Mice lacking GAD1 show neonatal mortality, but the human phenotype associated with GAD1 disruption is poorly characterized. Neuray et al. describe six patients with biallelic GAD1 mutations, presenting with early-infantile onset epilepsy, neurodevelopmental delay, muscle weakness and non-CNS manifestations.
Keywords: GAD1, epilepsy, neurodevelopmental delay, muscle weakness, cleft palate
Abstract
Gamma-aminobutyric acid (GABA) and glutamate are the most abundant amino acid neurotransmitters in the brain. GABA, an inhibitory neurotransmitter, is synthesized by glutamic acid decarboxylase (GAD). Its predominant isoform GAD67, contributes up to ∼90% of base-level GABA in the CNS, and is encoded by the GAD1 gene. Disruption of GAD1 results in an imbalance of inhibitory and excitatory neurotransmitters, and as Gad1−/− mice die neonatally of severe cleft palate, it has not been possible to determine any potential neurological dysfunction. Furthermore, little is known about the consequence of GAD1 disruption in humans. Here we present six affected individuals from six unrelated families, carrying bi-allelic GAD1 variants, presenting with developmental and epileptic encephalopathy, characterized by early-infantile onset epilepsy and hypotonia with additional variable non-CNS manifestations such as skeletal abnormalities, dysmorphic features and cleft palate. Our findings highlight an important role for GAD1 in seizure induction, neuronal and extraneuronal development, and introduce GAD1 as a new gene associated with developmental and epileptic encephalopathy.
Introduction
The neurotransmitter γ-aminobutyric acid (GABA) is one of the main inhibitory neurotransmitters deriving from glutamate (Cooper et al., 1996). It plays a critical signalling role in the nervous system and also in a number of non-neuronal cell types. The enzyme responsible for the conversion of glutamate into GABA is glutamate decarboxylase (GAD), and occurs in two isoforms GAD65 and GAD67, depending on its molecular weight (Kaufman et al., 1991). These isoforms are products of two different genes, GAD1 (encoding a 67 kDa molecular weight protein, GAD67) and GAD2 (encoding a 65 kDa molecular weight protein, GAD65). GAD67 is constitutively active and produces >90% of the base level GABA in the CNS, whilst GAD65 is transiently activated (Asada et al., 1997).
Animal studies have also shown a distinct role for GAD65 and GAD67. Gd65−/− mice are viable but show a higher susceptibility to seizure induction despite normal GABA levels (Asada et al., 1996), whereas Gad67−/− mice are characterized by neonatal death, severe cleft plate and respiratory failure. GAD activity and GABA concentration are also drastically reduced in Gad67−/− mice (Asada et al., 1997).
The severity of the Gad67−/− phenotype in animal models might suggest severe phenotypical manifestions in humans, yet there have been few reported families with GAD1 mutations (Lynex et al., 2004; Saito et al., 2010; Curley et al., 2011; Ruzicka et al., 2015; Magri et al., 2018). Previous reports have described seemingly unparalleled phenotypes, which include schizophrenia, autism spectrum disease and cerebral palsy, and functional studies are missing to confirm the pathogenicity of these reported mutations.
Here we report a series of six affected individuals with distinct phenotypical features from six unrelated families with bi-allelic mutations in the GAD1 gene (three carrying homozygous missense mutations, one carrying a homozygous frameshift variant, two compound heterozygous variants, and one harbouring a homozygus stop gain variant). All affected individuals presented with seizures, strongly impaired neurocognitive development, and reduced muscle tone of variable severity. Interestingly only one presented with cleft palate, which has been suggested to be one of the key features in the GAD1 animal models.
Material and methods
Patients and genetic analysis
Six patients from six unrelated families of Persian (Family A), Pakistani (Family B), African American (Family C), Sudanese (Family D), Egyptian (Family E) and Turkish ancestry (Family F) were identified through GeneMatcher (Sobreira et al., 2015) and enrolled in this study. The study was conducted according to the Declaration of Helsinki and with the approval of the institutional review boards of University College of London and participating centres. Genetic testing through whole exome sequencing (WES) was carried out in different research centres after informed consent was obtained from the parents or legal guardians of the studied subjects. Genomic DNA was extracted from peripheral blood obtained from the probands, parents, and unaffected siblings (when available). Exome sequencing and data analysis was performed as follows: Families A, B and D in the according centres as previously described (Monies et al., 2019; Dias et al., 2019), Family C through GeneDx (Retterer et al., 2016), and Families E and F at Centogene (Bauer et al., 2018). Potential candidate causal variants were subsequently confirmed by independent bi-directional Sanger sequencing. Detailed information is provided in the Supplementary material.
Data availability
The data that support the findings of this report are available from the corresponding author, upon reasonable request.
Results
Clinical manifestation
Four of the affected individuals were born from consanguineous parents (first or second cousins), and all were born at term following a normal pregnancy. A key clinical feature common to all affected individuals was early onset seizures (from 2 to 6 months), predominantly focal motor seizures with and without impaired awareness (two with additional epileptic spasms, four with focal non-motor seizures, and five with bilateral motor seizures). Seizures were pharmacologically controlled in three of six affected individuals, with three reported as drug-resistant. Drug regimens differed across the individuals. EEG at seizure onset showed a burst suppression pattern in two individuals, diffuse slowing with multifocal as well as generalized sharp waves in two, and hypsarrhythmia in two. Follow-up EEGs showed diffuse slowing of background activity or persistent epileptic activity (two of six). Cranial MRI was normal in all but two individuals, one showing slight ventricular enlargement and one moderate global atrophy.
The second common clinical feature was severe developmental delay. Most patients did not achieve any speech or non-verbal communication, only one was reported to have developed simple speech and basic perceptive language skills. This individual has remained seizure-free under medication. Of the more severely affected individuals, despite remaining seizure-free following pharmacological intervention, they still failed to accelerate in their intellectual development.
The third key feature we observed was a reduced muscle strength (five of six individuals) of varying severity ranging from slight muscle tone (one of five) to limited head control, inability to sit or crawl (four of six) and nasogastric tube dependence, due to dysphagia (two of six). While slight dysmorphic facial features were seen in four of six individuals (Table 1), only one presented with a cleft palate.
Table 1.
Clinical features of GAD1 patients
| Family | A (Patient III-2) | B (Patient III-4) | C (Patient II-1) | D (Patient II-4) | E (Patient III-3) | F (Patient II-2) |
|---|---|---|---|---|---|---|
| Sex/ ethnic origin | Female/ Persian | Male/ Pakistani | Male/ African American | Male/ Sudanese | Female/ Egyptian | Female/ Turkish |
| Consanguinity | Yes (double first cousins) | Yes | – | Yes (second cousins) | Yes (first cousins) | Yes (first cousins) |
| Age at first/last exam | 5 y/10 y 3 m | 6 m/7 y | 2 m/22 m | 2 m/18 m | 6 m/4 y | 1 m/3 y |
| Development | ||||||
| Milestones | Delayed in all milestones, simple speech at 4 y, walking delayed, no complex movements | Delayed in all milestones, sitting and crawling | Delayed in all milestones, no head control, no sitting, no speech | Severe delay, poor head control achieved at 18m, no sitting | Severe delay in all milestones, bed ridden | Severe delay in all milestones, no sitting or crawling |
| ID | Moderate | Severe | Severe | Severe | Severe | Severe |
| Vision/hearing | High myopia | Normal | NA | Normal | Moderate hearing impairment | High myopia |
| Dysmorphic facial features | Yes | Yes | – | Yes | Yes | – |
| Cleft palate | – | – | – | – | Yes (surgical correction) | – |
| Skeletal abnormalities | Clindodactyly, pes planus, scoliosis | Arthrogryposis of lower limbs | – | Short arms | Congenital hip dislocation and malformation | |
| Neurological examination | Mild hypotonia | Brisk DTR, stereotypic hand movements, oral automatisms | Mild hypotonia, spasticity in lower extremities, oropharyngeal dysphagia | Severe hypotonia, dysphagia (floppy epiglottis) | Severe hypotonia, hyporeflexia, dysphagia | Severe hypotonia |
| Epilepsy | ||||||
| Age at onset | 2 m | 6 m | 2 m | 2 m | 6 m | 2 m |
| Type(s) of seizure at onset | Focal non/motor with impaired awareness | Focal motor impaired awareness, bilateral tonic-clonic | Focal non/motor with impaired awareness, bilateral tonic clonic | Epileptic spasms | Bilateral tonic clonic | Focal ± to bilateral motor with impaired awareness |
| Seizure progression (age) | Controlled (10 y), last seizure at age 7 y | Controlled (7 y), last seizure at 5.5 y | Refractory (28 m) | Spasms continue, seizures controlled (18 m) | Refractory (4 y) | Partial control, 1 seizure/week |
| EEG at onset | Burst suppression | Multifocal and generalized epileptogenic activity | Burst suppression | Hypsarrhythmia | Generalized epileptogenic activity | Hypsarrhythmia |
| Follow-up EEG (age) | Normal (11 y) | Normal (7 y) | Slowing, multifocal epileptic discharges (28 m) | No epileptic abnormalities (18 m) | NA | Generalized epileptiform activity (4 m) |
| AEDs trialled | PB, VPA, CBZ, CLB | PB, VPA, CLZ | CBD, vigabatrin, ketogenic diet, CLZ, PB | steroids, TP, vigabatrin | LEV, CLZ, TP | PB, TP, LEV, CLZ, vigabatrin, steroids |
| Cardiovascular MRI (age) | Normal (5 y) | Prominent ventricular space (6 m) | Normal (2 m) | Normal (6 m) | Moderate global atrophy (1 y) | Normal (2 m) |
| Other features | Hydronephrosis, nephrocalcinosis, bilateral kidney stones | – | NG-tube dependent | Diastasis recti | Intermittent NG tube dependence | – |
Current antiepileptic drugs (AEDs) are highlighted in italics. CBD = cannabidiol; CBZ = carbamazepine; CLB = clobazam; CLZ = clonazepam; DTR = deep tendon reflexes; ID = intellectual disability; LEV = levetiracetam; NA = not applicable; NG = nasogastric; OXC = oxcarbazepine; PB = phenobarbital; TP = topiramate; VPA = valproic acid.
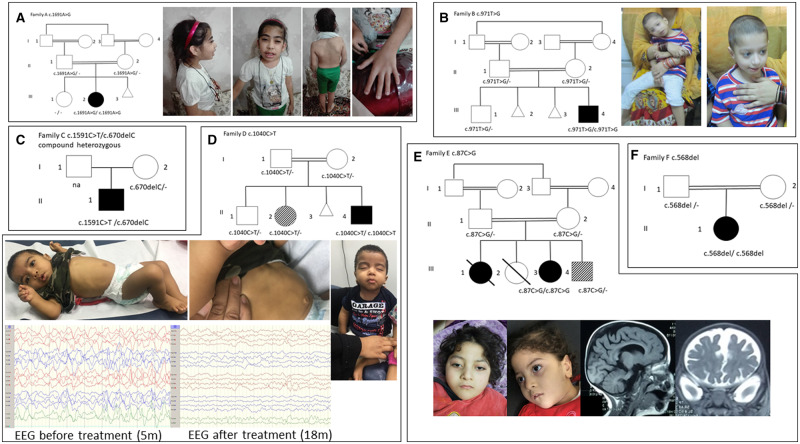
Other features that were observed without clear common elements included hirsutism, kidney stones, urogenital malformations, diastasis recti, reduced head circumference, clinodactyly, short arms, arthrogryposis of the lower limbs and congenital hip dislocation. Metabolic workups did not show any abnormalities. A detailed clinical summary of all affected individuals can be found in Table 1 along with images of key features and family pedigrees in Fig. 1.
Figure 1.
Pedigrees of the reported families and clinical pictures of GAD1 patients. (A) The female patient from Family A (Patient II-2) carries the homozygous c.1691A>G p.(Asn564Ser) variant and shows dysmorphic features with thick eyebrows, protruding ears, scoliosis, and long fingers with clinodactyly. (B) Patient from Family B (Patient III-4) harbours the c.971T>G p.(Phe324Cys) variant. He has slight dysmorphic features (wide mouth, thin upper lips, bitemporal narrowing and retrognathia). (C) Pedigree showing the segregation of the compound heterozygous variants c.1591C>T, c.1591C>T p.(Arg531*) and c.670delC p.(Leu224Serfs*5) in Family C. (D) Pedigree of Family D shows the segregation of the c.1040C>T p.(Thr347Met) variant. Patient II-4 carries the variant in homozygous state. He is severely hypotonic and shows severe dysmorphic features (infra-orbital creases, severely depressed nasal bridge, anteverted nares, prominent nasolabial folds). In addition, significant diastasis recti can be observed. His sister (Patient II-2) is heterozygous for the same variant and suffers from a different neurodevelopmental condition without seizures. (E) Patient III-2 from Family E harbours the c.87C>G (Tyr29*) variant, severely affected with dysmorphic facial features and global atrophy on cardiovascular MRI, one similarly affected sibling passed away without any genetic testing being performed, another sibling passed away only a few hours after birth, no phenotypical or genetic assessment could be carried out, and one sibling is alive with a different phenotype (sensoneural hearing loss, Hirschsprung disease). (F) Patient II-1 from Family F harbours the c.568del (Gln190Serfs*11) variant. His parents are both heterozygous carriers of the same variant. Empty and full symbols represent healthy and affected individuals, respectively. The symbol with diagonal lines indicates carrier status/different phenotype. The double line indicates consanguinity.
Genetic findings
Variants were prioritized in each family based on allele frequency <0.01%, predicted impact on protein function, and biological consistency. Potentially causal bi-allelic variants in GAD1 were identified in all affected individuals. The segregation of the variants with the clinical phenotype was confirmed by Sanger sequencing, which showed a recessive mode of inheritance. Detailed genetic results are provided in Table 2. All affected individuals carried ultrarare GAD1 variants, which were predicted to result in impaired protein function. Homozygous variants were identified in five families (Families A, B, D, E and F), whereas compound heterozygous variants were found in Family C (Table 2).
Table 2.
Frequency and predicted effect of the reported GAD1 variants
| GAD1 variant [NM_000817.2] | c.87C>G (p.Tyr29Ter) (Family E) | c.568delC (p.Gln190Serfs Ter11) (Family F) | c.670delC p.(Leu224Serfs*5) (Family C) | c.971T>G p.(Phe324Cys) (Family B) | c.1040C>T p.(Thr347Met) (Family D) | c.1591C>T p.(Arg531*) (Family C) | c.1691A>G p.(Asn564Ser) (Family A) |
|---|---|---|---|---|---|---|---|
| g. (hg19) | g.171678601C>G | g.171693323delC | g.171700586delC | g.171702542T>G | g.171704223C>T | g.171715383C>T | g.171716298A>G |
| Internal database | – | – | – | – | – | – | – |
| ExAC/GnomAD | – | – | – | 0.00000796 (2 het) | 0.00000795 (2 het) | 0.00000398 (1 het) | 0.00000398 (1 het) |
| GME | – | – | – | – | – | – | – |
| Iranome | – | – | – | – | – | – | – |
| Ensembl | – | – | – | – | – | – | – |
| SIFT | D- N/A | N/A | N/A | D (0.9125) | D (0.9125) | N/A | D (0.9125) |
| MutationTaster | DC (1) | DC (1) | DC (1) | DC (0.9768) | DC (1) | DC (1) | DC (1) |
| PolyPhen-2 | N/A | N/A | N/A | PD (0.978) | PD (1) | N/A | PD (1) |
| GERP score | 4.97 | 5.55 | 6.17 | 5.91 | 5.67 | 4.67 | 5.48 |
| CADD score | 35 | N/A | N/A | 28 | 29.1 | 43 | 26.1 |
| ACMG class | 5 (PVS1, PM2, PP4) | 5 (PVS1, PM2, PP3) | 5 (PVS1, PM2, PP3) | 3 (PM2, PP3) | 3 (PM2, PP3) | 5 (PVS1, PM2, PP3) | 3 (PM2, PP3) |
| GeneDx | 0 | 0 | 0/135 084 | 0 | 1/130 874 | 0/135 084 | 1/130 874 |
CADD = Combined Annotation Dependent Depletion; D = damaging; DC = disease causing; GeneDx = variant frequencies from the GeneDx database; GERP = Genomic Evolutionary Rate Profiling; GnomAD = Genome Aggregation Database; GME = Greater Middle East (GME) Variome Project; het = heterozygous; N/A = not applicable; PD = probably damaging; PM2 = Pathogenic Moderate 2; PP3 = Pathogenic Supporting 3; PVS1 = Pathogenic Very Strong 1; SIFT = Sorting Intolerant From Tolerant.
The affected proband from Family A carried a homozygous c.1691A>G, p.(Asn564Ser) variant, which is reported only once in the heterozygous state in gnomAD. This variant has a combined annotation dependent depletion (CADD) score of 26.1 and is predicted to be pathogenic by several bioinformatic prediction tools, including SIFT (score 0.9125), MutationTaster, and PolyPhen-2 (score 1) (Table 2). The proband from Family B harboured a homozygous c.971T>G, p.(Phe324Cys) variant, which causes the substitution of a phenylalanine residue at position 324 with cysteine. This position is not strictly conserved in other species, where leucine is found in place of phenylalanine. However, both phenylalanine and leucine belong to the class of the amino acids with long hydrophobic chains (Fig. 2A) and share several chemical features. This variant has been seen in the heterozygous state in gnomAD with a minor allele frequency of 0.00000796. It has a CADD score of 28 and is predicted damaging by all the prediction tools used (scores of 0.9125 and 0.978 for SIFT and PolyPhen-2, respectively). The proband from Family D carried a homozygous c.1040C>T, p.(Thr347Met) variant, which was reported twice in the heterozygous state in the gnomAD database. It was predicted damaging by both SIFT and PolyPhen-2 with high scores (0.9125 and 1, respectively). The CADD score for this variant was 29.1. Further in silico analysis predicted a reduction in protein stability for p.(Phe324Cys) and p.(Asn564Ser), in association with a break in H-bonds in the pyridoxal 5′-phosphate (PLP) binding domain for p.(Thr347Met). Both these changes might result in an impairment of the protein function due to the abnormal degradation or the decreased binding activity towards PLP, leading to a likely loss-of-function effect (Fig. 2B and Supplementary Table 1). In Family E, a homozygous stop gain variant c.87C>G, p.(Tyr29Ter) was identified. This variant is absent in all the queried population datasets. It is predicted damaging by SIFT (score not available) and disease-causing by Mutation Taster, with a CADD score of 35. This null variant likely causes a nonsense-mediated mRNA decay (NMD), leading to a complete loss-of-function. The proband from Family F carries a homozygous c.568delC p.(Gln190SerfsTer11) variant, which is absent in gnomAD and other population datasets queried. It is predicted damaging by MutationTaster (score 1) and classified as pathogenic according to the ACMG guidelines. Family C was the only family in which heterozygous variants were found: c.1591C>T, p.(Arg531*) and c.670delC, p.(Leu224Serfs*5). This individual’s mother was a carrier for c.670delC; however, the individual’s father was not available for genetic testing. The c.1591C>T, p.(Arg531*) variant results in a stop gain with a very high likely impact on protein function, as highlighted by a CADD score of 43. This is only reported in heterozygous state in the gnomAD database. Similarly, the frameshift variant c.670delC, p.(Leu224Serfs*5) is predicted to be disease-causing by MutationTaster and affects a very conserved residue, with a GERP score of 6.17. The null variants identified in Families B, E, and F may cause a premature termination of the transcript leading to a truncated protein or, alternatively, affected transcripts might be target of NMD.
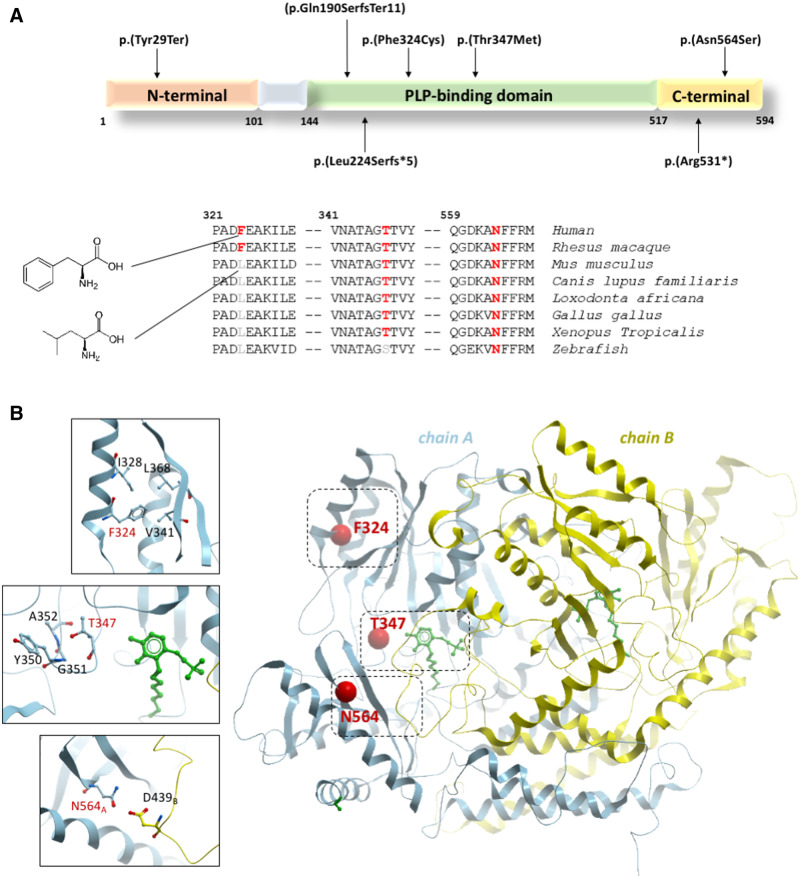
Figure 2.
Schematic and cartoon representation of GAD1. (A) Schematic representation of the GAD1 isoform GAD67 (NP_000808.2) with the pathogenic variants identified in this study. Of the six variants, four fall within the PLP-binding domain, a conserved region that is essential for the binding of the crucial cofactor pyridoxal 5′-phosphate (PLP). The remaining variants affect the C-terminal domain, which contains the catalytic site of the enzyme. Conservation status among different species is shown for the missense variants. (B) Cartoon representation of human GAD1 dimer (PDB: 2okj) with the two subunits in blue and yellow. Sites of the three missense mutations in this study are shown as red spheres, and close-up views of their nearby atomic environment are shown as insets. The PLP cofactor is shown in green.
Discussion
In humans, GAD1 encodes a 67-kDa protein, GAD67, which is the major contributor to GABA production in the CNS. As the major embryonic GAD isoform, it also plays a pivotal role in synaptogenesis and neuronal development (Asada et al., 1997; Soghomonian and Martin, 1998; Sgadò et al., 2011).
The 594 amino acid protein GAD67 is composed of a N-terminal domain involved in the generation of GAD65–GAD67 heterodimers and subcellular targeting, a C-terminal domain containing the catalytic site, and a central conserved domain binding PLP (Bosma et al., 1999; Martin et al., 2000; Lynex et al., 2004). This pyridoxal-dependent decarboxylase domain is essential for GAD67 function as GAD requires PLP as a cofactor to catalyze the generation of GABA from glutamate (Lernmark, 1996). Four of the seven variants identified in our families affect conserved residues in the PLP-binding domain, likely leading to loss-of-function (Fig. 1A). In particular, the two missense variants c.971T>G, p.(Phe324Cys) and c.1040C>T, p.(Thr347Met) probably cause impaired PLP binding, whereas the null variants c.568delC, p.(Gln190SerfsTer11) and c.670delC p.(Leu224Serfs*5) might result in a truncated protein or NMD. Similarly, the c.87C>G, p.(Tyr29Ter) variant localized to the N-terminal domain results in a premature stop codon, likely leading to NMD. The remaining two variants, c.1591C>T, p.(Arg531*) and c.1691A>G, p.(Asn564Ser), are localized to the C-terminal domain of the protein and are predicted to result in a complete and partial loss-of-function of the catalytic activity, respectively (Fig. 2B). According to gnomAD, GAD1 shows a moderate intolerance to missense variants (Z-score = 2.32; observed 217 and expected 336.5) and predicted loss-of-function variants (expected 33.1, observed 9). The likely negative impact of the missense variants on PLP-binding and C-terminal catalytic domains, together with the finding of four null pathogenic variants in our case series, supports the idea that the clinical phenotypes observed are likely due to a loss-of-function mechanism.
With regard to the possible genotype-phenotype correlation, there have been few clinical case reports of GAD1 mutations, with little overlap in clinical features (Lynex et al., 2004; Ruzicka et al., 2015; Magri et al., 2018). The six patients reported here, however, do show a clear phenotypical manifestation.
One of the main common features we observed was the occurrence of seizures at a young age (between 2 and 6 months of age). Since GAD1 is centrally involved in the production of GABA, GAD1 mutations likely lead to an imbalance of GABA in the brain. Previous studies have shown that abnormalities in GABAergic function play an important role in seizure induction (Olsen and Avoli, 1997). Dysfunction of the GABAergic system can be caused through either abnormal GABA synthesis (e.g. GAD dysfunction) or abnormal signalling (e.g. GABA receptor malfunction). Animal studies have shown that mutant mice lacking GAD or certain subunits of GABA-A receptors are prone to spontaneous epileptic seizures (Asada et al., 1996; Kash et al., 1997; DeLorey et al., 1998). It has also been shown that there is a reduction of GABAergic neurons in the epileptic brain, independent of the seizure’s aetiology supporting a conclusion that seizures themselves further decrease GABA release within the brain, causing further imbalance (Wang et al., 2011). This evidence suggests that there is a clear correlation between abnormalities within the GABAergic system and seizure occurrence. The occurrence of seizures early in life of our patients is therefore not a surprising phenotypical finding. However, previous data suggest that the intellectual development of newborns with epileptic encephalopathies is strongly dependent on seizure control, and seizure freedom usually leads to acceleration of intellectual development (Bombardieri et al., 2010). However, despite seizure control (seizure freedom achieved in three of six individuals), all patients described here still showed severe intellectual disability. This has been observed previously in other genetic conditions (Weckhuysen et al., 2012; Berecki et al., 2019), leading to the assumption that genetic defects themselves are influencing development and cognition, independently from seizure control. This is reflected in the ILAE’s terminology of ‘developmental and epileptic encephalopathies’, though a clear distinction between epileptic and developmental encephalopathy has been advised (Scheffer et al., 2017; Scheffer and Liao, 2020). Furthermore, some EEG abnormalities continued to be prominent after achieving seizure freedom. We therefore hypothesize that GAD1 mutations may also play an important role in intellectual development.
The reduced muscle tone and weakness, causing severe disability in four of six affected individuals was a surprising finding. There is a single case described in the literature of an individual with cerebral palsy and a GAD1 variant (Lynex et al., 2004). However, identification of the variant was based on autozygosity mapping, in which they identified a recessive locus of 5 cM located at 2q24-31.1. The investigators subsequently investigated the most interesting candidate in that region by sequencing the exons of GAD1 and identified a homozygous missense variant c.35C>G (p.Ser12Cys) (NM_013445.3) in the N-terminal domain of the protein (Lynex et al., 2004). This variant is rare in gnomAD (exomes allele frequency of 0.0000239) and absent in the homozygous state. However, the predictions on its pathogenicity are conflicting as it is predicted benign by MutationAssessor, DEOGEN2, and MetaLR. Furthermore, the 2q24-31.1 locus encompasses several other possible genes of interest (e.g. DYNC1I2, responsible for a neurodevelopmental disorder characterized by intellectual disability, spasticity, and neuroradiological anomalies), which were not investigated. The N-terminal domain of GAD67 is involved in the generation of GAD65–GAD67 heterodimers and subcellular targeting, and whilst we cannot rule out a possible impairment of the interaction of GAD67 with GAD65 or an alteration in GAD67 subcellular targeting as a result of this variant, we suggest that variants affecting the PLP-binding and C-terminal domain cause a more severe deficiency in GAD67 activity.
In addition, we also identified four null variants likely resulting in a loss-of-function. According to these observations and the consistency of the phenotype in our case series, we emphasize that GAD1 pathogenic variants should be considered the cause of a distinctive neurodevelopmental disorder instead of spastic cerebral palsy (Lynex et al., 2004). The clinical observation of muscle weakness is particularly interesting as it has not been described previously. Other GAD1-related diseases, such as antibody-mediated syndromes, have been associated with motor symptoms (Dayalu and Teener, 2012). However, these motor phenomena are usually linked to a hyperexcitability (increased muscle tone leading to rigidity, muscle spasms, and stiff person syndrome), as well as other neurological symptoms (e.g. Miller Fisher syndrome, eye movement disorders, cerebellar ataxia, epilepsy, limbic encephalitis, etc.) (Moersch and Woltman, 1956; Dalakas et al., 2000; Saiz et al., 2008; Tohid, 2016). Of note, in no case has muscle weakness been linked to GAD1 deficiency.
While Gad67−/− mice have been reported to die within the first hours of life due to a cleft palate (Asada et al., 1997; Condie et al., 1997), only one of our patients (Family E, Patient III-3) was born with a cleft palate (the same patient also showed congenital bilateral hip dislocation with shallow acetabulum, talipes equinovarus and hearing impairment.) Several studies have hypothesized that the development of a cleft palate is linked to reduced tongue movement during embryonic development and therefore secondary to CNS dysfunction (Iseki et al., 2007; Oh et al., 2010; Saito et al., 2010). Gad1 expression has also been shown in different non-neural tissues, such as the tail bud, limb mesenchyme, vibrissal placodes, and pharyngeal arches in various stages of embryonic development (Maddox and Condie, 2001). This observation has suggested a broader influence of GAD1 and GABA function on non-neural development (Maddox and Condie, 2001), supporting a possible primary role of GAD1 impaired function in the pathogenesis of non-neural defects. However, further studies are required to confirm this in humans.
In conclusion, this case series reports distinct phenotypical features caused by GAD1 variants, including early-infantile onset epilepsy, severe developmental delay and muscle weakness. Less consistent features include skeletal abnormalities and dysmorphic facial features, including cleft palate. Functional studies and larger clinical series will be necessary to further assess genotype-phenotype correlations for GAD1 variants.
Supplementary Material
Acknowledgements
We thank all families and collaborators for providing detailed clinical data and samples to conduct this study.
Funding
This study was supported by grants from The MRC (MR/S01165X/1, MR/S005021/1, G0601943), The National Institute for Health Research University College London Hospitals Biomedical Research Centre, Rosetree Trust, Ataxia UK, MSA Trust, Brain Research UK, Sparks GOSH Charity, Muscular Dystrophy UK (MDUK), Muscular Dystrophy Association (MDA USA). The families were collected as part of the SYNaPS Study Group collaboration funded by The Wellcome Trust and strategic award (Synaptopathies) funding (WT093205 MA and WT104033AIA). This research was conducted as part of the Queen Square Genomics group at University College London, supported by the National Institute for Health Research University College London Hospitals Biomedical Research Centre.
Competing interests
R.P. is an employee of GeneDx, Inc. S.K. and C.B. are employees of Centogene, AG.
Glossary
- GAD =
glutamate decarboxylase
Appendix 1
Consortia and network members involved in this study are listed below. The full details are available in the Supplementary material.
The Synaptopathies and Paroxysmal Syndromes (SYNaPS) Study Group
http://neurogenetics.co.uk/synaptopathies-synaps/
Stanislav Groppa, Blagovesta Marinova Karashova, Wolfgang Nachbauer, Sylvia Boesch, Larissa Arning, Dagmar Timmann, Bru Cormand, Belen Pérez-Dueñas, Gabriella Di Rosa, Jatinder S. Goraya, Tipu Sultan, Jun Mine, Daniela Avdjieva, Hadil Kathom, Dr Radka Tincheva, Selina Banu, Mercedes Pineda-Marfa, Pierangelo Veggiotti, Michel D. Ferrari, Alberto Verrotti, Giangluigi Marseglia, Salvatore Savasta, Mayte García-Silva, Alfons Macaya Ruiz, Barbara Garavaglia, Eugenia Borgione, Simona Portaro, Benigno Monteagudo Sanchez, Richard Boles, Savvas Papacostas, Michail Vikelis, Eleni Zamba Papanicolaou, Efthymios Dardiotis, Shazia Maqbool, Shahnaz Ibrahim, Salman Kirmani, Nuzhat Noureen Rana, Osama Atawneh, George Koutsis, Marianthi Breza, Salvatore Mangano, Carmela Scuderi, Eugenia Borgione, Giovanna Morello, Tanya Stojkovic, Massimi Zollo, Gali Heimer, Yves A. Dauvilliers, Pasquale Striano, Issam Al-Khawaja, Fuad Al-Mutairi, Hamed Sherifa.
Contributor Information
SYNaPS Study Group:
Stanislav Groppa, Blagovesta Marinova Karashova, Wolfgang Nachbauer, Sylvia Boesch, Larissa Arning, Dagmar Timmann, Bru Cormand, Belen Pérez-Dueñas, Gabriella Di Rosa, Jatinder S Goraya, Tipu Sultan, Jun Mine, Daniela Avdjieva, Hadil Kathom, Dr Radka Tincheva, Selina Banu, Mercedes Pineda-Marfa, Pierangelo Veggiotti, Michel D Ferrari, Alberto Verrotti, Giangluigi Marseglia, Salvatore Savasta, Mayte García-Silva, Alfons Macaya Ruiz, Barbara Garavaglia, Eugenia Borgione, Simona Portaro, Benigno Monteagudo Sanchez, Richard Boles, Savvas Papacostas, Michail Vikelis, Eleni Zamba Papanicolaou, Efthymios Dardiotis, Shazia Maqbool, Shahnaz Ibrahim, Salman Kirmani, Nuzhat Noureen Rana, Osama Atawneh, George Koutsis, Marianthi Breza, Salvatore Mangano, Carmela Scuderi, Eugenia Borgione, Giovanna Morello, Tanya Stojkovic, Massimi Zollo, Gali Heimer, Yves A Dauvilliers, Pasquale Striano, Issam Al-Khawaja, Fuad Al-Mutairi, and Hamed Sherifa
References
- Asada H, Kawamura Y, Maruyama K, Kume H, Ding R, Ji FY, et al. Mice lacking the 65 kDa isoform of glutamic acid decarboxylase (GAD65) maintain normal levels of GAD67 and GABA in their brains but are susceptible to seizures. Biochem Biophys Res Commun 1996; 229: 891–895. [DOI] [PubMed] [Google Scholar]
- Asada H, Kawamura Y, Maruyama K, Kume H, Ding RG, Kanbara N, et al. Cleft palate and decreased brain gamma-aminobutyric acid in mice lacking the 67-kDa isoform of glutamic acid decarboxylase. Proc Natl Acad Sci USA 1997; 94: 6496–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer P, Kandaswamy KK, Weiss MER, Paknia O, Werber M, Bertoli-Avella AM, et al. Development of an evidence-based algorithm that optimizes sensitivity and specificity in ES-based diagnostics of a clinically heterogeneous patient population. Genet Med 2018; 21: 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berecki G, Bryson A, Terhag J, Maljevic S, Gazina EV, Hill SL, et al. SCN1A gain of function in early infantile encephalopathy. Ann Neurol 2019; 85: 514–25. [DOI] [PubMed] [Google Scholar]
- Bombardieri R, Pinci M, Moavero R, Cerminara C, Curatolo P.. Early control of seizures improves long-term outcome in children with tuberous sclerosis complex. Eur J Paediatr Neurol 2010; 14: 146–9. [DOI] [PubMed] [Google Scholar]
- Bosma PT, Blázquez M, Collins MA, Bishop JD, Drouin G, Priede IG, et al. Multiplicity of glutamic acid decarboxylases (GAD) in vertebrates: molecular phylogeny and evidence for a new GAD paralog. Mol Biol Evol 1999; 16: 397–404. [DOI] [PubMed] [Google Scholar]
- Condie BG, Bain G, Gottlieb DI, Capecchi MR.. Cleft palate in mice with a targeted mutation in the gamma-aminobutyric acid-producing enzyme glutamic acid decarboxylase 67. Proc Natl Acad Sci USA 1997; 94: 11451–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JR, Bloom FE, Roth RH, editors. The biochemical basis of neuropharmacology. New York: New York: Oxford University Press; 1996. [Google Scholar]
- Curley AA, Arion D, Volk DW, Asafu-Adjei JK, Sampson AR, Fish KN, et al. Cortical deficits of glutamic acid decarboxylase 67 expression in schizophrenia: clinical, protein, and cell type-specific features. Am J Psychiatry 2011; 168: 921–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalakas MC, Fujii M, Li M, McElroy B.. The clinical spectrum of anti-GAD antibody-positive patients with stiff-person syndrome. Neurology 2000; 55: 1531–5. [DOI] [PubMed] [Google Scholar]
- Dayalu P, Teener JW.. Stiff Person syndrome and other anti-GAD-associated neurologic disorders. Semin Neurol 2012; 32: 544–9. [DOI] [PubMed] [Google Scholar]
- DeLorey TM, Handforth A, Anagnostaras SG, Homanics GE, Minassian BA, Asatourian A, et al. Mice lacking the beta3 subunit of the GABAA receptor have the epilepsy phenotype and many of the behavioral characteristics of Angelman syndrome. J Neurosci 1998; 18: 8505–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias CM, Punetha J, Zheng C, Mazaheri N, Rad A, Efthymiou S, et al. Homozygous missense variants in NTNG2, encoding a presynaptic netrin-G2 adhesion protein, lead to a distinct neurodevelopmental disorder. Am J Hum Genet 2019; 105: 1048–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iseki S, Ishii-Suzuki M, Tsunekawa N, Yamada Y, Eto K, Obata K.. Experimental induction of palate shelf elevation in glutamate decarboxylase 67-deficient mice with cleft palate due to vertically oriented palatal shelf. Birth Defect Res A Clin Mol Teratol 2007; 79: 688–95. [DOI] [PubMed] [Google Scholar]
- Kash SF, Johnson RS, Tecott LH, Noebels JL, Mayfield RD, Hanahan D, et al. Epilepsy in mice deficient in the 65-kDa isoform of glutamic acid decarboxylase. Proc Natl Acad Sci USA 1997; 94: 14060–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman DL, Houser CR, Tobin AJ.. Two forms of the gamma-aminobutyric acid synthetic enzyme glutamate decarboxylase have distinct intraneuronal distributions and cofactor interactions. J Neurochem 1991; 56: 720–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lernmark A. Glutamic acid decarboxylase–gene to antigen to disease. J Intern Med 1996; 240: 259–77. [DOI] [PubMed] [Google Scholar]
- Lynex CN, Carr IM, Leek JP, Achuthan R, Mitchell S, Maher ER, et al. Homozygosity for a missense mutation in the 67 kDa isoform of glutamate decarboxylase in a family with autosomal recessive spastic cerebral palsy: parallels with Stiff-Person Syndrome and other movement disorders. BMC Neurol 2004; 4: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox DM, Condie BG.. Dynamic expression of a glutamate decarboxylase gene in multiple non-neural tissues during mouse development. BMC Dev Biol 2001; 1: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magri C, Giacopuzzi E, La Via L, Bonini D, Ravasio V, Elhussiny MEA, et al. A novel homozygous mutation in GAD1 gene described in a schizophrenic patient impairs activity and dimerization of GAD67 enzyme. Sci Rep 2018; 8: 15470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DL, Liu H, Martin SB, Wu SJ.. Structural features and regulatory properties of the brain glutamate decarboxylases. Neurochem Int 2000; 37: 111–9. [DOI] [PubMed] [Google Scholar]
- Moersch FP, Woltman HW.. Progressive fluctuating muscular rigidity and spasm (“stiff-man” syndrome); report of a case and some observations in 13 other cases. Proc Staff Meet Mayo Clin 1956; 31: 421–7. [PubMed] [Google Scholar]
- Monies D, Abouelhoda M, Assoum M, Moghrabi N, Rafiullah R, Almontashiri N, et al. Lessons learned from large-scale, first-tier clinical exome sequencing in a highly consanguineous population. Am J Hum Genet 2019; 105: 879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh W-J, Westmoreland JJ, Summers R, Condie BG.. Cleft palate is caused by CNS dysfunction in Gad1 and Viaat knockout mice. PloS One 2010; 5: e9758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen RW, Avoli M.. GABA and epileptogenesis. Epilepsia 1997; 38: 399–407. [DOI] [PubMed] [Google Scholar]
- Retterer K, Juusola J, Cho MT, Vitazka P, Millan F, Gibellini F, et al. Clinical application of whole-exome sequencing across clinical indications. Genet Med 2016; 18: 696–704. [DOI] [PubMed] [Google Scholar]
- Ruzicka WB, Subburaju S, Benes FM.. Circuit- and diagnosis-specific DNA methylation changes at γ-aminobutyric acid-related genes in postmortem human hippocampus in schizophrenia and bipolar disorder. JAMA Psychiatry 2015; 72: 541–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Kakizaki T, Hayashi R, Nishimaru H, Furukawa T, Nakazato Y, et al. The physiological roles of vesicular GABA transporter during embryonic development: a study using knockout mice. Mol Brain 2010; 3: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiz A, Blanco Y, Sabater L, González F, Bataller L, Casamitjana R, et al. Spectrum of neurological syndromes associated with glutamic acid decarboxylase antibodies: diagnostic clues for this association. Brain 2008; 131: 2553–63. [DOI] [PubMed] [Google Scholar]
- Scheffer IE, Berkovic S, Capovilla G, Connolly MB, French J, Guilhoto L, et al. ILAE classification of the epilepsies: position paper of the ILAE Commission for Classification and Terminology. Epilepsia 2017; 58: 512–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffer IE, Liao J.. Deciphering the concepts behind “Epileptic encephalopathy” and “Developmental and epileptic encephalopathy”. Eur J Paediatr Neurol 2020; 24: 11–14. [DOI] [PubMed] [Google Scholar]
- Sgadò P, Dunleavy M, Genovesi S, Provenzano G, Bozzi Y.. The role of GABAergic system in neurodevelopmental disorders: a focus on autism and epilepsy. Int J Physiol Pathophysiol Pharmacol 2011; 3: 223–35. [PMC free article] [PubMed] [Google Scholar]
- Sobreira N, Schiettecatte F, Boehm C, Valle D, Hamosh A.. New tools for Mendelian disease gene identification: phenoDB variant analysis module; and GeneMatcher, a web-based tool for linking investigators with an interest in the same gene. Hum Mut 2015; 36: 425–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soghomonian JJ, Martin DL.. Two isoforms of glutamate decarboxylase: why? Trends Pharmacol Sci 1998; 19: 500–5. [DOI] [PubMed] [Google Scholar]
- Tohid H. Anti-glutamic acid decarboxylase antibody positive neurological syndromes. Neurosciences (Riyadh) 2016; 21: 215–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhan L, Zeng W, Li K, Sun W, Xu ZC, et al. Downregulation of hippocampal GABA after hypoxia-induced seizures in neonatal rats. Neurochem Res 2011; 36: 2409–16. [DOI] [PubMed] [Google Scholar]
- Weckhuysen S, Mandelstam S, Suls A, Audenaert D, Deconinck T, Claes LRF, et al. KCNQ2 encephalopathy: emerging phenotype of a neonatal epileptic encephalopathy. Ann Neurol 2012; 71: 15–25. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this report are available from the corresponding author, upon reasonable request.