Abstract
The recent wide-spread adoption of single cell profiling technologies has revealed that individual cancers are not homogenous collections of deregulated cells, but instead are comprised of multiple genetically and phenotypically distinct cell subpopulations that exhibit a wide range of responses to extracellular signals and therapeutic insult. Such observations point to the urgent need to understand cancer as a complex, adaptive system. Cancer systems biology studies seek to develop the experimental and theoretical methods required to understand how biological components work together to determine how cancer cells function. Ultimately, such approaches will lead to improvements in how cancer is managed and treated. In this review, we discuss recent advances in cancer systems biology approaches to quantify, model, and elucidate mechanisms of heterogeneity.
Introduction
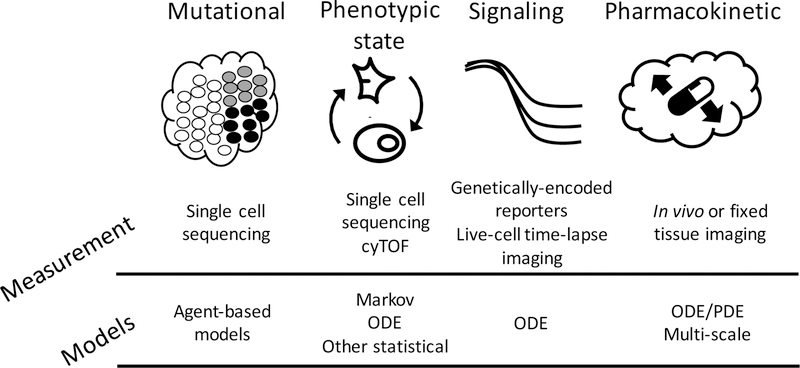
Cancers are marked by substantial genetic, epigenetic, and phenotypic heterogeneity. With the advent of single cell technologies, our ability to quantify cell-to-cell variability is advancing rapidly; however, the biological interpretation of these complex data is only at a nascent stage. A systems biology approach, in which quantitative technologies, comprehensive experimental measurements, and computational analyses are carefully integrated, is required to understand intratumoral heterogeneity and extract biological meaning. Moreover, computational frameworks will aid in the elucidation of the underlying biological mechanisms of heterogeneity. Such integrated approaches will ultimately lead to a deeper understanding of cancer and how to manage it to improve patient outcomes. Here, we cover recent advancements in cancer systems biology that have improved our understanding of heterogeneity in genetic, phenotypic, signaling, and pharmacokinetic realms (Figure 1).
Figure 1.
Classes of therapeutically-relevant heterogeneity. Variation may arise from cell-to-cell differences in mutational spectra, phenotype, cell signaling pathway activity, or pharmacokinetics. Measurement technologies and modeling approaches to study these various forms of heterogeneity are indicated.
Genetic heterogeneity
It is well-established that the initiation and progression of cancers depends on the acquisition of multiple driver mutations that activate oncogenic pathways and lead to the induction of cancer hallmarks1, 2. Moreover, the growth of tumors likely reflects a complex process of clonal evolution that ultimately results in considerable genetic heterogeneity within tumors3. A recent pan-cancer analysis of primary tumors revealed that the majority of cancers harbor multiple genetically distinct clones, and tumors with more clones harbored more driver mutations and showed greater nuclear heterogeneity4. Moreover, increased genetic heterogeneity is associated with poor outcome, indicating that measures of intratumoral heterogeneity may serve as useful biomarkers4. With the widespread adoption of single-cell sequencing technologies, we are poised to develop deeper and more precise views of intratumoral heterogeneity5.
Recent years have seen a substantial advancement in our understanding of genetic heterogeneity, particularly as it relates to therapeutic response. Using cell line model systems, Ramirez and colleagues studied drug-tolerant persister cells and found that they may provide a latent reservoir for the emergence of drug-resistance. Specifically, long-term drug exposure leads to survival and expansion of cells through a drug-tolerant state that can mediate genetically-driven resistance mechanisms6. Consistent with this idea, single-cell DNA and RNA sequencing of triple negative breast cancer patient samples profiled before and after treatment demonstrated that resistant genotypes were pre-existing and adaptively selected after treatment, while new transcriptional profiles were acquired by reprogramming in response to therapy7. Taken together, these studies further motivate the importance of understanding and managing intratumoral genetic heterogeneity. Laajala, et al (this issue) reviews systems biology approaches to study cancer genetic heterogeneity in greater depth.
Phenotypic heterogeneity
Cancers display substantial phenotypic heterogeneity and rapid adaptation in response to stimuli that occur on timescales that cannot be explained by genetic evolution or clonal selection, indicating a substantial contribution of the epigenome to cell-to-cell variation. A landmark study by Sharma and colleagues revealed that small sub-populations of cancer cells could mediate therapeutic resistance on time-scales too rapid to be explained by genetic evolution and selection8. Moreover, the acquisition of this drug-tolerant phenotype was found to be transient and result from chromatin changes, indicating a dynamic regulation of phenotypic heterogeneity which stands in contrast to long-term “hard-wired” genetic changes. In support of this hypothesis, a recent study of therapeutic response revealed substantial cell-to-cell transcriptional variability that involves infrequent, semi-coordinated transcription of a number of resistance markers in a small percentage of “jackpot” cells9. Such findings indicate that heterogeneity is mediated at multiple molecular levels and therefore requires integrated experimental approaches that can link across modalities at the single cell level.
Several groups have developed computational frameworks to shed light on the molecular basis and dynamic nature of phenotypic heterogeneity. Gupta and colleagues found that subpopulations of sorted phenotypically-distinct triple-negative breast cancer cells could return to equilibrium proportions over time10. This observation could be explained by a Markov model in which cells transition stochastically between phenotypically distinct states, indicating phenotypic plasticity even in the absence of an external pressure such as therapy. Building on this idea, Risom and colleagues used an ordinary differential equation (ODE) model to show that drug tolerant states can arise through differentiation state transitions rather than Darwinian selection of preexisting subpopulations11, 12. Such switching between differentiation states may enable cells to rapidly evade therapy. These studies highlight the importance of coupling experimental observation with computational frameworks to gain insight into the underlying mechanisms of cell-to-cell variation and plasticity.
The wide-spread adoption of single cell sequencing methods provides insights into the phenotypic state of hundreds to thousands of individual cells and has spurred novel analytics to aid in interpretation of these complex data13–15. In particular, some of the challenges of single cell sequencing include both technical artifacts related to differences in read-depth, dropouts (i.e., stochastic false-negative gene expression measurements), and variation in total transcript abundance16, 17. However, many of these technical challenges are being overcome with novel analytical approaches. For example, Azizi and colleagues developed the BISCUIT algorithm, a Bayesian approach for simultaneous clustering and imputing single cell RNAseq data13. Seurat is a suite of tools for analysis and visualization of single cell RNAseq data sets based on common sources of variation, which enables identification of shared populations across data sets; its major strengths are ease-of-use and extensive data visualization tools14. As new technologies develop, so too does the interest in integrating data from across various modalities to develop a complete picture of the biological mechanisms associated with heterogeneity18.
Ultimately, effective cancer therapy regimens will need to consider heterogeneity, and several methods to design therapies combatting cell-to-cell variation have emerged in recent years19–22. For example, DRUG-NEM is an integrated experimental-computational approach to rationally identify drug combinations that account for intratumoral heterogeneity19. This approach applies cyTOF (Mass Cytometry Time-of-Flight), a variant of flow cytometry that replaces fluorescence detection with metal ion mass spectrometry for greatly improved multiplexing23. cyTOF enables the simultaneous measurement of many intracellular and surface markers for hundreds of thousands of single cells in a sample23. The authors assess single-cell proteomic responses to individual drugs and then use a nested effects model to prioritize drug combinations that produce the maximal desired intracellular effects. Such nested effects models can identify common and unique signaling changes in response to drug treatments and this information can be used to optimize the minimum number of drugs that induce desired signaling changes24. Functional approaches such as these are quite advanced for blood cancers, where primary cancer cells are relatively accessible25.
Heterogeneity in cell signaling and pathway activity
While statistical models like those described above have refined our view of how cell-to-cell variability drives resistance, mechanistic information about relevant resistance pathways enables in silico experimentation of how individual molecular events influence overall behavior. Cells respond dynamically to treatment, and so genetically-encoded single cell reporters and in vitro live-cell time-lapse imaging, paired with dynamical systems models, have become a mainstay of studying this variability26–30. The continued development and use of these experimental and theoretical tools together demonstrate the power of this approach. From foundational studies of variability in TRAIL-induced apoptosis, the key elements of studying cell heterogeneity in specific pathways have remained largely unchanged31–34. Common to all these studies has been that cell-to-cell variability is explained by variation in the abundance of a few key proteins, rather than variation across many pathway components or intrinsic noise. Critically, computational models identified key components that both predict variation in phenotypic response across cells and pinpoint how to control this variability.
Imaging paired with dynamical systems analysis will continue to be used in studies of pathway activity variability and drug resistance. However, new experimental and computational tools will certainly expand the impact this pairing provides. Studies of pathway-specific cell-to-cell heterogeneity will always depend upon detailed knowledge of pathways and their relevant regulatory processes. Therefore, general improvements in our biological knowledge will expand the range of pathways and processes that can accurately be modeled. While imaging provides uniquely rich, dynamic data on single cell responses, it remains limited in the number, types, and throughput of molecular measurements one can perform. Methods to interrogate molecular state, such as endogenous protein tagging, will widen the accessible range of pathways and resistance mechanisms35. The number of measurements that can be multiplexed in single living cells is unlikely to considerably improve, and cell-to-cell variability dictates non-destructive techniques for sampling multiple measurements in the same cell. However, dynamic measurements can be paired with destructive measurements as an endpoint, and data can then be integrated to develop an understanding of the biological mechanisms that may be driving differences in dynamic responses36. Modeling developments to parameterize dynamical models on mixtures of population and individual cell measurements will be critical to enhance the value of these mixed measurements37. Finally, individual pathways operate within a larger context of broader molecular changes. Methods to integrate “phenotypic” and mechanistic cell variability will ultimately be necessary to recognize the simultaneous contributions of pathway-specific and global changes. These burgeoning improvements in measurement and modeling show that dynamical models are poised for expanded use in studies of cell signaling.
Spatial heterogeneity
Properties of the in vivo tumor environment introduce sources of phenotypic and molecular heterogeneity beyond those outlined above. In contrast to most in vitro models of cancer, tissues present variation in the extracellular matrix, resident host cells, and barriers to drug access38. Each of these can promote resistance; indeed, pharmacokinetic/pharmacodynamics (PK/PD) limitations have long been known to drive resistance across disease indications, particularly for tissues such as the central nervous system39. Spatial variation in these properties may co-exist in the same tumor and hinder drug function even in normally accessible tissues40.
In vivo and fixed tissue imaging modalities are uniquely powerful for capturing spatial variation in tumor properties. Mass spectrometry coupled to localized tissue sampling methods has recently enabled drug distribution imaging, alongside traditional immunohistochemistry techniques41. These methods also extend whole organism imaging methods in resolution and sensitivity to identify drug biodistribution within individual tissues. While limited to specific tissue sites, fluorescence intravital imaging has also provided a window into the spatial variation of tumors42. For example, even within a single patient, individual tumor nodes can have striking variation in immune infiltrates43. A continuing challenge will be the many length scales of spatial variation, from poor drug distribution across tissues to drug sequestration by neighboring cells44.
Pharmacokinetic models are widespread and accepted for modeling the diffusion and transport of molecules within cells and tissues. PK/PD models provide precise predictions linking molecular mechanism to cell response but are only accurate if built with a detailed understanding of the relevant properties to model. Randall et al recently integrated in vivo imaging of drug distribution and phosphoproteomic evaluation of signaling effect to reveal that intracranial EGFR inhibitor availability cooperates with other mechanisms of resistance in glioblastoma cells45. This integration is especially powerful for distinguishing between resistance driven by drug availability and other mechanisms.
Despite spatial effects being long-recognized as a source of intratumoral heterogeneity, more robust experimental tools and models are needed to help address how this variation influences cancer biology and treatment. In particular, the range of perturbations possible and the scale on which they can be performed is quite limited in the in vivo environment. In vitro methods to recapitulate critical features of the in vivo environment will be critical to enable mechanistic studies. For example, Wu et al. have applied a microfluidic device to study the interaction between breast cancer cell drug resistance and migration46. Others have demonstrated that hydrogel biomaterials can mimic the tumor extracellular matrix and enable detailed study of individual components47, 48. A key requirement of such mimetic systems is the inclusion of salient variables and features, which are not always well understood.
Future prospects for understanding and managing intratumoral heterogeneity
In reality, these distinct forms of heterogeneity likely operate to varying extents in parallel within a single tumor. Therefore, applying new findings from model systems will require methods to evaluate which forms of heterogeneity are most relevant to therapeutic response. Integrating methods to study these forms of heterogeneity will also reveal how they can impact each other. For example, Keren et al. recently showed that imaging cyTOF can provide both high dimensional and spatial information in breast tumors, and that the spatial organization of tumors can predict prognosis49. Forms of single-cell gene expression analysis50 that preserve spatial information are now also enabling similar analysis, along with highly-multiplexed immunofluorescence51, 52. These approaches afford assessment of multiple biomarkers in a single cell, all while maintaining spatial information about cell-to-cell variability and organizational structure. Of course, the studies described above have universally relied on a theoretical model tailored to the experimental system to understand the heterogeneity involved. As new experimental systems are developed, new computational models will need to be developed to leverage these data. Finally, a deep understanding of heterogeneity will ultimately require the integration of data across multiple physical and time scales, which can be achieved with multiscale modeling53.
Summary and conclusions
Genetic heterogeneity perhaps offers a glimpse into the future of how we will understand and manage other forms of intratumoral heterogeneity. Observation and characterization have given way to broad behavioral principles, such as that of clonal selection and expansion. Ultimately, all these forms of heterogeneity dynamically respond to treatment and therefore clinical management will require strategies to monitor patient’s tumors over time and in response to therapy 7, 54. An integrated cancer systems biology approach will provide the framework for understanding the molecular basis of heterogeneity and how to manage it clinically.
Table 1.
Experimental approaches for studying cancer heterogeneity
| Technique | Cost | Number of read-outs per cell | Spatial information | Temporal information |
|---|---|---|---|---|
| Single-cell sequencing | High | 10,000’s | No | No |
| seqFISH+ | Mid | 1,000’s | Yes | No |
| cyclicIF, mIHC | Low | 10’s | Yes | No |
| Intravital imaging | High | 10’s | Yes | Yes |
| Reporter molecules | Low | 1 | Yes | Yes |
Highlights.
Cell-cell variation manifests at multiple levels and scales in cancer
New profiling technologies are enabling quantitative assessment of heterogeneity
Integrated cancer systems biology approaches provide a mechanistic understanding of heterogeneity
Acknowledgements
This work was supported by National Cancer Institute grants U54 CA209988 (L.M.H.) U01-CA215709 (A.S.M.) and National Institute of Health grant DP5-OD01981 (A.S.M.) This work was also supported by the Jayne Koskinas Ted Giovanis Foundation for Health and Policy and the Breast Cancer Research Foundation, private foundations committed to critical funding of cancer research. The opinions, findings, conclusions or recommendations expressed in this material are those of the authors and not necessarily those of the Jayne Koskinas Ted Giovanis Foundation for Health and Policy, the Breast Cancer Research Foundation, the National Institutes of Health, or their respective directors, officers, or staffs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hanahan D & Weinberg RA Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B & Kinzler KW The Path to Cancer --Three Strikes and You’re Out. N Engl J Med 373, 1895–1898 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Marusyk A, Almendro V & Polyak K Intra-tumour heterogeneity: a looking glass for cancer? Nat Rev Cancer 12, 323–334 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Andor N et al. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat Med 22, 105–113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lawson DA, Kessenbrock K, Davis RT, Pervolarakis N & Werb Z Tumour heterogeneity and metastasis at single-cell resolution. Nat Cell Biol 20, 1349–1360 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramirez M et al. Diverse drug-resistance mechanisms can emerge from drug-tolerant cancer persister cells. Nat Commun 7, 10690 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim C et al. Chemoresistance Evolution in Triple-Negative Breast Cancer Delineated by Single-Cell Sequencing. Cell 173, 879–893 e813 (2018).* Single-cell DNA and RNAseq of triple negative breast cancers before and after treatment identifies pre-existing genomic alterations and transcriptional reprogramming of resistant signatures.
- 8.Sharma SV et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 141, 69–80 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shaffer SM et al. Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature 546, 431–435 (2017).** Cell-to-cell variability involves infrequent, semi-coordinated transcriptional changes in a very small percentage of “jack pot” cells that can mediate therapeutic resistance. The findings are related to the Luria-Delbruck model.
- 10.Gupta PB et al. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell 146, 633–644 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Chapman MP et al. Modeling differentiation-state transitions linked to therapeutic escape in triple-negative breast cancer. PLoS Comput Biol 15, e1006840 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Risom T et al. Differentiation-state plasticity is a targetable resistance mechanism in basal-like breast cancer. Nat Commun 9, 3815 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Azizi E et al. Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment. Cell 174, 1293–1308 e1236 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Butler A, Hoffman P, Smibert P, Papalexi E & Satija R Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420 (2018).* Seurat is a bioinformatics toolkit for analysis of large-scale single-cell RNAseq data sets.
- 15.van Dijk D et al. Recovering Gene Interactions from Single-Cell Data Using Data Diffusion. Cell 174, 716–729 e727 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiselev VY, Andrews TS & Hemberg M Challenges in unsupervised clustering of single-cell RNA-seq data. Nat Rev Genet 20, 273–282 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Nguyen QH, Pervolarakis N, Nee K & Kessenbrock K Experimental Considerations for Single-Cell RNA Sequencing Approaches. Front Cell Dev Biol 6, 108 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stuart T & Satija R Integrative single-cell analysis. Nat Rev Genet 20, 257–272 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Anchang B et al. DRUG-NEM: Optimizing drug combinations using single-cell perturbation response to account for intratumoral heterogeneity. Proc Natl Acad Sci U S A 115, E4294–E4303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brady SW et al. Combating subclonal evolution of resistant cancer phenotypes. Nat Commun 8, 1231 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Savage P et al. A Targetable EGFR-Dependent Tumor-Initiating Program in Breast Cancer. Cell Rep 21, 1140–1149 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Zhao B, Hemann MT & Lauffenburger DA Intratumor heterogeneity alters most effective drugs in designed combinations. Proc Natl Acad Sci U S A 111, 10773–10778 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spitzer MH & Nolan GP Mass Cytometry: Single Cells, Many Features. Cell 165, 780–791 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Markowetz F, Kostka D, Troyanskaya OG & Spang R Nested effects models for high-dimensional phenotyping screens. Bioinformatics (Oxford, England) 23, i305–312 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Kurtz SE et al. Molecularly targeted drug combinations demonstrate selective effectiveness for myeloid- and lymphoid-derived hematologic malignancies. Proc Natl Acad Sci U S A 114, E7554–E7563 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cohen-Saidon C, Cohen AA, Sigal A, Liron Y & Alon U Dynamics and variability of ERK2 response to EGF in individual living cells. Mol Cell 36, 885–893 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Kunkel MT, Ni Q, Tsien RY, Zhang J & Newton AC Spatio-temporal dynamics of protein kinase B/Akt signaling revealed by a genetically encoded fluorescent reporter. The Journal of biological chemistry 280, 5581–5587 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lahav G et al. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat Genet 36, 147–150 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Nelson DE et al. Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science 306, 704–708 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Regot S, Hughey JJ, Bajar BT, Carrasco S & Covert MW High-sensitivity measurements of multiple kinase activities in live single cells. Cell 157, 1724–1734 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen SH, Forrester W & Lahav G Schedule-dependent interaction between anticancer treatments. Science 351, 1204–1208 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paek AL, Liu JC, Loewer A, Forrester WC & Lahav G Cell-to-Cell Variation in p53 Dynamics Leads to Fractional Killing. Cell 165, 631–642 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roux J et al. Fractional killing arises from cell-to-cell variability in overcoming a caspase activity threshold. Molecular systems biology 11, 803 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spencer SL, Gaudet S, Albeck JG, Burke JM & Sorger PK Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature 459, 428–432 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Serebrenik YV, Sansbury SE, Kumar SS, Henao-Mejia J & Shalem O Efficient and flexible tagging of endogenous genes by homology-independent intron targeting. Genome Res 29, 1322–1328 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lane K et al. Measuring Signaling and RNA-Seq in the Same Cell Links Gene Expression to Dynamic Patterns of NF-kappaB Activation. Cell Syst 4, 458–469 e455 (2017).* Endpoint analyses paired with live-cell measurements provides both dynamic and global endpoint measurements in the same cells.
- 37.Yao J, Pilko A & Wollman R Distinct cellular states determine calcium signaling response. Molecular systems biology 12, 894 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tredan O, Galmarini CM, Patel K & Tannock IF Drug resistance and the solid tumor microenvironment. J Natl Cancer Inst 99, 1441–1454 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Hurst EW & Davies OL Studies on the blood-brain barrier. II. Attempts to influence the passage of substances into the brain. Br J Pharmacol Chemother 5, 147–164 (1950). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Provenzano PP et al. Enzymatic targeting of the stroma ablates physical barriers to treatment of pancreatic ductal adenocarcinoma. Cancer Cell 21, 418–429 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim M et al. Efficacy of the MDM2 Inhibitor SAR405838 in Glioblastoma Is Limited by Poor Distribution Across the Blood-Brain Barrier. Mol Cancer Ther 17, 1893–1901 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pittet MJ & Weissleder R Intravital imaging. Cell 147, 983–991 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cuccarese MF et al. Heterogeneity of macrophage infiltration and therapeutic response in lung carcinoma revealed by 3D organ imaging. Nat Commun 8, 14293 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arlauckas SP et al. In vivo imaging reveals a tumor-associated macrophage-mediated resistance pathway in anti-PD-1 therapy. Sci Transl Med 9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Randall EC et al. Integrated mapping of pharmacokinetics and pharmacodynamics in a patient-derived xenograft model of glioblastoma. Nat Commun 9, 4904 (2018).* Integrating both analysis of drug distribution and cell signaling can distinguish pharmacokinetics from other sources of cell heterogeneity.
- 46.Wu A et al. Cell motility and drug gradients in the emergence of resistance to chemotherapy. Proc Natl Acad Sci U S A 110, 16103–16108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwartz AD et al. A biomaterial screening approach reveals microenvironmental mechanisms of drug resistance. Integr Biol (Camb) 9, 912–924 (2017).* In vitro culture environments are used to explore the contribution of microenvironment on drug response and identify an effective drug combination across environments.
- 48.Xiao W et al. Brain-Mimetic 3D Culture Platforms Allow Investigation of Cooperative Effects of Extracellular Matrix Features on Therapeutic Resistance in Glioblastoma. Cancer Res 78, 1358–1370 (2018).* Biomaterial scaffolds enable detailed exploration of how individual ECM components modulate resistance development.
- 49.Keren L et al. A Structured Tumor-Immune Microenvironment in Triple Negative Breast Cancer Revealed by Multiplexed Ion Beam Imaging. Cell 174, 1373–1387 e1319 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eng CL et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature 568, 235–239 (2019).* SeqFISH+ is a novel technique that allows assessment of thousands of genes in single cells while preserving spatial context.
- 51.Lin JR, Fallahi-Sichani M & Sorger PK Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method. Nat Commun 6, 8390 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsujikawa T et al. Quantitative Multiplex Immunohistochemistry Reveals Myeloid-Inflamed Tumor-Immune Complexity Associated with Poor Prognosis. Cell Rep 19, 203–217 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Norton KA, Gong C, Jamalian S & Popel AS Multiscale Agent-Based and Hybrid Modeling of the Tumor Immune Microenvironment. Processes (Basel) 7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Navin N et al. Tumour evolution inferred by single-cell sequencing. Nature 472, 90–94 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]