Abstract
Liver organogenesis begins with hepatic precursors in the foregut endoderm, followed by hepatoblast specification, differentiation, outgrowth, and maturation for the formation of functional hepatocytes. Although several signaling pathways and critical factors that regulate liver specification, differentiation, and proliferation have been identified, little is known about how liver maturation is regulated. Here, we used a screen for mutations affecting liver development in zebrafish and identified a cq96 mutant that exhibits a specific defect in liver maturation. Results from positional cloning revealed that cq96 encodes an RNA-binding protein, Rbm15, which is an evolutionarily conserved Spen family protein and known to play a crucial role in RNA m6A modification, nuclear export, and alternative splicing. However, a function of Rbm15 in embryonic liver development has not been reported. We found that Rbm15 is specifically expressed in the liver after its differentiation. CRISPR/Cas9-mediated loss of rbm15 repressed hepatic maturation, but did not affect hepatoblast specification, differentiation, and hepatocyte proliferation and apoptosis. Additional experiments disclosed that the mTOR complex 1 (mTORC1) pathway is highly activated in rbm15-deficient hepatocytes. Moreover, rapamycin treatment partially restored normal hepatic gene expression as well as the nuclear location of the transcription factor Hnf4a. Taken together, these results reveal an unexpected role of Rbm15 in liver maturation.
Keywords: RNA-binding motif protein 15 (Rbm15), liver organogenesis, liver maturation, mTORC1, hepatocyte nuclear factor 4alpha (Hnf4a), RNA-binding protein, gene regulation, post-transcriptional regulation, zebrafish hepatocyte nuclear factor 4 (HNF-4)
The liver is the biggest digestive organ of the body and plays a central role in metabolism and systemic homeostasis. It mainly consists of hepatocytes, stellate cells, macrophages, sinusoidal endothelial cells, and cholangiocytes (1). The hepatocyte is the most populous cell of the liver; nearly 80% of hepatic cells are hepatocytes. It undertakes multiple physiological functions, such as bile secretion, glycol metabolism, lipid storage, protein secretion, and detoxification (2). Therefore, it is crucial to understand the mechanisms underlying liver development using animal models. Although the molecular regulation on liver organogenesis is starting to be unraveled, the critical factors to regulate liver development specifically have not been completely known.
In zebrafish, hepatogenesis can be divided into two steps. In the first stage, mesoderm around foregut endoderm induces hepatic precursor's proliferation and migration out from the gut endoderm and then specification to hepatoblast. Multiple layers of hepatoblast and the surrounding septum transversum form the liver bud. This progress lasts from 24 h post-fertilization (hpf) to 34 hpf (3). Mesoderm-derived FGF, WNT, and BMP signaling orchestrate this progress (4–6). Many transcription factors, such as GATA6, Hhex, and Foxas, are implicated in liver bud formation and hepatoblast specification (7–9). In the second stage, hepatoblast proliferates, differentiates, and maturates into hepatocytes and cholangiocytes, eventually forming a functional liver (2). In this progress, Wnt signaling is the key regulator during liver formation via increasing cell proliferation and repressing apoptosis (10, 11). The Hippo/YAP pathway is crucial for liver size control; hyper-YAP1 activity in the liver stimulates hepatocyte and cholangiocyte specification, whereas loss of YAP1 decreases hepatocyte survival and impairs biliary duct morphogenesis (12). Notch signaling and esr2b are also involved in hepatocyte differentiation and bile duct formation (13, 14). Numerous transcription factors govern cell fate choices. Hnf4a, Prox1, Hnf1a, and c/EBPa are important for hepatocyte differentiation and hepatic gene expression (15–18). Hnf1b and Hnf6 are responsible for bile duct formation (18). The mTOR pathway is also involved in liver development (19). Although many mechanisms underlying the hepatic fate decisions in vivo and in vitro have been reported, the key transcriptional factors that regulate hepatocyte maturation have been addressed less.
Rbm15 is an evolutionarily conserved Spen family protein involved in cell fate decisions. It can bind to RNA and recruit other proteins to specific binding sites to regulate post-transcriptional modification (20). In mice, RBM15 regulates megakaryocyte terminal differentiation by affecting alternative RNA splicing of a group of genes such as Gata1, C-mpl, Runx1, and Tal1, which are important for megakaryopoiesis (21). RBM15 can bind to specific sites in the intron of pre-mRNA, recruit SF3B1 to the splicing region, and facilitate alternative RNA splicing in different developmental stages (21). RBM15 can affect promoter activity of Notch target genes, such as Hes1, in a cell-specific manner. It can repress the expression of Notch-induced gene Hes1 in nonhematopoietic cells but enhance the Hes1 mRNA level in hematopoietic cell lines (22). RBM15 can also affect hematopoietic stem cell and megakaryocyte development via regulating c-myc expression (23). m6A is the most widespread RNA modification; the establishment of m6A relies on RBM15 (24, 25). RBM15 is also involved in RNA nuclear export (26). However, the role of Rbm15 in liver development is unknown.
Zebrafish has been developed into a popular model organism in recent years because of advantages of their transparent embryos developing outside of the mother, allowing constant visualization, and 69% of all zebrafish genes have a clear human orthologue (27). High genetic conservation among vertebrates makes zebrafish an excellent model system to study liver development and disease (27, 28). Here we used chemical mutagenesis to screen genes involved in liver development and got a specific liver maturation mutant, cq96. The mutant cq96 revealed normal liver specification, differentiation, and outgrowth, but the liver maturation was repressed. The development of the pancreas and intestine showed normal in cq96 mutant. Positional cloning and knockout experiments confirmed this mutation site located in gene rbm15. Deficiency of Rbm15 leads to up-regulation of the activity of mTORC1 signaling, and inhibition of the mTORC1 pathway by rapamycin can rescue liver maturation defect. Furthermore, the treatment of rapamycin enhanced the nuclear import of Hnf4a and restored hepatic gene expression. Our data revealed an unexpected function of rbm15 in liver development in zebrafish.
Results
The cq96 mutant confers a liver maturation defect
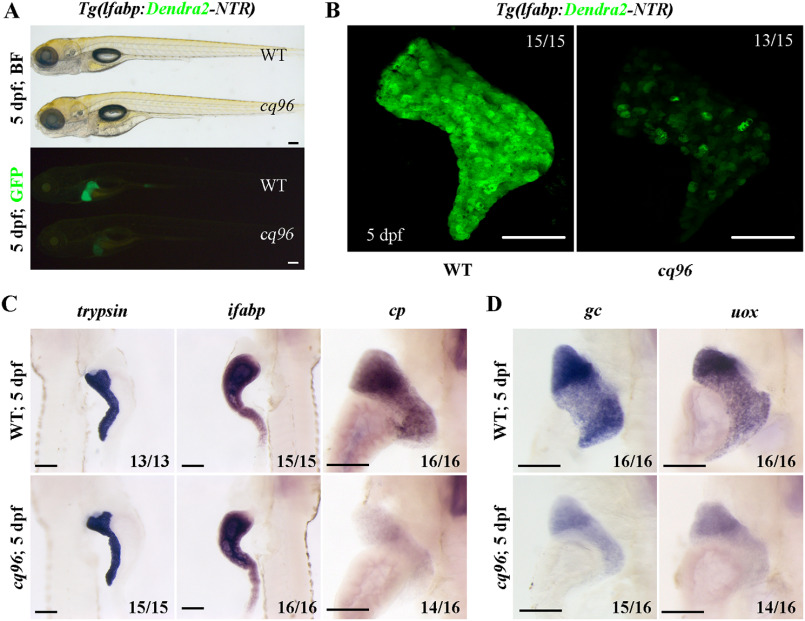
We have screened mutagenized zebrafish larvae using transgenic line Tg(lfabp:Dendra2-NTR) (29) and identified the cq96 mutant. At 5 dpf, this mutant showed normal body morphology but weaker Dendra2 expression (Fig. 1A). The confocal image exhibited relatively normal liver size but heterogeneous Dendra2 expression in the mutant (Fig. 1B). The expressions of digestive organ–specific markers trypsin (exocrine pancreas) and ifabp (intestine) were normal, but the hepatocyte-specific gene cp was repressed (Fig. 1C). The expressions of functional markers of hepatocytes such as uox, gc, and gys2 were repressed in the mutant (Fig. 1D and Fig. S1 (A and B)). These data indicate that the cq96 mutant specifically affects liver development and especially regulates liver maturation.
Figure 1.
Zebrafish cq96 mutants display aberrant liver maturation. The liver was labeled by Tg(lfabp:Dendra2-NTR)cq1. A, bright field and green fluorescence image of WT and cq96 mutant at 5 dpf showing the morphology and liver fluorescence expression. B, confocal images of WT and cq96 at 5 dpf revealing the liver size and morphology. C, whole-mount in situ hybridization (WISH) results showing the expressions of trypsin, ifabp, and cp in WT and cq96 mutant at 5 dpf. D, evaluating the liver function by WISH results of uox and gc in WT and cq96 mutant at 5 dpf. BF, bright field. Numbers indicate the proportion of larvae exhibiting the expression shown. Scale bars, 100 μm.
Zebrafish cq96 mutation site locates in gene rbm15
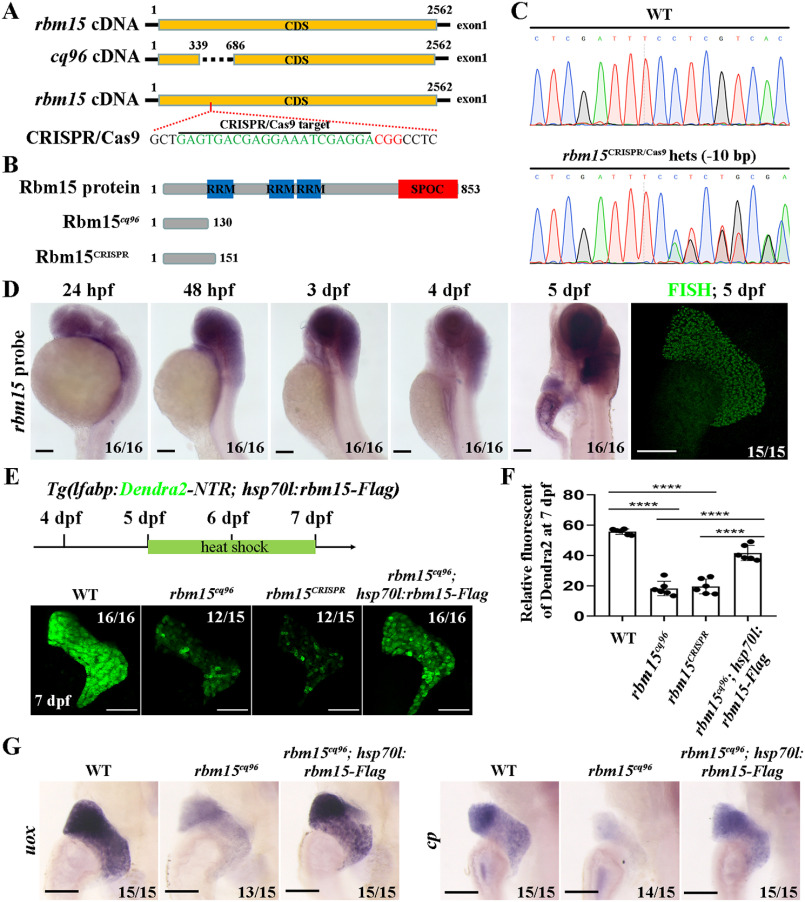
To determine the target gene of the cq96 mutant, we performed genome mapping and placed the cq96 mutation site locus to gene rbm15. The genomic sequencing result showed that there are 347 bp deleted in rbm15 exon 1, leading to translation into a truncated peptide (Fig. 2, A and B). To further confirm that rbm15 affects liver maturation, we generated a new rbm15 mutant by CRISPR/Cas9 that resembled the cq96 phenotype (Fig. 2, A–F). These results revealed that the mutation gene of cq96 is rbm15. Furthermore, rbm15 expressed in the liver region from 4 dpf and much stronger at 5 dpf (Fig. 2D). The weak expressions of Dendra2 and hepatic maturation markers such as uox and cp in cq96 mutant can be rescued by using Rbm15 overexpression line Tg(hsp70l:rbm15-Flagcq97) (Fig. 2, E–G). These results suggested that the mutation gene in cq96 is rbm15.
Figure 2.
The mutation gene in cq96 is rbm15. A and B, schematic drawing of the rbm15 locus, cq96, CRISPR/Cas9 targeting site (green), and the PAM sequence (red). C, genomic sequencing result of rbm15 mutation derived from the CRISPR/Cas9 knockout experiment. D, WISH and fluorescence in situ hybridization (FISH) results showing the expression pattern of rbm15 from 24 hpf to 5 dpf. E, heat shock transgenic line Tg(hsp70l:rbm15-Flagcq97) rescues liver defects of cq96; confocal images show the expression of Dendra2. F, the quantification of fluorescent intensity of Dendra2 in the liver. G, evaluating the rescue effects by detecting the expression of liver functional genes uox and gc via WISH. Numbers indicate the proportion of larvae exhibiting the expression shown. Asterisks indicate statistical significance: ****, p < 0.0001. Scale bars, 100 μm; error bars, S.D.
Zebrafish cq96 mutant affects liver maturation but not hepatoblast specification
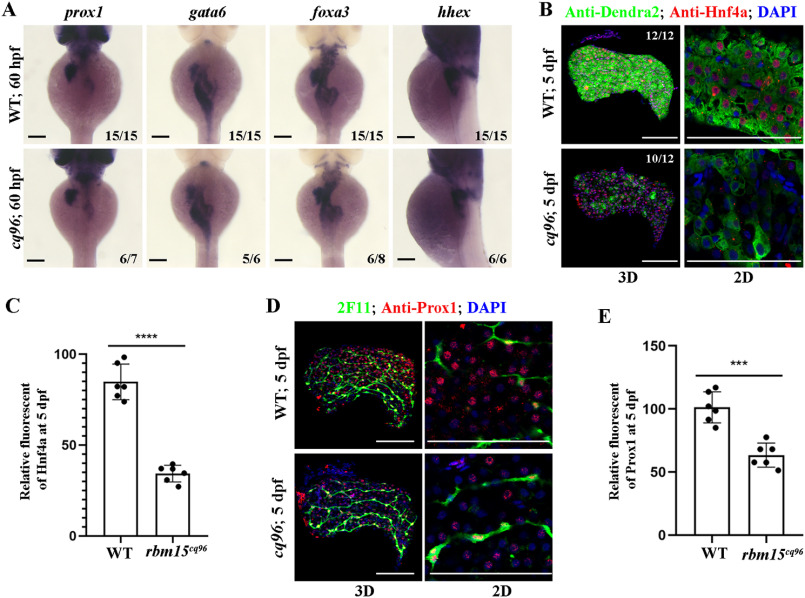
To evaluate hepatoblast specification in the rbm15 mutant, we assayed the expression of transcriptional factors important for hepatoblast formation and specification at 60 hpf. Interestingly, the expressions of prox1, gata6, hhex, and foxa3 were not greatly different between mutants and siblings (Fig. 3A). This means that liver specification was normal in the cq96 mutant. Then we performed antibody staining for hepatic factors Hnf4a and Prox1 at 5 dpf. The nuclear signal for Hnf4a and Prox1 was much weaker compared with siblings (Fig. 3, B–E). These results suggest that the cq96 mutant has liver maturation rather than liver specification defects.
Figure 3.
Hepatoblast specification is normal in the cq96 mutant. A, WISH results of gata6, prox1, foxa3, and hhex at 60 hpf revealing hepatic formation and specification in cq96 and WT. B, confocal images showing the antibody staining of Hnf4a and Dendra2 at 5 dpf in cq96 mutant. C, the quantification of Hnf4a fluorescent intensity in WT and cq96. D, confocal images showing the antibody staining of Prox1 and 2F11 in WT and mutant at 5 dpf. E, the quantification of Prox1 fluorescent intensity in WT and cq96. Numbers indicate the proportion of larvae exhibiting the expression shown. Asterisks indicate statistical significance: ***, p < 0.001; ****, p < 0.0001. Scale bars, 100 μm; error bars, S.D.
Loss of Rbm15 confers normal hepatic proliferation and apoptosis
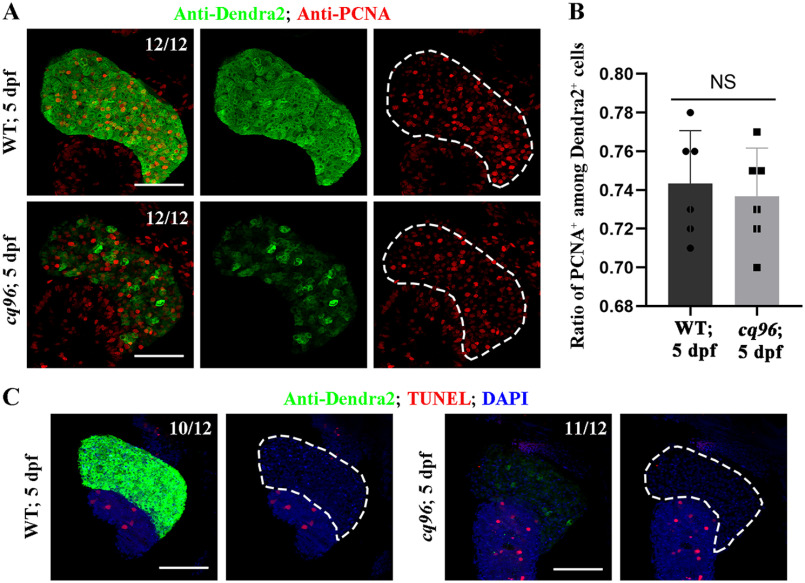
To investigate the underlying mechanism of liver development defect in cq96 mutant, we performed antibody staining for PCNA, which labeled cells outside of the G0 phase. We detected a similar ratio of PCNA and Dendra2 double-positive cells in siblings and mutants (Fig. 4, A and B). To assess whether cell death contributes to liver development defect in cq96 mutant, we performed a transferase-mediated dUTP nick-end labeling (TUNEL) assay on WT and cq96 at 5 dpf. Larval hepatocytes presented a low apoptotic index at 5 dpf, which was unchanged in cq96 mutant (Fig. 4C). Therefore, liver development defect in the cq96 mutant does not appear to be due to defective cell proliferation and apoptosis.
Figure 4.
Loss of Rbm15 has no effect on hepatic proliferation and apoptosis. A, antibody staining results of PCNA and Dendra2 showing hepatocytes outside of the G0 phase at 5 dpf. B, quantification of the percentage of the PCNA+ among Dendra2+ cells at 5 dpf in WT and rbm15−/−. TUNEL assay images showing the apoptosis of liver in WT and rbm15−/− at 5 dpf. Numbers indicate the proportion of larvae exhibiting the expression shown. NS, not significantly different; scale bars, 100 μm; error bars, S.D.
Inhibiting mTORC1 signaling pathway partially rescues the phenotype of cq96 mutant
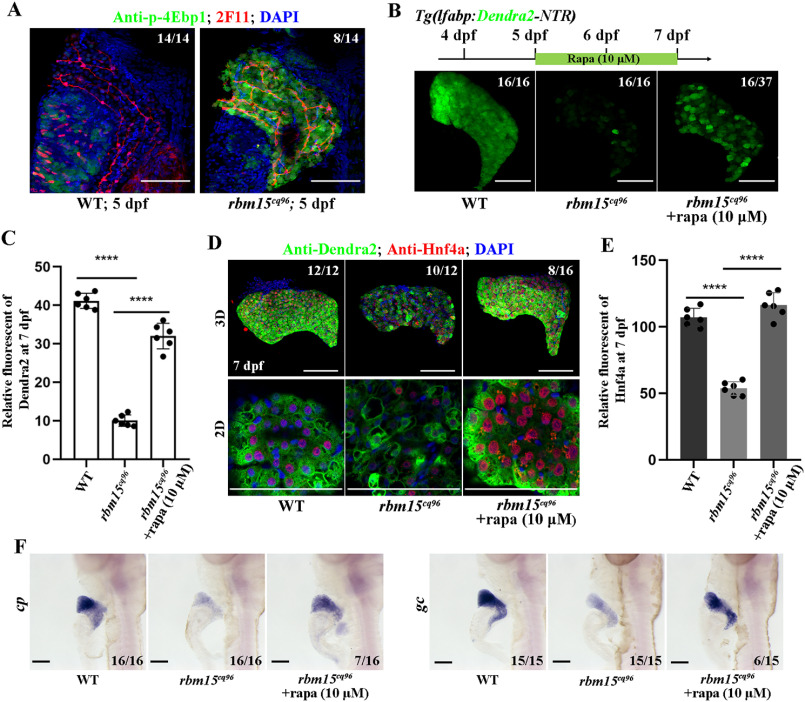
Hyper- and hypoactivated mTORC1 pathway will impair normal liver development (30, 31). To explore whether the loss of rbm15 will affect the mTORC1 signaling or not, we performed antibody staining to check the expression level of p-4Ebp1, which indicates the activity of mTORC1 signaling, and found that the rbm15−/− mutant larvae showed high p-4Ebp1 level in the liver (Fig. 5A). This indicates that the mTORC1 pathway was hyperactivated in the rbm15−/− liver. To further confirm that the liver maturation defect in the rbm15 mutant was caused by mTORC1 activation, we used 10 μm rapamycin to inhibit mTORC1 from 5 to 7 dpf. mTORC1 inhibition can partially rescue the developmental liver defect (Fig. 5 (B and C) and Fig. S2A). After rapamycin treatment, the protein level of Hnf4a in mutant hepatocyte was rescued (Fig. 5, D and E), but the mRNA level of hnf4a showed no big difference between WT, rbm15−/−, and sample groups (Fig. S2B). Furthermore, the expressions of hepatocyte-specific genes such as cp and gc were also restored after rapamycin treatment (Fig. 5F). These results indicate that inhibition of mTORC1 can partially rescue liver maturation defects in rbm15 mutant.
Figure 5.
Inhibiting the mTORC1 pathway partially rescues the liver maturation defects in rbm15cq96. A, confocal images (3D) showing the antibody staining of p-4Ebp1 and 2F11 to evaluate the activity of mTORC1 signal in hepatocytes at 5 dpf. B, rapamycin treatment and confocal images reflecting the rescue efficiency of rapamycin treatment. C, quantification of Dendra2 fluorescent intensity in WT, cq96, and rapamycin treatment. D, antibody staining results of Hnf4a and Dendra2 showing that rapamycin treatment affects the cellular and protein levels of Dnedra2 and Hnf4a. E, the quantification of Hnf4a fluorescent intensity in WT, cq96, and rapamycin treatment. F, WISH images showing the expression patterns of cp and gc after rapamycin treatment. rapa, rapamycin. The numbers indicate the proportion of larvae exhibiting the expression shown. Asterisks indicate statistical significance: ****, p < 0.0001. Scale bars, 100 μm; error bars, S.D.
Discussion
We have described a liver developmental mutant caused by rbm15 mutation, and inhibition of mTORC1 was an efficient strategy to rescue hepatic maturation defects. We showed that loss of Rbm15 specifically affected hepatic maturation, but not the developmental progress of intestine, pancreas. The mutant liver exhibited relative normal proliferation and apoptosis but weak hepatic gene expression. The two important hepatic transcriptional factors Hnf4a and Prox1 exhibited weak nuclear location, which could explain why rbm15 mutant showed hepatic maturation defects to some degree. Liver failure caused by rbm15 mutation also showed abnormal mTORC1 activation. Inhibition of mTORC1 can partially recover hepatic gene expression; this progress may rely on the enhancement of Hnf4a nuclear import.
Rbm15 is an important post-transcriptional regulator involved in RNA nuclear export, m6A modification, and alternative splicing (21, 25, 26). It is indispensable for megakaryocyte differentiation, and rbm15 defect can induce acute megakaryoblast leukemia (20). mTORC1 acts as a metabolic regulator important for cell growth and differentiation (19). Abnormal mTORC1 activation is associated with liver developmental defects and enhancement of liver damage (30, 32). Our finding first reveals that rbm15 is essential for hepatic maturation and that loss of rbm15-induced liver developmental failure partially depends on aberrant high mTORC1 activation.
We point out the importance of rbm15 in hepatic maturation, but we still do not know the target genes of rbm15. The relationship between rbm15 deficiency and high mTORC1 activity is still a mystery. RNA immunoprecipitation sequencing experiments will be indispensable to further answer these questions.
Experimental procedures
Ethics statement
All experimental protocols were approved by the Institute of Developmental Biology and Regenerative Medicine, Southwest University (Chongqing, China), and the methods were carried out in accordance with the approved guidelines. The zebrafish facility and study were approved by the Institutional Review Board of Southwest University (Chongqing, China). Zebrafish were maintained in accordance with the Guidelines of Experimental Animal Welfare from the Ministry of Science and Technology of the People's Republic of China (2006) and the Institutional Animal Care and Use Committee protocols from Southwest University (2007).
Zebrafish lines
Zebrafish (Danio rerio) AB strain-derived Tg(lfabp:Dendra2-NTR)cq1 was used as WT, and rbm15cq96 mutant was generated by ENU treatment. The IND line was used for mapping. These zebrafish lines were raised under standard conditions, and embryos/larvae for the experiment were treated with 0.003% PTU (Sigma) from 24 hpf.
CRISPR/Cas9-targeted rbm15 knockout
The CRISPR/Cas9 was carried out essentially as reported previously (33). The sequence for CRISPR RNA is shown in Fig. 2A. We used the following primers to identify the genotype of mutant: forward primer, 5′-GAATTCTGGCGGAGGAAGCA-3′; reverse primer, 5′-AAGCCGACCCAGTGCTAAC-3′.
Whole-mount in situ hybridization and fluorescent in situ hybridization
Whole-mount in situ hybridization and fluorescent in situ hybridization were based on a previous report (29) using antisense probes for hhex, gata6, foxa3, prox1, uox, gc, cp, hnf4a, and rbm15. Primers used for amplifying the rbm15 probe were 5′-GAGGCAGTTTACTTGAACAG-3′ (forward primer) and 5′-AAGCCGACCCAGTGCTAAC-3′ (reverse primer).
Antibody staining and TUNEL assay
Antibody staining and TUNEL assay were performed as described previously (29). The following antibodies were used: antibodies against Dendra2 (1:1000; AB821, Evrogen, Moscow, Russia), phospho-4E-BP1 (Thr-37/46) (1:500; catalog no. 2855, Cell Signaling), Hnf4a (1:200; sc-6556, Santa Cruz Biotechnology, Inc.), Prox1 (1:500; ab5475, Chemicon), 2F11 (1:1000; ab71826, Abcam, Cambridge, MA), and PCNA (1:1000; SAB2701819, Sigma).
Generation of transgenic line for rescue experiments
Full-length rbm15 cDNA was amplified by PrimeSTAR HS DNA Polymerase (Takara) and cloned into pBluescript vector. The full-length rbm15-Flag CDS was driven by hsp70l promoter, and the plasmid was injected into AB strain embryos to generate the transgenic line Tg(hsp70l:rbm15-Flag)cq97. Positive embryos showed cerulean expression in the eyes of the offspring.
Rapamycin treatment and heat shock
Embryos were treated with 10 μm rapamycin (Sangon Biotech, Shanghai, China) in PTU egg water from 5 to 7 dpf and replaced rapamycin solution every 24 h. The control group was treated with 0.2% DMSO. To induce rbm15 overexpression from Tg(hsp70l:rbm15-Flag)cq97, larvae were placed in egg water and then incubated in a 38.5 °C water bath for 30 min once a day from 5 to 7 dpf.
Data collection and analysis
All images were taken on a SteREO DiscoveryV20 microscope (Carl Zeiss, Germany) and LSM880 confocal microscope (Carl Zeiss). The intensities of fluorescent images were measured with ImageJ. The statistical analysis were performed with GraphPad Prism 8. Variation of individual data points was represented in S.D.
Data availability
All the data are contained within the article.
Supplementary Material
Acknowledgments
We are grateful to Prof. Lingfei Luo for guidance and assistance; Jinzi Chen and Kai Gang for discussions; and Qifen Yang, Rui Ni, and Xuemei Tang for technical assistance.
This article contains supporting information.
Author contributions—L. H. and J. H. conceptualization; L. H. and H. L. data curation; L. H. and H. L. formal analysis; L. H., H. L., and Z. C. methodology; L. H. writing-original draft; J. H. funding acquisition; J. H. writing-review and editing.
Funding and additional information—This work was supported by National Key R&D Program of China Grant 2019YFA0802703 (to J. H.) and National Natural Science Foundation of China Grants 31970784 and 31801214 (to J. H.).
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- hpf
- hours post fertilization
- dpf
- days post fertilization
- PTU
- 1-phenyl 2-thiourea
- WISH
- whole-mount in situ hybridization
- mTOR
- mechanistic target of rapamycin
- PCNA
- proliferating cell nuclear antigen
- TUNEL
- transferase-mediated dUTP nick-end labeling.
References
- 1. Berasain C., and Avila M. A. (2015) Regulation of hepatocyte identity and quiescence. Cell. Mol. Life Sci. 72, 3831–3851 10.1007/s00018-015-1970-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gordillo M., Evans T., and Gouon-Evans V. (2015) Orchestrating liver development. Development 142, 2094–2108 10.1242/dev.114215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Field H. A., Ober E. A., Roeser T., and Stainier D. Y. (2003) Formation of the digestive system in zebrafish. I. Liver morphogenesis. Dev. Biol. 253, 279–290 10.1016/S0012-1606(02)00017-9 [DOI] [PubMed] [Google Scholar]
- 4. Calmont A., Wandzioch E., Tremblay K. D., Minowada G., Kaestner K. H., Martin G. R., and Zaret K. S. (2006) An FGF response pathway that mediates hepatic gene induction in embryonic endoderm cells. Dev. Cell 11, 339–348 10.1016/j.devcel.2006.06.015 [DOI] [PubMed] [Google Scholar]
- 5. Shin D., Shin C. H., Tucker J., Ober E. A., Rentzsch F., Poss K. D., Hammerschmidt M., Mullins M. C., and Stainier D. Y. (2007) Bmp and Fgf signaling are essential for liver specification in zebrafish. Development 134, 2041–2050 10.1242/dev.000281 [DOI] [PubMed] [Google Scholar]
- 6. Ober E. A., Verkade H., Field H. A., and Stainier D. Y. R. (2006) Mesodermal Wnt2b signalling positively regulates liver specification. Nature 442, 688–691 10.1038/nature04888 [DOI] [PubMed] [Google Scholar]
- 7. Watanabe H., Takayama K., Inamura M., Tachibana M., Mimura N., Katayama K., Tashiro K., Nagamoto Y., Sakurai F., Kawabata K., Furue M. K., and Mizuguchi H. (2014) HHEX promotes hepatic-lineage specification through the negative regulation of eomesodermin. PLoS One 9, e90791 10.1371/journal.pone.0090791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee C. S., Friedman J. R., Fulmer J. T., and Kaestner K. H. (2005) The initiation of liver development is dependent on Foxa transcription factors. Nature 435, 944–947 10.1038/nature03649 [DOI] [PubMed] [Google Scholar]
- 9. Zhao R., Watt A. J., Li J., Luebke-Wheeler J., Morrisey E. E., and Duncan S. A. (2005) GATA6 is essential for embryonic development of the liver but dispensable for early heart formation. Mol. Cell Biol. 25, 2622–2631 10.1128/MCB.25.7.2622-2631.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tan X., Behari J., Cieply B., Michalopoulos G. K., and Monga S. P. (2006) Conditional deletion of β-catenin reveals its role in liver growth and regeneration. Gastroenterology 131, 1561–1572 10.1053/j.gastro.2006.08.042 [DOI] [PubMed] [Google Scholar]
- 11. Colletti M., Cicchini C., Conigliaro A., Santangelo L., Alonzi T., Pasquini E., Tripodi M., and Amicone L. (2009) Convergence of Wnt signaling on the HNF4α-driven transcription in controlling liver zonation. Gastroenterology 137, 660–672 10.1053/j.gastro.2009.05.038 [DOI] [PubMed] [Google Scholar]
- 12. Yimlamai D., Christodoulou C., Galli G. G., Yanger K., Pepe-Mooney B., Gurung B., Shrestha K., Cahan P., Stanger B. Z., and Camargo F. D. (2014) Hippo pathway activity influences liver cell fate. Cell 157, 1324–1338 10.1016/j.cell.2014.03.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zong Y., Panikkar A., Xu J., Antoniou A., Raynaud P., Lemaigre F., and Stanger B. Z. (2009) Notch signaling controls liver development by regulating biliary differentiation. Development 136, 1727–1739 10.1242/dev.029140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chaturantabut S., Shwartz A., Garnaas M. K., LaBella K., Li C. C., Carroll K. J., Cutting C. C., Budrow N., Palaria A., Gorelick D. A., Tremblay K. D., North T. E., and Goessling W. (2020) Estrogen acts via estrogen receptor 2b to regulate hepatobiliary fate during vertebrate development. Hepatology 10.1002/hep.31184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lüdtke T. H., Christoffels V. M., Petry M., and Kispert A. (2009) Tbx3 promotes liver bud expansion during mouse development by suppression of cholangiocyte differentiation. Hepatology 49, 969–978 10.1002/hep.22700 [DOI] [PubMed] [Google Scholar]
- 16. Hang H.-L., Liu X.-Y., Wang H.-T., Xu N., Bian J.-M., Zhang J.-J., Xia L., and Xia Q. (2017) Hepatocyte nuclear factor 4A improves hepatic differentiation of immortalized adult human hepatocytes and improves liver function and survival. Exp. Cell Res. 360, 81–93 10.1016/j.yexcr.2017.08.020 [DOI] [PubMed] [Google Scholar]
- 17. Sosa-Pineda B., Wigle J. T., and Oliver G. (2000) Hepatocyte migration during liver development requires Prox1. Nat. Genet. 25, 254–255 10.1038/76996 [DOI] [PubMed] [Google Scholar]
- 18. Odom D. T., Zizlsperger N., Gordon D. B., Bell G. W., Rinaldi N. J., Murray H. L., Volkert T. L., Schreiber J., Rolfe P. A., Gifford D. K., Fraenkel E., Bell G. I., and Young R. A. (2004) Control of pancreas and liver gene expression by HNF transcription factors. Science 303, 1378–1381 10.1126/science.1089769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chaturantabut S., Shwartz A., Evason K. J., Cox A. G., Labella K., Schepers A. G., Yang S., Acuna M., Houvras Y., Mancio-Silva L., Romano S., Gorelick D. A., Cohen D. E., Zon L. I., Bhatia S. N., et al. (2019) Estrogen activation of G-protein-coupled estrogen receptor 1 regulates phosphoinositide 3-kinase and mTOR signaling to promote liver growth in zebrafish and proliferation of human hepatocytes. Gastroenterology 156, 1788–1804.e13 10.1053/j.gastro.2019.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tran N. T., Su H., Khodadadi-Jamayran A., Lin S., Zhang L., Zhou D., Pawlik K. M., Townes T. M., Chen Y., Mulloy J. C., and Zhao X. (2016) The AS-RBM15 lncRNA enhances RBM15 protein translation during megakaryocyte differentiation. EMBO Rep. 17, 887–900 10.15252/embr.201541970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang L., Tran N. T., Su H., Wang R., Lu Y., Tang H., Aoyagi S., Guo A., Khodadadi-Jamayran A., Zhou D., Qian K., Hricik T., Cote J., Han X., Zhou W., et al. (2015) Cross-talk between PRMT1-mediated methylation and ubiquitylation on RBM15 controls RNA splicing. eLife 4, e07938 10.7554/eLife.07938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ma X., Renda M. J., Wang L., Cheng E. C., Niu C., Morris S. W., Chi A. S., and Krause D. S. (2007) Rbm15 modulates Notch-induced transcriptional activation and affects myeloid differentiation. Mol. Cell Biol. 27, 3056–3064 10.1128/MCB.01339-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Niu C., Zhang J., Breslin P., Onciu M., Ma Z., and Morris S. W. (2009) c-Myc is a target of RNA-binding motif protein 15 in the regulation of adult hematopoietic stem cell and megakaryocyte development. Blood 114, 2087–2096 10.1182/blood-2009-01-197921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Knuckles P., Lence T., Haussmann I. U., Jacob D., Kreim N., Carl S. H., Masiello I., Hares T., Villaseñor R., Hess D., Andrade-Navarro M. A., Biggiogera M., Helm M., Soller M., Bühler M., et al. (2018) Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m(6)A machinery component Wtap/Fl(2)d. Genes Dev. 32, 415–429 10.1101/gad.309146.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Patil D. P., Chen C. K., Pickering B. F., Chow A., Jackson C., Guttman M., and Jaffrey S. R. (2016) m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537, 369–373 10.1038/nature19342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zolotukhin A. S., Uranishi H., Lindtner S., Bear J., Pavlakis G. N., and Felber B. K. (2009) Nuclear export factor RBM15 facilitates the access of DBP5 to mRNA. Nucleic Acids Res. 37, 7151–7162 10.1093/nar/gkp782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Goessling W., and Sadler K. C. (2015) Zebrafish: an important tool for liver disease research. Gastroenterology 149, 1361–1377 10.1053/j.gastro.2015.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chu J., and Sadler K. C. (2009) New school in liver development: lessons from zebrafish. Hepatology 50, 1656–1663 10.1002/hep.23157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. He J., Lu H., Zou Q., and Luo L. (2014) Regeneration of liver after extreme hepatocyte loss occurs mainly via biliary transdifferentiation in zebrafish. Gastroenterology 146, 789–800.e8 10.1053/j.gastro.2013.11.045 [DOI] [PubMed] [Google Scholar]
- 30. Wang Z., Song J., Luo L., and Ma J. (2018) Loss of leucyl-tRNA synthetase B leads to ILFS1-like symptoms in zebrafish. Biochem. Biophys. Res. Commun. 505, 378–384 10.1016/j.bbrc.2018.09.133 [DOI] [PubMed] [Google Scholar]
- 31. He J., Yang Y., Zhang J., Chen J., Wei X., He J., and Luo L. (2017) Ribosome biogenesis protein Urb1 acts downstream of mTOR complex 1 to modulate digestive organ development in zebrafish. J. Genet. Genomics 44, 567–576 10.1016/j.jgg.2017.09.013 [DOI] [PubMed] [Google Scholar]
- 32. Zhu Q., Wang H., Jiang B., Ni X., Jiang L., Li C., Wang X., Zhang F., Ke B., and Lu L. (2018) Loss of ATF3 exacerbates liver damage through the activation of mTOR/p70S6K/HIF-1α signaling pathway in liver inflammatory injury. Cell Death Dis. 9, 910 10.1038/s41419-018-0894-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chang N., Sun C., Gao L., Zhu D., Xu X., Zhu X., Xiong J. W., and Xi J. J. (2013) Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 23, 465–472 10.1038/cr.2013.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data are contained within the article.