Abstract
The conserved region of influenza hemagglutinin (HA) stalk (or stem) has gained attention as a potent target for universal influenza vaccines1–5. Although the HA stalk region is relatively well conserved, the evolutionarily dynamic nature of influenza viruses6 raises concerns about the possible emergence of viruses carrying stalk escape mutation(s) under sufficient immune pressure. Here we show that immune pressure on the HA stalk can lead to expansion of escape mutant viruses in study participants challenged with a 2009 H1N1 pandemic influenza virus inoculum containing an A388V polymorphism in the HA stalk (45% wild type and 55% mutant). High level of stalk antibody titers was associated with the selection of the mutant virus both in humans and in vitro. Although the mutant virus showed slightly decreased replication in mice, it was not observed in cell culture, ferrets or human challenge participants. The A388V mutation conferred resistance to some of the potent HA stalk broadly neutralizing monoclonal antibodies (bNAbs). Co-culture of wild-type and mutant viruses in the presence of either a bNAb or human serum resulted in rapid expansion of the mutant. These data shed light on a potential obstacle for the success of HA-stalk-targeting universal influenza vaccines—viral escape from vaccine-induced stalk immunity.
Conserved regions of viral pathogens have long been targets for developing broadly protective vaccines and therapeutic antibodies. Such vaccines and antibodies are especially needed to mitigate threats from influenza A viruses (IAVs), which undergo rapid evolution and antigenic drift, leading to escape from pre-existing immunity induced by infection or vaccination7–9. The unpredictable emergences of antigenically novel pandemic influenza viruses, which, in the past, have resulted in millions of deaths globally10, could also ideally be prevented or mitigated by broadly protective (so-called ‘universal’) influenza vaccines11,12.
Candidate universal influenza vaccines that target the HA stalk, initially proposed as a ‘common neutralizing epitope’ more than two decades ago1, have shown protective efficacy in animal models2–5 and are currently being evaluated in clinical trials (for example, ClinicalTrials.gov identifiers NCT03275389, NCT03300050 and NCT03814720). Importantly, the HA stalk antibodies have been shown to be associated with protection against influenza virus in humans with varying magnitudes depending on study settings13–15. The HA stalk is relatively conserved across the 18 HA IAV subtypes16, presumably because 1) the HA stalk is under stricter structural constraints as it must undergo conformational changes during HA membrane fusion17, and 2) the HA stalk region is also under lower antigenic drift pressure owing to its immuno-subdominance to HA head epitopes18.
However, the HA stalk region clearly has plasticity. The stalk sequences of related HA subtypes are phylogenetically distinct19, and HA stalk broadly neutralizing monoclonal antibodies (bNAbs) do not bind stalk epitopes from different HA subtypes with equal affinity20. Indeed, viruses possessing stalk escape mutations have been experimentally generated under immune pressure by bNAbs21–25 or human serum26, raising concerns about emergence of escape mutants under immune pressure narrowly targeting the HA stalk. However, it remains unclear if immune pressure applied to the HA stalk13–15 could lead to the emergence of viable stalk escape mutants in humans.
This study examined selection of an HA stalk antibody-escape mutant virus in human influenza challenge participants inoculated with a well-characterized IAV stock that contained a mixed population of viruses with a nonsynonymous single-nucleotide polymorphism (SNP) in the HA stalk (A388V)27–29. This variant had arisen spontaneously in Vero cell passage during good manufacturing practice (GMP) manufacture of the challenge inoculum. We investigated association between pre-existing anti-HA stalk immunity and selection of the V388 mutant virus during human viral challenge. The effect of the mutation on HA stalk local structure was studied, and the mutant virus was characterized in vitro and in vivo with and without immune pressure. Selection dynamics between the wild-type and mutant viruses were evaluated in vitro under immune pressure by a bNAb and human challenge participant serum.
Results
Human challenge with an HA A388V polymorphic virus.
To investigate the relationship between pre-existing anti-HA stalk immunity and escape mutant virus selection during influenza challenge infection in humans, we analyzed pre-challenge serum for HA stalk antibody titers and post-challenge nasal wash samples for viral sequencing from 2009 H1N1 pandemic influenza (H1N1pdm) virus human challenge studies13,27,28, which showed that pre-challenge antibody titers against the HA head, HA stalk and neuraminidase (NA) were all correlates of protection for viral shedding, whereas only anti-NA titers were correlated with reduction in symptoms. In these prior studies, participants were challenged with an influenza virus inoculum containing a polymorphism in the HA stalk at position 388 (position 1 being initial methionine)—approximately 45% wild-type (A388) and 55% mutant (V388) HA stalk as measured by deep sequencing analysis29. The A388V polymorphism had arisen spontaneously from the clonal starting virus (A388) during the six passages in certified Vero cells used for its GMP manufacture27,29.
Association between pre-existing human serum anti-HA stalk immunity and A388V mutant virus expansion in challenge study participants.
To determine the viral selection outcomes (that is, whether a study participant sample showed predominantly wild-type or mutant virus after challenge), viruses were analyzed from nasal wash samples obtained on days 2–9 after challenge using a SNP assay that reliably detects the minor population (either A388 or V388) present at 10% or higher frequency (Extended Data Fig. 1). SNP results were obtained from 29 participants who were confirmed positive for post-challenge viral shedding by clinical molecular testing27,28,30 and whose nasal wash samples contained amplifiable viral RNA: 12 participants selected for the V388 mutant virus, eight selected for the A388 wild-type virus, and nine shed both wild-type and mutant viruses (Fig. 1a).
Fig. 1 |. Association between pre-existing anti-HA stalk immunity and mutant virus selection in humans.
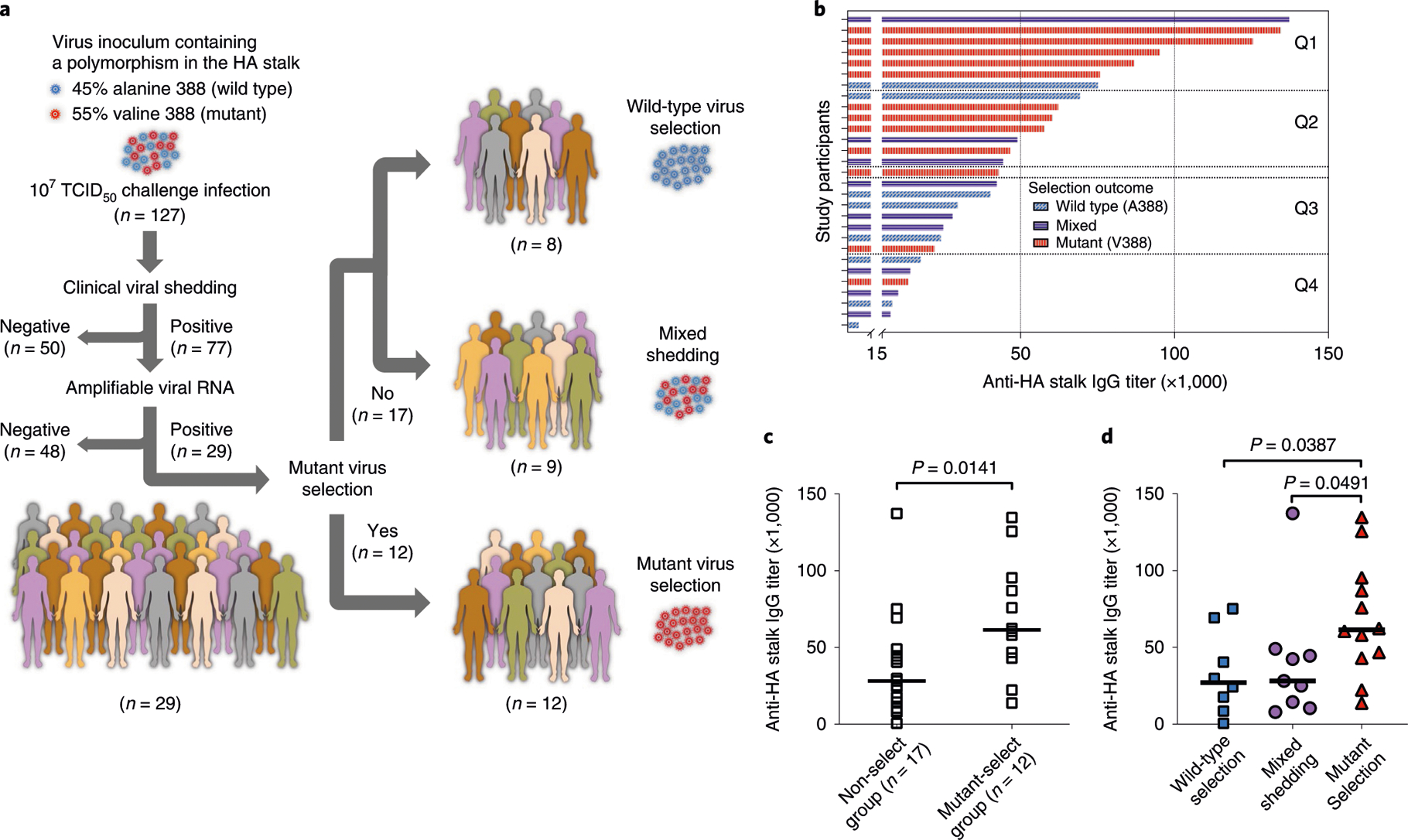
a, Study design. Influenza human challenge study participants were challenged using a virus stock containing a polymorphism in the HA stalk: 45% wild-type (A388) and 55% mutant (V388) virus. Viral RNA was extracted from post-challenge nasal wash samples and analyzed for selection outcomes (that is, whether the participants selected for the wild-type or mutant virus) by a SNP assay using a set of MGB-based TaqMan probes. b, Participants with a successful determination of the selection outcome were arranged in a descending manner by the participants’ pre-existing anti-HA stalk serum IgG titers measured using ELISA and divided into quartiles (Q1-Q4). Each bar represents an individual selection outcome by color (for example, a red bar represents a challenge study participant in which the mutant virus was selected during the viral challenge infection), and the length of each bar represents anti-HA stalk serum IgG titer of each participant. c, d, Participants were grouped depending on the selection outcome, and the stalk antibody titers from the mutant selection group were compared to other selection groups using a two-tailed nonparametric Mann-Whitney test. Median values are shown as a horizontal line.
Study participants with higher pre-challenge anti-HA stalk antibody titers (Fig. 1b) tended to select for the mutant virus (colored in red), as compared to participants with moderate to lower antibody levels. For example, five of seven participants in the highest quartile (Q1) selected for the mutant virus, but only one of seven in the lowest quartile (Q4) selected for the mutant virus. To investigate the association between the anti-HA stalk antibody levels and the mutant virus selection, participants were grouped into two categories by the selection outcomes: mutant-select group (V388 selection, n = 12) or non-select group (A388 selection or mixed shedding, n = 17). The pre-existing stalk antibody titers from the mutant-select group were significantly higher than that of the non-select group (P = 0.0141; Fig. 1c). When the study participants were divided into three groups, the mutant-select group still showed higher stalk antibody levels than the participants with wild-type selection or mixed shedding with statistical significance (Fig. 1d). Hemagglutination inhibition (HAI) titers, ratios between HA stalk antibodies and total HA antibodies and age of the participants were not associated with the mutant virus selection (Extended Data Fig. 2a–e). Interestingly, most of the study participants analyzed in this study had very low HAI titers (Extended Data Fig. 2a), suggesting the possibility that HA stalk antibodies, in the absence of other protective antibodies (for example, HAI antibodies), might drive selection for antibody-escape mutant viruses in humans.
Lack of effect of A388V stalk mutation on viral replication fitness and clinical disease.
Because HA stalk mutations have often been shown to decrease viral fitness23,25, we investigated the fitness of the A388V mutation using H1N1pdm viruses with A388 or V388 generated using a reverse genetics system. Replication kinetics of the viruses were evaluated in vitro using cell lines derived from different species (MDCK, canine origin; Vero, Chlorocebus sp. ‘African green, monkey origin; A549, human origin) and in vivo in mice and ferrets. Viral growth kinetics in vitro between the two viruses were indistinguishable (Fig. 2a–c). Although the mutant virus showed modestly (<ten-fold) decreased viral replication in mice with statistical significance (Fig. 2d), no decreased fitness was observed in ferrets (Fig. 2e). Additionally, secondary analysis of the human challenge study27,28 showed that clinical disease and shedding duration in participants who selected for the mutant virus were not different from study participants with other viral selection outcomes (Extended Data Fig. 2f–h).
Fig. 2 |. Effect of A388V stalk mutation on viral replicative fitness.
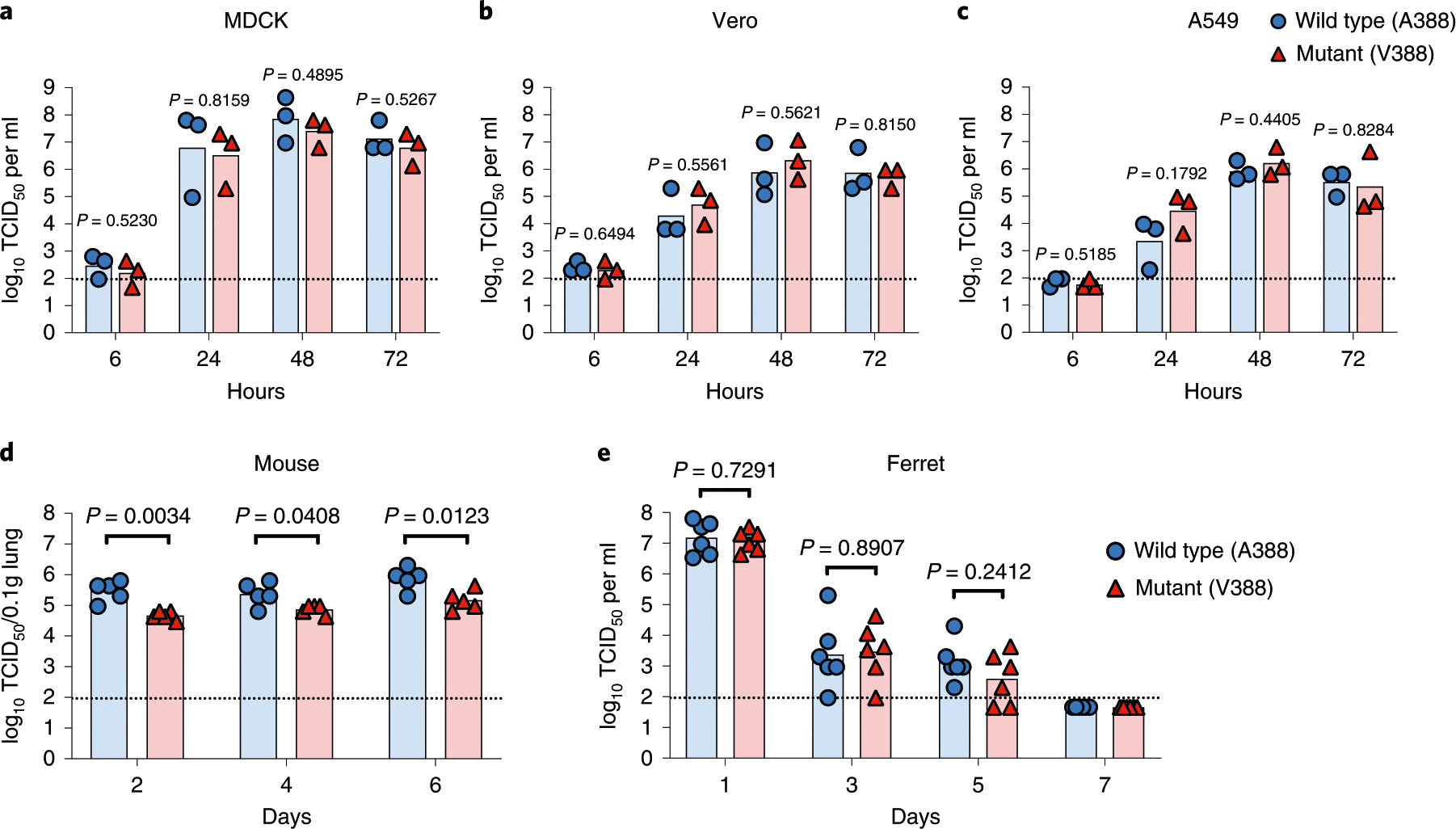
Wild-type (A388) or mutant (V388) H1N1pdm viruses were generated by reverse genetics and characterized for the viral replicative fitness. To measure in vitro fitness, (a) MDCK, (b) Vero and (c) A549 cells were infected at 0.001 MOI. Supernatants were collected at 6, 24, 48 and 72 h after infection, followed by titration in MDCK cells. Each symbol indicates an individual measurement. All conditions were independently replicated a total of three times. Bars represent geometric means. For measuring in vivo fitness, (d) 7–8-week-old female BALB/c mice were intranasally infected with 103 TCID50 of wild-type or mutant virus in a 50-μl inoculum; (e) 6–7-month-old female ferrets were intranasally infected with 105 TCID50 of wild-type or mutant virus in a 1-ml inoculum. Each symbol in d and e represents an individual animal. Viral loads were titrated in MDCK from (d) mice lung homogenate (n = 5) or (e) ferret nasal wash (n = 6). Bars represent geometric mean titers. Dashed lines show the detection limit of the TCID50 assay. An unpaired two-tailed Welch’s t-test was used to compare the level of viral growth between the wild-type and mutant virus. log10-transformed titers were used for the comparison.
A388V mutation induces a stalk conformational change that disrupts epitopes that bind different HA stalk monoclonal antibodies.
The effect of the A388V mutation on stalk epitopes was investigated by measuring binding levels of various bNAbs to full-length wild-type or mutant HAs. Six well-characterized bNAbs (CR6261 (refs. 20,31), CR9114 (ref. 32), FI6V3 (ref. 33), 70–1F02 (ref. 34), C179 (ref. 1) and CT149 (ref. 35)) all showed significant reduction in binding to the mutant HA compared to wild-type HA with varying degrees (Fig. 3a–f and Extended Data Figs. 3 and 4). FI6V3 showed the smallest reduction in binding of the bNAbs tested, suggesting that FI6V3 binding is less sensitive to the A388V mutation. Neutralizing globular head monoclonal antibodies EM-4C04 (ref. 34) and 2–12C36 retained binding to the A388V HA protein (Fig. 3g,h and Extended Data Fig. 3h). Notably, antibodies in human serum showed significantly decreased recognition of the mutant stalk construct (Fig. 3j and Extended Data Fig. 5), suggesting that the mutant virus would be less inhibited by human serum HA stalk antibodies.
Fig. 3 |. A significant conformational change to the HA stalk region induced by A388V mutation.
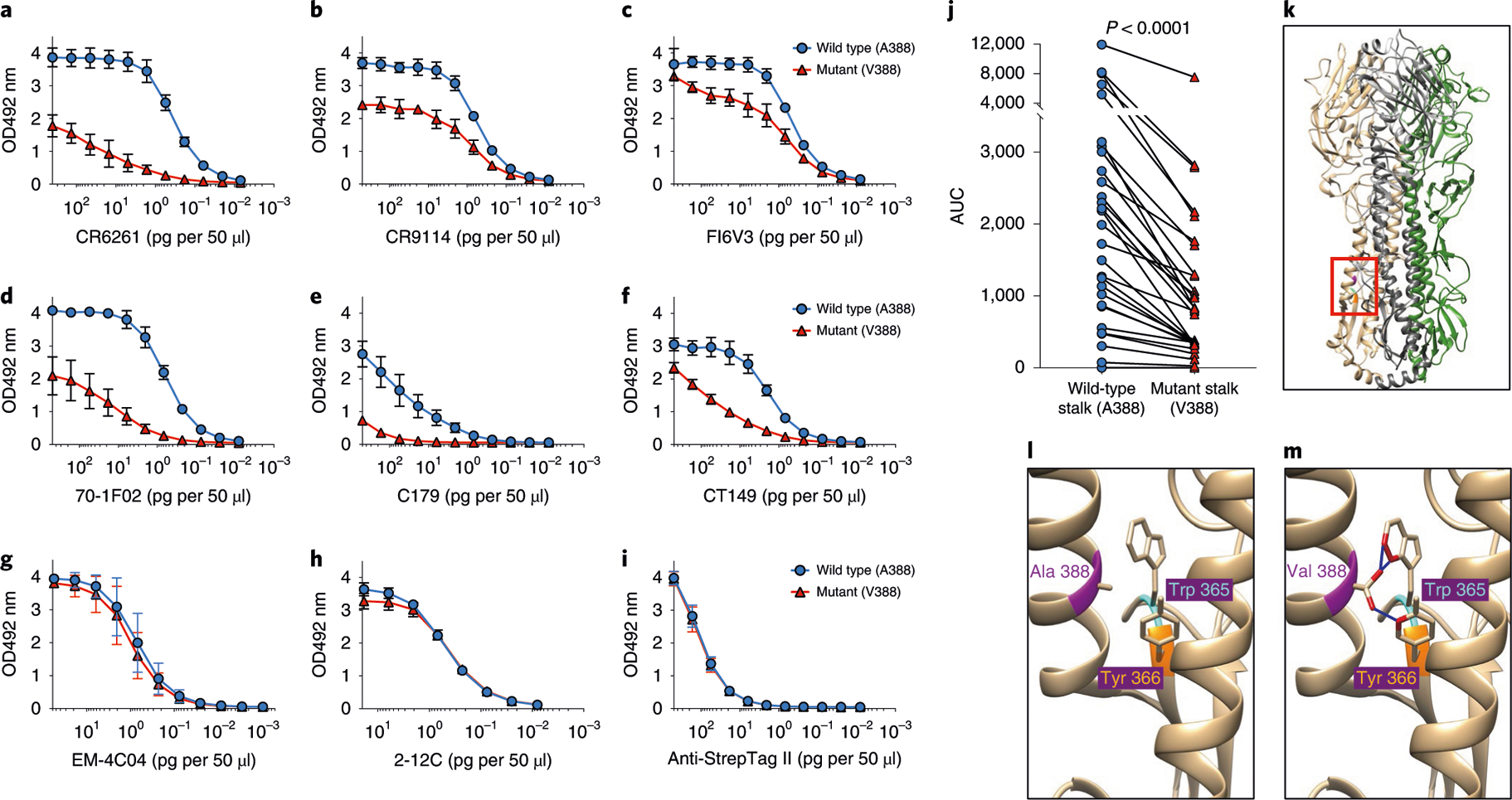
Full-length HA and HA stalk-only proteins with or without A388V mutation were used for ELISA to measure conformational changes introduced by the A388V mutation. Six bNAbs binding to the HA stalk—(a) CR6261, (b) CR9114, (c) FI6V3, (d) 70–1F02, (e) C179 and (f) CT149—were used to detect changes in the HA stalk structure using the full-length wild-type or mutant HAs. Neutralizing monoclonal antibodies that bind to the HA globular head, (g) EM-4C04 and (h) 2–12C, were used to show the structural integrity of the purified HA proteins. i, Anti-Strep-tag II antibody was used to show that equal amounts of wild-type or mutant HAs were used. Graphs show mean and s.d. from three independent measurements (a-i, n = 3). j, Decreased recognition of the mutant stalk by serum from the study participants (n = 29) is shown as assayed using stalk-only constructs (wild type or mutant) that measure only HA stalk-binding antibodies while excluding HA head-binding antibodies. A two-tailed paired t-test was used to compare the AUCs. A structural analysis was performed with UCSF Chimera using a previously published 2009 H1N1 influenza virus HA structure (PDB ID: 3LZG). k, Each monomer of the trimeric HA structure was colored in beige, gray and green. The location of A388V mutation in the short α-helix of the HA stalk structure is highlighted in pink. An area surrounding the A388V mutation (red box) was magnified for (l) wild-type HA (A388) and (m) mutant HA (V388) for clarity. A388V mutation and adjacent amino acids (Trp 365, cyan; Tyr 366, orange) that are predicted to undergo steric clash are shown. Molecules expected to experience steric clash are highlighted in red. Blue lines indicate the predicted clashes between molecules.
Structural modeling indicated that the A388V mutation does not interface directly with bNAbs but would likely cause steric clashes against the bulky aromatic side chains of W365 and Y366 on the β-hairpin structure of the HA stalk (Fig. 3k–m). Given the reductions in antibody binding to this region, it is probable that the α-helix of the HA stalk, the target of all known stalk bNAbs5, could bulge or rotate to accommodate the A388V mutation, resulting in structural changes. These results suggest that even a seemingly conserved stalk mutation (a methyl to an isopropyl side chain), and while positioned inter-helically, could nonetheless substantially affect the structure and potentially confer resistance against stalk antibodies.
Increased resistance of the A388V mutant to bNAbs.
Indeed, the mutant virus was significantly more resistant to CR6261 (refs. 20,31) (eight-fold) and CR9114 (ref. 32) (2.5-fold) neutralization compared to the wild-type virus in vitro (Fig. 4a,b). Effectiveness of FI6V3 (ref. 33) was maintained (Fig. 4c). The varying degree of decreased effectiveness of different bNAbs to the mutant virus was consistent with the differential changes in binding levels of the bNAbs (Fig. 3a–f and Extended Data Fig. 4). The mutant virus also showed higher resistance to prophylactically injected CR6261 in mice than the wild-type virus (Fig. 4d). Although all doses of CR6261 substantially inhibited both viruses when compared to untreated controls, the mutant virus replicated to significantly higher levels than wild-type virus in mice with 1.0 mg kg−1 and 30.0 mg kg−1 of CR6261 HA stalk monoclonal antibody (Fig. 4d).
Fig. 4 |. increased resistance to broadly neutralizing antibodies by A388V mutation and its rapid selection by immune pressure.
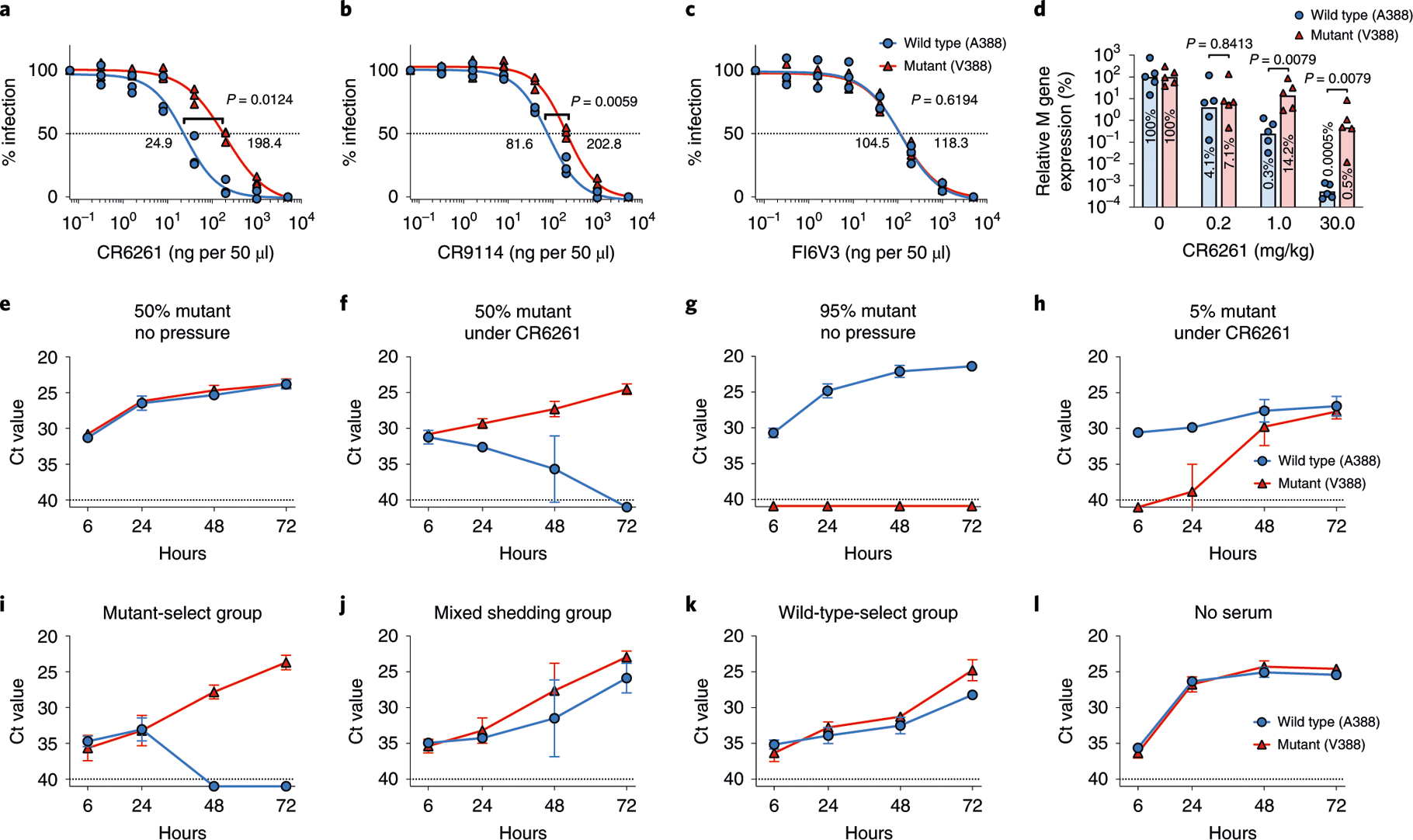
IC50 values of (a) CR6261, (b) CR9114 and (c) FI6V3 to the wild-type or mutant virus were measured three times independently (n = 3). An unpaired two-tailed Welch’s t-test was used to compare the IC50 values. d, 7–8-week-old female BALB/c mice were intranasally infected with 103 TCID50 of wild-type or mutant virus 24h after a prophylactic intraperitoneal injection of CR6261 antibody. At day 2 after infection, viral RNA levels from the mouse lung were quantified and compared to that of control mice not treated with antibodies. The level of viral RNA from control group was considered 100%. Each symbol represents an individual animal (n = 5). Bars represent geometric means. A two-tailed nonparametric Mann-Whitney test was used. Selection dynamics under immune pressure were measured by co-culturing the viruses with CR6261 antibody. The virus mixture, consisting of 50% wild-type and 50% mutant virus, was cultured in the (e) absence or (f) presence of CR6261. The mixture consisting of 95% wild-type and 5% mutant virus was cultured in the (g) absence or (h) presence of CR6261 antibody. Serum samples from each selection group (Fig. 1a) were pooled and used to place immune pressure. The viruses were mixed to represent the challenge virus inoculum used in the human challenge studies (45% wild type and 55% mutant) and co-cultured with pooled serum from each group: (i) mutant-select group, (j) mixed shedding group, (k) wild-type-select group and (l) without serum as a control. Data from e-l are presented as the Ct value from the SNP assay, and dashed lines show the Ct value limit (Ct 40) of the SNP assay. To generate graphs, undetected signals were assigned a Ct value of 41. Graphs show mean and s.d. from three independent experiments (n = 3).
Kinetics and characteristics of in vitro selection of the mutant virus by a bNAb.
To evaluate the effect of immune pressure on the selection of A388V mutant, wild-type and mutant viruses were mixed in different ratios (50:50 and 95:5) and cultured with or without immune pressure in vitro. A sub-neutralizing amount of CR6261 (approximately 95% maximal inhibitory concentration (IC95) for the wild type and IC75 for the mutant) was used to place the immune pressure. Although the initial ratios were well maintained without immune pressure (Fig. 4e,g), the growth kinetics of the viruses changed dramatically in the presence of the bNAb. When mixed in an equal ratio (50:50) and cultured under immune pressure, the mutant virus was rapidly expanded over the wild-type virus (Fig. 4f); when mixed at 5% under the same immune pressure, the mutant virus grew to levels similar to the wild-type virus in 72 h (Fig. 4h).
In vitro mutant virus selection by pre-existing human immunity.
Next, we measured the effect of immune pressure placed by the study participants that could be associated with the in-human selection shown in Fig. 1. Serum samples from each selection group (Fig. 1a) were pooled and used to assess immune pressure in vitro. A mixture of viruses (45% wild type and 55% mutant) representing the virus inoculum used in our human challenge studies was cultured with pooled serum from each group. Although serum from the mutant-select group rapidly selected for the mutant virus (Fig. 4i), serum from the non-mutant-select groups allowed both viruses to grow (Fig. 4j,k), similarly to the control without serum (Fig. 4l). Selection pressure from individual serum samples was also measured. Interestingly, the wild-type virus was not selected under serum pressure; however, only the mutant virus was favored or selected under several serum samples, most frequently (in five of seven) under those with high stalk antibody titers (Extended Data Fig. 6a, Q1). Serum with high stalk antibody titers (Q1) showed a significantly higher level of selection pressure than serum from the lowest quartile (Extended Data Fig. 6b). Moreover, the level of stalk antibody titers correlated well with the amount of selection pressure (Extended Data Fig. 6c). These results suggest that the mutant virus expansion that occurred in study participants with high stalk antibody levels (Fig. 1) was, at least in part, caused by selection pressure placed by pre-existing stalk immunity.
Discussion
In this study of H1N1 human challenge participants, selection of a stalk mutation that was associated with little to no loss of viral fitness or lessened clinical disease correlated with pre-challenge HA stalk serum antibody titers (Figs. 1 and 4 and Extended Data Fig. 6). This raises the possibility that immunity elicited by HA stalk-targeting influenza vaccines could result in expansion of HA stalk escape mutant viruses, in a manner analogous to the well-documented antigenic drift that occurs under population immune pressure at multiple HA and NA epitopes, possibly limiting the effectiveness of HA stalk-targeting ‘universal’ influenza vaccines.
The A388V stalk mutation of the 2009 H1N1pdm virus was first reported by Tan et al. in association with selection pressure applied using bNAb 6F12 (ref. 22). Anderson et al. further investigated the escape potential of the A388V mutation26. In the same study, an escape V41I mutant was generated under immune pressure by human serum26 without losing viral fitness, suggesting that influenza viruses are capable of escaping polyclonal immune pressure targeting the conserved HA stalk region. Doud et al.37 suggested that tolerated stalk mutations predominantly result in ‘modest’ (<ten-fold) effect on neutralization. Interestingly, both A388V and V41I37 mutations are also consistent with this hypothesis as they did not reduce, or only minimally reduced, viral fitness and had a less than ten-fold effect on neutralization. It will be important to investigate if these modest stalk mutation(s) would decrease vaccine efficacy and if stalk antibodies could be elicited in humans high enough to control mutations with these moderate effects.
The association between loss/retention of viral fitness and stalk mutations is not fully understood: some data show that stalk mutations are associated with loss of viral fitness, resulting in a decrease in viral pathogenicity in mice23,26 or an increase in sensitivity to anti-viral drugs25. However, an escape mutant that seemed to retain full viral fitness in vitro and in mice has also been reported26. The A388V mutation might cause subtle loss in viral fitness in humans that is below the threshold of detection in our small study and/or cause reduced transmissibility. However, it is important to note that around 40% of the study participants (12 of 29) selected for the V388 mutant virus after the viral challenge (Fig. 1), and only the mutant virus was selected under human serum, not the wild-type virus (Extended Data Fig. 6). This raises important questions about using current in vitro and in vivo models for measuring viral fitness because these experimental systems do not consider pre-existing immunity and might be less relevant for studying the complex characteristics of human IAV infection.
Although we showed that human intra-host expansion of the mutant virus occurred within a matter of days, selection characteristics on a population scale might not be easily predicted from this result. The emergence and establishment of an escape mutant would require multiple transmission events, in which the virus sequentially encounters different immune pressures, including pressures that select against it. Furthermore, it remains inconclusive whether selection for the V388 mutant virus that occurred in some study participants (Fig. 1), and in vitro under the participant serum (Fig. 4i and Extended Data Fig. 6), was driven by higher levels of stalk antibodies (Fig. 1c,d) or was due to fundamental differences in stalk antibody repertoire between the study participants. Further investigation of antibody repertoires on individual levels38 will be required to reveal underlying mechanisms of the mutant virus selection in humans. Moreover, although the present study was not designed to measure the effect of imprinting39 on HA stalk antibody responses and its possible role in the mutant virus selection, this will be important to examine in future studies.
To date, stalk escape mutations have been reported very infrequently in human IAV surveillance. For example, only five of approximately 25,000 human H1N1pdm HA sequences reported in the Influenza Research Database (https://www.fludb.org) carry the A388V mutation. However, these surveillance data suggest that stalk escape mutations such as A388V do arise spontaneously in nature. Possible explanations for the limited occurrence of stalk escape mutations include that current population-scale immune pressures to the HA stalk might not trigger or expand stalk escape mutations, which, in general, are not probable to confer evolutionary advantage under low immune pressure to them. Furthermore, the most frequently used system for global influenza virus surveillance (that is, passaging original clinical samples in MDCK cells or embryonated chicken eggs followed by Sanger consensus sequence analysis) might fail to detect these mutations while they are circulating at low level. Although stalk escape mutations have been infrequently reported, this study suggests that increased anti-HA stalk immunity without other types of antibodies (for example, HA inhibition and/or NA inhibition) can select for HA stalk escape mutants in people without apparent loss of fitness.
This study brings into question the generally supported concept40 that escape mutation(s) in the conserved HA stalk would not emerge in response to immune selective pressure. This concept derives from observations that the influenza HA stalk is under strict constraints owing to viral fitness costs imposed by mutations and from the hypothesis that broad human polyclonal immune responses would recognize multiple stalk epitopes. It is thought that these attributes would prevent viral escape in humans. Our study, however, demonstrated minimal to no loss of replicative fitness associated with the A388V escape mutation in vitro, in vivo and in humans. Moreover, human serum placed ‘biased, rather than broad, immune pressures that rapidly expanded the A388V mutant in vitro. High level of stalk antibody titers was also associated with the mutant virus selection both in humans and in vitro. Although our study suggests that V388 mutation would be substantially more resistant to multiple potent bNAbs, or to currently existing human serum antibodies in general, the neutralization efficacy of FI6V3 antibody was not affected by the A388V mutation. This suggests that current stalk vaccination strategies could be strengthened if it could induce specific types of stalk antibodies in the face of potential emergence of escape mutants.
In conclusion, this study sheds light on a potential obstacle for the success of universal influenza vaccines exclusively targeting the HA stalk. It will be important to investigate if new universal influenza vaccine strategies could generate a broader and higher antibody response than conventional vaccination or natural infection, to provide broader protection even against escape mutants. Further investigation is needed to understand the population-level dynamics of HA stalk immunity and viral mutation for HA stalk-targeting vaccine strategies to become truly ‘universal’ against ever-evolving influenza viruses.
Methods
Influenza human challenge study, sample collection, RNA extraction and determination of anti-HA stalk titers.
Clinical samples were obtained from previously performed influenza virus human challenge studies27,28. The studies were approved by the National Institute of Allergy and Infectious Diseases (NIAID) Institutional Review Board and performed at the National Institutes of Health (NIH) Clinical Center (ClinicalTrials.gov identifiers NCT01646138 and NCT01971255; see Life Sciences Reporting Summary). Written informed consent was obtained from all study participants. Volunteers were intranasally inoculated with 107 TCID50 of A/California/04/2009 (H1N1) virus (H1N1pdm) in a 1-ml inoculum as previously described27,28. The challenge virus stock was generated under a GMP and accumulated a polymorphism at position 388 (position 1 being initial methionine): 45% wild-type (A388) and 55% mutant (V388) HA measured by deep sequencing analysis29. Serum samples used for analyzing pre-existing immunity were collected right before the challenge inoculation. Nasal wash samples were collected daily after challenge until samples were confirmed negative for viral shedding for two consecutive days using BioFire FilmArray Respiratory Panel (BioFire Diagnostics)30. Nasal wash samples that were confirmed positive for influenza virus were selected for viral RNA isolation to be analyzed using the SNP assay (A388 versus V388). RNA isolation was performed using InnuPure C16 touch system (catalog no. 845-00020-2, Analytik Jena) with innuPREP Virus DNA/RNA Kit-IPC16 (catalog no. 845-IPP-7016096, Analytik Jena). RNA was extracted from 200 μl of nasal wash samples and resuspended in 100 μl of elution buffer. Anti-HA stalk IgG antibody titers were determined as previously described using serum samples from the study participants13.
SNP assay.
SNP (A388 versus V388) was determined using minor groove binder (MGB)-based TaqMan probes combined with multiplex quantitative polymerase chain reaction (qPCR). Primers and MGB probes were initially designed using Primer Express Software v3.0 (catalog no. 4363991, Thermo Fisher) and further modified to increase specificity. Primer and MGB probe sequences and concentrations used for the SNP assay are listed in Supplementary Table 1. The VIC-labeled TaqMan probe is designed to detect the wild-type (A388) sequence (gcc), and the FAM-labeled TaqMan probe is designed to detect the mutant (V388) sequence (gtc). The MGB probes were synthesized by Thermo Fisher. TaqMan Fast Virus 1-Step Master Mix (catalog no. 4444432, Thermo Fisher) was used for the SNP assay along with QuantStudio 6 Flex qPCR System (catalog no. 4485699, Thermo Fisher).
SNP determination of clinical nasal wash samples was done using the SNP assay after an initial one-step reverse transcription PCR (RT-PCR) amplification of the influenza HA stalk region. Primers listed in Supplementary Table 1 were used for the initial RT-PCR amplification. SuperScript IV One-Step RT-PCR System (catalog no. 12594100, Thermo Fisher) was used for high-fidelity RT-PCR amplification (cycle conditions: 53 °C for 10 min; 98 °C for 2 min; 35 cycles of 98 °C for 10 s, 60 °C for 10 s, 72 °C for 20 s; 72 °C for 5 min) following the manufacturer’s instruction using 5 ul of the viral RNA from clinical nasal wash samples purified as described above. One microliter of the RT-PCR product was used for SNP determination using TaqMan Fast Virus 1-Step Master Mix (catalog no. 4444432, Thermo Fisher) and presence/absence experiment mode of the QuantStudio 6 Flex qPCR System. The following cycle was used: 60 °C for 30 s (pre-read stage); 95 °C for 20 s; 40 cycles of 95 °C for 3 s, 65.5 °C for 30 s; 60 °C for 30 s (post-read stage). ΔRn for VIC (detecting wild type) and ΔRn for FAM (detecting mutant) were collected simultaneously and used to determine the presence or absence of the wild-type or mutant sequences. Rn was defined as the intensity of the reporter dye normalized to the passive reference ROX dye. ΔRn was defined as: Rn (post-PCR read) − Rn (pre-PCR read). RNA extracted from the wild-type and mutant H1N1pdm viruses was mixed at varying ratios (10:0, 9:1, 1:9 and 0:10) and processed identically along with the viral RNA from clinical nasal wash samples; the ΔRn values of the control RNA mixtures were used to determine the clinical sample polymorphism (Extended Data Fig. 1a–c). Specifically, the wild-type virus was considered to be present when the VIC ΔRn value of the sample was greater than that of the 1:9 control mixture (10% wild type). The mutant virus was considered to be present when the FAM ΔRn value of the sample was greater than that of the 9:1 mixture (10% mutant). When both viruses were present, the sample was considered to have mixed polymorphism (mixed shedding). The wild-type and mutant H1N1pdm viruses used to provide the control RNA were generated using a 12-plasmid reverse genetics system41 with all viral segments derived from the H1N1pdm virus. The SNP determination of clinical nasal wash samples was repeated three times (initial RT-PCR amplification as well as the following SNP determination) to ensure reliability of the data. Of those three repeats, each SNP (wild type or mutant) had to be detected at least twice to be counted toward the determination of the selection outcome; a SNP amplified only once out of three repeats was not counted. For example, when the wild-type sequence was detected three times and the mutant sequence was detected only once, the sample was considered as wild type only. Also, the SNP result from the latest time point was used to determine the selection outcome of a study participant. For example, when both viruses were detected from day 3 nasal wash, but only the mutant virus was detected from day 4 and 5 nasal wash, the study participant was considered to have selected for the mutant virus.
SNP assay was also used to track in vitro selection dynamics from using extracted RNA without the initial one-step RT-PCR amplification. RNA from virus culture media was extracted as described above using the InnuPure C16 touch system using 50 μl of virus culture media and resuspended in 50 μl of elution buffer. Two microliters of the extracted RNA was used for the SNP assay using the standard curve experiment mode of the QuantStudio 6 Flex qPCR System. The following cycle was used: 50 °C for 5 min; 95 °C for 20 s; 40 cycles of 95 °C for 3 s, 65.5 °C for 30 s. The results are reported as cycle threshold (Ct) values. The SNP assay, both for the clinical nasal wash samples and the in vitro selection experiment, was validated for accuracy under various conditions using the wild-type or mutant H1N1pdm viruses generated as described above. Viral RNA from wild-type and mutant viruses, isolated as described above, was mixed in varying ratios from 10:0 (0% mutant virus) to 0:10 (100% mutant virus) and diluted to represent varying viral loads: 103.0, 104.0 and 105.5 TCID50 per ml for the two-step SNP assay of clinical nasal wash samples (Extended Data Fig. 1a–c); 104.0, 105.5 and 107.0 TCID50 per ml for the SNP assay using RNA from virus cultures (Extended Data Fig. 1d–f).
Evaluation of viral fitness in vitro and in vivo.
Wild-type (A388) and mutant (V388) H1N1pdm viruses were generated using a reverse genetics system as described above and passaged twice in MDCK cells before being tested for viral fitness. To measure viral fitness in vitro, MDCK, Vero and A549 cells were seeded in 96-well plates and infected with 0.001 multiplicity of infection (MOI) of the wild-type or mutant viruses when cells were 90–100% confluent. After 60 min, virus inoculum was removed, and fresh virus growth media was added. For MDCK and Vero cells, the virus growth medium was prepared by adding Antibiotic-Antimycotic (catalog no. 15240062, Thermo Fisher) and 6- (1-tosylamido-2-phenyl) ethyl chloromethyl ketone (TPCK)-treated trypsin (catalog no. T1426, Millipore Sigma) at a final concentration of 1 μg ml−1 to DMEM (catalog no. 11995065, Thermo Fisher). For A549 cells, F-12K Medium (catalog no. 30–2004, ATCC) was used instead of DMEM. After 6, 24, 48 and 72 h, the supernatant was collected, clarified by centrifugation and titrated in MDCK cells using TCID50 assay, and the Reed-Muench method42 was used for the TCID50 calculation. For measuring viral fitness in mice, 7–8-week-old female BALB/c mice were intranasally infected with 103 TCID50 of wild-type (n = 15) or mutant (n = 15) virus in 50 μl of phosphate-buffered saline (PBS). At 2 (n = 5), 4 (n = 5) and 6 (n = 5) d post challenge (dpc), mice were euthanized using CO2, and lungs were harvested and homogenized in PBS using a tissue homogenizer (Omni International). Viral loads were titrated in MDCK cells using TCID50 assay as described above. For measuring viral fitness in ferrets, 6–7-month-old female ferrets were intranasally infected with 105 TCID50 of wild-type (n = 6) or mutant (n = 6) virus in 1 ml of PBS. At 1, 3, 5 and 7 dpc, ferrets were lightly anesthetized using isoflurane, and nasal washes were collected using 1 ml of PBS. Viral loads were titrated in MDCK cells using TCID50 assay as described above. All animal experiments were conducted under protocols approved by the Animal Care and Use Committee at the NIAID, NIH. See the Life Sciences Reporting Summary for further information on the in vivo study replication.
Production of full-length HA and HA stalk-only proteins.
Full-length wild-type (A388) and mutant (V388) H1N1pdm HA proteins were designed as previously described43 with a Strep-tag II instead of hexahistidine tag used in the reference study (Extended Data Fig. 7). The wild-type and mutant stalk-only constructs were designed based on a stalk-only construct that has been successfully used as a universal influenza vaccine candidate3 as well as to measure stalk-binding antibody levels in human serum13. Two minor modifications were added: 1) a carboxy-terminal T4 trimerization domain for increased stability43,44 and 2) Strep-tag II, instead of hexahistidine tag, for purification (Extended Data Fig. 7). Both the full-length HA and stalk-only constructs were produced in Sf9 insect cells using the Bac-to-Bac baculovirus expression system (catalog no. 10359016, Thermo Fisher) and purified using Strep-Tactin Sepharose (IBA) as previously described13. A Bicinchoninic Acid Protein Assay Kit (catalog no. 23225, Thermo Fisher) was used for protein quantification for both full-length HA and stalk-only constructs.
ELISA using full-length HAs.
To measure the effect of the A388V mutation on the HA structure, full-length wild-type or mutant HAs, produced as described above, were diluted in PBS (3 μg ml−1) and added to 96-well ELISA plates (50 μl per well) (catalog no. 456537, Thermo Fisher). The plates were incubated overnight at 4 °C followed by the addition of blocking buffer (1% bovine serum albumin (BSA) in PBS, 100 μl per well). After 30 min at room temperature, the plates were washed three times with wash buffer (0.05% Tween 20 in PBS). Six bNAbs that bind to HA stalk were used for the ELISA: CR6261 (refs. 20,31) (a gift from Janssen Pharmaceutica), CR9114 (ref. 32) (catalog no. PABX-119, Creative Biolabs), FI6V3 (ref. 33) (catalog no. PABL-214, Creative Biolabs), 70–1F02 (ref. 34) (a gift from Dr. Rafi Ahmed, Emory Vaccine Center), C179 (ref. 1) (catalog no. M145, Takara Bio) and CT149 (ref. 3) (catalog no. PABL-213, Creative Biolabs). Two neutralizing monoclonal HA head-binding antibodies, EM-4C04 (ref. 34) (a gift from Dr. Rafi Ahmed, Emory Vaccine Center) and 2–12C36 (a gift from Dr. Alain Townsend, University of Oxford), were used to show the structural integrity of the purified full-length HA proteins. Anti-Strep-tag II antibody (catalog no. Ab76949, Abcam) was used to confirm that equal amounts of wild-type or mutant HAs were used for ELISA. Antibodies were serially diluted in antibody diluent (1% BSA and 0.05% Tween 20 in PBS) and added to the washed plates. After incubation (room temperature, 2 h), the plates were washed three times, and 1:10,000 diluted horseradish peroxidase (HRP)-conjugated anti-human IgG antibody (catalog no. Ab 205630, Abcam) was added (100 μl per well) for CR6261, CR9114, FI6V3, 70–1F02, CT149, EM-4C04 and 2–12C. HRP-conjugated anti-mouse IgG antibody (catalog no. A28177, Thermo Fisher) and anti-rabbit IgG antibody (catalog no. A16110, Thermo Fisher) were 1:10,000 diluted and used for C179 and anti-Strep-tag II antibody, respectively. After incubation (room temperature, 1 h), the plates were washed six times followed by 30 min of room temperature incubation with HRP substrate solution (100 μl per well) prepared by adding a 10-mg o-phenylenediamine dihydrochloride tablet (catalog no. P8287, Millipore Sigma) to 20 ml of phosphate citrate buffer preparation (catalog no. P4922, Millipore Sigma). The reaction was stopped by adding 1 M sulfuric acid (100 μl per well), and the optical density was measured at 492 nm (OD492). Area under the curve (AUC) values were calculated using Prism8 software v.8.3.0 (GraphPad Software). The baseline for AUC calculation was set as 0.1 to exclude nonspecific signals from the AUC calculation. The OD492 of 0.1 is approximately two times the OD492 value from the control wells that were treated equally but without the primary antibodies.
ELISA using HA stalk-only constructs.
Wild-type and mutant stalk-only constructs were produced and purified as described above to specifically measure antibodies recognizing the mutant stalk in human serum while excluding head-binding antibodies. To measure the structural differences between the wild-type and mutant HA stalk-only construct, ELISA and the following AUC calculation was performed using serially diluted bNAbs (CR6261, CR9114, FI6V3, 70–1F02, C179 and CT149) as described above with one modification. The stalk-only constructs were diluted in PBS at 1 μg ml−1 for coating ELISA plates, rather than 3 μg ml−1. To show that the stalk-only A388V construct closely represents the stalk structure in the full-length A388V HA, the magnitude of the structural differences between the respective wild-type and mutant constructs was compared by ELISA and the following AUC calculation as described above. To measure the change in human serum’s ability to recognize the mutant stalk structure, the serum samples obtained from the influenza human challenge study were analyzed using ELISA. Pre-challenge serum samples were serially diluted (initial 1:200 dilution followed by four-fold serial dilution) and analyzed using ELISA and following AUC calculation as described above. Reciprocal dilutions (dilution factors) of the serum were used as x values for the AUC calculation.
Structural modeling.
A structural analysis was performed with UCSF Chimera45 using a previously published 2009 H1N1 influenza virus HA structure (PDB ID: 3LZG)46. Structural editing was performed using the Rotamers option in UCSF Chimera. Molecules expected to undergo steric clash upon A388V mutation were predicted using the default clash parameter in UCSF Chimera.
Flow cytometry.
MDCK cells were infected at 1 MOI of wild-type (A388) or mutant (V388) H1N1pdm viruses generated by reverse genetics. Twenty-four hours after infection, cell culture supernatant was discarded and cells were washed twice with PBS and treated with trypsin-EDTA (catalog no. 25200056, Thermo Fisher) for 10 min at 37 °C. The cells were then harvested, and the trypsin was neutralized by adding an equal volume of PBS supplemented with 5% fetal bovine serum. After centrifugation (300g for 5 min), the supernatant was discarded, and cells were fixed by resuspending in Fixative Solution (catalog no. R37814, Thermo Fisher) for 15 min at room temperature. Fixed cells were filtered using a 40-μm cell strainer (catalog no. 352340; Corning Life Sciences) to remove clumped cells. Filtered cells were spun (300g for 5 min), the supernatant was removed and the cells were washed twice with flow cytometry buffer (1% BSA, 0.1% sodium azide in PBS) using centrifugation (300g for 5 min) and resuspended in the flow cytometry buffer. Six stalk-binding bNAbs (CR6261, CR9114, FI6V3, 70–1F02, C179 and CT149) were conjugated with fluorescein isothiocyanate (FITC) using a FITC Conjugation Kit (catalog no. Ab 102884, Abcam). EM-4C04, a monoclonal antibody binding to the HA globular head, was conjugated with R-phycoerythrin (R-PE) using an R-PE Conjugation Kit (catalog no. Ab102918, Abcam). Anti-influenza nucleoprotein (NP) antibody (catalog no. MAB8257, Millipore Sigma) was conjugated with allophycocyanin (APC) using an APC Conjugation Kit (catalog no. Ab201807, Abcam) and used for gating. The estimated concentrations of the conjugated antibodies were 0.83 mg ml−1 for all antibodies. Cells were stained with an antibody mixture—one of the FITC-conjugated stalk-binding bNAbs (CR6261, CR9114, FI6V3, 70–1F02, C179 or CT149), the R-PE-conjugated HA head-binding antibody (EM-4C04) and the APC-conjugated NP antibody. For the staining, 0.5 × 106 cells were resuspended in 50 μl of the diluted antibody mixture (1:100 for stalk antibodies, 1:500 for EM-4C04 and 1:500 for NP antibody). Data were collected using BD LSR II (BD Biosciences) and FACSDiva software (version 6.2, BD Biosciences) and analyzed using FlowJo v.10.6.1 (BD Biosciences). The flow cytometry was independently repeated three times.
Measuring viral resistance to a broadly neutralizing antibody.
The effect of the A388V mutation on the increase of viral resistance to three bNAbs (CR6261, CR9114 and FI6V3) was measured using wild-type (A388) and mutant (V388) H1N1pdm viruses generated by reverse genetics. To measure the IC50 value of CR6261, CR9114 and FI6V3 to each virus, an ELISA-based microneutralization assay47 was performed using five-fold serially diluted bNAbs. The percentage of infection was normalized to the lowest antibody concentration, and the IC50 values were determined using a nonlinear least square regression model (variable slope, four parameters) using Prism8 software v.8.3.0 (GraphPad Software). To compare viral resistance in vivo, 7–8-week-old female BALB/c mice were intranasally infected with 103 TCID50 of wild-type or mutant virus in 50 μl of PBS 24 h after a prophylactic intraperitoneal injection of CR6261 antibody at 0.2 mg kg−1, 1.0 mg kg−1, 30.0 mg kg−1 or a saline control (200 μl per animal). To rule out the possibility of ex vivo neutralization of the viruses by CR6261 during tissue processing, viral titers were quantified using qPCR. Groups of five mice were euthanized at day 2 after challenge, lungs were collected and homogenized in Trizol and total RNA was isolated following the manufacturer’s protocol (Thermo Fisher) and purified using the RNeasy Mini Kit (Qiagen). Reverse transcription of total RNA was performed using the Superscript III First-Strand cDNA Synthesis Kit (catalog no. 18080051, Thermo Fisher) primed with an equal mix of oligo(dT) and the Uni12 influenza A specific primer: 5′-AGCRAAAGCAGG-3′. The influenza matrix (M) gene was quantified using primers and probe previously published48 (Supplementary Table 1). The average (n = 5) level of influenza M gene RNA (normalized to GAPDH; catalog no. 4352932E. Thermo Fisher) from CR6261-treated and infected mice was normalized to M gene RNA level in control mice (antibody untreated, infected mice) using the ΔΔCt method.
In vitro selection study.
The effect of monoclonal and polyclonal immune pressure on the selection of the mutant virus was measured by co-culturing the wild-type and mutant viruses under selection pressure by bNAb CR6261 or human serum. To measure the selection dynamics under CR6261, reverse genetics-generated wild-type and mutant H1N1pdm viruses were mixed in different ratios (50:50 and 95:5) while maintaining the combined viral titer at 2 × 105 TCID50 per ml. CR6261 antibody was diluted to a final concentration of 10 μg ml−1. Supplemented medium used for virus and CR6261 antibody dilution was made by supplementing 100 ml of DMEM (Thermo Fisher) with 10 ml of 7.5% BSA (catalog no. 15260037, Thermo Fisher), 1.1 ml of Antibiotic-Antimycotic (catalog no. 15240062, Thermo Fisher) and 110 μl of TPCK-treated trypsin (1 μg ml−1, Millipore Sigma). Diluted viruses at different ratios were mixed with an equal volume of diluted CR6261 antibody and incubated at 37 °C for 1 h and then added to MDCK cells at 90–100% confluency in 96-well plates (100 μl per well). Supernatants from three wells were pooled for analysis at 6, 24, 48 and 72 h after infection. Viral RNA was extracted and analyzed using the SNP assay as described above.
To assess the immune pressure placed by polyclonal human serum, serum samples were divided into three groups based on the selection outcomes according to the SNP assay results (Fig. 1). Equal volumes of serum from each group were pooled and used to place immune pressure. Reverse genetics-generated wild-type and mutant H1N1pdm viruses were mixed to represent the challenge virus used in the human challenge studies (45% wild type and 55% mutant) while maintaining the combined viral titer at 2 × 105 TCID50 per ml. The mixed viruses were incubated with an equal volume of 1:50-diluted human pooled serum. After incubation (37 °C, 1 h), the virus and antibody mixtures were added to MDCK cells at 90–100% confluency in six-well plates (800 μl per well). Supernatants were collected for analysis at 6, 24, 48 and 72 h after infection (70 μl per collection). Viral RNA was extracted, and the selection dynamics were analyzed using the SNP assay as described above. Individual serum samples were also analyzed for the selection pressure. Individual serum samples were 1:50 diluted and mixed with an equal volume of wild-type and mutant virus mixture (50:50, 2 × 105 TCID50 per ml combined viral titer) followed by incubation at 37 °C for 1 h and then added to MDCK cells at 90–100% confluency in six-well plates (800 μl per well). Supernatants were collected for analysis at 48 and 72 h after infection (70 μl per collection). Viral RNA was extracted using 50 μl of supernatant, and the selection dynamics were analyzed using the SNP assay as described above. Serum samples with a higher level of neutralizing antibodies, not allowing measurable viral growth, were further diluted at 1:100 or 1:200 to allow viruses to grow. A 1:200 dilution was used for the serum samples that neutralized viruses at 1:100. To investigate the association between the level of stalk antibodies and the level of immune pressure selecting for the mutant virus, mutant selection index of individual serum samples was calculated by the ΔΔCt method normalized to controls cultured without serum: ΔCt wild-type = Ct wild-type control − Ct wild-type with serum; ΔCt mutant = Ct mutant control − Ct mutant with serum; ΔΔCt = ΔCt mutant − ΔCt wild-type; mutant selection index = ΔΔCt × dilution factor (50, 100 or 200). All the in vitro selection studies were repeated three times independently.
Statistical analyses.
Anti-HA stalk antibody titers were compared between the mutant selection group and other groups using a nonparametric two-tailed Mann-Whitney test (Fig. 1c,d). A two-tailed unpaired Welch’s t-test was used to compare differences in in vitro and in vivo viral growth kinetics on the log10-transformed titers (Fig. 2). A two-tailed paired t-test was used to compare the recognition of the wild-type and mutant stalk by individual human serum using AUCs (Fig. 3j). A two-tailed unpaired Welch’s t-test was used to compare differences in IC50 values (Fig. 4a–c). IC50 curves were generated using a nonlinear least square regression model (variable slope, four parameters) using Prism8 software v.8.3.0 (GraphPad Software). A nonparametric two-tailed Mann-Whitney test was used for in vivo viral replication under CR6261 (Fig. 4d). The difference in HAI titers, ratios between HA stalk antibodies and total HA antibodies, age and birth year of the participants, days and number of symptoms and days of shedding between the mutant selection group and other groups were compared using a nonparametric two-tailed Mann-Whitney test. (Extended Data Fig. 2). The level of selection pressure (mutant selection index) between samples from different quartiles was compared using nonparametric one-way analysis of variance (Kruskal-Wallis test) and Dunn’s test as a post hoc test (Extended Data Fig. 6b). Correlation between the anti-stalk serum IgG titer and the mutant selection index was analyzed by calculating nonparametric two-tailed Spearman’s rank correlation analysis. The best-fit line was plotted using simple linear regression analysis. All statistical analyses were performed using Prism8 software v.8.3.0 (GraphPad Software).
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1 |. A Single Nucleotide Polymorphism (SNP) assay using minor groove binder (MGB)-based TaqMan probes for detecting wild-type (A388) or mutant (V388) genes in various conditions.
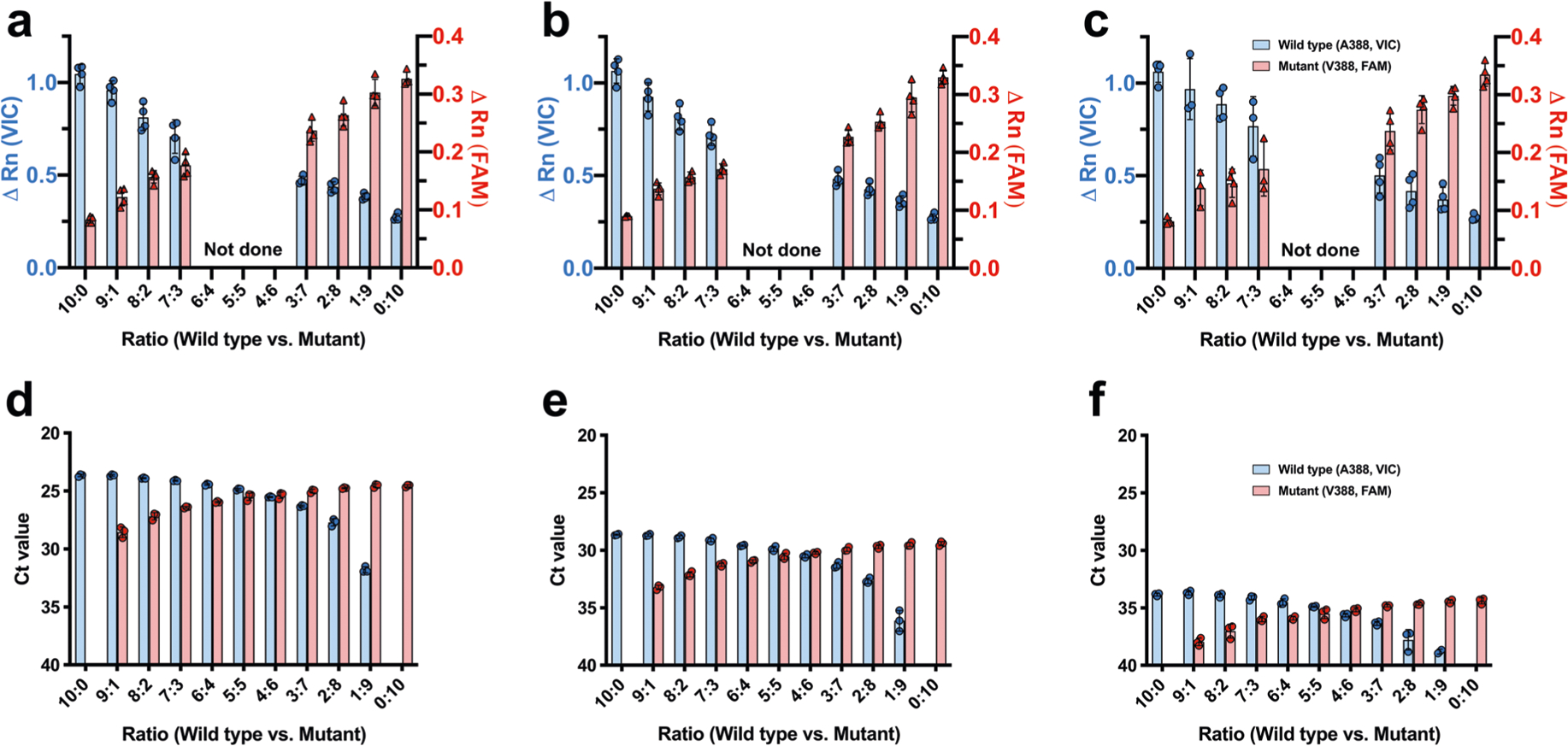
A SNP assay was developed for detecting wild-type (A388) or mutant (V388) HA genes in various conditions utilizing a set of Minor Groove Binder (MGB)-based TaqMan probes: VIC-labeled probe detecting the wild-type (A388) and a FAM-labeled probe detecting the mutant (V388). The SNP assay was validated using a mixed viral genome from the 2009 H1N1pdm wild-type (A388) and the mutant (V388) viruses. Viral RNA was mixed in varying ratios from 10:0 (0% mutant virus) to 0:10 (100% mutant virus). For the validation of two-step SNP assay for analyzing nasal wash samples from the human challenge study, mixed viral RNA was diluted to represent varying viral loads of (a) 105.5 TCID50/ml, (b) 104.0 TCID50/ml, and (c) 103.0 TCID50/ml. Prepared viral RNA was analyzed by the two-step SNP assay (see Methods). Blue and red bars indicate the ΔRn value of wild-type and mutant virus, respectively. Graphs show mean ΔRn value and standard deviation from 4 independent experiments (a-c, n = 4). For the validation of one-step SNP assay used to analyze selection dynamics in vitro, mixed viral RNA was diluted to represent varying viral loads of (d) 107.0 TCID50/ml, (e) 105.5 TCID50/ml, and (f) 104.0 TCID50/ml. Prepared viral RNA was analyzed by the one-step SNP assay (see Methods). Blue bars indicate the threshold cycle (Ct) values representing the amount of wild-type (A388) virus. Red bars represent the amount of mutant (V388) virus. Graphs show mean Ct value and standard deviation from 3 independent experiments (d-f, n = 3). Results show that the SNP assay reliably detects minor population, either wild-type or mutant, existing as low as 10% across the various viral loads.
Extended Data Fig. 2 |. Comparison of different parameters between study participants with different selection outcomes.
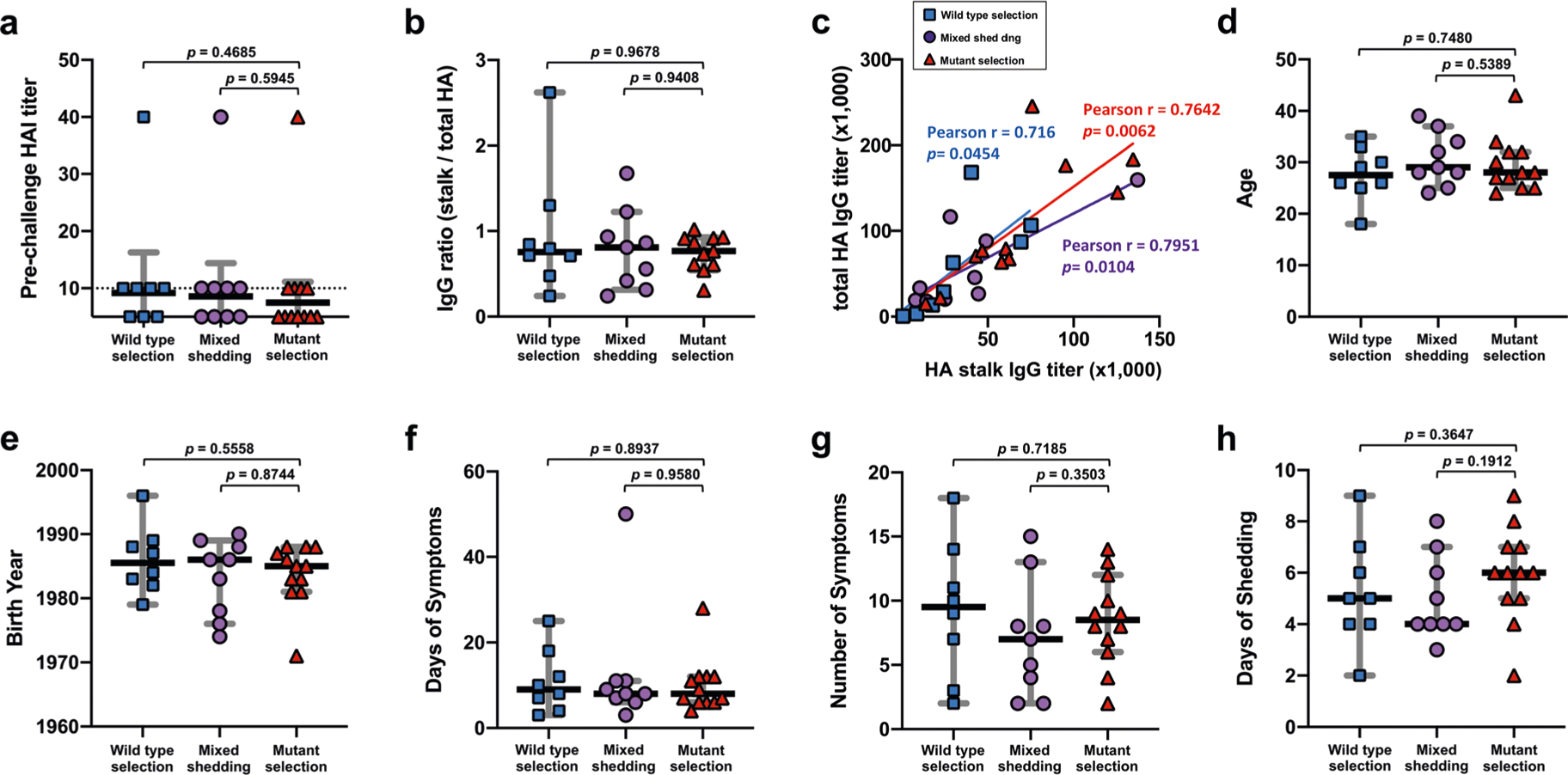
Different parameters were compared between groups to find possible correlates of the selection outcomes. a, Pre-challenge serum hemagglutination inhibition (HAI) titers were compared. Black horizontal lines and grey error bars represent geometric mean titer and 95% CI, respectively. Dashed line shows the detection limit. b, The fraction of HA stalk antibodies relative to total HA antibodies was compared. The ratio is a calculation of serum anti-stalk IgG and anti-full-length HA IgG titers measured by ELISA. c, Serum IgG titer data (anti-stalk and anti-total HA) generated in (b) were used to analyze the correlation between the stalk and total HA antibody titers in each selection group. A two-tailed Pearson’s correlation coefficient (Pearson’s r) was used for the analysis. The positive correlation between the stalk antibody titers and total HA titers may explain the lack of difference in stalk/total HA antibody ratio between groups seen in (b). To find a possible role of immunological imprinting on the selection outcomes, (d) age of the participants at the study enrollment and (e) birth year was compared. To find potential loss in viral fitness in humans caused by the A388V mutation, (f) days of symptoms, (g) number of symptoms, and (h) duration of shedding after the challenge infection were compared between groups. Black horizontal lines and grey error bars represent median value and 95% CI, respectively (b,d,e-h). A two tailed nonparametric Mann-Whitney test was used to compare parameters from the mutant selection group to other selection groups. Each symbol represents an individual study participant and their selection outcome (wild-type selection, n = 8; mixed shedding, n = 9; mutant selection, n = 12).
Extended Data Fig. 3 |. A significant conformational change to the HA stalk region induced by A388V mutation.
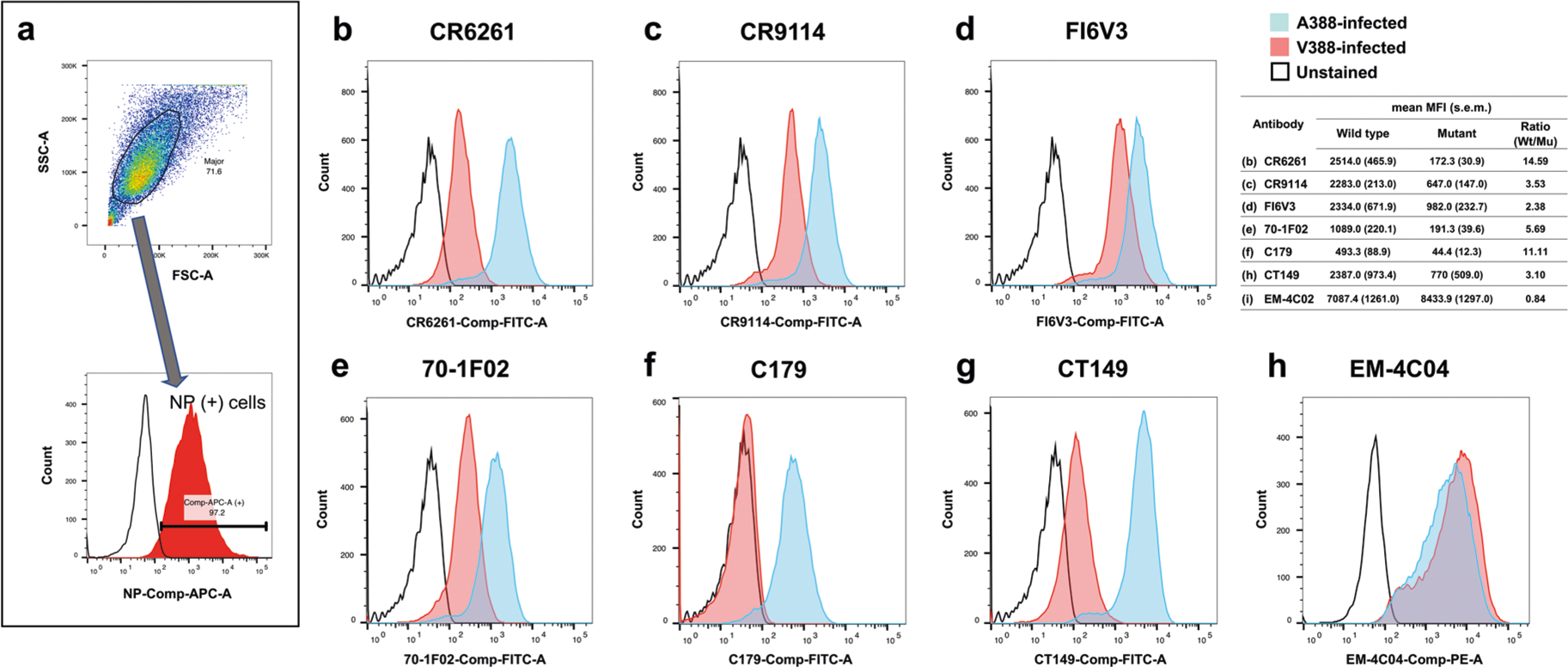
MDCK cells were infected with 1 multiplicity of infection (MOI) of wild-type (A388) or mutant (V388) H1N1pdm viruses generated by reverse genetics. 24 hours after infection, cells were harvested, and the expression of wild-type and mutant HA was measured by flow cytometry to evaluate the effect of the A388V mutation on the HA stalk epitopes. a, A representative example of the gating strategy. Dead cells and debris were excluded based on FSC/SSC cell dot plot. Anti-influenza nucleoprotein (NP) antibodies conjugated with allophycocyanin (APC) were used for gating. Only infected cells, expressing NP, were used for the analysis. Broadly neutralizing antibodies binding to the HA stalk, (b) CR6261, (c) CR9114, (d) FI6V3, (e) 70–1F02, (f) C179, and (g) CT149 were conjugated with fluorescein isothiocyanate (FITC). A monoclonal antibody that binds to the HA globular head, (h) EM-4C04, was conjugated with r-phycoerythrin (R-PE). Each stalk-binding antibody was mixed with the head-binding EM-4C04 antibody and NP antibody, and the antibody mixtures were used to stain cells expressing the wild-type or mutant HA. Histograms are colored differently to show different experimental groups: Blue - cells infected with wild-type (A388) virus; Pink - cells infected with mutant (V388) virus; Black - unstained cell control. Representative histograms from three independent experiments are shown. The summary table shows the average median fluorescence intensity (MFI) and standard error of mean (s.e.m.) of the three independent experiments.
Extended Data Fig. 4 |. Comparison between stalk-only and full-length HA construct.
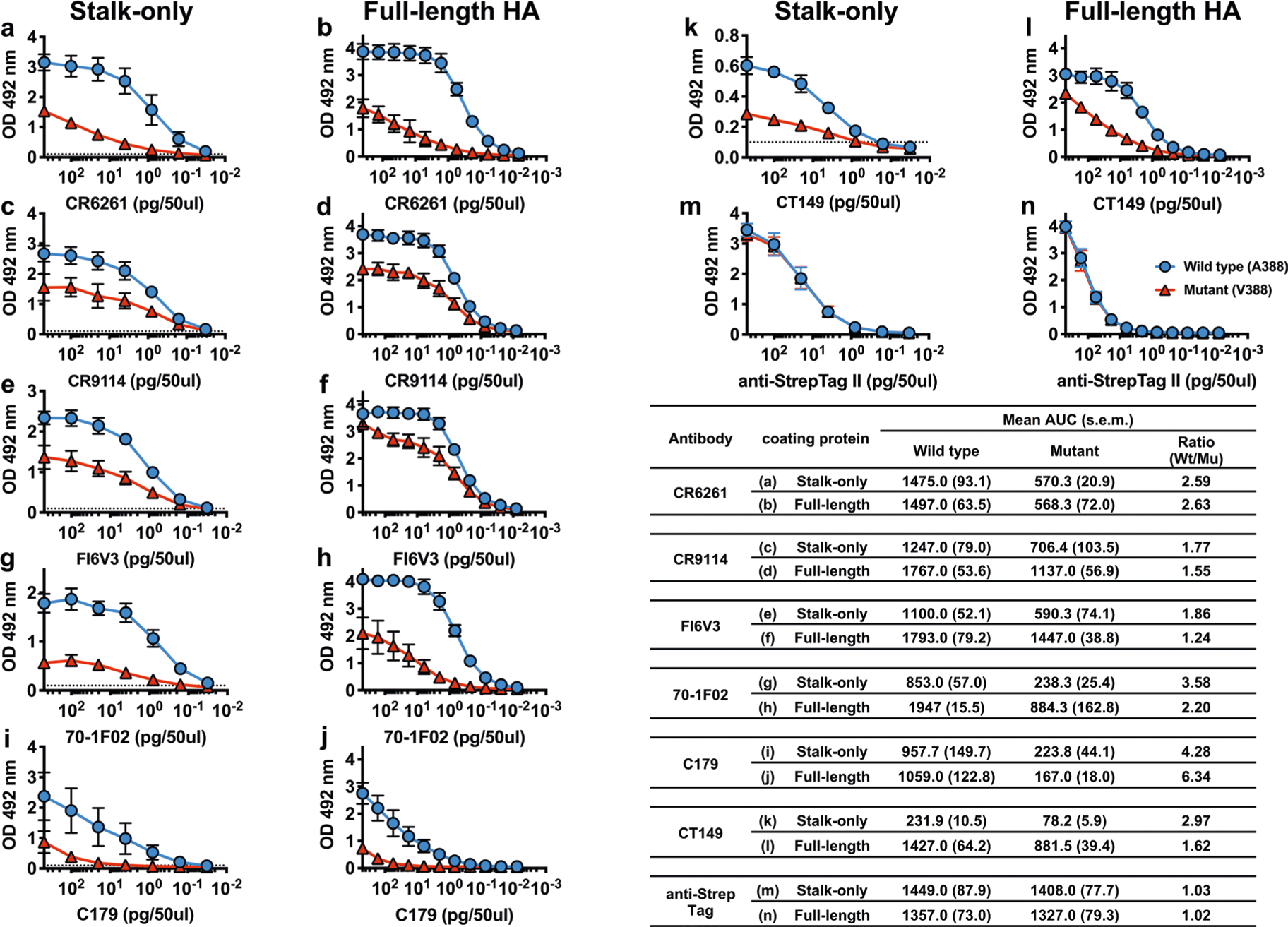
Stalk-only constructs with or without the A388V mutation (See Extended Data Fig. 7b for the amino acid sequence) were produced to measure the level of antibodies recognizing the mutant stalk in human serum while excluding head-binding antibodies (Fig. 3j). To confirm that the stalk-only construct with A388V mutation appropriately represents the natural A388V stalk structure, the level of decrease in broadly neutralizing monoclonal antibodies (bNAbs) binding was compared between the stalk-only construct and the full-length HA. ELISA was performed using serially diluted (a,b) CR6261, (c,d) CR9114, (e,f) FI6V3, (g,h) 70–1F02, (l,j) C179, and (k,l) CT149. (m,n) Anti-StrepTag II antibody was used to show that equal amounts of wild-type and mutant antigen were used for the analysis. The AUC was calculated using GraphPad Prism8 (v.8.3.0). The AUC for the full-length HAs (b,d,f,h,j,l) was calculated using the data from Fig. 3. Graphs show mean and standard deviations from three independent measurements. The summary table shows the mean OD492 values and standard error of mean (s.e.m.) of the three independent measurements. The comparison result (summarized in the table) shows that the A388V stalk-only construct closely represents the natural stalk structure of the full-length A388V HA.
Extended Data Fig. 5 |. AuC calculation using wile-type and mutant stalk-only construct.
Raw ELISA data used to generate Fig. 3j are shown. Stalk-only constructs with or without the A388V mutation (See Extended Data Fig. 7b for the amino acid sequence) were used to measure changes in the level of antibodies recognizing the wild-type (A388) of mutant (V388) stalk in human serum. twenty-nine pre-challenge serum samples from the influenza human challenge study participants were serially diluted and used for the ELISA. the AUC was calculated using graphPad Prism8 (v.8.3.0) with the baseline value of 0.1 (approximately 2 times the OD492 value from the control wells). Blue lines and numbers show the ELISA data and the AUC, respectively, obtained using the wild-type (A388) stalk construct. Red lines and numbers show the ELISA data and the AUC, respectively, obtained using the mutant (V388) stalk construct. graphs show mean and standard deviations from three independent measurements.
Extended Data Fig. 6 |. Association between the stalk antibody titers and selection pressure measured in vitro.
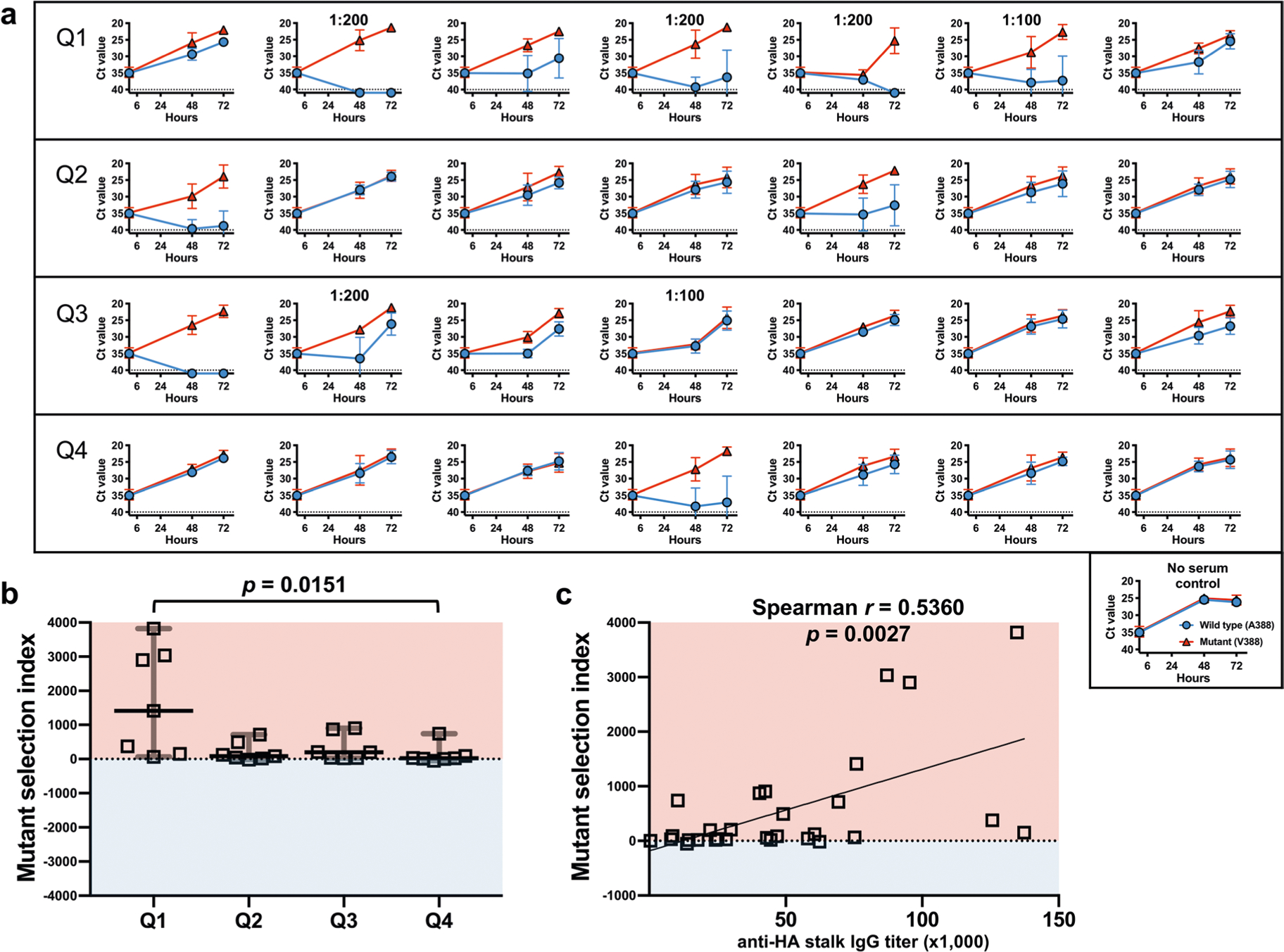
Selection pressure placed by individual human sera was measured using pre-challenge serum from the influenza human challenge study participants. a, An equal mixture of the wild-type (A388) and mutant (V388) virus was cultured with 1:50 diluted serum samples. Sera from Q1, Q2, Q3 and Q4 from Fig. 1 (n = 7 per quartile) were used. Culture supernatants were collected at 48 and 72 hours after infection followed by viral RNA extraction. The selection dynamics were measured using a Single Nucleotide Polymorphism (SNP) assay utilizing a set of Minor Groove Binder (MGB)-based TaqMan probes; VIC-labeled probe detects the wild-type (A388); FAM-labeled probe detects the mutant (V388). Data are presented as the threshold cycle (Ct) value from the SNP assay. Dashed lines show the Ct value limit (Ct 40) of the SNP assay. A Ct value of 41 was given to undetected signals to generate graphs. Error bars represent standard deviations from three independent experiments. The final dilution is noted on the individual graph if higher than 1:50. b, Mutant selection index was calculated based on data from (a) by ΔΔCt method using controls cultured without serum (see Methods). A mutant selection index higher than 0 (pink area) indicates a serum sample selected for the mutant virus. An index lower than 0 (blue area) indicates a serum sample selected for the wild-type virus. Horizontal lines show median values and error bars represent 95% CI. The indexes between samples from different quartiles were compared using nonparametric one-way analysis of variance (Kruskal-Wallis test) and Dunn’s test as a post-hoc test. c, Correlation between the anti-stalk serum IgG titer and the selection index of 29 sera samples were analyzed by calculating two-tailed Spearman’s rank correlation coefficient (Spearman r). The best-fit line was plotted using simple linear regression analysis. Statistical analyses were performed using GraphPad Prism8 (v.8.3.0).
Extended Data Fig. 7 |. Sequence of wild-type and mutant full-length HA or HA stalk-only constructs.

Amino acid sequences of (a) full-length HA and (b) stalk-only proteins with or without A388V mutation are shown. A388V mutation, HA trimerization domain, and StrepTag II sequence are highlighted. Consensus amino acid sequences are shown in dots.
Supplementary Material
Acknowledgements
This work was supported by the intramural research program of the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH). We thank the Comparative Medicine Branch (NIAID, NIH) for assistance with the animal studies and S. Hunsberger (Biostatistics Research Branch, NIAID, NIH) for the helpful discussion of statistical analyses performed in this study. We also thank Janssen Pharmaceutica, R. Ahmed (Emory Vaccine Center) and A. Townsend (University of Oxford) for generously sharing monoclonal antibodies.
Footnotes
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41591-020-0937-x.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Competing interests
All authors declare no competing financial or non-financial interests.
Additional information
Extended data is available for this paper at https://doi.org/10.1038/s41591-020-0937-x.
Supplementary information is available for this paper at https://doi.org/10.1038/s41591-020-0937-x.
References
- 1.Okuno Y, Isegawa Y, Sasao F & Ueda S A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J. Virol 67, 2552–2558 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krammer F, Pica N, Hai R, Margine I & Palese P Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J. Virol 87, 6542–6550 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Impagliazzo A et al. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science 349, 1301–1306 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Yassine HM et al. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat. Med 21, 1065–1070 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Andrews SF, Graham BS, Mascola JR & McDermott AB Is it possible to develop a “universal” influenza virus vaccine? Immunogenetic considerations underlying B-cell biology in the development of a pan-subtype influenza A vaccine targeting the hemagglutinin stem. Cold Spring Harb. Perspect. Biol 10, a029413 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rambaut A et al. The genomic and epidemiological dynamics of human influenza A virus. Nature 453, 615–619 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie H et al. H3N2 Mismatch of 2014–15 northern hemisphere influenza vaccines and head-to-head comparison between human and ferret antisera derived antigenic maps. Sci. Rep 5, 15279 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bush RM, Fitch WM, Bender CA & Cox NJ Positive selection on the H3 hemagglutinin gene of human influenza virus A. Mol. Biol. Evol 16, 1457–1465 (1999). [DOI] [PubMed] [Google Scholar]
- 9.Altman MO, Angeletti D & Yewdell JW Antibody immunodominance: the key to understanding influenza virus antigenic drift. Viral Immunol 31, 142–149 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taubenberger JK & Morens DM Influenza: the once and future pandemic. Public Health Rep. 125, 16–26 (2010). [PMC free article] [PubMed] [Google Scholar]
- 11.Morens DM & Taubenberger JK Making universal influenza vaccines: lessons from the 1918 pandemic. J. Infect. Dis 219, S5–S13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paules CI & Fauci AS Influenza vaccines: good, but we can do better. J. Infect. Dis 219, S1–S4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park JK, et al. Evaluation of preexisting anti-hemagglutinin stalk antibody as a correlate of protection in a healthy volunteer challenge with influenza A/H1N1pdm Virus. mBio 9, e02284–17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christensen SR, et al. Assessing the protective potential of H1N1 influenza virus hemagglutinin head and stalk antibodies in humans. J Virol 93, e02134–17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ng S et al. Novel correlates of protection against pandemic H1N1 influenza A virus infection. Nat. Med 25, 962–967 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirkpatrick E, Qiu X, Wilson PC, Bahl J & Krammer F The influenza virus hemagglutinin head evolves faster than the stalk domain. Sci. Rep 8, 10432 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu NC & Wilson IA A perspective on the structural and functional constraints for immune evasion: insights from influenza virus. J. Mol. Biol 429, 2694–2709 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andrews SF et al. Immune history profoundly affects broadly protective B cell responses to influenza. Sci. Transl. Med 7, 316ra192 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang R et al. Examining the hemagglutinin subtype diversity among wild duck-origin influenza A viruses using ethanol-fixed cloacal swabs and a novel RT-PCR method. Virology 375, 182–189 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Throsby M et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS ONE 3, e3942 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clementi N et al. A human monoclonal antibody with neutralizing activity against highly divergent influenza subtypes. PLoS ONE 6, e28001 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan GS et al. A pan-H1 anti-hemagglutinin monoclonal antibody with potent broad-spectrum efficacy in vivo. J. Virol 86, 6179–6188 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henry Dunand CJ et al. Preexisting human antibodies neutralize recently emerged H7N9 influenza strains. J. Clin. Invest 125, 1255–1268 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henry Dunand CJ et al. Both neutralizing and non-neutralizing human h7n9 influenza vaccine-induced monoclonal antibodies confer protection. Cell Host Microbe 19, 800–813 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chai N et al. Two escape mechanisms of influenza a virus to a broadly neutralizing stalk-binding antibody. PLoS Pathog. 12, e1005702 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson CS et al. Natural and directed antigenic drift of the H1 influenza virus hemagglutinin stalk domain. Sci. Rep 7, 14614 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Memoli MJ et al. Validation of the wild-type influenza A human challenge model H1N1pdMIST: an A(H1N1)pdm09 dose-finding investigational new drug study. Clin. Infect. Dis 60, 693–702 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Memoli MJ et al. Evaluation of antihemagglutinin and antineuraminidase antibodies as correlates of protection in an Influenza A/H1N1 virus healthy human challenge model. mBio 7, e00417–16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao Y et al. Deep sequencing of 2009 influenza A/H1N1 virus isolated from volunteer human challenge study participants and natural infections. Virology 534, 96–107 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poritz MA et al. FilmArray, an automated nested multiplex PCR system for multi-pathogen detection: development and application to respiratory tract infection. PLoS ONE 6, e26047 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ekiert DC et al. Antibody recognition of a highly conserved influenza virus epitope. Science 324, 246–251 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dreyfus C et al. Highly conserved protective epitopes on influenza B viruses. Science 337, 1343–1348 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Corti D et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333, 850–856 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Wrammert J et al. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J. Exp. Med 208, 181–193 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu Y et al. A potent broad-spectrum protective human monoclonal antibody crosslinking two haemagglutinin monomers of influenza A virus. Nat. Commun 6, 7708 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang KY et al. Focused antibody response to influenza linked to antigenic drift. J. Clin. Invest 125, 2631–2645 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doud MB, Lee JM & Bloom JD How single mutations affect viral escape from broad and narrow antibodies to H1 influenza hemagglutinin. Nat. Commun 9, 1386 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee J et al. Molecular-level analysis of the serum antibody repertoire in young adults before and after seasonal influenza vaccination. Nat. Med 22, 1456–1464 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Linderman SL & Hensley SE Antibodies with ‘Original Antigenic Sin’ properties are valuable components of secondary immune responses to influenza viruses. PLoS Pathog. 12, e1005806 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coughlan L & Palese P Overcoming barriers in the path to a universal influenza virus vaccine. Cell Host Microbe 24, 18–24 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Neumann G et al. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl Acad. Sci. USA 96, 9345–9350 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reed LJ & H. M A simple method of estimating fifty percent endpoints. Am. J. Epidemiol 27, 493–497 (1938). [Google Scholar]
- 43.Krammer F et al. A carboxy-terminal trimerization domain stabilizes conformational epitopes on the stalk domain of soluble recombinant hemagglutinin substrates. PLoS ONE 7, e43603 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stevens J et al. Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science 303, 1866–1870 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Pettersen EF et al. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Xu R et al. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science. 328, 357–360 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.WHO Global Influenza Surveillance Network. Manual for the Laboratory Diagnosis and Virological Surveillance of Influenza. (World Health Organization, 2011). [Google Scholar]
- 48.Shu B et al. Design and performance of the CDC real-time reverse transcriptase PCR swine flu panel for detection of 2009 A (H1N1) pandemic influenza virus. J. Clin. Microbiol 49, 2614–2619 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


