Abstract
Adenosine-to-inosine (A-to-I) editing in the seed sequence of microRNAs can shift the microRNAs’ targetomes and thus their function. Using public RNA-sequencing data, we identified 35 vasoactive microRNAs that are A-to-I edited. We quantified A-to-I editing of the primary (pri-)microRNAs in vascular fibroblasts and endothelial cells. Nine pri-microRNAs were indeed edited, and editing consistently increased under ischemia. We determined mature microRNA editing for the highest expressed microRNAs, i.e., miR-376a-3p, miR-376c-3p, miR-381-3p, and miR-411-5p. All four mature microRNAs were edited in their seed sequence. We show that both ADAR1 and ADAR2 (adenosine deaminase acting on RNA 1 and RNA 2) can edit pri-microRNAs in a microRNA-specific manner. MicroRNA editing also increased under ischemia in vivo in a murine hindlimb ischemia model and ex vivo in human veins. For each edited microRNA, we confirmed a shift in targetome. Expression of the edited microRNA targetomes, not the wild-type targetomes, was downregulated under ischemia in vivo. Furthermore, microRNA editing enhanced angiogenesis in vitro and ex vivo. In conclusion, we show that microRNA A-to-I editing is a widespread phenomenon, induced by ischemia. Each editing event results in a novel microRNA with a unique targetome, leading to increased angiogenesis.
Keywords: microRNA, A-to-I editing, ischemia, peripheral artery disease, angiogenesis, target gene regulation, ADAR, cardiovascular disease
Graphical Abstract
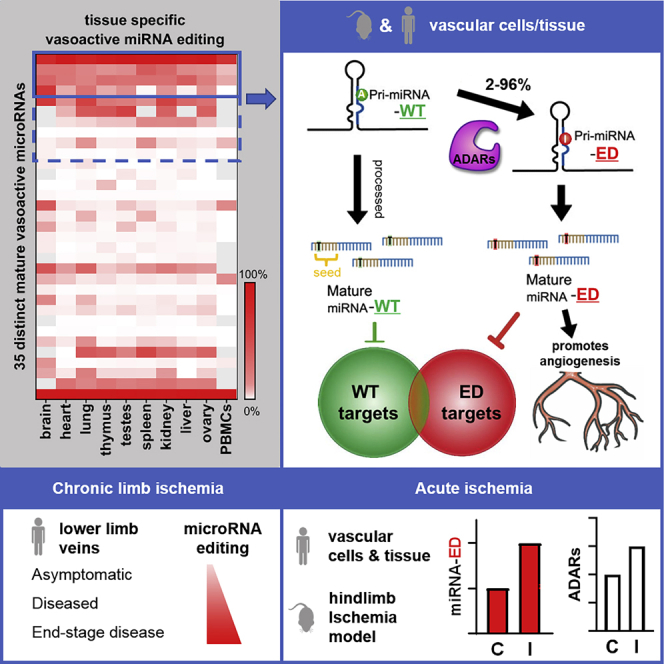
van der Kwast and colleagues demonstrate that a number of microRNAs with a known vascular function are edited by ADARs in vascular cells and tissue. MicroRNA editing consistently increased after limb ischemia and altered the microRNAs’ targets and function, ultimately promoting angiogenesis.
Introduction
MicroRNAs (miRNAs) play an important role in processes involved in cardiovascular disease, including neovascularization, atherosclerosis, hypertension, and aneurysm formation.1 MicroRNAs are short non-coding RNAs that inhibit translation of the mRNAs they target through partial complementary binding. In general, only the binding of nucleotides 2–8 from the 5′ end of the microRNA, a microRNA’s seed region, is required to induce target silencing.2 As a result, microRNAs can target hundreds of mRNAs, allowing them to regulate complex, multifactorial physiological and pathological processes, such as cardiovascular disease.3 Our group has shown that multiple microRNAs from a single microRNA gene cluster, located on the long arm of human chromosome 14 (14q32), are regulated under ischemia and directly affect restoration of blood flow to ischemic tissues.4
MicroRNAs are produced after a series of maturation steps of the primary transcript of microRNA genes (pri-microRNAs).2 Pri-microRNAs fold into hairpin-shaped, double-stranded RNA structures that are sequentially cleaved by ribonucleases Drosha and Dicer, yielding a microRNA duplex. Either side of the duplex can be incorporated into the RNA-induced silencing complex to become a functional mature microRNA, distinguished as either the microRNA on the 5′ or 3′ side of the pri-microRNA hairpin (microRNA-5p or microRNA-3p).2
However, similar to other RNA species, microRNA transcripts can also be modified at the nucleotide level. Adenosine-to-inosine (A-to-I) editing is the most prevalent RNA nucleotide modification that changes the sequence of the RNA molecule.5,6 The inosine preferentially binds to cytidine and is therefore interpreted as guanosine by the cellular machinery. This form of RNA editing is considered an essential post-transcriptional modification, which is regulated in a tissue- and context-specific manner.7 In mammals, A-to-I editing is catalyzed by either ADAR1 or ADAR2 (adenosine deaminase acting on RNA 1 and 2), which are abundantly expressed throughout the body.8
As ADARs target double-stranded RNA structures of at least 20 nt long, A-to-I editing of microRNAs occurs in the pri-microRNA stage.8 In fact, previous studies have suggested that at least 16% of all human pri-microRNAs contain sites that may be subject to A-to-I editing.9,10 Editing of a pri-microRNA can profoundly influence microRNA maturation,11 and several pri-microRNA editing events were shown to be associated with traits, including plasma high-density lipoprotein levels.12 However, if editing occurs in the seed sequences of either the microRNA-5p or microRNA-3p, editing can completely alter the mature microRNA’s target selection, resulting in regulation of a different set of target mRNAs, or “targetome”.13
Recently, we demonstrated that microRNA editing indeed plays a regulatory role in cardiovascular disease. We showed that the pri-microRNA of miR-487b-3p, a microRNA from the 14q32 cluster, is A-to-I edited in the seed sequence following ischemia.14 We found that the edited mature microRNA (ED-microRNA) indeed selects a completely different targetome than does the unedited “wild-type” microRNA (WT-microRNA). Because of this switch in the targetome, ED-miR-487b-3p promotes angiogenesis and neovascularization, whereas WT-miR-487b-3p does not. These findings demonstrated that microRNA editing can play an important role in the endogenous response to pathological stimuli such as ischemia. Whether vasoactive microRNAs besides miR-487b-3p are also subject to A-to-I editing in the vasculature, however, is still unknown.
Therefore, in this study, we aimed to identify vasoactive microRNAs that are robustly A-to-I edited in vascular cells and to examine whether the expression and editing of these microRNAs are regulated under ischemic conditions. Next, we aimed to validate our findings in a murine hindlimb ischemia (HLI) model and subsequently in human vascular tissue. Finally, we examined how editing affects the function of vascular microRNAs, specifically with regard to angiogenesis.
Results
Identification of Potential MicroRNA Editing Sites
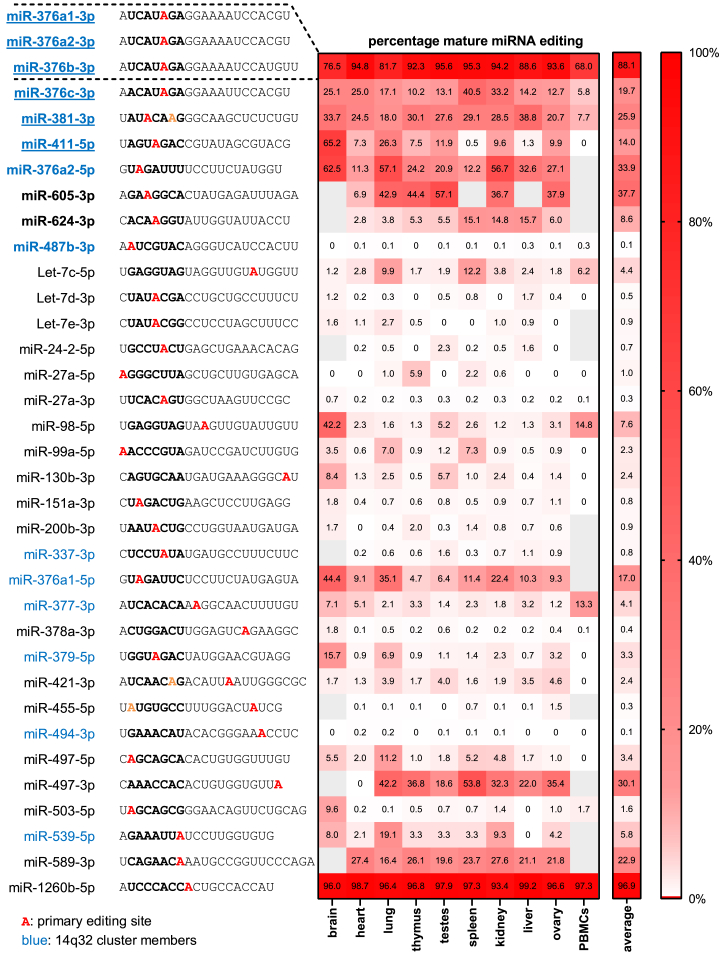
We identified microRNAs containing adenosines that can be subject to A-to-I editing by manual literature curation, in combination with reanalysis of public microRNA sequencing (microRNA-seq) datasets. In total, 60 “editable” mature microRNAs originating from 56 different microRNA genes were found to contain robust, context-dependent A-to-G mismatches, indicative of A-to-I editing (Table S1). 35 of these editable microRNAs (58%), of which the vast majority are located in the 14q32 locus (14 of 35, or 40%), could be linked to vascular functions. These 35 vasoactive microRNAs contain a total of 38 possible editing locations, which display a wide range of percentage of A-to-I editing across major human organs (Figure 1). Most of these potential vasoactive mature microRNA editing locations are within the dominantly expressed mature microRNA of the microRNA gene (25 of 38, 66%), and they are localized within the mature microRNA’s seed sequence (26 of 38, 68%).
Figure 1.
Vasoactive Mature MicroRNAs Containing A-to-G Mismatches Indicative of A-to-I Editing
Heatmap displaying the percentage of A-to-I editing in human tissues of the 35 vasoactive mature microRNAs that were identified to contain potential microRNA editing sites after manual literature curation in combination with reanalysis of public microRNA-seq datasets. The percentage of A-to-I editing was obtained by reanalysis of selected high-quality public datasets (n = 1) and quantification of percentage A-to-G mismatches, indicative of A-to-I editing. Due to their near-complete sequence homology, editing of miR-376a1-3p, miR-376a2-3p, and miR-376b-3p could not be calculated separately, so their overall percentage editing is presented instead. Gray squares indicate insufficient reads (<10). The location of the quantified editing within the microRNA’s sequence is highlighted in red, while the microRNA seed sequence is in bold. Adenosines highlighted in orange were edited to a lower extent than the microRNA’s primary editing site. MicroRNAs in blue are members of the 14q32 microRNA cluster.
MicroRNA Editing in Vascular Cells
Both endothelial cells and adventitial fibroblasts are known to play crucial roles in cardiovascular pathology and angiogenesis.15 Therefore, we determined whether these vasoactive microRNAs are also edited in human umbilical vascular endothelial cells (HUVECs) and human umbilical arterial fibroblasts (HUAFs), cultured either under normal conditions or a combination of hypoxia and serum starvation (hypoxia+starvation) to mimic ischemic conditions (Figure 2A). Measurements of HIF1A, VEGFA, and p53 expression and visual inspection of the cells validated that the culture conditions successfully induced hypoxia signaling and cell cycle arrest in both HUVECs and HUAFs (Figures S1A–S1D).
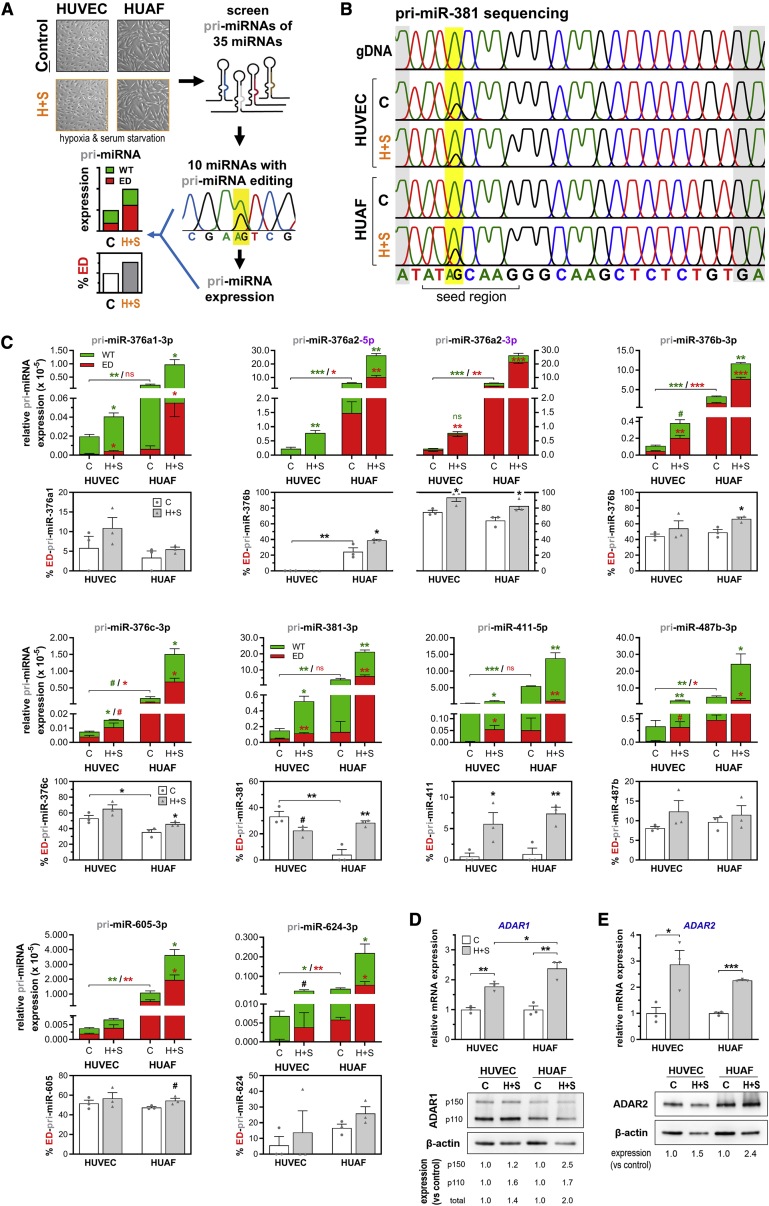
Figure 2.
Identification of A-to-I Editing within Vasoactive pri-MicroRNAs and Regulation of Expression and Editing under Conditions That Mimic Ischemia
(A) Schematic overview detailing the screening of pri-microRNA A-to-I editing in vascular cells. Human umbilical venous endothelial cells (HUVECs) and human umbilical arterial fibroblasts (HUAFs) were cultured separately under “control” conditions (C) or hypoxia+starvation (H+S) to mimic ischemia. Complementary DNA (cDNA) from these cells was subsequently used to screen the 35 selected vasoactive microRNAs for A-to-I editing by Sanger sequencing. The percentage of pri-microRNA editing quantified from sequencing chromatograms and qPCR quantification of total pri-microRNA expression were used to calculate relative expression of edited pri-microRNAs. (B) Representative chromatograms obtained by sequencing of pri-miR-381. A-to-I RNA editing sites were detected as an A-to-G change in the cDNA sequencing chromatogram, while being absent in the genomic DNA (gDNA) chromatogram. Location of editing within the microRNA’s seed region is highlighted in yellow. (C) Expression of unedited (WT) and edited (ED) pri-microRNA expression relative to U6 (top panels) and percentage pri-microRNA editing (bottom panels). (D and E) Fold change in relative ADAR1 (D) and ADAR2 (E) mRNA and protein expression upon culturing under hypoxia+starvation conditions. In the ADAR1 western blot, two isoforms are visible, known as p150 and p110. Expression of both ADARs was normalized to stable household genes RPLP0 or β-actin (for mRNA and protein expression, respectively) and expressed as fold change of the CAD group. All data are presented as mean ± SEM from three independent experiments performed with pooled cells from a total of 13 different umbilical cords. #p < 0.01, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, versus control condition unless otherwise indicated by a two-sided Student’s t test.
Screening of each of the selected pri-microRNAs revealed that 10 vasoactive microRNAs (29%), i.e., miR-376a1-3p, miR-376a2-5p and −3p, miR-376b-3p, miR-376c-3p, miR-381-3p, miR-411-5p, miR-605-3p, miR-624-3p, and miR-487b-3p, were indeed A-to-I edited in human vascular cells (Figures 2B and 2C). Except for pri-miR-605 and pri-miR-624, all edited pri-microRNAs are transcribed from the 14q32 microRNA mega-cluster. Each detected vascular pri-microRNA A-to-I editing location corresponds with one of the potential microRNA editing sites that we identified previously. Strikingly, each editing event is located within the microRNA’s seed sequence, and all edited microRNAs, except for miR-376a2-5p and miR-624-3p, represent the dominant microRNA of the precursor microRNA duplex, indicating that each editing event is potentially functionally significant. In fact, these edited seed sequences are different from any other validated microRNA seed sequence16 (TargetScan; http://www.targetscan.org/vert_72/, release 7.2; Table S2).
pri-MicroRNA Editing
Quantification of pri-microRNA editing and expression revealed that the rate of editing is regulated under hypoxia+starvation (Figure 2C). We observed that both pri-microRNA editing and expression generally increase under these culture conditions in both cell types, resulting in a consistent increase in the expression of the ED-pri-microRNA. An exception to the rule was pri-miR-381, for which the editing rate decreased under hypoxia+starvation from 33% to 22% (p < 0.1) in HUVECs. In HUAFs, however, pri-miR-381 editing was induced approximately 6-fold under hypoxia+starvation, as was the editing rate of pri-miR-411 (pri-miR-381, 4.3%–28%, p < 0.007; pri-miR-411, 1.3%–7.5%, p < 0.02). The baseline editing rate also differed between HUVECs and HUAFs; pri-miR-376c and pri-miR-381 showed a 1.5-fold and 6-fold higher editing rate in HUVECs compared to HUAFs, respectively, while pri-miR-376a2-5p was only edited in HUAFs (pri-miR-376c, 35.5% versus 53%, p < 0.03; pri-miR-381, 4.3% versus 27%, p < 0.007). Baseline pri-microRNA expression was consistently higher and was also induced stronger by hypoxia+starvation in HUAFs than in HUVECs. Combined, these results suggest that regulation of A-to-I editing under ischemic conditions is cell type-specific.
Consistent with these findings, hypoxia+starvation also induced the expression of A-to-I editing enzymes ADAR1 (both p150 and p110 isoforms) and ADAR2, both at the mRNA and protein level (Figures 2D and 2E). Protein levels of ADAR1 and ADAR2 increased more in hypoxic+starved HUAFs compared to hypoxic+starved HUVECs (at least 2-fold versus ∼1.5-fold induction, respectively). Regulation of ADAR expression in response to either serum starvation or hypoxia differed between HUVECs and HUAFs, indicating that ADARs are also regulated in a cell type-specific manner (Figures S1E and S1F).
Mature MicroRNA Editing
To determine whether these edited pri-microRNAs are processed into edited mature microRNAs, the four microRNAs with the highest expression were selected, i.e., miR-376a-3p (the mature microRNA produced by both pri-miR-376a1 and pri-miR-376a2), miR-376c-3p, miR-381-3p, and miR-411-5p (Figure S2A). Table S3 provides a comprehensive overview of all our subsequent findings organized per selected microRNA. Expression levels of WT-microRNA and ED-microRNA were quantified by version-specific quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) assays (Figure 3A). Due to technical limitations, ED-miR-376a-3p could not be distinguished from ED-miR-376b-3p, which has near complete sequence homology, resulting in quantification of their sum instead, i.e., ED-miR-376a+b-3p. miR-376b-3p is generally expressed at 20-fold lower levels than miR-376a-3p (Figure 3B). We found that each of the selected edited pri-microRNAs is indeed processed to an edited mature microRNA in both HUVECs and HUAFs. All of these ED-microRNAs, except for ED-miR-376c-3p, have a completely novel seed sequence (Figure 3A) and thus form novel mature microRNAs with a unique targetome.
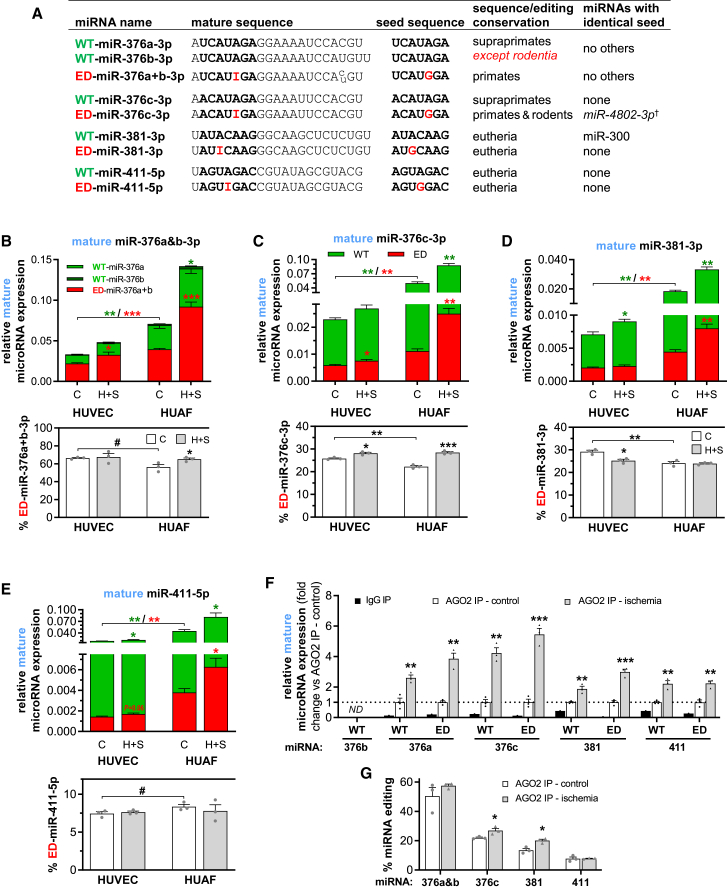
Figure 3.
Specific Quantification of Normal and Edited Mature MicroRNAs in Vascular Cells
(A) Unedited mature microRNAs (WT-microRNAs) and edited mature microRNAs (ED-microRNAs) were quantified using version-specific TaqMan qRT-PCR assays. Since inosines resulting from A-to-I editing are recognized as a G, the functional seed sequence of each microRNA is also highlighted and whether this seed is unique or shared by a different microRNA is noted. †Unvalidated microRNA according to TargetScan.16 (B–E) Relative mature WT-microRNA (green) and ED-microRNA (red) expression of (B) miR-376a&b-3p, (C) miR-376c-3p, (D) miR-381-3p, and (E) miR-411-5p in HUVECs and HUAFs cultured in control (C) conditions or hypoxia+starvation (H+S) to mimic ischemia (top panels) and the corresponding percentage of mature ED-microRNA (bottom panels). Relative microRNA expression was normalized to U6. (F) Relative expression of WT-microRNAs and ED-microRNAs in fractions after negative control IgG or AGO2 was immunoprecipitated from HUAFs cultured under control or hypoxia+starvation conditions. Data are expressed as fold change of the control AGO2 immunoprecipitation (IP). (G) Percentage editing measured in AGO2 IP fractions. All data are presented as mean ± SEM from three independent experiments performed with pooled cells from a total of 13 different umbilical cords. #p < 0.01, ∗∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, versus control condition unless otherwise indicated by a two-sided Student’s t test. Symbol color shows whether means of either WT or ED expression are compared.
Mature MicroRNA Editing under Ischemic Conditions
Baseline mature microRNA expression was consistently higher in HUAFs compared to HUVECs, but the percentage of mature ED-microRNAs was generally lower (Figures 3B–3E). The percentage of ED-miR-376a+b-3p and ED-miR-376c-3p was approximately 60% and 25%, respectively, a decrease compared to the percentage editing of pri-miR-376a2-3p and pri-miR-376c-3p in both cell types, suggesting certain edited pri-microRNAs are processed less efficiently than their WT counterparts. Despite this, increases in the percentage of miR-376a+b-3p and ED-miR-376c-3p during hypoxia+starvation still mirrored changes in the percentage of pri-microRNA editing. Moreover, consistent with pri-microRNA editing findings, only HUVEC mature miR-381-3p editing decreased with hypoxia+starvation. These aspects of ischemia also consistently induced total mature microRNA expression. As a result, expression of all mature ED-microRNAs also significantly increased under hypoxia+starvation in both HUVECs and HUAFs.
A microRNA has to be associated with an argonaute protein (AGO), the active part of the RNA-induced silencing complex, to be functional. To examine whether the ED-microRNAs are functional, we performed AGO2 immunoprecipitation (IP) on lysates from HUAFs cultured on either normal or hypoxia+starvation conditions and measured the expression of WT-microRNAs and ED-microRNAs in the IP fractions. Samples from normoxic cells showed enrichment of both WT- and ED-microRNAs in the AGO2 precipitate, compared to an immunoglobulin G (IgG) negative control, confirming that both variants of the four investigated microRNAs enter the cell’s microRNA machinery (Figure 3F). Conforming to their increased expression, AGO2 binding of both the WT-microRNA and ED-microRNA variants increased in hypoxic+starved cells. Furthermore, the percentage of microRNA editing increased in the AGO2 precipitates of hypoxic+starved versus normoxic cells for three of the four investigated microRNAs (Figure 3G).
ADAR1 and ADAR2 in MicroRNA Expression and Editing
To investigate the effect of ADAR1 and ADAR2 on microRNA expression and editing, we used short interfering RNAs (siRNAs) to knock down ADAR1 or ADAR2 expression in HUAFs (Figure S3A). Knockdown of either ADAR1 or ADAR2 decreased total pri-microRNA expression by 2-fold to more than 10-fold compared to control siRNA (Figure 4A). Furthermore, ADAR1 knockdown resulted in significant reduction of pri-microRNA editing for all examined pri-microRNAs except for pri-miR-376a2. Knockdown of ADAR2, however, only affected editing of pri-miR-376a1 and pri-miR-376a2.
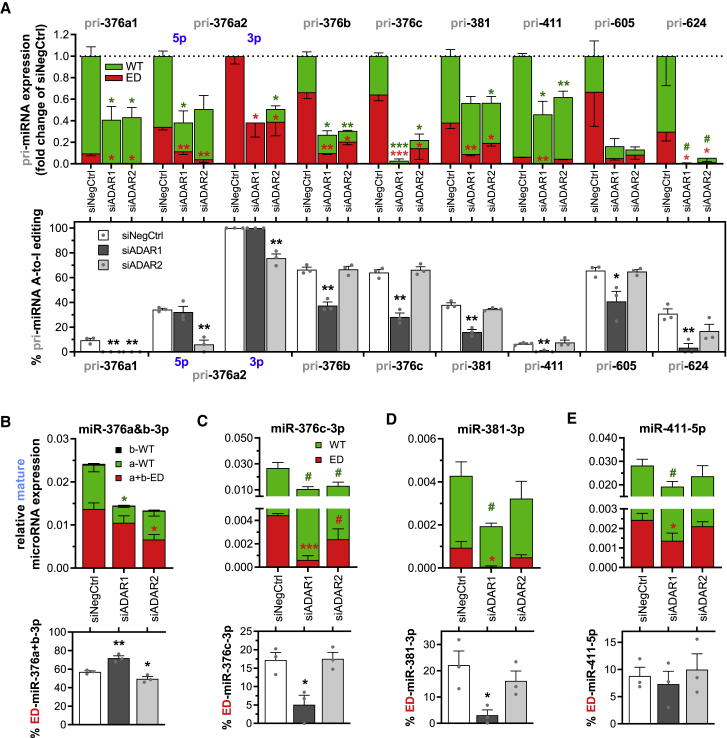
Figure 4.
Role of ADAR1 and ADAR2 in Vasoactive MicroRNA Editing and Maturation
(A–E) ADAR1 and ADAR2 were knocked down in HUAFs through transfection of an ADAR1-targeting or ADAR2-targeting siRNA (siADAR1 and siADAR2, respectively). The subsequent effects on microRNA expression and editing were analyzed as in Figures 2 and 3 and compared to transfection with a negative control siRNA (siNegCtrl). (A) pri-MicroRNA A-to-I editing and expression. (B–E) Mature (B) miR-376a&b-3p, (C) miR-376c-3p, (D) miR-381-3p, and (E) miR-411-5p expression relative to U6 (top panels) and the corresponding percentage of mature microRNA-ED (bottom panels). All data are presented as mean ± SEM from three independent experiments performed with pooled cells from 13 different umbilical cords. #p < 0.01, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, versus siNegCtrl or as indicated by a two-sided Student’s t test.
Consistent with these findings, mature microRNA expression and editing was similarly affected by the knockdown of ADARs (Figures 4B–4E). ADAR1 knockdown reduced mature microRNA expression as well as mature microRNA editing for all measured microRNAs, except for editing of miR-376a+b-3p, which increased instead. ADAR2 knockdown also decreased total mature microRNA expression but instead only decreased miR-376a+b editing. Our data suggest that both ADARs play a role in expression of these 14q32 microRNAs, while ADAR1 is responsible for A-to-I editing of all mature microRNAs except for miR-376a-3p.
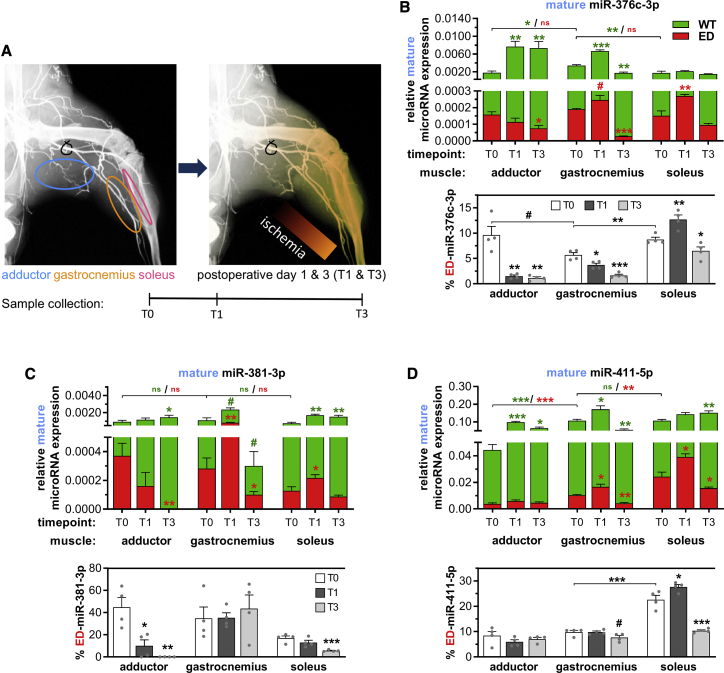
In Vivo MicroRNA Editing
We subjected C57BL/6 mice to HLI via single ligation of the left femoral artery and measured mature WT-microRNAs and ED-microRNAs in distinct whole-muscle tissues: the adductor, which remains relatively normoxic, and the gastrocnemius and the soleus, which become ischemic17 (Figure 5A). Of the four selected microRNAs, all editing events are conserved in mice except for miR-376a-3p. The seed sequence and editing are not conserved in the murine mmu-miR-376a-3p, and therefore mmu-miR-376a was excluded from all experiments using murine tissue. We observed differences in baseline microRNA expression and the percentage of ED-microRNA between muscle tissues (Figure 5B). Furthermore, expression of ED-miR-376c-3p, ED-miR-381-3p, and ED-miR-411-5p was increased in the gastrocnemius and soleus muscles 1 day (T1) after HLI compared to before surgery (T0), but not in the adductor muscle. In contrast, at day 3 after surgery (T3) ED-microRNA expression reduced to sub-T0 levels. Adar1 expression also increased only in the ischemic gastrocnemius and soleus muscles, while Adar2 expression was induced in all three muscles (Figures S3B–S3D). These results confirm that vasoactive microRNAs are edited in response to ischemia in vivo as well as in vitro.
Figure 5.
Mature MicroRNA Editing and Expression after Induction of Hindlimb Ischemia in Muscles Experiencing Ischemia or Increased Shear Stress
(A) Schematic representation of hindlimb ischemia induction by ligation of the femoral artery and the resulting ischemia downstream of the ligation site. Three different muscle tissues were harvested before surgery (T0) or 1 and 3 days after surgery (T1 and T3). While the gastrocnemius and soleus experience ischemia after surgery, the adductor remains relatively normoxic due to its more upstream anatomical location. (B–D) Mature (B) miR-376c-3p, (C) miR-381-3p, and (D) miR-411-5p expression relative to U6 (top panels) and the corresponding percentage of mature microRNA-ED (bottom panels) in the harvested muscles, presented as in Figure 3. All data are presented as mean ± SEM of muscles harvested from four different mice per time point. #p < 0.01, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, versus T0 unless otherwise indicated by a two-sided Student’s t test. Symbol color shows whether means of either WT or ED expression are compared.
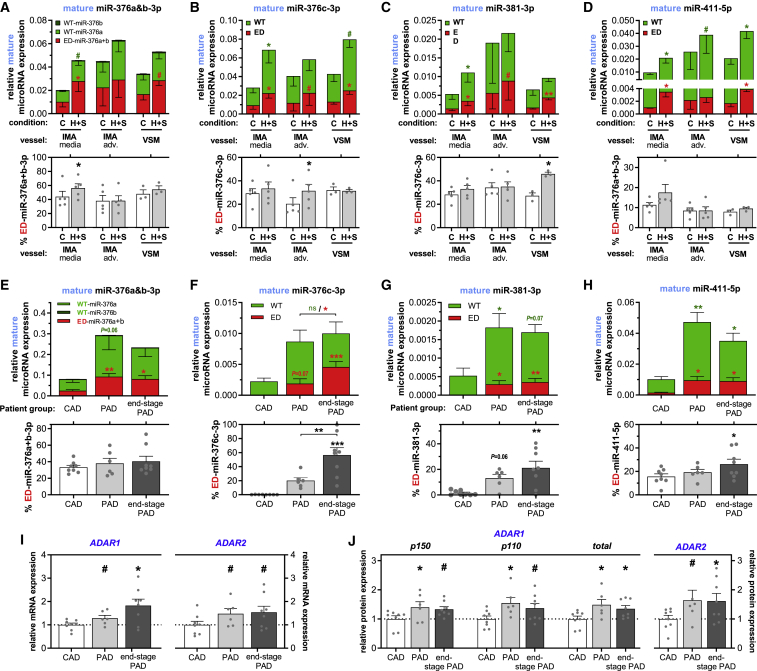
MicroRNA Editing in Human Tissues and Vascular Disease
To examine whether microRNA expression and editing in human vascular tissue responds similarly to ischemic conditions, we used human venae saphenae magnae (VSMs) and internal mammary arteries (IMAs), which were harvested during elective coronary bypass surgery on patients with coronary artery disease (CAD). After culturing the vessels ex vivo under control or hypoxia+starvation conditions for 24 h, expression and editing of all four selected microRNAs were measured. Prior to this, the IMAs were separated manually into the tunica adventitia and the tunica media/intima, while the VSMs were left intact. Consistent with our in vitro and in vivo ischemia models, expression of all ED-microRNAs increased significantly after hypoxia+starvation in all vessel segments, except for the adventitia of the IMAs, where trends toward increased expression were observed (Figures 6A–6D). As before, WT-microRNA expression was often also increased after hypoxia+starvation treatment. Nevertheless, a significant increase in the percentage of editing was observed in at least one of the vessel segments for miR-376a+b-3p, miR-376c-3p, and miR-411-5p (IMA adventitia, IMA media, and VSM, respectively).
Figure 6.
Mature MicroRNA Editing and Expression in Normoxic and Ischemic Human Vessels
(A) miR-376a+b-3p, (B) miR-376c-3p, (C) miR-381-3p, and (D) miR-411-5p expression and editing in internal mammary arteries (IMAs, n = 5) and venae saphenae magnae (VSMs, n = 3) cultured in ex vivo control (C) conditions or hypoxia+starvation (H+S) to mimic ischemia. Right before harvest, IMAs were separated manually into the tunica media/intima (media) and the tunica adventitia (adv.), whereas VSMs were left intact. #p < 0.01, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, versus its T0 by a paired two-sided Student’s t test. Symbol color shows whether means of either WT or ED expression are compared. (E) miR-376a+b-3p, (F) miR-376c-3p, (G) miR-381-3p, and (H) miR-411-5p expression and editing of the same microRNAs in lower leg vein (LLV) samples from three different patient groups with minimal to end-stage peripheral artery disease (PAD). Normoxic LLV samples (n = 8) from patients with coronary artery disease (CAD) rather than PAD undergoing coronary artery bypass surgery were compared to LLV samples (n = 6) from patients with severe PAD undergoing femoral artery to popliteal artery bypass surgery and to LLV samples (n = 8) from patients with end-stage PAD, undergoing lower limb amputation. All mature microRNA expressions are normalized to U6. (I and J) Relative ADAR1 and ADAR2 mRNA (I) and protein (J) expression in the LLV samples from different patient groups. Expression was normalized to stable household genes RPLP0 or β-actin (for mRNA and protein expression, respectively) and expressed as fold change of the CAD group. See Figure S4 for the western blots used for the ADAR protein quantification. For (E) and (J), all data are presented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, versus T0 unless otherwise indicated by one-way ANOVA.
Next, we examined microRNA A-to-I editing in patients with ischemic disease by comparing mature microRNA expression in lower leg vein (LLV) samples from patients with CAD but without clinically actionable peripheral artery disease (PAD) undergoing coronary artery bypass surgery to LLV samples from patients with severe PAD undergoing femoral artery to popliteal artery-bypass surgery and to LLV samples from patients with end-stage PAD, undergoing lower limb amputation.
We observed increased expression and editing in LLV tissues from PAD patients compared to CAD patients (Figures 6E–6H). Of the microRNAs examined, ED-microRNA expression increased by at least 3-fold, and only the percentage editing of miR-376a+b-3p remained largely unaffected. We also observed an increase in ADAR1 and ADAR2 mRNA and protein expression in the veins from both PAD and end-stage PAD patients (Figures 6I, 6J, and S4). These data suggest that microRNA editing is actively regulated in ischemic disease in humans.
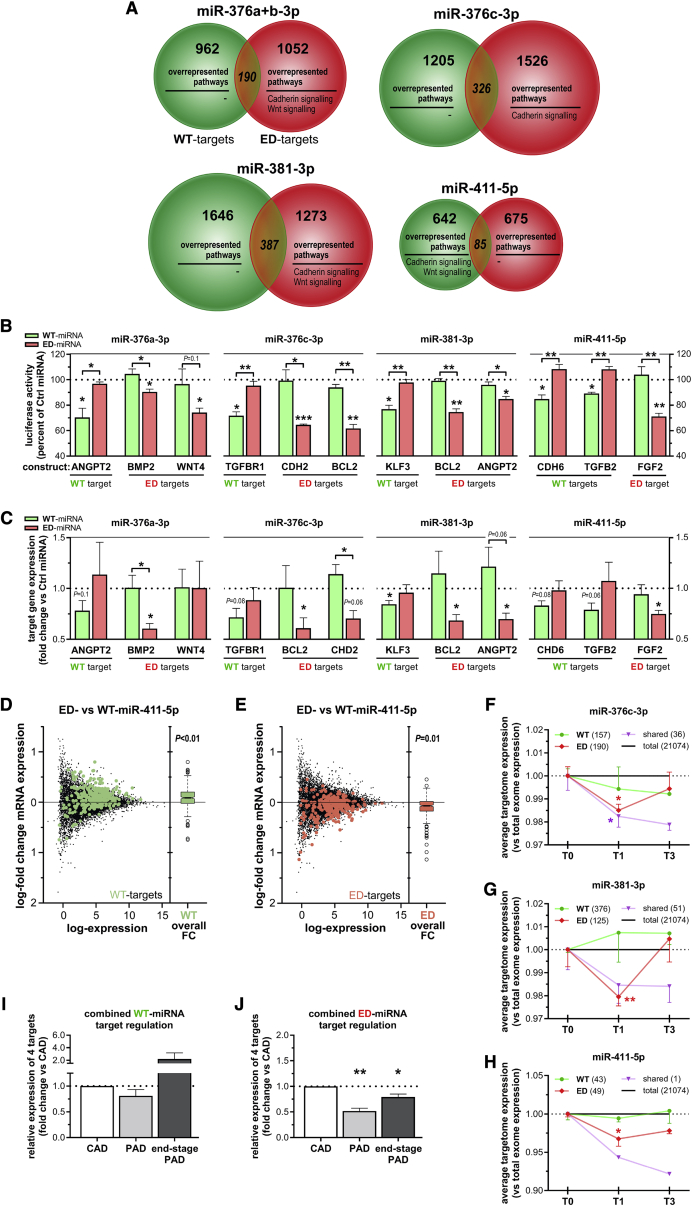
Editing Induces a Shift in MicroRNA Targetomes
Since editing of the microRNA resulted in a completely novel seed sequence for all but one microRNA, changes in putative targetomes of the edited microRNAs were determined using three distinct target prediction algorithms. We found that each WT-microRNA targetome had less than 25% overlap with its respective ED-microRNA targetome. The remaining overlap in targetomes was caused entirely by mRNAs containing two separate binding sites for both the WT-microRNA and the ED-microRNA (Figure 7A), rather than single sites that can bind both microRNA variants. In this analysis, the targetomes of miR-376a-3p and miR-376b-3p were combined, as they are nearly identical, both in microRNA sequence and in targetome (more than 95% overlap between individual WT targetomes and ED targetomes).
Figure 7.
Targetome Predictions and Validation of Target Sequence Binding and Endogenous Target Regulation
(A) Venn diagram of putative targetomes for WT-microRNA (green) and ED-microRNA (red) representing the putative targets that all three prediction algorithms indicated to be targeted. Within each putative targetome, significantly enriched pathways were displayed (see Table S4). (B) To examine the change in target binding induced by editing, luciferase assays were performed with endogenous microRNA binding sequences from vasoactive targets for either a WT-microRNA or a ED-microRNA as indicated. MicroRNA-specific target regulation was assessed by examining luciferase reporter activity after co-transfecting HeLa cells with a WT-microRNA mimic (green) or a ED-microRNA mimic (red), normalized to transfection of a non-targeting microRNA mimic (Ctrl microRNA). (C) Endogenous target regulation within HUAFs after transfection-mediated overexpression of microRNA mimics as indicated. For (B) and (C), data shown represent the averages of three independent experiments and are presented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, by a one-sample t test versus Ctrl microRNA or a two-sided Student’s t test to compare WT-microRNA versus ED-microRNA treatments. (D and E) The log fold changes (FCs) of mRNA expression calculated from RNA-seq data obtained after overexpression of ED-miR-411-5p compared to overexpression of WT-miR-411-5p in HUAFs. Data represent averages of three independent experiments and are visualized using mean-difference plots, highlighting the predicted target genes (with a 0.5 binding score threshold to minimize false positives; see Table S6) that were uniquely targeted by the WT-miR-411-5p (green, D) or ED-miR-411-5p (red, E). The overall distribution of the logFC per targetome is shown in the boxplots. Differential expression of each targetome was tested using ROAST.62Figure S6 shows target gene regulation by WT-miR-411-5p and ED-miR-411-5p compared to a non-targeting control microRNA. (F–H) Analysis of conserved targetome expression with a predicted binding score of >0.5 (see Table S7) in whole-muscle tissue of C57BL/6 mice subjected to HLI using whole-genome expression data obtained via microarray.21 Mean fold change in targetome expression of WT, ED, or shared targets (green, red, and purple, respectively) of miR-376c-3p (F), miR-381-3p (G), and miR-411-5p (H) targets after HLI. Per targetome, the number of conserved targets that were detected above array background levels is indicated between brackets. Targetome expression was normalized to expression of total number of genes detected (black line) and presented as mean ± SEM of at least three different mice per time point. ∗p < 0.05, ∗∗p < 0.01, versus whole-genome expression by a two-sided Student’s t test. (I and J) The combined average mRNA expression of the WT-microRNA targets (I) or ED-microRNA targets (J) validated in (B) and (C) within the LLV samples from three different patient groups (see Figure 6): CAD patients (LLVs without PAD, n = 8), PAD patients (LLVs suffering severe PAD, n = 6), and end-stage PAD patients (LLVs suffering end-stage PAD, n = 8). Combined average expression levels were calculated using the average expression of individual target genes shown in Figure S7 and are expressed as fold change of the CAD group. Data are presented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, by a one-sample t test.
Pathway enrichment analysis of each targetome using the PANTHER algorithm18 revealed that editing of the selected microRNAs shifts their targetome toward overrepresentation of cadherin signaling and/or Wnt signaling, with the exception of ED-miR-411-5p, which shifts away from overrepresentation of cadherin signaling and Wnt signaling (Figure 7A; Table S4).
Genes within each edited targetome could be linked to processes involved in the response to ischemia, including bone morphogenetic protein 2 (BMP2, target of ED-miR-376a+b-3p), cadherin-2 (CDH2, target of WT-miR-411-5p and ED-miR-376c-3p), and B cell lymphoma 2 (BCL2, target of ED-miR-376c-3p and ED-miR-381-3p) (Table S5).
Validation of Target mRNA Regulation
Next we set out to validate whether the single nucleotide change found in each seed sequence of ED-microRNA indeed causes a shift in target site selection. We performed dual-luciferase reporter gene assays using endogenous putative binding sequences from at least one vasoactive target per microRNA (Figure 7B). The luciferase activity of the WT-miR-381-3p binding site containing the Krüppel-like factor 3 (KLF3) sequence was indeed only repressed by WT-miR-381-3p to 77% ± 3% (p < 0.02) and not by ED-miR-381-3p (p = 0.7). Conversely, luciferase activity of the ED-miR-381-3p binding site containing BCL2 and angiopoietin-2 (ANGPT2) sequences were only inhibited by ED-miR-381-3p to 74% ± 2% (p < 0.001) and 85% ± 2% (p < 0.02), respectively. WT-microRNA- versus ED-microRNA-specific regulation of vasoactive target sequences was similarly validated for miR-376a-3p, miR-376c-3p, and miR-411-5p. These results confirm that each editing event results in a complete shift in target site recognition.
We then overexpressed either the WT-microRNA or ED-microRNA in HUAFs and examined endogenous target mRNA regulation (Figure 7C). Consistent with luciferase results, treatment with WT-miR-381-3p decreased endogenous KLF3 expression by 19% ± 6% (p = 0.05) but did not affect BCL2 and ANGPT2 expression (p = 0.6 and p = 0.4, respectively). Treatment with ED-miR-381-3p did not affect KLF3 expression (p = 0.6), while expression of both BCL2 and ANGPT2 were repressed by 30% (p > 0.04 for both). Similarly, overexpression of other WT-microRNAs consistently decreased WT targets only. Furthermore, treatment with ED-microRNAs successfully repressed only the ED targets. Only WNT4 did not appear to be affected by ED-miR-376a-3p in addition to WT-miR-376a-3p, possibly through indirectly being affected by regulation of other ED-miR-376a-3p targets. Overall, however, average vasoactive target repression by ED-microRNAs was stronger than average target repression by WT-microRNAs (p < 0.001, Figure S5E). This is in line with previous findings for WT-miR-487b-3p versus ED-miR-487b-3p and could be a consequence of the additional I-C bond (similar to a G-C bond) between ED-microRNAs and their target mRNAs, which is stronger than the A-U bonds between WT-microRNAs and their target mRNAs.14
To further validate our target predictions at a transcriptome-wide level, we compared the target gene regulation after overexpression of WT-miR-411-5p and ED-miR-411-5p using RNA-seq. miR-411-5p was chosen because it was highly expressed in lower limb vein (LLV) samples, its editing is highly conserved, and we have previously shown that other isoforms of this microRNA impact the cellular response to ischemia.19 We focused on the genes uniquely targeted by either WT-miR-411-5p and ED-miR-411-5p and filtered the predicted target genes based on using a binding score similar to or better than those of the targets that were confirmed with other in vitro assays (i.e., a binding score >0.5) as a threshold value to minimize the inclusion of false-positive putative targets.14,16,20 The numbers of predicted targets detected in the RNA-seq dataset are summarized in Table S6.
Consistent with our previous findings regarding target regulation, we found that overexpression of WT-miR-411-5p specifically led to a global downregulation of the WT targetome compared to a non-targeting control microRNA, while ED-miR-411-5p specifically downregulated the ED targetome (Figure S6). A direct comparison between ED-miR-411-5p and WT-miR-411-5p overexpression showed that the ED-miR-411-5p specifically decreases the ED targetome, while alleviating the repression of the WT targetome relative to WT-miR-411-5p overexpression (Figures 7D and 7E).
In Vivo Target Regulation
To examine target regulation in vivo after ischemia, we used whole-genome expression data obtained via microarray in whole-muscle tissue of C57BL/6 mice subjected to HLI.21 For this analysis, we took the human WT and ED targetomes of miR-376c-3p, miR-381-3p, and miR-411-5p (but not of miR-376a-3p due to lack of editing conservation). First, we determined which predicted human targets are conserved as predicted targets in mice. Next, we also filtered these predictions using a binding score threshold of 0.5 to minimize the inclusion of false-positive putative targets.14,16,20 The numbers of conserved predicted targets, as well as the fraction of predicted targets detected in the microarray dataset, are summarized in Table S7.
The average expression of all three ED-microRNAs’ targetomes were downregulated significantly 1 day after induction of ischemia (Figures 7F–7H). In contrast, the WT-microRNAs’ targetomes were not downregulated at any time point. Genes that are potentially targeted by both the ED-microRNAs and WT-microRNAs were also downregulated at both 1 and 3 days after induction of HLI.
Next we examined whether the human targets that we validated (see Figures 7B and 7C) were indeed regulated in the ischemic LLV samples from patients with PAD compared to the normoxic LLVs from patients with CAD. Targets of WT-microRNAs were not downregulated in the veins from patients with either PAD or end-stage PAD (Figure S7A). In contrast, ED-microRNA targets BCL2, BMP2, CHD2, and FGF2 showed decreased expression in veins from all PAD patients, compared to CAD patients (Figure S7B). A fifth ED-microRNA target, WNT4, was not expressed in these human LLVs. When combined into a single set of targets, the expression of the ED-microRNA targets was decreased significantly in both intermittent PAD patients (p = 0.001) and end-stage PAD patients (p = 0.03, Figure 7J), whereas the expression of the WT-microRNA target set was not regulated (Figure 7I).
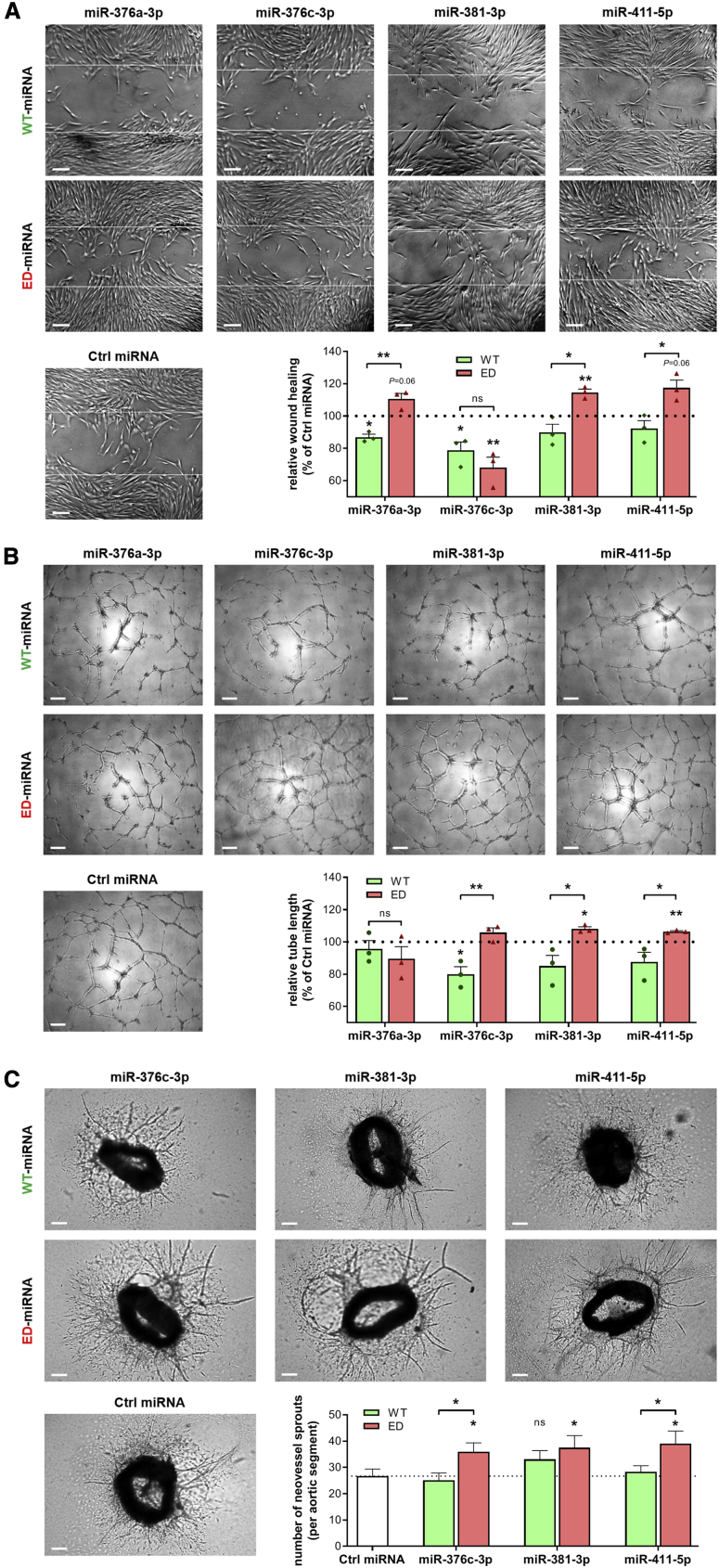
Functional Effects of MicroRNA Editing
Finally, the functional implications of the observed A-to-I editing events were examined using overexpression of either the WT-microRNA or ED-microRNA in three different functional assays. First we performed scratch-wound healing assays in HUAFs. We validated that transfection-mediated microRNA overexpression results in the desired changes in the percentage of microRNA editing (Figures S5A–S5D). Overexpression of all four WT-microRNAs reduced scratch-wound healing compared to the control. Treatment with ED-miR-376c-3p resulted in a comparable reduction in scratch-wound healing as with WT-miR-376c-3p. However, treatment with ED-miR-376a-3p, ED-miR-381-3p, and ED-miR-411-5p resulted in increased scratch-wound healing compared to the control and to their WT-microRNA counterparts (Figure 8A).
Figure 8.
Functional Effect of WT-MicroRNAs and ED-MicroRNAs on In Vitro and Ex Vivo Angiogenesis
(A) Representative images and quantification of scratch-wound healing after overexpression of a WT-microRNA mimic or ED-microRNA mimic in HUAFs relative to a non-targeting microRNA mimic (Ctrl microRNA). White lines indicate original scratch wound area. (B) Representative images and quantification of HUVEC tube formation after similar treatment with microRNA mimics as indicated. (A and B) Data are presented as mean ± SEM from three independent experiments performed with pooled cells from a total of 13 different umbilical cords. ∗p < 0.05, ∗∗p < 0.01, by a one-sample t test versus Ctrl microRNA or a two-sided Student t test to compare WT-microRNA versus ED-microRNA treatments. (C) Representative images and quantification of neovessel sprouting from aortic ring segments treated with microRNA mimics as indicated. Data are presented as mean ± SEM of at least 30 aortic segments per treatment, originating from 11 different mice. ∗p < 0.05, versus control condition unless otherwise indicated by a two-sided Student’s t test. Scale bars, 200 μm.
Next we studied the functional effects of ED-microRNAs in HUVECs by examining tube formation on Matrigel. We observed that overexpression of ED-miR-376c-3p, ED-miR-381-3p, and ED-miR-411-5p induced more HUVEC tube formation than did their WT-microRNA counterparts (Figure 8B).
Finally, we studied the effects of microRNA editing on complex neovessel growth by culturing murine aortic segments ex vivo. We found that, compared to control microRNA-mimic treatment, overexpression of ED-miR-376c-3p, ED-miR-381-3p, and ED-miR-411-5p induced the outgrowth of neovessel sprouts, while their WT-microRNA counterparts did not (Figure 8C).
Taken together, these three experimental setups demonstrate that ischemia-induced editing of vascular microRNAs enhances their angiogenic potential.
Discussion
In this study, we demonstrate that post-ischemic induction of pri-microRNA A-to-I editing is a widespread phenomenon in vascular cells, occurring in at least 10 vasoactive microRNAs (including miR-487b-3p).14 MicroRNA editing appears relatively pervasive in the vasculature, as pri-microRNA A-to-I editing was found in 29% of the 35 vasoactive microRNAs that we identified as editable in a context-dependent manner. For the four most prevalent microRNAs, i.e., miR-376a-3p, miR-376c-3p, miR-381-3p, and miR-411-5p, we demonstrate that induction of pri-microRNA editing by mimicking ischemic conditions also results in a significant increase in functional ED-microRNAs. The expression of these microRNAs was also increased after HLI in vivo and in veins from PAD patients. Strikingly, all the A-to-I editing events that we identified were located in the seed sequence of the microRNAs. We validated that seed-sequence editing indeed causes a shift in target regulation for each of the four most prevalent microRNAs, leading to pro-angiogenic functional changes in in vitro and ex vivo assays.
Our study shows an unprecedented number of microRNA editing events that are actively regulated in response to a pathological stimulus. Moreover, the newfound edited vascular microRNAs have a significantly higher percentage of editing at both baseline and under ischemic conditions compared to our initial discovery of ischemia-induced editing of miR-487b-3p. A study by Nigita et al.22 examined microRNA editing after hypoxia, albeit using a breast adenocarcinoma cell line and RNA-seq instead. However, the authors could not establish unidirectional regulation of microRNA editing or expression. In fact, they found only five microRNAs that displayed A-to-I editing at low percentages, and none of these was a microRNA that we identified as edited in vascular cells. A probable explanation for the differences in microRNA editing is the tissue and context specificity of A-to-I editing.7 This hypothesis is supported by the differences in microRNA expression and A-to-I editing that we found between different human organs, vascular cell types, and vessels (Figures 1, 3, and 5 respectively). Additionally, studies have shown that microRNA A-to-I editing is generally reduced in human cancer tissues, which could explain the low editing observed in the human cancer cell line.23,24
We observed that expression of both ED-pri-microRNAs and mature ED-microRNAs is consistently increased in cells cultured under hypoxia+starvation to mimic ischemic conditions. However, the increase in percentage of pri-microRNA editing did not always result in a similar increase in the percentage of mature microRNA editing, such as for miR-411-5p. Similarly, a previous study also observed differences between pri-miR-411-5p and mature miR-411-5p editing in human brains.25 These differences in editing rates support the principle that pri-microRNA editing can affect microRNA processing dynamics, resulting in a reduced processing efficiency compared to the unedited microRNA.11,26,27
A-to-I editing is directed by the enzymes ADAR1 and ADAR2. In the present study we showed that ischemia-induced vasoactive microRNA editing was consistently paired with increased expression of both ADARs in vitro and in vivo. ADAR1 and ADAR2 are also known to play editing-independent roles in microRNA biogenesis and maturation, however.28, 29, 30 We found that this was indeed the case for all of the examined microRNAs, as repression of either ADAR1 or ADAR2 also resulted in reduced expression of both WT and ED mature microRNAs. Additionally, knockdown of ADARs also resulted in reduced expression of the pri-microRNAs. Since these primary microRNAs were formed from spliced-out introns or intergenic transcripts that are usually degraded,31 our data suggest that ADARs may help preserve such pri-microRNA-containing transcripts and facilitate microRNA biogenesis.
However, where processing was influenced by both ADARs equally, editing of specific microRNAs was regulated by either ADAR1 or ADAR2 specifically. With the exception of pri-miR-376a1&2, and the previously reported pri-miR-487b,14 all vascular pri-microRNA editing depended solely on ADAR1. For pri-miR-376a2 and pri-miR-381-3p editing, these findings were in accordance with previous ADAR perturbation experiments on human cancer cell lines.27,32 For the rest of the vasoactive pri-microRNA editing, our results provide the first clear validation of their dependency on ADAR1 in human cells. Editing of pri-miR-376a2 was exclusively ADAR2-dependent. Alternatively, pri-miR-376a1 editing depends both on ADAR1 and ADAR2, similar to what we previously showed for editing of pri-miR-487b-3p.14 The mechanisms behind the selectivity of A-to-I editing of pri-microRNAs remain unclear, as ADAR1 and ADAR2 edit distinct sets of microRNAs without a strict target sequence specificity.8,33,34 Nevertheless, microRNA editing events are often strongly conserved, in contrast to editing events of other RNA species in the human transcriptome.24,35 Many of the microRNA-editing events that we observed are conserved across species, including miR-381-3p, miR-411-5p, and the miR-376 family, suggesting their biological importance.13,24
Indeed, all of the editing sites that we identified were located within the seed sequence of the microRNAs. Therefore, these editing events could all lead to a novel microRNA, the ED-microRNA, which inhibits a completely different set of target mRNAs. The seed sequence of each of the microRNAs that we found to be edited is different from any validated microRNA.16 This confirms that each of these A-to-I editing events produces an entirely new microRNA with a novel targetome. Through luciferase reporter gene assays and validation of endogenous targets we showed that seed-sequence editing of miR-376a-3p, miR-376c-3p, miR-381-3p, and miR-411-5p indeed completely shifts its target site selection (Figure 7). Finally, we demonstrated the relevance of ED-microRNAs in vivo by showing that the increased expression of the edited microRNAs in mouse muscles goes hand-in-hand with a decreased expression of ED-microRNA targets but not of WT-microRNA targets.
Interestingly, pathway enrichment analyses of the putative targetomes revealed that each microRNA editing event altered enrichment for cadherin signaling genes and, for three of the four microRNAs, also for Wnt signaling genes. Cadherins are known to play an important role in vascular cell function and angiogenesis, while Wnt signaling has been implicated to promote vascular remodeling and even cardiovascular regeneration.36,37 Using three different functional assays we confirmed that editing-induced targetome changes result in angiogenesis-associated functional changes. We assessed the effect of WT-microRNAs and ED-microRNAs on in vitro scratch-wound healing and tube formation, and ex vivo neovessel sprouting, and consistently found that ED-microRNAs had increased pro-angiogenic properties compared to the WT-microRNAs. These results are in line with the effects of editing of miR-487b-3p, which also enhanced in vitro and ex vivo angiogenesis.14
A technical limitation to current microRNA-editing studies is the fact that it is yet not possible to manipulate microRNA editing in vascular tissues specifically in vivo without also affecting the WT-microRNA expression or without large-scale off-target effects. An alternative in vivo experimental setup could be to quantify vascular ingrowth into subcutaneously injected Matrigel plugs mixed with synthetic microRNAs. However, the potential clinical relevance of microRNA editing has already been demonstrated in the field of oncology. miR-378a-3p editing was shown to prevent melanoma progression via regulation of PARVA expression,38 and miR-455 editing was shown to repress melanoma growth and reduce metastasis.39 Furthermore, Franzén et al.12 found that specific editing events can sometimes be associated with phenotypic traits, and Stellos et al.40 demonstrated the importance of A-to-I editing of the cathepsin S mRNA in cardiovascular disease. We provide new evidence herein that microRNA editing can play a direct role in cardiovascular disease. We demonstrate that vasoactive mature microRNAs are also edited in vivo in response to ischemia in a murine HLI model and in ex vivo-cultured human arteries and veins. Furthermore, we found that mature microRNA editing is significantly increased in limb veins from patients with PAD, compared to limb veins from patients with CAD (Figure 6). These results suggest that increased microRNA editing is clinically associated with PAD.
Our unbiased screening for editable microRNAs resulted in a striking enrichment for microRNAs from the 14q32 microRNA gene cluster. Indeed of the 10 pri-microRNAs that we confirmed to be edited, 8 microRNAs originate from this single gene cluster, including the 4 highest expressed microRNAs, i.e., miR-376a-3p, miR-376c-3p, miR-381-3p, and miR-411-5p. miR-487b-3p, which we previously reported to be edited, is also transcribed from the 14q32 locus. The 14q32 locus (12F1 in mice) encodes >50 microRNA genes, and we, and others, have previously shown that 14q32 microRNAs, as well as the other types of noncoding RNAs from this locus, play a vital regulatory role in many aspects of cardiovascular physiology and pathology.4,14,41, 42, 43, 44, 45, 46 Additionally, 14q32 microRNAs have been implicated in rapid placental growth during gestation by regulating capillary formation of the placenta’s labyrinth zone, which fits with the microRNAs’ regulatory roles in angiogenesis during adulthood.47 We now show that A-to-I editing of 14q32 microRNAs also contributes to the regulatory role of this locus in vascular remodeling.
In conclusion, we found widespread A-to-I editing of the seed sequence of multiple vasoactive microRNAs, resulting in novel mature microRNAs. Expression of each ED-microRNA was significantly increased after ischemia, and microRNA editing was also induced in ischemic veins from patients. Editing of miR-376a-3p, miR-376c-3p, miR-381-3p, and miR-411-5p causes a complete shift in the targetome of the microRNA. The ED-microRNAs were functionally different from the WT-microRNAs, and all ED-microRNAs enhanced certain angiogenic properties, compared to their wild-type counterparts. This study underlines the relevance of microRNA editing in the response to ischemia. Since the editing events identified herein represent only a small subset of the total pri-microRNA editing events,10 future studies are likely to uncover many more editing events relevant to cardiovascular disease.
Materials and Methods
Identification of Vasoactive MicroRNAs Containing A-to-I Editable Adenosines
To identify vasoactive microRNAs containing adenosines that can be subject to A-to-I editing in a context-specific manner, we combined manual literature curation with reanalysis of public microRNA-seq datasets.
We first searched the PubMed database for studies that identified microRNA editing events by analyzing high-throughput small RNA-seq datasets by searching for the search terms “microRNA” and “A-to-I OR inosine” in the titles and abstracts (performed December 2017). A total of eight studies were identified that fulfilled these criteria and none was excluded.10,23,27,32,48, 49, 50, 51 On average, these studies identified approximately 40 mature microRNAs with statistically significant A-to-I editing events. To ensure that microRNA candidates were confidently edited, we only included editing events that were identified in at least two different studies and/or were previously shown to be ADAR-dependent. When selecting microRNA candidates, no restrictions were applied to magnitude or tissue specificity of the editing to prevent excluding context-dependent editing events. This yielded a total of 60 confidently editable microRNAs (Table S1).
Next, the subset of “vasoactive” editable microRNAs was identified by selecting microRNAs that were linked to vascular biology in previous studies. This was done by searching the microRNAs in the PubMed database combined with the search term “cardiovascular OR vascular OR vessel OR endothelial OR angiogenesis” (within the title and abstract). MicroRNAs were considered vasoactive if at least one search result indicated that the microRNA was involved in or associated with cardiovascular function or disease (Table S1).
Finally, high-quality public microRNA deep-sequencing datasets were reanalyzed using the “miR-seq browser” function of the miRGator webtool (http://mirgator.kobic.re.kr/)52 to examine baseline prevalence and tissue specificity of selected microRNA editing in major human organs. The datasets used were the ones with highest read counts available: GSM548639 (whole brain), SRX050631 (heart), SRX050632 (lung), SRX050633 (thymus), SRX050634 (ovary), SRX050635 (testes), SRX050636 (spleen), SRX050637 (kidney), SRX050638 (liver), and GSM494810 (peripheral blood mononuclear cells [PBMCs]). The percentage of inferred editing was calculated per editable adenosine by dividing total number of reads containing a G mismatch in that position divided by the sum of the reads with an A or a G in that position. In both cases, 3′ templated isomiR reads were included since this does not alter the seed region and thus targets the same genes. miR-376a1-3p, miR-376a2-3p, and miR-376b-3p editing could not be accurately calculated separately due to their near-complete sequence homology, and was therefore expressed as percentage editing of miR-376a+b-3p.
The identification procedure of vasoactive microRNAs containing A-to-I editable adenosines is schematically summarized in Figure S8.
Isolation of Primary Vascular Cells from Human Umbilical Cords
Isolation and culturing of primary vascular human cells was performed as described previously.14
In brief, umbilical cords were collected from full-term pregnancies and used for isolation of either HUAFs or HUVECs.
For HUAF isolation, the tunica adventitia was removed from the umbilical artery and incubated overnight in serum-rich medium (DMEM GlutaMAX [Invitrogen, Gibco, Auckland, New Zealand], 10% heat-inactivated fetal bovine serum [FBS; PAA Laboratories, Pasching, Austria], 10% heat-inactivated human serum, 100 U of penicillin and 100 μg of streptomycin per mL [Lonza, Basel, Switzerland], and nonessential amino acids [Gibco, #11140050]). The next day the adventitia was incubated in a 2 mg/mL collagenase type II solution (Worthington, Lakewood, NJ, USA) at 37°C. The resulting cell suspension was filtered over a 70-μm cell strainer, pelleted, and resuspended and plated in HUAF culture medium (DMEM GlutaMAX [Invitrogen], 10% heat-inactivated FBS [PAA], and 100 U of penicillin and 100 μg of streptomycin per mL [Lonza]).
HUVECs were isolated from the umbilical veins by infusing a flushed vein with 0.75 mg/mL collagenase type II (Worthington Biochemical) and incubated at 37°C for 20 min. The cell suspension was collected and pelleted and resuspended in HUVEC culture medium (M199 [PAA], 10% heat-inactivated human serum [PAA], 10% heat-inactivated newborn calf serum [PAA], 100 U of penicillin and 100 μg of streptomycin per mL [Lonza]), 150 μg/mL endothelial cell growth factor [kindly provided by Dr. Koolwijk, VU Medical Center, Amsterdam, the Netherlands], and 5 U/mL heparin [LEO Pharma, Ballerup, Denmark]). HUVECs were cultured in plates coated with 10 μg/mL fibronectin (Sigma-Aldrich, Steinheim, Germany).
Cell Culture
Cells were cultured at 37°C in a humidified 5% CO2 environment. Culture medium was refreshed every 2–3 days. Cells were passed using trypsin-EDTA (Sigma-Aldrich) at 70%–80% confluency (HUAFs) or 90%–100% (HUVECs). HUAFs were used up to passage 5 and HUVECs up to passage 3. Stock solutions of isolated HUAFs and HUVECs up to passage 2 were stored at −180°C in DMEM GlutaMAX containing 20% FBS and 10% DMSO (Sigma-Aldrich). Experiments were performed with pooled cells isolated from 13 different donors.
In Vitro Acute Ischemia Model
For in vitro ischemia experiments, HUAFs and HUVECs were seeded in separate 12-well plates at 70.000 or 100.000 cell per well, respectively. After 24 h, medium was removed and cells were subjected to either control conditions (normal culture medium and ∼20% oxygen) or by mimicking ischemic conditions for an additional 24 h. Ischemic conditions were mimicked by culturing cells in starvation medium and hypoxia (1% oxygen). HUAF starvation medium consisted of DMEM GlutaMAX (Invitrogen) with 0.1% heat-inactivated FBS (PAA) and 100 U of penicillin and 100 μg of streptomycin per mL (Lonza). HUVEC starvation medium consisted of M199 (PAA), 10% heat-inactivated newborn calf serum (PAA), and 100 U of penicillin and 100 μg of streptomycin per mL (Lonza). At the end of the experiment, cells were washed with PBS and harvested with TRIzol reagent (Invitrogen).
RNA Isolation and cDNA Synthesis
Total RNA was isolated with TRIzol (Invitrogen), according to the manufacturer’s instructions. RNA concentration and purity were examined by NanoDrop (NanoDrop Technologies, Wilmington, DE, USA). For pri-microRNA experiments, DNase treatment was performed using RQ1 RNase-free DNase (Promega, Madison, WI, USA) according to the manufacturer’s instructions. Total complementary DNA (cDNA) was prepared using a high-capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s protocol.
Quantification of pri-MicroRNA editing by Sanger Sequencing
pri-MicroRNA-seq was performed as previously described.14 Genomic DNA (gDNA) was isolated from the interphase according to TRIzol reagent instructions to ensure observed cDNA sequencing variation was not due to SNPs. The pri-microRNA of each selected “editable” microRNA was individually amplified by PCR from HUAF gDNA and cDNA samples with GoTaq DNA polymerase (Promega) or HotStarTaq DNA polymerase (QIAGEN, Hilden, Germany) (for primer sequences, see Table S8). Gel electrophoresis of the product was performed, after which the correctly sized DNA band was excised and purified using a Wizard SV gel and PCR clean-up system (Promega). Amplified and purified pri-microRNA samples were submitted for Sanger sequencing to the Leiden Genome Technology Center (Leiden, the Netherlands) according to their instructions.
A-to-I editing presents itself as A-to-G substitutions on the resulting sequencing chromatograms. Therefore, the location of each genomic adenosine was analyzed for the presence of a secondary guanosine peak in the chromatogram from the cDNA samples. gDNA sequencing chromatograms were used to ensure A-to-G substitutions were cDNA-specific. The percentage pri-microRNA A-to-I editing was calculated as described previously.53 In summary, editing is equal to the height of the editing peak (the secondary G peak) expressed as a percentage of the combined heights of the overlapping A and G peaks.
Quantification of pri-MicroRNA and mRNA Expression
The expression of pri-microRNAs and mRNAs within cDNA samples was quantified by qPCR using QuantiTect SYBR Green (QIAGEN) on the ViiA7 real-time PCR system (Applied Biosystems). pri-MicroRNA expression was normalized against the stably expressed non-coding RNA U6. Pri-microRNA expression was combined with percentage editing to calculate individual expression of WT-pri-microRNA and ED-pri-microRNA. mRNA expression was measured with intron-spanning primers and normalized against RPLP0 mRNA expression, a household gene that remains stable under ischemic conditions40. All primer sequences are provided in Table S8.
ADAR1 and ADAR2 Protein Quantification by Western Blot
Protein was isolated from cell and human LLV lysates with TRIzol (Invitrogen), according to the manufacturer’s instructions. Total protein concentration was quantified by a Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific, Waltham, MA, USA), after which protein concentration was normalized to 1 μg/μL in Laemmli buffer (Bio-Rad, Hercules, CA, USA) containing 10% β-mercaptoethanol (Sigma-Aldrich).
Samples were heated to 95°C for 5 min and cooled before loading 1 μg of protein per lane in a 4%–15% Mini-PROTEAN TGX precast protein gel (Bio-Rad). Protein separation was performed in a vertical electrophoresis cell using premixed Tris/glycine/SDS running buffer (both Bio-Rad). Proteins were transferred onto a nitrocellulose membrane (GE Healthcare, Eindhoven, the Netherlands) by a wet transfer using premixed Tris/glycine transfer buffer (Bio-Rad). The membrane was blocked at room temperature in 5% non-fat dried milk in TBS-T (150 mM NaCl, 50mM Tris, 0.05% Tween 20 [Sigma-Aldrich]) and subsequently incubated overnight at 4°C with antibodies against ADAR1 (Abcam ab168809, 1:500 dilution), ADAR2 (Abcam ab64830, 1:500 dilution), or stable household protein β-actin (Abcam ab8226, 1:1,000 dilution), diluted in 5% non-fat dried milk in TBS-T. After multiples washes with TBS-T, the membrane was incubated at room temperature with anti-rabbit peroxidase-conjugated secondary antibody (31462, Thermo Fisher Scientific), diluted to 1:10,000 in 5% non-fat dried milk in TBS-T. Proteins of interest were revealed using SuperSignal West Pico Plus chemiluminescent substrate (Thermo Fisher Scientific) and imaged using the ChemiDoc touch imaging system (Bio-Rad Laboratories). ADAR1 and ADAR2 expression levels were quantified relative to stable household protein β-actin using ImageJ.
Quantification of Mature MicroRNA Expression and Editing
Specific quantification of WT-microRNA expression and ED-microRNA expression was performed using TaqMan qRT-PCR microRNA assays designed by and purchased from Applied Biosystems. WT-microRNA assays were predesigned while ED-microRNA assays were a custom-designed TaqMan small RNA assay specifically targeting the edited sequence where the inosine was replaced by a guanosine instead. Oligonucleotide sequences used for each custom ED-microRNA-specific qRT-PCR assay are presented in Table S9.
Expression quantification was performed according to the manufacturer’s instructions. Briefly, RNA was reverse transcribed into microRNA-specific cDNA using the TaqMan microRNA reverse transcription kit (Applied Biosystems). Samples were run in triplicate on a ViiA7 (Applied Biosystems). Relative expression of WT-microRNA and ED-microRNAs was calculated relative to noncoding household RNA U6. Amplification efficiency of all qRT-PCR kits was characterized using serial dilution (Figures S2B–S2E) and incorporated in expression calculations. The percentage of microRNA editing was calculated per mature microRNA by expressing ED-microRNA as a percentage of the combined expression of both WT-microRNA and ED-microRNA.
AGO2 IP and Quantification of Associated MicroRNAs
RNA-binding protein IP (RIP) was performed using the EZMagna RIP kit (Millipore). HUAFs were seeded in T75 flasks. After 24 h, medium was removed and cells were subjected to either control conditions (normal culture medium and ∼20% oxygen) or hypoxia+starvation conditions for an additional 24 h. Cells were then washed with cold PBS, trypsinized, and pelleted at 300 × g using a tabletop centrifuge. The cell pellet was then resuspended in 0.4% formaldehyde to crosslink RNA-protein complexes for 30 min on ice. Next, cells were pelleted and washed twice with cold PBS, after which the cell pellet was resuspended in complete RIP lysis buffer. Per RIP reaction, HUAF lysate from two T75 culture flasks were incubated with RIP buffer containing magnetic beads conjugated with antibodies against AGO2 (Abcam ab32381) and negative control rabbit control IgG (Millipore PP64B). After completing the RIP according to the manufacturer’s protocol, the samples were treated with proteinase K to digest protein, and RNA was isolated using TRIzol LS reagent (Invitrogen).
cDNA was made as described above. Finally, the expression of mature WT-microRNAs and ED-microRNAs in each RIP fraction was measured as described above to determine whether these microRNAs are indeed associated with AGO2 and whether hypoxia+starvation conditions also affect the amount of microRNA associated with AGO2.
siRNA-Mediated Knockdown of ADAR1 and ADAR2
Knockdown of ADAR1 and ADAR2 in HUAFs was performed as previously described.14 Briefly, HUAFs were seeded in 12-well plates, grown to 70% confluence, and then transfected using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s protocol, with a final concentration of 27.5 nM siRNA. siRNA sequences used were originally reported and validated by Stellos et al.40 and can be found in Table S10. After 48 h, cells were washed three times with PBS and total RNA was isolated and cDNA was synthesized as before. Expression of ADAR1, ADAR2, and RPLP0 was quantified to determine knockdown efficiency. Subsequent microRNA expression and editing analyses were performed as described above.
HLI Model
All animal experiments were approved by the Committee on Animal Welfare of the Leiden University Medical Center (Leiden, the Netherlands, approval reference no. 09163).
Adult male C57BL/6 mice, 8–12 weeks old (Charles River, Wilmington, MA, USA), were housed in groups of three to five animals, with free access to tap water and regular chow. The assignment of the mice to the experimental groups was conducted randomly. All animals were included in the study, and the definitions of inclusion and exclusion criteria as well as primary and secondary endpoints were not applicable.
Induction of HLI was performed as described previously.14 In brief, mice were anesthetized by intraperitoneal injection of midazolam (5 mg/kg, Roche Diagnostics, Almere, the Netherlands), medetomidine (0.5 mg/kg, Orion, Espoo, Finland), and fentanyl (0.05 mg/kg, Janssen Pharmaceuticals, Beerse, Belgium). Unilateral HLI was induced by electrocoagulation of the left femoral artery proximal to the superficial epigastric arteries. After surgery, anesthesia was antagonized with flumazenil (0.5 mg/kg, Fresenius Kabi, Bad Homburg vor der Höhe, Germany), atipamezole (2.5 mg/kg, Orion), and buprenorphine (0.1 mg/kg, MSD Animal Health, Boxmeer, the Netherlands). Mice were sacrificed by cervical dislocation, and the adductor and gastrocnemius muscles were excised en bloc and snap-frozen on dry ice before (T0) and at 1 and 3 days (T1 and T3, respectively) after induction of HLI. Muscle tissues were crushed with pestle and mortar, while using liquid nitrogen to preserve sample integrity. Tissue homogenates were stored at −80°C. Total RNA was isolated from tissue powder by standard TRIzol-chloroform extraction as before.
Collection of Surplus Human Artery and Vein Samples
All human artery and vein samples were collected at the Leiden University Medical Center. Collection, storage, and processing of the samples were performed in compliance with the Medical Treatment Contracts Act (WGBO, 1995) and the Code of Conduct for Health Research using Body Material (Good Practice Code, Dutch Federation of Biomedical Scientific Societies, 2002) and the Dutch Personal Data Protection Act (WBP, 2001).
Human VSMs and IMAs were harvested during elective coronary bypass surgery on patients with CAD. Only surplus tissue was collected. These samples were anonymized and no data were recorded that could potentially trace back to an individual’s identity. Vessels were left to rest overnight in culture medium (DMEM GlutaMAX with 10% heat-inactivated fetal calf serum and 100 U of penicillin and 100 μg of streptomycin per mL) at 37°C and 20% oxygen and subsequently cultured for 24 h, either at control conditions (20% oxygen and culture medium) or at hypoxia+starvation conditions (1% oxygen and fetal calf serum reduced to 0.5%). VSMs were left intact and snap-frozen directly. Before snap-freezing the IMAs, they were separated manually into the tunica adventitia and the tunica media/intima, while kept cold. Frozen tissues were crushed in liquid nitrogen and total RNA was isolated from tissue powder by standard TRIzol-chloroform extraction as described above.
Surplus LLV tissue samples were also collected during coronary bypass surgery and femoral artery to popliteal artery bypass surgery.
LLVs from eight patients with end-stage PAD were obtained directly after lower limb amputation. Inclusion criteria for the biobank were a minimum age of 18 years and lower limb amputation, excluding ankle, foot, or toe amputations. The exclusion criteria were suspected or confirmed malignancy and inability to give informed consent. Sample collection was approved by the Medical Ethics Committee of the Leiden University Medical Center (protocol no. P12.265), and written informed consent was obtained from these participants.
All human artery and vein samples were snap-frozen and stored at −80°C. Frozen tissues were crushed in liquid nitrogen and total RNA was isolated from tissue powder by standard TRIzol-chloroform extraction as described above.
In Silico Target Prediction and Pathway Enrichment Analysis
Putative human targetomes of each microRNA were determined using three distinct target prediction algorithms to reduce the number of false positives: TargetScan (http://www.targetscan.org/vert_72/),16 miRanda (http://www.microRNA.org),54 and Diana-MR-microT (http://diana.imis.athena-innovation.gr/DianaTools/index.php).20 TargetScan (version 6.2) and miRanda (2010 release) predictions were obtained through the miRmut2go webtool (http://compbio.uthsc.edu/miR2GO/home.php),55 whereas Diana-MR-microT predictions were obtained using its website. In the predictions, the inosine was replaced with guanosine for the ED-microRNA input sequence. No restrictions were applied for target prediction. Genes were only considered to be a particular microRNA’s putative target gene when each of the three target prediction algorithms identified them as a target.
For each targetome, the set of target genes of a particular microRNA, overrepresented pathways were identified using PANTHER pathway enrichment analysis (http://www.pantherdb.org/, version 11),18 as described previously.56
To identify putative target genes involved in the response to ischemia, several relevant gene ontology terms were selected, including “response to hypoxia,” “angiogenesis,” and “migration” (http://geneontology.org/) (Table S5). Putative target genes exclusively targeted by either the WT or ED variant of a particular microRNA were considered involved in the response to ischemia when they were associated with one or more of these terms.
Dual-Luciferase Reporter Gene Assays
Constructs
3′ UTR sequences containing one or more WT-microRNA or ED-microRNA binding sites from endogenous target genes were amplified from human cDNA using primers with a short extension containing cleavage sites for XhoI (5′ end) and NotI (3′ end) (Table S8). For BMP2, BCL2, and ANGPT2 (ED-miR-381-3p binding sequences only) endogenous binding sequences were purchased from Integrated DNA Technologies (IDT, Coralville, IA, USA) instead (Table S10).
Amplicons and synthetic sequences were digested with XhoI and NotI and cloned in between the XhoI and NotI cleavage sites of the PsiCHECK-2 vector (Promega) at the 3′ end of the coding region of the Renilla luciferase reporter gene. The sequence of each construct was confirmed using Sanger sequencing.
Sequences of the primers used are available in Table S8.
Luciferase Assays
HeLa cells were cultured at 37°C under 5% CO2 using DMEM (Gibco) with high glucose and stable l-glutamine, supplemented with 10% fetal calf serum and 100 U of penicillin and 100 μg of streptomycin per mL (Lonza). For experiments, HeLa cells were grown to 75%–80% confluence in white 96-well plates in their normal growth medium, at 37°C under 5% CO2. Lipofectamine 3000 (Invitrogen) in Opti-MEM (Gibco) was used, according to the manufacturer’s instructions, to transfect each well with 30 ng of PsiCHECK-2 vector containing endogenous microRNA binding sequences or the original empty vector. Cells were co-transfected with 2 pmol of miRCURY locked nucleic acid (LNA) microRNA mimic for either a WT-microRNA, ED-microRNA, or negative control microRNA (Qiagen) at a concentration of 10 nM. Firefly luciferase and Renilla luciferase were measured in cell lysates using a Dual-Luciferase reporter assay system (Promega) according to the manufacturer’s protocol on a Cytation 5 plate reader (BioTek, Winooski, VT, USA). Firefly luciferase activity was used as an internal control for cellular density and transfection efficiency. The luminescence ratios were corrected for differences in baseline vector luminescence observed in the vehicle-treated group and expressed as a percentage of scrambled control luminescence.
Displayed luciferase data represent the averages from three independent experiments.
Endogenous Transcript Regulation by WT-MicroRNAs or ED-MicroRNAs
Endogenous transcript regulation was examined by overexpression of WT-microRNA or ED-microRNA in HUAFs. HUAFs were seeded in 12-well plates at 70,000 cells per well and grown for 12 h in culture medium, after which the medium was replaced with starvation medium to synchronize cell cycle. After another 12 h, Lipofectamine RNAiMAX (Invitrogen) in Opti-MEM (Gibco) was used according to the manufacturer’s instructions to transfect each well with 1 pg of miRCURY LNA microRNA mimic for either a WT-microRNA, ED-microRNA, or negative control microRNA (Qiagen). After 12 h, transfection medium was replaced with new starvation medium. After 14 h (26 h total after transfection), cells were washed twice with PBS and harvested with TRIzol reagent, after which RNA was isolated.
To examine target mRNA expression of individual genes, total cDNA was prepared and target mRNA expression was measured by qPCR as described above. Target mRNA expression was normalized against RPLP0. The intron-spanning primers used can be found in Table S8. Displayed endogenous target regulations represent the averages from three independent experiments.
Targetome Regulation by WT-miR-411-5p and ED-miR-411-5p
For each independent miR-411-5p overexpression experiment, at least 500 ng of pooled total RNA was submitted to BGI Hong Kong for RNA-seq. BGI’s services included mRNA enrichment and purification using oligo(dT) selection, RNA fragmentation, reverse transcription, end repair, adaptor ligation, DNA nanoball synthesis, and finally sequencing on the DNBseq platform.
Raw reads were filtered by BGI Genomics to remove adaptor sequences, contaminations, and low-quality reads. Quality of the clean fastQ files was verified using FastQC version 0.11.8 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads were then mapped to the human genome using HISAT2 version 2.2.0 with the pre-compiled index GRCh38_snp_tran57 and subsequently assembled into transcripts using StringTie version 2.1.1.58 Transcript abundance was estimated per gene using Rsubread version 2.0.1.59 Read count normalization and filtering were done using edgeR, retaining all genes that had >1 counts per million (cpm) in three or more samples.60 Differential expression analysis was performed on voom-transformed log2 cpm values using limma.61 Finally, enrichment of the predicted targetomes for WT and ED miR-411-5p was tested using ROAST, a self-contained gene set test.62 p values were calculated by simulation, using 999 rotations. Data were visualized using limma’s plotMD and R’s built-in boxplot functions.
In Vivo Targetome Regulation after HLI
For each human targetome, except the unconserved miR-376a-3p, target genes that were also predicted to be target genes in mice according to the murine Diana-MR-microT algorithm (http://diana.imis.athena-innovation.gr/DianaTools/index.php)20 were considered the conserved murine targetome. On average, 62.5% of the human target genes were identified to be conserved targets in mice in this way (see Table S7).
To assess which conserved targetomes are regulated in response to HLI, we used a previously published whole-genome expression microarray dataset in which transcriptome expression was measured before and after HLI in the adductor muscle (Nossent et al.21). For each gene detected above background levels (21,074 out of 45,200), post-ischemic expression was calculated relative to its expression before HLI (T0) by calculating the 2Δlog2(measured gene intensity). Average putative targetome expression was determined by calculating the average post-ischemic change in expression of all genes within the particular targetome. Targetome expressions were compared to the average expression of all genes above the detection limit.
Effects of WT-MicroRNAs or ED-MicroRNAs on In Vitro Scratch-Wound Healing
Effects of WT-microRNA or ED-microRNA overexpression on scratch-wound healing of HUAFs were examined. HUAFs were seeded and transfected as described above. Transfection medium was removed after 12 h. Next, a p200 pipette tip was used to introduce a scratch wound across the diameter of each well. Subsequently, the cells were washed with sterile PBS and medium was replaced with new serum starvation medium. Three locations along the scratch wound were marked per well. The scratch wound at these sites was imaged at time 0 h and 14 h after scratch wound introduction using live phase-contrast microscopy (Axiovert 40C, Carl Zeiss, Oberkochen, Germany). After the 14-h time point, cells were washed three times in PBS and then lysed and harvested in TRIzol for RNA isolation as before. MicroRNA overexpression efficiency was validated by measuring microRNA expression as described above. For each imaged location, the area of the scratch wound at 0 h was superimposed on the 14-h scratch-wound area image. Scratch-wound healing was then determined per well as the newly covered scratch-wound area after 14 h using the wound-healing tool macro for ImageJ. Displayed scratch-wound healing represent the averages from three independent experiments.
Effects of WT-MicroRNAs or ED-MicroRNAs on HUVEC Tube Formation
HUVECs were seeded in 12-well plates in EBM-2 basal medium (CC-3156, Lonza) supplemented with EGM-2 SingleQuots supplements (CC-4176, Lonza). At 80% confluency, each well was transfected with 1 pg of miRCURY LNA microRNA mimics as described before, using Lipofectamine RNAiMAX (Invitrogen) in Opti-MEM (Gibco) according to the manufacturer’s instructions. After 24 h, the transfected HUVECs were detached using trypsin-EDTA (Sigma, Steinheim, Germany) and counted. Next, low-serum medium (EBM-2 basal medium [CC-3156] supplemented with 0.2% FBS and 1% GA-1000) was used to seed 15,000 cells per well in a 96-well plate, which was pre-coated with 50 μL/well of Geltrex extracellular matrix (A1413202, Gibco). After a 12-h incubation, images of each well were taken using live phase-contrast microscopy (Axiovert 40C, Carl Zeiss). The total length of the tubes formed was analyzed using the angiogenesis analyzer plugin for ImageJ.
Effects of WT-MicroRNAs or ED-MicroRNAs on Ex Vivo Angiogenesis
Mouse aortic ring assays were performed as described previously.4,63 In brief, six thoracic aortas were removed from 8- to 10-week-old mice, after which the surrounding fat and branching vessels were carefully removed and the aorta was flushed with Opti-MEM (Gibco). Aortic rings of ∼1 mm were cut and the rings from each mouse aorta were divided over 7 wells of 24-well plate groups for separate treatments. The different groups were then transfected overnight with 1 pg of miRCURY LNA microRNA mimics, using 1.5 μL of Lipofectamine RNAiMAX (Invitrogen) in fresh Opti-MEM (Gibco) with a total volume of 500 μL.
The next day, 96-well plates were coated with 75 μL of collagen matrix (collagen type I, Millipore) diluted in 1× DMEM (Gibco), and the pH was adjusted with 5 N NaOH. One aortic ring per well was embedded in the collagen matrix, for a total of at least 30 rings per pre-microRNA treatment. After letting the collagen solidify for an hour, 150 μL of Opti-MEM supplemented with 2.5% FBS (PAA, Austria), penicillin-streptomycin (PAA, Austria), and 30 ng/mL VEGF (R&D Systems) was added. Medium was refreshed every 2 days and was supplemented with microRNA mimics at a concentration of 100 nM, without transfection agent. Images of each embedded aortic ring and its neovessel outgrowth were made after 7 days using live phase-contrast microscopy (Axiovert 40C, Carl Zeiss). The number of neovessel sprouts was manually counted per aortic segment. Segments were excluded when they were too close to an obstacle (i.e., the wall of the well) or showed no outgrowth. Each neovessel emerging from the ring was counted as a sprout. Individual branches arising from each microvessel were counted as a separate sprout.
Statistical Analysis
All results are expressed as mean ± SEM. Normality of data obtained was examined using the Shapiro-Wilk’s normality test. Since all variables measured were continuous parameters, pairwise comparisons were tested using either t tests or one-way ANOVA. p values less than or equal to 0.05 were considered statistically significant.
Author Contributions
Conceptualization, R.V.C.T.v.d.K., A.Y.N., and P.H.A.Q.; Methodology, R.V.C.T.v.d.K., L.P., M.L.v.d.B., F.B., E.H.A.B.P., M.R.d.V., and A.Y.N.; Investigation, R.V.C.T.v.d.K., L.P., E.v.I., and E.H.A.B.P.; Resources, E.A.C.G., K.H.S., M.P., and M.R.d.V.; Writing – Original Draft, R.V.C.T.v.d.K.; Writing – Review & Editing, R.V.C.T.v.d.K., P.H.A.Q., and A.Y.N.; Visualization, R.V.C.T.v.d.K.; Supervision, R.V.C.T.v.d.K., P.H.A.Q., and A.Y.N.; Funding Acquisition, A.Y.N.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We thank the vascular and thoracic surgeons of the LUMC that helped with collecting the patient material. Furthermore, we acknowledge D.A.L. van den Homberg, J.S.A. van der Geest, E. Kalbus, and A.W. Meijer for technical support. This study was supported by a grant from the Dutch Heart Foundation, the Netherlands (Dr. E. Dekker Senior Postdoc, 2014T102), the LUMC Johanna Zaaijer Fund (2017), and the Austrian Science Fund FWF (Lise Meitner Grant, M 2578-B30).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.07.020.
Supplemental Information
References
- 1.Welten S.M., Goossens E.A., Quax P.H., Nossent A.Y. The multifactorial nature of microRNAs in vascular remodelling. Cardiovasc. Res. 2016;110:6–22. doi: 10.1093/cvr/cvw039. [DOI] [PubMed] [Google Scholar]
- 2.Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 3.Romaine S.P., Tomaszewski M., Condorelli G., Samani N.J. MicroRNAs in cardiovascular disease: an introduction for clinicians. Heart. 2015;101:921–928. doi: 10.1136/heartjnl-2013-305402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Welten S.M., Bastiaansen A.J., de Jong R.C., de Vries M.R., Peters E.A., Boonstra M.C., Sheikh S.P., La Monica N., Kandimalla E.R., Quax P.H., Nossent A.Y. Inhibition of 14q32 microRNAs miR-329, miR-487b, miR-494, and miR-495 increases neovascularization and blood flow recovery after ischemia. Circ. Res. 2014;115:696–708. doi: 10.1161/CIRCRESAHA.114.304747. [DOI] [PubMed] [Google Scholar]
- 5.Mallela A., Nishikura K. A-to-I editing of protein coding and noncoding RNAs. Crit. Rev. Biochem. Mol. Biol. 2012;47:493–501. doi: 10.3109/10409238.2012.714350. [DOI] [PubMed] [Google Scholar]
- 6.Nigita G., Veneziano D., Ferro A. A-to-I RNA editing: current knowledge sources and computational approaches with special emphasis on non-coding RNA molecules. Front. Bioeng. Biotechnol. 2015;3:37. doi: 10.3389/fbioe.2015.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tan M.H., Li Q., Shanmugam R., Piskol R., Kohler J., Young A.N., Liu K.I., Zhang R., Ramaswami G., Ariyoshi K., GTEx Consortium. Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group. Statistical Methods groups—Analysis Working Group. Enhancing GTEx (eGTEx) groups. NIH Common Fund. NIH/NCI. NIH/NHGRI. NIH/NIMH. NIH/NIDA. Biospecimen Collection Source Site—NDRI. Biospecimen Collection Source Site—RPCI. Biospecimen Core Resource—VARI. Brain Bank Repository—University of Miami Brain Endowment Bank. Leidos Biomedical—Project Management. ELSI Study. Genome Browser Data Integration &Visualization—EBI. Genome Browser Data Integration &Visualization—UCSC Genomics Institute, University of California Santa Cruz Dynamic landscape and regulation of RNA editing in mammals. Nature. 2017;550:249–254. doi: 10.1038/nature24041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishikura K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 2016;17:83–96. doi: 10.1038/nrm.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawahara Y., Megraw M., Kreider E., Iizasa H., Valente L., Hatzigeorgiou A.G., Nishikura K. Frequency and fate of microRNA editing in human brain. Nucleic Acids Res. 2008;36:5270–5280. doi: 10.1093/nar/gkn479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li L., Song Y., Shi X., Liu J., Xiong S., Chen W., Fu Q., Huang Z., Gu N., Zhang R. The landscape of miRNA editing in animals and its impact on miRNA biogenesis and targeting. Genome Res. 2018;28:132–143. doi: 10.1101/gr.224386.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang W., Chendrimada T.P., Wang Q., Higuchi M., Seeburg P.H., Shiekhattar R., Nishikura K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franzén O., Ermel R., Sukhavasi K., Jain R., Jain A., Betsholtz C., Giannarelli C., Kovacic J.C., Ruusalepp A., Skogsberg J. Global analysis of A-to-I RNA editing reveals association with common disease variants. PeerJ. 2018;6:e4466. doi: 10.7717/peerj.4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawahara Y., Zinshteyn B., Sethupathy P., Iizasa H., Hatzigeorgiou A.G., Nishikura K. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007;315:1137–1140. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Kwast R.V.C.T., van Ingen E., Parma L., Peters H.A.B., Quax P.H.A., Nossent A.Y. Adenosine-to-inosine editing of microRNA-487b alters target gene selection after ischemia and promotes neovascularization. Circ. Res. 2018;122:444–456. doi: 10.1161/CIRCRESAHA.117.312345. [DOI] [PubMed] [Google Scholar]
- 15.Newman A.C., Nakatsu M.N., Chou W., Gershon P.D., Hughes C.C. The requirement for fibroblasts in angiogenesis: fibroblast-derived matrix proteins are essential for endothelial cell lumen formation. Mol. Biol. Cell. 2011;22:3791–3800. doi: 10.1091/mbc.E11-05-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agarwal V., Bell G.W., Nam J.W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:4. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hellingman A.A., Bastiaansen A.J., de Vries M.R., Seghers L., Lijkwan M.A., Lowik C.W., Hamming J.F., Quax P.H. Variations in surgical procedures for hind limb ischaemia mouse models result in differences in collateral formation. Eur. J. Vasc. Endovasc. Surg. 2010;40:796–803. doi: 10.1016/j.ejvs.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 18.Mi H., Huang X., Muruganujan A., Tang H., Mills C., Kang D., Thomas P.D. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45(D1):D183–D189. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Kwast R., Woudenberg T., Quax P.H.A., Nossent A.Y. MicroRNA-411 and Its 5-IsomiR have distinct targets and functions and are differentially regulated in the vasculature under ischemia. Mol. Ther. 2020;28:157–170. doi: 10.1016/j.ymthe.2019.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paraskevopoulou M.D., Georgakilas G., Kostoulas N., Vlachos I.S., Vergoulis T., Reczko M., Filippidis C., Dalamagas T., Hatzigeorgiou A.G. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013;41:W169-73. doi: 10.1093/nar/gkt393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nossent A.Y., Bastiaansen A.J., Peters E.A., de Vries M.R., Aref Z., Welten S.M., de Jager S.C., van der Pouw Kraan T.C., Quax P.H. CCR7-CCL19/CCL21 axis is essential for effective arteriogenesis in a murine model of hindlimb ischemia. J. Am. Heart Assoc. 2017;6:e005281. doi: 10.1161/JAHA.116.005281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nigita G., Acunzo M., Romano G., Veneziano D., Laganà A., Vitiello M., Wernicke D., Ferro A., Croce C.M. MicroRNA editing in seed region aligns with cellular changes in hypoxic conditions. Nucleic Acids Res. 2016;44:6298–6308. doi: 10.1093/nar/gkw532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pinto Y., Buchumenski I., Levanon E.Y., Eisenberg E. Human cancer tissues exhibit reduced A-to-I editing of miRNAs coupled with elevated editing of their targets. Nucleic Acids Res. 2018;46:71–82. doi: 10.1093/nar/gkx1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warnefors M., Liechti A., Halbert J., Valloton D., Kaessmann H. Conserved microRNA editing in mammalian evolution, development and disease. Genome Biol. 2014;15:R83. doi: 10.1186/gb-2014-15-6-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jepson J.E., Reenan R.A. RNA editing in regulating gene expression in the brain. Biochim. Biophys. Acta. 2008;1779:459–470. doi: 10.1016/j.bbagrm.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 26.Tomaselli S., Galeano F., Alon S., Raho S., Galardi S., Polito V.A., Presutti C., Vincenti S., Eisenberg E., Locatelli F., Gallo A. Modulation of microRNA editing, expression and processing by ADAR2 deaminase in glioblastoma. Genome Biol. 2015;16:5. doi: 10.1186/s13059-014-0575-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alon S., Mor E., Vigneault F., Church G.M., Locatelli F., Galeano F., Gallo A., Shomron N., Eisenberg E. Systematic identification of edited microRNAs in the human brain. Genome Res. 2012;22:1533–1540. doi: 10.1101/gr.131573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ota H., Sakurai M., Gupta R., Valente L., Wulff B.E., Ariyoshi K., Iizasa H., Davuluri R.V., Nishikura K. ADAR1 forms a complex with Dicer to promote microRNA processing and RNA-induced gene silencing. Cell. 2013;153:575–589. doi: 10.1016/j.cell.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qi L., Song Y., Chan T.H.M., Yang H., Lin C.H., Tay D.J.T., Hong H., Tang S.J., Tan K.T., Huang X.X. An RNA editing/dsRNA binding-independent gene regulatory mechanism of ADARs and its clinical implication in cancer. Nucleic Acids Res. 2017;45:10436–10451. doi: 10.1093/nar/gkx667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawahara Y., Zinshteyn B., Chendrimada T.P., Shiekhattar R., Nishikura K. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep. 2007;8:763–769. doi: 10.1038/sj.embor.7401011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Houseley J., Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y., Xu X., Yu S., Jeong K.J., Zhou Z., Han L., Tsang Y.H., Li J., Chen H., Mangala L.S. Systematic characterization of A-to-I RNA editing hotspots in microRNAs across human cancers. Genome Res. 2017;27:1112–1125. doi: 10.1101/gr.219741.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bass B.L. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pinto Y., Cohen H.Y., Levanon E.Y. Mammalian conserved ADAR targets comprise only a small fragment of the human editosome. Genome Biol. 2014;15:R5. doi: 10.1186/gb-2014-15-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hermans K.C., Blankesteijn W.M. Wnt signaling in cardiac disease. Compr. Physiol. 2015;5:1183–1209. doi: 10.1002/cphy.c140060. [DOI] [PubMed] [Google Scholar]
- 37.Blaise S., Polena H., Vilgrain I. Soluble vascular endothelial-cadherin and auto-antibodies to human vascular endothelial-cadherin in human diseases: two new biomarkers of endothelial dysfunction. Vasc. Med. 2015;20:557–565. doi: 10.1177/1358863X15591201. [DOI] [PubMed] [Google Scholar]
- 38.Velazquez-Torres G., Shoshan E., Ivan C., Huang L., Fuentes-Mattei E., Paret H., Kim S.J., Rodriguez-Aguayo C., Xie V., Brooks D. A-to-I miR-378a-3p editing can prevent melanoma progression via regulation of PARVA expression. Nat. Commun. 2018;9:461. doi: 10.1038/s41467-018-02851-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shoshan E., Mobley A.K., Braeuer R.R., Kamiya T., Huang L., Vasquez M.E., Salameh A., Lee H.J., Kim S.J., Ivan C. Reduced adenosine-to-inosine miR-455-5p editing promotes melanoma growth and metastasis. Nat. Cell Biol. 2015;17:311–321. doi: 10.1038/ncb3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stellos K., Gatsiou A., Stamatelopoulos K., Perisic Matic L., John D., Lunella F.F., Jaé N., Rossbach O., Amrhein C., Sigala F. Adenosine-to-inosine RNA editing controls cathepsin S expression in atherosclerosis by enabling HuR-mediated post-transcriptional regulation. Nat. Med. 2016;22:1140–1150. doi: 10.1038/nm.4172. [DOI] [PubMed] [Google Scholar]
- 41.Nossent A.Y., Eskildsen T.V., Andersen L.B., Bie P., Brønnum H., Schneider M., Andersen D.C., Welten S.M., Jeppesen P.L., Hamming J.F. The 14q32 microRNA-487b targets the antiapoptotic insulin receptor substrate 1 in hypertension-induced remodeling of the aorta. Ann. Surg. 2013;258 doi: 10.1097/SLA.0b013e3182a6aac0. 743–751, discussion 752–753. [DOI] [PubMed] [Google Scholar]
- 42.Wezel A., Welten S.M., Razawy W., Lagraauw H.M., de Vries M.R., Goossens E.A., Boonstra M.C., Hamming J.F., Kandimalla E.R., Kuiper J. Inhibition of microRNA-494 reduces carotid artery atherosclerotic lesion development and increases plaque stability. Ann. Surg. 2015;262:841–847. doi: 10.1097/SLA.0000000000001466. [DOI] [PubMed] [Google Scholar]
- 43.Hakansson K.E.J., Goossens E.A.C., Trompet S., van Ingen E., de Vries M.R., van der Kwast R., Ripa R.S., Kastrup J., Hohensinner P.J., Kaun C. Genetic associations and regulation of expression indicate an independent role for 14q32 snoRNAs in human cardiovascular disease. Cardiovasc. Res. 2019;115:1519–1532. doi: 10.1093/cvr/cvy309. [DOI] [PubMed] [Google Scholar]
- 44.Welten S.M.J., de Jong R.C.M., Wezel A., de Vries M.R., Boonstra M.C., Parma L., Jukema J.W., van der Sluis T.C., Arens R., Bot I. Inhibition of 14q32 microRNA miR-495 reduces lesion formation, intimal hyperplasia and plasma cholesterol levels in experimental restenosis. Atherosclerosis. 2017;261:26–36. doi: 10.1016/j.atherosclerosis.2017.04.011. [DOI] [PubMed] [Google Scholar]
- 45.Boon R.A., Hofmann P., Michalik K.M., Lozano-Vidal N., Berghäuser D., Fischer A., Knau A., Jaé N., Schürmann C., Dimmeler S. Long noncoding RNA Meg3 controls endothelial cell aging and function: implications for regenerative angiogenesis. J. Am. Coll. Cardiol. 2016;68:2589–2591. doi: 10.1016/j.jacc.2016.09.949. [DOI] [PubMed] [Google Scholar]
- 46.Bijkerk R., Au Y.W., Stam W., Duijs J.M.G.J., Koudijs A., Lievers E., Rabelink T.J., van Zonneveld A.J. Long non-coding RNAs Rian and Miat mediate myofibroblast formation in kidney fibrosis. Front. Pharmacol. 2019;10:215. doi: 10.3389/fphar.2019.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Malnou E.C., Umlauf D., Mouysset M., Cavaillé J. Imprinted microRNA gene clusters in the evolution, development, and functions of mammalian placenta. Front. Genet. 2019;9:706. doi: 10.3389/fgene.2018.00706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chiang H.R., Schoenfeld L.W., Ruby J.G., Auyeung V.C., Spies N., Baek D., Johnston W.K., Russ C., Luo S., Babiarz J.E. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010;24:992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ekdahl Y., Farahani H.S., Behm M., Lagergren J., Öhman M. A-to-I editing of microRNAs in the mammalian brain increases during development. Genome Res. 2012;22:1477–1487. doi: 10.1101/gr.131912.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gong J., Wu Y., Zhang X., Liao Y., Sibanda V.L., Liu W., Guo A.Y. Comprehensive analysis of human small RNA sequencing data provides insights into expression profiles and miRNA editing. RNA Biol. 2014;11:1375–1385. doi: 10.1080/15476286.2014.996465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng Y., Li T., Ren R., Shi D., Wang S. Revealing editing and SNPs of microRNAs in colon tissues by analyzing high-throughput sequencing profiles of small RNAs. BMC Genomics. 2014;15(Suppl 9):S11. doi: 10.1186/1471-2164-15-S9-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cho S., Jang I., Jun Y., Yoon S., Ko M., Kwon Y., Choi I., Chang H., Ryu D., Lee B. MiRGator v3.0: a microRNA portal for deep sequencing, expression profiling and mRNA targeting. Nucleic Acids Res. 2013;41:D252–D257. doi: 10.1093/nar/gks1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eggington J.M., Greene T., Bass B.L. Predicting sites of ADAR editing in double-stranded RNA. Nat. Commun. 2011;2:319. doi: 10.1038/ncomms1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Betel D., Koppal A., Agius P., Sander C., Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bhattacharya A., Cui Y. miR2GO: comparative functional analysis for microRNAs. Bioinformatics. 2015;31:2403–2405. doi: 10.1093/bioinformatics/btv140. [DOI] [PubMed] [Google Scholar]
- 56.Mi H., Muruganujan A., Casagrande J.T., Thomas P.D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pertea M., Pertea G.M., Antonescu C.M., Chang T.C., Mendell J.T., Salzberg S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liao Y., Smyth G.K., Shi W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019;47:e47. doi: 10.1093/nar/gkz114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu D., Lim E., Vaillant F., Asselin-Labat M.L., Visvader J.E., Smyth G.K. ROAST: rotation gene set tests for complex microarray experiments. Bioinformatics. 2010;26:2176–2182. doi: 10.1093/bioinformatics/btq401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baker M., Robinson S.D., Lechertier T., Barber P.R., Tavora B., D’Amico G., Jones D.T., Vojnovic B., Hodivala-Dilke K. Use of the mouse aortic ring assay to study angiogenesis. Nat. Protoc. 2011;7:89–104. doi: 10.1038/nprot.2011.435. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.