaller than the MAE achieved with the other modeling strategies, and an R2 of 0.86, the second largest being 0.75 for EN. The best DL architecture had an input image with 0.3 of the original size, 5 encoder blocks, 64 nodes on the first fully connected layer, and 32 on the second. For MD, the DL approach again achieved the best performance, with a median R2 of 0.51 and MAE of 3.33 mm, which was on average 28% smaller than the MAE achieved by the other models. The best DL architecture had an input image with 0.2 of the original size, 5 encoder blocks, 64 nodes on the first fully connected layer, and 32 on the second. Regarding BF, DL achieved the best predictive performance with a median R2 of 0.45 and an MAE of 0.88 mm. Moreover, for BF, all the other approaches did perform very poorly when compared with DL. The best DL architecture had an input image with 0.2 of the original size, 5 encoder blocks, and 32 nodes on both fully connected layers.
Table 2.
Estimated CV MAE, MASE, RMSE, and R2 for BW, MD, and BF for the best multiple LM, PLS, EN, ANN, and DL image encoder
| Trait | Model | MAE | MASE | RMSE | R 2 |
|---|---|---|---|---|---|
| BW, kg | LM | 4.81 | 4.00 | 6.61 | 0.73 |
| PLS | 4.62 | 3.85 | 6.48 | 0.74 | |
| EN | 4.55 | 3.79 | 6.39 | 0.75 | |
| ANN | 5.00 | 4.16 | 6.83 | 0.70 | |
| DL | 3.26 | 2.69 | 4.56 | 0.86 | |
| MD, mm | LM | 4.10 | 6.30 | 5.16 | 0.35 |
| PLS | 4.36 | 6.67 | 5.37 | 0.30 | |
| EN | 4.12 | 6.32 | 5.12 | 0.31 | |
| ANN | 4.61 | 7.07 | 5.77 | 0.21 | |
| DL | 3.28 | 5.02 | 4.34 | 0.50 | |
| BF, mm | LM | 1.15 | 18.83 | 1.43 | 0.12 |
| PLS | 1.13 | 18.95 | 1.40 | 0.10 | |
| EN | 1.08 | 18.00 | 1.35 | 0.16 | |
| ANN | 1.20 | 19.69 | 1.52 | 0.10 | |
| DL | 0.80 | 13.56 | 1.11 | 0.45 |
Discussion
In the current work, we developed an automated CVS for the prediction of body composition (MD and BF) together with the improvement of the predictions of BW of pigs on barn conditions from past studies (Pezzuolo et al., 2018; Fernandes et al., 2019). Previous experimental results on the use of image features from 3D images of growing pigs for predicting BW have already shown the potential of CVS applications (Kongsro, 2014; Condotta et al., 2018). There are many advantages of using 3D images over 2D for the measurement of pig traits in barn conditions, such as easier image processing and removal of background, with less interference of background noise and light conditions (Kashiha et al., 2014; Kongsro, 2014), and the possibility for measurement of traits, such as body volume, shape, and gait analysis (Stavrakakis et al., 2014; Fernandes et al., 2019). However, most of those previous applications required some level of human processing of the images or manual measurement of the image features of interest. To the best of the authors’ knowledge, only Kashiha et al. (2014) and Fernandes et al. (2019) applied fully automated approaches for the prediction of BW in pigs. Also, most of the previous studies presented predictions for growing animals from different ages, with BW ranging from 15 to 50 kg (Kashiha et al., 2014) and 20 to 120 kg (Kongsro, 2014; Condotta et al., 2018); only Fernandes et al. (2019) presented results for finishing pigs (BW of 120 ± 12.4 kg). The predictive performance reported in previous studies ranged from 2% to 4% of the animals’ BW, while in the current study, it was of 2.7%, which is similar to what was observed in previous studies with the advantage that DL does not require any image preprocessing.
DL approaches have been successfully applied in many computer vision tasks in recent years, surpassing many traditional image analysis techniques (Goodfellow et al., 2016). A recent example was the use of DL for separation of fish body from fins and background from images acquired under challenging light conditions (Fernandes et al., 2020), achieving scores of intersection over union of 99, 90, and 64 for background, fish body, and fish fins, respectively. The main advantage of DL over the other techniques evaluated here is that there is no need for splitting the prediction into many steps, such as image processing, feature extraction, and finally prediction. Therefore, the model automatically searches for the best encoder features and weights that reduce the model loss (e.g., the mean squared error for Gaussian models). Moreover, it is important to notice that the input for the DL approach was only the images, while for the other approaches the input included not only the features extracted from the images but also sex and line information. Nevertheless, there was no significant improvement in the predictions for BW when considering these extra variables. To evaluate even further the impact of having animals from all three lines in the training, the selected DL architecture was retrained using only the data from two lines as a training set and validating the model in the remaining line (repeating this process three times). Moreover, for a fair comparison with a constant training set size, the DL approach was retrained also using a 3-fold CV with all three lines together (Table 3). This new set of analyses showed that, for BW, there was a decrease in model performance when training the model on two lines and validating it in another one. This result reinforces the hypothesis that the DL is accounting for differences across lines when animals from the three different lines are included in the training set.
Table 3.
Predictive MAE, MASE, RMSE, and R2 for the DL architecture for BW, MD, and BF for the three genetic lines evaluated on different validation schemes
| Trait | Validation Schemes1 | Line | MAE | MASE | RMSE | R 2 |
|---|---|---|---|---|---|---|
| BW, kg | Line | 1 | 4.86 | 4.22 | 6.34 | 0.68 |
| Line | 2 | 4.27 | 3.57 | 6.05 | 0.67 | |
| Line | 3 | 4.33 | 3.38 | 5.40 | 0.71 | |
| 3F-CV | 1 | 4.26 | 3.70 | 5.43 | 0.78 | |
| 3F-CV | 2 | 3.99 | 3.35 | 5.70 | 0.68 | |
| 3F-CV | 3 | 3.58 | 2.80 | 4.44 | 0.81 | |
| 5F-CV | 1 | 3.46 | 3.02 | 4.74 | 0.81 | |
| 5F-CV | 2 | 3.29 | 2.77 | 4.95 | 0.78 | |
| 5F-CV | 3 | 2.78 | 2.17 | 3.58 | 0.87 | |
| MD, mm | Line | 1 | 4.17 | 6.84 | 5.22 | 0.13 |
| Line | 2 | 5.20 | 7.63 | 6.43 | 0.05 | |
| Line | 3 | 4.04 | 6.17 | 4.95 | 0.10 | |
| 3F-CV | 1 | 4.45 | 7.08 | 5.40 | 0.10 | |
| 3F-CV | 2 | 5.15 | 7.75 | 6.44 | 0.04 | |
| 3F-CV | 3 | 4.04 | 6.17 | 5.00 | 0.08 | |
| 5F-CV | 1 | 3.39 | 5.50 | 4.38 | 0.36 | |
| 5F-CV | 2 | 3.57 | 5.27 | 4.83 | 0.39 | |
| 5F-CV | 3 | 3.04 | 4.62 | 4.00 | 0.41 | |
| BF, mm | Line | 1 | 0.97 | 15.15 | 1.22 | 0.20 |
| Line | 2 | 1.22 | 20.47 | 1.55 | 0.03 | |
| Line | 3 | 1.20 | 19.80 | 1.47 | 0.06 | |
| 3F-CV | 1 | 1.00 | 16.43 | 1.22 | 0.22 | |
| 3F-CV | 2 | 1.12 | 18.69 | 1.41 | 0.19 | |
| 3F-CV | 3 | 1.11 | 18.03 | 1.39 | 0.17 | |
| 5F-CV | 1 | 0.81 | 13.16 | 1.05 | 0.41 | |
| 5F-CV | 2 | 0.85 | 14.19 | 1.15 | 0.47 | |
| 5F-CV | 3 | 0.89 | 14.39 | 1.21 | 0.36 |
1Retaining one line for validation (Line), a 3-fold (3F-CV), and a 5-fold (5F-CV) CV.
Pig carcass composition is of great importance for farmers since packing plants tend to reward them according to lean muscle percentage (Engel et al., 2012). Moreover, information on individual pig growth and muscle and fat deposition can be used by farmers to improve feeding strategies, as feed costs account for approximately 60% to 70% of total pig production. Additionally, there is an increasing concern on fattening pigs emissions of phosphorus and nitrogen (Cadéro et al., 2018), and a well-balanced feed can reduce such costs and emissions. Therefore, the constant monitoring of pigs’ growth and body composition can be of utmost importance in the precision feeding of pigs, improving group homogeneity, reducing costs, as well as emissions of greenhouse gases, phosphorus, and protein intake and excretion (Pomar and Remus, 2019).
Imaging techniques such as visual image analysis, ultrasound, DXA, and computed tomography have been traditionally used in the evaluation of pig carcass composition within research conditions, with DXA and computed tomography presenting an overall higher accuracy than other techniques (Carabús et al., 2016). Previous studies evaluating ultrasound, computed tomography, and slaughter measurements of MD and BF showed that ultrasound measurements have correlations of 0.6 and 0.56 with the carcass measurements of MD and BF, respectively, while computed tomography has correlations of 0.48 to 0.67 for fat and 0.91 to 0.94 for lean meat (Font-i-Furnols et al., 2015; Lucas et al., 2017). However, there is little information in the literature regarding the use of CVS for the evaluation of live pig lean muscle and fat composition. The only other study found that evaluated the use of features from image analysis for the prediction of pig muscle and body fat composition (Doeschl-Wilson et al., 2005) reported predicted R2 of 0.31 and 0.19 for fat and 0.04 and 0.18 for prediction of lean on the foreloin and hindloin regions, respectively. In the same study, higher predictive accuracy was observed when including other sources of information, such as sex, BW, and BF. In the current study, it was not observed an improvement in the predictions when considering sex or line information. Moreover, the DL approach (which takes as input only the 3D image) presented MAE, MASE, and R2 for MD of 3.28 mm, 5.02%, and 0.50, respectively. Also, the median MAE, MASE, and R2 of the same method for BF were, respectively, of 0.84 mm, 13.62%, and 0.45. These results represent an interesting improvement in the application of CVS to the prediction of body composition in live finishing pigs. When comparing the effect of training size, the results show a decrease in performance for both MD and BF that followed the decrease in training dataset size from the 5-fold CV to the 3-fold CV scheme (Table 3). Also, the decrease was steeper for MD, for which the R2 went from around 0.39 to 0.07, while for BF, it went from 0.41 to 0.19. This result showing the importance of training size for DL is in accordance with what has been seen in other studies. As an example, Passafaro et al. (2020) evaluated the performance of DL and other methods on the genome-enabled prediction of BW in broilers and observed an increasing model performance as the training set size increased. The results from this and the previous work suggest that an increase in performance of the DL may be achieved with a bigger and richer dataset. Moving from the 3-fold CV (with all three lines) to the validation on line, a further decrease in performance was observed for BF, but for MD, the results of the validation on the lines were very similar to the 3-fold CV setting.
The BF and MD from the ultrasound measurements were virtually uncorrelated between themselves on this dataset. Nonetheless, both were positively correlated with BW. Therefore, one could question if the predictive approaches developed in this study could be better than using only BW for the prediction of MD or BF. By fitting a linear regression of MD on BW, we found predictive MAE of 4.84 mm, RMSE of 6.04, and R2 of 0.17, which is worse than the predictive accuracy of all the approaches evaluated. Similarly, for BF, we found higher values of MAE (1.29 mm) and RMSE (1.53 mm), and lower R2 (0.14) than what was observed for most of the approaches evaluated. Thus, even though BW is positively correlated with MD and BF, the DL is ultimately accounting for more information than only using BW as a predictor variable.
Regarding the correlations between BF or MD with the measurements extracted from the images, it is interesting to notice that they were generally mostly opposite. This trend was observed for length, widths, and the PSD. BF showed a higher correlation with the central PSDs measured around 225 to 300 degrees, while BW and MD had higher correlations with PSDs 1 to 25 and 335 to 360 degrees. Interestingly, the correlations observed are in accordance with the results from Peñagaricano et al. (2015), who investigated phenotypic/genetic causal networks of muscle and fat deposition in pigs. In their study, an antagonistic relationship existed between fat and muscle deposition, with BF having a negative causal effect on loin depth.
In the current study, single-trait models were developed for each trait of interest. Obviously, any field application of the current models would need to deploy at least three different predictive models, one for each trait. However, since there are patterns of association among the three traits, a future research direction would be to evaluate the predictive performance of models developed for the joint prediction of these traits. Another possible development would be using other methods for measurement of muscle and fat composition as ground truth (i.e., prediction targets) instead of ultrasound. Such measurements could be from DXA and computed tomography, which has been proven to provide more accurate measurements of muscle and fat composition, or even from direct carcass measurements.
In conclusion, a DL model using the raw 3D images as input provided higher prediction accuracy for BW, MD, and BF compared with the other methods evaluated. It was demonstrated that it is possible to predict MD and BF via CVS in a fully automated setting using 3D images from farm conditions, without the need for preprocessing images in steps such as image segmentation and feature extraction. Such finding is a key factor for optimizing data analysis workflows in CVS, where computational resources need to be efficiently used. Nevertheless, there is still room for improvement regarding the predictions of MD and BF, and additional research in this area is warranted.
Acknowledgments
This research was performed using the compute resources and assistance of the UW-Madison Center For High Throughput Computing (CHTC) in the Department of Computer Sciences. The CHTC is supported by UW-Madison, the Advanced Computing Initiative, the Wisconsin Alumni Research Foundation, the Wisconsin Institutes for Discovery, and the National Science Foundation and is an active member of the Open Science Grid, which is supported by the National Science Foundation and the U.S. Department of Energy’s Office of Science. The first author acknowledges financial support from the Coordination for the Improvement of High Education Personnel (CAPES), Brazil.
Glossary
Abbreviations
- ANN
artificial neural networks
- BF
back fat
- BW
body weight
- CV
cross-validation
- CVS
computer vision system
- DL
deep learning
- DXA
dual-energy x-ray absorptiometry
- EN
elastic networks
- LM
linear regression model
- MAE
mean absolute error
- MASE
mean absolute scaled error
- MD
muscle depth
- ML
multiple linear regression
- PFD
polar Fourier descriptors
- PIC
Pig Improvement Company
- PLS
partial least squares
- PSD
polar shape descriptors
- R2
squared correlation
- ReLU
rectified linear unit
- RMSE
root mean square error
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Literature Cited
- Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, Corrado G S, Davis A, Dean J, Devin M, . et al. 2015. Large-scale machine learning on heterogeneous distributed systems. TensorFlow. [accessed June 20, 2019]. Available from https://www.tensorflow.org/about/bib
- Alsahaf A, Azzopardi G, Ducro B, Hanenberg E, Veerkamp R F, and Petkov N. . 2019. Estimation of muscle scores of live pigs using a Kinect camera. IEEE Access. 7:52238–52245. doi: 10.1109/ACCESS.2019.2910986 [DOI] [Google Scholar]
- Benjamin M, and Yik. S. 2019. Precision livestock farming in swine welfare: a review for swine practitioners. Animals 9:133. doi: 10.3390/ani9040133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berckmans D. 2017. General introduction to precision livestock farming. Anim. Front. 7:6. doi: 10.2527/af.2017.0102 [DOI] [Google Scholar]
- Cadéro A, Aubry A, Brossard L, Dourmad J Y, Salaün Y, and Garcia-Launay F. . 2018. Modelling interactions between farmer practices and fattening pig performances with an individual-based model. Animal 12:1277–1286. doi: 10.1017/S1751731117002920 [DOI] [PubMed] [Google Scholar]
- Carabús A, Gispert M, and Font-i-Furnols M. . 2016. Imaging technologies to study the composition of live pigs: a review. Spanish J. Agric. Res. 14:e06R01. doi: 10.5424/sjar/2016143-8439 [DOI] [Google Scholar]
- Condotta I C F S, Brown-Brandl T M, Silva-Miranda K O, and Stinn J P. . 2018. Evaluation of a depth sensor for mass estimation of growing and finishing pigs. Biosyst. Eng 173: 11–18. doi: 10.1016/j.biosystemseng.2018.03.002 [DOI] [Google Scholar]
- Doeschl-Wilson A B, Green D M, Fisher A V, Carroll S M, Schofield C P, and Whittemore C T. . 2005. The relationship between body dimensions of living pigs and their carcass composition. Meat Sci. 70:229–240. doi: 10.1016/j.meatsci.2005.01.010 [DOI] [PubMed] [Google Scholar]
- Engel B, Lambooij E, Buist W G, and Vereijken P. . 2012. Lean meat prediction with HGP, CGM and CSB-Image-Meater, with prediction accuracy evaluated for different proportions of gilts, boars and castrated boars in the pig population. Meat Sci. 90:338–344. doi: 10.1016/j.meatsci.2011.07.020 [DOI] [PubMed] [Google Scholar]
- FAO. 2017. Livestock solutions for climate change. Available from http://www.fao.org/3/a-i8098e.pdf [Google Scholar]
- FAO. 2018. Shaping the future of livestock: sustainably, responsibly, efficiently. In: The 10th Global Forum for Food and Agriculture; Berlin FAO; p. 20 Available from http://www.fao.org/3/i8384en/I8384EN.pdf [Google Scholar]
- FASS. 2010. Guide for the care and use of agricultural animals in research and teaching. 3rd ed Champaign (IL): Federation of Animal Science Societies. [Google Scholar]
- Fernandes A F A, Dórea J R R, Fitzgerald R, Herring W, and Rosa G J M. . 2019. A novel automated system to acquire biometric and morphological measurements and predict body weight of pigs via 3D computer vision. J. Anim. Sci. 97:496–508. doi: 10.1093/jas/sky418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes A F A, Turra E M, de Alvarenga É R, Passafaro T L, Lopes F B, Alves G F O, Singh V, and Rosa G J M. . 2020. Deep learning image segmentation for extraction of fish body measurements and prediction of body weight and carcass traits in Nile tilapia. Comput. Electron. Agric. 170:105274. doi: 10.1016/j.compag.2020.105274 [DOI] [Google Scholar]
- Font-i-Furnols M, Carabús A, Pomar C, and Gispert M. . 2015. Estimation of carcass composition and cut composition from computed tomography images of live growing pigs of different genotypes. Animal. 9:166–178. doi: 10.1017/S1751731114002237 [DOI] [PubMed] [Google Scholar]
- Friedman J, Hastie T, and Tibshirani R. . 2010. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 33:1–22. [PMC free article] [PubMed] [Google Scholar]
- Goodfellow I, Bengio Y, and Courville A. . 2016. Deep learning. Cambridge (MS):MIT Press. [Google Scholar]
- Goodfellow I J, Warde-Farley D, Mirza M, Courville A, and Bengio Y. . 2013. Maxout networks [accessed April 3, 2005]. Available from https://arxiv.org/pdf/1302.4389v4.pdf
- Hastie T, Tibshirani R, and Friedman J. . 2009. The elements of statistical learning. 2nd ed New York (NY):Springer New York. [Google Scholar]
- Kashiha M A, Bahr C, Haredasht S A, Ott S Moons C P H, Niewold T A, Ödberg F O, and Berckmans D. . 2013. The automatic monitoring of pigs water use by cameras. Comput. Electron. Agric. 90:164–169. doi: 10.1016/j.compag.2012.09.015 [DOI] [Google Scholar]
- Kashiha M A, Bahr C, Ott S, Moons C P H, Niewold T A, Ödberg F O, and Berckmans D. . 2014. Automatic weight estimation of individual pigs using image analysis. Comput. Electron. Agric. 107:38–44. doi: 10.1016/j.compag.2014.06.003 [DOI] [Google Scholar]
- Kongsro J. 2014. Estimation of pig weight using a Microsoft Kinect prototype imaging system. Comput. Electron. Agric. 109:32–35. doi: 10.1016/j.compag.2014.08.008 [DOI] [Google Scholar]
- LeDell E, Gill N, Aiello S, Fu A, Candel A, Click C, Kraljevic T, Nykodym T, Aboyoun P Kurka M, . et al. 2019. R interface for “H2O.” [accessed June 17, 2005]. Available from https://cran.r-project.org/web/packages/h2o/h2o.pdf
- Lucas D, Brun A, Gispert M, Carabús A, Soler J, Tibau J, and Font-i-Furnols M. . 2017. Relationship between pig carcass characteristics measured in live pigs or carcasses with Piglog, Fat-o-Meat’er and computed tomography. Livest. Sci. 197:88–95. doi: 10.1016/J.LIVSCI.2017.01.010 [DOI] [Google Scholar]
- Maselyne J, Van Nuffel A, Briene P, Vangeyte J, De Ketelaere B, Millet S, Van den Hof J, Maes D, and Saeys W. . 2017. Online warning systems for individual fattening pigs based on their feeding pattern. Biosyst. Eng. 173:1–14. doi: 10.1016/j.biosystemseng.2017.08.006 [DOI] [Google Scholar]
- Mevik B-H, and Wehrens R. . 2007. The pls package: principal component and partial least squares regression in R. J. Stat. Softw. 18:1–23. doi:10.18637/jss.v018.i02 [Google Scholar]
- Microsoft 2014. Kinect SDK for Windows [accessed August 12, 2020]. Available from https://developer.microsoft.com/en-us/windows/kinect/. [Google Scholar]
- Murphy K P. 2012. Machine learning: a probabilistic perspective. Cambridge (MA): MIT Press. [Google Scholar]
- Passafaro T L, Lopes F B, Dorea J R R, Craven M, Breen V, Hawken R J, and Rosa G J M. . 2020. Would large dataset sample size unveil the potential of deep neural networks for improved genome-enabled prediction of complex traits? The case for body weight in broilers. BMC Genomics. doi: 10.21203/rs.2.22198/v2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñagaricano F, Valente B D, Steibel J P, Bates R O, Ernst C W, Khatib H, and Rosa G J. . 2015. Exploring causal networks underlying fat deposition and muscularity in pigs through the integration of phenotypic, genotypic and transcriptomic data. BMC Syst. Biol. 9:58. doi: 10.1186/s12918-015-0207-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezzuolo A, Guarino M, Sartori L, González L A, and Marinello F. . 2018. On-barn pig weight estimation based on body measurements by a Kinect v1 depth camera. Comput. Electron. Agric. 148:29–36. doi: 10.1016/J.COMPAG.2018.03.003 [DOI] [Google Scholar]
- Pomar C, and Remus A. . 2019. Precision pig feeding: a breakthrough toward sustainability. Anim. Front. 9:52–59. doi: 10.1093/af/vfz006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poudel R P K, Liwicki S, and Cipolla R. . 2019. Fast-SCNN: Fast Semantic Segmentation Network. [accessed February 21, 2005]. Available from http://arxiv.org/abs/1902.04502
- R Core Team 2017. R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing. [Google Scholar]
- Scholz A M, Bünger L, Kongsro J, Baulain U, and Mitchell A D. . 2015. Non-invasive methods for the determination of body and carcass composition in livestock: dual-energy X-ray absorptiometry, computed tomography, magnetic resonance imaging and ultrasound: Invited Review. Animal 9:1250–1264. doi: 10.1017/S1751731115000336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavrakakis S, Guy J H, Warlow O M E, Johnson G R, and Edwards S A. . 2014. Walking kinematics of growing pigs associated with differences in musculoskeletal conformation, subjective gait score and osteochondrosis. Livest. Sci. 165:104–113. doi: 10.1016/j.livsci.2014.04.008 [DOI] [Google Scholar]
- Terven J R, and Córdova-Esparza D M. . 2016. Kin2. A Kinect 2 toolbox for MATLAB. Sci. Comput. Program. 130:97–106. doi: 10.1016/j.scico.2016.05.009 [DOI] [Google Scholar]
- The MathWorks 2017. MATLAB Release 2017b. Available from https://www.mathworks.com/products/new_products/release2017b.html [Google Scholar]
- Venables W N, and Ripley B D. . 2002. Modern applied statistics with S. New York (NY):Springer-Verlag. [Google Scholar]
- Wu J, Tillett R, McFarlane N, Ju X, Siebert J P, and Schofield P. . 2004. Extracting the three-dimensional shape of live pigs using stereo photogrammetry. Comput. Electron. Agric. 44:203–222. doi: 10.1016/J.COMPAG.2004.05.003 [DOI] [Google Scholar]
- Zhang D, and Lu G. . 2002. Shape-based image retrieval using generic Fourier descriptor. Signal Process. Image Commun. 17:825–848. doi: 10.1016/S0923-5965(02)00084-X [DOI] [Google Scholar]
- Ostermeier G. C., Sargeant G. A., Yandell B. S., and Parrish J. J.. 2001. Measurement of bovine sperm nuclear shape using Fourier harmonic amplitudes. J. Androl. 22:584–594. doi: 10.1002/J.1939-4640.2001.B02218.X [DOI] [PubMed] [Google Scholar]