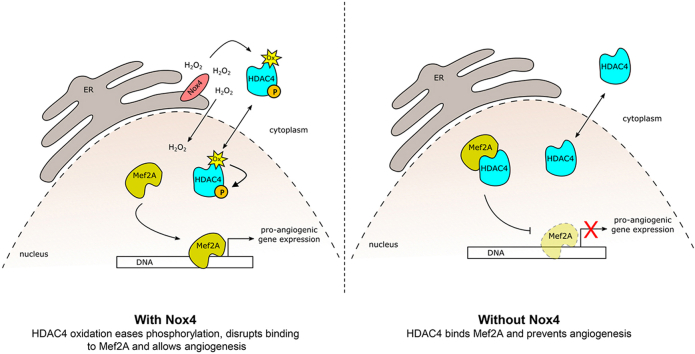
Abstract
NADPH oxidases produce reactive oxygen species that differ in localization, type and concentration. Within the Nox family only Nox4 produces H2O2 which can directly oxidize cysteine residues. With this post-translational modification, activity, stability, localization and protein-protein interactions of the affected protein is altered. Nox4 controls differentiation, cellular homeostasis and prevents inflammation. Therefore, is likely that epigenetic mechanisms contribute to the effects of Nox4. One group of epigenetic modifiers are class IIa histone deacetylases (HDACs). We hypothesize that Nox4-derived H2O2 oxidizes HDACs and analyzed whether HDACs can be differentially oxidized by Nox4. As an artificial system, we utilized HEK293 cells, overexpressing Nox4 in a tetracycline-inducible manner. HDAC4 was oxidized upon Nox4 overexpression. Additionally, Nox4 overexpression increased HDAC4 phosphorylation on Ser632. H2O2 disrupted HDAC4/Mef2A complex, which de-represses Mef2A. In endothelial cells such as HUVECs and HMECs, overexpression of HDAC4 significantly reduced tube formation. Overexpression of a redox insensitive HDAC4 had no effect on endothelial tube formation. Treatment with H2O2, induction of Nox4 expression by treatment of the cells with TGFβ and co-overexpression of Nox4 not only induced phosphorylation of HDAC4, but also restored the repressive effect of HDAC4 for tube formation, while overexpression of a redox dead mutant of Nox4 did not.
Taken together, Nox4 oxidizes HDAC4, increases its phosphorylation, and eventually ensures proper tube formation by endothelial cells.
Keywords: NADPH oxidase, Nox4, HDAC4
Graphical abstract
1. Introduction
Endothelial cells regulate vascular tone, vascularization processes, influence smooth muscle cell function and mediate immune as well as general inflammatory responses [6,11,31,34]. As such, endothelial dysfunction can contribute to the development of atherosclerosis, hypertension and coronary artery disease [15]. It has become clear that endothelial cells undergo permanent changes in their epigenetic programming, dependent on their metabolic/redox status [23].
Epigenetic programming includes alterations in DNA packing density, which among others is regulated via the acetylation-status of positively charged amino acids, namely lysine and arginine in histone tails. Acetyl transferases, such as p300, neutralize the positive charges by converting the amines into amides, which loosens the histone-DNA binding and eases transcription. In contrast, histone deacetylases remove acetyl groups, which tightens the histone-DNA binding and hinders transcription. In mammals, 18 HDACs sorted into four main classes. Out of those, class II HDACs (class IIa: HDACs 4, 5, 7 and 9 and class IIb: HDACs 6 and 10) appear to be redox-regulated via direct cysteine oxidation [2]. An interesting HDAC in endothelial cells is HDAC4. It maintains VEGFR2 expression as well as autophagy and it enhances angiotensin II-induced inflammation in vascular endothelial cells [5,14]. Besides histones, HDACs have other targets as well. Recently it was shown that fluid shear stress stimulates HDAC5 phosphorylation and its nuclear export in endothelial cells through a calcium/calmodulin-dependent pathway. This results in dissociation of HDAC5 and Mef2, which enhances Mef2 transcriptional activity, leading to the development of an anti-inflammatory phenotype of endothelial cells [38]. Phosphorylated HDAC5 in the nucleus therefore may play a protective role in inflammation. Further, overexpression of HDAC5 in vivo, as found in scleroderma, impaired the expression of key-angiogenic and fibrotic genes [35]. This finding is similar to the effect of Nox4 knock out, which results in a pro-inflammatory phenotype, in deficits in angiogenesis as well as in enhanced fibrosis [2,12,30].
Metabolic and especially redox regulation have become major fields of research within the last decade. NADPH oxidases are significant sources of reactive oxygen species. The family of NADPH oxidases is composed of seven members. Nox1, 2 and 3 are dependent on cytosolic subunits, the organizer, and activator. All subunits need to form a complex, which additionally contains the small G-protein Rac. The whole complex then allows formation of superoxide (•O2ˉ). Nox5 and DUOX1 and 2 produce •O2ˉ or H2O2 in a Ca2+-dependent manner. The only constitutively active NADPH oxidase is Nox4, which produces permanently small amounts of H2O2 and thereby facilitates long term processes such as differentiation [9]. Further, the H2O2-producing NADPH oxidase Nox4 is important for endothelial health, function and integrity [28]. In fact, Nox4 is expressed at high levels in endothelial cells [30]. Importantly, in endothelial cells as well as in the heart Nox4 elicits positive and protective effects [31,40].
Nox4 plays a pivotal role in angiogenesis via the maintenance of Hif-1 and consequently VEGFR2 expression [36]. Nox4 knock-out results in a pro-inflammatory phenotype, in deficits in angiogenesis as well as in enhanced fibrosis [2,12,30]. In vivo, HDAC4 contributes to the manifestation of cardiac hypertrophy after transverse aortic constriction (TAC) surgery [1,19], while Nox4 overexpression protects from TAC-induced hypertrophy [40]. We hypothesize that HDAC4 and Nox4 may have opposite roles and interact. Accordingly, we thought to investigate this potential interaction of Nox4 and class II HDACs.
2. Results
2.1. Endothelial HDAC4 is susceptible to H2O2-dependent oxidation and specifically oxidized by Nox4
HDAC expression may vary between certain endothelial cell lines or origins [35,41]. We analyzed for the expression of members of class IIa HDACs in endothelial cells (HUVECs and HMECs) as well as in the model system of HEK293 cells (Supplemental Fig. 1A–D). In all three cell lines, the most abundant HDAC was HDAC7, followed by HDAC4 and very little HDAC5, whereas HDAC9 was nearly at the detection limit.
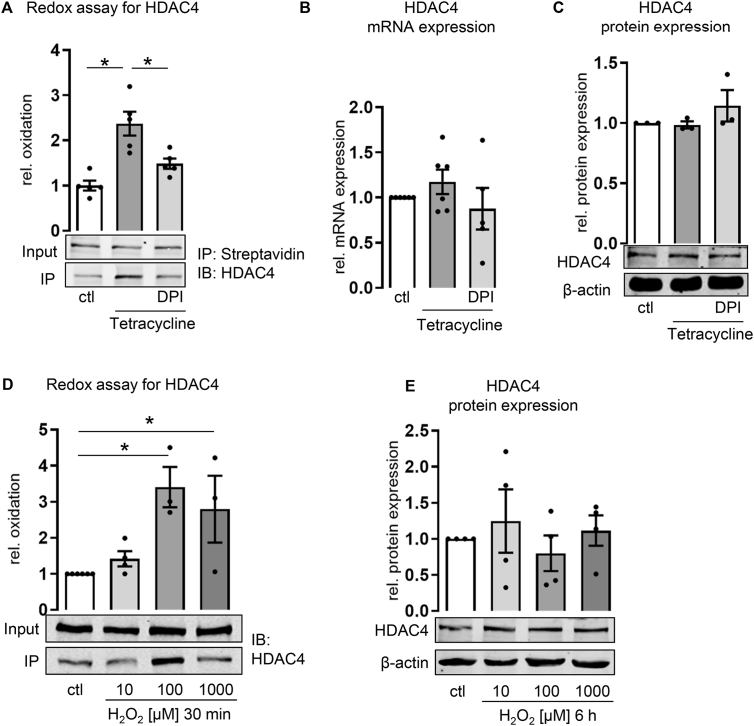
The NADPH oxidase Nox4 is an essential, direct, and constitutively active source of H2O2 in endothelial cells [30]. In order to analyze whether Nox4 is involved in oxidation of HDACs, an inducible Nox4 overexpression system in HEK293 cells was utilized (Nox4-tet-on cells). Tetracycline-dependent Nox4 overexpression was confirmed by qPCR and Western blot, while expression of other NADPH oxidases, such as Nox5 remained unaffected by tetracycline (Supplemental Fig. 2A+B). Tetracycline treatment and subsequent upregulation of Nox4 resulted in an increased catalase-sensitive H2O2 production, which was inhibited by the flavoprotein inhibitor DPI [17] (Supplemental Fig. 2C). In this work, DPI-treated cells served as a control, to show that the redox change in tetracycline-treated cells is likely due to a flavoenzyme-mediated ROS production and not due to some other effects tetracycline may have on the cells. Although we are aware of the many side effects DPI may have, it still is the most potent and reliable inhibitor of NADPH oxidases, with a fail-safe inhibition of ROS formation in the utilized system of Nox4 overexpression upon tetracycline treatment in HEK293 cells. Using this artificial system, we found that HDAC7 was not prone to Nox4-mediated redox modifications (Supplemental Fig. 2D). In contrast, HDAC5 was redox-modified (Supplemental Fig. 2E). In the DPI control, however, HDAC5 oxidation was highly variable and not significantly reduced. We conclude that other events related to tetracycline, rather than Nox4 derived ROS, elicit HDAC5 oxidation. In contrast, HDAC4 oxidation was enhanced when Nox4 was induced by tetracycline in Nox4-tet-on cells. DPI reduced Nox4-dependent HDAC4 oxidation (Fig. 1A). Neither mRNA nor protein expression of HDAC4 was altered by Nox4 overexpression (Fig. 1B&C). Further experiments were conducted to prove that the effects seen so far are specifically mediated by Nox4-derived H2O2 and do not represent a general oxidation of HDACs by ROS. Nox5 overexpressing HEK293 cells (Supplemental Fig. 3) were used to model the effect of plasma membrane bound NADPH oxidases and subsequent •O2ˉ production [33]. HEK293-Nox5 cells constitutively overexpress Nox5, which is inactive in normal cell culture conditions but can be activated by phorbol myristate acetate (PMA) treatment. Challenging HEK293-Nox5 with PMA induced superoxide production, while Nox4-induction in the Nox4-tet-on cells did not have such an effect (Supplemental Fig. 3B). Accordingly, in HEK293-Nox5 cells treated with PMA Prx3 was oxidized (Supplemental Fig. 3C), which serves as positive control for cellular effects of Nox5-derived superoxide, as it has been shown to be oxidized by Nox5 before [17]. Although Nox5-derived •O2ˉ technically is sufficient to oxidize target proteins, such as Prx3 in HEK-Nox5 cells, neither HDAC4 nor HDAC5 was oxidized in PMA-treated HEK-Nox5 cells (Supplemental Fig. 3D). Taken together, the data suggest HDAC4 to be the most likely redox target in endothelial cells. Indeed, HDAC4 can be oxidized by H2O2 in HUVECs and HMECs as shown by a modified BIAM (biotinylated iodoacetamide)-based redox-immunoprecipitation [17]. With the aid of this redox-IP, we were able to detect selectively reversible thiol oxidations. According to the concept of over-oxidation and irreversible modification by high concentrations of H2O2, BIAM labeling of HDAC4 was increased at 100 μM H2O2 and decreased when cells were treated with 1 mM H2O2 (Fig. 1D and Supplemental Fig. 4A). HDAC4 protein abundance in HUVECs and HMECs is not affected by H2O2 (Fig. 1E and Supplemental Fig. 4B).
Fig. 1.
Oxidation of HDAC4 upon Nox4 overexpression and treatment with H2O2. (A) BIAM switch redox assay (n = 5) (One-way ANOVA with Tukey post-hoc test, *p < 0.05), (B) relative mRNA expression analyzed by qPCR (n > 4) and (C) relative protein expression of HDAC4 analyzed by Western blot and normalized to β-actin (n = 3) in Nox4 tet-on cells with or without tetracycline (1 μg/ml, 24 h) and DPI (3 μM, 3 h). (D) BIAM switch redox assay (n > 3) (One-way ANOVA with Dunnett post-hoc test, *p < 0.05) and (E) relative protein expression of HDAC4 in HUVECs with H2O2 (10/100/1000 μM, 30 min or 6 h) (n = 4). Representative Western blots are depicted. All bar graphs show means ± SEM.
These data suggest HDAC4 to be specifically oxidized by Nox4-derived H2O2, which has no impact on the expression of HDAC4. We continued to analyze the consequences of Nox4-mediated HDAC4 oxidation.
2.2. Mef2A physically interacts with HDAC4, which is counteracted by H2O2 and Nox4 activity
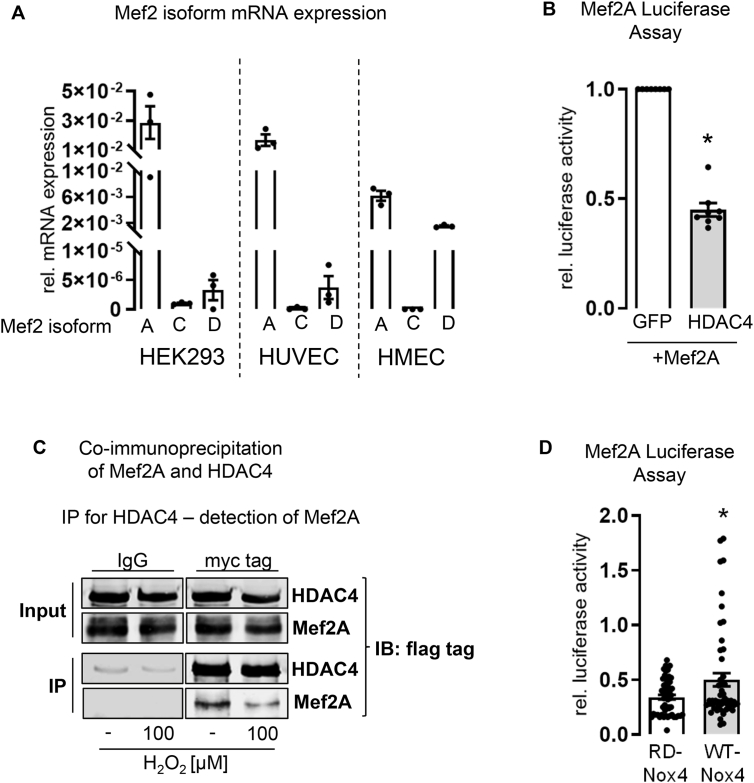
One major component of HDAC4-directed regulation of cellular processes is the repression of Mef2 transcription factors. Out of the four Mef2 isoforms (Mef2A-D), mainly Mef2A, C and D interact with HDAC4, which alters their physiological functions [22,37]. In contrast, any physiological consequences of Mef2Bs interaction with HDAC4 are uncertain [10]. Mef2A is the most abundant isoform in HEK293 cells, HMECs and HUVECs (Fig. 2A). Overexpression of HDAC4 in HEK293 cells impaired the activity of Mef2A as analyzed with the aid of a luciferase reporter gene assay (Fig. 2B). Importantly, HDAC4 physically interacts with Mef2A and H2O2 treatment disrupts this interaction (Fig. 2C). We therefore thought to analyze the effect of Nox4-derived H2O2 on Mef2A activity. We overexpressed a redox dead mutant of Nox4 (RD-Nox4; which does not produce H2O2), or wild type Nox4. If overexpressed together with HDAC4 and Mef2A, Nox4 was able to increase Mef2A activity, while RD-Nox4 was not (Fig. 2D). We conclude that Nox4-derived H2O2 oxidizes HDAC4, which disrupts the HDAC4/Mef2A complex and allows an increased Mef2A activity. Indeed, binding of HDAC4 to Mef2 in the nucleus inhibits Mef2s function [22]. Enhanced nuclear export of HDAC4 upon oxidation [19] may interfere with the repressive effect of the deacetylase on Mef2 and subsequently activate this transcription factor family. Indeed, when we analyzed for a correlation of Nox4 and Mef2A target genes in databases of human samples of endothelial cells, we found several of them to correlate with Nox4 in their expression (Supplemental Fig. 4C).
Fig. 2.
Analysis of Mef2A activity. (A) Basal mRNA expression of Mef2A, C and D in HEK293 cells, HUVECs and HMECs. Samples were analyzed by qPCR and normalized to β-actin (n = 3). (B) Mef2 luciferase reporter gene assay. HEK293 cells were transfected with the reporter plasmid 3xMef2-Luc and Mef2A together with either GFP as control or HDAC4, respectively. Luciferase activity was measured with luciferin and is illustrated relative to the GFP control (n = 8) (One sample t-test, *p < 0.05). (C) Co-immunoprecipitation of Mef2A and HDAC4. HEK293 cells were transfected with Mef2A-flag and flag-HDAC4-myc for 48 h. Cells were treated with or without H2O2 (100 μM, 30 min). IP was conducted with Protein G beads and IgG or myc tag antibodies. Western blots were stained for flag tag. (D) Mef2A luciferase reporter gene assay in HEK293 cells. Mef2A and HDAC4 were overexpressed together with Nox4 (WT-Nox4) or redox dead Nox4 mutant (RD-Nox4). Relative Mef2 activity is illustrated (n > 52) (Unpaired t-test, *p < 0.05). All bar graphs show means ± SEM.
2.3. HDAC4 phosphorylation is enhanced by H2O2 and Nox4 activity
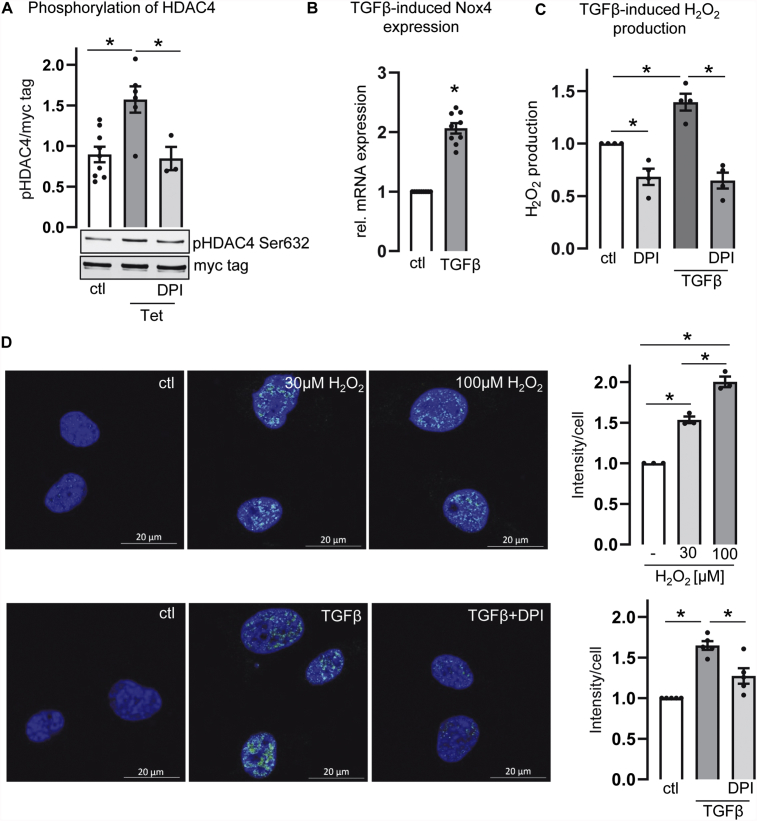
The inhibitory activity of class IIa HDACs on Mef2 isoforms is overcome by their phosphorylation on conserved serine residues [18]. Phosphorylation leads to the disruption of Mef2-HDAC complexes [21]. In the artificial system of tetracycline-inducible Nox4 expression in HEK-Nox4-tet-on cells, HDAC4 phosphorylation was increased upon Nox4 induction which was absent upon prevention of Nox4s activity by treatment of the cells with DPI (Fig. 3A). In endothelial cells treatment with increasing concentrations of H2O2 increased HDAC4-phosphorylation in a dose dependent manner (Fig. 3D). We also thought to analyze for the endogenous effect of Nox4 expression in endothelial cells. Endothelial cells endogenously express Nox4 and transforming growth factor (TGF) β is known to increase Nox4 expression in endothelial cells [25]. Therefore, we utilized TGFβ as an inducer of endogenous Nox4 expression. Indeed, TGFβ potently and robustly increased the expression of Nox4 as well as H2O2 production in HMECs (Fig. 3B&C). Further, TGFβ treatment increased the abundance of phosphorylated HDAC4 and prevention of Nox4 dependent ROS formation by DPI reduced the phosphorylation of HDAC4 back to the basal level (Fig. 3D).
Fig. 3.
Phosphorylation of HDAC4 . (A) Nox4-tet-on cells, were transfected with HDAC4-myc and treated with or without tetracycline (Tet) (1 μg/ml, 24 h) and DPI (3 μM, 3 h). Ratio of phosphorylated HDAC4 (pHDAC4) and total HDAC4 (myc tag) is depicted (n > 3). (One-way ANOVA with Tukey post-hoc test, *p < 0.05). One representative Western blot is depicted. (B) HMECs were treated with TGFβ (10 ng/ml, 24 h). Relative mRNA expression of Nox4 was analyzed by qPCR (n = 9) (Unpaired t-test, *p < 0.05). (C) Amplex Red assay in HMECs. H2O2 measurement of HMECs treated with TGFβ (10 ng/ml, 24 h) and/or DPI (3 μM, 3 h) (n = 4). (One-way ANOVA with Sidak post-hoc test, *p < 0.05). (D) Immunofluorescence staining for pHDAC4 (Ser632) in HMECs. Cells were stimulated with H2O2 (30/100 μM, 30 min) (n = 3) and TGFβ (10 ng/ml, 24 h) with or without DPI (3 μM, 24 h) (n = 5). Cells were fixed and stained with pHDAC4 (Ser632) (green). Nuclei were stained with DAPI (blue). (One-way ANOVA with Sidak post-hoc test, *p ≤ 0.05). All bar graphs show means ± SEM.
Our data suggest that Nox4 overexpression results in oxidation of HDAC4. This may ease HDAC4 phosphorylation at Ser632 and consequently, via disruption of the HDAC4/Mef2A complex, enhances Mef2A activity.
2.4. HDAC4 mediated inhibition of angiogenesis is rescued by H2O2 and Nox4 activity
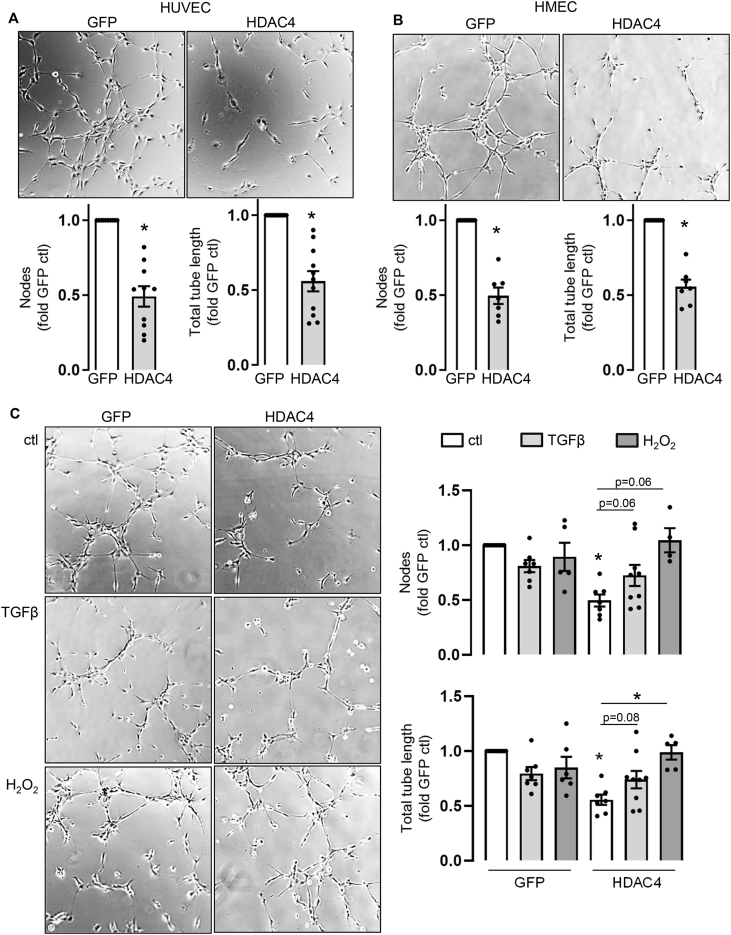
As an indicator of the physiological relevance of the data in this manuscript, we sought to investigate the effect of HDAC4 on angiogenesis. Active Mef2A, as well as phosphorylation of HDAC4, has been shown to enhance angiogenesis [16,27]. We first analyzed the effect of HDAC4 overexpression and we were able to verify that HDAC4 is a negative regulator of angiogenesis in HUVECs and HMECs (Fig. 4A and B). Treatment of HDAC4 overexpressing cells with H2O2 or TGFβ restored tube formation capacity (Fig. 4C and Supplemental Fig. 5A). Overexpression of Nox4 along with HDAC4 returned tube formation back to the basal level, while overexpressing a redox dead mutant of Nox4 (RD-Nox4) did not rescue the anti-angiogenic effect of HDAC4 overexpression (Supplemental Fig. 5B).
Fig. 4.
Tube formation assay with HDAC4 overexpression. (A) HUVECs were transfected with GFP or HDAC4. After 24 h cells were seeded on matrigel. (B + C) HMECs were transfected with GFP or HDAC4. After 24 h cells were treated with TGFβ (10 ng/ml) or H2O2 (30 μM) for 24 h and seeded on matrigel. Representative images after 4 h of tube formation are shown. Numbers of nodes were counted and total tube length was measured with an image angiogenesis analyzer (Carpentier, 2012) in ImageJ and summarized in the statistics (n > 4). All bar graphs show means ± SEM (One-sample t-test for GFP vs. HDAC4, One-way ANOVA with Sidak post-hoc test for TGFβ/H2O2 treatment *p ≤ 0.05).
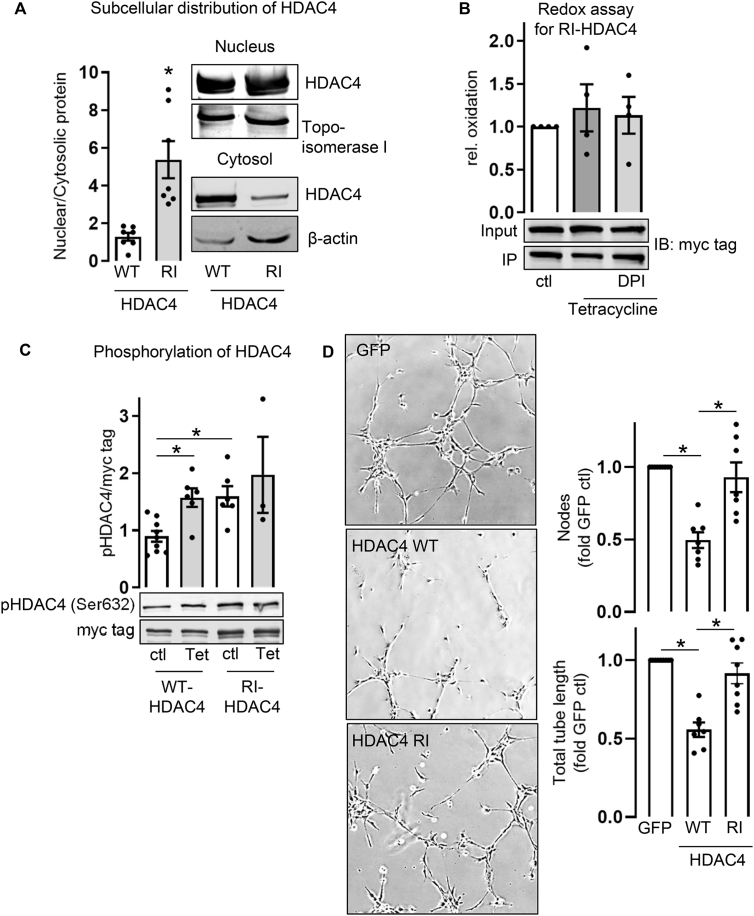
Oxidation is a prerequisite for HDAC4 to exit the nucleus, while phosphorylation ensures its accumulation in the cytosol [39]. We analyzed the effect of a redox-insensitive mutant of HDAC4 (C667/669A; RI-HDAC4). Upon overexpression in HEK293 cells, RI-HDAC4 accumulated substantially more in the nucleus than WT-HDAC4 does (Fig. 5A). Redox assays performed in Nox4-tet-on cells transfected with the RI-HDAC4 mutant did not provide reasonable evidence for a Nox4-mediated oxidation of RI-HDAC4 (Fig. 5B). Further, Nox4 did not induce RI-HDAC4s phosphorylation (Fig. 5C). Instead, RI-HDAC4 is already phosphorylated at a higher level than WT-HDAC4, for an unclear reason. Eventually, overexpression of RI-HDAC4 in HMECs did not reduce the angiogenic potential of these cells (Fig. 5D).
Fig. 5.
Effect of the C667/669A HDAC4 mutation. (A) Ratio of HDAC4 protein distribution between nucleus and cytosol. HEK293 cells were transfected with WT- or RI-HDAC4. Nuclear and cytosolic extracts were analyzed by Western blot (n = 7) (Unpaired t-test, *p < 0.05). (B) BIAM switch redox assay with Nox4-tet-on cells. Cells were transfected with RI-HDAC4 (C667/669A). After 24 h cells were treated with or without tetracycline (1 μg/ml, 24 h) and DPI (3 μM, 3 h). (C) Nox4-tet-on cells, were transfected with WT- or RI-HDAC4-myc and treated with or without tetracycline (Tet) (1 μg/ml, 24 h). Ratio of phosphorylated HDAC4 (pHDAC4) and HDAC4 (myc tag) is depicted (n > 3) (One-way ANOVA with Sidak post-hoc test, *p < 0.05). One representative Western blot is depicted. (D) HMECs were transfected with GFP, WT- or RI-HDAC4 for 48 h and seeded on matrigel. Representative images after 4 h of tube formation are shown. Numbers of nodes were counted and total tube length was measured with an image angiogenesis analyzer (Carpentier, 2012) in ImageJ and summarized in the statistics (n = 7). (One-way ANOVA with Tukey post-hoc test, *p ≤ 0.05). All bar graphs show means ± SEM.
We conclude that Nox4-mediated oxidation of HDAC4 is a prerequisite for HDAC4 to leave the nucleus, but also for the formation of the HDAC4/Mef2A complex. We conclude that HDAC4, oxidized by Nox4, has less capacity to inhibit Mef2A, which ensures endothelial tube formation.
3. Discussion
HDAC4 contributes to the manifestation of cardiac hypertrophy after TAC surgery [1,19], while Nox4 overexpression protects from TAC-induced hypertrophy [40]. Accordingly, we hypothesized that Nox4 may counteract the effects of HDAC4. Based on the finding of a high Nox4 expression in endothelial cells, we analyzed a potential connection of Nox4 and the histone deacetylase HDAC4 as well as its physiological relevance in endothelial cells. We provide evidence that Nox4-derived H2O2 oxidizes HDAC4and thereby eases the exit of HDAC4 from the nucleus. The exact localization of Nox4-mediated oxidation of HDAC4 remains to be resolved. Most prominently Nox4 is localized in the endoplasmic reticulum and the perinuclear membrane system [13]. As HDAC4 shuttles between cytosol and nucleus, it is possible that HDAC4 passes an area similar to a micro domain with high H2O2 concentration made by Nox4, a redox cloud [29]. In cardiac myocytes, oxidation of HDAC4 can be reversed by the reductase Trx1 and reduction of Cys667 and 669 in HDAC4 inhibits its nuclear export independently of its phosphorylation status [1]. Here we verify that oxidation is a prerequisite for HDAC4 to stay in the nucleus and show that Nox4 overexpression forces phosphorylation of HDAC4. Potentially both mechanisms regulate subcellular localization of HDAC4.
A mechanism how phosphorylation prevents nuclear accumulation of HDAC4 would be its binding to 14-3-3. Two models are discussed for this effect: Binding of 14-3-3 proteins could unmask a nuclear localization site (NLS) in HDAC4 and 5 [7] or expose a nuclear export site (NES) in HDAC5 [20]. Although it is uncertain, whether binding of 14-3-3 promotes nuclear export or prevents nuclear import, it is established that 14-3-3 binding controls the subcellular localization of class IIa HDACs. The Sadoshima lab published elegant studies showing that oxidation of HDAC4, facilitates nuclear export more than phosphorylation does. They found that phosphorylation is a follow up of oxidation. Accordingly, some conformational change may enable a better association of HDAC4 with its kinases. This however needs to be addressed in a separate study. Different to the work of Matsushima et al., 2013 performed in cardiac myocytes [19], we provide evidence that Nox4 overexpression in HEK293 as well as treatment of HMECs with H2O2 or TGFβ induces phosphorylation of HDAC4. We can only speculate, why this is so. The phosphatase PP2A dephosphorylates multiple serines including the 14-3-3 binding sites of HDAC4, which promotes HDAC4 nuclear import [24]. In fibrosarcoma cells the absence of Nox4 allows an accumulation of PP2A in the nucleus [11]. These findings open room for the following speculation concerning the situation in endothelial cells: In the absence of Nox4 PP2A dephosphorylates HDAC4, which accumulates in the nucleus, where it prevents angiogenesis. Indeed, one consequence of a Nox4 knock out is a reduced angiogenesis [12,36] and here we show that overexpression of HDAC4 itself prevents angiogenesis. Additionally in another work, utilizing HEK-tet-on cells for Nox4 overexpression, we identified the kinase MARK2 as a target of Nox4-mediated redox modification [17]. In HDAC7, MARK2 phosphorylates Ser155 as a prerequisite for the phosphorylation of the nearby 14-3-3 binding site in a signal-independent manner [4]. The authors of that study propose a signal-independent multisite hierarchical phosphorylation by a variety of kinases. This may also include Ser632, which was analyzed in this work. Although it is just speculation, oxidation of MARK2 could serve as an activator stimulus and facilitate HDAC4 phosphorylation. Alternatively, oxidized HDAC4 may interact easier with kinases of the CaMKII family.
A major function of endothelial cells is to perform angiogenesis. TGFβ can promote angiogenesis through multiple factors, including increased endothelial Nox4 expression and TGFβ-mediated tube formation of HUVECs which is abolished when Nox4 is inhibited [25]. TGFβ treatment elevates Nox4 expression in lung fibroblasts and leads to nuclear export of HDAC4 enabling myofibroblast differentiation [8]. Those findings are in line with the results of this study and therefore likely also to apply in endothelial cells Our results indicate that a TGFβ-induced Nox4 expression is likely to rescue the anti-angiogenic effect of HDAC4 overexpression in endothelial cells. In summary, here we show that Nox4-derived H2O2 oxidizes HDAC4, which forces its phosphorylation, which reverses HDAC4s inhibitory effect on angiogenesis. We further provide evidence that reduction of angiogenesis by overexpression of HDAC4 is abrogated by TGFβ treatment, likely based on a subsequent Nox4 expression.
4. Innovation
Overexpression of HDAC4 inhibits endothelial tube formation. Nox4-derived H2O2 specifically oxidizes HDAC4 and forces its phosphorylation. As a consequence Nox4 derived H2O2 can prevent the anti-angiogenic effect of HDAC4 overexpression in endothelial cells.
5. Materials and methods
5.1. Cell culture
Human embryonic kidney (HEK) 293 cells were purchased from ATCC (Manassas, USA) and used as an artificial model system. HEK293 cells overexpressing Nox4 in a tetracycline-inducible manner (Nox4-tet-on) were kindly provided by K.H. Krause, University of Geneva, Switzerland [32] as well as stably Nox5 overexpressing HEK293 cells (HEK293-Nox5) [26]. All HEK cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM), high glucose, GlutaMAX (Gibco, Carlsbad, USA), supplemented with 10% fetal calf serum (FCS, #S0115, Biochrom, Berlin, Germany), 1% MEM Non-essential Amino Acid Solution (100x), 1 mM Na-Pyruvat (both from Merck, Darmstadt, M7145, S8636) and gentamicin (50 μg/ml, Gibco). Human umbilical vein endothelial cells (HUVECs) were obtained from Lonza (#CC-2519, Lot No. 371074, 369146, 314457, 192485, 186864, 171772, Walkersville, USA) and PeloBiotech (#PB-CH-190-813, Lot No. QC-18P13F11, Planegg, Germany). HUVECs were cultured in dishes coated with gelatin in endothelial growth medium (EGM). EGM was composed of endothelial basal medium (EBM) supplemented with human recombinant epidermal growth factor (EGF), EndoCGS-Heparin (PeloBiotech), 8% FCS, penicillin (50 U/ml) and streptomycin (50 μg/ml) (#15140–122, Gibco). Human microvascular endothelial cells (HMECs) were purchased from Centers for Disease and Prevention (E−036-91/0, CDC, Atlanta, USA) and cultured in dishes coated with gelatin in HMEC-EGM, which consists of EGM with hydrocortisone (1 μg/ml, PeloBiotech).
5.2. Transfection
5.2.1. Lipofection of HEK293 cells and HMECs
HEK293 cells were transiently transfected with Lipofectamine 2000 reagent (#11668019, Thermo Fisher Scientific, Carlsbad, USA). Per 6 cm dish 50 μl transfection medium (HEK293 medium without antibiotics) including 1.5 μg DNA were mixed with 35 μl transfection medium including 3.5 μl Lipofectamine 2000. 35 μl transfection medium with the respective DNA amount were mixed with 17.5 μl transfection medium including 1.75 μl Lipofectamine 2000 per well in a 12-well plate. During 10 min incubation of the transfection mixture at room temperature, HEK293 cells were washed and the growth medium was replaced with 1.5 ml (6 cm dish) or 0.5 ml (12-well plate) transfection medium. The transfection mixture was then added dropwise and after 4 h the medium was changed to HEK293 growth medium.
HMECs were transiently transfected with Lipofectamine 3000 and P3000TM Enhancer Reagent (#L3000-015, Thermo Fisher Scientific). Per 3.5 cm dish 125 μl EBM were mixed with 5 μl Lipofectamine 3000 and another 125 μl EBM with 5 μl P3000TM Enhancer Reagent and 2 μg DNA. After 5 min incubation at room temperature both components were mixed and incubated for another 20 min at room temperature. The mixture was added dropwise to the dishes filled with HMECs in 1 ml EBM. After 4 h media was changed to HMEC-EGM.
5.2.2. Electroporation of HUVECs
HUVECs were transfected by electroporation using the Neon Transfection System (Thermo Fisher Scientific) according to the manufacturers’ protocol. Cells were then cultured in EBM with 8% FCS. Medium was changed to EGM after 6 h.
5.2.3. Overexpression plasmids
The following plasmids were used: GFP (#pEGFP C1, Clontech, Mountain View, USA), flag-WT- and flag-RI-HDAC4-myc (gift from Johannes Backs, Heidelberg, Germany), Mef2A-flag (#32970, Addgene, Cambridge, USA), Nox4 and an inactive Nox4 mutant (redox dead Nox4, RD-Nox4) in which Thr355 is mutated to lysine. 3XMef2-luc was a gift from Ron Prywes (Addgene plasmid #32967; http://n2t.net/addgene:32967; RRID:Addgene_32967).
5.3. H2O2 measurement with Amplex Red
Nox4-tet-on cells or HMECs were seeded on 12-well plates. Cells were washed with HEPES-Tyrode (HT) buffer (10 mM HEPES, 137 mM NaCl, 2.7 mM KCl, 1.8 mM CaCl2, 0.5 mM MgCl2, 5 mM glucose, 0.36 mM NaH2PO4). 300 μl of pre-warmed assay buffer (50 μl Amplex Red (#A12222, Invitrogen), 2 U/ml horseradish peroxidase (#P-6782, Sigma-Aldrich) in HT buffer) were added to the cells. Nox4-Tet-on cells were incubated for 3 min and HMECs for 30 min in the incubator. 200 μl were transferred to a 96-well plate and fluorescence was measured (Exc./Em. 540/585 nm). To calculate the catalase-sensitive portion fluorescence PEG-catalase (500 U/ml, #C4963, Sigma-Aldrich) was added to the assay buffer 30 min before starting the assay.
5.4. Superoxide measurement with cytochrome c
Nox4-tet-on and HEK293-Nox5 cells were seeded on 12-well plates. Cells were washed with PBS and resuspended in 1 ml HT buffer. Cells were counted and adjusted 4 × 106 cells/ml. 50 μl cell suspension were transferred to a 96-well plate and 200 μl assay buffer (20 μM cytochrome c (#C7752, Sigma-Aldrich) and 3000 U catalase (#C1345, Sigma-Aldrich)) were added. After incubation for 45 min at 37 °C, absorbance at 550 nm was measured. To calculate SOD-sensitive superoxide production every sample was subsequently measured including 500 U SOD (#S7571, Sigma-Aldrich) in the assay buffer.
5.5. Quantitative real-time polymerase chain reaction (qRT-PCR)
Isolation of mRNA was performed with the aid of an RNA-Mini-kit (Bio&Sell, Feucht, Germany) according to the manufacturer's protocol. Genomic DNA was digested with DNase I (#4536282001, Roche, Basel, Switzerland). For cDNA synthesis, Random hexamer primers (Promega, Madison, USA) together with Superscript III Reverse Transcriptase (Invitrogen, Darmstadt, Germany) were used. qRT-PCR was performed with the aid of an AriaMx qPCR cycler (Agilent Technologie, Santa Clara, USA). Primers are listed in Table 1 cDNA was detected using iQ™ SYBR® Green in a Supermix (BioRad, Hercules, USA). cDNA levels were calculated using the ΔΔct method and gene expression was normalized to β-actin.
Table 1.
qRT-PCR primers (human).
| Gene | Forward primer (5’→3′) | Reverse primer (5’→3′) |
|---|---|---|
| β-actin | AAAGACCTGTACGCCAACAC | GTCATACTCCTGCTTGCTGAT |
| HDAC4 | GAAACGAGCTTGATCCTCTCCCAG | GCTTCACGCCCACGGACAGCGAG |
| HDAC5 | TCTGAGGCTTGTGTCTCGGCTCTG | AGGCTCCTGCTCCATGGGCTCCTC |
| HDAC7 | TCTTCTGGGTAACAGGGTGGATC | ACCAGCTGCTCCGAGGGCCTATC |
| HDAC9 | TCAGGACCATCGTGAAGCCTGTG | TGCAAGTGGCTCCAGCTCATTTCC |
| Mef2A | TACACTAACCCAGGGAGTTC | TTGCACCAGTAGCTCCAATC |
| Mef2C | TACGAGGATTATGGATGAACG | GTGTACTTGAGAAGCACTTTG |
| Mef2D | TGCCCTAGGCCCAGCTTTC | ATGAACGGTCTGGGAACAGTG |
| Nox4 | TCCGGAGCAATAAGCCAGTC | CCATTCGGATTTCCATGACAT |
| Nox5 | GGTTTGTGAGCCTGCTGACTAA | GAAGCGGCCGTATTGAGC |
5.6. Western blot
Cells were washed with PBS, harvested by scratching them off the plate in RIPA lysis buffer containing Tris-HCl (50 mM), NaCl (150 mM), deoxycholate (1%), Triton X-100 (1%), sodium dodecyl sulfate (SDS) (0.1%), phenylmethylsulfonyl fluoride (PMSF, 1 mM), orthovanadate (OV, 2 mM), okadaic acid (OA, 10 nM) and protease inhibitor mix (PIM). If phosphorylation was investigated, cells were just washed quickly and shock frozen in liquid nitrogen before lysis. The lysate was centrifuged (17,000 g, 10 min, 4 °C) and the supernatant was collected. Protein concentration was determined using Bradford assay. Samples were boiled in Lämmli buffer under reducing conditions, separated by SDS-PAGE and transferred to a nitrocellulose membrane by Western blotting. Fluorescence-based detection of secondary antibodies was performed using the Odyssey CLx imaging system (Licor, Bad Homburg, Germany). Primary antibodies: HDAC4 (HEK293 cells: #sc-46672, Santa Cruz, Heidelberg, Germany; HUVECs and HMECs: #ab12172, Abcam, Cambridge, UK), phospho HDAC4 (#ab39408, Abcam), HDAC5 (#sc-133106, Santa Cruz), HDAC7 (#sc-74563, Santa Cruz), GFP (#632381, ClonTech), peroxiredoxin (Prx) 3 (#LF-PA0030, Abfrontier/Biomol, Hamburg, Germany), Mef2A (#ab32866, Abcam), phospho Mef2A (#ab30644, Abcam), Myc tag (#ab32, Abcam), Flag tag (#F3165, Sigma-Aldrich), Nox4 (#NB110-58851, Novus Biologicals, Wiesbaden, Germany), Topoisimerase I (#sc5342, Santa Cruz) and β-actin (#A1978, Sigma-Aldrich). Fluorescence-labeled secondary antibodies: IRDye® 680RD Donkey anti-Mouse, 680RD Donkey anti-Rabbit, 680RD Donkey anti-Goat, 800CW Donkey anti-Mouse, 800CW Donkey anti-Rabbit (#926–68072, #926–68073, #926–68074, #926–32212, #926–32213, Licor).
5.7. Redox-immunoprecipitation/BIAM switch assay
Alkylation of thiol groups was performed in living cells before harvest. N-ethylmaleimide (NEM, 50 mM, #4259, Sigma-Aldrich) was added to the medium and left for 5 min at room temperature. Cells were then washed with PBS containing 100 mM NEM and 500 μl trichloroacetic acid (TCA, 20%) were added. Cells were scratched off, transferred into a reaction tube, and incubated on ice for 15 min. Samples were centrifuged (13000 g, 30 min, 4 °C) and washed with 10% and then 5% TCA. The pellet was resuspended in denaturation buffer (pH 8.5, 100 mM Tris-HCl, 8 M Urea, 5 mM EDTA, 0.5% SDS) with NEM (25 mM) and incubated for 1 h at 37 °C. After addition of 800 μl acetone, proteins were precipitated over night at −20 °C. Samples were centrifuged (13000 g, 30 min, 4 °C) and washed twice with acetone. Then pellets were resuspended in denaturation buffer with 5 mM DTT for 5 min at 37 °C to reduce oxidized thiols, which were subsequently labeled by adding DB with EZ-Link Iodoacetyl-LC-Biotin (BIAM, 2 mg/ml, #21333, Thermo Fisher Scientific) for 1 h at 37 °C and 850 rpm. Proteins were precipitated by adding 800 μl acetone and stored over night at −20 °C. Samples were centrifuged (13000 g, 30 min, 4 °C) and washed twice with acetone. The pellet was resuspended with lysis buffer composed of Tris-HCl (50 mM), EDTA (5 mM), Triton X-100 (1%), SDS (1%), PMSF (1 mM), OV (2 mM), OA (10 nM) and PIM. After incubation for 1 h at 37 °C the protein amount was determined by Lowry assay. 500 μg protein were adjusted to 1 μg/μl with lysis buffer. Labeled proteins were pulled down with 100 μl Pierce™ Streptavidin Agarose beads (#20353, Thermo Fisher Scientific) slurry overnight at 4 °C. Beads were washed five times with lysis buffer and boiled with 1.5x Lämmli buffer. Samples were analyzed on Western blot [17].
5.8. Co-immunoprecipitation (Co-IP)
HEK293 cells were seeded on 6 cm dishes and transfected with 1 μg Mef2A and 0.5 μg WT- or RI-HDAC4 plasmids, respectively. Before harvesting cells were washed with PBS. Cells were scraped in lysis buffer (10 mM HEPES, 10 mM KCl,0.1 mM EDTA, 0.1 mM EGTA and 1% Nonidet P40) including OV, OA, PIM and PMSF. The lysate was sonificated three times for 10 s and 30 Hz. After that samples were centrifuged (15,000 g, 10 min, 4 °C) and the supernatant was collected in a new tube. Protein concentration was determined by Bradford assay. 1000 μg of protein were adjusted to a volume of 800 μl. Samples were pre-cleared for 30 min at 4 °C with 40 μl protein G beads (# 10004D, Thermo Fisher Scientific) which were washed before five times with lysis buffer. Pre-cleared lysates were separated from the beads and 4 μg of Mef2A-, myc tag- or IgG-antibody were added and incubated overnight at 4 °C. 40 μl protein G beads per sample were washed five times with lysis buffer and used for IP for 4 h at 4 °C. Beads were washed five times with lysis buffer and boiled for 5 min at 95 °C with 1.5x Lämmli buffer under reducing conditions. Samples were analyzed on Western blot.
5.9. Nuclear extraction
Cells were scratched off the plate in Hank's buffer and centrifuged (17,000 g, 1 min, 4 °C). The pellet was resuspended in nuclear extraction buffer containing HEPES (10 mM), KCl (10 mM), EDTA (0.1 mM), EGTA (0.1 mM), PMSF (1 mM) and PIM. After incubation on ice for 15 min, Nonidet P40 (1.5%) was added, samples were centrifuged (17,000 g, 1 min, 4 °C) and the cytosolic protein lysate (supernatant) was transferred into a new tube. The lysate and the pellet, containing nuclear proteins, were boiled in Lämmli buffer and used for Western blot.
5.10. Mef2 luciferase reporter assay
Promegas luciferase reporter assay was used to measure luciferase activity (#E1501, Promega, Madison, USA). Cells were seeded on 12-well plates and transfected for 48 h.1 μg 3xMef2-luc and 0.25 μg of the other plasmids were used for transfection. Cells were washed with PBS and scraped with 120 μl lysis buffer (1x passive lysis buffer (Promega) with PIM). Lysates were stored overnight at −80 °C. Samples were centrifuged (15,000 g, 1 min, 4 °C) and supernatant was transferred to a new tube. 20 μl were mixed with 100 μl luciferase assay reagent in a 96-well plate followed by measurement of luciferase activity.
5.11. Tube formation assay
Tube formation assays were performed in μ-Slide Angiogenesis coverslips (#81507, Ibidi, Planegg, Germany). Matrigel was thawed on ice over night. 10 μl of matrigel (#356231, Corning, Corning, USA) were added per well and allowed to polymerize for 30 min at 37 °C. HUVECs and HMECs were transfected and after 24 h cells were starved in EBM+0.1% BSA. Cells were, trypsinized and counted. 5000 HUVECs or 3000 HMECs were seeded onto the matrigel in 50 μl EBM with 1% FCS. After 4 h incubation in a cell culture incubator at 37 °C, 30 μl medium were replaced by 30 μl PBS with 4% PFA in order to fix the cells. Tube formation was analyzed by counting nodes and measuring total tube length using ImageJ [3].
5.12. Immunofluorescence
Immunofluorescence was performed in μ-Slide 8 Well chambers (#80826, Ibidi). 25,000 HMECs per well were seeded in EBM+0.1% BSA for the indicated periods. Then, cells were fixed in 2% PFA for 10 min and washed twice with PBS. To remove PFA PBS+2% glycine was added for 10 min and cells were washed twice with PBS afterwards. 0.05% Triton X-100 in PBS was used to permeabilize the cells for 10 min. PBS+3% BSA was added for 30 min to reduce unspecific binding. Phospho HDAC4 (#ab39408, Abcam) was diluted in PBS+3% BSA (1:100) and incubated over night at 4 °C. Cells were washed three times with PBS+0.3% Tween and once with PBS. Secondary antibody AlexaFluor488 donkey anti-rabbit (#A21206, Thermo Fisher Scientific) 1:500 in PBS+3% BSA was added for 30 min at room temperature. Cells were stained with DAPI for 10 min and washed three times with PBS+0.3% Tween and once with PBS. Images were taken with a confocal microscope LSM 510 Meta and analyzed with ImageJ.
5.13. Statistics
Statistical analysis was performed with GraphPad Prism 8. Means with standard error of the mean (SEM) are shown. The number of biological replicates is defined by “n”. Differences were considered as significant, if p < 0.05.
Funding sources
This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG) (to KS SFB815/TP1, SFB834/TPA2) and the Cardio-Pulmonary Institute, Germany (CPI), EXC 2026, Project ID: 390649896.
Declaration of competing interest
The authors declare that they have no conflicts to disclose.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101669.
Abbreviations
- Nox
NADPH oxidase
- HDAC
Histone deacetylase
- TGFβ
Transforming growth factor beta
- HUVEC
Human umbilical vein endothelial cells
- HEK293
Human embryonic kidney
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Ago T., Liu T., Zhai P., Chen W., Li H., Molkentin J.D., Vatner S.F., Sadoshima J. A redox-dependent pathway for regulating class II HDACs and cardiac hypertrophy. Cell. 2008;133:978–993. doi: 10.1016/j.cell.2008.04.041. [DOI] [PubMed] [Google Scholar]
- 2.Babelova A., Avaniadi D., Jung O., Fork C., Beckmann J., Kosowski J., Weissmann N., Anilkumar N., Shah A.M., Schaefer L., Schröder K., Brandes R.P. Role of Nox4 in murine models of kidney disease. Free Radic. Biol. Med. 2012;53:842–853. doi: 10.1016/j.freeradbiomed.2012.06.027. [DOI] [PubMed] [Google Scholar]
- 3.Carpentier G., Martinelli M., Courty J., Cascone I., editors. Proceedings of the ImageJ User and Developer Conference 2012. 24. - 26. Oktober 2012, Mondorf-Les-Bains, Luxembourg] CRP; Luxembourg: 2012. [Google Scholar]
- 4.Dequiedt F., Martin M., Blume J von, Vertommen D., Lecomte E., Mari N., Heinen M.-F., Bachmann M., Twizere J.-C., Huang M.C., Rider M.H., Piwnica-Worms H., Seufferlein T., Kettmann R. New role for hPar-1 kinases EMK and C-TAK1 in regulating localization and activity of class IIa histone deacetylases. Mol. Cell Biol. 2006;26:7086–7102. doi: 10.1128/MCB.00231-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Di, Xiao C., Long F., Su Z., Jia W., Qin M., Huang M., Wu W., Suguro R., Liu X., Zhu Y. HDAC4 regulates vascular inflammation via activation of autophagy. Cardiovasc. Res. 2018;114:1016–1028. doi: 10.1093/cvr/cvy051. [DOI] [PubMed] [Google Scholar]
- 6.Furchgott R.F., Zawadzki J.V. The obligatory role of endothelial cells in the relaxation of arterial smooth muscle by acetylcholine. Nature. 1980;288:373–376. doi: 10.1038/288373a0. [DOI] [PubMed] [Google Scholar]
- 7.Grozinger C.M., Schreiber S.L. Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc. Natl. Acad. Sci. U. S. A. 2000;97:7835–7840. doi: 10.1073/pnas.140199597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo W., Saito S., Sanchez C.G., Zhuang Y., Gongora Rosero R.E., Shan B., Luo F., Lasky J.A. TGF-beta1 stimulates HDAC4 nucleus to cytoplasm translocation and NADPH oxidase4-derived reactive oxygen species in normal human lung fibroblasts. Am. J. Physiol. Lung Cell Mol. Physiol.: ajplung.00256. 2017;2016 doi: 10.1152/ajplung.00256.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hahner F., Moll F., Schröder K. NADPH Oxidases in the differentiation of endothelial cells. Cardiovasc. Res. 2019;116:262–268. doi: 10.1093/cvr/cvz213. [DOI] [PubMed] [Google Scholar]
- 10.Han A., He J., Wu Y., Liu J.O., Chen L. Mechanism of recruitment of class II histone deacetylases by myocyte enhancer factor-2. J. Mol. Biol. 2005;345:91–102. doi: 10.1016/j.jmb.2004.10.033. [DOI] [PubMed] [Google Scholar]
- 11.Harris-Hooker S.A., Gajdusek C.M., Wight T.N., Schwartz S.M. Neovascular responses induced by cultured aortic endothelial cells. J. Cell. Physiol. 1983;114:302–310. doi: 10.1002/jcp.1041140308. [DOI] [PubMed] [Google Scholar]
- 12.Helfinger V., Henke N., Harenkamp S., Walter M., Epah J., Penski C., Mittelbronn M., Schröder K. The NADPH Oxidase Nox4 mediates tumour angiogenesis. Acta Physiol. 2016;216:435–446. doi: 10.1111/apha.12625. [DOI] [PubMed] [Google Scholar]
- 13.Helmcke I., Heumüller S., Tikkanen R., Schröder K., Brandes R.P. Identification of structural elements in Nox1 and Nox4 controlling localization and activity. Antioxidants Redox Signal. 2009;11:1279–1287. doi: 10.1089/ars.2008.2383. [DOI] [PubMed] [Google Scholar]
- 14.Hrgovic I., Doll M., Pinter A., Kaufmann R., Kippenberger S., Meissner M. Histone deacetylase inhibitors interfere with angiogenesis by decreasing endothelial VEGFR-2 protein half-life in part via a VE-cadherin-dependent mechanism. Exp. Dermatol. 2017;26:194–201. doi: 10.1111/exd.13159. [DOI] [PubMed] [Google Scholar]
- 15.Incalza M.A., D'Oria R., Natalicchio A., Perrini S., Laviola L., Giorgino F. Oxidative stress and reactive oxygen species in endothelial dysfunction associated with cardiovascular and metabolic diseases. Vasc. Pharmacol. 2018;100(1–19) doi: 10.1016/j.vph.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 16.Liu J., Zhou X., Li Q., Zhou S.-M., Hu B., Hu G.-W., Niu X., Guo S.-C., Wang Y., Deng Z.-F. Role of phosphorylated HDAC4 in stroke-induced angiogenesis. BioMed Res. Int. 2017;2957538 doi: 10.1155/2017/2957538. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Löwe O., Rezende F., Heidler J., Wittig I., Helfinger V., Brandes R.P., Schröder K. BIAM switch assay coupled to mass spectrometry identifies novel redox targets of NADPH oxidase 4. Redox Biol. 2019;21:101125. doi: 10.1016/j.redox.2019.101125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin M., Kettmann R., Dequiedt F. Class IIa histone deacetylases: regulating the regulators. Oncogene. 2007;26:5450–5467. doi: 10.1038/sj.onc.1210613. [DOI] [PubMed] [Google Scholar]
- 19.Matsushima S., Kuroda J., Ago T., Zhai P., Park J.Y., Xie L.-H., Tian B., Sadoshima J. Increased oxidative stress in the nucleus caused by Nox4 mediates oxidation of HDAC4 and cardiac hypertrophy. Circ. Res. 2013;112:651–663. doi: 10.1161/CIRCRESAHA.112.279760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKinsey T.A., Zhang C.L., Olson E.N. Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14-3-3 to histone deacetylase 5. Proc. Natl. Acad. Sci. U. S. A. 2000;97:14400–14405. doi: 10.1073/pnas.260501497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKinsey T.A., Zhang C.L., Olson E.N. Signaling chromatin to make muscle. Curr. Opin. Cell Biol. 2002;14:763–772. doi: 10.1016/s0955-0674(02)00389-7. [DOI] [PubMed] [Google Scholar]
- 22.Miska E.A., Karlsson C., Langley E., Nielsen S.J., Pines J., Kouzarides T. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J. 1999;18:5099–5107. doi: 10.1093/emboj/18.18.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Narayanan S., Loganathan G., Mokshagundam S., Hughes M.G., Williams S.K., Balamurugan A.N. Endothelial cell regulation through epigenetic mechanisms: depicting parallels and its clinical application within an intra-islet microenvironment. Diabetes Res. Clin. Pract. 2018;143:120–133. doi: 10.1016/j.diabres.2018.06.018. [DOI] [PubMed] [Google Scholar]
- 24.Paroni G., Cernotta N., Dello Russo C., Gallinari P., Pallaoro M., Foti C., Talamo F., Orsatti L., Steinkühler C., Brancolini C. PP2A regulates HDAC4 nuclear import. Mol. Biol. Cell. 2008;19:655–667. doi: 10.1091/mbc.E07-06-0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peshavariya H.M., Chan E.C., Liu G.S., Jiang F., Dusting G.J. Transforming growth factor-β1 requires NADPH oxidase 4 for angiogenesis in vitro and in vivo. J. Cell Mol. Med. 2014;18:1172–1183. doi: 10.1111/jcmm.12263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rezende F., Löwe O., Helfinger V., Prior K.-K., Walter M., Zukunft S., Fleming I., Weissmann N., Brandes R.P., Schröder K. Unchanged NADPH oxidase activity in nox1-nox2-nox4 triple knockout mice: what do NADPH-stimulated chemiluminescence assays really detect? Antioxidants Redox Signal. 2016;24:392–399. doi: 10.1089/ars.2015.6314. [DOI] [PubMed] [Google Scholar]
- 27.Sacilotto N., Chouliaras K.M., Nikitenko L.L., Lu Y.W., Fritzsche M., Wallace M.D., Nornes S., García-Moreno F., Payne S., Bridges E., Liu K., Biggs D., Ratnayaka I., Herbert S.P., Molnár Z., Harris A.L., Davies B., Bond G.L., Bou-Gharios G., Schwarz J.J., Val S de. MEF2 transcription factors are key regulators of sprouting angiogenesis. Genes Dev. 2016;30:2297–2309. doi: 10.1101/gad.290619.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schröder K. Vol. 104. Antioxid Redox Signal; 2018. Redox Control of Angiogenesis; pp. 447–452. [Google Scholar]
- 29.Schröder K. NADPH oxidase-derived reactive oxygen species: dosis facit venenum. Exp. Physiol. 2019;30:960–971. doi: 10.1113/EP087125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schröder K., Zhang M., Benkhoff S., Mieth A., Pliquett R., Kosowski J., Kruse C., Luedike P., Michaelis U.R., Weissmann N., Dimmeler S., Shah A.M., Brandes R.P. Nox4 is a protective reactive oxygen species generating vascular NADPH oxidase. Circ. Res. 2012;110:1217–1225. doi: 10.1161/CIRCRESAHA.112.267054. [DOI] [PubMed] [Google Scholar]
- 31.Schürmann C., Rezende F., Kruse C., Yasar Y., Löwe O., Fork C., van de Sluis B., Bremer R., Weissmann N., Shah A.M., Jo H., Brandes R.P., Schröder K. The NADPH oxidase Nox4 has anti-atherosclerotic functions. Eur. Heart J. 2015;36:3447–3456. doi: 10.1093/eurheartj/ehv460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Serrander L., Cartier L., Bedard K., Banfi B., Lardy B., Plastre O., Sienkiewicz A., Fórró L., Schlegel W., Krause K.-H. NOX4 activity is determined by mRNA levels and reveals a unique pattern of ROS generation. Biochem. J. 2007;406:105–114. doi: 10.1042/BJ20061903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Serrander L., Jaquet V., Bedard K., Plastre O., Hartley O., Arnaudeau S., Demaurex N., Schlegel W., Krause K.-H. NOX5 is expressed at the plasma membrane and generates superoxide in response to protein kinase C activation. Biochimie. 2007;89:1159–1167. doi: 10.1016/j.biochi.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 34.Thienel U., Loike J., Yellin M.J. CD154 (CD40L) induces human endothelial cell chemokine production and migration of leukocyte subsets. Cell. Immunol. 1999;198(87–95) doi: 10.1006/cimm.1999.1583. [DOI] [PubMed] [Google Scholar]
- 35.Tsou P.-S., Wren J.D., Amin M.A., Schiopu E., Fox D.A., Khanna D., Sawalha A.H. Histone deacetylase 5 is overexpressed in scleroderma endothelial cells and impairs angiogenesis via repression of proangiogenic factors. Arthritis & rheumatology (Hoboken, N.J.) 2016;68:2975–2985. doi: 10.1002/art.39828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vogel J., Kruse C., Zhang M., Schröder K. Nox4 supports proper capillary growth in exercise and retina neo-vascularization. J Physiol. 2015;593:2145–2154. doi: 10.1113/jphysiol.2014.284901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang A.H., Bertos N.R., Vezmar M., Pelletier N., Crosato M., Heng H.H., Th'ng J., Han J., Yang X.J. HDAC4, a human histone deacetylase related to yeast HDA1, is a transcriptional corepressor. Mol. Cell Biol. 1999;19:7816–7827. doi: 10.1128/mcb.19.11.7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang W., Ha C.H., Jhun B.S., Wong C., Jain M.K., Jin Z.-G. Fluid shear stress stimulates phosphorylation-dependent nuclear export of HDAC5 and mediates expression of KLF2 and eNOS. Blood. 2010;115:2971–2979. doi: 10.1182/blood-2009-05-224824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Z., Qin G., Zhao T.C. HDAC4: mechanism of regulation and biological functions. Epigenomics. 2014;6:139–150. doi: 10.2217/epi.13.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang M., Brewer A.C., Schröder K., Santos C.X.C., Grieve D.J., Wang M., Anilkumar N., Yu B., Dong X., Walker S.J., Brandes R.P., Shah A.M. NADPH oxidase-4 mediates protection against chronic load-induced stress in mouse hearts by enhancing angiogenesis. Proc. Natl. Acad. Sci. U. S. A. 2010;107:18121–18126. doi: 10.1073/pnas.1009700107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang R., Ge J. Proteinase-activated receptor-2 modulates ve-cadherin expression to affect human vascular endothelial barrier function. J. Cell. Biochem. 2017;118:4587–4593. doi: 10.1002/jcb.26123. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.