Summary
Stable genomic integration of exogenous transgenes is essential in neurodevelopmental and stem cell studies. Despite tools driving increasingly efficient genomic insertion with DNA vectors, transgenesis remains fundamentally hindered by the impossibility of distinguishing integrated from episomal transgenes. Here, we introduce an integration-coupled On genetic switch, iOn, which triggers gene expression upon incorporation into the host genome through transposition, thus enabling rapid and accurate identification of integration events following transfection with naked plasmids. In vitro, iOn permits rapid drug-free stable transgenesis of mouse and human pluripotent stem cells with multiple vectors. In vivo, we demonstrate faithful cell lineage tracing, assessment of regulatory elements, and mosaic analysis of gene function in somatic transgenesis experiments that reveal neural progenitor potentialities and interaction. These results establish iOn as a universally applicable strategy to accelerate and simplify genetic engineering in cultured systems and model organisms by conditioning transgene activation to genomic integration.
Keywords: genetic switch, somatic transgenesis, lineage tracing, mosaic analysis, DNA vectors, transposon systems, genomic integration, neural stem cells, genetic engineering
Graphical Abstract

Highlights
-
•
A gene expression switch powered by genomic integration
-
•
Accelerated readout of additive transgenesis with one or multiple vectors
-
•
Faithful lineage tracing and mosaic analysis by somatic transfection
-
•
Near-universal applicability in cultured cells and animal models
Kumamoto et al. introduce iOn, a genetic switch that conditions exogenous transgene expression to integration in the host cell genome by DNA transposition. This system radically simplifies stable transgenesis with one or multiple plasmid vectors, opening new options to genetically manipulate cells in cultured systems and model organisms.
Introduction
Gene transfer approaches enabling stable genomic insertion and expression of exogenous transgenes in host cells or organisms are central in biology. Constant progress in integrative vector systems facilitate their implementation for a growing range of purposes, such as investigation of gene function and regulation (Akhtar et al., 2013), genetic screens (Doench, 2018), stem cell study and engineering (Tewary et al., 2018), cell-based therapies (Hirsch et al., 2017), and emerging synthetic biology applications (Black et al., 2017, Ebrahimkhani and Ebisuya, 2019).
Neurobiology is one of the fields most profoundly impacted by these approaches. In cultured systems, additive transgenesis is widely used to derive cells homogenously expressing one or more genes of interest, a process instrumental to direct pluripotent stem cells toward specific neuronal lineages, decipher the mechanisms of their differentiation, and harness these cells to model neural pathologies (e.g., Kondo et al., 2017, Nehme et al., 2018, Yang et al., 2017). In vivo, somatic transgenesis approaches targeting neural progenitors have acquired major importance in neurodevelopmental studies by enabling the expression of lineage markers or experimental manipulation of gene function over the long-term (Cepko, 1988, Woodworth et al., 2017) without resorting to time-consuming germline modifications.
Since spontaneous integration occurs only at an extremely low rate, tools driving efficient genomic transgene insertion are essential to reach acceptable throughput in the above applications and implement more complex schemes involving multiple transgenes. Vectors adapted from genome-integrating RNA-based viruses have long been employed for this purpose. In particular, retroviruses capable of transducing dividing cells have been at the basis of seminal studies identifying the neuronal and glial output of embryonic neural progenitors with marker genes or sequence-based barcodes (e.g., Noctor et al., 2001, Price et al., 1987, Yu et al., 2012). However, viral vector production is burdensome, and their cargo capacity is inherently limited. Fast and convenient to assemble, store, and handle, DNA vectors have become the tool of choice in many gene transfer applications, thanks to the emergence of transposases and programmable endonucleases that drive random or site-specific integration, respectively (Ivics et al., 2009, Mikuni et al., 2016, Suzuki et al., 2016). Transposases provide the highest efficiency of integration, a process that they directly mediate, whereas endonucleases only indirectly stimulate transgene insertion through the host repair machinery upon target locus cleavage, with potential detrimental side effects (Haapaniemi et al., 2018). Transposon-based systems achieve high rates of integration with >1-kb-long sequences, can handle cargos up to 100 kb (Jung et al., 2016, Li et al., 2011), and are thus ideal for demanding applications requiring integration of one or multiple large transgenes comprised of a promoter and gene of interest (GOI). Beyond stable cell transfection in cultured systems, they advantageously replace transgenic animal lines in a growing number of in vivo applications, including fate mapping (Chen and LoTurco, 2012), multiplexed clonal analysis with combinatorial labels (Figueres-Oñate et al., 2016, García-Moreno et al., 2014, Loulier et al., 2014), assessment of gene function (Landrette and Xu, 2011, Serralbo et al., 2013), or direct screening of genes involved in developmental processes (Lu et al., 2018).
Integrative DNA vectors can be delivered in cultured cells by a number of transfection agents, and in vertebrate animal models, the location of neural progenitors along the lumen of the neural tube makes them accessible to electroporation-based approaches that yield strong expression in their neuronal and glial descendants. Alternatively, adeno-associated viruses (AAVs) offer a convenient way to directly introduce donor DNA in mature neurons for Cas9- or transposase-mediated genomic integration (Cammack et al., 2019, Gao et al., 2019, Nishiyama et al., 2017).
While molecular tools facilitating genomic integration have been the focus of considerable efforts, the design of transgenes carried by integrative DNA vectors has essentially remained the same since the beginning of the genetic engineering era; it simply follows the arrangement of endogenous transcriptional units where the GOI is positioned downstream of its promoter and is thus constitutively expressed by episomal vectors prior to integration. Consequently, genome-integration events cannot be distinguished from residual episomal transgenes. This is a major problem that universally affects stable transgenesis procedures with DNA vectors, making them considerably more complex than transient approaches. In cultured cells, week-long delays are required to eliminate episomes, usually in the presence of drugs and with inherent risks of genetic or epigenetic drift (Liang and Zhang, 2013, Merkle et al., 2017). In vivo, transient episomes confound the interpretation of somatic transgenesis experiments, as they can act on a progenitor while being absent in its descendants due to dilution by cell division. Thus, expression from integrated markers may not accurately reflect the past history of transgene expression in a cell. This essentially precludes the development of reliable functional mosaic analysis schemes based on exogenous DNA vectors applicable to probe cell-autonomous versus non-autonomous aspects of gene function over the long-term, until now involving complex genetic schemes and the establishment of transgenic animal lines (Pontes-Quero et al., 2017, Zong et al., 2005). Episomes may also cause leakage from cell-type-specific promoters and regulatory sequences (Inoue et al., 2017), preventing the development of somatic intersectional schemes based on Cre/lox recombination that offer the same reliability as transgenic animal lines. Moreover, in vitro as well as in vivo, integrative schemes aimed at assessing gene function are critically impeded by the burst of episomal expression that follows transfection, which may have harmful effects on cell behavior, identity, and viability (Batard et al., 2001).
To bypass the limitations of stable transfection mentioned above, there is a need for an approach through which large transgenes can be efficiently incorporated in the host genome and distinguished from episomal donor vectors. This could be achieved if transgene expression was suppressed in episomes and only switched on upon genomic integration. However, existing systems to control gene expression, such as those based on site-specific recombination (Dymecki and Kim, 2007), do not enable this.
Here, we introduce an integration-coupled On (iOn) gene expression switch through which genomic insertion of an initially silent transgene is accompanied by a rearrangement that triggers its expression. We present several implementations of this concept that efficiently couple transcriptional or translational gene activation to genomic integration. We demonstrate its advantages for highly efficient establishment of stable mouse and human pluripotent stem cell lines expressing multiple transgenes in vitro. In vivo, somatic transfection with iOn-based vectors constitutes a powerful alternative to additive transgenesis to drive constitutive or conditional expression of reporters and effectors. We applied this approach for lineage and functional mosaic analyses in chick embryos that enabled us to determine the clonal output of genetically defined progenitor subtypes in the developing retina and to uncover a homeostatic control of neurogenesis in the embryonic neural tube. These results establish iOn as an efficient strategy for direct readout of transgenesis that accelerates and facilitates genetic manipulations in neurodevelopmental and neural stem cell studies and is also applicable to virtually any cell type or species.
Results
Design and Validation of an Integration-Coupled Gene Expression Switch
To create the iOn switch (Figure 1), we took advantage of the piggyBac transposition system (Fraser et al., 1996), currently one of the most efficient tool for genomic integration of exogenous DNA (Ding et al., 2005), characterized by its very large cargo capacity (Jung et al., 2016, Li et al., 2011) and precise cut-and-paste mechanism mediating traceless transposon excision. In classic transposon vectors (Figure 1A, left), the transgene, framed by two antiparallel-oriented terminal repeats (TRs) recognized by the piggyBac transposase, is readily active prior to excision from the donor plasmid. We reasoned that placing the TRs in parallel rather than antiparallel orientation would create a situation in which transposase-mediated insertion in the host genome is accompanied by a rearrangement exploitable to trigger gene expression (Figure 1A, right).
Figure 1.
Principle and Validation of the iOn Switch
(A) Principle of gene transfer with a classic transposon (left) and iOn vector (right). While the former allows GOI expression from episomes, that from iOn vector is conditioned by transposase action that reunites the promoter (Prom) and GOI. Orange arrows: 5′ and 3′, transposon TRs; pA, transcription terminators; TTAA, PB transposition footprint.
(B) Validation in HEK293 cells 3 days after transfection with a classic transposon (PBCAG::RFP, left) and iOn vector (right). Top: epifluorescence images. Bottom: representative cytometry plots from cells transfected with PB/iOn vectors (red) versus control cells (gray).
(C) Time-course analysis of RFP expression with episomal, classic transposon, and iOn vectors. Values and error bars represent the mean and SEM of four replicates.
(D) Localization of a membrane-GFP (GFP-Kras) expressed from a classic transposon (left) or iOn vector (right) 3 days after transfection in HEK293 cells. This vector was designed with the translational LiOn switch presented in Figure 2.
(E) Clones established by sorting iOnCAG∞RFP-transfected cells display the sequence expected for precise junction between the promoter and GOI.
(F) Cells sorted based on iOnCAG∞RFP expression yield a high proportion of RFP-positive clones compared to PBCAG::RFP. Values and error bars represent the mean and SEM of three separate experiments. 1,078, 620, and 504 clones were assessed for PBCAG::RFP transfection without and with PBase and iOnCAG∞RFP, respectively (p < 0.0001 with χ2 test).
See also Figures S1 and S2.
We first designed transcriptional versions of iOn switch in which a promoter (Prom) and GOI, initially placed in opposite orientation, are reunited in a functional configuration by transposition (Figure 1A, right, and Figures S1A–S1D). We hereafter denote this transgene arrangement iOnProm∞GOI by opposition to a classic transposon driving constitutive expression noted PBProm::GOI. We tested different iOn vector designs using the broadly active CAG promoter and a red fluorescent protein (RFP) as GOI, which we assayed by transfection in HEK293 cells in presence or absence of piggyBac transposase (PBase) (Figures S1A–S1D). 3 days after transfection, PBase-dependent RFP expression was observed with all tested constructs, validating the iOn switch concept. We selected the transgene design with highest signal to noise ratio (Figure S1C). This construct, hereafter termed iOnCAG∞RFP, yielded efficient RFP expression with very low background transcription in absence of PBase (0.4%; Figure 1B, right).
Time-course experiments comparing the iOn vector with classic piggyBac-based and nonintegrative plasmids showed that the iOn switch achieves long-term GOI expression without the transient expression burst associated with vectors active in episomal form (Figure 1C). Instead, GOI expression gradually builds up, plateaus within a few days and remains in a narrower range compared to a classic transposon (Figures 1C, S1E, and S1F), a feature of high interest to reduce variability and improve protein localization in transfection assays. Indeed, in HEK293 cells transfected with an iOn vector expressing a farnesylated GFP (GFP-Kras), near-uniform membrane expression staining was observed 3 days after transfection, while signal from a classic transposon showed frequent clustering and overflowing of GFP from the membrane compartment (Figure 1D). Importantly, short- and long-term viability with iOn vectors were comparable to that of classic transposons (Figures S1G and S1H), and the switch was active in all cells tested, including HeLa and 3T3 (Figure S1I) as well as human and mouse pluripotent stem cells (see below).
To confirm that the iOn switch functions as expected, we performed the following tests. First, we verified that both TRs were necessary to power the switch using truncated iOnCAG∞RFP vectors lacking either the 5′ or 3′ piggyBac TR. As expected (Wang et al., 2014), transfection of these single-TR plasmids in HEK293 cells yielded only barely detectable fluorescence in very rare cells (Figure S2A). Second, to separately monitor each of the two vector “arms,” we designed an iOn plasmid bearing two distinct fluorescent protein (FP) markers upstream of each PB TR. 7 days after transfection, co-expression of these markers was observed in a vast majority of labeled cells (94% ± 3% SEM), indicating near-systematic co-integration of the two arms upon transposase action (Figure S2B). Finally, to verify whether the switch drives the predicted transgene rearrangement, we derived clones from individual fluorescent cells transfected with iOnCAG∞RFP in which we sequenced the junction between the promoter and GOI; all sequences demonstrated reunion of the two transgene elements with base-pair precision (Figure 1E; n = 6).
The latter approach also enabled us to compare the performance of iOn versus classic transposons for stable cell line establishment. Analysis of >500 clones showed that 95.78% (±0.73% SEM) remained RFP positive 10 days after sorting, indicating highly efficient integration of the transgene in the genome of founder cells and long-term maintenance of its expression (Figure 1F). Indeed, analysis of the few RFP-negative clones among 440 clones demonstrated that they had lost and not silenced the transgene (Figure S2C). By contrast, a classic PBCAG::RFP vector yielded only 55.18% (±5.07% SEM) of RFP-positive clones (Figure 1F). Similar to HEK cells, fluorescence-based clonal selection of mouse embryonic stem cells (ESCs) with the above GFP-Kras-expressing iOn vector demonstrated a high enrichment in integrative events compared to a classic transposon (Figure S2D). Thus, the dependence of the iOn switch on transposition enables efficient coupling of transgene activation and genomic integration.
Together, these results establish iOn as a tool for highly effective drug-free transgenesis through which genomic expression of GOIs can be assessed directly after transfection, without interfering episomal expression.
Improved Versions of iOn Vectors
Vectors based on the transcriptional version of the iOn switch, presented above, suppress most activity from episomes and shall suffice in a majority of uses, such as stable cell line establishment or assessment of gene function. In anticipation of highly sensitive applications, we nevertheless explored ways to achieve even tighter control of gene expression.
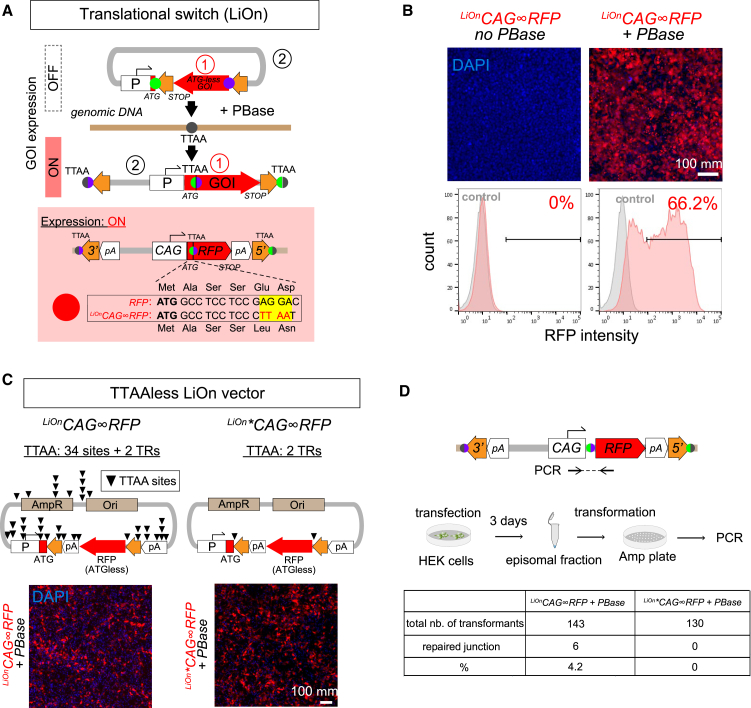
First, we sought to abolish the faint episomal leakiness observed with transcriptionally activated iOn vectors. Transcriptional background is a general problem with non-chromosomal DNA (Inoue et al., 2017), whether due to cryptic promoter activity or unavoidable “pervasive” transcription (Clark et al., 2011). To bypass this issue, we designed a leak-proof iOn (LiOn) switch in which both transcription and translation are blocked in absence of transposase (Figures 2A and 2B). In LiOn, the GOI open reading frame (ORF) is initially interrupted after the translational start and reconstituted upon PBase action, the piggyBac excision footprint (TTAA) being incorporated at a silent or neutral position (Figure 2A). A plasmid based on this design (hereafter denoted LiOnProm∞GOI) showed undetectable leakiness in absence of transposase (Figure 2B) and provided similar PBase-dependent expression compared to the transcriptional iOn, with the same reduced variability and low toxicity (Figures S1F and S1G).
Figure 2.
Improvement of the iOn Switch
(A) Principle of the leak-proof translational iOn (LiOn) switch. In episomes, the GOI is split in 5′ and 3′ portions that are reunited upon transposition with incorporation of the TTAA footprint at a silent position. Orange arrows: 5′ and 3′, transposon TRs; pA, transcription terminators.
(B) Validation of the LiOn switch. Top: epifluorescence views of HEK293 cells 3 days after transfection with a LiOnCAG∞RFP vector in presence and absence of PBase. Bottom: representative cytometry plots from cells transfected with the LiOn vector (red) versus control cells (gray).
(C) Design and validation of a “TTAA-less” LiOn vector (LiOn∗CAG∞RFP) devoid of PBase target sequences. Top: maps of the original LiOn vector (left) and LiOn∗CAG∞RFP plasmid (right) in which all TTAA sequences have been mutated (except in the two TRs). Bottom: control and mutated vectors show similar RFP expression 3 days after transfection in HEK293 cells.
(D) Assessment of episomal activation of the LiOnCAG∞RFP and LiOn∗CAG∞RFP vectors. Episomes purified from HEK cells 3 days after transfection were transformed in competent bacteria. PCR tests on 130 colonies grown from the TTAA-less LiOn vector did not reveal any activated episomes.
See also Figure S3.
Second, we sought to suppress any unwanted episomal activation of iOn vectors that may happen through self- (a.k.a. “suicidal”) integration, shown in previous work to occur at low frequency with classic transposon vectors (Wang et al., 2014). To avoid this effect, we synthesized a modified LiOn vector with punctual substitutions in all 34 PBase target sequences (TTAA) present in the backbone, promoter, and polyadenylation signals (Figure 2C), which we used to express RFP. In HEK293 cells, expression from this TTAA-less LiOn vector (denoted LiOn∗CAG∞RFP) in presence of PBase was comparable to that obtained with the non-edited LiOn version (Figure 2C, bottom). To evaluate the activation of the switch in nonintegrated plasmids, we then purified episomes from the transfected HEK cells, which we transformed in competent bacteria. While PCR tests performed on the resulting bacterial colonies revealed a low fraction (<5%) of activated episomes in cells transfected with the unmodified LiOn vector, activated plasmids were not observed with the TTAA-less LiOn plasmid (n = 130 colonies; Figure 2D). Further qPCR tests performed on the episomal fraction recovered from HEK cells also demonstrated a significant reduction of activated episomes with the TTAA-less strategy (Figure S3).
Thus, in cases requiring very tight transgene control, both the low PBase-independent transcriptional leakiness of iOn vectors and their rare PBase-dependent episomal activation can be efficiently controlled through the LiOn and TTAA-less iOn strategies, respectively.
High-Efficiency Multiplexed Stable Transfection with iOn Vectors
Having validated the iOn switch and its improved implementations, we next sought to extend its range of applications in cultured cells (Figure 3). Stable transfection with multiple independent transgenes is challenging with standard DNA vectors due to false positives resulting from episomal expression and the difficulty of combining orthogonal drug selection systems. The high enrichment for integrative events achieved with iOn bypasses these issues.
Figure 3.
Highly Efficient Multiplexed Stable Transfection with iOn Vectors
(A) 3 days after co-transfection in HEK293 cells of three LiOn plasmids expressing distinct FPs (EGFP, mRFP1, or IRFP670, 100 ng each, respectively coded as green, red, and blue), PBase-dependent expression is observed at similar levels for all markers.
(B) Dose dependence of expressed transgenes copy number. Co-transfection of 1 ng, 10 ng, and 100 ng of the three LiOnCAG∞FP plasmids results in increasingly complex colors that reveal activity of 1.1-6.9 transgene copies. Ternary graphs show RGB values from individual labeled cells. See also Figure S4C for transgene copy number estimation.
(C) Cell sorting of triple-labeled cells 2 days after transfection with the LiOn vectors yields a majority of clones co-expressing the three FPs, but only a minority with classic transposons (mean and SEM of three separate experiments; 158, 104, and 87 clones were assessed for each condition). χ2 test indicated significant differences among the three situations (p < 0.0001).
(D) Example of a human iPSC colony derived from cells co-transfected with the three-color LiOnCAG∞FP vectors, grown 45 days. All cells co-express the three FPs.
(E) Co-transfection of the three-color LiOnCAG∞FP vectors during human iPSC neuronal differentiation yields varied FP combinations reflecting their clonal organization. Inset: LiOnCAG∞RFP expression in iPSCs immunostained with the neuronal marker Tuj1 (green).
See also Figure S4.
To simultaneously identify multiple integration events, we created LiOn vectors expressing green and near-infrared FPs, forming a trichromatic set together with the abovementioned LiOnCAG∞RFP (Figures 3A, S4A, and S4B). Co-transfection of standard amounts of the three vectors in HEK293 cells resulted in varied FP combinations, revealing expression from multiple transgenes in a large fraction of cells (Figure 3A). Furthermore, the color palette expressed by the cells provided a way to quantify active transgene copy number, as done in former work (Kobiler et al., 2010; Figure S4C). Using this approach, we found that adjusting vector concentration enabled to fine tune this number from approximately one to seven transgene copies per labeled cell (Figures 3B and S4C; see STAR Methods).
We then applied the three-color LiOn vectors to establish stable cell lines bearing distinct transgenes. Strikingly, sorting and expansion of triple-fluorescent cells yielded a vast majority (80%) of clones co-expressing all three FPs 10 days after sorting (Figures 3C, S4D, and S4E) compared to only ~20% when using classic transposons, demonstrating the superior efficiency of iOn-based multiplexed transgenesis. In human induced pluripotent stem cells (iPSCs), clones co-expressing all three LiOnCAG∞FP vectors similarly maintained their expression at near-homogeneous levels over multiple passages (Figure 3D). The three-color LiOn transgenes also drove long-term expression during iPSC differentiation toward the neural lineage, with FP combinations providing a readout of clonal relationships (Figure 3E). Thus, iOn provides an efficient route for one-shot multiplexed transgenesis in cell culture models.
Additive Somatic Transgenesis and Cell Lineage Tracing with iOn
We next assessed the iOn switch in vivo (Figure 4). By efficiently coupling transgene integration and expression, iOn vectors have the potential to create a situation virtually equivalent to transgenic animal lines through simple transfection, without the shortcomings associated with expression from nonintegrated episomes.
Figure 4.
Cell Lineage Tracing and Conditional Expression by Additive Somatic Transgenesis with iOn
(A) Fate mapping in the mouse cerebral cortex. Left: co-electroporation of an iOnCAG∞RFP vector with PBase during neurogenesis (E12.5) yields streams of neurons migrating radially from the ventricular surface at E18.5, while an episome (CAG::GFP) only marks those born shortly after electroporation. No red labeling is observed in absence of PBase (inset). CP, cortical plate. Right: quantification of labeled cells confirms that iOn-labeled cells (RFP, RFP/GFP) occupy all cortical layers, while most cells bearing episomal labeling settle in intermediate layers. Values and error bars represent mean and SEM from 4 tissue sections.
(B) Longitudinal confocal views through E6 chick spinal cords electroporated at E2 with a classic transposon (top) or iOn vector (bottom) together with a control episome (CAG::GFP). The iOn vector homogenously labels radially migrating cells, while the classic transposon also strongly labels isolated neurons (white arrowheads), similar to the nonintegrating vector.
(C) LiOnCAG∞GFP electroporation with PBase in the embryonic mouse retina during neurogenesis (E14.5) labels all retinal layers at post-natal day 6 (P6), while expression from an episome (CAG::dsRed2) only marks ganglion cells, born shortly after electroporation (ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer).
(D) Multicolor clonal tracking. Radial view of a portion of neural retina (top) and en face views of the bipolar cell layer (BC; bottom) and retinal pigmented epithelium (RPE; right) from an E8 chicken embryo electroporated with triple-color LiOn vectors at E1.5. FP combinations identify clones.
See also Figure S5.
We tested this somatic cell transgenesis strategy through various assays in the developing vertebrate nervous system, where electroporation provides a convenient way to transfect neural progenitors located at the ventricular surface. In the mouse cerebral cortex, electroporation of a control episomal vector (CAG::GFP) only labeled neurons born at the time of the electroporation (embryonic day 12 [E12]) that migrated in intermediate layers due to rapid dilution in dividing progenitors (Figures 4A and S5A). In striking contrast, an iOnCAG∞RFP vector homogenously marked electroporated progenitors and all their radially migrating derivatives, including late-born upper-layer neurons and astrocytes, in a strict PBase-dependent manner. Further tests in the embryonic chicken spinal cord confirmed that iOn expression in progenitor cells, first detected 1 day after electroporation, was maintained in the neurons that they generated as these migrated away from the ventricular surface and differentiated in outer layers (Figures 4B, S5B, and S5C). Importantly, iOn vectors avoided the strong and irregular labeling of isolated neurons similar to that observed with episomal CAG::GFP that polluted patterns obtained with classic transposons, likely due to inheritance of multiple nonintegrated copies in cells born shortly after transfection, (Figure 4B, arrowheads). To verify whether the two types of vectors integrated within the same temporal frame in the genome of transfected progenitors, we compared the size of isolated clones marked by co-electroporated LiOnCAG∞GFP and PBCAG::RFP plasmids, focusing on the apical progenitor pool. The average number of progenitors per clone (measured by counting cells attached to the ventricular surface) did not differ between the iOn vector and the classic transposon (Figure S5D), indicating that they obeyed similar integration kinetics. As a third validation, we applied the iOn switch in the developing mouse retina, where similar to the above two models, iOn plasmid electroporation in progenitors at E14.5, during the first phase of neurogenesis, successfully labeled all cell layers, while an episomal vector only marked early-born retinal ganglion cells (Figure 4C).
As shown in vitro (Figure 3), the high efficiency of the iOn switch makes it an ideal candidate to implement complex schemes requiring multiple transgenes. We explored this potential by applying iOn vectors expressing different color markers to resolve cell lineage relationships in the nervous system (Figure S5E). In the chick neural retina, retinal pigmented epithelium, and spinal cord, FP combinations generated by co-transfecting the trichromatic LiOnCAG∞FP vectors presented in Figures 2A and S3A efficiently contrasted groups of cells that matched known clonal patterns (Fekete et al., 1994, Leber and Sanes, 1995; Figures 4D and S5F). Analysis of FP proportions expressed by individual retinal clones demonstrated their homogeneity among labeled cells, including those located in distinct layers, indicating that epigenetic silencing did not have a major influence on transgene expression (Figure S5G).
Thus, as is the case in cultured cells, the iOn switch functions with high efficiency in vivo to express one or multiple transgenes over the long-term.
Functional Mosaic Analysis with iOn
Beyond lineage tracing, the iOn strategy offers a means to create sustained experimental perturbations of specific cell signaling pathways (Figure 5). To test this possibility, we assembled a LiOn vector co-expressing RFP and the intracellular domain of the Notch receptor (NICD), a well-known regulator of neural progenitor fate (Pierfelice et al., 2011; Figure 5A). Electroporation of this plasmid in the embryonic chick spinal cord resulted in marked reduction of neurogenesis and expansion of progenitors compared to a LiOnCAG∞GFP control vector (Figures 5A and S6A), which are the effects anticipated in this model for Notch pathway activation (Hämmerle et al., 2011). Remarkably, these effects were still manifest 4 days after electroporation and were also augmented with respect to a transient perturbation.
Figure 5.
Functional Mosaic Analysis by Somatic Transgenesis
(A) Top: longitudinal confocal views through E5 chick spinal cords 3 days after electroporation of a LiOn vector expressing the Notch intracellular domain (LiOnCAG∞RFP-2A-NICD) compared to transient NICD overexpression (CAG::NICD-IRES-GFP) or control (LiOnCAG∞GFP). VZ, ventricular zone. Bottom: representative transverse section of E5 chick spinal cords electroporated with control (LiOnCAG∞GFP, green) and NICD-expressing LiOn vectors (LiOnCAG∞RFP-2A-NICD, red), immunostained for neuronal marker HuC/D (blue). Graph shows the percentage of HuC/D neurons among transfected cells with sustained versus transient NICD expression (CAG::NICD) and control cells (LiOnCAG∞RFP). Values and error bars show mean and SEM from distinct embryos. A Kruskal-Wallis test indicated significant difference between control and LiOnCAG∞NICD (p < 0.01).
(B) Longitudinal view through an E6 spinal cord co-electroporated with control (LiOnCAG∞GFP, green) and NICD-expressing LiOn vectors (LiOnCAG∞RFP-2A-NICD, red). Green cells migrate radially, while most red cells remain at the ventricular surface (dotted line).
(C) Non-cell-autonomous effects of NICD expression. Left: E6 chick spinal cord transverse sections co-electroporated at E2 with a GFP-expressing control LiOn vector and a LiOnCAG∞RFP (left) or LiOnCAG∞RFP-2A-NICD plasmid (right). Middle: quantification of the ratio between GFP+/RFP− and RFP+ neurons and ventricular zone cells. Values and error bars represent mean and SEM from distinct embryos (n ≥ 5). A Mann-Whitney test indicates significant difference between control and LiOnCAG∞NICD (p < 0.005). Right: summary. Increased neurogenic output from green cells in NICD-perturbed condition reveals a homeostatic interaction among progenitors.
See also Figure S6.
This experimental paradigm provided a unique opportunity to investigate the non-cell-autonomous effects of an extended maintenance of neural stem cells in the developing vertebrate nervous system (Figure 5C). To this aim, we generated color-coded mosaics by co-electroporating NICD-expressing (red) and control (green) iOn vectors in which we measured the neurogenic output of GFP+/RFP− (unperturbed) progenitors 4 days after the electroporation. Such mosaic analysis cannot be conclusively undertaken with classic integrative vectors active in episomal form, since cells unlabeled at the time of the analysis may have transiently expressed the transgene earlier on. Surprisingly, in the iOn mosaics, we observed that unperturbed spinal progenitors generated significantly expanded numbers of progeny (+314%, p < 0.005) compared to a control situation (Figure 5C). This non-cell-autonomous effect did not appear to be triggered by the apoptosis of NICD-expressing cells, since no activated caspase-3 was detected in labeled cells (Figure S6B). We thus conclude that neural stem cells of the embryonic spinal cord can modulate their output and compensate in a homeostatic manner for a reduction in the neurogenic activity of their neighbors.
Conditional Control of Transgene Expression in Somatic Transfection Experiments
Finally, iOn vectors are also of interest to assay and exploit transcriptional regulatory elements in a genomic configuration without the need to establish transgenic cell or animal lines. Cre/lox conditional labeling, widely applied in transgenic mice to target specific cell types, is so far largely incompatible with somatic transfection approaches due to leakage from episomes (Schick et al., 2019). We devised a LiOn vector that reconstitutes an interrupted Cre recombinase genes upon PBase action (Figure 6A, left). Strikingly, this LiOnCMV∞Cre plasmid presented no detectable leakiness in the absence of transposase when transfected in cells expressing a floxed reporter transgene, but it efficiently triggered Cre expression and recombination upon PBase action (Figure 6A, right).
Figure 6.
Intersectional Cre/lox Recombination and Analysis of the Output of Genetically Identified Neural Progenitors with iOn
(A) Cre/lox conditional expression. Left: LiOn vector in which full translation of Cre, initially blocked, is activated by PBase; inset: Cre-FLAG immunodetection after transfection of LiOnCMV∞Cre and PBase in HEK-RY cells stably expressing a CAG::RY reporter switching from RFP to YFP expression under Cre action. Right: strict PBase-dependent recombination is observed 3 days after LiOnCMV∞Cre transfection in HEK-RY cells (CAG::mTurquoise2: transfection control). Graph shows mean and SEM of replicates from three distinct experiments.
(B) Radial view through an E8 chick retina co-electroporated at E1.5 with a Cre-expressing LiOn vector driven by Atoh7 regulatory sequences (LiOnAtoh7∞Cre) and a Tol2CAG::RY transposon. Restricted recombination in the retinal ganglion (RGL) and outer nuclear (ONL) layers is observed. INL, inner nuclear layer.
(C) Multicolor clonal analysis of Atoh7+ progenitor output. Optical sections and radial views of an E8 chick retina electroporated at E1.5 with LiOnAtoh7∞Cre along with genome-integrating multicolor reporters (Tol2CAG::Nucbow and Tol2CAG::Cytbow). Left: 3D view of a retinal column containing labeled neurons. Middle: Four retinal columns in which clonal pairs or sister cells of a same type can be identified based on expression of identical color marker combinations (arrowheads). Some cells not included in same-type pairs are also observed (asterisks). Column 1 corresponds to that shown in the left panel. Right: quantification of the number of cells belonging to 1-, 2-, 3- or 4-cell clones within labeled photoreceptor (PR), horizontal cell (HC), amacrine cell (AC), and ganglion cells (RGC) in individual columns, consistent with a bias of Atoh7+ retinal progenitors to generate PRs, HCs, and ACs through terminal symmetric division patterns. Graph shows mean and SEM of 14 columns reconstructed from two distinct embryos.
See also Figure S7.
We then applied this LiOn Cre/lox switch to trace the fate of a subset of retinal progenitor cells (RPCs) defined by expression of the Atoh7 transcription factor. At the population level, Atoh7+ RPCs are known to be biased toward the generation of ganglion cells, for which they are essential (Wang et al., 2001), but they also generate much larger numbers of photoreceptors (PRs), horizontal cells (HCs), and amacrine cells (ACs) (Brzezinski et al., 2012, Feng et al., 2010). How this is accounted for at the individual progenitor level is unclear. Atoh7 regulatory sequences (Skowronska-Krawczyk et al., 2009), validated in separate experiments (Figure S7A), were incorporated in a LiOn Cre vector. Electroporation of this LiOnAtoh7∞Cre vector at early stages of chick retinogenesis (E1.5) drove expression in progenies restricted to the outer, amacrine, and ganglion layers of the retina at E8 and E14, while bipolar cells located in the inner nuclear layer (INL) were mostly not labeled, as confirmed by the rare occurrence of the bipolar marker Vsx2 in recombined versus nonrecombined cells (Figures 6B and S7B). This demonstrated tight control of Cre production by the Atoh7 element during retinal development and confirmed that chicken Atoh7+ progenitors are biased toward specific fates.
To determine the potency of individual Atoh7+ progenitors, we then combined the LiOnAtoh7∞Cre vector with stochastic multicolor lineage reporters encoded by Tol2 Cytbow and Nucbow transposons (Loulier et al., 2014; Figure 6C). This approach marked groups of retinal cells organized as columns interrupted at the level of the INL (Figure 6C, left), within which FP combinations further delineated clones whose cell type composition could be assigned based on layer position and morphology. Strikingly, at E8, these Atoh7+-derived clones mostly comprised only a single ganglion cell but frequently included pairs of nearby cells of a same type among photoreceptors, ACs, and HCs (Figures 6C and S7C; Table S1). This suggests that Atoh7 is expressed prior to mitosis in progenitors that generate an important fraction of these three types of neurons (at least 44% ± 4%, 36% ± 7%, and 30% ± 14% of their PR, AC, and HC output, respectively) through terminal symmetric divisions that result in identical fates.
Discussion
We introduce a novel expression strategy that efficiently couples transgene activation to integration in the genome of host cells. iOn vectors rely on an unconventional arrangement of their elements to reconstitute a split transcriptional unit by DNA transposition. Transcriptional and translational versions of the switch efficiently silence (and even completely abolish) transgene expression in absence of transposase. By conditioning GOI activation to transposition, the iOn switch fills a gap in the suite of tools currently available to control transgene activity. Contrary to classic DNA switches based on site-specific recombination (Jefferis and Livet, 2012), it does not require engineering of the host genome: iOn vectors are self-sufficient reagents applicable in virtually any cell or organism. By canceling expression from nonintegrated transgenes, iOn solves two general problems associated with stable transfection using DNA vectors: (1) the impossibility to identify transgenic cells directly after transfection due to episomes; and (2) the adverse effects of episomal expression, including leakiness (Inoue et al., 2017), toxicity (Batard et al., 2001), and risks of genetic and epigenetic drift during selection (Liang and Zhang, 2013). In effect, iOn provides most of the advantages of retro- and lentiviral vectors without their cargo restriction while offering the ease of use of DNA vectors and the capability of traceless excision of the piggyBac system (Fraser et al., 1996). Effective in all cell types and species tested here (human, mouse, and chick), iOn can easily be adapted to different GOIs and promoters. For this purpose, maps of all plasmids built for this study, including a CAG-driven iOn vector equipped with a multicloning site, are presented in Table S2.
The dependence of the iOn switch on transposition makes it prefectly suited to report stable transgenesis in varied contexts (Figure 7). In cultured cells, iOn essentially eliminates the need to select integration events through multiple rounds of cell division in stable transfection assays, making them as simple and rapid as transient approaches while considerably reducing variability. As demonstrated here (Figures 1, 2, and 3), the switch is ideal for rapid drug-free identification and sorting of stable integrants expressing one or multiple transgenes. In many uses, short-term transfection experiments with iOn vectors will provide information equivalent to stable cell lines on gene expression, regulation, and function. We anticipate that this will simplify and accelerate genetic screening procedures based on transposons (Kawakami et al., 2017). In vivo (Figures 4, 5, and 6), transfection with iOn vectors may substitute for additive transgenesis in most, if not all, of its applications, with more accurate readout compared to previous somatic transgenesis approaches that can be polluted by expression from episomal vectors (Chen et al., 2014, Loulier et al., 2014). In particular, somatic transfection with iOn vectors provides an efficient means to track cell lineage in a variety of model organisms including non-genetically tractable ones. iOn also provides a simple route for functional mosaic analysis using distinct FP markers to identify perturbed versus control cells (Figure 5), while implementing such color-based strategy usually requires complex, time-consuming genetic manipulations (Pontes-Quero et al., 2017, Zong et al., 2005). Critically in this approach, the coupling of transgene integration and expression achieved with iOn ensures that marker expression in a cell reflects that of its entire lineage. This enabled us to create sustained mosaic perturbations of neurogenesis that reveal an intriguing homeostatic interplay among progenitors in the embryonic neural tube that is capable of significantly influencing their output and reminiscent of that observed in some peripheral organs (Sharma et al., 2017, Stanger et al., 2007).
Figure 7.
Summary of iOn Potential Applications
iOn vectors enable fast and reliable analysis of genome integrative events shortly after transfection both in vitro (top) and in vivo (bottom).
We also demonstrate that accurate cell-type-specific Cre/lox recombination, until now largely restricted to transgenic animal lines, can be achieved using electroporated iOn transgenes (Figure 6). This opens the way to one-shot intersectional schemes based on somatic transfection to label and manipulate molecularly defined cells. As showed here, this approach can be advantageously combined with multicolor labeling (Weissman and Pan, 2015) to trace the lineage of genetically identified stem cells with clonal resolution. We applied it in chicken embryos to assess the output of fate-restricted retinal progenitors expressing the basic-helix-loop-helix (bHLH) transcription factor Atoh7. Frequent pairs of same-color cells observed among labeled PRs, HCs, and ACs, but not RGCs, support the idea that Atoh7+ progenitors produce the latter cell type by asymmetric divisions (Brzezinski et al., 2012) but generate an important fraction of the first three neuronal classes by symmetric-fated terminal divisions reminiscent of those observed with Drosophila ganglion mother cells (Holguera and Desplan, 2018). Thus, the lineage tree of chicken Atoh7+ progenitors varies depending on the identity of the cells that they generate, and some of them may be restricted to generate a single cell type among PRs, HCs, and ACs. These results extend former observations showing that a nonidentified fraction of retinal progenitors divide symmetrically to produce PRs or ACs in fish (He et al., 2012, Suzuki et al., 2013) and specific HC subtypes in chicken (Rompani and Cepko, 2008). Clonal analysis of genetically defined retinal progenitors has until now only been possible by resorting to a combination of transgenic mouse lines and targeted retroviral labeling (Hafler et al., 2012). Interestingly, another bHLH transcription factor, Olig2, defines retinal progenitors that undergo terminal divisions to produce a subset or neural types partially overlapping with that of Atoh7+ progenitors; however, the specific lineage patterns of the two populations appear to diverge as Olig2+ progenitors mostly generate ACs through heterotypic-fated divisions (Hafler et al., 2012).
Beyond the usages demonstrated here, we anticipate that somatic transgenesis with iOn will provide a platform for complex genetic schemes involving multiple transgenes, ranging from cellular barcoding to functional screening of effector combinations. Current advances in transposition systems also open exciting avenues to improve the iOn strategy. In particular, we anticipate that the switch may be combined with modified transposases enabling to restrict the duration of integration (Querques et al., 2019) or with recently discovered RNA-guided transposases to target specific loci (Klompe et al., 2019, Strecker et al., 2019).
In conclusion, by enabling to report DNA vector integration in host genome, iOn opens new avenues to genetically engineer cells in cultured systems and probe stem cell fate and regulation in the nervous system of model organisms or any tissue accessible to transfection.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-Tuj1 | BioLegend | Cat# MMS-435P; RRID:AB_2728521 |
| Rabbit anti-FLAG | Sigma | Cat# F7425; RRID:AB_439687 |
| Mouse anti-HuC/D (clone 16A11) | Thermo Fisher Scientific | Cat# A-21271; RRID:AB_221448 |
| Rabbit anti-activated Caspase 3 | Cell Signaling | Cat# 9661S; RRID:AB_2341188 |
| Alexa 647-conjugated phalloidin | Molecular Probes | Cat# A30107 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Lipofectamine 2000 | Invitrogen | 11668027 |
| paraformaldehyde (Antigenfix) | Diapath | F/P0014 |
| phosphate buffer saline | Sigma | 32129211 |
| Vectashield | Vector labs | H-1000; RRID:AB_2336789 |
| Vectashield with DAPI | Vector labs | H-1200; RRID:AB_2336790 |
| Trypan Blue | Sigma | T8154 |
| DAPI | Sigma | D9542 |
| Dulbecco’s Modification of Eagle’s Medium (DMEM) | Life technologies | 11965175 |
| E8 medium | Life technologies | A1517001 |
| Geltrex LDEV-Free, hESC-Qualified, Reduced Growth Factor Basement Membrane Matrix | Life technologies | A1413302 |
| Accutase | Life technologies | A1110501 |
| poly-L-ornithine | Sigma | P4957 |
| Laminin | Sigma | 23017-015 |
| Neurobasal Medium | Life technologies | 21103049 |
| Advanced DMEM/F-12 | Life technologies | 12634028 |
| B-27 Supplement (50X), minus vitamin A | Life technologies | 12587010 |
| N-2 Supplement (100X) | Life technologies | 17502048 |
| 2-Mercaptoethanol (50 mM) | Life technologies | 31350010 |
| GlutaMAX Supplement | Life technologies | 35050061 |
| Penicillin-Streptomycin (10,000 U/mL) | Life technologies | 15140122 |
| Opti-MEM® I Reduced Serum Medium (100ml) | Life technologies | 31985062 |
| Dreamfect | OZBioscience | DF40500 |
| Lipofectamine Stem Cell | Invitrogen | STEM00001 |
| Normal goat serum | Sigma | G9023 |
| Fetal Bovine Serum | Eurobio | CVFSVF00 01∗ |
| Fetal Bovine Serum | GIBCO | 10270-098 |
| Normal donkey serum | Jackson ImmunoResearch | 617-000-121 |
| Critical Commercial Assays | ||
| CloneAmp HiFi PCR Premix | Clontech | 639298 |
| Q5 high-fidelity DNA polymerase | NEB | M0491S |
| PowerUp SYBR Green Master Mix | Thermo Fisher Scientific | A25742 |
| NEBuilder® HiFi DNA Assembly Master Mix | NEB | E2621S |
| Nucleospin Tissue Kit | Macherey-Nagel | 740952.50 |
| Nucleospin NucleoSpin Kit | Macherey-Nagel | 740588.50 |
| XL2-Blue Ultracompetent Cells | Agilent Technologies | 50-125-016 |
| Experimental Models: Cell Lines | ||
| HEK293 cells | ATCC | 1573 |
| HeLa cells | ATCC | CCL-2 |
| NIH 3T3 cells | ATCC | 1658 |
| HEK RY reporter cells | This paper | N/A |
| hiPS cells: WTSIi008-A | EBiSC | RRID:CVCL_AH70 |
| ES cells: KH2 | Beard et al., 2006 | N/A |
| Experimental Models: Organisms/Strains | ||
| Mice (Swiss strain) | JANVIER LABS | N/A |
| Mice (C57BL6J strain) | JANVIER LABS | N/A |
| Chicken fertilized eggs (JA57 strain) | EARL Morizeau | N/A |
| Recombinant DNA | ||
| PB(1A)CAG∞RFP | This paper | N/A |
| PB(1B)CAG∞RFP | This paper | N/A |
| PB(2A)CAG∞RFP | This paper | N/A |
| iOnCAG∞RFP | This paper | Addgene #154014 |
| iOnCAG∞MCS | This paper | Addgene #154013 |
| LiOnCAG∞GFP | This paper | Addgene #154015 |
| LiOnCAG∞RFP | This paper | Addgene #154016 |
| LiOnCAG∞IRFP | This paper | Addgene #154017 |
| PBCAG::GFP | This paper | N/A |
| PBCAG::RFP | This paper | N/A |
| PBCAG::IRFP | This paper | N/A |
| LiOnCAG∞GFP-Kras | This paper | N/A |
| LiOn∗CAG∞RFP | This paper | Addgene #154019 |
| LiOnCMV∞Cre | This paper | Addgene #154018 |
| LiOnCAG∞RFP-2A-NICD | This paper | N/A |
| LiOnAtoh7∞Cre | This paper | N/A |
| LiOnAtoh7∞RFP | This paper | N/A |
| Tol2CAG::RY | This paper | N/A |
| Integration reporter | This paper | N/A |
| iOnCAG∞RFP (Δ5′TR) | This paper | N/A |
| iOnCAG∞RFP (Δ3′TR) | This paper | N/A |
| Software and Algorithms | ||
| ImageJ | NIH | https://imagej.nih.gov/ij/. RRID: SCR_003070; |
| Imaris | Bitplane | https://imaris.oxinst.com/packages. RRID: SCR_007370 |
| Adobe Photoshop | Adobe | https://www.adobe.com/products/photoshop.html. RRID: SCR_014199 |
| Adobe Illustrator | Adobe | https://www.adobe.com/fr/products/illustrator.html. RRID:SCR_010279 |
| R Project for Statistical Computing | The R Foundation | https://www.r-project.org/. RRID:SCR_001905 |
| Metamorph software | Molecular Devices | https://www.moleculardevices.com/ RRID: SCR_002368 |
| GraphPad prism | GraphPad Software | https://www.graphpad.com/scientific-software/prism/. RRID:SCR_002798 |
| Other | ||
| Arrayscan high-content system | Thermo Fisher Scientific | Arrayscan |
| FACS | Beckman Coulter | MoFlo Astrios cell sorter |
| FACS | Beckman Coulter | Astrios MoFlo EQ cell sorter |
| Sanger sequencing | Genewiz | N/A |
| Zeiss Discovery V20 | Zwiss | V20 |
| vibrating microtome | Leica | VT1000 |
| Cryostat | Thermo Fisher Scientific | Microm HM560 |
| Confocal microscope | Olympus | FV1000 |
| Epifluorescence microscope | Leica | DM6000 |
| EVOS FL auto inverted microscope | Life technologies | AMAFD1000 |
| Inverted automated microscope | Zeiss | Celldiscoverer 7 |
| Real-time PCR System | Thermo Fisher Scientific | 7500 Fast |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jean Livet (jean.livet@inserm.fr).
Materials Availability
iOn vectors have been deposited at the Addgene public plasmid repository under references #154013, #154014, #154015, #154016, #154017, #154018 and #154019. Detailed maps and sequences of all plasmids developed for this study are also available upon request.
Data and Code Availability
This study did not generate new datasets or codes.
Experimental Models and Subject Details
Cultured cells
Human embryonic kidney (HEK293T), HeLa and 3T3 cells were cultured in 10% fetal bovine serum in Dulbecco modified Eagle medium (DMEM, Life technologies).
Human induced pluripotent stem cells (iPS line WTSIi008-A, EBiSC, UK) were cultured in E8 medium (Life technologies) on Geltrex coating (Life technologies) and passaged with EDTA.
Mouse ES cells (C57BL/6 × 129/Sv, line KH2) (Beard et al., 2006) were cultured on primary embryonic fibroblasts feeder cells.
Mice
Swiss (for brain electroporation) and C57BL6J (for retinal electroporation) females (Janvier labs) were housed in a 12 hr light/12 hr dark cycle with free access to food, and animal procedures were carried out in accordance with institutional guidelines. Animal protocols were approved by the Charles Darwin animal experimentation ethical board (CEEACD/N°5). The date of the vaginal plug was recorded as embryonic day (E) 0.5 and the date of birth as postnatal day (P) 0.
Chicken embryos
JA57 chicken fertilized eggs were provided by EARL Morizeau (8 rue du Moulin, 28190 Dangers, France) and incubated at 38°C for the appropriate time in a FIEM incubator (Italy).
Method Details
DNA constructs
A schematized map of the plasmids designed for this study can be found in Table S2, along with restriction sites available to exchange GOIs and promoters. All iOn and control piggyBac vectors were assembled in a pUC57-mini plasmid backbone (Genscript Inc) using a combination of DNA synthesis (Genscript Inc), Gibson assembly (NEB) and standard restriction and ligation-based cloning. PCR for Gibson assembly was performed using CloneAmp HiFi PCR Premix (Clontech) and Q5 high-fidelity DNA polymerase (NEB). We used minimal piggyBac 5′ and 3′ TRs (Meir et al., 2011), with an additional 3 bp from the wild-type transposon in the 3′ TR as in Loulier et al. (2014). We drove GOI expression with the strong eukaryotic CAG (Niwa et al., 1991) and CMV promoters as well as a 2145 bp fragment regulating expression of the chicken Atoh7 gene (Skowronska-Krawczyk et al., 2009). GOIs were followed by a bovine growth hormone transcriptional terminator (pA1). In the final iOn vector design, a rabbit beta globin transcription terminator (pA2) was added upstream of the PB 3′TR to prevent cryptic episomal transcription. FPs used as GOI included RFP (mRFP1, Campbell et al., 2002), GFP (EGFP, Clontech) and IRFP (IRFP670, Shcherbakova and Verkhusha, 2013). In LiOn vectors, FP ORFs were split near the N terminus (Nt) in two opposite-oriented fragments that become reunited by transposition with incorporation of the TTAA footprint as indicated in Figures 2A and S4A. In the LiOnCMV∞Cre vector, the Cre recombinase ORF was separated in Nt and Ct portions as in (Jullien et al., 2003), with incorporation of the TTAA footprint at a silent position (Leu104) and addition of a FLAG epitope (DYKDDDDK) at the protein Ct. To limit expression of the Cre Nt fragment prior to transposition, its coding sequence was positioned in frame (through the PB 5′ TR) with a PEST degron (Li et al., 1998) followed by a translational stop. The membrane-restricted GFP was generated by adding a short Kras tethering sequence (Averaimo et al., 2016) at the Ct end of EGFP using annealed oligonucleotides. The TTAA-less LiOn∗CAG∞RFP vector was designed on the model of LiOnCAG∞RFP with punctual substitutions intended not to affect vector replication and expression. The 3-color integration reporter was constructed by inserting in LiOnCAG∞RFP two CMV-driven transcriptional units expressing mTurquoise2 and EYFP, 5′ of CAG and 3′ of the mRFP1 polyA, respectively. To assay Cre activity, we designed a floxed reporter (Tol2CAG::loxP-mCherry-loxP-EYFP, abbreviated as Tol2CAG::RY) in which expression switches from mCherry to EYFP upon recombination, framed with Tol2 transposition endfeet to enable genomic integration. The LiOnCAG∞RFP-2A-NICD vector was assembled by introducing a P2A cleavage sequence between the RFP and NICD ORFs to enable their co-expression. As non-integrative control vectors, we used a CAG::NICD-IRES-GFP plasmid (Rios et al., 2011). Other plasmids used in this study included CMV-driven vectors expressing Cre, mTurquoise2 (Goedhart et al., 2012) and IRFP670 (Shcherbakova and Verkhusha, 2013) as well as CAG-driven vectors producing EGFP, mCerulean, dsRed2 (Clontech), the Tol2 transposase (Kawakami and Noda, 2004) and an optimized piggyBac transposase (hyPBase, Yusa et al., 2011).
HEK293, HeLa and NIH 3T3 cell culture experiments
iOn and piggyBac plasmids were transfected in human HEK293, HeLa or mouse NIH 3T3 cells using cationic lipids. Except when otherwise noted, 1 × 105 cells/well were plated in a 24-well dish and transfected at day 1 with 100 ng iOn vector with or without 20 ng of PBase-expressing plasmid (CAG::hyPBase) using 0.7 μl of Lipofectamine 2000 reagent (Invitrogen). For triple-color labeling experiments, we used 100 ng/well of each LiOnCAG∞FP plasmid and 60 ng of PBase vector. To validate the LiOnCMV∞Cre transgene, 50 ng of the corresponding plasmid was co-transfected with 10 ng of PBase vector in a HEK293 cell line stably expressing the Tol2CAG::RY reporter. This line was established by successive use of Tol2 transposition, drug selection with G418 (300 μg/ml, Sigma) and picking of RFP-positive clones. In some experiments, 50 ng of non-integrative plasmid expressing an FP marker distinct from the iOn vector (CMV::mTurquoise2, CMV::IRFP or CAG::GFP) were applied as transfection control. For FACS analysis, transfections were performed in 6-cm dishes with scaled up concentrations. HEK293 cell viability after iOn plasmids transfection was assessed by dye exclusion with Trypan blue solution (0.4%, Sigma). FP expression was either assayed by flow cytometry, epifluorescence or confocal microscopy, or an Arrayscan high-content system (Thermo Fisher Scientific) (see below). For fixed observations, cells grown on 13 mm coverslips coated with collagen (50 μg/ml, Sigma) were immersed in 4% paraformaldehyde (PFA) in phosphate buffer saline (PBS) (Antigenfix, Diapath), rinsed in PBS and mounted in glycerol-based Vectashield mounting medium supplemented with DAPI (Vector labs). All images are representative of at least 3 independent experiments.
FACS analysis and selection of HEK293 clones
For FACS analysis, HEK293 cells grown on 6-cm dishes were dissociated three days after transfection, stained with DAPI and analyzed on a MoFlo Astrios cell sorter (Beckman Coulter) using the following laser lines: 405 nm (DAPI), 488 nm (GFP), 561 nm (RFP), 640 nm (IRFP). 10000 cells were analyzed for each condition; non-fluorescent controls were prepared from mock-transfected cells stained with DAPI. For clonal experiments, HEK293 cells were sorted as single cells two days after transfection. Selection windows were chosen to include most of the FP-positive population and exclude negative cells. For 3-color cell sorting, we first selected live dissociated cells and subsequently selected RFP+, IRFP+ cells within the GFP+ population. Cells were then plated as single cells in 96-well plates and grown for 7-10 days in 200 μL of 10% FBS/DMEM medium mixed 1:1 with filtrated HEK293-conditioned medium. FP expression was assayed by epifluorescence microscopy or Arrayscan High-Content imaging (see below). Some positive clones were expanded in larger dishes for sequencing. To this aim, genomic DNA was isolated from a confluent 3.5- or 10-cm dish with the Nucleospin Tissue Kit (Macherey-Nagel). The rearranged region between the promoter and GOIs (500-600 bp) was amplified using CloneAmp HiFi PCR premix (Clontech) followed by Sanger sequencing (Genewiz, UK).
Episome extraction and characterization
Extrachromosomal DNA was purified from HEK cells three days after transfection as in (Ziegler et al., 2004) using a NucleoSpin plasmid column (Macherey-Nagel). For the bacterial transformation assay, 5ng of episomal DNA was transformed into XL2-Blue Ultracompetent Cells (Stratagene); PCR was performed directly on colonies grow on Ampicillin using primers 5′GTGCTGGTTGTTGTGCTGTC3′ and 5′CAGCCTCATCTTGATCTCGCCCTTCA3′ located on each side of the TTAA footprint in activated LiOnCAG∞RFP transgenes. Quantitative PCR analysis was performed on purified episomes using an Applied Biosystems real-time PCR machine (7500 Fast System) with PowerUp SYBR green master mix following the manufacturer’s instructions. Results from PCR1 (primers 5′TAGAGCCTCTGCTAACCATGTTC3′ and 5′CGCACCTTGAAGCGCATGAAC3′) were normalized against a second PCR internal to RFP (primers 5′CCGACTACTTGAAGCTGTCCTT3′ and 5′GCTTCACCTTGTAGATGAACTCG3′, PCR2), Quantification of gene expression was based on the DeltaCt method in at least three independent biological experiments. Episomal DNA isolated from cells transfected with CAG::RFP was used as a reference. These qPCR measurements represented an overestimation of episomes due to unavoidable contamination by genomic DNA (Yu et al., 2009).
Human iPS cell transfection and differentiation
For iOn labeling of differentiating iPS cells, colonies were dissociated with Accutase (Life Technologies) and replated in 96-well plates coated with poly-L-ornithine (20 μg/ml, Sigma P4957) and laminin (3 μg/ml, Sigma 23017-015). On day 2, cells were transfected with Dreamfect (OZBioscience) according to the manufacturer’s instructions. Cells were then differentiated as spinal motor neurons, fixed on day 14 with 4% PFA and stained with Tuj1 antibody as previously done (Maury et al., 2015). For iPS line generation, WTSIi008-A iPS cells were plated and transfected with Lipofectamine Stem Cell reagent (Invitrogen) according to the manufacturer’s protocol. Transfected cells were isolated by manual or EDTA passages, and homogeneous colonies were obtained 18 days after transfection (4 passages).
Mouse ES cell transfection and clone selection
KH2 ES cells were transfected with LiOnCAG∞GFP-Kras and CAG::hyPBase plasmids (4:1 mass ratio) using Lipofectamine 2000 reagent. 48 hrs after transfection, GFP-positives cells (1.5%) were sorted using an Astrios MoFlo EQ cell sorter and plated at low density (103 cells/ 10-cm dish) on feeder cells. After eight days, GFP-positives clones were picked under a fluorescent stereomicroscope (Zeiss Discovery V20).
Mouse and chicken embryonic electroporation
In utero and in ovo electroporation in mouse and chicken embryos were performed as previously described (Loulier et al., 2014, Rebsam et al., 2009). A DNA mix containing 1-1.2 μg/μl of iOn vector, 0.5-1.2 μg/μl of non-integrative control plasmid and 0.2 μg/μl of CAG::hyPBase plasmid supplemented with fast green dye was injected with a glass capillary pipette into one lateral ventricle or one eye of E12.5 or E14.5 mice, or the optic cup or central spinal cord canal of E1.5 or E2 chick embryos, respectively. For multicolor labeling, the mix contained 1 μg/μl of each LiOnCAG∞FP vector and 0.6 μg/μl of PBase vector. Embryos were left to develop until sacrifice. Tissues were fixed in 4% PFA. Postnatal mouse brains were sectioned at 200-μm thickness with a vibrating microtome (VT1000, Leica), while mouse retinas, chick E6 spinal cords and E6-E8 retinas were flat-mounted on glass slides. Samples were mounted in Vectashield medium and imaged with epifluorescence or confocal fluorescence microscopy.
Immunostaining
For cell cultures: HEK293 cells plated on glass coverslips or iPS cells were fixed with 4% PFA, followed by washing in PBS and a 20-60 min blocking step at room temperature. Blocking solution for HEK293 and iPS cells respectively contained 10% normal goat serum (Sigma) or fetal bovine serum (Eurobio) and 0.5% or 0.2% Triton X-100 (Sigma). Cells were then incubated overnight at 4°C with primary antibody diluted in blocking solution (rabbit anti-FLAG, Sigma, 1:250 or mouse anti-Tuj1, Biolegend, 1:500). After washing in PBS and incubation with secondary antibody (Alexa 647 anti-goat IgG, 1:500, or Alexa 488 anti-goat IgG, 1:1000, Invitrogen) for 1 hr at room temperature, cells were washed again prior to mounting in Vectashield medium. For chicken spinal cords sections: embryos fixed for 1 hr in 4% PFA were equilibrated in 30% sucrose and embedded in TissueTek (Sakura), frozen on dry ice and stored at −80°C prior to cryostat sectioning (Microm HM560, 14 μm sections). After equilibration at room temperature, sections were washed in PBS before blocking in PBS-0.1% Triton-10% normal donkey serum (NDS) and overnight incubation with primary antibody (anti-HuC/D, Molecular Probes, 1:50, or anti-activated Caspase3, Cell Signaling, 1:100) in PBS-0.1% Triton-1% NDS. Following PBS washes, slides were incubated 1 hr with secondary antibody (Alexa 647 donkey anti-mouse or anti-rabbit, Invitrogen, 1:500) in the above buffer, washed and mounted with Vectashield medium. For whole chicken spinal cords: embryos were fixed overnight in 4% PFA. Spinal cords were dissected, washed in PBS and incubated with Alexa 647-conjugated phalloidin (Molecular Probes, 1:250) in PBS-0.1% Triton-1% NDS for 3 hr at room temperature. Following PBS washes, spinal cords mounted in Vectashield medium.
Fluorescence imaging and image analysis
Epifluorescence images were collected with a 10 × 0.6 NA or 20 × 0.7 NA objective on a Leica DM6000 microscope equipped with a VT1000 camera and separate filter cubes for GFP, RFP and IRFP. Confocal image stacks were acquired with 20 × 0.8 NA oil and 40 × 1.25 NA silicone objectives on an Olympus FV1000 microscope, using 440, 488, 515, 560, and 633 nm laser lines to excite CFP, GFP, YFP, RFP and IRFP/Alexa 647, respectively. For analysis with Arrayscan (Thermo Fisher Scientific), cells grown in 24- or 96-well plates were fixed 15 min with 4% PFA and stained with 300 nM DAPI prior imaging with the following laser lines: 386 nm (DAPI), 485 nm (GFP), 570 nm (RFP), and 650 nm (IRFP). Images of live ES cell clones were acquired using an EVOS FL auto inverted microscope (Life Technologies). Image analysis was performed with Fiji (Schindelin et al., 2012) and Imaris (Bitplane). Levels were uniformly adjusted across images with Adobe Photoshop.
Transgene copy number estimation
Images of HEK293 cells transfected with an equimolar mixture of the trichromatic LiOnCAG∞GFP/RFP/IRFP vectors (coded as green, red and blue, respectively) were acquired by confocal microscopy and the center of each labeled cell was manually pointed in Fiji. RGB values were extracted from 6 pixel-radius areas in each cell, and ternary diagrams showing the mean normalized values of the three channels were plotted as in Loulier et al., 2014. A threshold was determined for each channel based on single-color control transfections, which served to determine for each concentration of vector DNA the number of cells labeled with 1- 2- and 3-colors. Experimental data were approximated theoretically using randomized values of copy number frequencies for the interval 1-8 copies per cell following a Gaussian distribution, and each random set of values was used to calculate the expected numbers of cells for each of the three color-categories. This was achieved based on the probabilistic contribution of each copy number class and the frequency randomly assigned in each case, according to the formula:
where Nc is the number of single, two, or three-color cells; n, the copy number class; Fc, the frequency randomly attributed to each copy number class; N, the total number of evaluated cells for each observation; Pc, the probability of having one, two, or three-color cells in the respective copy number class. The theoretical approximation closest to experimental observations in terms of cell numbers was selected. A single Gaussian fit was applied on the chosen theoretical values to reveal the average copy number for each condition:
Quantification and Statistical Analysis
Statistical analysis
The number of samples analyzed is indicated in the figure legends. Statistical analyses were performed using R or GraphPad Prism software. Significance was assessed using χ2 (Figures 1F, 3C, S1H, and S2D) and Kruskal-Wallis tests (one-way ANOVA on ranks) (Figure 5A), Student t test (Figure S3), Welch’s t test (Figures S2B and S7B), and non-parametric Mann-Whitney U test (Figure 5C). Data represent mean ± SEM, ∗ p < 0.05; ∗∗ p < 0.01, ∗∗∗ p < 0.001.
FACS analysis
10000 cells were analyzed for each condition; non-fluorescent controls were prepared from mock-transfected cells stained with DAPI.
Acknowledgments
We thank X. Morin, G. Le Dréau, and G. Orieux for scientific discussions and comments on the manuscript; X. Nicol for the Kras sequence; J.-M. Matter for the Atoh7 promoter; C. Marcelle for the NICD-expressing plasmid; and all members of the Livet lab for assistance. We thank L. Riancho and P.-H. Commère from the Saint-Antoine and Institut Pasteur cytometry facilities and A. Potey, C. Condroyer, S. Fouquet, and the Institut de la Vision core facilities. This work was funded by fellowships from the Uehara Memorial Foundation and Naito Foundation to T.K., the French Ministry of Research (F.M. and C.V.), Fondation pour la Recherche Médicale (DBI20141231328), the European Research Council (ERC-CoG 649117), and Agence Nationale de la Recherche under contracts ANR-10-LABX-65 (Labex Lifesenses), ANR-18-IAHU-0001 (IHU FOReSIGHT), ANR-10-LABX-73-01, ANR-15-CE13-0010, ANR-19-CE11-0005, and ANR-19-CE16-0019. S.N. was funded by ATIP/Avenir and Association Française contre les Myopathies. A.R. was funded by ANR-12-BSV4-0002, Fondation NRJ, and Genespoir.
Author Contributions
J.L., R.B.-M., and T.K. conceived the iOn and LiOn switches. R.B.-M. performed the initial experiments, T.K. performed in vitro and chick spinal cord experiments, F.M. performed chick retina experiments, A.R. performed mouse retina experiments, T.K. and K.L. performed cortical electroporation, C.V. and S.N. performed iPSC experiments, and M.C.T. and S.V.-P. performed ESC experiments. M. Le and M. Lerat performed cell culture experiments and assisted with cloning and image analysis. T.K. and S.T. analyzed data. D.N. modeled copy-number.. T.K., S.T. and J.L. wrote the manuscript with input from all authors.
Declaration of Interests
J.L., T.K., R.B.-M., F.M., S.T., K.L., and M. Le have filed a patent application related to the technology described in this article (number EP18305623).
Published: June 18, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.neuron.2020.05.038.
Supplemental Information
References
- Akhtar W., de Jong J., Pindyurin A.V., Pagie L., Meuleman W., de Ridder J., Berns A., Wessels L.F.A., van Lohuizen M., van Steensel B. Chromatin position effects assayed by thousands of reporters integrated in parallel. Cell. 2013;154:914–927. doi: 10.1016/j.cell.2013.07.018. [DOI] [PubMed] [Google Scholar]
- Averaimo S., Assali A., Ros O., Couvet S., Zagar Y., Genescu I., Rebsam A., Nicol X. A plasma membrane microdomain compartmentalizes ephrin-generated cAMP signals to prune developing retinal axon arbors. Nat. Commun. 2016;7:12896. doi: 10.1038/ncomms12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batard P., Jordan M., Wurm F. Transfer of high copy number plasmid into mammalian cells by calcium phosphate transfection. Gene. 2001;270:61–68. doi: 10.1016/s0378-1119(01)00467-x. [DOI] [PubMed] [Google Scholar]
- Beard C., Hochedlinger K., Plath K., Wutz A., Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44:23–28. doi: 10.1002/gene.20180. [DOI] [PubMed] [Google Scholar]
- Black J.B., Perez-Pinera P., Gersbach C.A. Mammalian synthetic biology: engineering biological systems. Annu. Rev. Biomed. Eng. 2017;19:249–277. doi: 10.1146/annurev-bioeng-071516-044649. [DOI] [PubMed] [Google Scholar]
- Brzezinski J.A., 4th, Prasov L., Glaser T. Math5 defines the ganglion cell competence state in a subpopulation of retinal progenitor cells exiting the cell cycle. Dev. Biol. 2012;365:395–413. doi: 10.1016/j.ydbio.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammack A.J., Moudgil A., Lagunas T., Chen J., Vasek M.J., Shabsovich M., McCullough K., He J., Chen X., Hooda M. Transposon-mediated, cell type-specific transcription factor recording in the mouse brain. bioRxiv. 2019 doi: 10.1101/538504. [DOI] [Google Scholar]
- Campbell R.E., Tour O., Palmer A.E., Steinbach P.A., Baird G.S., Zacharias D.A., Tsien R.Y. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cepko C. Retrovirus vectors and their applications in neurobiology. Neuron. 1988;1:345–353. doi: 10.1016/0896-6273(88)90184-5. [DOI] [PubMed] [Google Scholar]
- Chen F., LoTurco J. A method for stable transgenesis of radial glia lineage in rat neocortex by piggyBac mediated transposition. J. Neurosci. Methods. 2012;207:172–180. doi: 10.1016/j.jneumeth.2012.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Maher B.J., LoTurco J.J. piggyBac transposon-mediated cellular transgenesis in mammalian forebrain by in utero electroporation. Cold Spring Harb. Protoc. 2014;2014:741–749. doi: 10.1101/pdb.prot073650. [DOI] [PubMed] [Google Scholar]
- Clark M.B., Amaral P.P., Schlesinger F.J., Dinger M.E., Taft R.J., Rinn J.L., Ponting C.P., Stadler P.F., Morris K.V., Morillon A. The reality of pervasive transcription. PLoS Biol. 2011;9 doi: 10.1371/journal.pbio.1000625. e1000625–discussion e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S., Wu X., Li G., Han M., Zhuang Y., Xu T. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122:473–483. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Doench J.G. Am I ready for CRISPR? A user’s guide to genetic screens. Nat. Rev. Genet. 2018;19:67–80. doi: 10.1038/nrg.2017.97. [DOI] [PubMed] [Google Scholar]
- Dymecki S.M., Kim J.C. Molecular neuroanatomy’s “three Gs”: a primer. Neuron. 2007;54:17–34. doi: 10.1016/j.neuron.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimkhani M.R., Ebisuya M. Synthetic developmental biology: build and control multicellular systems. Curr. Opin. Chem. Biol. 2019;52:9–15. doi: 10.1016/j.cbpa.2019.04.006. [DOI] [PubMed] [Google Scholar]
- Fekete D.M., Perez-Miguelsanz J., Ryder E.F., Cepko C.L. Clonal analysis in the chicken retina reveals tangential dispersion of clonally related cells. Dev. Biol. 1994;166:666–682. doi: 10.1006/dbio.1994.1346. [DOI] [PubMed] [Google Scholar]
- Feng L., Xie Z.H., Ding Q., Xie X., Libby R.T., Gan L. MATH5 controls the acquisition of multiple retinal cell fates. Mol. Brain. 2010;3:36. doi: 10.1186/1756-6606-3-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueres-Oñate M., García-Marqués J., López-Mascaraque L. UbC-StarTrack, a clonal method to target the entire progeny of individual progenitors. Sci. Rep. 2016;6:33896. doi: 10.1038/srep33896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser M.J., Ciszczon T., Elick T., Bauser C. Precise excision of TTAA-specific lepidopteran transposons piggyBac (IFP2) and tagalong (TFP3) from the baculovirus genome in cell lines from two species of Lepidoptera. Insect Mol. Biol. 1996;5:141–151. doi: 10.1111/j.1365-2583.1996.tb00048.x. [DOI] [PubMed] [Google Scholar]
- Gao Y., Hisey E., Bradshaw T.W.A., Erata E., Brown W.E., Courtland J.L., Uezu A., Xiang Y., Diao Y., Soderling S.H. Plug-and-play protein modification using homology-independent universal genome engineering. Neuron. 2019;103:583–597.e8. doi: 10.1016/j.neuron.2019.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Moreno F., Vasistha N.A., Begbie J., Molnár Z. CLoNe is a new method to target single progenitors and study their progeny in mouse and chick. Development. 2014;141:1589–1598. doi: 10.1242/dev.105254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedhart J., von Stetten D., Noirclerc-Savoye M., Lelimousin M., Joosen L., Hink M.A., van Weeren L., Gadella T.W.J., Jr., Royant A. Structure-guided evolution of cyan fluorescent proteins towards a quantum yield of 93% Nat. Commun. 2012;3:751. doi: 10.1038/ncomms1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haapaniemi E., Botla S., Persson J., Schmierer B., Taipale J. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat. Med. 2018;24:927–930. doi: 10.1038/s41591-018-0049-z. [DOI] [PubMed] [Google Scholar]
- Hafler B.P., Surzenko N., Beier K.T., Punzo C., Trimarchi J.M., Kong J.H., Cepko C.L. Transcription factor Olig2 defines subpopulations of retinal progenitor cells biased toward specific cell fates. Proc. Natl. Acad. Sci. USA. 2012;109:7882–7887. doi: 10.1073/pnas.1203138109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hämmerle B., Ulin E., Guimera J., Becker W., Guillemot F., Tejedor F.J. Transient expression of Mnb/Dyrk1a couples cell cycle exit and differentiation of neuronal precursors by inducing p27KIP1 expression and suppressing NOTCH signaling. Development. 2011;138:2543–2554. doi: 10.1242/dev.066167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Zhang G., Almeida A.D., Cayouette M., Simons B.D., Harris W.A. How variable clones build an invariant retina. Neuron. 2012;75:786–798. doi: 10.1016/j.neuron.2012.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch T., Rothoeft T., Teig N., Bauer J.W., Pellegrini G., De Rosa L., Scaglione D., Reichelt J., Klausegger A., Kneisz D. Regeneration of the entire human epidermis using transgenic stem cells. Nature. 2017;551:327–332. doi: 10.1038/nature24487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holguera I., Desplan C. Neuronal specification in space and time. Science. 2018;362:176–180. doi: 10.1126/science.aas9435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue F., Kircher M., Martin B., Cooper G.M., Witten D.M., McManus M.T., Ahituv N., Shendure J. A systematic comparison reveals substantial differences in chromosomal versus episomal encoding of enhancer activity. Genome Res. 2017;27:38–52. doi: 10.1101/gr.212092.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivics Z., Li M.A., Mátés L., Boeke J.D., Nagy A., Bradley A., Izsvák Z. Transposon-mediated genome manipulation in vertebrates. Nat. Methods. 2009;6:415–422. doi: 10.1038/nmeth.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferis G.S.X.E., Livet J. Sparse and combinatorial neuron labelling. Curr. Opin. Neurobiol. 2012;22:101–110. doi: 10.1016/j.conb.2011.09.010. [DOI] [PubMed] [Google Scholar]
- Jullien N., Sampieri F., Enjalbert A., Herman J.P. Regulation of Cre recombinase by ligand-induced complementation of inactive fragments. Nucleic Acids Res. 2003;31:e131. doi: 10.1093/nar/gng131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C.J., Ménoret S., Brusselle L., Tesson L., Usal C., Chenouard V., Remy S., Ouisse L.H., Poirier N., Vanhove B. Comparative analysis of piggyBac, CRISPR/Cas9 and TALEN mediated BAC transgenesis in the zygote for the generation of humanized SIRPA rats. Sci. Rep. 2016;6:31455. doi: 10.1038/srep31455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K., Noda T. Transposition of the Tol2 element, an Ac-like element from the Japanese medaka fish Oryzias latipes, in mouse embryonic stem cells. Genetics. 2004;166:895–899. doi: 10.1534/genetics.166.2.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K., Largaespada D.A., Ivics Z. Transposons as tools for functional genomics in vertebrate models. Trends Genet. 2017;33:784–801. doi: 10.1016/j.tig.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klompe S.E., Vo P.L.H., Halpin-Healy T.S., Sternberg S.H. Transposon-encoded CRISPR-Cas systems direct RNA-guided DNA integration. Nature. 2019;571:219–225. doi: 10.1038/s41586-019-1323-z. [DOI] [PubMed] [Google Scholar]
- Kobiler O., Lipman Y., Therkelsen K., Daubechies I., Enquist L.W. Herpesviruses carrying a Brainbow cassette reveal replication and expression of limited numbers of incoming genomes. Nat. Commun. 2010;1:146. doi: 10.1038/ncomms1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T., Imamura K., Funayama M., Tsukita K., Miyake M., Ohta A., Woltjen K., Nakagawa M., Asada T., Arai T. iPSC-based compound screening and in vitro trials identify a synergistic anti-amyloid β combination for Alzheimer’s disease. Cell Rep. 2017;21:2304–2312. doi: 10.1016/j.celrep.2017.10.109. [DOI] [PubMed] [Google Scholar]
- Landrette S.F., Xu T. Somatic genetics empowers the mouse for modeling and interrogating developmental and disease processes. PLoS Genet. 2011;7:e1002110. doi: 10.1371/journal.pgen.1002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leber S.M., Sanes J.R. Migratory paths of neurons and glia in the embryonic chick spinal cord. J. Neurosci. 1995;15:1236–1248. doi: 10.1523/JNEUROSCI.15-02-01236.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhao X., Fang Y., Jiang X., Duong T., Fan C., Huang C.C., Kain S.R. Generation of destabilized green fluorescent protein as a transcription reporter. J. Biol. Chem. 1998;273:34970–34975. doi: 10.1074/jbc.273.52.34970. [DOI] [PubMed] [Google Scholar]
- Li M.A., Turner D.J., Ning Z., Yusa K., Liang Q., Eckert S., Rad L., Fitzgerald T.W., Craig N.L., Bradley A. Mobilization of giant piggyBac transposons in the mouse genome. Nucleic Acids Res. 2011;39:e148. doi: 10.1093/nar/gkr764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G., Zhang Y. Genetic and epigenetic variations in iPSCs: potential causes and implications for application. Cell Stem Cell. 2013;13:149–159. doi: 10.1016/j.stem.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loulier K., Barry R., Mahou P., Le Franc Y., Supatto W., Matho K.S.K.S., Ieng S., Fouquet S., Dupin E., Benosman R. Multiplex cell and lineage tracking with combinatorial labels. Neuron. 2014;81:505–520. doi: 10.1016/j.neuron.2013.12.016. [DOI] [PubMed] [Google Scholar]
- Lu I.L., Chen C., Tung C.Y., Chen H.H., Pan J.P., Chang C.H., Cheng J.S., Chen Y.A., Wang C.H., Huang C.W. Identification of genes associated with cortical malformation using a transposon-mediated somatic mutagenesis screen in mice. Nat. Commun. 2018;9:2498. doi: 10.1038/s41467-018-04880-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maury Y., Côme J., Piskorowski R.A., Salah-Mohellibi N., Chevaleyre V., Peschanski M., Martinat C., Nedelec S. Combinatorial analysis of developmental cues efficiently converts human pluripotent stem cells into multiple neuronal subtypes. Nat. Biotechnol. 2015;33:89–96. doi: 10.1038/nbt.3049. [DOI] [PubMed] [Google Scholar]
- Meir Y.J.J., Weirauch M.T., Yang H.S., Chung P.C., Yu R.K., Wu S.C.Y. Genome-wide target profiling of piggyBac and Tol2 in HEK 293: pros and cons for gene discovery and gene therapy. BMC Biotechnol. 2011;11:28. doi: 10.1186/1472-6750-11-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle F.T., Ghosh S., Kamitaki N., Mitchell J., Avior Y., Mello C., Kashin S., Mekhoubad S., Ilic D., Charlton M. Human pluripotent stem cells recurrently acquire and expand dominant negative P53 mutations. Nature. 2017;545:229–233. doi: 10.1038/nature22312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikuni T., Nishiyama J., Sun Y., Kamasawa N., Yasuda R. High-throughput, high-resolution mapping of protein localization in mammalian brain by in vivo genome editing. Cell. 2016;165:1803–1817. doi: 10.1016/j.cell.2016.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehme R., Zuccaro E., Ghosh S.D., Li C., Sherwood J.L., Pietilainen O., Barrett L.E., Limone F., Worringer K.A., Kommineni S. Combining NGN2 programming with developmental patterning generates human excitatory neurons with NMDAR-mediated synaptic transmission. Cell Rep. 2018;23:2509–2523. doi: 10.1016/j.celrep.2018.04.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama J., Mikuni T., Yasuda R. Virus-mediated genome editing via homology-directed repair in mitotic and postmitotic cells in mammalian brain. Neuron. 2017;96:755–768.e5. doi: 10.1016/j.neuron.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H., Yamamura K., Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Noctor S.C., Flint A.C., Weissman T.A., Dammerman R.S., Kriegstein A.R. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409:714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- Pierfelice T., Alberi L., Gaiano N. Notch in the vertebrate nervous system: an old dog with new tricks. Neuron. 2011;69:840–855. doi: 10.1016/j.neuron.2011.02.031. [DOI] [PubMed] [Google Scholar]
- Pontes-Quero S., Heredia L., Casquero-García V., Fernández-Chacón M., Luo W., Hermoso A., Bansal M., Garcia-Gonzalez I., Sanchez-Muñoz M.S., Perea J.R. Dual ifgMosaic: a versatile method for multispectral and combinatorial mosaic gene-function analysis. Cell. 2017;170:800–814.e18. doi: 10.1016/j.cell.2017.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J., Turner D., Cepko C. Lineage analysis in the vertebrate nervous system by retrovirus-mediated gene transfer. Proc. Natl. Acad. Sci. USA. 1987;84:156–160. doi: 10.1073/pnas.84.1.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querques I., Mades A., Zuliani C., Miskey C., Alb M., Grueso E., Machwirth M., Rausch T., Einsele H., Ivics Z. A highly soluble Sleeping Beauty transposase improves control of gene insertion. Nat. Biotechnol. 2019;37:1502–1512. doi: 10.1038/s41587-019-0291-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebsam A., Petros T.J., Mason C.A. Switching retinogeniculate axon laterality leads to normal targeting but abnormal eye-specific segregation that is activity dependent. J. Neurosci. 2009;29:14855–14863. doi: 10.1523/JNEUROSCI.3462-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios A.C., Serralbo O., Salgado D., Marcelle C. Neural crest regulates myogenesis through the transient activation of NOTCH. Nature. 2011;473:532–535. doi: 10.1038/nature09970. [DOI] [PubMed] [Google Scholar]
- Rompani S.B., Cepko C.L. Retinal progenitor cells can produce restricted subsets of horizontal cells. Proc. Natl. Acad. Sci. USA. 2008;105:192–197. doi: 10.1073/pnas.0709979104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schick E., McCaffery S.D., Keblish E.E., Thakurdin C., Emerson M.M. Lineage tracing analysis of cone photoreceptor associated cis-regulatory elements in the developing chicken retina. Sci. Rep. 2019;9:9358. doi: 10.1038/s41598-019-45750-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serralbo O., Picard C.A., Marcelle C. Long-term, inducible gene loss-of-function in the chicken embryo. Genesis. 2013;51:372–380. doi: 10.1002/dvg.22388. [DOI] [PubMed] [Google Scholar]
- Sharma B., Ho L., Ford G.H., Chen H.I., Goldstone A.B., Woo Y.J., Quertermous T., Reversade B., Red-Horse K., Chen H.I. Alternative progenitor cells compensate to rebuild the coronary vasculature in Elabela- and Apj-deficient hearts. Dev. Cell. 2017;42:655–666.e3. doi: 10.1016/j.devcel.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbakova D.M., Verkhusha V.V. Near-infrared fluorescent proteins for multicolor in vivo imaging. Nat. Methods. 2013;10:751–754. doi: 10.1038/nmeth.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowronska-Krawczyk D., Chiodini F., Ebeling M., Alliod C., Kundzewicz A., Castro D., Ballivet M., Guillemot F., Matter-Sadzinski L., Matter J.-M. Conserved regulatory sequences in Atoh7 mediate non-conserved regulatory responses in retina ontogenesis. Development. 2009;136:3767–3777. doi: 10.1242/dev.033449. [DOI] [PubMed] [Google Scholar]
- Stanger B.Z., Tanaka A.J., Melton D.A. Organ size is limited by the number of embryonic progenitor cells in the pancreas but not the liver. Nature. 2007;445:886–891. doi: 10.1038/nature05537. [DOI] [PubMed] [Google Scholar]
- Strecker J., Ladha A., Gardner Z., Schmid-Burgk J.L., Makarova K.S., Koonin E.V., Zhang F. RNA-guided DNA insertion with CRISPR-associated transposases. Science. 2019;365:48–53. doi: 10.1126/science.aax9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S.C., Bleckert A., Williams P.R., Takechi M., Kawamura S., Wong R.O.L. Cone photoreceptor types in zebrafish are generated by symmetric terminal divisions of dedicated precursors. Proc. Natl. Acad. Sci. USA. 2013;110:15109–15114. doi: 10.1073/pnas.1303551110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Tsunekawa Y., Hernandez-Benitez R., Wu J., Zhu J., Kim E.J., Hatanaka F., Yamamoto M., Araoka T., Li Z. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature. 2016;540:144–149. doi: 10.1038/nature20565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewary M., Shakiba N., Zandstra P.W. Stem cell bioengineering: building from stem cell biology. Nat. Rev. Genet. 2018;19:595–614. doi: 10.1038/s41576-018-0040-z. [DOI] [PubMed] [Google Scholar]
- Wang S.W., Kim B.S., Ding K., Wang H., Sun D., Johnson R.L., Klein W.H., Gan L. Requirement for math5 in the development of retinal ganglion cells. Genes Dev. 2001;15:24–29. doi: 10.1101/gad.855301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wang J., Devaraj A., Singh M., Jimenez Orgaz A., Chen J.X., Selbach M., Ivics Z., Izsvák Z. Suicidal autointegration of sleeping beauty and piggyBac transposons in eukaryotic cells. PLoS Genet. 2014;10:e1004103. doi: 10.1371/journal.pgen.1004103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman T.A., Pan Y.A. Brainbow: new resources and emerging biological applications for multicolor genetic labeling and analysis. Genetics. 2015;199:293–306. doi: 10.1534/genetics.114.172510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodworth M.B., Girskis K.M., Walsh C.A. Building a lineage from single cells: genetic techniques for cell lineage tracking. Nat. Rev. Genet. 2017;18:230–244. doi: 10.1038/nrg.2016.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N., Chanda S., Marro S., Ng Y.-H.Y.-H., Janas J.A., Haag D., Ang C.E., Tang Y., Flores Q., Mall M. Generation of pure GABAergic neurons by transcription factor programming. Nat. Methods. 2017;14:621–628. doi: 10.1038/nmeth.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Hu K., Smuga-Otto K., Tian S., Stewart R., Slukvin I.I., Thomson J.A. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y.C., He S., Chen S., Fu Y., Brown K.N., Yao X.H., Ma J., Gao K.P., Sosinsky G.E., Huang K., Shi S.H. Preferential electrical coupling regulates neocortical lineage-dependent microcircuit assembly. Nature. 2012;486:113–117. doi: 10.1038/nature10958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusa K., Zhou L., Li M.A., Bradley A., Craig N.L. A hyperactive piggyBac transposase for mammalian applications. Proc. Natl. Acad. Sci. USA. 2011;108:1531–1536. doi: 10.1073/pnas.1008322108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler K., Bui T., Frisque R.J., Grandinetti A., Nerurkar V.R. A rapid in vitro polyomavirus DNA replication assay. J. Virol. Methods. 2004;122:123–127. doi: 10.1016/j.jviromet.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Zong H., Espinosa J.S., Su H.H., Muzumdar M.D., Luo L. Mosaic analysis with double markers in mice. Cell. 2005;121:479–492. doi: 10.1016/j.cell.2005.02.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate new datasets or codes.