Abstract
The phase separation occurring in a system of mutually interacting proteins that can bind on specific sites of a chromatin fiber is investigated here. This is achieved by means of extensive molecular dynamics simulations of a simple polymer model that includes regulatory proteins as interacting spherical particles. Our interest is particularly focused on the role played by phase separation in the formation of molecule aggregates that can join distant regulatory elements, such as gene promoters and enhancers, along the DNA. We find that the overall equilibrium state of the system resulting from the mutual interplay between binding molecules and chromatin can lead, under suitable conditions that depend on molecules concentration, molecule-molecule, and molecule-DNA interactions, to the formation of phase-separated molecular clusters, allowing robust contacts between regulatory sites. Vice versa, the presence of regulatory sites can promote the phase-separation process. Different dynamical regimes can generate the enhancer-promoter contact, either by cluster nucleation at binding sites or by bulk spontaneous formation of the mediating cluster to which binding sites are successively attracted. The possibility that such processes can explain experimental live-cell imaging data measuring distances between regulatory sites during time is also discussed.
Significance
Phase separation is a general physical mechanism that occurs in living cells at various levels and is fundamental for genome activity. Indeed, protein condensates mediate the interaction between distant regulatory elements along the chromatin chain and shape genome architecture. Correspondingly, the activity of the genes is strongly associated with this process. Remarkable experimental work has been recently done to investigate phase separation, and it is currently the object of intense research. Here, using polymer modeling and molecular dynamics simulations, we provide a systematic investigation of this fundamental process and explore its influence on the physical communication between regulatory elements along the chromatin chain, such as genes and enhancers.
Introduction
The formation of molecular aggregates through phase separation (PS) is a general physical mechanism for which there is an increasing amount of experimental evidence, highlighting its importance for the cell activity (1,2). Within the cell nucleus, PS occurs at many levels and leads to the formation of membraneless structures at different scales, ranging from micron-sized aggregates, such as nucleoli and Cajal bodies (1,2), to the few hundreds of nanometer droplets of transcriptional coactivators (3), mediator and RNA PolII clusters (4,5), and transcription factors (TFs) (6). Typically, phase-separated aggregates have round shapes, tend to coalesce if in spatial proximity, and exhibit a highly dynamic, liquid-like behavior, as highlighted by photobleaching experiments (3,5). Importantly, the formation of such condensates is deeply linked to the transcriptional activity of the genes because they mediate the contact with their distal regulatory elements (i.e., enhancers and superenhancers (7)). On the other hand, PS also plays a crucial role in shaping repressed heterochromatin through the formation of phase-separated condensates of the protein HP1 (8,9). Recently, it has been shown that the chromatin fiber can undergo PS, forming dense chromatin droplets, as shown in in vitro experiments under physiological conditions (10). Also, the phase-separated aggregates have been shown to have a complex, multilayered internal structure emerging from the different interactions among the molecules (11).
In contrast with prokaryotic cells, in which the organization of the DNA is much simpler and the mechanism of gene activation may involve the sliding of regulatory proteins along the DNA (12), in eukaryotic cells, the situation is much more complex, and the PS process plays an important role either for the spatial organization of chromatin within the nucleus or for gene regulation (13,14). Indeed, genome structure is intimately linked to the transcriptional activity of genes because a correct folding allows an efficient communication between genes and their distal enhancers (15), whereas if altered, they can cause severe diseases (16). To quantitatively investigate such three-dimensional (3D) architecture, classic polymer physics models (17,18) and mesoscale models (19) have been developed. Notably, they successfully explained general aspects of genomic structure and helped to better understand mechanistic principles that regulate chromatin folding, such as the formation of chromatin loops (20,21), the structure of megabase-sized human and murine loci (22, 23, 24), the impact of structural variants (25), and the structure of real loci at nucleosome level (26). Importantly, some of those models (27, 28, 29) rely on simple thermodynamic mechanisms and naturally envisage the formation of phase-separated molecular clusters necessary to mediate the contact between distant elements along the chromatin chain (30).
In this work, we use polymer physics and molecular dynamics simulations (28) to quantitatively investigate the relationship between the PS process and the formation of contacts between genes and enhancers. To this aim, we consider a simple model in which specific regulatory loci of the chromatin fiber can interact with diffusing multivalent molecules that in turn nonspecifically interact among themselves. Such interactions are known to exist for several proteins and are fundamental to promote PS events (2,31). From this point of view, the model is a generalization of previous models (32), in which the above interactions were typically overlooked.
By varying the parameters controlling the phase transition, which are the molecular concentration and the nonspecific affinity, we build the system’s phase diagram, extending the usual experimental approach typically focused only on the molecular concentration. The PS process is triggered by increasing the molecular concentration or their interaction affinity above threshold values. When the transition occurs and equilibrium is achieved, the structural properties of the molecular cluster can be changed by means of the control parameters. For very weak affinities, we find that the cluster exhibits dynamical properties, such as high exchange rates of particles and internal mobility, indicating a liquid-like nature of the molecular aggregate. Consistently with experiments (3,6), the formation of the phase-separated cluster is crucial to mediate a stable contact between the enhancer and its target gene. The predictions of the model are then compared with published experimental live-imaging data measuring distances between the Sox2 promoter and its superenhancer Sox2 control region (SCR) (33). On the other hand, genes and enhancers can act as a nucleation starting site and induce the PS. In general, the formation of molecular aggregates driven by PS and the interaction of the molecules with the genes (or enhancers) located along the chromatin chain act cooperatively, and their interplay determines transient and equilibrium properties of the system.
Methods
Model details
To investigate the interplay between the PS process and gene-enhancer dynamic, we use a simple polymer model, based on the strings-and-binders-switch polymer model (29) (also known as the TFs model (27)), in which a chromatin filament is modeled by a self-avoiding walk (SAW) string made of N beads, with some binding sites (bs) that can interact with binding factors (or simply binders) floating in the surrounding environment. Although the model envisages the possibility of different types of binders (25), the results discussed here are based, for sake of simplicity, on models with only one type of binder. The binders can interact with binding sites (bs) placed on the polymer, with an interaction affinity Eb-bs and a total concentration c. Furthermore, binders interact among themselves with an attractive multivalent interaction Eb-b. In all simulations, we use polymers with N = 200. The total number of bs is 6, arranged in two groups of three sites symmetrically located on the polymer with an average linear distance of 100 beads. To check the robustness of the results, we also used polymers with arrangements having four and two bs and found, in general, similar behaviors.
Molecular dynamics simulations details
The entire system (polymer beads and binders) is subject to thermal fluctuations at temperature T, so the particles obey the Langevin equation (34). Unless differently stated, beads and binders have same diameter σ and mass m that we set equal to 1 in dimensionless units (35). We use a purely repulsive Lennard-Jones (LJ) potential between any two particles to account for excluded volume effects, with length scale σ and energy scale ε measured in KBT units (35). Between any two adjacent beads of the polymer, we use a finitely extensible nonlinear elastic spring (35), with standard parameters (27,36,37) (length constant R0 = 1.6 σ and spring constant K = 30 KBT/σ2).
The interaction between beads that are bs representing genes or enhancers and binders, as well as the nonspecific interaction among the binders, was modeled by a short-range, truncated attractive LJ potential VLJ in the following form:
for r < Rint = 1.3 σ or 0 otherwise. The interaction affinities reported in the figures (named Eb-b and Eb-bs) are given by the minimum of VLJ and are controlled by ε (38). In our simulations, ε was sampled in the range (5.8–11.6) KBT for Eb-b and in the range (9–15) KBT for Eb-bs.
The Langevin equation is integrated using the online available LAMMPS package (39). The dynamics parameters are set to standard values (38); that is, friction coefficient ζ = 0.5, temperature T = 1, and integration time step dt = 0.012 (35,40), expressed in dimensionless units. The system is confined in a cubic simulation box with periodic boundary conditions, with edge size D = 30 σ for the simulations in Figs. 1, 2, and 4 and D = 60 σ, to minimize finite size effects for the simulations presented in Fig. 3.
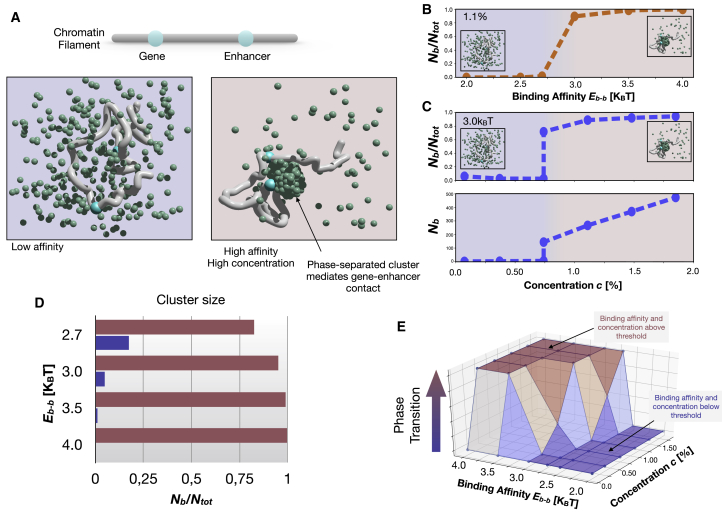
Figure 1.
Phase separation (PS) is a switch-like process mediating the formation of the gene-enhancer interaction. (A) The formation of a phase-separated cluster can mediate the contact between a gene and its enhancer. (B) The fraction of binders Nb/Ntot in the phase-separated cluster as a function of the binding affinity Eb-b among the binders at equilibrium. The concentration c is ∼1.1%. The transition occurs between 2.7 and 3.0 KBT. (C) The fraction of binders in the cluster as a function of the binder concentration c. The binding affinity is Eb-b = 3.0 KBT. At the transition point c ∼0.7%, in some cases, the phase-separated cluster could not form within the time window considered. (D) The horizontal red (or blue) bar shows the fraction of binders belonging (or not belonging) to the cluster for different binding affinities Eb-b (y axis) at high concentration (c is ∼1.9%). (E) A three-dimensional phase-diagram of the system. On the z axis, the fraction of times in which the system exhibited the phase transition for a fixed parameter configuration is reported. For all panels, interaction with bs Eb-bs is 3.1 KBT. Similar results are obtained in the absence of polymer. To see this figure in color, go online.
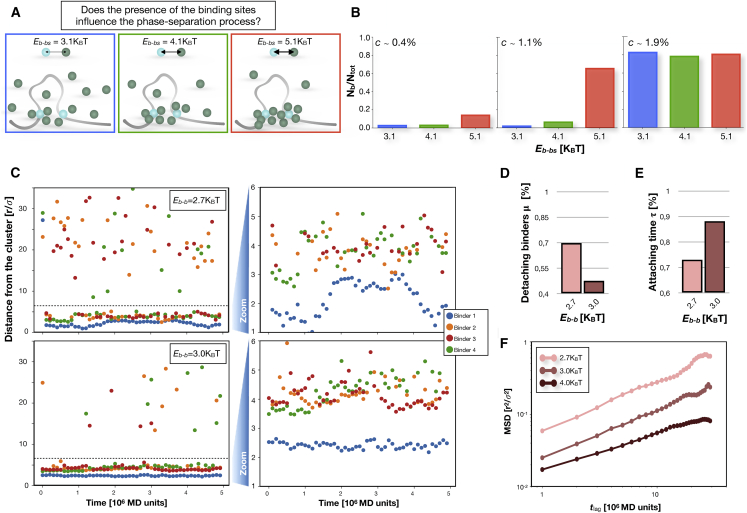
Figure 2.
Influence of the bs and structural properties of the phase-separated clusters. (A) The interaction between the bs and the binders influences the system properties by locally increasing the concentration around the bs. (B) Fraction of total binders forming the largest cluster Nb/Ntot for different concentrations c and Eb-bs, at the transition energy Eb-b = 2.7 KBT. When c is ∼1.1%, only at Eb-bs = 5.1 KBT does the system exhibits a macroscopic transition. (C) Left panels show distance between single binders and the center of the phase-separated cluster, as function of time, for two different values of Eb-b. Each color represents a different binder. The horizontal dashed line indicates an estimate of the cluster size. When the dot is above the line, the binder is escaped from the cluster. Right panels show zoom highlighting of the mobility of the binders when they move within the cluster. (D) The fraction of binders μ that detach from the cluster at least once in a characteristic time interval. For low affinities, the cluster exhibits highly dynamical properties because up to roughly 70% of the binders experience a detaching event. (E) The attaching time—that is, the average relative time spent by a single binder in the cluster—strongly depends on the binding affinity Eb-b. (F) The mean-square displacement (MSD; log-log scale) of the binders within the cluster, for different values of Eb-b. As the energy decreases, the binders have a higher mobility, and the cluster becomes more dynamic. For (C)–(F), the concentration used is c ∼1.5%. To see this figure in color, go online.
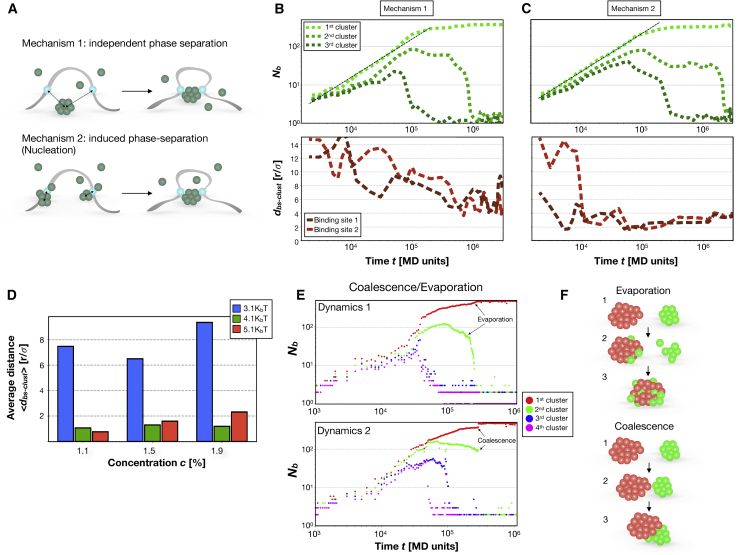
Figure 4.
Microscopic mechanisms of PS induction. (A) Possible mechanisms leading to PS and driving contact formation between regulatory elements. (B) The number of binders Nb composing the three main clusters (green curves). Note the linear increase during time. Below is shown the distance of the main clusters from the bs (dbs-clust, brown curves). Here, binding affinity Eb-bs = 3.1 KBT and concentration c is ∼1.5%. (C) shows the same as in (B), with Eb-bs = 5.1 KBT. (D) The average distance <dbs-clust> at early stage of the dynamics (see Methods) for different concentrations c and binding affinities Eb-bs. (E) The number of binders Nb of the first four largest clusters monitored over a highly time-resolved simulation. In dynamics 1, the gradual and continuous decrease of the top second cluster (green curve) and the gradual increase of the top first cluster (red curve) indicate an evaporation process. Conversely, in dynamics 2, the top second cluster drops and the top first cluster increases, respectively, in a discontinuous way, indicating the coalescence between the two (green plus red). Simulations were performed with Eb-b = 3.0 KBT, Eb-bs = 3.1 KBT, and c being ∼1.9%. (F) Schematic representation of evaporation and coalescence, the microscopic mechanisms that regulate PS dynamics. To see this figure in color, go online.
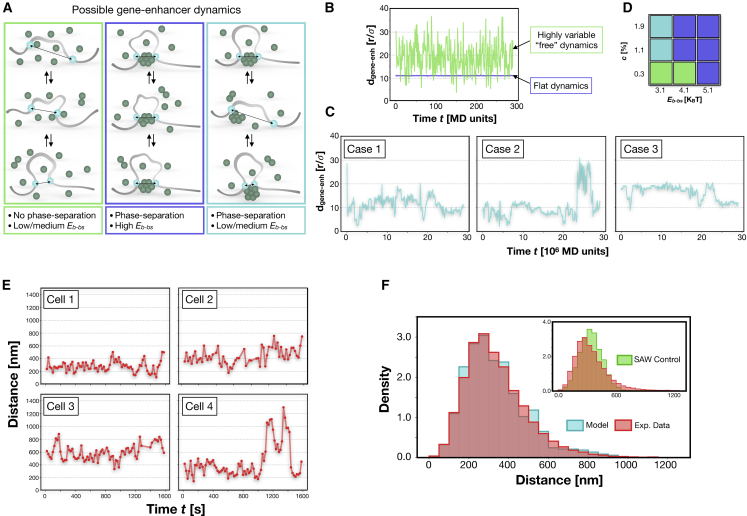
Figure 3.
The PS process produces different regimes of gene-enhancer contact dynamics. (A) The affinity between bs and binders influences the gene-enhancer contacts in time, and different dynamics emerge. (B) An example of gene-enhancer distance dynamics in which a stable contact is not formed (green curve, Eb-bs = 3.1 KBT, Eb-b = 3.0 KBT, and c is ∼0.3%, and there is no phase-separated cluster) and in which a highly stable contact (blue curve, Eb-bs = 5.1 KBT, Eb-b = 3.0 KBT, and c is ∼1.1%) mediated by a phase-separated cluster is formed. (C) Three examples of contact dynamics for intermediate affinity Eb-bs = 3.1 KBT (Eb-b = 3.0 KBT). Cases 1 and 2 are with c ∼1.1%, and case 3 is with c ∼1.9%. Here, the contact is stable, yet results are much more variable because the bs can move on the surface of the cluster and can also detach. (D) Diagram summarizing the different dynamic regimes and their corresponding parameters. (E) Examples of experimental single-cell gene-enhancer dynamics from Sox2 and its superenhancer SCR in mouse embryonic stem cells. Data are taken from (33). The profile results are very similar to the simulated dynamics in (C). (F) Comparison between the model (cyan) and experimental (red) distributions of distances. The distributions are consistent (Kolmogorov-Smirnov KS test, p-value > 0.01). The inset shows the comparison with the control distribution of the free SAW case (KS test, p-value = 10−24). To see this figure in color, go online.
Each polymer is initialized to a random SAW polymer configuration (35). The binders are uniformly distributed in the box as the simulation starts, with a concentration per volume unit c = (4πr3/3) × Ntot/D3, where Ntot is the total number of binders, and r is the radius of the binder. Note that c is linked to the molar concentration cm through the relation c = (cmσ3) × NA, where NA is the Avogadro number and σ is the physical length scale. In the figures, concentrations are rescaled by the geometric factor 4πr3/3 ∼ 0.5. To check the robustness of the results, we verified that a reduction of the binder size led to analogous results upon rescaling of other quantities (such as Ntot and D) so as to keep the binder concentration per volume unit c in the explored range of values.
For each parameter choice, we performed 10 independent simulations, which were equilibrated up to 20 × 107 timesteps so as to ensure the phase transition. Starting from the initial state, the configurations were sampled logarithmically in time and then, after 105 timesteps, taken every 105 timesteps, except for simulations presented in Fig. 4 E and Videos S1 and S2, in which configurations were sampled every 2 × 103 timesteps.
Clustering method
To obtain size distributions of binder aggregates, we use a standard iterative clustering method, in which the Euclidean distance among the binders is used as metric. To define the clusters, we set as threshold distance the cutoff Rint of the LJ interaction and defined as a cluster the set of particles whose mutual distance is smaller than Rint, using the Python package scipy.cluster. To avoid wrong counts in the cluster number, for each frame analyzed, we implemented iterative boundary corrections, in which the simulation box is replicated at each side and the configuration having the minimal number of clusters is considered. At equilibrium, when the fraction of binders Nb/Ntot in the main cluster is above 50%, then a PS event is considered. In the phase diagram of Fig. 1 E, the fraction of system realizations satisfying this condition is reported. When the system is close to the critical threshold, the time required to observe the full PS is longer. In these cases, some replicates do not exhibit the transition in the time window considered. Here, the fraction of realizations in which the transition occurs is less than 1.
Gene-enhancer distance dynamics
Distances dgene-enh are extracted from simulated 3D trajectories in equilibrium conditions; that is when the phase-separated cluster is formed. For each control parameter combination, distance distributions are obtained by considering 10 independent equilibrium trajectories. For sake of simplicity, each polymer binding region is made by three bs and has an average separation of 100 beads. Nevertheless, we verified that, by varying the separation and the number of bs, analogous dynamics regimes are found, as shown in Fig. S3, in which binding regions are made by one and three bs, respectively, separated by 50 beads.
Mixtures of systems using different concentrations are used for the comparison with experimental data. In Fig. 3 F, a 0.25:0.75 mixture in which c is ∼1.9% and c is ∼1.1%, respectively, is shown. By equating the experimental and model averages (24,41), we obtain the factor σ ∼29 nm that maps the dimensionless length scale in physical units. Because the genomic distance between Sox2 and SCR is ∼100 kb, the genomic content of each bead in our polymer results is ∼1 kb. Note that, in this case, equilibrium clusters count thousands of binders because simulations were performed with a bigger box to avoid finite size effects. Experimental trajectories are taken from (33) mouse embryonic stem cells.
As a characteristic biological time to compare with, we consider the average time between two contact events, defined when dgene-enh is shorter than the cluster diameter. For the parameter range explored, values of characteristic time roughly range in the interval 3–4 × 106 timesteps.
Equilibrium properties of the phase-separated cluster
The fraction of binders μ that detach from the cluster is defined as (Nb − Nin)/Nb, where Nb is the average number of binders in the cluster and Nin is the number of binders that never escapes from the cluster in a fixed time interval. A binder is considered detached if its distance from the cluster center is larger than the estimated size of the cluster. Analogously, the average attaching time τ spent by a single binder in the cluster in a time interval Δt is defined as <tin/Δt>, where tin is the number of times that the binder is found attached to the cluster and < > indicates average over the binders. In Fig. 2, D and E, the reported values are computed in a time interval Δt = 4.5 × 106 timesteps in equilibrium conditions in which they result in a plateau regime, as shown in Fig. S2 B.
The mean-square displacement (MSD) is calculated according to the standard formula , where d(t) is the distance from the center of the cluster at time t and Δt is the time interval considered. MSD is calculated and averaged over the binders that never escape from the cluster in the time interval Δt. A power-law fit MSD(tlag) = a × (tlag)α is performed over the first 10 tlag points. As for higher values of tlag, the number of configurations in the sum reduces, and the fluctuations of MSD are larger. For all the described quantities, ensemble averages are performed.
Dynamics of the cluster formation
To study the dynamics of the cluster formation, we consider system configurations from the initial state up to equilibrium states. For each configuration, we find the distribution of the cluster size Nb and select the largest ones. At each time, an ensemble average is performed, namely over the different replicas of the dynamical process.
The distance dbs-clust between one specific polymer bs and the clusters plotted in Fig. 4, B and C, is defined as mini([dist(bs,clusti)]), where “i” labels the first three major clusters ranked according their size Nb. Again, an ensemble average is performed. Estimation of the decay time for dbs-clust is made with an exponential fit dbs-clust(t) = a × exp(t/b) + c and returns b ∼1.3 × 105 and b ∼7.1 × 103, expressed in MD timesteps, for Eb-bs = 3.1 KBT and Eb-bs = 5.1 KBT, respectively.
Average <dbs-clust> in Fig. 4 D is defined as , where the sum runs over the first three timesteps with at least one cluster with Nb > 15, “i” labels the first three major clusters, and “j” labels the two bs.
Results and Discussion
The simulated system
To investigate the PS process mediating the contact between distant loci on the chromatin filament, we consider a model consisting of a polymer in which two distant binding regions are located. Such regions can attractively interact with particles (named “binders” in the following) floating in the surrounding environment. Biologically, the polymer represents a chromatin filament, and the bs represent the enhancer and the promoter of its target gene (Fig. 1), whereas the binders mimic the molecular factors that normally tie to chromatin in the cell nucleus (e.g., TFs). Importantly, binders can also attractively interact among each other in a nonspecific manner because it is known that TFs undergo weak attractive interactions through their intrinsically disordered regions (1,2), as highlighted by experimental evidence in vitro (3,6) and in vivo (5). Thus, the system is regulated by three main parameters: the binder concentration c, the interaction affinity between the binders and the DNA bs Eb-bs, and the interaction affinity among the binders Eb-b (see Methods). In general, for a fixed value Eb-bs, if c and Eb-b are above a certain threshold, a phase-separated cluster of binders can form and mediate a stable contact between distant bs (Fig. 1 A). Details about other specific parameters in the simulations, such as temperature and arrangement of the binding regions along the polymer, can be found in the Methods.
PS process occurs with a switch-like behavior
We first study the simple case with a weakly interacting polymer (i.e., with low Eb-bs values). Analogous results are obtained for the system in the absence of the polymer. To quantitatively study the PS process, we fix the binder concentration and vary the affinity Eb-b. To evaluate the thermodynamic state of the system, we consider the quantity Nb/Ntot, where Ntot is the total number of binders and Nb is the number of binders contained in the largest of the clusters that spontaneously may form (see Methods). The binding affinity with the polymer is set to Eb-bs = 3.1 KBT, where T is the temperature and KB the Boltzmann constant (see Methods). As shown in Fig. 1 B, if the interaction affinity Eb-b is below a transition value, no macroscopic cluster of binders is observed at equilibrium. Conversely, if the affinity is higher than an energetic threshold, a macroscopic phase-separated cluster is stable at equilibrium. The transition value is identified between Eb-b = 2.7 KBT and Eb-b = 3.0 KBT for the considered concentration c ≈ 1.1%, where c is expressed as volume fraction (see Methods).
Next, we fix the affinity at an above threshold value Eb-b = 3.0 KBT and vary the binder concentration c (Fig. 1 C, upper panel). Again, the system undergoes a transition and a phase-separated cluster is observed if c is above a threshold identified around c ≈ 0.7%. In Fig. 1 C (lower panel) we report also the absolute number Nb of binders that belong to the largest phase-separated cluster. In general, the number Nb depends both on c and Eb-b. For instance, by varying the affinity Eb-b from 2.7 to 4 KBT, Nb/Ntot ranges from roughly 80% to values higher than 95% (c ≈ 1.9%, Fig. 1 D). For the values of the affinity explored, which fall in the weak biochemical range, and the considered size of the system (see Methods), the minimal number of binders required for the formation of phase-separated clusters results in approximately hundreds, which is consistent with the number of molecules (200–400) found in in vivo stable condensates of PolII and mediator complexes (5).
All the discussed results are summarized in the phase diagram in Fig. 1 E, in which the z axis reports the fraction of system realizations that exhibit phase transition in the time window considered (see Methods). As previously specified, the affinity with polymer is Eb-bs = 3.1 KBT. A very similar phase diagram is found for the system made of only binders (Fig. S1 A).
The interaction with the polymer bs can macroscopically influence the system
Next, we investigate how the presence of the polymer with its bs influences the PS process. To this aim, we consider different values of Eb-bs ranging approximately from 3 to 5 KBT (Fig. 2 A) and study the equilibrium properties of the system by analyzing the cluster distribution (see Methods) in each considered condition.
We find that when the binding affinity Eb-b is very low (≤2.5 KBT), no macroscopic PS is observed for any value of Eb-bs and of the binder concentration (Fig. S1 B). Nevertheless, for high values of the concentration c, small clusters form whose size depends on Eb-bs, likely because of the presence of the bs that, for high-interaction affinities, produce a local increase in the binder density (as sketched in Fig. 1 A; Fig. S2 A). This effect becomes important and induces the macroscopic transition when the interaction energy Eb-b is increased to values proximal to the transition threshold in the absence of polymer or with a weakly interacting polymer (see Methods). Indeed, the presence of the polymer for large Eb-bs triggers the PS transition for sufficiently high values of c, even though the transition is not observed for lower values of Eb-bs and the same concentration c. This is shown in the bar plots in Fig. 2 B, where the fraction of binders in the largest cluster is reported for different values of concentration c and binding affinity Eb-bs.
It is interesting to stress that such macroscopic changes are induced by a very small perturbation to the system because the concentration of bs (around 0.02%; see Methods) is roughly two orders of magnitude lower than the binder concentration range explored. These results suggest that the combined action of the nonspecific interaction among the binders and their interaction with the bs on the polymer cooperatively influences the evolution of the system.
The phase-separated cluster exhibits a dynamical structure
In this section we focus on the structural properties of the clusters for different values of the control parameters. To this aim, we observe the behavior of the cluster by tracking in time, in equilibrium conditions, the distance of its binders from the cluster center (see Methods) by monitoring the respective trajectories. In Fig. 2 C, we show, as an example, such distance during time for four different binders, each tracked with a different color. From these tracks, two fundamental properties emerge. First, the phase-separated cluster is originated from a dynamic equilibrium because binders attach and detach from the cluster surface with a certain frequency (Fig. 2 C, left panels). Second, the cluster is characterized by a nontrivial internal mobility, with the single binders adsorbed inside the cluster moving for distances comparable with the size of the cluster. This last aspect is highlighted by the zoom plots (Fig. 2 C, right panels). Of course, binders are differently tied to the cluster, depending on the interaction affinity Eb-b, as shown for two values of the interaction affinity reported in the figure (Eb-b = 2.7 KBT, upper panels and Eb-b = 3.0 KBT, bottom panels).
More quantitatively, we calculate the fraction of binders μ that detach from the cluster within a fixed time interval (see Methods; Fig. S2 B). Interestingly, for low affinity (Eb-b = 2.7 KBT), we find that ∼70% of the binders escape at least one time (Fig. 2 D) over timescales comparable to the estimated characteristic time between two contact events involving the regulatory elements (see Methods). This implies that such highly dynamic exchange occurs as fast as typical biological processes, in agreement with the liquid-like nature of protein condensates having high exchange rates. Analogously, the estimated average value of attaching time (indicated with τ) (i.e., the relative time spent attached to the cluster within a fixed long observation time) results roughly 70% (Fig. 2 E). Such values vary sensibly upon the increase of Eb-b (e.g., μ and τ result in ∼50 and 90%, respectively, for Eb-b = 3.0 KBT). Finally, we focused on the internal mobility of the cluster and calculated the MSD (Fig. 2 F) of the binders that are always contained in the cluster by varying the lag time tlag (see Methods). The behavior is subdiffusive because we observe a rough behavior MSD of ∼(tlag)α, with α approximately ranging from 0.5 to 0.7 (see Methods), slightly decreasing with larger Eb-b.
Gene-enhancer interaction and cluster dynamics
Next, we investigate how the phase-separated cluster influences the interaction between the bs located along the polymer, representing an enhancer and a gene promoter on the chromatin filament. This is an interesting aspect of the model because in real cells, complex structural relationships exist between enhancers and target genes, whose features depend on the genomic region and its regulatory landscape (15). For instance, the contact can be invariant with respect to the transcriptional activity of the gene, such as the Shh gene with its enhancer ZRS (42), or it can be highly tissue-specific, such as the Pitx1 gene and its enhancer Pen (43). Many other biological examples are reviewed in (15).
In our simplified framework, we can easily study the equilibrium dynamics of the distance between the bs, as visually depicted in Fig. 3 A. We consider three possible scenarios: in the first case (highlighted in green) no phase-separated cluster is formed (that is, low concentration c or low-affinity Eb-b), and the affinity between binders and bs (Eb-bs) is not sufficiently high to mediate stable contacts. In this case, the bs come in spatial proximity very rarely, and interactions only occur as due to random fluctuations of their positions. A typical example of this “free” dynamics is shown in Fig. 3 B (green curve), in which the distance dgene-enh between bs is plotted against time. In the second case (highlighted in blue in Fig. 3 A), the phase-separated cluster mediates the contact between the bs with a very strong affinity and completely constrains their motion. The resulting distance dynamics (flat blue curve in Fig. 3 B) is very stable and practically constant over very long times. Of course, the distance value depends on the relative position of the bs when they start to interact with the cluster.
The third case (highlighted in cyan in Fig. 3 A) occurs when the phase-separated cluster interacts with the bs with an intermediate affinity Eb-bs. In Fig. 3 C, three examples of distance dynamics are shown that highlight the deep difference with the former cases. Indeed, here the single bs can detach from the cluster and the distance dgene-enh can increase in short time intervals. This dynamic behavior is compatible with the dynamic “kissing” model that has been proposed to explain experimental observations in which, for instance, the Esrrb gene is found to sporadically colocalize with the mediator cluster (5). Furthermore, the bs are much more mobile on the cluster surface. As a consequence, the resulting dynamics, although still influenced by the presence of the cluster, is richer than in the case of strong affinity. By acting on the binder concentration, it is also possible to expand the range of values of the equilibrium distances between the bs, as shown in the right-hand plot in Fig. 3 C (labeled as case 3), in which the higher concentration c ensures a larger equilibrium distance. We stress that the three different dynamic regimes are all observed in equilibrium conditions and naturally emerge by simply varying the control parameters (i.e., c, Eb-b, and Eb-bs).
The schematic diagram in Fig. 3 D summarizes the parameters corresponding to each of the described regimes. System details, such as the number of bs used to model the gene or the enhancer (see Methods), that biologically correspond to the regulatory landscape of the region under consideration can influence the values of the concentration and affinity that determine the kind of dynamic regime. Also, more realistic models would require the use of binders with different size because real proteins span different lengths (approximately in the range 5–30 nm (44)). However, we verified that, upon rescaling of the system parameters, similar results are found, and the overall dynamical behaviors described above remain qualitatively unchanged (Fig. S3; see Methods).
Gene-enhancer temporal dynamics
We compare, here, our results from the model dynamics with recently published imaging data (33), in which the distance between the Sox2 gene (chr3: 34, 548, 927-34, 551, 382, mm9) and its distal superenhancer SCR (chr3: 34653927–34660927, mm9) has been tracked in vivo in different murine tissues. In Fig. 3 E, we show four examples of such dynamics, each corresponding to a different individual cell. In making the comparison, we have considered only embryonic stem cells, in which the Sox2 gene is active through the contact with SCR (45), as also confirmed by high-resolution Hi-C data (46). As discussed in (33), from the experimental tracks, it is possible to appreciate the variety of behaviors that individual cells can exhibit, although they all belong to the same type. A first visual comparison with our results shows that the situation corresponding to the cyan dynamical regime in Fig. 3 A gives rise to distance dynamics (Fig. 3 C) that are qualitatively similar to the experimental ones (Fig. 3 E).
More quantitatively, we computed the histogram of the experimental distances and compared it with the same quantity obtained from our numerical simulations (Fig. 3 F; see Methods). To make a meaningful comparison, a suitable scaling of distances is used (see Methods). Similar to the experimental case, we see that the model distribution is not bimodal (33) (Hartigan’s dip test p-value > 0.1 in both cases) and is statistically compatible with the experiments (Kolmogorov-Smirnov KS test p-value > 0.01). On the contrary, the control distribution obtained from the “free” dynamics case is not able to describe the experiments (see inset of Fig. 3 F, KS test p-value = 10−24). Taken together, these results indicate that the formation of the phase-separated aggregate and its interaction with the gene/enhancer sites act in a cooperative manner to ensure a robust but dynamic contact that can sometimes be inhibited by thermal fluctuations.
In this comparison, the chromatin fiber is simply modeled with a uniform string of beads having a fixed genomic content (see Methods). In general, more details could also be taken into account, such as the heterogeneous genomic content along the chromatin experimentally observed in vivo (47), the above-mentioned size of the binders, and a more complex arrangement of bs.
Microscopic mechanisms underlying PS
In the previous sections, we showed that with sufficiently high-binding-affinity Eb-b, the presence of the bs along the polymer can induce a macroscopic transition that is not observed for lower affinities (Fig. 2, B and C). This observation prompted us to investigate the early microscopic dynamics leading to the formation of the phase-separated cluster and the role of the bs played in such process.
From our simulations, two main mechanisms emerge, as schematically depicted in Fig. 4 A. In the first mechanism, the PS is not initially influenced by the presence of the bs, and the cluster mediates the contacts only when it is already (or almost) formed (see Fig. 4 A, upper panel). In the second mechanism, bs induce the formation of small-sized clusters that, acting as nucleation sites, grow and eventually merge together, leading to the full phase transition with the formation of the macroscopic cluster (see Fig. 4 A, bottom panel). It is worth mentioning here that recent technological developments (CasDrop method (48)) allowed us to experimentally investigate such process and, in analogy with our findings, have highlighted that artificial protein aggregates, targeted to specific seeded loci, are able to pull in spatial proximity distant chromatin regions.
For a quantitative study of the above mechanisms, we have monitored the growth of the largest clusters by tracking their number of binders during time (green curves in Fig. 4, B and C; see Methods). In parallel, we have also monitored the average distance between such clusters and the bs (dbs-clust, brown curves in Fig. 4, B and C; see Methods). In this way, we tested whether the two processes are connected and, therefore, which mechanism regulates the system evolution. Note that, in any case, the cluster growth is well described by a linear increase in time, as shown by the black dashed line in Fig. 4, B and C. This is expected from classical results in statistical mechanics (49,50), as we will discuss below. We find that when Eb-bs is comparable or lower than Eb-b (around 3KBT in the considered case), PS is basically unaffected by the presence of the bs, as the distance dbs-clust decays on a timescale (Fig. 4 B, brown curves) when the cluster is already grown (Fig. 4 B, green curves). On the other hand, if Eb-bs ≫ Eb-b, the binder droplets tend to nucleate around the bs, as witnessed by the fact that the decay time of dbs-clust is approximately one order of magnitude shorter than in the previous case (brown curves, Fig. 4 C; see Methods). In this case, the contact between bs occurs when the cluster counts just a few binders (approximately tens of binders; Fig. 4 C, green curves). The results obtained with different model parameters are summarized in Fig. 4 D, in which the average distance between the bs and the major clusters at early times, well before the PS has been completed (Methods), is reported for different values of c and Eb-bs, Eb-b being ∼3 KBT. The described analysis, in which cluster growth and distance between bs and clusters are observed during time, could be a possible experimental strategy to understand the dynamics mechanism bridging distant chromatin regions in vivo.
Finally, we have investigated the microscopic process driving the formation of the phase-separated clusters. To this aim, we performed a highly time-resolved analysis (see Methods) of the growth of the cluster. In Fig. 4 E, two independent examples (labeled as 1 and 2) are reported, in which the number of binders Nb belonging to the major four clusters is tracked in time (see Methods). From these plots, it is possible to appreciate two different mechanisms occurring at microscopic level. In the upper plot (dynamics 1 in Fig. 4 E), the first and second clusters (red and green curves) regularly grow and compete until the red dominates and the green continuously decreases and eventually drops to negligible values. This behavior is the signature of an evaporation-condensation process, in which the green cluster undergoes a gradual loss of its binders that are adsorbed by the red one after traveling between the two droplets. Conversely, in the bottom plot (dynamics 2 in Fig. 4 E), the green curve suddenly disappears, whereas the red curve has a discontinuous jump, indicating that the two clusters merged together in a coalescence event. Alternatively, it is possible to appreciate the different processes by considering the entire distribution of the cluster size during time, which is dynamically shown in Videos S1 and S2. Evaporation-condensation and coalescence (schematically shown in Fig. 4 F) are therefore the two fundamental microscopic mechanisms that compete/cooperate to the overall PS process. From the theoretical point of view, it is well established (49, 50, 51, 52, 53) that in a 3D system both mechanisms lead to the linear increase of the volume of the aggregate, as indeed confirmed by our simulations (Fig. 4, B and C).
Biologically, coalescence is a well-known process for phase-separated protein aggregates and has been observed in several studies (2,3,5,48). On the other hand, transient small clusters have also been observed (e.g., PolII and Mediator) (5) and could be identified with the transient clusters shrinking because of evaporation. By experimental implementation of the described approach (e.g., by monitoring in time size and geometry of the clusters, as suggested in (31)) with live-imaging techniques at high temporal resolution (5), it would be possible to understand how the two processes are related in real cells and the conditions in which they occur.
Conclusions
In this work, we investigated the biophysical mechanisms behind the PS process that regulate the interaction between genes and enhancers by using an essential polymer model with bs simulating genomic regulatory elements and nonspecifically interacting multivalent binders modeling the proteins, such as TFs, that are present in the nuclear environment and are known to interact with chromatin. Typically, implementations of this polymer model, more commonly known as the strings-and-binders switch (29) or the TFs (27) model, do not take into account the weak nonspecific interaction among molecules, even though the formation of clusters spontaneously emerge as bridging-induced microphase separation (30).
We showed that such a model exhibits phase-transitions, leading to the formation of particle clusters, when the binding affinity among the molecules and molecular concentration are above threshold values. The structural properties of the molecular aggregates, as number of molecules, mobility, and exchange rate, depend on the system parameters and, upon certain conditions, can be highly dynamic in agreement with the liquid-like nature of protein condensates, experimentally highlighted by photobleaching experiments (3,30). By varying the interaction affinity between the binders and the bs of the polymer, that are genes or enhancers, it is possible to obtain gene-enhancer contact dynamics similar to live-imaging experimental data (33) and compatible with the “kissing” model proposed to explain colocalization data of protein clusters with the Esrrb gene (5). Conversely, the presence of the polymer with its bs can influence the PS process because it can be induced and catalyzed by a nucleation mechanism that can produce macroscopic effects. The dynamic and equilibrium properties of the system are therefore the result of a collective behavior emerging from the interplay between PS and interactions among the binding molecules, genes, and enhancers.
Naturally, the model can be generalized in different ways (e.g., by introducing more types of binders with different sizes and specific interactions, which have been shown to generate multilayered aggregates experimentally observed (11)), as well as more details accounting for the protein structure, such as the DNA binding domains and the intrinsically disordered regions (6). Also, more complex arrangements of bs along the polymer and better modeling the regulatory landscape of real genomic regions can be employed to investigate more accurately the associated chromatin architecture (23,24,41). Furthermore, the presence of a mechanoactive surrounding environment that influences and, in turn, is influenced by the formation of phase-separated clusters (48) is neglected in our study and could be implemented (e.g., with a dense viscoelastic matrix of polymers (54)). Nevertheless, although based on an essential model, our work provides a theoretical framework able to recapitulate many features of the PS mechanism and its interplay with the gene-enhancer communication.
Author Contributions
A.M.C., F.C., and M.S. designed the research project. A.M.C. run computer simulations and performed data analyses. A.M.C., F.C., and M.S. wrote the manuscript.
Acknowledgments
We thank Maria Barbi for interesting discussion and Simona Bianco for critical reading and helpful remarks.
A.M.C. acknowledges computer resources from ENEA CRESCO/ENEAGRID (55). We acknowledge funding from MIUR PRIN 2015K7KK8, “Statistical Mechanics and Complexity.”
Editor: Tamar Schlick.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2020.07.007.
Supporting Material
References
- 1.Banani S.F., Lee H.O., Rosen M.K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shin Y., Brangwynne C.P. Liquid phase condensation in cell physiology and disease. Science. 2017;357:eaaf4382. doi: 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- 3.Sabari B.R., Dall’Agnese A., Young R.A. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018;361:eaar3958. doi: 10.1126/science.aar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho W.-K., Jayanth N., Cisse I.I. RNA Polymerase II cluster dynamics predict mRNA output in living cells. eLife. 2016;5:e13617. doi: 10.7554/eLife.13617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho W.-K., Spille J.-H., Cisse I.I. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science. 2018;361:412–415. doi: 10.1126/science.aar4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boija A., Klein I.A., Young R.A. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell. 2018;175:1842–1855.e16. doi: 10.1016/j.cell.2018.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hnisz D., Shrinivas K., Sharp P.A. A phase separation model for transcriptional control. Cell. 2017;169:13–23. doi: 10.1016/j.cell.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strom A.R., Emelyanov A.V., Karpen G.H. Phase separation drives heterochromatin domain formation. Nature. 2017;547:241–245. doi: 10.1038/nature22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larson A.G., Elnatan D., Narlikar G.J. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature. 2017;547:236–240. doi: 10.1038/nature22822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibson B.A., Doolittle L.K., Rosen M.K. Organization of chromatin by intrinsic and regulated phase separation. Cell. 2019;179:470–484.e21. doi: 10.1016/j.cell.2019.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boeynaems S., Holehouse A.S., Gitler A.D. Spontaneous driving forces give rise to protein-RNA condensates with coexisting phases and complex material properties. Proc. Natl. Acad. Sci. USA. 2019;116:7889–7898. doi: 10.1073/pnas.1821038116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barbi M., Place C., Salerno M. A model of sequence-dependent protein diffusion along DNA. J. Biol. Phys. 2004;30:203–226. doi: 10.1023/B:JOBP.0000046728.51620.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dekker J., Mirny L. The 3D genome as moderator of chromosomal communication. Cell. 2016;164:1110–1121. doi: 10.1016/j.cell.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dixon J.R., Gorkin D.U., Ren B. Chromatin domains: the unit of chromosome organization. Mol. Cell. 2016;62:668–680. doi: 10.1016/j.molcel.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robson M.I., Ringel A.R., Mundlos S. Regulatory landscaping: how enhancer-promoter communication is sculpted in 3D. Mol. Cell. 2019;74:1110–1122. doi: 10.1016/j.molcel.2019.05.032. [DOI] [PubMed] [Google Scholar]
- 16.Spielmann M., Lupiáñez D.G., Mundlos S. Structural variation in the 3D genome. Nat. Rev. Genet. 2018;19:453–467. doi: 10.1038/s41576-018-0007-0. [DOI] [PubMed] [Google Scholar]
- 17.Fiorillo L., Bianco S., Chiariello A.M. A modern challenge of polymer physics: novel ways to study, interpret, and reconstruct chromatin structure. WIREs Comput. Mol. Sci. 2020;10:e1454. [Google Scholar]
- 18.Bianco S., Chiariello A.M., Nicodemi M. Computational approaches from polymer physics to investigate chromatin folding. Curr. Opin. Cell Biol. 2020;64:10–17. doi: 10.1016/j.ceb.2020.01.002. [DOI] [PubMed] [Google Scholar]
- 19.Portillo-Ledesma S., Schlick T. Bridging chromatin structure and function over a range of experimental spatial and temporal scales by molecular modeling. WIREs Comput. Mol. Sci. 2020;10:e1434. doi: 10.1002/wcms.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanborn A.L., Rao S.S.P., Aiden E.L. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl. Acad. Sci. USA. 2015;112:E6456–E6465. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fudenberg G., Imakaev M., Mirny L.A. formation of chromosomal domains by loop extrusion. Cell Rep. 2016;15:2038–2049. doi: 10.1016/j.celrep.2016.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buckle A., Brackley C.A., Gilbert N. Polymer simulations of heteromorphic chromatin predict the 3D folding of complex genomic loci. Mol. Cell. 2018;72:786–797.e11. doi: 10.1016/j.molcel.2018.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bianco S., Annunziatella C., Nicodemi M. Modeling single-molecule conformations of the HoxD region in mouse embryonic stem and cortical neuronal cells. Cell Rep. 2019;28:1574–1583.e4. doi: 10.1016/j.celrep.2019.07.013. [DOI] [PubMed] [Google Scholar]
- 24.Chiariello A.M., Bianco S., Nicodemi M. A dynamic folded hairpin conformation is associated with α-globin activation in erythroid cells. Cell Rep. 2020;30:2125–2135.e5. doi: 10.1016/j.celrep.2020.01.044. [DOI] [PubMed] [Google Scholar]
- 25.Bianco S., Lupiáñez D.G., Nicodemi M. Polymer physics predicts the effects of structural variants on chromatin architecture. Nat. Genet. 2018;50:662–667. doi: 10.1038/s41588-018-0098-8. [DOI] [PubMed] [Google Scholar]
- 26.Bascom G.D., Myers C.G., Schlick T. Mesoscale modeling reveals formation of an epigenetically driven HOXC gene hub. Proc. Natl. Acad. Sci. USA. 2019;116:4955–4962. doi: 10.1073/pnas.1816424116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brackley C.A., Taylor S., Marenduzzo D. Nonspecific bridging-induced attraction drives clustering of DNA-binding proteins and genome organization. Proc. Natl. Acad. Sci. USA. 2013;110:E3605–E3611. doi: 10.1073/pnas.1302950110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiariello A.M., Annunziatella C., Nicodemi M. Polymer physics of chromosome large-scale 3D organisation. Sci. Rep. 2016;6:29775. doi: 10.1038/srep29775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barbieri M., Chotalia M., Nicodemi M. Complexity of chromatin folding is captured by the strings and binders switch model. Proc. Natl. Acad. Sci. USA. 2012;109:16173–16178. doi: 10.1073/pnas.1204799109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brackley C.A., Marenduzzo D. Bridging-induced microphase separation: photobleaching experiments, chromatin domains and the need for active reactions. Brief. Funct. Genomics. 2020;19:111–118. doi: 10.1093/bfgp/elz032. [DOI] [PubMed] [Google Scholar]
- 31.Erdel F., Rippe K. Formation of chromatin subcompartments by phase separation. Biophys. J. 2018;114:2262–2270. doi: 10.1016/j.bpj.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scialdone A., Cataudella I., Nicodemi M. Conformation regulation of the X chromosome inactivation center: a model. PLoS Comput. Biol. 2011;7:e1002229. doi: 10.1371/journal.pcbi.1002229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alexander J.M., Guan J., Weiner O.D. Live-cell imaging reveals enhancer-dependent Sox2 transcription in the absence of enhancer proximity. eLife. 2019;8:e41769. doi: 10.7554/eLife.41769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allen M.P., Tildesley D.J. Oxford University Press; New York: 1989. Computer Simulation of Liquids. [Google Scholar]
- 35.Kremer K., Grest G.S. Dynamics of entangled linear polymer melts: a molecular-dynamics simulation. J. Chem. Phys. 1990;92:5057–5086. [Google Scholar]
- 36.Barbieri M., Xie S.Q., Pombo A. Active and poised promoter states drive folding of the extended HoxB locus in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 2017;24:515–524. doi: 10.1038/nsmb.3402. [DOI] [PubMed] [Google Scholar]
- 37.Annunziatella C., Chiariello A.M., Nicodemi M. Polymer models of the hierarchical folding of the Hox-B chromosomal locus. Phys. Rev. E. 2016;94:042402. doi: 10.1103/PhysRevE.94.042402. [DOI] [PubMed] [Google Scholar]
- 38.Annunziatella C., Chiariello A.M., Nicodemi M. Molecular dynamics simulations of the strings and binders switch model of chromatin. Methods. 2018;142:81–88. doi: 10.1016/j.ymeth.2018.02.024. [DOI] [PubMed] [Google Scholar]
- 39.Plimpton S. Fast parallel algorithms for short-range molecular dynamics. J. Comput. Phys. 1995;117:1–19. [Google Scholar]
- 40.Rosa A., Everaers R. Structure and dynamics of interphase chromosomes. PLoS Comput. Biol. 2008;4:e1000153. doi: 10.1371/journal.pcbi.1000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brackley C.A., Brown J.M., Marenduzzo D. Predicting the three-dimensional folding of cis-regulatory regions in mammalian genomes using bioinformatic data and polymer models. Genome Biol. 2016;17:59. doi: 10.1186/s13059-016-0909-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paliou C., Guckelberger P., Andrey G. Preformed chromatin topology assists transcriptional robustness of Shh during limb development. Proc. Natl. Acad. Sci. USA. 2019;116:12390–12399. doi: 10.1073/pnas.1900672116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kragesteen B.K., Spielmann M., Andrey G. Dynamic 3D chromatin architecture contributes to enhancer specificity and limb morphogenesis. Nat. Genet. 2018;50:1463–1473. doi: 10.1038/s41588-018-0221-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maeshima K., Kaizu K., Takahashi K. The physical size of transcription factors is key to transcriptional regulation in chromatin domains. J. Phys. Condens. Matter. 2015;27:064116. doi: 10.1088/0953-8984/27/6/064116. [DOI] [PubMed] [Google Scholar]
- 45.Li Y., Rivera C.M., Ren B. CRISPR reveals a distal super-enhancer required for Sox2 expression in mouse embryonic stem cells. PLoS One. 2014;9:e114485. doi: 10.1371/journal.pone.0114485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bonev B., Mendelson Cohen N., Cavalli G. Multiscale 3D genome rewiring during mouse neural development. Cell. 2017;171:557–572.e24. doi: 10.1016/j.cell.2017.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ricci M.A., Manzo C., Cosma M.P. Chromatin fibers are formed by heterogeneous groups of nucleosomes in vivo. Cell. 2015;160:1145–1158. doi: 10.1016/j.cell.2015.01.054. [DOI] [PubMed] [Google Scholar]
- 48.Shin Y., Chang Y.-C., Brangwynne C.P. Liquid nuclear condensates mechanically sense and restructure the genome. Cell. 2018;175:1481–1491.e13. doi: 10.1016/j.cell.2018.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bray A.J. Theory of phase-ordering kinetics. Adv. Phys. 1994;43:357–459. [Google Scholar]
- 50.Cugliandolo L.F. Coarsening phenomena. C. R. Phys. 2015;16:257–266. [Google Scholar]
- 51.Binder K., Stauffer D. Theory for the slowing down of the relaxation and spinodal decomposition of binary mixtures. Phys. Rev. Lett. 1974;33:1006–1009. [Google Scholar]
- 52.Ahmad S., Corberi F., Zannetti M. Aging and crossovers in phase-separating fluid mixtures. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2012;86:061129. doi: 10.1103/PhysRevE.86.061129. [DOI] [PubMed] [Google Scholar]
- 53.Corberi F., Cugliandolo L.F., Yoshino H. Growing length scales in aging systems. In: Berthier L., Biroli G., Bouchaud J.-P., Cipelletti L., van Saarloos W., editors. Dynamical Heterogeneities in Glasses, Colloids, and Granular Media. Oxford University Press; 2011. pp. 370–406. [Google Scholar]
- 54.Lucas J.S., Zhang Y., Murre C. 3D trajectories adopted by coding and regulatory DNA elements: first-passage times for genomic interactions. Cell. 2014;158:339–352. doi: 10.1016/j.cell.2014.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ponti G., Palombi F., Vita A. IEEE; 2014. The role of medium size facilities in the HPC ecosystem: the case of the new CRESCO4 cluster integrated in the ENEAGRID infrastructure. In 2014 International Conference on High Performance Computing & Simulation (HPCS) pp. 1030–1033. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.