Abstract
The Genera of Fungi series, of which this is the sixth contribution, links type species of fungal genera to their morphology and DNA sequence data. Five genera of microfungi are treated in this study, with new species introduced in Arthrographis, Melnikomyces, and Verruconis. The genus Thysanorea is emended and two new species and nine combinations are proposed. Kramasamuha sibika, the type species of the genus, is provided with DNA sequence data for first time and shown to be a member of Helminthosphaeriaceae (Sordariomycetes). Aureoconidiella is introduced as a new genus representing a new lineage in the Dothideomycetes.
Keywords: DNA barcodes, fungal systematics, ITS, LSU, new taxa
INTRODUCTION
This study focuses on five genera that form part of the Genera of Fungi project (www.generaoffungi.org; Crous et al. 2014a). The overall intention of this project is to revise and update the generic names of fungi, to provide DNA sequence data for them and to restudy or recollect their type species. In this study, we provide DNA sequence data for the unusual and poorly known genus Kramasamuha. Furthermore, the phylogenetic position of the genus Melnikomyces is clarified. We also resolve the taxonomy and phylogeny of Thysanorea and related Minimelanolocus species in the Herpotrichiellaceae. Additional new taxa are introduced based on morphological and DNA sequence data.
MATERIALS AND METHODS
Isolates
Freshly collected leaves and twigs were placed in damp chambers and treated as described by Castañeda-Ruiz et al. (2016). Protocols used for the collection and processing of soil samples are described in Giraldo et al. (2012, 2019) and Groenewald et al. (2018). After 1 wk of incubation on 2 % malt extract agar (MEA) supplemented with penicillin-G and streptomycin, individual colonies were transferred to MEA plates without antibiotics and incubated between 22–24 °C for 7–14 d, in order to obtain axenic cultures.
Colonies were sub-cultured onto 2 % potato dextrose agar (PDA), oatmeal agar (OA), MEA (Crous et al. 2019), autoclaved pine needles on 2 % tap water agar (PNA) (Smith et al. 1996), and incubated at 25 °C under continuous near-ultraviolet light to promote sporulation. Reference strains and specimens are maintained at the Westerdijk Fungal Biodiversity Institute (CBS Culture Collection and herbarium, respectively), Utrecht, The Netherlands or Coleção Octávio de Almeida Drummond (COAD), Viçosa, Brazil.
DNA isolation, amplification and analyses
Genomic DNA was extracted from fungal colonies growing on MEA using the Wizard® Genomic DNA purification kit (Promega, Madison, WI), following the manufacturer’s protocol. The primers V9G (de Hoog & Gerrits van den Ende 1998) or ITS5 (White et al. 1990) and LR5 (Vilgalys & Hester 1990) were used to amplify part of the nuclear rDNA operon (ITS) spanning the 3′ end of the 18S nrRNA gene, the first internal transcribed spacer (ITS1), the 5.8S nrRNA gene, the second ITS region (ITS2) and approximately 900 bp of the 5′ end of the 28S nrRNA gene. The primers ITS4 (White et al. 1990) and LR0R (Vilgalys & Hester 1990) were used as internal sequence primers to ensure high quality sequences over the entire length of the amplicon. Part of the 18S small subunit nrRNA gene (SSU) was amplified and sequenced for selected isolates using NS1 and NS4 (White et al. 1990). Amplification conditions followed those described by Cheewangkoon et al. (2008). Part of the actin gene (act) was amplified and sequenced for selected isolates using the primer set Act1/Act4 (Voigt & Wöstemeyer 2000). The software SeqMan Pro v. 13.0.0 (DNASTAR, Madison, WI) and Geneious v. 11.0.4 (Kearse et al. 2012; https://www.geneious.com) were used to obtain consensus sequences of each isolate. Blast searches using ITS and LSU sequences were performed for each isolate and the closest matches were retrieved from GenBank and included in the phylogenetic analyses. Multiple sequence alignments for individual genes were generated using the online version of MAFFT (http://mafft.cbrc.jp/alignment/software/). Subsequent phylogenetic analyses from individual and combined datasets were conducted using Maximum-likelihood (ML) performed on the CIPRES Science Gateway portal (Miller et al. 2012) using RAxML v. 8.2.10 (Stamatakis 2014). The default parameters were used, and bootstrap support (BS) was obtained using the rapid bootstrapping algorithm with the automatic halt option. A BS ≥ 95 % was considered as statistically significant. Sequence data were deposited in the GenBank/ENA public databases (Table 1) and the alignments and trees in TreeBASE (http://www.treebase.org).
Table 1.
GenBank accession numbers of taxa included in this study.
| Taxa | Isolates1 | Country | Substrate |
GenBank accession numbers2 |
References | ||
|---|---|---|---|---|---|---|---|
| ITS | LSU | act/SSU | |||||
| Dothideomycetes | |||||||
| Aliquandostipite khaoyaiensis | CBS 118232 | Thailand | Twig | – | GU301796.1 | – | Schoch et al. (2009) |
| SS3028 | Thailand | – | EF175649.1 | – | Campbell et al. (2007) | ||
| Alternaria tenuissima | CBS 918.96 | UK | Dianthus chinensis | – | KC584311.1 | – | Woudenberg et al. (2013) |
| Alysidiella parasitica | CBS 120088 | South Africa | Leaves Eucalyptus sp. | – | DQ923525.1 | – | Summerell et al. (2006) |
| Apiosporina collinsii | CBS 118973 | Canada | Amelanchier alnifolia | – | GU301798.1 | – | Schoch et al. (2009) |
| Arthrographis arxii | CBS 203.78 | India | Dung of herbivore | GQ272638.1 | AB213426.1 | HG316563.1/- | Murata et al. (2005), Kang et al. (2010), Giraldo et al. (2014a) |
| Arthrographis chlamydospora | CBS 135936 | USA | Human urine | HG004554.1 | HG004543.1 | HG316560.1/- | Giraldo et al. (2014a) |
| Arthrographis curvata | CBS 135933 | USA | Human nails | HG004557.1 | HG004539.1 | HG316557.1/- | Giraldo et al. (2014a) |
| CBS 135934 | Spain | River sediment | HG004556.1 | HG004542.1 | HG316558.1/- | Giraldo et al. (2014a) | |
| Arthrographis globosa | UTHSC 11-757 | USA | Bronchial wash | HG004553.1 | HG004541.1 | HG316561.1/- | Giraldo et al. (2014a) |
| Arthrographis grakistii | JW 22011 = CBS 145529 | The Netherlands | Soil | MN794359 | MN794336 | MN816497 | This study |
| JW 22015 | The Netherlands | Soil | MN794360 | MN794337 | MN816498 | This study | |
| JW 22019 | The Netherlands | Soil | MN794361 | MN794338 | MN816499 | This study | |
| JW 49011 | The Netherlands | Soil | MN794362 | MN794339 | MN816500 | This study | |
| JW 49012 | The Netherlands | Soil | MN794363 | MN794340 | MN816501 | This study | |
| JW 180011 | The Netherlands | Soil | MN794364 | MN794341 | MN816502 | This study | |
| JW 190014= CBS 145530 | The Netherlands | Soil | MN794365 | MN794342 | MN816503 | This study | |
| JW 199018 | The Netherlands | Soil | MN794366 | MN794343 | MN816504 | This study | |
| JW 209002 | The Netherlands | Soil | MN794367 | MN794344 | MN816505 | This study | |
| JW 209003 | The Netherlands | Soil | MN794368 | MN794345 | MN816506 | This study | |
| Arthrographis kalrae | CBS 693.77 | India | Sputum | AB116536.1 | AB116544.1 | HG316544.1/- | Xi et al. (2004), Giraldo et al. (2014a) |
| JW 21004 | The Netherlands | Soil | MN794369 | MN794346 | MN816507 | This study | |
| JW 21008 = CBS 145527 | The Netherlands | Soil | MN794370 | MN794347 | MN816508 | This study | |
| JW 21029 | The Netherlands | Soil | MN794371 | MN794348 | MN816509 | This study | |
| Arthrographis longispora | CBS 135935 | USA | Human foot | HG004555.1 | HG004540.1 | HG316559.1/- | Giraldo et al. (2014a) |
| JW 22007 = CBS 145528 | The Netherlands | Soil | MN794372 | MN794349 | MN816510 | This study | |
| Asterina chrysophylli | VIC 42823 | Brazil | Leaves Henriettea succosa | – | KP143738.1 | – | Guatimosim et al. (2015) |
| Asterina melastomatis | VIC 42822 | Brazil | Leaves Miconia sp. | – | NG_057055.1 | – | Guatimosim et al. (2015) |
| Asterotexis cucurbitacearum | PMA M141224 | Panama | Sechium edule | – | HQ610510.1 | – | Unpublished |
| VIC 24814 | Brazil | Leaves Cucurbita pepo | – | NG_057054.1 | – | Guatimosim et al. (2015) | |
| Aulographina eucalypti | CPC 12986 | Australia | Eucalyptus cloeziana | – | HM535600.1 | – | Cheewangkoon et al. (2012) |
| Aureoconidiella foliicola | CBS 145943 | South Africa | Leaves Syzygium cordatum | MN794373 | MN794350 | – | This study |
| Bezerromyces brasiliensis | URM7411 | Brazil | Tacinga inamoena | – | KX518623.1 | – | Bezerra et al. (2017) |
| Bezerromyces pernambucoensis | URM7412 | Brazil | Tacinga inamoena | – | KX518624.1 | – | Bezerra et al. (2017) |
| Blastacervulus eucalypti | CBS 124759 | Australia | Eucalyptus robertsonii subsp. hemisphaerica | – | GQ303302.1 | – | Cheewangkoon et al. (2009) |
| Blastacervulus robbenensis | CBS 124780 | Cyprus | Eucalyptus sp. | – | HM628777.1 | – | Cheewangkoon et al. (2012) |
| Botryosphaeria dothidea | CBS 115476 | Switzerland | Prunus sp. | – | DQ377852.1 | – | Crous et al. (2006b) |
| Brachiosphaera tropicalis | E192 | – | – | – | EF175653.1 | – | Campbell et al. (2007) |
| Byssosphaeria jamaicana | SMH 1403 | – | – | – | GU385152.1 | – | Mugambi & Huhndorf (2009a) |
| Byssosphaeria salebrosa | SMH 2387 | – | – | – | GU385162.1 | – | Mugambi & Huhndorf (2009a) |
| Capnodium coffeae | CBS 147.52 | Zaire | Berry Coffea robusta | – | MH868489.1 | – | Vu et al. (2019) |
| Cladoriella eucalypti | CBS 115899 | South Africa | Leaves Eucalyptus | – | EU040224.1 | – | Crous et al. (2007b) |
| Cladoriella kinglakensis | CPC 32730 | Australia | Leaves Eucalyptus | – | MG386126.1 | – | Crous et al. (2017) |
| Cladoriella paleospora | CBS 124761 | Australia | Leaves Eucalyptus | – | GQ303303.1 | – | Cheewangkoon et al. (2009) |
| Cladoriella rubrigena | CBS 124760 | Australia | Leaves Eucalyptus | – | MH874921.1 | – | Vu et al. (2019) |
| Cladoriella xanthorrhoeae | CBS 143398 | Australia | Leaves Xanthorrhoea sp. | – | NG_059054.1 | – | Crous et al. (2017) |
| Cladosporium halotolerans | CBS 127371 | Cuba | Human | – | MH875988.1 | – | Vu et al. (2019) |
| Cladosporium variabile | CBS 121636 | USA | Spinacia oleracea | – | MH874684.1 | – | Vu et al. (2019) |
| Clavatispora thailandica | MFLUCC 17-2237 | Thailand | Hevea brasiliensi | – | MH062960.1 | – | Unpublished |
| MFLUCC 10-0107 | Thailand | Dead stems | – | NG_058863.1 | – | Boonmee et al. (2014) | |
| Dibotryon morbosum | N/A | USA | Prunus sp. | – | EF114694.1 | – | Winton et al. (2007) |
| Diplodia mutila | CBS 431.82 | The Netherlands | Dead branches Fraxinus excelsior | – | DQ377863.1 | – | Crous et al. (2006b) |
| Dissoconium aciculare | CBS 204.89 | Germany | Astragalus | – | GU214419.1 | – | Crous et al. (2009a) |
| Eremomyces bilateralis | CBS 781.70 | USA | Dung of pack rat | HG004552.1 | HG004545.1 | HG316562.1/- | Giraldo et al. (2014a) |
| Fusicladium pini | CBS 463.82 | The Netherlands | Needle Pinus sylvestris | – | EU035436.1 | – | Crous et al. (2007c) |
| Fusicladium ramoconidii | CBS 462.82 | The Netherlands | Needle Pinus sp. | – | EU035439.1 | – | Crous et al. (2007c) |
| Gibbera conferta | CBS 191.53 | Switzerland | Vaccinium uliginosum | – | GU301814.1 | – | Schoch et al. (2009) |
| Gloniopsis arciformis | GKM L166A | – | – | – | GU323211.1 | – | Schoch et al. (2009) |
| Glonium circumserpens | CBS 123342 | Tasmania | Wood | – | FJ161208.1 | – | Boehm et al. (2009) |
| Glonium circumserpens | CBS 123343 | Tasmania | Saxicolous on limestone | – | FJ161200.1 | – | Boehm et al. (2009) |
| Helicomyces roseus | CBS 283.51 | Switzerland | Dead bark | – | AY856881.1 | – | Tsui et al. (2006) |
| Herpotrichia juniperi | AFTOL-ID 1608 | Switzerland | Juniperus nana | – | DQ678080.1 | – | Schoch et al. (2009) |
| Heteroconium eucalypti | CBS 120122 | Uruguay | Leaves Eucalyptus dunnii | – | DQ885893.1 | – | Crous et al. (2006a) |
| Hysterium angustatum | CBS 123334 | USA | Bark Pinus rigida | – | FJ161207.1 | – | Boehm et al. (2009) |
| Hysterium pulicare | ANM1455 | USA | – | – | GQ221904.1 | – | Mugambi & Huhndorf (2009b) |
| Hysteropatella clavispora | CBS 247.34 | USA | Salix sp. | – | AY541483.1 | – | Lumbsch et al. (2005) |
| Hysteropatella prostii | H.B. 9934b | Germany | Malus domestica | – | KT876980.1 | – | Unpublished |
| Jahnula appendiculata | SS2900 | Thailand | – | – | EF175654.1 | – | Campbell et al. (2007) |
| Jahnula aquatica | R68-1 | USA | – | – | EF175655.1 | – | Campbell et al. (2007) |
| Jahnula bipileata | F49-1 | USA | – | – | EF175657.1 | – | Campbell et al. (2007) |
| Jahnula seychellensis | SS2113.1 | Thailand | – | – | EF175665.1 | – | Campbell et al. (2007) |
| Leptoxyphium fumago | CBS 123.26 | Indonesia | Hibiscus tiliaceus | – | GU214430.1 | – | Crous et al. (2009a) |
| Macrophomina phaseolina | CBS 227.33 | – | Zea mays | – | DQ377906.1 | – | Crous et al. (2006b) |
| Melanomma pulvis-pyrius | CBS 124080 | France | Bark Salix caprea | – | GU456323.1 | – | Zhang et al. (2009a) |
| Melnikomyces thailandicus | CBS 145767 | Thailand | Soil | MN794374 | MN794351 | – | This study |
| Melnikomyces vietnamensis | CBS 136209 | Vietnam | Leaves | – | NG_058087.1 | – | Crous et al. (2014b) |
| Mycosphaerella punctiformis | CBS 113265 | The Netherlands | Dead leaves Quercus robur | – | DQ470968.1 | – | Spatafora et al. (2006) |
| Neocoleroa metrosideri | PDD107531 | New Zealand | Metrosideros excelsa | – | NG_059638.1 | – | Johnston & Park (2016) |
| Neofusicoccum mangiferae | CBS 118532 | Australia | Mangifera indica | – | NG_055730.1 | – | Crous et al. (2006b) |
| Neofusicoccum nonquaesitum | CBS 126655 | USA | Umbellularia californica | – | NG_058258.1 | – | Yang et al. (2017) |
| Ochroconis constricta | CBS 202.27 | USA | Soil | – | KF156147.1 | – | Samerpitak et al. (2014) |
| Ochroconis gamsii | CBS 239.78 | Sri Lanka | Leaf Caryota plumosa | – | NG057992.1 | – | Samerpitak et al. (2014) |
| Patellaria cf. atrata | BCC 28876 | Thailand | – | – | GU371828.1 | – | Schoch et al. (2009) |
| BCC 28877 | Thailand | – | – | GU371829.1 | – | Schoch et al. (2009) | |
| Phaeocryptopus gaeumannii | CBS 267.37 | Germany | Pseudotsuga menziesii | – | EF114698.1 | – | Winton et al. (2007) |
| Phaeotrichum benjaminii | CBS 541.72 | – | Dung of rodent | – | AY779311.1 | – | Lumbsch et al. (2005) |
| Phoma herbarum | CBS 567.63 | USA | Fruit Malus sylvestris | – | MH869982.1 | – | Vu et al. (2019) |
| Pirozynskiella laurisilvatica | CBS 138109 | Spain | Leaves Laurus sp. | – | NG_058462.1 | – | Hernández-Restrepo et al. (2017) |
| Psiloglonium simulans | CBS 206.34 | USA | Tilia sp. | – | FJ161178.1 | – | Boehm et al. (2009) |
| Rhexothecium globosum | CBS 955.73 | Egypt | Desert soil | MH860827.1 | HG004544.1 | – | Giraldo et al. (2014a), Vu et al. (2019) |
| Rhytidhysteron rufulum | CBS 306.38 | – | Pistacia chinensis | – | FJ469672.1 | – | Schoch et al. (2009) |
| Schizothyrium pomi | CBS 228.57 | Italy | – | – | EF134947.1 | – | Batzer et al. (2008) |
| CBS 486.50 | The Netherlands | Polygonum sachalinense | – | EF134948.1 | – | Batzer et al. (2008) | |
| Scolecobasidiella avellanea | CBS 772.73 | Somalia | Soil | – | EF204505.1 | – | Unpublished |
| Stemphylium herbarum | CBS 191.86 | India | Leaf Medicago sativa | – | JX681120.1 | – | Verkley et al. (2014) |
| Sympoventuria capensis | CBS 120136 | South Africa | Leaf litter Eucalyptus sp. | – | NG_057984.1 | – | Samerpitak et al. (2014) |
| Teratosphaeria destructans | CBS 111369 | Indonesia | Eucalyptus grandis | – | EU019287.2 | – | Crous et al. (2007a) |
| Teratosphaeria fibrillosa | CBS 121707 | South Africa | Leaves Protea sp. | – | KF902075.1 | – | Quaedvlieg et al. (2014) |
| Teratosphaeria stellenboschiana | CBS 116428 | South Africa | Leaf litter Eucalyptus sp. | – | EU019295.1 | – | Crous et al. (2007a) |
| Trichodelitschia bisporula | CBS 262.69 | The Netherlands | Dung of Rabbit | – | GU348996.2 | – | Schoch et al. (2009) |
| Tubeufia paludosa | CBS 245.49 | The Netherlands | Corylus avellana | – | MH856510.1 | – | Vu et al. (2019) |
| Uwebraunia commune | CBS 110747 | South Africa | Eucalyptus nitens | – | GQ852589.1 | – | Crous et al. (2009b) |
| Uwebraunia dekkeri | CBS 111282 | Zambia | Eucalyptus globulus | – | GU214425.1 | – | Crous et al. (2009b) |
| Venturia inaequalis | CBS 176.42 | France | – | – | GU348998.1 | – | Schoch et al. (2009) |
| Venturia populina | CBS 256.38 | Italy | Populus canadensis | – | GU323212.1 | – | Schoch et al. (2009) |
| Verruconis calidifluminalis | CBS 125818 | Japan | Hot spring effluent | – | NG_057985.1 | – | Samerpitak et al. (2014) |
| Verruconis gallopava | CBS 547.81 | New Zealand | – | – | KF156109.1 | – | Samerpitak et al. (2014) |
| CBS 437.64 | USA | Brain abscess Meleagris gallopavo | – | NG_58016.1 | – | Machouart et al. (2014) | |
| Verruconis thailandica | CBS 145768 | Thailand | Soil | MN794375 | MN794352 | – | This study |
| Verruconis verruculosa | CBS 119775 | Malaysia | Root Hevea species | – | KF282668.1 | – | Machouart et al. (2014) |
| Xiliomyces brasiliensis | URM7413 | Brazil | Tacinga inamoena | – | KX518625.1 | – | Bezerra et al. (2017) |
| Zasmidium cellare | CBS 146.36 | – | Wall in wine cellar | – | EU041878.1 | – | Arzanlou et al. (2007) |
| Eurotiomycetes | |||||||
| Aculeata aquatica | MFLUCC 11-0529 | Thailand | Submerged wood | MG922575.1 | MG922579.1 | –/MG922571.1 | Dong et al. (2018) |
| Capronia pilosella | AFTOL-ID 657 | – | – | DQ823099.1 | DQ823106.1 | –/DQ826737.1 | James et al. (2006) |
| Cladophialophora carrionii | CBS 160.54 | Australia | Man | FJ358234.1 | FJ358302.1 | –/AF050262.1 | Gueidan et al. (2008), Untereiner & Naveau (1999) |
| Cladophialophora minourae | CBS 556.83 | Japan | Decaying wood | FJ358235.1 | FJ358303.1 | –/AY251087.1 | Braun et al. (2003), Gueidan et al. (2008) |
| Cladophialophora parmeliae | CBS 129337 | Portugal | – | JQ342182.1 | – | –/JQ342180.1 | Diederich et al. (2013) |
| Cladophialophora subtilis | CBS 122642 | The Netherlands | Ice tea | NG_058961.1 | KX822283.1 | –/NR_111363.1 | Badali et al. (2008), Vasse et al. (2017) |
| Cyphellophora oxyspora | CBS 698.73 | Sri Lanka | – | KC455262.1 | KC455305.1 | –/KC455249.1 | Réblová et al. (2013) |
| Cyphellophora sessilis | CBS 243.85 | The Netherlands | Resin Picea abies | EU514700.1 | KC455308.1 | –/EU514700.1 | Untereiner et al. (2008), Réblová et al. (2013) |
| Exophiala jeanselmei | CBS 507.90 | Uruguay | Man | FJ358242.1 | FJ358310.1 | –/NR_111129.1 | Gueidan et al. (2008) |
| Exophiala nigra | dH 12,296 | – | – | FJ358244.1 | FJ358312.1 | – | Gueidan et al. (2008) |
| Exophiala pisciphila | CBS 537.73 | USA | Ictalurus punctatus | MH872483.1 | JN856018.1 | –/AF050272.1 | de Hoog et al. (2011), Untereiner & Naveau (1999), Vu et al. (2019) |
| AFTOL-ID 669 | – | – | DQ823101.1 | DQ823108.1 | –/DQ826739.1 | Gueidan et al. (2008) | |
| Exophiala salmonis | AFTOL-ID 671 | – | – | EF413609.1 | EF413608.1 | – | Geiser et al. (2006) |
| CBS 157.67 | Canada | Salmo clarkii | MH870616.1 | JN856020.1 | –/NR_121270.1 | de Hoog et al. (2011), Schoch et al. (2014), Vu et al. (2019) | |
| Exophiala xenobiotica | CBS 115831 | Germany | Browncol | FJ358246.1 | FJ358314.1 | –/AY857539.1 | Gueidan et al. (2008) |
| Fonsecaea monophora | CBS 102243 | FJ358247.1 | FJ358315.1 | –/EU938579.1 | Gueidan et al. (2008) | ||
| Melanoctona tectonae | MFLUCC 12-0389 | Thailand | Tectona grandis | KX258779.1 | KX258780.1 | –/KX258778.1 | Unpublished |
| Phialophora americana | AFTOL-ID 658 | – | – | FJ358226.1 | FJ358294.1 | – | Gueidan et al. (2008) |
| Phialophora verrucosa | AFTOL-ID 670 | – | – | EF413615.1 | EF413614.1 | – | Geiser et al. (2006) |
| Rhinocladiella anceps | AFTOL-ID 659 | – | – | DQ823102.1 | DQ823109.1 | –/DQ826740.1 | James et al. (2006) |
| Rhinocladiella anceps | CBS 181.65 | Canada | Soil | EU041862.1 | AY554292.1 | –/MH858534.1 | Arzanlou et al. (2007) |
| Thysanorea asiatica | MFLUCC 15-0237 | China | Submerged wood | KR215610.1 | KR215615.1 | –/KR215604.1 | Liu et al. (2015) |
| Thysanorea cantrelliae | CBS 145909 | USA | Unidentified twig | MN794376 | MN794353 | –/MN794382 | This study |
| Thysanorea curvata | MFLUCC 15-0259 | China | Submerged wood | KR215609.1 | KR215614.1 | –/KR215605.1 | Liu et al. (2015) |
| Thysanorea lotorum | CBS 235.78 | USA | Root Lotus corniculatus | MH872892.1 | – | –/MH861130.1 | Vu et al. (2019) |
| KUMCC 15-0206 | China | Submerged wood | KX789215.1 | – | –/KX789212.1 | Liu et al. (2015) | |
| Thysanorea melanica | MFLUCC 15-0415 | China | Submerged wood | KR215613.1 | KR215618.1 | –/KR215608.1 | Liu et al. (2015) |
| Thysanorea nonramosa | MFLUCC 17-2378 | Thailand | Wood | MH532970.1 | – | –/MH532971.1 | Wang et al. (2019) |
| Thysanorea obscura | MFLUCC 15-0416 | China | Submerged wood | KR215611.1 | KR215616.1 | –/KR215606.1 | Liu et al. (2015) |
| Thysanorea papuana | CBS 212.96 | Papua New Guinea | – | EU041871.1 | – | –/EU041814.1 | Arzanlou et al. (2007) |
| MFLUCC 15-0966 | Thailand | Submerged wood | MG922576.1 | MG922580.1 | –/MG922572.1 | Dong et al. (2018) | |
| Thysanorea rousseliana | CBS 126086 | Spain | Dead branches Quercus ilex | MH875246.1 | – | –/MH863784.1 | Vu et al. (2019) |
| Thysanorea seifertii | CBS 145910 | USA | Unidentified twig | MN794377 | MN794354 | –/MN794383 | This study |
| Thysanorea thailandensis | MFLUCC 15-0971 | Thailand | Submerged wood | MG922577.1 | MG922581.1 | –/MG922573.1 | Dong et al. (2018) |
| Thysanorea yunnanense | MFLUCC 15-0414 | Thailand | Submerged wood | KR215612.1 | KR215617.1 | –/KR215607.1 | Liu et al. (2015) |
| Veronaea botryosa | CBS 254.57 | Italy | Sansa olive | EU041873.1 | JN856021.1 | –/EU041816.1 | Arzanlou et al. (2007) |
| MFLUCC 11-0072 | Thailand | Submerged wood | MG922574.1 | MG922578.1 | –/MG922570.1 | Dong et al. (2018) | |
| Veronaea compacta | CBS 268.75 | South Africa | – | EU041876.1 | – | –/EU041819.1 | Arzanlou et al. (2007) |
| Veronaea japonica | CBS 776.83 | Japan | Dead bamboo culm | EU041875.1 | – | –/EU041818.1 | Arzanlou et al. (2007) |
| Sordariomycetes | |||||||
| Anthostomella sp. | SMH3101 | USA | – | – | AY780050.1 | – | Miller & Huhndorf (2005) |
| Camarops tubulina | SMH4614 | Denmark | – | – | AY346266.1 | – | Huhndorf et al. (2004) |
| Camarops ustulinoides | SMH1988 | USA | – | – | AY346267.1 | – | Huhndorf et al. (2004) |
| Chaetosphaeria ovoidea | SMH2605 | USA | – | – | AF064641.1 | – | Fernandez et al. (1999) |
| Coniochaeta discoidea | SANK 12878 | – | – | – | AY346297.1 | – | Huhndorf et al. (2004) |
| Coniochaetidium savoryi | TRTC 51980 | – | – | – | AY346276.1 | – | Huhndorf et al. (2004) |
| Cytospora ceratosperma | AR3426 | Austria | Quercus robur | – | AF408387.1 | – | Castlebury et al. (2002) |
| Diaporthe phaseolorum | FAU458 | USA | – | – | AY346279.1 | – | Huhndorf et al. (2004) |
| Echinosphaeria canescens | JHC97-006 | – | – | – | KF765604.1 | – | Miller et al. (2014) |
| SMH4666 | – | – | – | KF765605.1 | – | Miller et al. (2014) | |
| SMH4791 | – | – | – | AY436403.1 | – | Miller & Huhndorf (2004) | |
| TL5730 | – | – | – | AY436404.1 | – | Miller & Huhndorf (2004) | |
| Eutypa sp. | SMH3580 | Panama | Branch | – | AY346280.1 | – | Huhndorf et al. (2004) |
| Fusarium ambrosium | SMH1999 | – | – | – | AY780077.1 | – | Miller & Huhndorf (2005) |
| Helminthosphaeria carpathica | SMH3903 | – | – | – | KF765606.1 | – | Miller et al. (2014) |
| Helminthosphaeria cf. stuppea | JF04120 | – | – | – | KF765611.1 | – | Miller et al. (2014) |
| TL11998 | – | – | – | KF765612.1 | – | Miller et al. (2014) | |
| Helminthosphaeria clavariarum | SMH4609 | Denmark | Clavulina cristata | – | AY346283.1 | – | Huhndorf et al. (2004) |
| Helminthosphaeria corticiorum | JF04225 | – | – | – | KF765607.1 | – | Miller et al. (2014) |
| Helminthosphaeria odontiae | ANM928 | – | – | – | KF765610.1 | – | Miller et al. (2014) |
| Helminthosphaeria tomaculum | SMH2485 | – | – | – | KF765613.1 | – | Miller et al. (2014) |
| Helminthosphaeria triseptata | JF04015 | – | – | – | KF765614.1 | – | Miller et al. (2014) |
| Hilberina caudata | SMH1542 | – | – | – | KF765615.1 | – | Miller et al. (2014) |
| Hilberina munkii | SMH1531 | – | – | – | KF765616.1 | – | Miller et al. (2014) |
| Kramasamuha sibiki | CPC 35619 = CBS 146338 | Australia | Leaves Lophostemon confertus | MN794378 | MN794355 | – | This study |
| CPC 36725 = CBS 146339 | Malaysia | Needles Pinus tecunumanii | MN794379 | MN794356 | – | This study | |
| CBS 146133 = CPC 36153 | South Africa | Leaves Syzygium cordatum | MN794380 | MN794357 | – | This study | |
| COAD 2632 | Brazil | Leaves Hypericum innodorum | MN794381 | MN794358 | – | This study | |
| Lasiosphaeria ovina | SMH1538 | – | – | – | AF064643.1 | – | Fernandez et al. (1999) |
| Neurospora crassa | MUCL 19026 | – | – | – | AF286411.1 | – | Untereiner et al. (2001) |
| Ruzenia spermoides | ANM163 | – | – | – | KF765618.1 | – | Miller et al. (2014) |
| SMH4606 | – | – | – | AY436422.1 | – | Miller & Huhndorf (2004) | |
| SMH4655 | – | – | – | KF765619.1 | – | Miller et al. (2014) | |
| Sporoschisma hemipsila | SMH2125 | – | – | – | AY346292.1 | – | Huhndorf et al. (2004) |
| Synaptospora plumbea | ANM963 | – | – | – | KF765620.1 | – | Miller et al. (2014) |
| SMH3962 | – | – | – | KF765621.1 | – | Miller et al. (2014) | |
| Valsonectria pulchella | SMH1193 | – | – | – | AY346304.1 | – | Huhndorf et al. (2004) |
1 BCC: BIOTEC Culture Collection, National Center for Genetic Engineering and Biotechnology (BIOTEC), Bangkok, Thailand; CBS: Culture Collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands; COAD: Coleção Octávio de Almeida Drummond, Viçosa, Brazil; CPC: Culture Collection of Pedro Crous, Utrecht, The Netherlands; JW: Johanna Westerdijk Culture Collection, Utrecht, The Netherlands; MFLUCC: Mae Fah Luang University Culture Collection, Chiang Rai, Thailand; UTHSC: Fungus Testing Laboratory of the University of Texas Health Science Center at San Antonio, USA. For other acronyms see references.
2 LSU: Large subunit of the nrDNA; SSU: Small subunit of the nrDNA; ITS: internal transcribed spacer regions of the nrDNA and intervening 5.8S nrDNA; act: partial actin gene. Accession numbers of sequences newly generated in this study are indicated in bold.
Morphology
Slide preparations were mounted in lactic acid or water from colonies sporulating on the media previously mentioned. Observations were made with a Nikon SMZ1500 dissecting microscope and with a Nikon Eclipse Ni compound microscope using a DSRi2 digital camera (Nikon, Tokyo, Japan) and NIS-Elements imaging software v. 4.3. Colony characters and pigment production were noted after 1–2 wk of growth on MEA, PDA and OA incubated at 25 ºC under natural light. Colony colours (surface and reverse) were determined using the colour charts of Rayner (1970). Taxonomic novelties were deposited in MycoBank (www.MycoBank.org; Crous et al. 2004).
RESULTS
Phylogeny
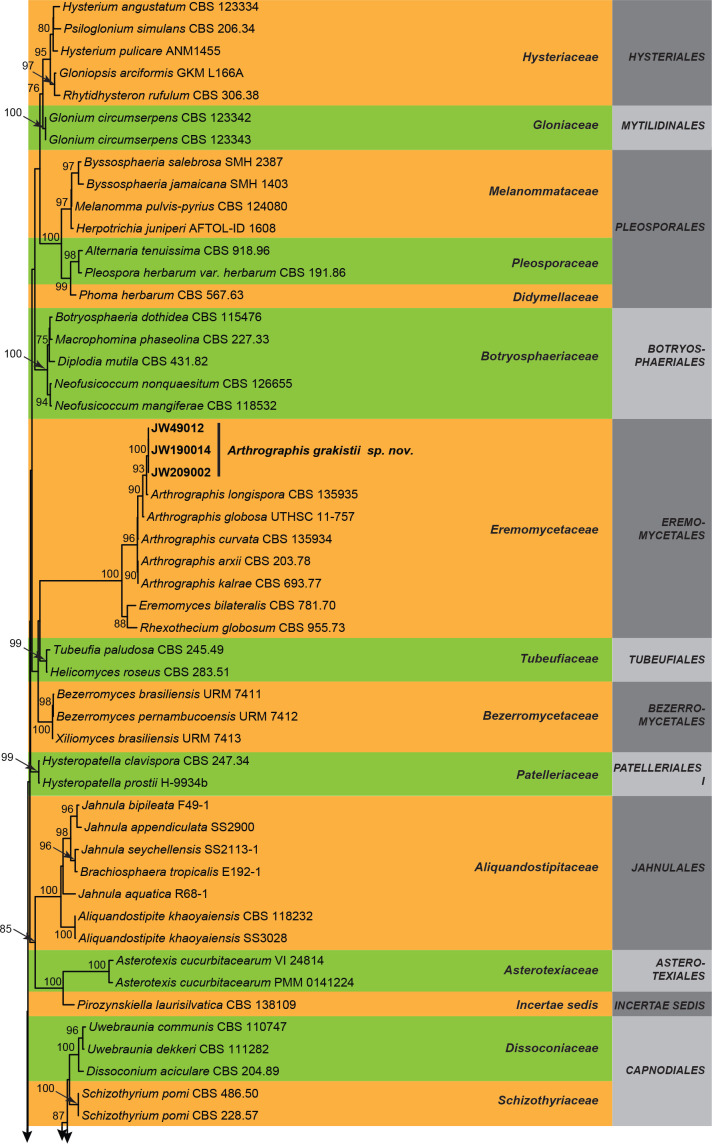
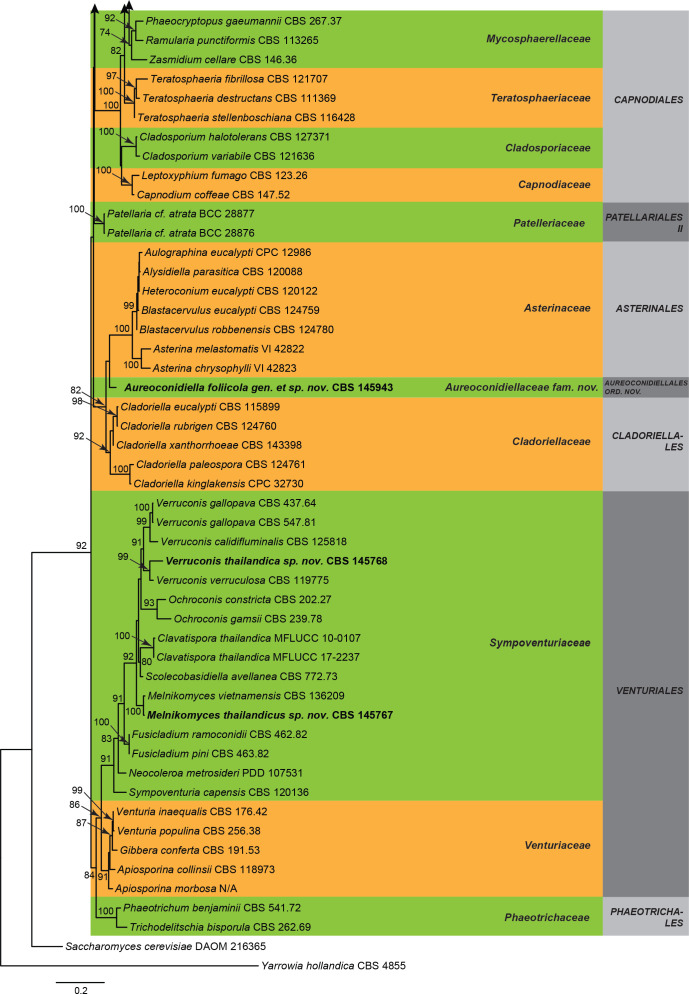
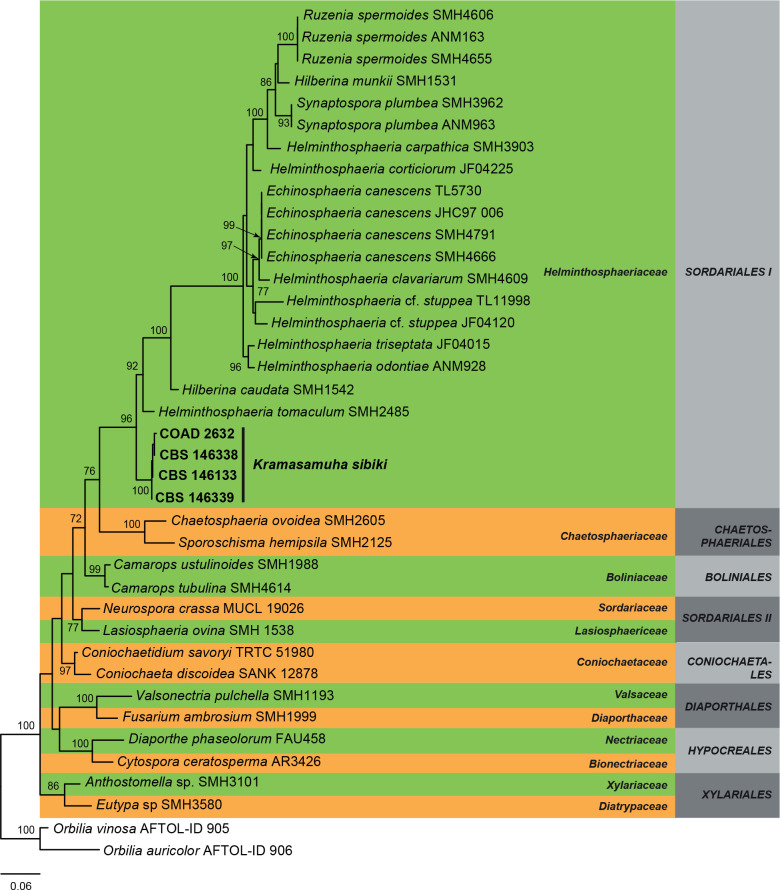
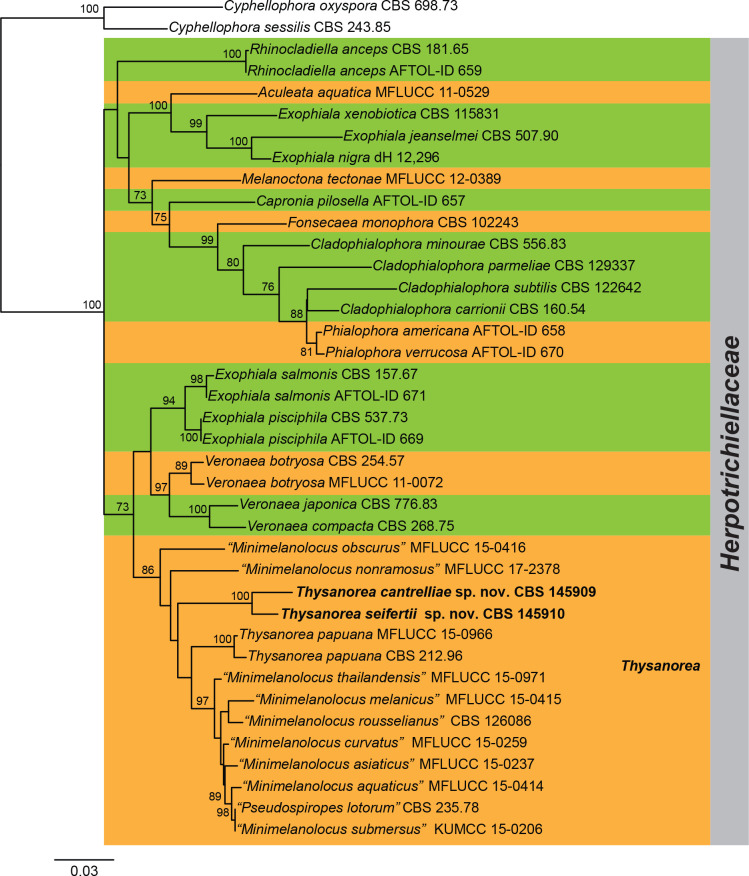
Three overview phylogenies were generated in this study. The first two of these were based on a partial alignment of LSU to provide the phylogenetic position of the treated genera and species within the Dothideomycetes (Fig. 1) and Sordariomycetes (Fig. 2). A third analysis was implemented for selected Herpotrichiellaceae genera based on a concatenated ITS/LSU/SSU alignment (Fig. 3). Other phylogenetic trees specific to the treated species are discussed in the notes for those taxa.
Fig. 1.
Maximum composite likelihood tree obtained from the RAxML analysis of the LSU sequence alignment of selected Dothideomycetes. Bootstrap support values above 70 % are shown at the nodes. Families and orders are indicated with coloured blocks to the right of the tree. Taxonomic novelties described in this study are indicated in boldface. The tree was rooted to Yarrowia hollandica (CBS 4855).
Fig. 2.
Maximum composite likelihood tree obtained from the RAxML analysis of the LSU sequence alignment of selected Sordariomycetes. Bootstrap support values above 70 % are shown at the nodes. Families and orders are indicated with coloured blocks to the right of the tree. Included strains described in this study are indicated in boldface. The tree was rooted to Orbilia vinosa (AFTOL-ID 905) and Orbilia auricolor (AFTOL-ID 906).
Fig. 3.
Maximum composite likelihood tree obtained from the RAxML analysis of the ITS/LSU/SSU sequence alignment of selected Herpotrichiellaceae. Bootstrap support values above 70 % are shown at the nodes. Taxonomic novelties described in this study are indicated in boldface. The tree was rooted to Cyphellophora oxyspora (CBS 698.73) and Cyphellophora sessilis (CBS 243.85).
The BLAST search results using the LSU and ITS sequences for the isolates CBS 145943, CBS 145767, CBS 145768, and JW showed that they were related to members of Cladoriellales and Asterinales, Venturiales and Eremomycetales in the Dothideomycetes. Consequently, the currently accepted taxa in those orders and other orders in the Dothideomycetes were included in our analyses (Fig. 1).
The overview phylogeny of Dothideomycetes (Fig. 1) revealed that in the Arthrographis subclade (96 % BS), the strains JW 49012, JW 190014, and JW 209002 grouped together in a separate clade representing a putative new species that is described below.
The genus Aureoconidiella is introduced to accommodate CBS 145943 that formed a lineage distinct from other genera, families and orders included in the analysis. A new family and order are introduced for this genus. Furthermore, the isolates CBS 145767 and CBS 145768 nested in the Sympoventuriaceae clade (91 % BS) within the Venturiales. They were closely related but different to Melnikomyces vietnamensis and Verruconis verruculosa, respectively. These isolates are consequently considered to represent putative new species in the genera Melnikomyces and Verruconis that are introduced below.
The overview phylogeny of Sordariomycetes (Fig. 2) revealed that four isolates, CBS 146133, CBS 146338, CBS 146339, and COAD 2632 of Kramasamuha sibika grouped together in a fully-supported terminal clade (100 % BS), related to Helminthosphaeriaceae (97 % BS).
The combined analysis of the ITS/LSU/SSU (Fig. 3) revealed that Minimelanolocus and Thysanorea cluster together in the same clade (86 % BS). Several species treated so far as belonging to Minimelanolocus, i.e. M. aquaticus, M. asiaticus, M. curvatus, M. melanicus, M. obscurus, M. rosselianus, M. submersus, and M. thailandensis, proved to be congeneric with Thysanorea and therefore new combinations are proposed to accommodate them. In addition, CBS 145909 and CBS 145910 formed a separate subclade (100 % BS) within Thysanorea, representing two putative new species.
Taxonomy
Aureoconidiellales Hern.-Restr. & Crous, ord. nov. MycoBank MB833918.
Description: See description of Aureoconidiella.
Type family: Aureoconidiellaceae Hern.-Restr. & Crous
Aureoconidiellaceae Hern.-Restr. & Crous, fam. nov. MycoBank MB833917.
Description: See description of Aureoconidiella.
Type genus: Aureoconidiella Hern.-Restr. & Crous
Aureoconidiella Hern.-Restr. & Crous, gen. nov. MycoBank MB833915.
Etymology: Name refers to the golden brown colour of its conidia.
Conidiophores macronematous, simple, septate, brown. Conidiogenous cells integrated, terminal, polyblastic, with thickened scars, brown to pale brown. Conidia globose to subglobose with apiculate base, initially subhyaline, golden brown at maturity, verrucose.
Type species: Aureoconidiella foliicola Hern.-Restr. & Crous
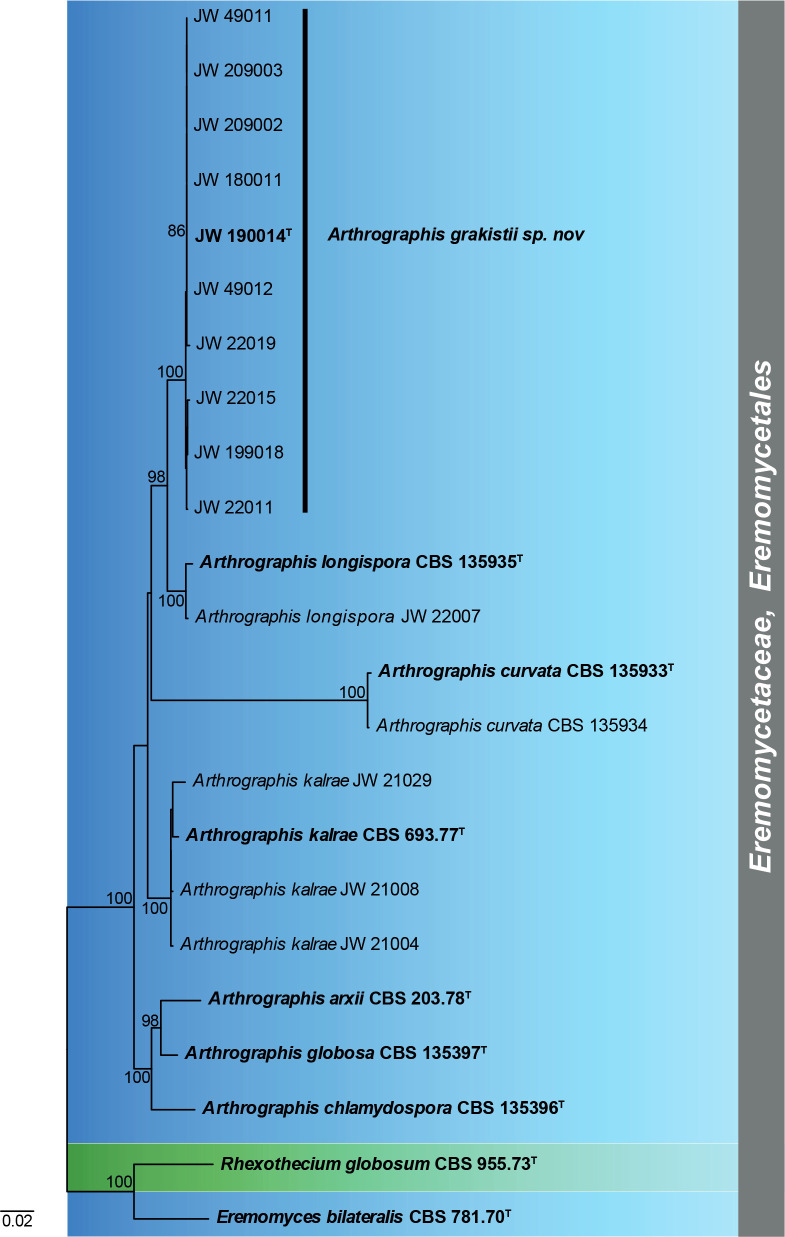
Aureoconidiella foliicola Hern.-Restr. & Crous, sp. nov. MycoBank MB833916. Fig. 4.
Fig. 4.
Aureoconidiella foliicola gen. et sp. nov. (CBS 145943). A. Conidiophores and conidia. B. Conidiophore with conidiogenous cell. C, D. Conidiogenous cells giving rise to conidia. E. Conidia. Scale bars: A = 20 μm, all others = 10 μm.
Etymology: The epithet “foliicola” refers to its habitat on a dead leaf.
Mycelium consisting of septate, smooth, brown, 1–2.5 μm wide hyphae. Conidiophores macronematous, simple, septate, brown, 40–85 × 3–5.5 μm. Conidiogenous cells integrated, terminal, polyblastic, with thickened scars, brown to pale brown, 25–53 × 3–4.5 μm. Conidia globose to subglobose with apiculate base, initially subhyaline, golden brown at maturity, verrucose, 5–8 μm diam, base 1–2 μm wide.
Culture characteristics: On MEA and OA surface cottony aerial mycelium Fawn, sepia to dark brick close to the agar, margin effuse, entire; reverse sepia to black.
Typus: South Africa, KwaZulu-Natal, Richards Bay, on living leaves of Syzygium cordatum (Myrtaceae), Jun. 2016, M.J. Wingfield (holotype CBS H-24099, culture ex-type CPC 36154 = CBS 145943).
Notes: This new lineage is introduced to accommodate a fungus characterised by unbranched conidiophores, cicatrised and sympodial conidiogenous cells with thickened scars, producing sub-globose, verruculose, and golden brown conidia. Other related lineages are those accommodating Asterinales and Cladoriellales (Fig. 1). However, they differ from those in the Aureoconidiellales based on the morphology of the asexual morphs. The Asterinales is mainly characterised by taxa that are coelomycetes with pycnothyrial conidiomata (Guatimosim et al. 2015, Jaklitsch et al. 2016). The Cladoriellales is a monotypic order related to cladosporium-like hyphomycetous fungi with conidia frequently remaining attached in long acropetal chains (Crous et al. 2006c, 2017).
Authors: M. Hernández-Restrepo, P.W. Crous and M.J. Wingfield
Arthrographis Sigler & J.W. Carmich., Mycotaxon 4: 359. 1976.
Synonym: [Arthrographis G. Cochet, Annls Parasit. Hum. Comp. 17: 97. 1939. (Nom. inval., Art. 39.1)]
Vegetative hyphae septate, hyaline, smooth- and thin-walled. Conidiophores macro- or micronematous, erect, simple or poorly branched, hyaline, smooth-walled. Conidiogenous hyphae simple or branched, thin-walled, forming septa basipetally to form arthroconidia released by schizolytic secession. Arthroconidia unicellular, cylindrical or cuboid, straight, subhyaline, thick- and smooth-walled. Synasexual morph trichosporiella-like, with conidia growing directly on undifferentiated hyphae, sessile, lateral, terminal, globose, subglobose or clavate, subhyaline, thin- and smooth-walled. Sexual morph not observed.
Type species: Arthrographis kalrae (R.P. Tewari & Macph.) Sigler & J.W. Carmich.
Arthrographis kalrae (R.P. Tewari & Macph.) Sigler & J.W. Carmich., Mycotaxon 4: 360. 1976.
Basionym: Oidiodendron kalrae R.P. Tewari & Macph., Mycologia 63: 603. 1971 [as ‘kalrai’].
Synonym: [Arthrographis langeronii G. Cochet (as ‘langeroni’), Annls Parasit. hum. comp. 17: 97. 1939. (Nom. inval., Art. 39.1)]
Descriptions and illustrations: Sigler & Carmichael (1976, 1983), Giraldo et al. (2014a).
Specimens examined: The Netherlands, Utrecht, isolated from soil, 2017, E. Kieviet (JW 21004 = CBS 145527), ibid. JW 21008, ibid. JW 21029.
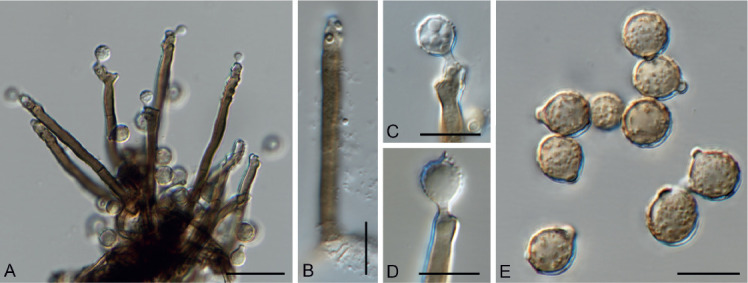
Arthrographis grakistii Giraldo López & Hern.-Restr., sp. nov. MycoBank MB833677. Fig. 5.
Fig. 5.
Arthrographis grakistii sp. nov. (CBS 145530). A. Poorly branched conidiophores and arthroconidia. B, C. Trichosporiella-like synasexual morph. Scale bars = 10 μm.
Etymology. Named after Ewan Grakist, who collected the soil sample. This species was discovered as part of a Citizen Science project in the Netherlands.
Vegetative hyphae septate, hyaline, smooth- and thin-walled, 1.5–2 μm wide. Conidiophores semi-macronematous or micronematous, erect, simple, hyaline, smooth-walled. Conidiogenous hyphae simple or branched, 1.5–2 μm wide, thin-walled, forming septa basipetally to form arthroconidia released by schizolytic secession. Arthroconidia unicellular, cylindrical or cuboid, straight, 2–4 × 2–3 μm, subhyaline, thick- and smooth-walled. Synasexual morph trichosporiella-like with conidia growing directly on undifferentiated hyphae, sessile, sometimes with a subcylindrical to clavate intercalary cell, lateral, terminal, globose, subglobose or clavate, 3–5 × 2–3 μm, subhyaline, thin- and smooth-walled. Sexual morph not observed.
Culture characteristics: Colonies at 25 °C after 14 d: on OA reaching 13–14 mm, flat, glabrous to floccose, surface and reverse buff. On MEA and PDA reaching 11–13 mm and 12–17 mm, respectively; flat or raised, dusty to cottony at centre, glabrous toward the periphery, buff to honey, reverse uncoloured. No growth at 37 °C.
Typus: The Netherlands, Utrecht Province, Wijk bij Duurstede, from garden soil, 2017, E. Grakist (holotype CBS H-23912, culture ex-type CBS 145530 = JW 190014).
Additional materials examined: The Netherlands, Utrecht Province, IJsselstein, from garden soil, 2017, J. Brus (JW 209002, JW 209003); from garden soil, 2017, R. de Bruyn (JW 180011); Utrecht, from garden soil, 2017, M. Wickham (JW 199018); Zeeland Province, Vlissingen, from garden soil, 2017, N. Penabad (CBS 145529 = JW 22011, JW 22015, JW 22019); Gelderland Province, Zaltbommel, from garden soil, 2017, K. & T. de Man (JW 49011, JW 49012).
Notes: Based on a BLAST search using the ITS and LSU loci several soil isolates (JW isolates listed in Table 1) were identified as belonging to Arthrographis. In order to confirm their identity at the species level, a combined analysis of the LSU/ITS/act loci was performed, including all members from Eremomycetaceae. The ML tree (Fig. 6) showed that three isolates (JW 21004, JW 21008 and JW 21029) grouped with the type species of A. kalrae (CBS 693.77) and one (JW 22007) with A. longispora (CBS 135935). However, most of the isolates formed a well-supported clade that represents the new species A. grakistii.
Fig. 6.
Maximum composite likelihood tree based on partial sequences from the LSU, ITS and act regions from all members of Eremomycetales. Colour boxes (blue and green) indicate the generic clades. Bootstrap support values above 70 % are shown at the nodes. Ex-type strains are in boldface. T = Ex-type.
The phylogenetic analyses showed that A. grakistii is closely related to A. longispora (Figs 1, 6). The latter species, however, has longer and narrower arthroconidia [5–10(–13) × 1–1.5 μm in A. longispora vs. 2–4 × 2–3 μm in A. grakistii] and does not produce the trichosporiella-like synasexual morph in culture (Giraldo et al. 2014a).
Morphologically, A. grakistii resembles A. kalrae and A. curvata in having cylindrical arthroconidia and a trichosporiella-like synasexual morph. Arthrographis kalrae and A. curvata are able to grow at 37 °C (Sigler & Carmichael 1983, Giraldo et al. 2014a), while A. grakistii does not grow at this temperature.
Authors: A. Giraldo López and M. Hernández-Restrepo
Kramasamuha Subram. & Vittal, Canad. J. Bot. 51: 1128. 1973.
Conidiophores erect, flexuous, solitary to fasciculate, arising from a swollen basal cell, which appears lobed due to rhizoids; medium brown, smooth, multi-septate, simple or branched, giving rise to parallel stipes, becoming paler towards apex, terminating in an acute conidiogenous cell. Conidiogenous cells monoblastic, pale brown, ampulliform, straight to curved, tapering to a truncate apex, thin-walled, solitary or in clusters, integrated or discrete, terminal and intercalary. Conidia solitary, smooth, septate, obovoid to pyriform; second cell from base thick-walled, dark brown, somewhat swollen, basal and apical cell subhyaline, with short narrow separating cell at base as remnant from conidiogenous cell.
Type species: Kramasamuha sibika Subram. & Vittal
Kramasamuha sibika Subram. & Vittal, Canad. J. Bot. 51: 1129. 1973. Fig. 7.
Fig. 7.
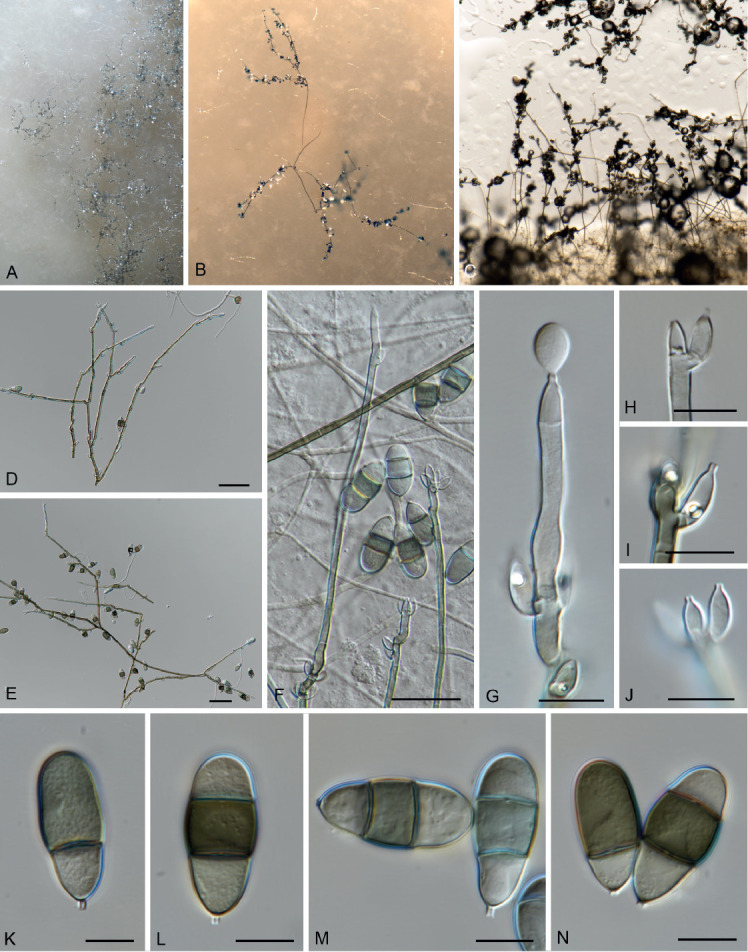
Kramasamuha sibika (CBS 146133). A–C. Conidiophores overview on OA. D–F. Conidiophores, conidiogenous cells and conidia. G. Conidiogenous cells and conidia. H–J. Conidiogenous cells. K–N. Conidia. Scale bars: D–F = 25 μm, all others = 10 μm.
Typus: India, Tamil Nadu, Chingleput district, Vandalur, on dead leaves of Gymnosporia emarginata (Celastraceae), 23 Apr. 1971, B.P.R. Vittal, Herb. MUBL 2153 (not seen).
Occurring on leaf litter. Conidiophores erect, flexuous, solitary to fasciculate, arising from a swollen basal cell, 10–14 μm diam, which appears lobed due to rhizoids; medium brown, thin-walled, smooth, multi-septate, septa 17–30 μm apart, up to 550 μm tall, 3–5 μm diam, unbranched or branched, giving rise to parallel stipes, becoming paler towards apex, terminating in an acute conidiogenous cell. Conidiogenous cells monoblastic, pale brown, ampulliform, straight to curved, tapering to a truncate apex, thin-walled, solitary or in clusters of 2–4, integrated or discrete, terminal and intercalary, 4–8 × 3–4 μm. Conidia solitary, smooth, (1–)2(–3)-septate, blastoconidia, obovoid to pyriform, apex obtuse, (18–)25–27(–34) × (10–) 11(–12) μm; second cell from base thick-walled, dark brown, somewhat swollen, basal and apical cell subhyaline, with short narrow separating cell at base as remnant from conidiogenous cell, 1–1.5 × 1 μm.
Culture characteristics: Colonies on OA with scarce aerial mycelium, cottony to velvety, grey olivaceous, submerged mycelium vinaceous, margin effuse, irregular, reverse vinaceous. Conidiophores, conidiogenous cells, and conidia very similar to those observed in natural substrate. Conidiogenous cells 4.5–9 × 2.5–4 μm. Conidia 14–32.5 × 9–14 μm, (0–)2(–3)-septate.
Materials examined: Australia, New South Wales, Mallanganee, on leaves of Lophostemon confertus (Myrtaceae), 17 Apr. 2018, A.J. Carnegie, CBS 146338. Brazil, state of Minas Gerais, Viçosa, on Hypericum innodorum (Hypericaceae) leaves bearing necrotic spots caused by Seimatosporium hypericinum (Pinaceae), 4 Jul. 2017, A.A. Colmán (VIC 47176, COAD 2632). Malaysia, on needles of Pinus tecunumanii, Oct. 2018, M.J. Wingfield, CBS 146339. South Africa, KwaZulu-Natal, Richards Bay, on living leaves of Syzygium cordatum (Myrtaceae), Jun. 2016, M.J. Wingfield, CBS 146133 = CPC 36153.
Notes: Kramasamuha resembles the genus Garnaudia and some species of Endophragmiella in having conidia with a short narrow separating cell at the base as remnant from the conidiogenous cell. However, species in these genera can be distinguished by the arrangement and colour of the conidiogenous cells. In Kramasamuha they are pale brown and solitary or in clusters along the conidiophores (Subramanian & Vittal 1973). In Garnaudia, the conidiogenous cells are brown, and verticillate in terminal branches (Borowska 1977), while in Endophragmiella the conidiogenous cells are hyaline and mainly solitary and terminal (Hughes 1979, Seifert et al. 2011). Kramasamuha is a monotypic genus originally described from India on Gymnosporia emarginata (Subramanian & Vittal 1973). However, the specimens examined here were from different substrates and continents, i.e. Lophostemon confertus in Australia, Hypericum×innodorum in Brazil, Pinus tecunumanii in Malaysia, and Syzygium cordatum in South Africa and can thus not serve as an epitype. This species has also been recorded from leaves on Feijoa sellowiana in New Zealand, and on Psidium guajava in Western Samoa (Landcare database 2019).
This is the first time that DNA sequence data has become available for K. sibika and shows that Kramasamuha is related to Helminthosphaeriaceae (Sordariomycetes, Fig. 2). Asexual morphs in Helminthosphaeria have been recognised as diplococcium-like (Samuels et al. 1997, Réblová 1999) with tretic conidiogenous cells, differing from the monoblastic conidiogenous cells observed in Kramasamuha (Seifert et al. 2011).
Authors: M. Hernández-Restrepo, P.W. Crous, M.J. Wingfield, A.A. Colmán, P.S.C. Mansur and R.W. Barreto
Melnikomyces Crous & U. Braun, Persoonia 32: 263. 2014.
Mycelium consisting of brown, septate, branched, smooth, thick-walled hyphae. Conidiophores subcylindrical, brown, smooth, erect, straight or geniculate, reduced to conidiogenous cells, or long, flexuous, multiseptate. Conidiogenous cells polyblastic, subcylindrical to subclavate, terminal or intercalary, brown, smooth, developing a rachis with numerous denticle-like loci. Conidia solitary, brown, verruculose, fusoid-ellipsoidal, 1-septate, ends sub-obtuse, released by rhexolytic secession. Chlamydospores terminal, globose to subglobose, in short chains, simple or branched, brown, smooth (modified from Crous et al. 2014b).
Type species: Melnikomyces vietnamensis Crous & U. Braun
Melnikomycesthailandicus Giraldo López, sp. nov. MycoBank MB833678. Fig. 8.
Fig. 8.
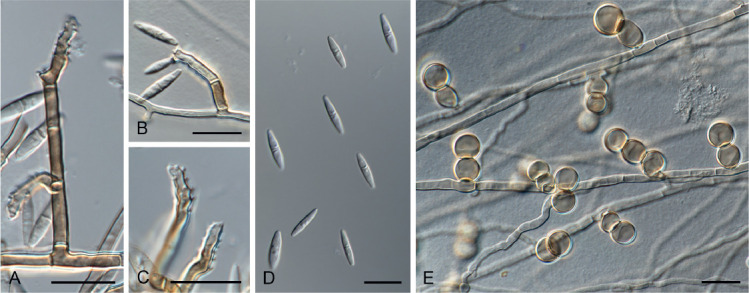
Melnikomyces thailandicus sp. nov. (CBS 145767). A–C. Conidiophores and denticulated conidiogenous cells. D. Conidia. E. Chlamydospores. Scale bars = 10 μm.
Etymology: Name refers to Thailand where the fungus was collected.
Mycelium consisting of brown, septate, branched, smooth, thick-walled, 2–2.5 μm diam hyphae. Conidiophores macronematous, arising directly from vegetative hyphae, erect, straight or slightly bent, simple, multiseptate, cylindrical, 14–37 × 2–3 μm, brown, paler apex, thick and smooth-walled. Conidiogenous cells integrated, terminal, polyblastic, brown to pale brown, sympodial, with long open denticles; denticles cylindrical, pale brown, up to 1 μm long. Conidia fusoid, ends subobtuse, 1-septate, solitary, subhyaline, smooth-walled, (8–)9.5–12(–13) × (2–)2.5(–3) μm. Chlamydospores lateral, globose to subglobose, in short and simple chains, light brown, thick-and smooth-walled, 5.5–10 μm diam.
Typus: Thailand, Nakhon Nayok Province, Mueang Nakhon Nayok district, Wang Takrai waterfall, N14.330023° E101.307168°, 64 m above sea level, from soil, 22 Jul. 2008, P.W. Crous (holotype CBS H-24236, culture ex-type CBS 145767).
Notes: The monotypic genus Melnikomyces was introduced by Crous and Braun (Crous et al. 2014b), based on M. vietnamensis collected from dry leaves in Vietnam. This species was treated as incertae sedis in the Chaetothyriales, Eurotiomycetes (Crous et al. 2014b). However, the results of this study show that it resides in the Sympoventuriaceae (Venturiales, Dothideomycetes), together with other genera producing septate conidia from denticulate conidiogenous cells, such as Ochroconis and Verruconis (Machouart et al. 2014, Samerpitak et al. 2014).
Melnikomyces thailandicus is the second species described in the genus, which differs morphologically from M. vietnamensis in having shorter conidiophores (10–60 μm long vs.14–37 μm long) and longer and narrower smooth-walled conidia (8–13 × (2–3) μm vs. 7–11 × 2.5–3.5 μm). Originally, M. vietnamensis was described with two types of conidiophores (Crous et al. 2014b), but in M. thailandicus one of the conidiophore types more closely resembles chlamydospores, as they appear to stay attached to the hyphae.
Authors: A. Giraldo López and P.W. Crous
Thysanorea Arzanlou, W. Gams & Crous, Stud. Mycol. 58: 80. 2007. emend.
Conidiophores micro- or macronematous, erect, simple or apically branched, sometimes proliferating percurrently in the apex, brown, smooth. Conidiogenous cells terminal or intercalary, polyblastic, smooth, brown at the base, paler towards the apex, subcylindrical, clavate to doliiform, sympodial, with crowded conidiogenous loci inconspicuous to slightly prominent, refractive to somewhat obscure, slightly thickened. Conidia solitary, oblong, obovoid, cylindrical, broadly fusiform to subpyriform, pale brown, smooth, with a narrowly truncate base and darkened hilum; conidial secession schizolytic. Synasexual morph: Conidiophores erect, simple, brown, smooth. Conidiogenous cells terminal, discrete, phialidic, subglobose to lageniform, with a balloon- to funnel-shaped collarette, brown, often in clusters at the apex of the conidiophores. Conidia solitary, subglobose to obovate, unicellular, hyaline, guttulate, smooth.
Type species: Thysanorea papuana (Aptroot) Arzanlou et al.
Notes: Thysanorea was established for a genus similar to Periconiella based on the branching pattern of the conidiophores (Arzanlou et al. 2007). However, recent studies have shown that those branching patterns depend on culture conditions, where those on natural substrates or in young cultures are not as prominently branched as previously described (Kirschner 2016, Wang et al. 2019). The generic description is emended here to include species with a phialidic synasexual morphs.
Phylogenetically, Thysanorea is closely related to Minimelanolocus in the Chaetothyriales (Fig. 3). However, the phylogenetic position of M. navicularis, the type species, remains unknown since DNA sequence data are not available for it, and the supposed phylogenetic position has been based on other species (Liu et al. 2015, Wang et al. 2019).
Morphologically, M. navicularis is characterised by terminal conidiogenous cells that produce navicular conidia with sub-hyaline polar cells and darker central cells (Castañeda-Ruiz 1987). They consequently differ from those species placed in Minimelanolocus based on DNA sequence data and in which the conidiogenous cells are terminal and intercalary, and the conidia are oblong, obovoid, cylindrical, broadly fusiform, and uniformly pale brown (Liu et al. 2015, Wang et al. 2019). In this regard, they would fit better with the generic concept of Thysanorea (Arzanlou et al. 2007, Wang et al. 2019). Based on these morphological differences and phylogenetic relationships, we propose new combinations for those species that have been shown as related to Thysanorea. The phylogenetic placement of other species for which DNA sequence data are not available, including M. navicularis must still be determined.
Thysanorea asiatica (H.Y. Su, et al.) Hern.-Restr. & Crous, comb. nov. MycoBank MB833919.
Basionym: Minimelanolocus asiaticus H.Y. Su, et al., Fungal Biol. 119: 1054. 2015.
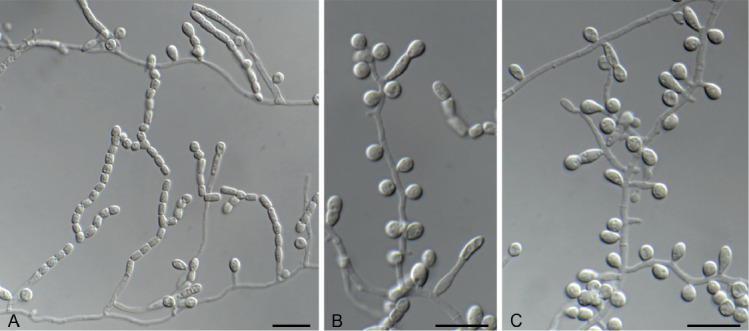
Thysanorea cantrelliae Hern.-Restr., R. van Doorn & Crous, sp. nov. MycoBank MB833914. Fig. 9.
Fig. 9.

Thysanorea cantrelliae sp. nov. (CBS 145909). A. Conidiophore and conidium. B, C. Conidiophores. D–G. Conidiogenous cells. H, I. Conidia. Scale bars = 10 μm.
Etymology: Named in honour of Sharon Cantrell, who was the organizer of the IMC 11 (2018) in Puerto Rico. This fungus was collected on a field trip held during the IMC 11.
Mycelium composed of hyaline to pale brown, septate, smooth, 1–2 μm wide hyphae. Conidiophores semi-micronematous, sometimes reduced to conidiogenous cells, simple, erect, straight or flexuous, cylindrical, pale brown, smooth, 8–31 × 2–3 μm. Conidiogenous cells holoblastic, polyblastic, mainly terminal, integrated, sympodial, pale brown, 7–22 × 2–3 μm. Conidia solitary, fusiform to acicular, straight or curved, (1–)3(–4)-septate, subhyaline to pale brown, smooth, 10–34 × 1.5–3 μm, apex acute, base truncate.
Culture characteristics: Colonies at 25 °C after 14 d: on OA reaching 22–25 mm, aerial mycelium moderate, cottony to floccose, olivaceous black, margin effuse entire; reverse black. On MEA and PDA reaching 22–30 mm, aerial mycelium abundant, cottony to floccose, olivaceous grey, black close to the agar, margin effuse, entire; reverse black.
Typus: USA, Puerto Rico, from unidentified twig, 20 Jul. 2018, M. Hernández-Restrepo (holotype CBS H-24100, culture ex-type CBS 145909).
Notes: Thysanorea cantrelliae clustered in a separate clade together with T. seifertii (Fig. 3). It can be distinguished from T. seifertii and other species in the genus by its acicular conidia (Fig. 9).
Thysanorea curvata (H.Y. Su et al.) Hern.-Restr. & Crous, comb. nov. MycoBank MB833921.
Basionym: Minimelanolocus curvatus H.Y. Su et al., Fungal Biol. 119: 1055. 2015.
Thysanorea lotorum (Morgan-Jones) Hern.-Restr. & Crous, comb. nov. MycoBank MB833922.
Basionym: Pseudospiropes lotorum Morgan-Jones, Mycotaxon 5: 481. 1977 [as ‘lotorus’]
Synonym: Nigrolentilocus lotorum (Morgan-Jones) R.F. Castañeda & Heredia, Cryptog. Mycol. 22: 15. 2001.
Minimelanolocus submersus Z.L. Luo et al., Fungal Diversity 80: 143. 2016.
Thysanorea melanica (H.Y. Su, et al.) Hern.-Restr. & Crous, comb. nov. MycoBank MB833923.
Basionym: Minimelanolocus melanicus H.Y. Su et al., Fungal Biol. 119: 1056. 2015.
Thysanorea nonramosa (X.D. Yu et al.) Hern.-Restr. & Crous, comb. nov. MycoBank MB833924.
Basionym: Minimelanolocus nonramosus X.D. Yu et al., Mycol. Progr. 18: 514. 2019.
Thysanorea obscura (Matsush.) Hern.-Restr. & Crous, comb. nov. MycoBank MB833925.
Basionym: Pseudospiropes obscurus Matsush., Matsushima Mycol. Mem. 3: 14. 1983.
Synonym: Minimelanolocus obscurus (Matsush.) R.F. Castañeda & Heredia, Cryptog. Mycol. 22:10. 2001.
Thysanorea rousseliana (Mont.) Hern.-Restr. & Crous, comb. nov. MycoBank MB833926.
Basionym: Helminthosporium rousselianum Mont., Ann. Sci. Nat., Sér. 3, Bot., 12: 300. 1849.
Synonyms: Pleurophragmium rousselianum (Mont.) S. Hughes, Canad. J. Bot. 36: 798. 1958.
Spiropes rousselianus (Mont.) de Hoog & Arx, Kavaka 1: 59. 1973.
Pseudospiropes rousselianus (Mont.) M.B. Ellis, More Dematiaceous Hyphomycetes: 221. 1976.
Minimelanolocus rousselianus (Mont.) R.F. Castañeda & Heredia, Cryptog. Mycol. 22: 10. 2001.
Thysanorea seifertii Hern.-Restr., R. van Doorn & Crous, sp. nov. MycoBank MB833920. Fig. 10.
Fig. 10.

Thysanorea seifertii sp. nov. (CBS 145910). A. Conidiophores and conidia. B, C. Conidiogenous cells and conidia. D. Conidia. E–J. Synasexual morph. E, F. Conidiophores. G–I. Conidiogenous cells and conidia. H. Conidia. Scale bars: G–J = 5 μm, all others = 10 μm.
Etymology: Named in honour of Prof. dr Keith A. Seifert, who was President of the International Mycological Association during the IMC 11 (2018) in Puerto Rico. This fungus was collected on a field trip during the IMC 11.
Mycelium composed of hyaline to pale brown, septate, smooth, 1–2 μm wide hyphae. Conidiophores mononematous, simple, erect, straight or flexuous, cylindrical, brown, paler towards the apex, smooth, 30–133 × 2–3 μm. Conidiogenous cells holoblastic, polyblastic, terminal or intercalary, integrated, sympodial, pale brown to brown, 7.5–46.5 × 2–3 μm. Conidia solitary, subcylindrical to clavate or oblong, straight or slightly curved, (0–)1–3(–4)-septate, pale brown, smooth, 7–15 × 1.5–3 μm, apex rounded, base darkened and truncated. Synasexual morph: Conidiophores micro- or macronematous, erect, straight or flexuous, cylindrical, brown, smooth, 10–51 × 2–4 μm. Conidiogenous cells enteroblastic, phialidic, arranged around the apex of the conidiophore, brown, subglobose to ampulliform, 2–4(–6) × 2–3 μm, with a balloon- to funnel-shaped collarette, 1–3 × 1–3 μm. Conidia solitary, subglobose to obovate, unicellular, hyaline, guttulate, smooth, 1–2 × 1–1.5 μm, base truncated.
Culture characteristics: Colonies at 25 °C after 14 d: on OA reaching 22–25 mm, aerial mycelium moderate, cottony to floccose, grey olivaceous to olivaceous black, margin effuse, entire; reverse black. On MEA and PDA reaching 18–20 mm, aerial mycelium moderate to abundant, cottony to floccose, olivaceous grey, black close to the agar, margin effuse, entire; reverse black.
Typus: USA, Puerto Rico, from unidentified twig, 20 Jul. 2018, M. Hernández-Restrepo (holotype CBS H-24101, culture ex-type CBS 145910).
Notes: Some of the conidia in T. seifertii resemble those of T. obscura in being 3-septate. However, conidia in T. seifertii are smaller than those of T. obscura (7–15 × 1.5–3 μm vs. 20–31 × 5–8 μm, Castañeda-Ruiz et al. 2001). Thysanorea seifertii is the only species in the genus known to produce a phialidic synasexual morph.
Thysanorea thailandensis (W. Dong et al.) Hern.-Restr. & Crous, comb. nov. MycoBank MB833927.
Basionym: Minimelanolocus thailandensis W. Dong et al., Mycol. Progr. 17: 622. 2018.
Thysanorea yunnanensis Hern.-Restr. & Crous, nom. nov. MycoBank MB833928.
Replaced synonym: Minimelanolocus aquaticus H.Y. Su et al., Fungal Biol. 119: 1049. 2015 [non Thysanorea aquatica W. Dong, H. Zhang & K.D. Hyde, 2018].
Etymology: The name refers to the Chinese Province of Yunnan where the fungus was collected.
Authors: M. Hernández-Restrepo, R. van Doorn and P.W. Crous
Verruconis Samerp. et al., Fungal Diversity 65: 117. 2014.
Mycelium consisting of septate, pale brown, smooth and thick-walled hyphae. Conidiophores differentiated, erect, straight or slightly bent, unbranched, pale brown. Conidiogenous cells mostly polyblastic, subcylindrical to narrowly mucronate, producing conidia sympodially on long open denticles; denticles cylindrical, pale brown, scattered at the apical third of the conidiogenous cell. Conidia two-celled, ellipsoidal, cylindrical or clavate, brown, verrucose or smooth-walled, released by rhexolytic secession. Sexual morph unknown.
Type species: Verruconis gallopava (W.B. Cooke) Samerp. & de Hoog
Verruconis thailandica Giraldo López & Crous, sp. nov. MycoBank MB833679. Fig. 11.
Fig. 11.
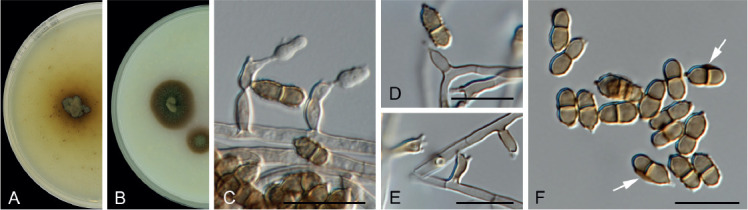
Verruconis thailandica sp. nov. (CBS 145768). A, B. Colonies on PDA and OA, respectively, at 25 °C after 14 d. C–E. Denticulated conidiogenous cells. F. Conidia with gelatinous brown sheath (arrow). Scale bars = 10 μm.
Etymology: The name refers to Thailand where the fungus was collected.
Mycelium consisting of septate, pale brown, smooth, thick-walled, 2–2.5 μm diam hyphae. Conidiophores differentiated, arising directly from vegetative hyphae, erect, straight or slightly bent, simple, 0–1-septate, subcylindrical, (3.6–)4.1–7.1(–9) × (1.3–)2(–2.3) μm, pale brown, thick- and smooth-walled, producing conidia sympodially on long open denticles; denticles cylindrical, pale brown, up to 1 μm long. Conidia abundant on OA and PCA, scarce on PDA, two-celled, broadly ellipsoidal with a protuberant hilum, constricted at the septum, (5–)5.8(–7) × (2.2–)2.6(–3.1) μm, brown, verrucose, thick-walled, sometimes with a wing-like gelatinous brown sheath, released by rhexolytic secession. Sexual morph not observed.
Cultural characteristics: Colonies at 25 °C after 14 d: on OA and PCA, flat, woolly at centre, glabrous at periphery, top and reverse sepia. On PDA raised, felty, top and reverse olivaceous with ochreous diffusible pigment.
Typus: Thailand, Nakhon Nayok Province, Mueang Nakhon Nayok district, Wang Takrai waterfall, N14.330023° E101.307168°, 64 m above sea level, from soil, 22 Jul. 2008, P.W. Crous (holotype CBS H-24237, culture ex-type CBS 145768).
Notes: The genus Verruconis (Sympoventuriaceae, Venturiales, Dothideomycetes) was established to accommodate thermophilic species segregated from Ochroconis (O. gallopava and O. calidifluminalis) and Scolecobasidium (S. verruculosum), which produce septate conidia from sympodially proliferating conidiophores, released by rhexolytic secession (Samerpitak et al. 2014). These species have been isolated from hot spring water, warm effluents or as soil saprobes (Yarita et al. 2007, Samerpitak et al. 2014, Giraldo et al. 2014b). However, the type species, V. gallopava has been reported as an opportunistic pathogen of humans and causing infections in other warm-blooded animals, mainly birds (Revankar & Sutton 2010, de Hoog et al. 2011).
Verruconis thailandica is phylogenetically related to V. verruculosa (Fig. 1), but the two species can be distinguished by the length of their conidiophores (up to 9 μm long in V. thailandica vs. up to 45 μm long in V. verruculosa) and conidia (up to 7 μm long in V. thailandica vs. up to 9 μm long in V. verruculosa; Samerpitak et al. 2014). Recently, three new species were added to the genus, namely V. panacis from Panax notoginseng (Zhang et al. 2018), V. hainanensis and V. pseudotricladiata from submerged decaying leaves (Qiao et al. 2019). These species clustered in a separate clade phylogenetically distant from V. thailandica. They can be distinguished by the presence of four-celled conidia in V. panacis and V. hainanensis, and branched Y-shaped conidia in V. pseudotricladiata (Zhang et al. 2018, Qiao et al. 2019).
Authors: A. Giraldo López and P.W. Crous
ACKNOWLEDGEMENTS
The authors thank the technical staff, A. van Iperen and T. Merkx for their valuable assistance with cultures.
REFERENCES
- Arzanlou M, Groenewald JZ, Gams W, et al. , (2007). Phylogenetic and morphotaxonomic revision of Ramichloridium and allied genera. Studies in Mycology 58: 57–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badali H, Gueidan C, Najafzadeh MJ, et al. , (2008). Biodiversity of the genus Cladophialophora. Studies in Mycology 61: 175–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer JC, Arias MM, Harrington TC, et al. , (2008). Four species of Zygophiala (Schizothyriaceae, Capnodiales) are associated with the sooty blotch and flyspeck complex on apple. Mycologia 100: 246–258. [DOI] [PubMed] [Google Scholar]
- Bezerra JDP, Oliveira R, Paiva L, et al. , (2017). Bezerromycetales and Wiesneriomycetales ord. nov. (class Dothideomycetes), with two novel genera to accommodate endophytic fungi from Brazilian cactus. Mycological Progress 16: 297–309. [Google Scholar]
- Boehm EW, Schoch CL, Spatafora JW. (2009). On the evolution of the Hysteriaceae and Mytilinidiaceae (Pleosporomycetidae, Dothideomycetes, Ascomycota) using four nuclear genes. Mycological Research 113: 461–479. [DOI] [PubMed] [Google Scholar]
- Boonmee S, Bhat JD, Maharachchikumbura SSN, et al. , (2014). Clavatispora thailandica gen. et sp. nov., a novel taxon of Venturiales (Dothideomycetes) from Thailand. Phytotaxa 176: 92–101. [Google Scholar]
- Borowska A. (1977). Garnaudia elegans gen. et sp. nov. and Endophragmiella tenera sp. nov., new dematiaceous hyphomycetes. Acta Mycologica 13: 169–174. [Google Scholar]
- Braun U, Crous PW, Dugan F, et al. , (2003). Phylogeny and taxonomy of Cladosporium-like hyphomycetes, including Davidiella gen. nov., the teleomorph of Cladosporium s. str. Mycological Progress 2: 3–18. [Google Scholar]
- Campbell J, Ferrer A, Raja HA, et al. , (2007). Phylogenetic relationships among taxa in the Jahnulales inferred from 18S and 28S nuclear ribosomal DNA sequences. Canadian Journal of Botany 85: 873–882. [Google Scholar]
- Castañeda-Ruiz RF. (1987). Fungi Cubenses II. Instituto de Investigaciones Fundamentales en Agricultura Tropical ‘Alejandro de Humboldt’, La Habana: 1–22 [Google Scholar]
- Castañeda-Ruiz RF, Heredia G, Gusmao LFP. (2016). Fungal diversity of central and south America. In: Biology of microfungi (De-Wei L, ed). Springer International Publishing, Switzerland: 197–218. [Google Scholar]
- Castañeda-Ruiz RF, Heredia G, Reyes, et al. , (2001). A revision of the genus Pseudospiropes and some new taxa. Cryptogamie Mycologie 22: 1–18. [Google Scholar]
- Castlebury LA, Rossman AY, Jaklitsch WJ, et al. , (2002). A preliminary overview of the Diaporthales based on large subunit nuclear ribosomal DNA sequences. Mycologia 94: 1017–1031. [PubMed] [Google Scholar]
- Cheewangkoon R, Crous PW, Hyde KD, et al. , (2008). Species of Mycosphaerella and related anamorphs on Eucalyptus leaves from Thailand. Persoonia 21: 77–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheewangkoon R, Groenewald JZ, Hyde KD, et al. , (2012). Chocolate spot disease of Eucalyptus. Mycological Progress 11: 61–69. [Google Scholar]
- Cheewangkoon R, Groenewald JZ, Summerell BA, et al. , (2009). Myrtaceae, a cache of fungal biodiversity. Persoonia 23: 55–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Braun U, Groenewald JZ. (2007a). Mycosphaerella is polyphyletic. Studies in Mycology 58: 1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Braun U, Schubert K, et al. , (2007b). Delimiting Cladosporium from morphologically similar genera. Studies in Mycology 58: 33–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Gams W, Stalpers JA, et al. , (2004). MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology 50: 19–22. [Google Scholar]
- Crous PW, Giraldo A, Hawksworth DL, et al. , (2014a). The Genera of Fungi: fixing the application of type species of generic names. IMA Fungus 5: 141–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Groenewald JZ, Wingfield MJ. (2006a) Heteroconium eucalypti. Fungal Planet No. 10. Centraalbureau voor Scimmelcultures, the Netherlands. [Google Scholar]
- Crous PW, Schoch CL, Hyde KD, et al. , (2009a). Phylogenetic lineages in the Capnodiales. Studies in Mycology 64: 17–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Schubert K, Braun U, et al (2007c). Opportunistic, human-pathogenic species in the Herpotrichiellaceae are phenotypically similar to saprobic or phytopathogenic species in the Venturiaceae. Studies in Mycology 58: 185–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Shivas RG, Quaedvlieg W, et al. , (2014b). Fungal Planet description sheets 214–280. Persoonia 32: 184–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Slippers B, Wingfield MJ, et al. , (2006b). Phylogenetic lineages in the Botryosphaeriaceae. Studies in Mycology 55: 235–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Summerell BA, Carnegie AJ, et al. , (2009b). Unravelling Mycosphaerella: do you believe in genera? Persoonia 23: 99–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Verkley GJM, Groenewald JZ. (2006c). Eucalyptus microfungi known from culture. 1. Cladoriella and Fulvoflamma genera nova, with notes on some other poorly known taxa. Studies in Mycology 55: 53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Verkley GJM, Groenewald JZ, et al. , (2019). Westerdijk Laboratory Manual Series 1: Fungal Biodiversity. Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands. [Google Scholar]
- Crous PW, Wingfield MJ, Burgess TI, et al. , (2017). Fungal Planet description sheets: 625–715. Persoonia 39: 270–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoog GS, Vicente VA, Najafzadeh MJ, et al. , (2011). Waterborne Exophiala species causing disease in cold-blooded animals. Persoonia 27: 46–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoog GS, Gerrits van den Ende AHG. (1998). Molecular diagnostics of clinical strains of filamentous Basidiomycetes. Mycoses 41: 183–189. [DOI] [PubMed] [Google Scholar]
- de Hoog GS, Guarro J, Gené J, et al. , (2011). Atlas of clinical fungi. CD-ROM version 3.1. CBS-KNAW Fungal Biodiversity Centre, Utrecht, the Netherlands. [Google Scholar]
- Diederich P, Ertz D, Lawrey JD, et al. , (2013). Molecular data place the hyphomycetous lichenicolous genus Sclerococcum close to Dactylospora (Eurotiomycetes) and S. parmeliae in Cladophialophora (Chaetothyriales). Fungal Diversity 58: 61–72. [Google Scholar]
- Dong W, Hyde KD, Bhat DJ, et al. , (2018). Introducing Aculeata aquatica gen. et sp. nov., Minimelanolocus thailandensis sp. nov. and Thysanorea aquatica sp. nov. (Herpotrichiellaceae, Chaetothyriales) from freshwater in northern Thailand. Mycological Progress 17: 617–629. [Google Scholar]
- Fernandez FA, Lutzoni FM, Huhndorf SM. (1999). Teleomorph-anamorph connections: the new pyrenomycetous genus Carpoligna and its Pleurothecium anamorph. Mycologia 91: 251–262. [Google Scholar]
- Geiser DM, Gueidan C, Miadlikowska J, et al. , (2006). Eurotiomycetes: Eurotiomycetidae and Chaetothyriomycetidae. Mycologia 98: 1053–1064. [DOI] [PubMed] [Google Scholar]
- Giraldo A, Gené J, Cano J, et al. , (2012). Two new species of Acremonium from Spanish soils. Mycologia 104: 1456–1465. [DOI] [PubMed] [Google Scholar]
- Giraldo A, Gené J, Sutton DA, et al. , (2014a). Phylogenetic circumscription of Arthrographis (Eremomycetaceae, Dothideomycetes). Persoonia 32: 102–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo A, Hernández-Restrepo M, Crous PW. (2019). New Plectosphaerellaceous species from Dutch garden soil. Mycological Progress 18: 1135–1154. [Google Scholar]
- Giraldo A, Sutton D, Samerpitak K, et al. , (2014b). Occurrence of Ochroconis and Verruconis species in clinical specimens from the United States. Journal of Clinical Microbiology 52: 4189–4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenewald M, Lombard L, de Vries M, et al. , (2018). Diversity of yeast species from Dutch garden soil and the description of six novel Ascomycetes. FEMS Yeast Research 18: 1–14. [DOI] [PubMed] [Google Scholar]
- Guatimosim E, Firmino AL, Bezerra JL, et al. , (2015). Towards a phylogenetic reappraisal of Parmulariaceae and Asterinaceae (Dothideomycetes). Persoonia 35: 230–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueidan C, Villasenor CR, de Hoog GS, et al. , (2008). A rock-inhabiting ancestor for mutualistic and pathogen-rich fungal lineages. Studies in Mycology 61: 111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Restrepo M, Gene J, Castañeda-Ruiz RF, et al. , (2017). Phylogeny of saprobic microfungi from Southern Europe. Studies in Mycology 86: 53–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes SJ. (1979). Relocation of species of Endophragmia auct. with notes on relevant generic names. New Zealand Journal of Botany 17: 139–188. [Google Scholar]
- Huhndorf SM, Miller AN, Fernandez FA. (2004). Molecular systematics of the Sordariales: the order and the family Lasiosphaeriaceae redefined. Mycologia 96: 368–387. [PubMed] [Google Scholar]
- Jaklitsch WM, Baral HO, Lücking R, et al (2016). Syllabus of plant families – A. Engler’s Syllabus der Pflanzenfamilien Part 1/2: Ascomycota, 13th edn Borntraeger, Stuttgart, Germany. [Google Scholar]
- James TY, Kauff F, Schoch CL, et al. , (2006). Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature 443: 818–822. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Sigler L, Lee J, et al. , (2010). Xylogone ganodermophthora sp. nov., an ascomycetous pathogen causing yellow rot on cultivated mushroom Ganoderma lucidum in Korea. Mycologia 102: 1167–1184. [DOI] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, et al. , (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner R. (2016). Revision of the morphology and biogeography of Thysanorea papuana. Mycosphere 7: 820–827. [Google Scholar]
- Landcare database (2019). New Zealand Fungi and Bacteria. https://nzfungi2.landcareresearch.co.nz/.
- Liu XY, Udayanga D, Luo ZL, et al. , (2015). Backbone tree for Chaetothyriales with four new species of Minimelanolocus from aquatic habitats. Fungal Biology 119: 1046–1062. [DOI] [PubMed] [Google Scholar]
- Lumbsch HT, Schmitt I, Lindemuth R, et al. , (2005). Performance of four ribosomal DNA regions to infer higher-level phylogenetic relationships of inoperculate euascomycetes (Leotiomyceta). Molecular Phylogenetics and Evolution 34: 512–524. [DOI] [PubMed] [Google Scholar]
- Machouart M, Samerpitak K, de Hoog GS, et al. , (2014). A multigene phylogeny reveals that Ochroconis belongs to the family Sympoventuriaceae (Venturiales, Dothideomycetes). Fungal Diversity 65: 77–88. [Google Scholar]
- Miller AN, Huhndorf SM. (2004). A natural classification of Lasiosphaeria based on nuclear LSU rDNA sequences. Mycological Research 108: 26–34. [DOI] [PubMed] [Google Scholar]
- Miller AN, Huhndorf SM. (2005). Multi-gene phylogenies indicate ascomatal wall morphology is a better predictor of phylogenetic relationships than ascospore morphology in the Sordariales (Ascomycota, Fungi). Molecular Phylogenetics and Evolution 35: 60–75. [DOI] [PubMed] [Google Scholar]
- Miller AN, Huhndorf SM, Fournier J. (2014) Phylogenetic relationships of five uncommon species of Lasiosphaeria and three new species in the Helminthosphaeriaceae (Sordariomycetes). Mycologia 106: 505–524. [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. (2012). The CIPRES science gateway: enabling high-impact science for phylogenetics researchers with limited resources. In: Proceedings of the 1st Conference of the Extreme Science and Engineering Discovery Environment: Bridging from the extreme to the campus and beyond: 1–8 Association for Computing Machinery, USA. [Google Scholar]
- Mugambi GK, Huhndorf SM. (2009a). Molecular phylogenetics of Pleosporales: Melanommataceae and Lophiostomataceae re-circumscribed (Pleosporomycetidae, Dothideomycetes, Ascomycota). Studies in Mycology 64: 103–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugambi GK, Huhndorf SM. (2009b). Parallel evolution of hysterothecial ascomata in ascolocularous fungi (Ascomycota, Fungi). Systematics and Biodiversity 7: 453–464. [Google Scholar]
- Murata Y, Sano A, Kamei K, et al. , (2005). Arthrographis kalrae isolated from the oral cavity of a house-hold dog. Program and Abstracts of the 49th Annual Meeting of the Japanese Society for Medical Mycology 49: 162. [Google Scholar]
- Qiao M, Tian W, Castañeda-Ruiz RF. (2019). Two new species of Verruconis from Hainan, China. MycoKeys 48: 41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaedvlieg W, Binder M, Groenewald JZ, et al. , (2014). Introducing the consolidated species concept to resolve species in the Teratosphaeriaceae. Persoonia 33: 1–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner RW. (1970). A mycological colour chart. British Mycological Society. Commonwealth Mycological Institute; Kew, Surrey. [Google Scholar]
- Réblová M. (1999). Teleomorph-anamorph connections in Ascomycetes 3. Three new lignicolous species of Helminthosphaericeae. Sydowia 51: 223–244. [Google Scholar]
- Réblová M, Untereiner WA, Réblová K. (2013). Novel evolutionary lineages revealed in the Chaetothyriales (fungi) based on multigene phylogenetic analyses and comparison of its secondary structure. PloS ONE 8: e63547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revankar SG, Sutton D. (2010). Melanized fungi in human disease. Clinical Microbiology Reviews 23: 884–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samerpitak K, Van der Linde E, Choi HJ, et al. , (2014). Taxonomy of Ochroconis, genus including opportunistic pathogens on humans and animals. Fungal Diversity 65: 89–126. [Google Scholar]
- Samuels GJ, Candoussau F, Magni JF. (1997). Fungicolous pyrenomycetes 1. Helminthosphaeria and the new family Helminthosphaeriaceae. Mycologia 89: 141–155. [Google Scholar]
- Schoch CL, Crous PW, Groenewald JZ, et al (2009). A class-wide phylogenetic assessment of Dothideomycetes. Studies in Mycology 64: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch CL, Robbertse B, Robert V, et al (2014). Finding needles in haystacks: linking scientific names, reference specimens and molecular data for Fungi. Database (Oxford): doi:10.1093/database/bau061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert K, Morgan-Jones G, Gams W, et al. , (2011). The genera of Hyphomycetes. CBS Biodiversity Series Vol. 9. Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands. [Google Scholar]
- Sigler L, Carmichael JW. (1976). Taxonomy of Malbranchea and some other Hyphomycetes with arthroconidia. Mycotaxon 4: 349–488. [Google Scholar]
- Sigler L, Carmichael JW. (1983). Redisposition of some fungi referred to Oidium microspermum and a review of Arthrographis. Mycotaxon 18: 495–507. [Google Scholar]
- Smith H, Wingfield MJ, Crous PW, et al. , (1996). Sphaeropsis sapinea and Botryosphaeria dothidea endophytic in Pinus spp. and Eucalyptus spp. in South Africa. South African Journal of Botany 62: 86–88. [Google Scholar]
- Spatafora JW, Sung GH, Johnson D, et al. , (2006). A five-gene phylogeny of Pezizomycotina. Mycologia 98: 1018–1028. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian CV, Vittal BPR. (1973). Three new Hyphomycetes from litter. Canadian Journal of Botany 51: 1127–1132. [Google Scholar]
- Summerell BA, Groenewald JZ, Carnegie AJ, et al. , (2006). Eucalyptus microfungi known from culture. 2. Alysidiella, Fusculina and Phlogicylindrium genera nova, with notes on some other poorly known taxa. Fungal Diversity 23: 323–350. [Google Scholar]
- Tsui CK, Sivichai S, Berbee ML. (2006). Molecular systematics of Helicoma, Helicomyces and Helicosporium and their teleomorphs inferred from rDNA sequences. Mycologia 98: 94–104. [DOI] [PubMed] [Google Scholar]
- Untereiner WA, Angus A, Reblova M, et al. , (2008). The systematics of the Phialophora verrucosa complex: new insights from B-tubulin, large subunit nuclear rDNA and ITS sequence data. Botany 86: 742–750. [Google Scholar]
- Untereiner WA, Debois V, Naveau FA. (2001). Molecular systematics of the ascomycete genus Farrowia (Chaetomiaceae). Canadian Journal of Botany 79: 321–333. [Google Scholar]
- Untereiner WA, Naveau FA. (1999). Molecular systematics of the Herpotrichiellaceae with an assessment of the phylogenetic positions of Exophiala dermatitidis and Phialophora americana. Mycologia 91: 67–83. [Google Scholar]
- Vasse M, Voglmayr H, Mayer V, et al (2017). A phylogenetic perspective on the association between ants (Hymenoptera: Formicidae) and black yeasts (Ascomycota: Chaetothyriales). Proceedings Biological Sciences 284: 1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkley GJ, Dukik K, Renfurm R, et al. , (2014). Novel genera and species of coniothyrium-like fungi in Montagnulaceae (Ascomycota). Persoonia 32: 25–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigalys R, Hester M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt K, Wöstemeyer J. (2000). Reliable amplification of actin genes facilitates deep-level phylogeny. Microbiological Research 155: 179–195. [DOI] [PubMed] [Google Scholar]
- Vu D, Groenewald M, de Vries M, et al. , (2019). Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Studies in Mycology 92: 135–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GN, Yu XD, Dong W, et al. , (2019). Freshwater hyphomycetes in Eurotiomycetes: a new species of Minimelanolocus and a new collection of Thysanorea papuana (Herpotrichiellaceae). Mycological Progress 18: 511–522. [Google Scholar]
- White TJ, Bruns T, Lee S, et al (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR protocols: a guide to methods and applications (Innis MA, Gelfand DH, Sninsky JJ, et al, eds). Academic Press, New York, USA: 315–322. [Google Scholar]
- Winton LM, Stone JK, Hansen EM, et al. , (2007). The systematic position of Phaeocryptopus gaeumannii. Mycologia 99: 240–252. [DOI] [PubMed] [Google Scholar]
- Woudenberg JH, Groenewald JZ, Binder M, et al. , (2013). Alternaria redefined. Studies in Mycology 75: 171–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi L, Fukushima K, Changming L, et al. , (2004). First case of Arthrographis kalrae ethmoid sinusitis and ophthalmitis in the people’s Republic of China. Journal of Clinical Microbiology 42: 4828–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Groenewald JZ, Cheewangkoon R, et al. , (2017). Families, genera, and species of Botryosphaeriales. Fungal Biology 121: 322–346. [DOI] [PubMed] [Google Scholar]
- Yarita K, Sano A, Murata Y, et al. , (2007). Pathogenicity of Ochroconis gallopava isolated from hot spring in Japan and a review of published reports. Mycopathologia 164: 135–147. [DOI] [PubMed] [Google Scholar]
- Zhang TY, Yu Y, Zhang MY, et al. , (2018). Verruconis panacis sp. nov., an endophyte isolated from Panax notoginseng. International Journal of Systematics and Evolutionary Microbiology 68: 2499–2503. [DOI] [PubMed] [Google Scholar]