Abstract
The qualified presumption of safety (QPS) was developed to provide a safety pre‐assessment within EFSA for microorganisms. Strains belonging to QPS taxonomic units (TUs) still require an assessment based on a specific data package, but QPS status facilitates fast track evaluation. QPS TUs are unambiguously defined biological agents assessed for the body of knowledge, their safety and their end use. Safety concerns are, where possible, to be confirmed at strain or product level, and reflected as ‘qualifications’. Qualifications need to be evaluated at strain level by the respective EFSA units. The lowest QPS TU is the species level for bacteria, yeasts and protists/algae, and the family for viruses. The QPS concept is also applicable to genetically modified microorganisms used for production purposes if the recipient strain qualifies for the QPS status, and if the genetic modification does not indicate a concern. Based on the actual body of knowledge and/or an ambiguous taxonomic position, the following TUs were excluded from the QPS assessment: filamentous fungi, oomycetes, streptomycetes, Enterococcus faecium, Escherichia coli and bacteriophages. The list of QPS‐recommended biological agents was reviewed and updated in the current opinion and therefore now becomes the valid list. For this update, reports on the safety of previously assessed microorganisms, including bacteria, yeasts and viruses (the latter only when used for plant protection purposes) were reviewed, following an Extensive Literature Search strategy. All TUs previously recommended for 2016 QPS list had their status reconfirmed as well as their qualifications. The TUs related to the new notifications received since the 2016 QPS opinion was periodically evaluated for QPS status in the Statements of the BIOHAZ Panel, and the QPS list was also periodically updated. In total, 14 new TUs received a QPS status between 2017 and 2019: three yeasts, eight bacteria and three algae/protists.
Keywords: safety, QPS, food and feed, bacteria, yeast, fungi, virus, algae
Summary
The European Food Safety Authority (EFSA) asked the Panel on Biological Hazards (BIOHAZ) to deliver a Scientific Opinion on the maintenance of the list of Qualified Presumption of Safety (QPS) biological agents. The QPS assessment was developed to provide a safety pre‐evaluation of microbiological agents notified to EFSA in the frame of an application dossier to support the risk assessments performed by EFSA's scientific Panels and Units. Strains belonging to QPS taxonomic units (TUs) still require an assessment based on a specific data package, but QPS status facilitates a fast track evaluation. The workflow diagrams of the QPS process are presented in this Opinion. QPS TUs are unambiguously defined biological agents assessed for the body of knowledge on the organisms, their safety and their end use. Identified safety concerns are, where possible, to be confirmed at strain or product level, and reflected as ‘qualifications’. Qualifications of QPS microorganisms need to be evaluated at strain level using the information provided in the respective dossiers. The lowest TU for which the QPS status is granted is the species level for bacteria, yeasts and protists/algae, and the family level for viruses. The BIOHAZ Panel was requested to undertake care of three specific tasks as mentioned in the terms of reference (ToR):
The first ToR requires the regular updating of the list of microbial agents being notified, in the context of technical dossiers to EFSA Units, for intentional use in feed and/or food or as sources of food and feed additives, enzymes and plant protection products (PPPs) for safety assessment. The list ‘Microbial species as notified to EFSA’ (https://doi.org/10.5281/zenodo.3607184 Appendix D of this opinion) compiles all microorganisms notified to EFSA from the beginning of the QPS exercise in 2007. Between October 2016 and September 2019, 328 notifications were received and included. From these, 185 were for feed additives, 78 for food enzymes, food additives and flavourings, 25 for novel foods and 40 for PPPs; 198 were bacteria, 84 filamentous fungi, 4 viruses and 34 yeasts, 6 protists/algae.
The second ToR concerns the revision of the TUs previously recommended for the QPS list and their qualifications based on any new information that has become available. This task has been covered by each of the Panel Statements published from June 2017. The current opinion summarises the results of the six Panel Statements published/prepared since then. For this update, reports on the safety of previously assessed microorganisms, including bacteria, yeasts and viruses (the latter only when used for plant protection purposes) were reviewed following an Extensive Literature Search strategy for which the protocol can be found in https://doi.org/10.5281/zenodo.3607190 (Appendix B) and the Search strategies in https://doi.org/10.5281/zenodo.3607193 (Appendix C). The list of the biological agents for which QPS status is recommended, prior to safety risk assessments being carried out by EFSA, was reviewed and updated by the current opinion and therefore becomes the current valid list (‘2019 QPS list’) available at the Knowledge Junction in Zenodo (https://doi.org/10.5281/zenodo.1146566, Appendix A). All TUs previously recommended for the ‘2016 QPS list’ had their status reconfirmed, as well as their qualifications. Relevant information from the extensive literature searches (ELS) includes case reports of human diseases. Several of the QPS‐TUs (e.g. Bifidobacterium species, Lactobacillus and Saccharomyces boulardii cerevisiae) are sporadically reported as causing infections in individuals with conditions that are recognised as predisposing to the acquisition of opportunistic infections. Previous use of the microorganisms as food supplements for humans, which does not fall under the remit of the QPS assessment, was reported in many of these cases.
The third ToR requires a (re)assessment of the suitability of TUs, notified to EFSA, that are not present in the previous ‘2016 QPS list’, for their inclusion in the updated list. The TUs related to the new notifications received since the 2016 QPS opinion were periodically evaluated, and their resulting QPS status published in the six Panel Statements published/prepared since then, and the ‘2016 QPS list’ was periodically updated.
Between 2017 and 2019, a total of 51 TUs were (re)assessed, and 14 are recommended for QPS status: three yeasts, eight bacteria and three algae/protists; Lactobacillus animalis, Lactobacillus parafarraginis and Zygosaccharomyces rouxii are recommended for full QPS status; Euglena gracilis, Aurantiochytrium limacinum, Tetraselmis chuii, Corynebacterium ammoniagenes, Cupriavidus necator, Komagataeibacter sucrofermentans and Yarrowia lipolytica are recommended for QPS status with the qualification ‘for production purposes only’. Bacillus velezensis is recommended for QPS status with the qualification ‘absence of toxigenic potential and absence of aminoglycoside production ability’; Parageobacillus thermoglucosidasius and Paenibacillus illinoisensis are recommended for QPS status with the qualification ‘for production purposes only’ and ‘absence of toxigenic potential’; Komagatella phaffii is recommended for QPS status with the qualification ‘when the species is used for enzyme production’.
During the 3‐year period of this QPS mandate, some aspects in relation to the application of QPS in safety assessments were clarified:
Based on the actual body of knowledge and/or the ambiguous taxonomic position, the following TUs were excluded from the QPS assessment: filamentous fungi, oomycetes, streptomycetes, Enterococcus faecium, Escherichia coli and bacteriophages.
In the case of Genetically Modified Microorganisms (GMM) for which the species of the recipient strain qualifies for QPS status, and for which the genetically modified state does not give rise to safety concerns, the QPS approach can be extended to genetically modified production strains.
The qualification ‘for production purpose only’ implies the absence of viable cells of the production organism in the final product, and can also be applied to food and feed products based on microbial biomass.
The QPS status of Corynebacterium glutamicum was confirmed with the qualification ‘extended to other production purposes’.
For yeasts, acquired antimicrobial resistance (AMR) genes are not of relevance, but susceptibility to antimycotic compounds used in human medicine should be proved when yeasts are used as viable organisms in the food and feed chains.
1. Introduction
1.1. Background and Terms of Reference as provided by EFSA
A wide variety of microorganisms are intentionally added at different stages into the food and feed chain. In the context of applications for market authorisation of these biological agents used either directly or as sources of food and feed additives, food enzymes, novel foods and plant protection products, EFSA is requested to assess their safety.
Several taxonomic units (TU) (usually species for bacteria, yeasts and protists/algae, families for viruses) have been included in the qualified presumption of safety (QPS) list either following notifications to EFSA or proposals made initially by stakeholders during a public consultation in 2005, even if they were not notified to EFSA (EFSA, 2005).1 The EFSA Scientific Committee reviewed the range and numbers of microorganisms likely to be the subject of an EFSA Opinion and in 2007 published a list of microorganisms recommended for the QPS list.2
In 2007, the Scientific Committee recommended that a QPS approach should provide a generic concept to prioritise and to harmonise safety risk assessment of microorganisms intentionally introduced into the food chain, in support of the respective Scientific Panels and EFSA Units in the frame of the market authorisations. The same Committee recognised that there would have to be continuing provision for reviewing and modifying the QPS list and in line with this recommendation, the EFSA Panel on Biological Hazards (BIOHAZ) took the prime responsibility for this and started reviewing annually the existing QPS list. In 2008, the first annual QPS update3 was published and EFSA's initial experience in applying the QPS approach included. The potential application of the QPS approach to microbial plant protection products was discussed in the 2009 update.4 Also, in 2009, bacteriophages were assessed and were not considered appropriate for the QPS list. After consecutive years of reviewing the existing scientific information, the filamentous fungi (2008 to 2013 update) and enterococci (2010–2013 update) were not recommended for the QPS list. The 2013 update5 of the recommended QPS list included 53 species of Gram‐positive non‐sporulating bacteria, 13 Gram‐positive spore forming bacteria (Bacillus species), one Gram‐negative bacterium,6 13 yeast species, and 3 virus families.
In 2014, the BIOHAZ Panel in consultation with the Scientific Committee, decided to change the revision procedure: the overall assessment of the taxonomic units previously recommended for the QPS list is no longer carried out annually but over a 3‐year period. From 2017, the search and revision of the possible safety concerns linked to those taxonomic units start to be done every 6‐month period. The update of the 2013 QPS list version (EFSA BIOHAZ Panel, 2013) was done in 2016 (EFSA BIOHAZ Panel, 2017) and the next update is included in this scientific Opinion of the BIOHAZ Panel adopted in December 2019.7 The QPS list of microorganisms has been maintained and frequently checked, based on the evaluation of extensive literature searches. In the mean time, and every 6 months, a Panel Statement, compiling the assessments for a QPS status of the microbial agents notified to EFSA requested by the Feed Unit, the Food Ingredients and Packaging (FIP) Unit, the Nutrition Unit or the Pesticides Unit, has been produced and published. In the follow‐up of the 2013 update5, the Scientific Committee agreed to exclude some biological groups (filamentous fungi, bacteriophages and Enterococcus faecium)8 notified to EFSA from the QPS assessment because it was considered unlikely that any taxonomical units within these groups would be granted the QPS status in the foreseeable future. Thus, the assessment of members of these biological groups needs to be done at a strain level, on a case by case basis, by the relevant EFSA Unit.
The QPS provides a generic safety pre‐assessment approach for use within EFSA that covers risks for human, animals and the environment. In the QPS concept, a safety assessment of a defined taxonomic unit is considered independently of any particular specific notification in the course of an authorisation process. The QPS concept does not address hazards linked to the formulation or other processing of the products containing the microbial agents and added into the food or feed chain. Although general human safety is part of the evaluation, specific issues connected to type and level of exposure of users handling the product (e.g. dermal, inhalation, ingestion) are not addressed. In the case Genetically Modified Microorganisms (GMM) for which the species of the recipient strain qualifies for the QPS status, and for which the genetically modified state does not give rise to safety concerns, the QPS approach can be extended to genetically modified production strains (EFSA BIOHAZ Panel, 2018a).9 Assessment of potential allergenicity to microbial residual components is beyond the QPS remit, however, if there is science‐based evidence for some microbial species it is reported. Where applicable, these aspects are evaluated, separately by the EFSA Panel responsible for assessing the notification. Antimicrobial resistance was introduced as a possible safety concern for the assessment of the inclusion of bacterial species in the QPS list published in 2008 QPS Opinion (EFSA, 2008).3 In the 2009 QPS Opinion (EFSA BIOHAZ Panel, 2009)4 a qualification regarding absence of antimycotic resistance for yeasts was introduced.
The Terms of Reference, as provided by EFSA are as follows:
ToR 1: Keep updated the list of biological agents being notified in the context of a technical dossier to EFSA Units such as Feed, Pesticides, Food Ingredients and Packaging (FIP) and Nutrition, for intentional use directly or as sources of food and feed additives, food enzymes and plant protection product for safety assessment.
ToR 2: Review taxonomic units previously recommended for the QPS list and their qualifications when new information has become available. The latter is based on a review of the updated literature aiming at verifying if any new safety concern has arisen that could require the removal of the taxonomic unit from the list, and to verify if the qualifications still efficiently exclude safety concerns.
ToR 3: (Re) assess the suitability of new taxonomic units notified to EFSA for their inclusion in the QPS list. These microbial agents are notified to EFSA and requested by the Feed Unit, the FIP Unit, the Nutrition Unit or by the Pesticides Unit.
1.2. Interpretation of the Terms of Reference
1.2.1. QPS definition
A wide variety of microorganisms are intentionally used at different stages of the food chain and are risk assessed in several EFSA areas; e.g. feed, food, pesticides, nutrition, on the basis of an application dossier to the European Commission. The qualified presumption of safety (QPS) assessment was developed to provide a safety pre‐assessment of microorganisms to support the risk assessments performed by EFSA's scientific panels. The lowest taxonomic unit (TU) for which the QPS status is granted is the species level for bacteria, yeasts and protists/algae, and family for viruses. The safety of unambiguously defined biological TUs and the body of knowledge on safety aspects are assessed.
In the case that scientific knowledge identifies a specific, or more generally applicable, hazard related to a TU, e.g. acquired antimicrobial resistance, which can be confirmed at the strain or product level, a ‘qualification’ to exclude that hazard maybe established. The subject of these qualifications in the microbial strain under investigation is evaluated by the EFSA Unit to which the application dossier has been allocated. Microorganisms belonging to bacterial, yeast and protists/algae species or virus families in the QPS list are still submitted to a safety assessment based on the individual data package, although with fewer requirements. The data required in each application have to confirm the unambiguous identification of the organism and the confirmation that the qualifications are met.
The BIOHAZ Panel confirmed that in the case of a Genetically Modified Microorganism (GMM) for which the species of the recipient strain qualifies for the QPS status, and for which the genetically modified state does not give rise to safety concerns, the QPS approach can be extended to genetically modified production strains (EFSA BIOHAZ Panel, 2018a).
In June 2018 (EFSA BIOHAZ Panel, 2018b), the BIOHAZ Panel clarified that the qualification ‘for production purpose only’ implies the absence of viable cells of the production organism in the final product and can also be applied to food and feed products based on microbial biomass.
1.2.2. Notification of microorganisms for QPS assessment
A QPS assessment is triggered upon receipt by EFSA of an application dossier seeking a market authorisation of a regulated product that requires a safety assessment of a microbial strain. After the establishment of the first QPS list, no new starter organisms used for food fermentation have been included because they are not subject to a notification to EFSA for market authorisation. Because the QPS status list only considers those microorganisms sent to EFSA in the frame of notifications for market authorisation, it is not exhaustive.
1.2.3. Decision of exclusion from QPS assessment
Some microbial groups are excluded from the QPS assessment based on an ambiguous taxonomic position, lack of a sufficient body of knowledge or the possession of potentially harming traits (e.g. pathogenicity, presence of virulence factors or production of biologically active toxic secondary metabolites), because it is considered unlikely that any TUs within these groups would be granted QPS status in the foreseeable future. Thus, the assessment of members of these biological groups needs to be done at a strain level, on a case‐by‐case basis, by the relevant EFSA Unit:
Filamentous fungi
While knowledge of fungal secondary metabolites has grown substantially, information on their toxic effects on humans and animals is evolving at a much slower rate. Therefore, it was decided that until further notice, filamentous fungi would be excluded from the QPS evaluations.
Bacteriophages
Bacteriophages were not considered appropriate for consideration as QPS organisms because: (i) the lowest level of phylogenetic TU should be the order Caudovirales (which includes 95% of all known phages), which is considered to be too wide; (ii) distinguishing between transducing and non‐transducing phages or whether they carry virulence factor determinants (encoding toxins, adhesins, antibiotic resistance, etc.) would involve thorough analysis of the genomes and has to be done at the individual phage level.
Streptomycetes
Streptomycetes are essentially non‐virulent, with the exception of some plant pathogens such a S. scabies. Genome sequencing has confirmed that all streptomycetes carry gene clusters for the production of secondary metabolites, which include antimicrobial compounds, depressors of the immune system and herbicides (Butaye et al., 2003). Many of these may select for antimicrobial resistance or being toxic. The Panel therefore decided to exclude the genus Streptomyces from future QPS evaluations.
Oomycetes – Pythium oligandrum
Pythium oligandrum is an oomycete used in plant protection products. Several factors lie behind the decision that the oomycetes collectively (Class Oomycota) are not considered eligible for QPS status: (i) They seem to be rarely used as components of food and feed fermentations; (ii) There is lack of experience of actively adding oomycetes in the food and feed chains (except for a few biocontrol agents); (iii) a high proportion of the known oomycete species are pathogens, particularly to plants; (iv) there is little information available about the production of toxins by oomycetes in general (Stam et al., 2013; Berger et al., 2016; Amaro et al., 2017; Kushawa et al., 2017). The Panel therefore decided to exclude oomycetes from future QPS evaluations.
Enterococcus faecium
The inappropriateness of granting a safety status to the species E. faecium has been recognised in several EFSA Opinions (EFSA, 2007, 2008) due to the pathogenic potential of some strains, as defined by their possession of putative and confirmed virulence markers (Freitas et al., 2018).The Panel confirmed the exclusion of this species from future QPS evaluations.
Escherichia coli
Many strains of E. coli are pathogens for humans and animals. In fact, numerous strains of this species are the leading urinary and intestinal bacterial pathogens in developed and underdeveloped countries, respectively. In addition, they are frequently a cause of sepsis and many other systemic infections.
The Panel decided to exclude this species from future QPS evaluations.
1.2.4. Deliverables produced in response to the Terms of reference
ToR 1: The notifications considered for each Panel Statement (from December 2016 until December 2019) have been published in each respective Appendix. The previous list (published with the QPS 2016 update Opinion) has been updated with the corresponding notifications received between October 2016 and September 2019 (see Appendix D, https://doi.org/10.5281/zenodo.3607184).
ToR 2: If required, a Panel Statement would have been published with an explanation of the reason that led to the exclusion of a TU or the change of a qualification. At the same time, the QPS Opinion from 2016 would have been properly changed and an erratum included. The work being developed in order to reply to this ToR is reflected in the current opinion.
ToR 3: The current opinion takes into consideration the outcome of several Panel Statements published since December 2016. The notifications received within that period and the respective evaluation for a QPS status of the associated TU have been included in Appendix D, together with the previous notifications and respective evaluations. The new recommendations for QPS status have been included in the current QPS list (Appendix A, https://doi.org/10.5281/zenodo.1146566).
1.3. Additional information
1.3.1. Implementation of the QPS approach in EFSA risk assessment
The EFSA risk assessments for regulated products are based on an application dossier sent to the individual EFSA Panels/Units by the respective services of the European Commission, the Applicant or a Competent Authority in a Member State (Figure 1).
Figure 1.
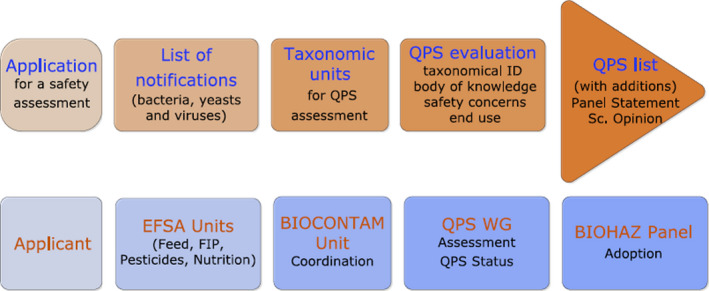
Workflow diagram describing how the QPS assessment is triggered by an application for market authorisation of a regulated product
The specific EFSA risk assessment areas involved in the assessment of regulated products that may involve the use of microorganisms are:
Feed additives safety assessment area
The EFSA Unit responsible for this area (Feed Unit) applies the QPS evaluation on the assessment of biological agents intended for use as feed additives or as a source of a feed additive, as defined in Regulation (EC) 1831/200310. The opinions of the EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP) dealing with QPS recommended microorganisms consider that no assessment of safety for the target species, consumer and the environment is required, provided that the microorganism is unambiguously identified and the existing qualifications are met. Since QPS assessments are made independently of the dose, the FEEDAP Panel concludes for QPS organisms that unless a specific provision relating to dose is included in the ‘qualification’ for a given TU, safety is presumed (EFSA FEEDAP Panel, 2018).
Pesticides safety assessment area
The EFSA Unit responsible for this area (Pesticide Peer Review Unit) organises the peer review of the microorganisms that are submitted for approval under Regulation (EC) No 1107/200911. In this regulation, data are required at strain level, including investigations of effects on human health and on non‐target organisms (in the environment), assessment of residues in or on treated crops, and information on the fate and behaviour of the plant protection product in the environment after application. The need for an environmental risk assessment is a generic qualification for all TUs evaluated for use as plant protection agents. Considering the extensive data requirements of the current legislation, the QPS evaluation will be limited to the safety evaluation of the exposure of humans and livestock to microorganisms or their residues (e.g. toxins) via food or feed. It is noted that non‐dietary exposure during or after the application of the plant protection product represents a set of situations not normally covered by the QPS assessment. In addition, environmental risk assessment as defined by the regulation cannot be considered to be covered by the QPS assessment, since the application of the organisms to agricultural or horticultural fields or protected cropping systems before harvest(s) triggers an assessment of risk for a variety of non‐target organisms covering a wide range of taxonomic and functional groups.
Microorganisms recommended for the QPS list and proposed as plant protection products under the Council Directive 91/414/EC12 were often exempted from certain data requirements, such as oral toxicity data. As an example, the QPS recommendation of the Baculoviridae family was used during the peer review of several species of baculoviruses (EFSA, 2012a,b).
Food Ingredients and Packaging safety assessment area
The EFSA Unit responsible for this area (FIP Unit) applies the QPS evaluation of those specific microbial TUs used for the production of food enzymes in agreement with the QPS approach that entered EU law with the publication of a Commission Implementing Regulation (EU) No 562/201213 amending Commission Regulation (EU) No 234/201114 with regard to specific data required for risk assessment of food enzymes. If the microorganism used in the production of a food enzyme qualifies for the QPS approach, the safety assessment of the food enzyme would not need specific toxicological test data. However, if residues, impurities or degradation products linked to the total food enzyme production process (production, recovery and purification) could give rise to concern, toxicological data may be requested. In the same legislative framework, the QPS approach can also be applied for the risk assessment of food flavourings and food additives produced from microbial sources, including genetically modified microorganisms where the parental strain fulfils the qualifications for QPS status (EFSA BIOHAZ Panel, 2018a).
Nutrition safety assessment area
The tasks of the Panel on Nutrition, Novel Foods and Food Allergens (NDA) include the safety assessment of novel foods (NF) that fall under Regulation (EU) 2015/2283.15 Novel foods (NF) are foods and food ingredients that have not been used for human consumption to a significant degree in the EU before 15 May 1997. Following this regulation, two different procedures are followed: 1. safety assessment of traditional foods (TF) from third countries based on a history of safe food use is requested by European Commission for comments to all the Member States (MS) and for safety assessment by EFSA in a time frame of 6 months; 2. all other applications are safety assessed by EFSA within 9 months.
If an NF (including TFs) consists of, contains, or is produced by, a microorganism which has been granted QPS status, the NDA Panel would not question the safety of that microorganism (regarding toxin production, infectivity or pathogenicity in general). Toxicological tests (e.g. genotoxicity and subchronic toxicological studies) may still be needed depending on the available data, not because of concerns about the microorganism used, but to address potential toxicity arising from other components (raw materials, reagents, residuals, contamination) and the production process. Also, assessments that result in QPS status not being granted have been shown to be useful for the NDA Panel, because in such cases, the NDA Panel gets useful information from the QPS outcome regarding reported infections, pathogenicity, toxin production etc.
QPS process general workflows
The QPS provides a generic safety pre‐assessment approach for use within EFSA. A QPS assessment is triggered by EFSA receiving an application dossier seeking a market authorisation of a regulated product that requires a safety assessment of a microbial strain.
Strains of TUs with QPS status still require an assessment based on the data included in the application sent to EFSA. Whenever foreseen by the specific regulatory framework, a fast track evaluation can be done with fewer requirements in relation to the risks that might be associated with the microorganism (see Figure 2). From 2014, the process includes the publication of a QPS Panel Statement every 6 months (Figure 3) and a QPS Opinion every 3 years (Figure 4). Because only those microorganisms sent to EFSA in the frame of notifications for market authorisation are considered for the QPS status, it is important to stress that the QPS list is not exhaustive.
Figure 2.
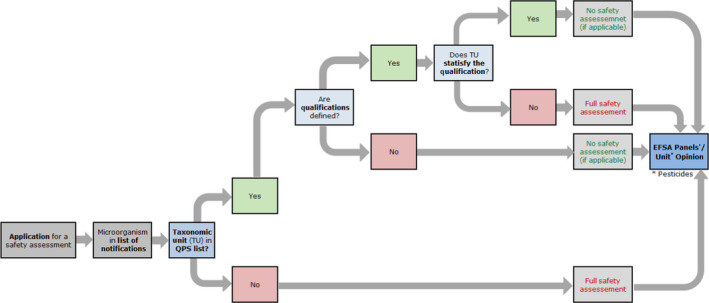
- QPS: Qualified Presumption of Safety.
Figure 3.
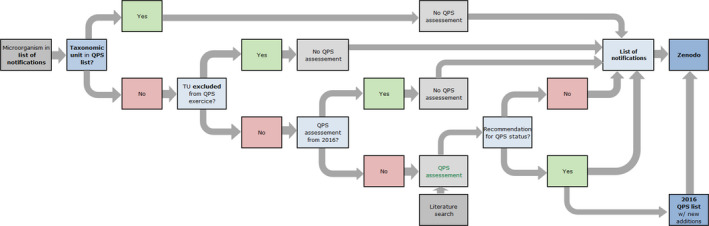
- BIOHAZ: Biological Hazards Panel; QPS: Qualified Presumption of Safety.
Figure 4.
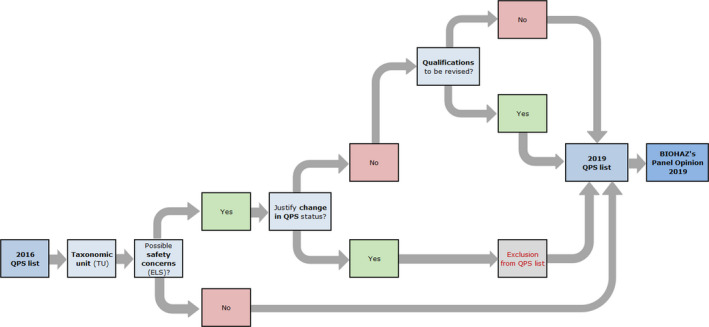
- BIOHAZ: Biological Hazards Panel; QPS: Qualified Presumption of Safety.
Each QPS Panel Statement contains the evaluations of the new notifications for microorganisms submitted for possible QPS status. It also contains a screening of the literature published during the previous 6‐month period concerning possible new safety concerns related to the TUs already included in the QPS list. The data identified in the literature are used to decide whether any TU may or may not remain in the QPS list, and whether any qualifications need to be revised.
Since 2016, the literature update has been performed by extensive literature searches (ELS). This Opinion contains an updated QPS list and summarises the main results of the 3‐year ELS on the QPS TUs, together with an update of the process for granting QPS Status.
The three flowcharts below illustrate how the QPS approach is incorporated in the EFSA safety assessment independently of the particularities for each specific risk assessment area:
The first flowchart (Figure 2) describes how EFSA Units incorporate the QPS status of a certain TU into the safety assessment process for a microorganism notified through an application for market authorisation. Possible qualifications of QPS microorganisms need to be evaluated by the EFSA Unit based on the information provided in the respective dossiers. The specific safety assessment is included in the EFSA Unit's Opinion and reference to the QPS status of the TU notified and eventual qualifications are included in that Opinion.
The second flowchart (Figure 3) describes how the evaluation of a newly notified TU, not in the QPS list, is included in each BIOHAZ Panel Statement. EFSA Units update the ‘List of notifications’ (Figure 1), and for each 6‐month period, includes them in an appendix of the ongoing Panel Statement. EFSA checks the respective TUs and chooses which are to be considered for the QPS status assessment. If a new TU receives a QPS recommendation (and possible qualifications), it is included in the valid QPS list.
The third flowchart (Figure 4) describes how the QPS list is maintained and the QPS Opinion is prepared. The QPS Opinion contains an update of the QPS list and the results of the 3‐year ELS on the existing QPS taxonomic units (TUs), together with an update of the QPS process.
1.3.2. Summary of the BIOHAZ Panel statements adopted between June 2017 and December 2019
In response to ToR1, the list of biological agents being notified to EFSA has been updated. Between October 2016 and September 2019, 328 notifications were received, of which 185 were for feed additives, 78 for food enzymes, food additives and flavourings, 25 for novel foods and 40 for PPP (see Table 2). With regard to the type of microorganisms, 196 were bacteria, 84 filamentous fungi, 1 oomyctes, 4 viruses, 34 yeasts, 6 algae/protists and 1 bacteriophage.
Table 2.
Notifications received by biological group and agent from October 2016 until September 2019, per Panel Statements (from part 6 to part 11)
| Panel StatementBiological group | 6 | 7 | 8 | 9 | 10 | 11 | Grand total |
|---|---|---|---|---|---|---|---|
| Algae | 2 | 4 | 6 | ||||
| Aurantiochytrium limacinum | 1 | 1 | |||||
| Euglena gracilis | 1 | 1 | |||||
| Phaeodactylum tricornutum | 1 | 1 | |||||
| Schizochytrium sp. | 2 | 2 | |||||
| Tetraselmis chuii | 1 | 1 | |||||
| Bacteria | 18 | 1 | 2 | 3 | 6 | 10 | 40 |
| Bacillus circulans | 3 | 3 | |||||
| Bacillus velezensis | 1 | 1 | |||||
| Burkholderia ubonensis | 1 | 1 | |||||
| Corynebacterium ammoniagenes | 1 | 1 | |||||
| Corynebacterium casei | 1 | 1 | |||||
| Corynebacterium stationis | 1 | 1 | |||||
| Cupriavidus necator | 1 | 1 | |||||
| Escherichia coli | 7 | 7 | |||||
| Parageobacillus thermoglucosidasius | 1 | 1 | |||||
| Gluconobacter frateurii | 1 | 1 | |||||
| Hyphomicrobium denitrificans | 1 | 1 | |||||
| Kitasatospora paracochleata | 1 | 1 | |||||
| Komagataeibacter sucrofermentans | 1 | 1 | |||||
| Lactobacillus animalis | 1 | 1 | |||||
| Lactobacillus parafarraginis | 1 | 1 | |||||
| Microbacterium foliorum | 1 | 1 | |||||
| Mycobacterium setense | 1 | 1 | |||||
| Paenibacillus lentus | 1 | 1 | |||||
| Paenibacillus illinoisensis | 1 | 1 | |||||
| Paracoccus carotinifaciens | 1 | 1 | |||||
| Protaminobacter rubrum | 1 | 1 | |||||
| Pseudomonas amyloderamosa | 1 | 1 | |||||
| Pseudomonas fluorescens | 1 | 1 | |||||
| Rhodococcus aetherovorans | 1 | 1 | |||||
| Rhodococcus ruber | 1 | 1 | |||||
| Sphingomonas elodea | 1 | 1 | |||||
| Pantoea ananatis | 1 | 1 | |||||
| Streptomyces cinnamonensis | 1 | 1 | |||||
| Streptomyces mobaerensis | 1 | 1 | |||||
| Streptomyces netropsis | 1 | 1 | |||||
| Streptomyces rubiginosus | 1 | 1 | |||||
| Streptomyces violaceoruber | 1 | 1 | |||||
| Yeast | 1 | 1 | 3 | 5 | |||
| Hamamotoa singularis | 1 | 1 | |||||
| Kodamaea ohmeri | 1 | 1 | |||||
| Komagatella phaffii | 1 | 1 | |||||
| Yarrowia lipolytica | 1 | 1 | |||||
| Zygosaccharomyces rouxii | 1 | 1 | |||||
| Grand Total | 18 | 2 | 3 | 3 | 8 | 17 | 51 |
In response to ToR3, these biological agents were (re)assessed for their suitability for inclusion in the updated QPS list. From the 328 notifications, 146 biological agents already had a QPS status and were not further evaluated, nor were other 131 notifications: 84 filamentous fungi and 4 of Enterococcus faecium, which were excluded from QPS consideration following a recommendation of the QPS 2013 update (EFSA BIOHAZ Panel, 2013). Similarly, no assessments were triggered by the 27 notifications of E. coli (bacterium) and 4 of Streptomyces spp., excluded in the Panel Statement adopted in December 2016 (EFSA BIOHAZ Panel, 2017), 1 notification of a bacteriophage, excluded in the Panel Statement adopted in December 2017 (EFSA BIOHAZ Panel, 2018a) and Sphingomonas paucimobilis which has already been evaluated in a previous Panel Statement (EFSA BIOPHAZ Panel, 2019a). Furthermore, it was agreed not to include 10 notifications from Pesticides Unit as the respective dossiers (including the literature review) had not been received (8 of Bacillus thuringiensis, 1 of Pseudomonas sp. and 1 of an Oomycetes). The remaining 51 notifications were considered for the assessment of the suitability of the respective TUs for inclusion in the QPS list. From these 51, 40 were bacteria, 5 were yeasts and 6 were protists/algae (see Table 2).
The assessment of the respective TUs was published in six Panel statements, adopted every 6 months between June 2017 and December 2019.
Table 1.
Notifications received by type of risk assessment area and by biological group from October 2016 until September 2019 (included in one of six Panel Statements, from part 6 to part 11)
| Risk assessment area | Not evaluated | Evaluated | Total | |
|---|---|---|---|---|
| Biological group | Already QPS | Excluded in QPS | ||
| Feed additives | 115 | 53 | 17 | 185 |
| Bacteria | 90 | 21 | 16 | 127 |
| Bacteriophages | 0 | 1 | 0 | 1 |
| Filamentous fungi | 0 | 31 | 0 | 31 |
| Yeasts | 25 | 0 | 1 | 26 |
| Novel foods | 4 | 9 | 12 | 25 |
| Algae | 0 | 0 | 6 | 6 |
| Bacteria | 1 | 8 | 5 | 14 |
| Filamentous fungi | 0 | 1 | 0 | 1 |
| Yeasts | 3 | 0 | 1 | 4 |
| Plant protection products | 9 | 30 | 1 | 40 |
| Bacteria | 5 | 10 | 1 | 16 |
| Filamentous fungi | 0 | 19 | 0 | 19 |
| Oomycetes | 0 | 1 | 0 | 1 |
| Viruses | 4 | 0 | 0 | 4 |
| Food enzymes, food additives and flavourings | 18 | 39 | 21 | 78 |
| Bacteria | 17 | 6 | 18 | 41 |
| Yeasts | 0 | 31 | 3 | 34 |
| Filamentous fungi | 1 | 2 | 0 | 3 |
| Total | 146 | 131 | 51 | 328 |
In total, 14 new TUs received a QPS status between 2017 and 2019: 3 yeasts, 8 bacteria and 3 algae/protists (Table 3).
Table 3.
New QPS recommendations per TU group and TU, for notifications received from October 2016 until September 2019
| TU group/TU | QPS status | Qualification |
|---|---|---|
| Protists/Algae | ||
| Euglena gracilis | Yes | For production purposes only |
| Aurantiochytrium limacinum | Yes | For production purposes only |
| Tetraselmis chuii | Yes | For production purposes only |
| Bacteria | ||
| Bacillus velezensis | Yes | Absence of toxigenic potential and absence of aminoglycoside production ability |
| Corynebacterium ammoniagenes | Yes | For production purposes only |
| Cupriavidus necator | Yes | For production purposes only |
| Parageobacillus thermoglucosidasius | Yes | For production purposes only and absence of toxigenic potential |
| Komagataeibacter sucrofermentans | Yes | For production purposes only |
| Lactobacillus animalis | Yes | |
| Lactobacillus parafarraginis | Yes | |
| Paenibacillus illinoisensis | Yes | For production purposes only and absence of toxigenic potential |
| Yeast | ||
| Komagatella phaffii | Yes | When the species is used for enzyme production |
| Yarrowia lipolytica | Yes | Qualification for production purposes only |
| Zygosaccharomyces rouxii | Yes | |
2. Data and methodologies
In reply to ToR 2, concerning the revision of the TUs previously recommended for the QPS list and their qualifications, an extensive literature search (ELS) was conducted as described in Section 2.2.1 and Appendices B and C.
In reply to ToR 3, (re)assessment of the suitability of TUs notified within the time period covered by this QPS mandate (from October 2016 until September 2019) was carried out. Relevant databases such as PubMed, Web of Science, CasesDatabase, CAB Abstracts or Food Science Technology Abstracts (FSTA) and Scopus were searched for possible new safety concerns. For evaluations of new TUs, details on the search strategy, search keys and approach followed are described in each Panel Statement.
2.1. Data
The QPS assessment is carried out considering the following pillars:
Taxonomic aspects;
Body of knowledge;
Safety concerns in relation to virulence/pathogenicity;
Safety for the environment.
2.1.1. Taxonomic identification
The TU for which the QPS status is granted is the species for bacteria, yeast and protists/algae, and the family for viruses. Only unambiguously defined biological TUs are considered for inclusion in the QPS list. Microbial taxonomy is a very dynamic discipline, recently supported mainly by phylogenetic analysis of housekeeping genes and whole genome relatedness (e.g. ANI, phylogenomics). The resulting reclassifications of microorganisms will lead to necessary adaptations in the QPS list, which are updated in the successive QPS Statements.
Bacterial taxonomy
Taxonomic identity is based on the internationally accepted classification, overseen by the International Committee on Systematics of Prokaryotes. The nomenclature of bacteria and the nomenclatural changes as cited in the Approved Lists of Bacterial Names or validly published in the International Journal of Systematic Bacteriology or in the International Journal of Systematic and Evolutionary Microbiology are reported in the website List of Prokaryotic Names with Standing in Nomenclature (LPSN) (Parte, 2018).
Fungal taxonomy
The nomenclature and taxonomy of fungi, including yeasts, is covered by the International Code of Nomenclature for algae, fungi and plants (ICN) (Turland et al., 2018). The most recent authoritative taxonomy of yeasts was published in 2011 (Kurtzman et al., 2011).
Virus taxonomy
The taxonomy and nomenclature of viruses are the responsibility of the International Committee on Taxonomy of Viruses (ICTV, 2018). Updates are made annually, based on proposals by Study Groups and after adoption by the Executive Committee. These updates form the 10th Report of the ICTV and are available through the ICTV website (https://talk.ictvonline.org/taxonomy/vmr/). The most recent update is from 2018 (ICTV, 2018). Two orders and 20 families of plant viruses have been recognised by the ICTV (ICTV, 2018). Two families (Alphaflexviridae, 49 species and Potyviridae, 160 species) and one insect virus family (Baculoviridae, 55 species) contain viruses notified to EFSA.
A species is the lowest taxon recognised by the ICTV and is based on a consensus sequence of a mixture of genotypes. An isolate or strain is a mixture of genotypes with certain biological characteristics. In the case of baculoviruses, a species is based on a consensus sequence with > 95% sequence homology (Wennmann et al., 2019).
Protists/Algae taxonomy
For protists/algae taxonomy, the Catalogue of Life (https://www.catalogueoflife.org) and the Global Biodiversity Information Facility (https://www.gbif.org) are used as basis for the assessment.
2.1.2. Body of Knowledge
The body of knowledge is one of the pillars of the QPS evaluation and is investigated based on the scientific literature. This includes peer‐reviewed papers published in journals and books that appear in scientific literature databases. To evaluate if the body of knowledge is sufficient to grant a TU, the QPS status several aspects are taken into account, such as the amount of available scientific knowledge indicating a certain degree of exposure of humans and animals through food and feed use.
Aspects on the ecology of the organism are also taken into account. This includes the distribution of the TU in natural environments (e.g. in the gut of humans, wild and farmed animals, and in the plant ecosystem) and their colonisation ability and routes for dispersal. The body of knowledge includes also the history of use of a TU in the agro‐food chain or in other sectors (e.g. biotechnological or medical applications). For this, information on the direct use of viable cells (e.g. as feed additives, food starter cultures, novel foods, probiotic or plant protection products), the use for production purposes (e.g. production of amino acids, biomass, enzymes, vitamins and polysaccharides) or its use in biotechnological or medical applications is examined. When detection in food or feed microbial community is reported, its presence as spontaneous contaminant vs. as main fermentative agent is considered.
2.1.3. Safety concerns in relation to pathogenicity and virulence
TUs assessed for the QPS list should not represent a hazard to human and animal health when used in the food or feed chain.
Relevant information includes case reports of human diseases, particularly infections or human intoxications linked to the TU under assessment. Additional important information is whether any negative impacts are confined to affected persons with conditions favouring opportunistic infections, for example, immunosuppression, and whether transmission occurred through food or other routes (e.g. medical devices). Studies indicating the presence of virulence factors (e.g. toxins and enzymes that may contribute to the pathogenicity of the microorganism) in the TU are also relevant for identification of potential safety concerns.
Several of the QPS‐TUs (e.g. Bifidobacterium species, Lactobacillus and Saccharomyces boulardii cerevisiae) are sporadially reported as causing infections in individuals with recognised predisposing conditions for the acquisition of opportunistic infections e.g. cardiovascular conditions favouring endocarditis, populations in the extreme lower or upper age spectrum or other conditions which can lead to impairment of the immunological system, such as patients submitted to transplants, undergoing cancer therapy, with physical trauma or tissue damage, or HIV patients. Moreover, gastrointestinal tract‐related conditions with mucosal impairment can also be predisposing factors for infections. Previous use of the microorganisms as food supplements for humans was reported in many of these cases. The living microorganism used as a food supplement does not fall under the remit of the QPS assessment. Nevertheless, QPS assessment will continue to take into consideration these reports, extracting relevant information whenever justified.
Assessment of allergenicity to microbial residual components is beyond the QPS assessment remit; nevertheless, if there is science‐based evidence for some microbial species related to well‐defined clinical cases, this is taken into consideration. Although general human safety is part of the evaluation, specific issues connected to exposure of users handling the product (e.g. dermal, inhalation, ingestion) are not addressed.
Reports of infection, intoxication or other diseases caused by the assessed TU on livestock domesticated and wild animals are also a relevant set of information for identifying potential safety concerns. As with safety concerns for humans, whether diseases are acquired through exposure via feed or other routes (e.g. wounds, inhalation) is also relevant information.
2.1.4. Safety for the environment
The assessment of environmental safety considers information on the natural presence of the TU in the microbiota of humans and animals, and the wider environment, and if its use is expected to pose additional risks to these different environments.
2.1.5. Qualifications
In the case that scientific knowledge identifies a specific safety concern related to a TU or more generally applicable to a group of TUs, which can be tested at the strain or product level, a ‘qualification’ to exclude that safety concern maybe established.
Generic qualification on antimicrobial resistance for bacteria
The absence of acquired genes coding for resistance to antimicrobials relevant for humans and animals in QPS recommended bacterial TUs is a generic qualification. The verification that a specific bacterial strain, notified to a certain Panel, fulfils the qualification of the absence of acquired antimicrobial resistance (AMR) genes is conducted by the specific EFSA Unit/Panel to which the notification was assigned. Within the framework of EFSA activities, the use of interpretative criteria and methods to define and monitor AMR have been harmonised and are reflected in EFSA's guidance documents (e.g. EFSA FEEDAP Panel, 2018).
Generic qualification on antimycotic resistance for viable yeasts
For yeasts, acquired AMR genes are not of relevance, but susceptibility to antimycotic compounds used in human medicine should be proved if the yeasts are to be used as viable organisms in the food and feed chains.
In order to better understand the prevalence and impact of antimycotic resistance of yeasts used in the feed chain, as well as on the mechanisms and genetic processes associated with it, a special investigation was undertaken, including consultation with a hearing expert in the field, that confirmed the usefulness of the previous qualification.
One main difference from bacteria is that in fungi, horizontal gene transfer between strains or species is not considered to play a significant role for development and transmission of antimycotic resistance. Resistance in fungi typically evolves due to mutations caused by exposure to antifungals in the patient or the environment (Perlin et al., 2016; Morio et al., 2017). For QPS yeasts occasionally reported to occur in opportunistic infections (D. hansenii, K. marxianus, L. jadinii, S. cerevisiae, W. anomalus), there are occasional reports of resistance or reduced susceptibility to antimycotics (see sections on the specific species below).
The verification that a specific yeast strain, used as a viable organism in the food or feed chains, fulfils the qualification of the absence of antimycotic resistance has to be conducted by the specific EFSA Unit/Panel to which the notification was assigned.
Qualification on toxigenic potential for Bacillus spp.
Several Bacillus species are included in the QPS list with the qualification ‘absence of toxigenic activity’. This is based on the observation that some strains among the Bacillus species on the QPS list have caused food‐borne disease in the past. Technical guidance to identify toxic compounds among Bacillus species has been elaborated and updated by EFSA (EFSA FEEDAP Panel, 2018). The application of the qualification should permit identification of this safety concern among strains of the QPS Bacillus species. It is the purpose of the regular update of the QPS list to verify that no other relevant safety concerns have been identified for the QPS species of Bacillus.
Qualification for production purposes only
The qualification ‘for production purpose only’ applies to TUs used for the biosynthesis of specific products for the food chain and subject to a specific authorisation (e.g. feed additives – vitamins, amino acids, polysaccharides and enzymes – and food processing enzymes). For most of the TUs used for production, data are lacking on the direct exposure of humans and animals, while there is a long history of use of their fermentation products in the food chain. This qualification implies the absence of viable production organisms in the final product and is also applicable to food products based on the non‐viable biomass of the microorganism (EFSA BIOHAZ Panel, 2018b).
Generic qualification for environmental risk assessment of microbial plant protection products
In Regulation (EC) No 1107/2009.5 data requirements are described that applicants must address in relation to the environmental risk assessment. The need for this environmental risk assessment is indicated in the QPS evaluation as a qualification ‘environmental risk assessment for use as microbial plant protection product at strain level following the requirements of the current legislation’. This qualification is generic for all microorganisms applied as microbial plant protection products.
2.2. Methodologies
2.2.1. Review of the scientific literature
The aim of the Extensive Literature Search (ELS) carried out in response to ToR 2 (review of the recommendations for the QPS list and specific qualifications) was to identify any publicly available studies reporting on safety concerns for humans, animals or the environment caused by organisms that have QPS status, since the publication of the previous QPS review in 2016 (i.e. publications from June 2016 to June 2019). For a detailed protocol of the process and search strategies, refer to Appendices B and C.
Within this time frame, a total of five ELS exercises have been run, with searches made for the following periods of publication
From June 2016 to June 2017
From July to December 2017
From January to June 2018
From July to December 2018
From January to June 2019
The summary of the results obtained for the data retrieved for each of these periods was published within the respective Panel Statement:
Panel Statement part 6/7
Panel Statement part 8
Panel Statement part 9
Panel Statement part 10
Panel Statement part 11
After removal of duplicates, records were submitted to the title screening step, which led to the exclusion of 18,618 of them. The remaining 1,608 records were found eligible for the Title and abstract screening step, which led to the exclusion of 704 of these. Of the 904 articles that finally reached the Article evaluation step (full text), 241 were considered to be relevant for the QPS project. The flow of records from their identification by the different search strategies (as reported in Appendix C) to their consideration as potentially relevant papers for QPS, is shown in Table 4.
Table 4.
Flow of records by search strategy, per taxonomic unit and per BIOHAZ Panel Statement
| Species | Title screening step | Title/abstract screening step | Article evaluation step (screening for potential relevance)(a) | Article evaluation step (identification of potential safety concerns)(b) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No papers | Stat 7 | Stat 8 | Stat 9 | Stat 10 | Stat 11 | Total | Stat 7 | Stat 8 | Stat 9 | Stat 10 | Stat 11 | Total | Stat 7 | Stat 8 | Stat 9 | Stat 10 | Stat 11 | Total | Stat 7 | Stat 8 | Stat 9 | Stat 10 | Stat 11 | Total |
| Bacteria | 3,977 | 2,592 | 1,750 | 2,244 | 1,833 | 12,396 | 445 | 65 | 57 | 258 | 65 | 890 | 82 | 29 | 23 | 224 | 31 | 389 | 41 | 16 | 14 | 11 | 14 | 96 |
| Bacillus spp. | 1,325 | 1,107 | 537 | 804 | 183 | 3,956 | 54 | 16 | 3 | 199 | 15 | 287 | 16 | 8 | 2 | 199 | 9 | 234 | 2 | 5 | 1 | 2 | 6 | 16 |
| Bifidobacterium spp. | 347 | 204 | 168 | 206 | 270 | 1,195 | 167 | 11 | 17 | 21 | 14 | 230 | 17 | 2 | 6 | 3 | 6 | 34 | 12 | – | 1 | 1 | 1 | 15 |
| Carnobacterium divergens | – | – | – | – | – | 1 | – | – | – | – | 1 | |||||||||||||
| Corynebacterium glutamicum | 73 | 42 | 47 | 45 | 39 | 246 | 8 | 3 | – | – | – | 11 | 1 | – | – | – | – | 1 | 1 | – | – | – | – | 1 |
| Gluconobacter oxydans | 394 | 155 | 132 | 115 | 164 | 960 | 2 | 1 | 2 | 1 | 1 | 7 | – | – | – | – | – | – | – | – | – | – | – | – |
| Xanthomonas campestris | – | 1 | – | – | 1 | – | – | – | – | – | – | |||||||||||||
| Lactobacillus spp. | 874 | 565 | 426 | 555 | 620 | 3,040 | 154 | 14 | 13 | 22 | 23 | 226 | 32 | 6 | 7 | 12 | 8 | 65 | 13 | 6 | 7 | 3 | 2 | 31 |
| Lactococcus lactis | 316 | 152 | 152 | 173 | 165 | 958 | 9 | 3 | 9 | 6 | 2 | 29 | 5 | 2 | 4 | 4 | 2 | 17 | 3 | – | 4 | 3 | – | 10 |
| Leuconostoc spp. | 151 | 62 | 44 | 68 | 81 | 406 | 18 | 8 | 9 | 5 | 2 | 42 | 6 | 5 | 3 | 4 | 1 | 19 | 5 | 4 | 1 | 1 | 1 | 12 |
| Microbacterium imperiale | – | – | – | – | – | – | – | – | – | – | – | |||||||||||||
| Oenococcus oeni | 78 | 37 | 43 | 24 | 39 | 221 | 9 | – | – | 1 | 1 | 11 | – | – | – | – | 1 | 1 | – | – | – | – | 1 | 1 |
| Pasteuria nishizawae | – | – | – | – | – | – | – | – | – | – | – | |||||||||||||
| Pediococcus spp. | 245 | 137 | 126 | 146 | 166 | 820 | 10 | 4 | 1 | 2 | 3 | 20 | 1 | 3 | – | 2 | 2 | 8 | 1 | – | – | 1 | 1 | 3 |
| Propionibacterium spp. | 64 | 49 | 30 | 27 | 27 | 197 | 1 | 1 | 1 | – | – | 3 | 1 | – | – | – | – | 1 | 1 | – | – | – | – | 1 |
| Streptococcus thermophilus | 110 | 82 | 45 | 81 | 79 | 397 | 13 | 4 | 2 | 1 | 4 | 24 | 3 | 3 | – | – | 2 | 8 | 2 | 1 | – | – | 2 | 5 |
| Viruses | 193 | 98 | 74 | 108 | 99 | 572 | 15 | 5 | 1 | – | 5 | 26 | – | 3 | – | – | 5 | 8 | – | – | – | – | – | – |
| Alphaflexiviridae | 57 | 28 | 26 | 39 | 41 | 191 | 5 | – | – | – | 2 | 7 | – | – | – | – | 2 | 2 | – | – | – | – | – | – |
| Baculoviridae | 136 | 70 | 48 | 69 | 58 | 381 | 10 | 5 | 1 | – | 3 | 19 | – | 3 | – | – | 3 | 6 | – | – | – | – | – | – |
| Yeasts | 2,833 | 1,500 | 1,210 | 1,358 | 357 | 7,258 | 240 | 72 | 58 | 273 | 49 | 692 | 121 | 52 | 35 | 268 | 31 | 507 | 42 | 31 | 21 | 30 | 21 | 145 |
| Debaryomyces hansenii (anamorph=Candida famata) | 2,833 | 1,500 | 1,210 | 1,358 | 357 | 7,258 | 240 | 72 | 58 | 273 | 49 | 692 | 121 | 52 | 35 | 268 | 31 | 507 | 9 | 7 | 5 | 10 | 5 | 36 |
| Kluyveromyces marxianus (anamorph= Candida kefyr) | 15 | 15 | 13 | 16 | 13 | 72 | ||||||||||||||||||
| Candida pelliculosa (synonymus = Pichia anomala, teleomorph = Wickerhamomyces anomalus) | 4 | 6 | 2 | 6 | – | 18 | ||||||||||||||||||
| Candida utilis (teleomorph = Lindnera jadinii) | 3 | – | – | 1 | – | 4 | ||||||||||||||||||
| Hanseniaspora uvarum | – | – | 1 | 1 | – | 2 | ||||||||||||||||||
| Saccharomyces cerevisiae including Saccharomyces boulardii | 13 | 9 | 5 | 8 | 7 | 42 | ||||||||||||||||||
| Kluyveromyces lactis (anamorph=Candida spherica) | – | – | – | 1 | 1 | 2 | ||||||||||||||||||
| Schizosaccharomyces pombe | – | – | – | 1 | – | 1 | ||||||||||||||||||
| Total | 7,003 | 4,190 | 3,034 | 3,710 | 2,289 | 20,226 | 700 | 142 | 116 | 531 | 119 | 1,608 | 203 | 84 | 58 | 492 | 67 | 904 | 83 | 47 | 35 | 41 | 35 | 241 |
| Excluded | 6,303 | 4,048 | 2,918 | 3,179 | 2,170 | 18,618 | 497 | 58 | 58 | 39 | 52 | 704 | 120 | 37 | 23 | 451 | 31 | 662 | ||||||
Full text phase. To be excluded: references not in English, without a full text PDF, not dealing with the TU or without any safety concern described.
Reference with a safety concern described. To be excluded: references with a methodological problem (related to identity confirmation method, reliability of source attribution, misuse of microorganims, predisposing factors in exposed subjects).
2.2.2. Consultation on antimycotic resistance
In order to better understand the prevalence and impact of antimycotic resistance of yeasts used in the feed chain, as well as on the mechanisms and genetic processes associated with it, a special investigation (by consultation with a hearing expert in the field) was made that confirmed the usefulness of the previous qualification.
3. Assessment
The following section includes the re‐evaluation of the TUs included in the QPS list published in the 2016 QPS Opinion (ToR2), and the assessment of the new TUs corresponding to the microorganisms notified to EFSA under the frame of an application for market authorisation for a possible inclusion in the list (ToR3). The QPS approach has been applied in the same way and based on the four main pillars in reply to both ToRs, as described in Section 2.1.
The TUs included in the 2016 QPS list were re‐evaluated periodically and published in each of the Panel Statements adopted between June 2017 and December 2019. As explained in Section 2.2.1, this assessment was based on ELS.
For the (re)evaluation of the TUs corresponding to the microorganisms notified to EFSA between October 2016 and September of 2019, the available literature databases were searched for possible safety concerns and analysed according to experts’ knowledge and the outcomes have been included in the respective Panel Statements. These TUs, newly added to the QPS list, were not included in the ELS revisions as only the ones that were already present in the 2016 QPS opinion were considered for this exercise.
The results of both evaluations are summarised below.
3.1. Gram‐positive non‐sporulating bacteria
3.1.1. QPS Bifidobacterium species and Carnobacterium divergens
A search for papers potentially relevant for the QPS consideration of Bifidobacterium spp. and Carnobacterium divergens provided 1,195 references. The analysis of their titles left 230 articles; 34 articles were found relevant at the level of screening for potential relevance. After screening the entire papers, 14 of them were finally discarded because they did not deal with safety concerns, did not concern this TU or were not in English. Fifteen papers were analysed in detail for the potential safety concern identified only for QPS Bifidobacterium.
Bifidobacterium species with QPS status
A total of 15 papers for QPS Bifidobacterium species were selected for further analysis. From these, 10 were excluded because no safety concerns were identified, or methodological shortcomings were found. The five remaining articles were considered for further analysis. In four of these articles, the cases reported were presenting severe underlying conditions (cancer, preterm infants with health problems, alcoholic cirrhosis, etc.) predisposing them to infections by some QPS Bifidobacterium spp. (B. longum, B. breve, etc.) (Esaiassen et al., 2016, 2017; Sato et al., 2016; Wilson and Ong, 2017). The paper of Martinez et al. (2018) was kept in the final analysis because the authors characterised a novel gene conferring erythromycin and clindamycin resistance that should be verified at the strain level of QPS Bifidobacterium spp.
All safety concerns were linked to patients with severe underlying health conditions or to an immunocompromised status, and therefore, the QPS status of Bifidobacterium species on the list is not changed.
Carnobacterium divergens
Only one article arrived to the final stage but does not describe a safety concern related to Carnobacterium divergens, so no new safety concern was found. Consequently, the QPS status of C. divergens is not changed.
3.1.2. Corynebacterium ammoniagenes
This is a new taxonomic unit evaluated from notifications received from October 2016 and now included in the QPS list. The evaluation was published in a recent BIOHAZ Panel Statement (EFSA BIOPHAZ Panel, 2019b).
Corynebacterium ammoniagenes is a species with standing in nomenclature. It was first described by Cooke and Keith (1927) as Brevibacterium ammoniagenes, a urea‐splitting bacterium isolated from the human intestinal tract. It was transferred to the genus Corynebacterium by Collins (1987).
C. ammoniagenes is used for the industrial production of nucleotides, nucleosides and riboflavin (Koizumi et al., 2000; Serrano et al., 2017). C. ammoniagenes derived single‐cell protein can also be used as a non‐conventional protein source in animal diets, such as for broilers (An et al., 2018) and growing pigs (Wang et al., 2013), without any negative effects on blood, bone characteristics or meat quality (An et al., 2018).
No information was found in relation to pathogenicity of the organism, including when it was used as single cell protein for feed (Oliveira et al., 2017).
C. ammoniagenes can be recommended for the QPS list with the qualification ‘for production purposes only’.
3.1.3. Corynebacterium glutamicum
A search for papers potentially relevant for the QPS consideration of Corynebacterium glutamicum provided 246 references. The analysis of their titles left 11 articles. Only one paper reached the final selection phase (full text), but it did not deal with any safety concerns, so the QPS status of C. glutamicum is not changed.
In parallel to the standard procedure for assessing a TU for a possible QPS status and for the maintenance of a QPS status, it was decided to run a complementary reassessment for another specific end use of this TU as the QPS qualification ‘only applies when the species is used for amino acid production’ (EFSA BIOPHAZ Panel, 2019b). The QPS status of C. glutamicum is confirmed with the qualification extended to other production purposes.
3.1.4. Lactobacillus species
Lactobacillus spp.
A search for papers potentially relevant for the QPS consideration of any of the 37 Lactobacillus species included in the list provided 3040 references. The analysis of their titles left 226 articles and of their titles/abstracts left 65 articles. Revision of their full texts allowed selection of 31 reports that raised safety concerns, all of which described human pathological processes. The claimed aetiological agents comprise L. acidophilus (Cohen et al., 2016; Haghighat and Crum‐Cianflone, 2016; Hubbard et al., 2018), L. animalis (Somayaji et al., 2016), L. casei (Passera et al., 2016; Vanichanan et al., 2016; Pailhories et al., 2017; Stroupe et al., 2017; de Seynes et al., 2018), L. coryniformis (Datta et al., 2017), L. delbrueckii (Chaini et al., 2016; Maillet et al., 2018), L. gasseri (Chaini et al., 2016; Elikowski et al., 2017; Esquibel et al., 2017), L. paracasei (Harding‐Theobald and Maraj, 2018; Kao et al., 2018; Kato et al., 2016; Pararajasingam and Uwagwu, 2017), L. plantarum (Biesiada et al., 2018), L. rhamnosus (Felekos et al., 2016; Molinaro et al., 2016; Aaron et al., 2017; Norena et al., 2017; Boumis et al., 2018; Kane et al., 2018; Koyama et al., 2018; Naqvi et al., 2018; Nayeem et al., 2018; Zeba et al., 2018) and L. salivarius (Garcia Carretero et al., 2018; Wang et al., 2017).
The pathological processes reported included endocarditis (10 cases), bacteraemia/sepsis (8 cases), abdominal, including liver, abscesses (4 cases), pulmonary infection (4 cases), meningoencephalitis, spondylodiscitis and prosthetic joint, urinary tract and genital infections (one case each).
In three reports (Hubbard et al., 2018; Kao et al., 2018; Nayeem et al., 2018), no information was provided on how the causal microorganism was undet, while in several others (Cohen et al., 2016; Haghighat and Crum‐Cianflone, 2016; Elikowski et al., 2017; Biesiada et al., 2018; Harding‐Theobald and Maraj, 2018; de Seynes et al., 2018; Zeba et al., 2018), phenotypical methods for identification were used, which are not considered completely reliable for lactobacilli.
The articles involved single cases of infection in patients that suffered from predisposing illnesses such as metastatic lung (Biesiada et al., 2018) and pancreas (Nayeem et al., 2018) tumours, congenital heart problems (Norena et al., 2017), haemorrhagic telangiectasia (Boumis et al., 2018), alcoholic cirrhosis (Harding‐Theobald and Maraj, 2018), anastomotic leak from bariatric surgery (Garcıa Carretero et al., 2018), chronic obstructive pulmonary disease, caries and uncontrolled diabetes (Hubbard et al., 2018; Pailhoriès et al., 2017) or were immunocompromised due to untreated AIDS and cirrhosis (Haghighat and Crum‐Cianflone, 2016), prematurity (Molinaro et al., 2016) or had received a bone marrow transplant (Koyama et al., 2018).
Based on the available evidence as described above (the safety concerns identified were considered to be linked to severe underlying health conditions or to immunocompromised people or had methodological problems in the identification of the strain), the QPS status of the lactobacilli involved in the reported cases and, by extension, of all others included in the QPS list, is not changed.
Lactobacillus animalis
This is a new taxonomic unit evaluated from notifications received from October 2016 and now included in the QPS list. The full evaluation was published in a previous BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2017).
Lactobacillus animalis strains have been isolated from the oral cavity and the gastrointestinal tract of animals (Dent and Williams, 1982) and from kimchi, a traditional Korean fermented vegetable dish (Nam et al., 2011). The strain type is L. animalis NCDO 2425. L. animalis is an obligate homofermentative organism that produces mainly L‐(+) lactic acid and is closely related to Lactobacillus acidophilus and L. ruminis. DNA base composition is between 41.3 and 44.4% G + C. A search for the body of knowledge on L. animalis was undertaken in the Web of Science Core collection (search strings in Appendix A) and a total of nine papers were retrieved, with one considered relevant for QPS (Dent and Williams, 1982). Another literature search was performed in PubMed, 289 papers were identified, from which nine were further considered.
L. animalis strain TMW 1.971 has been shown to improve the water holding and gas retention ability of gluten‐free doughs by the production of exopolysaccharides (Rühmkorf et al., 2012, 2013). Effects of L. animalis strain LA4 on the composition and the metabolism of the intestinal microbiota in dogs indicate that it might be considered as a potential probiotic for dogs (Biagi et al., 2007). L. animalis DPC6134 (Hayes et al., 2007) generated peptides with angiotensin‐converting enzyme inhibitory activity from bovine caseinate containing media, with the potential to reduce blood pressure and antihypertensive effects. Bacteriocin production has been characterised for L. animalis strain TSU4 (Sahoo et al., 2015).
The genome sequence of L. animalis P38, isolated from the caecal content of chickens (Rezvani et al., 2016), of L. animalis 381‐IL‐28, a component of a multistrain commercial food biopreservative (Sturino et al., 2014) and of L. animalis KCTC 3501, isolated from kimchi (Nam et al., 2011), have been determined and not found to harbour any genes encoding known virulence factors.
A case of chronic hip prosthetic joint infection caused by L. animalis has been described (Somayaji et al., 2016). This occurred in a 70‐year‐old patient, 5 years after a transient bacteraemia by the same organism as deduced through whole genome sequencing of both causal agents. The patient presented a medical history of type 2 diabetes mellitus and pancreatic cancer.
The species L. animalis is a component of the bacterial communities that colonise the oral cavity and gastrointestinal tract of diverse animal species, and is also commonly used as a starter for fermented vegetables. A single case of human infection by the organism has been reported, but it was linked to life‐compromising predisposing factors. It is therefore concluded that L. animalis does not pose a health risk for the consumer. Consequently, QPS status can be granted for this species.
Lactobacillus parafarraginis
This new taxonomic unit was evaluated from notifications received since October 2016 and is now included in the QPS list. The full evaluation has been published in a previous BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2020).
Lactobacillus parafarraginis is a valid species name according to the List of Prokaryotic Names with Standing in Nomenclature. It was first described upon isolation from Shochu compost (shochu is a sake‐derived distilled beverage) (Endo and Okada, 2007) and belongs to the L. buchneri group of heterofermentative lactobacilli (Salvetti et al., 2018).
L. parafarraginis is used in the fermentation of food and feed. Consequently, it is frequently consumed by humans and livestock. There are no reports of safety concerns.
L. parafarraginis is recommended for inclusion in the QPS list.
3.1.5. Lactococcus lactis
A search for papers potentially relevant for the QPS consideration of Lactococcus lactis provided 958 references. Analysis of their titles left 29 articles and of their abstracts, 17. Full text revision of these allowed the selection of 10 papers that raised safety concerns.
Of these, two dealt with animal infections; one reported bovine mastitis (Rodrigues et al., 2016) and the other involved farmed Alosa‐Alosa fish (Wünnemann et al., 2018). The remaining papers described pathological processes in humans; four related to endocarditis (Mansour et al., 2016; Chen et al., 2018; Georgountzos et al., 2018; Tato Rodriguez et al., 2018), two to oral lesions (Kabore et al., 2018; Mussano et al., 2018) and the other two to liver abscesses (Fragkiadakis et al., 2017) and cholangitis (Shimizu et al., 2019). In two of the reports (Fragkiadakis et al., 2017; Georgountzos et al., 2018), no indication is provided on how the identification of the microorganism was done, while in the other two (Chen et al., 2018; Kabore et al., 2018), phenotypical methods for identification of the aetiological agent, which are known not to be reliable for L. lactis, were used. Finally, Mansour et al. (2016) report repetitive negative blood cultures and a single PCR‐positive determination, which suggests contamination rather than aetiology. Rodrigues et al. (2016) found a higher proportion of L. lactis in milk from mastitic than from healthy cows which, in itself, does not imply causality. The two oral inflammation articles described polymicrobial infections and concomitant isolation of well‐known pathogens, which make doubtful the L. lactis aetiology of the lesions. Predisposing conditions for opportunistic infection were detected in several of the reports dealing with human infections; these comprise pernicious anaemia, autoimmune atrophic gastritis and severe periodontitis (Fragkiadakis et al., 2017), previous cerebral haemorrhage, coronary heart disease and Alzheimer's disease (Chen et al., 2018), valve replacement and aortocoronary by‐pass (Tato Rodriguez et al., 2018) and insertion of a catheter to allow bile secretion after blocking by a cholangiocarcinoma (Shimizu et al., 2019). Finally, the fish studied by Wünnemann et al. (2018) had been recently captured from the wild and kept in an overpopulated tank under very low oxygen concentrations, conditions described by the authors as very stressful.
Based on the available evidence as described above (all safety concerns identified were considered linked to severe underlying health conditions or to immunocompromised status people or had methodological problems in the identification of the strain), the QPS status of L. lactis is not changed.
3.1.6. Leuconostoc species and Microbacterium imperiale
The search for papers potentially relevant for the QPS consideration of Leuconostoc and Microbacterium imperiale provided 406 references. The analysis of their titles left 42 articles. From these, 19 papers were screened for a possible safety concern. After screening the entire papers, four of them were discarded because they did not deal with safety concerns, did not concern this TU or were not in English. Twelve were analysed in detail for information on the potential safety concern identified.
Leuconostoc species
Twelve articles described safety concerns. Four papers on Leuconostoc mesenteroides dealt with nosocomial infections of patients that suffered predisposing conditions. Franco‐Cendejas et al. (2017) refer to a case of acute infection of a knee prosthesis associated with L. mesenteroides 3 years after surgery. The isolated strain was identified using both phenotypic tests and molecular analyses. The authors proposed that the patient's previous upper respiratory tract infection, which caused hyperpermeability and the subsequent bacterial entrance into the bloodstream, may be the origin of the L. mesenteroides infection. In another case, L. mesenteroides was isolated from the blood of a 50‐day‐old baby hospitalised with diarrhoea, and presenting with catheter‐related septicaemia. In this latter study, only a phenotypic identification procedure was performed (Karbuz et al., 2017). For another paper (Ananieva et al., 2017), the identification was achieved using biochemical methods, and the last article concerned the AMR of a small number of L. mesenteroides strains (Cai et al., 2017).
Two cases of L. pseudomesenteroides were described – one catheter‐related sepsis in which the patient was successfully treated with antibiotic lock therapy (Ho et al., 2016) and another paper on bacteraemia in a patient with acute lymphoblastic leukaemia (Ino et al., 2016) without any indication of the identification procedures.
Another case involved a 44‐year‐old woman with acute myeloid leukaemia under myelosuppression, who had bacteraemia caused by L. lactis (Matsuda et al., 2017).
All safety concerns were considered to be linked to severe underlying health conditions, or patients were immunocompromised, or there were methodological problems in the identification of the strain, and therefore, there is no need to change the QPS recommendation of L. pseudomesenteroides and of other Leuconostoc species included in the QPS list.
Microbacterium imperiale
No articles progressed to the level of screening for potential safety concerns for Microbacterium imperiale, so no new safety concerns were identified. Consequently, the QPS status of M. imperiale is not changed, and nor is the qualification ‘QPS applies for production purposes only’.
3.1.7. Oenococcus oeni and Pasteuria nishizawae
The search for papers potentially relevant for the QPS consideration of Oenococcus oeni and Pasteuria nishizawae provided 221 references. The analysis of their titles left 11 articles for further consideration. One, related to O. oeni, was analysed in full with reference to the potential safety concern identified.
Oenococcus oeni
As described above, one article was analysed in detail, but no potential safety concern was identified. Consequently, the QPS status of O. oeni is not changed.
Pasteuria nishizawae
As described above, no article arrived to the final screening stage, so no new safety concerns were identified. Consequently, the QPS status of P. nishizawae is not changed.
3.1.8. Pediococcus species
The search for papers potentially relevant for the QPS consideration of Pediococcus spp. provided 820 references. Analysis of their titles left 20 articles and of their abstracts, 8. After screening the papers that emerged from the different steps considered, only three papers (Han et al., 2016; Chen et al., 2018; Thumu and Halami, 2019) were analysed in full for potential safety concerns, but they did not relate to food‐borne infections, and/or had methodological problems in the identification of the isolate, so they did not give rise to any new safety concerns. Consequently, the QPS status of Pediococcus spp. is not changed.
3.1.9. Dairy propionic acid bacteria – Propionibacterium species
The search for papers potentially relevant for the QPS consideration of Propionibacterium spp. provided 197 references. The analysis of their titles left three articles. After screening the entire papers, only one paper (Giok, 2016) reached the final selection phase, but it does not refer to a food‐borne disease and had methodological identification problems. Consequently, the QPS status of Propionibacterium spp. is not changed.
3.1.10. Streptococcus thermophilus
The search for papers potentially relevant for the QPS consideration of Streptococcus thermophilus provided 397 references. The analysis of their titles left 24 articles. After following the screening procedure, a total of eight arrived to the full‐text phase. From these, five papers arrived to the final stage (Cohen et al., 2016; Yu et al., 2016; Florez and Mayo, 2017; Yang and Yu, 2019; Wardill et al., 2019). They were fully analysed for potential safety concerns, but they did not describe food‐borne infections and/or had methodological problems in the identification, so they did not provide information on any new safety concerns. Therefore, the QPS status of S. thermophilus is not changed.
3.2. Gram‐positive spore‐forming bacteria
3.2.1. Bacillus species with QPS status and Geobacillus stearothermophilus
The search for papers potentially relevant for the QPS consideration of QPS Bacillus spp. and Geobacillus stearothermophilus provided 3,956 references. The analysis of their titles left 287 articles; 234 papers reached the full‐text phase and were analysed in‐depth for their potential relevance. After screening the entire papers, 250 were finally discarded because they did not deal with safety concerns, did not concern this TU or were not in English. Sixteen articles described possible safety concerns.
Bacillus species with QPS status
Sixteen papers were analysed in depth. Eleven had methodological problems with regard to the identification procedures (Danilova et al., 2017; Garcia‐Ramon et al., 2017; Osman et al., 2018; Joshi et al., 2019; Shah et al., 2019a,b; Li et al., 2019; Tsonis et al., 2018), source attribution (Allam et al., 2017; Joshi et al., 2019, Shah et al., 2019a,b) or were not food related (Aydin et al., 2018).
Four papers described human cases where immunosuppression or particular disease features explain the Bacillus spp. clinical manifestations: (i) one paper (Kim et al., 2018) describes a case of pyometra in an immunosuppressed dog, (ii) one paper (Crisafulli et al., 2019) described pleuritic lesions caused by B. megaterium in an old patient with underlying disease, (iii) a case of a 5‐year‐old immunocompetent patient with a deep skin abscess due to B. licheniformis and related to a retained plant thorn (Yuste et al., 2016), (iv) cases due to infection with B. flexus hospitalised in a burns unit (Ucar et al., 2016);
The paper of Gu et al. (2019) reports the analysis of the whole genome sequence of a Bacillus strain indicating a low degree of homology with some virulence determinants of specific pathogens. Moreover, this strain was shown to have haemolitic activity. This pathogenicity feature can be detected by the cytotoxicity test that is required by the current qualification for all Bacillus spp.
Two papers were not considered because they dealt with intrinsic AMR, not associated with any known genetic element able to mobilise resistance genes (Glenwright et al., 2017; Jeong et al., 2017) and with strain‐specific acquired antimicrobial resistance genes (Jeong et al., 2017).
These papers described opportunistic infections linked to specific predisposing factors and do not suggest a risk for consumers or animals via exposure through the food and feed chains, had methodological shortcomings on source attribution or on strain identification, and therefore, there is no change in the status of the Bacillus species included in the QPS list nor in the qualification ‘absence of toxigenic activity’.
Bacillus velezensis
This is a new taxonomic unit that is now included in the QPS list. The evaluation is published in a previous BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2020).
B. velezensis was first described by Ruiz‐García et al. (2005), and is considered a bacterial species with standing in nomenclature (LPSN bactero.net).
The natural habitats of B. velezensis are the soil, the rhizosphere and the marine environment. It has been involved in the fermentation of foods such as kimchi and fermented soybean paste. This species has been used as a plant growth promoting rhizobacterium, in the biological control of plant pathogens and mycotoxigenic fungi and in the detoxification of mycotoxins. Moreover, studies describe the use of B. velezensis as a probiotic in chickens and in fish, being able to control fish bacterial pathogens. This species produces compounds of biotechnological interest, such as β‐glucanases, L‐asparaginase and surfactins.
No association of B. velezensis to intoxication or infection has been reported in humans or animals.
A strain of this species, isolated from a marine environment and identified by 16S rRNA gene analysis, was shown to produce an antimicrobial substance that, based on structural analysis, is classified as an aminoglycoside (Pournejati et al., 2019). A recent study (Agersø et al., 2018) addressed the minimal inhibitory concentration (MIC) distribution and the presence of genes coding for antimicrobial resistance in five Bacillus species, including B. velezensis. The tetracycline efflux gene, tet(L) was found in strains with reduced tetracycline susceptibility but not in susceptible strains.
B. velezensis can be recommended for the QPS list with the qualification ‘absence of toxigenic potential and absence of aminoglycoside production ability’.
Geobacillus stearothermophilus
No article arrived at the level of screening of potential safety concerns for Geobacillus stearothermophilus, so no new safety concern was identified. Consequently, the QPS status of G. stearothermophilus is not changed; nor is the qualification ‘absence of toxigenic activity’.
3.2.2. Paenibacillus illinoisensis
This is a new taxonomic unit that is now included in the QPS list. The evaluation was published in a previous BIOHAZ Panel Statement (EFSA BIOPHAZ Panel, 2019a).
Paenibacillus illinoisenis, previously known as Bacillus circulans, group 6, was described by Shida et al. (1997). It is a valid species with standing in nomenclature.
P. illinoisensis was isolated from the rhizosphere of soil and characterised for its siderophore‐producing capacity, promoting iron absorption by plants (Liu et al., 2017a,b). Strains of P. illinoisensis were reported to secrete cyclodextrin gluconotransferase (Doukyu et al., 2003; Lee et al., 2013, chitinases (Jung et al., 2008) and enzymes degrading methane (Jhala et al., 2014).
P. illinoisensis can be recommended for QPS with the specific qualifications for production purposes and absence of toxigenic potential.
3.2.3. Parageobacillus thermoglucosidasius
This is a new taxonomic unit that is now included in the QPS list. The full evaluation was published in a BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2020).
Parageobacillus thermoglucosidasius is the valid species name with standing in nomenclature (Oren and Garrity, 2019). The basonym is Bacillus thermoglucosidasius (Suzuki et al., 1983); Geobacillus thermoglucosidasius can be used as homotypic synonym; the name Geobacillus thermoglucosidans (Coorevits et al., 2012) has not been accepted as a correct name.
Parageobacillus thermoglucosidasius is a facultative, anaerobic, thermophilic bacterium which is frequently isolated from high temperature environments including hot springs (Brumm et al., 2015) and compost (Brumm et al., 2016; Sung et al., 2002; Fong et al., 2006). The body of knowledge is mainly related to its biotechnological potential for fermentation of plant biomasses (Iwazaki et al., 2018) to generate bio‐ethanol (Zhou et al., 2016) and biohydrogen (Mohr et al., 2018). Also of interest is the production of (heterologous) thermostable enzymes for various industrial applications (Holland et al., 2019) and the biomineralisation potential of this species (Murai and Yoshida, 2016).
Parageobacillus thermoglucosidasius has been frequently found as a spontaneous contaminant in dairy powder products and is isolated in biofilms from the dairy processing industry (Zhao et al., 2013, 2018).
P. thermoglucosidasius can be recommended for QPS list with the qualification ‘for production purposes only’ and the absence of toxigenic potential.
3.3. Gram‐negative bacteria
3.3.1. Gluconobacter oxydans and Xanthomonas campestris
A search of papers potentially relevant for the QPS consideration of Gluconobacter oxidans and Xanthomonas campestris provided 960 references. The analysis of their titles left seven articles. After screening the abstracts, six were discarded because they did not deal with safety concerns. One arrived to the full text phase but did not reached the final selection phase.
Gluconobacter oxydans
As no paper describing a safety concern was found, the QPS status of G. oxydans is not changed, and neither is the qualification ‘QPS only applies when the species is used for vitamin production’.
Xanthomonas campestris
One paper arrived at the full text phase (Sundin and Wang, 2018) dealing with antibiotic resistance in plant‐pathogenic bacteria, but no safety concerns were described. Therefore, the QPS status of X. campestris was not changed, and neither was the qualification ‘QPS only applies when the species is used for the production of xanthan gum’.
3.3.2. Komagataeibacter sucrofermentans
This is a new taxonomic unit that is now included in the QPS list. An evaluation was published in a previous BIOHAZ Panel Statement (EFSA BIOPHAZ Panel, 2019a).
The bacterial species K. sucrofermentans (Validation List nr. 149, IJSM 2013, 63, 1‐5) was previously named Acetobacter xylinus subsp. sucrofermentans (Toyosaki et al., 1996) and Gluconacetobacter sucrofermentans (Cleenwerck et al., 2010). The species is clearly described based on a polyphasic approach (Cleenwerck et al., 2010).
K. sucrofermentans strains are characterised by their ability to produce large amounts of cellulose from sucrose in agitated cultures (Cleenwerck et al., 2010). Searching PubMed database for this species delivered 11 hits, all concerning the cellulose production capacity. In Asia, cellulose has traditionally been produced from the fermentation of coconut waste water by K. sucrofermentans, and used in food.
K. sucrofermentans (A. xylinus subsp. sucrofermentans) can be proposed for the QPS list with the qualification ‘QPS applies for production purposes only’.16
3.3.3. Cupriavidus necator
This new taxonomic unit was evaluated from notifications received after October 2016, and is now included in the QPS list. The evaluation was published in a recent BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2020).
Cupriavidus necator was first described by Makkar and Casida (1987) and is confirmed by DNA–DNA hybridization to be the validated species name with standing in nomenclature (Vandamme and Coenye, 2004). Members of the species were formerly named as Alcaligenes eutrophus, Ralstonia eutropha or Wautersia eutropha (Vaneechoutte et al., 2004). These are Gram‐negative bacteria belonging to the family Burkholderiaceae and the class β–proteobacteria. The whole genome sequence of C. necator strain NH9 and another set of Cupriavidus and Ralstonia strains confirmed the clear delineation of both genera (Gan, 2019; Moriuchi et al., 2019).
C. necator has been reported to prey upon a wide range of Gram‐negative and Gram‐positive bacteria (Seccareccia et al., 2016). C. necator is used as source of polyhydroxybutyrate (PHB) which can be used for the production of bioplastics after recovery from the cell cytoplasm (Aramvash et al., 2015) or by using the dried biomass (Kunasundari et al., 2013). PHBs and the dried biomass of C. necator can be used for its antimicrobial, insecticidal and antiviral activities based on the degradation by bacteria of PHB into β‐Hydroxybutyrate (van Hung et al., 2019). C. necator has been genetically modified to produce several compounds such as isopropanol, hydrocarbons, methyl ketones, free fatty acids, alkanes etc. (Marc et al., 2017).
Cupriavidus necator can be recommended for QPS status with the qualification ‘for production purposes only’.
3.4. Yeasts
Fungi are unique among living organisms because they may have two valid names. The primary name is based on the sexual state or teleomorph, but a second valid name may be based on the asexual state or anamorph. This redundancy of names developed because for many fungi teleomorphs have not been found, or it has not been clear that a particular teleomorph is the same species as a particular anamorph (Kurtzman et al., 2011). In the screened scientific reports on yeasts, either the teleomorph or anamorph names (and sometimes both) are used. Thus, all synonyms or alternative names are included in the literature searches. In the following summaries of the evaluations of the yeast species, the teleomorph name is preferentially used, but for clarity the anamorph name (when known), and synonyms, are also mentioned.
A search of papers potentially relevant for the QPS consideration of the yeast species included in the QPS list provided 7258 references. The analysis of their titles left 692 articles and of their abstracts 507. Three hundred and sixty‐two of these were immediately excluded because they were not in English, or because they were not dealing with safety concerns.
Collectively, the ELS identified 145 articles referring to different yeast species with QPS status (see Appendix D), of which 72 referred to Kluyveromyces marxianus (anamorph=Candida kefyr), 42 to Saccharomyces cerevisiae including Saccharomyces boulardii, 36 to Debaryomyces hansenii (anamorph=Candida famata), 18 to Wickerhamomyces anomalus (anamorph=Candida pelliculosa), 4 to Lindnera jadinii (anamorph=Candida utilis), 2 for Hanseniaspora uvarum. 2 for Kluyveromyces lactis (anamorph=Candida sperica) and 1 for Schizosaccharomyces pombe. For the other yeast species with QPS status, no relevant studies were identified through the ELS.
From these 145 articles, 93 described a potential safety concern. From these 93, 70 had some type of methodological problem identified, either due to weaknesses in the methodology used for identity confirmation of the microorganism (55 articles), or a lack of information regarding the source attribution (3 articles), or uncertainty about whether an infection had actually been diagnosed (40 articles).The remaining 23 articles had no methodological problems, and safety concerns were identified in 15 articles after careful reading. These safety concerns are discussed in the sections below, dedicated to the individual yeast TUs.
3.4.1. Candida cylindracea
C. cylindracea belongs to the Ogataea clade of the Ascomycetous yeasts (Kurtzman et al., 2011; Daniel et al., 2014). The species was described by Yamada and Machida (1962) and validated by Meyer and Yarrow (1998). No synonym names have been used. Only the anamorphic form is known and described. The type strain for C. cylindracea – CBS 6330 – is also marketed under other designations, e.g. DSMZ 2031 (online) and ATCC 14930 (online). Unfortunately, in the literature on lipase‐producing yeasts, the C. cylindracea type strain has at times been referred to as Candida rugosa (e.g. Benjamin and Pandey (1998); Takaç et al. (2010)). This has caused some confusion since C. cylindracea and C. rugosa are two well‐defined species, not closely related phylogenetically (Kurtzman et al., 2011). It is also unfortunate since C. rugosa is considered an emerging, opportunistic yeast (Miceli et al., 2011). However, identification according to molecular methods can easily separate the two species. It is therefore recommended that the species identity of lipase‐producing strains of Candida is confirmed by using such methods.
No references related to possible concerns for human or animal safety, or other related aspects, were identified, and therefore, the QPS status is not changed. The qualification ‘QPS only applies when the species is used for enzyme production’ is also unchanged.
3.4.2. Debaryomyces hansenii
The anamorph form of D. hansenii is Candida famata. Based on Nguyen et al. (2009), Betancourt et al. (2016) concluded that C. famata should not be considered the anamorphic form of D. hansenii. However, Nguyen et al. proposed that a special riboflavin overproducing form of C. famata, also referred to as Candida famata var. flareri, did not belong to the same species as D. hansenii, while C. famata var. famata should be considered to belong to the same species as D. hansenii. Additionally, Kurtzman et al. (2011) supported the view that C. famata should be considered the anamorph of D. hansenii. Since clinical reports usually do not distinguish between the varieties of C. famata/D. hansenii, it is not possible to say to what extent the flareri variety might be involved. But obviously there is reason to closely follow developments in the taxonomy of D. hansenii and related species.
Seventeen references related to possible safety concerns or relevant aspects of the body of knowledge were identified in the ELS, of which 16 used the name C. famata and one the name D. hansenii.
Most of the studies had some type of methodological problem identified and thus had to be given less weight in the evaluation. Most often there were weaknesses in the methods used for species identification.
Taverna et al. (2019) compared different methods to further identify 38 clinical yeast isolates, tentatively identified as C. famata/D. hansenii by morphological and physiological growth tests methods, from a collection in Argentina. Three different molecular methods and MALDI‐TOF MS were largely convergent and showed that more than half of the 38 isolates actually did not belong to the D. hansenii complex, but to the Candida guilliermondii complex. This suggests that studies relying on morphological and biochemical/physiological identification methods are likely to exaggerate the frequency of D. hansenii in human infections, and reinforces the need to use molecular approaches for correct identification of this species. Karapetsa et al. (2019) claim to be the first to report septic shock due to D. hansenii in an immunocompetent subject, although the patient was characterised as showing ‘immunoparalysis’. The young male had serious injuries after a car accident and was admitted to an intensive care unit. Predisposing conditions included a central venous catheter, recurrent bacterial infections and prolonged use of antibiotics. The patient recovered from the fungal infection after treatment with amphotericin B.
Sonmez and Erbas (2017) identified Candida spp. isolates involved in mycotic mastitis in cattle and investigated susceptibility to antimycotics. Three isolates (12%) were identified as D. hansenii, and all were susceptible to ketoconazole but resistant to fluconazole, miconazole and amphotericin B. However, species identification was only based on physiological growth tests and is therefore uncertain. Lo et al. (2017) found that four isolates of D. hansenii from fresh fruit were susceptible to both the antimycotics tested, fluconazole and triadimenol. Taverna et al. (2019) reported that their eight isolates of D. hansenii all had comparatively low MIC values (i.e. high susceptibility) to all eight tested antimycotics. Espinel‐Ingroff et al. (2019) present MIC distributions and epidemiological cut‐off values for D. hansenii for four triazole antimycotics.
In conclusion, relatively few studies reported isolation of D. hansenii in clinical samples, and in most of them, the species identification was uncertain. No studies reported infection in humans without predisposing factors. In retrospective studies of clinical isolate collections, the prevalence of D. hansenii was generally low compared to other yeasts. No studies reported any increased prevalence of antimycotic resistance. In conclusion, no information was obtained to indicate the need for a change in QPS status.
3.4.3. Hanseniaspora uvarum
The anamorph form of H. uvarum is Kloeckera apiculata. The species name has not been changed since the 2016 QPS opinion.
One reference relating to possible concerns for human or animal safety, or other related aspects, was identified.
Siavoshi et al. (2018) claim that common food‐borne yeasts, among them H. uvarum, can harbour the bacterial pathogen Helicobacter pylori as an intracellular parasite or commensal. Thereby yeasts might possibly function as a vector for transmission of this bacterium. Several earlier studies on possible H. pylori association with yeasts have been published by the same research group, but no studies on this subject by any other laboratory or group could be found. While some studies have reported an association of H. pylori with free‐living amoebae (reviewed by Quaglia and Dambrosio, 2018), more evidence is needed before any firm conclusions can be drawn about the possibility that food‐borne yeasts might function as a vector for H. pylori.
No references indicating any new possible concerns for human or animal safety, or other related aspects, were identified. Therefore, the QPS status of H. uvarum is unchanged.
3.4.4. Kluyveromyces lactis
The anamorph form of K. lactis is Candida spherica. The species name has not been changed since the 2016 Opinion.
No references related to possible concerns for human or animal safety, or other related aspects, were identified. Therefore, its QPS status is not changed. The qualification ‘for production purposes only’ is also unchanged.
3.4.5. Kluyveromyces marxianus
The anamorph form of K. marxianus is Candida kefyr. The species name has not been changed since the 2016 Opinion.
In total, 40 references related to possible safety concerns, or relevant aspects of the body of knowledge, were identified in the ELS. All 40 studies were retrieved when using the anamorph name C. kefyr. The majority of these 40 studies had some type of methodological problem identified and thus had to be given less weight in the evaluation. Most often there were weaknesses in the methods used for identifying the microorganism, or a lack of information regarding the source attribution, or uncertainty about whether or not an infection had actually been diagnosed.
The ability of K. marxianus/C. kefyr to cause opportunistic infections in humans with predisposing disease conditions has received increased attention in recent years. For instance, Charsizadeh et al. (2018a,b,c) reported the isolation of K. marxianus from patients with suspected candidiasis in neonatal and paediatric intensive care units, although the prevalence of K. marxianus compared to other fungal species was only 1–2%. Jahanshiri et al. (2018) reported that K. marxianus constituted 5% of the isolates from cancer patients with oropharyngeal candidiasis in a hospital in Iran. Ghajari et al. (2018) reported that one out of 31 yeast isolates from women with suspected vulvovaginal candidiasis was K. marxianus. This is a low prevalence, it is not entirely clear that this isolate caused an infection, and there is no information regarding whether or not there were any predisposing factors. The review of Benedict et al., 2016 lists an earlier study by Pineda et al. (2012), which reports a case of bloodstream infection in a pregnant woman and her twin fetuses. All three survived the infection after successful antimycotic therapy. The mother became pregnant by in vitro fertilisation and delivered the twins by caesarean section at 29 weeks of gestation. The two bloodstream isolates (from the mother and one of the twins) were identified as two different strains of K. marxianus by ITS sequencing and RAPD. No further details on the identification were reported. The mechanism by which the yeast gained access to the placenta and bloodstreams is not clear, and a link to intake of dairy products containing K. marxianus is only hypothesised.
A few studies reported antimycotic susceptibility of clinical K. marxianus isolates. Nagy et al. (2018) demonstrated differences in susceptibility of planktonic and biofilm cells of K. marxianus to several antimycotics. Farmakiotis and Kontoyiannis (2017) reviewed antimycotic non‐susceptibility in pathogenic and opportunistic yeasts and concluded that there is an increasing trend for resistance in K. marxianus. Another review focused on multidrug resistance in pathogenic Candida spp. (Colombo et al., 2017), but also highlighted K. marxianus as an emerging opportunistic pathogen. Salse et al. (2019) presented epidemiological cut‐off values for K. marxianus that may be useful to identify isolates with potential resistance to antimycotics. Espinel‐Ingroff et al. (2019) present MIC distributions and epidemiological cut‐off values for D. hansenii for four triazole antimycotics. One study reported divergent results of the E‐test and Vitek 2 methods for determining antimycotic susceptibility in K. marxianus (Alfouzan et al., 2017), although there were methodological uncertainties regarding how the selection and identification of the studied isolates were performed. Reales‐Calderon et al. (2016) reviewed information regarding the acquisition and development of resistance to antimycotics in yeasts and other fungi, although no specific information was given regarding K. marxianus.
Karstrup et al. (2017) identified a yeast‐like organism in uterine lavage fluids from three cows with slight signs of inflammation but no mastitis, as K. marxianus. The organism could not be cultured but was identified using PCR and sequencing.
There is no doubt that K. marxianus/C. kefyr can behave as an opportunistic fungus in humans with predisposing factors. Reports where it has been unambiguously shown that food intake of K. marxianus is the cause of infectious disease in otherwise healthy individuals do not exist. Therefore, its QPS status remains unchanged.
3.4.6. Komagataella pastoris
The anamorph of K. pastoris is not described. The previous name of this species is Pichia pastoris. The species name has not been changed since the 2016 QPS opinion.
No references related to possible concerns for human or animal safety, or other related aspects, were identified. Therefore, its QPS status does not change. The qualification ‘QPS only applies when the species is used for enzyme production’ is unchanged.
3.4.7. Komagataella phaffi
This is a new taxonomic unit, evaluated from notifications received since October 2016 and now included in the QPS list. The evaluation was published in a previous BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2018a).
The anamorph of Komagataella phaffii is not described. K. phaffii is closely related to Komagataella pastoris, a species with a QPS status, from which it was separated (Kurtzman, 2005). The three species of the genus Komagataella, K. pastoris, K. phaffii and K. pseudopastoris show no differences in standard fermentation and growth tests. Consequently, it is recommended that the species be separated based on differences in D1/D2 26S rRNA gene sequences or on differences in restriction patterns of SSU rRNA (Kurtzman et al., 2011).
In total, 24 studies were identified (see Appendix A) and screened, dealing with the properties of the species as a protein expression and model organism. K. phaffii is a sibling species of K. pastoris (Naumov et al., 2013). In the literature, it has been described as being used for the same purpose as K. pastoris; that is for the production of heterologous proteins (Chessa et al., 2017).
There is very little information about the ecology of K. phaffii, but at least some strains have a similar ecology to K. pastoris, since both species have been isolated from sap fluxes in trees (Kurtzman et al., 2011).
There is no information available about any potential safety concerns regarding K. phaffii. However, reports on the safety of K. pastoris as production organism also have relevance for K. phaffii because this was changed, on the basis of taxonomic position, to the species K. phaffii.
The species K. phaffii, a sibling species of K. pastoris, can be recommended for the QPS list with the qualification ‘QPS only when the species is used for enzyme production’.
3.4.8. Lindnera jadinii
The anamorph form of L. jadinii is Candida utilis. Synonyms of this species are Pichia jadinii, Hansenula jadinii and Torulopsis utilis. The species name has not been changed since the 2016 QPS opinion.
Three references related to possible safety concerns or relevant aspects of the body of knowledge were identified in the ELS. All three studies were retrieved when using the anamorph name C. utilis.
Two studies reported L. jadinii in human clinical samples. Yagmur et al. (2016) isolated yeasts from postmortem specimens from 1309 cases with suspected fungal infection in Turkey. L. jadinii was reported in low prevalence (three isolates, =3%). Identification was only by physiological and morphological properties and it is very uncertain whether the putative L. jadinii strains actually caused infection. In a retrospective study by Kim et al. (2016), a relatively high number of ‘Candida’ isolates (304 isolates, =12%) from clinical samples from patients in a hospital in South Korea were L. jadinii. All patients had underlying disease and the most common source of L. jadinii was a urinary catheter.
Watanasrisin et al. (2016) characterised the ABC‐transporters in L. jadinii. These transporters can be involved in the development of resistance to antimycotics, and knowledge about their structure and function can facilitate the development of novel antimycotic substances.
Few studies reported isolation of L. jadinii/C. utilis in clinical samples and no studies reported infection in humans without predisposing factors. Prevalence was generally low compared to other yeasts isolated from collections of clinical samples. No studies reported increased prevalence of antimycotic resistance. In conclusion, no information was retrieved to indicate a change in the QPS status, nor in the qualification ‘QPS only when the species is used for enzyme production’.
3.4.9. Ogataea angusta
The anamorph form of O. angusta is not described. A synonym of this species is Pichia angusta. The species name has not been changed since the 2013 QPS opinion.
No references related to possible concerns for human or animal safety, or other related aspects, were identified. Therefore, its QPS status, and the qualification ‘QPS only when the species is used for enzyme production’ is unchanged.
3.4.10. Saccharomyces cerevisiae/species
The anamorph form of S. cerevisiae is not described. A synonym of this species is Saccharomyces boulardii. The species name has not been changed since the 2016 QPS opinion.
In total, five references were identified in the ELS, after exclusion of the ones from which methodological problems were identified. From these five references, only one described an infection associated with S. cerevisiae, an osteomyelitis after surgical reconstruction following serious physical injury of an arm in an adult when working in a bakery. The infection was cured after antimycotic treatment (Seng et al., 2016, ELS 3245).
Espinel‐Ingroff et al. (2019) presents MIC distributions and epidemiological cut‐off values for S. cerevisiae for four triazole antimycotics. Pérez‐Cantero et al. (2019) evaluated the in vitro activity of nine antifungal compounds against S. cerevisiae and they also studied in vivo efficacy of the three antifungals showing the highest in vitro activity by using a murine model of systemic infection.
These new reports of Saccharomyces cerevisiae did not add any new information that would change the QPS status of this species. These new reports also confirm the previous qualifications, that the consumption of Saccharomyces boulardii (synonym of S. cerevisiae) by patients with fragile health may be considered as the possible origin of the infection, although the use of microorganisms intended to be used as ‘probiotic’ for humans as a health claim does not fall under the remit of the QPS assessment. These new reports also confirm the previous QPS qualifications; the absence of resistance to antimycotics used for medical treatment of yeast infections in cases where viable cells are added to the food or feed chain and inability to grow above 37°C. Therefore, its QPS status, and the qualifications ‘Absence of resistance to antimycotics used for medical treatment of yeast infections in cases where viable cells are added to the food or feed chain’ is not changed. In the case of Saccharomyces cerevisiae, this qualification applies for yeast strains able to grow above 37°C.
3.4.11. Schizosaccharomyces pombe
There are no synonym names in common use for this species, and the name has not been changed since 2016.
No references related to possible concerns for human or animal safety, or other related aspects, were identified. Therefore, its QPS status and qualification are unchanged.
3.4.12. Wickerhamomyces anomalus
The anamorph form of W. anomalus is Candida pelliculosa. Synonyms of this species are Hansenula anomala, Pichia anomala and Saccharomyces anomalus. The species name has not been changed since the 2016 QPS opinion.
Nine references related to possible concerns for human or animal safety, or other related aspects, were identified. Seven references related to possible safety concerns, or relevant aspects of the body of knowledge, were selected for further investigation. These papers reported proper identification of the W. anomalus/C. pelliculosa strain.
The studies reported W. anomalus/C. pelliculosa in human clinical samples with low prevalence in bloodstream infection (Kumar et al., 2017; Liu et al., 2017a,b; Suhr et al., 2017; Tan et al., 2016) or in immunosuppressed patients or in intensive care units (Fernandez‐Ruiz et al., 2017; Jung et al., 2017; Tejan et al., 2017).
W. anomalus was always a minor fraction of the isolates, and there were no indications that exposure may have been through the food‐borne route. No studies reported infection in healthy, non‐hospitalised subjects or signs of increased prevalence of antimycotic resistance. Therefore, its QPS status is not changed. The qualifications ‘QPS only applies when the species is used for enzyme production’ and ‘Absence of resistance to antimycotics used for medical treatment of yeast infections in cases where viable cells are added to the food or feed chain’ are also unchanged.
3.4.13. Yarrowia lipolytica
This is a new taxonomic unit evaluated from notifications received since October 2016, and now included in the QPS list. The evaluation was published in a previous BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2018b).
Yarrowia lipolytica and Candida lipolytica are the teleomorph and anamorph names of the same species.
The species is widespread in nature, as seen from the sources of isolation. Substrates high in lipids are a common source of this species. Y. lipolytica has several physiological properties of industrial significance. The species is well known for the production of proteases and lipases. It is also a widely reported contaminant in dairy and meat products, and is common on raw poultry products and in traditional sausages (Kurtzman et al., 2011).
Hazen (1995) paid special attention to C. lipolytica, since the species had been implicated as the cause of human infection in one study, and it was therefore considered an ‘emerging pathogen’. Pfaller and Diekema (2004) reported four C. lipolytica isolates among 6,082 Candida spp. isolates (i.e. less than 0.1%) from human bloodstream infections. Shin et al. (2000) reported a temporal outbreak of hospital‐acquired infections with C. lipolytica in five patients during 3 months in a paediatric ward in a hospital in Korea. All patients had suppressed immune systems and all but one had a catheter fitted. All recovered from the infection after chemotherapy. Tumbarello et al. (1996) reported the presence of C. lipolytica in one out of 64 HIV patients with oral candidiasis. A comprehensive review by Groenewald et al. (2014) concluded that all described human infections with this species have occurred in immunocompromised patients with underlying disease, and that the majority were catheter‐related. Antifungal therapy invariably resulted in clearance of the pathogen. In the last 5 years, several reports have confirmed that C. lipolytica can behave as an opportunistic pathogen (e.g.: Trabelsi et al., 2015; Abbes et al., 2017; Boyd et al., 2017).
Based on the available information, Y. lipolytica is a commonly occurring species in many habitats/environments. It may behave as an opportunistic pathogen for immunocompromised patients, especially for those who are using catheters. Yarrowia lipolytica is recommended for the QPS list with the qualification ‘QPS applies for production purpose only’.
3.4.14. Xanthophyllomyces dendrorhous
The anamorph form of X. dendrorhous is Phaffia rhodozyma. The species name has not been changed since the 2016 QPS opinion.
No references related to possible concerns for human or animal safety, or other related aspects, were identified. Therefore, the QPS status is unchanged, as is the qualification.
3.4.15. Zygosaccharomyces rouxii
This new taxonomic unit was evaluated following notifications received since October 2016 and it is now included in the QPS list. The evaluation was published in a recent BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2020).
The genus Zygosaccharomyces is a member of the Saccharomycetaceae family and most closely related to Torulaspora, Zygotorulaspora, Vanderwaltozyma and Tetrapisispora. Six species are present in this genus (Z. bailii, Z. bisporus, Z. kombuchaensis, Z. lentus, Z. mellis and Z. rouxii). Z. rouxii is considered the neotype of the genus.
Z. rouxii is typically found in highly osmotic habitats. Strains of Z. rouxii have been isolated from a wide variety of sources, including cane sugar, chocolate syrup, concentrated black grape must, honey, jam, maple syrup, marmalade, marzipan, miso, red wine, salted beans, soft drinks and soy sauce (for a review see Kurtzman et al., 2011). Z. rouxii is used to ferment a number of salted, oriental fermented foods, the best known being soy sauce and miso. This species is also important in the early stages in the manufacture of balsamic vinegar.
Z. rouxii is primarily a spoilage yeast of high‐sugar or high‐salt foods, such as sugar syrups, candied fruit and soy sauce.
Z. rouxii is recommended for inclusion in the QPS list.
3.5. Protists/Algae
3.5.1. Aurantiochytrium limacinum
This is a new taxonomic unit evaluated from notifications received since October 2016 and now included in the QPS list. The evaluation was published in a recent BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2020).
Aurantiochytrium limacinum is a marine protozoan, belonging to the genus Aurantiochytrium, composed of unicellular eukaryotes, belonging to the order Thraustochytrida, class Labyrinthulea within the phylum Bigyra (Catalogue of Life, online). The taxonomic identification is mainly based on life cycle and developmental stages. The whole genome sequence of strain CCTCC M209059 has been determined (Ji et al., 2015). Shizochytrium limacinum is considered to be a synonym (Catalogue of Life, online). A. limacinum is most often called a microalga, although it is autotrophic and not photosynthetic.
A. limacinum strains are known to produce large amounts of docosahexaenoic acid/docosapenaenoic acid (DHA/DPA), eicosapentaenoic acid (EPA), astaxanthin and β‐carotene (Liang et al., 2011, Du et al., 2019, Ye et al., 2015, Zhang et al., 2017; Bindea et al., 2018). It may also produce peptides with antioxidant activity (Hu et al., 2019) and can be used for the production of biofuel (Xu et al., 2018). It is able to grow on saline waste water (e.g. demineralisation water from cheese whey) (Humhal et al., 2017), cull potato (Chi et al., 2007) and biodiesel‐derived crude glycerol (Ethier et al., 2011) for biomass production. A combined effect of the probiotic Lactococcus lactis and the prebiotic A. limacinum biomass, fed to fish, induced positive effects on their growth and immunity (Sun et al., 2019). A. limacinum biomass is commercialised and was successfully used as alternative for fish oil in feeding laying hens for the enrichment of table eggs with n‐3 fatty acids (Kralik et al., 2019).
No safety concerns were reported related to A. limacinum.
The species Aurantiochytrium limacinum is recommended for QPS status with the qualification ‘for production purposes only’.
3.5.2. Euglena gracilis
This new taxonomic unit was evaluated following notifications received since October 2016 and it is now included in the QPS list. The evaluation was published in a recent BIOHAZ Panel Statement (EFSA BIOPHAZ Panel, 2019b).
E. gracilis belongs to the genus Euglena, phototrophic euglenoid flagellates possessing complex chloroplasts. The taxonomy of E. gracilis and closely related species has not been amended so far, based on molecular phylogenetic analyses (Zakryś et al., 2017). The whole genome of the E. gracilis strain Z1 was recently sequenced (Ebenezer et al., 2019).
There are many scientific papers published on this TU. E. gracilis is found in many freshwater habitats, especially in shallow eutrophic ponds. The species is able to synthesise biotechnologically relevant compounds such as polyunsaturated fatty acids, vitamins, b‐glucans and tyrosine, used in cosmetics and food supplements. E. gracilis is also used for bioremediation of heavy metals in contaminated water and as a toxicity bioindicator (Krajčovič et al., 2015).
E. gracilis biomass, generally based on dried cells, is used as a feed additive in aquaculture and in animal feed as well as in human food (Suzuki, 2017). Food products containing E. gracilis are marketed in Japan as cookies, cereal bars and nutritional drinks.
Using dried preparations of non‐viable E. gracilis, no genotoxicity was observed in bacterial reverse mutation and mammalian micronucleus tests. Moreover, subchronic toxicity tests in rats did not show any adverse effect and a no‐observed‐adverse‐effect‐level (NOAEL) of 50,000 ppm was determined (Simon et al., 2016). Literature searches did not provide any evidence of any safety concerns for human or animal health related to any use of E. gracilis.
Euglena gracilis may be recommended for the QPS list with the qualification ‘QPS applies for production purposes only’.
3.5.3. Tetraselmis chuii
This is a new taxonomic unit, evaluated from notifications received since October 2016, and it is now included in the QPS list. The evaluation was published in a recent BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2020).
Tetraselmis chuii (also sometimes spelled Tetraselmis chui) is a unicellular, planktonic microalga belonging to the phylum Chlorophyta (green algae) and family Chlorodendraceae. Members of the genus (currently around 25 species) have four flagella arranged in two pairs and are capable of active movement. The taxonomy of the genus has been described based mainly on morphological and ultrastructural information. A few studies employed rDNA sequencing for species identification in limited collections of Tetraselmis strains (Lee and Hur, 2009; Arora et al., 2013; González et al., 2015); however, systematic taxonomic studies of the genus combining morphological and molecular approaches are lacking.
The species was first described in the 1950s (Butcher, 1959) but has since then been found in phytoplankton communities in marine and brackish environments around the world. An interesting ecological observation is that the functional chloroplast of T. chuii can be retained within the cell in some ciliates that graze the alga and thereby possibly give the ciliate a nutritional supplement by photosynthesis (McManus et al., 2018).
T. chuii is cultured and refined in large‐scale facilities and has a long history of use as an efficient feed in the aquaculture industry (Camus et al., 2009; Galimany et al., 2014). It has a high nutritional value (Tibbetts et al., 2015) and is of considerable interest for biotechnological production of useful compounds, e.g. essential fatty acids, antioxidants (carotenoids and phenolic compounds), starch and bulk lipids and oils (Araujo et al., 2011; Custódio et al., 2012; Gifuni et al., 2018). Genetic tools for transformation of T. chuii have been developed (Úbeda‐Mínguez et al., 2015). The species has also been used as a test organism in toxicity assays of different types of pollutants (Debelius et al., 2009; Prata et al., 2018; Davarpanah and Guilhermino, 2019.).
The alga T. chuii has also found use in human food, based on its content of antioxidants (Widowati et al., 2017). Dried biomass of T. chuii has been authorised in the EU as a novel food and food supplement (Commission Implementing Regulation (EU) 2017/247017), and it is sold under the name TetraSOD®.
No safety concern was identified. Cerezuela et al. published a series of papers (2012a,b,c, 2013) reporting the effects of different diets, including supplements of T. chuii, on expression of genes related to intestinal and immune functions in sea bream (Sparus aurata L.). However, no information is given about any effects of the diets on the growth or health status of the fish. One study investigated the toxicity of freeze‐dried biomass of T. chuii in a rat model (Mantecón et al., 2019). Different doses of T. chuii had no effect on growth rate, and no clinical signs or effects on blood parameters, organ weights or histopathology were observed.
Tetraselmis chuii is recommended for the QPS list ‘for production purposes only’.
3.6. Viruses used for plant protection
A number of viruses for use to control plant pests have been recommended as candidates for QPS. The first category encompasses ‘mild strains’ of plant viruses used to mitigate the effects of infection with ‘severe strains’ of the same virus species, the latter causing severe disease, e.g. in tomato and squash. The viruses notified to EFSA are members of two well‐characterised plant virus families, the Alphaflexviridae (Order Tymovirales) and Potyviridae. The second category consists of baculoviruses (Family Baculoviridae) that kill specific species of pest insects.
3.6.1. Alphaflexiviridae and Potyviridae
A search of papers, published between 2016 and 2019 and potentially relevant for the consideration QPS status relating to Alphaflexviridae and Potyviridae provided 191 references. A total of two papers reached the title/abstract selection phase but did not arrive to the final phase of the selection.
Alphaflexiviridae
No papers reached the final selection phase, so no new safety concern was identified. Therefore, the QPS recommendation on members of the Alphaflexviridae can be maintained, and the family Alphaflexviridae is the lowest TU with QPS status.
Potyviridae
Two papers (Cong et al., 2019; Gachara and Wisser, 2018) reached the final selection phase, but no new safety concern was described. No references related to possible concerns for human or animal safety, or other related aspects, were identified, so the QPS recommendation on members of the Potyviridae family can be maintained, and the family Potyviridae is the lowest TU with QPS status.
3.6.2. Insect viruses
Baculoviridae
A search for papers published between 2016 and 2019, and potentially relevant for the QPS consideration of Baculoviridae, provided 381 references. Three papers (Lacey, 2017; Maciel‐Vergara and Ros, 2017; van Oers et al., 2017) were evaluated but did not deal with safety concerns. Zhao et al. (2019) reported an in‐depth study on the fate and consequences of baculovirus infection by intravenous infection of rats. This is an unusual route for interaction of baculoviruses and vertebrate hosts; normally vertebrates only ingest baculoviruses with food. Nevertheless, no adverse effects on pathology or animal health were observed. Charon et al. (2019) describe the regulatory framework for agrochemicals (including baculoviruses) and promote the inclusion of baculoviruses as a low risk substance. Nan et al. (2019) note the prion‐like properties of one gene product (Late Essential Factor 10) related to the insect pathology of baculoviruses. This has no consequences for the safety of baculoviruses as a biocontrol agent of insect pests for vertebrates.
Therefore, the QPS recommendation of the members of the Baculoviridae family is not changed and the family Baculoviridae is the lowest TU with QPS status.
4. Conclusions
Answer to the terms of reference (ToR):
ToR 1: Keep updated the list of biological agents being notified in the context of a technical dossier to EFSA Units (such as Feed, Pesticides, Food Ingredients and Packing, and Nutrition) for intentional use in feed and/or food or as sources of food and feed additives, enzymes and plant protection products for safety assessment.
The list of biological agents notified in the context of technical dossiers was updated. Three hundred and twenty‐eight notifications were received between October 2016 and September 2019, of which, 185 were from feed additives, 78 from food enzymes, food additives and flavourings, 25 from novel foods, and 40 from PPPs; 198 were bacteria, 84 filamentous fungi, 4 viruses and 34 yeasts and 6 protists/algae.
The list ‘Microbial species as notified to EFSA’ (https://doi.org/record/zenodo.3607184), Appendix D of this opinion, compiles all the microorganisms notified to EFSA from the beginning of the QPS exercise in 2007.
ToR 2: Review taxonomic units previously recommended for the QPS list and their qualifications (especially the qualification regarding antimicrobial resistance) when new information has become available. Update the information provided in the previous opinion where appropriate.
This task has been covered by each of the Panel Statements published from June 2017. The current opinion summarises the results of the six Panel Statements published/prepared since then. All TUs that had been previously recommended for the QPS list in the 2016 Opinion were reviewed and confirmed. The information in the previous opinion was updated and the qualifications were also confirmed. The updated list (‘2019 QPS list’) is available on the EFSA Knowledge Junction community18 in Zenodo (https://doi.org/10.5281/zenodo.1146566, Appendix A).
Relevant information from the ELS includes case reports of human diseases. Several of the QPS‐TUs (e.g. Bifidobacterium species, Lactobacillus and Saccharomyces boulardii cerevisiae) are sporadically reported as causing infections in individuals with recognised predispositions for the acquisition of opportunistic infections. Previous use of the microorganisms as food supplements for humans, which does not fall under the remit of the QPS assessment, was reported in many of these cases.
During the 3‐year period of this QPS mandate, some aspects in relation to the application of QPS in safety assessments were clarified:
Based on the actual body of knowledge and/or an ambiguous taxonomic position, the following TUs were excluded from the QPS assessment: filamentous fungi, oomycetes, streptomycetes, Enterococcus faecium, Escherichia coli and bacteriophages (EFSA BIOHAZ Panel et al., 2017b).
In the case of Genetically Modified Microorganisms (GMM) for which the species of the recipient strain qualifies for the QPS status, and for which the genetically modified state does not give rise to safety concerns, the QPS approach can be extended to genetically modified production strains (EFSA BIOHAZ Panel et al., 2018a).
The qualification ‘for production purpose only’ implies the absence of viable cells of the production organism in the final product, and can also be applied to food and feed products based on microbial biomass (EFSA BIOHAZ Panel et al., 2018b).
The QPS status of Corynebacterium glutamicum was confirmed with the qualification extended to other production purposes.
For yeasts, acquired AMR genes are not of relevance, but susceptibility to antimycotic compounds used in human medicine should be proved in circumstances where the yeasts are used as viable organisms in the food and feed chain.
ToR 3: (Re) assess the suitability of taxonomic units notified to EFSA not present in the current QPS list for their inclusion in that list.
Six Panel statements have been published periodically (approximately every 6 months) in order to assess the suitability of new TU notified to EFSA and to update the list with those biological agents that were recommended for the QPS list. From those 328 notifications, 131 biological agents already had a QPS status and were not further evaluated, neither were the notifications of 84 filamentous fungi and 4 of Enterococcus faecium, excluded from QPS consideration following a recommendation of the QPS 2013 update, the 27 notifications of E. coli (bacterium) and 4 of Streptomyces spp., excluded in the Panel Statement adopted in December 2016, 1 notification of a bacteriophage, excluded in the Panel Statement adopted in December 2017 and Sphingomonas paucimobilis which has already been evaluated in a previous Panel Statement. Furthermore, it was agreed not to include 10 notifications from the Pesticides Unit as the respective dossiers (including the literature review) had not yet been received (8 of Bacillus thuringiensis, 1 of Pseudomonas sp. and 1 of an Oomycetes). The remaining 51 notifications were considered for the assessment of the suitability of the respective TUs for inclusion in the QPS list. From these 51, 40 were bacteria, 5 yeasts and 6 protists/algae. Of these, 14 new TUs received a QPS status: 3 yeasts, 8 bacteria and 3 algae/protists:
Lactobacillus animalis, Lactobacillus parafarraginis and Zygosaccharomyces rouxii are recommended for the QPS status; Euglena gracilis, Aurantiochytrium limacinum, Tetraselmis chuii, Corynebacterium ammoniagenes, Cupriavidus necator, Komagataeibacter sucrofermentans and Yarrowia lipolytica are recommended for the QPS status with the qualification ‘for production purposes only’; Bacillus velezensis is recommended for the QPS status with the qualification ‘absence of toxigenic potential and absence of aminoglycoside production ability’; Paenibacillus illinoisensis and Parageobacillus thermoglucosidasius are recommended for the QPS status with the qualification ‘for production purposes only’ and absence of toxigenic potential; Komagatella phaffii is recommended for QPS status with the qualification ‘when the species is used for enzyme production’.
5. Recommendations
Studies to assess the potential for virulence of QPS‐TU are recommended to be intensified, using whole genome sequencing and experimental tests, to identify any specific factors that might contribute to their pathogenicity.
The verification that specific yeast strains, used as viable organisms in the food and feed chain, fulfil the qualification of ‘absence of antimycotic resistance’ has to be conducted by the specific EFSA Unit/Panel to which the notification was assigned. Specific guidance needs to be developed.
A generic qualification for Plant Protection Products ‘environmental risk assessment for use as microbial plant protection product should be assessed at strain level following the requirements of the current legislation’.
Glossary
- Anamorph
namesecond valid name of a fungi based on the asexual state reproductive state (morphologically)
- Basonym
the earliest validly published name of a taxon
- Synonymous name/Homotypic synonym
have the same type (specimen) and the same taxonomic rank.
- Teleomorph name
primary name of a fungi based on the sexual reproductive state (morphologically)
Abbreviations
- AMR
Antimicrobial resistance
- BIOHAZ
EFSA Panel on Biological Hazards
- ELS
Extensive literature search
- FEEDAP
EFSA Panel on Additives and Products of Substances used in Animal Feed
- FIP
Food Ingredients and Packaging
- FSTA
Food Science Technology Abstracts
- GMM
Genetically modified microorganisms
- ICN
International Code of Nomenclature
- ICTV
International Commission on the Taxonomy of Viruses
- IJSEM
International Journal of Systematic and Evolutionary Microbiology
- MALDI‐TOF MS
Matrix‐Assisted Laser Desorption/ionisation Time‐of‐Flight Mass Spectrometry
- MIC
Minimal inhibitory concentration values
- NDA
EFSA Panel on Dietetic Products, Nutrition and Allergies
- NF
Novel Food
- QPS
Qualified Presumption of Safety
- PCR
Polymerase Chain Reaction
- PCR‐RFLP
Polymerase chain reaction‐restriction fragment length polymorphism
- PECO
Population Exposure Comparator Outcome
- PPP
Plant protection product
- ToR
Term of reference
- TF
Traditional food
- TU
Taxonomic unit
Appendix A – The 2019 updated list of microorganisms with QPS status
1.
The list of QPS status recommended biological agents (EFSA BIOHAZ Panel, 2020) is being maintained in accordance with the self‐task mandate of the BIOHAZ Panel (2020–2022). Possible additions to this list are included around every 6 months, with the first Panel Statement adopted in June 2020 and the last Panel Statement planned for adoption in December 2022. These additions are published as updates to the Scientific Opinion (EFSA BIOHAZ Panel, 2020) and as supporting information linked to every Panel Statement available on the EFSA Knowledge Junction community on Zenodo at: https://doi.org/10.5281/zenodo.1146566
Appendix B – Extensive literature search, relevance screening and article evaluation for the maintenance and update of list of QPS‐recommended biological agents intentionally added to the food or feed chain as notified to EFSA
1.
This extensive literature search (ELS) protocol used in the context of the EFSA mandate on the list of QPS‐recommended biological agents intentionally added to the food or feed is available on the EFSA Knowledge Junction community on Zenodo at: https://doi.org/10.5281/zenodo.3607190
Appendix C – Search strategies
1.
The search strategies for each taxonomic unit (TU), i.e. the string for each TU and the search outcome, are available on the EFSA Knowledge Junction community on Zenodo at: https://doi.org/10.5281/zenodo.3607193
Appendix D – Microbial species as notified to EFSA until September 2019
1.
Appendix D contains the list of ‘Microbial species as notified to EFSA’, and compiles all microorganisms notified to EFSA from the beginning of the QPS exercise in 2007 – available on the EFSA Knowledge Junction community on Zenodo at: https://doi.org/10.5281/zenodo.3607184
Suggested citation: EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards) , Koutsoumanis K, Allende A, Alvarez‐Ordóñez A, Bolton D, Bover‐Cid S, Chemaly M, Davies R, De Cesare A, Hilbert F, Lindqvist R, Nauta M, Peixe L, Ru G, Simmons M, Skandamis P, Suffredini E, Cocconcelli PS, Fernández Escámez PS, Maradona MP, Querol A, Suarez JE, Sundh I, Vlak J, Barizzone F, Correia S and Herman L, 2020. Scientific Opinion on the update of the list of QPS‐recommended biological agents intentionally added to food or feed as notified to EFSA (2017–2019). EFSA Journal 2020;18(2):5966, 56 pp. 10.2903/j.efsa.2020.5966
Requestor: EFSA
Question number: EFSA‐Q‐2016‐00684
Former Panel members: Ana Allende, Declan Bolton, Marianne Chemaly, Robert Davies, Pablo Salvador Fernáandez Escáamez, Rosina Girones, Lieve Herman, Konstantinos Koutsoumanis, Roland Lindqvist, Birgit Nørrung, Antonia Ricci, Lucy Robertson, Giuseppe Ru, Moez Sanaa, Marion Simmons, Panagiotis Skandamis, Emma Snary, Niko Speybroeck, Benno Ter Kuile, John Threlfall and Helene Wahlström.
Current Panel members: Ana Allende, Avelino Alvarez‐Ordóñez, Declan Bolton, Sara Bover‐Cid, Marianne Chemaly, Robert Davies, Alessandra De Cesare, Lieve Herman, Friederike Hilbert, Kostas Koutsoumanis, Roland Lindqvist, Maarten Nauta, Luisa Peixe, Giuseppe Ru, Marion Simmons, Panagiotis Skandamis and Elisabetta Suffredini.
Acknowledgements: The Panel wishes to thank the EFSA staff members: Margarita Aguillera‐Gomez, Jaime Aguillera, Wolfgang Gelbmann, Rosella Brozzi, Katrin Bote, Leng Heng, Annamaria Rossi, Patricia Romero and Frédérique Istace for the support provided to this scientific output. The Panel also wants to thank Prof. Malcolm Richardson for the information provided on antimycotic resistance in September of 2018.
Adopted: 12 December 2019
Amendment: The hyperlinks sending to Appendix A on Zenodo were corrected on pages 3, 10, 41 and 56, and the surname of Annamaria Rossi has also been corrected in the ‘Acknowledgements’ paragraph. These editorial corrections do not materially affect the contents or outcome of this scientific output. To avoid confusion, the older version has been removed from the EFSA Journal, but is available on request, as is a version showing all the changes made.
Erratum: On pages 3 in the Summary, and on pages 18, 30 and 41, the reference to the qualification of B. velezensis has been corrected from ‘absence of aminoglycoside production including the genes encoding it’ to ‘absence of aminoglycoside production ability’. To avoid confusion, the older version of the output has been removed from the EFSA Journal but is available on request as is a version showing all the changes made.
Amended: 15 May 2020
Notes
Opinion of the Scientific Committee on a request from EFSA related to a generic approach to the safety assessment by EFSA of microorganisms used in food/feed and the production of food/feed additives. EFSA Journal 2005;226, 1–12.
Introduction of a Qualified Presumption of Safety (QPS) approach for assessment of selected microorganisms referred to EFSA ‐ Opinion of the Scientific Committee. EFSA Journal 2007;587, 1–16.
Scientific Opinion of the Panel on Biological Hazards on a request from EFSA on the maintenance of the list of QPS microorganisms intentionally added to food or feed. EFSA Journal 2008;923, 1–48.
Scientific Opinion of the Panel on Biological Hazards (BIOHAZ) on the maintenance of the list of QPS microorganisms intentionally added to food or feed (2009 update). EFSA Journal 2009;7(12):1431, 92 pp. https://doi.org/10.2903/j.efsa.2009.1431
EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), 2013. Scientific Opinion on the maintenance of the list of QPS biological agents intentionally added to food and feed (2013 update). EFSA Journal 2013;11(11):3449, 107 pp. https://doi.org/10.2903/j.efsa.2013.3449
Gluconobacter oxydans.
References updated from original mandate.
The TU was corrected from the original mandate: ‘enterococci’. It is only referred to Enterococcus faecium, the only species which was evaluated for a possible QSP status.
Sentence included, correcting the previous sentence from the original mandate: ‘Genetically modified microorganisms are similarly not taken into account’.
Regulation (EC) No 1831/2003 of the European Parliament and of the Council of 22 September 2003 on additives for use in animal nutrition. OJ L 268, 18.10.2003, p. 29–43.
Regulation (EC) No 1107/2009 of the European Parliament and of the Council of 21 October 2009 concerning the placing of plant protection products on the market and repealing Council Directives 79/117/EEC and 91/414/EEC. OJ L 309, 24.11.2009, p. 1–49.
Council Directive of 15 July 1991 concerning the placing of plant protection products on the market (91/414/EEC). OJ L 230, 19.8.1991, p. 1–32.
Commission Implementing Regulation (EU) No 562/2012 of 27 June 2012 amending Commission Regulation (EU) No 234/2011 with regard to specific data required for risk assessment of food enzymes. OJ L 168, 28.6.2012, p. 21–23.
Commission Regulation (EU) No 234/2011 of 10 March 2011 implementing Regulation (EC) No 1331/2008 of the European Parliament and of the Council establishing a common authorisation procedure for food additives, food enzymes and food flavourings. OJ L 64, 11.3.2011, p. 15–24.
Regulation (EU) 2015/2283 of the European Parliament and of the Council of 25 November 2015 on novel foods, amending Regulation (EU) No 1169/2011 of the European Parliament and of the Council and repealing Regulation (EC) No 258/97 of the European Parliament and of the Council and Commission Regulation (EC) No 1852/2001. OJ L 327, 11.12.2015, p. 1–22.
Commission Implementing Regulation (EU) 2017/2470 establishing the Union list of novel foods in accordance with Regulation (EU) 2015/2283 of the European Parliament and of the Council on novel foods. OJ L 351, 30.12.2017, p. 72–201.
The Knowledge Junction is a curated, open repository for the exchange of evidence and supporting materials used in food and feed safety risk assessments ‐ https://zenodo.org/communities/efsa-kj/
References
- Aaron JG, Sobieszczyk ME, Weiner SD, Whittier S and Lowy FD, 2017. Lactobacillus rhamnosus endocarditis after upper endoscop. Open Forum Infectious Diseases, 4, ofx085 10.1093/ofid/ofx085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbes S, Amouri I, Trabelsi H, Neji S, Sellami H, Rahmouni F, Makni F, Rebai T and Ayadi A, 2017. Analysis of virulence factors and in vivo biofilm‐forming capacity of Yarrowia lipolytica isolated from patients with fungemia. Medical Mycology, 55, 193–202. 10.1093/mmy/myw028 [DOI] [PubMed] [Google Scholar]
- Agersø Y, Stuer‐Lauridsen B, Bjerre K, Jensen M, Johansen E, Bennedsen M, Brockmann E and Nielsen B, 2018. Antimicrobial susceptibility testing and tentative epidemiological cutoff values for five Bacillus species relevant for use as animal feed additives or for plant protection. Applied and Environmental Microbiology, 84, 10.1128/AEM.01108-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfouzan W, Al‐Enezi T, AlRoomi E, Sandhya V, Chandy R and Khan ZU, 2017. Comparison of the VITEK 2 antifungal susceptibility system with Etest using clinical isolates of Candida species. Revista Iberoamericana de Micología, 34, 171–174. [DOI] [PubMed] [Google Scholar]
- Allam NAT, Sedky D and Mira EK, 2017. The clinical impact of antimicrobial resistance genomics in competition with she‐camels recurrent mastitis metabolomics due to heterogeneous Bacillus licheniformis field isolates. Veterinary World, 10, 1353–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaro TMMM, Thilliez GJA, Motion GB and Huitema E, 2017. A perspective on CRN proteins in the genomics age: evolution, classification, delivery and function revisited. Frontiers in Plant Science, 8, 99. 10.3389/fpls.2017.00099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An B‐K, Choi Y‐I, Kang C‐W and Lee K‐W, 2018. Effects of dietary Corynebacterium ammoniagenes‐derived single cell protein on growth performance, blood and tibia bone characteristics, and meat quality of broiler chickens. Journal of Animal and Feed Sciences, 27, 140–147. 10.22358/jafs/91966/2018 [DOI] [Google Scholar]
- Ananieva MM, Faustova MO, Basarab IO and Loban GA, 2017. Kocuria rosea, kocuria kristinae, leuconostoc mesenteroides as caries‐causing representatives of oral microflora. Wiadomosci lekarskie (Warsaw Poland, 1960), 70, 296–298. [PubMed] [Google Scholar]
- Aramvash A, Shahabi Z, Aghjeh S and Ghafari M, 2015. Statistical physical and nutrient optimization of bioplastic polyhydroxybutyrate production by Cupriavidus necator. International Journal of Environmental Science Technology, 12, 2307 10.1007/s13762-015-0768-3 [DOI] [Google Scholar]
- Araujo GS, Matos LJBL, Gonçalves LRB, Fernandes FAN and Farias WRL, 2011. Bioprospecting for oil producing microalgal strains: evaluation of oil and biomass production for ten microalgal strains. Bioresource Technology, 102, 5248–5250. 10.1016/j.biortech.2011.01.089 [DOI] [PubMed] [Google Scholar]
- Arora M, Anil AC, Leliaert F, Delany J and Mesbahi E, 2013. Tetraselmis indica (Chlorodendrophyceae, Chlorophyta), a new species isolated from salt pans in Goa, India. European Journal of Phycology, 48, 61–78. 10.1080/09670262.2013.768357 [DOI] [Google Scholar]
- Aydin ZGG, Aydemir D, Arslan EA, Ozkaya E, Kamasak T, Sahin S, Guvercin AR, Yazar U, Arslan E, Cakir E and Cansu A, 2018. Evaluation of ventriculoperitoneal shunt infections in children. Journal of Pediatric Infection, 12, 147–152. [Google Scholar]
- Benedict K, Chiller TM and Mody RK, 2016. Invasive fungal infections acquired from contaminated food or nutritional supplements: a review of the literature. Foodborne Pathogens and Disease, 13, 343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin S and Pandey A, 1998. Candida rugosa lipases: molecular biology and versatility in biotechnology. Yeast, 14, 1069–1087. [DOI] [PubMed] [Google Scholar]
- Berger H, Yacoub A, Gerbore J, Grizard D, Rey P, Sessitsch A and Compant S, 2016. Draft genome sequence of biocontrol agent Pythium oligandrum strain Po37, an oomycota. Genome Announcements, 4, e00215‐16. 10.1128/genomeA.00215-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt AAS, Sibaja Alvarez P, Arguedas Camacho R and Guevara Espinoza E, 2016. Candida famata mediastinitis. A rare complication of open heart surgery. Case Report and Brief Review, IDCases, 5, 37–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biagi G, Cipollini I, Pompei A, Zaghini G and Matteuzzi D, 2007. Effect of a Lactobacillus animalis strain on composition and metabolism of the intestinal microflora in adult dogs. Veterinary Microbiology, 124, 160–165. 10.1016/j.vetmic.2007.03.013 [DOI] [PubMed] [Google Scholar]
- Biesiada G, Krycinska R, Czepiel J, Stazyk K, Kedzierska J and Garlicki A, 2018. Meningoencephalitis caused by Lactobacillus plantarum ‐ case report. The International Journal of Neuroscience, 1–9. [DOI] [PubMed] [Google Scholar]
- Bindea M, Rusu B, Rusu A, Trif M, Leopold LF, Dulf F and Vodnar DC, 2018. Valorification of crude glycerol for pure fractions of docosahexaenoic acid and β‐carotene production by using Schizochytrium limacinum and Blakeslea trispora. Microbial Cell Factories, 17, 97 10.1186/s12934-018-0945-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boumis E, Capone A, Galati V, Venditti C and Petrosillo N, 2018. Probiotics and infective endocarditis in patients with hereditary hemorrhagic telangiectasia: a clinical case and a review of the literature. BMC Infectious Diseases, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd AS, Wheless L, Brady BG and Ellis D, 2017. Cutaneous Yarrowia lipolytica infection in an immunocompetent woman. JAAD Case Reports, 3, 219–221. 10.1016/j.jdcr.2017.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumm PJ, Land ML and Mead DA, 2015. Complete genome sequence of Geobacillus thermoglucosidasius C56‐YS93, a novel biomass degrader isolated from obsidian hot spring in Yellowstone National Park. Standards in Genomic Sciences, 10, 73–73. 10.1186/s40793-015-0031-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumm PJ, Land ML and Mead DA, 2016. Complete genome sequences of Geobacillus sp. WCH70, a thermophilic strain isolated from wood compost. Stand Genomic Sciences, 11, 33 10.1186/s40793-016-0153-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butaye P, Devriese LA and Haesebrouck F, 2003. Antimicrobial growth promoters used in animal feed: effects of less well known antibiotics on gram‐positive bacteria. Clinical Microbiology Reviews, 16, 175–188. 10.1128/cmr.16.2.175-188.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher RW, 1959. An Introductory Account of the Smaller Algae of British Coastal Waters: Part I: Introduction and Chlorophyceae. Great Britain: Ministry of Agriculture, Fisheries and Food, Fish Investigation IV (Part 1). H. M. Stationery Office; 74 pp. [Google Scholar]
- Cai TLQ, Xiang W, Zhang Q, Chen G and Cai Y, 2017. Antibiotic resistance evaluation and resistance gene profile of epibiotic lactic acid bacteria on red bell peppers used for Sichuan pickle fermentation. Food Science, China, 38, 27–33. 10.7506/spkx1002-6630-201702005 [DOI] [Google Scholar]
- Camus T, Zeng C and McKinnon AD, 2009. Egg production, egg hatching success and population increase of the tropical paracalanid copepod, Bestiolina similis (Calanoida: Paracalanidae) fed different microalgal diets. Aquaculture, 297, 169–175. 10.1016/j.aquaculture.2009.09.018 [DOI] [Google Scholar]
- Catalogue of Life , online. Catalogue of Life: 2019 Annual Checklist. Available online: http://www.catalogueoflife.org/annual-checklist/2019/details/species/id/fd4c2685b651e6d01f3b9dad65707003
- Cerezuela R, Fumanal M, Tapia‐Paniagua ST, Meseguer J, Morinigo MA and Esteban MA, 2012a. Histological alterations and microbial ecology of the intestine in gilthead seabream (Sparus aurata L.) fed dietary probiotics and microalgae. Cell Tissue Research, 350, 477–489. 10.1007/s00441-012-1495-4 [DOI] [PubMed] [Google Scholar]
- Cerezuela R, Guardiola FA, Gonzalez P, Meseguer J and Esteban MA, 2012b. Effects of dietary Bacillus subtilis, Tetraselmis chuii, and Phaeodactylum tricornutum, singularly or in combination, on the immune response and disease resistance of sea bream (Sparus aurata L.). Fish Shellfish Immunology, 33, 342–349. 10.1016/j.fsi.2012.05.004 [DOI] [PubMed] [Google Scholar]
- Cerezuela R, Guardiola FA, Meseguer J and Esteban MA, 2012c. Enrichment of gilthead seabream (Sparus aurata L.) diet with microalgae: effects on the immune system. Fish Physiology and Biochemistry, 38, 1729–1739. 10.1007/s10695-012-9670-9 [DOI] [PubMed] [Google Scholar]
- Cerezuela R, Meseguer J and Esteban MA, 2013. Effects of dietary inulin, Bacillus subtilis and microalgae on intestinal gene expression in gilthead seabream (Sparus aurata L.). Fish Shellfish Immunology, 34, 843–848. 10.1016/j.fsi.2012.12.026 [DOI] [PubMed] [Google Scholar]
- Chaini E, Chainis ND, Ioannidis A, Magana M, Nikolaou C, Papaparaskevas J, Liakata M‐V, Katopodis P, Papastavrou L, Tegos GP and Chatzipanagiotou S, 2016. Pneumonia and Pleural Empyema due to a Mixed Lactobacillus spp. infection as a possible early esophageal carcinoma signature. Frontiers in Medicine, 3, 42–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charon M, Robin D and Marchand PA, 2019. The major interest for crop protection of agrochemical substances without maximum residue limit (MRL). Biotechnologie Agronomie Societe Et Environnement, 23, 22–29. [Google Scholar]
- Charsizadeh A, Mirhendi H, Nikmanesh B, Eshaghi H, Rahmani M, Farhang A, Bakhshi H and Makimura K, 2018a. Candidemia in children caused by uncommon species of Candida. Archives of Pediatric Infectious Diseases, 6, e11895. [Google Scholar]
- Charsizadeh A, Mirhendi H, Nikmanesh B, Eshaghi H and Makimura K, 2018b. Microbial epidemiology of candidaemia in neonatal and paediatric intensive care units at the Children's Medical Center, Tehran. Mycoses, 61, 22–29. 10.1111/myc.12698 [DOI] [PubMed] [Google Scholar]
- Charsizadeh A, Nikmanesh B, Ahmadi B, Jalalizand N, Jafari Z, Rahmani M, Kordbacheh P and Mirhendi H, 2018c. Frequency of Candida species isolated from patients in children's Medical Center, Tehran, Iran. Archives of Pediatric Infectious Diseases, 6, e62410 10.5812/pedinfect.11895 [DOI] [Google Scholar]
- Chen F, Zhang Z and Chen J, 2018. Infective endocarditis caused by Lactococcus lactis subsp. lactis and Pediococcus pentosaceus: a case report and literature review. Medicine (Baltimore), 97, e13658 10.1097/md.0000000000013658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chessa R, Landolfo S, Ciani M, Budroni M, Zara S, Ustun M, Cakar ZP and Mannazzu I, 2017. Biotechnological exploitation of Tetrapisispora phaffii killer toxin: heterologous production in Komagataella phaffii (Pichia pastoris). Applied Microbiology and Biotechnology, 101, 1–12. 10.1007/s00253-016-8050-2 [DOI] [PubMed] [Google Scholar]
- Chi Z, Hu B, Liu Y, Frear C, Wen Z and Chen S, 2007. Production of omega‐3 polyunsaturated fatty acids from cull potato using an algae culture process. Applied Biochemistry Biotechnology, 137–140, 805–815. 10.1007/s12010-007-9099-2 [DOI] [PubMed] [Google Scholar]
- Cleenwerck I, De Vos P and De Vuyst L, 2010. Phylogeny and differentiation of species of the genus Gluconacetobacter and related taxa based on multilocus sequence analyses of housekeeping genes and reclassification of Acetobacter xylinus subsp. sucrofermentans as Gluconacetobacter sucrofermentans (Toyosaki et al. 1996) sp. nov., comb. nov. International Journal of Systematic and Evolutionary Microbiology, 60, 2277–2283. 10.1099/ijs.0.018465-0 [DOI] [PubMed] [Google Scholar]
- Cohen SA, Woodfield MC, Boyle N, Stednick Z, Boeckh M and Pergam SA, 2016. Incidence and outcomes of bloodstream infections among hematopoietic cell transplant recipients from species commonly reported to be in over‐the‐counter probiotic formulations. Transplant Infectious Disease, 18, 699–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins MD, 1987. Transfer of Brevibacterium ammoniagenes (Cooke and Keith) to the Genus Corynebacterium as Corynebacterium ammoniagenes comb. nov. International Journal of Systematic and Evolutionary Microbiology, 37, 442–443. 10.1099/00207713-37-4-442 [DOI] [Google Scholar]
- Colombo AL, e Almeida Jr JN and Guinea J, 2017. Emerging multidrug‐resistant Candida species. Current Opinion in Infectious Diseases, 30, 528–538. [DOI] [PubMed] [Google Scholar]
- Cong QQ, Wang Y, Liu J, Lan YF, Guo ZK, Yang JG, Li XD and Tian YP, 2019. Evaluation of Potato virus X mild mutants for cross protection against severe infection in China. Virology Journal, 16, 36 10.1186/s12985-019-1143-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke JV and Keith HR, 1927. A type of urea‐splitting bacterium found in the human intestinal tract. Journal of Bacteriology, 13, 315–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coorevits A, Dinsdale AE, Halket G, Lebbe L, De Vos P, Van Landschoot A and Logan NA, 2012. Taxonomic revision of the genus Geobacillus: emendation of Geobacillus, G. stearothermophilus, G. jurassicus, G. toebii, G. thermodenitrificans and G. thermoglucosidans (nom. corrig., formerly ‘thermoglucosidasius’); transfer of Bacillus thermantarcticus to the genus as G. thermantarcticus comb. nov.; proposal of Caldibacillus debilis gen. nov., comb. nov.; transfer of G. tepidamans to Anoxybacillus as A. tepidamans comb. nov.; and proposal of Anoxybacillus caldiproteolyticus sp. nov. International Journal of Systematic and Evolutionary Microbiology, 62, 1470–1485. 10.1099/ijs.0.030346-0 [DOI] [PubMed] [Google Scholar]
- Crisafulli E, Aredano I, Valzano I, Burgazzi B, Andrani F and Chetta A, 2019. Pleuritis with pleural effusion due to a Bacillus megaterium infection. Respirology Case Reports, 7, e00381 10.1002/rcr2.381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custódio L, Justo T, Silvestre L, Barradas A, Duarte CV, Pereira H, Barreira L, Rauter AP, Alberício F and Varela J, 2012. Microalgae of different phyla display antioxidant, metal chelating and acetylcholinesterase inhibitory activities. Food Chemistry, 131, 134–140. 10.1016/j.foodchem.2011.08.047 [DOI] [Google Scholar]
- Daniel HM, Lachance MA and Kurtzman CP, 2014. On the reclassification of species assigned to Candida and other anamorphic ascomycetous yeast genera based on phylogenetic circumscription. Antonie van Leeuwenhoek, 106, 67–84. 10.1007/s10482-014-0170-z [DOI] [PubMed] [Google Scholar]
- Danilova IV, Toymentseva AA, Baranova DS and Sharipova MR, 2017. The genetic mechanism of resistance to antibiotics in Bacillus pumilus 3‐19 strain. Bionanoscience, 7, 88–91. [Google Scholar]
- Datta P, Gupta V, Mohi GK, Chander J and Janmeja AK, 2017. Lactobacillus coryniformis causing pulmonary infection in a patient with metastatic small cell carcinoma: case report and review of literature on lactobacillus pleuro‐pulmonary infections. Journal of Clinical and Diagnostic Research, 11, DE1–DE5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davarpanah E and Guilhermino L, 2019. Are gold nanoparticles and microplastics mixtures more toxic to the marine microalgae Tetraselmis chuii than the substances individually? Ecotoxicology and Environmental Safety, 181, 60–68. 10.1016/j.ecoenv.2019.05.078 [DOI] [PubMed] [Google Scholar]
- Debelius B, Forja JM, DelValls Á and Lubián LM, 2009. Toxicity and bioaccumulation of copper and lead in five marine microalgae. Ecotoxicology and Environmental Safety, 72, 1503–1513. 10.1016/j.ecoenv.2009.04.006 [DOI] [PubMed] [Google Scholar]
- Dent VE and Williams RAD, 1982. Lactobacillus animalis sp. nov., a new species of lactobacillus from the alimentary canal of animals. Zentralblatt für Bakteriologie Mikrobiologie und Hygiene: I. Abt. Originale C: Allgemeine, Angewandte und ökologische Mikrobiologie, 3, 377–386. 10.1016/S0721-9571(82)80018-5 [DOI] [Google Scholar]
- Doukyu N, Kuwahara H and Aono R, 2003. Isolation of Paenibacillus illinoisensis that produces cyclodextrin glucanotransferase resistant to organic solvents. Bioscience, Biotechnology, and Biochemistry, 67, 334–340. 10.1271/bbb.67.334 [DOI] [PubMed] [Google Scholar]
- Du H, Liao X, Gao Z, Li Y, Lei Y, Chen W, Chen L, Fan X, Zhang K, Chen S, Ma Y, Meng C and Li D, 2019. Effects of Methanol on Carotenoids as Well as Biomass and Fatty Acid Biosynthesis in Schizochytrium limacinum B4D1. Applied and Environmental Microbiology, 85, e01243‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebenezer TE, Zoltner M, Burrell A, Nenarokova A, Novák Vanclová AMG, Prasad B, Soukal P, Santana‐Molina C, O'Neill E, Nankissoor NN, Vadakedath N, Daiker V, Obado S, Silva‐Pereira S, Jackson AP, Devos DP, Lukeš J, Lebert M, Vaughan S, Hampl V, Carrington M, Ginger ML, Dacks JB, Kelly S and Field MC, 2019. Transcriptome, proteome and draft genome of Euglena gracilis. BMC Biology, 17, 11 10.1186/s12915-019-0626-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA (European Food Safety Authority), 2005. Opinion of the Scientific Committee on a request from EFSA related to a generic approach to the safety assessment by EFSA of microorganisms used in food/feed and the production of food/feed additives. EFSA Journal 2005;3 (6):226, 12 pp. 10.2903/j.efsa.2005.226 [DOI] [Google Scholar]
- EFSA (European Food Safety Authority), 2007. Introduction of a Qualified Presumption of Safety (QPS) approach for assessment of selected microorganisms referred to EFSA ‐ Opinion of the Scientific Committee. EFSA Journal 2007;5 (12):587, 16 pp. 10.2903/j.efsa.2007.587 [DOI] [Google Scholar]
- EFSA (European Food Safety Authority), 2008. The maintenance of the list of QPS microorganisms intentionally added to food or feed ‐ Scientific Opinion of the Panel on Biological Hazards. 2008;6 (12):923, 48 pp. 10.2903/j.efsa.2008.923 [DOI] [Google Scholar]
- EFSA (European Food Safety Authority), 2012a. Conclusion on the peer review of the pesticide risk assessment of the active substance Adoxophyes orana granulovirus. EFSA Journal 2012;10 (4):2654, 32 pp. 10.2903/j.efsa.2012.2654 [DOI] [Google Scholar]
- EFSA (European Food Safety Authority), 2012b. Conclusion on the peer review of the pesticide risk assessment of the active substance Cydia pomonella granulovirus (CpGV). EFSA Journal 2012;10 (4):2655, 40 pp. 2655 10.2903/j.efsa.2012.2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), 2009. Scientific Opinion on the maintenance of the list of QPS microorganisms intentionally added to food or feed (2009 update). EFSA Journal 2009;7 (12):1431, 92 pp. e01431 10.2903/j.efsa.2009.1431 [DOI] [Google Scholar]
- EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), 2013. Scientific Opinion on the maintenance of the list of QPS biological agents intentionally added to food and feed (2013 update). EFSA Journal 2013;11 (11):3449, 107 pp. e03449 10.2903/j.efsa.2013.3449 [DOI] [Google Scholar]
- EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), Ricci A, Allende A, Bolton D, Chemaly M, Davies R, Girones R, Herman L, Koutsoumanis K, Lindqvist R, Nørrung B, Robertson L, Ru G, Sanaa M, Simmons M, Skandamis P, Snary E, Speybroeck N, Ter Kuile B, Threlfall J, Wahlström H, Cocconcelli PS, Klein G, Prieto Maradona M, Querol A, Peixe L, Suarez JE, Sundh I, Vlak JM, Aguilera‐Gómez M, Barizzone F, Brozzi R, Correia S, Heng L, Istace F, Lythgo C and Fernández Escámez PS, 2017. Scientific Opinion on the update of the list of QPS‐recommended biological agents intentionally added to food or feed as notified to EFSA. EFSA Journal 2017;15 (3):4664, 178 pp. 10.2903/j.efsa.2017.4664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), Ricci A, Allende A, Bolton D, Chemaly M, Davies R, Girones R, Koutsoumanis K, Lindqvist R, Nørrung B, Robertson L, Ru G, Fernández Escámez PS, Sanaa M, Simmons M, Skandamis P, Snary E, Speybroeck N, Ter Kuile B, Threlfall J, Wahlström H, Cocconcelli PS, Peixe L, Maradona MP, Querol A, Suarez JE, Sundh I, Vlak J, Barizzone F, Correia S and Herman L, 2018a. Update of the list of QPS‐recommended biological agents intentionally added to food or feed as notified to EFSA 7: suitability of taxonomic units notified to EFSA until September 2017. EFSA Journal 2018;16 (1):5131, 43 pp. 10.2903/j.efsa.2018.5131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), Ricci A, Allende A, Bolton D, Chemaly M, Davies R, Fernández Escámez PS, Girones R, Koutsoumanis K, Lindqvist R, Nørrung B, Robertson L, Ru G, Sanaa M, Simmons M, Skandamis P, Snary E, Speybroeck N, Ter Kuile B, Threlfall J, Wahlström H, Cocconcelli PS, Peixe L, Maradona MP, Querol A, Suarez JE, Sundh I, Vlak J, Barizzone F, Correia S and Herman L, 2018b. Update of the list of QPS‐recommended biological agents intentionally added to food or feed as notified to EFSA 8: suitability of taxonomic units notified to EFSA until March 2018. EFSA Journal 2018;16(7):5315, 42 pp. 10.2903/j.efsa.2018.5315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), Koutsoumanis K, Allende A, Álvarez‐Ordóñez A, Bolton D, Bover‐Cid S, Chemaly M, Davies R, Hilbert F, Lindqvist R, Nauta M, Peixe L, Ru G, Simmons M, Skandamis P, Suffredini E, Cocconcelli PS, Fernández Escámez PS, Maradona MP, Querol A, Suarez JE, Sundh I, Vlak J, Barizzone F, Correia S and Herman L, 2019a. Update of the list of QPS‐recommended biological agents intentionally added to food or feed as notified to EFSA 9: suitability of taxonomic units notified to EFSA until September 2018. EFSA Journal 2019;17 (1):5555, 46 pp. 10.2903/j.efsa.2019.5555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), Koutsoumanis K, Allende A, Alvarez‐Ordóñez A, Bolton D, Bover‐Cid S, Chemaly M, Davies R, De Cesare A, Hilbert F, Lindqvist R, Nauta M, Peixe L, Ru G, Simmons M, Skandamis P, Suffredini E, Cocconcelli PS, Fernández Escámez PS, Maradona MP, Querol A, Suarez JE, Sundh I, Vlak J, Barizzone F, Correia S and Herman L, 2019b. Update of the list of QPS‐recommended biological agents intentionally added to food or feed as notified to EFSA 10: suitability of taxonomic units notified to EFSA until March 2019. EFSA Journal 2019;17 (7):5753, 79 pp. 10.2903/j.efsa.2019.5753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), Koutsoumanis K, Allende A, Alvarez‐Ordóñez A, Bolton D, Bover‐Cid S, Chemaly M, Davies R, De Cesare A, Hilbert F, Lindqvist R, Nauta M, Peixe L, Ru G, Simmons M, Skandamis P, Suffredini E, Cocconcelli PS, Fernández Escámez PS, Maradona MP, Querol A, Suarez JE, Sundh I, Vlak JM, Barizzone F, Correia S and Herman L, 2020. Scientific Opinion on the update of the list of QPS‐recommended biological agents intentionally added to food or feed as notified to EFSA (2017‐2019). EFSA Journal 2020;18(2):5966, 10.2903/j.efsa.2020.5966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA FEEDAP Panel (EFSA Panel on Additives and Products or Substances used in Animal Feed), Rychen G, Aquilina G, Azimonti G, Bampidis V, Bastos ML, Bories G, Chesson A,Cocconcelli PS, Flachowsky G, Gropp J, Kolar B, Kouba M, López‐Alonso M, López Puente S, Mantovani A,Mayo B, Ramos F, Saarela M, Villa RE, Wallace RJ, Wester P, Glandorf B, Herman L, Kärenlampi S,Aguilera J, Anguita M, Brozzi R and Galobart J, 2018. Guidance on the characterisation of microorganisms used as feed additives or as production organisms. EFSA Journal 2018;16(3):5206, 24 pp. 10.2903/j.efsa.2018.5206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elikowski W, Malek‐Elikowska M, Lisiecka M, Bodora A, Wisniewska K and Oko‐Sarnowska Z, 2017. Lactobacillus gasseri endocarditis on the aortic valve bioprosthesis ‐ a case report. Polski merkuriusz lekarski: organ Polskiego Towarzystwa Lekarskiego, 43, 220–223. [PubMed] [Google Scholar]
- Endo A and Okada S, 2007. Lactobacillus farraginis sp. nov. and Lactobacillus parafarraginis sp. nov., heterofermentative lactobacilli isolated from a compost of distilled shochu residue. The International Journal of Systematic and Evolutionary Microbiology, 57, 708–712. 10.1099/ijs.0.64618-0 [DOI] [PubMed] [Google Scholar]
- Esaiassen E, Cavanagh P, Hjerde E, Simonsen GS, Stoen R and Klingenberg C, 2016. Bifidobacterium longum subspecies infantis bacteremia in 3 extremely preterm infants receiving probiotics. Emerging Infectious Diseases, 22, 1664–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esaiassen E, Hjerde E, Cavanagh JP, Simonsen GS and Klingenberg C and Norwegian Study Group on Invasive Bifidobacterial Infections , 2017. Bifidobacterium bacteremia ‐ clinical characteristics and a genomic approach to assess pathogenicity. Journal of Clinical Microbiology, 55, 2234–2248. 10.1128/jcm.00150-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinel‐Ingroff A, Turnidge J, Alastruey‐Izquierdo A, Botterel F, Canton E, Castro C, Chen YC, Chen Y, Chryssanthou E, Dannaoui E, Garcia‐Effron G, Gonzalez GM, Govender NP, Guinea J, Kidd S, Lackner M, Lass‐Flörl C, Linares‐Sicilia MJ, López‐Soria L, Magobo R, Pelaez T, Quindós G, Rodriguez‐Iglesia MA, Ruiz MA, Sánchez‐Reus F, Sanguinetti M, Shields R, Szweda P, Tortorano A, Wengenack NL, Bramati S, Cavanna C, DeLuca C, Gelmi M, Grancini A, Lombardi G, Meletiadis J, Negri CE, Passera M, Peman J, Prigitano A, Sala E and Tejada M, 2019. Method‐dependent epidemiological cutoff values for detection of triazole resistance in Candida and Aspergillus species for the sensititre yeastone colorimetric broth and etest agar diffusion methods. Antimicrobial Agents and Chemotherapy, 63, e01651–01618. 10.1128/AAC.01651-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquibel A, Dababneh AS and Palraj BR, 2017. Lactobacillus gasseri Causing Bilateral Empyema. Case Reports in Infectious Diseases, 2017, 4895619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ethier S, Woisard K, Vaughan D and Wen Z, 2011. Continuous culture of the microalgae Schizochytrium limacinum on biodiesel‐derived crude glycerol for producing docosahexaenoic acid. Bioresource Technology, 102, 88–93. 10.1016/j.biortech.2010.05.021 [DOI] [PubMed] [Google Scholar]
- Farmakiotis D and Kontoyiannis DP, 2017. Epidemiology of antifungal resistance in human pathogenic yeasts: current viewpoint and practical recommendations for management. International Journal of Antimicrobial Agents, 50, 318–324. [DOI] [PubMed] [Google Scholar]
- Felekos I, Lazaros G, Tsiriga A, Pirounaki M, Stavropoulos G, Paraskevas J, Toutouza M and Tousoulis D, 2016. Lactobacillus rhamnosus endocarditis: an unusual culprit in a patient with Barlow's disease. Hellenic Journal of Cardiology: HJC = Hellenike kardiologike epitheorese, 57, 445–448. [DOI] [PubMed] [Google Scholar]
- Fernandez‐Ruiz M, Guinea J, Puig‐Asensio M, Zaragoza O, Almirante B, Cuenca‐Estrella M, Aguado JM, Candipop Project G and Gemicomed R, 2017. Fungemia due to rare opportunistic yeasts: data from a population‐based surveillance in Spain. Medical Mycology, 55, 125–136. [DOI] [PubMed] [Google Scholar]
- Florez AB and Mayo B, 2017. Antibiotic resistance‐susceptibility profiles of streptococcus thermophilus isolated from raw milk and genome analysis of the genetic basis of acquired resistances. Frontiers in Microbiology, 8, 2608 10.3389/fmicb.2017.02608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong JC, Svenson CJ, Nakasugi K, Leong CT, Bowman JP, Chen B, Glenn DR, Neilan BA and Rogers PL, 2006. Isolation and characterization of two novel ethanol‐tolerant facultative‐anaerobic thermophilic bacteria strains from waste compost. Extremophiles, 10, 363–372. 10.1007/s00792-006-0507-2 [DOI] [PubMed] [Google Scholar]
- Fragkiadakis K, Ioannou P, Barbounakis E and Samonis G, 2017. Intra‐abdominal abscesses by Lactococcus lactis ssp cremoris in an immunocompetent adult with severe periodontitis and pernicious anemia. IDCases, 7, 27–29. 10.1016/j.idcr.2016.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco‐Cendejas R, Colin‐Castro CA, Hernandez‐Duran M, Lopez‐Jacome LE, Ortega‐Pena S, Ceron‐Gonzalez G, Vanegas‐Rodriguez S, Mondragon‐Eguiluz JA and Acosta‐Rodriguez E, 2017. Leuconostoc mesenteroides periprosthetic knee infection, an unusual fastidious Gram‐positive bacteria: a case report. BMC Infectious Diseases, 17, 227 10.1186/s12879-017-2315-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas AR, Tedim AP, Novais C, Coque TM and Peixe L, 2018. Distribution of putative virulence markers in Enterococcus faecium: towards a safety profile review. Journal of Antimicrobial Chemotherapy, 73, 306–319. 10.1093/jac/dkx387 [DOI] [PubMed] [Google Scholar]
- Gachara S and Wisser RJ, 2018. Synthetic biology for plant viral diagnostics: application to maize lethal necrosis disease. Phytopathology, 108, 15.28876210 [Google Scholar]
- Galimany E, Rose JM, Alix J, Dixon MS and Wikfors GH, 2014. Responses of the ribbed mussel, Geukensia demissa, to the harmful algae Aureococcus anophagefferens and Heterosigma akashiwo. Journal of Molluscan Studies, 80, 123–130. 10.1093/mollus/eyt055 [DOI] [Google Scholar]
- Gan HM, 2019. Commentary: complete genome sequence of 3‐chlorobenzoate‐degrading bacterium cupriavidus necator NH9 and reclassification of the strains of the genera cupriavidus and ralstonia based on phylogenetic and whole‐genome sequence analyses. Frontiers in Microbiology, 10 10.3389/fmicb.2019.02011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia Carretero R, Regodon Dominguez M, Ruiz Bastian M and Lopez Lomba M, 2018. Lactobacillus salivarius infection as a postoperative complication after bariatric surgery. Enfermedades Infecciosas Y Microbiologia Clinica, 36, 60–61. [DOI] [PubMed] [Google Scholar]
- Garcia‐Ramon DC, Berry C, Tse C, Fernandez‐Fernandez A, Osuna A and Vilchez S, 2017. The parasporal crystals of Bacillus pumilus strain 15.1: a potential virulence factor? Microbial Biotechnology, 11, 302–316. 10.1111/1751-7915.12771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgountzos G, Michopoulos C, Grivokostopoulos C, Kolosaka M, Vlassopoulou N and Lekkou A, 2018. Infective endocarditis in a young adult due to Lactococcus lactis: a case report and review of the literature. Case Reports in Medicine, 5091456 10.1155/2018/5091456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghajari A, Lotfali E, Ahmadi NA, Fassihi PN, Shahmohammadi N, Ansari S, Norouzi M and Arab‐Mazar Z, 2018. Isolation of different species of candida in patients with vulvovaginal candidiasis from Damavand, Iran. Archives of Clinical Infectious Diseases, 13, e59291 10.5812/archcid.59291 [DOI] [Google Scholar]
- Gifuni I, Olivieri G, Pollio A and Marzocchella A, 2018. Identification of an industrial microalgal strain for starch production in biorefinery context: the effect of nitrogen and carbon concentration on starch accumulation. New Biotechnology, 41, 46–54. 10.1016/j.nbt.2017.12.003 [DOI] [PubMed] [Google Scholar]
- Giok FX, 2016. Antimicrobial resistance in direct‐fed microbial preparations used in cattle. Master of Science thesis, Department of Diagnostic Medicine and Pathobiology, College of Veterinary Medicine. Kansas State University; 91 pp. [Google Scholar]
- Glenwright H, Pohl S, Navarro F, Miro E, Jimenez G, Blanch AR and Harwood CR, 2017. The identification of intrinsic chloramphenicol and tetracycline resistance genes in members of the Bacillus Cereus group (sensu lato). Frontiers in Microbiology, 7, 2122 10.3389/fmicb.2016.02122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González MA, Aguayo PA, Inostroza I, Castro PA, Fuentes GA and Gómez PI, 2015. Ultrastructural and molecular characterization of Tetraselmis strains (Chlorodendrophyceae, Chlorophyta) isolated from Chile. Gayana ‐ Botanica, 72, 47–57. 10.4067/S0717-66432015000100007 [DOI] [Google Scholar]
- Groenewald M, Boekhout T, Neuvéglise C, Gaillardin C, van Dijck PWM and Wyss M, 2014. Yarrowia lipolytica: safety assessment of an oleaginous yeast with a great industrial potential. Critical Reviews in Microbiology, 40, 187–206. 10.3109/1040841X.2013.770386 [DOI] [PubMed] [Google Scholar]
- Gu H‐J, Sun Q‐L, Luo J‐C, Zhang J and Sun L, 2019. A first study of the virulence potential of a Bacillus Subtilis isolate from deep‐sea hydrothermal vent. Frontiers in Cellular and Infection Microbiology, 9 10.3389/fcimb.2019.00183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghighat L and Crum‐Cianflone NF, 2016. The potential risks of probiotics among HIV‐infected persons: Bacteraemia due to Lactobacillus acidophilus and review of the literature. International Journal of STD & AIDS, 27, 1223–1230. [DOI] [PubMed] [Google Scholar]
- Han A, Mehta J and Pauly RR, 2016. Septic shock secondary to a urinary tract infection with Pediococcus Pentosaceus. Missouri Medicine, 113, 179–181. [PMC free article] [PubMed] [Google Scholar]
- Harding‐Theobald E and Maraj B, 2018. Spontaneous bacterial peritonitis due to Lactobacillus paracasei in Cirrhosis. Case Reports in Gastrointestinal Medicine. 10.1155/2018/5714053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes M, Stanton C, Slattery H, Sullivan O, Hill C, Fitzgerald GF and Ross RP, 2007. Casein fermentate of Lactobacillus animalis; DPC6134 contains a range of novel propeptide angiotensin‐converting enzyme inhibitors. Applied and Environmental Microbiology, 73, 4658 10.1128/AEM.00096-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen KC, 1995. New and emerging yeast pathogens. Clinical Microbiology Reviews, 8, 462 10.1128/CMR.8.4.462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J, Jolliff JC and Heidari A, 2016. Antibiotic lock therapy for leuconostoc pseudomesenteroides catheter‐related bacteremia. American Journal of the Medical Sciences, 352, 229–230. [DOI] [PubMed] [Google Scholar]
- Holland ATN, Danson MJ and Bolhuis A, 2019. Inhibition of extracellular proteases improves the production of a xylanase in Parageobacillus thermoglucosidasius. BMC Biotechnology, 19, 17 10.1186/s12896-019-0511-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Yang X, Wu Q, Li L, Wu Y, Chen S, Li R and Ren J, 2019. Purification and identification of antioxidant peptides from schizochytrium limacinum hydrolysates by consecutive chromatography and electrospray ionization‐mass spectrometry. Molecules, 24 10.3390/molecules24163004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard J, Jariwala B, Hill A, Gega A and Palesty JA, 2018. A new Bacterium, Lactobacillus acidophilus, Causing Necrotizing Fasciitis. American Surgeon, 84, e61–e63. [PubMed] [Google Scholar]
- Humhal T, Kastanek P, Jezkova Z, Cadkova A, Kohoutkova J and Branyik T, 2017. Use of saline waste water from demineralization of cheese whey for cultivation of Schizochytrium limacinum PA‐968 and Japonochytrium marinum AN‐4. Bioprocess and Biosystems Engineering, 40, 395–402. 10.1007/s00449-016-1707-5 [DOI] [PubMed] [Google Scholar]
- van Hung N, Bossier P, Hong NTX, Ludeseve C, Garcia‐Gonzalez L, Nevejan N and De Schryver P, 2019. Does Ralstonia eutropha, rich in poly‐? hydroxybutyrate (PHB), protect blue mussel larvae against pathogenic vibrios? Journal of Fish Diseases, 4, 777‐787. 10.1111/jfd.12981 [DOI] [PubMed]
- ICTV (International Committee on Taxonomy of Viruses), 2018. The ICTV Report #2018b. Virus Taxonomy: The Classification and Nomenclature of Viruses. Available online: https://talk.ictvonline.org/ictv-reports/ictv_online_report/
- Ino K, Nakase K, Suzuki K, Nakamura A, Fujieda A and Katayama N, 2016. Bacteremia due to Leuconostoc pseudomesenteroides in a patient with acute Lymphoblastic Leukemia: case report and review of the literature. Case Reports in Hematology, 2016, 7648628–7648628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwazaki S, Hirai H, Hamaguchi N and Yoshida N, 2018. Isolation of levoglucosan‐utilizing thermophilic bacteria. Scientific Reports, 8, 4066 10.1038/s41598-018-22496-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahanshiri Z, Manifar S, Moosa H, Asghari‐Paskiabi F, Mahmoodzadeh H, Shams‐Ghahfarokhi M and Razzaghi‐Abyaneh M, 2018. Oropharyngeal candidiasis in head and neck cancer patients in Iran: species identification, antifungal susceptibility and pathogenic characterization. Journal de Mycologie Médicale, 28, 361–366. [DOI] [PubMed] [Google Scholar]
- Jeong D‐W, Jeong M and Lee J‐H, 2017. Antibiotic susceptibilities and characteristics of Bacillus licheniformis isolates from traditional Korean fermented soybean foods. LWT, 75, 565–568. 10.1016/j.lwt.2016.10.001 [DOI] [Google Scholar]
- Jhala YK, Vyas RV, Shelat HN, Patel HK, Patel HK and Patel KT, 2014. Isolation and characterization of methane utilizing bacteria from wetland paddy ecosystem. World Journal of Microbiology & Biotechnology, 30, 1845–1860. 10.1007/s11274-014-1606-3 [DOI] [PubMed] [Google Scholar]
- Ji X‐J, Mo K‐Q, Ren L, Li G‐L, Huang J‐Z and Huang H, 2015. Genome sequence of Schizochytrium sp. CCTCC M209059, an effective producer of docosahexaenoic acid‐rich lipids. Genome Announcements, 3 10.1128/genomea.00819-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi S, Udani S, Sen S, Kirolikar S and Shetty A, 2019. Bacillus Clausii septicemia in a pediatric patient after treatment with probiotics. The Pediatric infectious disease journal, 38, e228–e230. 10.1097/INF.0000000000002350 [DOI] [PubMed] [Google Scholar]
- Jung W‐J, Mabood F, Souleimanov A, Park R‐D and Smith DL, 2008. Chitinases produced by Paenibacillus illinoisensis and Bacillus thuringiensis subsp. pakistani degrade Nod factor from Bradyrhizobium japonicum. Microbiological Research, 163, 345–349. 10.1016/j.micres.2006.06.013 [DOI] [PubMed] [Google Scholar]
- Jung J, Moon YS, Yoo JA, Lim J‐H, Jeong J and Jun J‐B, 2017. Investigation of a nosocomial outbreak offungemia caused by Candida pelliculosa (Pichia anomala) in a Korean tertiary care center. Journal of Microbiology, Immunology, and Infection, 51, 794–801. 10.1016/j.jmii.2017.05.005 [DOI] [PubMed] [Google Scholar]
- Kabore WAD, Dembele R, Bagre TS, Konate A, Boisrame S, Chevalier V, Konsem T, Traore AS and Barro N, 2018. Characterization and antimicrobial susceptibility of Lactococcus lactis isolated from endodontic infections in Ouagadougou, Burkina Faso. Dentistry Journal, 6, pii:E69. 10.3390/dj6040069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane AF, Bhatia AD, Denning PW, Shane AL and Patel RM, 2018. Routine supplementation of Lactobacillus rhamnosus GG and risk of necrotizing enterocolitis in very low birth weight infants. Journal of Pediatrics, 195, 73–79.e2. 10.1016/j.jpeds.2017.11.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao B‐Z, Lin H‐J, Chen M‐Y, Wu C‐S, Lin S‐T, Lee M‐H, Lai Y‐X and Hu P‐J, 2018. Lactobacillus paracasei as cause of liver abscess: case report. Journal of Gastroenterology and Hepatology, 33, 433–433. [Google Scholar]
- Karapetsa M, Tsolaki V, Arabatzis M, Petinaki E, Velegraki A and Zakynthinos E, 2019. Septic shock due to Candida famata (Debaryomyces hansenii) candidemia in an ICU immunocompetent trauma‐patient. Journal of Infection and Public Health, 12, 594–597. 10.1016/j.jiph.2018.12.015 [DOI] [PubMed] [Google Scholar]
- Karbuz A, Kocabas BA, Yalman A, Kuloglu Z, Aysev AD, Ciftci E and Ince E, 2017. Catheter related leuconostoc mesenteroides bacteremia: a rare case and review of the literature. Journal of Pediatric Research, 4, 35–38. [Google Scholar]
- Karstrup CC, Aalbaek B, Klitgaard K, Jensen TK, Pedersen HG and Agerholm JS, 2017. Colonization of the bovine uterus by Candida kefyr. Acta Veterinaria Scandinavica, 59, 61 10.1186/s13028-017-0329-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Funabashi N, Takaoka H, Kohno H, Kishimoto T, Nakatani Y, Matsumiya G and Kobayashi Y, 2016. Lactobacillus paracasei endocarditis in a consumer of probiotics with advanced and severe bicuspid aortic valve stenosis complicated with diffuse left ventricular mid‐layer fibrosis. International Journal of Cardiology, 224, 157–161. [DOI] [PubMed] [Google Scholar]
- Kim G‐Y, Jeon J‐S and Kim JK, 2016. Isolation frequency characteristics of Candida species from clinical specimens. Mycobiology, 44, 99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MK, Yoon HY, Lee MH and Kim JH, 2018. Canine pyometra associated with Bacillus species: a case report. Veterinarni Medicina, 63, 143–149. [Google Scholar]
- Koizumi S, Yonetani Y, Maruyama A and Teshiba S, 2000. Production of riboflavin by metabolically engineered Corynebacterium ammoniagenes. Applied Microbiology and Biotechnology, 53, 674–679. [DOI] [PubMed] [Google Scholar]
- Koyama S, Fujita H, Shimosato T, Kamijo A, Ishiyama Y, Yamamoto E, Ishii Y, Hattori Y, Hagihara M, Yamazaki E, Tomita N and Nakajima H and Yokohama Cooperative Study Group for Hematology (YACHT) , 2018. Septicemia from Lactobacillus rhamnosus GG, from a probiotic enriched Yogurt, in a patient with autologous stem cell transplantation. Probiotics and Antimicrobial Proteins, 11, 295–298. 10.1007/s12602-018-9399-6 [DOI] [PubMed] [Google Scholar]
- Krajčovič J, Matej V and Schwartzbach SD, 2015. Euglenoid flagellates: a multifaceted biotechnology platform. Journal of Biotechnology, 202, 135–145. 10.1016/j.jbiotec.2014.11.035 [DOI] [PubMed] [Google Scholar]
- Kralik Z, Kralik G, Grčević M, Hanžek D and Margeta P, 2019. Microalgae Schizochytrium limacinum as an alternative to fish oil in enriching table eggs with n‐3 polyunsaturated fatty acids. Journal of the Science of Food and Agriculture, n/a. 10.1002/jsfa.10052 [DOI] [PubMed] [Google Scholar]
- Kumar A, Roy P, Rai G, Das S and Ansari MA, 2017. Cyberlindnera fabianii and Wickerhamomyces anomalous fungemia in newborns: an experience from a North Indian tertiary‐care centre. Indian Journal of Medical Specialities, 8, 131–133. [Google Scholar]
- Kunasundari B, Murugaiyah V, Kaur G, Maurer FH and Sudesh K, 2013. Revisiting the single cell protein application of Cupriavidus necator H16 and recovering bioplastic granules simultaneously. PLoS One, 8:e78528. 10.1371/journal.pone.0078528 [DOI] [PMC free article] [PubMed]
- Kurtzman CP, 2005. Description of Komagataella phaffii sp. nov. and the transfer of Pichia pseudopastoris to the methylotrophic yeast genus Komagataella. International Journal of Systematic and Evolutionary Microbiology, 55, 973–976. 10.1099/ijs.0.63491-0 [DOI] [PubMed] [Google Scholar]
- Kurtzman CP, Fell JW and Boekhout T, 2011. The Yeasts, A Taxonomic Study, 5th Edition Elsevier, London: 2354 pp. [Google Scholar]
- Kushwaha SK, Vetukuri RR and Grenville‐Briggs LJ, 2017. Draft genome sequence of the mycoparasitic oomycete Pythium oligandrum strain CBS 530.74. Genome Announcements, 5, e00346‐17. 10.1128/genomea.00346-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey LA, 2017. Entomopathogens Used as Microbial Control Agents. Microbial Control of Insect and Mite Pests: From Theory to Practice, 3–12. [Google Scholar]
- Lee HJ and Hur Sb, 2009. Genetic relationships among multiple strains of the genus tetraselmis based on partial 18S rDNA sequences. Algae, 24, 205–212. 10.4490/ALGAE.2009.24.4.205 [DOI] [Google Scholar]
- Lee YS, Zhou Y, Park DJ, Chang J and Choi YL, 2013. beta‐cyclodextrin production by the cyclodextrin glucanotransferase from Paenibacillus illinoisensis ZY‐08: cloning, purification, and properties. World Journal of Microbiology & Biotechnology, 29, 865–873. 10.1007/s11274-012-1241-9 [DOI] [PubMed] [Google Scholar]
- Li C, Li XX, Xiao XM, Di Zhai S, Shi LW and Ge QG, 2019. Five cases of Bacillus Licheniformis bacteremia in immunocompetent hosts receiving Bacillus Licheniformis Capsules: the safety of probiotics in critical patients is worthy of further disscussion. Gastroenterology, 156, S460. [Google Scholar]
- Liang Y, Garcia RA, Piazza GL and Wen Z, 2011. Nonfeed Application of Rendered Animal Proteins for Microbial Production of Eicosapentaenoic Acid by the Fungus Pythium irregulare . Journal of Agricultural and Food Chemistry, 59, 11990–11996. 10.1021/jf2031633 [DOI] [PubMed] [Google Scholar]
- Liu D, Yang Q, Ge K, Hu X, Qi G, Du B, Liu K and Ding Y, 2017a. Promotion of iron nutrition and growth on peanut by Paenibacillus illinoisensis and Bacillus sp. strains in calcareous soil. Brazilian Journal of Microbiology, 48, 656–670. 10.1016/j.bjm.2017.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W‐L, Lai C‐C, Li M‐C, Wu C‐J, Ko W‐C, Hung Y‐L, Tang H‐J and Hsueh P‐R, 2017b. Clinical manifestations of candidemia caused by uncommon Candida species and antifungal susceptibility of the isolates in a regional hospital in Taiwan, 2007–2014. Journal of Microbiology, Immunology, and Infection, 52, 612–619. 10.1016/j.jmii.2017.08.007 [DOI] [PubMed] [Google Scholar]
- Lo H‐J, Tsai S‐H, Chu W‐L, Chen Y‐Z, Zhou Z‐L, Chen H‐F, Lee C‐F and Yang Y‐L, 2017. Fruits as the vehicle of drug resistant pathogenic yeasts. Journal of Infection, 75, 254–262. [DOI] [PubMed] [Google Scholar]
- Maciel‐Vergara G and Ros VID, 2017. Viruses of insects reared for food and feed. Journal of Invertebrate Pathology, 147, 60–75. [DOI] [PubMed] [Google Scholar]
- Maillet F, Passeron A, Podglajen I, Ranque B and Pouchot J, 2018. Lactobacillus delbrueckii urinary tract infection in a male patient. Médecine et Maladies Infectieuses, 49, 226–228. 10.1016/j.medmal.2018.11.006 [DOI] [PubMed] [Google Scholar]
- Makkar NS and Casida LE, 1987. Cupriavidus necator gen. nov., sp. nov.; a Nonobligate Bacterial Predator of Bacteria in Soil. International Journal of Systematic and Evolutionary Microbiology, 37, 323–326. 10.1099/00207713-37-4-323 [DOI] [Google Scholar]
- Mansour B, Habib A, Asli N, Geffen Y, Miron D and Elias N, 2016. A case of infective endocarditis and pulmonary septic emboli caused by Lactococcus lactis. Case Reports in Pediatrics, 2016, 1024054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantecón L, Moyano R, Cameán AM and Jos A, 2019. Safety assessment of a lyophilized biomass of Tetraselmis chuii (TetraSOD®) in a 90‐day feeding study. Food and Chemical Toxicology, 133, 110810 10.1016/j.fct.2019.110810 [DOI] [PubMed] [Google Scholar]
- Marc J, Grousseau E, Lombard E, Sinskey AJ, Gorret N and Guillouet SE, 2017. Over expression of GroESL in Cupriavidus necator for heterotrophic and autotrophic isopropanol production. Metabolic Engineering, 42, 74–84. 10.1016/j.ymben.2017.05.007 [DOI] [PubMed] [Google Scholar]
- Martinez N, Luque R, Milani C, Ventura M, Banuelos O and Margolles A, 2018. A gene homologous to rRNA methylase genes confers erythromycin and clindamycin resistance in bifidobacterium breve. Applied and Environmental Microbiology, 84, pii:e02888–17. 10.1128/AEM.02888-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda K, Koya J, Toyama K, Ikeda M, Arai S, Nakamura F, Okugawa S, Moriya K and Kurokawa M, 2017. A therapeutic benefit of daptomycin against glycopeptide‐resistant gram‐positive cocci bloodstream infections under neutropenia. Journal of Infection and Chemotherapy, 23, 788–790. [DOI] [PubMed] [Google Scholar]
- McManus GB, Liu W, Cole RA, Biemesderfer D and Mydosh JL, 2018. Strombidium rassoulzadegani: a model species for chloroplast retention in oligotrich ciliates. Frontiers in Marine Science, 5 10.3389/fmars.2018.00205 [DOI] [Google Scholar]
- Meyer SA and Yarrow D, 1998. Validation of the names of three Candida species. Mycotaxon, 66, 99–101. [Google Scholar]
- Miceli MH, Diaz JA and Lee SA, 2011. Emerging opportunistic yeast infections. The Lancet Infectious Diseases, 11, 142–151. 10.1016/s1473-3099(10)70218-8 [DOI] [PubMed] [Google Scholar]
- Mohr T, Aliyu H, Kuchlin R, Zwick M, Cowan D, Neumann A and de Maayer P, 2018. Comparative genomic analysis of Parageobacillus thermoglucosidasius strains with distinct hydrogenogenic capacities. BMC Genomics, 19, 880 10.1186/s12864-018-5302-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinaro M, Aiazzi M, La Torre A, Cini E and Banfi R, 2016. Lactobacillus Rhamnosus sepsis in a preterm infant associated with probiotic integrator use: a case report. Recenti Progressi in Medicina, 107, 485–486. [DOI] [PubMed] [Google Scholar]
- Morio F, Jensen RH, Le Pape P and Arendrup MC, 2017. Molecular basis of antifungal drug resistance in yeasts. International Journal of Antimicrobial Agents, 50, 599–606. 10.1016/j.ijantimicag.2017.05.012 [DOI] [PubMed] [Google Scholar]
- Moriuchi R, Dohra H, Kanesaki Y and Ogawa N, 2019. Complete genome sequence of 3‐chlorobenzoate‐degrading bacterium cupriavidus necator nh9 and reclassification of the strains of the genera cupriavidus and ralstonia based on phylogenetic and whole‐genome sequence analyses. Frontiers in Microbiology, 10 10.3389/fmicb.2019.00133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai R and Yoshida N, 2016. Effect of doping with metals, silicate, and phosphate ions on fluorescence properties and morphology of calcite single crystals synthesized in geobacillus thermoglucosidasius parent colonies. Journal of Microbial and Biochemical Technology, 8, 100–106. 10.4172/1948-5948.1000270 [DOI] [Google Scholar]
- Mussano F, Ferrocino I, Gavrilova N, Genova T, Dell'Acqua A, Cocolin L and Carossa S, 2018. Apical periodontitis: preliminary assessment of microbiota by 16S rRNA high throughput amplicon target sequencing. Bmc Oral Health, 18, 55 10.1186/s12903-018-0520-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy F, Bozo A, Toth Z, Daroczi L, Majoros L and Kovacs R, 2018. In vitro antifungal susceptibility patterns of planktonic and sessile Candida kefyr clinical isolates. Medical Mycology, 56, 493–500. [DOI] [PubMed] [Google Scholar]
- Nam S‐H, Choi S‐H, Kang A, Kim D‐W, Kim RN, Kim A, Kim D‐S and Park H‐S, 2011. Genome sequence of Lactobacillus animalis; KCTC 3501. Journal of Bacteriology, 193, 1280 10.1128/JB.01505-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan H, Chen H, Tuite MF and Xu X, 2019. A viral expression factor behaves as a prion. Nature Communications, 10, 359 10.1038/s41467-018-08180-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi SSB, Nagendra V and Hofmeyr A, 2018. Probiotic related Lactobacillus rhamnosus endocarditis in a patient with liver cirrhosis. IDCases, 13, e00439 10.1016/j.idcr.2018.e00439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumov G, Naumova E, Tyurin O and Kozlov D, 2013. Komagataella kurtzmanii sp. nov., a new sibling species of Komagataella (Pichia) pastoris based on multigene sequence analysis. Antonie van Leeuwenhoek, 104 10.1007/s10482-013-9956-7 [DOI] [PubMed] [Google Scholar]
- Nayeem M, Firdous N, Dang M‐T, Hafeez W and Arsene C, 2018. When the good becomes the bad: a case of lactobacillus rhamnosus septicemia unrelated to probiotic use. American Journal of Gastroenterology, 113, S1220–S1220. [Google Scholar]
- Nguyen HV, Gaillardin C and Neuveglise C, 2009. Differentiation of Debaryomyces hansenii and Candida famata by rRNA gene intergenic spacer fingerprinting and reassessment of phylogenetic relationships among D. hansenii, C. famata, D. fabryi, C. flareri (=D. subglobosus) and D. prosopidis: description of D. vietnamensis sp. nov. closely related to D. nepalensis. FEMS Yeast Research, 9, 641–662. 10.1111/j.1567-1364.2009.00510.x [DOI] [PubMed] [Google Scholar]
- Norena I, Cabrera‐Marante O and Fernandez‐Ruiz M, 2017. Endocarditis due to Lactobacillus rhamnosus in a patient with bicuspid aortic valve: potential role for the consumption of probiotics? Medicina Clinica, 149, 181–182. 10.1016/j.medcli.2017.03.021 [DOI] [PubMed] [Google Scholar]
- van Oers MM, Bateman KS and Stentiford GD, 2017. Viruses of invertebrates related to the food‐chain. Journal of Invertebrate Pathology, 147, 1–3. [DOI] [PubMed] [Google Scholar]
- Oliveira A, Oliveira LC, Aburjaile F, Benevides L, Tiwari S, Jamal SB, Silva A, Figueiredo HCP, Ghosh P, Portela RW, De Carvalho Azevedo VA and Wattam AR, 2017. Insight of genus corynebacterium: ascertaining the role of pathogenic and non‐pathogenic species. Frontiers in Microbiology, 8, 1937 10.3389/fmicb.2017.01937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren A and Garrity GM, 2019. Notification that new names of prokaryotes, new combinations, and new taxonomic opinions have appeared in volume 69, part 3 of the IJSEM. International Journal of Systematic and Evolutionary Microbiology, 69, 1529–1530. 10.1099/ijsem.0.003174 [DOI] [PubMed] [Google Scholar]
- Osman KM, Kappell AD, Orabi A, Al‐Maary KS, Mubarak AS, Dawoud TM, Hemeg HA, Moussa IMI, Hessain AM, Yousef HMY and Hristova KR, 2018. Poultry and beef meat as potential seedbeds for antimicrobial resistant enterotoxigenic Bacillus species: a materializing epidemiological and potential severe health hazard. Scientific Reports, 8, 11600 10.1038/s41598-018-29932-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pailhoriès H, Sanderink D, Abgueguen P and Lemarie C, 2017. A neglected pathogen responsible for deep infections: a case report of spondylodiscitis due to Lactobacillus sp. Médecine et Maladies Infectieuses, 47, 302–303. 10.1016/j.medmal.2017.03.006 [DOI] [PubMed] [Google Scholar]
- Pararajasingam A and Uwagwu J, 2017. Lactobacillus: the not so friendly bacteria. BMJ Case Reports. 10.1136/bcr-2016-218423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parte AC, 2018. LPSN – list of prokaryotic names with standing in nomenclature (bacterio.net), 20 years on. International Journal of Systematic and Evolutionary Microbiology, 68, 1825–1829. 10.1099/ijsem.0.002786 [DOI] [PubMed] [Google Scholar]
- Passera M, Pellicioli I, Corbellini S, Corno M, Vailati F, Bonanomi E and Farina C, 2016. Lactobacillus casei subsp rhamnosus septicaemia in three patients of the paediatric intensive care unit. Journal of Hospital Infection, 94, 361–362. [DOI] [PubMed] [Google Scholar]
- Perlin DS, Shor E and Zhao Y, 2016. Update on Antifungal Drug Resistance. Current Clinical Microbiology Reports, 2, 84–95. 10.1007/s40588-015-0015-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez‐Cantero A, Thomson P, Paredes K, Guarro J and Capilla J, 2019. Antifungal susceptibility of Saccharomyces cerevisiae and therapy in a murine model of disseminated infection. Revista Iberoamericana de Micología, 36, 37–40. 10.1016/j.riam.2018.04.004 [DOI] [PubMed] [Google Scholar]
- Pfaller MA and Diekema DJ, 2004. Twelve years of fluconazole in clinical practice: global trends in species distribution and fluconazole susceptibility of bloodstream isolates of Candida. Clinical Microbiology & Infection, 10(Suppl. 1), 11–23. 10.1111/j.1470-9465.2004.t01-1-00844.x [DOI] [PubMed] [Google Scholar]
- Pineda C, Kaushik A, Kest H, Wickes B and Zauk A, 2012. Maternal sepsis, chorioamnionitis, and congenital Candida kefyr infection in premature twins. The Pediatric Infectious Disease Journal, 31, 320–322. 10.1097/INF.0b013e31823eee1a [DOI] [PubMed] [Google Scholar]
- Pournejati R, Gust R and Karbalaei‐Heidari HR, 2019. An aminoglycoside antibacterial substance, S‐137‐R, produced by newly isolated Bacillus velezensis strain RP137 from the Persian Gulf. Current Microbiology, 76, 1028–1037. 10.1007/s00284-019-01715-7 [DOI] [PubMed] [Google Scholar]
- Prata JC, Lavorante B, MDC BSMM and Guilhermino L, 2018. Influence of microplastics on the toxicity of the pharmaceuticals procainamide and doxycycline on the marine microalgae Tetraselmis chuii. Aquatic Toxicology, 197, 143–152. 10.1016/j.aquatox.2018.02.015 [DOI] [PubMed] [Google Scholar]
- Quaglia NC and Dambrosio A, 2018. Helicobacter pylori: a foodborne pathogen? World Journal of Gastroenterology, 24, 3472–3487. 10.3748/wjg.v24.i31.3472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reales‐Calderon JA, Molero G, Gil C and Martinez JL, 2016. The fungal resistome: a risk and an opportunity for the development of novel antifungal therapies. Future Medicinal Chemistry, 8, 1503–1520. [DOI] [PubMed] [Google Scholar]
- Rezvani M, Mendoza M, Koci MD, Daron C, Levy J and Hassan HM, 2016. Draft genome sequences of Lactobacillus animalis strain P43 isolated from chicken cecum. Genome Announcements, 4, e01229–01216. 10.1128/genomeA.01229-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues MX, Lima SF, Higgins CH, Canniatti‐Brazaca SG and Bicalho RC, 2016. The Lactococcus genus as a potential emerging mastitis pathogen group: a report on an outbreak investigation. Journal of Dairy Science, 99, 9864–9874. [DOI] [PubMed] [Google Scholar]
- Rühmkorf C, Jungkunz S, Wagner M and Vogel RF, 2012. Optimization of homoexopolysaccharide formation by lactobacilli in gluten‐free sourdoughs. Food Microbiology, 32, 286–294. 10.1016/j.fm.2012.07.002 [DOI] [PubMed] [Google Scholar]
- Rühmkorf C, Bork C, Mischnick P, Rübsam H, Becker T and Vogel RF, 2013. Identification of Lactobacillus curvatus TMW 1.624 dextransucrase and comparative characterization with Lactobacillus reuteri TMW 1.106 and Lactobacillus animalis TMW 1.971 dextransucrases. Food Microbiology, 34, 52–61. 10.1016/j.fm.2012.11.002 [DOI] [PubMed] [Google Scholar]
- Ruiz‐García C, Béjar V, Martínez‐Checa F, Llamas I and Quesada E, 2005. Bacillus velezensis sp. nov., a surfactant‐producing bacterium isolated from the river Vélez in Málaga, southern Spain. International Journal of Systematic and Evolutionary Microbiology, 55, 191–195. 10.1099/ijs.0.63310-0 [DOI] [PubMed] [Google Scholar]
- Sahoo TK, Jena PK, Patel AK and Seshadri S, 2015. Purification and molecular characterization of the novel highly potent bacteriocin TSU4 produced by Lactobacillus animalis TSU4. Applied Biochemistry and Biotechnology, 177, 90–104. 10.1007/s12010-015-1730-z [DOI] [PubMed] [Google Scholar]
- Salse M, Gangneux JP, Cassaing S, Delhaes L, Fekkar A, Dupont D, Botterel F, Costa D, Bourgeois N, Bouteille B, Houze S, Dannaoui E, Guegan H, Charpentier E, Persat F, Favennec L, Lachaud L and Sasso M, 2019. Multicentre study to determine the Etest epidemiological cut‐off values of antifungal drugs in Candida spp. and Aspergillus fumigatus species complex. Clinical Microbiology & Infection, 25, 1546–1552. 10.1016/j.cmi.2019.04.027 [DOI] [PubMed] [Google Scholar]
- Salvetti E, Harris HMB, Felis GE and O'Toole PW, 2018. Comparative genomics of the genus lactobacillus reveals robust phylogroups that provide the basis for reclassification. Applied and Environment Microbiology, 84, 10.1128/aem.00993-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Uchida T, Kuwana S, Sasaki K, Watanabe T, Saito J and Kawaji T, 2016. Bacteremia induced by Bifidobacterium breve in a newborn with cloacal exstrophy. Pediatrics International, 58, 1226–1228. [DOI] [PubMed] [Google Scholar]
- Seccareccia I, Kovacs AT, Gallegos‐Monterrosa R and Nett M, 2016. Unraveling the predator‐prey relationship of Cupriavidus necator and Bacillus subtilis. Microbiological Research, 192, 231–238. 10.1016/j.micres.2016.07.007 [DOI] [PubMed] [Google Scholar]
- Seng P, Cerlier A, Cassagne C, Coulange M, Legre R and Stein A, 2016. Saccharomyces cerevisiae osteomyelitis in an immunocompetent baker. IDCases, 5, 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano A, Sebastián M, Arilla‐Luna S, Baquedano S, Herguedas B, Velázquez‐Campoy A, Martínez‐Júlvez M and Medina M, 2017. The trimer interface in the quaternary structure of the bifunctional prokaryotic FAD synthetase from Corynebacterium ammoniagenes. Scientific Reports, 7, 404–404. 10.1038/s41598-017-00402-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Seynes C, Dutronc H, Cremer P and Dupon M, 2018. Lactobacillus casei prosthetic joint infection. Médecine et Maladies Infectieuses, 48, 422–423. 10.1016/j.medmal.2018.04.390 [DOI] [PubMed] [Google Scholar]
- Shah MM, Miringu G, Wada A, Kaneko S and Ichinose Y, 2019a. Case report: Bacillus pumilus‐Caused Bacteremia in a patient with food poisoning. American Journal of Tropical Medicine and Hygiene, 100, 688–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah TA, Lee CC, Orts WJ and Tabassum R, 2019b. Biological pretreatment of rice straw by ligninolytic Bacillus sp. strains for enhancing biogas production. Environmental Progress & Sustainable Energy, 38, e13036 10.1002/ep.13036 [DOI] [Google Scholar]
- Shida O, Takagi H, Kadowaki K, Nakamura LK and Komagata K, 1997. Transfer of Bacillus alginolyticus, Bacillus chondroitinus, Bacillus curdlanolyticus, Bacillus glucanolyticus, Bacillus kobensis, and Bacillus thiaminolyticus to the genus Paenibacillus and emended description of the genus Paenibacillus. International Journal of Systematic Bacteriology, 47, 289–298. 10.1099/00207713-47-2-289 [DOI] [PubMed] [Google Scholar]
- Shimizu A, Hase R, Suzuki D, Toguchi A, Otsuka Y, Hirata N and Hosokawa N, 2019. Lactococcus lactis cholangitis and bacteremia identified by MALDI‐TOF mass spectrometry: a case report and review of the literature on Lactococcus lactis infection. Journal of Infection and Chemotherapy: Official Journal of the Japan Society of Chemotherapy, 25, 141–146. 10.1016/j.jiac.2018.07.010 [DOI] [PubMed] [Google Scholar]
- Shin JH, Kook H, Shin DH, Hwang TJ, Kim M, Suh SP and Ryang DW, 2000. Nosocomial cluster of Candida lipolytica fungemia in pediatric patients. European Journal of Clinical Microbiology and Infectious Diseases, 19, 344–349. 10.1007/s100960050491 [DOI] [PubMed] [Google Scholar]
- Siavoshi F, Sahraee M, Ebrahimi H, Sarrafnejad A and Saniee P, 2018. Natural fruits, flowers, honey, and honeybees harbor Helicobacterpylori‐positive yeasts. Helicobacter, 23, e12471 10.1111/hel.12471 [DOI] [PubMed] [Google Scholar]
- Simon RR, Vo TD and Levine R, 2016. Genotoxicity and subchronic toxicity evaluation of dried Euglena gracilis ATCC PTA‐123017. Regulatory Toxicology and Pharmacology, 80, 71–81. 10.1016/j.yrtph.2016.06.007 [DOI] [PubMed] [Google Scholar]
- Somayaji R, Lynch T, Powell JN and Gregson D, 2016. Remote transient Lactobacillus animalis bacteremia causing prosthetic hip joint infection: a case report. BMC Infectious Diseases, 16, 634 10.1186/s12879-016-1980-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonmez M and Erbas G, 2017. Isolation and identification of Candida spp. from mastitis cattle milk and determination of antifungal susceptibilities. International Journal of Veterinary Science, 6, 104–107. [Google Scholar]
- Stam R, Jupe J, Howden AJM, Morris JA, Boevink PC, Hedley PE and Huitema E, 2013. Identification and characterisation CRN effectors in Phytophtora capsici shows modularity and functional diversity. PLoS ONE, 8, e59517, 10.1371/journal.pone.0059517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroupe C, Pendley J, Isang E and Helms B, 2017. Persistent bacteremia secondary to delayed identification of Lactobacillus in the setting of mitral valve endocarditis. IDCases, 10, 132–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturino JM, Rajendran M and Altermann E, 2014. Draft genome sequence of Lactobacillus animalis 381‐IL‐28. Genome Announcements, 2, e00478–00414. 10.1128/genomeA.00478-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhr MJ, Gomes‐Neto JC, Banjara N, Florescu DF, Mercer DF, Iwen PC and Hallen‐Adams HE, 2017. Epidemiological investigation of Candida species causing bloodstream infection in paediatric small bowel transplant recipients. Mycoses, 60, 366–374. [DOI] [PubMed] [Google Scholar]
- Sun Y, Xiang Y, He M, Zhang X, Wang S, Guo W, Liu C, Cao Z and Zhou Y, 2019. Evaluation of Lactococcus lactis HNL12 combined with Schizochytrium limacinum algal meal in diets for humpback grouper (Cromileptes altivelis). Fish & Shellfish Immunology, 94, 880–888. 10.1016/j.fsi.2019.09.059 [DOI] [PubMed] [Google Scholar]
- Sundin GW and Wang N, 2018. Antibiotic resistance in plant‐pathogenic bacteria. Annual Review of phytopathology, 56, 161–180. 10.1146/annurev-phyto-080417-045946 [DOI] [PubMed] [Google Scholar]
- Sung MH, Kim H, Bae JW, Rhee SK, Jeon CO, Kim K, Kim JJ, Hong SP, Lee SG, Yoon JH, Park YH and Baek DH, 2002. Geobacillus toebii sp. nov., a novel thermophilic bacterium isolated from hay compost. International Journal of Systematic and Evolutionary Microbiology, 52, 2251–2255. 10.1099/00207713-52-6-2251 [DOI] [PubMed] [Google Scholar]
- Suzuki S, 2017. Large‐Scale Cultivation of Euglena. Advances in Experimental Medicine and Biology, 979, 285–293. 10.1007/978-3-319-54910-1_14 [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Kishigami T, Inoue K, Mizoguchi Y, Eto N, Takagi M and Abe S, 1983. Bacillus thermoglucosidasius sp. nov., a new species of obligately Thermophilic Bacilli. Systematic and Applied Microbiology, 4, 487–495. 10.1016/S0723-2020(83)80006-X [DOI] [PubMed] [Google Scholar]
- Takaç S, Ünlü AE and Erdem B, 2010. Oxygen transfer strategy modulates the productions of lipase and esterase enzymes by Candida rugosa. Journal of Molecular Catalysis B: Enzymatic, 64, 150–154. 10.1016/j.molcatb.2009.07.005 [DOI] [Google Scholar]
- Tan TY, Hsu LY, Alejandria MM, Chaiwarith R, Chinniah T, Chayakulkeeree M, Choudhury S, Chen YH, Shin JH, Kiratisin P, Mendoza M, Prabhu K, Supparatpinyo K, Tan AL, Xuan Thi P, Thi Thanh Nga T, Gia Binh N, Mai Phuong D, Van An H, Su Minh Tuyet N, Thanh Binh T and Van Hung P, 2016. Antifungal susceptibility of invasive Candida bloodstream isolates from the Asia‐Pacific region. Medical Mycology, 54, 471–477. [DOI] [PubMed] [Google Scholar]
- Tato Rodriguez R, Guzman Figueroa DM, Trigo Daporta M and Garcia Campello M, 2018. Fever in an 80‐year‐old male carrying biologic aortic prosthesis endocarditis due to Lactococcus lactis subsp lactis. Journal of Clinical Microbiology, 56, e03357–15. 10.1128/JCM.03357-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna CG, Córdoba S, Vivot M, Szusz W, Vivot W, Bosco‐Borgeat M‐E and Davel G, 2019. Reidentification and antifungal susceptibility profile of Candida guilliermondii and Candida famata clinical isolates from a culture collection in Argentina. Medical Mycology, 57, 314–323. 10.1093/mmy/myy038 [DOI] [PubMed] [Google Scholar]
- Tejan N, Singla N, Guglani V, Gombar S and Chander J, 2017. Comparison of incidence, risk factors, outcome and antifungal susceptibility between Candida albicans and non‐albicans Candida species in patients of candidemia in intensive care setting. International Journal of Pharmaceutical Sciences and Research, 8, 4891–4898. [Google Scholar]
- Thumu SCR and Halami PM, 2019. Heterogeneity of macrolide‐lincosamide‐streptogramin phenotype & conjugal transfer of erm(B) in Pediococcus pentosaceus. Indian Journal of Medical Research, 149, 270–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbetts SM, Milley JE and Lall SP, 2015. Chemical composition and nutritional properties of freshwater and marine microalgal biomass cultured in photobioreactors. Journal of Applied Phycology, 27, 1109–1119. 10.1007/s10811-014-0428-x [DOI] [Google Scholar]
- Toyosaki H, Kojima Y, Tsuchida T, Hoshino K, Yamada Y and Yoshinaga F, 1996. Acetobacter xylinum subsp. sucrofermentans subsp. nov. in Validation of the publication of new names and new combinations previously effectively published outside the IJSB, List no. 58. International Journal of Systematic and Evolutionary Microbiology, 46, 836–837. [Google Scholar]
- Trabelsi H, Chtara K, Khemakhem N, Neji S, Cheikhrouhou F, Sellami H, Guidara R, Makni F, Bouaziz M and Ayadi A, 2015. Fungemia caused by Yarrowia lipolytica. Mycopathologia, 179, 437–445. 10.1007/s11046-015-9859-4 [DOI] [PubMed] [Google Scholar]
- Tsonis I, Karamani L, Xaplanteri P, Kolonitsiou F, Zampakis P, Gatzounis G, Marangos M and Assimakopoulos SF, 2018. Spontaneous cerebral abscess due to Bacillus subtilis in an immunocompetent male patient: a case report and review of literature. World Journal of Clinical Cases, 6, 1169–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbarello M, Caldarola G, Tacconelli E, Morace G, Posteraro B, Cauda R and Ortona L, 1996. Analysis of the risk factors associated with the emergence of azole resistant oral candidosis in the course of HIV infection. Journal of Antimicrobial Chemotherapy, 38, 691–699. 10.1093/jac/38.4.691 [DOI] [PubMed] [Google Scholar]
- Turland NJ, Wiersema JH, Barrie FR, Greuter W, Hawksworth DL, Herendeen PS, Knapp S, Kusber W‐H, Li D‐Z, Marhold K, May TW, McNeill J, Monro AM, Prado J, Price MJ and Smith GFe, 2018. International Code of Nomenclature for algae, fungi, and plants (Shenzhen Code) adopted by the Nineteenth International Botanical Congress Shenzhen, China, July 2017. Regnum Vegetabile 159. Available online: https://www.iapt-taxon.org/nomen/main.php/ [Accessed: 31 July 2018]. Glashütten, Koeltz Botanical Books. 10.12705/code.2018 [DOI]
- Úbeda‐Mínguez P, Chileh T, Dautor Y, García‐Maroto F and Alonso DL, 2015. Tools for microalgal biotechnology: development of an optimized transformation method for an industrially promising microalga—Tetraselmis chuii. Journal of Applied Phycology, 27, 223–232. 10.1007/s10811-014-0306-6 [DOI] [Google Scholar]
- Ucar AD, Ergin OY, Avci M, Ari A, Yildirim M and Erkan N, 2016. Bacillus flexus outbreak in a tertiary burn center. Burns, 42, 948–949. [DOI] [PubMed] [Google Scholar]
- Vandamme P and Coenye T, 2004. Taxonomy of the genus Cupriavidus: a tale of lost and found. International Journal of Systematic and Evolutionary Microbiology, 54, 2285–2289. 10.1099/ijs.0.63247-0 [DOI] [PubMed] [Google Scholar]
- Vaneechoutte M, Kämpfer P, De Baere T, Falsen E and Verschraegen G, 2004. Wautersia gen. nov., a novel genus accommodating the phylogenetic lineage including Ralstonia eutropha and related species, and proposal of Ralstonia [Pseudomonas] syzygii (Roberts et al. 1990) comb. nov. International Journal of Systematic and Evolutionary Microbiology, 54, 317–327. 10.1099/ijs.0.02754-0 [DOI] [PubMed] [Google Scholar]
- Vanichanan J, Chavez V, Wanger A, De Golovine AM and Vigil KJ, 2016. Carbapenem‐resistant Lactobacillus intra‐abdominal infection in a renal transplant recipient with a history of probiotic consumption. Infection, 44, 793–796. [DOI] [PubMed] [Google Scholar]
- Wang JP, Kim JD, Kim JE and Kim IH, 2013. Amino acid digestibility of single cell protein from Corynebacterium ammoniagenes in growing pigs. Animal Feed Science and Technology, 180, 111–114. 10.1016/j.anifeedsci.2012.12.006 [DOI] [Google Scholar]
- Wang H‐K, Teng L‐J, Chen Y‐C, Du S‐H and Hsueh P‐R, 2017. Lactobacillus salivarius empyema with respiratory failure. Journal of Microbiology Immunology and Infection, 50, 923–925. [DOI] [PubMed] [Google Scholar]
- Wardill HR, Tissing WJE, Kissow H and Stringer AM, 2019. Animal models of mucositis: critical tools for advancing pathobiological understanding and identifying therapeutic targets. Current Opinion in Supportive and Palliative Care, 13, 119–133. [DOI] [PubMed] [Google Scholar]
- Watanasrisin W, Iwatani S, Oura T, Tomita Y, Ikushima S, Chindamporn A, Niimi M, Niimi K, Lamping E, Cannon RD and Kajiwara S, 2016. Identification and characterization of Candida utilis multidrug efflux transporter CuCdr1p. FEMS Yeast Research, 16 10.1093/femsyr/fow042 [DOI] [PubMed] [Google Scholar]
- Wennmann JT, Keilwagen J and Jehle JA, 2019. Baculovirus Kimura two‐parameter species demarcation criterion is confirmed by the distances of 38 core gene nucleotide sequences. Journal of General Virology, 99, 1307–1320. 10.1099/jgv.0.001100 [DOI] [PubMed] [Google Scholar]
- Widowati I, Zainuri M, Kusumaningrum HP, Susilowati R, Hardivillier Y, Leignel V, Bourgougnon N and Mouget J‐L, 2017. Antioxidant activity of three microalgae Dunaliella salina, Tetraselmis chuii and Isochrysis galbana clone Tahiti. IOP Conference Series: Earth and Environmental Science, 55, 012067 10.1088/1755-1315/55/1/012067 [DOI] [Google Scholar]
- Wilson HL and Ong CW, 2017. Bifidobacterium longum vertebrodiscitis in a patient with cirrhosis and prostate cancer. Anaerobe, 47, 47–50. [DOI] [PubMed] [Google Scholar]
- Wünnemann H, Eskens U, Prenger‐Berninghoff E, Ewers C and Lierz M, 2018. Lactococcus lactis, causative agent of an endocarditis valvularis and parietalis thromboticans in the allis shad, Alosa alosa (L.) [published online ahead of print, 2018 May 28]. Journal of Fish Diseases, 41, 1207–1215. 10.1111/jfd.12813 [DOI] [PubMed] [Google Scholar]
- Xu Y‐P, Duan P‐G, Wang F and Guan Q‐Q, 2018. Liquid fuel generation from algal biomass via a two‐step process: effect of feedstocks. Biotechnology for Biofuels, 11, 83 10.1186/s13068-018-1083-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagmur G, Sav H, Ziyade N, Elgormus N, Sen S, Bilgi EA, Atan Y, Buyuk Y and Kiraz N, 2016. Evaluation of virulence factors and antifungal susceptibility in yeast isolates from postmortem specimens. Journal of Forensic Sciences, 61, 1000–1006. [DOI] [PubMed] [Google Scholar]
- Yamada K and Machida H, 1962. Studies on the production of lipase by microorganisms. Part I. The selection and identification of a new strain (in Japanese). Nippon Nogeikagaku Kaishii, 36, 858–860. [Google Scholar]
- Yang C and Yu T, 2019. Characterization and transfer of antimicrobial resistance in lactic acid bacteria from fermented dairy products in China. Journal of Infection in Developing Countries, 13, 137–148. [DOI] [PubMed] [Google Scholar]
- Ye C, Qiao W, Yu X, Ji X, Huang H, Collier JL and Liu L, 2015. Reconstruction and analysis of the genome‐scale metabolic model of Schizochytrium limacinum SR21 for docosahexaenoic acid production. BMC Genomics, 16, 799. 10.1186/s12864-015-2042-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Jiang X, Li L, Wang H, Zuo S, Zhang M, Qi Z and Yu M, 2016. Antimicrobial resistance and resistance genes in lactic acid bacteria isolated from commercial yogurt Food Science. China, 37, 131–136. [Google Scholar]
- Yuste JR, Franco SE, Sanders C, Cruz S, Fernández‐Rivero ME and Mora G, 2016. Bacillus licheniformis as a cause of a deep skin abscess in a 5‐year‐old girl: an exceptional case following a plant thorn injury. Journal of Microbiology, Immunology and Infection, 49, 819–821. 10.1016/j.jmii.2014.08.031 [DOI] [PubMed] [Google Scholar]
- Zakryś B, Milanowski R and Karnkowska A, 2017. Evolutionary origin of Euglena. Advances in Experimental Medicine and Biology, 979, 3–17. 10.1007/978-3-319-54910-1_1 [DOI] [PubMed] [Google Scholar]
- Zeba F, Yirerong J, Assali M, Tewary G and Noska A, 2018. A Double Whammy: Lactobacillus acidophilus Bacteremia and subsequent Lactobacillus rhamnosus prosthetic valve infective endocarditis in an elderly diabetic patient. Rhode Island Medical Journal, 101, 32–35. [PubMed] [Google Scholar]
- Zhang K, Li H, Chen W, Zhao M, Cui H, Min Q, Wang H, Chen S and Li D, 2017. Regulation of the docosapentaenoic acid/docosahexaenoic acid ratio (DPA/DHA Ratio) in Schizochytrium limacinum B4D1. Applied Biochemistry and Biotechnology, 182, 67–81. 10.1007/s12010-016-2311-5 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Caspers MPM, Metselaar KI, de Boer P, Roeselers G, Moezelaar R, Nierop Groot M, Montijn RC, Abee T and Kort R, 2013. Abiotic and microbiotic factors controlling biofilm formation by thermophilic sporeformers. Applied and Environmental Microbiology, 79, 5652 10.1128/AEM.00949-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Kumar M, Caspers MPM, Nierop Groot MN, van der Vossen J and Abee T, 2018. Short communication: growth of dairy isolates of Geobacillus thermoglucosidans in skim milk depends on lactose degradation products supplied by Anoxybacillus flavithermus as secondary species. Journal of Dairy Science, 101, 1013–1019. 10.3168/jds.2017-13372 [DOI] [PubMed] [Google Scholar]
- Zhao M, Li S, Zhou Q, Zhou D, He N and Qian Z, 2019. Safety evaluation of microbial pesticide (HaNPV) based on PCR method. Frontiers of Chemical Science and Engineering, 13, 377–384. [Google Scholar]
- Zhou J, Wu K and Rao CV, 2016. Evolutionary engineering of Geobacillus thermoglucosidasius for improved ethanol production. Biotechnology and Bioengineering, 113, 2156–2167. 10.1002/bit.25983 [DOI] [PubMed] [Google Scholar]


