Abstract
Astrotactin 1 (ASTN1) is known to serve a physiological role in neuronal migration; however its role in liver cancer remains to be determined. In the present study, ASTN1 levels were lower in liver cancer tissues compared with those in matching normal tissue. ASTN1 levels were negatively associated with microscopic vascular invasion, advanced clinical stage and a less favorable prognosis in patients with hepatocellular carcinoma (HCC). Furthermore, ASTN1 overexpression in a liver cancer cell line reduced the migratory and invasive capacity of the cells. Based on bioinformatics analysis, ASTN1 levels were negatively associated with the Wnt signaling pathway. In addition, ASTN1 downregulated the protein expression levels of β-catenin, T-cell factor (TCF)1, TCF4, Jun proto-oncogene (C-jun), Myc proto-oncogene (C-myc), cyclooxygenase-2 (COX2), metalloproteinase (MMP)2, MMP9 and vascular endothelial growth factor (VEGF) protein levels, indicative of suppression of Wnt signaling. Furthermore, XAV939-induced Wnt signaling suppression reversed the ASTN1-mediated inhibition of invasion and migration in cells. Overexpression of ASTN1 in xenografts reduced cancer development as well as Wnt signaling. TIMER analysis showed that ASTN1 expression was negatively correlated with B cell, macrophage and neutrophil infiltrating levels in HCC. Together, the results of the present study showed that ASTN1 reduced the migratory and invasive capacity of liver cancer cells, potentially served as a candidate biomarker for diagnosis and prediction of the prognosis of HCC, and was associated with immune infiltration. Understanding the underlying mechanisms of action of ASTN1 may facilitate the development of novel strategies for prevention and treatment of liver cancer.
Keywords: astrotactin 1, liver cancer, tumorigenicity, Wnt/β-catenin signal transduction pathway, tumor immune infiltrate
Introduction
Hepatocellular carcinoma (HCC) is the most common type of liver cancer, and ranks fifth in terms of mortality rates worldwide amongst all types of cancer, and its morbidity rates have been increasing on a yearly basis (1). The outcomes of patients with HCC have improved significantly over the past decades, which has been ascribed to the development of improved local therapeutic techniques and resection criteria; nonetheless, the prognosis of patients with HCC remains poor, and the rates of distant metastasis and local recurrence are also high (2–5). It is well documented that dysregulated gene expression serves an important role during the development and progression of cancer (6–8). Certain cancer markers have been identified for predicting the prognosis of surgical resection outcomes; however, they may not always exhibit the best predictive capacity (9,10). Therefore, identifying novel cancer markers is required to improve prediction of clinical outcomes. Additionally, understanding the mechanisms of action at the molecular level underlying the involvement of predictive biomarkers may highlight potential novel targets for the treatment of patients with liver cancer.
Long non-coding RNAs (lncRNAs) are non-coding RNAs that are 2.2 kb in length. Hox transcript antisense RNA (HOTAIR) is a lncRNA encoded by the HOX gene cluster at the HOXC site (11). HOTAIR is upregulated in HCC compared with that in non-cancerous tissues, and exerts its function through the activation of the Wnt/β catenin signal transduction pathway and is associated with metastasis (12). In vitro, it has been demonstrated that HOTAIR promotes migration and invasion of HCC cells (13,14). Furthermore, gene expression profiling has shown that HOTAIR knockdown results in upregulated astrotactin 1 (ASTN1) expression (15). ASTN1 is a neuronal adhesion molecule, which is required for the physiological migration of granule cells during brain development (16). However, its expression and functions within tumor tissues remain largely unknown, and additional studies are required to examine the role of ASTN1 in migration and invasion in liver cancer.
The results of the present study showed that ASTN1 expression was decreased in liver cancer tissues compared with that noted in matched adjacent normal liver tissues. ASTN1 expression was upregulated or silenced to examine its functions in human liver cancer cells. Overexpression of ASTN1 reduced liver cancer invasion and migration through suppression of the Wnt signaling pathway. Additionally, ASTN1 expression was associated with HCC immune infiltration. Taken together, the results of the present study suggest that ASTN1 may be used as a diagnostic and prognostic marker in patients with liver cancer.
Materials and methods
Patients and tissue specimens
In the present study 145 consecutive HCC cases (mean age, 47 years; age range, 22–75 years; 128 male and 17 female patients) undergoing surgical resection at Sun Yat-sen University Cancer Center during 2004 were immunohistochemically analyzed. Their clinical data were acquired through reviewing medical records. Informed consent was obtained from all patients, and the study was approved by the Ethics Committee of Sun Yat-sen University Cancer Center. Each case was followed once every month for the first 6 months postoperatively, and once every 3 months thereafter. This study was ended on December 31, 2017. Magnetic resonance imaging (MRI) or computed tomography (CT) was used to confirm tumor relapse or metastasis. Overall survival (OS) was deemed as the duration from the date of surgical resection to the date of death, whereas recurrence-free survival (RFS) was the duration from the date of surgical resection to the date of metastasis or relapse.
Reverse transcription-quantitative (RT-q)PCR
Total RNA was extracted from cells or tissues using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol, and subsequently treated with DNase. The RNA was reverse transcribed into cDNA using a RevertAid First-Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc.). The thermocycling conditions was as follows: Pre-degeneration at 95°C for 10 min, denaturation at 95°C for 5 sec; annealing at 60°C for 30 sec; followed by 40 cycles, and extension at 60°C for 30 sec. qPCR was performed using SYBR-Green qPCR Master mix (Thermo Fisher Scientific, Inc.) on an ABI 7300 system (Applied Biosystem; Thermo Fisher Scientific, Inc.). The sequences of the primers used were: ASTN1 forward, 5′-CAATCTCTTCAATGGCTACAC-3′ and reverse, 5′-TCCTTTCCTCCAATCATCTAC-3′; and GAPDH forward, 5′-AATCCCATCACCATCTTC-3′ and reverse 5′-AGGCTGTTGTCATACTTC-3′. GAPDH was used as the loading control. Experiments were repeated three times. The 2−ΔΔCq method was used to normalize target gene mRNA expression to GAPDH mRNA levels (17).
Bioinformatics analysis
An HCC dataset including 374 tumor and 50 non-carcinoma tissues was acquired from The Cancer Genome Atlas project (TCGA; tcga-data.nci.nih.gov/tcga/). ASTN1 levels between tumor and adjacent non-carcinoma tissues were compared using a Student's t-test. The ASTN1 levels between tumor and non-tumor tissues were compared in the GEO datasets, GSE22058 (18) and GSE14520 (19), using Integrative Molecular Database of Hepatocellular Carcinoma (HCCDB) (20). ASTN1 expression in various types of cancer was determined using the Oncomine database (https://www.oncomine.org/) with the selection criteria set as a significance threshold of P≤0.0001 and a fold change of 2. UALCAN (ualcan.path.uab.edu/index.html) was used to show ASTN1 expression and patient clinical stage information based on gene expression. Survival analysis based on ASTN1 expression was performed using Kaplan-Meier-plotter (21). In addition, the pathways associated with ASTN1 were identified based on TCGA HCC dataset using gene set enrichment analysis (GSEA) using broad.mit.edu/gsea as described previously (22). Using Tumor Immune Estimation Resource (TIMER), ASTN1 expression was analyzed across a range of different types of cancer and immune infiltration in HCC was estimated (23). The abundance of six immune infiltrates (B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages and dendritic cells) were estimated, and their association with ASTN1 expression were analyzed using Spearman's rank correlation coefficient analysis. Scatter diagrams were drawn to show the correlation between immune-related gene expression and ASTN1 expression.
Cell culture
HepG2, Hep3B, HCCLM3, Huh7, SKHEP1 and MIHA cells were provided by The Cell Bank of the Type Culture Collection of the Chinese Academy of Sciences, and cultured in DMEM supplemented with 10% FBS (Invitrogen, Thermo Fisher Scientific, Inc.), in the absence of antibiotics with 5% CO2 at 37°C and 99% relative humidity.
Preparation and transfection of lentivirus
The lentiviral vector pLKO.1-puro (Addgene, Inc.) containing one of the three anti-ASTN1 short hairpin RNAs (sh)RNAs or the negative control (shNC) was transfected into cells. The sequences of the shRNAs were: shASTN1−1, 5′-GCCAGAGAAAGCGGATCAA-3′; shASTN1−2, 5′-GCAACTGCCAGATGGTCTT-3′; and shASTN1−3, 5′-CCTGGAACCTGACACCATT-3′. Human ASTN-1 cDNA was inserted into a pLVX-puro lentiviral vector (Addgene, Inc.) using the BamHI and EcoRI sites. Subsequently, Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) was used to transfect 293T cells with the lentiviral vector and packaging plasmids according to the manufacturer's protocol, to produce the lentiviruses. After 48 h of transfection, the medium from transfected cells were collected, mixed and filtered. The liver cancer cells were subjected to specific lentiviral infection using the filtered medium.
Western blotting
Antibodies against ASTN1 (dilution 1:1,000; product code ab154522), T-cell factor (TCF)1 (dilution 1:1,000; product code ab183862), TCF4 (dilution 1:1,000; product code ab130014), C-jun (Jun proto-oncogene) (dilution 1:5,000; product code ab40766), C-myc (Myc proto-oncogene) (dilution 1:1,000; product code ab32072), cyclooxygenase-2 (COX2) (dilution 1:500; product code ab23672), metalloproteinase (MMP)2 (dilution 1:1,000; cat. no. ab97779), MMP9 (dilution 1:500; product code ab119906), vascular endothelial growth factor (VEGF) (dilution 1:300; product code ab1316) (all from Abcam), β-catenin (dilution 1:1,000; cat. no. 8480) and GAPDH (dilution 1:2,000; cat. no. 5174) (both from Cell Signaling Technology, Inc.) were used for western blotting. RIPA lysis buffer (JRDUN) supplemented with a protease inhibitor cocktail (Sigma Aldrich; Merck KGaA), was used to lyse tissues or cells. A bicinchoninic acid protein assay kit (Thermo Fisher Scientific, Inc.) was used to measure protein concentration. A total of ~25 µg protein lysate loaded on a 10 or 12% SDS gel, resolved using SDS-PAGE and transferred to nitrocellulose membranes (EMD Millipore). Membranes were incubated with the primary antibodies overnight at 4°C, and subsequently incubated with a horseradish peroxidase (HRP)-labeled secondary antibody (dilution 1:1,000; cat. no. A0208 from Beyotime Institute of Biotechnology) for 1 h at room temperature. Enhanced chemiluminescence reagent (EMD Millipore) was used to visualize the signals.
Transwell and wound healing assays
Transwell assays were performed to determine the migratory and invasive capacity of the cells. In the migration assay, the transduced cells were plated in 24-well plates. A total of 24 h later, cells were serum starved overnight, followed by trypsin digestion; 200 µl serum-free medium containing 5×104 cells was added to the upper chamber of a Transwell insert and 700 µl culture medium supplemented with 10% FBS was added to the lower chamber. The cells were incubated for 24 h at 37°C. Subsequently, the cells which had migrated were fixed using 4% paraformaldehyde and stained using 0.5% crystal violet, and the number of cells stained was counted under a microscope. All tests were performed three times. Similarly, invasion assays were performed using a similar method to that of migration assay, with the addition of 80 µl Matrigel® (Corning, Inc.) to the upper chamber of the Transwell insert prior to addition of cells.
For the wound healing assays, liver cancer cells were cultured in wells, and once confluent, the monolayer was scratched using a 10-µl pipette tip. Subsequently, the cells were cultured using FBS-free DMEM. An inverted microscope (Olympus IX73) was used to observe the migration of cells at 0, 12 and 24 h post-scratching.
Tumor xenograft experiments in vivo
Animal studies were performed in accordance with guidelines released by the Experimental Animal Care Commission of Sun Yat-sen University Cancer Center. Brief, 12 BALB/c nude mice (4–5 weeks) (Shanghai SLAC Laboratory Animal Co., Ltd.) were raised under specific pathogen-free conditions with a 12 h light/dark cycle, and ad libitum access to water and food. Establishment of tumor-bearing nude mice was routinely accomplished through tail-vein injections of HCCLM3 cells infected with the ASTN1-expressing or control vector virus (1×107 cells in 100 µl PBS). A total of 4 weeks after cell transplantation, the mice were sacrificed and the number of lung metastatic foci were counted. The tumors were extracted and subjected to hematoxylin and eosin (H&E) and immunofluorescence staining. The 5-µm paraffin sections were stained with H&E for histological evaluation based on standard pathological methods. In the immunofluorescence assay, the 5-µm sections were prepared from the paraffin-embedded tissue blocks, followed by deparaffinization and rehydration according to a standard protocol. After antigen retrieval using citrate buffer solution (pH 6.0), tissues were incubated with anti-ASTN1 (Santa-Cruz Biotechnology, Inc.; cat. no. sc-514299) and anti-β-catenin (Abcam; cat. no. ab32572) overnight at 4°C. Subsequently, anti-mouse and anti-rabbit horseradish peroxidase (HRP)-conjugated fluorescent dye-labeled secondary antibodies (cat. nos. A0428 and A0423; Beyotime Institute of Biotechnology) were used to label and visualize target protein expression in tissues.
Immunohistochemistry (IHC) analysis
For IHC, 5-µm sections prepared from formalin-fixed and paraffin-embedded tissue blocks were used for analysis. Briefly, the paraffin-embedded sections were deparaffinized using xylene and rehydrated using a series of solutions of decreasing alcohol concentrations. Subsequently, tissues were placed in boiling citrate buffer for antigen retrieval, and 3% H2O2 was used to block endogenous peroxidase activity. Tissues were incubated overnight with anti-ASTN1 antibody (dilution, 1:150; cat. no. ab140533; Abcam) at 4°C. The following day, the sections were washed and incubated using a HRP-polymer anti-Rabbit IHC kit (Maixin) at room temperature, and developed using DAB HRP Color Development kit (Maixin), followed by hematoxylin counter-staining. Quantification of ASTN1 expression was performed as described in our previous study (9).
Statistical methods
GraphPad Prism (GraphPad Software, Inc.) was used for all statistical analyses. Differences between two groups were compared using a Student's t-test. Difference between multiple groups were compared using a one-way ANOVA with post hoc contrasts by Tukey test. The area under the receiver operating characteristic curve (AUC) was calculated using the pROC package in R. A χ2 test was used to analyze the association between ASTN1 expression levels and the clinicopathological characteristics. The OS was compared between patients with high and low ASTN1 expression levels using a log-rank test and Kaplan-Meier analysis. Furthermore, univariate and multivariate Cox proportional hazards regression models were used to determine survival-related factors. P<0.05 was considered to indicate a statistically significant difference.
Results
ASTN1 expression is downregulated in HCC tissues relative to the matching adjacent normal tissue
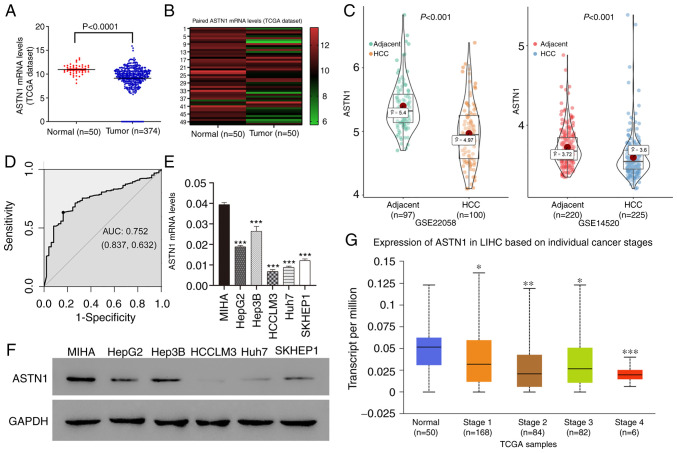
ASTN1 levels within human HCC tissues were measured based on the TCGA dataset. ASTN1 expression levels were downregulated in HCC tissues (n=374) compared with the normal liver tissue (n=50) (9.14±0.14 vs. 10.95±0.14; P<0.0001; Fig. 1A). Furthermore, ASTN1 expression in the 50 paired HCC and adjacent non-carcinoma tissue samples showed that ASTN1 mRNA levels were lower in tumor tissues compared with that in the paired non-carcinoma HCC tissues (9.74±1.64 vs. 10.95±1.02; P<0.0001; Fig. 1B). Downregulated expression of ASTN1 in HCC was further validated in the GSE22058 (P=8.600×10−11) and GSE14520 (P=2.480×10−7) datasets (Fig. 1C). ASTN1 differential expression between HCC and adjacent samples in HCCDB is shown in Fig. S1. ASTN1 protein and mRNA expression levels were analyzed in 5 liver cancer cell lines as well as in the MIHA normal liver cell line through western blotting and RT-qPCR. ASTN1 levels were significantly lower in the liver cancer cells compared with the normal liver cell line (Fig. 1E and F). In addition, receiver operating characteristic curve analysis suggested that ASTN1 was a potential diagnostic marker for HCC, with an AUC of 0.752 (Fig. 1D), and lower ASTN1 levels were associated with advanced patient clinical stage (Fig. 1G).
Figure 1.
Downregulated ASTN1 levels in human liver cancer tissue samples are associated with advanced clinical stage. (A) ASTN1 expression was detected in HCC as well as normal liver tissues based on the TCGA HCC dataset. (B) ASTN1 mRNA expression levels were detected in HCC tissues as well as paired adjacent non-cancerous tissues. (C) Further validation of ASTN1 expression using GEO datasets. (D) Diagnostic ability of ASTN1 in HCC. ASTN1 (E) mRNA and (F) protein expression levels in 5 liver cancer cell lines as well as the MIHA normal liver cell line. (G) Low ASTN1 levels were associated with advanced clinical stage. *P<0.05, **P<0.01, ***P<0.001 vs. normal control. ASTN1, astrotactin 1; TCGA, The Cancer Genome Atlas; HCC, hepatocellular carcinoma; GEO, Gene Expression Omnibus; AUC, area under the curve.
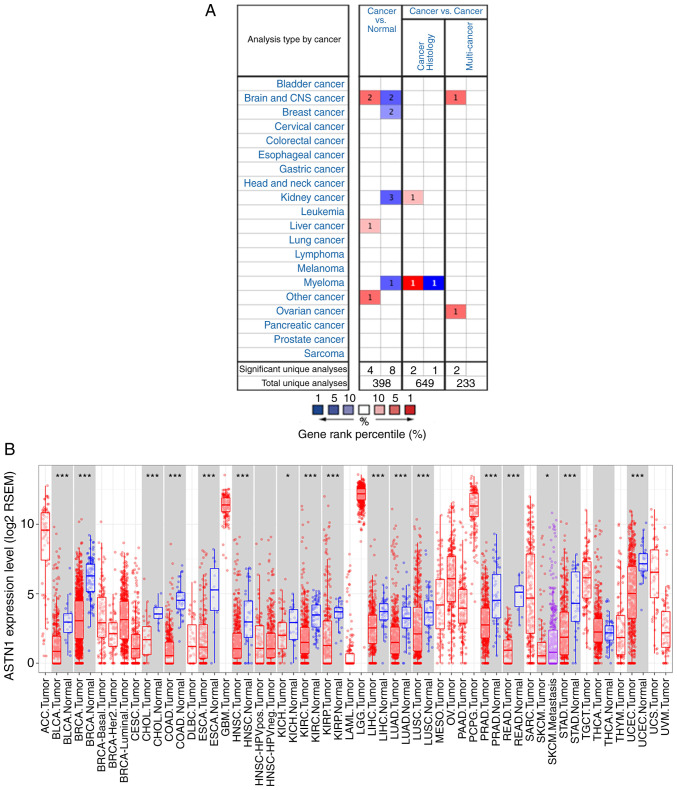
The Oncomine database showed that the ASTN1 expression was higher in two brain and CNS cancer studies, one liver cancer study [relatively small population using a micro-array with lack of validation (GSE6764)], and one other cancer study; low in two brain and CNS cancer studies, two breast cancer studies, three kidney cancer studies and one myeloma cancer study (Fig. 2A). The differential expression of ASTN1 across all TCGA cancer cases in TIMER is shown in Fig. 2B.
Figure 2.
ASTN1 expression in different types of cancer. (A) ASTN1 expression in different types of cancer based on the Oncomine database. (B) TCGA database ASTN1 expression levels in TIMER. *P<0.05, ***P<0.001. ASTN1, astrotactin 1; TCGA, The Cancer Genome Atlas; TIMER, Tumor Immune Estimation Resource.
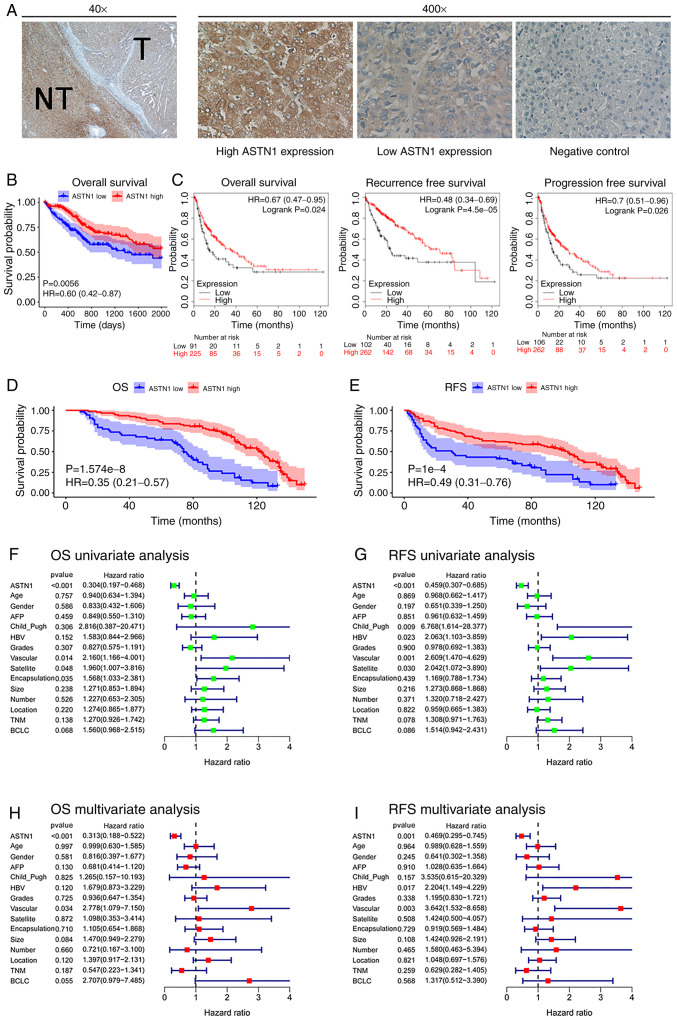
Associations between ASTN1 expression with clinicopathological parameters and prognosis in patients with HCC
The associations between ASTN1 expression levels with the clinicopathological parameters were analyzed. Further, the ASTN1 levels in 145 HCC samples were also detected using IHC (Fig. 3A). The 145 HCC patients were split into two groups, based on ASTN1 expression; 92 patients had levels of expression (score ≥6), and 53 patients had a low (score <6) ASTN1 levels. ASTN1 levels were significantly associated with the presence of microscopic satellite nodules (P=0.018), microscopic vascular invasion (P=0.010), tumor grade (P=0.003), encapsulation (P<0.001), Barcelona Clinic Liver Cancer (BCLC) stage (P=0.024) and Tumor-Node-Metastasis stage (P=0.007) as shown in Table I.
Figure 3.
Association of ASTN1 levels with HCC prognosis. (A) Immunohistochemistry staining of HCC tissues. T, tumor; NT, nontumor tissue. (B) Kaplan-Meier survival curves based on expression of ASTN1. (C) Kaplan-Meier-plotter was used to further validate the differences in survival based on ASTN1 expression. Low ASTN1 levels predicted a (D) less favorable OS and (E) RFS. Univariate analysis of (F) OS and (G) RFS. Multivariate analysis of (H) OS and (I) RFS. ASTN1, astrotactin 1; HCC, hepatocellular carcinoma; OS, overall survival; RFS, recurrence-free survival.
Table I.
Association of ASTN1 with the clinicopathological variables of the HCC patients.
| ASTN1 level | ||||
|---|---|---|---|---|
| Clinicopathological parameters | Case (n=145) | Low (n=53) | High (n=92) | P-value |
| Age (years) | 0.424 | |||
| <45 | 73 | 29 | 44 | |
| ≥45 | 72 | 24 | 48 | |
| Sex | 0.338 | |||
| Male | 128 | 45 | 83 | |
| Female | 17 | 8 | 9 | |
| AFP (µg/l) | 0.805 | |||
| <400 | 103 | 37 | 66 | |
| ≥400 | 42 | 16 | 26 | |
| Child_Pugh score | 0.132 | |||
| 5 | 143 | 51 | 92 | |
| 6 | 2 | 2 | 0 | |
| HBV history | 0.248 | |||
| No | 20 | 5 | 15 | |
| Yes | 125 | 48 | 77 | |
| Grades of differentiation | 0.003 | |||
| Low | 26 | 17 | 9 | |
| Medium | 99 | 30 | 69 | |
| High | 20 | 6 | 14 | |
| Microscopic vascular invasion | 0.010 | |||
| Absent | 128 | 42 | 86 | |
| Present | 17 | 11 | 6 | |
| Microscopic satellite nodules | 0.018 | |||
| Absent | 134 | 45 | 89 | |
| Present | 11 | 8 | 3 | |
| Encapsulation | <0.001 | |||
| Intact | 77 | 15 | 62 | |
| Destructed | 68 | 38 | 30 | |
| Tumor size (cm) | 0.264 | |||
| <5 | 88 | 29 | 59 | |
| ≥5 | 57 | 24 | 33 | |
| Tumor number | >0.999 | |||
| Single | 132 | 48 | 84 | |
| Multiple | 13 | 5 | 8 | |
| Tumor location | 0.526 | |||
| Left lobe | 43 | 15 | 28 | |
| Right lobe | 73 | 35 | 62 | |
| Both lobes | 5 | 3 | 2 | |
| TNM stage | 0.007 | |||
| I | 113 | 35 | 78 | |
| II | 23 | 12 | 11 | |
| III | 9 | 6 | 3 | |
| BCLC stage | 0.024 | |||
| A | 127 | 43 | 84 | |
| B | 15 | 7 | 8 | |
| C | 3 | 3 | 0 | |
ASTN, astrotactin 1; AFP, α-fetoprotein; HBV, hepatitis B virus; BCLC, Barcelona Clinic Liver Cancer.
Prognosis and survival analysis was performed using the 365 cases from TCGA. Increased ASTN1 mRNA expression levels was associated with improved OS rates (P=0.0056; Fig. 3B). KM-plotter was used to validate the differences in OS based on ASTN1 expression, which showed that OS, RFS and progression-free survival were significantly different in patients with HCC cases between low and high ASTN1 expression level groups (Fig. 3C). Low ASTN1 expression was associated with shorter survival times. To further verify the above findings, the ASTN1 levels among the recruited 145 HCC patients were detected. The results suggested that patients with increased ASTN1 expression levels had higher OS (P<0.0001; Fig. 3D) as well as RFS rates (P<0.0001; Fig. 3E). According to results of univariate Cox regression analysis, ASTN1, microscopic vascular invasion, microscopic satellite nodules, and encapsulation were all associated with OS rates (Fig. 3F). Additionally, ASTN1, Child_Pugh, microscopic satellite nodule, microscopic vascular invasion and HBV history were all associated with RFS (Fig. 3G). To determine whether ASTN1 was an independent factor for predicting patient prognosis, multivariate analysis was performed on ASTN1 expression levels and clinicopathological characteristics. According to the Cox proportional hazards regression analysis, ASTN1 and microscopic vascular invasion were independent risk factors of OS (Fig. 3H). Additionally, ASTN1, HBV history and microscopic vascular invasion were independent risk factors affecting patients RFS (Fig. 3I). Taken together, the above results suggest that, ASTN1 may serve as an independent biomarker for prediction of OS and RFS in patients with HCC [hazard ratio (HR), 0.313; 95% confidence interval (CI), 0.188–0.522; HR, 0.469; 95% CI, 0.295–0.745, respectively]. These results suggest that ASTN1 may exhibit clinical value for the diagnosis and prognosis of patients with HCC.
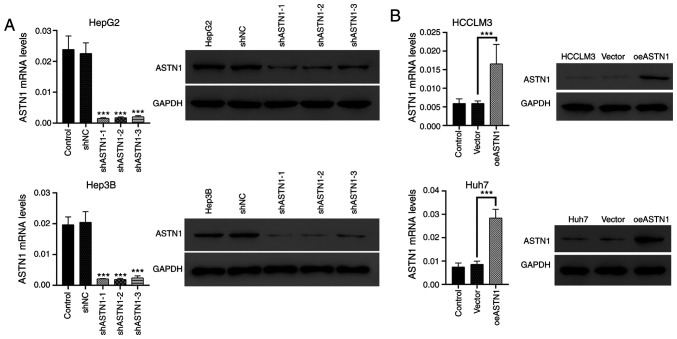
Effects of knockdown and overexpression of ASTN1 levels in liver cancer cells
ASTN1 levels were significantly decreased in liver cancer cells, particularly in the HCCLM3 and Huh7 cells. To further investigate the role of ASTN1 in liver cancer, lentiviral transfection was performed to overexpress ASTN1 (oeASTN1) in HCCLM3 and Huh7 cells, and to knock down expression (shASTN1 1–3) in HepG2 and Hep3B cells which exhibited higher levels of ASTN1 expression. Following transfection for 48 h, RT-qPCR and western blotting were performed. Transfection with control shNC or the empty vector did not affect ASTN1 expression levels (Fig. 4A). To knockdown ASTN1 expression, shASTN1 1–3 were transfected into cells, and this resulted in a significant knockdown in ASTN1 expression in HepG2 and Hep3B cells; shASTN1-3 exhibited the lowest knockdown efficiency and was thus not used in subsequent experiments. ASTN1 mRNA and protein expression levels were significantly upregulated in the ASTN1 viral transfected cells compared with the untransfected and empty vector virus-transfected cells (Fig. 4B).
Figure 4.
Knockdown and overexpression of ASTN1 expression in liver cancer cells. (A) HepG2 and Hep3B cells were subjected to ASTN1 shRNA (shASTN1-1-3) and shNC lentiviral infection. The protein levels of ASTN1 were detected following viral infection for 48 h. ***P<0.001 vs. with shNC. (B) ASTN1 was overexpressed in HCCLM3 and Huh7 cells using lentiviral transfection, and the protein levels of ASTN1 were detected following viral infection for 48 h. Untransfected cells were used as the negative control. ***P<0.001. ASTN1, astrotactin 1; shNC, short hairpin negative control; Vector, empty vector transfected cells; oeASTN1, ASTN1-overexpressing vector.
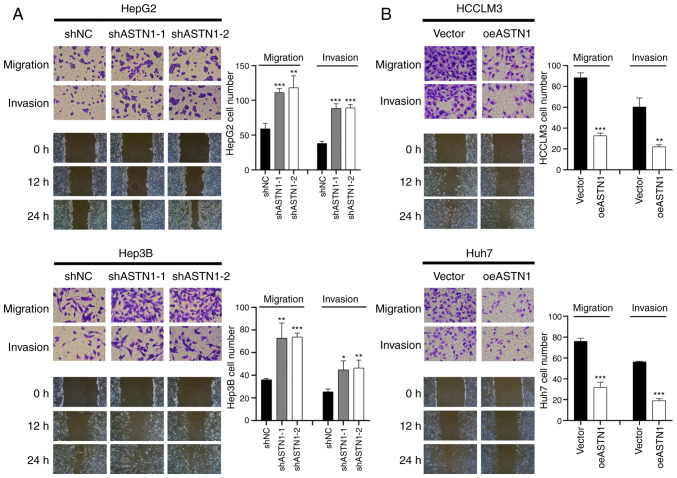
ASTN1 suppresses the migratory and invasive capacities of liver cancer cells
Transwell invasion and migration assays were performed to determine the role of ASTN1 in the invasive and migratory capacities of liver cancer cells. Knockdown of ASTN1 expression in HepG2 and Hep3B cells significantly increased the migratory and invasive capacity of the cells (Fig. 5A). Compared with the controls, overexpression of ASTN1 in HCCLM3 and Huh7 cell lines resulted in a significant downregulation in migration and invasion (Fig. 5B). These results suggest that ASTN1 suppressed the migratory and invasive capacity of liver cancer cells.
Figure 5.
ASTN1 reduces the migratory and invasive capacities of liver cancer cells. Transwell migration and invasions assays were performed to determine the migratory and invasive capacities of HepG2 and Hep3B cells following (A) ASTN1 silencing or (B) ASTN1 overexpression in HCCLM3 and Huh7 cells. *P<0.05, **P<0.01, ***P<0.001 vs. the shNC or Vector group. ASTN1, astrotactin 1; shNC, short hairpin negative control; Vector, empty vector transfected cells; oeASTN1, ASTN1-overexpressing vector.
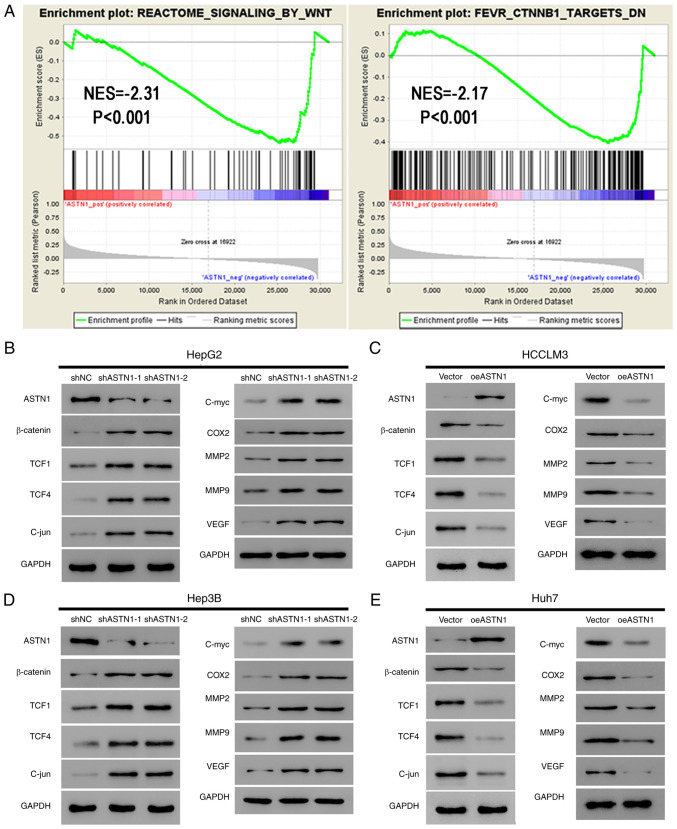
ASTN1 suppresses the Wnt signal transduction pathway in liver cancer
Pathways associated with ASTN1 were evaluated among TCGA HCC samples using GSEA. ASTN1 levels were negatively associated with REACTOME_SIGNALING_BY_WNT and FEVR_CTNNB1_TARGETS_DN (Fig. 6A), suggesting ASTN1 potentially affected HCC migration and invasion through the Wnt/β-catenin signaling pathway.
Figure 6.
ASTN1 suppresses the Wnt signaling pathway in liver cancer cells. (A) As suggested by gene set enrichment analysis, ASTN1 was negatively associated with the Wnt signal transduction pathway and CTNNB1 in TCGA HCC samples. Expression of the primary components of the Wnt signaling pathway (β-catenin, TCF1, TCF4 and C-jun) and the downstream effectors (C-myc, COX2, MM2, MMP9, and VEGF) of the Wnt signaling pathway were assessed by western blotting in (B) HepG2 cells and (D) Hep3B cells following ASTN1 silencing, and in (C) HCCLM3 cells and (E) Huh7 cells following ASTN1 overexpression. HCC, hepatocellular carcinoma; ASTN1, astrotactin 1; shNC, short hairpin negative control; Vector, empty vector transfected cells; oeASTN1, ASTN1-overexpressing vector; TCGA, The Cancer Genome Atlas; TCF, T-cell factor; COX2, cyclooxygenase-2; MMP, metalloproteinase; VEGF, vascular endothelial growth factor.
Activation of the Wnt signaling pathway induces invasion and proliferation of cells, which may serve a vital role in enhancing carcinogenesis. In the present study, ASTN1 levels were negatively associated with the Wnt signal transduction pathway; therefore, ASTN1 was hypothesized to inhibit the Wnt signaling activity. Western blotting was performed to measure the expression levels of the primary components (β-catenin, TCF1, TCF4 and C-jun) and the downstream effectors (C-myc, COX2, MMP2, MMP9 and VEGF) of the Wnt signaling pathway in liver cancer cells overexpressing ASTN1 or with ASTN1 expression knocked down. Knockdown of ASTN1 expression in HepG2 and Hep3B cells resulted in a significant upregulation in β-catenin, TCF1, TCF4, C-jun, C-myc, COX2, MM2, MMP9 and VEGF protein expression levels (Fig. 6B and D). Overexpression of ASTN1 in HCCLM3 and Huh7 cells resulted in the opposite effect (Fig. 6C and E).
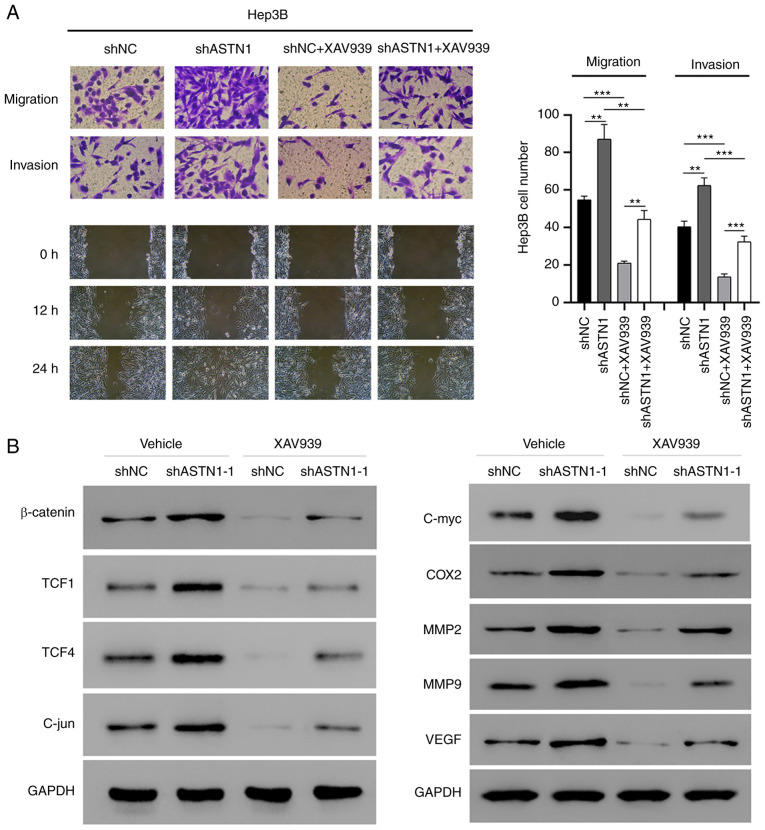
Wnt signaling mediates the effect of ASTN1 on migration and invasion
To determine the role of Wnt signal transduction in mediating the effect of ASTN1 on migration and invasion of cells, ASTN1-silenced Hep3B cells were treated with XAV939 to inhibit the Wnt signal transduction pathway (Fig. 7). ASTN1 silencing significantly increased migration and invasion of cells, whereas XAV939 treatment suppressed these effects. Furthermore, western blot analysis showed that β-catenin, TCF1, TCF4, C-jun, C-myc, COX2, MM2, MMP9 and VEGF protein expression levels were notably decreased in the Hep3B cells with ASTN1 expression knocked down when treated with XAV939. These results further verify the role of the Wnt signaling pathway in the effects of ASTN1 on migration and invasion.
Figure 7.
The Wnt signal transduction pathway regulates the effects of ASTN1 on the migratory and invasive capacities of cells. Hep3B cells were transfected with ASTN1-shRNA or shNC. Cells were treated with XAV939 after viral infection for 24 h. (A) Migratory and invasive capacities of Hep3B cells with ASTN1 expression silenced following XAV939 treatment were determined using Transwell assays. **P<0.01, ***P<0.001. (B) Western blot analysis of the key components and the downstream effectors of the Wnt signal transduction pathway. ASTN1, astrotactin 1; sh, short hairpin; NC, negative control; TCF, T-cell factor; COX2, cyclooxygenase-2; MMP, metalloproteinase; VEGF, vascular endothelial growth factor.
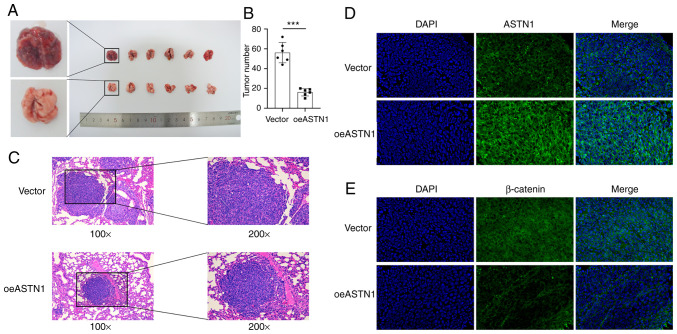
ASTN1 inhibits tumorigenesis of HCC cells in vivo
To determine the role of ASTN1 levels in tumorigenesis, HCCLM3 cells overexpressing ASTN1 (test group; oeASTN1) or the cells transfected with an empty vector virus (control group; vector), were injected into nude mice via the tail vein. The results suggested that, ASTN1 overexpression exhibited significantly slower tumor growth compared with the vector group (Fig. 8A). After 4 weeks, the number (Fig. 8B) and size (Fig. 8C) of tumors were significantly reduced in mice injected with ASTN1 overexpressing cells compared with the vector group. Furthermore, counterstaining of ASTN1 and β-catenin was evaluated using immunofluorescence staining in tissues (Fig. 8D and E), which indicated that ASTN1 expression was upregulated, whereas β-catenin expression was downregulated in the test group compared with the control group. These results suggest that overexpression of ASTN1 notably reduced HCC tumor development and metastasis in vivo, and this may have been mediated though inhibition of the Wnt signaling pathway.
Figure 8.
ASTN1 suppresses the carcinogenesis of HCC cells in vivo. Nude mice (n=6) were injected with ASTN1-overexpressing HCCLM3 cells (oeASTN1) or empty vector transfected cells. A total of 4 weeks after injection, the xenografts were collected and counted. (A) Representative images of the tumors generated by empty vector (top panel) and ASTN1-overexpressing (bottom panel) cells. (B) Mean counts of two groups of mice were quantified. Less tumors were formed in the ASTN1-overexpressing HCC cells compared with the empty vector transfected cells. *P<0.05, ***P<0.001. (C) Tumor sections stained with hematoxylin and eosin showed the presence of smaller tumor sizes in the ASTN1-overexpressing group. Immunofluorescence staining was performed to assess (D ASTN1 and (E) β-catenin expression in xenografts. ASTN1 expression was increased and β-catenin expression was decreased in the ASTN1-overexpressing group. ASTN1, astrotactin 1; HCC, hepatocellular carcinoma.
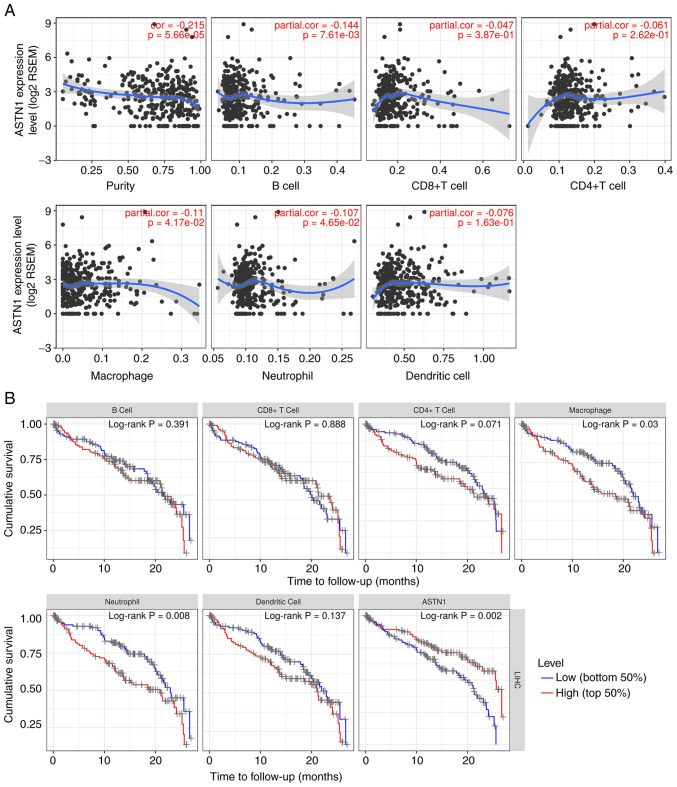
ASTN1 is correlated with immune infiltration
The correlation between ASTN1 expression and immune infiltration levels were investigated using TIMER. The results showed that ASTN1 expression was significantly negatively correlated with tumor purity, B cell, macrophage and neutrophil infiltration (P<0.05; Fig. 9A). Patients with low macrophage (P=0.030) or neutrophil counts (P=0.008), or increased ASTN1 levels (P=0.002) exhibited improved survival rates (Fig. 9B). As shown in Table II, the correlations between ASTN1 expression and immune marker genes were also analyzed. The detailed correlation diagram results are presented in Figs. S2 and S3.
Figure 9.
ASTN1 is correlated with immune infiltration. (A) Correlation between ASTN1 expression and tumor purity together with six immune infiltrates (B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages and dendritic cells). (B) Prognostic analysis of ASTN1 expression and six immune infiltrates. ASTN1, astrotactin 1.
Table II.
Correlation analysis between ASTN1 and immune cell markers in TIMER.
| Correlation without adjustment | Correlation adjusted by purity | ||||
|---|---|---|---|---|---|
| Cell types | Gene markers | Correlation | P-value | Correlation | P-value |
| CD8+ T cells | CD8A | 0.034182 | 0.511591 | 0.081896 | 0.128411 |
| CD8B | 0.015941 | 0.759587 | 0.063422 | 0.239343 | |
| T cells (general) | CD3D | −0.12757 | 0.013933 | −0.10127 | 0.059859 |
| CD3E | 0.007557 | 0.884658 | 0.034257 | 0.52537 | |
| CD2 | −0.03618 | 0.487164 | −0.00988 | 0.854771 | |
| B cells | CD19 | 0.014771 | 0.776747 | 0.039349 | 0.465652 |
| CD79A | 0.11551 | 0.026093 | 0.13686 | 0.010818 | |
| Monocytes | CD86 | −0.04087 | 0.432495 | −0.01351 | 0.802259 |
| CD115 (CSF1R) | 0.043919 | 0.398952 | 0.065139 | 0.226833 | |
| TAMs | CCL2 | 0.180094 | 0.000491 | 0.191626 | 0.000337 |
| CD68 | −0.07161 | 0.168719 | −0.05022 | 0.351714 | |
| IL10 | 0.002457 | 0.962381 | 0.023704 | 0.660375 | |
| M1 macrophages | INOS (NOS2) | 0.126491 | 0.01477 | 0.131225 | 0.01458 |
| IRF5 | −0.16654 | 0.001285 | −0.16246 | 0.002436 | |
| COX2 (PTGS2) | 0.26749 | 1.69E-07 | 0.27122 | 3.01E-07 | |
| M2 macrophages | CD163 | 0.136422 | 0.008511 | 0.161701 | 0.002554 |
| VSIG4 | 0.035601 | 0.494211 | 0.054388 | 0.313091 | |
| MS4A4A | 0.050425 | 0.332751 | 0.066037 | 0.220477 | |
| Neutrophils | CD66b (CEACAM8) | 0.003447 | 0.94725 | −0.00637 | 0.906081 |
| CD11b (ITGAM) | −0.17579 | 0.000671 | −0.16203 | 0.002503 | |
| CCR7 | 0.1447 | 0.005231 | 0.147673 | 0.005923 | |
| Natural killer cells | KIR2DL1 | 0.081523 | 0.116985 | 0.102597 | 0.05658 |
| KIR2DL3 | 0.042478 | 0.41462 | 0.053668 | 0.319551 | |
| KIR2DL4 | −0.06845 | 0.18832 | −0.02314 | 0.667981 | |
| KIR3DL1 | 0.073519 | 0.157592 | 0.089658 | 0.095901 | |
| KIR3DL2 | 0.056232 | 0.28001 | 0.097365 | 0.070477 | |
| KIR3DL3 | −0.00972 | 0.851954 | 0.010157 | 0.850686 | |
| KIR2DS4 | 0.01597 | 0.759156 | 0.024581 | 0.648643 | |
| Dendritic cells | HLA-DPB1 | 0.01705 | 0.743422 | 0.047378 | 0.37963 |
| HLA-DQB1 | −0.01022 | 0.844391 | 0.020789 | 0.699987 | |
| HLA-DRA | 0.01886 | 0.717289 | 0.049302 | 0.360557 | |
| HLA-DPA1 | 0.032908 | 0.527457 | 0.056162 | 0.297547 | |
| BDCA-1 (CD1C) | 0.188423 | 0.000262 | 0.181648 | 0.000687 | |
| BDCA-4 (NRP1) | 0.193742 | 0.000173 | 0.219065 | 3.95E-05 | |
| CD11c (ITGAX) | −0.06498 | 0.211775 | −0.04713 | 0.382092 | |
| Th1 | T-bet (TBX21) | 0.111911 | 0.031157 | 0.127942 | 0.017264 |
| STAT4 | −0.03855 | 0.45914 | −0.03675 | 0.495677 | |
| STAT1 | −0.06341 | 0.223052 | −0.05128 | 0.341569 | |
| IFN-γ (IFNG) | −0.11173 | 0.031431 | −0.0763 | 0.156743 | |
| TNF-α (TNF) | −0.00913 | 0.860897 | −0.00176 | 0.973948 | |
| Th2 | GATA3 | 0.050592 | 0.331143 | 0.076333 | 0.156542 |
| STAT6 | 0.148376 | 0.004181 | 0.167407 | 0.00178 | |
| STAT5A | 0.045432 | 0.382891 | 0.082603 | 0.125135 | |
| IL13 | −0.01168 | 0.822557 | −0.00147 | 0.978293 | |
| Tfh | BCL6 | 0.136255 | 0.008592 | 0.123545 | 0.02153 |
| IL21 | −0.00174 | 0.973357 | −0.01194 | 0.824809 | |
| Th17 | STAT3 | 0.067729 | 0.19304 | 0.095733 | 0.075343 |
| IL17A | 0.076524 | 0.141253 | 0.069255 | 0.198757 | |
| Treg | FOXP3 | 0.030033 | 0.564174 | 0.016873 | 0.754475 |
| CCR8 | −0.0099 | 0.849268 | −0.00339 | 0.949862 | |
| STAT5B | 0.288155 | 1.59E-08 | 0.295426 | 2.13E-08 | |
| TGFβ (TGFB1) | 0.061255 | 0.239205 | 0.083081 | 0.122961 | |
| T cell exhaustion | PD-1 (PDCD1) | −0.0909 | 0.080357 | −0.05511 | 0.306696 |
| CTLA4 | −0.13813 | 0.007714 | −0.10896 | 0.042819 | |
| LAG3 | −0.0897 | 0.08444 | −0.05067 | 0.347395 | |
| TIM-3 (HAVCR2) | −0.11253 | 0.030226 | −0.08619 | 0.109528 | |
| GZMB | 0.008927 | 0.863935 | 0.037332 | 0.488851 | |
ASTN1, astrotactin 1; TIMER, Tumor Immune Estimation Resource; TAM, tumor-associated macrophage; Th, T helper cell; Tfh, follicular helper T cell; Treg, regulatory T cell.
Discussion
HOTAIR is a lncRNA located on chromosome 12q13.13 at the HOXC site which is associated with cancer progression (24). Numerous studies examining the biological functions of HOTAIR during the progression of HCC have suggested that expression of HOTAIR is upregulated in hepatocellular carcinoma (HCC) tissues relative to the normal tissues, and that HOTAIR may serve as a potential marker of HCC (25). Inspired by the bioinformatics analysis on genes associated with invasiveness targeted by HOTAIR silencing, astrotactin 1 (ASTN1) has been identified as one of the top most upregulated genes (15). Therefore, ASTN1 is speculated to be associated with liver cancer progression. To assess this hypothesis, ASTN1 expression was examined using the TCGA HCC dataset as well as in patients who visited the Sun Yat-sen University Cancer Center. According to the results of the present study, ASTN1 expression was significantly decreased in HCC tissues compared with non-carcinoma tissues. Using Oncomine gene expression data we found that ASTN1 was slightly increased in HCC when compared with the normal liver. However, the Oncomine result should be interpreted with caution since GSE6764: i) included a relatively small population (n=45), ii) was based on microarray instead of RNA sequencing, and iii) had lack of validation. Additionally, downregulated ASTN1 expression levels in HCC were associated with a lower tumor grade, microscopic vascular invasion, microscopic satellite nodule, destructed encapsulation, advanced TNM stage, advanced BCLC stage and a less favorable OS. These results highlight the potential of ASTN1 as a candidate marker for diagnosis and prognostic prediction in patients with HCC.
There are only a few existing studies that have examined the effects of ASTN1 on tumor development, and only one study has examined its role in small lung cancer patients (26). Using exome sequencing, Iwakawa et al (27) analyzed 44 cancer tissues from 38 cases of small cell lung cancer in a Japanese cohort, as well as 38 normal controls, and there were notable ASTN1 gene mutations identified, highlighting the potential of ASTN1 during cancer development. The involvement of ASTN1 in suppressing the migration of neuronal cells has garnered increasing attention over the last decades (28–30). Yi et al (31) reported that miR-sc3 enhanced the proliferation and migration of Schwann cells through targeting of ASTN1. Horn et al (28) demonstrated that neuron migration and neuron-glia attachment were determined by neural cadherin (CDH2) expression in glial cells, and ASTN1 enhanced cell migration through directly interacting with CDH2 in neurons and glial cells. CDH2 and ASTN1 formed a complex at the migration junction, which served a critical role in the glial-mediated neuronal migration. In the present study, cases with ASTN1 downregulation were associated with increased risks of microscopic vascular invasion and advanced clinical stage; therefore, the effects of ASTN1 on liver cancer migration and invasion were examined. The role of ASTN1 in the migratory and invasive capacity of liver cancer cells was assessed in the present study through regulating the expression of ASTN1 using lentiviral transfection. To the best of our knowledge, the present study is the first to show that ASTN1 may be considered a tumor-suppressor gene in liver cancer.
At present, a number of signal transduction pathways have been shown to participate in regulating the invasive and metastatic capacity of liver cancer cells. Of these, the Wnt/β-catenin signal transduction pathway is a classical pathway, which serves a critical role in embryonic development as well as self-renewal of adult tissues (32). When abnormally activated, this pathway results in abnormal cell growth as well as malignant transformation (33). Approximately 30% of HCC patients show abnormally high levels of Wnt/β-catenin signal transduction pathway activity (34–36). The abnormal activation of the Wnt/β-catenin signaling pathway within HCC enhances cell proliferation and increases resistance to sorafenib (37), and suppressing the Wnt/β-catenin signaling pathway reduces the tumorigenic potential (38). This suggests that the excessive activation of the Wnt/β-catenin signal transduction pathway facilitates HCC tumorigenesis. Therefore, it is essential to explore the related mechanism of the hyperactivation of the Wnt/β-catenin signal transduction pathway within liver cancer, which may assist in the development of targeted therapeutics for preventing tumor relapse. Nonetheless, the role of ASTN1 in mediating the Wnt signal transduction pathway remains unclear. In the present study, the GSEA results based on TCGA dataset suggested that the ASTN1 levels were negatively associated with the Wnt signal transduction pathway. Consistent with this, the Wnt/β-catenin signal transduction pathway was also found to be excessively activated in human liver cancer tissues. Therefore, these results suggest that ASTN1 functioned via the Wnt/β-catenin signal transduction pathway and downregulated several of the downstream genes of this pathway. Overexpression of ASTN1 resulted in a significant downregulation in β-catenin, TCF1, TCF4, C-jun, C-myc, COX2, MM2, MMP9 and VEGF protein expression levels, which are the primary signaling components or effectors downstream of the Wnt signaling pathway, and are involved in the development of liver cancer. Treatment with XAV939, an inhibitor of the Wnt/β-catenin signaling pathway, significantly increased the migratory and invasive capacity of ASTN1-overexpressing cells. Taken together, these results suggest that the Wnt signaling transduction pathway regulated the functions of ASTN1 with regards to the invasive and migratory capacities of cells.
Recent advances in checkpoint inhibition therapy have renewed investigative interest into tumor immunotherapy. The present study also predicted the association between ASTN1 expression and different immune infiltration levels in HCC. The results showed that the expression of ASTN1 in HCC was significantly associated with increased infiltration of B cells, macrophages and neutrophils. ASTN1 and marker genes of M1 macrophages were also significantly correlated. It has been shown that SPON2 may inhibit the metastasis of HCC through the recruitment of M1 macrophages (39). The Il-6/STAT3 pathway mediates the polarization of M1 macrophages in HCC (40). These results suggest the potential functional mechanism underlying ASTN1-mediated regulation of M1 macrophages in HCC, although further validation is required to confirm this hypothesis. Additionally, the correlation between ASTN1 and cancer immune infiltrates requires further investigation.
In conclusion, downregulated ASTN1 expression was associated with a less favorable prognosis in patients with HCC. In addition, overexpression of ASTN1 in liver cancer cells reduced the migratory and invasive capacity of the cells, and reduced the activity of the Wnt signal transduction pathway. ASTN1 was shown to be correlated with immune infiltrates in HCC. Therefore, it is of crucial significance to examine the underlying mechanism of action of ASTN1 to develop novel preventative and therapeutic strategies for treatment of liver cancer.
Supplementary Material
Acknowledgements
Not applicable.
Glossary
Abbreviations
- ASTN1
astrotactin 1
- HCC
hepatocellular carcinoma
- TCGA
The Cancer Genome Atlas
- GEO
Gene Expression Omnibus
- TIMER
Tumor Immune Estimation Resource
- HOTAIR
Hox Transcript Antisense RNA
- GSEA
gene set enrichment analysis
- OS
overall survival
- RFS
recurrence-free survival
- shRNA
short hairpin RNA
- mRNA
messenger RNA
- IHC
immunohistochemistry
- RT-qPCR
reverse transcription-quantitative PCR
- AUC
area under the roc curve
- HR
hazard ratio
- CI
confidence interval
Funding
The present study was supported by the National Natural Science Foundation of China (grant no. 81801804) and Guangdong Provincial People's Hospital Project 2017 (2017bq05).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
QFC, FS, PW, and WL conceived and designed the experiments. QFC, FS, TH, and CH performed the experiments and collected the data. QFC, FS, TH, CH and LS analyzed the data. QFC, PW, and WL wrote the paper. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
The study of clinical samples was approved by the Ethics Committee of Sun Yat-sen University Cancer Center and was performed in accordance with the principles embodied in the Declaration of Helsinki. All patients provided written informed consent to participate in the study. All animal procedures were approved by the Ethics Committee of Sun Yat-sen University Cancer Center.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Dasgupta P, Henshaw C, Youlden DR, Clark PJ, Aitken JF, Baade PD. Global trends in incidence rates of primary adult liver cancers: A systematic review and meta-analysis. Front Oncol. 2020;10:171. doi: 10.3389/fonc.2020.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen QF, Jia ZY, Yang ZQ, Fan WL, Shi HB. Transarterial chemoembolization monotherapy versus combined transarterial chemoembolization-microwave ablation therapy for hepatocellular carcinoma tumors ≤5 cm: A propensity analysis at a single center. Cardiovasc Intervent Radiol. 2017;40:1748–1755. doi: 10.1007/s00270-017-1736-8. [DOI] [PubMed] [Google Scholar]
- 3.Shen L, Zeng Q, Guo P, Huang J, Li C, Pan T, Chang B, Wu N, Yang L, Chen Q, et al. Dynamically prognosticating patients with hepatocellular carcinoma through survival paths mapping based on time-series data. Nat Commun. 2018;9:2230. doi: 10.1038/s41467-018-04633-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen QF, Xia JG, Li W, Shen LJ, Huang T, Wu P. Examining the key genes and pathways in hepatocellular carcinoma development from hepatitis B virus-positive cirrhosis. Mol Med Rep. 2018;18:4940–4950. doi: 10.3892/mmr.2018.9494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen QF, Wu PH, Huang T, Shen LJ, Huang ZL, Li W. Efficacy of treatment regimens for advanced hepatocellular carcinoma: A network meta-analysis of randomized controlled trials. Medicine (Baltimore) 2019;98:e17460. doi: 10.1097/MD.0000000000017460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang ZL, Li W, Chen QF, Wu PH, Shen LJ. Eight key long non-coding RNAs predict hepatitis virus positive hepatocellular carcinoma as prognostic targets. World J Gastrointest Oncol. 2019;11:983–997. doi: 10.4251/wjgo.v11.i11.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen QF, Li W, Wu P, Shen L, Huang ZL. Alternative splicing events are prognostic in hepatocellular carcinoma. Aging (Albany NY) 2019;11:4720–4735. doi: 10.18632/aging.102085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen QF, Huang T, Si-Tu QJ, Wu P, Shen L, Li W, Huang Z. Analysis of competing endogenous RNA network identifies a poorly differentiated cancer-specific RNA signature for hepatocellular carcinoma. J Cell Biochem. 2020;121:2303–2317. doi: 10.1002/jcb.29454. [DOI] [PubMed] [Google Scholar]
- 9.Huang T, Chen QF, Chang BY, Shen LJ, Li W, Wu PH, Fan WJ. TFAP4 promotes hepatocellular carcinoma invasion and metastasis via activating the PI3K/AKT signaling pathway. Dis Markers. 2019;2019:7129214. doi: 10.1155/2019/7129214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeon Y, Kim H, Jang ES, Hong S, Kim JW, Yoon YS, Cho JY, Han HS, Jeong SH. Expression profile and prognostic value of glypican-3 in post-operative South Korean hepatocellular carcinoma patients. APMIS. 2016;124:208–215. doi: 10.1111/apm.12491. [DOI] [PubMed] [Google Scholar]
- 11.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao JZ, Li J, Du JL, Li XL. Long non-coding RNA HOTAIR is a marker for hepatocellular carcinoma progression and tumor recurrence. Oncol Lett. 2016;11:1791–1798. doi: 10.3892/ol.2016.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geng YJ, Xie SL, Li Q, Ma J, Wang GY. Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression. J Int Med Res. 2011;39:2119–2128. doi: 10.1177/147323001103900608. [DOI] [PubMed] [Google Scholar]
- 14.Yang T, He X, Chen A, Tan K, Du X. LncRNA HOTAIR contributes to the malignancy of hepatocellular carcinoma by enhancing epithelial-mesenchymal transition via sponging miR-23b-3p from ZEB1. Gene. 2018;670:114–122. doi: 10.1016/j.gene.2018.05.061. [DOI] [PubMed] [Google Scholar]
- 15.Ono H, Motoi N, Nagano H, Miyauchi E, Ushijima M, Matsuura M, Okumura S, Nishio M, Hirose T, Inase N, Ishikawa Y. Long noncoding RNA HOTAIR is relevant to cellular proliferation, invasiveness, and clinical relapse in small-cell lung cancer. Cancer Med. 2014;3:632–642. doi: 10.1002/cam4.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng C, Heintz N, Hatten ME. CNS gene encoding astrotactin, which supports neuronal migration along glial fibers. Science. 1996;272:417–419. doi: 10.1126/science.272.5260.417. [DOI] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Burchard J, Zhang C, Liu AM, Poon RT, Lee NP, Wong KF, Sham PC, Lam BY, Ferguson MD, Tokiwa G, et al. microRNA-122 as a regulator of mitochondrial metabolic gene network in hepatocellular carcinoma. Mol Syst Biol. 2010;6:402. doi: 10.1038/msb.2010.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roessler S, Jia HL, Budhu A, Forgues M, Ye QH, Lee JS, Thorgeirsson SS, Sun Z, Tang ZY, Qin LX, Wang XW. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70:10202–10212. doi: 10.1158/0008-5472.CAN-10-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lian Q, Wang S, Zhang G, Wang D, Luo G, Tang J, Chen L, Gu J. HCCDB: A database of hepatocellular carcinoma expression atlas. Genomics Proteomics Bioinformatics. 2018;16:269–275. doi: 10.1016/j.gpb.2018.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Menyhárt O, Nagy Á, Győrffy B. Determining consistent prognostic biomarkers of overall survival and vascular invasion in hepatocellular carcinoma. R Soc Open Sci. 2018;5:181006. doi: 10.1098/rsos.181006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS. TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77:e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ezponda T, Licht JD. Molecular pathways: Deregulation of histone h3 lysine 27 methylation in cancer-different paths, same destination. Clin Cancer Res. 2014;20:5001–5008. doi: 10.1158/1078-0432.CCR-13-2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan SX, Tao QF, Wang J, Yang F, Liu L, Wang LL, Zhang J, Yang Y, Liu H, Wang F, et al. Antisense long non-coding RNA PCNA-AS1 promotes tumor growth by regulating proliferating cell nuclear antigen in hepatocellular carcinoma. Cancer Lett. 2014;349:87–94. doi: 10.1016/j.canlet.2014.03.029. [DOI] [PubMed] [Google Scholar]
- 26.Chang H. Cleave but not leave: Astrotactin proteins in development and disease. IUBMB Life. 2017;69:572–577. doi: 10.1002/iub.1641. [DOI] [PubMed] [Google Scholar]
- 27.Iwakawa R, Kohno T, Totoki Y, Shibata T, Tsuchihara K, Mimaki S, Tsuta K, Narita Y, Nishikawa R, Noguchi M, et al. Expression and clinical significance of genes frequently mutated in small cell lung cancers defined by whole Exome/RNA sequencing. Carcinogenesis. 2015;36:616–621. doi: 10.1093/carcin/bgv026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horn Z, Behesti H, Hatten ME. N-cadherin provides a cis and trans ligand for astrotactin that functions in glial-guided neuronal migration. Proc Natl Acad Sci USA. 2018;115:10556–10563. doi: 10.1073/pnas.1811100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Behesti H, Fore TR, Wu P, Horn Z, Leppert M, Hull C, Hatten ME. ASTN2 modulates synaptic strength by trafficking and degradation of surface proteins. Proc Natl Acad Sci USA. 2018;115:E9717–E9726. doi: 10.1073/pnas.1809382115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson PM, Fryer RH, Fang Y, Hatten ME. Astn2, a novel member of the astrotactin gene family, regulates the trafficking of ASTN1 during glial-guided neuronal migration. J Neurosci. 2010;30:8529–8540. doi: 10.1523/JNEUROSCI.0032-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yi S, Wang S, Zhao Q, Yao C, Gu Y, Liu J, Gu X, Li S. miR-sc3, a novel microRNA, promotes Schwann cell proliferation and migration by targeting Astn1. Cell Transplant. 2016;25:973–982. doi: 10.3727/096368916X690520. [DOI] [PubMed] [Google Scholar]
- 32.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 33.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 34.Lachenmayer A, Alsinet C, Savic R, Cabellos L, Toffanin S, Hoshida Y, Villanueva A, Minguez B, Newell P, Tsai HW, et al. Wnt-pathway activation in two molecular classes of hepatocellular carcinoma and experimental modulation by sorafenib. Clin Cancer Res. 2012;18:4997–5007. doi: 10.1158/1078-0432.CCR-11-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, Calderaro J, Bioulac-Sage P, Letexier M, Degos F, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44:694–698. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kan Z, Zheng H, Liu X, Li S, Barber TD, Gong Z, Gao H, Hao K, Willard MD, Xu J, et al. Whole-genome sequencing identifies recurrent mutations in hepatocellular carcinoma. Genome Res. 2013;23:1422–1433. doi: 10.1101/gr.154492.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Y, Ye X, Zhang JB, Ouyang H, Shen Z, Wu Y, Wang W, Wu J, Tao S, Yang X, et al. PROX1 promotes hepatocellular carcinoma proliferation and sorafenib resistance by enhancing β-catenin expression and nuclear translocation. Oncogene. 2015;34:5524–5535. doi: 10.1038/onc.2015.7. [DOI] [PubMed] [Google Scholar]
- 38.Yamashita T, Ji J, Budhu A, Forgues M, Yang W, Wang HY, Jia H, Ye Q, Qin LX, Wauthier E, et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology. 2009;136:1012–1024. doi: 10.1053/j.gastro.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang YL, Li Q, Yang XM, Fang F, Li J, Wang YH, Yang Q, Zhu L, Nie HZ, Zhang XL, et al. SPON2 promotes M1-like macrophage recruitment and inhibits hepatocellular carcinoma metastasis by distinct integrin-Rho GTPase-Hippo pathways. Cancer Res. 2018;78:2305–2317. doi: 10.1158/0008-5472.CAN-17-2867. [DOI] [PubMed] [Google Scholar]
- 40.Yin Z, Ma T, Lin Y, Lu X, Zhang C, Chen S, Jian Z. IL-6/STAT3 pathway intermediates M1/M2 macrophage polarization during the development of hepatocellular carcinoma. J Cell Biochem. 2018;119:9419–9432. doi: 10.1002/jcb.27259. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.